- Fisheries College, Guangdong Ocean University, Zhanjiang, China
Neuroparsins (NP) are small-size cysteine-rich neuropeptides first discovered in insects. They are known to be involved in insect reproduction. In this study, we have cloned two neuroparsin cDNAs (i.e., MrNP1 and MrNP2) from the freshwater shrimp Macrobrachium rosenbergii. The two neuroparsins consist of 12 cysteines, which is characteristic of the neuroparsin family. These cysteines are arranged in identical relative positions that form 6-disulfide bonds. MrNP1 and MrNP2 are most similar to the corresponding neuroparsin counterparts of the shrimp Macrobrachium nipponense. Phylogenetic study results suggested that MrNP1 and MrNP2 are closely related to MnNP1 and MnNP3, respectively. Also, an additional MrNP gene similar to MnNP2 is expected to exist in M. rosenbergii. The MrNP1 expression level is the highest in the ovary, and MrNP2 expression is higher in the brain and heart of the females. In addition, during the ovary maturation cycle, MrNP1 expression in the hepatopancreas is highest in stage V; in the ovary it is variable. MrNP2 expression in the hepatopancreas and ovary is the highest in stage II and stage I, respectively. In vivo and in vitro bioassay experiment results indicate that MrNP1 and MrNP2 recombinant proteins can stimulate the expression of the MrVg gene. In contrast, silencing of MrNP1 and MrNP2 genes would suppress MrVg, VgR, and CyclinB gene expressions. The results indicate that the products of both genes can stimulate vitellogenesis by up-regulating the MrVg gene expression. Results from their difference in expression patterns indicate that they might have different regulatory roles in vitellogenin synthesis. Since gene silencing of either MrNP1 or MrNP2 affected the expression of the other NP, we have hypothesized that coordinated regulatory action between MrNP1 and MrNP2 may be necessary for the normal vitellogenesis in M. rosenbergii.
Introduction
Insects and crustaceans belong to the subphylum of Arthropoda. They share many similar structural/anatomical features. For example, they share many similar/identical hormones that regulate similar or different biological processes (Veenstra, 2016). These hormones include ecdysteroids, juvenoids, CHH-family neuropeptides, and neuroparsins (Chung et al., 2010). The first neuroparsin (NP) was isolated from the pars intercerebralis-corpora cardiaca complex of the locust Locusta migratoria (Girardie et al., 1987; Moreau et al., 1988; Girardie et al., 1989). Neuroparsin is involved in the regulation of mineral, water, and reproduction. In addition to their roles in reproduction, they are involved in promoting nerve growth, green coloration of insect larvae, anti-diuretic, adipokinetic hormone (AKH) regulation of carbohydrates and lipids, and behavioral regulation of locust phase migration. Therefore, they are also known as multi-effect neuropeptides (Fournier et al., 1987; Girardie et al., 1987; Nogaro et al., 1995; Claeys et al., 2010; Claeys et al., 2006).
In recent years, with the rapid development of RNA and genomic DNA sequencing technology, many neuroparsin-like cDNAs have been identified from different crustaceans including Litopenaeus vannamei, Scylla paramamosain, lobster Homarus americanus, M. rosenbergii, Carcinus maenas, and Panulirus argus (Badisco et al., 2007; Ma et al., 2010; Marco et al., 2014; Bao et al., 2015; Christie et al., 2015; Andrew, 2020; Kyei et al., 2020; Liu et al., 2020). Biochemically, crustacean neuroparsins are poly-cysteine-rich compounds. They consist of 12 cysteine residues that form six disulfide bonds for tertiary structure formation. They share some similarities with the neuroparsin of the locust and the ovary ecdysteroidogenic hormone (OEH) of mosquitoes (Brown et al., 1998), the N-terminal end of the growth factor-binding protein region of the vertebrate and molluscan insulin-like growth factor-binding protein and single insulin-binding domain protein (Kyei et al., 2020). The major characteristic of neuroparsin is that the spacing of Cys residues is conserved for different groups of cysteine-rich neuropeptides (Badisco et al., 2007; Tanaka, 2021). Different neuroparsins were initially identified as anti-gonadotropic peptides from the pars intercerebralis-corpora cardiaca complex of the migratory locust, Locusta migratoria, and further studies revealed the pleiotropic activities of these peptides. Discovered in crustaceans quite late as compared to insects, neuroparsins have also been found in many Decapoda in recent years. Similar to insects, the crustacean neuroparsins are reported to have gonad regulatory function. For example, in the shrimp M. ensis, in vivo gene silencing of neuroparsins could cause a significant decrease in vitellogenin transcript level in the hepatopancreas and ovary (Yang et al., 2014). In Marsupenaeus japonicus, in vivo injection of recombinant neuroparsin could increase the expression of MjVg (Tsutsui et al., 2020).
The freshwater shrimp M. rosenbergii is one of the most important freshwater crustaceans cultured in China. There is a rapid increase in demand for high-quality shrimp fry for aquaculture. In this study, we have reported the characterization of two neuroparsins and investigated the role of the two neuroparsins in the regulation of vitellogenesis. Because the amount of culture has increased, the need for shrimp seedling has also increased greatly in recent years. The production of high-quality healthy seedings has become the bottleneck for the further increase in M. rosenbergii culture. Therefore, the study on the reproductive physiology mechanism of M. rosenbergii is of great significance for understanding their reproduction and for better development of contemporary farming techniques. Recently, we have identified two neuroparsin cDNA sequences from the transcriptome of M. rosenbergii. We have used gene cloning techniques to obtain two neuroparsin open reading frame (ORF) sequences and have studied their gene expression patterns in the early stages of female shrimp gonad development and the level of mRNA expression in the hepatopancreas (Hp) and ovary (Ov) during different developmental cycles. Using RNAi interference technology, the regulatory role of neuroparsin in the expression of the major egg yolk protein of M. rosenbergii (vitellogenin, Vg) is explored. It is hoped that these data can provide background information for the role of the neuroparsins in shrimp.
Material and Methods
Animals
Female M. rosenbergii used in the study were obtained from the shrimp-breeding research facility of Guangdong Ocean University, Zhanjiang, Guangdong, China. They were temporarily cultured in a small container (i.e., 1 M3) for 2–3 days. About 50% of water was changed daily, and shrimps were fed thrice daily with pellet diets. The reproduction status of the animal was determined before the experiment following the reported methods (Chang and Shih, 1995; Yang et al., 2000); shrimps at different ovarian development stages (stage I–V) in this study are listed in Table 1.
RNA Preparation, cDNA Synthesis, and Gene Cloning
Based on the anatomical structure and external appearance of the ovary, the developmental conditions of the shrimps were determined (Chang and Shih, 1995). Various tissues including the hepatopancreas (Hp), eyestalk (Es), brain (Br), stomach (St), thoracic ganglion (Tg), intestine (Int), heart (He), and ovary (Ov) were dissected and extracted for total RNA using a spin-column-based TransZol Up Plus RNA preparation kit (TransGen Biotech, Peking, China). After elution of the total RNA from the column, the quality of total RNA was analyzed by 1.5% agarose gel electrophoresis and the concentration was determined by a NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific, Inc., Waltham, MA, USA). High-quality total RNAs were used for first-stranded cDNA synthesis using the 5× All-In-One RT MasterMix cDNA synthesis kit (ABM, Vancouver, Canada). The cDNA synthesis condition followed the program as follows: 25°C for 10 min, 42°C for 15 min, 85°C for 5 min. At the end of the synthesis, the cDNA was diluted 20× for real-time quantitative PCR (qRT-PCR) analysis or for use (1 μl) directly in semiquantitative RT-PCR. The PCR reaction in general consists of 20 µl: 10 µl 2× PCR mix, 0.5 µl cDNA, 1 µl primer (forward and reverse 1 µl, respectively), and 7 µl free water, and the PCR condition was 95°C for 3 min, 95°C for 30 s, 58°C for 30 s (MrNP1 and MrNP2 have the same temperature), 72°C for 30 s for 34 cycles, and 72°C for 10 min. At the end of the PCR, the PCR products were analyzed by 1.5% agarose gel. The amplified target cDNA in the gel was cut and purified by a spin-column based purification kit, subcloned and the DNA sequence was determined. Obtaining the cDNA of containing tagetart band by a spin column-based gel purification kit, for gene cloning.
Bioinformatic Characterization of MrNPs
The program SingnalP5.0 (http://www.cbs.dtu.dk/services/SignalP/) was used to identify the signal peptide region. The pI and pMW of NPs were estimated using the pI/pMW program from Expasy (http://web.expasy.org/protparam/). For alignment analysis, several invertebrate neuroparsin molecules were obtained through the BLAST program in the NCBI database (https://blast.ncbi.nlm.nih.gov/Blast.cgi) similarity search and analyzed using the program ClustalX (http://www.ebi.ac.uk/Tools/msa/clustalo/). All primers were designed using the Primer 5.0 program, and EF-1α was used as internal reference gene (Table 2). MEGA6.0 (megasoftware.net) was used to build the phylogenetic tree with the neighbor-joining method (NJ) with confidence intervals set at 1,000 bootstrap replications.
