
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mar. Sci., 05 September 2022
Sec. Marine Conservation and Sustainability
Volume 9 - 2022 | https://doi.org/10.3389/fmars.2022.967435
This article is part of the Research TopicAquatic One Health—The Intersection of Marine Wildlife Health, Public Health, and Our OceansView all 10 articles
E. coli is a species of enteric bacteria found in the intestinal tract of humans and animals that can persist in the environment and contaminate food. Anthropogenic activity has led to pathogenic E. coli from humans and animals contaminating environments through the discharge of fecal wastes in sewage and agricultural runoff. While anthropogenic sources of E. coli have been described in terrestrial and freshwater environments, gaps remain in scientific knowledge about E. coli diversity in marine environments and the risk to human and animal health. This study aims to fill in some of the knowledge gaps on the diversity of E. coli in marine ecosystems, including: 1) describe the spatial variation of the E. coli sequence types (STs) found in the study region; 2) describe available information on E. coli STs from marine environments in terms of known relationships to determine if the isolates were related to human, animal, environment strains or novel. We analyzed a dataset of 332 E. coli isolates from the Salish Sea ecosystem, comprising 196 multi-locus sequence types. Sample sources included marine water near shellfish beds, marine wildlife, river otters, and a small number of marine water sites near beaches and freshwater samples from creeks into the Salish Sea. ST10 was the most frequent ST (n=12) and was found in multiple locations and sample types. For the identified STs, we searched metadata for E. coli STs in EnteroBase, an international E. coli database. Additional information on E. coli STs was derived from searches of published studies in PubMed. We found that diversity varied between different regions of the study area, with the greatest diversity found in an area which has partially treated wastewater outflows. A higher diversity of STs associated with animals was found in an area near were animals are raised. Many of the STs identified have been associated with virulence in humans. For a number of identified STs, no references could be found in either PubMed or EnteroBase. These findings support the importance of further studies to understand the relevance of marine E. coli to human and wildlife health.
While E. coli is part of the normal flora of warm-blooded mammals’ intestines, it can be a significant pathogen, causing severe disease inside and outside of the intestinal tract (Centers for Disease Control and Prevention, 2014). There is tremendous genetic diversity among E. coli, and Multi-locus Sequencing Typing (MLST) has been used to partition E. coli into genetically related groups by assigning sequence types (STs) based on allelic variation within specific housekeeping genes (Ibarz Pavón and Maiden, 2009). Since its inception, MLST has been used to characterize the epidemiologic behavior of various clones of E. coli (Jolley et al., 2018; Vingino et al., 2021).
There are multiple routes by which E. coli enter water systems. Animal manure is often used for fertilizer in crop agriculture, thus contaminating vegetables and the surrounding soil (Centers for Disease Control and Prevention, 2022). Once the manure is applied to fields, bacteria can make their way to groundwater or wash into nearby bodies of water from rain, flooding, or inadequate containment of animal waste (McEwen and Collignon, 2018). The bacteria may ultimately end up in the marine environment. Wastewater treatment plants (WWTPs) are able to dump partially treated effluent, containing viable bacteria, directly into ocean waters. Raw sewage is released into the environment due to flooding, line breaks, and power outages, resulting in polluting water and environmental sources (Environmental Protection Agency, 2021). The illegal discharge of ship ballast water also contributes to bacterial contamination in marine waters with enteric bacteria, including strains of E. coli O157:H7 (Ruiz et al., 2000; Altug et al., 2012).
This study aims to fill in some of the knowledge gaps on the diversity of E. coli in marine ecosystems. The study analyzes data on previously collected isolates of E. coli from marine water (primarily around commercial shellfish operations) and wildlife samples while comparing the diversity of STs across four quadrants of the Salish Sea (north, south, central, and Strait of Juan de Fuca) (Norman et al., 2021; Vingino et al., 2021)(Figure 1). We simultaneously compared the STs from our dataset to metadata about associated hosts for each ST appearing in EnteroBase, a public database of E. coli scientific knowledge (https://enterobase.warwick.ac.uk/) and the published scientific literature in PubMed. We hypothesized that there would be variability in the diversity of the STs within E. coli isolated from the Salish Sea across the region, with more human-associated E. coli in a quadrant of the sound with less treated sewage and more animal-associated E. coli in a quadrant near livestock farming. Our study aim was to test this hypothesis and the objective was to look for diversity of the E. coli between the quadrants.
