- 1Aquatic Environmental Biotechnology & Nanotechnology Division, (AEBN) Division, Indian Council of Agricultural Research (ICAR)- Central Inland Fisheries Research Institute (ICAR-CIFRI), Barrackpore, India
- 2Indian Council of Agricultural Research - Central Inland Fisheries Research Institute (ICAR-CIFRI), Barrackpore, India
- 3College of Fisheries, Central Agricultural University (CAU), Lembucherra, India
- 4Laboratory of Marine Biology, Kyushu University, Fukuoka, Japan
- 5AquaBioResource Innovation Center (ABRIC), Kyushu University, Fukuoka, Japan
Fisheries and aquaculture are the fastest-growing food-producing sector and rapidly becoming an important element for the global food security since they are the primary source of seafood and high animal protein in the human diet. Genome editing offers new possibilities such as the clustered regularly interspaced palindromic repeats (CRISPR)/CRISPR-associated protein (Cas9) technology, which has the potential to accelerate the sustainable genetic improvement in fisheries and aquaculture. The CRISPR/Cas9 system has four key components, namely, target DNA, Cas9, the protospacer adjacent motif sequence, and the guide RNA or single-guide RNA. CRISPR/Cas is cheaper, easier, and more precise than the other genome editing technologies and can be used as a new breeding technology in fisheries and aquaculture to solve the far-reaching challenges. The attributes like high fecundity, external fertilization, short generation interval, the established method of breeding, and the larval rearing of most aquaculture species have advantages for CRISPR/Cas9 genome editing applications. CRISPR/Cas9 has recently been applied to the traits valued in some aquaculture species (almost >20 species), targeting the main traits of traditional genetic improvement initiatives like growth, disease resistance, reproduction, sterility, and pigmentation. Genome editing can fast forward the breeding process with precision where changes occur in the targeted genes. The probability of desired changes occurring and passing the trait in the next generation is high, so it takes 1-3 generations to establish a breed. Moreover, CRISPR/Cas genome editing rapidly introduces favorable changes by disrupting genes with targeted minor changes, in contrast to transgenesis, which introduces foreign genes into the host genome and thereby alleviates major public concerns on safety. Although the CRISPR/Cas technology has a tremendous potential, there are several technical challenges and regulatory and public issues concerning the applications in fisheries and the aquaculture breeding sector. Nonetheless, the exciting point in the CRISPR/Cas9 genome editing is that two CRISPR-edited fish, namely, red sea bream and tiger puffer developed by the Kyoto-based startup got approval and are now on the market for sale, and another fish, FLT-01 Nile tilapia developed by the AquaBounty, is not classified under genetically modified organism regulatory. However, there is still a way to go before it revolutionizes and becomes viable in commercial aquaculture as the new breeding technology for aquaculture-important traits and species.
Introduction
Food security has become a major global challenge with an increasing population and a growing demand for the animal protein. Aquaculture, as the leading source of high-quality animal protein for human nutrition, has a significant role to play in the global food security (FAO, 2020). Gene editing offers new possibilities, and applying it in the aquaculture sector can be a smart way to reach the full potential and enable a more affordable access to nutrient-rich foods throughout the world (Hallerman, 2021). At first, zinc finger nucleases (ZFNs) and transcription activator–like endonuclease (TALEN) techniques were used for genome editing. At present, the most novel method of genome editing, the clustered regularly interspaced palindromic repeats (CRISPR) system, dominates all over the world (Jinek et al., 2012; Doudna and Charpentier, 2014), and it is much cheaper and efficient for targeted genome editing at new sites compared to ZFN and TALEN (Boch and Bonas, 2010). The CRISPR/Cas system has been applied to several plant, animal, and fish species by many researchers at an unprecedented pace and shows no signs of slowing down. CRISPR/Cas editing is an easy and powerful tool as a new breeding technology including for aquaculture and has recently been applied to the traits valued in few aquaculture species; however, there is still a long way to go before it becomes viable in commercial aquaculture. Here, we will discuss the current understanding of the CRISPR/Cas genome editing technology, the present status, challenges, and future directions in aquaculture applications.
What Is CRISPR and How Does It Work?
CRISPR is an amazing technology with a fast, easy, and efficient high-tech method of editing the genomes and allows for targeted genetic alterations such as deleting, substituting, or adding the DNA or switching genes on or off without adding the exogenous genes (Urnov et al., 2010). CRISPR (clustered regularly interspaced short palindromic repeat) sequences were discovered in the Escherichia coli genome in 1987, and later in 2007, their function as defense against bacteriophages was elucidated. Genome editing technology type II CRISPR/Cas9 has been developed from the Streptococcus pyogenes CRISPR-associated protein (Cas9)–adaptive immune system, a simple natural defense strategy found in many bacteria. Bacteria use CRISPR-derived RNA (crRNA) and the Cas protein system to cope and catalog the numerous virus threats and foreign invaders by maintaining a sort of genetic library within their DNA that contains bits of gene sequences from the past encounters (CRISPR 101: A Desktop Resource, www.addgene.org). Through sequence homology, crRNAs guide a Cas nuclease to the specified exogenous genetic material; then, the CRISPR/Cas complex binds to the foreign DNA and cleaves it to destroy the invader. In a native CRISPR/Cas system, Cas9 is guided to its target sites with the aid of two RNAs: the (CRISPR RNA) crRNA, which defines the genomic target for Cas9, and the transactivating crRNA (tracrRNA), which acts as a scaffold linking the crRNA to Cas9 and facilitates the processing of mature crRNAs from pre-crRNAs. While native CRISPR/Cas systems have a variety of enzymes responsible for processing foreign DNA, for CRISPR genome editing, the only protein required is the Cas9 endonuclease or a variant thereof. The CRISPR/Cas system was first demonstrated for targeted in vitro DNA cleavage in 2012 by Jinek et al. and CRISPR/Cas-based genome editing described in the cell culture in 2013 by Cong et al., 2013 and Mali et al., 2013 Then, within a very short span of time, the CRISPR/Cas technology has been developed at a truly astonishing pace with much work directed toward the development of new applications, increasing specificity and multiplexibility in various species and targeted genome modifications in various model systems (Hwang et al., 2013; Mali et al., 2013). Further, as an improvement in most CRISPR/Cas genome editing systems, the two-component system described previously, crRNA and tracrRNA, are fused into a guide RNA (gRNA) or single-guide RNA (sgRNA) to efficiently create target mutations (Jinek et al., 2012; Hwang et al., 2013; Mali et al., 2013). The gRNA/sgRNA is composed of two parts: CRISPR RNA (crRNA) at the 5′ end for the recognition of the target sequence and a trans-activating crRNA (tracrRNA) at the 3′ end, which enables interaction with Cas9 to activate the endonuclease activity. The CRISPR/Cas system can be used for the editing any desired target genomic region if the sequence is unique compared to the rest of the genome and is located adjacent to protospacer-adjacent motif (PAM) sequences (NGG), which are recognized by the gRNA-containing complimentary sequences to the target site. Cas9 endonuclease binding to the target genomic locus is mediated both by the target sequence contained within the guide RNA and a species-specific short PAM sequence (typically 3–5 base pairs) and are cleaved by gRNA and the nuclease Cas9 complex (Kleinstiver et al., 2016). The enzymatic activity of Cas9 cleaves both strands of genomic DNA, resulting into a double-strand break (DSB), and then, DNA damage repair responses are initiated to join the broken ends (Ota et al., 2014). The DSB can be repaired by two major DNA repair pathways, namely, non-homologous end joining (NHEJ) and homology-directed repair (HDR). The NHEJ pathway repairs two adjacent strands of DNA that are error-prone and usually induce unpredictable insertion or deletions (indels) and substitutions (Segev-Hadar et al., 2021). Additionally, if a repair template is present, another pathway HDR can be used to insert the desired mutations for precise gene editing (Figure 1).
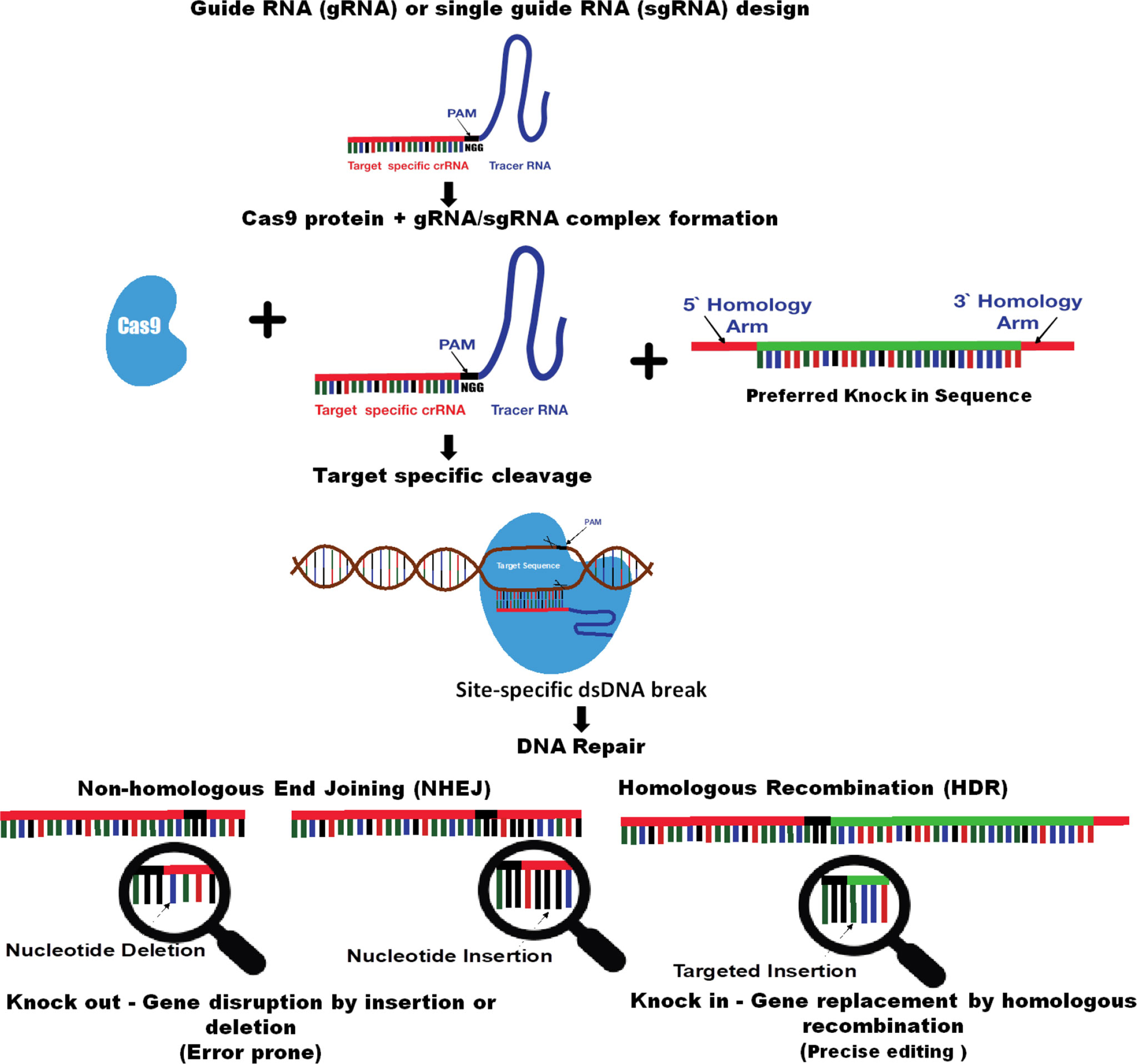
Figure 1 Schematic diagram of CRIPSR/Cas–mediated genome editing technology (adapted from Mohapatra and Chakraborty, 2021). Guide RNA (gRNA) or single-guided RNA (sgRNA) consists of a CRISPR RNA (crRNA) sequence that is specific to the targeted DNA and a transactivating crRNA (tracrRNA) sequence that interacts with the Cas9 protein. The target sequence contained within the gRNA or sgRNA has a species-specific short protospacer adjacent motif (PAM) sequence (–NGG). gRNA/ sgRNA guides the Cas9 endonuclease to the targeted genomic locus; then, the CRISPR/ Cas9 complex binds and cleaves it. Cas9-mediated site-specific DNA cleavage is directed by complementary between the gRNA/ sgRNA and the target sequence upon the presence of a PAM sequence on the opposite strand. The enzymatic activity of Cas9 cleaves both strands of genomic DNA, resulting in a double-strand break (DSB). A CRISPR-Cas induced DSB is repaired by either non-homologous end-joining (NHEJ) or homologous recombination (HDR). NHEJ is more efficient than HDR but is error prone and may induce unpredictable indels, whereas HDR can provide a precise gene modification.
