
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mar. Sci. , 22 July 2022
Sec. Marine Fisheries, Aquaculture and Living Resources
Volume 9 - 2022 | https://doi.org/10.3389/fmars.2022.946182
This article is part of the Research Topic The Transmission and Prevention of Infectious Diseases in Aquatic Animals View all 7 articles
The leucine-rich repeat (LRR) motif is evolutionarily conserved in many pattern recognition receptors. Compared to the reported LRR proteins with multiple functional domains, the role of LRR-only proteins merely containing LRR motifs remain largely unexplored. In this study, two LRR-only proteins, PtLRR1 and PtLRR2, were identified from the swimming crab Portunus trituberculatus. Five LRR motifs with a consensus sequence LxxLxxLxLxxNxL were found in their encoded peptides. Both PtLRR1 and PtLRR2 were dominantly expressed in the hepatopancreas and showed a time-dependent response post bacteria and virus stimulation. The recombinant PtLRR1 could bind to various PAMPs, including LPS, PGN, and GLU. PtLRR1 and PtLRR2 displayed different regulatory activities in inducing the expression of inflammation and proPO system-related genes. Knockdown of PtLRR2 led to the decreased expression of the tested cytokines and adapter, while PtLRR1 knockdown enhanced the expression of serine proteases, serine protease homologues, and proPO genes. In addition, knockdown of PtLRR1 or PtLRR2 reduced the clearance activity of Vibrio but upregulated the expression levels of AMPs and key genes of Toll, IMD, and JNK pathways. These results suggest that PtLRR1 and PtLRR2 could act as potential immune receptors and regulate antibacterial immunity in crab.
Leucine-rich repeat (LRR) proteins constitute a large protein superfamily widely found in microorganisms, plants, and animals (Kobe and Kajava, 2001). These LRR proteins usually have 2–45 LRR motifs and can be divided into two protein groups, LRR-only proteins merely containing LRR motifs and proteins containing LRR motifs and other motifs (Huang et al., 2011; Xu, 2011). Most LRR proteins are involved in multiple physiological processes through LRR motifs, for example, protein–ligand or protein–protein interaction, signal transduction, cell adhesion, and host immune defense (Kobe and Kajava, 2001; Kędzierski et al., 2004 ).
The LRR motif was firstly identified in the leucine-rich alpha 2-glycoprotein from human plasma (Takahashi et al., 1985). It consists of 20–30 amino acid residues with the highly conserved sequence LxxLxLxxN/CxL (Bella et al., 2008). Multiple LRR motifs form an LRR domain adopting a horseshoe-shaped structure to mediate the protein–protein interactions (Kędzierski et al., 2004). The LRR domain is a common feature of many pattern recognition receptors (PRRs), such as NBS (nucleotide-binding site)-LRR proteins, Toll-like receptors (TLRs), NOD-like receptors (NLRs), and variable lymphocyte receptors (VLRs) (Ng and Xavier, 2011; Matsushima et al., 2019). Plant NBS-LRR proteins are involved in the recognition of pathogen effector avirulence (AVR) proteins (DeYoung and Innes, 2006). TLRs and NLRs play crucial roles in immune recognition of PAMPs from pathogens and activation of specific signaling pathways, such as NF-κB and MAPK pathways (Kawai and Akira, 2009; Valanne et al., 2011).
Apart from the above LRR proteins, LRR-only proteins merely containing LRR motifs remain uncharacterized functionally in invertebrates. There were several LRR-only proteins found in crustaceans, such as PmLRR with 16 LRR motifs from black tiger shrimp Penaeus monodon (Sriphaijit and Senapin, 2007), SpLRR with 17 LRR motifs from mud crab Scylla paramamosain (Cao et al., 2013), and SsLRR with 17 LRR motifs from S. serrata (Vidya et al., 2016). These LRR-only proteins contained more than 15 LRR motifs, and their expression levels could be significantly changed after stimulation with bacteria or virus. In addition, MjLRRC1 with 19 LRR motifs from kuruma shrimp Marsupenaeus japonicus could regulate the expression of some antimicrobial peptide (AMP) genes for bacterial clearance in stomach (Shi et al., 2017). In scallop, two LRR-only proteins CfLRRop-1 and 3 identified from Chlamys farreri were reported to bind various PAMPs, and CfLRRop-1 and 2 might act as a pro-inflammatory factor that induce the release of TNF-α in scallop hemocytes (Wang et al., 2016a; Wang et al., 2016b).
Compared with the reports in shrimp and scallop, the roles of the LRR-only proteins in crabs remain largely unknown. In the present study, two novel LRR-only proteins containing five LRR motifs, designed as PtLRR1 and PtLRR2, were identified and characterized from the swimming crab Portunus trituberculatus. The expression patterns of PtLRR1 and PtLRR2 of tissue distribution and after pathogen challenge were detected. The recombinant protein (rPtLRR1) was tested for the PAMP-binding ability. In the case of PtLRR1 or PtLRR2 knockdown by small interfering RNA (siRNA), the roles of PtLRR1 and PtLRR2 in regulating the expression of cytokines, proPO system-related genes, and AMPs were investigated, and the effects of promoting the clearance activity of bacteria were detected.
Live healthy crabs (110 ± 10 g) were obtained from an aquatic product market in Qingdao, Shandong Province, China. The crabs were transported to the laboratory and temporarily cultured at 12°C–14°C aerated seawater for 1 week. During the period, the crabs were fed with clam meat once daily, and the seawater in the tank was replaced daily after 3 h of feeding. The samples of the eyestalk, hemocytes, muscle, gill, hepatopancreas, intestine, stomach, and testis from five healthy crabs were obtained for tissue distribution analysis.
For the immune stimulation experiment, Vibrio parahaemolyticus, Staphylococcus aureus, and white spot syndrome virus (WSSV) were prepared and diluted in PBS at the concentration of 3 × 108 CFU/ml, 3 × 108 CFU/ml, and 3.2 × 108 copies/ml, respectively. Crabs were randomly divided into four groups including one control and three immune stimulation groups, and each group contained 70 individuals. In the immune stimulation groups, crabs were injected with 100 μl V. parahaemolyticus, S. aureus or WSSV, respectively. Crabs injecting with PBS served as the control group. Hepatopancreas was collected from crabs at 0, 3, 6, 12, 24, 48, and 72 h post injection. Each sampling contained five biologically repeats.
Total RNA was extracted from various tissues and cells using RNAiso Plus Reagent (TaKaRa). The first-strand cDNA was synthesized using the PrimeScript™ II 1st cDNA Synthesis Kit (TaKaRa). Based on the unigene sequences, primers (PtLRR1-F and PtLRR1-R, PtLRR2-F1 and PtLRR2-R1, PtLRR2-F2 and PtLRR2-R2, Table 1) were designed to amplify the open reading frames of PtLRR1 or PtLRR2. The PCR was performed in a 25-μl reaction volume containing 9.5 μl sterile distilled H2O, 12.5 μl of 2× Accurate Taq Master Mix (Accurate Biotechnology, Changsha, China), 1 μl of each primer (10 μmol l-1), and 1 μl of DNA template (approximately 50 ng). The PCR amplification procedure was as follows: 94°C for 30 s, followed by 35 cycles at 98°C for 10 s, 55°C for 30 s, and 72°C for 1 min, and finally 72°C for 2 min. The primers (3P2, PtLRR1-3′-1 and PtLRR2-3′-1, 3P4, PtLRR1-3′-2 and PtLRR2-3′-2, Table 1) were designed to clone the 3′ ends of PtLRR1 and PtLRR2 by RACE. The full length of PtLRR1 and PtLRR2 was obtained by overlap extension.
Sequence similarity analysis was analyzed with the BLAST algorithm (http://blast.ncbi.nlm.nih.gov/Blast.cgi). The SignalP 4.0 program was used to predict the signal peptide (http://www.cbs.dtu.dk/services/SignalP). The ExPASy ProtParam tool was applied to predict the isoelectric point (pI) and molecular weight (Mw) (http://prosite.expasy.org/). Multiple alignments of LRR motifs in PtLRR1 and PtLRR2 were performed with ClustalW (http://www.clustal.org) and illustrated using WebLogo application 3.7.4 (http://weblogo.threeplusone.com/). A phylogenetic tree was constructed using the neighbor-joining (NJ) method by MEGA 7.0 software. Tertiary protein structures of PtLRR1 and PtLRR2 were predicted using the SWISS-MODEL program (https://swissmodel.expasy.org/).
The expression patterns of PtLRR1 and PtLRR2 were determined by the specific primers PtLRR1-RTF and PtLRR1-RTR, PtLRR2-RTF and PtLRR2-RTR (Table 1). The cDNA product was diluted 20-fold in deionized water. PCR reaction was performed in a 10-µl reaction system, containing 2.28 µl of sterile distilled H2O, 3.33 µl of 2× SYBR Premix Ex Taq (TaKaRa), 0.13 µl of 50× ROX Reference Dye, 0.13 µl of each primer (10 µmol l−1), and 4 µl of the diluted cDNA. The PCR program was 95°C for 30 s, followed by 40 cycles of 95°C for 5 s and 60°C for 35 s. Each sample was run in triplicate. The relative expression levels of PtLRR1 and PtLRR2 were calculated by the 2-ΔΔCt method using the β-actin gene as internal standardization. Data were analyzed via one-way ANOVA and Duncan’s test using SPSS 16.0, and the difference was considered significant if P values are less than 0.05 and 0.01.
PtLRR1_ReF and PtLRR1_ReR (Table 1) with restriction enzymes were used to obtain the sequence encoding the mature peptide of PtLRR1. The amplified PCR fragments digested with restriction enzymes (Bam HI and Xho I) and subcloned into the pET-32a expression vector with the 6× His tag. The plasmid pET32a-PtLRR1 and pET-32a (empty vector) were transformed into E. coli BL21 (DE3) cells (Transgen, Beijing, China) for IPTG-induced expression. The recombinant PtLRR1 protein (rPtLRR1) and the negative control (rTrx) were purified under denaturing conditions using TALON metal affinity resin (Clontech, Shiga, Japan) following the instructions. The obtained proteins were concentrated with Amicon Ultra Centrifugal Filter, and the concentrations were measured by BCA Protein Assay Kit (Beyotime, Shanghai, China). After separation on an 8%–15% SDS-PAGE gel, the proteins were observed with Coomassie Brilliant Blue.