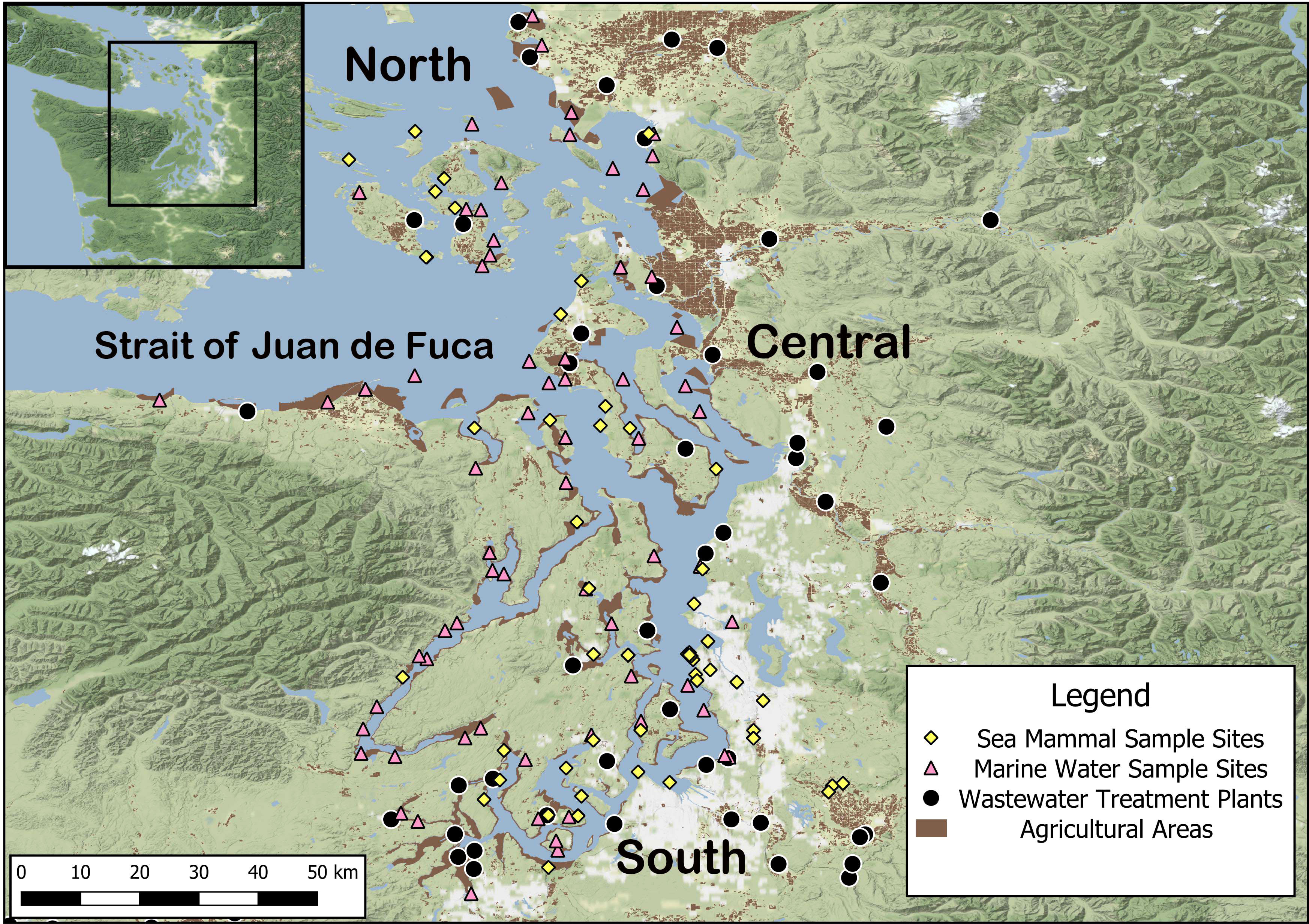
Figure 1 All sample locations in the Salish Sea are for water and marine mammals and anthropogenic sources.
The Salish Sea is an international inland sea that stretches from Campbell River, British Columbia, to Olympia, Washington, encompassing the Puget Sound and San Juan Islands. The Salish Sea has a complex system of estuaries and interconnecting marine waterways. The coastline spans ~ 7,470 km. It is home to ~37 species of animals, 172 birds, and 250 fish species (The Seadoc Society, 2022). The Strait of Juan de Fuca is the primary direct access to the Pacific Ocean from the Salish Sea. According to the State of Washington Department of Ecology [Puget Sound Nutrient Source Reduction Project. Volume 1: Model Updates and Bounding Scenarios (wa.gov)], the Salish Sea area is home to about 4.5 million people (2019). Approximately 99 WWTPs line the shores of the Salish Sea. Seventy-nine WWTPs are in the United States and twenty on the Canadian side (2019). WWTPs near Vancouver and Vancouver Island, British Columbia, dump partially treated wastewater into the Salish Sea (Johannessen et al., 2015). In the counties which border the Salish Sea (Whatcom, Skagit, Snohomish, King, Pierce, Thurston, Mason, Jefferson, and Clallam), pasture and hay make up the most agricultural land use behind shellfish harvesting (Agricultural Land Use | Washington State Department of Agriculture) (See map 1).
The E. coli isolates in this study were collected as part of previously published studies (Norman et al., 2021; Vingino et al., 2021). In those studies, five hundred fifty-one isolates of E. coli were obtained from various samples from the Salish Sea ecosystem. From these, 332 E. coli isolates were further studied with whole genome sequencing. These included isolates from marine water samples near shellfish beds (n=238), marine water samples near local Seattle beaches (n=3), freshwater samples of estuaries entering into the Salish Sea (n=5), fecal samples from river otters (Lontra canadensis) (n=24), rectal swabs, and fecal samples from both dead and alive seals (Phoca vituline) (n=53), fecal sampling from dead porpoise (Phocoena phocoena) (n=7), and samples isolated from English sole (Parophrys vetulus) (n=2) (Table S1). Differences in sampling numbers are due to the previous study (Vingino et al., 2021) assessing for antibiotic resistance in the E. coli isolates. While 332 isolates were sent for whole-genome sequencing (WGS) and MLST, not all were able to be sent for antibiotic resistance testing. The previous study by Vingino et al. (2021) used 305 isolates, while the current study uses all 332 whole genome sequences (Table S1).