Although the CRISPR/Cas9 genome editing system has many advantages compared to its counterparts, there are still practical challenges in CRISPR experiments, such as optimal sgRNA designing, which is the most important task (Cui et al., 2018; Hanna and Doench, 2020; Luo et al., 2022). The sgRNA consists of a custom-designed crRNA sequence fused to a tracrRNA, and there is a challenge in designing the sgRNA with high on-target activity and low off-target effects that can affect the efficacy and specificity of the genome-editing results (Mojica and Rodriguez-Valera, 2016; Luo et al., 2022). Plenty of research has been done for improving the sgRNA efficiency and specificity (Doench et al., 2016; Hanna and Doench, 2020), and many sgRNA design tools have been developed to support the CRISPR/Cas9 genome editing such as E-CRISP (http://www.e-crisp.org/E-CRISP/) (Heigwer et al., 2014), ZiFiT (https://zifit.partners.org/ZiFiT/) (Hwang et al., 2013; Mali et al., 2013), CHOPCHOP (https://chopchop.cbu.uib.no) (Montague et al., 2014; Labun et al., 2016), CRISPOR (http://crispor.org/) (Montague et al., 2014; Labun et al., 2016), CRISPRscan (http://www.crisprscan.org) (Moreno-Mateos et al., 2015), CCTop (https://crispr.cos.uni-heide) (Stemmer et al., 2015), and CRISPRdirect (http://crispr.dbcls.jp/) (Naito et al., 2015). In aquaculture and farmed fish studies, ZiFit, CRISPRScan, and CRISPOR were the most frequently used sgRNA-designing tools.Assessing the genome editing outcome is also another critical step in CRISPR experiments (Luo et al., 2022). CRISPR/Cas9 is suitable for multiple genome modifications since it consists of two components, sgRNA and Cas9 nuclease, which make it very easy to inject multiple sgRNAs with the nuclease Cas9 in various types of embryos (Urnov et al., 2010; Ota et al., 2014). There are several methods to detect the indels such as CelI assay (Miller et al., 2007) and high-resolution melt analysis (HRMA) (Bassett et al., 2013) for single-locus indels and the heteroduplex mobility assay (HMA) for multiple genome modifications (Ota et al., 2013; Ota et al., 2014). For the genome-editing result assessment, direct (Sanger) sequencing of PCR products can be used, which can be analyzed by TIDE (http://tide.nki.nl.) (Brinkman et al., 2018) and ICE (Conant et al., 2022; Luo et al., 2022), or else, next-generation sequencing (NGS) can be used with tools like CRISPResso2 (Clement et al., 2019), CRISPR RGEN, and CRISPR Cas-Analyzer (http://www.rgenome.net/) (Park et al., 2017; Luo et al., 2022).
Based on crRNA processing and further action, the CIRSPR/Cas system can be classified into 3 types (Khan, 2019):
CRISPR/Cas9 has already been widely applied to genome editing in a great range of organisms as model species, including the nematode (Caenorhabditis elegans) (Tzur et al., 2013), fruit fly (Drosophila) (Bassett et al., 2013), zebrafish (Danio rerio) (Hwang et al., 2013), rat (Rattus norvegicus) (Li et al., 2013), mouse (Mus musculus) (Wang et al., 2013), and monkey (Macaca fascicularis) (Niu et al., 2014) with efficiencies similar to or surpassing those obtained by ZFNs and TALENs.
CRISPR/Cas9 Genome-Editing Alternatives and Their Implications
The field of genome-editing technologies is rapidly evolving, and in the recent years, several genome-editing technologies have been developed. There are several genome-editing techniques that evolved such as the homing endonuclease system, a protein-based nuclease system like meganucleases, ZFNs, TALENs, and RNA-DNA system CRISPR/Cas (Khan, 2019). The core technologies commonly used in genome editing are (i) ZFNs (ii) TALENs (iii) CRISPR/Cas. ZFNs (zinc-finger nucleases), and TALENs (transcription activator–like effector nucleases), genome-editing technologies, utilize the dimer FokI nuclease for genome targeting. ZFNs are fusions between a custom-designed Cys2-His2 zinc-finger protein and a FokI endonuclease cleavage domain (Kim et al., 1996; Urnov et al., 2010). Each ZFN is typically composed of 3 or 4 zinc-finger domains, where each individual domain is composed of ∼30 amino acid residues. ZFNs function as dimers and the dimerization of the ZFN proteins is mediated by the FokI cleavage domain, which recognizes and cuts DNA within a 5–7 base pair spacer sequence (Smith et al., 2000). The major concern that prevents wider applications of ZFNs is off-target cleavage and undesired mutations (Pattanayak et al., 2011). Compared to ZFNs, TALENs exhibit reduced off-target effects and provides better specificity. TALENs are the fusions of transcription activator-like (TAL) proteins and a FokI nuclease and almost follow the same principle as ZFNs. TAL proteins are composed of 33–34 amino acid repeating motifs with two variable positions that have a strong recognition for specific nucleotides. TALENs recognize the target sites that consist of two TALE DNA-binding sites that flank a 12–20 base pair spacer sequence recognized by the FokI cleavage domain (Yang and Huang, 2019; Li et al., 2020). However, TALENs face major technical hurdles for cloning the repeat TALE arrays in the designing of the large-scale identical repeat sequences. Although ZFN and TALEN technologies have improved the capacity for genome editing, both of these techniques have limitations. Nevertheless, the newest developed CRISPR/Cas system employed a simple RNA-programmable method to edit the genome that can be used to generate gene knockouts (via insertion/deletion) or knockins (via HDR) (Gaj et al., 2016; Li et al., 2020). In contrast, ZFNs and TALENs use large fusion proteins consisting of a DNA-binding domain to recognize DNA and the FokI nuclease catalytic domain; the CRISPR/Cas system uses a single-guide RNA (sgRNA) that directs the Cas9 nuclease to a specific/targeted genomic location for disrupting the gene (Jinek et al., 2012; Mohapatra and Chakraborty, 2021). In addition, the protospacer adjacent motif (PAM, NGG) immediately downstream of the target sequence determines the specificity of this system. Moreover, CRISPR/Cas9 has the capacity to edit the multiple genes simultaneously, producing conditional alleles and generating tagged proteins with high efficiency. Thereafter, the CRISPR/Cas system shows superiority in convenience, efficiency, specificity, and versatility over ZFNs and TALENs; hence, it is widely utilized and revolutionized the genome editing (Khan, 2019; Mohapatra and Chakraborty, 2021). Very recently, a group of researchers from Kyushu University Japan have reported the pentatricopeptide repeat (PPR) protein–based genome editing technology (Yagi et al., 2013). Pentatricopeptide repeat (PPR) proteins constitute an interesting protein family and are distinguished by the presence of tandem-degenerate PPR motifs. The PPR technology can edit both DNA and RNA, which is very powerful, and in the future, this can revolutionize the gene editing technology. Although the PPR technology has not been used in aquaculture yet, it has a potential for future genome editing in the aquaculture field.
Advances of CRISPR/Cas-Genome Editing in Aquaculture and Fisheries
The CRISPR/Cas9 genome editing technology holds a great promise for disease control and molecular breeding in aquaculture. Unlike traditional transgenesis, which involves the transfer of a gene from one organism to other concerning the public safety regulations of genetically modified organisms, CRISPR/Cas genome editing allows specific, targeted, and often minor changes to the genome of the species of interest, and it does not introduce any foreign DNA, thereby alleviating major public concerns on the safety of GMOs (Okoli et al., 2021). The provision of the in vitro synthesis of sgRNA allows for multiplex CRISPR/Cas9 for editing at multiple sites on the targeted gene and also for the higher probability of a gene knockout event. CRISPR/Cas has truly revolutionized genetics in a variety of model organisms and is also employed in various non-model organisms including important aquaculture species. The CRISPR/Cas system has emerged as a new breeding technology to promote sustainable genetic improvement including for aquaculture because of the high efficiency and flexibility of gene targeting without the requirement for exogenous gene integration (Blix et al., 2021; Hallerman, 2021). Genome editing can fast-forward the breeding process with precision where changes occur in the targeted genes and there is a probability for the desired changes to occur. Passing the changing trait in the next generation is high, so it takes 1–3 generations to establish a breed.
On the contrary, in a selective breeding program, changes are random where the probability of desired changes to occur is low and a lengthy process for many generations to establish a breed. The main categories by which the applications of genome editing can be employed to improve aquaculture production and sustainability are fixing alleles at the existing QTL (quantitative trait locus), targeted introgression-by-editing accessing alleles from different strains or species, and creating de novo variants based on knowledge of the trait (Gratacap et al., 2019). The life history and culture attributes like high fecundity, short generation time, external fertilization of eggs, and embryonic and larval development outside the mother of most aquaculture species, a well- established method of artificial spawning and larval rearing for many cultured species have an advantage for the genome editing research and applications over farmed terrestrial animals.
The CRISPR/Cas genome editing experiment in farmed fish required a few necessary steps (Figure 2). First, the target gene has to be selected after searching the genome database of candidate species. The sgRNA has to be designed with the help of sgRNA-designing tools, and then, the oligo syntheses of sgRNA proof has to be done. To conduct genome editing, different approaches like the recombinant vector (containing sgRNA designed according to the targeted DNA sequence and cas9 protein sequence) or alternatively the cloning-free PCR approach (PCR to amplify the gRNA template for transcription) were employed and can be directly transfected into cells, and/or the lentivirus CRISPR system can used for in vitro gene editing. For target-specific cleavage, in vivo, sgRNA and the cas9 mixture need to be delivered to the newly fertilized embryo at a one-cell stage by microinjection, electroporation, and so on. The microinjection of the CRISPR/Cas9 complex (sgRNAs and the cas9 mixture) into newly fertilized eggs at the one-cell stage of development is the preferred procedure to induce in vivo mutations in aquaculture species. To induce heritable modifications through gene editing, it must happen very early in the development, ideally at the one-cell stage so that they can be incorporated in its germ line, hence the need for specialized methods like microinjection to deliver the CRISPR/Cas complex on a microscopic scale. Nevertheless, other procedures to deliver the CRISPR/Cas9 complex into one-cell fertilized eggs or cultured cell are lentivirus- mediated methods, electroporation, and ribonucleoprotein complexes (Okoli et al., 2021; Yang et al., 2022). Mutants should be carefully screened using sequence analysis, and off-target affects should be assessed. At the F0 generation, mutants contain various mosaic patterns of mutation, which might affect the further clear distinction of associated phenotypes. So, off-target free candidate founder mutants have to be crossed with the wild population, and the heterozygous population of specific mutants should be raised. The F1 heterozygotes should be intercrossed to obtain the homozygous F2 progeny. The final step is the selection and application stage, which includes the selection of phenotypes with CRISPR-induced mutations, and the establishment of new varieties with improved values in aquaculture (Luo et al., 2022). It is recommended that all phenotypic analyses should be conducted using at least the F3 generation of homozygous progeny.