The PAMP binding activity of rPtLRR1 was determined by ELISA (Zhang et al., 2008). Lipopolysaccharide (LPS, Yuanye, China), peptidoglycan (PGN, Yuanye, China), and glucan (GLU, Yuanye, China) were dissolved in carbonate-bicarbonate buffer (50 mmol l−1, pH 9.6), respectively. Then, 100 μl of the PAMP solution (20 μg) was added to the 96-well plate (Costar/Corning, Tewksbury, MA, USA) and placed overnight at 4°C. After washing by PBS-T (pH 7.5), the plate was blocked by PBS containing 3% BSA for 1 h at 37°C and then washed with PBS-T for three times. One hundred microliters of diluted rPtLRR1 was added to wells to bind PAMPs in the presence of 0.1 mg ml−1 BSA and 5 mmol l−1 CaCl2. Moreover, the serial dilution of rTrx was used as a negative control. The mouse anti-His tag antibody (Transgen, China) as primary antibody and the goat-anti-mouse Ig-HRP conjugate (Transgen, China) as secondary antibody could be diluted 1:1,000 from 1 mg ml−1 BSA in PBS. For each well, 100 µl of primary antibody was incubated for 1 h at 37°C, unbound primary antibodies were washed away with PBS-T buffer, and then 100 µl of secondary antibody was added and incubated at 37°C for 1 h. After washing the excess antibody, the chromogenic reaction was performed using EL-TMB chromogenic kit (Sangon, Shanghai, China). The absorbance was measured at 450 nm. Each experiment was repeated in triplicate. The ELISA index (EI) was calculated as follows: EI = ODsample/cutoff, where the cutoff was mean OD values of negative controls + three SDs per point. Each sample was run in triplicate. Samples with EI > 1.0 were considered to be positive.
The sequence-specific PtLRR1-siRNA(5′-CCUACACUACAU CAACACC-3′) and PtLRR2-siRNA(5′TACTTGTGCTGGCA ACGACTT-3′) were designed to knock down the expression of PtLRR1 or PtLRR2. The iRNA of EGFP(5′CAGCCACAACGU CUAUAUC-3′) was synthesized as the control group. The siRNAs of PtLRR1 and PtLRR2 was synthesized using invitro transcription T7 kit (Takara, Japan) according to the manufacturer’s instructions. The synthesized siRNAs were diluted with 0.1 mol l-1 PBS to a final concentration of 2.0 μg μl-1 before injection.
Crabs (100 g ± 5 g) were randomly divided into three groups with 10 individuals in each group. Each individual of the experimental groups were injected with 100 μl PtLRR1-siRNA or PtLRR2-siRNA. The crabs were injected with 100 μl PBS or 100 μl siRNA-EGFP as blank group and control group, respectively. Each crab was injected with 100 μl siRNA or PBS for the first time, and the same amount of siRNA or PBS was injected at 24 h after the first injection. At 48 h after the first injection of siRNA, the hepatopancreas from crabs in each group was separately isolated and frozen in liquid nitrogen for RNA extraction and cDNA synthesis. The amount of injected siRNA was chosen according to our preexperiment and the RNAi assay in the swimming crab (Su et al., 2020). Then the RNA interference efficiency of PtLRR1 and PtLRR2 was determined using qRT-PCR. The experiments were biologically repeated five times.
The transcripts of inflammation-related genes PtIL-17, PtIL-16, PtTRAF6, and PtLITAF; six proPO system-related genes PtcSP1-3, PtSPH, PtPPAF, and PtproPO; eight AMP genes PtALF1-7 and PtCrustin1-3; and five genes PtTLR, PtMyD88, PtRelish, PtPelle, and PtJNK of Toll, IMD, and JNK pathways were examined in the hepatopancreas of PtLRR1 or PtLRR2-knockeddown crabs. The amplification efficiencies of the above genes were within 96%–105%.
V. parahaemolyticus was confirmed as the main causative agent of cultured crustaceans. V. parahaemolyticus was selected in this assay and cultured in TSB medium at 28°C to the logarithmic growth stage. After centrifugation, the bacteria were washed with PBS for two times and diluted to 3 × 106 CFU ml-1. For the bacterial clearance test, the PtLRR1 or PtLRR2-knockdown crabs were injected with V. parahaemolyticus. Five crabs of each group were injected with 100 μl diluted V. parahaemolyticus. At 0, 6, 12, and 24 h postinjection, 50 μl of hemolymph was extracted from each crab. The hemolymph of five crabs at each time point was mixed and diluted 10-fold in PBS, and then 100 μl of the diluted hemolymph was plated on LB medium. After culture at 28°C for 16 h, bacterial colonies on the plate were counted. The assay was repeated three times.
The full-length cDNA sequences of PtLRR1 and PtLRR2 obtained by RACE were 1,601 and 1,026 bp in length, respectively. PtLRR1 contained a 28-bp 5′-untranslated region (UTR), a 966-bp open reading frame (ORF), and a 607-bp 3′-UTR with the polyadenylation signal AATAA and poly (A) tail (Figure 1A). PtLRR1 encoded a protein of 301 amino acid residues that contained a signal peptide (from M1 to A23) and five LRR motifs (from S73 to A95, from L120 to Y142, from L143 to P166, from L167 to F188, from L189 to D212). The molecular weight of mature PtLRR1 was 34.20 kDa, and the theoretical isoelectric point was 4.61.
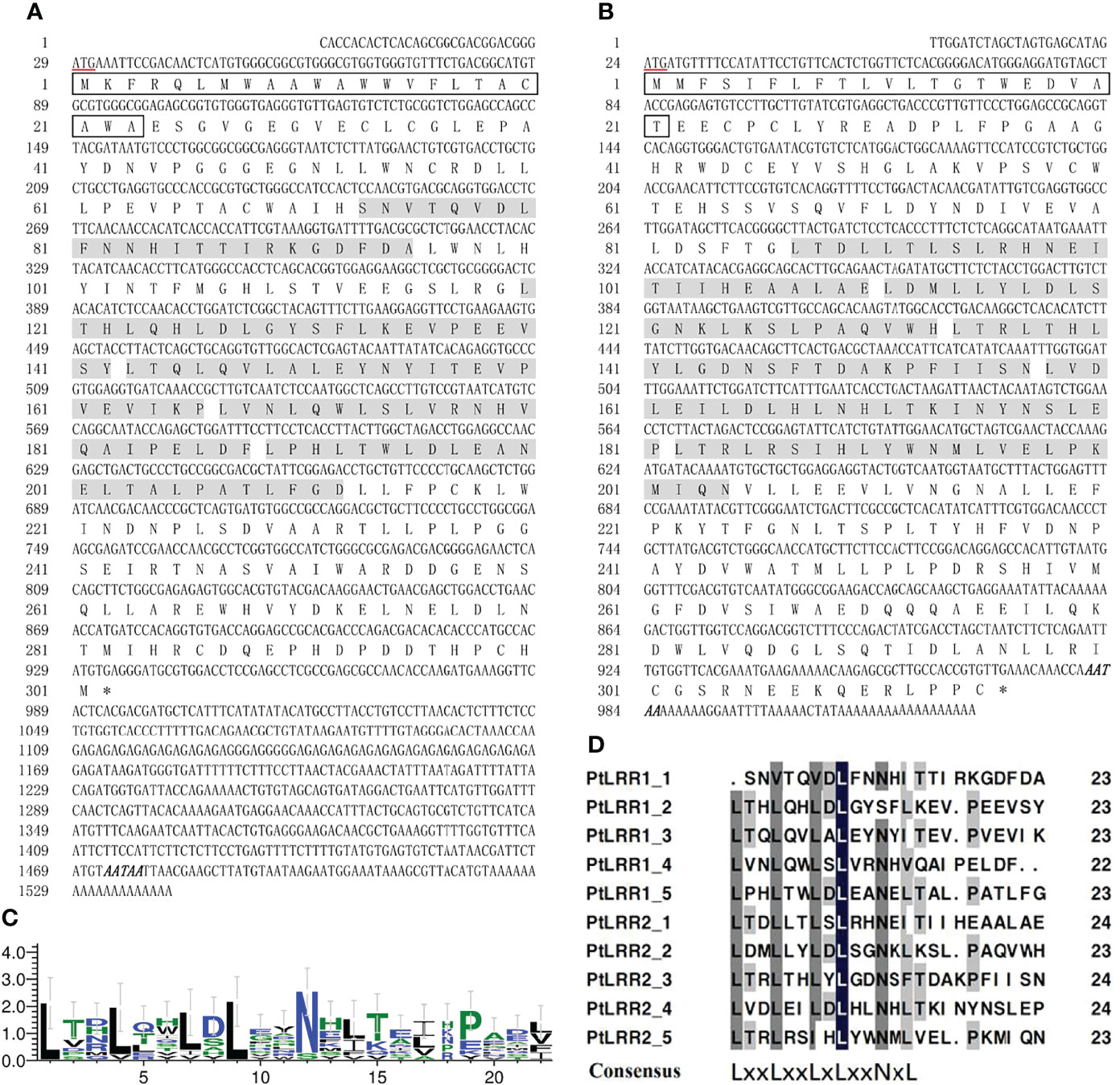
Figure 1 Sequence characterization of PtLRR1 and PtLRR2. (A) cDNA and protein sequences of PtLRR1. (B) cDNA and protein sequences of PtLRR2. The start codon is underlined in red. The stop codon is marked with an asterisk. The signal peptide is boxed in black. The LRR motifs are shaded with gray. The polyadenylation signal is bolded and italicized. (C) The consensus sequence of LRR motifs in PtLRR1 and PtLRR2. (D) Multiple alignments of LRR motifs in PtLRR1 and PtLRR2. L indicates leucine or bulky, non-polar residues, x indicates any amino acid, N refers to asparagine.
