
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mar. Sci. , 15 July 2022
Sec. Marine Biotechnology and Bioproducts
Volume 9 - 2022 | https://doi.org/10.3389/fmars.2022.939716
This article is part of the Research Topic Sustainable Production of Marine Natural Products - From Discovery to Application View all 9 articles
 Shitong Liang1†
Shitong Liang1† Xuewei Yang1*†
Xuewei Yang1*† Xingyu Zhu1
Xingyu Zhu1 Muhammad Ibrar1
Muhammad Ibrar1 Liangxu Liu1
Liangxu Liu1 Siting Li1,2
Siting Li1,2 Xia Li3
Xia Li3 Tian Tian4
Tian Tian4 Shuangfei Li1*
Shuangfei Li1*Docosahexaenoic acid (DHA) has attracted attention from researchers because of its pharmacological and nutritional importance. Currently, DHA production costs are high due to fermentation inefficiency; however, improving DHA yield by metabolic engineering in thraustochytrids is one approach to reduce these costs. In this study, a high-yielding (53.97% of total fatty acids) DHA production strain was constructed by disrupting polyunsaturated fatty acid beta-oxidation via knockout of the 2,4-dienyl-CoA reductase (DECR) gene (KO strain) in Aurantiochytrium sp. Slight differences in cell growth was observed in the wild-type and transformants (OE and KO), with cell concentrations in stationary of 2.65×106, 2.36×106 and 2.56×106 cells mL-1 respectively. Impressively, the KO strain yielded 21.62% more neutral lipids and 57.34% greater DHA production; moreover, the opposite was observed when overexpressing DECR (OE strain), with significant decreases of 30.49% and 64.61%, respectively. Furthermore, the KO strain showed a prolonged DHA production period with a sustainable increase from 63 to 90 h (170.03 to 203.27 mg g−1 DCW), while that of the wildtype strain decreased significantly from 150.58 to 140.10 mg g−1 DCW. This new approach provides an advanced proxy for the construction of sustainable DHA production strains for industrial purposes and deepens our understanding of the metabolic pathways of Aurantiochytrium sp.
Docosahexaenoic acid (DHA, C22:6) is an omega-3 polyunsaturated fatty acid (PUFA) that exists in a wide range of organisms, from unicellular microorganisms to mammals (Abedi and Sahari, 2014; Snetselaar et al., 2021). Recently, DHA has garnered attention because of its pharmacological and nutritional activities, such as enhancing brain and retina development (Bazan, 2005) and ability to reduce hypertension (Boyer-Diaz et al., 2020), atherosclerosis (Liu et al., 2016), schizophrenia (Harper et al., 2011), hypertriglyceridemia (Kelley et al., 2009), and cancer risk (Newell et al., 2017). Industrial-scale DHA production uses fish oils, although the primary producers are mainly thraustochytrids and microalgae (Raghukumar, 2008; Gupta et al., 2012). Overfishing and global warming are causing a decline in fish populations, which will result in a disturbance to the DHA demand and supply flow chain. The fatty acid profiles of microalgae are stable and vegetarian algal oil has excellent bioequivalence and safety, is a suitable alternative for DHA production (Arterburn et al., 2007; Rahmawaty et al., 2013). The omega-3 PUFA market was valued at approximately 4.3 billion USD in 2019 (Aasen et al., 2016); however, a gradual increase in water temperature due to climate change may result in a loss of up to 58% of globally available DHA by 2100 (Colombo et al., 2020). Thus, the development of a sustainable production method for DHA via genetically engineered marine microorganisms is crucial.
Marine protists are regarded as one of the most promising sources for omega-3 PUFAs, including DHA and eicosapentaenoic acid (EPA) (Abdel-Mawgoud and Stephanopoulos, 2018; Xu et al., 2020), owing to their high growth rate, excellent production density, and minimal occupation of farmland. Strains such as Schizochytrium mangrovei SM3 (Pleissner et al., 2013), Thraustochytrium sp. ATCC 20889 (Jiang et al., 2004), and Ulkenia TC 010 (Chang et al., 2012) have shown great potential, with contents of 24.00%, 26.00%, and 37.50% DHA of the total fatty acids (TFAs), respectively (Fossier Marchan et al., 2018; Morabito et al., 2019). Aurantiochytrium sp. is a promising DHA-producing candidate among microalgae, accumulating up to 54.0% DHA of total fatty acids (TFAs) (Dellero et al., 2018). In Aurantiochytrium sp., the fatty acid synthase (FAS) route and the polyketide synthase (PKS) pathway are both involved in DHA synthesis (Metz et al., 2001). The FAS process uses a cascade enzymatic reaction of elongase and desaturase to generate fatty acids (Morabito et al., 2019), whereas the PKS pathway follows dehydration and isomerization employing fatty acid acyl intermediates for carbon chain elongation (Gupta et al., 2012). As DHA is a secondary metabolite that can be utilized as a carbon source, the accumulation of DHA in the cell is affected by anabolism and catabolism (beta-oxidation) (Sprecher, 2002). The beta-oxidation of DHA mainly occurs in peroxisome and mitochondria, consisting of four reaction cycles, namely oxidation, hydration, dehydrogenation, and thiolysis (Schulz, 1991). Unlike the beta-oxidation of saturated fatty acids (SFAs), the catabolism of DHA involves an oxidative and reductive reaction on the double bond at position four (Sprecher, 2002). To oxidize DHA, 2,4-dienoyl-CoA reductase (DECR) is necessary for reducing 2-trans-4,7,10,13,16,19-heptaenoyl-CoA, followed by the isomerization of 3,7,10,13,16,19-hexenoyl-CoA (Sprecher, 2002) and 2-trans-7,10,13,16,19-hexenoyl-CoA formation (shown in Figure 1A). This process clearly indicates the crucial role of DECR in DHA beta-oxidation.
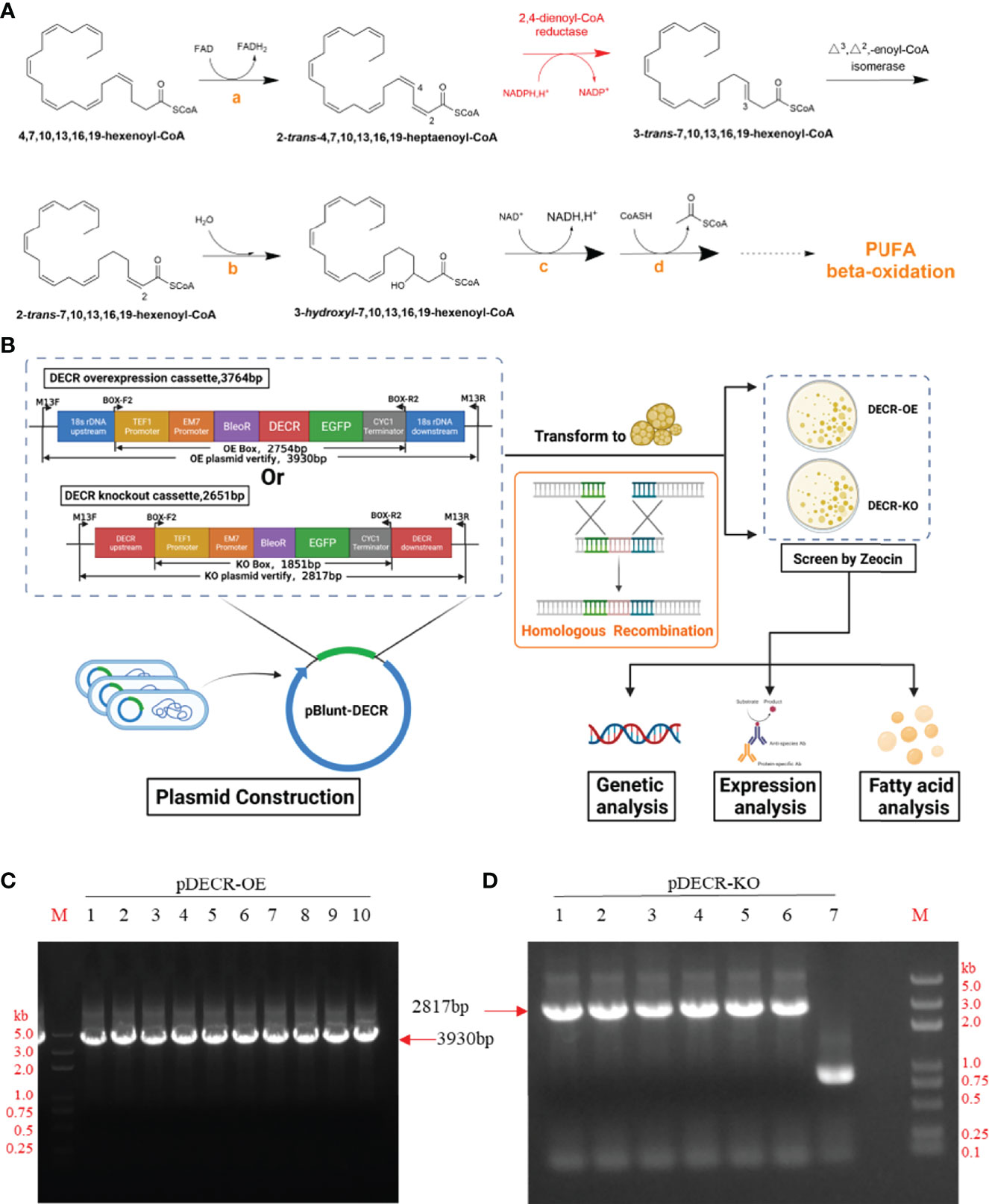
Figure 1 Pathway, approach, and plasmid identification. (A) The pathway of DHA beta-oxidation. The orange letters indicate enzymes involved in standard beta-oxidation: acetyl-CoA dehydrogenase (a), enoyl-CoA hydratase (b), hydroxyacyl-CoA dehydrogenase (c), and beta-ketothiolase (d), respectively. Created with Chemdraw 20.0. (B) Approach to study the effect of DECR on fatty acids. The upstream and downstream homology arms of the overexpression (OE) cassette contain 18s rDNA, those of the knockout (KO) cassette contain DECR. After transformation, the transformants of DECR-OE and DECR-KO were screened with Zeocin and analyzed. Created with BioRender.com. (C) and (D) Identification of the pBlunt plasmid containing DECR overexpression (OE) and knockout (KO) cassettes in E. coli with the primer pair M13F/M13R by PCR, respectively. (C) The 3930 bp band indicates the amplification of the DECR overexpression cassette. Numbers 1 to 10 represent the colony numbers of the OE plasmids in E.coli.(D) The 2817 bp band indicates the amplification of the DECR knockout cassette. Numbers 1 to 7 represent the colony numbers of the corresponding plasmids in E.coli. M, DL5000 DNA Marker.