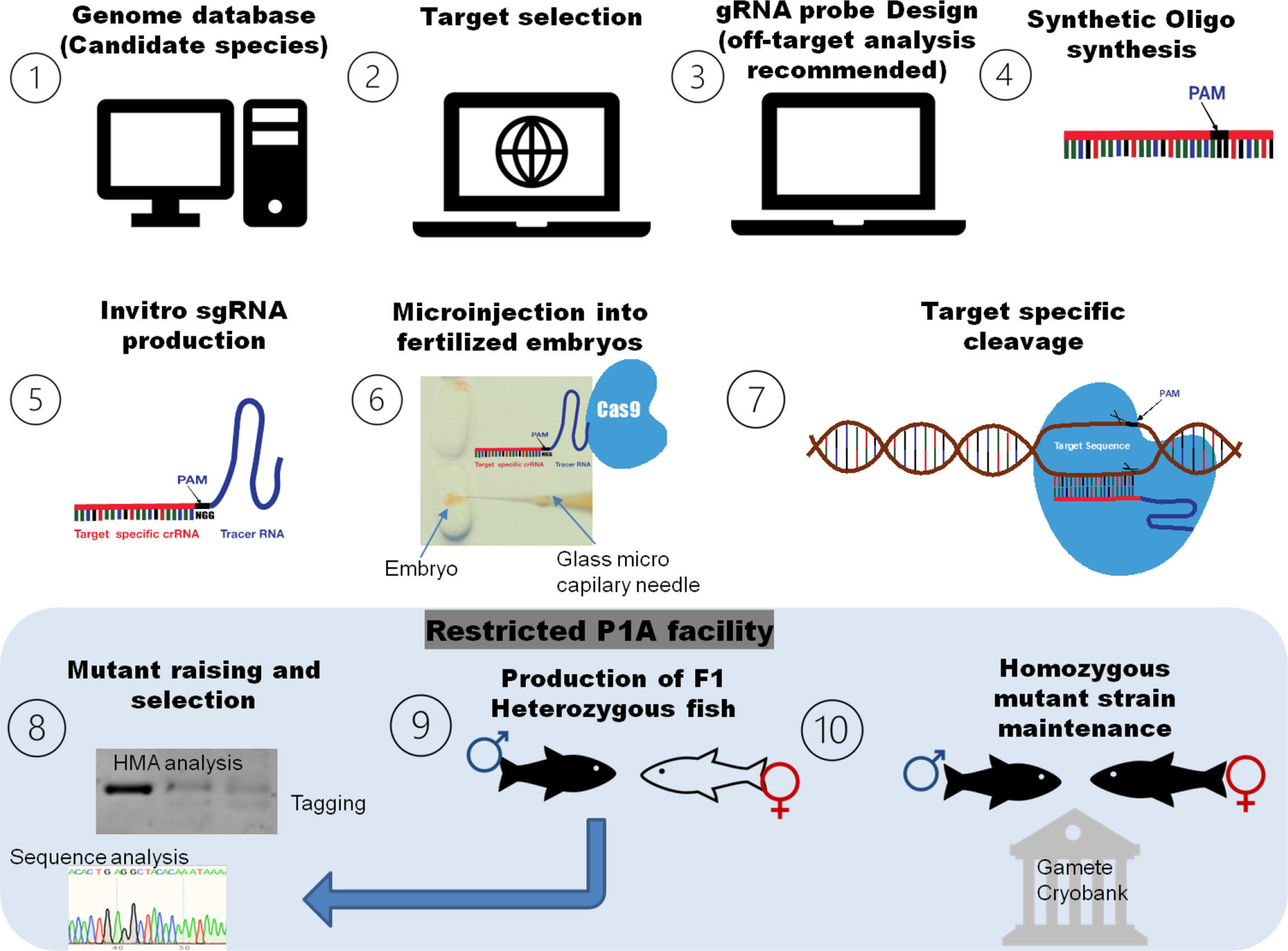
Figure 2 A simple roadmap of general methodology for CRIPSR/Cas genome editing in aquaculture and fisheries. The target gene has to be selected after searching the genome database of candidate species. The sgRNA has to be designed with the help of sgRNA-designing tools, and then, the sgRNA oligo has to be synthesized. For target-specific cleavage, the sgRNA and cas9 mixture needs to be delivered to the newly fertilized embryo at a one-cell stage by microinjection or similar methods. The final step is the assessing the genome-editing results and application stage that includes mutagenesis analysis, the selection of mutants, crossing with wild population and production of a specific mutant line, the evaluation of CRISPR-induced mutation associated phenotyp(s), and the establishment of new varieties with improved values in aquaculture.
Current Status of CRISPR/Cas Genome Editing in Aquaculture and Fisheries
The CRISPR/Cas technology offers new possibilities, and studies on genome editing in aquaculture species confirmed that the CRISPR/Cas editing system is easy, highly efficient, has lower off-target tendencies, and is able to generate long fragment deletions. Recently, the CRISPR/Cas9 system was studied on gene functions and breeding in vivo and/or in the cell lines of several major aquaculture species (almost over 20 species), namely, Atlantic salmon; Nile tilapia; red sea bream; channel and southern catfish; rainbow trout; blunt snout bream; and farmed carps like Rohu, grass, and common carp, as well as Pacific oyster, Atlantic killifish, fighting fish, Chinese tongue sole, olive flounder, oriental and white prawn, and other species (Gratacap et al., 2019; Blix et al., 2021). To date, plenty of CRISPR research in aquaculture reflects more traditional genetic improvement initiatives like growth, disease resistance, reproduction, sterility, and coloration patterns in fishes (Table 1). The most targeted genes in the genome-editing experiments of cultured fishes are myostatin (mstn), which is a key regulator for skeletal muscle growth and fertility, and sex-related genes like dead end (dnd) and doublesex- and mab-3-related transcription factor (dmrt), and several other genes like solute carrier family 45 member 2 (slc45a2) involved in the pathways of pigment synthesis and genes related to immunity and diseases like Toll-like receptor (TLR22) (Hallerman, 2021).
Growth and Productivity
Genetic improvement programs may have multiple breeding goals; growth enhancement and a rapid growth rate are universally valued traits. The CRISPR genome-editing technique has been used for growth enhancement by targeting the myostatin (mstn) gene in several cultured fishes (Khalil et al., 2017; Kishimoto et al., 2018; Hallerman, 2021). Myostatin is a member of the transforming growth factor β (TGFβ) superfamily, and it is a key regulator of skeletal muscle growth (Sun et al., 2020b). The expression of mstn is linked with the double-muscled phenotype in cattle and other animals. CRISPR techniques have been used for growth enhancement by disabling the mstn gene in many fishes like the common carp (Cyprinus carpio) (Zhong et al., 2016), channel catfish (Ictalurus punctatus) (Khalil et al., 2017), red sea bream (Pagrus major) (Kishimoto et al., 2018), olive founder (Paralichthys olivaceus) (Kim et al., 2019), pacific oyster (Crassostrea gigas) (Yu et al., 2019), blunt snout bream (Megalobrama amblycephala) (Sun et al., 2020b), and Nile tilapia (Oreochromis niloticus) developed by the US-based company AquaBounty. The Mstn knockout red sea bream has been reported to significantly increase its muscle mass (16% increase of skeletal muscle), short body length, and better feed efficiency, resulting in better overall growth, which was not observed in other fishes (Kishimoto et al., 2018; Ohama et al., 2020). Recently, the CRISPR/Cas9 system was used for mstn-targeted disruption to obtain the first growing mud loach (Misgurnus anguillicaudatus) (Tao et al., 2021). Apart from the mstn gene, CRISPR is also used for growth-associated traits in the tiger puffer (Takifugu rubripe) by disrupting the leptin receptor gene that controls the appetite, causing fish to eat more (Kishimoto et al., 2018; Shimbun, 2021). Very recently, Japan has approved the sales of two CRISPR-edited fish, for example, tiger puffer and red sea bream, which grow faster than their counterpart. The growth hormone (GH)–insulin-like growth factor (IGF) is a positive regulator of growth, whereas IGF-binding proteins (IGFBP) binds to IGF and acts as a negative regulator of somatic growth. Hence, CRISPR/Cas9 was used to disrupt the IGFBP-2b gene in rainbow trout (Oncorhynchus mykiss) (Cleveland et al., 2018). To increase the per capita productivity via suppressing a cannibalistic behavior, we produced AVTR (alginin vasotosin receptor) V1a2 knockout chub mackerel (Scomber japonicus) (Ohga and Matsuyama, 2021) and found that, apart from the desired phenotype, they also showed docility and reduced oxygen demand (unpublished data). Such an alternative approach toward increasing productivity through genome editing might improve the situations of aquaculture sustainability in the future.
Disease Resistance
Disease resistance is another important trait for commercial production, and the genetic improvement of disease resistance remains a high breeding priority. Genome-editing experiments have already been investigated for addressing the improvement of immunity and disease resistance. A targeted disruption of the Toll-like receptor TLR22 gene has been done in farmed Indian major carp Rohu (Labeo rohita) that can be used as a model system to investigate the role of TLR22 against pathogenic dsRNA viruses, bacteria, and parasites, for example, fish lice (Chakrapani et al., 2016). As an effective approach to control the hemorrhagic disease induced by grass carp reovirus (GCRV), CRISPR/Cas9 was used to knock out the (Ctenopharyngodon idella) grass carp Junctional adhesion molecule-A (JAM-A) gene and in vitro resistance evaluated against GCRV (Ma et al., 2018). In the channel, catfish (Ictalurus punctatus) targeted the altered immunity toll/interleukin 1 receptor domain-containing adapter molecule (TICAM1) gene involved in the signaling pathway initiated by TLR3 and disease susceptibility–related gene rhamnose binding lectin (RBL) (Elaswad et al., 2018). Sun et al. (2020) demonstrated the function of the carotenoid isomerooxygenase (EcNinaB-X1) gene in the immune defense of Exopalaemon carinicauda by performing CRISPR/Cas9-mediated gene mutagenesis. EcNinaB-X1-Knock out prawns (E. carinicauda) had lower mortality than wild-type after the bacterial challenge. In another study, CRISPR/Cas9 HDR was used to integrate (knock-in) an exogenous alligator cathelicidin gene (which exhibits a broad-spectrum antimicrobial activity) into a targeted non-coding region of the channel catfish genome (Simora et al., 2020). Since many genes are involved in the complex traits like disease resistance and growth, many of the genes might require editing to attain the desired phenotypes; hence, problems arise in the genome editing. Significant work will be required for the application of CRISPR to determine which genes and versions should be targeted through editing.
Sterility and Reproductive Confinement
The escape of fishes from aquaculture facilities may cause environmental, ecological, and genetic impacts, especially if they are a non-native species. Creating sterile animals for aquaculture is desirable to profit the industry to prevent the introgression with wild stock for conserving the genetic integrity and to avoid the negative production consequences of early maturation. Additionally, in a genetic improvement program, reproductive confinement protects the interest of the breeder investment. In this context, CRISPR/Cas9 was used to develop new sterility models in Atlantic salmon (Salmo salar) fish by knocking out a germ cell-specific candidate, dead end (dnd) gene, for producing germ cell-free salmon (Wargelius et al., 2016). Germ cell-free Atlantic salmon remained immature and did not undergo puberty (Kleppe et al., 2017). In another study, sterile channel catfish (Ictalurus punctatus) were produced by editing the luteinizing hormone (LH) gene using a modified ZNF technology with electroporation (Qin et al., 2016). Along with the genome editing technique for producing sterile fish in breeding programs, the knock-in technique also need to be developed, which could lead to the production of an inducible “on-off” system for sterility. Nevertheless, some prior knock-in work with model fish species medaka and zebrafish suggests that restoring fertility for breeding stocks might be possible (Zhang et al., 2015; Nagasawa et al., 2019). In a similar investigation, we, at Kyushu university, employed platinum TALEN to knockout the LH gene Japanense anchovy (Engraulis japonicus) and could reverse the LH-KO mediated “immature phenotype” effects by gonadotropin [HCG (human chorionic gonadotropin)] administration even at the F0 generation (unpublished data). A strategy for recovering the reproductive ability is needed for salmon lacking germ cells to be useful for breeding and aquaculture, and a rescue approach is reported for producing germ cells in dnd knockout fish (Güralp et al., 2020). Sterlet (Acipenser ruthenus), which has the fastest reproductive cycle, can be used for the surrogate production of critically endangered and late age-at-maturity sturgeon fish. CRISPR/Cas9 was used to the knockout dnd1 gene and prepares a sterile Sterlet host for the surrogate production of the late-maturing and large sturgeon species (Chen et al., 2018).