PtLRR2 contained a 23-bp 5′-UTR, a 948-bp ORF, and a 55-bp 3′-UTR with the polyadenylation signal AATAA and poly (A) tail (Figure 1B). PtLRR2 encoded a protein of 315 amino acid residues including a signal peptide (from M1 to T21) and five LRR motifs (from L87 to E110, from L111 to H133, from L134 to N157, from L158 to P181, from L182 to N204). The mature PtLRR2 was estimated to be 36.00 kDa, and the theoretical isoelectric point was 4.66. The sequences of PtLRR1 and PtLRR2 were submitted to GenBank under the accession numbers of ON367496 and ON367497.
The LRR motifs of PtLRR1 and PtLRR2 contained 22 or 23 residues, and all conformed to a consensus sequence LxxLxxLxLxxNxL except the first and second LRR motifs of PtLRR1 (Figures 1C, D). The 3D-model analysis revealed that the potential tertiary structures of both PtLRR1 and PtLRR2 could form a typical horseshoe (U-shaped) structure (Figure 2).
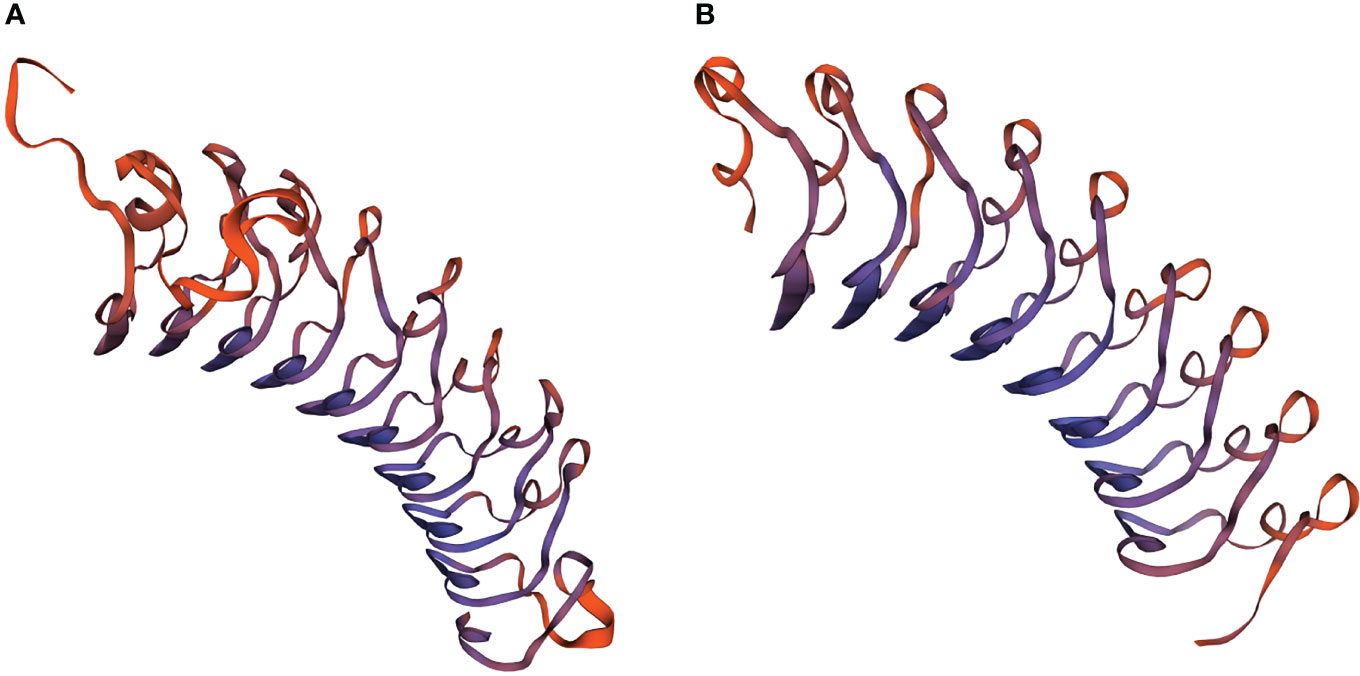
Figure 2 The potential spatial structures of (A) PtLRR1 and (B) PtLRR2 predicted by the SWISS-MODEL program.
The mRNA expression levels of PtLRR1 and PtLRR2 were detected in all the examined tissues (Figure 3). The highest expression of PtLRR1 was found in hepatopancreas, which was 34,875.03-fold of that in hemocytes. The moderate expression of PtLRR1 was detected in the testis and eyestalk, and the lower expression was observed in hemocytes, stomach, and gill. The PtLRR2 transcript was also mainly expressed in the hepatopancreas which was 84,071.99-fold of that in hemocytes, while the relative expression levels in muscle, intestine and testis were much lower.
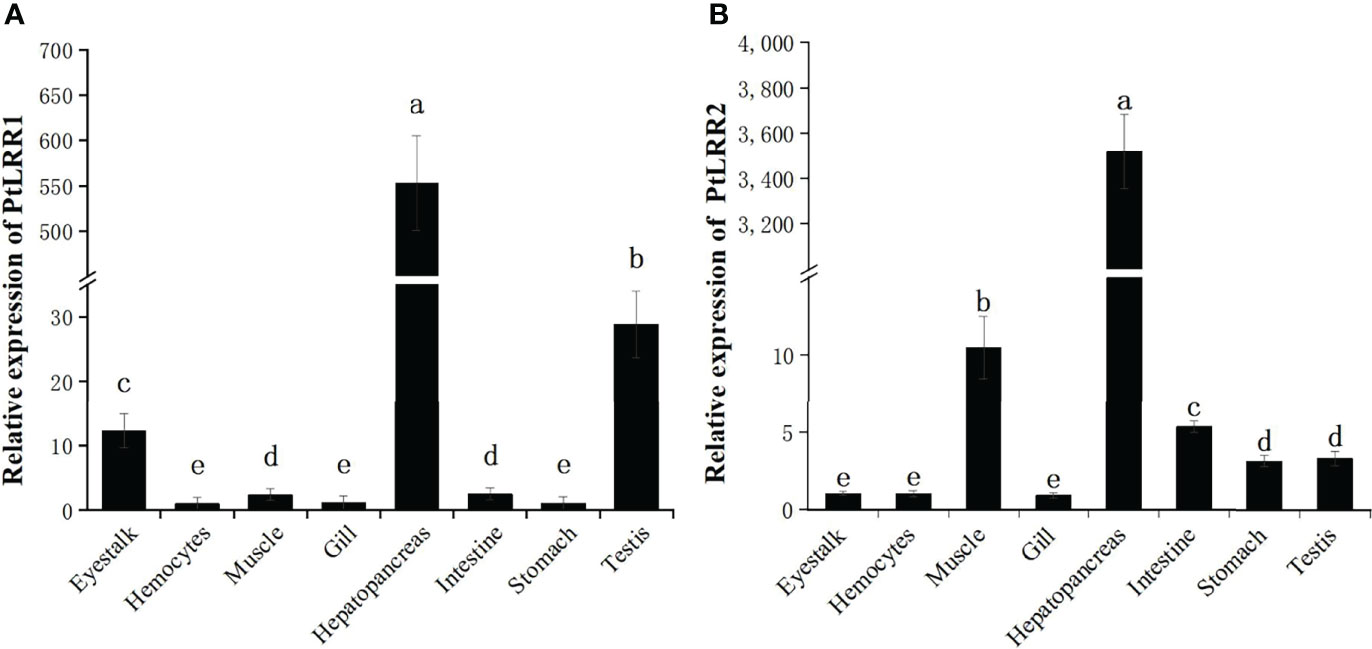
Figure 3 Tissue distribution of (A) PtLRR1 and (B) PtLRR2. Data are shown as mean ± S.D. (n = 5) and different letters indicate a significant difference at P < 0.05 or P < 0.01.
The temporal expression of PtLRR1 and PtLRR2 in the hepatopancreas was investigated post V. parahaemolyticus, S. aureus, and WSSV stimulation (Figure 4). After being challenged with WSSV, the expression of PtLRR1 could be rapidly upregulated at 6 h, peaked at 12 h (8.21-fold to control, P < 0.01), and recovered to the control level after 24 h postinjection (Figure 4A). PtLRR1 expression was only increased and peaked at 12 h (4.21-fold to control, P < 0.01) in the S. aureus injection group. PtLRR1 showed a slow response to V. parahaemolyticus, and its expression was peaked until 24 h after injection (4.37-fold to control, P < 0.01). Then, PtLRR1 expression decreased but was 2.00-fold higher than that in the control group (P < 0.05).
