
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mar. Sci., 30 June 2022
Sec. Marine Biology
Volume 9 - 2022 | https://doi.org/10.3389/fmars.2022.930156
This article is part of the Research TopicEchinoderms: Biology, Ecology and ExploitationView all 17 articles
A correction has been applied to this article in:
Corrigendum: Sea urchins in acute high temperature and low oxygen environments: The regulatory role of microRNAs in response to environmental stress
 Lingshu Han1
Lingshu Han1 Yanglei Wu2
Yanglei Wu2 Pengfei Hao2
Pengfei Hao2 Beichen Ding2
Beichen Ding2 Yuanxin Li2
Yuanxin Li2 Wenpei Wang2
Wenpei Wang2 Xianglei Zhang2
Xianglei Zhang2 Chuang Gao2
Chuang Gao2 Heng Wang2
Heng Wang2 Luo Wang2
Luo Wang2 Weijie Zhang2
Weijie Zhang2 Yaqing Chang2
Yaqing Chang2 Dewen Ding1
Dewen Ding1 Jun Ding2*
Jun Ding2*Strongylocentrotus intermedius is an economically valuable sea urchin species in China. However, its growth and survival are severely constrained by ocean warming and the hypoxia that often accompanies high water temperatures. MicroRNAs (miRNAs) are important regulators of gene expression in response to environmental change. In this study, high-throughput RNA sequencing was used to investigate changes in miRNA expression in S. intermedius under heat (25°C), hypoxia (2 mg/L O2), and combined heat and hypoxia stresses. Twelve small RNAs libraries were constructed and 17, 14, and 23 differentially expressed miRNAs (DEMs) were identified in the heat, hypoxia, and combined stress groups (P<0.05), respectively. Gene ontology and Kyoto Encyclopedia of Genes and Genomes pathway functional analyses of putative target genes of the DEMs suggested that these miRNAs were important in basal metabolism, apoptosis, oxidative stress, and immune-related pathways. By co-analysis with published transcriptome data, key DEMs (miR-193, miR-184, miR-133, miR-125, miR-2008) and their key target genes (EGF3, ABCB4, CYCL, PAN2, CALN) were identified. Quantitative real-time PCR analysis of the expression of 10 DEMs and their key target genes confirmed the RNA sequencing results. These results provide information on gene expression regulation of the molecular mechanisms underlying the response of S. intermedius to multi-cause environmental stresses.
Global warming has profoundly affected the nearshore and marine environments. The resulting environmental changes include increased seawater surface temperature, acidification, increased hypoxic zones, and extreme weather, which seriously threaten the reproduction of marine fishery species and the sustainable development of aquaculture (Blencowe, 2006; Xu et al., 2018; García Molinos, 2020). Among them, water temperature is the most fundamental and widespread ecological (environmental) factor that affects the survival of marine organisms (Pinsky et al., 2019). Increased seawater temperatures have been shown to impact the growth, metabolism, reproduction, and development of marine animals (García-Echauri et al., 2020; Gouda and Agatsuma, 2020; Strøm et al., 2020). Hypoxia caused by high temperatures, nutrient enrichment, water pollution, and high-density farming affects wild and farmed aquatic animals (Vaquer-Sunyer and Duarte, 2008; Breitburg et al., 2018). Dissolved oxygen levels of <2.8 mg/L (equivalent to 2 mL O2/L or 91.4 mM) are considered hypoxic. Very high summer temperatures and depleting aquaculture resources lead to hypoxic conditions that can be fatal or sub-lethal for sea urchins and can cause mass mortality (Riedel et al., 2014). Under hypoxic stress, significant changes in the oxygen consumption rate (Tomi et al., 2011) and the expression of immune and metabolic-related genes were found to occur in sea urchins (Suh et al., 2014; Hao et al., 2022). Therefore, hypoxia is another important factor that limits the survival and growth of sea urchins.
Sea urchins are a model organism for embryological development and a globally crucial marine fishery resource species (Nesbit et al., 2019). Among them, Strongylocentrotus intermedius, which has fast growth and excellent qualities, is China’s main sea urchin culture species, accounting for >90% of the total sea urchin culture in China (Chang et al., 2016). However, S. intermedius is a cold-water species that is sensitive to temperature changes. The high temperatures in the northern summers of 2017 and 2020 led to high mortality rates (up to 80%) of S. intermedius cultured in the Shandong and Liaoning provinces (Ding and Chang, 2020). Furthermore, between 1978 and 2014, the monthly average dissolved oxygen content in the Bohai Sea declined (Shi, 2016). These patterns suggest that environmental factors will seriously threaten the sustainable development of the aquaculture industry. Echinoderms have specific molecular biological mechanisms to respond to stress, such as histone modification, transcription, translation, and post-translational modification (Branco et al., 2013; Huo et al., 2018; Huo et al., 2021).
Various non-coding RNAs are known to be involved in post-transcriptional regulation. Among them, microRNAs (miRNAs) are small non-coding RNAs with about 18-25 nucleotides, which are endogenous regulatory RNAs found mainly in eukaryotes. Most miRNAs are transcribed from DNA sequences into primary miRNA, and processed into precursor RNAs by a series of nucleases to obtain mature miRNAs. The mature miRNAs are assembled into RNA-induced silencing complexes that recognize the target mRNAs through complementary base pairing and instruct the silencing complex to degrade or inhibit the translation of the target mRNAs according to the different degrees of complementation (Fabian et al., 2010; Fu, 2014; Zgheib et al., 2017). MiRNAs play irreplaceable roles in cell proliferation and differentiation, cell apoptosis, gene expression, restoration of homeostasis, and target recognition (Bartel, 2009; Leung and Sharp, 2010; Noman et al., 2017; Tian et al., 2019a; Tian et al., 2019b). Many studies have shown that miRNAs are activated in animals under environmental stress. For example, miR-210-5p and miR-92b-3p were highly activated in sea cucumber in response to the environmental pressures of hypoxia and high temperature (Huo et al., 2021). Sun et al. (2019) reported that miR-122, miR-15b-5p, miR-30b, miR-20a-5p, and miR-7b helped maintain the energy requirements of common carp (Cyprinus carpio) under high temperature stress by regulating glycolysis. In sea cucumbers under salinity stress, miR-2008 and miR-92a were shown to respond by regulating proteins and phospholipids (Tian et al., 2019b). In Chinese shrimp (Fenneropenaeus chinensis), novel-mir-76 and novel-mir-193 responded to high pH stress by regulating lipid metabolism, amino acid metabolism, and carbon metabolism pathways (Li et al., 2019). Multiple environmental factors can affect organisms in natural environments. In a previous study (Hao et al., 2022), we performed transcriptome-wide gene expression profiling by RNA sequencing (RNA-seq) of S. intermedius under short-term high temperature, hypoxia, and combined stresses. We found that exposure to the combined stresses resulted in a two-factor additive effect at the transcriptome level and had a broader effect on immune-related pathways in the S. intermedius than a single stress had (Hao et al., 2022). However, no reports on the involvement of miRNAs in the regulatory mechanism of sea urchins under acute environmental stresses have been published so far.
In this study, we used RNA-seq technology to screen and identify differentially expressed miRNAs (DEMs) under high temperature, low oxygen, and combined stresses. We analyzed the obtained DEMs jointly with published transcriptome data to further explore the molecular mechanisms of S. intermedius in response to multiple environmental stresses. The results provide information for selecting and breeding resistant sea urchins and provide a theoretical basis for healthy sea urchin culture.
Healthy 1-years-old S. intermedius (average test diameter: 35.74 ± 1.35 mm; average test height: 16.34 ± 1.27 mm; weight: 19.25 ± 0.37 g) were bought from Dalian Haibao Fishery Co., Ltd. (38°91′25′′N” 121°60′25′′E) in Dalian, Liaoning Province, China, then immediately transported to the Key Laboratory of Mariculture and Stock Enhancement in North China’s Sea, Ministry of Agriculture and Rural Affairs, Dalian Ocean University. All the S. intermedius were temporarily raised in the same environment for 14 days before the experiment. The breeding conditions are as follows: temperature, 15 - 16 °C; salinity, 31.22 - 31.36 ppt; pH, 8.15 - 8.25; and oxygen content, 8 - 9 mg/L. Sea urchins were fasted for 48 h before the start of the experiment to expel the intestinal contents.
According to the changes in temperature and dissolved oxygen in the Yellow Sea and the Bohai Sea in the past ten years (Song et al., 2020) and previous experiments (Hao et al., 2022), this study selected 25°C and 2 mg/L oxygen as the conditions of temperature and hypoxia stress. Healthy S. intermedius were randomly cultured in high temperature (HT group, 25 °C, 8 mg/L O2), low oxygen (LO group, 15 °C, 2 mg/L O2), combined stress of heat and hypoxia (HL group, 25 °C, 2 mg/L O2) and control group (NC group, 15 °C, 8 mg/L O2), using 6 S. intermedius per group. The stress experiment was carried out in the automatic temperature control system and the dissolved oxygen control system (Figure 1). The temperature control system’s temperature deviation was less than 0.5 °C, and the oxygen concentration deviation of the dissolved oxygen control system was less than 0.5 mg/L (Huo et al., 2018). Previous experiments showed that individuals died at 12 hours of combined stress, and transcriptome sequencing revealed significant changes in genes related to sea urchin immunity and metabolism at 12 hours of acute stress (Hao et al., 2022). Therefore, we selected 12 h as the time of stress. After 12 h, 6 sea urchins in each group were dissected on ice to absorb the coelomic fluid and centrifuged at 3000 rpm for 10 min at 4 °C. The coelomocyte of S. intermedius were collected and immediately stored in an -80 °C for further experiments.