Expression Study of MrNP1 and MrNP2
Gene-specific primers for neuroparsin (MrNPI, MrNP2), vitellogenin (Vg, GenBank: ADK55596.1), vitellogenin receptor (VgR, GenBank: ADK55596.1), cyclin B (GenBank: ADP95148.1), elongation factor (EF-1α), and β-actin are listed in Table 2 (i.e., qNP1F, qNP1R, qNP2F, qNP2R, qVgF, qVgR, qVgRF, qVgRR, qCBF, qCBR). qRT-PCR was performed using the SYBR qPCR kit (Vazyme, Nanjing, China). For the 20-μl reaction, it consisted of 10 µl SYBR Premix Ex Taq, 1 µl cDNA, and 1 µl of each forward and reverse primer (10 µM). PCR was performed in a real-time PCR machine (Bio-Rad CFX® 96 system, Bio-Rad, USA), and the conditions were 95°C denature and 35 cycles of 95°C 30 s, annealing at 55°C for 30 s and denature at 72°C for 30 s. A further 10 min at 72°C was included for complete product extension. The results were determined using 2-ΔΔCt and further analyzed by statistical methods. The sample size N = 5 and the experiment were triplicated.
Functional Study by Recombinant Protein Technology and RNA Interference
Production of His-tagged recombinant protein: For rMrNP1, a pair of restriction enzyme sequence-linked primers (Table 2) were used to amplify the mature peptide sequence of MrNP1 from the full-length cDNA. After PCR, the product was digested with the restriction Kpn I and BamH I, respectively, and the digested fragments were ligated into Kpn I and BamH I cut sites of the linearized pET-32a protein expression vector. The ligated product was transformed into DH5a E. coli bacterial cells. For protein expression, the positive clone (i.e., rMrNP1 plasmid) was transformed into a BL21 cell. Positive clones were induced by isopropyl β-d-1-thiogalactopyranoside (IPTG) for fusion protein generation. After optimization for incubation time, IPTG, and temperature, mass production was performed in 500 ml of LB medium buffer using the determined parameters. Mass production and subsequent purification of the his-tagged recombinant protein followed the procedure as described before. Similar procedures were used for the production of recombinant protein MrNP2. Primer sequences are found in Table 2.
For double-strand RNA (dsRNA) production, forward and reverse primers (i.e., T7MrNP1F (Table 2), T7MrNP1R, T7MrNP1F, and T7MrNP2R containing T7 adapters) were designed using the program Primer 5.0 to synthesize dsNP1 and dsNP2 with MrNP1 and MrNP2 clones as templates. The PCR reaction in general consists of 20 μl: 10 μl 2× PCR mix and 0.5 μl cDNA, and the PCR reaction used the following procedure: 95°C for 3 min; 34 cycles of 95°C for 30 s, 63°C (T7NP1) or 60°C (T7NP2) for 30 s, 72°C for 30 s, and 72°C for 10 min. The purified product was the template for dsRNA production using the OneScribe T7 Synthesis Kit (ABM, Vancouver, Canada). The control double-strand RNA included the dsRNA for the Tiger Frog virus (dsTFV), as described previously (Tiu and Chan, 2007), and it has no effect on M. rosenbergii in this study.
Adult females at the early stage of gonadal development were selected for in vivo injection study. The experimental group was injected with 50 μl injection volume containing 2 μg/g of double-stranded RNA (i.e., dsMrNP1 and dsMrNP2), and the control group was injected with the same volume of dsTFV (Tiu and Chan, 2007). All animals were placed in the same culture environment for 48 h until sampling. Shrimps were dissected for collecting the hepatopancreas and ovary for total RNA preparation. The interference efficiency of dsRNA and gonad maturation conditions were determined for monitoring the expression of related genes (i.e., MrNP1, MrNP2, vitellogenin, VgR, and CyclinB). For in vitro RNAi and recombinant protein study, a the nutrient medium M199 (Sigma, USA) was sterile filtered with a 0.45-μM filter unit. Culture medium (1.5 ml) was added to each well of the 12-well culture plate. The hepatopancreas and ovary were dissected and cut to small fragments of similar size (i.e., 8 cm3). They were first rinsed separately in 1× cold PBS and placed in a well containing the recombinant proteins or dsRNA and control 1× cold PBS or control dsTFV. The culture plate was placed on an orbital shaker and incubated at 25°C. After 3 h, for the in vitro recombinant protein experiment, the tissue pieces were extracted for crude protein for SDS-PAGE analysis (Okuno et al, 2002). In the experiment of adding dsRNA in vitro, the tissue pieces were extracted for total RNA using the TRIzol RNA preparation kit, and total RNA was extracted to determine the RNA interference efficiency and changes in the expression of related genes. Both experiments were repeated five times (i.e., N = 5).
Results
Characterization of the Shrimp Neuroparsins
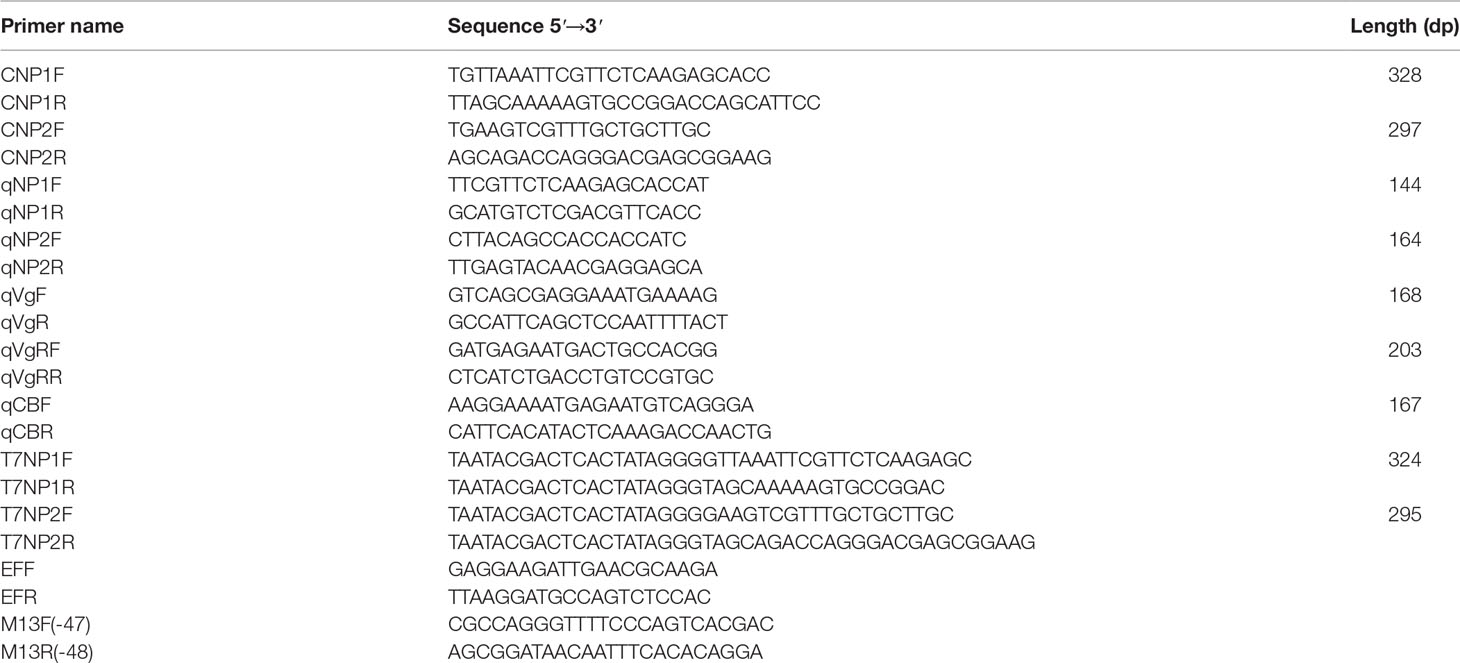
Two different neuroparsin-like cDNAs (i.e., MrNP1 and MrNP2) were cloned from the eyestalk of the freshwater shrimp M. rosenbergii. The open reading frame of MrNP1 was 303 bp encoding for a peptide of 100 amino acid residues. The signal peptide was from AA1 to AA26, and the mature peptide contains 74 aas in size (Figure 1). For pI and pMW, MrNP1 is predicted to have theoretical pI and pMW values of 7.34/10,862.6 and 6.28/8014.9, respectively. The open reading frame of MrNP2 was 300 bp encoding for a peptide of 99 amino acid residues. The signal peptide was from AA1 to AA24, and the mature peptide contains 75 AAs (Figure 1). The pro-peptide and mature peptide of MrNP2 are predicted to have a theoretical pI/pMW of 7.96/10,808.56 and pI/MW of 8.06/8150.3, respectively. Although MrNP1 and MrNP2 share low sequence homology in the signal peptide region, they both consist of 12 cysteine residues forming six disulfide bridges and the spacing of these cysteines is identical, which suggests that they share a similar tertiary structure (Figure 2A).

Figure 1 Deduced amino acid of the partial cDNAs of MrNP1 (left) and MrNP2 (right). One letter amino acid code is shown. The two partial cDNAs consist of all the coding/translation information. Underlined parts represent mature peptide.