Samples of marine water were taken in a coordinated manner by the Washington State Department of Health as part of their shellfish monitoring program (Vingino et al., 2021). To our knowledge, no samples were collected after significant rainfall or flooding events which may have influenced the amount of E. coli in the area. The current study does not measure the survivability of E. coli but rather detection by culture at the time of sampling. The E. coli came from the study by Vingino et al. (2021) and included marine waters samples provided by the Washington Department of Health collected from GPS-located shellfish beds as part of a fecal coliform analysis using the Environmental Protection Agency modified A-1 method (Rice et al., 2012). In addition marine water samples near beaches were collected 15 cm below the water surface. These marine water samples were diluted 1:10 (10 mL marine water and 90 mL deionized sterile water) (2021). The freshwater samples from Piper’s Creek and a beaver pond adjacent to marine beaches (Golden Garden Park, Seattle, WA) were used directly and both the marine water by beaches and freshwater samples were processed using Colilert Standard Quanti-Tray2000® (IDEXX Laboratories, Westbrook, ME, USA) at the University of Washington Laboratory.
Opportunistic sampling was performed on deceased harbor seals or porpoises via fecal swab obtained during necropsy or by rectal swab, and obtained from the study by Norman et al. (2021). Swabs were placed into Amies transport medium without charcoal, refrigerated, and shipped to a reference lab (Phoenix Laboratory, Mukilteo, WA, USA [Zoeitis References Laboratory] (Norman et al., 2021). Fecal samples from alive seals were obtained by the Washington Department of Fish and Wildlife (WDFW) from various dock sites in the Salish Sea (Vingino et al., 2021). River otter fecal sampling was collected along the Green-Duwamish River in Washington at six otter latrine locations. The samples were iced and transported to the University of Washington laboratory and processed using eosin methylene blue agar, and verified with biochemical testing (Vingino et al., 2021). More specific details have been described previously by Norman et al. (2021) and Vingino et al. (2021).
As described in the study by Vingino et al. (2021), whole-genome sequencing (WGS) was done in cooperation with the Washington Department of Health in conjunction with the FDA GenomeTrakr Project (ID 283914 - BioProject - NCBI) using Illumina (Illumina, San Diego, CA,USA) (2021). The DNA was extradite using Qiagen DNeasy Blood and Tissue Kit (Qiagen Sciences Inc. Germantown, MD USA) or MagnaPure 96 with Roche DNA and viral nucleic acid small volume kits (Roche Pharmaceuticals, Indianapolis, IND USA). From that sequence data, MLST analysis was able to be performed to determine the ST for each isolate. NCBI accession numbers for the isolates in that study have previously been described by Vingino et al. (2021). Additional NCBI accession numbers used for this study are as follows: [SAMN] 13392847, 14137889, 14214489, 15182309, 15182314, 15182317, 15344669, 15483653. [SRR] 12424353, 12643355, 12618578, 12618581, 12643343, 12643347, 12643348, 12643351, 12643352, 12643360, 12643362, 12643364, 12643366-12643368, 12643370, 12663941.
EnteroBase version 1.1.3 is an online public and voluntary repository of scientific data regarding E. coli. Researchers contribute data to the database, including results of laboratory analysis and metadata, including sequence type. Other metadata for accessions include the source (animal, human or environmental), source type, source details, country of origin, and laboratory analytic results. The data are not centrally curated, so there is potential for data heterogeneity. Despite these limitations, EnteroBase data have been used for epidemiologic investigation of E. coli (Zhou et al., 2020).
The database was accessed on 8/11/2021 (https://enterobase.warwick.ac.uk/) to retrieve and download data for investigating the 196 STs identified in this study. A total of 153,428 entries existed in EnteroBase at the time of the query. Of the 153,428 entries, 47,319 were for the 196 STs found in this study. Entries for all 196 STs were found in the database. Information about the source (human, animal, environmental) for isolates was categorized based on available information found in EnteroBase using the listed source of the isolates: 1) found in human, animal, and environment, 2) found in animals, 3) found in humans, 4) found in the environment, 5) in animals and environment, 6) in humans and environment, and 7) in humans and animals (Table S2). STs with ten or fewer entries in EnteroBase were coded as “novel.” Attention was made to any reference in the data about an ST regarding a documented marine water source, marine animals, shellfish, and fish.
We noted instances when the source niche and the source type did not match (e.g., the source niche was listed as human, but the source type entered listed as ovine/goat). Additionally, the source was not entered for every isolate. While every attempt was made to identify and mitigate these inconsistencies, some source information for isolates may be inaccurate. It is also possible that isolates in this study have been recovered from other sources but were not listed. Only a few isolates were reported by pathogen type (e.g., STEC, ExPEC, and EHEC). Thus, we may have under-identified pathogenic potential in our isolates.
