- Red Sea Research Center (RSRC), King Abdullah University of Science and Technology (KAUST), Thuwal, Saudi Arabia
Solar ultraviolet B radiation (UVB) has recently been described as a relevant stressor in fish confined to aquaculture cages. In gilthead seabream (Sparus aurata), UVB exposure resulted in decreased growth, epidermal sloughing, increased oxidative stress in the skin, and induced changes in behavior, physiology, and immune system. Several molecular responses should accompany such detrimental effects; however, little is known in fish about the overall UVB-mediated changes at the transcriptional level. Thus, this study aimed to investigate the effects of UVB exposure on the global gene expression profiles of S. aurata skin through transcriptome analysis. S. aurata juveniles were exposed for 43 days to two experimental groups: 1) UVB (daily dose, 6 kJ m-2; representing levels between 5 and 7 m depth); 2) Unirradiated treatment, used as a control. The comparison of skin transcriptomes between the control and UVB treatments revealed 845 differentially expressed genes (580 up-regulated and 265 down-regulated). The reliability of the transcriptome analysis was confirmed by qRT-PCR for selected genes. Functional annotation and PPI analyses revealed that genes related to the immune system and inflammatory response, cell cycle regulation, proteasome, proteolysis, and oxidative stress might be involved in the response to UVB exposure. In contrast, UVB exposure inhibited the expression of several genes related to growth factor activity, cell growth and differentiation, and pigmentation. p53 signaling pathway was enriched in fish exposed to UVB. Moreover, pathways involved in the immune system and inflammatory response (cytokine-cytokine receptor interaction, RIG-I-like receptor signaling pathway, and Toll-like receptor signaling pathway) were also enriched in the skin of UVB-exposed fish. UVB-induced skin damage and a high level of infiltration of immune-related cells were confirmed through histopathological examination. Together, our results provide noteworthy insights into the molecular changes in fish after long-term exposure to UVB. These findings will help in the future to identify biomarkers of fish reared in offshore aquaculture systems in oligotrophic and highly transparent waters.
1 Introduction
Solar ultraviolet radiation (UVR) reaching the earth’s surface is a well-recognized natural abiotic stressor for aquatic organisms (Williamson et al., 2019). Although the ultraviolet B (UVB, 280-315 nm) represents only a low percentage of the total UVR reaching the water surface, more harmful effects are generally attributed to this band (Häder et al., 2015; Neale et al., 2021). In fish, commonly reported impacts of UVB exposure include (1) increased mortality (Fukunishi et al., 2006; Sucré et al., 2012), (2) development of body abnormalities (Nuñez et al., 2012; Vásquez et al., 2016), (3) growth reduction (Vitt et al., 2017; Alves et al., 2021), (4) behavioral changes (Pulgar et al., 2015; Valiñas and Walter Helbling, 2016), (5) immune system suppression (Markkula et al., 2005; Jokinen et al., 2011), (6) metabolic and physiological changes (Alemanni et al., 2003; Sharma et al., 2010), and (7) DNA damage and increased oxidative stress (Carrasco-Malio et al., 2014; Braun et al., 2016; Alves and Agustí, 2021).
In fish, the direct contact between the skin and UVR exposure makes it one of the most vulnerable tissues to UVB-induced damage. Previous studies reported that fish skin undergoes noteworthy structural, morphological, and functional changes due to UVR exposure, including hyperpigmentation, epidermal sloughing, modifications on the mucous layer, hypertrophy and hyperplasia, inflammation, and high susceptibility to fungal infections (Bullock and Coutts, 1985; Blazer et al., 1997; Häkkinen et al., 2004; Sayed et al., 2007; Kazerouni and Khodabandeh, 2011; Manek et al., 2012; Sayed et al., 2013). Fish skin plays a protective role against UVR exposure (Fabacher and Little, 1995; Blazer et al., 1997; Fabacher and Little, 1998). Research shows that epidermal mucous can be abundant in UV-absorbing compounds, such as the mycosporine-like amino acids (MAAs), (Braun et al., 2016; Zamzow and Losey, 2002). Moreover, the presence of specialized epidermal cells can also contribute to the photoprotective role of fish skin. Club cells can promote the healing process in the damaged skin after UVB exposure (Blazer et al., 1997; Chivers et al., 2007). Thus, the aforementioned changes in the skin can activate significant changes at the transcriptional level. Few studies addressed the changes in the fish skin global gene expression after UVB exposure. For example, the disassembled dermal skin structure observed in zebrafish exposed to UVB was associated with changes in the expression of extracellular matrix-associated genes. While collagen and fibronectin genes were down-regulated, several matrix metallopeptidase proteolytic genes involved in procollagen synthesis inhibition were up-regulated (Chen et al., 2020). Transcriptome analysis on the UVB-exposed Xiphophorus Sp-Couch hybrid, which displays an enhanced macromelanophore pigmentation, revealed enhanced mRNA expression levels of melanin synthesis and transport-related genes in the skin (Lu et al., 2015). Moreover, a recent RNA-seq study showed that UVB exposure (to 8-32 kJ m-2) in southern platyfish (Xiphophorus maculatus) activates, in the skin, the expression of genes related to the immune system (e.g., inflammatory response, activation of complement, mucous secretion), cell restructuring and cytoskeleton remodeling (Yang et al., 2014).
Gilthead seabream (Sparus aurata), a widely cultured Mediterranean fish species, has gained importance in the last years for the Saudi Arabia offshore aquaculture scenario in the Red Sea (FAO, 2013; Khan et al., 2018), which is characteristic of oligotrophic and transparent waters, and high underwater UVB levels (Overmans and Agustí, 2019; Overmans and Agustí, 2020). We recently reported that exposure to natural underwater UVB daily doses of 6 kJ m−2 d−1 for 43 days resulted in lesions in the skin (e.g., typical sunburn and epidermal sloughing) but also remarkable behavioral, metabolic, and physiological changes, as well as increased oxidative stress (Alves et al., 2020; Alves & Agustí, 2021). With the aim to understand whether the responses and tissue damage previously reported in S. aurata towards exposure to UVB are accompanied by overall transcriptional changes, the present study addressed the global gene expression profiles after long-term exposure to natural underwater UVB levels (43 days, daily dose, 6 kJ m-2). Hence, using the next-generation sequencing (NGS) technology, differential gene expression analysis was performed on the skin, followed by functional annotation, including gene ontology and pathways enrichment analyses. Finally, UVB-induced damage in the skin was confirmed through histopathological examination.
2 Material and methods
2.1 Fish, UVB exposure, and sampling
Experimental design, including fish husbandry and UVB exposure daily dose selection, is detailly described in Alves et al. (2020). Briefly, S. aurata juveniles were obtained from Tharawat Seas Company (Ar Rayis, Saudi Arabia) in April 2018. Fish were kept in quarantine for 30 days at CMOR laboratory facilities. After the quarantine period, fish were transferred to the experimental tanks (300 L rectangular glass tanks; 40 juveniles per tank; body weight: 32 ± 4 g) and maintained in a flow-through system with a constant water flow rate (0.3 m3 h−1). After 2 weeks of acclimation, juveniles were selected for two experimental groups: 1) Unirradiated treatment, used as a control; and 2) UVB treatment (defined as a moderate daily dose in Alves et al., 2020, 0.21 W m-2, 6.1 kJ m-2 d-1) representative of the underwater levels typical of oligotrophic waters (Garcia-Corral et al., 2015). The daily UVB dose used in this experiment corresponded to the UVB levels between 5 and 7 m depth in the central Red Sea (depending on the seasonal variability of underwater UVB radiation). Overmans and Agustí (2020) observed in 2018 an average daily UVB doses of 9 and 5 kJ m-2 at 4 and 6 m depth, respectively. For example, at 4 m, an average daily UVB dose of 6 kJ m-2 was observed between January and March. Conversely, the same daily UVB doses were registered at 6 m depth between May and August (Overmans and Agustí, 2020). Furthermore, observed similar levels at several Red Sea farms (Alves et al., 2020). Both experimental groups were tested in duplicate. Fish were daily exposed to UVB (lighting system with TL20W/12RS UVB Broadband lamps wrapped with neutral mosquito layers, see Alves et al., 2020 for full details) for 8 h to obtain the required daily doses. UVB irradiance was measured daily in the tanks using an underwater PMA2100 data-logging radiometer (Solar LightTM, USA) fitted to a PMA2106 Digital Non-Weighted UVB Sensor (Solar LightTM, 280-320 nm, peak sensitivity at 312 nm). Before starting the experiment, undetectable UVC and negligible UVA doses were confirmed inside the tanks (see Alves et al., 2021). Environmental parameters were daily monitored (temperature 27.0 ± 0.8°C, oxygen saturation 92.9 ± 6.1%, pH 8.1 ± 0.1, salinity 40.5 ± 0.1), and fish were maintained in a photoperiod of 12 h: 12 h light/dark using daylight tubes FH039W Reef White 10000 (Aqua Medic, Germany). Fish were manually fed 3-5% of body weight two times a day with a commercial diet, with the following proximal composition: 94.4% dry matter, 52.5% crude protein, 13.3% crude fat, and 21.0 MJ kg−1 gross energy. The experiment duration was 43 days (UVB treatment, absolute dose, 261 kJ m−2).
At the end of the experiment, sixteen fish from each treatment (n = 8 per tank) were randomly sampled and euthanized in an overdose of tricaine methanesulfonate solution (MS222, 150-200 mg L-1; Sigma-Aldrich, United States). After cervical sectioning, the skin was collected from each fish from the same body region (after the end of the operculum and just below the dorsal fin). For the transcriptome analysis (n = 8 per treatment), seabream juveniles were kept on a cold plate (0-4°C), and the skin was immediately sampled, incubated in RNA later (Life Technologies, Carlsbad, USA) at 4°C for 24 h, and then stored at -20°C until further analysis. For the histopathology analysis (n = 8 per treatment), the skin was fixed in 10% neutral buffered formalin (formaldehyde-phosphate buffer, pH 7.2) at 4°C overnight with gentle agitation. After fixation, samples were washed in phosphate buffer and preserved in 75% ethanol until further use.
2.2 Total RNA extraction, Illumina library preparation, and sequencing
Total RNA was isolated from the skin (30-40 mg) of control and UVB-exposed fish. RNA extraction and purification were optimized by combining both TRIzol® (Sigma, USA) and RNeasy® PowerLyzer® Tissue & Cells Kit (Qiagen, Germany). Briefly, tissues preserved in RNA later were weighted, transferred directly to 2.8 mm ceramic beads tubes (Qiagen PowerLyzer kit) containing 1 ml of chilled TRIzol, and homogenized in the TissueLyser II (Qiagen, Germany) for 2 min at 25 Hz (2-3 cycles, with a pause on ice for 20 seconds between cycles). Skin homogenates were transferred to new microcentrifuge tubes, and solvent extraction was performed following the TRIzol manufacturer’s protocol. The aqueous phase was transferred to a new microcentrifuge, and total RNA was purified using the RNeasy PowerLyzer Tissue & Cells Kit following the manufacturer’s instructions. 10 μg of total RNA was treated with a Turbo DNA-free kit (Invitrogen, USA) and purified with the RNeasy® PowerClean ® Pro Cleanup Kit (Qiagen, Germany), preventing contamination with genomic DNA. RNA concentration and purity were determined using the Qubit™ RNA BR Assay Kit (Thermo Scientific, USA) and Nanodrop 2000c (Thermo Scientific, USA), respectively. OD 260/280 and OD 260/230 ranged between 2.08-2.12 and 2.15-2.22, respectively. The integrity and concentration were verified with an Agilent 2100 Bioanalyzer (Agilent Technologies, USA). For all the samples, RIN (RNA integrity number) values were equal to or above 8.
Four biological replicates from each treatment (Control, UVB) were selected for library preparation and RNA sequencing was based on the highest RNA quality and integrity scores. A total of eight libraries were prepared from 1 μg of total RNA using the Illumina TruSeq™ Stranded mRNA LT Sample Prep Kit (Illumina, USA) according to the manufacturer’s instructions. The quality of each library was confirmed using the Agilent 2100 Bioanalyzer (Agilent Technologies, USA). All the eight RNA-seq libraries were pooled (average size, 337-407 bp inserts). After generating the clusters, libraries were sequenced on the Illumina HiSeq 4000 platform (Illumina, USA), to create paired-end reads (2 x 150 bp). Image analysis and base calling were done using the Illumina pipeline. The RNA sequencing was performed at the KAUST Bioscience Core Labs.
2.3 Bioinformatics analyses
2.3.1 Quality assessment, reads mapping, gene quantification, and identification of differentially expressed genes (DEGs)
Raw reads quality assessment was performed with FastQC v.011.7 software (Andrews, 2010). FASTQ files were processed with the BBDuk tool from the BBMap suite (Bushnell B, http://sourceforge.net/projects/bbmap/) to remove short and low-quality reads and Illumina adaptors and phiX sequences. Any residual rRNA sequences were removed by mapping the reads to the Sparus aurata ribosomal RNA using BBMap and retaining the unmapped set of reads. The remaining data analysis was performed using OmicsBox 1.3.11 (Biobam Bioinformatics, 2019). Clean and trimmed reads from the 8 libraries were then mapped to the Sparus aurata reference genome assembly (NCBI fSpaAur1.1, SC - July 2019, RefSeq GCF_900880675.1, https://www.ncbi.nlm.nih.gov/genome/11609?genome_assembly_id=648533; Ensembl annotation file of genomic features, fSpaAur1.1 GCA_900880675.1, http://ftp.ensembl.org/pub/release-103/gff3/sparus_aurata/), using the fast RNA-Seq read mapper algorithm STAR-2pass v.2.7.3 (Spliced Transcript Alignment to a Reference), (Dobin et al., 2012). Read mapping was performed using the following settings: overhang - 150 bp, sort by coordinate, min. intron length - 20 bp, max. intron length - 1,000,000 bp, max. # of mismatches - 999, and max. # of multiple alignments - 200. Only reads mapped unambiguously to a single genomic feature were considered. HTSeq v.0.9.0 tool (Anders et al., 2014) was used to count the number of reads mapped for each gene. For each sorted SAM file, gene quantification was performed using default settings (overlap mode - union, minimum mapping quality - 10; non-strand specific). Using the g:Profiler web server (https://biit.cs.ut.ee/gprofiler/gost, Raudvere et al., 2019), the top 500 highly expressed genes in both conditions were used to identify the abundant gene ontology (GO) terms associated with biological processes in the skin transcriptomes.
Differentially expressed genes (DEGs) were identified using the software package edgeR v.3.24.3 from the Bioconductor project (Robinson et al., 2010). Comparisons were performed for skin exposed to UVB (43 days of exposure, n = 4 for each tissue) against their corresponding unexposed control treatment (n = 4 for each tissue). A CPM (count-per-million) filter of 1 was established to exclude genes with low counts across libraries. Data normalization was performed with the TMM (weighted trimmed mean of M-values) method and a GLM (Likelihood Ratio Test), based on fitting negative binomial Generalized Linear Models with the Cox-Reid dispersion estimates, was applied to detect DEGs. Genes showing a false discovery rate (FDR) < 0.05 and the absolute value of the log2 fold change ≥ 1 (up-regulated) or ≤ -1 (down-regulated) were defined as DEGs.
