
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mar. Sci. , 17 June 2022
Sec. Global Change and the Future Ocean
Volume 9 - 2022 | https://doi.org/10.3389/fmars.2022.921877
This article is part of the Research Topic The Adaptation and Response of Aquatic Animals in the Context of Global Climate Change View all 18 articles
Background: Understanding how organisms respond and adapt to environmental changes is central to evolutionary biology. As a sessile organism that has adapted to life in estuaries and intertidal zones, the eastern oyster Crassostrea virginica can tolerate wide fluctuations in temperature and salinity and survive for weeks out of water. To understand the molecular mechanisms underlying the remarkable stress tolerance of the eastern oyster, we studied the transcriptomic changes induced by exposure to air and cold stress. Eastern oysters were maintained for 7 days under four conditions, namely, in seawater (normal) at 22°C, in air at 22°C, in seawater at 5°C and in air at 5°C, and then sampled for RNA sequencing.
Results: Transcriptomic analysis revealed that many genes involved in cell cycle progression and DNA replication were downregulated in oysters exposed to air and cold, which indicates that stress inhibits cell division. Exposure to air at 22°C induced a concerted inhibition of apoptosis through the upregulation of expanded inhibitors of apoptosis and the downregulation of caspases. Interactions between TNF and NF-κB signalling implied a reduction in the inflammatory response and immune functions. Key genes for proline production, fatty acid synthesis and chromosomal proteins were upregulated during exposure to low temperatures, which suggested that proline accumulation, energy conservation, and epigenetic modification of chromosomes are important for coping with cold stress. The upregulation of melatonin, FMRFamide, and neural acetylcholine receptors indicate the significance of the neurohormonal regulation of homeostasis.
Conclusion: These results show that air exposure and cold stress alter the expression of key genes for cell division, apoptosis, proline accumulation, fatty acid metabolism, neurohormonal signalling, and epigenetic modifications, suggesting regulation of these processes plays an important role in the stress response of the eastern oyster and possibly other marine molluscs. This study provides new insights into molecular mechanisms of stress response that are essential for understanding the adaptive potential of marine organisms under climate change.
The mechanism through which organisms respond and adapt to environmental shifts is a fundamental question of evolutionary biology. As climate change causes significant shifts in the oceanic environment, understanding its effects on marine organisms and ecosystems is critical. The eastern oyster (Crassostrea virginica) is a marine bivalve widely distributed along the Atlantic coast of America, ranging from the Gulf of St. Lawrence in Canada to the Gulf of Mexico and extending to West Indies, and an important fishery and aquaculture species in the US and Canada. Due to its adaptation to estuarine and intertidal environments across a wide latitudinal range, the eastern oyster is remarkably resilient and can tolerate wide fluctuations in salinity and temperature as well as long periods of exposure to air. In the intertidal zone, the eastern oyster may face icy conditions in the winter and 45°C in the summer (Galtsoff, 1964; Shumway, 1996). This species can survive well for at least a month during exposure to air at 4°C (Kawabe et al., 2010). This remarkable resilience makes the eastern oyster an interesting model for studying stress adaptation.
Among all environmental challenges, prolonged exposure to air may be very stressful for oysters and can cause heavy mortalities during summer months (Malek, 2010; Clements et al., 2018). As observed in field experiments, increases in intertidal air exposure increase disease prevalence and mortality and reduce growth (Malek, 2010). Genomic analyses have provided some insights into the molecular mechanisms of the stress response and adaptation of oysters. Transcriptomic analyses of the eastern oyster have identified genes and pathways related to the immune responses of the species and its responses to salinity and crude oil (Eierman and Hare, 2014; McDowell et al., 2014; Zhang et al., 2014a; López-Landavery et al., 2019; Modak and Gomez-Chiarri, 2020). Transcriptomic studies of Pacific oysters Crassostrea gigas subjected to thermal, salinity, air exposure and heavy metal stresses have shown that air exposure induces transcriptional changes in the largest number of genes (Zhang et al., 2012). These changes include strong upregulation of the expression of heat shock proteins (HSPs) and inhibitor of apoptosis (IAPs), which suggests that these proteins play key roles in stress response and adaptation (Zhang et al., 2012; Guo et al., 2015). A proteomic study of Pacific oysters under desiccation stress revealed increases in ubiquinone synthesis, reductions in reactive oxygen species (ROS), and downregulation of the expression of proteins involved in metabolism, ion transport, DNA replication and protein synthesis (Zhang et al., 2014b). In the eastern oyster, air exposure induces the upregulation and alternative splicing of alternative oxidases that participate in ROS removal (Liu and Guo, 2017).
Few studies have examined the relationship between air exposure and the innate immune system. Allen and Burnett (2008) found that hypoxia and high temperatures reduce the ability of haemocytes to kill bacteria in C. gigas under air exposure. Air exposure may result in reduced oxygen levels in oysters or hypoxia. Alvarez and Friedl (1992) reported that hypoxia and anoxia have no effects on phagocytosis and particle clearance in the eastern oyster, whereas another study (Boyd and Burnett, 1999) in the same species showed that the respiratory burst function (ROS production) of haemocytes is reduced under hypoxic conditions. Environmental stressors exert immunosuppressive effects in Pinctada imbricata (Kuchel et al., 2010). The eastern oyster populations are seriously impacted by two lethal diseases, MSX caused by Haplosporidium nelsoni and Dermo caused by Perkinsus marinus (Guo and Ford, 2016). The effects of air exposure on the immune function of the eastern oyster are unclear.
Compared with air exposure, cold exposure is a relatively mild stressor for oysters. The LT50 (median lethal time to reach 50% mortality) values for oysters exposed to air at temperatures of 4°C, 15°C and 20°C are 47.8, 15.9 and 12.2 days, respectively (Kawabe et al., 2010). The regulation of energy storage and consumption is crucial for the survival of animals during long periods of cold conditions (Storey and Storey, 1990). During cold periods, bivalves can enter a state of metabolic rate depression (MRD) (Saucedo et al., 2004; Xiao et al., 2014). An analysis of the physiological states of oysters at low temperatures has suggested that the metabolic pathways in oysters change after one week of exposure (Kawabe et al., 2010), although most of the regulatory networks at the protein and transcriptomic levels are largely unknown.
The availability of the C. virginica genome and high-throughput next-generation sequencing technology has allowed accurate measurement of transcriptional levels of all genes, which provided a valuable tool for studying molecular mechanisms of stress response in oysters (Zhang et al., 2012; Gómez-Chiarria et al., 2015; Guo et al., 2015; Guo and Ford, 2016). In this study, we used the reference genome (GCA_002022765.4 C_virginica-3.0) and transcriptomic sequencing to characterize transcriptional changes in eastern oysters subjected to air exposure and cold stress and thus elucidate the molecular mechanisms or regulatory pathways underlying stress response in this species.
Hatchery-produced eastern oysters of the same age (18 months) and similar shell lengths (5–8 cm) were used in this study. The oysters were obtained from the Haskin Shellfish Research Laboratory, Rutgers University, Port Norris, New Jersey, USA. The oysters were maintained in ambient seawater at 9°C ~ 12°C and then acclimated to 22°C for 3 days for this experiment in April. To elicit a strong response to chronic air exposure and cold stress, oysters were exposed to air and cold (5°C) for 7 days. The low temperature of 5°C was selected because it is the lowest possible temperature above freezing that can be stably maintained in the lab. The duration of 7 days was selected based on a previous study (Zhang et al., 2012), where oysters exposed to 5°C for 7 days and to air for 7-9 days exhibited the strongest responses. Although 7-day exposure is rare in the field (only experienced by oysters at extremely high tidal positions), this extreme length helps to identify key genes responding to air exposure stress. Also, eastern oysters are often stored in cold rooms without water for 1-2 weeks before being consumed live. Taking all these factors into account, we subjected oysters to 7 days of air exposure. In winter, the coldest month in New Jersey is January when the average temperature ranged from -6.7°C to 3.3°C. The oysters may be covered by ice for several days at a time in the field. The low temperate tested in this study is quite common for eastern oysters in NJ and the northeastern US. The average ambient temperature in April was 10°C. The control oysters were maintained in seawater at 22°C with heaters, an optimal temperature that is routinely used for oyster conditioning in our lab. Accordingly, the oysters were divided into 4 groups (7 per group): one group was maintained in seawater at 22°C with aeration and served as the control group (hereafter the 22SW group), one group was exposed to air in an incubator with temperature set at 22°C (the 22AE group), one group was maintained in seawater at 5°C (the 5SW group), and one group was exposed to air at 5°C (the 5AE group). The cold treatment was conducted in a cold storage room with both water and air temperature set at 5°C. The temperature decreased from 22°C to 5°C over approximately one day. The oysters exposed to air were covered with a wet towel to keep them moist, and the towel was rewetted daily. The oysters kept in seawater were not fed to ensure consistency with those exposed to air. The salinity of the seawater was 32 parts per thousand.
After 7 days of treatment, 6 oysters were sampled from each group. Gill tissue from each oyster was flash-frozen in liquid nitrogen and used for transcriptome sequencing. Gills were used for this study because gills are the organ needed for respiration and are important for maintaining homeostasis. Total RNA was extracted from the gill tissue using TRIzol reagent (Invitrogen, USA) following the manufacturer’s protocol. The quantity and quality of the extracted total RNA were assessed with a NanoDrop 2000 (Thermo Scientific, USA) and agarose gel electrophoresis. For each group, RNA was extracted from six oysters, and equal amounts of RNA from each oyster were combined into one sample. A total of 3 μg of RNA per group was used for library preparation.
