- 1Natural Product Informatics Research Center, Korea Institute of Science and Technology, Gangneung, South Korea
- 2Smart Farm Research Center, Korea Institute of Science and Technology, Gangneung, South Korea
- 3Biological and Genetic Resources Assessment Division, National Institute of Biological Resources, Incheon, South Korea
Streptomyces, the largest genus in Actinobacteria, has been known as a chemically prolific bacterial group producing pharmaceutically important small molecules. Various endeavors have been made to discover novel secondary metabolites from strains inhabiting diverse environmental niches. In our course of collecting bacterial strains to discover biologically active molecules, a marine-derived Streptomyces sp. V17-9 was isolated from a seagrass collected from a beach on Côn Đảo, Vietnam. Phylogenetic and genomic analyses suggested the possibility that this strain might form a new taxonomic group with a few closely related unclassified strains. The genome sequence of strain V17-9 was predicted to have 20 putative biosynthetic gene clusters. A chemical investigation identified amino acid derivatives (N-acetyltryptamine, N-acetyltyramine, and 6-prenyltryptophol) and siderophores (desferrioxamine E and spoxazomicin A) from culture extracts, linking gene clusters with actual productions. In particular, prenylated indole compounds were enhanced in production as part of metabolic conversion under supplement with ferric ions. Sequence similarity networks for indole and siderophore gene clusters showed their diversity and complexity in the genus Streptomyces. Phylogenomic analysis of gene cluster for 6-prenyltryptophol suggested strains of genomic potential for production of such compounds. They also suggested how these gene clusters may have shaped the biosynthesis of natural products. Chemotaxonomic profiling coupled with genome analysis would provide new insights into comparative studies on Actinobacteria producing prenylated indoles and siderophores.
1 Introduction
Actinobacteria are common soil inhabitants and are known to be involved in organic matter recycling (Bhatti et al., 2017). They have also been well known as a chemically prolific bacterial group producing pharmaceutically important small molecules (Takahashi and Nakashima, 2018). However, it has become harder to discover novel molecules from soil-dwelling Actinobacteria, mainly challenged by frequent re-isolation of known compounds (Zotchev, 2012). Accordingly, numerous endeavors have been made to isolate taxonomically underrepresented microbes from non-terrestrial niches, for example, marine-derived bacteria, endophytes, and insect symbionts, as potential sources of new secondary metabolites (Lam, 2006; Janso and Carter, 2010; Madden et al., 2013; Matsumoto and Takahashi, 2017). As a major group of Actinobacteria, the genus Streptomyces has been prodigiously studied because of its versatility to produce diverse secondary metabolites for new antibiotics or drug candidates and adaptability to inhabit ecosystems including the marine environment. In particular, marine Streptomyces have been highlighted in their wide range of genetic diversity and potential for mining novel biosynthetic gene clusters (BGCs) (Xu et al., 2019).
The similarity of 16S rDNA sequences has been widely used as a criterion for species demarcation of bacterial species (Coenye and Vandamme, 2004; Konstantinidis and Tiedje, 2007). However, the genetic complexity of this genus has been demonstrated by highly variable average nucleotide identity (ANI) and the presence/absence of orthologs even in identical strains based on 16S rDNA sequence analysis. In addition, differences between phylogenetic measures and metabolic characteristics including BGCs suggested that 16S rDNA sequence-based studies might be insufficient to resolve diversity in the genus Streptomyces (Chevrette et al., 2019). Moreover, a comparative genomics survey provided that only 5.83% of orthologous clusters among 17 Streptomyces spp. belonged to the core genome, while 33.95% and 60.22% of them turned out to be dispensable and unique genomes, respectively. These two large groups of genes were thought to render their adaptability to diverse environmental niches, reflecting a high degree of genomic diversity (Kim et al., 2015). Meanwhile, intragenus diversity in secondary metabolism was reported in Streptomyces albus and Streptomyces rimosus (Seipke, 2015; Park and Andam, 2019). Furthermore, remarkable differences in abundance and diversity were found in the distribution of BGCs in members of the same species (Sottorff et al., 2019; Belknap et al., 2020), emphasizing the importance of chemotaxonomic resolution consisting of phylogenetic, genomic, and chemical evidence.
During the course of collecting bacterial strains to discover biologically active molecules, a marine bacterial strain V17-9 was isolated from seagrass growing at a beach on Côn Đảo, Vietnam. The 16S rRNA gene sequence of strain V17-9 showed high similarity with that of Streptomyces ardesiacus. S. ardesiacus was initially classified as a subspecies of Actinomyces diastaticus, later renamed Streptomyces diastaticus subsp. ardesiacus and recently reclassified as an independent species (Komaki and Tamura, 2020). Prior to the taxonomic clarification, there have been previous studies reporting the production of secondary metabolites, such as oligomycin, rimocidin, phenazinolines, amylostatin XG, urdamycins, and CE-108 from S. diastaticus (Fukuhara et al., 1982; Seco et al., 2004; Yang et al., 2010; Li et al., 2015; Gui et al., 2018). These results further highlight the importance of an ensemble approach consisting of phylogenetic, genomic, and chemical profiling.
Here, we report chemical profiles and the complete genome sequence of a marine-derived Streptomyces sp. V17-9. Phylogenomic analysis revealed that strain V17-9 may form a new taxon with unclassified strains from marine environments. The chemical investigation identified endogenous production of small molecules in strain V17-9. In particular, the production of prenylated indole products was dependent on the presence of ferric ions, suggesting a way of awakening silent or cryptic gene clusters. Gene clusters for prenylated indoles and siderophores were searched in 2,061 Streptomyces genome sequences, assessing the biosynthetic potential of such compounds. Chemical profiling coupled with genomic analysis would provide new insights in studies of silent clusters and comparative genomics of microbes producing prenylated indoles and siderophores.
2 Materials and methods
2.1 Isolation of marine bacteria
Seagrass samples, collected on the beach of Côn Đảo, Vietnam (8°41′21.8″N 106°37′42.4″E), were rinsed three times with sterile seawater in order to remove surface contamination. A 1.0-g sample was triturated with sterile seawater and spread on the entire surface of Marine Agar (peptone 5.0 g, yeast extract 1.0 g, MgSO4 0.1 g, KH2PO4 0.1 g, and agar 18.0 g dissolved in 500 ml of seawater and 500 ml of distilled water, pH 7.0–7.2). After incubation at 28°C for 24 h, all colonies with different pigmentations and morphologies were picked out. The isolated colonies were repeatedly streaked to obtain pure cultures and stored at −80°C in Marine Broth supplemented with 30% (v/v) glycerol for further studies. A marine-derived bacterium, strain V17-9, was isolated from a rhizome of the seagrass Thalassia sp. and grown as orange-colored colonies with representative morphology of Actinobacteria.
2.2 Genomic DNA preparation and sequencing
The strain was cultivated at the TSB medium for 3 days. The cells, obtained after centrifugation at 12,000 rpm for 2 min, were extracted using SolGent™ Genomic DNA Prep Kit following the manufacturer’s instructions (SolGent, Daejeon, Republic of Korea). The genome of strain V17-9 was sequenced by PacBio RS II technology (Macrogen, Inc., Seoul, Republic of Korea). The sequencing generated 135,225 reads with a total of 1,413,328,505 bases.
2.3 Analyses for species identification
Species identification was performed by using the consensus 16S rRNA gene sequence (1,515 bp) obtained from the genome. The sequence was analyzed by using the EZBioCloud 16S database (Yoon et al., 2017). Hits from the database were obtained and subjected to phylogenetic analysis. A phylogenetic tree based on the 16S rDNA sequences was created by the maximum likelihood method under the Tamura 3‐parameter with a discrete gamma distribution and invariable sites model (T92+G+I) using MEGA X (Kumar et al., 2018). The value of DNA–DNA relatedness was obtained by calculation of digital DNA–DNA hybridization (dDDH) under the recommended setting of the Genome-to-Genome Distance Calculator (GGDC v3.0) (Meier-Kolthoff et al., 2013; Meier-Kolthoff et al., 2022). The OrthoANI values were calculated by the Orthologous Average Nucleotide Identity Tool (v1.40) (Lee et al., 2016). OrthoANI and dDDH calculations were visualized as a scatter plot by using ggplot2, ggExtra, and ggthemes packages (Wickham, 2016; Arnold, 2021; Attali and Baker, 2022) in R (v4.1.2) (R Core Team, 2021).
2.4 Genome assembly, annotation, and bioinformatics
The reads were assembled by using the Hierarchical Genome Assembly Process (HGAP v3.0) (Chin et al., 2013). Completeness of the genome assembly was assessed by analyzing the Actinobacteria (odb9) dataset of Benchmarking Universal Single-Copy Orthologs (BUSCO v3.0.2) (Simão et al., 2015). Genome annotation was conducted by using Prokka (v1.13) (Seemann, 2014). RNA genes were predicted by RNAmmer (v1.2) (Lagesen et al., 2007). Genomic features, the predicted genes, and the distribution of BGCs were visualized in a circular plot by Circos (v0.69-9) (Krzywinski et al., 2009). Functional annotation of the predicted genes was carried out by eggNOG-mapper (Huerta-Cepas et al., 2019). Prediction of biosynthetic gene clusters was performed by using antiSMASH (v5.1.2) (Blin et al., 2019). Sequences of antiSMASH-predicted BGCs were analyzed by biosynthetic gene similarity clustering and prospecting engine (BiG-SCAPE) (Navarro-Muñoz et al., 2020). The resulting networks of indole and siderophore BGCs were visualized by using Cytoscape (v3.9.1) (Shannon et al., 2003). Protein sequences of a cluster for prenylated indole products were obtained from the gene prediction of strain V17-9. The sequence of a tryptophanase (WP_014061276.1) was obtained from the genome sequence of Streptomyces violaceusniger Tu 4113. They were searched against 2,061 Streptomyces genome sequences (Table S1) by using TBLASTN. In order to visualize the result with other 2,061 Streptomyces genomes, a phylogenomic tree was constructed by the standalone version of CVTree (Qi et al., 2004). The length of K for CVTree was set to be 6 since it was suggested to be optimal for bacterial phylogeny (Zuo et al., 2010). The significance of homology for the proteins was shown with the phylogenomic tree by using Graphical Phylogenetic Analysis (GraPhlAn v1.1.4) (Asnicar et al., 2015). Synteny diagrams for gene clusters were created by using BLASTN (v2.2.31+) and visualized by Easyfig (v2.2.5) (Sullivan et al., 2011).