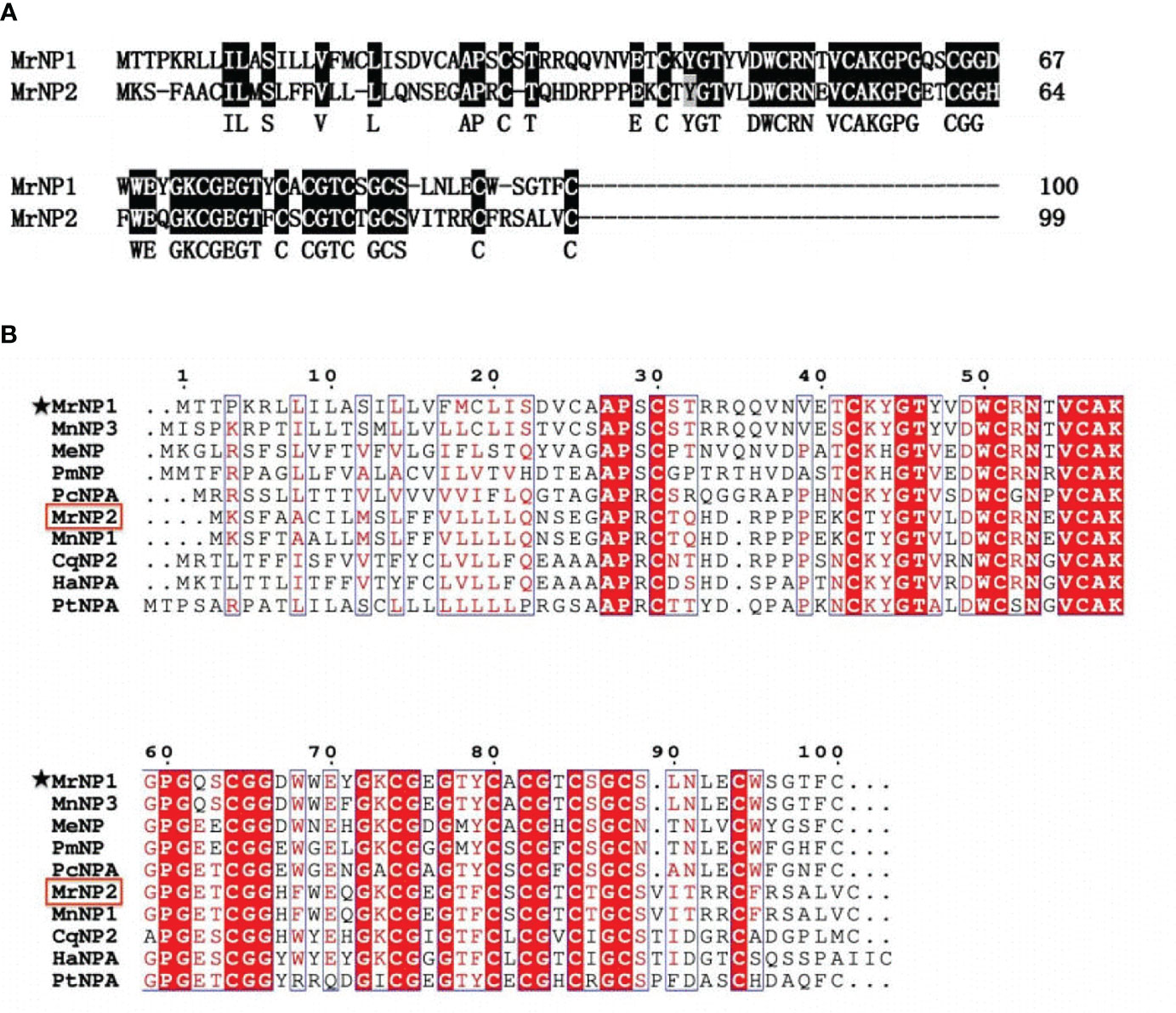
Figure 2 (A) Alignment of MrNP1 and MrNP2. (B) Multiple-sequence alignment of selected neuroparsins from different arthropods including the insects and crustaceans. These sequences are as follows: M. nipponense NP3 (GenBank# QBG05398.1); Metapenaeus ensis NP (GenBank# AHX39208.1); Penaeus monodon NP (GenBank# ALO17555.1); Procambarus clarkii NPA (GenBank# XP_045591875.1); Macrobrachium nipponense NP1 (GenBank# QBG05396.1); Culex quinquefasciatus NP2 (GenBank# AWK57531); Homarus americanus NPA (GenBank# XP042239197.1); Portunus trituberculatus NPA (GenBank# XP045122305.1). M. rosenbergii neuroparsin 1 and neuroparsin 2 (MrNP1, MrNP2) were marked with an asterisk and a box, respectively.
Multiple-sequence alignment results (Figure 2B) indicate that MrNP1 shared the highest amino acid sequence identity with the neuroparsin 3 precursor of M. nipponense (MnNP3, 84.2%), followed by that of the shrimp M. ensis neuroparsin (MeNP, 55%), Penaeus monodon neuroparsin (PmNP, 55%), the crayfish Procambarus clarkii neuroparsin A-like (PcNPA, 50.5%), the neuroparsin A-like of the lobster Homarus americanus (HaNPA, 44%), the neuroparsin A of the crab Portunus trituberculatus (PtNPA, 44%), and M. nipponense neuroparsin 1 (MnNP1, 42.4%), and the lowest homology (39%) with Cherax quadricarinatus neuroparsin 2 (CqNP2), while MrNP2 shared the highest amino acid sequence identity with MnNP1 (97%), followed by CqNP2 (64%), HaNPA (64.7%), PtNPA (63.8%) and PcNPA (46.5%), MnNP3 (42.4%), MeNP (41.4%), and the lowest homology (38.4%) with PmNP.
Based on the amino acid sequence of the neuroparsin-like and other cysteine-rich peptides from vertebrates and invertebrates, the phylogenetic tree result indicates that neuroparsins and other cysteine-rich peptides were clustered into three major groups: i.e., the crustacean NPs, insect NPs, and other cysteine-rich peptides. The first group consisted of crustacean neuroparsins containing MrNP1 and MrNP2 which are evolutionarily related. Group 2, the neuroparsins of insects, includes NP1, NP2, NP3, and NP4 of S. paramamosain and L. migratoria NPA and OEH protein of Acedes aegypti and Culex quinquefasciatus. The single insulin-binding domain (SIBD) protein of invertebrates and insulin-like growth factor-binding protein (IGFBP) of the cysteine-rich peptides were clustered into group 3 (Figure 3).
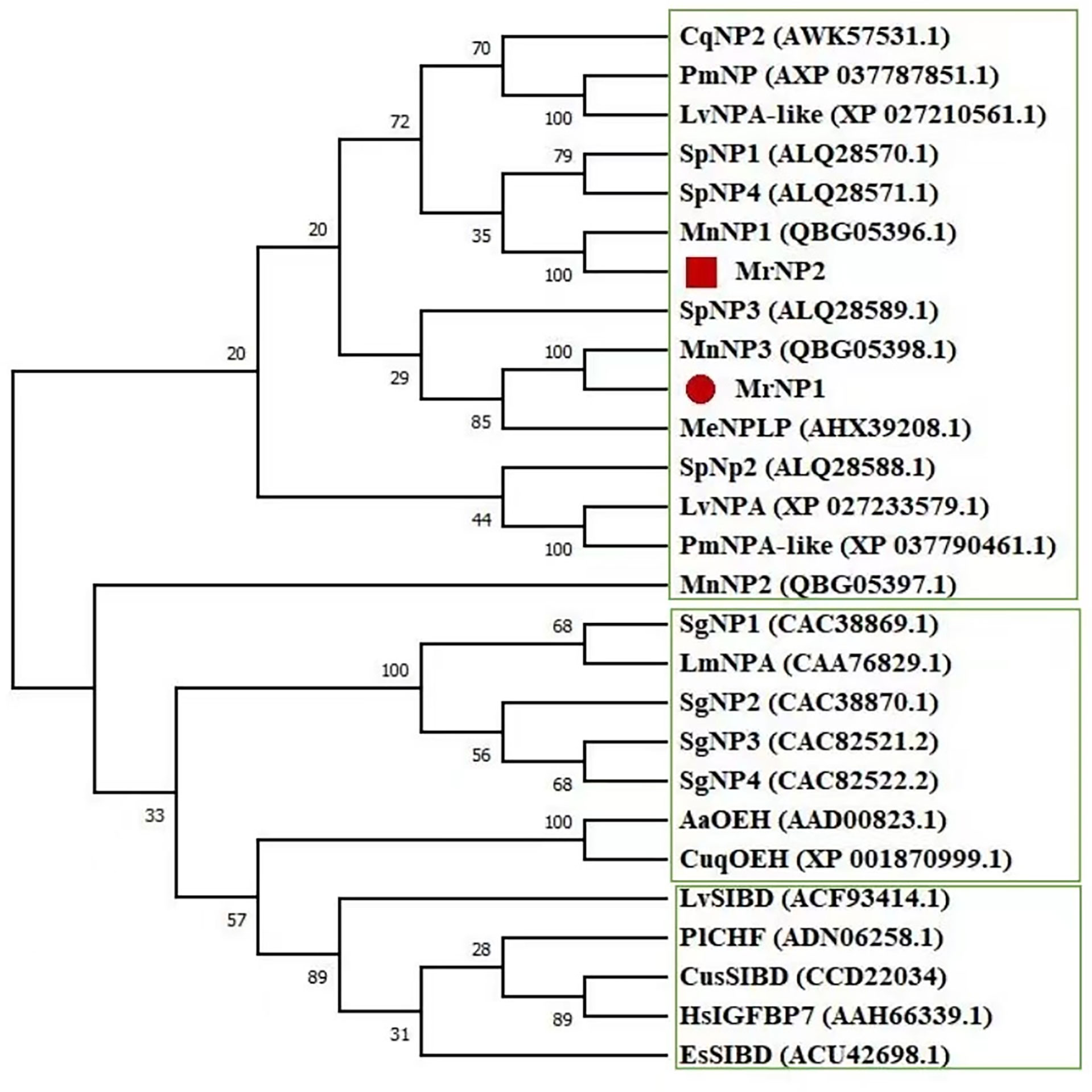
Figure 3 Phylogenetic tree analysis of the M. rosenbergii neuroparsins (MrNP1, MrNP2) with other cysteine-rich related peptides from the crustaceans, insects, and other species. The sequences include Penaeus monodon NPA-like (GenBank# XP037790461.1); Litopenaeus vannamei NPA (GenBank# XP027233579.1); the locust Schistocerca gregaria NP1 (GenBank# CAC38869.1); S. gregaria NP2 (GenBank# CAC38870.1); S. gregaria NP3 (GenBank# CAC82521.2); S. gregaria NP4 (GenBank# CAC82522.2); L. migratoria LmNPA (GenBank# CAA76829.1); Acedes aegypti AeOEH (GenBank# AAD00823.1); Culex quinquefasciatus CuqOEH (GenBank# XP_001870999.1); the crab S. paramamosain SpNP1 (GenBank# ALQ28588.1); S. paramamosain SpNP2 (GenBank# ALQ28570.1); S. paramamosain SpNP3 (GenBank# ALQ28589.1); S. paramamosain SpNP4 (GenBank# ALQ28571.1); M. nipponense MnNP1 (GenBank# QBG05396.1); M. nipponense MnNP2 (GenBank# QBG05397.1); M. nipponense MnNP3 (GenBank# QBG05398.1); M. ensis MeNP (GenBank# AHX39208.1); L. vannamei single IB domain protein (LvSIBD, GenBank# ACF93414.1); Pacifastacus leniusculus crustacean hematopoietic factor (PlCHF, GenBank# ADN06258.1); crab Eriocheir sinensis single insulin binding domain protein (EsSIBD, GenBank# ACU42698.1); spider Cupiennius salei SIBD (CusSIBD, GenBank# CCD22034); Homo sapiens insulin-like growth factor-binding protein 7 (HsIGFBP7,GenBank# AAH66339.1).
