- 1Key Laboratory of Freshwater Fish Reproduction and Development (Ministry of Education), College of Fisheries, Southwest University, Chongqing, China
- 2Key Laboratory of Aquatic Biodiversity and Conservation of Chinese Academy of Sciences, Institute of Hydrobiology, Chinese Academy of Sciences, Wuhan, China
Rock carp, Procypris rabaudi, is a vulnerable carp endemic to the upper reaches of the Yangtze River and included in the National Key Protected Wildlife in 2021 as a second-class aquatic animal. Evaluating the genetic makeup of released individuals before a restocking activity is carried out is essential, and a molecular marker with simple, rapid, and universal characteristics will be helpful to the evaluation. In this study, the genetic diversity and structure of rock carp from two representative hatcheries [Yibin (YB) and Wanzhou (WZ)] and a section of the upper Yangtze [Zhuyang (ZY)] were investigated using three mtDNA markers to select one marker instead of genetic evaluation of release. The results of three mtDNA markers revealed basically the same, indicating that the level of genetic diversity in rock carp was low, and there was significant genetic differentiation between the ZY and YB. Except for Cyt b–labeled YZ (0.81) and D-loop–labeled WZ (0.59), most of the haplotypic diversity values (h) were below 0.5, the nucleotide diversity values (π) of each group were lower than 0.5 × 10-2, and the haplotype number of rock carp is 1 to 4. Among the three mtDNA markers selected, D-loop marker detected higher diversity, more haplotypes, and private haplotypes, and significant differences between the YB and WZ. The results in this study pointed out the importance of pre-release genetic evaluation and the urgency of protecting the genetic diversity of rock carp, and the D-loop marker was preferentially selected in the pre-release genetic evaluation of fish. Hatchery release is the main strategy for the recovery of rock carp populations, similar to more than 20 endemic fish species in the upper Yangtze River. This study has guiding significance for the protection and restoration of other endemic fishes in the Yangtze River by hatchery release.
Introduction
Rock carp, Procypris rabaudi, is an endemic and vulnerable Cyprinid fish in the upper Yangtze River (Yang et al., 2009). The fish is demersal, inhabits mainly slow-flowing areas with rocky substrate, and spawns under rocky shelters in rushing streams (Wang et al., 2015). Rock carp had relatively extensive distribution records in the mainstem of the upper Yangtze and its tributaries, such as the Jinsha River, Jialing River, and Minjiang River (Yang et al., 2009; Wang et al., 2015). The upper mainstem and its tributaries are typical canyon rivers with steep gradients, staggered shoals, and rapids (Cheng et al., 2015). Hence, the fish have extensive distribution in the upper Yangtze, and its suitable habitats may be limited. A lot of dams have been or are being constructed in the upper Yangtze, such as 10 cascade dams in the middle and lower reaches of the Jinsha River, which have dramatically degraded the habitats of the fish (Cheng et al., 2015). Combining other anthropogenic effects including the construction of shipping channels, water pollution, and overfishing, wild populations of rock carp have dramatically declined and become extremely rare in its native habitats (Yang et al., 2009; Cheng et al., 2011; Wang et al., 2015). As a result, the fish has been listed as a second-class aquatic animal in the National List of Key Protected Wild Animals in 2021 (Zhang et al., 2022). Furthermore, many endemic fishes in the upper Yangtze River, such as Acipenser dabryanus, Percocypris pingi, and Myxocyprinus asiaticus, have similar threats to rock carp. Population conservation and restoration of these endemic fishes have greatly been concerning.
Hatchery release is one of the major techniques for fish stock enhancement and has been practiced as a major strategy for population conservation and restoration of endemic fishes in the upper Yangtze River (Cheng et al., 2011; Yang and Wei, 2021; Zhang et al., 2022). A total of 2.77 × 107 and 1.65 × 107 endemic fishes were resealed into the upper Yangtze River in replenishment activities in Sichuan province and Chongqing city only in 2019 and 2020, respectively (Fishery Supervision and Administration Office of the Yangtze River Basin, 2019; 2020). Rock carp are the main component of these released endemic fish. For example, at least 2.29 × 107 rock carp were released by replenishment activities in Wanzhou district, Chongqing city, between 2013 and 2017 (Zhang et al., 2022). However, when the release of individuals with substantially different genetic backgrounds contributes to gene flow in wild populations, hatchery release may have potentially adverse effects such as loss of genetic diversity, alteration in genetic structure, and a decrease in effective population size (Cross, 1999; Bert et al., 2001; Grant et al., 2017). To rock carp, Cheng et al. (2011) found lower genetic diversity in hatchery individuals and hybrids with released individuals in the wild population. Zhang et al. (2022) demonstrated the genetic effects of hatchery release, which resulted in a substantial change in the genetic structure of the wild population and the threat of inbreeding decline of this vulnerable fish. Both studies used different microsatellite loci to compare population genetics between wild and hatchery groups of rock carp (Cheng et al., 2011; Zhang et al., 2022). Microsatellite marker can reveal abundant genetic information for analysis, is laborious and time-consuming, and needs a specific primer for loci amplification (Liu and Cordes, 2004; Portnoy and Heist, 2012). Evaluating the genetic makeup of released individuals and their genetic effects on wild counterparts prior to restocking activity is critical to ensuring population conservation and restoration of endemic fishes in the upper Yangtze River, and a molecular marker with simple, rapid, and universal characteristics will be helpful to the evaluation of pre-released.