2.5 Phenotypic characterization and chemotaxonomy
To determine the optimal temperature and pH for growth, the strain was incubated for 7 days in a TSB or ISP2 medium containing 3% sea salts at temperatures of 10°C, 15°C, 20°C, 25°C, 28°C, 37°C, and 45°C and pH values ranging from 1 to 11 (in 2 pH unit intervals, adjusted by 1 N HCl or NaOH). Growth in various concentrations of NaCl (0%–12%, w/v, in 1% increments) was evaluated after 7 days of incubation in TSB. Biochemical characteristics were evaluated using API ZYM Biochemical Test Kit (bioMérieux, Marcy l’étoile, France) according to the manufacturer’s instructions. The morphological feature was observed under a field emission scanning electron microscope (Inspect F, FEI, Hillsboro, OR, USA). The use of sole carbon in the strain was examined on basal salt media fortified with 3% sea salt supplemented with 1% of the carbon source (Shirling and Gottlieb, 1966). Cellular fatty acids were prepared by extraction and methylation and then analyzed by a 6890N gas chromatography system (Agilent Technologies, Santa Clara, CA, USA) using the Sherlock Microbial Identification software package (version 6.1) (Sasser, 1990).
2.6 General spectrometric experiment
NMR spectra were recorded with a Varian 500 MHz Superconducting FT-NMR system or a JEOL 400 MHz NMR system. Electrospray ionization–mass spectrometry (ESI-MS) spectra were obtained on an Agilent Technologies 1200 series/Agilent 6120 Quadrupole LC/MS with a Phenomenex Luna® C18(2) 5 μm (4.6 × 150 mm) column. High-resolution ESI-MS (HR-ESI-MS) spectra were obtained by using a Q Exactive™ Hybrid Quadrupole-Orbitrap™ Mass Spectrometer with ACQUITY UPLC® BEH 1.7 μm C18 130 Å (2.1 × 100 mm) column. Preparative high-performance liquid chromatography (HPLC) was performed using a Luna® 10 μm C8(2) 100 Å (10 × 250 mm) column.
2.7 Mass cultivation
The strain was cultivated at the TSB medium containing 3% sea salts for 3 days under a shaking incubator at 200 rpm. To isolate secondary metabolites from fermentation, seed culture was prepared with 30 ml of TSB broth for 3 days at 30°C. The seed culture was inoculated in a 4-L culture of A1SS medium (starch 10.0 g/L, peptone 4.0 g/L, yeast extract 2.0 g/L, and sea salt 30.0 g/L) and incubated for 7 days at 30°C at 200 rpm.
2.8 Isolation of secondary metabolites from culture extract
The culture supernatant was extracted with 6 L of ethyl acetate and concentrated in a rotary evaporator. Ethyl acetate extract was carried out by flash column chromatography using gradient elution with 20%, 40%, 60%, 80%, and 100% MeOH–water mixture, respectively. Compounds (1–3) were isolated directly from 40% MeOH fraction using preparative HPLC (Luna® 10 μm C18(2) 100 Å, 250×21.2 mm, 8 ml/min) with isocratic mobile phase (20% acetonitrile–water). We obtained pure compounds 1 (2.5 mg, tR 15 min), 2 (1.7 mg, tR 18 min), and 3 (1.5 mg, tR 25 min). In addition, 80% MeOH fraction was applied to preparative HPLC using the same column under a gradient condition of 20% to 100% MeCN for 1 h at a 2 ml/min flow rate, and two compounds (4 and 5) were eluted at 20 and 40 min, respectively. In addition, compounds 1–5 were identified on the 1H NMR spectrum comparisons with previously reported data along with HR-MS data (Itoh et al., 2003; Schumacher et al., 2003; Stritzke et al., 2004).
2.8.1 N-Acetyltryptamine (1)
ESI-MS m/z 203.1 [M + H]+, 225.1 [M + Na]+; HR-ESI-MS m/z 203.1180 for C12H15ON2 ([M + H]+, Δ0.52 ppm), 225.1000 for C12H14ON2Na ([M + Na]+, Δ 0.76 ppm); UV (MeCN) λmax: 220 nm, 280 nm; 1H NMR (500 MHz, CD3OD) δ 7.55 (d, J = 7.9 Hz, 1H), 7.32 (d, J = 8.1 Hz, 1H), 7.07 (m, 2H), 7.00 (t, J = 7.5 Hz, 1H), 3.46 (t, J = 7.4 Hz, 2H), 2.93 (t, J = 7.1 Hz, 2H), and 1.91 (s, 3H).
2.8.2 Desferrioxamine E (2)
ESI-MS m/z 601 [M + H]+, 301.3[M+2H]2+; HR-ESI-MS m/z 623.3370 for C27H48O9N6Na ([M + Na]+, Δ −0.84 ppm); UV (MeCN) λmax: 210 nm; 1H NMR (500 MHz, CD3OD) δ 3.61 (t, J = 6.6 Hz, 1H), 3.17 (t, J = 6.6 Hz, 1H), 2.78 (t, J = 7.0 Hz, 1H), 2.47 (t, J = 7.1 Hz, 1H), 1.63 (p, J = 6.9 Hz, 1H), 1.52 (p, J = 6.9 Hz, 1H), and 1.32 (p, J = 7.8 Hz, 1H).
2.8.3 N-Acetyltyramine (3)
ESI-MS m/z 180.1 [M + H]+, 202.1 [M + Na]+; HR-ESI-MS m/z 180.1020 for C10H14O2N ([M + H]+, Δ 0.30 ppm), 202.0839 for C10H13NO2Na ([M + Na]+, Δ0.21 ppm); UV (MeCN) λmax: 240 nm, 280 nm; 1H NMR (500 MHz, CD3OD) δ 7.02 (dt, 1H), 6.70 (dt, 1H), 3.33 (d, J = 7.3 Hz, 1H), 2.68 (t, J = 7.4 Hz, 1H), and 1.90 (s, 1H).
2.8.4 6-Prenyltryptophol (4)
ESI-MS m/z 230.1 [M + H]+, 251.1 [M+Na]+; HR-ESI-MS m/z 230.1533 for C15H20ON ([M + H]+, Δ −2.96 ppm); UV (MecN) λmax: 228 nm, 282 nm; 1H NMR (400 MHz, CD3OD) δ 7.39 (dd, J = 8.0, 0.7 Hz, 1H), 7.08 (dd, J = 1.4, 0.8 Hz, 1H), 6.96 (d, J = 0.9 Hz, 1H), 6.80 (dd, J = 8.2, 1.5 Hz, 1H), 5.33 (dddt, J = 7.4, 5.8, 2.9, 1.4 Hz, 1H), 3.77 (t, J = 7.3 Hz, 2H), 3.38 (d, J = 7.4 Hz, 2H), 2.91 (td, J = 7.3, 0.9 Hz, 2H), and 1.72 (t, J = 1.3 Hz, 6H).
2.8.5 Spoxazomicin A (5)
HR-ESI-MS m/z 336.1364 for C16H22N3O3S ([M + H]+, −3.74 ppm); 1H NMR (500 MHz, CD3OD) δ 7.64 (dd, J = 7.8, 1.8 Hz, 1H), 7.38 (m, 1H), 6.92, 6.87, 4.59 (m, 1H), 4.54 (m, 1H), 4.40 (dd, J = 8.4, 6.8 Hz, 1H), 4.21 (d, J = 4.8 Hz, 1H), 3.46 (dt, J = 3.3, 1.7 Hz), 3.20 (overlapped), 3.11 (p, J = 1.7 Hz, 1H), 2.96 (t, J = 5.5 Hz, 1H), 2.84 (dd, J = 11.1, 8.3 Hz, 1H), 2.52 (d, J = 1.4 Hz, 3H), and 1.96 (d, J = 2.5 Hz, 3H).
3 Results and discussion
3.1 Phenotypic and chemotaxonomic characteristics of strain V17-9
Streptomycetes were well known for mold-like colony morphology with aerial and substrate mycelium (Tresner et al., 1961). However, strain V17-9 exhibited a glossy bald appearance due to its inability to form a sporulating aerial mycelium when grown on solid media such as marine agar, ISP2, TSA, and A1SS. As for culture and growth characteristics, strain V17-9 grew at a temperature ranging from 15°C to 37°C (optimum, 28°C), and the pH range for growth was pH 5.0 to 9.0 (optimum, pH 7.0). The concentration of NaCl supporting growth was 0%–10% (optimal, 5%). The color of mycelium was observed as white on the ISP2 medium, and the scanning electron micrograph identified that the mycelium of V17-9 was rectiflexible with rugose ornamented spores (Figure S1 and Table S2).
Based on the API ZYM test, strain V17-9 was positive for alkaline phosphatase, esterase (C4), esterase lipase (C8), lipase (C14), leucine arylamidase, valine arylamidase, trypsin, α-chymotrypsin, acid phosphatase, naphthol-AS-BI-phosphohydrolase, β-galactosidase, α-glucosidase, N-acetyl-β-glucosaminidase, and α-mannosidase. Negative activity was observed for cystine arylamidase, α-galactosidase, β-glucuronidase, β-glucosidase, and α-fucosidase.
The cellular fatty acids in strain V17-9 consisted of the predominant fatty acids, (>10%) including C16:0 iso (39.75%), C15:0 anteiso (13.30%), and 14:0 iso (11.11%), and minor fatty acids (>1%) such as 16:1 iso H (6.60%), 16:0 (5.66%), 17:0 anteiso (5.52%), 15:0 iso (5.30%), 17:1 anteiso w9c (3.33%), 17:0 iso (2.31%), 18:0 (2.11%), and 17:0 cyclo (1.31%). This fatty acid profile was in accordance with that of the genus Streptomyces comprising complex mixtures of saturated, iso-, and anteiso-fatty acids (Kämpfer, 2012).
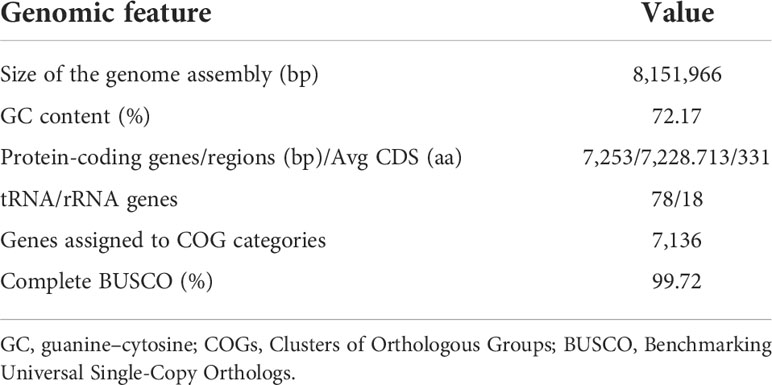
3.2 Genome sequence and identification of strain V17-9
The complete genome sequence was assembled in a single chromosome of 8,151,966 bp with 72.17% of guanine–cytosine (GC) content. Gene prediction revealed 7,253 protein-coding, 78 tRNA, and 18 rRNA genes from the genome sequence (Table 1 and Figure 1). A total of 7,136 predicted proteins were classified into 21 Clusters of Orthologous Groups (COGs). The majority of proteins, 45.38% of the total hits, were assigned as the ones of unknown function and only with general functional prediction. Among the rest of the hits, the largest functional category was genes involved in transcription (595), followed by carbohydrate transport and metabolism (457), amino acid transport and metabolism (408), energy production and conversion (335), signal transduction mechanisms (317), and inorganic ion transport and metabolism (291). In addition, 153 genes were predicted to be involved in the biosynthesis of secondary metabolites, transport, and catabolism, suggesting potential for the production of bioactive small molecules (Table S3).
