- 1Department of Integrative Marine Ecology, Stazione Zoologica Anton Dohrn, Napoli, Italy
- 2Department of Veterinary Medicine, Università di Bari Aldo Moro, Valenzano, Italy
- 3Istituto Nazionale di Oceanografia e di Geofisica Sperimentale, Oceanography Division – Osservatorio Geofisico Sperimentale (OGS), Trieste, Italy
- 4Consiglio Nazionale delle Ricerche, Istituto di Scienze Marine (CNR-ISMAR), Venezia, Italy
- 5Institut de Ciències del Mar, Consejo Superior de Investigaciones Scientificas (CSIC), Barcelona, Spain
The Northern Adriatic Sea is a key area of the Mediterranean Sea, strongly affected by freshwater inputs, mainly from the Po River, which bring high amounts of nutrients as well as organic and inorganic particles. Free-living and particle-attached prokaryotes were characterized by 16S rRNA gene amplicon sequencing of size-fractionated samples collected during a diatom bloom in this area. The diversity of free-living and particle-attached prokaryotic assemblages was investigated with the aim to understand how the microbial communities are structured in the two fractions and whether specific microbial groups are associated to one lifestyle or the other. The results highlight a diverse prokaryotic community dominated by Proteobacteria, Bacteroidetes, and, remarkably, Firmicutes. Taxa within Firmicutes and Alphaproteobacteria are identified as the main particle-attached indicators by LEfSe, while members of Bacteroidetes and Gammaproteobacteria were representative of the free-living lifestyle, although they were also usually found as particle-attached. Collectively, the results suggest that both the free-living and the particle-attached lifestyles are a complex combination of specialization and adaptation to local conditions.
Introduction
The aggregation of organic and inorganic matter is an important process in the functioning of aquatic ecosystems. Aggregate formation occurs through the collision and adhesion to particles of inorganic and organic particles, including bacteria, phytoplankton, feces, detritus, and bio-minerals (Alldredge and Silver, 1988; Simon et al., 2002; Thornton, 2002), ranging in size from a few microns to many centimeters (Alldredge and Silver, 1988; Passow et al., 2012). These aggregates host a great diversity, and include different microorganisms, such as bacteria, protists, and viruses, as well as gels, colloids, and cell debris (Azam and Malfatti, 2007). It has been hypothesized that bacteria and microzooplankton growing on aggregates, may build an area of enriched organic and inorganic nutrients around the aggregates (Hoppe, 1981). Also, microorganisms maximize their position within such nutrient field, leading to the formation of microbial clusters in their vicinity (Azam and Ammerman, 1984). Biddanda and Pomeroy (1988) called this microenvironment the “phytoplankton-derived detritosphere”, which is similar to the “phycosphere” concept used by Bell and Mitchell (1972) to define the microscopic region rich in organic molecules that surrounds phytoplankton cells. The phycosphere represents the key interface where tight interactions between algae and other organisms are controlled by exuded chemicals (Seymour et al., 2017) and act as hot spots of high microbial abundance and activity (Smith et al., 1992; Azam et al., 1994) as well as of biogeochemical processes. These hot spots are also the site of aggregation–disaggregation processes determining the fate of this detritus in the water column (Biddanda and Pomeroy,1988).
The interactions that occur between phytoplankton and bacteria are crucial in global carbon and nutrient cycles (Amin et al., 2015; Christie-Oleza et al., 2017; Seymour et al., 2017; Mühlenbruch et al., 2018; Fu et al., 2020). Phytoplankton are responsible for half of Earth’s photosynthesis, and heterotrophic marine bacteria process 40% to 50% of this ocean fixed carbon (Moran et al., 2016; Fu et al., 2020). Heterotrophic bacteria, in addition to using organic carbon, also recycle inorganic nutrients, such as nitrogen and phosphorus, directly affecting phytoplankton dynamics (Amin et al., 2012; Buchan et al., 2014; Amin et al., 2015; Eigemann et al., 2022). It is well known that prokaryotes and phytoplankton are closely linked in coastal marine environments, with correlations frequently observed between their biomasses (Fuhrman et al., 1980; Gasol and Duarte, 2000), and species-specific interactions, suggesting that prokaryotes, especially bacteria, can play a major role in controlling phytoplankton dynamics (Furuki and Kobayashi, 1991; Imai et al., 1993; Yoshinaga et al., 1995; Yoshinaga et al., 1997). Indeed, some prokaryotes are capable of stimulating or inhibiting phytoplankton growth (Ferrier et al., 2002), killing or lysing phytoplankton (Mayali and Azam, 2004), or altering their physiology (Gallacher et al., 1997). During phytoplankton growth, organic carbon is made available to prokaryotes, and both healthy and dying algal cells can release large amounts of dissolved organic matter into their surroundings (Myklestad, 1995; Thornton, 2014; Christie-Oleza et al., 2017). The metabolites released by phytoplankton include several chemical classes, such as carboxylic acids, amino acids, carbohydrates, C1 compounds, organic sulfur compounds (Thornton, 2014; Durham et al., 2015; Landa et al., 2017), and secondary metabolites (Ribalet et al., 2007). The metabolites present in a given phycosphere depend on the phytoplankton taxonomy and physiology (Allen et al., 2008; Barofsky et al., 2009; Becker et al., 2014; Landa et al., 2017) as is well known that different types of phytoplankton might generate distinct types of organic matter (Sarmento et al., 2013; Moran et al., 2022), and this could explain why some prokaryotes might prefer some specific phytoplankters.
Depending on their relation with the particulate organic and inorganic matter present in the environment, prokaryotes have often been classified into two types of communities: free-living (FL) or particle-attached (PA) (Crump et al., 1999; Simon et al., 2002). Their respective niches are generally operationally defined according to the pore size of the filter used to collect them (Milici et al., 2016). Filters with larger pore size (generally larger than 3 μm) collect particles, micro-eukaryotes, and prokaryotes (attached or aggregated) associated with the larger items, while the truly planktonic or free-living prokaryotes are typically captured on 0.1- or 0.2-μm filters. Currently, the PA and FL communities have been reported to harbor different groups of microbial lineages in various marine environments including coastal or surface water (Mohit et al., 2014; Zhang et al., 2016; Mestre et al., 2017a; Mestre et al., 2017b), bathypelagic water (Wilkins et al., 2013; Salazar et al., 2016; Milici et al., 2017), or extreme depths of hadalpelagic habitats (>6,000 m) (Eloe et al., 2011; Tarn et al., 2016).
Some coastal marine plankton sites are highly affected by freshwater inputs that bring high concentration of nutrients and many organic and inorganic particles. How the microbial communities in the FL and the PA fractions of coastal sites are structured and whether they differ from, e.g., those of oligotrophic plankton sites are a matter of relevance to understand the dynamics of microbial communities and their possible preference for one or the other lifestyle. A powerful and reproducible aid that helps us better understand microbial diversity comes from the development of high-throughput sequencing (HTS) technologies, along with a metabarcoding approach, e.g., the large-scale analyses of biodiversity through the amplification and sequencing of marker genes (in this case, 16S rDNA), providing an efficient way to conduct the biodiversity monitoring of the whole ecosystem (Creer et al., 2010; Zhang et al., 2020). Metabarcoding has revolutionized our capacity to gather biodiversity data from environmental samples (Malviya et al., 2016) and transformed our understanding of microbial diversity (Pedrós-Alió et al., 2018).
In this study, the distribution and structure of the prokaryotic community in the Northern Adriatic Sea is investigated as this area is highly dynamic, under the alternating influence of eutrophic low-salinity waters originating from the Po River, and oligotrophic high-salinity water from the central Adriatic (Orlić et al., 2013; Steiner et al., 2019; Steiner et al., 2020). The Northern Adriatic Sea is also exposed to various anthropogenic impacts such as different types of wastewater pollution and agricultural and urban runoff. These inputs can introduce allochthonous organic matter and microbes into the environment (Orel et al., 2021). It is reported that the abundance, composition, and activity of microbial communities in this area depend, to a large extent, on river discharges (Celussi et al., 2010; Mozetič et al., 2012), and the Northern Adriatic Sea receives significant freshwater inputs from several rivers, such as Adige, Brenta, Piave, and Po, along the northwestern coast of Italy. Russo and Artegiani (1996) estimated that about 20% of Mediterranean freshwater inputs occur in the Northern Adriatic Sea. The Po River is the main river in Italy for length and water load and the second freshwater contributor to the Mediterranean Sea (1,500 m3/s; Raicich, 1994), discharging high amounts of particles (both inert and living) and fueling high primary productivity (Sournia, 1973). The haline stratification segregates phytoplankton at surface, triggering and controlling phytoplankton blooms, especially in the frontal area of the river delta (Mangoni et al., 2008). In general, it has been observed that total phytoplankton abundances decreased over the past two decades with a significant shift in 2003 (Cozzi and Giani, 2011; Djakovac et al., 2012; Bernardi Aubry et al., 2021), probably due to the reduction in the total load of freshwater discharge and related nutrient loads.
Given the great variability of oceanographic conditions in the Northern Adriatic Sea, including the large effects of river inputs with high particle load, and the recurrent presence of large diatom populations, free-living prokaryotes (FL) were characterized and compared to those attached to particles (PA), during a diatom bloom using HTS of the 16S RNA gene. The starting hypothesis is that specific prokaryotic communities would be associated with either debris from the river, or particles including phytoplankton, considering the diatom bloom present at the time. This hypothesis is supported by previous surveys in the same area and season (e.g., Catalano et al., 2014), reporting that the detrital fraction in the upper mixed layer and the deep mixed layer represents 25%–29% of the total POC. Comparing communities in these two fractions provides information on potential actors in the aggregation processes that often have provoked mucilage events in the area and also on the potential impact of harmful organisms that may thrive and aggregate on particles, considering that this is the most productive area of the Mediterranean Sea (Salgado-Hernanz et al., 2022).
Materials and Methods
Sample Collection
From March 14 to 21, 2017, an oceanographic research cruise (Project INTERNOS 2017) was conducted in the Northern Adriatic Sea with R/V Minerva Uno. Fourteen stations were sampled (Figure 1), using a rosette sampler equipped with 10-L Niskin bottles and a CTD probe (SBE 911, Sea-Bird Electronics, USA) with fluorometer ECO FLNTU to measure conductivity, temperature, depth, and fluorescence. Samples for phytoplankton abundance, flow cytometry, and 16S rRNA gene sequencing were collected from the same Niskin bottles. Duplicate samples for flow cytometry (1 ml each) were placed in 2.0-ml cryovials, fixed with a mix of glutaraldehyde (0.05% final concentration) and paraformaldehyde (1% final concentrations) for 10 min in the dark, frozen in liquid nitrogen, and stored at −80°C until analysis (Marie et al., 1999). Microphytoplankton samples were collected at stations 1, 4, 7, and 10 and disposed in 500-ml dark glass bottles, fixed with buffered 37% formaldehyde (1.6% final concentration), and counted following the Utermöhl method (Edler and Elbrächter, 2010). For 16S sequencing, 1 L of seawater was filtered sequentially through 47-mm Millipore polycarbonate filters with pore sizes 3.0 μm and 0.22 μm (Whatman-Nucleopore and Millipore-GSWP, respectively) using a Swinnex filter holder system connected to a vacuum pressure pump, to collect free-living microbes (FL, between 0.2 and 3.0 µm in size) and those attached to particles (PA, >3.0 µm), respectively. The filters were disposed in cryovials, frozen in liquid nitrogen and stored at −80°C until DNA extraction.
