- 1Key Laboratory of Mariculture and Enhancement, Marine Fishery Research Institute of Zhejiang Province, Zhoushan, China
- 2Marine and Fisheries Research Institute, Zhejiang Ocean University, Zhoushan, China
- 3Fisheries College, Zhejiang Ocean University, Zhoushan, China
Investigations have demonstrated a strong and positive association between dietary intact phospholipid (PL) inclusion and aquatic larval growth, nevertheless, the precise molecular mechanism underlying PL inclusion on growth performance has not been well elucidated. This study aimed to investigate the effects of dietary soybean lecithin (SL) inclusion on growth performance, liver metabolism, resistance to hypoxia stress, and potential molecular mechanisms in rock bream (Oplegnathus fasciatus) larvae. Four types of equal-protein and equal-lipid content microdiets (MDs) were formulated with graded levels of SL to achieve phospholipid levels of (PLs, dry matter) 3.84% (SL0), 6.71% (SL4), 9.38% (SL8), and 12.21% (SL12). Rock bream larvae (25 days post-hatching) were fed the respective MDs for 30 days with three replicates. We found that dietary SL inclusion promoted growth performance, survival rate, and stress resistance to hypoxia stress. The increased dietary SL inclusion improved intestinal structure, as shown by the increased perimeter ratio, muscular thickness, and mucosal fold height of the mid-intestinal tissue. Moreover, a high SL inclusion diet (SL12) increased the activity of the key lipolysis-related enzyme (lipase [LP]) in liver tissue but decreased the activity of amino acid catabolism-related enzymes (aspartate aminotransferase [AST] and alanine aminotransferase [ALT]). RNA sequencing results in liver tissue revealed that the SL12 diet increased the transcriptional level of fatty acid activation-related genes (acsl6 and acsbg2), phospholipid catabolism-related genes (acat2, lpin2, and crls), and amino acid synthesis-related genes (gs, csb, aldh18a1, and oct), but decreased the expression of amino acid catabolism-related gene gprt2. Notably, the SL12 diet significantly increased the expression of ribosome biogenesis-related genes (pes1, nop56, nop58, and rpf2) in liver tissue. The ribosome protein-related pathways were the most enriched pathways mapped in the GO database. Collectively, this study demonstrated the necessity of dietary SL for survival, growth performance, promotion of mid-intestinal morphology, and hypoxia stress during the rock bream larval stage. The SL-induced growth performance promotion was likely attributed to increasing nutrient acquisition by intestinal morphology improvement and to increasing SL catabolism and thereby sparing amino acids for protein synthesis.
Introduction
Larval rearing is a bottleneck for aquaculture development. To obtain a large quantity and high-quality larvae, a microdiet (MD) with rich and comprehensive nutrients is indispensable for larval rearing. With numerous studies focused on MD phospholipids (PLs), the inclusion of intact PLs has been recognized to improve culture performance, survival, and stress resistance as well as reduce the occurrence of deformities (Tocher et al., 2008).
Progress has been made to elucidate the limited ability for endogenous de novo biosynthesis of PLs during the early life stage of marine fish larvae (Carmona-Antoñanzas et al., 2015). Nevertheless, the precise molecular mechanism underlying PL inclusion on growth performance in the early life stage of fish has not been well elucidated. A series of proposed plausible hypotheses have been proposed to explain the possible mechanism. One of the possible explanations for the requirement for PLs is that PL inclusion increases lipid and fatty acid transportation from enterocytes to the rest of the body through lipoprotein synthesis, as PLs are essential for lipoprotein assembly (Tocher et al., 2008). The beneficial effects of PL inclusion were generally believed to act on intestinal tissue (De Santis et al., 2015). The mid-intestine is the main tissue for nutrient digestion and absorption. A well-developed intestinal structure is important for nutrient digestion, absorption, and subsequent nutrient acquisition. However, there has been a debate about the effects of intact PL inclusion on intestinal morphology at the fish larval stage. Feeding with a PL-deficient diet led to intestinal lumen epithelial damage, whereas high PL inclusion increased goblet cell quantity and therefore aided digestion in Arctic char (Salvelinus alpinus L.) larvae (Olsen et al., 1999). However, dietary PL source and level showed no significant effect on the mid-intestinal enterocyte height of Atlantic salmon (Salmo salar) larvae (Taylor et al., 2015). Our recent study discovered that the high inclusion level of dietary SL led to intestinal morphology development in yellow drum (Nibea albiflora) larvae (Tan et al., 2022). Further studies are warranted to confirm whether dietary PL inclusion can improve intestinal morphology.
After exogenous PLs are absorbed, their biochemical metabolism primarily occurs in liver tissue. Liver tissue plays a central role in the synthesis, metabolism, and redistribution of carbohydrates, proteins, and lipids. Controversial results have been obtained across aquatic species in response to dietary PLs. A previous study on large yellow croaker (Larimichthys crocea) noted that increased PL content significantly increased the activity of lipase, the key enzyme for lipid catabolism (Cai et al., 2016). Further studies have elucidated that PLs decrease excessive lipid accumulation by inhibiting fatty acid uptake and lipid synthesis, together with promoting lipid export at the transcriptional level (Cai et al., 2017). Consistently, PL inclusion decreased hepatic lipid content in blunt snout (Megalobrama amblycephala) fingerlings (Li et al., 2015) and juvenile hybrid snakehead (Channa argus×Channa maculata) (Lin et al., 2018). However, a recent study on hybrid grouper (Epinephelus fuscoguttatus ♀ × E. lanceolatus ♂) reported that dietary PL inclusion increased hepatic crude lipid and triglyceride contents in a dose-dependent manner, possibly by regulating the synthesis and hydrolysis of triglycerides (Huang et al., 2021). Despite this being the case, most of the studies in the field of PLs have only focused on lipid metabolism but fail to explain the growth promotion effect of dietary PL inclusion.
The over-exploitation of wild fisheries decreased the availability of phospholipid from fish meal and fish oil. The sustainable, alternative to marine products is plant-based, but these contain very little phospholipid in comparison with marine products. Crude soybean oil contains around 1.5-3.1% phospholipids that are removed to become the by-product, SL (Tocher et al., 2008). SL is widely used in diet manufacture and the supplementation of SL in larvae MD has attracted much attention. Individual growth, more precisely protein retention, is closely related to lipid metabolism (Francis and Turchini, 2017). Based on the finding that dietary PLs can serve as a preferential source of energy in the larval stage, the larvae could decrease the amino acid flux into the tricarboxylic acid (TCA) cycle and thereby spare precious amino acids for body constitution. We propose a hypothesis that the growth promotion effect of high dietary SL inclusion was partially attributed to the protein-sparing effect. To our knowledge, there is no relevant research focusing on protein or amino acid metabolism regarding dietary SL inclusion in fish larvae. The mechanistic investigation and systematic understanding of how dietary SL level affects growth performance and liver tissue metabolism in fish larvae are still in the preliminary stage.
Unique molecular identifiers (UMIs) RNA sequencing has provided a method to accurately qualify mRNA duplicates because they have identical alignment coordinates and identical UMI sequences (Alejo and Tafalla, 2011). Rock bream (Oplegnathus fasciatus) is a vital commercial species in the East China Sea. Due to overexploitation and environmental pressures, rock bream stocks have experienced a drastic decline in recent years. To date, the strategy for improving their survival and growth is still unclear. This has hindered the industrialization and commercialization of this species. Therefore, the overall purpose of the present study was to investigate the effects of dietary SL inclusion on growth performance and the underlying molecular mechanism using UMI-RNA sequencing. The present study could contribute to the development of the rock bream larval MD. Moreover, this study could provide a better understanding of how dietary SL affects growth performance at the larval stage.
Materials and Methods
All animal experiments were conducted according to the Guide for the Care and Use of Laboratory Animals formulated by the Ministry of Science and Technology of China. The study was approved by the Committee on Ethics of Animal Experiments of Zhejiang Ocean University and was performed in compliance with the ethical principles and standards of Frontiers in Marine Science. Prior to sampling, the larvae were anesthetized with 10 mg L−1 tricaine methanesulfonate (MS-222, Sigma, USA) until the respiratory opercula stopped moving.
Chemical Analysis
The crude protein, lipid, moisture, and ash contents of the ingredients, larval body, and MDs were determined according to the methods of the Association of Official Analytical Chemists (AOAC, 2016). The MDs were placed in an oven at 105°C for 7 hours to a constant weight. Crude protein content was determined using the Kjeldahl method with an automatic digester (KjelFlex K-360, BUCHI, Switzerland). Ether extraction was used to determine crude lipids using a Soxtec System HT (Soxtec 2055, FOSS Tecator, Sweden). Ash content was determined by burning off the organic matter in a muffle furnace at 550°C for 6 hours.
Total lipid isolation and purification procedures were the same as described previously description (Folch et al., 1957). Briefly, the lipids were extracted by homogenizing the freeze-dried samples with 2:1 chloroform-methanol (v/v) and filtering the homogenate. Afterward, the filtrate was freed from these substances by being placed in contact with over 5-fold its volume of water. The lipid solvent was evaporated under nitrogen gas and desiccated in a vacuum for at least 16 hours. The fatty acid methyl esters (FAMEs) were prepared using the procedures in a previous description (Zuo et al., 2012). Fatty acids were eventually reported as mol % of total identified fatty acids. The amino acid content of MDs and the larval body was analyzed as described previously (Wu et al., 2019).
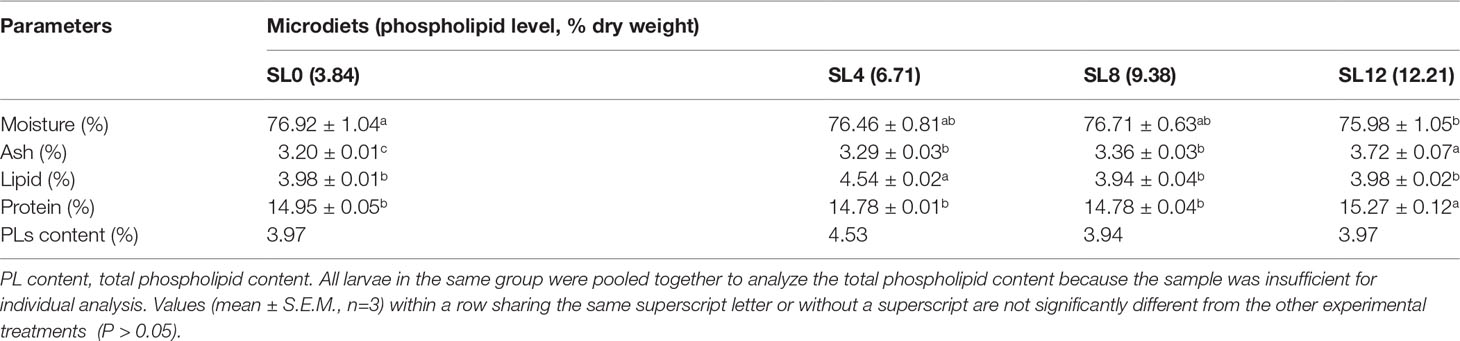
The PL content of MDs and the larval body was determined using the molybdenum blue colorimetry method. Briefly, total lipids were extracted as described in the fatty acid profile analysis. Dietary PLs from the extracted lipids were burned to phosphorus pentoxide and further oxidized to phosphoric acid by hydrochloric acid. Phosphoric acid and sodium molybdate formed sodium phosphomolybdate, which was further reduced to molybdenum blue by hydrazine sulfate. A colorimetric analysis was performed using a visible spectrophotometer at 650 nm wavelength (Biochrom Ultrospec 2100 pro, England). The number of larval bodies was not sufficient to be analyzed for each individual tank; therefore, the PL content determination was pooled together for each treatment.
