
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mar. Sci., 12 September 2022
Sec. Global Change and the Future Ocean
Volume 9 - 2022 | https://doi.org/10.3389/fmars.2022.971796
This article is part of the Research TopicThe Adaptation and Response of Aquatic Animals in the Context of Global Climate ChangeView all 18 articles
 Junxia Mao
Junxia Mao Xiaofang Huang
Xiaofang Huang Hongyan Sun
Hongyan Sun Xin Jin
Xin Jin Wenjuan Guan
Wenjuan Guan Jiahui Xie
Jiahui Xie Yiying Wang
Yiying Wang Xubo Wang
Xubo Wang Donghong Yin
Donghong Yin Zhenlin Hao
Zhenlin Hao Ying Tian
Ying Tian Jian Song
Jian Song Jun Ding*
Jun Ding* Yaqing Chang*
Yaqing Chang*High temperature and hypoxia greatly threaten marine life and aquaculture. Scallops, a diverse and ecologically important group of high economic value, mostly thrive in fluctuating environments, and are vulnerable to environmental stress. In the present study, the molecular response mechanism of scallops to a combination of environmental stressors was investigated via transcriptome analysis of the gill tissues in three scallop species, the Yesso scallop (Patinopecten yessoensis), Zhikong scallop (Chlamys farreri) and bay scallop (Argopecten irradians) that were exposed to transient heat, hypoxia and a combination thereof. The Yesso scallop had the most differentially expressed genes (DEGs) compared with the other two scallop species, indicating the highest sensitivity of the Yesso scallop to environmental stress. With increased temperature and decreased dissolved oxygen, the number of DEGs was greatly increased in the three scallop species, indicative of the enhancement in gene expression regulation in scallops in response to severe environmental changes. Heat and hypoxia had a synergistic effect on scallops. GO and KEGG enrichment analysis of DEGs under different stressors revealed overlapping molecular mechanisms of response in scallops following exposure to heat and hypoxia. Several immune and apoptosis-related pathways were highly enriched in the upregulated DEGs of the three scallops, suggesting that immune system activation and apoptosis promotion occurred in scallops in response to environmental stress. Heat shock proteins (HSPs) were significantly upregulated under heat and hypoxia, which likely assisted in correct protein folding to facilitate the adaption of the scallops to the altered environment. Additionally, the HIF-1 signaling pathway—the key pathway associated with hypoxia response—was triggered by extremely acute environmental changes. Comparative transcriptome analysis revealed 239 positively selected genes among the different scallops, including those involved in immune system and environmental adaptation, suggesting a long-term mechanism of environmental adaptation. The present study provides new insights into the molecular response mechanism in scallops to multiple environmental stressors and improves our understanding of the adaptive mechanisms of marine organisms under changing global climate conditions.
Water temperature and dissolved oxygen (DO) are two important environmental factors for marine organisms. The changing global climate has increased the ocean temperatures, which in turn has led to a decline in the DO levels in coastal zones (Vaquer-Sunyer and Duarte, 2008; Keeling et al., 2010; Vaquer-Sunyer and Duarte, 2011; Doney et al., 2012). High ocean temperatures have been proven to be a major driver of hypoxia (Vaquer-Sunyer and Duarte, 2011). Over the past decades, global warming and marine eutrophication have increased the number of coastal hypoxic zones, as well as their extension, severity and duration, which has been a major threat to coastal ecosystems, globally (Vaquer-Sunyer and Duarte, 2011; Sun et al., 2021). High temperature and hypoxia decrease the feeding, growth, reproduction and even survival of marine species (Levin et al., 2009; Hoegh-Guldberg and Bruno, 2010; Doney et al., 2012; Gobler et al., 2014; Li et al., 2020). Particularly, hypoxia caused by high temperatures resulted in high mortality in benthic marine organisms, leading to a major loss of marine biodiversity and also impacting aquaculture (Breitburg, 2002; Vaquer-Sunyer and Duarte, 2011). High temperatures in summer and hypoxia due to global warming, algal blooms and high-density aquaculture have led to frequent outbreaks of disease and the large-scale death of aquatic species, resulting in huge economic losses (Kemp et al., 2009; Pörtner, 2010; Pörtner, 2012; Stevens and Gobler, 2018). Thus, uncovering the molecular response mechanism of marine life to environmental stress (heat and hypoxia) is of priority concern to better comprehend the effects of the changing climate on marine organisms. Recently, the adaptive responses of marine organisms to heat and hypoxia have been widely investigated at molecular, metabolic, and cellular levels. However, most studies focused on only a single stressor (Truebano et al., 2010; Artigaud et al., 2014; Huo et al., 2019; Sun et al., 2020; Sun et al., 2021), whereas the two environmental stressors (heat and hypoxia) often affect marine life concurrently (Parthasarathy et al., 1992; Pörtner, 2010). Although the physiological effects of combined environmental stressors have been reported in some marine species (Pörtner and Knust, 2007; Pörtner and Farrell, 2008; Stevens and Gobler, 2018), the molecular response mechanism in marine organisms, including scallops, remains largely unknown.
Scallops, a diverse group of animals, consist of more than 300 extant marine species distributed worldwide. They play an important role in the structure and function of local benthic ecosystems (Brand, 2016). As filter-feeding bivalves, they are recognized for their ability to mitigate eutrophication, improve light availability, and recycle nutrients and organic matter, which reduces the risk of algal blooms in the coastal ecosystems (Officer et al., 1982; Cerrato et al., 2004; Gobler et al., 2005; Carroll et al., 2008; Wall et al., 2008). Additionally, many scallop species are of great economic value and support both commercial fisheries and mariculture. However, most of the commercially valuable species live and are farmed in shallow water, where environmental conditions are fluctuating, making scallops vulnerable to heat and hypoxia stresses (Guo and Luo, 2016; Stevens and Gobler, 2018). Therefore, elucidating the response and adaptive mechanism of scallops to heat, hypoxia and multiple environmental stressors is imperative to understanding the ecology of scallops and improving aquaculture strategies. Studies have shown that individual or combined environmental stressors could have different physiological effects on bivalve species with different environmental sensitivities (Stevens and Gobler, 2018). However, comparisons of molecular response mechanisms of different scallop species remain poorly studied, limiting our knowledge of their evolutionary adaptation to environmental changes.
Particularly, the Yesso scallop (Patinopecten yessoensis), Zhikong scallop (Chlamys farreri) and bay scallop (Argopecten irradians) are the three most important aquaculture scallop species in northern China with varying degrees of temperature tolerance. The Yesso scallop is a cold-water species that is naturally distributed along the coastlines of northern Japan, the far east of Russia, and the northern Korean Peninsula (Kosaka, 2016). The tolerable temperature range of the Yesso scallop is 4 to 23°C with the optimum temperature for growth ranging from 10 to 15°C (Kosaka, 2016). Since it was introduced into China in the 1980s, the aquaculture of the Yesso scallop has been growing rapidly due to its large size (Guo and Luo, 2016). However, the high summer water temperatures of the Bohai Sea, which can reach 25 to 28°C, are a challenge (Guo and Luo, 2016). The Zhikong scallop is a local species of China, which thrives mainly in the southern Liaoning Peninsula and the north and south coasts of the Shandong Peninsula (Guo and Luo, 2016). Although the Zhikong scallop can tolerate a wide range of water temperatures (about -1.5°C to 30°C, with rapid growth at temperatures ranging from 16 to 18°C), growth decreases sharply when the water temperature exceeds 23°C (Yang et al., 1999; Guo and Luo, 2016). The large-scale mortality of cultured individuals has occurred each summer in most areas of northern China since 1996 (Xiao et al., 2005). In contrast to Yesso and Zhikong scallops, the bay scallop is a native of the east coast of the United States, occurring from Cape Cod to around New Jersey and Maryland (Clarke, 1965; Shumway and Castagna, 1994). It was successfully introduced into China in 1982 and can tolerate higher temperatures (the tolerable temperature range of adults is about -1 to 31°C with the optimum temperature for growth ranging from 18 to 28°C), growing fast in summer (Wang et al., 2011; Robinson et al., 2016). In addition to being impacted by the high temperatures, these three scallop species often suffer from hypoxia caused by environmental changes and intensive culture (Chen et al., 2007; Li et al., 2020; Silina, 2019; Gurr et al., 2021). For example, a decline in the growth rate of a Yesso scallop population in the coastal waters of Amur Bay off Vladivostok was found to be due to the lower oxygen saturation of water (Silina, 2019). A large-scale mortality event for the Zhikong scallop was caused by severe hypoxia resulting from eutrophication (Chen et al., 2007). Low DO also significantly increases the heartbeat rates and respiration rates of the bays scallop during hypoxia (Gurr et al., 2021; Yang et al., 2021). Therefore, to elucidate the molecular response mechanism of scallops with different thermal tolerance to combined environmental stressors, transcriptome analysis of the three scallops exposed to heat, hypoxia and heat plus hypoxia was first performed in the present study. Next, comparative transcriptome analysis was performed and positively selected genes were identified. This study provides new insights into the molecular response mechanism of scallops under multiple environmental stressors and improves our understanding of the adaptive mechanism of marine organisms in response to a changing global climate.
To elucidate the response mechanism of scallops to high temperature and hypoxia, a total of 63 RNA-seq libraries were constructed to analyze the gill tissues of the Yesso scallop, Zhikong scallop, and bay scallop under normal and stressed conditions (heat or/and hypoxia). After eliminating adaptors and low-quality reads, a total of 3023.51 million reads (~424.64 Gb) with an average of 47.99 million reads (~6.74 Gb) for each sample were obtained, and the detailed sequencing information for each library is presented in Supplementary Table S1 and summarized in Table 1. A de novo assembly was separately carried out for each species using the Trinity method to construct the reference sequences for the analysis of differential gene expression and comparative transcriptome analysis. Finally, a total of 94,592, 112,109 and 83,299 unigenes with an average length of 1351.12 bp, 1174.67 bp and 1254.20 bp were obtained for the Yesso scallop, Zhikong scallop and bay scallop, respectively. The assembly information for the three scallops is summarized in Supplementary Table S2. The unigenes for each scallop were annotated by searching the sequences against the Nr, Swiss-Prot, GO, KOG, eggNOG and KEGG databases using BLASTX with a cut-off Evalue ≤ 1e-5 to describe their functions at different levels. The annotation information is summarized in Supplementary Table S3 and Figures S1-3.
The expression of the assembled unigenes was analyzed via the FPKM method. Analysis of differential gene expression was conducted in the three individual scallop species exposed to heat stress (HIG1: 22°C, HIG2: 25°C) and under normal (NOR: 15°C) conditions. At 22°C, there were 5231 differentially expressed genes (DEGs) detected in the Yesso scallop (PY_HIG1 vs PY_NOR), 3838 DEGs in the Zhikong scallop (CF_HIG1 vs CF_NOR) and 3227 DEGs in the bay scallop (AI_HIG1 vs AI_NOR) (Figure 1). After the temperature was increased to 25°C, a total of 8713 unigenes in the Yesso scallop (PY_HIG2 vs PY_NOR), 4018 unigenes in the Zhikong scallop (CF_HIG2 vs CF_NOR), and 3882 unigenes in the bay scallop (AI_HIG2 vs AI_NOR) were found be differentially expressed (Figure 1). Overall, the number of DEGs in the three scallops increased as the temperature increased. The Yesso scallop had more DEGs than the other two scallops.
