- 1Key Laboratory of Freshwater Aquatic Genetic Resources, Ministry of Agriculture, Shanghai Ocean University, Shanghai, China
- 2Shanghai Engineering Research Center of Aquaculture, Shanghai Ocean University, Shanghai, China
- 3Shanghai Collaborative Innovation Center for Cultivating Elite Breeds and Green-culture of Aquaculture Animals, Shanghai, China
Cyclin dependent kinase 6 (CDK6) is a serine/threonine kinase that plays important roles in cell cycle progression and differentiation. In this study, full-length cDNA of Hc-CDK6 was obtained from freshwater pearl mussels (Hyriopsis cumingii, Hc) with 3´,5´ rapid-amplification of cDNA ends (RACE). The Hc-CDK6 expression profiles were analyzed with quantitative real-time PCR and in situ hybridization. The function of the Hc-CDK6 gene was studied with both RNA interference (RNAi) and overexpression in H. cumingii. Hc-CDK6 was found to encode 331 amino acids and to have a CDK4/6-like serine/threonine kinase catalytic structural domain. In terms of the amino acid sequence, the protein Hc-CDK6 was most closely related to its homolog in Crassostrea gigas, with a similarity of 75.23%. Hc-CDK6 was expressed in all examined tissues (adductor, foot, visceral mass, gill, outer mantle, inner mantle and gonads), and the highest expression was observed in the gonads (P<0.05). The relative expression of Hc-CDK6 increased during embryonic development, and was higher in the blastocyst and gastrulation stages, which were characterized by rapid division and differentiation. Hc-CDK6 showed hybridization signals in all parts of the mantle. After knockdown of Hc-CDK6 through RNAi, a significant decrease in CDK6 expression was found, and the percentage of cells in G0/G1 significantly increased. Overexpression of Hc-CDK6 in mantle cells increased the proliferation of cultured cells (P<0.05). Hc-CDK6 appeared to promote the cell cycle in H. cumingii, and overexpression of Hc-CDK6 promoted mantle cell proliferation. The functional study of this gene may provide new ideas for solving the problem of slow proliferation of shellfish cells in in vitro culture.
1 Introduction
Cell division is the most fundamental developmental process in unicellular or multicellular organisms, and the cell cycle describes the entire process of cell division from the end of the last mitosis to the completion of the next mitosis (Jacobs, 1992). This process is tightly regulated by cyclins and cyclin dependent kinases (CDKs) (Satyanarayana and Kaldis, 2009; Lim and Kaldis, 2013). CDKs play important roles in cell cycle checkpoints in eukaryotic cells, and cells entering S-phase must meet the basic requirements for CDK activation (Massague, 2004).
Each CDK has a structural domain consisting of a cyclin-binding domain and a T-loop motif that together participate in CDK activation. During the cell cycle, CDK protein amounts are essentially unchanged, and these kinases interact with different cyclins and form active complexes (Pines and Hunter, 1991; Malumbres et al., 2009; Suryadinata et al., 2010). CDKs are catalytic subunits that are activated by the corresponding cyclins. The CDK6-Cyclin D complex regulates cell proliferation through the G1-S checkpoint (Pietenpol and Stewart, 2002). This complex has kinase-dependent and independent functions and is a hub for the integration of signals affecting transcription, proliferation and differentiation (Tigan et al., 2016). Current research on CDK6 focuses on tumor and cancer-related aspects. Overexpression of CDK6 is often used as a biomarker for tumor diagnosis, and its overactivation leads to uncontrolled cell proliferation (Baba et al., 2014). Given the role of CDK6 in cell cycle regulation, many studies have achieved antitumor effects by inhibiting the activity of CDK6 (Schaer et al., 2018). In aquatic organisms, CDK6 has been less studied, and only cloning and tissue expression analysis of CDK6 in Scophthalmus maximus have been performed; however, further study of its function is lacking (Guo et al., 2016). The function of CDK6 in aquatic organisms remains poorly understood.
H. cumingii is an important freshwater pearl mussel: 95% of the freshwater pearls worldwide are produced by H. cumingii, which has high economic and research value (Wang et al., 2021a). In production, defective pearls such as tail pearls and specially shaped pearls may be formed through the influence of nuclear insertion technology, pearl breeding and other factors (Pan and Wen, 2002). The mantle is an important site not only for pearl formation but also for making mantle saibos. Some researchers have studied the cultivation of pearls with free mantle cells instead of mantle saibos, and have found that the number of defective pearls produced by mantle free cells significantly decreases, but the activity and proliferation of cells affect the formation of pearls (Qian et al., 2002; Jin et al., 2011; Li et al., 2014). In cell cycle regulation, late G1 and G2/M limited points phases determine whether a cell is in a proliferative or quiescent state (Fu et al., 1999; Pietenpol and Stewart, 2002). It is reported that with the increase in mantle cells with culture time, the number of G0/G1 phase cells increases significantly, thus indicating that, over the course of culture, some cells might not be able to pass the late G1 limited point, and the cell proliferation activity decreases (Cao et al., 2020). The important regulatory role of CDK6 in the G1 phase of the cell cycle is crucial for enhancing the proliferation of mantle cells in vitro.
In this study, we identified Hc-CDK6 in H. cumingii, and analyzed their expression in different tissues and developmental stages of embryo. The effect of Hc-CDK6 silencing and overexpression on the expression of Hc-CCND2 and distribution of the cell cycle were investigated. These results will be helpful to understand the regulation of the shellfish cell cycle. and provide a novel approach for improving the slow proliferation rate of shellfish cells in in vitro cultures.
2 Materials and Methods
2.1 Animals and Tissue Samples
Healthy 1-year-old mussels (H. cumingii, 15 individuals, 6–7 cm shell length) were collected from a farm in Wuyi (Zhejiang, China) in March, 2021.
Cut off the adductor from selected mussels, the tissues were sampled respectively. The samples used in the experiment included the adductor, foot, gill, outer mantle, inner mantle and gonads. Embryo samples were sampled from adult mussels’ gill tissue at 1 day, 3 day, 5 day, 7 day, 9 day, 10 day, including the cleavage stage, blastula stage, gastrula stage, and glochidium. All samples were immediately frozen in liquid nitrogen and stored at - 80°C (Wang et al., 2021b).
For suspension cell preparation, the adductors were cut off from selected mussels with a scalpel, and the mantle and gill tissues were repeatedly washed with sterile distilled water. The outer mantle was then separated and cut into small pieces (1 mm2). After digestion in 0.25% trypsin at 37°C for 20 min, excess tissue mass was filtered through a 300 mesh strainer. The cell suspension was counted with a hemocytometer, the cell number was adjusted to 5×105 to 1×106 mL-1 used 1640 medium, and cultured in a constant temperature incubator at 25°C. The medium formula is 500 μL sodium pyruvate, 500 μL nonessential amino acid, 5 mL fetal bovine serum and 44 mL of 1640 basal medium.
