
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mar. Sci. , 26 October 2022
Sec. Aquatic Microbiology
Volume 9 - 2022 | https://doi.org/10.3389/fmars.2022.985756
Micro/macro plastics are ubiquitous and are emerging agents causing many ecological problems in marine environments. Plastics carry various fouling organisms, including harmful microorganisms, that could potentially have ecological impacts on the marine environment and eventually human health. In this study, marine micro/macro plastics drifting at multiple locations in the Jeju Strait and around Jeju Island, Korea, were collected and analyzed. DNA metabarcoding and scanning electron microscopy were used to characterize the species composition of the attached eukaryotic microbial communities, with a special emphasis on harmful or toxic dinoflagellates, fungi, and parasites. A total of 1,035 eukaryotic microbial operational taxonomic units (OTUs) were identified from plastics and surrounding seawater samples. Two types of samples shared half of the eukaryotic OTUs, and of which approximately 12% were unique to plastic samples. These included 68 Archaeplastida (54.0%), 45 Stramenopiles (35.7%), 12 Opisthokonta (9.5%), and 3 Alveolata (2.4%) OTUs. The harmful or toxic dinoflagellates observed on the plastic surface were Alexandrium, Coolia, Dinophysis, Heterocapsa, Karlodinium, Noctiluca, Ostreopsis, Prorocentrum, Scrippsiella, and Tripos. The most dominant parasite community of the plastisphere was represented by fungi (42% of all parasite OTUs), followed by Oomycota (33%), Stramenopiles (14%), and dinoflagellates (12%). Nonetheless, this study demonstrates that the eukaryotic communities are considerably shared between the plastisphere and seawater. This implies plastics serve as a new ecological habitat in the sea and could function as dispersal vectors that facilitate the spread of harmful eukaryotic species and parasites.
Plastic particles in oceans are becoming one of the most pressing ecological concerns of this century (Law and Thompson, 2014). Marine plastics occur in a variety of size, i.e., macro- (> 5 mm), micro- (0.001 – 5 mm), and nanoplastics (< 0.001 mm) (Andrady, 2011). Microplastic particles can be further subdivided into ‘primary’ microplastics that are already small in size before entering the environment (Browne et al., 2011; Cole et al., 2011; Eerkes-Medrano et al., 2015) and ‘secondary’ microplastics that result from the breakdown of larger macroplastics, such as bottles, plastic bags, clothes, and fishing nets, by a combination of physical, biological, and chemical processes (Thompson et al., 2004; Ryan et al., 2009). The major cause of plastics entering the oceans is the direct introduction from terrestrial environments (Hammer et al., 2012).
Once introduced in marine environments, plastics could have deleterious effects on marine organisms; these include entanglement and ingestion by various marine organisms from zooplankton to fish, seabirds, sea turtles, and cetaceans (Laist, 1997; Eriksson and Burton, 2003; Gall and Thompson, 2015). When ingested, microplastics can result in irritation and injuries to the digestive tract and can also cause several problems associated with energy reallocation, reproductive success, and offspring performance (Browne et al., 2008; Von Moos et al., 2012; Sussarellu et al., 2016).
Apart from the deleterious direct and indirect effects of plastics on marine organisms, plastic-associated microbial communities, referred to as “plastisphere” (Zettler et al., 2013), have also received considerable attention. Micro/macro plastics can be readily colonized by various marine organisms such as virus, bacteria, microalgae, macroalgae, and invertebrates, and provide new habitats for them (Kiessling et al., 2015; De Tender et al., 2015; Gerritse et al., 2020; Asiandu et al., 2021; Moresco et al., 2021). Through microscopic observations or next-generation amplicon sequencing (NGS), several studies have shown the presence of a variety of prokaryotic and eukaryotic microbial communities (ranging from cyanobacteria, coccolithophores, diatoms to dinoflagellates) on plastic particles (Briand et al., 2012; Carson et al., 2013; Zettler et al., 2013; Oberbeckmann et al., 2014; Reisser et al., 2014; De Tender et al., 2017; Ibabe et al., 2020). Notably, these plastics-associated microbial communities have been reported to occasionally contain high numbers of pathogenic bacteria such as Vibrio spp., Campylobacter spp., and Pseudomonas spp., as well as harmful or toxic microalgae such as diatoms and dinoflagellates (Masó et al., 2003; Zettler et al., 2013; Reisser et al., 2014; Masó et al., 2016; Silva et al., 2019; Bowley et al., 2021). Under such conditions, plastic particles enriched with harmful microorganisms may threaten biodiversity and ecosystem health in a given marine environment, especially when transported by physical forces such as ocean currents and winds (Barnes, 2002; Gregory, 2009).
Jeju Island and the adjacent Jeju Strait are located in the southern part of the Korean Peninsula. They are known to be affected by the Tsushima Warm Current flowing northeast through the Korea Strait and its branch, i.e., the Jeju Warm Current, flowing in a clockwise direction; both originate from the Kuroshio Current (Lie and Cho, 2016). Due to this circulation pattern of the ocean currents, this area can act as a gateway with a large potential to influence the southern coastal areas around Korea, for example, in terms of the introduction of harmful microorganisms associated with drifting plastic particles. While limited studies have reported the abundance, occurrence, and distribution of plastic types in coastal waters around Korea (Lee et al., 2013; Eo et al., 2018; Jang et al., 2020; Kwon et al., 2020), none have addressed the eukaryotic microbial community attached to plastic debris from Korean coastal waters. By using both scanning electron microscopy (SEM) and DNA metabarcoding, the present study aimed to (1) characterize the plastic-associated eukaryotic microbial communities on plastic particles, emphasizing on potentially harmful or toxic dinoflagellates, fungi, and eukaryotic parasites, and (2) compare these communities with those in the surrounding seawater.
A total of 42 plastic and surrounding seawater samples were collected from 13 sites around Jeju Island and the adjacent Jeju Strait from March to June 2019 (Figure 1 and Supplementary Table S1). At coastal sites (stations 1, 6 – 13), macroplastics only were collected, due to the difficulty of deploying a manta net for microplastic sampling. By comparison, macroplastics were not observed at the Strait sampling stations (stations 2 – 5) and microplastics only were collected using a manta net. For the collection of microplastic samples, sampling was performed during the research cruise at four stations (stations 2 – 5) using a manta net with a 330 μm pore size. The research vessel’s speed was approximately 2.5 knots during sampling, and the manta net was towed alongside the vessel for 30 min at each station. Sorting of microplastics collected by the manta net was carried out as follows: once the manta net was back to research vessel, it was first rinsed with autoclaved seawater from the outside to inside direction to collect all the materials captured by the net in the sample bucket. Then, the bucket was detached from the manta net and the samples were filtered using a sieve to remove particles greater than 5 mm in size. Next, the filtered samples (smaller than 5 mm) were poured into a wide tray. A small number of microplastics concentrated in this way were large (0.33 – 5 mm) enough to be identified by a naked eye. Therefore, aliquots of the collected microplastic samples were transferred to a 50 mL conical tube (Corning Inc., Corning, NY) with 2% glutaraldehyde (final concentration) using a forcep for SEM analysis. The collected microplastic samples were once again confirmed under a stereomicroscope (SteREO Discovery.V8, Carl Zeiss Inc., Germany) to make sure if the materials were really plastics before further experiments were carried out in the laboratory. Some aliquots were transferred to a 5 mL cryogenic vial (SPL Life Sciences, Pocheon, Korea) with 2 mL STE buffer (100 mM NaCl, 10 mM Tris-HCl, 1 mM EDTA, pH 8.0) and preserved in liquid nitrogen for DNA metabarcoding. Seawater samples were collected simultaneously using Niskin bottles at each station and filtered onto the polycarbonate membrane filters with a diameter of 47 mm and pore size of 0.2 μm (Pall Corporation, New York, United States).
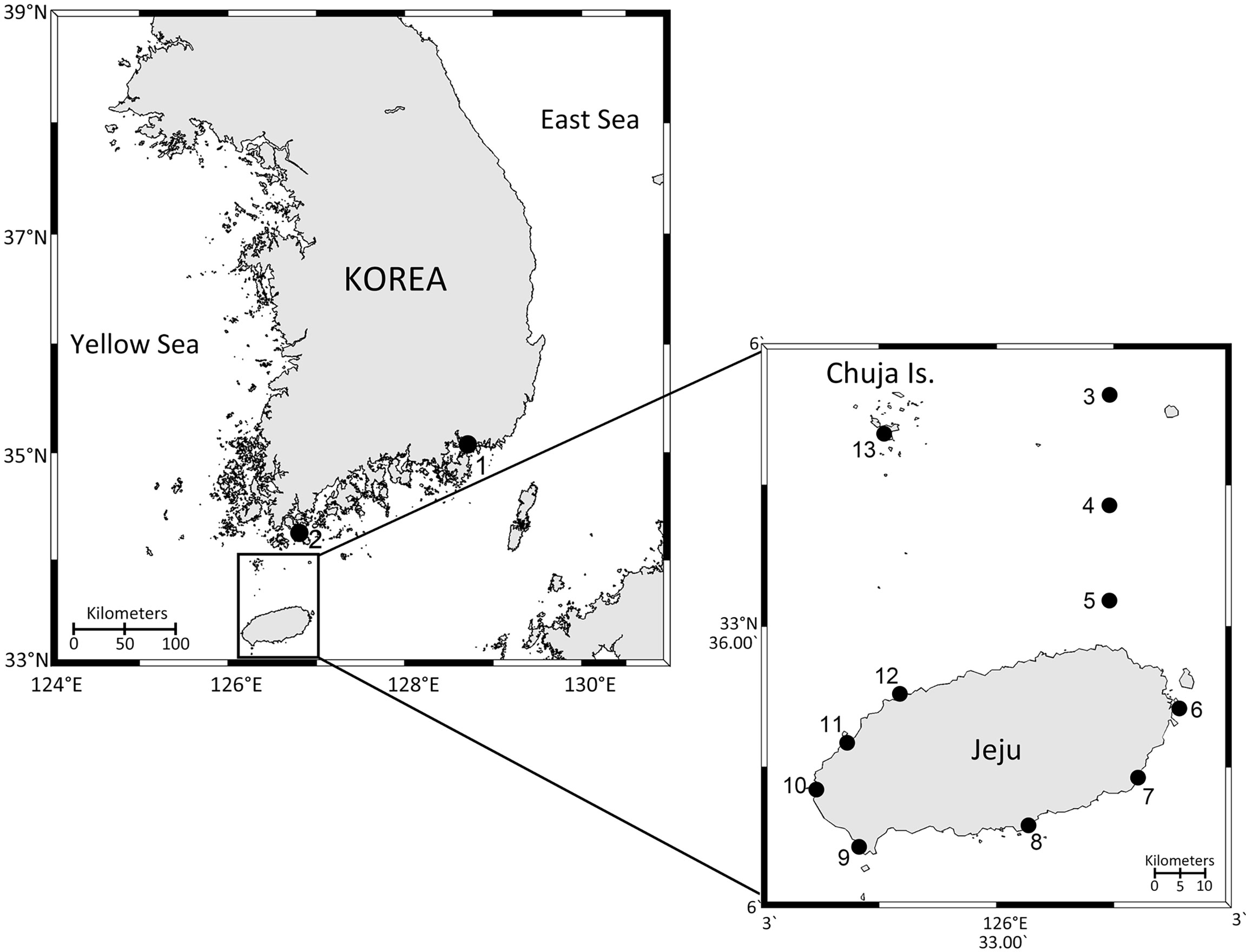
Figure 1 Map of the study area showing the sampling sites. 1, Jangcheon; 2, W1; 3, W4; 4, W6; 5, W8; 6, Seongsan; 7, Pyoseon; 8, Hahyo; 9, Unjin; 10, Jagunae; 11, Gemneung; 12, Aewol; 13, Sinyang. Circles 2 – 5 are microplastic sampling sites. Circles 1 and 6 – 13 are macroplastic sampling sites.