Three coelomocyte samples were selected from every group of sea urchins. The extraction, quality testing, and purification of total RNA were performed according to Wang’s method (Wang et al., 2022). The experimental procedure was performed following the standard steps provided by Illumina, including library preparation and sequencing experiments. The Small RNA sequencing library was prepared using TruSeq Small RNA Sample Prep Kits (Illumina, San Diego, USA). The Illumina HiSeq 2500 (LC Sciences, Houston, Texas, USA) platform was used to sequence the constructed library, and the reading length was 1×50 bp on a single end. Data analysis was based on this literature (Huo et al., 2021). The obtained sequences with 18-25 nt were used to perform BLAST analysis in miR Base 22.0 database (http://microrna.sanger.ac.uk) to identify miRNAs and examine their variations.
To identify DEMs between treatment group (HT, LO, and HL) and control group (NC), the expression levels of miRNAs in the whole library were normalized, and then DEMs were screened using Student’s T-test. P<0.05 was considered significant. A heatmap was constructed using TBtools and clustered by row scale. Target genes of miRNAs were predicated by using Target Finder (http://www.targetfinder.org) and were subjected to the enrichment analysis of functions and pathways by GO and KEGG database (P-value<0.05).
Association analysis of DEM and differentially expressed genes (DEGs) were based on published transcriptome data (Database number: PRJNA776993). All possible positively and negatively correlated miRNA-mRNA pairs were predicted using ACGT101-CORR1.1. Based on the comprehensive analysis of DEGs and DEMs, we screened the negatively associated miRNA-mRNA pairs and constructed interaction networks using Cytoscape 2.8.3 software (http://www.cytoscape.org/).
To validate the results of RNA-seq, DEMs and target DEGs were randomly selected for real-time fluorescence quantitative PCR (qRT-PCR). Total RNA extraction and reverse transcription were performed by referring to Han et al. (Han et al., 2021). 18s rRNA and U6 RNA were used as internal reference gene for qRT-PCR, which was conducted with the LightCycler96 Real-time System (Roch, Switzerland) and followed by the manufacturer’s instructions of the FastStart Essential DNA Green Master (Roch, Switzerland). The PCR primers were designed and synthesized by Shanghai Sangon Biotech and the primer sequences are shown in Table 1. The qRT-PCR was performed in a 20 μL reaction sample containing 2 μL of cDNA, 10 μL of FastStart Essential DNA Green Master, 6 μL of ddH2O water, and 1 μL (10 mM) of each primer. The reaction conditions were followed by 40 cycles of 95°C for 30 s, 95°C for 5 s, 60°C for 32 s, 95°C for 15 s, 60°C for 60 s, 95°C for 15 s and 60°C for 15 s. Compared with the control gene, fold-change of expression levers was determined using the 2−ΔΔCT method (Livak and Schmittgen, 2001), both in miRNA and mRNA PCR amplification. Statistical analysis was performed using SPSS software version 19.0 (IBM, Armonk, NY, USA). The data are expressed as the mean ± standard error of the mean (SEM) (n=3). In the results of gene expressions, significant differences (P<0.05) for each variable were first detected using the one-way ANOVA test between different groups, followed by Tukey’s HSD test.
We constructed small RNA libraries from S. intermedius and sequenced them on an Illumina HiSeq2500 platform to identify the miRNAs involved in heat, hypoxia, and combined stresses. Twelve libraries were established from the four experimental groups. A total of 12,049,632 ± 1,609,813 (NC), 9,169,509 ± 1,182,352 (HT), 12,077,449 ± 2,083,876 (LO), and 12,214,860 ± 1,271,035 (HL) raw reads were obtained. After removing low-quality reads, we mapped the clean reads against the Rfam database, which contains rRNA, tRNA, snRNA, snoRNA families, and Repbase to filter unwanted sequences and obtain valid small RNA data. A total of 7,846,735 ± 1,822,577 (NC), 5,549,774 ± 3,009,597 (HT), 8,869,277 ± 1,415,005 (LO), and 9,613,111 ± 885,961 (HL) valid reads were obtained (Table 2). The length distribution statistics for the total number of filtered valid reads showed that most of the reads were 22-nt (Supplementary Figure 1).
The bioinformatic analysis identified 140 miRNAs in the four experimental groups; 70 known and 70 novel miRNAs (Table 3). By co-expression analysis of the four groups (three comparison groups), a total of 70 (55 co-expressed, HT 1, NC 14), 78 (61 co-expressed, LO 9, NC 8) and 82 (67 co-expressed, HL 13, NC 2) miRNAs were identified in the HT vs NC, LO vs NC, and HL vs NC comparisons, respectively (Figure 2).
We identified 17, 14, and 23 DEMs in the HT vs NC, LO vs NC, and HL vs NC comparisons, respectively, by analyzing the RNA-seq data. Among the 17 DEMs in the HT vs NC comparison, 7 were up-regulated and 10 were down-regulated. Among the 14 DEMs in the LO vs NC comparison, 8 were up-regulated and 6 were down-regulated. Among the 23 DEMS in the HL vs NC comparison, 14 were up-regulated and 9 were down-regulated (Supplementary Table 1). The heat map (Figure 3) shows that 42 of the DEMs in the three comparisons were significantly differentially expressed (P <0.05), and four of them (PC-3p-56283_77, lva-miR-2008-5p, lva-miR-219-5p_R-1, lva-miR-9-5p_R+1) were significantly different in all three comparison groups.
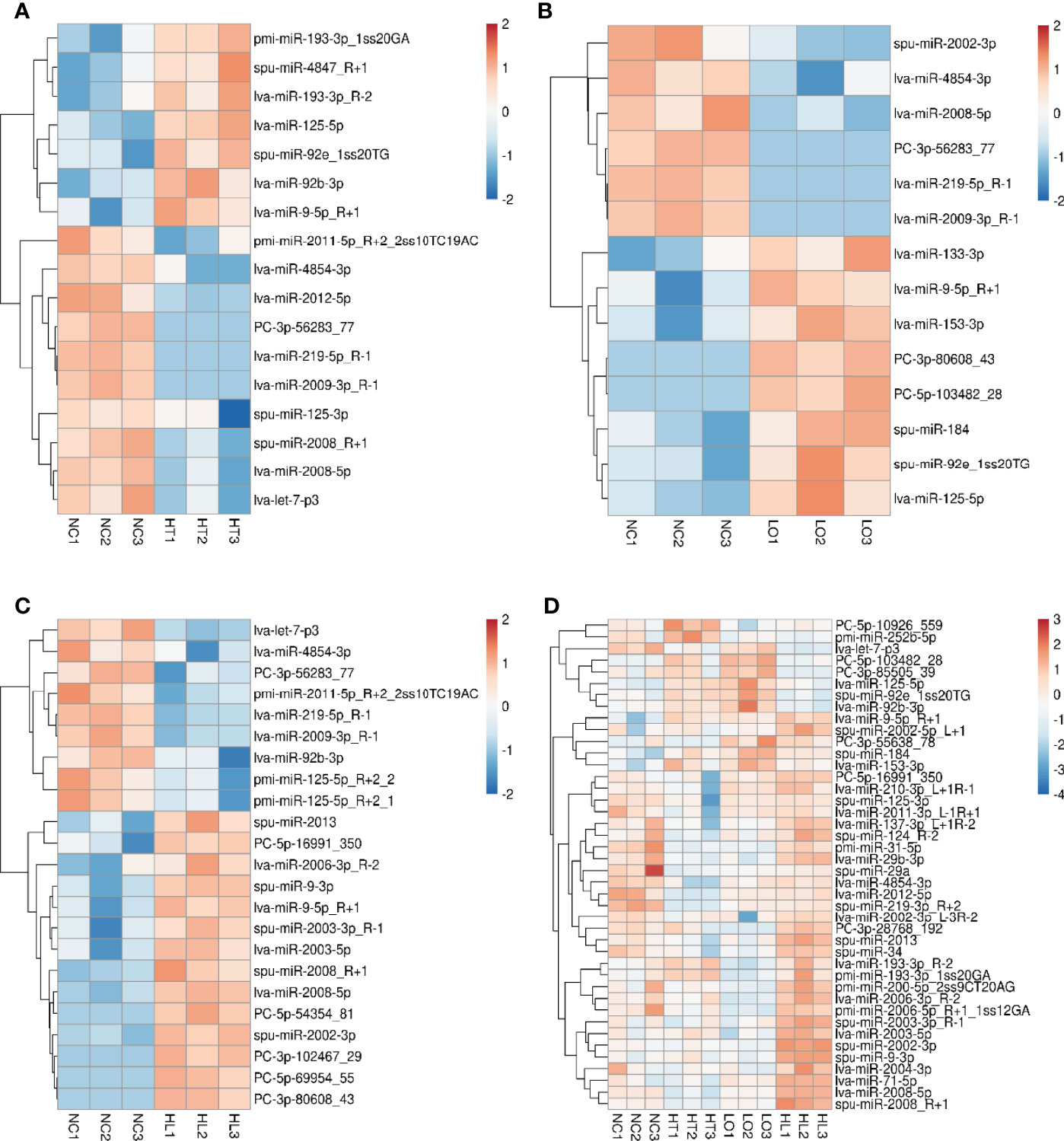
Figure 3 Differentially expressed miRNAs (DEMs) in the four groups. (A) HT vs NT, (B) LO vs NT, (C) HL vs NT, (D) DEMs (non-repetitive) in the pairwise comparison among the three treatments (P < 0.05). All red rectangles indicate higher levels of miRNAs, and blue rectangles indicate lower levels of miRNAs. Color scale bar represents log2FoldChange.
