
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mar. Sci. , 16 August 2022
Sec. Marine Ecosystem Ecology
Volume 9 - 2022 | https://doi.org/10.3389/fmars.2022.980695
This article is part of the Research Topic Degradation, Ecological Restoration and Adaptive Management of Estuarine Wetlands under Intensifying Global Changes View all 20 articles
Soil salinization has been one of the main causes of ecosystem degradation in many estuarine wetlands under global climate changes, but it remains unclear how salinization shifts the phenotypic variability and genetic diversity of the foundation plant species in estuarine wetlands. To reveal the effects of salinization on natural populations of foundation plant species, we investigated the intraspecific variation of Phragmites australis using five functional traits (shoot height, leaf length, panicle length, seed number per panicle, and mass per seed) and ten microsatellite markers in the five sites across the Yellow River Delta. The salinity was indicated by electrical conductivity, and the reproductive strategy was estimated by the ratio of panicle length to shoot height. The linear models showed that the shoot height, leaf length, and panicle length had significantly negative correlations to soil salinity, while the mass per seed had a significantly positive correlation to soil salinity. However, there were no significant relationships between the seed number per panicle or reproductive ratio and soil salinity. The genetic diversity within populations was high in all sites (He > 0.5), but the genetic differentiation between populations was very weak (FST from 0.0074 to 0.0212), which suggested that there was a strong genetic flow among populations. Genetic structure analyses showed two phylogenetic groups of P. australis distributed in four of five surveyed sites across the Yellow River Delta. Our study also found significant phylogenetic signals in the leaf length and mass per seed, suggesting a substantial role of phylogenetic relationship (technically, neutral genetic relatedness) in intraspecific variation and salt adaptation of P. australis. Our study provides novel insight into the adaptative responses of the foundation plant species to soil salinization from individual traits to population genetics and offers significant implications for ecological restoration and adaptive management of saline lands in estuarine wetlands.
Soil salinization is an increasingly serious problem due to extreme drought events and sea-level rise caused by global climate change and high-intensity land use (Yu et al., 2014; Zhou et al., 2022). The area of saline land has increased from 800 million hectares in 1991 to about 1 billion hectares of soil worldwide (Hopmans et al., 2021) and continues to expand at a speed of 2.00×106 hectares per year (Abbas et al., 2013). Estuarine wetland, one of the most sensitive ecosystems to salinization, can provide protection from windstorms, fishery production, biodiversity maintenance, and the prevention of biological invasion (Barbier, 2013). Soil salinization has resulted in vegetation decline, biodiversity reduction, and ecosystem degradation in many estuarine wetlands worldwide (Day et al., 2000; Herbert et al., 2015). Understanding the vegetation response to soil salinization in estuarine wetlands is significant for predicting the ecological effects of soil salinization and making adaptive management of the estuarine wetland in the future.
Soil salinization could directly affect the physiological processes and deeply shift the ecological strategies of plant individuals. There have been numerous methods to characterize ecological strategies, such as the competitor, stress-tolerator, ruderal (CSR) strategy (Grime, 1977), leaf economics spectrum (Wright et al., 2004), reproductive strategy (Doust, 1989), and seed size-number trade-off (Venable, 1992). One of the most important dimensions of reproductive strategy is that plants tend to allocate more resources early to reproductive growth when the environment becomes extreme (Metz et al., 2020). Seed size-number trade-off suggested the plants could give their reproductive efforts to provide large seeds that could allow them to have a high survival probability in the stressful conditions under low resource availability, or they could spread their reproductive efforts among many small seeds to maximize adaptability or dispersal under high resource availability or serious disturbance (Coomes and Grubb, 2003). However, there are few studies on plant ecological strategies under the influence of salinity currently at a population level within species (Agrawal, 2020).
Genetic diversity, containing adaptive and neutral diversity within populations or species, plays an important role in the maintenance and adaptation of plants in different degraded habitats (Holderegger et al., 2006). Most molecular markers presently used in population genetics are usually considered neutral, such as AFLPs (amplified fragment length polymorphisms) and microsatellites. The neutral diversity can demonstrate genetic processes, including gene flow and drift at the population level (Holderegger et al., 2006), but it may also indicate the natural selection when the neutral diversity is observed to change with environmental factors because these neutral loci could also be associated to adaptive traits or specific phylogenetic groups. Low genetic diversity is caused by genetic drift and inbreeding, which usually symbolizes an unhealthy population growth rate. A high level of genetic diversity can ensure the adaptive potential of natural populations in the face of rapidly changing environmental pressures. For example, the research on Zostera marina suggested that a high level of genetic diversity could enhance the resistance to disturbance (Hughes and Stachowicz, 2004). The changes in genetic diversity under environmental shifts can help reveal the potential driving forces that promote population differentiation and adaptation, and predict how the population responds to the changing environment (Gao et al., 2012). Gene flow between populations could reduce genetic differentiation. Recently, phylogenetic analysis has been a popular method to link the evolutionary process with the plant ecological features among different species. The phenomenon that close phylogenetic relatives have similar traits is usually termed the phylogenetic signal of the traits (Blomberg and Garland, 2002; Collyer et al., 2022).
Phragmites australis (Cav.) Trin. ex Steud. (common reed) is a significant wetland foundation species widely distributed worldwide (Eller et al., 2017). Phragmites australis has several distinct phylogeographical groups (Saltonstall, 2016) and ploidy levels ranging from 3× to 12× (Clevering and Lissner, 1999), which give them a high adaptive potential to environmental shifts. As a pseudohalophyte, P. australis could grow in freshwater and slightly saline wetlands (Eller et al., 2017). High salinity usually hinders the shoot growth, photosynthetic rate, and final total biomass of P. australis (Achenbach et al., 2013; Naidoo, 2021). To survive saline conditions, P. australis tends to allocate more resources to reproductive traits to ensure their offspring’s survival or escape (Meyerson et al., 2000; Farrer et al., 2021). The salt tolerance of P. australis may be attributed to the geographic and phylogenetic origin, but not the ploidy level (Achenbach et al., 2013; Lambertini et al., 2020). However, little is known about how soil salinization affects the intraspecific variation of P. australis populations combined with different phylogenetic groups.
The Yellow River Delta is one of the three largest estuarine deltas in China, located in the alluvial plain at the mouth of the Yellow River, near the Bohai Sea. Salinization in the Yellow River Delta has become a troubling issue due to seawater, groundwater evaporation, and storm-tide disasters (Yu et al., 2014; Wang et al., 2017). The Yellow River Delta has varied saline conditions in the small-scale environment, and P. australis are widely distributed in the Yellow River Delta, providing an ideal system for salinization’s ecological effects on P. australis. The previous studies have well documented the responses of P. australis in morphological traits and genetic diversity indicators to saline conditions (Guo et al., 2003; Gao et al., 2012; Zhou et al., 2021). Our recent studies have indicated that there are two distinct lineages (phylogenetic groups) of P. australis, i.e., tetraploid lineage O and octoploid lineage P (Liu et al., 2022b) in the Yellow River Delta, and lineage O might be more tolerant to salinization (Liu et al., 2021b). However, these studies reported few traits related to propagation, such as seed size and number, and did not link the trait variability to genetic relatedness between different genotypes.
