- 1School of Environmental and Geographical Sciences, University of Nottingham Malaysia Campus, Semenyih, Selangor, Malaysia
- 2Institute of Biology, College of Science, University of the Philippines Diliman, Quezon City, Philippines
- 3Centre of Advanced Study in Marine Biology, Faculty of Marine Sciences, Annamalai University, Portonovo, India
- 4Institute of Biological Sciences, Faculty of Science, Universiti Malaya, Kuala Lumpur, Malaysia
- 5Center of Excellence for Mangrove, Universitas Sumatera Utara, Medan, Indonesia
- 6University Institute of Fisheries and Aquaculture, University Cheikh Anta Diop, Dakar, Senegal
- 7Dept. of Fisheries Biology and Genetics, Faculty of Fisheries, Aquaculture & Marine Science, Sher-e-Bangla Agricultural University, Dhaka, Bangladesh
- 8Iriomote Station, Tropical Biosphere Research Center, University of the Ryukyus, Uehara, Taketomi, Yaeyama, Okinawa, Japan
- 9College of Marine and Allied Sciences, Mindanao State University at Naawan, Naawan, Misamis Oriental, Philippines
- 10Department of Botany and Zoology, Stellenbosch University, Stellenbosch, South Africa
- 11Department of Zoology, Faculty of Science, University of Peradeniya, Peradeniya, Sri Lanka
- 12Department of Marine Science, Faculty of Science, Chulalongkorn University, Bangkok, Thailand
- 13Institute for Tropical Biology and Conservation, Universiti Malaysia Sabah, Kota Kinabalu, Sabah, Malaysia
Species detection using environmental DNA (eDNA) is a biomonitoring tool that can be widely applied to mangrove restoration and management. Compared to traditional surveys that are taxa-specific and time-consuming, eDNA metabarcoding offers a rapid, non-invasive and cost-efficient method for monitoring mangrove biodiversity and characterising the spatio-temporal distribution of multiple taxa simultaneously. General guidelines for eDNA metabarcoding are well-established for aquatic systems, but habitat-specific guidelines are still lacking. Mangrove habitats, as priority ecosystems for restoration in Southeast Asia, present unique prospects and challenges in these regards. Environmental DNA metabarcoding can be used to (1) track functional recovery in ecological restoration, (2) prioritise conservation areas, (3) provide early warning for threats, (4) monitor threatened taxa, (5) monitor response to climate change, and (6) support community-based restoration. However, these potential applications have yet been realized in Southeast Asia due to (1) technical challenges, (2) lack of standardised methods, (3) spatio-temporal difficulties in defining community, (4) data limitations, and (5) lack of funding, infrastructure and technical capacity. Successful implementation of eDNA metabarcoding in mangrove restoration activities would encourage the development of data-driven coastal management and equitable conservation programs. Eventually, this would promote Southeast Asia’s shared regional interests in food security, coastal defence and biodiversity conservation.
1 Introduction
Mangroves provide various ecosystem services and socio-economic benefits (Hochard et al., 2019; Spalding and Parrett, 2019; Friess et al., 2020). Mangroves are also recognized for their roles in climate change adaptation and mitigation (CCAM; Zari et al., 2019) and biodiversity conservation (da Rosa and Marques, 2022). The largest (32.2%; 43,767 km2; Spalding and Leal, 2021) and most diverse mangroves (> 50 species; Spalding, 2010) are found in Southeast Asia (SEA). Although mangrove degradation rate declined globally in the last 10 years, SEA remains a hotspot for mangrove loss (Spalding and Leal, 2021; Bhowmik et al., 2022), with Myanmar and the Philippines experiencing the highest rate of loss (Long et al., 2014; Estoque et al., 2018). Aquaculture is the main driver of loss in most SEA countries, though other drivers could be country-specific, e.g. rice farms in Myanmar, and oil palms in Malaysia and Indonesia (Richards & Friess, 2016). When mangroves are lost or degraded, the delivery of ecosystem services and biodiversity conservation are expected to decline (Sannigrahi et al., 2020).
Mangroves and SEA are therefore priority ecosystems and regions, respectively, for global CCAM and biodiversity conservation (Bunting et al., 2022). Mangrove restoration is advocated as a nature-based solution (NbS; Zimmer et al., 2022) and an integral component of the United Nations Decade on Ecosystem Restoration (Su et al., 2021). The region has already implemented several restoration programs since the 1990s (e.g. Walters, 2003; Ilman et al., 2011; Hamdan et al., 2014) although the efficacy was not systematically assessed and reported (Lee et al., 2019). Generally, when mangroves are restored, the vegetation and sediment conditions are expected to improve with age of the stands (Salmo et al., 2013), followed by improvements in the faunal assemblages and associated food web indicators (Salmo et al., 2017; Then et al., 2021; Basyuni et al., 2022). As more researchers move away from simple metrics of forest cover to assess the efficacy of restoration programs, regular monitoring and documentation of biodiversity becomes more important in mangrove restoration ecology (da Rosa and Marques, 2022). In this regard, conventional biodiversity monitoring approaches (e.g. faunal sampling, transect and plot techniques) remain vital to undoubtedly provide foundational empirical data but are time consuming and expensive (Taddeo and Dronova, 2018).