Expression of MrNP1 and MrNP2 in Female M. rosenbergii
QRT-PCR results showed that the MrNP1 expression level was the highest in the ovary (Ov), followed by the hepatopancreas (Hp), and it is only moderately expressed in the eyestalk (Figure 4A). In contrast, MrNP2 transcript levels were higher in the heart and brain. It is moderately expressed in the thoracic ganglion and intestine, and only traces of transcripts were detected in the gill, eyestalk, and hepatopancreas (Figure 4B). Since the ovary (Ov) and hepatopancreas (Hp) are major sites for vitellogenesis, we also studied the expression of these two genes during the gonad maturation cycle of M. rosenbergii. In the Hp, MrNP1 expression was high in stage V (GSIa = 7.03 ± 0.48) and the MrNP2 expression level is the highest in stage II (GSIa = 1.48 ± 0.38) (Figures 5A, B); in the Ov, the MrNP1 transcript level is variable but the expression level of MrNP2 is the highest in stage I (GSIa = 0.49 ± 0.03) (Figures 5C, D).
Figure 4 QRT-PCR expression study of (A) MrNP1 and (B) MrNP2 in various tissues of female M. rosenbergii. Tissues used in the study are eyestalk (Es), gill (Gi), brain (Br), stomach (St), thoracic ganglion (Tg), intestine (Int), heart (Ht), hepatopancreas (Hp), and ovary (Ov). Values are relative expression level ± standard deviation (SD) (N = 5). Different letters indicate significant differences at P < 0.05 computed with ANOVA and the Tukey–Kramer test.
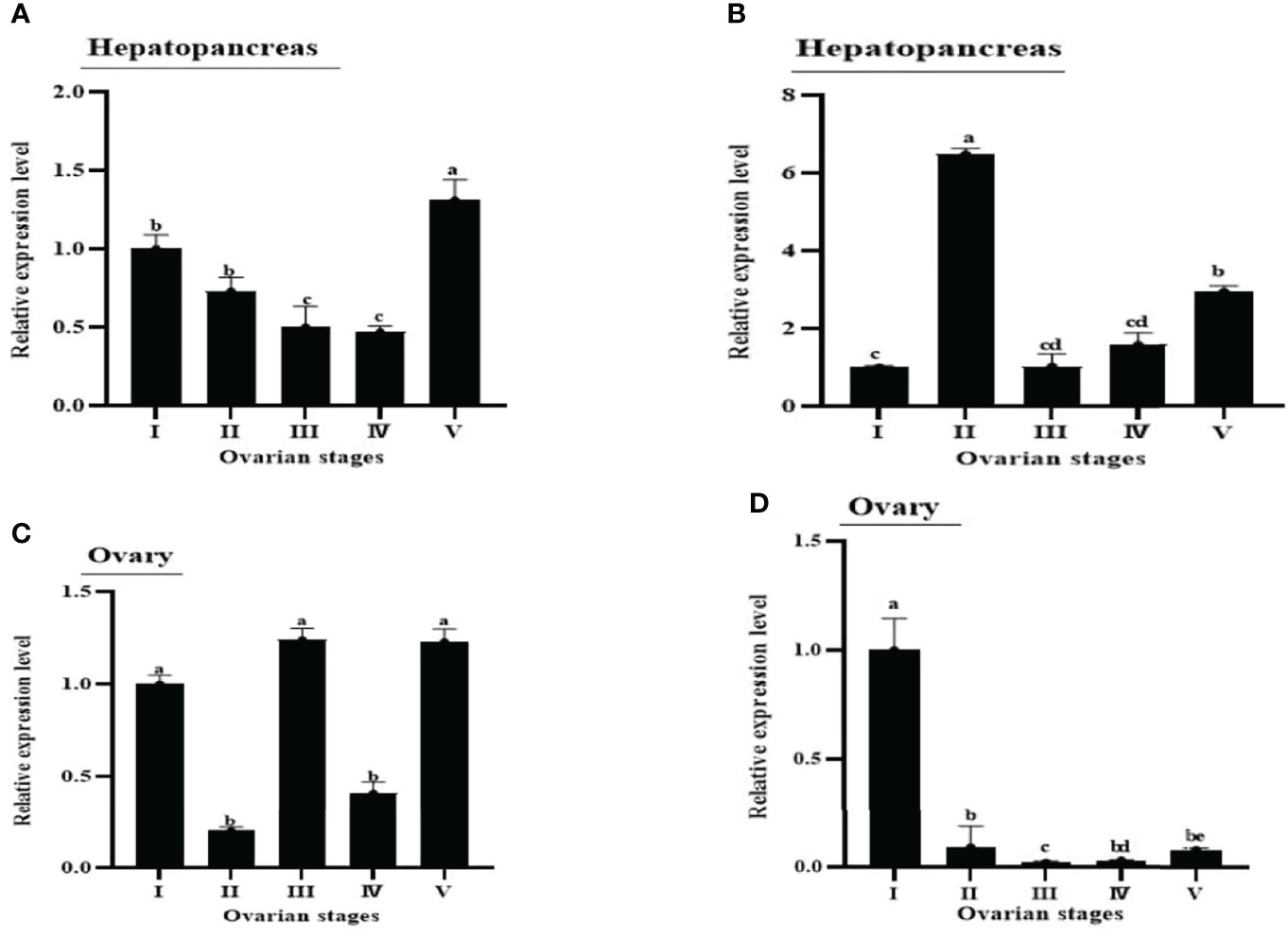
Figure 5 Figure 5 | Expression patterns of MrNP1 and MrNP2 in (A, B) hepatopancreas and (C, D) ovary in female M. rosenbergii in different reproductive stages (i.e., I, II, III, IV, V). Data shown are means of relative expression levels ± SD. Different letters indicate significant differences at P < 0.05 computed with ANOVA and the Tukey–Kramer test.
RNAi and Recombinant Protein Assay to Study the Function of MrNP1 and MrNP2 in Vitellogenesis
Females at the early (stage II) to early-middle (stages II–III) stages of the ovary maturation were used for in vivo and in vitro recombinant protein and RNAi bioassay experiments. Shrimp were selected at the early (stage II) for in vivo dsRNA injection studies, and they were sacrificed for 48 h after dsRNA injections. The injection of dsMrNP1 and dsMrNP2 had no observable harmful effects as there was no mortality in any of the groups. For dsMrNP1-injected shrimps, MrNP1 transcript levels in the hepatopancreas and ovary had decreased significantly. Also, there was a significant reduction in MrNP2 transcript level in the hepatopancreas (P < 0.05) but not in the ovary. The injection of dsMrNP2 had little effect on MrNP1 transcript level in the ovary, but the level has reduced to only 30% in the hepatopancreas as compared to the control. Interestingly, the hepatopancreas and ovary expression of the MrNP1 transcript and MrNP2 was reduced significantly (Figures 6A, B). The gene knockdown of MrNP1 and MrNP2 has caused a significant reduction in MrVg transcript levels in the hepatopancreas and ovary (Figure 6C). Also, the ovary vitellogenin receptor transcript level was reduced significantly. However, the injection of dsMrNP1 and dsMrNP2 had little effect on the transcript levels of CyclinB (Figures 6D, E).
