- 1Key Laboratory of Livestock Infectious Diseases in Northeast China, Ministry of Education, Shenyang Agricultural University, Shenyang, China
- 2Aquaculture Department, College of Animal Science and Veterinary Medicine, Shenyang Agricultural University, Shenyang, China
Enterospora artemiae, an obligate intracellular parasitic microsporidium, severely affects the development of Chinese grass shrimp (Palaemonetes sinensis) aquaculture. Currently, no effective drugs or vaccines are available for treatment. To improve the diagnosis and prevention of microsporidia infection in P. sinensis, two recombinase polymerase amplification (RPA) detection methods (visualized by electrophoresis [RPA-AGE] and a colloidal gold lateral flow dip-strip [RPA-LFD], respectively) were established based on the E. artemiae S8 serine protease gene. RPA-AGE showed optimal amplification at 37°C for 30 min, and amplification by RPA-LFD was completed in 10 min at 37°C and produced detection results within 5 min. Regarding specificity, both methods showed specific amplification of E. artemiae but not of other pathogens. Regarding sensitivity, the minimum detection limit for both RPA-AGE and RPA-LFD was 4.7 copies/μL. Using 30 clinical samples, the 70%-positive rate was lower than that of fluorescence quantitation, but accuracy was improved compared with conventional polymerase chain reaction-based amplification (56.7%). Our RPA-AGE and RPA-LFD methods showed high specificity and sensitivity, with short detection time. In particular, the RPA-LFD method can be used for simple on-site detection of E. artemiae in P. sinensis farms without the requirement of experimental equipment, which can facilitate the prevention and control of this microsporidial disease.
1 Introduction
The Chinese grass shrimp Palaemontes sinensis (Decapoda, Palaemonidae) is a small crustacean with high consumer preference in China. Moreover, P. sinensis is of ornamental value and shows environmental tolerance (Oonuki et al., 2010; Jiang et al., 2016; Bao et al., 2017; Bao et al., 2018), making it a promising species for large-scale artificial culture. However, with the rapid development of aquaculture, diseases of P. sinensis have emerged (Li et al., 2018; Jiang et al., 2020; Cao et al., 2022). In recent years, many farmed shrimp have become infected with a microsporidium. Jiang et al. (2020) identified Enterocytospora artemiae as the main parasite in the hepatopancreas of P. sinensis based on molecular biology and electron microscopy. After severe infection, the typical body transparency of the affected shrimp decreases, and the hepatopancreas and muscles show white turbidity. Large numbers of microsporidia may be observed in the hepatopancreas by smear observation (Jiang et al., 2020).
The high infection rate of E. artemiae severely limits the healthy development of rice field and pond cultures of P. sinensis in northern China. E. artemiae is an obligate intracellular parasitic fungus belonging to a branch of the Enterospora-like clade (Rode et al., 2013), and there are currently no effective drugs for treatment. Therefore, effective and rapid identification of this pathogen and avoiding transmission are the most effective preventive measures. At present, detection of E. artemiae mainly relies on histological and molecular methods (Rode et al., 2013; Jiang et al., 2021). However, histological detection requires the presence of a large number of parasites or characteristic lesions in the tissues and organs. Therefore, infection may be easily missed or misjudged when the pathogen load is low or when tissue samples are not processed adequately. Polymerase chain reaction (PCR)-based diagnosis has been commonly used for these infections because of its simplicity and rapid detection. Quantitative PCR (qPCR) and nested PCR are the two most commonly used methods for detecting E. artemiae (Rode et al., 2013; Jiang et al., 2021). However, these approaches depend on the availability of thermocyclers and thus testing is typically limited to a laboratory. In addition, these methods are time-consuming, laborious, and require specific skills and technical expertise (Table 1).
Recombinase polymerase amplification (RPA) is a newly developed and commonly applied nucleic acid amplification method that can help achieve the miniaturization of detection equipment and thus facilitate simple on-site detection (Moore and Jaykus, 2017; Wand et al., 2018; Ma et al., 2019). The underlying principle is the formation of a protein–DNA complex by the combination of recombinase and primers, which then recognizes homologous sequences in double-stranded DNA and initiates DNA amplification. The important advantages of this technology include the convenience of amplification under constant temperature, fast reaction speed, and limited requirements for hardware (Daher et al., 2016; Lillis et al., 2016; Lobato and O'Sullivan, 2018). RPA can also be combined with a lateral flow dip-strip (LFD), which is used to detect the product after target amplification using biotin-labeled primers and carboxyl fluorescein-labeled probes. For this approach, the amplification products are coated with gold nanoparticles with a carboxyl fluorescein antibody and the detection line is coated with a biotin antibody. When the reaction solution enters the test strip, the amplification products with carboxyl fluorescein and biotin form biotin antibody–nucleic acid–gold nanoparticle complexes, leading to the development of color on the detection line through antigen–antibody binding (Zhang et al., 2019; Wang et al., 2022). Detection results can be visually assessed with the naked eye within 5 min (Table 1).