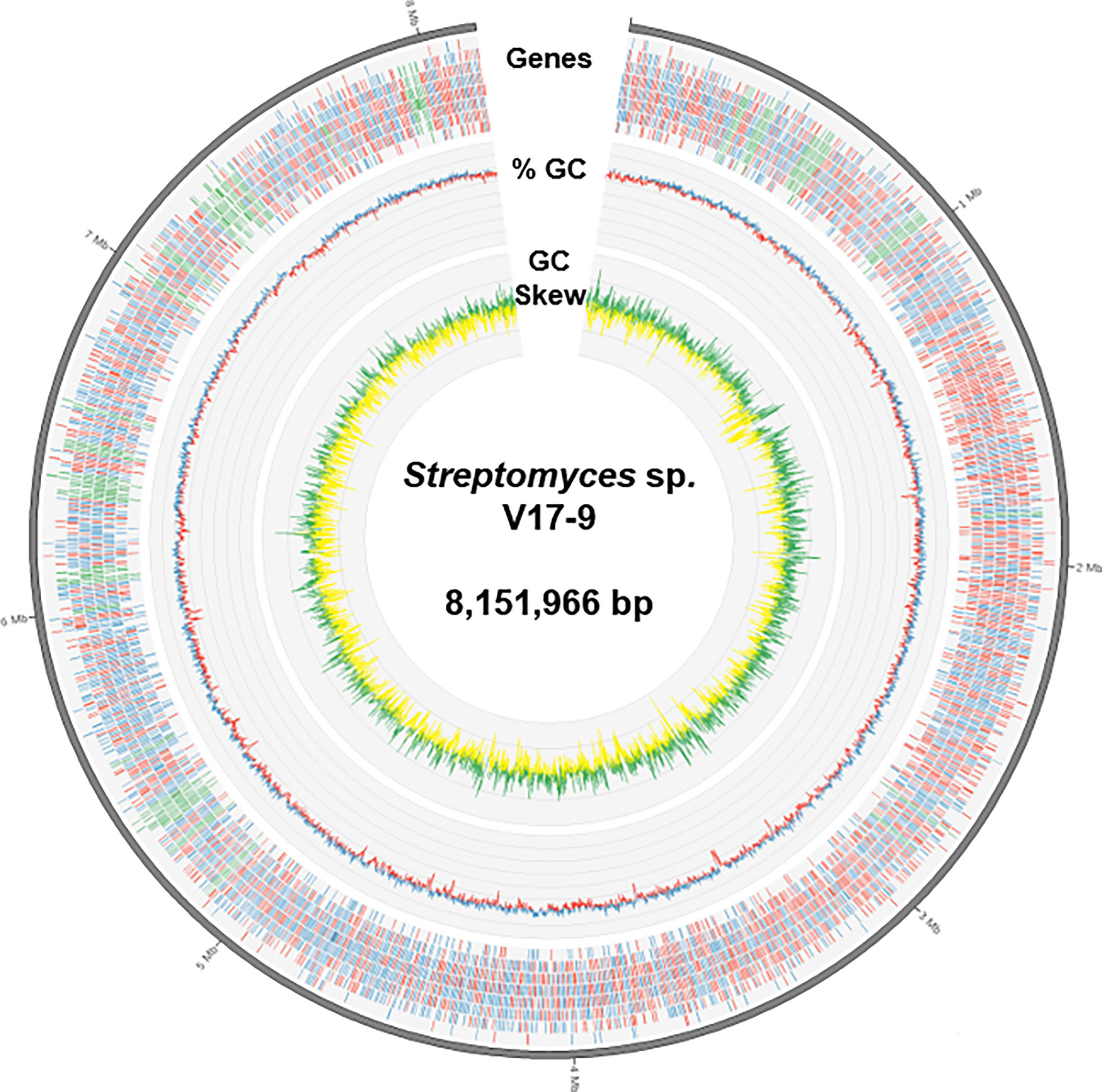
Figure 1 A diagram representing genomic features of Streptomyces sp. V17-9. From the outermost track to the center: i) the predicted genes (blue/red for forward/reverse strand and green for BGCs), ii) GC content (blue/red for above/below average), and iii) GC skew (green, >0; yellow, <0). BGCs, biosynthetic gene clusters; GC, guanine–cytosine.
Analysis of 16S rRNA gene sequences was performed in order to identify strain V17-9. The consensus sequence of the six 16S rRNA genes predicted from the genome showed sequence identities of 99.17%, 99.17%, and 99.14% against that of S. ardesiacus NRRL B-1773T, Streptomyces coelicoflavus NBRC 15399T, and Streptomyces hyderabadensis OU-40T, respectively (Table S4). The phylogenetic tree of the 16S rDNA sequences showed that strain V17-9 was clustered as a sister taxon with S. ardesiacus NRRL B-1773T, with S. coelicoflavus NBRC 15399T in a basal position (Figure S2). A sequence identity cutoff value of 97%–98% for 16S rRNA gene sequences has been accepted for species identification (Stackebrandt and Goebel, 1994; Yarza et al., 2008). Despite the high 16S rDNA sequence similarity, bacterial strains were often classified as different species according to additional analyses (Antony-Babu et al., 2017; Kusuma et al., 2020; Saygin et al., 2020; Maiti and Mandal, 2021). Thus, genome-based analyses, including dDDH and OrthoANI calculations, were performed on the 2,061 Streptomyces genomes. There are criteria including 95%–96% by OrthoANI and 70% dDDH for bacterial species delineation (Auch et al., 2010; Lee et al., 2016); however, all closely related type strains based on the 16S rDNA sequence analysis showed lower values than the recommended thresholds (Figure 2, Figure S3, and Table S5). In addition, the distribution of genes in predicted BGCs showed intricate patterns among strain V17-9 and 10 genomes shown in Table S5, suggesting that strain V17-9 might be genetically distinct from these species (Figure S4). Phenotypic and chemotaxonomic differences among close relatives based on the 16S rDNA sequence analysis further suggested that strain V17-9 might be a new species of Streptomyces (Figure S1 and Table S2). On the other hand, there were seven genomes showing OrthoANI > 95% as well as dDDH > 70% (Figure 2). Five of them were quite closely related to the V17-9 showing OrthoANI values greater than 98%. Interestingly, all these seven strains were collected from marine environments or the vicinity of the sea (Table S6). The results suggested that strain V17-9 might build a new taxon with these unclassified strains from marine environments.

Figure 2 Distribution of OrthoANI and dDDH values between strain V17-9 and the other Streptomyces genomes. Pairwise calculations of OrthoANI (x-axis) and dDDH (y-axis) are shown as a scatter plot. The gray box highlights the seven strains showing OrthoANI > 95% and dDDH > 70% compared to strain V17-9. The top 10 strains showing the highest 16S rDNA sequence similarities are shown as red dots. See Table S1 and Table S5 for more details.
3.3 Genome mining-based identification of secondary metabolites of strain V17-9
The complete genome sequence of strain V17-9 was scanned for BGCs in order to evaluate the genetic potential of strain V17-9 for producing bioactive secondary metabolites. The result revealed the presence of 20 BGCs, including four terpene clusters, two non-ribosomal peptide synthetases (NRPSs), three siderophores, and four polyketide synthases (PKSs; types I, II, and III). Fourteen of the 20 clusters showed significant similarity (>40%), whereas the rest only had negligible or no similarities to the known clusters (Table S7). The result may indicate the genetic potential of strain V17-9 for producing physiologically required metabolites, such as ectoine to survive in extreme osmotic stresses, germicidin to act as an autoregulatory inhibitor of spore germination, undecylprodigiosin, melanin, isorenieratene and carotenoid to protect against oxidative stress, and siderophores (coelichelin, coelibactin, and desferrioxamine) to chelate metal ions and transport them into the cells (Petersen et al., 1993; Bursy et al., 2008; Chu et al., 2010; Rao et al., 2017). Meanwhile, BGCs responsible for the production of substances isolated from the phylogenetic neighbors were not found in the genome, e.g., urdamycins, rimodicin, and CE-108 in S. diastaticus (Seco et al., 2004; Gui et al., 2018) and thiazostatin and butyloactol in S. ardesiacus (Komaki et al., 2018). It may imply that strain V17-9 would be far different from S. ardesiacus and S. diastaticus based not only on genetic compositions but also on molecular and chemical taxonomies.
3.4 Isolation and structural elucidation of secondary metabolites from culture extract
To isolate secondary metabolites, strain V17-9 was grown in an A1SS liquid medium and extracted with ethyl acetate. Diverse compound peaks were observed from the culture extract according to LC-MS data. Two of these peaks showed similar UV absorption spectra with 2-hydroxyphenylthiazoline, a family of iron-chelating non-ribosomal peptide natural products along with indole molecules with representative UV absorption spectrum via our in-house UV library (Figure 3 and Figure S5). The siderophore-like compounds were not produced along with germicidins when V17-9 was cultivated in a ferric ion-supplemented medium (A1SS + 50 μg/ml of FeCl3), which revealed their siderophore characteristics. Meanwhile, the production of hydrophobic indole compounds detected at 20–25 min of retention times was increased when ferric ions were supplied (Figure 3). This might provide a new strategy to induce a cryptic BGC for the ones producing iron-chelating molecules by supplying required metal ions. Subsequent flash column chromatography provided a rich fraction containing siderophores and indole compounds. After additional preparative HPLC experiments, five pure compounds were obtained. For elucidating the structure of the isolated compounds, the NMR analysis and high-resolution MS experiment were carried out. The 1H NMR was consistent with the corresponding spectral data of the N-acetyltryptamine (1), desferrioxamine E (2), N-acetyltyramine (3), 6-prenyltryptophol (4), and spoxazomicin A (5), supporting the HR-MS data (Figure 3C and Figure S5A). Except for the five identified compounds, there was an additional one with the identical molecular formula and UV absorption spectra to spoxazomicin A (5) in the culture extract. It is likely to be spoxazomicin B, because spoxazomicin A was originally isolated along with its stereoisomer spoxazomicin B (Inahashi et al., 2011).