In this study, we investigated functional traits and microsatellites of P. australis in the Yellow River Delta to uncover the following scientific questions: (i) how salinity alters the trait trade-off strategies in natural P. australis populations; (ii) whether the genetic diversity is related to soil salinity; and (iii) whether the evolutionary history limits the intraspecific functional trait related to salt adaptation. Our research could help understand the evolutionary mechanism of foundation plant species under soil salinization, and offer implications for ecosystem conservation and restorations in estuarine wetlands.
In October 2020, five populations of P. australis in different habitats were investigated in the Yellow River Delta (37.76° -38.05°N, 118.67° -119.17°E) (Figure 1). We investigated about 20 P. australis shoots of each population and recorded the habitat type and coexistence of plant species. To reduce the probability of sampling the same individuals, the distance between sampled shoots was at least 50 m. We recorded the geographical information of each shoot with a GPS instrument and measured the soil electrical conductivity of each reed at a 5-10 cm depth using an EC 110 Meter portable conductometer (Spectrum Technologies, Georgia, America). A soil with EC greater than or equal to 4 dS m-1 at 25°C is defined as saline soil (Richards, 1954).
We measured shoot height (SH) and leaf length (LL) for all sampled shoots and panicle length (PL) for the shoots with panicles. Shoot height was measured from the soil surface to shoot tip point (usually panicle or leaf tip, sometimes stem tip). We selected the largest leaf to measure LL from all complete, healthy, and mature leaves for each shoot. In sites wu01 and wu04, the seed number per panicle (SN) and mass per seed (SM) were also estimated. We counted the seed number from about a quarter of seed weight for each panicle to calculate SM and then SN. A ratio of panicle length to shoot height was used to quantify the reproductive allocation ratio (R). Leaves on the upper part of each plant were harvested and placed in a plastic bag with silica gel, and transported to the laboratory.
DNA was extracted with the improved CTAB plant DNA extraction method from leaves conserved in silica gel. Leaves frozen in liquid nitrogen were ground into powder using 3 mm steel beads in a JXFSTPRP-24 automatic sample rapid grinder (Shanghai Jingxin Industrial Development Co., Ltd., Shanghai, China). 900 μL of 65°C preheated 2% CTAB extract was added to each tube, and the mixture was vortexed vigorously. Tubes were incubated at 60°C for one hour and were put under vortex movement every 15 min. These tubes were centrifuged at 12000 rpm for 30 seconds, and the supernatant was transferred to new centrifuge tubes. 600 μL 25:24:1 phenol/chloroform/isoamyl alcohol was added to each tube, and these were mixed slowly. Then tubes were centrifuged at 12000 rpm for 10 min, and the supernatant was again transferred to new tubes. An equal volume of 24:1 chloroform/isoamyl alcohol was added to each tube, and these were mixed slowly. Then tubes were centrifuged at 12000 rpm for 10 min, and the supernatant was again retained. Approximate 1.5 μL RNase was added to each tube and kept at 37°C for one hour, followed by extraction with an equal volume of 24:1 chloroform/isoamyl alcohol. Then tubes were centrifuged at 12000 rpm for 10 min, and the supernatant was again retained. About 0.6 volume of cold isopropanol was added and mixed by inversion. These tubes were placed at room temperature for 20 min for precipitation. After precipitation, the tubes were centrifuged for 5 min at 10000 rpm, and the supernatant was poured off carefully. Then the pellet was washed twice using 600 µl cold 70% ethanol. After the pellet was dried completely, it was suspended in 50-100 μL TE buffer (or sterile water) and stored at - 20°C. Its quality was analyzed with a 1% agarose gel electrophoresis and Micro-Spectrophotometer (YIMA, Xi’an, China).
To infer the lineages of sampled P. australis in a large evolutionary background, we added the individuals from Lianyungang (LYG) and Tangshan (TS) to our microsatellite analysis, which were identified as lineage P for LYG and O for TS with chloroplast fragments and nuclear microsatellites in a large sample from China (Liu et al., 2022b). Ten pairs of primers developed by Saltonstall (2003) and Yu et al. (2016) were used to amplify the microsatellite polymorphism of P. australis (n = 96). We used five primer sets (set1 for PaGT4 and PaGT9; set2 for PaGT12 and PaGT13; set3 for PaGT14 and PaGT16; set4 for PaGT21 and PaGT22; and set5 for Phra315 and Phra522), and each forward primer had a fluorochrome tag (FAM, HEX, or TAMRA) on its 5’ tail. To ensure the method’s repeatability, 10% of randomly chosen samples had two replicates—the reaction volume and reaction procedure we used by Liu et al. (2022b). Then the product was sequenced by the AB13730XL sequencer, and the data file was imported into GeneMapper 2.2.0 for allele identification.
To evaluate the genetic diversity, we calculated the number of alleles (Na), Shannon’s information index (I), expected heterozygosity (He), and Bruvo distances using the R package polysat (v1.7-5). The FST as the measure of differentiation between population pairs was estimated using R package polysat. Genetic structure was visualized by principal coordinate analysis (PCoA) and hierarchical clustering with function cmdscale and hclust in R software. A Bayesian clustering approach with STRUCTURE 2.3.4 (Pritchard et al., 2000; Falush et al., 2007) was used to assign individuals to clusters (K) based on their genotypes. We used the admixture model and ran STRUCTURE ten times for each K (from 1 to 7) with 20,000 burn-in iterations followed by 100,000 Markov Chain Monte Carlo iterations. The most likely K was inferred with the ΔK method of Evanno et al. (2005) using the online tool structure harvest. Finally, the output was averaged and visualized by CLUMPP and DISTRUCT 1.1.
The coefficient of variation (CV) was calculated to express the variation between plant traits under variable electrical conductivity. We performed Shapiro-Wilk test for normality of plant traits with function shapiro.test in R package stats, and Levene’s test for homogeneity of variance among different populations or lineages with function leveneTest in R package car. Data were log-transformed to achieve approximate normality if necessary. When the data have approximate normality and equal variance, we used one-way ANOVA with function aov in R package stats to test the significant difference among different groups and then used the Duncan post-hoc test to complete the multiple comparisons. Otherwise, we used Kruskal-Wallis test to finish multiple comparisons with function kruskal in R package agricolae. The results of multiple comparisons were visualized by boxplots using R package graphics.