Fishes exhibit a diversity of sex-determination systems, and these are not well characterized in aquaculture species (Mank and Avise, 2009). In addition, fishes also exhibit sexual plasticity since multiple factors including the genetic makeup or environmental factors (such as temperature, light, hormones, and pH) regulate the process indicating the involvement of epigenetic regulation in sexual plasticity (Kikuchi et al., 2020; Zhou et al., 2021). Rapid advances of sequencing and the genome editing technology such as CRISPR/cas9 have accelerated the understanding of the steroidogenic pathway, hormonal regulation, and molecular mechanisms of fish sex determination and differentiation (Li and Wang, 2017). The CRISPR/cas9 gene editing of single- or multiple-gene mutation helps to elucidate the precise roles of the different paralogs in sex determination and differentiation. Until now, functional studies of steroidogenic enzymes and sex steroid receptors of different mutant lines of star1, star2, cyp11a1, cyp17a1, cyp17a2, cyp11c1, cyp19a1a, and cyp19a1b have been constructed by gene editing in model fish species like zebrafish, medaka, and tilapia (Tang et al., 2018; Yan et al., 2019; Alward et al., 2020; Shu et al., 2020; Zhang et al., 2020; Nishiike et al., 2021; Zhou et al., 2021). Genome-editing techniques were used for understanding the sex determination in tilapia as a good model for studying the fish sex determination/reproduction and have contributed toward the development of reproductively confined aquaculture stocks. CRISPR/Cas9 was employed to knock out the doublesex- and mab-3-related transcription factor (dmrt6 and dmrt1) and forkhead box L2 (foxl2), Nanos C2HC-Type Zinc Finger (nanos2 and nanos3), steroidogenic factor-1 (Sf- 1), gonadal soma-derived factor (Gsdf), Wilms tumor 1 (Wt1), and eukaryotic translation elongation factor 1A (eEF1A) gene in (Oreochromis niloticus) Nile tilapia (Li et al., 2014; Xie et al., 2016; Chen et al., 2017; Jiang et al., 2017). Very recently, CRISPR/Cas9 was employed to produce the all-female common carp (Cyprinus carpio L.) population through the knockout of the Cytochrome P450 17A1 (cyp17a1) gene (Zhai et al., 2022).
Omega-3 Production
The health benefits of fish oil omega-3 long chain (≥C20) polyunsaturated fatty acids (n-3 LC-PUFAs), particularly EPA (eicosapentaenoic acid, 20:5n-3) and DHA (docosahexaenoic acid, 22:6n-3), are well documented and established as good fatty acids for human health. Since humans have a limited capacity for the endogenous synthesis of LC-PUFA (EPA and DHA), dietary supplementation remains the best way to meet the requirements. Fishes including Atlantic salmon are the primary sources of omega-3 LC-PUFAs (EPA and DHA) for the human diet. The capacity for LC-PUFA synthesis in any species depends on the complementary activities of fatty acyl desaturases (Fads2) and elongases of very- long-chain fatty acid (Elovls). An understanding of endogenous synthesis and the regulation of LC-PUFAs is required in in vivo functional studies. Datsomor et al. (2019) used the CRISPR/Cas9 editing of Δ5 and Δ6 desaturases, which impairs Δ8-desaturation and docosahexaenoic acid synthesis in Atlantic salmon (Salmo salar). In a subsequent study, Datsomor et al. (2019) used the CRISPR/Cas9 editing of elovl2 in Atlantic salmon, which inhibits the elongation of PUFA, demonstrating the key roles of elovl2 in PUFA synthesis and also suggests that sterol regulatory element binding protein-1 (Srebp-1) is a main regulator of endogenous PUFA synthesis in Atlantic salmon.
Pigmentation
Coloration and color patterns in fishes are one of the important considerations in some aquaculture species, especially in ornamental trade. In fish, 6 kinds of pigment cells were identified (Fujii, 2000), while different fish species may have different sets of pigment cell types, and many genes that affect fish pigmentation can also affect the color pattern development. With the recent development of genome-editing tools like CRISPR/Cas9 methods for studying, these pigmentation genes have enriched the understanding and more colors of mutant fish have been obtained (Kimura et al., 2014). In Nile tilapia (Oreochromis niloticus), 25 genes related to pigmentation were involved in melanogenesis, pteridine metabolism, and the carotenoid absorption, and cleavage pathways were edited via CRISPR/Cas9. Among them, 13 genes had a phenotype in both F0 and F2 generations. Four types of pigment cells were identified in wild-type tilapia and, together with various naturally and artificially induced color gene mutants, can serve tilapia as a model system for studying color patterns in teleost fishes (Wang et al., 2021). In some cases, removing genes for pigmentation is also used as a tool to trace successful genome editing, as those individuals that are genetically changed will be without pigment, rendering a white or yellow color. The CRISPR/Cas9 system was used to knock out two genes, tyrosinase (tyr) and solute carrier family 45, member 2 (slc45a2), involved in pigmentation in Atlantic salmon and observed a graded range of phenotypes, ranging from a complete lack of pigmentation (albino) to partial loss and normal pigmentation (Hege Straume et al.; Edvardsen et al., 2014). The CRISPR/Cas9 system was used to target the tyr gene, a key enzyme in melanin synthesis and pigmentation, in two economically important fish species in Asia, loach (Paramisgurnus dabryanus) (Xu et al., 2019) and white crucian carp (Carassius auratus cuvieri) (Liu et al., 2019), which showed different degrees of pigmentation to clear albino. CRISPR/Cas was used to disrupt the HDL receptor/Scavenger receptor B1 (Scarb1); Scarb1-like genes involved in carotenoid metabolism and Gch1 (GTP cyclohydrolase 1) gene in the pteridine pathway resulted in the regional red skin fading into white color in ornamental common carp (Du et al., 2021). In another study, two Agouti signaling protein genes (ASIP 1 and ASIP 2) were disrupted via the CRISPR/Cas9 system and by which, black patches disappeared in the Oujiang color common carp (Chen et al., 2019). Recently, there is an increasing market demand for red tilapia that are the hybrids of different tilapia species or the red variant of the highly inbred Nile tilapia, and this red-color phenotype is unstable, resulting in a non-uniform coloration in appearance or dark-red color blotches that reduce the market value. Hence, the CRISPR/Cas9 system was used to target the slc45a2 gene in melanin biosynthesis and produces an albino variant of Nile tilapia with red skin and eyes (Segev-Hadar et al., 2021). Additionally, researchers are utilizing slc family genes to improve the visual confirmation of knockout fish or strain, and in such a research, Pandey et al. (2021) showed that slc2415 mutants showed reduced melanin pigmentation in the retina and bodies of Kawakawa (Euthynus affinis), highlighting toward a future where the visual recognition of a genome-edited stock will be possible.
Other Aquaculture-Important Studies
Since fertilization is internal, crustaceans are difficult to transform; however, the CRISPR/Cas technology has also been demonstrated in crustaceans. Two potential new techniques, the receptor-mediated ovary transduction of cargo (ReMOT control) and electroporation, can be used for delivering CRISPR/Cas9 components into cultured crustacean species like model crustaceans Daphnia pulex and the decapod Exopalaemon carinicauda (Xu et al., 2020). CRISPR/Cas9 editing has not yet been reported in one of the important groups of aquaculture species shrimp (Penaeus sp.) that may be partly due to practical limitations of internal fertilization. Nevertheless, the microinjection method was developed for the ridgetail white prawn Exopalaemon carinicauda and the CRISPR/Cas9 system was demonstrated to knock out the chitinase 4 (EcChi4) gene (Gui et al., 2016).
Challenges in Fisheries and Aquaculture Applications
The CRISPR/Cas technology has a tremendous potential for aquaculture applications such as assessing the gene functions and the improvement of important commercial traits (Figure 3). However, several technical hurdles and ethical issues concerning the applications in aquaculture breeding still exist. Risk assessment, regulatory approval, and public/consumer acceptance are necessary for the commercialization of the CRISPR products.
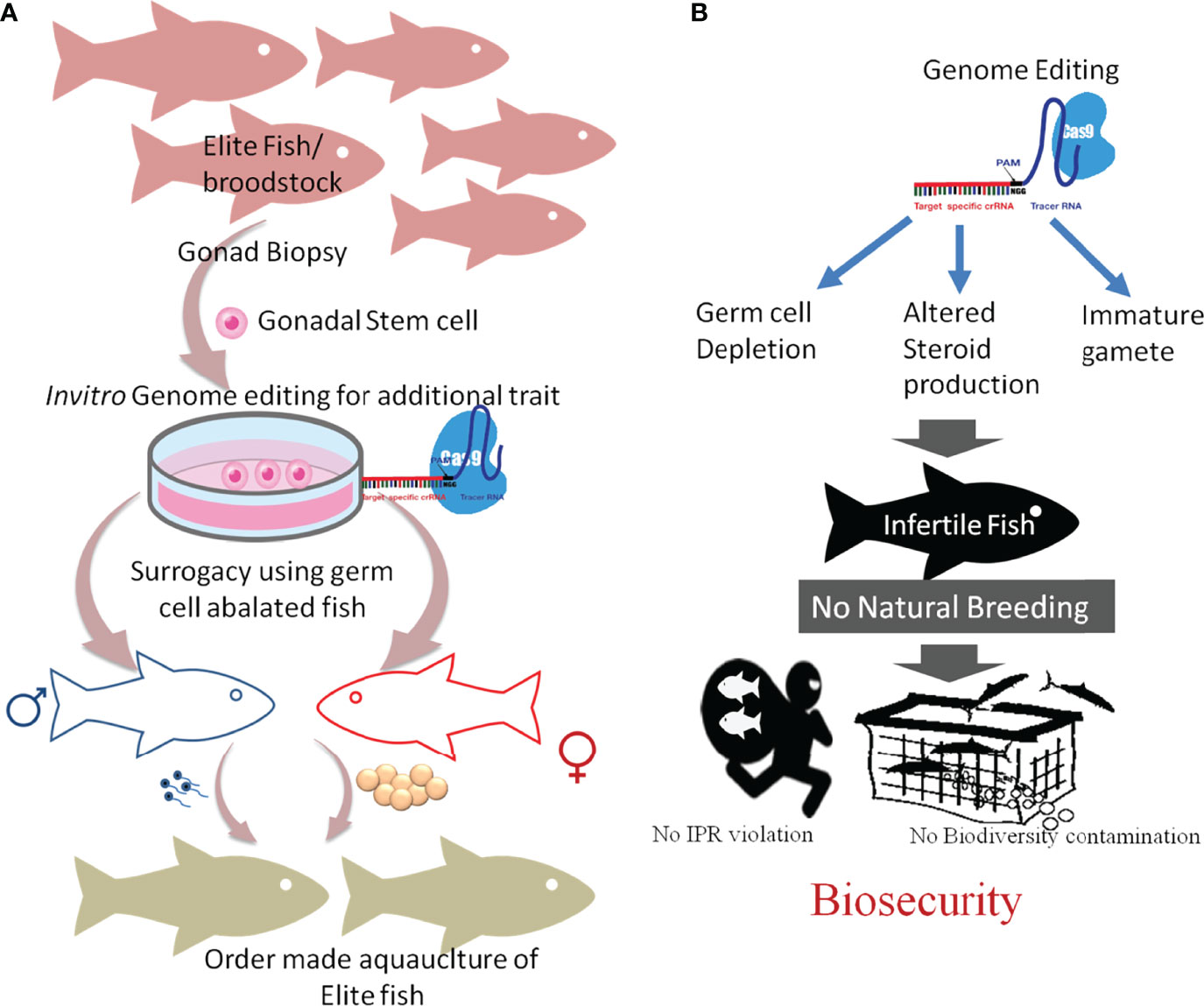
Figure 3 Future of genome editing in aquaculture and fisheries. It is assumed that genome editing will be largely adopted in aquaculture to improve its sustainability. To produce a genome-edited stock, germline mutation is pivotal. (A) The right panel showed a gonadal stem cell [germ cell precursor cells or gamete-producing cells (GSC)] from various high-performing (elite) fish will be collected, cultured, and modified using genome editing. The in vitro genome- edited GSC, which carries the mutation, when transplanted into sterilized (germ cell-less or abalated) host will produce male and female gametes, depending on the sex of the host. The mixing of these gametes will produce a specific superior strain. (B) Illustration showing the application of GSC-based ordermade strain production in infertile fish production, controlled aquaculture, and biosecurity.
