- 1Ministry of Education Key Laboratory of Marine Genetics and Breeding, College of Marine Life Sciences, Ocean University of China, Qingdao, China
- 2Key Laboratory of Tropical Aquatic Germplasm of Hainan Province, Sanya Oceanographic Institution, Ocean University of China, Sanya, China
- 3Qingdao Dayi Haichuang Biotechnology Co., Ltd, Qingdao, China
Intertidal biota is subjected to significant fluctuations in salinity. Urechis unicinctus, a typical species thriving in intertidal sediments, have relatively simple anatomy structure and therefore could be considered as an ideal species for salinity acclimation. Moreover, due to the high nutrients, U. unicinctus has become an emerging aquaculture species in China. In this study, we investigated the effects of salinity on the growth performance, coelomic fluid biochemical indices, antioxidant enzyme system, Na+/K+-ATPase, and non-specific immune enzyme activities, as well as intestinal microbiota composition of this species in an 8-week experiment. The results indicated U. unicinctus is a euryhaline species that can tolerate salinities from 15‰ to 40‰ and demonstrated the highest growth performance at 30‰. Physiological characteristic analyses of coelomic fluid and intestines indicated that the salinity range of 15‰ to 30‰ is more suitable, while 10‰ and 40‰ salinity ranges seem unsuitable for juvenile U. unicinctus. Furthermore, intestinal microbiota analysis indicated salinity had a significant effect on the composition structure. KEGG pathway analysis indicated that antioxidant related metabolic pathways and amino acid metabolic pathways may play important roles in gut microbiota under salinity stress. Comprehensive analysis of intestinal bacteria and enzyme activities indicated Unidentified Rhodobacteraceae, Vibrio, and Shimia may play important roles in high salinity acclimation, while Legionellaceae may act as important microbiota in low salinity acclimation. Moreover, Ruminococcus, Oscillospira, Lachnospiraceae, Clostridiales, and S24-7 showed negative correlations with the antioxidant and nonspecific immune enzymes, indicating that these bacteria can be considered as important candidates of probiotics for U. unicinctus aquaculture industry. The findings of this study will extend our understanding of the effects of salinity on the growth and health of U. unicinctus and contribute to a better understanding of the salinity acclimation strategies of organisms in intertidal zones.
Introduction
Salinity is a critical environmental factor for aquatic organisms. Due to rainstorm or freshwater input, a marine organism living in an intertidal zone may experience changes in salinity over the course of a tidal cycle (Montagna et al., 2018). The instability of salinity challenges the homeostasis of organisms. Salinity can affect the distribution (Kneib, 1984; Conde et al., 2013), physiology (Zhang et al., 2014; Falconer et al., 2019; Yang et al., 2022), and reproduction (Deschaseaux et al., 2010) of intertidal species, which also employ physiological strategies to maintain function in rapid salinity fluctuation. Therefore, intertidal species, especially sedentary species and benthic animals in intertidal environments have drawn increasing attention for salinity acclimation investigation (Falconer et al., 2019; Yang et al., 2022).
Salinity is a fundamental factor affecting the normal growth of aquatic organisms. Salinity alterations can significantly affect physiological activity of aquatic organisms, such as osmolality regulation, antioxidant enzyme system, as well as non-specific immune enzyme activities including Na+/K+-ATPase, superoxide dismutase (SOD), acid phosphatase (ACP), and alkaline phosphatase (ALP) (Kültz, 2015; Liu et al., 2017; Tian et al., 2020; Yang et al., 2022). Rapid salinity changes result in the generation of excessive reactive oxygen species (ROS) in marine species (Lushchak, 2011). SOD reduces ROS into H2O2, which can be further reduced into H2O by other antioxidant enzymes (Shen, 2007). An over-production of ROS also leads to a permeabilization of lysosomal membranes as demonstrated by the release of lysosomal proteolytic enzymes such ACP and ALP to the cytosol (Chien et al., 2003; Wang et al., 2012). In addition, blood biochemical parameters are often used to evaluate health status (Edsall, 1999; Li et al., 2011). Several studies indicate that salinity has a significant effect on blood physiological indices of aquatic organisms (Jarvis and Ballantyne, 2003; Xu et al., 2008; Zarejabad et al., 2010; Tian et al., 2020; Yang et al., 2022). Conversely, the intestines of aquatic organisms play significant roles in osmotic regulation by dynamically regulating water/salt absorption and ion exchange under salinity stress (Giffard-Mena et al., 2006).
As a component contributing to host life activities, the intestinal microbial composition may be variously affected by the presence of environmental water. The intestinal microbiota plays a critical role in regulating many biological functions of the hosts, including nutritional provisioning, immune reaction, and metabolic homeostasis (Gomez and Balcazar, 2008; Sullam, 2012). At present, salinity stress has been mainly investigated on the aquatic organism itself, whereas the influence of salinity stress on host–microbe interaction remains in its infancy. Only several studies, in fish and shrimp, have focused on the effects of salinity on the intestinal microbiota composition and function (Eichmiller et al., 2016; Dehler et al., 2017; Hou et al., 2020; Lai et al., 2020; Tian et al., 2020). To extend the understanding of the intestinal microbiota of how host species respond to environmental variations, more investigation will be necessary to fully understand the important role of intestinal microbiota in host salinity acclimation.
The Echiura worm Urechis unicinctus mainly inhabits U-shaped burrows in the intertidal zone of China, Russian, Korea, and Japan (Li et al., 1995). U. unicinctus has high nutritional value and is rich in essential human amino acids, glycosaminoglycan saccharides, and polypeptides (Li et al., 2000; Zhang & Li, 2011; Bi et al., 2013; Cui et al., 2021). Moreover, U. unicinctus polypeptides have been reported to enhance immune functions and inhibit tumor in mice (Zhao et al., 2008; Sung et al., 2008; Niu and Chen, 2016). At present, the natural resources of U. unicinctus have declined sharply because of overfishing in China (Li et al., 2017). U. unicinctus has become an emerging aquaculture species in China. Conversely, U. unicinctus is a filter feeding animal which can filter algae and organic matter in water. U. unicinctus live in U-shaped burrows in the mud- and sandflats of coastal areas and do not compete for a niche with other species. Moreover, U. unicinctus demonstrate high sulfide metabolism (Ma et al., 2011; Ma et al., 2012; Zhang et al., 2021; Liu et al., 2022) which could improve sulfur-containing coastal substrates. Based on these advantages, U. unicinctus have potential application in pond polyculture with other species such as Litopenaeus vannamei, Stichopus japonicus, as well as marine fish (Zhao and Wang, 2018; Chen et al., 2019). Taken together, the artificial breeding is urgently needed in order to meet the increasing consumer demand of human and polyculture application. At present, U. unicinctus breeding technology is still nascent (Xu et al., 2016). Therefore, investigation of the environmental condition for the breeding of juveniles is very important for the U. unicinctus aquaculture industry.
Salinity is an important environmental factor in U. unicinctus cultivation, especially in pond polyculture with other species such as euryhaline organism L. vannamei. To date, investigations on the effects of salinity on U. unicinctus have mainly focused on the adult, including the suitable salinity from 24‰-35‰ (Li et al., 1998) as well as digestive enzyme activity and antioxidant enzyme activity variation for 96 h under different salinities (15‰, 20‰, 25‰, and 30‰) (Xu et al., 2017; Zhu et al., 2019), while studies on the juveniles is only limited to growth and survival rate (Zheng et al., 2006; Li et al., 2019; Sun et al., 2020). A previous study of marine leeches revealed the juvenile was more sensitive to salinity stress than the adult (Kua et al., 2014). Chinese mitten crab also demonstrated different responses between adults and juveniles (Yang et al., 2022; Zhao et al., 2016). Conversely, the investigation of the influence of salinity on juveniles is important for the U. unicinctus aquaculture industry. Therefore, to extend the understanding of the influence of salinity on U. unicinctus cultivation, we investigated the effects of salinity on the growth, coelomic fluid biochemical indices, antioxidant enzyme system, nonspecific immunity, Na+/K+-ATPase activities, and intestinal microbiota composition of U. unicinctus juveniles in an 8-week experiment. The findings of this study will provide suitable salinity conditions for the rationalization of breeding techniques and thereby contribute to the development of the U. unicinctus aquaculture. Meanwhile, the comprehensive analysis of physicochemical indices and intestinal microbiota of this study could also provide new insight in salinity acclimation for aquatic organism.
Materials and Methods
Specimens and Experimental Design
U. unicinctus juveniles (mean weight, 533.3 ± 121.3 mg) were supplied by Qingdao Dayi HC Biotechnology Co (Qingdao, China). The juveniles were cultivated temporarily in three 200-L tanks with 500 juveniles per tank for one week before different salinity treatments (17 ± 0.2°C, 30 ± 0.2‰). After acclimatization, the juveniles were randomly grouped into 15 aquariums (0.25 m long and 0.4 wide) with 20 mesh screened sand at a height of 0.05 m and 0.25 m water depth. Previous studies indicated U. unicinctus juveniles could grow well in 20.0‰ and 30.0‰ salinity, while 40.0‰ and 10‰ were the salinity limit (Zheng et al., 2006; Li et al., 2019; Sun et al., 2020). To further investigate the salinity effect on U. unicinctus juveniles, 40.0‰, 30.0‰, 20.0‰, 15‰, and 10‰ salinity was selected for this study. The aquariums were divided into five different salinity groups: 40.0‰, 30.0‰, 20.0‰, 15‰, and 10‰, with three replicates for each group. Salinity levels were obtained by artificial seawater, which was prepared by seawater crystal (Sea Salt Science and Technology Co., Ltd., Qingdao, China), and measured with an optical refractometer (American Optical Corporation, USA) for salinity. The breeding density was 1,000 per cubic meter of water. Water was renewed 50% every three days and metabolic waste was siphoned daily. The environmental conditions during the different salinity cultivation were as follows: pH 7.8-8.5, water temperature 16.5-20°C, and dissolved oxygen 5.5-7.4 mg/L. The juveniles were reared for 8 weeks and fed 2% of their body weight, twice daily, with a manually mixed diet (Spirulina and Saccharomyces cerevisiae at a ratio of 9:1).
Sample Collection and Preparation
At the end of the 8 week experimental period, the survival rate, final weight, and body wall weight of the U. unicinctus juveniles, in the different salinity groups, were recorded. Ten juveniles were randomly collected from each aquarium. After dissecting for the collection of coelomic fluids, the intestine samples were aseptically removed from the juvenile. Then, the intestine and coelomic fluid samples were obtained. After being frozen in liquid nitrogen, all the samples were stored at −80°C for subsequent analysis.
Determining Coelomic Fluid Biochemical Indices and Enzymatic Activities
Biochemical indices, including total protein, glucose (Glu), triglycerides (TG), total cholesterol (T-Cho), as well as ionic concentrations including Na+ and Cl− in the coelomic fluid of U. unicinctus juveniles were detected following the manufacturer’s commercial assay kit protocol (Nanjing Jiancheng, Nanjing, China). The cat. no. of the kits for total protein, glucose, T-Cho, TG, and Na+ and Cl− were A045-3, A154-1-1, A111-1-1, A110-1-1, C002-1-1 and C003-2, respectively.
The activities of superoxide dismutase (SOD), total antioxidant capacity (T-AOC), the content of malondialdehyde (MDA), acid phosphatase (ACP), alkaline phosphatase (ALP), and Na+/K+-ATPase in the coelomic fluids and intestines were measured using commercial assay kits (Nanjing Jiancheng, Nanjing, China). The cat. no. of the kits for SOD, T-AOC, MDA, ACP, ALP, and Na+/K+-ATPase were A001-3, A015-2-1, G0109W G0903W, G0904W, and G0859W, respectively.The above coelomic fluid biochemical index and enzymatic activity were measured using 1510 Enzyme-labelled meter (Thermo Fisher Scientific, Waltham, MA, USA).
Intestinal Microbiota Analysis
Total genomic DNA of intestine samples were extracted using the Fast DNA SPIN extraction kits (MP Biomedicals, Santa Ana, CA, USA), following the manufacturer’s instructions. DNA quality and concentration were checked by agarose gel electrophoresis and a NanoDrop ND-1000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA).
PCR amplification of the bacterial 16S rRNA genes V3–V4 region was performed using the forward primer 338F (5’-ACTCCTACGGGAGGCAGCA-3’) and the reverse primer 806R (5’-GGACTACHVGGGTWTCTAAT-3’). The PCR components contained 5 μl of Q5 reaction buffer (5×), 5 μl of Q5High-Fidelity GC buffer (5×), 2 μl (2.5 mM) of dNTPs, and 1 μl (10 uM) of each Forward and Reverse primer, 0.25 μl of Q5 High-Fidelity DNA Polymerase (5U/μl), 2 μl of DNA Template, and 8.75 μl of ddH2O. PCR amplicons were purified by Agencourt AMPure Beads (Beckman Coulter, Indianapolis, IN) and quantified using the PicoGreen dsDNA Assay Kit (Invitrogen, Carlsbad, CA, USA). After the individual quantification step, amplicons were pooled into equal amounts and pair-end 2×300 bp sequencing was performed using the Illlumina NovaSeq 6000 platform at the Shanghai Personal Biotechnology Co., Ltd (Shanghai, China).
