- 1Department of Molecular Biotechnology, Graduate School of Advanced Sciences of Matter, Hiroshima University, Higashihiroshima, Japan
- 2Research Institute for Science and Engineering, Waseda University, Tokyo, Japan
- 3Department of Chemistry and Biochemistry, Graduate School of Advanced Science and Engineering, Waseda University, Tokyo, Japan
- 4Department of Environmental Science, Hankuk University of Foreign Studies, Yongin, South Korea
- 5Li Dak Sum Yip Yio Chin Kenneth Li Marine Biopharmaceutical Research Center, College of Food and Pharmaceutical Sciences, Ningbo University, Ningbo, China
- 6Ningbo Institute of Marine Medicine, Peking University, Ningbo, China
- 7Department of Civil and Environmental Engineering, Graduate School of Engineering, Hiroshima University, Higashihiroshima, Japan
- 8Graduate School of Advanced Science and Engineering, Hiroshima University, Higashihiroshima, Japan
- 9Unit of Biotechnology, Graduate School of Integrated Sciences for Life, Hiroshima University, Higashihiroshima, Japan
Most of the microbes from natural habitats cannot be cultivated with standard cultivation in laboratory, and sponge-associated microbes are no exception. We used two advanced methods based on a continuous-flow bioreactor (CF) and in situ cultivation (I-tip) to isolate previously uncultivated marine sponge-associated bacteria. We also characterized the physiological properties of the isolates from each method and attempted to clarify the mechanisms operating in each cultivation method. A greater number of novel bacteria were isolated using CF and in situ cultivation compared to standard direct plating (SDP) cultivation. Most isolates from CF cultivation were poor growers (with lower specific growth rates and saturated cell densities than those of isolates from SDP cultivation), demonstrating that it is effective to carry out pre-enrichment cultivation targeting bacteria that are less competitive on conventional cultivation, especially K-strategists and bacterial types inhibited by their own growth. Isolates from in situ cultivation showed a positive influence on cell recovery stimulated by chemical compounds in the extract of sponge tissue, indicating that some of the bacteria require a “growth initiation factor” that is present in the natural environment. Each advanced cultivation method has its own distinct key mechanisms allowing cultivation of physiologically and phylogenetically different fastidious bacteria for cultivation compared with conventional methods.
Introduction
The vast number of microbial species that remain uncultivated is sometimes referred to as the mysterious dark-matter of the microbial world (Rinke et al., 2013). Although there is argument over the proportion of uncultivated microorganisms in nature (Martiny, 2019; Steen et al., 2019; Martiny, 2020), culture independent surveys have demonstrated high diversity of uncultivated microbial species (Handelsman, 2004; Locey and Lennon, 2016; Hofer, 2018; Lloyd et al., 2018). Cultivation and isolation of microorganisms are essential steps to enable the discovery of natural products with biotechnological potential. Therefore, solving this microbial uncultivability challenge is a prerequisite for bioprospecting of microbial resources.
The mechanisms underlying why most microbes cannot grow using standard cultivation methods are still unclear. A significantly low proportion (generally less than 1%) of plated microbes readily form visible colonies on standard agar plates, thus leading to plate count anomalies (Staley and Konopka, 1985; Amann et al., 1995). However, no studies have led to a full resolution of the phenomenon. Some investigations have focused on modifying the nutrient composition of media and incubation conditions to overcome the limitations of conventional methods. Modifying growth conditions by adding specific organic or inorganic compounds to media (Bruns et al., 2002; Vartoukian et al., 2016), extending incubation times (Davis et al., 2005), lowering nutrient concentrations (Janssen et al., 2002), modifying the preparation of agar media (Tanaka et al., 2014; Kawasaki and Kamagata, 2017; Kato et al., 2018), and the use of alternative gelling agents (Tamaki et al., 2005) has led to increased microbial recovery, but most of the postulated extant microbes in the natural environment remain uncultivated. This suggests that microbial uncultivability cannot simply be explained by the unfitness of specific strains to certain culture conditions such as media composition, gas composition, temperature, or pH (Stewart, 2012). It might be the case that other essential factors that can more critically affect the growth of fastidious microbes are absent from the artificial conditions in these cultivation methods.
In this study, we used two advanced cultivation methods based on a continuous-flow bioreactor (CF) and in situ cultivation to successfully cultivate previously uncultivated bacteria associated with marine sponges. The advanced cultivation method using a CF bioreactor has been applied to enrichment culture of specific microbial targets that play a vital ecological role in nitrogen cycling but were previously uncultivated such as nitrite-oxidizing bacteria, ammonium oxidation (anammox) bacteria, and the methanogenic microbial community in sea floor sediments (Isaka et al., 2006; Imachi et al., 2011; Fujitani et al., 2013). The distinctive feature of this system is the use of porous materials such as polyester nonwoven fabric or polyurethane sponge as carrier material in the bioreactor to provide adequate pore space in the medium, providing an enlarged surface area for growth and a longer cell residence time. In addition, the reactor can maintain low substrate concentrations and high flow rate conditions. The use of the continuous-flow bioreactor is expected to facilitate enrichment of slow/poor growing microbial types which do not grow well under the batch-type cultivation methods. However, the effectiveness of this method to cultivate diverse previously uncultivated microbial types (non-targeted approach for novel species) is still unknown. We expected that use of the CF cultivation before standard cultivation on agar plates would lead to the enrichment of diverse slow and/or poor growing sponge-associated bacteria that have been difficult to isolate using the standard direct plating (SDP) cultivation method.
We also employed one in situ cultivation method, the I-tip, to isolate previously uncultivated sponge-associated bacteria. Previous studies using in situ cultivation methods aiming to better simulate the natural environment in a variety of environments such as activated sludge, sediments, soils, alkaline soda lakes, sponges, and thermal springs, have shown success for isolating novel microbes (Bollmann et al., 2007; Aoi et al., 2009; Jung et al., 2013; Jung et al., 2014; Jung et al., 2016; Jung et al., 2018; Chaudhary et al., 2019). Recently we clarified some of the principles of in situ cultivation methods including the presence of growth initiation factors (signaling-like compounds) in natural environments that can stimulate bacterial resuscitation from a nongrowing state (Jung et al., 2021). Therefore, we expected that in situ cultivation methods allow the isolation of sponge-associated bacteria that were considered previously non-growing using standard cultivation methods.
In the present work, we applied those two advanced cultivation methods to a marine sponge, Theonella swinhoei, to isolate previously uncultivated bacterial species. Marine sponges have been identified as rich sources of bioactive secondary metabolites of biotechnological interest for their antiviral, antitumoral, antimicrobial and cytotoxic properties (Koopmans et al., 2009; Mehbub et al., 2014; Anjum et al., 2016). Many studies have suggested that symbiotic microbes in marine sponges produce some of these bioactive metabolites (Brantley et al., 1995; Bewley et al., 1996; Wang, 2006). However, these talented producers remain unexplored and unavailable because the microbes associated with the sponges cannot be easily cultivated (Wang, 2006).
The main objectives of this study were to use two different advanced cultivation methods to access the hidden diversity of marine sponge-associated bacteria, isolate previously uncultivated species and identify some of the key factors for successful isolation previously uncultivated bacteria. We hypothesized that each advanced method would incorporate different key factors for successful isolation of fastidious bacteria. To evaluate the hypothesis, we compared patterns of phylogenetic diversity between isolates from two advanced cultivation methods and compared their cultures with the standard agar plating method. In addition, the physiological characteristics were compared among the isolates from each cultivation method to identify some possible mechanisms operating in each cultivation method.
Results
Identification of isolated bacteria
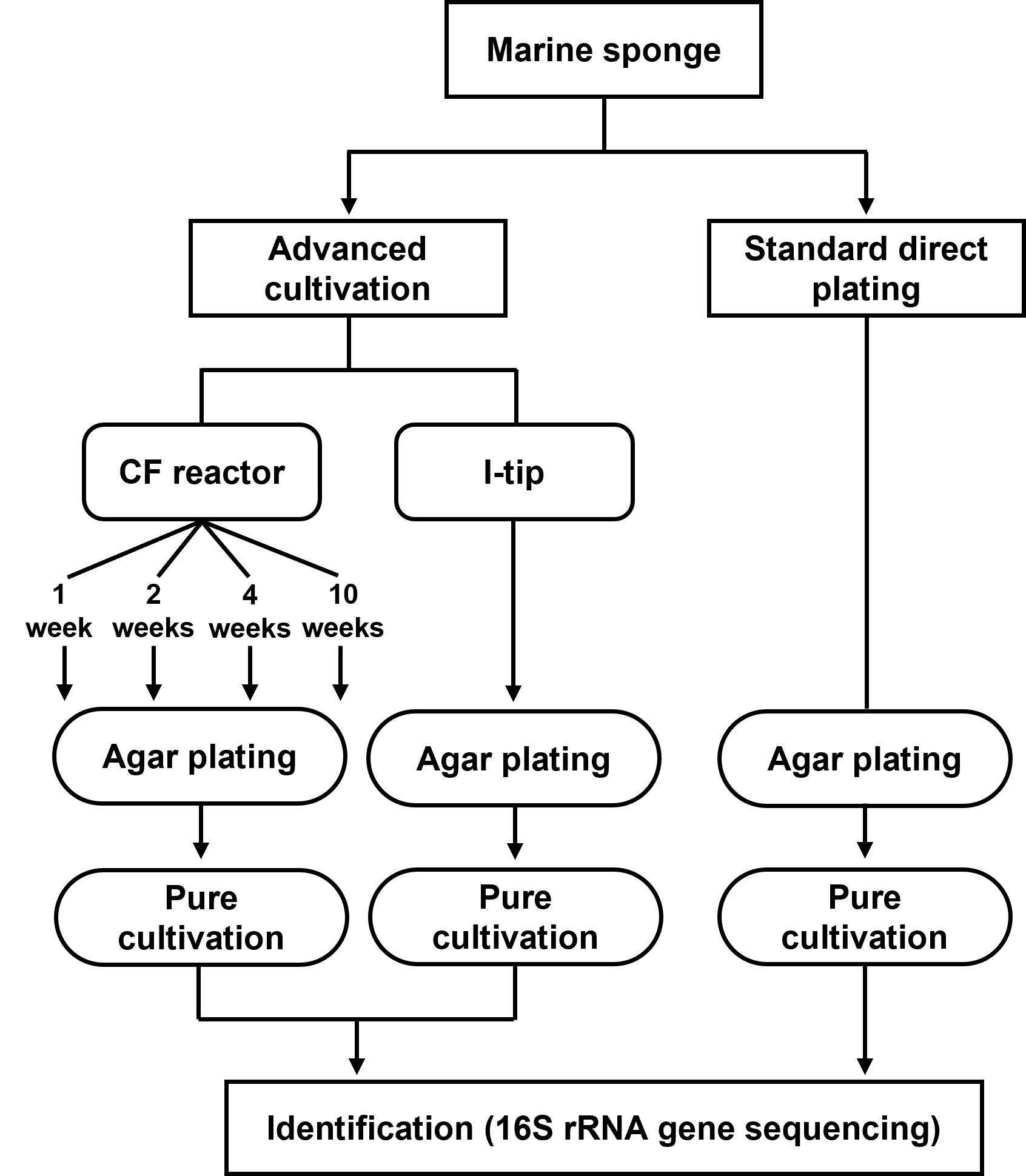
We applied two advanced cultivation and standard direct plating methods to a marine sponge (Figures 1, 2). The sequencing results of 60 isolates for each cultivation method (in total 120 isolates) were compared. The CF cultivation resulted in isolates representing 31 species (defined as OTUs composed of 16S rRNA gene sequences sharing over 97% identity) from four taxonomic groups (Actinobacteria, Firmicutes, Alphaproteobacteria, and Gammaproteobacteria; Figure 3, Table S1). The in situ cultivation (I-tip) resulted in isolates representing 25 species from three taxonomic groups (Bacteroidetes, Alphaproteobacteria, and Gammaproteobacteria; Table S2).
