
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
METHODS article
Front. Mar. Sci. , 23 September 2022
Sec. Marine Conservation and Sustainability
Volume 9 - 2022 | https://doi.org/10.3389/fmars.2022.961095
This article is part of the Research Topic Contributions of Zoos and Aquariums to the Advancement of Marine Science View all 14 articles
 Ashleigh M. Westphal1*
Ashleigh M. Westphal1* C-Jae C. Breiter1,2
C-Jae C. Breiter1,2 Sarah Falconer1
Sarah Falconer1 Najmeh Saffar3
Najmeh Saffar3 Ahmed B. Ashraf3
Ahmed B. Ashraf3 Alysa G. McCall4
Alysa G. McCall4 Kieran McIver4
Kieran McIver4 Stephen D. Petersen1
Stephen D. Petersen1Successful conservation efforts often require novel tactics to achieve the desired goals of protecting species and habitats. One such tactic is to develop an interdisciplinary, collaborative approach to ensure that conservation initiatives are science-based, scalable, and goal-oriented. This approach may be particularly beneficial to wildlife monitoring, as there is often a mismatch between where monitoring is required and where resources are available. We can bridge that gap by bringing together diverse partners, technologies, and global resources to expand monitoring efforts and use tools where they are needed most. Here, we describe a successful interdisciplinary, collaborative approach to long-term monitoring of beluga whales (Delphinapterus leucas) and their marine ecosystem. Our approach includes extracting images from video data collected through partnerships with other organizations who live-stream educational nature content worldwide. This video has resulted in an average of 96,000 underwater images annually. However, due to the frame extraction process, many images show only water. We have therefore incorporated an automated data filtering step using machine learning models to identify frames that include beluga, which filtered out an annual average of 67.9% of frames labelled as “empty” (no beluga) with a classification accuracy of 97%. The final image datasets were then classified by citizen scientists on the Beluga Bits project on Zooniverse (https://www.zooniverse.org). Since 2016, more than 20,000 registered users have provided nearly 5 million classifications on our Zooniverse workflows. Classified images are then used in various researcher-led projects. The benefits of this approach have been multifold. The combination of machine learning tools followed by citizen science participation has increased our analysis capabilities and the utilization of hundreds of hours of video collected each year. Our successes to date include the photo-documentation of a previously tagged beluga and of the common northern comb jellyfish (Bolinopsis infundibulum), an unreported species in Hudson Bay. Given the success of this program, we recommend other conservation initiatives adopt an interdisciplinary, collaborative approach to increase the success of their monitoring programs.
Modern threats to species and habitats demand multi-faceted and collaborative solutions that engage a diverse set of stakeholders and expertise. Given the complexities of many conservation challenges, several authors have proposed solutions that incorporate multiple disciplines, collaborative frameworks, and diverse perspectives to find solutions that fit each issue (Meffe and Viederman, 1995; Naiman, 1999; Dick et al., 2016). Unprecedented shifts in global climate, habitat loss, and the rapid loss of biodiversity emphasizes the need for interdisciplinary approaches now (Dick et al., 2016; Berger et al., 2019). There are a number of successful examples that integrate multiple disciplines to enhance conservation actions for habitats and land use (e.g., reducing deforestation in Amazonia, Hecht, 2011; climate change risk within land and water management, Lanier et al., 2018; interdisciplinary approach to preventing the biodiversity loss, Nakaoka et al., 2018) as well as protecting individual species (grizzly bear Ursus arctos, Rutherford et al., 2009; hippopotamus Hippopotamus amphibius Sheppard et al., 2010). The nature of interdisciplinary solutions is that many solutions may exist, therefore, documenting and sharing successful methods increases the opportunities for tailored solutions to be created for other species and habitats (Pooley et al., 2013; Dick et al., 2016).
Monitoring is an essential part of many successful conservation programs as it allows for adaptive strategies and rapid detection of changes. However, monitoring often receives limited support after the initial conservation action and can be difficult to prioritize when a species or habitat is not currently facing a threat. There is a growing emphasis on improving monitoring efforts to maximize conservation outcomes, exemplified by initiatives put forward by the International Union for Conservation of Nature (IUCN) Species Monitoring Specialist Group (Stephenson, 2018). However, conservation monitoring is difficult and often limited by high costs, lack of trained personnel, or low detectability of target species (McDonald-Madden et al., 2010; Linchant et al., 2015; Rovang et al., 2015). Remote locations pose an even greater challenge as even when resources are available, access to the study site may impede proper monitoring. Therefore, incorporating new disciplines and novel technologies into monitoring efforts can increase their effectiveness, scope, and reliability.
Citizen science has been recognized for increasing the capacity of monitoring projects (Chandler et al., 2017). Citizen science, also referred to as community science or participatory science, involves participants contributing to the scientific process through observations, indexing, and even analyzing data (Conrad and Hilchey, 2011). The benefits of citizen science are multifold for both researchers and participants. Harnessing the efforts of volunteers allows researchers to expand their data processing capabilities and can provide data resources for researching and developing machine learning tools (Swanson et al., 2015; Willi et al., 2019; Anton et al., 2021). Additionally, citizen science allows volunteers to meaningfully contribute to scientific endeavors, aiding in increasing scientific literacy and trust in scientific processes (Tulloch et al., 2013). There have been notable examples of citizen science projects resulting in effective conservation management, including detecting population declines of monarch butterflies (Danaus plexippus; Schultz et al., 2017), implementing policies to protect British breeding birds (Greenwood, 2003), and monitoring killer whale (Orcinus orca) populations (Towers et al., 2019).
Rapid improvement in passive monitoring and data storage technology have also contributed to monitoring efforts by dramatically increasing the size and quality of conservation datasets. Additionally, technological advances have expanded the variety and availability of non- or minimally invasive means of monitoring (Marvin et al., 2016; Stephenson, 2018). There are a growing number of examples where passive acoustic monitoring (Wrege et al., 2017; Wijers et al., 2019), satellite or remote sensing (Luque et al., 2018; Ashutosh and Roy, 2021), and drone monitoring (Burke et al., 2019; Lopez and Mulero-Pazmany, 2019; Harasyn et al., 2022) are used to increase monitoring effort while remaining cost-effective and minimally invasive to the study species. Large datasets collected using these methods provide conservation scientists and practitioners with opportunities to explore new research questions; however, manually processing these datasets can become extremely time-consuming, costly, and may increase the likelihood of human error. Passive monitoring systems need to be implemented in tandem with solutions that allow rapid processing to ensure these datasets can be effectively used to maximize benefit.
Machine learning, a subset of the broader term of artificial intelligence, is an emerging tool that can address the challenges of processing large datasets for conservation projects (Lamba et al., 2019). Deep learning models, a part of machine learning, and in particular convolutional neural networks (CNN), are able to automatically learn useful feature representations at various levels of pixel organization (Krizhevsky et al., 2017). As a result, CNNs have shown highly accurate results for image recognition and object detection in a variety of applications (Simonyan and Zisserman, 2015; He et al., 2016; Krizhevsky et al., 2017). Biologists and land managers have already begun to employ these models in a variety of contexts, including detecting, identifying, and counting a variety of wildlife species in camera trap images (Norouzzadeh et al., 2018; Tabak et al., 2019; Willi et al., 2019) and underwater video (Siddiqui et al., 2018; Lopez-Vazquez et al., 2020; Anton et al., 2021). Effectively applying machine learning tools like deep learning models to automate essential but time-consuming classifying tasks can greatly reduce the labor and time-costs of initial data processing. However, deep learning in conservation is still a relatively novel application and there is the opportunity to develop this discipline further.
The need for monitoring Arctic species and ecosystems is growing as northern regions are being disproportionately affected by climate change (Dunham et al., 2021). At the same time, studying Arctic species remains particularly challenging due to extreme weather, remoteness, and limited infrastructure (Høye, 2020). This is compounded when studying Arctic marine mammals as they migrate long distances, can be distributed widely across the seascape, and are submerged and out of view of researchers for most of their life (Simpkins et al., 2009). There have been successful non-invasive monitoring projects in the Arctic using developing technology, for example the use of environmental DNA (eDNA; Lacoursière-Roussel et al., 2018), acoustic monitoring (Marcoux et al., 2017), or drone surveliance (Eischeid et al., 2021); however, each of these methods used exclusively can be inhibited by cost, access to the habitat or species, and may provide limited information about the species of interest. Thus, Arctic species and ecosystems can particularly benefit from interdisciplinary, collaborative approaches to enhance monitoring efforts. In fact, an international collaborative framework for studying Arctic marine mammals specifically recognized the need for multi-disciplinary studies to enhance monitoring in these regions (Simpkins et al., 2009).
Here, we describe a successful multi-year interdisciplinary, collaborative project for monitoring beluga whales (Delphinapterus leucas) and their marine ecosystem in the Churchill River estuary near Churchill, Manitoba, Canada. We use the terms “interdisciplinary” and “collaborative” intentionally, as our study integrates multiple disciplines to reach a cohesive goal and involves contributions from several stakeholders. This interdisciplinary approach requires collaboration with environmental organizations, biologists and wildlife professionals, research scientists, educators, and community members. Key objectives for this research project include: 1) Developing a multi-year photo database of beluga within the Churchill River estuary, 2) identifying individual beluga using distinctive markings, and 3) monitoring beluga and broader ecosystem health. We will provide detail on the methods we employed as well as the numerous outcomes of this approach. We hope that this project demonstrates another example of a successful interdisciplinary, collaborative monitoring strategy, while providing a framework for other researchers to implement.