The Quantitative Insights into Microbial Ecology (QIIME2,2019.4) pipeline was employed to process the sequencing data (Caporaso et al., 2010). Briefly, raw sequencing reads with exact matches to the barcodes were assigned to respective samples and identified as valid sequences. The low-quality sequences were filtered through following criteria (Chen and Jiang, 2014): sequences that had a length of <150 bp, sequences that had average Phred scores of <20, sequences that contained ambiguous bases, and sequences that contained mononucleotide repeats of >8 bp. Paired-end reads were assembled using FLASH (Magoc and Salzberg, 2011). After chimera detection, the remaining high-quality sequences were clustered into operational taxonomic units (OTUs) at 97% sequence identity by UCLUST (Edgar, 2010). A representative sequence was selected from each OTU using default parameters. OTU taxonomic classification was conducted by BLAST searching the representative sequences set against the Greengenes Database (DeSantis et al., 2006) using the best hit (Altschul et al., 1997). An OTU table was further generated to record the abundance of each OTU in each sample and the taxonomy of these OTUs. OTUs containing less than 0.001% of total sequences across all samples were discarded. To minimize the difference of sequencing depth across samples, an averaged, rounded rarefied OTU table was generated by averaging 100 evenly resampled OTU subsets under the 90% of the minimum sequencing depth for further analysis. Sequence data analyses were mainly performed using QIIME2 and R packages (v3.3.1). OTU-level alpha diversity indices, such as Chao1, observed species, Shannon, and Simpson index were calculated using the OTU table in QIIME2. A Venn diagram was obtained to visualize the shared and unique OTUs among different salinity groups using R package (Zaura et al., 2009). Rarefaction curves of intestinal bacterial in different salinity groups were generated to estimate diversity and richness. Beta diversity analysis was performed to investigate the structural variation of microbial communities across different salinity groups using UniFrac distance metrics (Lozupone and Knight, 2005; Lozupone et al., 2007) and was visualized via principal coordinate analysis (PCoA) (Ramette, 2007). Taxa abundances at the phylum, class, order, family, and genus levels were statistically compared among different salinity groups by Metastats (White et al., 2009) and visualized as violin plots. Krona analysis, a visualization tool that allows intuitive exploration of relative abundances in different taxa was applied (Ondov et al., 2011). Microbial functions were predicted by PICRUSt2 based on high-quality sequences of intestinal bacterial in different salinity groups (Douglas et al., 2020). Then, the MetaCyc (https://metacyc.org/) database was used for KEGG pathway prediction.
Statistical Analysis
Significant differences in the growth performance, coelomic fluid physiological indices, enzyme activities, antioxidant capacity, and intestinal microbiota alpha diversity index among U. unicinctus juveniles under different salinity were determined using one-way analysis of variance followed by Tukey’s multiple-range tests, using the SPSS package (version 16.0) at a significance level of P < 0.05. The correlations between the enzyme activities and intestinal microbes of U. unicinctus juveniles were determined by the Spearman coefficient.
Results and Discussion
Effects of Salinity on Growth Performance
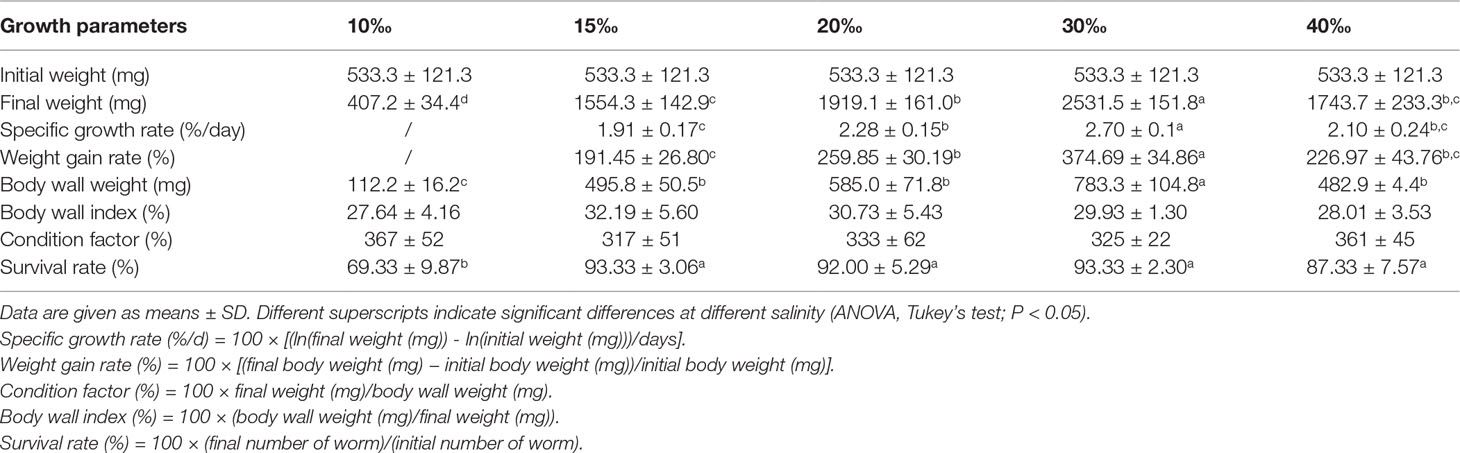
To illustrate the effect of salinity on U. unicinctus growth, U. unicinctus juveniles were reared for 8 weeks at five different salinities (10‰, 15‰, 20‰, 30‰, and 40‰). The results showed salinity had a significant effect on U. unicinctus growth (Table 1). U. unicinctus juveniles in the 30‰ group showed the best growth performance with the highest final weight (FW) (2531.5 ± 151.8 mg), specific growth rate (SGR) (2.70 ± 0.13%/day), and body wall weight (BWW) (783.3 ± 104.8 mg), followed by 20‰ (FW1919.1 ± 161.0 mg, SGR2.28 ± 0.15%/day, BWW585.0 ± 71.8 mg), 40‰ (FW1743.7 ± 233.3 mg, SGR2.10 ± 0.24%/day, BWW482.9 ± 4.4 mg), 15‰ (FW1554.3 ± 142.9 mg, SGR1.91 ± 0.17%/day, BWW495.8 ± 50.5 mg) groups, and finally the 10‰ group (FW407.2 ± 34.4 mg, BWW112.2 ± 16.2 mg) not even growing in the 8 week feeding trial. A previous study reported that the adult U. unicinctus has suitable salinity ranging from 24.8‰ to 35.8‰ (Li et al., 1998) and thus, our results further implied 30‰ is possibly the universal salinity for juvenile and adult U. unicinctus. In addition, no significant difference was observed in body wall index and condition factors. Moreover, the survival rate was significantly lower in the 10‰ group with 69.33 ± 9.87% compared to the other salinity groups. The results indicate that 30‰ is the best salinity condition for U. unicinctus growth, while U. unicinctus could grow well at 15-40‰ salinity, indicating that the species is a euryhaline organism. The trend was similar with the previous feeding trial report of U. unicinctus juveniles with different salinities (Zheng et al., 2006; Li et al., 2019; Sun et al., 2020). In addition, the survival rate (93.33%), weight gain rate, and specific growth rate of U. unicinctus juveniles in this study were higher than the previous reports with survival rates from 65.37% to 82.67% at 30‰ salinity (Chen et al., 2020; Chen et al., 2021) which may result from the different diet of the feeding trial. The diet in this study was composed of Spirulina and Saccharomyces cerevisiae which was more suitable for U. unicinctus juvenile growth than the diet of sea mud and powders of Sargassum, Mytilusedulis, and Spirulina in the previous report (Chen et al., 2020; Chen et al., 2021).
Effects of Salinity on Coelomic Fluid Biochemical Indices
The anatomy of the U. unicinctus is relatively simple, which can be roughly divided into body wall, coelomic fluid, and intestine (Li et al., 1998). The coelomic fluid is composed of coelomic fluid cells and blood, which act as the circulatory system and play an important role in environment adaption (Arp et al., 1995; Ma et al., 2005). To further investigate the effect of salinity on U. unicinctus growth, the coelomic fluid biochemical indices, including total protein, glucose, triglycerides, total cholesterol, Na+, and Cl- was determined after 8 weeks of cultivation at five different salinities (10‰, 15‰, 20‰, 30‰, and 40‰). The results showed that salinity had a significant effect on coelomic fluid biochemical indices (Figure 1). The salinity of the 20‰ and 30‰ groups exhibited higher total protein content (14.97 mg/mL and 14.71 mg/mL, respectively), followed by the 15‰ group (12.60 mg/mL), and finally the 40‰ and 10‰ groups (4.87mg/mL and 2.56 mg/mL, respectively) were relative lower (Figure 1A). The total protein level is considered a valuable indicator for the general health and stress status of aquatic animals (Magnadottir, 2006). This result is consistent with other studies where serum total protein content remarkably declined with increasing or decreasing salinity (Kelly and Woo, 1999; Martínez-Álvarez et al., 2002). Glucose is a major energy source for the body to help cope with osmotic stress and the body can enhance glucose to provide the energy under hypo-osmotic or hyper-osmotic conditions (Long et al., 2017). Moreover, lipid metabolism, including triglyceride and cholesterol, is also an important energy source for aquatic animals (Long et al., 2017). The glucose and triglyceride content in the 15‰, 20‰ and 30‰ groups were significantly high compared to those in the 10‰ and 40‰ groups (Figures 1B, C). Total cholesterol content continuously increased from 10‰ (0.03 mmol/L) to 30‰ (1.22 mmol/L) and a significant decrease in the 40‰ group (0.28 mmol/L) is shown in Figure 1D. The results indicate that U. unicinctus juveniles, under hypo-hyper osmotic conditions, increased energy demand for osmoregulation then decreased energy allocated to other biological processes, resulting in lower growth performance especially in the 10‰ group. Osmotic regulation is mainly achieved by Na+ and Cl− regulation (Chen and Chia, 1997; Henry et al., 2002). The concentration of Na+ and Cl- was increased with salinity enhancement from 10‰ to 40‰ in U. unicinctus coelomic fluid. This trend indicates the passive entry of Na+ and Cl- into the worm which is consistent with other aquatic animals (Tian et al., 2020; Yang et al., 2022).
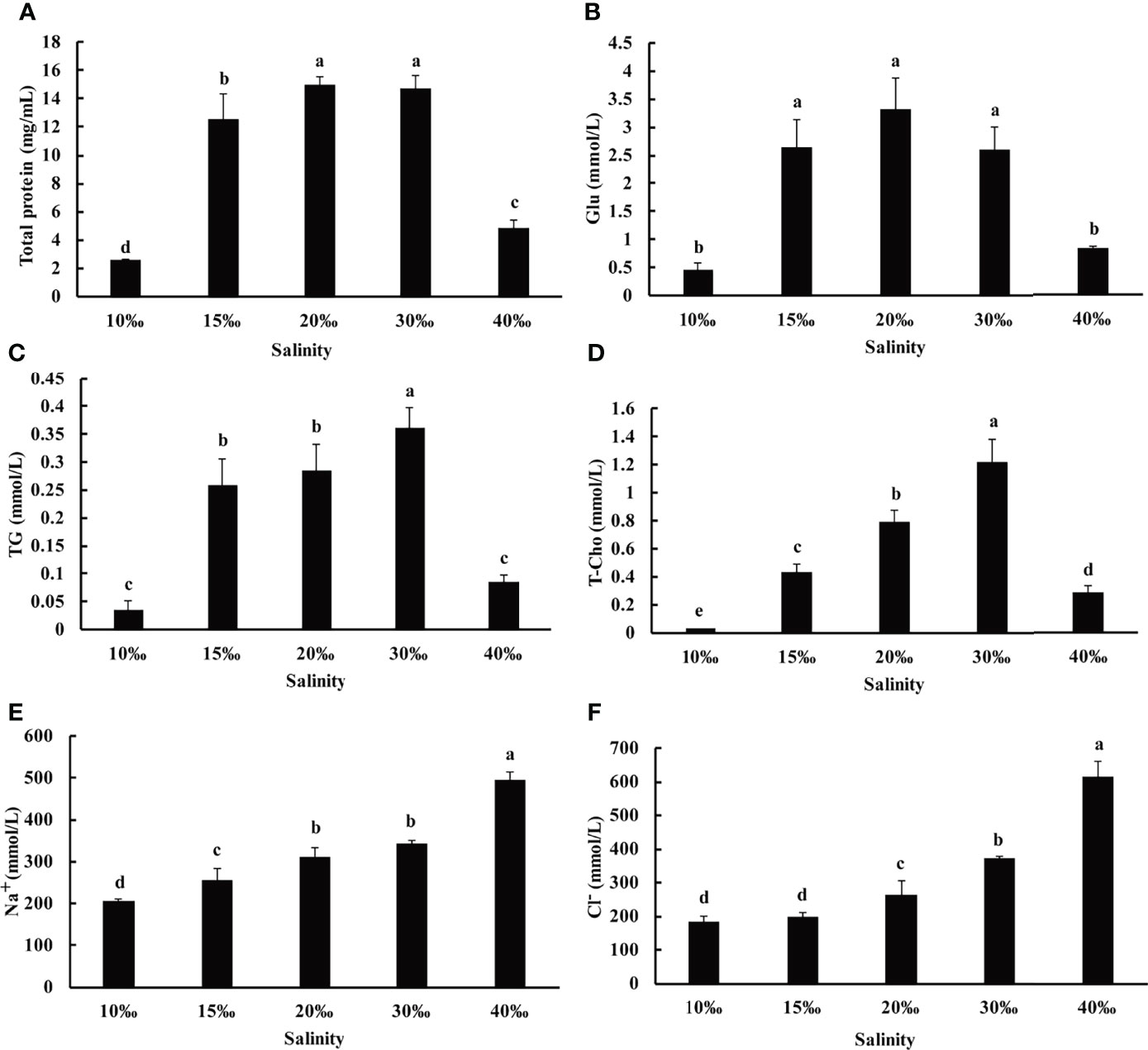
Figure 1 The contents of total protein (A), glucose (Glu, B), triglycerides (TG, C), total cholesterol (T-Cho, D), Na+(E) and Cl-(F) in the coelomic fluid samples of U. unicinctus juvenile reared in different salinities for 8 weeks. The data are mean values of three replicates + SD (n = 3). Different letters indicate significant differences at different salinity (ANOVA, Tukey’s test; P < 0.05).
