- 1Fisheries College, Guangdong Ocean University, Zhanjiang, China
- 2Guangdong Provincial Key Laboratory of Aquatic Animal Disease Control and Healthy Culture & Key Laboratory of Control for Diseases of Aquatic Economic Animals, Guangdong Higher Education Institutes, Zhanjiang, China
- 3Institute of Shenzhen, Guangdong Ocean University, Shenzhen, China
- 4Shenzhen Public Seawater Economic Animal Seedling Health Evaluation Public Technology Platform, Shenzhen, China
Offshore industries and resources are the links between humans and the marine ecosystem. Several risk factors are associated with beach activities. The risk of Vibrio infection at bathing beaches is often overlooked as there are virtually no regulations regarding them. In this study, we investigated the current prevalence of pathogenic Vibrio spp. on the main beaches of Shenzhen. The risk indicator levels of pathogenic Vibrio species obtained were analyzed using 17 virulence genes mainly associated with Vibrio species and are responsible for several ailments and infections. A total of 60 Vibrio strains were isolated and identified by morphological observation, evolutionary tree alignment, and biochemical testing. There was a high abundance of Vibrio in the seawater. Also, a positive correlation was observed between the presence of virulence genes and the exhibition of high pathogenicity after artificially infecting fish with some of the virulent Vibrio species. In the infection experiment, it was observed that all the zebrafish infected with MEDF7 (Vibrio alginolyticus) and JSW-YELLOW (Vibrio harveyi) died a day after injection, with varying degrees of abdominal enlargement and skin ulceration. The mortality rates for strains with medium risk and low risk were 65% and 45%, respectively. Lethal dose 50 (LD50) values were 5.67 ×104 CFU/fish, 3.72 ×105 CFU/fish, and 4.31 ×105 CFU/fish for the high-risk, medium-risk, and low-risk strains of zebrafish, respectively. The results of the antibiotic sensitivity test showed that all the six experimental strains, except JSW-YELLOW, were resistant to doxycycline and neopenicillin. In summary, our study first identified and evaluated the pathogenicity of Vibrio in the Shenzhen beach baths, serving as a scientific benchmark for Vibrio risk prevention and control as well as guidance for Vibrio diagnosis through virulence factor detection and risk classification.
Introduction
Bathing beaches are of economic, social, and cultural importance. They provide many jobs for coastal residents and the agriculture industry. China’s coastal tourism market is rapidly expanding, with revenues predicted to exceed RMB 180.86 billion by 2019 (Yu et al., 2020). According to a study conducted by Choe in 2021, 56% of Chinese travelers plan to travel within China in 2020, with beaches being the most popular destination (Choe, 2021). Shenzhen is accelerating its development as a global center of the ocean, with millions of tourists expected every year.
Human infection with vibriosis occurs frequently (Vugia et al., 2009). Pathogenic marine microorganisms come into close contact with human beings through bathing beaches, water sports bases, mariculture animals, marine aquatic products, and estuaries, which cause major public health incidents (Zulkifli et al., 2009). A new study shows that even frequently monitored beaches with very low fecal index concentrations are at risk of swimming-related illness (Tomenchok et al., 2020). Several species of Vibrionaceae, including Vibrio harveyi, Vibrio vulnificus, Vibrio parahaemolyticus, and Vibrio alginolyticus, are mostly associated with health problems in marine animals (Fahmy et al., 2021). Groups at higher risk of infection include young children, people over the age of 64, and people with chronic illnesses or weak immune systems (Tomenchok et al., 2020). According to the United States Centers for Disease Control and Prevention, the incidence of Vibrio infections has increased dramatically since 2010 (Vugia et al., 2009). Common symptoms of vibriosis include chills, nausea, diarrhea, and fever. In severe cases of infection, it can cause conjunctivitis, otitis media, etc. (Eiler et al., 2006; Li et al., 2005; Hsieh et al., 2007).
The pathogenesis of Vibrio has been a hot topic of research, and virulence factors encoded by virulence genes are the main factors in the pathogenicity of Vibrio (Schroeder et al., 2017). Five major virulence factors have been identified: capsular polysaccharide, adhesion factor, cytotoxin, lipopolysaccharide, and flagellum (Wu et al., 2008; Daniela et al., 2013). Vibrio infects and damages the host through these virulence factors, allowing pathogens to attach, enter, establish and multiply while avoiding the host’s immune defense system (Darshanee Ruwandeepika et al., 2012). In addition, Vibrio can acquire atypical virulence genes, which increase pathogenicity (Deng et al., 2020). It has been found that these atypical virulence genes can be acquired by horizontal gene transfer (HGT) from the environment and/or other bacteria (Soucy et al., 2015). Global climate change, antibiotics, heavy metals, and environmental eutrophication can increase the pathogenicity and resistance of pathogens by affecting horizontal gene transfer (HGT) (Sun et al., 2015; Hu et al., 2016). Shenzhen belongs to the subtropical monsoon climate, with a long summer and short winter, a mild climate, sufficient sunshine, and abundant rainfall. The average annual temperature is 23.3°C, which is very favorable for the growth of Vibrio (Ferchichi et al., 2020). According to the investigation, vibriosis occurs worldwide, usually in summer and autumn, with a high incidence at 25–32°C and a rapid epidemic over 28°C (Jin et al., 2004). Multiple regional outbreaks of vibriosis in Alaska (McLaughlin et al., 2005), Denmark (Dalsgaard etal., 1999), and Spain (Martinez-Urtaza et al., 2005) support the hypothesis that vibriosis epidemics are influenced by rising global temperatures. Furthermore, some studies have shown that Shenzhen’s coastal waters have a certain degree of heavy metal pollution (Huang et al., 2017). Therefore, investigating and studying Vibrio virulence genes (especially atypical virulence genes) is very meaningful for Vibrio control.
Antibiotics are widely used to combat bacterial infections in aquaculture. It was estimated that in 2013, 54,000 tons of antibiotics were excreted into the environment by humans in China, an amount that has gradually increased over the following years (Q.-Q. Zhang et al., 2015). Antibiotics can be transmitted directly to humans through the consumption of contaminated food or water (Duarte et al., 2022). In addition, repeated exposure to antibiotics by pathogenic microorganisms has led to a decrease or even loss of susceptibility to the drugs, resulting in reduced efficacy or even failure of the drugs against drug-resistant bacteria (Letchumanan et al., 2015). Antibiotic resistance has become one of the most serious threats to global aquaculture and public health, and the level of resistance of Vibrio to antibiotics is on the increase (Larsson & Flach, 2021). As a result, determining the antibiotic resistance of pathogens in a specific region is essential for developing local antibiotic use policies and programs.
For the first time, we investigated the abundance of Vibrio in Shenzhen sea bathing beaches as well as the distribution of virulence genes in 60 strains of Vibrio. This study is important for preventing and controlling vibriosis in aquaculture and protecting public health.
Materials and methods
Ethics statement
All animal studies were carried out according to the Guangdong Ocean University’s Animal Ethics Committee Guide for the Care and Use of Laboratory Animals. Guangdong Ocean University’s Animal Ethics Committee approved the animal research (Zhanjiang, China).
Study area
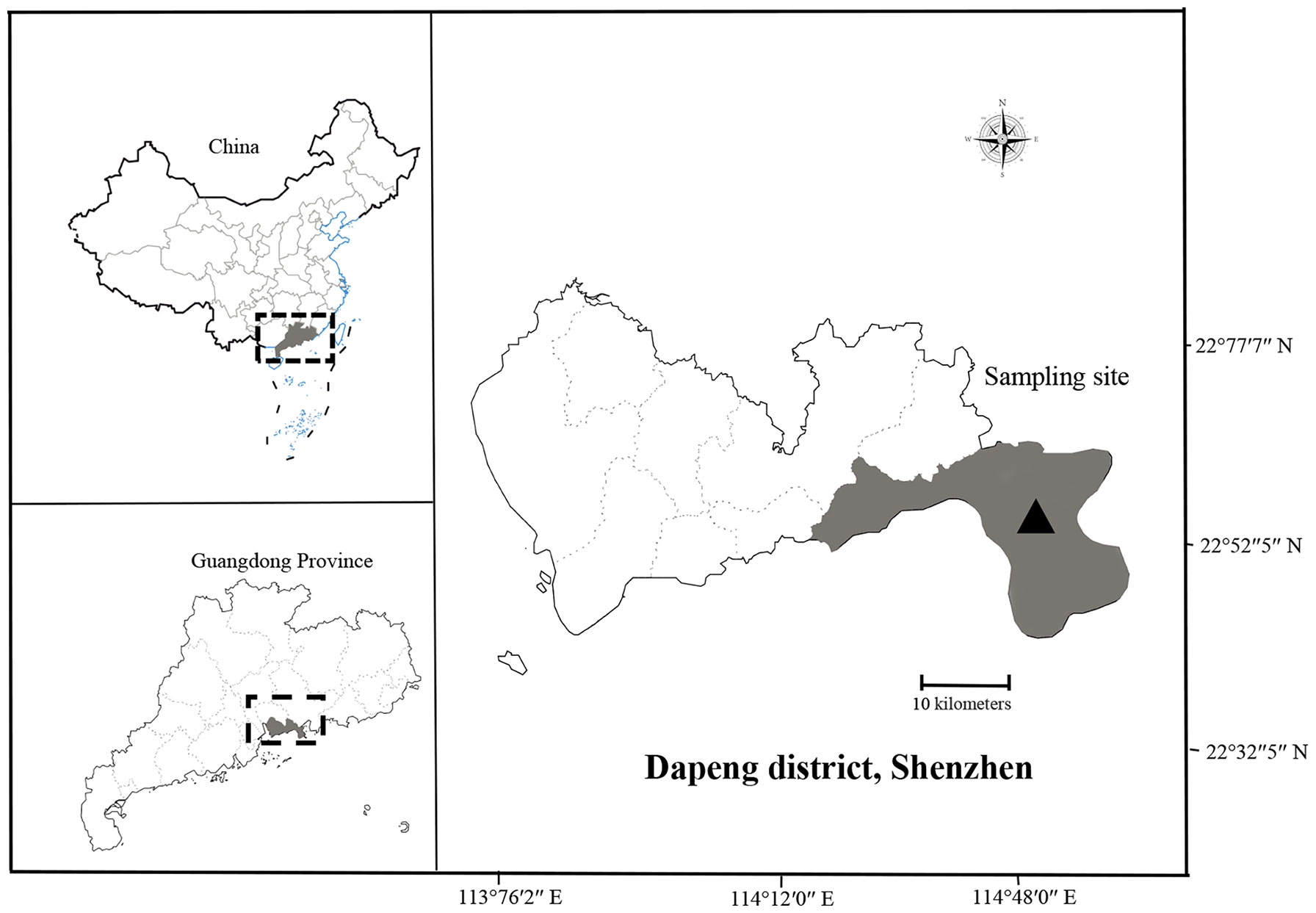
The study area was the main bathing beach in Shenzhen, China. Shenzhen is located at longitude 113 degrees 46 ‘to 114 degrees 37’ east and latitude of 22 degrees 27 ‘to 22 degrees 52’ north, south of Guangdong, and on the east bank of the Pearl River Estuary, with Daya Bay and Dapeng Bay in the east, the Pearl River Estuary and Lingdingyang in the west (Figure 1).