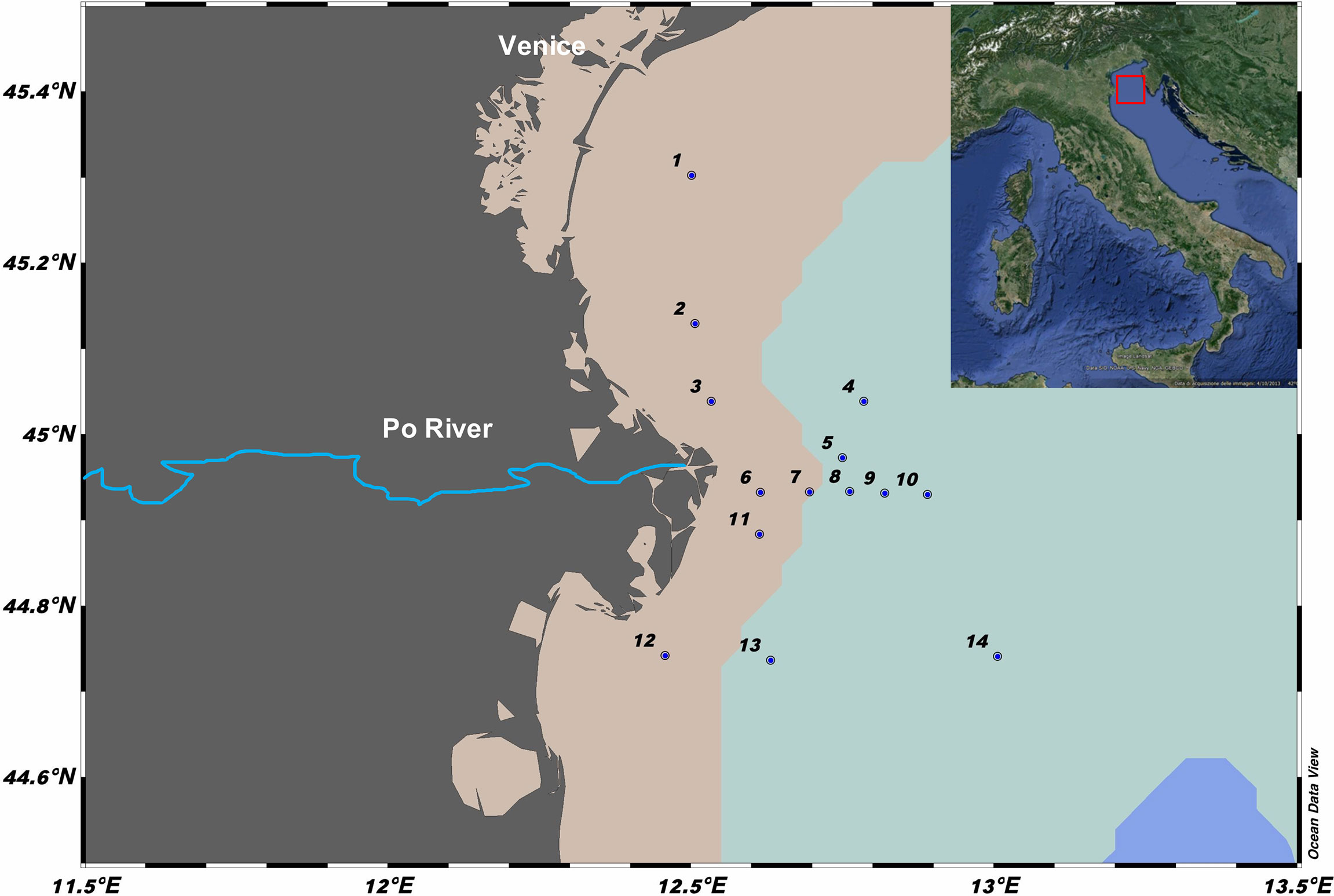
Figure 1 Map of the study area in the Northern Adriatic Sea (Mediterranean Sea) showing the 14 stations sampled in 2017. Created by ODV version 5.1.7. The different shades on the map represent the delimitation of 20-m (pink) and 60-m (light blue) isobaths.
Flow Cytometry
Total prokaryotic abundances were determined using a BD FACSVerse flow cytometer (BD Biosciences, Franklin Lake, USA) equipped with a 488-nm laser excitation and the standard filter setup. Analysis of heterotrophic prokaryotes was performed after thawing and staining the samples with SYBRGreen I (Molecular Probes, Inc., USA as described in Balestra et al., 2011). Heterotrophic prokaryotes were discriminated from other particles or background based on side scatter and by their high green fluorescence from SYBRGreen I, also used to discriminate HNA (High Nucleic Acid content) from LNA (Low Nucleic Acid content) prokaryotic cells (Gasol and del Giorgio, 2000). Data acquisition was performed using software FACSuite (BD Biosciences, Franklin Lake, USA) while file analysis was carried out using the FCS Express 6 Flow v 6.06.0025 software (DeNovo Software, Glendale, USA).
DNA Extraction and Sequencing
DNA was extracted from the filters as described in Boström et al. (2004). DNA concentration purity was measured using an ND-1000UV-Vis Nano-Drop spectrophotometer (ThermoFisher, UK). The extracted DNA samples were sent to the Integrated Microbiome Resource of Dalhousie University (Canada) for PCR amplification and Illumina MiSeq 2x300 bp sequencing. Raw sequences were deposited in the Sequence Read Archive (SRA) of the National Center for Biotechnology Information (NCBI) under BioProject #PRJNA834028.
The V4–V5 hypervariable region of the 16S rRNA gene was amplified using the 515-Y (5′ GTGYCAGCMGCCGCGGTAA) and 926R (5′ CCGYCAATTYMTTTRAGTTT) primers (Parada et al., 2016). After primer removal, raw sequencing reads were submitted to the QIIME2 pipeline (Bolyen et al., 2019). Amplicon sequence variants (ASVs) were identified through the DADA2 strategy within QIIME2 (Callahan et al., 2016), with standard parameters. For the taxonomic affiliation of the ASVs, a reference database was first created within QIIME2 by trimming the SILVA database v132 (Quast et al., 2012) to the region amplified by the used sequencing primers and training a Naive-Bayes classifier on the subset. This classifier was then applied to the representative ASVs for taxonomic assignment. ASVs assigned to chloroplasts, mitochondria, or “unknown” (i.e., that could not be classified at the kingdom level) were removed and excluded from further analyses.
Data Treatment and Diversity Analyses
The generated ASV table was normalized with a random subsampling at the lowest number of reads present in the samples (1,000 reads) and used to calculate alpha- and beta-diversity and the taxonomic description of the dataset. The analyses were performed with the phyloseq (McMurdie and Holmes, 2013), vegan (Oksanen et al., 2013), and ggplot2 (Wickham, 2010) R packages, within R-software (version 3.6.3). For alpha-diversity, rarefaction curves were generated and estimators of alpha-diversity including observed ASVs (richness), Shannon, and Chao indexes were calculated. For beta-diversity, Hellinger transformation was applied to the normalized abundance table and then the dissimilarity matrix was computed using the Bray–Curtis index (function vegdist). The Bray–Curtis dissimilarity matrix was then used to perform ordination analyses [hierarchical clustering and non-metric multidimensional scaling (NMDS)]. Analysis of similarities (ANOSIM test) (Clarke, 1993) was used for testing differences between the FL and PA prokaryotic communities. PerMANOVA (adonis function in vegan) was applied to detect significant associations between environmental parameters and prokaryotic community structure. Venn diagrams were built in order to calculate the shared and exclusive ASVs in the FL and PA communities (Oliveros, 2007–2015) (available at http://bioinfogp.cnb.csic.es/tools/venny/index.html). Finally, linear discriminant analysis (LDA) effect size (LEfSe) analysis (Segata et al., 2011) was used to identify the features (e.g., the taxa) that most likely would explain differences between classes (FL vs. PA), with a stringent threshold LDA score of 4.0. The taxa that violate a null hypothesis of no difference between classes were considered to be biomarkers for the taxon considered (Segata et al., 2011). Using the microeco R package (Liu et al., 2021), the metabolic or other ecologically relevant functions of the taxa were predicted using Functional Annotation of Prokaryotic Taxa (FAPROTAX) database, which extrapolates putative functional profiles based on the literature of cultured taxa (Louca et al., 2016).
R Codes used in this study are available in the Supplementary Material.
Results
Sampled Area
Surface temperatures were higher in the area influenced by the Po River (stt. 5, 6, 8) associated with lower salinities and relatively higher prokaryotic abundances (Table 1). The highest fluorescence-derived chlorophyll a with an average of 3.95 ± 2.02 µg L−1 were reported in stations 3 and 4, just above the mouth of the Po River due to a diatom bloom (mainly Chaetoceros spp.) present in the area. In fact, diatoms accounted for 81% of total microphytoplankton abundance, on average, with a peak of 7.49 × 103 cells ml−1 (95% of total) at station 4 (Table 1), and within diatoms, Chaetoceros was always the dominant genus.
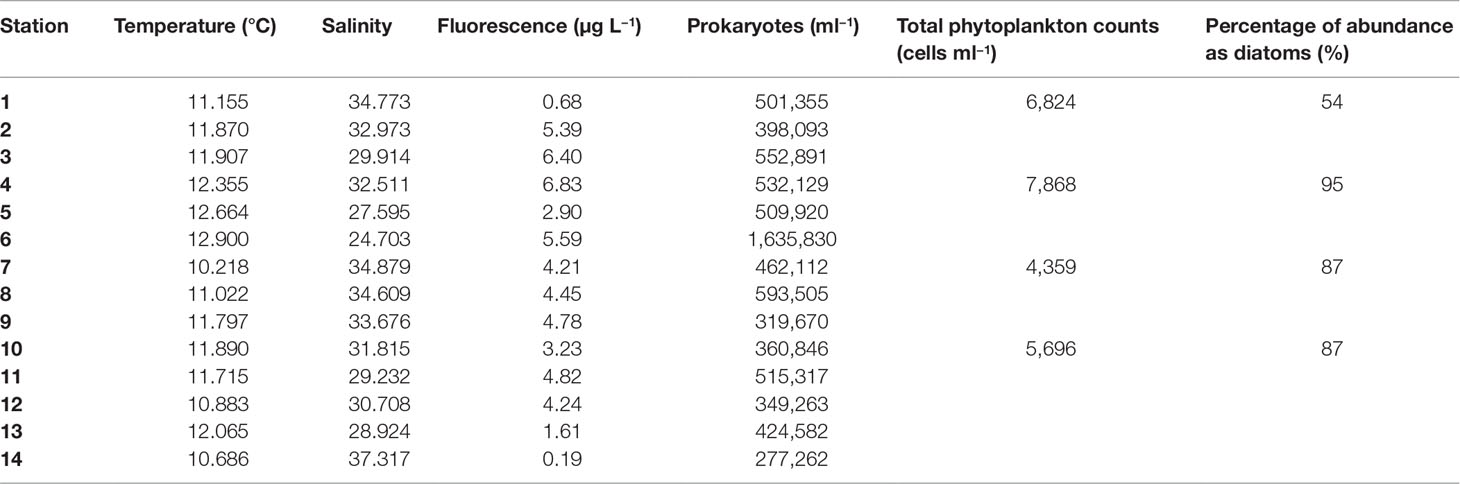
Table 1 Surface values of temperature, salinity, fluorescence levels, total heterotrophic prokaryote abundance (cells ml−1), total phytoplankton abundance, and diatom percentage in 2017 in the Northern Adriatic Sea.
Prokaryotic Diversity and Community Composition
The total (PA+FL) clean dataset contained 44,425 total reads corresponding to 2,293 ASVs with PA including 20,619 reads (1,516 ASVs) and FL having 23,806 reads (1,410 ASVs). After normalization, a total of 2,124 ASVs were left, with 25.4% of them shared between FL and PA samples, 40.2% exclusive of the PA community, and 34.4% exclusive of FL communities (Figure 2).