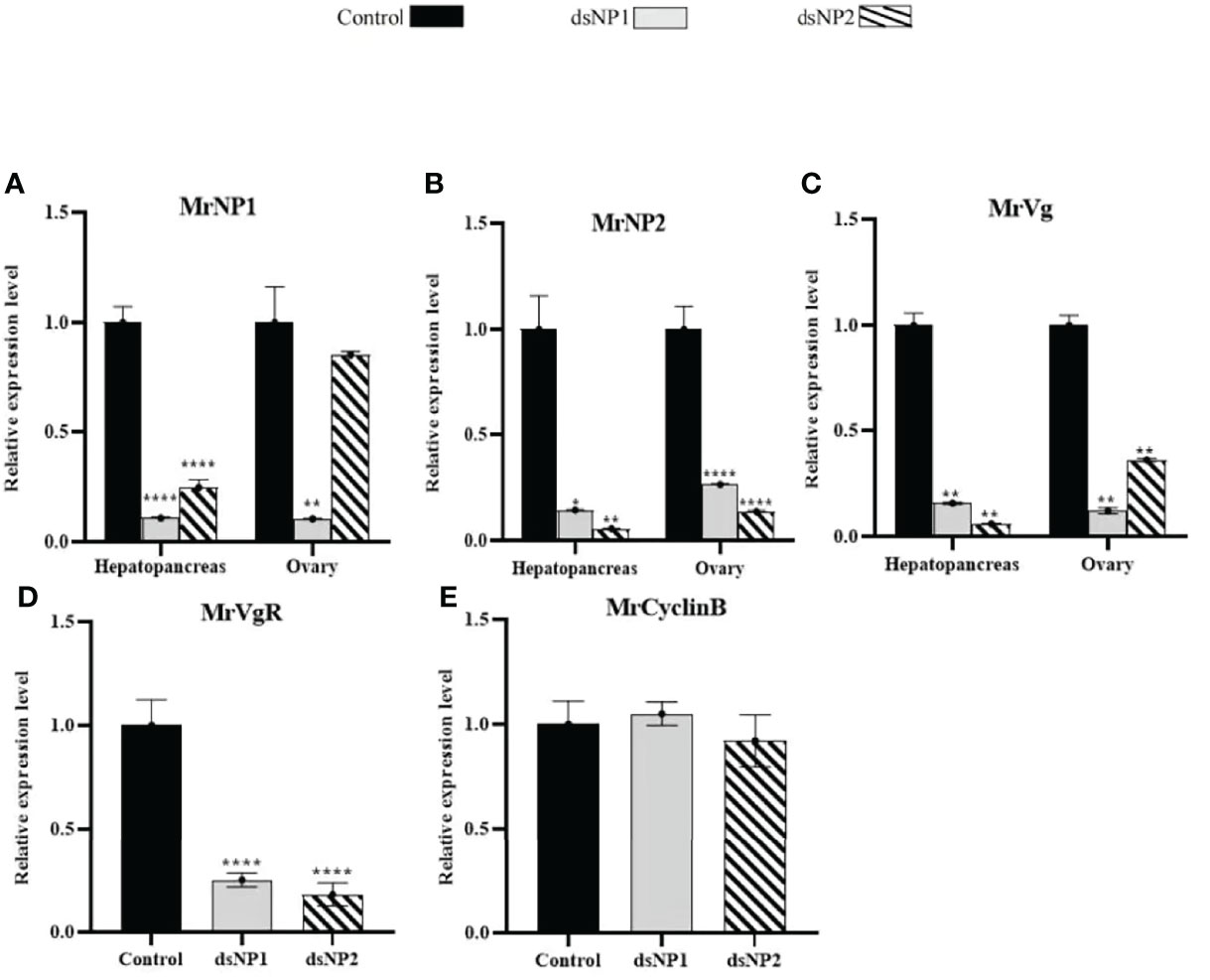
Figure 6 In vivo RNA interference of dsMrNP1 and dsMrNP2. Expression of (A) MrNP1, (B) MrNP2, and (C) MrVg after shrimps were injected with dsTFV (black bar), dsMrNP1 (gray bar) and dsMrNP2 (diagonally stripe bar). Expression of VgR (D) and CyclinB (E) in the ovary (data shown are means of relative expression levels ± SD. One-way ANOVA method was used in the statistical analysis . The significance levels are: * , (P < 0.05); **,(P < 0.01); ***, (P < 0.005) ; and ****,(P<0.001).
The in vitro RNAi results were similar to those of the in vivo RNAi experiment with minor differences (Figures 7A, B, D, E). The change in transcript levels of MrNP1, MrNP2, MrVg, and MrVgR in dsMrNP1- and dsMrNP2-treated tissues was similar in the in vitro and in vivo experiments. However, in the hepatopancreas the expression level of MrVg was unaffected after dsRNA treatments (Figure 7C).
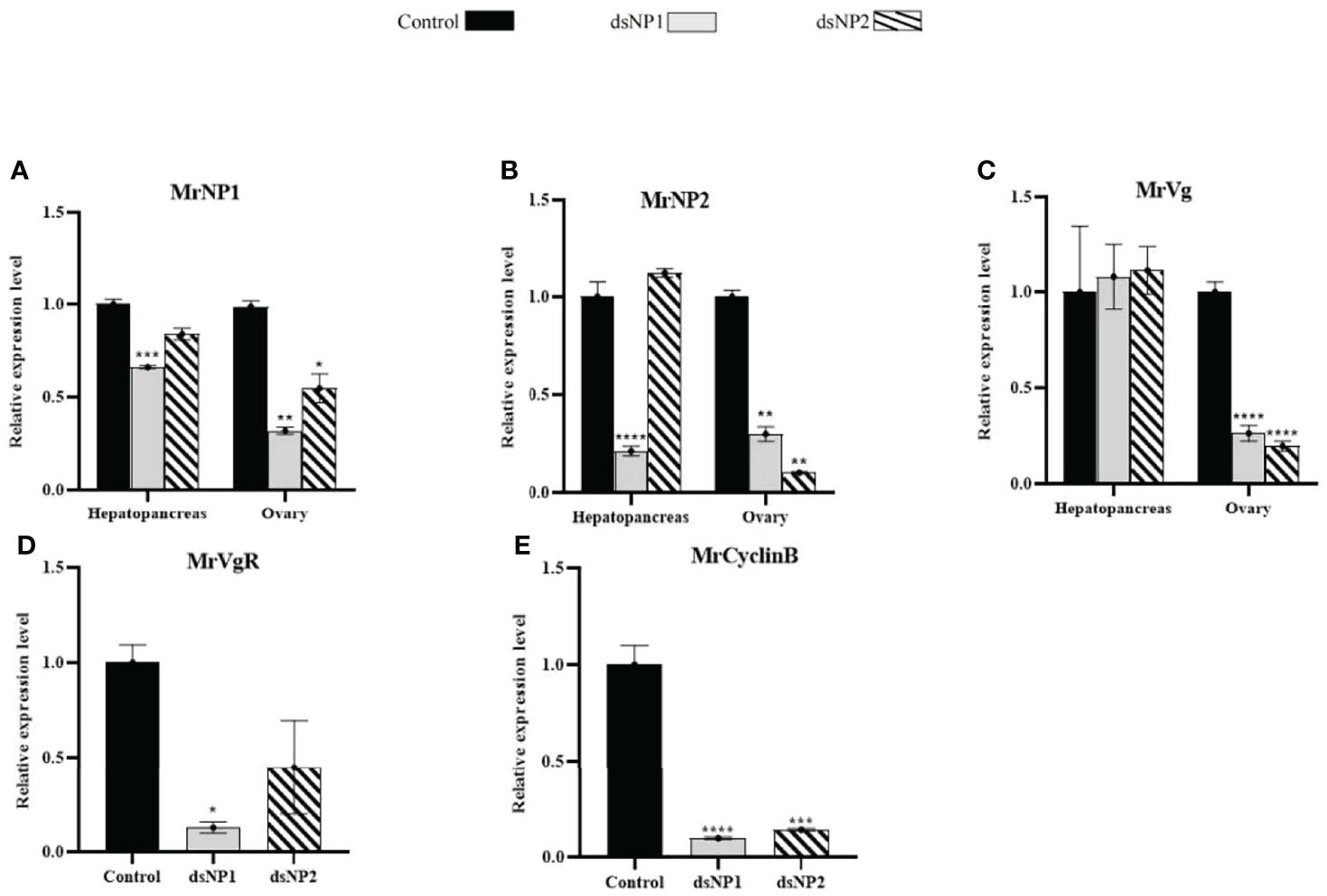
Figure 7 In vitro RNA interference of dsMrNP1 and dsMrNP2. Expression of (A) MrNP1, (B) MrNP2, and (C) MrVg after shrimp hepatopancreas and ovary fragments were treated with dsTFV (black bar), dsMrNP1 (gray bar), and dsMrNP2 (diagonally stripe bar); expression of VgR (D) and CyclinB (E) after RNAi. Data were mean relative expression levels ± SD. One-way ANOVA method was used in the statistical analysis. The significance levels are: * , (P < 0.05); **, (P < 0.01); ***, (P < 0.005); and ****, (P<0.001).
Since the expressions of MrNP1 and MrNP2 are important for the expression of MrVg, we extend the in vitro functional study by treating the hepatopancreas and ovary fragments with recombinant proteins for MrNP1 and MrNP2. Compared to the control, hepatopancreas and ovary fragments treated with recombinant proteins for MrNP1 and MrNP2 express a higher level of MrVg (the densitometry results in Supplement 1). The effect existed in the ovary of early stage (stage II) and also in ovaries of further matured stages II to III (stages II–III) (Figure 8).
