- 1Climate Change Cluster, University of Technology Sydney, Ultimo, NSW, Australia
- 2Centre for Marine Science and Innovation, University of New South Wales Sydney, Kensington, NSW, Australia
- 3School of Life Sciences, University of Technology Sydney, Ultimo, NSW, Australia
- 4Australian Institute of Marine Science, Townsville, QLD, Australia
Reef-building corals are among the largest producers of dimethylsulfoniopropionate (DMSP), an essential compound in marine biogeochemical cycles. DMSP can be catabolised in coral mucus by a wide diversity of coral-associated bacteria, where it can either be demethylated, leading to the incorporation of sulfur and carbon into bacterial biomass – or cleaved by lyases, releasing the climatically-active gas dimethyl sulfide (DMS). It has been demonstrated that thermal stress increases DMSP concentrations in many coral species, however the effect of increased DMSP availability on coral-associated bacteria has not been explored. Here we performed thermal stress experiments to examine how changes in DMSP availability impact bacterial degradation pathways in the mucus of Acropora millepora. DMSP concentrations increased with temperature, reaching a maximum of 177.3 μM after 10 days of heat stress, which represents the highest concentration of DMSP recorded in any environment to date. Bacterial communities in coral mucus were significantly different from the surrounding seawater, yet they did not vary significantly between temperature or time. However, during thermal stress, when DMSP concentrations increased, a significant increase in the abundance of both the demethylation gene dmdA and the cleavage gene dddP were recorded. Importantly, our results show that for the highest DMSP concentrations recorded (above 30 μM), the cleavage pathway became more abundant than the demethylation pathway. This suggests that under high DMSP concentrations characteristic of heat stress, a larger fraction of the DMSP pool in the coral mucus is likely catabolised through the DMS-producing cleavage pathway.
Introduction
Dimethylsulfoniopropionate (DMSP) is a critical compound in the marine sulfur and carbon cycles (Sievert et al., 2007) and an essential chemical currency in microbial interactions (Kiene et al., 2000). Global DMSP production is likely to exceed one billion tons of sulfur per year (Howard et al., 2006), but the distribution of this compound in the ocean is not homogenous. Although DMSP is present in seawater at low nanomolar concentrations (oceanic average: 16.91 ± 22.17 nM; see Kettle et al., 1999), specific marine environments have been identified as hotspots, such as highly productive polar waters (Trevena et al., 2003), or coral reefs (Hill et al., 1995). Within tropical coral reefs, large concentrations of DMSP have been recorded in many benthic organisms, including macroalgae (Van Alstyne et al., 2007), giant clams (Hill et al., 2000), soft corals (Haydon et al., 2018) and reef-building corals (Broadbent et al., 2002), and their microalgal symbionts (phylum Dinoflagellata; Caruana and Malin, 2014). DMSP concentrations in these different benthic organisms can vary by more than two orders of magnitude (Broadbent et al., 2002; Tapiolas et al., 2013), but to date the highest concentration ever measured (54 µM; or more than three orders of magnitude higher than the global seawater average) was recorded from the mucus of corals from the genus Acropora (Broadbent and Jones, 2004).
Coral mucus is a viscous mixture secreted by specialised epithelial cells, forming a coating over the polyps that is important for many aspects of coral biology (Meikle et al., 1988; Bythell and Wild, 2011). The surface mucus layer protects corals against desiccation at low tide (Brown and Bythell, 2005), as well as sudden changes in environmental conditions (e.g., temperature, salinity) (Piggot et al., 2009), while allowing gas and metabolite exchanges (Bythell and Wild, 2011), it is key to feeding and cleansing processes (Brown and Bythell, 2005) and acts as a significant input of carbon to reef waters that sustains other benthic organisms (Wild et al., 2004). The chemical composition of the mucus is variable between coral species (Ducklow and Mitchell, 1979; Meikle et al., 1988), but one commonality is the presence of large glycoproteins giving the mucus its gel-like texture (Bythell and Wild, 2011). Coral mucus has also been described as the first line of defence against pathogens (Ritchie, 2006a; Shnit-Orland and Kushmaro, 2009), as it is densely populated by specific bacterial communities (Garren and Azam, 2010), some of which can degrade DMSP (Raina et al., 2009; Frade et al., 2015).
DMSP is an important nutrient source for marine bacteria, contributing significantly to their sulfur (up to 95%; Zubkov et al., 2004) and carbon (up to 15%; Simó et al., 2002) requirements. Degradation of DMSP by bacteria occurs through two major pathways producing either methanethiol (MeSH; demethylation) or dimethyl sulfide (DMS; cleavage) as end products (Reisch et al., 2011; Sun et al., 2016). The demethylation pathway is estimated to catabolise approximately 70% of dissolved DMSP (Reisch et al., 2011), leading to the assimilation of both carbon and sulfur into bacterial biomass (Kieber et al., 1996; Kiene et al., 1999; Simó, 2001). The gene encoding the first enzymatic step of this pathway was discovered in an Alphaproteobacterium from the Roseobacter clade and termed dmdA (Howard et al., 2006). The second degradation route is the cleavage pathway which leads to the release of DMS and is suspected to degrade the remaining 30% of dissolved DMSP. Eight different DMSP lyases, termed ddd+ have been identified so far (Zhang et al., 2019), among which dddP (Todd et al., 2009) is one of the most prevalent in the marine environment. Many marine bacteria harbour both degradation pathways; however, the environmental factors that dictate which pathway is used have remained hypothetical for more than 20 years (Kiene et al., 2000; Simó, 2001). Understanding which pathway marine bacteria preferentially use under specific conditions is critical because it directly affects how much DMS is produced and ultimately released into the atmosphere.
A long standing hypothesis in the DMSP field proposed that external DMSP concentrations regulate which degradation pathway is usedby bacteria (Kiene et al., 2000; Simó, 2001). According to this hypothesis, when DMSP availability is low (or if there is a high bacterial sulfur demand), most DMSP is expected to be catabolised through demethylation. On the other hand, when DMSP availability is high, the less costly cleavage pathway is likely used (Reisch et al., 2011). Recent experimental evidence confirmed that external DMSP concentrations dictate the relative expression of the two pathways with an increase in DMSP cleavage (leading to DMS formation) measured near the surface of coral-associated microalgae, where DMSP concentrations are the highest (Gao et al., 2020). A previous study comparing different coral species also revealed that when DMSP concentrations were high, a lower relative abundance of the gene encoding the first step of the demethylation pathway (dmdA) was present in bacterial communities (Frade et al., 2015). Environmental stressors, such as heat, are known to cause an increase in DMSP concentrations in some corals (e.g., members of the Acropora genus; see Raina et al., 2013; Deschaseaux et al., 2014; Gardner et al., 2017). However, we do not know the effect that a sudden rise in DMSP availability may have on coral-associated bacterial communities and on the pathways they use to catabolise this compound.
Here we investigated how variations in DMSP concentrations caused by heat stress affect the abundance of bacterial genes involved in the demethylation and cleavage pathways in Acropora millepora, a reef building coral widespread throughout the Indo Pacific. To achieve this goal, we simultaneously measured: i) DMSP concentrations; ii) bacterial community structure; and iii) prevalence of DMSP-degrading genes during thermal stress in the mucus of A. millepora.
Methods
Coral collection and experimental set-up
Five colonies of Acropora millepora were collected from Heron Island lagoon in the southern Great Barrier Reef, Australia (151°55´E, 23°26´S) and acclimatised at ambient seawater temperature (27°C) in a flow-through aquaria system under 50% shaded sunlight (daily average of 600 µmol photons m-2 s-1) for 3 days at 27 ± 0.5°C. Each colony was then split in half and divided between the control and the treatment tanks (1 tank per colony). Experimental tanks (30 L) were set up in a shaded flow-through aquaria with constant flow (5 L min-1) of lagoon seawater (approx. 27 ± 0.5°C). The ambient light intensity was measured every 5 min using PAR loggers (Odyssey) and temperature was recorded every 10 min (Thermochron, Australia). For the thermal stress treatment tanks (n = 5), temperature was increased 1°C per day over 5 days from the ambient temperature of 27°C to reach the target temperature of 32°C and then held for a further 2 days. Physiological parameters (FV/FM and ΔF/FM'; see below) were recorded daily for the duration of the experiment. Coral mucus, coral host and seawater samples were collected over 4 time points; T0 (day 3), T1 (day 6), T2 (day 8) and T3 (day 10) (Figure 1), for chlorophyll a, cell density, surface area, quantification of DMSP/DMSO, 16S rRNA gene sequencing and identification of dmdA and dddP degradation genes.
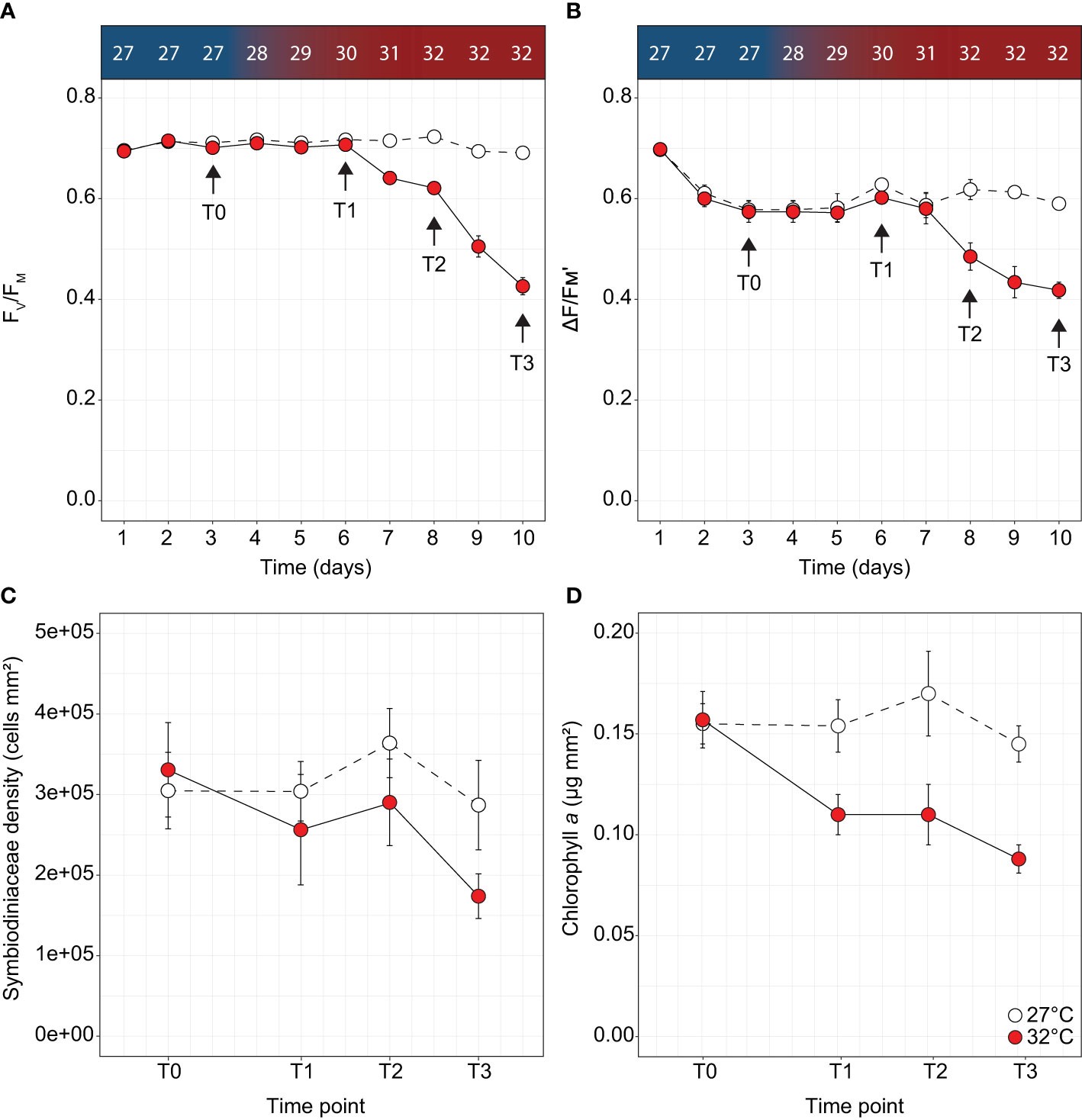
Figure 1 Physiological parameters measured over time for Acropora millepora under thermal stress including (A) maximum quantum yield of photosystem II (FV/FM), (B) effective quantum yield of photosystem II (ΔF/FM'), (C) Symbiodiniaceae density and (D) chlorophyll a for the control (white circles) and treatments (red circles). Letters indicate significant differences between timepoints for the treatment. Temperature increases indicated along the top panel for corresponding days and sampling timepoints. Averages (± SE) are shown (n = 4-5).
