- 1Key Laboratory of Tropical & Subtropical Fishery Resource Application & Cultivation, Ministry of Agriculture and Rural Affairs, Key Laboratory of Aquatic Animal Immune Technology of Guangdong Province, Pearl River Fisheries Research Institute, Chinese Academy of Fishery Science, Guangzhou, China
- 2Maoming Branch, Guangdong Laboratory for Lingnan Modern Agriculture, Maoming, China
Nile tilapia (Oreochromis niloticus) is one of the most important commercial freshwater fish in China, and dietary transition occurs in their different life stages. The gut microbiota is important to host health. The relationships among the diet, gut microbiota, and development of tilapia are not well known. In the present study, we attempted to understand how diet is associated with microbiota community dynamics during the development of tilapia. The first experiment was performed under standard laboratory feeding operation to determine the effect of diet transition on intestinal microbiota. In the second experiment, tilapia were fed with Artemia or plant-based dry (PBD) food from the fish started feeding to their late juvenile stage (90 days post-fertilization). The results in the first experiment showed that feeding habit transition in juvenile fish had a low effect on the microbiota of the tilapia intestine. In the second experiment, plant-based food negatively affected the survival rate and intestinal development of tilapia. The phylum Planctomycetes was dominant in juvenile fish fed PBD food. The phylum Fusobacteria was dominant in the juvenile fish fed Artemia. At the genus level, Gemmobacter, Pirellula, and Planctomyces, belonging to the phylum Planctomycetes, were significantly abundant in the guts of fish fed the PBD food diet. Cetobacterium of the phylum Fusobacteria was dominant in juvenile fish fed Artemia. Thus, we can conclude that diet types have a great effect on the microbiota of tilapia intestine in their early life stages. The intestinal microflora of tilapia was established in juvenile tilapia, approximately 2 months after hatching. Our results provide useful information for the experimental design of studies on the microbial community of the tilapia gut. We suggest that modulation of gut microbiota of tilapia could be performed in their early life.
Introduction
The intestinal microbiota is considered an additional ‘organ’ of fish, which has beneficial effects on nutrition, development, and immunity (Ghanbari et al., 2015; Wang et al., 2018). These are of vital importance regarding fish health. Many researchers have reported that the community structure and function of the fish intestine are related to their diet (Ringø et al., 2016; Wei et al., 2018; Villasante et al., 2019). However, the gut microbiota could be modulated by dietary manipulations as well as by host factors, environmental factors, microbial factors, and individual variations (Ringø et al., 2016). To reduce these effects on the gut microbiota, we conducted research on the intestinal microbial community of a single large sibling all-male group of Nile tilapia (Oreochromis niloticus) throughout development.
Nile tilapia are one of the major fish in tropical and subtropical freshwater (Negassa and Prabu, 2008). Due to their high tolerance to environmental conditions and omnivorous feeding habits, tilapia have become one of the most important commercial fish in several countries throughout the globe, particularly China (Fishery Bureau, 2020). Tilapia have been recorded to undergo dietary transition (Le Roux, 1956; Ibrahim et al., 2015; Tesfahun and Temesgen, 2018). The fry prefer zooplankton (Le Roux, 1956). Juvenile tilapia depend on animal-based food, e.g., zooplankton and insect larvae, whereas adults feed mainly on macrophytes followed by phytoplankton (Tesfahun and Temesgen, 2018). Previous research found that eggs, rearing water, and live feed are possible sources of the intestinal microbiota of fish larvae (Wang et al., 2018). A study on zebrafish showed that fish development affected the intestinal microbial community (Stephens et al., 2016). The intestinal microbiota of blunt snout bream (Megalobrama amblycephala) was influenced by feed transition across development (Wei et al., 2018). Thus, diet and physiology over fish development probably interact, and both are involved in the establishment of intestine microbiota.
During the last decade, to reduce the use of fish meal and oil, sustainable alternative protein sources have been increasingly in demand in the aquaculture industry (Morais et al., 2012; Hansen and Hemre, 2013; Ringø et al., 2016). In the present study, we attempted to determine how diet-associated microbial communities change with the development of tilapia to reveal the possibility of using a plant-based diet for tilapia culture throughout their entire life. One experiment was performed under standard operating tilapia husbandry practices (Fujimura and Okada, 2007) to understand the effect of the diet transition on the gut microbiota, in which tilapia were fed with Artemia until 40 days post-fertilization (dpf). Then, dry pelleted food with fish meal was added to the diet to avoid interindividual variance in growth, and in that case, bigger fish could feed on both Artemia and dry pelleted food, while smaller fish feed on Artemia. After 60 dpf, tilapia were fed dry pelleted food with fish meal. In another experiment, to determine the effect of different diets on the establishment of gut microbiota, tilapia were fed with artemia or plant-based dry (PBD) food from fish started feeding to 90 dpf. The results will provide information on how diet affects the microbiota of tilapia intestine across development, and novel insights into the modulation of the gut microbiome of tilapia.
Materials and Methods
Experimental Design and Sample Collection
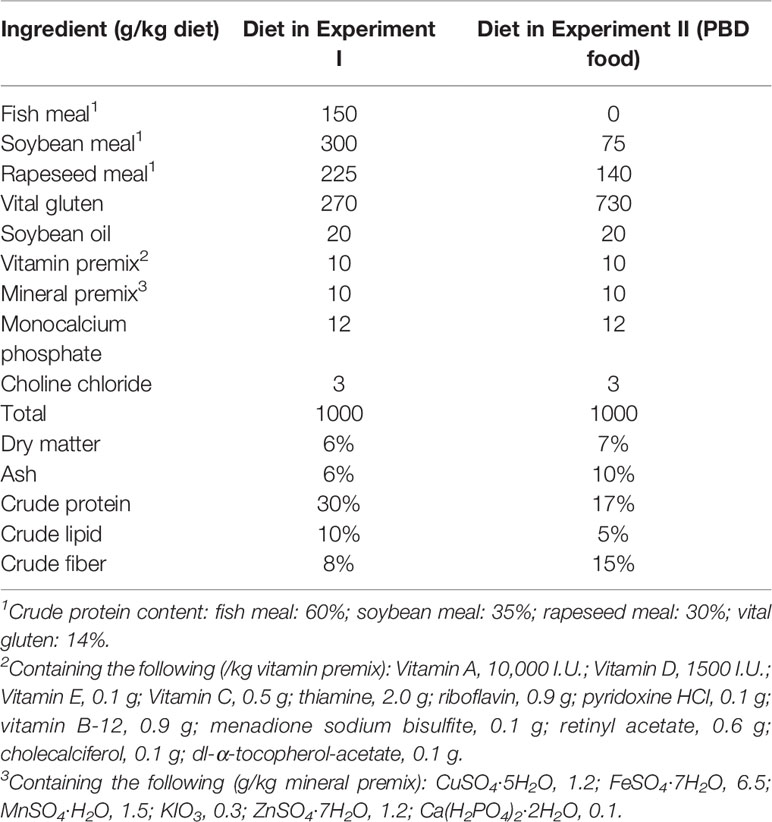
Experiment I: To reduce the potential impacts of host genotypic variation, we used offspring from a single mating pair in the experiment. All-male tilapia were obtained by crossing YY super-male tilapia with XX females, in which the genotypes of females and males were confirmed by sex-related molecular markers developed in our laboratory. The intestinal microbiota of offspring of a pair of adult Nile tilapia parents at multiple stages in their development was analyzed using high-throughput 16S rRNA gene sequencing. The offspring (all male) were cultured in glass tanks (45 cm× 35 cm× 35 cm) with three replicate tanks and 120 fish per tank. The fish were raised in a way reflecting commonly operated tilapia husbandry practices for indoor experiments (Fujimura and Okada, 2007) in an recycling aquaculture system with a water exchange rate of 6 m3/h. Tilapia were fed with Artemia from 11 dpf to 59 dpf. From 40 dpf, dry pelleted food was added to their diet to avoid interindividual variance in growth. From 60 dpf, tilapia were fed with totally dry pelleted food. Artemia was offered twice a day (1 nauplii/ml water volume) from 11 dpf (Drossou et al., 2006), and the amount was increased to 2 and 4 nauplii/ml water volume from 20 and 30 dpf. Dry feed was offered twice a day with 5% of the mean wet body weight from 40 dpf, and the amount was increased to 10% of the mean wet body weight from 60 dpf. Live Artemia were hatched from Artemia salina egg (Fengnian Aquaculture Co., Ltd, Tianjin, China) at 28°C for 24 h. Dry pelleted food (Nanhai Jieda Feed Co., Ltd., Foshan, China) contained 30% protein. During the whole experiment, water temperature was 27.5 ± 0.5°C. pH was 7.20 ± 0.10. Light: dark period was 12L:12D. Total nitrogen (TN), ammonia nitrogen (NH4-N), and nitrite nitrogen (NO2-N) content of water of the system was 0.22 ± 0.05 mg/L, 0.10 ± 0.03 mg/L, and 0.01 ± 0.002 mg/L. TN, NH4-N, and NO2-N levels in water were determined weekly using the spectrophotometric method according to Chinese Environmental Quality Standards HJ 636-2012, HJ 535-2009 and GB 7493-87. The dissolved oxygen (DO) concentration of water was 5.90 ± 0.90 mg/L. DO and pH were measured by multi-parameter system (HQ2100, Hach, USA). Tilapia and tank water were sampled at multiple time points to catch vital nutritional transitions: when fish relied on egg yolk for nutrition (11 dpf, before feeding), fish were fed Artemia (15, 20, and 40 dpf), dry pelleted food was added to the fish diet (60 dpf), and fish were fed dry pelleted food (90 dpf; Figure 1A). We sampled the intestines of multiple fish at each time point, resulting in 24 fish per time point for ages 11 and 20 dpf and 9 fish per time point for ages 40 to 90 dpf. The ingredients of the dry pelleted food diet are shown in Table 1.