To quantify the relationships between P.australis traits and electrical conductivity, we performed a general linear model and linear mixed model with R package nlme, and calculated the AIC, R2, and P of each model to determine which model’s description was more appropriate. Spearman correlation analysis was performed to test the relationships between each traits. The output was visualized by the pheatmap function in the pheatmap package of R. The trait records with missing values were deleted. The relationship between electrical conductivity and genetic diversity was performed by the general linear model, and the fitting function was tested by joint hypotheses test and coefficient of determination.
To determine the phylogenetic signals of continuous traits, we calculated Pagel’s λ (Pagel, 1999) and Blomberg’s K (Blomberg and Garland, 2002) with R package phytools (Revell, 2012). Brownian motion (BM) can represent the random process of evolution and is applied as a random walk model in these estimations. The λ is a value between 0 and 1. λ = 0 indicates no phylogenetic dependence, and λ = 1 indicates that trait variation depends on the phylogeny (Freckleton et al., 2002). Blomberg’s K was tested with 10,000 randomization simulations. K = 0 indicates no significant signal, and K > 1 suggests stronger similarities among closely related lineages than expected under BM (Blomberg et al., 2003).
The soil electrical conductivity varied from 1.83 ± 1.14 dS m-1(wu02) to 9.75 ± 6.38 dS m-1 (wu04) (dS m-1, deciSiemens per metre; 1 dS m-1 ~ 0.55 ppt, as the main salt type is NaCl in the Yellow River Delta). The reed stands in site wu01 resulted from secondary succession from farmland abandoned due to a recent storm surge several years ago and held at least nine plant species. Site wu02 was a monodominant stand of P. australis with a few Suaeda salsa individuals in a shallow river (abandoned course of the Yellow River). Site wu01 and wu02 had slightly saline soils (Table 1). The saline soils were found in site wu03 (meadow patches in oil field) and wu05 (an intertidal zone near the Yellow River), and the hypersaline soils were found in site wu04 (an intertidal zone with Spartina alterniflora invasion) (Table 1). However, all sites had large EC variances (Figure 1; CV of EC in Table 1).
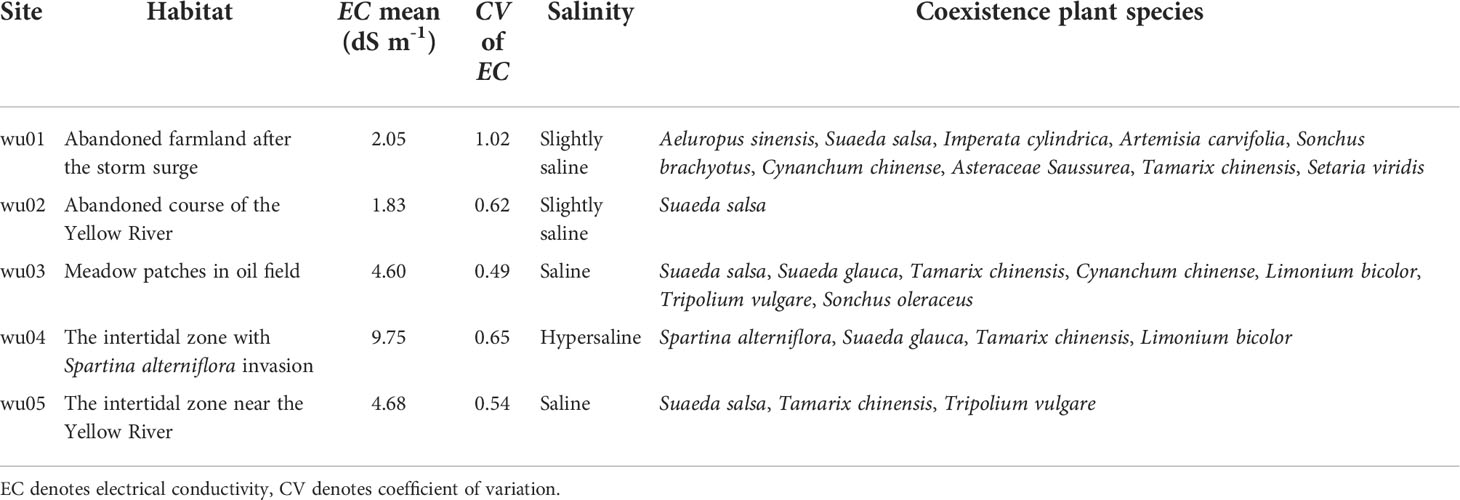
Table 1 The habitat information of sampled Phragmites australis populations in the Yellow River Delta.
There were significant differences in shoot height, leaf length, reproductive allocation, and mass per seed among populations (Figure 2; Table S1), but were no significant differences in panicle length and seed number per panicle (Figure 2, P > 0.05). Shoot height (175.95 ± 45.11 cm) and leaf length (26.40 ± 8.28 cm) were highest in the slightly saline soils (wu02) but lowest (118.65 ± 43.69 cm, 17.75 ± 6.73 cm) in the hypersaline soils (wu04) (Figure 2). The panicle length ranged from 18.53 ± 5.26 cm (wu04, hypersaline soils) to 23.67 ± 2.21 cm (wu02, slightly saline soils). In the wu02 population (slightly saline soils), the reproductive allocation was the lowest (0.11 ± 0.02), and in the wu01 population (slightly saline soils) was the highest (0.16 ± 0.02) (Figure 2).

Figure 2 Plant functional traits (A, shoot height; B, leaf length; C, panicle length; D, reproductive allocation, i.e., the ratio of panicle length to shoot height; E, log-transformed seed number per panicle; F, log-transformed mass per seed) of Phragmites australis from different populations. Different letters represent significant differences. In the box diagram, the middle horizontal line is the median, and the box frame is the interquartile distance (IQR). Each whisker extends to the furthest data point in each wing that is within 1.5 times the IQR, data points further than 1.5IQR are marked with a dot. ns represents no significance; · P < 0.1; *P < 0.05; **P < 0.01; ***P < 0.001.
The variation of shoot height was highest in the hypersaline soil (Table 2, CVSH = 0.37 in wu04 population) but lowest in the slightly saline soil (Table 2, CVSH = 0.16 in wu01 population). The variation of leaf length was highest in the hypersaline soil (Table 2, CVLL = 0.38 in wu04 population) but lowest in the saline soil (Table 2, CVLL = 0.18 in wu05 population). The variation of panicle length was highest in the hypersaline soil (Table 2, CVPL = 0.28 in wu04 population) but lowest in the slightly saline soil (Table 2, CVPL = 0.09 in wu02 population).
