- 1Key Laboratory of South China Sea Fishery Resources Exploitation and Utilization, South China Sea Fisheries Research Institute (CAFS), Guangzhou, China
- 2College of Fisheries and Life Science, Shanghai Ocean University, Shanghai, China
- 3South China Sea Fisheries Research Institute, Chinese Academy of Fishery Sciences, Tropical Fisheries Research and Development Center, Sanya Tropical Fisheries Research Institute, Sanya, China
Glutaredoxins (Grxs) are glutathione-dependent oxidoreductases that belong to the thioredoxin (Trx) superfamily and are an essential part of the redox system in living organisms. However, there is a serious lack of sequence information and functional validation associated with Grxs in crustaceans. In this study, a new Grx gene (PmGrx5) was identified and characterized in black tiger shrimp (Penaeus monodon). The full-length cDNA of PmGrx5 is 787 bp and consists of 114 bp 5′-UTR, 232 bp 3′-UTR, and 441 bp ORF, encoding a hypothetical protein of 146 amino acids. The putative PmGrx5 protein is 16.27 kDa with a theoretical isoelectric point of 5.90. Sequence alignment showed that PmGrx5 had the highest amino acid sequence homology with Grx5 from Penaeus vannamei at 98.63% and clustered with Grx5 from other crustaceans. Quantitative real-time PCR (qRT-PCR) analysis showed that PmGrx5 was expressed in all tissues examined, with a higher expression in the testis, stomach, lymphoid organ, and gill. PmGrx5 was continuously expressed during development, with the highest expression in zoea I. Ammonia-N stress and bacterial infection both differentially upregulated PmGrx5 expression in the hepatopancreas and gill. In addition, when PmGrx5 was inhibited, the expression of some other antioxidant enzymes was upregulated at the beginning of ammonia-N stress, but as the stress time increased, the expression of antioxidant enzymes was inhibited, the expression of apoptotic genes was increased, and the GSH content was significantly reduced. Inhibition of PmGrx5 led to a greater risk of oxidative damage in shrimp. In addition, the relationship between SNPs in exons of the PmGrx5 gene and tolerance to ammonia-N stress was identified and analysed. A total of nine SNPs were successfully identified, eight of which were significantly associated with ammonia and nitrogen stress tolerance trait in shrimp (P < 0.05). The present study shows that PmGrx5 is involved in redox regulation and plays an important role in shrimp resistance to marine environmental stresses. Meanwhile, this study will provide a basis for molecular marker breeding in shrimp.
Introduction
Black tiger shrimp (Penaeus monodon) is currently the third most cultured shrimp in the world, accounting for approximately 8% of total crustacean farming production and is an important part of the mariculture economic industry (FAO, 2020). However, due to the deteriorating marine environment, especially in coastal areas, black tiger shrimp farming is often exposed to viral attacks and bacterial infections, resulting in high mortality rates and huge economic losses (Li et al., 2018). A good marine environment is a prerequisite for the sustainable development of the mariculture industry. Adverse environmental factors can upset the original physiological balance in organisms, and prolonged environmental stress can cause irreversible damage to organs and tissues and even lead to the death of organisms (Dunier and Siwicki, 1994; Chen et al., 1994; Chen and Chen, 2002). Ammonia-N is one of the most important environmental factors in mariculture, and ammonia-N stress is the most common environmental challenge faced by mariculture organisms (Liu et al., 2021). Ammonia-N stress usually causes oxidative stress in crustaceans, which disrupts redox homeostasis in crustaceans and ultimately leads to the production of excess reactive oxygen species (ROS). ROS are a class of single-electron reduction products of oxygen atoms in the body, mainly produced during mitochondrial oxidative phosphorylation and during cellular defense against exogenous compounds, which cause oxidative damage to biological macromolecules such as proteins and deoxyribonucleotides (Nathan and Cunningham-Bussel, 2013). Crustaceans can use a variety of redox systems in their bodies to eliminate excess ROS in order to protect themselves (Bu et al., 2017). Therefore, the study of redox systems is fundamental and necessary for mariculture management and shrimp breeding.