2.2 Amplification of Full-Length cDNA
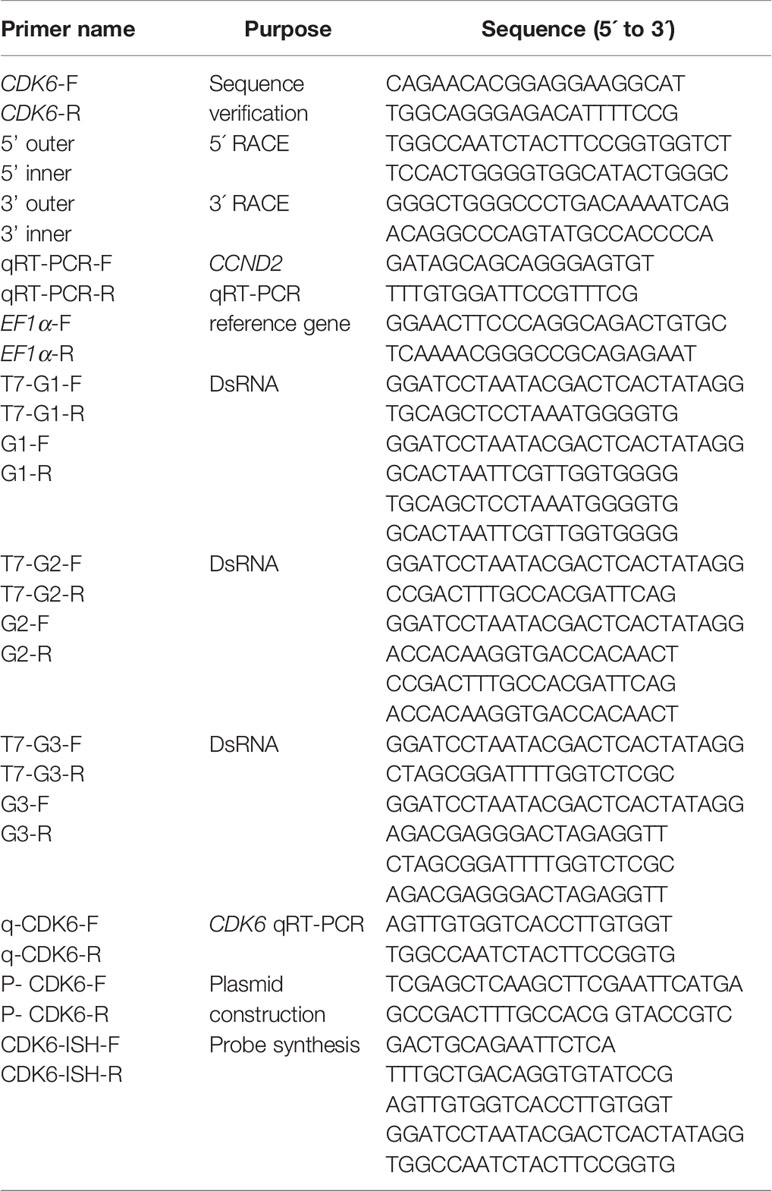
Total RNA was extracted with RNAiso Plus (TaKaRa, Japan). The RNA concentration and quality were detected with a spectrophotometer (Thermo Fisher Scientific, USA) and 1% agarose gel, respectively. Partial cDNA sequences of Hc-CDK6 were obtained from the transcriptome of H. cumingii (unpublished). Primers were designed with NCBI Primer BLAST (https://www.ncbi.nlm.nih.gov/tools/primer-blast/) for sequence verification. After reverse transcription of RNA (TransGen Biotech, China), the obtained cDNA was used for sequence verification.
The 3’ and 5’ ends of Hc-CDK6 cDNA were amplified through the RACE method with a SMARTer® RACE 5´/3´ Kit (Takara Bio, USA). The primers were designed on the basis of partial Hc-CDK6 sequences verified with an NCBI primer Blast search (Table 1). The PCR product was purified with a Gel Extraction Kit (TaKaRa, Japan) according to the manufacturer’s instructions, then incubated with pMD19-T (TaKaRa, Japan) at 16°C for 4 h. Subsequently, ten µL of the above incubation products was transformed into DH5α cells. PCR positive clones were selected and subjected to sequencing with 3730xlDNAAnalyzer by GENEWIZ, Inc (Suzhou). The 3’ and 5’ sequences were spliced to obtain the full length cDNA.
2.3 Sequence and Phylogenetic Analysis
Prediction of Hc-CDK6 amino acid sequences was performed with the NCBI ORF finder online tool (https://www.ncbi.nlm.nih.gov/orffinder/). Prediction of Hc-CDK6 amino acid signal peptides was performed with SignalP4.1 (http://www.cbs.dtu.dk/services/SignalP/). Hc-CDK6 phosphorylation sites were analyzed using NetPhos 3.1 (http://www.cbs.dtu.dk/services/NetPhos/), and isoelectric points and molecular weights were predicted with the Compute pI/Mw tool (https://web.expasy.org/compute_pi/). Multiple comparisons of amino acid sequences and phylogenetic tree construction (neighbor-joining method, 1000 bootstrap replicates) were performed in Bioedit and MEGA7.0, respectively.
2.4 Expression Analysis of Hc-CDK6 by qRT-PCR
The RNA extraction method for frozen samples was as described in section 2.2. The manufacturer’s instructions for PrimeScript™ RT Master Mix (TaKaRa, Japan) were followed for reverse transcription of RNA samples. The information on quantitative primers and reference primers used to examine tissues and embryo expression levels is shown in Table 1. Hc-CDK6 relative expression were detected with the CFX96 system (Bio-Rad, USA), each sample was performed on three technical replicates. The qRT-PCR reaction conditions followed the TB Green® Fast qPCR Mix instructions (TaKaRa, Japan). Relative expression of Hc-CDK6 was calculated with the 2-ΔΔCT method.
2.5 In Situ Hybridization
The cDNA fragments were synthesized with probe synthetic primers (Table 1), then purified and subjected to in vitro transcription (Transgen, China). The probes were labeled with DIG RNA Labeling Mix. Healthy H. cumingii (1 year old) were selected, and the mantle tissue (5×8 mm) was cut and fixed in 4% paraformaldehyde for 18 h. The tissue was then dehydrated in alcohol, rendered transparent in xylene and finally embedded in paraffin. A paraffin slicer was used to slice sections with a thickness of 3 μm. The in situ hybridization procedure was performed according to the in situ hybridization kit instructions (Boster, USA). Samples were observed and photographed under a microscope.
2.6 Synthesis of dsRNA In Vitro
According to the sequence of the ORF region of Hc-CDK6, three pairs of primers approximately 500 bp in length were designed with NCBI Primer BLAST. We added a T7 sequence to the 5’ end of each primer pair. The primers were amplified and purified to obtain the target sequence. The in vitro transcription of target sequences was performed according to the instruction manual (TransGen Biotech, China). The dsRNA was obtained as previously described (Wang et al., 2021). After purification, dsRNAs were diluted to working concentration (300 ng/µL) with RNase/DNase-free water.
2.7 Knockdown of the CCND2 Gene In Vivo by RNAi
Sixty healthy 1-year-old H. cumingii (average weight 55 ± 2 g) were randomly assigned to four groups (15 individuals/group). Three groups were randomly selected as experimental groups to be injected with 100 μL of the corresponding dsRNA. The negative control group was injected with 100 μL of RNase/DNase-free per mussel. Gill tissue was collected from each group at three time points post-injection (n=5, at 24 h, 48 h and 72 h). The tissue collection and preservation methods were as described in section 2.1, and the RNA extraction and reverse transcription methods were as described in sections 2.2 and 2.4, respectively.
After RNAi, we used flow cytometry to analyze cell cycle phases. Briefly, gill cells were collected from mussels after RNAi. The gill tissue suspension cell collection method was as described in section 2.1. Cells were collected after removal of culture medium, and washed and fixed according to the manufacturer’s instructions for PI/RNase Staining Buffer (BD, USA). The cells were then resuspended in 500 µL PI/RNase solution (BD, USA) for 15–20 min in the dark in a 37°C incubator. Samples were analyzed with an Accuri C6 Plus Cytometer (BD Biosciences, USA). The numbers of cells in the different cell cycle phases were calculated.
2.8 Overexpression of Hc-CDK6 Gene in Mantle Cells
The ORF region of the Hc-CDK6 gene was amplified by PCR, and the purified PCR product was used in plasmid construction (Table 1). The restriction enzyme site was EcoR I (GAATTC). The pcDNA3.1 vector (Clontech, USA) was digested with EcoR I. The purified PCR product was ligated into the pcDNA3.1 vector with a Seamless Cloning Kit (Beyotime Biotechnology, China), and the plasmids were transformed into DH5α cells. Finally, single clones were selected for sequence identification (GENEWIZ, China).
In the transient transfection experiments, pcDNA3.1-CDK6 plasmids (2.5 μg) were transfected with Lipo8000 (Beyotime, China) according to the user manual. Cell growth state was observed under an inverted microscope (Olympus, Japan) during cell culture, and pictures were taken. Cell proliferation was assessed in normal mantle cells and Hc-CDK6 overexpressing mantle cells with CCK-8 dye (Beyotime, China). Analysis of the cell cycle after transfection was performed by flow cytometry.
2.9 Statistical Analysis
All data were analyzed with one-way analysis of variance. Data are expressed as mean ± SE. Multiple comparisons between groups were analyzed with Duncan’s multiple range tests. A value of P<0.05 was considered significant. OriginPro 9.1 (OriginLab, Massachusetts, USA) was used to plot bar charts.
3 Results
3.1 Obtained and Characterization of Hc-CDK6 cDNA Clones
The complete cDNA sequence of Hc-CDK6 was obtained with RACE, was deposited in GenBank accession number OM572509. The full length cDNA was 1,818 bp, the ORF was 996 bp, and the 5’UTR and 3’UTR were 228 bp and 594 bp, respectively (Figure 1). A typical polyadenylation signal sequence was found in the 3’UTR. The ORF of the Hc-CDK6 cDNA encoded 331 aa.