2,4-Dienoyl-CoA reductase (DECR) is a peroxisomal NADPH coenzyme-dependent oxidoreductase, converting 2,4-dienoyl-CoA into 3-enoyl-CoA (Gurvitz et al., 1997). During the peroxisomal beta-oxidation of arachidonic acid (AA), delta-3,5-delta-2,4-dienoyl-CoA isomerase and 2,4-dienoyl-CoA reductase are required to remove double bonds from odd-numbered carbons (Luthria et al., 1995). At present, DECR has been identified and cloned in mammals (Shirley and Murphy, 1990; Koivuranta et al., 1994; Das et al., 2000), plants (Goepfert et al., 2005), fungus (Dommes et al., 1983; Gurvitz et al., 1997; Mastalski et al., 2020), bacteria (Dommes and Kunau, 1984; He et al., 1997; Ogawa et al., 2020), and parasites (Semini et al., 2020). The bacterium-like peroxisomal 2,4-dienoyl-CoA reductase gene (DECR) mutant of Leishmania spp. could not oxidize unsaturated fatty acids (UFAs) and consequently accumulated the intermediate 2,4-decadienoyl-CoA (Semini et al., 2020). The knockout of bacterial-FadH encoding 2,4-dienoyl-CoA reductase in Shewanella livingstonensis Ac10 reduced the conversion rate of DHA to EPA by 86.00% (Ogawa et al., 2020). DECR was considered to participate in controlling the balance between SFAs and UFAs. The deletion of DECR in LNCaP cells resulted in higher PUFAs, particularly AA and DHA (Blomme et al., 2020). In addition to the enzymes required for saturated fatty acid beta-oxidation, DECR might have an auxiliary effect on the beta-oxidation of DHA (Schulz and Kunau, 1987). Although previous literature on lipid enhancement focused mainly on the disruption of lipid homeostasis through the loss of DECR in mammal animals (Mäkelä et al., 2019; Blomme et al., 2020), there are a lack of data on the regulatory effects of DECR on the PUFA yields in the primary producer thraustochytrids.
To explore a new proxy to improve DHA production yield and efficiency, an engineered strain (KO strain) was constructed by disrupting PUFA beta-oxidation via knocking out 2,4-dienyl-CoA reductase (DECR) gene that has been detected in Aurantiochytrium sp. SZU445 genome (Zhu et al., 2020). To clarify the effects of DECR on the accumulation of PUFAs in Aurantiochytrium sp., the DECR overexpressed (OE) strain was also constructed. The fatty acid profiles and yields (neutral lipids, total FAs, total UFAs, and DHA) in various growth stages were investigated to explore how DECR regulates the flux of PUFA metabolism. This aim of this study was to produce a foundation for the use of genetic engineering to construct an efficient oleaginous protist to achieve high yields of DHA.
Aurantiochytrium sp. SZU445 was obtained by UV mutagenesis of Aurantiochytrium sp. PKU#Mn16 (Liu et al., 2020). The culture conditions were same as those in our previously reported study (Zhu et al., 2020). To measure growth curves, the strain was inoculated in M4 medium at 23°C in a shaker with a speed of 200 rpm (IS-RDS6, CRYSTAL, Suzhou, China). Natural seawater containing 1 g L−1 yeast extract, 15 g L−1 agar, 20 g L−1 glucose, 0.025 g L−1 monopotassium phosphate, 0.3 mg mL−1 zeocin, and 1.5 g L−1 peptone served as the transformant screening medium.
The expression vector pREMI-Z (Addgene ID 59527) containing TEF1 promoter, the BleoR gene, and CYC1 terminator was used in this study (Mukaiyama et al., 2002). The vector was constructed by inserting the fusion expression of enhanced green fluorescent protein (EGFP) gene after BleoR. The fusion between BleoR and eGFP was generated by “ggtggtggt”, with six His tags used after the eGFP gene for purification. The sequential connection of TEF1 promoter–EM7 promoter–BleoR–EGFP–CYC1 terminator as the basic frame (BF) (Additional file 1: Figure S1) was optimized by thraustochytrids codons and synthesized by GENERAL BIOL company (Anhui, China). The basic frame (BF) was amplified from pBluescript SK (+) using KOD DNA polymerase (KFX-101B, TOYOBO, Osaka, Japan) with the primer pair BOX-F2/BOX-R2 (Additional file 2: Table S1). The rDNA is highly conserved, with multiple copies, and feasible as an insertion site for homologous recombination (HR) (Amador et al., 2000; Cheng et al., 2011). The sequence of Aurantiochytrium sp. SZU445 18s rDNA (GenBank: MT232522.1) (Liu et al., 2020; Zhu et al., 2020) was amplified with the primer pair 18S-F/18S-R. The sequence of DECR was amplified from Aurantiochytrium sp. SZU445 genome with primers DECR-F/DECR-R (Additional file 2:Table S1).
For the construction of the DECR overexpression cassette, the 505 bp 18s rDNA upstream and 505 bp 18s rDNA downstream were amplified with primers OEP1-F/OEP1-R and OEP5-F/OEP5-R, respectively (Figure 1B). DECR in the OE-P3 was amplified with primers OEP3-F/OEP3-R (Additional file 2: Table S1). The OE-P2 containing TEF1-EM7-BleR was amplified with primers OEP2-F/OEP2-R, whereas the OE-P4 containing EGFP-CYC1 was amplified with primers OEP4-F/OEP4-R. The overexpression cassette was assembled from the OE-P1, OE-P2, OE-P3, OE-P4, and OE-P5 sequences by overlap polymerase chain reaction (PCR) with KOD DNA polymerase. On both sides of the basic frame (BF), 800 bp DECR homologous flanks for the DECR gene knockout were inserted (Figure 1B). The 400 bp DECR upstream and 400bp DECR downstream homologous flank were amplified from the DECR gene with the primer pairs KOP1-F/KOP1-R and KOP3-F/KOP3-R, respectively. The KO-P2 was amplified with the primer pair KOP2-F/KOP2-R using BF as a template. The knockout cassette was assembled as the KO-P1, KO-P2, and KO-P3 sequence by overlapping PCR with KOD DNA polymerase. The DECR overexpression (OE) and knockout (KO) cassette were ligated with pEASY-Blunt (TransGen Biotech, Beijing, China) and transformed into DH5-alpha E. coli (named as pDECR-OE and pDECR-KO, respectively). The fragments were confirmed from single colonies by PCR with the primer pair M13F/M13R and sequenced by Sangon Biotech Company (Shanghai, China).
After verifying the construct by sequencing, the linear OE and KO cassette DNA for transforming Aurantiochytrium sp. strains was amplified by PCR from the construct with the primer pairs OEP1-F/OEP5-R and KOP1-F/KOP3-R, respectively. Linear OE and KO cassettes were transferred to Aurantiochytrium sp. SZU445, following the established protocol (Cheng et al., 2011) with some modification. Briefly, 5–10 μg linear DNA and 80 μL cells were added into the 0.2-cm cuvette, followed by an electroporation pulse (1500 V, 200 Ω, and 50 μF). Following electroporation, the solution was recovered in 1 mL M4 liquid medium and incubated at 23°C, with shaking at 200 rpm, for 24 h.
The transformants were selected by plating on M4 solid medium with 0.3 mg mL−1 Zeocin at 23°C. The transformants genome was extracted with yeast genome extraction kit (D2500, Omega, USA) and identified by PCR using primers BOX-F2/BOX-R2 (Additional file 2: Table S1). The green fluorescence of eGFP was used to determine the overexpressed and knockout mutants via laser scanning with a confocal microscope (LSM 710 NLO, ZEISS, Germany). A 488 nm argon laser line was used as the excitation source, and the detector was set at 509 nm for eGFP mutants.
Strains were inoculated in a 250-mL flask containing 100 mL of culture medium using a 10% (v/v) inoculation ratio. To analyze biomass and construct growth curves, we controlled the initial cell density of starting culture to be 2 × 104 cells mL-1. Triplicate cell samples were collected from each flask at regular intervals. The biomass was expressed as the number of cells counted using a hemocytometer microscope (DM500, Leica, Germany).
Neutral lipids were stained with Nile red (Rhawn, Shanghai, China) and analyzed using a fluorescent microplate reader (excitation at 488 nm and emission at 592 nm) (Synergy H1, Bio-Tek, USA) (Chen et al., 2009; Wang et al., 2020). One milliliter of collected cell culture broth with different growth phases was stained with 10 μL Nile red (0.1 mg mL−1 in acetone). The cells were incubated in an orbital shaker at 200 rpm in the dark for 20 min at 23°C before confocal microscopy (LSM 710 NLO, ZEISS, Germany). From the test samples, blank controls (cells incubated without Nile red stain) were subtracted. Data were expressed as the Nile red fluorescence ratio of transformants (OE and KO) to WT.
The cultured cells were collected by centrifugation at 6000 rpm for 10 min followed by freeze-drying for 72 h using a freeze dryer (10N, SCIENTZ, Ningbo, China) (Liu et al., 2020). Then, 500 mg of freeze-dried cells was extracted with chloroform: methanol (ratio 2:1 v/v) in a Soxhlet extractor (AG-SXT-06, OUGE, Shanghai, China) at 65°C for 60 h. After extraction, the samples were prepared according to our previously reported method (Li et al., 2020). Crude total lipids were collected after solvent evaporation at 65°C for 15 min. The fatty acid methyl esters (FAMEs) were obtained by adding 4 mL of 4% sulfuric acid in methanol to crude total lipids at 65°C for 1 h. The FAMEs were then extracted twice with 2 mL n-hexane and deionized water. Subsequently, n-hexane was volatilized in a nitrogen stream to obtain methyl esterified fatty acids (MEFs). The MEFs were dissolved in 1 mL of dichloromethane for analysis by gas chromatography–mass spectrometry (GC-MS, 7890-5975 Agilent, Santa Clara, CA, USA).