The Churchill River estuary (58.737138, -94.202522) is one of three major estuaries along the coastline of western Hudson Bay where large annual aggregations of the Western Hudson Bay beluga population occur (Figure 1; Matthews et al., 2017). This beluga population migrate seasonally from wintering areas in Hudson and Davis Strait to southern coastal regions and estuaries in western Hudson Bay (NAMMCO, 2018).The Churchill River estuary is approximately 13 kilometers in length and 3 km wide at high tide (Kuzyk et al., 2008). Hudson Bay experiences a complete annual freeze-thaw cycle, with ice formation in the Churchill River estuary typically beginning in October-November and break-up in May-June (Kuzyk et al., 2008). Large inputs of freshwater mix with the marine waters of Hudson Bay to produce an ecologically rich area that supports a complex food web. The diversity and abundance of animals in the area has given rise to a thriving tourism industry based in the town of Churchill that attracts over 530,000 people annually (Greenslade, 2018).
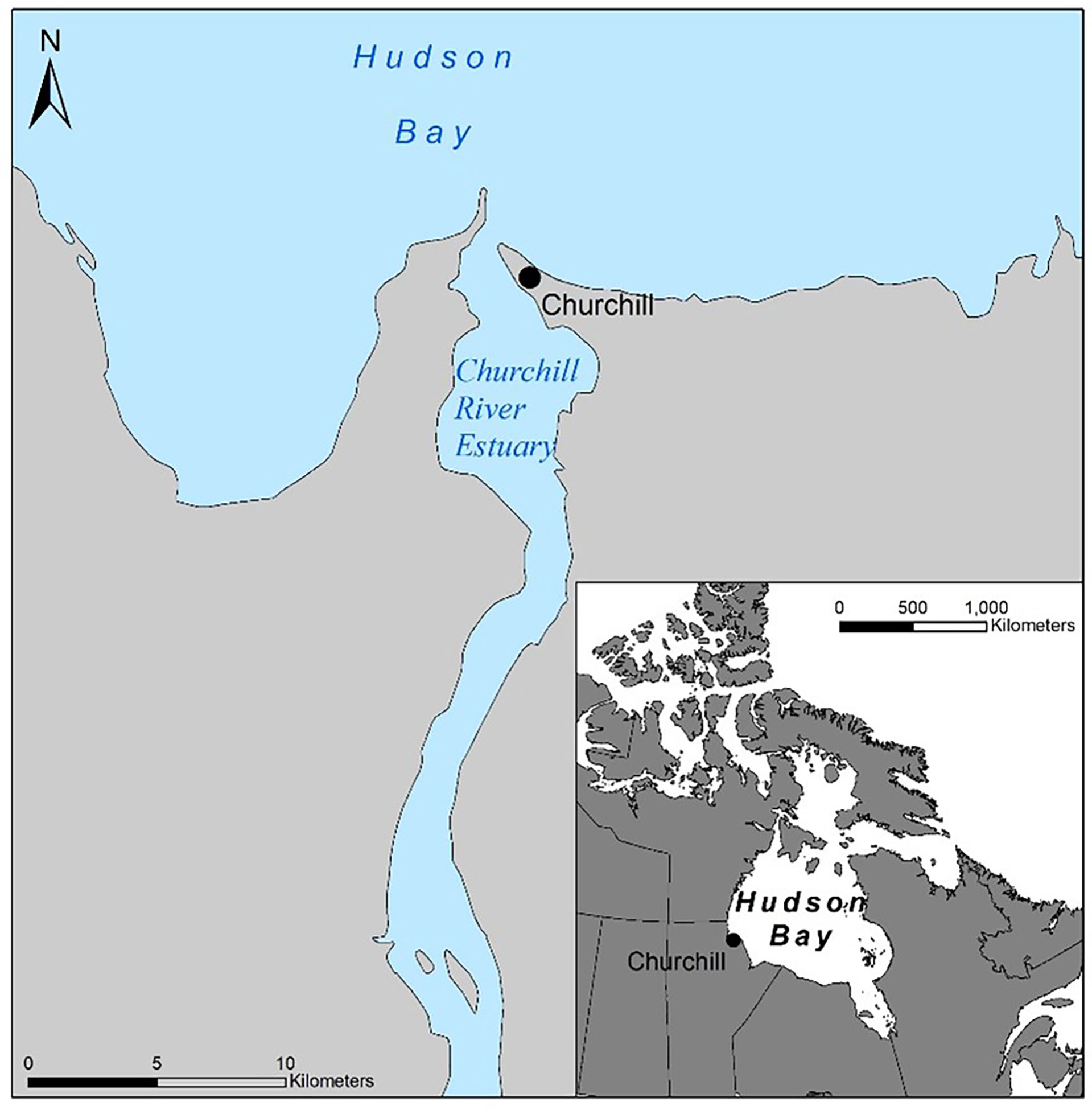
Figure 1 Map of the Churchill River estuary in northern Manitoba, Canada. Base maps were obtained from the Government of Manitoba – Manitoba Land Initiative (Retrieved from: mli2.gov.mb.ca/mli_data/index.html).
Beluga whales are a highly social, medium-sized species of toothed whale and the only living member of their genus. They are endemic to higher latitudes within the Northern Hemisphere and found throughout the circumpolar Arctic and sub-Arctic (NAMMCO, 2018). Beluga whales are an ice-adapted species; they lack a dorsal fin, permitting them to surface between ice flows and minimizing heat loss (O’Corry-Crowe, 2008). Globally, many populations of beluga are threatened by direct and indirect effects of climate change. Indirect effects include increased human activity in the north with accompanying increases in noise (Halliday et al., 2019; Vergara et al., 2021) and pollution (Moore et al., 2020), while direct effects include changing sea ice conditions, shifting prey availability, and increased exposure to novel pathogens (Lair et al., 2016).
Polar Bears International, a non-profit conservation organization, and explore.org, an online multimedia platform, manages field data collection which has occurred since 2016 in the Churchill River estuary and adjacent waters of Hudson Bay. The observation platform is either a 5.8 meter aluminum hull boat or a 1.2 m inflatable hull boat with an underwater camera mounted to the stern or side, respectively. The boat operates for approximately four hours each day from two hours before high tide to two hours after high tide. Trips occur daily from early to mid-July until the first week of September, weather dependent, every year.
The vessel and camera equipment underwent several changes and upgrades since the initiation of the project; however, basic setup remains effectively the same and, anecdotally, the behavior of whales around the boat has not changed. The setup includes an underwater camera capturing video footage below the surface, a hydrophone which records beluga vocalizations, and microphones on-board for the vessel operator and onboard guests to commentate during the tour. The vessel operator’s objective is to maneuver the boat into the vicinity of whales without disturbing or altering their behavior and remain in that area either by drifting or idling to maintain position. Views of the whales on the camera are entirely reliant on whales choosing to approach and/or follow the boat and are permitted each year by Fisheries and Oceans Canada (FWI-ACC-2018-59, FWI-ACC-2019-17, FWI-ACC-2020-13, FWI-ACC-2021-23).
Live-stream video broadcast on explore.org1 occurs for the entirety of the 4-hour trip each day. Additionally, when possible the video is archived for further analysis. Remote volunteers from explore.org continually monitor the video/audio feeds, notifying the vessel operator if technical issues arise during the live-stream. The vessel operator navigates the boat in the areas of the Churchill River estuary and adjacent Hudson Bay where Wi-Fi signal strength can be maintained. During each tour the vessel operator records GPS tracks of their movements.
Our photo dataset is created by two means: 1) images taken by viewers watching the live-stream on explore.org, which we will refer to as snapshots, and 2) still frames extracted from raw video footage, hereafter referred to as frames. During the live-stream, moderators and viewers watch and collect snapshots of beluga or other objects in the water. At the end of the field season, all snapshots and video are shared with researchers from Assiniboine Park Zoo, in Winnipeg, Manitoba, Canada. Frames from the video footage are subsampled at a rate of one frame for every three seconds (2016-2020) or one frame per second (2021). Frames are extracted using the av (Ooms, 2022) package in R (R Core Team, 2021).
Snapshots and frames are classified as part of the Beluga Bits2 citizen science research project created and managed by Assiniboine Park Zoo researchers. The Beluga Bits project is hosted on the Zooniverse platform. Beluga Bits was developed specifically for Zooniverse to engage citizen scientists around the world in answering questions about the life history, social structure, health, and threats of beluga whales inhabiting the Churchill River estuary. We launched the project on Zooniverse in 2017 and recruited citizen scientists primarily through the platform, social media, and Zoo-based educational programs and presentations.
