- 1Graduate Institute of Pharmacognosy, College of Pharmacy, Taipei Medical University, Taipei, Taiwan
- 2National Museum of Marine Biology & Aquarium, Pingtung, Taiwan
- 3Graduate Institute of Medical Sciences, College of Medicine, Taipei Medical University, Taipei, Taiwan
- 4Department of Dermatology, Taipei Medical University-Shuang Ho Hospital, New Taipei City, Taiwan
- 5Department of Dermatology, School of Medicine, College of Medicine, Taipei Medical University, Taipei, Taiwan
- 6International Ph.D. Program for Cell Therapy and Regeneration Medicine, Taipei Medical University, College of Medicine, Taipei, Taiwan
- 7Department of Pharmaceutical Biology, German University in Cairo, Cairo, Egypt
- 8Department of Pharmacognosy, Faculty of Pharmacy, Ain-Shams University, Cairo, Egypt
- 9Department of Food Science, College of Human Ecology, Fu Jen Catholic University, New Taipei City, Taiwan
- 10Department of Marine Biotechnology and Resources, National Sun Yat-sen University, Kaohsiung, Taiwan
- 11PhD Program in Pharmaceutical Biotechnology, Fu Jen Catholic University, New Taipei City, Taiwan
- 12Cell Physiology and Molecular Image Research Center, Wan Fang Hospital, Taipei Medical University, Taipei, Taiwan
- 13PhD Program in Clinical Drug Development of Herbal Medicine, College of Pharmacy, Taipei Medical University, Taipei, Taiwan
- 14Traditional Herbal Medicine Research Center, Taipei Medical University Hospital, Taipei, Taiwan
In-depth analysis of metabolomics diversity of marine species through advanced mass spectrometric analysis is one of the most promising new tools for the development of marine drugs against mild and life-threatening diseases. Neurofibromas are a common type of tumor in the peripheral nervous system. Currently, there are very limited treatment options for neurofibromas. In our course of exploring potential therapeutic agents for neurofibroma treatment, the multi-informative molecular networking (MIMN) approach was proposed. The MIMNs of the Lendenfeldia sp. sponge extract and sub-fractions were established according to their inhibitory activity against several inflammatory chemokines (CCL3, CCL4, CCL5, CXCL1, CXCL8, and CXCL10) in neurofibroma cell line hTERT-NF1-ipNF95.11b-C (CRL-3390). The visualized MIMN revealed the anti-inflammatory potential of scalarane-enriched fractions, and the follow-up annotation and isolation led to the identification of a scalarane, 24-methyl-12,24,25-trioxoscalar-16-en-22-oic acid (2). Our results revealed that the most abundant scalarane (2) dominated the anti-chemokine effect of Lendenfeldia sp. extract together with other scalaranes, indicating the potential application of sponge-derived scalaranes to be developed as therapeutic agents for neurofibromas.
Introduction
Recent advances in spectroscopic analysis led to the development of many applied strategies and techniques for the discovery of novel secondary metabolites from marine sources. Among these techniques are the “MS/MS molecular networking (MN) approaches” (Watrous et al., 2012) that help in the secondary metabolites identification as well as the “spectroscopic coupled bioassay screening strategies” (Faleschini et al., 2019) that aid in the identification of the most potent natural components. These techniques drew most of the scientific community’s attention and led to significant advances in bioactive metabolites discovery (Allard et al., 2016; Caesar et al., 2018; Woo et al., 2019; Olivon et al., 2020; Peng et al., 2021). MN is a visual computational strategy that can be intuitively implemented using LC-MS/MS data (Watrous et al., 2012). The visual computational strategy of MN is based on comparing the theoretical to raw MS/MS spectra to establish a relative network. Similar structures are clustered together with similar MS/MS spectra. Unlike traditional mass spectrometry techniques that focus on the annotation of known metabolites, MN is more focused on exposing the structural relationships between metabolites with visualized “clusters”, which is conducive to the systematic and comprehensive study of metabolites (Kang et al., 2019; Beniddir et al., 2021; Yu et al., 2022). In the current study, we took the advantage of mastering these complex spectroscopic techniques, to design a useful multi-informative molecular networking (MIMN) platform for analyzing and refining the valuable information in structural diversity and bioactivities of marine sponges. Moreover, based on this metabolomic information, we will be able to suggest the lead chemical skeletons with the most significant bioactive potential through primary fractionations, bioassays, and bioactive score calculations.
Neurofibroma is a type of nerve sheath tumor developed in the peripheral nervous system. It is the most prevalent tumor associated with the tumor predisposition syndrome neurofibromatosis type 1 (NF1), an autosomal dominant genetic disorder (Gutmann et al., 2017). NF1 patients are associated with germline monoallelic NF1 mutation, and their somatic cells could acquire the potential to give rise to a tumor when the only normal NF1 allele is mutated. This process is known as NF1 loss of heterozygosity (Gutmann et al., 2017).
The tumor cells of neurofibroma are derived from NF1 mutant Schwann cells. Neurofibroma is featured by a heterogeneous and complex tumor microenvironment; among the microenvironmental cells, immune cells are emphasized for their critical contributions to neurofibroma development and progression (Staser et al., 2012; Jiang et al., 2021). In mice, Nf1-/- Schwann cells require the support of immune cells to give rise to a neurofibroma. Furthermore, the Nf1 status of microenvironmental immune cells determines their ability to sustain neurofibroma development; the Nf1-/- Schwann cells are driven to form a tumor by blood cells with Nf1+/- but not Nf1+/+ status (Yang et al., 2008). This is critical evidence showing the indispensable contribution from immune cells to neurofibroma tumorigenesis and highlights Nf1 status (Table S1).
