- 1Aquafeed Research Center, National Institute of Fisheries Science, Pohang, Republic of Korea
- 2Department of Fisheries Biology, Pukyong National University, Busan, Republic of Korea
- 3Department of Biotechnology, Pukyong National University, Busan, Republic of Korea
- 4Division of Fisheries Life Sciences, Pukyong National University, Busan, Republic of Korea
The necessity for additional macroalgal biomass is warranted amid the ongoing expansion of abalone aquaculture. In the current study, an in-depth investigation of the food value of Sargassum horneri as a replacement for Undaria pinnatifida on growth, body composition, body histology, gut microbiota, and the taste sensory profile of abalone Haliotis discus hannai was carried out. After a 12-week feeding trial, the results indicated that 25% replacement (SH25) induced the highest shell-growth performance and best feed utilization values, although there was no significant difference in weight-related growth parameters with the basal (no inclusion of S. horneri) and 50% replacement (SH50) diets. Interestingly, the highest protein content was detected in abalone fed the basal diet followed by SH25 and SH50. The high replacement of S. horneri in the diet showed signs of structural integrity loss in the digestive gland. The results from the pyrosequencing of the genomic DNA from the gut of the abalone indicated an insignificant difference in α-diversity parameters and only a clear boundary was observed between SH0 and SH100 diets based on principal coordinate analysis. The bacterial genera Psychrilobacter, Vibrio, and Mycoplasma predominantly occupied the gut of the abalone, although there is a difference in their relative abundance. The taste screening experiment through the use of an electronic tongue showed a comparable taste profile of the abalone and is independent of the diets given. The findings of the present study showed that S. horneri, up to 50% replacement of macroalgal ingredients, can be utilized in feed formulation for H. discus hannai. These, in turn, contribute to the expanding knowledge on the utilization of biofouling and non-traditional macroalgae for sustainable abalone aquaculture practices.
1 Introduction
Sargassum horneri (Turner) C. Agardh is a large brown seaweed endemic to north-northwest Pacific coasts, particularly in China, Japan, and Korea (Pang et al., 2009; Su et al., 2017; Kim et al., 2018). S. horneri are important ecological habitats playing crucial roles such as nursery sites and food for aquatic animals in the marine ecosystem (Smetacek and Zingone, 2013; Wang et al., 2014; Kim et al., 2018). It has been shown to have potential as a functional biomaterial for pharmaceutical applications (Kim et al., 2018) and has been used as a traditional medicine in East Asian nations (Sanjeewa et al., 2017). Sargassum species produce large blooms or golden tides (Smetacek and Zingone, 2013) and can negatively affect nearby ecosystems and aquaculture farms, and become a nuisance on the coastlines. In Korea, this species mainly inhabits the coasts of Jeju Island and South Jeolla Province (Hwang et al., 2016; Kim et al., 2018) and large-scale blooms that destroyed seaweed and abalone culture sites has been reported (Hwang et al., 2016; Byeon et al., 2019). Additionally, Jeju island’s fish harbors and shores are spoiled by shore-washed, decaying gulfweed that emits noxious hydrogen sulfide (Hwang et al., 2016). The lack of use for direct human consumption is also seen as one of the causes why S. horneri natural population tends to go beyond reasonable quantity. Alternative ways of using this species have thus been suggested to minimize the harmful effects of gulfweed bloom, with a focus on its potential as an ingredient in aquafeed formulations and to establish application in aquaculture industry (Zhang et al., 2017; Zhu et al., 2017; Ansary et al., 2019a; Ansary et al., 2019b).
Abalone, an algivore marine gastropod mollusk, is an economically-important aquaculture species worldwide. In recent years, the abalone production trend is increasing globally (Cook, 2016; Cook, 2019) and Korea is one of the major suppliers in the market (Cook, 2014). There are about 90 species of abalone (Bullon et al., 2022), six of which are found in Korean waters but aquaculture production dominantly comes from one species, the disk or Japanese abalone Haliotis discus hannai (Park and Kim, 2013; Cook, 2016). Large portions of abalone culture farms are located in the Wando County of South Jeolla Province comprising approximately 500,000 sea cages in the early 2010s (Park and Kim, 2013). The majority of produced abalone is consumed locally and some are exported to Japan, China, the USA, and Taiwan (Cook, 2016). Abalones are herbivorous organisms with a distinct shift in feeding pattern across life stages, from being planktivorous consuming microalgae during larval and post-larval stages and shifting to macroalgal diets starting at the juvenile stage (Takami and Kawamura, 2003; Bullon et al., 2022). The formulated feeds for abalone are sourced both from terrestrial and aquatic environments. These include the use of traditional feed ingredients such as cereal crops, fishmeal, fish oil, and seaweeds (Kirkendale et al., 2010; Bullon et al., 2022). Bullon et al. (2022) did a compilated review of research reports on the utilization of both common or traditional and non-traditional macroalgal food source in the formulated feed for various abalone species, wherein positive animal feed responses were described. In addition, studies emphasizing the substitution potential of various animal, bacterial and plant proteins in the formulated diet have been reported as well to have a beneficial effect on the animal (Cho, 2010, Wu et al., 2022; Yu et al., 2022). Nevertheless, macroalgae, the natural feed of abalone is essential in diets to increase feed intake and to improve digestibility and meat yield (Dlaza et al., 2008; Kemp et al., 2015; Nel et al., 2017) and can reach up to 40% of the total feed composition. In Korea, fresh macroalgae Saccharina japonica and Undaria pinnatifida are predominantly used over formulated diets as abalone feed (Park and Kim, 2013) primarily due to ease of management in abalone farms (Lee et al., 2016; Jang et al., 2018). However, these species are bounded with several problems including availability as only exclusively harvestable during the winter season (Lee et al., 2016), increasing market price (Jang et al., 2018), and unsustainable in the long run as also being farmed for biopharmaceutical application and human consumption (Hwang and Park, 2020). It is further known from previous studies that over a single macroalga, a combined macroalgae in the diets offers well-balanced nutrition and improves immunity and subsequently faster growth performance in a range of abalone species (Dang et al., 2011; Viera et al., 2011; O'Mahoney et al., 2014; Ansary et al., 2019b; Sun et al., 2021). As a result, earlier studies affirmed the necessity of using additional seaweed species as a source of macroalgal biomass for abalone aquaculture (Mulvaney et al., 2013; Jang et al., 2018; Ansary et al., 2019a).
The potential of fouling macroalgae as a feed for abalone has been reported (Alcantara and Noro, 2006; Zhang et al., 2010; Dang et al., 2011; Mulvaney et al., 2013; Ansary et al., 2019b). Particularly, Ansary et al. (2019a) conducted an earlier study on the replacement of U. pinnatifida by S. horneri on abalone, but with some similarities and differences in the current study. The similitudes include utilization of juvenile abalone and analysis of somatic growth and soft-body composition of the animal. The differences include percentage replacement of S. horneri (up to 20% versus 40% in this study) and culture system (cage in raceway system versus tank in flow-through system in this study). The works of Ansary et al. (2019a); Ansary et al. (2019b) provided baseline information on the utilization of S. horneri as additional macroalgal food for abalone, but nevertheless recommended that further research is required before commercial-scale application. Hence, in order to better understand the impact of S. horneri as a dietary replacement in formulated feed, the current study examined growth (morphological growth, feed utilization, body composition, body histology), gut microbiota, and taste sensory profile of commercially-important abalone Haliotis discus hannai.
2 Materials and methods
2.1 Experimental diets
The feed formulation and proximate composition of diets are shown in Table 1. The experimental diets were formulated with fish meal, corn gluten meal and defatted soybean meal as protein, wheat flour and dextrin as carbohydrate and fish oil as lipid sources. The formulated feed containing 40% macroalgae U. pinnatifida with 0% inclusion of S. horneri (SH0) was designated as a basal diet. Other experimental diets were prepared by using S. horneri (SH) to replace 25%, 50%, 75% and 100% of U. pinnatifida named SH25, SH50, SH75, and SH100, respectively. The formulated diets were produced using a pellet extruder at Ewha Oil and Fat Industry Co. Ltd. (Busan, Korea) and the resulting feed is an extruded, smooth round-edge flakes with surface area of 10 mm2. All prepared diets were stored in -20°C throughout the experiment.
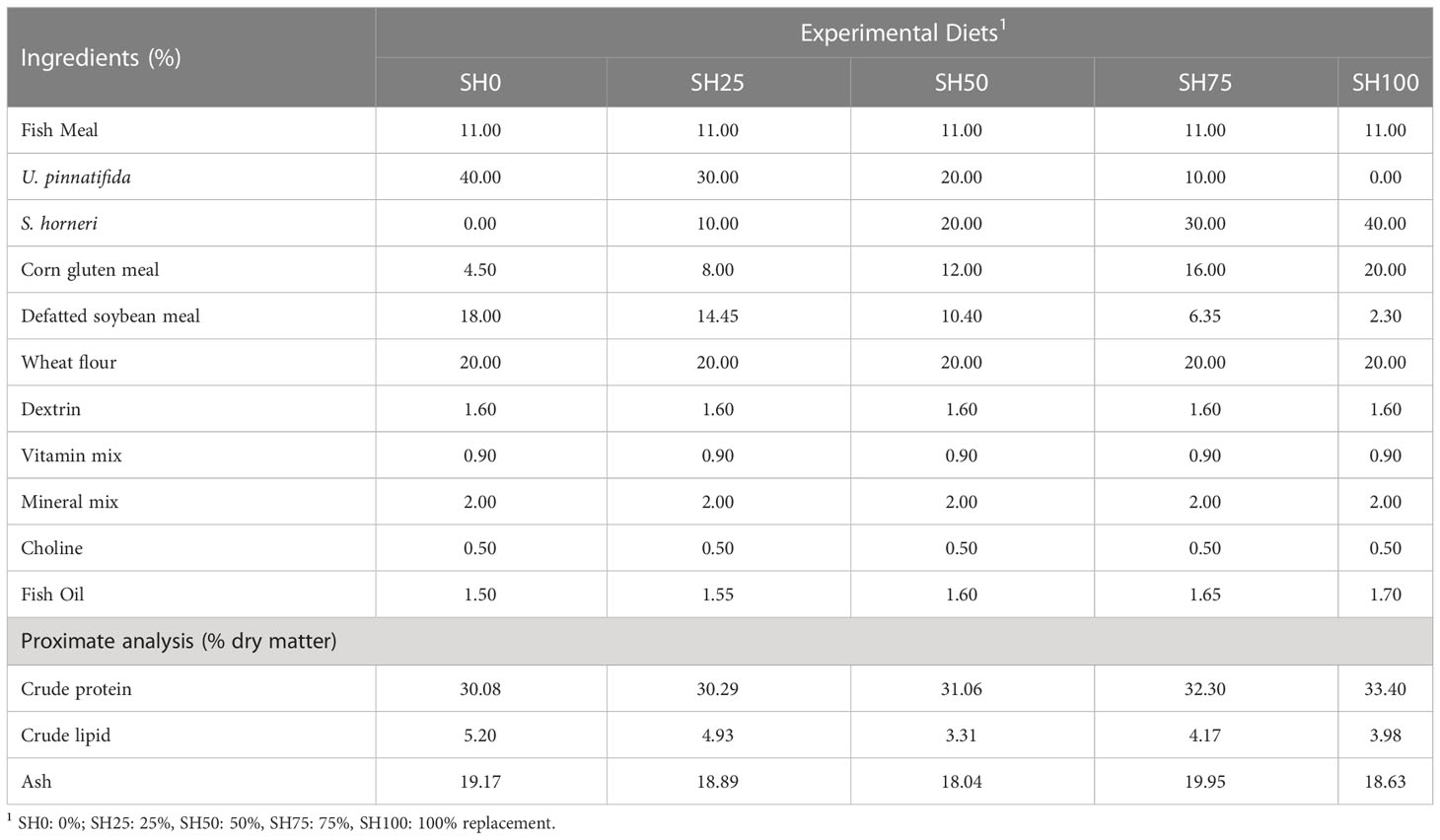
Table 1 Feed formulation and proximate composition of the abalone diets with varying Sargassum horneri inclusion levels (% dry matter).