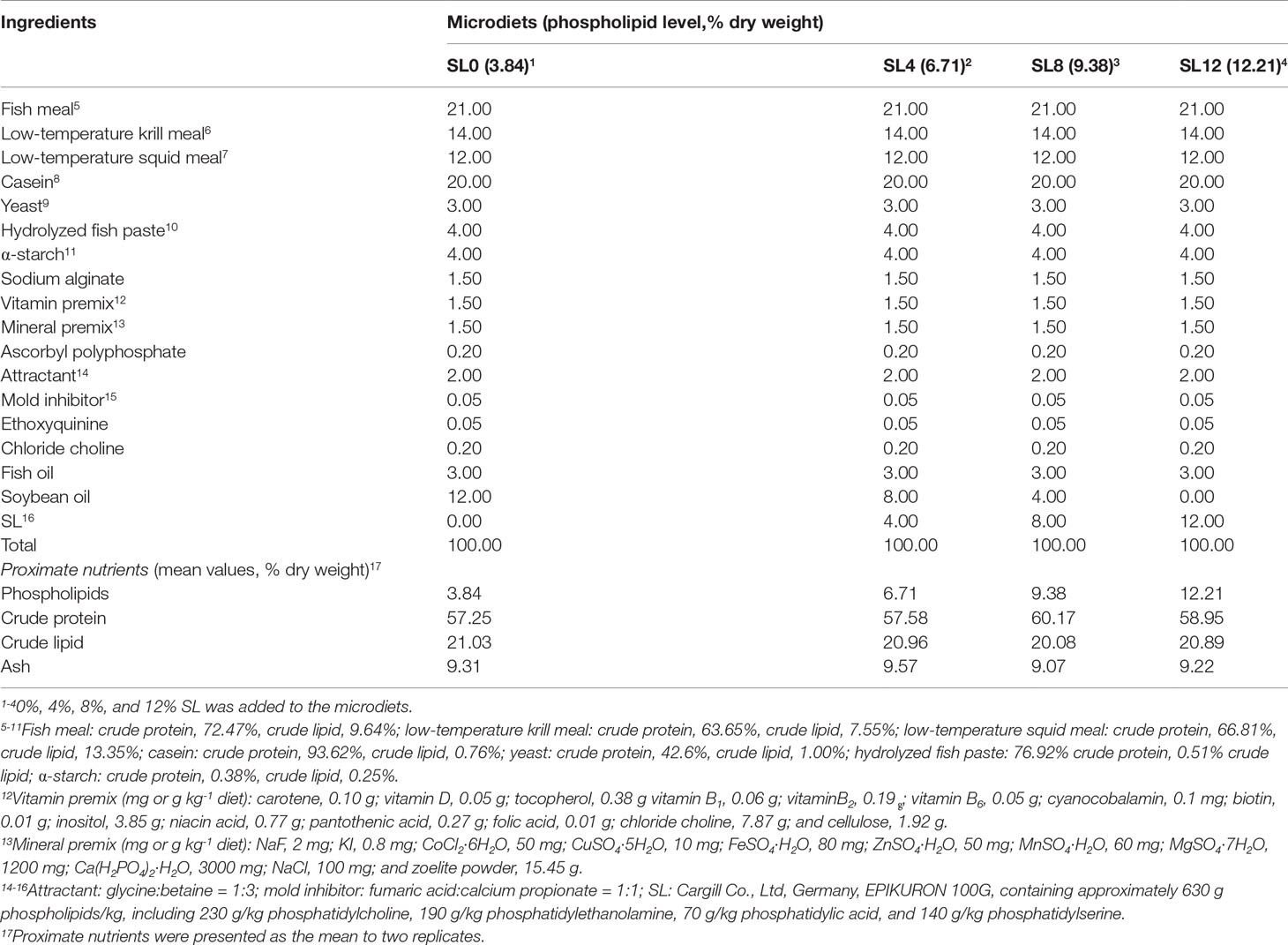
Microdiet Preparation
The MD formulation was primarily based on our previous study by Tan et al. (2022). Fish meal, low-temperature krill meal, low-temperature squid meal, and casein meal were the main protein sources. Fish oil, soybean oil, and SL served as the main lipid sources (Table 1). MDs were incorporated with increasing SL content (Cargill, Germany): 0% (SL0), 4% (SL4), 8% (SL8), and 12% (SL12). The diet formulation was as follows: protein sources, yeast, hydrolyzed fish paste, α-starch, sodium alginate, vitamin premix, mineral premix, ascorbyl polyphosphate, attractant, and mold inhibitor were mashed into a fine powder and passed through a 125-μm mesh. These ingredients were thoroughly mixed according to the proportion and then mixed with the remaining ingredients, including fish oil, soybean oil, SL, ethoxyquinine, and chloride choline solution, to make a dough. Strips of 1.0 mm diameter were processed in an F-26-type screw plodder (South China University of Technology, China) and then ripened for approximately 3 hours in an oven at 60°C. The strips were then smashed in a granulating machine (South China University of Technology, China) and further sifted into three different particle sizes: over 178 μm, 125 to 178 μm, and under 125 μm. The prepared MDs were packed and stored at −20°C until use.
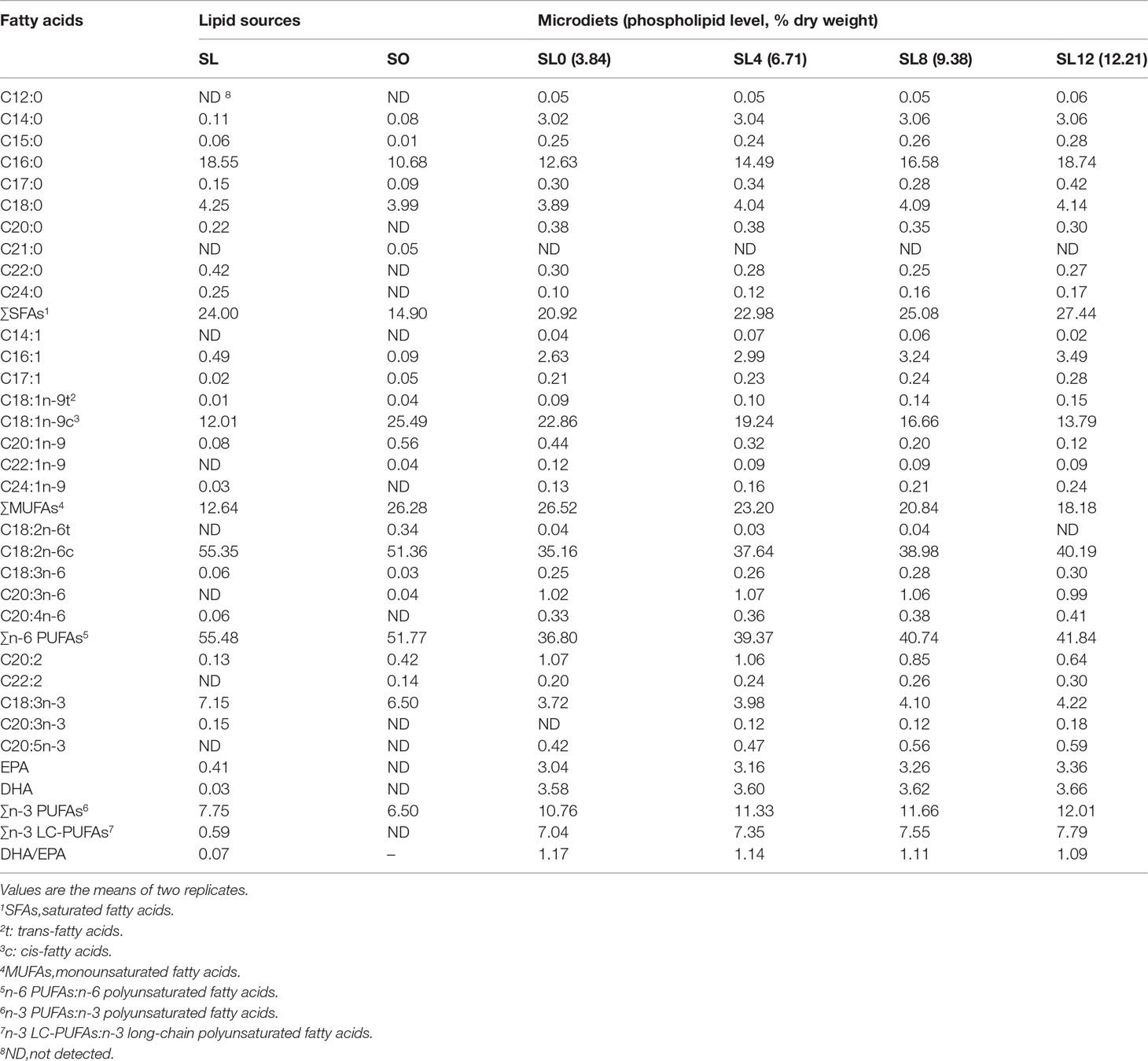
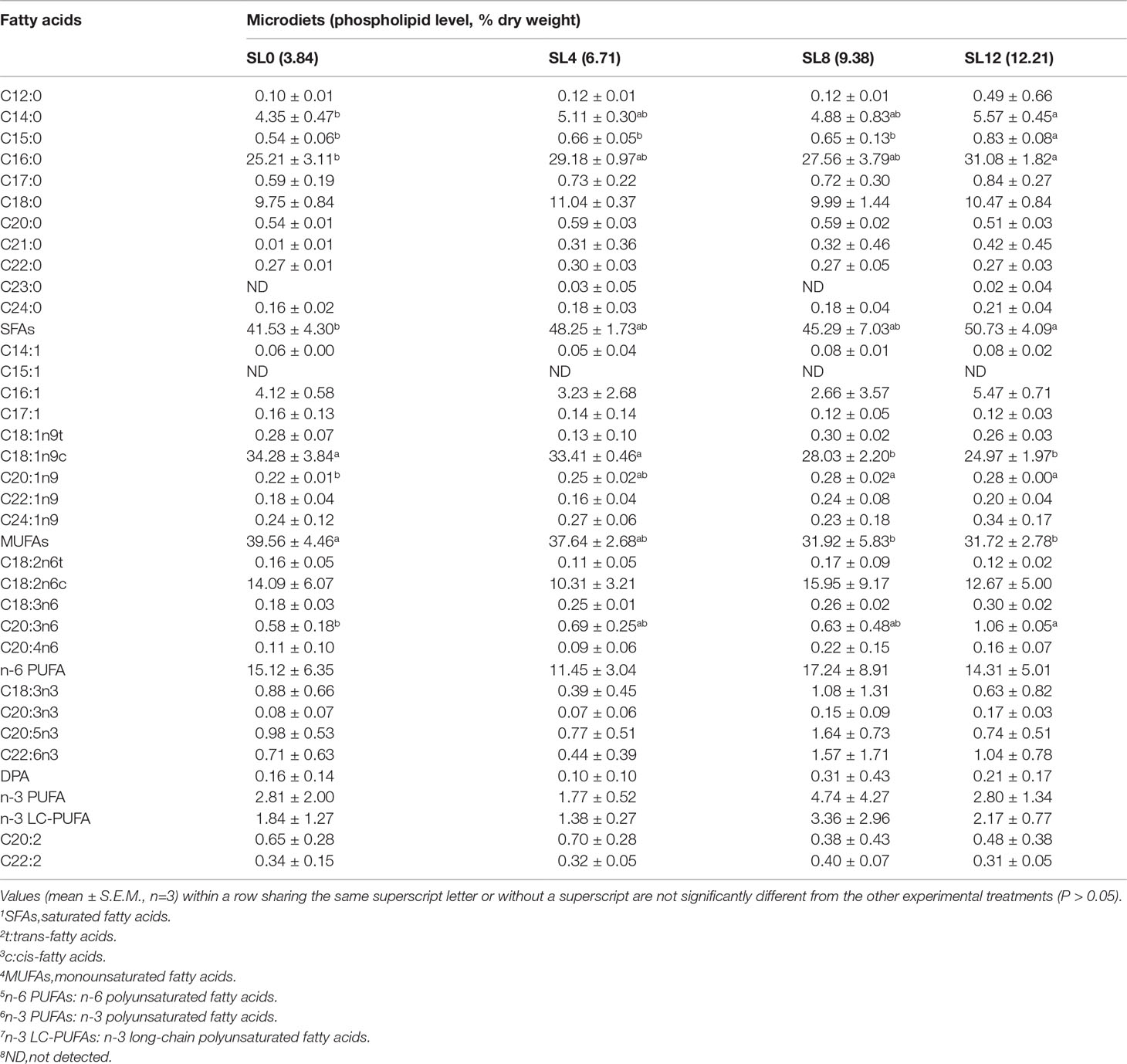
The PL contents (dry matter) were 3.84% (SL0), 6.71% (SL4), 9.38% (SL8), and 12.21% (SL12). The decrease in soybean oil content in MDs was compensated for by SL inclusion (Table 1). Composition analyses of the dietary lipid sources and MDs indicated only a trace amount of docosahexaenoic acid (DHA, C22:6n-3) and eicosapentaenoic acid (EPA, C20:5n-3) in SL, with similar fatty acid profiles obtained among the MDs (Table 2).
Larval Rearing
The larvae were reared in a 40-m3 pool after hatching on Xixuan Island (Zhoushan, Zhejiang, China). The larvae were fed rotifers (Brachionus rotundiformis) from 3 days post-hatching (dph) (mouth opening) to 20 dph and a mix of rotifers with Artemia nauplii from 20 to 22 dph. To acclimatize the experimental larvae, 1800 larvae (150 larvae per tank, 25 dph) were transferred to a flowing water system with water temperatures of 20-23°C, a salinity of 28 ± 1 ‰, and dissolved oxygen of over 6.0 mg per liter of seawater. To wean the larvae onto an MD, they were fed a mix of Artemia nauplii and MD for an additional 3 days prior to the feeding trial (22-24 dph). During the acclimation period, dead larvae were siphoned out, and fresh individuals were added to maintain a fixed quantity of larvae in each tank. During the feeding trial, larvae (initial body weight [IBW] 15.1 mg; initial body length [IBL] 1.01 cm) were hand-fed with the MDs every 2 hours from 06:00 to 20:00 (7 times a day). The amount of water volume in each tank was doubled every day (flow rate ~0.5 L/min). Using the siphon method, the tank was cleaned twice daily to remove feces, residual bait, and corpses. The larval corpses were carefully identified and recorded. From 0 to 10 days of larval rearing, the larvae were fed MD particles under 125 μm in size. Then, the larvae were fed with MDs of larger particle sizes.