Sequencing libraries were generated using an NEBNext® Ultra™ RNA Library Prep Kit for Illumina® (NEB, USA) following the manufacturer’s recommendations, and unique index codes were utilised to distinguish the samples. Briefly, mRNA was purified from total RNA using poly-T oligo-attached magnetic beads. Fragmentation was performed using divalent cations under elevated temperature in NEBNext First Strand Synthesis Reaction Buffer (5X). First-strand cDNA was synthesized using random hexamer primers and M-MuLV reverse transcriptase (RNase H-). Second-strand cDNA synthesis was performed using DNA polymerase I and RNase H. The remaining overhangs were converted into blunt ends by exonuclease/polymerase. After adenylation of the 3’ ends, NEBNext adaptors with hairpin loop structures were ligated to the fragments to prepare them for hybridization. To preferentially select cDNA fragments with a length of 150~200 bp, the library fragments were purified with an AMPure XP System (Beckman Coulter, USA). The size-selected, adaptor-ligated cDNA was then incubated with 3 μl of USER Enzyme at 37°C for 15 min and then at 95°C for 5 min before PCR. PCR was performed with Phusion High-Fidelity DNA polymerase, universal PCR primers and the Index (X) Primer. The PCR products were purified (AMPure XP System), and the library quality was assessed with an Agilent Bioanalyzer 2100 system. Clustering of the index-coded samples was performed using a cBot Cluster Generation System with a TruSeq PE Cluster Kit v3-cBot-HS (Illumina) according to manufacturer’s instructions. After cluster generation, the libraries were sequenced on an Illumina HiSeq 2000 platform to generate 100-bp paired-end reads.
Raw sequence data in fastq format were first processed through in-house Perl scripts. Clean reads were obtained by removing reads containing adapters or poly-N sequences (for which the proportion of N bases was > 10%) and reads containing more than 50% of low quality (Q-value ≤ 20) bases. The C. virginica reference genome and gene model annotation files were downloaded from NCBI (https://www.ncbi.nlm.nih.gov/genome/398). An index of the reference genome was built, and clean paired-end reads were mapped to the reference genome using HISAT2.2.4 (Kim et al., 2015) with the “-rna-strandness RF” setting and default settings for other parameters.
The mapped reads of each sample were assembled with StringTie v1.3.1 (Pertea et al., 2015; Pertea et al., 2016) using a reference-based approach. For each transcription region, fragments per kilobase of transcript per million mapped reads (FPKM) values were calculated to quantify transcript abundance of each gene using StringTie software. Differential expression between two samples was analysed with edgeR (Robinson et al., 2010; McCarthy et al., 2012). P-values were obtained using the exactTest function with normalized counts and assuming a negative binomial distribution. Correction for multiple testing was performed by applying the Benjamini–Hochberg method to the p-values to control the false discovery rate (FDR) (Benjamini and Hochberg, 1995). To reduce false positives due to the lack of replication, a fold-change threshold was also applied. Genes with a false discovery rate (FDRs) < 0.001 and absolute fold change ≥ 2 were considered DEGs. A hierarchical cluster analysis of the top 1000 DEGs was performed to assess transcriptional variations among oysters exposed to different stresses using in-house R scripts.
Gene Ontology (GO) enrichment analysis of the DEGs was performed with the GOseq R package (Young et al., 2010). We used the Kyoto Encyclopedia of Genes and Genomes (KEGG) Orthology-Based Annotation System (KOBAS) (Mao et al., 2005) to test the significance of the enrichment of DEGs in KEGG (Kanehisa et al., 2008) pathways. KOBAS uses the whole genome as the default background distribution. For each pathway that occurs in a geneset, it counts the number of genes in the geneset that belong to the pathway and compares it against the whole-genome background. The calculating formula of P-value is:
where N is the number of all genes that with KEGG annotation, n is the number of DEGs in N, M is the number of all genes annotated to specific pathways, and m is number of DEGs in M. The calculated p-values were corrected for FDR (Benjamini and Hochberg, 1995) with FDR ≤ 0.05 as the threshold. Pathways meeting this condition were defined as significantly enriched pathways in DEGs.
Before combining RNA from 6 oysters for library construction, 2 µg of total RNA from each oyster was removed and separately reverse-transcribed into cDNA for qPCR validation. Complementary DNA (cDNA) was prepared using a FastQuant RT kit (with gDNase) (Tiangen, Beijing, China).
We selected eight genes for qPCR validation rather arbitrarily to cover genes with different expression levels. Most of the eight genes were significantly expressed in at least one treatment group, but not all of them were DEGs, such as EC-SOD (Extracellular superoxide dismutaseare). They were chosen to show the correlation between the read counts obtained by RNA-seq and the relative expression levels obtained by qPCR. Some genes were abundantly expressed (KLF5) and some were not (EEF2). At the same time, they may or may not be key genes or in important pathways analyzed. Primers for the eight genes were designed with Primer-BLAST online (http://www.ncbi.nlm.nih.gov/tools/primer-blast/) (Supplementary Table 2). The expression of the selected genes and the housekeeping gene β-actin (Ivanina et al., 2010) was quantified by qPCR using a 7500 Fast Real-time PCR System (Applied Biosystems, USA). The cycle threshold (CT) values of each gene were normalized to those of β-actin, and the relative amount of mRNA specific to each of the selected genes was calculated using the 2−ΔΔCt method (Livak and Schmittgen, 2001). Validation analyses were performed by comparing the read count of each gene obtained from the RNA-seq data (adjusted by edgeR software using one scaling normalized factor) with the mean 2−ΔΔCt value from the qPCR results.
A total of 247,921,390 clean paired-end reads were generated from the four groups of oysters, and these numbers ranged from 55.2 to 67.9 million per group (Supplementary Table 1). The raw sequence data have been submitted to NCBI under the accession numbers SRR5643669, SRR5643668, SRR5643671 and SRR5643670. For all the groups, approximately 80% of the reads were successfully mapped to the reference genome, covering 84.30% to 86.73% of the genes (Supplementary Table 1). For the eight genes selected for validation (Supplementary Table 2), a highly significant correlation (p < 0.0001, r = 0.8553) was found between the read counts obtained by RNA-seq and the relative expression levels obtained by qPCR (Supplementary Figure 1), which indicated that the RNA-seq data are reliable due to the pooling of six individuals and a high read coverage. Further, we applied more stringent selection criteria (FDR < 0.001 plus fold-change ≥ 2) to DEG identification to compensate for the lack of sequencing replication, and identified affected pathways based on concerted regulation of multiple protagonist and antagonist genes (see below) so that the conclusions are not affected by random false positives. Nevertheless, it should be noted that the inclusion of independent samples or sample pools would increase the statistical power and detect more DEGs with small fold changes that could not be identified in this study. Additionally, findings of this study should be viewed with caution due to the lack of treatment replicates. Finally, the response to stress is a temporal process, and our observations are limited to the response at day 7, which may differ from the responses at other time points.
Differential expression analysis revealed large numbers of DEGs among the four groups. Compared with the control condition (22SW), exposure to air at 22°C resulted in the largest number of DEGs (2488), which suggested that air at 22°C is the harshest of the tested conditions for the eastern oyster (Figure 1A). Exposure to air at 5°C produced 1969 DEGs, and exposure to seawater at 5°C produced 1802 DEGs, which indicated that these conditions are relatively less stressful. The two conditions at 5°C shared 987 DEGs (Figure 1B). Under all three stress conditions, a higher number of genes were downregulated than upregulated. These results indicate that all three stress conditions depressed the transcriptional activities of the eastern oyster and that air exposure at 22°C was the most stressful. This finding is consistent with the results obtained for the Pacific oyster, in which air exposure also induces the highest number (4420) of DEGs (Zhang et al., 2012).
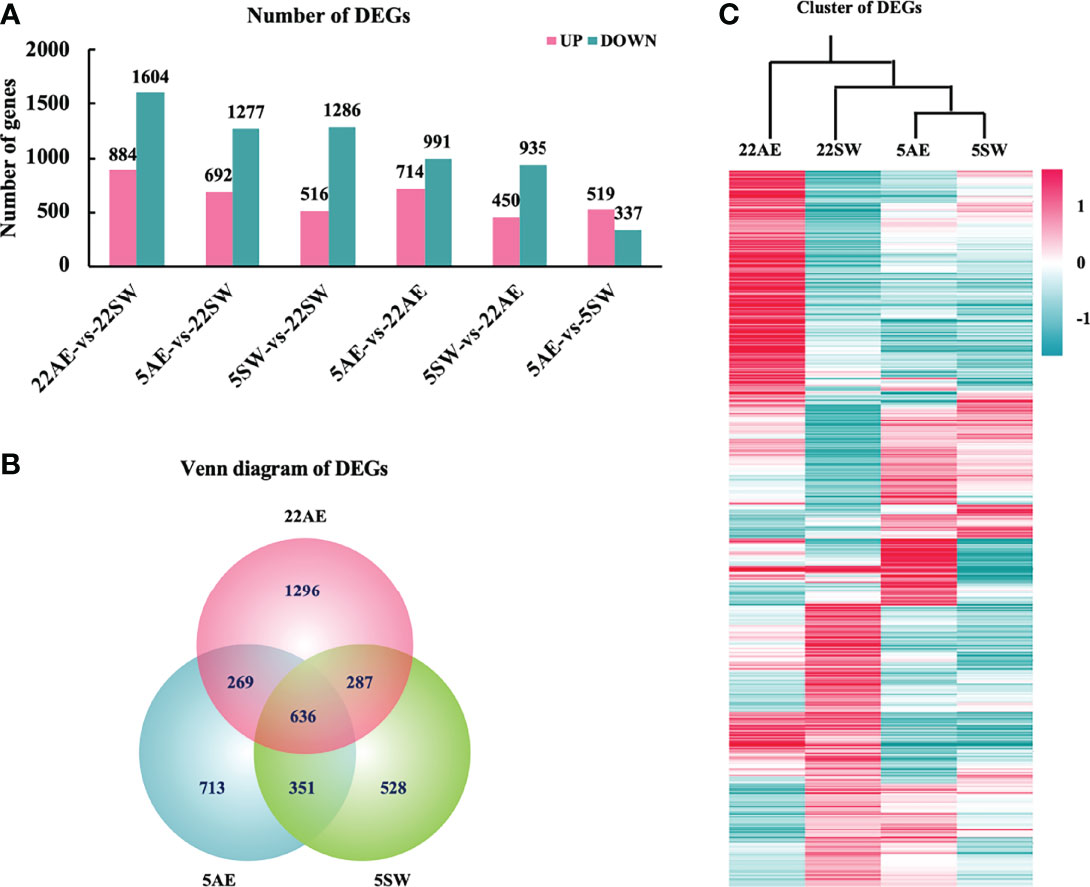
Figure 1 Differentially expressed genes (DEGs) among the four experimental groups of eastern oysters. (A), Number of DEGs between pairs of four groups: seawater at 22°C (22SW, control), exposure to air at 22°C (22AE), seawater at 5°C (5SW) and exposure to air at 5°C (5AE); (B), Venn diagram of DEGs between the three stress conditions and the control 22SW condition; (C), hierarchical clustering of the expression of the top 1000 DEGs in the four groups. The colour range indicates the log10normalized (read count+1) values from high (red, the maximum value is 8.14) to low (green, the minimum value is -6.82).