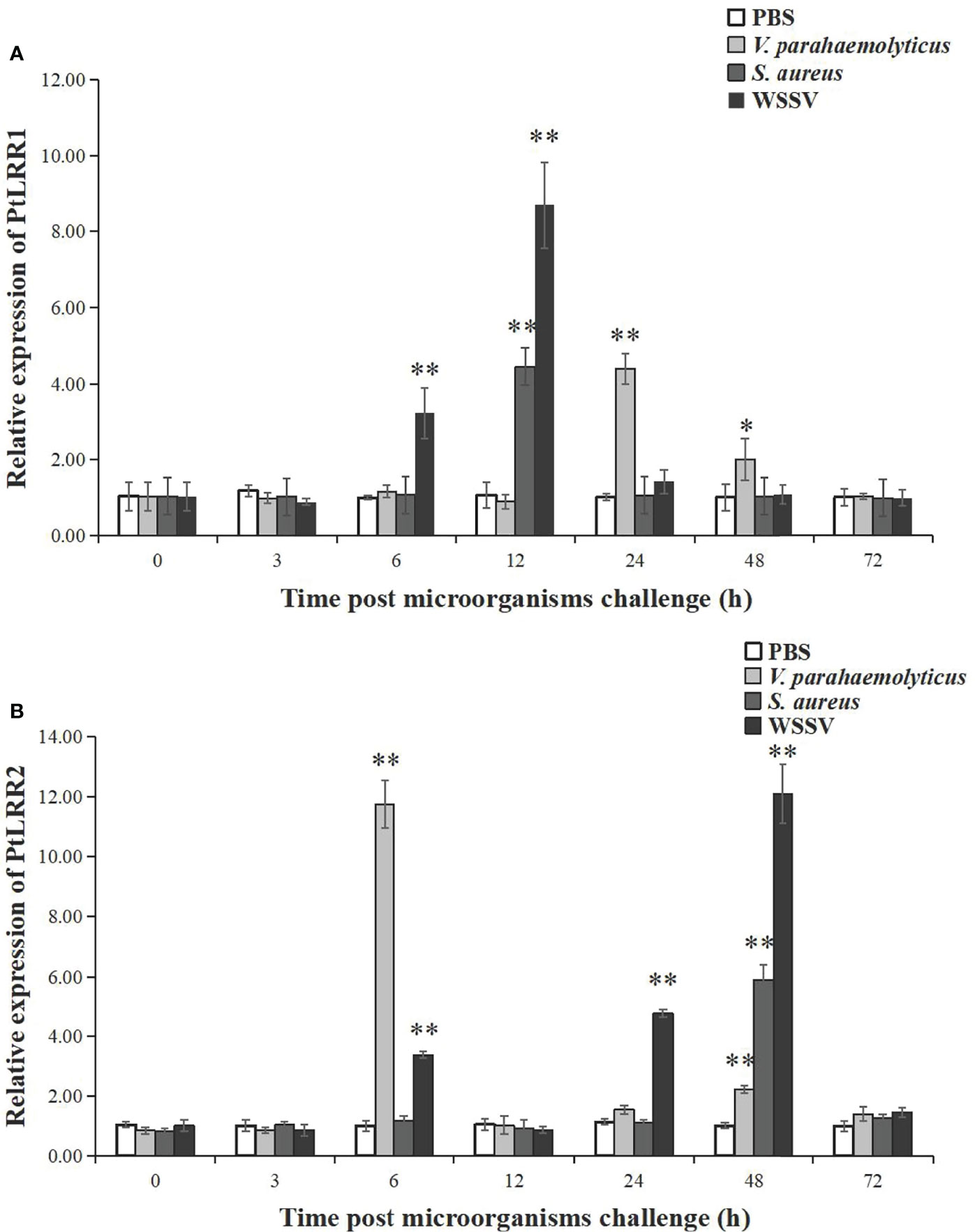
Figure 4 Relative expression of (A) PtLRR1 and (B) PtLRR2 after challenge with V. parahaemolyticus (light grey bars), S. aureus (gray bars), and WSSV (black bars). Data are represented as mean ± S.D. (n = 5). Asterisks indicate the significant differences between the experiment group and the control group at the same sampling point (*P < 0.05, **P < 0.01).
As shown in Figure 4B, the expression of PtLRR2 could be significantly increased and peaked at 6 h (11.75-fold to control, P < 0.01) after being challenged with V. parahaemolyticus. As time progressed, the expression level of PtLRR2 was dropped and reached a second peak at 48 h post V. parahaemolyticus injection (2.19-fold to control, P < 0.01). After WSSV injection, PtLRR2 expression was increased to the first peak at 6 h postinjection (3.35-fold to control, P < 0.01), then decreased at 12 h and reached the maximum level at 48 h postinjection, which was 12.03-fold to that of the control (P < 0.01). The expression of PtLRR2 was only significantly upregulated at 48 h (5.86-fold to control, P < 0.01) post S. aureus injection.
The recombinant PtLRR1 protein was successfully expressed as inclusion bodies using an E. coli expression system (Figure 5). Consistent with the predicted molecular weight, rPtLRR1 had a distinct band with a molecular mass of approximately 47.44 kDa revealed by SDS-PAGE. Moreover, rTrx was also successfully expressed as a control and was detected to be 20.5 kDa. The obtained rPtLRR1 and rTrx were at the concentrations of 3.64 and 2.14 mg ml-1, respectively.
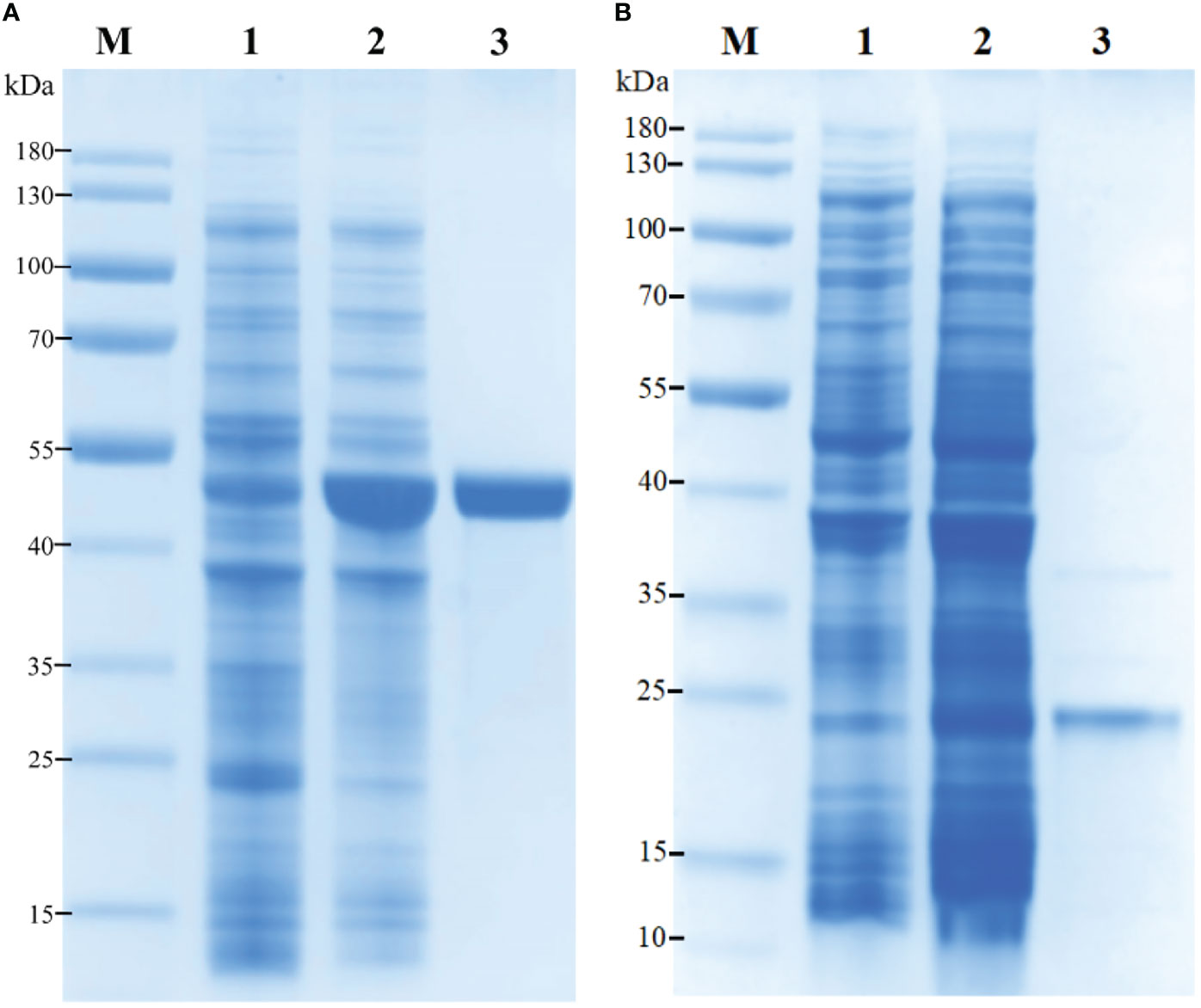
Figure 5 Recombinant expression and purification of (A) rPtLRR1 and (B) rTrx. M, protein marker (kDa); lane 1, total protein of E. coli BL21 (DE3) with recombinant plasmid of pET32a-PtLRR1 or pET32a; lane 2, total protein of E. coli with recombinant plasmid of pET32a-PtLRR1 or pET32a after induction; lane 3, purified recombinant protein.
The binding activity of rPtLRR1 toward various PAMPs was detected by ELISA (Figure 6). The EI values for LPS, PGN, and GLU were above 1.0 when rPtLRR1 was at the concentrations of 4, 8, and 4 μg ml-1, and the binding activity was dose-dependent. At the same protein concentration, rPtLRR1 displayed relatively higher binding affinity to GLU than to PGN and LPS. As the control, rTrx showed no binding effect toward the tested PAMPs.