The roles of the identified DEMs in regulating the expression and function of their target genes were predicted by gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway functional enrichment analysis. A total of 69,912 target genes were predicted for the 17 DEMs identified in the HT vs NC comparison, and 12,825 of them were annotated to 9,879 GO terms under the three main GO categories: biological process (BP), cellular component (CC), and molecular function (MF). Under BP, biological process, signal transduction, and positive regulation of transcription by RNA polymerase II were the three most enriched terms (P <0.05). Under CC, integral component of membrane, membrane, and plasma membrane were the three most enriched terms. Under MF, calcium ion binding, serine-type endopeptidase activity, and GTP binding were the three most enriched terms (Figure 4A and Supplementary Table 2). In addition, 5,710 target genes were annotated to 388 KEGG pathways. Five of these pathways were significantly enriched, namely Glycosaminoglycan biosynthesis - chondroitin sulfate/dermatan sulfate (ko00532), Neuroactive ligand–receptor interaction (ko04080), mTOR signaling pathway (ko04150), FoxO signaling pathway (ko04068), and Drug metabolism - cytochrome P450 (ko00982) (Figure 5A and Supplementary Table 3).
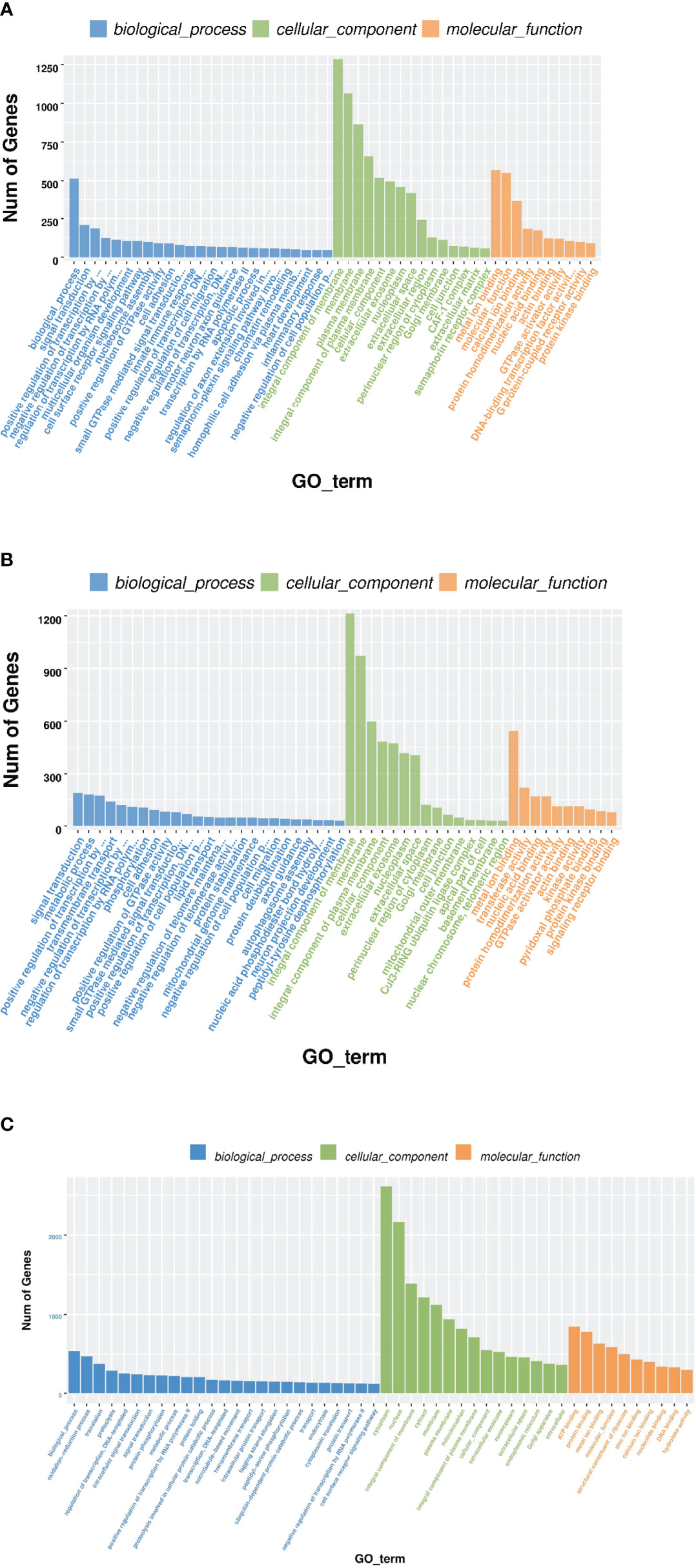
Figure 4 GO enrichment of target genes of DEMs. The x-axis is the rich factor, which indicates the proportion of genes in total genes in a GO term. The y-axis is the gene functional classification of GO. Various colors of plots indicate different values of −log 10 (P-value). Plot diameter represents target gene numbers in a GO term. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.). (A) HT VS NT, (B) LO vs NT, (C) HL vs NT.
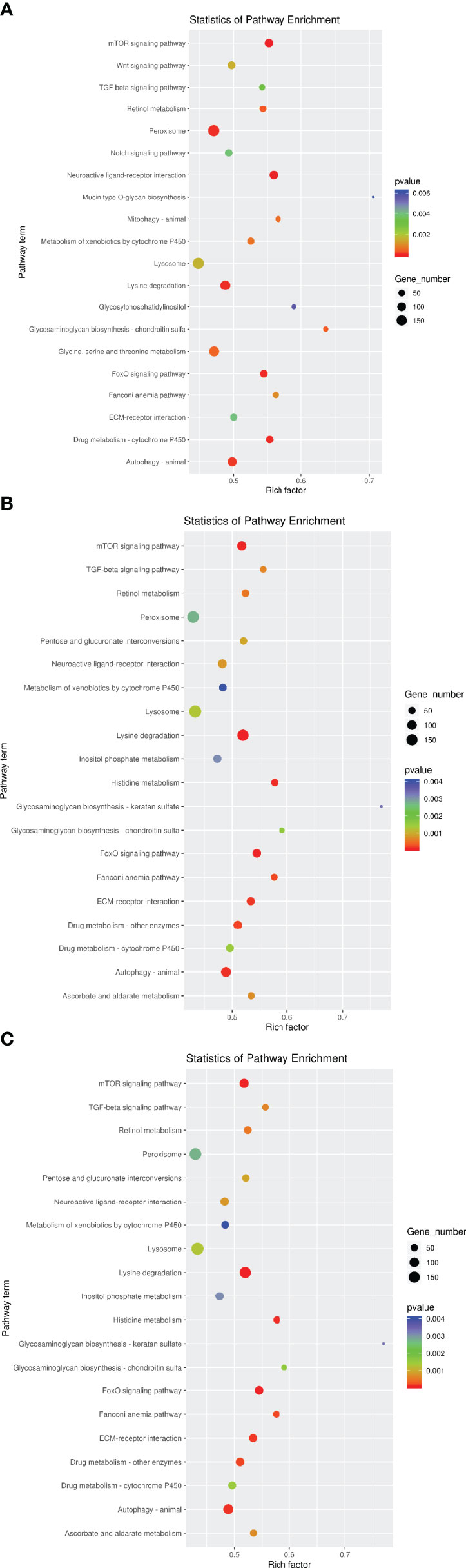
Figure 5 KEGG enrichment of target genes of DEMs. The x-axis is the rich factor, which means that the proportion of target genes in total genes in a KEGG term. The y-axis is the gene functional classification of KEGG. Various colors of plots indicate different values of −log 10 (P-value). Plot diameter represents target gene numbers in a KEGG term. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article). (A) HT VS NT, (B) LO vs NT, (C) HL vs NT.
A total of 56,715 target genes were predicted for the 14 DEMs identified in the LO vs NC comparison, and 12,217 of them were annotated to 9,592 GO terms. Under BP and CC, the three most enriched terms were the same as those for the HT vs NC comparison. Under MF, metal ion binding, transferase activity, and nucleic acid binding were the three most enriched terms (Figure 4B). A total of 5,471 target genes were annotated to 383 KEGG pathways. Four of these pathways were the most significantly enriched, namely Histidine metabolism, mTOR signaling pathway, ECM-receptor interaction, and FoxO signaling pathway (Figure 5B).