The three stress conditions differed among the types of DEGs induced. Although the three conditions shared 636 DEGs, they also had large numbers of unique DEGs: 1296 in 22AE, 713 in 5AE and 528 in 5SW (Figure 1B). Among the four groups, the expression profiles obtained for 5AE and 5SW were most similar but differed significantly from that obtained for 22AE, as indicated by the clustering heatmap (Figure 1C). As low temperature tends to reduce metabolic activity, oysters might be less susceptible to air exposure at low temperature than at normal or high temperatures. All DEGs revealed by pairwise comparisons are shown in Supplementary Table 3.
Enrichment analysis revealed that the following KEGG pathways were overrepresented in the DEGs between the 22AE and 22SW control groups: DNA replication, cell cycle control, retinol metabolism, apoptosis, nuclear factor kappa B (NF-κB) signalling, lipid metabolism, peroxisome and tumour necrosis factor (TNF) signalling (P-value < 0.05, Table 1). The main processes affected in the 5SW and 5AE groups were lipid metabolism and DNA replication. The only GO term significantly enriched in DEGs between 22AE and 5AE was DNA metabolism (0006259), and these DEGs included genes involved in DNA replication and cell cycle control. Some of the significantly enriched pathways and processes are highlighted below. For brevity, the gene IDs from the reference genome were shortened by omitting the string “OC1111”, which is shared by all the genes (i.e., L00001 for LOC111100001).
One of the most significantly affected processes in the eastern oyster under stress is cell division, as evidenced by the identification of large number of DEGs related to DNA replication and cell cycle control under all three stressors at day 7, particularly in the 22AE group (Table 1). Cell cycle progression, a process that is conserved in metazoans, is regulated by cyclin-dependent kinases (CDKs) and their specific cyclin partners, positive and negative regulators, and downstream targets (Morgan, 1997). In this study, cyclin A (L22860, L22939), cyclin D (L03611), cyclin E (L32839, L99092), and CDK2 (L21435), which are expressed at the G1 phase and determine whether cells proceed through the G1 phase into the S phase, were all downregulated under air exposure and cold stress (Supplementary Table 4 and Figure 2). During the S phase, DNA replication starts with the building of replication complexes, where minichromosome maintenance (MCM) proteins are loaded onto chromatin to “licence” DNA for replication (Blow and Laskey, 1988). Six MCM2-7 complex genes (L09564, L24297, L35545, L31693, L03623, L36042), the associated component MCMBP (L00880) and CDC7 (L22674), which can phosphorylate MCM2 and MCM3, were significantly downregulated in oysters under the 22AE conditions (Supplementary Table 4). All six genes under cold stress showed reduced expression, although the reduction in the expression of some of the genes was not significant. Once replication complexes are built and activated, a series of replicating forks form along the chromosome, generating bubbles that eventually fuse together to complete DNA replication (Alberts et al., 2010). Genes associated with the replication protein A complex, such as replication protein A (L20888), DNA polymerase (L12959, L30346, L37485, L30057, L26184, L28010), DNA primase (L26192, L21347) and DNA clamp proteins (RFC and PCNA, L08767, L31355, L32381, L34900, L22897, L35799, L20673), were downregulated (Supplementary Table 4). Genes encoding cyclins and CDKs that promote the progression of cells from the G2 phase to the M phase, such as G2/mitotic-specific cyclin-A (L22860, L22939) and M-phase inducer phosphatase (CDC25A, L32091, L32548, L35070), which function as dosage-dependent inducers of mitotic progression (Nilsson and Hoffmann, 2000), were also downregulated (Figure 2). Other genes, such as CDK7 (L25401), involved in transcription initiation and DNA repair were also downregulated. These results strongly suggest that exposure of the eastern oyster to air and cold stress suppresses the transcription of key genes needed for cell cycle progression or cell division, which is crucial for growth and immune functions. The discovery of the downregulation of key genes needed for cell division provides molecular support for the well-known negative effects of stress on growth and the immune response (Malek, 2010; Guo et al., 2015; Guo and Ford, 2016; Clements et al., 2018; La Peyre et al., 2018).
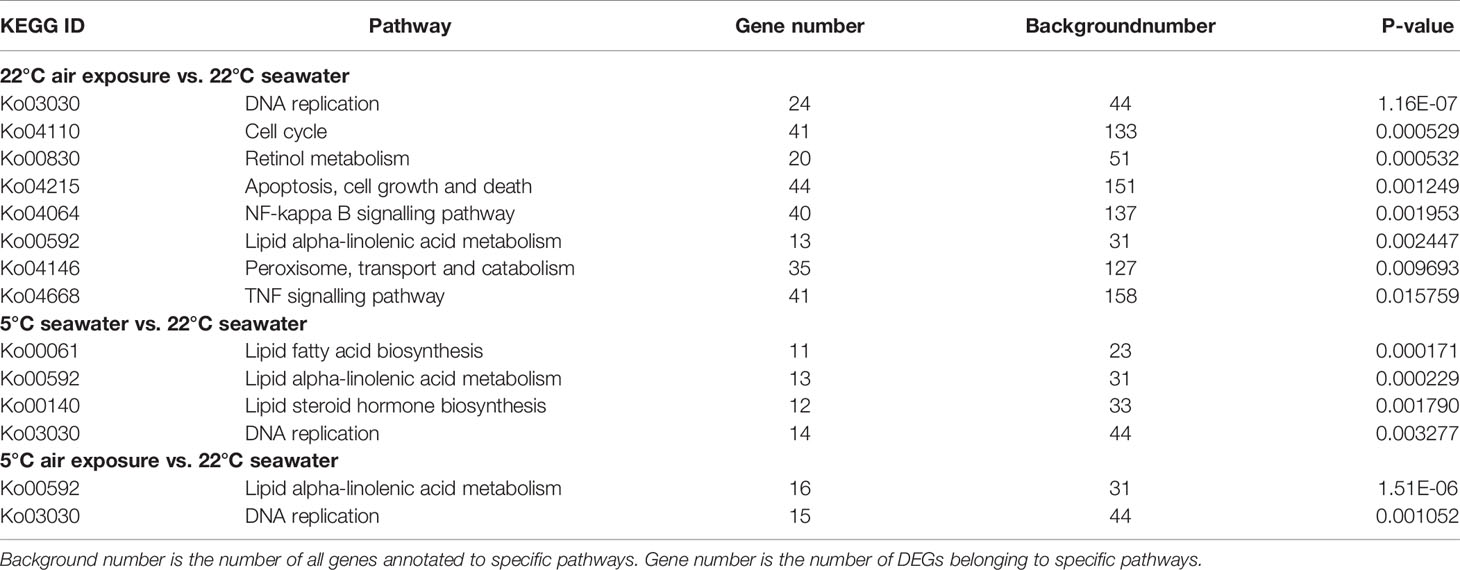
Table 1 KEGG pathways enriched in differentially expressed genes induced by stressors in the eastern oyster.
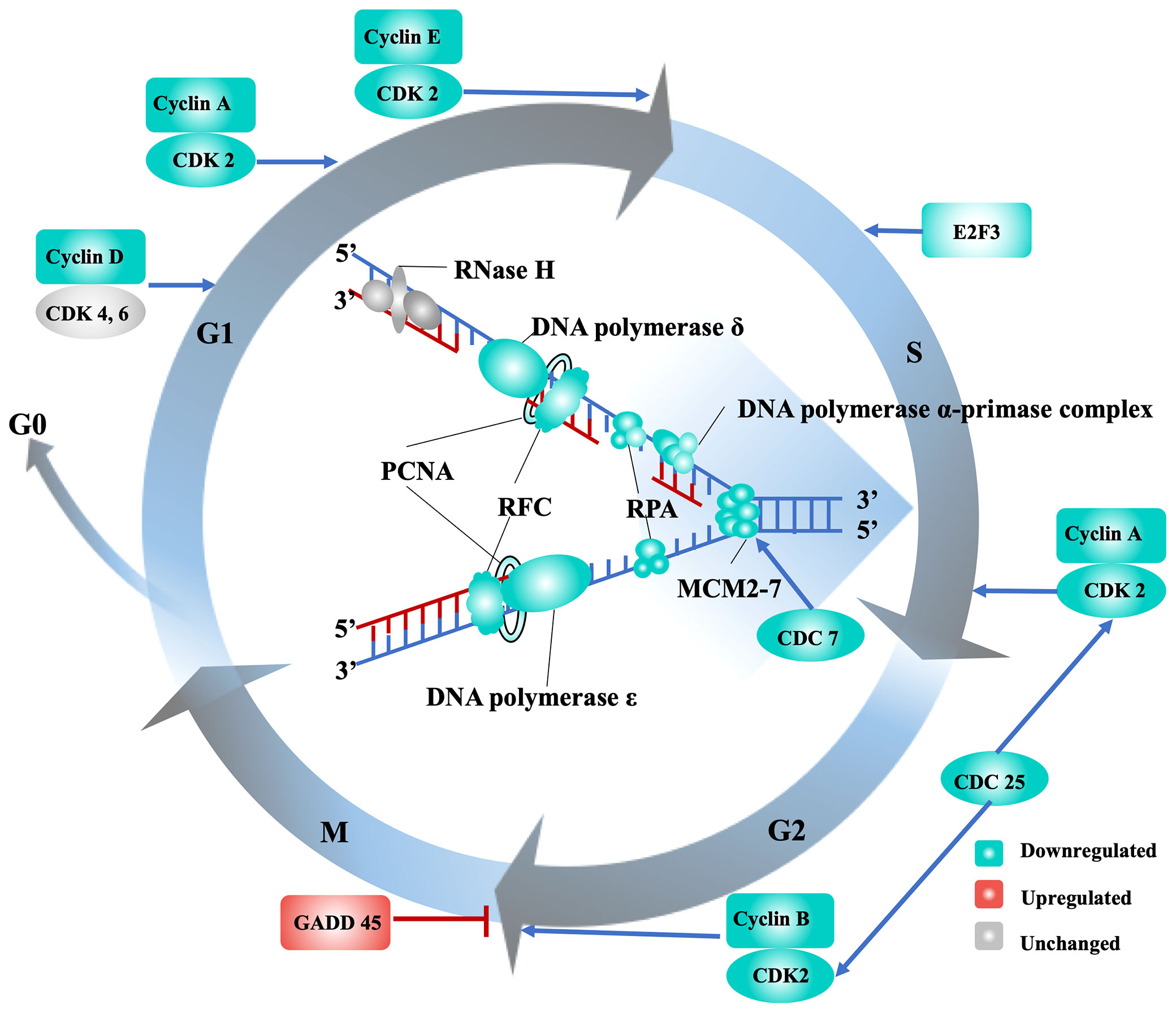
Figure 2 Transcriptional regulation of the cell cycle and DNA replication in eastern oysters under stress. The gene names are the following: CDC, cell division cycle-related protein kinase; CDK, cyclin-dependent kinase; E2F, transcription factor E2F3; GADD 45, growth arrest and DNA damage-inducible protein 45; MCM2-7, minichromosome maintenance proteins; PCNA, proliferating cell nuclear antigen; RFC, replication factor C; RPA, replication protein A.