Effects of Salinity on Antioxidant Enzyme System
To further investigate the effect of salinity on U. unicinctus, several oxidative stress related indices including SOD, T-AOC, and MDA of U. unicinctus intestine and coelomic fluid reared in different salinity levels were determined in this study (Figure 2). The salinity of the 10‰ and 40‰ groups (144.65 U/mg and 102.67 U/mg, respectively) exhibited higher SOD activity compared to other groups in the intestine of U. unicinctus (Figure 2A), whereas the 10‰ and 40‰ groups (141.63 U/mL and 1063.66 U/mL, respectively) exhibited relative lower SOD activity than the other groups in coelomic fluid (Figure 2B). The T-AOC results demonstrated the similar trend with SOD activity in the U. unicinctus intestine and coelomic fluid exposed to different salinity treatments (Figures 2C, D). There was no significant difference in the MDA content in U. unicinctus intestine exposed to different salinity treatments (Figure 2E). In U. unicinctus coelomic fluid, the 10‰ group exhibited relatively lower MDA content (2.08 U/mL) compared to that in other groups (Figure 2F).
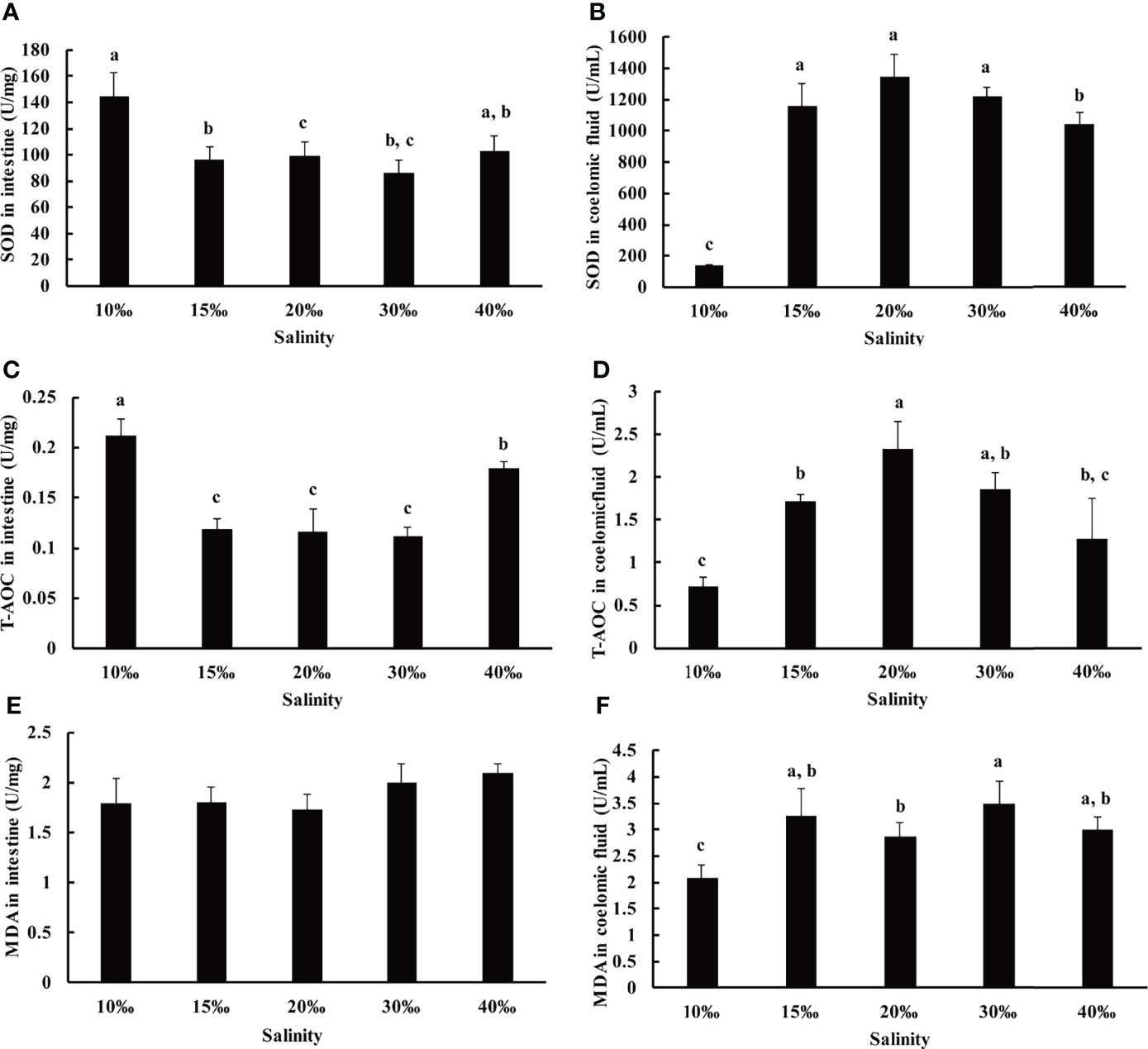
Figure 2 Oxidative stress related indices in the intestine and coelomic fluid of U. unicinctus juvenile reared in different salinities for 8 weeks. Superoxide dismutase (SOD, A, B), total antioxidant capacity (T-AOC, C, D) and malondialdehyde (MDA, E, F). The data are mean values of three replicates + SD (n = 3). Different letters indicate significant differences at different salinity (ANOVA, Tukey’s test; P < 0.05).
It has been reported that the alterations in salinity can induce ROS generation and result in oxidative stress in aquatic organisms (Lushchak, 2011). SOD is an important antioxidant enzyme in scavenging superoxide anion free radicals (Shen, 2007). Total antioxidant capacity (TAC) is an analyte frequently used to assess the antioxidant status of biological samples and can evaluate the antioxidant response against the free radicals produced in the samples. T-AOC reflects the total antioxidant capacity including enzymatic systems, such as SOD, CAT and GST, as well as non-enzymatic systems, such as amino acids, vitamins, and metalloproteins (Rubio et al., 2016). MDA is one of the main products of membrane lipid peroxidation which often occurs under stress, reflecting the degree of biological damage under stress (Lepage et al., 1991). In this study, SOD activity and T-AOC increased significantly in the 10‰ and 40‰ groups of U. unicinctus intestine, indicating 10‰ and 40‰ salinity were not suitable conditions for U. unicinctus juvenile growth. Under those conditions, we speculate salinity stress induces oxygen free radical production in intestines, destroying the balance of active oxygen metabolism and then, results in the enhancement of SOD activity and T-AOC in intestine. The result is consistent with the reports in other aquatic animals (Shen, 2007; Yang et al., 2022). No significant difference in the MDA content indicated salinity stress did not induced membrane lipid peroxidation in U. unicinctus intestine. The oxidative stress related indices variation trend in coelomic fluid was different with that in U. unicinctus intestine. U. unicinctus intestine is divided into three parts according to the region including foregut, midgut and hindgut. Midgut is the main peptic organ and hindgut functions as a type of water lung similar with gill in other aquatic organisms (Arp et al., 1995; Li et al., 1995). SOD activity and T-AOC increased significantly in 10‰ and 40‰ groups suggest intestine may act as important functional organ in salinity stress. In contrast, SOD activity and T-AOC decreased significantly in 10‰ and 40‰ groups in coelomic fluid suggest coelomic fluid may act as effector organ in salinity stress. As the significant deceased MDA content in coelomic fluid of 10‰ group compared to that in other groups, the reason may lie in the abnormal growth state in the conditions.
Effects of Salinity on Nonspecific Immune System
Besides the antioxidant system being involved in salinity stress, the nonspecific immune system also plays an important role to repair the damage caused by salinity stress. ACP and ALP are two important phosphatases that are crucial for metabolism and the nonspecific immune system (Chien et al., 2003; Wang et al., 2012). To further investigate the effect of salinity on U. unicinctus, ACP and ALP activity was investigated in this study (Figure 3). The result showed ACP and ALP activities were significantly increased in the 10‰ (12.84 U/L and 408.77 U/L, respectively) and 40‰ (17.13 U/L and 353.68 U/L, respectively) groups compared to other groups in the intestine (Figures 3A, C). In the coelomic fluid, ACP and ALP activity significantly decreased in the 10‰ (30.15 U/L and 27.04 U/L, respectively) and 40‰ (88.18 U/L and 101.93 U/L, respectively) groups compared to other groups (Figures 3B, D). It is worth noting that the ACP and ALP variation trend is similar with that in SOD and T-AOC. The results indicated the antioxidant system and nonspecific immune system simultaneously activate response to salinity stress in the U. unicinctus intestine. The result is consistent with the previous report in aquatic organisms (Yang et al., 2022). The ACP and ALP results also support that intestine and coelomic fluid act as a functional and effector organ in U. unicinctus stress, respectively.
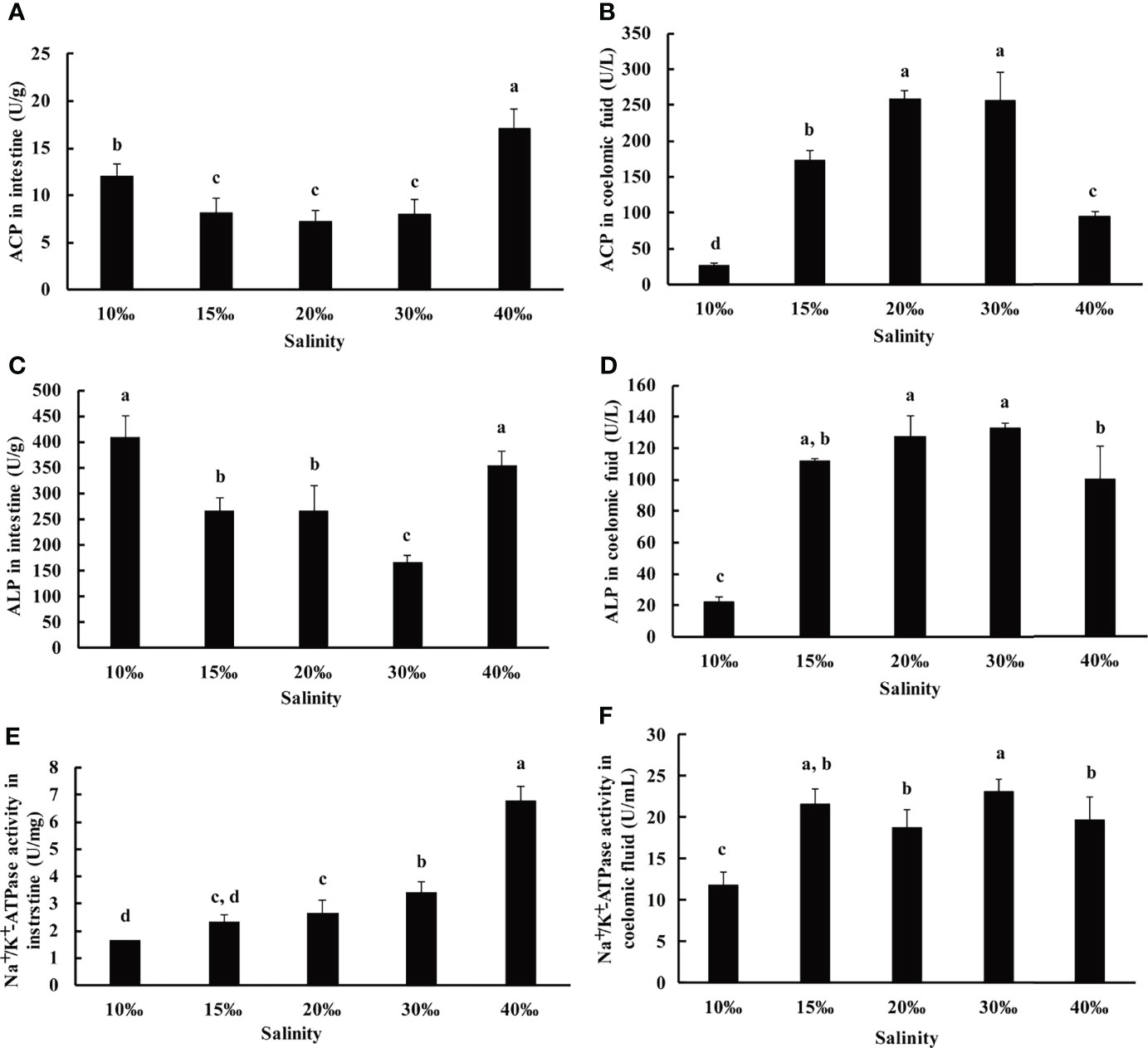
Figure 3 The immune indices and Na+/K+-ATPase activity in the intestine and coelomic fluid (A, B: ACP; C, D: ALP; E, F: Na+/K+-ATPase) of U. unicinctus juvenile reared in different salinities for 8 weeks. The data are mean values of three replicates + SD (n = 3). Different letters indicate significant differences at different salinity (ANOVA, Tukey’s test; P < 0.05).