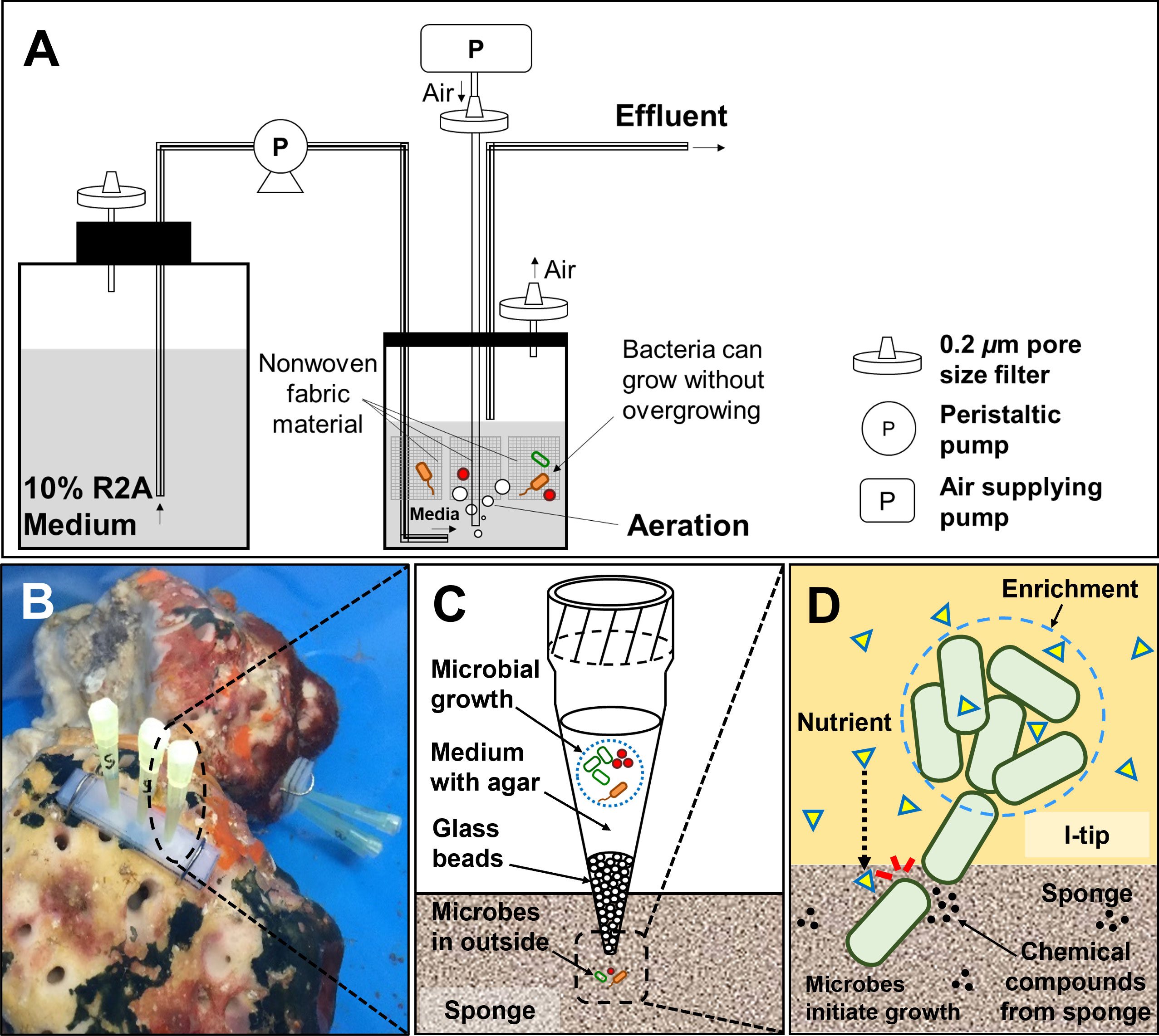
Figure 2 Structure and principle of CF (continuous-flow bioreactor) cultivation (A). Schematic showing the in situ view (B), structure (C) and principle (D) of I-tip.
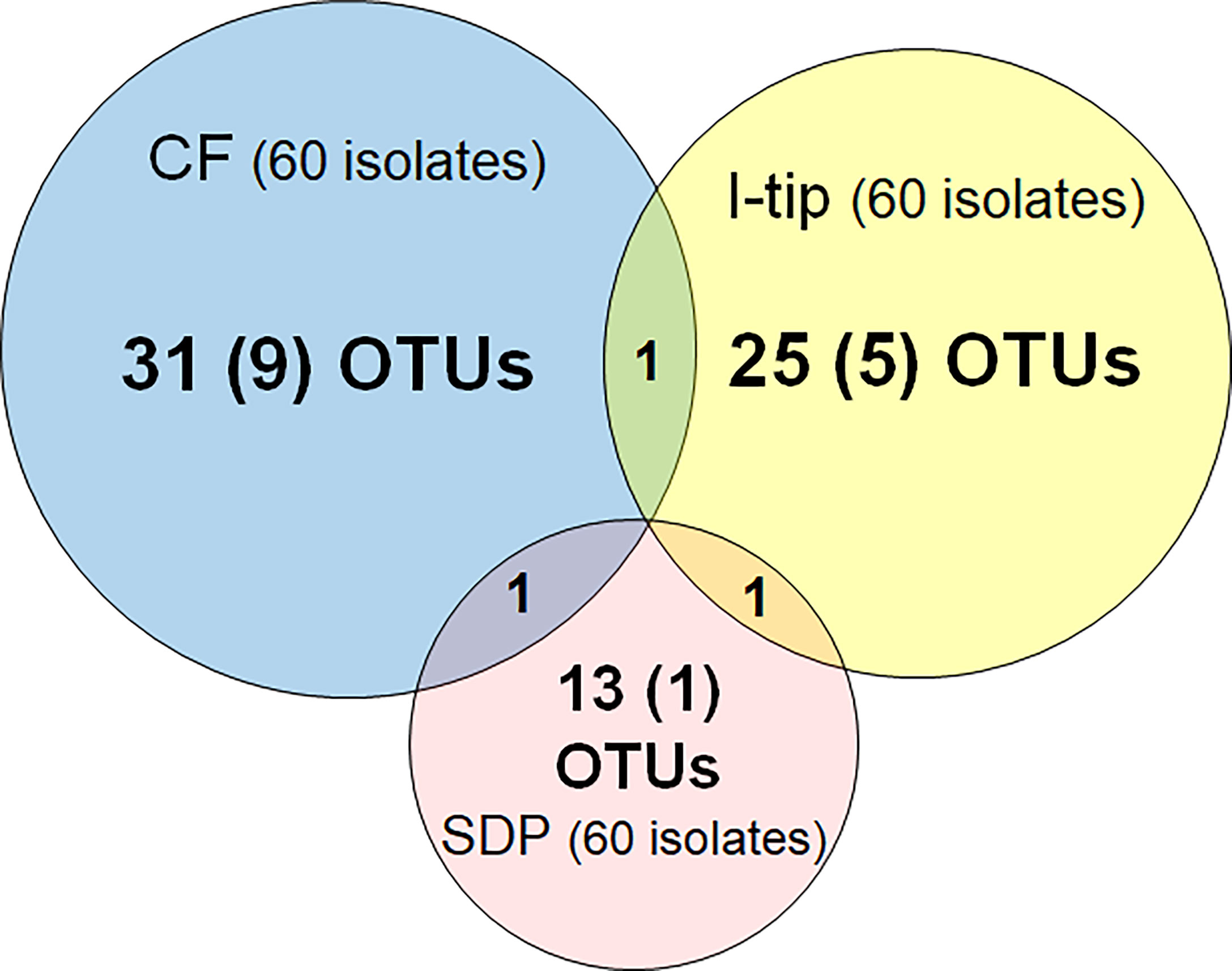
Figure 3 Overlap among culture collections obtained using three different cultivation approaches. Values in the center of each circle represent the total number of different isolated species (defined as OTUs composed of 16S rRNA gene sequences sharing over 97% identity) by method; values in parentheses represent the numbers of novel species (defined as a strain with ≤97% 16S rRNA similarity to the closest known relative among cultivated isolates); values in the overlapping areas represent the numbers of co-isolated species. Venn diagrams were made with Venn Diagram Plotter v. 1.5.5228.29250 (Pacific Northwest National Laboratory http://www.pnl.gov/; http://omics.pnl.gov/).
We refer to isolates from SDP cultivation performed in the same series of experiments (Jung et al., 2021) using the same sample. SDP cultivation resulted in isolates representing 13 species from three taxonomic groups (Bacteroidetes, Alphaproteobacteria and Gammaproteobacteria; Figure 3, Table S3). Shannon-Weaver diversity indexes (effective number of species) of isolated bacterial species from CF, I-tip and SDP were 21.9, 19.1 and 10.8, respectively. These results showed that advanced cultivation methods yielded higher diversity among the isolates at the species level than among those obtained via SDP cultivation. In addition, few common species were found between the three cultivation methods. (Figure 3). There were no species common among the isolates from all three cultivation methods.
The ratios of candidates for novel species, defined as a strain with ≤97% 16S rRNA similarity to the closest known relative among cultivated isolates, were different between the advanced cultivation (I-tip and CF) and SDP methods. Only one SDP isolate (1.6%) was a novel species, while 23% (14 isolates belonging to 5 species) and 45% (27 isolates belonging to 10 species) of I-tip and CF isolates, respectively, were novel species (Figures 3, 4, Tables S1-S3). None of these novel species have more than 95% 16S rRNA similarity to the cultivated isolates obtained from marine sponges in GenBank (www.ncbi.nlm.nih.gov) databases.