The general linear models across all sites showed that the soil electrical conductivity was negatively correlated to shoot height, leaf length, and panicle length but positively correlated to mass per seed (Figure 3, P < 0.01). There was no significant correlation between electrical conductivity and reproductive allocation or seed number per panicle (Figure 3, P > 0.05). The general linear models separately performed in 5 populations revealed some conflicting results among sites (Figure S1), indicating a strong site effect on the correlations between traits and salinity. The linear mixed models with site as the random factor found the positive correlations of EC to reproductive allocation (AIC = -264.62) and to mass per seed (AIC = -58.95) but the negative correlations to shoot height (AIC = 1049.47), leaf length (AIC = 678.68), panicle length (AIC = 390.14) and seed number per panicle (AIC = 636.17) (Figure 4; Table S2).

Figure 3 Relationships between Phragmites australis traits (A, shoot height; B, leaf length; C, panicle length; D, reproductive allocation, i.e. the ratio of panicle length to shoot height; E, seed number per panicle; F, mass per seed) and soil electrical conductivity across five populations based on general linear models. **P < 0.01; ***P < 0.001.
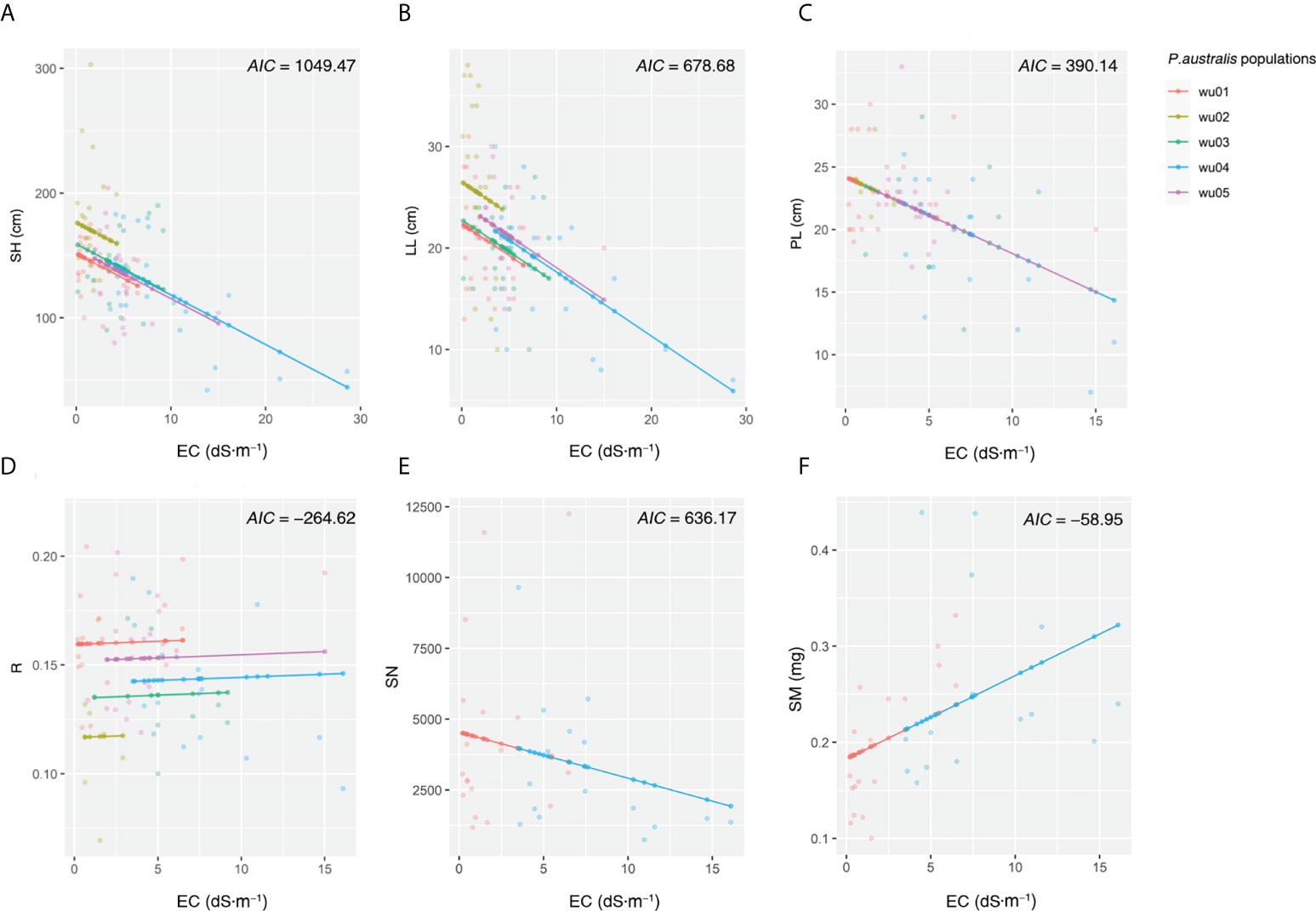
Figure 4 Relationships between Phragmites australis traits (A, shoot height; B, leaf length; C, panicle length; D, reproductive allocation, i.e., the ratio of panicle length to shoot height; E, seed number per panicle; F, mass per seed) and electrical conductivity in five populations based on linear mixed models with the random factor of site.
According to Spearman correlation analysis between traits (Figure S2), the shoot height of P. australis had the significant positive correlations with the leaf length (r = 0.50, P < 0.001), panicle length (r = 0.57, P < 0.001) and seed number per panicle (r = 0.56, P < 0.01). The panicle length had the significant positive correlations with the leaf length (r = 0.53, P < 0.001) and seed number per panicle (r = 0.61, P < 0.001).
The number of alleles (Na) varied from 5.90 (wu03, saline soils) to 9.00 (wu01, slightly saline soils). Shannon’s information index (I) varied from 2.94 (wu02, wu03, wu04, and wu05) to 3.00 (wu01, slightly saline soils), and the expected heterozygosity (He) varied from 0.56 (wu03, saline soils) to 0.70 (wu02 and wu04) (Table 2, Figure S3). There were no significant correlations between genetic diversity indicators and soil EC (Figure S3). Pairwise genetic differentiation (FST) values ranged between 0.0074 (wu01 and wu02) and 0.0212 (wu03 and wu04) (Table 3).
