- 1Oceans Institute, The University of Western Australia, Crawley, WA, Australia
- 2School of Biological Sciences, University of Western Australia, Crawley, WA, Australia
- 3Cold Spring Harbor Laboratory, Simons Center for Quantitative Biology, Cold Spring Harbor, New York, NY, United States
- 4Institute of Marine Research, His, Norway
- 5National Marine Science Centre, New South Wales Fisheries, Coffs Harbour, NSW, Australia
- 6National Marine Science Centre, Southern Cross University, Coffs Harbour, NSW, Australia
Genetic diversity can influence resilience and adaptative capacity of organisms to environmental change. Genetic diversity within populations is largely structured by reproduction, with the prevalence of asexual versus sexual reproduction often underpinning important diversity metrics that determine selection efficacy. Asexual or clonal reproduction is expected to reduce genotypic diversity and slow down adaptation through reduced selection efficacy, yet the evolutionary consequences of clonal reproduction remain unclear for many natural populations. Here, we examine the genomic consequences of sympatric sexual (haplodiplontic) and clonal morphs of the kelp Ecklonia radiata that occur interspersed on reefs in Hamelin Bay, Western Australia. Using genome-wide single nucleotide polymorphisms, we confirm significant asexual reproduction for the clonal populations, indicated by a significantly lower number of multi-locus lineages and higher intra-individual diversity patterns (individual multi-locus heterozygosity, MLH). Nevertheless, co-ancestry analysis and breeding experiments confirmed that sexual reproduction by the clonal morph and interbreeding between the two morphs is still possible, but varies among populations. One clonal population with long-term asexuality showed trends of decreased selection efficacy (increased ratio non- vs. synonymous gene diversities). Yet, all clonal populations showed distinct patterns of putative local adaptation relative to the sexual morph, possibly indicating maladaptation to local environmental conditions and high vulnerability of this unique clonal morph to environmental stress.
Introduction
Global climate change is threatening ecosystems worldwide (Pecl et al., 2017) with rising temperatures and increasing frequency and intensity of extreme climatic events (Oliver et al., 2019; IPCC, 2021) resulting in range contractions, species loss and decoupling of entire ecosystems (Vergés et al., 2014; Wernberg et al., 2016; Pecl et al., 2017; Hastings et al., 2020; Pinsky et al., 2020). The resilience of species to changing environments is significantly underpinned by genetic diversity (Wernberg et al., 2018; Morikawa and Palumbi, 2019; Becheler et al., 2020; Hoban et al., 2021) because the higher the intraspecific genetic diversity, the more likely a species is to contain beneficial genetic variation that can be selected for, allowing adaption to change. Understanding patterns of genetic diversity is therefore crucial to assess species’ resilience to climate change especially for foundation species that support entire ecosystems (Vranken et al., 2021).
Genetic diversity is primarily created and maintained by genetic recombination involved in sexual reproduction. Although sexual reproduction includes significant higher genetic and energetic costs than asexual reproduction, it is considered more evolutionarily advantageous because it creates more variable offspring which facilitates faster adaptation to changing environments (Red Queen Hypothesis) (Becks and Agrawal, 2012; Bell, 2019). In contrast, where there is no independent segregation of alleles and recombination, as in asexual or clonal reproduction, selection efficacy is predicted to be reduced (Muller, 1932; Muller, 1964; Hill and Robertson, 1966; Hartfield, 2016). This is because the increased linkage among loci in clones inhibits selection from working independently on each locus, resulting in selection interference and decreased selection efficacy (Otto, 2021). This, in turn, can lead to a higher accumulation of deleterious mutations (Muller’s ratchet, Muller, 1964) and a slower adaptation rate in asexual individuals. Despite disadvantages, there exist several forms of asexual reproduction that are widespread across numerous taxa (De Meeûs et al., 2007), but often occurring in marginal habitats or areas where abiotic factors reduce the relative benefit of sexual reproduction (Silvertown, 2008). Because theory predicts that only infrequent sex is required to negate selection interference (Hartfield, 2016), periodic gene flow with sexual lineages has been assumed to occur for clonal lineages to survive extended periods of time. The mode of reproduction within populations can thus have a profound effect on the evolutionary trajectory and adaptation capacity of a species (Holsinger and Weir, 2009), though the relative consequences of reproducing asexually vs. sexually remains poorly understood for many natural populations.
Kelp are important habitat formers or foundation species in the marine environment and have a complex haplodiplontic lifecycle where the macroscopic diploid (sporophyte) alternates with a microscopic haploid (gametophyte) (Figure 1A). However, several kelp species can also reproduce asexually, for example by endomitosis, where haploid gametophytes produce diploid sporophytes without fertilisation [e.g. Lessonia nigrescens; Oppliger et al., 2007)], or by vegetative reproduction by fragmentation by the gametophytes (Destombe and Valeria Oppliger, 2011). In addition, vegetative reproduction of kelp sporophytes can happen through expansion of the holdfast through rhizoids (e.g. Laminaria sinclarii; Demes and Graham, 2011), stolons (e.g. Ecklonia stolonifera, Macrocystis pyrifera; Murúa et al., 2017; Hayashi et al., 2020), or less frequently from the thallus frond (e.g. E. radiata, E. stolonifera; Coleman and Wernberg, 2018; Hayashi et al., 2020). In algae, the presence of unique asexual lineages has often been linked to specific environmental conditions where asexual reproduction is thought to be more advantageous than sex and promote survival and enhance recovery (Pacheco-Ruíz et al., 2005; Tatarenkov et al., 2005; Demes and Graham, 2011; Murúa et al., 2017). However, the ecology and vulnerability to climate change of these unique asexual lineages remains poorly understood.
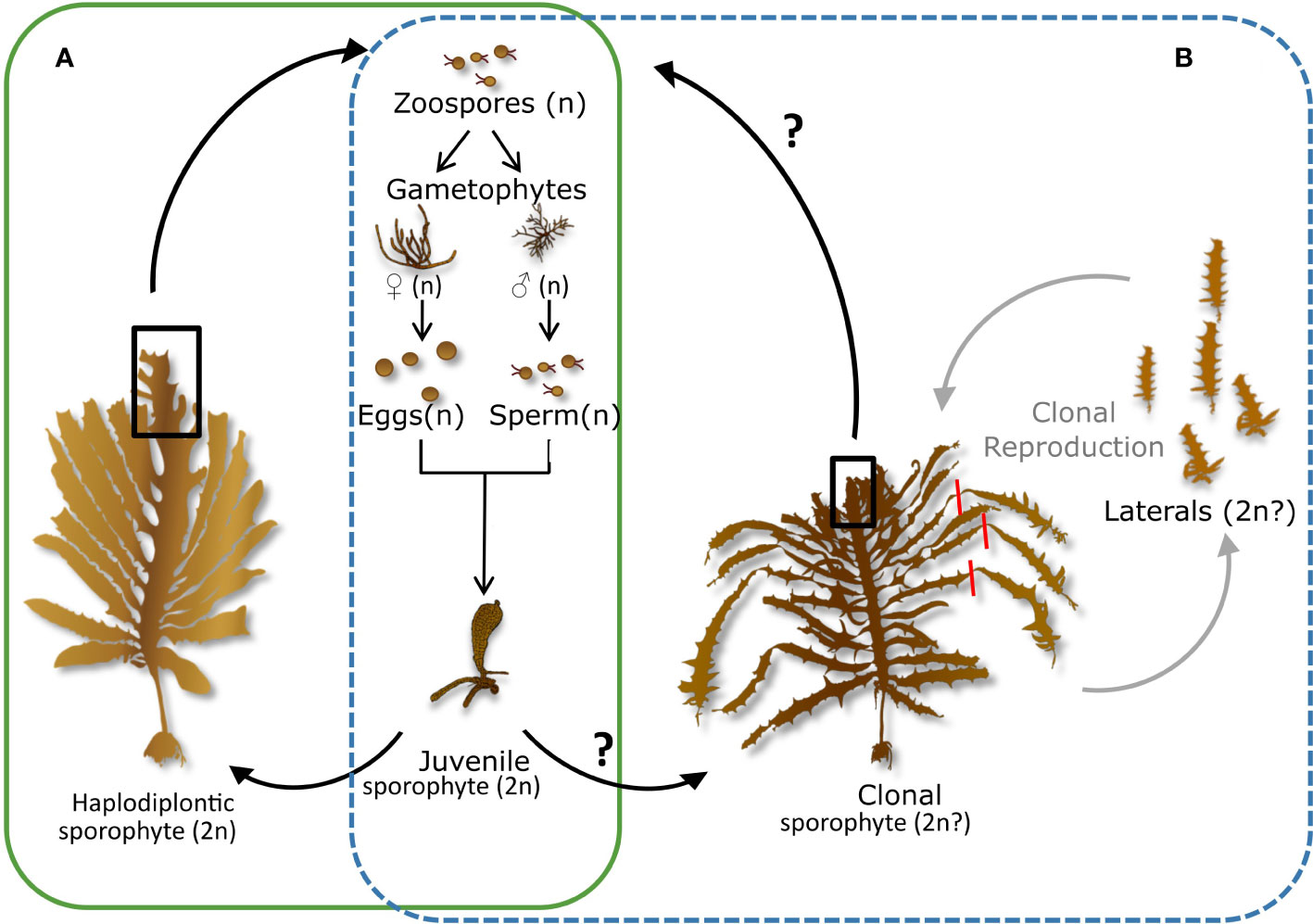
Figure 1 Life cycle of Ecklonia radiata. (A) normal sexual haplodiplontic life cycle. The macroscopic sporophyte (2n) produces zoospores (n) through meiosis that develop into male and female gametophytes (n). Male and female gametophytes produce sperm (n) and eggs (n), respectively. The sperm can fertilise the egg and a zygote will develop into a sporophyte (2n). (B) Life cycle of the unique clonal morphotype observed in Hamelin Bay, Western Australia. Clonal reproduction happens through haptera at the distal end of the laterals that allow attachment to the seafloor. When laterals attach to the seafloor and tear off the ‘parent’ individual, a new sporophyte can be formed via clonal reproduction.
The kelp E. radiata is one of the dominant foundation species of Australia’s Great Southern reef [GSR; Bennett et al. (2016); Wernberg et al. (2019a)] but has undergone major declines due to climate change (Wernberg, 2021). E. radiata reproduces through a sexual haplodiplontic life cycle (Figure 1A) but a unique asexual morph has been reported from the Cape Leeuwin area, Western Australia, where a clonal morph grows sympatrically, interspersed and under the same environmental conditions as the ‘normal’ haplodiplontic morph (Coleman and Wernberg, 2018) (Figure 2). This clonal morph is morphologically distinguishable (more slender) and forms many club-like haptera at the distal end of most primary laterals (up to 20) that can each attach to the seafloor (Figure 1B, Figure 2) (Coleman and Wernberg, 2018). Once these attached laterals tear off the parent thallus, they are assumed to grow into a new individual (Lindauer, 1945) (Figure 1B). What is particularly unique about this mode of reproduction is that it requires a total reversal in polarity, with the distal end of the thallus becoming the base of the new individual (Lindauer, 1945). Although previous research confirmed widespread asexual reproduction and revealed slightly reduced genetic diversity in the clonal morph (Coleman and Wernberg, 2018), much about the ecology and vulnerability to climate change of this clonal morph remains unknown. Here, we advance understanding of the ecology and consequences of asexual reproduction in this species using a combination of breeding experiments and ddRAD (double digest restriction-site associated DNA) sequencing. Specifically, we test for the effect of clonal reproduction on selection efficacy and putative adaptation to a changing climate and provide essential information needed to develop management plans.