Photochemical efficiency of PSII
Using a MiniPAM (Walz GmbH, Effeltrich, Germany; MI: 8, Gain: 5, SI: 10, SW: 0.8s) chlorophyll a fluorescence was measured daily at midday (effective quantum yield of PSII; ΔF/FM') and just after sunset (maximum quantum yield of PSII; FV/FM) to monitor photo-physiological stress of each colony, with 2 technical replicate measurements from distinct branches. At T3, a DivingPAM (Walz GmbH, Effeltrich, Germany; MI: 8 Gain: 7 SI: 8 SW: 0.8s) was used to conduct rapid light curves (RLC; see Ralph and Gademann, 2005) in the first hour of daylight, using the same settings as above. The RLC protocol consisted of a low-light acclimated (c.a. 5 µmol photons m-2 s-1) FV/FM measurement to initiate the RLC, followed by ΔF/FM' measurements under actinic light at eight 30 second intervals of increasing intensity (98, 162, 240, 325, 480, 610, 971, and 1,359 µmol photons m-2 s-1). The saturating irradiance (EK), an estimate of the irradiance where PSII transitions from a light-limited to a light-saturated state, and the maximum photochemical efficiency of PSII (FQ/FM(max)) were described using equation 1 (Hennige et al., 2008) where E = irradiance.
To detect whether utilisation of alternative pathways of excitation energy dissipation differed across temperature treatments, the extent of light dependant photochemical quenching ([1 – C], equation 2) and non-photochemical quenching ([1 – Q], equation 3) were calculated at each step of the RLC (Suggett et al., 2015; Nitschke et al., 2018).
Cell density, chlorophyll a and surface area
Coral tissue was removed from the skeleton of coral fragments using an airgun in 5 mL FSW (0.2 μm). The tissue slurry was concentrated via centrifugation at 3000 g for 10 min. The algal pellets were resuspended in 5 mL FSW (0.2 µm), and homogenised. A 3 mL subsample was then centrifuged at ~3,600 g for 4 min and resuspended in 3 mL of 90% acetone and left at 4°C in the dark for 24 h before spectrophotometric chlorophyll determination. Concentrations of chlorophyll a were calculated using the equations from Ritchie (2006b). The remaining 2 mL subsample was used for cell density measurements using a haemocytometer (n = 8). The bare coral skeletons were dried, and the surface area of each coral fragment was calculated using the paraffin wax technique (Stimson and Kinzie, 1991; Veal et al., 2010).
Sample collection
Coral mucus was collected by exposing the colony to air for 2 min (prompting mucus production from the epithelial cells and the gastrovascular cavity; see Sweet et al., 2011). Coral colonies were held inverted to allow excess seawater and mucus to drip off the branches. After 2 min air exposure, each colony was dipped back into the water for 1 min, removed from the seawater, and then held inverted in the air for a further 15 s to allow excess water to drip from the branches. The resulting mucus secreted during air exposure was then collected from the branch tips using a sterile 10 mL syringe with a 21″G sterile needle. For each colony, 4-5 mL mucus was collected in a falcon tube, of which 2 mL was immediately added to 4 mL analytical grade methanol (95%), sealed with parafilm and stored at -20°C in the dark for nuclear magnetic resonance (NMR) spectroscopy. An additional 2 mL of coral mucus was filtered using a glass filter tower (Cole-Parmer Instrument Company, USA) onto membrane filter (Whatman; 25 mm × 0.2 µm pore size), then the filter was snap frozen in liquid nitrogen and stored in cryovials at -80°C for 16S rRNA gene amplicon sequencing and quantitative PCR (qPCR) analyses. To sample the coral host, a 3 cm fragment was removed from each colony using sterile bone cutters; the fragment tip was discarded, and the middle section added to 3 mL methanol in a falcon tube, sealed with parafilm and stored in the dark at -20°C for NMR analysis. Water samples were also taken at each time-point (n = 3), where 1 L was collected from each tank in a large glass Schott bottle at 20 cm distances from the coral colony and filtered using a vacuum pump (Capex 8C, Charles Austen Pumps Ltd, Surrey, UK) onto 45 mm × 0.45 µm filters (Whatman). The filters were snap frozen and stored in cryovials at -80°C.
Quantification of DMSP and DMSO
Coral host fragments and mucus samples in methanol were stored at -20°C until processing at the Australian Institute of Marine Science, Townsville, Australia (AIMS). Coral fragments were further extracted using sonication on ice (bath at 40 kHz) in 1 mL of HPLC-grade methanol for 5 min. The two extracts from the coral host were pooled and dried overnight (8 h) using a concentrator (Savant SpeedVac SC210A, Thermo Scientific, USA). The coral mucus samples were also dried overnight (14 h) in the concentrator, then in a freeze drier (Dynavac FD12 Perogon Technologies, Australia) for a further 4 h to ensure complete dryness. The dried extracts from the host and mucus were resuspended in a mixture of deuterated methanol (CD3OD; 750 µL) and deuterium oxide (D2O; 250 µL), vortexed to solubilise the compounds and then centrifuged for 10 min to pellet the debris. An 800 µL aliquot of the particulate free extract was transferred into a 5 mm NMR tube (Norell 509-UP) and analysed immediately by 1H NMR following the method described in Tapiolas et al. (2013). Spectra were recorded on a 600 MHz NMR spectrometer (Bruker Avance, Germany) with a TXI cryoprobe, using the Bruker TOPSPIN 2.1 software, and referenced using CD3OD (δH 3.31). Concentrations of DMSP and DMSO were quantified using the ERETIC method (electronic reference to access in vivo concentrations; Akoka and Trierweiler, 2002), which electronically generates an external reference signal that was calibrated using a 2 mM stock solution of acrylate. After calibration, the concentration of each target compound was determined by comparing the signal intensities of well resolved non-exchangeable protons: (CH3)2SCH2CH2CO2 at δ2.95 ppm for DMSP and (CH3)2SO at δ2.73 ppm for DMSO (in the coral host and mucus samples) in a 0.20 ppm window against the intensity of the reference signal through signal integration (Akoka and Trierweiler, 2002). Coral skeletons remaining after extraction were soaked in 10% bleach overnight, dried at 60°C in an oven and used for surface area measurements using the paraffin wax technique (Stimson and Kinzie, 1991; Veal et al., 2010). The surface area of each fragment was then used to normalise NMR data. The coral mucus samples were normalised to the volume of mucus originally collected in methanol (2 mL).
DNA extraction, amplicon sequencing and processing
DNA was extracted from filters used to collect mucus and water samples using methods modified from Schauer et al. (2000). Briefly, lysis buffer (0.5 mL) and 75 µL lysozyme (100 mg/mL) were added to cryovials containing the filters and incubated for 1 h at 37°C. Subsequently, 100 µL SDS (25%) and 10 µL Proteinase K (20 mg/mL) were added and incubated for 1 hr at 55°C. The recovered lysate (600 µL) was extracted twice using equal volumes of phenol:chloroform:isoamyl alcohol (25:24:1, pH8), followed by chloroform:isoamyl alcohol (24:1) and centrifuged for 30 min at 4°C and 16,000g. DNA was precipitated from the recovered aqueous phase (top layer) with 500 µL ice cold isopropanol, left in the dark for 15 min and centrifuged at 16,000g for 30 min. The pellet was rinsed with 500 µL of 70% ethanol before being dried in a concentrator for 10 min to ensure all ethanol was removed. The remaining DNA pellet was resuspended in 40 µL sterile milliQ, and the purity of the DNA was assessed using a NanoDrop spectrophotometer (Thermo Scientific, USA). Aliquots were stored at -20°C until use.
To examine the composition of bacterial assemblages, the 16S rRNA gene was amplified using 27F/519R (V1-V3 region) primers. PCR was carried out using the HotStarTaq Plus Master Mix Kit (Qiagen, USA) using the following conditions: (i) 3 min at 94°C; (ii) 28 cycles, with each cycle consisting of 30 s at 94°C, 40 s at 53°C, and 1 min at 72°C; (iii) a final elongation step of 5 min at 72°C. PCR products were checked in 2% agarose gels, and samples were pooled in equal proportions based on molecular weight and DNA concentrations. Pooled samples were purified using Ampure XP beads (Beckman-Coulter, USA) and processed using the Illumina TruSeq DNA library protocol. Sequencing was performed on an Illumina MiSeq platform (2 × 300 cycles; Molecular Research LP; Shallowater, TX, USA).