Glutaredoxins (Grxs), also known as thioltransferase, are heat-stable, small molecular weight enzymes that are an essential part of the redox system in living organisms (Holmgren et al., 2005). In the classification of proteins, Grxs are classified as a thioredoxin (Trx) superfamily (Carvalho et al., 2006). On the one hand, the members of the Trx superfamily all have a similar tertiary structure. The typical Trx structure is a centrally located four β-strands surrounded by three α-helices (Pan and Bardwell, 2006). However, the folding pattern of Grxs is subtly different from the typical Trx structure: its central four β-strands are surrounded by five α-helices (Chi et al., 2018). On the other hand, Grxs and Trxs have many of the same or similar functions in living organisms. The main role of Grxs is to use glutathione (GSH) to maintain redox homeostasis in living organisms. Glutathione (GSH) is a small molecular peptide consisting of the amino acids glutamate, cysteine, and glycine and is the main determinant of the redox state of cells. Classically, Grxs regulate the glutathionylation or deglutathionylation of proteins in an organism, thus keeping the GSH to GSSG ratio within normal limits. Because Grxs are required to recognize only the glutathione fraction of the substrate and not the substrate itself, Grxs are more flexible than Trxs in terms of substrate selection and reaction catalysis mechanism (Holmgren, 1979). Grxs were first identified in 1976 in Escherichia coli with a deficiency in Trx activity (Holmgren, 1979). Depending on the sequence of the central active sites, typical Grxs can be divided into two types: the dithiol Grxs, which contain Cys-Pro-Tyr-Cys as the catalytic sites; and the monothiol Grxs, which contain Cys-Gly-Phe-Ser as the catalytic sites and contain only one N-terminal cysteine residue. The monothiol Grxs can be further classified into two types: one type contains only a Grx structural domain and the other type contains a Trx structural domain at its N-terminal end with multiple Grx structural domains attached (Lillig et al., 2008). Notably, the second monothiol Grxs are currently only found in eukaryotes (Eklund et al., 1984; Couturier et al., 2009). The mechanism of action of the two Grxs differs somewhat, but both can use cysteine to accomplish reversible regulation of protein glutathionylation (Lillig et al., 2008).
In recent years, as studies have progressed, more other types of Grxs have been discovered. Plants are now known to possess the greatest quantities and types of Grxs, with the active site Cys-Cys-X-X being unique to plants (Verma et al., 2021). Grxs have been shown to be involved in a number of physiological processes in plants, mainly developmental regulation, stress resistance, and environmental adaptation (Xing et al., 2005; Cheng, 2008; Wang et al., 2009), and play an important role in plant growth and resistance to adverse factors. In addition, Grxs of photosynthetic organisms possess more complex structures and more diverse types. For example, a new type of complex Grx linking multiple proteins at the N-terminus has been identified in cyanobacteria (Mondal et al., 2019). The types and quantities of Grxs in animals are far less than in plants, but they still play a very crucial role. In mammals, Grxs have been reported to be involved in iron–sulfur cluster coordination, antioxidative stress, cell growth, and apoptosis (Hiroaki et al., 2003; Potamitou and Arne, 2004; Bräutigam et al., 2013). As Grxs have been found to function in a variety of human pathological processes, their development as drug targets is equally of interest (Levin et al., 2018). In aquatic organisms, Grxs have been reported to have important effects on embryonic development, brain and heart development, and innate immunity (Bräutigam et al., 2011; Bräutigam et al., 2013; Berndt et al., 2014; Omeka et al., 2019). However, few studies on Grxs in crustaceans have been reported. In this study, the full-length cDNA of Grx5 (PmGrx5) from P. monodon was cloned for the first time and analyzed for its mRNA expression during development and in different tissues. To determine whether PmGrx5 is involved in other physiological activities, we observed the expression of PmGrx5 after injection with various pathogenic bacteria and under ammonia-N stress, as well as physiological changes after PmGrx5 was disturbed. In addition, we screened for the SNP markers of PmGrx5 associated with the ammonia-N stress tolerance trait. This study aims to lay the foundation for further exploring the role of Grx5 in natural immune regulation and resistance to ammonia-N stress.