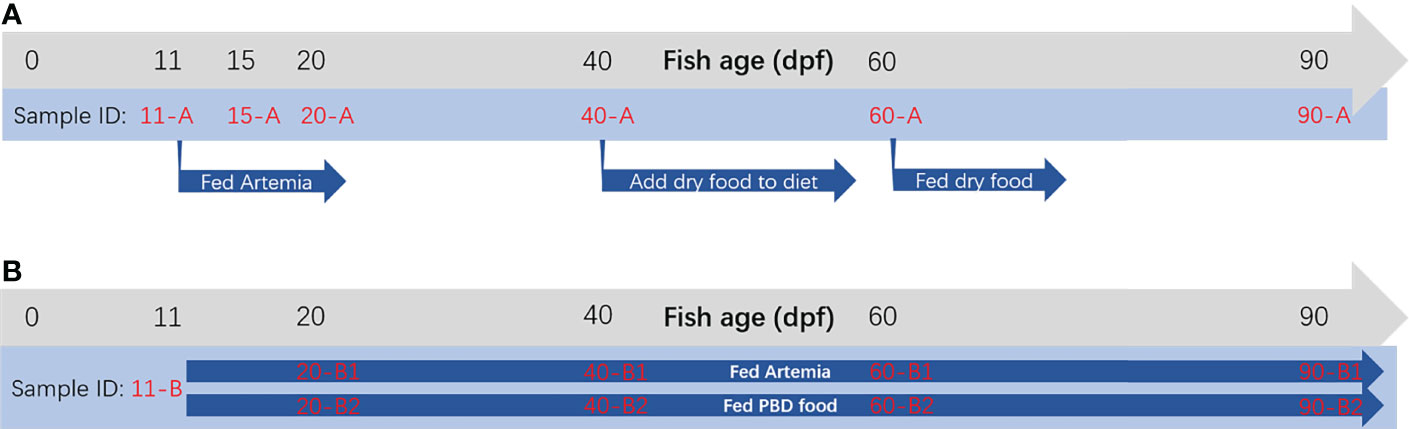
Figure 1 Experimental design. (A) Experiment I studied the effect of diet transition on the gut microbiota of tilapia. (B) Experiment II studied the effects of diet on the intestinal microbiota of tilapia after the first feeding. (A) 11-A, 15-A, 20-A, 40-A, 60-A and 90-A: tilapia at 11, 15, 20, 40, 60 and 90 dpf in Experiment I. (B) 11-B: tilapia at 11 dpf in Experiment II. 20-B1, 40-B1, 60-B1, and 90-B1: tilapia at 20, 40, 60 and 90 dpf fed Artemia in Experiment II. 20-B2, 40-B2, 60-B2, and 90-B2: tilapia at 20, 40, 60 and 90 dpf fed PBD food in Experiment II. The same applies below.
Experiment II: Offspring (all male) from a single mating pair were used in the experiment. The offspring were split evenly among six glass tanks (40 cm× 32 cm× 32 cm), with three replicate tanks for each group and 80 fish per tank. The fish in group one were fed Artemia, and the fish in group two were fed PBD food from 11 dpf (Figure 1B). The ingredients of the PBD food are shown in Table 1. The PBD food was grounded with mortar and pestle and sieved to a proper particle size according to the fish size. Feed was offered twice a day with 10% of the mean wet body weight. Water temperature was 26.50 ± 2.10°C. pH was 7.40 ± 0.20. Light: dark period was 12L:12D. Tank water was changed every week. TN, NH4-N, and NO2-N content of water of the system was 0.45 ± 0.12 mg/L, 0.30 ± 0.10 mg/L, and 0.02 ± 0.005 mg/L during the whole experiment. DO of tank water was 5.20 ± 0.50 mg/L. Fish were sampled at 11 dpf (before feeding), 20 dpf, 40 dpf, 60 dpf, and 90 dpf. The intestine of 24 fish per time point for ages 11 and 20 dpf and nine fish per time point for ages 40 to 90 dpf were sampled. The mortality of fish was recorded.
Tilapia from each time point were sampled from the facility before they were fed between 9 and 10 a.m. of the sampling day. The intestines were put into 2 ml screw cap tubes, frozen in liquid N2 and preserved at -80°C in a refrigerator before DNA extraction. Two hundred ml of water per tank was sampled. Water samples were filtered through a 0.22 μm cellulose nitrate filter. The filter was preserved at -80°C in a refrigerator for DNA extraction.
All tilapia experiments were performed in conformity with the Ethical Committee for Animal Experiments of Pearl River Fisheries Research Institute, Chinese Academy of Fishery Sciences, China. All tilapia experiments followed the guidelines of the Animal Welfare Council of China.
Intestinal Histology
Fish samples at 20 dpf and 40 dpf in Experiment II were collected and maintained in 4% formalin-buffered saline solution. Then, the whole fish samples were prepared for paraffin embedding, stained with Harris hematoxylin and eosin (HE), and sealed with a neutral gum. Observation of the morphological structure of the intestine was conducted under a photomicroscope (Nikon YS100, Japan).
Illumina Library Preparation and Sequencing
DNA from the samples was isolated using Quick-DNA™ Universal Kits (Zymo Research, USA) following the manufacturer’s instructions. To study the structure and diversity of the bacterial communities in samples, a protocol reported previously was used (Caporaso et al., 2010). PCR amplifications were performed with the 515f/806r primer set (5-GTGCCAGCMGCCGCGGTAA-3′ and 5-GGACTACHVGGGTWTCTAAT-3, accordingly) that amplifies the V4 region of the 16S rRNA gene. DNA was amplified according to a protocol reported previously (Magoč and Salzberg, 2011). Sequencing was performed on an Illumina Hiseq2500 PE250 platform.
FLASH constructed pairs of reads from the original DNA fragments (Edgar, 2013). Sequencing reads were assigned to each sample following the unique barcode of each sample. Sequences were analyzed with the QIIME2 and UPARSE pipeline. The OTUs sequence was annotated based on SILVA138 SSUrRNA database (http://www.arb-silva.de/), and the community composition of each sample was calculated according to the obtained taxonomic information.