FAMEs were analyzed using the HP-5MS GC column (30.0 m × 250 µm, I.D. × 0.25 µm film thickness, Agilent) with a maximum temperature of 350°C. The inlet temperature was set to 250°C. Helium was used as a carrier gas. Constant pressure mode was used with a split ratio of 10:1. One microliter of each FAME sample was injected into the column. The column temperature was increased from 60°C to 180°C at a constant rate of 25°C per minute, then to 240°C at a rate of 3°C min−1, held for 1 min at 240°C, and then finally was raised to 250°C at a constant rate of 5°C per minute. The mass spectrum was collected by full scan mode detection. Fatty acids were identified by using National Institute of Standards and Technology (NIST) mass spectral library. As an internal standard, nonadecanoic acid (Solarbio, Beijing, China) was employed according to Li et al. (Li et al., 2020), and the fatty acid content was determined by comparison of internal standard peak areas. All samples used were analyzed in triplicate.
All experiments were performed in triplicate, and the data were presented as the mean and standard deviation (SD). GraphPad Prism (version 8.0.2) was used for data analysis. Two-way ANOVA and Duncan’s multiple range test at the p < 0.05 (confidence level) were used to determine differences between groups. Different numbers of asterisks (*) above each columns indicate significant differences at p < 0.05.
To generate DECR overexpression and knockout mutants in Aurantiochytrium sp. SZU445, we transformed cells with expression cassettes that harbored a Zeocin antibiotic resistance cassette with 800–1010 bp flanks homologous to the upstream and downstream ends of the insertion site locus for fragment exchange (Figure 1B). The 1010 bp 18s rDNA homologous flanks were designed to replace 577 bp of the 1786 bp long 18s rDNA gene with an overexpression cassette containing Zeocin resistance, DECR, and eGFP. Simultaneously, the 800 bp DECR homologous flanks were designed to replace 100 bp of the 900 bp long DECR gene with a knockout cassette containing the eGFP gene and zeocin resistance. The overexpression cassette (total length 3764 bp) and knockout cassette (2651 bp) were constructed and identified with the primer pair M13F/M13R by PCR (Figures 1C, D). Finally, the cassette was sequenced to verify its completeness and correctness.
The resistance screening experiment (Additional file 1: Figure S2) showed that 0.3 mg mL−1 Zeocin resulted in cell death of Aurantiochytrium sp. SZU445 (wildtype, WT). Thus, 0.3 mg mL−1 Zeocin was used for resistance screening throughout the experiments. The PCR-amplified linear overexpression and knockout cassettes were successfully transformed into WT cells via electroporation. The DECR overexpression transformants (hereinafter referred to as OE) and knockout transformants (hereinafter referred to as KO) were selected on solid M4 medium with 0.3 mg mL−1 Zeocin (Figure 2A). The Zeocin-resistant strains were confirmed via PCR using primers (BOX-F2/BOX-R2) that bound to the fragment between the TEF1 promoter and the CYC1 terminator in the expression cassette. The expected PCR amplicon sizes of the OE and KO mutant colonies were 2754 bp and 1851 bp, respectively (Figure 2B). A partial fragment alignment of the sequencing peak map of the transformant genome is shown in Additional file 1: Figure S3. The visualization of eGFP green fluorescence in the transformants (OE and KO) compared with WT confirmed the successful expression (Figure 2C).
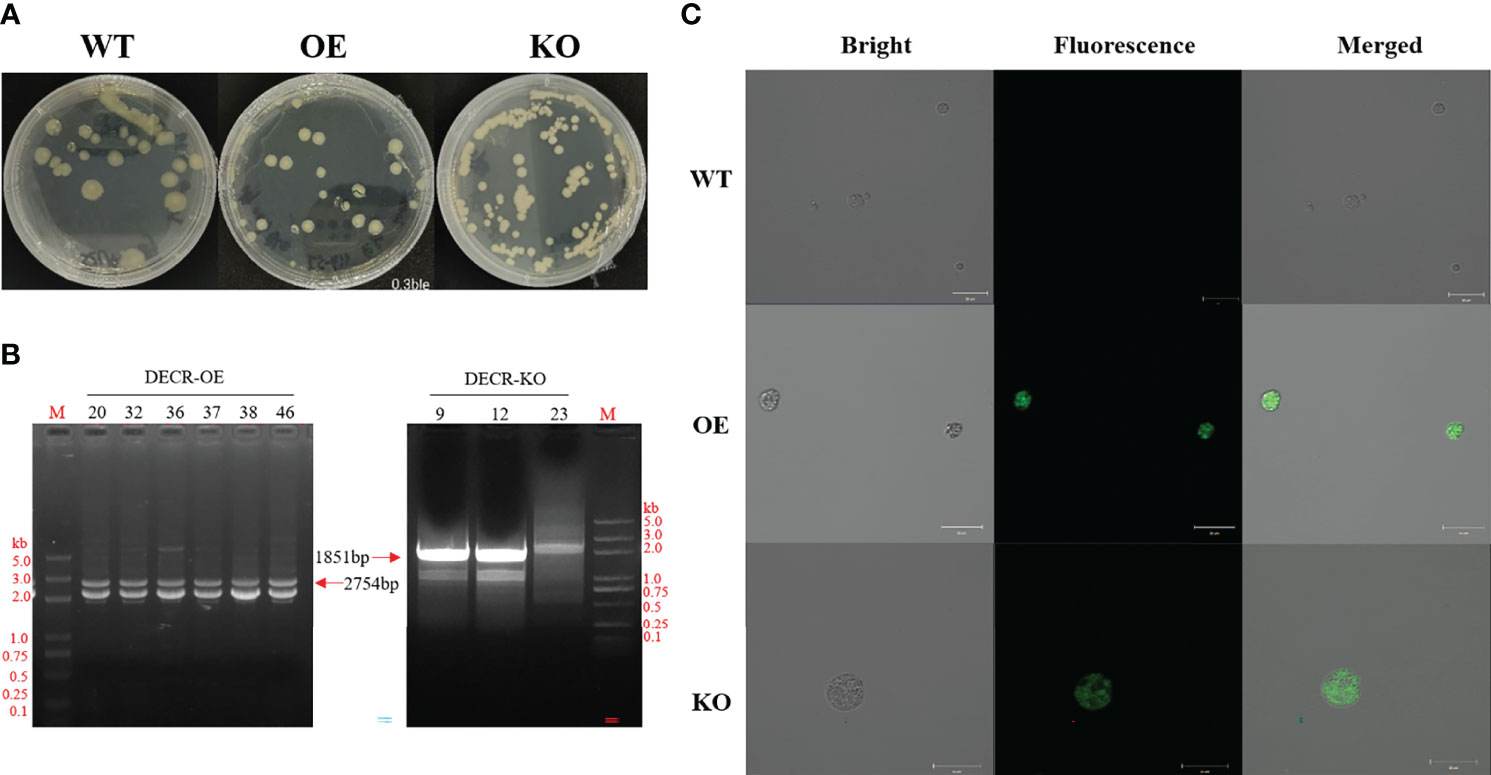
Figure 2 Obtaining and identifying the transformants. (A) Screening transformants through Zeomycin resistance (0.3 mg mL−1). Yellow clones were visible after 24–48 h, and transformants were kept in an M4 plate containing 0.3 mg mL−1 Zeomycin. Left, wildtype (WT) grown on an M4 solid plate. Centre and right, DECR overexpression (OE) and knockout (KO) mutants grown on M4 plates containing 0.3 mg mL−1 bleomycin, respectively. (B) PCR verification of transgenic Aurantiochytrium sp. SZU445 with OE-overlap and KO-overlap. Specific fragments of 2754 bp and 1851 bp were amplified from the genome of OE and KO transformants, respectively; 20, 32, 36, 37, 38, and 46 are the transformants with the OE cassette, and 9, 12, 23 are the transformants with the KO cassette. M, DL 5000 DNA Marker. (C) Confocal microscope observations of the green fluorescence produced by EGFP expression in OE and KO mutants. WT has no autofluorescence. Scale bar: 20 μm.
To explore the effect of DECR expression level on the growth of Aurantiochytrium sp. SZU445, the growth curves of wildtype (WT) and transformant (OE and KO) strains were compared. As shown in Figure 3A, the growth patterns were almost identical, with minor differences between transformants and the WT strain suggesting that knockout of DECR did not influence normal growth. After incubation for 22 h, the cell concentration was higher in the WT strain (by 1.03–1.26 times) than in the transformants, although the KO strains had 1.08 times higher growth than the OE strain. From the growth curves of all the three strains, the growth period can divide as follows: after inoculation, the cultures strains were in the lag phase for 0–12 h, in the log phase for 12–38 h, in the exponential phase for 38–50 h, in the stationary phase for 50–72 h; beyond 72 h, growth declined (the declining phase). Five periods (lag, log, exponential, stationary and declined phase) were represent respectively by different time points (12, 24, 42, 63 and 90 h) for follow-up researches.