At present, detection of P. sinensis microsporidia in aquafarms requires transporting the collected samples to a laboratory in order to complete the time-consuming testing procedures, which is also financially costly. Thus, to overcome the limitations of current PCR-based detection methods, in this study, we developed two RPA detection methods of P. sinensis, visualized by electrophoresis (RPA-AGE) and using the LFD test strip (RPA-LFD), respectively. The current PCR detection method of microsporidia mainly uses ribosomal DNA as the target, which can also produce non-specific amplification in similar species. To improve the specificity and sensitivity of our RPA methods, we selected the S8 serine protease gene. Serine protease is an important hydrolase with serine as the active center, catalyzing and hydrolyzing the specific amino acids of the substrate. The S8 serine protease family has three unique catalytically active sites (aspartic acid, histidine, and serine residues) and one substrate-binding site. Although these active domains are highly conserved, the complete amino acid sequences are more diverse, with less than 20% similarity for some protein sequences. Therefore, S8 serine protease gene is considered to be suitable for target gene detection (Beers et al., 2004; Laskar et al., 2011).
Our new RPA methods can provide an important practical advance to achieve the rapid diagnosis of E. artemiae in the field, which is of considerable importance for the prevention and control of this microsporidia disease in Chinese grass shrimp.
2 Materials and methods
2.1 Sample collection
P. sinensis shrimp were purchased from three farms in Panjin City, Liaoning province, China, which were transported to the aquaculture laboratory of Shenyang Agricultural University. The shrimp were temporarily cultured in three 300-L circular aquaria for seven days during which they were fed artificial compound feed twice, and one third of the water was exchanged each day. After this period, 10 shrimp of each farm were randomly selected to determine the E. artemiae prevalence using different methods. During the sampling process, live shrimp were anesthetized by placing them on ice for 5 min, and the hepatopancreas was quickly removed and placed in a centrifuge tube containing 100% alcohol for detection and analysis.
2.2 DNA extraction
DNA was extracted from the hepatopancreas of P. sinensis using a Tiangen Marine Animal DNA Kit (Tiangen Biotech Co. Ltd., Beijing, China), according to the manufacturer’s instructions. The concentration of extracted DNA was measured using an ultra-micro spectrophotometer (K5500, Beijing Kaiao Technology Development Co., Ltd., Beijing, China) and was stored at –20°C until analysis.
2.3 Primer design
Three pairs of RPA primers were designed based on the S8 serine protein gene (GenBank accession number: ON182063) and according to the instructions of the DNA constant-temperature rapid amplification kit (WeiFang Amp Future Biotech Co., Ltd., WeiFang, China), using Premier 5.0 software. The most suitable primers (Table 2) were determined through testing. Real-time fluorescence qPCR and conventional PCR detection primers and reaction procedures were performed as described by Jiang et al. (2021).
2.4 Preparation of the plasmid standard
An S8 serine protein gene fragment (681 bp, obtained by simple genome sequencing) of E. artemiae was amplified through PCR. The amplification product was recovered using FastPure Gel DNA Extraction Mini Kit (Vazyme, Nanjing, China), and the DNA fragment was integrated in a PMD™19-T Vector (Takara, Shiga, Japan). Subsequently, the recombinant plasmid was transferred into E. coli DH5α cells (Sangon, Shanghai, China) and the cells were cultured on LB plates containing X-Gal, IPTG, and Ampicillin at 37°C overnight. Next, the positive cells were picked out for LB liquid culture after identification of positive clones by sequencing with universal primer M13 (Table 2). Then, plasmids were extracted using a FastPure Plasmid Mini Kit (Vazyme). The concentration of the RPA recombinant plasmid was 152 ng/μL and the copy number of the plasmid was 4.7 × 1010 copies/μL. The prepared plasmid was stored at –20°C.
2.4.1 RPA-AGE
DNA was amplified using a constant-temperature rapid amplification kit (electrophoresis-type; Weifang Amp Future Biotech Co., Ltd.). A 50-μL reaction system was used, including buffer A (29.4 μL), 2 μL upstream primer (10 μM), 2 μL downstream primer (10 μM), ddH2O (10.1 μL), 200 ng DNA template, and buffer B (2.5 μL). The other main mixtures were prepared in the form of dried enzyme particles in freeze-dried reaction tubes. To optimize the reaction conditions, four reaction times (10, 20, 30, and 40 min) and five reaction temperatures (25, 30, 37, 39, and 42°C) were tested. The amplified products were extracted with 50 μL solution (phenol:chloroform:isoamyl alcohol = 25:24:1). Five microliters of the supernatant was collected after centrifugation for 5 min at 7,780 g, which was then added to 2 μL of 6× sample buffer solution. The mixed solution was analyzed by 1.5% agarose gel electrophoresis at 120 V for 30 min.