Cell cycle arrest may be induced by DNA damage, and cells attempt to repair this damage during cell cycle arrest (Pucci et al., 2000). In this study, genes that are induced by DNA damage, such as the growth arrest and DNA damage-inducible protein GADD45 (L18431, L18444, L18682, L21109), which is induced by p53 and arrests the cell cycle at the G2/M checkpoint (Papa et al., 2004), were upregulated. High-mobility group proteins (L09677, L09703, L12806, L17359, L25762, L26226), which respond to DNA damage and oxidative stress in yeast (Reeves and Adair, 2005; Lange et al., 2008), were also upregulated. In addition, sestrin1 (L03315), which is induced by the p53 tumour suppressor protein and has the ability to arrest cell growth and decrease ROS production, was also upregulated (Sun et al., 2014). It is widely accepted that p53 mediates cell cycle arrest, apoptotic cell death, and/or cellular senescence (Li et al., 2012). Although the transcription level of p53 did not change significantly under stress in this study, it is possible that p53 activity, a key event in the cell cycle arrest process, is controlled by a diverse array of posttranslational modifications (Kruse and Gu, 2009). Additionally, because this study only provides a snapshot in time, p53 and its regulators may be differentially expressed at other time points. It should be noted that in addition to DNA damage repair, stress and nutritional limitations may also lead to cell cycle arrest and growth arrest (Yanagida et al., 2011; Rossi and Antonangeli, 2015).
Although the underlying mechanisms are not completely defined, cell cycle arrest is an active response to stress that enables organisms to survive under challenging environmental conditions. The results of this study demonstrate that the exposure of eastern oyster to air and cold stress suppresses the transcription of genes promoting cell cycle progression, which may lead to cell cycle arrest and in turn affect growth, immune responses and other biological processes. Growth retardation inferred from cell cycle arrest is consistent with the results from field experiments conducted by Malek (2010), who found that the growth of eastern oysters decreased with increases in the duration of intertidal air exposure, and La Peyre et al. (2018), who found that a lower growth rate among oysters that experienced daily air exposure. Air exposure in the field may also reduce the time dedicated to feeding and increase the use of glycogen storage, which contribute to reduced growth.
Exposure to air at 22°C triggers the inhibition of apoptosis and related regulatory pathways. Inhibitors of apoptosis (IAPs) play important roles in promoting cell survival. IAP family members inhibit programmed cell death by binding to and/or directly inhibiting caspases (LaCasse et al., 1998). The C. gigas genome exhibits an expanded IAP family (48 genes), members of which are highly expressed after air exposure and pathogen infection, which indicates that IAPs play crucial roles in anti-apoptotic processes in oysters (Zhang et al., 2012; Green et al., 2015). We identified 58 IAPs in the eastern oyster genome; 8 and 4 of these IAPs were highly and significantly upregulated and slightly downregulated, respectively, in the 22AE group, resulting in an overall upregulation of IAPs (Supplementary Table 4). Furthermore, genes encoding caspases, CASP7 (L3317, L2489, L24891) and CASP10 (L04823, L33185, L33189), were slightly downregulated in the 22AE group. These results suggest the existence of a concerted effort to inhibit apoptosis in oysters under stress (Figure 3 and Supplementary Table 4).
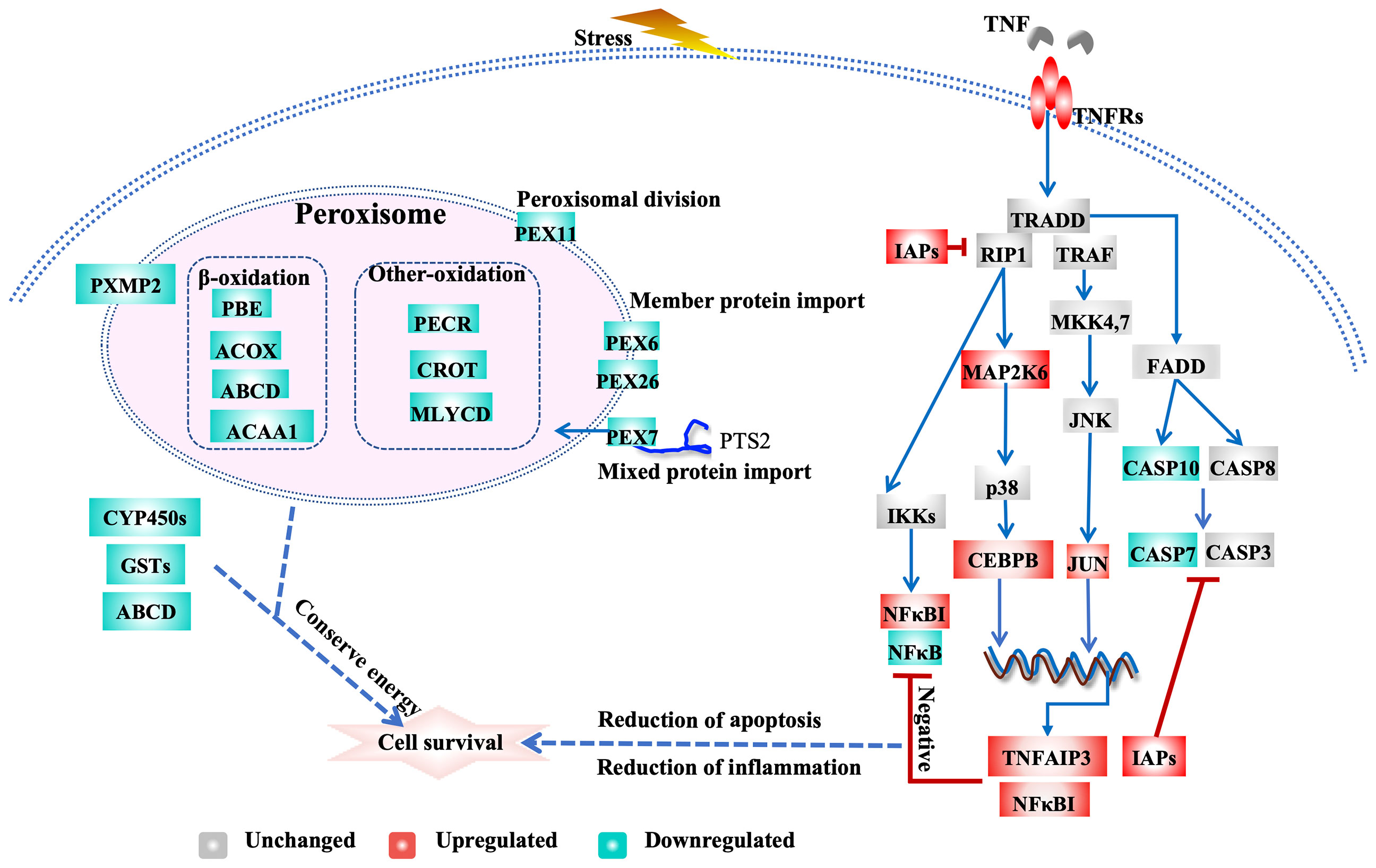
Figure 3 Transcriptional regulation of apoptosis, TNF signalling and peroxisomal activity in eastern oysters during exposure to air. The key genes are the following: ABCD, ATP-binding cassette subfamily D; ACAA1, 3-ketoacyl-CoA thiolase A, peroxisomal; ACOX1, peroxisomal acyl-coenzyme An oxidase 1; NUDT19, nucleoside diphosphate-linked moiety X motif 19; JUN, Jun proto-oncogene, AP-1 transcription factor subunit; CASP7, caspase-7; CASP10, caspase-10; CEBP, CCAAT/enhancer-binding protein; CYP450s, cytochrome P450s; CROT, peroxisomal carnitine O-octanoyltransferase; GSTs, glutathione S-transferases; IAP, inhibitor of apoptosis; TNFR, tumour necrosis factor receptor; MAP2K6, dual specificity mitogen-activated protein kinase 6; NFKBIA, NF-kappa-B inhibitor alpha-like; NFKB, nuclear factor NF-kappa-B p105; PEX7, peroxisomal targeting signal 2 receptor; PEX26, peroxisome assembly protein 26; PEX6, peroxisome assembly factor 2; PEX11, peroxisomal membrane protein 11; PXMP2, peroxisomal membrane protein 2; PECR, peroxisomal trans-2-enoyl-CoA reductase-like; PIK3, phosphatidylinositol-4,5-bisphosphate 3-kinase; TNFAIP3, tumour necrosis factor alpha-induced protein 3.