Image data on Beluga Bits is processed through a hierarchical series of workflows, where initial workflows are used to filter images into subsequent workflows of increasing specificity. All images are first processed as part of a “General Photo Classification” workflow, where participants annotate images by quality, whether a beluga is present, and content before being filtered into more question-driven workflows. We design our Zooniverse workflows to guide citizen scientists through the image classification process and provide the tools necessary for accurate assessment. Participants are presented with a tutorial for each workflow that provides written instructions and visual examples demonstrating how to complete the tasks asked of them. Moreover, task responses frequently include an option that participants can select if they are uncertain. Additional resources that are not workflow-specific are available as information pages, FAQs, or “talk” forum discussions.
To assess the reliability and accuracy of citizen scientist classification, we used images classified within the “General Photo Classification” workflow. We included images collected between 2016-2021 and considered each year of data collection separately to account for differences that field conditions (e.g., water color and clarity) and/or workflow structure (i.e., providing Yes/No responses vs. placing markers on individual beluga) may have had on agreement among participants. Each image was classified by a minimum of 10 participants. We evaluated the agreement among participants using a Fleiss’ kappa (Fleiss, 1971) measure implemented in the irr package (Gamer et al., 2019) in R. We then considered the accuracy of participant responses by comparing them with classifications provided by researchers. We created a subset of images that had been classified by both a researcher and participants, filtering for images where 70% or more participants agreed on whether or not a beluga was present. This resulted in a dataset of 1,936 images. We then compared classifications provided by researchers with the aggregated response from participants using a confusion matrix (caret package, Kuhn, 2022) in R.
Extracting frames from the video allows us to greatly expand our dataset; however, it produces a large number of images that do not contain species of interest. Anecdotally, this lack of beluga images reduces the level of participation of citizen scientists during the image classification steps. Therefore, to increase data processing efficiency and maintain participant interest, we developed convolutional neural networks to sort frames that contain beluga whales from empty (just water) images.
We collected images to train the deep learning model from the “General Photo Classification” workflow. Each photo was seen by 10 participants. A curated, balanced set of 12,678 images where 100% of participants agreed on presence or absence of beluga was selected to train and test the model. We used cross-validation to evaluate the generalization of our trained models. Cross-validation involves dividing the dataset into partitions (commonly referred to as folds), wherein multiple models are trained and evaluated by letting different folds assume the role of training and validation sets (Bishop, 2006). We carried out the two-fold cross validation by randomly shuffling the dataset into two subsets of equal size, designated as D0 (first fold) and D1 (second fold). We then trained deep learning models on D0 and validated it using D1, followed by training on D1 and validating on D0. To establish a deep learning baseline for classifying image frames with and without beluga, we selected three established CNN architectures for testing. We implemented our code in Python (using PyTorch framework; Paszke et al., 2019). All the models were trained on the computing platform with the following specifications: Intel Core i9 10th generation processor with 256 GB RAM, and 48 GB NVIDIA Quadro RTX 8000 GPU. For training, we used the following parameters: a stochastic gradient descent optimizer with momentum of 0.9, 50 epochs, learning rate of 0.001, and batch size of 64 (Goodfellow et al., 2016).
CNN architectures, including AlexNet (Krizhevsky et al., 2017), VGG-16 (Simonyan and Zisserman, 2015), and ResNet50 (He et al., 2016) were used. Deep neural networks can either be trained for the task from scratch or models that have been pre-trained on a large publicly available dataset, such as ImageNet, can be used to improve the training accuracy and speed of new models (Deng et al., 2009). The latter approach is referred to as transfer learning (Ribani and Marengoni, 2019). The extracted features of the images on the pre-trained network can be transferred to the new task and do not have to be learned again. In our work, we have employed transfer learning with two of the architectures, AlexNet and ResNet50. For VGG-16, we incorporated attention mechanisms to further improve the performance (Jetley et al., 2018). Attention mechanisms in deep neural networks are a class of methods through which the neural network can learn to pay attention to certain parts of the image based on the context and the task.
AlexNet (Krizhevsky et al., 2017) is composed of five convolutional layers followed by max pooling and three fully connected layers (Figure 2). It has the fewest number of layers compared to the other two architectures, VGG-16 and ResNet-50, while it uses larger receptive fields.
The second architecture we trained was VGG-16 combined with trainable attention modules (Figure 3). VGG architectures have deeper networks, but with smaller filters. This enhances their representational powers to implement nonlinear functions (Simonyan and Zisserman, 2015). This can assist the networks in discriminating between different classes more efficiently. Additionally, we applied an attention mechanism to help the model focus on critical and salient regions most pertinent to the object of interest (Jetley et al., 2018). We can gain insight into where the model focuses when it makes predictions by looking at the attention estimator output of the image frames, paying particular attention to images the network struggles to identify or incorrectly assigned class labels (Figure 3).
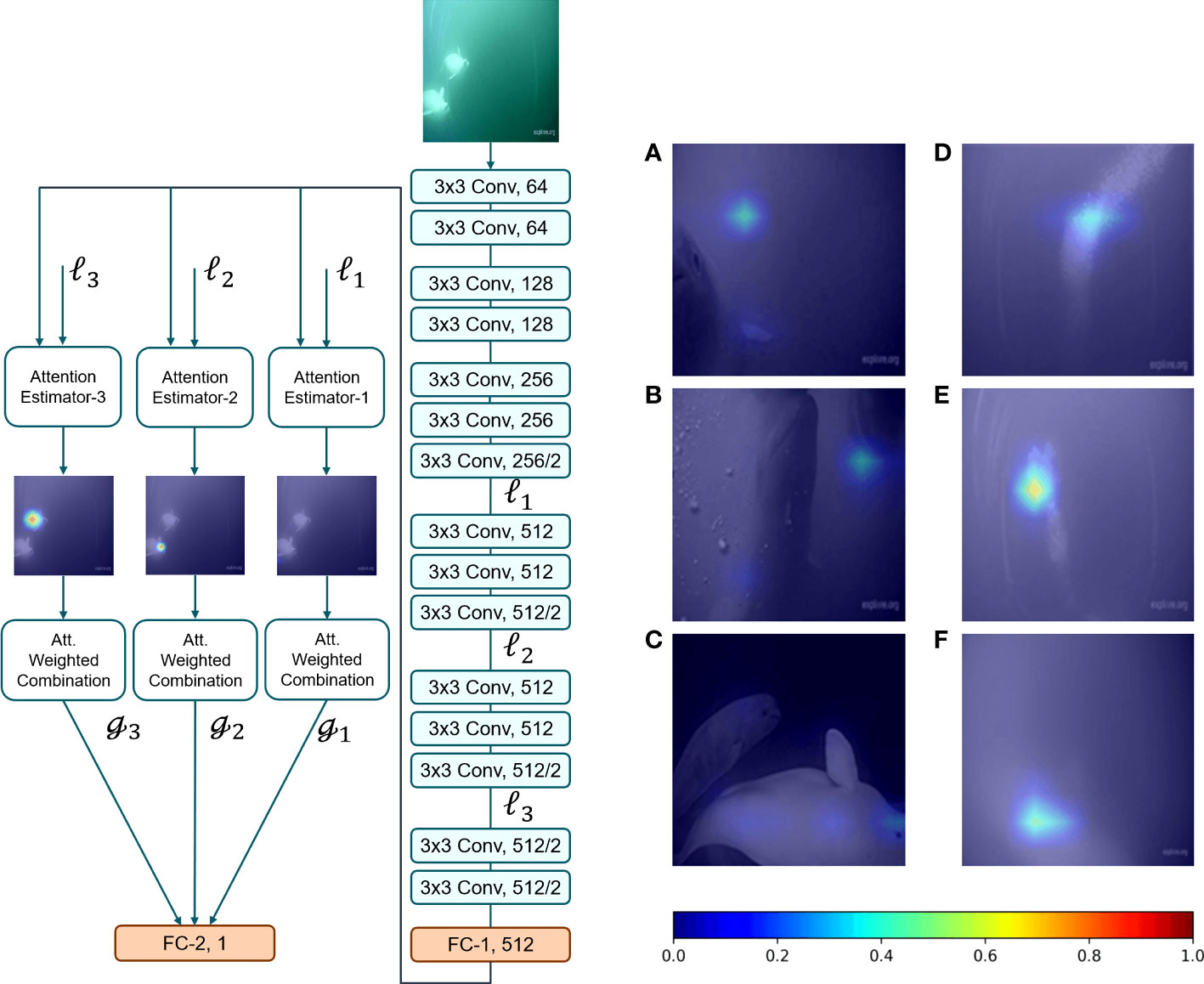
Figure 3 Left: The architecture of VGG-16 with the attention module. Right: Panels (A–C) show correctly classified images that contain beluga and (D–F) show incorrectly classified images. The colour bar defines regions with varying attention values, where the blue and red extremes signify lower and higher attention values, respectively.
The attention estimator masks shown above demonstrate that the neural network learns to pay attention to only those regions of the image which contain beluga whales. Right: Panels A-C are the example frames that contained belugas and were predicted correctly, but were challenging for the network. Panels D-F are the example frames that were incorrectly classified by the network, in this instance, bubbles were classified as beluga. The color bar defines regions with varying attention values, where the blue and red ends of the spectrum signify lower and higher attention values, respectively.