Amino acid analysis indicated that Aa 13–300 contained the catalytic structural domain of a CDK4/6-like serine/threonine kinase, and amino acid residues bound to D-type cyclins are present in this structural domain (Figure 2). Predictions indicated the absence of signal peptide sequences and transmembrane structures in this protein. The secondary structure of the protein comprised 30.21% α-helix, 17.22% extended strand and 52.57% random coil. The predicted molecular weight and theoretical isoelectric point were 37773.18 Da and 5.81, respectively.
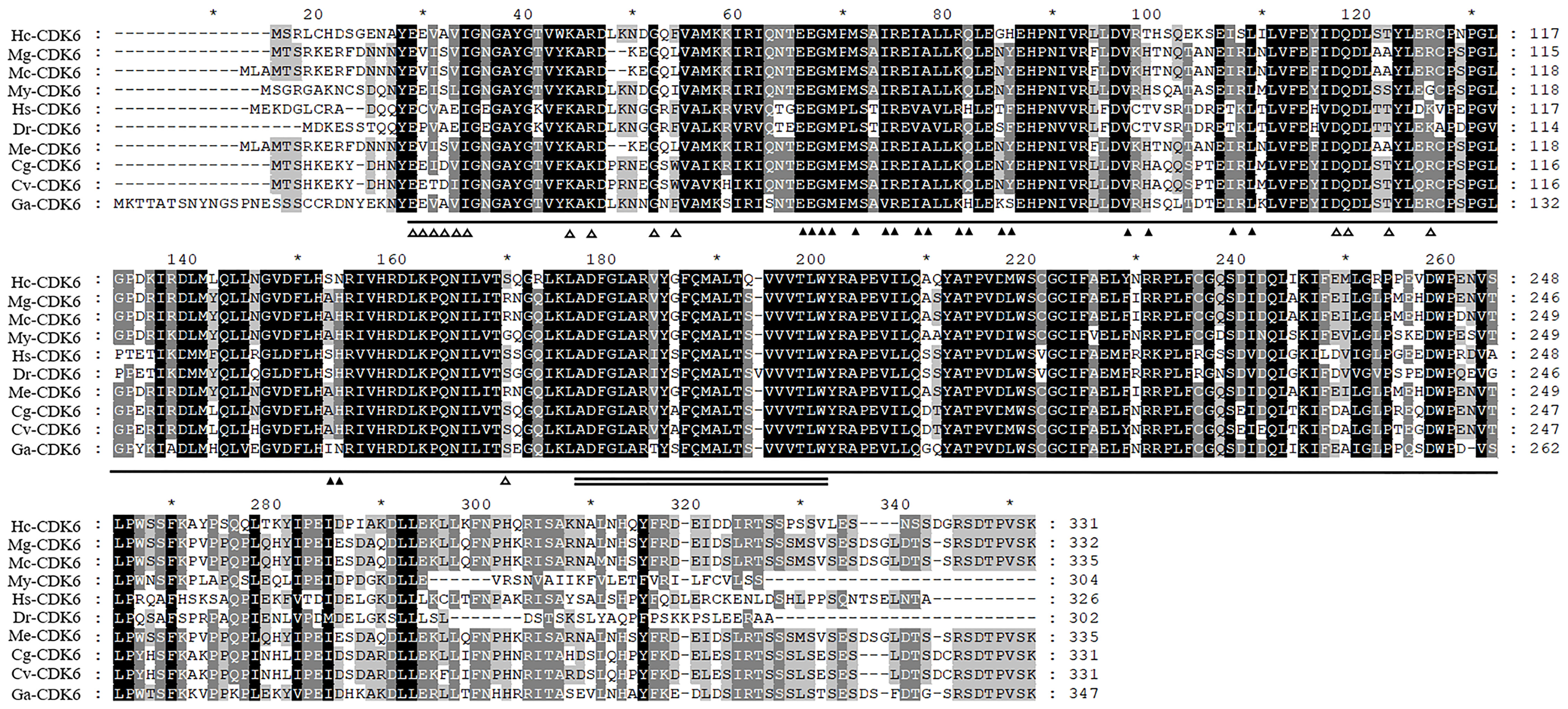
Figure 2 Alignment analysis of Hc-CDK6 with homologous sequences in other species. ▲Indicates the amino acid residue of CDK binding to CCND, △indicates the amino acid residue of CDK binding to INK4, a horizontal line indicates the catalytic domain of cyclin-dependent kinase 4 and 6-like serine/threonine kinases of CDK6, and the double solid line indicates the activation loop (A-loop). GenBank accession numbers: Mytilus galloprovincialis (Mg), VDI04144.1; Mytilus coruscus (Mc), CAC5413630.1; Mizuhopecten yessoensis (My), XP_021356142.1; Homo sapiens (Hs), NP_001138778.1; Danio rerio (Dr), NP_001137525.1; Crassostrea gigas (Cg), XP_011426627.1; Crassostrea virginica (Cv), XP_022322423.1; Gigantopelta aegis (Ga), XP_041371365.1; Mytilus edulis (Me), CAG2221073.1. Black, conserved amino acid residue; gray, analogous residues. * Indicates that starting from the 10th amino acid, every 20 amino acids are marked with a *.
Amino acid comparison indicated 75% sequence identity with Mizuhopecten yessoensis CDK6, 75.23% with Crassostrea gigas CDK6 and 74.61% with Crassostrea virginica CDK6 (Figure 2). The phylogenetic tree was constructed with MEGA7.0 (Figure 3), and the Hc-CDK6 amino acid sequence was compared with other available CDK6 sequences. The phylogenetic tree showed that Hc-CDK6 clustered with other marine bivalve sequences.
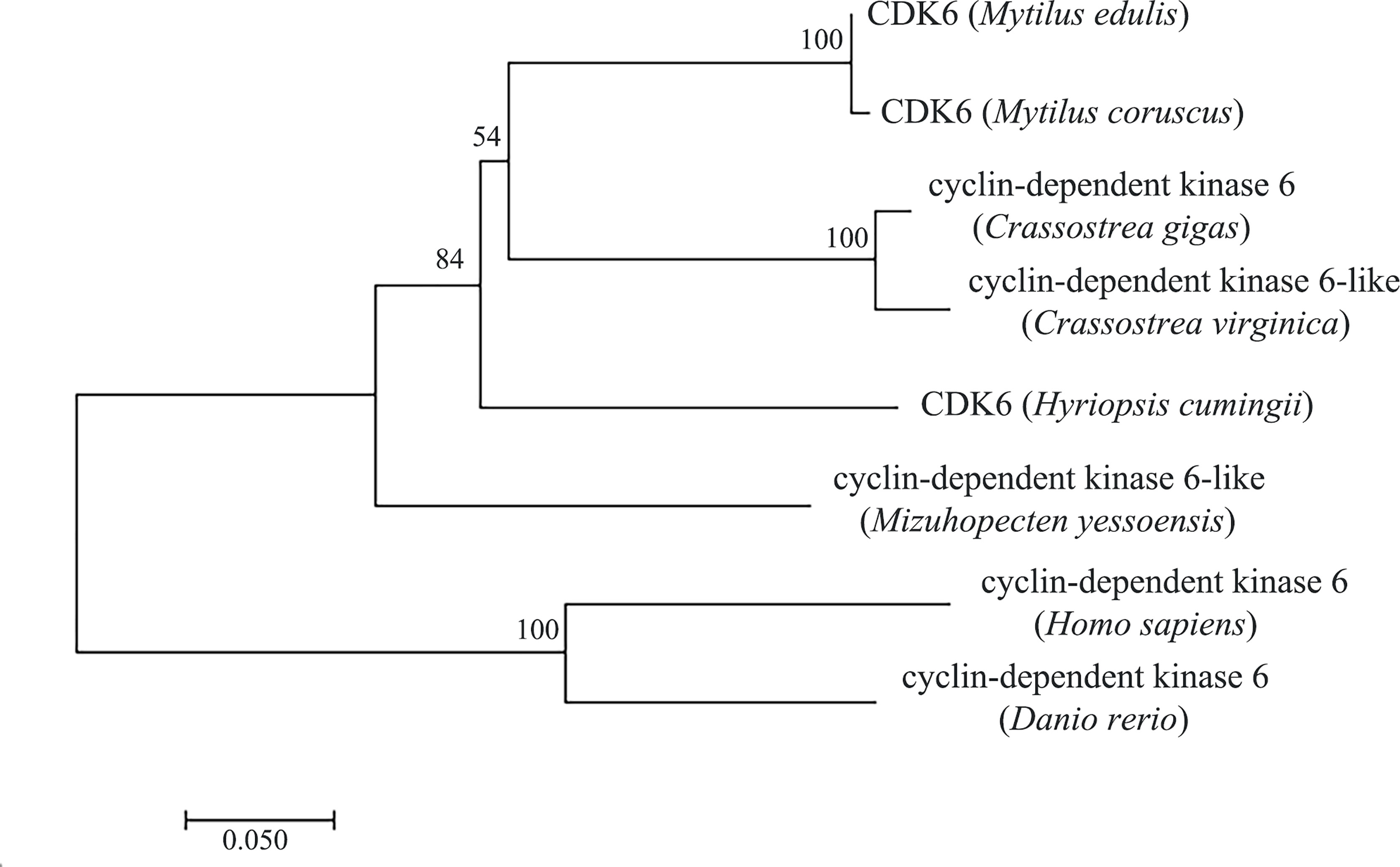
Figure 3 NJ tree of the Hc-CDK6 amino acid sequence with CDK6 sequences from other species. GenBank accession numbers are as in Figure 2, the number on the node indicates the confidence value of the test for 1000 Bootstrap repetitions.