Technical Challenges
- CRISPR editing requires a very clear and robust knowledge of the genetic background and genomic sequences. Although many important fish and aquatic species (>70) have been sequenced (Wargelius, 2019), they are still less compared to the total number of aquaculture (>600) species existing (FAO, 2020). Additionally, for many species, genome sequences were not well assembled and annotated; thus, the functions are still unknown for most of the genes (Sundaram et al., 2017). In aquaculture species, the genes that are known to be associated with important traits, such as growth, disease resistance, and robustness, are still very limited, which hampers in the CRISPR/Cas applications in aquaculture. Hence, an improved knowledge of the genome sequences and trait-related genes will help in the designing of gRNAs specific to a single targeted region.
- For the rapid selection of optimal gRNAs, CRISPR/Cas constructs better to be tested in the cell culture before in vivo editing (Gratacap et al., 2020; Yang et al., 2022). However, a problem arises since for aquaculture research, there is a short fall of well-characterized and suitable cell lines for many species of interest. In addition, for many crustacean and molluscan species, there are no well-established immortalized cell lines. Furthermore, various aspects of in vitro genome editing need to be optimized in aquaculture species. Therefore, developing such platforms will make genome-wide screening approaches a more realistic possibility in major aquaculture species.
- CRISPR/Cas construct needs to be delivered into fertilized eggs at the one-cell stage in fish (Güralp et al., 2020; Yang et al., 2022). The timing for the development of the fertilized egg to first cell division varies from species to species in fish; therefore, it is essential to know the time duration from fertilization to the first cell division so that the critical time for genome editing is not missed. Microinjection is labor intensive and needs a specific microinjection platform that is costly. The microinjection of a large number of eggs within a short time slot poses a challenge. Moreover, for species like certain shrimp, access to newly fertilized embryos is difficult. Therefore, the alternative methods of delivery of CRISPR/Cas9 have to be developed and tested for breeding purposes, for example, electroporation, sperm-mediated transfer, the microinjection of unfertilized ova, and editing primordial germ cells.
- The egg membrane makes a low microinjection success rate for oviparous fish, and in ovoviviparous fishes, there is no established gene editing platform at present. Moreover, in some fish species, the egg membrane/shell is soft and in some species, it is very hard (e.g., tilapia), so the standardization of the needle type and injection is essential (Okoli et al., 2021).
- Teleost fish experienced a whole genome duplication (WGD) event, and in the case of salmon, this WGD is expanded with the salmon-specific 4th round (Ss4R) (Glasauer and Neuhauss, 2014). Duplication hinders the CRISPR-editing efficiency and a comparison between genes with various copies in the genome could be performed to elucidate this issue (Cleveland et al., 2018; Datsomor et al., 2019b; Gratacap et al., 2019).
- In aquatic species where CRISPR/Cas9 has already been applied, the optimization of methods is required to maximize the editing efficiency, minimize off-target effects, and reduce the problem of mosaicism. Off-target editing can result in a non-specific and unintended modification to the genome that may also lead to unwanted impacts on the organism, and it requires a more careful assessment including both off-targeting in the genome and the potential risk related to food safety. Post-editing screening for unexpected mutations is necessary; however, because of the natural genetic variation in different families and strains, post-editing detection becomes complicated (Blix et al., 2021).
Regulatory and Consumer Acceptance
Government regulation and public acceptance are the greatest challenge for CRISPR/Cas genome editing in aquaculture. The CRISPR technology has the potential to improve aquaculture production; however, public and regulatory acceptance are the key to its potential being realized (Okoli et al., 2021). CRISPR/Cas-editing applications remain mostly at the research stage, and in aquaculture breeding programs, it is still limited. This occurs mainly due to the fact that the common public has not been convinced yet for the genome-edited products and the global regulatory environment on genome editing remains uncertain. CRISPR/as genome editing is different from the genetically modified organisms (GMOs). In GMOs, new-foreign genes combine into the original DNA, whereas CRISPR genome editing does not introduce any foreign DNA into genome of interest species. Rather, it is a small-controlled change to the original DNA that improves the targeted trait by removing the inferior alleles (Yang et al., 2022). This means that the GMOs product/fish is not a creature that would exist in nature, whereas CRISPR-edited product/fish where the original genes are partly altered similar to the selective breeding process, thus shortening the mutation evolution process that occurs in nature over many generations of breeding. However, still there is a debate going on whether genome-editing approaches should be considered separately than the GMOs. There are no specific and harmonized guidelines for regulatory acceptance yet available for the genome-edited products. Nevertheless, different regulatory authorities throughout the world including the EU, Argentina, Brazil, Australia, New Zealand, Canada, the United States, and Norway have started discussions on how to regulate products arising from the new CRISPR editing techniques (Eckerstorfer et al., 2019). The process of regulatory acceptance and guidelines may vary considerably in different countries where in some countries, genome-edited products are not considered as GMOs and hence bio-safety screening is not required, whereas some countries will consider for safety screening.
CRISPR genome editing products will be relatively easy for the consumer acceptance compared to GMOs, since CRISPR/Cas technology-derived products are called “genome edited” rather than “genetically modified,” this changes in nomenclature itself will save many adverse response which happened earlier for the GMOs (Figure 4). For promoting the consumer acceptance, public must be communicated well how and why CRISPR/Cas was applied and as well as for its value system, for example, producing healthy and quality food and increasing animal welfare, its safety, and sustainability points (Hallerman, 2021).
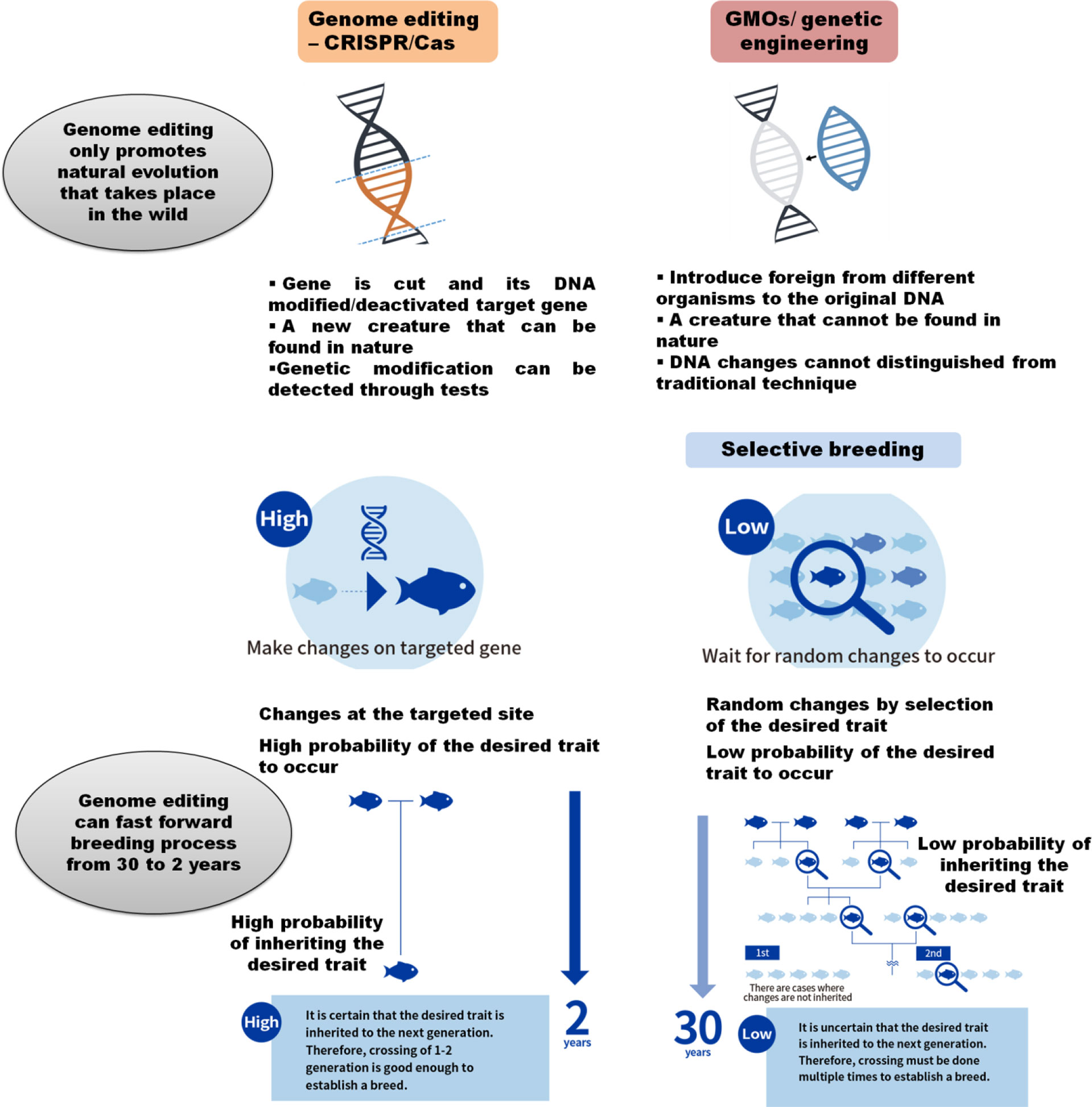
Figure 4 CRISPR/Cas as high-speed new breeding technology in aquaculture: CRISPR/Cas genome editing promotes natural evolution that naturally takes place in the wild and can fast forward the genetic improvement process at a high speed with minor targeted changes in the genome (Adapted from https://regional.fish/en/genome/).
Commercialization
The world’s first CRISPR/Cas genome-edited fish are now on the market for sale and reached to the consumer. Japan has approved the two CRISPR-edited fish: red sea bream and tiger puffer both developed by the Kyoto-based startup Regional Fish Institute with Kyoto University and Kindai University. Approval has been done from the Ministry of Health, Labour and Welfare (MHLW) and the Ministry of Agriculture, Forestry and Fisheries (MAFF) of Japan after completing the national application process (Japan embraces CRISPR-edited fish, 2022). The fishes were CRISPR edited to grow bigger and have more edible parts (1.2 times higher red sea bream and 1.9 times higher tiger puffer fish than the conventional counterpart). In Japan, GMO products/food that contain foreign genes must undergo safety screening, but CRISPR edited was exempt from this step since it was developed without the addition of foreign genes, so it is not genetically modified, but a change that can happen in nature. As a food, it is safe and has no negative impact on biodiversity. Another aquatic product has been submitted to regulatory review is FLT-01 Nile tilapia developed by the AquaBounty Company (Hallerman, 2021; https://www.fishfarmingexpert.com/article/aquabounty-gets-argentina-go-ahead-for-edited-tilapia/). According to AquaBounty, the CRISPR-edited Nile tilapia has led to increased muscle mass, a significant improvement in the fillet yield and feed conversion than its unedited counterpart. Since FLT-01 Nile tilapia fish does not contain any foreign DNA or a new combination of genetic material that would warrant the regulation as genetically modified in Argentina, unlike transgenic AquAdvantage salmon developed by the same company, it is not covered by the definition of a regulated article under Res. 763 under the Cartagena Protocol for Biosafety. Under Argentine Resolution 173/15-New Breeding Techniques, this fish is not classified as a GMO and Brazil made a similar determination for this genome-edited fish in 2019.