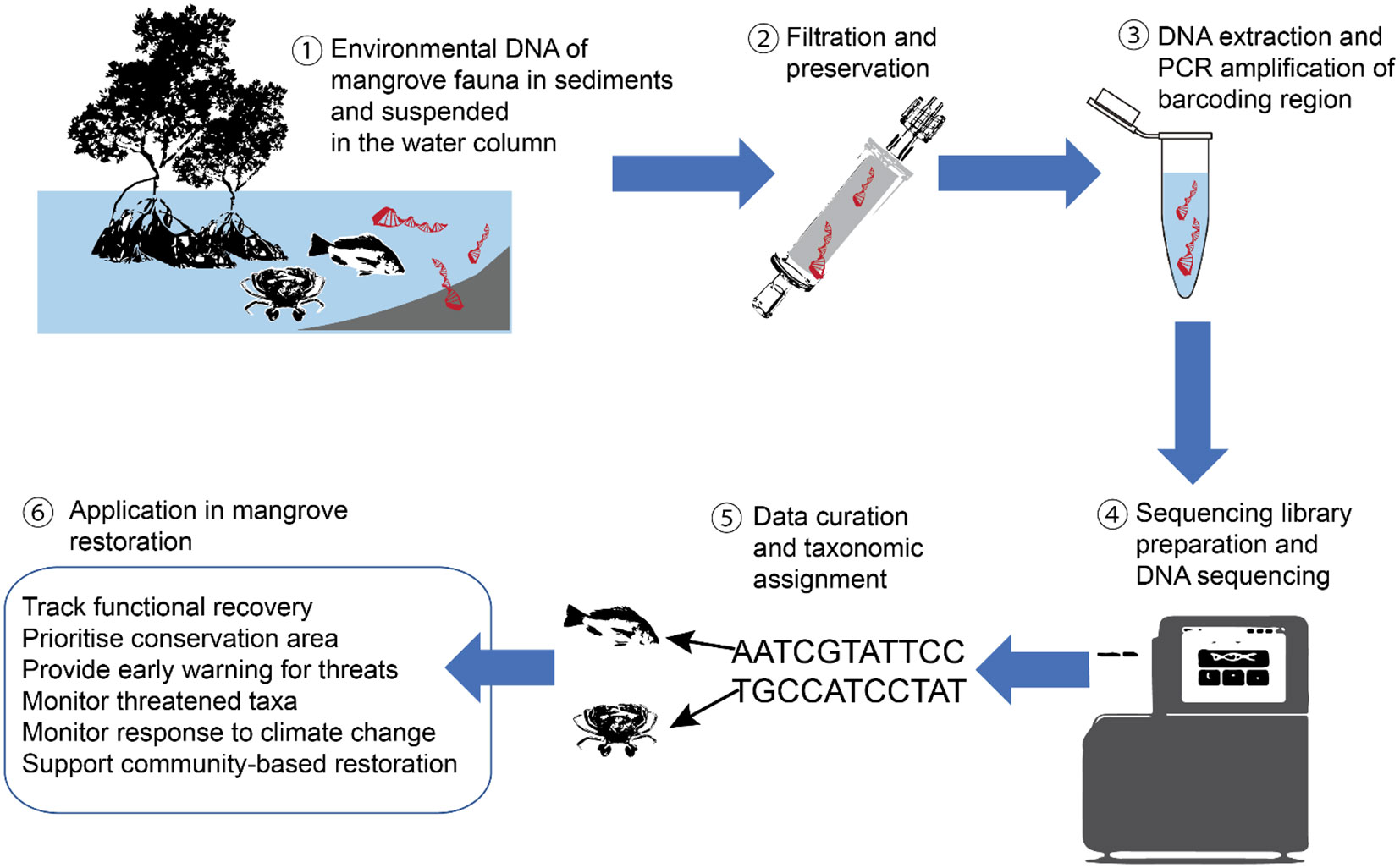
The development of environmental DNA (eDNA) provides an opportunity for biodiversity documentation that can complement conventional biodiversity monitoring techniques (e.g. Oka et al., 2021; Polanco Fernández et al., 2021). The term eDNA refers to organismal DNA and extra-organismal DNA sourced from marine, aquatic, aerial and terrestrial environmental samples, such as water and sediments (Rodriguez-Ezpeleta et al., 2021). Environmental DNA metabarcoding is a novel method of assessing biodiversity from a wide taxonomic group using remnant DNA from environmental samples (Ruppert et al., 2019; see Figure 1 for the workflow of eDNA metabarcoding). The utilisation of eDNA metabarcoding has revolutionised biodiversity research in that (1) it is faster than conventional biodiversity assessment using morphological identification, making it efficient and relatively cheap, (2) the sampling process is simple, non-destructive, and non-invasive, (3) it can detect rare, cryptic, and elusive species, (4) it enables the early detection of imminent, significant environmental change, (5) it is standardised and reproducible across different life stages and environment, and (6) it allows for the simultaneous biodiversity assessment for a wide range of organisms (Beng & Corlett, 2020). Despite its increased popularity in aquatic and marine ecosystems (Bessey et al., 2021), eDNA metabarcoding is still not widely adapted yet in SEA due to several key technical and practical limitations. In this mini-review, we synthesize the viability, potentials, challenges and future prospects of applying eDNA metabarcoding approaches in mangrove restoration in SEA. We align our recommendations to the broader aims of the UN’s Decade on Ecosystem Restoration and highlight the role of eDNA metabarcoding in advancing mangrove restoration research in SEA.
2 Environmental DNA applications for mangrove restoration
2.1 Track functional recovery in ecological restoration
The success of mangrove restoration to date is often measured by forest cover recovery using remote sensing and quadrat/transect surveys (Giri, 2016). This is because measuring the recovery of all ecosystem components (and thus the full ecosystem function – Bosire et al., 2008) is more challenging, especially the recovery of mangrove biota such as macrofaunal, meiofaunal and microbial communities. Often, excavation, careful taxonomic identification, and abundance/biomass quantification, are needed to obtain meaningful results. Thus, assessing restoration success can often be time-consuming, resource-intensive, and subjective due to the limited availability of expertise. Environmental DNA metabarcoding would allow for faster, accurate and cost-efficient data collection at a large number of sites, including healthy and degraded ones (Wang et al., 2019; Saenz-Agudelo et al., 2022), and for tracking community-wide shifts in response to recovery (but see quantification limitation in Section 3.4). The possibility to standardise sampling protocols across time and user groups allows for an accurate chronosequence of species succession. Furthermore, the high resolution of eDNA metabarcoding data can improve understanding of fine-scale movement of taxa or connectivity between systems. Environmental DNA also allows for the detection of rare species, and the accurate identification of specimens that would be difficult through morphological examination alone, e.g. cryptic species (Lim et al., 2016; Mennesson et al., 2018) or juvenile/larval stages (Marshall and Stepien, 2019).
2.2 Prioritise conservation area
Evidence from aquatic and marine ecosystems have demonstrated the usefulness of eDNA metabarcoding in informing protected area design. Significant differences in fish species composition among coral reefs with different levels of protection in Lombok Island (Indonesia) was observed using eDNA metabarcoding (Gelis et al., 2021). Environmental DNA techniques can also be used in the conservation management of priority species, especially in monitoring populations and understanding habitat boundaries. For example, in blacktip reef sharks, single-species eDNA surveys using real-time PCR detected spatio-temporal changes in abundance that are comparable to extensive fishing surveys and acoustic telemetry (Postaire et al., 2020), highlighting the potential of eDNA for monitoring shark populations and understanding their habitat boundaries. These eDNA approaches can be applied to the mangrove habitat to understand the movement ecology of priority species and the connectivity across habitats. Such information on metapopulation dynamics will aid in the prioritisation and design of conservation areas and the selection of restoration sites.
2.3 Provide early warning for threats
Environmental DNA metabarcoding allows for the early detection of elusive and invasive species at low densities, which is otherwise challenging to monitor using conventional techniques (Loeza-Quintana et al., 2021). Environmental DNA has been used to monitor the invasive Burmese python Python bivittatus in South Florida (Piaggio et al., 2014) and the Atlantic Charru mussel Mytella strigata in Singapore (Ip et al., 2021). Also, significant changes in indicator copepod species like Paracalanus indicus and Hexanauplia detected by eDNA metabarcoding can be used to monitor heat waves and their impacts on aquatic ecosystems (Berry et al., 2019). Interestingly, the recent development of 60 species-specific real-time, or quantitative, PCR (qPCR) assays for invasive, threatened, and exploited freshwater vertebrates and invertebrates in North America (Hernandez et al., 2020) paved the way forward for other aquatic ecosystems, e.g. mangroves, that are constantly under threat. In mangroves, invasive plants (e.g. carrotwood) and harmful algae could cause significant loss of biodiversity and damage to ecosystem function. Thus, early detection and regular monitoring of these species using eDNA metabarcoding and even species-specific qPCR assays is crucial to ensure successful ecological restoration.