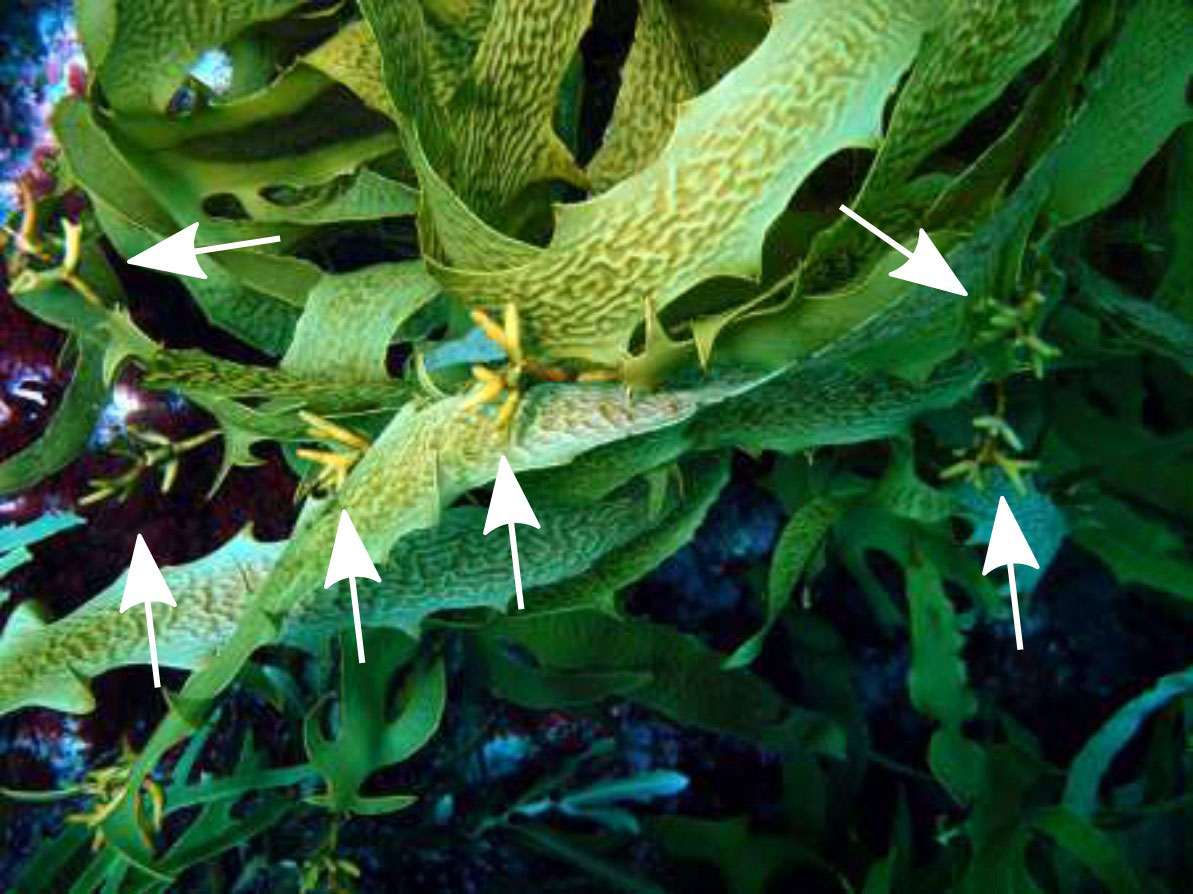
Figure 2 Ecklonia radiata from Hamelin Bay, Western Australia. The ‘normal’ haplodiplontic morph and the unique clonal morph with haptera at the end of every lateral (white arrow) grow interspersed on the same reef. Picture by TW.
We predicted higher heterozygosity and allelic diversity, but a lower genotypic diversity, in clonal individuals than in sexual individuals due to the lack of allele segregation and the accumulation of mutations (Balloux et al., 2003). As previous research suggested the possibility of sexual reproduction and admixture of the asexual and haplodiplontic morph (Coleman and Wernberg, 2018), we also tested for long-term asexuality, and co-ancestry among individuals to identify gene flow among haplodiplontic and clonal populations. Further, if admixture among the clonal and haplodiplontic morph is possible, we investigated whether it can lift the potential negative consequences of clonal reproduction. To confirm that sexual reproduction by the clonal morph and interbreeding with the sexual morph was possible, as well as to determine whether asexually produced sporophytelaterals developed into the asexual morph, we conducted a laboratory breeding and an aquarium experiment. Finally, we identified candidate SNPs associated with clonality to explore the genetic basis of this unique reproduction mode.
Material and methods
Field sampling, DNA extraction, library preparation, SNP calling and filtering
Adult sporophytes (stage 3 cf. Kirkman, 1981) were collected for both the haplodiplontic (sporophytes without haptera on the laterals) and the clonal morph (sporophytes with club-like structures on laterals, e.i. haptera by carefully detaching the holdfast from the sea floor. For the clonal morph, also the haptera at the end of the laterals that were attached to the seafloor were carefully removed, so plants were not damaged. Sporophytes were collected in 2018 and 2019 from two sites, four km apart in Hamelin Bay (34°13’48.79”S; 115° 0’52.70”E), Western Australia, with the two morphs growing interspersed on the same reefs. At every site, samples were haphazardly collected at 10 m depth in ~50 m2 and from distinct thalli at least 1 m apart. Clean tissue samples for DNA extraction were snap frozen and stored at -80°C until further processing.
For every sample, total genomic DNA was extracted and cleaned as in (Vranken et al., 2021). For seven haplodiplontic individuals a technical replicate was taken and for these individuals DNA was extracted twice. After quality control, 200 ng of DNA per sample was used to prepare ddRAD (double digestive restriction associated DNA) libraries as in (Vranken et al., 2021). Final libraries were pooled and each sent for paired-end sequencing using the Illumina XTEN at the Kinghorn Centre for Clinical Genomics’ Sequencing Facility (Darlinghurst, NSW, Australia).
SNP calling was performed as in Vranken et al. (2021). To retain only high quality SNPs, vcftools 0.1.15 (Danecek et al., 2011) was used to first remove all indels, multiallelic sites and all individuals containing > 75% missing data. Further, a minimum depth of coverage (DP) of 3 reads per allele and a maximum coverage of 2x mean sequencing depth, and a minimum allele frequency of 0.02 was required, and a maximum missingness of 10% per site. Pilots using lower levels of maximum missingness per sites (1 and 2%) did not change the number of MLLs detected (see further). Only SNPs with a maximum heterozygosity of 80%, and SNPs present in 3/4 sites, and in at least 80% of the individuals were included using Stacks (Catchen et al., 2013). Resulting in a final dataset of 5597 SNPs with multiple SNPs per RAD locus or a dataset with 4231 SNPs with max 1 SNP per locus.
Clonal validation, genotypic richness and long-term clonality
To confirm and assess levels of clonality, clonal or multi-locus lineages (MLLs) (Arnaud-Haond et al., 2007) and clonal richness statistics were estimated using the dataset with multiple SNPs per RAD locus. Individuals from the same MLL will not have the exact same genotype due to sequencing errors and somatic mutations (Arnaud-Haond et al., 2007). To set the distance threshold to define distinct MLLs two distance-based methods were implemented. First, the maximum proportion of allelic differences (i.e. bitwise distance) found between technical replicates of the haplodiplontic morph (d = 0.019), calculated with bitwise.dist in the R package poppr (Kamvar et al., 2014; Kamvar et al., 2015), was used as a threshold. Second, GENODIVE v 3.05 (Meirmans, 2020) was used to calculate a frequency distribution of pairwise allelic distances calculated among all sampled individuals (without technical replicates). The primary small hump of the bimodal distribution was assumed to represent mutations and sequencing errors between individuals from the same MLL, the second peak to represent distances between individuals from different MLL. The maximum of the first hump was used to define MLL (Figure S2) (Meirmans and Van Tienderen, 2004). Because MLL can also be observed when sexual reproduction is present due to low genetic variation or when the number of genetic markers used is insufficient to detect the present variation, we tested for clonal population structure with a permutation test implemented in GENODIVE. The permutation test was based on corrected Nei’s diversity index, alleles were randomised over individuals within populations for 999 permutations, assuming random mating.
Clonal richness (R) was calculated as R = (G-1)/(N-1) with G as the unique number of identified MLLs and N the number of samples analysed (Dorken and Eckert, 2001; Arnaud-Haond et al., 2007) and the Pareto Beta index to describe the distribution of the clonal memberships (high b value for high evenness, low b value for skewed distribution with dominance of a few MLLs).
As clonal reproduction may persist over extended periods of time, accumulation of mutation at different alleles of the same locus within one individual will occur (i.e. Meselson effect) (Hartfield, 2016), leading to an increase of intraindividual heterozygosity. Moreover, when heterozygous sites are fixed throughout the genome and shared among multiple clonal populations, one single shared origin and long-term clonality is suggested (Wachi et al., 2021). But when no such sites are present, multiple origins or the presence of sexual reproduction are supported (Wachi et al., 2021, Figure S3). To test whether the clonal populations have persisted over extended periods of time and have a shared origin, individual multilocus heterozygosity (MLH) was first calculated and compared among morphs and sampling populations using InbreedR (Stoffel et al., 2016) and a pairwise Wilcoxon test with Bonferroni p-value correction. Subsequently, the number of shared fixed heterozygous SNPs per sampling site were calculated using a custom python script (https://github.com/ascheben/phd_scripts/blob/master/scripts/find_fixed_sites_by_pop_in_vcf.py).
Genetic diversity and population structure
Using the dataset with only one SNP per RAD tag, genetic diversity within each sampling site was estimated as mean expected heterozygosity (He), mean observed heterozygosity (Ho), and average inbreeding coefficient (FIS) in the R package Hierfstat (Goudet and Jombart, 2022). Significance of FIS from 0 was tested by applying 9999 permutations and calculating the 99% confidence interval. As variance of FIS and distribution patterns of FIS values across loci are thought to be slightly more informative in cases of partial clonality than average FIS (Reynes et al., 2021; Stoeckel et al., 2021), these metrics were also calculated. Linkage disequilibrium was assessed by calculating the standard index of association (r ̄d) as implemented in poppr v2.8.3 (Kamvar et al., 2014). We tested for departure from random association between loci by performing 999 permutations. Individual multilocus heterozygosity (MLH) was calculated using the R-package InbreedR (Stoffel et al., 2016), and MLH was compared among sampling populations with a pairwise Wilcoxon rank sum test with Bonferroni p-value correction. All diversity metrics were calculated before and after clone correction (retaining only one sample from each set of clones).
To assess population structure and admixture among clonal and haplodiplontic populations, we used the Bayesian model-based approach implemented in the program fineRADstructure (Malinsky et al., 2018) to estimate co-ancestry among all samples. fineRADstructure offers especially high resolution in inference of recent shared ancestry by using multiple SNPs per RAD loci and nearest-neighbour haplotype relationships. First, the haplotype output from populations (Stacks) was converted to a fineRADpainter input file, using Stacks2fineRAD.py script included and allowing max 10 SNPs per locus. As recommended, RAD loci were ordered according to linkage disequilibrium using the fineRADstructure script sampleLD.R to ensure the correct estimation of the maximum number of clusters by RADpainter v0.3.2 which subsequently estimates the co-ancestry matrix (Malinsky et al., 2018). To assign individuals, fineSTRUCTURE v4.1.0 was used with 100,000 burn-in iterations, 100,000 sample iterations and thinning every 1000th generation. fineSTRUCTURE was also used to run a simple tree-building algorithm using 10,000 burn-in generations. The resulting co-ancestry matrix, MCMC output and the coalescence tree were all visualised through R. The effect of missing data on the final output was verified by comparing the final output with a missing-data-PCA (Malinsky et al., 2018). The final output was used to infer a number of clusters and explore relationships among clusters. Additionally, using the clone-corrected dataset with only one SNP per RAD tag pairwise FST between sampling sites was estimated using the R-package Stampp (Pembleton et al., 2013).
Putative pattern of environmental adaptation and selection efficacy
To test whether clonal kelps have a similar pattern of putative adaptation to the local environment as the haplodiplontic morph, we performed a Discriminant Analysis of Principle Components (DAPC) to compare allele frequencies of 174 loci previously defined as putatively linked to environmental variables (i.e. mean diffuse attenuation, annual mean temperature, annual mean temperature range) in Vranken et al. (2021). DAPC was performed using the R-package adegenet (Jombart, 2008) for the best fitting number of clusters K = 4, as defined by minimal cross entropy (CE) and using the optimal number of principal components as defined by cross validation. Loci contributions were used to identify the most important loci driving the observed allelic changes between the morphotypes. Annotations of the putative environmental linked loci were taken from Vranken et al. (2021). Further, to test whether clonal populations accumulate more deleterious alleles due to relaxed selection, we calculated the polymorphism ratio πN/πS with πN and πS as the mean population gene diversity for nonsynonymous and synonymous loci. As selection efficacy is decreasing, πN/πS is expected to increase. Nonsynonymous (SNP changing protein sequence) and synonymous SNPs (coding SNP not changing protein sequence) were first identified by mapping the stacks loci to our annotated reference genome using bowtie v1.3.1 (Langmead et al., 2009), annotating SNPs with using SNPeff v5.0e (Cingolani et al., 2012), and then calculating gene diversities per locus per populations with Stacks. A Kruskal-Wallis rank sum test and subsequent pairwise Wilcoxon test with Bonferroni p-value adjustment were applied to test if gene diversities of nonsynonymous and synonymous SNPs differed among populations.