Materials and Methods
Experimental Shrimps
All shrimps used in this study were from the experimental base of the South China Sea Fisheries Research Institute in Shenzhen (Guangdong, China) and from a mixed culture pool of different families. The shrimps used for RNA interference weighed 7.01 ± 0.8 g and were 5.12 ± 0.69 cm in length, the shrimps used for the analysis of SNPs weighed 1.71 ± 0.31 g and were 2.53 ± 0.32 cm in length, while the rest of the experimental shrimps weighed 15.08 ± 1.20 g and were 12.37 ± 0.57 cm in length. We kept the shrimps in plastic cylinders filled with aerated filtered seawater under salinity- (29‰), temperature- (26°C ± 2°C), and pH-controlled (7.5–7.8) conditions for 1 week. During the temporary feeding period, we fed the shrimps once a day with compound feed (8.32% moisture, 48.65% crude protein, 5.70% crude fat, and 11.89% ash) until 24 h before treatment.
Sample Collection
To test the PmGrx5 expression in different tissues, 14 kinds of tissues, namely, the testis, ovary, brain, eyestalk nerve, thoracic nerve, abdominal nerve, epidermis, gill, heart, muscle, stomach, intestine, hepatopancreas, and lymphoid organ, were dissected from three random healthy untreated shrimps. To test the PmGrx5 expression during the developmental period, we collected samples from a total of 14 different developmental stages, from oosperm to post-larvae (Gürel, 2005). All the above samples were preserved in the RNAlater solution (Ambion, USA) at 4°C for 24 h and then moved to −80°C until used.
Ammonia-N Stress
To test the PmGrx5 expression under ammonia-N stress, first, we determined the 96-h median lethal concentration (96-h LC50) and the safe concentration (SC) through an acute ammonia-N stress pre-experiment. A total of 180 shrimps were used for the pre-experiment. In a plastic cylinder of 200 L filtered seawater (salinity 29‰, temperature 26°C ± 2°C, pH 7.5–7.8) aerated continuously using an air stone, 30 shrimps per cylinder were placed separately. The seawater was replenished daily. Six concentrations of ammonia-N, i.e., 0, 20, 40, 60, 80, and 100 mg/L, were prepared by applying NH4Cl per cylinder to seawater until the required concentration was reached. Mortality was registered every 3 h during the pre-experiment. After completion, 96-h LC50 and SC were computed by linear regression, which were 29.47 and 2.95 mg/L, respectively (Li et al., 2012; Zhou et al., 2017).
The shrimp sizes and experimental conditions of the formal experiment were similar to those of the pre-experiment. Experimental shrimps were randomly divided into three treatments (control, 96-h LC50, and SC), each having three replicates with 30 shrimps per replicate. During the experiment, no feed was supplied to the shrimps. Shrimp survival was monitored and reported every 3 h, and the dead shrimps were instantly pulled from the cylinders. At 0, 3, 6, 12, 24, 48, 72, and 96 h after ammonia-N stress, three shrimps were chosen for each duplication. Since the gill is the first barrier to the water environment and the hepatopancreas is the most important detoxification and metabolic organ of the shrimp (Rőszer, 2014; Rowley, 2016), these were chosen as representative target organs to detect the expression of PmGrx5 under ammonia-N stress. The hepatopancreas and gills of the shrimps were immediately dissected and preserved for 24 h at 4°C in the RNAlater solution (Ambion, USA) and then moved to −80°C until used.
Bacteria Challenge Experiment In Vivo
To test the PmGrx5 expression after the immune challenge, the experimental shrimps were randomly divided into four treatments (control, Staphylococcus aureus, Vibrio harveyi, and Vibrio anguillarum) with three replicates per treatment and 25 shrimps per replicate. The Key Laboratory of South China Sea Fishery Resources Extraction & Utilization supplied those three kinds of bacteria. According to a previous study, 100 μl sterile phosphate buffer solution (PBS, pH 7.4) was injected into the shrimps in the control group, and 100 μl (1.0 × 108 CFU/ml) of the corresponding bacterial solution was injected into the shrimps in the other three treatment groups, respectively (Qin et al., 2019). Each shrimp in the second abdominal segment was inserted intramuscularly. Shrimp survival was monitored and reported every 3 h, and the dead shrimps were instantly pulled from the cylinders. From each replication, three shrimps were picked at 0, 3, 6, 12, 24, 48, and 72 h after the injection. In addition, the hepatopancreas is the most important immune and phagocytic organ of the shrimp (Rőszer, 2014). In P. monodon, it has been found that after phagocytosis of pathogenic Vibrio cell fragments, hepatopancreatic cells can release bacterial antigens into the hemolymph (Alday et al., 2002). We, therefore, chose the gill and the hepatopancreas as representative target organs to detect PmGrx5 under bacterial infection. The hepatopancreas and gills of the shrimps were immediately dissected and preserved for 24 h at 4°C in the RNAlater solution (Ambion, USA) and then moved to −80°C until used.