Sample Collection
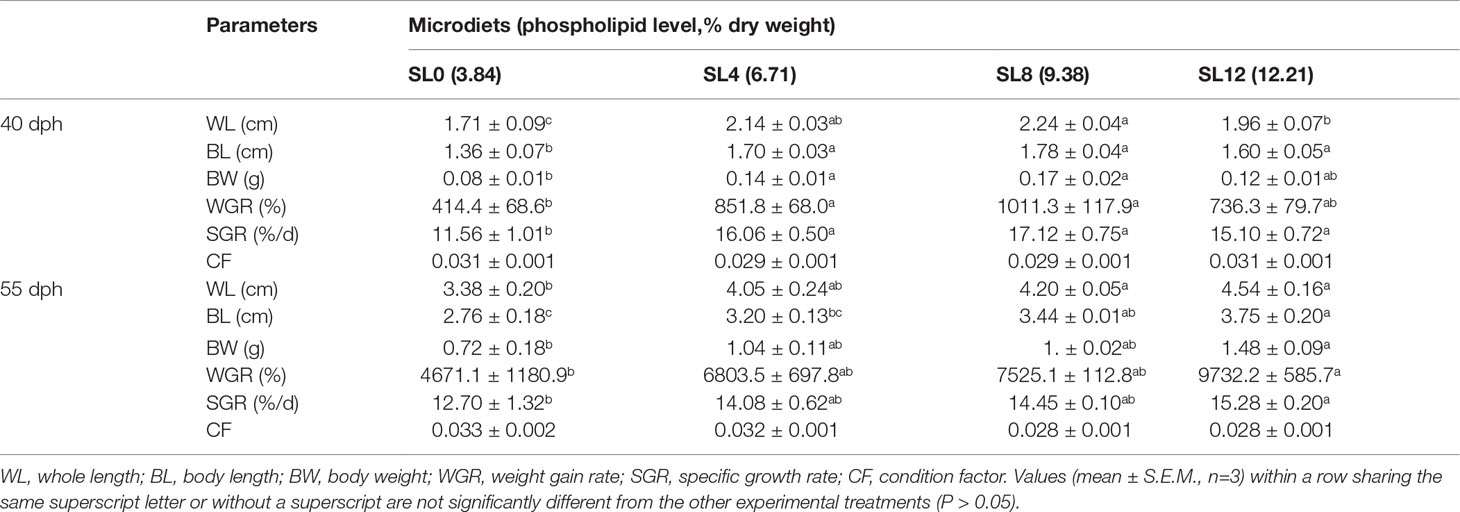
Prior to the beginning of the feeding trial, 50 larvae were randomly collected, drained of seawater, and weighed to obtain the total weight, and then the total length of each larva was measured and recorded. In the middle of the feeding trial (15-day rearing, larvae 40-dph), seven larvae per tank were randomly collected to measure the whole length (WL, length from the front of the mouth to the end of the caudal fin), body length (BL, length from the front of the mandible to the root of the caudal fin), and body weight (BW).
At the end of the feeding trial (larvae 55-dph), the mid-intestinal tissues of 2 larvae pre-tank were randomly collected under a dissecting microscope and stored in a 4% paraformaldehyde (PFA) solution for hematoxylin and eosin (H&E) section analysis. Twenty larvae were reserved per tank for the air exposure challenge, and the WL, BL, and BW of the remaining larvae were measured and recorded individually. The liver tissue of ten larvae per tank was randomly collected and pooled together in 1.5 mL microtubes (Biotech, China). These hepatic samples were immediately frozen in liquid nitrogen and stored at −80°C before enzyme activity analysis. After washing with phosphate-buffered saline (PBS), the liver tissue of three larvae per tank from larvae fed SL0 and SL12 were randomly collected and pooled (n=3 for each treatment), and tissues were stored in RNAlater (Thermo Fisher Scientific, USA) at −20°C until total RNA extraction. The remaining sampled larval bodies were collected by tank and stored at −20°C until proximate composition analysis and fatty acid and amino acid profile analysis.
Hypoxia Stress Challenge
After the sampling was conducted, the rest of the fish (20 larvae per tank) were fed with corresponding MDs for an additional two days. The primary experiments were conducted to determine the appropriate time for the hypoxia stress challenge. At the beginning of the hypoxia stress challenge, 20 larvae from each tank were captured by a mesh net. The larvae were exposed to air for an accurate 5 minutes and then returned to their original tank. Over the next 24 hours, the dead fish were collected, and body parameters were recorded. After completing the aforementioned steps, all live larval bodies were weighed (BW), measured (WL and BL), collected, and stored at −20°C for the analysis of larval body PL content.
Intestinal Structure by H&E Section Analysis
Mid-intestinal samples were sliced transversely into 6 μm sections and stained with H&E according to a previously described method (Baeverfjord and Krogdahl, 1996; Dimitroglou et al., 2009). A Panoramic 250 FLASH digital pathology system (3DHISTECH, Hungary) was adopted to image the slices. The mucosal fold height, muscular thickness, and perimeter ratio of the intestinal tissue slices were further analyzed according to previously described methods (Penn et al., 2011). The mucosal fold height, perimeter ratio, and muscular thickness were determined from 24 slices (6 slices for each treatment).
Enzyme Activity Analysis
To analyze hepatic metabolism enzyme activities, samples were homogenized with cold PBS (0.1 M, pH 7.0, 4°C) at a proportion of 1:9 (m/v). Afterward, the homogenate was centrifuged at 4°C at 3000 ×g for 10 minutes. The sediment was discarded, and the supernatant was stored at − 80°C until analysis. Using commercial kits (a total protein quantitative assay kit, alanine aminotransferase [ALT] kit, aspartate aminotransferase [AST] kit, and lipase [LP] assay kit, Nanjing Jiancheng Bioengineering Institute, China), the total protein content and ALT, AST, and LP enzyme activities were analyzed following the manufacturer’s instructions. To normalize the enzyme activity, the tissue protein concentration was utilized.
UMI-RNA-Seq and Bioinformatics Analysis
The total RNA of the liver tissue from larvae fed a PL-insufficient diet (SL0) and PL-sufficient diet (SL12) was extracted using TRIzol reagent (Invitrogen, USA). RNA degradation was monitored in a 1% agarose gel, and RNA purity was measured using a NanoPhotometer® spectrophotometer (Implen, USA). The RNA concentration was determined using a Qubit® RNA Assay Kit with Qubit® 2.0 Fluorometer (Life Technologies, USA). RNA integrity was measured using the RNA Nano 6000 Assay Kit of the Agilent Bioanalyzer 2100 system (Agilent Technologies, USA). Clustering and sequencing were performed with the help of the Experimental Department of Novogene Co., Ltd. (Beijing, China). All libraries were sequenced using the Illumina HiSeq 2500 sequencing platform (Illumina, USA). The library preparation for RNA-seq, sequence quality assessment and assembly, annotation, and differential expression gene (DEG) analyses were as described in detail in our previous publication with some differences (Xu et al., 2018). In the synthesis of second-strand cDNA, a UMI label and sequencing adaptor were added to the cDNA duplicates. After RNA sequencing, UMI reads were mapped to the genome sequences of the rock bream. The DEG cutoff was set as an adjusted P (q) value < 0.05 and fragments per kilobase per million (FPKM) fold change > 2. Detailed information on the sequencing data and mapping are summarized in Table S1.
DEG Validation by Real-Time Quantitative PCR
Total RNA was extracted from identical samples used for Illumina sequencing. Reversed cDNA was synthesized using a Prime Script RT Reagent Kit with a gDNA eraser (Takara, Japan) following the manufacturer’s instructions. The gene expression profiles of the 12 target genes were validated by real-time quantitative PCR (qRT–PCR). The primers used are summarized in Table S2. PCR was performed using an ABI Real-Time PCR machine (ABI Step One Plus, ABI, USA). The program settings used were the same as those described in our previous study (Wu et al., 2019). Briefly, SYBR® Premix Ex Taq (TaKaRa, Japan) was used for RT-qPCR. Each PCR reaction system consisted of 10.0 μl SYBR® Premix Ex Taq, 0.4 μl of each forward and reverse primer (10 μM), 0.4 μl ROX Reference Dye, 2 μl cDNA, and 6.8 μl dH2O in a total volume of 20 μl. Then, RT-qPCR was performed on a StepOnePlus™ Real-Time PCR machine (Applied Biosystems, USA). The standard procedure for amplification was as follows: 95°C for 30 s and 40 cycles of 95°C for 5 s, 57°C for 30 s. β-actin and g6pdh were used as internal controls for the normalization of gene expression levels. Expression levels were calculated using the comparative CT method (Livak & Schmittgen, 2001). The relative expression level of target genes was calculated by dividing the copy number of target genes by the geometric mean copy number of the two reference genes (Vandesompele et al., 2002).
Calculations and Data Statistics
The following variables were calculated:
Final body weight (FBW, g) = Wt/NtSurvival rate (%) = 100 × Nt/N0Survival rate after hypoxia stress (%) = 100 × larvae survival quantity/number of larvae for the stress testSpecific growth rate (SGR, %/d) = 100 × [Ln (Wt/Nt) – Ln (W0/N0)]/tWeight gain rate (WGR, %) = 100 × (Wt − W0)/W0Condition factor (CF, %) = 100 × (BW, g)/(BL, cm3) where Wt and W0 are the sums of the final and initial larval BWs, respectively; Nt and N0 are the final and initial larval quantities in each tank, respectively; and t is the experimental duration in days. All data were processed using GraphPad version 9.0.0 for Windows (USA, www.graphpad.com) and are presented as the mean ± standard error of the mean (S.E.M.). The normality and homogeneity of variances of the data were confirmed using the Levene test. The Kruskal–Wallis test was performed when the data were not homogeneous. Otherwise, one-way ANOVA with Tukey’s HSD test was used to assess significant differences among treatments at a significance level of P < 0.05. The DEG association networks were predicted by String (string-db.org) and further subjected to Cytoscape software (www.cytoscape.org) (Shannon et al., 2003). We performed a multidimensional correlation analysis using the R packages ggplot2, linkET, and dplyr. A schematic representation of the effects of the PL-rich diet (SL12) on hepatic metabolic and physiological responses was created with “BioRender.com”.
Results
Larval Growth Performance
At the end of 15 days of rearing (larvae 40-dph), the statistical results showed that the WL, BL, BW, WGR, and SGR of the larvae fed the SL4 and SL8 diets were significantly higher than those of larvae fed the SL0 diet (P < 0.05). The larvae fed the SL12 diet showed significantly higher WL, BL, and SGR than those fed the SL0 diet (P < 0.05). There was no significant difference in CF among treatments (P > 0.05) (Table 3).
At the end of the feeding trial (larvae 55-dph), the larvae fed the SL12 diet showed significantly higher WL, BL, BW, WGR, and SGR than those fed the SL0 diet (P < 0.05). The WL and BL were significantly higher in the larvae fed the SL8 diet than in the larvae fed the SL0 diet (P < 0.05). There was no significant difference in the aforementioned parameters between the larvae fed the SL4 and SL0 diets (P > 0.05). There was also no significant difference in CF among treatments (P > 0.05) (Table 3).
Larval Body Proximate Nutrient, Fatty Acid, and Amino Acid Profiles
The increased dietary SL inclusion significantly increased larval whole-body ash and protein contents (P < 0.05). The whole-body lipid content was significantly higher in the larvae fed the SL4 diet than in the rest of the groups (P < 0.05). The larval whole-body PL content reached comparable values among groups (Table 4).
The dietary fatty acid composition was roughly reflected by the composition of larval bodies. Significantly increased saturated fatty acids (SFAs) and decreased monosaturated fatty acids (MUFAs) proportions were observed in the larvae with increasing dietary SL inclusion (P < 0.05). No significant differences in n-6 polyunsaturated fatty acids (PUFAs), n-3 PUFAs, or n-3 long-chain polyunsaturated fatty acids (LC-PUFAs) were observed among the groups (P > 0.05) (Table 5).