FST based loci differences
To identify loci significantly different among morphs, per loci FST and a subsequent Fisher’ exact test with p-value correction were performed using Stacks to assess which loci significantly differentiate between the two morphs (Aumer et al., 2019). To account for differences in sample sizes between morphs, one unique sample per MLL for the clonal morph was compared with a similar number of randomly chosen haplodiplontic individuals. Loci with p< 0.0001 were visually inspected using TASSEL5 (Bradbury et al., 2007) to identify alleles fully segregating between the two morphs. Further, candidate SNPs were mapped to the current draft genome with bowtie and functionally annotated based on the current E. radiata gene models using SNPeff. Protein function of coding regions were identified using homology based on blastp analysis.
Zoospore release, zoospore germination and culture set-up
To test the capacity of sexual (haplodiplontic) reproduction of the clonal and haplodiplontic morph and confirm whether interbreeding is a possible, microscopic life-stages were cultured in the laboratory and a breeding experiment was performed. During April 2019, fifteen clean and healthy individuals of the haplodiplontic and clonal morph were selected from clonal and haplodiplontic site 2 and prepared for zoospore extraction following Alsuwaiyan et al. (2019). Briefly, tissue discs were cut out of the top part of the central lamina using a PVC corer, softly scraped with a razor blade, washed with autoclaved filtered seawater, briefly sterilized in iodine solution, again washed with filtered autoclaved seawater (0.2 µm) and patted dry with clean tissues. Tissue material was then placed between moist tissue paper and transported on ice. Once back in the lab, after 20 h, tissue material of all processed thalli was immersed in 100 ml of filtered-autoclaved seawater to release zoospores in separate sterile beakers. After 20 minutes, 10 ml of spore solutions was transferred to a 10 ml test tube with 10 drops of 100% ethanol. However, no zoospore release could be observed. For the next 12h,spore release was monitored without success (possibly spores were too small to be seen) and all tissue material was removed from the beakers, and all prepared solution was poured in two sterile petri dishes per individual to test for sporadic zoospore presence and capacity for spore germination and development into gametophytes. After a 12 h settlement period, the water was renewed with autoclaved Provasoli enriched seawater (PES) (Provasoli, 1968) and added germanium oxide (GeO2) (Lewin, 1966). Petri dishes were incubated at 18°C, with a 12:12 light:dark photoperiod, 6 ± 3 µmol photons m-2 s-1 red light. Petri dishes were checked for gametophyte development and received a PES medium change every two weeks. Presence or absence of gametophyte development was recorded per individual. As soon as gametophytes were observed, pictures were taken for observation, sex ratio evaluation and gametophyte growth rate (See below). After 2 months, light intensities were gradually increased to 20 µmol photons m-2 s-1 red light. Using tweezers and a light microscope, male and female gametophytes were isolated and put in new petri dishes to create single sex cultures. Male and female gametophytes were distinguished based on morphology; females have bigger cells and are more coarsely branched whereas males have thinner filaments and are more finely branched. Only gametophytes with clear morphological distinction were selected for culture set up. As gametophyte biomass accumulated, gametophytes were transferred to 250 ml sterile culture flasks. Gametophytes were grown in a vegetative state under the above condition until enough material for a crossing experiment was available (> 1 year) (Figure S1).
Gametophyte measurements: Sex ratio, length and growth
Sex ratio was estimated for four haplodiplontic individuals and one clonal individual (only one asexual individual produced viable gametophytes), 12 weeks after zoospore release, when male and female gametophytes were big enough to distinguish sexes. Male and female occurrences were recorded by photographing ten field of views using the 10x objective for the haplodiplontic morph and the 40x objective for the clonal morph and counting 50 random gametophytes. Sex ratio was calculated as the total number of males per total of gametophytes. Deviance from 50:50 male:female ratio was assessed per individual with an exact binomial test.
Length and growth rate were estimated for three haplodiplontic individuals and one clonal individual and compared between morphs. To estimate gametophyte length, 20 field of views were photographed for one replicate dish per parent individual, 7 weeks after spore release, using the 10x objective for the haplodiplontic and the 40x objective clonal morph. The software ImageJ was used to measure 20 random gametophytes per individual. Welsh’s t-test was performed to test for differences in gametophyte length between the two morphs, after confirming normality. To estimate growth, pictures were taken a second time, 10 weeks after spore release. Differences in gametophyte growth rate between morphs was tested by applying a linear regression on gametophyte lengths per morph and testing difference between slopes using a t-test in MedCalc (Software, 2020). Results were visualised in R.
Crossing experiment: Reproductive success, sporophyte length, sporophyte relative growth rate and density change
To conduct a controlled breeding experiment, healthy single sex male and female gametophytes cultures were first broken down into smaller fragments using a blender. To obtain equal gametophytes size among cultures, gametophytes were filtered at 60 and 30 μm and eluted in PES, at a mean density of ± 1000 fragments per ml. Male and female cultures from four haplodiplontic individuals and male and female cultures from one clonal individual were crossed following a full diallel mating design (all parents are crossed to all others in every combination including direct crosses, reciprocal crosses and selfing), resulting in both inter- and intra-morph crosses or five treatments (Figure S1).
- ♀ clonal x ♂ clonal - selfing
- ♀ clonal x ♂ haplodiplontic
- ♀ haplodiplontic x ♂ clonal
- ♀ haplodiplontic x ♂ haplodiplontic - outcrossing
- ♀ haplodiplontic x ♂ haplodiplontic - selfing
Because only one clonal individual produced gametophytes, there was no intra-morph outcrossing for the clonal morph. For every cross 2 x 9 ml of gametophytes PES solution was aliquoted to a sterile 60 mm diameter Petri dish and replicated four times. Petri dishes were transferred to the red-light culture conditions as above to let the gametophytes recover. After three days, Petri dishes were transferred to incubators set at 19.5°C, 25 μmol photons m-2 s-1 white light, 12:12 light:dark photoperiod. PES was renewed weekly. The Petri dishes were examined weekly for four consecutive weeks under a microscope and seven random fields of view were photographed.
Reproductive success (RS) was estimated as RS = s/fg with s as the number juvenile sporophytes, and fg as all female gametophytes observed. For sporophyte length measures, the five largest juvenile sporophytes from every replicate dish were selected and measured. We avoided random sampling because it is likely to choose sporophytes that grew from eggs developed at various dates, thereby misguiding true growth rate trends (Kain and Jones, 1965). Sporophyte relative growth rate (RGR) was calculated (ln L2 – ln L1)/(t2 – t1) with L1 and L2 sporophyte lengths at week 3 (t1) and 4 (t2). The percentage change in sporophyte density was estimated by counting the number of healthy sporophytes at week 4 (t2) relative to the density at week 3 (t1). Because there were only gametophytes of one clonal parent individual, which made the experimental design heavily unbalanced, and because the data did not follow a normal distribution, a non-parametric Kruskal-Wallis test, using the stats R-package, was performed to test the effect of the different crossings on reproductive success, sporophyte length, sporophyte RGR and sporophyte density change. A pairwise Wilcoxon rank sum test with Bonferroni p-value adjustment for multiple testing was performed with the stats R package, to test for significant differences among crossing treatments.
Growth capacity of asexual laterals - confirming the mechanism of asexual reproduction in the clonal morphotype
To test growth capacity of clonal laterals, between two to seven healthy laterals were selected from 10 clonal adults, attached to tiles and put in aquaria at ± 17 °C and 12:12 light:dark cycle. After 8 days of acclimatising, the laterals were hole punched 2 cm above the proximate end of the haptera. Survival, growth and development of new haptera on secondary laterals were recorded after 69 days. RGR of the laterals was calculated as above where L1 and L2 are lengths between the hole and proximate end of the haptera at times t1 and t2 (weeks). Difference in survival and growth rate of laterals sourced from different parent individuals was tested with respectively a Chi-Square test and a one-way ANOVA after confirming assumptions of normality and homogeneity of variances.
Results
Validation of clonality, genotypic richness and long-term clonality
In total, the same 69 MLLs were identified by both methods with a total of 15 clonal MLLs and 54 haplodiplontic MLLs. There were three MLLs within the clonal population at site 1 (clonal site 1 hereafter) and 12 MLLS within the clonal population at site 2 (clonal site 2 hereafter). For the haplodiplontic morph, there were 30 MLLs at site 1 (haplodiplontic site 1 hereafter) and 24 MLLs in the haplodiplontic populations at site 2 (haplodiplontic site 2 hereafter). For the clonal morph, we found a low clonal richness (R = 0.06), with one particularly big MLL including 22 samples (Beta = 0.70) in clonal site 1. In contrast, clonal site 2 had a high clonal richness (R = 0.58) with five unique MLLs, including only one sample and all other MLLs only two or three individuals (Beta = 2.10). For the haplodiplontic morph, all MLLs were unique and represented only one sample and no clonality was found (R = 1). No MLLs were shared across sampling sites or morphotypes. The null hypothesis of random mating was rejected for both clonal populations, confirming clonal reproduction, but accepted for both haplodiplontic populations, confirming sexual reproduction (Table S1).
Clonal individuals have a significantly higher MLH compared to all haplodiplontic individuals (p < 0.001) (Figure 3A). However, only MLH of clonal site 2 (Mdn 0.128 ± 0.002) was significantly higher than MLH in both populations of the haplodiplontic morph (haplod1: Mdn 0.091 ± 0.004, haplod2: Mdn 0.104 ± 0.002) (Figure 3A). MLH of clonal site 1 was only significantly (clonal1: Mdn 0.103 ± 0.006) different from overall MLH of haplodiplontic site 1 (haplod1) (Figure 3A).