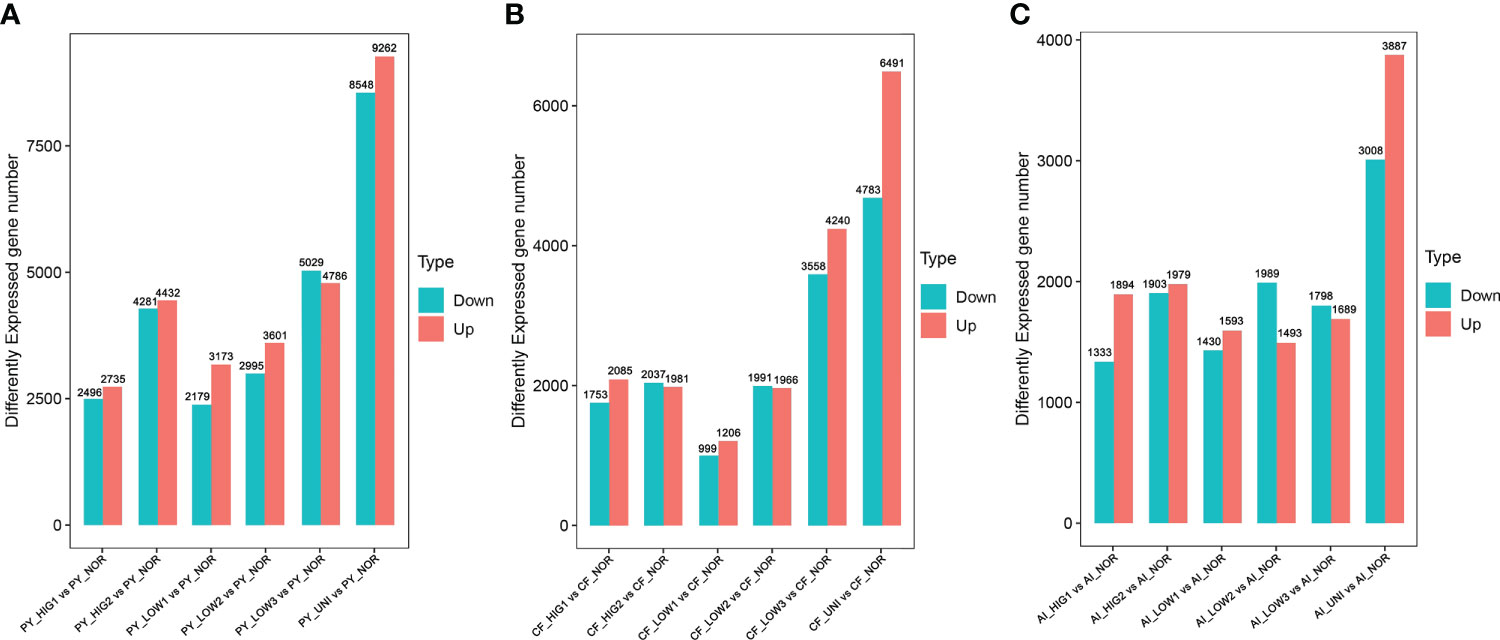
Figure 1 Number of DEGs for the Yesso scallop (A), Zhikong scallop (B), and bay scallop (C) exposed to heat, hypoxia and heat combined with hypoxia. (PY, Patinopecten yessoensis; CF, Chlamys farreri; AI, Argopecten irradians; HIG1 and HIG2, heat stress at 22 and 25°C, respectively; LOW1-LOW3, hypoxia at DO levels of 6 mg/L, 4 mg/L, and 2 mg/L, respectively; UNI, heat plus hypoxia at 25°C and the 2 mg/L DO; NOR, normal conditions at 15°C and 8 mg/L DO).
GO enrichment analysis was performed to elucidate the functions of the DEGs. At 22°C and 25°C, a total of 288 and 444 GO terms in the Yesso scallop (Tables S4, 5), 165 and 179 GO terms in the Zhikong scallop (Tables S6, 7), and 192 and 177 GO terms in the bay scallop (Tables S8, 9) were significantly enriched, respectively. Among these GO terms, many immune response-related functions—such as ‘immune response’, ‘innate immune response’, and ‘regulation of immune response’ —and apoptosis-related functions—such as ‘regulation of apoptotic process’ and ‘regulation of necroptotic process’ —were enriched at both temperatures for the three scallop species. GO functions related to heat response, such as ‘cellular response to heat’, ‘regulation of cellular response to heat’, and ‘Hsp 70 (heat shock protein 70) protein binding’, were significantly enriched at 25 °C, indicating the obvious cellular response processes to severe heat stress. Additionally, protein processing-related GO functions, such as ‘response to unfolded protein’, ‘protein refolding’, ‘chaperone cofactor-dependent protein refolding’, ‘misfolded protein binding’, and ‘regulation of proteolysis’, were much more enriched at the higher temperature. This was indicative of heat damage to the structures of many proteins in cells and the organisms’ activated response mechanisms to repair or eliminate these abnormal proteins. Additionally, GO functions associated with the oxidative stress response, such as ‘regulation of cellular response to oxidative stress’ and ‘response to hypoxia’, were also enriched at higher temperatures, which probably resulted from the hypoxia caused by high temperature. Overall, the above related GO terms were found much more enriched in the Yesso scallop, as highlighted in Supplementary Tables S4-S9.
KEGG enrichment analysis was further performed for all DEGs as well as up- and downregulated DEGs to identify the important molecular pathways involved in heat stress. The results showed that, at 22°C and 25°C, a total of 40 and 76 pathways in the Yesso scallop, 19 and 24 pathways in the Zhikong scallop, and 17 and 14 pathways in the bay scallop were significantly enriched for all the DEGs, respectively. Notably, the most significantly enriched pathways were those related to immune response and apoptosis, which were significantly upregulated at two heat stresses in the three scallops (Figure 2). However, many more related pathways were found in the Yesso scallop (Supplementary Tables S10-S15). Among these pathways, ‘TNF (tumor necrosis factor) signaling pathway’, ‘Toll and Imd (immune deficiency) signaling pathway’, ‘NF (nuclear factor)-kappa B signaling pathway’, ‘NOD (nucleotide oligomerization domain)-like receptor signaling pathway’, ‘Apoptosis-multiple species’ and ‘Necroptosis’ were simultaneously enriched at both heat stress groups for the three scallop species (Figure 2; Tables S10-S15). The pathways of ‘Protein processing in endoplasmic reticulum’ and ‘Ubiquitin mediated proteolysis’, which were involved in protein processing, were also significantly enriched in the three scallop species. ‘Ubiquitin mediated proteolysis’ was upregulated in all the heat-treated groups of the three scallop species; and ‘Protein processing in endoplasmic reticulum’ was upregulated in both two treated groups of the Yesso scallop and HIG2 group of the Zhikong scallop, but downregulated in HIG2 group of the bay scallop (Tables S10-S15; Figure S4). With the temperature increase, the ‘HIF-1 signaling pathway’ was significantly upregulated in the Yesso and bay scallops at 25°C (Tables S11, 15).
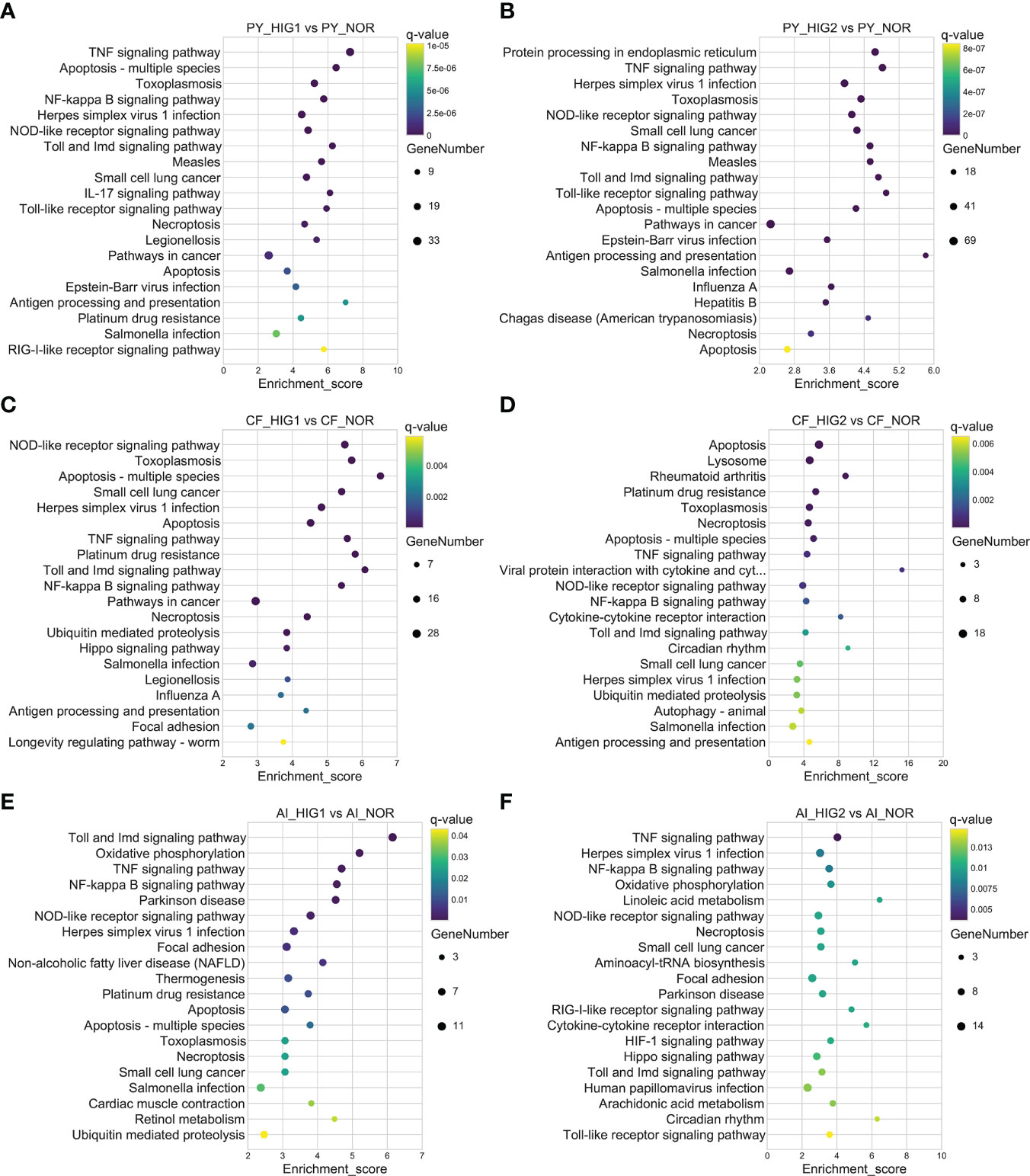
Figure 2 Top 20 significantly enriched KEGG pathways of upregulated DEGs in the Yesso scallop (A, B), Zhikong scallop (C, D), and bay scallop (E, F) exposed to different heat stressors. (PY, Patinopecten yessoensis; CF, Chlamys farreri; AI, Argopecten irradians; HIG1 and HIG2, heat stress at 22 and 25°C, respectively; NOR, normal conditions at 15°C and 8 mg/L DO).
Changes in transcriptomic expression in the three scallop species exposed to different levels of hypoxia (LOW1: 6 mg/L, LOW2: 4 mg/L, LOW3: 2 mg/L) were then determined by comparison with scallops under normal conditions (NOR: 8 mg/L). A total of 5552, 6596, and 9815 DEGs were identified in the Yesso scallop at DO concentrations of 6 mg/L (PY_LOW1 vs PY_NOR), 4 mg/L (PY_LOW2 vs PY_NOR), and 2 mg/L (PY_LOW3 vs PY_NOR), respectively (Figure 1A). In the Zhikong scallop, a total of 2205, 3957 and 7828 DEGs were detected at DO concentrations of 6 mg/L (CF_LOW1 vs CF_NOR), 4 mg/L (CF_LOW2 vs CF_NOR), and 2 mg/L (CF_LOW3 vs CF_NOR), respectively (Figure 1B). In the bay scallop, the number of DEGs were 3023, 3482 and 3487 at different DO levels (AI_LOW1 vs AI_NOR, AI_LOW2 vs AI_NOR and AI_LOW3 vs AI_NOR) (Figure 1C). Under hypoxia stress, the Yesso scallop exhibited the most DEGs among the three scallop species, followed by the Zhikong scallop, and the bay scallop. Generally, the number of DEGs increased with the decrease in DO among the three scallop species, but especially in the Yesso and Zhikong scallops.
GO enrichment analysis of the DEGs in the three scallop species showed that a total of 319, 353 and 527 GO terms in the Yesso scallop (Tables S16-S18), 89, 163 and 390 GO terms in the Zhikong scallop (Tables S19-S21), and 204, 196 and 228 GO terms in the bay scallop (Tables S22-S24) were significantly enriched under different levels of hypoxia stress (LOW1 to LOW3). A variety of functions were affected. Among these GO terms, functions related to immune response, apoptosis, protein folding, and hypoxia response (highlighted with different colors in Tables S16-S24) were significantly enriched in three scallop species, especially in the Yesso scallop. Some related GO functions were also found in the Zhikong and bay scallops, but fewer than in the Yesso scallop.