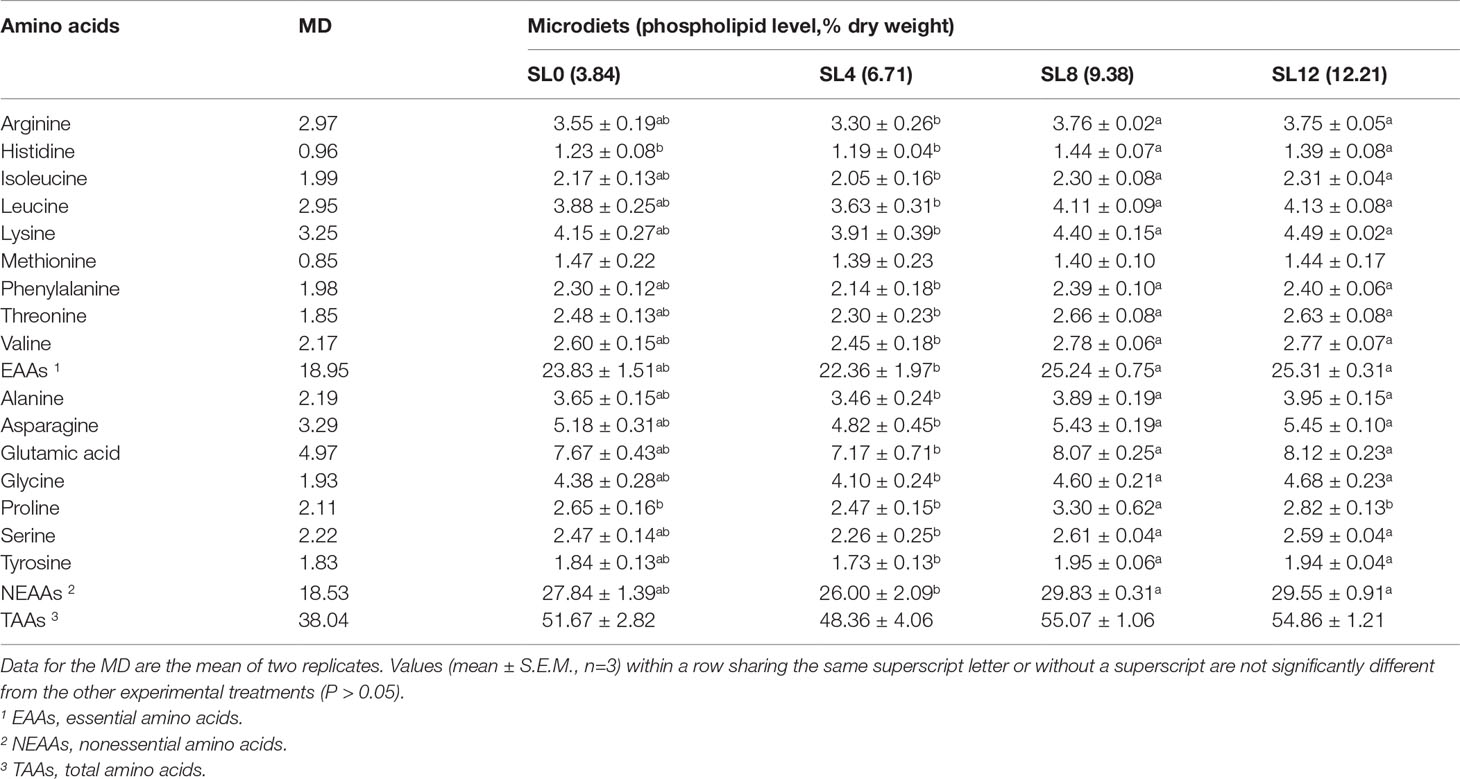
The larval whole-body amino acid content showed an increasing trend with dietary PL content increasing from 3.84% to 12.21% (Table 6). The essential amino acids (EAAs) and nonessential amino acids (NEAAs) contents of the larval body were found to be significantly higher in the SL12 and SL8 groups than in the SL4 group (P < 0.05) (Table 6).
Larval Survival, Resistance to Hypoxia Stress, and Hepatic Metabolism-Related Enzyme Activities
The siphon method was adopted to collect corpses, and the corpse quantity was carefully recorded twice daily (Figure 1A). At the end of the feeding trial, the survival rates were significantly higher in larvae fed the SL8 (P <0.01) and SL12 (P <0.05) diets than in the larvae fed the SL0 diet. Similarly, the resistance to hypoxia test showed that the high dietary inclusion of SL (SL8 and SL12 groups) increased the survival rate (P<0.05) (Figure 1B).
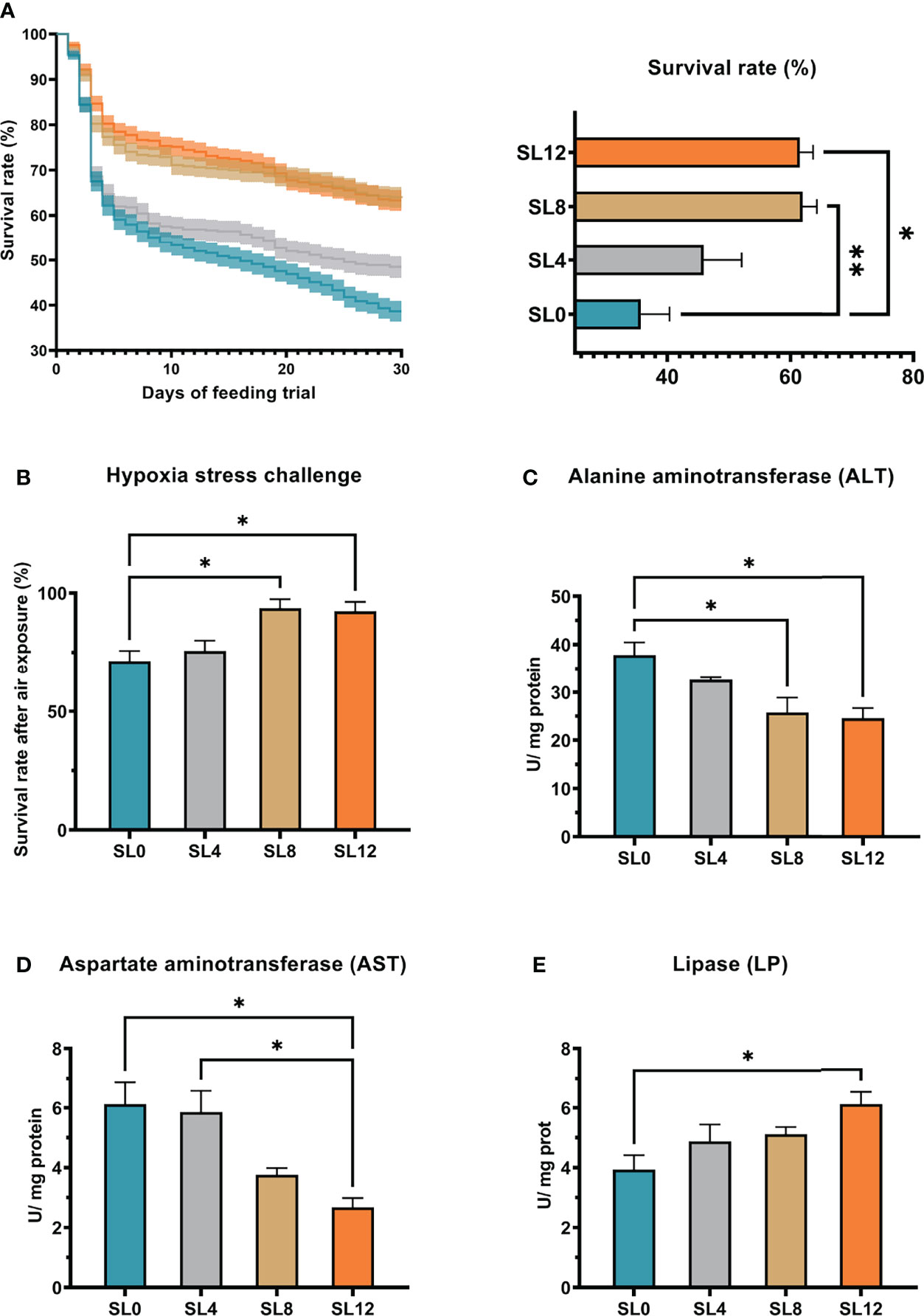
Figure 1 Rock bream larval survival rate, stress resistance to hypoxia stress, and liver tissue metabolism-related enzyme activities. (A) The rock bream larval survival rate was recorded during the feeding trial daily, and final statistical analyses are presented in the right panel. (B) Larval stress resistance to air exposure for 5 minutes. (C-E) Larval liver tissue alanine aminotransferase (ALT) Aspartate aminotransferase (AST) and lipase (LP) enzyme activities correspond to dietary soybean lection inclusion levels. *P < 0.05, **P < 0.01, otherwise, no significant difference (P > 0.05).
Results of metabolism-related enzyme activity in liver tissue indicated that the SL12 and SL8 diets significantly decreased hepatic ALT activity (Figure 1C) (P<0.05). Moreover, feeding with the SL12 diet significantly decreased the hepatic AST activity compared with that of the larvae fed the SL0 or SL4 diet (Figure 1D) (P<0.05). In contrast, the larvae fed the SL12 diet showed significantly higher hepatic LP activity than the larvae fed the SL0 diet (Figure 1E) (P <0.05).
Larval Mid-Intestinal Structure Analysis
Mid-intestinal histological sections in 55-dph larvae revealed improved intestinal structure with increased SL inclusion (Figure 2). Statistical analysis of the mid-intestinal structure of 55-dph larvae demonstrated that insufficient inclusion of dietary SL (SL0) significantly decreased the mucosal fold height, muscular thickness, and perimeter ratio (P < 0.05). The SL12 group showed the best intestinal structure among the groups.
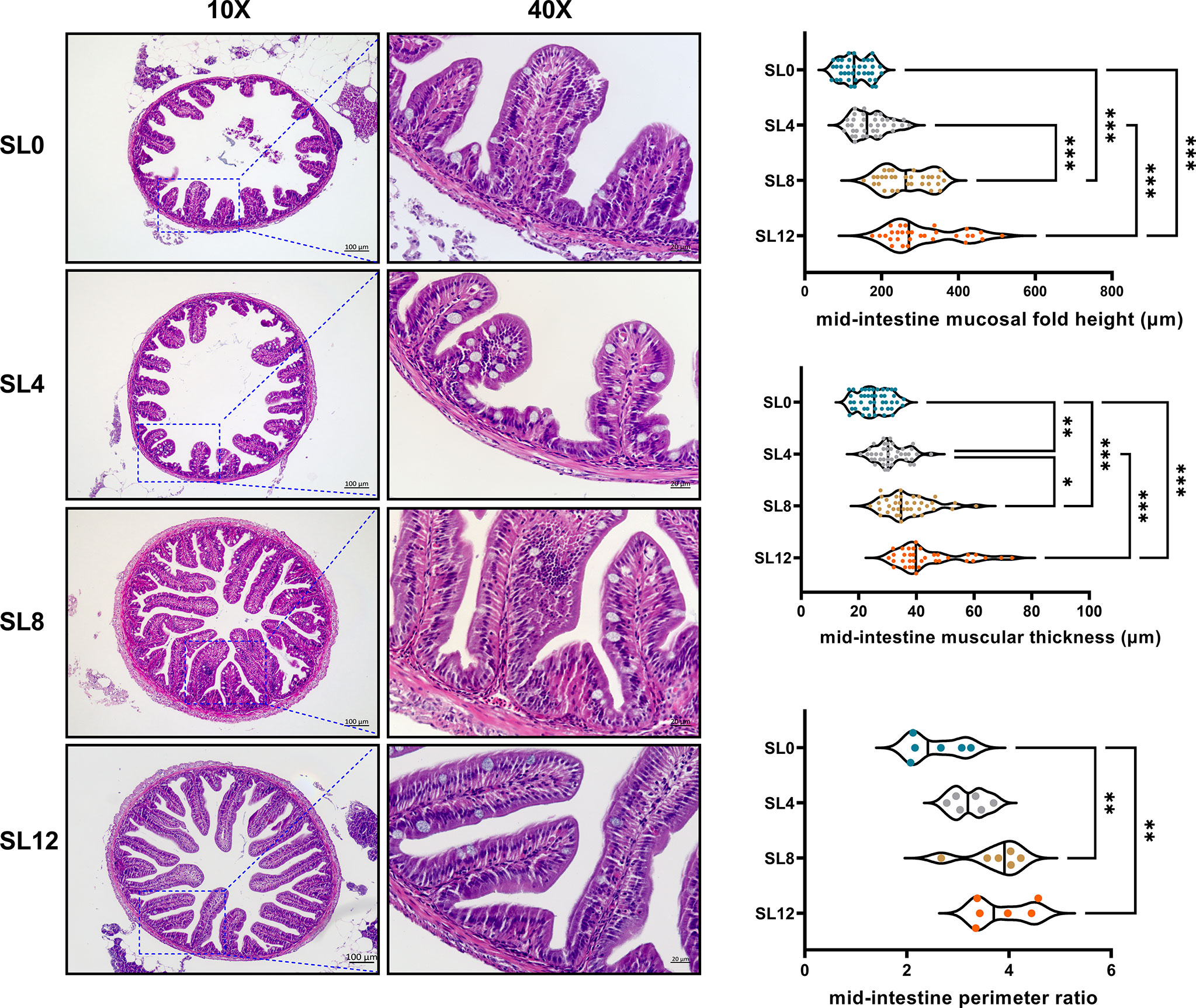
Figure 2 Longitudinal paraffin section of the mid-intestine. A total of 24 slices with 6 slices per treatment were observed. The perimeter ratio, intestinal muscular thickness, and mucosal fold height are presented by a violin plot. *P < 0.05, **P < 0.01, ***P < 0.001; otherwise, no significant difference(P > 0.05)
Liver Tissue RNA-Seq Quality Control, Mapping, and Bioinformatics Analysis
Clean reads were mapped on the annotated genome of rock bream, and an average of 96.88% of the UMI reads could be successfully mapped, of which an average of 89.42% were uniquely mapped (Table S1).