Exposure to air at 22°C affected signalling pathways such as the TNF signalling and NF-kappa B signalling pathways (Table 1 and Supplementary Table 4). These pathways play important roles in the responses to biotic and abiotic stresses in the Pacific oyster (Zhang et al., 2015) and other molluscs, such as the European flat oyster Ostrea edulis (Martín-Gómez et al., 2012), disk abalone Haliotis discus (De Zoysa et al., 2009), and Chinese scallop Chlamys farreri (Li et al., 2009). Depending on which downstream genes are activated, the TNF signalling pathway can induce apoptosis or necroptosis or promote cell survival (Baud and Karin, 2001; Sabio and Davis, 2014). TNF signalling may induce apoptosis via TNFα-TNFR-(TRADD/RIP1/TRAF2 complex)-FADD-CASP signalling (Rath and Aggarwal, 1999). However, the upregulation of IAPs and the downregulation of CASPs may have a negative feedback effect on TNF signalling. The TNF signalling pathway induces inflammation via TNFα-TNFR-(TNFR1 complex I)-TAB/TAK1-IKK-NF-κB signalling (Bantel and Schulze-Osthoff, 2012). The major pathway for the systemic inflammatory responses depends on NF-κB. The strong upregulation of NF-κB inhibitor alpha-like genes (NFκBIA, L31108, L31219) and the downregulation of NF-κB (L21590) suggest that the NF-κB signalling pathway is inhibited by stress (De Martin et al., 1993). Notably, genes encoding TNF alpha-induced protein 3 (TNFAIP3, L04432, L34417) (Figure 3 and Supplementary Table 4) induced by TNF were highly upregulated, and these genes inhibit NF-κB activation and TNF-mediated apoptosis and thereby negatively regulate downstream cell death signalling (Song et al., 1996; Jiang et al., 1999). The prostaglandin E receptor 4 (EP4, L17914, L20936, L18192, L22415) interacts with prostaglandin E receptor 4-associated protein (EPRAP) to inhibit the activation of NF-κB (Minami et al., 2008). Other genes that negatively regulate the inflammatory response, such as suppressor of cytokine signalling (SOCS2, L17924), were also upregulated, which could limit the cytokine response by inhibiting Janus kinase 2 (JAK2) activity (Rico-Bautista et al., 2006; Posselt et al., 2011). These processes inhibit apoptosis through TNF signalling pathways and attenuate the inflammatory response by inhibiting NFκB activation. Inhibition of the inflammatory response may lead to weakened immune responses, which explains the susceptibility and mortality of oysters under stress (Malek, 2010; Guo et al., 2015; Guo and Ford, 2016; Clements et al., 2018). In addition, Jun proto-oncogene (JUN, L20138) and CEBP-beta (CEBPB, L36304), which regulate a number of cellular processes, including differentiation, proliferation, apoptosis and the immune response, were upregulated, which indicated their potential role in the stress response and the complex crosstalk among signalling pathways.
Overall, these transcriptomic changes indicate that an orchestrated response involving the upregulation of expanded IAPs and the downregulation of caspases through interactions with TNF and NF-κB signalling work in concert to help stressed oyster cells evade apoptosis, whereas the downregulation of NF-κB signalling may suppress inflammatory responses and impair immune functions.
The key genes and pathways involved in xenobiotic biotransformation and lipid metabolism were affected during exposure to air at 22°C (Table 1). The xenobiotic biotransformation system helps oysters cope with environmental contaminants (Zhang et al., 2016). Genes that are known to be important for xenobiotic biotransformation, such as cytochrome P450s (CYP450s, 11 genes), myeloperoxidase (MPO, L099927, L01491, L01639, L01744), GSTs (L02975, L04070), and ATP-binding cassette subfamily D member (L30560) (Supplementary Table 4), were significantly downregulated under 22AE conditions.
Genes related to fatty acid synthesis, particularly the key gene fatty acid synthase (FAS, L18924, L21707), were highly upregulated under 22AE conditions. Moreover, genes playing key roles in fatty acid oxidation in peroxisomes, such as peroxisomal acyl-coenzyme A oxidase 1 (ACOX1, L13840, L15085, L32545, MSTRG.2967), which is the first enzyme in the fatty acid beta-oxidation pathway that produces hydrogen peroxide (Inestrosa et al., 1979), were downregulated. The 3-ketoacyl-CoA thiolase (ACAA1, L15744, L15745), which catalyses the final step in the β-oxidation cycle, was also downregulated. The upregulation and downregulation of these key genes were also observed in the groups exposed to cold stress, albeit at a small magnitude, which suggested similar regulation of fatty acid metabolism under cold stress (Supplementary Table 4). Maintaining the energy balance is critical for stress tolerance (Sokolova et al., 2012), and our results indicate the involvement of lipid metabolism in coping with air exposure and cold stress. Additionally, organisms in cold environments often have more unsaturated fatty acids for membrane fluidity (Dey et al., 1993).
Peroxisomes play particularly important roles in lipid metabolism, ether-phospholipid biosynthesis, and reactive oxygen species (ROS) metabolism (Fransen et al., 2012; Jo and Cho, 2019). Moreover, peroxisomes are signalling platforms for antiviral innate immune responses (Dixit et al., 2010). More peroxisomal genes were downregulated during exposure to air at 22°C than under cold stress conditions. Proteins that control peroxisome assembly, division, and inheritance, such as peroxins or peroxisome biogenesis factors (PEXs, L03004, L08788, L31586, L17608, L28390, L35112) (Ma et al., 2011), were downregulated (Figure 3 and Supplementary Table 4), which indicated that peroxisome biogenesis is downregulated during exposure to air at 22°C.
The downregulation of xenobiotic biotransformation and peroxisome-related genes may decrease fatty acid oxidation (Mayer et al., 1995), whereas the upregulation of fatty acid synthase increases the de novo synthesis of fatty acids; altogether, these effects alter the fatty acid composition. These results all indicate that fatty acid metabolism plays a critical role in mediating the responses of oysters to air exposure.
The proline accumulation pathway was activated in oysters under cold stress (Figure 4). The synthesis of proline is mediated by two pathways: the L-glutamic acid pathway and the ornithine pathway (Verbruggen and Hermans, 2008). Delta-1-pyrroline-5-carboxylate synthase (P5CS, L36221), a key enzyme responsible for the synthesis of L-glutamate 5-semialdehyde from L-glutamate in steps 1 and 2 of the subpathway, was significantly upregulated (Figure 4 and Supplementary Table 4). Arginase (ARG, L30979), which transforms arginine to ornithine, and ornithine aminotransferase (OAT, L36290), which catalyses the transformation of ornithine into L-glutamate 5-semialdehyde, were both upregulated by cold stress. The enzyme pyrroline-5-carboxylate reductase (PYCR2, L28747, PYCR3, L28753), which catalyses the last step in proline biosynthesis, was also upregulated. Together, these results suggest that the proline accumulation pathway is activated or upregulated under cold stress. The accumulation of proline has been observed in oysters under hyperosmotic stress (Meng et al., 2013) and is a key mechanism for cold tolerance in plants (Szabados and Savoure, 2010). The activation of the proline accumulation pathway observed in this study implies that a large proline pool is important for the survival of oysters under cold stress conditions.
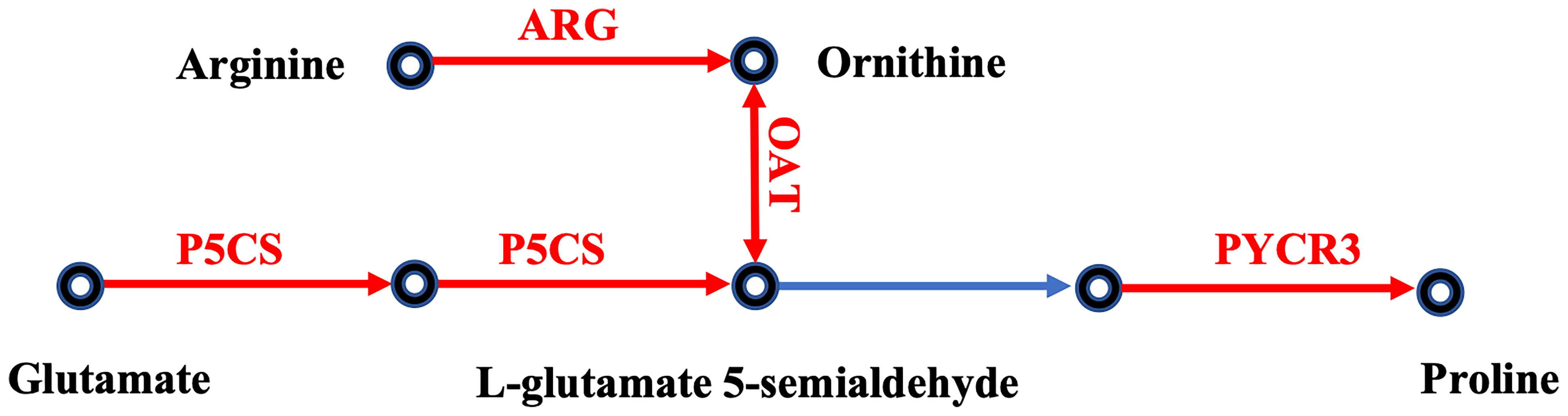
Figure 4 Effects of cold stress on the proline accumulation pathway in eastern oysters. The genes in red exhibit upregulated expression. The gene names are the following: P5CS, delta-1-pyrroline-5-carboxylate synthase; PYCR, pyrroline-5-carboxylate reductase; ARG, arginase; OAT, ornithine aminotransferase.