2.2 Feeding trial
The feeding trial was conducted at Aquafeed Research Center, National Institute of Fisheries Science (ARC-NIFS) in Pohang, South Korea. Post-larval abalone was sourced from a commercial farm in Wando, South Jeolla Province, and transferred to the culture facility of ARC-NIFS. Abalones were set to acclimatize to the local conditions for two weeks and fed a commercial diet (Trueone Abalone, Kyongwon Feed, Dangjin, South Korea) before the commencement of the study. Fifteen 200 L-capacity cylindrical tanks (corresponding to five treatment diets replicated thrice) were used to contain the abalone. A black, W-shaped PVC material (n=2 per tank, 33 × 18 × 4 cm) was used as a shelter for the abalone. A total of 1500 abalone were stocked, 100 individuals per tank with an initial weight (IW) of 8.0 ± 0.02 g and initial shell length (ISL) of 37.50 ± 0.04 mm. These culture tanks are continuously supplied by fresh sand-filtered seawater via a flow-through system. The amount of feed was calculated at 1.5% total biomass per tank, adjusted monthly, and given once daily at 4:30 PM. Tanks were cleaned daily to remove feces and uneaten feeds. The experiment lasted for 12 weeks from April to June 2021.
2.3 Sample collection
At the end of the feeding trial, abalones were starved for 24 h before sampling. Abalones were counted, collectively weighted to the nearest 0.01 g, and shell length was measured to the nearest 0.01 mm to calculate the survival rate, growth performance and feed utilization efficiency. The growth was indicated by the increase in body weight shown as weight gain rate and weight specific growth rate and increase in shell length shown as daily increment in shell length. The feed utilization values were calculated as the conversion of feed input and nutrients into somatic growth (specific formulas are shown in section 2.8 in this article). A random sampling of three abalone individuals per diet group were used for histological analysis, biochemical analysis and taste screening analysis, and five individual samples for the genomic analysis of the gut bacterial population. For body composition analysis, whole soft tissues were carefully shucked from the shell using a spatula. For histological examination, an incision in the midsection of the abalone body was made to obtain the digestive gland from the stomach/intestine part (Schaefer et al., 2013) and fixed in Bouin’s solution. The whole gastrointestinal tract and adductor muscle were similarly separated from the body for microbial genomic sequencing and taste profile analysis, respectively. All samples were snap-frozen in liquid nitrogen and stored at -80°C until analysis.
2.4 Analysis of feed and whole-body composition
The proximate composition of the formulated diets (n=2) and abalone soft-body tissues (n=3 per diet group) were analyzed following standard procedures (AOAC, 1995). The samples were constantly weighed in an oven at 105°C to determine moisture content. Crude protein (CP) was analyzed using Kjeldahl nitrogen analyzer (Kjeltec™ 8400, Foss, Denmark) and calculated as N × 6.25 after digestion of standard acid. Crude lipid was extracted from the samples using Soxhlet extraction method utilizing hexane as solvent and detected using Soxtec™ 2043 Fat Extraction System (Foss, Denmark). Ash was measured after sample combustion in a muffle furnace at 600°C for 4 hours.
2.5 Histology
Foot and digestive gland tissues (n=3 per diet group) were dehydrated through a series of increasing ethanol concentrations, washed with xylene, embedded in paraffin, and sectioned at 6 µm using a microtome. Sectioned tissues were floated onto glass slides and left to dry overnight in a slide warmer. Digestive gland tissues were stained with Hematoxylin (Dako, CA, USA) and Eosin (Sigma, MO, USA) (H&E). All stained tissues were mounted using canada balsam (Junsei, Tokyo, Japan), examined under a light microscope (SEX41; Olympus, Tokyo, Japan) and images were captured using Motic Image Plus 2.0 ML (Motic, Xiamen, China).
2.6 Gut microbiota analysis
The abalone gut samples (n=5 per diet group) were subjected to total bacterial DNA extraction using FavorPrep™ Tissue Genomic Extraction Mini Kit (Favorgen Biotech Corp., Taiwan). The concentration and quality of extracted DNA were determined by DropSense 96 (Trinean) microplate reader and gel electrophoresis. Then, a sequencing library was built targeting the V3 and V4 hypervariable regions of the microbial 16S rRNA gene. These regions were amplified by KAPA HiFi HotStart ReadyMix (KAPA Biosystems, Wilmington, MA, USA) and PCR products were purified using Agencourt AMPure XP system (Beckman Coulter Genomics, Brea, CA, USA). PCR analysis was run using region-specific primers shown to have compatibility with Illumina index and sequencing adapters (forward primer: 5′-TCGTCGGCAGCGTCAGATGTGTATAAGGACAGTCGTCGGCAGCGTCAGATGTGTATAAGAGACAGCCTACGGGNGGCWGCAG-3′, reverse primer: 5′-GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGGTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGGACTACHVGGTATCTAATCC-3′). The PCR conditions were as follows 95°C for 3 min, and 25 cycles of 95°C for 30 sec, 55°C for 30 sec, and 72°C for 30 sec, with a final elongation at 72°C for 5 min. Subsequently, purified PCR products were visualized using 2% gel electrophoresis and quantified with a Qubit dsDNA HS Assay Kit (Thermo Scientific) on a Qubit 3.0 fluorometer. Sequencing was conducted on Miseq system (Illumina) with 300 bp paired-end reads. The 16S rRNA raw reads were trimmed using cutadapt (ver. 1.9). Operational taxonomic units (OTUs) with 97% similarity cut-off were used and compared with known sequences in the database of the 16S rRNA gene using BLAST search (https://blast.ncbi.nlm.nih.gov). The alpha diversity indices were calculated based on richness and diversity parameters while the beta diversity index is shown as a result of principal coordinate analysis (PCoA) based on UniFrac metrics among diets performed by Quantitative Insights into Microbial Ecology 2 (QIIME2, https://qiime2.org/) (Shannon, 1948; Simpson, 1949; Chao and Bunge, 2002; Hamady et al., 2010). The raw sequence reads were deposited in NCBI with accession number PRJNA889551.
2.7 Taste sensory analysis using an electronic tongue
An electronic tongue (e-tongue) (ASTREE II, Alpha MOS, France) was used to determine the taste sensory profile of the abalone. The e-tongue is composed of three parts: a sampling system or autosampler, a sensor array or system, and an analyzer or data processing system (Jayasundar et al., 2021; Wang et al., 2021). The autosampler consists of 16 sample holders (fitted to hold a standard 100 mL-capacity beaker) and a movable arm. The sensor system is composed of seven cross-inductance sensors, a reference electrode (Ag/AgCl) and a stirrer all attached to the movable arm. Sensors PKS, CPS, ANS, and SCS are general-purpose sensors, whereas AHS, CTS, and NMS corresponds to sourness, saltiness, and umami, respectively. The data processing system is a chemometric software package used to analyze and evaluate signals from the sensors. It is composed of programable sequences used to run the analysis and statistical packages to analyze results. The procedures employed in the use of e-tongue were strictly followed as per the manufacturer’s instructions. Samples were measured after reaching constant potential for each of the sensors passing conditioning and calibration steps. A rinsing step was done during post-cycle analysis.
The adductor muscle (AM) (n=3 per diet group), together with the foot muscle is primarily consumed in abalone (Porturas Olaechea et al., 1993) and thus used to determine the taste sensory profile in this study. During analysis, frozen AM was thawed, weighed, and homogenized in an ice-cold bath for 2-3 mins. Homogenates were centrifuged at 10,000 rpm for 8 mins at 4°C to obtain supernatants. Then, the supernatant was mixed with purified water to obtain a 100 ppm solution in a 100 mL-capacity beaker. The beakers containing sample solutions were placed in the autosampler corresponding to the sequence made to read five times.
2.8 Data calculation and statistical analysis
Growth performance and feed utilization parameters were calculated as follows:
Daily increment in shell length (DISL, μm/day) = [final shell length (mm) - initial shell length (mm)]/days × 1000.
Weight gain rate (WGR, %) = [(final weight (g) - initial weight (g))/initial weight (g)] × 100.
Weight specific growth rate (WSGR, %) = [(Ln final weight (g) - Ln initial weight (g))/days of culture] × 100.
Feed conversion ratio (FCR) = feed intake (g)/[final weight (g) – initial weight (g)].
Feed efficiency rate (FE, %) = [(final weight (g) – initial weight (g))/feed intake (g)] × 100
Protein efficiency ratio (PER) = [(final weight (g) – initial weight (g))/protein given]
Survival rate (SR, %) = (final number of abalone/initial number of abalone) × 100.
All growth performance, feed utilization, whole-body proximate composition data, and bacterial alpha indices were analyzed using SPSS version 26 (IBM, USA). Homogeneity of variances was assessed using Levene’s test of homogeneity before proceeding to the One-way Analysis of Variance (ANOVA) analysis. If statistical significance is detected, Duncan posthoc test was performed to determine the difference among treatment means. All groups were analyzed at 95% confidence level. All data are presented as mean ± standard error (SE).