3.2 Expression Profiles of Hc-CDK6 in Different Tissues and Embryonic Periods
The expression level of the Hc-CDK6 gene in H. cumingii tissues was detected with qRT-PCR (Figure 4A). Hc-CDK6 was distributed in the gonads, gills, adductor, foot, outer mantle and inner mantle. The expression of Hc-CDK6 was highest in the gonads, then in the foot and gills, and was lowest in the adductor; the level in the outer mantle was higher than that in the inner mantle (P<0.05).
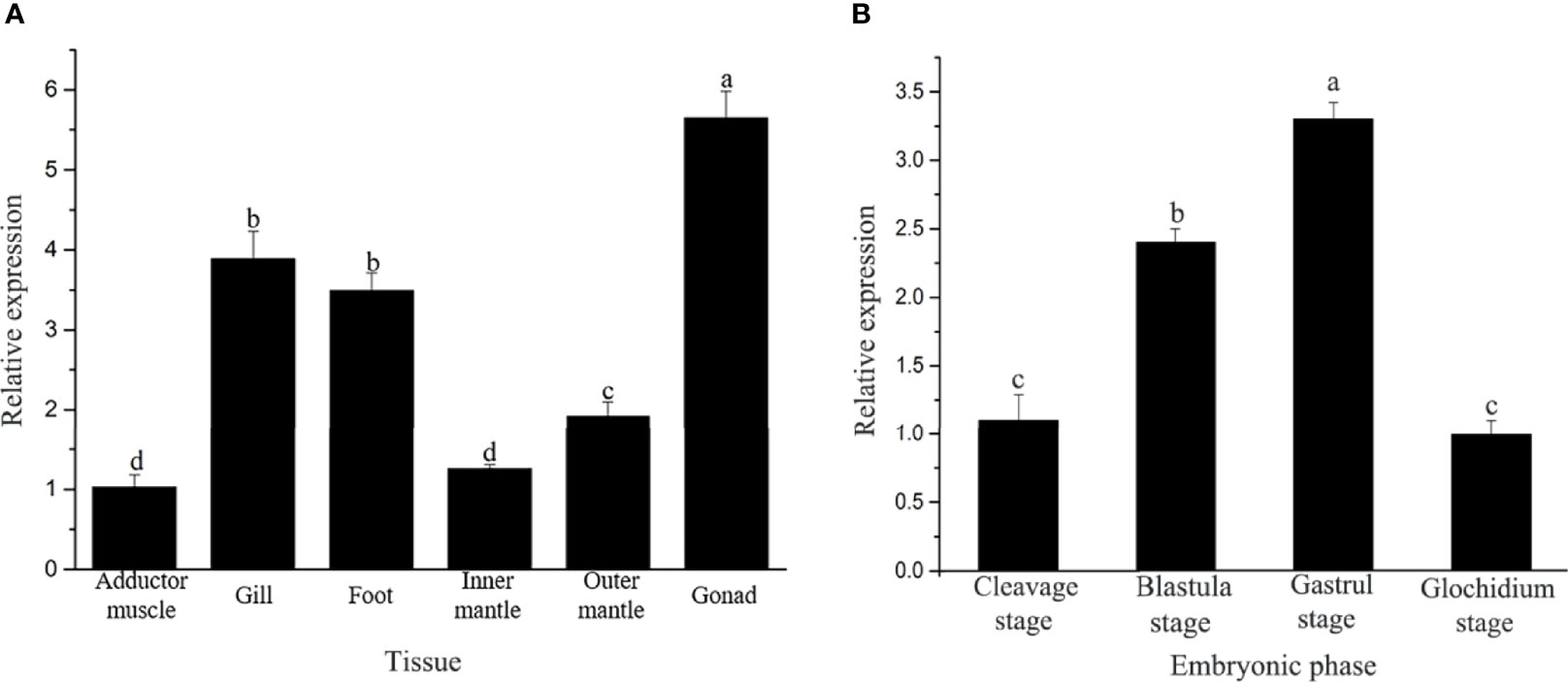
Figure 4 (A) Analysis of relative Hc-CDK6 expression levels in different tissues. (B) Hc-CDK6 expression levels during different embryonic periods. Values with different letters indicate P < 0.05.
The qRT-PCR showed that Hc-CDK6 was expressed in different embryonic stages. The expression of Hc-CDK6 first increased and then decreased with embryonic development, and the relative expression was highest in the gastrula stage, then decreased in the glochidium stage (Figure 4B).
3.3 Localization of Hc-CDK6 in the Mantle
In situ hybridization was performed to explore the localization of Hc-CDK6 in the mantle tissue. Figures 5A, B shows the in situ hybridization of the experimental and control groups, respectively. Compared with the control, CDK6 showed positive hybridization signals in all parts of the mantle, on the outer fold as well as on the middle fold (at the arrow), and CCND2 showed positive hybridization signals on the ventral mantle, the outer fold as well as the middle fold (Figure 5C, at the arrow), whereas no hybridization signals were present in the control.
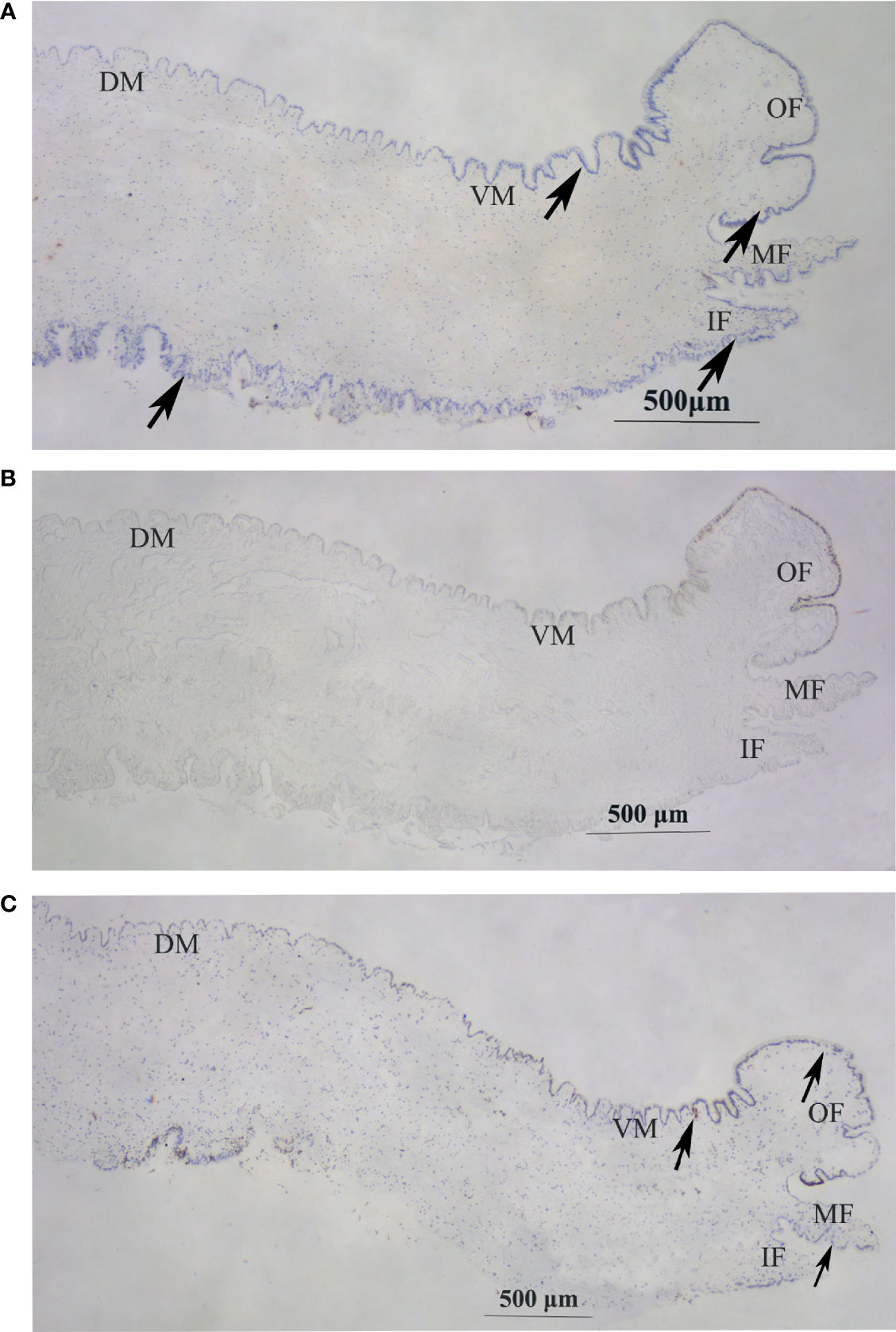
Figure 5 In situ hybridization in the mantle of H. cumingii. (A) Signal of Hc-CDK6 in the experimental groups. (B) Micrograph of the control groups. (C) Signal of Hc-CCND2 in experimental groups. The arrow indicates the position of the signal. OF, outer fold; MF, middle fold; IF, inner fold; DM, dorsal mantle; VM, ventral mantle.