The main immune cell types observed in the neurofibromas microenvironment are mast cells and macrophages (Prada et al., 2013; Liao et al., 2018). The data from the genetically engineered mouse models showed that mast cells and macrophages are attracted to neurofibroma tumors by SCF/KIT and CXCL10/CXCR3 signaling respectively, and the chemokine/cytokine are derived from the tumor cells (Liao et al., 2018; Fletcher et al., 2019). Taken together along with the fact that immune cells are essential drivers for neurofibroma tumorigenesis (Yang et al., 2008), we proposed compounds with the potential to downregulate the chemokine/cytokine expressions in neurofibroma cells might be developed as anti-neurofibroma candidates. Therefore, we designed a useful strategy to analyze and define the valuable information on structural diversity and anti-neurofibroma activity of marine invertebrates. In the current study, the Lendenfeldia sp. sponge was screened for anti-chemokine activity. Multi-informative molecular networking (MIMN) approach was carried out to simultaneously map the chemical and biological profiles based on the reduction of CXCL10 expression and some other chemokines, including CCL3, CCL4, CCL5, CXCL1, and CXCL8. These data demonstrated a potent anti-inflammatory ability of the sclaranes identified from Lendenfeldia sp. sponge and suggested a neurofibroma therapeutic potential by targeting the tumor cell-initiated inflammatory microenvironment.
Materials and Methods
Animal Material
The original specimen of Lendenfeldia sp. was collected via scuba diving off the coast of Southern Taiwan in April of 2019. The sponge was aquacultured in a seawater cylinder (four tons) with a cooler to control temperature (25–28°C) and a LED lamp (9–12 h light support per day) at the National Museum of Marine Biology & Aquarium (Pingtung, Taiwan). The sponge samples were collected and freeze-dried in September of 2021 before extraction.
Extraction, Fractionation, and Isolation
The freeze-dried Lendenfeldia sp. material was minced and extracted three times with a 1:1 mixture of dichloromethane (CH2Cl2) and methanol (MeOH). The crude extract was evaporated under reduced pressure to afford a residue for further fractionation. A Shimazu LC-2050 HPLC system (Shimazu, Kyoto, Japan) was utilized to profile the crude Lendenfeldia sp. extract. Liquid chromatography was performed using a Galaksil EF-C18-H (5 μm, 120 Å, 10 × 250 mm, C18) column (Galak Chromatography, Jiangsu, China). The mobile phase was prepared by mixing MeOH (A) and water (W, containing 0.1% formic acid) gradient sequence: 0–10 min, 65–70% A; 10–45 min, 70–80% A; 45–55 min, 80–100% A; 55–70 min, 100% A. The flow rate was fixed at 2 mL/min, the column temperature was maintained at 40°C, and the detection wavelengths were set from 190 to 500 nm. To prepare the fractionation sample, the sample was dissolved in methanol and filtered through a 0.45 μm membrane filter before loading into the column (with concentration of 1 mg/μL). The sample injection was implemented manually with a 20 μL volume per injection. Further fractionation was performed based on the retention time.
The fractions BRW7 was selected for the further produced in large scale and isolation due to its anti-chemokine potential. The purifications of scalarane 2 from BRW7 were executed using the isocratic elution [MeOH–water (75:25, v/v)]. The identification of isolated compounds was completed based on the comparison between the obtained spectra [nuclear magnetic resonance (NMR) and mass (MS) spectra] and previous reported ones (please check the spectra from Supplementary Materials: Table S2, Figures S1–S12). The purity of 2 was further qualitatively analyzed (Figure S13) using UPLC.
MS/MS Non-Targeted Fragment Ions Collection Using Ultra-Performance Liquid Chromatography Quadrupole Time-of-Flight Mass Spectrometry
The MS2 data collection was carried out based on a Waters SYNAPT G2 LC/Q-TOF (Waters Corporation, Milford, MA, USA) system. The chromatographic separation before the MS spectra was performed using a C18 column of Waters Acquity UPLC BEH (Waters, 1.7 µm, 2.1 mm × 100 mm). The mobile phase was prepared with a MeCN (A, containing 0.1% formic acid)/water (W, containing 0.1% formic acid) gradient sequences as follows: 0–1 min, 5% A; 1–16 min, 5–99.5% A; 16–26 min, 99.5% A; 26–26.1 min, 99.5–5% A; 26.1–28 min, 5% A. The flow rate was set up at 0.4 mL/min, and the temperature of the column part was maintained at 40°C in the oven. The extract (5 mg) was dissolved in 1 mL of methanol (5,000 ppm) and filtered through a 0.45 μm membrane filter. The sample injection was executed automatically with a 5 μL volume per injection. The non-targeted MS1 and MS2 data were collected within the range of m/z 100–2000. The automated data-dependent acquisition (DDA) approach was applied in the MS2 scans, and the non-targeted selections of 5 precursor ions were fragmented with ramping of the collision energy from 10−50 V. The acquired MS data were finalized by Waters MassFragment software (MassLynx4.1, Waters, MA, USA).
GNPS-Based Molecular Networking Analysis
A GNPS web-based platform (https://gnps.ucsd.edu) was applied to analyze and output the MS/MS molecular networking data (January 27th of 2022). The MS/MS spectra were window-filtered according to the top 5 strongest ion peaks in the ± 50 Da window throughout the spectrum. A network was then created in which linkages between nodes were filtered by a cosine value above 0.70 and at least four matched peaks. Then the appeared nodes in the network were annotated based on the experimental MS2 fragmentations of isolates. The molecular network was visualized and was laid out using Cytoscape 3.8.2 (Cytoscape 3.8.2, NRNB, CA, USA).
Bioactivity Score Calculation
The multi-informative molecular networking (MIMN) presented in the current study revealed the bioactivity, distribution, and putative chemical class of specific precursor ion nodes, representing the bioactive probability of a group of compounds clustered in MIMN. In order to quantify this chemical structure-relevant bioactive probability, the bioactivity scores were calculated based on their bioactivity levels together with relative abundances (pie chart of the precursor nodes).
Bioactivity scores (BS) = anti-chemokine level (1: inhibition rate ≥ 50%; 0: inhibition rate < 50%) x relative abundances (%)
Neurofibroma Cell Culture
The neurofibroma cell line hTERT-NF1-ipNF95.11b-C (CRL-3390) was obtained from the American Type Culture Collection (ATCC, Manassas, VA, USA). CRL-3390 cells were cultured in Dulbecco’s Modified Eagle Medium (DMEM) with high glucose (Gibco, Thermo Fisher Scientific, Waltham, MA, USA), and were supplemented with 10% fetal bovine serum, L-glutamine, penicillin-streptomycin, and sodium pyruvate (HyClone Laboratories Inc., UT, USA). CRL-3390 cells were maintained in an incubator at 37°C with 5% CO2 and were passaged every 3-4 days. CRL-3390 neurofibroma cells were cultured in a 6-well or a 96-well plate for qPCR or MTT assay. When the cells reached about 60-70% confluence, the culture media were replaced by compound-containing media for the drug treatment.