Bacterial isolates
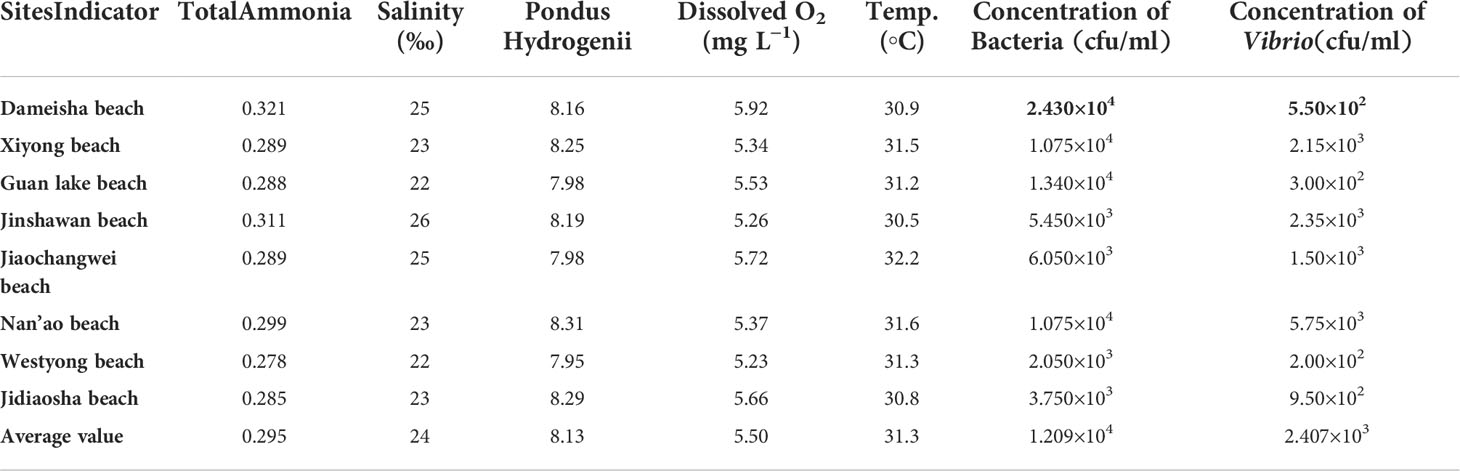
The experimental samples were collected from Shenzhen’s main 8 bathing beaches (Table 3). The samples collected are coastal water. Three replicates were set for each sampling site, and a total of 24 samples were collected and obtained. In this experiment, 200 μL of seawater (each sample) was inoculated on a TCBS solid plate for isolation and cultured at 28°C for 18 h. The dominant colonies were selected with repeated scribing separation to obtain the pure cultured strains. The same volume of 30% glycerol was added for mixed preservation and stored at -80°C.
DNA extraction
Bacterial DNA was extracted using the TaKaRa MiniBEST Bacteria Genomic DNA Extraction Kit Ver. 3.0 from Dalian Bao Biological Co. and stored at -20°C.
16S rRNA and gram stain
The 16S rRNA primers used are shown in Table 1. The PCR was performed in 25 μL reactions containing 12.0 μL of Premix Taq (TaKaRa Taq Version 2.0 plus dye) (Takara, Japan), 1.0 μL of each primer (10 μM), 1.0 μL of template DNA (20 ng/L), and 10.0 μL of sterilized water. The amplification was performed in an automatic thermal cycler (Bio-Rad, USA) as follows: initial denaturation at 95°C for 5 min; 35 cycles of denaturation at 95 °C for 30 s, annealing at 55°C for 30 s, and extension at 72°C for 60 s/kb; and final extension at 72°C for 10 min. The gram stain was used using the HB8278 Gram Stain Kit (Hopebiol Co., Ltd., Qingdao, China).
Biochemical tests
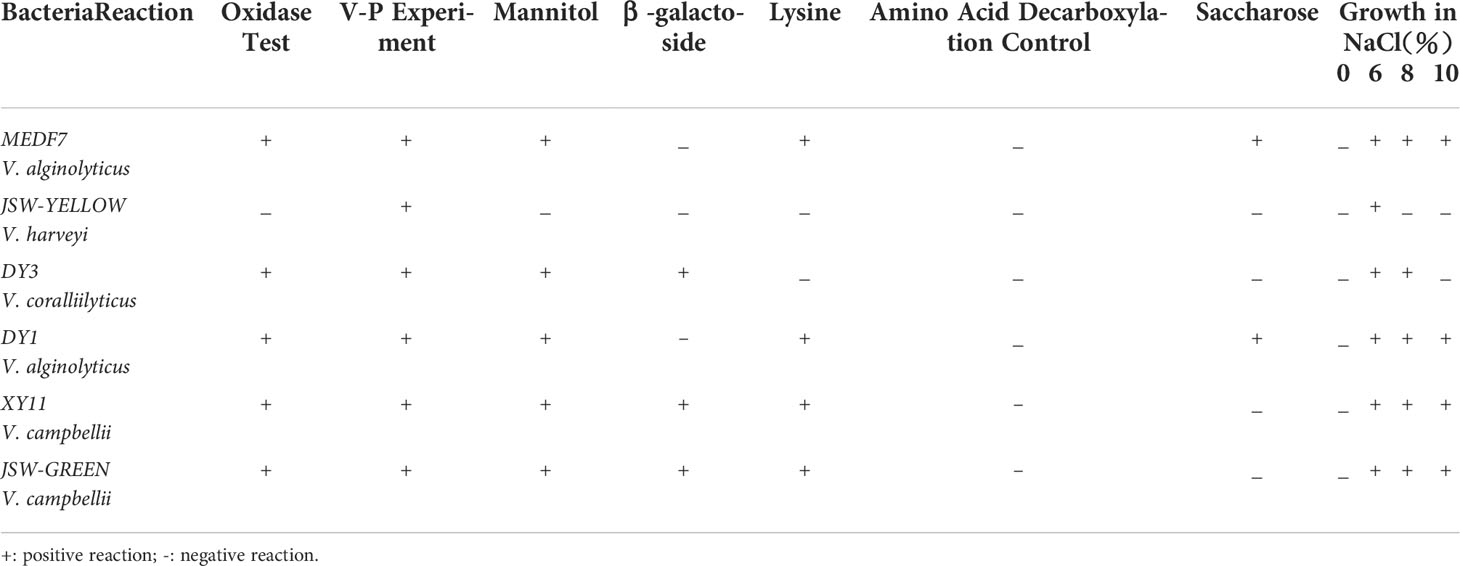
Biochemical tests were performed using a GYZ-9V (070070) biochemical identification kit for Vibrio bacteria (Huankai Microorganism Reagent Co., Ltd., China). Presumptive tests included glucose, arabinose, mannose, sucrose, lysine, arginine dihydrolase, peptone water, and the Voges-Proskauer test. Growth conditions with incubation at 25°C for 24–48 hours with 0%, 3%, 6%, 8%, and 10% NaCl (Table 2). The pathogens were analyzed and identified by referring to the “Manual of Common Bacterial System Identification” and the “Manual of Berger’s Bacterial Identification” (ninth edition) (Floyd et al., 2020).