KEGG enrichment analysis revealed that, at different degrees of hypoxia (LOW1, LOW2, and LOW3), a total of 47, 42 and 91 pathways in the Yesso scallop, 38, 45 and 51 pathways in the Zhikong scallop, and 31, 18 and 11 pathways in the bay scallop, were significantly enriched, respectively. Like in response to heat stress, the most significantly enriched pathways in the three scallop species were those related to immune response and apoptosis. This was most pronounced in the Yesso scallop, with all pathways upregulated simultaneously in all the three hypoxia groups, including ‘TNF signaling pathway’, ‘Toll and Imd signaling pathway’, ‘Toll-like receptor signaling pathway’, ‘NF-kappa B signaling pathway’, ‘NOD-like receptor signaling pathway’, ‘IL (interleukin)-17 signaling pathway’, ‘RIG (retinoic acid-inducible gene)-I-like receptor signaling pathway’, ‘Apoptosis’, ‘Apoptosis-multiple species’, and ‘Necroptosis’ (Figures 3A-C; Figures S5A-C; Tables S25-S27). In the Zhikong scallop, some of these immune and apoptosis-related pathways were downregulated or both up- and downregulated (Figures 3D-F; Figures S5D-F; Tables S28-30). In the bay scallop, some of these pathways were upregulated in the LOW1 and LOW2 groups but not in the LOW3 group, except for the downregulated pathway of ‘Apoptosis’ (Figures 3G-I; S5G-I; Tables S31-S33). Additionally, protein processing-related pathways of ‘Protein processing in endoplasmic reticulum’ and ‘Ubiquitin mediated proteolysis’ were upregulated at all hypoxia levels for the Yesso scallop and at most for the Zhikong scallop, but were not enriched in the bay scallop except for the upregulatation of ‘Ubiquitin mediated proteolysis’ in the LOW2 group. Following decreased DO, the ‘HIF-1 signaling pathway’ was found to be significantly upregulated in the severe hypoxia groups of the three scallop species, i.e., the LOW3 group of the Yesso and Zhikong scallops and the LOW2 and LOW3 groups of the bay scallop (Figure 3). DNA replication- and repair-associated pathways, such as ‘DNA replication’, ‘Mismatch repair’, ‘Base excision repair’, and ‘Nucleotide excision repair’, were also found to be significantly enriched, but were downregulated, in severe hypoxia groups of the Yesso (LOW2 and LOW3) and Zhikong scallops (LOW3) (Figure S5).

Figure 3 Top 20 significantly enriched KEGG pathways of upregulated DEGs in the Yesso scallop (A–C), Zhikong scallop (D–F), and bay scallop (G–I) exposed to different levels of hypoxia stress. (PY, Patinopecten yessoensis; CF, Chlamys farreri; AI, Argopecten irradians; LOW1-LOW3, hypoxia stress associated with DO levels of 6 mg/L, 4 mg/L, and 2 mg/L, respectively; NOR, normal conditions at 15°C and 8 mg/L DO).
The regulation of gene expression in the three scallop species following exposure to a combination of heat hypoxia (UNI: 25 °C and 2 mg/L) was analyzed. A total of 17810 (PY_UNI vs PY_NOR), 11174 (CF_UNI vs CF_NOR) and 6885 unigenes were differentially expressed in the Yesso, Zhikong, and bay scallops, respectively (Figure 1). This was much higher than that induced by each stressor individually. GO enrichment analysis of the DEGs showed that a total of 481, 501, and 280 GO terms were significantly enriched in the Yesso, Zhikong and bay scallops, respectively (Tables S34-S36), which covered a variety of functions, including those related to immune response, apoptosis, and stress response (highlighted with different colors in Tables S34-S36). KEGG enrichment analysis showed that a total of 84, 50 and 25 pathways were significantly enriched in the Yesso, Zhikong, and bay scallops, respectively. As expected, the pathways involved in immune response, apoptosis, and protein processing were most significantly enriched in the three scallop species All were upregulated under combined stress, with significant upregulation of ‘TNF signaling pathway’, ‘Toll and Imd signaling pathway’, ‘NF-kappa B signaling pathway’, ‘NOD-like receptor signaling pathway’, ‘Apoptosis’, ‘Apoptosis-multiple species’, ‘Necroptosis’, as well as ‘Ubiquitin mediated proteolysis’ and ‘HIF-1 signaling pathway’, in the three scallop species (Tables S37-S39; Figure 4). Like the downregulated pathways of the severe hypoxia groups, DNA replication- and repair-associated pathways were mainly enriched in the downregulated DEGs of the Yesso and Zhikong scallops (Figure S6).
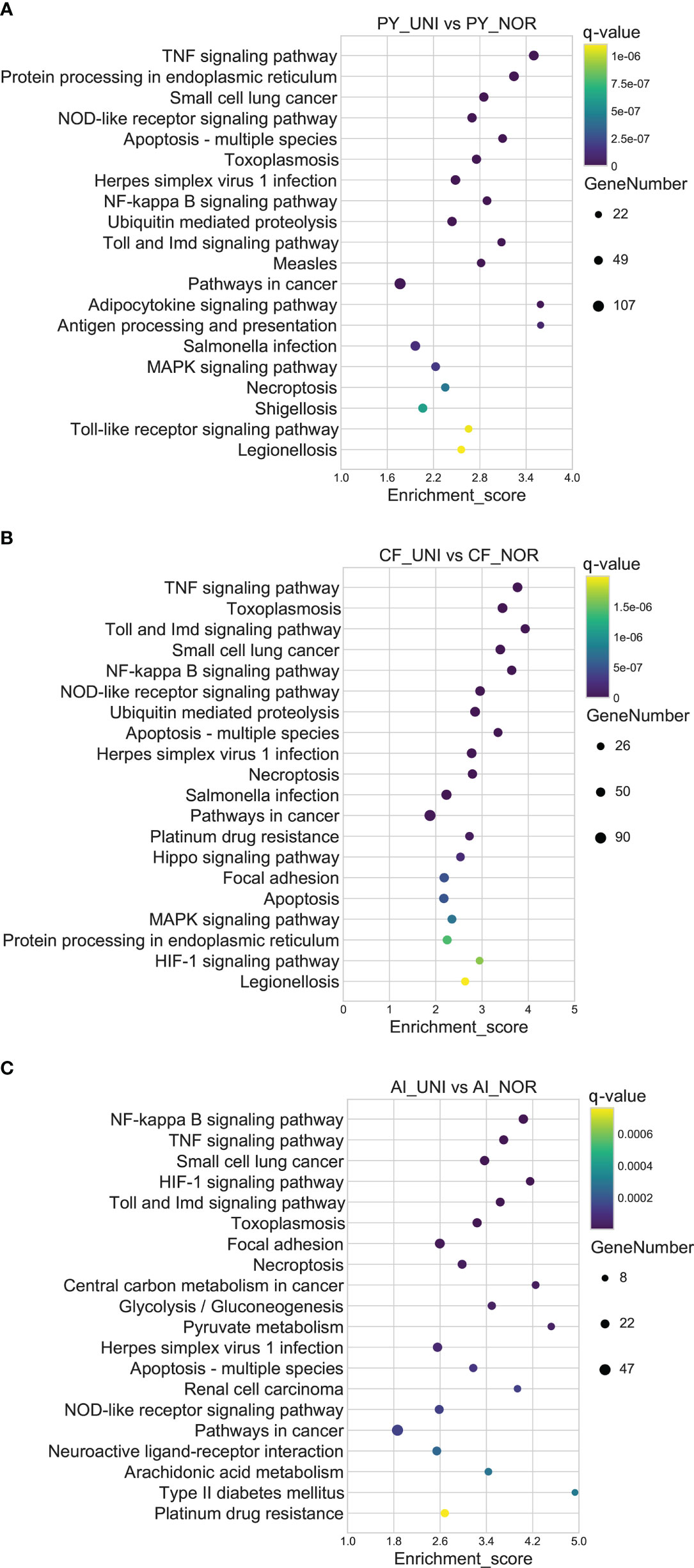
Figure 4 Top 20 significantly enriched KEGG pathways of upregulated DEGs in the Yesso scallop (A), Zhikong scallop (B), and bay scallop (C) exposed to heat and hypoxia. (PY, Patinopecten yessoensis; CF, Chlamys farreri; AI, Argopecten irradians; UNI, heat + hypoxia stress at 25°C and 2 mg/L DO; NOR, normal conditions at 15°C and 8 mg/L DO).
Finally, a Venn analysis of the DEGs was performed in all the treatment groups (HIG1, HIG2, LOW1, LOW2, LOW3, and UNI). Following exposure to heat and hypoxia, the number of non-redundant DEGs in the three scallops was 14019, 10787, and 6466 for the Yesso, Zhikong, and bay scallops, respectively (Figures 7A-C). Additionally, 722, 71, and 164 DEGs were shared by all the separate treatment groups of the Yesso, Zhikong, and bay scallops (Figure 5). Due to the relatively few shared DEGs—including several non-annotated ones—that were identified in the Zhikong and bay scallops, a significant enrichment of GO functions and pathways was mainly observed in the Yesso scallop. A total of 91 GO terms, including those related to immune response, apoptosis, heat and hypoxia response were significantly enriched in the Yesso scallop (Table S40). KEGG enrichment analysis showed 51 significantly enriched pathways, including some associated with immune response and apoptosis (Table S41). Furthermore, among the enriched GO functions and pathways for the Yesso scallop, several key candidate genes involved in stress response were detected, including TNF receptor-associated factor 3, TNF receptor superfamily member 11B, TNF receptor superfamily member 27, HSP 70B2, Heat shock cognate 71 kDa protein, HSP 90-beta, HSP 90 co-chaperone Cdc37, Putative tyrosinase-like protein-3, Cell division cycle and apoptosis regulator protein 1, and Cytochrome P450 2C38 (Figure 6).

Figure 5 Venn diagrams representing DEGs in the Yesso scallop (A), Zhikong scallop (B), and bay scallop (C) under stress. (PY, Patinopecten yessoensis; CF, Chlamys farreri; AI, Argopecten irradians; HIG1 and HIG2, heat stress at 22 and 25°C; LOW1-LOW3, hypoxia stress induced by DO levels of 6 mg/L, 4 mg/L, and 2 mg/L, respectively; UNI, stress induced by heat and hypoxia at 25°C and 2 mg/L DO; NOR, normal conditions at 15°C and 8 mg/L DO).