Many E. coli STs are commensal and do not have pathogenic potential to cause disease (Centers for Disease Control and Prevention, 2014). While other E. coli STs are associated with one or more pathogenic types, which can cause varying degrees of illness in humans and animals (Jolley et al., 2018). The pathogenic types examined included: 1) Shiga toxin-producing E. coli (STEC), 2) verocytoxic E. coli (VTEC), and 3) enterohemorrhagic E. coli (EHEC), which can produce a toxin that causes various diseases with associated mortality (CDC, 2014). E. coli (EIEC) causes intestinal inflammation and bloody diarrhea similar to STEC (Belotserkovsky and Sansonetti, 2018). Enterotoxigenic E. coli (ETEC) causes diarrheal illness and is the leading cause of traveler diarrhea and illness in low-income countries, especially among children (Centers for Disease Control and Prevention, 2014). Enteroaggregative E. coli (EAEC) is an emerging foodborne pathogen that may cause acute or chronic diarrhea (Kaur et al., 2010), while Enteropathogenic E. coli (EPEC) is a common cause of diarrheal illness in children in developing countries (Ochoa and Contreras, 2011). Extraintestinal Pathogenic E. coli (ExPEC) can cause E. coli infections outside the intestinal tract, including urinary tract and bloodstream infections. It is also the second most common cause of meningitis in neonates (Russo and Johnson, 2003). Uropathogenic E. coli (UPEC) is a subset of extraintestinal pathogenic E. coli, causing only urinary tract infections (Spurbeck and Mobley, 2013). EnteroBase was queried for mention of those pathogenic types listed above. We additionally coded ST types as to whether they had been associated with STEC, EPEC, EAEC, UPEC, ExPEC, EIEC, ETEC, and EHEC strains. The number of pathogenic types for each ST was identified and added to create a “pathogen score” for a particular ST (Table S3).
ST distribution across different areas of the Salish Sea was performed using QGIS version 3.22. The Salish Sea was divided in to 4 quadrants for analysis (north, central, south and Strait of Juan de Fuca) (Figure 1). Maps were created to demonstrate the field collection sites and anthropogenic E. coli sources. We also used maps to give a visualization of the distribution of ST diversity. Metadata for WWTP in the Puget Sound area were obtained from the U.S. Geological Survey (USGS Water Mission Area NSDI Node). Metadata for agricultural areas were obtained from the Washington State Department of Agriculture (Agricultural Land Use | Washington State Department of Agriculture) (Figure 1).
PubMed (PubMed (nih.gov) was searched between July and September of 2021 to determine the number of citations in the entire PubMed database for specific STs. The following search terms were utilized: (“Escherichia coli”[Mesh : NoExp] OR “E coli”[TIAB] AND (“ST (number)” OR “Sequence type (number) “ OR “ST (number)”[TIAB]). The Result was categorized as follows:
0 articles found; 1-10 articles found; 11-25 articles found; 26-50 articles found; 51-100 articles found; 101-200 articles found; 201-500 articles found; 501-1000 articles found, and >1000 articles found.
Statistical analysis of E. coli diversity was performed using Shannon diversity index, Shannon equitable index (evenness), and Hutcheson t-test to characterize diversity of E. coli in the four quadrants of the Salish Sea region. The Shannon diversity index quantifies biodiversity so it can be compared to other locations or communities, but statistical significance cannot be inferred from these index values. The Hutcheson t-test is a modified version of a t-test that specifically compares the indices of the Shannon diversity index so that comparisons can be made between the diversity of two samples (Data Analytics, 2019). These statistical measures were used to test the null hypotheses 1) diversity measures of STs are similar across the four quadrants of the Puget Sound and there would be no spatial variation, 2) there are no geographic differences in diversity measures of isolates with source metadata derived from EnteroBase, Source metadata from EnteroBase were used to characterize isolates according to likely sources (human, animal, environmental). Utilizing these same tests, diversity among the quadrants for STs with pathogenic potential was also assessed. Data analysis was performed using R studio version 3.6.2.
Amongst the 332 E. coli isolates, 196 different STs were identified by MLST. A total of 238 isolates were obtained from marine water (72%), which included 159 STs. An additional 86 isolates with 56 different STs were from animals; seal (n=17), deceased seal (n=36), porpoise (n=7), sole (n=2), and river otter (n=24), while 33 different STs were found in animals only (Figure 1) Five isolates were from freshwater samples (ST297, ST405, ST616, ST681, ST2557), and three were from samples of marine water by beaches (ST58, ST117, ST1308). Only two STs (ST10 and ST720) were found in marine water in all four quadrants of the marine water in the Salish Sea. The most common ST found was ST10, with twelve isolates (3.6% of the total), followed by ST162, with eight isolates (2.4%). Two STs had seven isolates each (ST297, ST372). Three STs had six isolates found (ST117, ST967, and ST 1079). Most STs identified were represented by a single isolate (n=139). The remaining (n=43) E. coli isolates had a total of 2-5 isolates for each ST (Table S2).