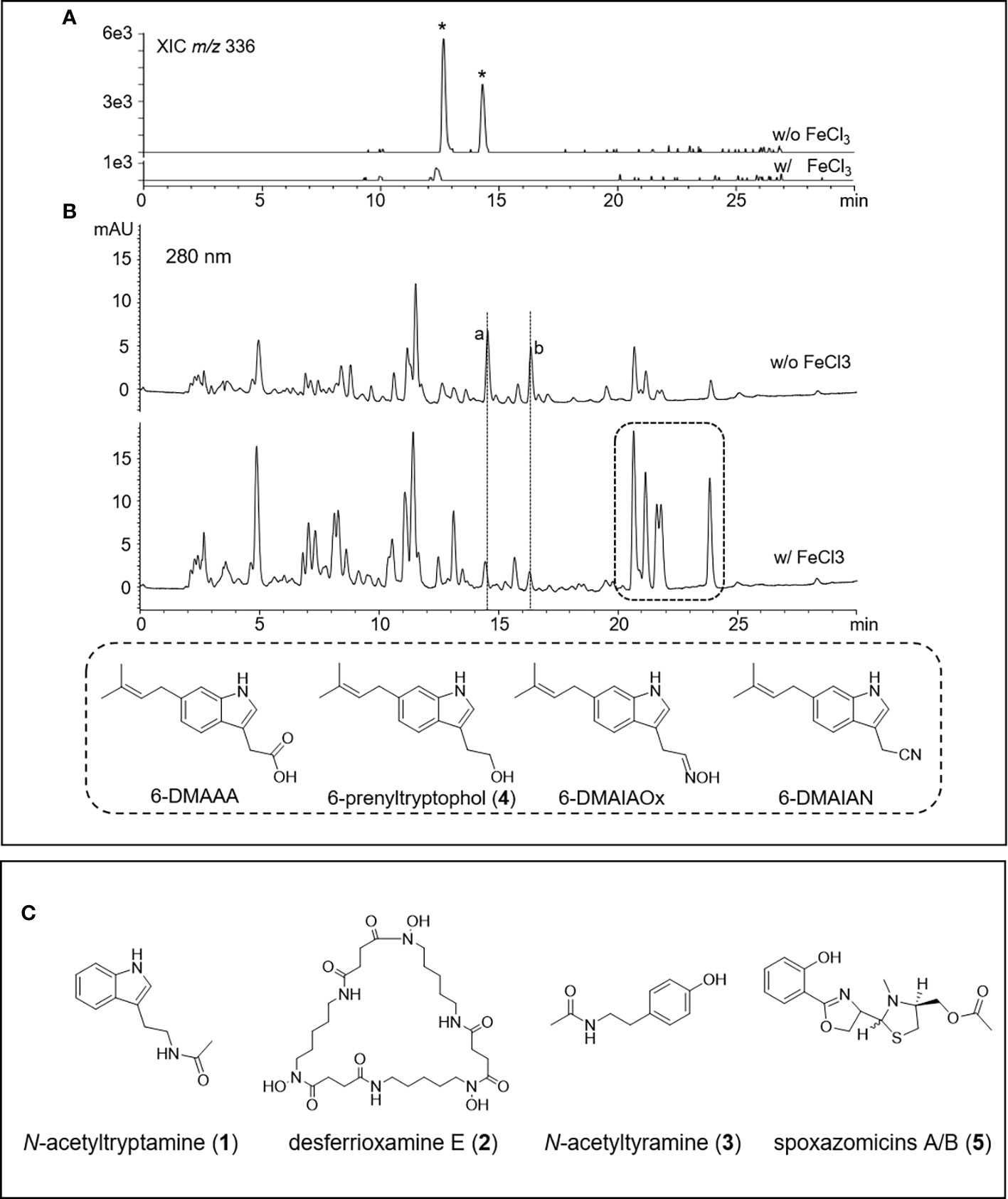
Figure 3 HPLC-MS chromatograms of V17-9 culture extracts of normal culture and ferric ion supplied culture and isolated compounds from V17-9. (A) When ferric ions were supplied as FeCl3, two compounds labeled with an asterisk with identical molecular ions (m/z 336) were depleted and subsequently identified as spoxazomicins A and B after isolation and purification. (B) Along with spoxazomicins, additional two compounds with the same UV absorption spectrum (a, b) disappeared under cultivation supplied with FeCl3, and indole compounds (6-dimethylallylindole-3-acetic acid (6-DMAAA, monaprenylindole A), 6-dimethylallylindole-3-acetonitrile (6-DMAIAN), and 6-dimethylallylindole-3-acetaldoxime (6-DMAIAOx) in the dashed box increased. Peaks were observed at similar retention times, but the peaks were different molecules from a and b based on UV and mass spectra. The compounds corresponding to a and b were expected as germicidins based on their UV absorption spectrum and mass-to-charge ratio (m/z). (C) Chemical structures of the compounds isolated from the culture extract of V17-9, N-acetyltryptamine (1), desferrioxamine E (2), N-acetyltyramine (3), and spoxazomicins A and B (5).
N-Acetyltryptamine and N-acetyltyramine are amino acid-derived amine compounds isolated from diverse biological sources including plants, fungi, and bacteria and represented assorted biological activities covering antiviral to antithrombotic agents (Nonno et al., 1999; Lee et al., 2017; Shen and Li, 2020). Spoxazomicins as trypanosomicides had been isolated from endophytic actinobacterium, Streptosporangium oxazolinicum K07-0460T, and a marine isolate Streptomyces olivaceus FXJ8.012 (Inahashi et al., 2011; Liu et al., 2013). Although it was predicted as 2-hydroxyphenylthiazoline by chemo-profiling with the in-house UV library, spoxazomicin A possessed 2-hydroxyphenyloxazine moiety found at microbial siderophores such as oxachelins, amychelins, and mycobactin-T (Wang et al., 2014). Desferrioxamine E is a cyclic hydroxamate siderophore, produced by diverse bacteria with antitumor and antineoplastic activities (Wang et al., 2014). A cytotoxic indolic metabolite, 6-prenyltryptophol, was isolated along with cyanide and oxime derivatives from the marine Streptomyces sp. BL-49-58-005 (López et al., 2003).
3.5 Correlation between biosynthetic gene clusters from genome sequence and identified compounds
Among the isolated compounds, biosynthetic pathways for 6-prenyltryptophol and spoxazomicins have not been identified in full (Rudolf et al., 2021); thus, BGCs responsible for their production were searched in the genome sequence of strain V17-9. Indole prenyltransferase (IPT) is one of the key players in the biosynthesis of prenylated indole products (Tanner, 2015). One of the predicted BGCs, Region 1 (Table S7), had a putative gene encoding IPT (KE639_00345) with an aromatic prenyltransferase domain (IPR033964). In addition, the sequence showed significant similarity with proteins contributing production of prenylated indole products, for example, SCO7467 of Streptomyces coelicolor A3(2) (80.21%) (Ozaki et al., 2013), PriB of Streptomyces sp. RM-5-8 (64.20%) (Elshahawi et al., 2017), and IptA of Streptomyces sp. SN-593 (62.61%) (Takahashi et al., 2010). Interestingly, IPT genes in Streptomyces were often accompanied by a conservon consisting of four genes, a sensor-like histidine kinase gene (cvnA), two hypothetical protein genes (cvnB and cvnC), and a gene encoding ATP/GTP-binding protein (cvnD), which might be a membrane-associated heterocomplex similar to a G-protein-coupled regulatory system in Eukaryotes (Komatsu et al., 2006). Previous genetic and biochemical investigations have shown that cvn1 operon, one of 13 conservons in S. coelicolor A3(2), regulates antibiotic biosynthesis as well as morphological and physiological development (Takano et al., 2011). Although these conservons were commonly found in actinobacterial genomes, it was unusual that the conservon was located within a specific BGC for secondary metabolites. CvnA (KE639_00350 in strain V17-9) orthologs with E-value < 1e−10 were found in only four BGCs including 5-isoprenylindole-3-carboxylate β-D-glycosyl ester, cyslabdan, raimonol, and frankiamicin (Ogasawara et al., 2015; Ikeda et al., 2016; Elshahawi et al., 2017) from Minimum Information about a Biosynthetic Gene cluster (MIBiG) database covering 1,923 secondary metabolite clusters. This may suggest that a certain conservon is involved in activation or repression to regulate the expression of a specific BGC by converting the environmental signal into an intracellular response. In the case of V17-9, metal ions might act as an extracellular signal, because the prenylated indole molecules were produced by a supplement of FeCl3. In addition, genes encoding flavin monooxygenase (FMO), tryptophanase, and cytochrome P450 (CYP450) were often found in the adjacent region of a prenyltransferase gene and the conservon. In Region 1, FMO and CYP450 genes were located along with the conservon and a putative gene encoding prenyltransferase. It was reported that an indole prenyltransferase (SCO7467) and an FMO (SCO7468) were responsible for the production of 5-dimethylallylindole-3-acetonitrile (5-DMAIAN) and 5-dimethylallylindole-3-acetaldoxime (5-DMAIAOx), which was structurally close to 6-prenyltryptophol (Ozaki et al., 2013). In strain V17-9, prenylated indole-3-acetonitrile and indole-3-acetaldoxime predicted as 6-DMAIAN and 6-DMAIAOx were also observed from the culture along with prenylated indole-3-acetic acid corresponding to monaprenylindole A (Figure 3B and Figure S5). Therefore, 6-prenyltryptophol would be a product of the gene cluster in Region 1 (Table S7). In addition, it was suggested that 6-prenyltryptophol could also be synthesized by nitrilase reaction and reduction after 6-DMAIAN (Ozaki et al., 2013).
Since spoxazomicin A was structurally related to non-ribosomal peptide siderophore pyochelin produced from Pseudomonas aeruginosa, it was inferred that spoxazomicin A was synthesized by NRPS consisting of three adenylation (A) domains for 2,3-dihydroxybenzoate (DHB), serine, and cysteine along with a methyltransferase (MT) gene to catalyze N-methylation at a thiazoline moiety. Among the 20 predicted BGCs, only Region 17 was predicted to produce coelibactin, which contained an A domain for DHB along with additional A domains for three cysteine and an MT gene (Table S7). Oxazoline moiety of spoxazomicin A would be synthesized by A domain predicted as cysteine according to NRPSsp and NRPSpredictor2 (Röttig et al., 2011; Prieto et al., 2012), in a similar way that methyloxazoline moiety of coelibactin was produced by A domain for cysteine, not threonine. Therefore, spoxazomicin A might be synthesized by a side product or a shunt product of the coelibactin BGC through modular skipping for the second heterocycle of coelibactin.