Statistical Analysis
Alpha diversity was calculated using Simpson and Shannon indices with the VEGAN package in R (version: 3.6.0). Weighted UniFrac distances were calculated in GUniFrac to evaluate beta diversity. Principal coordinate analysis (PCoA) was conducted for beta diversity with the R (version: 3.6.0) package Ape. A phylogenetic tree was constructed using FastTree (version 2.1). Discriminatory analysis of taxonomic groups among different groups was conducted by a Kruskal-Wallis test with R (version: 3.6.0).
Results
Diet Affected the Dominant Microbial Taxa of the Intestinal Microbiota of Tilapia
The tilapia intestinal microbiota was characterized under diet changes during the larval to juvenile stages in Experiment I. Diet and environment were held constant during the early juvenile stages (15, 20, and 40 dpf). Dry food was added to the diet after 40 dpf to avoid individual variance in growth, and then fish of a mid-juvenile stage (60 dpf) were sampled. Tilapia were fed only dry food after 60 dpf, and after that, fish of a late-juvenile stage (90 dpf) were added to compare gut microbiota (Figure 1). In Experiment II, tilapia were divided into two groups, and within each group, diet and environment were kept constant during the whole experiment. In both experiments, sequencing depth was all > 50000 sequences per sample. The Shannon and Simpson diversity indices showed that larvae before feeding (11 dpf) had higher bacterial diversity than those at other stages, and the bacterial diversity decreased with the development of tilapia (Figure 2). In Experiment I, significant differences were found between 11 and 20 dpf of both the Shannon and Simpson diversity indices (p < 0.05).
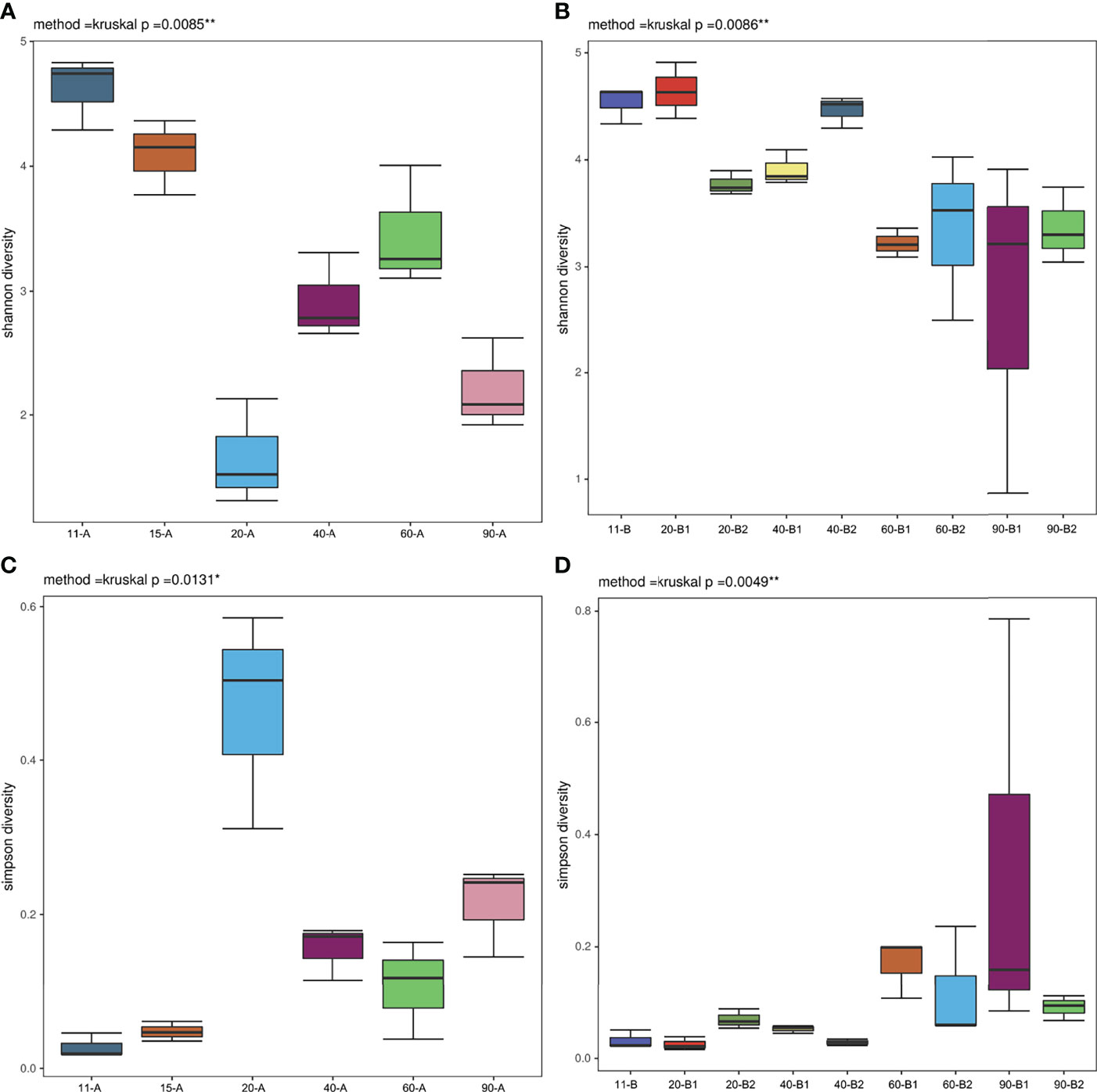
Figure 2 Alpha diversity measures at the OTU level. (A) Shannon index for Experiment I (n=3), (B) Shannon index for Experiment II (n=3), (C) Simpson index for Experiment I (n=3) and (D) Simpson index for Experiment II (n=3). *p < 0.05, **p < 0.01.
These changes in community diversity were accompanied by large changes in the phylum-level composition of fish before feeding (11 dpf) and fish fed different diets (15, 20, 40, 60, and 90 dpf), with especially significant differences in the composition of Planctomycetes in Experiment I (Figure 3A). Planctomycetes were the most abundant phylum of bacteria in larval fish (11 dpf), followed by the mid-juvenile fish (60 dpf) in Experiment I, and were particularly abundant in larval intestines along with water samples. In Experiment II, the relative composition of the phylum Planctomycetes also decreased after fish started feeding (20 dpf), except in one fish sample that was fed with PBD food (Figure 3B). The relative abundance of the phyla Planctomycetes and Proteobacteria was higher in juvenile fish (40, 60 and 90 dpf) in the PBD-food feeding group than in the Artemia feeding group. The relative abundance of Fusobacteria increased after fish started feeding (15 dpf) in Experiment I and showed a marked increase in the juvenile fish (60 and 90 dpf) of the Artemia feeding group in Experiment II. We noticed a high abundance of Fusobacteria OTUs within those intestines (20, 40, 60, and 90 dpf in Experiment I; 60 and 90 dpf in Experiment II), with the large proportion of these OTUs (> 98%) being classified in the genus Cetobacterium. In Experiment I, Proteobacteria and Bacteroidetes were abundant in all Artemia and water samples, which did not always show high abundance in intestinal samples (Figure 3A).