The final architecture tested in our study was ResNet-50, a fifty-layer deep convolutional neural network known as a residual network (He et al., 2016; Figure 4). A deeper network increases the efficiency of learning a more complex function; however, as a network goes deeper, performance will eventually drop due to vanishing gradient problems. Skip connection has been introduced in residual networks to address this.
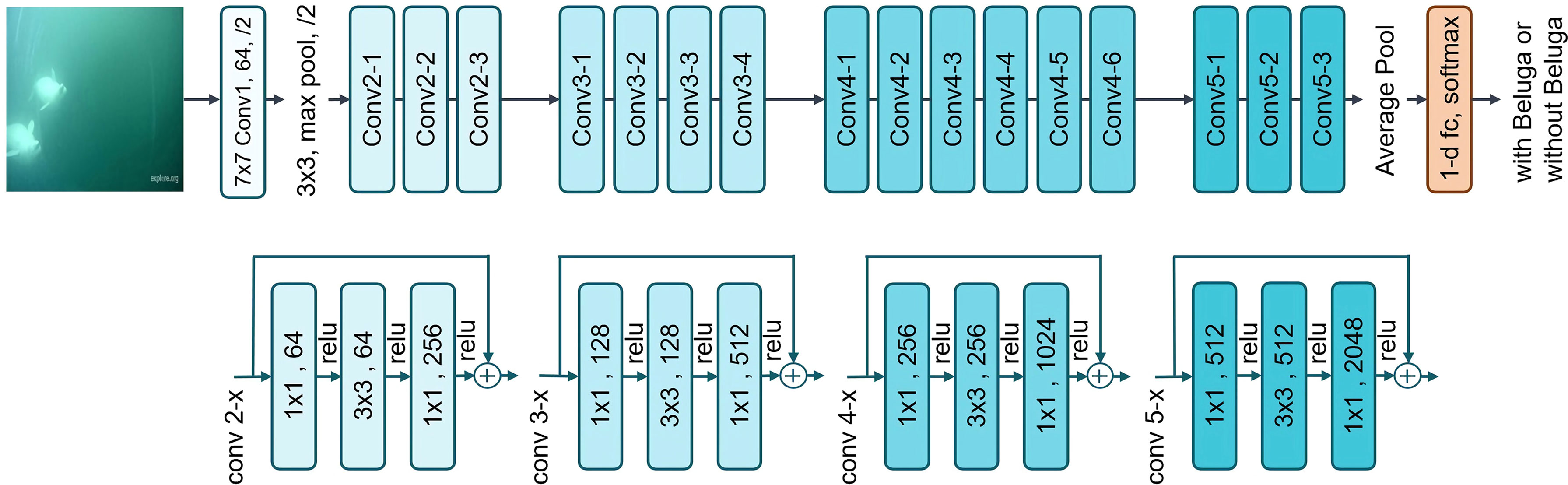
Figure 4 Schematic of a ResNet-50 architecture with skip connections, adding the input of each convolution block to its output.
Once a final model was developed, we used it to classify whether frames did or did not contain a beluga on new image datasets. Images in which the model was at least 90% confident a beluga was present were processed and uploaded to the Beluga Bits project on Zooniverse.
After the initial photo processing using deep learning, our team has addressed more specific questions about belugas and their environment using curated photo datasets. Many of these questions were based on initial observations and comments made by citizen scientists and workflow testers in earlier forms of the project. In 2019, the “General Photo Classification” workflow was updated to ask participants to answer questions related to what parts of the whale are in view. For example, the question “Can you see the underside of any beluga in this photo” identified images that could later be used to determine sex by genitalia. In this workflow, users were also asked “Do any of the beluga in this photo have major wounds or identifiable marks?”, which would later be used to identify individuals based on unique markings. Additionally, after a number of observations from citizen scientists of jellyfish (Cnidaria) and jellies (Ctenophora) in the photographs, a targeted workflow called “Is that jellyfish?” was created in 2020 to assist researchers in counting and identifying jellyfish and jelly species within the estuary. Researchers initially created the workflow and provided educational materials to record three species (Lion’s Mane Cyanea capillata, moon jellyfish Aurelia aurita, and Arctic comb jelly Mertensia ovum) that had been confirmed within the estuary.
Live-stream viewership and engagement on explore.org varied across the years of the project (Table 1). The first year of the project saw its highest number of participants with 276 unique usernames contributing snapshots for the Beluga Bits project. In 2020, poor water clarity and technical difficulties likely contributed to lower engagement, with just 30 users contributing snapshots. Participation on Beluga Bits has grown continually since its launch on Zooniverse (Figure 5).
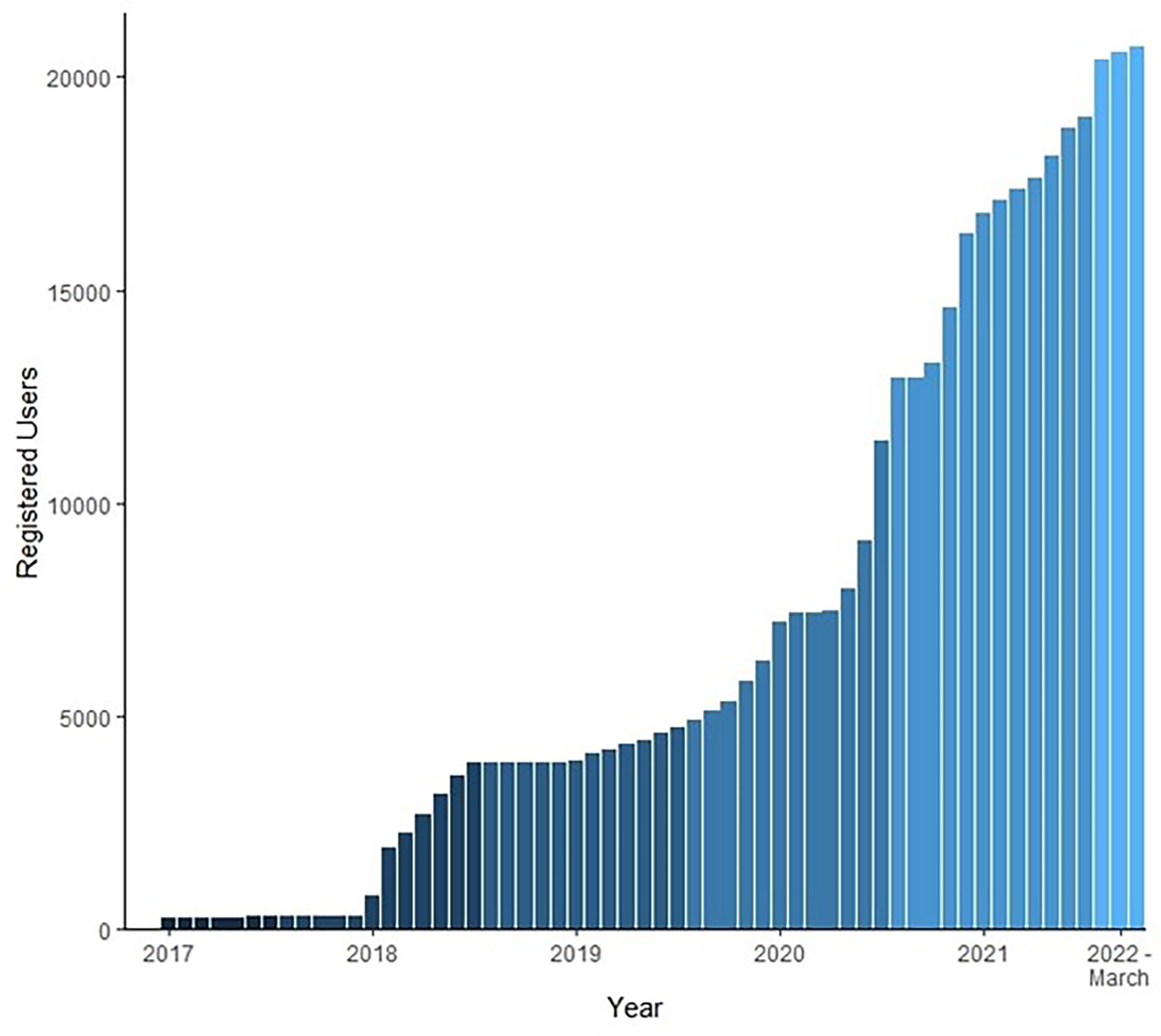
Figure 5 Total registered users contributing to Beluga Bits since the project’s inception. Bars represent cumulative users per month per year. Beluga Bits project officially launched on Zooniverse in April, 2017.
We extracted frames from video in each year of the project; however, the amount of video available to be subsampled varied each season for logistical and technical reasons. On average, 67.9% (range: 55.1-77.4%) of frames were designated as empty (i.e., did not contain beluga) and were removed from the dataset. The remaining frames and snapshots were uploaded to the Beluga Bits project on Zooniverse.
Agreement among participants was stronger in more recent years of the project (Table 2). Frames collected during years which had generally higher water clarity (2016, 2019, and 2021) did not necessarily have stronger levels of agreement compared to those which had lower clarity (2017, 2018, and 2020).