2.3.2 Functional annotation: gene ontology, pathways, and enrichment analysis
Functional annotation and enrichment analysis were performed using annotated DEGs (up and down-regulated) lists. Gene ontology (GO) enrichment analysis (BP, biological processes; MF, molecular function; CC, cellular component) was carried out by the FatiGO package (Al-Shahrour et al., 2004) using OmicsBox 1.3.11 (Biobam Bioinformatics, 2019). After retrieving the GO terms for all the DEGs, over and under-represented GO terms were identified using Fisher’s exact test and applying a False Discovery Rate (FDR) adjusted P-value of 0.05. Enriched GO terms for each category were also uploaded to the REViGO Web server (Supek et al., 2011) to summarize them by removing redundant GO terms (semantic similarity of 0.5), and then visualized using the CirGO (Circular Gene Ontology) Python software package (Kuznetsova et al., 2019).
KEGG (Kyoto Encyclopedia of Genes and Genomes) and Reactome pathways analysis and enrichment were performed for each up and down-regulated DEGs list using DAVID software (https://david.ncifcrf.gov/), (Huang et al., 2009; Sherman et al., 2022) and g:Profiler a web server (https://biit.cs.ut.ee/gprofiler/gost), (Raudvere et al., 2019). To obtain gene IDs compatible with both DAVID and g:Profiler, Ensembl zebrafish (Danio rerio) gene IDs were retrieved from the corresponding gilthead seabream Ensembl ID entries using the BioMart data-mining tool (http://www.ensembl.org/biomart/martview/). DAVID analyses were performed with the following settings: gene count = 2, ease = 0.1, and P-value of 0.05. Using the g:Profiler, significantly enriched pathways were obtained after FDR correction using a cutoff of 0.05. Dot plot pathway enrichment maps were constructed using the ggplot2 R package (Wickham, 2011).
Combined enrichment analysis (BP and KEGG/Reactome pathways) was performed using the Metascape tool (https://metascape.org/, accessed in July 2022; Zhou et al., 2019) under default parameters: minimum overlap of 3 genes, P-value of 1, and enrichment factor of 1.5. Metascape was also used to construct a protein-protein interaction (PPI) network through String and BioGrid modules under the default parameters, followed by MCODE (Molecular Complex Detection) algorithm analysis to identify the connected network components. After enrichment analysis, the top 3 annotations (BP GO terms, KEGG/Reactome pathways; defined by the P-value) were used to characterize the functional description of each MCODE complex. Both combined enrichment and PPI analyses were performed after retrieving corresponding Ensembl zebrafish (Danio rerio) gene IDs.
2.4 RNA-seq data validation: Quantitative real-time PCR (qRT-PCR)
To verify accuracy and reliability of DEGs identification through RNA‐seq, six genes from those showing differential gene expression were selected and validated using RT-qPCR: beta-2-microglobulin-like (ENSSAUG00010016139, b2ml), glutathione peroxidase 1-like (ENSSAUG00010007839, gpx1), carboxypeptidase A1-like (ENSSAUG00010008967, cpa1), superoxide dismutase [Cu-Zn] (ENSSAUG00010015634, sod3), insulin-like growth factor 1 (ENSSAUG00010015109, igf1), and galactose-specific lectin nattectin-like (ENSSAUG00010004346, ntcl), (Supplementary Table S1). Genes selection considered: (1) different abundance levels, (2) different fold change, and (3) relevance based on previous data and the analysis of functional enrichment. Specific primers for the target genes were designed using Beacon design 8.21 and Primer Premier 6.25 (Premier Biosoft Int., CA, USA).
Identical RNA samples were used for Illumina sequencing and the qRT-PCR analyses. First-strand cDNA synthesis was carried out for each sample using 600 ng of DNase treated RNA primed with 100 ng of random hexamers and the SuperScriptTM III First-Strand Synthesis SuperMix kit (Thermo Fisher, CA, USA) following the manufacturer’s instructions. qPCR reactions were carried out in triplicate for a final reaction volume of 25 μl using 9 ng of cDNA, 200 nM of the specific primers, and Power SYBR® Green PCR Master Mix (Thermo Fisher, CA, USA). QuantStudio™ 3 Real-Time PCR System thermocycle was used to conduct the qPCR reactions with the following cycling conditions: 10 min at 95°C, 40 cycles of 15 sec at 95°C and 1 min at the optimal annealing temperature for the specific primers (60, 62 or 64°C, depending on the primers used, Supplementary Table S1), and followed by a final melt curve between 60 and 95°C. The melt curve confirmed a single product’s amplification and the dissociation curves in all reactions for each gene. Negative controls included the absence of cDNA, and RT control (reverse transcriptase was omitted from the reaction) were performed for each target gene. All amplicons were analyzed by electrophoresis and sequenced to confirm the size of the amplified targets and the specificity of the reactions. qPCR results were analyzed using the QuantStudio™ Design and Analysis Desktop software (Thermo Fisher, CA, USA).
For each gene, purified PCR products were used to generate standard curves relating the amplification cycle to the initial template quantity in copy number (initial concentration, 108 copies amplicon μl-1, series dilutions of 1:10), as previously described in Viera et al. (2011). qPCR efficiencies and R2 are indicated in Supplementary Table S1. qPCR results were normalized by dividing the respective copy number in the cDNA samples by the geometric mean of the reference genes. Three candidate reference genes previously described in Costa et al. (2017) and Riera-Heredia et al. (2018) were tested: 40S ribosomal protein S18 (rps18), ribosomal protein L27A (rpl27a), and actin beta (actb). The geometric mean of the rps18 and rpl27a was used to normalize the expression of b2ml, gpx1, cpa1, sod3, igf1, and ntcl between control and UVB skin samples. The relative gene expression expressed as log2 fold change in UVB skin samples was compared with the transcriptome data.
2.5 Histopathological changes
After fixation, skin samples (n = 4 fish/treatment) were dehydrated in graded ethanol series (70-100%), saturated in xylene, and impregnated and embedded in low melting point paraffin wax (Surgipath Paraplast, Leica, Germany) using an automated tissue processor (ASP300S, Leica, Germany). Before tissue processing, samples were decalcified overnight in 0.5 M ethylenediaminetetraacetic acid (EDTA, pH 8). Serial 5 μm sections were obtained using a rotary microtome (Leica RM2125) mounted on polylysine coated slides (VWR, 631-0107, Lutterworth, UK), dried overnight at 37°C, cooled to room temperature, and stored until further use. Several sections per individual were stained using hematoxylin and eosin. Stained sections were analyzed using a microscope (DM6000B, Leica, Germany) equipped with a digital camera (DFC7000 T, Leica, Germany). Digital images were processed and analyzed using Leica Application Suite X (LAS X, Leica, Germany).
3 Results
3.1 Illumina sequencing: Quality assessment and reads mapping
A total of 575,410,677 paired-end reads of 150 bp were generated with the HiSeq 4000 platform. An average of 71,926,335 raw reads was generated for each biological replicate in the skin libraries. All reads exceeded Q30 with an average quality Q score of 37, and the average GC content in all samples was 50%. After adaptor sequence trimming and ambiguous reads, and any residual rRNA removal, more than 99% of raw reads in each library were used for mapping to the seabream genome (Supplementary Table S2). The average number of mapped reads in the control and UVB skin libraries were 66,136,102 and 62,976,757, which accounted for 90.9% and 89.5% of total reads, respectively. Of all 25,222 coding genes annotated in the S. aurata genome
(http://asia.ensembl.org/Sparus_aurata/Info/Annotation), 17,947 and 17,747 were detected in all the Control and UVB skin samples, respectively (Supplementary Table S2). Supplementary Figure 1 shows the most enriched GO terms associated with biological processes for the top 500 highly expressed genes in the skin transcriptomes. Translation, biosynthetic and metabolic processes were dominant GO biological processes terms. Further, and as expected, the top 500 most expressed genes were involved in actin filament synthesis and organization and anatomical structure development, concomitant with the structural role played by the skin tissue (Supplementary Figure S1).
3.2 Differential gene expression analysis
Differentially expressed genes (DEGs) in response to UVB exposure were identified in the skin using the following criteria: FDR < 0.05 and log2 fold change ≥ 1 (up-regulated) and FDR < 0.05 log2 fold change ≤ -1 (down-regulated). A total of 845 DEGs were identified in the skin of UVB-exposed fish (compared to control), (Figures 1A, B). The complete list of DEGs is described in Supplementary Table S3. Hierarchical clustering within the heatmaps showed that control and UVB treatments represent separated clusters in the skin transcriptomes. For each treatment, transcriptional expression patterns of DEGs were similar between the four biological replicates (Figure 1C). A total of 580 genes were up-regulated in the skin of fish exposed to UVB, while 265 genes were down-regulated. The top up-regulated genes in the skin of fish exposed to UVB were associated with the immune system, inflammatory response, and leucocytes activation (ifi27l2, rtp3, pbp, clec4e, ifi44l, cfp, lag3, cd209e, cxorf21), cellular defense response (lgals3bp), protein ubiquitination (Probable E3 ubiquitin-protein ligase ARI5, rnf213a), and double-stranded DNA break repair (trex2). The top down-regulated genes are related to development, cell component organization, anatomical structural development and receptor activity (inhbb, ar, scube1, col28a2a), inflammatory processes (ccl20, scube1), oxidative stress (sod3b), RNA and protein metabolic processes (dbpb, mmel1, ciarta), and regulation of transcription (sinhcaf). A total of 57 identified DEGs corresponded to uncharacterized proteins (45 up-regulated, 12 down-regulated), (Supplementary Table S3).
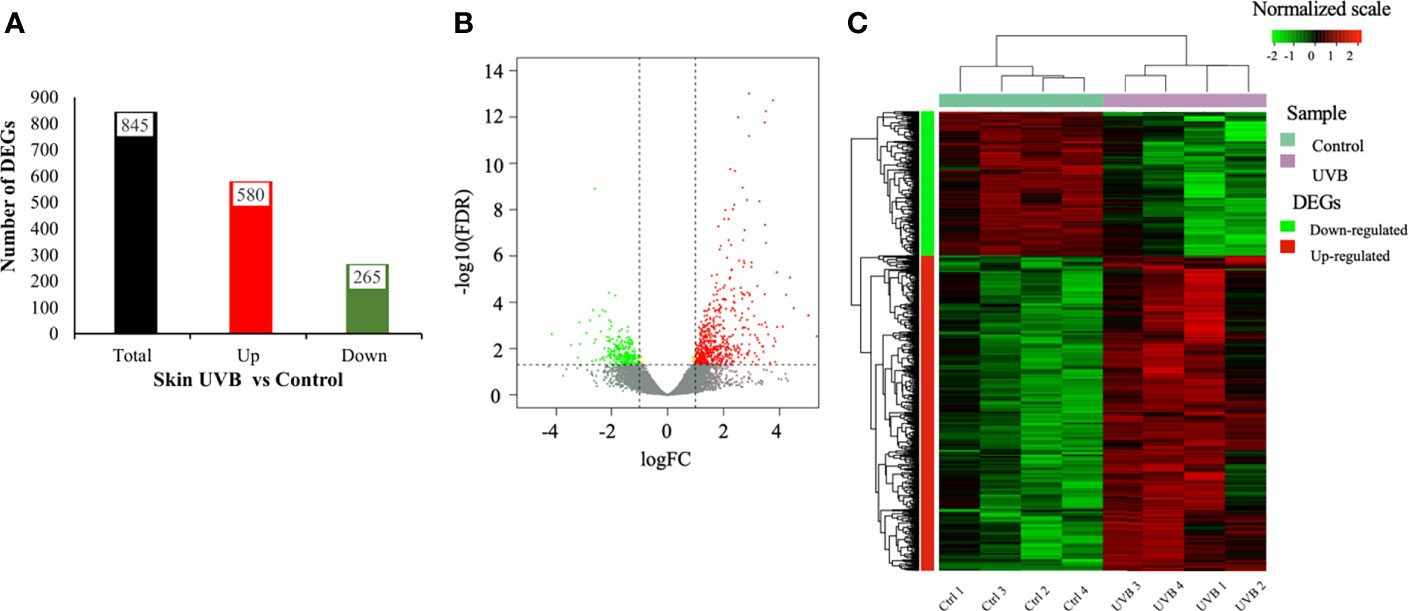
Figure 1 DEGs identified in the skin (A-C) of S. aurata exposed to UVB. (A) Number of DEGs; (B) Volcano plot of distribution patterns for differentially expressed genes in UVB exposure and control treatments. Each dot represents one gene. Red dots represent up-regulated genes, and green dots represent down-regulated genes in UVB-exposed fish. Grey dots represent genes showing no significant changes in expression between control and UVB (FDR > 0.05 or -log10(FDR) < 1.301). Yellow dots represent genes exhibiting differential expression (FDR < 0.05 or -log10(FDR) > 1.301) but with log fold change (FC) between -1 and 1; (C) Heatmap showing the hierarchical clustering analysis of genes differentially expressed regulated by UVB exposure (four biological replicates per treatment). The analysis was conducted based on log2 FC values of the DEGs between UVB and control treatments. Columns represent biological replicates, and rows represent genes. Color bar showed log2 FC values and corresponding colors (red, black, and green for up-regulated, no significant changes, and down-regulated, respectively).
3.3 Functional annotation of DEGs
To obtain insights into the function of the DEGs identified in the skin transcriptomes. Enriched GO terms and pathways associated with up-regulated genes were designated over-represented in UVB-exposed fish. In contrast, enriched GO terms associated with down-regulated genes were deemed to be over-represented in control fish.
Out of the 845 genes that showed differential gene expression in the skin, 773 (91.5%, 513 up-regulated, and 260 down-regulated) had a successful GO term assignment. Enrichment analysis revealed a total of 783 enriched GO terms in the skin. The number of enriched GO terms was higher for up-regulated genes (652 terms: 561 BP, 18 MF, 73 CC) than for down-regulated genes (131 terms: 10 BP, 116 MF, 5 CC), and the lists of all enriched GO terms are described in Supplementary Tables S4-6 and summarized in Figure 2 and Supplementary Figures S2-3). At least 50% of the over-represented BP in the skin of fish exposed to UVB was related to the immune system (innate and adaptive immune responses) and inflammatory response. Several BP associated with cytokine response were also over-represented. The second-highest proportion of the enriched BP terms in the UVB treatment was related to the cell cycle. It included “mitotic sister chromatid segregation”, “cell division”, “chromosome organization”, “DNA packaging”, and “spindle checkpoint”. Under-represented BP terms in the skin of UVB-exposed fish (enriched in control) included signal transduction, cell surface signaling, and pigmentation (Figure 2 and Supplementary Table S4). In the skin of UVB-exposed fish, over-represented MF terms were mainly associated with receptor, peptidase, protein kinase, and transferase activities. In addition, several enriched terms were related to the immune response (e.g., “immune receptor activity”, “cytokine receptor activity”, “MHC protein binding”). A high number of molecular functions were under-represented in fish exposed to UVB. They were associated with growth factor activity and transcription. MF terms related to hormone receptors and metallopeptidase activities were also enriched in the control fish. Moreover, the “DNA (6-4) photolyase activity” term was also under-represented in the skin of UVB-exposed fish (Supplementary Figure S2 and Supplementary Table S5). For the CC, more than 60% of the enriched terms in the skin of UVB-exposed fish were associated with the cytoskeleton, chromosome, and DNA packing complex. Several over-represented CC terms (> 20%) were related to the immune system (e.g., “MHC protein complex”, “immunoglobulin complex”, and “B cell receptor complex”). Some CC terms related to the proteasome complex were also enriched in this treatment. Under-represented CC terms in the skin of UVB-exposed fish included annotations related to pigmentation, cell periphery, and extracellular region (Supplementary Figure S3 and Supplementary Table S6).