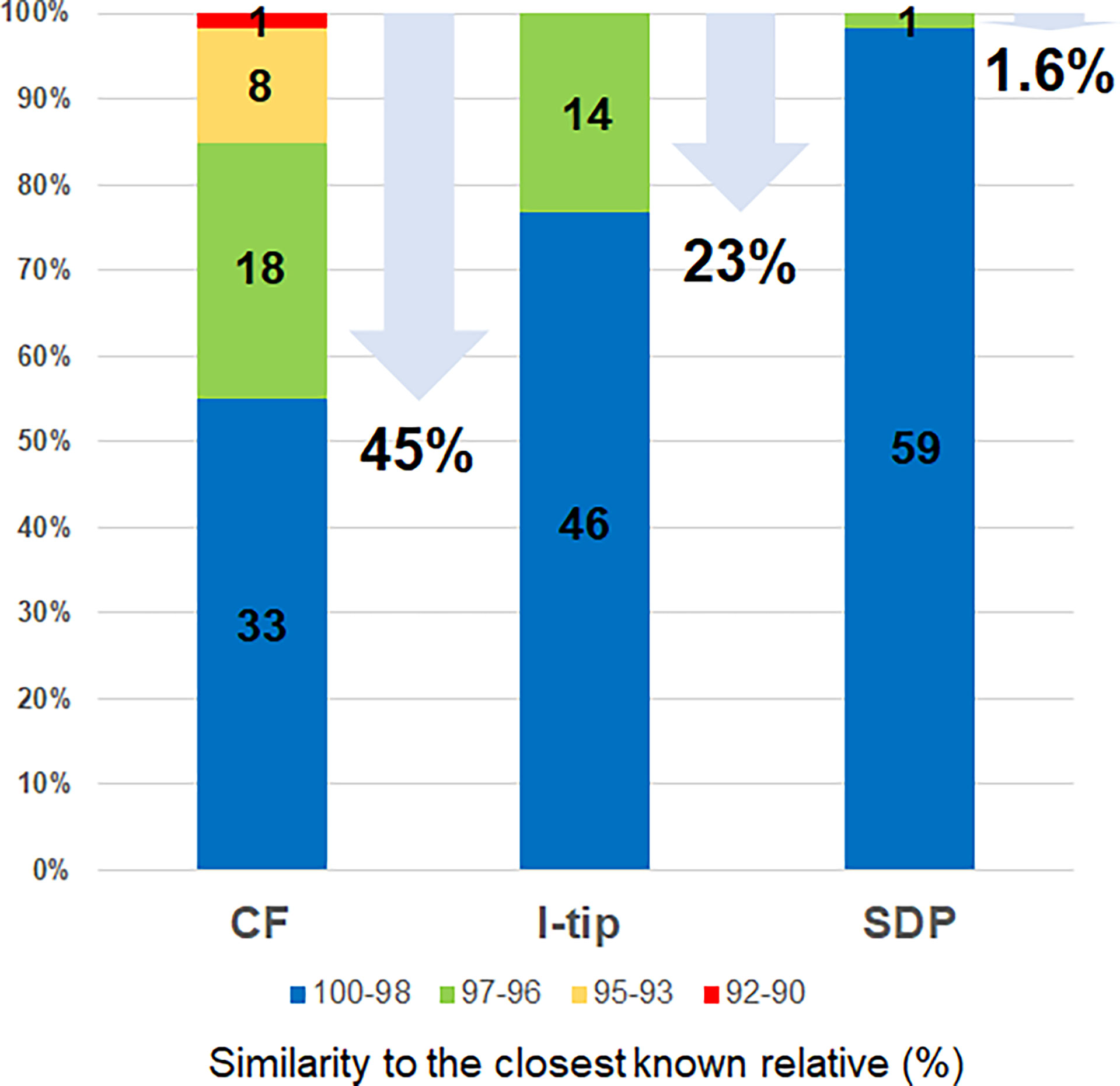
Figure 4 Similarity of isolates from each cultivation method based on 16S rRNA gene to the closest known relative in GenBank databases. Numbers in the bar graphs represent OTUs numbers in each similarity level and numbers under arrows represent the ratio of novel species, defined as a strain ≤97% 16S rRNA similarity to the closest known relative.
Bacterial community composition determined by the culture-independent method
The microbial community compositions of the marine sponge (Theonella swinhoei) samples were analyzed by Illumina-MiSeq sequencing based on the 16S rRNA gene. A total of 114,624 reads with a median length of 252 base pairs (bp) (V4~533–786 bp) assigned to 1625 OTUs were obtained from the sample. In total 16 taxonomic groups, Acidobacteria, Actinobacteria, Bacteroidetes, Chlamydiae, Chloroflexi, Cyanobacteria, Firmicutes, Nitrospirae, Planctomycetes, Alpha-, Beta-, Delta- Epsilon- Gammaproteobacteria, Spirochaetes and Verrucomicrobia were observed in the sponge sample (Figure 5). The isolates from the culture-dependent methods belonged to 5 groups, Actinobacteria, Bacteroidetes, Firmicutes, Alpha-, and Gammaproteobacteria.
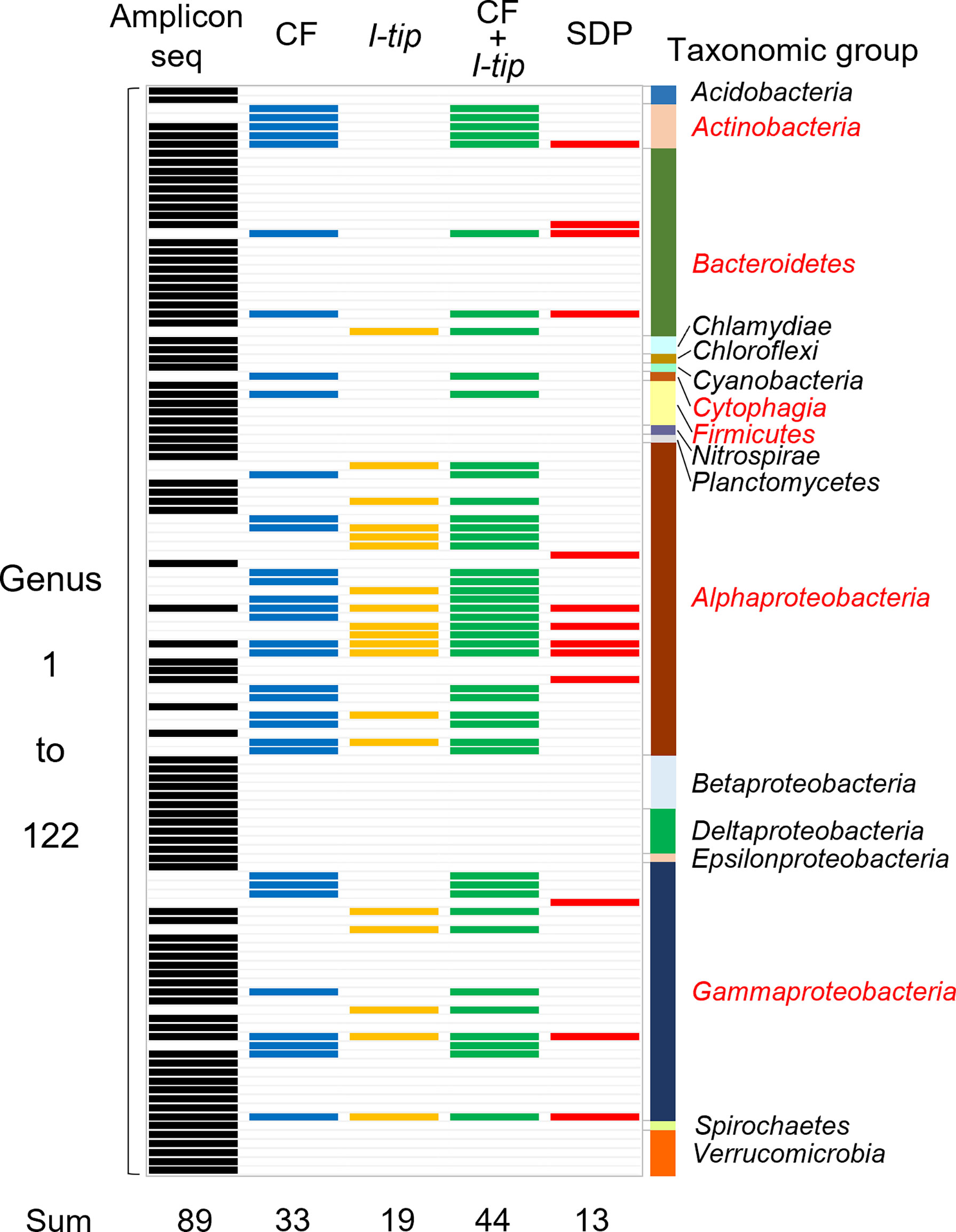
Figure 5 Band chart showing the comparison of microbial community compositions at the genera level among the sponge microbial community (amplicon sequencing data), isolates from CF, I-tip, and SDP cultivations. A total of 122 genera were detected from culture-independent and dependent approaches. Numbers at the bottom indicate number of genera detected with each method. Those are 89 from the amplicon sequencing data (black-colored bands), 33 from CF isolates (blue-colored bands), 19 from I-tip isolates (yellow-colored bands), 44 from sum of CF and I-tip isolates (green-colored bands), and 13 from SDP isolates (red-colored bands). Bar graph on the right indicates the taxonomic group to which those genera belong. The red letters for taxonomic group’s name represent the group that also isolated by the culture-dependent methods.
We also analyzed the 16S rRNA sequences from Illumina-MiSeq sequencing at the genus level and compared the results with those of culture-dependent approaches (Figure 5). For CF cultivation, we used results of isolates from 1, 2, 3 and 4 weeks of CF incubation. A total of 89 genera were identified in the environmental samples. On the other hand, a total of 44 genera appeared in the culture collection for advanced cultivations (33 and 19 in CF and I-tip cultivation, respectively), while the SDP isolates belonged to only 13 genera (Figure 5). Among 44 genera obtained from advanced cultivations, 13 genera appeared in the sponge sample but other 31 genera were obtained only from the culture collection.
Time course of changes in diversity and novelty of isolates from CF
To characterize the phylogenetic variation of isolates from CF over time in the continuous flow bioreactor, 30 isolates were obtained at each sub-cultivation step (at 1, 2, 4 and 10 weeks). Changes in bacterial community composition, diversity and proportion of novel species of isolates from CF cultivation over time were observed (Figure 6). The relative abundance of isolated bacteria at the phylum level dramatically shifted over time. After 1 week of CF cultivation, isolates belonging to Bacteroidetes, Alphaproteobacteria and Gammaproteobacteria were cultivated. The community composition at the phylum level was exactly the same as that from SDP cultivation (Table S3), but dramatically shifted at 2 weeks of CF cultivation.