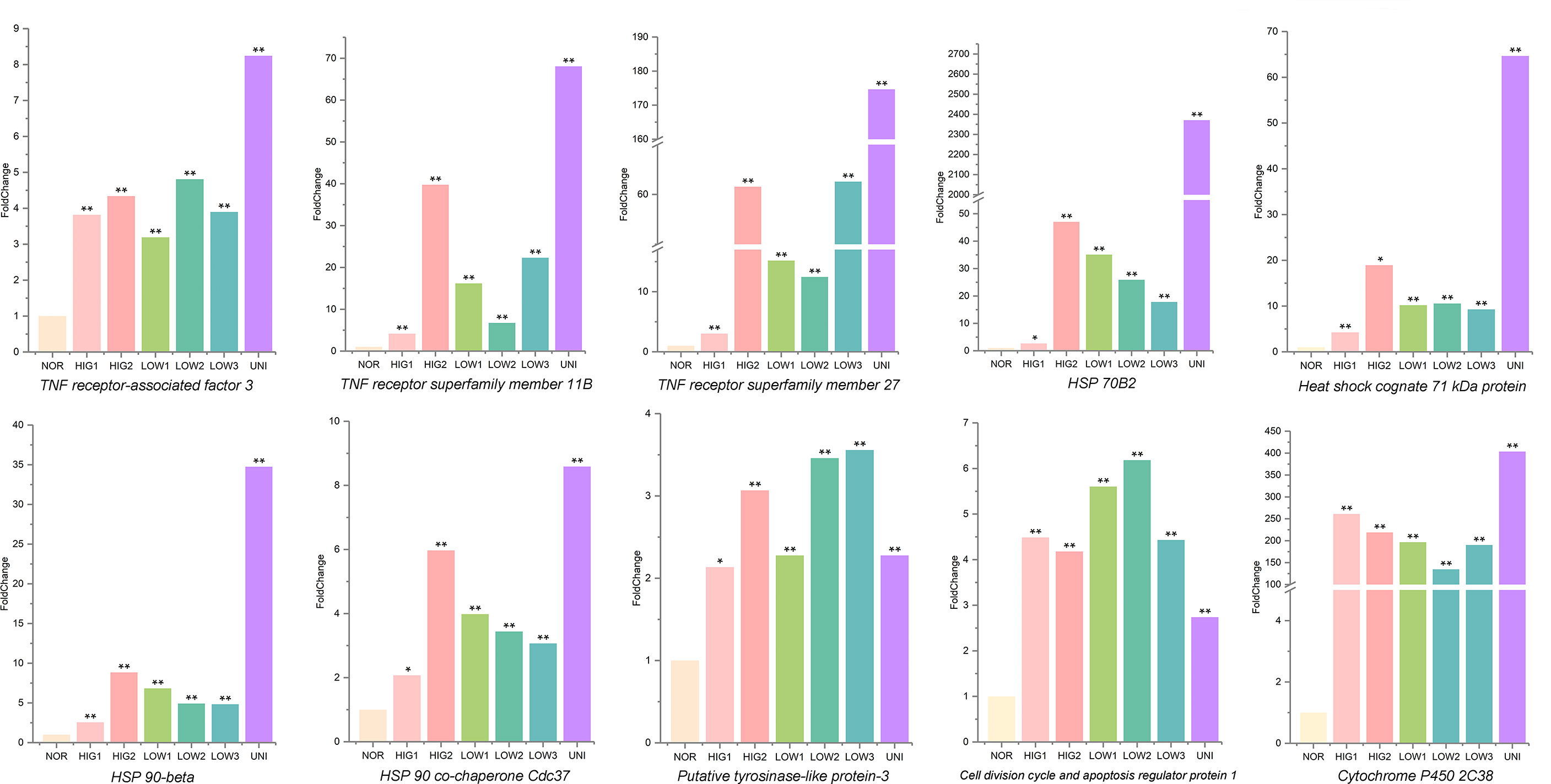
Figure 6 Levels of mRNA expression associated with key candidate genes involved in stress response in the Yesso scallop. (Values marked with an asterisk indicate significant differences, *p-value < 0.05, **p-value < 0.01; HIG1and HIG2, heat stress at 22 and 25°C; LOW1-LOW3, hypoxia stress at DO levels of 6 mg/L, 4 mg/L, and 2 mg/L; UNI, stress due to heat and hypoxia at 25°C and 2 mg/L DO; NOR, normal conditions at 15°C and 8 mg/L DO).
The transcriptome sequences of the king scallop (Pecten maximus) reported by Kenny et al. (2020) were used because at least four species were required for phylogenetic analysis. A total of 5693 single-copy orthologs were detected with the CDS sequences of the four scallop species. Phylogenetic analysis of these orthologs indicated clusters of Yesso and Zhikong scallops, followed by the bay scallop and king scallop, which was consistent with their species phylogeny (Figure 7A). The color of each branch indicated the dN/dS value of the ‘super gene’ and was generated by directly connecting all the orthologs of each species. The distribution of dN/dS in each species is shown in Figure 7B. Overall, the dN/dS value of the Yesso scallop was the largest among the three scallop species, followed by the Zhikong and bay scallops, suggesting a relatively rapid evolutionary speed of the Yesso scallop compared with that of the other two scallops. A total of 239 positively selected genes were identified using the branch-site model of the PAML software (p < 0.05). These genes may be associated with positive selection. GO enrichment analysis of these positively selected genes indicated enrichment of 44 GO functions (level 2), including ‘response to stimulus’ and ‘immune system process’ (Figure 7C). KEGG enrichment showed that pathways related to ‘immune system’, ‘infectious disease’ and ‘environmental adaptation’ were also significantly enriched (Figure 7D). Positively selected genes that were possibly involved in stress adaptation are listed in Table 2.

Figure 7 Comparative transcriptome analysis of different scallops. (A) Phylogenetic analysis of different scallop species with single-copy orthologs. The color of the branch indicates the dN/dS value of the ‘super gene’ connected by all orthologs of the species. (B) Distribution of dN/dS values in different scallops. (C) GO enrichment analysis of positively selected genes. (D) KEGG enrichment analysis of positively selected genes.
High temperature and hypoxia have become common stress factors that pose a great threat to marine life and aquaculture. Although some studies have explored the influence of each of these stressors on aquatic animals individually, the combined effects of the two stressors remain largely unknown, especially in scallops. In the present study, three scallop species—Yesso, Zhikong and bay scallops— were exposed to short-time heat and hypoxia, both individually and combined. Then, an analysis of transcriptome regulation was performed to elucidate the molecular response mechanism of the scallops under environmental stress, especially under combined stress. The key molecular pathways of scallops involved in the response of the scallops to heat, hypoxia and heat plus hypoxia were obtained. Comparative transcriptome analysis among the different scallop species was also performed to reveal an evolutionary mechanism of the scallops’ adaptation to environmental changes. This study provides insights into the response mechanisms of scallops exposed to multiple environmental stressors and improves our understanding of adaptive mechanisms in marine organisms in response to a changing global climate.
Environmental stress can damage macromolecules within cells, such as DNA and proteins, which induces a response mechanism by the organisms to maintain cellular homeostasis (Halliwell and Cross, 1994; Visick and Clarke, 1995; Chen and Jin, 2019). Accumulated DNA damages and unrepaired abnormal proteins will activate apoptosis or cell death (Franco et al., 2009; Erguler et al., 2013). In the present study, heat, hypoxia, and their combination resulted in high levels of protein disorder. Many protein folding-related GO functions—such as “protein folding”, “chaperone-mediated protein folding”, “protein refolding”, “chaperone cofactor-dependent protein refolding”, “response to unfolded protein”, “unfolded protein binding”, and “misfolded protein binding”—were significantly enriched in the three scallop species, especially the Yesso scallop, in response to the different environmental stressors. However, it appeared that DNA damages mainly occurred under acute hypoxia stress, as DNA replication and repair-related pathways—such as “DNA replication”, “mismatch repair”, “base excision repair”, and “nucleotide excision repair”—were significantly downregulated in the acute hypoxia groups of the Yesso (LOW 2, LOW3 and UNI) and Zhikong scallops (LOW3 and UNI). These effects would lead to a high mutation ratio (Artigaud et al., 2015). Acute hypoxia probably caused high levels of reactive oxygen species (ROS) in scallops, which would lead to DNA damage and, ultimately, cell death (de Almeida et al., 2007).
Exposure to single stressors—increased temperature and decreased DO—increased the number of DEGs in the three scallop species. This was consistent with results for the hard clam (Mercenaria mercenaria); that is, the number of DEGs increased with an increase in the severity of heat and hypoxia stresses (Hu et al., 2022). Heat often accompanies hypoxia, as high temperatures decrease the solubility of oxygen, while increasing the metabolic and respiratory rates of organisms, thus enhancing oxygen requirements in warmer water that holds less DO (Vaquer-Sunyer and Duarte, 2011). However, the combined impact of heat and hypoxia on scallops was unclear before this study. Multiple stressors interact in unpredictable ways (Stevens and Gobler, 2018). Environmental stressors are more likely to act synergistically or antagonistically than additively (Crain et al., 2008). In the present study, the number of DEGs in the three scallop species following exposure to combined stress (heat plus hypoxia) was significantly higher than for either single stressor and was also greater than the summation of non-redundant DEGs under heat and hypoxia individually. This was suggestive of a synergistic effect of the two stressors. GO and KEGG enrichment analysis revealed the simultaneous enrichment of multiple functions and pathways related to immune response, apoptosis, and protein processing in response to different stressors (Figure 8). This suggested shared molecular responses, that is, activation of the immune system and promotion of apoptosis, to heat and hypoxia in scallops. Similar results were reported in small abalone (Haliotis diversicolor), suggesting a similar molecular response to thermal stress and hypoxia (Zhang et al., 2014).
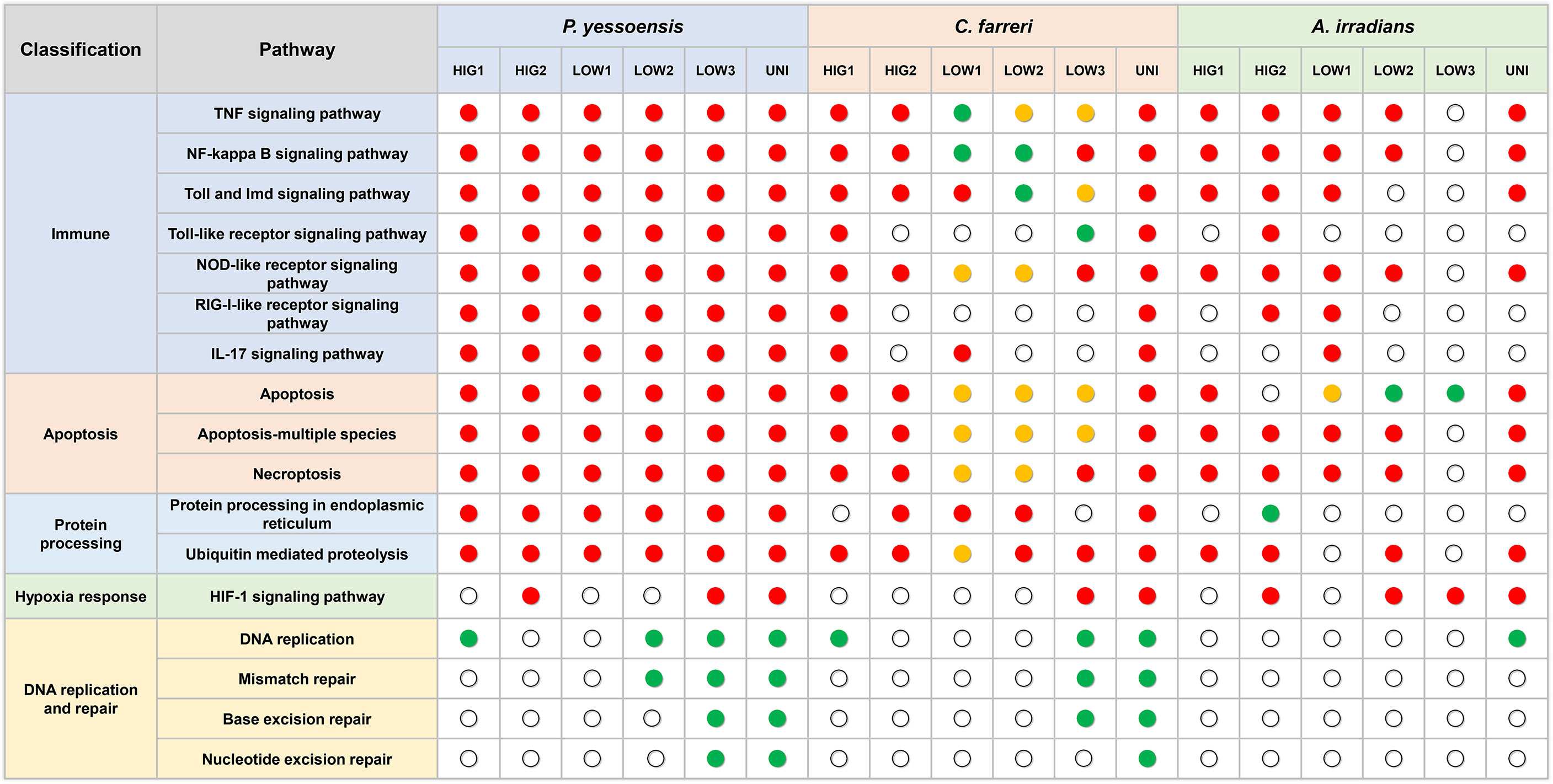
Figure 8 Key pathways in the three scallop species in response to different kinds of environmental stresses. Red dot represent enriched pathways in upregulated DEGs, green dots represent enriched pathways in downregulated DEGs, and yellow dots represent enriched pathways in both up- and downregulated DEGs.