Based on the FPKM method, a total of 427 significant DEGs, including 126 up- and 301 downregulated DEGs, were detected in the SL0 versus SL12 groups (Figure 3A). Among the DEGs, some protein metabolism-related genes (gpt2, gs, and pro1) as well as lipid metabolism-related genes (acsbg2, acsl6, crls, lpin2, acsbg2, pemt, and mgll), were observed. The heatmap (Figure 3B) presented special selected DEGs involved in metabolism.
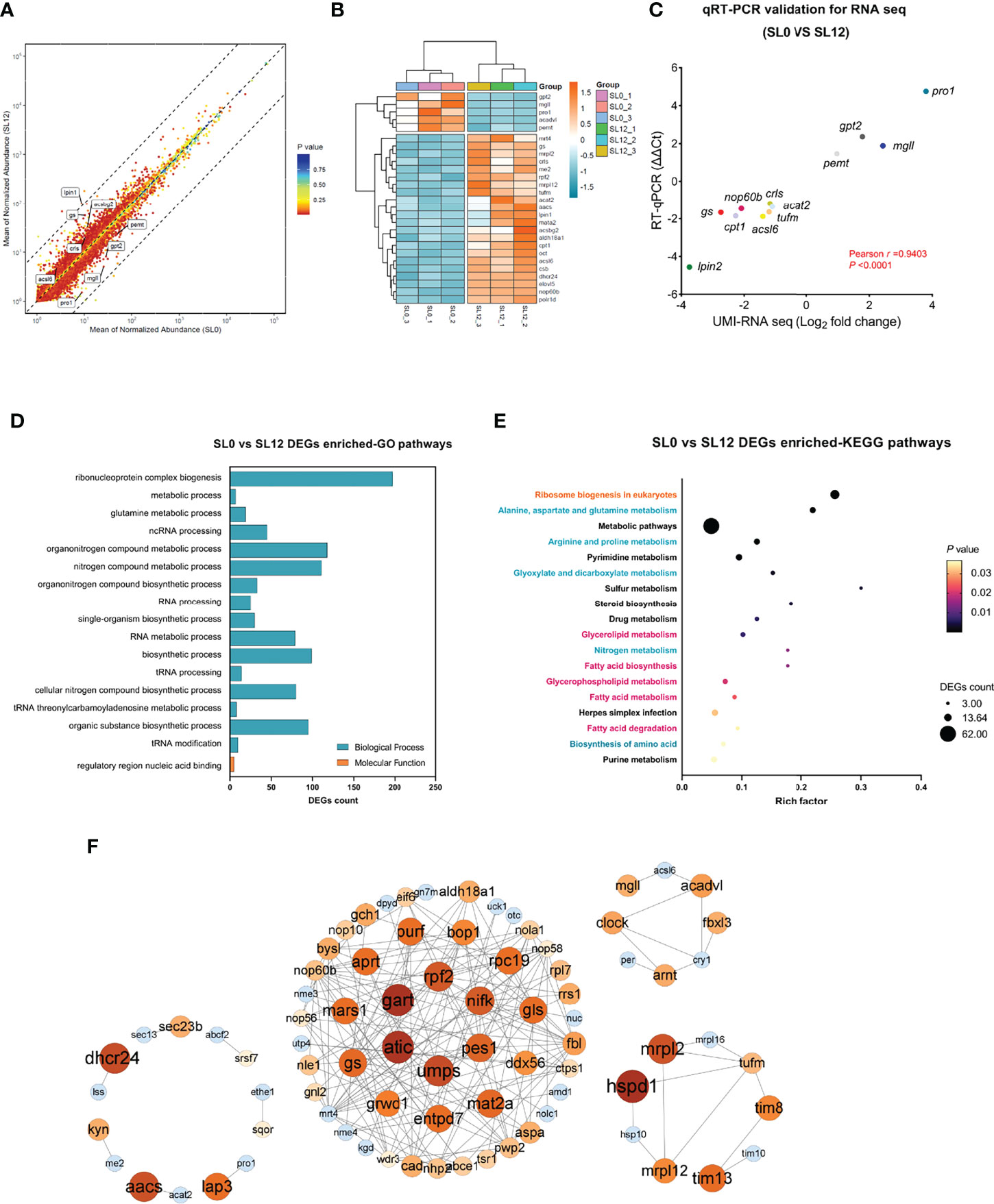
Figure 3 Transcriptome analysis of larval liver tissue corresponding to dietary phospholipid levels. (A) Volcano plot of detected gene expression levels between the SL0 and SL12 groups. A total of 427 DEGs were detected. (B) Heatmap presentation for partial DEGs between treatments. The block color diversity characterizes the gene expression levels. (C) UMI-RNA-seq DEG validation by qRT–PCR. (D, E) DEG-enriched GO and KEGG pathways between groups. (F) Potential association network for predicted DEGs constructed using Cytoscape software with high confidence.
To validate the DEGs identified by RNA-seq analysis, we performed qRT–PCR on 12 selected genes (Figure 3C). The qRT–PCR analysis revealed that all genes shared the same expression tendencies as those from the transcriptome data (ΔΔCt versus log2 fold change). Pearson’s correction analysis indicated that the r-value was 0.9403 between these two methods, and the P-value was below 0.0001. The qRT–PCR results confirmed the credibility of the UMI-RNA-seq data in this study.
All DEGs were subjected to Gene Ontology (GO) analysis, and the results showed strong enrichment of DEGs in two main categories: biological processes and molecular functions (Figure 3D). Among them, ribonucleoprotein complex biogenesis, metabolic process, and glutamine metabolic process were the most enriched GO pathways. The top 18 Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways are shown in Figure 3E. Ribosome biogenesis in eukaryotes was the KEGG pathway with the highest rich factor, with the involvement of protein metabolism-related pathways (in blue) and lipid metabolism-related pathways (purplish-red character), and some other pathways (in black).
The DEG association network was predicted using STRING with high confidence, and the interaction network was constructed by Cytoscape software. Figure 3F left panel shows a cluster illustrating some lipid metabolism-related gene interactions, such as dhcr24 and aacs, as well as some protein metabolism-related gene interactions, such as lap3 and pro1. The middle panel primarily highlights a clustering illustrating ribosome protein biosynthesis-related gene interactions and their interactions with nucleotide metabolism. The genes aprt, gart, atic, and umps translate key proteins that are involved in adenosine synthesis, while the genes pes1, nop56, nop58, and rpf2 translate key proteins for ribosome biogenesis, maturation, and assembly. The upright panel shows a clustering illustrating lipid metabolism gene interactions, such as acadvl, mgll, and acsl6. The bottom right panel shows a clustering illustrating genes encoding mitochondrial ribosome proteins (tufm, mrpl2, mrSL12, and mrpl16) interacting with genes encoding mitochondrial inner membrane proteins (tim8, tim10, and tim13).
Multidimensional Correlation
As shown in Figure 4, the correlation analysis between gene expression and nutritional parameters showed that the survival rate was significantly positively correlated with growth performance (WGR) and body ash content (P < 0.05). The whole-body ash content was positively correlated with the whole-body protein content (P<0.001) and TAA content (P<0.01) but negatively correlated with the MUFAs proportion (P<0.05), and lipid and fatty acid metabolism-related genes, as well as protein and amino acid metabolism-related genes, were significantly correlated with the whole-body ash content (Mantel’s r≥0.9, P<0.05). Notably, the whole-body protein content was significantly negatively correlated with the MUFAs proportion, and protein and amino acid metabolism-related genes were significantly correlated with the whole-body protein content (Mantel’s r≥0.4, P<0.01). Ribosome biogenesis-related gene expression was closely related to the whole-body EAA (Mantel’s P<0.01) and TAA (Mantel’s r≥0.9, P<0.01) contents.
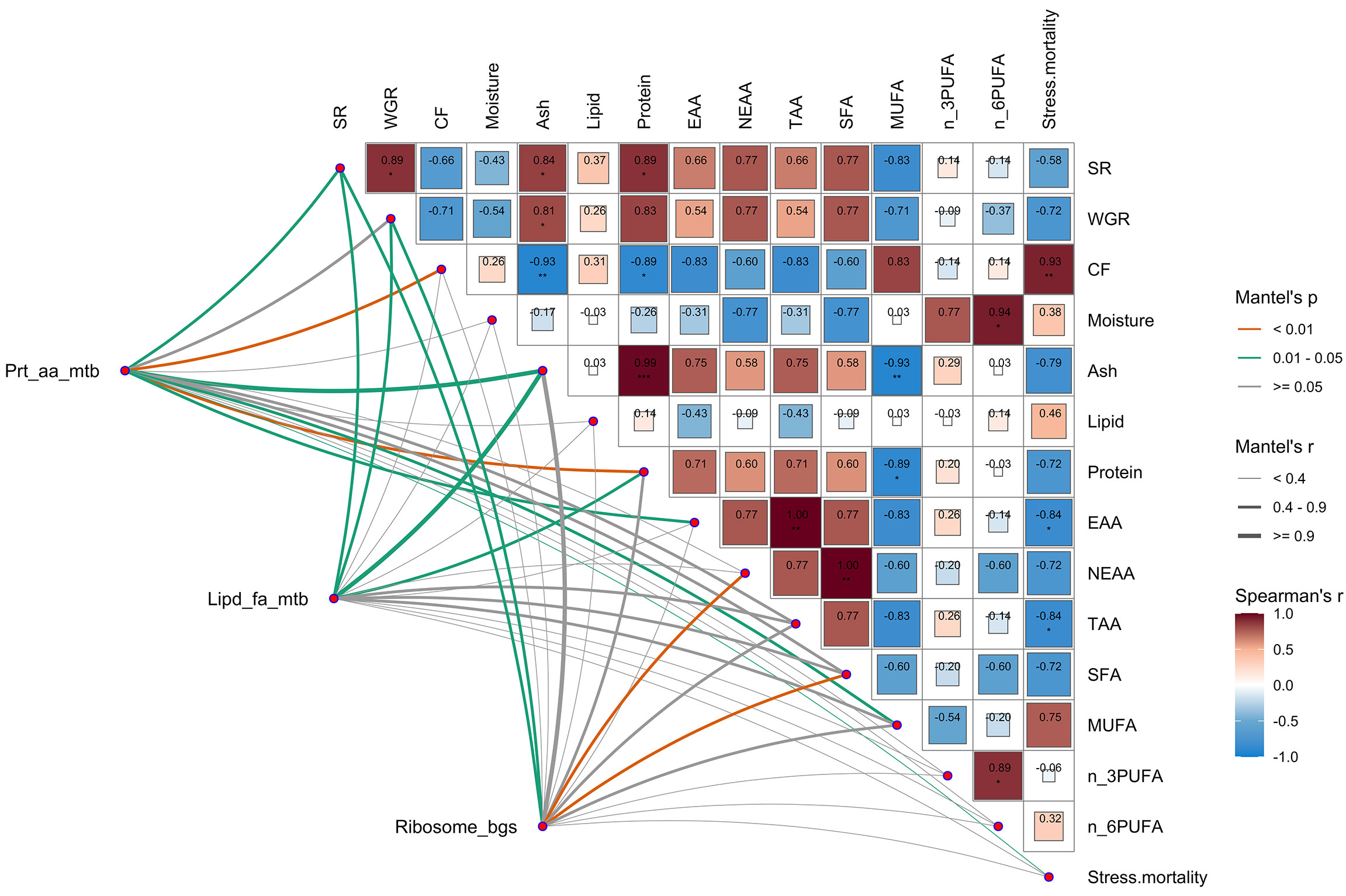
Figure 4 Multidimensional correlation analysis between gene expression and larval nutrition parameters. Nutritional parameters: survival rate (SR), weight gain rate (WGR), condition factor (CF), larval body moisture, ash, lipid, protein, essential amino acid content (EAA), nonessential amino acid content (NEAAs), total amino acid content (TAAs), saturated fatty acids (SFAs), monounsaturated fatty acids (MUFAs), n-3 polyunsaturated fatty acids (n_3PUFAs), n-6 polyunsaturated fatty acids (n_6PUFAs) and stress resistance to hypoxia stress (stress mortality). Gene expression: hepatic protein and amino acid metabolism-related gene expression, including pro1, mata2, gs, oct, aldh18a1, csb, and gpt2 (Prt_aa_mtb); hepatic lipid and fatty acid metabolism-related gene expression, including acat2, crls, me, aacs, lpin2, elovl5, mgll, acsbg2, acsl6, and pemt (Lipd_fa_mtb); hepatic ribosome biogenesis-related gene expression, including rpf2, polr1d, nop60b, mrpl2, mrSL12, mrpl16, nop56, nop58, bop1 and mrt4 (Ribosome_bgs). Pairwise comparisons of nutritional parameters are presented, with a color gradient and color block size denoting Pearson’s correlation coefficients. Related gene expression levels were related to the characterized parameters by Mantel’s tests. The edge width and color correspond to Mantel’s r statistic and Mantel’s p statistic for the corresponding distance correlations, respectively.