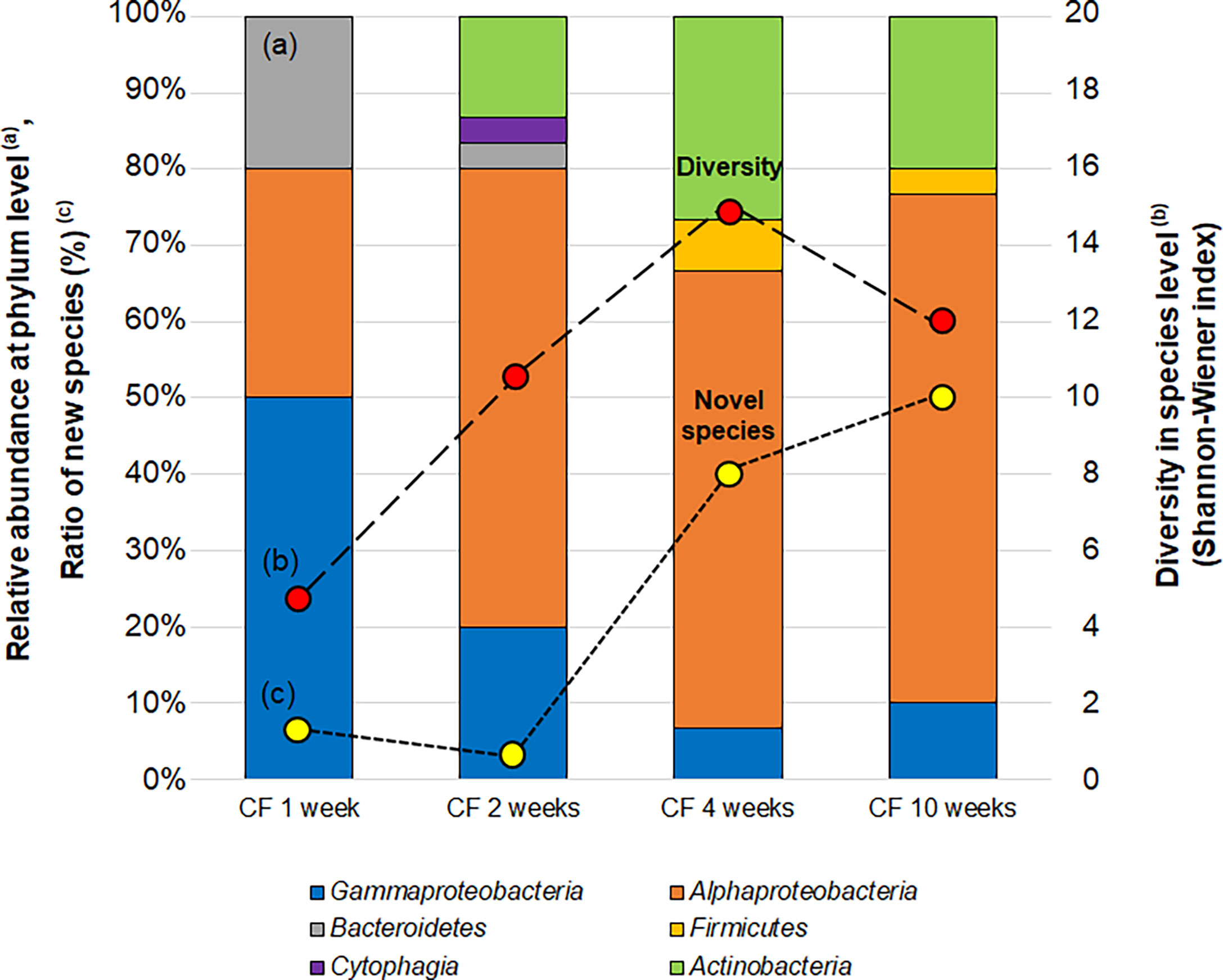
Figure 6 Changes in the bacterial community (a; bar graph), diversity (b; red circle) and proportion of novel species (c; yellow circle) of isolates CF (continuous-flow bioreactor) cultivation at 1, 2, 4 and 10 weeks. Bacterial community (A) shown as relative abundance at phylum level (class level for alpha, Gamma and Epsilon proteobacteria group). Diversities of isolates (B) are compared by the effective number of species from Shannon-Wiener index, exp (). Novel species (C) were defined as a strain ≤97% 16S rRNA similarity to the closest known relative.
The diversity at the species level (Shannon-Wiener index) and novelty of isolates (the ratio of novel species among the isolates) also shifted over time (Figure 6). The diversity index (effective number of species) was 4.7 at 1 week increased to 14.9 and 12.0 at 4 and 10 weeks, respectively. While the ratio of novel species among the isolates at 1 week was 6.7% (2/30), the proportion increased to 40% (12/30) and 50% (15/30) at 4 and 10 weeks, respectively.
More than 70% of the organic compounds fed into the reactor were consumed, resulting in a low total organic carbon concentration (TOC) in the reactor (below 30 mg-C/L during most of the incubation) throughout the experiment (Figure S1).
Growth characteristics of isolated species
For further investigation of the growth characteristics (specific growth rate, saturated cell density and growth curve type), we selected 25, 15 and 11 strains obtained from CF, I-tip and SDP cultivations, respectively, representing identical species of all identified isolates in each cultivation method except some species which had failed to be sub-cultivated during further experiments.
The specific growth rates and saturated cell density of selected strains from CF, I-tip, and SDP cultivation are plotted in Figure 7 (we hereafter define those selected strains as CF, I-tip, SDP strains, respectively). Most of the patterns for the CF strains were distinct from the I-tip, and SDP strains, as the specific growth rates and saturated cell density (carrying capacity) of most CF strains (20/25, 80%) were significantly lower than those of the I-tip, and SDP strains (optical density [OD] at 600 nm less than 0.005 h-1 and 0.115, respectively), except for five strains (belonging to 5 distinct OTUs).
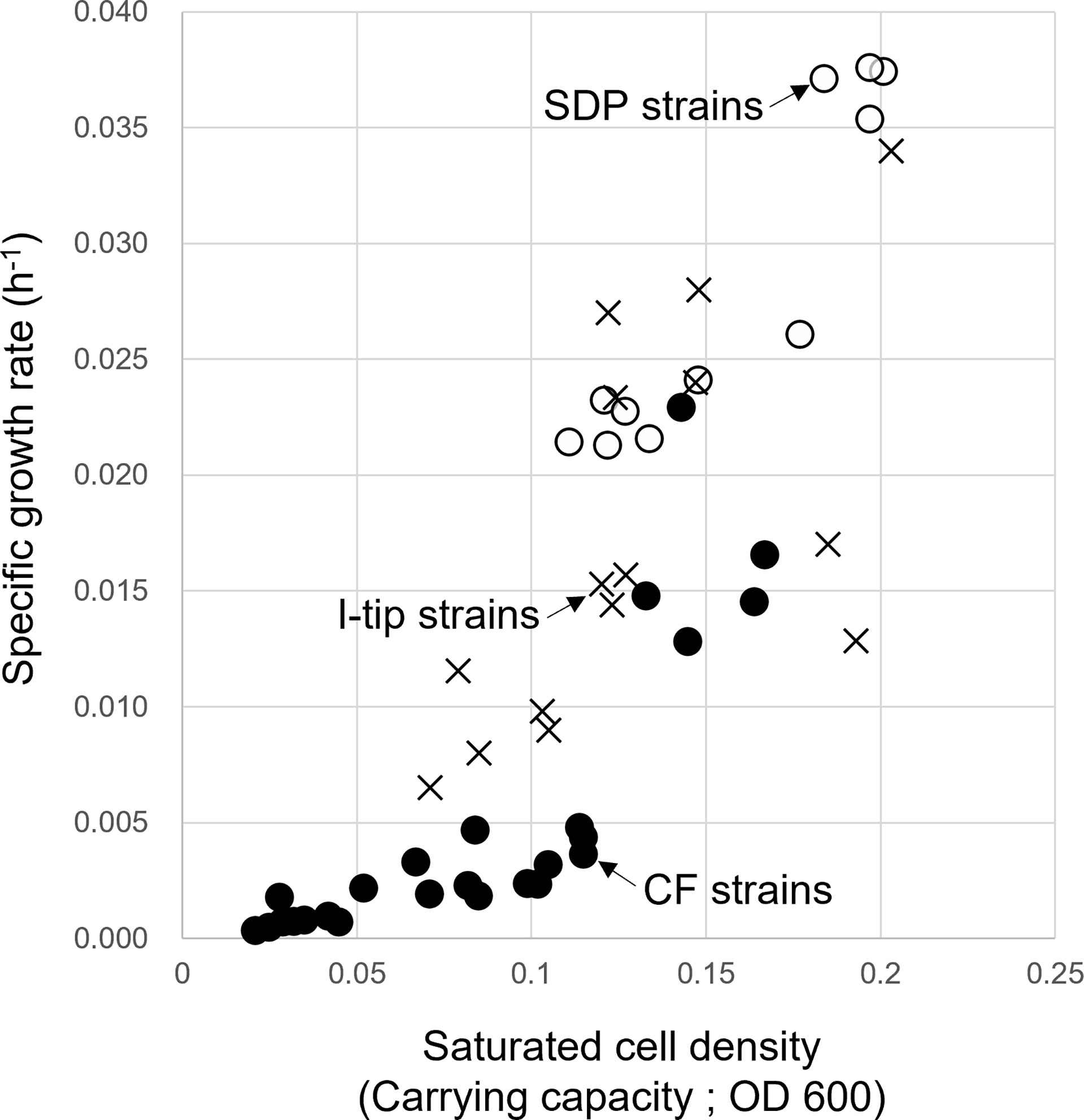
Figure 7 Specific growth rate and saturated cell density of CF (continuous-flow bioreactor; filled circle), I-tip (cross), and SDP (standard direct plating; open circle) strains. Data for specific growth rate and saturated cell density of SDP strains have already been reported (Jung et al., 2021).
The type of growth curve for CF strains also differed between that of strains from other cultivation methods (Table S4, Figure S2). Most CF strains (19/25, 75%) showed concave down growth curves, whereas all I-tip (15/15) and SDP strains (11/11) had concave up growth curves (logistic growth curve).
Colony size
The average colony size among the cultivation methods was compared using the Kruskal-Wallis procedure followed by post-hoc Dunn’s tests (Weitzman et al., 2018) (Figure 8). These comparisons indicated that the average colony diameter of CF strains was significantly smaller than that for I-tip and SDP strains (Kruskal-Wallis Rank Test; df =2, P< 0.00001; Figure 8).