2.4.2 RPA-LFD
DNA was amplified using a constant-temperature rapid amplification kit (colloidal gold lateral flow dip-strip type) (Weifang Amp Future Biotech Co., Ltd.). The reaction mixture (50 μL) was the same as that described above for RPA-AGE, with addition of the probe (0.6 μL 10 μM; Table 2) and using 9.5 µL ddH2O. Other main mixtures were prepared in freeze-drying reaction tubes in the form of dried enzyme particles. Three reaction times (5, 10, and 20 min) and five reaction temperatures (25, 30, 37, 39, and 42°C) were tested to optimize the reaction conditions. After the reaction, 10 μL of the amplification product was added to a centrifuge tube containing 190 μL ddH2O. The diluted reaction product (50 μL) was added to the sample-adding hole of an LFD test strip, and the test results were recorded within 5 min.
2.5 RPA specificity test
DNA from the hepatopancreas of infected and healthy P. sinensis was used as positive and negative controls, respectively, and microsporidia (Enterocytozoon hepatopenaei, Hepatospora eriocheir, and Microsporidia sp.), white spot syndrome virus, and fungi (Metschnikowia bicuspidata and Wicherhamomyces anomalus), which are routinely maintained in our laboratory, were used for specificity verification.
2.6 RPA sensitivity test
The sensitivity of RPA-AGE and RPA-LFD was tested using recombinant plasmids at concentrations ranging from 4.7 × 105 to 4.7 copies/μL, and detection limits were determined based on whether the respective target bands appeared separately.
2.7 Detection of clinical samples
RPA-AGE, RPA-LFD, qPCR, and PCR were used to detect E. artemiae in 30 samples of P. sinensis collected from the three aquafarms. The qPCR and PCR reaction systems were used as described by Jiang et al. (2021). The qPCR was performed using an Applied Biosystems Quantum Studio 3 Real-Time Quantitative PCR device (Thermo Fisher Scientific, Waltham, MA, USA).