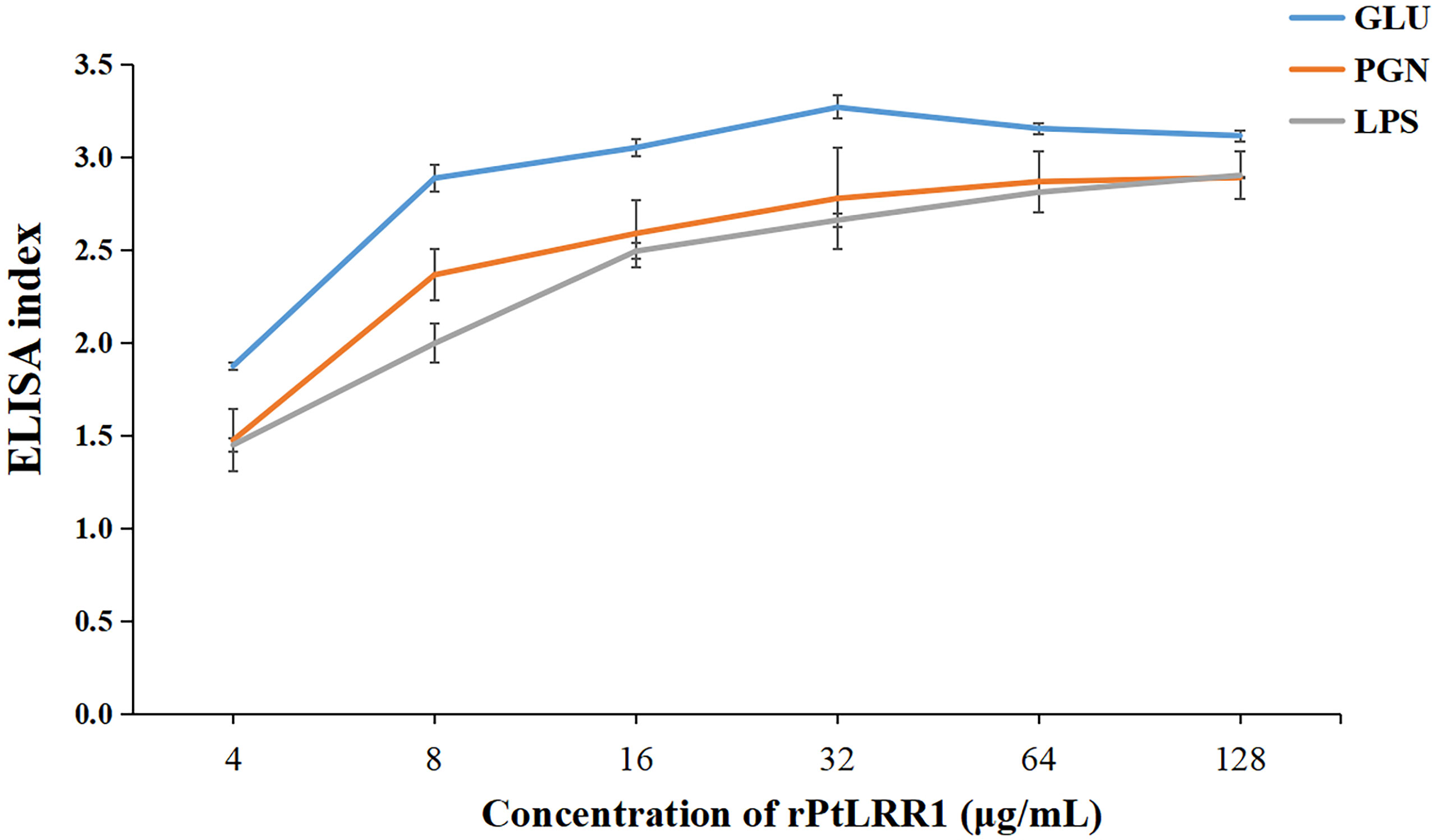
Figure 6 Binding activities of rPtLRR1 to PAMPs. EI values > 1.0 were considered positive. Data are shown as mean ± S.D. (n = 3). LPS, lipopolysaccharide; PGN, peptidoglycan; GLU, glucan.
The siRNA-induced RNAi was performed to knock down the expression of PtLRR1 or PtLRR2. The expression of PtLRR1 and PtLRR2 in the hepatopancreas was significantly down-regulated in crabs injected with PtLRR1-siRNA or PtLRR2-siRNA compared to that in the EGFP-siRNA group. The knockdown efficiency of PtLRR1 and PtLRR2 was 76.31% or 75.35%, respectively (Figure 7).
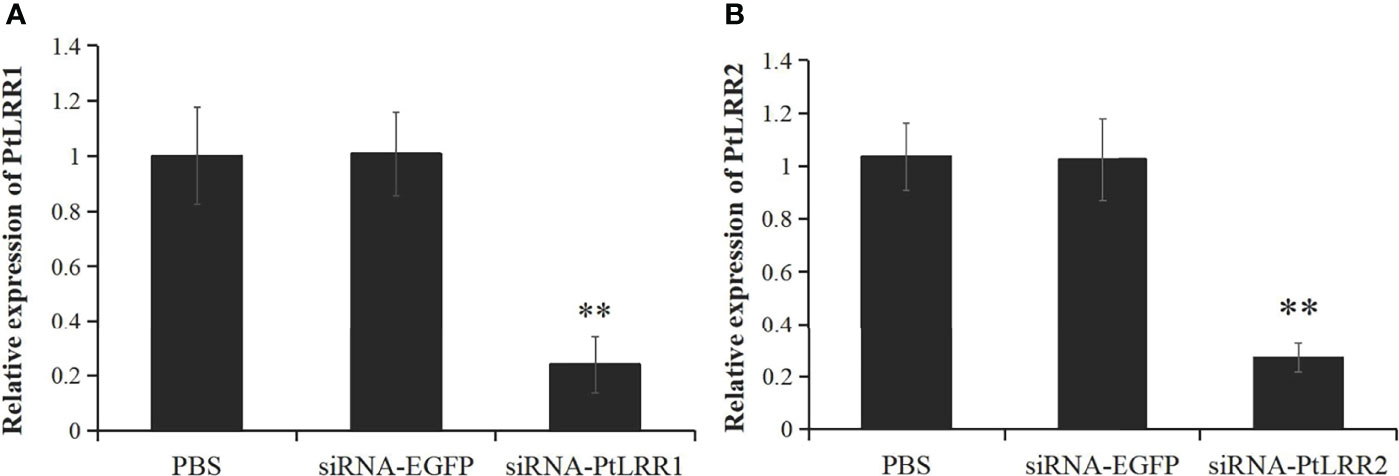
Figure 7 The knockdown efficiency of (A) PtLRR1 and (B) PtLRR2 in crab hepatopancreas by 48 h post siRNA first injection. Data are represented as mean ± S.D. (n = 5). Significant differences across control are indicated with two asterisks (P < 0.01).
After PtLRR1 gene knockdown, the transcription of inflammation-related genes PtIL-16 and PtLITAF, proPO system-related genes PtcSP1-3, PtSPH, PtPPAF, and PtproPO, and AMPs PtALF1-3, PtALF5, PtALF6, and PtCrustin1-3 were obviously upregulated compared with the control group (Figures 8A–C). On the contrary, the expression levels of inflammation-related genes PtIL-17 and PtTRAF6 and five genes of the Toll, IMD, and JNK signaling pathways PtTLR, PtMyD88, PtPelle, PtRelish, and PtJNK were significantly downregulated (Figures 8A–D).
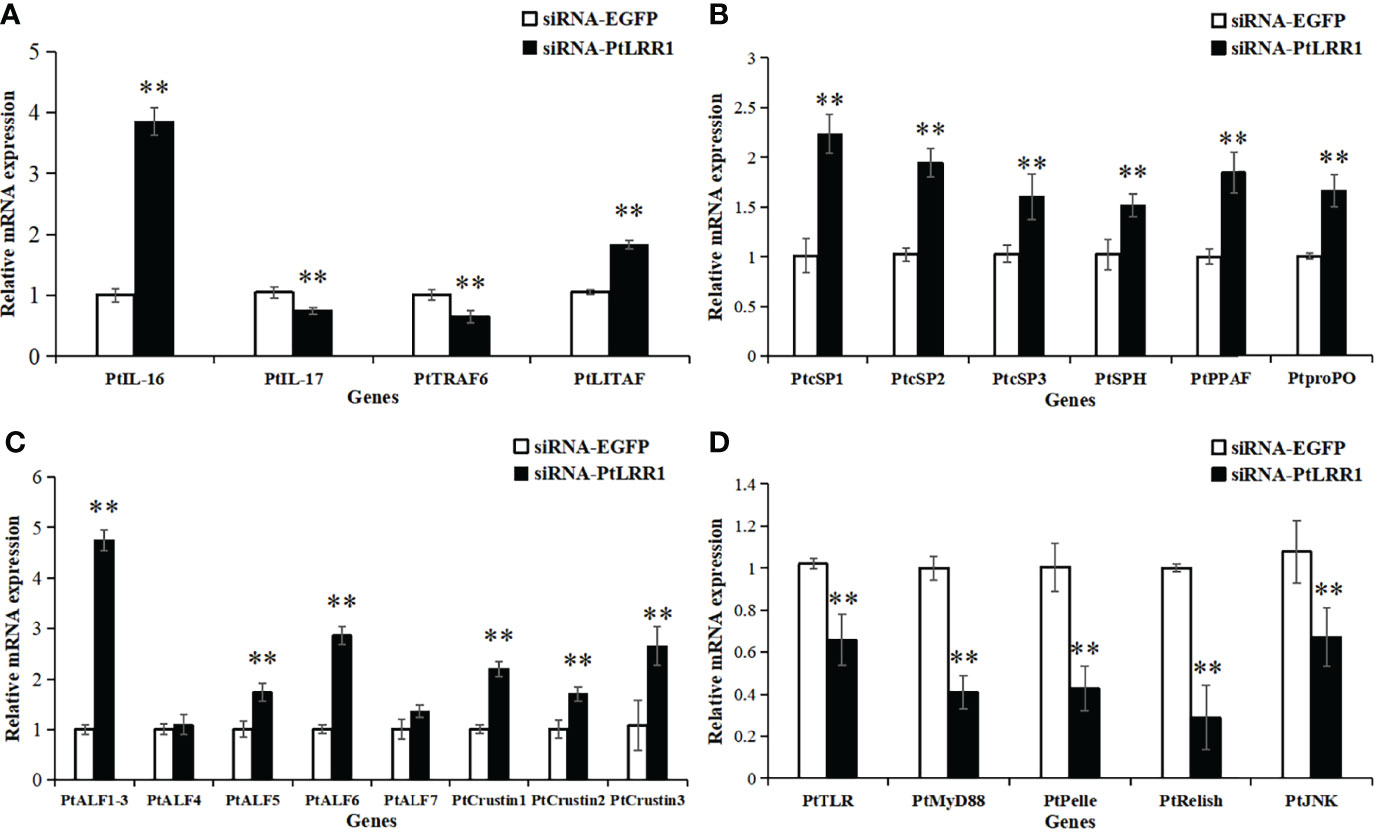
Figure 8 Expression pattern of immune genes after PtLRR1 RNAi. The immune genes included (A) inflammation-related genes, (B) proPO system-related genes, (C) AMPs, and (D) five genes of the Toll, IMD, and JNK pathways. Data are shown as mean ± S.D. (n = 5). Asterisks indicate the significant differences compared with control at the same sampling point (*P < 0.05, **P < 0.01). The genes are denoted as follows: PtIL-17 (interleukin-17, POR|c47537_g2), PtIL-16 (interleukin-16, POR|c99420_g1), PtTRAF6 (tumor necrosis factor receptor-associated factor 6, AKD94181.1) and PtLITAF (LPS induced TNF-α factor, XP_04112677.1), PtcSP1-3 (clip domain serine protease, JF412648, JF412649, and JF412650), PtSPH (serine protease homologue, JF412651), PtPPAF (proPO-activating factor, GQ914996), PtproPO (FJ215871), PtALF1-3 (anti-lipopolysaccharide factor, HM627757, HM627758, and GQ165621), PtALF4-7 (JF756050-JF756053), PtCrustin1-3 (FJ612106, JQ728435, and JQ728425), PtTLR (Toll-like receptor, KR108027.1), PtMyD88 (myeloid differentiation factor 88, KM521426.2), PtPelle (POR|c93425_g1), PtRelish (MF624027.1), and PtJNK (c-Jun N-terminal kinase, POR|c99916_g3).