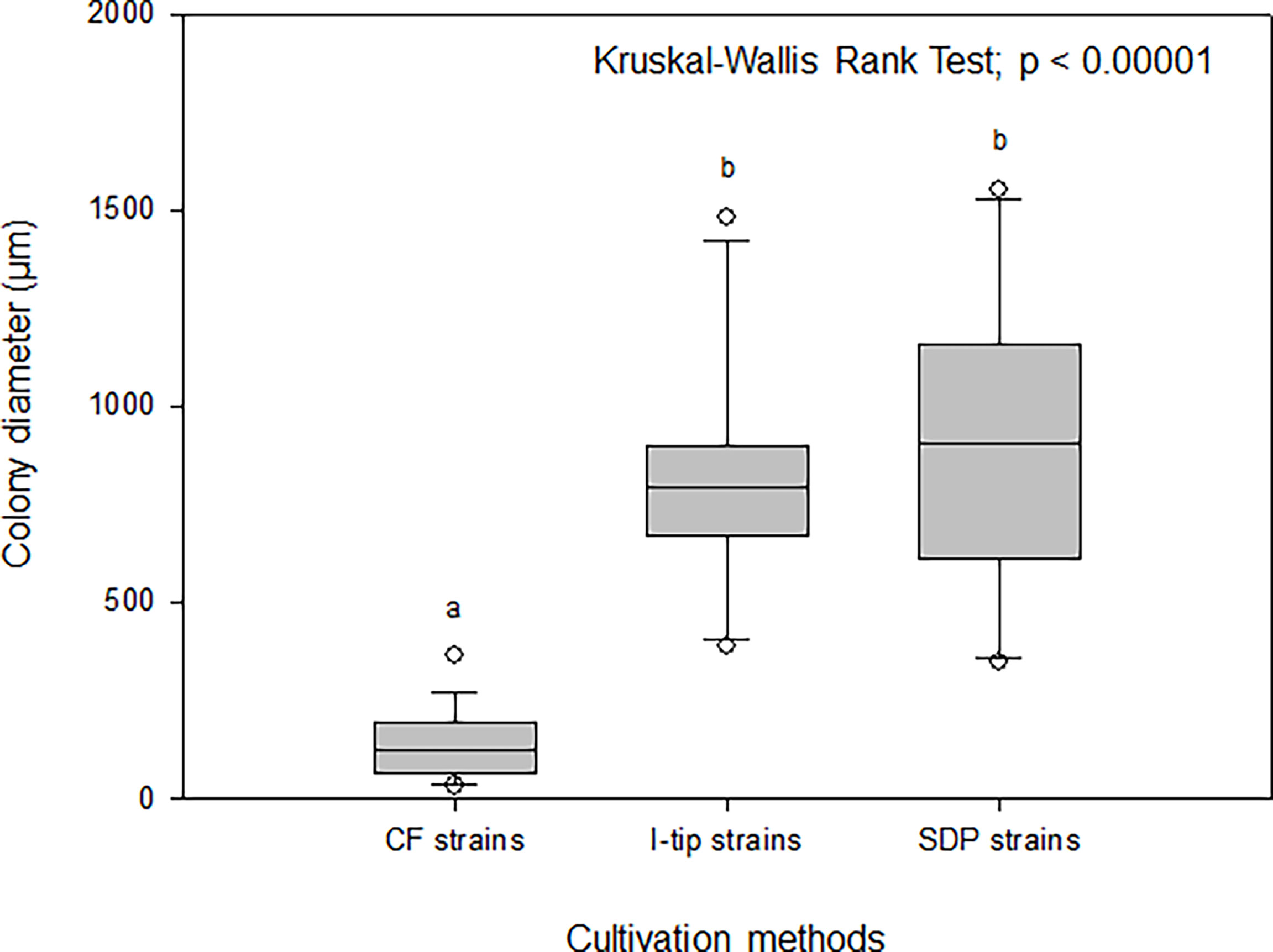
Figure 8 Box-and-whisker plots of colony diameter for CF (continuous-flow bioreactor), I-tip and SDP (standard direct plating) strains. The line inside each box indicates the median value. Lines extending from the boxes represent minimum and maximum values excluding outliers. Closed circles indicate the mean. Average colony diameter of each selected strain was measured by randomly selected 20 colonies on agar plate. Average colony diameter was compared among cultivation methods using the Kruskal-Wallis procedure. Letters above boxes indicate significant differences using post-hoc Dunn’s tests.
Effect of sponge extract on cell recovery
The colony formation efficiency ratios between the two culture conditions (media with and without the sponge extract) were calculated for selected CF, I-tip and SDP strains to examine the effect of the sponge extract on starvation recovery. The effect of sponge extract on the cell recovery rate on agar media was clearly different between the I-tip and SDP strains (Figure 9). The addition of sponge extract positively affected the recoveries of all I-tip strains. Especially two strains, I14 and I17, had more than 5 times colony numbers on the agar media with the sponge extract compared with the colony numbers on the media with no sponge extract. In contrast, no SDP strains were positively affected or rather negatively affected by the sponge extract addition, except for two strains that showed a slight positive effect (Figure 9). Note that the closest species for one SDP strain (SA7) showing a positive effect was Pseudovibrio denitrificans, which was commonly found in I-tip isolates (I9; Table S2).
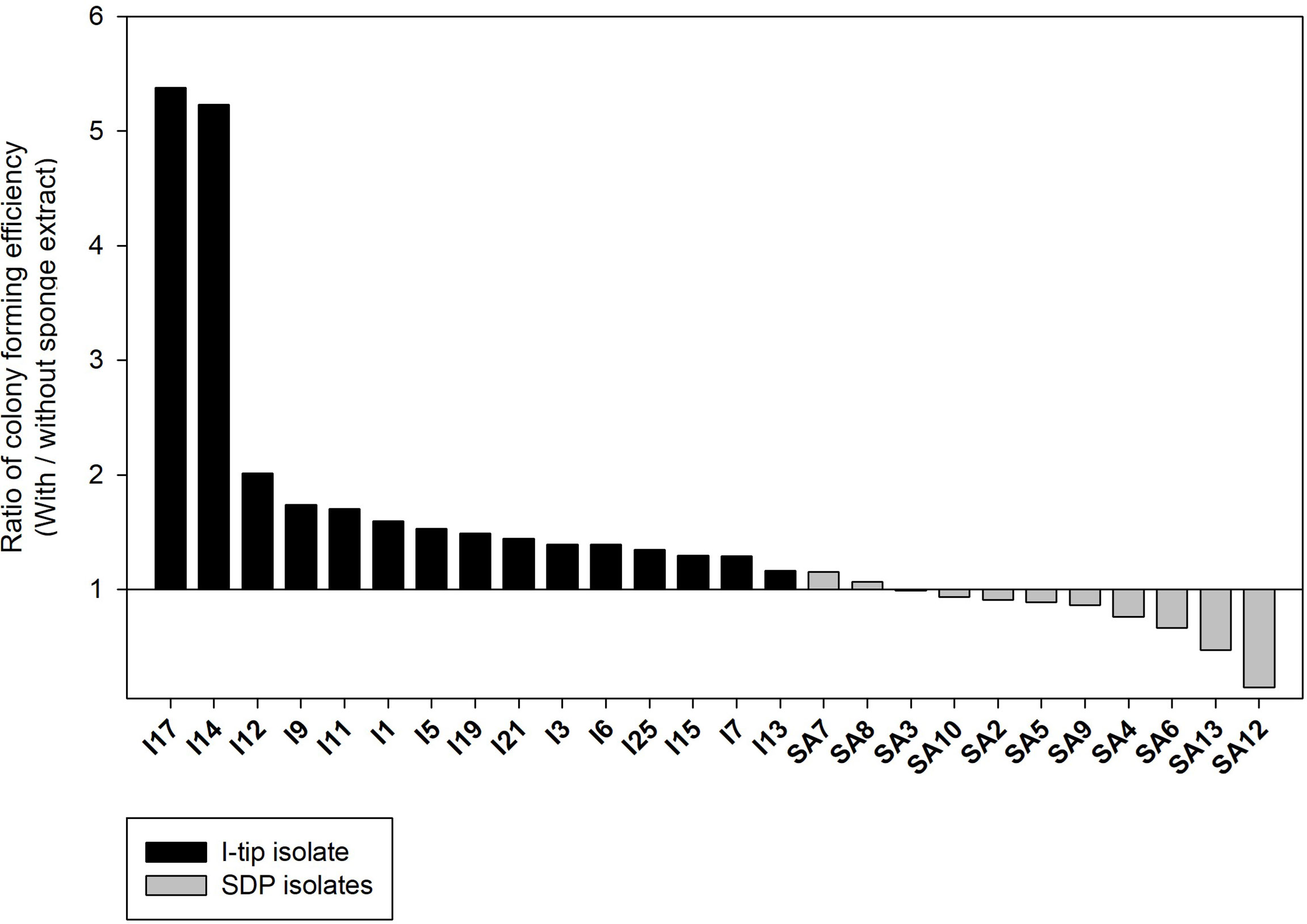
Figure 9 The effect of sponge extract on cell recovery rate of I-tip and SDP (standard direct plating) strains. The ratio of colony forming numbers between the two culture conditions (colony numbers on the agar medium with the sponge extract/colony numbers on the agar medium without the sponge extract) was calculated for each selected strain. The ratio was calculated using the average value from triplicate measurements (n=3) of the CFUs on each agar plate. Data for SDP strains have already been reported (Jung et al., 2021).
Discussion
How does CF cultivation allow cultivation of fastidious bacterial types?
A variety of cultivation efforts have made use of several alternative techniques to isolate previously uncultivated sponge-associated microbes, and those approaches yielded an increased novelty of sponge isolates and improved cultivability rates up to 14% in some cases (Sipkema et al., 2011). However, most postulated extant microbes in sponges remain uncultivated; thus, further efforts to discover such novel microbes are needed (Wang, 2006).
In this study, CF cultivation enabled obtaining a different culture collection that was larger and more novel than that obtained by SDP cultivation (Figure 4, Table S1). Identified CF isolates shared only one species in common with isolates from I-tip and SDP cultivation, respectively (Figure 3). This indicates that CF cultivation method has its own selectivity, which is different from that of the I-tip and SDP methods. CF cultivation allows enrichment of physiologically different bacterial types (Figure 7) and resulting isolation of phylogenetically different bacterial types. Furthermore, more novel species were obtained from the sample with a longer period of CF cultivation (Figure 6), suggesting that uncultivated bacterial types unique to CF cultivation require a certain length of time to be enriched in the reactor.
As described earlier, we expected that CF cultivation results in the growth of specific bacterial types that grow poorly on agar plates, which increases the efficiency of isolating previously uncultivated bacterial types. The specific growth rates of CF strains were much lower than those of strains from other methods (Figure 7), indicating that CF cultivation is suitable for the relatively slow-growing bacteria. The nonwoven fabric material used in the CF method was effective in retaining bacterial biomass in the bioreactor under the high flow rate condition, and enriching slow-growing microbes.
Furthermore, medium was continuously provided and completely mixed by aeration in the reactor and continuously discharged to keep the low concentration of the substrate in the CF reactor. Unlike batch or batch-fed type enriched cultivation, the low substrate concentration prevents overgrowth of specific bacterial types, especially r-strategists (higher growth rate under substrate unlimited conditions). This is effective to enrich K-strategists (competitive under substrate limited conditions, and usually slow growers in conventional cultivation methods).