Environment stress has been shown to significantly alter the immunity of several marine organisms (Chen et al., 2007; Malagoli et al., 2007; Sun et al., 2020). Immune regulation can be activated by the exposure to environmental stressors to increase the immune defenses of organisms in adverse environments (Huo et al., 2019; Zhang et al., 2019; Nie et al., 2020). In the current study, several innate immune-related GO functions and pathways were significantly enriched in the three scallop species following exposure to heat, hypoxia, and heat plus hypoxia. Most of these pathways were upregulated and included in the most significantly enriched pathways, which indicated the activation of the immune response of the scallops to the different environmental stresses. The TNF signaling pathway, NF-kappa B signaling pathway, Toll and Imd signaling pathway, and NOD-like receptor signaling pathway were significantly upregulated under different levels of heat stress and the combination of heat and hypoxia in all the three scallop species, and were significantly enriched in all the hypoxia groups of the Yesso scallop (all upregulated) and at least one hypoxia group of the Zhikong and bay scallops (Figure 8). These findings indicated the important role of these pathways in the immune response of the scallops to heat, hypoxia and heat plus hypoxia.
Similar immune pathways were also detected in response to environmental stressors in several other bivalves. For example, both the NF-kappa B signaling pathway and TNF signaling pathway were significantly enriched in ark shells (Scapharca subcrenata) for DEGs under heat and hypoxia stresses (Ning et al., 2021). In the Manila clam (Ruditapes philippinarum), KEGG analyses of the DEGs showed that the TNF signaling pathway, NOD-like receptor signaling pathway, RIG-I-like receptor signaling pathway, and NF-kappa B signaling pathway were significantly enriched under hypoxia stress (Nie et al., 2020), and the TNF signaling pathway and NF-kappa B signaling pathway were highly enriched after aerial exposure stress (Nie et al., 2020). Similar results have also been found in hard clams under aerial exposure; that is, the NF-kappa B signaling pathway, NOD-like receptor signaling pathway, and RIG-I-like receptor signaling pathway were significantly enriched (Zhou et al., 2021). Therefore, mollusks may have developed sophisticated immune regulatory mechanisms to sense the environmental changes and orchestrate appropriate signaling pathways to facilitate adaptation in the marine environment.
The TNF family members and their receptors are important pleiotropic cytokines that mediated inflammatory signaling in infectious diseases (Wajant et al., 2003). TNF can coordinate with Imd, Toll, and JNK pathways to regulate the innate immune response through controlling the capacity of phenoloxidase and the expression of antimicrobial peptide genes synergistically (Tang et al., 2019), while the NF-kappa B signaling pathway is crucial for mediating cellular TNF responses (Tanji and Ip, 2005). NF-kappa B is prominently involved in the regulation of immune and inflammatory responses, typically by inducing antiapoptotic gene expression to promote cell survival (Mattson and Meffert, 2006). NOD-like receptors have emerged as pivotal intracellular sensors in innate and adaptive immunity, which can integrate positive and negative incoming signals and stimulate other signaling pathways, such as the NF-kappa B signaling pathway (Shaw et al., 2010; Liu et al., 2019). Therefore, these immune pathways do not function alone. The simultaneous regulation of these pathways in the three scallop species under different environmental stressors is also indicative that the pathways can interact to protect organisms from stresses. Among these pathways, the TNF signaling pathway may represent a key environmental response mechanism in the Yesso scallop, given the significant upregulation of several TNF receptor superfamily genes (TNF receptor-associated factor 3, TNF receptor superfamily member 11B, TNF receptor superfamily member 27) in all the stressed groups of the Yesso scallop. Additionally, TNF is also a crucial regulator of the generation of ROS, and high ROS concentrations in cells ultimately lead to DNA damage and cell death (Blaser et al., 2016).
Apoptosis, or programmed cell death, plays a vital role in organ development, tissue homeostasis regulation, elimination of abnormal or damaged cells, and the immune response (Jacobson et al., 1997; Ameisen, 2002; Sokolova, 2009). Environmental stressors are well known to induce apoptotic cell death (Franco et al., 2009). In the present study, the apoptotic GO functions and pathways were highly enriched for DEGs in all the three scallop species following exposure to heat, hypoxia, and heat plus hypoxia, indicating the key role of apoptosis in response to environmental stress. Furthermore, these apoptosis-related pathways, such as “Apoptosis”, “Apoptosis-multiple species”, and “Necroptosis”, were significantly upregulated and mostly involved in the top enriched pathways, suggesting the induction of apoptosis under environmental stress. Similar conclusions were also obtained for the Manila clam in response to hypoxia and aerial exposure stresses (Nie et al., 2020; Nie et al., 2020), the king scallop in response to aeration stress (Pauletto et al., 2018), the Pacific oyster (Crassostrea gigas) in response to heat stress (Yang et al., 2017) and the Pacific abalone (H. discus hannai) in response to hypoxia stress (Shen et al., 2019).
Apoptosis can be induced via the unfolded protein response (UPR), which is a major signaling cascade controlling the quality of protein folding in the endoplasmic reticulum (ER) (Hetz, 2012; Erguler et al., 2013; Sun et al., 2020). If the unfolded or misfolded proteins in cells are not repaired within a certain period, the UPR triggers cellular apoptosis (Sun et al., 2020). In the present study, many protein folding-related GO functions were significantly enriched, suggesting that an active UPR occurred in scallop gill cells in response to environmental changes. Protein quality control is critical in maintaining cellular homeostasis under environmental stress, and the ER has quality control systems that ensure the correct folding of proteins (Ellgaard et al., 1999). In this study, the pathway of “protein processing in endoplasmic reticulum” was significantly upregulated in most of the Yesso and Zhikong scallop treatment groups; this would recognize and selectively direct misfolded proteins to be either refolded or degraded. “Ubiquitin mediated proteolysis”, which plays an important role in degrading irreparable proteins (Hampton, 2002), was another significantly upregulated pathway in nearly all of the treated groups of the three scallop species. This indicated that abnormal proteins induced by environmental stress in scallops may be eliminated by ubiquitin-mediated proteolysis. Similar conclusions have also been reported in the hard clam, with these same two pathways being the most significantly enriched under heat and hypoxia stresses. These results suggest that bivalves employ a strict quality control system to guarantee correct protein folding and the elimination of irreparable proteins, thereby alleviating cytotoxicity and maintaining cell homeostasis under environmental stress (Hu et al., 2022). However, even in the presence of a quality control system, incorrectly folded proteins will accumulate in the ER under severe stress (Ma and Hendershot, 2004). Therefore, as exposure to heat or hypoxia may induce considerable protein disorder in scallops, apoptosis could eventually result despite the attempts at maintaining homeostasis via the UPR.
HSPs are highly conserved proteins that act as biomarkers of environmental stress (Kregel, 2002; Sørensen et al., 2003). Most studies on HSPs, such as HSPs 70 and 90, have mainly focused on thermal tolerance in mollusks; however, these genes are widely involved in several other environmental stresses responses, including hypoxia and aerial exposure (Delaney and Klesius, 2004; Mohindra et al., 2015; Huo et al., 2019; Nie et al., 2020; Nie et al., 2020). In the hard clam, the elevated expression of two HSP 90 genes and some HSP 70 members was induced by heat and hypoxia stresses (Hu et al., 2022). In the present study, the GO functions of “HSP 70 protein binding” were significantly enriched in the HIG2, LOW1, LOW2, LOW3, and UNI groups of the Yesso scallop and the UNI group of the Zhikong scallop. The GO functions of “HSP binding” were significantly enriched in the LOW1, LOW2, and LOW3 groups of the bay scallop and the LOW1 group of the Yesso scallop. Furthermore, for DEGs shared by all stressed groups in the Yesso scallop, the “HSP binding” GO functions were also highly enriched, and the involved genes HSP 70B2, Heat shock cognate 71 kDa protein, HSP 90-beta, and Hsp 90 co-chaperone Cdc37 were significantly upregulated in all the heat, hypoxia, and heat plus hypoxia groups. These results suggested that HSPs play an important role in response to different types of environmental stress in scallops. HSPs have extensive functions associated with apoptosis, including enhanced cell survival by inhibiting apoptosis (Roberts et al., 2010; Artigaud et al., 2015). Acting as molecular chaperones, HSPs can catalyze protein folding and refolding, stabilize normal proteins to prevent degeneration, eliminate irreversibly damaged proteins, and maintain cellular homeostasis. HSP 90 and HSP 70 are two major chaperone classes (Frydman, 2001; Sørensen et al., 2003; Liu et al., 2019). Therefore, given the highly enriched protein folding and apoptosis-related functions and pathways in the present study, it is likely that HSPs inhibited apoptosis by interacting with abnormal proteins to facilitate the adaption of scallops to environmental changes.
The ability of marine organisms to adapt to changing levels of DO is important for their survival. Several studies have suggested that the modulation of oxygen homeostasis in animals greatly depends on HIF-1 signaling. This induces similar biochemical and physiological responses in different organisms, including oxygen sensor mobilization and oxygen transport, to facilitate cellular or tissue adaptation to low oxygen levels (Semenza, 1999; Zhang et al., 2017). The important role of the HIF-1 signaling pathway in environmental stress tolerance and resistance has been reported in some mollusks. In the Pacific oyster, the expression of HIF-α was significantly increased by air exposure and heat shock (Kawabe and Yokoyama, 2012). The HIF-1 signaling pathway was significantly enriched in the Manila clam and small abalone under hypoxia stress (Sun et al., 2019; Nie et al., 2020). In the Yesso scallop, Kotsyuba reported that the activity of HIF-1α depended on the duration of anoxia and the temperature (Kotsyuba, 2017). In the present study, the HIF-1 signaling pathway was significantly enriched for the upregulated DEGs under severe heat and hypoxia. The pathway was significantly enriched in the HIG2 groups of the Yesso and bay scallops, and the LOW3 and UNI groups of all the three scallop species, suggesting that the HIF-1 signaling pathway was triggered by extreme and acute environmental changes. Thus, the activation of the HIF-1 signaling pathway in scallops depends on the intensity of stress. A few GO functions related to the hypoxia response were also significantly enriched at increased temperatures, suggesting that the activation of the HIF-1 signaling pathway under acute heat stress was probably due to a sharp decrease in DO at high temperatures.
Different numbers of DEGs in the three scallop species were induced by individual and combined stressors, which were associated with their susceptibility to environmental stress. This is perhaps the first study to simultaneously analyze the gene expression regulation induced by environmental stress in more than one bivalve species. In the present study, many more DEGs were observed in the Yesso scallop compared with the other two scallop species in response to both the individual and combined stressors. This suggested the higher sensitivity of the Yesso scallop to environmental changes (heat and hypoxia), followed by the Zhikong scallop, and then the bay scallop. Consistent with the DEGs numbers, more immune response and apoptosis-related GO functions and pathways were enriched in the Yesso scallop. This indicated that the Yesso scallop likely enhanced its gene expression related to the biological processes of immune response and apoptosis in response to environmental stresses. As a tropical species, the bay scallop has a higher thermal tolerance than the other two scallop species. Therefore, the environmental stresses used in this study were likely mild for the bay scallop, resulting in the least DEGs being induced. It is probable that fewer cytotoxic effects—such as DNA and proteins damages—were caused by the environmental stressors in the bay scallop: the DNA repair-related pathways and the protein processing in endoplasmic reticulum pathway were virtually unaffected this species following exposure to any environmental stressor but were down- or upregulated in the other two scallop species (Figure 8). As a result, fewer molecular pathways—such as immune response-related pathways—were induced in the bay scallop in response to environmental changes.
The extent of impacts of environmental stressors varies among taxa, depending on their physiological strategies, life stages, and motility, but also depending on the capacity of microevolutionary processes to increase the resistance of organisms (Vaquer-Sunyer and Duarte, 2011). Intriguingly, different dN/dS ratios, indicating different evolutionary ratio, were discovered among the three scallop species and were consistent with their sensitivity to environmental stress. The highest dN/dS ratio was for the Yesso scallop, followed by the Zhikong and then the bay scallops, This result indicated that scallops with higher environmental sensitivity may experience ongoing accelerated evolution to enable the species to better cope with the changing environment.
Bivalves are mostly weak-swimming or stationary filter-feeders. Many live in intertidal zones or shallow waters where there are wide fluctuations in environmental conditions. Thus, bivalves may have evolved mechanisms such as high genetic diversity and expansion of key stress response genes to adapt to diverse and highly variable environments (Guo et al., 2015). A growing body of evidence shows that environmental stress can affect genome stability in eukaryotes (Galhardo et al., 2007). For instance, hypoxia can suppress DNA repair pathways and cause an increase in mutagenesis in mammalian cells (Yuan et al., 2000; Mihaylova et al., 2003), and yeast regulates mutagenesis in response to environmental stress (Shor et al., 2013). In mollusks, downregulated genes involved in DNA replication and/or repair were detected in the king scallop and Pacific abalone under different environmental stresses (Artigaud et al., 2015; Shen et al., 2019). Consistently, pathways associated with DNA replication and repair were also suppressed in the Yesso and Zhikong scallops under severe hypoxia and combined stresses, which indicated that these individuals might experience increased mutation rates under different environmental stresses (Artigaud et al., 2015).