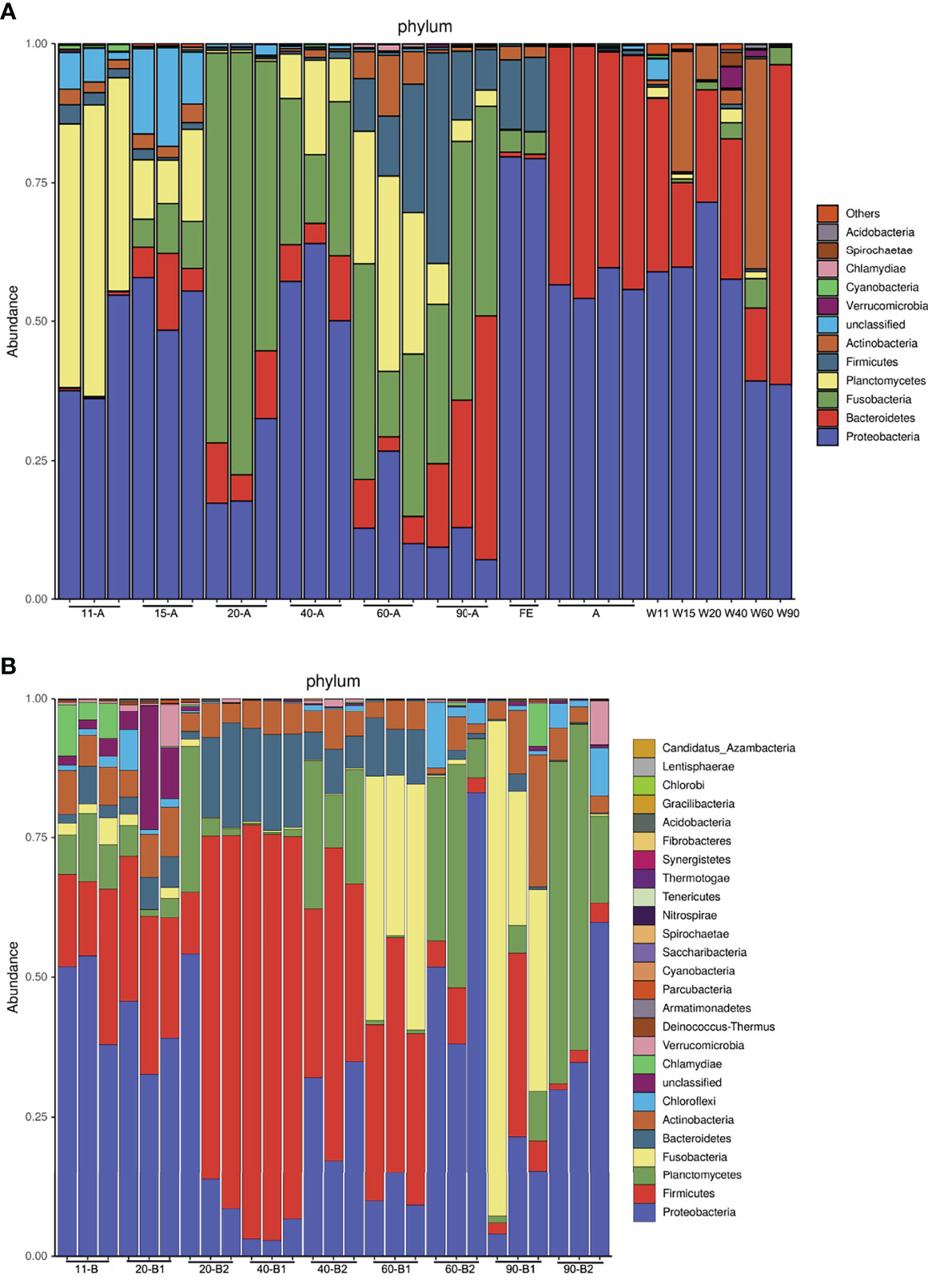
Figure 3 Taxonomic composition of the microbiota community at the phylum level. (A) Experiment I and (B) Experiment II. (A): 11-A, 15-A, 20-A, 40-A, 60-A and 90-A represent tilapia at 11, 15, 20, 40, 60 and 90 dpf with three duplicates; FE represents dry feed sampled when tilapia at 40 and 60 dpf; A represents Artemia sampled when tilapia at 15 and 20 dpf with two duplicates; W11, W15, W20, W40, W60 and W90 represent water sampled when tilapia at 11, 15, 20, 40, 60 and 90 dpf. (B): 11-B represents tilapia at 11 dpf; 20-B1, 40-B1, 60-B1, and 90-B1 repesent tilapia at 20, 40, 60 and 90 dpf fed Artemia; 20-B2, 40-B2, 60-B2, and 90-B2 represent tilapia at 20, 40, 60 and 90 dpf fed PBD food, all with three duplicates.
Nine genera, including Bacillus, Bosea, Cetobacterium, Gemmata, Gemmobacter, Paracoccus, Planctomyces, Plesiomonas, and Singulisphaera, were identified as core microbiota of the larval and juvenile tilapia intestine in both experiments. In the present study, core microbiota were genus identified in all the tested samples, whose relative abundance was more than 1% in at least one sample (Table 2). Additionally, 11 and 23 core genera were found in the intestines of samples in Experiment I and Experiment II, respectively.
Variation in Microbial Community Composition
Despite the genetic similarity of the tilapia, the composition of microbial intestinal communities displayed a variation among fish since the start of feeding, as calculated by the UniFrac distance (Figures 4A, B). In Experiment I, communities associated with individual tilapia showed a higher similarity to communities associated with tilapia of the same age than they were to those associated with tilapia of different ages (Figure 4C). In Experiment II, most of the fish fed the same diet clustered together (Figure 4D).
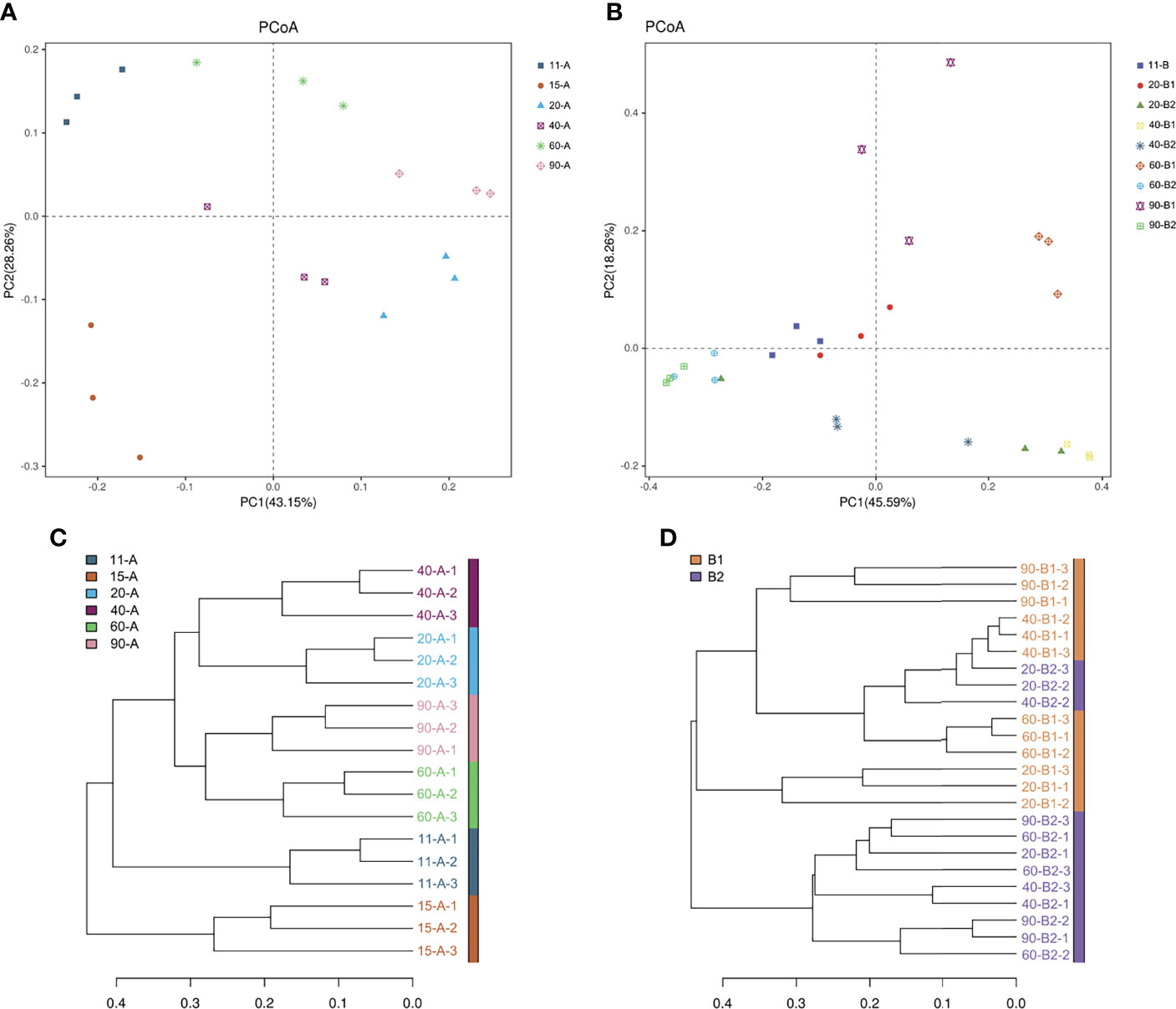
Figure 4 Beta diversity measures at the OTU level. (A) PCoA plot using beta-weighted UniFrac of Experiment I. (B) PCoA plot using beta-weighted UniFrac of Experiment II. (C) Clustering of intestinal samples from Experiment I according to the weighted UniFrac distance metric. (D) Clustering of intestinal samples from Experiment II according to the weighted UniFrac distance metric.