Nine genes for histone H4 and four genes for histone H2B were highly upregulated under cold stress, particularly under 5SW conditions. The upregulation of some but not all histone subunits was also observed in the Pacific oyster under stress (Zhang et al., 2012). Canonical histones are synthesized during the S phase for the packaging of newly synthesized DNA. Because genes that promote cell division were downregulated under stress, the upregulation of some histone genes is intriguing and begs other explanations. Moving, ejecting or restructuring nucleosomes is one of the strategies for maintaining chromatin stability (Cedar and Bergman, 2009; Clapier and Cairns, 2009). The high-mobility groups (HMGs) of proteins (L09677, L09703, L12806, L17359, L25762, L26226), another group of major chromatin architectural components of eukaryotes, were also highly upregulated under cold stress (Supplementary Table 4). Although preliminary and to be confirmed through further studies, these results indicate that genes encoding chromosomal proteins are upregulated under cold stress, probably for repairing or stabilizing DNA under stress or as epigenetic modifications that regulate transcription. In plants, stress induces dramatic epigenetic changes, such as DNA methylation, histone modifications and changes in the composition of different histone variants, which regulate gene expression and enable stress responses and memory (Chinnusamy and Zhu, 2009). We hypothesize that the upregulation of the expression of certain chromosomal structural protein genes observed in this study is involved in the epigenetic modifications of chromatin that are needed for regulating the transcriptional response to stress and facilitating epigenetic memory in oysters.
Several genes encoding hormone and neurotransmitter receptors were upregulated in oysters under air exposure and cold stress conditions. The melatonin receptor (MTNR, L03709, L04987, L03710) and the FMRFamide receptor (FMRFAR, L22789, L22537, L21307) were upregulated under all three conditions, particularly under 22AE conditions (Supplementary Table 4). FMRFamide and FMRFamide-related neuropeptides are abundant in invertebrates and have several important functions. FMRFamide-related peptides (FaRPs) in gastropods are involved in movement, feeding, defecation, and reproduction (López-Vera et al., 2008). Melatonin is involved in circadian and seasonal timing (Reppert et al., 1994; Dubocovich et al., 2003) and was highly upregulated under all stresses in this study. In addition, genes encoding atrial natriuretic peptide receptor A and neuronal acetylcholine receptor were also upregulated under 5AE conditions. The up regulation of these hormone and neurotransmitter receptors may be critical for activating physiological responses or inducing physiological state transitions as in a circadian system shift (Ottaviani and Franceschi, 1996; Fuentes-Pardo et al., 2013; Lenz et al., 2015). In C. gigas, many GPCR genes, including those encoding prostaglandin E2 receptors, FMRFamide receptors, cholecystokinin receptors, melatonin receptors, prolactin-releasing peptide receptors, adenosine receptors and dopamine D2-like receptors, are upregulated by OsHV-1 infection (He et al., 2015). It has been shown that neuronal acetylcholine receptors are greatly expanded in bivalve molluscs and are important for adaptation to stationary life under variable environments (Jiao et al., 2019).
The remarkable resilience of the eastern oyster is not well understood at the molecular level. The results obtained in this study clearly demonstrate that exposure to air and cold stress induce significant changes in the transcription of large numbers of genes that are likely crucial for the response and tolerance of the eastern oyster to stress. The observed suppression of genes promoting cell cycle progression suggests that cell division may be arrested in oysters under stress. The upregulation of anti-apoptosis genes and the downregulation of pro-apoptosis genes and regulatory pathways indicate a concerted effort to evade cell death. The inhibition of key signalling pathways that activate the immune response suggests that immune defence is impaired by stress. Receptors for hormones and neurotransmitters were found to be upregulated in response to stress. The upregulation of genes for proline synthesis and chromosome structural proteins indicates that proline accumulation and epigenetic modifications of chromosomes are important for coping with cold stress. Although it is well known that stress depresses growth and immune responses, which often lead to disease outbreaks and mass summer mortalities in molluscs (Guo and Ford, 2016), the underlying molecular mechanisms are not well understood. By revealing the downregulation of genes involved in cell cycle progression and the inflammatory response under stress, this study provides transcriptomic evidence for the negative effects of stress on growth and immune functions. This study reveals several previously undescribed responses to stress in oysters and other molluscs. The genes and pathways implicated in this study provide useful resources for further studies on the stress response and adaptation in molluscs and other marine invertebrates. Knowing how stress affects growth and immune response at molecular levels is essential for understanding stress impact and assessing the adaptive potential of marine organisms under climate change. Oysters are economically important and support major aquaculture industries worldwide. The sustainable development of the oyster aquaculture industry depends on genetic improvement of cultured stocks (Guo, 2021). The candidate genes and their polymorphisms can be used for marker-assisted selection for the breeding of more resilient oysters.
XG and CL conceived and designed the study. CL performed the experiments and analysed the data. CL, XG and HW interpreted the data and wrote the manuscript. All authors have read and approved the manuscript. All authors contributed to the article and approved the submitted version.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, PRJNA378873.
This work was partly supported by grants from the National Science Foundation of China (No. 41906083), the Taishan Overseas Scholar Project of Shandong, the US National Oceanic and Atmospheric Agency (NA18NMF4720321) and the Department of Agriculture Animal Health (NJ32920). The statements, findings, conclusions, and recommendations are those of the authors and do not necessarily reflect the views of the National Oceanic and Atmospheric Administration or the Department of Commerce.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We thank Baoyu Huang for helping with the sampling.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2022.921877/full#supplementary-material
ABCD, ATP-binding cassette subfamily D; ACAA1, 3-ketoacyl-CoA thiolase A, peroxisomal; ACOX1, peroxisomal acyl-coenzyme A oxidase 1; ARG, arginase; CASP, caspase; CDC, cell division cycle-related protein kinase; CDK, cyclin-dependent kinase; CEBP, CCAAT/enhancer-binding protein; CROT, peroxisomal carnitine O-octanoyltransferase; CYP450, cytochrome P450; E2F, transcription factor E2F3; ER, endoplasmic reticulum; GADD 45, growth arrest and DNA damage-inducible protein 45; GST, glutathione S-transferase; IAP, inhibitor of apoptosis; JUN, AP-1 transcription factor; MAP2K6, dual specificity mitogen-activated protein kinase 6; MCM2-7, minichromosome maintenance proteins; NFKB, nuclear factor NF-kappa-B p105; NFKBIA, NF-kappa-B inhibitor alpha-like; NUDT19, nucleoside diphosphate-linked moiety X motif 19; OAT, ornithine aminotransferase; P5CS, delta-1-pyrroline-5-carboxylate synthase; PCNA, proliferating cell nuclear antigen; PECR, peroxisomal trans-2-enoyl-CoA reductase-like; PEX, peroxin protein; PIK3, phosphatidylinositol-4,5-bisphosphate 3-kinase; PYCR, pyrroline-5-carboxylate reductase; RFC, replication factor C; ROS, reactive oxygen species; RPA, replication protein A; TNF, tumour necrosis factor; TNFAIP3, tumour necrosis factor alpha-induced protein 3; TNFR, tumour necrosis factor receptor.
Alberts B., Johnson A., Lewis J., Raff M., Roberts K., Walter P. (2002). “The Initiation and Completion of DNA Replication in Chromosomes,” in Molecular Biology of the Cell 4th edition. (New York, NY: Garland Science), 377–392.
Allen S. M., Burnett L. E. (2008). The Effects of Intertidal Air Exposure on the Respiratory Physiology and the Killing Activity of Hemocytes in the Pacific Oyster, Crassostrea Gigas (Thunberg). J. Exp. Mar. Biol. Ecol. 357, 165–171. doi: 10.1016/j.jembe.2008.01.013
Alvarez M. R., Friedl F. E. (1992). Effects of a Fungicide on In Vitro Hemocyte Viability, Phagocytosis and Attachment in the American Oyster, Crassostrea Virginica. Aquaculture 107, 135–140. doi: 10.1016/0044-8486(92)90058-S
Bantel H., Schulze-Osthoff K. (2012). Mechanisms of Cell Death in Acute Liver Failure. Front. Physiol. 3. doi: 10.3389/fphys.2012.00079
Baud V., Karin M. (2001). Signal Transduction by Tumor Necrosis Factor and its Relatives. Trends Cell Biol. 11, 372–377. doi: 10.1016/S0962-8924(01)02064-5
Benjamini Y., Hochberg Y. (1995). Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. B Methodol. 57, 289–300. doi: 10.1111/j.2517-6161.1995.tb02031.x
Blow J. J., Laskey R. A. (1988). A Role for the Nuclear Envelope in Controlling DNA Replication Within the Cell Cycle. Nature 332, 546–548. doi: 10.1038/332546a0
Boyd J. N., Burnett L. E. (1999). Reactive Oxygen Intermediate Production by Oyster Hemocytes Exposed to Hypoxia. J. Exp. Biol. 202 (22), 3135–3143. doi: 10.1242/jeb.202.22.3135
Cedar H., Bergman Y. (2009). Linking DNA Methylation and Histone Modification: Patterns and Paradigms. Nat. Rev. Genet. 10, 295–304. doi: 10.1038/nrg2540
Chinnusamy V., Zhu J. K. (2009). Epigenetic Regulation of Stress Responses in Plants. Curr. Opin. Plant Biol. 12, 133–139. doi: 10.1016/j.pbi.2008.12.006
Clapier C. R., Cairns B. R. (2009). The Biology of Chromatin Remodeling Complexes. Annu. Rev. Biochem. 78, 273–304. doi: 10.1146/annurev.biochem.77.062706.153223
Clements J. C., Davidson J. D. P., McQuillan J. G., Comeau L. A. (2018). Increased Mortality of Harvested Eastern Oysters (Crassostrea Virginica) is Associated With Air Exposure and Temperature During a Spring Fishery in Atlantic Canada. Fish. Res. 206, 27–34. doi: 10.1016/j.fishres.2018.04.022
De Martin R., Vanhove B., Cheng Q., Hofer E., Csizmadia V., Winkler H., et al. (1993). Cytokine-Inducible Expression in Endothelial Cells of an I Kappa B Alpha-Like Gene is Regulated by NF Kappa B. EMBO J. 12, 2773–2779. doi: 10.1002/j.1460-2075.1993.tb05938.x
Dey I., Buda C., Wiik T., Halver J. E., Farkas T. (1993). Molecular and Structural Composition of Phospholipid Membranes in Livers of Marine and Freshwater Fish in Relation to Temperature. Proc. Natl. Acad. Sci. U.S.A. 90, 7498–7502. doi: 10.1073/pnas.90.16.7498
De Zoysa M., Jung S., Lee J. (2009). First Molluscan TNF-α Homologue of the TNF Superfamily in Disk Abalone: Molecular Characterization and Expression Analysis. Fish Shellfish Immunol. 26, 625–631. doi: 10.1016/j.fsi.2008.10.004
Dixit E., Boulant S., Zhang Y., Lee A. S., Odendall C., Shum B., et al. (2010). Peroxisomes are Signaling Platforms for Antiviral Innate Immunity. Cell 141, 668–681. doi: 10.1016/j.cell.2010.04.018
Dubocovich M. L., Rivera-Bermudez M. A., Gerdin M. J., Masana M. I. (2003). Molecular Pharmacology, Regulation and Function of Mammalian Melatonin Receptors. Front. Biosci. 8, d1093–d1108. doi: 10.2741/1089
Eierman L. E., Hare M. P. (2014). Transcriptomic Analysis of Candidate Osmoregulatory Genes in the Eastern Oyster Crassostrea Virginica. BMC Genom. 15, 503. doi: 10.1186/1471-2164-15-503
Fransen M., Nordgren M., Wang B., Apanasets O. (2012). Role of Peroxisomes in ROS/RNS-Metabolism: Implications for Human Disease. Biochim. Biophys. Acta 1822, 1363–1373. doi: 10.1016/j.bbadis.2011.12.001
Fuentes-Pardo B., Barriga-Montoya C., de la O-Martínez A. (2013). “"Melatonin as a Circadian Hormone in Invertebrates,",” in New Developments in Melatonin Research. Eds. Acuna-Castroviejo D., Rusanova I., Escames G. (New York, NY: Nova Science Publishers, Inc), 27–38.