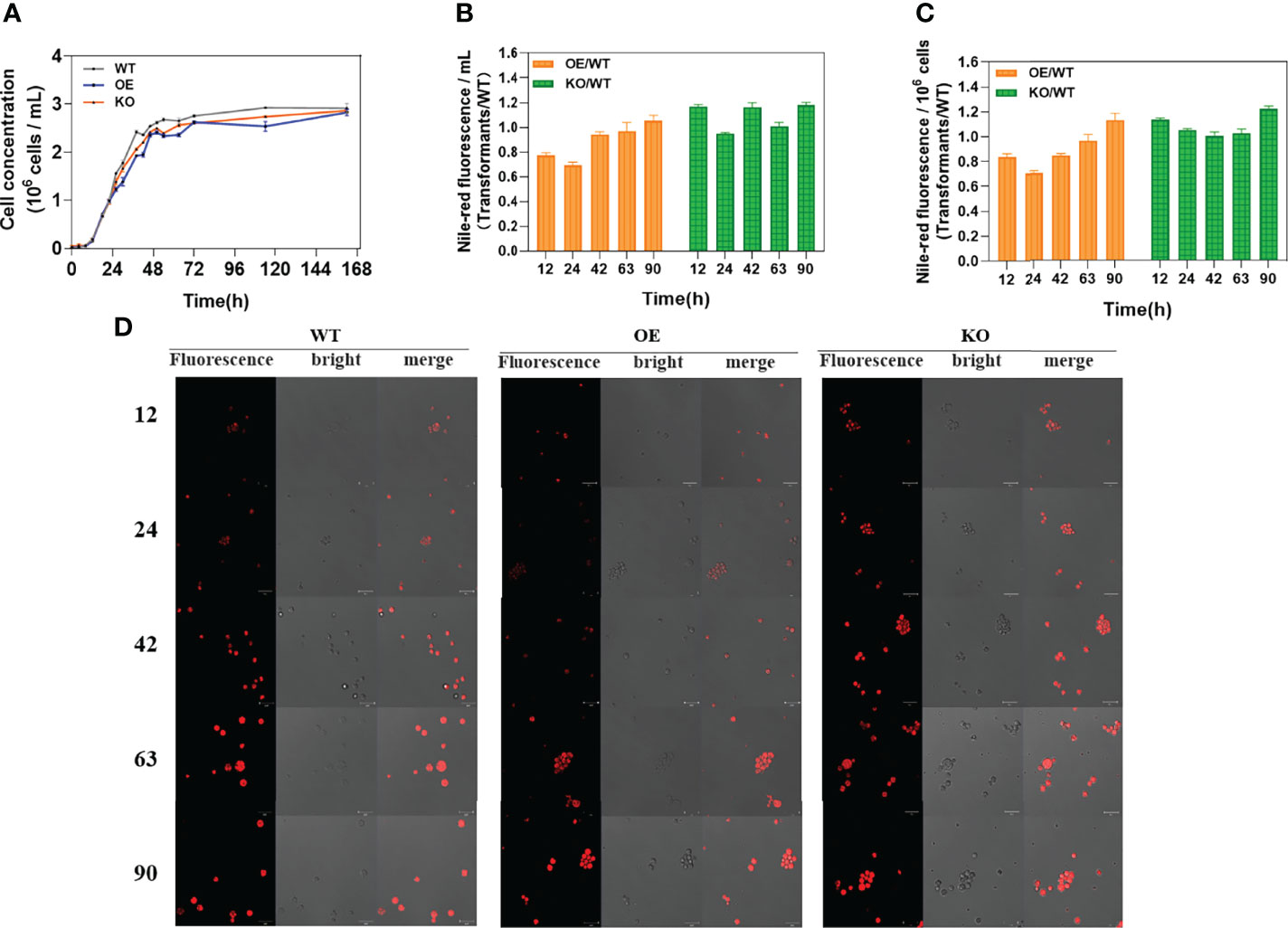
Figure 3 Growth curves and determination of neutral lipids. (A) The growth curves of wildtype (WT) strain, DECR overexpression (OE), and DECR knockout (KO) transformants were determined over a total period of 162 h. (B) Nile red fluorescence ratio of transformants (OE and KO) to wildtype (WT) per milliliters of strain solution. (C) Nile red fluorescence ratio of transformants (OE and KO) to wildtype (WT) per 106 cells. (D) Nile red fluorescence observation of wild-type (WT) strain, DECR overexpression (OE), and DECR knockout (KO) transformants in five various periods. All data were collected from three independent experiments. Scale bar: 20 μm. Values are the mean ± standard deviation.
The effect of DECR manipulation of neutral lipids was evaluated by assay of Nile red fluorescence in five growth phases. A significant difference was observed between the transformants and WT strains (Figure 3D). Shown as Figures 3B, 3C, the Nile red relative fluorescence (RFU) intensity of the KO strain was higher than that of the WT, whereas that of OE strain was lower than that of the WT in all observations. The RFU intensities of KO cells were the highest among all three strains, indicating the crucial effect of DECR on the accumulation of neutral lipids. The accumulation of neutral lipids in KO strain was 1.22-fold (p < 0.01) higher than WT strain at 90 h for 106 cells. At the same culture volume and cell concentration, the KO strains produced up to 1.23 and 1.48 times more neutral lipids than the OE strains, respectively, indicating its major potential to be applied as the nutrient additive.
Fatty acids at different growth stages (12, 24, 42, 63, and 90 h) were extracted and analyzed by GC-MS. Two predominant compositions (PA (C16:0) and DHA (C22:6)) for KO, OE, and WT were observed at various stages (Figures 4A–E). SFAs, especially PA, showed increased accumulation in the early growth stage (before 42 h). However, the UFAs, especially DHA, started to accumulate after 42 h accompanied by the decrease of SFAs at the same time period. In the exponential period (42 h) (Figure 4C), the abundance of DHA (C22:6) for the KO strain increased by 135.87% (p < 0.0001) and 17.09% (p < 0.05) compared with the OE and WT strains, respectively. Meanwhile, the content of PA (C16:0) for the KO strain decreased by 56.29% (p < 0.01) and 172.46% (p < 0.0001) compared with the WT and OE strain, respectively. This decrease indicated that DHA formation (C22:6) by consuming PA (C16:0) as a substrate was more efficient in the KO strains (Lee et al., 2015). Furthermore, the content of DHA (C22:6) in KO strains was significantly higher (51.46%–53.98%) than that of WT in the stationary (63 h) (Figure 4D) and declining phases (90 h) (Figure 4E), with increases of up to 1.52-fold (p < 0.01) and 1.57-fold (p < 0.001), respectively. Knockout of DECR substantially enhanced the DHA percentage (up to 54.00%) in the TFAs, while overexpressing DECR (OE strain) facilitated the beta-oxidation of PUFAs, leading to a lower proportion of DHA (36.43%–37.14%). These observations indicated that knockout of DECR is a practical approach for increasing the DHA percentage (of TFAs) in genetically engineered strains.
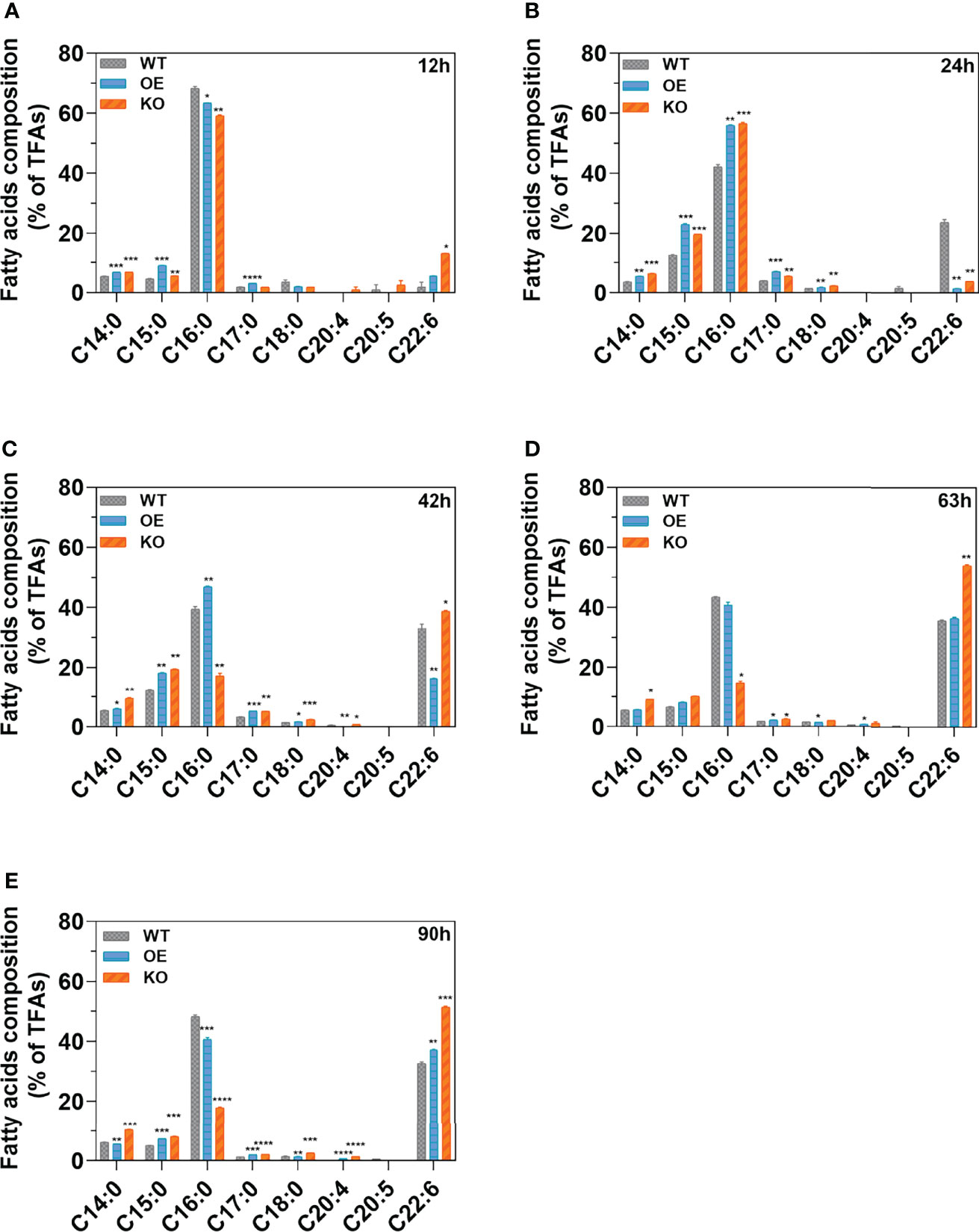
Figure 4 Fatty acid composition in various growth stages. (A–E) indicate the fatty acid composition of the wildtype (WT) strain, DECR overexpression (OE), DECR and knockout (KO) transformants detected by GC-MS after 12 h, 24 h, 42 h, 63 h, and 90 h, respectively. Data are shown as the percentage of total fatty acids (TFAs). C14:0, tetradecanoic acid; C15:0, pentadecanoic acid; C16:0, hexadecanoic acid; C17:0, heptadecanoic acid; C18:0, octadecanoic acid; C20:4, eicosatetraenoic acid; C20:5 eicosapentaenoic acid; C22:6, docosahexaenoic acid. Significant difference between wildtype strain (WT) and transformants (OE and KO) indicated at the p < 0.05 (*), p < 0.01 (**), p < 0.001 (***), or p < 0.0001 (****) level. All data are expressed as the mean ± S.D. of three independent experiments.