Table 2 Summary of agreement among citizen scientist responses as measured by Fleiss’ K (Fleiss, 1971).
Participants had an accuracy of 70.4% (95% CI: 68.3%, 72.4%) and moderate agreement (K = 0.396, Landis and Koch, 1977) when compared with researcher responses (Table 3). Citizen scientists were better at correctly determining when beluga were not present (74.2% accuracy) compared to when they were (65.4%).

Table 3 Confusion matrix comparing the aggregated responses of citizen scientists with researcher responses for a subset of images.
Three CNN architectures were trained and tested for the task of classifying whether frames did or did not contain beluga whales. All the models performed well on both the training and testing datasets. We report two performance metrics: 1) the classification accuracy, with confusion matrices, and 2) the Area Under Receiver Operating Characteristics curve (ROC-AUC). The ROC-AUC did not substantially differ between the models, all with an Area Under Curve (AUC) of greater than 0.99 for the testing data and 1.00 for the training data (Table 4). Among all networks that we used, the best performance was achieved by VGG-16 architecture using attention modules with an AUC of 0.99 and the class accuracies of 97.48% (with beluga) and 98.22% (without beluga) (Figure 6).
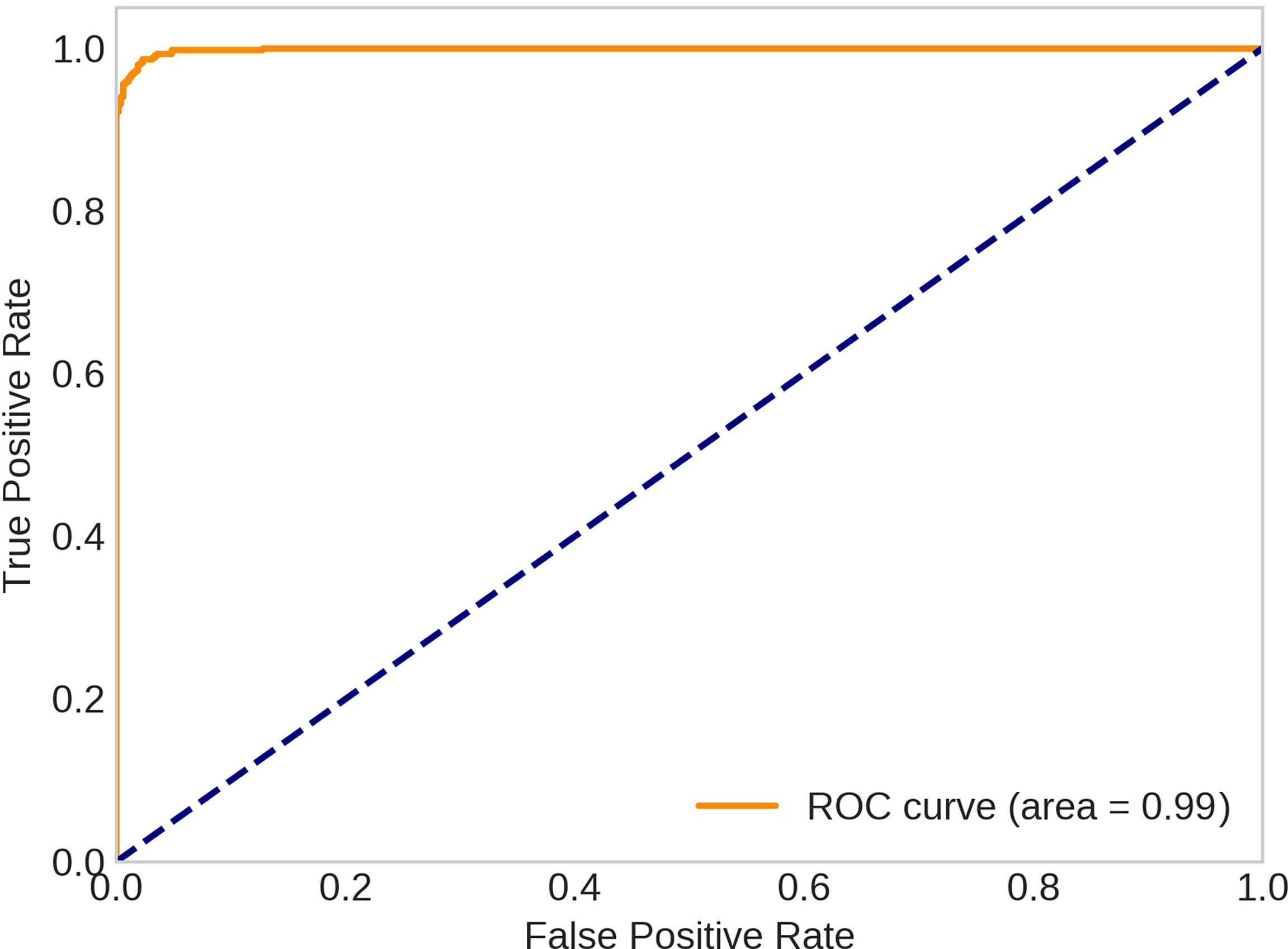
Figure 6 ROC curves for VGG-16 with attention model (trained on the first fold of the dataset and tested on the second fold).
Using remarks from citizen scientists from the “talk” forum discussions on Zooniverse, there have been a number of interesting observations noted about beluga and their environment. Often, these observations were explored further through discussions with project partners and experts in their fields. For example, citizen scientists highlighted a photo of a whale from 2016 that had a series of dots along the right side of its dorsal ridge. This same whale was later resighted by researchers in both 2019 and 2021 during boat-based observations because of this distinctive dotted lesion at the same location on its body. Likewise, in 2020 a participant commented on a photo of a beluga that had distinctive markings that they suspected was a result of an injury due to a boat propeller. After consulting with researchers studying wild beluga, it was determined that this whale’s scars were created by a previously attached satellite-tracking device and the observation subsequently published in Ryan et al. (2022).
Soon after the launch of the “Is that a Jellyfish?” workflow, citizen scientists highlighted images containing species that did not match the descriptions of those we had previously identified. These discussions, with consultation with jellyfish researchers, ultimately confirmed the presence of two jellyfish and three jelly species in the estuary that had been captured in our images. Two of these jelly species had not been noted in the original workflow materials, the first being the melon comb jelly (Beroe cucumis), a comb jelly that has been observed across northern Atlantic and Pacific coastlines. The second species, the common northern comb jelly (Bolinopsis infundibulum), has not previously been photo-documented or recorded within Hudson Bay to the best of our knowledge.
Other than these important observations from the photo database that were revealed by citizen scientists, there are a number of other observations that have been indicated to researchers that require further exploration and may dictate future research objectives such as the high incidence of curved flippers, wounding around the mouth, interesting skin and moulting patterns, and the timing and presence of calves with fetal folds.
Our results demonstrate the value that an interdisciplinary approach has brought to this project, particularly in the integration of wildlife biology, citizen science, and computer science. In collaborating with project partners and citizen scientists, we were able to collect thousands of images each year to index and classify. Overall, citizen scientists had a classification accuracy of 70.4% (95% CI: 68.3%, 72.4%) when determining the presence or absence of beluga in frames. Agreement on presence and absence of beluga did vary from year to year, with a higher agreement in more recent years. This variation may be due in part to improvements to workflow structure over time, as well as varying water quality amoung years. Of the three deep learning architectures tested, the VGG-16 architecture had the highest class accuracy of 97.48% (with beluga) and 98.22% (without beluga). The two other architectures tested also had high class accuracies, all of which were over 96%. Our project demonstrates that large-scale image sorting of underwater images of marine life is viable through deep learning models. This could be the first of many such applications towards wildlife research, where building efficient and engaging processes allow for increased focus on research questions and monitoring. Ultimately, this will aid in supporting wildlife management and conservation.
While the CNN models were more accurate than citizen scientists in predicting whether belugas were present, this does not discredit the value of these participants in our project. For instance, although the CNN models performed well in determining the presence or absence of beluga, there are more complicated questions we are addressing on Zooniverse. This includes questions that would be challenging for machine learning models to correctly identify, such as classifying unique markings on beluga. Moreover, interactions between participants and researchers on Zooniverse through forum discussions have led to crucial discoveries and project developments (i.e., new jellyfish species, the previously tagged individual). Finally, the aggregated responses from citizen scientists were necessary for testing and training the computer models. This data integration among disciplines highlights an aspect of this approach that we consider essential to the success of our project. For our project, multiple disciplines have been integrated with each other, leading to the development of a shared framework. Citizen scientist classifications would be necessary for any further model development for this project and, in turn, the output from the models could create more engaging workflows for participants in the future. We suggest that other projects that are considering interdisciplinary methodologies consider how their disciplines and partnerships can build on each other cohesively to maximize success.