Galtsoff P. S. (1964). “The American Oyster, Crassostrea Virginica Gmelin,” in Fishery Bulletin (Washington, D.C.: US Government Printing Office), 1–480.
Gómez-Chiarria M., Warrenb W. C., Guo X., Proestoud D. (2015). Developing Tools for the Study of Molluscan Immunity: The Sequencing of the Genome of the Eastern Oyster, Crassostrea Virginica. Fish Shellfish Immunol. 46, 2–4. doi: 10.1016/j.fsi.2015.05.004
Green T. J., Rolland J. L., Vergnes A., Raftos D., Montagnani C. (2015). OsHV-1 Countermeasures to the Pacific Oyster's Anti-Viral Response. Fish Shellfish Immunol. 47, 435–443. doi: 10.1016/j.fsi.2015.09.025
Guo X. (2021). “Genetics in Shellfish Culture,” in Molluscan Shellfish Aquaculture: A Practical Guide. Ed. Shumway S. (Essex, UK: 5m Books Ltd), 393–413.
Guo X., Ford S. E. (2016). Infectious Diseases of Marine Molluscs and Host Responses as Revealed by Genomic Tools. Philos. Trans. R. Soc Lond. B Biol. Sci. 371, 20150206. doi: 10.1098/rstb.2015.0206
Guo X., He Y., Zhang L., Lelong C., Jouaux A. (2015). Immune and Stress Responses in Oysters With Insights on Adaptation. Fish Shellfish Immunol. 46, 107–119. doi: 10.1016/j.fsi.2015.05.018
He Y., Jouaux A., Ford S. E., Lelong C., Sourdaine P., Mathieu M., et al. (2015). Transcriptome Analysis Reveals Strong and Complex Antiviral Response in a Mollusc. Fish Shellfish Immunol. 46, 131–144. doi: 10.1016/j.fsi.2015.05.023
Inestrosa N. C., Bronfman M., Leighton F. (1979). Detection of Peroxisomal Fatty Acyl-Coenzyme A Oxidase Activity. Biochem. J. 182, 779–788. doi: 10.1042/bj1820779
Ivanina A. V., Sokolov E. P., Sokolova I. M. (2010). Effects of Cadmium on Anaerobic Energy Metabolism and mRNA Expression During Air Exposure and Recovery of an Intertidal Mollusk Crassostrea Virginica. Aquat. Toxicol. 99, 330–342. doi: 10.1016/j.aquatox.2010.05.013
Jiang Y., Woronicz J. D., Liu W., Goeddel D. V. (1999). Prevention of Constitutive TNF Receptor 1 Signaling by Silencer of Death Domains. Science 283, 543–546. doi: 10.1126/science.283.5401.543
Jiao Y., Cao Y., Zheng Z., Liu M., Guo X. (2019). Massive Expansion and Diversity of Nicotinic Acetylcholine Receptors in Lophotrochozoans. BMC Genom. 20, 937. doi: 10.1186/s12864-019-6278-9
Jo D. S., Cho D. H. (2019). Peroxisomal Dysfunction in Neurodegenerative Diseases. Arch. Pharm. Res. 42, 393–406. doi: 10.1007/s12272-019-01131-2
Kanehisa M., Araki M., Goto S., Hattori M., Hirakawa M., Itoh M., et al. (2008). KEGG for Linking Genomes to Life and the Environment. Nucleic Acids Res. 36, D480–D484. doi: 10.1093/nar/gkm882
Kawabe S., Takada M., Shibuya R., Yokoyama Y. (2010). Biochemical Changes in Oyster Tissues and Hemolymph During Long-Term Air Exposure. Fish. Sci. 76, 841–855. doi: 10.1007/s12562-010-0263-1
Kim D., Langmead B., Salzberg S. L. (2015). HISAT: A Fast Spliced Aligner With Low Memory Requirements. Nat. Methods 12, 357–360. doi: 10.1038/nmeth.3317
Kruse J. P., Gu W. (2009). Modes of P53 Regulation. Cell 137, 609–622. doi: 10.1016/j.cell.2009.04.050
Kuchel R. P., Raftos D. A., Nair S. (2010). Immunosuppressive Effects of Environmental Stressors on Immunological Function in Pinctada Imbricata. Fish Shellfish Immunol. 29, 930–936. doi: 10.1016/j.fsi.2010.07.033
LaCasse E. C., Baird S., Korneluk R. G., MacKenzie A. E. (1998). The Inhibitors of Apoptosis (IAPs) and Their Emerging Role in Cancer. Oncogene 17, 3247–3259. doi: 10.1038/sj.onc.1202569
Lange S. S., Mitchell D. L., Vasquez K. M. (2008). High Mobility Group Protein B1 Enhances DNA Repair and Chromatin Modification After DNA Damage. Proc. Natl. Acad. Sci. U.S.A. 105, 10320–10325. doi: 10.1073/pnas.0803181105
La Peyre J. F., Casas S. M., Supan J. E. (2018). Effects of Controlled Air Exposure on the Survival, Growth, Condition, Pathogen Loads and Refrigerated Shelf Life of Eastern Oysters. Aquac. Res. 49, 19–29. doi: 10.1111/are.13427
Lenz O., Xiong J., Nelson M. D., Raizen D. M., Williams J. A. (2015). FMRFamide Signaling Promotes Stress-Induced Sleep in Drosophila. Brain Behav. Immun. 47, 141–148. doi: 10.1016/j.bbi.2014.12.028
Li T., Kon N., Jiang L., Tan M., Ludwig T., Zhao Y., et al. (2012). Tumor Suppression in the Absence of P53-Mediated Cell-Cycle Arrest, Apoptosis, and Senescence. Cell 149, 1269–1283. doi: 10.1016/j.cell.2012.04.026
Li L., Qiu L., Song L., Song X., Zhao J., Wang L., et al. (2009). First Molluscan TNFR Homologue in Zhikong Scallop: Molecular Characterization and Expression Analysis. Fish Shellfish Immunol. 27, 625–632. doi: 10.1016/j.fsi.2009.07.009
Liu M., Guo X. (2017). A Novel and Stress Adaptive Alternative Oxidase Derived From Alternative Splicing of Duplicated Exon in Oyster Crassostrea Virginica. Sci. Rep. 7, 10785. doi: 10.1038/s41598-017-10976-w
Livak K. J., Schmittgen T. D. (2001). Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
López-Landavery E. A., Amador-Cano G., Alejandri N., Ramirez-Álvarez N., Montelongo I., Díaz F., et al. (2019). Transcriptomic Response and Hydrocarbon Accumulation in the Eastern Oyster (Crassostrea Virginica) Exposed to Crude Oil. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 225, 108571. doi: 10.1016/j.cbpc.2019.108571
López-Vera E., Aguilar M. B., Heimer de la Cotera E. P. (2008). FMRFamide and Related Peptides in the Phylum Mollusca. Peptides 29, 310–317. doi: 10.1016/j.peptides.2007.09.025
Ma C., Agrawal G., Subramani S. (2011). Peroxisome Assembly: Matrix and Membrane Protein Biogenesis. J. Cell Biol. 193, 7–16. doi: 10.1083/jcb.201010022
Malek J. C. (2010). The Effects of Intertidal Exposure on Disease, Mortality, and Growth of the Eastern Oyster, Crassostrea Virginica (Maryland, College Park, MD: College Park, MD: ProQuest Dissertations Publishing, University of Maryland).
Mao X., Cai T., Olyarchuk J. G., Wei L. (2005). Automated Genome Annotation and Pathway Identification Using the KEGG Orthology (KO) as a Controlled Vocabulary. Bioinformatics 21, 3787–3793. doi: 10.1093/bioinformatics/bti430
Martín-Gómez L., Villalba A., Abollo E. (2012). Identification and Expression of Immune Genes in the Flat Oyster Ostrea Edulis in Response to Bonamiosis. Gene 492, 81–93. doi: 10.1016/j.gene.2011.11.001
Mayer J. M., Testa B., Roy-de Vos M., Audergon C., Etter J. C. (1995). “"Interactions Between the In Vitro Metabolism of Xenobiotics and Fatty Acids,",” in Toxicology in Transition. Eds. Degen G. H., Seiler J. P., Bentley P. (Berlin, Heidelberg: Springer), 499–513.