3 Results
3.1 Growth performance and feed utilization
The growth performance and feed utilization of abalone-fed varying inclusion levels of SH are shown in Table 2. Except for the initial data and survival rate, all parameters calculated were significantly influenced by experimental diets (P < 0.05). In terms of shell length growth, the inclusion of 25% SH recorded the highest FL and DISL with a remarkable difference in SH50, SH75, and SH100 inclusion (P < 0.05) but without a difference to abalone-fed SH0 (P > 0.05). Meanwhile, FW, WGR, and WSGR of abalone fed SH0, SH25 and SH50 did not significantly differ from each other (P > 0.05) and significantly lower values were calculated in SH75 and SH100 (P < 0.05). Significantly higher FCR and lower FE in SH75 and SH100 than in other experimental diets (P < 0.05) were also calculated. In terms of protein efficiency ratio, SH0, SH25, and SH50 were statistically similar (P > 0.05) among each other and SH100 had the lowest PER among experimental diets (P < 0.05). No mortality was recorded during the conduct of the feeding trial.
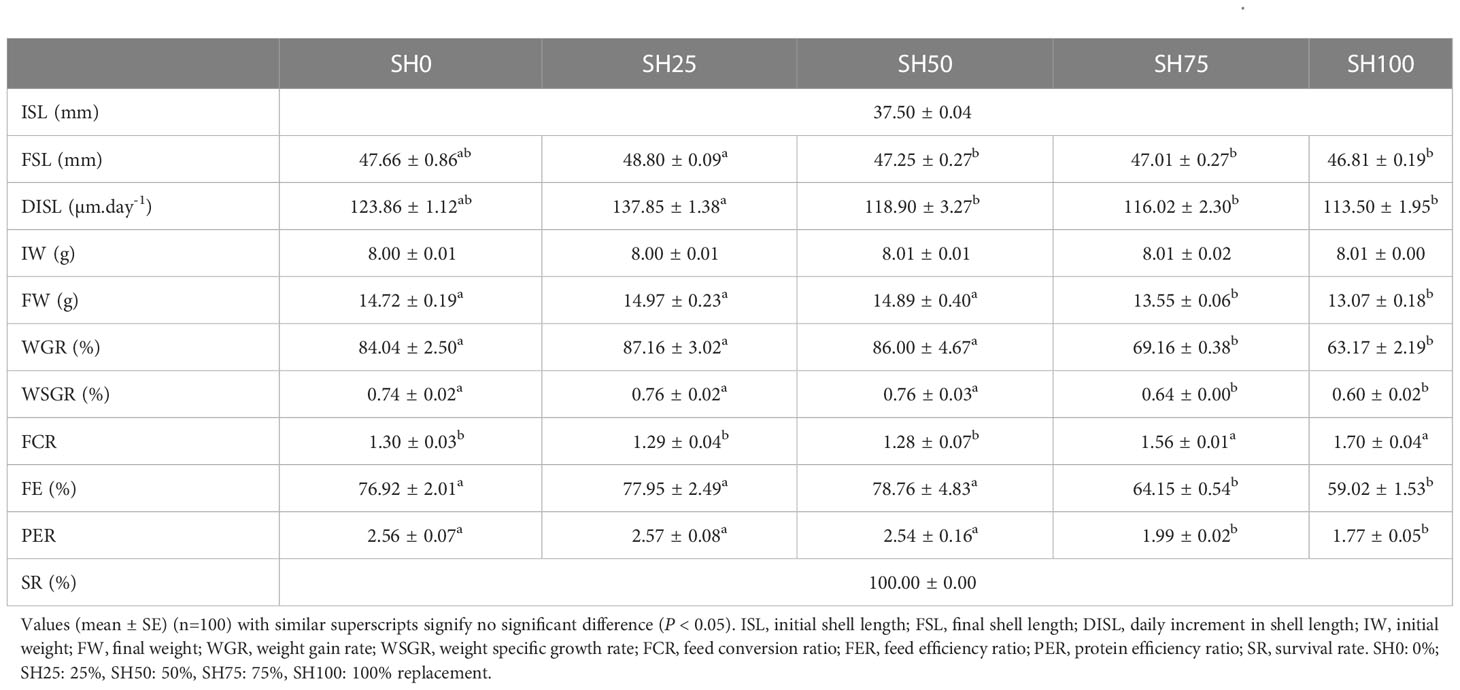
Table 2 Effects of Sargassum horneri inclusion levels on growth performance and feed utilization of abalone Haliotis discus hannai.
3.2 Soft-body composition
The soft-body composition of abalone fed varying inclusion levels of SH is shown in Table 3. The moisture content and crude lipid content of abalone were insignificantly influenced by experimental diets (P > 0.05). The crude protein content of abalone is significantly highest in SH0 (P < 0.05) followed by SH25 and SH50.
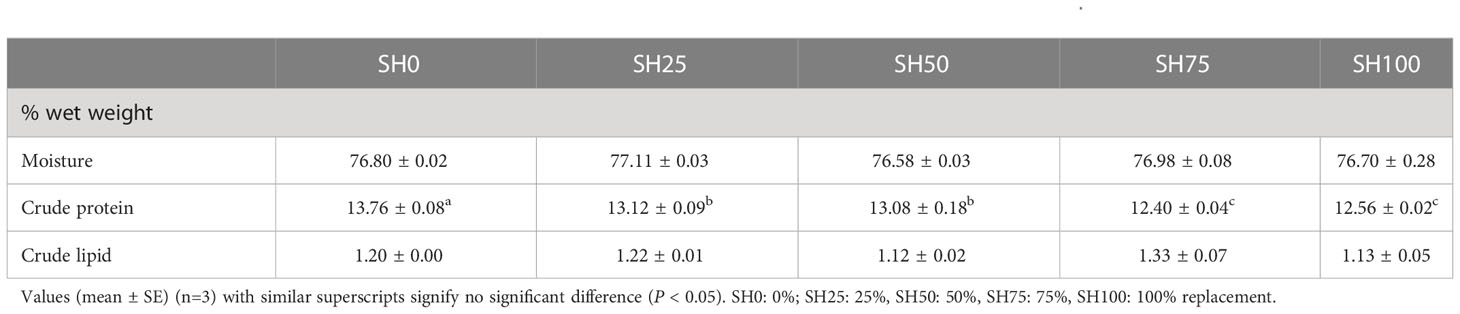
Table 3 Effects of Sargassum horneri inclusion levels on the body composition of abalone Haliotis discus hannai. .
3.3 Histology
The histological analysis of the digestive gland of abalone revealed a series of well-arranged digestive tubules, well-developed columnar epithelial layer, and digestive and basophilic cells with clear cell boundaries and visible nuclei. During ocular examination of histological samples, no discernible structural differences observed on abalone fed SH0, SH25 and SH50 diets. However, signs of digestive tubule degeneration, sloughing of the epithelial layer due to loss of structural integrity, and the nuclei of digestive cells becoming less prominent were observed in abalone fed the SH75 diet and more pronounced degradation in SH100-fed abalone (Figures 1A–E). The prominent structural integrity of the digestive tubule in the SH25-fed abalone is similarly presented (Figure 1F). In all of the experimental diets, the histological analysis of the abalone foot showed a prominent division between epithelial and muscular layers where mucous cells are sporadically situated. Additionally, no discernible difference was observed in the pedal histology of abalone-fed all experimental diets (data not shown).
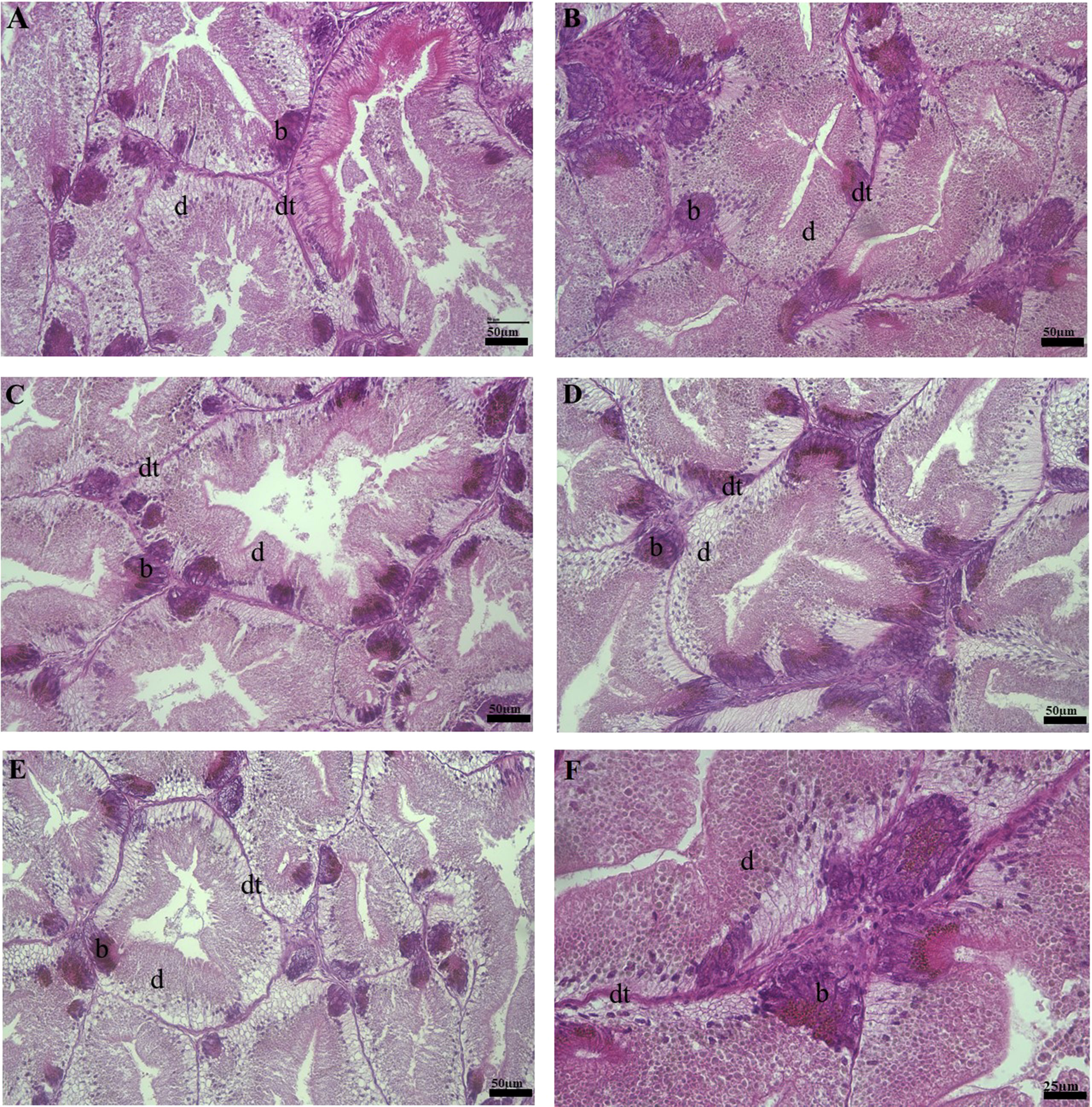
Figure 1 Histological photographs of abalone digestive gland fed experimental diets of varying dietary replacement of Sargassum horneri. Hematoxylin and Eosin staining method. (A) 0% (B) 25% (C) 50% (D) 75% (E) 100% replacement. 200× magnification. Scale bar = 50 µm. (F) Digestive gland of an abalone fed SH25 demonstrating structural integrity of the digestive tubule. 400× magnification. Scale bar = 25 µm. d, digestive cell; b, basophilic cell; dt, digestive tubule.