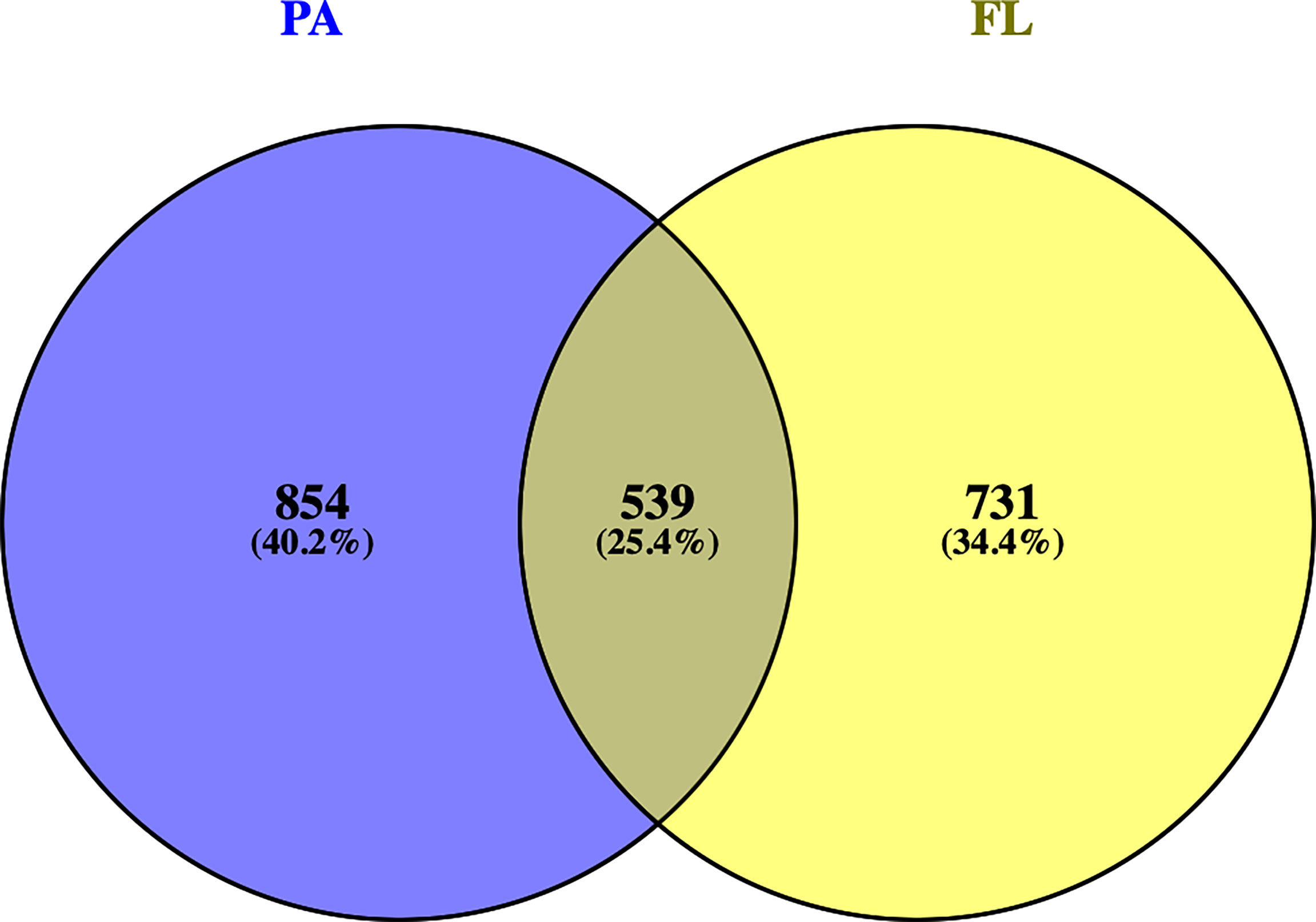
Figure 2 Venn diagram showing the ASV distribution in each of the two fractions: FL (Free-Living, yellow) and PA (Particle-Attached, blue), and those shared by both fractions.
Wilcoxon tests highlighted significantly higher values of alpha-diversity (Observed ASVs, Shannon, and Chao1) in the FL than in the PA communities (Figure 3). In addition, alpha-diversity values within FL were more homogeneous (interquartile range = 0.2) than in PA samples (interquartile range = 0.7).
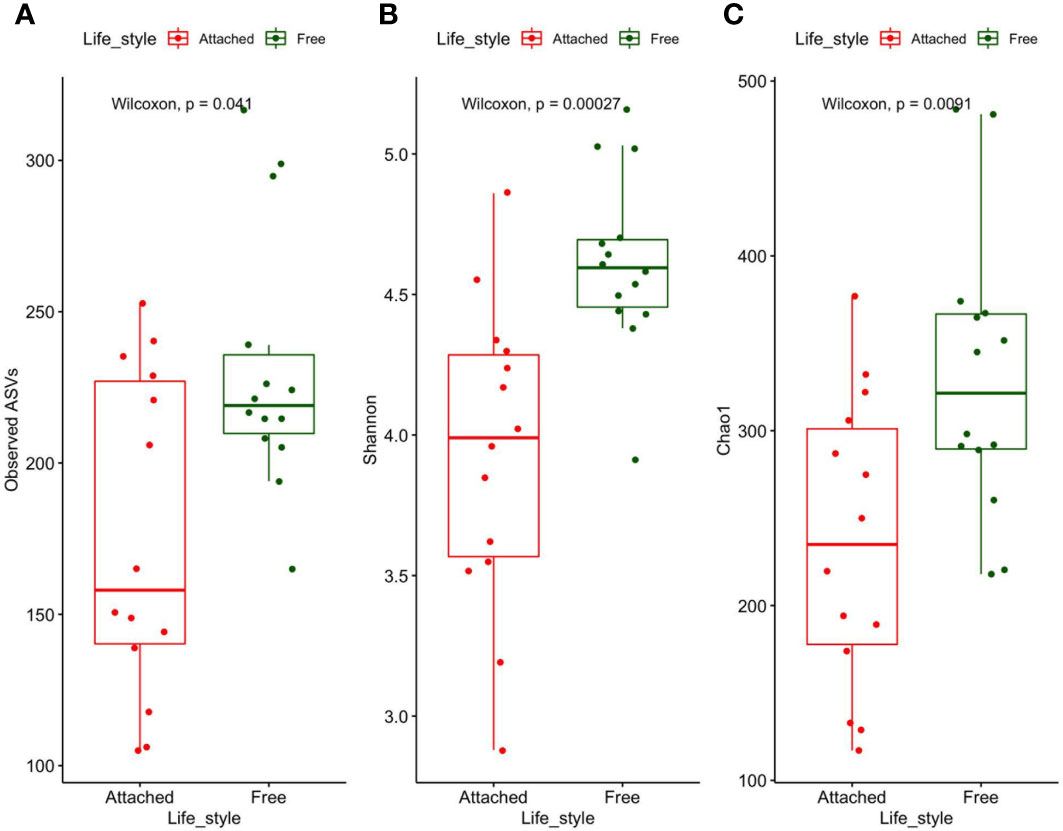
Figure 3 Number of observed ASVs (A), Shannon diversity (B), and Chao I richness (C) in FL (green) and PA (red) prokaryotes. Upper and lower lines correspond to the first and third quartile of the distribution of values, while the horizontal lines indicate the median values.
A clear separation between FL and PA samples was observed by cluster analysis (Figure 4), with the FL samples grouped into one homogeneous cluster and the PA samples, instead, separated into two distinct subgroups, one closer to the FL communities (I) grouping five samples and the other including nine samples mainly located far from the Po River (II). Cluster I was characterized by lower salinity and higher fluorescence values than the other stations (Table 1), suggesting a less selective environment. The distinction between FL and PA was confirmed by the ANOSIM test (R = 0.711, p-value ≤ 0.001) and also by the PERMANOVA analysis, which identified a significant correlation only with salinity (p-value ≤ 0.05), among all environmental parameters. Unfortunately, alpha-diversity was not significantly different between the two clusters (p > 0.5 for all indexes).
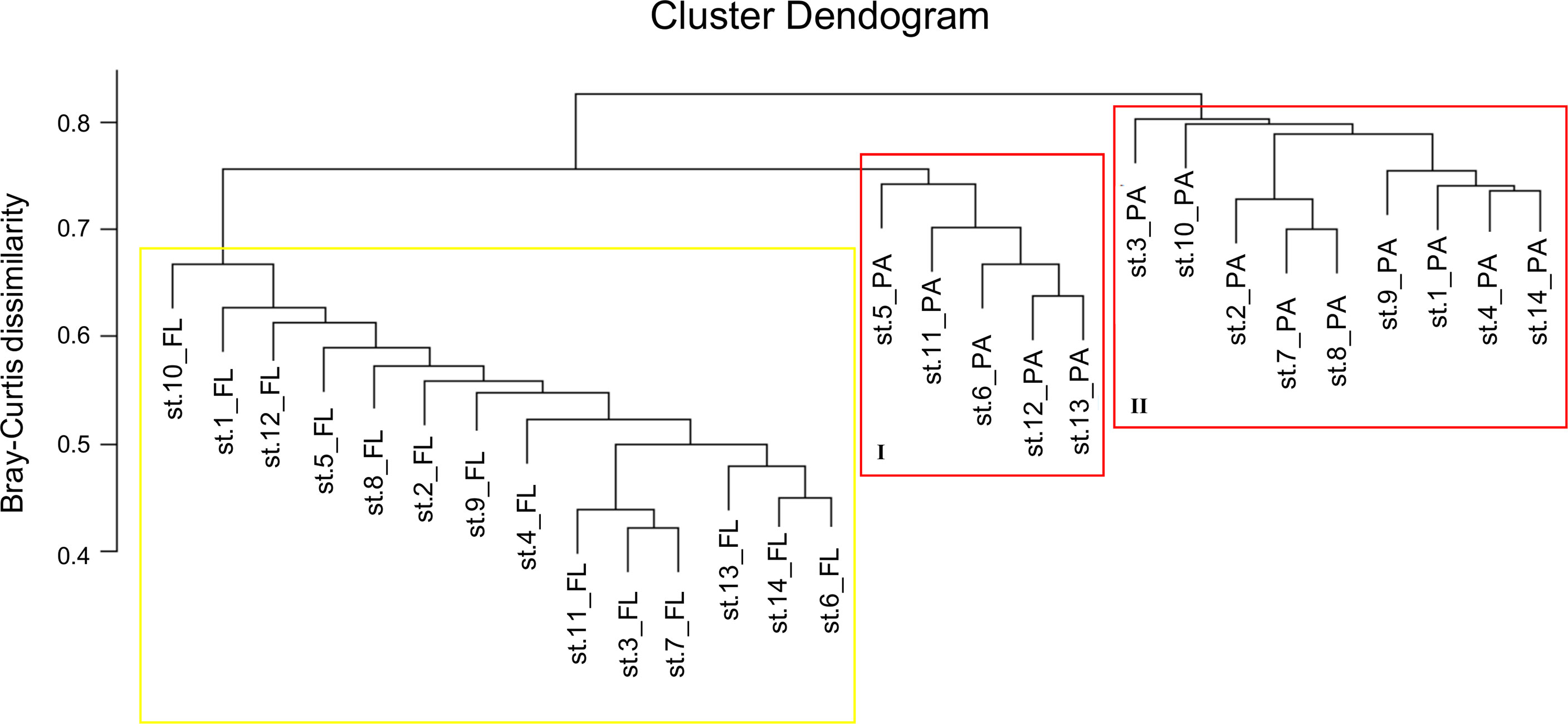
Figure 4 Hierarchical clustering based on a Bray–Curtis dissimilarity matrix. The yellow square delimitates FL communities. The red squares delimitate PA prokaryotic communities.
At the phylum level, the global dataset (FL+PA) was dominated by Proteobacteria, Bacteroidetes, and Firmicutes, representing 52.0%, 28.6%, and 7.4% of the total, respectively. Firmicutes, Planctomycetes, and Acidobacteria showed higher relative abundances in the PA community, with 5.2% (out of 7.4% total), 1.8% (out of 2.7% total), and 0.7% (out of 1.0% total), respectively. Within the PA fraction, Firmicutes were twice as abundant in cluster II compared to cluster I (10.3% vs. 5.3%).
Cyanobacteria were present in both fractions but with very low contributions (1.4% of FL plus PA) (Figure 5). The total of Archaea sequences was less than 1% and therefore they were grouped into “< 1% abund.”.