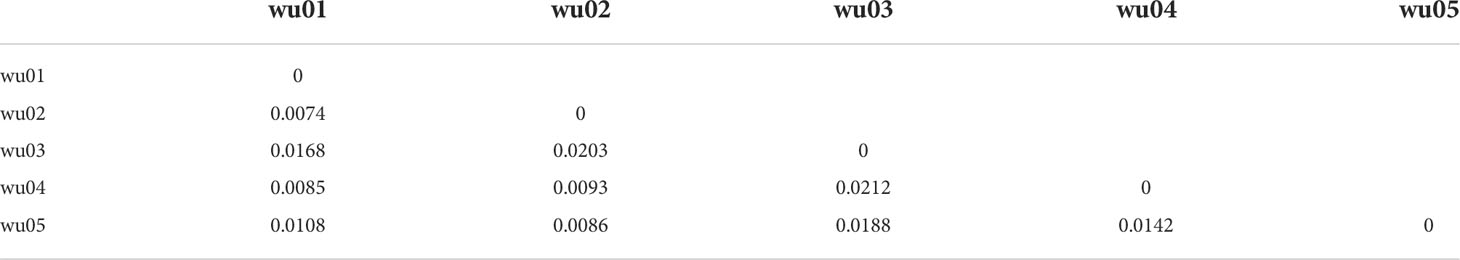
Table 3 Pairwise genetic differentiation (Fst) matrix between five Phragmites australis populations.
The first and the second axes of PCoA explained 21.52% and 8.72% of the total variance. The PCoA showed two clusters separated by the first axis (Figure 5A). The UPGMA clustering and Bayesian methods also supported these two genetic clusters (Figures 5B–D). Since the LYG and TS populations used in this study were regarded as the lineage P and O, respectively, the two genetic clusters in the Yellow River Delta were reasonable to be inferred into these two lineages. According to the results of these three methods, we determined 77 genotypes of lineage O and 19 genotypes of lineage P in populations from the Yellow River Delta. There were no significant difference of soil EC between two lineages (t-test, P = 0.188).

Figure 5 Microsatellite-based genetic structure of Phragmites australis populations in the Yellow River Delta, Lianyungang (LYG, lineage P), and Tangshan (TS, lineage O). (A) Principal coordinate analysis. (B) UPGMA clustering. Blue indicates lineage P, and orange indicates lineage O. (C) ΔK-value at different K. (D) STRUCTURE plot at K = 2. A thin line depicts each individual, and its color depends on its partitioning into the K clusters.
According to the Pagel’s λ, the leaf length had a significant phylogenetic signal (λ = 0.639, P < 0.05), the mass per seed had a marginally significant phylogenetic signal (λ = 0.488, P = 0.069). Shoot height, panicle length, reproductive allocation, and seed number per panicle did not have any phylogenetic signal (Table 4). The Blomberg’s K demonstrated that the leaf length and mass per seed had a significant phylogenetic signal (K > 0.80, P < 0.05), but other traits did not (Table 4). In addition, lineage O has a larger panicle length and a larger seed number per panicle but a lower mass per seed than lineage P (Figure 6; Table S3). The leaf length of lineage O was marginally smaller than lineage P (Figure 6B; P = 0.072).

Figure 6 Plant functional trait comparisons (A, shoot height; B, leaf length; C, panicle length; D, reproductive allocation, i.e., the ratio of panicle length to shoot height; E, log-transformed seed number per panicle; F, log-transformed mass per seed) between lineage O and P of Phragmites australis. Different letters represent significant differences. In the box diagram, the middle horizontal line is the median, and the box frame is the interquartile distance (IQR). Each whisker extends to the furthest data point in each wing that is within 1.5 times the IQR, data points further than 1.5 IQR are marked with a dot. ns represents no significance; · P < 0.1; *P < 0.05; **P < 0.01.
Consistent with the previous study in the Yellow River Delta (Zhou et al., 2021), P. australis in the saline habitats is likely to adopt a stress-tolerator strategy that plants reduce shoot height (Figure 3A), leaf size (Figure 3B), and other morphological traits to survival in stressful environments. The study in perennial grass Lolium perenne revealed that frost-and drought-sensitive populations had high growth rates, wide leaves, and erect growth habits, which helped them deal with interspecific and intraspecific competition in suitable conditions (Dolferus, 2014), and similar trade-off between salinity tolerance and competitive ability also exists in P. australis (Qi et al., 2018; Zhou et al., 2021). Competitor strategy that plants invest more resources in the rapid growth of large organs to outcompete neighbors is advantageous in highly productive habitats with low stress and disturbance (Vasseur et al., 2018). In the recently abandoned farmland (site wu01), the interspecific competition might be the main biological pressure, since there were many other plant species (Table 1) and the P. australis stands were preliminarily established after the recent storm surge. In other sites, the environmental filtering might be the key driver shaping the community assemble and trait variation.
Besides tolerant and competitive traits, reproductive traits also play a significant role in the functional responses of plants to soil salinization. Perennial plants usually give priority to sexual reproduction in variable and stressful environments (Yang and Kim, 2018), but the relationship between salinity and reproductive allocation was not significant in the sampled P. australis populations (Figure 3D). Sexual reproduction is considered uncommon in mature reed beds, but it seems crucial for the establishment in new sites (Albert et al., 2015). Rhizome-based asexual reproduction allows plants spread rapidly and farther (Douhovnikoff and Hazelton, 2014). However, our study found a seed size-number trade-off: the panicle length and seed number were decreased with salinization (Figures 4C, E) while the mass per seed increased (Figure 4F). Smaller seeds can save the cost of high seed production, but seedlings from larger seed plants have more competitiveness (Jakobsson and Eriksson, 2000; Leishman, 2001). Our results suggested that the reproduction investment of P. australis was used to provide large seeds that could tolerate saline habitats and ensure a high survival probability in extreme conditions.
Although soil salinization is dynamic over time and depth, our data showed significant correlations between functional traits and one-time measurement of topsoil EC with a probe method. On the one hand, the results suggest the functional response of P. australis is so robust that the measurement uncertainty of EC in our study cannot affect the statistical results. On the other hand, there might be a significant correlation between soil ECs in different depths and stages. Previous investigations of soil salinity in the Yellow River Delta have well documented the significant positive correlation in soil salinity between different soil depths (Yu et al., 2014; Wang et al., 2017). Since the elevation in natural micro-topography is the major factor in soil salinity at the fine scale (Wang et al., 2017), the rainfall or tidal inundation might not change the difference among different sites over time. The coexistent plant records near every sample also support the reliability of our salinity estimations. For example, in site wu01, nine coexisting plant species were observed, but Suaeda salsa was almost the only coexistent plant where the EC was higher than 1.00 dS m-1. However, continuous observation in multiple depths and composition analysis of salt ions are needed for the further understanding of the mechanisms underpinning the functional response of wetland plants to different salinization conditions.