Indeed, previous research successfully demonstrated the enrichment of anaerobic ammonium oxidation (anammox) bacteria, which were hardly cultivated even in enrichment and still not yet purely isolated (Date et al., 2009) and the selective enrichment of slow growing nitrite oxidizing bacteria Nitrospira (Fujitani et al., 2013; Fujitani et al., 2014). In this study, we used CF cultivation for the first time to isolate diverse marine sponge-associated bacteria, rather than targeting single species.
However, the favorable conditions for slow-growing bacteria are likely not the only mechanism for isolating a unique bacterial culture collection, because extending the incubation time of SDP cultivation did not affect microbial composition, diversity and novelty of isolates (Figure S3). Other factors in CF cultivation, such as the removal of metabolic waste from bacteria being cultured and difference in medium (solid vs liquid) can explain the result here.
The maximum growth (saturated cell density) of CF strains shown in their growth curves were remarkably lower than those of I-tip and SDP strains (Figure 7). Furthermore, by classifying the type of growth curve for each strain for the different cultivation methods, additional information on the growth characteristics was gained. Most CF strains had the growth curve classified as a concave down type, whereas none of the I-tip and SDP strains showed this type of growth curve but were classified as concave up (logistic growth curve) (Table S4, Figure S2). These results indicate that the growth of CF isolates was suppressed even if their cell concentrations were comparatively low. In other words, it suggests that most strains from CF cultivation are bacterial species that are more sensitive to metabolites or metabolic by-products and, as a result, those strains are less competitive in batch cultivation. This is also suggested by the observation that the average colony diameter of CF strains. was significantly smaller than that of I-tip and SDP strains (Figure 8). The flow-through conditions in the CF reactor minimizes this negative effect and allow enrichment of less competitive bacterial species under the standard cultivation condition. This concept has been previously presumed but not fully explored (Imachi et al., 2011).
How does in situ cultivation (I-tip) allow cultivation of fastidious bacterial types?
The I-tip method produced a richer collection of more novel strains that are fastidious for cultivation with the SDP method (Figure 3, 4). This is likely because the first step of the method, in situ cultivation, mimics natural conditions and allows for growth of those species that are only active in the environment (Jung et al., 2014). What mechanisms yield a different culture collection that differs entirely from the culture collection from the standard cultivation method?
We have developed and utilized various types of in situ cultivation methods to isolate microbes from a variety of environmental samples (Aoi et al., 2009; Jung et al., 2013; Jung et al., 2014; Jung et al., 2016; Jung et al., 2018). From these previous studies, two key principles of in situ cultivation can be identified. The first is that, if microbes are grown in situ, e.g., in a diffusion chamber, the cultivation results in higher recovery and increased novelty of isolates (Jung et al., 2013; Jung et al., 2014; Jung et al., 2016; Jung et al., 2018). The second is the observation that some microbes, those which initiate growth and thrive during in situ incubation, continue their growth even during the sub-cultivation step ex situ. Therefore, we hypothesized that some factors facilitating bacterial resuscitation in natural environments arouse inactive cells and enrich them during in situ cultivation, which results in recovery of fastidious bacterial species that cannot be cultivated by using standard cultivation.
This hypothesis was supported by the results of this study, and also those obtained using a diffusion chamber with the same marine sponge samples as in the present study (Jung et al., 2021). The results of the cell recovery experiment clearly support the hypothesis (Figure 9). Most of I-tip strains showed a positive effect on cell recovery using chemical compounds in sponge tissue. Therefore, I-tip cultivation led to isolation of a different bacterial culture collection that could not be cultivated with SDP cultivation. In contrast, most SDP strains showed a negative effect on cell growth with the sponge extract, likely due to some antibacterial metabolites in sponges such as secondary metabolites or antimicrobials from Actinomycetes (Bewley et al., 1996; Schmidt et al., 2000), or high concentration of trace toxic compounds which can accumulate in sponge tissue (Keren et al., 2016). Those bacterial types probably grow rapidly on rich media typical in standard cultivation methods (see growth rate result of SDP strains; Figure 7), and often have negative effects on the growth of more slowly growing oligotrophs (Jung et al., 2013), which results in culture collections with skewed species distributions (Jung et al., 2018).
Adding the sponge extract at higher concentrations than that in the original experimental condition (e.g., 1%, which was 10 times higher than the original experiment condition) strongly inhibited growth even for isolates from I-tip cultivation (data not shown). This might be one reason why SDP cultivation with sponge extract media (including 4% of sponge extract) did not result in isolating diverse and novel microbial species. Although media containing 1-10% of sponge extract have been widely used in several previous studies for isolating sponge-associated microbes (Selvin et al., 2009; Sipkema et al., 2011; Steinert et al., 2014; Esteves et al., 2016), our results suggest that adding lower concentrations of sponge extract (0.1% in this study) is advantageous in isolating a variety of microorganisms.
We performed the same experiment with CF strains, and compared the results with that of I-tip (Figure S4). Unlike for I-tip strains, the addition of the sponge extract did not measurably affect the cell recovery of strains from CF cultivation.
Bacterial community comparison of the culture-dependent and -independent methods
Comparing the bacterial composition of the 16S rRNA gene from the culture-independent method and that of the culture collections indicates that some bacterial groups that cannot be cultured by standard cultivation can be isolated with advanced cultivation approaches (Figure 5). The advanced cultivation methods, CF and I-tip, led to the isolation of several bacterial groups at the genus level that were not cultivated in the SDP method. On the other hand, there are several genera obtained from culture-dependent approaches (31 and 4 in advanced cultivations and SDP cultivation, respectively) that does not appear in the sponge samples observed by Illumina MiSeq sequencing analysis. These bacterial species are probably present in extremely low numbers in the sponge tissue, and may not be detected in a 16S rRNA gene clone library. This hypothesis is supported by several previous studies about analysis of the diversity in the clone library from sponge and other environmental samples (Sipkema et al., 2009; Sipkema et al., 2011). Some isolates that exclusively exist in culture-dependent methods may correspond to these “hidden” OTUs in the clone library.
Conclusion and perspectives
The results presented here show the utility of advanced cultivation methods to cultivate diverse previously uncultivated sponge-associated bacteria, including 1) combined use of a CF cultivation system and 2) combined use of in situ cultivation (I-tip), prior to agar plating as an isolation step. Furthermore, by demonstrating advanced approaches to microbial cultivation and comparing physiological properties of isolates from different approaches, we clearly identified two key points focused on the efficient cultivation of previously uncultivated microbes. Those are (1) targeting bacterial types that are less competitive (poorly or slow growing) on batch type cultivation, especially K-strategists and bacterial types inhibited by themselves, and (2) focusing on bacterial types which require a signaling-like factor for restarting to grow (turning on the switch to grow). Achieving this provides crucial insights into microbial uncultivability that can be applied to build new cultivation strategies. In addition, newly discovered microbes are candidates as sources for valuable secondary metabolites.
Nevertheless, there are still microbial species in marine sponges that have been represented in culture-independent methods but were not represented in our culture collection (Figure 5). In addition to the mechanisms explored in this study, there are likely several other important factors operating to cultivate uncultivated microbial types. Continued advances in cultivation methods together with improved understanding of the key factors for cultivating previously uncultivated microbes are needed.
Materials and methods
Sample collection
Marine sponges (Theonella swinhoei) were collected by scuba diving near Okino Island, (Kochi Prefecture, Japan) in August 2015 at a depth of approximately 15-20 m. Sponges were kept cool in an insulated box with sea water and transported to the aquarium and laboratory for the experiment.
Media
We used 1/10 diluted Reasoner’s 2A (R2A) media (10% of the manufacturer’s suggested concentration, Nihon Seiyaku, Japan) for the advanced cultivations, which were adapted from previous study isolating wide variety of sponge-associated bacteria (Jung et al., 2014), and used four different media for sub cultivation in order to cover diverse microbial species: (1) 1/10 diluted R2A medium; (2) marine medium (Difco, Franklin Lakes, NJ, USA); (3) fish extract media (0.2 g fish extract, 0.1 g yeast extract per liter); (4) sponge extract medium (0.01 g peptone, 40 ml aqueous sponge extract per liter). The sponge extract was prepared by mixing homogenized sponge tissue and sterile distilled water including 3.5% of artificial sea salt at a 1:1 (vol/vol) ratio, vortexing for 1 min, centrifugation (10 min, 8,000 rpm), and filter sterilization using a 0.2-μm pore size filter. All media except marine media were supplemented with 3.5% artificial sea salt, SEALIFE (Marine Tech, Tokyo, Japan).
CF cultivation
The overall experimental design of the cultivation experiments is shown in Figure 1. The CF with porous polyester nonwoven fabric (0.5 cm thickness; Japan Vilene, Tokyo, Japan) was set up to maintain the bacteria in an active state without overgrowing (Figure 2A), according to Fujitani et al. (2013). The bioreactor volume was 500 ml and maintained at 20°C. The reactor was aerated to provide oxygen. Due to the aeration, the liquid phase was completely mixed. A total of 200 ml of homogenized sponge tissue was used as inoculum. To prepare the inoculum, marine sponge tissue (about 150 g) was rinsed three times with sterile artificial sea water and pulverized using a sterile mortar and pestle with the same volume of sterile artificial sea water. The 1/10 diluted R2A including 3.5% of artificial sea salt was supplied from the medium chamber by a peristaltic pump at a velocity of 500 ml per day (hydraulic retention time of 24 h). Therefore, any media overflowing was automatically discharged from the bioreactor in order to reduce the concentration of metabolic by- products and overgrown biomass such as inactive cells, debris, and extracellular polymeric substances. The total organic carbon concentration in the bioreactor was measured during the experiment at intervals of three to seven days with a TOC analyzer (Shimadzu, TOC-500, Kyoto, Japan) to monitor conditions in the bioreactor.
Biomass from the nonwoven fabric material was sub-cultured on agar plate at 1, 2, 4 and 10 weeks of CF cultivation to examine temporal changes in the bacterial community. For sub-culturing, a piece of fabric material was randomly selected and retrieved from the reactor. One gram of fabric was added to 10 ml of sterile water including 3.5% of artificial sea salt and vortexed for 10 min. We then applied the homogenizer by controlling the power at the low level (less than 30% of the maximum power of the machine, Ultrasonic Homogenizer NR-50M, Microtec, Japan) for 10 seconds at a frequency of 20 kHz to dislodge biofilm and the associated bacteria from the surface of fabric material. Liquid with detached bacteria was diluted and inoculated on media described above with 1.5% agar. After one-week incubation at 20°C, 30 colonies were randomly selected for bacterial identification. The isolates identified from 4 and 10 weeks of CF cultivation (total 60) were used as the final isolates to compare among the cultivation methods.