3 Results
3.1 Primers and reaction conditions
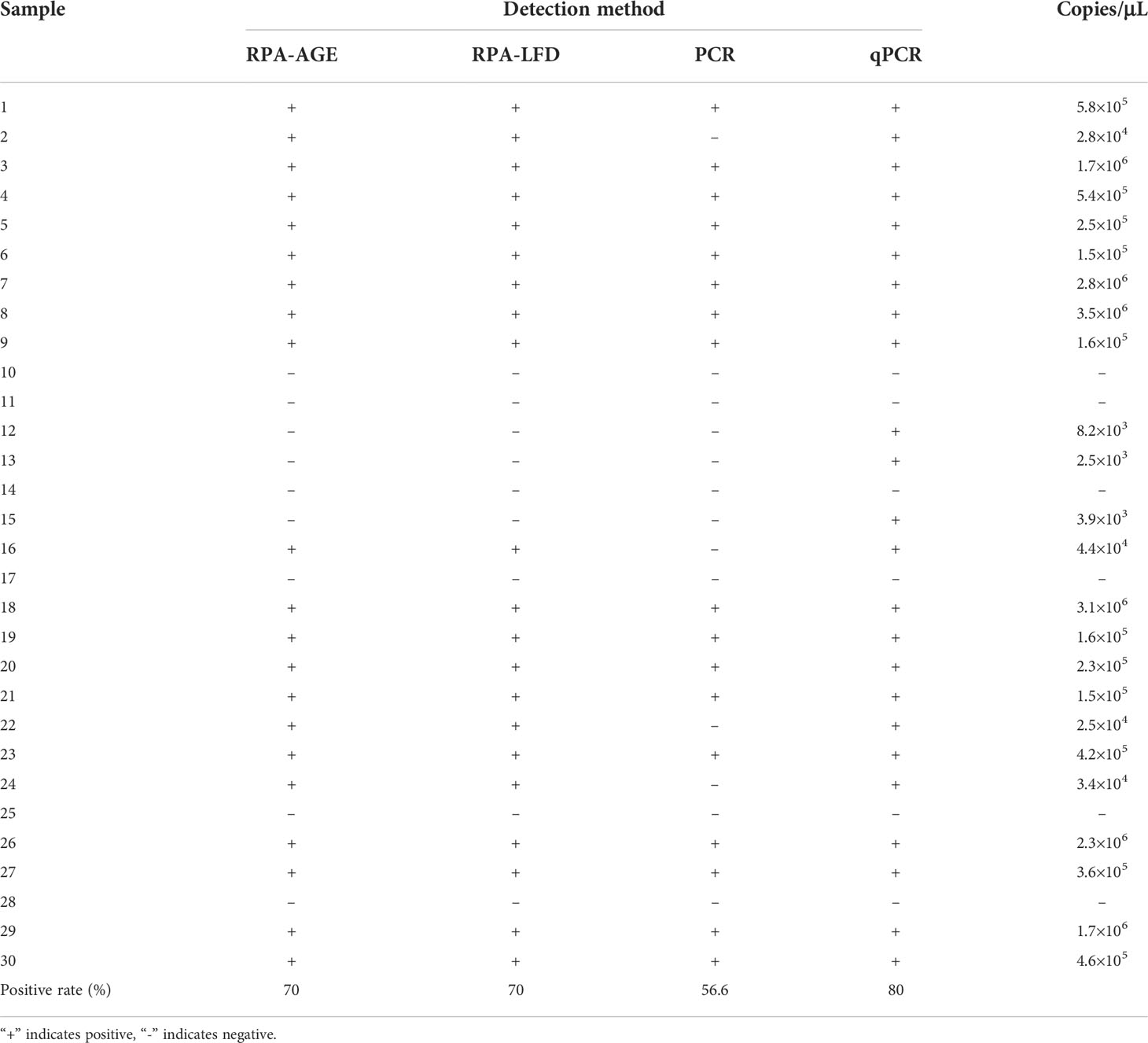
The RPA-AGE reactions were incubated for 30 min at different temperatures (25, 30, 37, 39, and 42°C). At 25°C, a visible amplification band was observed (Figure 1A); however, the gel strip bands were more pronounced at 37, 39, and 42°C. Thus, we selected 37°C as the optimal reaction time, since this is a more convenient temperature to achieve for on-site testing. To optimize the reaction time, the reaction was performed at 37°C for 10, 20, 30, and 40 min. The amplification intensity was the highest at 30 min (Figure 1B), which was the reaction time used in subsequent tests.
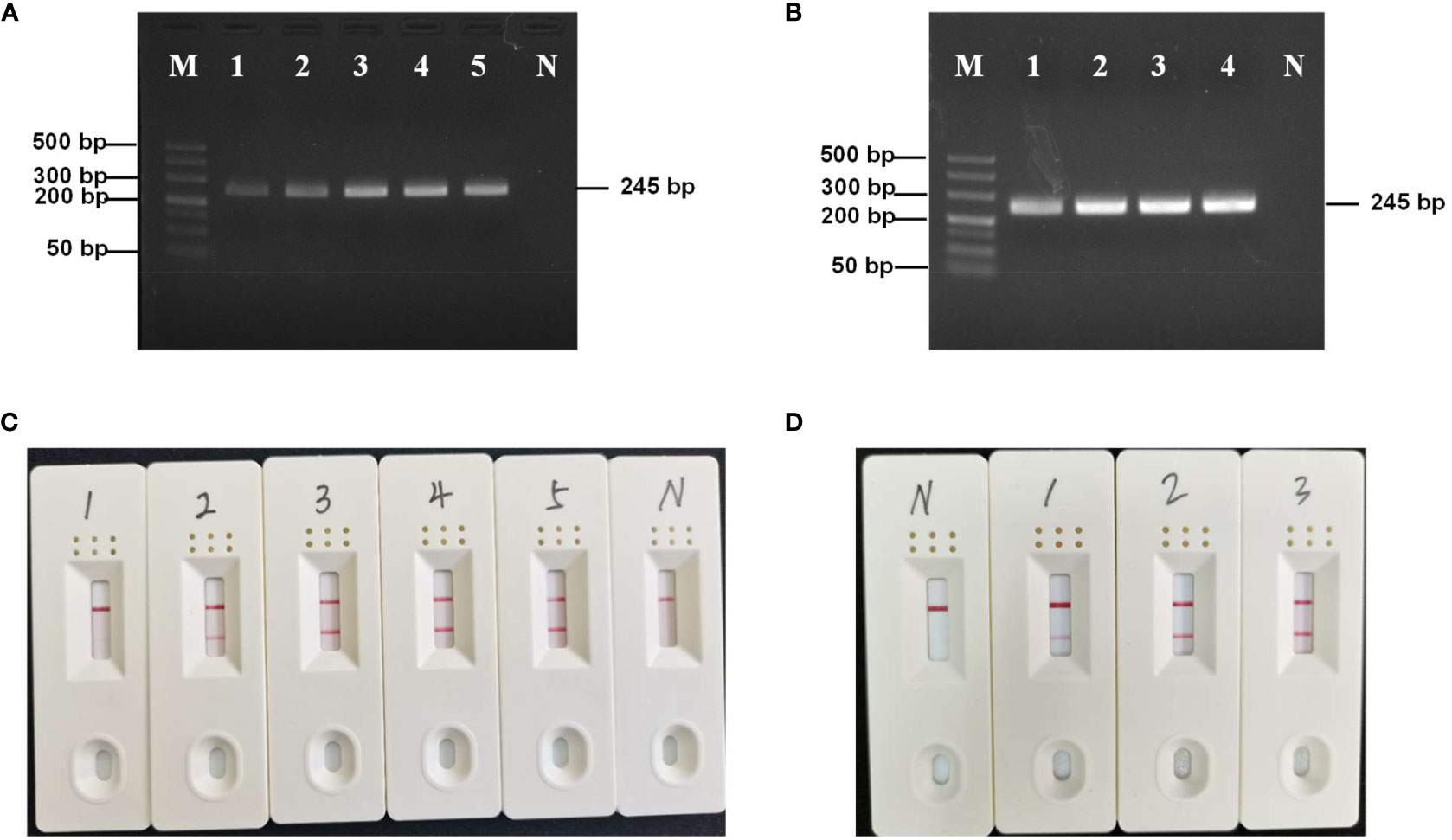
Figure 1 (A) Optimizing the RPA-AGE reaction temperature. M: DL 500 Marker; N: negative control; lines 1–5: 25°C, 30°C, 37°C, 39°C, and 42°C, respectively. (B) Optimizing the RPA-AGE reaction time. M: DL 500 Marker; N: negative control; 1: 10 min; 2: 20 min; 3: 30 min; 4: 40 min. (C) Optimizing the RPA-LFD reaction temperature. N: negative control; 1: 25°C; 2: 30°C; 3: 37°C; 4: 39°C; 5: 42°C. (D) Optimizing the RPA-LFD reaction time. N: negative control; 1: 5 min; 2: 10 min; 3: 20 min.
The RPA-LFD reactions were incubated for 10 min at different temperatures (25, 30, 37, 39, and 42°C). A visible positive signal was observed at 30°C (Figure 1C) and the strongest signal was obtained at 37°C (Figure 1C). To optimize the reaction time, the reaction was performed at 37°C for 5, 10, and 20 min; at 10 min, the amplification intensity was high (Figure 1D). Thus, 37°C and 10 min were the optimized conditions used for subsequent tests.
3.2 RPA specificity