3.6 Distribution of prenylated indole biosynthetic gene clusters in the genus Streptomyces
According to previous studies, bacterial indole derivatives have diverse biological functions, including biofilm formation, motility, virulence, and antibiotic resistance (Melander et al., 2014). Prenylation of aromatic compounds is also known to affect the biological activity of chemical scaffolds, such as reduced plasticity of the cellular membrane (Botta et al., 2005) and signaling between bacteria by quorum sensing inhibition (Paguigan et al., 2019). To assess genetic diversity in the biosynthesis of such compounds, a sequence similarity network was constructed by using 657 predicted indole BGCs. The indole BGCs were grouped into 147 gene cluster families (GCFs) and further clustered into nine gene cluster clans (GCCs) (Figure 4). The indole cluster predicted in strain V17-9 was included in the largest component (dashed box in Figure 4), suggesting that it was one of the most frequently found types of indole clusters. In GCC478, 117 out of 124 BGCs were the first neighbors of the indole BGC of V17-9, showing their close genetic relatedness. Meanwhile, IPT gene-containing clusters have been classified into two types: those with a tryptophanase gene and those with an FMO gene (Ozaki et al., 2013) (Figure 5A). It was also demonstrated that the two types of clusters were capable of producing prenylated indole products (Ozaki et al., 2013). These clusters showed several combinations of genes encoding IPT, FMO, CYP450, and tryptophanase (Trpase), in addition to the conservon comprising four genes (cvnA–cvnD) (Figure S6). It was hypothesized that these are key enzymes responsible for the production of prenylated indole compounds. Protein sequences encoded by the key genes found in the indole BGC of strain V17-9 and a Trpase of S. violaceusniger Tu 4113 (WP_014061276.1) were searched against 2,061 Streptomyces genome sequences. Hits to the FMO (KE639_00344) were the least conserved throughout the genomes, showing a median E-value of 3.81e−23. Meanwhile, 492 out of 2,061 genomes had hits to the IPT (KE639_00345) with E-value < 1.0e−8, where 101 of them showed extremely high homology (E-value = 0). In addition, 491 out of 492 genomes had hits to all the seven proteins (KE639_00344–KE639_00350), supporting that IPT is a key enzyme of this cluster (Table S8).
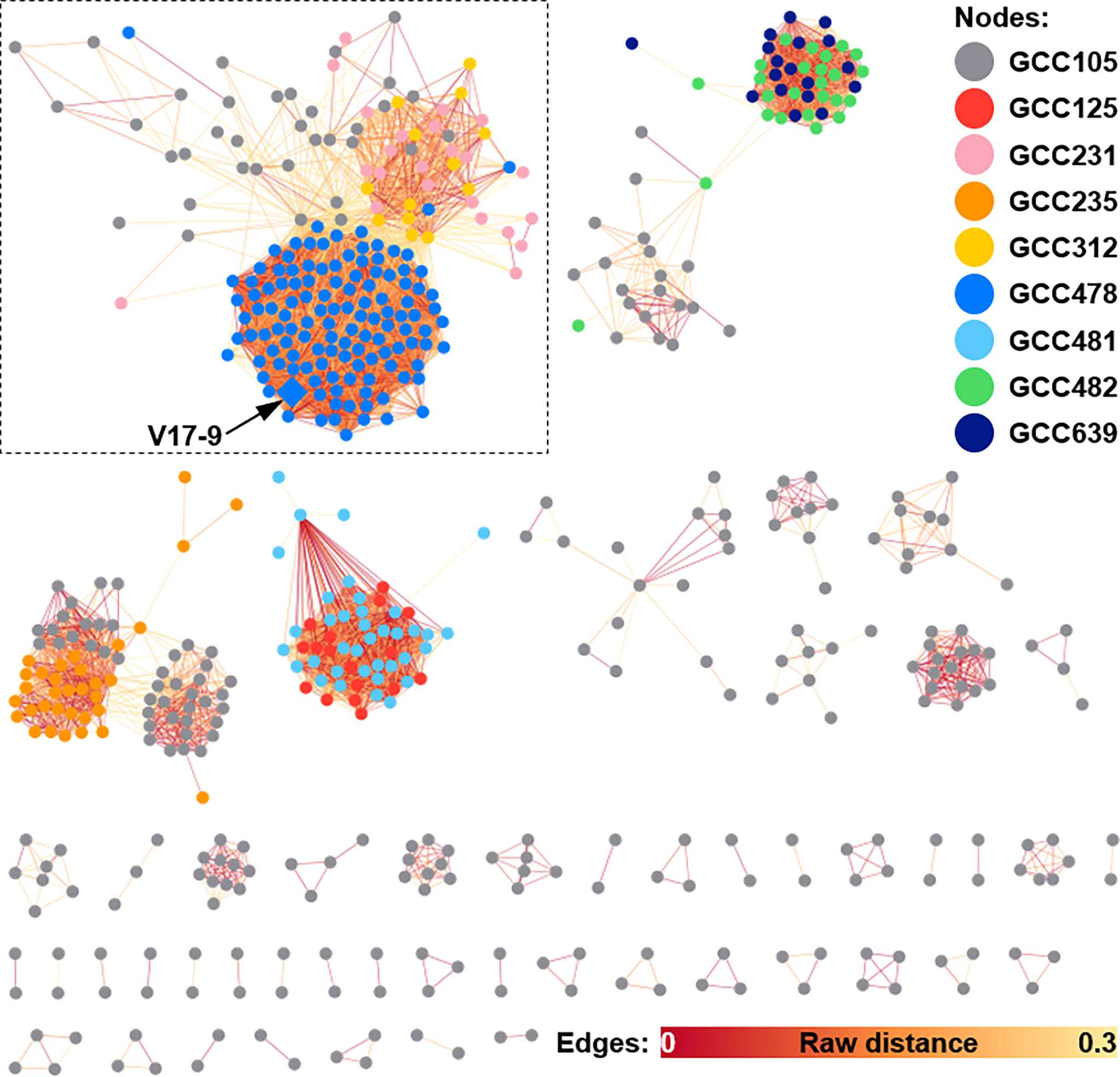
Figure 4 Sequence similarity network of indole BGCs generated by BiG-SCAPE. A total of 578 indole BGCs, excluding 79 singletons, from 2,062 Streptomyces genomes are shown as a similarity network. Nodes are filled with the nine different colors by the gene family clans (GCCs) they belong to. Edges are shown in the color gradient denoting the raw distance retrieved from the BiG-SCAPE analysis. The largest component is highlighted with a dashed box. The BGC identified from the V17-9 is shown as a diamond and indicated by an arrow. BGCs, biosynthetic gene clusters.
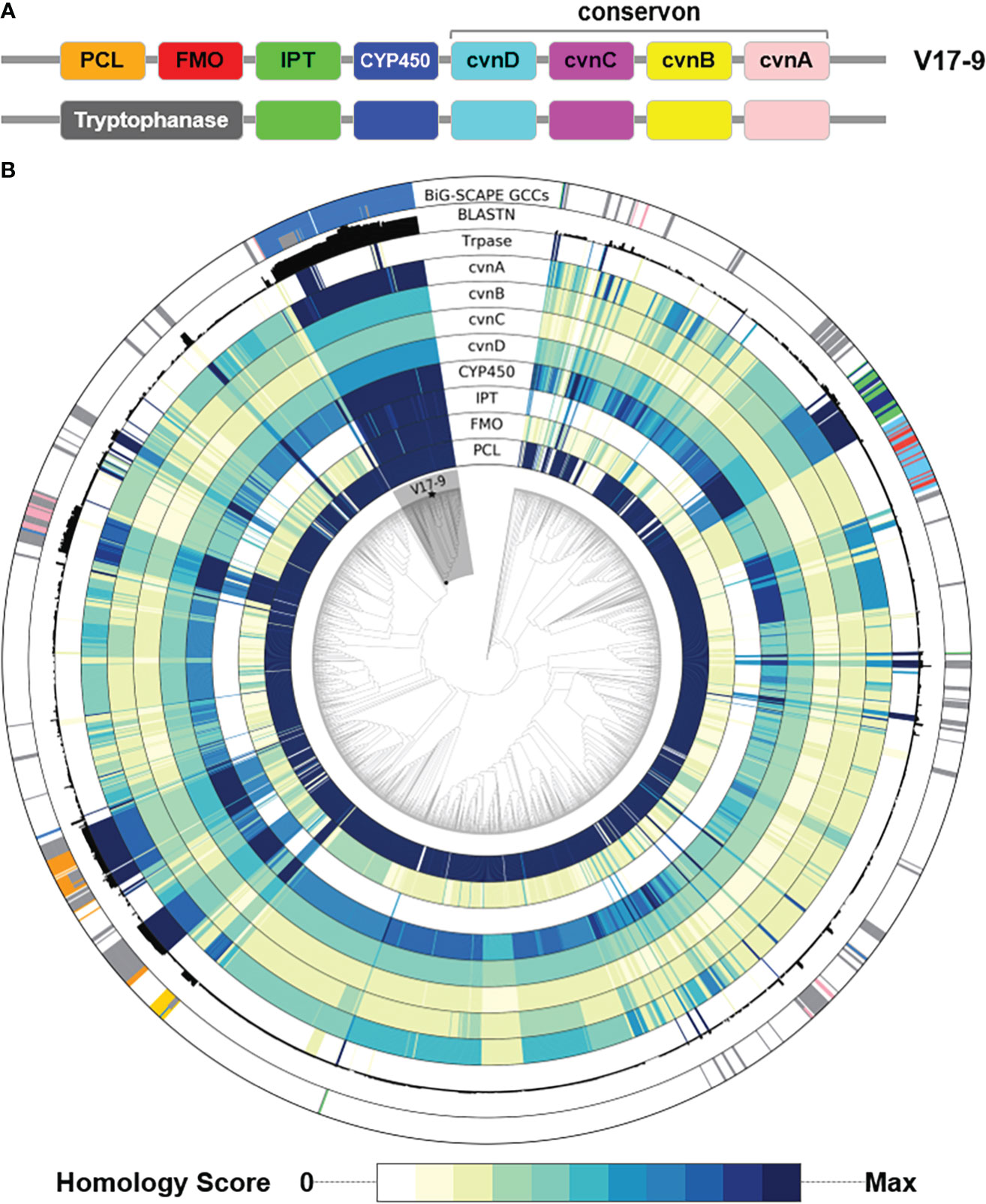
Figure 5 Distribution of key genes involved in the biosynthesis of prenylated indoles over a phylogenomic tree of 2,062 Streptomyces genome sequences. (A) The two types of gene structure. (B) For the nine tracks labeled with protein names, homology of each protein in the cluster was shown in the color gradient from white to dark navy. The more intense the color, the greater the homology with the corresponding query sequence. Homology score was calculated by –log10(E-value), where E-value of 0 was replaced with 1e−200 to avoid infinity. BLASTN track shows the sum of bit scores of hits with Evalue < 1e−20. The most peripheral track shows clan assignment from the BiG-SCAPE analysis shown in Figure 4. A monophyletic group including the V17-9 is shaded in gray. Strain V17-9 is indicated with a filled star mark at the terminal node. PCL, phenylacetate-CoA ligase; FMO, flavin monooxygenase; IPT, indole prenyltransferase; CYP450, cytochrome P450; Trpase, tryptophanase.