3.4 Knockdown of Hc-CDK6 Induces Cell Cycle Arrest in Gill Cells
The three interfering chains were synthesized for screening, and the qRT-PCR results of the mantle and gill tissues after the injection of the three interfering chains showed that the expression of the three interfering chains was significantly lower in the gill tissues than that in the control group after RNAi (P<0.05). The highest knockdown rates were 76.3%, 88.4% and 51.7% in the gills on the 2nd day (Figure 6A). In the mantle tissues, the expression of Hc-CDK6 significantly differed from that in the control group on the 2nd day (P<0.05), but the knockdown efficiency was lower than that in gill tissues (Figure 6B). Interfering chain 2, which had the highest knockdown rate, was selected for RNAi, and gill tissue was selected for downstream gene expression detection and flow cytometric analysis.
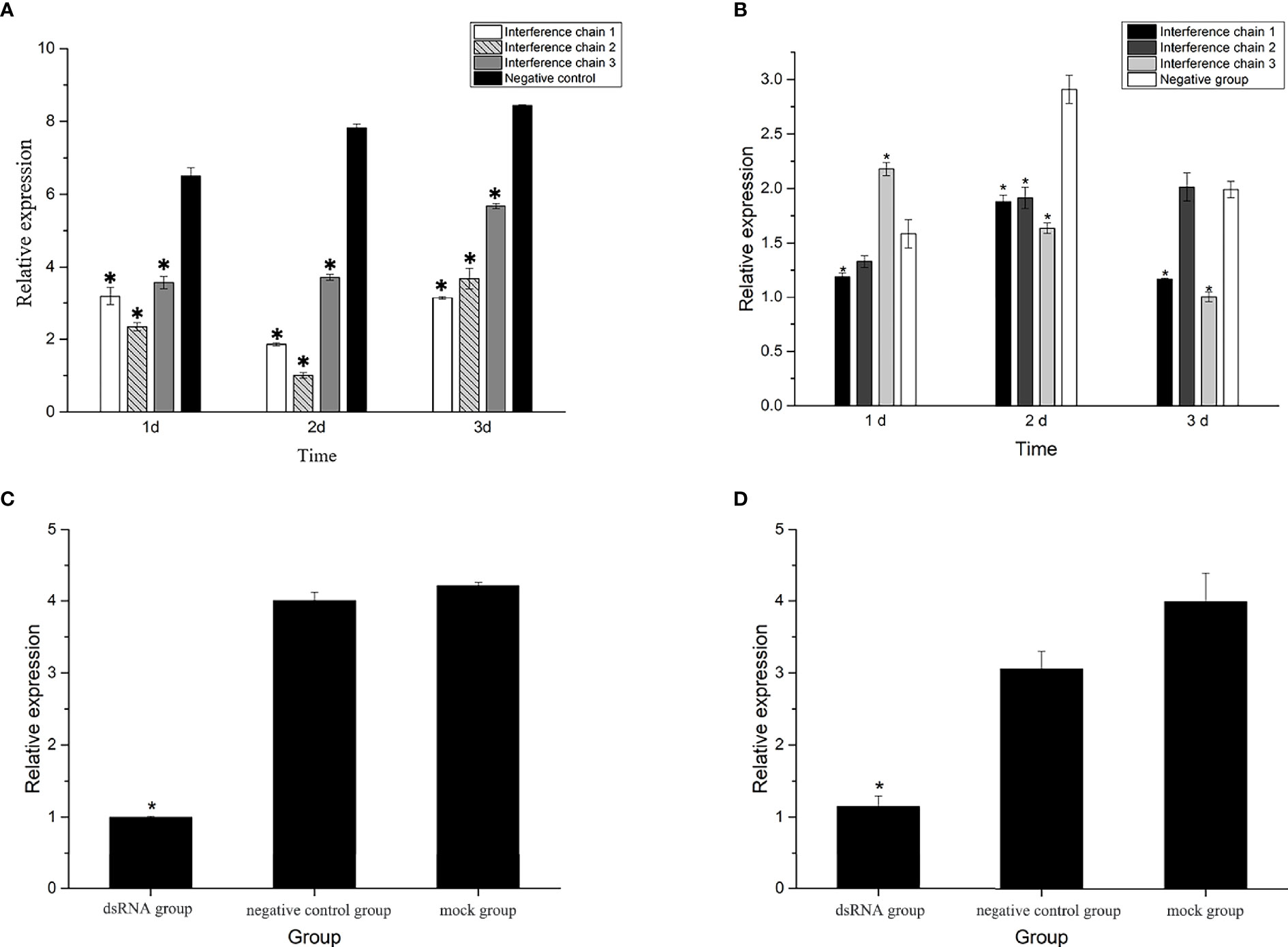
Figure 6 (A) Relative expression of Hc-CDK6 in the gills after RNAi at three time points. (B) Relative expression of Hc-CDK6 in the mantle after RNAi at three time points. (C) Relative expression of Hc-CDK6 in group 2, and the negative control and blank groups. (D) Relative expression of Hc-CCND2 in group 2, and the negative control and blank groups. *Significance level (P < 0.05).
After RNAi, the relative expression of Hc-CCND2 (Figure 6D) and Hc-CDK6 (Figure 6C) was significantly lower than that in the control groups (P<0.05). Pearson analysis showed a significant positive correlation between Hc-CCND2 and Hc-CDK6 relative expression after knockdown (P<0.05).
The percentage of G0/G1 phase cells increased significantly, and the percentage of S phase cells decreased significantly after Hc-CDK6 RNAi (P<0.05, Figure 7). The cell proliferation index was calculated and analyzed, and also showed a significant decrease after RNAi (P<0.05).