For macroplastic sampling, pieces of plastic floating on the sea surface were collected using a hand-made net in the coastal area of Jeju Island and Jangcheon harbor (Figure 1 and Supplementary Table S1). After collection, half of the samples were cut into several small pieces and transferred into a 50 mL conical tube with 2% glutaraldehyde (final concentration) for SEM analysis. The remaining samples were gently scraped using a cell scraper (SPL Life Sciences, Pocheon, Korea) to detach the microbial community, rinsed with autoclaved fresh seawater using a squeeze bottle, and filtered onto the polycarbonate membrane filters as mentioned above. In addition, 1 L of surrounding seawater samples were taken and filtered as described above. During sample collection, in situ seawater temperature and salinity at each site were measured using a Yellow Springs Instrument (YSI; YSI Inc, Ohio, USA).
In the laboratory, environmental DNA was extracted according to the protocol used by Choi et al. (2015) with some modifications. The extracted environmental DNA samples were used as a template to amplify the nuclear SSU rDNA V4 regions. The polymerase chain reaction (PCR) amplification was performed using the TA-Reuk454FWD1 and TAReukREV3 primer set (Stoeck et al., 2010). The PCR was conducted in 25 μL volume containing 2.5 μL 10X Taq polymerase buffer, 0.5 μL of dNTP (10 mM), 1 μL of each primer, and 0.125 μL of Diastar-Taq DNA polymerase (Solgent Co., Daejeon, Korea). The reactions were conducted using a C1000 Touch Thermal Cycler (Bio-Rad, CA, USA) and run following the PCR program: the initial denaturing step was at 95°C for 2 min, then 35 cycles of 20 s at 95°C, 40 s at 56°C, and 30 s at 72°C, followed by a final extension step of 5 min at 72°C. The second-round PCR for attaching Illumina barcodes was conducted using the index primers. Each final PCR product was purified using a LaboPassTM PCR Purification Kit (COSMO Genetech, Seoul, Korea). DNA quantification was performed using a Qubit 3.0 Fluorometer (Thermo Fisher Scientific, Waltham, USA), and identical quantities of each DNA were pooled and sequenced using the Illumina MiSeq platform (ChunLab, Seoul, Korea).
The raw sequence data is available in the NCBI Sequence Read Archive under the accession number Bioproject ID PRJNA854290. Demultiplexing of the amplicon data set and removal of the barcodes was done by using Mothur v.1.36.1 (Schloss et al., 2009). Briefly, paired-end reads were joined using the default settings of the “make.contigs” command. The quality of the data was controlled by removing ambiguous base pairs, and excessively short (< 300 bp) and long (> 550 bp) sequences as well as removing homopolymers (> 8). Potential chimeras were conducted via Uchime in Mothur (Edgar, 2016), and chimeric sequences were removed. Finally, 1,239,593 clean reads were obtained, and all were clustered and aligned. Taxonomic classification of sequences was conducted in Mother with a SILVA SSU Ref database (release 128). Among the classified sequences, sequences assigned to the superkingdoms of ‘Bacteria,’ ‘Virus,’ ‘Archaea,’ and ‘Unclassified’ groups were excluded. To investigate the biodiversity of marine eukaryotic microorganisms, the classified OTUs were matched with the World Register of Marine Species (WoRMS; http://www.marinespecies.org) database and the AlgaeBase (https://www.algaebase.org).
To assess structural similarities in the eukaryotic microbial communities between plastic and seawater samples, a non-metric multidimensional scaling (NMDS) plot was generated based on Bray–Curtis dissimilarities. This provides measures of community composition differences between samples based on OTU counts, regardless of the taxonomic assignment. The NMDS plots, analysis of similarity (ANOSIM) test, rarefaction curves, and alpha-diversity indices for all samples were obtained using the R Studio using vegan R package version 3.6.2. Three diversity indices, namely, Shannon index, Simpson’s index, and the number of observed OTUs, were computed for each sample. Subsequently, a t-test was performed to evaluate whether the alpha-diversity of the eukaryotic microbial community from plastics was significantly different from that obtained from seawater samples. A p-value of less than 0.05 was considered statistically significant for all tests.
The fixed microplastics and the scraped samples from macroplastic were used for SEM studies. The scraped samples from macroplastic were filtered onto Isopore membrane filters (0.8 μm pore size; Millipore, Cork, Ireland). The samples were washed in distilled water for 1 h and dehydrated in a graded ethanol series (25, 50, 75, 90, and 99%) for 15 min at each step, and then rinsed three times in absolute ethanol at each 15 min interval. The samples were critical point dried in liquid CO2 using a Leica EM CPD300 (Leica Microsystems, Germany). Samples were subsequently mounted on SEM stubs with carbon tape, sputter-coated with platinum, and examined with an SEM (model Genesis-1000, EmCrafts, Korea) scanning electron at 15 kV.
When micro/macro plastic samples were collected from sampling sites, sea surface temperature ranged from 11 °C in March at station 2 (sampling site W1) to 21.1 °C in June at station 6 (sampling site W6) (Figure 1 and Supplementary Table S1). The range of salinity was from 29.3 to 35.4.
A total of 1,239,593 raw rDNA sequence reads and 1,035 OTUs were obtained from the 42 samples. From each sample, the raw rDNA sequence reads obtained ranged from 10 to 78,272 (on average 28,858 reads) and obtained OTUs ranged from 6 to 237 (on average 108 OTUs) (Supplementary Figure S1). Large differences in the number of eukaryotic OTUs were detected between samples obtained from plastic and surrounding seawater. The number of eukaryotic OTUs from the seawater samples (N = 908) was larger than that from the plastic samples (N = 663) (Figure 2). A total of 536 eukaryotic OTUs were shared between the two samples, and only 127 eukaryotic OTUs were plastic-specific.
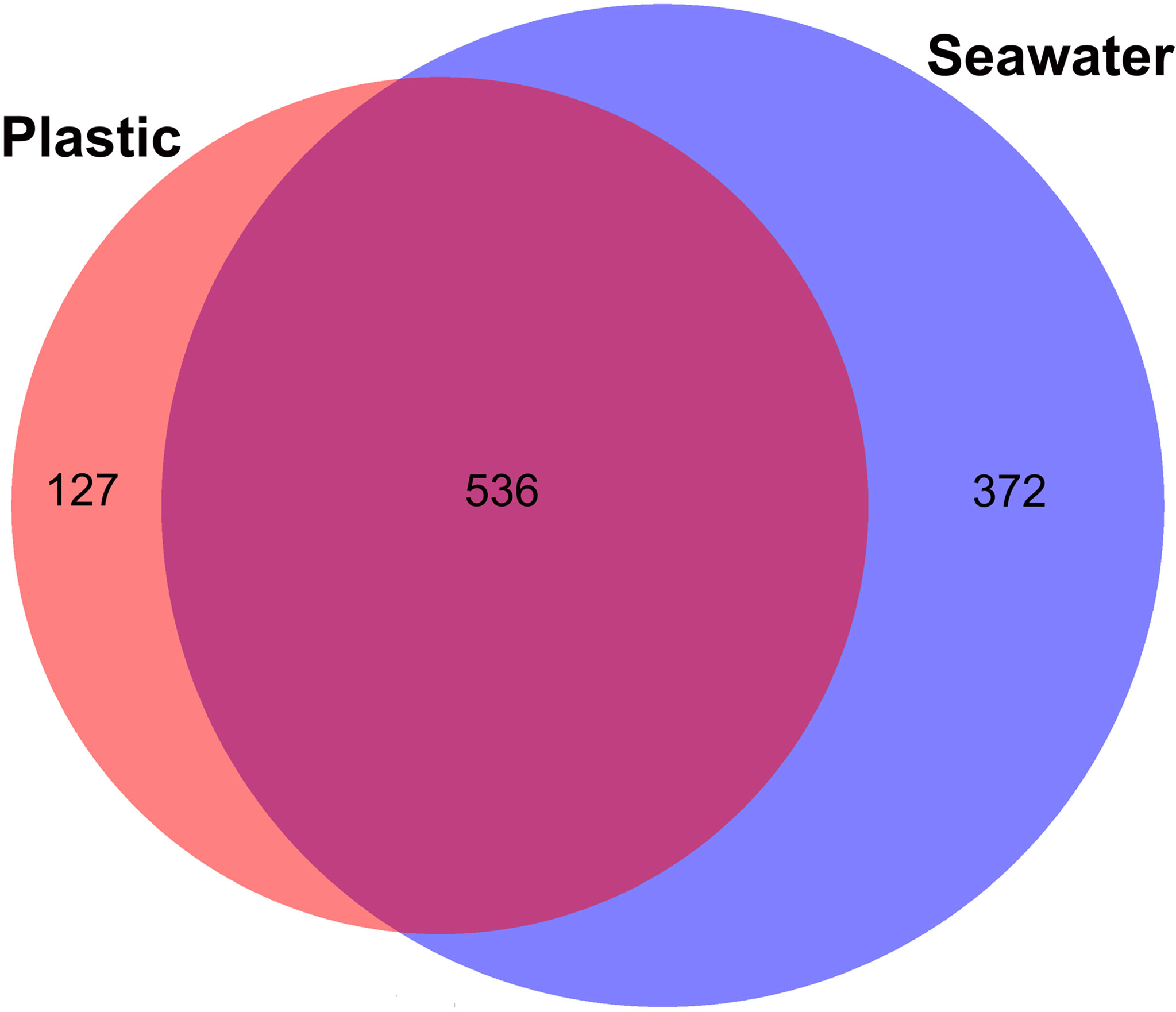
Figure 2 Eukaryotic operational taxonomic units (OTUs) shared between plastics and seawater samples.
The NMDS plot generated based on Bray–Curtis dissimilarities revealed no clear separation of eukaryotic microbial community structure between plastic and seawater samples (Figure 3). The result of the ANOSIM test also showed that community differences between plastic and seawater samples was not statistically significant (R = 0.1797, p-value = 0.0005). In contrast, the ANOSIM statistical tests revealed differences between coastal and strait sampling sites (R = 0.7067, p-value = 0.0001).