Mitochondrial (mtDNA) markers are frequently used to reveal genetic diversity and structure, population origins and mixing, and hybrid introgression of fishes (Jackson et al., 2012). This has been attributed to its practical advantages such as simpler operation, universal primer, and quick and comparable results, in part due to faster accumulating of sequence divergence in vertebrate than in nuclear DNA (Brown, 1985; Liu and Cordes, 2004). Common mtDNA markers applied in fish include the cytochrome oxidase I (CO I), cytochrome b (Cyt b), and the control region (D-loop). Among these markers, the D-loop is a non-coding region and exhibits generally higher genetic variations relative to coding regions such as the CO I and Cyt b (Keeney and Heist, 2006; Jackson et al., 2012; Portnoy and Heist, 2012). Furthermore, the genetic diversity and structure of rock carp are relatively clear (Cheng et al., 2011; Song et al., 2014; Zhang et al., 2022). In general, wild populations of the fish showed a moderate level of genetic diversity and no obvious geographical distribution patterns in the Yangtze River (He et al., 2008; Zhu et al., 2008; Zhang et al., 2020); hatchery-raised individuals showed lower genetic diversity and significant genetic differentiation with wild populations and among different hatcheries (Cheng et al., 2011; Yue et al., 2021). In addition, impacts of hatchery release on the genetic makeup of wild rock carp in the upper Yangtze have already been demonstrated (Cheng et al., 2011; Zhang et al., 2022).
Hatchery releases have not always achieved the anticipated population conservation and restoration of endemic fishes in the Yangtze River (Wei, 2020; Xie, 2020; Yang and Wei, 2021). One reason may be that many individuals in released fishes are genetically inferior to wild individuals (Cheng et al., 2011; Grant et al., 2017). A molecular marker with practical advantages will be helpful to evaluate rapidly genetic makeup of released individuals before restocking activity which is important to improve the restocking plan and to predict genetic effects on wild counterparts. In this study, samples of rock carp were collected from representative hatcheries and the upper reaches of the Yangtze River, and the genetic diversity and structure of sampling groups were investigated using three mtDNA markers. The aim of this study was to prioritize mtDNA markers for pre-released genetic evaluation of rock carp, which would also be helpful to hatchery release protect the population conservation and restoration of other endemic fishes in the upper Yangtze River.
Materials and Methods
A total of 21 adult individuals as a wild population (ZY) were collected from fishers located in the Zhuyang section of the upper Yangtze River in 2020 (Figure 1). ZY section is a main habitat and spawning ground of the fish and is an important part of the National Natural Reserve Areas of Rare and Special Fishes in the upper reaches of the Yangtze River (Cheng et al., 2011). Two released groups of the fish were sampled from the Yibin hatchery of Sichuan province (YB) and Wanzhou hatchery of Chongqing city (WZ) in 2020. Fin specimens were collected from 30 adult individuals for each hatchery. The Yibin hatchery collected the original broodstock of rock carp from the Zhuyang section in the 1990s and produced the majority of released rock carp for Sichuan province, China (Cheng et al., 2011). The Wanzhou hatchery is a major supplied hatchery for released activities of Chongqing city; however, the information on the original broodstock of the Wanzhou hatchery is unknown (Zhang et al., 2022). All specimens were collected and fixed immediately with 95% ethanol and stored at −20°C in the laboratory.
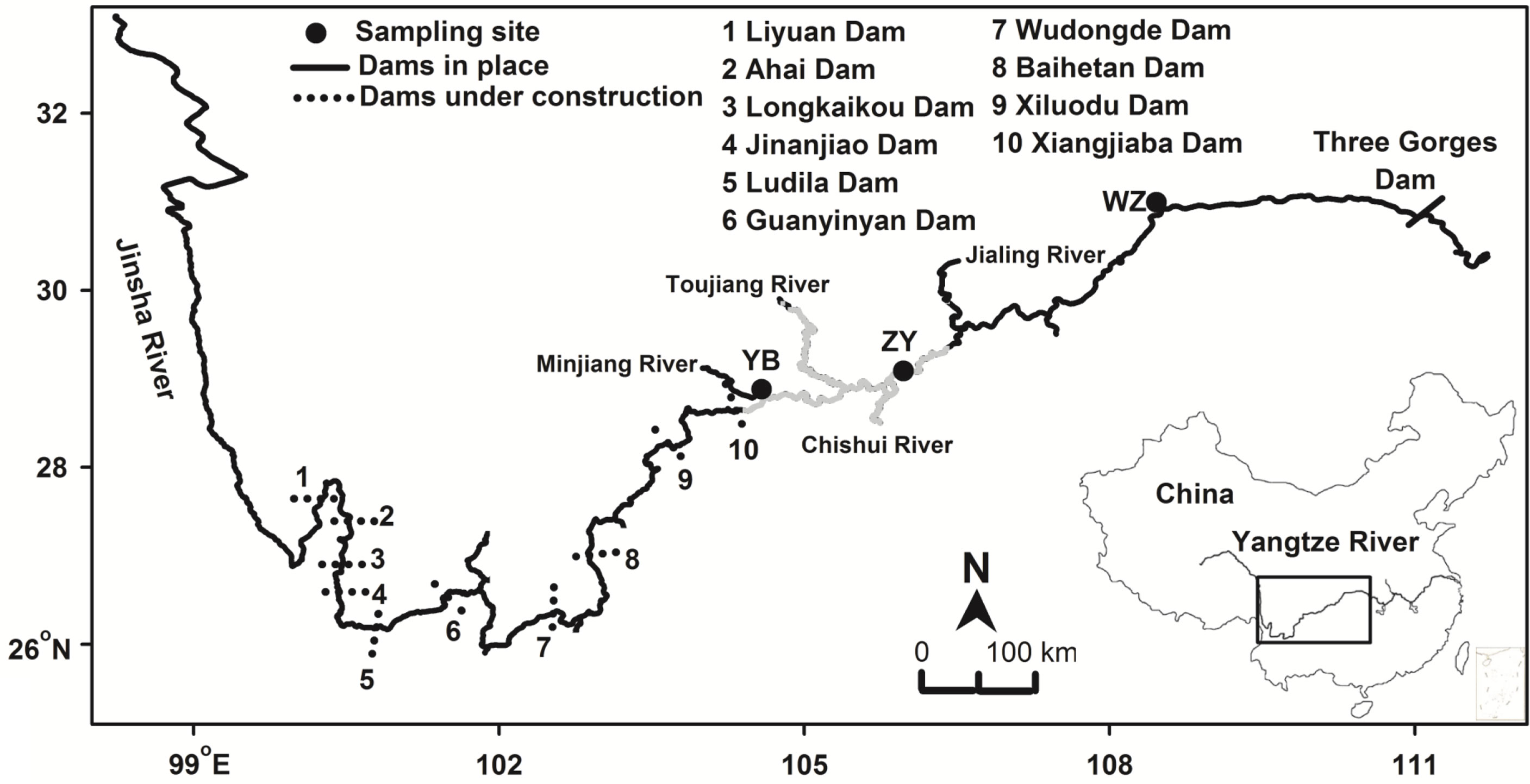
Figure 1 Sampling sites of Procypris rabaudi for the wild population (ZY), Yibin hatchery (YB), and Wanzhou hatchery (WZ) in the upper Yangtze River. Main section of the National Natural Reserve Areas of Rare and Special Fishes of the Upper Yangtze River is shown using gray, and main tributaries, the Three Gorges Dam, and 10 cascade dams in the middle and lower reaches of the Jinsha River are located.