Evolutionary pressures of various kinds have often been hypothesized to cause active and rapid evolutionary changes, which occasionally generate fitter mutants and potentially accelerate adaptive evolution (Galhardo et al., 2007). Positive selection is a form of natural selection that influences the process by which new advantageous genetic variants sweep across populations (Wang et al., 2010). In Tibetan Schizothoracinae fish (Gymnocypris przewalskii), significant signals of positive selection were discovered on genes controlling innate immunity (Tong et al., 2017). In oysters, some members of expanded immune-related gene families diverged in function at different temperatures and salinities or assumed new roles in abiotic stress responses (Guo et al., 2015), and a positive selection pressure on immune-related genes has also been documented (Yu et al., 2011). In the present study, a total of 239 positively selected genes were identified among four different scallop species, including those involved in immune system and environmental adaptation such as the toll-like, toll-like receptor, c-type lectin, cytochrome c oxidase, and cytochrome P450 genes. The selection of these genes represents an important adaptive mechanism in response to the long-term environmental changes. Further study is needed to investigate the molecular functions of these genes that are associated with environmental adaptation.
In the present study, a transcriptome analysis of the gill tissues of three scallop species—the Yesso, Zhikong, and bay scallops—was performed following short-term exposure to heat, hypoxia, and heat plus hypoxia. The molecular response mechanism of the scallops under environmental stress, especially under combined stress was explored. The DEGs under different types of stress were determined and important functional pathways in environmental stress were identified via GO and KEGG analyses. Comparative transcriptome analysis was performed and positively selected genes were identified, suggesting possible mechanisms of long-term environmental adaptation. The present study provides new insights into the molecular response mechanisms of scallops exposed to combined environmental stressors and improves our understanding of adaptive mechanisms in marine organisms in response to the changing global climate.
Healthy Yesso, Zhikong, and bay scallops with an average shell size of 71.33 ± 0.72 mm, 63.07 ± 0.85 mm and 54.87 ± 1.13 mm, respectively, were obtained from culture populations in the Lvshun Sea area in Dalian, Liaoning Province. The scallops were then cultured under laboratory conditions in the Key Laboratory of Mariculture & Stock Enhancement in North China’s Sea for one week, with filtered and aerated seawater at 15°C, and a twice-daily mixed algal diet of Chlorella sp. and Spirulina platensis. The individual scallop species were divided into the following groups: normal control (NOR) at a water temperature of 15°C and adequate DO (8 mg/L); heat stress at a water temperature of 22°C (HIG1) and 25°C (HIG2) under adequate DO (8 mg/L); hypoxia stress with decreased DO at 6 mg/L (LOW1), 4 mg/L (LOW2), and 2 mg/L (LOW3) at a water temperature of 15°C; and combined stress (UNI) at a water temperature of 25°C and low DO of 2 mg/L. Although the three types of scallops thrive at different temperature ranges, 15 °C is within the optimum temperature range of all the three scallop species and was selected as the control temperature. The temperatures of 22 °C and 25 °C were to indicate the tolerance of the Yesso scallop. The lowest DO of 2 mg/L would be stressful but not lethal for the scallops and has been used in previous studies on hypoxia stress in scallops (Chen et al., 2007; Stevens and Gobler, 2018). The three levels of DO were set as low (6 mg/L), moderate (4 mg/L), and high (2 mg/L) levels of hypoxia compared with the control group of 8 mg/L.
A total of 105 individuals of each species with similar size and good activity were divided into the above seven groups and cultured in 70 L tanks. There were three replicates for each group. For heat stress, the temperature was controlled by a thermostatic heating rod (RS Electrical®, China). For hypoxia stress, the scallops were cultured in closed tanks in which the DO was controlled by pumping nitrogen or air into the seawater with adjustment of the nitrogen flow to maintain the DO level (Sun et al., 2020; Nie et al., 2020). The temperature and DO levels were constantly monitored with a thermometer and O2 sensing probe (YSI ProPlus, USA), respectively, and adjusted whenever necessary. All the scallops from each group were exposed to stress for 36 h. Due to the use of short-term stress treatments in the present study, to avoid the effects of feeding on the culture environment and physiology of the scallops, the scallops were not fed during the experiment. Following treatment, the gill tissues of scallops from each group were sampled, immediately frozen in liquid nitrogen, and stored at -80°C. Three individuals from different tanks of each group (three biological replicates) were selected for transcriptome sequencing. The experimental design is presented in Figure 9. No specific permits were needed for the described field studies. All the Yesso, Zhikong, and bay scallops utilized in this study were cultured marine species that are commercially available and are not endangered or protected species. All experiments were conducted in accordance with the regulations of the local and central governments.
The total RNA of each sample was isolated with the RNAprep pure tissue kit (Tiangen, China) according to the manufacturer’s protocol. The quantity and quality of total RNA were determined using an NV3000 micro-spectrophotometer (Vastech, US), and the integrity was determined by agarose gel electrophoresis. The mRNA was purified from total RNA using oligo(dT) magnetic beads. The RNA-seq libraries were constructed using the TruSeq Stranded mRNA LTSample Prep Kit (Illumina, San Diego, CA, USA) following the manufacturer’s instructions. The quality of the libraries was evaluated using the Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA). Finally, a total of 63 libraries were subjected to 150 bp paired-end sequencing on the Illumina Hiseq X TEN sequencing platform.
Raw sequencing data were processed utilizing Trimmomatic (Bolger et al., 2014) to remove reads containing poly-N and low-quality reads. The high-quality clean reads of each scallop species were combined and used for de novo transcriptome assembly using the Trinity software (version: 2.4) (Grabherr et al., 2011) with the paired-end method and a kmer size of 32. The longest transcript was selected as the unigene based on the similarity and length of a sequence for subsequent analysis. The functions of the unigenes in each scallop species were separately annotated by aligning the unigene sequences to the NCBI non-redundant protein (Nr) database, Swiss-Prot database, Gene Ontology (GO) database, Eukaryotic Orthologous Groups (KOG) protein database and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway database (Kanehisa et al., 2007) using BLASTX with an E-value of 10-5.
The expression of unigenes was determined by mapping the reads from each sample to the assembled references using Bowtie 2 (Langmead and Salzberg, 2012) and the fragments per kilobase per million mapped reads (FPKM) method, which eliminated the effects of gene length and sequencing depth on the calculation of gene expression. Differentially expressed genes (DEGs) in stressed and normal groups were identified by DESeq (Anders and Huber, 2012). The negative binomial distribution hypothesis was tested to determine the significance of differential expression. A fold change > 2 or < 0.5 and unadjusted p-value < 0.05 was set as the threshold for statistically significant differential expression. GO and KEGG enrichment analysis of DEGs were carried out using the clusterProfiler package in R (Yu et al., 2012) based on the hypergeometric distribution test. GO functions or pathways with q-values (calibrated p-values via the Benjamini-Hochberg method) < 0.05 and gene numbers ≥ 3 were considered to be enriched.
The transcripts of the three scallop species and the king scallop (Kenny et al., 2020) were subjected to comparative transcriptome analysis. The transcript coding region sequences (CDS) were predicted by ESTScan software (Iseli et al., 1999). The CDS were then translated into amino acid sequences via standard genetic coding. The orthologous gene clusters were constructed with the CDS sequences of four scallop species using OrthoMCL software (Li et al., 2003) with BLASTP (E-value < 10-10). Single-copy genes with clear one-to-one orthologs between species were used for subsequent analysis. The putative orthologous pairs were then aligned using MUSCLE software (Edgar, 2004) with the default parameters. The polygenetic tree of the four scallops was constructed using RAxML software (Stamatakis, 2014) using the maximum likelihood (ML) method. The dN, dS, and dN/dS values of the orthologs were calculated using the CodeML program of the PAML package (Yang, 2007). The positively selected genes were identified using the branch-site model of PAML with the threshold p-value < 0.05. GO and KEGG enrichment analyses were performed to determine the function of these positively selected genes using the method described in the previous section.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, PRJNA786240.
Ethical review and approval was not required for the animal study because no specific permits were needed for the described field studies. All the Yesso, Zhikong and bay scallops utilized in this study were cultured marine species available commercially, and were not part of an endangered or protected species. All experiments were conducted in accordance with the regulations of the local and central government.
JD and YC conceived and designed the experiments. XH, XJ, WG, JX, YW, DY, and JS prepared the samples. XH, HS, XJ, WG, and JX performed the experiments. JM, XH, XW, ZH, and YT analyzed the data. JM wrote the paper. JD, ZH, and YC reviewed and revised the paper. and JM, JD, and YC acquired the funding. All authors contributed to manuscript revision, read, and approved the submitted version.
This project was supported by the Science and Technology Foundation of Dalian (2021JB11SN035), the University Innovation Team and Innovation Talent Support Plan of Liaoning Province (LT2019003), Dalian High-level Talent Innovation Support Program (2019RQ143), Scientific Research Fund of Liaoning Provincial Department of Education (LJKZ0705), and the National Natural Science Foundation of China (31702342).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2022.971796/full#supplementary-material
Ameisen J. C. (2002). On the origin, evolution, and nature of programmed cell death: a timeline of four billion years. Cell Death Differentiation 9 (4), 367–393. doi: 10.1038/sj.cdd.4400950
Anders S., Huber W. (2012). Differential expression of RNA-seq data at the gene level-the DESeq package Vol. 10 (Heidelberg, Germany: European Molecular Biology Laboratory (EMBL), f1000research.
Artigaud S., Lacroix C., Pichereau V., Flye-Sainte-Marie J.. (2014). Respiratory response to combined heat and hypoxia in the marine bivalves Pecten maximus and Mytilus spp. Comp. Biochem. Physiol. Part A: Mol. Integr. Physiol. 175, 135–140. doi: 10.1016/j.cbpa.2014.06.005
Artigaud S., Richard J., Thorne M. A. S., Lavaud R., Flye-Sainte-Marie J., Jean F., et al. (2015). Deciphering the molecular adaptation of the king scallop (Pecten maximus) to heat stress using transcriptomics and proteomics. BMC Genomics 16, 1–14. doi: 10.1186/s12864-015-2132-x
Blaser H., Dostert C., Mak T. W., Brenner D.. (2016). TNF and ROS crosstalk in inflammation. Trends Cell Biol. 26 (4), 249–261. doi: 10.1016/j.tcb.2015.12.002
Bolger A. M., Lohse M., Usadel B. (2014). Trimmomatic: a flexible trimmer for illumina sequence data. Bioinformatics 30 (15), 2114–2120. doi: 10.1093/bioinformatics/btu170
Brand A. R. (2016). “Scallop ecology: distributions and behaviour,” in Scallops: biology, ecology and aquaculture. Eds. Shumway S., Parsons G. J. (Amsterdam, Netherlands:Elsevier), pp:469–pp:533.