Discriminatory Taxa at the Genus Level
To identify taxonomic groups associated with the feed of tilapia, a nonparametric test of significance implemented by Kruskal-Wallis was used. The top 20 genera identified as discriminatory taxa are shown in Figure 5. In Experiment I, 15 dpf age tilapia, which had begun feeding on Artemia, showed a large presence of the genus Cetobacterium. In Experiment II, this genus showed high relative abundance in the Artemia feeding group for 60 and 90 dpf tilapia (all p < 0.05). Genus Planctomyces was present in 11 dpf age tilapia in both experiments. Its relative abundance in the 60 dpf age tilapia whose feed had added dry food was high in Experiment I, which was significantly higher than in the 15, 20, 40, and 90 dpf age fish (all p < 0.05). The relative abundance of the genus Planctomyces was higher in all the PBD groups than in the Artemia-feeding group for all the age tilapia, with significant differences between the two groups of 60 dpf age (p < 0.05). Gemmobacter and Pirellula were significantly more abundant in fish fed the PBD food diet for all age tilapia. For Pirellula, its relative abundance was significantly higher in the PBD-feeding group for 90 dpf fish (p < 0.05). If only the effect of diet was taken into account, Cetobacterium was significantly higher in the Artemia-feeding group than the PBD-feeding group (p < 0.01) (Figure 5C). While several genera, including Rhodobacter, Gemmobacter, Pirellula, Planctomyces, and Gemmata were significantly higher in the PBD-feeding group (all p < 0.05).
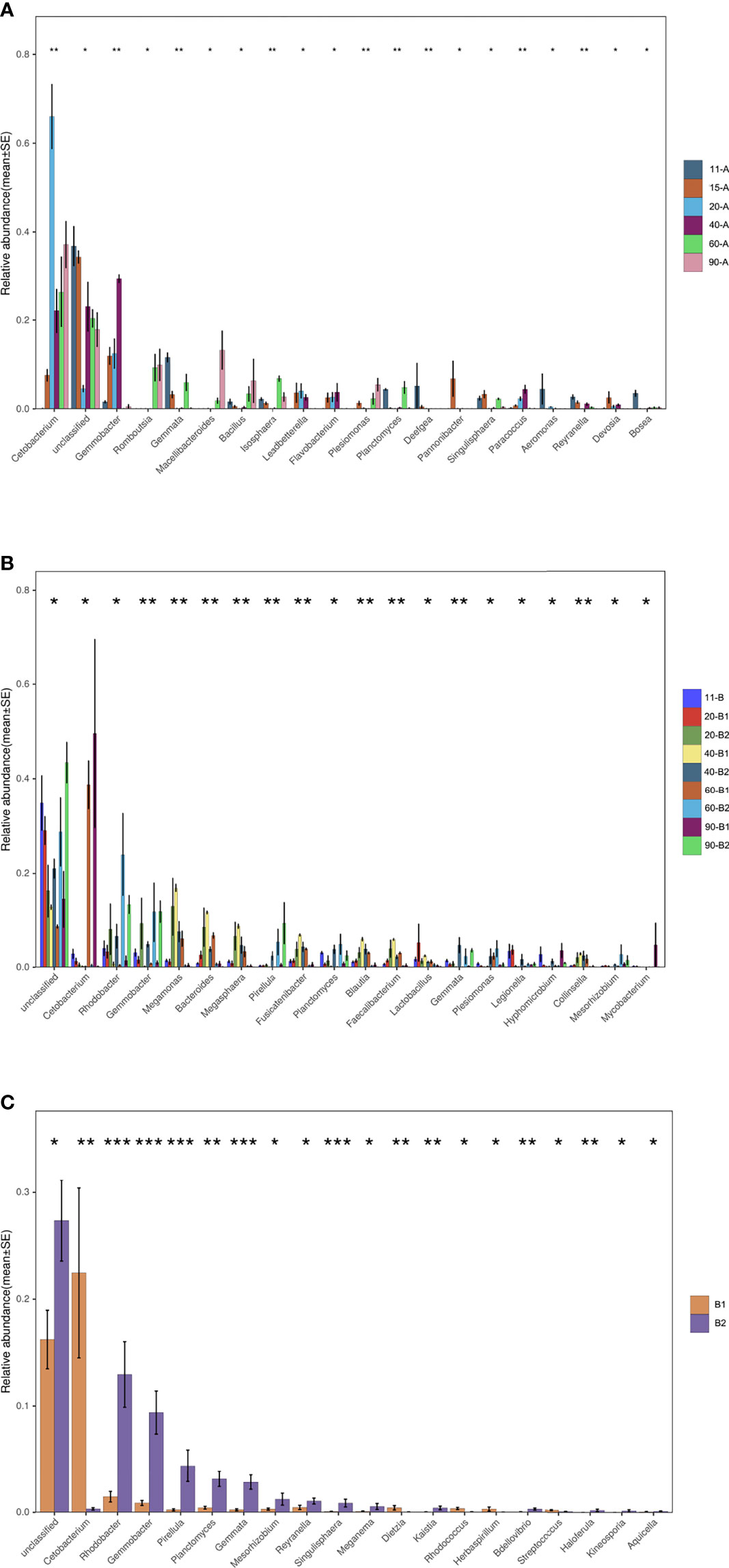
Figure 5 Discriminatory taxa of top 20 at the genus level. (A) Experiment I (n=3), (B) Experiment II (n=3) comparing all samples, and (C) Experiment II comparing between the two diet groups (n=12). *p < 0.05, **p < 0.01, ***p < 0.001.
Intestinal Structure of 20 dpf and 40 dpf Tilapia
Microphotographs showed that smooth muscle layers were easily separated from the lamina propria, and villi were damaged in the fish intestine of the PBD-feeding group in Experiment II (Figure 6).