Total DNA was extracted using an animal genomic DNA extraction kit (Sangon, Shanghai, China). DNA quality and concentration were determined by agarose gel electrophoresis and spectrophotometer (Eppendorf, Germany). For partial sequences of the mtDNA CO I marker, universal primers were FishF1 and FishR1, and amplification was performed following Ward et al. (2005). For partial sequences of the mtDNA Cyt b marker, universal primers were L14724 and H15915, and amplification was performed following Xiao et al. (2001). For partial sequences of the mtDNA D-loop marker, universal primers were MitD1-F and MitD1-R, and amplification was performed following Dong et al. (2014). Products of polymerase chain reaction (PCR) products were detected by agarose gel electrophoresis and purified and sequenced by a commercial sequencing company (BGI, Shenzhen, China).The Clustal X software (version 1.83) was used to edit and align sequences of the three markers (Thompson et al., 1997). The MEGA 5.0 software translated sequences into amino acids to check for sequencing errors and pseudogene presences for the mtDNA CO I and Cyt b markers (Tamura et al., 2011). The DnaSP v5.1 software calculated standard genetic diversity indices for sequences of the three mtDNA markers such as the haplotype number (k), haplotypic diversity (h), average pairwise sequence differences (П), and nucleotide diversity (π) (Librado and Rozas, 2009). The ARLEQUIN v3.1 software estimated genetic differentiation among the three sampling groups through mean pairwise differences (NST) for each marker (Excoffier et al., 2005), and p < 0.05 was used to screen out the significant differences between different populations. The TCS v1.21 software constructed a minimum-spanning haplotype network to evaluate relationships between sampling groups and the distribution of haplotypes for each marker using a statistical parsimony procedure with 95% connection limits (Clement et al., 2000). The DnaSP v5.1 software also evaluated the historical dynamics of the population through the mismatch distribution analysis and implemented Tajima’s D test and Fu’s Fs statistic for the neutrality test in the wild population. The ARLEQUIN v3.1 software assessed the goodness of fit by the raggedness index (r) and the sum of square deviations (SSD) between the observed and expected mismatch with 1,000 parametric bootstrap replicates for the wild population.
Results
Partial sequences of the mtDNA CO I, Cyt b, and D-loop markers were extracted from about 900, 997, and 637 base pairs for furthermore analysis, respectively. No indels, sequencing errors, and pseudogenes were detected from the CO I and Cyt b sequences. The D-loop sequence showed relatively higher variations and diversities than the CO I and Cyt b sequences. Haplotypes were detected in four (GenBank: 2562447) from the CO I sequences, five (GenBank: 2563182) from the Cyt b sequences, and six (GenBank: 2562496) from the D-loop sequences. Variable sites, parsimony informative sites, and G+C content were 7, 5, and 45.0% in the CO I sequence; 6, 4, and 41.7% in the Cyt b sequences; and 22, 8, and 27.8% in the D-loop sequences, respectively. Haplotypic and nucleotide diversity indices were 0.31 and 0.16 × 10−2 in the CO I sequence, 0.37 and 0.79 × 10−3 in the Cyt b sequences, and 0.39 and 0.35 × 10−2 in the D-loop sequences, respectively.
Among the sampling groups, the k, h, П, and π values of the CO I sequences were lowest in the YB and similar between the ZY and the WZ and ranged from 1 to 4, 0.00 to 0.47, 0.00 to 2.00, and 0.00 to 0.22 × 10−2, respectively (Table 1). The diversity indices also were lowest in the YB and highest in the ZY, and the k, h, П, and π values ranged from 1 to 4, 0.00 to 0.81, 0.00 to 1.90, and 0.00 to 0.19 × 10−2, respectively (Table 1). However, the genetic diversity indices of the D-loop regions were relatively higher than the other two markers’ sequences in all groups and ranged from 2 to 4, 0.11 to 0.59, 1.26 to 2.67, and 0.20 × 10−2 to 0.42 × 10−2, respectively (Table 1).
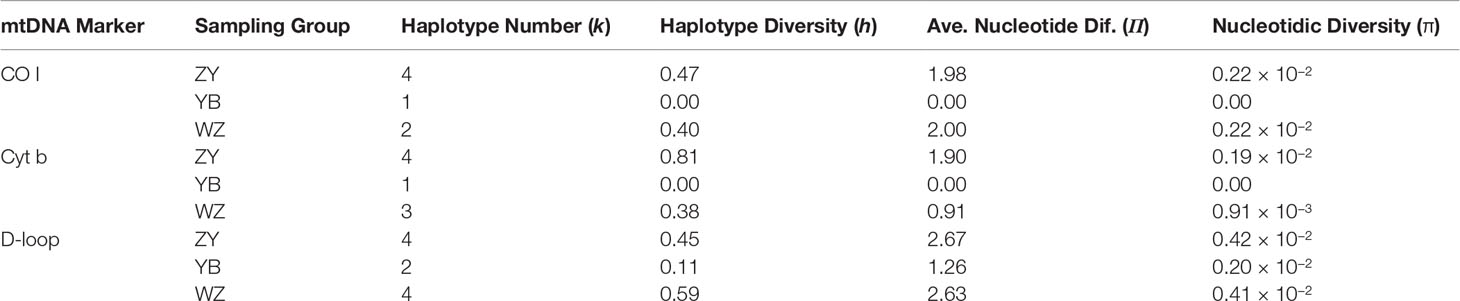
Table 1 Genetic diversity indices for partial sequences of the mtDNA CO I, Cyt b, and D-loop markers in the wild population (ZY), Yibin hatchery (YB), and Wanzhou hatchery (WZ) of Procypris rabaudi.