Raw data was demultiplexed and primers were removed using cutadapt (Martin, 2011). The open-source software package DADA2 (Callahan et al., 2016) was used to produce a table of chimera-free amplicon sequence variants (ASVs). ASVs were taxonomically classified using the RDP Classifier against the SILVA v.138 (August, 2020) reference database (Quast et al., 2013). A phylogenetic tree was generated by sequence alignment with MAFFT (Katoh and Standley, 2013) and the tree was produced with FastTreeMP (Price et al., 2009) on the CIPRES v3.3 Science Gateway (Miller et al., 2010). ASV count and taxonomic data were imported using the phyloseq package v.1.28 (McMurdie and Holmes, 2013) for analysis in R v3.6.1. Sequences classified as chloroplast, mitochondria, eukaryota, and archaea, as well as known common reagent contaminants (Sheik et al., 2018), were removed before further analysis. Singleton ASVs, together with one seawater sample containing <1,000 reads, were removed from the dataset. Overall, the dataset comprised 17 samples (9 for mucus and 8 for seawater), resulting in 414,856 sequences with a mean length of 294 bp. After quality filtration, exclusion of chimeras, and specific retention of bacterial sequences, 1,873 ASVs (Supplementary Table 1) were obtained from the 17 samples. Samples were rarefied to 5,099 to account for the variability in sequencing depth between samples. The mean good’s coverage score was 99.81% ± 0.38 for the dataset indicating the sequencing depth was adequate to capture the majority of diversity in the samples.
Statistical analyses of the 16S rRNA gene sequencing data
Amplicon sequence variants (ASVs) were analysed with Phyloseq v1.28 (McMurdie and Holmes, 2013), Vegan v2.5.6 (Oksanen et al., 2007) and rstatix v0.7.0 (Kassambara, 2021). Alpha diversity indices (chao1 richness and the observed number of ASVs) were computed and a one-way ANOVA was run through rstatix for each diversity index after checking for homogeneity and normality using Levene’s and Shapiro’s tests, respectively. A principal coordinates analysis (PCoA) was used to visualise dissimilarities in microbial communities between sample types (mucus and seawater) and treatments (27°C and 32°C) using weighted unifrac distances, which considers the relative abundance of each ASV and integrates phylogenetic distance (Lozupone et al., 2011).
To statistically analyse differences in microbial community structure, ASV counts were Hellinger transformed to reduce the effects of numerically large values from very abundant taxa and weighted unifrac distances were computed. Permutational multivariate analysis of variance (PERMANOVA; n = 9,999 permutations, adonis/adonis2 function in Vegan) was performed to test for significant differences between mucus and seawater samples and homogeneity of dispersions around group centroids (i.e., variation) was assessed using PERMDISP (betadisper function in Vegan). Data were further analysed using a two-factor nested PERMANOVA to assess differences between temperature treatment as a fixed factor (two levels: 27°C and 32°C) and time point (two levels: T0 and T3) nested within sample type (mucus and seawater). Stacked bar graphs were plotted using ggplot2 v3.3.5 to phylum level and represent those bacterial taxa with a relative abundance above 1% across all samples.
DMSP degrading predictions of the bacterial communities
To determine if thermal stress induced an increase in the relative abundance of taxa harbouring the demethylation pathway (dmdA) or the cleavage pathway (dddP), we functionally predicted the abundance of these two genes in the bacterial communities from the mucus T3 samples. Briefly, the mucus samples were rarefied to 9,182 (lowest read depth for the mucus samples), and all ASVs were compared to genomes harbouring dmdA and dddP (Supplementary Table 2) derived from KEGG (Kyoto Encyclopedia of Genes and Genomes) and NCBI (National Center for Biotechnology Information). Each ASV with a query coverage ≥ 90% and a similarity ≥ 95% with the 16S rRNA gene of one of the dmdA or dddP-harbouring genomes was considered as a putative DMSP degrader.
Amplification and identification of dmdA and dddP genes
Quantitative PCR (qPCR) was used to determine the total abundance of bacterial 16S rRNA genes and genes involved in marine DMSP cycling. Gene abundance was quantified using an automated Liquid Handling robot (epMotion 5075l) on a Bio-Rad CFX Touch Real-Time PCR Detection System. A range of dmdA subclades were initially screened (A/1, A/2, B/4, C/2, D/1 and D/3, E/2; Varaljay et al., 2010) using qPCR, but only a few successfully amplified in the samples (A/1, D/3, E/2), the most successful being dmdA A/1. All sample plates included a triplicate, six-point calibration curve constructed from a known amount of amplicon DNA measured by Qubit (according to the manufacturer’s instructions), followed by five successive 10-fold dilutions and negative template controls of nuclease-free water.
Absolute quantification of the bacterial 16S rRNA gene was performed using the 16S rRNA specific primers BACT1369F (5’ – CGGTGAATACGTTCYCGG – 3’) and PROK1492R (5’ – GGWTACCTTGTTACGACTT – 3’) and a TaqMan probe TM1389F (5’ – CTTGTACACACCGCCCGTC – 3’) (Suzuki et al., 2000; Supplementary Table 3) with following cycling conditions: 95°C for 3 minutes followed by 39 cycles of 95°C for 30 s and 56°C for 60 s with an extension at 72°C for 30 s. Each individual PCR reaction volume was 5 µL and contained 2.5 µL iTaq Universal probes SMX (Bio-Rad), 0.2 µL of each forward and reverse 16S rRNA gene specific primers (10 µM), 0.1 µL of Taqman Probe Mix, TM1389F (5’ – CTTGTACACACCGCCCGTC – 3’) (10 µM), 1 µL of nuclease-free water and 1 µL of template DNA. The relative abundance of bacterial DMSP-degrading genes was acquired by normalising their copy numbers to the copy number of bacterial 16S rRNA gene, however, it should be noted that some bacterial genomes have multiple copies of the 16S rRNA gene (Cui et al., 2015).
Absolute quantification of the genes encoding DMSP catabolism were performed using primers for the DMSP cleavage gene dddP_874F (5’ – AAYGAAATWGTTGCCTTTGA – 3’) and dddP_971R (5’ – GCATDGCRTAAATCATATC – 3’) (Levine et al., 2012) and DMSP demethylation gene dmdA A/1-spFP (5’ – ATGGTGATTTGCTTCAGTTTCT – 3’) and A/1-spRP (5’ – CCCTGCTTTGACCAACC – 3’) (Varaljay et al., 2010; Supplementary Table 3). Analysis of DMSP degradation genes were performed on three technical replicates of the following mixture: 2.5 µL of 2 × Sensifast SYBR Hi-ROX mastermix, 0.2 µL of forward primer (10 µM initial), 0.2 µL of reverse primer (10 µM initial), 0.1 µL nuclease-free water and 2 µL of DNA template (diluted between 1:1 and 1:20). Amplification of the DMSP cleavage gene dddP and demethylation gene dmdA A/1, consisted of an initial denaturation step of 95°C for 5 min, followed by 40 cycles of 95°C for 30 s, 41°C (dddP) and 53°C (dmdA) for 30 s and 72°C for 30 s. To differentiate target amplicons from non-specific products, a dissociation melt curve was generated after each reaction.
Statistical analyses of the qPCR data
Quantitative PCR data was analysed with rstatix v0.7.0 (Kassambara, 2021) in R v3.6.1. Two-way repeated measures ANOVAs were run for the mucus datasets to determine the interaction between temperature treatment (27°C and 32°C) and time point (T0, T1, T2 and T3) after checking for homogeneity and normality using Levene’s and Shapiro’s tests, respectively. Tukey’s post-hoc tests were used to determine differences between groups. Pairwise comparisons, using t-tests, were then run for each timepoint, for the coral host and mucus samples. Throughout the manuscript, all reported values are mean ± SE, unless mentioned otherwise.
Results
Thermal bleaching responses and a sharp increase in coral mucus DMSP concentrations
Measures of Symbiodiniaceae PSII photochemical efficiency, cell densities, and chlorophyll content indicate A. millepora corals subjected to 32°C were in the early stages of a thermal bleaching response. Ten days after the onset of thermal stress, dark-adapted maximum quantum yields decreased by 39% relative to controls (FV/FM; Two-way ANOVA; F9,180 = 53.35, p < 0.001; Supplementary Table 4 and Figure 1A) from 0.694 ± 0.007 (day 1) to 0.426 ± 0.017 (day 10). In addition, a 38% decrease in effective quantum yield was also recorded (ΔF/FM'; F8,162 = 5.67, p < 0.001; Supplementary Table 4 and Figure 1B) from 0.698 ± 0.005 (day 1) to 0.434 ± 0.031 (day 10). In RLCs performed at day 10, no difference in EK was observed between treatments (Figure S1A), however FQ/FM(max) was significantly different, with decreases from 0.72 ± 0.01 to 0.66 ± 0.01 in thermally stressed corals (One-way ANOVA; F1,8 = 12.64, p < 0.05, Figure S1B and Supplementary Table 4). No differences were observed in [1 - C]; however, stressed corals exhibited significantly increased utilisation of [1 - Q] at all RLC irradiance steps (One-way ANOVA, p < 0.05 for all irradiances, Supplementary Table 4 and Figure S1C), indicating increased reliance on non-photochemical quenching. Although no difference in Symbiodiniaceae density was recorded between treatments (Figure 1C), chlorophyll a in thermally stressed corals decreased by 43% from 0.157 ± 0.014 µg chl a mm-2 to 0.088 ± 0.007 µg chl a mm-2 (Two-way ANOVA; F3,19 = 5.72, p = 0.007, Supplementary Table 4 and Figure 1D).
At the beginning of the thermal stress experiment (T0), DMSP concentrations in the coral mucus secreted after air exposure averaged 4.24 ± 1.35 µM across both treatments but started to sharply increase when the temperature reached 32°C by T2 (Figure 2A) to 71.23 ± 12.12 µM. By T3 this value reached 83.15 ± 27.87 µM, corresponding to a 15.6-fold increase over the controls (F3,32 = 7.031, p < 0.001, Supplementary Table 4 and Figure 2A). We also measured a 2.9-fold increase for DMSO (F3,32 = 3.954, p < 0.02, Supplementary Table 4 and Figure 2B), from 0.79 ± 0.15 µM at T0 to 1.77 ± 0.34 µM at T3 under elevated temperature. Conversely, in the coral host, no difference was detected for DMSO (p = 0.570; Supplementary Table 4 and Figure 2D) between control and thermally-stressed samples, while DMSP concentrations significantly differed between control and treatment by T2, and were 2.7-fold higher in thermally-stressed samples by T3 (F3,32 = 3.85, p = 0.018, Supplementary Table 4 and Figure 2C).