Effects of Salinity on Osmotic Regulation
As antioxidant and nonspecific immune system are the indirect influence of salinity, osmotic regulation is the direct response for salinity stress. Na+/K+-ATPase plays an important role in osmotic regulation (Onken and Putzenlechner, 1995). To further investigate the salinity effect on U. unicinctus, the Na+/K+-ATPase activity was determined in the intestine and coelomic fluid after 8 weeks of cultivation at five different salinities (10‰, 15‰, 20‰, 30‰, and 40‰). The results showed that the Na+/K+-ATPase activity was increased with salinity enhancement in the intestine which demonstrated the highest level in the 40‰ group (6.80 U/mg) and the lowest level in the 10‰ group (1.65 U/mg) (Figure 3E). The result is consistent with the findings of other aquatic organisms (Feng et al., 2013; Zhao et al., 2016; Yang et al., 2022). The results also indicated the intestine play important role in osmotic regulation in U. unicinctus. In coelomic fluid, the Na+/K+-ATPase activity was not increased with the salinity enhancement (Figure 3F), suggesting coelomic fluid did not play an important role in osmotic regulation. Taken together, based on antioxidant and nonspecific immunity, as well as osmotic regulation results, we suggest the intestine plays a significant role in salinity acclimation in U. unicinctus. The result is also consistent with the previous reports, which have shown that the intestine plays a critical role in osmotic regulation in aquatic organisms (Evans, 2010; Takei et al., 2014).
Effects of Salinity on Intestinal Microbiota Composition
Intestine microbes have been proved to play roles in many physiological processes such as nutritional provisioning, immune reaction, and metabolic homeostasis (Gomez and Balcazar, 2008; Sullam, 2012). To investigate the changes in microbial communities in various salinity environments, the intestine microbes in U. unicinctus were analyzed by 16S rRNA gene sequencing. After being filtered and denoised, a total of 996,561 sequences were retained out of a total of 1,180,529 clean sequencing reads. The microbial diversity of the intestine microbes were classified by operational taxonomic units (OTUs). Of the OTUs recognized in the LS10 (843), LS15 (1636), LS20 (2157), NS30 (1938), and HS40 (2794) groups, only 47 were shared among the five salinities (Figure 4A), indicating salinity may have a significant effect on U. unicinctus intestinal microbiota. Alpha diversity was applied to assess the complexity of species diversity in the U. unicinctus intestine (Schlos set al., 2009). Rarefaction curves analysis indicated that the sequencing data of intestinal microbiota in different salinity group was sufficient (Figure 4B). The alpha diversity analysis including Chao1, observed species, Shannon, and Simpson indices indicated that species richness and diversity was lower in the 10‰ group compared to other groups (P < 0.05) (Table 2). PCoA was performed to analyze the difference in the intestinal microbiota compositions under different salinities (Figure 4C). Based on the PCA plot, 30‰ and 40‰ salinity percentages were separated with other treatments by PC1 (36.2% of the total variant), whereas PC2 (22.9%) further exhibited the separation of 30‰ and 40‰, indicating that salinity had a significant effect on the intestinal microbiota composition in the U. unicinctus juveniles.
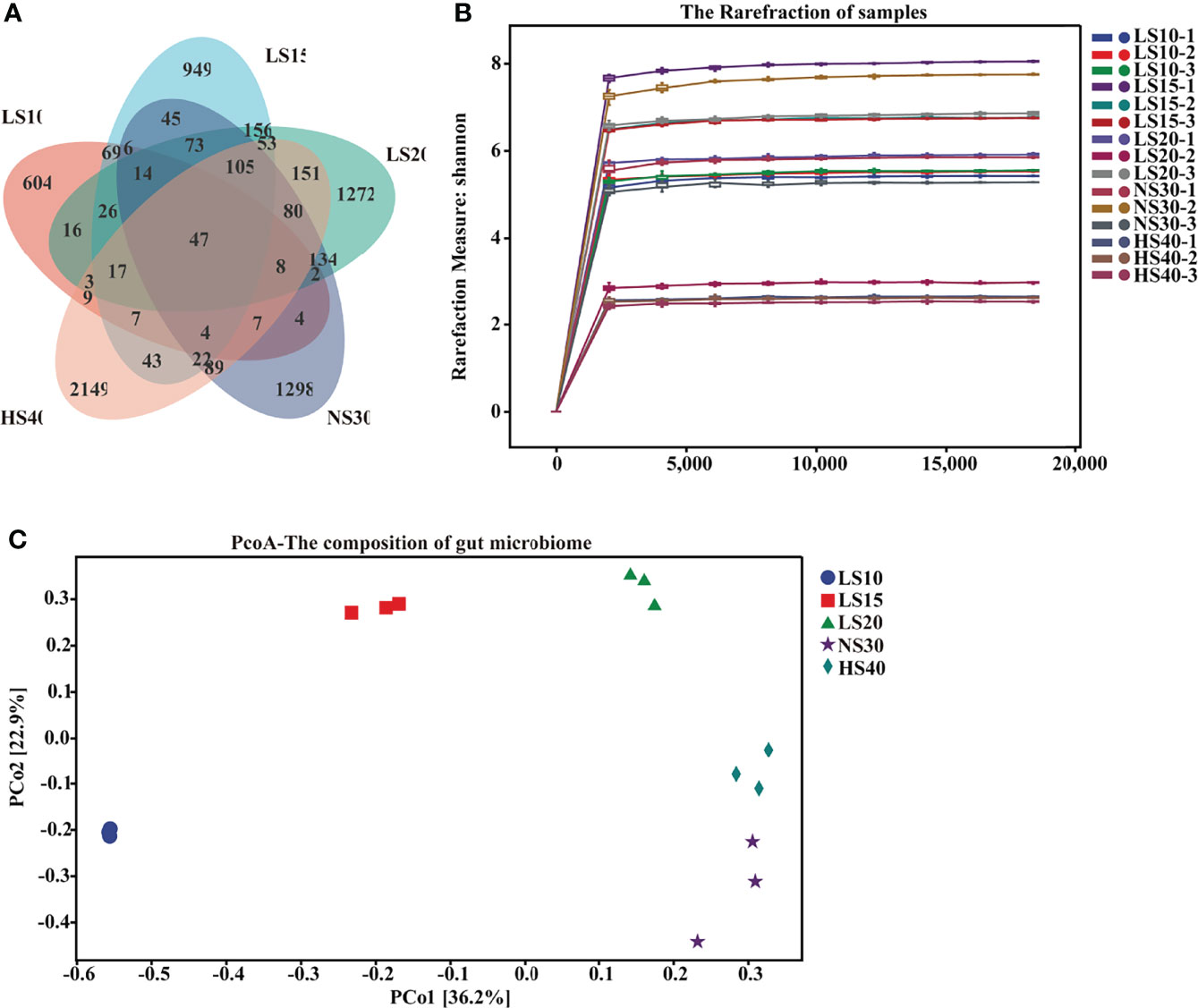
Figure 4 Changes in diversity of U. unicinctus intestine microbial species upon salinity stress. (A) Venn diagram of OTUs detected in the gut of 10‰ (LS10), 15‰ (LS15), 20‰ (LS20), 30‰ (NS30) and 40‰ (HS40) salinity. Different colors represent different groups. (B) Rarefaction curves of different salinity groups based on the Shannon indices. (C) Principal coordinate analysis (PCoA) of different salinity groups based on the Bray-Curtis distance.
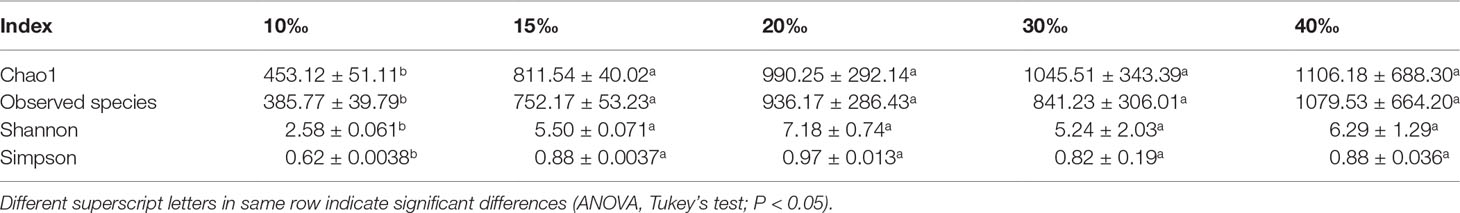
Table 2 Alpha diversity analysis of the intestinal bacteria of U. unicinctus exposed to different salinities.
To further investigate the variation of intestinal microbial composition in different salinity groups, a comparative analysis of the intestinal microbial communities from the phylum to genus levels was conducted (Supplementary Figure S1). The relative abundance of Firmicutes and Bacteroidetes was higher in the 15‰ (8.96% and 5.13%), 20‰ (22.57% and 10.35%), 30‰ (13.95% and 7.82%) and 40‰ (4.51% and 5.08%) groups than the 10‰ group (0.61% and 0.24%) (Supplementary Figure S1A). Firmicutes was also identified as a major constituent in Atlantic salmon (Rudi et al., 2018). At the class level, Gammaproteobacteria, Alphaproteobacteria, Clostridia, and Bacteroidia was the main class (top 80%) in the intestinal microbiota of U. unicinctus in the five different salinity groups (Supplementary Figure S1B). The relative abundance of Gammaproteobacteria microflora was significantly higher in the 10‰ group (91.39%) than in the 15‰ (26.59%), 20‰ (9.44%), 30‰ (52.16%) and 40‰ (14.76%) groups (P < 0.05), whereas the relative abundance of Alphaproteobacteria (5.09%) and Clostridia (0.40%) was lower in this group. Alphaproteobacteria and Gammaproteobacteria had been also identified as major class in the guts of numerous aquatic species (Lai et al., 2020; Chen et al., 2021), indicating these classes of bacteria may play important roles for nutritional provision and environment adaptation for marine organisms. At the order, family, and genus levels, the variation in the intestinal microbiota of U. unicinctus was more significant than those in the phylum and class levels (Supplementary Figure S1).
At the order level, the relative abundance of Legionellales was significantly higher in the 10‰ group than other groups, which comprise more than 90% in this group (Supplementary Figure S1C). Legionellales are bacteria linked to waterborne disease for humans (Morton et al., 1986; van Loenhout et al., 2012). The high relative abundance of Legionellales in the 10‰ group (91.17%) may have resulted in the lower growth compared to the other salinity groups. The Vibrionales was the major order in the 30‰ group (50.59%), while it altered the salinity results in the decreased, to almost non-existant, low salinity groups. The result is consistent with the previous report in shrimp (Cao et al., 2020; Hou et al., 2020; Tian et al., 2020), in which low salinity treatment resulted in decreased Vibrionales abundance in the gut. In addition, in the 40‰ group, the relative abundance of Rhodobacterale (46.76%) was significantly higher than other groups (P < 0.05). As some bacteria in Rhodobacterale are more tolerant to high salinity (Hansen and Veldkamp, 1973; Imhoff, 2001; Hülsen et al., 2019), we deduced that the increased population in the 40‰ group may due to this reason. Conversely, some Rhodobacterale, such as Rhodobacter capsulatus are widely used as probiotics in feed additive to improve the immunity of aquatic organisms. Considering the increased abundance in the 40‰ group, Rhodobacterale may play an important role in improving the high salinity acclimation for U. unicinctus. The family analysis results were consistent with the order level (Supplementary Figures S1C, S1D, S2). Due to the unclassified Legionellales (25% of the total) and Rhodobacterale (13% of the total) at the genus level (Supplementary Figure S2) which comprise most microbial communities in the 10‰ and 40‰ groups (Supplementary Figure S3), only the 30‰ group identified more than 50% intestinal microbial communities (Supplementary Figure S1E). To investigate the variation of intestinal microbial composition under salinity stress at the genus level more comprehensively, the unclassified genus was also analyzed in this study. Similar methods have been used in microbial composition analysis (Chen et al., 2020). The results indicated that the dominant genera of U. unicinctus intestinal microbiota from the 10‰ group were unidentified Legionellaceae (89.67%) and Labrenzia (3.95%); 15‰ and 20‰ groups with Labrenzia (35.93% and 25.77%), unidentified Legionellaceae (25.67% and 7.00%) and unidentified Rhodobacteraceae (7.67% and 10.97%); 30‰ group with Vibrio (50.59%), Shimia (8.98%), and unidentified Rhodobacteraceae (7.33%), whereas in the 40‰ group, the dominant genera were unidentified Rhodobacteraceae (41.33%), Vibrio (11.62%) and Labrenzia (7.73%) (Figure 5). Horizontal clustering of abundance at the genus level indicated the composition of intestinal microbial species in the U. unicinctus from the 15‰, 20‰, and 30‰ groups were more similar than those in the 40‰ and 10‰ groups, which was consistent with the variations in physiological characteristics.