2.4 Monitor threatened taxa
Mangroves in SEA are key habitats of multiple threatened species such as the dugong (Dugong dugon), proboscis monkey (Nasalis larvatus), roughnose stingray (Pastinachus solocirostris) and tiger tail seahorse (Hippocampus comes). Reliable monitoring of these organisms is crucial for data-driven conservation actions but remains a challenge owing to the lack of standardised methods, elusiveness of the species, and dependence on species experts (Thomsen et al., 2012). Environmental DNA has various proven applications in monitoring specific threatened taxa, including uncovering previously unrecorded elasmobranch species in Singapore’s waters (Ip et al., 2021), mapping the distribution of endangered European eel (Weldon et al., 2020) and documenting rare and threatened sharks and rays across eastern Indonesia (Moore et al., 2021). This is accelerated by the development of universal primers that allows for metabarcoding and multi-taxa detection, e.g. the reptile primers simultaneously detected the vulnerable flatback turtle and the Indo-Australian water snake, which inhabit mangrove forests in SEA (West et al., 2021). Furthermore, species detection by eDNA can now include qPCR, whereby the presence and even abundance of a species can be quantitatively estimated based on the eDNA concentration (Weltz et al., 2017). Species detection by qPCR of eDNA samples is still less expensive than traditional surveys, and represent a highly repeatable and sensitive method for behaviorally elusive species (Qu & Stewart, 2019). The presence and recovery of threatened taxa can thus be used as an important metric in monitoring mangrove restoration success.
2.5 Monitor response to climate change
Environmental DNA metabarcoding is useful in tracking the response of mangrove communities to climate change. For example, a 5-year eDNA metabarcoding survey demonstrated a significant seasonal change in meroplankton communities, including fish, molluscs, and cnidarians, especially after the 2015 strong El Niño event (Djurhuus et al., 2020). In 2021, UNESCO launched a global eDNA metabarcoding expedition to study species vulnerability to climate change at marine World Heritage sites (UNESCO, 2021). As successful restoration includes resilience towards climate change, similar eDNA metabarcoding approaches can be applied to mangrove ecosystems to understand climatic response and facilitate adaptation.
2.6 Support community-based restoration
The benefits of involving local communities in restoration efforts and monitoring are three-fold (Schmitt and Duke, 2015; Miya et al., 2022): (1) shared ownership ensures multi-stakeholder support and continued protection of the site, (2) payment for restoration efforts contribute towards local livelihoods, and, (3) increased participation allows for more detailed data collection. However, the lack of taxonomic expertise and the need to maintain a standardised protocol present challenges to increase community involvement in restoration (Eger et al., 2022). The use of eDNA metabarcoding addresses these limitations as community volunteers or employees are able to collect the eDNA metabarcoding sample with minimal training (Miya et al., 2022), and samples are then analysed by taxon experts and molecular researchers who can easily provide a comprehensive species list. This rapid assessment by local communities across a finer temporal and spatial scale (Agersnap et al., 2022) could also allow for early detection of invasive species (Larson et al., 2020) and changes in coastal communities or targeted species (Biggs et al., 2015). In addition, it is important to sustain the collaboration between molecular ecologists and local stakeholders through the Adoption of Translational Molecular Ecology that enhances consensus on objectives, methods, and outcomes of environmental management projects (Aylagas et al., 2020). Establishing a sustained dialogue among stakeholders is key to accelerating the adoption of molecular-based approaches for marine monitoring and assessment (Aylagas et al., 2020).
3 Challenges
3.1 Technical challenges
The eDNA metabarcoding workflow – from sample collection, preservation, amplification, sequencing to bioinformatic analysis – presents multiple technical challenges. As eDNA metabarcoding work involves detecting minute amounts of degraded DNA from environmental samples, the sample collection process is therefore prone to contamination, from sampling instruments, lab supplies (e.g. collection tubes), boats and other field gear (e.g. boots). Therefore, field decontamination before sampling is paramount to ensure sample independence (Goldberg et al., 2016). The most widely-adopted approach to capture eDNA is by filtration, which is highly dependent on factors like pH, amount of suspended particles, filter pore size, and filtered water volume (Majaneva et al., 2018; Wong et al., 2020). For the turbid estuarine waters characteristic of mangroves in SEA, high concentrations of suspended organic and inorganic particles are associated with higher eDNA abundance but also rapid clogging of filters. Sample preservation is key, as the degradation of eDNA accelerated by the warm humid tropical climate results in shorter DNA fragments (Ruppert et al., 2019) that reduce PCR amplification success. Also, studies in relatively remote areas can pose logistical challenges in sample preservation. Higher levels of PCR inhibitions are also linked to turbid waters (Kumar et al., 2022); some inhibitors, such as humic acid, fulvic acid, tannic acid, and hematin, are naturally excreted in the environment from animals or plants (Sato et al., 2017; Minegishi et al., 2019; Uchii et al., 2019). In highly biodiverse tropical mangroves, successful co-amplification of eDNA strongly depends on suitable design primer(s) that is (are) specific and sensitive (Coissac et al., 2012). These challenges would have to be overcome to facilitate the standardisation of experimental protocol.
3.2 Lack of standardised methods
The standardisation of methodology is necessary to ensure comparability across studies. Taxon detection using eDNA metabarcoding is highly dependent on environmental variables (Stewart, 2019; Blabolil et al., 2021). Taxa groups may differ based on the types of waters sampled in the mangroves (Majaneva et al., 2018; Jerde et al., 2019) – eDNA from estuarine waters will likely reflect both surface and bottom estuarine fauna such as fish and prawns but interstitial or pore water may better represent terrestrially-associated fauna, e.g. gastropods. Other sources such as aquatic sediments may contain more eDNA of fish as compared to water samples (Turner et al., 2015). The lack of consensus on best practices for collection and analysis (Goldberg et al., 2015) ultimately prevents the development of a universal eDNA protocol for SE Asian mangroves and reduces the comparability of biodiversity studies (Fonseca, 2018).
3.3 Spatio-temporal difficulties in defining community
The terrestrial, aquatic and intertidal fauna found in mangroves are diverse in life history strategies, and may use mangroves for part of or throughout their lifespans. Mobile taxa such as fish may use mangrove creeks and estuaries exclusively or opportunistically in conjunction with other adjacent habitats, and may move upstream even into more freshwater zones (Krumme, 2004; Russell and McDougall, 2005). Hence, taxon detection depends on multiple factors, e.g. tidal condition, monsoons, salinity gradient, and stratification of the water column (Figure 2). Furthermore, understanding processes that govern both eDNA release from focal mangrove fauna and removal of eDNA in the intertidal mangrove environment, i.e. eDNA sources and sinks respectively, are critical (Stewart, 2019) but remain poorly documented. The dynamic spatio-temporal heterogeneity in faunal utilisation of mangroves also poses a problem of scale in terms of delineating the sampling boundaries for different ecological and ecosystem applications. Due to the bio-geomorphological complexity of mangrove habitats, the species distribution data could be influenced by the microhabitat, estuarine position and extent, biogeographic region and mangrove type (Figure 2), in addition to being confounded by human influences (e.g. the presence of introduced or invasive species). Hence, it is important that eDNA metabarcoding in mangroves includes sampling at multiple spatial scales and checklist records of native species to allow for better understanding of species distribution. Consensus in sampling protocols at congruent scales is needed to facilitate comparative studies across mangroves in SEA.