Extraction of RNA and Synthesis of cDNA
For the quantification experiments, the total RNA of all samples obtained above was harvested according to the instructions of the HiPure Fibrous RNA Plus Kit (Megan, China). In addition, the total RNA of 14 different tissues was harvested in conjunction with the instructions of the RNeasy Mini Kit (Qiagen, Germany) for cloning the full-length cDNA of PmGrx5. The purity and quantity of total RNA were calculated by measuring the ultraviolet absorbance ratio at 260/280 nm using a NanoDrop 2000 device (Thermo Scientific, USA). Agarose gel electrophoresis (1%) was used to assess the integrity of total RNA. Total RNA for two different purposes was immediately synthesized into the corresponding cDNA according to the instructions of the Evo M-MLV RT Kit with gDNA Clean for qPCR (AG, China) and HiScript-TS 5′/3′ RACE Kit (Vazyme, China), respectively. The synthetic cDNA was preserved at −80°C until used.
Cloning the Full-Length cDNA of PmGrx5
The partial fragment of PmGrx5 was filtered and determined by the NCBI database BLAST on the basis of the cDNA library of P. monodon in our laboratory. The primers were constructed by Premier 6.0 based on the identified fragment, and the PmGrx5 open reading frame (ORF) was expanded by PCR technology. The PCR program was run in three steps: 95°C for 3 min; 35 cycles of 95°C for 15 s, 55°C for 15 s, and 72°C for 15 s; and completed in an ultimate duration of 5 min at 72°C. In 25 μl of the reaction mixture consisting of 1 μl of forward primer, 1 μl of reverse primer, 1 μl of cDNA template, 12.5 μl of double-distilled water, and 12.5 μl of 2× Taq Plus Master Mix II (Vazyme, China), PCR amplification was done.
The 3′ and 5′ end nested PCR primers were constructed by Premier 6.0 according to the PmGrx5 ORF. The first-round PCR program was run in three steps: 95°C for 3 min; 35 cycles of 95°C for 30 s, 60°C for 30 s, and 72°C for 1 min; and completed in an ultimate duration of 10 min at 72°C. In 50 μl of the reaction mixture consisting of 2 μl of a mixture of UPM long primers and short primers, 2 μl of first-round specific primer, 3 μl of cDNA template, and 43 μl of Mix Green (Tsingke, China), the first-round PCR amplification was done. The second-round PCR program was run in three steps: 95°C for 3 min; 35 cycles of 95°C for 30 s, 55°C for 30 s, and 72°C for 1 min; and completed in an ultimate duration for 10 min at 72°C. In 50 μl of the reaction mixture consisting of 2 μl of NUP, 2 μl of second-round specific primer, 3 μl of first-round PCR product, and 43 μl of Mix Green (Tsingke, China), the second-round PCR amplification was done.
All PCR products were cloned into pEASY®-T1 Cloning Vector (TransGen, China), sequencing a positive monoclonal colony. Lastly, the RACE technology obtained the full-length cDNA of PmGrx5. RuiBiotech (Guangzhou, China) was responsible for the synthesis of all primers and the sequencing of all PCR products mentioned above. All the primers above are shown in Table 1.