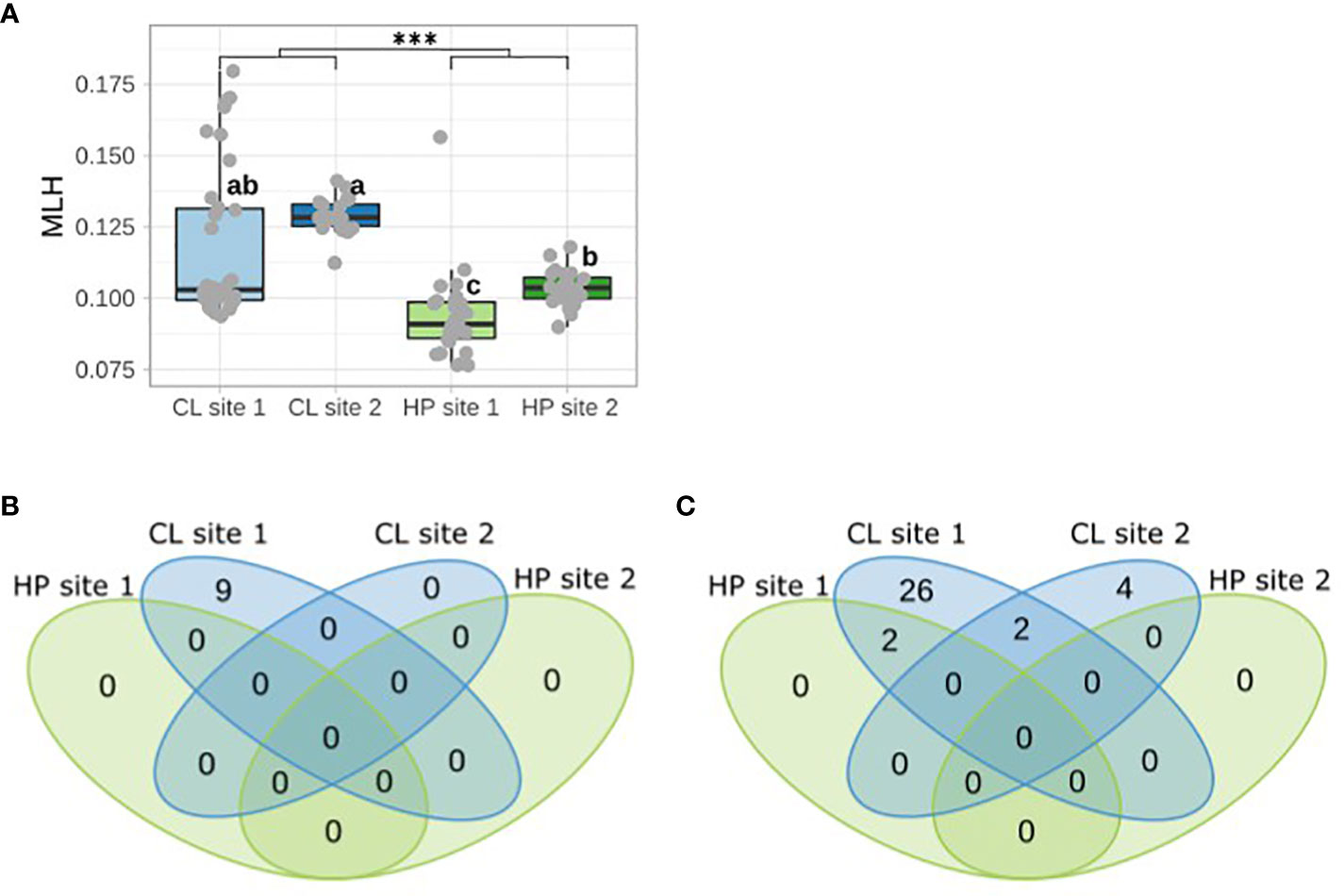
Figure 3 (A) Individual multilocus heterozygosity (MLH) per sampling population represented with a Box and Whisker plot. Grey dots indicate MLH per individual. *** indicates a significant difference among morphs (Kruskal rank sum test, p < 0.001). Lowercase letters indicate significant differences among sampling populations (pairwise Wilcoxon rank sum test, p < 0.05). (B) Number of fixed heterozygous sites and (C) almost fixed heterozygous sites among populations. Almost fixed heterozygous are sites with >85% of heterozygous sites. HP refers to haplodiplontic, CL to clonal.
There were only shared fixed heterozygous sites present at clonal site 1, suggesting all clonal individuals at site 1 have a single shared origin and long-term clonality (Figure 3B). The number of almost fixed heterozygous sites was also the highest for clonal site 1 followed by clonal site 2 (Figure 3C).
Genetic diversity and population structure
Populations of the clonal morph showed higher expected and observed heterozygosity than populations of the haplodiplontic morph both before and after clone correction (Table 1). Before clone correction, there was a significant departure from panmixia for clonal site 1, indicated with a small, but significant, negative average FIS of -0.027, but not for clonal site 2 (Table 1). However, distributions of FIS indicated both for clonal site 1 and 2 a substantial proportion of negative per locus FIS values (Figure S4) and a higher FIS variance than the haplodiplontic morph (Table 1). After clone correction both clonal populations indicated a deficit of heterozygotes, indicated by a significant positive FIS of 0.100 and 0.036. Before clone correction, there was a significant linkage disequilibrium for clonal site 1 of 0.213, which resolved after clone correction. For clonal site 2, there was significant linkage disequilibrium before and after clone correction of respectively 0.027 and 0.002 (Table 1). As no clones were observed within the haplodiplontic populations, clone correction was not done. There was departure from panmixia in both haplodiplontic populations, indicated by a small, but significant, positive FIS of 0.065 and 0.025 (Table 1). Distributions of both haplodiplontic site 1 and 2 indicted less negative per locus FIS values (Figure S4) and lower variance than the clonal (Table 1). Significant but low linkage disequilibrium was observed for haplodiplontic site 1 (0.007) but not for haplodiplontic site 2 (0.001) (Table 1).
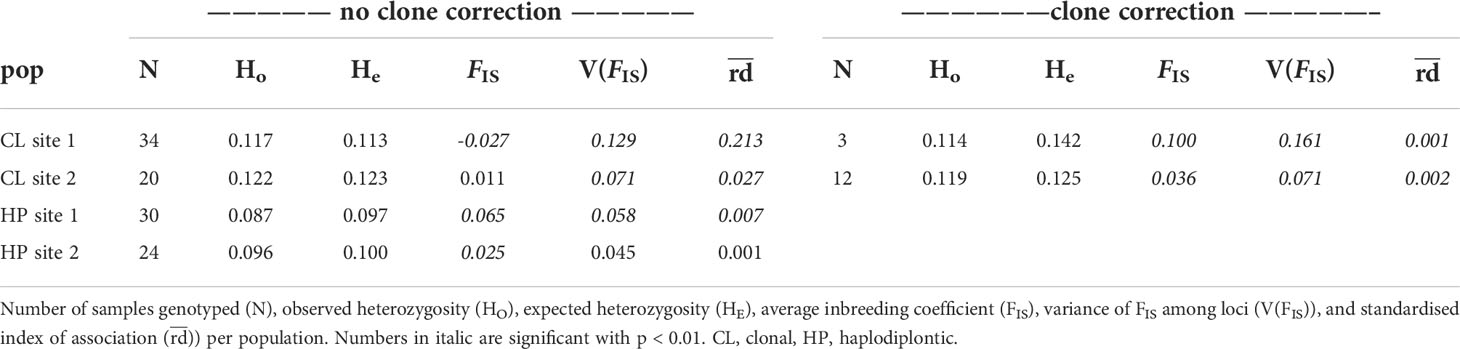
Table 1 Genetic diversity within Hamelin Bay populations of Ecklonia radiata before and after clone correction.
The clustered co-ancestry matrix indicated three clusters, two for the clonal morph (Figure 4, cluster 1 and 3) and one for the haplodiplontic morph (Figure 4, cluster 2). Clustering of the clonal morph did not correspond to sampling locations. Twenty-two individuals of clonal site 1 shared more co-ancestry with three individuals from clonal site 2 (Figure 4, cluster 1) than the remaining clonal individuals from their own sampling site. The second cluster of the clonal morph (Figure 4, cluster 3), indicated profound substructure. Interestingly, one of the subclusters (Figure 4, cluster 3A) indicated a substantial co-ancestry (67 loci) among clonal individuals from different sites, suggesting dispersal of clonal morphs between sampling sites. All individuals from clonal site 2 indicate substantially more shared co-ancestry with the haplodiplontic morph than individuals of clonal site 1, indicating possible hybridisation between the two morphotypes at site 2. The individuals from the haplodiplontic morph from both the sampling sites cluster together, indicating homogenous geneflow among haplodiplontic individuals. Pairwise FST analysis indicated the biggest genetic differentiation between clonal site 1 and both the haplodiplontic populations, suggesting little admixture among clonal individuals at site1 and the haplodiplontic individuals (Figure 5). Clonal site 2 on the other hand showed much lower genetic differentiation with the haplodiplontic individuals, indicating much more admixture among the clonal individuals at site 2 and the haplodiplontic individuals. The genetic differentiation between clonal site 1 and clonal site 2 is bigger than between clonal site 2 and the haplodiplontic populations (Figure 5). The haplodiplontic populations showed almost no genetic differentiation (Figure 5).
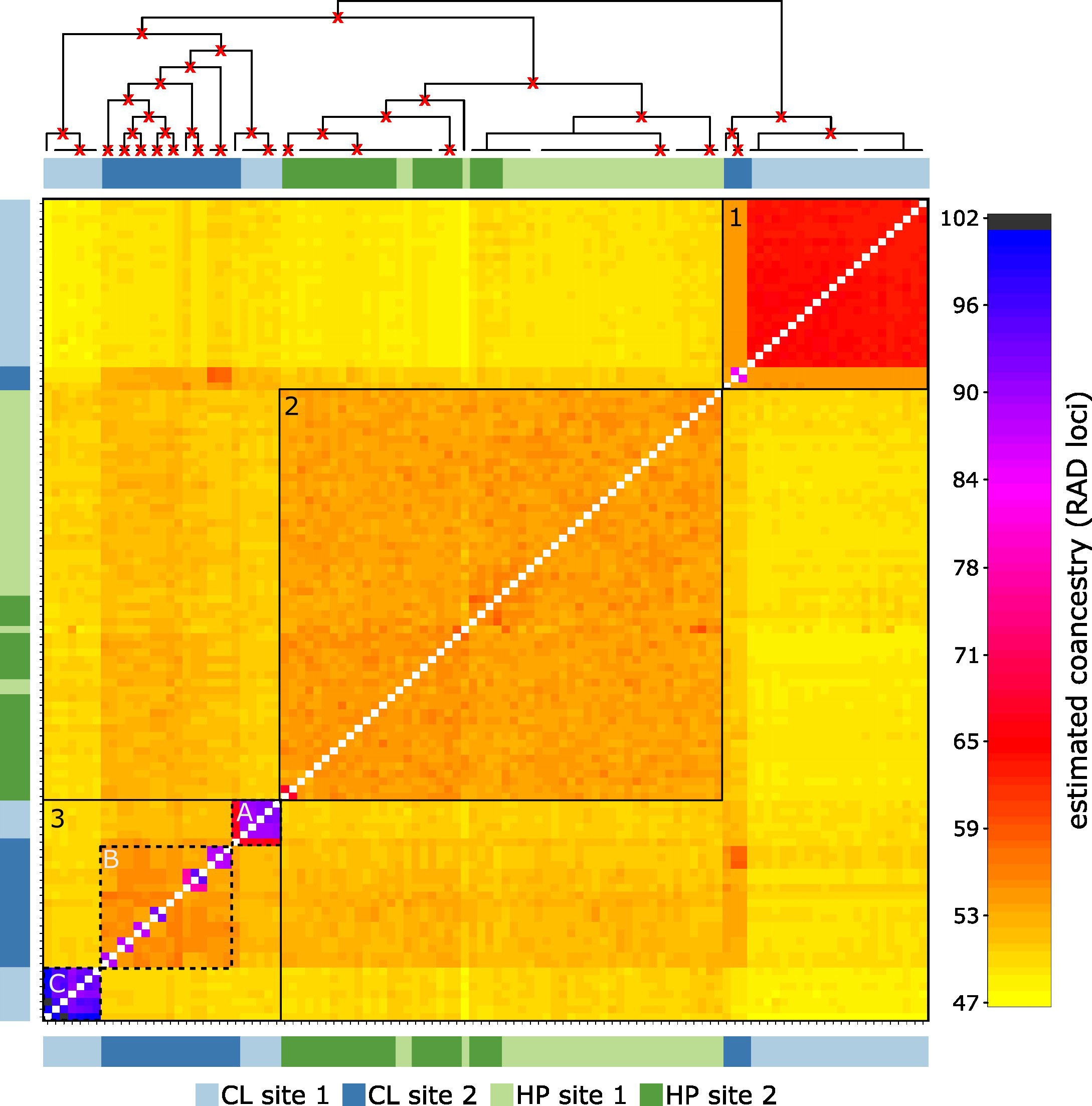
Figure 4 Clustered co-ancestry matrix among all individuals of Ecklonia radiata samples at Hamelin Bay, estimated using fineRADstructure. The heatmap indicates variation in estimated pairwise co-ancestry among all genotyped individuals using the colour scale on the right. The dendrogram on the top indicates the clustering of individuals based on the pairwise matrix of co-ancestry coefficients, bootstraps values ≥ 0.90 are indicated with a red cross on the dendogram. Black numbered squares indicate main clusters. Dashed lines indicate subclusters. Legends on the left and below indicate sampling population with CL referring to clonal and HP to haplodiplontic individuals.