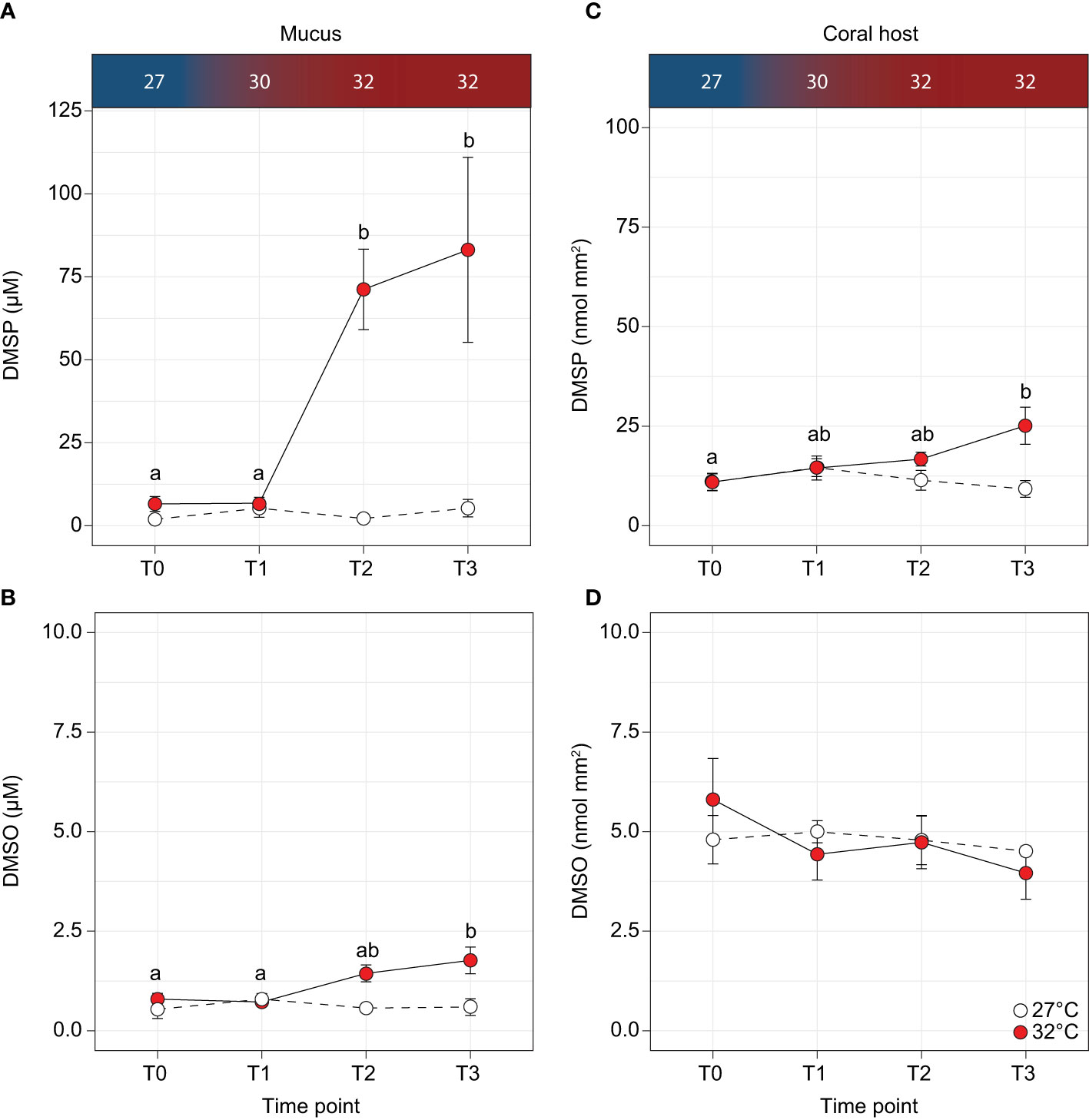
Figure 2 Concentrations of (A, C) dimethylsulphoniopropionate (DMSP) and (B, D) dimethylsulphoxide (DMSO) in the Acropora millepora mucus and coral host for the control (white circles) and temperature treatments (red circles). Letters indicate significant differences between timepoints for the treatments. Temperatures are indicated along the top panel for corresponding sampling time-points. Averages (± SE) are shown (n = 5).
Bacterial community structure differs across mucus and seawater
Both the observed number of ASVs and their richness (Chao1) were significantly higher in the mucus samples (observed: 201.56 ± 45.97; Chao1: 206.24 ± 48.33) compared with the surrounding seawater (observed: 77.70 ± 15.21, [F1,15 = 5.83, p = 0.029]; Chao1: 77.86 ± 15.19, [F1,15 = 5.78, p = 0.030]; Figure 3A). The overall microbial community structure of the two environments (coral mucus and seawater) also showed limited overlap (principal coordinates analysis (PCoA); Figure 3B). Mucus samples were more tightly clustered than the seawater samples; however, group dispersion was not significantly different (PERMDISP, p = 0.959, Figures 3B, C). Microbial community composition significantly varied between sample types (PERMANOVA, F = 3.85, R2 = 0.20, p < 0.005), and between sample types when partitioned between timepoints and temperature treatments (PERMANOVA, F = 2.49, R2 = 0.21, p = 0.011). However, no differences in microbial community composition in mucus samples were detected between temperatures (PERMANOVA, F = 0.76, R2 = 0.05, p = 0.613) or timepoints (PERMANOVA, F = 1.62, R2 = 0.10, p = 0.115), and variance was similar between groups for both (PERMDISP, p = 0.590 and p = 0.415, respectively).
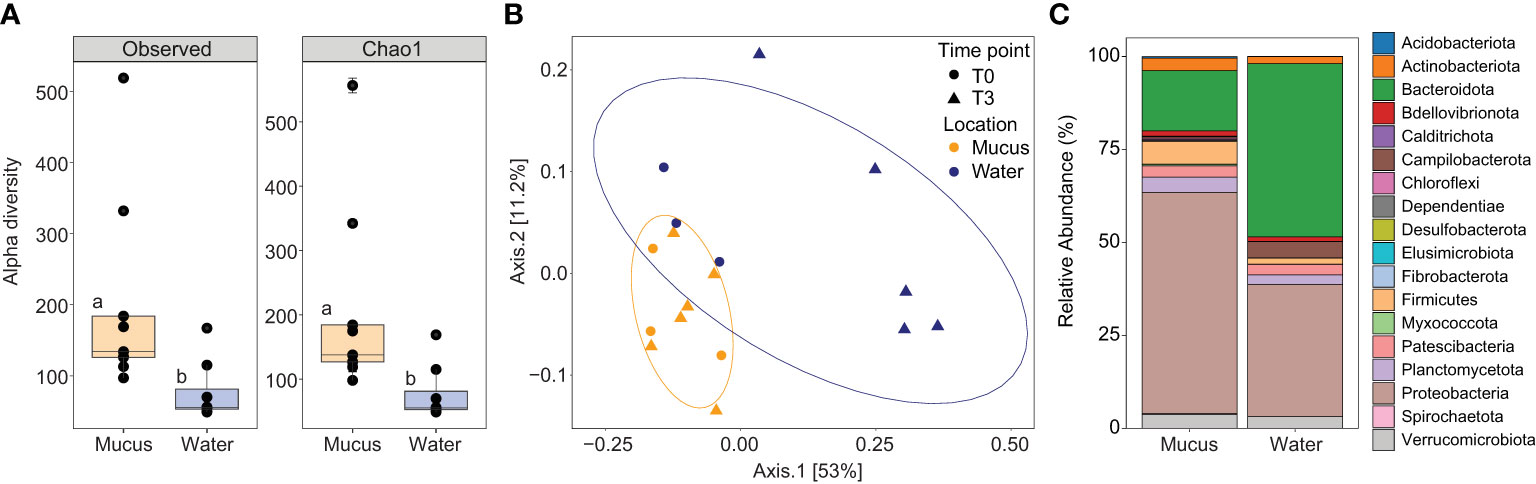
Figure 3 (A) Alpha diversity indices including the number of observed ASVs and species richness (Chao1) for the mucus (orange boxes), and seawater (blue boxes) samples. Data was rarefied to the smallest sample size depth of 5,099. Letters indicate post-hoc groupings. (B) Principal coordinate analysis (PCoA) based on weighted unifrac distance of the 16S rRNA gene amplicon sequences for the mucus (orange symbols) and seawater (dark blue symbols) at timepoint T0 (circle symbols) and T3 (triangle symbols). Ellipses show 80% confidence intervals. (C) Relative abundance of bacterial taxa plotted at the phylum level for the mucus and seawater samples, showing total relative abundance of each phylum >1%.
The mucus samples were dominated by Proteobacteria from the Gammaproteobacteria (40.08% ± 5.04%), Alphaproteobacteria (19.40% ± 2.67) and Bacteroidia (16.10% ± 3.66) classes (Figure 3C). More specifically, three families represented 34.35% of the communities: Rhodanobacteraceae (20.63% ± 2.71%), Flavobacteriaceae (8.49% ± 2.27) and Rhodobacteraceae (5.23% ± 1.51; Supplementary Table 5A and Figure 3C). Conversely, the seawater samples were dominated by Bacteroidia (47%), Alphaproteobacteria (26%), and Gammaproteobacteria (10%; Figure 3C and Supplementary Table 5B). A predictive analysis was carried out on the samples collected during the last time point of the experiment (T3; day 10) to determine the taxonomic composition of DMSP degraders in coral mucus, together with their relative abundance in response to the large increase in DMSP concentrations recorded in the thermally stressed colonies. According to our analysis, an average of 5% of the bacterial communities harboured the demethylation gene dmdA in the control samples, the most abundant genera belonging to the Pelagibacter, Phaeobacter and Cognatishimia (Supplementary Table 2 and Figure 4). Importantly, the relative abundance of the bacteria putatively harbouring dmdA significantly increased in the heat-stressed samples to reach an average of 6.7% of the communities (t-test; t = -2.86, df = 4, p = 0.045). In comparison, 5.4% of the communities putatively harboured the cleavage gene dddP, with the Gammaproteobacteria Caballeronia, Acinetobacter and the Alphaproteobacteria Phaeobacter being the most abundant. However, heat stress did not significantly affect the relative abundance of dddP in the communities (t-test; t = -1.18, df = 4, p = 0.302; Figure 4).
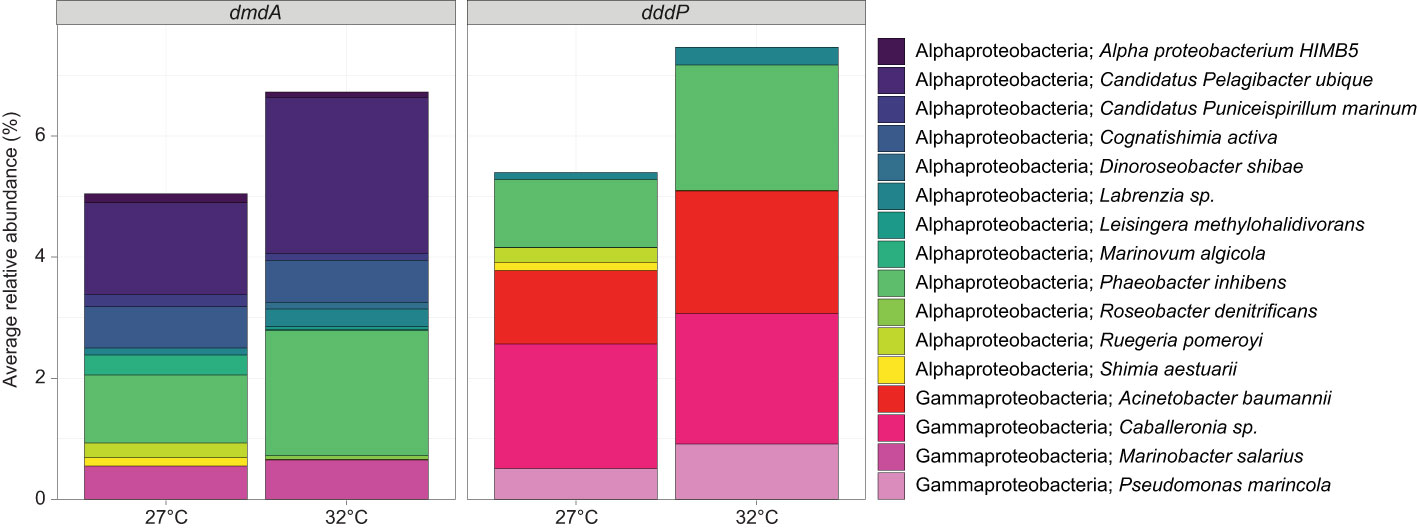
Figure 4 Average relative abundance (%) of ASVs that were taxonomically assigned to bacteria (>95% similarity) harbouring the DMSP-degrading genes dmdA and/or dddP for mucus samples at timepoint T3, for the control (27°C) and temperature (32°C) treatments. Data were rarefied to the smallest sample size depth of 9,182.