Detection of virulence genes
To evaluate the distribution of virulence genes in Vibrio species, 8 typical V. harveyi virulence genes (vhpA, vhhA, vhhB, luxR, chiA, vhml, vhs, and toxRVh), the V. vulnificus hemolysin gene (vvh), the V. parahaemolyticus virulence genes (trh), the V. anguillarum virulence genes (flaB, flaC), the V. alginolyticus hemolysin gene serine protease, and 5 V. cholera typical virulence genes (toxR, toxS, tcpA, ctxA, and zot) were analyzed with PCR. The PCR was performed in 25 μL reactions containing 12.0 μL of Premix Taq (TaKaRa Taq Version 2.0 plus dye) (Takara, Japan), 1.0 μL of each primer (10 μM), 1.0 μL of template DNA (20 ng/L), and 10.0 μL of sterilized water. The amplification was performed in an automatic thermal cycler (Bio-Rad, USA) as follows: initial denaturation at 95 °C for 5 min; 33 cycles of denaturation at 95 °C for 30 s, annealing at the annealing temperature for 30 s, and extension at 72°C for 60 s/kb; and final extension at 72°C for 10 min. The PCR amplification products were assessed by 1.0% agarose gel electrophoresis. Following electrophoresis, gels were photographed under UV illumination.
Pathogenicity test
A pathogenicity test was performed according to Saleh and Hela (Saleh et al., 2021) for a single randomly selected isolate from each recovered bacterial species to ensure the virulence of Danio rerio. Three hundred and twenty-two fish were used for pathogenicity testing. Groups were randomly divided into 10 fish in each group. The experimental group was intramuscularly injected with 10 μL bacterial solution per tail (2 × 108 CFU/fish). The control group was injected with 1×PBS in the same way. The mortality of fish within 7 days was recorded until the death was stable. The median lethal dose was calculated using the Finney method (Finney, 1985). Feeding was restricted for 24 hours before infection and resumed at 12 hours post-infection. All fish groups were kept under observation for 7 days for abnormal clinical signs, postmortem lesions, and mortality rates.
Clinical examination
Infected fish were examined to determine any external abnormalities. The challenged fish were observed twice daily throughout the experiment period according to the method described by Austin and Austin (Austin & Austin, 2016).
Histopathological investigation
Infected fish were fixed in tissue fixative fluid. Samples were sent to Seville Biotechnology Co., Ltd. for case section observation. The histopathological examination was performed as described by Suvarna and Layton (Suvarna et al., 2019).
Antimicrobial resistance testing
Two strains were selected from three groups of high, medium, and low-risk of virulence gene distribution for antibiotic resistance testing, respectively. Strains MEDF7, JCW-YELLOW, DY1, DY3, JCW-Green, and XY11 were investigated for doxycycline (DOX, 30 µg/disk), chloramphenicol (CHL, 30 µg/disk), ciprofloxacin (CIP, 5 µg/disk), trimethoprim-sulfamethoxazole (T/S, 23.75/1.25 µg/disk), neopenicillin (NEO, 30 µg/disk), macrodantin (MAC, 300 µg/disk), gentamicin (GEN, 10 µg/disk), rifampicin (RIF, 5 µg/disk), meropenem (MER, 10 µg/disk), florfenicol (FLO, 10 µg/disk), ampicillin (AMP, 10 µg/disk), streptomycin (STR, 10 µg/disk), and erythromycin (ERY, 15 µg/disk). All the bacterial isolates were tested for resistance or sensitivity to different antibiotics using the standard disc diffusion method (Kirby Bauer test). For the disc diffusion assay, bacteria were grown between 18 and 24 h on Mueller-Hinton agar and then suspended in a 0.85% sterile physiological saline solution adjusted to a 0.5 McFarland turbidity standard, corresponding to 108 CFU/ml. Antimicrobial discs were gently fixed into the agar surface by fine forceps. The agar plates were incubated at 35°C for 24 h; Escherichia coli ATCC 25922 was used as a control strain. The inhibition zone was measured to the nearest mm using a digital caliper and interpreted according to breakpoints mentioned by the Clinical and Laboratory Standards Institute (CLSI) (2016) and shown in Table 6.
Results
Physiochemical water parameters
Water parameters were in the acceptable range. The average value of dissolved oxygen was higher than the optimum concentration (5 mg L−1); the temperature was about 31.3°C the concentration of heterotrophic bacteria was 1.209×104 CFU/ml; the concentration of Vibrio was about 2.407×103 CFU/ml as presented in Table 3.
Isolation of Vibrio species from coastal water
A total of 60 Vibrio strains were collected from coastal water in Shenzhen Beach in September 2021. These bacterial species accounted for approximately 70% of the dominant clones. Among them, 18 V. harveyi, 14 V. campbellii, 5 V. owenii, 3 V. coralliilyticus, 3 V. sinaloensis, 3 V. mediterranei, 2 V. ponticus, 2 V. orientalis, 2 V. alginolyticus, 2 V. aestivus, 2 V. tubiashii, 1 V. alfarensis, 1 V. parahaemolyticus, 1 V. hangzhouensis, and 1 V. rotiferianus were identified, accounting for 30%, 23.33%, 8.33%, 5%, 5%, 5%, 3.33%, 3.33%, 3.33%, 3.33%, 3.33%, 1.67% 1.67%, 1.67% and 1.67% of the total strains, respectively (Figure 2). The 16S rRNA phylogenetic tree clustered with the corresponding reference strain and was separate from other reference strains (Figure 3); the biochemical reactions of 6 Vibrio strains were detected (Table 3); the gram stain of V. alginolyticus (MEDF7) showed a negative, short rod-shaped, and slightly curved (Figure 4).
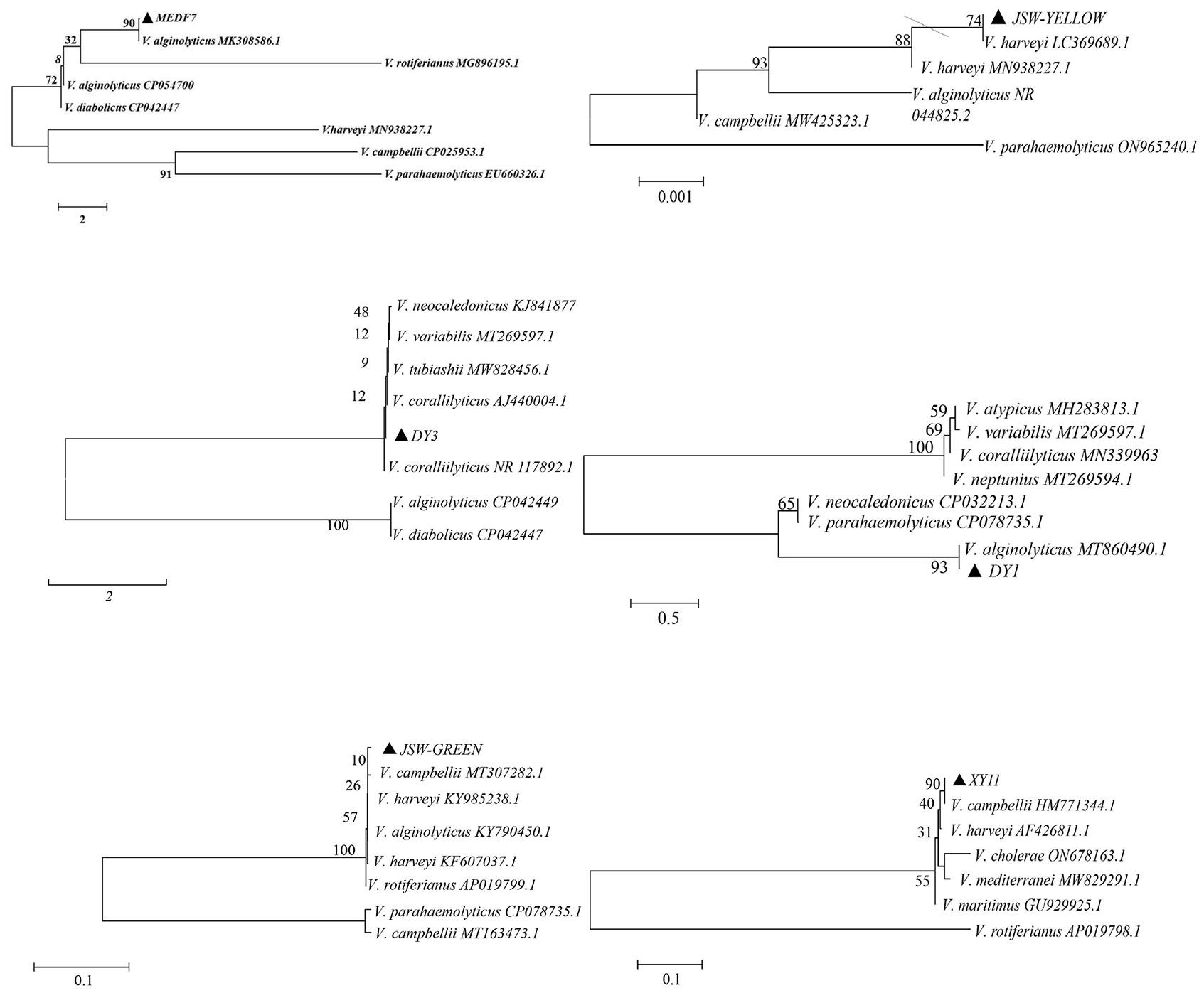
Figure 3 The 16S rRNA phylogenetic tree. Phylogenetic tree construction using MEGA6.0 and neighbor-Joining method (1000-fold bootstrap).