3.4 Gut microbiota profile
The relative abundance of gut microbial communities of abalone-fed experimental diets is shown in Figure 2. At the phylum level, regardless of diets, the top three phyla that predominated the gut of abalone are Fusobacteria, Tenericutes, and Proteobacteria, albeit difference in abundance is visible. The abalone fed of the diet SH0, SH100, and SH50 had the highest relative abundance of these bacterial phyla, in respective order. Meanwhile, Psychrilyobacter, Vibrio, and Mycoplasma genera predominantly occupied the gut of the abalone. Although the gut bacterial composition of the abalone-fed experimental diets was similar, there is a visible difference in the relative abundance of these bacterial florae. The α-diversity indices of the intestinal bacterial communities of abalone fed varying SH inclusion are shown in Table 4. The bacterial richness (ACE, CHAO) and diversity (Shannon and Simpson) index analyses revealed a range of values corresponding to experimental diets; however, no significant difference was calculated (P > 0.05). The PCoA based on the weighted UniFrac metrics is shown in two- and three-dimensional space (Figures 3A, B). In 3-dimensional coordinate analysis, PC1 explains 57.12% variances, PC2 explains 30.12% variances and PC3 explains 8.71% variances totaling to 96.13% total variability. In the two-dimensional analysis, PC1 (57.21%) and PC2 (30.12%) explains 87.24% total variability. The high percentages of total system variance (96.13% and 87.14%) mean high expression of microbial diversity information from the total set of data generated in the analysis. The overlapping of samples would indicate a similar bacterial composition and nonoverlapping would suggest otherwise. In beta diversity, no overlapping was observed and a clear boundary between experimental diets is found only between SH0 and SH100.
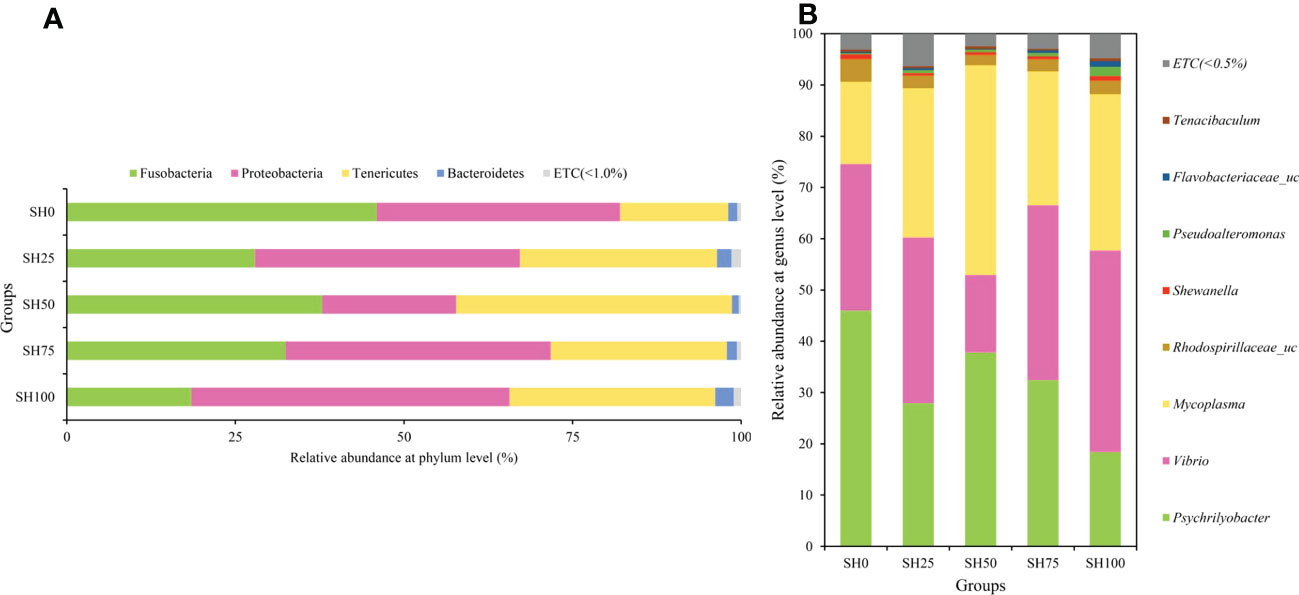
Figure 2 Relative abundance of microbial communities in the gut of abalone fed varying inclusion of Sargassum horneri in (A) phylum and (B) genus level. SH0: 0%, SH25: 25%, SH50: 50%, SH75: 75%, SH100: 100% replacement.
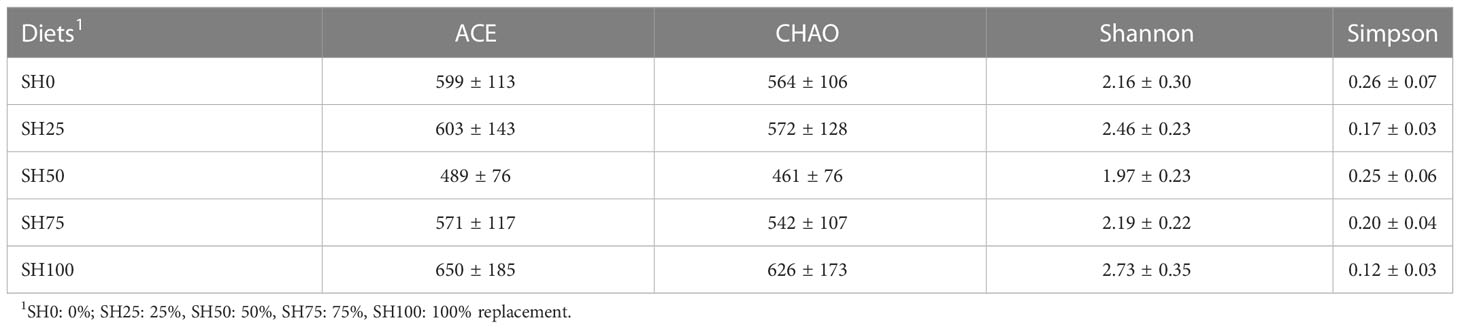
Table 4 Effect of Sargassum horneri replacement rate on α-diversity of the gut bacterial communities of abalone Haliotis discus hannai..
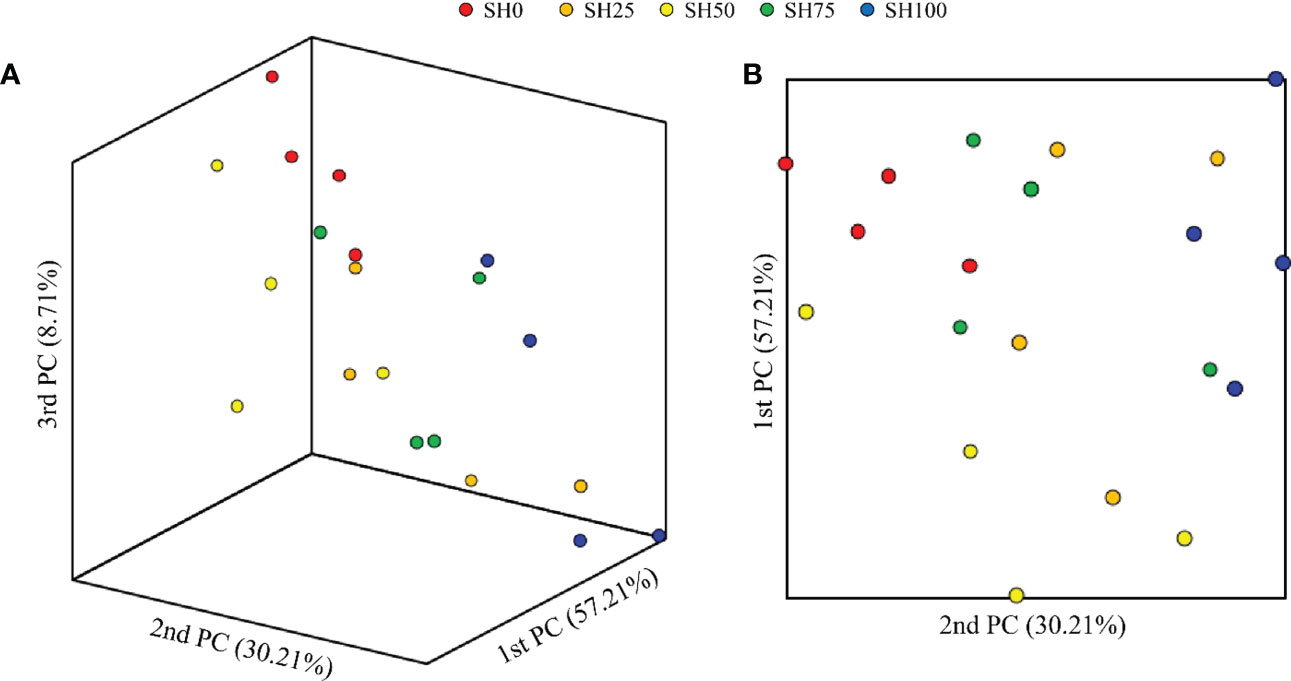
Figure 3 Principal Coordinate Analysis (PCoA) based on the weighted unifrac metrics of bacterial operational taxonomic units among different experimental diets. (A) 3-dimensional space. (B) 2-dimensional space. SH0: 0%, SH25: 25%, SH50: 50%, SH75: 75%, SH100: 100% replacement.