A total of 79,746 target genes were predicted for the 23 DEMs identified in the HL vs NC comparison, and 14,343 of them were annotated to 9,958 GO terms. Under BP, biological process, oxidation-reduction process, and signal transduction were the three most enriched terms (P <0.05). Under CC, integral component of membrane, plasma membrane, and mitochondrion were the three most enriched terms. Under MF, metal ion binding, zinc ion binding, and transferase activity were the three most enriched terms (P <0.05) (Figure 4C). Unlike the KEGG pathway annotations of the single stresses, the KEGG pathway annotations of the target genes of the 23 DEMs obtained for the combined stresses were mostly enriched in metabolic pathways, such as Glycosaminoglycan biosynthesis, Lysine degradation, Peroxisome, and Valine, leucine and isoleucine degradation and Fatty acid elongation (P <0.05) (Figure 5C).
The pre-constructed transcriptome library and previously identified DEGs (Hao et al., 2022) were used for co-analysis with the target genes of the DEMs. We identified 15 DEMs that interacted with 33 DEGs under high temperature stress by association analysis. Among them, the expression of 24 DEM–DEG pairs was negatively correlated and the expression of 32 DEM–DEG pairs was positively correlated. We also found 14 DEMs that interacted with 171 DEGs under hypoxia stress; 139 were negatively correlated DEM–DEG pairs and 108 were positively correlated DEM–DEG pairs. Under the combined stresses, 23 DEMs interacted with 102 DEGs; 69 were negatively correlated DEM–DEG pairs and 79 were positively correlated DEM–DEG pairs (Supplementary Table 4). The predicted DEM–DEG network map is shown in Figure 6. Meanwhile, we found that the key DEM in high-temperature stress was lva-miR-193-3p_R-2, whose predicted target genes were DHX33 and CSTF1; the key DEM in hypoxic stress was spu-miR-92e_1ss20TG, with nine essential target genes predicted to be negatively regulated, namely ZNFX1, Pola2, DDB_ G0276821, ACSM3, ctps1, PLOD1, GRID2IP, Pole, and FAU; the key DEM was lva-miR-9- in combined stresses, whose had 28 key negatively regulated target genes, mainly SOD, PAN2, and PRPF.
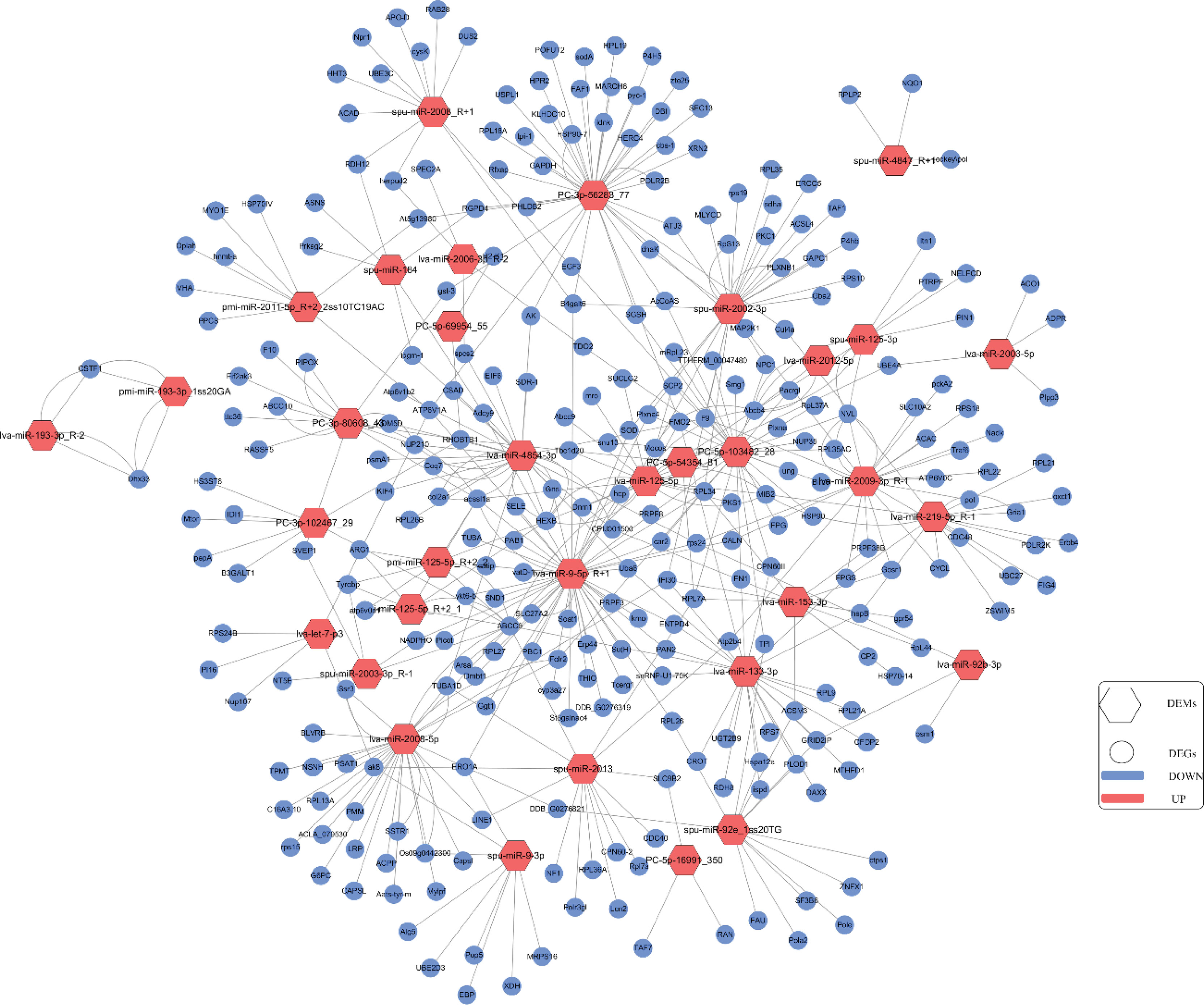
Figure 6 The network of DEMs and their target DEGs was constructed using Cytoscape 3.3.0. Hexagon is miRNA, circle is target DEGs, red means up-regulated expression, blue means down-regulated expression.
A qRT-PCR analysis was performed to validate the RNA-seq results. A total of 10 DEGs (including ABCB4, CYCL, EGF3, ZNFX1, CALN) and 10 DEMs (such as miR-193, miR-184, miR-133, miR-125, miR-2008) were selected for the qRT-PCRs. The results of the qRT-PCR analysis were consistent with the results obtained by the RNA-seq analysis and showed similar expression trends (Figure 7). In addition, the relative expressions of ABCB4, KIF4, ZNFX1, EGF3, DHX33, and PTRPF were significantly increased in high-temperature stress, and CALN, CUL4A, and PAN2 were up-regulated in hypoxic stress; CYCL genes showed a decreasing trend in high temperature and hypoxic stress, but their expressions were increased under combined stress.
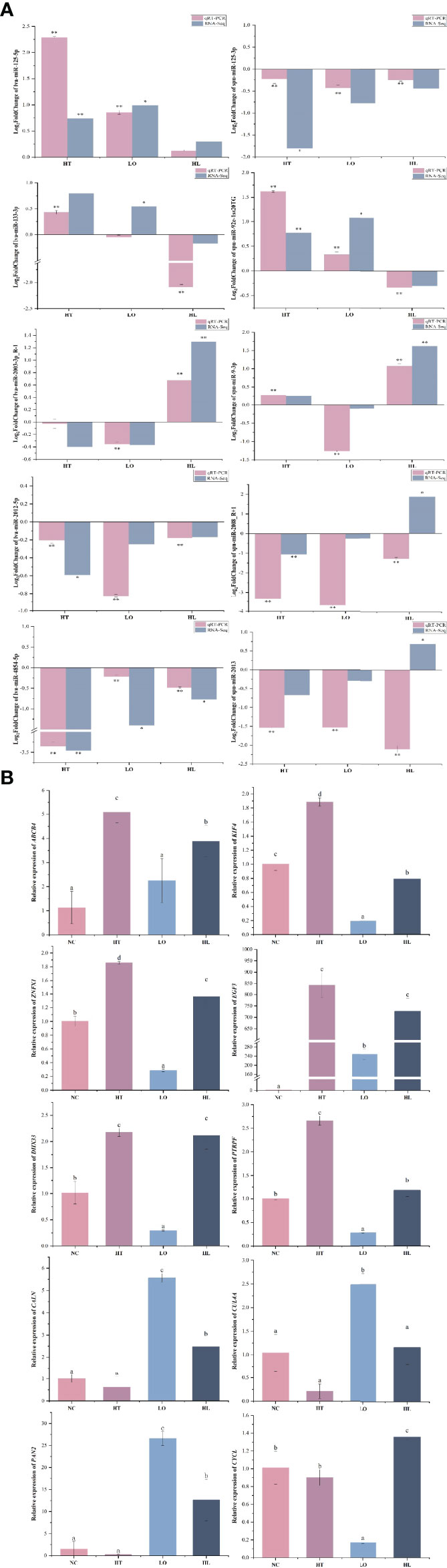
Figure 7 qRT-PCR verification of DEMs (A) and target DEGs (B). Each vertical bar represents the Mean ± SD (n=3), U6 and18s rRNA were used as a reference miRNA/gene. *Significant differences at P<0.05 vs control (NT). **Highly significant differences at P<0.01 vs control (NT). Letters above the bars indicate significant differences at P<0.05.