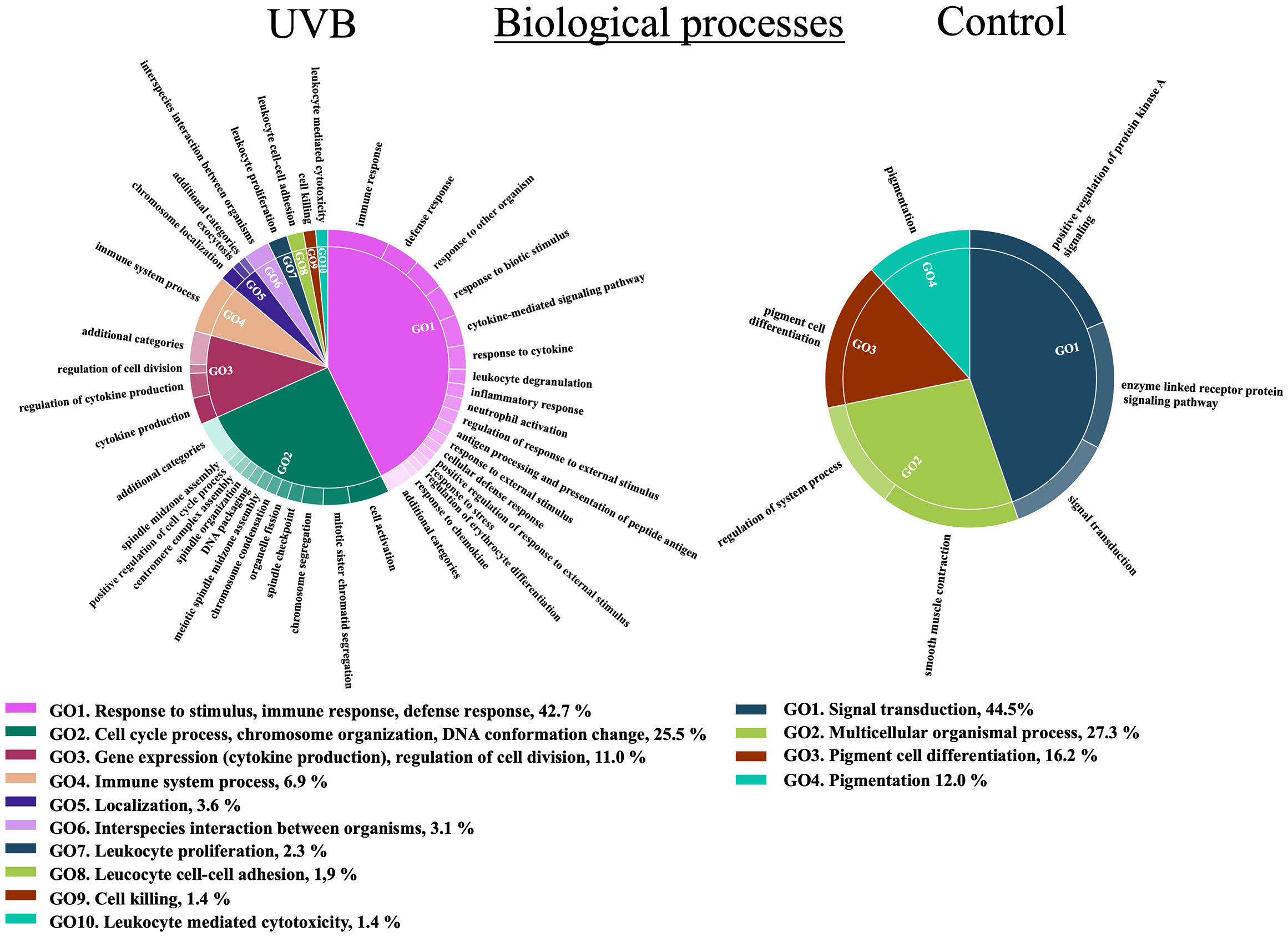
Figure 2 Functional representation of the enriched Biological Processes (BP) terms in the S. aurata skin of UVB (up-regulated genes) and control (down-regulated genes) treatments. Over-represented BP terms were summarized and grouped by hierarchical clustering after removing the most redundant terms using REViGO based on semantic similarity of 0.5, and visualized with CirGO. Parent GO terms and their respective proportions are represented below the CirGO image plots.
A total of 10 enriched KEGG pathways were enriched in the skin. Proteasome (8 genes), lysosome (10 genes), phagosome (10 genes), and cell cycle (9 genes) were the top four most significantly enriched pathways in the skin of UVB-exposed fish, according to their p-value (p < 0.01). Several pathways related to the immune system and inflammatory response were enriched in the skin of UVB-exposed fish, such as cytokine-cytokine receptor interaction (8 genes), RIG-I-like receptor signaling pathway (5 genes), and Toll-like receptor signaling pathway (7 genes). Furthermore, p53 signaling (6 genes) and pyrimidine metabolism (7 genes) pathways were also enriched in UVB treatment. The melanogenesis pathway (6 genes) was enriched in the skin of control fish (under-represented in the UVB treatment), (Figure 3). 62 enriched Reactome pathways were retrieved in the skin, 60 in the UVB treatment, while only 2 pathways were enriched in control (Supplementary Table S7). During the Reactome pathway enrichment analysis, reactions can be considered pathways “steps” and were recognized as pathways, as shown in Supplementary Figure S4. The top three enriched pathway categories in the skin of UVB-exposed fish are related to the cell cycle (35 genes), immune system (26 genes), and signal transduction (38 genes). Pathways involved in DNA replication, homeostasis, metabolism, metabolism of RNA, and transport of small molecules were also enriched in the UVB treatment (Supplementary Table S7). Two Reactome pathways related to signal transduction were enriched in the skin of control fish (Supplementary Figure S4; Supplementary Table S7).
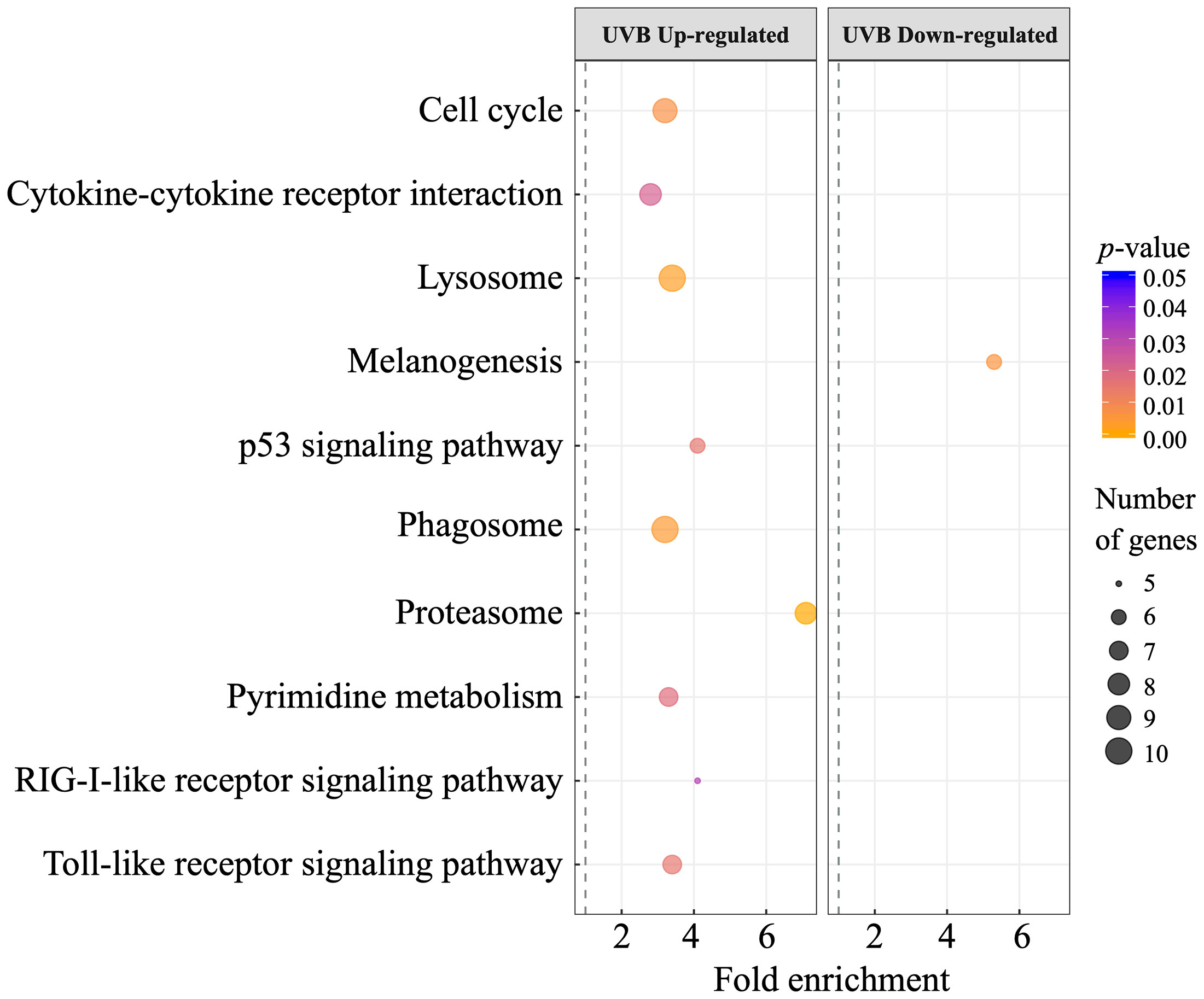
Figure 3 Dot plot showing the enriched KEGG pathways (p < 0.05) of DEGs (left, up-regulated; right, down-regulated) in S. aurata skin after UVB exposure. The vertical axis represents the enriched pathways, and the horizontal axis represents the rich factor of the enriched pathways. The size and color of dots represent the gene number and the range of p-values, respectively. Fold enrichment is the ratio of differentially expressed gene number enriched in the pathway to the total gene number in a particular pathway.
Combined enrichment analysis using Metascape showed that the top 20 enriched GO terms and pathways in up-regulated genes were related to the cell cycle and immune system (Supplementary Figure S5). The top 3 enrichment results in the down-regulated genes were circadian rhythm, regulation of blood pressure, and regulation of organismal processes. Melanogenesis, G alpha (z) signaling, and PI5P, PP2A and IER3 Regulate PI3K/AKT signaling were the enriched pathways in the list of down-regulated genes (Supplementary Figure S6). PPI networks for up- and down-regulated genes were obtained using Metascape (Figure 4). In the up-regulated genes PPI, 95 nodes and 280 edges were included. The top 3 functional enriched terms for each MCODE are identified in Supplementary Figure S7. From the 11 clusters identified in the PPI network for the up-regulated genes, 9 were functionally annotated. Genes from MCODE1, MCODE3, and MCODE10 were mainly associated with the cell cycle. The genes of MCODE2 and MCODE7 were involved in the immune system. Biological processes related to cell motility and Signaling by Interleukins pathway were well represented in the genes from MCODE2. The genes in MCODE7 were mainly correlated with the Cytokine-cytokine receptor interaction pathway. Proteasome and lysosome pathways were represented in the MCODE4 and MCODE8, respectively. Clustered genes in MCODE5 and MCODE9 were related to metabolic pathways (Figure 4A). Only two MCODES were functionally enriched in the list of down-regulated genes and are involved in regulating phosphorylation, signaling molecules, and interaction processes (Figure 4B).
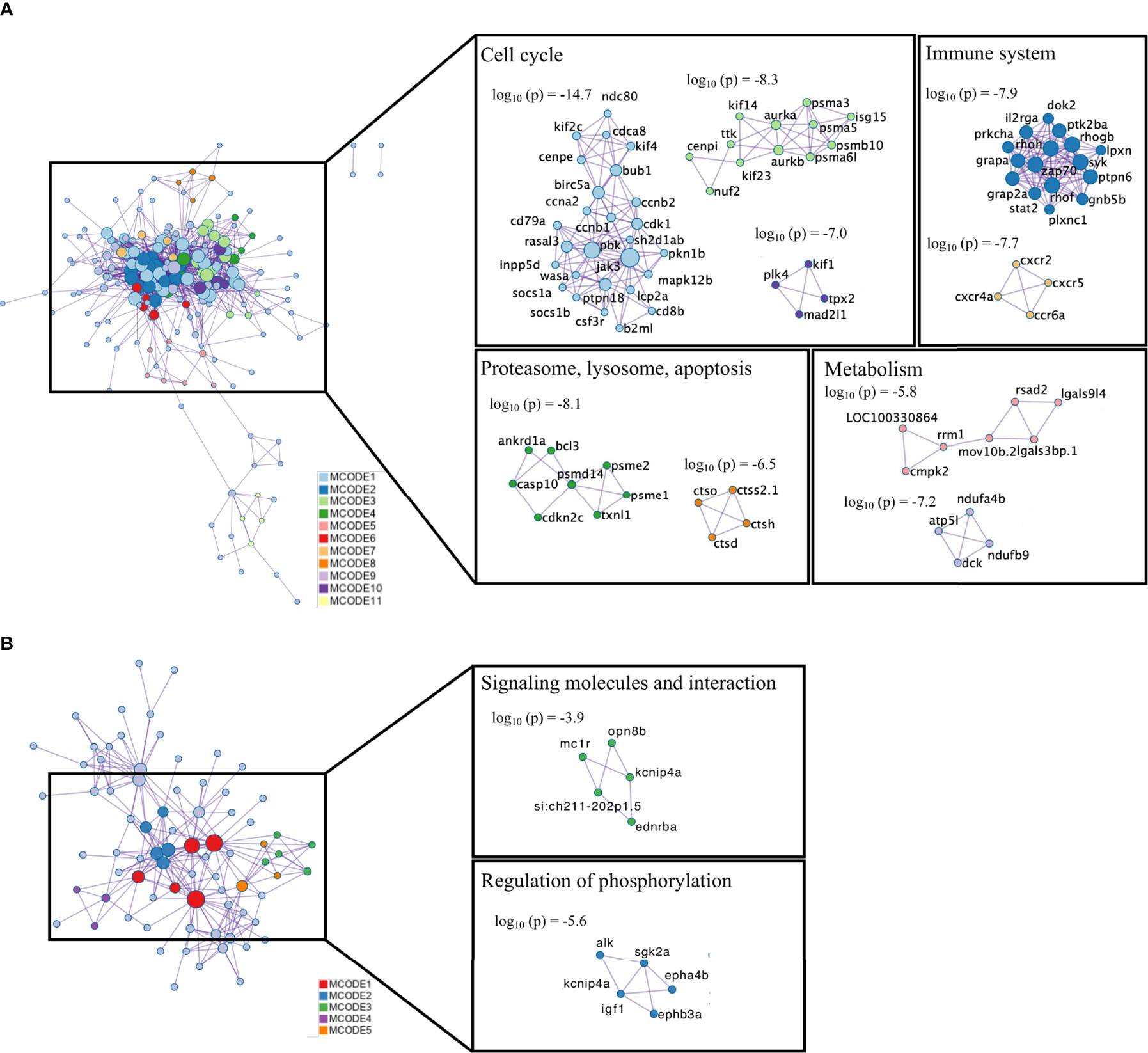
Figure 4 Protein-protein interaction (PPI) network and MCODE complexes identified in the list of DEGs (A) up-regulated; (B) down-regulated). PPI analysis was performed with Metascape, and MCODE complexes are colored by their identities. Top-three functional enriched terms for each MCODE complex are shown in Supplementary Figure S9.