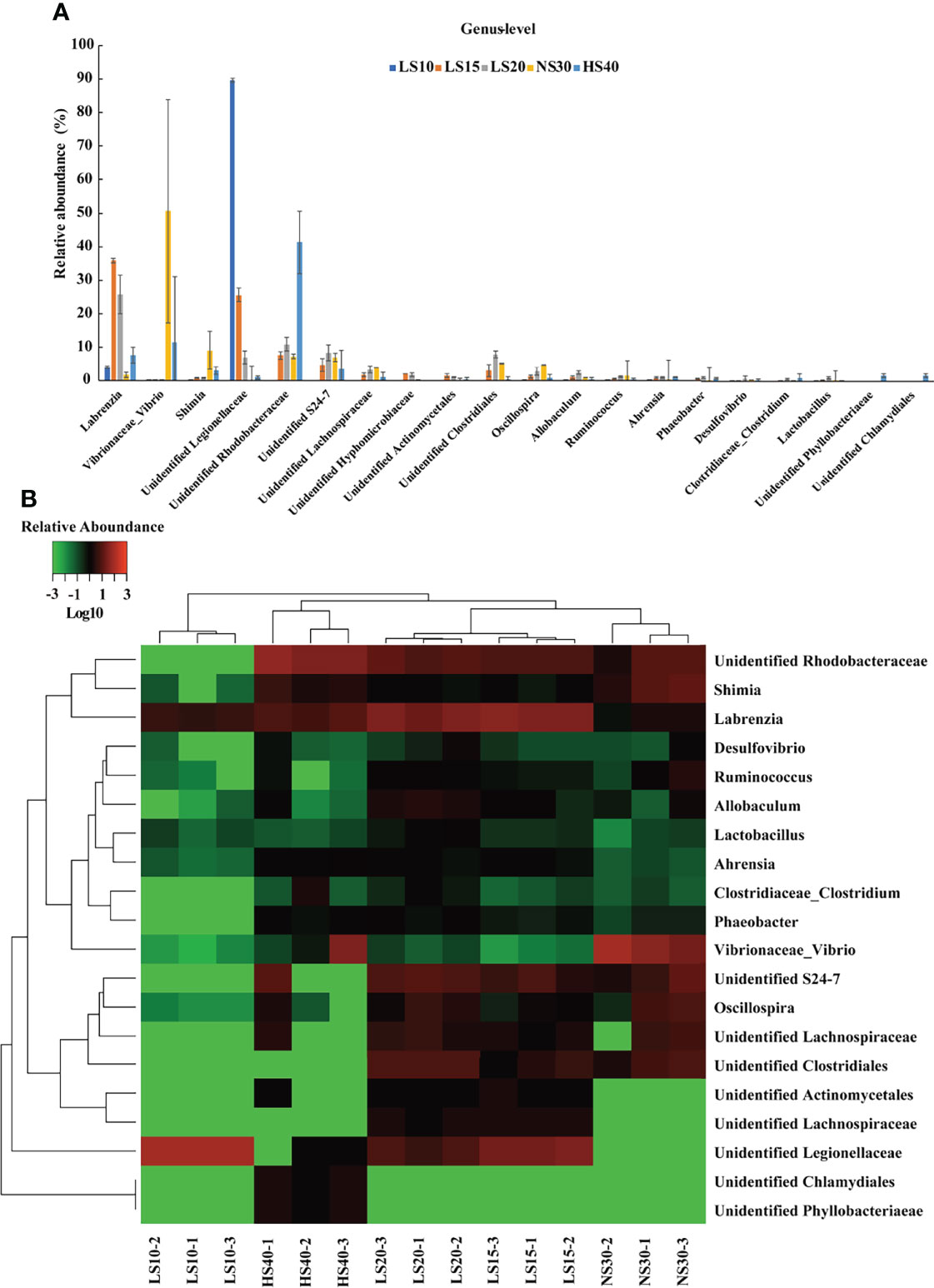
Figure 5 Changes of U. unicinctus intestine microbial species at the genus level in different salinity groups. (A) Relative abundance of the intestine microbiota at the genus level in 10‰ (LS10), 15‰ (LS15), 20‰ (LS20), 30‰ (NS30), and 40‰ (HS40) salinity groups. The data are mean values of three replicates ± SD (n = 3). (B) Heat map indicating the abundance of each bacterium at the genus level. Red represents a higher abundance. Green indicates a lower abundance.
Effects of Salinity on Intestinal Microbiota Functional Structure
Besides composition structure, salinity can also change the intestinal microbiota function of U. unicinctus juveniles. Based on PICRUSt2 analysis, we found that cofactor, prosthetic group, electron carrier, vitamin biosynthesis, nucleoside and nucleotide biosynthesis, amino acid biosynthesis, as well as fatty acid and lipid biosynthesis were the highest 4 abundant function pathways (Supplementary Figure S4), indicating these pathways may play important roles in nutritional provision in the gut (Dehler et al., 2017; Lai et al., 2020).
Intestinal microbiota functioning differently in different salinity groups was also observed, especially in the 10‰ group which showed significantly different functions compared with the other groups (Supplementary Figure S5). To further investigate the difference KEGG pathways, the up-regulated and down-regulated pathways in different salinity group compared to the 30‰ group were analyzed. The up-regulated KEGG pathways analysis showed that the 10‰ group included 89 unique pathways compared to other groups, while 20 pathways were included in all salinity groups (Figure 6A; Supplementary Table S1). The down-regulated KEGG pathways analysis showed that the 10‰ group included 24 unique pathways compared to other groups, whereas 10 pathways were included in all salinity groups (Figure 6B; Supplementary Table S2). The KEGG analysis results indicated the 10‰ group showed significant difference compared with the other groups, which was consistent with the microbial composition and functional structure analysis results, as well as with the growth performance. Furthermore, we found antioxidant related metabolic pathways including NAD biosynthesis II, mycothiol biosynthesis, factor 420 biosynthesis, and ergothioneine biosynthesis I (bacteria) (Ying, 2008; Gurumurthy et al., 2013; Cumming et al., 2014) were enriched in the up-regulated 20 pathways in all salinity stress groups (Figure 6C). As alterations in salinity can induce ROS generation and result in oxidative stress in the host (Lushchak, 2011), we suggest the up-regulated antioxidant metabolic pathways may play important roles in salinity acclimation of the host and bacteria themselves. In addition, amino acid metabolic pathways including the superpathway of ornithine degradation and L-tryptophan degradation to 2-amino-3-carboxymuconate semialdehyde, as well as aromatic biogenic amine degradation (bacteria), were also enriched (Figure 6C). Considering that free amino acids play important roles in osmoregulation of aquatic organisms (Lee et al., 2017; Yang et al., 2022), we suggest the up-regulated amino acid metabolic pathways in intestinal microbiota of U. unicinctus play an important role in assisting the host and bacteria under altered salinity environments. The result is consistent with the previous report in Atlantic salmon, which found amino acid metabolism was significantly differently abundant between freshwater and sea water fish bacterial communities (Dehler et al., 2017). Together, we conclude that the shift of bacterial composition will change the intestinal microbiota function under salinity stress and finally benefit the salinity acclimation of U. unicinctus.
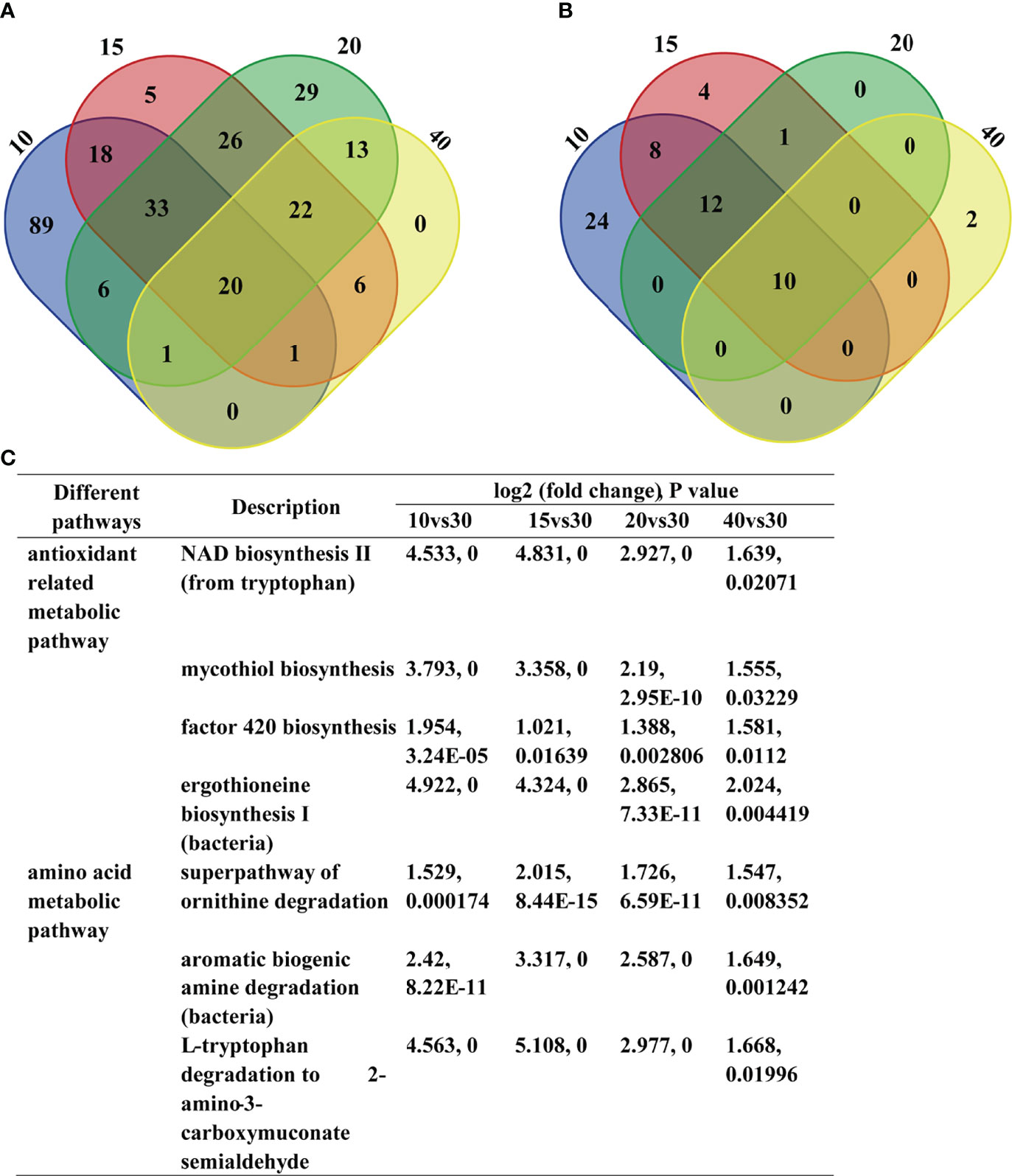
Figure 6 Changes of U. unicinctus intestine microbial function in different salinity groups. (A) Venn diagram showing the overlap between the up-regulated KEGG pathways in different salinity groups compare to 30‰ group (P < 0.05). (B) Venn diagram showing the overlap between the down-regulated KEGG pathways in different salinity groups compare to 30‰ group (P < 0.05). (C) Antioxidant related metabolic pathways and amino acid metabolic pathways in different salinity groups compare to 30‰ group.
Correlation Analysis Between the Intestinal Bacteria and the Physiological Characteristics
To further investigate the relationship between enzyme activities and the intestinal microbiota, the Spearman correlation was analyzed. The results indicated several intestinal enzyme activities were significantly correlated with the relative abundances of some intestinal bacteria genera among all test samples (Figure 7). The abundance of Ruminococcus, Oscillospira, and unidentified Lachnospiraceae were negatively correlated with the activity of SOD (P < 0.05), ALP (P < 0.05) and T-AOC (P < 0.01). Significant negative correlations were also found in the relative abundance of the unidentified Clostridiales and unidentified S24-7 with ACP activity and T-AOC (P < 0.05). In addition, the relative abundance of Shimia and Vibrio genus were positively correlated with the activity of Na+/K+-ATPase (P < 0.05), whereas unclassified Legionellaceae was negatively correlated with Na+/K+-ATPase activity (P < 0.05). Vibrio and Shimia are widely distribute in marine sediment and aquatic organism intestine (Cao et al., 2020; Hou et al., 2020; Tian et al., 2020; Zhu et al., 2021). Especially, Vibrio is a famous and widely distributed pathogen in aquatic organisms (Cao et al., 2020; Hou et al., 2020; Tian et al., 2020). Legionellaceae are bacteria linked to waterborne diseases in humans (Morton et al., 1986; van Loenhout et al., 2012). In this study, the Vibrio and Shimia were mainly detected in high salinity 30‰ and 40‰ groups, while Legionellaceae were most abundant (approximately 90%) in the low salinity 10‰ group. Given that the specific distribution and correlation with Na+/K+-ATPase activity, we suggest Vibrio and Shimia may play important roles in high salinity acclimation, while Legionellaceae may act as important gut microbiota in low salinity acclimation for U. unicinctus juveniles. Previous reports suggest the pathogens in the host intestine can create ecological niches that facilitate their expansion and advantages to outcompete commensals (Thiennimitr et al., 2011; Mallon et al., 2015). For example, the pathogen Salmonella enterica demonstrated a growth advantage in an inflamed gut due to its ability to respire ethanolamine, which is released from host tissue (Mallon et al., 2015). High salinity culture resulted in the increased abundance of Vibrio in the gut and had also been reported in shrimp (Hou et al., 2020) and three-spined stickleback (Milligan-Myhre et al., 2016). In this study, we demonstrated some pathogens may play a significant role in salinity acclimation for the host.