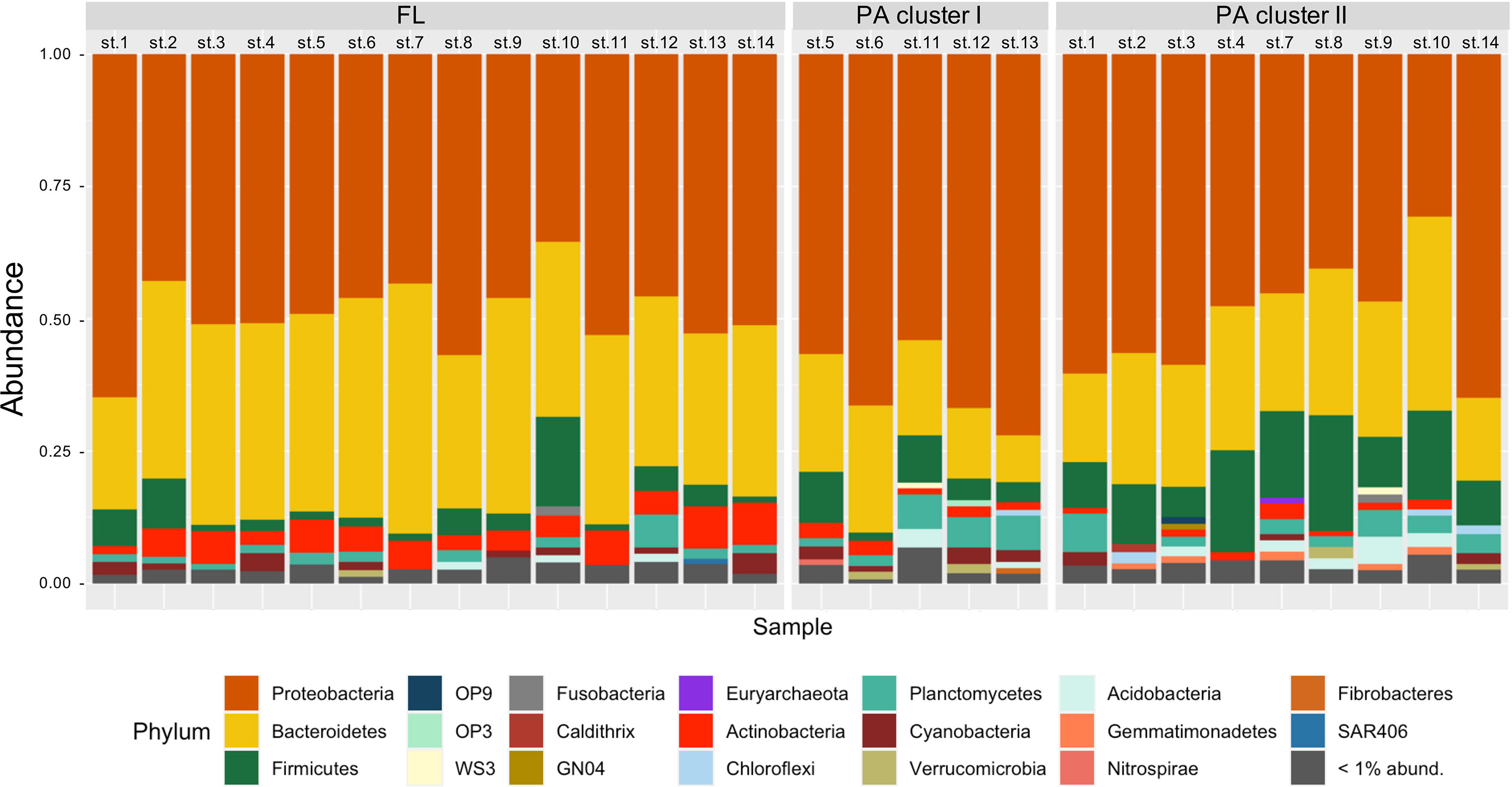
Figure 5 Taxonomic composition (at the phylum level) of FL and PA prokaryotic communities (left and right bars, respectively) at each station. The phyla with abundance < 1% are grouped and represented as “< 1% abund.”, in gray. Station names are on top.
At the family level, Phyllobacteriaceae, Rhodobacteriaceae, and Flavobacteriaceae, as well as Bacteroidaceae, were the most abundant families, with 16.3%, 13.9%, 13.5%, and 6.4% of the total reads, respectively. Of these, 84.0% of Flavobacteriaceae were present in the FL fraction, while 72.3% of Phyllobacteriaceae and 70.0% of Bacteroidaceae were present in the PA fraction. The Rhodobacteriaceae were present in both fractions with 53.9% in the FL and 46.1% in the PA community.
In order to identify taxa that could be considered as indicators of PA or FL lifestyle, LEfSe analysis showed Alphaproteobacteria (Phyllobacterium genus), Firmicutes (Clostridiales order), and the class Bacteroidia (Bacteroides genus of the Bacteroidaceae family) as PA indicators, with other Bacteroidetes (particularly Polaribacter, Sediminicola, and Flavobacterium of the Flavobacteriaceae family), Gammaproteobacteria (OM60 and Alteromonadaceae families), and Actinobacteria (Candidatus Aquiluna genus of the Microbacteriaceae family) as representatives of the FL lifestyle (Figure 6).
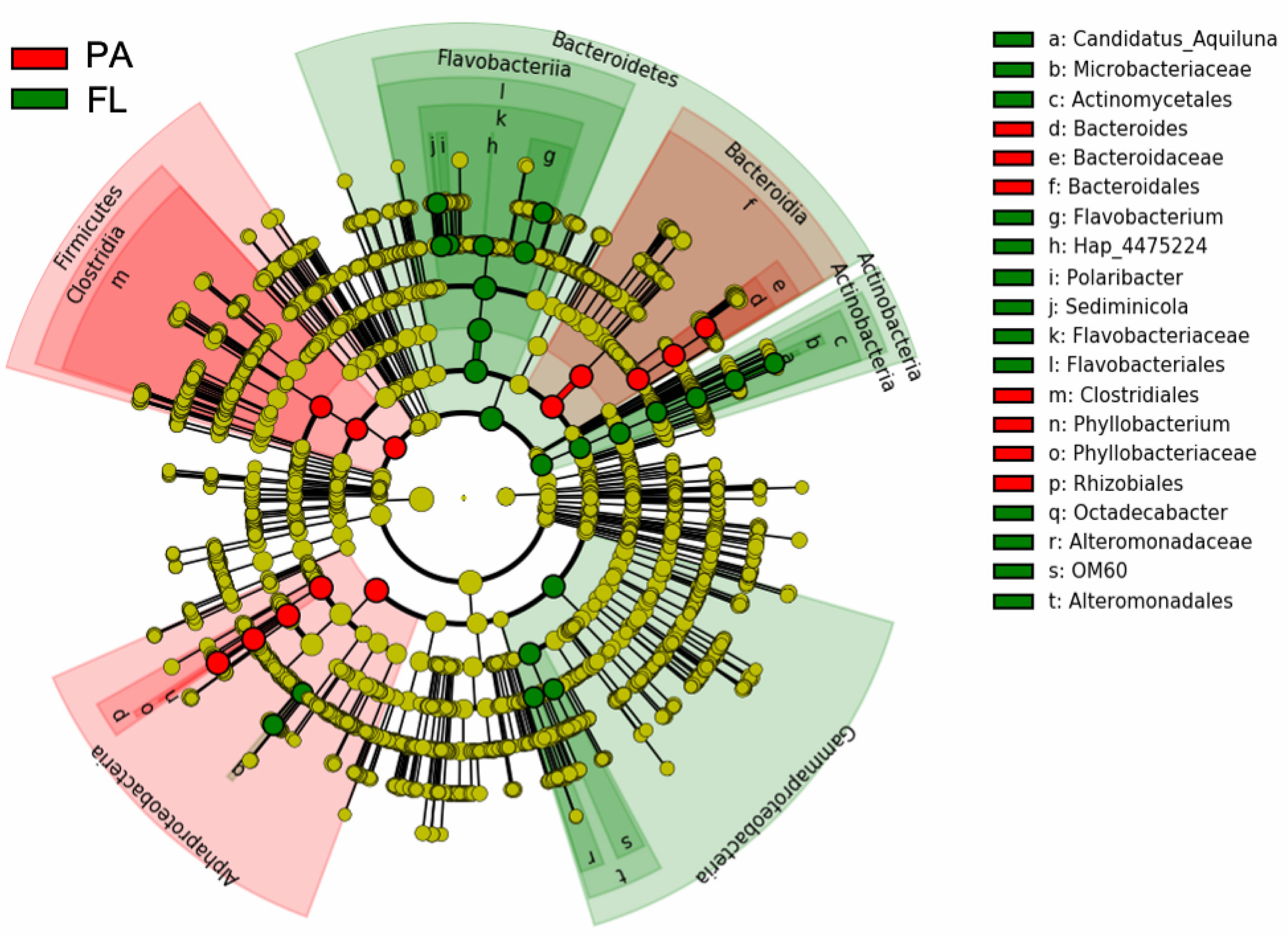
Figure 6 LEfSe analyses (LDA >4; p = 0.05) showing the taxonomic groups that statistically differentiate FL (green) and PA (red) communities using the prokaryotic ASVs table. Taxa with non-significant differences are presented as yellow circles and the diameter of the circles is proportional to relative abundance. Circles represent taxonomic levels from phyla (innermost ring) to genus (outermost ring). On the right, the list of taxa represented in the taxonomic hierarchical cladogram.
A total of 65 functional groups were identified using FAPROTAX, accounting for 42% of all ASVs. Predominant functional groups included aerobic and anaerobic chemoheterotrophy (55%), followed by C fermentation (8.6%) (Table S1 and Figure 7). However, functional signals showed some differences between the FL and PA communities (Figure 7), with the FL bacteria showing a dominance of aerobic chemoheterotrophy and the PA community showing a prevalence of anaerobic chemoheterotrophy and fermentation functions. Within the PA community, the samples were grouped into two main clusters (I and II) that are distinguished by small differences in fermentation and aerobic as well as anaerobic chemoheterotrophy functions.
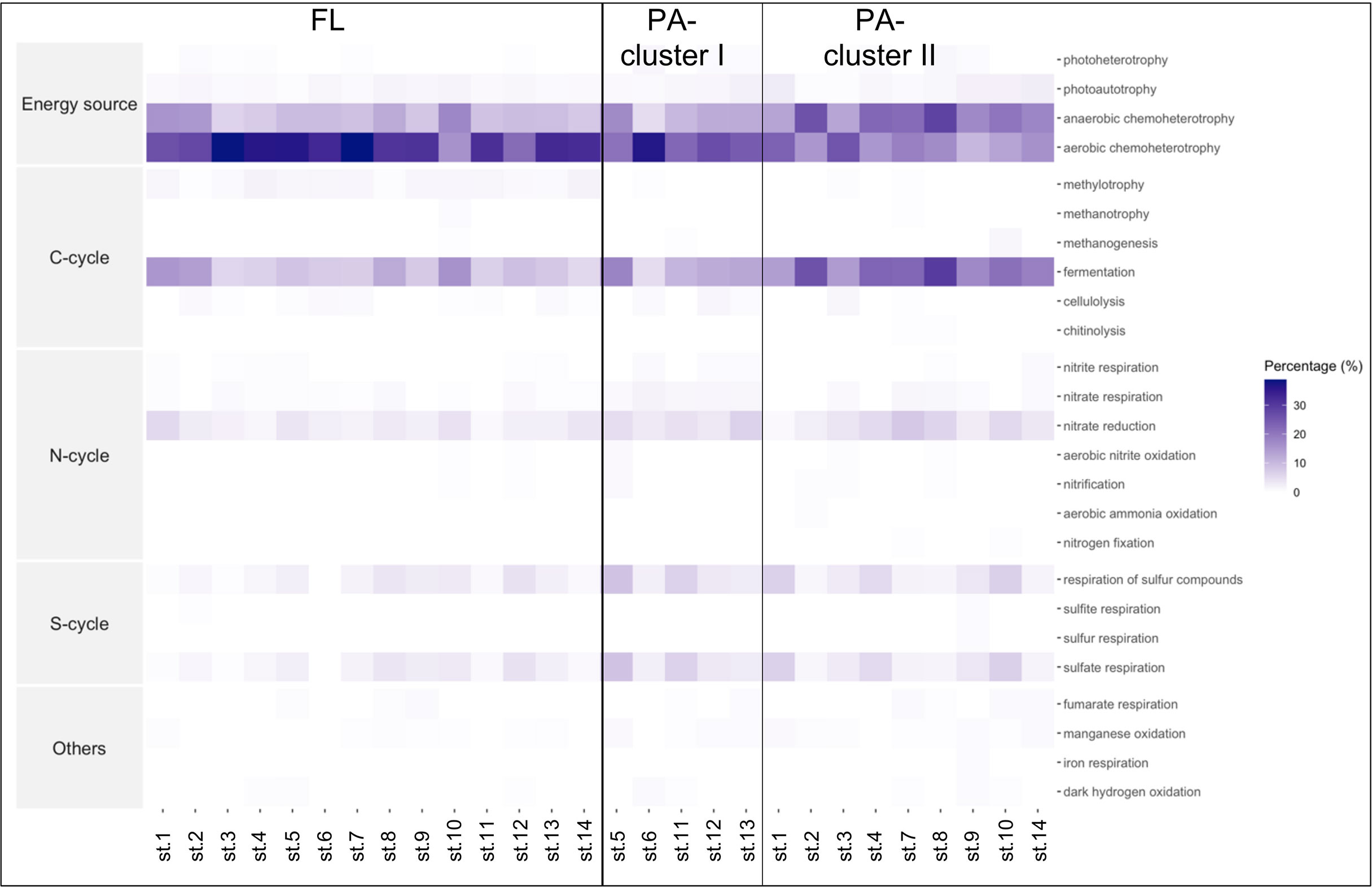
Figure 7 Heatmap of the function distributions generated by FAPROTAX. The columns correspond to the samples and the rows correspond to the percentage of functional groups predicted within each sample. On the left, the Free-Living (FL) samples, and on the right, the Particle-Attached (PA) samples separated in the 2 clusters.