Many aquatic organisms are located in environmentally sensitive coastal zones or estuaries, which are extremely vulnerable to climate change. Water temperature and dissolved oxygen are the most important environmental factors that affect the survival of S. intermedius. MiRNAs are known to modulate gene expression and have critical roles in many biological processes, including cell proliferation, apoptosis, differentiation, cell cycle progression, and organ development (Tse et al., 2016; Biggar and Storey, 2018). In this study, miRNA libraries of sea urchins under heat, hypoxia, and combined stresses were constructed, sequenced by RNA-seq, and analyzed together with published transcriptome data to investigate the molecular mechanisms of sea urchins in response to different environmental conditions. The miRNAs were mainly distributed at 22-nt long, which is consistent with a previous study of S. intermedius (Zhan et al., 2018). A total of 140 miRNAs were identified; 70 were known and 70 were newly identified miRNAs. We identified 17, 14, and 23 DEMs in the HT, LO, and HL comparisons with NC, respectively, and found that they were involved in multiple key biological processes related to biosynthesis, metabolism, immunity, and signaling transduction by functional enrichment analysis.
These biological processes may be associated with major changes in the predicted target genes. We found changes in lysosomal pathways related to translation that were similar to the results of Huo et al. (2018). Regarding signal transduction, for the DEMs in the HT vs NC comparison, the mTOR and Wnt signaling pathways were among the enriched pathways; for the DEMs in the LO vs NC comparison, the FoxO signaling and Fanconi anemia pathways were among the enriched pathways; and for the DEMs in the HL vs NC comparison, the Hippo signaling and TGF-β signaling pathways were among the enriched pathways. Interestingly, the KEGG pathway annotations Lysine degradation, Peroxisome, Glycerolipid metabolism, and TGF-β signaling pathway were assigned to targent genes of DEMs in all three comparisons. Small amounts of lysine ingested by animals have been shown to cause triglyceride accumulation and significantly inhibit growth and development (Forster and Ogata, 1998; Ahmed and Khan, 2004). Peroxisomes are involved in multiple metabolic processes, including fatty acid oxidation, ether lipid synthesis, and reactive oxygen species (ROS) metabolism. Recent studies suggest that peroxisomes are critical mediators of cellular responses to various forms of stress, including oxidative stress, hypoxia, starvation, cold exposure, and noise (He et al., 2021a). Jain et al. discovered regulators of cell fitness in high and low oxygen conditions which led to the identification of an essential role for peroxisomes in metabolic adaptation to hypoxic stress (Jain et al., 2020). TGF-β signaling is a multifunctional pathway that controls cell proliferation, cell differentiation and tissue homeostasis (Hata and Chen, 2016). A study has found that TGF-β sensu stricto signaling plays an essential role in the formation of the embryonic skeleton of this sea urchin (Sun and Ettensohn, 2017). This finding together with the changes in the multi-metabolic and apoptosis-multi-species pathways, suggest that the metabolic system of sea urchins will be affected and apoptosis will occur under environmental stresses, in addition, multiple signal transduction pathways may work together to activate the same defense response. The DEMs were also involved in biosynthesis and metabolism-related pathways such as “Glucose metabolism”, “Serotonin metabolism”, and “Galactose metabolism”. These results indicates that sea urchins respond to the adverse effects of environmental stress through a variety of miRNA regulatory mechanisms.
MiRNA sequences are usually partially or totally matched with specific regions of the target genes. This interaction results in endonuclease digestion or translation inhibition, thereby negatively regulating the expression of the target gene. Therefore, identifying target genes that are differentially expressed is critical for understanding the biological functions of the encoded proteins.
MiR-193 plays an important role in cell growth, proliferation, and apoptosis. MiR-193 down-regulates the insulin growth factor 2 gene (IGF2) to induce cell proliferation and migration, which affect the angiogenic process (Yi et al., 2017). We found that miR-193 expression was up-regulated and its target genes DHX33 (putative ATP-dependent RNA helicase DHX33) and CSTF1 (cleavage stimulation factor subunit 1) were down-regulated under high temperature stress. DHX33 is a nucleoprotein involved in cell cycle regulation (Zhang et al., 2011), and CSTF1 encodes a nuclear protein that contains a ribonucleoprotein (RNP)-type RNA binding domain at its N-terminal end, which is essential for mRNA cleavage and polyadenylation. When DNA-damaged cells undergo oxidative stress, CSTF1 can bind to the BARD1/BRCA1 ubiquitin ligase heterodimer, thereby inhibiting 3′ end processing (Fontana et al., 2017). This finding and the regulation of alternative polyadenylation in development, differentiation, and neuronal activation suggest that 3′ end processing can be regulated in response to physiological and pathological stimuli. Therefore, we hypothesized that DNA was damaged under high temperature stress and miR-193a negatively regulated DHX33 and CSTF1 to promote apoptosis.
The expression of miRNA let-7 was also found to be down-regulated under high temperature stress. Members of the let-7 family are significantly elevated under high temperature stimulation, and are involved in regulating cell growth, differentiation, apoptosis, and metabolism (Gibadulinova et al., 2020; Tristán-Ramos et al., 2020; Huo et al., 2021). In the current study, the expression of lva-let-7-p3 was significantly down-regulated under heat stress and combined heat and hypoxic stresses. Lva-let-7-p3 may regulate potential target genes, including PI16 (peptidase inhibitor 16), ATP6V1A (V-type proton ATPase catalytic subunit A isoform X4), and Nup107 (nuclear pore complex protein Nup107 isoform X2).
MiR-184 was shown to be an important regulator of stem cell proliferation and growth (Liu et al., 2015), and its overexpression led to apoptosis and its suppression led to an increase in cell numbers (Foley et al., 2010; Liu et al., 2010). Previous studies have shown that miR-184 overexpression inhibited autophagy and exacerbated oxidative damage, and miR-184 also negatively regulated Wnt signaling in vivo and in vitro (Takahashi et al., 2015). MiR-184 was found to inhibit gene expression in human trabecular meshwork cell cytotoxicity, apoptosis, and the extracellular matrix by targeting the hypoxia-inducible factor HIF-1αin vivo, and it also exhibited angiostatic properties by regulating signaling pathways such as the Akt, TNF-α, and VEGF signaling pathways (Park et al., 2017). Furthermore, reduced expression of miR-184 was shown to inhibit cell growth through the CDC25A-dependent Notch signaling pathway (Cao et al., 2020).
MiR-133 is a key regulator of muscle proliferation and myocardial differentiation and is associated with cell proliferation and apoptosis (Chen et al., 2006; Uchida et al., 2013). The target of miR-133 was shown to be epidermal growth factor (EGF), and when EGF was down-regulated by miR-133, it inhibited its downstream signaling pathways, including the MAPK and AKT signaling pathways (Hernes et al., 2004; Dimauro et al., 2014). MiR-133 was also found to be an important component of the apoptotic pathway. In this study, we found that miR-133 expression was down-regulated under hypoxic stress and that its target genes HSP70, EGF, PAN2, and ABCC9 were up-regulated. In addition, the functional enrichment analysis of miRNA target genes under hypoxic stress showed that most of the genes were related to basal metabolism. Therefore, we hypothesized that miR-133 inhibited apoptosis by regulating HSP70, EGF, and other genes when sea urchins were exposed to hypoxia stress, and that metabolism-related genes were mobilized to maintain vital metabolism processes.
We found that the expression of lva-miR-125-5p was remarkably up-regulated in sea urchins under hypoxic conditions. Previous studies of miR-125-5p focused mainly on its role in regulating cell migration, apoptosis, immunity, proliferation, and cancer (Fassan et al., 2013; Natalia et al., 2018; Xu et al., 2019; Zheng et al., 2019; He et al., 2021b). In zebrafish (Danio rerio), the main target gene for hypoxia response was HIF-1a, which up-regulated the expression of miR-125 under hypoxic conditions (He et al., 2017). However, no up-regulation of HIF genes was detected in the present study. Previous studies have demonstrated that under hypoxic stress, the expression of HIF in the brain of the Wuchang bream (Megalobrama amblycephala) did not differ significantly from that of the control group (Shen et al., 2010). It is speculated that different species may have different hypoxia tolerance thresholds or species-specific differences in oxygen requirements. It might be duo to the short duration of the stress treatment in this study, we plan to further investigate the response of sea urchins under long-term hypoxic stress in future work. MiR-125 was shown to inhibit the expression of the mitofusin Mfn1 and reduce the disordered growth of pulmonary arterial smooth muscle cells under hypoxic conditions, as well as protect pulmonary blood vessels from mitochondrial dysfunction and abnormal remodeling (Ma et al., 2017). These findings provided a theoretical foundation for successful lung organ care. In this study, the potential target genes that may be regulated by lva-miR-125-5p included ZFP708 (zinc finger protein 708-like), ALDH (aldehyde dehydrogenase), KMT2A (lysine methyltransferase 2A), and MAP3K7 (mitogen-activated protein kinase kinase kinase 7). In the Gene Expression Omnibus (GEO) Profiles database (Tanya et al., 2007), ZFP708, ALDH, and MAP3K7 are recorded as being involved in the regulation of organisms in a hypoxic environment. These findings suggest that these genes may be involved in the response of sea urchins to low oxygen environments.