McCarthy D. J., Chen Y., Smyth G. K. (2012). Differential Expression Analysis of Multifactor RNA-Seq Experiments With Respect to Biological Variation. Nucleic Acids Res. 40, 4288–4297. doi: 10.1093/nar/gks042
McDowell I. C., Nikapitiya C., Aguiar D., Lane C. E., Istrail S., Gomez-Chiarri M. (2014). Transcriptome of American Oysters, Crassostrea Virginica, in Response to Bacterial Challenge: Insights Into Potential Mechanisms of Disease Resistance. PLoS One 9, e105097. doi: 10.1371/journal.pone.0105097
Meng J., Zhu Q., Zhang L., Li C., Li L., She Z., et al. (2013). Genome and Transcriptome Analyses Provide Insight Into the Euryhaline Adaptation Mechanism of Crassostrea Gigas. PLoS One 8, e58563. doi: 10.1371/journal.pone.0058563
Minami M., Shimizu K., Okamoto Y., Folco E., Ilasaca M. L., Feinberg M. W., et al. (2008). Prostaglandin E Receptor Type 4-Associated Protein Interacts Directly With NF-κb1 and Attenuates Macrophage Activation. J. Biol. Chem. 283, 9692–9703. doi: 10.1074/jbc.M709663200
Modak T. H., Gomez-Chiarri M. (2020). Contrasting Immunomodulatory Effects of Probiotic and Pathogenic Bacteria on Eastern Oyster, Crassostrea Virginica, Larvae. Vaccines (Basel) 8, 588. doi: 10.3390/vaccines8040588
Morgan D. O. (1997). Cyclin-Dependent Kinases: Engines, Clocks, and Microprocessors. Annu. Rev. Cell Dev. Biol. 13, 261–291. doi: 10.1146/annurev.cellbio.13.1.261
Nilsson I., Hoffmann I. (2000). Cell Cycle Regulation by the Cdc25 Phosphatase Family. Prog. Cell Cycle Res. 4, 107–114. doi: 10.1007/978-1-4615-4253-7_10
Ottaviani E., Franceschi C. (1996). The Neuroimmunology of Stress From Invertebrates to Man. Prog. Neurobiol. 48, 421–440. doi: 10.1016/0301-0082(95)00049-6
Papa S., Zazzeroni F., Bubici C., Jayawardena S., Alvarez K., Matsuda S., et al. (2004). Gadd45β Mediates the NF-κb Suppression of JNK Signalling by Targeting MKK7/JNKK2. Nat. Cell Biol. 6, 146–153. doi: 10.1038/ncb1093
Pertea M., Kim D., Pertea G. M., Leek J. T., Salzberg S. L. (2016). Transcript-Level Expression Analysis of RNA-Seq Experiments With HISAT, StringTie and Ballgown. Nat. Protoc. 11, 1650–1667. doi: 10.1038/nprot.2016.095
Pertea M., Pertea G. M., Antonescu C. M., Chang T. C., Mendell J. T., Salzberg S. L. (2015). StringTie Enables Improved Reconstruction of a Transcriptome From RNA-Seq Reads. Nat. Biotechnol. 33, 290–295. doi: 10.1038/nbt.3122
Posselt G., Schwarz H., Duschl A., Horejs-Hoeck J. (2011). Suppressor of Cytokine Signaling 2 is a Feedback Inhibitor of TLR-Induced Activation in Human Monocyte-Derived Dendritic Cells. J. Immunol. 187, 2875–2884. doi: 10.4049/jimmunol.1003348
Pucci B., Kasten M., Giordano A. (2000). Cell Cycle and Apoptosis. Neoplasia 2, 291–299. doi: 10.1038/Sj.Neo.7900101
Rath P. C., Aggarwal B. B. (1999). TNF-Induced Signaling in Apoptosis. J. Clin. Immunol. 19, 350–364. doi: 10.1023/A:1020546615229
Reeves R., Adair J. E. (2005). Role of High Mobility Group (HMG) Chromatin Proteins in DNA Repair. DNA Repair (Amst.) 4, 926–938. doi: 10.1016/j.dnarep.2005.04.010
Reppert S. M., Weaver D. R., Ebisawa T. (1994). Cloning and Characterization of a Mammalian Melatonin Receptor That Mediates Reproductive and Circadian Responses. Neuron 13, 1177–1185. doi: 10.1016/0896-6273(94)90055-8
Rico-Bautista E., Flores-Morales A., Fernández-Pérez L. (2006). Suppressor of Cytokine Signaling (SOCS) 2, A Protein With Multiple Functions. Cytokine Growth Factor Rev. 17, 431–439. doi: 10.1016/j.cytogfr.2006.09.008
Robinson M. D., McCarthy D. J., Smyth G. K. (2010). Edger: A Bioconductor Package for Differential Expression Analysis of Digital Gene Expression Data. Bioinformatics 26, 139–140. doi: 10.1093/bioinformatics/btp616
Rossi M. N., Antonangeli F. (2015). Cellular Response Upon Stress: P57 Contribution to the Final Outcome. Mediat. Inflamm. 2015, 259325. doi: 10.1155/2015/259325
Sabio G., Davis R. J. (2014). TNF and MAP Kinase Signalling Pathways. Semin. Immunol. 26, 237–245. doi: 10.1016/j.smim.2014.02.009
Saucedo P. E., Ocampo L., Monteforte M., Bervera H. (2004). Effect of Temperature on Oxygen Consumption and Ammonia Excretion in the Calafia Mother-of-Pearl Oyster, Pinctada Mazatlanica (Hanley 1856). Aquaculture 229, 377–387. doi: 10.1016/S0044-8486(03)00327-2
Shumway S. E. (1996). “Natural Environmental Factors,” in The Eastern Oyster Crassostrea Virginica. Eds. Kennedy V. S., Newell R. I. E., Eble A. F. (Maryland, College Park, MD: Maryland Sea Grant College), 467–513.
Sokolova I. M., Frederich M., Bagwe R., Lannig G., Sukhotin A. A. (2012). Energy Homeostasis as an Integrative Tool for Assessing Limits of Environmental Stress Tolerance in Aquatic Invertebrates. Mar. Environ. Res. 79, 1–15. doi: 10.1016/j.marenvres.2012.04.003
Song H. Y., Rothe M., Goeddel D. V. (1996). The Tumor Necrosis Factor-Inducible Zinc Finger Protein A20 Interacts With TRAF1/TRAF2 and Inhibits NF-kappaB Activation. Proc. Natl. Acad. Sci. U.S.A. 93, 6721–6725. doi: 10.1073/pnas.93.13.6721
Storey K. B., Storey J. M. (1990). Metabolic Rate Depression and Biochemical Adaptation in Anaerobiosis, Hibernation and Estivation. Q. Rev. Biol. 65, 145–174. doi: 10.1086/416717
Sun G., Xue R., Yao F., Liu D., Huang H., Chen C., et al. (2014). The Critical Role of Sestrin 1 in Regulating the Proliferation of Cardiac Fibroblasts. Arch. Biochem. Biophys. 542, 1–6. doi: 10.1016/j.abb.2013.11.011
Szabados L., Savoure A. (2010). Proline: A Multifunctional Amino Acid. Trends Plant Sci. 15, 89–97. doi: 10.1016/j.tplants.2009.11.009
Verbruggen N., Hermans C. (2008). Proline Accumulation in Plants: A Review. Amino Acids 35, 753–759. doi: 10.1007/s00726-008-0061-6
Xiao B. C., Li E. C., Du Z. Y., Jiang R. L., Chen L. Q., Yu N. (2014). Effects of Temperature and Salinity on Metabolic Rate of the Asiatic Clam Corbicula Fluminea (Müller 1774). SpringerPlus 3, 455. doi: 10.1186/2193-1801-3-455
Yanagida M., Ikai N., Shimanuki M., Sajiki K. (2011). Nutrient Limitations Alter Cell Division Control and Chromosome Segregation Through Growth-Related Kinases and Phosphatases. Philos. Trans. R. Soc Lond. B Biol. Sci. 366, 3508–3520. doi: 10.1098/rstb.2011.0124
Young M. D., Wakefield M. J., Smyth G. K., Oshlack A. (2010). Gene Ontology Analysis for RNA-Seq: Accounting for Selection Bias. Genome Biol. 11, R14. doi: 10.1186/gb-2010-11-2-r14
Zhang G., Fang X., Guo X., Li L., Luo R., Xu F., et al. (2012). The Oyster Genome Reveals Stress Adaptation and Complexity of Shell Formation. Nature 490, 49–54. doi: 10.1038/nature11413
Zhang L., Li L., Guo X., Litman G. W., Dishaw L. J., Zhang G. (2015). Massive Expansion and Functional Divergence of Innate Immune Genes in a Protostome. Sci. Rep. 5, 8693. doi: 10.1038/srep08693
Zhang G., Li L., Meng J., Qi H., Qu T., Xu F., et al. (2016). Molecular Basis for Adaptation of Oysters to Stressful Marine Intertidal Environments. Annu. Rev. Anim. Biosci. 4, 357–381. doi: 10.1146/annurev-animal-022114-110903
Zhang L., Li L., Zhu Y., Zhang G., Guo X. (2014a). Transcriptome Analysis Reveals a Rich Gene Set Related to Innate Immunity in the Eastern Oyster (Crassostrea Virginica). Mar. Biotechnol. 16, 17–33. doi: 10.1007/s10126-013-9526-z
Keywords: stress, transcriptome, cell cycle control, apoptosis, growth, immune function, epigenetic modification
Citation: Li C, Wang H and Guo X (2022) Regulation of the Cell Cycle, Apoptosis, and Proline Accumulation Plays an Important Role in the Stress Response of the Eastern Oyster Crassostrea Virginica. Front. Mar. Sci. 9:921877. doi: 10.3389/fmars.2022.921877
Received: 16 April 2022; Accepted: 24 May 2022;
Published: 17 June 2022.
Edited by:
Juan D. Gaitan-Espitia, The University of Hong Kong, Hong Kong, SAR ChinaReviewed by:
Joanna Griffiths, University of California, Davis, United StatesCopyright © 2022 Li, Wang and Guo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ximing Guo, eGd1b0Boc3JsLnJ1dGdlcnMuZWR1
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.