3.4 Relevant differentially expressed genes
To help interpret the results, relevant DEGs were summarized, which included genes previously identified as responsive to UVR exposure (Table 1 and Supplementary Tables S8, S9). Table 1 resumes only the candidate DEGs showing a log2 FC ≥ 2 (fold change ≥ 4, up-regulated in UVB) or log2 FC ≤ -2 (fold change ≤ 4, down-regulation in UVB). Several genes related to cell cycle control and arrest were up-regulated. Genes annotated with the GO BP term “DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest” were up-regulated in the skin of UVB-treated fish. Several genes involved in the resolution of sister chromatid cohesion and chromosome maintenance (e.g., ndc80, nuf2, spc24, knl1, cenpf, cenpi, cenpl, cenpt) were also up-regulated in the skin of UVB-treated fish (Table 1 and Supplementary Table S8). Genes involved in the immune system response, including innate and adaptive immune systems components, were well represented in the DEGs list. Overall, the majority of the innate immune components were up-regulated in the skin of UVB-exposed fish, such as interleukins, associated receptors, chemokines and, toll-like receptors, interferon regulatory factors (IRFs) and induced proteins. Additionally, several adaptive immune system components were also up-regulated in the skin of UVB-exposed fish. A general up-regulation was observed in the genes identified in the enriched pathway “Phagosome” such as the cathepsin S-like (ctss), coronin-1A-like (coro1a), integrin beta-2-like (itgb2), and neutrophil cytosolic factors (ncf1, ncf2, ncf4). Several genes involved in the inflammatory response were modulated by UVB exposure (Table 1 and Supplementary Table S8).
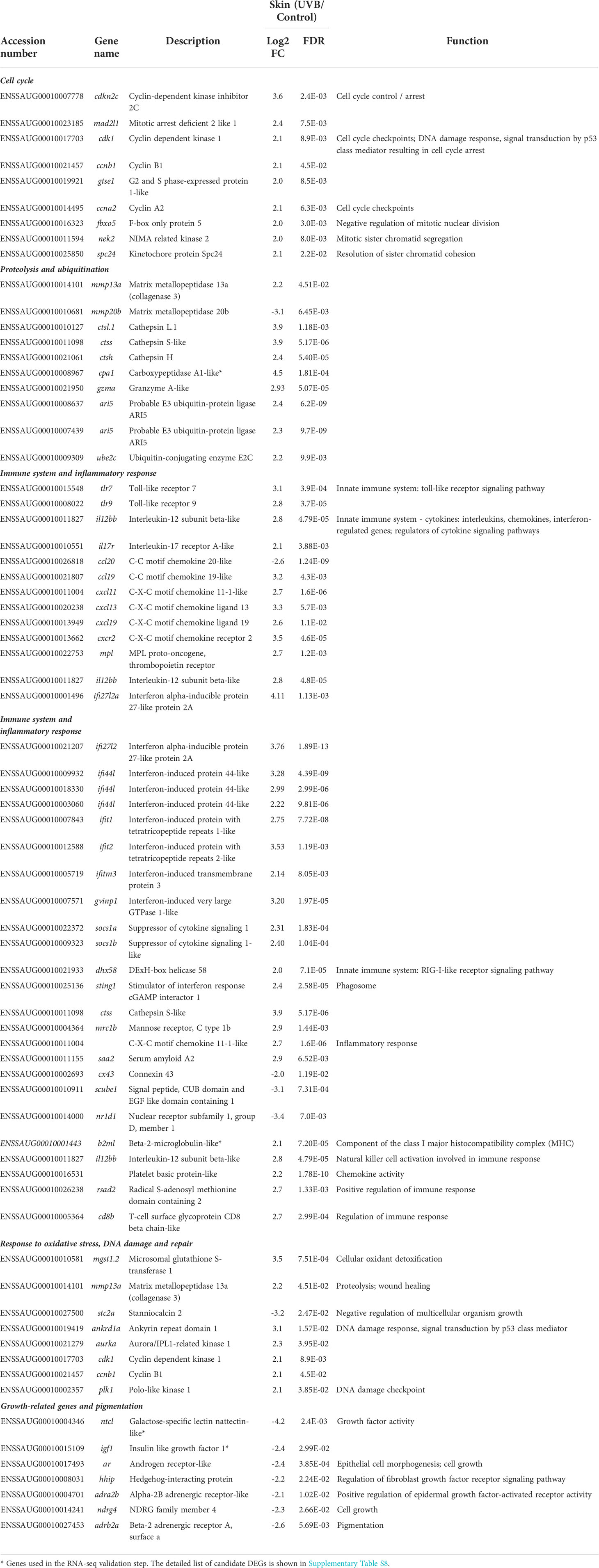
Table 1 Expression of selected candidate DEGs (UVB/Control; log2 Fold change, FC) related to cell cycle, proteolysis, immune response, oxidative stress, DNA damage and repair, growth, pigmentation, mucous production, angiogenesis, and cell adhesion/junction in S. aurata skin following UVB exposure (Selected up-regulated genes - log2 FC ≥ 2, FDR < 0.05; selected down-regulated genes - log2 FC ≥ 2, FDR < 0.05).
Seven genes belonging to the proteasome complex (psme1, psme2, psma1, psma3, psma5, psmb10, and psmd14) were up-regulated in response to UVB exposure. These genes were mapped in several enriched pathways related to the cell cycle, immune system, and signal transduction (Supplementary Figure S5). Several genes involved in protein ubiquitination were overexpressed in the skin after UVB exposure. Moreover, the expression of several proteolytic enzymes, including cathepsins (7 genes), granzymes (2 genes), and matrix metalloproteinases (4 genes), were also modulated by UVB exposure in the S. aurata skin. For example, Cathepsin L.1 (ctsl.1, fold change – 15x) and cathepsin S-like (ctss, fold change -15x) were strongly up-regulated in the skin of UVB-exposed fish (Table 1 and Supplementary Table S8). The expression of several genes involved in the protection of cells from oxidative damage, such as mgst1.2, gpx1, txnl1, hmox1a, and gss was induced in S. aurata skin after UVB exposure. In contrast, a slight down-regulation was observed in the superoxide dismutase [Cu-Zn] (sod3b), as well as in the forkhead box O6 (foxo6a), an essential transcription factor that regulates oxidative stress response (Table 1 and Supplementary Table S8). No significant changes were observed in the other important antioxidant enzymes (e.g., catalase, superoxide dismutase 1, superoxide dismutase 2, glutathione S-transferase 3-like, glutathione S-transferase Mu 3-like), neither in most genes involved in the oxidative stress regulatory mechanisms (Supplementary Table S10). The expression of several genes involved in the cellular response to DNA damage stimulus (ankrd1a, aurka, bl3, plk1, pclaf, top2a, rbbp6, irf7) was induced after UVB exposure. No significant changes between control and UVB treatments were observed in the photoreactive repair enzyme CPD photolyase (cpdp) expression. However, reduced mRNA levels of cryptochrome circadian regulator 5 (cry5) gene, which enables DNA (6-4) photolyase activity, were observed in UVB-exposed fish (Table 1 and Supplementary Table S8).
Growth factor activity-related DEGs identified in the S. aurata skin following UVB exposure were generally down-regulated. At least, six genes were annotated with the GO term “Growth factor activity” including tgfb3, fgf7, fgf16, and csf1b. Galactose-specific lectin nattectin-like (ntcl) and insulin-like growth factor 1 (igf1) were also down-regulated after UVB exposure. Several regulators of cell and multicellular organism growth (ar, adra2b, igfbp5b, sgk2a, wfdc1) were also down-regulated in UVB-exposed fish. In addition, transcripts abundance of genes related to pigmentation, mucous production, angiogenesis and cell adhesion decreased following UVB exposure (Table 1 and Supplementary Table S8).
3.5 DEGs validation using qRT-PCR
The reliability of the RNA-seq results was confirmed by qRT-PCR and is shown in Supplementary Figure S8. Six DEGs were selected for the qRT-PCR validation including b2ml, gpx1, cpa1, sod3, igf1, and ntcl. The expression of b2ml, gpx1, and cpa1 increased in the skin of UVB-exposed fish, while a decrease was observed in sod3, igf1 and ntcl. As shown in Supplementary Figure S8, the expression levels of each gene detected by qRT-PCR were consistent with those obtained in the RNA-seq results.
3.6 Histopathology
The typical organization of S. aurata skin was observed in control fish: a thicker and multicellular epidermis layer overlays the dermis and the adipose tissue of the hypodermis (Figures 5A–F). Overall, UVB exposure resulted in structural and morphological changes in the skin (Figures 5A–F, 6). An inflammatory response was observed with significant infiltration of immune-related cells (e.g., lymphocytes, granulocytes), particularly in the loose dermis (stratum spongiosum). This inflammatory infiltration with immune-related cells was not limited only to the surrounding area of the scale pocket but also to other areas through this skin layer (Figures 5C–E and Figures 6E–I). Moreover, some infiltration of immune-related cells was also observed in the stratum compactum of the dermis (Figures 5F, Figures 6J, K).
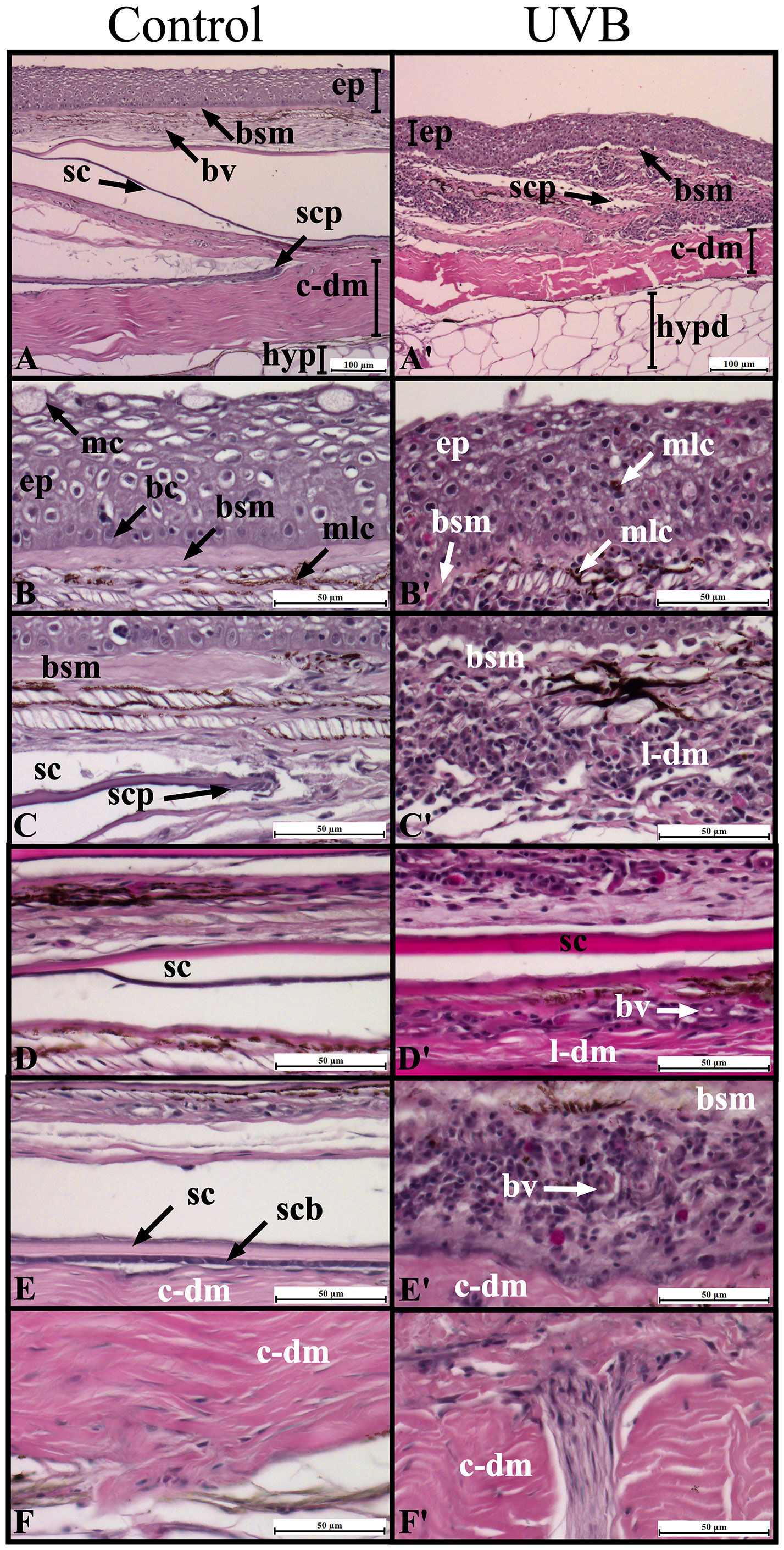
Figure 5 Morphological characterization of Sparus aurata skin in Control (A-F) and after UVB exposure (A’-F’) using histological sections stained with hematoxylin and eosin. Images are grouped according to skin layers: epidermis (B-B’), dermis stratum spongiosum (C-E, C’-E’), and dermis stratum compactum (F-F’). The basal cell layer and the collagen enriched basement membrane separate the epidermis from the dermis (B). Scales are enclosed in scale pockets in the stratum spongiosum (loose dermis), and their posterior regions are projected to the epidermis (C–E). The stratum compactum of the dermis is composed of a thicker layer of collagen fibers and underlies the stratum spongiosum (F). ep, epidermis; c-dm, compact dermis; l-dm, loose dermis; hypd, hypodermis; mc, goblet/mucus cells; bc, basement membrane; bsm, basement membrane; mlc, melanocytes; sc, scale; scp, scale pocket; scb, scleroblasts; bv, blood vessel.