Future Perspectives and Conclusions
Genome editing technology offers great promise for functional genomics and gene therapy. The outcomes of CRISPR/Cas genome editing are promising and have potential to speed up the aquaculture breeding process. One thing is certain that majority of the expected genome-edited aquaculture products for commercialization will be developed through the CRISPR/Cas technology. However, its application for genetic improvement in aquaculture is just in its infancy. So far, the major research efforts in aquaculture are based on the traditional genetic improvement initiatives like growth, production, and reproduction. In the future, more precise and complex genome editing needs to be performed at multiple loci/chromosomal locations to improve the complex traits and there is also a need to include other economically important traits and diversify the aquaculture species. The continuous improvement of the CRISPR/Cas9 technique and its application in aquaculture as a new breeding technology will revolutionize the sector by increasing the production and quality. Moreover, many solutions in aquaculture are expected in the near future (disease resistance, sterile breeding and targeted therapy, etc.) that could not have been possible to solve before by the traditional ways. For CRISPR/Cas to play a bigger role in aquaculture as new breeding technology, there is a need for the identification and functional annotation of the trait-related genes and signal pathways. Nevertheless, substantial success has been observed in CRISPR/Cas editing in Atlantic salmon and tilapia fish species, and the derivable knowledge from this can be transferable to other fish species. Accordingly, Atlantic salmon and tilapia can be employed as aquaculture model fish species to initiate the optimization of aquatic protocols and assess the off-target effects. It is also relevant to ensure that the process of genome editing is sustainable with regard to environmental interests and animal welfare. At last, the key for CRISPR/Cas genome editing success in aquaculture relies on the subject to favorable regulatory, public, and consumer acceptance.
Author Contributions
SR has conceptualized, designed and written the original draft; VK reviewed and edited the manuscript, TC and SM have reviewed and edited the figure and have given the expertise inputs for improving the manuscript; JP, SM, BB, and BD have reviewed and edited the manuscript. All authors contributed to edit the manuscript and agreed to the published version of the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors are thankful to the Director ICAR-Central Inland Fisheries Research Institute (ICAR-CIFRI) for the support. The authors appreciate the help of Mr. Issei Yahiro for modification of Figure 3. Funds for JSPS Kakenhi (grant no: JP22K05832), Japan were used in the research. We would like to acknowledge all the researchers whose references were used for building the manuscript and reviewers for the inputs into the manuscript.
References
Alward B. A., Laud V. A., Skalnik C. J., York R. A., Juntti S. A., Fernald R. D. (2020). Modular Genetic Control of Social Status in a Cichlid Fish. Proc. Natl. Acad. Sci. U. S. A. 117, 28167–28174. doi: 10.1073/pnas.2008925117
Bassett A., Tibbit C., Ponting C., Liu J. (2013). Highly Efficient Targeted Mutagenesis of Drosophila With the CRISPR/Cas9 System. Cell Rep. 4, 220–228. doi: 10.1016/j.celrep.2013.06.020
Blix T. B., Dalmo R. A., Wargelius A., Myhr A. I. (2021). Genome Editing on Finfish: Current Status and Implications for Sustainability. Rev. Aquac. 13, 2344–2363. doi: 10.1111/raq.12571
Boch J., Bonas U. (2010). Xanthomonas AvrBs3 Family-Type III Effectors: Discovery and Function. Annu. Rev. Phytopathol. 48, 419–436. doi: 10.1146/annurev-phyto-080508-081936
Brinkman E. K., Kousholt A. N., Harmsen T., Leemans C., Chen T., Jonkers J., et al. (2018). Easy Quantification of Template-Directed CRISPR/Cas9 Editing. Nucleic Acids Res. 46, e58–e58. doi: 10.1093/nar/gky164
Chakrapani V., Patra S. K., Panda R. P., Rasal K. D., Jayasankar P., Barman H. K. (2016). Establishing Targeted Carp TLR22 Gene Disruption via Homologous Recombination Using CRISPR/Cas9. Dev. Comp. Immunol. 61, 242–247. doi: 10.1016/j.dci.2016.04.009
Chen J., Jiang D., Tan D., Fan Z., Wei Y., Li M., et al. (2017). Heterozygous Mutation of Eef1a1b Resulted in Spermatogenesis Arrest and Infertility in Male Tilapia, Oreochromis Niloticus. Sci. Rep. 7, 1–15. doi: 10.1038/srep43733
Chen H., Wang J., Du J., Si Z., Yang H., Xu X., et al. (2019). ASIP Disruption via CRISPR/Cas9 System Induces Black Patches Dispersion in Oujiang Color Common Carp. Aquaculture 498, 230–235. doi: 10.1016/j.aquaculture.2018.08.057
Chen J., Wang W., Tian Z., Dong Y., Dong T., Zhu H., et al. (2018). Efficient Gene Transfer and Gene Editing in Sterlet (Acipenser Ruthenus). Front. Genet. 9. doi: 10.3389/fgene.2018.00117
Clement K., Rees H., Canver M. C., Gehrke J. M., Farouni R., Hsu J. Y., et al. (2019). CRISPResso2 Provides Accurate and Rapid Genome Editing Sequence Analysis. Nat. Biotechnol. 37, 224–226. doi: 10.1038/s41587-019-0032-3
Cleveland B. M., Yamaguchi G., Radler L. M., Shimizu M. (2018). Editing the Duplicated Insulin-Like Growth Factor Binding Protein-2b Gene in Rainbow Trout (Oncorhynchus Mykiss). Sci. Rep. 8. doi: 10.1038/s41598-018-34326-6
Conant D., Hsiau T., Rossi N., Oki J., Maures T., Waite K., et al. (2022). Inference of CRISPR Edits From Sanger Trace Data. Cris. J. 5, 123–130. doi: 10.1089/crispr.2021.0113
Cong L., Ran F., Cox D., Lin S., Barretto R., Habib N., et al. (2013). Multiplex Genome Engineering Using CRISPR/Cas Systems. Sci. (80-. ) 339, 819–823. doi: 10.1126/science.1231143
Cui Y., Xu J., Cheng M., Liao X., Peng S. (2018). Review of CRISPR/Cas9 sgRNA Design Tools. Interdiscip. Sci. Comput. Life Sci. 10, 455–465. doi: 10.1007/s12539-018-0298-z
Dan C., Lin Q, Gong G., Yang T., Xiong S., Xiong Y., et al (2018). A Novel PDZ Domain-Containing Gene Is Essential for Male Sex Differentiation and Maintenance in Yellow Catfish (Pelteobagrus fulvidraco). Sci. Bull. 63, 1420–1430. doi: 10.1016/j.scib.2018.08.012.
Datsomor A. K., Olsen R. E., Zic N., Madaro A., Bones A. M., Edvardsen R. B., et al. (2019a). CRISPR/Cas9-Mediated Editing of Δ5 and Δ6 Desaturases Impairs Δ8-Desaturation and Docosahexaenoic Acid Synthesis in Atlantic Salmon (Salmo Salar L.). Sci. Rep. 9, 16888. doi: 10.1038/s41598-019-53316-w
Datsomor A. K., Zic N., Li K., Olsen R. E., Jin Y., Vik J. O., et al. (2019b). CRISPR/Cas9-Mediated Ablation of Elovl2 in Atlantic Salmon (Salmo Salar L.) Inhibits Elongation of Polyunsaturated Fatty Acids and Induces Srebp-1 and Target Genes. Sci. Rep. 9, 7533. doi: 10.1038/s41598-019-43862-8
Dehler C. E., Lester K., Della Pelle G., Jouneau L., Houel A., Collins C., et al. (2019). Viral Resistance and IFN Signaling in STAT2 Knockout Fish Cells. J. Immunol. 203, 465–475. doi: 10.4049/jimmunol.1801376.
Doench J. G., Fusi N., Sullender M., Hegde M., Vaimberg E. W., Donovan K. F., et al. (2016). Optimized sgRNA Design to Maximize Activity and Minimize Off-Target Effects of CRISPR-Cas9. Nat. Biotechnol. 34, 184–191. doi: 10.1038/nbt.3437
Doudna J., Charpentier E. (2014). The New Frontier of Genome Engineering With CRISPR–Cas9. Sci. (80-. ). 346, 1258096. doi: 10.1126/science.1258096
Du J., Chen H., Mandal B. K., Wang J., Shi Z., Lu G., et al. (2021). HDL Receptor/Scavenger Receptor B1-Scarb1 and Scarb1-Like Mediate the Carotenoid-Based Red Coloration in Fish. Aquaculture 545. doi: 10.1016/j.aquaculture.2021.737208
Eckerstorfer M. F., Dolezel M., Heissenberger A., Miklau M., Reichenbecher W., Steinbrecher R. A., et al. (2019). An EU Perspective on Biosafety Considerations for Plants Developed by Genome Editing and Other New Genetic Modification Techniques (nGMs). Front. Bioeng. Biotechnol. 7. doi: 10.3389/fbioe.2019.00031
Edvardsen R. B., Leininger S., Kleppe L., Skaftnesmo K. O., Wargelius A. (2014). Targeted Mutagenesis in Atlantic Salmon (Salmo Salar L.) Using the CRISPR/Cas9 System Induces Complete Knockout Individuals in the F0 Generation. PloS One 9, e108622. doi: 10.1371/journal.pone.0108622
Elaswad A., Khalil K., Cline D., Page-McCaw P., Chen W., Michel M., et al. (2018). Microinjection of CRISPR/cas9 Protein Into Channel Catfish, Ictalurus Punctatus, Embryos for Gene Editing. J. Vis. Exp. 2018, 1–12. doi: 10.3791/56275
FAO (2020). The State of World Fsheries and Aquaculture 2020: Sustainability in Action (Rome: FAO). Available at: 10.4060/ca9229en.