Discussion
Numerous investigations have demonstrated a strong and positive association between dietary intact PL inclusion and aquatic larval growth performance as well as stress resistance (Zhao et al., 2013; Gao et al., 2014; Zhang et al., 2022). Consistent with these results, larvae fed the high SL content MDs showed significantly higher growth performance and hypoxia stress resistance than those fed the SL0 MD. To investigate the potential mechanism underlying the growth promotion effect of dietary SL inclusion, mid-intestinal morphology, body nutrient composition, the activity of metabolism-related enzymes, and RNA sequencing in liver tissues were conducted. All DEGs have been listed as a supplemental table (Table S3). A schematic representation of the effects of the PL-enriched (SL12) diet on liver tissue metabolic and physiological responses is presented in Figure 5.
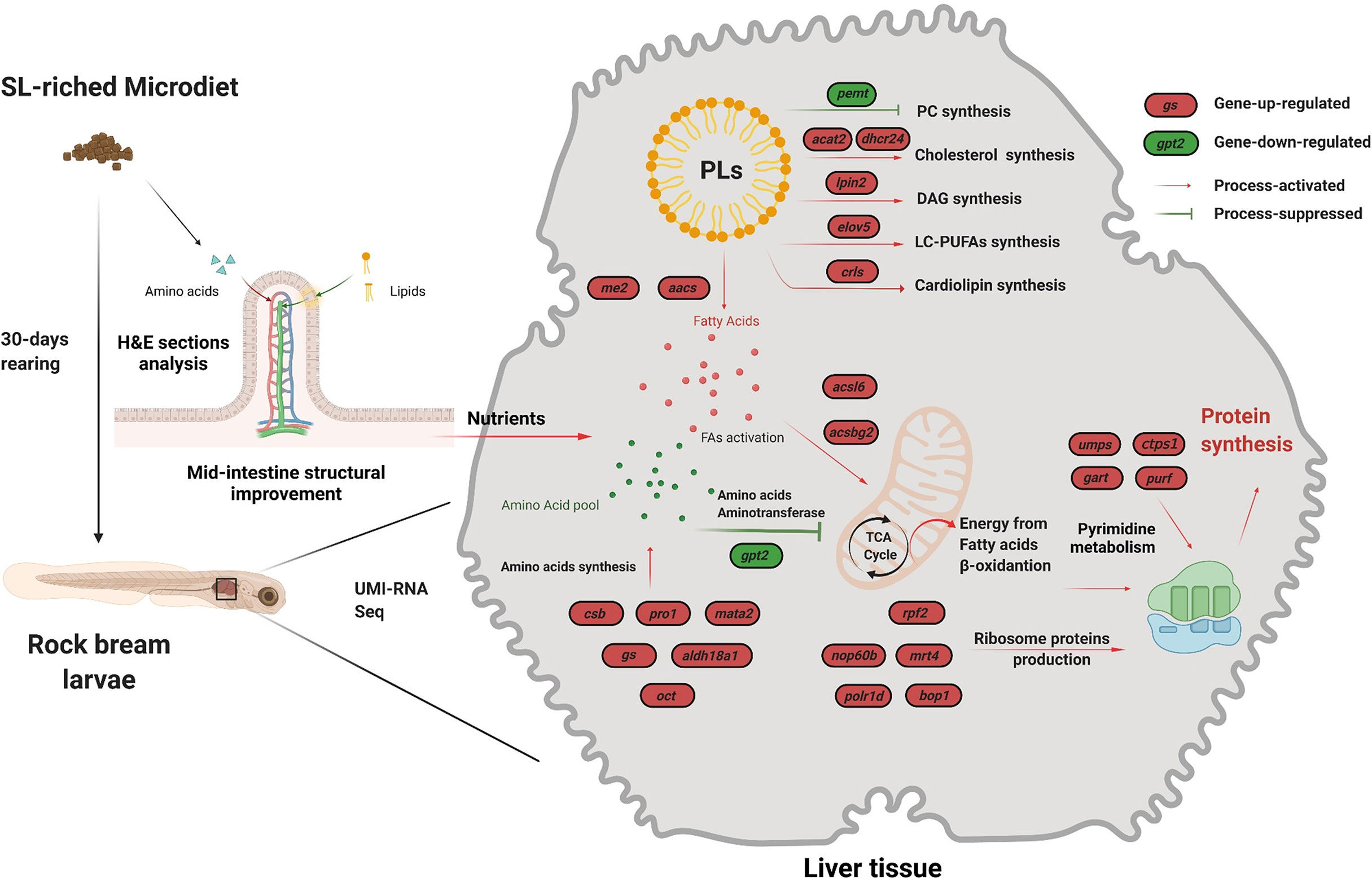
Figure 5 Schematic representation of the effects of the PL-rich (SL12) diet on intestine morphology and liver tissue molecular response. Transcriptome analysis was used to elucidate the potential molecular mechanisms underlying the observed results. The red ovals indicate upregulated genes, and the green ovals indicate downregulated genes. The SL12 diet improved the mid-intestine structure and thereby increased nutrient acquisition. The UMI-RNA sequencing results indicated that the SL12 diet could increase the de novo synthesis of lipids other than phosphatidylcholine. The SL12 diet increased energy production from fatty acid β-oxidation by fatty acid activation but decreased amino acid aminotransferase activity in the TCA cycle. The SL12 diet increased amino acid synthesis and ribosome production-related gene expression.
Dietary Phospholipids Improved Intestinal Morphology
Studies have revealed the beneficial effects of PLs in diet emulsification (Koven et al., 1993), fatty acid absorption (Fontagné et al., 2000; Hadas et al., 2003), digestive enzyme activities (Gisbert et al., 2005), intestinal lipid transportation (Lin et al., 2018) and microbiota composition (Wang et al., 2021). However, no consensus has been reached about the relationship between dietary intact PLs and intestinal morphology at the fish larval stage. A previous study on Atlantic salmon larvae argued that there was no significant difference in mid-intestinal enterocyte height among dietary treatments (Taylor et al., 2015). Moreover, the height of enterocytes was higher in carp (Cyprinus carpio L.) larvae that were fed the PL-deficient diet than in larvae fed a phosphatidylcholine-enriched diet (Fontagné et al., 1998). In contrast, the PL-deficient diet was reported to cause epithelial damage in the intestinal lumen of Arctic char larvae (Olsen et al., 1999). In line with that, our recent study in yellow drum larvae found that the high inclusion of dietary PLs induced increased fatty acid β-oxidation, which provided energy for the proliferation of enterocytes and eventually improved intestinal morphology (Tan et al., 2022). The rapid proliferation of enterocytes requires a considerable amount of energy, and dietary PL inclusion can serve as a preferential energy source during larval growth and development (Penn et al., 2011). In addition, fatty acids and choline from PL catabolism were indispensable for the synthesis of new cellular structures (Sargent et al., 1999). In the present study, the mid-intestine perimeter ratio, muscular thickness, and mucosal fold height were observed to significantly increase with dietary soybean lection inclusion. Therefore, the improved mid-intestine morphology could probably contribute to exogenous nutrient acquisition in larvae fed diets with high SL contents. The possible molecular mechanism was likely caused by the significantly increased mitochondrial fatty acid oxidation and energy production, as mitochondrial fatty acid β-oxidation key genes (ppara and cpt1) in larvae fed a high soybean lectin inclusion diet were significantly enhanced (Carmona-Antoñanzas et al., 2015; Cai et al., 2016; Tan et al., 2022).
SL-Enriched Diets Increased Phospholipid Catabolism
In this study, the activity of LP, a key lipolysis enzyme in the liver was significantly enhanced by the high inclusion of SLs; in addition, the expression level of fatty acid activation-related genes (acsl6 and acsbg2) was significantly increased. These results were in accordance with the findings in some other aquatic species that dietary SL inclusion increased lipid catabolism (Cahu et al., 2003; Niu et al., 2008; Azarm et al., 2013; Cai et al., 2016; Zhang et al., 2022). Larvae fed a high PL diet have increased energy production because PLs can serve as a preferential source of energy in the larval stage (Tocher et al., 2008). ACSBG2 and ACSL6 can both catalyze the conversion of long-chain fatty acids and activate diverse SFAs, MUFAs, and PUFAs to their active form acyl-CoA for degradation via β-oxidation (Pei et al., 2006; Nakahara et al., 2012). The increased gene expression of ascl6 and acsbg2 in the liver of the SL12 group indicated the increased activation of fatty acid conversion and thereby increased fatty acid flux to β-oxidation. Corresponding to the increase in fatty acid β-oxidation, the expression levels of mitochondrial biogenesis-related genes (mrpl2, mrSL12, and mrpl16), as well as mitochondrial intermembrane chaperone-related genes (tim8, tim10, and tim13), were significantly upregulated. These mitochondrial biogenesis-related genes encoding 39S ribosomal proteins are essential for mitochondrial construction (Amunts et al., 2015). At a high confidence level, DEG interactions (Figure 3F) indicated that genes encoding mitochondrial biogenesis and mitochondrial intermembrane chaperones interacted with tufm, which was required for mitochondrial protein biosynthesis and maintenance of mitochondrial DNA (Christian and Spremulli, 2012).
The increased dietary SL content seems to have little effect on the whole-body PL content. Extra dietary SLs could lead to a crowding-out effect, characterized as increasing PL catabolism and increasing the biosynthesis of other lipids. Correspondingly, RNA sequencing results indicate that the SL12 diet significantly increased the expression of some other lipid anabolism-related genes, such as cholesterol synthesis-related acat2 (An et al., 2006) and dhcr24 (Waterham et al., 2001), diacylglycerol synthesis-related lpin2 (Donkor et al., 2007), LC-PUFA synthesis-related elov5 (Monroig et al., 2022), and diphosphatidylglycerol (cardiolipin) synthesis-related crls (Lu et al., 2006). These results were in accordance with the findings in hybrid grouper, in which dietary PL inclusion increased hepatic crude lipid and triglyceride by regulating the synthesis and hydrolysis of triglyceride (Huang et al., 2021). Moreover, upregulated mgll expression in the SL12 group indicated that an SL-enriched diet enhances the conversion of monoacylglycerides to free fatty acids and glycerol (Taschler et al., 2011) and thereby increases the fatty acids supply for lipid biosynthesis. Notably, high dietary SL content significantly decreased the expression of pemt. PEMT is the key enzyme that catalyzes the three sequential steps of the methylation pathway of phosphatidylcholine biosynthesis (Tocher et al., 2008). This result acts in cooperation with the whole-body PL content, and the mechanism could be the negative feedback loop in response to high dietary PL content. In summary, a high soybean lection content improves fatty acid flux to energy production and increases PL catabolism to other lipids, such as cholesterol, diacylglycerol, LC-PUFAs, and diphosphatidylglycerol.
SL-Enriched Diets Decreased Amino Acid Catabolism
AST and ALT are the key enzymes mediating amino acid catabolism through the transamination of amino acids into TCA cycle precursors (Bibiano Melo et al., 2006). A previous study demonstrated that the activities of ALT and AST were negatively correlated with protein efficiency (He et al., 2010). A study in largemouth bass (Micropterus salmoides) larvae reported that the inclusion of marine protein hydrolysates significantly decreased the activities of AST and ALT accompanied by a growth promotion effect (Dai et al., 2020). In the present study, the activities of the AST and ALT enzymes in liver tissue were observed to be significantly higher in the larvae fed the SL0 diet than in the larvae fed the SL12 diet. A possible explanation for this result could be that the increased of AST and ALT activities enhanced the flux of amino acids into the TCA cycle for ATP production in the SL0 group. In accordance with the decreased aminotransferase enzyme activities, the RNA sequencing results showed that the gpt2 (encoding alanine aminotransferase 2) expression level in the liver tissue of larvae fed the SL12 diet was significantly decreased. Interestingly, the expression of amino acid synthesis-related genes was significantly increased. These genes include glutamine synthesis-related gs (Eelen et al., 2018), cystathionine synthesis-related csb (Casique et al., 2013), proline synthesis-related aldh18a1 (Baumgartner et al., 2000), S-adenosylmethionine synthesis-related mata2 and L-citrulline-synthesis-related oct (Couchet et al., 2021). As a consequence, the increased amino acid synthesis and decreased amino acid catabolism in the SL12 group could be attributed to the increased whole-body protein and amino acid contents in larvae. The multidimensional correlation analysis indicated that the whole-body protein content was significantly negatively correlated with the whole-body MUFAs proportion, while protein and amino acid metabolism-related genes were significantly correlated with the whole-body protein content (Mantel’s r≥0.4, P<0.01). The multidimensional correlation analysis supports our hypothesis that larval whole-body protein content was regulated by fatty acid and amino acid metabolism processes.