A key objective of the Beluga Bits project was to create a long-term photo database of beluga within the Churchill River estuary to identify distinctive markings that would identify individual beluga, potential threats to belugas, and changes in the ecosystem. Here we have had several successes capitalizing on observations from citizen scientists. Scars or other markings on animals can be used to identify individuals, but they have also been used to provide insights into broader ecosystem health and emerging threats (Aguirre and Lutz, 2004; LaDue et al., 2021). Identifying wounds and scars can provide information about interactions beluga have within their environment and threats they may be encountering across their range, for example vessel strikes or infectious agents. The whale that was originally sighted in 2016 was identified using a distinct dot-like scarring pattern, referred to as a “morse-code” lesion (Le Net, 2018). Similar lesions have been attributed to a pox-like viral skin infection in other beluga populations (Krasnova et al., 2015, Le Net, 2018) and can persist on the skin for several years (Krasnova et al., 2015). Epidemiological studies have not found evidence that poxvirus infections induce high mortality in affected cetacean populations; however, it has been suggested that neonates or calves could be at higher risk of mortality, potentially affecting population dynamics (Van Bressem et al., 1999). Additionally, in other beluga populations poxvirus lesions are considered indicators of various pollutants (Krasnova et al., 2015), therefore monitoring the presence and prevalence of these infections could provide insight into the environmental conditions individuals are encountering.
An additional success was sighting a previously satellite-tagged beluga whale based on a citizen scientist observation. Satellite-tagging within the Greater Hudson Bay area has occurred infrequently and resighting a tagged whale is rare. Confirming this previously tagged whale within our dataset resulted in a collaboration on a larger project examining previously tagged whales in both Western Hudson Bay and Cumberland Sound populations (Ryan et al., 2022). This resighting provided insights into tag loss, wound healing, and the long-term impacts of tagging for these animals. These become important considerations when planning monitoring activities using satellite-tags on cetaceans in the future. Ultimately, resighting individual whales over their lifespan can provide valuable insight into the process of wound healing (Krasnova et al., 2015; McGuire et al., 2021), long-term changes in health (Krasnova et al., 2015; McGuire et al., 2021), and degree of site fidelity (McGuire et al., 2020). By resighting known whales we are able to gain insight about the use of this estuary and life history.
The applications of citizen science can extend beyond single species to multi-species (Swanson et al., 2015) and broader ecosystem monitoring (Gouraguine et al., 2019). An objective that emerged from the Beluga Bits project has been to monitor the health of the Churchill River estuary ecosystem using jellyfish as an indicator species. Jellyfish have previously been considered indicators for ecosystem disturbances given their propensity to thrive in disturbed marine environments (Lynam et al., 2011; Brodeur et al., 2016). Moreover, many jellyfish species are expanding their ranges as climate change results in warmer waters (Hay, 2006). This may be of particular importance for Arctic oceans, as jellyfish species have been documented increasing in occurrence and abundance in these areas (Attrill et al., 2007; Purcell et al., 2010; Geoffroy et al., 2018). Some researchers are advocating for increased monitoring of jellyfish species, as invasive jellyfish can have significant impacts to ecosystems (Brodeur et al., 2016). Therefore, detecting new jelly and jellyfish species in the Hudson Bay ecosystem could provide critical insight into the potential population or community-level changes that may be occurring within the estuary or surrounding waters.
Despite numerous successes already in this project, there are some limitations we are hoping to address in the future, or are taking steps to address now. Viewership on explore.org does vary from year to year and can be hard to predict. However, frame sampling the live-video ensures that regardless of viewership we are capturing all frames of interest. This will likewise increase the likelihood of capturing useful images in years with poor water quality. To that effect, we are also looking to develop our image capturing technology (such as implementing a 360 degree camera) to maximize the quality of photos regardless of water quality. We have also seen participation on Beluga Bits grow every year since the project began and we believe implementing new workflows and new photo data every year will maintain engagement. Additionally, although we have established relationships with veterinarians, facilities with animals in care and researchers, we are limited in the number of parties and disciplines we work with. We would like to expand our network further, for example, local Indigenous communities and knowledge keepers hold intimate knowledge and expertise of beluga and their environment. This is an essential perspective on how we understand and conserve beluga within Hudson Bay. Additionally, although our data has led to important insights into beluga biology in the estuary, the further we develop our data capabilities, the more opportunities we have to inform conservation and management decision-making more directly. For example, linking GPS locations with underwater images may provide insights into how different groups of beluga use specific areas of the estuary. Georeferencing detections of beluga mothers and calves could inform guidelines for speed zones or temporary restrictions within the estuary to minimize impacts on vulnerable parts of the population (also suggested by Malcolm and Penner, 2011). There are numerous avenues still to explore by expanding the reach of the project into other adjacent areas and building connections with nearby communities, knowledge keepers, and researchers.
As the project grows its reach and research capabilities, there is great potential to continue expanding this network to incorporate disciplines and perspectives. For example, understanding the origins and healing processes of injuries and infections can be highly informative for conservation and management. We have established relationships with veterinarians, facilities with animals in care, and researchers to share images of distinct injuries and markings to learn more about them
We have described a successful interdisciplinary, collaborative monitoring project focusing on beluga whales and the Churchill River estuary ecosystem. This project has been built by bringing together concepts and tools from multiple disciplines with collaborations from diverse partnerships to produce a comprehensive approach. We have demonstrated the successes of applying such an approach to produce innovative applications for emerging technologies, increase data collection capabilities, provide critical insights, and engage a broad audience to participate in conservation research. As we have shown, we can bring together diverse perspectives, partners, technologies, and resources to expand monitoring efforts and deliver tools where they are needed most.
The data that support the findings of this study are available from the corresponding author upon reasonable request.
The animal study was reviewed and approved by Fisheries and Oceans Canada, Government of Canada.
AW, CB, SF, and SP contributed to the conception and design of the study. SP designed the Beluga Bits project on Zooniverse. AM and KM collected field data, managed the live stream on explore.org, and contributed manuscript sections. AW, CB, SF, and SP organized and managed the image database. NS and AA designed and tested deep learning models and contributed manuscript sections. AW and NS performed statistical analysis. AW, SF, and SP wrote the first draft of the manuscript. All authors contributed to manuscript revision, read, and approved the submitted version.
This work was supported by the Assiniboine Park Conservancy, the RBC Environmental Donations Fund, Churchill Northern Studies Centre’s Northern Research Fund, Calm Air, and Mitacs Accelerate program
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Aguirre A. A., Lutz P. L. (2004). Marine turtles as sentinels of ecosystem health: Is fibropapillomatosis an indicator? EcoHealth. 1, 275–83. doi: 10.1007/s10393-004-0097-3
Anton V., Germishuys J., Bergström P., Lindegarth M., Obst M. (2021). An open-source, citizen science and machine learning approach to analyze subsea movies. Biodivers Data J. 9, e60548. doi: 10.3897/BDJ.9.e60548
Ashutosh S., Roy P. S. (2021). Three decades of nationwide forest cover mapping using Indian remote sensing satellite data: A success story of monitoring forests for conservation in India. J. Indian Soc. Remote Sens. 49, 61–70. doi: 10.1007/s12524-020-01279-1
Attrill M. J., Wright J., Edwards M. (2007). Climate-related increases in jellyfish frequency suggest a more gelatinous future for the North Sea. Limnol Oceanogr. 52, 480–485. doi: 10.4319/lo.2007.52.1.0480
Berger C., Bieri M., Bradshaw K., Brümmer C., Clemen T., Hickler T., et al. (2019). Linking scales and disciplines: An interdisciplinary cross-scale approach to supporting climate-relevant ecosystem management. Clim Change. 156, 139–150. doi: 10.1007/s10584-019-02544-0
Brodeur R. D., Link J. S., Smith B. E., Ford M. D., Kobayashi D. R., Jones T. T. (2016). Ecological and economic consequences of ignoring jellyfish: A plea for increased monitoring of ecosystems. Fisheries 41, 630–637. doi: 10.1080/03632415.2016.1232964
Burke C., Rashman M., Wich S., Symons A., Theron C., Longmore S. (2019). Optimizing observing strategies for monitoring animals using done-mounted thermal infrared cameras. Inter J. Remote Sen. 40, 439–467. doi: 10.1080/01431161.2018.1558372
Chandler M., See L., Copas K., Bonde A. M. Z., López B. C., Danielsen F., et al. (2017). Contribution of citizen science towards international biodiversity monitoring. Biol. Cons. 213, 280–294. doi: 10.1016/j.biocon.2016.09.004
Conrad C. C., Hilchey K. G. (2011). A review of citizen science and community-based environmental monitoring: Issues and opportunities. Environ. Monit. Assess. 176, 273–291. doi: 10.1007/s10661-010-1582-5
Core Team R. (2021). R: A language and environment for statistical computing (Vienna, Austria: R Foundation for Statistical Computing). Available at: https://www.R-project.org/.