Bioinformatics Analysis of PmGrx5
The ORF and its corresponding amino acid sequences of PmGrx5 were predicted using the ORF Finder (https://www.ncbi.nlm.nih.gov/orffinder). The nucleotide sequences and the protein sequences were analyzed using BLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi?CMD=Web&PAGE_TYPE=BlastHome). Protein domains were predicted using SMART (http://smart.embl-heidelberg.de/). Signal peptides were predicted using the SignalP-5.0 Server (http://www.cbs.dtu.dk/services/SignalP/). The O-glycosylation sites were predicted using the YinOYang 1.2 Server (http://www.cbs.dtu.dk/services/YinOYang/). The N-glycosylation sites were predicted using the NetNGlyc 1.0 Server (http://www.cbs.dtu.dk/services/NetNGlyc/). The phosphorylation sites were predicted using the NetPhos 3.1 Server (http://www.cbs.dtu.dk/services/NetPhos/). The N-myristoylation sites were predicted using the Motif Scan (https://myhits.sib.swiss/cgi-bin/motif_scan). The protein secondary structure was predicted using SOPMA (https://npsa-prabi.ibcp.fr/cgi-bin/npsa_automat.pl?page=/NPSA/npsa_sopma.html). The protein tertiary structure was predicted using the SWISS-MODEL (https://swissmodel.expasy.org/). Molecular mass and theoretical isoelectric point were predicted using Expasy (https://web.expasy.org/compute_pi/). The nucleotide and protein sequences of Grx5 from different species were downloaded from the NCBI database (https://www.ncbi.nlm.nih.gov/). With the ClustalX 1.83 software, multiple sequence alignment was conducted. The phylogenetic tree with 1,000 bootstrap trial replicates was built by the MEGA-X software. The Grx protein interaction networks were built by the STRING database (https://cn.string-db.org/).
mRNA Expression by Quantitative Real-Time PCR Analysis
The expression levels of PmGrx5 and related genes in all samples collected in this experiment were detected by quantitative real-time PCR (qRT-PCR). The qRT-PCR primers were constructed by Premier 6.0. In P. monodon, elongation factor 1α (EF-1α), as a housekeeping gene, has been used as an internal control for all qRT-PCR experiments to normalize the cDNA quantity (Arun et al., 2002; Soonthornchai et al., 2010; Amornrat and Montip, 2010). EF-1α was used as the reference gene in this study. The PmEF-1α gene from the genomic sequence was deposited in GenBank (MG775229.1). All primers were synthesized by RuiBiotech (Guangzhou, China) and shown in Table 1. A Roche LightCycler® 480II was used for the qRT-PCR. The program was run in six steps: 95°C for 30 s, 40 cycles of 94°C for 5 s and 60°C for 30 s, 95°C for 5 s, 60°C for 1 min, 95°C for 1 min, and completed in an ultimate duration for 30 s at 50°C. In 12.5 μl of the reaction mixture consisting of 0.5 μl of forward primer, 0.5 μl of reverse primer, 1 μl of cDNA template (50 ng/μl), 6.25 μl of SYBR Green Premix Pro Taq (AG, China), and 4.25 μl of double-distilled water, PCR amplification was done. The PCR data were analyzed using the 2−ΔΔCT method. Statistical analyses were conducted using IBM SPSS Statistics 26.0. One-way ANOVA, followed by Tukey’s multiple range test, estimated the statistical differences. When P <0.05, the difference was considered significant. The test data were shown as mean ± standard deviation (SD).
Ammonia-N Stress on PmGrx5-Interfered Shrimps
The double-stranded RNA (dsRNA) for PmGrx5 (dsGrx5) and the green fluorescent protein (GFP, as a non-specific negative control) gene (dsGFP) were synthesized using the T7 RiboMAX Express RNAi kit (Promega, USA). The primers used to synthesize the dsRNA for the PmGrx5 and the GFP gene are shown in Table 1. The purity and quantity of the dsRNA were calculated by measuring the ultraviolet absorbance ratio at 260/280 nm using a NanoDrop 2000 device (Thermo Scientific, USA). Agarose gel electrophoresis (1%) was used to assess the integrity of the dsRNA. The synthetic dsRNA was preserved at −80°C until used.
First, the PmGrx5 expression was measured by qRT-PCR to determine the interference efficiency of dsGrx5. The concentration of dsRNA was diluted to 1 μg/μl using PBS buffer before the in-vivo injection. We divided the experimental shrimps into two groups of three replicates each, with five shrimps in each replicate. We injected dsGrx5 or dsGFP dilution (3 μg per gram of shrimp weight) at the second ventral segment of each shrimp. The dsGFP-injected shrimps were the control group and the dsGrx5-injected shrimps were the experimental group. Due to the importance of the hepatopancreas and the multiple previous reports on the use of the hepatopancreas as the target organ for RNA interference in crustaceans (Alday et al., 2002; Rőszer, 2014; Rowley, 2016; Zheng et al., 2018; Zheng et al., 2021; Jie et al., 2022), therefore, in this study, 24 h after injection, we selected three shrimps from each replicate and immediately collected the shrimp hepatopancreas, following the same procedure provided in the mRNA Expression by Quantitative Real-Time PCR Analysis section.