Quantitative Polymerase Chain Reaction
For qPCR assay, the CRL-3390 cells were treated with compound-containing media for 24 h. At the time of cell harvesting, the media were aspirated and were gently washed with phosphate-buffered saline. The attached cells were lysed by 1 ml of TRIzol Reagent (Invitrogen, Waltham, MA, USA) for RNA extraction. A total of 1 µg RNA was subjected to reserve transcription by the iScript cDNA Synthesis Kit (Bio-Rad, Hercules, CA, USA). The cDNA was used as a template for qPCR with the iTaq Universal SYBR Green Supermix (Bio-Rad, Hercules, CA, USA) in a StepOnePlus Real-Time PCR System (Applied Biosystems, Waltham, MA, USA). All the above procedures were performed according to the manufacturer’s protocols. The primer sequences used for this study were listed in Table 1.
MTT Cell Viability Assay
The MTT assay is a colorimetric assay to determine the cellular metabolic activity by analyzing the NAD(P)H-dependent cellular oxidoreductase enzymes. We used the MTT assay as a readout to determine any potential cytotoxicity to the CRL-3390 neurofibroma cells from the tested compounds. Briefly, 5,000 cells in 100 µl media were plated in each well of a 96-well plate. When the cell density reached 60-70 confluence, the media were replaced by drug-containing media. The cells were treated with each compound for 24 or 48 h. For the MTT assay, each well was added with 20 µl of 2.5 mg/ml MTT and was incubated for 30 min at 37°C. The media were then sucked away and the purple crystals in cells were lysed by 100 µl DMSO followed by 20 min incubation. The relative cell viability was determined by the light absorbance at 570 nm.
Statistics
The qPCR and MTT data were presented by mean ± SEM from 3-5 independent samples. The statistics were performed by a two-tailed Student’s t-test with the Prism (GraphPad, San Diego, CA, USA).
Results and Discussion
Multi-Informative Molecular Networking Platform for Anti-Neurofibroma Lead Discovery
To facilitate the discovery of anti-neurofibroma leads from the Lendenfeldia sp. sponge, we designed a workflow based on chemical molecular networking (MN) and anti-chemokine assessments (Figure 1). First, a primary chromatographic separation and collection were performed to obtain fractions from the crude extract. Then the chemical and bioactive profiles of these fractions were constructed based on the MS/MS MN analysis and anti-neurofibroma assessments, respectively. The followed-up clustering, classification, and annotation were performed through the Global Natural Product Social (GNPS) MN platform. Then bioactive probabilities of each identified compounds or specific chemical clusters were calculated based on their bioactivity levels together with relative abundances. Moreover, the target isolation was designed and executed based on the results from the visualized multi-informative molecular networking (MIMN). Finally, the ultimate correlation between bioactivities, single compounds, fractions, and crude extracts was generalized to suggest the lead single compounds or chemical skeletons with the most significant anti-neurofibroma potential.
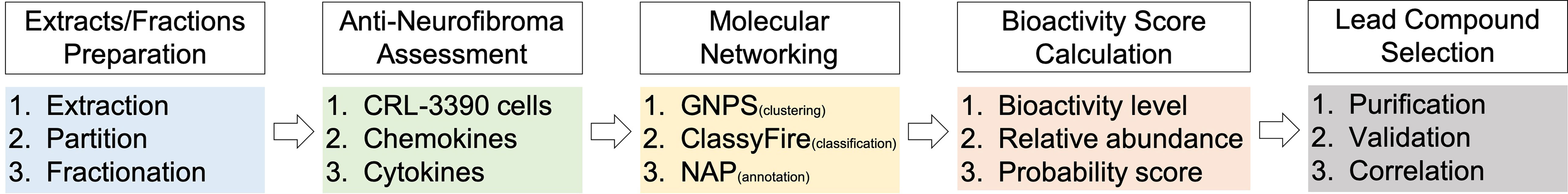
Figure 1 The multi-informative molecular networking (MIMN) workflow for discovering anti-neurofibroma leads.
Establishing a High-Performance Liquid Chromatography-Based Anti-Chemokine Profile
The fractionation methods with the traditional column chromatography usually deal with problems of high content overlap that reduces the accuracy of bioassay-guided isolation toward searching for anti-neurofibroma biomarkers. Herein, high-performance liquid chromatography (HPLC) was employed to assure the distinction of the tested fractions during the bioassay. In the current work, the chromatogram retention time-based fractionation of the extract of a Lendenfeldia sp. sponge was performed within 70 min. The collected eleven fractions (BRW1–11, at concentrations of 1 and 4 μg/mL) were further subjected to a quantitative polymerase chain reaction (qPCR) assay with the CRL-3390 cells to scheme a high-resolution anti-chemokine profile against CCL3, CCL4, CCL5, CXCL1, CXCL8, and CXCL10 (Figure 2A). Among all tested fractions, BRW5, 7, 9, and 10 showed inhibitory activity against most of the chemokines at a concentration of 4 μg/ml (Figure 2B). When treating CRL-3390 cells by BRW fractions at 1 μg/ml, we observed that CXCL10 mRNA expression was suppressed by the BRW1 (11%), BRW7 (70%), BRW8 (37%), BRW9 (34%), BRW10 (32%), and BRW11 (64%). Of note, the BRW7 (p = 0.0012) is the most potent fraction to significantly inhibit CXCL10 gene expression, implying the presence of major biomarkers in BRW7. To assess the chemokines inhibitory properties, the viability of CRL-3390 cells treated with these fractions was evaluated using MTT cell proliferative assay at various concentrations (2, 4, and 8 μg/ml) (Figure 3). Since BRW10 and 11 showed cytotoxicity at 4, and 8 μg/ml, the bioactivity of these two fractions was not considered nor was discussed in the current study.