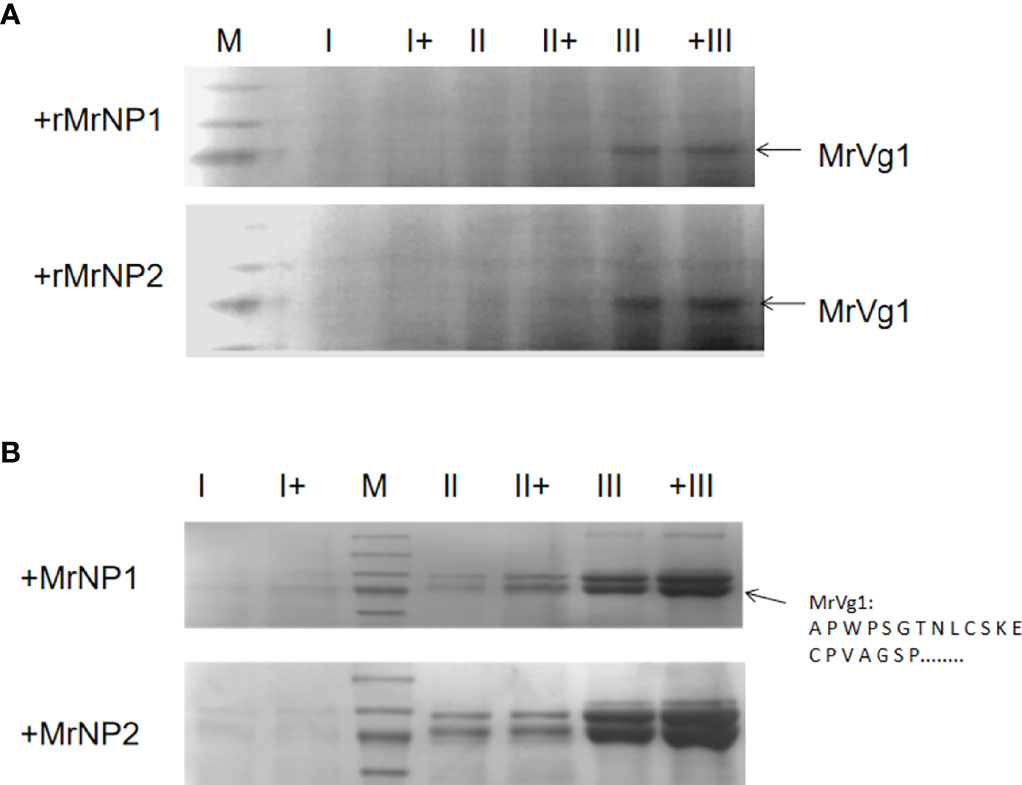
Figure 8 Analysis of hepatopancreas (A) and ovary (B) tissue protein by 7% SDS-PAGE after in vitro treatment with recombinant protein for MrNP1 and MrNP2 (N = 3). Representative Coomassie Blue-stained gel after SDS-PAGE. Each lane represents protein extracted from the from medium. Protein in gel (78 kDa, arrow) was excised, and the amino acid sequence was determined by amino acid sequencing and confirmed to be MrVg as previously (Okuno et al., 2002). Hepatopancreas from the early (stage I), middle (stage II), and late (stage III) stages of ovary maturation were either incubated with (+) or without the recombinant proteins (i.e., rMrNP1 and rMrNP2).
Discussion
Compared with other neuropeptide hormones, neuroparsins are more recently discovered cysteine-rich signaling biomolecules in Arthropoda. Sequence comparison and multiple alignments of neuroparsins with other selected cysteine-rich peptides revealed that there are significant sequence similarities among arthropod neuroparsins, crustacean hematopoietic factor, single insulin-binding domain protein of vertebrate, and mollusk insulin-like growth factor-binding proteins (IGFBP) (Kleijn and Herp, 1998). Therefore, we agreed with that neuroparsins might interact with endogenous insulin-related peptides (Badisco et al., 2007). Lm-NPA was initially identified from corpora cardiaca of the locust based on its inhibiting role in vitellogenesis. Later in Aedes aegypti, a neuroparsin ortholog was identified and named ovary ecdysis hormone (i.e., OEH) (Brown et al., 1998; Matsumoto et al., 1989). Although the OEH shared only 29% amino acid sequence similarity to Lm-NP, it has eight cysteine residues spaced in a conserved identical position similar to that of Lm-NP, which is considered to be a major characteristic of the NP family (Tanaka, 2021). Also, most of these cysteine sites are concentrated in the mature peptide region, and there are 12 cysteines in them.
Three different NPs were identified in M. nipponense (Qiao et al., 2020). Despite that M. rosenbergii MrNP1 and MrNP2 share only 56% amino acid identity, the comparison between M. rosenbergii MrNP1 and M. nipponense MnNP3 and the comparison between M. rosenbergii MrNP2 and M. nipponense MnNP1 show high degrees of amino acid sequence identity (97%), which suggests that they should have a strong structure–function relationship. From an evolutionary standpoint, the presence of a third NP in M. nipponense suggested that a third MrNP3 that shared a high sequence identity with NP2 of M. nipponense may exist in M. rosenbergii. Further experiments are needed to confirm or identify this NP. The locust neuroparsin gene produces five different transcripts, of which only one coding for the neurohormone was identified from the corpora cardiaca. The existences of multiple-neuroparsin genes have been reported in some insects including the beg bug Cimex lectularius (GenBank# Q034340.1) (Predel et al., 2018). In locust, the organically reported LmNPB was actually derived from the alternative splicing of the LmNP giving rise only to the Lm-NPA transcript. Considering high sequence similarity with the crab NP (Sp-NP1), the two NPs found in M. rosenbergii may also have a similar function of promoting the development of gonads (Liu et al., 2020).
The development of ovary in M. rosenbergii is artificially divided into five stages based on the amount of the major yolk protein vitellin present in the oocytes (Chang and Shih, 1995; Ching and Tung, 2011). During the early phase of ovary maturation, vitellogenin, the precursor of vitellin, was produced in large quantity (Meusy, 1980). The understanding of how vitellogenin synthesis is regulated in the early stage of maturation is valuable as its knowledge may contribute to the development of a technique to stimulate ovary maturation in hatchery production. Therefore, M. rosenbergii at the early stage of development was used in this study. The hepatopancreas and the ovary are the major non-neuronal tissues that express NP in female M. rosenbergii. However, the expression patterns of M. rosenbergii MrNP1 and MrNP2 are different. In the ovary, MrNP1 expression is the highest in stage III, while the MrNP2 expression level is the highest in stage I. Because of the difference in the expression patterns of MrNP1 and MrNP2, it is likely that the main regulation of Vg by the two MrNPs occurred in different developmental stages.
After vitellogenin is synthesized in the Hp, Vg produced in the hepatopancreas is secreted into the hemolymph and transported to the oocytes (Chen and Raikhel, 1996). In the oocytes, Vg interacts with VgR to form a complex and then enters the cytoplasm through endocytosis. Therefore, an up-regulated Vg production, in general, is accompanied by an increase in expression of VgR in the ovary (Tiu et al., 2008). CyclinB is an important maturation factor (i.e., MPF) involved in cell division and is highly expressed in the ovary in oocyte formation. In rainbow trout (Oncorhynchus mykiss) (Feng et al., 2020), Cyclin B1 was absent in vitellogenic oocytes but present in young previtellogenic and mature oocytes and showed a strong relationship with vitellogenesis (Draetta et al., 1989; Qiu et al., 2008). In the crab, Eriocheir sinensis, it was reported that CyclinB is involved in the oogenesis during ovary maturation (Fang and Qiu, 2009). To further understand and explore the function of two NP genes in M. rosenbergii, the expressions of the Vg, VgR, and CyclinB genes were analyzed as indicators. The results of the RNAi in vivo and in vitro experiments were monitored by the gene knockdown of MrNP1 and MrNP2 genes and also the expression of the reproduction-related marker gene. The in vivo and in vitro knockdown of MrNP1 and MrNP2 genes was highly effective as the gene expression levels of the MrNP1 and MrNP2 were reduced significantly. As in other decapods, vitellogenin in M. rosenbergii is produced in the hepatopancreas during vitellogenesis. After synthesis, Vg is transported through the hemolymph to the ovarian tissue for further processing and storage. Our experimental results show that it is very likely that neuroparsin can directly affect the synthesis and storage of Vg in M. rosenbergii. Therefore, contrasting vitellogenin stimulation and inhibition results have been reported in crustaceans, which is consistent with our experimental results; the expression of the VgR gene in the ovary decreases significantly. Knockdown of MrNP1 and MrNP2 genes can significantly reduce the expression of VgR genes, indicating that MrNP1 and MrNP2 genes may participate in the processing of Vg. There is a significant difference between the amount and the control group. The CyclinB gene mainly plays a role in the process of oocyte mitosis. During the entire vitellogenesis period, the CyclinB gene expression is the highest during the prophase of vitellogenesis; therefore, knockdown of the NP1 and NP2 genes have no effect in CyclinB expression.
In locusts, the deposition of NP can inhibit the growth of oocytes and production of Vg (Girardie et al., 1998; Badisco et al., 2011). In mosquitoes, OEH can increase the expression of the Vg gene (Riehle et al., 2002; Gulia-Nuss et al., 2012). In Scylla paramamosain, NP could down-regulate the expression of the Vg gene (Liu et al., 2020), while the expression of the Vg gene decreased significantly after injection of dsNP in female M. ensis (Yang et al., 2014). These results show that the function of the NP gene may be species-specific. In this study, we found that knockdown of MrNP1 and MrNP2 could decrease the expression of the Vg gene in both hepatopancreas and ovary, and addition of recombinant protein could increase the expression of Vg. At this stage, the reason for the lack of change in MrVg expression after RNAi by dsMrNP1 and dsMrNP2 is unknown. However, since there was no significant change in Vg expression in these animals, the results reaffirmed that NP2 was involved in Vg regulation.