SL-Enriched Diets Increased Ribosome Biogenesis
Individual growth, more precisely protein retention, primarily occurs in the ribosome. The ribosome is the essential unit of all living organisms that executes protein synthesis and therefore fuels cell growth and cell proliferation. The biogenesis of ribosomes requires abundant nutrients and ATP; therefore, the synthesis of ribosomes is tightly controlled (Piazzi et al., 2019). In this study, the SL12 diet significantly increased ribosome biogenesis-related gene expression in the liver tissue, characterized as rpf2, pes1, nop56, nop58, nop60b, mrt4, polr1d, and bop1. The proteins RPF2, PES1, NOP56, NOP58, NOP60B, MRT4, and BOP1 are essential elements for ribosome biogenesis, maturation, and assembly processes (Hayano et al., 2003; Rohrmoser et al., 2007; Michalec et al., 2010; Sloan et al., 2013). The DEG-enriched GO and KEGG pathways revealed that ribosome protein synthesis-related pathways were the most enriched pathways. These results indicated that the SL12 diet significantly increased ribosome biogenesis, which was in accordance with the increased whole-body protein content. It was inferred that a high content of amino acids together with abundant ribosomes led to increased protein synthesis in larvae fed the SL12. In line with this, the multidimensional correlation analysis indicated that the whole-body EAA and TAA contents were closely related to ribosome biogenesis-related gene expression.
Surprisingly, the protein-to-protein interaction analysis by Cytoscape software indicated that nucleotide metabolism-related genes (aprt, ctps1, gart, atic, and umps), especially pyrimidine and purine metabolism-related genes, closely interacted with ribosome protein biosynthesis-related }gene expression. The protein synthesis process not only requires amino acids and ribosomes but also requires templates for that process. Nucleotide metabolism plays a role in supplying messenger RNAs, transfer RNA and ribosomal RNA, as well as precursor substances for energy carriers (ATP and GTP) (Lane & Fan, 2015). Meanwhile, nucleotide metabolism is closely related to amino acid metabolism, and nucleotides and amino acids can be transformed into each other. Further study is warranted to reveal the role of nucleotide metabolism in the process.
Conclusions
Feeding trials and subsequent transcriptome analysis revealed the effects of dietary SL content on survival rate, growth performance, intestinal structure in rock bream larvae, as well as the underlying molecular mechanism. The conclusions are summarized as follows:(1) The PL-enriched diet (SL12) increased the growth performance and resistance to hypoxia in rock bream at the larval stage.(2) Dietary SL improved mid-intestine morphology.(3) The PL-enriched diet could increase phospholipid catabolism and amino acid synthesis-related gene expression but decreased amino acid catabolism-related gene expression.(4) The PL-enriched diet increased ribosome biogenesis- related gene expression.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below:
https://www.ncbi.nlm.nih.gov/bioproject/PRJNA848209
Ethics Statement
The animal study was reviewed and approved by Animal Research and Ethics Committees of Ocean University of Zhejaing Province.
Author Contributions
PT: conceptualization, data curation, formal analysis, funding acquisition, project administration, supervision, software, validation, roles/writing – original draft, and writing – review and editing. PZ: data curation. LZ: data curation. WZ: data curation. LW: data curation. RC: data curation. QZ: formal analysis. DX: funding acquisition, project administration, supervision. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Key R&D Program of China [grant number: 2020YFD0900804], the Zhejiang Provincial Natural Science Foundation of China (grant number: LQ20C190008) and the Science and Technology Program of Marine Fisheries Research Institute of Zhejiang Province (HYS-CZ-003).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The hydrolyzed fish paste was kindly provided by Kenon Biotechnology Co., Ltd. (Weifang, China).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2022.942259/full#supplementary-material
References
Alejo A., Tafalla C. (2011). Chemokines in Teleost Fish Species. Dev. Comp. Immunol. 35 (12), 1215–1222. doi: 10.1016/j.dci.2011.03.011
Amunts A., Brown A., Toots J., Scheres S. H. W., Ramakrishnan V. (2015). Ribosome. The Structure of the Human Mitochondrial Ribosome. Science 348 (6230), 95–98. doi: 10.1126/science.aaa1193
An S., Cho K. H., Lee W. S., Lee J. O., Paik Y. K., Jeong T. S. (2006). A Critical Role for the Histidine Residues in the Catalytic Function of Acyl-CoA:cholesterol Acyltransferase Catalysis: Evidence for Catalytic Difference Between ACAT1 and ACAT2. FEBS Lett. 580 (11), 2741–2749. doi: 10.1016/j.febslet.2006.04.035
AOAC (2016). Official Methods of Analysis. 20th ed (Rockville, MD) Association of Official Analytical Chemists.
Azarm H. M., Kenari A. A., Hedayati M. (2013). Effect of Dietary Phospholipid Sources and Levels on Growth Performance, Enzymes Activity, Cholecystokinin and Lipoprotein Fractions of Rainbow Trout (Oncorhynchus Mykiss) Fry. Aquaculture. Res. 44 (4), 634–644. doi: 10.1111/j.1365-2109.2011.03068.x
Baeverfjord G., Krogdahl A. (1996). Development and Regression of Soybean Meal Induced Enteritis in Atlantic Salmon, Salmo Salar L., Distal Intestine: A Comparison With the Intestines of Fasted Fish. J. Fish. Dis. 19 (5), 375–387. doi: 10.1046/j.1365-2761.1996.d01-92.x
Baumgartner M. R., Hu C. A., Almashanu S., Steel G., Obie C., Aral B., et al. (2000). Hyperammonemia With Reduced Ornithine, Citrulline, Arginine and Proline: A New Inborn Error Caused by a Mutation in the Gene Encoding Delta(1)-Pyrroline-5-Carboxylate Synthase. Hum. Mol. Genet. 9 (19), 2853–2858. doi: 10.1093/hmg/9.19.2853
Bibiano Melo J. F., Lundstedt L. M., Meton I., Baanante I. V., Moraes G. (2006). Effects of Dietary Levels of Protein on Nitrogenous Metabolism of Rhamdia Quelen (Teleostei: Pimelodidae). Comp. Biochem. Physiol. A. Mol. Integr. Physiol. 145 (2), 181–187. doi: 10.1016/j.cbpa.2006.06.007
Cahu C. L., Zambonino Infante J. L., Barbosa V. (2003). Effect of Dietary Phospholipid Level and Phospholipid:Neutral Lipid Value on the Development of Sea Bass (Dicentrarchus Labrax) Larvae Fed a Compound Diet. Br. J. Nutr. 90 (1), 21–28. doi: 10.1079/bjn2003880
Cai Z., Feng S., Xiang X., Mai K., Ai Q. (2016). Effects of Dietary Phospholipid on Lipase Activity, Antioxidant Capacity and Lipid Metabolism-Related Gene Expression in Large Yellow Croaker Larvae (Larimichthys Crocea). Comp. Biochem. Physiol. B. Biochem. Mol. Biol. 201, 46–52. doi: 10.1016/j.cbpb.2016.06.007
Cai Z., Mai K., Ai Q. (2017). Regulation of Hepatic Lipid Deposition by Phospholipid in Large Yellow Croaker. Br. J. Nutr. 118 (12), 999–1009. doi: 10.1017/s000711451700294x
Carmona-Antoñanzas G., Taylor J. F., Martinez-Rubio L., Tocher D. R. (2015). Molecular Mechanism of Dietary Phospholipid Requirement of Atlantic Salmon, Salmo Salar, Fry. Biochim. Biophys. Acta (BBA). -. Mol. Cell Biol. Lipids 1851 (11), 1428–1441. doi: 10.1016/j.bbalip.2015.08.006
Casique L., Kabil O., Banerjee R., Martinez J. C., De Lucca M. (2013). Characterization of Two Pathogenic Mutations in Cystathionine Beta-Synthase: Different Intracellular Locations for Wild-Type and Mutant Proteins. Gene 531 (1), 117–124. doi: 10.1016/j.gene.2013.08.021
Christian B. E., Spremulli L. L. (2012). Mechanism of Protein Biosynthesis in Mammalian Mitochondria. Biochim. Biophys. Acta 1819 (9-10), 1035–1054. doi: 10.1016/j.bbagrm.2011.11.009
Couchet M., Breuillard C., Corne C., Rendu J., Morio B., Schlattner U., et al. (2021). Ornithine Transcarbamylase - From Structure to Metabolism: An Update. Front. Physiol. 12. doi: 10.3389/fphys.2021.748249
Dai M., Li S. L., Fu C. F., Qiu H. J., Chen N. S. (2020). The Potential Role of Marine Protein Hydrolyzates in Elevating Nutritive Values of Diets f or Largemouth Bass, Micropterus Salmoides. Front. Mar. Sci. 7. doi: 10.3389/fmars.2020.00197
De Santis C., Taylor J. F., Martinez-Rubio L., Boltana S., Tocher D. R., (2015). Influence of Development and Dietary Phospholipid Content and Composition on Intestinal Transcriptome of Atlantic Salmon (Salmo Salar). PloS One 10 (10), e0140964. doi: 10.1371/journal.pone.0140964
Dimitroglou A., Merrifield D. L., Moate R., Davies S. J., Spring P., Sweetman J., et al. (2009). Dietary Mannan Oligosaccharide Supplementation Modulates Intestinal Microbial Ecology and Improves Gut Morphology of Rainbow Trout, Oncorhynchus Mykiss (Walbaum). J. Anim. Sci. 87 (10), 3226–3234. doi: 10.2527/jas.2008-1428
Donkor J., Sariahmetoglu M., Dewald J., Brindley D. N., Reue K. (2007). Three Mammalian Lipins Act as Phosphatidate Phosphatases With Distinct Tissue Expression Patterns. J. Biol. Chem. 282 (6), 3450–3457. doi: 10.1074/jbc.M610745200
Eelen G., Dubois C., Cantelmo A. R., Goveia J., Bruning U., DeRan M., et al. (2018). Role of Glutamine Synthetase in Angiogenesis Beyond Glutamine Synthesis. Nature 561 (7721), 63–69. doi: 10.1038/s41586-018-0466-7
Folch J., Lees M., Sloane Stanley G. H. (1957). A Simple Method for the Isolation and Purification of Total Lipides From Animal Tissues. J. Biol. Chem. 226 (1), 497–509. doi: 10.1016/S0021-9258(18)64849-5
Fontagné S., Burtaire L., Corraze G., Bergot P. (2000). Effects of Dietary Medium-Chain Triacylglycerols (Tricaprylin and Tricaproin) and Phospholipid Supply on Survival, Growth and Lipid Metabolism in Common Carp (Cyprinus Carpio L.) Larvae. Aquaculture 190 (3-4), 289–303. doi: 10.1016/s0044-8486(00)00400-2
Fontagné S., Geurden I., Escaffre A.-M., Bergot P. (1998). Histological Changes Induced by Dietary Phospholipids in Intestine and Liver of Common Carp (Cyprinus Carpio L.) Larvae. Aquaculture 161 (1-4), 213–223. doi: 10.1016/s0044-8486(97)00271-8
Francis D. S., Turchini G. M. (2017). Retro-Engineering the Protein Sparing Effect to Preserve N-3 LC-PUFA From Catabolism and Optimise Fish Oil Utilisation: A Preliminary Case Study on Juvenile Atlantic Salmon. Aquaculture 468, 184–192. doi: 10.1016/j.aquaculture.2016.10.013