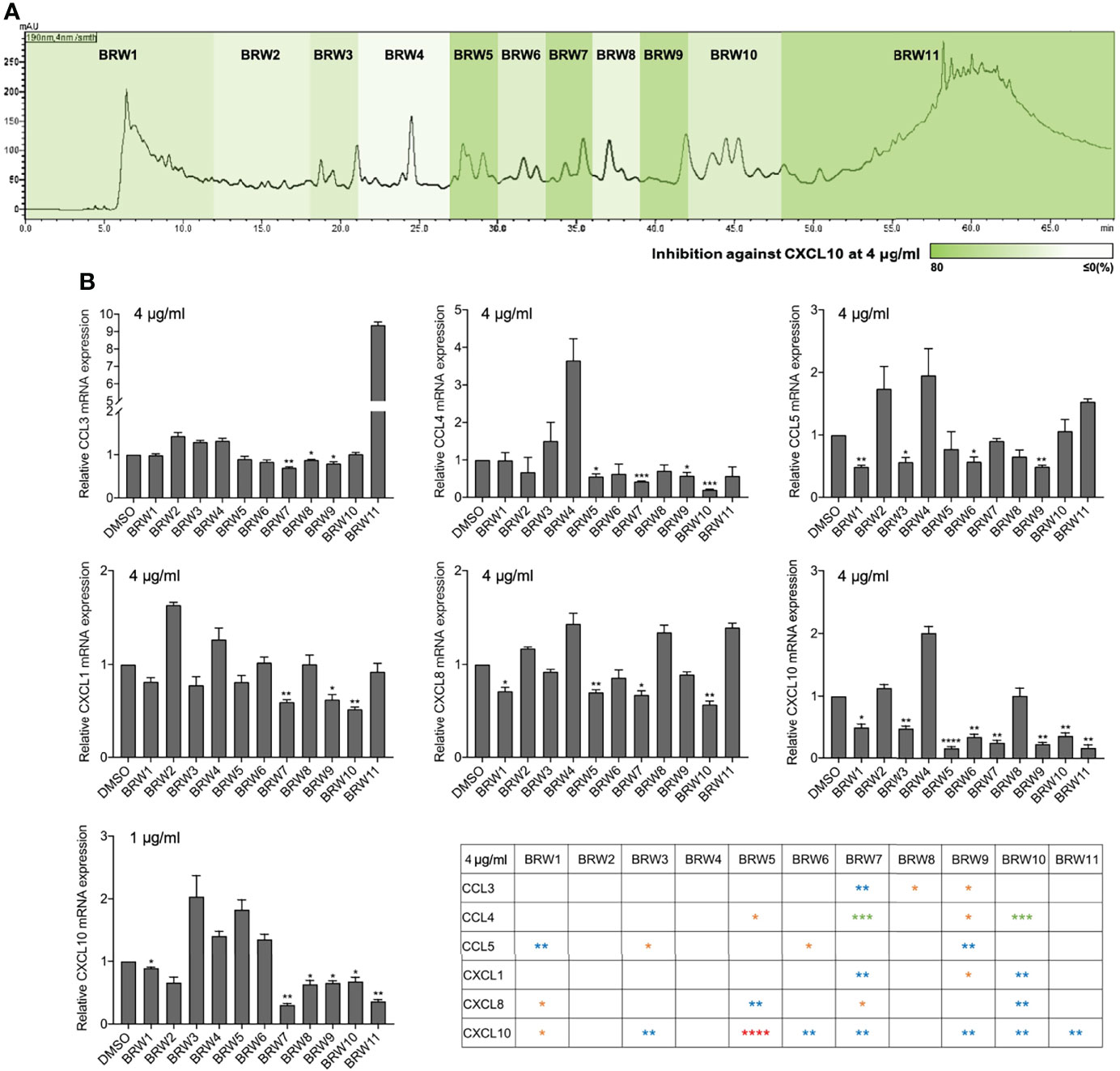
Figure 2 The anti-neurofibroma assessment of the Lendenfeldia sp. sponge extract. (A) The high-performance liquid chromatography (HPLC) bioassay profiling was performed using the retention time-based fractionation approach together with (B) the assessment of chemokine inhibition (1 and 4 μg/ml) in neurofibroma cells CRL-3390. Results are presented as mean ± S.E.M. (n = 3). *p < 0.05, **p < 0.01, *** p < 0.001, ****p < 0.0001 compared with the control (DMSO) on the reduction samples.
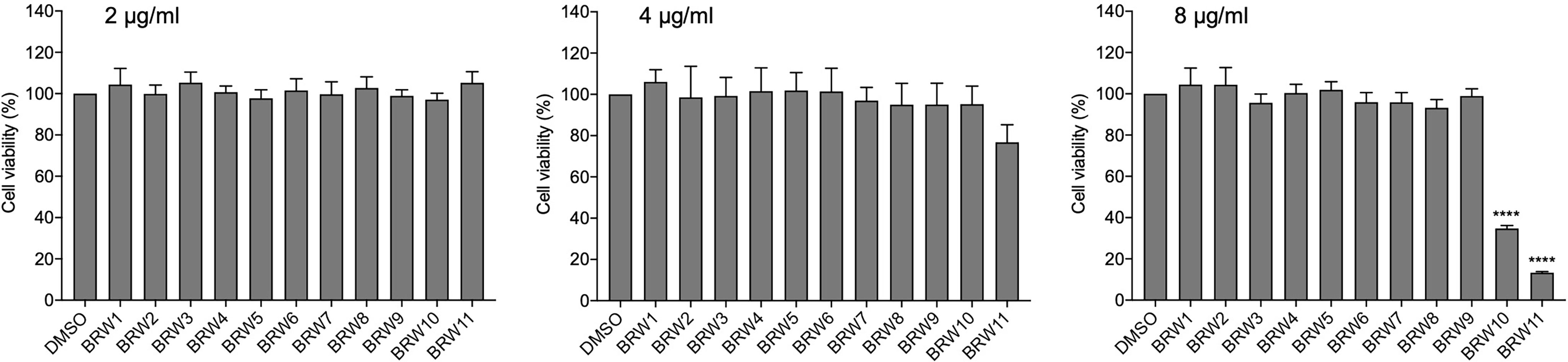
Figure 3 The CRL-3390 neurofibroma cell viability at various concentrations (2, 4, and 8 μg/ml) of BRW1–11 treatments. Results are presented as mean ± S.E.M. (n = 5). ****p < 0.0001 compared with the control (DMSO).