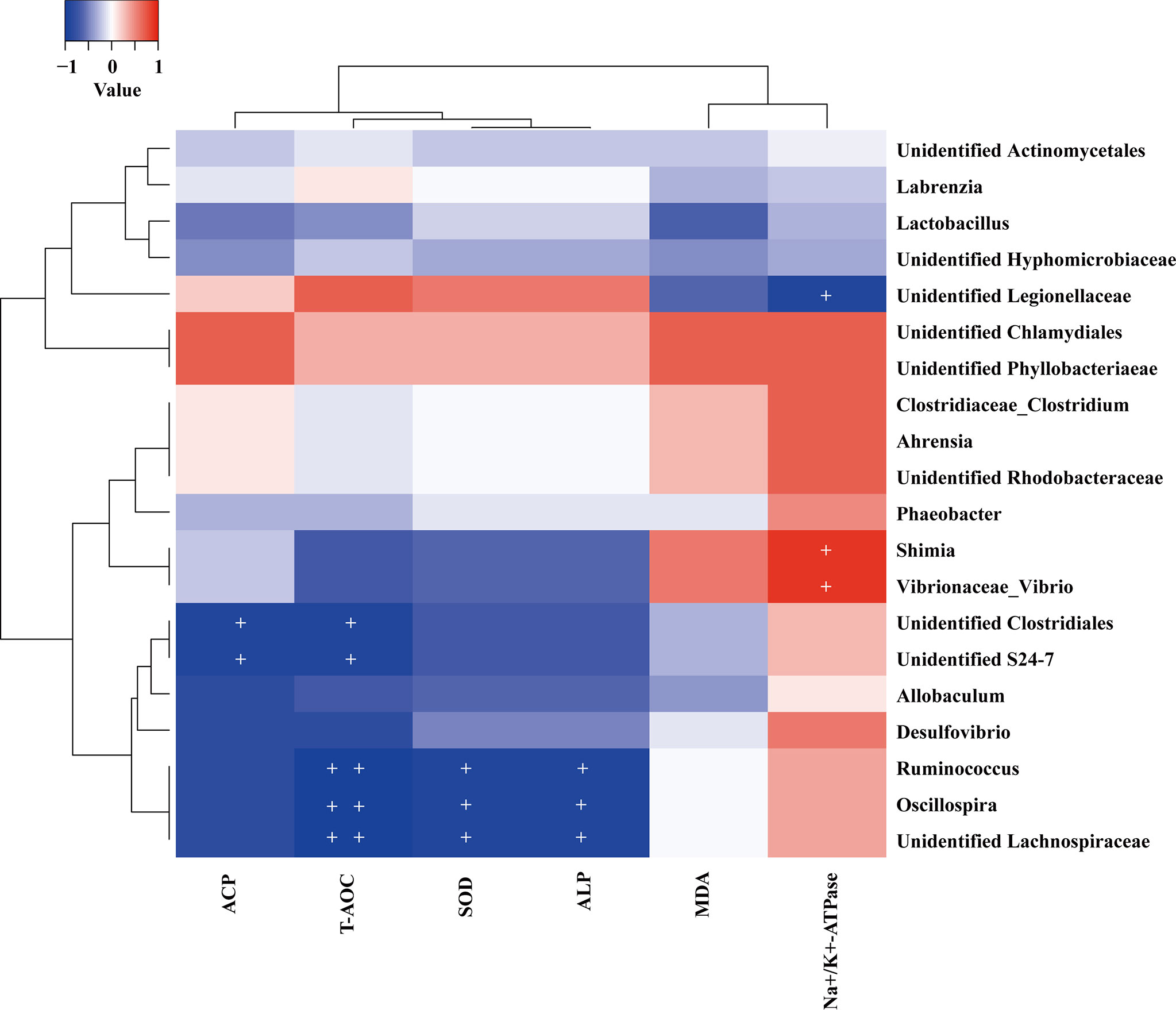
Figure 7 Heat map showing Spearman correlation between the abundance of the top 20 genus intestinal bacteria and the physiological characteristics in the U. unicinctus juveniles. + indicates that P < 0.05 and ++ indicates that P < 0.01.
Moreover, some genus such as Ruminococcus, Oscillospira, Lachnospiraceae, Clostridiales, and S24-7 caught our attentions due to their negative correlations with the antioxidant and nonspecific immune indices. The abundance of Ruminococcus is increased in irritable bowel syndrome (IBS) patients (Han et al., 2021). Oscillospira is an enigmatic bacterial genus that has never been cultured. Oscillospira is positively associated with leanness and health and negatively associated with inflammatory diseases in humans (Konikoff and Gophna, 2016). Oscillospira probably produces the short-chain fatty acid butyrate and is suggested as an important candidate for the next generation of probiotics. Besides Oscillospira, Lachnospiraceae, and Clostridiales could also produce the anti-inflammatory short chain fatty acid butyrate (Atarashi et al., 2011; Cabrera et al., 2022). Butyrate has attracted much attention due to its beneficial effects on the host, such as trophic and anti-inflammatory effects on epithelial cells (Inan et al., 2000) and has been proved to mediate host-microbial crosstalk in gut immune homeostasis (Hori et al., 2013; Furusawa et al., 2015). In addition, S24-7 has been also suggested to be involved in host microbe interactions (Palm et al., 2014; Bunker et al., 2015). In this study, the negative correlations of Ruminococcus, Oscillospira, Lachnospiraceae, Clostridiales, and S24-7 with antioxidant and nonspecific immune indices including SOD, ALP, ACP, and T-AOC which suggest these bacteria may play important roles in U. unicinctus salinity acclimation and could be considered as important candidates of probiotics for the U. unicinctus breeding industry.
Conclusion
In this study, we confirmed that U. unicinctus belongs to euryhaline organisms which can tolerate salinity of 15‰ to 40‰ and indicates the highest growth performance at 30‰. Physiological characteristic analyses of coelomic fluid and intestine indicated that the range of salinities from 15‰ to 30‰ were more suitable, while 10‰ and 40‰ salinities were not suitable for U. unicinctus juveniles. Furthermore, the intestinal microbiota analysis indicated salinity had a significant effect on the composition structure and functional structure in the U. unicinctus juveniles. The shift of microbiota may influence various metabolism-dependent pathways, such as antioxidant related metabolic pathways and amino acid metabolism pathways, to assist the host and bacteria in salinity acclimation. Comprehensive analysis of intestinal bacteria and the enzyme activities indicated unidentified Rhodobacteraceae, Vibrio, and Shimia may play important roles in high salinity acclimation, while Legionellaceae may act as important gut microbiota in low salinity acclimation. Given that the negative correlations with the antioxidant and nonspecific immune enzymes, Ruminococcus, Oscillospira, Lachnospiraceae, Clostridiales, and S24-7 may also play important roles in U. unicinctus salinity acclimation and could be considered important probiotic candidates for the U. unicinctus breeding industry.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: NCBI [accession: PRJNA822550].
Ethics Statement
This study was carried out in accordance with the recommendations of Animal Research and Ethics Committees of Ocean University of China.
Author Contributions
QZ: Methodology, Investigation. YW, JC, YL, FZ, DL, MW, and DK: Investigation. ZC: Resources. ZZ: Supervision, Funding acquisition. YM: Conceptualization, Writing – original draft, Writing – review & editing, Supervision, Funding acquisition. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the key research & development plan of Hainan province (ZDYF2021XDNY180), National Natural Science Foundation of China (31372506, 32170373), Shandong province science outstanding Youth Fund (ZR2020YQ20), China Postdoctoral Science Foundation (2020M680095) and Qingdao postdoctoral application research project.
Conflict of Interest
Author ZC was employed by Qingdao Dayi HC Biotechnology Co., Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2022.912023/full#supplementary-material
References
Altschul S. F., Madden T. L., Schaffer A. A., Zhang J. H., Zhang Z., Miller W., et al. (1997). Gapped BLAST and PSI-BLAST: A New Generation of Protein Database Search Programs. Nucleic Acids Res. 25 (17), 3389–3402. doi: 10.1093/nar/25.17.3389
Arp A. J., Menon J. G., Julian D. (1995). Multiple Mechanisms Provide Tolerance to Environmental Sulfide in Urechis Caupo. Integr. Comp. Biol. 35, 132–144. doi: 10.1093/icb/35.2.132
Atarashi K., Tanoue T., Shima T., Imaoka A., Kuwahara T., Momose Y., et al. (2011). Induction of Colonic Regulatory T Cells by Indigenous Clostridium Species. Science 331, 337–341. doi: 10.1126/science.1198469
Bi Q., Chu J., Feng Y., Jiang Z., Han B., Liu W. (2013). Purification and Characterization of a New Serine Protease With Fibrinolytic Activity From the Marine Invertebrate, Urechis Unicinctus. App. Biochem. Biotechnol. 170 (3), 525–540. doi: 10.1007/s12010-013-0168-4
Bunker J. J., Flynn T. M., Koval J. C., Shaw D. G., Meisel M., McDonald B. D., et al. (2015). Innate and Adaptive Humoral Responses Coat Distinct Commensal Bacteria With Immunoglobulin A. Immunity 43 (3), 541–553. doi: 10.1016/j.immuni.2015.08.007
Cabrera S. M., Coren A. T., Pant T., Ciecko A. E., Jia S., Roethle M. F., et al. (2022). Probiotic Normalization of Systemic Inflammation in Siblings of Type 1 Diabetes Patients: An Open-Label Pilot Study. Sci. Rep. 12, 3306. doi: 10.1038/s41598-022-07203-6
Cao Q., Najnine F., Han H., Wu B., Cai J. (2020). BALOs Improved Gut Microbiota Health in Postlarval Shrimp (Litopenaeus Vannamei) After Being Subjected to Salinity Reduction Treatment. Front. Microbiol. 11. doi: 10.3389/fmicb.2020.01296
Caporaso J. G., Kuczynski J., Stombaugh J., Bittinger K., Bushman F. D., Costello E. K., et al. (2010). QIIME Allows Analysis of High-Throughput Community Sequencing Data. Nat. Methods 7, 335–336. doi: 10.1038/nmeth.f.303
Chen J. C., Chia P. G. (1997). Osmotic and Ionic Concentrations of Scylla Serrata (Forskal) Subjected to Different Salinity Levels. Comp. Biochem. Physiol. A Physiol. 117 (2), 239–244. doi: 10.1016/S0300-9629(96)00237-X
Chen H., Jiang W. (2014). Application of High-Throughput Sequencing in Understanding Human Oral Microbiome Related With Health and Disease. Front. Microbiol. 5. doi: 10.3389/fmicb.2014.00508
Chen W. B., Zhang S. S., Sun Y., Tian B., Song L. J., Xu Y., et al. (2021). Effects of Substrate on the Physiological Characteristics and Intestinal Microbiota of Echiura Worm (Urechis Unicinctus) Juveniles. Aquaculture 530, 735710. doi: 10.1016/j.aquaculture.2020.735710
Chen W. B., Zhang S. S., Sun Y., Xu Y., Song L. J., Tian B., et al. (2020). Effects of Stocking Density on the Growth Performance, Physiological Response and Intestinal Microbiota of Juvenile Echiura Worms (Urechis Unicinctus). Aquac. Res. 00, 1–10. doi: 10.1111/are.14741
Chen X. L., Zhang L. M., Xu C. X., Yin X. H., Wei B. (2019). Industrialized Three-Dimensional Ecological Polyculture Experiment of Urechis Unicinctus and Stichopus Japonicus. Hebei Fisheries 6, 8–9. doi: 10.3969/j.issn.1004-6755.2019.06.004
Chien Y. H., Pan C. H., Hunter B. (2003). The Resistance to Physical Stresses by Penaeus Monodon Juveniles Fed Diets Supplemented With Astaxanthin. Aquaculture 216 (1-4), 177–191. doi: 10.1016/S0044-8486(02)00056-X
Conde A., Novais M., Domínguez J. (2013). Intertidal Macrofauna and Environmental Stress at a Riverine-Marine Boundary. Mar. Environ. Res. 92, 1–9. doi: 10.1016/j.marenvres.2013.07.002