In the PtLRR2-knockdown crabs, the expression levels of inflammation-related genes PtIL-16, PtIL-17, PtTRAF6, and PtLITAF; proPO system-related PtSP2 and PtproPO; and four genes of the Toll or JNK signaling pathways PtTLR, PtMyD88, PtPelle, and PtJNK were remarkably suppressed (Figures 9A, B–D). Otherwise, the expression levels of proPO system-related genes PtSP3, PtSPH, and PtPPAF, and most AMPs PtALF1-6, PtCrustin1, and PtCrustin3, and PtRelish in the IMD signaling pathway were significantly increased (Figures 9−D).
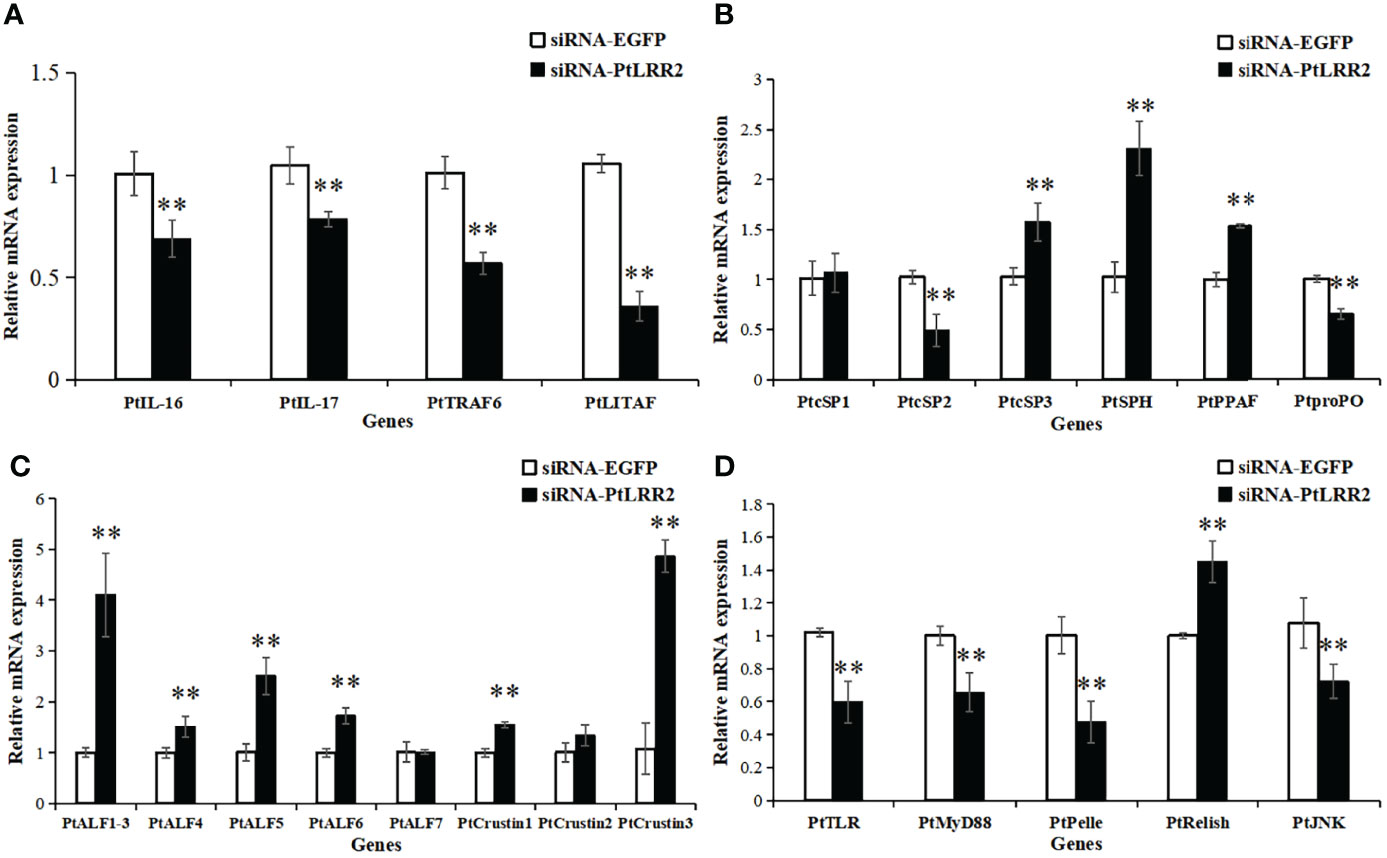
Figure 9 Expression pattern of immune genes after PtLRR2 RNAi. The immune genes included (A) inflammation-related genes, (B) proPO system-related genes, (C) AMPs, and (D) five genes of the Toll, IMD, and JNK pathways. Data are shown as mean ± S.D. (n = 5). Asterisks indicate the significant differences compared with control at the same sampling point (*P < 0.05, **P < 0.01).
To study the immune protection of PtLRR1 and PtLRR2 against V. parahaemolyticus infection, the bacterial clearance activity was tested in PtLRR1-knockdown or PtLRR2-knockdown crabs. Compared to the siRNA-EGFP crabs, the viable bacterial numbers in the siRNA-PtLRR1 or PtLRR2-knockdown crabs were found to significantly increase in the hemocytes from 1 to 12 h (Figure 10). The highest bacterial counts in the PtLRR1-knockdown or PtLRR2-knockdown crabs were found after 6 h post V. parahaemolyticus infection, which were 9.40-fold or 7.92-fold higher amounts to the siRNA-EGFP control (P < 0.01), respectively.
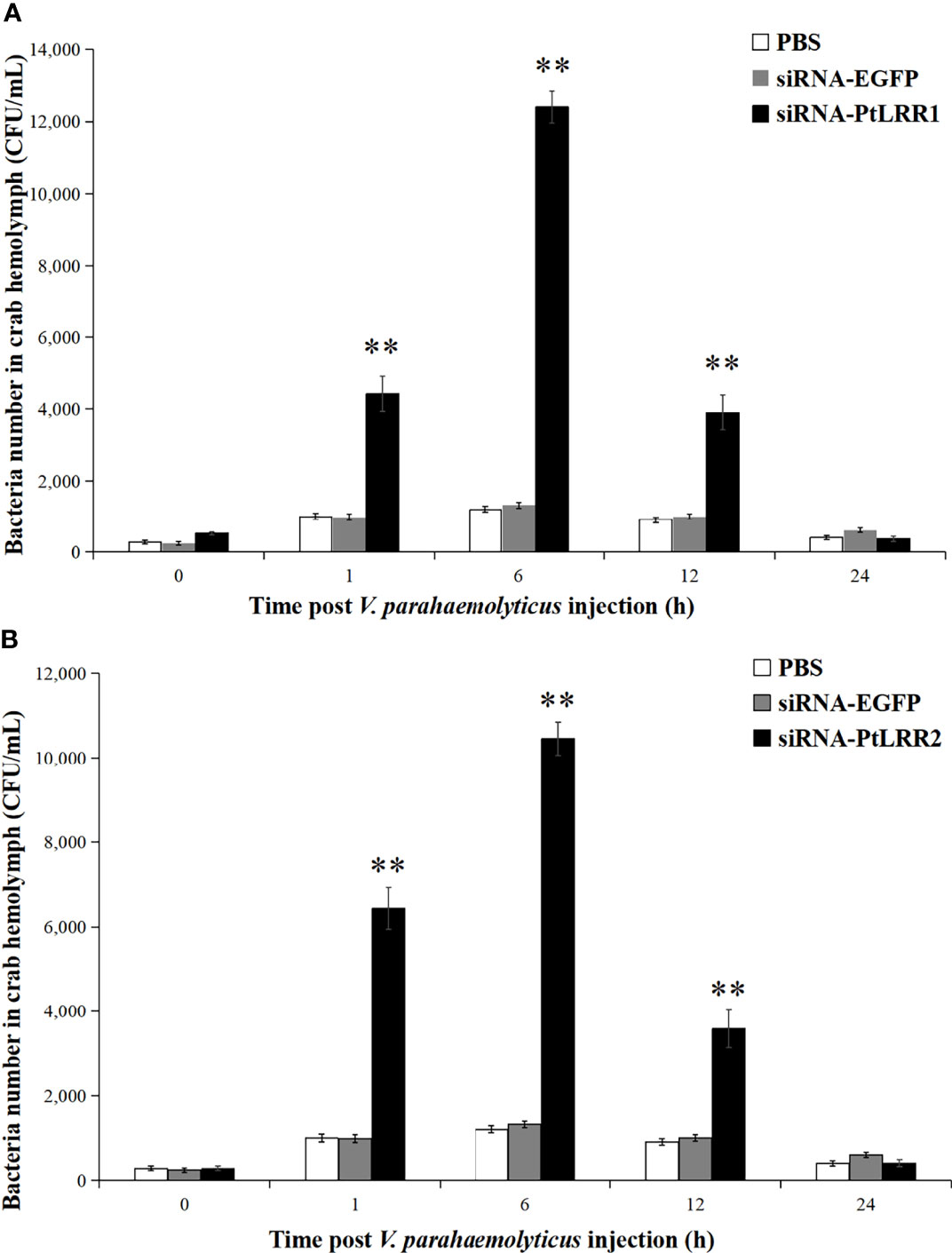
Figure 10 Bacterial clearance activity in (A) PtLRR1-knockdown and (B) PtLRR2-knockdown crabs. The number of bacteria on LB medium were counted and recorded. Data are represented as mean ± S.D. (n = 5). Asterisks indicate the significant differences between the experiment group and the control.