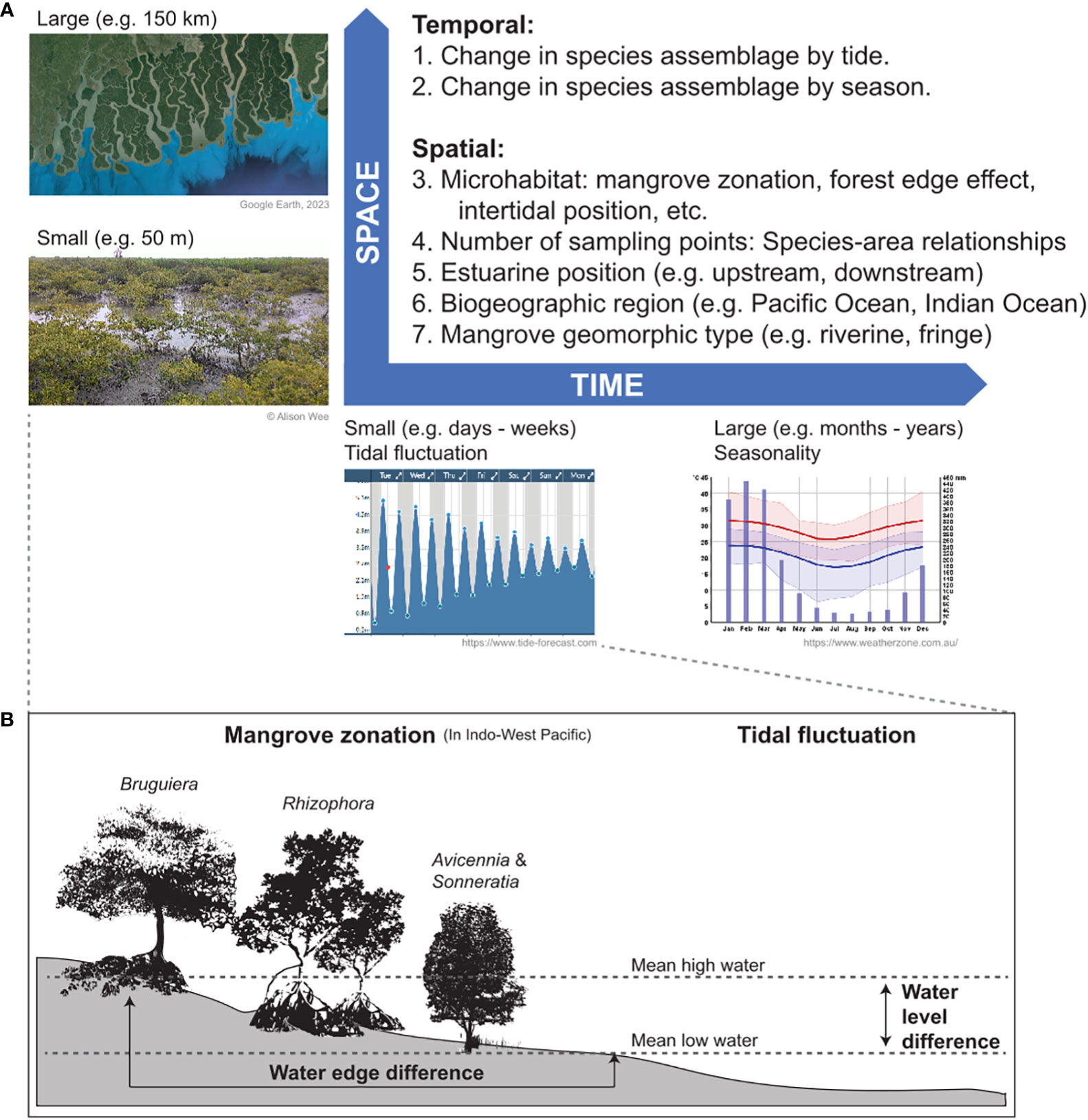
Figure 2 (A) Key examples of spatio-temporal challenges of designing eDNA metabarcoding studies in mangroves, (B) mangrove zonation and tidal fluctuation are two examples of the spatio-temporal challenges in mangrove eDNA metabarcoding study design.
3.4 Data limitations
A comprehensive reference database is lacking for many highly speciose but poorly studied mangrove fauna (Rajpar and Zakaria, 2014; Jerde et al., 2021). Across all taxa, fishes (especially freshwater fish) have the most comprehensive reference database, e.g. FISH-BOL (Ward et al., 2009). However, the available reference databases are mostly region-specific and have limited transferability across ecosystems and ecoregions of the world. The absence of a region-specific reference database has confined eDNA metabarcoding research in SEA to single-species study on ecologically significant taxa, e.g. highly invasive bivalve (Xia et al., 2018). As species identification using eDNA metabarcoding is only as good as the reference database (Thomsen & Sigsgaard, 2019), establishing a regional, comprehensive database is the necessary step forward for the application of eDNA metabarcoding as a routine biomonitoring tool.
Environmental DNA metabarcoding holds great promise in determining relative species abundance and/or biomass. To date, abundance/or biomass estimation has been shown feasible for select taxa in ‘controlled’ environments such as aquaria and in selected natural environments (see review by Rourke et al., 2022). For this to be feasible, correlations must be established between eDNA concentrations and abundance and/or biomass across taxa (Fonseca, 2018). Further work will be essential to overcome eDNA data limitations beyond documentation of species presence.
3.5 Lack of funding, infrastructure and capacity building
Although eDNA metabarcoding can be more cost-effective than traditional approaches in highly biodiverse regions (Bálint et al., 2018), it remains relatively cost-prohibitive in local SEA currencies, especially when many molecular supplies come from developed countries like the USA and Germany. Advances in eDNA biodiversity applications to date are primarily confined to high-income developed countries (Rourke et al., 2022). This is further hampered by limitations in funding, molecular laboratories and infrastructure that can support the post-sampling analyses. The molecular component poses a steep learning curve, and coupled with the lack of user-friendly reference databases, hampers the uptake of eDNA metabarcoding within SEA. Knowledge transfer and capacity building training that promote hands-on experiences in field sampling, laboratory and data analysis, as well as mutual academic exchanges to develop the best practice, are necessary.