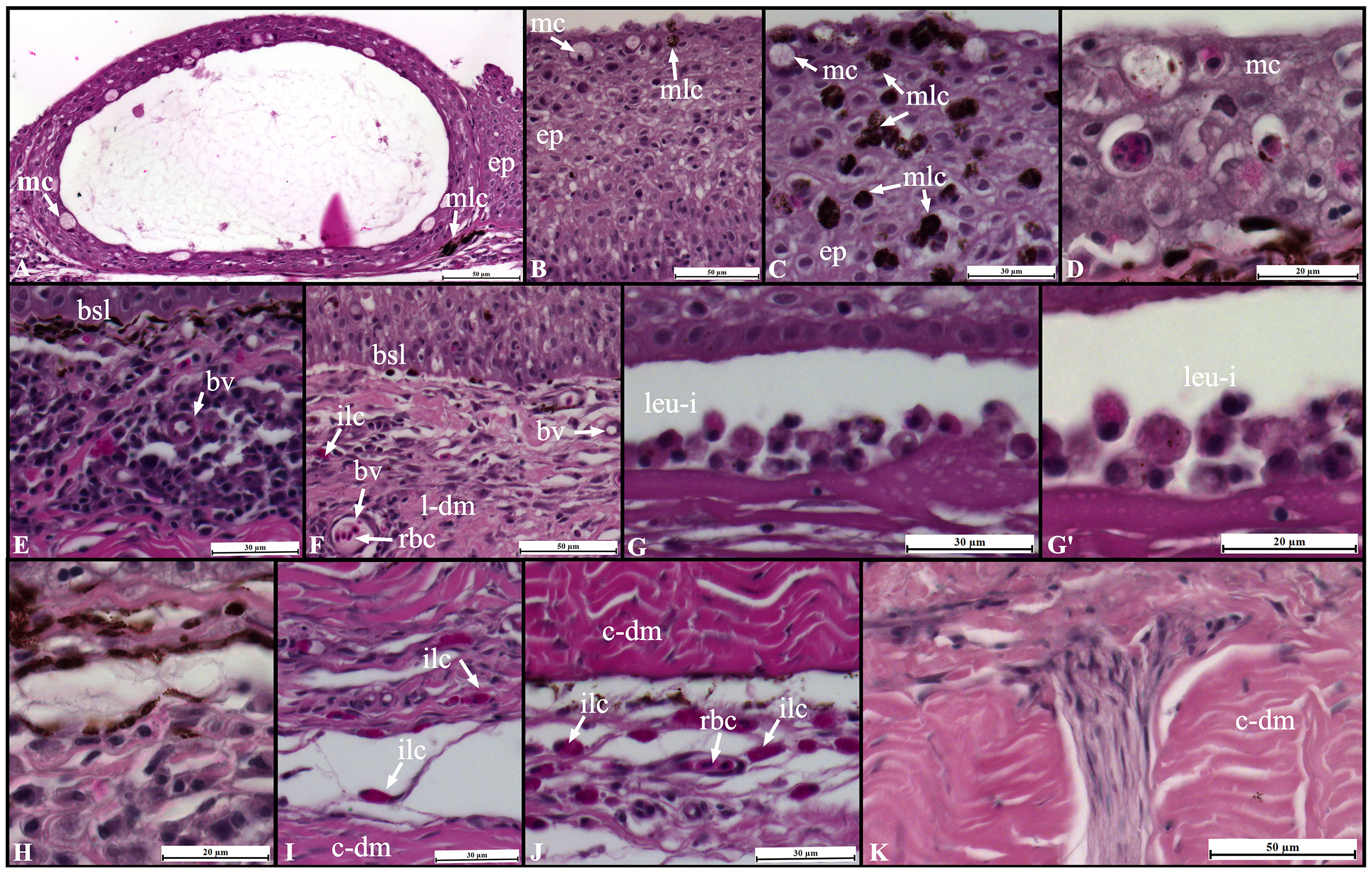
Figure 6 Changes in Sparus aurata skin morphology following UVB exposure: epidermis (A–D), dermis stratum spongiosum (E–I), and dermis stratum compactum (J, K). ep, epidermis; mc, goblet/mucous cells; mlc,melanin-containing cells; bv, blood vessel; bsl, basal-cell layer; ilc, immune-like cells; rbc, red blood cells; leu-i, leucocytes infiltration; l-dm, loose dermis; c-dm, compact dermis.
In the epidermis, the main changes included: loss of cellular organization, particularly in the basal layer epithelial cells; edema, hyperplasia, and sloughing/loss of the epidermis in the most severe cases; decreases in the abundance of mucous producing cells (goblet cells); intraepidermal blisters; agglomeration of melanin-containing cells (Figures 5A, B, 6A–D). In the dermo-epidermal junction, a decrease in the thickness of the basement membrane and loss of scales were also observed in UVB-exposed fish (Figures 5B, C, 6E, F). No major changes were observed in the hypodermis (data not shown).
4 Discussion
Transcriptome-wide changes as a response to UVB exposure have been widely studied in the skin of mammals (Enk et al., 2006; Sun et al., 2016; Xiao et al., 2021), but in fish remain poorly unexplored. Here, we described for the first time the transcriptional responses in the skin of S. aurata after long-term exposure to UVB. Our results revealed a higher number of differentially expressed genes (845 DEGs: 580 up-regulated, 365 down-regulated). Furthermore, GO/pathways enrichment, and PPI analyses demonstrated that biological processes and pathways related to cell cycle control/arrest, proteolysis, cell signaling, immune system, and inflammatory responses were enriched after UVB exposure. In contrast, a significant inhibition in genes related to growth, pigmentation, and cell adhesion/junction occurred. Our results are in agreement with previous transcriptomic studies. For example, leukocyte activation, defense inflammatory response, cell cycle, DNA damage, and apoptosis are some of the enriched pathways identified in human keratinocytes exposed to UVB (Shen et al., 2016). In addition, single exposure of human keratinocytes to UVR down-regulates the expression of genes involved in cell cycle progression. Conversely, repetitive exposure induced metabolism and cell signaling changes (Marais et al., 2017). UVB exposure also triggers the expression of genes associated with keratinization and apoptosis, followed by genes involved in inflammation and immune system activation (Bustamante et al., 2020).
In the present study, functional analyzes revealed that the cell cycle pathway was undoubtedly enriched in the skin of UVB-exposed fish. Concerning the general increase in the expression of genes involved in the cell cycle, we highlight the up-regulation observed in several key regulatory genes (e.g., cdkn2c, bub1, mad2l1, socs1a, socs1b, gtse1, ttk). These regulatory genes are important components of the cell cycle checkpoints. It has been demonstrated that activation of the cell cycle checkpoints and subsequent G1/S and G2/M cell cycle arrest acts as a mechanism of response to DNA damage induced by UVB, preventing inappropriate cell division (Bolognia et al., 1994; Latonen et al., 2001; Liu et al., 2007; Placzek et al., 2007; Granados-López et al., 2021). For example, we observed that cdkn2c was up-regulated in the skin of UVB-exposed fish in the order of 12-fold. This cell growth regulator is a cyclin-dependent kinase inhibitor that leads the G1 cell cycle arrest due to its interaction with the cyclin-dependent kinases CDK4 and CDK6, inhibiting the action of cyclin D (Cánepa et al., 2007; Lim and Kaldis, 2013). Furthermore, the upregulation of bub1 and mad2l1 suggests that there was a response to the changes in the maintenance of chromosomal and intrachromosomal stability following UVB exposure. These two genes are involved in the spindle assembly checkpoint. They have putative role in cell cycle arrest, and bub1 has also been described as involved in the DNA damage response (Percy et al., 2000; Yang et al., 2012; Vleugel et al., 2015; Foijer et al., 2017; Raaijmakers et al., 2018). Likewise, several genes involved in the DNA damage response were up-regulated in the skin of UVB- exposed fish. For example, as mentioned above, plk1 expression increased in the skin after UVB exposure. Plk1 has different roles during the DNA damage response, including the DNA checkpoint activation/maintenance and DNA repair and recovery (Hyun et al., 2014). The PCNA-associated factor was also up-regulated in the skin following UVB exposure, and Turchi et al. (2009) showed that transcription factor 3 (ATF3) and proliferating cell nuclear antigen (PCNA)-associated factor KIAA0101/p15PAF are key elements to trigger the DNA repair mechanisms in human keratinocytes. Genes involved in the resolution of sister chromatid cohesion and separation of sister chromatids (aurka, aurkb, birc5a, plk1, plk4, spdl1, ttk) were found to be up-regulated after UVB exposure. Actually, increased expression of these mitotic-related kinases after UVB exposure can result in the significant arrest of the cell cycle at the G2/M phase or induction of apoptosis, as previously suggested by Shukla et al. (2005). In addition, the transition from G2 to M is generally delayed when gtse1 is overexpressed (Monte et al., 2003; Monte et al., 2004). The higher abundance of transcripts of gtse1 observed in the UVB-exposed fish reinforces the importance of these regulatory elements in cell cycle checkpoints after exposure to UVB.
We recently suggested that modulation in the immune system responses can occur in fish after long-term exposure to UVB (Alves et al., 2020; Alves et al., 2021). Remarkably, in our study, significant transcriptional changes observed in the skin after UVB exposure were significantly enriched with GO terms and pathways related to the immune system and inflammatory responses. In agreement with our results, Patra et al. (2019) revealed that DEGs in UVB-exposed mice skin were enriched with several immune-related biological processes and pathways, including defense response, leukocyte migration, chemokine-mediated signaling, and Toll-like receptor signaling pathway. The absorption of UV photons by skin chromophores stimulates an innate immune response by increasing inflammatory mediators, followed by an infiltration of leucocytes, including neutrophils, monocytes, and T helper cells. These inflammatory mediators include interleukins, chemokines, interferons (IFNs), and antimicrobial peptides (AMPs), (Hart and Norval, 2018). Our results suggest that there was a stimulation of the innate immune response in seabream through an activated pro-inflammatory environment in the skin. In addition to the transcriptional activation of several inflammatory mediators and their receptors (e.g., il12b, ccl4, ccr6, cxcr2, irf7, ifi272l, nkl, b2ml), we also observed a generalized infiltration of leucocytes at the different layers of the skin after UVB exposure.
Furthermore, we suggest that increased transcription levels of il12b occurred to stimulate the repair of damaged DNA by activating DNA repair mechanisms (Schwarz et al., 2002). This gene is essential to prevent UVB-mediated immunosuppression, and interleukin-12-deficient mice are more susceptible to UVR-induced skin tumors than wild-type mice (Schwarz et al., 2002; Meeran et al., 2006a; Meeran et al., 2006b). Additionally, the PPI analysis also evidenced the importance of cytokine signaling in the immune system in the seabream skin after UVB exposure. Here, we show that UVB modulated the expression of several Toll-like receptors (TLRs) in the S. aurata. TLRs also play a vital role in the innate immune system, particularly in pathogen recognition and defense (Rebl et al., 2009; Takeda et al., 2003). Three of the TLRs genes up-regulated in the skin of UVB-exposed fish (tlr2, tlr7, tlr9) were previously linked to the mechanisms involved in the DNA repair induced by UVB exposure (reviewed by Kim and He, 2014). In addition, limited inflammatory responses in the skin were observed in TLR2-deficient mice exposed for 6 weeks to UVB (Park et al., 2014). Lawrence et al. (2019) suggested that under UVR exposure, immune mediators can activate dendritic-like cells and generate B and T regulatory cells, which can have immunosuppressive actions locally and systemically. Thus, UVB exposure may decrease the efficiency of immunological responses to pathogens. Consequently, an increase in microbial load can lead to a higher susceptibility to severe infections (Cramp et al., 2014; Markkula et al., 2007; Subramani et al., 2015). We have no direct evidence of these B- and T- immunosuppressive cells in the skin of UVB-exposed fish; however, genes involved in B- and T-cells differentiation, migration, and activation were overrepresented in the UVB fish (rhoh, zap70, syk, grap2a, il2rga), as evidenced by GO enrichment and PPI analyses.
In this study, many genes from the proteasome complex were up-regulated in the skin after UVB exposure. The proteasome pathway was one of the top enriched pathways, and PPI analysis suggested an interaction between proteasome elements with genes involved in the cell cycle and immune system. The proteasome proteolytic system is responsible for degrading oxidized and misfolded proteins, contributing to the cellular defense against oxidative stress (Dahlmann, 2007; Jung and Grune, 2008). Consistent with the present results, increased expression of several subunits of the proteasome system was observed in human epithelial keratinocytes irradiated even at lower doses of UVR (Sesto et al., 2002; Howell et al., 2004; Perluigi et al., 2010). Likewise, increased ubiquitinated protein levels have been associated with UVB exposure (Chen et al., 2020). We identified several up-regulated genes related to protein ubiquitination in the skin of UVB-exposed fish. We suggest that S.aurata uses the proteasome-ubiquitin degradation mechanism to eliminate oxidized and damaged proteins in the skin, counteracting the UVB-mediated damage (Perluigi et al., 2010). Unfortunately, the current study did not determine levels of ubiquitinated proteins or protein carbonylation.
We also showed that UVB exposure induced the expression of several genes with proteolytic activity (cathepsins, MMPs, and granzymes). For instance, we observed an increase in transcripts abundance of seven cathepsin genes. Cathepsins L and S (ctsl, ctss) were among the top up-regulated genes in the skin of fish exposed to UVB. Previous studies have demonstrated that UVR can regulate mRNA and protein levels of cathepsins, the most abundant group of proteases in lysosomes (Assefa et al., 2003; Tyrell, 2012; Ciazynska et al., 2018; Wang et al., 2019). Similarly, exposure to UVB (0.3 kJ m-2) in zebrafish resulted in increased transcriptional levels of cathepsin S and B (ctssb.1, ctsba), (Chen et al., 2020). Cathepsins L and S (ctsl2, ctss) genes were also up-regulated in cultured human keratinocytes exposed to UVB 30-50 mJ cm-2 (Lee et al., 2005; Shen et al., 2016). Previous work has confirmed that cathepsin L may be involved in the apoptotic-signaling pathway in UV-irradiated keratinocytes through the cleavage of pro-caspase 8 in the cytosol. However, this cleavage mechanism still needs to be confirmed. Additionally, recent studies demonstrated that cathepsin D has an essential role in initiating the caspase cascade through the direct activation of caspase 8 in human keratinocytes and neutrophils (Conus et al., 2012; Appelqvist et al., 2013). Interestingly, both cathepsin (ctsd) and caspase 8-like genes were up-regulated in the S. aurata skin after UVB exposure, and PPI enrichment analysis showed that a gene cluster including (ctso, ctsl.1, ctsh, and ctsd) was associated with lysosome and apoptosis. Previous studies showed that both UVA and UVB can stimulate the production of granzyme B (GZMB) in human keratinocytes, and these cells acquired cellular cytotoxicity capacity after UVB exposure. The ability of keratinocytes to degrade extracellular matrix components after UVB exposure can be supported by GZMB (Hernandez-Pigeon et al., 2006; Hernandez-Pigeon et al., 2007). In S. aurata, the expression of gzma and gzmb was previously detected in the leucocytes. Their expression and activity profiles suggested that the innate CMC occurs mainly due to the action of GZMA (Chaves-Pozo et al., 2019). The unmistakable infiltration of immune-related cells observed in the skin UVB-treated fish can also justify the increase observed in the expression of gzma and gzmb. Further studies should be conducted to explore the possible role of these genes on the degradation of extracellular matrix components in the S. aurata skin under long-term UVB exposure.