Discussion
In our study, prokaryotic abundances were highest near the Po River delta with a general decrease from the coast to offshore, confirming their elevated concentrations in meso to eutrophic areas (Shiah, 1999; Šilović et al., 2012; Paterson et al., 2013). All the bacterial diversity patterns observed indicated a clear distinction between FL and PA prokaryotic communities as reported in various marine environments including coastal or surface water (Mohit et al., 2014; Zhang et al., 2016), bathypelagic water (Wilkins et al., 2013; Salazar et al., 2016; Milici et al., 2017), or extreme depths of hadalpelagic habitats (>6,000 m) (Eloe et al., 2011; Tarn et al., 2016). Higher diversity in FL than in PA was observed in this study. Although the question of which fraction contains more diversity has been extensively investigated both in the Mediterranean Sea and in other regions, no trend attributable to geographic location, trophic conditions or other factors could be found. In the Mediterranean Sea, some authors found more diversity in the FL fraction (Acinas et al., 1999; Ghiglione et al., 2007), while others, in the PA fraction (Crespo et al., 2013; Mestre et al., 2017b). The same is true for other areas (e.g., Kellogg and Deming, 2009) reporting higher diversity in the FL prokaryotes, with others finding the opposite (e.g., Zhang et al., 2007; Eloe et al., 2011).
Within the FL and PA lifestyles, the FL communities sampled were more homogeneous in diversity and functional profiles than the PA communities, which were, instead, split into two subgroups (see Figure 4). Although a separation of stations based on distance from the Po River or salinity values was statistically supported, the stations of cluster II of PA were located farther away from the Po River. This suggests that factors such as freshwater influence, presence of different particles, and nutrients create different substrates to which PA communities can attach in different ways, likely generating variability of the prokaryotic communities associated to particles.
Although the rarefaction procedure may have affected the absolute value of alpha-diversity, in our study, the communities of the FL fraction show higher signal of diversity and richness than the PA communities. This is likely to be explained by the diatom bloom occurring during sampling, which attracted PA taxa associated or specialized to exploit diatom-derived organic carbon, such as Rhodobacteraceae (Roseobacter and Sulfitobacter) and Bacteroidetes (Flavobacterium), reducing diversity in the PA fraction.
The general taxonomic composition and the LEfSe analysis suggest the preference of specific taxa for either the FL or the PA lifestyle. However, while some previous studies reported a high degree of specialization in different bacterioplankton groups (DeLong et al., 1993; Acinas et al., 1999; Moeseneder et al., 2001; Allgaier et al., 2007; Smith et al., 2013), others observed significant overlap between the FL and PA communities (Hollibaugh et al., 2000; Ghiglione et al., 2007; Crespo et al., 2013). Taxa found in both fractions could be able to switch their lifestyles from PA to FL and vice versa (Crespo et al., 2013; Li et al., 2015; Mestre et al., 2018). It is reasonable to expect that PA prokaryotes can also be capable of surviving freely, so they can colonize new particles (Pedrós-Alió and Brock, 1983; Ghiglione et al., 2007). In marine sites similar to the area sampled in this study, Alphaproteobacteria, Gammaproteobacteria, and Bacteroidetes are present and even dominant in both PA and FL fractions (Yung et al., 2016), although only a finer taxonomical detail can elucidate real differences or similarities.
The Bacteroidaceae family (mostly genus Bacteroides), the Alphaproteobacteria class (particularly the genus Phyllobacterium), and phylum Firmicutes (order Clostridiales) were selected by LEfSe as PA indicators. These taxa were predicted by FAPROTAX analysis to provide anaerobic chemoheterotrophy and C fermentation as expected from anaerobic microenvironments, such as marine particles, guts of zooplankton, or others (Bickel and Tang, 2014). In particular, members belonging to the Bacteroidia class, within the Bacteroidaceae family, are frequently found in soils and freshwater, and are considered specialized in the degradation of high-molecular-weight organic matter, i.e., proteins and carbohydrates (Thomas et al., 2011). These groups are also found to attach to plastic debris (Lee et al., 2008; Jain and Krishnan, 2017). In addition, in Korean coastal waters, Chun et al. (2021) reported an increase of Bacteroidia during diatom (e.g., Chaetoceros, Pseudo-nitzschia, Leptocylindrus, Thalassiosira, and Skeletonema) blooms, probably related to their ability in degradation of phytoplankton-derived organic matter, which are further used by other bacteria as carbon sources (Williams et al., 2013). Our results support these observations, confirming the ecological role of Bacteroidia as an important group in the PA fraction of coastal areas, especially in the presence of diatom blooms. The other taxa with a PA lifestyle preference were genus Phyllobacterium (Alphaproteobacteria) and the Firmicutes phylum (order Clostridiales). Members of the Phyllobacteriaceae family are reported to thrive in soil and water and/or to be associated with plants (Willems, 2014), and Firmicutes represent large components of microbial communities in soils (Roesch et al., 2007; Youssef and Elshahed, 2009), the members of which have also been reported as feces-associated bacteria in coastal waters (Basili et al., 2020). Within Firmicutes, members of the Lachnospiraceae family, and genera Streptococcus and Ruminococcus are typical sewage-associated prokaryotes (Oberbeckmann et al., 2015). In our dataset, Firmicutes differentiate between clusters, with members of Clostridia class (Blautia and Dorea genera in the Lachnospiraceae family and Oscillospira, Ruminococcus, and Faecalibacterium genera of the Ruminococcaceae family) and members of Bacilli (Enterococcus, Lactobacillus, and Streptococcus genera) particularly abundant in cluster II, indicating anaerobic chemoheterotrophy and fermentation activity that may mark their terrestrial origin and/or untreated sewage water discharged into the Po River or directly into the sea.
Members of Flavobacteria (Bacteroidetes), instead, show a preference for the FL lifestyle, despite the fact that they are often reported as associated with phytoplankton and/or other particles (Kirchman, 2002; Karrasch et al., 2003; Teeling et al., 2012). In our data, Flavobacterium and Polaribacter were associated to aerobic chemohetrotrophy function, likely related to the presence of high levels of nutrients and organic matter (Kirchman, 2002; Elifantz et al., 2007), as expected for the Po river discharge area. Other groups, such as Gammaproteobacteria and Actinobacteria, were selected by LEfSe as FL indicators. Within Gammaproteobacteria, the Alteromonadales and OM60 families were predicted to have aerobic chemoheterotrophy function, and were found to be associated to FL fraction, contrary to what has been reported in other studies (Zhang et al., 2007; Fuchsman et al., 2011; Gomez-Consarnau et al., 2019), where they are usually considered as typical PA prokaryotes. It is well known that most Gammaproteobacteria have high nutrient requirements (Schattenhofer et al., 2009) and they live attached to particles as a strategy to prevent nutrient limitation (Crespo et al., 2013). However, this was hardly the case of this study, as the Po River is a large source of nutrients, possibly allowing for their presence in the FL fraction. The presence of genus Candidatus Aquiluna of the Microbacteriaceae family (Actinobacteria phylum) is a remarkable indicator of the FL fraction. This genus was initially proposed as an indigenous freshwater species (Hahn, 2009), but has also been reported in the coastal marine environment (Šilović et al., 2012; Elifantz et al., 2013). Its presence in the sampled area could therefore be considered a marker of river influence and terrestrial discharges in general.
Interestingly, FAPROTAX analysis revealed different functional patterns not only between FL and PA communities but also among the two PA clusters, with cluster I showing prevalence of aerobic chemoheterotrophy, similar to FL communities (e.g., Flavobacteriaceae members), as compared to the strong anaerobic signal found in cluster II. This has implications for the ecological adaptation abilities to different sources and kinds of organic matter, which reverberates on the functioning and efficiency of the microbial loop in the two areas. This outcome also suggests that the PA lifestyle is heterogeneous and can include specialized taxa, especially in cluster II, together with other, more generalist, able to shift between lifestyles and functions. This contrasting evidence observed for some taxa may suggest that for a better definition of taxa lifestyle or preferences for different types of particles or substrates, a finer taxonomic level is needed, which is not always possible with metabarcoding data, as it does not differentiate between ecotypes.
In conclusion, our study confirms different taxonomic profiles between PA and FL prokaryotes and, coupled with functional diversity, provides new insights into the potential drivers shaping the two types of communities including biotic and abiotic factors as well as anthropic pressures. Despite the relatively high number of studies published describing the prokaryotic components of PA and FL communities in coastal ocean areas, further studies including the full seasonal cycle and the effect of environmental fluctuations would allow researchers to elucidate the complexity of the factors governing the community structure of particle-attached and free-living prokaryotic communities. Metagenomic and metatranscriptomic profiles would also be key to better assess taxonomical and associated functional diversity of these two prokaryotic community types.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/ - BioProject ID PRJNA834028; accession numbers for single samples are SAMN28028918- SAMN28028941.
Author Contributions
AT and RC conceived the research. AT performed the experiments, produced and analyzed the data, and prepared figures and tables under the supervision and following advice from RP, RC, and JG. CB, RP, and MB were involved in sampling, sample processing, data analysis, and interpretation. AT and RC wrote the draft manuscript. All authors were involved in writing, and read and approved the final version of the manuscript.