4 Future perspectives
For eDNA metabarcoding to fully realise its potential as a biomonitoring tool in mangrove restoration in SEA, several key organisational, technical and logistical improvements will need to be implemented. First, cohesive international and regional collaboration is necessary to overcome the data limitations, enable capacity building and facilitate the much-needed development of a comprehensive reference database. Successful endeavours have been undertaken at the national level, e.g. the Atlas of Living Australia (hosted by CSIRO) and the Biodiversity of Singapore (hosted by the Lee Kong Chian Natural History Museum) initiatives, whereby concerted efforts among government agencies, scientists and citizens have led to the establishment of a well-curated biodiversity inventory supplemented by DNA barcoding. To advance eDNA metabarcoding in mangrove habitats with pantropical distribution, transboundary cooperation is essential, and hence such efforts should be emulated at an international level. International collaboration will benefit not only the development of the reference database, but also encourages technical exchanges and the sharing of facilities and resources. All of this will be crucial for developing countries in SEA and elsewhere, which are commonly underfunded for molecular biodiversity research. Long term funding, preferably from multiple sources, would ensure the continuity of the collaborative platform. One key example is the “Global analyses of mangrove ecosystem by eDNA metabarcoding” international research project supported by the Japan Society for the Promotion of Science (JSPS) Core-to-Core, which enabled capacity-building through seminars and workshops and eventually led to spin-off projects funded by regional, multinational sources. One of the spin-offs being the international collaborative research project between Japan, Philippines, and Indonesia on the “Application of eDNA metabarcoding in faunal biodiversity assessment of Indo-Pacific mangroves vulnerable to climate change” under the East Asia Science and Innovation Area Joint Research Program (e-ASIA JRP), supported by public co-funding from the East Asia Summit (EAS) member countries.
Second, explorative studies are essential in standardising the methodology, developing the collaborative platform, and building the baseline data for biomonitoring. Regular meetings among the regional mangrove eDNA metabarcoding community are needed to coordinate and refine a common methodology for local use. A standardised, freely available experimental manual, e.g. the Environmental DNA Sampling and Experiment Manual (The eDNA Society, 2019), would be an ideal starting point, upon which technical improvements to overcome excessive sediments in samples and PCR inhibitors can be based. To address the difficulties in defining the sampling scale, it is important to first conduct baseline studies that sample widely and across various spatio-temporal and forest configurations. This would include sampling across tidal regimes, monsoonal seasons, intertidal zones, water depths, salinity gradients, mangrove habitat types (e.g. fringe, estuarine), spatial scales and levels of anthropogenic impacts. Following this, a collaborative platform for inventorizing the eDNA samples would be necessary to facilitate the comparison of biodiversity across spatio-temporal configurations. For example, the ANEMONE DB (https://db.anemone.bio/) based in Tohoku University provides targeted inventorizing of MiFish eDNA metabarcoding surveys in the Pacific Ocean and the WilderLab platform (https://www.wilderlab.co.nz/) showcased publicly available eDNA data in New Zealand. Such a data repository would facilitate the sharing of eDNA data that can eventually inform decision-making on sampling design and workload planning. In addition, the initial studies would provide essential baseline data for future biomonitoring efforts.
In summary, this review provides an essential, practical guide to scientists, policymakers, conservation practitioners and mangrove forest managers in implementing eDNA metabarcoding as a biomonitoring tool in mangrove restoration programs. The implementation of eDNA metabarcoding would encourage the development of data-driven coastal management and equitable conservation programs. Such advancement is especially needed in SEA as it comprises coastal nations with shared coastal resources, threats and ecosystem restoration goals. Hence, international collaboration and capacity building in mangrove eDNA metabarcoding are crucial to promote the region’s interests in food security, coastal defense and biodiversity conservation.
Author contributions
All authors contributed to the conceptualization, literature review and writing. All authors contributed to the article and approved the submitted version.
Funding
This study is an output from the 2nd International Workshop for Mangrove Biodiversity Studies by eDNA Metabarcoding supported by the Japan Society for the Promotion of Science (JSPS) Core-to-Core Program (JPJSCCB20200007). The work is also supported by the Japan Science and Technology Agency (JST) SICORP (e-ASIA JPMJSC21E5 and AJ-CORE JPMJSC21E5), and the JSPS KAKENHI Fund for the Promotion of Joint International Research (21KK0115). AW, AT and TK are supported by the Fundamental Research Grant Scheme FRGS 2021 (FRGS/1/2021/WAB11/UNIM/02/1) awarded by the Ministry of Higher Education Malaysia. AW, AT, MB and TK are supported by the Collaborative Regional Research Programme 2020 (CRRP2020-SP1103-Wee) awarded by the Asia-Pacific Network for Global Change Research. SS was supported by the Foreign Researcher Invitation Program at the University of the Ryukyus. MB was supported by Basic Research Collaboration Scheme 2022-2024 (e-Asia Joint Research Program) (079/UN5.2.3.1/PPM/KP-DRTPM/L/2022) from the Ministry of Education, Culture, Research and Technology of Government of Indonesia.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Agersnap S., Sigsgaard E. E., Jensen M. R., Avila M. D. P., Carl H., Møller P. R., et al. (2022). A national scale “BioBlitz” using citizen science and eDNA metabarcoding for monitoring coastal marine fish. Front. Mar. Sci. 9. doi: 10.3389/fmars.2022.824100
Aylagas E., Borja A., Pochon X., Zaiko A., Keeley N., Bruce K., et al. (2020). Translational molecular ecology in practice: Linking DNA-based methods to actionable marine environmental management. Sci. Total Environ. 744, 140780. doi: 10.1016/j.scitotenv.2020.140780
Bálint M., Nowak C., Márton O., Pauls S. U., Wittwer C., Aramayo J. L., et al. (2018). Accuracy, limitations and cost efficiency of eDNA-based community survey in tropical frogs. Mol. Ecol. Resour. 18, 1415–1426. doi: 10.1111/1755-0998.12934
Basyuni M., Bimantara Y., Cuc N. T., Balke T., Vovides A. G. (2022). Macrozoobenthic community assemblage as key indicator for mangrove restoration success in north Sumatra and aceh, Indonesia. Restor. Ecol. 30, e13614. doi: 10.1111/rec.13614
Beng K. C., Corlett R. T. (2020). Applications of environmental DNA (eDNA) in ecology and conservation: Opportunities, challenges and prospects. Biodivers. Conserv. 29, 2089–2121. doi: 10.1007/s10531-020-01980-0
Berry T. E., Saunders B. J., Coghlan M. L., Stat M., Jarman S., Richardson A. J., et al. (2019). Marine environmental DNA biomonitoring reveals seasonal patterns in biodiversity and identifies ecosystem responses to anomalous climatic events. PLos Genet. 15, e1007943. doi: 10.1371/journal.pgen.1007943
Bessey C., Neil Jarman S., Simpson T., Miller H., Stewart T., Kenneth Keesing J., et al. (2021). Passive eDNA collection enhances aquatic biodiversity analysis. Commun. Biol. 4, 236. doi: 10.1038/s42003-021-01760-8
Bhowmik A. K., Padmanaban R., Cabral P., Romeiras M. M. (2022). Global mangrove deforestation and its interacting social-ecological drivers: A systematic review and synthesis. Sustainability 14 (8), 4433. doi: 10.3390/su14084433