Under the combined stresses, spu-miR-2002-3p, which was highly expressed under hypoxic stress, was significantly up-regulated. This miRNA has not been reported in this context in previous studies. We found that under hypoxic stress, 14, 30, 5, 25, 39, and 11 target genes of spu-miR-2002-3p were enriched in neuroactive ligand–receptor interaction, lysine degradation, biosynthesis of unsaturated fatty acids, valine, leucine and isoleucine degradation, lysosome, and tryptophan metabolism KEGG pathways, respectively. The neuroactive ligand–receptor interaction signaling pathway involves all the receptors and ligands on the plasma membrane associated with intracellular and extracellular signaling pathways (Lauss et al., 2007). Under hypoxic stress, the cells pass through the surface of the sea urchin, then the receptors interact with extracellular ligands to trigger a series of metabolic changes. These processes may support the response of sea urchins to high temperature and low oxygen stress to protect them from damage.
We also identified miR-2008 and miR-9, which were significantly expressed under all three conditions. MiR-2008 was identified as a regulator of sea cucumber skin ulcer syndrome outbreaks by deep sequencing, and its target gene is the toll like receptor TLR3 (Li et al., 2012; Zhou et al., 2018). The expression of miR-2008 was also found to be up-regulated in sea cucumber under hypoxic stress. In the present study, miR-2008 expression was up-regulated under all three stress conditions, and the key target genes were EGF3, ABCC9, TUBA, and ACAD. We hypothesized that when S. intermedius is damaged by environmental stresses, the expression of genes related to apoptosis and immunity is significant. Our results support the view that miRNA–target gene interactions are complex, and a single miRNA can regulate multiple target genes simultaneously, and a gene can be regulated by multiple miRNAs simultaneously (Agarwal et al., 2015; Lan et al., 2016; Lai et al., 2022; Miao et al., 2022). The mechanism of the S. intermedius response to environmental stress is also complex. Compared with the effects of a single-factor stress, under multi-factor stress conditions, S. intermedius suffers an increased degree of organismal damage and responds comprehensively through metabolic and immune pathways. The qRT-PCR results further imply that there may be meaningful targeted regulatory interactions between the candidate DEMs and DEGs. In future research, the targeted regulatory interactions of the putative DEM–DEG pairs need to be confirmed by additional in vivo and in vitro experiments (e.g., dual luciferase reporter analysis, gain or loss of function analysis, and western blotting).
In this study, we report the miRNAs profiles of the sea urchin S. intermedius under high temperature, low oxygen, and combined stresses, and identified 17, 14, and 23 DEMs, respectively. By co-analysis with published transcriptome data, key DEMs (miR-193, miR-184, miR-133, miR-125, miR-2008) and their key target genes (EGF3, ABCC9, TUBA, PAN2, CALN) were identified. We found that the S. intermedius defense responses were activated by multiple miRNAs that regulate multiple signal transduction pathways in response to the adverse effects of environmental stress. Furthermore, under the combined heat and hypoxic stresses, the effects of both these factors were superimposed. The expression of target genes associated with apoptosis and oxidative stress was up-regulated, implying transcriptional regulation is a comprehensive response in sea urchins in the face of global climate change. Our results provide information on gene expression regulation of the molecular mechanisms underlying the response of S. intermedius to multi-cause environmental stress and provide a theoretical basis for healthy sea urchin reproduction.
The datasets presented in this study can be found in online repositories. The data presented in the study are deposited in the NCBI repository, accession number PRJNA828018.
JD, DD, and YC: conceptualization and resources. JD and BD: conceived and designed the experiment. BD, PH, and XZ: performed the experiment. PH, YL, and WW: data curation. LH, PH and YW: analyzed the data. WZ, HW, LW and CG: contributed reagents/materials/analysis tools. YW and LH: writing—original draft preparation. LH: writing—review and editing. JD: funding acquisition. All authors have read and agreed to the published version of the manuscript.
This work was financially supported by Liaoning Department of Natural Resources Promotion of Marine Economic Development Project No.84[2021], High-level Talent Support Grant for Innovation in Dalian [2020RD03], and Liaoning Province Higher Education Innovation Team and Innovative Talent Support Program Project [LT2019003].
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We thank Margaret Biswas, PhD, from Liwen Bianji (Edanz) (www.liwenbianji.cn/) for editing the English text of a draft of this manuscript.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2022.930156/full#supplementary-material
MiRNA, MicroRNA; DEGs, Different expression genes; DEMs, Different expression microRNA; GO, Gene Ontology; BP, Biological processes; MF, Molecular functions; CC, Cellular components; KEGG, Kyoto Encyclopedia of Genes and Genomes; qRT-PCR; Quantitative Real-time PCR.
Agarwal V., Bell G. W., Nam J. W., Bartel D. P. (2015). Predicting Effective microRNA Target Sites in Mammalian mRNAs. Elife 4, e05005. doi: 10.7554/eLife.05005
Ahmed I., Khan M. A. (2004). Dietary Lysine Requirement of Fingerling Indian Major Carp, Cirrhinus Mrigala (Hamilton). Aquaculture 235, 499–511. doi: 10.1016/j.aquaculture.2003.12.009
Bartel D. P. (2009). MicroRNAs: Target Recognition and Regulatory Functions. Cell 136, 215–233. doi: 10.1016/j.cell.2009.01.002
Biggar K. K., Storey K. B. (2018). Functional Impact of microRNA Regulation in Models of Extreme Stress Adaptation. J. Mol. Cell Biol. 10, 93–101. doi: 10.1093/jmcb/mjx053
Blencowe B. J. (2006). Alternative Splicing: New Insights From Global Analyses. Cell 126, 37–47. doi: 10.1016/j.cell.2006.06.023
Branco P. C., Borges J. C., Santos M. F., Jensch Junior B. E., Da Silva J. R. (2013). The Impact of Rising Sea Temperature on Innate Immune Parameters in the Tropical Subtidal Sea Urchin Lytechinus Variegatus and the Intertidal Sea Urchin Echinometra Lucunter. Mar. Environ. Res. 92, 95–101. doi: 10.1016/j.marenvres.2013.09.005
Breitburg D., Levin L. A., Oschlies A., Grégoire M., Chavez F. P., Conley D. J., et al. (2018). Declining Oxygen in the Global Ocean and Coastal Waters. Science 359, eaam7240. doi: 10.1126/science.aam7240
Cao Q., Xu W., Chen W., Peng D., Liu Q., Dong J., et al. (2020). MicroRNA-184 Negatively Regulates Corneal Epithelial Wound Healing via Targeting CDC25A, CARM1, and LASP1. Eye Vis. (Lond) 7, 35. doi: 10.1186/s40662-020-00202-6
Chang Y., Tian X., Zhang W., Han F., Chen S., Zhou M., et al. (2016). Family Growth and Survival Response to Two Simulated Water Temperature Environments in the Sea Urchin Strongylocentrotus Intermedius. Int. J. Mol. Sci. 17, 1356. doi: 10.3390/ijms17091356
Chen J. F., Mandel E. M., Thomson J. M., Wu Q., Callis T. E., Hammond S. M., et al. (2006). The Role of microRNA-1 and microRNA-133 in Skeletal Muscle Proliferation and Differentiation. Nat. Genet. 38, 228–233. doi: 10.1038/ng1725