3.5 Taste sensory characteristics
The multi-variate principal component analysis (PCA) plot (Figures 4A, B), taste screening radar (Figure 5) and response signals on the seven sensors (data not shown) were used to explain differences in the taste sensory characteristics of abalone fed experimental diets. In the two-dimensional space, PC1 reads 71.25% variability and PC2 reads 23.54% variability for a total of 94.79% variability. In the three-dimensional space, PC1, PC2 and PC3, respectively explains 71.25%, 23.54%, 4.52% variability totaling to 99.31% variability. The high percentages of total system variance (94.79% and 99.31%) mean high expression of sensor scores from the total set of scores generated in the analysis. Taste can be separated into two groups based on the shape and clustering created by scores in the PCA plots. SH0, SH25, and SH50 make up one group, while SH75 and SH100 make up the other. Nevertheless, the negative value of the discrimination index (-265) revealed that the overall taste characteristics of abalone are nearly similar and unlikely to have a significant difference from each other. In the taste screening matrix, relative signal intensities varied widely in general-purpose sensors but only a slight deviation was observed in umami, saltiness and sourness sensors. These signal data made it possible to sort the specific taste of the abalone meat. The relative signal intensities of the various abalone AM samples varied across each of the seven taste sensors, and all of the samples exhibited similar responses with minimal deviation in every sensor. The PCA analysis, taste screening radar and sensor values allowed overall discrimination of taste sensory profile of abalone following various S. horneri inclusion in the diet.
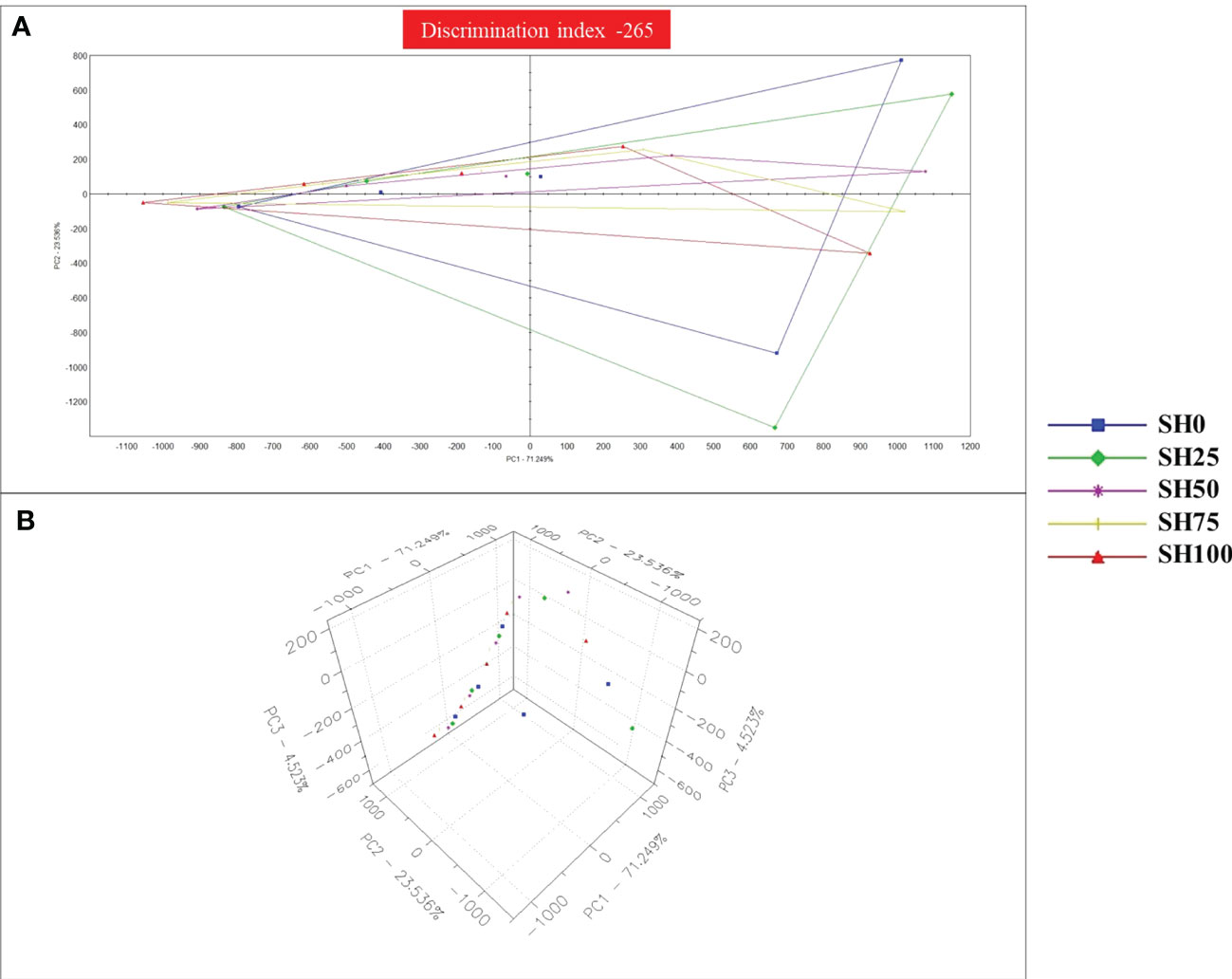
Figure 4 Principal Component Analysis (PCA) plot of the response signals on the electronic tongue by abalone’s adductor muscle as affected by Sargassum horneri inclusion levels in diets. (A) 2-dimensional space. (B) 3-dimensional space. SH0: 0%, SH25: 25%, SH50: 50%, SH75: 75%, SH100: 100% replacement.
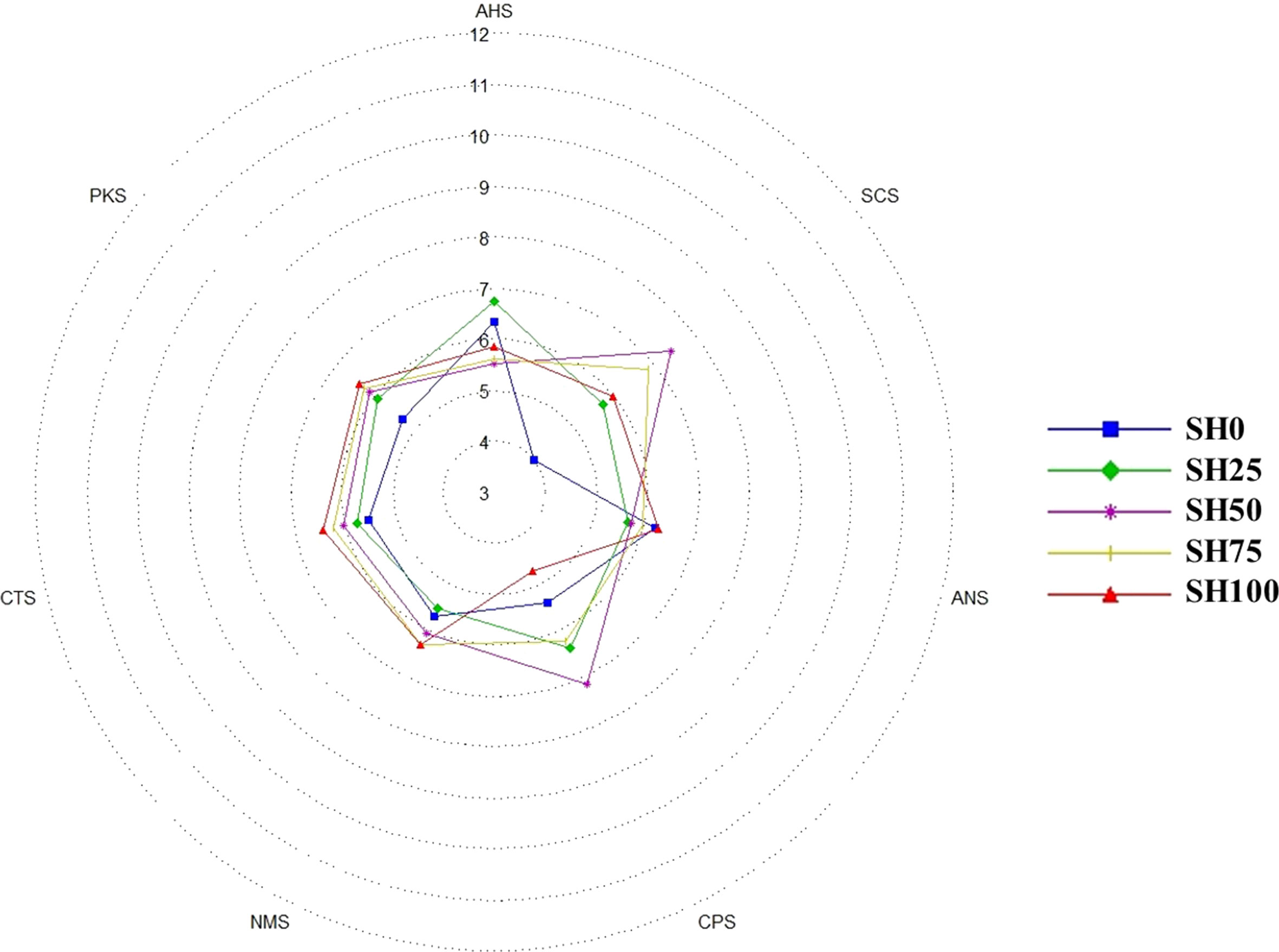
Figure 5 Radar map of relative taste intensity of abalone meat using the e-tongue for different Sargassum horneri inclusion levels in diets. SH0: 0%, SH25: 25%, SH50: 50%, SH75: 75%, SH100: 100% replacement.
4 Discussion
Abalone as an herbivorous marine organism primarily consumes seaweed as source of energy and overall nutrition (Bautista-Teruel et al., 2011), thus macroalgae are vital ingredient in feed formulation for the animal. In the present study, the highest shell growth (FSL and DISL) was recorded in SH25, and shell weight growth (FW, WGR, and WSGR) of up to SH50 is higher than in the control diet. These results present a significant increase in body growth of abalone after feeding dietary inclusion of SH up to a maximal level. This can be attributed to more balanced nutrition available to the organism compared to a mono-macroalgal-based formulated diet, correspondingly with the results of other studies (Viera et al., 2011; Yu et al., 2014; Bansemer et al., 2016a; Jang et al., 2018; Ansary et al., 2019a). The highly suggested reason is that a single species would compensate for low or absent nutritional components of each other, which would result in better attainment of the animal’s nutritional needs (Kemp et al., 2015). Additionally, the ideal combination of macroalgal diets would offer abalone a well-balanced amino acid and overall nutritional profiles (Bullon et al., 2020). The availability of essential amino acids (EAA) is known to positively correlate with better growth (Mai et al., 1994; Bansemer et al., 2016b) and although we were not able to determine the amino acid profile of the diet and the animal, the addition of S. horneri would be expected to improve EAA profile and growth performance. In the study of Ansary et al. (2019a), up to 60% inclusion of SH was reported to have the best beneficial effect on the growth of abalone, and higher would result to less growth performance. In the present study, 75% and 100% of SH had an adverse effect on the growth performance of abalone. These presents the low value of SH when given in high amounts or given in full replacement. Zhu et al. (2017), in their study on white spotted snapper Lutjanus stellatus, reported that a high replacement ratio of SH in the diet reduced the digestive enzyme activities of the animal. Moreover, species under the genus Sargassum are also known to exhibit anti-nutritional activity (Øverland et al., 2018) and Zhu et al. (2017) discussed that non-starch polysaccharides (NSPs), a type of anti-nutritional factors from seaweed produce digestive stress that negatively affected protein digestibility consequently affecting growth. These factors thus could partly explain the overall inferior growth of abalone-fed SH75 and SH100 diets.