Biggs J., Ewald N., Valentini A., Gaboriaud C., Dejean T., Griffiths R. A., et al. (2015). Using eDNA to develop a national citizen science-based monitoring programme for the great crested newt (Triturus cristatus). Biol. Conserv. 183, 19–28. doi: 10.1016/j.biocon.2014.11.029
Blabolil P., Harper L. R., Říčanová Š., Sellers G., Di Muri C., Jůza T., et al. (2021). Environmental DNA metabarcoding uncovers environmental correlates of fish communities in spatially heterogeneous freshwater habitats. Ecol. Indic. 126, 107698. doi: 10.1016/j.ecolind.2021.107698
Bosire J. O., Dahdouh-Guebas F., Walton M., Crona B. I., Lewis Iii R. R., Field C., et al. (2008). Functionality of restored mangroves: a review. Aquat. Bot. 89, 251–259. doi: 10.1016/j.aquabot.2008.03.010
Bunting P., Rosenqvist A., Hilarides L., Lucas R. M., Thomas N. (2022). Global mangrove watch: Updated 2010 mangrove forest extent (v2. 5). Remote Sens. 14, 1034. doi: 10.3390/rs14041034
Coissac E., Riaz T., Puillandre N. (2012). Bioinformatic challenges for DNA metabarcoding of plants and animals. Mol. Ecol. 21, 1834–1847. doi: 10.1111/j.1365-294X.2012.05550.x
da Rosa C. M., Marques M. C. M. (2022). How are biodiversity and carbon stock recovered during tropical forest restoration? supporting the ecological paradigms and political context involved. J. Nat. Conserv. 65, 126115. doi: 10.1016/j.jnc.2021.126115
Djurhuus A., Closek C. J., Kelly R. P., Pitz K. J., Michisaki R. P., Starks H. A., et al. (2020). Environmental DNA reveals seasonal shifts and potential interactions in a marine community. Nat. Commun. 11, 254. doi: 10.1038/s41467-019-14105-1
Eger A. M., Earp H. S., Friedman K., Gatt Y., Hagger V., Hancock B., et al. (2022). The need, opportunities, and challenges for creating a standardized framework for marine restoration monitoring and reporting. Biol. Conserv. 266, 109429. doi: 10.1016/j.biocon.2021.109429
Estoque R. C., Myint S. W., Wang C., Ishtiaque A., Aung T. T., Emerton L., et al. (2018). Assessing environmental impacts and change in myanmar's mangrove ecosystem service value due to deforestation, (2000–2014). Glob. Change Biol. 24, 5391–5410. doi: 10.1111/gcb.14409
Fonseca V. G. (2018). Pitfalls in relative abundance estimation using eDNA metabarcoding. Mol. Ecol. Resour. 18, 923–926. doi: 10.1111/1755-0998.12902
Friess D. A., Yando E. S., Wong L. W., Bhatia N. (2020). Indicators of scientific value: An under-recognised ecosystem service of coastal and marine habitats. Ecol. Indic. 113, 106255. doi: 10.1016/j.ecolind.2020.106255
Gelis E. R. E., Kamal M. M., Subhan B., Bachtiar I., Sani L. M. I., Madduppa H. (2021). Environmental biomonitoring of reef fish community structure with eDNA metabarcoding in the coral triangle. Environ. Biol. Fishes 104, 887–903. doi: 10.1007/s10641-021-01118-3
Giri C. (2016). Observation and monitoring of mangrove forests using remote sensing: Opportunities and challenges. Remote Sens. 8, 783. doi: 10.3390/rs8090783
Goldberg C. S., Strickler K. M., Pilliod D. S. (2015). Moving environmental DNA methods from concept to practice for monitoring aquatic macroorganisms. Biol. Conserv. 183, 1–3. doi: 10.1016/j.biocon.2014.11.040
Goldberg C. S., Turner C. R., Deiner K., Klymus K. E., Thomsen P. F., Murphy M. A., et al. (2016). Critical considerations for the application of environmental DNA methods to detect aquatic species. Methods Ecol. Evolution. 7, 1299–1307. doi: 10.1111/2041-210X.12595
Hamdan O., Aziz H. K., Hasmadi I. M. (2014). L-band ALOS PALSAR for biomass estimation of matang mangroves, Malaysia. Remote Sens. Environ. 155, 69–78. doi: 10.1016/j.rse.2014.04.029
Hernandez C., Bougas B., Perreault-Payette A., Simard A., Côté G., Bernatchez L. (2020). 60 specific eDNA qPCR assays to detect invasive, threatened, and exploited freshwater vertebrates and invertebrates in Eastern Canada. Environ. DNA 2 (3), 373–386. doi: 10.1002/edn3.89
Hochard J. P., Hamilton S., Barbier E. B. (2019). Mangroves shelter coastal economic activity from cyclones. Proc. Natl. Acad. Sci. USA 116, 12232–12237. doi: 10.1073/pnas.1820067116
Ilman M., Wibisono I. T. C., Suryadiputra I. N. N. (2011). State of the art information on mangrove ecosystems in Indonesia. Wetlands International-Indonesia Programme Bogor. 58. Available at: https://indonesia.wetlands.org/ (accessed 20 Feb 2023).
Ip Y. C. A., Tay Y. C., Chang J. J. M., Ang H. P., Tun K. P. P., Chou L. M., et al. (2021). Seeking life in sedimented waters: Environmental DNA from diverse habitat types reveals ecologically significant species in a tropical marine environment. Environ. DNA 3, 654–668. doi: 10.1002/edn3.162
Jerde C. L., Mahon A. R., Campbell T., McElroy M. E., Pin K., Childress J. N., et al. (2021). Are genetic reference libraries sufficient for environmental DNA metabarcoding of Mekong river basin fish? Water 13, 1767. doi: 10.3390/w13131767
Jerde C. L., Wilson E. A., Dressler T. L. (2019). Measuring global fish species richness with eDNA metabarcoding. Mol. Ecol. Resour. 19, 19–22. doi: 10.1111/1755-0998.12929
Krumme U. (2004). Patterns in tidal migration of fish in a Brazilian mangrove channel as revealed by a split-beam echosounder. Fish. Res. 70, 1–15. doi: 10.1016/j.fishres.2004.07.004
Kumar G., Farrell E., Reaume A. M., Eble J. A., Gaither M. R. (2022). One size does not fit all: Tuning eDNA protocols for high-and low-turbidity water sampling. Environ. DNA 4, 167–180. doi: 10.1002/edn3.235
Larson E. R., Graham B. M., Achury R., Coon J. J., Daniels M. K., Gambrell D. K., et al. (2020). From eDNA to citizen science: Emerging tools for the early detection of invasive species. Front. Ecol. Environ. 18, 194–202. doi: 10.1002/fee.2162
Lee S. Y., Hamilton S., Barbier E. B., Primavera J., Lewis R. R. (2019). Better restoration policies are needed to conserve mangrove ecosystems. Nat. Ecol. Evol. 3, 870–872. doi: 10.1038/s41559-019-0861-y
Lim H., Zainal Abidin M., Pulungan C. P., de Bruyn M., Mohd Nor S. A. (2016). DNA Barcoding reveals high cryptic diversity of the freshwater halfbeak genus Hemirhamphodon from sundaland. PLos One 11, e0163596. doi: 10.1371/journal.pone.0163596
Loeza-Quintana T., Crookes S., Li P. Y., Reid D. P., Smith M., Hanner R. H. (2021). Environmental DNA detection of endangered and invasive species in kejimkujik national park and historic site. Genome 64, 172–180. doi: 10.1139/gen-2020-0042
Long J., Napton D., Giri C., Graesser J. (2014). A mapping and monitoring assessment of the philippines' mangrove forests from 1990 to 2010. J. Coast. Res. 30, 260–271. doi: 10.2112/JCOASTRES-D-13-00057.1
Majaneva M., Diserud O. H., Eagle S. H., Boström E., Hajibabaei M., Ekrem T. (2018). Environmental DNA filtration techniques affect recovered biodiversity. Sci. Rep. 8, 4682. doi: 10.1038/s41598-018-23052-8
Marshall N. T., Stepien C. A. (2019). Invasion genetics from eDNA and thousands of larvae: A targeted metabarcoding assay that distinguishes species and population variation of zebra and quagga mussels. Ecol. Evol. 9, 3515–3538. doi: 10.1002/ece3.4985
Mennesson M. I., Bonillo C., Feunteun E., Keith P. (2018). Phylogeography of eleotris fusca (Teleostei: Gobioidei: Eleotridae) in the indo-pacific area reveals a cryptic species in the Indian ocean. Conserv. Genet. 19, 1025–1038. doi: 10.1007/s10592-018-1063-x
Minegishi Y., Wong M. K. S., Kanbe T., Araki H., Kashiwabara T., Ijichi M., et al. (2019). Spatiotemporal distribution of juvenile chum salmon in otsuchi bay, iwate, Japan, inferred from environmental DNA. PLos One 14, e0222052. doi: 10.1371/journal.pone.0222052
Miya M., Sado T., Oka S. I., Fukuchi T. (2022). The use of citizen science in fish eDNA metabarcoding for evaluating regional biodiversity in a coastal marine region: A pilot study. Metabarcoding Metagenom. 6, e80444. doi: 10.3897/mbmg.6.80444
Moore A. M., Jompa J., Tassakka A. C. M. A. R., Yasir I., Ndobe S., Umar W., et al. (2021). Sharks and rays (Chondrichthyes) around Banggai Island, Banggai MPA indonesia: biodiversity data from an environmental DNA pilot study. AACL Bioflux 14, 725–745.