Dimauro I., Grasso L., Fittipaldi S., Fantini C., Mercatelli N., Racca S., et al. (2014). Platelet-Rich Plasma and Skeletal Muscle Healing: A Molecular Analysis of the Early Phases of the Regeneration Process in an Experimental Animal Model. PLoS One 9, e102993. doi: 10.1371/journal.pone.0102993
Ding J., Chang Y. Q. (2020). Research Progress in Conservation and Utilization of Economic Echinoderm: A Review. J. Dalian Ocean Univ. 35, 645–656. (in Chinese)
Fabian M. R., Sonenberg N., Filipowicz W. (2010). Regulation of mRNA Translation and Stability by microRNAs. Annu. Rev. Biochem. 79, 351–379. doi: 10.1146/annurev-biochem-060308-103103
Fassan M., Pizzi M., Realdon S., Balistreri M., Guzzardo V., Zagonel V., et al. (2013). The HER2-Mir125a5p/Mir125b Loop in Gastric and Esophageal Carcinogenesis. Hum. Pathol. 44, 1804–1810. doi: 10.1016/j.humpath.2013.01.023
Foley N. H., Bray I. M., Tivnan A., Bryan K., Murphy D. M., Buckley P. G., et al. (2010). MicroRNA-184 Inhibits Neuroblastoma Cell Survival Through Targeting the Serine/Threonine Kinase AKT2. Mol. Cancer 9, 83. doi: 10.1186/1476-4598-9-83
Fontana G. A., Rigamonti A., Lenzken S. C., Filosa G., Alvarez R., Calogero R., et al. (2017). Oxidative Stress Controls the Choice of Alternative Last Exons via a Brahma-BRCA1-CstF Pathway. Nucleic Acids Res. 45, 902–914. doi: 10.1093/nar/gkw780
Forster I., Ogata H. Y. (1998). Lysine Requirement of Juvenile Japanese Flounder Paralichthys Olivaceus and Juvenile Red Sea Bream Pagrus Major. Aquaculture 161, 131–142. doi: 10.1016/S0044-8486(97)00263-9
Fu X. D. (2014). Non-Coding RNA: A New Frontier in Regulatory Biology. Natl. Sci. Rev. 1, 190–204. doi: 10.1093/nsr/nwu008
García-Echauri L. L., Liggins G., Cetina-Heredia P., Roughan M., Coleman M. A., Jeffs A. (2020). Future Ocean Temperature Impacting the Survival Prospects of Post-Larval Spiny Lobsters. Mar. Environ. Res. 156, 104918. doi: 10.1016/j.marenvres.2020.104918
García Molinos J. (2020). Global Marine Warming in a New Dimension. Nat. Ecol. Evol. 4, 16–17. doi: 10.1038/s41559-019-1037-5
Gibadulinova A., Bullova P., Strnad H., Pohlodek K., Jurkovicova D., Takacova M., et al. (2020). CAIX-Mediated Control of LIN28/let-7 Axis Contributes to Metabolic Adaptation of Breast Cancer Cells to Hypoxia. Int. J. Mol. Sci. 21, 4299. doi: 10.3390/ijms21124299
Gouda H., Agatsuma Y. (2020). Effect of High Temperature on Gametogenesis of the Sea Urchin Strongylocentrotus Intermedius in the Sea of Japan, Northern Hokkaido, Japan. J. Exp. Mar. Biol. Ecol. 525, 151324. doi: 10.1016/j.jembe.2020.151324
Han L., Quan Z., Han B., Ding B., Ding J. (2021). Molecular Characterization and Expression of the SiUCP2 Gene in Sea Urchin Strongylocentrotus Intermedius. J. Oceanol. Limnol. 39, 1523–1537. doi: 10.1007/s00343-020-0181-8
Hao P., Ding B., Han L., Xie J., Wu Y., Jin X., et al. (2022). Gene Expression Patterns of Sea Urchins (Strongylocentrotus Intermedius) Exposed to Different Combinations of Temperature and Hypoxia. Comp. Biochem. Physiol. Part D Genomics Proteomics 41, 100953. doi: 10.1016/j.cbd.2021.100953
Hata A., Chen Y. G. (2016). TGF-β Signaling From Receptors to Smads. Cold Spring Harb. Perspect. Biol. 8, a022061. doi: 10.1101/cshperspect.a022061
He A. Y., Dean J. M., Lodhi I. J. (2021a). Peroxisomes as Cellular Adaptors to Metabolic and Environmental Stress. Trends Cell Biol. 31, 656–670. doi: 10.1016/j.tcb.2021.02.005
He Y., Huang C. X., Chen N., Wu M., Huang Y., Liu H., et al. (2017). The Zebrafish miR-125c is Induced Under Hypoxic Stress via Hypoxia-Inducible Factor 1α and Functions in Cellular Adaptations and Embryogenesis. Oncotarget 8, 73846–73859. doi: 10.18632/oncotarget.17994
Hernes E., Fosså S. D., Berner A., Otnes B., Nesland J. M. (2004). Expression of the Epidermal Growth Factor Receptor Family in Prostate Carcinoma Before and During Androgen-Independence. Br. J. Cancer 90, 449–454. doi: 10.1038/sj.bjc.6601536
He W., Zhang N., Lin Z. (2021b). MicroRNA-125a-5p Modulates Macrophage Polarization by Targeting E26 Transformation-Specific Variant 6 Gene During Orthodontic Tooth Movement. Arch. Oral. Biol. 124, 105060. doi: 10.1016/j.archoralbio.2021.105060
Huo D., Sun L., Ru X., Zhang L., Lin C., Liu S., et al. (2018). Impact of Hypoxia Stress on the Physiological Responses of Sea Cucumber Apostichopus Japonicus: Respiration, Digestion, Immunity and Oxidative Damage. PeerJ 6, e4651. doi: 10.7717/peerj.4651
Huo D., Sun L., Sun J., Zhang L., Liu S., Su F., et al. (2021). Sea Cucumbers in a High Temperature and Low Dissolved Oxygen World: Roles of miRNAs in the Regulation of Environmental Stresses. Environ. Pollut. 268, 115509. doi: 10.1016/j.envpol.2020.115509
Jain I. H., Calvo S. E., Markhard A. L., Skinner O. S., To T. L., Ast T., et al. (2020). Genetic Screen for Cell Fitness in High or Low Oxygen Highlights Mitochondrial and Lipid Metabolism. CELL 181, 716–727. doi: 10.1016/j.cell.2020.03.029
Lai K. P., Tam N. Y. K., Chen Y., Leung C. T., Lin X., Tsang C. F., et al. (2022). miRNA–mRNA Integrative Analysis Reveals the Roles of miRNAs in Hypoxia-Altered Embryonic Development- and Sex Determination-Related Genes of Medaka Fish. Front. Mar. Sci. 8. doi: 10.3389/fmars.2021.736362
Lan C., Chen Q., Li J. (2016). Grouping miRNAs of Similar Functions via Weighted Information Content of Gene Ontology. BMC Bioinf. 17, 507. doi: 10.1186/s12859-016-1367-0
Lauss M., Kriegner A., Vierlinger K., Noehammer C. (2007). Characterization of the Drugged Human Genome. Pharmacogenomics 8, 1063–1073. doi: 10.2217/14622416.8.8.1063
Leung A. K., Sharp P. A. (2010). MicroRNA Functions in Stress Responses. Mol. Cell 40, 205–215. doi: 10.1016/j.molcel.2010.09.027
Li C., Feng W., Qiu L., Xia C., Su X., Jin C., et al. (2012). Characterization of Skin Ulceration Syndrome Associated microRNAs in Sea Cucumber Apostichopus Japonicus by Deep Sequencing. Fish Shellfish Immunol. 33, 436–441. doi: 10.1016/j.fsi.2012.04.013
Liu X., Fu B., Chen D., Hong Q., Cui J., Li J., et al. (2015). miR-184 and miR-150 Promote Renal Glomerular Mesangial Cell Aging by Targeting Rab1a and Rab31. Exp. Cell Res. 336, 192–203. doi: 10.1016/j.yexcr.2015.07.006
Liu C., Teng Z. Q., Santistevan N. J., Szulwach K. E., Guo W., Jin P., et al. (2010). Epigenetic Regulation of miR-184 by MBD1 Governs Neural Stem Cell Proliferation and Differentiation. Cell Stem Cell 6, 433–444. doi: 10.1016/j.stem.2010.02.017
Livak K. J., Schmittgen T. D. (2001). Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Li Z., Wang J., He Y., Hu S., Li J. (2019). Comprehensive Identification and Profiling of Chinese Shrimp (Fenneropenaeus Chinensis) microRNAs in Response to High pH Stress Using Hiseq2000 Sequencing. Aquac. Res. 50, 14269. doi: 10.1111/are.14269
Ma C., Zhang C., Ma M., Zhang L., Zhang L., Zhang F., et al. (2017). MiR-125a Regulates Mitochondrial Homeostasis Through Targeting Mitofusin 1 to Control Hypoxic Pulmonary Vascular Remodeling. J. Mol. Med. (Berl) 95, 977–993. doi: 10.1007/s00109-017-1541-5
Miao B.-B., Niu S.-F., Wu R.-X., Liang Z.-B., Zhai Y. (2022). The MicroRNAs-Transcription Factors-mRNA Regulatory Network Plays an Important Role in Resistance to Cold Stress in the Pearl Gentian Grouper. Front. Mar. Sci. 8. doi: 10.3389/fmars.2021.824533
Natalia M. A., Alejandro G. T., Virginia T. J., Alvarez-Salas L. M. (2018). MARK1 is a Novel Target for miR-125a-5p: Implications for Cell Migration in Cervical Tumor Cells. Microrna 7, 54–61. doi: 10.2174/2211536606666171024160244