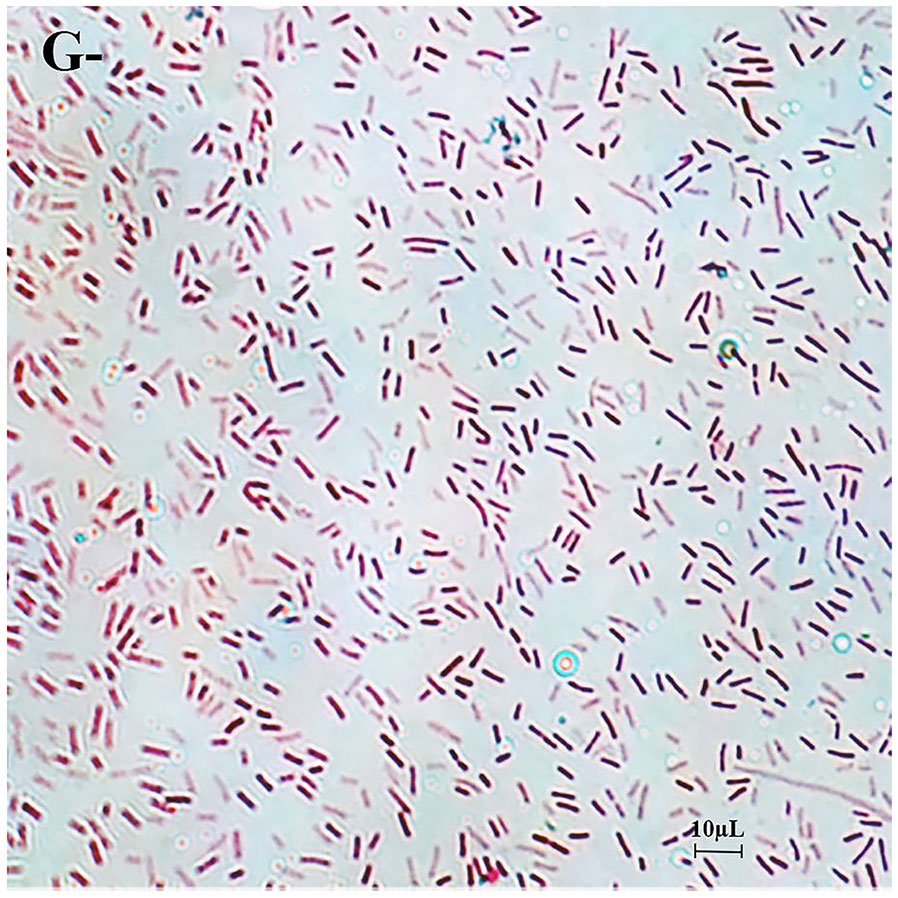
Figure 4 The gram stain. Gram stain shows negative, short rod-shaped bacteria. The scale used in the Figure is 10μm.
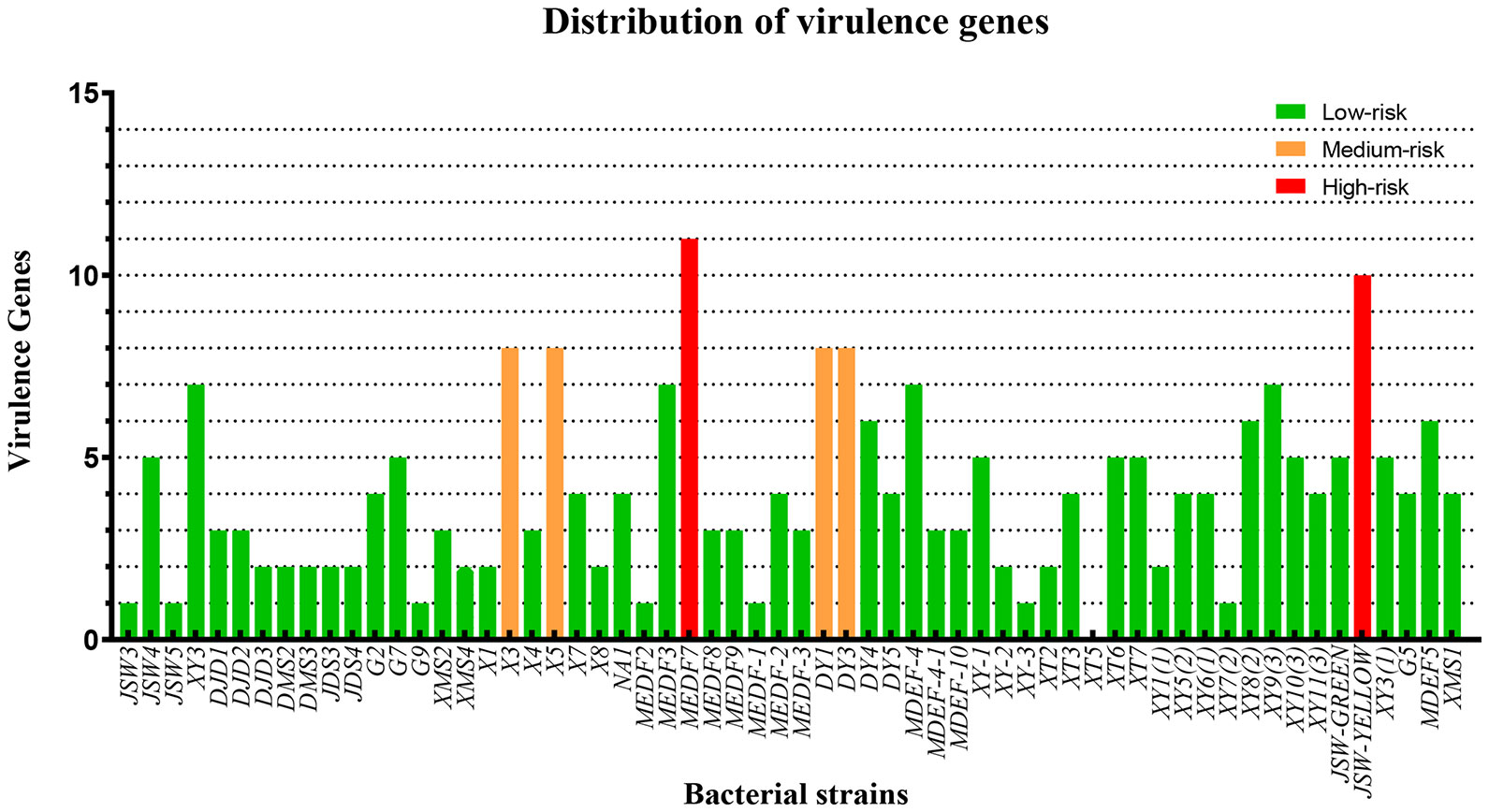
Distribution of virulence genes in Vibrio isolates
The isolated Vibrio strains were identified by the risk classification of virulence genes for 17 common Vibrio virulence genes. Isolates carrying virulence genes over 10 (≥ 60%) were considered high-risk strains, and isolates carrying virulence genes less than 7 (≤ 40%) were considered low-risk strains. Those carrying 8-9 virulence genes were classified as medium-risk strains. PCR analysis showed that MEDF7 and JSW-YELLOW were high-risk strains with 11 and 10 virulence genes, respectively. Strains DY1, DY3, X1 and X3 were medium-risk strains with 8 virulence genes, respectively. The remaining strains contained 1–7 virulence genes and were low-risk strains. In 60 Vibrio strains, the detection rate of pathogenic genes was 5.00%–71.67% (Figure 5), and the one-time detection rate was: serine protease, 71.67%; chiA, 56.67%; luxR, 53.33%; flaC, 31.67%; toxRVh, 26.67%; flaB, 18.33%; flaA, 16.67%; vhhB, 16.67%; toxR, 15%; toxS, 15% and vvh, 15%. Six atypical virulence genes, luxR, toxR, vhhA, flaA, chiA, and trh were detected in the high-risk strain MEDF7, accounting for 54.54%. Four atypical virulence genes vvh, flaA, flaC, and tcpA were detected in strain JSW-YELLOW, accounting for 40% (Supplementary Tables).