A high level of diversity of STs was found amongst all marine water samples in each of the four quadrants of the Salish Sea (Figure 1). The Strait of Juan de Fuca quadrant had the greatest richness of diversity among STs found there (n=51) with a Shannon diversity index (H=3.852), followed closely by the south quadrant (H=3.827). The north quadrant contained the least diversity (H=3.718). Using the Hutcheson t-test to compare the Shannon diversity index values of the Strait of Juan de Fuca and the north quadrant, there was no statistical difference in the diversity of the two quadrants (p=0.13). Of those STs found in animals and the environment, the central quadrant contained the greatest diversity (H=2.398), while the south quadrant was least diverse (H=0.95). The difference in diversity was statistically significant (p=0.0018). Of the isolates only found in the environment, the south quadrant had the highest level of diversity (H=1.7), while the north and Strait of Juan de Fuca had the least (H=1.38). There was a statistically significant higher diversity of these environmental isolates in the southern quadrant compared to the northern quadrant and Strait of Juan de Fuca (p=0.02). Of the isolates coded as novel (n=62), the Strait of Juan de Fuca contained the highest amount of diversity (H=2.75), while the south quadrant had the least diversity (H=2.39). This difference was also statistically significant (p=0.009) (Table 1). Of those STs previously described as found in humans and the environment (n=8), humans only (n=2), humans and animals (n=4), and animal only (n=2), the small sample size did not allow for statistical analysis of diversity.
Of the 196 STs found in this study, 82 (41.8%) had no published articles retrieved on a PubMed search, suggesting that these STs are rarely identified and/or reported. An additional 91 STs had only 1-10 citations available (46%). Only one ST (ST131), had greater than 1,000 articles retrieved (Figure 2). In contrast, in EnteroBase, only 47 (24%) had ¾ five entries, and 15 (7.7%) had ¾ ten entries in EnteroBase. Compared to source data from EnteroBase, 129 (65.8%) STs from this study were found in humans, animals, and the environment. Of the remaining STs, 21(10.7%) were identified in the environmental sources only, 30 (15.3%) were identified in both environmental and animal sources, eight had known sources in humans and environment, four were found in humans and animals, two were known to be found in humans only, and two were known animal sources. (Figure S1). All quadrants of the study region contained STs that were associated with animal sources only. In the northern quadrant, 84% of all STs were associated with animals (n=38), in comparison to 85% in the southern quadrant (n=41), 88% in the central quadrant (n=43), and 78% in the Strait of Juan de Fuca (n=40). STs associated with human sources were also isolated in the four quadrants, 76% (n=34) in the north, in comparison to 70% (n=34) in the central, 78% (n=38) in the south, and 86% (n=44) in the Strait of Juan de Fuca. For those isolates found in humans and the environment (n=8), five isolates were found in the Strait of Juan de Fuca. The two human-only isolates were obtained in dead seals. The two animal-only isolates were found in the Strait of Juan de Fuca and the southern quadrant. The four isolates found in humans and animals were found in two deceased seals, the central and the southern quadrants.
Five STs stood out when compared to known sources in EnteroBase (ST5869, ST1065, ST2164, ST9001, and ST11343) which had no previous evidence of occurrence in marine water or sea mammals. STs found in the current study were compared to available source data in EnteroBase; four isolates STs were found in marine water; twenty-two isolates STs were found in marine fish, eleven STs in marine mammals, and forty STs in shellfish (Table 2).
The 196 STs found in this study were queried in EnteroBase to determine if there were known STEC/VTEC, EPEC, EAEC, ExPEC, EIEC, ETEC, EHEC, or UPEC isolates associated with these STs. Thirty-five of the STs found in our study were UPEC strains. ST10 was the most abundant ST found in this study (n=12) and was associated with STEC/VTEC, EPEC, EAEC, ExPEC, and ETEC strains. Six STs (ST10, ST58, ST101, ST155, ST297, ST1248) were associated with STEC strains, ten EPEC strains, three EIEC strains, 19 EAEC, 28 ExPEC strains, seven ETEC, and three EHEC (Table S3). In those STs with at least one pathogen type, the south quadrant contained the highest diversity (H=2.955), while the central quadrant contained the least diversity (H=2.58). All quadrants sampled for marine water, freshwater, seals (alive and deceased), porpoise, and sole had at least one pathogen associated ST strain. EnteroBase was queried for pathogenic type STs with sources found in marine mammals and shellfish, with known disease-causing strains. There were 21 STs with pathogenic potential found in shellfish and 8 STs found in marine animals with pathogenic potential (Table S3).
This study describes a high degree of E. coli ST diversity in a marine environment as evidenced by Shannon diversity index scores of >3 in the marine water samples from the four quadrants of the Salish Sea. Five of 62 novel STs [5869, 1065, 11343, 9001, 2164] have not previously been reported in marine and animal environments in published literature. Within the quadrants, there was a high degree of diversity in E. coli STs, including novel isolates, those with known environmental sources, environmental and/or animal sources. There was not a high number of isolates of human origin in the northern quadrant which was surprising given the outflow of partially treated sewage. There was higher diversity of STs associated with animals in the central quadrant where there was proximity to animal agriculture. Evenness across the four quadrants was consistently close to one. This finding is not surprising given the high proportion of identified STs represented by one isolate; thus, there is a similar abundance, suggesting there is no dominant strain present in the quadrants at the sampling time. There was no concentrated variation in the distribution of STs. ST10 was the most abundant isolate found (n=12), and it was isolated in all four quadrants of the marine water and in a river otter E. coli.