Characterizing the Bioactive Chemical Classes Using Tandem Mass Spectroscopy and MIMN Approach
Tandem MS is the most commonly used platform in the holistic profiling of natural products because it provides meaningful information related to the structural characterization of complex mixtures. Recently, the tandem MS-derived MN approach has been used in studies on natural products derived from microorganisms, marine organisms, fungi, plants, and other biological sources (Allard et al., 2016; Caesar et al., 2018). To clarify the relationships between the chemical diversity and anti-chemokine properties among these sponge-derived fractions, we combined the MN approach with bioactivity data to characterize the potential anti-neurofibroma constituents in terms of their chemical classes from the fractions and extract the Lendenfeldia sp. sponge.
First, an ultra-performance liquid chromatography quadrupole time-of-flight mass spectrometry (UPLC-QTOF-MS) was applied for the MS2 fragmentation collection in data dependent analysis (DDA) mode. The acquired MS2 data was then subjected to the Global Natural Product Social (GNPS) MN platform (https://gnps.ucsd.edu) for fragmentation similarity analysis. The compounds with similar chemical skeletons were clustered and visualized using Cytoscape software. In the constructed MIMN (Figure 4A), the pie charts were colored according to their CXCL10 inhibition (or rough distribution in each fraction). The compounds annotations and chemical classifications were carried out through GNPS database (or our in-house library) matches and ClassyFire software, which led to the putative identification of several major chemical classes (prenol lipids, benzenes, steroids, carboxylic acids, and organooxygen compounds) (Figure 4B) and four scalararane-type sesterterpenoids (Figures S1–S12): felixin B (1), 24-methyl-12,24,25-trioxoscalar-16-en-22-oic acid (2), 12α-acetoxy-22-hydroxy-24-methyl-24-oxoscalar-16-en-25-al (3), and 16-β-hrydroxy-24-methyl-12,24-dioxo-scalaran-25-al (4). These results suggested the prolific chemical diversity of scalarane-type metabolites (classified as prenol lipids). The anti-CXCL10 potential of these scalaranes was revealed due to their high average bioactivity scores [cluster A (83), B (85), and C (72); (Table S3)] presented in those clusters.
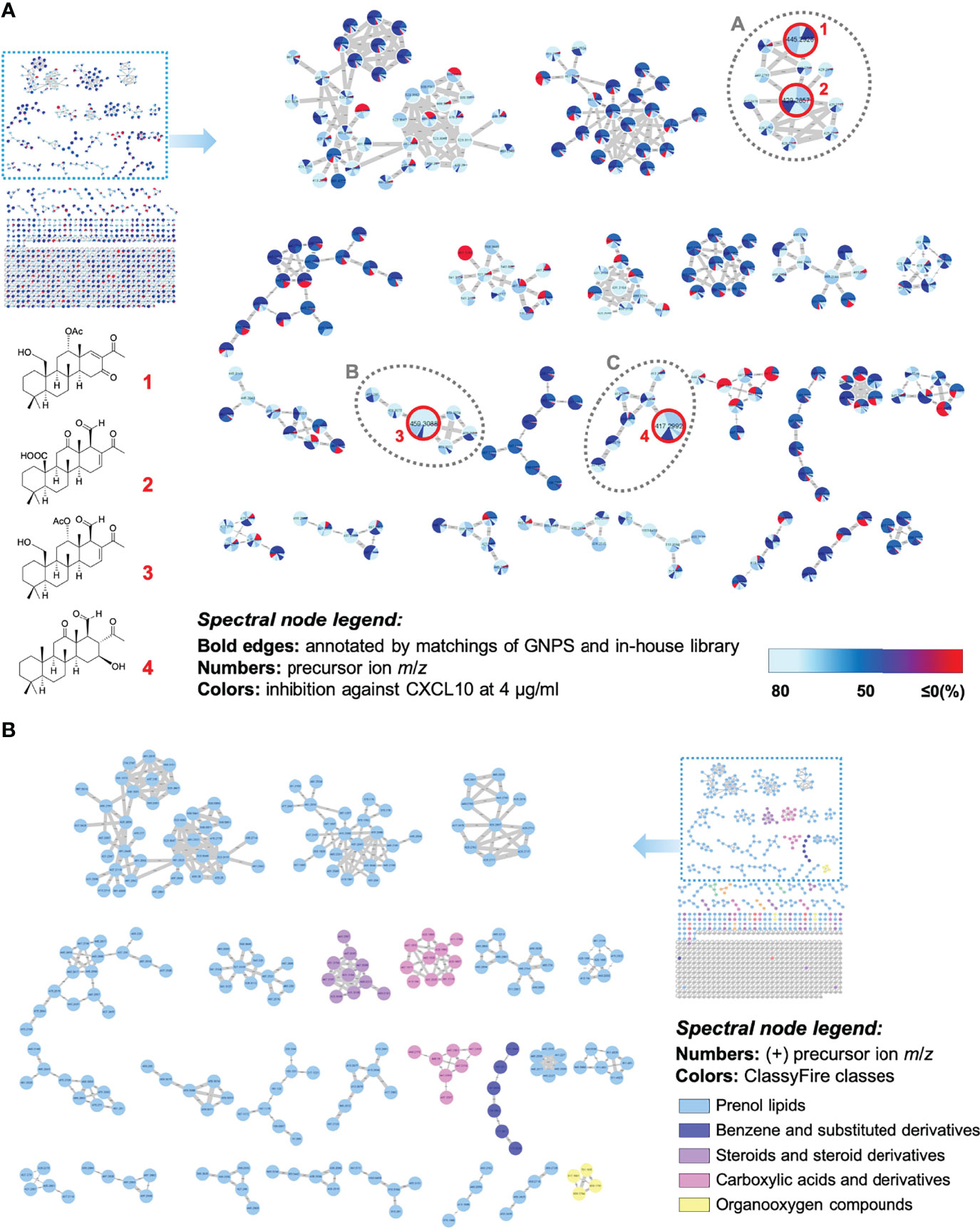
Figure 4 The multi-informative MN illustrated the relationships between scalarane diversity and anti-neurofibroma property in the Lendenfeldia sp. sponge extract. (A) The constructed MIMN is based on the inhibition of CXCL10 expression (4 μg/ml) in CRL-3390 cells; (B) the molecular network spectral nodes are colored according to ClassyFire classes.