Pathogenicity test
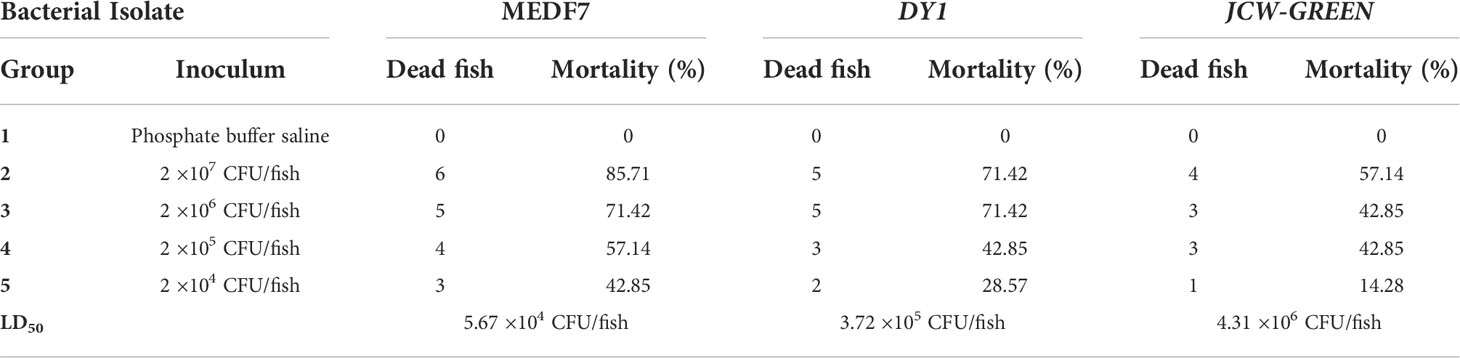
Strains MEDF7, JCW-YELLOW, DY1, DY3, JCW-GREEN, and XY11 were selected for the zebrafish infection experiment. Zebrafish began to die 10 hours after infection with MEDF7 and JSW-YELLOW strains (2 × 108 CFU/fish). All the zebrafish infected with the MEDF7 strain died within 24 hours (2 × 108 CFU/fish), and the mortality rate was about 70% within 7 days in the medium-risk strain group and 50% in the low-risk strain group (Table 4). MEDF7, DY1, and JSW-GREEN strains had a lethal dose of 50 (LD50) to zebrafish of 5.67 ×104 CFU/fish, 3.72 ×105 CFU/fish, and 4.31 ×106 CFU/fish, respectively (Table 5).
Clinical and pathological examination
The first clinical sign observed in naturally and experimentally infected fish was a decrease in feed intake. The infected zebrafish got darker and accumulated at the water surface, gasping for air, particularly in the afternoon. Diseased fish showed hemorrhagic ulcerations on the external body surface and eroded hemorrhagic pectoral and pelvic fins (Figure 6). Internally, the diseased fish showed congested and hemorrhagic internal organs mainly, while the gill filaments and intestinal tract were damaged, and inflammation increased (Figure 7).
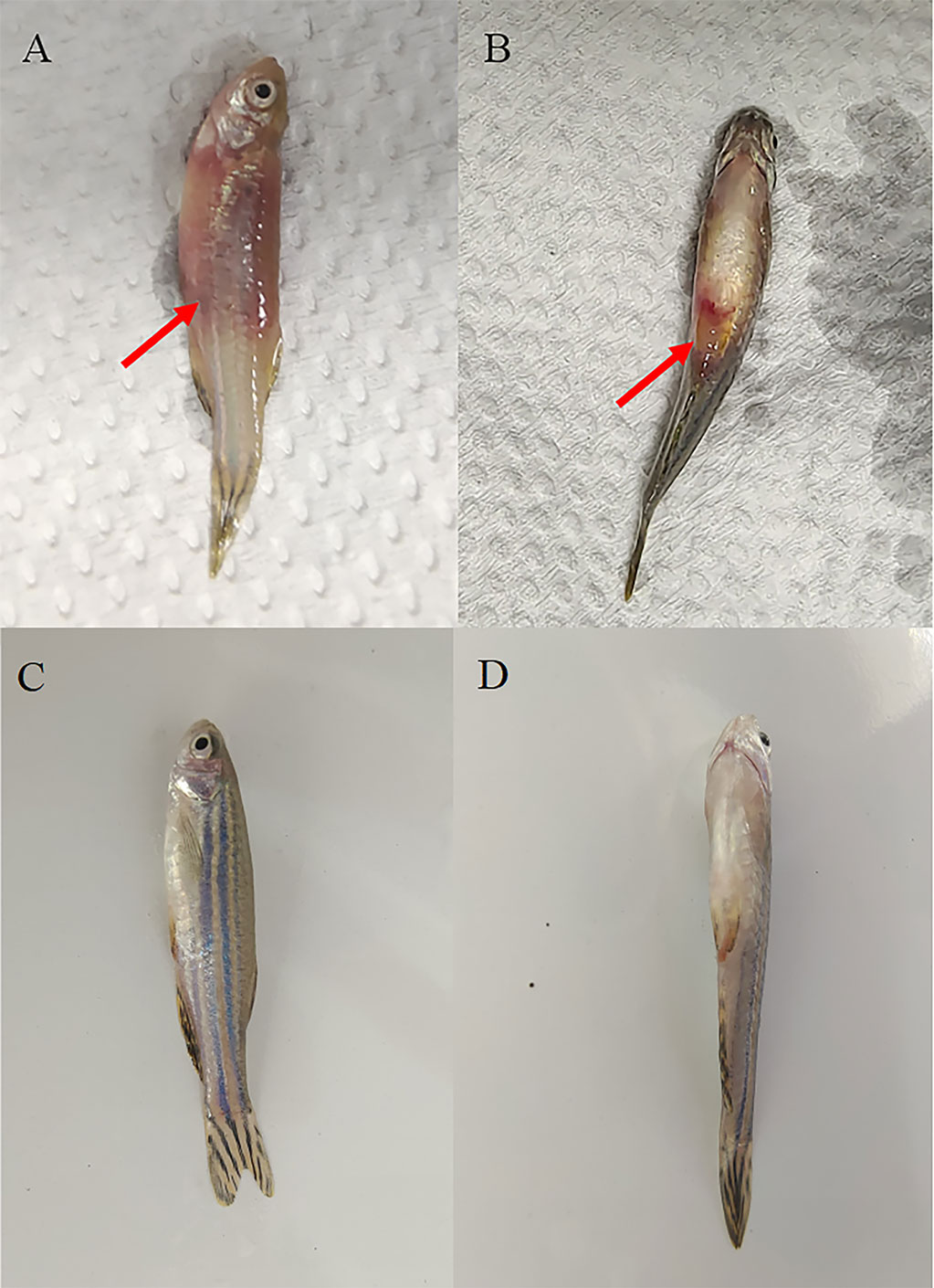
Figure 6 Clinical signs of Vibrio infection. Symptoms of Danio rerio fingerlings experimentally infected with MEDF7 (red arrow). (A, B): Diseased fish group; (C, D): Healthy fish group.
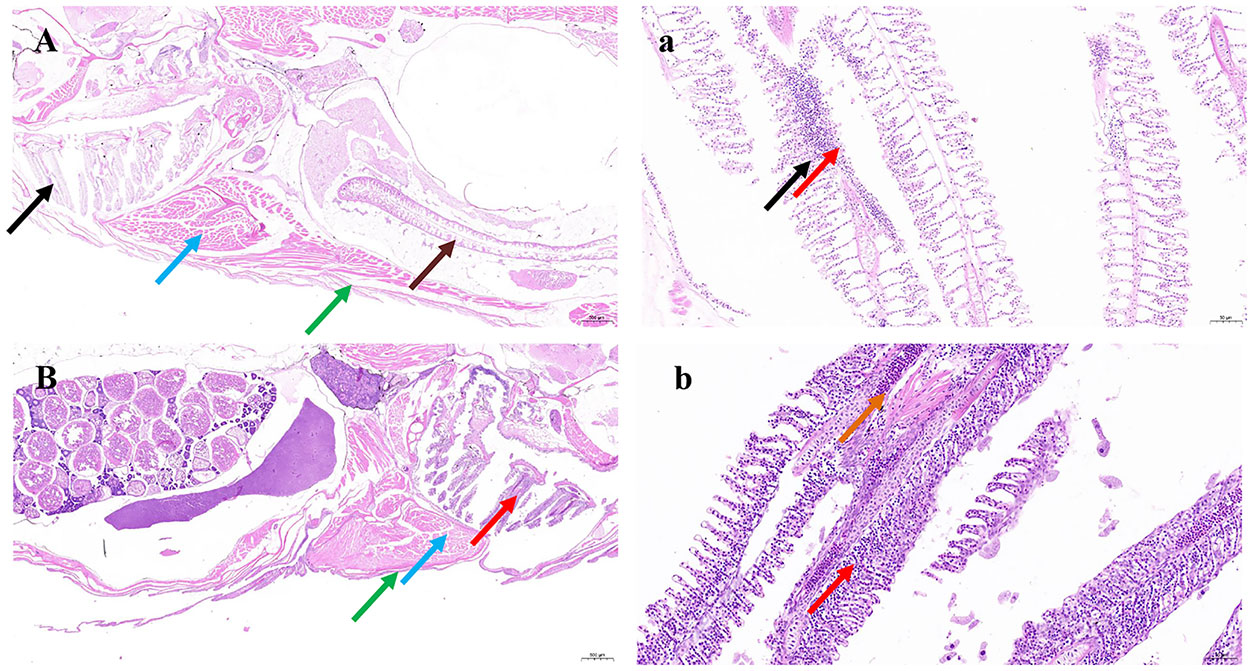
Figure 7 Pathological tissue section results. Tissue sections of Danio rerio fingerlings experimentally infected with MEDF7. (A-a): Necrosis foci are occasionally seen in the gill, with some gill filament necrosis, cell nuclei fragmentation or dissolution (black arrow), accompanied by inflammatory cell dot infiltration (red arrow); Intestinal epithelial exfoliation and lamina propria exposure (brown arrow); Note the regular arrangement of muscle fibers (blue arrow); There are more mucus cells in the skin (green arrow),H&E, X =2.0-20.0. (B-b): The gill filaments are short and thick with extravasation (orange arrows) and more inflammatory cells (red arrows). Loose .