Feed utilization is reflected in the growth performance of abalone. In the present study, FCR, FE, and PER were recorded similarly in 25% and 50% inclusion levels of S. horneri. This suggests that up to 50% replacements had satisfactory feed utilization values compared to higher replacement ratios. A lower FCR (close to 1) would mean higher feed efficiency. O'Mahoney et al. (2014) reported that a higher feed efficiency would mean better food potential for abalone. The increasing feed proportion (up to 50%) of S. horneri in the experimental diets improved PER of abalone which is indicative of increased protein availability, as previously stipulated in the study of Kemp et al. (2015).
One of the factors that affect the growth of the abalone is the transformation of feed nutrients to the body tissues (e.g. protein deposition) (Bansemer et al., 2016a; Bullon et al., 2022). The moisture and lipid content of the abalone soft-body was not affected by any dietary inclusion of S. horneri. This conforms with the previous study by Ansary et al. (2019a). The results showed that the increased dietary inclusions of S. horneri were associated with a decreasing trend in body protein. This is in contrast with the study of Ansary et al. (2019a) that reported no distinct trend between S. horneri inclusion and body protein content of abalone. Based on feed formulation, SH75 and SH100 diets had relatively higher protein content compared to other diets, yet the abalone ingested in these diets had significantly lower protein content, and can be partly because of their low feed intake and utilization. Conversely, the possible accumulation of NSPs could also result in reduced digestibility of protein (Zhu et al., 2017). Interestingly, the abalone fed basal diet had the highest protein content among experimental diets. The protein content and feed utilization values (FCR, FE, and FCR) in SH0 are nearly similar in SH25 and SH50, so to discuss based on these factors alone would be inadequate and unconvincing. In the current paper, we also report the relative abundance of bacterial composition wherein the highest abundance of the bacterial genus Psychrilyobacter was detected in SH0-fed abalone. The bacterium Psychrilyobacter was recently shown to be a prominent protein degrader in necromass (Pelikan et al., 2021) and these bacteria might significantly help in protein catabolism in the organisms making them rapidly assimilated by the animal, which, in turn, could result in high nutrient deposition.
The digestive gland of abalone is made up of columnar epithelial cells arranged into branching tubules and contains secretory or basophilic cells and digestive gland cells. These cells play roles in the production and accumulation of digestive enzymes and subsequently function during feed digestion, nutrient absorption, food storage, and excretion (Taïeb, 2001; Johnston et al., 2005; Zarai et al., 2011; Dennis et al., 2021). It is previously reported that dietary ingredients can affect digestive gland morphology and composition of abalone. For example, full replacement of fish meal with soybean meal in the diet produces vacuole accumulation and gradual dissolving of digestive cell nuclei (Yu et al., 2022) and the digestive cells of kelp Egregia menziesii-fed juvenile red abalone H. rufescens have an abundant presence of proteoglycan granules, which were correlated with faster growth performance (Serviere-Zaragoza et al., 2016). In the present study, morphological alterations might partly affect digestive enzyme secretion and negatively influenced nutrient absorption in the abalone-fed SH75 and SH100 diets. Similarly, digestive tissue degradation is proof of compromised nutrition from the limited ability to ingest food (Johnston et al., 2005).
Microorganisms are known to play a significant function during digestion, nutrient absorption, and immunity in a wide range of aquatic fauna (Tanaka et al., 2003; Pérez et al., 2010; Zhao et al., 2018). Among others, food is one of the exogenous factors that affect gut microflora and subsequently affects the overall health and growth of the animal (Tanaka et al., 2003; Cicala et al., 2018). In the present study, the varying levels of inclusion of SH seemed to affect bacterial abundance but not the complexity of bacterial communities. The wide range of scores calculated in the richness estimates indicators (ACE, CHAO) and diversity estimates indicators (Shannon and Simpson) suggests heterogeneity in both the number of species present and their abundance. The PCoA analyses revealed nonoverlapping of bacterial taxonomic units, and distances within and among experimental diets are showing the complex and varied microbial communities of abalone.
The most abundant microbial phyla in the present study are Fusobacteria, Tenericutes, and Proteobacteria. These phyla are known to inhabit the intestine of abalone in abundance as shown in previous studies. Choi et al. (2021) showed that 89.29% of the total bacterial population of large abalone is composed of these phyla. Nel et al. (2017) reported similar bacteria phyla that predominantly occupied the gut of abalone H. midae fed kelp Ecklonia maxima. Wu et al. (2022) reported Firmicutes and Proteobacteria are the most abundant phyla in H. discuss hannai fed various inclusion of corn meal as a substitute to fish meal. The high relative abundance of Psychrilyobacter, Vibrio, and Mycoplasma at the genus level is also consistent with the known bacterial genus that predominantly inhabits the intestine of the abalone (Cicala et al., 2018; Gobet et al., 2018; Choi et al., 2021; Wu et al., 2022). The positive association between Psychrilyobacter and abalone growth was reported in fast-growing abalone highlighting the possible role in degrading undigested oligopolysaccharides (Choi et al., 2021). Recently, this bacterial genus was shown to be an excellent protein degrader in necromass and might help in protein catabolism in the animal (Pelikan et al., 2021), however, the exact function of this bacteria in abalone needs to be further studied. Mycoplasma is abundant in H. fulgens and H. corrugata (Cicala et al., 2018) and H. discus hannai (Wu et al., 2022), and may play essential roles in the metabolic functions of the animal such as catabolism of complex substances such as glycans, proteins, and oligosaccharides (Wang et al., 2016; Cicala et al., 2018). The bacterial species in the genus Vibrio function in the degradation of complex polysaccharides by producing exogenous digestive enzymes and also helps in nutrient digestibility (Sawabe et al., 2003; Tanaka et al., 2016; Nel et al., 2017; Choi et al., 2021). The high abundance of these bacterial genera may have partly contributed to the food digestion and nutrient absorption and subsequently to the growth of the abalone. The high relative composition of Psychrilobacter in SH0 and low in SH100 is thus hypothesized to affect nutrient utilization and indirectly induce growth performance. The highest growth rate observed can be attributed to a more balanced bacterial composition in the SH25 diet.
One of the factors that significantly affect the taste sensory quality of cultured animals is feed (Brown et al., 2008; Smit et al., 2010; Bansemer et al., 2016b). For example, Chiou and Lai (2002) described a taste preference of cooked abalone-fed artificial diets over those fed with macroalgae Gracilaria sp. Smit et al. (2010) showed that an apparent difference in color, texture, and aroma intensity in raw abalone was attributed to the diets consumed. Preece (2006) showed no distinguishing flavor preference for abalone fed on two commercial diets and between cultivated and wild abalone. The contrasting relationship between feed and abalone taste profile in published works could be explained by the use of human resources as trained panelists to assess the taste quality of seafood, which is posed by several practical and economic limitations. These include intrinsic subjectivity of how the panel members perceived the product, the scarce number of trained panelists, and the cost involved in the training (Smit et al., 2010; Marx et al., 2021). These, in turn, may result in inferences that are inconsistent and challenging to reproduce (Mabuchi et al., 2018). In the current study, we present the first report on establishing the link between dietary ingredient in formulated feed and taste sensory attributes of abalone meat using an e-tongue. The e-tongue is a sensor-based technology that mimics how the human tongue perceived the five basic taste sensations namely - bitter, salty, sour, sweet, and umami (Marx et al., 2021). The application of e-tongue on taste screening of seafood has been studied, albeit scarce. For instance, Zhang et al. (2012) identified flavor peptides from puffer fish (Takifugu obscurus). Mabuchi et al. (2018) noted taste differences among white-flesh fish species in terms of sourness, acidic bitterness, umami, and saltiness, and elucidated the role played by phosphoric acid in determining taste attributes. Hu et al. (2020) detected similar taste profiles of different fish soups sourced from the different body regions (back, abdomen and tail) of silver carp (Hypophthalmichthys molitrix). In the present study, e-tongue was used to discriminate the taste profile of abalone concerning the dietary replacement of S. horneri in the diet. The negative value of the discrimination index and overlapping of sensor values in the two-dimensional PCA would give evidence that abalone has similar taste characteristics and is overall independent of the amount of S. horneri inclusion in the diet. Additionally, sensor values of AHS, CTS, and NMS that correspond to sourness, saltiness, and umami flavors in the taste screening matrix are clustered closely and improbably suggest a significant difference in taste. The similar response signals in each of the seven sensors further cemented that abalone fed experimental diets had a comparable taste profile. Collectively considered, an e-tongue is a useful instrument for identifying the flavor characteristics of abalone meat and that a single macroalgal food source is unlikely to significantly alter the overall taste of abalone. This agrees with the study of Mørkøre et al. (2007) wherein a single dietary ingredient (100% fish oil and 40% replaced by soybean oil) showed no overall significant preference in Atlantic cod (Gadus morhua).
Conclusion
In this study, we confirmed the food value of SH as additional macroalgal biomass for abalone. The growth was enhanced, the gut was similarly occupied by microbial communities and no distinct change in taste profile was recognized. Taken together, S. horneri can be included in the feed formulation for abalone aquaculture up to 50% of macroalgal content without significantly impairing normal physiological processes and meat characteristics.
Data availability statement
The datasets presented in this article are not readily available because NONE. Requests to access the datasets should be directed to Y-HC,dW5pY2hvaUBwa251LmFjLmty.
Ethics statement
The experiment was performed following the guidelines of the Korean Association for Laboratory Animals (approval no. 18-0680, 2018).
Author contributions
S-WH: methodology, investigation, conceptualization, writing-original draft. JC: investigation, formal analysis, writing-original draft. SL: investigation. J-HL: formal analysis, investigation. S-JP: formal analysis, resources. W-JJ: formal analysis. Y-HC: supervision, writing-review and editing, funding acquisition. All authors contributed to the article and approved the submitted version.