MIMN-Guided Isolation and Validation of Anti-Neurofibroma Leads
After completing the comprehensive analysis of the anti-chemokine chemical skeletons, figuring out the biomarker and the most potent lead compound became the primary topic for the future development of Lendenfeldia sp. sponge as clinical anti-neurofibroma agents. Therefore, another MIMN was drafted based on the inhibition of CXCL10 expression (1 μg/ml) in CRL-3390 cells (Figure 5A). This multi-informative MIMN spectroscopic map indicated that the scalaranes from BRW7 possessed the most significant anti-CXCL10 potential. The most abundant annotated scalarane, 24-methyl-12,24,25-trioxoscalar-16-en-22-oic acid (2) was suggested to be the major bioindicator (Figure 5B). Then a series of isolation and purification experiments were conducted to probe compound 2 from BRW7. The ultimate validation of the anti-neurofibroma activity of 2 was achieved through the qPCR assay against CCL3 (55%), CCL4 (27%), CCL5 (74%), CXCL1 (24%), CXCL8 (35%), and CXCL10 (61%) expressions in CRL-3390 cells (Figure 5A). Among these genes inhibited by 2, we would like to highlight that the CCL5 (p = 0.0024) and CXCL10 (p = 0.0092) are the most highly suppressed chemokines with reliable statistical significance. These results confirmed the indicative role of scalarane (2) in the anti-neurofibroma activity of the Lendenfeldia sp. sponge extract.
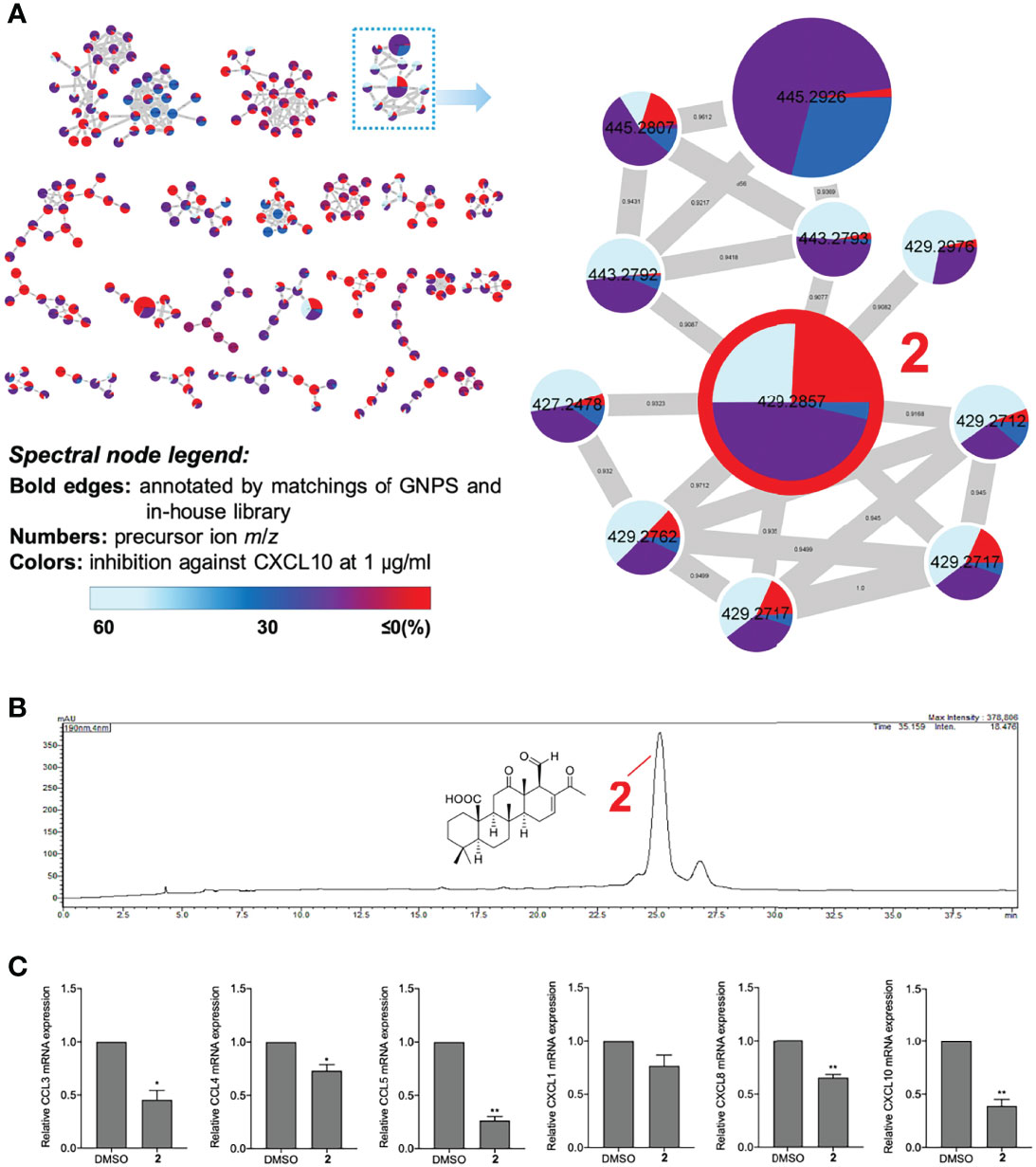
Figure 5 The visualized MIMN revealed the most potent anti-chemokine scalarane, 24-methyl-12,24,25-trioxoscalar-16-en-22-oic acid (2). (A) The MIMN was established according to the inhibition of CXCL10 expression (1 μg/ml) in CRL-3390 cells. (B) The isocratic HPLC-PDA chromatogram of BRW7 at 190 nm. (C) The assessment of chemokine inhibition (1 μM). Results are presented as mean ± S.E.M. (n = 3). *p < 0.05, **p < 0.01 compared with the control (DMSO).