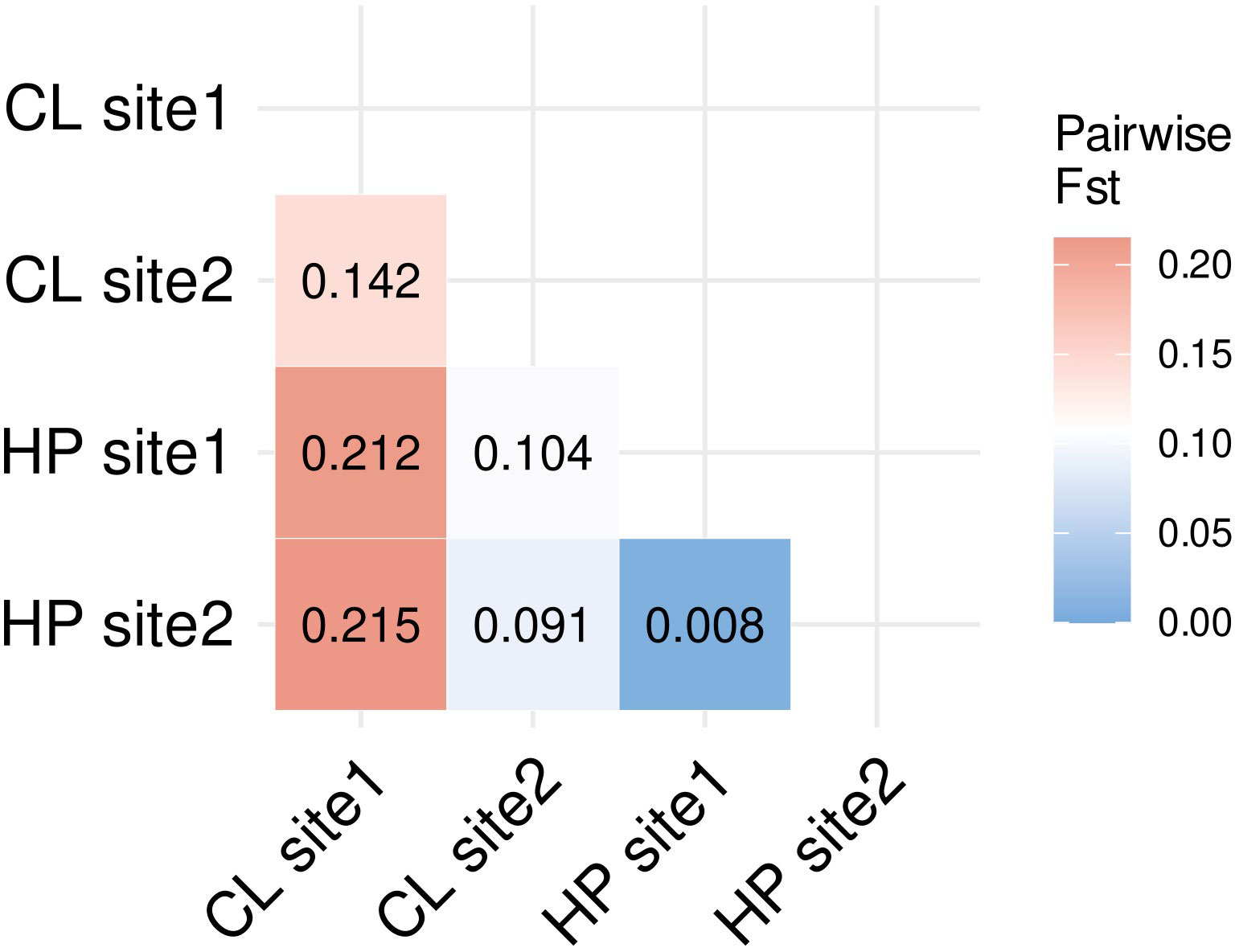
Figure 5 Pairwise FST estimates among all sampling sites using clone-corrected data. CL and HP refer to clonal and haplodiplontic respectively.
Putative pattern of environmental adaptation and selection efficacy
DAPC analysis indicated different trends in putative environment-linked loci for the clonal and haplodiplontic morph, possibly indicating maladaptation of the clonal morph (Figure 6A). The individuals from the clonal morph formed three distinct clusters of mixed sampling sites while all individuals from the haplodiplontic morph from both site 1 and site 2 cluster together (Figure 6A). Five loci (11920_39, 12012_31, 19346_48, 31079_35, 66811_33) contribute significantly to the first discriminant function LD1 and the difference among the clonal and haplodiplontic morphotype (Figure S5). No annotations were available for these loci. The clonal population at site 1 had the highest πN/πS ratio, suggestive of relaxed selection (Figure 6B). This high πN/πS ratio for clonal site 1 was driven by a higher, but non-significantly different, average non-synonymous gene diversity than the other populations (H(3) = 7.1, p = 0.068) (Figure 6C). Notably, clonal site 2 had the lowest πN/πS value, suggesting high selection efficacy, driven by a relatively higher synonymous gene diversity. Synonymous gene diversity was significantly different among populations (H(3) = 13.3, p = 0.004), with clonal site 2 and haplodiplontic site 1 significantly different from only each other (p = 0.003) (Figure 6D).
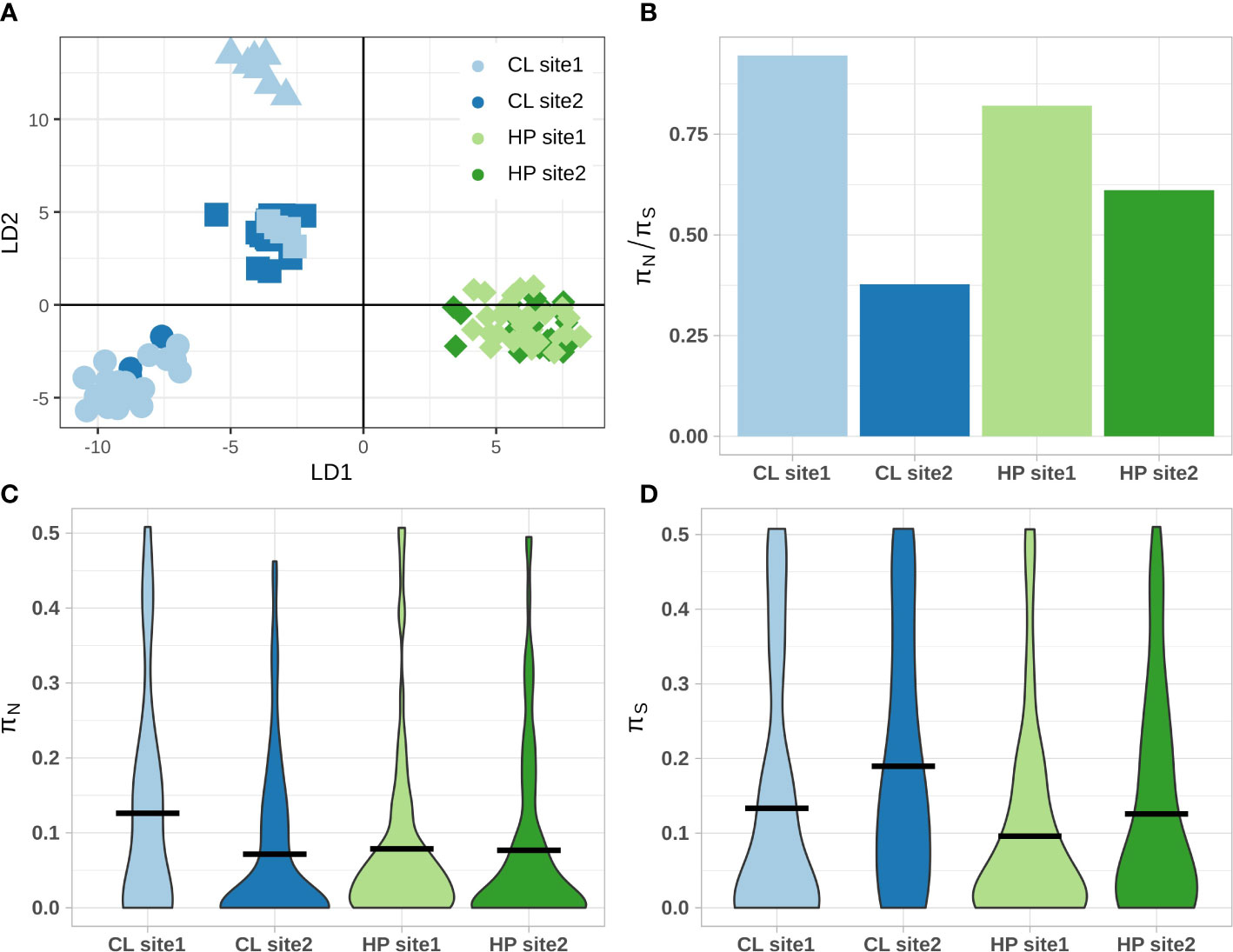
Figure 6 Putative trends of selection, polymorphism ratio and non- and synonymous gene diversities for all individuals per sampling population. (A) DAPC cluster analysis for putative environment-linked loci (174 SNPs) for all sampled populations, only the first 2 discriminant functions are shown, shape of data points refers to the genetic cluster (K = 4). (B) average polymorphism ratio πN/πS. (C) Nonsynonymous (πN) and (D) synonymous (πS) gene diversities, the black horizontal line indicates average population gene diversity (π). Colours indicate sampling populations in all panels. CL and HP stand for clonal and haplodiplontic respectively.
FST based loci differences
We identified 49 SNPs as significantly different between morphs, of which 19 SNPs were included in the coding regions of a known gene, although no missense mutations were observed. Two gene models could be linked to known proteins (Endo-1,3-beta-glucanase, family GH81, Leucyl aminopeptidase) with potential function in cell cycling such as fertilisation, seed germination, somatic embryogenesis, and meiosis (Table S2). Other putative functions of proteins linked to gene models were related to diverse physiological processes, secondary metabolites, defence mechanisms and immune response (Table S2).
Gametophyte measurements: germination, sex ratio, length and growth rate
Gametophytes developed for only one (7%) of the clonal individuals from which spores were released, whereas 12 (80%) of the haplodiplontic individuals had gametophyte development. Although the clonal morph somewhat developed males and females with distinct morphology similar to the haplodiplontic morph (males; thinner filaments, finely branched, females; broader filaments, coarsely branched, Figure 7), they also developed gametophytes looking like clumps of cells without much branching (Figure 7A). Mean sex ratio for the haplodiplontic morph was balanced (mean 0.56 ± 0.03) but the clonal morph displayed a significant female (0.22) bias as indicated by comparison to the binomial law, although given difficulties in identification of males and females for the clonal morph, this result should be interpreted with caution. Gametophyte length 7 weeks after spore release was significantly smaller for the clonal morph (t(43) = -19.0, p < 0.05). Growth rate differed significantly among the two morphs (t(156) = -3.70, p < 0.001) (Figure 8A); gametophytes from the clonal morph did not grow (average growth of -0.3068 ± 0.5483 µm/day) compared to the haplodiplontic morph which grew at 3.9890 ± 0.6427 µm/day.
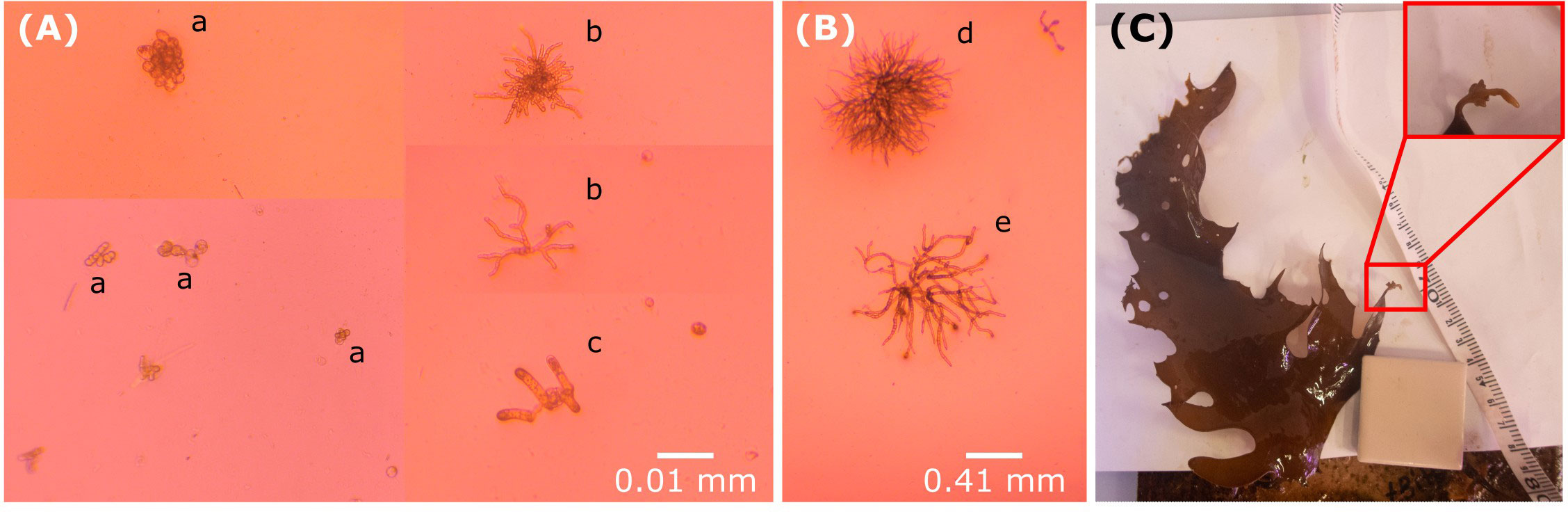
Figure 7 Examples of Ecklonia radiata in Hamelin Bay, Western Australia. (A) Gametophytes developed from spores from a clonal sporophyte and (B) a ‘normal’ haplodiplontic sporophyte. a) gametophyte cell clumps without branching, b) male gametophyte, c) female gametophyte, d) male gametophyte, e) female gametophyte. (C) Lab grown clonal lateral (cut from parent clonal plants) growing into a new adult individual with the formation of its own new haptera at the top of its own secondary lateral. Pictures taken one month after start of experiment.