The effects of DECR expression on total fatty acids (TFAs), omega-3 polyunsaturated fatty acids (n-3 PUFAs), and DHA was analyzed at various growth stages and the results are shown in Figure 5. In Aurantiochytrium sp., the predominant composition of n-3 PUFAs was DHA, accounting for 94.37%–98.41% in the WT, OE, and KO strains. Thus, the trend in DHA accumulation was quite similar to n-3 PUFAs in all strains. The WT strain had the highest total fatty acid production among three strains, followed by OE (Figure 5A). However, the n-3 PUFAs yield of the KO strain (208.96 mg g−1) was significantly higher (1.47-fold) than that in the WT strain, whereas that of the OE strain was reduced by 13.03% at 90 h (Figure 5B). These results validate the fatty acid profile analysis (Figure 4). The content of saturated fatty acids was higher in the WT and OE strains, while unsaturated fatty acids in KO strain predominates. The KO strain showed a dramatic increase, achieving 203.268 mg g−1 (90 h) for the DHA yield over the whole growth profile, which is 67.55% and 45.10% more than that of the OE (121.318 mg g−1) and WT (140.092 mg g−1) strain (Figure 5C). Interestingly, Figure 5C shows that the DHA accumulation in the KO strain exhibited a sustainable growth until 90 h, whereas the DHA yield in the OE and WT strains peaked at 63 h and decreased by 7.07% and 6.97% at 90 h, respectively. For the OE and WT strains, the DHA tended to decompose and was consumed from 63 h, which led to a lower final yield of DHA at 90 h. However, for the KO strain, the PUFA beta-oxidation was disrupted by DECR knockout; thus, DHA continuously accumulated until 90 h (19.55% increase compared with 63 h) with a higher final yield (1.68-fold and 1.45-fold more than the OE and WT strains). From these results, it can be projected that the fermentation period can be prolonged to continuously accumulate DHA by knocking out DECR to interrupt PUFA consumption in the late growth stage, thus achieving a higher yield.
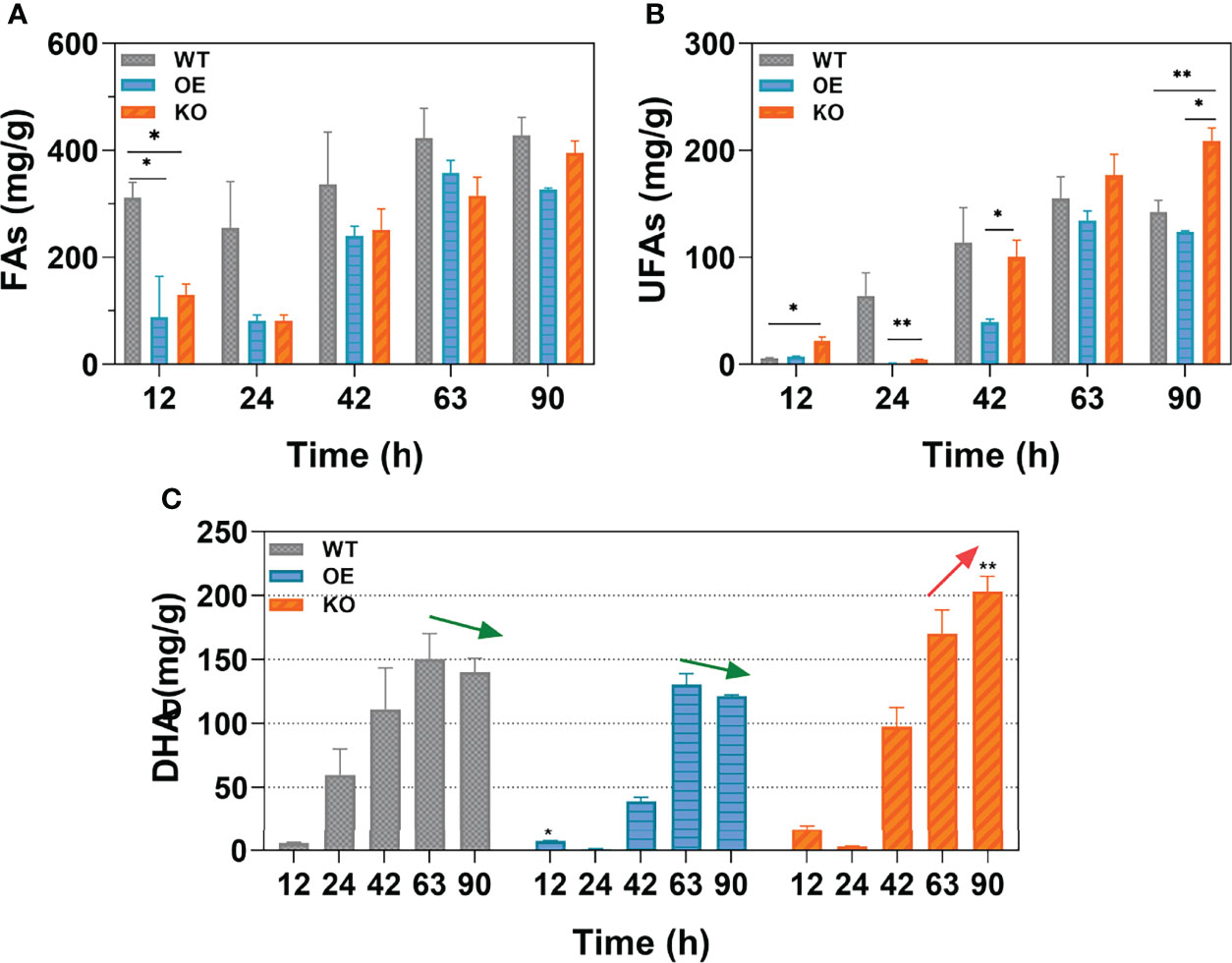
Figure 5 The yield of different types of fatty acids. (A-C) are the contents of total fatty acids (TFAs), unsaturated fatty acids (UFAs), and docosahexaenoic acid (DHA) respectively. Data shown are the weight of fatty acids per gram dry cells weight (DCW). Arrows (green and red) indicate the trend in DHA yield from 63 h to 90 h. Statistically significant differences between wildtype and transformants (OE and KO) were indicated at the p < 0.05 (*) and p < 0.01 (**) level. Values are the mean ± standard deviation.
The major neutral storage lipids in thraustochytrids are triacylglycerols (TAGs), which act as intracellular storage molecules for sterols, free fatty acids, and diacylglycerols (DAGs), as well as acting as a source of energy (Athenstaedt and Daum, 2006; Czabany et al., 2007; Jain et al., 2007; Du et al., 2021). The homeostasis of neutral lipids is maintained by hydrolases and synthetases, and the stored TAGs are hydrolyzed first into free fatty acids for energy consumption (Morabito et al., 2019; You et al., 2020). TAG accumulation is triggered under conditions of nitrogen deficiency at the end of growth (Yang et al., 2013; Dellero et al., 2018). The substantial physical connection between peroxisomes and liposomes allows lipolysis within liposomes to be coupled to peroxisomal fatty acid oxidation (Binns et al., 2006). Therefore, it is reasonable to speculate that the disruption of peroxisome-located DECR (Dommes et al., 1981) impedes the beta-oxidation of fatty acids, which leads to the continuous increase in PUFA content and inhibition of the hydrolytic tendency of TAGs, thereby accumulating TAGs. By disrupting DECR, the reaction progresses towards the synthesis of TAGs owing to the accumulation of hydrolysates, which thus enhances the accumulation of neutral lipids (Figure 6).
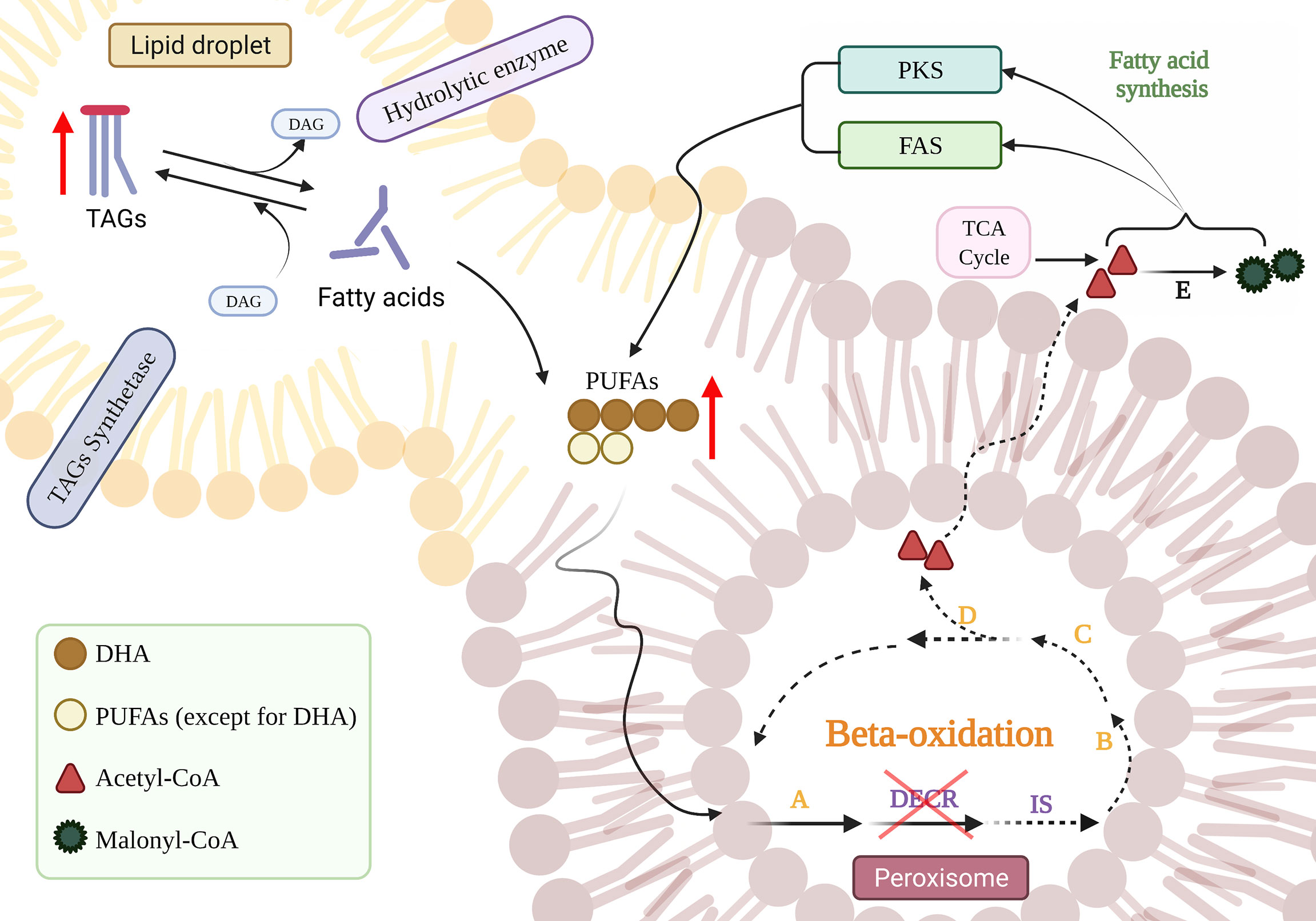
Figure 6 The effect of DECR disruption on the accumulation of PUFAs. The blue downward arrow implies that DECR was knocked down. Red upward arrows indicate increased product accumulation and interrupted lines indicate diminished responses. The orange letters refer to enzymes involved in standard beta-oxidation: acetyl-CoA dehydrogenase (A), enoyl-CoA hydratase (B), hydroxyacyl-CoA dehydrogenase (C), and beta-ketothiolase (D), respectively. DECR, 2,4-dienoyl-CoA reductase; IS, delta-3,5-delta-2,4-dienoyl-CoA isomerase; DAG, diacylglycerol; TAG, triacylglycerol; PKS, polyketide synthase; FAS, fatty acid synthase; TCA, tricarboxylic acid. Created with BioRender.com.