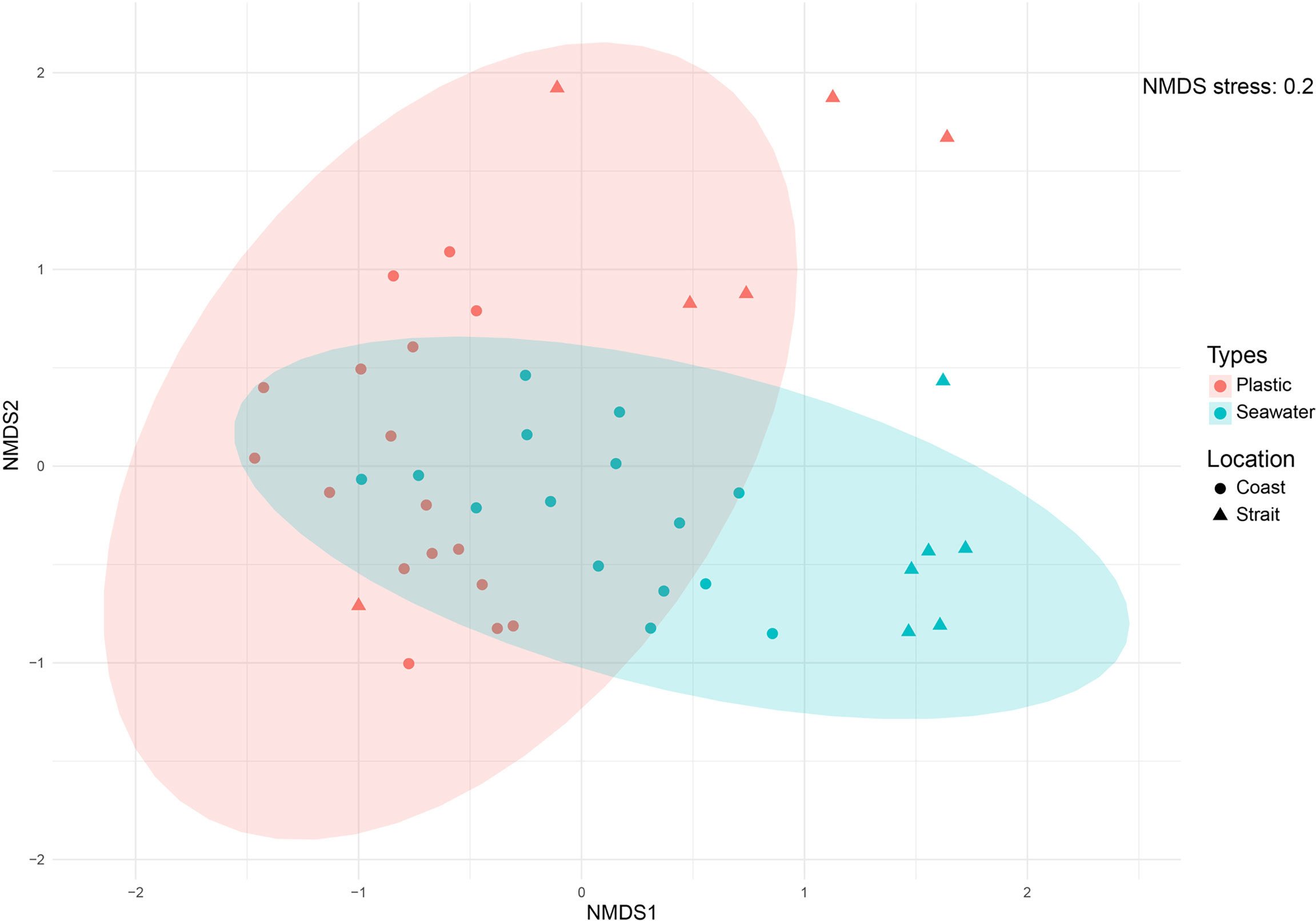
Figure 3 Non-metric multidimensional scaling (NMDS) ordination plots of Bray-Curtis dissimilarities for all samples across sample categories (i.e., sample types and sampling sites). Analysis was based on operational taxonomic units (OTUs) data metrics.
Several rarefaction curves based on the OTUs present in our dataset did not reach the theoretical asymptotic shape (Supplementary Figure S2). The alpha-diversity of the eukaryotic microbial communities associated with the plastic and seawater samples was measured in terms of the number of observed OTUs per sample (richness), Shannon index, and Simpson’s index (evenness) (Figure 4 and Supplementary Figure S3). The key measure of alpha-diversity, including richness was significantly different between plastic and seawater samples (p-value < 0.05; Figure 4). Overall sample diversity, based on Shannon and Simpson indices (Figure 4), was higher in the plastic samples. However, the difference was only significant for the Simpson’s index (p-value < 0.05). The average alpha-diversity scores for all metrics, except for the observed species, were higher for the plastic samples than for the seawater samples (Supplementary Figure S3).
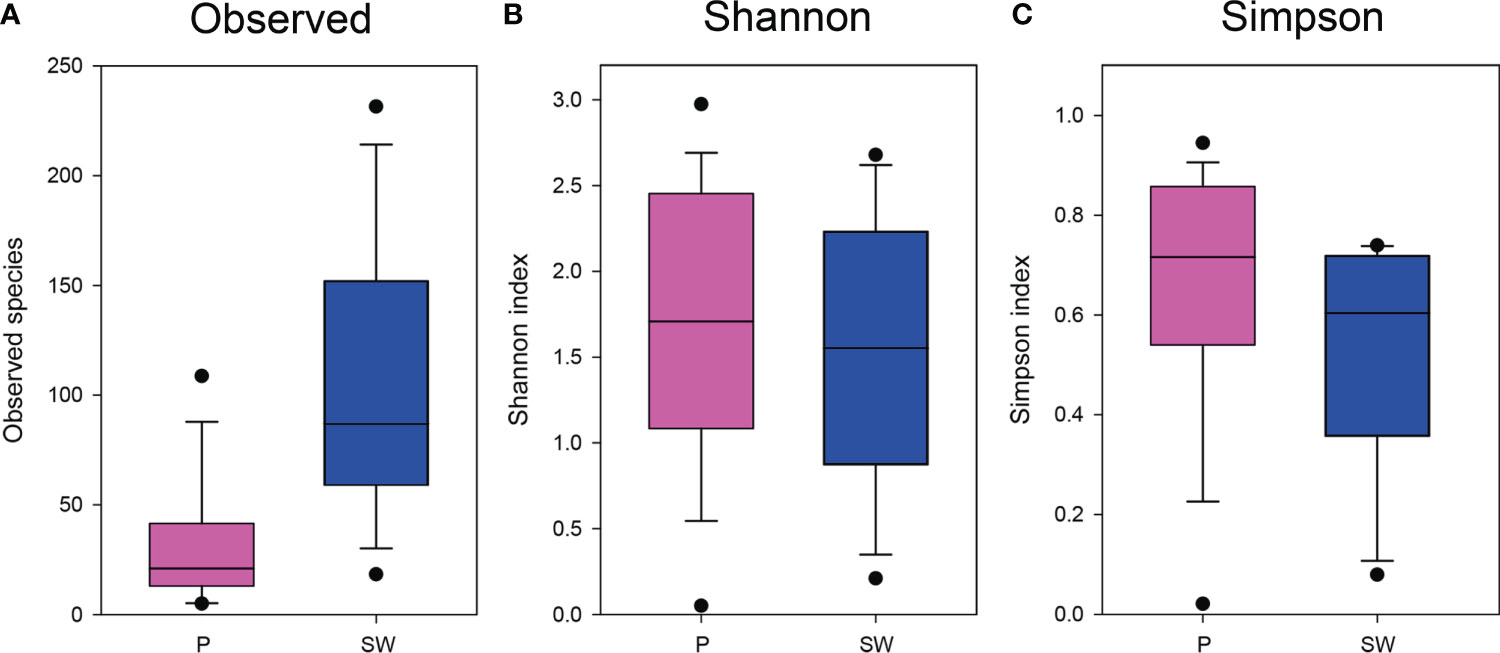
Figure 4 Boxplot of alpha diversity indices for eukaryotic taxa calculated with operational taxonomic units (OTUs) data and arranged according to sample types. (A), Observed species; (B), Shannon indices; (C), Simpson indices. Pink box = plastic samples; Blue box = seawater samples.
The 1,035 eukaryotic OTUs identified comprised five supergroups: 347 OTUs from Archaeplastida (33.5%), 12 OTUs from Hacrobia (1.2%), 317 OTUs from Stramenopiles (30.6%), 311 OTUs from Alveolata (30%), and 48 OTUs from Opisthokonts (4.6%). These five eukaryotic supergroups were composed of 14 phyla (1 NA), 41 classes, 105 orders (3 NA), 166 families (7 NA), and 222 genera, including the “not available (NA)” taxa. The eukaryotic microbial community composition showed differences between plastic and seawater samples regarding the percentages of reads and OTUs (Figures 5, Figure 6). When represented by the percentage of reads, the most abundant group in plastic samples was Archaeplastida (68.3%), followed by Stramenopiles (21.4%), Opisthokonta (7.6%), Alveolata (2.6%), and Hacrobia (0.003%) (Figure 5). By percentage of OTUs, the most abundant group in plastic samples was Archaeplastida (40.7%), followed by Stramenopiles (36.5%), Alveolata (16.4%), Opisthokonta (5.6%), and Hacrobia (0.5%) (Figure 5). The total OTUs obtained from the plastic samples (N = 663) were comprised of 13 phyla, 37 classes, 74 orders (2 NA), 115 families (3 NA), and 150 genera. In the case of seawater samples, when represented by the percentage of reads, the most abundant group was Archaeplastida (46.7%), followed by Alveolata (35.1%), Stramenopiles (15.4%), Hacrobia (1.8%), and Opisthokonta (1%) (Figure 5). Based on the percentage of OTUs, the most abundant group in seawater samples was Alveolata (31.7%), followed by Archaeplastida (28.8%), Stramenopiles (28%), Hacrobia (7.5%), and Opisthokonta (3.9%) (Figure 5). The total OTUs from seawater samples (N = 908) were comprised of 13 phyla, 45 classes, 95 orders (3 NA), 150 families (7 NA), and 210 genera.
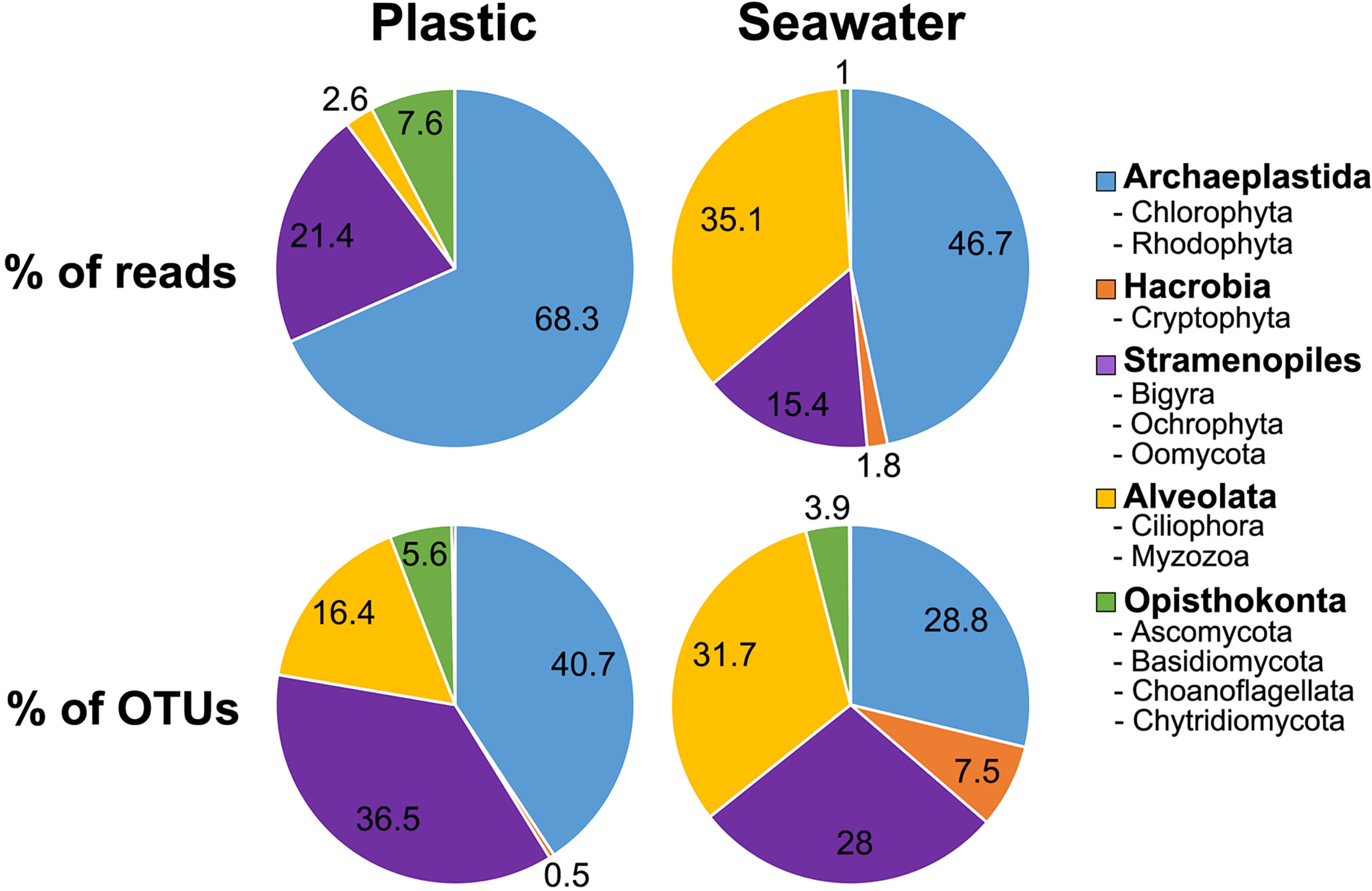
Figure 5 Pie charts illustrating the relative proportion of the number of operational taxonomic units (OTUs) and the number of reads in the taxonomic supergroups detected in plastic and seawater samples.