The abundance of DMSP degrading genes increase in coral mucus under thermal stress
To go beyond the functional predictions based on 16S rRNA gene similarities, we quantified the abundance of both dmdA A/1 and dddP in coral mucus throughout the experiment. The demethylation gene dmdA A/1 significantly increased in abundance in the thermally stressed fragments between T2 and T3 (ANOVA; p = 0.0144; Supplementary Table 2 and Figure 5A). At T3, the abundance of dmdA A/1 (per 16S rRNA gene copy) was 1.5-fold higher in the mucus from fragments exposed to 32°C compared to the controls (F3,23 = 4.89, p = 0.009; Supplementary Table 2 and Figure 5A). The abundance of the cleavage gene dddP in coral mucus exhibited a similar but more pronounced increase with temperature, with an 8.6-fold increase recorded between T1 and T2 (ANOVA; p < 0.001; Supplementary Table 2 and Figure 5B). At T2, the abundance of the dddP gene was 2.95-fold higher in the thermally stressed fragments than the controls. This difference slightly decreased by T3, with 2.3-fold more dddP gene copies in the thermally stressed fragments compared with the controls.
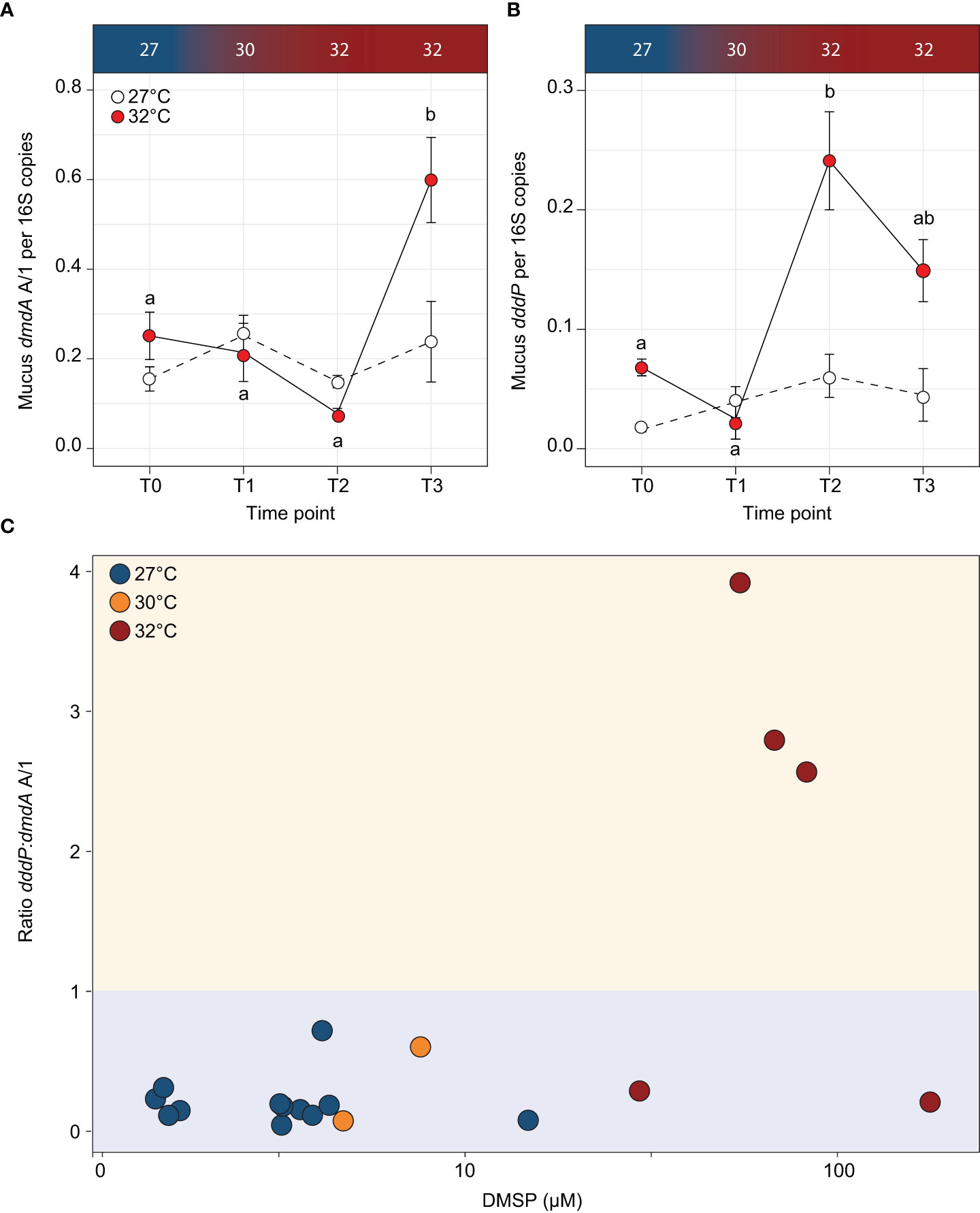
Figure 5 Abundance of DMSP degrading genes (A) dmdA A/1 and (B) dddP per 16S copies for each timepoint for the control (white circles) and temperature treatments (red circles) in the Acropora millepora mucus. Letters indicate significant differences between timepoints for the treatments. Averages (± SE) are shown (n = 3-5). (C) Ratio of dddP:dmdA A/1 (cleavage:demethylation pathway) to concentration of DMSP in Acropora millepora mucus for experimental temperatures 27°C (T0; blue circles), 30°C (T1; orange circles) and 32°C (T2 and T3; red circles). Ratio >1 (yellow shading) indicates when the cleavage pathway is higher than demethylation. When the ratio of dddP:dmdA is <1 (blue shading), the demethylation pathway is higher than the cleavage pathway. Temperature treatments are indicated along the top panel for corresponding sampling timepoints. DMSP concentration on the x-axis is on a log10 scale.
To determine if DMSP concentrations affected the prevalence of a specific pathway, we calculated the ratio of dddP:dmdA A/1 in each sample collected during the experiment (Figure 5C). For DMSP concentrations under 29.4 µM, the dddP:dmdA ratio was consistently less than 1, indicating that the demethylation pathway was more abundant in the mucus microbiome than the cleavage pathway. However, when DMSP concentrations were above 29.4 µM, 75% of the samples had a ratio larger than 1, which means that the cleavage pathway became more abundant than demethylation. Based on these data, a significant association was identified between DMSP concentrations and dddP:dmdA ratio (Fisher’s exact test; p = 0.002; Figure 5C).
Discussion
DMSP is a key molecule in the marine sulfur cycle (Sievert et al., 2007), which is produced in large amounts by reef-building corals (Broadbent et al., 2002; Broadbent and Jones, 2004). Thermal stress is known to cause sudden increases in DMSP concentrations that have been linked to its antioxidant capabilities (Raina et al., 2013; Deschaseaux et al., 2014; Gardner et al., 2017), yet the subsequent response of coral-associated microbes that can catabolise this compound remains undefined. Our study aimed to identify how the increased availability of DMSP in Acropora millepora mucus during an ecologically relevant thermal stress affected the abundance of DMSP degrading bacteria and the genes they use to degrade this molecule. As predicted, elevated seawater temperature resulted in order of magnitude increase in DMSP concentration in the mucus layer of A. millepora. We subsequently identified putative DMSP degraders in the mucus secreted after air exposure and confirmed that the abundance of genes mediating the two DMSP degradation pathways increased with higher DMSP concentrations. Notably, at the highest DMSP concentrations recorded, the cleavage gene causing DMS production became more abundant than the demethylation gene, suggesting that a greater proportion of DMSP is catabolised through this route during heat stress, which has potentially significant implications given the climatic importance of DMS.
Elevated seawater temperature caused reductions in PSII photochemical efficiency (FV/FM) and chlorophyll content without a loss of Symbiodiniaceae cells. This, along with the increased reliance on non-photochemical quenching, indicates a nascent bleaching response where light energy is in excess and the integrity of the photosynthetic electron transport chain is compromised (Rochaix, 2011). A reduction in FV/FM is typical of thermally stressed Acropora symbionts at our sampling site (Heron Island) (Fisher et al., 2012; Gardner et al., 2017; Nitschke et al., 2018), which belong to Cladocopium of the C3 radiation (LaJeunesse et al., 2003; Fisher et al., 2012; Gardner et al., 2017). Cumulative thermal stress likely overwhelmed Symbiodiniaceae photosystem repair mechanisms (Takahashi et al., 2009), alternative electron flow pathways (e.g., the mehler reaction, see Roberty et al., 2014), and antioxidant systems (Gardner et al., 2017), leading to net production of reactive oxygen species (ROS) (Lesser, 2006; Lesser, 2011). Elevated cellular ROS emissions are characteristic of temperature sensitive Symbiodiniaceae cells (Suggett et al., 2008; Goyen et al., 2017; Buerger et al., 2020), and ROS leakage into host cells has been proposed as one of the mechanisms responsible for Cnidarian-Symbiodiniaceae dysbiosis (Lesser, 1997; Lesser, 2006; Lesser, 2011).