Cui Q. M., Zhao Z. Y., Gao T., Yuan C. Y. (2021). Effects of Glycosaminoglycan From Urechis Unicinctus on ADP-Induced Platelet Calcium and Membrane Glycoprotein Expressions in Rats. Acta Haematologia 144 (1), 44–47. doi: 10.1159/000505769
Cumming B. M., Lamprecht D. A., Wlls R. M., Saini V., Mazorodze J., Steyn A. J. (2014). The Physiology and Genetics of Oxidative Stress in Mycobacteria. Microbiol. Spectr. 2 (3), 1–22. doi: 10.1128/microbiolspec.MGM2-0019-2013
Dehler C. E., Secombes C. J., Martin S. A. (2017). Environmental and Physiological Factors Shape the Gut Microbiota of Atlantic Salmon Parr (Salmo Salar L.). Aquaculture 467, 149–157. doi: 10.1016/j.aquaculture.2016.07.017
DeSantis T. Z., Hugenholtz P., Larsen N., Rojas M., Brodie E. L., Keller K., et al. (2006). Greengenes, a Chimera-Checked 16S rRNA Gene Database and Workbench Compatible With ARB. App. Environ. Microbiol. 72 (7), 5069–5072. doi: 10.1128/AEM.03006-05
Deschaseaux E. S. M., Taylor A. M., Maher W. A., Davis A. R. (2010). Cellular Responses of Encapsulated Gastropod Embryos to Multiple Stressors Associated With Climate Change. J. Exp. Mar. Biol. Ecol. 383, 130–136. doi: 10.1016/j.jembe.2009.12.013
Douglas G. M., Maffei V. J., Zaneveld J. R., Yurgel S. N., Brown J. R., Taylor C. M., et al. (2020). PICRUSt2 for Prediction of Metagenome Functions. Nat. Biotechnol. 38, 685–688. doi: 10.1038/s41587-020-0548-6
Edgar R. C. (2010). Search and Clustering Orders of Magnitude Faster Than BLAST. Bioinformatics 26 (19), 2460–2461. doi: 10.1093/bioinformatics/btq461
Edsall C. C. (1999). A Blood Chemistry Profile for Lake Trout. J. Aquat. Anim. Health 11, 81–86. doi: 10.1577/1548-8667(1999)011<0081:ABCPFL>2.0.CO;2
Eichmiller J. J., Hamilton M. J., Staley C., Sadowsky M. J., Sorensen P. W. (2016). Environment Shapes the Fecal Microbiome of Invasive Carp Species. Microbiome 4, 44. doi: 10.1186/s40168-016-0190-1
Evans T. G. (2010). Co-Ordination of Osmotic Stress Responses Through Osmosensing and Signal Transduction Events in Fishes. J. Fish Biol. 76, 1903–1925. doi: 10.1111/j.1095-8649.2010.02590.x
Falconer T. R. L., Marsden I. D., Hill J. V., Glover C. N. (2019). Does Physiological Tolerance to Acute Hypoxia and Salinity Change Explain Ecological Niche in Two Intertidal Crab Species? Conserv. Physiol. 7 (1), coz086. doi: 10.1093/conphys/coz086
Feng G. P., Liu J., Zhuang P., Zhang L. Z., Wang R. F., Liu J. Y. (2013). Effects of Salinity on Osmo-Ionic Regulation and Enzyme Activities in Mature Female Eriocheir Sinensis. Mar. Fish 35 (4), 468–473. doi: 10.13233/j.cnki.mar.fish.2013.04.015
Furusawa Y., Obata Y., Fukuda S., Endo T. A., Nakato G., Takahashi D., et al. (2015). A New View of the Fish Gut Microbiome: Advances From Next-Generation Sequencing. Aquaculture 448, 464–475. doi: 10.1016/j.aquaculture.2015.06.033
Giffard-Mena I., Charmantier G., Grousset E., Aujoulat F., Castille R. (2006). Digestive Tract Ontogeny of Dicentrarchus Labrax: Implication in Osmoregulation. Dev. Growth Differ. 48, 139–151. doi: 10.1111/j.1440-169X.2006.00852.x
Gomez G. D., Balcazar J. L. (2008). A Review on the Interactions Between Gut Microbiota and Innate Immunity of Fish. FEMS Immunol. Med. Microbiol. 52, 145–154. doi: 10.1111/j.1574-695X.2007.00343.x
Gurumurthy M., Rao M., Mukherjee T., Rao S. P. S., Boshoff H. I., Dick T., et al. (2013). A Novel F(420) -Dependent Anti-Oxidant Mechanism Protects Mycobacterium Tuberculosis Against Oxidative Stress and Bactericidal Agents. Mol. Microbiol. 87 (4), 744–755. doi: 10.1111/mmi.12127
Hansen T., Veldkamp H. (1973). Rhodopseudomonas Sulfidophila, Nov. Spec., a New Species of the Purple Nonsulfur Bacteria. Arch. Microbiol. 92 (1), 45–58. doi: 10.1007/BF00409510
Han L. J., Zhao L., Zhou Y., Yang C., Xiong T., Lu L., et al. (2021). Altered Metabolome and Microbiome Features Provide Clues in Understanding Irritable Bowel Syndrome and Depression Comorbidity. ISME J. 16 (4), 983–996. doi: 10.1038/s41396-021-01123-5
Henry R. P., Garrelts E. E., McCarty M. M., Towle D. W. (2002). Differential Induction of Branchial Carbonic Anhydrase and NA+/K+ ATPase Activity in the Euryhaline Crab, Carcinus Maenas, in Response to Low Salinity Exposure. J. Exp. Zool. 292, 595–603. doi: 10.1002/jez.10075
Hori S., Ohara O., Morita T., Koseki H., Kikuchi J., Honda K., et al. (2013). Commensal Microbe-Derived Butyrate Induces the Differentiation of Colonic Regulatory T Cells. Nature 504, 446–450. doi: 10.1038/nature12721
Hou D., Zhou R., Zeng S., Wei D., Deng X., Xing C., et al. (2020). Intestine Bacterial Community Composition of Shrimp Varies Under Low- and High-Salinity Culture Conditions. Front. Microbiol. 11. doi: 10.3389/fmicb.2020.589164
Hülsen T., Hsieh K., Batsrone D. J. (2019). Saline Wastewater Treatment With Purple Phototrophic Bacteria. Water Res. 160, 259–267. doi: 10.1016/j.watres.2019.05.060
Imhoff J. F. (2001). True Marine and Halophilic Anoxygenic Phototrophic Bacteria. Arch. Microbiol. 176 (4), 243–254. doi: 10.1007/s002030100326
Inan M. S., Rasoulpour R. J., Yin L., Hubbard A. K., Rosenberg D. W., Giardina C. (2000). The Luminal Short-Chain Fatty Acid Butyrate Modulates NF-kB Activity in a Human Colonic Epithelial Cell Line. Gastroenterology 118, 724–734. doi: 10.1016/s0016-5085(00)70142-9
Jarvis P. L., Ballantyne J. S. (2003). Metabolic Responses to Salinity Acclimation in Juvenile Shortnose Sturgeon Acipenser Brevirostrum. Aquaculture 219, 891–909. doi: 10.1016/S0044-8486(03)00063-2
Kelly S. P., Woo N. Y. S. (1999). The Response of Sea Bream Following Abrupt Hypoosmotic Exposure. J. Fish Bio. 55, 732–750. doi: 10.1111/j.1095-8649.1999.tb00714.x
Kneib R. T. (1984). Patterns of Invertebrate Distribution and Abundance in the Intertidal Salt Marsh: Causes and Questions. Estuar. Coast. 7, 392–412. doi: 10.2307/1351621
Konikoff T., Gophna U. (2016). Oscillospira: A Central, Enigmatic Component of the Human Gut Microbiota. Trends Microbiol. 24 (7), 523–524. doi: 10.1016/j.tim.2016.02.015
Kua B. C., Choong F. C., Leaw Y. Y. (2014). Effect of Salinity and Temperature on Marine Leech, Zeylanicobdella Arugamensis (De Silva) Under Laboratory Conditions. J. Fish Dis. 37, 201–207. doi: 10.1111/jfd.12087
Kültz D. (2015). Physiological Mechanisms Used by Fish to Cope With Salinity Stress. J. Exp. Biol. 218, 1907–1914. doi: 10.1242/jeb.118695
Lai K. P., Lin X., Tam N., Ho J. C. H., Wong M. K. S., Gu J., et al. (2020). Osmotic Stress Induces Gut Microbiota Community Shift in Fish. Environ. Microbiol. 22 (9), 3784–3802. doi: 10.1111/1462-2920.15150
Lee S. H., Lee M. C., Puthumana J., Park J. C., Kang S. J., Hwang D. S., et al. (2017). Effects of Salinity on Growth, Fatty Acid Synthesis, and Expression of Stress Response Genes in the Cyclopoid Copepod Paracyclopina Nana. Aquaculture 470, 182–189. doi: 10.1016/j.aquaculture.2016.12.037
Lepage G., Munoz G., Champagne J., Roy C. C. (1991). Preparative Steps Necessary for the Accurate Measurement of Malondialdehyde by High-Performance Liquid Chromatography. Anal. Biochem. 197 (2), 277–283. doi: 10.1016/0003-2697(91)90392-7
Li H. H., Liu X., Sun N., Li H. Y. (2019).  Effects of Salinity and Sediment on Growth and Survival of Urechis Unicinctus Juvenile. Hunan Agr. Sci. 4, 85–88. doi: 10.16498/j.cnki.hnnykx.2019.004.022
Li N., Song S. L., Tang Y. Z. (1995). The Life History of the Urechis Unicinctus (Von. Draschi). Shandong Fisheres 12 (6), 24–27.
Li N., Song S. L., Tang Y. Z. (2000). Analysis of Composition and Content of Aminoacid in the Bodywall Muscle Urechis Unicinctus. Shandong Fisheres 17 (5), 26–27.
Li N., Song S. L., Tang Y. Z., Wang B. G. (1998). Study on Culture and Enhancement Biology of Urechis Unicinctus. Shandong Fisheres 15 (1), 11–14.
Li F., Sun T., Ji Y., Wang L., Yu H. (2017). Advances in Studies on the Biology and Ecology of Urechis Unicinctus. Mar. Sci. 41 (10), 125–131.