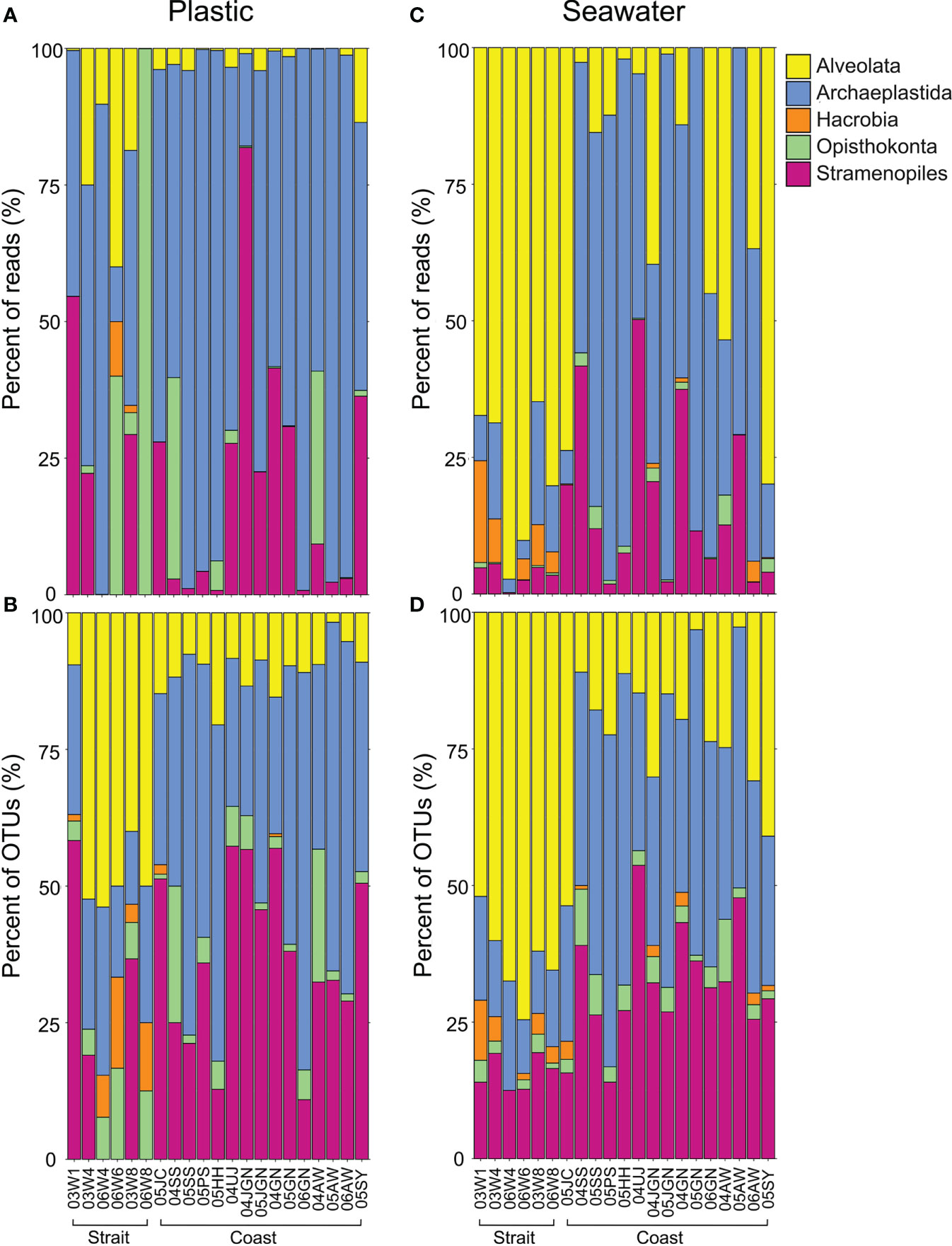
Figure 6 Bar graphs illustrating the relative proportion of the number of reads and the number of operational taxonomic units (OTUs) in the taxonomic supergroups detected in plastic and seawater samples. (A, B), plastic samples; (C, D), seawater samples.
There were also marked differences in the eukaryotic microbial community between plastic and seawater samples from within sampling sites and inter-sampling sites (i.e., coastal vs. strait) (Figure 6). In the case of plastic samples from the strait, the supergroup Archaeplastida occupied more than two-thirds of the number of reads for all of the eukaryotic microbial community (41%), followed by Opisthokonta (24%) and Stramenopiles (18%) (Figure 6A). Similar results were observed for the coastal plastic samples; the most abundant group was Archaeplastida (71%), followed by Stramenopiles (20%) and Opisthokonta (5%) (Figure 6A). However, when represented by the percentages of OTUs for individual samples (Figure 6B), Alveolata (43%) was the most abundant group in strait plastic samples, followed by Archaeplastida (23%) and Stramenopiles (19%). For coastal plastic samples, almost half of the eukaryotic diversity was Archaeplastida (46%), with Stramenopiles accounting for 37% and Alveolata accounting for 10% (Figure 6B). In the case of seawater samples, Alveolata (78% and 64%) was the most dominant eukaryotic community in strait samples. However, Archaplastida (55% and 41%) was the dominant in coastal samples in terms of both reads and OTUs (Figures 6C, D).
To determine the number of species (as OTUs) associated exclusively with plastic, the eukaryotic microbial communities not associated with plastic samples were excluded. Of 663 OTUs from plastic samples, 127 OTUs (19%) were detected only in the plastics samples (Figure 2). These consisted of 67 Archaeplastida (54.0%), 45 Stramenopiles (35.7%), 12 Opisthokonta (9.5%), and 3 Alveolata (2.4%) OTUs. The plastic-specific OTUs belonged to four supergroups mentioned above, with 8 phyla, 15 classes (1 NA), 27 orders (2 NA), 38 families (3 NA), and 40 genera, including NA taxa. The most abundant phylum was Chlorophyta (41.3%), followed by Ochrophyta (31.7%) and Rhodophyta (12.7%), and the other groups were less than 10% (Table 1).
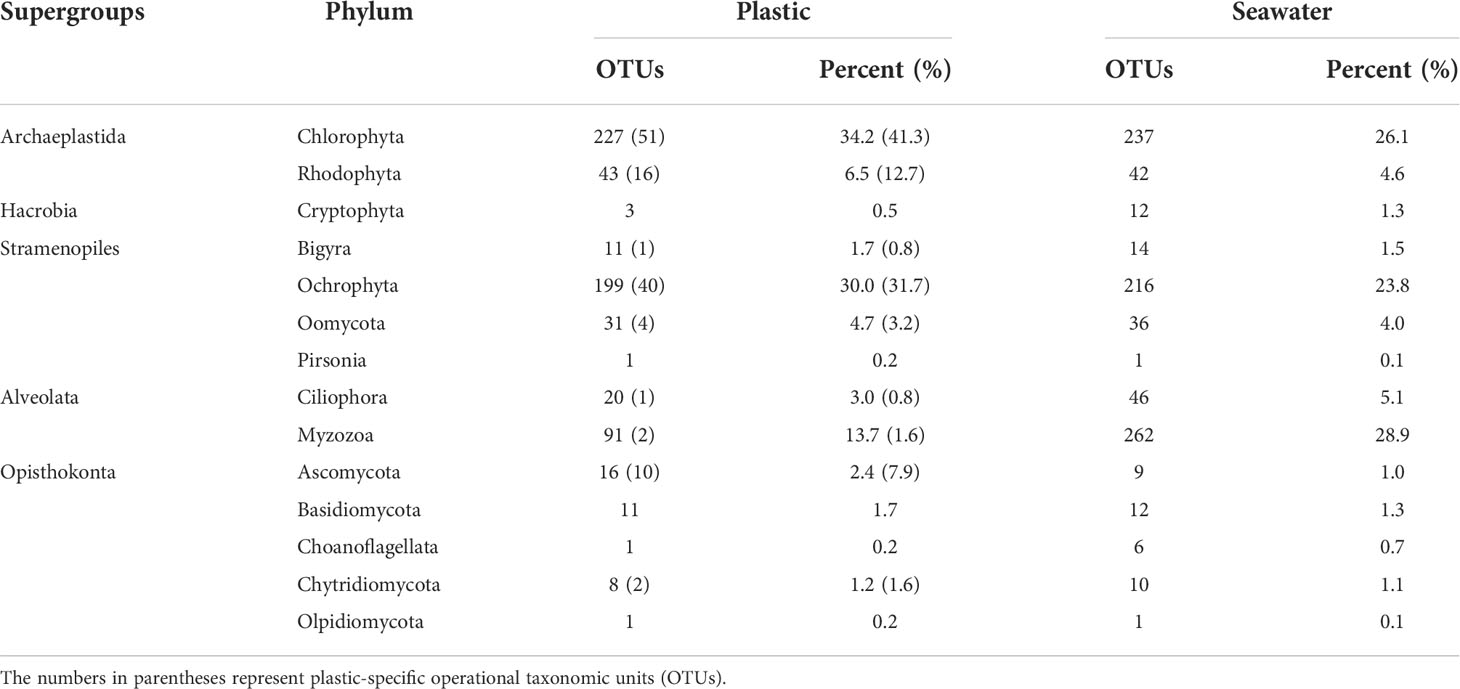
Table 1 The number of eukaryotic operational taxonomic units (OTUs) identified from plastic and seawater samples according to phylum.
While the Myzozoa OTUs ranged from 1 – 122 (mean = 23) in each sample, the number of reads varied from 1 – 26,112 (mean = 4,679) (data not shown). The 264 Myzozoa OTUs from all the samples were segregated into 2 classes, 13 orders, 38 families, and 53 genera. In the plastic samples, the 91 OTUs were segregated into 2 classes, 12 orders, 23 families, and 30 genera. Peridiniales (39%) and Gymnodiniales (34%) were the most abundant orders in the plastic samples (Figure 7A) and showed a similar abundance in communities from both the plastic and seawater samples (Figure 7). Suessiales, Syndiniales, and Thoracosphaerales prevailed in seawater (Figure 7B). The difference between the sampling sites (e.g., coastal and strait) was also observed. Although sparse, Blastodiniales (0.1%) and Noctilucales (0.04%) were only present in coastal plastic samples (04GN and 05JC, respectively; Figure 7A). In contrast, Pyrocystales (0.3%) was only present in strait plastic samples (03W1; Figure 7A).