Breitburg D. (2002). Effects of hypoxia, and the balance between hypoxia and enrichment, on coastal fishes and fisheries. Estuaries 25, 767–781. doi: 10.1007/BF02804904
Carroll J., Gobler C. J., Peterson B. J. (2008). Resource-restricted growth of eelgrass in new York estuaries: light limitation, and alleviation of nutrient stress by hard clams. Mar. Ecol. Prog. Ser. 369, 51–62. doi: 10.3354/meps07593
Cerrato R. M., Caron D. A., Lonsdale D. J., Rose J. M., Schaffner R. A.. (2004). Effect of the northern quahog Mercenaria mercenaria on the development of blooms of the brown tide alga aureococcus anophagefferens. Mar. Ecol. Prog. Ser. 281, 93–108. doi: 10.3354/meps281093
Chen D., Jin C. (2019). Histone variants in environmental-stress-induced DNA damage repair. Mutat. Res. 780, 55–60. doi: 10.1016/j.mrrev.2017.11.002
Chen J., Mai K., Ma H., Wang X., Deng D., Liu X., et al. (2007). Effects of dissolved oxygen on survival and immune responses of scallop (Chlamys farreri Jones et Preston). Fish Shellfish Immunol. 22 (3), 272–281. doi: 10.1016/j.fsi.2006.06.003
Chen M., Yang H., Delaporte M., Zhao S., Xing K.. (2007). Immune responses of the scallop Chlamys farreri after air exposure to different temperatures. J. Exp. Mar. Biol. Ecol. 345 (1), 52–60. doi: 10.1016/j.jembe.2007.01.007
Crain C. M., Kroeker K., Halpern B. S. (2008). Interactive and cumulative effects of multiple human stressors in marine systems. Ecol. Lett. 11 (12), 1304–1315. doi: 10.1111/j.1461-0248.2008.01253.x
de Almeida E. A., Bainy A. C. D., de Melo Loureiro A. P., Martinez G. R., Miyamoto S., Onuki J., et al. (2007). Oxidative stress in Perna perna and other bivalves as indicators of environmental stress in the Brazilian marine environment: antioxidants, lipid peroxidation and DNA damage. Comp. Biochem. Physiol. Part A: Mol. Integr. Physiol. 146 (4), 588–600. doi: 10.1016/j.cbpa.2006.02.040
Delaney M. A., Klesius P. H. (2004). Hypoxic conditions induce Hsp70 production in blood, brain and head kidney of juvenile Nile tilapia Oreochromis niloticus (L.). Aquaculture 236 (1-4), 633–644. doi: 10.1016/j.aquaculture.2004.02.025
Doney S. C., Ruckelshaus M., Emmett Duffy J., Barry J. P., Chan F., English C. A., et al. (2012). Climate change impacts on marine ecosystems. Annu. Rev. Mar. Sci. 4, 11–37. doi: 10.1146/annurev-marine-041911-111611
Edgar R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32 (5), 1792–1797. doi: 10.1093/nar/gkh340
Ellgaard L., Molinari M., Helenius A. (1999). Setting the standards: quality control in the secretory pathway. Science 286 (5446), 1882–1888. doi: 10.1126/science.286.5446.1882
Erguler K., Pieri M., Deltas C. (2013). A mathematical model of the unfolded protein stress response reveals the decision mechanism for recovery, adaptation and apoptosis. BMC Syst. Biol. 7 (1), 1–18. doi: 10.1186/1752-0509-7-16
Franco R., Sánchez-Olea R., Reyes-Reyes E. M., Panayiotidis M. I.. (2009). Environmental toxicity, oxidative stress and apoptosis: Ménage à trois. Mutation 674 (1-2), 3–22. doi: 10.1016/j.mrgentox.2008.11.012
Frydman J. (2001). Folding of newly translated proteins in vivo: the role of molecular chaperones. Annu. Rev. Biochem. 70 (1), 603–647. doi: 10.1146/annurev.biochem.70.1.603
Galhardo R. S., Hastings P. J., Rosenberg S. M. (2007). Mutation as a stress response and the regulation of evolvability. Crit. Rev. Biochem. Mol. Biol. 42 (5), 399–435. doi: 10.1080/10409230701648502
Gobler C. J., DePasquale E. L., Griffith A. W., Baumann H.. (2014). Hypoxia and acidification have additive and synergistic negative effects on the growth, survival, and metamorphosis of early life stage bivalves. PLoS One 9 (1), e83648. doi: 10.1371/journal.pone.0083648
Gobler C. J., Lonsdale D. J., Boyer G. L. (2005). A review of the causes, effects, and potential management of harmful brown tide blooms caused by Aureococcus anophagefferens (Hargraves et sieburth). Estuaries 28, 726–749. doi: 10.1007/BF02732911
Grabherr M. G., Haas B. J., Yassour M., Levin J. Z., Thompson D. A., Amit I., et al. (2011). Trinity: reconstructing a full-length transcriptome without a genome from RNA-seq data. Nat. Biotechnol. 29 (7), 644. doi: 10.1038/nbt.1883
Guo X., He Y., Zhang L., Lelong C., Jouaux A.. (2015). Immune and stress responses in oysters with insights on adaptation. Fish shellfish Immunol. 46 (1), 107–119. doi: 10.1016/j.fsi.2015.05.018
Guo X., Luo Y. (2016). “Scallops and scallop aquaculture in China,” in . scallops: biology, ecology and aquaculture. Eds. Shumway S., Parsons G. J. (Amsterdam, Netherlands:Elsevier), pp: 937–952.
Gurr S. J., Dwyer I. P., Goleski J., Lima F. P., Seabra R., Gobler C. J.. (2021). Acclimatization in the bay scallop Argopecten irradians along a eutrophication gradient: insights from heartbeat rate measurements during a simulated hypoxic event. Mar. Freshw. Behav. Physiol. 54, 23–49. doi: 10.1080/10236244.2020.1867477
Halliwell B., Cross C. E. (1994). Oxygen-derived species: their relation to human disease and environmental stress. Environ. Health Perspect. 102 (suppl 10), 5–12. doi: 10.1289/ehp.94102s105
Hampton R. Y. (2002). ER-associated degradation in protein quality control and cellular regulation. Curr. Opin. Cell Biol. 14 (4), 476–482. doi: 10.1016/S0955-0674(02)00358-7
Hetz C. (2012). The unfolded protein response: controlling cell fate decisions under ER stress and beyond. Nat. Rev. Mol. Cell Biol. 13 (2), 89–102. doi: 10.1038/nrm3270
Hoegh-Guldberg O., Bruno J. F. (2010). The impact of climate change on the world’s marine ecosystems. Science 328 (5985), 1523–1528. doi: 10.1126/science.1189930
Hu Z., Feng J., Song H., Zhou C., Yu Z., Yang M., et al. (2022). Mechanisms of heat and hypoxia defense in hard clam: Insights from transcriptome analysis. Aquaculture 549, 737792. doi: 10.1016/j.aquaculture.2021.737792
Huo D., Sun L., Zhang L., Ru X., Liu S., Yang X., et al. (2019). Global-warming-caused changes of temperature and oxygen alter the proteomic profile of sea cucumber Apostichopus japonicus. J. Proteomics 193, 27–43. doi: 10.1016/j.jprot.2018.12.020
Iseli C., Jongeneel C. V., Bucher P. (1999). ESTScan: a program for detecting, evaluating, and reconstructing potential coding regions in EST sequences. ISMB. 99, 138–148.
Jacobson M. D., Weil M., Raff M. C. (1997). Programmed cell death in animal development. Cell 88 (3), 347–354. doi: 10.1016/S0092-8674(00)81873-5
Kanehisa M., Araki M., Goto S., Hattori M., Hirakawa M., Itoh M., et al. (2007). KEGG for linking genomes to life and the environment. Nucleic Acids Res. 36 (suppl_1), D480–D484. doi: 10.1093/nar/gkm882
Kawabe S., Yokoyama Y. (2012). Role of hypoxia-inducible factor α in response to hypoxia and heat shock in the pacific oyster Crassostrea gigas. Mar. Biotechnol. 14 (1), 106–119. doi: 10.1007/s10126-011-9394-3
Keeling R. F., Körtzinger A., Gruber N. (2010). Ocean deoxygenation in a warming world. Annu. Rev. Mar. Sci. 2, 199–229. doi: 10.1146/annurev.marine.010908.163855
Kemp W. M., Testa J. M., Conley D. J., Gilbert D., Hagy J. D.. (2009). Temporal responses of coastal hypoxia to nutrient loading and physical controls. Biogeosciences 6 (12), 2985–3008. doi: 10.5194/bg-6-2985-2009
Kenny N. J., McCarthy S. A., Dudchenko O., James K., Betteridge E., Corton C., et al. (2020). The gene-rich genome of the scallop Pecten maximus. GigaScience 9 (5), giaa037. doi: 10.1093/gigascience/giaa037
Kosaka Y. (2016). “Scallop fisheries and aquaculture in Japan,” in scallops: biology, ecology and aquaculture. Eds. Shumway S., Parsons G. J. (Amsterdam, Netherlands:Elsevier), pp: 891–936.
Kotsyuba E. P. (2017). Hypoxia-inducible factor 1α in the central nervous system of the scallop Mizuhopecten yessoensis jay, 1857 (Bivalvia: Pectinidae) during anoxia and elevated temperatures. Russian J. Mar. Biol. 43 (4), 293–301. doi: 10.1134/S1063074017040071
Kregel K. C. (2002). Invited review: heat shock proteins: modifying factors in physiological stress responses and acquired thermotolerance. J. Appl. Physiol. 92 (5), 2177–2186. doi: 10.1152/japplphysiol.01267.2001
Langmead B., Salzberg S. L. (2012). Fast gapped-read alignment with bowtie 2. Nat. Methods 9 (4), 357. doi: 10.1038/nmeth.1923
Levin L. A., Ekau W., Gooday A. J., Jorissen F., Middelburg J. J., Naqvi S. W.A., et al. (2009). Effects of natural and human-induced hypoxia on coastal benthos. Biogeosciences 6 (10), 2063–2098. doi: 10.5194/bg-6-2063-2009
Li L., Stoeckert C. J., Roos D. S. (2003). OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 13 (9), 2178–2189. doi: 10.1101/gr.1224503
Liu Y., Li L., Huang B., Wang W., Zhang G.. (2019). RNAi based transcriptome suggests genes potentially regulated by HSF1 in the pacific oyster Crassostrea gigas under thermal stress. BMC Genomics 20 (1), 1–15. doi: 10.1186/s12864-019-6003-8
Liu P., Lu Z., Liu L., Li R., Liang Z., Shen M., et al. (2019). NOD-like receptor signaling in inflammation-associated cancers: from functions to targeted therapies. Phytomedicine 64, 152925. doi: 10.1016/j.phymed.2019.152925
Li Q., Zhang F., Wang M., Li M., Sun S.. (2020). Effects of hypoxia on survival, behavior, and metabolism of Zhikong scallop Chlamys farreri Jones et Preston 1904. J. Ocean. Limnol. 38(2), 351–363. doi: 10.1007/s00343-019-9074-0.
Ma Y., Hendershot L. M. (2004). ER chaperone functions during normal and stress conditions. J. Chem. Neuroanat. 28 (1-2), 51–65. doi: 10.1016/j.jchemneu.2003.08.007
Malagoli D., Casarini L., Sacchi S., Ottaviani E.. (2007). Stress and immune response in the mussel mytilus galloprovincialis. Fish Shellfish Immunol. 23 (1), 171–177. doi: 10.1016/j.fsi.2006.10.004
Mattson M. P., Meffert M. K. (2006). Roles for NF-κB in nerve cell survival, plasticity, and disease. Cell Death Differentiation 13 (5), 852–860. doi: 10.1038/sj.cdd.4401837
Mihaylova V. T., Bindra R. S., Yuan J., Yuan J., Campisi D., Narayanan L., Jensen R., et al. (2003). Decreased expression of the DNA mismatch repair gene Mlh1 under hypoxic stress in mammalian cells. Mol. Cell. Biol. 23 (9), 3265–3273. doi: 10.1128/MCB.23.9.3265-3273.2003
Mohindra V., Tripathi R. K., Yadav P., Singh R. K., Lal K. K.. (2015). Hypoxia induced altered expression of heat shock protein genes (Hsc71, Hsp90α and Hsp10) in Indian catfish, Clarias batrachus (Linnaeus, 1758) under oxidative stress. Mol. Biol. Rep. 42 (7), 1197–1209. doi: 10.1007/s11033-015-3855-0
Nie H., Jiang K., Li N., Yan X.. (2020). Transcriptomic analysis of Ruditapes philippinarum under aerial exposure and reimmersion reveals genes involved in stress response and recovery capacity of the Manila clam. Aquaculture 524, 735271. doi: 10.1016/j.aquaculture.2020.735271
Nie H., Wang H., Jiang K., Yan X.. (2020). Transcriptome analysis reveals differential immune related genes expression in Ruditapes philippinarum under hypoxia stress: potential HIF and NF-κB crosstalk in immune responses in clam. BMC Genomics 21 (1), 1–16. doi: 10.1186/s12864-020-6734-6
Ning J., Zou D., Lu X., Cao W., Chen M., Liu B., et al. (2021). Transcriptomic analyses provide insights into the adaptive responses to heat stress in the ark shells, Scapharca subcrenata. Comp. Biochem. Physiol. Part D: Genomics Proteomics 38, 100813. doi: 10.1016/j.cbd.2021.100813
Officer C. B., Smayda T. J., Mann R. (1982). Benthic filter feeding: a natural eutrophication control. Mar. Ecol. Prog. Ser. 9, 203–210. doi: 10.3354/meps009203
Parthasarathy A., Srinivasan S., Appleby A. J., Martin C. R.. (1992). Temperature dependence of the electrode kinetics of oxygen reduction at the platinum/Nafion® interface-a microelectrode investigation. J. Electrochemical Soc. 139 (9), 2530. doi: 10.1149/1.2221258
Pauletto M., Di Camillo B., Miner P., Huvet A., Quillien V., Milan M., et al. (2018). Understanding the mechanisms involved in the high sensitivity of Pecten maximus larvae to aeration. Aquaculture 497, 189–199. doi: 10.1016/j.aquaculture.2018.07.059
Pörtner H. O. (2010). Oxygen-and capacity-limitation of thermal tolerance: a matrix for integrating climate-related stressor effects in marine ecosystems. J. Exp. Biol. 213 (6), 881–893. doi: 10.1242/jeb.037523
Pörtner H. O. (2012). Ntegrating climate-related stressor effects on marine organisms: unifying principles linking molecule to ecosystem-level changes. Mar. Ecol. Prog. Ser. 470, 273–290. doi: 10.3354/meps10123
Pörtner H. O., Farrell A. P. (2008). Physiology and climate change. Science, 322(5902)690–692. doi: 10.1126/science.1163156
Pörtner H. O., Knust R. (2007). Climate change affects marine fishes through the oxygen limitation of thermal tolerance. science 315 (5808), 95–97. doi: 10.1126/science.1135471
Roberts R. J., Agius C., Saliba C., Bossier P., Sung Y. Y.. (2010). Heat shock proteins (chaperones) in fish and shellfish and their potential role in relation to fish health: a review. J. fish Dis. 33 (10), 789–801. doi: 10.1111/j.1365-2761.2010.01183.x
Robinson S. M. C., Parsons G. J., Davidson L. A., Shumway S. E., Blake N. J.. (2016). “Scallop aquaculture and fisheries in Eastern north America,” in Scallops: biology, ecology and aquaculture. Eds. Shumway S., Parsons G. J. (Amsterdam, Netherlands:Elsevier), pp: 737–779.