Fujii R. (2000). The Regulation of Motile Activity in Fish Chromatophores. Pigment Cell Res. 13, 300–319. doi: 10.1034/j.1600-0749.2000.130502.x
Gan R. H., Wang Y., Li Z., Yu Z. X., Li X. Y., Tong J. F., et al. (2021). Functional Divergence of Multiple Duplicated Foxl2 Homeologs and Alleles in a Recurrent Polyploid Fish. Mol. Biol. Evol. 38, 1995–2013. doi: 10.1093/molbev/msab002
Gaj T., Sirk S. J., Shui S.-L., Liu J. (2016). Genome-Editing Technologies: Principles and Applications. Cold Spring Harb. Perspect. Biol. 8, a023754. doi: 10.1101/cshperspect.a023754
Glasauer S. M. K., Neuhauss S. C. F. (2014). Whole-Genome Duplication in Teleost Fishes and Its Evolutionary Consequences. Mol. Genet. Genomics 289, 1045–1060. doi: 10.1007/s00438-014-0889-2
Gratacap R. L., Regan T., Dehler C. E., Martin S. A. M., Boudinot P., Collet B., et al. (2020). Efficient CRISPR/Cas9 Genome Editing in a Salmonid Fish Cell Line Using a Lentivirus Delivery System. BMC Biotechnol. 20. doi: 10.1186/s12896-020-00626-x
Gratacap R. L., Wargelius A., Edvardsen R. B., Houston R. D. (2019). Potential of Genome Editing to Improve Aquaculture Breeding and Production. Trends Genet. 35, 672–684. doi: 10.1016/j.tig.2019.06.006
Gui T., Zhang J., Song F., Sun Y., Xie S., Yu K., et al. (2016). CRISPR/Cas9-Mediated Genome Editing and Mutagenesis of EcChi4 in Exopalaemon Carinicauda. G3 Genes Genomes Genet. 6, 3757–3764. doi: 10.1534/g3.116.034082
Güralp H., Skaftnesmo K. O., Kjærner-Semb E., Straume A. H., Kleppe L., Schulz R. W., et al. (2020). Rescue of Germ Cells in Dnd Crispant Embryos Opens the Possibility to Produce Inherited Sterility in Atlantic Salmon. Sci. Rep. 10, 1–12. doi: 10.1038/s41598-020-74876-2
Hallerman E. (2021). Genome Editing in Cultured Fishes. CABI Agric. Biosci. 2. doi: 10.1186/s43170-021-00066-3
Hanna R. E., Doench J. G. (2020). Design and Analysis of CRISPR–Cas Experiments. Nat. Biotechnol. 38, 813–823. doi: 10.1038/s41587-020-0490-7
Hege Straume A., Kjaerner-Semb E., Skaftnesmo K. O., Güralp H., Lillico S., Wargelius A., et al. A Refinement to Gene Editing in Atlantic Salmon Using Asymmetrical Oligonucleotide. doi: 10.1101/2021.02.08.430296
Heigwer F., Kerr G., Boutros M. (2014). E-CRISP: Fast CRISPR Target Site Identification. Nat. Methods 11, 122–123. doi: 10.1038/nmeth.2812
hu T., Zhai G., Pradhan A., Olsson P.-E., Yin Z. (2020). Zebrafish Cyp17a1 Knockout Reveals That Androgen-Mediated Signaling Is Important for Male Brain Sex Differentiation. Gen. Comp. Endocrinol. 295, 113490. doi: 10.1016/j.ygcen.2020.113490
Hwang W. Y., Fu, Y., Reyon D., Morgan L. M. S. Q. T., Sander J. D., Peterson R. T., et al. (2013). Efficient Genome Editing in Zebrafish Using a CRISPR-Cas System. Nat. Biotechnol. 31, 227–229. doi: 10.1038/nbt.2501
Jiang D., Chen J., Fan Z., Tan D., Zhao J., Shi H., et al. (2017). CRISPR/Cas9-Induced Disruption of Wt1a and Wt1b Reveals Their Different Roles in Kidney and Gonad Development in Nile Tilapia. Dev. Biol. 428, 63–73. doi: 10.1016/j.ydbio.2017.05.017
Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J., Charpentier E. (2012). A Programmable Dual-RNA-Guided DNA Endonuclease in Adaptive Bacterial Immunity. Sci. 337(6096), 816−821. doi: 10.1126/science.1225829
Khalil K., Elayat M., Khalifa E., Daghash S., Elaswad A., Miller M., et al. (2017). Generation of Myostatin Gene-Edited Channel Catfish (Ictalurus Punctatus) via Zygote Injection of CRISPR/Cas9 System. Sci. Rep. 7, 1–12. doi: 10.1038/s41598-017-07223-7
Khan S. H. (2019). Genome-Editing Technologies: Concept, Pros, and Cons of Various Genome-Editing Techniques and Bioethical Concerns for Clinical Application. Mol. Ther. - Nucleic Acids 16, 326–334. doi: 10.1016/j.omtn.2019.02.027
Kikuchi M., Nishimura T., Ishishita S., Matsuda Y., Tanaka M. (2020). Foxl3, a Sexual Switch in Germ Cells, Initiates Two Independent Molecular Pathways for Commitment to Oogenesis in Medaka. Proc. Natl. Acad. Sci. U. S. A. 117, 12174−12181 doi: 10.1073/pnas.1918556117
Kim Y. G., Cha J., Chandrasegaran S. (1996). Hybrid Restriction Enzymes: Zinc Finger Fusions to Fok I Cleavage Domain. Proc. Natl. Acad. Sci. U. S. A. 93, 1156–1160. doi: 10.1073/pnas.93.3.1156
Kim J., Cho J. Y., Kim J.-W., Kim H.-C., Noh J. K., Kim Y.-O., et al. (2019). CRISPR/Cas9-Mediated Myostatin Disruption Enhances Muscle Mass in the Olive Flounder Paralichthys Olivaceus. Aquaculture 512, 734336. doi: 10.1016/j.aquaculture.2019.734336
Kim J., Cho J. Y., Kim J. W., Kim D. G., Nam B. H., Kim B. S., et al. (2021). Molecular Characterization of Paralichthys Olivaceus Maf1 and its Potential Role as an Anti-Viral Hemorrhagic Septicaemia Virus Factor in Hirame Natural Embryo Cells. Int. J. Mol. Sci. 22, 1–15. doi: 10.3390/ijms22031353
Kimura T., Nagao Y., Hashimoto H., Yamamoto-Shiraishi Y., Yamamoto S., Yabe T., et al. (2014). Leucophores are Similar to Xanthophores in Their Specification and Differentiation Processes in Medaka. Proc. Natl. Acad. Sci. U. S. A. 111, 7343–7348. doi: 10.1073/pnas.1311254111
Kishimoto K., Washio Y., Yoshiura Y., Toyoda A., Ueno T., Fukuyama H., et al. (2018). Production of a Breed of Red Sea Bream Pagrus Major With an Increase of Skeletal Muscle Muss and Reduced Body Length by Genome Editing With CRISPR/Cas9. Aquaculture 495, 415–427. doi: 10.1016/j.aquaculture.2018.05.055
Kleinstiver B. P., Prew M. S., Tsai S. Q., Topkar V., Nguyen T., Zheng Z., et al. (2016). Engineered CRISPR-Cas9 Nucleases With Altered PAM Specificities. Nature 523, 481–485. doi: 10.1038/nature14592
Kleppe L., Andersson E., Skaftnesmo K. O., Edvardsen R. B., Fjelldal P. G., Norberg B., et al. (2017). Sex Steroid Production Associated With Puberty Is Absent in Germ Cell-Free Salmon. Sci. Rep. 7, 1–11. doi: 10.1038/s41598-017-12936-w
Labun K., Montague T. G., Gagnon J. A., Thyme S. B., Valen E. (2016). CHOPCHOP V2: A Web Tool for the Next Generation of CRISPR Genome Engineering. Nucleic Acids Res. 44, W272–W276. doi: 10.1093/nar/gkw398
Li D., Qiu Z., Shao Y., Chen Y., Guan Y., Liu M., et al. (2013). Heritable Gene Targeting in the Mouse and Rat Using a CRISPR-Cas System. Nat. Biotechnol. 31, 681–683. doi: 10.1038/nbt.2661
Li Q., Shao G., Ding Y., Xu L., Shao J., Ao J., et al. (2021). Effective CRISPR/Cas9-Based Genome Editing in Large Yellow Croaker (Larimichthys crocea). Aquac. Fish. doi: 10.1016/j.aaf.2021.04.008
Liu Q., Qi Y., Liang Q., Song J., Liu J., Li W., et al. (2019). Targeted Disruption of Tyrosinase Causes Melanin Reduction in Carassius Auratus Cuvieri and its Hybrid Progeny. Sci. China Life Sci. 62, 1194–1202. doi: 10.1007/s11427-018-9404-7
Li M., Wang D. (2017). Gene Editing Nuclease and its Application in Tilapia. Sci. Bull. 62, 165–173. doi: 10.1016/j.scib.2017.01.003
Li H., Yang Y., Hong W., Huang M., Wu M., Zhao X. (2020). Applications of Genome Editing Technology in the Targeted Therapy of Human Diseases: Mechanisms, Advances and Prospects. Signal Transduction Targeting Ther. 5. doi: 10.1038/s41392-019-0089-y
Li M., Yang H., Zhao J., Fang L., Shi H., Li M., et al. (2014). Efficient and Heritable Gene Targeting in Tilapia by CRISPR/Cas9. Genetics 197, 591–599. doi: 10.1534/genetics.114.163667
Luo M., Wang J., Dong Z., Wang C., Lu G. (2022). CRISPR-Cas9 sgRNA Design and Outcome Assessment: Bioinformatics Tools and Aquaculture Applications. Aquac. Fish. 7, 121–130. doi: 10.1016/j.aaf.2021.10.002
Ma J., Fan Y., Zhou Y., Liu W., Jiang N., Zhang J., et al. (2018). Efficient Resistance to Grass Carp Reovirus Infection in JAM-A Knockout Cells Using CRISPR/Cas9. Fish Shellfish Immunol. 76, 206–215. doi: 10.1016/j.fsi.2018.02.039
Mali P., Yang L., Esvelt K., Aach J., Guell M., DiCarlo J., et al. (2013). RNA-Guided Human Genome Engineering via Cas9. Sci. (80-. ). 339, 823–826. doi: 10.1126/science.1232033
Mank J. E., Avise J. C. (2009). Evolutionary Diversity and Turn-Over of Sex Determination in Teleost Fishes. Sex Dev. 3, 60–67. doi: 10.1159/000223071
Miller J. C., Holmes M. C., Wang J., Guschin D. Y., Lee Y.-L., Rupniewski I., et al. (2007). An Improved Zinc-Finger Nuclease Architecture for Highly Specific Genome Editing. Nat. Biotechnol. 25, 778–785. doi: 10.1038/nbt1319
Mohapatra S., Chakraborty T. (2021). Advances in Fisheries Biotechnology. Eds. Kumar P.P., Janmejay P. (Singapore: Springer International) doi: 10.1007/978-981-16-3215-0
Mojica F. J. M., Rodriguez-Valera F. (2016). The Discovery of CRISPR in Archaea and Bacteria. FEBS J. 283, 3162–3169. doi: 10.1111/febs.13766
Montague T. G., Cruz J. M., Gagnon J. A., Church G. M., Valen E. (2014). CHOPCHOP: A CRISPR/Cas9 and TALEN Web Tool for Genome Editing. Nucleic Acids Res. 42, W401–W407. doi: 10.1093/nar/gku410
Moreno-Mateos M. A., Vejnar C. E., Beaudoin J.-D., Fernandez J. P., Mis E. K., Khokha M. K., et al. (2015). CRISPRscan: Designing Highly Efficient sgRNAs for CRISPR-Cas9 Targeting In Vivo. Nat. Methods 12, 982–988. doi: 10.1038/nmeth.3543
Nagasawa K., Ishida M., Octavera A., Kusano K., Kezuka F., Kitano T., et al. (2019). Novel Method for Mass Producing Genetically Sterile Fish From Surrogate Broodstock via Spermatogonial Transplantation. Biol. Reprod. 100, 535–546. doi: 10.1093/biolre/ioy204
Naito Y., Hino K., Bono H., Ui-Tei K. (2015). CRISPRdirect: Software for Designing CRISPR/Cas Guide RNA With Reduced Off-Target Sites. Bioinformatics 31, 1120–1123. doi: 10.1093/bioinformatics/btu743
Nishiike Y., Miyazoe D., Togawa R., Yokoyama K., Nakasone K., Miyata M., et al. (2021). Estrogen Receptor 2b Is the Major Determinant of Sex-Typical Mating Behavior and Sexual Preference in Medaka. Curr. Biol. 31, 1699–1710.e6. doi: 10.1016/j.cub.2021.01.089
Niu Y., Shen B., Cui Y., Chen Y., Wang J., Wang L., et al. (2014). Generation of Gene-Modified Cynomolgus Monkey via Cas9/RNA-Mediated Gene Targeting in One-Cell Embryos. Cell 156, 836–843. doi: 10.1016/j.cell.2014.01.027
Ohama M., Washio Y., Kishimoto K., Kinoshita M., Kato K. (2020). Growth Performance of Myostatin Knockout Red Sea Bream Pagrus Major Juveniles Produced by Genome Editing With CRISPR/Cas9. Aquaculture 529, 735672. doi: 10.1016/j.aquaculture.2020.735672
Ohga H., Matsuyama M. (2021). Effects of LPXRFamide Peptides on Chub Mackerel Gonadotropin Secretion†. Biol. Reprod. 105, 1179–1188. doi: 10.1093/biolre/ioab130
Okoli A. S., Blix T., Myhr A. I., Xu W., Xu X. (2021). Sustainable Use of CRISPR/Cas in Fish Aquaculture: The Biosafety Perspective. Transgenic Res. doi: 10.1007/s11248-021-00274-7
Ota S., Hisano Y., Ikawa Y., Kawahara A. (2014). Multiple Genome Modifications by the CRISPR/Cas9 System in Zebrafish. Genes to Cells 19, 555–564. doi: 10.1111/gtc.12154
Ota S., Hisano Y., Muraki M., Hoshijima K., Dahlem T. J., Grunwald D. J., et al. (2013). Efficient Identification of TALEN-Mediated Genome Modifications Using Heteroduplex Mobility Assays. Genes Cells 18, 450–458. doi: 10.1111/gtc.12050
Pandey D., Matsubara T., Saito T., Kazeto Y., Gen K., Sakuma T., et al (2021). TALEN-Mediated Gene Editing of slc24a5 (Solute Carrier Family 24, Member 5) in Kawakawa, Euthynnus affinis. J. Mar. Sci. Eng. 9. doi: 10.3390/jmse9121378
Park J., Childs L., Kim D., Hwang G.-H., Kim S., Kim S.-T., et al. (2017). Digenome-Seq Web Tool for Profiling CRISPR Specificity. Nat. Methods 14, 548–549. doi: 10.1038/nmeth.4262
Pattanayak V., Ramirez C. L., Joung J. K., Liu D. R. (2011). Revealing Off-Target Cleavage Specificities of Zinc-Finger Nucleases by In Vitro Selection. Nat. Methods 8, 765–770. doi: 10.1038/nmeth.1670
Qin Z., Li Y., Su B., Cheng Q., Ye Z., Perera D. A., et al. (2016). Editing of the Luteinizing Hormone Gene to Sterilize Channel Catfish, Ictalurus Punctatus, Using a Modified Zinc Finger Nuclease Technology With Electroporation. Mar. Biotechnol. 18, 255–263. doi: 10.1007/s10126-016-9687-7
Segev-Hadar A., Slosman T., Rozen A., Sherman A., Cnaani A., Biran J. (2021). Genome Editing Using the CRISPR-Cas9 System to Generate a Solid-Red Germline of Nile Tilapia (Oreochromis Niloticus). Cris. J. 4, 583–594. doi: 10.1089/crispr.2020.0115
Shimbun Y. (2021). Kyoto Frm Puts Genome-Edited Tiger Pufer on the Table (Japan: Japan News). Available at: https://the-japan-news.com/news/article/000793605.