Funding
This is a contribution of RITMARE Flagship Project by Italian MIUR DTA.AD008. AT was supported by a Ph.D. fellowship funded by the Stazione Zoologica Anton Dohrn (Open University-Stazione Zoologica Anton Dohrn Ph.D. Program).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank Stefania Finotto (CNR-ISMAR, Venice) for the phytoplankton counts, Jessica Titocci, and the captain and crew of R/V Minerva Uno for the sampling. André Comeau from IMR is kindly acknowledged for his support in the sequencing.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2022.912528/full#supplementary-material
References
Acinas S. G., Antón J., Rodríguez-Valera F. (1999). Diversity of Free-Living and Attached Bacteria in Offshore Western Mediterranean Waters as Depicted by Analysis of Genes Encoding 16s rRNA. Appl. Environ. Microbiol. 65, 514–522. doi: 10.1128/AEM.65.2.514-522.1999
Alldredge A. L., Silver M. W. (1988). Characteristics, Dynamics and Significance of Marine Snow. Prog. Oceanogr. 20, 41–82. doi: 10.1016/0079-6611(88)90053-5
Allen A. E., Laroche J., Maheswari U., Lommer M., Schauer N., Lopez P. J, et al. (2008). Whole-Cell Response of the Pennate Diatom Phaeodactylum Tricornutum to Iron Starvation. Proc. Natl. Acad. Sci. U. S. A. 105 (30), 10438–10443. doi: 10.1073/pnas.0711370105
Allgaier M., Brückner S., Jaspers E., Grossart H. P. (2007). Intra- and Inter-Lake Variability of Free-Living and Particle-Associated Actinobacteria Communities. Environ. Microbiol. 9, 2728–2741. doi: 10.1111/j.1462-2920.2007.01385.x
Amin S. A., Hmelo L. R., van Tol H. M., Durham B. P., Carlson L. T., Heal K. R., et al. (2015). Interaction and Signalling Between a Cosmopolitan Phytoplankton and Associated Bacteria. Nature 522 (7554), 98–101. doi: 10.1038/nature14488
Amin S. A., Parker M. S., Armbrust E. F. (2012). Interactions Between Diatoms and Bacteria. Microbiol. Mol. Biol. Rev. 76, 667–684. doi: 10.1128/MMBR.00007-12
Azam F., Ammerman J. W. (1984). “Cycling of Organic Matter by Bacterioplankton in Pelagic Marine Ecosystems: Microenvironmental Considerations,” in Flows of Energy and Materials in Marine Ecosystems. Ed. Fasham M. J. R. (Boston, MA: Springer US), 345–360. doi: 10.1007/978-1-4757-0387-0_14
Azam F., Malfatti F. (2007). Microbial Structuring of Marine Ecosystems. Nat. Rev. Microbiol. 5, 782–791. doi: 10.1038/nrmicro1747
Azam F., Smith D. C., Steward G. F., Hagström Å. (1994). Bacteria-Organic Matter Coupling and Its Significance for Oceanic Carbon Cycling. Microbial Ecol. 28, 167–179. doi: 10.1007/BF00166806
Balestra C., Alonso-Sáez L., Gasol J. M., Casotti R. (2011). Group-Specific Effects on Coastal Bacterioplankton of Polyunsaturated Aldehydes Produced by Diatoms. Aquat. Microb. Ecol. 63, 123–131. doi: 10.3354/ame01486
Barofsky A., Vidoudez C., Pohnert G. (2009). Metabolic Profiling Reveals Growth Stage Variability in Diatom Exudates. Limnol. Oceanogr. Methods 7, 382–390. doi: 10.4319/lom.2009.7.382
Basili M., Quero G. M., Giovannelli D., Manini E., Vignaroli C., Avio C. G., et al. (2020). Major Role of Surrounding Environment in Shaping Biofilm Community Composition on Marine Plastic Debris. Front. Mar. Sci. 7. doi: 10.3389/fmars.2020.00262
Becker J. W., Berube P. M., Follett C. L., Waterbury J. B., Chisholm S. W., Delong E. F., et al. (2014) Closely Related Phytoplankton Species Produce Similar Suites of Dissolved Organic Matter. Front. Microbiol. 5, 1–14. doi: 10.3389/fmicb.2014.00111
Bell W., Mitchell R. (1972). Chemotactic and Growth Responses of Marine Bacteria to Algal Extracellular Products. Biol. Bull. 143, 265–277. doi: 10.2307/1540052
Bernardi Aubry F., Acri F., Finotto S., Pugnetti A. (2021). Phytoplankton Dynamics and Water Quality in the Venice Lagoon. Water 13, 2780. doi: 10.3390/w13192780
Bickel S. L., Tang K. W. (2014). Carbon Substrate Usage by Zooplankton-Associated Bacteria, Phytoplankton-Associated Bacteria, and Free-Living Bacteria Under Aerobic and Anaerobic Conditions. Mar. Biol. 161, 2233–2242. doi: 10.1007/s00227-014-2501-z
Biddanda B. A., Pomeroy L. R. (1988). Microbial Aggregation and Degradation of Phytoplankton-Derived Detritus in Seawater. Microbial Metabolism. Mar. Ecol. Prog. Ser. 42, 79–88.
Bolyen E., Rideout J. R., Dillon M. R., Bokulich N. A., Abnet C. C., Al-Ghalith G. A., et al. (2019). Reproducible, Interactive, Scalable and Extensible Microbiome Data Science Using QIIME 2. Nat. Biotechnol. 37, 852–857. doi: 10.1038/s41587-019-0209-9
Boström K. H., Simu K., Hagström Å., Riemann L. (2004). Optimization of DNA Extraction for Quantitative Marine Bacterioplankton Community Analysis: DNA Extraction From Bacterioplankton. Limnol. Oceanogr. Methods 2, 365–373. doi: 10.4319/lom.2004.2.365
Buchan A., LeCleir G. R., Gulvik C. A., Gonzalez J. M. (2014). Master Recyclers: Features and Functions of Bacteria Associated With Phytoplankton Blooms. Nat. Rev. Microbiol. 12, 686–698. doi: 10.1038/nrmicro3326
Callahan B. J., McMurdie P. J., Rosen M. J., Han A. W., Johnson A. J. A., Holmes S. P. (2016). DADA2: High-Resolution Sample Inference From Illumina Amplicon Data. Nat. Methods 13, 581–583. doi: 10.1038/nmeth.3869
Catalano G., Azzaro M., Bastianini M., Bellucci L. G., Bernardi Aubry F., Bianchi F., et al. (2014). The Carbon Budget in the Northern Adriatic Sea, a Winter Casestudy. J. Geophys. Res.: Biogeosci. 119, 1399–1417. doi: 10.1002/2013JG002559
Celussi M., Bussani A., Cataletto B., Del Negro P. (2010). Assemblages’ Structure and Activity of Bacterioplankton in Northern Adriatic Sea Surface Waters: A 3-Year Case Study. FEMS Microbiol. Ecol. 75 (1), 77–88. doi: 10.1111/j.1574-6941.2010.00997.x
Christie-Oleza J. A., Sousoni D., Lloyd M., Armengaud J., Scanlan D. J. (2017). Nutrient Recycling Facilitates Long-Term Stability of Marine Microbial Phototroph-Heterotroph Interactions. Nat. Microbiol. 2, 1–10. doi: 10.1038/nmicrobiol.2017.100
Chun S.-J., Cui Y., Baek S. H., Ahn C.-Y., Oh H.-M. (2021). Seasonal Succession of Microbes in Different Size-Fractions and Their Modular Structures Determined by Both Macro- and Micro-Environmental Filtering in Dynamic Coastal Waters. Sci. Total Environ. 784, 147046. doi: 10.1016/j.scitotenv.2021.147046
Clarke K. R. (1993). Non-Parametric Multivariate Analyses of Changes in Community Structure. Austral Ecol. 18, 117–143. doi: 10.1111/j.1442-9993.1993.tb00438.x
Cozzi S., Giani M. (2011). River Water and Nutrient Discharges in the Northern Adriatic Sea: Current Importance and Long Term Changes. Continental Shelf Res. 31, 1881–1893. doi: 10.1016/j.csr.2011.08.010
Creer S., Fonseca V. G., Porazinska D. L., Giblin-Davis R. M., Sung W., Power D. M., et al. (2010). Ultrasequencing of the Meiofaunal Biosphere: Practice, Pitfalls and Promises. Mol. Ecol. 19, 4–20. doi: 10.1111/j.1365-294X.2009.04473.x
Crespo B. G., Pommier T., Fernández-Gómez B., Pedrós-Alió C. (2013). Taxonomic Composition of the Particle-Attached and Free-Living Bacterial Assemblages in the Northwest Mediterranean Sea Analyzed by Pyrosequencing of the 16S rRNA. Microbiology Open 2, 541–552. doi: 10.1002/mbo3.92
Crump B. C., Armbrust E. V., Baross J. A. (1999). Phylogenetic Analysis of Particle-Attached and Free-Living Bacterial Communities in the Columbia River, Its Estuary, and the Adjacent Coastal Ocean. Appl. Environ. Microbiol. 65, 3192–3204. doi: 10.1128/AEM.65.7.3192-3204.1999
Crump B. C., Hopkinson C. S., Sogin M. L., Hobbie J. E. (2004). Microbial Biogeography Along an Estuarine Salinity Gradient: Combined Influences of Bacterial Growth and Residence Time. Appl. Environ. Microbiol. 70, 1494–1505. doi: 10.1128/aem.70.3.1494-1505.2004
DeLong E. F., Franks D. G., Alldredge A. L. (1993). Phylogenetic Diversity of Aggregate-Attached vs. Free-Living Marine Bacterial Assemblages. Limnol. Oceanogr. 38, 924–934. doi: 10.4319/lo.1993.38.5.0924
Djakovac T., Degobbis D., Supić N., Precali R. (2012). Marked Reduction of Eutrophication Pressure in the Northeastern Adriatic in the Period 2000–2009. Estuar. Coast. Shelf Sci. 115, 25–32. doi: 10.1016/j.ecss.2012.03.029
Durham B. P., Sharma S., Luo H., Smith C. B., Amin S. A., Bender S. J., et al. (2015). Cryptic Carbon and Sulfur Cycling Between Surface Ocean Plankton. Proc. Natl. Acad. Sci. U. S. A. 112 (2), 453–457. doi: 10.1073/pnas.1413137112
Edler L., Elbrächter M. (2010). “The Utermöhl Method for Quantitative Phytoplankton Analysis,” in Microscopic and Molecular Methods for Quantitative Phytoplankton Analysis. Eds. Karlson B., Cusack C. K., Bresnan E. (Paris: UNESCO), 13–20.
Eigemann F., Rahav E., Grossart H. P., Aharonovich D., Sher D., Vogts A., et al. (2022). Phytoplankton Exudates Provide Full Nutrition to a Subset of Accompanying Heterotrophic Bacteria via Carbon, Nitrogen and Phosphorus Allocation. Environ. Microbiol. 24, 2467–2483. doi: 10.1111/1462-2920.15933
Elifantz H., Dittel A., Cottrell M., Kirchman D. (2007). Dissolved Organic Matter Assimilation by Heterotrophic Bacterial Groups in the Western Arctic Ocean. Aquat. Microb. Ecol. 50, 39–49. doi: 10.3354/ame01145
Elifantz H., Horn G., Ayon M., Cohen Y., Minz D. (2013). Rhodobacteraceae are the Key Members of the Microbial Community of the Initial Biofilm Formed in Eastern Mediterranean Coastal Seawater. FEMS Microbiol. Ecol. 85, 348–357. doi: 10.1111/1574-6941.12122
Eloe E. A., Shulse C. N., Fadrosh D. W., Williamson S. J., Allen E. E., Bartlett D. H. (2011). Compositional Differences in Particle-Associated and Free-Living Microbial Assemblages From an Extreme Deep-Ocean Environment: Microbial Diversity in the Puerto Rico Trench. Environ. Microbiol. Rep. 3, 449–458. doi: 10.1111/j.1758-2229.2010.00223.x
Ferrier M., Martin J. L., Rooney-Varga J. N. (2002). Stimulation of Alexandrium Fundyense Growth by Bacterial Assemblages From the Bay of Fundy. J. Appl. Microbiol. 92, 706–716. doi: 10.1046/j.1365-2672.2002.01576.x
Fuchsman C. A., Kirkpatrick J. B., Brazelton W. J., Murray J. W., Staley J. T. (2011). Metabolic Strategies of Free-Living and Aggregate-Associated Bacterial Communities Inferred From Biologic and Chemical Profiles in the Black Sea Suboxic Zone. FEMS Microbiol. Ecol. 78, 586–603. doi: 10.1111/j.1574-6941.2011.01189.x
Fuhrman J. A., Ammerman J. W., Azam F. (1980). Bacterioplankton in the Coastal Euphotic Zone: Distribution, Activity and Possible Relationships With Phytoplankton. Mar. Biol. 60, 201–207. doi: 10.1007/BF00389163
Furuki M., Kobayashi M. (1991). Interaction Between Chattonella and Bacteria and Prevention of This Red Tide. Mar. pollut. Bull. 23, 189–193. doi: 10.1016/0025-326X(91)90673-G
Fu H., Uchimiya M., Gore J., Moran M. A. (2020). Ecological Drivers of Bacterial Community Assembly in Synthetic Phycospheres. Proc. Natl. Acad. Sci. 117.7, 3656–3662. doi: 10.1073/pnas.1917265117
Gallacher S., Flynn K. J., Franco J. M., Brueggemann E. E., Hines H. B. (1997). Evidence for Production of Paralytic Shellfish Toxins by Bacteria Associated With Alexandrium Spp. (Dinophyta) in Culture. Appl. Environ. Microbiol. 63, 239–245. doi: 10.1128/aem.63.1.239-245.1997
Gasol J. M., Del Giorgio P. A. (2000). Using Flow Cytometry for Counting Natural Planktonic Bacteria and Understanding the Structure of Planktonic Bacterial Communities. Sci. Mar. 64, 197–224. doi: 10.3989/scimar.2000.64n2197
Gasol J. M., Duarte C. M. (2000). Comparative Analyses in Aquatic Microbial Ecology: How Far do They Go? FEMS Microbiol. Ecol. 31, 99–106. doi: 10.1111/j.1574-6941.2000.tb00675.x
Ghiglione J. F., Mevel G., Pujo-Pay M., Mousseau L., Lebaron P., Goutx M. (2007). Diel and Seasonal Variations in Abundance, Activity, and Community Structure of Particle-Attached and Free-Living Bacteria in NW Mediterranean Sea. Microb. Ecol. 54, 217–231. doi: 10.1007/s00248-006-9189-7
Gómez-Consarnau L., Needham D. M., Weber P. K., Fuhrman J. A., Mayali X. (2019). Influence of Light on Particulate Organic Matter Utilization by Attached and Free-Living Marine Bacteria. Front. Microbiol. 10. doi: 10.3389/fmicb.2019.01204
Hahn M. W. (2009). Description of Seven Candidate Species Affiliated With the Phylum Actinobacteria, Representing Planktonic Freshwater Bacteria. Internation J. Of Systematic And Evolutionary Microbiol. 59, 112–117. doi: 10.1099/ijs.0.001743-0
Hollibaugh J., Wong P., Murrell M. (2000). Similarity of Particle-Associated and Free-Living Bacterial Communities in Northern San Francisco Bay, California. Aquat. Microb. Ecol. 21, 103–114. doi: 10.3354/ame021103
Hoppe H. G. (1981). Blue-Green Algae Agglomeration in Surface Water: A Microbiotope of High Bacterial Activity. Kieler Meeresforsch 5, 291–303.