The distribution of the presence of indole BGCs was scattered in a number of phyletic groups (the outermost track in Figure 5B). Diverse gene structures (Figure S6) and patchy distribution (Figure 5B) might imply that indole products might be one of the ancestral chemical traits. Overall, the strong homology of the key protein sequences coincided with the presence of predicted indole BGCs. In addition, members of a monophyletic group including strain V17-9 showed significant homology to IPT, FMO, CYP450, and the four conservon components (Figure 5). The clade was comprised of 121 Streptomyces spp., and 115 of them contained BGCs that were the first neighbors to the indole BGC of V17-9. Nucleotide sequence homology of the indole BGC of V17-9 was higher in genome sequences of these strains than the others, showing that they share highly identical BGCs for indole production (Figure 5). Interestingly, 16 of the 121 Streptomyces genomes had multiple indole BGCs: one homologous to that of the V17-9 and the other containing a gene encoding carboxylesterase domain (PF00135)-containing protein. It might be relevant for their ecological fitness in certain habitats.
3.7 Distribution of siderophore biosynthetic gene clusters in the genus Streptomyces
Bacteria often secrete siderophores to acquire iron as a trace element for catalyzing redox reactions. Furthermore, siderophores may render ecological dynamics and mediate interactions with eukaryotic hosts (Kramer et al., 2020). In strain V17-9, the production of the indole compounds was activated by supplying ferric ions, mediated by siderophores (Figure 3). To explore the diversity of siderophore BGCs, a network was constructed based on sequence similarity (Figure 6). A total of 5,368 BGCs were grouped into 385 GCFs, and these were clustered into 32 GCCs. Three siderophore BGCs predicted in strain V17-9 belong to each of the three largest components, suggesting that they may produce the most common siderophore scaffolds. The distribution of the 32 GCCs over the phylogenomic tree showed complicated patterns (Figure S7). GCC profiles of siderophore BGCs for 54 genomes were the same as those of strain V17-9. In addition, 45 out of them were located in the clade comprising 121 strains, which showed high homology to the indole BGC of strain V17-9 (Figure S7). It may suggest that a certain combination of siderophores has been co-evolved and has worked in concert with BGCs to produce natural products for environmental fitness.
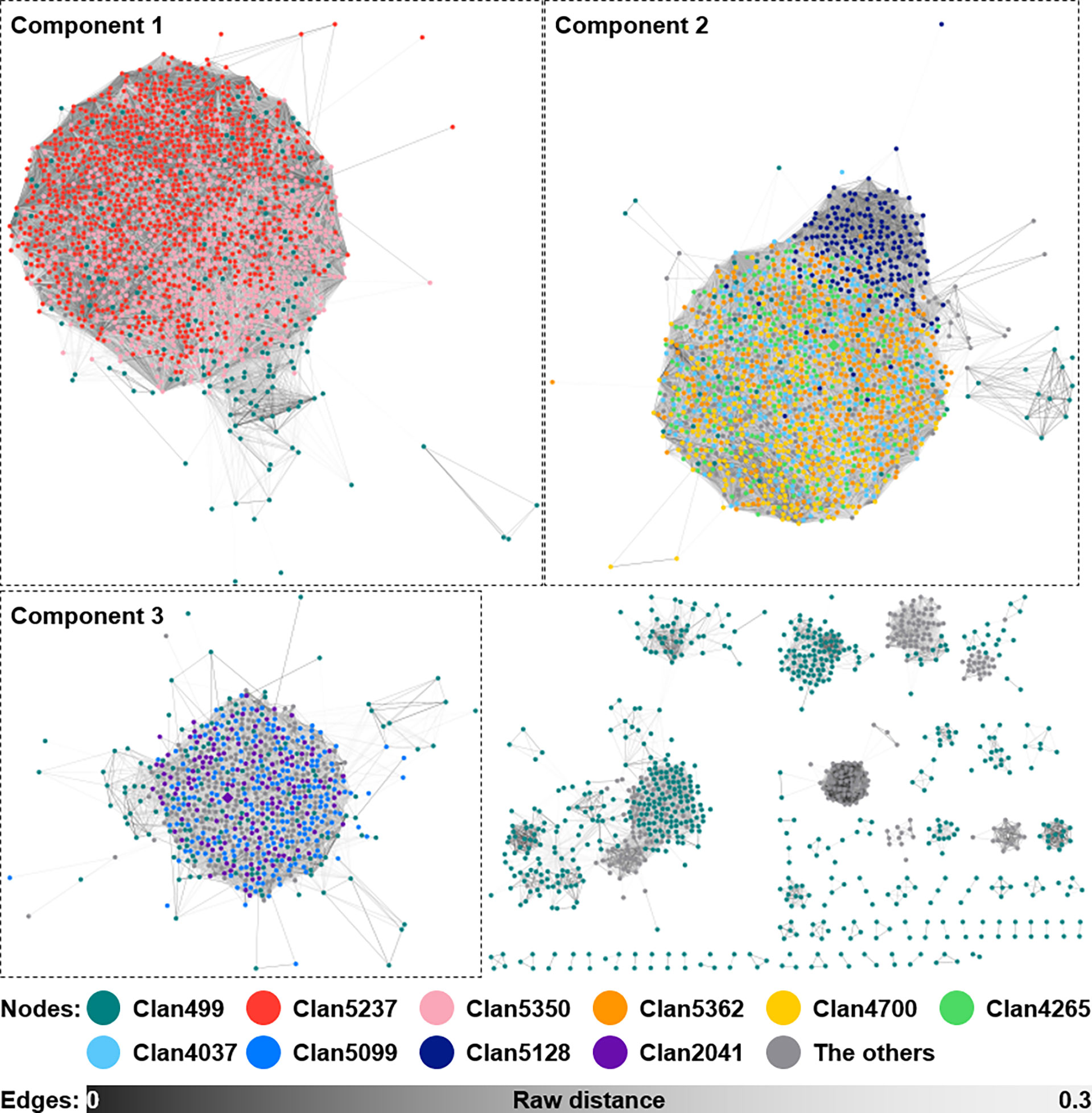
Figure 6 Sequence similarity network of siderophore BGCs predicted from the 2,062 Streptomyces genomes. A total of 5,217 siderophore BGCs, excluding 151 singletons, from 2,062 Streptomyces genomes are shown as a similarity network. Nodes belonging to the top 10 largest clans are filled with different colors as shown in the figure. Edges are shown in the color gradient denoting the raw distance retrieved from the BiG-SCAPE analysis. The top three largest components, to which BGCs of strain V17-9 belonged, are shown with the dashed boxes. BGCs, biosynthetic gene clusters.
4 Conclusion
The marine-derived Streptomyces sp. V17-9 was isolated and taxonomically analyzed with both the 16S rRNA genes and genome sequences. Based on its genomic relatedness with neighboring strains, V17-9 might construct a novel species with unclassified strains such as Streptomyces sp. SID10362, S. diastaticus deep-sea A18, and Streptomyces sp. H23. In addition, the prediction of BGCs for secondary metabolites proposed that this marine-derived microbe has substantial potential to produce small molecules. Our chemical investigation showed the biogenesis of siderophore compounds (spoxazomicins and desferrioxamine E) and amino acid derivatives (6-prenyltryptophol, N-acetyltryptamine, and N-acetyltyramine) from V17-9. In particular, prenylated indole compounds including 6-prenyltryptophol were induced when biosynthesis of siderophore was no longer needed due to the external supplement of ferric ions. Moreover, BGCs for prenylated indole compounds were well conserved in the specific clades including V17-9 among the Streptomyces strains. Phylogenomic analysis of gene structures of such BGCs showed that putative producers of prenylated indoles were mainly grouped into a few clades, suggesting that these molecules might serve as a chemotaxonomic marker in the genus Streptomyces. Genomes with the same GCC profiles for indole and siderophore BGCs suggested that they might have been co-evolved to work together in the production of natural products. Chemotaxonomic profiling coupled with genome analysis would provide new insights into comparative studies on Actinobacteria producing amino acid derivatives and siderophores.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material. The complete genome sequence of Streptomyces sp. strain V17-9 was deposited in NCBI GenBank under accession number CP074111 (BioProject: PRJNA725373). Cytoscape session files for the networks of indole and siderophore BGCs shown in Figure 4 and 6 can be accessed at https://doi.org/10.5281/zenodo.6582750 and https://doi.org/10.5281/zenodo.6582754, respectively.
Author contributions
D-EK: formal analysis, investigation, validation, writing—original draft, and writing—review and editing. S-CH: formal analysis, investigation, software, visualization, writing—original draft, and writing—review and editing. YY: resources and writing—review and editing. JC: data curation, formal analysis, investigation, methodology, software, supervision, visualization, writing—original draft, and writing—review and editing. J-SP: conceptualization, formal analysis, investigation, methodology, supervision, validation, writing—original draft, and writing—review and editing. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by an intramural grant from the Korea Institute of Science and Technology (2Z06663) and a grant from the Ministry of Oceans and Fisheries, Republic of Korea (20210641).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2022.959690/full#supplementary-material
References
Antony-Babu S., Stien D., Eparvier V., Parrot D., Tomasi S., Suzuki M. T. (2017). Multiple Streptomyces species with distinct secondary metabolomes have identical 16S rRNA gene sequences. Sci. Rep.7, 11089. doi: 10.1038/s41598-017-11363-1
Arnold J. B. (2021). "ggthemes: Extra themes, scales and geoms for 'ggplot2'". Available at: https://cran.r-project.org/web/packages/ggthemes/index.html.
Asnicar F., Weingart G., Tickle T. L., Huttenhower C., Segata N. (2015). Compact graphical representation of phylogenetic data and metadata with GraPhlAn. PeerJ3, e1029. doi: 10.7717/peerj.1029
Attali D., Baker C. (2022). "ggExtra: Add marginal histograms to 'ggplot2', and more 'ggplot2' enhancements".