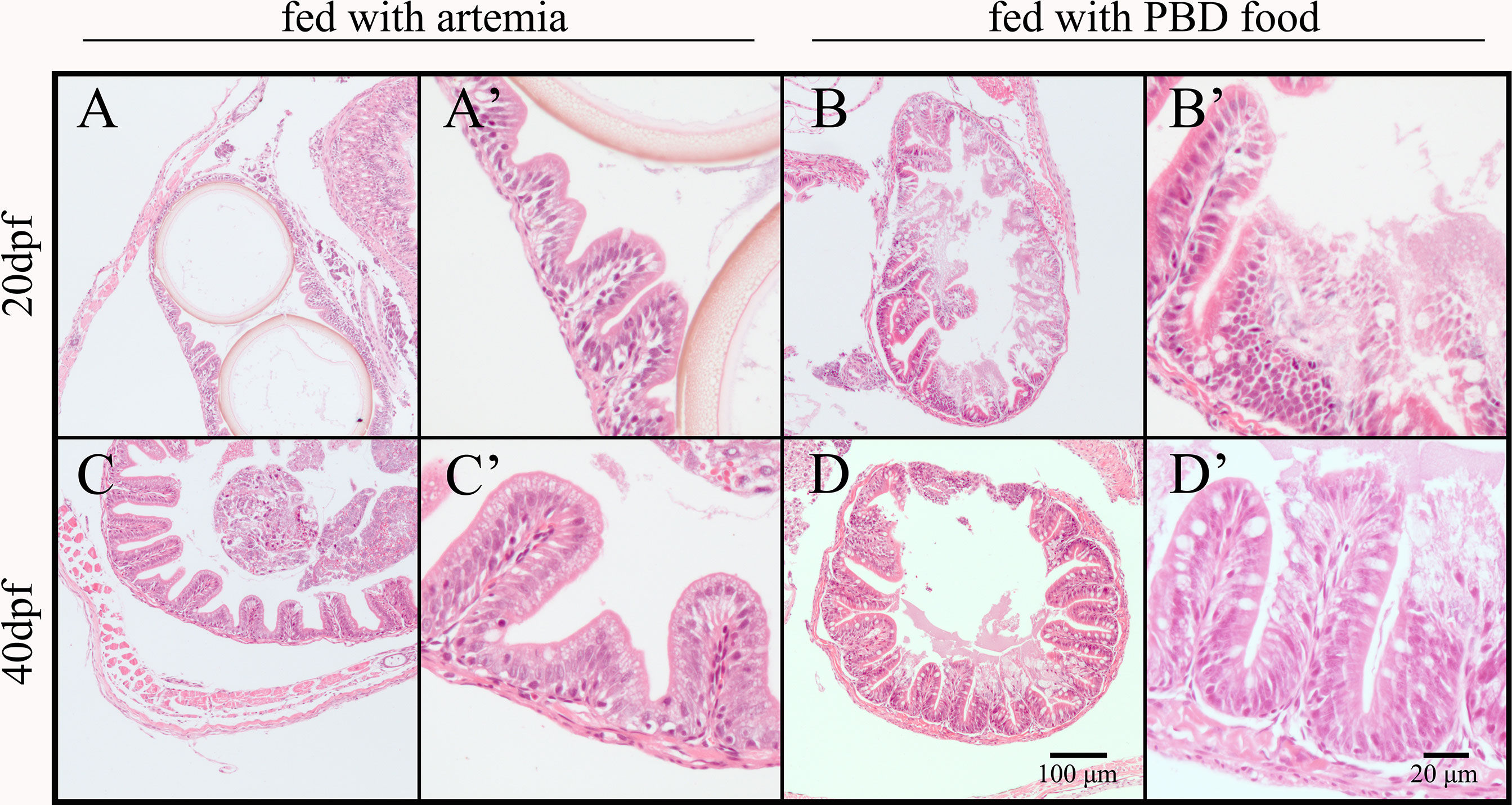
Figure 6 Intestinal morphology of Nile tilapia fed different diets (mid-intestine). (A) 20 dpf fish fed Artemia (10×); (A’) 20 dpf fish fed Artemia (40×); (B) 20 dpf fish fed PBD food (10×); (B’) 20 dpf fish fed PBD food (40×); (C) 40 dpf fish fed Artemia (10×); (C’) 40 dpf fish fed Artemia (40×); (D) 40 dpf fish fed PBD food (10×); (D’) 40 dpf fish fed PBD food (40×).
Cumulative Overall Survival Rate
No death of fish was recorded in Experiment I. In Experiment II, no death of fish was recorded in the Artemia-feeding group. The fish in the PBD-feeding group died from 23 dpf until 41 dpf (Figure 7). The cumulative overall survival rates from 41 dpf to 90 dpf in the three PBD-feeding tanks were 93.57%, 94.29%, and 95.00%, respectively. The cumulative overall survival rate of the Artemia-feeding group was significantly higher than that of PBD-feeding group (p < 0.05).
Discussion
Under the standard laboratory feeding operation of tilapia, the microbiota composition of fish changed with the development of fish even when the feed and environment remained constant (15 dpf, 20 dpf, and 40 dpf), suggesting that fish physiological development has impacts on the microbiota. This is consistent with research on zebrafish (Stephens et al., 2016). They suggested that the main drivers of changes in the microbiota were morphological changes during development. For tilapia, the ratio of intestine length to body length increases with the development of fish (Hu and Zhang, 1983). When the water temperature was approximately 27°C, tilapia larvae developed to juvenile at 12 dpf (Fujimura and Okada, 2007). The ratio of intestinal length to body length was 1.02, 1.34, and 1.53 for tilapia at 14, 18, and 23 dpf, respectively, with four curvatures. Then, the intestine of tilapia developed into five curvatures at 24 dpf. The intestine of 57 dpf juveniles developed seven curvatures. The preferred food of pond tilapia changed with the development of the fish intestine, from zooplankton to plant-based food. In the present research, the microbiota composition of tilapia fed Artemia changed with development as well, although similar results were not found in tilapia fed with PBD food. We found that tilapia fed with PBD food died from 23 dpf to 41 dpf, suggesting that PBD food was not completely suitable for tilapia during the early juvenile stages. This result was supported by the unhealthy intestinal structure of fish at 20 dpf and 40 dpf in the PBD food group. We found that the microbiota of fish intestines did not simply reflect the microbiota in their habitat or feed but was closely related to species-specific behavior, metabolism, and immunity, which were linked to both their development and diet (Stephens et al., 2016; Zhang et al., 2018). The intestinal microflora of tilapia becomes established at the early juvenile stage, approximately 2 months after hatching. Similar results were reported for goldfish (Carassius auratus) (Sugita et al., 1988).
To understand the effect of diet on the developmental stage-specific gut microbiota composition and diversity, tilapia were fed two different diets since they started feeding. We observed major compositional shifts in the two diet groups. The main intestinal microbiota of tilapia larvae before feeding at the phylum level included Proteobacteria, Firmicutes, Actinobacteria, Planctomycetes, Fusobacteria and Bacteroidetes. The relative abundance of Proteobacteria was high (38.0%-53.8%) in the two experiments. Fusobacteria became the dominant phylum for 90 dpf fish under standard laboratory feeding operation and under Artemia feeding condition. Planctomycetes was the dominant phylum for 90 dpf fish fed with PBD food. Similarly, research on the first-feeding impact on the intestinal microbiota of rainbow trout (Oncorhynchus mykiss) showed that the plant-based diet favored the phylum Firmicutes, whereas the marine-based diet favored the phylum Proteobacteria (Ingerslev et al., 2014). In research on the effect of feeding a high carbohydrate diet to juvenile Atlantic salmon (Salmo salar) on intestinal microbiota composition, the phylum Planctomycetes was significantly increased (Villasante et al., 2019). They suggested that the effect of diet on the structure of gut microbiota was low. It is possible that juvenile fish possess a gut microbiome that is more resistant to dietary changes. In the present research, we also found that feeding habit transition (60 dpf and 90 dpf) had little impact on the gut microbiota composition of juvenile tilapia under standard laboratory feeding operation.