I-tip in situ cultivation
The I-tip method allows both microbes and chemical compounds to enter from the environment, thereby better simulating the natural environment (Figure 2). Because a micropipette tip is used as main element of the I-tip device, the sharp end can be easily installed in invertebrates such as sponges. The I-tip was prepared as described previously (Jung et al., 2014). In brief, 200μL pipette tips were prepared (Eppendorf, No. 022493022, Hamburg, Germany) with the bottom part of the tip filled with acid-washed glass beads (60 to 100 μm in diameter, Sigma-Aldrich, No. G1145, St. Louis, MO, USA) to prevent invasion of larger organisms. Then 70 μl of sterilized 1/10 diluted R2A medium containing 0.7% (wt/vol) agar was added to the tip for bacterial growth. After autoclaving the assembly, the wide end of the tip was aseptically sealed with a waterproof adhesive (Cemedain, Tokyo, Japan) to prevent contamination from air and water.
For in situ cultivation using the I-tip method, sponges were kept in aquaria at the Takehara marine station of Hiroshima University (Takehara, Japan). The aquarium was cylindrical with a holding capacity of ca. 43 L (Ø = 0.35 m, d = 0.45 m), and seawater was continuously supplied. To install I-tip, silicon supporting plates (w = 3.5 cm × 1.2 cm, d = 0.6 cm) were placed on the surface of a sponge, and six I-tips (three per plate) were inserted into the specimen through the silicon plate to a depth of 0.3 cm (Figure 2D). After one week of in situ incubation, I-tips were retrieved from the sponges and then transported to the laboratory. The wide end of each I-tip was cut aseptically with a blade, and then the agar with microbes was removed using a sterile loop. The agar material from each tip was added to 1 ml of sterilized water including 3.5% of artificial sea salt, homogenized with a sterile stick, vortexed, diluted with sterile water including 3.5% artificial sea salt, and sub-cultured on media as described above with 1.5% agar for incubation at 20°C. After one week, 60 colonies were selected randomly and pure cultured for further experiments.
Identification of isolates
Taxonomic identification was carried out by sequencing 643 to 782 long fragments of the 16S rRNA gene. The colony material was used directly as a template for PCR. The 16S rRNA gene was amplified using the universal primer as follows; 27F (5’-AGAGTTTGATCCTGGCTCAG-3’) and 1492R (5’-GGTTACCTTGTTACGACTT-3’) with a KOD FX Neo system (Toyobo, Osaka, Japan), then purified with fast gene purification kit (Nippon Genetics, Tokyo, Japan). The purified PCR products were sequenced using a commercial sequencer (Takara Bio, Shiga, Japan) by fluorescent dye terminator sequencing. The sequences were compared to those available in GenBank (www.ncbi.nlm.nih.gov) databases using the MEGA program (MEGA software, Tempe, AZ, USA) to determine their closest relatives and their similarity.
DNA extraction and amplicon sequencing targeting the 16S rRNA gene
To compare the cultivated bacterial diversity with the microbial molecular signatures in the host sponge, 16S amplicon sequencing was performed based on the 16S rRNA genes. After transferring the marine sponges to the laboratory, samples were washed three times with DNA-free water and homogenized. Genomic DNA from the homogenized tissue was extracted using a FastDNA spin kit for soil (MP Biomedicals, Irvine, CA, USA) per the manufacturer’s guidelines. The extracted genomic DNA was amplified using the primers, 341F and 805R, with a Hifi hot start ready mix PCR (Kapa Biosystems, Wilmington, MA, USA), then purified using a fast gene purification kit (Nippon Genetics, Tokyo, Japan). DNA was sequenced by Hokkaido System Science (Sapporo, Japan) using an Illumina MiSeq System (Illumina, San Diego, CA, USA). Each read was assigned taxonomically using Quantitative Insights Into Microbial Ecology (QIIME) software (Kuczynski et al., 2011).
Selection of strains for further experiments
For further investigation, we selected 25, 15 and 11 strains from CF, I-tip and SDP cultivation methods, respectively, except for a few lost isolates. Each strain represents each identified OTU within the group of isolates derived from the same cultivation method.
Growth characteristics of isolated strains
Growth curves of the selected strains were analyzed to compare bacterial growth among the strains from each cultivation. Diluted R2A medium (5 mL) was inoculated with 5-20 μL of cell suspension, in triplicate, from pure liquid cultures grown, then incubated at 20°C with shaking at 150 rpm. The optical density (OD) was measured at 600 nm using a spectrophotometer (DR 3900, HACH, USA). Growth curves from OD 600 values were fitted using the nonlinear regression function with logistic model of Sigma plot software (Systat Software Inc). The specific growth rate (μ), representing the maximum rate of change, was calculated by the change in OD 600, and the saturated cell density (maximum growth) was determined by its maximum value on the fitted growth curve.
We also classified the type of growth curve for the strains into two categories. One type was a concave up growth curve (logistic growth curve) that showed exponential growth during the log phase, and the other was a concave down growth curve with the growth increments decreasing and become slower. To determine the type of growth curve, we used the values in fitted growth curves with logistic model of each tested strain above. Then these values were examined on a log scale to determine whether exponential growth occurred during log phase (concave up growth curve) or not (concave down growth curve).
Colony size of isolated strains
Colony size on agar medium (1/10 R2A) was compared among selected strains for each cultivation method. The average colony diameter of each strain was measured for selected 20 colonies on the agar plate under a digital microscope (KH-8700, Hirox Europe, France). For that, we used colonies of pure cultured strains on agar plate that grew four weeks after inoculation, and randomly selected 20 colonies. The colony diameter was compared among cultivation methods using the Kruskal-Wallis procedure and post-hoc Dunn’s tests were carried out on each pair of groups to test for significant differences between the groups (30).
Effect of sponge extract on cell recovery
The colony formation efficiency ratios between the two culture conditions (media with and without the sponge extract) were calculated for selected strains from CF and I-tip cultivation, and compared with the results for those strains from SDP cultivation to evaluate the effect of chemical compounds on cell recovery. First, one strain per OTU (defined at 97% 16S rRNA gene sequence identity) was pure cultured in 5 ml of 1:10 diluted R2A broth with 3.5% artificial sea salt at 20°C, then the liquid culture was diluted with artificial sea water (1:100) followed by incubation at 5°C for 3 days to inactivate microbes. As test strains after the pre-cultivation were diluted in 100-fold with artificial seawater, this process made microbial cells starved under low temperature, and this condition was enough for the strains from marine sponge to show a different recovery ratio according to two conditions, with and without sponge extract in a previous study (Jung et al., 2021). After 3 days, the liquid culture serial dilutions were inoculated in triplicate on two types of agar media: 1:10-diluted R2A agar medium with 0.1% (vol/vol) sponge extract and the same medium without the sponge extract. The sponge extract was added to the medium before autoclaving as we confirmed that autoclaved sponge extract remains a key function and worked for our purpose in a previous study (Jung et al., 2021). After 5 days of incubation at 20°C, the colony number ratios between the two cultivation methods (with and without the sponge extract) were calculated for each strain.
Nucleotide sequence accession numbers
Newly determined sequence data have been deposited in GenBank (www.ncbi.nlm.nih.gov) under accession numbers MK697035 to MK697065 for isolates from CF, MK689993 to MK690017 for isolates from I-tip, and MK674856 to MK674892 for isolates from SDP cultivation. The data obtained by Illumina-MiSeq sequencing (microbial community analysis) have been deposited in the DNA Data Bank of Japan (DDBJ) Sequence Read Archive (DRA) under accession number DRA008671.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author contributions
DJ, YN and YA conceived and designed the study. DJ and KM performed the laboratory experiments. DJ, JO, SH, TK, AO and YA analyzed the data and compiled the results. DJ and YA wrote the manuscript draft. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by JSPS KAKENHI Grant Numbers JP17F17098, JP19H02873, JP25630383, 26709063, JP18K19181. DJ was supported by the Japan Society for the Promotion of Science (JSPS) as an International Research Fellow of the JSPS. DJ and SH were supported by the National 111 Project of China (D16013).