Auch A. F., von Jan M., Klenk H. P., Göker M. (2010). Digital DNA-DNA hybridization for microbial species delineation by means of genome-to-genome sequence comparison. Stand. Genomic Sci.2 (1), 117–134. doi: 10.4056/sigs.531120
Belknap K. C., Park C. J., Barth B. M., Andam C. P. (2020). Genome mining of biosynthetic and chemotherapeutic gene clusters in Streptomyces bacteria. Sci. Rep. 10 (1), 2003. doi: 10.1038/s41598-020-58904-9
Bhatti A. A., Haq S., Bhat R. A. (2017). Actinomycetes benefaction role in soil and plant health. Microb. Pathog.111, 458–467. doi: 10.1016/j.micpath.2017.09.036
Blin K., Shaw S., Steinke K., Villebro R., Ziemert N., Lee S. Y., et al. (2019). antiSMASH 5.0: updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res.47 (W1), W81–W87. doi: 10.1093/nar/gkz310
Botta B., Vitali A., Menendez P., Misiti D., Delle Monache G. (2005). Prenylated flavonoids: Pharmacology and biotechnology. Curr. Medicinal Chem.12 (6), 713–739. doi: 10.2174/0929867053202241
Bursy J., Kuhlmann A. U., Pittelkow M., Hartmann H., Jebbar M., Pierik A. J., et al. (2008). Synthesis and uptake of the compatible solutes ectoine and 5-hydroxyectoine by Streptomyces coelicolor A3(2) in response to salt and heat stresses. Appl. Environ. Microbiol.74 (23), 7286–7296. doi: 10.1128/AEM.00768-08
Chevrette M. G., Carlos-Shanley C., Louie K. B., Bowen B. P., Northen T. R., Currie C. R. (2019). Taxonomic and metabolic incongruence in the ancient genus Streptomyces. Front. Microbiol.10, 2170. doi: 10.3389/fmicb.2019.02170
Chin C. S., Alexander D. H., Marks P., Klammer A. A., Drake J., Heiner C., et al. (2013). Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat. Methods10 (6), 563–569. doi: 10.1038/nmeth.2474
Chu B. C., Garcia-Herrero A., Johanson T. H., Krewulak K. D., Lau C. K., Peacock R. S., et al. (2010). Siderophore uptake in bacteria and the battle for iron with the host; a bird's eye view. Biometals23 (4), 601–611. doi: 10.1007/s10534-010-9361-x
Coenye T., Vandamme P. (2004). Use of the genomic signature in bacterial classification and identification. Syst. Appl. Microbiol.27 (2), 175–185. doi: 10.1078/072320204322881790
Elshahawi S. I., Cao H. N., Shaaban K. A., Ponomareva L. V., Subramanian T., Farman M. L., et al. (2017). Structure and specificity of a permissive bacterial c-prenyltransferase. Nat. Chem. Biol.13 (4), 366–368. doi: 10.1038/nchembio.2285
Fukuhara K., Murai H., Murao S. (1982). Amylostatins, other amylase inhibitors produced by Streptomyces diastaticus subsp. Amylostaticus no. 2476. Agric. Biol. Chem.46 (8), 2021–2030. doi: 10.1080/00021369.1982.10865392
Gui C., Liu Y. N., Zhou Z. B., Zhang S. W., Hu Y. F., Gu Y. C., et al. (2018). Angucycline glycosides from mangrove-derived Streptomyces diastaticus subsp. SCSIO GJ056. Mar. Drugs16 (6), 185. doi: 10.3390/md16060185
Huerta-Cepas J., Szklarczyk D., Heller D., Hernández-Plaza A., Forslund S. K., Cook H., et al. (2019). eggNOG 5.0: a hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic Acids Res.47 (D1), D309–D314. doi: 10.1093/nar/gky1085
Ikeda H., Shin-ya K., Nagamitsu T., Tomoda H. (2016). Biosynthesis of mercapturic acid derivative of the labdane-type diterpene, cyslabdan that potentiates imipenem activity against methicillin-resistant Staphylococcus aureus: cyslabdan is generated by mycothiol-mediated xenobiotic detoxification. J. Ind. Microbiol. Biotechnol.43 (2-3), 325–342. doi: 10.1007/s10295-015-1694-6
Inahashi Y., Iwatsuki M., Ishiyama A., Namatame M., Nishihara-Tsukashima A., Matsumoto A., et al. (2011). Spoxazomicins a-c, novel antitrypanosomal alkaloids produced by an endophytic actinomycete, Streptosporangium oxazolinicum K07-0460T. J. Antibiot.64 (4), 303–307. doi: 10.1038/ja.2011.16
Itoh T., Kinoshita M., Aoki S., Kobayashi M. (2003). Komodoquinone a, a novel neuritogenic anthracycline, from marine streptomyces sp. KS3. J. Nat. Prod.66 (10), 1373–1377. doi: 10.1021/np030212k
Janso J. E., Carter G. T. (2010). Biosynthetic potential of phylogenetically unique endophytic actinomycetes from tropical plants. Appl. Environ. Microbiol.76 (13), 4377–4386. doi: 10.1128/AEM.02959-09
Kämpfer P. (2012). “Genus i. streptomyces waksman and henrici 1943, 339AL emend. Witt and stackebrandt 1990, 370 emend. Wellington, stackebrandt, sanders, wolstrup and Jorgensen 1992, 159,” in Bergey’s manual of systematic bacteriology, part b, 2nd edn, vol. Vol 5 . Eds. Goodfellow M., Kämpfer P., Busse H.-J., Trujillo M. E., Suzuki K.-I., Ludwig W., Whitman W. B. (New York: Springer), 1455–1767.
Kim J. N., Kim Y., Jeong Y., Roe J. H., Kim B. G., Cho B. K. (2015). Comparative genomics reveals the core and accessory genomes of Streptomyces species. J. Microbiol. Biotechnol.25 (10), 1599–1605. doi: 10.4014/jmb.1504.04008
Komaki H., Sakurai K., Hosoyama A., Kimura A., Igarashi Y., Tamura T. (2018). Diversity of nonribosomal peptide synthetase and polyketide synthase gene clusters among taxonomically close Streptomyces strains. Sci. Rep.8, 6888. doi: 10.1038/s41598-018-24921-y
Komaki H., Tamura T. (2020). Reclassification of Streptomyces diastaticus subsp. ardesiacus, Streptomyces gougerotii and Streptomyces rutgersensis. Int. J. Syst. Evol. Microbiol.70 (7), 4291–4297. doi: 10.1099/ijsem.0.004287
Komatsu M., Takano H., Hiratsuka T., Ishigaki Y., Shimada K., Beppu T., et al. (2006). Proteins encoded by the conservon of Streptomyces coelicolor A3(2) comprise a membrane-associated heterocomplex that resembles eukaryotic G protein-coupled regulatory system. Mol. Microbiol.62 (6), 1534–1546. doi: 10.1111/j.1365-2958.2006.05461.x
Konstantinidis K. T., Tiedje J. M. (2007). Prokaryotic taxonomy and phylogeny in the genomic era: advancements and challenges ahead. Curr. Opin. Microbiol.10 (5), 504–509. doi: 10.1016/j.mib.2007.08.006
Kramer J., Özkaya Ö., Kümmerli R. (2020). Bacterial siderophores in community and host interactions. Nat. Rev. Microbiol.18 (3), 152–163. doi: 10.1038/s41579-019-0284-4
Krzywinski M., Schein J., Birol I., Connors J., Gascoyne R., Horsman D., et al. (2009). Circos: an information aesthetic for comparative genomics. Genome Res.19 (9), 1639–1645. doi: 10.1101/gr.092759.109
Kumar S., Stecher G., Li M., Knyaz C., Tamura K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol.35 (6), 1547–1549. doi: 10.1093/molbev/msy096
Kusuma A. B., Nouioui I., Klenk H. P., Goodfellow M. (2020). Streptomyces harenosi sp. nov., a home for a gifted strain isolated from Indonesian sand dune soil. Int. J. Syst. Evol. Microbiol.70 (9), 4874–4882. doi: 10.1099/ijsem.0.004346
Lagesen K., Hallin P., Rødland E. A., Staerfeldt H. H., Rognes T., Ussery D. W. (2007). RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res.35 (9), 3100–3108. doi: 10.1093/nar/gkm160
Lam K. S. (2006). Discovery of novel metabolites from marine actinomycetes. Curr. Opin. Microbiol.9 (3), 245–251. doi: 10.1016/j.mib.2006.03.004
Lee I., Kim Y. O., Park S. C., Chun J. (2016). OrthoANI: An improved algorithm and software for calculating average nucleotide identity. Int. J. Syst. Evol. Microbiol.66, 1100–1103. doi: 10.1099/ijsem.0.000760
Lee W., Kim M. A., Park I., Hwang J. S., Na M., Bae J. S. (2017). Novel direct factor xa inhibitory compounds from Tenebrio molitor with anti-platelet aggregation activity. Food Chem. Toxicol.109, 19–27. doi: 10.1016/j.fct.2017.08.026
Li Y. Q., Han L., Rong H., Li L. Y., Zhao L. X., Wu L. X., et al. (2015). Diastaphenazine, a new dimeric phenazine from an endophytic Streptomyces diastaticus subsp. ardesiacus. J. Antibiot.68 (3), 210–212. doi: 10.1038/ja.2014.124
Liu N., Shang F., Xi L., Huang Y. (2013). Tetroazolemycins a and b, two new oxazole-thiazole siderophores from deep-sea Streptomyces olivaceus FXJ8.012. Mar. Drugs11 (5), 1524–1533. doi: 10.3390/md11051524
López J. M. S., Insua M. M., Baz J. P., Puentes J. L. F., Hernández L. M. C. (2003). New cytotoxic indolic metabolites from a marine Streptomyces. J. Nat. Prod.66 (6), 863–864. doi: 10.1021/np0204444
Madden A. A., Grassetti A., Soriano J. A. N., Starks P. T. (2013). Actinomycetes with antimicrobial activity isolated from paper wasp (Hymenoptera: Vespidae: Polistinae) nests. Environ. Entomol.42 (4), 703–710. doi: 10.1603/EN12159
Maiti P. K., Mandal S. (2021). Streptomyces cupreus sp. nov., an antimicrobial producing actinobacterium isolated from Himalayan soil. Arch. Microbiol. 203, 1601–1609. doi: 10.1007/s00203-020-02160-y
Matsumoto A., Takahashi Y. (2017). Endophytic actinomycetes: promising source of novel bioactive compounds. J. Antibiot.70 (5), 514–519. doi: 10.1038/ja.2017.20
Meier-Kolthoff J. P., Auch A. F., Klenk H. P., Göker M. (2013). Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinf.14, 60. doi: 10.1186/1471-2105-14-60
Meier-Kolthoff J. P., Carbasse J. S., Peinado-Olarte R. L., Göker M. (2022). TYGS and LPSN: a database tandem for fast and reliable genome-based classification and nomenclature of prokaryotes. Nucleic Acids Res.50 (D1), D801–D807. doi: 10.1093/nar/gkab902
Melander R. J., Minvielle M. J., Melander C. (2014). Controlling bacterial behavior with indole-containing natural products and derivatives. Tetrahedron70 (37), 6363–6372. doi: 10.1016/j.tet.2014.05.089
Navarro-Muñoz J. C., Selem-Mojica N., Mullowney M. W., Kautsar S. A., Tryon J. H., Parkinson E. I., et al. (2020). A computational framework to explore large-scale biosynthetic diversity. Nat. Chem. Biol.16 (1), 60–68. doi: 10.1038/s41589-019-0400-9
Nonno R., Pannacci M., Lucini V., Angeloni D., Fraschini F., Stankov B. M. (1999). Ligand efficacy and potency at recombinant human MT2 melatonin receptors: evidence for agonist activity of some mt1-antagonists. Br. J. Pharmacol.127 (5), 1288–1294. doi: 10.1038/sj.bjp.0702658
Ogasawara Y., Yackley B. J., Greenberg J. A., Rogelj S., Melançon C. E. (2015). Expanding our understanding of sequence-function relationships of type II polyketide biosynthetic gene clusters: Bioinformatics-guided identification of frankiamicin a from frankia sp. EAN1pec. PloS One10 (4), e0121505. doi: 10.1371/journal.pone.0121505
Ozaki T., Nishiyama M., Kuzuyama T. (2013). Novel tryptophan metabolism by a potential gene cluster that is widely distributed among actinomycetes. J. Biol. Chem.288 (14), 9946–9956. doi: 10.1074/jbc.M112.436451
Paguigan N. D., Rivera-Chávez J., Stempin J. J., Augustinović M., Noras A. I., Raja H. A., et al. (2019). Prenylated diresorcinols inhibit bacterial quorum sensing. J. Nat. Prod.82 (3), 550–558. doi: 10.1021/acs.jnatprod.8b00925
Park C. J., Andam C. P. (2019). Within-species genomic variation and variable patterns of recombination in the tetracycline producer Streptomyces rimosus. Front. Microbiol.10. doi: 10.3389/fmicb.2019.00552
Petersen F., Zähner H., Metzger J. W., Freund S., Hummel R. P. (1993). Germicidin, an autoregulative germination inhibitor of Streptomyces viridochromogenes NRRL b-1551. J. Antibiot.46 (7), 1126–1138. doi: 10.7164/antibiotics.46.1126
Prieto C., García-Estrada C., Lorenzana D., Martin J. F. (2012). NRPSsp: non-ribosomal peptide synthase substrate predictor. Bioinformatics28 (3), 426–427. doi: 10.1093/bioinformatics/btr659
Qi J., Wang B., Hao B. I. (2004). Whole proteome prokaryote phylogeny without sequence alignment: A K-string composition approach. J. Mol. Evol.58 (1), 1–11. doi: 10.1007/s00239-003-2493-7
Rao M. P. N., Xiao M., Li W. J. (2017). Fungal and bacterial pigments: Secondary metabolites with wide applications. Front. Microbiol.8, 1113. doi: 10.3389/fmicb.2017.01113
R Core Team (2021). "R: A language and environment for statistical computing" (Vienna, Austria: R Foundation for Statistical Computing).