Deng J., Dong W., Socher R., Li L.-J., Li K., Fei-Fei L. (2009). “ImageNet: a Large-scale hierarchical image database. IEEE conference on computer vision and pattern recognition,” in IEEE Computer society conference on computer vision and pattern recognition, 248–255. doi: 10.1109/CVPR.2009.5206848
Dick M., Rous A. M., Nguyen V. M., Cooke S. J. (2016). Necessary but challenging: multiple disciplinary approaches to solving conservation problems. FACETS. 1, 67–82. doi: 10.1139/facets-2016-0003
Dunham K. D., Tucker A. M., Koons D. N., Abebe A., Dobson S. F., Grand J. B. (2021). Demographic responses to climate change in a threatened Arctic species. Ecol. Evol. 11, 10627–10643. doi: 10.1002/ece3.7873
Eischeid I., Soininen E. M., Assmann J. J., Ims R. A., Madsen J., Pedersen Å.Ø., et al. (2021). Disturbance mapping in Arctic tundra improved by a planning workflow for drone studies: Advancing tools for future ecosystem monitoring. Remote Sens. 13, 4466. doi: 10.3390/rs13214466
Fleiss J. L. (1971). Measuring nominal scale agreement among many raters. Psychol. Bull. 76, 378–382. doi: 10.1037/h0031619
Gamer M., Lemon J., Singh I. F. P. (2019) Irr: Various coefficients of interrater reliability and agreement. Available at: https://CRAN.R-project.org/package=irr.
Geoffroy M., Berge J., Majaneva S., Johnsen G., Langbehn T. J., Cottier F., et al. (2018). Increased occurrence of the jellyfish Periphylla periphylla in the European high Arctic. Polar Biol. 41, 2615–2619. doi: 10.1007/s00300-018-2368-4
Gouraguine A., Moranta J., Ruiz-Frau A., Hinz H., Reñones O., Ferse S. C. A., et al. (2019). Citizen science in data and resource-limited areas: A tool to detect long-term ecosystem changes. PLoS One 14, e0210007. doi: 10.1371/journal.pone.0210007
Greenslade B. (2018). Tourism industry in Churchill taking hit since rail line wash-out last spring. Global News. https://globalnews.ca/news/3982336/tourism-industry-in-churchill-taking-hit-since-rail-line-wash-out-last-spring/.
Greenwood J. J. D. (2003). The monitoring of British breeding birds: A success story for conservation science? Sci. Total Environ. 310, 221–230. doi: 10.1016/S0048-9697(02)00642-3
Høye T. T. (2020). Arthropods and climate change – arctic challenges and opportunities. Curr. Opin. Insect Sci. 41, 40–45. doi: 10.1016/j.cois.2020.06.002
Halliday W. D., Scharffenberg K., Whalen D., MacPhee S. A., Loseto L. L., Insley S. J. (2019). The summer soundscape of a shallow-water estuary used by beluga whales in the western Canadian Arctic. Arct. Sci. 6, 361–383. doi: 10.1139/as-2019-0022
Harasyn M. L., Chan W. S., Ausen E. L., Barber D. G. (2022). Detection and tracking of belugas, kayaks and motorized boats in drone video using deep learning. Drone Syst. Appl. 10, 77–96. doi: 10.1139/juvs-2021-0024
Hay S. (2006). Marine ecology: Gelatinous bells may ring change in marine ecosystems. Curr. Biol. 16, R679–R682. doi: 10.1016/j.cub.2006.08.010
Hecht S. B. (2011). From eco-catastrophe to zero deforestation? Interdisciplinarities, politics, environmentalisms and reduced clearing in Amazonia. Environ. Conserv. 39, 4–19. doi: 10.1017/S0376892911000452
He K., Zhang X., Ren S., Sun J. (2016). “Deep residual learning for image recognition,” in Proceedings of the IEEE computer society conference on computer vision and pattern recognition. doi: 10.1109/CVPR.2016.90
Jetley S., Lord N. A., Lee N., Torr P. H. S. (2018). “Learn to pay attention,” in 6th international conference on learning representations, ICLR 2018 - conference track proceedings. arXiv. doi: 10.48550/ARXIV.1804.02391
Krasnova V., Chernetsky A., Russkova O. (2015). Skin defects in the beluga whale Delphinapterus leucas (Pallas, 1776) from the Solovetsky gathering, as revealed by photo-identification analysis. Russ J. Mar. Biol. 41, 372–383. doi: 10.1134/S1063074015050077
Krizhevsky A., Sutskever I., Hinton G. E. (2017). ImageNet classification with deep convolutional neural networks. Commun. ACM. 60, 84–90. doi: 10.1145/3065386
Kuhn M. (2022). Caret: Classification and regression training. Available at: https://CRAN.R-project.org/package=caret.
Kuzyk Z. A., Macdonald R. W., Granskog M. A., Scharien R. K., Galley R. J., Michel C., et al. (2008). Sea Ice, hydrological, and biological processes in the Churchill River estuary region, Hudson Bay. Estuar. Coast. Shelf Sci. 77, 369–384. doi: 10.1016/j.ecss.2007.09.030
Lacoursière-Roussel A., Howland K., Normandeau E., Grey E. K., Archambault P., Deiner K., et al. (2018). eDNA metabarcoding as a new surveillance approach for coastal Arctic biodiversity. Ecol. Evol. 8, 7763–7777. doi: 10.1002/ece3.4213
LaDue C. A., Vandercone R. P. G., Kiso W. K., Freeman E. W. (2021). Scars of human–elephant conflict: Patterns inferred from field observations of Asian elephants in Sri Lanka. Wildl Res. 48, 540–553. doi: 10.1071/WR20175
Lair S., Measures L. N., Martineau D. (2016). Pathologic findings and trends in mortality in the beluga (Delphinapterus leucas) population of the St. Lawrence Estuary, Quebec, Canada, from 1983 to 2012. Vet. Pathol. 53, 22–36. doi: 10.1177/0300985815604726
Lamba A., Cassey P., Segaran R. R., Koh L. P. (2019). Deep learning for environmental conservation. Curr. Biol. 29, R977–R982. doi: 10.1016/j.cub.2019.08.016
Landis J. R., Koch G. G. (1977). The measurement of observer agreement for categorical data. Biometrics 33, 159–174. doi: 10.2307/2529310
Lanier A. L., Drabik J. R., Heikkila T., Bolson J., Sukop M. C., Watkins D. W., et al. (2018). Facilitating integration in interdisciplinary research: lessons from a south Florida water, sustainability, and climate project. Enviro. Manage. 62, 1025–1037. doi: 10.1007/s00267-018-1099-1
Le Net R. (2018). Épidémiologie et pathologie des dermatopathies chez les bélugas (Delphinapterus leucas) de l’estuaire du Saint-Laurent (Montreal, Canada: [Montréal (QC)]: Université de Montréal).
Linchant J., Lisein J., Semeki J., Lejeune P., Vermeulen C. (2015). Are unmanned aircraft systems (UASs) the future of wildlife monitoring? A review of accomplishments and challenges. Mamm. Rev. 45, 239–252. doi: 10.1111/mam.12046
López J. J., Mulero-Pázmány M. (2019). Drones for conservation in protected areas: Present and future. Drones 3, 1–23. doi: 10.3390/drones3010010
Lopez-Vazquez V., Lopez-Guede J. M., Marini S., Fanelli E., Johnsen E., Aguzzi J. (2020). Video image enhancement and machine learning pipeline for underwater animal detection and classification at cabled observatories. Sensors 20, 726. doi: 10.3390/s20030726
Luque S., Pettorelli N., Vihervaara P., Wegmann M. (2018). Improving biodiversity monitoring using satellite remote sensing to provide solutions towards the 2020 conservation targets. Methods Ecol. Evol. 9, 1784–1786. doi: 10.1111/2041-210x.13057
Lynam C. P., Lilley M. K. S., Bastian T., Doyle T. K., Beggs S. E., Hays G. C. (2011). Have jellyfish in the Irish Sea benefited from climate change and overfishing? Glob. Change Biol. 17, 767–782. doi: 10.1111/j.1365-2486.2010.02352.x
Malcolm C., Penner H. (2011). “Behaviour of belugas in the presence of whale watching vessels in Churchill, Manitoba and recommendations for local beluga watching activities,” in Polar tourism: Environmental, political and social dimensions. Eds. Maher P. T., Stewart E. J., Luck M. (New York, NY: Cognizant Communications), 54–79.
Marcoux M., Ferguson S. H., Roy N., Bedard J. M., Simard Y. (2017). Seasonal marine mammal occurrence detected from passive acoustic monitoring in Scott Inlet, Nunavut, Canada. Polar Biol. 40, 1127–1138. doi: 10.1007/s00300-016-2040-9
Marvin D. C., Koh L. P., Lynam A. J., Wich S., Davies A. B., Krishnamurthy R., et al. (2016). Integrating technologies for scalable ecology and conservation. Glob. Ecol. Conserv. 7, 262–275. doi: 10.1016/j.gecco.2016.07.002
Matthews C. J. D., Watt C. A., Asselin N. C., Dunn J. B., Young B. G., Montsion L. M., et al. (2017). “Estimated abundance of the Western Hudson Bay beluga stock from the 2015 visual and photographic aerial survey,” in DFO Can. Sci. Advis. Sec. Res. Doc. 2017/061, v + 20.