Our findings of high diversity of E. coli in a marine environment are consistent with published studies of freshwater environments that have reported higher E. coli diversity utilizing the Shannon diversity index. A high degree of E. coli diversity was found in freshwater wells in Nigeria, which the authors speculated was due to multiple sources of contamination, including proximity to septic tanks and erosion (Odetoyin et al., 2022). Chandran and Mazumder (2015) described a high level of E. coli diversity, with seasonal variations, within a freshwater lake on Vancouver Island, British Columbia, Canada. The authors had previously described greater E. coli diversity in animal and avian hosts than in human hosts (Chandran and Mazumder, 2013; Chandran and Mazumder, 2014). They hypothesized that the high level of diversity and seasonality variation of E. coli in the 2015 study was due to the variations in animal populations in the area as the lake was in a forested watershed. This lake is located near the Strait of Georgia, which connects to the north quadrant of the Salish Sea. The Salish Sea is home to multiple species of animals and birds that depend on the marine ecosystem (SeaDoc, 2021). This may be one reason for the overall high diversity and differences in diversity in the quadrants when comparing isolates with known sources of animal and environment and environmental-only isolates.
In the current study, isolates derived from water samples from the Strait of Juan de Fuca had the greatest ST richness. Five of the eight isolates previously associated with humans and the environment were also sampled in this region. The Strait of Juan de Fuca had the highest Shannon diversity index score, followed very closely by the southern quadrant, however, there was no statistical significance in the amount of diversity between the Strait and the quadrant with the lowest score (north). Interestingly, the Strait of Juan de Fuca also had the most diversity of novel STs as compared to the other quadrants. Though it is important to note the other quadrants had high diversity with index scores greater than two when assessing the biodiversity for the novel isolates. The northern quadrant had the fewest total number of isolates identified and the diversity did not differ significantly from the other quadrants for any of the categories investigated. This is somewhat surprising due to the presence of multiple untreated sewage outfalls in Vancouver and Victoria, British Columbia (Krogh et al., 2017). We had hypothesized due to the presence of untreated sewage, there would be a higher level of diversity and more human isolates in this quadrant. One possible explanation is that tidal currents and winds can carry the sewage westward into the Strait of Juan de Fuca, which may account for the variation found in this region (Krogh et al., 2018).
The central quadrant had the most diversity in STs found in animals and the environment, and this difference was statistically significant compared to diversity in the other quadrants. This quadrant had the second-highest diversity index score for environmental isolates. Isolates of STs with known pathogenic potential were found in all sample types analyzed, though the greatest diversity was found in samples from the southern quadrant of the Salish Sea. In those STs with known environmental sources, the highest diversity was in the southern quadrant, followed closely by the central quadrant. It is unclear why the southern quadrant had the most diversity. The diversity of STs may be influenced by the density of marine animal populations found in the Vingino et al. study (2021) and the presence of WWTPs in the southern quadrant, similar to the findings of Chandran and Mazumder (2015).
In addition to the high diversity of E. coli STs in this study, there were also interesting findings in regard to the novel STs. For example, E. coli ST5869, found in marine water, had entries in EnteroBase listing human and animal sources, including animals from China, Belgium (Brussels), and the United States (Tennessee), human sources from Cambodia and the Netherlands, and an avian source in Kenya. Scholarly articles have previously described this ST as being found in food sources in Germany and Belgium (Pauly et al., 2021; Garcia-Graells et al., 2020), animals treated in a veterinary clinic in Switzerland (Schmitt et al., 2021), and humans in Nigeria (Jesumirhewe et al., 2020). We could not find a previous report of this ST in a marine environment.
While E. coli ST1065 and ST11343 were associated only with human sources in EnteroBase, they were found in necropsy samples from dead seals in the current study. In EnteroBase, ST1065 was mentioned to have human source in an isolate from France and from the United States (Washington). In the expanded literature review, one article linked ST1065 to cattle (Isiko et al., 2015). We did not find any previous case reports of this ST in a marine animal. No scholarly articles were found regarding ST11343, even in an expanded search from multiple databases. While this ST was mentioned in EnteroBase as being isolated from human sources, mention of its occurrence in animals has apparently not been reported previously.
E. coli ST9001, isolated from a live seal in the current study, was referenced as human and environment in EnteroBase with sources from North America (not further specified) from humans and water. E. coli ST2164 isolated from a river otter in the current study was referenced in the environment only in EnteroBase with sources from Canada and Massachusetts and water sources from Florida and Washington. Neither E. coli ST9001 nor E. coli ST2164 were found in an expanded literature search in multiple databases. We did not find previous reports of these STs in animals.
There were several strengths of this study including how sampling of animals were done. For example most of the animals were from deceased marine animals which was opportunistic. In addition there was no human contact with the otters and alive seals while collecting fecal samples (Vingino et al., 2021), thus there was no harm or stress caused to marine mammals during the collection of samples. Although the lack of human isolates for comparison, the current study expanded our knowledge using a large collection of E. coli isolates from the Salish Sea including water and animal samples.