Funding
This research was supported by grant from National Institute of Fisheries Science, Republic of Korea (R2023037).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Øverland M., Mydland L. T., Skrede A. (2018). Marine macroalgae as sources of protein and bioactive compounds in feed for monogastric animals. J. Sci. Food Agric. 99, 13–24. doi: 10.1002/jsfa.9143
Alcantara L., Noro T. (2006). Growth of the abalone Haliotis diversicolor (Reeve) fed with macroalgae in floating net cage and plastic tank. Aquac. Res. 37, 708–717. doi: 10.1111/j.1365-2109.2006.01484.x
Ansary M., Rahman W., Baek S. I., Jeong H. S., Lee K. W., Cho S. H., et al. (2019b). Substitution effect of the combined fouling macroalgae Ulva australis and Sargassum horneri for Undaria pinnatifida in formulated diets on growth and body composition of juvenile abalone (Haliotis discus, reeve 1846) subjected to air exposure stressor. J. Appl. Phycol. 31, 3245–3254. doi: 10.1007/s10811-019-01812-x
Ansary M., Rahman W., Jeong H. S., Lee K. W., Kim H. S., Kim J., et al. (2019a). The effect of substituting Undaria pinnatifida in formulated feeds with Sargassum horneri on growth and body composition of juvenile abalone (Haliotis discus, reeve 1846). J. Appl. Phycol. 31, 2125–2132. doi: 10.1007/s10811-018-1672-2
AOAC (1995). “Official methods of analysis of AOAC international,” in Official analytical chemists, 16th (Arlington, VA, USA: AOAC International), 1141.
Bansemer M. S., Qin J. G., Harris J. O., Duong D. N., Currie K. L., Howarth G. S., et al. (2016a). Dietary inclusions of dried macroalgae meal in formulated diets improve the growth of greenlip abalone (Haliotis laevigata). J. Appl. Phycol. 28, 3645–3658. doi: 10.1007/s10811-016-0829-0
Bansemer M. S., Qin J. G., Harris J. O., Howarth G. S., Stone D. A. (2016b). Nutritional requirements and use of macroalgae as ingredients in abalone feed. Rev. Aquac. 8 (2), 121–135. doi: 10.1111/raq.12085
Bautista-Teruel M. N., Koshio S. S., Ishikawa M. (2011). Diet development and evaluation for juvenile abalone, Haliotis asinina linne: Lipid and essential fatty acid levels. Aquac. 312, 172–179. doi: 10.1016/j.aquaculture.2011.01.004
Brown M. R., Sikes A. L., Elliott N. G., Tume R. K. (2008). Physicochemical factors of abalone quality: a review. J. Shellfish Res. 27 (4), 835–842. doi: 10.2983/0730-8000
Bullon N., Seyfoddin A., Alfaro A. C. (2022). The role of aquafeeds in abalone nutrition and health: A comprehensive review. J. World Aquac Soc, 1–25. doi: 10.1111/jwas.12883
Byeon S. Y., Oh H. J., Kim S., Yun S. H., Kang J. H., Park S. R., et al. (2019). The origin and population genetic structure of the ‘golden tide’ seaweeds, Sargassum horneri, in Korean waters. Sci. Rep. 9, 1–13. doi: 10.1038/s41598-019-44170-x
Chao A., Bunge J. (2002). Estimating the number of species in a stochastic abundance model. Biometrics 58 (3), 531–539. doi: 10.1111/j.0006-341X.2002.00531.x
Chiou T. K., Lai M. M. (2002). Comparison of taste components in cooked meats of small abalone fed different diets. Fish. Sci. 68 (2), 388–394. doi: 10.1046/j.1444-2906.2002.00437.x
Cho S. H. (2010). Effect of fishmeal substitution with various animal and/or plant protein sources in the diet of the abalone Haliotis discus hannai ino. Aquac. Res. 41 (10), e587–e593. doi: 10.1111/j.1365-2109.2010.02561.x
Choi M. J., Oh Y. D., Kim Y. R., Lim H. K., Kim J. M. (2021). Intestinal microbial diversity is higher in pacific abalone (Haliotis discus hannai) with slower growth rates. Aquac. 537, 736500. doi: 10.1016/j.aquaculture.2021.736500
Cicala F., Cisterna-Céliz J. A., Moore J. D., Rocha-Olivares A. (2018). Structure, dynamics and predicted functional role of the gut microbiota of the blue (Haliotis fulgens) and yellow (H. corrugata) abalone from Baja California sur, Mexico. PeerJ 6, e5830. doi: 10.7717/peerj.5830
Cook P. A. (2014). The worldwide abalone industry. Mod. Econ. 5, 1181–1186. doi: 10.4236/me.2014.513110
Cook P. A. (2016). Recent trends in worldwide abalone production. J. Shellfish Res. 35, 581–583. doi: 10.2983/035.035.0302
Cook P. A. (2019). Worldwide abalone production statistics. J. Shellfish Res. 38, 401–404. doi: 10.2983/035.038.0222
Dang V. T., Li Y., Speck P., Benkendorff K. (2011). Effects of micro and macroalgal diet supplementations on growth and immunity of greenlip abalone, Haliotis laevigata. Aquac. 320, 91–98. doi: 10.1016/j.aquaculture.2011.08.009
Dennis M. M., Molnár K., Kriska G., Lőw P. (2021). “Mollusca: Gastropoda,” in Invertebrate histology. Ed. LaDouceur E. E. B. Hoboken, New Jersey, USA. Wiley-Blackwell 87–132. doi: 10.1002/9781119507697.ch4
Dlaza T. S., Maneveldt G. W., Viljoen C. (2008). Growth of post-weaning abalone Haliotis midae fed commercially available formulated feeds supplemented with fresh wild seaweed. Afr. J. Mar. Sci. 30, 199–203. doi: 10.2989/AJMS.2008.30.1.22.472
Gobet A., Mest L., Perennou M., Dittami S. M., Caralp C., Coulombet C., et al. (2018). Seasonal and algal diet-driven patterns of the digestive microbiota of the European abalone Haliotis tuberculata, a generalist marine herbivore. Microbiome 6 (1), 1–14. doi: 10.1186/s40168-018-0430-7
Hamady M., Lozupone C., Knight R. (2010). Fast UniFrac: facilitating high-throughput phylogenetic analyses of microbial communities including analysis of pyrosequencing and PhyloChip data. ISME J. 4 (1), 17–27. doi: 10.1038/ismej.2009.97
Hu Z., Tong Y., Manyande A., Du H. (2020). Effective discrimination of flavours and tastes of Chinese traditional fish soups made from different regions of the silver carp using an electronic nose and electronic tongue. Czech J. Food Sci. 38 (2), 84–93. doi: 10.17221/103/2018-CJFS
Hwang E. K., Lee S. J., Ha D. S., Park C. S. (2016). Sargassum golden tides in the shinan-gun and jeju island, Korea. Korean J. Fish Aquat Sci. 49, 689–693. doi: 10.5657/KFAS.2016.0689
Hwang E. K., Park C. S. (2020). Seaweed cultivation and utilization of Korea. Algae 35, 107–121. doi: 10.4490/algae.2020.35.5.15
Jang B., Kim P. Y., Kim H. S., Lee K. W., Kim H. J., Choi D. G., et al. (2018). Substitution effect of sea tangle (ST) (Laminaria japonica) with tunic of sea squirt (SS) (Halocynthia roretzi) in diet on growth and carcass composition of juvenile abalone (Haliotis discus, reeve 1846). Aquac. Nutri. 24, 586–593. doi: 10.1111/anu.12593
Jayasundar R., Singh A., Kumar D. (2021). Challenges in using electronic tongue to study rasa of plants: I. finding the right tool for the right job. J. Ayurveda Integr. Med. 12 (2), 234–237. doi: 10.1016/j.jaim.2020.12.011
Johnston D., Moltschaniwskyj N., Wells J. (2005). Development of the radula and digestive system of juvenile blacklip abalone (Haliotis rubra): potential factors responsible for variable weaning success on artificial diets. Aquac. 250, 341–355. doi: 10.1016/j.aquaculture.2005.03.012
Kemp J. O. G., Britz P. J., Agüero P. T. (2015). The effect of macroalgal, formulated and combination diets on growth, survival and feed utilisation in the red abalone Haliotis rufescens. Aquac. 448, 306–314. doi: 10.1016/j.aquaculture.2015.06.016
Kim H. S., Sanjeewa K. K., Fernando I. P., Ryu B., Yang H. W., Ahn G., et al. (2018). A comparative study of Sargassum horneri Korea and China strains collected along the coast of jeju island south Korea: its components and bioactive properties. Algae 33, 341–349. doi: 10.4490/algae.2018.33.11.15
Kirkendale L., Robertson-Andersson D., Winberg P. C. (2010) Review on the use and production of algae and manufactured diets as feed for sea-based abalone aquaculture in Victoria. Available at: http://ro.uow.edu.au/smfc/7/ (Accessed June 15, 2021).
Lee K. W., Kim H. S., Yun A., Choi D. G., Jang B. I., Kim H. J., et al. (2016). Effect of the formulated diets on performance and resistance of juvenile abalone [Haliotis discus (Reeve 1846)] subjected to various stress conditions. J. Shellfish Res. 35, 481–491. doi: 10.2983/035.035.0221
Mabuchi R., Ishimaru A., Tanaka M., Kawaguchi O., Tanimoto S. (2018). Metabolic profiling of fish meat by GC-MS analysis, and correlations with taste attributes obtained using an electronic tongue. Metabolites 9 (1), 1–11. doi: 10.3390/metabo9010001
Mai K., Mercer J. P., Donlon J. (1994). Comparative studies on the nutrition of two species of abalone, Haliotis tuberculata l. and Haliotis discus hannai ino: II. amino acid composition of abalone and six species of macroalgae with an assessment of their nutritional value. Aquac. 128 (1-2), 115–130. doi: 10.1016/0044-8486(94)90107-4
Marx Í.M., Veloso A. C., Casal S., Pereira J. A., Peres A. M. (2021). “Sensory analysis using electronic tongues,” in Innovative food analysis. Ed. Galanakis C. M. (London:England: Academic Press), 323–343.