The neurofibroma tumor is well recognized for its heterogeneous tumor microenvironment, especially in the immune cells (Yang et al., 2008; Prada et al., 2013; Brosseau et al., 2018; Liao et al., 2018). In this study, we strategically explored the anti-inflammatory potential of compounds extracted from the Lendenfeldia sp. sponge (Peng et al., 2020a; Peng et al., 2020b) in neurofibroma cells by analyzing their chemokine expressions after the compound treatment. Our results indicated that eight out of the eleven BRW fractions and the identified scalarane (2) demonstrated potent activities against the expression of CXCL10, a key tumor derived-molecule mediating the tumor-sustaining macrophage chemotaxis (Prada et al., 2013; Fletcher et al., 2019). This finding suggested the potential of 2 as well as other scalarane-type sesterterpenoids to serve as natural products to manage clinical manifestations caused by the overexpression of CXCL10, such as tumorous diseases (Liu et al., 2011b), autoimmune diseases (Lee et al., 2009), and infectious diseases (Liu et al., 2011a). These scalarane-type sesterpenoids have drawn much attention due to the recent findings as potent anti-inflammatory agents (Peng et al., 2020a; Chakraborty and Francis, 2021; Peng et al., 2021). However, research has been carried out herein to illustrate for the first time the anti-neurofibroma potential of these sponge-derived scalaranes. In addition to CXCL10, we also found that chemokines CCL3, CCL4, CCL5, CXCL1, and CXCL8 can be suppressed by certain extracts from the Lendenfeldia sp. sponge, and a single molecule 2 targeting all except CXCL1. These chemokines have also been implicated in the development of diseases (Raman et al., 2011). The data emphasized the anti-inflammatory activity of these sponge-derived natural products by downregulating chemokine expressions. Most current anti-chemokine approaches focus on developing small molecules against chemokine receptors (Mollica Poeta et al., 2019). Our data in this study revealed a relative uncommon success to directly downregulate chemokine expressions by a series of scalarane-enriched natural products extracted from the Lendenfeldia sp. sponge.
Conclusion
The breakthrough development of marine medicinal resources will be achieved through the application of new strategies and advanced spectroscopic technologies such as the novel multi-informative molecular networking (MIMN) application demonstrated in our current study. Through this promising approach, a significant anti-chemokine property of scalarane-type sesterterpenoids from the Lendenfeldia sp. sponge extract was suggested. Further biomarker investigation revealed a promising scalarane lead, 24-methyl-12,24,25-trioxoscalar-16-en-22-oic acid (2). Based on this achievement, the MIMN strategy will be employed to address holistically chemical-biological correlations and causations, which can be applied for future new drug discoveries.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
Author Contributions
K-HL, C-PL, and H-JW conceived and designed the experiments. B-RP, P-JS, and J-HS performed the sample collections and extraction. L-YC and C-HS carried out the fractionation and MS/MS analytical works and molecular networking data layout. The pharmacological experiments were executed by G-YL and C-PL. K-HL and C-PL contributed reagents and analysis tools. L-YC, B-RP, G-YL, K-HL, ME-S, C-PL, and H-JW participated in data interpretation, wrote and revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by a grant from the Ministry of Science and Technology, Taiwan (MOST 110-2320-B-038-034, 110-2320-B-038-014-MY2); Ministry of Education, Taiwan DP2-110-21121-01-N-12-01~03; and from Taipei Medical University (TMU109-AE1-B15, TMU109-AE1-B07).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at:https://www.frontiersin.org/articles/10.3389/fmars.2022.930736/full#supplementary-material
References
Allard P. M., Peresse T., Bisson J., Gindro K., Marcourt L., Pham V. C., et al. (2016). Integration of Molecular Networking and In-Silico MS/MS Fragmentation for Natural Products Dereplication. Anal. Chem. 88, 3317–3323. doi: 10.1021/acs.analchem.5b04804
Beniddir M. A., Kang K. B., Genta-Jouve G., Huber F., Rogers S., van der Hooft J. J. J. (2021). Advances in Decomposing Complex Metabolite Mixtures Using Substructure- and Network-Based Computational Metabolomics Approaches. Nat. Prod. Rep. 38, 1967–1993. doi: 10.1039/D1NP00023C
Brosseau J. P., Liao C. P., Wang Y., Ramani V., Vandergriff T., Lee M., et al. (2018). NF1 Heterozygosity Fosters De Novo Tumorigenesis But Impairs Malignant Transformation. Nat. Commun. 95014. doi: 10.1038/s41467-018-07452-y
Caesar L. K., Kellogg J. J., Kvalheim O. M., Cech R. A., Cech N. B. (2018). Integration of Biochemometrics and Molecular Networking to Identify Antimicrobials in Angelica Keiskei. Planta Med. 84, 721–728. doi: 10.1055/a-0590-5223
Chakraborty K, Francis P., and . (2021). Hyrtioscalaranes A and B, Two New Scalarane-Type Sesterterpenes From Hyrtios Erectus with Anti-Inflammatory and Antioxidant Effects. Nat. Prod. Res. 35, 5559–5570. doi: 10.1080/14786419.2020.1795854
Faleschini M. T., Maier A., Fankhauser S., Thasis K., Hebeisen S., Hamburger M., et al. (2019). A FLIPR Assay for Discovery of GABAA Receptor Modulators of Natural Origin. Planta Med. 85, 925–933. doi: 10.1055/a-0921-7602