This study had several limitations. The EnteroBase is a public database not centrally curated and, as such, relies on the users to enter accurate information. In addition, STs, especially those seeming to be novel, may be underreported as the database relies on human entry and is not auto populated by lab detection software. The literature review and counting of articles associated with each ST were performed with PubMed only. Additional articles may be found on different scholarly publication databases. This study aimed not to search multiple databases for each ST, but to give a broad overview of available literature in an extensive, primarily peer-reviewed database associated with each. Of note, an expanded literature review utilizing multiple databases was performed for E. coli with ST5869, ST1065, ST11343, ST9001, and ST2164 as described above to identify whether our reporting was novel. Finally, because this study utilized a previously collected dataset, the assumption is made that the data contained in the dataset is accurate and without errors. Samples for dead seals and porpoises collected for the original dataset were opportunistic and taken postmortem.
To our knowledge, this study is the most extensive characterization of E. coli diversity in a marine environment to date. The diversity found may be due to many changes in the Salish Sea and surrounding environment but determining these factors was beyond the scope of the current project. However, even given these limitations, the study can serve as a blueprint for future studies of E. coli in marine ecosystems including both the water and the animals living in and near marine environments around the world.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: Genome Trakr Project ID 283914-BioProject-NCBI.
JG did all the literature searches, data analysis and writing. MR, PR, and SW were involved with the original paper that generated the data set, the direction of the study and in the writing of the current manuscript. All authors contributed to the article and approved the submitted version.
Funding for the original data collection came from the SeaDoc Foundation. No funding was available for the current manuscript.
We would like to acknowledge Alexandria Vingino and Stephanie Norman’s previous published work from this dataset. SeaDoc provided the funding for the original study sampling. As previously acknowledged in the aforementioned authors studies, WA DOH for processing samples. Thank you to Dr. Daniel Enquobahrie for reviewing the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2022.967435/full#supplementary-material
Altug G., Sevan G., Cardak M., Ciftci P., Kalkan S.. (2012). The occurrence of pathogenic bacteria in some ships' ballast water incoming from various marine regions to the Sea of marmara, Turkey. Mar. Environ. Res. 81, 35–42. doi: 10.1016/j.marenvres.2012.08.005
Belotserkovsky I., Sansonetti P. (2018). Shigella and enteroinvasive Escherichia coli. Curr. Topics Microbiol. Immunol. 416, 1–26. doi: 10.1007/82_2018_104
Centers for Disease Control and Prevention (2014) E. coli. questions and answers (CDC) (Accessed March 26, 2022).
Centers for Disease Control and Prevention (2022). Antibiotic/Antimicrobial resistance (AR/AMR). where resistance spreads: Water, soil, environment. (Accessed April 4, 2022)
Chandran A., Mazumder A. (2013). Prevalence of diarrhea-associated virulence genes and genetic diversity in Escherichia coli isolates from fecal material of various animal hosts. Appl. Environ. Microbiol. 79 (23), 7371–7380. doi: 10.1128/AEM.03949-13
Chandran A., Mazumder A. (2014). Occurrence of diarrheagenic virulence genes and genetic diversity in Escherichia coli isolates from fecal material of various avian hosts in British Columbia, Canada. Appl. Environ. Microbiol. 80 (6), 1933–1940. doi: 10.1128/AEM.03738-14
Chandran A., Mazumder A. (2015). Pathogenic potential, genetic diversity, and population structure of Escherichia coli strains isolated from a forest-dominated watershed (Comox lake) in British Columbia, Canada. Appl. Environ. Microbiol. 81 (5), 1788–1798. doi: 10.1128/AEM.02653-13
Data Analytics (2019). Statistics for ecologists. comparing ecological diversity. Stat Ecologists Exercises. Dataanalytics.org.uk (Accessed May 27, 2022)
Environmental Protection Agency (2021) LEARN: What affects beach health (US EPA) (Accessed April 2, 2022).
Garcia-Graells C., Berbers B., Verhaegen B., Vanneste K., Marchal K., Roosens. N., et al. (2020). First detection of a plasmid located carbapenem resistant blaVIM-1 gene in E. coli isolated from meat products at retail in Belgium in 2015. Int. J. Food Microbiol. 324, 108624. doi: 10.1016/j.ijfoodmicro.2020.108624
Ibarz Pavón A. B., Maiden M. C. (2009). Multilocus sequence typing. Methods Mol. Biol. 551, 129–140. doi: 10.1007/978-1-60327-999-4_11
Isiko J., Khaitsa M., Bergholz T. M. (2015). Novel sequence types of non−O157 shiga toxin-producing Escherichia coli isolated from cattle. Lett. Appl. Microbiol. 60 (6), 552–557. doi: 10.1111/lam.12404