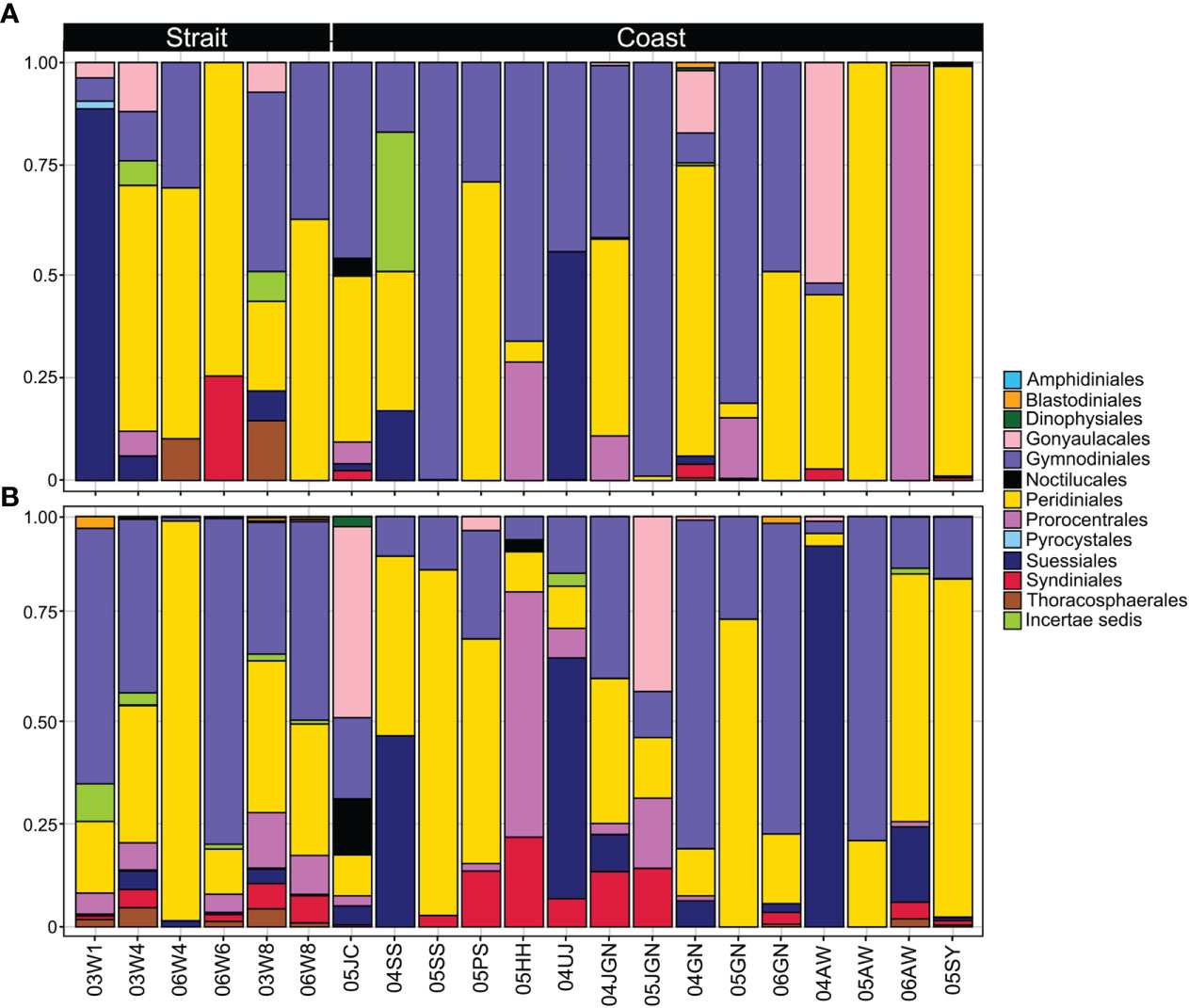
Figure 7 Relative abundance of dinoflagellate orders. (A), plastic samples; (B), seawater samples at the different sampling sites (from straits and coastal).
When ranked based on the number of the reads in all plastic samples, the most abundant dinoflagellate OTUs were Pentapharsodinium sp. (OTU21), followed by Lebouridinium glaucum (c.f. Katodinium glaucum; OTU16) and Karlodinium veneficum (OTU78) (Figure 8). Several benthic/epiphytic dinoflagellates (e.g., Coolia and Ostreopsis) were attached to the plastic (Figures 8, 9). Additionally, the diverse bloom-forming or toxic/harmful dinoflagellates were detected in plastic samples, although their abundances were relatively low. Among the 91 dinoflagellate OTUs obtained from plastic samples, 36 were potentially harmful (approximately 40%); these included members of genera Alexandrium, Coolia, Dinophysis, Heterocapsa, Karlodinium, Noctiluca, Ostreopsis, Prorocentrum, Scrippsiella, and Tripos (Figures 8, 9). Among those, Ostreopsis was detected only in the plastic samples.
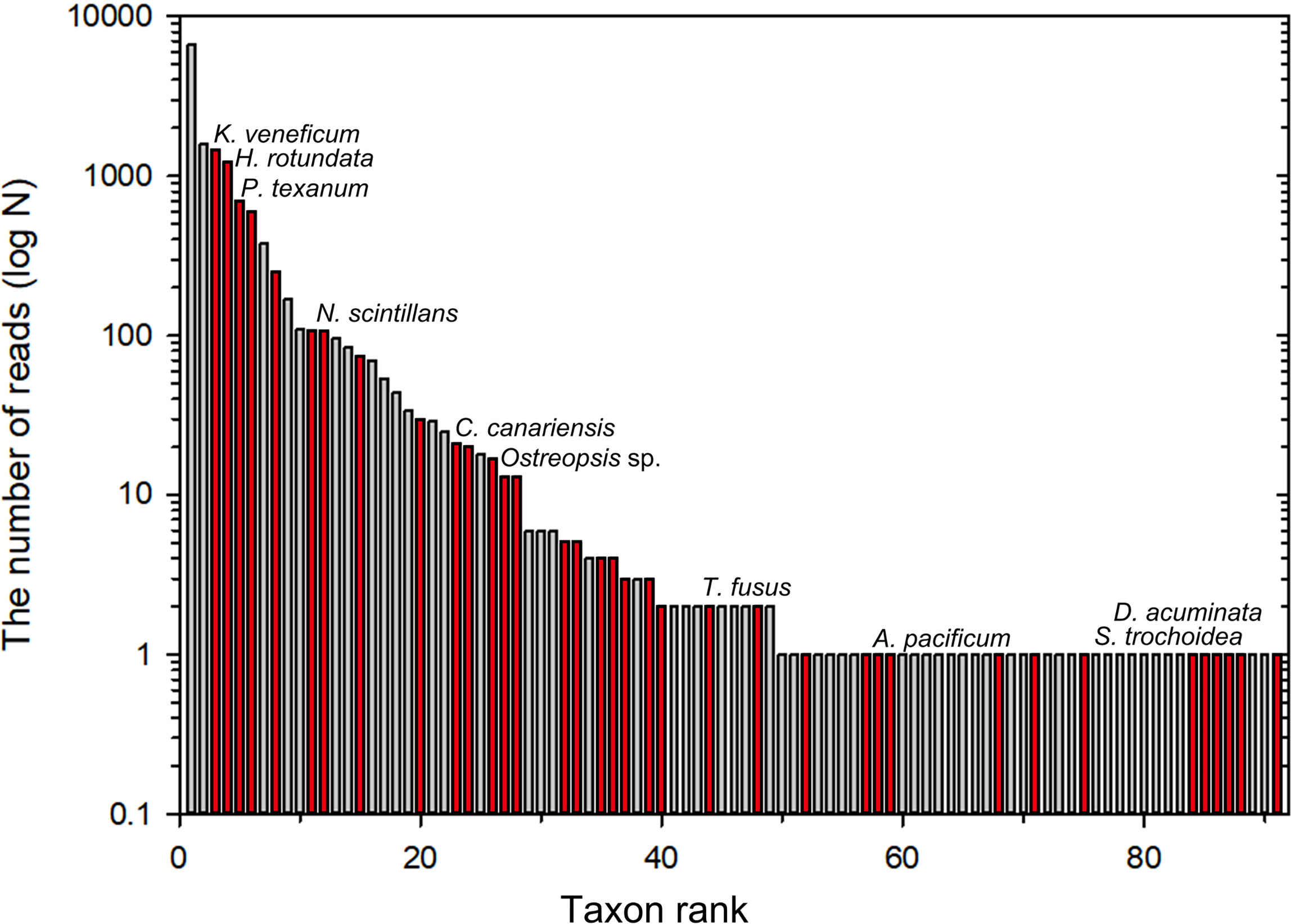
Figure 8 Bar graphs showing order by the number of reads of dinoflagellate operational taxonomic units (OTUs) detected in plastic samples. Red bars represent harmful (bloom-forming) or toxic dinoflagellates.
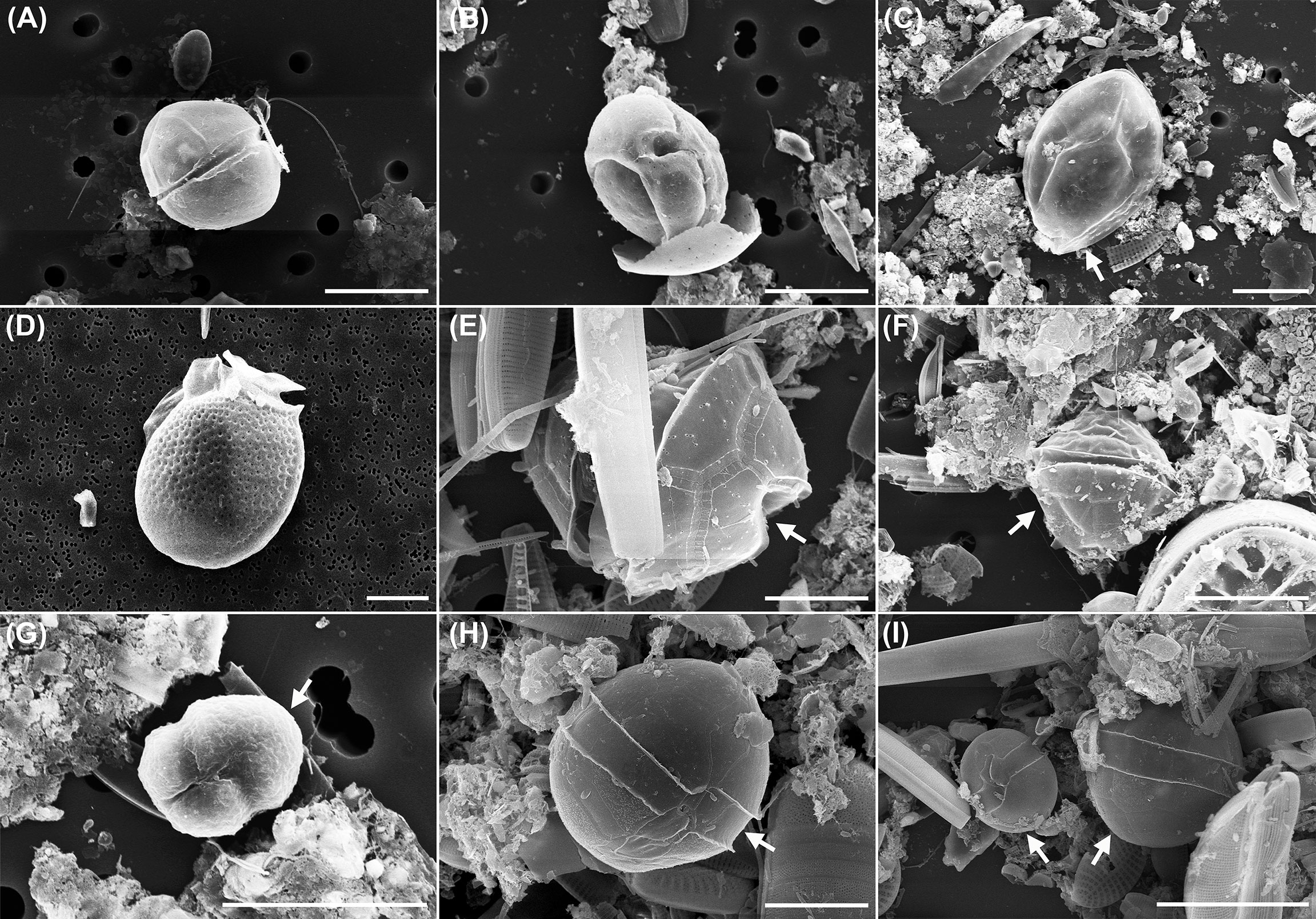
Figure 9 Dinoflagellates on plastic surfaces. (A, B) Coolia sp.; (C) Ostreopsis sp.; (D), Dinophysis acuminata; (E), Pentapharsodinium sp.; (F–I) Unidentified dinoflagellates. Scale bars: A–D, F, G, I = 20 µm; E, H = 10 µm. Note that the dinoflagellate images were obtained either by placing a sample scraped from the macroplastics surface on a membrane filter paper (A–G) or directly from the macroplastic surfaces (H, I). Dinoflagellates are indicated by arrows in C, E–I.