Previous research reported that anaerobic metabolism existed among all major Planctomycetes lineages and that Planctomycetes used fermentations of carbohydrates for growth and survival under anaerobic conditions (Elshahed et al., 2007). An increase in these microbial communities fermenting digestible carbohydrates under anaerobic conditions in the intestine was found in tilapia fed the PBD food diet. Similarly, at the genus level, Gemmobacter, Pirellula, and Planctomyces were significantly more abundant in fish fed the PBD food diet. They are members of Planctomycetes and have been described to be capable of carbohydrate fermentation (Schlesner and Stackebrandt, 1986; Ward et al., 2015; Hao et al., 2016). Pirellula was a dominant genus for tilapia (Zheng et al., 2019) and cyprinid fishes such as bighead carp (Hypophthalmichthys nobilis) and common carp (Cyprinus carpio) (Sakalli et al., 2018; Luo et al., 2021). Gemmobacter was revealed to be dominant in the gut of freshwater fish, including Nile tilapia (Deng et al., 2022) and common carp (Zhang et al., 2020). Gemmobacter were significantly enriched in the intestinal microbiota of blunt snout bream after plants were added to the diet (Wei et al., 2018). The increase in the relative abundance of Gemmobacter in fish fed PBD food could be due to their ability to degrade carbohydrates (Hao et al., 2016).
Cetobacterium, belonging to the phylum Fusobacteria, was a core microbe in the freshwater fish intestine (Sugita et al., 1991; Roeselers et al., 2011; Deng et al., 2022) and was involved in the fermentative metabolism of peptides and amino acids (Finegold et al., 2003). Under standard laboratory feeding conditions, Cetobacterium was the dominant genus from 20 dpf to 90 dpf. Cetobacterium was dominant in 60 dpf and 90 dpf fish fed Artemia. In research on gut microbiota composition in blunt snout bream during feeding habit transition, Cetobacterium was considered a microbiological marker of intestinal microbiota for zooplankton-based diet feeding (Wei et al., 2018). In previous research, it was shown that the dominant food of 24 dpf juvenile tilapia in ponds is zooplankton (Hu and Zhang, 1983). The proportion of phytoplankton increases with fish development. Under standard laboratory feeding operation, we added dry food to the fish diet beginning at 40 dpf. However, Cetobacterium was still dominant after fish were fed dry food. This could be because of the ability of Cetobacterium to utilize glucose (Wang et al., 2021). Previous research also found that Cetobacterium was a core microbiota of wild juvenile Nile tilapia, although they preferred plant-based food at that age (Tesfahun and Temesgen, 2018; Bereded et al., 2020; Bereded et al., 2021). Additionally, Cetobacterium was the main genus constituting the phylum Fusobacteria. This explained the high relative abundance of Fusobacteria in the intestine for 90 dpf fish under the standard laboratory feeding condition and under the Artemia-feeding condition.
We conclude that diet has a strong effect on the microbiota of tilapia intestine in the early life of fish. The intestinal microflora of tilapia is established approximately 2 months after hatching. Plant-based food negatively affected the survival rate and intestinal development of tilapia. The relative composition of the phyla Planctomycetes and Proteobacteria was higher in juvenile fish in the PBD food group than in the Artemia group. The relative abundance of Fusobacteria was higher in the juvenile fish of the Artemia group than in those of the PBD food group. Feeding habit transition in juvenile tilapia had a low effect on the microbiota of the intestine under standard laboratory feeding conditions. Our results provide useful information for future studies on the microbiota in the tilapia gut. We suggest that modulation of gut microbiota of tilapia could be performed in their early life.
Data Availability Statement
The data presented in the study are deposited in the NCBI Sequence Read Archive repository, accession number PRJNA832485.
Ethics Statement
The animal study was reviewed and approved by Ethical Committee for Animal Experiments of Pearl River Fisheries Research Institute, Chinese Academy of Fishery Sciences, China.
Author Contributions
MW contributed to the experimental design, data analysis, and primary writing. ZF and ZZ contributed to fish trials and sampling. MY and ZL contributed to sampling and data analysis. XK and FG contributed to manuscript reviewing and proofing. JC and ML contributed to the experimental design and manuscript review. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by National Natural Science Foundation of China (32102818), funds from China Agriculture Research System of MOF and MARA (CARS-46), Guangdong Basic and Applied Basic Research Foundation (2019A1515111046), Guangdong Provincial Key R&D Program (2021B0202020001), and Independent Research and Development Projects of Maoming Laboratory (2021ZZ007).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
This study was supported by National Natural Science Foundation of China (32102818), funds from China Agriculture Research System of MOF and MARA (CARS-46), Guangdong Basic and Applied Basic Research Foundation (2019A1515111046), Guangdong Provincial Key R&D Program (2021B0202020001), and Independent Research and Development Projects of Maoming Laboratory (2021ZZ007).
References
Bereded N. K., Abebe G. B., Fanta S. W., Curto M., Waidbacher H., Meimberg H., et al. (2021). The Impact of Sampling Season and Catching Site (Wild and Aquaculture) on Gut Microbiota Composition and Diversity of Nile Tilapia (Oreochromis Niloticus). Biology 10, 180. doi: 10.3390/biology10030180
Bereded N. K., Curto M., Domig K. J., Abebe G. B., Fanta S. W., Waidbacher H., et al. (2020). Metabarcoding Analyses of Gut Microbiota of Nile Tilapia (Oreochromis Niloticus) From Lake Awassa and Lake Chamo, Ethiopia. Microorganisms 8, 1040. doi: 10.3390/microorganisms8071040
Caporaso J. G., Kuczynski J., Stombaugh J. (2010). QIIME Allows Analysis of High-Throughput Community Sequencing Data. Nat. Methods 7, 335–336. doi: 10.1038/nmeth.f.303
Deng Y., Verdegem M., Eding E. H., Kokou F. (2022). Effect of Rearing Systems and Dietary Probiotic Supplementation on the Growth and Gut Microbiota of Nile Tilapia (Oreochromis Niloticus) Larvae. Aquaculture 546, 737297. doi: 10.1016/j.aquaculture.2021.737297
Drossou A., Ueberschär B., Rosenthal H., Herzig K.-H. (2006). Ontogenetic Development of the Proteolytic Digestion Activities in Larvae of Oreochromis Niloticus Fed With Different Diets. Aquaculture 256 (1-4), 479–488. doi: 10.1016/j.aquaculture.2006.01.038
Edgar R. C. (2013). UPARSE: Highly Accurate OTU Sequences From Microbial Amplicon Reads. Nat. Methods 10, 996–998. doi: 10.1038/nmeth.2604
Elshahed M. S., Youssef N. H., Luo Q., Najar F. Z., Roe B. A., Sisk T. M., et al. (2007). Phylogenetic and Metabolic Diversity of Planctomycetes From Anaerobic, Sulfide- and Sulfur-Rich Zodletone Spring, Oklahoma. Appl. Environ. Microbiol. 73, 4707–4716. doi: 10.1128/AEM.00591-07
Finegold S. M., Vaisanen M. L., Molitoris D. R., Tomzynski T. J., Song Y., Liu C., et al. (2003). Cetobacterium Somerae Sp. Nov. From Human Feces and Emended Description of the Genus Cetobacterium. Syst. Appl. Microbiol. 26, 177–181. doi: 10.1078/072320203322346010
Fishery Bureau, Ministry of Agriculture and Rural Affairs, China (2020). China Fisheries Yearbook (Beijing: Chinese Agriculture Express).
Fujimura K., Okada N. (2007). Development of the Embryo, Larva and Early Juvenile of Nile Tilapia Oreochromis Niloticus (Pisces: Cichlidae). Developmental Staging System. Dev. Growth Differ. 49, 301–324. doi: 10.1111/j.1440-169X.2007.00926.x
Ghanbari M., Kneifel W., Domig K. J. (2015). A New View of the Fish Gut Microbiome: Advances From Next-Generation Sequencing. Aquaculture 448, 464–475. doi: 10.1016/j.aquaculture.2015.06.033
Hansen A. C., Hemre G. I. (2013). Effects of Replacing Fish Meal and Oil With Plant Resources in on-Growing Diets for Atlantic Cod Gadus Morhua L. Aquac. Nutr. 19, 641–650. doi: 10.1111/anu.12078
Hao L., Bize A., Conteau D., Chapleur O., Courtois S., Kroff P., et al. (2016). New Insights Into the Key Microbial Phylotypes of Anaerobic Sludge Digesters Under Different Operational Conditions. Water Res. 102, 158–169. doi: 10.1016/j.watres.2016.06.014
Hu M., Zhang Z. (1983). A Study on the Development of Digesitive System and Feeding Habit of Fry and Juvenile of Tilapia Nilotica. J. Fish China 7, 207–217.