Activating antioxidant defense mechanisms is essential to prevent or mitigate the reactive oxygen species (ROS) generated by UVB and the resulting tissue damage (Cadenas, 1997). Although no enriched BP or pathways were associated with the oxidative stress response, UVB induced the expression of several antioxidant components, including gpx1, gss, hmox1a and txnl1. Increased gene expression or enzyme activity of GPx due to UVB exposure has been described in several organisms, from invertebrates such as copepods and crustaceans to mammals (Leccia et al., 2001; Kim et al., 2015; Hollmann et al., 2015). Furthermore, UVR can induce the expression of thioredoxin-1 in human/mouse melanocytes, keratinocytes, and fibroblasts (Funasaka and Ichihashi, 1997; Didier et al., 2001; Mustacich et al., 2004). Enhanced heme oxygenase 1 (HO-1) gene expression protects skin cells from UVA-induced damage (Allanson and Rieve, 2004; Xiang et al., 2011). Nevertheless, we suggest the activation of these antioxidant defenses was not efficient or sufficient to reduce the UVB-generated ROS, as evidenced by the general damage and morphological changes observed in the skin, as well as the notably high LPO levels (> 650%) in UVB-exposed fish (previously reported in Alves and Agustí, 2021).
With reference to the down-regulated genes in the seabream skin after UVB exposure, it is worth mentioning the inhibition observed in the expression of genes related to growth factor activity, including igf1, tgfb3, fgf7 fgf16, and csf1b. The growth inhibition, as well as changes in metabolism, reported in our recent work (Alves et al., 2020), can be connected with the reduced mRNA levels of igf1 (Reinecke et al., 2005). It is true that down-regulation of igf1 can occur in fish under stress conditions (Salas-Leiton et al., 2010; Jia et al., 2016). Transcriptional inhibition of fibroblast growth factors (fgf7, fgf16) may impair tissue repair and remodeling in UVB-damaged skin (Itoh and Ornitz, 2004; Yun et al., 2010). Kovac et al. (2009) showed that FGF7 is involved in the reduction of intracellular ROS levels after UVB exposure. If fgf7 has the same role in fish skin following UVB exposure, its down-regulation in our study can justify the increased LPO levels that resulted from the UVB-generate ROS. In agreement with our results, reduced transcriptional levels of transforming growth factor β due to UVB exposure was previously demonstrated in human cultured cells. Choi et al. (2007) demonstrated that UVB (50-800 mJ cm-2) inhibits the expression of tfgb1 in fibroblasts. On the contrary, in situ hybridization revealed that mRNA levels of tfgb1 and tgfb3 increased in the human epidermis and dermis following UVB exposure (Quan et al., 2002).
Genes related to focal adhesion were also down-regulated in fish following UVB exposure. Our results indicate that UVB may have affected focal adhesion dynamics in S. aurata skin, as previously demonstrated in human keratinocytes irradiated with UVB (Liu et al., 2007). In addition, other studies showed that UVB can down-regulate the expression of cell-adhesion molecules and integrins (itga6a, melanocytes; itga4a, Langerhans cells), affecting cell migration (Krengel et al., 2005; Hamakawa et al., 2006). Here, we observed decreased mRNA levels of itga11a in the skin after exposure to UVB. In fish, itga11a was up-regulated during skin regeneration (Huang et al., 2021). Several genes involved in pigmentation were down-regulated in the skin of UVB-exposed fish, and the melanogenesis pathway and biological processes related to pigmentation were underrepresented. In addition, the epidermis and basement membrane in some of the UVB-exposed fish were significantly damaged (data not shown). Moreover, the external lesions identified in UVB-exposed fish (desquamation, generalized epidermal sloughing) may also justify the down-regulation of pigmentation-related genes following UVB exposure. In fish, the skin mucous layer is the first line of defense against pathogens, and goblet cells are responsible for releasing mucous granules abundant in mucins (Elliott, 2011; Long et al., 2013; Gomez et al., 2013; Esteban and Cerezuela, 2015). Although no goblet cell counting was performed, our histology results suggest a decrease in the abundance of goblet cells in the skin of fish exposed to UVB. Moreover, a reduction in the abundance of muc5AC (secreted-gel forming mucin) transcript was observed following UVB exposure. Reduced mucous production cells have been reported as one of the adverse effects of UVB exposure in fish (Kaweewat and Hofer, 1997; Manek et al., 2012; Sucré et al., 2012).
5 Conclusion
In summary, we described for the first time the transcriptional and morphological changes occurring in the skin of S. aurata after long-term exposure to UVB. Several UVB-induced lesions were identified in the skin, particularly an infiltration of immune-related cells resulting from an inflammatory response. Simultaneously, many differentially expressed genes related to the immune and inflammatory responses (interleukins, chemokines, interferon-induced proteins, Toll-like receptors) were observed in the skin of UVB-exposed fish. The cell cycle pathway was one of the top enriched pathways observed after UVB exposure. Several key genes related to cell cycle regulation, particularly those involved in the cell cycle arrest (cdkn2c, bub1, mad2l1, plk1, gtse1), were up-regulated in UVB-exposed fish.
The transcription of genes related to proteolytic activity, including cathepsins, MMPs, and granzymes, was induced after UVB exposure. These genes may be involved in cell differentiation, cell cycle regulation, stress signaling, and inflammatory response as a consequence of UVB-induced damage. Moreover, UVB exposure induced the activation of several antioxidant components (gpx1, hmox1a, txnl1) and some genes involved in the DNA damage response. In contrast, UVB exposure inhibited the expression of several genes related to growth factor activity, cell growth and differentiation, and pigmentation. Finally, this study provides noteworthy insights into the molecular changes in fish skin with long-term exposure to UVB. To sum up, our data can be used in the future to identify potential biomarkers and explore the mechanisms underlying the molecular changes occurring in fish reared in offshore aquaculture systems in oligotrophic and highly transparent waters.
Data availability statement
All the raw Illumina sequencing reads were deposited in the NCBI SRA repository under the BioProject (PRJNA848258). The accession numbers for each dataset are SRR19631938, SRR19631937, SRR19631936, SRR19631943, SRR19631942, SRR19631941, SRR19631940, and SRR19631939. https://www.ncbi.nlm.nih.gov/bioproject/848258.
Ethics statement
The animal study was reviewed and approved by the KAUST Institutional Animal Care and Use Committee, and was in accordance with Saudi Arabia’s National Committee of Bioethics (NCBE) code of practice.
Author contributions
SA and RA designed the study. RA conducted the experiments and performed the transcriptome analysis, including library preparation and data analysis, qPCR, and histological analyses. RA and SA contributed to the critical interpretation of the data and drafted the manuscript. Both authors contributed to the critical reading and approved the final version of the manuscript.
Funding
This work was supported by the King Abdullah University of Science and Technology under the baseline funding to SA.
Acknowledgments
We acknowledge the KAUST Coastal and Marine Resources Core Lab (CMOR) for their support during the UVB exposure experiment and the KAUST Bioscience Core Lab (BCL) for their help with the NGS data generation. We would also like to thank Dr. Chakkiath Paul Antony (RSRC, KAUST) for his advice and comments during the bioinformatic analyses. Dr. Chakkiath kindly helped with the raw reads quality assessment, validation of each step during the analysis with the OmicsBox software, and during the generation of the enrichment analysis figures. We would like to thank our collaborators from Beacon Development Company (KAUST), Jorge F. Alarcon, Dr. Asaad H. Mahamed, Dr. Abdulaziz Al Suwailem, Dr. Joseph Leopoldo Q. Laranja, Muhammad Danial A. Nor Azli, and Nurhisham Razali for their assistance with seabream juveniles transport from the fish farms to the lab and maintenance during the quarantine period. Seabream juveniles were kindly provided by Tharawat Seas Company (Ar Rayis, Saudi Arabia).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2022.966654/full#supplementary-material
References
Al-Shahrour F., Díaz-Uriarte R., Dopazo J. (2004). FatiGO: A web tool for finding significant associations of Gene Ontology terms with groups of genes. Bioinformatics. 20, 578–580. doi: 10.1093/bioinformatics/btg455
Alemanni M. E., Lozada M., Zagarese H. E. (2003). Assessing sublethal effects of ultraviolet radiation in juvenile rainbow trout (Oncorhynchus mykiss). Photochem. Photobiol. Sci. 2, 867–870. doi: 10.1039/b301564e
Allanson M., Reeve V. E. (2004). Immunoprotective UVA (320-400 nm) irradiation upregulates heme oxygenase-1 in the dermis and epidermis of hairless mouse skin. J. Invest. Dermatol. 122, 1030–1036. doi: 10.1111/j.0022-202X.2004.22421.x
Alves R. N., Agustí S. (2021). Oxidative stress in tissues of gilthead seabream (Sparus aurata) and European seabass (Dicentrarchus labrax) juveniles exposed to ultraviolet-b radiation. J. Photochem. Photobiol. 8, 100070. doi: 10.1016/j.jpap.2021.100070
Alves R. N., Justo M. S. S., Laranja J. L. Q., Alarcon J. F., Al Suwailem A., Agustí S. (2021). Exposure to natural ultraviolet b radiation levels has adverse effects on growth, behavior, physiology, and innate immune response in juvenile European seabass (Dicentrarchus labrax). Aquaculture 533, 736215. doi: 10.1016/j.aquaculture.2020.736215
Alves R. N., Mahamed A. H., Alarcon J. F., Al Suwailem A., Agustí S. (2020). Adverse effects of ultraviolet radiation on growth, behavior, skin condition, physiology, and immune function in gilthead seabream (Sparus aurata). Front. Mar. Sci. 7, 1–20. doi: 10.3389/fmars.2020.00306
Anders S., Pyl P. T., Huber W. (2014). HTSeq — a Python framework to work with high-throughput sequencing data. Bioinformatics 31 (2), 166–169. doi: 10.1101/002824
Andrews S. (2010). FastQC: A quality control tool for high throughput sequence data [Online]. Available online at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/
Appelqvist H., Wäster P., Eriksson I., Rosdahl I., Öllinger K. (2013). Lysosomal exocytosis and caspase-8-mediated apoptosis in UVA-irradiated keratinocytes. J. Cell Sci. 126, 5578–5584. doi: 10.1242/jcs.130633
Assefa Z., Garmyn M., Vantieghem A., Declercq W., Vandenabeele P., Vandenheede J. R., et al. (2003). Ultraviolet b radiation-induced apoptosis in human keratinocytes: Cytosolic activation of procaspase-8 and the role of bcl-2. FEBS Lett. 540, 125–132. doi: 10.1016/S0014-5793(03)00238-2
BioBam Bioinformatics. (2019). OmicsBox: Bioinformatics made easy (version 1.3.11). BioBam Bioinformatics, Valencia, Spain. Available from: https://www.biobam.com/omicsbox.
Blazer V. S., Fabacher D. L., Little E. E., Ewing M. S., Kocan K. M. (1997). Effects of ultraviolet-b radiation on fish: Histologic comparison of a UVB-sensitive and a UVB-tolerant species. J. Aquat. Anim. Health 9, 132–143. doi: 10.1577/1548-8667(1997)009<0132:EOUBRO>2.3.CO;2
Bolognia J. L., Sodi S. A., Chakraborty A. K., Fargnoli M. C., Pawelek J. M. (1994). Effects of ultraviolet irradiation on the cell cycle. Pigment Cell Res. 7, 320–325. doi: 10.1111/j.1600-0749.1994.tb00634.x
Braun C., Reef R., Siebeck U. E. (2016). Ultraviolet absorbing compounds provide a rapid response mechanism for UV protection in some reef fish. J. Photochem. Photobiol. B Biol. 160, 400–407. doi: 10.1016/j.jphotobiol.2016.04.020
Bullock A. M., Coutts R. R. (1985). The impact of solar ultraviolet-radiation upon the skin of rainbow-trout, salmo gairdneri Richardson, farmed at high-altitude in Bolivia. J. Fish Dis. 8, 263–272. doi: 10.1111/j.1365-2761.1985.tb00942.x
Bustamante M., Hernandez-Ferrer C., Tewari A., Sarria Y., Harrison G. I., Puigdecanet E., et al. (2020). Dose and time effects of solar-simulated ultraviolet radiation on the in vivo human skin transcriptome. Br. J. Dermatol. 182, 1458–1468. doi: 10.1111/bjd.18527
Cadenas E. (1997). Basic mechanisms of antioxidant activity. BioFactors 6, 391–397. doi: 10.1002/biof.5520060404
Cánepa E. T., Scassa M. E., Ceruti J. M., Marazita M. C., Carcagno A. L., Sirkin P. F., et al. (2007). INK4 proteins, a family of mammalian CDK inhibitors with novel biological functions. IUBMB Life 59, 419–426. doi: 10.1080/15216540701488358
Carrasco-Malio A., Díaz M., Mella M., Montoya M. J., Miranda A., Landaeta M. F., et al. (2014). Are the intertidal fish highly resistant to UV-b radiation? a study based on oxidative stress in girella laevifrons (Kyphosidae). Ecotoxicol. Environ. Saf. 100, 93–98. doi: 10.1016/j.ecoenv.2013.07.030
Chaves-Pozo E., Valero Y., Lozano M. T., Rodríguez-Cerezo P., Miao L., Campo V., et al. (2019). Fish granzyme a shows a greater role than granzyme b in fish innate cell-mediated cytotoxicity. Front. Immunol. 10, 1–14. doi: 10.3389/fimmu.2019.02579
Chen R. Y., Lin C. J., Liang S. T., Villalobos O., Villaflores O. B., Lou B., et al. (2020). UVB irradiation induced cell damage and early onset of junbb expression in zebrafish. Animals 10, 1–16. doi: 10.3390/ani10061096
Chivers D. P., Wisenden B. D., Hindman C. J., Michalak T. A., Kusch R. C., Kaminskyj S. G., et al. (2007). Epidermal a’larm substance’ cells of fishes maintained by non-alarm functions: Possible defense against pathogens, parasites and UVB radiation. Proc. R. Soc Ser. B. 274, 2611–2619. doi: 10.1098/rspb.2007.0709
Choi C. P., Kim Y. I., Lee J. W., Lee M. H. (2007). The effect of narrowband ultraviolet B on the expression of matrix metalloproteinase-1, transforming growth factor-beta1 and type I collagen in human skin fibroblasts. Clin. Exp. Dermatol. 32 (2), 180–185. doi: 10.1111/j.1365-2230.2006.02309.x.