Simora R. M. C., Xing D., Bangs M. R., Wang W., Ma X., Su B., et al. (2020). CRISPR/Cas9-Mediated Knock-in of Alligator Cathelicidin Gene in a Non-Coding Region of Channel Catfish Genome. Sci. Rep. 10, 1–14. doi: 10.1038/s41598-020-79409-5
Smith J., Bibikova M., Whitby F. G., Reddy A. R., Chandrasegaran S., Carroll D. (2000). Requirements for Double-Strand Cleavage by Chimeric Restriction Enzymes With Zinc Finger DNA-Recognition Domains. Nucleic Acids Res. 28, 3361–3369. doi: 10.1093/nar/28.17.3361
Stemmer M., Thumberger T., Del Sol Keyer M., Wittbrodt J., Mateo J. L. (2015). CCTop: An Intuitive, Flexible and Reliable CRISPR/Cas9 Target Prediction Tool. PloS One 10, 1–11. doi: 10.1371/journal.pone.0124633
Straume A. H., Kjærner-Semb E., Skaftnesmo K.O., Güralp H., Lillico S., Wargelius A., et al (2021). Single Nucleotide Replacement in the Atlantic Salmon Genome using CRISPR/Cas9 and Asymmetrical Oligonucleotide Donors. BMC Genomics 22, 563. doi: 10.1186/s12864-021-07823-8
Sundaram A., Tengs T., Grimholt U. (2017). Issues With RNA-Seq Analysis in Non-Model Organisms: A Salmonid Example. Dev. Comp. Immunol. 75, 38–47. doi: 10.1016/j.dci.2017.02.006
Sun Y., Yan C., Liu M., Liu Y., Wang W., Cheng W., et al. (2020a). CRISPR/Cas9-Mediated Deletion of One Carotenoid Isomerooxygenase Gene (EcNinaB-X1) From Exopalaemon Carinicauda. Fish Shellfish Immunol. 97, 421–431. doi: 10.1016/j.fsi.2019.12.037
Sun Y., Zheng G.-D., Nissa M., Chen J., Zou S.-M. (2020b). Disruption of Mstna and Mstnb Gene Through CRISPR/Cas9 Leads to Elevated Muscle Mass in Blunt Snout Bream (Megalobrama Amblycephala). Aquaculture 528, 735597. doi: 10.1016/j.aquaculture.2020.735597
Tang H., Chen Y., Wang L., Yin Y., Li G., Guo Y., et al. (2018). Fertility Impairment With Defective Spermatogenesis and Steroidogenesis in Male Zebrafish Lacking Androgen Receptor. Biol. Reprod. 98, 227–238. doi: 10.1093/biolre/iox165
Tao B., Tan J., Chen L., Xu Y., Liao X., Li Y., et al. (2021). CRISPR/Cas9 System-Based Myostatin-Targeted Disruption Promotes Somatic Growth and Adipogenesis in Loach, Misgurnus Anguillicaudatus. Aquaculture 544, 737097. doi: 10.1016/j.aquaculture.2021.737097
Tzur Y. B., Friedland A. E., Nadarajan S., Church G. M., Calarco J. A., Colaiácovo M. P. (2013). Heritable Custom Genomic Modifications in Caenorhabditis Elegans via a CRISPR-Cas9 System. Genetics 195, 1181–1185. doi: 10.1534/genetics.113.156075
Urnov F., Rebar E., Holmes M., Zhang H., Gregory P. (2010). Genome Editing With Engineered Zinc Finger Nucleases. Nat. Rev. Genet. 11, 636–646. doi: 10.1038/nrg2842
Wang C., Baoyue L., Li T., Xu G. L. M., Liu X., Tao W., et al. (2021). Nile Tilapia: A Model for Studying Teleost Color Patterns. J. Hered. 112, 469–484. doi: 10.1093/jhered/esab018 Special Issue Article doi: 10.1093/jhered/esab018
Wang H., Yang H., Shivalila C. S., Dawlaty M. M., Cheng W. (2013). One-Step Generation of Mice Carrying Mutations in Multiple Genes by CRISPR/Cas-Mediated Genome Engineering. Cell 153, 910–918. doi: 10.1016/j.cell.2013.04.025
Wargelius A. (2019). Application of Genome Editing in Aquatic Farm Animals: Atlantic Salmon. Transgenic Res. 28, 101–105. doi: 10.1007/s11248-019-00163-0
Wargelius A., Leininger S., Skaftnesmo K. O., Kleppe L., Andersson E., Taranger G. L., et al. (2016). Dnd Knockout Ablates Germ Cells and Demonstrates Germ Cell Independent Sex Differentiation in Atlantic Salmon. Sci. Rep. 6, 1–8. doi: 10.1038/srep21284
Xie Q. P., He X., Sui Y. N., Chen L. L., Sun L. N., Wang D. S. (2016). Haploinsufficiency of SF-1 Causes Female to Male Sex Reversal in Nile Tilapia, Oreochromis Niloticus. Endocrinology 157, 2500–2514. doi: 10.1210/en.2015-2049
Xu X., Cao X., Gao J. (2019). Production of a Mutant of Large-Scale Loach Paramisgurnus Dabryanus With Skin Pigmentation Loss by Genome Editing With CRISPR/Cas9 System. Transgenic Res. 28, 341–356. doi: 10.1007/s11248-019-00125-6
Xu S., Pham T. P., Neupane S. (2020). Delivery Methods for CRISPR/Cas9 Gene Editing in Crustaceans. Mar. Life Sci. Technol. 2. doi: 10.1007/s42995-019-00011-4
Yagi Y., Hayashi S., Kobayashi K., Hirayama T., Nakamura T. (2013). Elucidation of the RNA Recognition Code for Pentatricopeptide Repeat Proteins Involved in Organelle RNA Editing in Plants. PLoS One 8. doi: 10.1371/journal.pone.0057286
Yan T., Cai Y., He J., Zhang Q., Wang X., Zhang S., et al. (2019). Characterization and Expression Profiles of Cyp19a1a in the Schizothoracine Fish Schizothorax Prenanti. Tissue Cell 58, 70–75. doi: 10.1016/j.tice.2019.04.008
Yang G., Huang X. (2019). Methods and Applications of CRISPR/Cas System for Genome Editing in Stem Cells. Cell Regen. 8, 33–41. doi: 10.1016/j.cr.2019.08.001
Yang Z., Yu Y., Tay Y. X., Yue G. H. (2022). Genome Editing and its Applications in Genetic Improvement in Aquaculture. Rev. Aquac. 14, 178–191. doi: 10.1111/raq.12591
Yu H., Li H., Li Q., Xu R., Yue C., Du S. (2019). Targeted Gene Disruption in Pacific Oyster Based on CRISPR/Cas9 Ribonucleoprotein Complexes. Mar. Biotechnol. 21, 301–309. doi: 10.1007/s10126-019-09885-y
Zhai G., Shu T., Chen K., Lou Q., Jia J., Huang J., et al. (2022). Successful Production of an All-Female Common Carp (Cyprinus Carpio L.) Population Using Cyp17a1-Deficient Neomale Carp. Engineering 8, 181–189. doi: 10.1016/j.eng.2021.03.026
Zhang Z., Lau S. W., Zhang L., Ge W. (2015). Disruption of Zebrafish Follicle-Stimulating Hormone Receptor (Fshr) But Not Luteinizing Hormone Receptor (Lhcgr) Gene by TALEN Leads to Failed Follicle Activation in Females Followed by Sexual Reversal to Males. Endocrinol. (United States) 156, 3747–3762. doi: 10.1210/en.2015-1039
Zhang Q., Ye D., Wang H., Wang Y., Hu W., Sun Y. (2020). Zebrafish Cyp11c1 Knockout Reveals the Roles of 11-Ketotestosterone and Cortisol in Sexual Development and Reproduction. Endocrinology 161, bqaa048. doi: 10.1210/endocr/bqaa048
Zhong Z., Niu P., Wang M., Huang G., Xu S., Sun Y., et al. (2016). Targeted Disruption of Sp7 and Myostatin With CRISPR-Cas9 Results in Severe Bone Defects and More Muscular Cells in Common Carp. Sci. Rep. 6, 1–14. doi: 10.1038/srep22953
Zhou L., Li M., Wang D. (2021). Role of Sex Steroids in Fish Sex Determination and Differentiation as Revealed by Gene Editing. Gen. Comp. Endocrinol. 313, 113893. doi: 10.1016/j.ygcen.2021.113893
Keywords: genome editing, CRISPR/Cas, aquaculture, knock out, CRISPR technology, fisheries
Citation: Roy S, Kumar V, Behera BK, Parhi J, Mohapatra S, Chakraborty T and Das BK (2022) CRISPR/Cas Genome Editing—Can It Become a Game Changer in Future Fisheries Sector? Front. Mar. Sci. 9:924475. doi: 10.3389/fmars.2022.924475
Received: 25 April 2022; Accepted: 06 June 2022;
Published: 14 July 2022.
Edited by:
Dong Han, Institute of Hydrobiology (CAS), ChinaReviewed by:
Xiaodan Wang, East China Normal University, ChinaGang Zhai, University of Chinese Academy of Sciences, China
Copyright © 2022 Roy, Kumar, Behera, Parhi, Mohapatra, Chakraborty and Das. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Basanta Kumar Das, YmFzYW50YWt1bWFyZEBnbWFpbC5jb20=; Bijay Kumar Behera, YmVoZXJhYmsxOEB5YWhvby5jby5pbg==; Tapas Chakraborty, dGFwYXNfY2hAYWdyLmt5dXNodS11LmFjLmpw
 Suvra Roy
Suvra Roy Vikash Kumar
Vikash Kumar Bijay Kumar Behera
Bijay Kumar Behera Janmejay Parhi
Janmejay Parhi Sipra Mohapatra4,5
Sipra Mohapatra4,5 Tapas Chakraborty
Tapas Chakraborty Basanta Kumar Das
Basanta Kumar Das