Imai I., Ishida Y., Hata Y. (1993). Killing of Marine Phytoplankton by a Gliding Bacterium Cytophaga Sp., Isolated From the Coastal Sea of Japan. Mar. Biol. 116, 527–532. doi: 10.1007/BF00355470
Jain A., Krishnan K. P. (2017). Differences in Free-Living and Particle-Associated Bacterial Communities and Their Spatial Variation in Kongsfjorden, Arctic. J. Basic Microbiol. 57, 827–838. doi: 10.1002/jobm.201700216
Janse I., Zwart G., van der Maarel M., Gottschal J. (2000). Composition of the Bacterial Community Degrading Phaeocystis Mucopolysaccharides in Enrichment Cultures. Aquat. Microb. Ecol. 22, 119–133. doi: 10.3354/ame022119
Karrasch B., Ullrich S., Mehrens M., Zimmermann-Timm H. (2003). Free and Particle-Associated Extracellular Enzyme Activity and Bacterial Production in the Lower Elbe Estuary, Germany. Acta Hydrochim. Hydrobiol. 31, 297–306. doi: 10.1002/aheh.200300505
Kellogg C., Deming J. (2009). Comparison of Free-Living, Suspended Particle, and Aggregate-Associated Bacterial and Archaeal Communities in the Laptev Sea. Aquat. Microb. Ecol. 57, 1–18. doi: 10.3354/ame01317
Kirchman D. L. (2002). The Ecology of Cytophaga–Flavobacteria in Aquatic Environments. FEMS Microbiol. Ecol. 39, 91–100. doi: 10.1111/j.1574-6941.2002.tb00910.x
Kraus R., Supić N., Precali R. (2016). Factors Favouring Phytoplankton Blooms in the Northern Adriatic: Towards the Northern Adriatic Empirical Ecological Model. Ocean Sci. 12, 19–37. doi: 10.5194/os-12-19-2016
Lage O. M., Bondoso J. (2011). Planctomycetes Diversity Associated With Macroalgae: Planctomycetes-Macroalgae Diversity. FEMS Microbiol. Ecol. 78, 366–375. doi: 10.1111/j.1574-6941.2011.01168.x
Landa M., Burns A. S., Roth S. J., Moran M. A. (2017). Bacterial Transcriptome Remodeling During Sequential Co-Culture With a Marine Dinoflagellate and Diatom. Isme J. (12), 2677–2690. doi: 10.1038/ismej.2017.117
Lee J.-W., Nam J.-H., Kim Y.-H., Lee K.-H., Lee D. H. (2008). Bacterial Communities in the Initial Stage of Marine Biofilm Formation on Artificial Surfaces. J. Microbiol. 46, 174–182. doi: 10.1007/s12275-008-0032-3
Liu C., Cui Y., Li X., Yao M. (2021). Microeco: An R Package for Data Mining in Microbial Community Ecology. FEMS Microbiol. Ecol. 97 (2), fiaa255. doi: 10.1093/femsec/fiaa255
Li J., Wei B., Wang J., Liu Y., Dasgupta S., Zhang L., et al. (2015). Variation in Abundance and Community Structure of Particle-Attached and Free-Living Bacteria in the South China Sea. Deep Sea Res. Part II: Topical Stud. Oceanography 122, 64–73. doi: 10.1016/j.dsr2.2015.07.006
Louca S., Parfrey L. W., Doebeli M. (2016). Decoupling Function and Taxonomy in the Global Ocean Microbiome. Science 353, 1272–1277. doi: 10.1126/science.aaf4507
Mühlenbruch M., Grossart H., Eigemann F., Voss M. (2018). Mini-Review: Phytoplankton-Derived Polysaccharides in the Marine Environment and Their Interactions With Heterotrophic Bacteria. Environ. Microbiol. 20, 2671–2685. doi: 10.1111/1462-2920.14302
Malviya S., Scalco E., Audic S., Vincent F., Veluchamy A., Poulain J., et al. (2016). Insights Into Global Diatom Distribution and Diversity in the World’s Ocean. Proc. Natl. Acad. Sci. U. S. A. 113 (11), E1516–E1525. doi: 10.1073/pnas.1509523113
Mangoni O., Modigh M., Mozetič P., Bergamasco A., Rivaro P., Saggiomo V. (2008). Structure and Photosynthetic Properties of Phytoplankton Assemblages in a Highly Dynamic System, the Northern Adriatic Sea. Estuarine Coast. Shelf Sci. 77, 633–644. doi: 10.1016/j.ecss.2007.10.023
Marie D., Brussaard C. P. D., Thyrhaug R., Bratbak G., Vaulot D. (1999). Enumeration of Marine Viruses in Culture and Natural Samples by Flow Cytometry. Appl. Environ. Microbiol. 65, 45–52. doi: 10.1128/AEM.65.1.45-52.1999
Mayali X., Azam F. (2004). Algicidal Bacteria in the Sea and Their Impact on Algal Blooms1. J. Eukaryotic Microbiol. 51, 139–144. doi: 10.1111/j.1550-7408.2004.tb00538.x
McMurdie P. J., Holmes S. (2013). Phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLoS One 8, e61217. doi: 10.1371/journal.pone.0061217
Mestre M., Borrull E., Sala M. M., Gasol J. M. (2017a). Patterns of Bacterial Diversity in the Marine Planktonic Particulate Matter Continuum. ISME J. 11, 999–1010. doi: 10.1038/ismej.2016.166
Mestre M., Ferrera I., Borrull E., Ortega-Retuerta E., Mbedi S., Grossart H. P., et al. (2017b). Spatial Variability of Marine Bacterial and Archaeal Communities Along the Particulate Matter Continuum. Mol. Ecol. 26, 6827–6840. doi: 10.1111/mec.14421
Mestre M., Ruiz-González C., Logares R., Duarte C. M., Gasol J. M., Sala M. M. (2018). Sinking Particles Promote Vertical Connectivity in the Ocean Microbiome. Proc. Natl. Acad. Sci. U. S. A. 115, E6799–E6807. doi: 10.1073/pnas.1802470115
Milici M., Deng Z. L., Tomasch J., Decelle J., Wos-Oxley M. L., Wang H., et al. (2016). Co-Occurrence Analysis of Microbial Taxa in the Atlantic Ocean Reveals High Connectivity in the Free-Living Bacterioplankton. Front. Microbiol. 7. doi: 10.3389/fmicb.2016.00649
Milici M., Vital M., Tomasch J., Badewien T. H., Giebel H. A., Plumeier I., et al. (2017). Diversity and Community Composition of Particle-Associated and Free-Living Bacteria in Mesopelagic and Bathypelagic Southern Ocean Water Masses: Evidence of Dispersal Limitation in the Bransfield Strait: Bacteria in the Deep Southern Ocean. Limnol. Oceanogr. 62, 1080–1095. doi: 10.1002/lno.10487
Moeseneder M. M., Winter C., Herndl G. J. (2001). Horizontal and Vertical Complexity of Attached and Free-Living Bacteria of the Eastern Mediterranean Sea, Determined by 16S rDNA and 16S rRNA Fingerprints. Limnol. Oceanogr. 46, 95–107. doi: 10.4319/lo.2001.46.1.0095
Mohit V., Archambault P., Toupoint N., Lovejoy C. (2014). Phylogenetic Differences in Attached and Free-Living Bacterial Communities in a Temperate Coastal Lagoon During Summer, Revealed via High-Throughput 16s rRNA Gene Sequencing. Appl. Environ. Microbiol. 80, 2071–2083. doi: 10.1128/AEM.02916-13
Moran M. A., Ferrer-González F. X., Fu H., Nowinski B., Olofsson M., Powers M. A., et al. (2022). The Ocean’s Labile DOC Supply Chain. Limnol. Oceanogr. 7, 1007–1021. doi: 10.1002/lno.12053
Moran M. A., Kujawinski E. B., Stubbins A., Fatland R., Aluwihare L. I., Buchan A., et al. (2016). Deciphering Ocean Carbon in a Changing World. Proc. Natl. Acad. Sci. U. S. A. 113 (12), 3143–3151. doi: 10.1073/pnas.1514645113
Mozetić P., Francé J., Kogovšek T., Malej A. (2012). Plankton Trends and Community Changes in a Coastal Sea (Northern Adriatic): Bottom-Up vs. Top-Down Control in Relation to Environmental Drivers. Estuar. Coast. Shelf Sci. 115, 138–148. doi: 10.1016/j.ecss.2012.02.009
Myklestad S. M. (1995). Release of Extracellular Products by Phytoplankton With Special Emphasis on Polysaccharides. Sci. Total Environ. 165, 155–164. doi: 10.1016/0048-9697(95)04549-G
Oberbeckmann S., Loder M. G. J., Labrenz M. (2015). Marine Microplastic-Associated Biofilms - a Review. Environ. Chem. 12 (5), 551–562. doi: 10.1071/EN15069
Oksanen J., Blanchet F., Kindt G. R., Legendre P., Minchin P. R., O’Hara R. B. (2013). Vegan: Community Ecology Package. Available at: https://cran.r-project.org, https://github.com/vegandevs/vegan.
Oliveros J. C. (2007-2015). Venny. An Interactive Tool for Comparing Lists With Venn’s Diagrams. Available at: https://bioinfogp.cnb.csic.es/tools/venny/index.html.
Orel N., Fadeev E., Klun K., Ličer M., Tinta T., Turk V. (2021). Bacterial Indicators Are 671 Ubiquitous Members of Pelagic Microbiome in Anthropogenically Impacted Coastal Ecosystem. Front. Microbiol. 12, 765091. doi: 10.3389/fmicb.2021.765091
Orlić S., Najdek M., Supić N., Ivančić I., Fuks D., Blažina M., et al. (2013). Structure and Variability of Microbial Community at Transect Crossing a Double Gyre Structure (North-Eastern Adriatic Sea). Aquat. Microb. Ecol. 69, 193–203. doi: 10.3354/ame01631
Parada A. E., Needham D. M., Fuhrman J. A. (2016). Every Base Matters: Assessing Small Subunit rRNA Primers for Marine Microbiomes With Mock Communities, Time Series and Global Field Samples: Primers for Marine Microbiome Studies. Environ. Microbiol. 18, 1403–1414. doi: 10.1111/1462-2920.13023
Passow U., Ziervogel K., Asper V., Diercks A. (2012). Marine Snow Formation in the Aftermath of the Deepwater Horizon Oil Spill in the Gulf of Mexico. Environ. Res. Lett. 7, 35301. doi: 10.1088/1748-9326/7/3/035301
Paterson H., Heel K., Waite A. (2013). A Warm-Core Eddy Linking Shelf, Leeuwin Current and Oceanic Waters Demonstrated by Near-Shelf Distribution Patterns of Synechococcus Spp. And Prochlorococcus Spp. In the Eastern Indian Ocean. Mar. Freshw. Res. 64, 1011. doi: 10.1071/MF12271
Pedrós-Alió C., Acinas S., Logares R., Massana R. (2018). “Marine Microbial Diversity as Seen by High Throughput Sequencing,” in Microbial Ecology of the Oceans, 3rd ed. Eds. Gasol J., Kirchman D. (Hoboken: John Wiley & Sons), 47–98.