The neutral genetic diversity can demonstrate processes such as natural selection, migration, and dispersal (Holderegger et al., 2006). In the previous studies, there was a high genetic diversity within P. australis populations in the Yellow River Delta (Gao et al., 2012; Liu et al., 2021b). We also found a very high level of genetic diversity in all populations except the population in the oil field (site wu03) and no significant relationships between the genetic diversity and salinity. The low genetic diversity of P. australis in site wu03 might be the very strong selection of genotypes due to the high-intensity disturbance in saline habitats during the oil production activities. Soil contamination from oil pollutants and drilling muds are well known to have negative impacts on plant and animal growth and survival, including reproductive investments. The intermediate disturbance might increase the genetic diversity by providing chances for external seed establishment, but extreme disturbance, stressful conditions or their combination could prevent the establishment of some sensitive genotypes (e.g., lineage P in wu03). Combined with previous studies, we could conclude that P. australis could always keep a high genetic diversity level under habitat fragmentation (Liu et al., 2021a), annual winter mowing (Kuprina et al., 2022), human management (Paul et al., 2011), and hydrographic modification (Naugžemys et al., 2021), but the synergistic impacts of natural stressors and human disturbances might impair the genetic diversity of P. australis populations.
Although our results did not find a significant relationship between genetic diversity and soil salinity due to the limited population size, previous studies found high salinity could decrease the genetic diversity of P. australis in the Yellow River Delta (Guo et al., 2003; Gao et al., 2012). The selection target has been identified as the homozygote of allele 203 at locus PaGT9 (Gao et al., 2012) and the tetraploid lineage O (Liu et al., 2021b). In this study, the PCoA and FST results suggested little differentiation and frequent gene flow between populations (Figure 5, Table 3), suggesting that there are few gene exchange barriers among populations in the Yellow River Delta and all populations might constitute a single metapopulation, which is similar to that found in the southern Baltic Sea (Kuprina et al., 2022). The mixed populations with two lineages also supported that limited diffusion might unlikely lead to the spatial genetic structure of P. australis population in the Yellow River Delta.
Niche conservatism and adaptive radiation play a central role in ecological diversification, and these two processes can be regarded as both ends of the spectrum for phenotypic evolution (Ackerly, 2009; Münkemüller et al., 2012). Niche conservatism represents a process of slow evolution, and adaptive radiation represents a process of rapid adaptation to environmental shifts. The phylogenetic conservation of plant traits such as leaf size and seed mass has been well analyzed among different species (Ackerly, 2009; Zanetti et al., 2020; Liu et al., 2022a), but to the best of our knowledge, our study is one of the first studies to apply the phylogenetic approach for trait evolution among so closely related lineages within a species (Agrawal, 2020). Within P. australis, we found a significant phylogenetic signal of the leaf length and the mass per seed (Table 4). There are many challenges for the interpretation of this method within species. The first concern is the certainty of our phylogenetic trees because it is a more difficult task to reconstruct phylogenetic history at lower taxonomic levels. The frequent, even free, gene communications among different individuals will mislead the phylogenetic reconstruction and alter the evolutionary implications of phylogenetic signal analysis at the individual level. Nevertheless, it could be a possible alternative to population Qst-Fst comparison when not enough samples are available, or populations are hard to define.
Phylogenetic conservatism of leaf length and the mass per seed might be mainly ascribed to the genetic differentiation between two lineages of P. australis. The ploidy difference has hindered the hybridization and introgression between these two lineages (Liu et al., 2022b), and the adaptive gene related to functional traits might also be fixed in a specific lineage (Gao et al., 2012; Liu et al., 2021a). The larger leaf length and seed mass of lineage P provided more evidence for the phylogenetic conservatism of leaf length and the mass per seed. The result of leaf length comparison is well consistent with our observation in the common garden (Liu et al., 2021b), which highlights the role of genetic factors in intraspecific variation of leaf size in the natural populations of P. australis. Previous studies suggest that lineage O of P. australis is more salt tolerant than lineage P (Lambertini et al., 2020; Liu et al., 2021b), but our study revealed that lineage P has a larger mass per seed which might facilitate its offspring survival and growth in saline conditions. Seed germination experiments for genotypes from two lineages are needed for screening genetic candidates for survival in higher salinity conditions. However, the effect of phenotypic plasticity cannot be ignored in the natural variation of plant traits at the individual level (Agrawal, 2020). The high level of phenotypic plasticity of both lineages allows them to survive in different saline habitats across nearly all sites in this study.
Our study revealed the high functional and genetic variation of P. australis in different saline habitats of the Yellow River Delta. The results indicated that salinization could reduce the vegetative growth but increase investment in some reproductive traits like mass per seed to ensure the seed vigor in stressful habitats. Moreover, the phylogenetic signal of mass per seed implied a phylogenetic limitation on salt adaptation due to the strong fixation of functional traits related to salt tolerance in P. australis lineages. Our study provides novel insights into the adaptation of P. australis to soil salinization from the aspects of traits, genetics, and phylogeny, and could be helpful in restoration management in estuarine wetlands under global environmental changes.
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
YW, LL, and WG conceived the idea and designed the study. YW, LL, and MY contributed to the field survey, sample collection, and molecular laboratory work. LL and YW analyzed and interpreted the data. YW and LL drafted the manuscript, and all authors participated in manuscript modifications and gave final approval for publication.
This work is supported by the National Natural Science Foundation of China (No. 32100304; 31970347), Open Project Fund of Key Laboratory of Ecological Prewarning, Protection and Restoration of Bohai Sea, Ministry of Natural Resources (No. 2022101), Postdoctoral Innovation Project of Shandong, and Postdoctoral Research Project of Qingdao.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2022.980695/full#supplementary-material
Abbas A., Khan S., Hussain N., Hanjra M. A., Akbar S. (2013). Characterizing soil salinity in irrigated agriculture using a remote sensing approach. Phys. Chem. Earth 55-57, 43–52. doi: 10.1016/j.pce.2010.12.004
Achenbach L., Eller F., Nguyen L. X., Brix H. (2013). Differences in salinity tolerance of genetically distinct Phragmites australis clones. AoB Plants 5, plt019. doi: 10.1093/aobpla/plt019
Ackerly D. (2009). Conservatism and diversification of plant functional traits: Evolutionary rates versus phylogenetic signal. Proc. Natl. Acad. Sci. U.S.A. 106, 19699–19706. doi: 10.1073/pnas.0901635106
Agrawal A. A. (2020). A scale-dependent framework for trade-offs, syndromes, and specialization in organismal biology. Ecology 101 (2), e02924. doi: 10.1002/ecy.2924
Albert A., Brisson J., Belzile F., Turgeon J., Lavoie C. (2015). Strategies for a successful plant invasion: the reproduction of Phragmites australis in north-eastern north America. J. Ecol. 103 (6), 1529–1537. doi: 10.1111/1365-2745.12473
Barbier E. B. (2013). Valuing ecosystem services for coastal wetland protection and restoration: Progress and challenges. Resources 2 (3), 213–230. doi: 10.3390/resources2030213
Blomberg S. P., Garland T. (2002). Tempo and mode in evolution: Phylogenetic inertia, adaptation and comparative methods. J. Evol. Biol. 15 (6), 899–910. doi: 10.1046/j.1420-9101.2002.00472.x
Blomberg S. P., Garland T., Ives A. R. (2003). Testing for phylogenetic signal in comparative data: Behavioral traits are more labile. Evolution 57 (4), 717–745. doi: 10.1111/j.0014-3820.2003.tb00285.x
Clevering O. A., Lissner J. (1999). Taxonomy, chromosome numbers, clonal diversity and population dynamics of Phragmites australis. Aquat. Bot. 64 (3-4), 185–208. doi: 10.1016/S0304-3770(99)00059-5
Collyer M. L., Baken E. K., Adams D. C. (2022). A standardized effect size for evaluating and comparing the strength of phylogenetic signal. Methods Ecol. Evol. 13 (2), 367–382. doi: 10.1111/2041-210X.13749
Coomes D. A., Grubb P. J. (2003). Colonization, tolerance, competition and seed-size variation within functional groups. Trends Ecol. Evol. 18 (6), 283–291. doi: 10.1016/S0169-5347(03)00072-7
Day J. W., Psuty N. P., Perez B. C. (2000). “The role of pulsing events in the functioning of coastal barriers and wetlands: Implications for human impact, management and the response to sea level rise,” in Concepts and controversies in tidal marsh ecology. Eds. Weinstein M. P., Kreeger D. A. (Dordrecht: Springer Netherlands), 633–659.