Jesumirhewe C., Springer B., Allerberger F., Ruppitsch W.. (2020). Whole genome sequencing of extended-spectrum β-lactamase genes in enterobacteriaceae isolates from Nigeria. PloS One 15 (4), e0231146. doi: 10.1371/journal.pone.0231146
Johannessen S. C., Macdonald R. W., Burd B., van Roodselaar A., Bertold S.. (2015). Local environmental conditions determine the footprint of municipal effluent in coastal waters: a case study in the strait of Georgia, British Columbia. Sci. Total Environ. 508, 228–239. doi: 10.1016/j.scitotenv.2014.11.096
Jolley K. A., Bray J. E., Maiden M. C. J. (2018). Open-access bacterial population genomics: BIGSdb software, the PubMLST.org website and their applications. Wellcome Open Res. 3, 124. doi: 10.12688/wellcomeopenres.14826.1
Kaur P., Chakraborti A., Asea A. (2010). Enteroaggregative Escherichia coli: An emerging enteric food borne pathogen. Interdiscip. Perspect. Infect. Dis. 2010, 254159. doi: 10.1155/2010/254159
Krogh J., Ianson D., Hamme R. C., Lowe C. J.. (2018). Risks of hypoxia and acidification in the high energy coastal environment near Victoria, canada's untreated municipal sewage outfalls. Mar. pollut. Bull. 133, 517–531. doi: 10.1016/j.marpolbul.2018.05.018
Krogh J., Lyons S., Lowe C. J. (2017). Pharmaceuticals and personal care products in municipal wastewater and the marine receiving environment near Victoria Canada. Front. Mar. Sci. 4, 2–18. doi: 10.3389/fmars.2017.00415
McEwen S., Collignon P. (2018). “Antimicrobial resistance: A one health perspective”. In Antimicrobial Resistance in Bacteria from Livestock and Companion Animals, 6 (2), 521–47. Washington, DC, USA.:ASM Press. doi: 10.1128/9781555819804.ch25
Norman S. A., Lambourn D. M., Huggins J. L., Gaydos J. K., Dubpernell S., Berta S., et al. (2021). Antibiotic resistance of bacteria in two marine mammal species, harbor seals and harbor porpoises, living in an urban marine ecosystem, the salish Sea, Washington state, USA. Oceans (Basel Switzerland) 2 (6), 86–104. doi: 10.3390/oceans2010006
Ochoa T. J., Contreras C. A. (2011). Enteropathogenic Escherichia coli infection in children. Curr. Opin. Infect. Dis. 24 (5), 478–483. doi: 10.1097/QCO.0b013e32834a8b8b
Odetoyin B., Ogundipe O., Onanuga A. (2022). Prevalence, diversity of diarrhoeagenic Escherichia coli and associated risk factors in well water in ile-ife, southwestern Nigeria. One Health Outlook 4 (1). doi: 10.1186/s42522-021-00057-4
Pauly N. N., Hammerl J. A., Grobbel M., Käsbohrer A., Tenhagen B. A., Malorny B., et al. (2021). Identification of a blaVIM-1-Carrying IncA/C2 multiresistance plasmid in an Escherichia coli isolate recovered from the German food chain. Microorg. (Basel) 9 (1), 29. doi: 10.3390/microorganisms9010029
QGIS.org (2022). QGIS geographic information system (QGIS Association). Available at: http://www.qgis.org.
Rice E. W., Baird R. B., Eaton A. D., Clesceri L. S.. (2012). Standard methods for examination of water and wastewater. 22th ed (Washington DC, USA: American Public Health Association).
Ruiz G. M., Rawlings T. K., Dobbs F. C, Drake L. A., Mullady T., Huq A., et al. (2000). Global spread of microorganisms by ships. Nat. (London) 408 (6808), 49–50. doi: 10.1016/S1286-4579(03)00049-2
Russo T., Johnson J. (2003). Medical and economic impact of extraintestinal infections due to Escherichia coli : Focus on an increasingly important endemic problem. Microbes Infect. 5 (5), 449–456. doi: 10.1016/S1286-4579(03)00049-2
Schmitt K., Kuster S., Zurfluh K., Jud R. S., Sykes J. E., Stephan R., et al. (2021). Transmission chains of extended-spectrum beta-Lactamase-Producing enterobacteriaceae at the companion animal veterinary clinic-household interface. Antibiotics (Basel) 10 (2), 171. doi: 10.3390/antibiotics10020171
Spurbeck R. R., Mobley H. L. T. (2013). “Uropathogenic escherichia coli,” in Escherichia coli: Pathotypes and principles of pathogenesis, 2nd ed, 275–304. Elsevier Inc. doi: 10.1016/B978-0-12-397048-0.00009-7
The SeaDoc Society (2022) About the salish Sea (SeaDoc Society) (Accessed 5/17/2022). About the Salish Sea.
Vingino A., Roberts M. C., Wainstein M., West J., Norman S. A., Lambourn D., et al. (2021). Surveillance for antibiotic-resistant E. coli in the salish Sea ecosystem. Antibiotics (Basel) 10 (10), 1201. doi: 10.3390/antibiotics10101201
Washington State Department of Ecology (2019). Puget sound nutrient source reduction project. Model. Updates Bounding Scenarios 1. Puget Sound Nutrient Source Reduction Project.
Keywords: E. coli, microbial diversity, marine mammals, EnteroBase, multilocus sequence types
Citation: Grunwald JL, Rabinowitz P, Weissman SJ and Roberts MC (2022) Diversity of Escherichia coli found in the Salish Sea. Front. Mar. Sci. 9:967435. doi: 10.3389/fmars.2022.967435
Received: 12 June 2022; Accepted: 05 August 2022;
Published: 05 September 2022.
Edited by:
Ayanna Carla N. Phillips Savage, Virginia Tech, United StatesReviewed by:
Chui Wei Bong, University of Malaya, MalaysiaCopyright © 2022 Grunwald, Rabinowitz, Weissman and Roberts. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Marilyn C. Roberts, bWFyaWx5bnJAdXcuZWR1
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.