Antimicrobial resistance
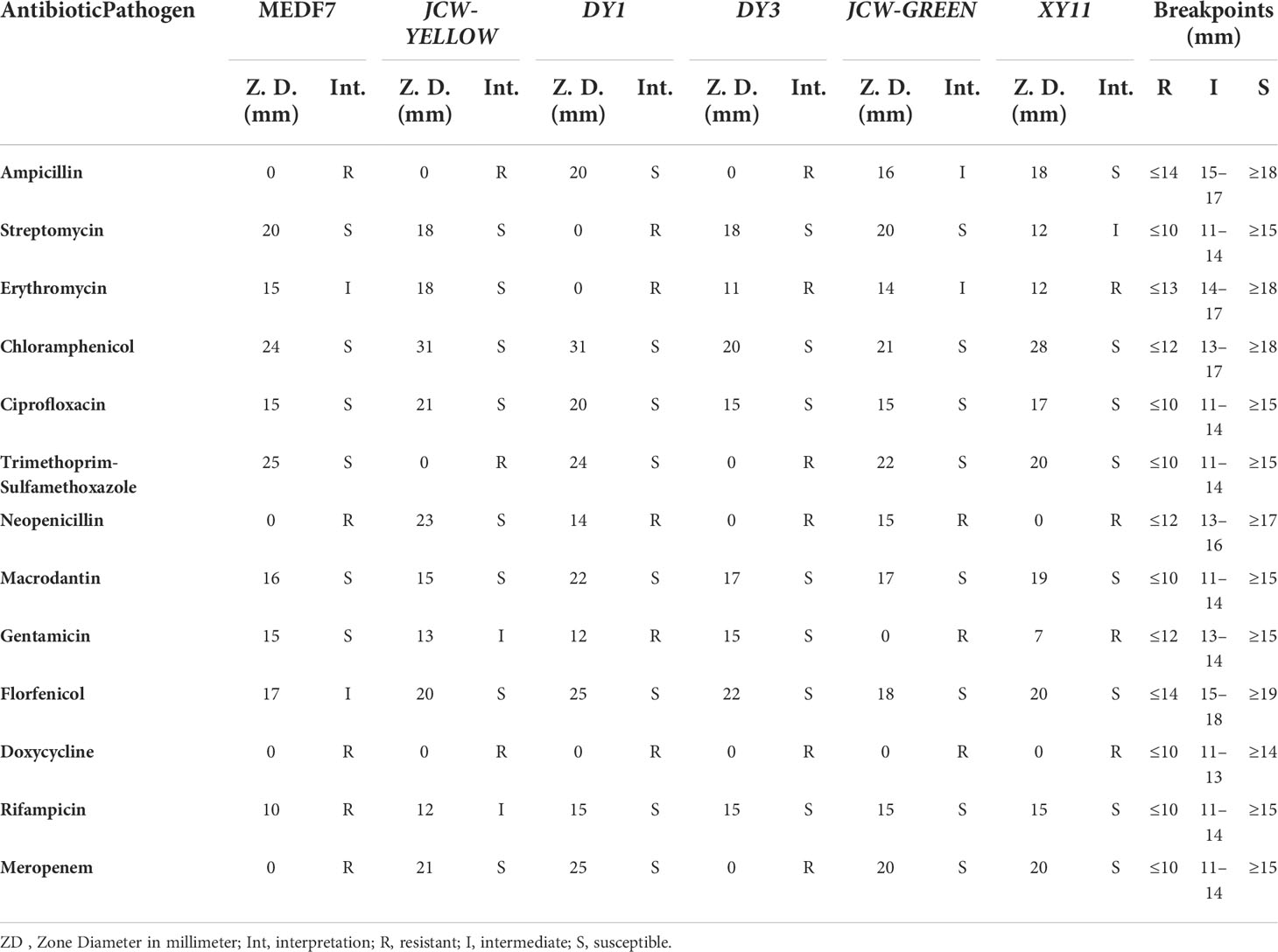
The antibiotic resistance results showed that the strain MEDF7 was resistant to ampicillin, neopenicillin G, doxycycline, rifampicin, meropenem, streptomycin, and flubenicol; JSW-YELLOW was resistant to ampicillin, trimethoprim-sulfamethoxazole, and doxycycline; DY1 was resistant to streptomycin, erythromycin, and neopenicillin; DY3 was resistant to ampicillin, erythromycin, and trimethoprim-sulfamethoxazole; JSW-GREEN was resistant to gentamicin, and doxycycline; XY11 was resistant to erythromycin, neopenicillin, gentamicin, and doxycycline. The 6 Vibrio strains except JSW-YELLOW were all resistant to doxycycline and neopenicillin (Table 6).
Discussion
With the rise of global temperature and the aggravation of offshore pollution, the pathogenicity and antibiotic resistance of Vibrio bacteria are increasing, and the existence of pathogenic Vibrio bacteria in the ocean is attracting more and more attention because of their ability to cause systemic infection, resulting in fish death and even human disease (Md Yasin et al., 2019).
Vibriosis outbreaks have been widely reported in the summer (Baker-Austin et al., 2016). In this study, the average temperature of the seawater was suitable for the growth of Vibrio. The abundance of Vibrio was high and accounted for 19.91% of the heterotrophic bacteria in the seawater. Several 15 Vibrio species and 60 Vibrio strains were discovered. The most prevalent strain found was V. harveyi, which accounted for 30% of the total. V. harveyi, V. alginolyticus, V. rotiferianus, V. campbellii, and V. parahaemolyticus were among the isolates, with 36 isolates belonging to the Harveyi clade, which is the most important pathogen clade in aquatic organisms (60%) (Darshanee Ruwandeepika et al., 2012). The Harveyi clade, particularly V. harveyi, prefers warm temperatures (Bossart, 2007; Igbinosa & Okoh, 2008), and is prevalent in marine fish farming in China, resulting in substantial losses (Deng et al., 2020). Warming temperatures can also directly induce the expression of virulence genes (Bouza & Cercenado, 2002). Guijarro et al. reported that when the temperature rose from 24 to 27℃, virulence factors involved in motility, host degradation, secretion, antimicrobial resistance, and transcriptional regulation were increased 16 times in V. corallilyticus (Guijarro et al., 2015). Yuan et al. mentioned that the risk of Vibrio infection in Shenzhen was higher (Yuan & Yuan, 2014), posing a danger to the people and fishery industry in the city. In order to assess bacterial pathogenicity, virulence factors must be identified. These factors allow bacteria to infect and damage hosts (Md Yasin et al., 2019). Include potential toxins, accessory colonization factors, outer membrane proteins, proteases, hemolysins, hemagglutinins, and a capsular polysaccharide, all of which could help Vibrio species survive and reproduce in the host (Shinoda, 2005; Gutierrez et al., 2013; Yokochi et al., 2013). Serine protease had the maximum one-time detection rate (71.67%), followed by chiA (56.67%), and luxR (53.33%). Extracellular protease: Serine protease is a virulence factor found in many pathogenic organisms. It is an essential hydrolytic enzyme. Vibrio species produce a variety of extracellular hydrolytic enzymes that are important in the invasiveness and establishment of infections (Yuan & Yuan, 2014). Lee and Cheng found a serine protease that was toxic to CHO, Hela, and Vero cells and could degrade various host proteins, resulting in tissue bleeding and death (Lee et al., 2002). Furthermore, the quorum-sensing regulator luxR, and the chitinase chiA were all discovered in V. harveyi, which was in line with previous findings (Xiandong et al., 2017; Zhu et al., 2017). In the high-risk strain MEDF7, six atypical virulence factors, luxR, toxR, vhhA, flaA, chiA, and trh, were found, accounting for 54.54%. These findings suggest that virulence genes are transferred among Vibrio species and that acquiring atypical virulence genes would increase Vibrio’s virulence to hosts (Deng et al., 2020). In aquatic environments, Vibrio species are a significant reservoir of potential virulence and bacterial DNA material exchange (Igbinosa, 2015). The emergence of new pathogenic Vibrio from non-harmful species may be possible (Balakrishnan et al., 2019). Moreover, all six experimental strains caused gill filament necrosis and increased the number of inflammatory cells in zebrafish. Especially the MEDF7 strain, which has a high mortality rate and can cause death in a short time. MEDF7 has the lowest LD50 (5.67 ×104 CFU/fish). It is shown that, to some extent, the superposition of virulence genes, especially the increase in the number of atypical virulence genes, has an important effect on the pathogenicity of Vibrio. This is consistent with the conclusion of related studies (Deng et al., 2020).
Antibiotics are widely used to prevent or treat bacterial diseases in aquaculture. Their indiscriminate use has led to an increase in antibiotic resistance in bacteria. The yearly use of raw antibiotic ingredients in China is estimated at about 180,000 t (R. Zhang et al., 2012). Therefore, monitoring the spread of antibiotic resistance is particularly important for assessments involving Vibrio species (Letchumanan et al., 2015). According to Wenguang and Xiong, the presence of potentially resistant and pathogen-associated taxonomic groups (Acinetobacter, Arcobacter, and Clostridium) may indicate a risk to human health (Xiong et al., 2015). In our study, the six strains of Vibrio were almost resistant to neopenicillin (β-lactams) and doxycycline (tetracycline). β-lactams and tetracycline are commonly used in animals as feed additives or veterinary drugs to promote growth and prevent disease (Zhao et al., 2009). For example, extensive intrinsic resistance to doxycycline and neopenicillin has been reported (Saha et al., 2020). This may be related to the use of biofertilizers, which contain high levels of residual antibiotic compounds, resistance factors, and drug-resistant bacteria (Aedo et al., 2014). Even the Chinese Ministry of Agriculture and Rural Affairs has banned the use of antibiotics in the aquaculture industry. Most antibiotics, including these banned antibiotics are commonly accessible and inexpensive (Wang et al., 2014), and their discharge from aquaculture activities is not currently regulated. This study focused on and investigated the drug resistance of pathogenic Vibrio in Shenzhen bathing beaches.