Moreover, the neutral lipid content of DECR-overexpressing strain (OE) was 0.70 times that of wild-type at 24 h (Figure 3C). Phospholipids, one of the main components of biological membranes, are synthesized at the expense of storing lipid TAG during the exponential growth phase (Pascual et al., 2013). when TAG is decomposed to generate one molecule of DAG to synthesize phospholipids, and at the same time generate one molecule of free fatty acid (FFA) (Czabany et al., 2007). We speculate that Overexpression of DECR breaks down more free fatty acids, and thus OE strains accelerate TAG decomposition due to continuous consumption of DAG and FFA products during exponential growth.
DHA (C22:6) enrichment may help thraustochytrids achieve improved membrane buoyancy and increased antioxidant capacity (Jain et al., 2007). The consumption of greater amounts of n-3 PUFAs (such as DHA) in the diet enhances the synthesis of cardioprotective and anti-inflammatory/pro-resolving lipid mediators generated from n-3 PUFAs (Ostermann et al., 2019). The accumulation of DHA in thraustochytrids is dynamic, determined by the net levels of synthesis and degradation (Figure 6). Fatty acids are usually utilized as an energy source for the initial growth period, whereas DHA is rapidly synthesized from the organic carbon present in the external medium via the PKS and FAS pathways in the stationary phase (Morabito et al., 2019). After nutrients are depleted, the DHA accumulated during the stationary phase becomes a readily available energy source during the declining phase as a result of beta-oxidation (Jain et al., 2007). The above results suggest that the disruption of DECR extends the DHA accumulation period (Figure 5C). DHA accumulation reached a peak in the stationary phase (63 h) in the WT and OE strains but continued to grow steadily in the declining phase (90 h) in KO strains. DECR is an essential enzyme required for the beta-oxidation of PUFAs (Sprecher, 2002); the disruption of DECR blocks beta-oxidation by eliminating the double bond at C4, thus reducing the utilization of DHA as an energy source. Although acetyl-CoA acts as a byproduct obtained from the degradation of other fatty acids, it is also used for DHA synthesis. The rate of DHA synthesis is higher than DHA decomposition, so the KO strain keeps accumulating DHA, even in the declining phase. The WT and OE strains tend to degrade DHA during starvation in the declining phase (Coutteau and Mourente, 1997; Jain et al., 2007), resulting in a slight reduction in accumulation (6.97% and 7.07%, respectively, compared to the stationary phase). In contrast, the KO strain appeared to prolong the accumulation period of DHA, possibly producing more DHA during the fermentation process; thus, this is a potential replacement for fish oil as a dietary supplement for n-3 PUFAs.
Other functions of DECR were also reported in the previous researches, including inhibition of gluconeogenesis (Miinalainen et al., 2009), antibacterial activity (Semini et al., 2020), and antitumor (Blomme et al., 2020; Gajewski et al., 2020). In the current study, we achieved high DHA accumulation and prolonged accumulation time by blocking beta-oxidation via the downregulation of DECR in Aurantiochytrium sp. In addition to enzymes on the beta-oxidation pathway, the PPAR pathway (Tahri-Joutey et al., 2021) and regulators (Xu et al., 2020) can be used to regulate the beta-oxidation. In further research, it is vital to apply this genetic engineering strategy to other enzymes in regulating the rate of beta-oxidation pathway, to explore their great potential to improve the accumulation of PUFAs. Moreover, production of DHA using thraustochytrids may be more sustainable than farming fish and cheaper than oilseeds crops (Salem and Eggersdorfer, 2015). The DHA yield enhancement in thraustochytrids obtained by various strategies thus far has been 97.10% in Schizochytrium sp. by multi-stage continuous fermentation (Guo et al., 2018), 83.20% via atmospheric and room temperature plasma mutagenesis of Aurantiochytrium sp. (Wang et al., 2022), and 1.18-fold by co-culture of Schizochytrium sp. and Rhodotorula glutinis (Sahin et al., 2018). By DECR knockout, we report a 46.80% increase in DHA accumulation in Aurantiochytrium sp. Thus, our genetic engineering strategy can be combined with other approaches to construct an efficient strain for DHA synthesis on an industrial scale and provide more sustainable n-3 PUFA supplements.
In this study, using homologous recombination to construct the transformants DECR knockout (KO) and overexpression (OE), the mechanism through which DECR affected the omega-3 PUFAs was thoroughly explored. By disrupting DECR expression, the DHA accumulation period was significantly prolonged (from 63 h to 90 h) with enhanced yields of neutral lipids, omega-3 PUFAs, and DHA up to 1.22-, 1.47-, and 1.45-fold compared to WT, respectively. In contrast, DECR overexpression led to the significant inhibition of FA production, with neutral lipids, omega-3 PUFAs, and DHA decreased by up to 30.52%, 65.38%, and 65.61%, respectively. This study confirmed the essential role of DECR in the accumulation of omega-3 PUFAs, especially DHA, and proposed a new strategy for the pursuit of sustainable renewable biofuels through marine protists.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
XY, SFL and ShiL designed the research experiments and wrote the paper. XZ, LL, SiL and ShiL performed the experiments and analyzed the data. ShiL, SFL, MI, XL, TT, and XY supervised the study, coordinated the writing and edited the drafts of the article. ShiL and XY contributed equally to this work. All authors contributed to the article and approved the submitted version.
This research was supported by the National Key Research and Development Project (2018YFA0902500), the Natural Science Foundation of Guangdong Province (2021A1515012557), Shenzhen Fundamental Research Projects (JCYJ20180507182405562), Joint R&D Project of Shenzhen-Hong Kong Innovation (SGLH20180622152010394), Hong Kong Innovation and Technology Commission TCFS (GHP/087/18SZ), Natural Science Foundation of Shenzhen (KQJSCX2018032809806045), Shenzhen Special Fund for Agricultural Development (Fishery) Project (2021-928), Shenzhen sustainable development project (2021N048).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We would like to thank Shenzhen University Testing Center for sharing equipment.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2022.939716/full#supplementary-material
Aasen I. M., Ertesvag H., Heggeset T. M., Liu B., Brautaset T., Vadstein O., et al. (2016). Thraustochytrids as Production Organisms for Docosahexaenoic Acid (DHA), Squalene, and Carotenoids. Appl. Microbiol. Biotechnol. 100 (10), 4309–4321. doi: 10.1007/s00253-016-7498-4
Abdel-Mawgoud A. M., Stephanopoulos G. (2018). Simple Glycolipids of Microbes: Chemistry, Biological Activity and Metabolic Engineering. Synthetic. Syst. Biotechnol. 3 (1), 3–19. doi: 10.1016/j.synbio.2017.12.001
Abedi E., Sahari M. A. (2014). Long-Chain Polyunsaturated Fatty Acid Sources and Evaluation of Their Nutritional and Functional Properties. Food Sci. Nutr. 2 (5), 443–463. doi: 10.1002/fsn3.121
Amador E., Martín J. F., Castro J. M. (2000). A Brevibacterium Lactofermentum 16S rRNA Gene Used as Target Site for Homologous Recombination. FEMS Microbiol. Lett. 185 (2), 199–204. doi: 10.1111/j.1574-6968.2000.tb09062.x
Arterburn L. M., Oken H. A., Hoffman J. P., Bailey-Hall E., Chung G., Rom D., et al. (2007). Bioequivalence of Docosahexaenoic Acid From Different Algal Oils in Capsules and in a DHA-Fortified Food. Lipids 42 (11), 1011. doi: 10.1007/s11745-007-3098-5
Athenstaedt K., Daum G. (2006). The Life Cycle of Neutral Lipids: Synthesis, Storage and Degradation. Cell Mol. Life Sci. 63 (12), 1355–1369. doi: 10.1007/s00018-006-6016-8
Bazan N. G. (2005). Neuroprotectin D1 (NPD1): A DHA-Derived Mediator That Protects Brain and Retina Against Cell Injury-Induced Oxidative Stress. Brain Pathol. 15 (2), 159–166. doi: 10.1111/j.1750-3639.2005.tb00513.x
Binns D., Januszewski T., Chen Y., Hill J., Markin V. S., Zhao Y., et al. (2006). An Intimate Collaboration Between Peroxisomes and Lipid Bodies. J. Cell Biol. 173 (5), 719–731. doi: 10.1083/jcb.200511125
Blomme A., Ford C. A., Mui E., Patel R., Ntala C., Jamieson L. E., et al. (2020). 2, 4-Dienoyl-CoA Reductase Regulates Lipid Homeostasis in Treatment-Resistant Prostate Cancer. Nat. Commun. 11 (1), 1–17. doi: 10.1038/s41467-020-16126-7
Boyer-Diaz Z., Morata P., Aristu-Zabalza P., Gibert-Ramos A., Bosch J., Gracia-Sancho J. (2020). Oxidative Stress in Chronic Liver Disease and Portal Hypertension: Potential of DHA as Nutraceutical. Nutrients 12 (9), 2627. doi: 10.3390/nu12092627
Chang K. J. L., Dunstan G. A., Abell G. C., Clementson L. A., Blackburn S. I., Nichols P. D., et al. (2012). Biodiscovery of New Australian Thraustochytrids for Production of Biodiesel and Long-Chain Omega-3 Oils. Appl. Microbiol. Biotechnol. 93 (5), 2215–2231. doi: 10.1007/s00253-011-3856-4
Cheng R.-B., Lin X.-Z., Wang Z.-K., Yang S.-J., Rong H., Ma Y. (2011). Establishment of a Transgene Expression System for the Marine Microalga Schizochytrium by 18S rDNA-Targeted Homologous Recombination. World J. Microbiol. Biotechnol. 27 (3), 737–741. doi: 10.1007/s11274-010-0510-8
Chen W., Zhang C., Song L., Sommerfeld M., Hu Q. (2009). A High Throughput Nile Red Method for Quantitative Measurement of Neutral Lipids in Microalgae. J. Microbiolog. Methods 77 (1), 41–47. doi: 10.1016/j.mimet.2009.01.001
Colombo S. M., Rodgers T. F. M., Diamond M. L., Bazinet R. P., Arts M. T. (2020). Projected Declines in Global DHA Availability for Human Consumption as a Result of Global Warming. Ambio 49 (4), 865–880. doi: 10.1007/s13280-019-01234-6
Coutteau P., Mourente G. (1997). Lipid Classes and Their Content of N-3 Highly Unsaturated Fatty Acids (HUFA) in Artemia Franciscana After Hatching, HUFA-Enrichment and Subsequent Starvation. Mar. Biol. 130 (1), 81–91. doi: 10.1007/s002270050227
Czabany T., Athenstaedt K., Daum G. (2007). Synthesis, Storage and Degradation of Neutral Lipids in Yeast. Biochim. Biophys. Acta 1771 (3), 299–309. doi: 10.1016/j.bbalip.2006.07.001
Das A. K., Uhler M. D., Hajra A. K. (2000). Molecular Cloning and Expression of Mammalian Peroxisomaltrans-2-Enoyl-Coenzyme A Reductase cDNAs. J. Biol. Chem. 275 (32), 24333–24340.