Pedros-Alio C., Brock T. D. (1983). The Impact of Zooplankton Feeding on the Epilimnetic Bacteria of a Eutrophic Lake. Freshw. Biol. 13, 227–239. doi: 10.1111/j.1365-2427.1983.tb00673.x
Quast C., Pruesse E., Yilmaz P., Gerken J., Schweer T., Yarza P., et al. (2012). The SILVA Ribosomal RNA Gene Database Project: Improved Data Processing and Web-Based Tools. Nucleic Acids Res. 41, D590–D596. doi: 10.1093/nar/gks1219
Raicich F. (1994). A Note on the Flow Rates of the Adriatic Rivers, Tech. Rep., RF 2/94 (Consiglio Nazionale delle Ricerche, Trieste: Istituto Sperimentale Trieste), 6 pp.
Rawat S. R., Männistö M. K., Bromberg Y., Häggblom M. M. (2012). Comparative Genomic and Physiological Analysis Provides Insights Into the Role of Acidobacteria in Organic Carbon Utilization in Arctic Tundra Soils. FEMS Microbiol. Ecol. 82, 341–355. doi: 10.1111/j.1574-6941.2012.01381.x
Ribalet F., Berges J. A., Ianora A., Casotti R. (2007). Growth Inhibition of Cultured Marine Phytoplankton by Toxic Algal-Derived Polyunsaturated Aldehydes. Aquat. Toxicol. 85, 219–227. doi: 10.1016/j.aquatox.2007.09.006
Roesch L. F. W., Fulthorpe R. R., Riva A., Casella G., Hadwin A. K. M., Kent A. D., et al. (2007). Pyrosequencing Enumerates and Contrasts Soil Microbial Diversity. ISME J. 1, 283–290. doi: 10.1038/ismej.2007.53
Salazar G., Cornejo-Castillo F. M., Benítez-Barrios V., Fraile-Nuez E., Álvarez-Salgado X. A., Duarte C. M., et al. (2016). Global Diversity and Biogeography of Deep-Sea Pelagic Prokaryotes. ISME J. 10, 596–608. doi: 10.1038/ismej.2015.137
Salgado-Hernanz P. M., Regaudie-de-Gioux A., Antoine D., Basterretxea G. (2022). Pelagic Primary Production in the Coastal Mediterranean Sea: Variability, Trends, and Contribution to Basin-Scale Budgets. Biogeosciences 19, 47–69. doi: 10.5194/bg-19-47-2022
Sarmento H., Romera-Castillo C., Lindh M., Pinhassi J., Sala M. M., Gasol J. M., et al. (2013). Phytoplankton Species-Specific Release of Dissolved Free Amino Acids and Their Selective Consumption by Bacteria. Limnol. Oceanogr. 58, 1123–1135. doi: 10.4319/lo.2013.58.3.1123
Schattenhofer M., Fuchs B. M., Amann R., Zubkov M. V., Tarran G. A., Pernthaler J. (2009). Latitudinal Distribution of Prokaryotic Picoplankton Populations in the Atlantic Ocean. Environ. Microbiol. 11, 2078–2093. doi: 10.1111/j.1462-2920.2009.01929.x
Segata N., Izard J., Waldron L., Gevers D., Miropolsky L., Garrett W. S., et al. (2011). Metagenomic Biomarker Discovery and Explanation. Genome Biol. 12, R60. doi: 10.1186/gb-2011-12-6-r60
Seymour J. R., Amin S. A., Raina J.-B., Stocker R. (2017). Zooming in on the Phycosphere: The Ecological Interface for Phytoplankton–Bacteria Relationships. Nat. Microbiol. 2, 17065. doi: 10.1038/nmicrobiol.2017.65
Shiah F. K. (1999). Die1 Cycles of Heterotrophic Bacterioplankton Abundance and Production in the Ocean Surface Waters. Aquat Microb. Ecol. 17 (3), 239– 246. doi: 10.3354/ame017239
Šilović T., Balagué V., Orlić S., Pedrós-Alió C. (2012). Picoplankton Seasonal Variation and Community Structure in the Northeast Adriatic Coastal Zone. FEMS Microbiol. Ecol. 82, 678–691. doi: 10.1111/j.1574-6941.2012.01438.x
Simon M., Grossart H., Schweitzer B., Ploug H. (2002). Microbial Ecology of Organic Aggregates in Aquatic Ecosystems. Aquat. Microb. Ecol. 28, 175–211. doi: 10.3354/ame028175
Smith D. C., Simon M., Alldredge A. L., Azam F. (1992). Intense Hydrolytic Enzyme Activity on Marine Aggregates and Implications for Rapid Particle Dissolution. Nature 359, 139–142. doi: 10.1038/359139a0
Smith M. W., Zeigler Allen L., Allen A. E., Herfort L., Simon H. M. (2013). Contrasting Genomic Properties of Free-Living and Particle-Attached Microbial Assemblages Within a Coastal Ecosystem. Front. Microbiol. 4. doi: 10.3389/fmicb.2013.00120
Sournia A. (1973). The Primary Productivity in the Mediterranean Sea. Essai mise jour. Bull. Etud. Commun. Medit. Monaco. 5, 1–128.
Steiner P. A., Geijo J., Fadeev E., Obiol A., Sintes E., Rattei T., et al. (2020). Functional Seasonality of Free-Living and Particle-Associated Prokaryotic Communities in the Coastal Adriatic Sea. Front. Microbiol. 11. doi: 10.3389/fmicb.2020.584222
Steiner P. A., Sintes E., Simó R., De Corte D., Pfannkuchen D. M., Ivančić I., et al. (2019). Seasonal Dynamics of Marine Snow-Associated and Free-Living Demethylating Bacterial Communities in the Coastal Northern Adriatic Sea. Environ. Microbiol. Rep. 11 (5), 699–707. doi: 10.1111/1758-2229.12783
Tarn J., Peoples L. M., Hardy K., Cameron J., Bartlett D. H. (2016). Identification of Free-Living and Particle-Associated Microbial Communities Present in Hadal Regions of the Mariana Trench. Front. Microbiol. 7. doi: 10.3389/fmicb.2016.00665
Teeling H., Fuchs B. M., Becher D., Klockow C., Gardebrecht A., Bennke C. M., et al. (2012). Substrate-Controlled Succession of Marine Bacterioplankton Populations Induced by a Phytoplankton Bloom. Science 336, 608–611. doi: 10.1126/science.1218344
Thomas F., Hehemann J.-H., Rebuffet E., Czjzek M., Michel G. (2011). Environmental and Gut Bacteroidetes: The Food Connection. Front. Microbio. 2. doi: 10.3389/fmicb.2011.00093
Thornton D. C. O. (2002). Diatom Aggregation in the Sea: Mechanisms and Ecological Implications. Euro. J. Phycol. 37, 149–161. doi: 10.1017/S0967026202003657
Thornton D. C. (2014). Dissolved Organic Matter (DOM) Release by Phytoplankton in the Contemporary and Future Ocean. Eur. J. Phycology 49(1), 20–46. doi: 10.1080/09670262.2013.875596
Trano A. C. (2021). Exploring Marine Bacterial Communities With a Focus on Bacteria Attached to Particles, The Open University.
Wilkins D., van Sebille E., Rintoul S. R., Lauro F. M., Cavicchioli R. (2013). Advection Shapes Southern Ocean Microbial Assemblages Independent of Distance and Environment Effects. Nat. Commun. 4, 2457. doi: 10.1038/ncomms3457
Willems A. (2014). “The Family Phyllobacteriaceae,” in The Prokaryotes. Eds. Rosenberg E., DeLong E. F., Lory S., Stackebrandt E., Thompson F. (Berlin, Heidelberg: Springer Berlin Heidelberg), 355–418. doi: 10.1007/978-3-642-30197-1_298
Williams T. J., Wilkins D., Long E., Evans F., DeMaere M. Z., Raftery M. J., et al. (2013). The Role of Planktonic Flavobacteria in Processing Algal Organic Matter in Coastal East Antarctica Revealed Using Metagenomics and Metaproteomics. Environ. Microbiol. 15 (5), 1302–1317. doi: 10.1111/1462-2920.12017
Yoshinaga I., Kawai T., Ishida Y. (1997). Analysis of Algicidal Ranges of the Bacteria Killing the Marine Dinoflagellate Gymnodinium Mikimotoi Isolated From Tanabe Bay, Wakayama Pref., Japan. Fish. Sci. 63, 94–98. doi: 10.2331/fishsci.63.94
Yoshinaga I., Kawai T., Takeuchi T., Ishida Y. (1995). Distribution and Fluctuation of Bacteria Inhibiting the Growth of a Marine Red Tide Phytoplankton Gymnodinium Mikimotoi in Tanabe Bay (Wakayama Pref., Japan). Fish. Sci. 61, 780–786. doi: 10.2331/fishsci.61.780
Youssef N. H., Elshahed M. S. (2009). Diversity Rankings Among Bacterial Lineages in Soil. ISME J. 3, 305–313. doi: 10.1038/ismej.2008.106
Yung C. M., Ward C. S., Davis K. M., Johnson Z. I., Hunt D. E. (2016). Insensitivity of Diverse and Temporally Variable Particle-Associated Microbial Communities to Bulk Seawater Environmental Parameters. Appl. Environ. Microbiol. 82, 3431–3437. doi: 10.1128/AEM.00395-16
Zhang R., Liu B., Lau S. C. K., Ki J.-S., Qian P. Y. (2007). Particle-Attached and Free-Living Bacterial Communities in a Contrasting Marine Environment: Victoria Harbor, Hong Kong: Particle-Attached and Free-Living Bacterial Communities. FEMS Microbiol. Ecol. 61, 496–508. doi: 10.1111/j.1574-6941.2007.00353.x
Zhang Y., Pavlovska M., Stoica E., Prekrasna I., Yang J., Slobodnik J. (2020). Holistic Pelagic Biodiversity Monitoring of the Black Sea via eDNA Metabarcoding Approach: From Bacteria to Marine Mammals. Environ. Int. 135, 105307. doi: 10.1016/j.envint.2019.105307
Zhang Y., Xiao W., Jiao N. (2016). Linking Biochemical Properties of Particles to Particle-Attached and Free-Living Bacterial Community Structure Along the Particle Density Gradient From Freshwater to Open Ocean: Linking POM to Particle-Attached and Free-Living Bacteria. J. Geophys. Res. Biogeosci. 121, 2261–2274. doi: 10.1002/2016JG003390
Keywords: prokaryotic communities, particles, phycosphere, prokaryotic diversity, Northern Adriatic Sea, 16S rRNA
Citation: Trano AC, Piredda R, Balestra C, Bastianini M, Gasol JM and Casotti R (2022) Diversity of Free-Living and Particle-Attached Prokaryotes in a River-Influenced Coastal Area of the Northern Adriatic Sea. Front. Mar. Sci. 9:912528. doi: 10.3389/fmars.2022.912528
Received: 04 April 2022; Accepted: 30 May 2022;
Published: 07 July 2022.
Edited by:
Antje Wichels, Alfred Wegener Institute Helmholtz Centre for Polar and Marine Research (AWI), GermanyReviewed by:
Tinkara Tinta, National Institute of Biology, SloveniaDaniel P. R. Herlemann, Estonian University of Life Sciences, Estonia
Copyright © 2022 Trano, Piredda, Balestra, Bastianini, Gasol and Casotti. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Raffaella Casotti, cmFmZmFlbGxhLmNhc290dGlAc3puLml0
 Anna Chiara Trano
Anna Chiara Trano Roberta Piredda2
Roberta Piredda2 Josep M. Gasol
Josep M. Gasol Raffaella Casotti
Raffaella Casotti