Conclusions
The warm climate conditions in Shenzhen provide favorable conditions for the growth and reproduction of Vibrio, and the high abundance of Vibrio in the coastal water leads to a high potential incidence of vibriosis. In addition, the transfer of atypical virulence factors and antibiotic resistance has led to the increasing pathogenic capacity of Vibrio, which poses a great threat to aquaculture and human health. The results of our study will provide theoretical support for the prevention and control of Vibrio coastal water in Shenzhen, and the virulence gene detection classification will offer indications for the risk prevention of Vibrio coastal water in Shenzhen. It will provide scientific guidance on the management of coastal ecology and sustainable development of the Shenzhen region.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author contributions
ZH: bacterial isolation and identification, pathogenicity and challenge tests, and antimicrobial susceptibility tests, wrote the manuscript, and analyzed and processed data. ZH, JL, and MA: review and editing. ZH and YL designed the study. YL and JC: funds and experimental reagents. All authors read and agreed to the final version.
Funding
This project was supported by JCYJ20180507183240459, through the Shenzhen Science and Technology Project. Projects subsidized by special fouds for Science Technology Innovation and Industrial Development of Shenzhen Dapeng New District, Grand No. PT202101-24.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2022.980593/full#supplementary-material
References
Aedo S., Ivanova L., Tomova A., Cabello F. C. (2014). Plasmid-related quinolone resistance determinants in epidemic Vibrio parahaemolyticus, uropathogenic escherichia coli, and marine bacteria from an aquaculture area in Chile. Microbial Ecol. 68 (2), 324–328. doi: 10.1007/s00248-014-0409-2
Austin B., Austin D. A. (2016). Bacterial fish pathogens enterobacteriaceae representatives. 323–396. Springer International Publishing Switzerland, Springer, Cham
Bai F., Pang L., Qi Z., Chen J., Austin B., Zhang X.-H. (2008). Distribution of five Vibrio virulence-related genes among Vibrio harveyi isolates. The Journal of General and Applied Microbiology 54 (1), 71–78. doi: 10.2323/jgam.54.71
Baker-Austin C., Triñanes J., Salmenlinna S., Löfdahl M., Siitonen A., Taylor N., et al. (2016). Heat wave–associated vibriosis, Sweden and Finland 2014. Emerging Infect. Dis. 22 (7), 1216–1220. doi: 10.3201/eid2207.151996
Balakrishnan M., Lawrance A., Thadikamala S., Das A., Vinithkumar N. V., Kirubagaran R., et al. (2019). Studies on diversity of Vibrio sp. and the prevalence of hapA, tcpI, st, rtxA&C, acfB, hlyA, ctxA, ompU and toxR genes in environmental strains of Vibrio cholerae from port Blair bays of south Andaman, India. Mar. Pollut. Bull. 144, 105–116. doi: 10.1016/j.marpolbul.2019.05.011
Bossart G. (2007). Emerging diseases in marine mammals: from dolphins to manatees. Microbe Magazine 2, 544–549. doi: 10.1128/microbe.2.544.1
Bouza E., Cercenado E. (2002). Klebsiella and enterobacter: Antibiotic resistance and treatment implications. Semin. Respir. infections 17, 215–230. doi: 10.1053/srin.2002.34693
Choe J. (2021). Asia Pacific travel and tourism report. Springer International Publishing, Switzerland, Springer Cham
Conejero M., Hedreyda C. (2003). Isolation of partial toxR gene of Vibrio harveyi and design of toxRtargeted PCR primers for species detection. Journal of Applied Microbiology. 95 (3), 602–611. doi: 10.1046/j.1365-2672.2003.02020.x
Conejero M. J. U., Hedreyda C. T. (2004). PCR detection of hemolysin (vhh) gene in Vibrio harveyi. J. General Appl. Microbiol. 50 (3), 137–142. doi: 10.2323/jgam.50.137
Dalsgaard I., Høi L., Siebeling R. J., Dalsgaard A. (1999). Indole-positive Vibrio vulnificus isolated from disease outbreaks on a Danish eel farm. Dis. Aquat. organisms 35 (3), 187–194. doi: 10.3354/dao035187
Daniela C., Hasan N. A., Anwar H., Colwell R. R. (2013). Distribution and dynamics of epidemic and pandemic Vibrio parahaemolyticus virulence factors. Front. Cell. Infection Microbiol. 3 (97). doi: 10.3389/fcimb.2013.00097
Darshanee Ruwandeepika H. A., Sanjeewa Prasad Jayaweera T., Paban Bhowmick P., Karunasagar I., Bossier P., Defoirdt T. (2012). Pathogenesis, virulence factors and virulence regulation of vibrios belonging to the harveyi clade. Rev. Aquacult 4 (2), 59–74. doi: 10.1111/j.1753-5131.2012.01061.x
Deng Y., Liwen X., Chen H., Liu S., Guo Z., Cheng C., et al. (2020). Prevalence, virulence genes, and antimicrobial resistance of Vibrio species isolated from diseased marine fish in south China. Sci. Rep. 10, 14329. doi: 10.1038/s41598-020-71288-0
Duarte A., Rodrigues S., Afonso A., Nogueira A., Coutinho P. (2022). Antibiotic resistance in the drinking water: Old and new strategies to remove antibiotics, resistant bacteria, and resistance genes. Pharmaceuticals 15 (4), 393. doi: 10.3390/ph15040393
Eiler A., Johansson M., Bertilsson S. (2006). Environmental influences on Vibrio populations in northern temperate and boreal coastal waters (Baltic and skagerrak seas). Appl. Environ. Microbiol. 72 (9), 6004–6011. doi: 10.1128/AEM.00917-06
Fahmy H., Gobarah D., Helmy S., Mahfouz N., Abou Zeid M., Moustafa E., et al. (2021). Phenotypic and molecular characterization of Vibrio species isolated from fish markets in Egypt. 72 (2), 2817–2024.
Ferchichi H., St-Hilaire A., Ouarda T., Levesque B. (2020). Impact of the future coastal water temperature scenarios on the risk of potential growth of pathogenic Vibrio marine bacteria. Estuarine Coast. Shelf Sci. 250, 107094. doi: 10.1016/j.ecss.2020.107094
Finney D. J. (1985). The median lethal dose and its estimation. Arch. Toxicol. 56 (4), 215–218. doi: 10.1007/bf00295156
Floyd K., Lee C., Xian W., Nametalla M., Valentine A., Crair B., et al. (2020). C-di-GMP modulates type IV MSHA pilus retraction and surface attachment in Vibrio cholerae. Nat. Commun. 11 (1), 1549. doi: 10.1038/s41467-020-15331-8
Guijarro J., Cascales D., Garcia Torrico A., Garcia M., Mendez J. (2015). Temperature-dependent expression of virulence genes in fish-pathogenic bacteria. Front. Microbiol. 6. doi: 10.3389/fmicb.2015.00700
Gutierrez C., Klein S., Lovell C. (2013). High frequency of virulence factor genes tdh, trh, and tlh in Vibrio parahaemolyticus strains isolated from a pristine estuary. Appl. Environ. Microbiol. 79 (7), 2247–2252. doi: 10.1128/AEM.03792-12
Hsieh J. L., Fries J. S., Noble R. T. (2007). Vibrio and phytoplankton dynamics during the summer of 2004 in a eutrophying estuary. Ecol. Appl. 17 (5), S102–S109. doi: 10.1890/05-1274.1
Huang F., Xu Y., Tan Z., Wu Z., Xu H., Shen L., et al. (2017). Assessment of pollutions and identification of sources of heavy metals in sediments from west coast of shenzhen, China. Environ. Sci. Pollut. Res. 25 (4), 3647–3656. doi: 10.1007/s11356-017-0362-y
Hu H. W., Wang J. T., Li J., Li J. J., Ma Y. B., Chen D., et al. (2016). Field-based evidence for copper contamination induced changes of antibiotic resistance in agricultural soils. Environ. Microbiol. 18, 13. doi: 10.1111/1462-2920.13370
Igbinosa E. (2015). Detection and antimicrobial resistance of Vibrio isolates in aquaculture environments: Implications for public health. Microbial Drug resistance 22 (3), 238–245. doi: 10.1089/mdr.2015.0169
Igbinosa E., Okoh A. (2008). Emerging Vibrio species: an unending threat to public health in developing countries. Res. Microbiol. 159, 495–506. doi: 10.1016/j.resmic.2008.07.001
Jin S., Wang G., Zhao Q., Zheng T., Chen Y. (2004). Epidemiology of vibriosis in large yellow croaker pseudosciaena crocea (Richardson) in marine cage culture. Fish. Sci. 24, 17–19.