Of the 1035 OTUs, 129 OTUs belonged to eukaryotic fungi and parasites. They belonged to 6 different phyla and 11 orders, which are known to infect a wide range of hosts. Among them, the 86 parasite OTUs obtained from plastic samples occupied 8% of total OTUs and 13% of plastic OTUs. According to the references, they consisted of a total of four groups (Figure 10). In the parasite community of the plastisphere, the most dominant group was represented by fungi (42% of all parasite OTUs), followed by Oomycota (33%), Stramenopiles (14%), and dinoflagellates (12%). The fungi, which belong to the supergroup Opisthokonta, were composed of Ascomycota (44%), Basidiomycota (31%), and Chytridiomycota (25%) (Figure 10). Oomycota included five orders, namely Anisolpodiales, Haliphthorales, Leptomitales, Peronosporales, and Pontismatales, the latter being the most abundant (46%). Stramenopiles included four orders, namely Labyrinthulales, Oblongichytriales, Thraustochytriales, and Pirsonia-clade, with the highest number of OTUs belonging to the Thraustochytriales (42%). In the dinoflagellate group, most OTUs were Syndiniales-Group II, which includes the genus Amoebophrya. Dinophycean parasites Blastodinium and Dissodinium were also detected in lower read abundances (30%).
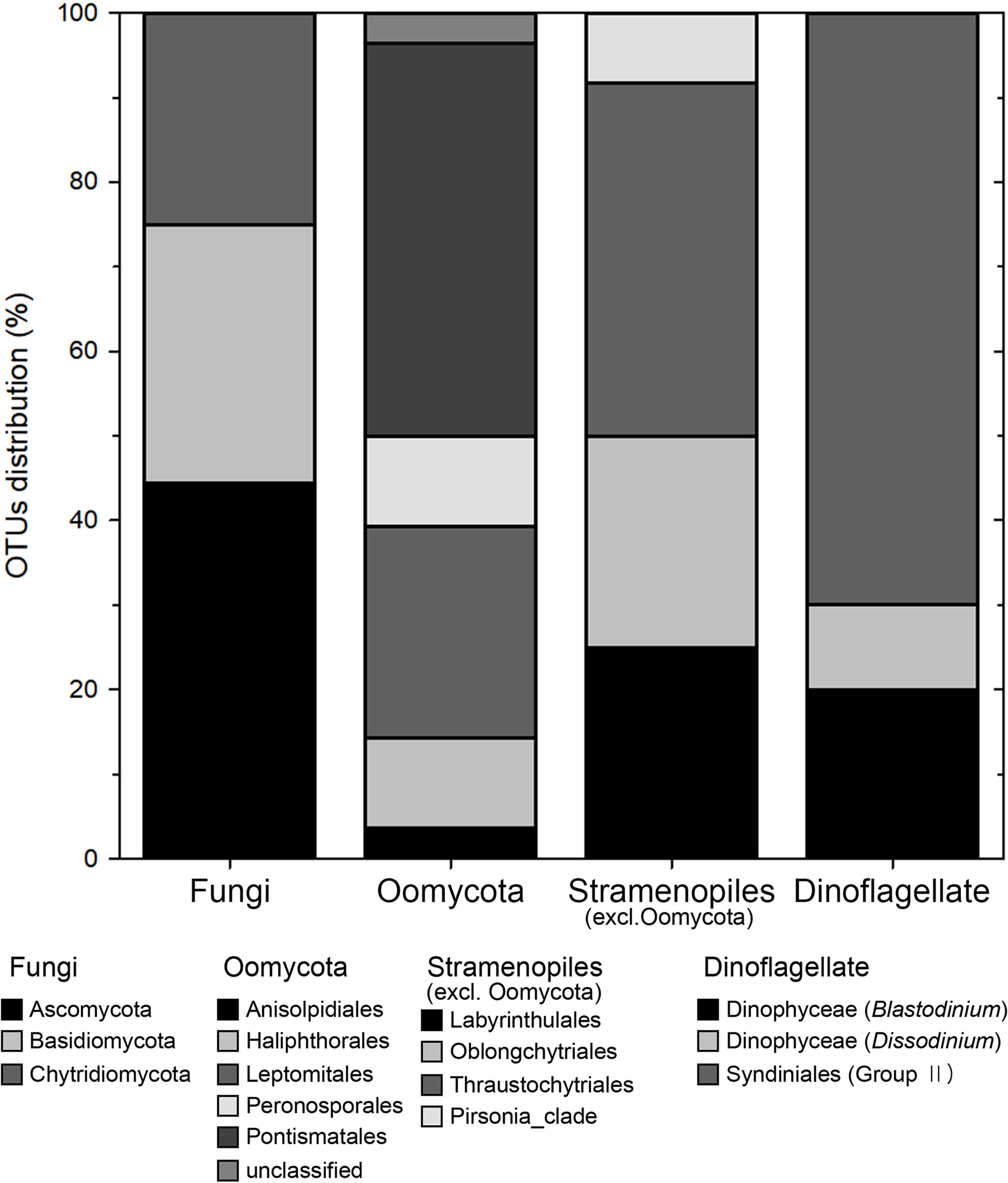
Figure 10 Stacked bar graphs representing eukaryotic parasite/pathogen groups. Each bar represents the relative proportion of operational taxonomic units (OTUs) distributions in each group.
The major findings from the present study were that (1) micro/macro plastics and surrounding seawater shared half of the eukaryotic microbial community composition in terms of the eukaryotic OTUs, and (2) the plastics harbored a variety of fungi, toxic or harmful dinoflagellates, and eukaryotic parasites.
Unlike some previous studies that have shown different bacterial community compositions on plastic compared to that in the surrounding seawater (Zettler et al., 2013; Amaral-Zettler et al., 2015; De Tender et al., 2015), the NMDS plot and ANOSIM test in this study showed no significant differences in eukaryotic microbial community composition between all plastic and seawater samples. Indeed, those two types of samples shared many eukaryotic OTUs (52% out of the 1,035 OTUs obtained in this study). This result indicates that the shared eukaryotic OTUs occurring on the plastics originate from the surrounding seawater or may be released from the plastics to the surrounding seawaters, acting as a vector to transport the eukaryotic microorganisms. Both imply the presence of active eukaryotic microbial interactions between the drifting plastics and surrounding seawater. Nonetheless, a notable difference in diversity patterns was found between the two types of samples. While the average number of observed species (richness) was much higher in seawater than on plastics, plastic samples showed greater evenness than the seawater samples. This result suggests that the eukaryotic microbial community on the plastics in the study area was not dominated by a few species, probably due to the high competition for the limited space resource available on the plastics.
Another interesting observation concerning eukaryotic diversity is that plastic samples collected from the strait and coast, showed a distinct difference in eukaryotic microbial community composition in terms of the percentage of OTUs; while Stramenopiles (40%) dominated the eukaryotic microbial community composition in strait plastic samples, both Archaeplastida (42%) and Stramenopiles (42%) were the most abundant in coastal plastic samples. In the present study, the supergroup Archaeplastida mainly consisted of macroalgae, which distribute worldwide, especially in shallow tropical and sub-tropical waters (Hanisak and Samuel, 1987; Mattio et al., 2008; Mattio and Payri, 2011), as well as release spores or seeds for widespread dispersal in the environment (Norton, 1992). Therefore, macroalgae spores or seeds are likely to easily adhere to the floating plastic surface in coastal areas where macroalgae species are relatively abundant. This may in part explain why Archaeplastida dominated the eukaryotic microbial community along with Stramenopiles in coastal plastic samples. Recently, De Tender et al. (2017) investigated the taxonomic composition of bacterial and fungal communities on various plastics at both a harbor and an offshore location in the Belgian part of the North Sea and showed the difference in bacterial and fungal colonization on plastic debris between the two different sampling sites. This and the present studies suggest that sampling locations could greatly influence the composition of microbial communities on floating plastic debris.
Apart from the sampling locations, other factors can also influence the colonization of plastic in the ocean, including plastic types (e.g., PE, PP, PS, and polyolefin particles), particle size and shape, time of the year (season), exposure time (e.g., biofilm age), environmental conditions such as nutrient concentration and hydrography (e.g., Oberbeckmann et al., 2014; Amaral-Zettler et al., 2015; Oberbeckmann et al., 2016; Kettner et al., 2019; He et al., 2022). Plastic types might contribute to the difference observed in diversity and composition of the microbial community on plastics collected from the strait and coast in the present study, as already shown in previous studies (e.g., Zettler et al., 2013; De Tender et al., 2015; Debroas et al., 2017). Unfortunately, however, their relative contributions associated with plastic types to the eukaryotic microbial community cannot be quantitatively assessed in this study because the present study did not distinguish plastic types. On the other hand, it seems unlikely that the time of year could cause the difference observed in this study between strait and coastal plastics, as samples analyzed in this study were collected over a limited period (i.e., April – June). Rather, plastic size and exposure time likely contribute to the observed difference between the two sample locations. In the present study, microplastics were collected using a manta net with a 330 μm pore size from the strait, and macroplastics were manually collected using a hand-made net from coastal sites. Given the different sampling tools employed to obtain plastic samples, this difference in plastic size suggests that while macroplastics have been recently introduced to sampling sites, microplastics may have been present in the ocean for longer. Thus, the recently introduced macroplastics are likely to harbor Archaeplastida (mostly Rhodophyta and Chlorophyta in this study) which is the most abundant at coastal sampling sites.