Significant genetic differences were detected between the YB and other groups and not found between the ZY and WZ by the three mtDNA markers (p < 0.05) (Table 2). The D-loop marker also detected significant genetic differences between the YB and WZ (p < 0.05) (Table 2). The minimum-spanning haplotype network of the three mtDNA markers indicated a segregating trend between the wild population and hatchery groups (Figure 2). Among four haplotypes of rock carp detected by the CO I marker, the ZY processed two private (Hap 2 and Hap 4) and shared Hap 3 with the WZ and Hap 1 with the YB and WZ, and similar haplotype networks among the three sampling groups are also detected by the Cyt b and D-loop markers (Figure 2). Furthermore, the D-loop marker detected more private haplotypes among the groups than the other two markers, which showed the three groups had, respectively, one private haplotype (Figure 2).
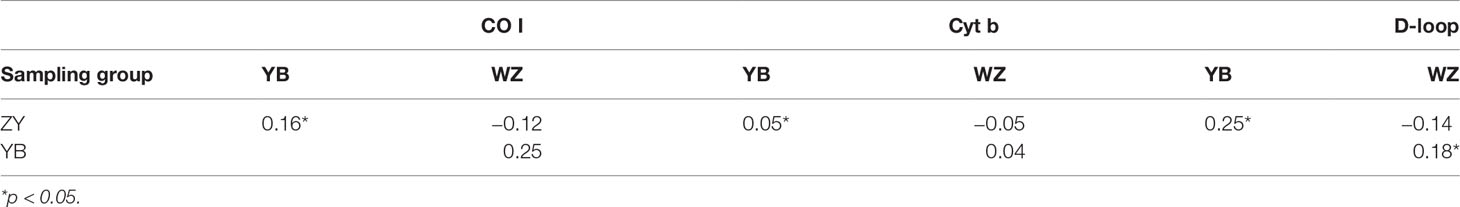
Table 2 Pairwise differences (NST) among the wild population (ZY), Yibin hatchery (YB), and Wanzhou hatchery (WZ) of Procypris rabaudi using partial sequences of the mtDNA CO I, Cyt b, and D-loop markers.
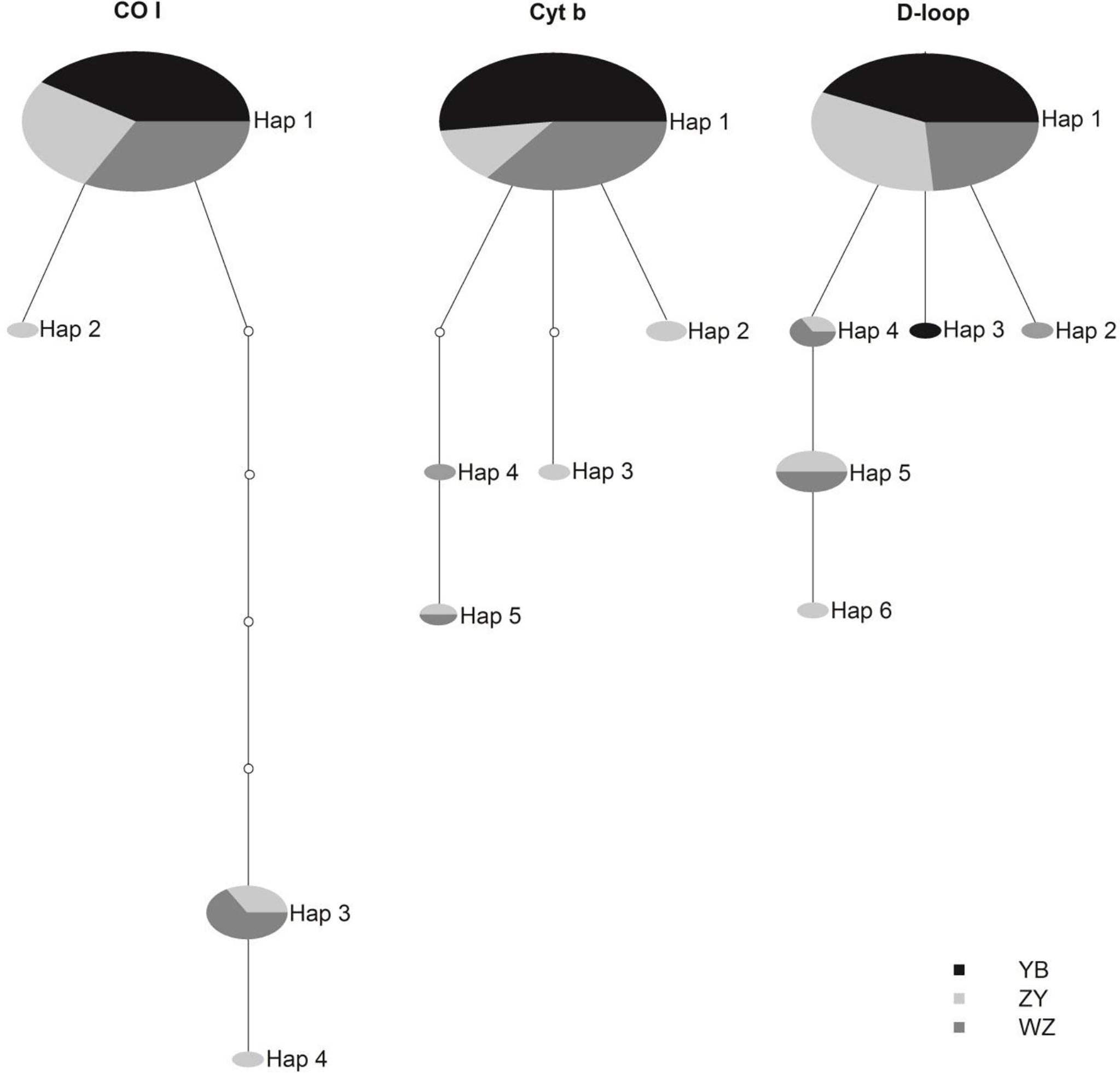
Figure 2 Statistical parsimony network based on haplotype frequencies of the mtDNA CO I, Cyt b, and D-loop markers among the wild population (ZY), Yibin hatchery (YB), and Wanzhou hatchery (WZ) of Procypris rabaudi.