Dellero Y., Cagnac O., Rose S., Seddiki K., Cussac M., Morabito C., et al. (2018). Proposal of a New Thraustochytrid Genus Hondaea Gen. Nov. And Comparison of its Lipid Dynamics With the Closely Related Pseudo-Cryptic Genus Aurantiochytrium. Algal. Res. 35, 125–141. doi: 10.1016/j.algal.2018.08.018
Dellero Y., Rose S., Metton C., Morabito C., Lupette J., Jouhet J., et al. (2018). Ecophysiology and Lipid Dynamics of a Eukaryotic Mangrove Decomposer. Environ. Microbiol. 20 (8), 3057–3068. doi: 10.1111/1462-2920.14346
Dommes V., Baumgart C., Kunau W. H. (1981). Degradation of Unsaturated Fatty Acids in Peroxisomes. Existence of a 2,4-Dienoyl-CoA Reductase Pathway. J. Biol. Chem. 256 (16), 8259–8262. doi: 10.1016/s0021-9258(19)68833-2
Dommes P.a., Dommes V., Kunau W. (1983). Beta-Oxidation in Candida Tropicalis. Partial Purification and Biological Function of an Inducible 2, 4-Dienoyl Coenzyme A Reductase. J. Biol. Chem. 258 (18), 10846–10852. doi: 10.1016/S0021-9258(17)44352-3
Dommes V., Kunau W. (1984). 2, 4-Dienoyl Coenzyme A Reductases From Bovine Liver and Escherichia Coli. Comparison of Properties. J. Biol. Chem. 259 (3), 1781–1788. doi: 10.1016/S0021-9258(17)43476-4
Du F., Wang Y. Z., Xu Y. S., Shi T. Q., Liu W. Z., Sun X. M., et al. (2021). Biotechnological Production of Lipid and Terpenoid From Thraustochytrids. Biotechnol. Adv. 48, 107725. doi: 10.1016/j.biotechadv.2021.107725
Fossier Marchan L., Lee Chang K. J., Nichols P. D., Mitchell W. J., Polglase J. L., Gutierrez T. (2018). Taxonomy, Ecology and Biotechnological Applications of Thraustochytrids: A Review. Biotechnol. Adv. 36 (1), 26–46. doi: 10.1016/j.biotechadv.2017.09.003
Gajewski T., Higgs E., Li S., Flood B., Hatogai K. (2020). 239 Decr2 Loss Promotes Resistance of Tumor Cells to Immunotherapy by Affecting CD8+ T Cell-Regulated Tumor Ferroptosis. J. ImmunoTher. Cancer 8 (Suppl 3), A142. doi: 10.1136/jitc-2020-SITC2020.0239
Goepfert S., Vidoudez C., Rezzonico E., Hiltunen J. K., Poirier Y. (2005). Molecular Identification and Characterization of the Arabidopsis Δ3, 5, Δ2, 4-Dienoyl-Coenzyme A Isomerase, a Peroxisomal Enzyme Participating in the β-Oxidation Cycle of Unsaturated Fatty Acids. Plant Physiol. 138 (4), 1947–1956. doi: 10.1104/pp.105.064311
Guo D.-S., Ji X.-J., Ren L.-J., Yin F.-W., Sun X.-M., Huang H., et al. (2018). Development of a Multi-Stage Continuous Fermentation Strategy for Docosahexaenoic Acid Production by Schizochytrium Sp. Biores. Technol. 269, 32–39. doi: 10.1016/j.biortech.2018.08.066
Gupta A., Barrow C. J., Puri M. (2012). Omega-3 Biotechnology: Thraustochytrids as a Novel Source of Omega-3 Oils. Biotechnol. Adv. 30 (6), 1733–1745. doi: 10.1016/j.biotechadv.2012.02.014
Gurvitz A., Rottensteiner H., Kilpelainen S. H., Hartig A., Hiltunen J. K., Binder M., et al. (1997). The Saccharomyces Cerevisiae Peroxisomal 2,4-Dienoyl-CoA Reductase is Encoded by the Oleate-Inducible Gene SPS19. J. Biol. Chem. 272 (35), 22140–22147. doi: 10.1074/jbc.272.35.22140
Harper K. N., Hibbeln J. R., Deckelbaum R., Quesenberry C. P. Jr., Schaefer C. A., Brown A. S. (2011). Maternal Serum Docosahexaenoic Acid and Schizophrenia Spectrum Disorders in Adult Offspring. Schizophr. Res. 128 (1–3), 30–36. doi: 10.1016/j.schres.2011.01.009
He X. Y., Yang S. Y., Schulz H. (1997). Cloning and Expression of the fadH Gene and Characterization of the Gene Product 2, 4-Dienoyl Coenzyme A Reductase From Escherichia Coli. Eur. J. Biochem. 248 (2), 516–520. doi: 10.1111/j.1432-1033.1997.00516.x
Jain R., Raghukumar S., Sambaiah K., Kumon Y., Nakahara T. (2007). Docosahexaenoic Acid Accumulation in Thraustochytrids: Search for the Rationale. Mar. Biol. 151 (5), 1657–1664. doi: 10.1007/s00227-007-0608-1
Jiang Y., Fan K.-W., Tsz-Yeung Wong R., Chen F. (2004). Fatty Acid Composition and Squalene Content of the Marine Microalga Schizochytrium Mangrovei. J. Agric. Food Chem. 52 (5), 1196–1200. doi: 10.1021/jf035004c
Kelley D. S., Siegel D., Fedor D. M., Adkins Y., Mackey B. E. (2009). DHA Supplementation Decreases Serum C-Reactive Protein and Other Markers of Inflammation in Hypertriglyceridemic Men. J. Nutr. 139 (3), 495–501. doi: 10.3945/jn.108.100354
Koivuranta K., Hakkola E., Hiltunen J. (1994). Isolation and Characterization of cDNA for Human 120 kDa Mitochondrial 2, 4-Dienoyl-Coenzyme A Reductase. Biochem. J. 304 (3), 787–792. doi: 10.1042/bj3040787
Lee D., Jeong D.-E., Son H. G., Yamaoka Y., Kim H., Seo K., et al. (2015). SREBP and MDT-15 Protect C. Elegans From Glucose-Induced Accelerated Aging by Preventing Accumulation of Saturated Fat. Genes Dev. 29 (23), 2490–2503. doi: 10.1101/gad.266304.115
Li S. T., Hu Z. L., Yang X. W., Li Y. (2020). Effect of Nitrogen Sources on Omega-3 Polyunsaturated Fatty Acid Biosynthesis and Gene Expression in Thraustochytriidae Sp. Mar. Drugs 18 (12), 17. doi: 10.3390/md18120612
Liu L. X., Hu Z. L., Li S. F., Yang H., Li S. T., Lv C. H., et al. (2020). Comparative Transcriptomic Analysis Uncovers Genes Responsible for the DHA Enhancement in the Mutant Aurantiochytrium Sp. Microorganisms 8 (4), 19. doi: 10.3390/microorganisms8040529
Liu L., Hu Q., Wu H., Xue Y., Cai L., Fang M., et al. (2016). Protective Role of N6/N3 PUFA Supplementation With Varying DHA/EPA Ratios Against Atherosclerosis in Mice. J. Nutr. Biochem. 32, 171–180. doi: 10.1016/j.jnutbio.2016.02.010
Luthria D. L., Baykousheva S. P., Sprecher H. (1995). Double Bond Removal From Odd-Numbered Carbons During Peroxisomal Beta-Oxidation of Arachidonic Acid Requires Both 2,4-Dienoyl-CoA Reductase and Delta 3,5, Delta 2,4-Dienoyl-CoA Isomerase. J. Biol. Chem. 270 (23), 13771–13776. doi: 10.1074/jbc.270.23.13771
Mäkelä A. M., Hohtola E., Miinalainen I. J., Autio J. A., Schmitz W., Niemi K. J., et al. (2019). Mitochondrial 2, 4-Dienoyl-CoA Reductase (Decr) Deficiency and Impairment of Thermogenesis in Mouse Brown Adipose Tissue. Sci. Rep. 9 (1), 1–16. doi: 10.1007/s13399-021-02147-9
Mastalski T., Brinkmeier R., Platta H. W. (2020). The Peroxisomal PTS1-Import Defect of PEX1- Deficient Cells Is Independent of Pexophagy in Saccharomyces Cerevisiae. Int. J. Mol. Sci. 21 (3), 867. doi: 10.3390/ijms21030867
Metz J. G., Roessler P., Facciotti D., Levering C., Dittrich F., Lassner M., et al. (2001). Production of Polyunsaturated Fatty Acids by Polyketide Synthases in Both Prokaryotes and Eukaryotes. Science 293 (5528), 290–293. doi: 10.1126/science.1059593
Miinalainen I. J., Schmitz W., Huotari A., Autio K. J., Soininen R., Ver Loren van Themaat E., et al. (2009). Mitochondrial 2,4-Dienoyl-CoA Reductase Deficiency in Mice Results in Severe Hypoglycemia With Stress Intolerance and Unimpaired Ketogenesis. PLoS Genet. 5 (7), e1000543. doi: 10.1371/journal.pgen.1000543