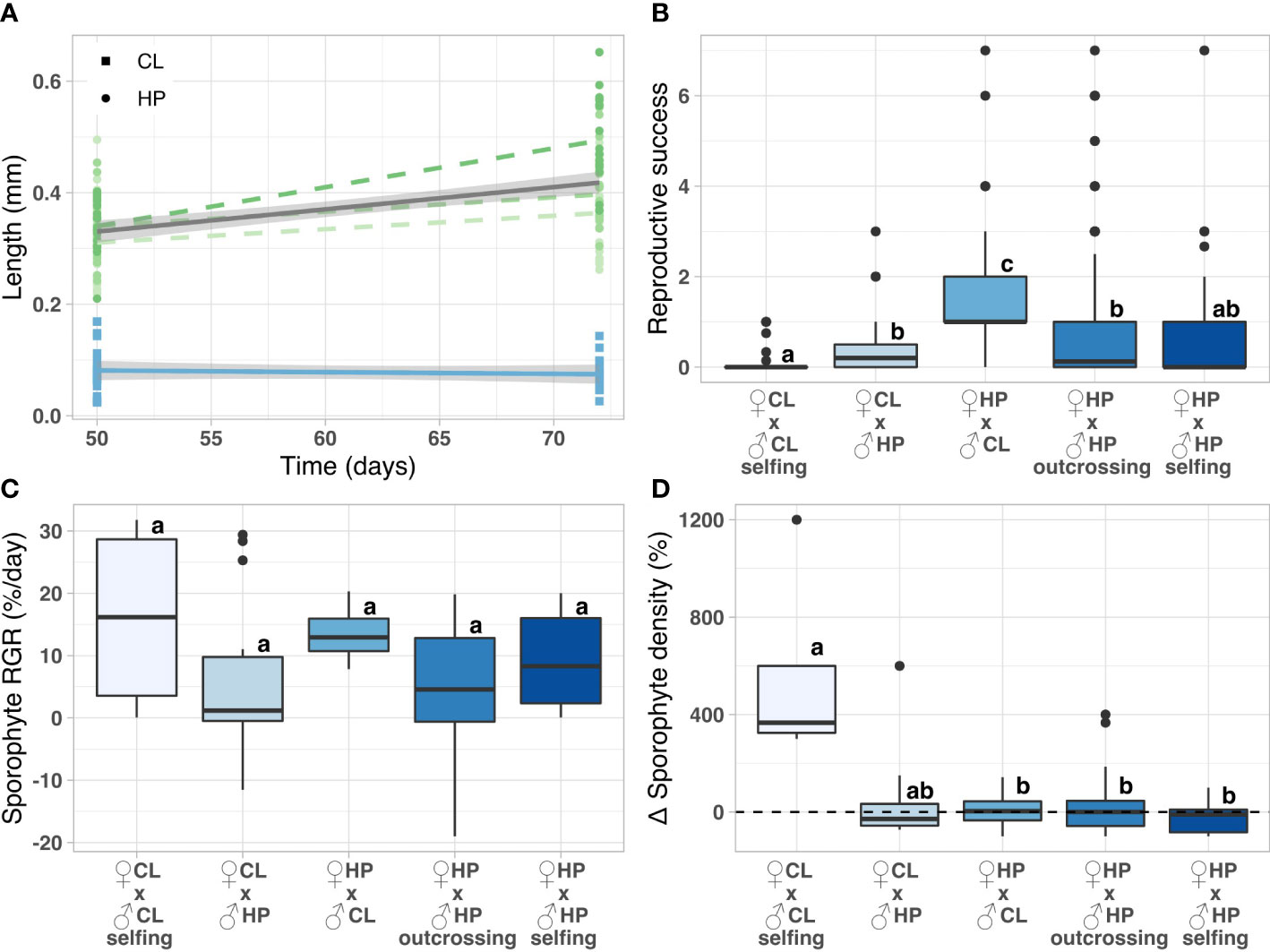
Figure 8 (A) Linear regression of gametophyte length measured 7 and 10 weeks after spore release for the haplodiplontic (HP) and clonal morph (CL). (B–D) Sporophyte measurements for the crossing experiment. (B) reproductive success (number of sporophytes per total number of female gametophytes), (C) sporophyte relative growth rate (RGR), (D) percentage sporophyte density change (t2 relative to t1). All measurements were taken 3 (t1) or 4 (t2) weeks after the start of the experiment. Lowercase letters indicate significant differences among sampling populations (pairwise Wilcoxon rank sum test, p < 0.05). CL and HP stand for clonal and haplodiplontic respectively.
Crossing experiment: Reproductive success, sporophyte length, sporophyte relative growth rate and density change
Reproductive success (H(4) = 65.19, p < 0.001) and sporophyte length (H(4) = 16.23, p = 0.003) were significantly different among crossing treatments (Figure 8B, Table 2). Sporophyte RGR was not significantly different among crossing treatments (H(4) = 9.14, p = 0.058) (Figure 8C, Table 2). However, overall there was much variation and differences among treatments were hard to interpret. Sporophyte density change differed significantly among crossing treatments (H(4) = 11.1, p = 0.026) (Figure 8D, Table 2). The clonal selfing treatment had a density increase of 367 ± 269% (Mdn ± SE), which was significantly higher than all other treatments (p < 0.05), except not significantly for treatment female clonal x male haplodiplontic (p = 0.08, Mdn -28.6 ± 54.9%, Figure 8D), indicating a delayed increase in sporophytes or a reproduction time lag for clonal selfing treatment. As only well-formed gametophytes from the clonal morphotype were selected (no clump-like gametophytes) to build cultures and perform this crossing experiment, reproductive success and sporophytes fitness may be overestimated compared to natural conditions. Further, due to the lack of replication in the clonal morph, results of these experiments should be interpreted with caution. Nevertheless, this experiment still provides a qualitative confirmation that sexual reproduction in the clonal morph and interbreeding among the two morphs is possible.
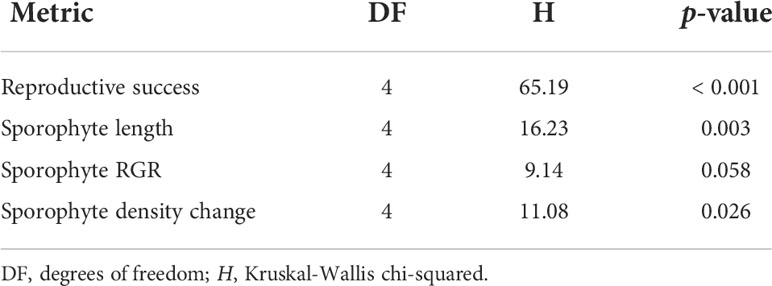
Table 2 Summary of Kruskal-Wallis rank sum test of differences in sporophyte measurements (reproductive success (number of sporophytes per number of female gametophytes), length, relative growth rate (RGR), density) among different crossings.
Growth capacity of asexual laterals - confirming the mechanism of asexual reproduction in the clonal morphotype
Fifty five percent of the clonal laterals survived for 10 weeks independent of parent ID (X214 = 14.67, p > 0.05). Mean growth rate of the clonal laterals was 11.07 ± 0.51% per week and was also independent of the parent individual (F10,10 = 5.22, p > 0.05). The lateral that grew the most (i.e., 15.81% per week) was the only lateral that developed a new haptera on a secondary lateral at the end of the 10-week period (Figure 7C).
Discussion
Understanding genetic diversity and reproductive mechanisms of species is key to predicting their adaptive capacity and resilience to environmental changes. Here, we investigated the genomic and ecological consequences of clonal reproduction in a unique clonal morph of kelp, which grows sympatrically with the haplodiplontic morph. Asexual reproduction by the clonal morph through the development of haptera at the tips of the laterals was confirmed by both an aquarium experiment and genomics. Yet, our results indicated that sexual reproduction by the clonal morph and interbreeding with the sexual morph is also possible. High genetic differentiation with the sexual morph and long-term clonality were established in one of the two clonal populations, together with a trend of decreased selection efficacy. Patterns of putative adaptive variation, however, deviated from the sexual morph in all clonal populations, indicating possible maladaptation and high vulnerability to environmental changes in the clonal morph.
Clonal reproduction can reduce genotypic diversity and maintain heterozygosity through the lack of independent allele segregation (Balloux et al., 2003; Hartfield, 2016). Moreover, in long-term clonality, mutations will accumulate at different sites at each allele over time, further increasing heterozygosity and creating high intra-individual diversity (Meselson effect) (Hartfield, 2016). Relative to the sexual populations, we identified significantly fewer MLLs, higher levels of heterozygosity, more negative per locus FIS values,and elevated levels of linkage disequilibrium in the clonal populations, indicating asexual reproduction. Moreover, the test for clonal population structure confirmed that the observed genotypic structure cannot be explained by sexual reproduction, even when genetic diversity is limited. Notably, the observed pattern of fixed heterozygous sites in clonal site 1 suggest that the individuals from clonal site 1 had a common evolutionary origin and have long since evolved individually.
Despite the fact that gametophytes from the clonal morph developed differently and slower, the crossing experiment indicated that sexual reproduction by the clonal morph and interbreeding between the clonal and haplodiplontic morph is possible. This was further supported with substantial admixture and limited genetic differentiation (low FST) between the sexual morph and clonal site 2, indicating substantial interbreeding between the morphs at clonal site 2. Conversely, in clonal site 1 there was limited admixture, high genetic differentiation from the sexual populations (high FST) and proof of long-term clonality (fixed heterozygous sites), indicating absence of sexual reproduction in clonal site 1. Hence, the frequency of sexual reproduction and interbreeding with the sexual morph likely varies among clonal lineages.
Underwater kelp forests are threatened by climate change and the increase of extreme climate events (Wernberg et al., 2019b). Extreme events can cause mortality, range shifts, new ecosystem states (Vergés et al., 2014; Wernberg et al., 2016; Arafeh-Dalmau et al., 2019; Filbee-Dexter et al., 2020) and drive changes or losses in genetic diversity (Becheler et al., 2020; Coleman et al., 2020a; Gurgel et al., 2020), which may impact the adaptability and vulnerability of these kelp forests to climate change (Wernberg et al., 2018). Reproductive mode can have a profound effect in buffering and creating resilience against extreme events (Becheler et al., 2020). For example, compared to sexual populations, clonal populations of the red algae Agarophyton chilense were more resilient to genetic impoverishment immediately after a catastrophic earthquake event, although long-term clonality reduced the chances of long-term survival after the environmental changes (Becheler et al., 2020). Asexual or clonal individuals are predicted to have lower selection efficacy and adaptive potential due to selection interference caused by high linkage among loci (Jaron et al., 2021; Otto, 2021). Although, theory predicts that only infrequent sex is needed to counter these negative effects (Hartfield, 2016; Becheler et al., 2020). We observed signals of decreased selection efficacy (higher πN/πS) in clonal site 1, where we also found proof of long-term asexuality. In the more admixed clonal site 2, where sexual reproduction and interbreeding with the sexual morph is more frequent, selection efficacy was high (lower πN/πS), confirming that occasional sex reduces the negative evolutionary effects of clonal reproduction. In contrast, relative to the sexual morph, both clonal populations showed a distinct pattern of putative local adaptation, suggesting possible maladaptation to current environmental conditions, lower selection efficacy and potential high vulnerability of the clonal morph to environmental changes. Further research using manipulative experiments linking to fitness measurements and better genomic resources (e.g. a fully annotated genome) are needed to further unravel the adaptive capacity and vulnerability of the clonal morph to environmental stressors.