Ibrahim A. N., Noll M. S., Valenti W. C. (2015). Zooplankton Capturing by Nile Tilapia, Oreochromis Niloticus (Teleostei: Cichlidae) Throughout Post-Larval Development. Zoologia 32, 469–475. doi: 10.1590/s1984-46702015000600006
Ingerslev H. C., Jørgensen L. V. G., Strube M. L., Larsen N., Dalsgaard I., Boye M., et al. (2014). The Development of the Gut Microbiota in Rainbow Trout (Oncorhynchus Mykiss) Is Affected by First Feeding and Diet Type. Aquac. Nutr. 424-425, 24–34. doi: 10.1016/j.aquaculture.2013.12.032
Le Roux P. (1956). Feeding Habits of the Young of Four Species of Tilapia. S. Afr. J. Sci. 53, 33–37.
Luo M., An R., Fu J., Wan S., Zhu W., Wang L., et al. (2021). Comparative Analysis of the Gut Microbiota in Bighead Carp Under Different Culture Patterns. J. Appl. Microbiol. 132, 1357–1369. doi: 10.1111/jam.15248.
Magoč T., Salzberg S. L. (2011). FLASH: Fast Length Adjustment of Short Reads to Improve Genome Assemblies. Bioinformatics 27, 2957–2963. doi: 10.1093/bioinformatics/btr507
Morais S., Silva T., Cordeiro O., Rodrigues P., Guy D. R., Bron J. E., et al. (2012). Effects of Genotype and Dietary Fish Oil Replacement With Vegetable Oil on the Intestinal Transcriptome and Proteome of Atlantic Salmon (Salmo Salar). BMC Genomics 13, 1–21. doi: 10.1186/1471-2164-13-448
Negassa A., Prabu P. C. (2008). Abundance, Food Habits, and Breeding Season of Exotic Tilapia Zillii and Native Oreochromis Niloticus L. Fish Species in Lake Zwai, Ethiopia. Maejo Int. J. Sci. Tech. 2, 345–359.
Ringø E., Zhou Z., Vecino J. L. G., Wadsworth S., Romero J., Krogdahl Å., et al. (2016). Effect of Dietary Components on the Gut Microbiota of Aquatic Animals. A Never-Ending Story? Aquacult. Nutr. 22, 219–282. doi: 10.1111/anu.12346
Roeselers G., Mittge E. K., Stephens W. Z., Parichy D. M., Cavanaugh C. M., Guillemin K., et al. (2011). Evidence for a Core Gut Microbiota in the Zebrafish. ISME J. 5, 1595–1608. doi: 10.1038/ismej.2011.38
Sakalli S., Giang P. T., Burkina V., Zamaratskaia G., Rasmussen M. K., Bakal T., et al. (2018). The Effects of Sewage Treatment Plant Effluents on Hepatic and Intestinal Biomarkers in Common Carp (Cyprinus Carpio). Sci. Total Environ. 635, 1160–1169. doi: 10.1016/j.scitotenv.2018.04.188
Schlesner H., Stackebrandt E. (1986). Assignment of the Genera Planctomyces and Pirella to a New Family Planctomycetaceae Fam. Nov. And Description of the Order Planctomycetales Ord. Nov. Syst. Appl. Microbiol. 8, 174–176. doi: 10.1016/S0723-2020(86)80072-8
Stephens W. Z., Burns A. R., Stagaman K., Wong S., Rawls J. F., Guillemin K., et al. (2016). The Composition of the Zebrafish Intestinal Microbial Community Varies Across Development. ISME J. 10, 644–654. doi: 10.1038/ismej.2015.140
Sugita H., Miyajima C., Deguchi Y. (1991). The Vitamin B 12 - Producing Ability of the Intestinal Microflora of Freshwater Fish. Aquaculture 92, 267–276. doi: 10.1016/0044-8486(91)90028-6
Sugita H., Tsunohara M., Ohkoshi T., Deguchi Y. (1988). The Establishment of an Intestinal Microflora in Developing Goldfish (Carassius Auratus) of Culture Ponds. Microb. Ecol. 15, 333–344. doi: 10.1007/BF02012646
Tesfahun A., Temesgen M. (2018). Food and Feeding Habits of Nile Tilapia Oreochromis Niloticus (L.) in Ethiopian Water Bodies: A Review. Int. J. Fish Aquat Stud. 6, 43–47.
Villasante A., Ramírez C., Catalán N., Opazo R., Dantagnan P., Romero J. (2019). Effect of Dietary Carbohydrate-to-Protein Ratio on Gut Microbiota in Atlantic Salmon (Salmo Salar). Animals 9, 89. doi: 10.3390/ani9030089
Wang A. R., Ran C., Ringø E., Zhou Z. G. (2018). Progress in Fish Gastrointestinal Microbiota Research. Rev. Aquacult. 10, 626–640. doi: 10.1111/raq.12191
Wang A., Zhang Z., Ding Q., Yang Y., Bindelle J., Ran C., et al. (2021). Intestinal Cetobacterium and Acetate Modify Glucose Homeostasis via Parasympathetic Activation in Zebrafish. Gut Microbes 13, 1–15. doi: 10.1080/19490976.2021.1900996
Ward N. L., Staley J. T., Schmidt J. M. (2015). Bergey’s Manual of Systematics of Archaea and Bacteria, Planctomyces (New Jersey: John Wiley & Sons).
Wei J., Guo X., Han L., Chen Y., Wang W. (2018). The Variation Profile of Intestinal Microbiota in Blunt Snout Bream (Megalobrama Amblycephala) During Feeding Habit Transition. BMC Microbiol. 18, 99. doi: 10.1186/s12866-018-1246-0
Zhang Z., Li D., Refaey M. M., Xu W., Rong T., Li L. (2018). Host Age Affects the Development of Southern Catfish Gut Bacterial Community Divergent From That in the Food and Rearing Water. Front. Microbiol. 9, 495. doi: 10.3389/fmicb.2018.00495
Zhang Y., Zhang P., Shang X., Lu Y., Li Y. (2020). Exposure of Lead on Intestinal Structural Integrity and the Diversity of Gut Microbiota of Common Carp. Comp. Biochem. Phys. C 239, 108877. doi: 10.1016/j.cbpc.2020.108877
Keywords: microbiota, intestine, tilapia, development, diet
Citation: Wang M, Fan Z, Zhang Z, Yi M, Liu Z, Ke X, Gao F, Cao J and Lu M (2022) Effects of Diet on the Gut Microbial Communities of Nile Tilapia (Oreochromis niloticus) Across Their Different Life Stages. Front. Mar. Sci. 9:926132. doi: 10.3389/fmars.2022.926132
Received: 22 April 2022; Accepted: 10 May 2022;
Published: 14 June 2022.
Edited by:
Piotr Rzymski, Poznan University of Medical Sciences, PolandReviewed by:
Yao Zheng, Chinese Academy of Fishery Sciences, ChinaHany M. R. Abdel-Latif, Alexandria University, Egypt
Mahmoud A. O. Dawood, Kafrelsheikh University, Egypt
Copyright © 2022 Wang, Fan, Zhang, Yi, Liu, Ke, Gao, Cao and Lu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jianmeng Cao, Y2Fvamlhbm1lbmdAYWxpeXVuLmNvbQ==; Maixin Lu, bXgtbHVAMTYzLmNvbQ==
†These authors have contributed equally to this work
 Miao Wang
Miao Wang Zijian Fan1†
Zijian Fan1† Mengmeng Yi
Mengmeng Yi Zhigang Liu
Zhigang Liu Fengying Gao
Fengying Gao Jianmeng Cao
Jianmeng Cao