Fletcher J. S., Wu J., Jessen W. J., Pundavela J., Miller J. A., Dombi E., et al. (2019). Cxcr3-Expressing Leukocytes are Necessary for Neurofibroma Formation in Mice. JCI Insight 4, e98601 doi: 10.1172/jci.insight.98601
Gutmann D. H., Ferner R. E., Listernick R. H., Korf B. R., Wolters P. L., Johnson K. J. (2017). Neurofibromatosis Type 1. Nat. Rev. Dis. Primers 3, 17004. doi: 10.1038/nrdp.2017.4
Jiang C., Mckay R. M., Le L. Q. (2021). Tumorigenesis in Neurofibromatosis Type 1: Role of the Microenvironment. Oncogene 40, 5781–5787. doi: 10.1038/s41388-021-01979-z
Kang K. B., Ernst M., van der Hooft J. J. J., Da Silva R. R., Park J., Medema M. H., et al. (2019). Comprehensive Mass Spectrometry-Guided Phenotyping of Plant Specialized Metabolites Reveals Metabolic Diversity in the Cosmopolitan Plant Family Rhamnaceae. Plant J. 98, 1134–1144. doi: 10.1111/tpj.14292
Lee E. Y., Lee Z. H., Song Y. W. (2009). CXCL10 and Autoimmune Diseases. Autoimmun. Rev. 8, 379–383. doi: 10.1016/j.autrev.2008.12.002
Liao C. P., Booker R. C., Brosseau J. P., Chen Z., Mo J., Tchegnon E., et al. (2018). Contributions of Inflammation and Tumor Microenvironment to Neurofibroma Tumorigenesis. J. Clin. Invest. 128, 2848–2861. doi: 10.1172/JCI99424
Liu M., Guo S., Hibbert J. M., Jain V., Singh N., Wilson N. O., et al. (2011a). CXCL10/IP-10 in Infectious Diseases Pathogenesis and Potential Therapeutic Implications. Cytokine Growth Factor Rev. 22, 121–130. doi: 10.1016/j.cytogfr.2011.06.001
Liu M., Guo S., Stiles J. K. (2011b). The Emerging Role of CXCL10 in Cancer (Review). Oncol. Lett. 2, 583–589. doi: 10.3892/ol.2011.300
Mollica Poeta V, Massara M, Capucetti A, Bonecchi R., ., ., and . (2019). Chemokines and Chemokine Receptors: New Targets for Cancer Immunotherapy. Front. Immunol. 10, 379. doi: 10.3389/fimmu.2019.00379
Olivon F., Retailleau P., Desrat S., Touboul D., Roussi F., Apel C., et al. (2020). Isolation of Picrotoxanes From Austrobuxus Carunculatus Using Taxonomy-Based Molecular Networking. J. Nat. Prod. 83, 3069–3079. doi: 10.1021/acs.jnatprod.0c00636
Peng B. R., Lai K. H., Chang Y. C., Chen Y. Y., Su J. H., Huang Y. M., et al. (2020a). Sponge-Derived 24-Homoscalaranes as Potent Anti-Inflammatory Agents. Mar. Drugs 18, 434 doi: 10.3390/md18090434
Peng B. R., Lai K. H., Chen Y. Y., Su J. H., Huang Y. M., Chen Y. H., et al. (2020b). Probing Anti-Proliferative 24-Homoscalaranes From a Sponge Lendenfeldiasp. Mar. Drugs 18, 76 doi: 10.3390/md18020076
Peng B. R., Lai K. H., Lee G. H., Yu S. S., Duh C. Y., Su J. H., et al. (2021). Scalarane-Type Sesterterpenoids From the Marine Sponge Lendenfeldia Sp. Alleviate Inflammation in Human Neutrophils. Mar. Drugs 19, 561 doi: 10.3390/md19100561
Prada C. E., Jousma E., Rizvi T. A., Wu J., Dunn R. S., Mayes D. A., et al. (2013). Neurofibroma-Associated Macrophages Play Roles in Tumor Growth and Response to Pharmacological Inhibition. Acta Neuropathol. 125, 159–168. doi: 10.1007/s00401-012-1056-7
Raman D, Sobolik-Delmaire T, Richmond A., ., and . (2011). Chemokines in Health and Disease. Exp. Cell Res. 317, 575-589. doi: 10.1016/j.yexcr.2011.01.005
Staser K., Yang F. C., Clapp D. W. (2012). Pathogenesis of Plexiform Neurofibroma: Tumor-Stromal/Hematopoietic Interactions in Tumor Progression. Annu. Rev. Pathol. 7, 469–495. doi: 10.1146/annurev-pathol-011811-132441
Watrous J., Roach P., Alexandrov T., Heath B. S., Yang J. Y., Kersten R. D., et al. (2012). Mass Spectral Molecular Networking of Living Microbial Colonies. Proc. Natl. Acad. Sci. U. S. A. 109, E1743–E1752. doi: 10.1073/pnas.1203689109
Woo S., Kang K. B., Kim J., Sung S. H. (2019). Molecular Networking Reveals the Chemical Diversity of Selaginellin Derivatives, Natural Phosphodiesterase-4 Inhibitors From Selaginella Tamariscina. J. Nat. Prod. 82, 1820–1830. doi: 10.1021/acs.jnatprod.9b00049
Yang F. C., Ingram D. A., Chen S., Zhu Y., Yuan J., Li X., et al. (2008). Nf1-Dependent Tumors Require a Microenvironment Containing Nf1+/– and C-Kit-Dependent Bone Marrow. Cell 135, 437–448. doi: 10.1016/j.cell.2008.08.041
Keywords: neurofibromas, multi-informative molecular networking, anti-inflammation, Lendenfeldia sp., scalarane
Citation: Chen L-Y, Peng B-R, Lai G-Y, Weng H-J, El-Shazly M, Su C-H, Su J-H, Sung P-J, Liao C-P and Lai K-H (2022) Chemometric-Guided Exploration of Marine Anti-Neurofibroma Leads. Front. Mar. Sci. 9:930736. doi: 10.3389/fmars.2022.930736
Received: 28 April 2022; Accepted: 06 June 2022;
Published: 13 July 2022.
Edited by:
Wolfram Brück, University of Applied Sciences and Arts of Western Switzerland, SwitzerlandReviewed by:
Chung-Kuang Lu, National Research Institute of Chinese Medicine, TaiwanJinshan Tang, Jinan University, China
Copyright © 2022 Chen, Peng, Lai, Weng, El-Shazly, Su, Su, Sung, Liao and Lai. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chung-Ping Liao, Y2h1bmdwaW5nbGlhb0B0bXUuZWR1LnR3; Kuei-Hung Lai, a3VlaWh1bmdsYWlAdG11LmVkdS50dw==
†These authors have contributed equally to this work
 Lo-Yun Chen
Lo-Yun Chen Bo-Rong Peng
Bo-Rong Peng Guan-Yi Lai3†
Guan-Yi Lai3† Hao-Jui Weng
Hao-Jui Weng Mohamed El-Shazly
Mohamed El-Shazly Jui-Hsin Su
Jui-Hsin Su Ping-Jyun Sung
Ping-Jyun Sung Chung-Ping Liao
Chung-Ping Liao Kuei-Hung Lai
Kuei-Hung Lai