Nesbit K. T., Fleming T., Batzel G., Pouv A., Rosenblatt H. D., Pace D. A., et al. (2019). The Painted Sea Urchin, Lytechinus Pictus, as a Genetically-Enabled Developmental Model. Methods Cell Biol. 150, 105–123. doi: 10.1016/bs.mcb.2018.11.010
Noman A., Fahad S., Aqeel M., Ali U., Amanullah, Anwar S., et al. (2017). miRNAs: Major Modulators for Crop Growth and Development Under Abiotic Stresses. Biotechnol. Lett. 39, 685–700. doi: 10.1007/s10529-017-2302-9
Park J. K., Peng H., Yang W., Katsnelson J., Volpert O., Lavker R. M. (2017). miR-184 Exhibits Angiostatic Properties via Regulation of Akt and VEGF Signaling Pathways. FASEB J. 31, 256–265. doi: 10.1096/fj.201600746R
Pinsky M. L., Eikeset A. M., Mccauley D. J., Payne J. L., Sunday J. M. (2019). Greater Vulnerability to Warming of Marine Versus Terrestrial Ectotherms. Nature 569, 108–111. doi: 10.1038/s41586-019-1132-4
Riedel B., Pados T., Pretterebner K., Schiemer L., Steckbauer A., Haselmair A., et al. (2014). Effect of Hypoxia and Anoxia on Invertebrate Behaviour: Ecological Perspectives From Species to Community Level. Biogeosciences 11, 1491–1518. doi: 10.5194/bg-11-1491-2014
Shen R. J., Jiang X. Y., Pu J. W., Zou S. M. (2010). HIF-1alpha and -2alpha Genes in a Hypoxia-Sensitive Teleost Species Megalobrama Amblycephala: cDNA Cloning, Expression and Different Responses to Hypoxia. Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 157, 273–280. doi: 10.1016/j.cbpb.2010.06.013
Shi Q. (2016). Spatio-Temporal Mode for Inter-Annual Change of Dissolved Oxygen and Apparent Oxygen Utilization in Summer Bohai Sea. J. Appl. Oceanogr. 35, 243–255. (in Chinese)
Song G., Zhao L., Chai F., Liu F., Li M., Xie H. X. (2020). Summertime Oxygen Depletion and Acidification in Bohai Sea, China. Front. Mar. Sci. 7. doi: 10.3389/fmars.2020.00252
Strøm J. F., Thorstad E. B., Rikardsen A. H. (2020). Thermal Habitat of Adult Atlantic Salmon Salmo Salar in a Warming Ocean. J. Fish Biol. 96, 327–336. doi: 10.1111/jfb.14187
Suh S. S., Hwang J., Park M., Park S. Y., Ryu T. K., Lee S., et al. (2014). Hypoxia-Modulated Gene Expression Profiling in Sea Urchin (Strongylocentrotus Nudus) Immune Cells. Ecotoxicol. Environ. Saf. 109, 63–69. doi: 10.1016/j.ecoenv.2014.08.011
Sun Z., Ettensohn C. A. (2017). TGF-β Sensu Stricto Signaling Regulates Skeletal Morphogenesis in the Sea Urchin Embryo. Dev. Biol. 421, 149–160. doi: 10.1016/j.ydbio.2016.12.007
Sun J., Liu Q., Zhao L., Cui C., Wu H., Liao L., et al. (2019). Potential Regulation by miRNAs on Glucose Metabolism in Liver of Common Carp (Cyprinus Carpio) at Different Temperatures. Comp. Biochem. Physiol. Part D Genomics Proteomics 32, 100628. doi: 10.1016/j.cbd.2019.100628
Takahashi Y., Chen Q., Rajala R. V. S., Ma J. X. (2015). MicroRNA-184 Modulates Canonical Wnt Signaling Through the Regulation of Frizzled-7 Expression in the Retina With Ischemia-Induced Neovascularization. FEBS Lett. 589, 1143–1149. doi: 10.1016/j.febslet.2015.03.010
Tanya B., Troup D., Wilhite S. E., Pierre L., Dmitry R., Carlos E., et al. (2007). NCBI GEO: Mining Tens of Millions of Expression Profiles—Database and Tools Update. Nucleic Acids Res. 35, 760–765. doi: 10.1093/nar/gkl887
Tian Y., Li G., Bu X., Shen J., Tao Z., Chen L., et al. (2019a). Changes in Morphology and miRNAs Expression in Small Intestines of Shaoxing Ducks in Response to High Temperature. Mol. Biol. Rep. 46, 3843–3856. doi: 10.1007/s11033-019-04827-2
Tian Y., Shang Y., Guo R., Chang Y., Jiang Y. (2019b). Salinity Stress-Induced Differentially Expressed miRNAs and Target Genes in Sea Cucumbers Apostichopus Japonicus. Cell Stress Chaperones 24, 719–733. doi: 10.1007/s12192-019-00996-y
Tomi S., Stankovi S., Lucu Č. (2011). Oxygen Consumption Rate and Na+/K+-ATPase Activity in Early Developmental Stages of the Sea Urchin Paracentrotus Lividus Lam. Helgoland Mar. Res. 65, 431–434. doi: 10.1007/s10152-011-0261-4
Tristán-Ramos P., Rubio-Roldan A., Peris G., Sánchez L., Amador-Cubero S., Viollet S., et al. (2020). The Tumor Suppressor microRNA Let-7 Inhibits Human LINE-1 Retrotransposition. Nat. Commun. 11, 5712. doi: 10.1038/s41467-020-19430-4
Tse A. C. K., Rubio-Roldan A., Li J. W., Wang S. Y., Chan T. F., Lai K. P., et al. (2016). Hypoxia alters testicular functions of marine medaka through microRNAs regulation. Aquatic Toxicology 180, 266–273. doi: 10.1016/j.aquatox.2016.10.007
Uchida Y., Chiyomaru T., Enokida H., Kawakami K., Tatarano S., Kawahara K., et al. (2013). MiR-133a Induces Apoptosis Through Direct Regulation of GSTP1 in Bladder Cancer Cell Lines. Urol. Oncol. 31, 115–123. doi: 10.1016/j.urolonc.2010.09.017
Vaquer-Sunyer R., Duarte C. M. (2008). Thresholds of Hypoxia for Marine Biodiversity. Proc. Natl. Acad. Sci. U.S.A. 105, 15452–15457. doi: 10.1073/pnas.0803833105
Wang H., Zhao W., Liu X., Gang D., Zuo R., Han L., et al. (2022). Comparative Lipidome and Transcriptome Provide Novel Insight Into Polyunsaturated Fatty Acids Metabolism of the Sea Urchin. Front. Mar. Sci. 9. doi: 10.3389/fmars.2022.777341
Xu X., Lai Y., Zhou W., Hua Z. (2019). Lentiviral Delivery of a shRNA Sequence Analogous to miR-4319/miR-125-5p Induces Apoptosis in NSCLC Cells by Arresting G2/M Phase. J. Cell Biochem. 120, 14017–14027. doi: 10.1002/jcb.28676
Xu Y., Ramanathan V., Victor D. G. (2018). Global Warming Will Happen Faster Than We Think. Nature 564, 30–32. doi: 10.1038/d41586-018-07586-5
Yi F., Shang Y. G., Li B., Dai S. L., Wu W., Cheng L., et al. (2017). MicroRNA-193-5p Modulates Angiogenesis Through IGF2 in Type 2 Diabetic Cardiomyopathy. Biochem. Biophys. Res. Commun. 491, 876–882. doi: 10.1016/j.bbrc.2017.07.108
Zgheib C., Hodges M. M., Hu J., Liechty K. W., Xu J. (2017). Long non-Coding RNA Lethe Regulates Hyperglycemia-Induced Reactive Oxygen Species Production in Macrophages. PLoS One 12, e0177453. doi: 10.1371/journal.pone.0177453
Zhang Y., Forys J. T., Miceli A. P., Gwinn A. S., Weber J. D. (2011). Identification of DHX33 as a Mediator of rRNA Synthesis and Cell Growth. Mol. Cell. Biol. 31, 4676–4691. doi: 10.1128/MCB.05832-11
Zhan Y., Li Y., Cui D., Pei Q., Sun J., Zhang W., et al. (2018). Identification and Characterization of microRNAs From the Tube Foot in the Sea Urchin Strongylocentrotus Intermedius. Heliyon 4, e00668. doi: 10.1016/j.heliyon.2018.e00668
Zheng X., Wu Z., Xu K., Qiu Y., Su X., Zhang Z., et al. (2019). Interfering Histone Deacetylase 4 Inhibits the Proliferation of Vascular Smooth Muscle Cells via Regulating MEG3/miR-125a-5p/IRF1. Cell Adh. Migr. 13, 41–49. doi: 10.1080/19336918.2018.1506653
Keywords: strongylocentrotus intermedius, microRNA, global climate change, high temperature stress, hypoxia
Citation: Han L, Wu Y, Hao P, Ding B, Li Y, Wang W, Zhang X, Gao C, Wang H, Wang L, Zhang W, Chang Y, Ding D and Ding J (2022) Sea Urchins in Acute High Temperature and Low Oxygen Environments: The Regulatory Role of microRNAs in Response to Environmental Stress. Front. Mar. Sci. 9:930156. doi: 10.3389/fmars.2022.930156
Received: 27 April 2022; Accepted: 26 May 2022;
Published: 30 June 2022.
Edited by:
Mark Botton, Fordham University, United StatesReviewed by:
Lusheng Xin, Chinese Academy of Fishery Sciences (CAFS), ChinaCopyright © 2022 Han, Wu, Hao, Ding, Li, Wang, Zhang, Gao, Wang, Wang, Zhang, Chang, Ding and Ding. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jun Ding, ZGluZ2p1bjE5NzMxMTE5QGhvdG1haWwuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.