Morabito C., Bournaud C., Maes C., Schuler M., Aiese Cigliano R., Dellero Y., et al. (2019). The Lipid Metabolism in Thraustochytrids. Prog. Lipid Res. 76, 101007. doi: 10.1016/j.plipres.2019.101007
Mukaiyama H., Oku M Fau - Baba M., Baba M Fau - Samizo T., Samizo T Fau - Hammond A. T., Hammond At Fau - Glick B. S., Glick Bs Fau - Kato N., et al. (2002). Paz2 and 13 Other PAZ Gene Products Regulate Vacuolar Engulfment of Peroxisomes During Micropexophagy. Genes to cells: devoted to Mol. Cell. Mech. 7 (1), 75–90. doi: 10.1046/j.1356-9597.2001.00499.x
Newell M., Baker K., Postovit L. M., Field C. J. (2017). A Critical Review on the Effect of Docosahexaenoic Acid (DHA) on Cancer Cell Cycle Progression. Int. J. Mol. Sci. 18 (8), 1784. doi: 10.3390/ijms18081784
Ogawa T., Hirose K., Yusuf Y., Kawamoto J., Kurihara T. (2020). Bioconversion From Docosahexaenoic Acid to Eicosapentaenoic Acid in the Marine Bacterium Shewanella Livingstonensis Ac10. Front. Microbiol. 11. doi: 10.3389/fmicb.2020.01104
Ostermann A. I., West A. L., Schoenfeld K., Browning L. M., Walker C. G., Jebb S. A., et al. (2019). Plasma Oxylipins Respond in a Linear Dose-Response Manner With Increased Intake of EPA and DHA: Results From a Randomized Controlled Trial in Healthy Humans. Am. J. Clin. Nutr. 109 (5), 1251–1263. doi: 10.1093/ajcn/nqz016
Pascual F., Soto-Cardalda A., Carman G. M. (2013). PAH1-Encoded Phosphatidate Phosphatase Plays a Role in the Growth Phase- and Inositol-Mediated Regulation of Lipid Synthesis in Saccharomyces Cerevisiae. J. Biol. Chem. 288 (50), 35781–35792. doi: 10.1074/jbc.M113.525766
Pleissner D., Lam W. C., Sun Z., Lin C. S. K. (2013). Food Waste as Nutrient Source in Heterotrophic Microalgae Cultivation. Biores. Technol. 137, 139–146. doi: 10.1016/j.biortech.2013.03.088
Raghukumar S. (2008). Thraustochytrid Marine Protists: Production of PUFAs and Other Emerging Technologies. Mar. Biotechnol. 10 (6), 631–640. doi: 10.1007/s10126-008-9135-4
Rahmawaty S., Charlton K., Lyons-Wall P., Meyer B. J. (2013). Dietary Intake and Food Sources of EPA, DPA and DHA in Australian Children. Lipids 48 (9), 869–877. doi: 10.1007/s11745-013-3812-4
Sahin D., Altindag U. H., Tas E. (2018). Enhancement of Docosahexaenoic Acid (DHA) and Beta-Carotene Production in Schizochytrium Sp. Using Symbiotic Relationship With Rhodotorula Glutinis. Process Biochem. 75, 10–15. doi: 10.1016/j.procbio.2018.09.004
Salem N. Jr., Eggersdorfer M. (2015). Is the World Supply of Omega-3 Fatty Acids Adequate for Optimal Human Nutrition? Curr. Opin. Clin. Nutr. Metab. Care 18 (2), 147–154. doi: 10.1097/MCO.0000000000000145
Schulz H. (1991). Beta Oxidation of Fatty Acids. Biochim. Biophys. Acta (BBA)-Lipids Lipid Metab. 1081 (2), 109–120. doi: 10.1016/0005-2760(91)90015-A
Schulz H., Kunau W.-H. (1987). Beta-Oxidation of Unsaturated Fatty Acids: A Revised Pathway. Trends Biochem. Sci. 12, 403–406. doi: 10.1016/0968-0004(87)90196-4
Semini G., Paape D., Blume M., Sernee M. F., Peres-Alonso D., Calvignac-Spencer S., et al. (2020). Leishmania Encodes a Bacterium-Like 2,4-Dienoyl-Coenzyme A Reductase That Is Required for Fatty Acid β-Oxidation and Intracellular Parasite Survival. mBio 11 (3), e01057–e01020. doi: 10.1128/mBio.01057-20
Shirley M., Murphy R. (1990). Metabolism of Leukotriene B4 in Isolated Rat Hepatocytes. Involvement of 2,4-Dienoyl-Coenzyme A Reductase in Leukotriene B4 Metabolism. J. Biol. Chem. 265 (27), 16288–16295. doi: 10.1016/S0021-9258(17)46220-X
Snetselaar L. G., de Jesus J. M., DeSilva D. M., Stoody E. E. (2021). Dietary Guidelines for Americans 2020–2025: Understanding the Scientific Process, Guidelines, and Key Recommendations. Nutr. Today 56 (6), 287. doi: 10.1097/NT.0000000000000512
Sprecher H. (2002). The Roles of Anabolic and Catabolic Reactions in the Synthesis and Recycling of Polyunsaturated Fatty Acids. Prostaglandins Leukotrienes Essential Fatty Acids 67 (2), 79–83. doi: 10.1054/plef.2002.0402
Tahri-Joutey M., Andreoletti P., Surapureddi S., Nasser B., Cherkaoui-Malki M., Latruffe N. (2021). Mechanisms Mediating the Regulation of Peroxisomal Fatty Acid Beta-Oxidation by PPARalpha. Int. J. Mol. Sci. 22 (16), 8969. doi: 10.3390/ijms22168969
Wang Q., Jin W., Han W., Song K., Chen Y., Chen C., et al (2022). Enhancement of DHA Production From Aurantiochytrium Sp. By Atmospheric and Room Temperature Plasma Mutagenesis Aided With Microbial Microdroplet Culture Screening. Biomass Conversion Biorefinery 12(1), 1–12 doi: 10.1007/s133Xu99-021-02147-9
Wang X., Liu S. F., Qin Z. H., Balamurugan S., Li H. Y., Lin C. S. K. (2020). Sustainable and Stepwise Waste-Based Utilisation Strategy for the Production of Biomass and Biofuels by Engineered Microalgae. Environ. pollut. 265, 114854. doi: 10.1016/j.envpol.2020.114854
Xu X., Huang C., Xu Z., Xu H., Wang Z., Yu X. (2020). The Strategies to Reduce Cost and Improve Productivity in DHA Production by Aurantiochytrium Sp.: From Biochemical to Genetic Respects. Appl. Microbiol. Biotechnol. 104 (22), 9433–9447. doi: 10.1007/s00253-020-10927-y
Xu T., Lim Y. T., Chen L., Zhao H., Low J. H., Xia Y., et al. (2020). A Novel Mechanism of Monoethylhexyl Phthalate in Lipid Accumulation via Inhibiting Fatty Acid Beta-Oxidation on Hepatic Cells. Environ. Sci. Technol. 54 (24), 15925–15934. doi: 10.1021/acs.est.0c01073
Yang Z.-K., Niu Y.-F., Ma Y.-H., Xue J., Zhang M.-H., Yang W.-D., et al. (2013). Molecular and Cellular Mechanisms of Neutral Lipid Accumulation in Diatom Following Nitrogen Deprivation. Biotechnol. Biofuels 6 (1), 67. doi: 10.1186/1754-6834-6-67
You J., Mallery K., Mashek D. G., Sanders M., Hong J., Hondzo M. (2020). Microalgal Swimming Signatures and Neutral Lipids Production Across Growth Phases. Biotechnol. Bioengineering 117 (4), 970–980. doi: 10.1002/bit.27271
Keywords: omega-3 polyunsaturated fatty acid, docosahexaenoic acid (DHA), PUFA beta-oxidation, 2,4-dienoyl-CoA reductase (DECR), Aurantiochytrium sp.
Citation: Liang S, Yang X, Zhu X, Ibrar M, Liu L, Li S, Li X, Tian T and Li S (2022) Metabolic Engineering to Improve Docosahexaenoic Acid Production in Marine Protist Aurantiochytrium sp. by Disrupting 2,4-Dienoyl-CoA Reductase. Front. Mar. Sci. 9:939716. doi: 10.3389/fmars.2022.939716
Received: 09 May 2022; Accepted: 20 June 2022;
Published: 15 July 2022.
Edited by:
Thomas Bartholomäus Brück, Technical University of Munich, GermanyCopyright © 2022 Liang, Yang, Zhu, Ibrar, Liu, Li, Li, Tian and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xuewei Yang, eWFuZ3h3QHN6dS5lZHUuY24=; Shuangfei Li, c2ZsaUBzenUuZWR1LmNu
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.