Liu Z. F., Gao X. Q., Yu J. X., Qian X. M., Xue G. P., Zhang Q. Y., et al. (2017). Effects of Different Salinities on Growth Performance, Survival, Digestive Enzyme Activity, Immune Response, and Muscle Fatty Acid Composition in Juvenile American Shad (Alosa Sapidissima). Fish Physiol. Biochem. 43, 761–773. doi: 10.1007/s10695-016-0330-3
Liu D. W., Qin Z. Q., Wei M. K., Kong D. X., Zheng Q. J., Bai S. M., et al. (2022). Genome-Wide Analyses of Heat Shock Protein Superfamily Provide New Insights on Adaptation to Sulfide-Rich Environments in Urechis Unicinctus (Annelida, Echiura). Int. J. Mol. Sci. 23,2715. doi: 10.3390/ijms23052715
Li Z. H., Zlabek V., Velisek J., Grabic R., Machova J., Kolarova J., et al. (2011). Acute Toxicity of Carbamazepine to Juvenile Rainbow Trout (Oncorhynchus Mykiss): Effects on Antioxidant Responses, Hematological Parameters and Hepatic EROD. Ecotox. Environ. Safe. 74, 319–327. doi: 10.1016/j.ecoenv.2010.09.008
Long X., Wu X., Zhao L., Ye H., Cheng Y., Zeng C. (2017). Effects of Salinity on Gonadal Development, Osmoregulation and Metabolism of Adult Male Chinese Mitten Crab, Eriocheir Sinensis. PLoS One 12 (6), e0179036. doi: 10.1371/journal.pone.0179036
Lozupone C. A., Hamady M., Kelley S. T., Knight R. (2007). Quantitative and Qualitative Beta Diversity Measures Lead to Different Insights Into Factors That Structure Microbial Communities. Appl. Environ. Microb. 73 (5), 1576–1585. doi: 10.1128/AEM.01996-06
Lozupone C., Knight R. (2005). UniFrac: A New Phylogenetic Method for Comparing Microbial Communities. Appl. Environ. Microb. 71 (12), 8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005
Lushchak V. I. (2011). Environmentally Induced Oxidative Stress in Aquatic Animals. Aquat. Toxicol. 101 (1), 13–30. doi: 10.1016/j.aquatox.2010.10.006
Ma Z. J., Bao Z. M., Kang K. H., Yu L., Zhang Z. F. (2005). The Changes of Three Components in Coelomic Fluid of Urechis Unicinctus Exposed to Different Concentrations of Sulfide. Chin. J. Oceanol. Limnol. 23, 104–109. doi: 10.1007/BF02845150
Magnadóttir B. (2006). Innate Immunity of Fish (Overview). Fish Shellfish Immunol. 20 (2), 137–151. doi: 10.1016/j.fsi.2004.09.006
Magoc T., Salzberg S. L. (2011). FLASH: Fast Length Adjustment of Short Reads to Improve Genome Assemblies. Bioinformatics 27 (21), 2957–2963. doi: 10.1093/bioinformatics/btr507
Mallon C. A., Elsas J. D., Salles J. F. (2015). Microbial Invasions: The Process, Patterns, and Mechanisms. Trends Microbiol. 23, 719–729. doi: 10.1016/j.tim.2015.07.013
Martínez-Álvarez R. M., Hidalgo M. C., Domezain A., Morales A. E., García-Gallego M. (2002). Physiological Changes of Sturgeon Acipenser Naccarii Caused by Increasing Environmental Salinity. J. Exp. Biol. 205, 3699–3706. doi: 10.1242/jeb.205.23.3699
Ma Y. B., Zhang Z. F., Shao M. Y., Kang K. H., Tan Z., Li J. L. (2011). Sulfide: Quinone Oxidoreductase From Echiuran Worm Urechis Unicinctus. Mar. Biotechnol. 13, 93–107. doi: 10.1007/s10126-010-9273-3
Ma Y. B., Zhang Z. F., Shao M. Y., Kang K. H., Tan Z., Shi X. L., et al. (2012). Response of Sulfide:Quinone Oxidoreductase to Sulfide Exposure in the Echiuran Worm. Mar. Biotechnol. 14 (2), 245–251. doi: 10.1007/s10126-011-9408-1
Milligan-Myhre K, Small C. M., Mittge E. K., Agarwal M., Currey M., Cresko W. A., et al(2016)Innate Immune Responses to Gut Microbiota Differ Between Oceanic and Freshwater Threespine Stickleback Populations. Dis. Model Mech9187198doi: 1242/dmm.021881
Montagna P. A., Hu X., Palmer T. A., Wetz M. (2018). Effect of Hydrological Variability on the Biogeochemistry of Estuaries Across a Regional Climatic Gradient. Limnol. Oceanogr. 63, 2465–2478. doi: 10.1002/lno.10953
Morton S., Bartlett C. L. R., Bibby L. F., Hutchinson D. N., Dyer J. V., Dennis P. J. (1986). Outbreak of Legionnaires’ Disease From a Cooling Water System in a Power Station. Br. J. Ind. Med. 43, 630–635. doi: 10.1136/oem.43.9.630
Niu R., Chen X. (2016). Full-Length cDNA, Prokaryotic Expression, and Antimicrobial Activity of UuHb-F-I From Urechis Unicinctus. BioMed. Res. Int. 2, 1–8. doi: 10.1155/2016/5683026
Ondov B. D., Bergman N. H., Phillippy A. M. (2011). Interactive Metagenomic Visualization in a Web Browser. BMC Bioinf. 12, 385. doi: 10.1186/1471-2105-12-385
Onken H., Putzenlechner M. (1995). A V-Atpase Drives Active, Electrogenic and Na+-Independent Cl- Absorption Across the Gills of Eriocheir Sinensis. J. Exp. Biol. 198 (3), 767–774. doi: 10.1242/jeb.198.3.767
Palm N. W., de Zoete M. R., Cullen T. W., Barry N. A., Stefanowski J., Hao L., et al. (2014). Immunoglobulin A Coating Identifies Colitogenic Bacteria in Inflammatory Bowel Disease. Cell 158 (5), 1000–1010. doi: 10.1016/j.cell.2014.08.006
Ramette A. (2007). Multivariate Analyses in Microbial Ecology. FEMS Microbiol. Ecol. 62 (2), 142–160. doi: 10.1111/j.1574-6941.2007.00375.x
Rubio C. P., Hernández-Ruiz J., Martinez-Subiela S., Tvarijonaviciute A., Ceron J. J. (2016). Spectrophotometric Assays for Total Antioxidant Capacity (TAC) in Dog Serum: An Update. BMC Vet. Res. 12, 166. doi: 10.1186/s12917-016-0792-7
Rudi K., Angell I. L., Pope P. B., Vik J. O., Sandve S. R., Snipen L. G. (2018). Stable Core Gut Microbiota Across the Freshwater-to-Saltwater Transition for Farmed Atlantic Salmon. Appl. Environ. Microb. 84, e01974–e01917. doi: 10.1128/AEM.01974-17
Schloss P. D., Westcott S. L., Ryabin T., Hall J. R., Hartmann M., Hollister E.B., et al. (2009). Introducing mothur: Open-Source, Platform-Independent, Community-Supported Software for Describing and Comparing Microbial Communities. Appl. Environ. Microbiol. 75, 7537–7541. doi: 10.1128/AEM.01541-09
Shen Y. H. (2007). Effect of Salinity Stress on the Activities of Phenoloxidase and Superoxide Dismutase of the Serum, Tissue and Organ of Mud Crab, Scylla Serrata. J. Oceanography Taiwan Strait 4, 569–575.
Sullam K. E. (2012). Environmental and Ecological Factors That Shape the Gut Bacterial Communities of Fish: A Meta-Analysis. Mol. Ecol. 21, 3363–3378. doi: 10.1111/j.1365-294X.2012.05552.x
Sun F. Y., Gai S. S., Ji Y., Liu F., Chen K. W. (2020). Effects of Different Salinity on Survival Rate and Behavioral Response of Urechis Unicinctus. Jiangsu Agr. Sci. 48 (22), 178–185. doi: 10.15889/j.issn.1002-1302-2020.22.034
Sung W. S., Park S. H., Lee D. G. (2008). Antimicrobial Effect and Membrane-Active Mechanism of Urechis Tachykinins, Neuropeptides Derived From Urechis Unicinctus. FEBS Lett. 582 (16), 2463–2466. doi: 10.1016/j.febslet.2008.06.015
Takei Y., Hiroi J., Takahashi H., Sakamot T. (2014). Diverse Mechanisms for Body Fluid Regulation in Teleost Fishes. Am. J. Physiol. Reg. I. 307, R778–R792. doi: 10.1152/ajpregu.00104.2014
Thiennimitr P., Winter S. E., Winter M. G., Xavier M. N., Tolstikov V., Huseby D. L., et al. (2011). Intestinal Inflammation Allows Salmonella to Use Ethanolamine to Compete With the Microbiota. P. Nat. Acad. Sci. U.S.A. 108, 17480–17485. doi: 10.1073/pnas.1107857108
Tian L., Tan P., Yang L., Zhu W. L., Xu D. D. (2020). Effects of Salinity on the Growth, Plasma Ion Concentrations, Osmoregulation, non-Specific Immunity, and Intestinal Microbiota of the Yellow Drum (Nibea Albiflora). Aquaculture 528, 735470. doi: 10.1016/j.aquaculture.2020.735470
van Loenhout J. A. F., Paget W. J., Vercoulen J. H., Wijkmans C. J., Hautvast J. L. A., van der Velden K. (2012). Assessing the Long-Term Health Impact of Q-Fever in the Netherlands: A Prospective Cohort Study Started in 2007 on the Largest Documented Q-Fever Outbreak to Date. BMC Infect. Dis. 12, 2–7. doi: 10.1186/1471-2334-12-280
Wang R. F., Zhuang P., Feng G. P., Zhang L. Z., Huang X. R., Jia X. Y. (2012). Osmotic and Ionic Regulation and Na+/K+-ATPase, Carbonic Anhydrase Activities in Mature Chinese Mitten Crab, Eriocheir Sinensis H. Milne Edwards 1853 (Decapoda, Brachyura) Exposed to Different Salinities. Crustaceana 85 (12-13), 1431–1447. doi: 10.1163/15685403-00003125
White J. R., Nagarajan N., Pop M. (2009). Statistical Methods for Detecting Differentially Abundant Features in Clinical Metagenomic Samples. PloS Comput. Biol. 5 (4), e1000352. doi: 10.1371/journal.pcbi.1000352
Xu L. W., Feng J., Guo Z. X., Lin H. Z., Guo G. X. (2008). Effect of Salinity on Hematology and Gill Na+/K+-ATPase Activity of Juvenile Cobia, Rachycentron Canadum Linnaeus. Mar. Environ. Sci. 27, 602–606.
Xu X. H., Zhu J. X., Huo W., Xiao M., Chen Y. Q. (2016). The Artificial Breeding and Aquatics Technique of Urechis Unicinctus. Sci. Fish Farming 2, 53–55.
Xu X. H., Zhu X. Y., Que Y. J., Xu G. C., Xu J. T., Huo W., et al. (2017). Effects of Ph, Water Temperature and Salinity on Activities of Intestinal Digestive Enzymes and Serum Lysozyme in Echiuran Worm Urechis Unicinctus. Fish. Sci. 36 (2), 138–142. doi: 10.16378/j.cnki.1003-1111.2017.02.003
Yang Z. G., Zhu L. L., Zhao X. J., Cheng Y. X. (2022). Effects of Salinity Stress on Osmotic Pressure, Free Amino Acids, and Immune-Associated Parameters of the Juvenile Chinese Mitten Crab, Eriocheir Sinensis. Aquaculture 549, 737776. doi: 10.1016/j.aquaculture.2021.737776
Ying W. H. (2008). NAD+/NADH and NADP+/NADPH in Cellular Functions and Cell Death: Regulation and Biological Consequences. Antioxid. Redox Signal. 10 (2), 179–206. doi: 10.1089/ars.2007.1672
Zarejabad A. M., Jalali M. A., Sudagar M., Pouralimotlagh S. (2010). Hematology of Great Sturgeon (Huso Huso Linnaeus 1758) Juvenile Exposed to Brackish Water Environment. Fish Physiol. Biochem. 36, 655–659. doi: 10.1007/s10695-009-9339-1
Zaura E., Keijser B. J. F., Huse S. M., Crielaard W. (2009). Defining the Healthy Core Microbiome of Oral Microbial Communities. BMC Microbiol. 9, 12. doi: 10.1186/1471-2180-9-259
Zhang H. Y., Ceung S. G., Shin P. K. S. (2014). The Larvae of Congeneric Gastropods Showed Differential Responses to the Combined Effects of Ocean Acidification, Temperature and Salinity. Mar. pollut. Bull. 79, 39–46. doi: 10.1016/j.marpolbul.2014.01.008
Zhang X. M., Li K. J. (2011). Research Progress on Biology and Physiology of Urechis Unicinctus. J. Henan Agr. Sci. 40 (11), 26–29. doi: 10.15933/j.cnki.1004-3268.2011.11.041
Zhang T. T., Qin Z. K., Liu D. W., Wei M. K., Fu Z., Wang Q., et al. (2021). A Novel Transcription Factor MRPS27 Up-Regulates the Expression of Sqr, a Key Gene of Mitochondrial Sulfide Metabolism in Echiuran Worm Urechis Unicinctus. Comp. Biochem. Physiol. C 243, 108997. doi: 10.1016/j.cbpc.2021.108997
Zhao H., Han B. Q., Liu W. S., Wang C. H. (2008). Antitumor Effect and Immunomodulation of Polypeptides From Urechis Unicinctus in Mice. Chin. J. Nat. Med. 6 (4), 302–306. doi: 10.3724/SP.J.1009.2008.00302
Zhao L., Long X. W., Wu X. G., He J., Shi Y. H., Zhang G. Y., et al. (2016). Effects of Water Salinity on Osmoregulation and Physiological Metabolism of Adult Male Chinese Mitten Crab Eriocheir Sinensis. Acta Hydrobiologica Sin. 40 (1), 27–34. 10.7541/2016.4
Zhao Y. H., Wang Z. J. (2018). High Quality and Efficient Culture Model of Litopenaeus Vannamei-Optimal Polyculture Density of Urechis Unicinctus.Test Summary 2018,1006–3188.
Zheng Y., Bai H. J., Wang Y. P. (2006). Tolerance of Urechis Unicinctus Juveniles to Temperature, Salinity and Ph Value. Fisheries Sci. 25 (10), 513–516. doi: 10.16378/j.cnki.1003-1011.2006.10.008
Zhu S. T., Chen C., Cheng Y. P., Guo C. B., Peng N., Zhang W. Y., et al. (2021). Shimia Sediminis Sp. Nov., a Bacterium Isolated From Marine Sediment in the East China Sea. Int. J. Syst. Evol. Microbiol. 71, 4782. doi: 10.1099/ijsem.0.004782
Keywords: Urechis unicinctus, salinity acclimation, growth performance, osmoregulation, intestinal microbiota, immune response
Citation: Zheng Q, Wang Y, Chen J, Li Y, Zhao F, Liu D, Wei M, Kong D, Chen Z, Zhang Z and Ma Y (2022) Effects of Salinity on the Growth, Physiological Characteristics, and Intestinal Microbiota of the Echiura Worm (Urechis unicinctus). Front. Mar. Sci. 9:912023. doi: 10.3389/fmars.2022.912023
Received: 03 April 2022; Accepted: 30 May 2022;
Published: 12 July 2022.
Edited by:
Yun-Zhang Sun, Jimei University, ChinaReviewed by:
Shuyan Chi, Guangdong Ocean University, ChinaJack Chi-Ho Ip, Hong Kong Baptist University, Hong Kong SAR, China
Copyright © 2022 Zheng, Wang, Chen, Li, Zhao, Liu, Wei, Kong, Chen, Zhang and Ma. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yubin Ma, bWF5dWJpbkBvdWMuZWR1LmNu
 Qiaojun Zheng
Qiaojun Zheng Yunjian Wang1
Yunjian Wang1 Yubin Ma
Yubin Ma