Dolferus R. (2014). To grow or not to grow: A stressful decision for plants. Plant Sci. 229, 247–261. doi: 10.1016/j.plantsci.2014.10.002
Douhovnikoff V., Hazelton E. L. G. (2014). Clonal growth: Invasion or stability? a comparative study of clonal architecture and diversity in native and introduced lineages of Phragmites australis (Poaceae). Am. J. Bot. 101 (9), 1577–1584. doi: 10.3732/ajb.1400177
Doust J. L. (1989). Plant reproductive strategies and resource allocation. Trends Ecol. Evol. 4 (8), 230–234. doi: 10.1016/0169-5347(89)90166-3
Eller F., Skálová H., Caplan J. S., Bhattarai G. P., Burger M. K., Cronin J. T., et al. (2017). Cosmopolitan species as models for ecophysiological responses to global change: The common reed Phragmites australis. Front. Plant Sci. 8, 1833. doi: 10.3389/fpls.2017.01833
Evanno G., Regnaut S., Goudet J. (2005). Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol. Ecol. 14 (8), 2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x
Falush D., Stephens M., Pritchard J. K. (2007). Inference of population structure using multilocus genotype data: Dominant markers and null alleles. Mol. Ecol. Notes 7 (4), 574–578. doi: 10.1111/j.1471-8286.2007.01758.x
Farrer E. C., Birnbaum C., Waryszak P., Halbrook S. R., Brady M. V., Bumby C. R., et al. (2021). Plant and microbial impacts of an invasive species vary across an environmental gradient. J. Ecol. 109 (5), 2163–2176. doi: 10.1111/1365-2745.13629
Freckleton R. P., Harvey P. H., Pagel M. (2002). Phylogenetic analysis and comparative data: A test and review of evidence. Am. Nat. 160 (6), 712–726. doi: 10.1086/343873
Gao L., Tang S., Zhuge L., Nie M., Zhu Z., Li B., et al. (2012). Spatial genetic structure in natural populations of Phragmites australis in a mosaic of saline habitats in the Yellow River Delta, China. PloS One 7 (8), e43334. doi: 10.1371/journal.pone.0043334
Grime J. P. (1977). Evidence for the existence of three primary strategies in plants and its relevance to ecological and evolutionary theory. Am. Nat. 111 (982), 1169–1194. doi: 10.1086/283244
Guo W., Wang R., Zhou S., Zhang S., Zhang Z. (2003). Genetic diversity and clonal structure of Phragmites australis in the Yellow River Delta of China. Biochem. Syst. Ecol. 31 (10), 1093–1109. doi: 10.1016/S0305-1978(03)00032-2
Herbert E. R., Boon P., Burgin A. J., Neubauer S. C., Franklin R. B., Ardón M., et al. (2015). A global perspective on wetland salinization: Ecological consequences of a growing threat to freshwater wetlands. Ecosphere 6 (10), 206. doi: 10.1890/ES14-00534.1
Holderegger R., Kamm U., Gugerli F. (2006). Adaptive vs. neutral genetic diversity: Implications for landscape genetics. Landscape Ecol. 21 (6), 797–807. doi: 10.1007/s10980-005-5245-9
Hopmans J. W., Qureshi A. S., Kisekka I., Munns R., Grattan S. R., Rengasamy P., et al. (2021). "Critical knowledge gaps and research priorities in global soil salinity," in advances in agronomy (the United States: Academic Press), 1–191.