RPA specificity was tested using other microsporidia, fungi, and a virus. Only the hepatopancreatic DNA of E. artemiae-infected shrimp showed a bright single target band (Figure 2A), indicating that the RPA primer was highly specific for this species. In addition, in RPA-LFD amplification, only E. artemiae-positive samples produced a positive band in the LFD, whereas no positive bands were produced from the other pathogens (Figure 2B), suggesting high specificity.
Figure 2 (A) Specificity of the RPA-AGE assay. M: DL 1000 Marker; P: Enterocytospora artemiae; 1: Enterocytozoon hepatopenaei; 2: Hepatospora eriocheir; 3: Microsporidia sp.; 4: Metschnikowia bicuspidata; 5: white spot syndrome virus; 6: Wicherhamomyces anomalus; N: negative control. (B) Specificity of the RPA-LFD assay. P: Enterocytospora artemiae; N: negative control; 1: Enterocytozoon hepatopenaei; 2: Hepatospora eriocheir; 3: Microsporidia sp.; 4: Metschnikowia bicuspidata; 5: white spot syndrome virus; 6: Wicherhamomyces anomalus. (C) Sensitivity of RPA-AGE to gradient dilution of the recombinant plasmid. M: DL 500 Marker; N: negative control; 1: 4.7 × 105 copies/µL; 2: 4.7 × 104 copies/µL; 3: 4.7 × 103 copies/µL; 4: 4.7 × 102 copies/µL; 5: 4.7 × 10 copies/µL; 6: 4.7 copies/µL. (D) Sensitivity of RPA-LFD to gradient dilution of the recombinant plasmid. N: negative control; 1: 4.7 × 105 copies/µL; 2: 4.7 × 104 copies/µL; 3: 4.7 × 103 copies/µL; 4: 4.7 × 102 copies/µL; 5: 4.7 × 10 copies/µL; and 6: 4.7 copies/µL.
3.3 RPA-AGE and RPA-LFD sensitivity
To test RPA-AGE and RPA-LFD sensitivity, a 4.7 × 105–4.7 copies/μL gradient of recombinant plasmid was used as a template for amplification. The minimum detection limit of both RPA-AGE and RPA-LFD was 4.7 copies/μL (Figures 2C, D).
3.4 Detection of clinical samples
Thirty shrimp samples from three farms were analyzed by RPA-AGE, RPA-LFD, PCR, and qPCR. The positive rate of RPA-AGE and RPA-LFD (21/30, 70.0% for both) was lower than that of qPCR (24/30, 80.0%), but higher than that of PCR (17/30, 56.7%) (Table 3).