Coral mucus contains high concentrations of DMSP under normal conditions (Broadbent and Jones, 2004). In the present study, concentrations measured in the temperature treatments averaged 83 µM, more than 1.5-fold higher than previously reported for Acropora. In addition, increased temperature led to the highest concentrations yet reported in any environment (177 µM). Our observations of Symbiodiniaceae photosystem stress and concurrent increases in DMSP concentrations in the coral host aligns well with its putative role in photosystem repair (Archer et al., 2010) and also corroborate the ROS-detoxifying role of DMSP proposed for Acropora (Raina et al., 2013; Deschaseaux et al., 2014; Jones and King, 2015; Gardner et al., 2017) and demonstrated for other species (Sunda et al., 2002). Mucus has long been thought to play an essential role in a coral’s ability to adapt to environmental change (Reshef et al., 2006). The exceptionally high concentrations of DMSP measured here, together with a nutrient-rich cocktail of proteins, polysaccharides and lipids, make coral mucus an ideal environment for microbial growth (Wild et al., 2004; Tremblay et al., 2011), and as such, the maintenance of a healthy coral microbiome.
We found little overlap in the identity of the taxonomy of the bacterial communities between mucus and seawater samples, consistent with previous work showing that mucus harbour specific taxa (Rohwer et al., 2001; Frias-Lopez et al., 2002; Rohwer et al., 2002). The two most abundant classes of bacteria present in mucus samples were Alphaproteobacteria and Gammaproteobacteria, known to harbour DMSP-degrading genes (Howard et al., 2008; Varaljay et al., 2010). We matched the 16S rRNA gene of bacteria present in the mucus samples to genomes that harbour dmdA and dddP, to identify and quantify putative DMSP degraders in the control and heat-stressed samples. The majority of the dmdA-harbouring bacteria identified in mucus belong to the Alphaproteobacteria class and include many representatives of the Roseobacter clade, such as Phaeobacter, Ruegeria, Shimia, or Marinovum. These Roseobacters are commonly associated with reef-building corals (Huggett and Apprill, 2019; Luo et al., 2021; Kuek et al., 2022), and akin to their interaction with phytoplankton (Seyedsayamdost et al., 2011), their relationships with corals can range from mutualistic (Sharp et al., 2015; Freire et al., 2019; Miura et al., 2019) to pathogenic under environmental stress (Casey et al., 2015; Pollock et al., 2017). In addition, the oligotrophic bacterium Pelagibacter was also present among the dmdA-harbouring bacteria identified in coral mucus. Given that this DMSP-degrading bacterium is the most abundant microorganism in seawater (Giovannoni, 2017), and mucus viscosity is known to trap particles from seawater (Wild et al., 2004), is it possible that some of these oligotrophs get also trapped in mucus. Conversely, the majority of dddP-harbouring bacteria belonged to the Gammaproteobacteria class, most notably members of the genus Caballeronia. The functional role of this genus is unknown in corals, but it is involved in mutualisms with plants (South et al., 2021) and insects (Mendiola et al., 2022). Although our approach enabled us to identify some of the DMSP degraders in coral mucus, our estimated proportion of DMSP degraders in the mucus (less than 10% of the total community) is most likely an underestimation, since our conservative functional assignment relies on a small number of genomes harbouring ratified or orthologous dmdA/dddP genes, but disregards the many homologous sequences that may be functional.
The abundance of the cleavage and demethylation genes in bacterial communities is an indicator of how DMSP is being catabolised. This is important because the demethylation pathway shunts the sulfur moiety towards the synthesis of amino acids (e.g., methionine; see Howard et al., 2006; Reisch et al., 2011), while the cleavage leads to the production of the climatically active DMS (Curson et al., 2011). Quantification of both dmdA and dddP genes using qPCR revealed that their abundance in coral mucus (per 16S rRNA gene copy) increased (1.5-fold and 3-fold increases, respectively) with DMSP concentrations during thermal stress. In addition, when DMSP concentrations increased beyond 30 µM in mucus, the cleavage gene dddP became more abundant than the demethylation gene dmdA (ratio of dddP:dmdA A/1 above 1). DMSP concentrations have long been hypothesised to be the key determinant of which of the two DMSP degradation pathways is preferentially used (Kiene et al., 2000; Simó, 2001). Recent laboratory results on a model bacterium confirmed that at low to medium concentrations, most of the DMSP is degraded via demethylation, but at elevated concentrations (>10 μM) a shift occurs toward cleavage (Gao et al., 2020). Although we did not investigate the expression of these pathways, we found that a similar threshold exists for the abundance of these genes in coral mucus. Therefore, our results suggest that under high DMSP concentrations, such as the ones recorded in coral mucus under thermal stress, a larger proportion of DMSP may be converted to DMS.
Here we measured the largest DMSP concentrations ever recorded in any environment in the mucus of the reef-building coral Acropora millepora. Thermal stress caused sharp increases in both DMSP, and the proportion of bacterial genes involved in its degradation. In addition, our results suggest that when DMSP concentrations are high, a greater proportion of DMSP is converted to DMS by bacteria, which corroborates the increase in DMS concentrations previously measured in corals under various environmental stressors (Deschaseaux et al., 2014). Stronger convection forces occurring in tropical regions result in fast transport of volatile gases, such as DMS (Randel and Jensen, 2013), which means that shifts in DMSP-degradation route used by coral-associated bacteria may have important effects on atmospheric chemistry.
Data availability statement
The raw sequencing data used in this study is available in the NCBI Sequence Read Archive under BioProject number PRJNA816931. Scripts to reproduce these analyses are available on GitHub (https://github.com/StephGardner/Mucus-DMSP). Additional information is available in the supplementary information.
Ethics statement
Corals were collected under the Great Barrier Reef Marine Park Authority permit G14/36977.1 issued to KP.
Author contributions
SG, KP and J-BR designed the research. SG, MN, KP, and J-BR carried out the experiment. CM provided NMR expertise. JO’B performed qPCR. SG, MN and JB-R performed sample and data analysis. SG, J-BR and MN wrote the first draft of the manuscript. All authors edited the final manuscript. All authors contributed to the article and approved the submitted version.
Funding
SG was supported by an Australian Postgraduate Award, and research funding was provided by the Climate Change Cluster and the School of Life Sciences, University of Technology Sydney, a PADI Foundation Grant and a Danielle Simmons Research Award from the Australian Coral Reef Society. Corals were collected under the Great Barrier Reef Marine Park Authority permit G14/36977.1 issued to KP. J-BR was supported by an Australian Research Council Future Fellowship (FT210100100).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2022.912862/full#supplementary-material
References
Akoka S., Trierweiler M. (2002). Improvement of the ERETIC method by digital synthesis of the signal and addition of a broadband antenna inside the NMR probe. Instrum. Sci. Technol. 30 (1), 21–29. doi: 10.1081/CI-100108768
Archer S. D., Ragni M., Webster R., Airs R. L., Geider R. J. (2010). Dimethyl sulfoniopropionate and dimethyl sulfide production in response to photoinhibition in emiliania huxleyi. Limnol. Oceanogr. 55 (4), 1579–1589. doi: 10.4319/lo.2010.55.4.1579
Broadbent A. D., Jones G. B. (2004). DMS and DMSP in mucus ropes, coral mucus, surface films and sediment pore waters from coral reefs in the great barrier reef. Mar. Freshw. Res. 55 (8), 849–855. doi: 10.1071/MF04114
Broadbent A. D., Jones G. B., Jones R. J. (2002). DMSP in corals and benthic algae from the great barrier reef. Estuar. Coast. Shelf Sci. 55 (4), 547–555. doi: 10.1006/ecss.2002.1021
Brown B. E., Bythell J. C. (2005). Perspectives on mucus secretion in reef corals. Mar. Ecol. Prog. Ser. 296, 291–309. doi: 10.3354/meps296291
Buerger P., Alvarez-Roa C., Coppin C. W., Pearce S. L., Chakravarti L. J., Oakeshott J. G., et al. (2020). Heat-evolved microalgal symbionts increase coral bleaching tolerance. Sci. Adv. 6 (20), 2498. doi: 10.1126/sciadv.aba2498
Bythell J. C., Wild C. (2011). Biology and ecology of coral mucus release. J. Exp. Mar. Biol. Ecol. 408 (1), 88–93. doi: 10.1016/j.jembe.2011.07.028
Callahan B. J., McMurdie P. J., Rosen M. J., Han A. W., Johnson A. J. A., Holmes S. P. (2016). DADA2: High-resolution sample inference from illumina amplicon data. Nat. Methods 13, 581. doi: 10.1038/nmeth.3869
Caruana A. M. N., Malin G. (2014). The variability in DMSP content and DMSP lyase activity in marine dinoflagellates. Prog. Oceanogr. 120, 410–424. doi: 10.1016/j.pocean.2013.10.014
Casey J. M., Connolly S. R., Ainsworth T. D. (2015). Coral transplantation triggers shift in microbiome and promotion of coral disease associated potential pathogens. Sci. Rep. 5 (1), 11903. doi: 10.1038/srep11903
Cui Y., Suzuki S., Omori Y., Wong S. K., Ijichi M., Kaneko R., et al. (2015). Abundance and distribution of dimethylsulfoniopropionate degradation genes and the corresponding bacterial community structure at dimethyl sulfide hot spots in the tropical and subtropical pacific ocean. Appl. Environ. Microbiol. 81 (12), 4184–4194. doi: 10.1128/aem.03873-14
Curson A. R. J., Todd J. D., Sullivan M. J., Johnston A. W. B. (2011). Catabolism of dimethylsulphoniopropionate: microorganisms, enzymes and genes. Nat. Rev. Microbiol. 9 (12), 849–859. doi: 10.1038/nrmicro2653
Deschaseaux E. S. M., Jones G. B., Deseo M. A., Shepherd K. M., Kiene R. P., Swan H. B., et al. (2014). Effects of environmental factors on dimethylated sulfur compounds and their potential role in the antioxidant system of the coral holobiont. Limnol. Oceanogr. 59 (3), 758–768. doi: 10.4319/lo.2014.59.3.0758
Ducklow H. W., Mitchell R. (1979). Composition of mucus released by coral reef coelenterates. Limnol. Oceanogr. 24 (4), 706–714. doi: 10.4319/lo.1979.24.4.0706
Fisher P. L., Malme M. K., Dove S. (2012). The effect of temperature stress on coral-symbiodinium associations containing distinct symbiont types. Coral Reefs 31, 473–485. doi: 10.1007/s00338-011-0853-0
Frade P. R., Schwaninger V., Glasl B., Sintes E., Hill R. W., Simó R., et al. (2015). Dimethylsulfoniopropionate in corals and its interrelations with bacterial assemblages in coral surface mucus. Environ. Chem. 13 (2), 252–265. doi: 10.1071/EN15023
Freire I., Gutner-Hoch E., Muras A., Benayahu Y., Otero A. (2019). The effect of bacteria on planula-larvae settlement and metamorphosis in the octocoral rhytisma fulvum fulvum. PloS One 14 (9), e0223214. doi: 10.1371/journal.pone.0223214
Frias-Lopez J., Zerkle A. L., Bonheyo G. T., Fouke B. W. (2002). Partitioning of bacterial communities between seawater and healthy, black band diseased, and dead coral surfaces. Appl. Environ. Microbiol. 68 (5), 2214–2228. doi: 10.1128/AEM.68.5.2214-2228.2002
Gao C., Fernandez V. I., Lee K. S., Fenizia S., Pohnert G., Seymour J. R., et al. (2020). Single-cell bacterial transcription measurements reveal the importance of dimethylsulfoniopropionate (DMSP) hotspots in ocean sulfur cycling. Nat. Commun. 11 (1), 1942. doi: 10.1038/s41467-020-15693-z
Gardner S. G., Raina J.-B., Nitschke M. R., Nielsen D. A., Stat M., Motti C. A., et al. (2017). A multi-trait systems approach reveals a response cascade to bleaching in corals. BMC Biol. 15 (1), 117. doi: 10.1186/s12915-017-0459-2
Garren M., Azam F. (2010). New method for counting bacteria associated with coral mucus. Appl. Environ. Microbiol. 76 (18), 6128–6133. doi: 10.1128/AEM.01100-10
Giovannoni S. J. (2017). SAR11 bacteria: The most abundant plankton in the oceans. Annu. Rev. Mar. Sci. 9 (1), 231–255. doi: 10.1146/annurev-marine-010814-015934
Goyen S., Pernice M., Szabó M., Warner M. E., Ralph P. J., Suggett D. J. (2017). A molecular physiology basis for functional diversity of hydrogen peroxide production amongst symbiodinium spp. (Dinophyceae). Mar. Biol. 164 (3), 46. doi: 10.1007/s00227-017-3073-5
Haydon T. D., Seymour J. R., Suggett D. J. (2018). Soft corals are significant DMSP producers in tropical and temperate reefs. Mar Biol. 165, 109. doi: 10.1007/s00227-018-3367-2
Hennige S. J., Smith D. J., Perkins R., Consalvey M., Paterson D. M., Suggett D. J. (2008). Photoacclimation, growth and distribution of massive coral species in clear and turbid waters. Mar. Ecol. Prog. Ser. 369, 77–88. doi: 10.3354/meps07612
Hill R. W., Dacey J. W. H., Edward A. (2000). Dimethylsulfoniopropionate in giant clams (Tridacnidae). Biol. Bull. 199 (2), 108–115. doi: 10.2307/1542870
Hill R. W., Dacey J. W. H., Krupp D. A. (1995). Dimethylsulfoniopropionate in reef corals. Bull. Mar. Sci. 57 (2), 489–494.