During the early stage of ovary maturation, it was estimated that 90% of Vg was synthesized in the hepatopancreas in M. rosenbergii (Li, 2012). Unlike the in vivo injection study, in vitro treatment of the hepatopancreas with dsMrNP1 did not affect the expression of MrVg. The results suggest that MrNP1 may not directly be involved in the synthesis of Vg. After the synthesis of Vg by the hepatopancreas, it is transported from the hemolymph to the ovary and binds to VgR to form a Vg/VgR complex. The Vg/VgR complex enters the oocytes cytoplasm via endocytosis (Meusy, 1980).
The presence of SpNP1 in the hemolymph has been reported in the crab S. paramamosain through Western blot, which indicates that SpNP1 serves as an endocrine factor in the regulation of physiological activities. In vitro experiments have further shown that the mRNA level of vitellogenin in the hepatopancreas notably decreases following administration of recombinant SpNP1, while the mRNA levels of the vitellogenin receptor and cyclin B in the ovary showed no significant differences. In S. paramamosain, in vitro experiments confirmed that recombinant SpNP1 inhibited the expression of Vg, but it had no effect on the expression of the VgR gene in the ovary (Liu et al., 2020). However, in M. rosenberg in an in vivo study showed that knockdown of MrNP1/2 could significantly decrease the expression of the VgR gene. Therefore, MrNP1 and MrNP2 play important roles in the transportation of Vg from hepatopancreas to ovary.
The oocytes’ meiotic competence and maturation occur at the termination of vitellogenesis. In this physiological process, cyclin B acts as a regulatory subunit of the maturation-promoting factor and plays a key role in the regulation of meiotic resumption (Okano-Uchida et al., 1998); the maturation-promoting factor controls the G2/M checkpoint in eukaryotic cell division cycles and induces oocytes to enter the M phase and develop into meiosis maturation (Basu et al., 2004; Tomy et al., 2016). Thus, a high level of cyclin B in the ovary is necessary for the maturation of oocytes (Phinyo et al., 2013). Moreover, researchers found that cyclin B has high expression at both the previtellogenesis and late vitellogenesis stages (Phinyo et al., 2013; Tomy et al., 2016; Xie et al., 2001). The result of the in vivo injection experiment showed that the expression of the cyclin B gene had no obvious change, while significant decreases were seen after the addition of dsMrNP1/2 in vitro incubating experiments. These results indicated that cyclin B could be regulated by MrNP1 and MrNP2 in the individual ovary, while in M. rosenbergii at the early vitellogenesis stage, the expression of cyclin B may be influenced by some unknown factors so as not to be decreased with the knockdown of MrNP1/2. According to this result, and combining with the results of the characteristics of sequences, patterns of expression, and experiments of injection and incubation, we concluded that both MrNP1 and MrNP2 can promote the synthesis of vitellogenin in M. rosenbergii, which might have different regulatory roles in the regulatory mechanism. However, the relationship between MrNPs and CyclinB gene is still unclear and needs further study.
Conclusion
In brief, the complete ORF sequences of MrNP1 and MrNP2 in M. rosenbergii have been cloned and confirmed as members of the neuroparsin family. The expression patterns of MrNP1 and MrNP2 were different, which indicate different regulatory roles. RNAi and overexpression experiments were carried out to identify the roles of MrNP1 and MrNP2 in the development of vitellogenin within M. rosenbergii in the early stage of the ovary. MrNP1 and MrNP2 may act as stimulators of early ovarian development via promoting the expression of the MrVg gene, MrVgR gene, and MrCyclinB gene. The current study expands our understanding of the regulatory mechanism of ovary in M. rosenbergii and provides valuable information for regulating ovarian maturation in the aquaculture of M. rosenbergii. However, the differences with regard to regulatory mechanisms observed for MrNP1 and MrNP2 in M. rosenbergii need further exploration and will help to better understand the developmental mechanism of the ovary in crustaceans.
Data Availability
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
Conceptualization, SC; methodology, SC; software, CW; validation, LS and WW; formal analysis, CA, SC, and WW; investigation, CA; resources, CW; writing—original draft preparation, CA, CW; writing—review and editing, CW and SC; visualization, LS; supervision, WW, LS; project administration, SC; funding acquisition, SC. All authors contributed to the article and approved the submitted version.
Funding
This research was funded by the Guangdong Provisional Research Grant (#2014B020202014).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2022.917274/full#supplementary-material
References
Andrew E. C. (2020). Identification of Putative Neuropeptidergic Signaling Systems in the Spiny Lobster, Panulirus Argus. Invertebrate Neurosci. 20 (1), 1–12. doi: 10.1007/s10158-020-0235-9
Badisco L., Claeys I., Loy T. V., Hiel M. V., Franssens V., Simonet G., et al. (2007). Neuroparsins, a Family of Conserved Arthropod Neuropeptides. Gen. Comp. Endocrinol. 153, 64–71.
Badisco L., Marchal E., Wielendaele P. V., Verlinden H., Vleugels R., Broeck J. V. (2011). RNA Interference of Insulin-Related Peptide and Neuroparsins Affects Vitellogenesis in the Desert Locust Schistocerca Gregaria. Peptides 32, 573–580. doi: 10.1016/j.peptides.2010.11.008
Bao C., Yang Y., Huang H., Ye H. (2015). Neuropeptides in the Cerebral Ganglia of the Mud Crab, S. Paramamosain: Transcriptomic Analysis and Expression Profiles During Vitellogenesis. Sci. Rep. 5, 17055. doi: 10.1038/srep17055
Basu D., Navneet A. K., Dasgupta S., Bhattacharya S. (2004). Cdc2-Cyclin B-Induced G2 to M Transition in Perch Oocytes is Dependent on Cdc25. Biol. Reprod. 71, 894–900. doi: 10.1095/biolreprod.104.029611
Brown M. ,. R., Graf R., Swiderk K. M., Fendley D., Stracker T. H., Champagne D. E., et al. (1998). Identification of a Steroidogenic Neurohormone in Female Mosquitoes. J. Bio. Chem. 273, 3967–3971. doi: 10.1074/jbc.273.7.3967
Chang C. F., Shih T. W. (1995). Reproductive Cycle of Ovarian Development and Vitellogenin Profies in the Freshwater Prawns, Macrobrachium Rosenbergii. Invertebr. Reprod. Dev. 27, 11–20. doi: 10.1080/07924259.1995.9672429
Chen J. S., Raikhel A. S. (1996). Subunit Cleavage of Mosquito Pro-Vitellogenin by a Subtilisin-Like Convertase. Proc. Natl. Acad. Sci. U.S.A. 93, 6186–6190.
Ching F. C., Tung W. S. (2011). Reproductive Cycle of Ovarian Development and Vitellogenin Profiles in the Freshwater Prawns, Macrobrachium Rosenbergii. Invertebr. Reprod. Dev. 27, 11–20. doi: 10.1080/07924259.1995.9672429
Christie A. E., Chi M., Lameyer T. J., Pascual M. G., Shea D. N., Stanhope M. E., et al. (2015). Neuropeptidergic Signaling in the American Lobster Homarus Americanus: New Insights From High-Throughput Nucleotide Sequencing. PLos One 10, e0145964. doi: 10.1371/journal.pone.0145964
Chung J. S., Zmora N., Katayama H., Tsutsui N. (2010). Crustacean Hyperglycemic Hormone (CHH) Neuropeptides Family: Functions, Titer, and Binding to Target Tissues. Gen. Comp. Endocrinol. 166, 447–454. doi: 10.1016/j.ygcen.2009.12.011
Claeys I., Breugelmans B., Simonet G., Franssens V., Broeck J. V. (2006). Regulation of Schistocerca Gregaria Neuroparsin Transcript Levels by Juvenile Hormone and 20-Hydroxyecdysone. Arch. Insect Biochem. Physiol. 62, 107–115. doi: 10.1002/arch.20127
Claeys G., Simonet B., Breugelmans S., Van S. V., Franssens F., Sas A., et al. (2010). Quantitative Real-Time RT-PCR Analysis in Desert Locusts Reveals Phase Dependent Differences in Neuroparsin Transcript Levels. Insect Mol. Biol. 14, 415–422. doi: 10.1111/j.1365-2583.2005.00572.x
Draetta G., Luca F., Westendorf J., Brizuela L., Ruderman J., Beach D. (1989). Cdc2 Protein Kinase I Complexed With Both Cyclin A and B: Evidence for Proteolytic Inactivation of MPF. Cell 56, 829–838. doi: 10.1016/0092-8674(89)90687-9
Fang J. J., Qiu G. F. (2009). Molecular Cloning of Cyclin B Transcript With an Unusually Long 3’ Untranslation Region and its Expression Analysis During Oogenesis in the Chinese Mitten Crab, Eriocheir Sinensis. Mol. Bio. Rep. 36, 1521–1529. doi: 10.1007/s11033-008-9346-9
Feng H., Dong Y. T., Liu X., Qiu G. F. (2020). Cyclin B Protein Undergoes Increased Expression and Nuclear Relocation During Oocytes Meiotic Maturation of the Freshwater Prawn Macrobrachium Rosenbergii and the Chinese Mitten Crab Eriocheir Sinensis. Gene 758, 144955. doi: 10.1016/j.gene.2020.144955
Fournier B., Herault J. P., Proux J. (1987). Antidiuretic Factor From the Nervous Corpora Cardiaca of the Migratory Locust: Improvement of an Existing In Vitro Bioassay. Gen. Com. Endocrinol. 68, 49–56. doi: 10.1016/0016-6480(87)90059-1
Girardie J., Boureme D., Couillaud F., Tamarelle M., Girardie A. (1987). Anti-Juvenile Effect of Neuroparsin a, a Neuroprotein Isolated From Locust Corpora Cardiaca. Insect Biochem. 17, 977–983. doi: 10.1016/0020-1790(87)90106-5
Girardie J., Girardie A., Huet J. C., Pernollet J. C. (1989). Amino Acid Sequence of Locust Neuroparsins. FEBS Lett. 245, 4–8. doi: 10.1016/0014-5793(89)80179-6
Girardie J., Huet J. C., Atay-Kadiri Z., Ettaouil S., Delbecque J. P., Fournier B., et al. (1998). Isolation, Sequence Determination, Physical and Physiological Characterization of the Neuroparsins and Ovary Maturing Parsins of Schistocerca Gregaria. Insect Biochem. Mol. Biol. 28, 641–650. doi: 10.1016/s0965-1748(98)00053-8
Gulia-Nuss M., Eum J. H., Strand M. R., Brown M. R. (2012). Ovary Ecdysteroidogenic Hormone Activates Egg Maturation in the Mosquito Georgecraigius Atropalpus After Adult Eclosion or a Blood Meal. J. Exp. Biol. 215, 3758–3767. doi: 10.1242/jeb.074617
Kleijn D., Herp F. V. (1998). Involvement of the Hyperglycemic Neurohormone Family in the Control of Reproduction in Decapod Crustaceans. Inverterb. Reprod. Dev. 33, 263–272. doi: 10.1080/07924259.1998.9652637
Kyei A. B., Wang C. G., Sun C. B., Wang W., Chan S. (2020). Crustacean Neuroparsins-a Mini-Review. Gene 732, 144361. doi: 10.1016/j.gene.2020.144361
Li Y. Y. (2012). Study on Site of Vitellogenin Synthesis in the Shrimp Litopenaeus Vannamei and Macrobrachium Rosenbergii. J. Fisheries China. 36, 1025–1027. doi: 10.3724/SP.J.1231.2012.27893
Liu J., Liu A., Liu F., Huang H., Ye H. (2020). Role of Neuroparsin 1 in Vitellogenesis in the Mud Crab, S Paramamosain. Gen. Com. Endocrinol. 285, 113248.