Processes underlying this unique mode of clonal reproduction in E. radiata remain elusive. Previously, it has been suggested that development of this clonal morph was triggered by environmental cues (Rothman et al., 2015) or wounding or abrasion (McCook and Chapman, 1992). However, Coleman and Wernberg (2018) argued that clonal reproduction is likely genetically defined and heritable due to the abundant production of haptera on the apex of the laterals (> 50% of the laterals, pers. observation), the vastly different morphologies among the morphs, and the localised extent of clonal reproduction despite the sympatric distribution of the sexual and clonal morphs interspersed on the same reef. Indeed, a breeding experiment in E. stolonifera suggested that comparable clonal reproduction from the thallus is heritable (Hayashi et al., 2020). Clonal reproduction from the thallus in E. stolonifera is however slightly different from that in E. brevipes. In E. stolonifera shoots can develop from multiple pinnae on the serration of the laterals, while in E. radiata a maximum of one haptera is formed at the tip of each lateral. Yet both clonal reproduction mechanisms require reversal of cell polarity and may be underpinned by the same genetic basis. We identified 49 candidate loci that are significantly different among morphs. These candidate loci might possibly underly clonal reproduction or be linked to genes underlying clonal reproduction. Links were found with proteins linked to cell cycling function but also to tyrosine decarboxylase which has been mentioned in the auxin biosynthesis process (Bogaert et al., 2019). Auxin influences algal development and cell polarity and may also underlie development of the clonal haptera (Ramakrishna and De Smet, 2014; Coleman and Wernberg, 2018). Currently, sporophytes resulting from all crosses performed here are being grown in controlled environments until phenotyping (presence/absence of the haptera) can be conducted. Together with further genomic studies, this will allow an understanding of the heritability of this unique clonal mode of reproduction and the genomic basis of polarity reversal in E. radiata.
Underwater forests are under threat from global change which is outpacing the capacity of kelp forests to adapt (Vranken et al., 2021; Wood et al., 2021). Preadapting threatened populations by either assisted adaptation, gene editing or priming might be a possible solution to protect these habitats (Coleman and Goold, 2019; Coleman et al., 2020b; Jueterbock et al., 2021). Clonality is a trait that has been extensively used and researched for agronomics and it is generally accepted that introduction of clonal reproduction or apomixis in agronomically important crops will revolutionize agriculture (Pupilli and Barcaccia, 2012). One of the major benefits of clonal reproduction is that it can assure immediate fixation of a beneficial genotype or trait in one generation (Savidan, 2000). In this sense, the trait of clonal reproduction could be highly beneficial in future management programs for kelp forests, to quickly spread preadapted (e.g. heat resilient) individuals in vulnerable populations. Gene editing could be used to engineer ‘super kelps’ that are both highly resilient to environmental changes and reproduce clonally to ensure such traits are fixed within populations. As future-proofing techniques can hold great potential for kelp restoration and conservation practices (Coleman et al., 2020b), further research focusing on manipulative experiments and identifying candidate genes useful for genetic engineering of clonality will be necessary.
Data availability statement
The datasets presented in this study can be found on Figshare: https://doi.org/10.6084/m9.figshare.19772866.v1.
Author contributions
SV, MC and TW conceptualized the study. SV and TW conducted fieldwork. SV performed all molecular and culture lab work. AS performed SNP calling and annotation. SV analysed the data. All authors contributed to the article and approved the submitted version.
Funding
This research was supported by the Australian Research Council through DP160100114 and DP200100201 granted to TW and MC and by a Holsworth Wildlife Research Endowment Grant of The Ecological Society of Australia granted to SV. SV and AS were supported by IPRS and RTP scholarships awarded by the Australian Government
Acknowledgments
We thank S. Bell, H. Denham and J. Ranson for assistance with the practical work. We thank A. Severn-Ellis for advice in the molecular lab and L. Thomas for constructive discussions about the analyses.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2022.921912/full#supplementary-material
References
Alsuwaiyan N. A., Mohring M. B., Cambridge M., Coleman M. A., Kendrick G. A., Wernberg T. (2019). A review of protocols for the experimental release of kelp (Laminariales) zoospores. Ecol. Evol. 9 (14), 8387–8398. doi: 10.1002/ece3.5389
Arafeh-Dalmau N., Montaño-Moctezuma G., Martinez J. A., Beas-Luna R., Schoeman D. S., Torres-Moye G. (2019). Extreme marine heatwaves alter kelp forest community near its equatorward distribution limit. Front. Mar. Sci. 6, 499. doi: 10.3389/fmars.2019.00499
Arnaud-Haond S., Duarte C. M., Alberto F., Serrao E. A. (2007). Standardizing methods to address clonality in population studies. Mol. Ecol. 16 (24), 5115–5139. doi: 10.1111/j.1365-294X.2007.03535.x
Aumer D., Stolle E., Allsopp M., Mumoki F., Pirk C. W. W., Moritz R. F. A. (2019). A single SNP turns a social honey bee (Apis mellifera) worker into a selfish parasite. Mol. Biol. Evol. 36 (3), 516–526. doi: 10.1093/molbev/msy232
Balloux F., Lehmann L., de Meeûs T. (2003). The population genetics of clonal and partially clonal diploids. Genetics 164 (4), 1635–1644. doi: 10.1093/genetics/164.4.1635
Becheler R., Guillemin M. L., Stoeckel S., Mauger S., Saunier A., Brante A., et al. (2020). After a catastrophe, a little bit of sex is better than nothing: Genetic consequences of a major earthquake on asexual and sexual populations. Evolutionary Appl. 13 (8), 2086–2100. doi: 10.1111/eva.12967
Becks L., Agrawal A. F. (2012). The evolution of sex is favoured during adaptation to new environments. PloS Biol. 10 (5), e1001317. doi: 10.1371/journal.pbio.1001317
Bell G. (2019). The masterpiece of nature: the evolution and genetics of sexuality (London: Routledge).
Bennett S., Wernberg T., Connell S. D., Hobday A. J., Johnson C. R., Poloczanska E. S. (2016). The ‘Great southern reef’: social, ecological and economic value of australia’s neglected kelp forests. Mar. Freshw. Res. 67 (1), 47–56. doi: 10.1071/MF15232
Bogaert K. A., Blommaert L., Ljung K., Beeckman T., De Clerck O. (2019). Auxin function in the brown alga dictyota dichotoma. Plant Physiol. 179 (1), 280–299. doi: 10.1104/pp.18.01041
Bradbury P. J., Zhang Z., Kroon D. E., Casstevens T. M., Ramdoss Y., Buckler E. S. (2007). TASSEL: Software for association mapping of complex traits in diverse samples. Bioinformatics 23 (19), 2633–2635. doi: 10.1093/bioinformatics/btm308
Catchen J., Hohenlohe P. A., Bassham S., Amores A., Cresko W. A. (2013). Stacks: an analysis tool set for population genomics. Mol. Ecol. 22 (11), 3124–3140. doi: 10.1111/mec.12354
Cingolani P., Platts A., Wang L. L., Coon M., Nguyen T., Wang L., et al. (2012). A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of drosophila melanogaster strain w1118; iso-2; iso-3. Fly 6 (2), 80–92. doi: 10.4161/fly.19695
Coleman M. A., Goold H. (2019). Harnessing synthetic biology for kelp forest conservation. J. Phycol. 55 (4), 745–751. doi: 10.1111/jpy.12888
Coleman M. A., Minne A. J. P., Vranken S., Wernberg T. (2020a). Genetic tropicalisation following a marine heatwave. Sci. Rep. 10 (1), 1–11. doi: 10.1038/s41598-020-69665-w
Coleman M. A., Wernberg T. (2018). Genetic and morphological diversity in sympatric kelps with contrasting reproductive strategies. Aquat. Biol. 27, 65–73. doi: 10.3354/ab00698
Coleman M. A., Wood G., Filbee-Dexter K., Minne A. J., Goold H. D., Verges A., et al. (2020b). Restore or redefine: future trajectories for restoration. Front. Mar. Sci. 7, 237. doi: 10.3389/fmars.2020.00237
Danecek P., Auton A., Abecasis G., Albers C. A., Banks E., DePristo M. A., et al. (2011). The variant call format and VCFtools. Bioinformatics 27 (15), 2156–2158. doi: 10.1093/bioinformatics/btr330
De Meeûs T., Prugnolle F., Agnew P. (2007). Asexual reproduction: Genetics and evolutionary aspects. Cell. Mol. Life Sci. 64 (11), 1355. doi: 10.1007/s00018-007-6515-2
Demes K. W., Graham M. H. (2011). Abiotic regulation of investment in sexual versus vegetative reproduction in the clonal kelp laminaria sinclairii (Laminariales, phaeophyceae). J. phycol. 47 (3), 463–470. doi: 10.1111/j.1529-8817.2011.00981.x
Destombe C., Valeria Oppliger L. (2011). Male Gametophyte fragmentation in laminaria digitata: A life history strategy to enhance reproductive success. CBM-Cahiers Biologie Mar. 52 (4), 385.
Dorken M. E., Eckert C. G. (2001). Severely reduced sexual reproduction in northern populations of a clonal plant, decodonverticillatus (Lythraceae). J. Ecol. 89 (3), 339–350. doi: 10.1046/j.1365-2745.2001.00558.x
Filbee-Dexter K., Wernberg T., Grace S., Thormar J., Fredriksen S., Narvaez C., et al. (2020). Marine heatwaves and the collapse of marginal North Atlantic kelp forests. Sci. Rep. 10 (1), 1–11. doi: 10.1038/s41598-020-70273-x
Goudet J., Jombart T. (2022). Hierfstat: Estimation and Tests of Hierarchical F-Statistics. R package version 0. 5–11
Gurgel C. F. D., Camacho O., Minne A. J., Wernberg T., Coleman M. A. (2020). Marine heatwave drives cryptic loss of genetic diversity in underwater forests. Curr. Biol. 30 (7), 1199–1206. doi: 10.1016/j.cub.2020.01.051
Hartfield M. (2016). Evolutionary genetic consequences of facultative sex and outcrossing. J. Evolutionary Biol. 29 (1), 5–22. doi: 10.1111/jeb.12770
Hastings R. A., Rutterford L. A., Freer J. J., Collins R. A., Simpson S. D., Genner M. J. (2020). Climate change drives poleward increases and equatorward declines in marine species. Curr. Biol. 30 (8), 1572–1577. doi: 10.1016/j.cub.2020.02.043
Hayashi Y. M., Notoya M., Yotsukura N. (2020). Abnormal asexual reproduction of thalli of ecklonia stolonifera (Laminariales, Phaeophyceae) off the coast of Nakanoshima in the Oki Islands, Japan. Botanica Marina 63 (3), 247–52. doi: 10.1515/bot-2018-0086
Hill W. G., Robertson A. (1966). The effect of linkage on limits to artificial selection. Genet. Res. 8 (3), 269–294. doi: 10.1017/S0016672300010156
Hoban S., Bruford M., Funk W. C., Galbusera P., Griffith M. P., Grueber C. E., et al. (2021). Global commitments to conserving and monitoring genetic diversity are now necessary and feasible. BioScience 71 (9), 964–976. doi: 10.1093/biosci/biab054
Holsinger K. E., Weir B. S. (2009). Genetics in geographically structured populations: Defining, estimating and interpreting FST. Nat. Rev. Genet. 10 (9), 639–650. doi: 10.1038/nrg2611
IPCC (2021). “Summary for policymakers,” in Climate change 2021: The physical science basis. contribution of working group I to the sixth assessment report of the intergovernmental panel on climate change. Eds. Masson-Delmotte V., Zhai P., Pirani A., Connors S. L., Peían C., Berger S., Caud N., Chen Y., Goldfarb L., Gomis M. I., Huang M., Leitzell K., Lonnoy E., Matthews J. B. R., Maycock T. K., Waterfield T., Yelekçi O., Zhou RYu &B. (Geneva: Cambridge University Press, In Press).