Howard E. C., Henriksen J. R., Buchan A., Reisch C. R., Bürgmann H., Welsh R., et al. (2006). Bacterial taxa that limit sulfur flux from the ocean. Science 314 (5799), 649–652. doi: 10.1126/science.1130657
Howard E. C., Sun S., Biers E. J., Moran M. A. (2008). Abundant and diverse bacteria involved in DMSP degradation in marine surface waters. Environ. Microbiol. 10 (9), 2397–2410. doi: 10.1111/j.1462-2920.2008.01665.x
Huggett M. J., Apprill A. (2019). Coral microbiome database: Integration of sequences reveals high diversity and relatedness of coral-associated microbes. Environ. Microbiol. Rep. 11 (3), 372–385. doi: 10.1111/1758-2229.12686
Jones G. B., King S. (2015). Dimethylsulphoniopropionate (DMSP) as an indicator of bleaching tolerance in scleractinian corals. J. Mar. Sci. Eng. 3 (2), 444–465. doi: 10.3390/jmse3020444
Kassambara A. (2021). "rstatix: Pipe-friendly framework for basic statistical tests". https://CRAN.R-project.org/package=rstatix
Katoh K., Standley D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. evol. 30 (4), 772–780. doi: 10.1093/molbev/mst010
Kettle A. J., Andreae M. O., Amouroux D., Andreae T. W., Bates T. S., Berresheim H., et al. (1999). A global database of sea surface dimethylsulfide (DMS) measurements and a procedure to predict sea surface DMS as a function of latitude, longitude, and month. Global Biogeochem. Cycles 13 (2), 399–444. doi: 10.1029/1999GB900004
Kieber D. J., Jiao J., Kiene R. P., Bates T. S. (1996). Impact of dimethylsulphide photochemistry on methyl sulphur cycling in the equatorial pacific ocean. J. Geophys. Res.: Oceans 101 (C2), 3715–3372. doi: 10.1029/95JC03624
Kiene R. P., Linn L. J., Bruton J. A. (2000). New and important roles for DMSP in marine microbial communities. J. Sea Res. 49 (3-4), 209–224. doi: 10.1016/S1385-1101(00)00023-X
Kiene R. P., Linn L. J., González J., Moran M., Bruton J. A. (1999). Dimethylsulfoniopropionate and methanethiol are important precursors of methionine and protein-sulfur in marine bacterioplankton. Appl. Environ. Microbiol. 65 (10), 4549–4558. doi: 10.1128/AEM.65.10.4549-4558.1999
Kuek F. W. I., Motti C. A., Zhang J., Cooke I. R., Todd J. D., Miller D. J., et al. (2022). DMSP production by coral-associated bacteria. Front. Mar. Sci 9. doi: 10.3389/fmars.2022.869574
LaJeunesse T. C., Loh W. K. W., van Woesik R., Hoegh-Guldberg O., Schmidt G. W., Fitt W. K. (2003). Low symbiont diversity in southern great barrier reef corals, relative to those of the Caribbean. Limnol. Oceanogr. 48 (5), 2046–2054. doi: 10.4319/lo.2003.48.5.2046
Lesser M. P. (1997). Oxidative stress causes coral bleaching during exposure to elevated temperatures. Coral Reefs 16, 187–192. doi: 10.1007/s003380050073
Lesser M. P. (2006). Oxidative stress in marine environments: Biochemistry and physiological ecology. Annu. Rev. Physiol. 68, 253–278. doi: 10.1146/annurev.physiol.68.040104.110001
Lesser M. P. (2011). “Coral bleaching: causes and mechanisms,” in Coral reefs: an ecosystem in transition, 1st ed. Eds. Dubinsky Z., Stambler N. (New York: Springer), 405–419.
Levine N. M., Varaljay V. A., Toole D. A., Dacey J. W. H., Doney S. C., Moran M. A. (2012). Environmental, biochemical and genetic drivers of DMSP degradation and DMS production in the Sargasso Sea. Environ. Microbiol. 14 (5), 1210–1223. doi: 10.1111/j.1462-2920.2012.02700.x
Lozupone C., Lladser M. E., Knights D., Stombaugh J., Knight R. (2011). UniFrac: an effective distance metric for microbial community comparison. ISME J.: Multidiscip. J. Microb. Ecol. 5 (2), 169–172. doi: 10.1038/ismej.2010.133
Luo D., Wang X., Feng X., Tian M., Wang S., Tang S.-L., et al. (2021). Population differentiation of rhodobacteraceae along with coral compartments. ISME J. 15 (11), 3286–3302. doi: 10.1038/s41396-021-01009-6
Martin M. (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal 17(1), 10–12. doi: 10.14806/ej.17.1.200
McMurdie P. J., Holmes S. (2013). Phyloseq: An r package for reproducible interactive analysis and graphics of microbiome census data. PloS One 8 (4), e61217. doi: 10.1371/journal.pone.0061217
Meikle P., Richards G. N., Yellowlees D. (1988). Structural investigations on the mucus from six species of coral. Mar. Biol. 99 (2), 187–193. doi: 10.1007/BF00391980
Mendiola S. Y., Stoy K. S., DiSalvo S., Wynn C. L., Civitello D. J., Gerardo N. M. (2022). Competitive exclusion of phytopathogenic serratia marcescens from squash bug vectors by the gut endosymbiont caballeronia. Appl. Environ. Microbiol. 88 (1), e0155021. doi: 10.1128/aem.01550-21
Miller M. A., Pfeiffer W., Schwartz T. (2010). "Creating the CIPRES science gateway for inference of large phylogenetic trees in proceedings of the gateway computing environments workshop (GCE)” (New Orleans, Los Angeles), 1–8. doi: 10.1109/GCE.2010.5676129
Miura N., Motone K., Takagi T., Aburaya S., Watanabe S., Aoki W., et al. (2019). Ruegeria sp. strains isolated from the reef-building coral galaxea fascicularis inhibit growth of the temperature-dependent pathogen vibrio coralliilyticus. Mar. Biotechnol. 21 (1), 1–8. doi: 10.1007/s10126-018-9853-1
Nitschke M. R., Gardner S. G., Goyen S., Fujise L., Camp E. F., Ralph P. J., et al. (2018). Utility of photochemical traits as diagnostics of thermal tolerance amongst great barrier reef corals. Front. Mar. Sci. 5 (45). doi: 10.3389/fmars.2018.00045
Oksanen J., Blanchet F. G., Friendly M., Kindt R., Legendre P., McGlinn D., et al (2019). vegan: Community Ecology Package. R package version 2.5-6. https://CRAN.R-project.org/package=vegan
Piggot A. M., Fouke B. W., Sivaguru M., Sanford R. A., Gaskins H. R. (2009). Change in zooxanthellae and mucocyte tissue density as an adaptive response to environmental stress by the coral, montastraea annularis. Mar. Biol. 156 (11), 2379–2389. doi: 10.1007/s00227-009-1267-1
Pollock F. J., Wada N., Torda G., Willis B. L., Bourne D. G., Kostka J. E. (2017). White syndrome-affected corals have a distinct microbiome at disease lesion fronts. Appl. Environ. Microbiol. 83 (2), e02799–e02716. doi: 10.1128/AEM.02799-16
Price M. N., Dehal P. S., Arkin A. P. (2009). FastTree: Computing Large minimum-evolution trees with profiles instead of a distance matrix. Mol. Biol. Evol. 26, 1641–1650. doi: 10.1093/molbev/msp077
Quast C., Pruesse E., Yilmaz P., Gerken J., Schweer T., Yarza P., et al. (2013). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41 (Database issue), D590–D596. doi: 10.1093/nar/gks1219
Raina J.-B., Tapiolas D. M., Foret S., Lutz A., Abrego D., Ceh J., et al. (2013). DMSP biosynthesis by an animal and its role in coral thermal stress response. Nature 502 (7473), 677–680. doi: 10.1038/nature12677
Raina J. B., Tapiolas D., Willis B. L., Bourne D. G. (2009). Coral-associated bacteria and their role in the biogeochemical cycling of sulfur. Appl. Environ. Microbiol. 75 (11), 3492–3501. doi: 10.1128/aem.02567-08
Ralph P. J., Gademann R. (2005). Rapid light curves: A powerful tool to assess photosynthetic activity. Aquat. Bot. 82, 222–237. doi: 10.1016/j.aquabot.2005.02.006
Randel W. J., Jensen E. J. (2013). Physical processes in the tropical tropopause layer and their roles in a changing climate. Nat. Geosci. 6 (3), 169–176. doi: 10.1038/ngeo1733
Reisch C. R., Moran M. A., Whitman W. B. (2011). Bacterial catabolism of dimethylsulfoniopropionate (DMSP). Front. Microbiol. 2. doi: 10.3389/fmicb.2011.00172
Reshef L., Koren O., Loya Y., Zilber-Rosenberg I., Rosenberg E. (2006). The coral probiotic hypothesis. Environ. Microbiol. 8 (12), 2068–2073. doi: 10.1111/j.1462-2920.2006.01148.x
Ritchie K. B. (2006a). Regulation of microbial populations by coral surface mucus and mucus-associated bacteria. Mar. Ecol. Prog. Ser. 322, 1–14. doi: 10.3354/meps322001
Ritchie R. J. (2006b). Consistent sets of spectrophotometric chlorophyll equations for acetone, methanol and ethanol solvents. Photosyn. Res., 89 (1), 27–41. doi: 10.1007/s11120-006-9065-9
Roberty S., Bailleul B., Berne N., Franck F., Cardol P. (2014). PSI mehler reaction is the main alternative photosynthetic electron pathway in symbiodinium sp., symbiotic dinoflagellates of cnidarians. New Phytol. 204 (1), 81–91. doi: 10.1111/nph.12903