Ciążyńska M., Bednarski I. A., Wódz K., Kolano P., Narbutt J., Sobjanek M., et al. (2018). Proteins involved in cutaneous basal cell carcinoma development. Oncol. Lett. 16, 4064–4072. doi: 10.3892/ol.2018.9126
Conus S., Pop C., Snipas S. J., Salvesen G. S., Simon H. U. (2012). Cathepsin d primes caspase-8 activation by multiple intra-chain proteolysis. J. Biol. Chem. 287, 21142–21151. doi: 10.1074/jbc.M111.306399
Costa R. A., Cardoso J. C., Power D. M. (2017). Evolution of the angiopoietin-like gene family in teleosts and their role in skin regeneration. BMC Evol. Biol. 17, 14. doi: 10.1186/s12862-016-0859-x.
Cramp R. L., Reid S., Seebacher F., Franklin C. E. (2014). Synergistic interaction between UVB radiation and temperature increases susceptibility to parasitic infection in a fish. Biol. Lett. 10, 20140449. doi: 10.1098/rsbl.2014.0449
Dahlmann B. (2007). Role of proteasomes in disease. BMC Biochem. 8, 1–12. doi: 10.1186/1471-2091-8-S1-S3
Didier C., Pouget J. P., Cadet J., Favier A., Béani J. C., Richard M. J. (2001). Modulation of exogenous and endogenous levels of thioredoxin in human skin fibroblasts prevents DNA damaging effect of ultraviolet a radiation. Free Radic. Biol. Med. 30, 537–546. doi: 10.1016/S0891-5849(00)00502-5
Dobin A., Davis C. A., Schlesinger F., Drenkow J., Zaleski C., Jha S., et al. (2012). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29 (1), 15–21. doi: 10.1093/bioinformatics/bts635
Elliott D. G. (2011). “Functional morphology of the integumentary system in fishes,” in Encyclopedia of fish physiology: From gene to environment. Ed. Farrell A. P. (San Diego, CA: Academic Press), 476–488.
Enk C. D., Jacob-Hirsch J., Gal H., Verbovetski I., Amariglio N., Mevorach D., et al. (2006). The UVB-induced gene expression profile of human epidermis in vivo is different from that of cultured keratinocytes. Oncogene 25, 2601–2614. doi: 10.1038/sj.onc.1209292
Esteban M. A., Cerezuela R. (2015). “Fish mucosal immunity: skin,” in Mucosal health in aquaculture. Eds. Beck B. H., Peatman E. (San Diego, CA: Academic Press), 67–92.
Fabacher D. L., Little E. E. (1995). Skin component may protect fishes from ultraviolet-b radiation. Environ. Sci. pollut. Res. 2, 30–32. doi: 10.1007/BF02987508
Fabacher D. L., Little E. E. (1998). Photoprotective substance occurs primarily in outer layers of fish skin. Environ. Sci. pollut. Res. 5, 4–6. doi: 10.1007/BF02986366
Foijer F., Albacker L. A., Bakker B., Spierings D. C., Yue Y., Xie S. Z., et al. (2017). Deletion of the MAD2L1 spindle assembly checkpoint gene is tolerated in mouse models of acute T-cell lymphoma and hepatocellular carcinoma. Elife 6, 1–22. doi: 10.7554/eLife.20873
Fukunishi Y., Masuda R., Yamashita Y. (2006). Ontogeny of tolerance to and avoidance of ultraviolet radiation in red sea bream pagrus major and black sea bream acanthopagrus schlegeli. Fish Sci. 72, 356–363. doi: 10.1111/j.1444-2906.2006.01157.x
Funasaka Y., Ichihashi M. (1997). The effect of ultraviolet b induced adult T cell leukemia-derived Factor/Thioredoxin (ADF/TRX) on survival and growth of human melanocytes. Pigment Cell Res. 10, 68–73. doi: 10.1111/j.1600-0749.1997.tb00469.x
Garcia-Corral L. S., Martinez-Ayala J., Duarte C. M., Agustí S. (2015). Experimental assessment of cumulative temperature and UV-B radiation effects on Mediterranean plankton metabolism. Front. Mar. Sci. 2, 48. doi: 10.3389/fmars.2015.00048
Gomez D., Sunyer J. O., Salinas I. (2013). The mucosal immune system of fish: the evolution of tolerating commensals while fighting pathogens. Fish Shellfish Immunol. 35, 1729–1739. doi: 10.1016/j.fsi.2013.09.032
Granados-López A. J., Manzanares-Acuña E., López-Hernández Y., Castañeda-Delgado J. E., Fraire-Soto I., Reyes-Estrada C. A., et al. (2021). Uvb inhibits proliferation, cell cycle and induces apoptosis via p53, e2f1 and microtubules system in cervical cancer cell lines. Int. J. Mol. Sci. 22, 1–14. doi: 10.3390/ijms22105197
Häder D. P., Williamson C. E., Wängberg SÅ, Rautio M., Rose K. C., Gao K., et al. (2015). Effects of UV radiation on aquatic ecosystems and interactions with other environmental factors. Photochem. Photobiol. Sci. 14, 108–126. doi: 10.1039/c4pp90035a
Häkkinen J., Vehniäinen E., Oikari A. (2004). High sensitivity of northern pike larvae to UV-b but no UV-photoinduced toxicity of retene. Aquat. Toxicol. 66, 393–404. doi: 10.1016/j.aquatox.2003.11.001
Hamakawa M., Sugihara A., Okamoto H., Horio T. (2006). Ultraviolet b radiation suppresses langerhans cell migration in the dermis by down-regulation of a4 integrin. Photodermatol. Photoimmunol. Photomed. 22, 116–123. doi: 10.1111/j.1600-0781.2006.00187.x
Hart P. H., Norval M. (2018). Ultraviolet radiation-induced immunosuppression and its relevance for skin carcinogenesis. Photochem. Photobiol. Sci. 17, 1872. doi: 10.1039/C7PP00312A
Hernandez-Pigeon H., Jean C., Charruyer A., Haure M. J., Baudouin C., Charveron M., et al. (2007). UVA Induces granzyme b in human keratinocytes through MIF: Implication in extracellular matrix remodeling. J. Biol. Chem. 282, 8157–8164. doi: 10.1074/jbc.M607436200
Hernandez-Pigeon H., Jean C., Charruyer A., Haure M. J., Titeux M., Tonasso L., et al. (2006). Human keratinocytes acquire cellular cytotoxicity under UV-b irradiation: Implication of granzyme b and perforin. J. Biol. Chem. 281, 13525–13532. doi: 10.1074/jbc.M512694200
Hollmann G., Ferreira G de J., Geihs M. A., Vargas M. A., Nery L. E. M., Leitão Á, et al. (2015). Antioxidant activity stimulated by ultraviolet radiation in the nervous system of a crustacean. Aquat. Toxicol. 160, 151–162. doi: 10.1016/j.aquatox.2015.01.008
Howell B. G., Wang B., Freed I., Mamelak A. J., Watanabe H., Sauder D. N. (2004). Microarray analysis of UVB-regulated genes in keratinocytes: Downregulation of angiogenesis inhibitor thrombospondin-1. J. Dermatol. Sci. 34, 185–194. doi: 10.1016/j.jdermsci.2004.01.004
Huang Z., Ma B., Guo X., Wang H., Ma A., Sun Z., et al. (2021). Comparative transcriptome analysis of the molecular mechanism underlying the golden red colour in mutant Taiwanese loach. Aquaculture 543, 736979. doi: 10.1016/j.aquaculture.2021.736979
Huang D. W., Sherman B. T., Lempicki R. A. (2009). Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4 (1), 44–57. doi: 10.1038/nprot.2008.211
Hyun S. Y., Hwan H. I., Jang Y. J. (2014). Polo-like kinase-1 in DNA damage response. BMB Rep. 47, 249–255. doi: 10.5483/BMBRep.2014.47.5.061
Itoh N., Ornitz D. M. (2004). Evolution of the fgf and fgfr gene families. Trends Genet. 20, 563–569. doi: 10.1016/j.tig.2004.08.007
Jia R., Liu B. L., Feng W. R., Han C., Huang B., Lei J. L. (2016). Stress and immune responses in skin of turbot (Scophthalmus maximus) under different stocking densities. Fish Shellfish Immunol. 55, 131–139. doi: 10.1016/j.fsi.2016.05.032
Jokinen I. E., Salo H. M., Markkula E., Rikalainen K., Arts M. T., Browman H. I. (2011). Additive effects of enhanced ambient ultraviolet b radiation and increased temperature on immune function, growth and physiological condition of juvenile (parr) Atlantic salmon, salmo salar. Fish Shellfish Immunol. 30, 102–108. doi: 10.1016/j.fsi.2010.09.017
Jung T., Grune T. (2008). The proteasome and its role in the degradation of oxidized proteins. IUBMB Life 60, 743–752. doi: 10.1002/iub.114
Kaweewat K., Hofer R. (1997). Effect of UV-b radiation on goblet cells in the skin of different fish species. J. Photochem. Photobiol. B. 41, 222–226. doi: 10.1016/S1011-1344(97)00104-8
Kazerouni E. G., Khodabandeh S. (2011). Ionocyte immunolocalization and the effects of ultraviolet radiation on their abundance and distribution in the alenins of Caspian sea salmon, salmo trutta caspius. Cell J. 13, 45–54. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3652540/.
Khan A. Q., Aldosari F., Hussain S. M. (2018). Fish consumption behavior and fish farming attitude in kingdom of Saudi Arabia (KSA). J. Saudi Soc Agric. Sci. 17, 195–199. doi: 10.1016/j.jssas.2016.04.003
Kim I. Y., He Y. Y. (2014). Ultraviolet radiation-induced non-melanoma skin cancer: Regulation of DNA damage repair and inflammation. Genes Dis. 1, 188–198. doi: 10.1016/j.gendis.2014.08.005
Kim B. M., Rhee J. S., Lee K. W., Kim M. J., Shin K. H., Lee S. J., et al. (2015). UV-B radiation-induced oxidative stress and p38 signaling pathway involvement in the benthic copepod tigriopus japonicus. Comp. Biochem. Physiol. Part C Toxicol. Pharmacol. 167, 15–23. doi: 10.1016/j.cbpc.2014.08.003
Kovac D., Raffa S., Flori E., Aspite N., Briganti S., Cardinali G., et al. (2009). Keratinocyte growth factor down-regulates intracellular ROS production induced by UVB. J. Dermatol. Sci. 54, 106–113. doi: 10.1016/j.jdermsci.2009.01.005
Krengel S., Stark I., Geuchen C., Knoppe B., Scheel G., Schlenke P., et al. (2005). Selective down-regulation of the a6-integrin subunit in melanocytes by UVB light. Exp. Dermatol. 14, 411–419. doi: 10.1111/j.0906-6705.2005.00295.x
Kuznetsova I., Lugmayr A., Siira S., Rackham O. (2019). Filipovska. CirGO: an alternative circular way of visualising gene ontology terms. BMC Bioinf. 20, 84. doi: 10.1186/s12859-019-2671-2
Latonen L., Taya Y., Laiho M. (2001). UV-Radiation induces dose-dependent regulation of p53 response and modulates p53-HDM2 interaction in human fibroblasts. Oncogene 20, 6784–6793. doi: 10.1038/sj.onc.1204883
Lawrence K. P., Young A. R., Diffey B. L., Norval M. (2019). The impact of solar ultraviolet radiation on fish: Immunomodulation and photoprotective strategies. Fish Fish 02, 1–16. doi: 10.1111/faf.12420
Leccia M.-T., Yaar M., Allen N., Gleason M., Gilchrest B. A. (2001). Solar simulated irradiation modulates gene expression and activity of antioxidant enzymes in cultured human dermal fibroblasts. Exp. Dermatol. 10, 272–279. doi: 10.1034/j.1600-0625.2001.100407.x
Lee K. M., Lee J. G., Seo E. Y., Lee W. H., Nam Y. H., Yang J. M., et al. (2005). Analysis of genes responding to ultraviolet b irradiation of HaCaT keratinocytes using a cDNA microarray. Br. J. Dermatol. 152, 52–59. doi: 10.1111/j.1365-2133.2005.06412.x
Lim S., Kaldis P. (2013). Cdks, cyclins and CKIs: Roles beyond cell cycle regulation. Dev. 140, 3079–3093. doi: 10.1242/dev.091744
Liu S., Mizu H., Yamauchi H. (2007). Molecular response to phototoxic stress of UVB-irradiated ketoprofen through arresting cell cycle in G2/M phase and inducing apoptosis. Biochem. Biophys. Res. Commun. 364, 650–655. doi: 10.1016/j.bbrc.2007.10.046
Long Y., Li Q., Zhou B., Song G., Li T., Cui Z. (2013). De novo assembly of mud loach (Misgurnus anguillicaudatus) skin transcriptome to identify putative genes involved in immunity and epidermal mucus secretion. PloS One 8 (2), 1–14. doi: 10.1371/journal.pone.0056998
Lu Y., Bowswell M., Bowswell W., Yang K., Schartl M., Walter R. B. (2015). Molecular genetic response of xiphophorus maculatus-x. couchianus interspecies hybrid skin to UVB exposure. Comp. Biochem. Physiol. Part - C Toxicol. Pharmacol. 178, 86–92. doi: 10.1016/j.cbpc.2015.07.011
Manek A. K., Ferrari M. C. O., Sereda J. M., Niyogi S., Chivers D. P. (2012). The effects of ultraviolet radiation on a freshwater prey fish: Physiological stress response, club cell investment, and alarm cue production. Biol. J. Linn. Soc 105, 832–841. doi: 10.1111/j.1095-8312.2011.01829.x
Marais T. L. D., Kluz T., Xu D., Zhang X., Gesumaria L., Matsui M. S., et al. (2017). Transcription factors and stress response gene alterations in human keratinocytes following solar simulated ultra violet radiation. Sci. Rep. 7, 1–13. doi: 10.1038/s41598-017-13765-7
Markkula S. E., Karvonen A., Salo H., Valtonen E. T., Jokinen E. I. (2007). Ultraviolet b irradiation affects resistance of rainbow trout (Oncorhynchus mykiss) against bacterium yersinia ruckeri and trematode diplostomum spathaceum. Photochem. Photobiol. 83, 1263–1269. doi: 10.1111/j.1751-1097.2007.00165.x
Markkula S. E., Salo H. M., Immonen A. K., Jokinen E. I. (2005). Effects of short- and long-term ultraviolet b irradiation on the immune system of the common carp (Cyprinus carpio). Photochem. Photobiol. 81, 595. doi: 10.1562/2004-07-13-RA-231.1
Meeran S. M., Mantena S. K., Katiyar S. K. (2006a). Prevention of ultraviolet radiation - induced immunosuppression by (-)-epigallocatechin-3-gallate in mice is mediated through interleukin 12-dependent DNA repair. Clin. Cancer Res. 12 (7), 2272–2280. doi: 10.1158/1078-0432.CCR-05-2672
Meeran S. M., Mantena S. K., Meleth S., Elmets C. A., Katiyar S. K. (2006b). Interleukin-12-deficient mice are at greater risk of UV radiation-induced skin tumors and malignant transformation of papillomas to carcinomas. Mol. Cancer Ther. 5, 825–832. doi: 10.1158/1535-7163.MCT-06-0003