Similar results in historical dynamics of the wild population were revealed by the three mtDNA markers, which failed to reject the raw hypothesis of population inflation and neutrality theory. The mismatch distribution analysis showed multimodal distribution (Figure 3). The goodness of fit and neutrality tests did not detect a significant departure from the raw hypothesis, except for the mtDNA CO I and D-loop–labeled markers SSDs (p < 0.05) (Table 3).
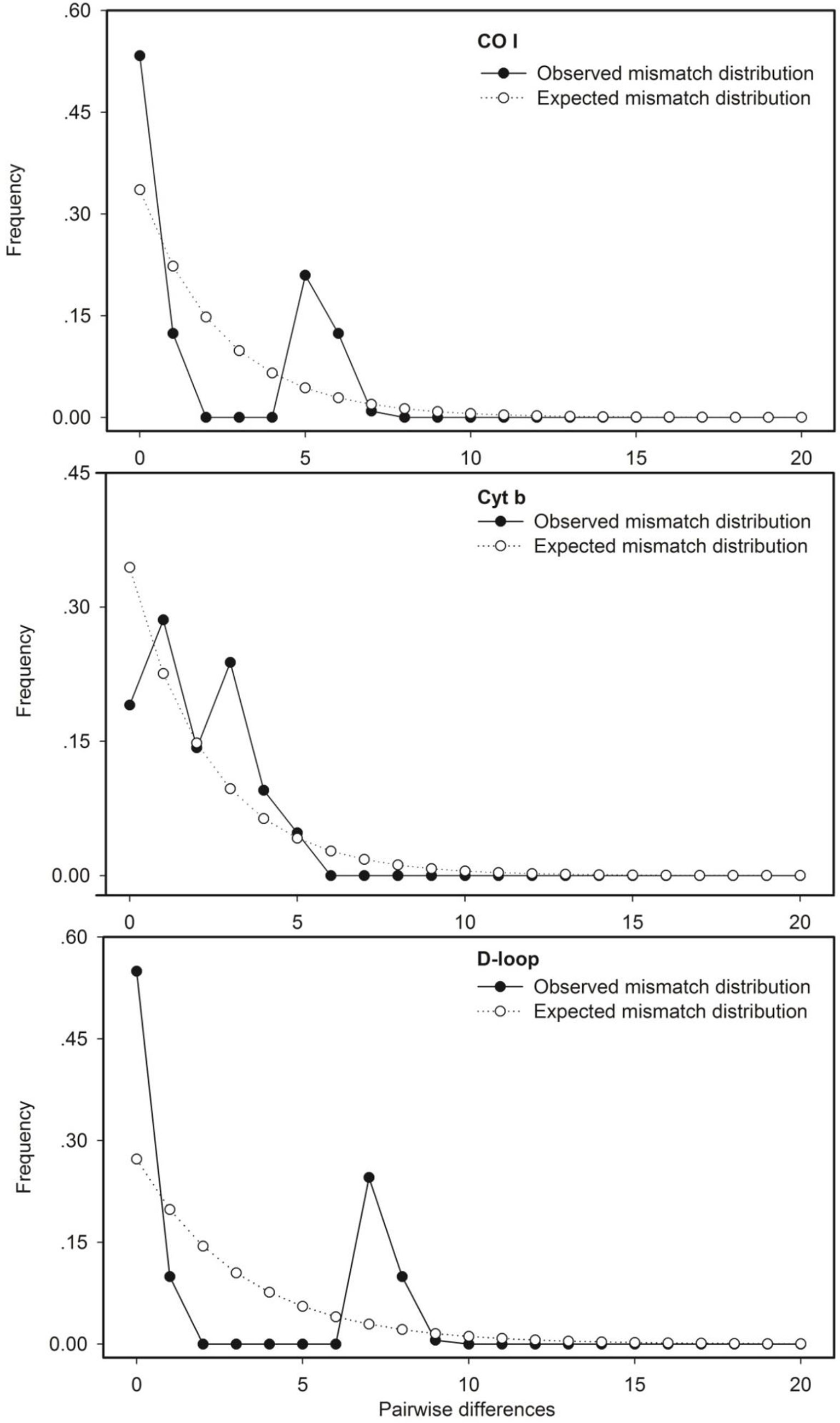
Figure 3 Mismatch distribution of partial sequences of the mtDNA CO I, Cyt b, and D-loop markers for the wild population of Procypris rabaudi.

Table 3 Goodness of fit and neutrality tests for partial sequences of the mtDNA CO I, Cyt b, and D-loop markers in the wild population of Procypris rabaudi.
Discussion
The three mtDNA markers revealed consistent results and indicated a low level of genetic diversity in rock carp and significant genetic differentiation between the wild population and the YB hatchery group. These results pointed out the importance of genetic evaluation of pre-released and the urgency in protecting the genetic diversity of rock carp. Among the three mtDNA markers, the D-loop marker showed higher genetic variations and more effective detection in genetic makeups among different groups of rock carp, which may be preferred in pre-released genetic evaluation for the fish. Hatchery release is a major strategy for population conservation and restoration of rock carp, similar to over 20 endemic fishes in the upper Yangtze River (Yin and Zhang, 2008). Application of pre-released genetic evaluation is suggested for protecting the genetic diversity of rock carp and avoiding genetic risks of hatchery-raised fish released based on our results. This study also is instructive for hatchery release for population conservation and restoration of other endemic fishes in the upper Yangtze River.