Acknowledgments
The authors would like to acknowledge S. Iwasaki at Takehara Station (Fisheries Research Station), Hiroshima University for his support on operation of the aquarium.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2022.963277/full#supplementary-material
References
Amann R. I., Ludwig W., Schleifer K. H. (1995). Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59, 143–169. doi: 10.1128/mr.59.1.143-169.1995
Anjum K., Abbas S. Q., Shah S. A. A., Akhter N., Batool S., Hassan S. S. U. (2016). Marine sponges as a drug treasure. Biomol. Ther. 24, 347–362. doi: 10.4062/biomolther.2016.067
Aoi Y., Kinoshita T., Hata T., Ohta H., Obokata H., Tsuneda S. (2009). Hollow-fiber membrane chamber as a device for in situ environmental cultivation. Appl. Environ. Microbiol. 75, 3826–3833. doi: 10.1128/AEM.02542-08
Bewley C. A., Holland N. D., Faulkner D. J. (1996). Two classes of metabolites from Theonella swinhoei are localized in distinct populations of bacterial symbionts. Experientia 52, 716–722. doi: 10.1007/BF01925581
Bollmann A., Lewis K., Epstein S. S. (2007). Incubation of environmental samples in a diffusion chamber increases the diversity of recovered isolates. Appl. Environ. Microbiol. 73, 6386–6390. doi: 10.1128/AEM.01309-07
Brantley S. E., Molinski T. F., Preston C. M., DeLong E. F. (1995). Brominated acetylenic fatty acids from xestospongia sp., a marine sponge bacteria association. Tetrahedron 51, 7667–7672. doi: 10.1016/0040-4020(95)00405-W
Bruns A., Cypionka H., Overmann J. (2002). Cyclic AMP and acyl homoserine lactones increase the cultivation efficiency of heterotrophic bacteria from the central Baltic Sea. Appl. Environ. Microbiol. 68, 3978–3987. doi: 10.1128/AEM.68.8.3978-3987.2002
Chaudhary D. K., Khulan A., Kim J. (2019). Development of a novel cultivation technique for uncultured soil bacteria. Sci. Rep. 9, 6666. doi: 10.1038/s41598-019-43182-x
Date Y., Isaka K., Ikuta H., Sumino T., Kaneko N., Yoshie S., et al (2009). Microbial diversity of anammox bacteria enriched from different types of seed sludge in an anaerobic continuous-feeding cultivation reactor. J. Biosci. Bioeng. 107, 281–286. doi: 10.1016/j.jbiosc.2008.11.015
Davis K. E. R., Joseph S. J., Janssen P. H. (2005). Effects of growth medium, inoculum size, and incubation time on culturability and isolation of soil bacteria. Appl. Environ. Microbiol. 71, 826–834. doi: 10.1128/AEM.71.2.826-834.2005
Esteves A. I. S., Amer N., Nguyen M., Thomas T. (2016). Sample processing impacts the viability and cultivability of the sponge microbiome. Front. Microbiol. 7, 499. doi: 10.3389/fmicb.2016.00499
Fujitani H., Aoi Y., Tsuneda S. (2013). Selective enrichment of two different types of Nitrospira-like nitrite-oxidizing bacteria from a wastewater treatment plant. Microbes Environ. 28 (2), 236–243. doi: 10.1264/jsme2.ME12209
Fujitani H., Ushiki N., Tsuneda S., Aoi Y. (2014). Isolation of uncultured Nitrospira. Environ. Microbiol. 16, 3030–3040. doi: 10.1111/1462-2920.12248
Handelsman J. (2004). Metagenomics: application of genomics to uncultured microorganisms. Microbiol. Mol. Biol. Rev. 68, 669–685. doi: 10.1128/MMBR.68.4.669-685.2004
Hofer U. (2018). The majority is uncultured. Nat. Rev. Microbiol. 16, 716–717. doi: 10.1038/s41579-018-0097-x
Imachi H., Aoi K., Tasumi E., Saito Y., Yamanaka Y., Saito Y., et al. (2011). Cultivation of methanogenic community from subseafloor sediments using a continuous-flow bioreactor. ISME J. 5, 1913–1925. doi: 10.1038/ismej.2011.64
Isaka K., Date Y., Sumino T., Yoshie S., Tsuneda S. (2006). Growth characteristic of anaerobic ammonium-oxidizing bacteria in an anaerobic biological filtrated reactor. Appl. Microbiol. Biotechnol. 70, 47–52. doi: 10.1007/s00253-005-0046-2
Janssen P. H., Yates P. S., Grinton B. E., Taylor P. M., Sait M. (2002). Improved culturability of soil bacteria and isolation in pure culture of novel members of the divisions Acidobacteria, Actinobacteria, Proteobacteria, and Verrucomicrobia. Appl. Environ. Microbiol. 68, 2391–2396. doi: 10.1128/AEM.68.5.2391-2396.2002
Jung D., Aoi Y., Epstein S. S. (2016). In situ cultivation allows for recovery of bacterial types competitive in their natural environment. Microbes Environ. 31, 456–459. doi: 10.1264/jsme2.ME16079
Jung D., Machida K., Nakao Y., Kindaichi T., Ohashi A., Aoi Y. (2021). Triggering growth via growth initiation factors in nature: a putative mechanism for in situ cultivation of previously uncultivated microorganisms. Front. Microbiol. 12, 537194. doi: 10.3389/fmicb.2021.537194
Jung D., Seo E., Epstein S. S., Joung Y., Han J., Parfenova V. V., et al. (2014). Application of a new cultivation technology, I-tip, for studying microbial diversity in freshwater sponges of Lake Baikal, Russia. FEMS Microbiol. Ecol. 90, 417–423. doi: 10.1111/1574-6941.12399
Jung D., Seo E., Epstein S. S., Joung Y., Yim J. H., Lee H., et al. (2013). A new method for microbial cultivation and its application to bacterial community analysis in Buus Nuur, Mongolia. Fundam. Appl. Limnol. 182, 171–181. doi: 10.1127/1863-9135/2013/0391
Jung D., Seo E., Owen J. S., Aoi Y., Yong S., Lavrentyeva E. V., et al. (2018). Application of the filter plate microbial trap (FPMT), for cultivating thermophilic bacteria from thermal springs in Barguzin area, eastern Baikal, Biosci. Biotechnol. Biochem. 82, 1624–1632. doi: 10.1080/09168451.2018.1482194
Kato S., Yamagishi A., Daimon S., Kawasaki K., Tamaki H., Kitagawa W., et al. (2018). Isolation of previously uncultured slow-growing bacteria by using a simple modification in the preparation of agar media. Appl. Environ. Microbiol. 84, e00807–e00818. doi: 10.1128/AEM.00807-18
Kawasaki K., Kamagata Y. (2017). Phosphate-catalyzed hydrogen peroxide formation from agar, gellan, and κ-carrageenan and recovery of microbial cultivability via catalase and pyruvate. Appl. Environ. Microbiol. 83, e01366–e01317. doi: 10.1128/AEM.01366-17
Keren R., Lavy A., Ilan M. (2016). Increasing the richness of culturable arsenic-tolerant bacteria from Theonella swinhoei by addition of sponge skeleton to the growth medium. Microb. Ecol. 71, 873–886. doi: 10.1007/s00248-015-0726-0
Koopmans M., Martens D., Wijffels H. R. (2009). Towards commercial production of sponge medicines. Mar. Drugs 7, 787–802. doi: 10.3390/md7040787
Kuczynski J., Stombaugh J., Walters W. A., González A. A., Caporaso J. G, Knight R. (2011). Using QIIME to analyze 16S rRNA gene sequences from microbial communities. Curr. Protoc. Bioinformatics 10 10, 7. doi: 10.1002/0471250953.bi1007s36
Lloyd K. G., Steen A. D., Ladau J., Yin J., Crosby L. (2018). Phylogenetically novel uncultured microbial cells dominate earth microbiomes. mSystems 3, e00055–e00018. doi: 10.1128/mSystems.00055-18
Locey K. J., Lennon J. T. (2016). Scaling laws predict global microbial diversity. Proc. Natl. Acad. Sci. U.S.A. 113, 5970–5975. doi: 10.1073/pnas.1521291113
Martiny A. C. (2019). High proportions of bacteria are culturable across major biomes. ISME J. 13, 2125–2128. doi: 10.1038/s41396-019-0410-3
Martiny A. C. (2020). The ‘1% culturability paradigm’ needs to be carefully defined. ISME J. 14, 10–11. doi: 10.1038/s41396-019-0507-8
Mehbub F. M., Lei J., Franco C., Zhang W. (2014). Marine sponge derived natural products between 2001 and 2010: Trends and opportunities for discovery of bioactives. Mar. Drugs 12, 4539–4577. doi: 10.3390/md12084539
Rinke C., Schwientek P., Sczyrba A., Ivanova N. N., Anderson I. J., Cheng J., et al. (2013). Insights into the phylogeny and coding potential of microbial dark matter. Nature 499, 431–437. doi: 10.1038/nature12352
Schmidt E. W., Obraztsova A. Y., Davidson S. K., Faulkner D. J., Haygood M. G. (2000). Identification of the antifungal peptide-containing symbiont of the marine sponge Theonella swinhoei as a novel δ-proteobacterium, “Candidatus Entotheonella palauensis”. Mar. Biol. 136, 969–977. doi: 10.1007/s002270000273
Selvin J., Gandhimathi R., Kiran G. S., Priya S. S., Ravji T. R., Hema T. A. (2009). Culturable heterotrophic bacteria from the marine sponge Dendrilla nigra: isolation and phylogenetic diversity of actinobacteria. Helgol. Mar. Res. 63, 239–247. doi: 10.1007/s10152-009-0153-z
Sipkema D., Holmes B., Nichols S. A., Blanch H. W. (2009). Biological characterisation of Haliclona (?gellius) sp.: sponge and associated microorganisms. Microbial. Ecol. 58, 903–920. doi: 10.1007/s00248-009-9534-8
Sipkema D., Schippers K., Maalcke W. J., Yang Y., Salim S., Blanch H. W. (2011). Multiple approaches to enhance the cultivability of bacteria associated with the marine sponge Haliclona (gellius) sp. Appl. Environ. Microbiol. 77, 2130–2140. doi: 10.1128/AEM.01203-10
Staley J. T., Konopka A. (1985). Measurement of in situ activities of nonphotosynthetic microorganisms in aquatic and terrestrial habitats. Annu. Rev. Microbiol. 39, 321–346. doi: 10.1146/annurev.mi.39.100185.001541
Steen A. D., Crits-Christoph A., Carini P., DeAngelis K. M., Fierer N., Lloyd K. G., et al. (2019). High proportions of bacteria and archaea across most biomes remain uncultured. ISME J. 13, 3126–3130. doi: 10.1038/s41396-019-0484-y
Steinert G., Whitfield S., Taylor M. W., Thoms C., Schupp P. J. (2014). Application of diffusion growth chambers for the cultivation of marine sponge-associated bacteria. Mar. Biotechnol. 16, 594–603. doi: 10.1007/s10126-014-9575-y
Stewart E. J. (2012). Growing unculturable bacteria. J. Bacteriol. 194, 4151–4160. doi: 10.1128/JB.00345-12
Tamaki H., Sekiguchi Y., Hanada S., Nakamura K., Nomura N., Matsumura M., et al. (2005). Comparative analysis of bacterial diversity in freshwater sediment of a shallow eutrophic lake by molecular and improved cultivation-based techniques. Appl. Environ. Microbiol. 71, 2162–2169. doi: 10.1128/AEM.71.4.2162-2169.2005
Tanaka T., Kawasaki K., Daimon S., Kitagawa W., Yamamoto K., Tamaki H., et al. (2014). A hidden pitfall in the preparation of agar media undermines microorganism cultivability. Appl. Environ. Microbiol. 80, 7659–7666. doi: 10.1128/AEM.02741-14
Vartoukian S. R., Adamowska A., Lawlor M., Moazzez R., Dewhirst F. E., Wade W. G. (2016). In vitro cultivation of “Unculturable” oral bacteria, facilitated by community culture and media supplementation with siderophores. PloS One 11, e0146926. doi: 10.1371/journal.pone.0146926
Wang G. (2006). Diversity and biotechnological potential of the sponge-associated microbial consortia. J. Ind. Microbiol. Biotechnol. 33, 545–551. doi: 10.1007/s10295-006-0123-2
Keywords: uncultured microbes, cultivation, continuous-flow bioreactor, slow-growing bacteria, in situ cultivation, I-tip cultivation
Citation: Jung D, Machida K, Nakao Y, Owen JS, He S, Kindaichi T, Ohashi A and Aoi Y (2022) Cultivation of previously uncultured sponge-associated bacteria using advanced cultivation techniques: A perspective on possible key mechanisms. Front. Mar. Sci. 9:963277. doi: 10.3389/fmars.2022.963277
Received: 07 June 2022; Accepted: 12 August 2022;
Published: 30 August 2022.
Edited by:
Tony Gutierrez, Heriot-Watt University, United KingdomReviewed by:
Dagmar Hajkova Leary, United States Naval Research Laboratory, United StatesCristiane Cassiolato Pires Hardoim, São Paulo State University, Brazil
Copyright © 2022 Jung, Machida, Nakao, Owen, He, Kindaichi, Ohashi and Aoi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yoshiteru Aoi, eW9zaGl0ZXJ1YW9pQGhpcm9zaGltYS11LmFjLmpw
†Present addresses: Dawoon Jung, Li Dak Sum Yip Yio Chin Kenneth Li Marine Biopharmaceutical Research Center, College of Food and Pharmaceutical Sciences, Ningbo University, Ningbo, China;
Ningbo Institute of Marine Medicine, Peking University, Ningbo, China
 Dawoon Jung
Dawoon Jung Koshi Machida
Koshi Machida Yoichi Nakao
Yoichi Nakao Jeffrey S. Owen
Jeffrey S. Owen Shan He
Shan He Tomonori Kindaichi
Tomonori Kindaichi Akiyoshi Ohashi
Akiyoshi Ohashi Yoshiteru Aoi
Yoshiteru Aoi