Gao J., Koshio S., Wang W. M., Li Y., Huang S. Q., Cao X. J. (2014). Effects of Dietary Phospholipid Levels on Growth Performance, Fatty Acid Composition and Antioxidant Responses of Dojo Loach Misgurnus Anguillicaudatus Larvae. Aquaculture 426, 304–309. doi: 10.1016/j.aquaculture.2014.02.022
Gisbert E., Villeneuve L., Zambonino-Infante J. L., Quazuguel P., Cahu C. L. (2005). Dietary Phospholipids are More Efficient Than Neutral Lipids for Long-Chain Polyunsaturated Fatty Acid Supply in European Sea Bass Dicentrarchus Labrax Larval Development. Lipids 40 (6), 609–618. doi: 10.1007/s11745-005-1422-0
Hadas E., Koven W., Sklan D., Tandler A. (2003). The Effect of Dietary Phosphatidylcholine on the Assimilation and Distribution of Ingested Free Oleic Acid (18:1n-9) in Gilthead Seabream (Sparus Aurata) Larvae. Aquaculture 217 (1-4), 577–588. doi: 10.1016/s0044-8486(02)00431-3
Hayano T., Yanagida M., Yamauchi Y., Shinkawa T., Isobe T., Takahashi N. (2003). Proteomic Analysis of Human Nop56p-Associated Pre-Ribosomal Ribonucleoprotein Complexes. Possible Link Between Nop56p and the Nucleolar Protein Treacle Responsible for Treacher Collins Syndrome. J. Biol. Chem. 278 (36), 34309–34319. doi: 10.1074/jbc.M304304200
He Y., Song H. L., Li G. X., Xu J., Gao B. X., Yu T., et al. (2010). The Relationship of CPS-I, OCT and Hepatic Encephalopathy. Zhonghua. Gan. Zang. Bing. Za. Zhi. 18 (9), 699–702. doi: 10.3760/cma.j.issn.1007-3418.2010.09.013
Huang Y., Xu J., Sheng Z., Chen N., Li S. (2021). Integrated Response of Growth Performance, Fatty Acid Composition, Antioxidant Responses and Lipid Metabolism to Dietary Phospholipids in Hybrid Grouper (Epinephelus Fuscoguttatus ♀ × E. Lanceolatus ♂) Larvae. Aquaculture 541, 736728. doi: 10.1016/j.aquaculture.2021.736728
Koven W. M., Kolkovski S., Tandler A., Kissil G. W., Sklan D. (1993). The Effect of Dietary Lecithin and Lipase, as a Function of Age, on N-9 Fatty Acid Incorporation in the Tissue Lipids of Sparus Aurata Larvae. Fish. Physiol. Biochem. 10 (5), 357–364. doi: 10.1007/bf00004502
Lane A. N., Fan T. W. (2015). Regulation of Mammalian Nucleotide Metabolism and Biosynthesis. Nucleic Acids Res. 43 (4), 2466–2485. doi: 10.1093/nar/gkv047
Li Y., Gao J., Huang S. (2015). Effects of Different Dietary Phospholipid Levels on Growth Performance, Fatty Acid Composition, PPAR Gene Expressions and Antioxidant Responses of Blunt Snout Bream Megalobrama Amblycephala Fingerlings. Fish. Physiol. Biochem. 41 (2), 423–436. doi: 10.1007/s10695-014-9994-8
Lin S. M., Li F. J., Yuangsoi B., Doolgindachbaporn S. (2018). Effect of Dietary Phospholipid Levels on Growth, Lipid Metabolism, and Antioxidative Status of Juvenile Hybrid Snakehead (Channa Argus × Channa Maculata). Fish. Physiol. Biochem. 44 (1), 401–410. doi: 10.1007/s10695-017-0443-3
Livak K. J., Schmittgen T. D. (2001). Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25 (4), 402–408. doi: 10.1006/meth.2001.1262
Lu B., Xu F. Y., Jiang Y. J., Choy P. C., Hatch G. M., Grunfeld C., et al. (2006). Cloning and Characterization of a cDNA Encoding Human Cardiolipin Synthase (Hcls1). J. Lipid Res. 47 (6), 1140–1145. doi: 10.1194/jlr.C600004-JLR200
Michalec B., Krokowski D., Grela P., Wawiórka L., Sawa-Makarska J., Grankowski N., et al. (2010). Subcellular Localization of Ribosomal P0-Like Protein MRT4 is Determined by its N-Terminal Domain. Int. J. Biochem. Cell Biol. 42 (5), 736–748. doi: 10.1016/j.biocel.2010.01.011
Monroig Ó., Shu-Chien A. C., Kabeya N., Tocher D. R., Castro L. F. C. (2022). Desaturases and Elongases Involved in Long-Chain Polyunsaturated Fatty Acid Biosynthesis in Aquatic Animals: From Genes to Functions. Prog. Lipid Res. 86, 101157. doi: 10.1016/j.plipres.2022.101157
Nakahara K., Ohkuni A., Kitamura T., Abe K., Naganuma T., Ohno Y., et al. (2012). The Sjögren-Larsson Syndrome Gene Encodes a Hexadecenal Dehydrogenase of the Sphingosine 1-Phosphate Degradation Pathway. Mol. Cell 46 (4), 461–471. doi: 10.1016/j.molcel.2012.04.033
Niu J., Liu Y. J., Tian L. X., Mai K. S., Yang H. J., Ye C. X., et al. (2008). Effects of Dietary Phospholipid Level in Cobia (Rachycentron Canadum) Larvae: Growth, Survival, Plasma Lipids and Enzymes of Lipid Metabolism. Fish. Physiol. Biochem. 34 (1), 9–17. doi: 10.1007/s10695-007-9140-y
Olsen R. E., Myklebust R., Kaino T., Ringø E. (1999). Lipid Digestibility and Ultrastructural Changes in the Enterocytes of Arctic Char (Salvelinus Alpinus L.) Fed Linseed Oil and Soybean Lecithin. Fish. Physiol. Biochem. 21 (1), 35–44. doi: 10.1023/a:1007726615889
Pei Z., Jia Z., Watkins P. A. (2006). The Second Member of the Human and Murine Bubblegum Family is a Testis- and Brainstem-Specific Acyl-CoA Synthetase. J. Biol. Chem. 281 (10), 6632–6641. doi: 10.1074/jbc.M511558200
Penn M. H., Campbell P., Krogdahl Å. (2011). High Level of Dietary Pea Protein Concentrate Induces Enteropathy in Atlantic Salmon (Salmo Salar L.). Aquaculture 310 (3-4), 267–273. doi: 10.1016/j.aquaculture.2010.10.040
Piazzi M., Bavelloni A., Gallo A., Faenza I., Blalock W. L. (2019). Signal Transduction in Ribosome Biogenesis: A Recipe to Avoid Disaster. Int. J. Mol. Sci. 20 (11). 2718 doi: 10.3390/ijms20112718
Rohrmoser M., Hölzel M., Grimm T., Malamoussi A., Harasim T., Orban M., et al. (2007). Interdependence of Pes1, Bop1, and WDR12 Controls Nucleolar Localization and Assembly of the PeBoW Complex Required for Maturation of the 60S Ribosomal Subunit. Mol. Cell Biol. 27 (10), 3682–3694. doi: 10.1128/mcb.00172-07
Sargent J., McEvoy L., Estevez A., Bell G., Bell M., Henderson J., et al. (1999). Lipid Nutrition of Marine Fish During Early Development: Current Status and Future Directions. Aquaculture 179 (1-4), 217–229. doi: 10.1016/s0044-8486(99)00191-x
Shannon P., Markiel A., Ozier O., Baliga N. S., Wang J. T., Ramage D., et al. (2003). Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res. 13 (11), 2498–2504. doi: 10.1101/gr.1239303
Sloan K. E., Bohnsack M. T., Watkins N. J. (2013). The 5s RNP Couples P53 Homeostasis to Ribosome Biogenesis and Nucleolar Stress. Cell Rep. 5 (1), 237–247. doi: 10.1016/j.celrep.2013.08.049
Tan P., Zhu W., Zhang P., Wang L., Chen R., Xu D. (2022). Dietary Soybean Lecithin Inclusion Promotes Growth, Development, and Intestinal Morphology of Yellow Drum (Nibea Albiflora) Larvae. Aquaculture. 559, 738446. doi: 10.1016/j.aquaculture.2022.738446
Taschler U., Radner F. P., Heier C., Schreiber R., Schweiger M., Schoiswohl G., et al. (2011). Monoglyceride Lipase Deficiency in Mice Impairs Lipolysis and Attenuates Diet-Induced Insulin Resistance. J. Biol. Chem. 286 (20), 17467–17477. doi: 10.1074/jbc.M110.215434
Taylor J. F., Martinez-Rubio L., del Pozo J., Walton J. M., Tinch A. E., Migaud H., et al. (2015). Influence of Dietary Phospholipid on Early Development and Performance of Atlantic Salmon (Salmo Salar). Aquaculture 448, 262–272. doi: 10.1016/j.aquaculture.2015.06.012
Tocher D. R., Bendiksen E.Å., Campbell P. J., Bell J. G. (2008). The Role of Phospholipids in Nutrition and Metabolism of Teleost Fish. Aquaculture 280 (1-4), 21–34. doi: 10.1016/j.aquaculture.2008.04.034
Vandesompele J., De Preter K., Pattyn F., Poppe B., Van Roy N., De Paepe A., et al. (2002). Accurate Normalization of Real-Time Quantitative RT-PCR Data by Geometric Averaging of Multiple Internal Control Genes. Genome Biol. 3 (7), RESEARCH0034. doi: 10.1186/gb-2002-3-7-research0034
Wang S., Han Z., Turchini G. M., Wang X., Fang Z., Chen N., et al. (2021). Effects of Dietary Phospholipids on Growth Performance, Digestive Enzymes Activity and Intestinal Health of Largemouth Bass (Micropterus Salmoides) Larvae. Front. Immunol. 12. doi: 10.3389/fimmu.2021.827946
Waterham H. R., Koster J., Romeijn G. J., Hennekam R. C., Vreken P., Andersson H. C., et al. (2001). Mutations in the 3beta-Hydroxysterol Delta24-Reductase Gene Cause Desmosterolosis, an Autosomal Recessive Disorder of Cholesterol Biosynthesis. Am. J. Hum. Genet. 69 (4), 685–694. doi: 10.1086/323473
Wu X., Wang L., Xie Q., Tan P. (2019). Effects of Dietary Sodium Butyrate on Growth, Diet Conversion, Body Chemical Compositions and Distal Intestinal Health in Yellow Drum (Nibea Albiflora, Richardson). Aquaculture. Res. 51 (1), 69–79. doi: 10.1111/are.14348
Xu D., You Q., Chi C., Luo S., Song H., Lou B., et al. (2018). Transcriptional Response to Low Temperature in the Yellow Drum (Nibea Albiflora) and Identification of Genes Related to Cold Stress. Comp. Biochem. Physiol. Part D. Genomics Proteomics 28, 80–89. doi: 10.1016/j.cbd.2018.07.003
Zhang F., Ning Y. C., Yuan R., Ding J., Chang Y. Q., Zuo R. T. (2022). Effects of Soya Lecithin Addition on the Growth, Antioxidant Capacity, Gonad Development and Nutritional Quality of Adult Sea Urchin (Strongylocentrotus Intermedius). Aquaculture. Rep. 22, 100990. doi: 10.1016/j.aqrep.2021.100990
Zhao J., Ai Q., Mai K., Zuo R., Luo Y. (2013). Effects of Dietary Phospholipids on Survival, Growth, Digestive Enzymes and Stress Resistance of Large Yellow Croaker, Larmichthys Crocea Larvae. Aquaculture 410-411, 122–128. doi: 10.1016/j.aquaculture.2013.05.018
Zuo R., Ai Q., Mai K., Xu W., Wang J., Xu H., et al. (2012). Effects of Dietary N-3 Highly Unsaturated Fatty Acids on Growth, Nonspecific Immunity, Expression of Some Immune Related Genes and Disease Resistance of Large Yellow Croaker (Larmichthys Crocea) Following Natural Infestation of Parasites (Cryptocaryon Irritans). Fish. Shellfish. Immunol. 32 (2), 249–258. doi: 10.1016/j.fsi.2011.11.005
Keywords: soybean lecithin, larvae, growth performance, intestinal morphology, liver tissue, fatty acid metabolism
Citation: Tan P, Zhang P, Zhang L, Zhu W, Wang L, Chen R, Zhu Q and Xu D (2022) Effects of Soybean Lecithin on Growth Performance, Intestine Morphology, and Liver Tissue Metabolism in Rock Bream (Oplegnathus fasciatus) Larvae. Front. Mar. Sci. 9:942259. doi: 10.3389/fmars.2022.942259
Received: 12 May 2022; Accepted: 13 June 2022;
Published: 11 July 2022.
Edited by:
Min Jin, Ningbo University, ChinaReviewed by:
Lingyu Li, Umeå, University, SwedenHongyan Tian, Yancheng Institute of Technology, China
Songlin Li, Shanghai Ocean University, China
Copyright © 2022 Tan, Zhang, Zhang, Zhu, Wang, Chen, Zhu and Xu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dongdong Xu, eHVkb25nMDU4MEAxNjMuY29t
 Peng Tan1,2
Peng Tan1,2 Ligai Wang
Ligai Wang Ruiyi Chen
Ruiyi Chen Dongdong Xu
Dongdong Xu