4 Discussion
Chinese grass shrimp have become an economically important freshwater aquaculture species in China because of their high nutritional value and delicious taste. Two main pathogens of P. sinensis have been reported to date, the isopod parasite Tachaea chinensis (Li et al., 2018) and the microsporidium E. artemiae, with the latter showing a considerably higher rate of infection. E. artemiae is the most destructive pathogen of P. sinensis, which severely affects the development of aquaculture for this shrimp (Jiang et al., 2020). Microsporidia are considered fungi, which are broadly parasitic on terrestrial organisms and aquatic animals, including shrimp and crab. Since the identification of Nosema bombycis in the mid-19th century, more than 1,500 species of 187 genera of microsporidia have been identified, with more than 60 genera capable of infecting aquatic crustaceans and 16 genera capable of infecting shrimp (Wang et al., 2013; Weiss and Becnel, 2014; Bojko et al., 2020). E. artemiae can infect not only Chinese grass shrimp but also Artemia. The size of E. artemiae is 1.9 × 1.1 μm, and thus it cannot be observed with the naked eye (Jiang et al., 2020). In cases of severe infection, microscopic examination can be performed using a hepatopancreatic smear, which is not possible during initial infection. Tissue sections and electron microscopy can be used to identify E. artemiae; however, these approaches require considerable time and examination by trained experts, which is not conducive to rapid and accurate identification under field conditions. Several rapid detection methods for microsporidia have been tested. For example, Zhao et al. (2020) detected E. hepatopenaei spores using the calcofluor white method, and Wang et al. (2020) used fluorescent brightener 28 staining to detect E. hepatopenaei. Although these methods can be used to observe microsporidia, they are not informative with respect to the species of microsporidia. Moreover, these methods require specialized technical devices such as a fluorescence microscope. RPA has been widely used for the detection of bacterial, viral, fungal, and parasitic pathogens in aquatic animals, food safety, agriculture, and other fields in recent years (Jaroenram and Owens, 2014; Gao et al., 2017; Yang et al., 2020; Ma et al., 2021). Therefore, in this study, we established RPA-AGE and RPA-LFD detection methods for the rapid and accurate identification of E. artemiae.
We designed primers for E. artemiae detection based on the S8 serine protease gene. We found that RPA reliably and exclusively detected E. artemiae, suggesting high specificity; thus, contamination by other pathogens, including other microsporidium species, does not pose a problem for this assay. Regarding reaction conditions, RPA amplified the target gene fragment at 30°C and showed clear detection results at 37°C. This temperature is easy to achieve without the need for specialized equipment. The RPA-LFD assay showed the best results at a reaction time of 10 min. This method is thus suitable for rapid on-site detection. The reaction time of the RPA-AGE method was longer than that of RPA-LFD, and the best result was obtained after 30 min. However, compared with PCR, the reaction time of the RPA-AGE method is considerably shorter, and it can be executed at a constant temperature of 37°C, without the need for a thermocycler. This result is in line with those of Zhou et al. (2020), showing that RPA of E. hepatopenaei produced good amplification and specificity at 30°C for 40 min. Sensitivity evaluation of the present study showed that the RPA-AGE and RPA-LFD methods detected E. artemiae present at the lowest concentration of 4.7 copies/µL, which was significantly better than the detection limit of conventional PCR using the ribosomal protein S9 gene (2.2 × 102 copies/µL) (Jiang et al., 2021). We confirmed that the sensitivity of this method is higher than that of conventional PCR for clinical sample detection. The positivity rate of RPA in 30 samples was 70%, which was higher than that of conventional PCR (56.7%). Although the positive rate detected by the RPA method was slightly lower than that of qPCR, this method nonetheless meets the detection requirements of field testing.
In conclusion, RPA-AGE and RPA-LFD detection of E. artemiae can achieve amplification at a constant temperature of 37°C. In particular, RPA-LFD can be completed within 10 min, and the detection results can be assessed within 5 min. Moreover, there is no need for special devices, and the sensitivity was higher than that of conventional PCR methods. This method showed high specificity and efficient detection in clinical samples, and is thus suitable for the on-site detection of E. artemiae on farms without specialized laboratory equipment.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/genbank/, ON182063.
Ethics statement
The animal study was reviewed and approved by Animal Experiments Ethics Committee of Shenyang Agricultural University (permit no. SYXK 2020041503).
Author contributions
HJ, JL, and QC were involved in designing the experiment and wrote the manuscript. JL, JB, CF, and HJ performed the majority of the experiments, data analysis, and interpretation. YX, QH, and XL assisted in sample collection and primer screening. All authors contributed to the article and approved the submitted version.