A low level of genetic diversity in rock carp was revealed by the mtDNA markers, which may be related to the genetic impacts of released hatchery-raised individuals. Most of the h values were below 0.5, except for the YZ (0.81) by the Cyt b marker and the WZ (0.59) by the D-loop marker; the π values of all groups were below 0.5 × 10−2, and the k values ranged from 1 to 4 in rock carp. All these results showed low genetic diversity, which was consistent with the recent results of rock carp (Zhang et al., 2020; Yue et al., 2021). The h and π values of the D-loop marker were 0.41 and 0.13 × 10−2 and 0.40 and 0.13 × 10−2 for two released hatcheries of Chongqing city (Yue et al., 2021), and the h and π values of the Cyt b marker were 0.63 and 0.14 × 10−2 and 0.46 and 0.10 × 10−2 for wild populations from the Wanzhou section and the Chishui River of the upper Yangtze, respectively (Zhang et al., 2020). Compared with earlier results of wild populations, our results showed a sharp decrease in genetic diversity in the last decade with the implementation of the mass-scale release of hatchery fish. The h and π values of the D-loop marker ranged from 0.60 to 0.97 with a mean of 0.86 and from 0.25 × 10−2 to 1.60 × 10−2 with a mean of 0.79 × 10−2 for eight wild populations collected from 2002 to 2006 (Song et al., 2014). Research studies before 2010 also showed high genetic diversity of rock carp, such as three wild populations with average h = 0.86 using the D-loop marker and two wild populations with average h = 0.88 using the Cyt b marker (He et al., 2008; Zhu et al., 2008). The research using microsatellite markers had identified the negative effects of hatchery release on the genetic diversity and structure of rock carp (Cheng et al., 2011; Zhang et al., 2022). Combined with other research, our results may be indicated that hatchery release is likely to important reason for the sharp decrease in genetic diversity of rock carp, which is consistent with the reference of Zhang et al. (2020). Genetic diversity is closely associated with population adaptation and fitness and is important to the long-term persistence of a species (Schindler et al., 2010; Grant et al., 2017). Thus, our results pointed out the importance of pre-released genetic evaluation and the urgency of protecting the genetic diversity of rock carp.
Significant genetic differences were detected between the ZY and YB, which were consistent with the results using microsatellite markers and cautioned the genetic impacts of released hatchery-raised rock carp (Cheng et al., 2011). The observed segregating trend of the haplotype network between the wild population and hatchery groups also supported the results of population differentiation. Previous research showed that no genetic differentiation and obvious geographical distribution patterns were found in wild populations of the fish using the mtDNA markers (He et al., 2008; Song et al., 2014; Zhang et al., 2020). However, recent research demonstrated genetic differentiation between the Wanzhou population and other natural populations due to long-term released activities of hatchery-raised rock carp in Wanzhou, Chongqing city (Zhang et al., 2022). Furthermore, the YB, the same broodstock in Cheng et al. (2011), showed extremely low genetic diversity and dramatically homogenizing of genetic variability in the last decade. Broodstock is generally small for economic efficiency to reduce the costs in hatchery practices, which inevitably produces a cohort of individuals that differs genetically from the wild population (Araki et al., 2007; Araki et al., 2008; Grant et al., 2017). Hybridizations with released hatchery-raised individuals like the YB have the potential to lower the fitness of wild counterparts. In addition, our results in historical dynamics indicated that the wild population complied with the neutrality theory and held relatively stable. Protecting the genetic resources of wild populations is an important mission in implementing stock restoration (Teletchea and Fontaine, 2014; Grant et al., 2017). Combined with this research, our results underscored the importance of broodstock management in the released hatchery. Broodstock management, especially effective broodstock size for rock carp, is central to successful hatchery release and then stock restoration (Grant et al., 2017). The Ryman–Laikre model with its extensions can be used to estimate appropriate broodstock size (Ryman and Laikre, 1991; Waples et al., 2016), which should be applied in broodstock management of released hatcheries, especially in the YB for rock carp. A careful evaluation of the genetic makeup of released individuals before restocking activity is needed to avoid the genetic impacts of released hatchery-raised rock carp.
The D-loop marker results revealed higher genetic variations and more effective detection of the genetic makeups of different groups of rock carp, which may be preferred in pre-release genetic assessment for the fish. Compared with other mtDNA markers, the D-loop marker detected higher diversity, more haplotypes and private haplotypes, and significant differences between the YB and WZ. As the non-coding region in mtDNA, the D-loop marker generally exhibits higher genetic variations and the fastest evolution rate relative to coding regions such as the CO I and Cyt b markers (Keeney and Heist, 2006; Jackson et al., 2012; Portnoy and Heist, 2012). In addition, the D-loop marker was also the most widely used molecular marker in rock carp and had comprehensively depicted the genetic background of two hatchery groups and all wild populations across the natural range of the fish (Song et al., 2014; Zhang et al., 2020; Yue et al., 2021). It is impossible to implement a risk-free program for population conservation and restoration (Waples and Drake, 2004). On the basis of our results, pre-released genetic evaluation is proposed for protecting the genetic diversity of rock carp and for avoiding the genetic risks of released hatchery-raised individuals. The D-loop marker with its practical advantages may be preferred in the evaluation for rock carp.
Conclusion
Rock carp (Procypris rabaudi) is included in the National Key Protected Wildlife in 2021 as a second-class aquatic animal. Hatchery release is the main strategy for the recovery of rock carp populations. In this study, we presented that the D-loop marker was preferentially selected in the pre-release genetic evaluation from three mtDNA markers; the D-loop marker detected higher diversity, more haplotypes, and private haplotypes, and significant differences between the two hatcheries: YB and WZ. This study pointed out the importance of pre-release genetic evaluation and the urgency of protecting the genetic diversity of rock carp. Hatchery release has been practiced for more than 20 endemic fishes including rock carp, and the results of this study are also instructive to hatchery release for population conservation and restoration of other endemic fishes in the upper Yangtze River.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material.