In the present study, two novel LRR-only proteins named PtLRR1 and PtLRR2 were identified from P. trituberculatus. Both PtLRR1 and PtLRR2 contained five LRR motifs and shared the consensus signature sequence LxxLxLxxNxL with other LRR proteins. Another conserved amino acids “Lxx” were found in the front of the consensus sequence of the LRR motifs of PtLRR1 and PtLRR2, which is similar to that observed in the LRR motifs of MjLRRC1 from M. japonicus (Shi et al., 2017) and SpLRR from S. paramamosain (Cao et al., 2013). These structural features indicate that PtLRR1 and PtLRR2 are conformed to be the typical LRR-only proteins.
Consistent with that reported in CfLRRop-1 and 2 from scallop C. farreri (Wang et al., 2016a; Wang et al., 2016b), PtLRR1 and PtLRR2 transcripts were dominantly expressed in the hepatopancreas. However, it was quite different from the tissue expression patterns of the most reported crustacean LRR-only proteins. For example, PmLRR from shrimp P. monodon was primarily detected in hemocytes, intestine, and lymphoid organ but weakly expressed in the hepatopancreas (Sriphaijit and Senapin, 2007). SpLRR from crab S. paramamosain was highly expressed in the gill, heart, hemocytes, stomach, and intestine while it was not detected in the hepatopancreas (Cao et al., 2013). The hepatopancreas in crustaceans is an integrated organ of immunity and metabolism, which is involved in the initiation of the immune response and the clearance of pathogens (Roszer, 2014). Moreover, the expression of PtLRR1 and PtLRR2 increased significantly after being challenged by bacteria and virus. These expression patterns together suggest that PtLRR1 and PtLRR2 could modulate in the immune defense in the hepatopancreas of swimming crab. The PtLRR1 transcript was upregulated rapidly at 6 h and peaked at 12 h after the injection of WSSV, while post V. parahaemolyticus challenge, the expression of PtLRR1 was induced and increased significantly until 24 h, suggesting that PtLRR1 might respond to invading WSSV more sensitively than V. parahaemolyticus. PtLRR2 was increased rapidly and significantly at 6 h after V. parahaemolyticus challenge and upregulated significantly at 6, 24, and 48 h after WSSV injection, suggesting that PtLRR2 might be an inducible acute factor against invading V. parahaemolyticus and provide long-lasting protection against WSSV infection. These diversities of the temporal expression patterns also indicate that PtLRR1 and PtLRR2 might have different operating times against pathogens.
The LRR proteins are thought to play an important role in immune recognition (Bell et al., 2003; Kędzierski et al., 2004). We tried to express the recombinant proteins of both PtLRR1 and PtLRR2 in the E. coli expression system; however, PtLRR2 was failed to express due to its high hydrophobic property. The successfully expressed rPtLRR1 could bind to LPS, GLU, and PGN in a dose-dependent manner, which is similar to those reported in MjLRRC1 from shrimp M. japonicus (Shi et al., 2017), and CfLRRop-1 and CfLRRop-3 from scallop C. farreri (Wang et al., 2016a; Wang et al., 2016b). This finding suggests that PtLRR1 could act as a PRR to recognize various PAMPs and initiate the downstream immune response in crab.
Some LRR-containing proteins have been reported to regulate the inflammatory cascade in immune cells by inducing the release of inflammatory cytokines. For example, TLR could contribute to inflammatory responses by inducing the expression of TNF-α and IL-1β (Telepnev et al., 2003). Transmembrane receptor LRRC19 could induce the expression of proinflammatory cytokine IL-8 (Chai et al., 2009). In this study, knockdown of PtLRR1 or PtLRR2 led to the decreased expression of PtIL-17 and PtTRAF6. Similarly, the expression of TNF-α was significantly increased after the cultured scallop hemocytes incubated with the recombinant CfLRRop-1 or 2 (Wang et al., 2016a; Wang et al., 2016b). PtLRR2 could also positively regulate the expression of cytokines PtIL-16 and PtLITAF, while PtLRR1 displayed the opposite regulatory activity, suggesting the different roles of PtLRR1 and PtLRR2 in regulation of some cytokines. These results indicate that PtLRR1 and PtLRR2 might be associated with modulation of genes involved in inflammatory response.
The proPO-activating system, an important innate immune response in arthropods, is initiated by the specific recognition of pathogenic PAMPs (Cerenius and Söderhäll, 2004). Several studies in insects have shown that there is a cross talk between the Toll pathway and the proPO system (Ligoxygakis et al., 2002; Cerenius et al., 2008). TLRs in the Toll pathway could regulate the proPO system by several serine proteinases and serine protease inhibitors (Kan et al., 2008; An et al., 2009; Dudzic et al., 2019). Our results firstly reported that LRR-only protein could also have a potential function in regulating the proPO system. Knockdown of PtLRR1 resulted in the significant increase in the tested proPO system-related genes, suggesting that PtLRR1 might be a negative regulator of the proPO system. It is inconsistent with those reported in MrToll1 and MrToll3 from prawn Macrobrachium rosenbergii (Li et al., 2020; Li et al., 2021). Also, PstLRR1 and PtLRR2 displayed different roles in this process, as PtLRR2 could positively regulate the expression of PtcSP2 and PtproPO.
Previous studies of LRR-only proteins, such as MjLRRC1 (Shi et al., 2017) and CfLRRop-7 (Wang et al., 2019), could positively regulate the expression of AMP genes. However, after PtLRR1 and PtLRR2 knockdown, the expression of most AMP genes was significantly increased. The Toll, IMD, and JNK pathways are the main regulators of AMP synthesis in Drosophila (De Gregorio et al., 2002; Kallio et al., 2005). We further investigated the expression of key genes in these signaling pathways and found that the expression of most genes was downregulated in PtLRR1 or PtLRR2-knockdown crabs. These inconsistency results between AMP expression and AMP-related pathways were also observed in PtCLec2 from P. trituberculatus (Liu et al., 2021). These data suggest that PtLRR1 and PtLRR2 might participate in AMP regulation by inducing multiple signaling pathways. In addition, both PtLRR1 and PtLRR2 could enhance the clearance of V. parahaemolyticus, which is similar to the bacterial clearance activity found in MjLRRC1 from shrimp M. japonicus (Shi et al., 2017) and LvLRRm from Litopenaeus vannamei (Zhang et al., 2020). Therefore, PtLRR1 and PtLRR2 could play important roles in antibacterial immune reaction of swimming crab.
In conclusion, two LRR-only proteins PtLRR1 and PtLRR2 were identified and characterized from P. trituberculatus. PtLRR1 and PtLRR2 were similar in structure, tissue distribution, bacterial clearance activity, and regulation of the AMP expression; nevertheless, they were different in expression after pathogen stimulation, as well as activities in inducing the expression of inflammation and proPO system-related genes. The PtLRR1 transcript displayed a quicker response to WSSV infection, while PtLRR2 increased more intensely after V. parahaemolyticus challenge. The recombinant PtLRR1 exhibited broad-spectrum binding activities against PAMPs. PtLRR1 or PtLRR2 knockdown could significantly change the expression levels of cytokines, proPO system-related genes, and AMPs. PtLRR2 could positively regulate the expression of inflammation-related genes. PtLRR1 might be a negative regulator of the proPO system. All these results indicate that PtLRR1 and PtLRR2 could function as novel PRRs to modulate innate immune signaling pathways in crab.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/genbank/, ON367496 and ON367497.
YL designed the experiments. AZ performed the experiments and analyzed the data. YL and AZ wrote the manuscript. AZ and NG participated in the sample collection. YL, SL, and FL participated in the data discussion and interpretation. All authors contributed to the article and approved the submitted version.