Ma M., Gard A. L., Xiang F., Wang J., Davoodian N., Lenz P. H., et al. (2010). Combining in Silico Transcriptome Mining and Biological Mass Spectrometry for Neuropeptide Discovery in the Pacific White Shrimp Litopenaeus Vannamei. Peptides 31, 27–43. doi: 10.1016/j.peptides.2009.10.007
Marco H. G., Anders L., Gerad G. (2014). cDNA Cloning and Transcript Distribution of Two Novel Members of the Neuroparsin Peptide Family in a Hemipteran Insect (Nezara Viridula) and a Decapod Crustacean (Jasus Lalandii). Peptides 53, 97–105. doi: 10.1016/j.peptides.2013.10.013
Matsumoto S., Brown M. R., Suzuki A., Lea A. O. (1989). Isolation and Characterization of Ovarian Ecdysteroidogenic Hormones From the Mosquito, Aedes Aegypti. Insect Biochem. 19, 651–656. doi: 10.1016/0020-1790(89)90100-5
Meusy J. J. (1980). Vitellogenin, the Extraovarian Precursor of the Protein Yolk in Crustacea: A Review. Reprod. Nutr. Dev. 20, 1–21. doi: 10.1051/rnd:19800101
Moreau R., Gourdoux L., Girardie J. (1988). Neuroparsin: A New Energetic Neurohormone in the African Locust. Arch. Insect. Biochem. Physiol. 8, 135–145. doi: 10.1002/arch.940080207
Nogaro M., Radallah D., Guillon G., Fournier B. (1995). Hplc Analysis of Inositol Phosphate Isomers Induced by Neuroparsin in Rectal Epithelial Cells of the African Locust: Role of External Calcium. Insect Biochem. Mol. Biol. 25, 559–567. doi: 10.1016/0965-1748(94)00094-X
Okano-Uchida T., Sekiai T., Lee K. S., Okumura E., Tachibana K., Kishimoto T. (1998). In Vivo Regulation of Cyclin A/Cdc2 and Cyclin B/Cdc2 Through Meiotic and Early Cleavage Cycles in Starfish. Dev. Biol. 197, 39–53. doi: 10.1006/dbio.1998.8881
Okuno A., Yang W. J., Jayasankar V., Saido-Sakanaka H., Huong D. T., Jasmani S., et al. (2002). Deduced Primary Structure of Vitellogenin in the Giant Freshwater Prawn, Macrobrachium Rosenbergii, and Yolk Processing During Ovarian Maturation. J. Exp. Zool. 292, 417–429. doi: 10.1002/jez.10083
Phinyo M., Visudtiphole V., Roytrakul S., Phaonakrop N., Jarayabhand P., Klinbunga S. (2013). Characterization and Expression of Cell Division Cycle 2 (Cdc2) mRNA and Protein During Ovarian Development of the Giant Tiger Shrimp Penaeus Monodon. Gen. Comp. Endocrinol. 193, 103–111. doi: 10.1016/j.ygcen.2013.07.012
Predel R., Neupert S., Derst C., Reinhardt K., Wegener C. (2018). Neuropeptidomics of the Bed Bug Cimex Lectularius. J. Proteome. Res. 17, 440–454. doi: 10.1021/acs.jproteome.7b00630
Qiao H., Xiong Y., Jiang S., Zhang W., Fu H. (2020). Three Neuroparsin Genes From Oriental River Prawn, Macrobrachium Nipponense, Involved in Ovary Maturation. Biotech 10, 537. doi: 10.1007/s13205-020-02531-8
Qiu G.-F., Ramachandra R., Yao J. B. (2008). Molecular Characterization and Expression Profiles of Cyclin B1, B2 and Cdc2 Kinase During Oogenesis and Spermatogenesis in Rainbow Trout (Oncorhynchus Mykiss). Anim. Reprod. science. 105, 209–225. doi: 10.1016/j.anireprosci.2007.03.005
Riehle M. A., Garczynski S. F., Crim J. W., Hill C. A., Brown M. R. (2002). Neuropeptides and Peptide Hormones in Anopheles Gambiae. Science 298, 172–175. doi: 10.1126/science.1076827
Tanaka Y. (2021). “Neuroparsin-ScienceDirect,” in Handbook of Hormones, (Amsterdam, Netherlands: Elsevier) 2nd ed., vol. 2. , 761–763. doi: 10.1016/B978-0-12-820649-2.00204-7
Tiu S. H., Benzie J., Chan S. M. (2008). From Hepatopancreas to Ovary: Molecular Characterization of a Shrimp Vitellogenin Receptor Involved in the Processing of Vitellogenin. Biol. Reprod. 79, 66–74. doi: 10.1095/biolreprod.107.066258
Tiu S. H. K., Chan S. M. (2007). The Use of Recombinant Protein and RNA Interference Approaches to Study the Reproductive Functions of a Gonad-Stimulating Hormone From the Shrimp Metapenaeus Ensis. FEBS J. 274, 4385–4395. doi: 10.1111/j.1742-4658.2007.05968.x
Tomy S., Saikrithi P., James N., Balasubramanian C. P., Panigrahi A., Otta S. K., et al. (2016). Serotonin Induced Changes in the Expression of Ovarian Gene Network in the Indian White Shrimp, Penaeus Indicus. Aquaculture 452, 239–246. doi: 10.1016/j.aquaculture.2015.11.003
Tsutsui N., Kobayashi Y., Izumikawa K., et al. (2020). Transcriptomic Analysis of the Kuruma Prawn Marsupenaeus Japonicus Reveals Possible Peripheral Regulation of the Ovary. Front. Endocrinol. 11. doi: 10.3389/fendo.2020.0054
Veenstra J. A. (2016). Similarities Between Decapod and Insect Neuropeptidomes. PeerJ 4, e2043. doi: 10.7717/peerj.2043
Xie J., Wen J. J., Chen B., Gui J. F. (2001). Differential Gene Expression in Fully-Grown Oocytess Between Gynogenetic and Gonochoristic Crucian Carps. Gene 271, 109–116. doi: 10.1016/S0378-1119(01)00491-7
Yang S. P., He J. G., Sun C. B., Chan S. F. (2014). Characterization of the Shrimp Neuroparsin (MeNPLP): RNAi Silencing Resulted in Inhibition of Vitellogenesis. FEBS Open Bio. 4, 976–986. doi: 10.1016/j.fob.2014.09.005
Keywords: neuroparsin, vitellogenin, Macrobrachium rosenbergii, reproduction, cysteine-rich protein
Citation: Ao CM, Shi LL, Wang W, Wang CG and Chan SF (2022) Neuroparsin 1 (MrNP1) and Neuroparsin 2 (MrNP2) Are Involved in the Regulation of Vitellogenesis in the Shrimp Macrobrachium rosenbergii. Front. Mar. Sci. 9:917274. doi: 10.3389/fmars.2022.917274
Received: 11 April 2022; Accepted: 30 May 2022;
Published: 20 July 2022.
Edited by:
Heinrich Dircksen, Stockholm University, SwedenReviewed by:
Isam Khalaila, Ben-Gurion University of the Negev, IsraelWenying Shen, Shaoxing University, China
Copyright © 2022 Ao, Shi, Wang, Wang and Chan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Siuming F. Chan, c2l1bWluZzU3M0BzaW5hLmNvbQ==; Cheng Gui Wang, bG9uZ3Nob3JlQDE2My5jb20=
 Chun Mei Ao
Chun Mei Ao Li Li Shi
Li Li Shi Wei Wang
Wei Wang Cheng Gui Wang
Cheng Gui Wang Siuming F. Chan
Siuming F. Chan