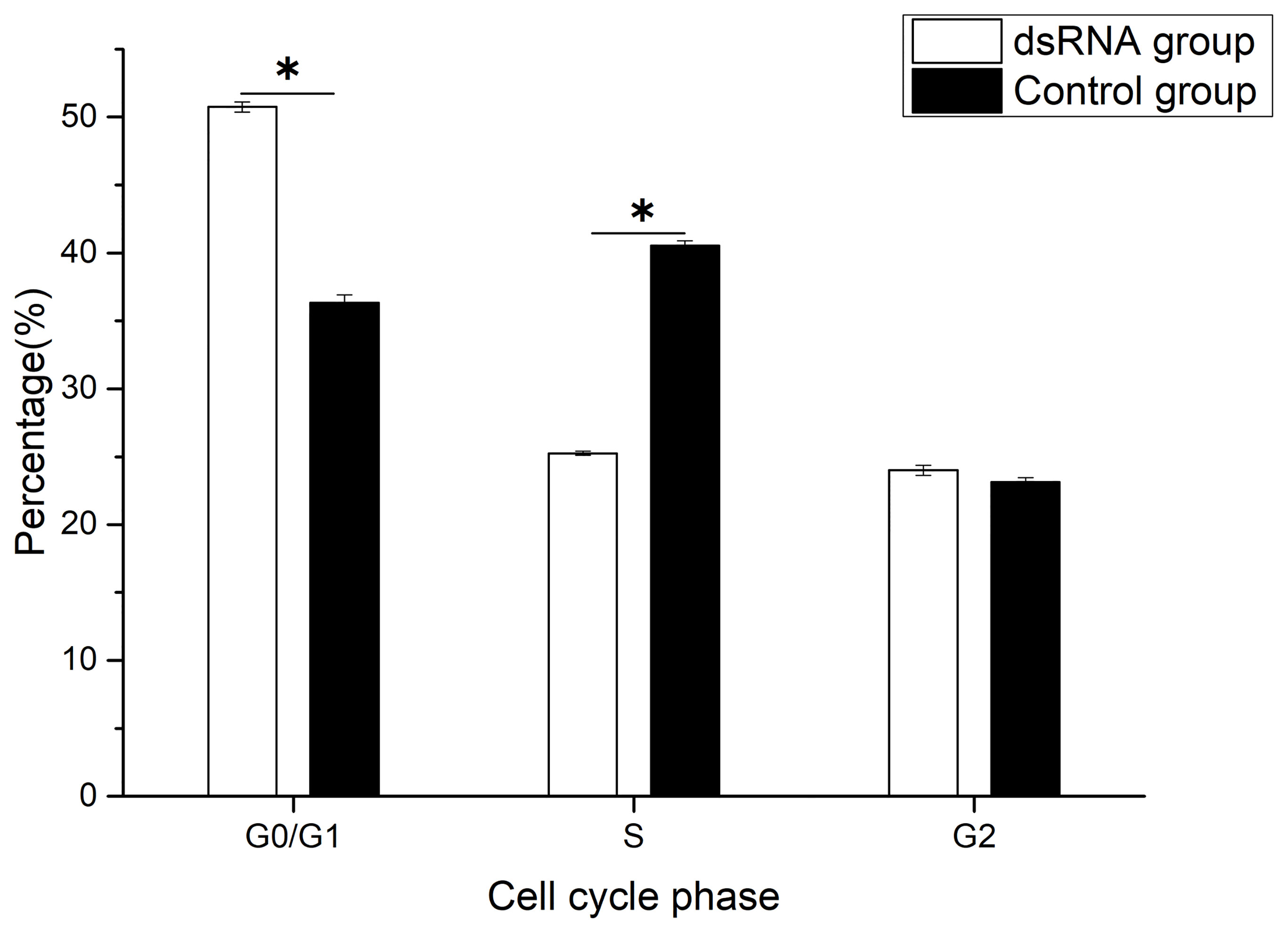
Figure 7 Cell cycle phase distribution of gill cells in Hyriopsis cumingii after Hc-CDK6 RNAi, *Significant difference (P < 0.05), Maximum value of vertical axis is 100%.
3.5 Overexpression of Hc-CDK6 Promotes Mantle Cell Proliferation
After transfection, no significant difference was observed in cell number compared with that in the control group (P>0.05, Figure 8), after analysis with a hemocytometer. Cell proliferation viability assays showed that the cell proliferation viability was significantly higher (P<0.05) than that in the control group at the fourth day of culture after transfection (Figure 9).
Figure 8 (A) Adherent mantle cells in the pcDNA3.1-CDK6 transfection group. (B) Mantle cells in the control group.
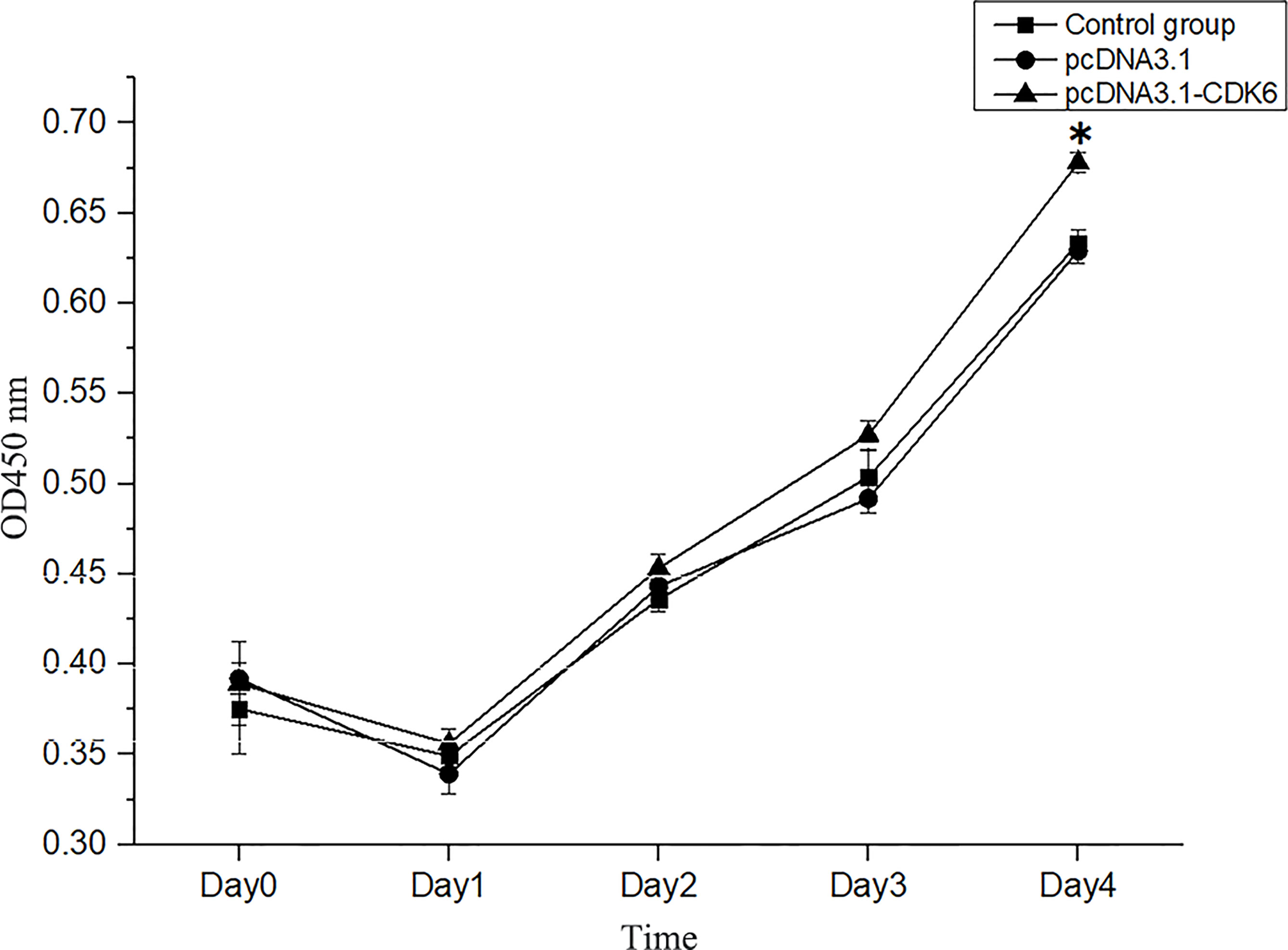
Figure 9 Cell viability assays of transfected and normal H. cumingii mantle cells cultured for 0 d, 1 d, 2 d, 3 d and 4 d. *Indicates a significant difference.
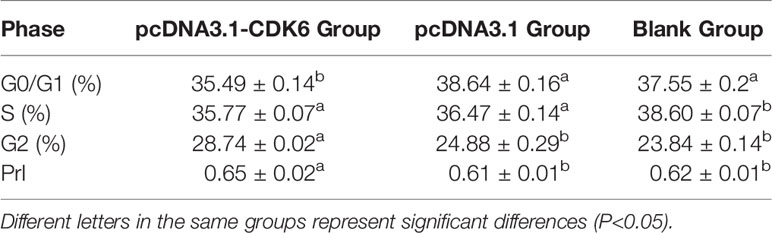
The cell cycle percentage after plasmid transfection was analyzed with flow cytometry. After pcDNA3.1-CDK6 plasmid transfection, the percentage of G1 phase cells in the recombinant plasmid group was significantly lower than that in the blank and empty plasmid groups (P<0.05), and the calculated PrI was significantly higher in the recombinant plasmid group than the control groups (P<0.05, Table 2).