Despite the first report on the occurrence of diatoms and hydroids on plastics collected from the Sargasso Sea surface by Carpenter and Smith Jr. (1972), substantial attention was not paid to the plastisphere community until the start of this century (Masó et al., 2003). Masó et al. (2003) have reported the occurrence of several harmful dinoflagellates such as Alexandrium taylori, Coolia sp., and Ostreopsis sp. on plastic surfaces collected in coastal waters of the Mediterranean Sea. However, subsequent studies have mostly focused on reporting the presence of various pathogenic bacteria such as Vibrio and other microorganisms such as members of Campylobacteraceae, Aeromonas salmonicida, Arcobacter spp., and Pseudomonas alcaligenes (De Tender et al., 2015; Kirstein et al., 2016; Oberbeckmann et al., 2016; Frère et al., 2018; Jiang et al., 2018; Curren and Leong, 2019; Bowley et al., 2021), and research on eukaryotic microorganisms (including eukaryotic pathogens) is still limited. The present study demonstrated that various harmful or toxic dinoflagellates (e.g., Alexandrium, Coolia, Dinophysis, Karlodinium, Scrippsiella, and Ostreopsis) were frequently observed on plastics collected in our study area. These results are similar to those reported in previous studies that reported the presence of harmful dinoflagellates such as Alexandrium, Coolia, Ostreopsis, and diatom Pseudo-nitzschia attached to plastic surfaces in coastal waters of the Mediterranean Sea (Masó et al., 2003; Masó et al., 2016; Casabianca et al., 2019). This indicates that the plastisphere community should be considered a worldwide environmental concern, not restricted to a given local area. Furthermore, in the present study, several harmful or toxic dinoflagellates (e.g., Alexandrium, Dinophysis, Karlodinium, and Scrippsiella), which are generally regarded as planktonic, were frequently found on the plastics, further supporting the presence of active eukaryotic microbial interactions between the drifting plastics and surrounding seawaters. Thus, the floating plastics may act as a vector for these harmful (e.g., toxic and bloom-forming) species (Masó et al., 2003), facilitating their spread into new habitats. It is interesting to observe the subtropical benthic dinoflagellates such as Coolia and Ostreopsis on the drifting plastics from the present study area. Lee and Park (2020) recently reported that the geographic distribution of the benthic dinoflagellate Ostreopsis is expanding in coastal waters around the Korean peninsula through a phylogeographic analysis based on Internal Transcribed Spacer (ITS) and 5.8S regions of rDNA sequences, although the dispersal mechanism was not fully understood. The result from the present study suggests that drifting plastics could be an important alternative vector concerning the expansion of such benthic dinoflagellates in Korean coastal waters.
Along with several harmful dinoflagellates, another notable finding from this study was to reveal the presence of diverse fungi and eukaryotic parasites attached to the plastics. So far, limited studies have reported the presence of considerable fungal OTUs attached to marine plastics through a metabarcoding approach (De Tender et al., 2015; Oberbeckmann et al., 2016; De Tender et al., 2017; Debroas et al., 2017; Jiang et al., 2018). Similar to previous studies, however, fungal OTUs could not be identified to the species level in this study and, thus, the taxonomic resolution offered is little. Nonetheless, it is interesting to note that fungal OTUs belonging to Ascomycota and Basidiomycota predominated (75%) the fungal community on plastics in this study, indicating that these fungal groups might be the preferred candidates for isolation and cultivation for their potential use in plastic degradation. In addition, the presence of abundant chytrid and Pontismatales OTUs (Oomycota) on plastics in this study suggests that floating plastics may act as hotspots to transport vectors capable of infecting phytoplankton (in particular, diatoms) and rhodophytes (Buaya and Thines, 2020), respectively.
Parasitic dinoflagellates were also detected on plastics, although in low abundance. Most OTUs (7 out of 10) of the parasitic dinoflagellates belonged to the Syndiniales group II, which includes the genus Amoebophrya. However, five OTUs were grouped with environmental sequences, of which the hosts are unknown. Interestingly, two OTUs (OTU138 and 576) conformed to Amoebophrya sp. ex Cochlodinium polykrikoides (KF791347) occurred around the coast of Yeosu, Korea (Kim and Park, 2014). Despite the absence of Cochlodinium polykrikoides on the plastics, it was surprising to detect the two Amoebophrya sequences infecting the dinoflagellate host, as the formation of resting cysts in Amoebophrya species was unknown. Thus, this result indicates that Amoebophrya infecting the dinoflagellates may form a resting cyst on plastics at its free-living stage (i.e., zoospore) as a survival strategy during the absence of its host. On the other hand, Pennino et al. (2020) recently reported a positive relationship between the ingestion of microplastics in fish and parasitism in the northwestern Mediterranean. They found that approximately 60% occurrence of microplastics was detected inside the gut of fish Sardina pilchardus (European sardine) and Engraulis encrasicolus (European anchovy). Further, fish with microplastics in their digestive system also had parasites such as larvae, trematodes, and nematodes and generally had a worse overall body condition. The parasitic dinoflagellate genus Blastodinium, detected on plastics in the present study, is known to live in the intestines of marine copepods (Skovgaard and Salomonsen, 2009; Skovgaard et al., 2012), although the mechanism of infection remains unknown. Further studies are needed to test whether the copepod could be infected by ingesting a microplastic to which the Blastodinium parasite is attached, and if microplastic ingestion could act as a vector for transmission of Blastodinium infection in copepods.
In conclusion, our results demonstrate the presence of diverse eukaryotic microbial communities, including harmful or toxic dinoflagellates, fungi, oomycetes, and parasitic dinoflagellates, on floating plastic surfaces collected from Jeju Island and Jeju Strait in southern coastal areas of Korea. As suggested for pathogenic bacteria in previous studies, the drifting plastics can also act as mediums for transporting diverse eukaryotic microorganisms, including harmful dinoflagellates and parasites or pathogens. On the other hand, the plastics highly enriched with those eukaryotic microorganisms might increase their sinking rates, enabling them to be transported to the deep sea environment at a much higher speed rate. Testing whether the parasitic infection through microplastic ingestion occurs at a higher trophic level remains an open question for future studies.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
All authors listed have made a substantial, direct and intellectual contribution to the work and approved the manuscript for publication.
This study was supported by the National Research Foundation of Korea (research grant NRF-2016R1A6A1A03012647).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2022.985756/full#supplementary-material
Supplementary Figure 1 | The number of operational taxonomic units (OTUs) at each sample. Blue line, seawater samples; Red line, plastic samples.
Supplementary Figure 2 | Rarefaction curves for each sample, relating the number of operational taxonomic units (OTUs) detected depending on the sequencing effort.
Supplementary Figure 3 | Box plots representing three alpha-diversity estimators (observed species, Shannon and Simpson indices) for eukaryotic microbial community calculated with operational taxonomic units (OTUs) data and arranged according to sampling sites (strait, coast) and sampling time. Pink circles = plastic samples; Blue circles = seawater samples.
Supplementary Figure 4 | Diatoms on plastic surfaces. (A), Navicula sp.; (B), Cymbela sp.; (C), Lyrella sp.; (D), Arachnoidiscus sp.; (E), Licomphora sp.; (F), Chaetoceros sp.; (G), Cylindrotheca closterium; (H), Achnanthes sp.; (I), Grammatophora sp.; (J), Leptocylindrus sp.; (K), Cocconeis sp.; (L), Microplastic covered with pennate diatoms of genus Synedropsis. Scale bars: A = 5 µm; B, C, G–J = 20 µm; D–F, K, L = 50 µm.
Amaral-Zettler L. A., Zettler E. R., Slikas B., Boyd G. D., Melvin D. W., Morrall C. E., et al. (2015). The biogeography of the plastisphere: Implications for policy. Front. Ecol. Environ. 13, 541–546. doi: 10.1890/150017
Andrady A. L. (2011). Microplastics in the marine environment. Mar. pollut. Bull. 62, 1596–1605. doi: 10.1016/j.marpolbul.2011.05.030
Asiandu A. P., Wahyudi A., Sari S. W. (2021). A review: Plastics waste biodegradation using plastics-degrading bacteria. J. Environ. Treat Techniques 9, 148–157. doi: 10.47277/JETT/9(1)157
Barnes D. K. (2002). Invasions by marine life on plastic debris. Nature 416, 808–809. doi: 10.1038/416808a
Bowley J., Baker-Austin C., Porter A., Hartnell R., Lewis C. (2021). Oceanic hitchhikers–assessing pathogen risks from marine microplastic. Trends Microbiol. 29, 107–116. doi: 10.1016/j.tim.2020.06.011
Briand J.-F., Djeridi I., Jamet D., Coupé S., Bressy C., Molmeret M., et al. (2012). Pioneer marine biofilms on artificial surfaces including antifouling coatings immersed in two contrasting French Mediterranean coast sites. Biofouling 28, 453–463. doi: 10.1080/08927014.2012.688957
Browne M. A., Crump P., Niven S. J., Teuten E., Tonkin A., Galloway T., et al. (2011). Accumulation of microplastic on shorelines woldwide: sources and sinks. Environ. Sci. Technol. 45, 9175–9179. doi: 10.1021/es201811s
Browne M. A., Dissanayake A., Galloway T. S., Lowe D. M., Thompson R. C. (2008). Ingested microscopic plastic translocates to the circulatory system of the mussel, mytilus edulis (L.). Environ. Sci. Technol. 42, 5026–5031. doi: 10.1021/es800249a
Buaya A. T., Thines M. (2020). An overview on the biology and phylogeny of the early-diverging oomycetes. Philipp J. Syst. Biol. 14, 1–20. doi: 10.26757/pjsb2020a14004
Carpenter E. J., Smith J. K. (1972). Plastics on the Sargasso Sea surface. Science 175, 1240–1241. doi: 10.1126/science.175.4027.1240
Carson H. S., Nerheim M. S., Carroll K. A., Eriksen M. (2013). The plastic-associated microorganisms of the north pacific gyre. Mar. pollut. Bull. 75, 126–132. doi: 10.1016/j.marpolbul.2013.07.054
Casabianca S., Capellacci S., Giacobbe M. G., Dell’aversano C., Tartaglione L., Varriale F., et al. (2019). Plastic-associated harmful microalgal assemblages in marine environment. Environ. pollut. 244, 617–626. doi: 10.1016/j.envpol.2018.09.110
Choi D. H., Park K.-T., An S. M., Lee K., Cho J.-C., Lee J.-H., et al. (2015). Pyrosequencing revealed SAR116 clade as dominant dddP-containing bacteria in oligotrophic NW pacific ocean. PloS One 10, e0116271. doi: 10.1371/journal.pone.0116271
Cole M., Lindeque P., Halsband C., Galloway T. S. (2011). Microplastics as contaminants in the marine environment: A review. Mar. pollut. Bull. 62, 2588–2597. doi: 10.1016/j.marpolbul.2011.09.025