Ethics Statement
The animal study was reviewed and approved by Institutional Animal Care and Use Committee(IACUC) of Southwest University. Written informed consent was obtained from the owners for the participation of their animals in this study.
Author Contributions
WH, methodology, genetic analysis and writing-original draft. ZD performed the field and laboratory work. TM, performed the field and laboratory work. HY, performed the field and laboratory work. ZC, performed the field and laboratory work. FC, conceptualization, supervision, writing-review & editing. All authors contributed to the article and approved the submitted version.
Funding
This research was supported by the Fundamental Research Funds for the Central Universities (XDJK2019C025), the National Natural Science Foundation of Chongqing (cstc2020jcyj-msxmX0438), and the National Natural Science Foundation of China (32071651).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher
References
Araki H., Berejikian B. A., Ford M. J., Blouin M. S. (2008). Fitness of Hatchery-Reared Salmonids in the Wild. Evol. Appl. 1, 342–355. doi: 10.1111/j.1752-4571.2008.00026.x
Araki H., Cooper B., Blouin M. S. (2007). Genetic Effects of Captive Breeding Cause a Rapid, Cumulative Fitness Decline in the Wild. Science 318, 100–103. doi: 10.1126/science.1145621
Bulletin on Hydrobiological Resources and Habitat in the Yangtze River Basin. (2020) Fishery Supervision and Administration Office of the Yangtze River Basin. http://www.cjyzbgs.moa.gov.cn/.
Bert T. M., Tringali M. D.and , Baker J. (2001).“Considerations for Sustainable Aquaculture, Biodiversity, and Ecosystem Processes a Genetics Perspective, ” in: Promoting Global Innovation of Agricultural Science and Technology and Sustainable Agriculture DevelopmentEd. ICAST Organizing Committee (Beijing: Proceedings of the International Conference on Agriculture Science and Technology), –, 238254
Brown W. M. (1985). “The Mitochondrial Genome of Animals, ” in Molecular Evolutionary Genetics. Ed. MacIntyre R. J., 95–130. Plenum. (New York: Springer), 95–130.
Cheng F., Li W., Castello L., Murphy B. R., Xie S. G. (2015). Potential Effects of Dam Cascade on Fish: Lessons From the Yangtze River. Rev. Fish Biol. Fisher. 25, 569–585. doi: 10.1007/s11160-015-9395-9
Cheng F., Wu Q. J., Liu M., Radhakrishnan K. V., Murphy B. R., Xie S. G. (2011). Impacts of Hatchery Release on Genetic Structure of Rock Carp Procypris Rabaudi in the Upper Yangtze River, China. Fish. Sci. 77, 765–771. doi: 10.1007/s12562-011-0383-2
Clement M., Posada D., Crandall K. A. (2000). TCS: A Computer Program to Estimate Gene Genealogies. Mol. Ecol. 9, 1657–1659. doi: 10.1046/j.1365-294x.2000.01020.x
Cross T. F. (1999). “Genetic Considerations in Enhancement and Ranching of Marine and Anadromous Species,” in Stock Enhancement and Sea Ranching. Ed. M Kenneth, Kitada S., Blankenship H. L., Svasand T. (Oxford: Blackwell), 37–48.
Dong X. P., Mu S. M., Zhou N., Kang X. J., Lou Q., Bai J. J. (2014). Structure Analysis of mtDNA D-Loop Region and the Genetic Diversity of Chanan Argu in Different Populations. J. Fish. China 38 (9), 1277–1285. doi: 10.3724/SP.J. 1231.2014.49331
Excoffier L., Laval G., Schneider S. (2005). Arlequin Ver. 3.0: An Integrated Software Package for Population Genetics Data Analysis. Evol. Bioinform. 1, 47–50. doi: 10.1177/117693430500100003
Grant W. S., Jasper J., Bekkevold D., Adkison M. (2017). Responsible Genetic Approach to Stock Restoration, Sea Ranching and Stock Enhancement of Marine Fishes and Invertebrates. Rev. Fish Biol. Fish. 27, 615–649. doi: 10.1007/s11160-017-9489-7
He Y. F., Tang Q. Y., Wang J. W., Liu H. Z., Tan D. Q. (2008). Population Differentiation of Procypris Rabaudi (Tchang) in the Upper Reaches of the Yangtze River Revealed by Mitochondrial DNA and RAPD Markers. Genes Genom. 30, 223–233. Available from https://www.researchgate.net/publication/259782818
Jackson A. J., Laikre L., Baker C. S., Kendall C. K. (2012). Guidelines for Collecting and Maintaining Archives for Genetic Monitoring. Conserv. Genet. Res. 4 (2), 527–536. doi: 10.1007/s12686-011-9545-x
Keeney D. B., Heist E. J. (2006). Worldwide Phylogeography of the Blacktip Shark (Carcharhinus Limbatus) Inferred From Mitochondrial DNA Reveals Isolation of Western Atlantic Populations Coupled With Recent Pacific Dispersal. Mol. Eco. 15, 3669–3679. doi: 10.1111/j.1365-294X.2006.03036.x
Librado P., Rozas J. (2009). DnaSP V5: A Software for Comprehensive Analysis of DNA Polymorphism Data. Bioinformatics 25 (11), 1451–1452. doi: 10.1093/bioinformatics/btp187
Liu Z. J., Cordes J. F. (2004). DNA Marker Technologies and Their Applications in Aquaculture Genetics. Aquaculture 238, 1–37. doi: 10.1016/j.aquaculture.2004.05.027
Portnoy D. S., Heist E. J. (2012). Molecular Markers: Progress and Prospects for Understanding Reproductive Ecology in Elasmobranchs. J. Fish Biol. 80, 1120–1140. doi: 10.1111/j.1095-8649.2011.03206.x