Mørkøre T., Netteberg C., Johnsson L., Pickova J. (2007). Impact of dietary oil source on product quality of farmed Atlantic cod, Gadus morhua. Aquac. 267, 236–247. doi: 10.1016/j.aquaculture.2007.01.033
Mulvaney W. J., Winberg P. C., Adams L. (2013). Comparison of macroalgal (Ulva and grateloupia spp.) and formulated terrestrial feed on the growth and condition of juvenile abalone. J. Appl. Phycol. 25, 815–824. doi: 10.1007/s10811-013-9998-2
Nel A., Pletschke B. I., Jones C. L. W., Kemp J., Robinson G., Britz P. J. (2017). Effects of kelp Ecklonia maxima inclusion in formulated feed on the growth, feed utilisation and gut microbiota of south African abalone Haliotis midae. Afr. J. Mar. Sci. 39, 183–192. doi: 10.2989/1814232X.2017.1338203
O'Mahoney M., Rice O., Mouzakitis G., Burnell G. (2014). Towards sustainable feeds for abalone culture: evaluating the use of mixed species seaweed meal in formulated feeds for the Japanese abalone, Haliotis discus hannai. Aquac. 430, 9–16. doi: 10.1016/j.aquaculture.2014.02.036
Pang S. J., Liu F., Shan T. F., Gao S. Q., Zhang Z. H. (2009). Cultivation of the brown alga Sargassum horneri: sexual reproduction and seedling production in tank culture under reduced solar irradiance in ambient temperature. J. Appl. Phycol. 21, 413–422. doi: 10.1007/s10811-008-9386-5
Park C. J., Kim S. Y. (2013). Abalone aquaculture in Korea. J. Shellfish Res. 32, 17–19. doi: 10.2983/035.032.0104
Pelikan C., Wasmund K., Glombitza C., Hausmann B., Herbold C. W., Flieder M., et al. (2021). Anaerobic bacterial degradation of protein and lipid macromolecules in subarctic marine sediment. ISME J. 15, 833–847. doi: 10.1038/s41396-020-00817-6
Pérez T., Balcázar J. L., Ruiz-Zarzuela I., Halaihel N., Vendrell D., De Blas I., et al. (2010). Host–microbiota interactions within the fish intestinal ecosystem. Mucosal Immunol. 3, 355–360. doi: 10.1038/mi.2010.12
Porturas Olaechea R., Ushio H., Watabe S., Takada K., Hatae K. (1993). Toughness and collagen content of abalone muscles. Biosci. Biotechnol. Biochem. 57, 6–11. doi: 10.1271/bbb.57.6
Preece M. A. (2006). Sensory qualities of the new Zealand abalone, Haliotis iris, reared in offshore structures on artificial diets. N.Z.J. Mar. Freshwater Res. 40, 2, 223–226. doi: 10.1080/00288330.2006.9517416
Sanjeewa K. A., Lee J. S., Kim W. S., Jeon Y. J. (2017). The potential of brown-algae polysaccharides for the development of anticancer agents: An update on anticancer effects reported for fucoidan and laminarin. Carbohydr. Polym. 177, 451–459. doi: 10.1016/j.carbpol.2017.09.005
Sawabe T., Setoguchi N., Inoue S., Tanaka R., Ootsubo M., Yoshimizu M., et al. (2003). Acetic acid production of Vibrio halioticoli from alginate: a possible role for establishment of abalone–V. halioticoli association. Aquac. 219, 671–679. doi: 10.1016/S0044-8486(02)00618-X
Schaefer E. N., Harris J. O., Howarth G. S., Bansemer M. S., Stone D. A. J. (2013). Comparative histological changes in the greenlip abalone Haliotis laevigata gastrointestinal tract in response to water temperature, different dietary protein levels, and animal age. J. Shellfish Res. 32, 131–141. doi: 10.2983/035.032.0119
Serviere-Zaragoza E., Pérez-Estrada C. J., Aldana Aranda D. (2016). Status of the digestive gland and feed index in juvenile green abalone Haliotis fulgens fed rehydrated macroalgae. Aquac. Nutr. 22, 767–775. doi: 10.1111/anu.12295
Shannon C. E. (1948). A mathematical theory of communication. Bell Syst. Tech. J. 27 (3), 379–423. doi: 10.1002/j.1538-7305.1948.tb01338.x
Smetacek V., Zingone A. (2013). Green and golden seaweed tides on the rise. Nature 504, 84–88. doi: 10.1038/nature12860
Smit A. J., Robertson-Andersson D. V., Bolton J. J. (2010). The effect of macroalgal and compound feeds on the sensory quality of cultivated south African abalone, Haliotis midae Linnaeus (Mollusca, Gastropoda). Aquac. Nutr. 16 (6), 590–603. doi: 10.1111/j.1365-2095.2009.00696.x
Su L., Shan T., Pang S., Li J. (2017). Analyses of the genetic structure of Sargassum horneri in the yellow Sea: implications of the temporal and spatial relations among floating and benthic populations. J. Appl. Phycol. 30, 1417–1424. doi: 10.1007/s10811-017-1296-y
Sun L., Guo Y., Ma S., Fan W., Liu Y., Liu D., et al. (2021). Replacement of dietary kelp meal with three macroalgae sources on the growth performance, immune responses and anti-stress capacity of abalone Haliotis discus hannai. J. Appl. Phycol. 33, 4051–4065. doi: 10.1007/s10811-021-02592-z
Taïeb N. (2001). Distribution of digestive tubules and fine structure of digestive cells of Aplysia punctata (Cuvier 1803). J. Molluscan Stud. 67, 169–182. doi: 10.1093/mollus/67.2.169
Takami H., Kawamura T. (2003). Dietary changes in the abalone, Haliotis discus hannai, and relationship with the development of the digestive organ. Jpn. Agric. Res. Q. 37 (2), 89–98. doi: 10.6090/jarq.37.89
Tanaka R., Shibata T., Miyake H., Mori T., Tamaru Y., Ueda M., et al. (2016). Temporal fluctuation in the abundance of alginate-degrading bacteria in the gut of abalone Haliotis gigantea over 1 year. Aquac. Res. 47 (9), 2899–2908. doi: 10.1111/are.12740
Tanaka R., Sugimura I., Sawabe T., Yoshimizu M., Ezura Y. (2003). Gut microflora of abalone Haliotis discus hannai in culture changes coincident with a change in diet. Fish. Sci. 69, 951–958. doi: 10.1046/j.1444-2906.2003.00712.x
Viera M. P., De Vicose G. C., Gómez-Pinchetti J. L., Bilbao A., Fernandez-Palacios H., Izquierdo M. S. (2011). Comparative performances of juvenile abalone (Haliotis tuberculata coccinea reeve) fed enriched vs non-enriched macroalgae: Effect on growth and body composition. Aquac. 319, 423–429. doi: 10.1016/j.aquaculture.2011.07.024
Wang G., Sun J., Liu G., Wang L., Yu J., Liu T., et al. (2014). Comparative analysis on transcriptome sequencings of six Sargassum species in China. Acta Oceanol. Sin. 33, 37–44. doi: 10.1007/s13131-014-0439-0
Wang Y., Huang J. M., Wang S. L., Gao Z. M., Zhang A. Q., Danchin A., et al. (2016). Genomic characterization of symbiotic mycoplasmas from the stomach of deep-sea isopod Bathynomus sp. Environ. Microbiol. 18 (8), 2646–2659. doi: 10.1111/1462-2920.13411
Wang Z., Li J., Hong X., Han X., Liu B., Li X., et al. (2021). Taste masking study based on an electronic tongue: The formulation design of 3D printed levetiracetam instant-dissolving tablets. Pharm. Res. 38 (5), 831–842. doi: 10.1007/s11095-021-03041-9
Wu Z., Yu X., Guo J., Fu Y., Guo Y., Pan M., et al. (2022). Effects of replacing fish meal with corn gluten meal on growth performance, intestinal microbiota, mTOR pathway and immune response of abalone Haliotis discus hannai. Aquacul. Rep. 23, 101007. doi: 10.1016/j.aqrep.2022.101007
Yu S. N., Duan Z. L., Huang B., Wang Y. L., Wang X. B., Li Y., et al. (2014). Effects of different diet combinations on the growth of juvenile abalone (Haliotis asinina Linnaeus). Anim. Feed Sci. Tech. 194, 106–112. doi: 10.1016/j.anifeedsci.2014.04.018
Yu X., Wu Z., Guo J., Fu Y., Luo K., Guo Y., et al. (2022). Replacement of dietary fish meal by soybean meal on growth performance, immunity, anti-oxidative capacity and mTOR pathways in juvenile abalone Haliotis discus hannai ino. Aquac. 551, 1–12. doi: 10.1016/j.aquaculture.bulo.737914
Zarai Z., Boulais N., Karray A., Misery L., Bezzine S., Rebai T., et al. (2011). Immunohistochemical localization of hepatopancreatic phospholipase A2 in Hexaplex trunculus digestive cells. Lipids Health Dis. 10, 1–9. doi: 10.1186/1476-511X-10-91
Zhang J., Shang D., Wang W., Jiang Z., Xue S., Fang J. (2010). The potential for utilizing fouling macroalgae as feed for abalone Haliotis discus hannai. Aquac. Res. 41, 1770–1777. doi: 10.1111/j.1365-2109.2009.02478.x
Zhang L., Zhao C., Shi D., Hu W., Wei J., Chang Y. (2017). Gulfweed Sargassum horneri is an alternative diet for aquaculture of juvenile sea urchins Strongylocentrotus intermedius in summer. Aquac. Int. 25, 905–914. doi: 10.1007/s10499-016-0088-8
Zhang M. X., Wang X. C., Liu Y., Xu X. L., Zhou G. H. (2012). Isolation and identification of flavour peptides from puffer fish (Takifugu obscurus) muscle using an electronic tongue and MALDI-TOF/TOF MS/MS. Food Chem. 135 (3), 1463–1470. doi: 10.1016/j.foodchem.2012.06.026
Zhao J., Ling Y., Zhang R., Ke C., Hong G. (2018). Effects of dietary supplementation of probiotics on growth, immune responses, and gut microbiome of the abalone Haliotis diversicolor. Aquac. 493, 289–295. doi: 10.1016/j.aquaculture.2018.05.011.
Keywords: Sargassum horneri, abalone aquaculture, Haliotis discus hannai, growth, gut microbiota, taste profile
Citation: Hur S-W, Cadangin J, Lee S, Lee J-H, Park S-J, Jang W-J and Choi Y-H (2023) Dietary replacement of Undaria pinnatifida by Sargassum horneri in feed formulation for abalone Haliotis discus hannai: Effect on growth, gut microbiota, and taste sensory profile. Front. Mar. Sci. 10:1053240. doi: 10.3389/fmars.2023.1053240
Received: 25 September 2022; Accepted: 09 January 2023;
Published: 01 February 2023.
Edited by:
Eduardo Almansa, Spanish Institute of Oceanography (IEO), SpainReviewed by:
Ana Pombo, Center for Marine and Environmental Sciences (MARE- IPLeiria), PortugalFucun Wu, Institute of Oceanology, Chinese Academy of Sciences (CAS), China
Copyright © 2023 Hur, Cadangin, Lee, Lee, Park, Jang and Choi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Youn-Hee Choi, dW5pY2hvaUBwa251LmFjLmty
†These authors have contributed equally to this work and share first authorship
 Sang-Woo Hur1†
Sang-Woo Hur1† Josel Cadangin
Josel Cadangin Seunghan Lee
Seunghan Lee Won-Je Jang
Won-Je Jang Youn-Hee Choi
Youn-Hee Choi