Curren E., Leong S. C. Y. (2019). Profiles of bacterial assemblages from microplastics of tropical coastal environments. Sci. Total Environ. 655, 313–320. doi: 10.1016/j.scitotenv.2018.11.250
Debroas D., Mone A., Ter Halle A. (2017). Plastics in the north Atlantic garbage patch: a boat-microbe for hitchhikers and plastic degraders. Sci. Total Environ. 599, 1222–1232. doi: 10.1016/j.scitotenv.2017.05.059
De Tender C. A., Devriese L. I., Haegeman A., Maes S., Ruttink T., Dawyndt P. (2015). Bacterial community profiling of plastic litter in the Belgian part of the north Sea. Environ. Sci. Technol. 49, 9629–9638. doi: 10.1021/acs.est.5b01093
De Tender C., Devriese L. I., Haegeman A., Maes S., Vangeyte J., Cattrijsse A., et al. (2017). Temporal dynamics of bacterial and fungal colonization on plastic debris in the north Sea. Environ. Sci. Technol. 51 (13), 7350–7360. doi: 10.1021/acs.est.7b00697
De Tender C., Schlundt C., Devriese L., Mincer T., Zettler E., Amaral-Zettler L. (2017). A review of microscopy and comparative molecular-based methods to characterize “Plastisphere” communities. Analytical Methods 9, 2132–2143. doi: 10.1039/C7AY00260B
Edgar R. C. (2016). UCHIME2: improved chimera prediction for amplicon sequencing. BioRxiv 074252. doi: 10.1101/074252
Eerkes-Medrano D., Thompson R. C., Aldridge D. C. (2015). Microplastics in freshwater systems: a review of the emerging threats, identification of knowledge gaps and prioritisation of research needs. Water Res. 75, 63–82. doi: 10.1016/j.watres.2015.02.012
Eo S., Hong S. H., Song Y. K., Lee J., Lee J., Shim W. J. (2018). Abundance, composition, and distribution of microplastics larger than 20 μm in sand beaches of south Korea. Environ. pollut. 238, 894–902. doi: 10.1016/j.envpol.2018.03.096
Eriksson C., Burton H. (2003). Origins and biological accumulation of small plastic particles in fur seals from macquarie island. AMBIO: A J. Hum. Environ. 32, 380–384. doi: 10.1579/0044-7447-32.6.380
Frère L., Maignien L., Chalopin M., Huvet A., Rinnert E., Morrison H., et al. (2018). Microplastic bacterial communities in the bay of Brest: Influence of polymer type and size. Environ. pollut. 242, 614–625. doi: 10.1016/j.envpol.2018.07.023
Gall S. C., Thompson R. C. (2015). The impact of debris on marine life. Mar. pollut. Bull. 92, 170–179. doi: 10.1016/j.marpolbul.2014.12.041
Gerritse J., Leslie H. A., De Tender C. A., Devriese L. I., Vethaak A. D. (2020). Fragmentation of plastic objects in a laboratory seawater microcosm. Sci. Rep. 10, 1–16. doi: 10.1038/s41598-020-67927-1
Gregory M. R. (2009). Environmental implications of plastic debris in marine settings–entanglement, ingestion, smothering, hangers-on, hitch-hiking and alien invasions. Philos. Trans. R. Soc. B: Biol. Sci. 364, 2013–2025. doi: 10.1098/rstb.2008.0265
Hammer J., Kraak M. H., Parsons J. R. (2012). Plastics in the marine environment: the dark side of a modern gift. Rev. Environ. Contamination Toxicol., 220, 1–44. doi: 10.1007/978-1-4614-3414-6_1
Hanisak M. D., Samuel M. A. (1987). “Growth rates in culture of several species of sargassum from Florida, USA,” in Twelfth international seaweed symposium (Dordrecht: Springer), 399–404. doi: 10.1007/978-94-009-4057-4_59
He S., Jia M., Xiang Y., Song B., Xiong W., Cao J., et al. (2022). Biofilm on microplastics in aqueous environment: Physicochemical properties and environmental implications. J. Hazardous Materials 424, 127286. doi: 10.1016/j.jhazmat.2021.127286
Ibabe A., Rayon F., Martinez J. L., Garcia-Vazquez E. (2020). Environmental DNA from plastic and textile marine litter detects exotic and nuisance species nearby ports. PloS One 15, e0228811. doi: 10.1371/journal.pone.0228811
Jang M., Shim W. J., Cho Y., Han G. M., Song Y. K., Hong S. H. (2020). A close relationship between microplastic contamination and coastal area use pattern. Water Res. 171, 115400. doi: 10.1016/j.watres.2019.115400
Jiang P., Zhao S., Zhu L., Li D. (2018). Microplastic-associated bacterial assemblages in the intertidal zone of the Yangtze estuary. Sci. Total Environ. 624, 48–54. doi: 10.1016/j.scitotenv.2017.12.105
Kettner M. T., Oberbeckmann S., Labrenz M., Grossart H. P. (2019). The eukaryotic life on microplastics in brackish ecosystems. Front. Microbiol. 10. doi: 10.3389/fmicb.2019.00538
Kiessling T., Gutow L., Thiel M. (2015). “Marine litter as habitat and dispersal vector,” in Marine anthropogenic litter (Cham: Springer), 141–181.
Kim S., Park M. G. (2014). Amoebophrya spp. from the bloom-forming dinoflagellate Cochlodinium polykrikoides: Parasites not nested in the “Amoebophrya ceratii complex”. J. Eukaryotic Microbiol. 61, 173–181. doi: 10.1111/jeu.12097
Kirstein I. V., Kirmizi S., Wichels A., Garin-Fernandez A., Erler R., Löder M., et al. (2016). Dangerous hitchhikers? evidence for potentially pathogenic Vibrio spp. on microplastic particles. Mar. Environ. Res. 120, 1–8. doi: 10.1016/j.marenvres.2016.07.004
Kwon O. Y., Kang J.-H., Hong S. H., Shim W. J. (2020). Spatial distribution of microplastic in the surface waters along the coast of Korea. Mar. pollut. Bull. 155, 110729. doi: 10.1016/j.marpolbul.2019.110729
Laist D. W. (1997). Impacts of marine debris: entanglement of marine life in marine debris including a comprehensive list of species with entanglement and ingestion records, in Marine debris (New York, NY: Springer), 99–139. doi: 10.1007/978-1-4613-8486-1_10
Law K. L., Thompson R. C. (2014). Microplastics in the seas. Science 345, 144–145. doi: 10.1126/science.1254065
Lee J., Hong S., Song Y. K., Hong S. H., Jang Y. C., Jang M., et al. (2013). Relationships among the abundances of plastic debris in different size classes on beaches in south Korea. Mar. pollut. Bull. 77, 349–354. doi: 10.1016/j.marpolbul.2013.08.013
Lee B., Park M. G. (2020). Distribution and genetic diversity of the toxic benthic dinoflagellate genus Ostreopsis in Korea. Harmful Algae 96, 101820. doi: 10.1016/j.hal.2020.101820
Lie H. J., Cho C. H. (2016). Seasonal circulation patterns of the yellow and East China seas derived from satellite-tracked drifter trajectories and hydrographic observations. Prog. Oceanogr. 146, 121–141. doi: 10.1016/j.pocean.2016.06.004
Masó M., Fortuño Alós J. M., De Juan S., Demestre M. (2016). Microfouling communities from pelagic and benthic marine plastic debris sampled across Mediterranean coastal waters. Scientia Marina 80, 117–127. doi: 10.3989/scimar.04281.10A
Masó M., Garcés E., Camp J. (2003). Drifting plastic debris as a potential vector for dispersing harmful algal bloom (HAB) species. Scientia Marina 67 (1), 107–111. doi: 10.3989/scimar.2003.67n1107
Mattio L., Dirberg G., Payri C. E., Andréfouët S. (2008). “Diversity, biomass and distribution pattern of sargassum beds in the south West lagoon of new Caledonia (South pacific),” in Nineteenth international seaweed symposium (Dordrecht: Springer), 361–373. doi: 10.1007/978-1-4020-9619-8_45
Mattio L., Payri C. E. (2011). 190 years of Sargassum taxonomy, facing the advent of DNA phylogenies. Botanical Rev. 77 (1), 31–70. doi: 10.1007/s12229-010-9060-x
Moresco V., Oliver D. M., Weidmann M., Matallana-Surget S., Quilliam R. S. (2021). Survival of human enteric and respiratory viruses on plastics in soil, freshwater, and marine environments. Environ. Res. 199, 111367. doi: 10.1016/j.envres.2021.111367
Norton T. A. (1992). Dispersal by macroalgae. Br. Phycological J. 27 (3), 293–301. doi: 10.1080/00071619200650271
Oberbeckmann S., Loeder M. G., Gerdts G., Osborn A. M. (2014). Spatial and seasonal variation in diversity and structure of microbial biofilms on marine plastics in northern European waters. FEMS Microbiol. Ecol. 90, 478–492. doi: 10.1111/1574-6941.12409
Oberbeckmann S., Osborn A. M., Duhaime M. B. (2016). Microbes on a bottle: substrate, season and geography influence community composition of microbes colonizing marine plastic debris. PloS One 11, e0159289. doi: 10.1371/journal.pone.0159289
Pennino M. G., Bachiller E., Lloret-Lloret E., Albo-Puigserver M., Esteban A., Jadaud A., et al. (2020). Ingestion of microplastics and occurrence of parasite association in Mediterranean anchovy and sardine. Mar. pollut. Bull. 158, 111399. doi: 10.1016/j.marpolbul.2020.111399
Reisser J., Shaw J., Hallegraeff G., Proietti M., Barnes D. K., Thums M., et al. (2014). Millimeter-sized marine plastics: a new pelagic habitat for microorganisms and invertebrates. PloS One 9, e100289. doi: 10.1371/journal.pone.0100289
Ryan P. G., Moore C. J., Van Franeker J. A., Moloney C. L. (2009). Monitoring the abundance of plastic debris in the marine environment. Philos. Trans. R. Soc. B: Biol. Sci. 364, 1999–2012. doi: 10.1098/rstb.2008.0207
Schloss P. D., Westcott S. L., Ryabin T., Hall J. R., Hartmann M., Hollister E. B., et al. (2009). Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75, 7537–7541. doi: 10.1128/AEM.01541-09
Silva M. M., Maldonado G. C., Castro R. O., De Sá Felizardo J., Cardoso R. P., Dos Anjos R. M., et al. (2019). Dispersal of potentially pathogenic bacteria by plastic debris in guanabara bay, RJ, Brazil. Mar. pollut. Bull. 141, 561–568. doi: 10.1016/j.marpolbul.2019.02.064
Skovgaard A., Karpov S. A., Guillou L. (2012). The parasitic dinoflagellates Blastodinium spp. inhabiting the gut of marine, planktonic copepods: morphology, ecology, and unrecognized species diversity. Front. Microbiol. 3. doi: 10.3389/fmicb.2012.00305
Skovgaard A., Salomonsen X. M. (2009). Blastodinium galatheanum sp. nov.(Dinophyceae) a parasite of the planktonic copepod Acartia negligens (Crustacea, calanoida) in the central Atlantic ocean. Eur. J. Phycol. 44, 425–438. doi: 10.1080/09670260902878743
Stoeck T., Bass D., Nebel M., Christen R., Jones M. D., Breiner H. W., et al. (2010). Multiple marker parallel tag environmental DNA sequencing reveals a highly complex eukaryotic community in marine anoxic water. Mol. Ecol. 19, 21–31. doi: 10.1111/j.1365-294X.2009.04480.x
Sussarellu R., Suquet M., Thomas Y., Lambert C., Fabioux C., Pernet M. E. J., et al. (2016). Oyster reproduction is affected by exposure to polystyrene microplastics. Proc. Natl. Acad. Sci. 113, 2430–2435. doi: 10.1073/pnas.1519019113
Thompson R. C., Olsen Y., Mitchell R. P., Davis A., Rowland S. J., John A. W., et al. (2004). Lost at sea: where is all the plastic? Science 304, 838–838. doi: 10.1126/science.1094559
Von Moos N., Burkhardt-Holm P., KoüHler A. (2012). Uptake and effects of microplastics on cells and tissue of the blue mussel Mytilus edulis l. after an experimental exposure. Environ. Sci. Technol. 46, 11327–11335. doi: 10.1021/es302332w
Keywords: fungi, plastisphere, metabarcoding, pathogen, vector
Citation: Lee B and Park MG (2022) Drifting marine plastics as new ecological habitats for harmful eukaryotic microbial communities in Jeju Strait, Korea. Front. Mar. Sci. 9:985756. doi: 10.3389/fmars.2022.985756
Received: 04 July 2022; Accepted: 13 October 2022;
Published: 26 October 2022.
Edited by:
Kuoping Chiang, National Taiwan Ocean University, TaiwanReviewed by:
Ruei-Feng Shiu, National Taiwan Ocean University, TaiwanCopyright © 2022 Lee and Park. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Myung Gil Park, bXBhcmtAY2hvbm5hbS5hYy5rcg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.