Jaron K. S., Bast J., Nowell R. W., Ranallo-Benavidez T. R., Robinson-Rechavi M., Schwander T. (2021). Genomic features of parthenogenetic animals. J. Heredity 112 (1), 19–33. doi: 10.1093/jhered/esaa031
Jombart T. (2008). Adegenet: a r package for the multivariate analysis of genetic markers. Bioinformatics 24 (11), 1403–1405. doi: 10.1093/bioinformatics/btn129
Jueterbock A., Minne A. J., Cock J. M., Coleman M. A., Wernberg T., Scheschonk L., et al. (2021). Priming of marine macrophytes for enhanced restoration success and food security in future oceans. Front. Mar. Sci 8, 658485. doi: 10.3389/fmars.2021.658485
Kain J. M., Jones M. N. (1965). Aspects of the biology of laminarja hyperborea IV. growth of early sporophytes. J. Mar. Biol. Assoc. United Kingdom 45 (1), 129–143. doi: 10.1017/S0025315400004033
Kamvar Z. N., Brooks J. C., Grünwald N. J. (2015). Novel r tools for analysis of genome-wide population genetic data with emphasis on clonality. Front. Genet. 6, 208. doi: 10.3389/fgene.2015.00208
Kamvar Z. N., Tabima J. F., Grünwald N. J. (2014). Poppr: an r package for genetic analysis of populations with clonal, partially clonal, and/or sexual reproduction. PeerJ 2, e281. doi: 10.7717/peerj.281
Kirkman H. (1981). The first year in the life history and the survival of the juvenile marine macrophyte, ecklonia radiata (Turn.) j. agardh. J. Exp. Mar. Biol. Ecol. 55 (2-3), 243–254. doi: 10.1016/0022-0981(81)90115-5
Langmead B., Trapnell C., Pop M., Salzberg S. L. (2009). Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10 (3), 1–10. doi: 10.1186/gb-2009-10-3-r25
Lewin J. (1966). Silicon metabolism in diatoms. v. germanium dioxide, a specific inhibitor of diatom growth. Phycologia 6 (1), 1–12. doi: 10.2216/i0031-8884-6-1-1.1
Lindauer V. W. (1945). Note on the brown alga ecklonia brevipes j. Ag. Trans. R. Soc. New Z. 75, 394–397.
Malinsky M., Trucchi E., Lawson D. J., Falush D. (2018). RADpainter and fineRADstructure: Population inference from RADseq data. Mol. Biol. Evol. 35 (5), 1284–1290. doi: 10.1093/molbev/msy023
McCook L., Chapman A. R. (1992). Vegetative regeneration of fucus rockweed canopy as a mechanism of secondary succession on an exposed rocky shore. Botanica Marina 35, 35–46. doi: 10.1515/botm.1992.35.1.35
Meirmans P. G. (2020). Genodive version 3.0: Easy-to-use software for the analysis of genetic data of diploids and polyploids. Mol. Ecol. Resour. 20 (4), 1126–1131. doi: 10.1111/1755-0998.13145
Meirmans P. G., Van Tienderen P. H. (2004). GENOTYPE and GENODIVE: two programs for the analysis of genetic diversity of asexual organisms. Mol. Ecol. Notes 4 (4), 792–794. doi: 10.1111/j.1471-8286.2004.00770.x
Morikawa M. K., Palumbi S. R. (2019). Using naturally occurring climate resilient corals to construct bleaching-resistant nurseries. Proc. Natl. Acad. Sci. 116 (21), 10586–10591. doi: 10.1073/pnas.1721415116
Muller H. J. (1964). The relation of recombination to mutational advance. Mutat. Research/Fundamental Mol. Mech. Mutagenesis 1 (1), 2–9. doi: 10.1016/0027-5107(64)90047-8
Murúa P., Müller D. G., Patiño D. J., Westermeier R. (2017). Giant kelp vegetative propagation: Adventitious holdfast elements rejuvenate senescent individuals of the macrocystis pyrifera “integrifolia” ecomorph. J. phycol. 53 (1), 230–234. doi: 10.1111/jpy.12493
Oliver E. C., Burrows M. T., Donat M. G., Sen Gupta A., Alexander L. V., Perkins-Kirkpatrick S. E., et al. (2019). Projected marine heatwaves in the 21st century and the potential for ecological impact. Front. Mar. Sci. 6, 734. doi: 10.3389/fmars.2019.00734
Oppliger L. V., Correa J. A., Peters A. F. (2007). Parthenogenesis in the brown alga lessonia nigrescens (Laminariales, phaeophyceae) from central Chile 1. J. Phycol. 43 (6), 1295–1301. doi: 10.1111/j.1529-8817.2007.00408.x
Otto S. P. (2021). Selective interference and the evolution of sex. J. Heredity 112 (1), 9–18. doi: 10.1093/jhered/esaa026
Pacheco-Ruíz I., Zertuche-González J., Espinoza-Avalos J. (2005). The role of secondary attachment discs in the survival of chondracanthus squarrulosus (Gigartinales, rhodophyta). Phycologia 44 (6), 629–631. doi: 10.2216/0031-8884(2005)44[629:TROSAD]2.0.CO;2
Pecl G. T., Araújo M. B., Bell J. D., Blanchard J., Bonebrake T. C., Chen I. C., et al. (2017). Biodiversity redistribution under climate change: Impacts on ecosystems and human well-being. Science 355 (6332), eaai9214. doi: 10.1126/science.aai9214
Pembleton L. W., Cogan N. O., Forster J. W.. (2013). StAMPP: an R package for calculation of genetic differentiation and structure of mixed–ploidy level populations. Mol. Ecol. Resour. 13:946–52. doi: 10.1111/1755-0998.12129
Pinsky M. L., Selden R. L., Kitchel Z. J. (2020). “Climate-Driven Shifts in Marine Species Ranges: Scaling from Organisms to Communities,” in Ann. Rev. Mar. Sci 12, 153–179. doi: 10.1146/annurev-marine-010419-010916
Provasoli L. (1968). “Media and prospects for the cultivation of marine algae,” in Cultures and collections of algae. Eds. Watanabe A., Hattori A. (Hakone, Japan: Japanese Society Plant Physiology), 63–75
Pupilli F., Barcaccia G. (2012). Cloning plants by seeds: inheritance models and candidate genes to increase fundamental knowledge for engineering apomixis in sexual crops. J. Biotechnol. 159 (4), 291–311. doi: 10.1016/j.jbiotec.2011.08.028
Ramakrishna P., De Smet I. (2014). “Evolutionary aspects of auxin signalling,” in Auxin and its role in plant development (Vienna: Springer), 265–290.
Reynes L., Thibaut T., Mauger S., Blanfuné A., Holon F., Cruaud C., et al. (2021). Genomic signatures of clonality in the deep water kelp laminaria rodriguezii. Mol. Ecol. 30 (8), 1806–1822. doi: 10.1111/mec.15860
Rothman M. D., Mattio L., Wernberg T., Anderson R. J., Uwai S., Mohring M. B., et al. (2015). A molecular investigation of the genus ecklonia (Phaeophyceae, laminariales) with special focus on the southern hemisphere. J. phycol. 51 (2), 236–246. doi: 10.1111/jpy.12264
Savidan Y. (2000). Apomixis: genetics and breeding. Plant Breed. Rev. 18, 13–86. doi: 10.1002/9780470650158.ch2
Silvertown J. (2008). The evolutionary maintenance of sexual reproduction: Evidence from the ecological distribution of asexual reproduction in clonal plants. Int. J. Plant Sci. 169 (1), 157–168. doi: 10.1086/523357
Stoeckel S., Porro B., Arnaud-Haond S. (2021). The discernible and hidden effects of clonality on the genotypic and genetic states of populations: Improving our estimation of clonal rates. Mol. Ecol. Resour. 21 (4), 1068–1084. doi: 10.1111/1755-0998.13316
Stoffel M. A., Esser M., Kardos M., Humble E., Nichols H., David P., et al. (2016). inbreedR: an r package for the analysis of inbreeding based on genetic markers. Methods Ecol. Evol. 7 (11), 1331–1339. doi: 10.1111/2041-210x.12588
Tatarenkov A., Bergström L., Jönsson R. B., Serrão E. A., Kautsky L., Johannesson K. (2005). Intriguing asexual life in marginal populations of the brown seaweed fucus vesiculosus. Mol. Ecol. 14 (2), 647–651. doi: 10.1111/j.1365-294X.2005.02425.x
Vergés A., Steinberg P. D., Hay M. E., Poore A. G., Campbell A. H., Ballesteros E., et al. (2014). The tropicalization of temperate marine ecosystems: climate-mediated changes in herbivory and community phase shifts. Proc. R. Soc B 281 (1789), 20140846. doi: 10.1098/rspb.2014.0846
Vranken S., Wernberg T., Scheben A., Severn-Ellis A., Batley J., Philipp B. E., et al. (2021). Genotype-environment mismatch of kelp forests under climate change. Mol. Ecol. 30 (15), 3730–3746. doi: 10.1111/mec.15993
Wachi N., Gau J. J., Fujie S., Fukano K., Maeto K. (2021). Genomic population structure of sympatric sexual and asexual populations in a parasitic wasp, meteorus pulchricornis (Hymenoptera: Braconidae), inferred from six hundred single-nucleotide polymorphism loci. Mol. Ecol. 30 (7), 1612–1623. doi: 10.1111/mec.15834
Wernberg T. (2021). “Marine heatwave drives collapse of kelp forests in Western Australia,” in Ecosystem collapse and climate change (Cham: Springer), 325–343.
Wernberg T., Bennett S., Babcock R. C., de Bettignies T., Cure K., Depczynski M., et al. (2016). Climate-driven regime shift of a temperate marine ecosystem. Science 353 (6295), 169–172. doi: 10.1126/science.aad8745
Wernberg T., Coleman M. A., Babcock R. C., Bell S. Y., Bolton J. J., Connell S. D., et al. (2019a). Biology and ecology of the globally significant kelp ecklonia radiata. Oceanogr. Mar. Biol.: Annu. Rev. 57, 265–324. doi: 10.1201/9780429026379-6
Wernberg T., Coleman M. A., Bennett S., Thomsen M. S., Tuya F., Kelaher B. P. (2018). Genetic diversity and kelp forest vulnerability to climatic stress. Sci. Rep. 8 (1), 1851. doi: 10.1038/s41598-018-20009-9
Wernberg T., Krumhansl K., Filbee-Dexter K., Pedersen M. F. (2019b). “Chapter 3 - status and trends for the world’s kelp forests,” in World seas: an environmental evaluation (Second edition). Ed. Sheppard C. (London: Academic Press), 57–78.
Keywords: Ecklonia radiata, E. brevipes, clonal reproduction, vegetative reproduction, adaptation, genetic diversity
Citation: Vranken S, Scheben A, Batley J, Wernberg T and Coleman MA (2022) Genomic consequences and selection efficacy in sympatric sexual versus asexual kelps. Front. Mar. Sci. 9:921912. doi: 10.3389/fmars.2022.921912
Received: 16 April 2022; Accepted: 27 September 2022;
Published: 17 October 2022.
Edited by:
Zi-Min Hu, Yantai University, ChinaReviewed by:
Ricardo T. Pereyra, University of Gothenburg, SwedenTifeng Shan, Institute of Oceanology, Institute of Oceanology (CAS), China
Copyright © 2022 Vranken, Scheben, Batley, Wernberg and Coleman. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sofie Vranken, c29maWVtdnJhbmtlbkBnbWFpbC5jb20=; Melinda Ann Coleman, bWVsaW5kYS5jb2xlbWFuQGRwaS5uc3cuZ292LmF1
 Sofie Vranken
Sofie Vranken Armin Scheben
Armin Scheben Jacqueline Batley
Jacqueline Batley Thomas Wernberg
Thomas Wernberg Melinda Ann Coleman
Melinda Ann Coleman