Sørensen J. G., Kristensen T. N., Loeschcke V. (2003). The evolutionary and ecological role of heat shock proteins. Ecol. Lett. 6 (11), 1025–1037. doi: 10.1046/j.1461-0248.2003.00528.x
Semenza G. L. (1999). Regulation of mammalian O2 homeostasis by hypoxia-inducible factor 1. Annu. Rev. Cell Dev. Biol. 15 (1), 551–578. doi: 10.1146/annurev.cellbio.15.1.551
Shaw P. J., Lamkanfi M., Kanneganti T. D. (2010). NOD-like receptor (NLR) signaling beyond the inflammasome. Eur. J. Immunol. 40 (3), 624–627. doi: 10.1002/eji.200940211
Shen Y., Huang Z., Liu G., Ke C., You W.. (2019). Hemolymph and transcriptome analysis to understand innate immune responses to hypoxia in pacific abalone. Comp. Biochem. Physiol. Part D: Genomics Proteomics 30, 102–112. doi: 10.1016/j.cbd.2019.02.001
Shor E., Fox C. A., Broach J. R. (2013). The yeast environmental stress response regulates mutagenesis induced by proteotoxic stress. PLoS Genet. 9 (8), e1003680. doi: 10.1371/journal.pgen.1003680
Shumway S. E., Castagna M. (1994). Scallop fisheries, culture and enhancement in the united states. Memoirs Queensland Museum 36 (2), 283–298.
Silina A. V. (2019). A yesso scallop population exposed to climate-induced and anthropogenic habitat changes in amur bay, Sea of Japan. Oceanology 59, 75–85. doi: 10.1134/S000143701901020X
Sokolova I. M. (2009). Apoptosis in molluscan immune defense. Invertebrate Survival J. 6 (1), 49–58.
Stamatakis A. (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30 (9), 1312–1313. doi: 10.1093/bioinformatics/btu033
Stevens A. M., Gobler C. J. (2018). Interactive effects of acidification, hypoxia, and thermal stress on growth, respiration, and survival of four north Atlantic bivalves. Mar. Ecol. Prog. Ser. 604, 143–161. doi: 10.3354/meps12725
Sun X., Tu K., Li L., Wu B., Wu L., Liu Z., et al. (2021). Integrated transcriptome and metabolome analysis reveals molecular responses of the clams to acute hypoxia. Mar. Environ. Res. 168, 105317. doi: 10.1016/j.marenvres.2021.105317
Sun Y., Zhang X., Wang Y., Day R., Yang H., and Zhang Z., et al. (2019). Immunity-related genes and signaling pathways under hypoxic stresses in Haliotis diversicolor: a transcriptome analysis. Sci. Rep. 9 (1), 1–15. doi: 10.1038/s41598-019-56150-2
Sun J. L., Zhao L. L., Liao L., Tang X., Cui C., Liu Q., et al. (2020). Interactive effect of thermal and hypoxia on largemouth bass (Micropterus salmoides) gill and liver: Aggravation of oxidative stress, inhibition of immunity and promotion of cell apoptosis. Fish shellfish Immunol. 98, 923–936. doi: 10.1016/j.fsi.2019.11.056
Tang T., Li W., Wang X., Wu Y., Liu F.. (2019). A house fly TNF ortholog eiger regulates immune defense via cooperating with toll and imd pathways. Dev. Comp. Immunol. 90, 21–28. doi: 10.1016/j.dci.2018.08.016
Tanji T., Ip Y. T. (2005). Regulators of the toll and imd pathways in the drosophila innate immune response. Trends Immunol. 26 (4), 193–198. doi: 10.1016/j.it.2005.02.006
Tong C., Fei T., Zhang C., Zhao K. (2017). Comprehensive transcriptomic analysis of Tibetan schizothoracinae fish Gymnocypris przewalskii reveals how it adapts to a high altitude aquatic life. BMC evolutionary Biol. 17 (1), 1–11. doi: 10.1186/s12862-017-0925-z
Truebano M., Burns G., Thorne M. A. S., Hillyard G., Peck L. S., Skibinski D. O.F., et al. (2010). Transcriptional response to heat stress in the Antarctic bivalve laternula elliptica. J. Exp. Mar. Biol. Ecol. 391, 65–72. doi: 10.1016/j.jembe.2010.06.011
Vaquer-Sunyer R., Duarte C. M. (2008). Thresholds of hypoxia for marine biodiversity. Proc. Natl. Acad. Sci. 105 (40), 15452–15457. doi: 10.1073/pnas.0803833105
Vaquer-Sunyer R., Duarte C. M. (2011). Temperature effects on oxygen thresholds for hypoxia in marine benthic organisms. Global Change Biol. 17 (5), 1788–1797. doi: 10.1111/j.1365-2486.2010.02343.x
Visick J. E., Clarke S. (1995). Repair, refold, recycle: how bacteria can deal with spontaneous and environmental damage to proteins. Mol. Microbiol. 16 (5), 835–845. doi: 10.1111/j.1365-2958.1995.tb02311.x
Wajant H., Pfizenmaier K., Scheurich P. (2003). Tumor necrosis factor signaling. Cell Death Differentiation 10 (1), 45–65. doi: 10.1038/sj.cdd.4401189
Wall C. C., Peterson B. J., Gobler C. J. (2008). Facilitation of seagrass zostera marina productivity by suspension-feeding bivalves. Mar. Ecol. Prog. Ser. 357, 165–174. doi: 10.3354/meps07289
Wang C., Liu B., Li J., Liu S., Li J., Hu L., et al. (2011). Introduction of the Peruvian scallop and its hybridization with the bay scallop in China. Aquaculture 310 (3-4), 380–387. doi: 10.1016/j.aquaculture.2010.11.014
Wang J. R., Wei Y. M., Deng M., Nevo E., Yan Z. H., Zheng Y. L.. (2010). The impact of single nucleotide polymorphism in monomeric alpha-amylase inhibitor genes from wild emmer wheat, primarily from Israel and golan. BMC Evolutionary Biol. 10 (1), 1–13. doi: 10.1186/1471-2148-10-170
Xiao J., Ford S. E., Yang H., Zhang G., Zhang F., Guo X.. (2005). Studies on mass summer mortality of cultured zhikong scallops (Chlamys farreri Jones et Preston) in China. Aquaculture 250, 602–615. doi: 10.1016/j.aquaculture.2005.05.002
Yang Z. (2007). PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24 (8), 1586–1591. doi: 10.1093/molbev/msm088
Yang C., Gao Q., Wang L., Zhou Z., Gong C., et al. (2017). The transcriptional response of the pacific oyster Crassostrea gigas against acute heat stress. Fish Shellfish Immunol. 68, 132–143. doi: 10.1016/j.fsi.2017.07.016
Yang B., Gao X., Zhao J., Liu Y., Xie L., Lv X., et al. (2021). Summer deoxygenation in a bay scallop (Argopecten irradians) farming area: The decisive role of water temperature, stratification and beyond. Mar. pollut. Bull. 173, 113092. doi: 10.1016/j.marpolbul.2021.113092
Yang H. S., Zhang T., Wang J., Wang P., He Y., Zhang F.. (1999). Growth characteristics of Chlamys farreri and its relation with environmental factors in intensive raft-culture areas of sishiliwan bay, yantai. J. Shellfish Res. 18 (1), 71–83.
Yuan J., Narayanan L., Rockwell S., Glazer P. M.. (2000). Diminished DNA repair and elevated mutagenesis in mammalian cells exposed to hypoxia and low pH. Cancer Res. 60 (16), 4372–4376.
Yu H., He Y., Wang X., Zhang Q., Bao Z., Guo X.. (2011). Polymorphism in a serine protease inhibitor gene and its association with disease resistance in the eastern oyster (Crassostrea virginica gmelin). Fish shellfish Immunol. 30 (3), 757–762. doi: 10.1016/j.fsi.2010.12.015
Yu G., Wang L. G., Han Y., He Q. Y.. (2012). clusterProfiler: an r package for comparing biological themes among gene clusters. Omics: J. Integr. Biol. 16 (5), 284–287. doi: 10.1089/omi.2011.0118
Zhang X., Huang Y., Cai X., Zou Z., Wang G., Wang S., et al. (2014). Identification and expression analysis of immune-related genes linked to Rel/NF-κB signaling pathway under stresses and bacterial challenge from the small abalone Haliotis diversicolor. Fish shellfish Immunol. 41 (2), 200–208. doi: 10.1016/j.fsi.2014.08.022
Zhang X., Shi J., Sun Y., Habib Y. J., Yang H., Zhang Z., et al. (2019). Integrative transcriptome analysis and discovery of genes involving in immune response of hypoxia/thermal challenges in the small abalone Haliotis diversicolor. Fish Shellfish Immunol. 84, 609–626. doi: 10.1016/j.fsi.2018.10.044
Zhang G., Zhao C., Wang Q., Gu Y., Li Z., Tao P., et al. (2017). Identification of HIF-1 signaling pathway in Pelteobagrus vachelli using RNA-seq: effects of acute hypoxia and reoxygenation on oxygen sensors, respiratory metabolism, and hematology indices. J. Comp. Physiol. B 187 (7), 931–943. doi: 10.1007/s00360-017-1083-8
Keywords: scallops, heat, hypoxia, transcriptome, response mechanism
Citation: Mao J, Huang X, Sun H, Jin X, Guan W, Xie J, Wang Y, Wang X, Yin D, Hao Z, Tian Y, Song J, Ding J and Chang Y (2022) Transcriptome analysis provides insight into adaptive mechanisms of scallops under environmental stress. Front. Mar. Sci. 9:971796. doi: 10.3389/fmars.2022.971796
Received: 17 June 2022; Accepted: 22 August 2022;
Published: 12 September 2022.
Edited by:
Juan D. Gaitan-Espitia, The University of Hong Kong, Hong Kong SAR, ChinaReviewed by:
Jesús Antonio López-Carvallo, Center for Scientific Research and Higher Education in Ensenada (CICESE), MexicoCopyright © 2022 Mao, Huang, Sun, Jin, Guan, Xie, Wang, Wang, Yin, Hao, Tian, Song, Ding and Chang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jun Ding, ZGluZ2p1bjExMTlAZGxvdS5lZHUuY24=; Yaqing Chang, Y2hhbmdsYWJAaG90bWFpbC5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.