This work was supported by the National Key R&D Program of China (2018YFD0901304), National Natural Science Foundation of China (41776159 and 31772880), and China Agriculture Research System of MOF and MARA.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
An C., Ishibashi J., Ragan E. J., Jiang H., Kanost M. R. (2009). Functions of Manduca Sexta Hemolymph Proteinases HP6 and HP8 in Two Innate Immune Pathways. J. Biol. Chem. 284, 19716–19726. doi: 10.1074/jbc.M109.007112
Bella J., Hindle K. L., McEwan P. A., Lovell S. C. (2008). The Leucine-Rich Repeat Structure. Cell Mol. Life Sci. 65, 2307–2333. doi: 10.1007/s00018-008-8019-0
Bell J. K., Mullen G. E., Leifer C. A., Mazzoni A., Davies D. R., Segal D. M. (2003). Leucine-Rich Repeats and Pathogen Recognition in Toll-Like Receptors. Trends Immunol. 24, 528–533. doi: 10.1016/S1471-4906(03)00242-4
Cao J., Lin Z., Zhang Y., Zhang X., Li S., Zhang N., et al. (2013). Cloning and Characterization of the SpLRR cDNA From Green Mud Crab, Scylla paramamosain. Fish. Shellfish. Immunol. 34, 129–135. doi: 10.1016/j.fsi.2012.10.018
Cerenius L., Lee B. L., Söderhäll K. (2008). The proPO-System: Pros and Cons for its Role in Invertebrate Immunity. Trends Immunol. 29, 263–271. doi: 10.1016/j.it.2008.02.009
Cerenius L., Söderhäll K. (2004). The Prophenoloxidase-Activating System in Invertebrates. Immunol. Rev. 198, 116–126. doi: 10.1016/0305-0491(84)90217-7
Chai L., Dai L., Che Y., Xu J., Liu G., Zhang Z., et al. (2009). LRRC19, a Novel Member of the Leucine-Rich Repeat Protein Family, Activates NF-kappaB and Induces Expression of Proinflammatory Cytokines. Biochem. Biophys. Res. Commun. 388, 543–548. doi: 10.1016/j.bbrc.2009.08.043
De Gregorio E., Spellman P. T., Tzou P., Rubin G. M., Lemaitre B. (2002). The Toll and Imd Pathways are the Major Regulators of the Immune Response in Drosophila. Embo. Journal. 21, 2568–2579. doi: 10.1093/emboj/21.11.2568
DeYoung B. J., Innes R. W. (2006). Plant NBS-LRR Proteins in Pathogen Sensing and Host Defense. Nat. Immunol. 7, 1243–1249. doi: 10.1038/ni1410
Dudzic J. P., Hanson M. A., Iatsenko I., Kondo S., Lemaitre B. (2019). More Than Black or White: Melanization and Toll Share Regulatory Serine Proteases in Drosophila. Cell Rep. 27, 1050–1061. doi: 10.1016/j.celrep.2019.03.101
Huang S., Wang X., Yan Q., Guo L., Yuan S., Huang G., et al. (2011). The Evolution and Regulation of the Mucosal Immune Complexity in the Basal Chordate Amphioxus. J. Immunol. 186, 2042–2055. doi: 10.4049/jimmunol.1001824
Kędzierski L., Montgomery J., Curtis J., Handman E. (2004). Leucine-Rich Repeats in Host-Pathogen Interactions. Arch. Immunol. Ther. Ex. 52, 104–112.
Kallio J., Leinonen A., Ulvila J., Valanne S., Ezekowitz R. A., Ramet M. (2005). Functional Analysis of Immune Response Genes in Drosophila Identifies JNK Pathway as a Regulator of Antimicrobial Peptide Gene Expression in S2 Cells. Microbes Infect. 7, 811–819. doi: 10.1016/j.micinf.2005.03.014
Kan H., Kim C. H., Kwon H. M., Park J. W., Roh K. B., Lee H., et al. (2008). Molecular Control of Phenoloxidase-Induced Melanin Synthesis in an Insect. J. Biol. Chem. 283, 25316–25323. doi: 10.1074/jbc.M804364200
Kawai T., Akira S. (2009). The Roles of TLRs, RLRs and NLRs in Pathogen Recognition. Int. Immunol. 21, 317–337. doi: 10.1093/intimm/dxp017
Kobe B., Kajava A. V. (2001). The Leucine-Rich Repeat as a Protein Recognition Motif. Curr. Opin. Struc. Biol. 11, 725–732. doi: 10.1016/s0959-440x(01)00266-4
Ligoxygakis P., Pelte N., Ji C., Leclerc V., Duvic B., Belvin M., et al. (2002). A Serpin Mutant Links Toll Activation to Melanization in the Host Defence of Drosophila. EMBO J. 21, 6330–6337. doi: 10.1093/emboj/cdf661
Liu Y., Su Y., Zhang A., Cui Z. (2021). A C-Type Lectin Lighly Expressed in Portunus trituberculatus Intestine Functions in AMP Regulation and Prophenoloxidase Activation. Antibiotics. (Basel). 10,541. doi: 10.3390/antibiotics10050541
Li Y., Xu Q., Gepan Y. Y., Liu H., Su J., Dai X. L. (2021). Molecular Cloning, Characterization and RNA Interference Assay of Two Toll-Like Receptors in Giant Freshwater Prawn, Macrobrachium rosenbergii. Aquac. Rep. 20, 100643. doi: 10.1016/j.aqrep.2021.100643
Li Y., Yuan W., Xu Q., Liu H., Dai X. L. (2020). The Regulation of Immune Responses Against White Spot Syndrome Virus or Vibrio Alginolyticus in Toll-Like Receptors Silenced Giant Freshwater Prawn (Macrobrachium rosenbergii). Fish. Shellfish. Immunol. 107, 84–94. doi: 10.1016/j.fsi.2020.10.003
Matsushima N., Takatsuka S., Miyashita H., Kretsinger R. H. (2019). Leucine Rich Repeat Proteins: Sequences, Mutations, Structures and Diseases. Protein Pept. Lett. 26, 108–131. doi: 10.2174/0929866526666181208170027
Ng A., Xavier R. J. (2011). Leucine-Rich Repeat (LRR) Proteins Integrators of Pattern Recognition and Signaling in Immunity. Autophagy 7, 1082–1084. doi: 10.4161/auto.7.9.16464
Roszer T. (2014). The Invertebrate Midintestinal Gland (“Hepatopancreas”) is an Evolutionary Forerunner in the Integration of Immunity and Metabolism. Cell Tissue Res. 358, 685–695. doi: 10.1007/s00441-014-1985-7
Shi X. Z., Feng X. W., Sun J. J., Zhao X. F., Wang J. X. (2017). Leucine-Rich Repeats Containing Protein Functions in the Antibacterial Immune Reaction in Stomach of Kuruma Shrimp Marsupenaeus japonicus. Fish. Shellfish. Immunol. 61, 130–137. doi: 10.1016/j.fsi.2016.12.029
Sriphaijit T., Senapin S. (2007). High Expression of a Novel Leucine-Rich Repeat Protein in Hemocytes and the Lymphoid Organ of the Black Tiger Shrimp Penaeus monodon. Fish. Shellfish. Immunol. 22, 264–271. doi: 10.1016/j.fsi.2006.06.001
Su Y., Liu Y., Gao F., Cui Z. (2020). A Novel C-Type Lectin With a YPD Motif From Portunus trituberculatus (PtCLec1) Mediating Pathogen Recognition and Opsonization. Dev. Comp. Immunol. 106, 103609. doi: 10.1016/j.dci.2020.103609
Takahashi N., Takahashi Y., Putnam F. W. (1985). Periodicity of Leucine and Tandem Repetition of a 24-Amino Acid Segment in the Primary Structure of Leucine-Rich Alpha-2-Glycoprotein of Human-Serum. P. Natl. Acad. Sci. U.S.A. 82, 1906–1910. doi: 10.1073/pnas.82.7.1906
Telepnev M., Golovliov I., Grundstrom T., Tarnvik A., Sjostedt A. (2003). Francisella tularensis Inhibits Toll-Like Receptor-Mediated Activation of Intracellular Signalling and Secretion of TNF-Alpha and IL-1 From Murine Macrophages. Cell. Microbiol. 5, 41–51. doi: 10.1046/j.1462-5822.2003.00251.x
Valanne S., Wang J. H., Ramet M. (2011). The Drosophila Toll Signaling Pathway. J. Immunol. 186, 649–656. doi: 10.4049/jimmunol.1002302
Vidya R., Makesh M., Purushothaman C. S., Chaudhari A., Gireesh-Babu P., Rajendran K. V. (2016). Report of Leucine-Rich Repeats (LRRs) From Scylla serrata: Ontogeny, Molecular Cloning, Characterization and Expression Analysis Following Ligand Stimulation, and Upon Bacterial and Viral Infections. Gene 590, 159–168. doi: 10.1016/j.gene.2016.06.026
Wang M. Q., Wang L. L., Guo Y., Yi Q. L., Song L. S. (2016a). An LRR-Only Protein Representing a New Type of Pattern Recognition Receptor in Chlamys farreri. Dev. Comp. Immunol. 54, 145–155. doi: 10.1016/j.dci.2015.09.006
Wang M. Q., Wang B. J., Liu M., Jiang K. Y., Wang L. (2019). A Novel LRR-Only Protein Mediates Bacterial Proliferation in Hemolymph Through Regulating Expression of Antimicrobial Peptides in Mollusk Chlamys farreri. Dev. Comp. Immunol. 92, 223–229. doi: 10.1016/j.dci.2018.11.013
Wang M. Q., Wang L. L., Xin L. S., Wang X. D., Wang L., Xu J. C., et al. (2016b). Two Novel LRR-Only Proteins in Chlamys farreri: Similar in Structure, Yet Different in Expression Profile and Pattern Recognition. Dev. Comp. Immunol. 59, 99–109. doi: 10.1016/j.dci.2016.01.013
Zhang H., Li S., Wang F., Xiang J., Li F. (2020). Identification and Functional Study of an LRR Domain Containing Membrane Protein in Litopenaeus vannamei. Dev. Comp. Immunol. 109, 103713. doi: 10.1016/j.dci.2020.103713
Keywords: Portunus trituberculatus, Leucine-rich repeat, pattern recognition receptor, RNA interference, signaling pathway
Citation: Zhang A, Liu Y, Guo N, Li S and Li F (2022) Two LRR-Only Proteins Involved in Antibacterial Defense and Prophenoloxidase System of Swimming Crab Portunus trituberculatus. Front. Mar. Sci. 9:946182. doi: 10.3389/fmars.2022.946182
Received: 17 May 2022; Accepted: 17 June 2022;
Published: 22 July 2022.
Edited by:
JInghui Fang, Chinese Academy of Fishery Sciences (CAFS), ChinaCopyright © 2022 Zhang, Liu, Guo, Li and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yuan Liu, bGl1eXVhbkBxZGlvLmFjLmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.