Oka S. I., Doi H., Miyamoto K., Hanahara N., Sado T., Miya M. (2021). Environmental DNA metabarcoding for biodiversity monitoring of a highly diverse tropical fish community in a coral reef lagoon: Estimation of species richness and detection of habitat segregation. Environ. DNA 3, 55–69. doi: 10.1002/edn3.132
Piaggio A. J., Engeman R. M., Hopken M. W., Humphrey J. S., Keacher K. L., Bruce W. E., et al. (2014). Detecting an elusive invasive species: A diagnostic PCR to detect b urmese python in f lorida waters and an assessment of persistence of environmental DNA. Mol. Ecol. Resour. 14, 374–380. doi: 10.1111/1755-0998.12180
Polanco Fernández A., Marques V., Fopp F., Juhel J. B., Borrero-Pérez G. H., Cheutin M. C., et al. (2021). Comparing environmental DNA metabarcoding and underwater visual census to monitor tropical reef fishes. Environ. DNA 3, 142–156. doi: 10.1002/edn3.140
Postaire B. D., Bakker J., Gardiner J., Wiley T. R., Chapman D. D. (2020). Environmental DNA detection tracks established seasonal occurrence of blacktip sharks (Carcharhinus limbatus) in a semi-enclosed subtropical bay. Sci. Rep. 10, 11847. doi: 10.1038/s41598-020-68843-0
Qu C., Stewart K. A. (2019). Evaluating monitoring options for conservation: comparing traditional and environmental DNA tools for a critically endangered mammal. Sci. Nat. 106, 9. doi: 10.1007/s00114-019-1605-1
Rajpar M. N., Zakaria M. (2014). “Mangrove fauna of Asia,” in Mangrove ecosystems of Asia. Eds. Faridah-Hanum I., Latiff A., Hakeem K. R., Ozturk M. (New York, NY: Springer), 153–197. doi: 10.1007/978-1-4614-8582-7_8
Richards D. R., Friess D. A. (2016). Rates and drivers of mangrove deforestation in southeast asia 2000–2012. Proc. Natl. Acad. Sci. U.S.A. 113, 344–349. doi: 10.1073/pnas.1510272113
Rodriguez-Ezpeleta N., Morissette O., Bean C. W., Manu S., Banerjee P., Lacoursière-Roussel A., et al. (2021). Trade-offs between reducing complex terminology and producing accurate interpretations from environmental DNA: Comment on “Environmental DNA: What's behind the term?” by pawlowski et al 2020. Mol. Ecol. 30 (19), 4601–4605. doi: 10.1111/mec.15942
Rourke M. L., Fowler A. M., Hughes J. M., Broadhurst M. K., DiBattista J. D., Fielder S., et al. (2022). Environmental DNA (eDNA) as a tool for assessing fish biomass: A review of approaches and future considerations for resource surveys. Environ. DNA 4, 9–33. doi: 10.1002/edn3.185
Ruppert K. M., Kline R. J., Rahman M. S. (2019). Past, present, and future perspectives of environmental DNA (eDNA) metabarcoding: A systematic review in methods, monitoring, and applications of global eDNA. Glob. Ecol. Conserv. 17, e00547. doi: 10.1016/j.gecco.2019.e00547
Russell D. J., McDougall A. J. (2005). Movement and juvenile recruitment of mangrove jack, Lutjanus argentimaculatus (Forsskål), in northern Australia. Mar. Freshw. Res. 56, 465–475. doi: 10.1071/MF04222
Saenz-Agudelo P., Delrieu-Trottin E., DiBattista J. D., Martínez-Rincon D., Morales-González S., Pontigo F., et al. (2022). Monitoring vertebrate biodiversity of a protected coastal wetland using eDNA metabarcoding. Environ. DNA 4, 77–92. doi: 10.1002/edn3.200
Salmo S. G., Lovelock C., Duke N. C. (2013). Vegetation and soil characteristics as indicators of restoration trajectories in restored mangroves. Hydrobiologia 720, 1–18. doi: 10.1007/s10750-013-1617-3
Salmo S. G., Tibbetts I., Duke N. C. (2017). Colonization and shift of mollusc assemblages as a restoration indicator in planted mangroves in the Philippines. Biodivers. Conserv. 26, 865–881. doi: 10.1007/s10531-016-1276-6
Sannigrahi S., Zhang Q., Pilla F., Joshi P. K., Basu B., Keesstra S., et al. (2020). Responses of ecosystem services to natural and anthropogenic forcings: A spatial regression based assessment in the world's largest mangrove ecosystem. Sci. Total. Environ. 715, 137004. doi: 10.1016/j.scitotenv.2020.137004
Sato H., Sogo Y., Yamanaka H. (2017). Usefulness and limitations of sample pooling for environmental DNA metabarcoding of freshwater fish communities. Sci. Rep. 7, 14860. doi: 10.1038/s41598-017-14978-6
Schmitt K., Duke N. C. (2015). “Mangrove management, assessment and monitoring,” in Tropical forestry handbook. Eds. Köhl M., Pancel L. (Berlin, Heidelberg: Springer), 1–29. doi: 10.1007/978-3-642-41554-8_126-1
Spalding M. D., Leal M. (2021). The state of the world’s mangroves 2021. Global Mangrove Alliance. 79. Available online at: https://www.mangrovealliance.org/mangrove-forests/ (accessed 20 Feb 2023).