Hughes A. R., Stachowicz J. J. (2004). Genetic diversity enhances the resistance of a seagrass ecosystem to disturbance. Proc. Natl. Acad. Sci. U.S.A. 101 (24), 8998–9002. doi: 10.1073/pnas.0402642101
Jakobsson A., Eriksson O. (2000). A comparative study of seed number, seed size, seedling size and recruitment in grassland plants. Oikos 88 (3), 494–502. doi: 10.1034/j.1600-0706.2000.880304.x
Kuprina K., Seeber E., Schnittler M., Landeau R., Lambertini C., Bog M. (2022). Genetic diversity of common reed in the southern Baltic Sea region-is there an influence of disturbance? Aquat. Bot. 177, 103471. doi: 10.1016/j.aquabot.2021.103471
Lambertini C., Guo W. Y., Ye S. Y., Eller F., Guo X., Li X. Z., et al. (2020). Phylogenetic diversity shapes salt tolerance in Phragmites australis estuarine populations in East China. Sci. Rep. 10 (1), 17645. doi: 10.1038/s41598-020-74727-0
Leishman M. R. (2001). Does the seed size/number trade-off model determine plant community structure? An assessment of the model mechanisms and their generality. Oikos 93 (2), 294–302. doi: 10.1034/j.1600-0706.2001.930212.x
Liu L., Wang J., Ma X., Li M., Guo X., Yin M., et al. (2021a). Impacts of the yellow river and qingtongxia dams on genetic diversity of Phragmites australis in ningxia plain, China. Aquat. Bot. 169, 103341. doi: 10.1016/j.aquabot.2020.103341
Liu H., Ye Q., Simpson K. J., Cui E., Xia J. (2022a). Can evolutionary history predict plant plastic responses to climate change? N Phytol 235(3)1260-1271. doi: 10.1111/nph.18194
Liu L. L., Yin M. Q., Guo X., Wang J. W., Cai Y. F., Wang C., et al. (2022b). Cryptic lineages and potential introgression in a mixed-ploidy species (Phragmites australis) across temperate China. J. Syst. Evol. 60 (2), 398–410. doi: 10.1111/jse.12672
Liu L. L., Yin M. Q., Guo X., Yu X. N., Song H. J., Eller F., et al. (2021b). The river shapes the genetic diversity of common reed in the Yellow River Delta via hydrochory dispersal and habitat selection. Sci. Total Environ. 764, 144382. doi: 10.1016/j.scitotenv.2020.144382
Metz J., Lampei C., Bäumler L., Bocherens H., Dittberner H., Henneberg L., et al. (2020). Rapid adaptive evolution to drought in a subset of plant traits in a large-scale climate change experiment. Ecol. Lett. 23 (11), 1643–1653. doi: 10.1111/ele.13596
Meyerson L. A., Saltonstall K., Windham L., Kiviat E., Findlay S. (2000). A comparison of Phragmites australis in freshwater and brackish marsh environments in North America. Wetlands Ecol. Manage. 8 (2), 89–103. doi: 10.1023/A:1008432200133
Münkemüller T., Lavergne S., Bzeznik B., Dray S., Jombart T., Schiffers K., et al. (2012). How to measure and test phylogenetic signal. Methods Ecol. Evol. 3 (4), 743–756. doi: 10.1111/j.2041-210X.2012.00196.x
Naidoo G. (2021). Salt tolerance of the African haplotype of Phragmites australis (Poaceae). Afr. J. Ecol. 59 (3), 724–734. doi: 10.1111/aje.12876
Naugžemys D., Lambertini C., Patamsytė J., Butkuvienė J., Khasdan V., Žvingila D. (2021). Genetic diversity patterns in phragmites australis populations in straightened and in natural river sites in Lithuania. Hydrobiologia 848 (14), 3317–3330. doi: 10.1007/s10750-021-04606-w
Pagel M. (1999). Inferring the historical patterns of biological evolution. Nature 401 (6756), 877–884. doi: 10.1038/44766
Paul J., Kirk H., Freeland J. (2011). Genetic diversity and differentiation of fragmented reedbeds (Phragmites australis) in the United Kingdom. Hydrobiologia 665 (1), 107–115. doi: 10.1007/s10750-011-0608-5
Pritchard J. K., Stephens M., Donnelly P. (2000). Inference of population structure using multilocus genotype data. Genetics 155 (2), 945–959. doi: 10.1093/genetics/155.2.945
Qi M., Sun T., Xue S., Yang W., Shao D., Martinez-Lopez J. (2018). Competitive ability, stress tolerance and plant interactions along stress gradients. Ecology 99 (4), 848–857. doi: 10.1002/ecy.2147
Revell L. J. (2012). Phytools: An r package for phylogenetic comparative biology (and other things). Methods Ecol. Evol. 3 (2), 217–223. doi: 10.1111/j.2041-210X.2011.00169.x
Richards L. A. (1954). Diagnosis and improvement of saline and alkali soils. AIBS Bull. 4 (3), 14–14. doi: 10.1093/aibsbulletin/4.3.14-a
Saltonstall K. (2003). Microsatellite variation within and among North American lineages of Phragmites australis. Mol. Ecol. 12 (7), 1689–1702. doi: 10.1046/j.1365-294X.2003.01849.x
Saltonstall K. (2016). The naming of Phragmites haplotypes. Biol. Invasions 18 (9), 2433–2441. doi: 10.1007/s10530-016-1192-4
Vasseur F., Sartori K., Baron E., Fort F., Kazakou E., Segrestin J., et al. (2018). Climate as a driver of adaptive variations in ecological strategies in Arabidopsis thaliana. Ann. Bot. 122 (6), 935–945. doi: 10.1093/aob/mcy165
Venable D. L. (1992). Size-number trade-offs and the variation of seed size with plant resource status. Am. Nat. 140 (2), 287–304. doi: 10.1086/285413
Wang Z., Zhao G., Gao M., Chang C. (2017). Spatial variability of soil salinity in coastal saline soil at different scales in the Yellow River Delta, China. Environ. Monit. Assess. 189 (2), 80. doi: 10.1007/s10661-017-5777-x
Wright I. J., Reich P. B., Westoby M., Ackerly D. D., Baruch Z., Bongers F., et al. (2004). The worldwide leaf economics spectrum. Nature 428 (6985), 821–827. doi: 10.1038/nature02403
Yang Y. Y., Kim J. G. (2018). The optimal balance between sexual and asexual reproduction in variable environments: A systematic review. J. Ecol. Environ. 40 (1), 1–18. doi: 10.1007/s12665-013-2980-0
Yu S., Zhang Y. X., Ren Y. L., Sun Q. X. (2016). Isolation and characterization of microsatellite loci for Phragmites australis. J. Genet. 93, E89–E92. doi: 10.1007/s12041-013-0278-3
Yu J. B, Li Y. Z., Han G. X., Zhou D., Fu Y. Q., Guan B., et al (2014). The spatial distribution characteristics of soil salinity in coastal zone of the Yellow River Delta. Environmental Earth Sciences 72(2), 589–599. doi: 10.1007/s12665-013-2980-0
Zanetti M., Dayrell R. L. C., Wardil M. V., Damasceno A., Fernandes T., Castilho A., et al. (2020). Seed functional traits provide support for ecological restoration and ex situ conservation in the threatened Amazon ironstone outcrop flora. Front. Plant Sci. 11 (599496). doi: 10.3389/fpls.2020.599496
Zhou D. Y., Ni Y. H., Yu X. N., Lin K. X., Du N., Liu L. L., et al. (2021). Trait-based adaptability of Phragmites australis to the effects of soil water and salinity in the Yellow River Delta. Ecol. Evol. 11 (16), 11352–11361. doi: 10.1002/ece3.7925
Keywords: coastal wetland, soil salinization, adaptation, intraspecific variation, ecological strategy
Citation: Wu Y, Liu L, Yin M and Guo W (2022) Phylogenetic relationship and soil salinity shape intraspecific trait variability of Phragmites australis in the Yellow River Delta. Front. Mar. Sci. 9:980695. doi: 10.3389/fmars.2022.980695
Received: 28 June 2022; Accepted: 28 July 2022;
Published: 16 August 2022.
Edited by:
Junhong Bai, Beijing Normal University, ChinaReviewed by:
Patrick Biber, University of Southern Mississippi, United StatesCopyright © 2022 Wu, Liu, Yin and Guo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Weihua Guo, d2hndW9Ac2R1LmVkdS5jbg==; Lele Liu, bGl1bGVsZUBzZHUuZWR1LmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.