4 Discussion
CDK4/6 and CDK2 are the first kinases activated by mitogenic signals that release cells from the G0 block (Park and Krause, 1999), which regulate cell cycle progression (Kozar and Sicinski, 2005). The amino acid residues that bind D-type cyclins and INK4 were also found in the amino acid sequence of Hc-CDK6, thus indicating that this kinase is a typical CDK family protein, in agreement with findings reported by Guo Jiawei et al. (Guo et al., 2016). Amino acid similarity comparison indicated that this domain was highly similar to those in other species. Thus, Hc-CDK6 might function similarly to homologs in other species and co-regulate the G1-S phase transition with D-type cyclins and CKIs. Phylogenetic tree analysis indicated that the H. cumingii CDK6 sequence clustered with those of marine bivalves such as oysters and scallops, and was furthest from those of vertebrates, thus indicating that Hc-CDK6 is highly evolutionarily conserved.
CDK6 plays an important role in cell cycle progression and differentiation (Ericson et al., 2003; Musgrove et al., 2011). The expression of CDK6 was highest in tissues with fast growth and development, such as the gonads in Scophthalmus maximus (Guo et al., 2016). This finding is consistent with the expression characteristics of H. cumingii CDK6 in this study. The similarity in tissue expression between Hc-CDK6 and Hc-CCND2 (on the basis of previous experiments) also suggested their synergistic role in the cell cycle process. The cleavage and blastocyst stages involve rapid division of embryonic cells, and gastrulation is a process of regular migration, arrangement and differentiation of embryonic cells (Eyal-Giladi and Kochav, 1976; Lin et al., 2010). The increase in CDK6 expression during embryonic development also indicates that this protein is closely associated with the rapid division and differentiation of early embryonic cells, in agreement with the results of Guo Jiawei et al. (Guo et al., 2016).
In situ hybridization of the mantle showed different hybridization signal sites of Hc-CDK6 and Hc-CCND2. CCND2 is closely associated with cell proliferation, and quantitative and in situ hybridization results indicated that the outer mantle is more viable than the inner mantle. CDK is the core regulatory molecule of the cell cycle. CDK expression tends to be stable during the cell cycle, and it is activated by the corresponding cyclins, which are expressed periodically, and then participate in cell cycle regulation (Jacobs, 1992; Malumbres and Barbacid, 2009; Lim and Kaldis, 2013). In this study, the extensive hybridization signals of CDK6 showed that mantle cells had proliferation potential, and its different localization and expression characteristics from those of CCND2 also indicated that cells in different locations in the mantle may vary in their proliferation states. CCND2 and CDK6 are closely related in the cell cycle, and their different localization and expression characteristics must be further studied.
H. cumingii are shellfish with an open-tube circulatory system, and dsRNA can circulate into various tissues within the mussel after injection through the adductor muscle. D-type cyclin associated CDK4/6 are activated and are important for the progression of the G1 cell cycle phase (Suryadinata et al., 2010). The cyclin D-Cdk4/6 complex phosphorylates Rb1 family proteins, thus resulting in the synthesis of S-phase factors required for cytokinesis (Martinez-Alonso and Malumbres, 2020). CCND2 and CDK6 genes were associated with G1-S phase regulation of the cell cycle (Dong et al., 2018). Quantitative results indicated that Hc-CCND2 gene expression levels were significantly decreased in H. cumingii after knockdown of Hc-CDK6 gene, indicating that there was a close correlation between Hc-CCND2 and Hc-CDK6, consistent with higher organisms (Malumbres et al., 2004). Knockdown of the Hc-CDK6 gene altered the distribution of cell cycle phases, as detected by flow cytometry: G0 cells increased, and S phase cells decreased, thus indicating cell cycle arrest in G1/S phase. CDK6 is involved in the regulation of G1 phase, and its silencing slows cell cycle progression (Deng et al., 2017). In tumor cells, silencing of CDK6 inhibits proliferation and hinders cycle progression (Whiteway et al., 2013; Zan et al., 2018; Su et al., 2019; Li et al., 2020). This indicates that the correct expression of CDK6 is necessary for normal cell proliferation.
The cell cycle is controlled by the synthesis of a single cyclin, and the subsequent activation of CDKs and inactivation of a series of specific Cyclin-CDK complexes at specific cell stages. D-type cyclins and CDK4/6 mainly play roles in the transformation of G1 and promote cell processes. Many studies have shown that overexpression of CCND promoted cell viability and proliferation. In previous studies, increased upstream signal transduction of CCND2 or overexpression of CDK6 has been found to lead to CCND2 accumulation and cell division (Fiaschi-Taesch et al., 2010; Baba et al., 2014). Our previous results showed that highly expressed CCND2 participated in the regulation of the cell cycle and led to the proliferation of mantle cells. Chen et al. have found that the overexpression of CDK6-CCND3 in mouse fibroblasts enhanced their proliferative potential (Chen et al., 2003). CDK6 must be activated with D-type cyclin to function. In this experiment, no difference was observed in cell proliferation activity 3 days after overexpression. D-type cyclin might not have reached the threshold of activating CDK6 activity and thus could not promote cell cycle progression. On the fourth day after transfection, the proliferative activity of cells significantly increased, and CCND2 accumulated and reached the threshold for activating CDK6. CDK6 activation promoted cell cycle progression, but the mechanism underlying this regulation by overexpression of CDK6 must be further explored.
In summary, the expression of Hc-CDK6 played an important role in the transformation of G1/S in the cell cycle. After treatment with Hc-CDK6 RNAi, cells were arrested in G0/G1. Overexpression of Hc-CDK6 promoted the proliferation of mantle cells. This study enriched research on the cell cycle in shellfish, providing a basis for further study on cell cycle regulation in shellfish. However, the mechanism of Hc-CDK6 regulated cell cycle has not been identified. Therefore, further study will be necessary for pathway analysis to explore mechanisms of cell cycle regulatory gene interactions.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/genbank/, OM572509.
Ethics Statement
The animal study was reviewed and approved by Ethics Committee of Shanghai Ocean University.
Author Contributions
SF: performed the experiments and data collection, data analysis and writing; XL: collected the review literature and participated in experimental design; HW: collected the review literature and participated in experiments; WL: performed analysis and experimental design; ZB: revised and submitted the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by the National Natural Science Foundation of China (31872565), the China Agriculture Research System of MOF and MARA and the Program of Shanghai Academic Research Leader (19XD1421500).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Baba Y., Watanabe M., Murata A., Shigaki H., Miyake K., Ishimoto T., et al. (2014). LINE-1 Hypomethylation, DNA Copy Number Alterations, and CDK6 Amplification in Esophageal Squamous Cell Carcinoma. Clin. Cancer Res. 20, 1114–1124. doi: 10.1158/1078-0432.CCR-13-1645
Cao Y. X., Lu A. L., Li W. J., Shi Z. Y., Tao J. K. (2020). Proliferative Capacity and Biomineralization of Mantle Cells in Hyriopsis Cumingii of Different Ages. J. Fish. Sci. China 27, 516–523.
Chen Q., Lin J., Jinno S., Okayama H. (2003). Overexpression of Cdk6-Cyclin D3 Highly Sensitizes Cells to Physical and Chemical Transformation. Oncogene 22, 992–1001. doi: 10.1038/sj.onc.1206193
Deng M., Zeng C., Lu X., He X., Zhang R., Qiu Q., et al. (2017). miR-218 Suppresses Gastric Cancer Cell Cycle Progression Through the CDK6/Cyclin D1/E2F1 Axis in a Feedback Loop. Cancer Lett. 403, 175–185. doi: 10.1016/j.canlet.2017.06.006
Dong P., Zhang C., Parker B. T., You L., Mathey-Prevot B. (2018). Cyclin D/CDK4/6 Activity Controls G1 Length in Mammalian Cells. PloS One 13, e0185637. doi: 10.1371/journal.pone.0185637
Ericson K. K., Krull D., Slomiany P., Grossel M. J. (2003). Expression of Cyclin-Dependent Kinase 6, But Not Cyclin-Dependent Kinase 4, Alters Morphology of Cultured Mouse Astrocytes. Mol. Cancer Res. 1, 654–664. doi: 10.1002/mpo.10269
Eyal-Giladi H., Kochav S. (1976). From Cleavage to Primitive Streak Formation: A Complementary Normal Table and a New Look at the First Stages of the Development of the Chick. I. General Morphology. Dev. Biol. 49, 321–337. doi: 10.1016/0012-1606(76)90178-0
Fiaschi-Taesch N. M., Salim F., Kleinberger J., Troxell R., Cozar-Castellano I., Selk K., et al. (2010). Induction of Human Beta-Cell Proliferation and Engraftment Using a Single G1/S Regulatory Molecule, Cdk6. Diabetes 59, 1926–1936. doi: 10.2337/db09-1776
Fu X. Y., Shen Z. H., Chao S. L. (1999). Cell Cycle, Restriction Point, Checkpoints. Chin. Bull. Life Sei. 11, 31–33.