Funding
This study was funded by China Agriculture Research System of MOF and MARA (CARS-48); Liaoning Key Technologies R&D Project (2021JH1/10400040); Shenyang Science and Technology Mission Project (21-116-3-38); Liaoning Province Department of Education fund item (LSNQN202002).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Bao J., Jiang H., Cheng H., Yu Y., Li X. (2017). Effect of ammonia nitrogen on acute toxicity and non-specific immune parameters of Palaemonetes sinensis. Acta Hydrobiologica Sinica 41 (3), 516–522. doi: 10.13292/j.1000-4890.201709.006
Bao J., Li X., Yu H., Jiang H. (2018). Respiratory metabolism responses of Chinese mitten crab, Eriocheir sinensis and Chinese grass shrimp, Palaemonetes sinensis, subjected to environmental hypoxia stress. Front. Physiol. 9. doi: 10.3389/fphys.2018.01559
Beers E. P., Jones A. M., Dickerman A. W. (2004). The S8 serine, C1A cysteine and A1 aspartic protease families in arabidopsis. Phytochemistry 65 (1), 43–58. doi: 10.1016/j.phytochem.2003.09.005
Bojko J., Behringer D. C., Moler P., Stratton C., Reisinger L. (2020). A new lineage of crayfish-infecting microsporidia: The Cambaraspora floridanus n. gen. n. sp. (Glugeida: Glugeidae) complex from Floridian freshwaters (USA). J. Invertebrate Pathology. 171, 107345. doi: 10.1016/j.jip.2020.107345
Cao G., Bao J., Feng C., Li X., Lang Y., Xing Y., et al. (2022). First report of metschnikowia bicuspidata infection in Chinese grass shrimp (Palaemonetes sinensis) in China. Trans. Emerg. Dis. 69, 3133–3141. doi: 10.1111/tbed.14460
Daher R. K., Stewart G., Boissinot M., Bergeron M. G. (2016). Recombinase polymerase amplification for diagnostic applications. Clin. Chem. 62 (7), 947–958. doi: 10.1373/clinchem.2015.245829
Gao W., Huang H., Zhang Y., Zhu P., Yan X., Fan J., et al. (2017). Recombinase polymerase amplification-based assay for rapid detection of Listeria monocytogenes in food samples. Food analytical Methods 10 (6), 1972–1981. doi: 10.1007/s12161-016-0775-0
Jaroenram W., Owens L. (2014). Recombinase polymerase amplification combined with a lateral flow dipstick for discriminating between infectious Penaeus stylirostris densovirus and virus-related sequences in shrimp genome. J. virological Methods 208, 144–151. doi: 10.1016/j.jviromet.2014.08.006
Jiang H., Bao J., Cong Y., Shen X., Pan D., Yu Y., et al. (2016). Temperature tolerance and the effects of temperature on respiratory metabolism of Palaemonetes sinensis. Chin. J. Ecol. 35 (8), 2202–2207. doi: 10.13292/j.1000-4890.201608.002
Jiang H., Bao J., Liu J., Chen Y., Feng C., Li X., et al. (2021). Development of a quantitative PCR method for specific and quantitative detection of Enterocytospora artemiae, a microsporidian parasite of Chinese grass shrimp (Palaemonetes sinensis). Front. Mar. Science. 8. doi: 10.3389/fmars.2021.730569
Jiang H., Chen Y., Bao J., Li X., Feng C., Xing Y., et al. (2020). Isolation of the parasite Enterocytospora artemiae from Chinese grass shrimp (Palaemonetes sinensis)–first report in Asia. Front. Cell. Infection Microbiol. 10. doi: 10.3389/fcimb.2020.580088
Laskar A., Rodger E. J., Chatterjee A., Mandal C. (2011). Modeling and structural analysis of evolutionarily diverse S8 family serine proteases. Bioinformation 7 (5), 239. doi: 10.6026/97320630007239
Lillis L., Siverson J., Lee A., Cantera J., Parker M., Piepenburg O., et al. (2016). Factors influencing recombinase polymerase amplification (RPA) assay outcomes at point of care. Mol. Cell Probes. 30 (2), 74–78. doi: 10.1016/j.mcp.2016.01.009
Li Y., Xu W., Li X., Jiang H., She Q., Han Z., et al. (2018). Comparative transcriptome analysis of Chinese grass shrimp (Palaemonetes sinensis) infected with isopod parasite Tachaea chinensis. Fish Shellfish Immunol. 82, 153–161. doi: 10.1016/j.fsi.2018.08.013
Lobato I. M., O'Sullivan C. K. (2018). Recombinase polymerase amplification: Basics, applications and recent advances. Trac Trends analytical Chem. 98, 19–35. doi: 10.1016/j.trac.2017.10.015
Ma C., Fan S., Wang Y., Yang H., Qiao Y., Jiang G., et al. (2021). Rapid detection of Enterocytozoon hepatopenaei infection in shrimp with a real-time isothermal recombinase polymerase amplification assay. Front. Cell. infection Microbiol. 11. doi: 10.3389/fcimb.2021.631960