Spalding M., Parrett C. L. (2019). Global patterns in mangrove recreation and tourism. Mar. Policy 110, 103540. doi: 10.1016/j.marpol.2019.103540
Stewart K. A. (2019). Understanding the effects of biotic and abiotic factors on sources of aquatic environmental DNA. Biodivers. Conserv. 28, 983–1001. doi: 10.1007/s10531-019-01709-8
Su J., Friess D. A., Gasparatos A. (2021). A meta-analysis of the ecological and economic outcomes of mangrove restoration. Nat. Commun. 12, 5050. doi: 10.1038/s41467-021-25349-1
Taddeo S., Dronova I. (2018). Indicators of vegetation development in restored wetlands. Ecol. Indic. 94, 454–467. doi: 10.1016/j.ecolind.2018.07.010
The eDNA Society (2019) Environmental DNA sampling and experiment manual version 2.1. Available at: http://ednasociety.org/eDNA_manual_Eng_v2_1_3b.pdf (Accessed August 30, 2022).
Then A. Y. H., Adame M. F., Fry B., Chong V. C., Riekenberg P. M., Mohammad Zakaria R., et al. (2021). Stable isotopes clearly track mangrove inputs and food web changes along a reforestation gradient. Ecosystems 24, 939–954. doi: 10.1007/s10021-020-00561-0
Thomsen P. F., Kielgast J. O. S., Iversen L. L., Wiuf C., Rasmussen M., Gilbert M. T. P., et al. (2012). Monitoring endangered freshwater biodiversity using environmental DNA. Mol. Ecol. 21, 2565–2573. doi: 10.1111/j.1365-294X.2011.05418.x
Thomsen P. F., Sigsgaard E. E. (2019). Environmental DNA metabarcoding of wild flowers reveals diverse communities of terrestrial arthropods. Ecol. Evol. 9, 1665–1679. doi: 10.1002/ece3.4809
Turner C. R., Uy K. L., Everhart R. C. (2015). Fish environmental DNA is more concentrated in aquatic sediments than surface water. Biol. Conserv. 183, 93–102. doi: 10.1016/j.biocon.2014.11.017
Uchii K., Doi H., Okahashi T., Katano I., Yamanaka H., Sakata M. K., et al. (2019). Comparison of inhibition resistance among PCR reagents for detection and quantification of environmental DNA. Environ. DNA 1, 359–367. doi: 10.1002/edn3.37
UNESCO (2021) UNESCO Launches global eDNA project to study vulnerability of species to climate change at marine world heritage sites. Available at: https://ioc.unesco.org/news/unesco-launches-global-edna-project-study-vulnerability-species-climate-change-marine-world (Accessed August 30, 2022).
Walters B. B. (2003). People and mangroves in the Philippines: fifty years of coastal environmental change. Environ. Conserv. 30, 293–303. doi: 10.1017/S0376892903000298
Wang P., Yan Z., Yang S., Wang S., Zheng X., Fan J., et al. (2019). Environmental DNA: an emerging tool in ecological assessment. Bull. Environ. Contam. Toxicol. 103, 651–656. doi: 10.1007/s00128-019-02720-z
Ward R. D., Hanner R., Hebert P. D. (2009). The campaign to DNA barcode all fishes, FISH-BOL. J. fish Biol. 74 (2), 329–356. doi: 10.1111/j.1095-8649.2008.02080.x
Weldon L., O’Leary C., Steer M., Newton L., Macdonald H., Sargeant S. L. (2020). A comparison of European eel Anguilla anguilla eDNA concentrations to fyke net catches in five Irish lakes. Environ. DNA 2, 587–600. doi: 10.1002/edn3.91
Weltz K., Lyle J. M., Ovenden J., Morgan J. A. T., Moreno D. A., Semmens J. M. (2017). Application of environmental DNA to detect an endangered marine skate species in the wild. PLos One 12 (6), e0178124. doi: 10.1371/journal.pone.0178124
West K. M., Heydenrych M., Lines R., Tucker T., Fossette S., Whiting S., et al. (2021). Development of a 16S metabarcoding assay for the environmental DNA (eDNA) detection of aquatic reptiles across northern Australia. Mar. Freshw. Re. doi. doi: 10.1071/MF20288
Wong M. K. S., Nakao M., Hyodo S. (2020). Field application of an improved protocol for environmental DNA extraction, purification, and measurement using sterivex filter. Sci. Rep. 10, 21531. doi: 10.1038/s41598-020-77304-7
Xia Z., Zhan A., Gao Y., Zhang L., Haffner G. D., MacIsaac H. J. (2018). Early detection of a highly invasive bivalve based on environmental DNA (eDNA). Biol. Invasions 20, 437–447. doi: 10.1007/s10530-017-1545-7
Zari M. P., Kiddle G. L., Blaschke P., Gawler S., Loubser D. (2019). Utilising nature-based solutions to increase resilience in pacific ocean cities. Ecosyst. Serv. 38, 100968. doi: 10.1016/j.ecoser.2019.100968
Keywords: biodiversity, conservation, environmental DNA, metabarcoding, monitoring, rehabilitation
Citation: Wee AKS, Salmo III SG, Sivakumar K, Then AY-H, Basyuni M, Fall J, Habib KA, Isowa Y, Leopardas V, Peer N, Artigas-Ramirez MD, Ranawana K, Sivaipram I, Suleiman M and Kajita T (2023) Prospects and challenges of environmental DNA (eDNA) metabarcoding in mangrove restoration in Southeast Asia. Front. Mar. Sci. 10:1033258. doi: 10.3389/fmars.2023.1033258
Received: 31 August 2022; Accepted: 13 February 2023;
Published: 27 February 2023.
Edited by:
Nathan Knott, NSW Government, AustraliaReviewed by:
Eva Aylagas, King Abdullah University of Science and Technology, Saudi ArabiaMatt Rees, NSW Government, Australia
Copyright © 2023 Wee, Salmo III, Sivakumar, Then, Basyuni, Fall, Habib, Isowa, Leopardas, Peer, Artigas-Ramirez, Ranawana, Sivaipram, Suleiman and Kajita. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Alison K. S. Wee, a2ltc2hhbi53ZWVAbm90dGluZ2hhbS5lZHUubXk=; Tadashi Kajita, a2FqaXRhQG1haWwucnl1ZGFpLmpw
 Alison K. S. Wee
Alison K. S. Wee Severino G. Salmo III
Severino G. Salmo III Kannan Sivakumar
Kannan Sivakumar Amy Y-H Then
Amy Y-H Then Mohammad Basyuni
Mohammad Basyuni Jean Fall6
Jean Fall6 Kazi Ahsan Habib
Kazi Ahsan Habib Venus Leopardas
Venus Leopardas Nasreen Peer
Nasreen Peer Maria D. Artigas-Ramirez
Maria D. Artigas-Ramirez Itchika Sivaipram
Itchika Sivaipram Monica Suleiman
Monica Suleiman Tadashi Kajita
Tadashi Kajita