Ryman N., Laikre L. (1991). Effects of Supportive Breeding on the Genetically Effective Population Size. Conserv. Biol. 5, 325–329. doi: 10.1111/j.1523-1739.1991.tb00144.x
Schindler D., Hilborn R., Chasco B., Boatright C., Quinn T., Rogers L., et al. (2010). Population Diversity and the Portfolio Effect in an Exploited Species. Nature 465, 609–613. doi: 10.1038/nature09060
Song J., Hou F. X., Zhang X. Y., Yue B. S., Song Z. B. (2014). Mitochondrial Genetic Diversity and Population Structure of a Vulnerable Freshwater Fish, Rock Carp (Procypris Rabaudi) in Upper Yangtze River Drainage. Biochem. Syst. Ecol. 55, 1–9. doi: 10.1016/j.bse.2014.02.008
Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. (2011). MEGA5: Molecular Evolutionary Genetics Analysis Using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Mol. Biol. Evol. 28, 2731–2739. doi: 10.1093/molbev/msr121
Teletchea F., Fontaine P. (2014). Levels of Domestication in Fish: Implications for the Sustainable Future of Aquaculture. Fish 15, 181–195. doi: 10.1111/faf.12006
Thompson J. D., Gibson T. J., Plewniak F., Jeanmougin F., Higgins D. G. (1997). The Clustal X Windows Interface: Flexible Strategies for Multiple Sequences Alignment Aided by Analysis Tools. Nucleic Acids Res. 25, 4876–4882. doi: 10.1093/nar/25.24.4876
Wang T., Gao X., Wang J., Jakovlic I., Dan S. G., Liu H. Z. (2015). Life History Traits and Implications for Conservation of Rock Carp Procypris Rabaudi Tchang, an Endemic Fish in the Upper Yangtze River, China. Fish. Sci. 81, 515–523. doi: 10.1007/s12562-015-0872-9
Waples R. S., Drake J. 2004”Risk-benefit Consideration for Marine Stock Enhancement: a Pacific Salmon Perspective,”in Stock Enhancement and Sea Ranchinged Leber K. M., Kitada S., Blankenship H. L.and , Svasand T. (Oxford Blackwell)160–306
Ward R. D., Zemlak T. S., Bronwyn H., Last P. R., Hebert P. D. N. (2005). DNA Barcoding Australia’s Fish Species. Philos. Trans. R. Soc Lond. B. Biol. Sci. 360, 1847–1857. doi: 10.1098/rstb.2005.1716
Wei Q. W. (2020). Conservation of Chinese Sturgeon (Acipenser Sinensis) Based on its Life History: Dilemma and Breakthrough. J. Lake Sci. 32 (5), 1297–1319. doi: 10.18307/2020.0509
Xiao W. H., Zhang Y. P., Liu H. Z. (2001). Molecular Systematics of Xenocyprinae (Teleostei: Cyprinidae): Taxonomy, Biogeography, and Coevolution of a Special Group Restricted in East Asia. Mol. Phylogenet. Evol. 18, 163–173. doi: 10.1006/mpev.2000.0879
Yang L., Mayden R. L., Cai Y. (2009). Threatened Fishes of the World: Procypris Rabaudi (Tchang 1930) (Cyprinidae). Environ. Biol. Fish. 84, 275–276. doi: 10.1007/s10641-008-9417-8
Yang H. L., Wei Q. W. (2021). Suppressive and Active Protective Actions in Aquatic Wildlife Conservation. J. Lake Sci. 33 (1), 1–10. doi: 10.18307/2021.0102
Yin Z. Q., Zhang S. Y. (2008). Thoughts About Released and Proliferation of Fisheries Resources in China. Fish Sin. 3, 9–11. doi: 10.3969/j.issn.1002-6681.2008.03.009
Yue H. M., Ruan R., Cao H., Zhou L., Jiang W., Li S., et al. (2021). Analysis on Genetic Diversity of Cultured Rock Carp (Procypris Rabaudi) From Chongqing City Based on the mtDNA D-Loop Sequences. Acta Hydrobiol. Sin. 45 (4), 846–850. doi: 10.7541/2021.2020.082
Zhang X. Y., Ouyang M., Zhang F. T., Wang J. W. (2022). Study on the Genetic Structure of Wild and Hatchery Populations of Procypris Rabaudi Tchang, an Endemic Fish in the Upper Yangtze River. Fish Res. 245, 106134. doi: 10.1016/j.fishres.2021.106134
Zhang X. Y., Zhang F. T., Yao F. C., Wang J. W. (2020). Study on Genetic Diversity and Population Historical Dynamics of Procypris Rabaudi (Tchang) Endemic in the Upper Yangtze River. Acta Hydrobiol. Sin. 44, 330–338. doi: 10.7541/2020.040
Zhu C. K., Zhang Q. Z., Juan Y., Luo F. (2008). Analysis of Genetic Diversity of the Mitochondrial DNA D-Loop Control Region of Rock Carp (Procypris Rabaudi (Tchang)) in the Three Gorges Reservoir. J. Southwest Univ. 30, 126–133. doi: 10.13718/j.cnki.xdzk.2008.12.012
Keywords: Cyt b, CO I, D-loop, hatchery release, population differentiation, rock carp
Citation: He W, Dong Z, Ma T, Yan H, Chen Z, Yao W and Cheng F (2022) Three Mitochondrial Markers Reveal Genetic Diversity and Structure of Rock Carp (Procypris rabaudi) Endemic to the Upper Yangtze: Implications for Pre-release Genetic Assessment. Front. Mar. Sci. 9:939745. doi: 10.3389/fmars.2022.939745
Received: 09 May 2022; Accepted: 06 June 2022;
Published: 13 July 2022.
Edited by:
Pengfei Cheng, Ningbo University, ChinaReviewed by:
Shengjie Ren, Queensland University of Technology, AustraliaZhenhua Ma, Chinese Academy of Fishery Sciences (CAFS), China
Copyright © 2022 He, Dong, Ma, Yan, Chen, Yao and Cheng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Fei Cheng, Y2hlbmdmZWlAaWhiLmFjLmNu
 Wenping He
Wenping He Zhiling Dong1
Zhiling Dong1