Ma L., Zeng F., Huang B., Zhu Y., Wu M., Xu F., et al. (2019). Point-of-care diagnostic assay for rapid detection of porcine deltacoronavirus using the recombinase polymerase amplification method. Transboundary Emerging Diseases. 66 (3), 1324–1331. doi: 10.1111/tbed.13155
Moore M. D., Jaykus L. A. (2017). Recombinase polymerase amplification: a promising point-of-care detection method for enteric viruses. Future Virology. 12 (8), 421–429. doi: 10.2217/fvl-2017-0034
Oonuki T., Suzuki N., Akiyama N. (2010). Annual reproductive cycle of the female Palaemonetes sinensis recorded for the first time in a pond of hamamatsu city, shizuoka prefecture, Japan. Aquaculture Science. 58 (4), 509–516. doi: 10.11233/aquaculturesci.58.509
Rode N. O., Landes J., Lievens E. J., Flaven E., Segard A., Jabbour-Zahab R., et al. (2013). Cytological, molecular and life cycle characterization of Anostracospora rigaudi n. g. n. sp. and Enterocytospora artemiae n. g. n. sp. two new microsporidian parasites infecting gut tissues of the brine shrimp artemia. Parasitology 140 (09), 1168–1185. doi: 10.1017/S0031182013000668
Wand N. I. V., Bonney L. C., Watson R. J., Graham V., Hewson R. (2018). Point-of-care diagnostic assay for the detection of zika virus using the recombinase polymerase amplification method. J. Gen. virology. 99 (8), 1012. doi: 10.1099/jgv.0.001083
Wang H., Dong J., Zhang T., Wang F., Yang R., Zhang Y., et al. (2022). A novel rapid detection of senecavirus a using recombinase polymerase amplification (RPA) coupled with lateral flow (LF) dipstrip. Analytical Biochem. 646, 114627. doi: 10.1016/j.ab.2022.114627
Wang L., Lv Q., He Y., Gu R., Zhou B., Chen J., et al. (2020). Integrated qPCR and staining methods for detection and quantification of Enterocytozoon hepatopenaei in shrimp Litopenaeus vannamei. Microorganisms 8 (9), 1366. doi: 10.3390/microorganisms8091366
Wang T. C., Nai Y. S., Wang C. Y., Solter L. F., Hsu H. C., Wang C. H., et al. (2013). A new microsporidium, Triwangia caridinae gen. nov., sp. nov. parasitizing fresh water shrimp, Caridina formosae (Decapoda: Atyidae) in Taiwan. J. invertebrate pathology. 112 (3), 281–293. doi: 10.1016/j.jip.2012.12.014
Weiss L. M., Becnel J. J. (2014). Microsporidia: Pathogens of opportunity. 1st ed. (Ames, USA: John Wiley & Sons, Inc). doi: 10.1002/9781118395264.ch17
Yang X., Zhao P., Dong Y., Shen X., Shen H., Li J., et al. (2020). An improved recombinase polymerase amplification assay for visual detection of Vibrio parahaemolyticus with lateral flow strips. J. Food Science. 85 (6), 1834–1844. doi: 10.1111/1750-3841.15105
Zhang T. T., Liu M. Z., Yin R. H., Yao L. Q., Liu B. S., Chen Z. L. (2019). Rapid and simple detection of Glaesserella parasuis in synovial fluid by recombinase polymerase amplification and lateral flow strip. BMC veterinary Res. 15 (1), 1–7. doi: 10.1186/s12917-019-2039-x
Zhao R. H., Gao W., Qiu L., Chen X., Dong X., Li C., et al. (2020). A staining method for detection of Enterocytozoon hepatopenaei (EHP) spores with calcofluor white. J. Invertebrate Pathology. 172, 107347. doi: 10.1016/j.jip.2020.107347
Keywords: Palaemonetes sinensis, Enterocytospora artemiae, RPA detection, S8 serine protease gene, on-site detection
Citation: Jiang H, Liu J, Bao J, Xing Y, Feng C, Hu Q, Li X and Chen Q (2022) Rapid detection of Enterocytospora artemiae in Chinese grass shrimp (Palaemonetes sinensis) through isothermal recombinase polymerase amplification. Front. Mar. Sci. 9:945809. doi: 10.3389/fmars.2022.945809
Received: 26 July 2022; Accepted: 04 October 2022;
Published: 18 October 2022.
Edited by:
Ji Hyung Kim, Gachon University, South KoreaReviewed by:
Susheel Kumar Sharma, ICAR Research Complex for NEH Region, IndiaParin Chaivisuthangkura, Srinakharinwirot University, Thailand
Copyright © 2022 Jiang, Liu, Bao, Xing, Feng, Hu, Li and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qijun Chen, cWlqdW5jaGVuNzU5QHN5YXUuZWR1LmNu
 Hongbo Jiang
Hongbo Jiang Jinghui Liu1
Jinghui Liu1 Jie Bao
Jie Bao Qingbiao Hu
Qingbiao Hu Qijun Chen
Qijun Chen