Guo J. W., Zhu X. P., Wu Z. H., Song Z. C., Fan Z. F., Tan X. G., et al. (2016). Cloning and Expression Analysis of Cdk1 and Cdk6 in Scophthalmus Maximus. Marine Sci. 40, 11–21.
Jacobs T. (1992). Control of the Cell Cycle. Dev. Biol. 153, 1–15. doi: 10.1016/0012-1606(92)90087-W
Jin Y. L., Shi Z. Y., Li W. J., Hao Y. Y., Qiang G. (2011). Improvement on Mantle Cell Culture and Technique for Large Nucleated Pearl Producing in Hyriopsis Cumingii. J. Shanghai Ocean Univ. 20, 705–711.
Kozar K., Sicinski P. (2005). Cell Cycle Progression Without Cyclin D-CDK4 and Cyclin D-CDK6 Complexes. Cell Cycle 4, 388–391. doi: 10.4161/cc.4.3.1551
Li F., Jiang Z., Shao X., Zou N. (2020). Downregulation of lncRNA NR2F2 Antisense RNA 1 Induces G1 Arrest of Colorectal Cancer Cells by Downregulating Cyclin-Dependent Kinase 6. Digest. Dis. Sci. 65, 464–469. doi: 10.1007/s10620-019-05782-5
Lim S., Kaldis P. (2013). Cdks, Cyclins and CKIs: Roles Beyond Cell Cycle Regulation. Development 140, 3079–3093. doi: 10.1242/dev.091744
Lin Y. Q., Geng J. G., Yang X. F., Wang L. J. (2010). Cell Cycle Control During the Early Stages of Chick Embryo Development. J. Guangdong Pharm. Univ. 26, 648–652. doi: 10.3969/j.issn.1006-8783.2010.06.025
Li Q., Shi Z. Y., Li W. J., Huang K., Qi X. X. (2014). Impact of Invitro Optimization and Invivo Implantation Culture on the Growth of Hyriopsis Cumingii Mantle Cells. J. Fish. Sci. China 21, 225–234. doi: 10.3724/SP.J.1118.2014.00225
Malumbres M., Barbacid M. (2009). Cell Cycle, CDKs and Cancer: A Changing Paradigm. Nat. Rev. Cancer 9, 153–166. doi: 10.1038/nrc2602
Malumbres M., Harlow E., Hunt T., Hunter T., Lahti J. M., Manning G., et al. (2009). Cyclin-Dependent Kinases: A Family Portrait. Nat. Cell Biol. 11, 1275–1276. doi: 10.1038/ncb1109-1275
Malumbres M., Sotillo R., Santamaria D., Galan J., Cerezo A., Ortega S., et al. (2004). Mammalian Cells Cycle Without the D-Type Cyclin-Dependent Kinases Cdk4 and Cdk6. Cell 118, 493–504. doi: 10.1016/j.cell.2004.08.002
Martinez-Alonso D., Malumbres M. (2020). Mammalian Cell Cycle Cyclins. Semin. Cell Dev. Biol. 107, 28–35. doi: 10.1016/j.semcdb.2020.03.009
Musgrove E. A., Caldon C. E., Barraclough J., Stone A., Sutherland R. L. (2011). Cyclin D as a Therapeutic Target in Cancer. Nat. Rev. Cancer 11, 558–572. doi: 10.1038/nrc3090
Pan B., Wen Z. (2002). Farming and Processing of Good Large-Circular Freshwater Pearl. Modern Fish. Inf. 17, 15–17. doi: 10.3969/j.issn.1004-8340.2002.02.004
Park M., Krause M. W. (1999). Regulation of Postembryonic G1cell Cycle Progression in Caenorhabditis Elegansby a Cyclin D/CDK-Like Complex. Development 126, 4849–4860. doi: 10.1242/dev.126.21.4849
Pietenpol J. A., Stewart Z. A. (2002). Cell Cycle Checkpoint Signaling: Cell Cycle Arrest Versus Apoptosis. Toxicology, 181–182, 475–481. doi: 10.1016/S0300-483X(02)00460-2
Pines J., Hunter T. (1991). Cyclin-Dependent Kinases: A New Cell Cycle Motif? Trends Cell Biol. 1, 117–121. doi: 10.1016/0962-8924(91)90116-Q
Qian W. P., Xu Z. R., Zhang M. X., Shi M. Q. (2002). Study on Culturing Pearl Sacs In Vitro and Injection Method for Pearl Producing. Acta Agricult. Zhejiangensis 14, 82–86. doi: 10.3969/j.issn.1004-1524.2002.02.004
Satyanarayana A., Kaldis P. (2009). Mammalian Cell-Cycle Regulation: Several Cdks, Numerous Cyclins and Diverse Compensatory Mechanisms. Oncogene 28, 2925–2939. doi: 10.1038/onc.2009.170
Schaer D. A., Beckmann R. P., Dempsey J. A., Huber L., Forest A., Amaladas N., et al. (2018). The CDK4/6 Inhibitor Abemaciclib Induces a T Cell Inflamed Tumor Microenvironment and Enhances the Efficacy of PD-L1 Checkpoint Blockade. Cell Rep. 22, 2978–2994. doi: 10.1016/j.celrep.2018.02.053
Su H., Chang J., Xu M., Sun R., Wang J. (2019). CDK6 Overexpression Resulted From Microrna320d Downregulation Promotes Cell Proliferation in Diffuse Large Bcell Lymphoma. Oncol. Rep. 42, 321–327. doi: 10.3892/or.2019.7144
Suryadinata R., Sadowski M., Sarcevic B. (2010). Control of Cell Cycle Progression by Phosphorylation of Cyclin-Dependent Kinase (CDK) Substrates. Biosci. Rep. 30, 243–255. doi: 10.1042/BSR20090171
Tigan A. S., Bellutti F., Kollmann K., Tebb G., Sexl V. (2016). CDK6-A Review of the Past and a Glimpse Into the Future: From Cell-Cycle Control to Transcriptional Regulation. Oncogene 35, 3083–3091. doi: 10.1038/onc.2015.407
Wang Y., Chen Y., Cao M., Wang X., Wang G., Li J. (2021a). Identification of Wnt2 in the Pearl Mussel Hyriopsis Cumingii and its Role in Innate Immunity and Gonadal Development. Fish Shellfish Immunol. 118, 85–93. doi: 10.1016/j.fsi.2021.08.022
Wang Y., Wang X., Ge J., Wang G., Li J. (2021b). Identification and Functional Analysis of the Sex-Determiner Transformer-2 Homologue in the Freshwater Pearl Mussel, Hyriopsis Cumingii. Front. Physiol. 12, 704548. doi: 10.3389/fphys.2021.704548
Whiteway S. L., Harris P. S., Venkataraman S., Alimova I., Birks D. K., Donson A. M., et al. (2013). Inhibition of Cyclin-Dependent Kinase 6 Suppresses Cell Proliferation and Enhances Radiation Sensitivity in Medulloblastoma Cells. J. Neuro Oncol. 111, 113–121. doi: 10.1007/s11060-012-1000-7
Keywords: Hyriopsis cumingii, Hc-CDK6, cell cycle, RNAi, overexpression
Citation: Feng S, Li X, Wang H, Li W and Bai Z (2022) Identification and Functional Analysis of the G1 Phase Cyclin Dependent Kinase Gene Hc-CDK6 in Pearl Mussels (Hyriopsis cumingii). Front. Mar. Sci. 9:921726. doi: 10.3389/fmars.2022.921726
Received: 16 April 2022; Accepted: 06 May 2022;
Published: 01 June 2022.
Edited by:
Xiaotong Wang, Ludong University, ChinaReviewed by:
Gen Hua Yue, Temasek Life Sciences Laboratory, SingaporeZhongming Huo, Dalian Ocean University, China
Copyright © 2022 Feng, Li, Wang, Li and Bai. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: ZhiYi Bai, enliYWlAc2hvdS5lZHUuY24=; WenJuan Li, d2psaUBzaG91LmVkdS5jbg==
†These authors have contributed equally to this work
 ShangLe Feng1†
ShangLe Feng1† XueNan Li
XueNan Li He Wang
He Wang WenJuan Li
WenJuan Li ZhiYi Bai
ZhiYi Bai