Monte M., Benetti R., Buscemi G., Sandy P., Del Sal G., Schneider C. (2003). The cell cycle-regulated protein human GTSE-1 controls DNA damage-induced apoptosis by affecting p53 function. J. Biol. Chem. 278, 30356–30364. doi: 10.1074/jbc.M302902200
Monte M., Benetti R., Collavin L., Marchionni L., Del Sal G., Schneider C. (2004). hGTSE-1 expression stimulates cytoplasmic localization of p53. J. Biol. Chem. 279, 11744–11752. doi: 10.1074/jbc.M311123200
Mustacich D., Wagner A., Williams R., Bair W., Barbercheck L., Stratton S. P., et al. (2004). Increased skin carcinogenesis in a keratinocyte directed thioredoxin-1 transgenic mouse. Carcinogenesis 25, 1983–1989. doi: 10.1093/carcin/bgh195
Neale R. E., Barnes P. W., Robson T. M., Neale P. J., Williamson C. E., Zepp R. G., et al. (2021). Environmental effects of stratospheric ozone depletion, UV radiation, and interactions with climate change: UNEP environmental effects assessment panel, update 2020 (Springer International Publishing). doi: 10.1007/s43630-020-00001-x
Nuñez E. T., Sobrino C., Neale P. J., Ceinos R. M., Du S. J., Rotllant J. (2012). Molecular response to ultraviolet radiation exposure in fish embryos: implications for survival and morphological development. Photochem. Photobiol. 88, 701–707. doi: 10.1111/j.1751-1097.2012.01088.x
Overmans S., Agustí S. (2019). Latitudinal gradient of UV attenuation along the highly transparent red sea basin. Photochem. Photobiol. 95, 1267–1279. doi: 10.1111/php.13112
Overmans S., Agustí S. (2020). Unraveling the seasonality of UV exposure in reef waters of a rapidly warming (Sub-)tropical Sea. Front. Mar. Sci. 7. doi: 10.3389/fmars.2020.00111
Park H. S., Jin S. P., Lee Y., Oh I. G., Lee S., Kim J. H., et al. (2014). Toll-like receptor 2 mediates a cutaneous reaction induced by repetitive ultraviolet b irradiation in C57/BL6 mice in vivo. Exp. Dermatol. 23, 591–595. doi: 10.1111/exd.12477
Patra V. K., Wagner K., Arulampalam V., Wolf P. (2019). Skin microbiome modulates the effect of ultraviolet radiation on cellular response and immune function. iScience 15, 211–222. doi: 10.1016/j.isci.2019.04.026
Percy M. J., Myrie K. A., Neeley C. K., Azim J. N., Ethier S. P., Petty E. M. (2000). Expression and mutational analyses of the human MAD2L1 gene in breast cancer cells. Genes Chromosom. Cancer 29, 356–362. doi: 10.1002/1098-2264(2000)9999:9999<::AID-GCC1044>3.0.CO;2-N
Perluigi M., Di Domenico F., Blarzino C., Foppoli C., Cini C., Giorgi A., et al. (2010). Effects of UVB-induced oxidative stress on protein expression and specific protein oxidation in normal human epithelial keratinocytes: A proteomic approach. Proteome Sci. 8, 1–14. doi: 10.1186/1477-5956-8-13
Placzek M., Przybilla B., Kerkmann U., Gaube S., Gilbertz K. P. (2007). Effect of ultraviolet (UV) a, UVB or ionizing radiation on the cell cycle of human melanoma cells. Br. J. Dermatol. 156, 843–847. doi: 10.1111/j.1365-2133.2007.07795.x
Pulgar J., Lagos P., Maturana D., Valdés M., Aldana M., Pulgar V. M. (2015). Effect of UV radiation on habitat selection by girella laevifrons and graus nigra (Kyphosidae). J. Fish Biol. 86, 812–821. doi: 10.1111/jfb.12566
Quan T. H., He T. Y., Kang S., Voorhees J. J., Fisher G. J. (2002). Ultraviolet irradiation alters transforming growth factor β/Smad pathway in human skin in vivo. J. Invest. Dermatol. 119, 499–506. doi: 10.1046/j.1523-1747.2002.01834.x
Raaijmakers J. A., van Heesbeen R. G. H. P., Blomen V. A., Janssen L. M. E., van Diemen F., Brummelkamp T. R., et al. (2018). BUB1 is essential for the viability of human cells in which the spindle assembly checkpoint is compromised. Cell Rep. 22, 1424–1438. doi: 10.1016/j.celrep.2018.01.034
Raudvere U., Kolberg L., Kuzmin I., Arak T., Adler P., Peterson H., et al. (2019). g:Profiler: a web server for functional enrichment analysis and conversions of gene list Update). Nucleic Acids Res. 47, 191–198. doi: 10.1093/nar/gkz369
Rebl A., Goldammer T., Seyfert H. M. (2009). Toll-like receptor signaling in bony fish. Vet Immunol Immunopathol. 134 (3-4), 139–150. doi: 10.1016/j.vetimm.2009.09.021
Reinecke M., Björnsson B. T., Dickhoff W. W., McCormick S. D., Navarro I., Power D. M., et al. (2005). Growth hormone and insulin-like growth factors in fish: Where we are and where to go. Gen. Comp. Endocrinol. 142, 20–24. doi: 10.1016/j.ygcen.2005.01.016
Riera-Heredia N., Martins R., Mateus A.P., Costa R.A., Gisbert E., Navarro I., et al. (2018). Temperature responsiveness of gilthead sea bream bone an in vitro and in vivo approach. Scientific Reports. 8 (1), 11211. doi: 10.1038/s41598-018-29570-9.
Robinson M. D., McCarthy D. J., Smyth G. K. (2010). edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. doi: 10.1093/bioinformatics/btp616
Salas-Leiton E., Anguis V., Martín-Antonio B., Crespo D., Planas J. V., Infante C., et al. (2010). Effects of stocking density and feed ration on growth and gene expression in the Senegalese sole (Solea senegalensis): Potential effects on the immune response. Fish Shellfish Immunol. 28, 296–302. doi: 10.1016/j.fsi.2009.11.006
Sayed A. E. H., Abdel-Tawab H. S., Hakeem S. S. A., Mekkawy I. A. (2013). The protective role of quince leaf extract against the adverse impacts of ultraviolet-a radiation on some tissues of clarias gariepinus (Burchel). J. Photochem. Photobiol. B 119, 9–14. doi: 10.1016/j.jphotobiol.2012.11.006
Sayed A. E. H., Ibrahim A. T., Mekkawy I. A. A., Mahmoud U. M. (2007). Acute effects of ultraviolet-a radiation on African catfish clarias gariepinus (Burchel). J. Photochem. Photobiol. B 89, 170–174. doi: 10.1016/j.jphotobiol.2007.09.010
Schwarz A., Ständer S., Berneburg M., Böhm M., Kulms D., van Steeg H., et al. (2002). Interleukin-12 suppresses ultraviolet radiation-induced apoptosis by inducing DNA repair. Nat. Cell Biol. 4, 26–31. doi: 10.1038/ncb717
Sesto A., Navarro M., Burslem F., Jorcano J. L. (2002). Analysis of the ultraviolet b response in primary human keratinocytes using oligonucleotide microarrays. Proc. Natl. Acad. Sci. U S A. 99, 2965–2970. doi: 10.1073/pnas.052678999
Sharma J. G., Rao Y. V., Kumar S., Chakrabarti R. (2010). Impact of UV-B radiation on the digestive enzymes and immune system of larvae of Indian major carp Catla catla. Int. J. Radiat. Biol. 86, 181–186. doi: 10.3109/09553000903419312
Shen Y., Kim A. L., Du R., Liu L. (2016). Transcriptome analysis identifies the dysregulation of ultraviolet target genes in human skin cancers. PloS One 11, 1–14. doi: 10.1371/journal.pone.0163054
Sherman B. T., Hao M., Qiu J., Jiao X., Baseler M. W., Lane H. C., et al. (2022). DAVID: a web server for functional enrichment analysis and functional annotation of gene listUpdate). Nucleic Acids Res. 23, 216–221. doi: 10.1093/nar/gkac194
Shukla Y., Reagan-Shaw S. R., Ahmad N. (2005). Ultraviolet radiation causes induction of mitotic kinases polo like kinase (Plk1) and aurora kinases -a and -b in HaCaT keratinocytes and SKH-1 hairless mouse skin: Relevance for skin carcinogenesis. Proc. Amer. Assoc. Cancer Res. 46, 1338–1339. https://aacrjournals.org/cancerres/article/65/9_Supplement/1338/524537.
Subramani P. A., Hameed B., Michael R. D. (2015). Effect of UV-b radiation on the antibody response of fish: implication on high altitude fish culture. J. Photochem. Photobiol. B. 143, 1–4. doi: 10.1016/j.jphotobiol.2014.12.021
Sucré E., Vidussi F., Mostajir B., Charmantier G., Lorin-Nebel C. (2012). Impact of ultraviolet-b radiation on planktonic fish larvae: Alteration of the osmoregulatory function. Aquat. Toxicol. 109, 194–201. doi: 10.1016/j.aquatox.2011.09.020
Sun X., Kim A., Nakatani M., Shen Y., Liu L. (2016). Distinctive molecular responses to ultraviolet radiation between keratinocytes and melanocytes. Exp. Dermatol. 25, 708–713. doi: 10.1111/exd.13057
Supek F., Bošnjak M., Škunca N., Šmuc T. (2011). REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS One 6, e21800. doi: 10.1371/journal.pone.0021800.
Takeda K., Kaisho T., Akira S. (2003). Toll-like receptors. Annual. Rev. Immunol. 21, 335–376. doi: 10.1146/annurev.immunol.21.120601.141126
Turchi L., Fareh M., Aberdam E., Kitajima S., Simpson F., Wicking C., et al. (2009). ATF3 and p15PAF are novel gatekeepers of genomic integrity upon UV stress. Cell Death Differ. 16, 728–737. doi: 10.1038/cdd.2009.2
Tyrrell R. M. (2012). Modulation of gene expression by the oxidative stress generated in human skin cells by UVA radiation and the restoration of redox homeostasis. Photochem. Photobiol. Sci. 11, 135–147. doi: 10.1039/C1PP05222E
Valiñas M. S., Walter Helbling E. (2016). Metabolic and behavioral responses of the reef fish patagonotothen cornucola to ultraviolet radiation: Influence of the diet. J. Exp. Mar. Bio Ecol. 474, 180–184. doi: 10.1016/j.jembe.2015.10.011
Vásquez P., Llanos-Rivera A., Castro L. R., Fernandez C. (2016). UV Radiation effects on the embryos of anchoveta (Engraulis ringens) and common sardine (Strangomera bentincki) off central Chile. Mar. Freshw. Res. 67, 195–209. doi: 10.1071/MF14038
Vieira F. A., Gregorio S. F., Ferraresso S., Thorne M. A., Costa R., Milan M., et al. (2011). Skin healing and scale regeneration in fed and unfed sea bream, Sparus auratus. BMC Genomics. 12, 490. doi: 10.1186/1471-2164-12-490
Vitt S., Rahn A. K., Drolshagen L., Bakker T. C. M., Scharsack J. P., Rick I. P. (2017). Enhanced ambient UVB light affects growth, body condition and the investment in innate and adaptive immunity in three-spined sticklebacks (Gasterosteus aculeatus). Aquat. Ecol. 51, 499–509. doi: 10.1007/s10452-017-9632-5
Vleugel M., Hoek T. A., Tromer E., Sliedrecht T., Groenewold V., Omerzu M., et al. (2015). Dissecting the roles of human BUB1 in the spindle assembly checkpoint. J. Cell Sci. 128, 2975–2982. doi: 10.1242/jcs.169821
Wang P. W., Hung Y. C., Lin T. Y., Fang J. Y., Yang P. M., Chen M. H., et al. (2019). Comparison of the biological impact of UVA and UVB upon the skin with functional proteomics and immunohistochemistry. Antioxidants 8(12), 1–20. doi: 10.3390/antiox8120569
Williamson C. E., Neale P. J., Hylander S., Rose K. C., Figueroa F. L., Robinson S. A., et al. (2019). The interactive effects of stratospheric ozone depletion, UV radiation, and climate change on aquatic ecosystems. Photochem. Photobiol. Sci. 18, 717–746. doi: 10.1039/C8PP90062K
Xiang Y., Liu G., Yang L., Zhong J. L. (2011). UVA-Induced protection of skin through the induction of heme oxygenase-1. Biosci. Trends. 5, 239–244. doi: 10.5582/bst.2011.v5.6.239
Xiao T., Chen Y., Song C., Xu S., Lin S., Li M., et al. (2021). Possible treatment for UVB-induced skin injury: Anti-inflammatory and cytoprotective role of metformin in UVB-irradiated keratinocytes. J. Dermatol. Sci. 102, 25–35. doi: 10.1016/j.jdermsci.2021.02.002
Yang K., Boswell M., Walter D. J., Downs K. P., Gaston-Pravia K., Garcia T., et al. (2014). UVB-induced gene expression in the skin of xiphophorus maculatus jp 163 b. Comp. Biochem. Physiol. Part - C Toxicol. Pharmacol. 163, 86–94. doi: 10.1016/j.cbpc.2014.01.008
Yang C., Wang H., Xu Y., Brinkman K. L., Ishiyama H. (2012). The kinetochore protein Bub1 participates in the DNA damage response. DNA Repair (Amst) 11 (2), 185–191. doi: 10.1016/j.dnarep.2011.10.018
Yun Y. R., Won J. E., Jeon E., Lee S., Kang W., Jo H., et al. (2010). Fibroblast growth factors: Biology, function, and application for tissue regeneration. J. Tissue Eng. 1, 1–18. doi: 10.4061/2010/218142
Zamzow J. P., Losey G. S. (2002). Ultraviolet radiation absorbance by coral reef fish mucus: photo-protection and visual communication. Environ. Biol. Fish 63, 41–47. doi: 10.1023/A:1013846816869
Keywords: UVB, seabream, transcriptomics, skin, cell cycle regulation, immune system, proteolysis, oxidative stress
Citation: Alves RN and Agustí S (2022) Transcriptional changes in the gilthead seabream (Sparus aurata) skin in response to ultraviolet B radiation exposure. Front. Mar. Sci. 9:966654. doi: 10.3389/fmars.2022.966654
Received: 11 June 2022; Accepted: 02 September 2022;
Published: 20 September 2022.
Edited by:
Rafael Magno Costa Melo, Federal University of Minas Gerais (UFMG), BrazilReviewed by:
Ali Reza Khansari, Universitat Autònoma de Barcelona, SpainLei Wei, Ludong University, China
Copyright © 2022 Alves and Agustí. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ricardo N. Alves, cmljYXJkby5hbHZlc0BrYXVzdC5lZHUuc2E=
 Ricardo N. Alves
Ricardo N. Alves Susana Agustí
Susana Agustí