Röttig M., Medema M. H., Blin K., Weber T., Rausch C., Kohlbacher O. (2011). NRPSpredictor2-a web server for predicting NRPS adenylation domain specificity. Nucleic Acids Res.39, W362–W367. doi: 10.1093/nar/gkr323
Rudolf J. D., Alsup T. A., Xu B., Li Z. (2021). Bacterial terpenome. Nat. Prod. Rep.38 (5), 905–980. doi: 10.1039/d0np00066c
Sasser M. (1990). Identification of bacteria by gas chromatography of cellular fatty acids. MIDI technical note 101 (Newark, DE: Microbial ID Inc).
Saygin H., Ay H., Guven K., Cetin D., Sahin N. (2020). Streptomyces cahuitamycinicus sp. nov., isolated from desert soil and reclassification of Streptomyces galilaeus as a later heterotypic synonym of Streptomyces bobili. Int. J. Syst. Evol. Microbiol.70 (4), 2750–2759. doi: 10.1099/ijsem.0.004103
Schumacher R. W., Talmage S. C., Miller S. A., Sarris K. E., Davidson B. S., Goldberg A. (2003). Isolation and structure determination of an antimicrobial ester from a marine sediment-derived bacterium. J. Nat. Prod.66 (9), 1291–1293. doi: 10.1021/np020594e
Seco E. M., Pérez-Zúñiga F. J., Rolón M. S., Malpartida F. (2004). Starter unit choice determines the production of two tetraene macrolides, rimocidin and CE-108, in Streptomyces diastaticus var. 108. Chem. Biol.11 (3), 357–366. doi: 10.1016/j.chembiol.2004.02.017
Seemann T. (2014). Prokka: rapid prokaryotic genome annotation. Bioinformatics30 (14), 2068–2069. doi: 10.1093/bioinformatics/btu153
Seipke R. F. (2015). Strain-level diversity of secondary metabolism in Streptomyces albus. PloS One10 (1), e0116457. doi: 10.1371/journal.pone.0116457
Shannon P., Markiel A., Ozier O., Baliga N. S., Wang J. T., Ramage D., et al. (2003). Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res.13 (11), 2498–2504. doi: 10.1101/gr.1239303
Shen S., Li W. (2020). The inhibitory effects of metabolites from Bacillus pumilus on potato virus y and the induction of early response genes in Nicotiana tabacum. AMB Express10 (1), 152. doi: 10.1186/s13568-020-01089-1
Shirling E. T., Gottlieb D. (1966). Methods for characterization of Streptomyces species. Int. J. Syst. Bacteriol.16 (3), 313–340. doi: 10.1099/00207713-16-3-313
Simão F. A., Waterhouse R. M., Ioannidis P., Kriventseva E. V., Zdobnov E. M. (2015). BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics31 (19), 3210–3212. doi: 10.1093/bioinformatics/btv351
Sottorff I., Wiese J., Lipfert M., Preußke N., Sönnichsen F. D., Imhoff J. E. (2019). Different secondary metabolite profiles of phylogenetically almost identical Streptomyces griseus strains originating from geographically remote locations. Microorganisms7 (6), 166. doi: 10.3390/microorganisms7060166
Stackebrandt E., Goebel B. M. (1994). Taxonomic note: A place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int. J. Syst. Evol. Microbiol.44 (4), 846–849. doi: 10.1099/00207713-44-4-846
Stritzke K., Schulz S., Laatsch H., Helmke E., Beil W. (2004). Novel caprolactones from a marine streptomycete. J. Nat. Prod.67 (3), 395–401. doi: 10.1021/np030321z
Sullivan M. J., Petty N. K., Beatson S. A. (2011). Easyfig: a genome comparison visualizer. Bioinformatics27 (7), 1009–1010. doi: 10.1093/bioinformatics/btr039
Takahashi Y., Nakashima T. (2018). Actinomycetes, an inexhaustible source of naturally occurring antibiotics. Antibiotics7 (2), 45. doi: 10.3390/antibiotics7020045
Takahashi S., Takagi H., Toyoda A., Uramoto M., Nogawa T., Ueki M., et al. (2010). Biochemical characterization of a novel indole prenyltransferase from streptomyces sp. SN-593. J. Bacteriol.192 (11), 2839–2851. doi: 10.1128/JB.01557-09
Takano H., Hashimoto K., Yamamoto Y., Beppu T., Ueda K. (2011). Pleiotropic effect of a null mutation in the cvn1 conservon of Streptomyces coelicolor A3(2). Gene477 (1-2), 12–18. doi: 10.1016/j.gene.2011.01.005
Tanner M. E. (2015). Mechanistic studies on the indole prenyltransferases. Nat. Prod. Rep.32 (1), 88–101. doi: 10.1039/c4np00099d
Tresner H. D., Davies M. C., Backus E. J. (1961). Electron microscopy of Streptomyces spore morphology and its role in species differentiation. J. Bacteriol.81 (1), 70–80. doi: 10.1128/jb.81.1.70-80.1961
Wang W. F., Qiu Z. Q., Tan H. M., Cao L. X. (2014). Siderophore production by actinobacteria. Biometals27 (4), 623–631. doi: 10.1007/s10534-014-9739-2
Xu L., Ye K. X., Dai W. H., Sun C., Xu L. H., Han B. N. (2019). Comparative genomic insights into secondary metabolism biosynthetic gene cluster distributions of marine Streptomyces. Mar. Drugs17 (9), 498. doi: 10.3390/md17090498
Yang P. W., Li M. G., Zhao J. Y., Zhu M. Z., Shang H., Li J. R., et al. (2010). Oligomycins a and c, major secondary metabolites isolated from the newly isolated strain Streptomyces diastaticus. Folia Microbiol.55 (1), 10–16. doi: 10.1007/s12223-010-0002-0
Yarza P., Richter M., Peplies J., Euzeby J., Amann R., Schleifer K. H., et al. (2008). The all-species living tree project: A 16S rRNA-based phylogenetic tree of all sequenced type strains. Syst. Appl. Microbiol.31 (4), 241–250. doi: 10.1016/j.syapm.2008.07.001
Yoon S. H., Ha S. M., Kwon S., Lim J., Kim Y., Seo H., et al. (2017). Introducing EzBioCloud: a taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int. J. Syst. Evol. Microbiol.67 (5), 1613–1617. doi: 10.1099/ijsem.0.001755
Zotchev S. B. (2012). Marine actinomycetes as an emerging resource for the drug development pipelines. J. Biotechnol.158 (4), 168–175. doi: 10.1016/j.jbiotec.2011.06.002
Keywords: Streptomyces, secondary metabolites, comparative genomics, biosynthesis, marine isolate
Citation: Kim D-E, Hong S-C, Yang Y, Choi J and Park J-S (2022) Chemical and genomic analyses of a marine-derived Streptomyces sp. V17-9 producing amino acid derivatives and siderophores. Front. Mar. Sci. 9:959690. doi: 10.3389/fmars.2022.959690
Received: 02 June 2022; Accepted: 11 July 2022;
Published: 02 August 2022.
Edited by:
Ahmed M Sayed, AlMaaqal University, IraqReviewed by:
Hilal Ay, Ondokuz Mayıs University, TurkeyPatrycja Golinska, Nicolaus Copernicus University in Toruń, Poland
Copyright © 2022 Kim, Hong, Yang, Choi and Park. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jaeyoung Choi, amFleW91bmcuY2hvaUBraXN0LnJlLmty; Jin-Soo Park, amluc29vcGFya0BraXN0LnJlLmty
†Present address: Da-Eun Kim, Green-Bio Research Facility Center, Institutes of Green-Bio Science and Technology, Seoul National University, Pyeongchang, South Korea
‡These authors have contributed equally to this work and share first authorship
 Da-Eun Kim1†‡
Da-Eun Kim1†‡ Sung-Chul Hong
Sung-Chul Hong Jaeyoung Choi
Jaeyoung Choi Jin-Soo Park
Jin-Soo Park