Rochaix J.-D. (2011). Regulation of photosynthetic electron transport. Biochim. Biophys. Acta (BBA) - Bioenerg. 1807 (3), 375–383. doi: 10.1016/j.bbabio.2010.11.010
Rohwer F., Breitbart M., Jara J., Azam F., Knowlton N. (2001). Diversity of bacteria associated with the Caribbean coral montastraea franksi. Coral Reefs 20 (1), 85–91. doi: 10.1007/s003380100138
Rohwer F., Seguritan V., Azam F., Knowlton N. (2002). Diversity and distribution of coral-associated bacteria. Mar. Ecol. Prog. Ser. 243, 1–10. doi: 10.3354/meps243001
Schauer M., Massana R., Pedrós-Alió C. (2000). Spatial differences in bacterioplankton composition along the Catalan coast (NW Mediterranean) assessed by molecular fingerprinting. FEMS Microbiol. Ecol. 33 (1), 51–59. doi: 10.1111/j.1574-6941.2000.tb00726.x
Seyedsayamdost M. R., Case R. J., Kolter R., Clardy J. (2011). The Jekyll-and-Hyde chemistry of phaeobacter gallaeciensis. Nat. Chem. 3 (4), 331–335. doi: 10.1038/nchem.1002
Sharp K. H., Sneed J. M., Ritchie K. B., McDaniel L., Paul V. J. (2015). Induction of larval settlement in the reef coral porites astreoides by a cultivated marine roseobacter strain. Biol. Bull. 228 (2), 98–107. doi: 10.1086/BBLv228n2p98
Sheik C. S., Reese B. K., Twing K. I., Sylvan J. B., Grim S. L., Schrenk M. O., et al. (2018). Identification and removal of contaminant sequences from ribosomal gene databases: Lessons from the census of deep life. Front. Microbiol. 9. doi: 10.3389/fmicb.2018.00840
Shnit-Orland M., Kushmaro A. (2009). Coral mucus-associated bacteria: a possible first line of defense. FEMS Microbiol. Ecol. 67 (3), 371–380. doi: 10.1111/j.1574-6941.2008.00644.x
Sievert S. M., Kiene R. P., Schulz-Vogt H. N. (2007). The sulfur cycle. Oceanography 20 (2), 117–123. doi: 10.5670/oceanog.2007.55
Simó R. (2001). Production of atmospheric sulfur by oceanic plankton: biogeochemical, ecological and evolutionary links. Trends Ecol. Evol. 16 (6), 287–294. doi: 10.1016/S0169-5347(01)02152-8
Simó R., Archer S. D., Pedrós-Alió C., Gilpin L., Stelfox-Widdicombe C. E. (2002). Coupled dynamics of dimethylsulfoniopropionate and dimethylsulfide cycling and the microbial food web in surface waters of the north Atlantic. Limnol. Oceanogr. 47 (1), 53–61. doi: 10.4319/lo.2002.47.1.0053
South K. A., Nordstedt N. P., Jones M. L. (2021). Identification of plant growth promoting rhizobacteria that improve the performance of greenhouse-grown petunias under low fertility conditions. Plants 10 (7), 1410. doi: 10.3390/plants10071410
Stimson J., Kinzie R. A. (1991). The temporal pattern and rate of release of zooxanthellae from the reef coral Pocillopora damicornis (Linnaeus) under nitrogen-enrichment and control conditions. J. Exp. Mar. Biol. Ecol. 153, 63–74. doi: 10.1016/S0022-0981(05)80006-1
Suggett D. J., Goyen S., Evenhuis C., Szabo M., Pettay D. T., Warner M. E., et al. (2015). Functional diversity of photobiological traits within the genus symbiodinium appears to be governed by the interaction of cell size with cladal designation. New Phytol. 208 (2), 370–381. doi: 10.1111/nph.13483
Suggett D. J., Warner M. E., Smith D. J., Davey P., Hennige S., Baker N. R. (2008). Photosynthesis and production of hydrogen peroxide by Symbiodinium (pyrrhophyta) phylotypes with different thermal tolerances. J. Phycol. 44 (4), 948–956. doi: 10.1111/j.1529-8817.2008.00537.x
Sunda W., Kieber D. J., Kiene R. P., Huntsman S. (2002). An antioxidant function for DMS and DMSP in marine algae. Nature 418, 317–320. doi: 10.1038/nature00851
Sun J., Todd J. D., Thrash J. C., Qian Y., Qian M. C., Temperton B., et al. (2016). The abundant marine bacterium pelagibacter simultaneously catabolizes dimethylsulfoniopropionate to the gases dimethyl sulfide and methanethiol. Nat. Microbiol. 1, 16065. doi: 10.1038/nmicrobiol.2016.65
Suzuki M. T., Taylor L. T., DeLong E. F. (2000). Quantitative analysis of small-subunit rRNA genes in mixed microbial populations via 5′-nuclease assays. Appl. Environ. Microbiol. 66 (11), 4605–4614. doi: 10.1128/AEM.66.11.4605-4614.2000
Sweet M. J., Croquer A., Bythell J. C. (2011). Bacterial assemblages differ between compartments within the coral holobiont. Coral Reefs 30 (1), 39–52. doi: 10.1007/s00338-010-0695-1
Takahashi S., Whitney S. M., Badger M. R. (2009). Different thermal sensitivity of the repair of photodamaged photosynthetic machinery in cultured symbiodinium species. Proc. Natl. Acad. Sci. 106 (9), 3237–3242. doi: 10.1073/pnas.0808363106
Tapiolas D. M., Raina J. B., Lutz A., Willis B. L., Motti C. A. (2013). Direct measurement of dimethylsulfoniopropionate (DMSP) in reef-building corals using quantitative nuclear magnetic resonance (qNMR) spectroscopy. J. Exp. Mar. Biol. Ecol. 443, 85–89. doi: 10.1016/j.jembe.2013.02.037
Todd J. D., Curson A. R. J., Dupont C. L., Nicholson P., Johnston A. W. B. (2009). The dddP gene, encoding a novel enzyme that converts dimethylsulfoniopropionate into dimethyl sulfide, is widespread in ocean metagenomes and marine bacteria and also occurs in some ascomycete fungi. Environ. Microbiol. 11 (6), 1376–1385. doi: 10.1111/j.1462-2920.2009.01864.x
Tremblay P., Weinbauer M. G., Rottier C., Guérardel Y., Nozais C., Ferrier-Pagès C. (2011). Mucus composition and bacterial communities associated with the tissue and skeleton of three scleractinian corals maintained under culture conditions. J. Mar. Biol. Assoc. United Kingdom. 91 (03), 649–657. doi: 10.1017/S002531541000130X
Trevena A. J., Jones G., Wright S., Enden R. (2003). Profiles of dimethylsulphoniopropionate (DMSP), algal pigments, nutrients, and salinity in the fast ice of prydz bay, Antarctica. Sch. Environ. Sci. Manage. Pap. 108, 3145. doi: 10.1029/2002JC001369
Van Alstyne K. L., Koellermeier L., Nelson T. A. (2007). Spatial variation in dimethylsulfoniopropionate (DMSP) production in ulva lactuca (Chlorophyta) from the northeast pacific. Mar. Biol. 150 (6), 1127–1135. doi: 10.1007/s00227-006-0448-4
Varaljay V. A., Howard E. C., Sun S., Moran M. A. (2010). Deep sequencing of a dimethylsulfoniopropionate-degrading gene (dmdA) by using PCR primer pairs designed on the basis of marine metagenomic data. Appl. Environ. Microbiol. 76 (2), 609–617. doi: 10.1128/AEM.01258-09
Veal C. J., Holmes G., Nunez M., Hoegh-Guldberg O., Osborn J. (2010). A comparative study of methods for surface area and three dimensional shape measurement of coral skeletons. Limnol. Oceanogr. 8, 241–253. doi: 10.4319/lom.2010.8.241
Wild C., Huettel M., Klueter A., Kremb S. G., Rasheed M. Y. M., Jørgensen B. B. (2004). Coral mucus functions as an energy carrier and particle trap in the reef ecosystem. Nature 428 (6978), 66–70. doi: 10.1038/nature02344
Zhang X.-H., Liu J., Liu J., Yang G., Xue C.-X., Curson A. R. J., et al. (2019). Biogenic production of DMSP and its degradation to DMS–their roles in the global sulfur cycle. Sci. China Life Sci. 62 (10), 1296–1319. doi: 10.1007/s11427-018-9524-y
Keywords: microbiome, coral mucus associated bacteria, DMSP-degrading genes, thermal stress, Acropora millepora, dimethylsulfoniopropionate (DMSP)
Citation: Gardner SG, Nitschke MR, O’Brien J, Motti CA, Seymour JR, Ralph PJ, Petrou K and Raina J-B (2022) Increased DMSP availability during thermal stress influences DMSP-degrading bacteria in coral mucus. Front. Mar. Sci. 9:912862. doi: 10.3389/fmars.2022.912862
Received: 05 April 2022; Accepted: 15 August 2022;
Published: 09 September 2022.
Edited by:
Graham Barry Jones, Southern Cross University, AustraliaReviewed by:
Stephen D. Archer, Bigelow Laboratory For Ocean Sciences, United StatesJohn Bythell, Newcastle University, United Kingdom
Copyright © 2022 Gardner, Nitschke, O’Brien, Motti, Seymour, Ralph, Petrou and Raina. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Stephanie G. Gardner, c3RlcGhhbmllLmdhcmRuZXJAc3lkbmV5LmVkdS5hdQ==
 Stephanie G. Gardner
Stephanie G. Gardner Matthew R. Nitschke
Matthew R. Nitschke James O’Brien
James O’Brien Cherie A. Motti
Cherie A. Motti Justin R. Seymour
Justin R. Seymour Peter J. Ralph
Peter J. Ralph Katherina Petrou
Katherina Petrou Jean-Baptiste Raina
Jean-Baptiste Raina