Larsson J., Flach C.-F. (2021). Antibiotic resistance in the environment. Nat. Rev. Microbiol. 20, 1–13. doi: 10.1038/s41579-021-00649-x
Lee C.-Y., Cheng M.-F., Yu M.-S., Pan M.-J. (2002). Purification and characterization of a putative virulence factor, serine protease, from Vibrio parahaemolyticus. FEMS Microbiol. Lett. 209, 31–37. doi: 10.1016/S0378-1097(02)00477-9
Letchumanan V., Yin W.-F., Lee L.-H., Chan K.-G. (2015). Prevalence and antimicrobial susceptibility of Vibrio parahaemolyticus isolated from retail shrimps in Malaysia. Front. Microbiol. 6. doi: 10.3389/fmicb.2015.00033
Li C., Lin Q., Zhang H., Cai W., Huang H., Dai M. (2005). Ecological characteristics of phytoplankton in caged culturing waters of dapengao in daya bay. J. Agro-Environment Sci. 24, 784–789.
Martinez-Urtaza J., Simental L., Velasco D., DePaola A., Ishibashi M., Nakaguchi Y., et al. (2005). Pandemic Vibrio parahaemolyticus O3:K6, Europe. Emerging Infect. Dis. 11 (8), 1319–1320. doi: 10.3201/eid1108.050322
McLaughlin J., Depaola A., Bopp C., Martinek K., Napolilli N., Allison C., et al. (2005). Outbreak of Vibrio parahaemolyticus gastroenteritis associated with alaskan oysters. New Engl. J. Med. 353, 1463–1470. doi: 10.1056/NEJMoa051594
Md Yasin I., Al-saari N., Mohamad A., Fathin-Amirah M., Mohd Aris A., Kasai H., et al. (2019). Vibriosis in fish: A review on disease development and prevention. J. Aquat. Anim. Health 31, 3–22. doi: 10.1002/aah.10045
Rivera I., Chun J., Huq A., Sack R., Colwell R. (2001). Genotypes Associated with Virulence in Environmental Isolates of Vibrio cholerae. Applied and Environmental Microbiology 67 (6), 2421–2429. doi: 10.1128/AEM.67.612
Saha S., Halder M., Mookerjee S., Palit A. (2020). Preponderance of multidrug-resistant, toxigenic, and thermotolerant enteropathogenic bacteria in raw and cooked seafood of indo-gangetic basin and associated health risks. J. Aquat. Food Product Technol. 29, 1–12. doi: 10.1080/10498850.2020.1813858
Saleh N., Helal M., Ali N., Abbas E. (2021). Effects of using vital wheat gluten in practical diets on growth, intestinal histopathology, proinflammation-related gene expression, and resistance of white seabream (Diplodus sargus) to staphylococcus epidermidis infection. Aquaculture 537, 736508. doi: 10.1016/j.aquaculture.2021.736508
Schroeder M., Brooks B. D., Brooks A. E. (2017). The complex relationship between virulence and antibiotic resistance. Genes 8 (1), 39.
Sechi L., Duprè I., Deriu A., Fadda G., Zanetti S. (2000). Distribution of Vibrio cholerae virulence genes among different Vibrio species isolated in Sardina, Italy. Journal of Applied Microbiology 88 (3), 475–481. doi: 10.1046/j.1365-2672.2000.00982.x
Shinoda S. (2005). Pathogenic factors of vibrios with special emphasis on Vibrio vulnificus. Yakugaku zasshi J. Pharm. Soc. Japan 125, 531–547. doi: 10.1002/chin.200544294
Soucy S. M., Huang J., Gogarten J. P. (2015). Horizontal gene transfer: building the web of life. Nat. Rev. Genet. 16 (8), 472–482. doi: 10.1038/nrg3962
Sun M., Ye M., Wu J., Feng Y., Wan J., Tian D., et al. (2015). Positive relationship detected between soil bioaccessible organic pollutants and antibiotic resistance genes at dairy farms in nanjing, Eastern China. Environ. Pollut. 206, 421–428. doi: 10.1016/j.envpol.2015.07.022
Tada J., Ohashi T., Nishimura N., Shirasaki Y., Ozaki H., Fukushima S., et al. (1992). Detection of the thermostable direct hemolysin gene (tdh) and the thermostable direct hemolysin-related hemolysin gene (trh) of Vibrio parahaemolyticus by polymerase chain reaction. Mol. Cell. Probes 6 (6), 477–487. doi: 10.1016/0890-8508(92)90044-x
Tomenchok L., Gidley M., Mena K., Ferguson A., Solo-Gabriele H. (2020). Children’s abrasions in recreational beach areas and a review of possible wound infections. Int. J. Environ. Res. Public Health 17, 4060. doi: 10.3390/ijerph17114060
Vugia D., Cronquist A., Cartter M., Tobin-D'Angelo M., Blythe D., Smith K., et al. (2009). Preliminary foodnet data on the incidence of infection with pathogens transmitted commonly through food 10 states 2008. MMWR Morb. Mortal. Wkly. Rep. 58, 333–337.
Wang D., Sui Q., Zhao W., Lü S., Qiu Z., Yu G. (2014). Pharmaceutical and personal care products in the surface water of China: A review. Chin. Sci. Bull. (Chinese Version) 59, 743. doi: 10.1360/972013-370
Wu H.-J., Wang A. H. J., Jennings M. P. (2008). Discovery of virulence factors of pathogenic bacteria. Curr. Opin. Chem. Biol. 12 (1), 93–101. doi: 10.1016/j.cbpa.2008.01.023
Xiandong X., Liu K., Shifeng W., Guo W., Xie Z., Zhou Y. (2017). Identification of pathogenicity, investigation of virulent gene distribution and development of a virulent strain-specific detection PCR method for Vibrio harveyi isolated from hainan province and guangdong province, China. Aquaculture 468 (1), 226–234. doi: 10.1016/j.aquaculture.2016.10.015
Xiong W., Sun Y., Zhang T., Ding X., Li Y., Wang M., et al. (2015). Antibiotics, antibiotic resistance genes, and bacterial community composition in fresh water aquaculture environment in China. Microbial Ecol. 70 (2), 425–432. doi: 10.1007/s00248-015-0583-x
Yokochi N., Tanaka S., Matsumoto K., Oishi H., Tashiro Y., Yoshikane Y., et al. (2013). Distribution of virulence markers among Vibrio vulnificus isolates of clinical and environmental origin and regional characteristics in Japan. PLS One 8, e55219. doi: 10.1371/journal.pone.0055219
Yuan Y. M., Yuan M. (2014). Detection and analysis the contamination of Vibrio vulnificus from environment in shenzhen. Henan J. Prev. Med. 25 (1), 16–18.
Yu D., Rutty M., Scott D., Li S. (2020). A comparison of the holiday climate index:beach and the tourism climate index across coastal destinations in China. Int. J. Biometeorol 65 (2), 741–748. doi: 10.1007/s00484-020-01979-w
Zhang Q.-Q., Ying G.-G., Pan C.-G., Liu Y.-S., Zhao J.-L. (2015). Comprehensive evaluation of antibiotics emission and fate in the river basins of China: Source analysis, multimedia modeling, and linkage to bacterial resistance. Environ. Sci. Technol. 49 (11), 6772–6782. doi: 10.1021/acs.est.5b00729
Zhang R., Zhang G., Tang J., Xu W., Li J., Liu X., et al. (2012). Levels, spatial distribution and sources of selected antibiotics in the East river (Dongjiang), south China. Aquat. Ecosystem Health Manage. 15, 210–218. doi: 10.1080/14634988.2012.689576
Zhao L., Hua D., Wang H. (2009). Residues of veterinary antibiotics in manures from feedlot livestock in eight provinces of China. Sci. Total Environ. 408, 1069–1075. doi: 10.1016/j.scitotenv.2009.11.014
Zhu Z., Dong C., Weng S., He J. (2017). The high prevalence of pathogenic Vibrio harveyi with multiple antibiotic resistance in scale drop and muscle necrosis disease of the hybrid grouper, epinephelus fuscoguttatus (♀) × e. lanceolatus (♂), in China. J. Fish Dis. 41 (4), 589–601. doi: 10.1111/jfd.12758
Keywords: pathogenicity, virulence gene, antibiotic resistance, risk classification, Shenzhen
Citation: Huang Z, Anokyewaa MA, Wang J, Jian J and Lu Y (2022) Pathogenicity and antibiotic resistance analysis of Vibrio species found in coastal water at mainly beach of Shenzhen, China. Front. Mar. Sci. 9:980593. doi: 10.3389/fmars.2022.980593
Received: 28 June 2022; Accepted: 11 August 2022;
Published: 05 September 2022.
Edited by:
Jin Zhou, Tsinghua University, ChinaReviewed by:
Sang Wha Kim, College of Veterinary Medicine, South KoreaXiaojun Zhang, Yangzhou University, China
Copyright © 2022 Huang, Anokyewaa, Wang, Jian and Lu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yishan Lu, ZmlzaGRpc0AxNjMuY29t
 Ziwei Huang
Ziwei Huang Melody Abena Anokyewaa
Melody Abena Anokyewaa Junlin Wang
Junlin Wang Jichang Jian
Jichang Jian Yishan Lu
Yishan Lu