McDonald-Madden E., Baxter P. W. J., Fuller R. A., Martin T. G., Game E. T., Montambault J., et al. (2010). Monitoring does not always count. Trends Ecol. Evol. 25, 547–550. doi: 10.1016/j.tree.2010.07.002
McGuire T. L., Boor G. K. H., McClung J. R., Stephens A. D., Garner C., Shelden K. E. W., et al. (2020). Distribution and habitat use by endangered Cook Inlet beluga whales: Patterns observed during a photo-identification study, 2005–2017. Aquat. Conserv. 30, 2402–2427. doi: 10.1002/aqc.3378
McGuire T., Stephens A. D., McClung J. R., Garner C., Burek-Huntington K. A., Goertz C. E. C., et al. (2021). Anthropogenic scarring in long-term photo-identification records of Cook Inlet beluga whales, Delphinapterus leucas. Mar. Fish Rev. 82, 20–40. doi: 10.7755/MFR.82.3-4.3
Meffe G. K., Viederman S. (1995). Combining science and policy in conservation biology. Wildl Soc. Bull. 23, 327–332. http://www.jstor.org/stable/3782936.
Moore R. C., Loseto L., Noel M., Etemadifar A., Brewster J. D., MacPhee S., et al. (2020). Microplastics in beluga whales (Delphinapterus leucas) from the Eastern Beaufort Sea. Mar. pollut. Bull. 150, 1–7. doi: 10.1016/j.marpolbul.2019.110723
Naiman R. J. (1999). A perspective on interdisciplinary science. Ecosystems 2, 292–295. doi: 10.1007/s100219900078
Nakaoka M., Sudo K., Namba M., Shibata H., Nakamura F., Ishikawa S., et al. (2018). TSUNAGARI: A new interdisciplinary and transdisciplinary study toward conservation and sustainable use of biodiversity and ecosystem services. Ecol. Res. 33, 35–49. doi: 10.1007/s11284-017-1534-4
NAMMCO (2018). Report of the NAMMCO global review of monodontids. 13-16 march 2017 (Hillerød, Denmark: North Atlantic Marine Mammal Commission).
Norouzzadeh M. S., Nguyen A., Kosmala M., Swanson A., Palmer M. S., Packer C., et al. (2018). Automatically identifying, counting, and describing wild animals in camera-trap images with deep learning. Proc. Natl. Acad. Sci. 115, E5716–E5725. doi: 10.1073/pnas.1719367115
O’Corry-Crowe G. (2008). Climate change and the molecular ecology of Arctic marine mammals. Ecol. App. 18, S56–S76. doi: 10.1890/06-0795.1
Ooms J. (2022). Av: Working with audio and video in r. Available at: https://CRAN.R-project.org/package=av.
Paszke A., Gross S., Massa F., Lerer A., Bradbury J., Chanan G., et al. (2019). “PyTorch: An imperative style, high-performance deep learning library,” in Advances in neural information processing systems (Red Hook, NY, USA:Curran Associates, Inc), 8024–8035.
Pooley S. P., Mendelsohn J. A., Milner-Gulland E. J. (2013). Hunting down the chimera of multiple disciplinarity in conservation science. Cons. Biol. 28, 22–32. doi: 10.1111/cobi.12183
Purcell J. E., Hopcroft R. R., Kosobokova K. N., Whitledge T. E. (2010). Distribution, abundance, and predation effects of epipelagic ctenophores and jellyfish in the western Arctic Ocean. Deep-Sea Res. II: Top. Stud. Oceanogr. 57, 127–135. doi: 10.1016/j.dsr2.2009.08.011
Ribani R., Marengoni. M. (2019). “A survey of transfer learning for convolutional neural networks,” in 32nd SIBGRAPI Conference on Graphics, Patterns and Images Tutorials (SIBGRAPI-T), 47–57. doi: 10.1109/SIBGRAPI-T.2019.00010
Rovang S., Nielsen S. E., Stenhouse G. (2015). In the trap: Detectability of fixed hair trap DNA methods in grizzly bear population monitoring. Wildl Biol. 21, 68–79. doi: 10.2981/wlb.00033
Rutherford M. B., Gibeau M. L., Clark S. G., Chamberlain E. C. (2009). Interdisciplinary problem solving workshops for grizzly bear conservation in Banff National Park, Canada. Policy Sci. 42, 163–187. doi: 10.1007/s11077-009-9075-5
Ryan K. P., Petersen S. D., Ferguson S. H., Breiter C. C., Watt C. A. (2022). Photographic evidence of tagging impacts for two beluga whales from the Cumberland Sound and western Hudson Bay populations. Arct Sci. Just-IN. doi: 10.1139/AS-2021-0032
Schultz C. B., Brown L. M., Pelton E., Crone E. E. (2017). Citizen science monitoring demonstrates dramatic declines of monarch butterflies in western North America. Biol. Conserv. 214, 343–346. doi: 10.1016/j.biocon.2017.08.019
Sheppard D. J., Moehrenschlager A., McPherson J., Mason J. (2010). Ten years of adaptive community-governed conservation: Evaluating biodiversity protection and poverty alleviation in a West African hippopotamus reserve. Environ. Conserv. 37, 270–282. doi: 10.1017/S037689291000041X
Siddiqui S. A., Salman A., Malik M. I., Shafait F., Mian A., Shortis M. R., et al. (2018). Automatic fish species classification in underwater videos: Exploiting pre-trained deep neural network models to compensate for limited labeled data. ICES J. Mar. Sci. 75, 374–389. doi: 10.1093/icesjms/fsx109
Simonyan K., Zisserman A. (2015). Very deep convolutional networks for large-scale image recognition (3rd International Conference on Learning Representations, ICLR 2015: San Diego, CA, USA, May 7-9, 2015). Conference Track Proceedings. doi: 10.48550/ARXIV.1409.1556
Simpkins M., Kovacs K. M., Laidre K., Lowry L. (2009). A framework for monitoring Arctic marine mammals - findings of a workshop sponsored by the U.S. Marine Mammal Commission and U.S. Fish and Wildlife Service, Valencia, CAFF International Secretariat, CAFF CBMP Report No. 16.
Stephenson P. J. (2018). A global effort to improve species monitoring for conservation. Oryx 52, 409–415. doi: 10.1017/S0030605318000509
Swanson A., Kosmala M., Lintott C., Simpson R., Smith A., Packer C. (2015). Snapshot Serengeti, high-frequency annotated camera trap images of 40 mammalian species in an African savanna. Sci. Data 2, 150026. doi: 10.1038/sdata.2015.26
Tabak M. A., Norouzzadeh M. S., Wolfson D. W., Sweeney S. J., Vercauteren K. C., Snow N. P., et al. (2019). Machine learning to classify animal species in camera trap images: Applications in ecology. Methods Ecol. Evol. 10, 585–590. doi: 10.1111/2041-210X.13120
Towers J. R., Sutton G. J., Shaw T. J. H., Malleson M., Matkin D., Gisborne B., et al. (2019). Photo-identification catalogue, population status, and distribution of Bigg’s killer whales known from coastal waters of British Columbia, Canada. Can. Tech Rep. Fish Aquat. Sci. 3311, vi + 299.
Tulloch A. I. T., Possingham H. P., Joseph L. N., Szabo J., Martin T. G. (2013). Realizing the full potential of citizen science monitoring programs. Biol. Conserv. 165, 128–138. doi: 10.1016/j.biocon.2013.05.025
Van Bressem M.-F., Van Waerebeek K., Raga J. A. (1999). A review of virus infections of cataceans and the potential impact of morbilliviruses, poxviruses and papillomaviruses on host population dynamics. Dis. Aquat. Org. 38, 53–65. doi: 10.3354/dao038053
Vergara V., Wood J., Lesage V., Ames A., Mikus M. A., Michaud R. (2021). Can you hear me? Impacts of underwater noise on communication space of adult, sub-adult and calf contact calls of endangered St. Lawrence belugas (Delphinapterus leucas). Polar Res. 40, 5521. doi: 10.33265/polar.v40.5521
Wijers M., Loveridge A., Macdonald D. W., Markham A. (2019). CARACAL: A versatile passive acoustic monitoring tool for wildlife research and conservation. Bioacoustics 30, 41–57. doi: 10.1080/09524622.2019.1685408
Willi M., Pitman R. T., Cardoso A. W., Locke C., Swanson A., Boyer A., et al. (2019). Identifying animal species in camera trap images using deep learning and citizen science. Methods Ecol. Evol. 10, 80–91. doi: 10.1111/2041-210X.13099
Keywords: citizen science, machine learning, Cnidara, Ctenophora, conservation, deep learning, beluga whale (Delphinapterus leucas), wildlife monitoring
Citation: Westphal AM, Breiter C-JC, Falconer S, Saffar N, Ashraf AB, McCall AG, McIver K and Petersen SD (2022) Citizen science and machine learning: Interdisciplinary approach to non-invasively monitoring a northern marine ecosystem. Front. Mar. Sci. 9:961095. doi: 10.3389/fmars.2022.961095
Received: 03 June 2022; Accepted: 22 August 2022;
Published: 23 September 2022.
Edited by:
Laura Airoldi, University of Padova Chioggia Hydrobiological Station, ItalyCopyright © 2022 Westphal, Breiter, Falconer, Saffar, Ashraf, McCall, McIver and Petersen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ashleigh M. Westphal, YXdlc3RwaGFsQGFzc2luaWJvaW5lcGFyay5jYQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.