After determining that the interference efficiency of dsRNA was significant, we again divided the experimental shrimps into two groups with three replicates of 40 shrimps each. Injections were carried out as above. Twenty-four hours after the injection, the PmGrx5-interfered and dsGFP-injected shrimps were subjected to 48 h of acute ammonia-N stress using 96-h LC50. Sampling was done at 0, 3, 6, 12, 24, and 48 h post-stress. At each time point, we selected three shrimps from each replicate and immediately collected the shrimp hepatopancreas: one part was stored in RNAlater solution (Ambion, USA) at 4°C for 24 h and then moved to −80°C until used, and the other part was stored directly in liquid nitrogen until used.
Similar to the steps provided in the Extraction of RNA and Synthesis of cDNA and mRNA Expression by Quantitative Real-Time PCR Analysis sections, the same procedures were followed to analyze the relative expression levels of various related genes such as PmTrx, PmPrx1, PmCAT, PmCYC, and PmIAP in PmGrx5-interfered shrimps and dsGFP-injected shrimps. The specific primers for the above genes are shown in Table 1. As Grx5 is a glutathione-dependent oxidoreductase, samples stored directly in liquid nitrogen were assayed using reduced glutathione (GSH) assay kit (Nanjing Jiancheng Bioengineering Institute, China). Three replicates of each sample were assayed and statistically analyzed according to the kit’s instructions.
Correlation Analysis Between the SNPs of PmGrx5 and Ammonia-N Stress Tolerance Trait
Seven hundred healthy juvenile shrimps were selected for the ammonia-N stress experiment (using the same method provided in the Ammonia-N Stress section). The first 70 shrimps to die were considered to be the sensitive group and the last 70 shrimps to die and those still alive were considered to be the resistant group. Samples were stored in ethanol and used to extract the DNA. The DNA was extracted from the samples using the MagPure Tissue/Blood DNA LQ Kit (Megan, China).
The NCBI database was used to predict the exonic region of PmGrx5. Based on the results, Premier 6.0 was used to design the primers that span the exonic regions to amplify the DNA sequence of the samples. The PCR reaction system consisted of the following: 1 µl of forward primer, 1 µl of reverse primer, 2 µl of DNA template, and 21 µl of Mix Green (Tsingke, China). The PCR procedure was as follows: firstly, 98°C for 2 min, followed by 35 cycles of 98°C for 10 s, 55°C for 15 s, and 72°C for 1 min, and finally 72°C for 10 min. Tsingke Biotechnology was responsible for the synthesis of all the above primers and the sequencing of the PCR products (Guangzhou, China), and the above primers are shown in Table 1.
The DNA sequences of each sample were examined using DNAMAN and Chromas to identify the SNPs of PmGrx5. Genetic parameters were calculated using WPS Office, including observed heterozygosity (Ho), expected heterozygosity (He), effective allele number (Ne), minimum allele frequency (MAF), polymorphism information content (PIC), and Hardy–Weinberg equilibrium (HWE). The χ2 test using SPSS 26.0 was used to analyze the correlation between the SNPs of PmGrx5 and ammonia-N stress tolerance trait, and P <0.05 was considered to be a significant difference.
Results
Bioinformatics Results for PmGrx5
As seen in Figure 1A, the full-length cDNA of PmGrx5 (GenBank accession number: ON086315) was 787 bp, which included 114 bp 5′-UTR, 232 bp 3′-UTR, a poly-A tail, and 441 bp ORF, encoding a 146 amino acid putative protein. The putative PmGrx5 protein was 16.27 kDa with a theoretical isoelectric point of 5.90. The putative PmGrx5 protein contained only one Grx structural domain. The active center sequence of the PmGrx5 domain was C-G-F-S (54–57 aa), which was a typical characteristic of the currently known Grx5 proteins. These features suggested that PmGrx5 was a new member of the monothiol Grxs. According to the putative secondary structure, the proportions of α-helix, extended strand, β-turn, and random coil were 45.21%, 8.90%, 8.90%, and 36.99%, respectively. The putative tertiary structure of PmGrx5 is shown in Figure 1B.
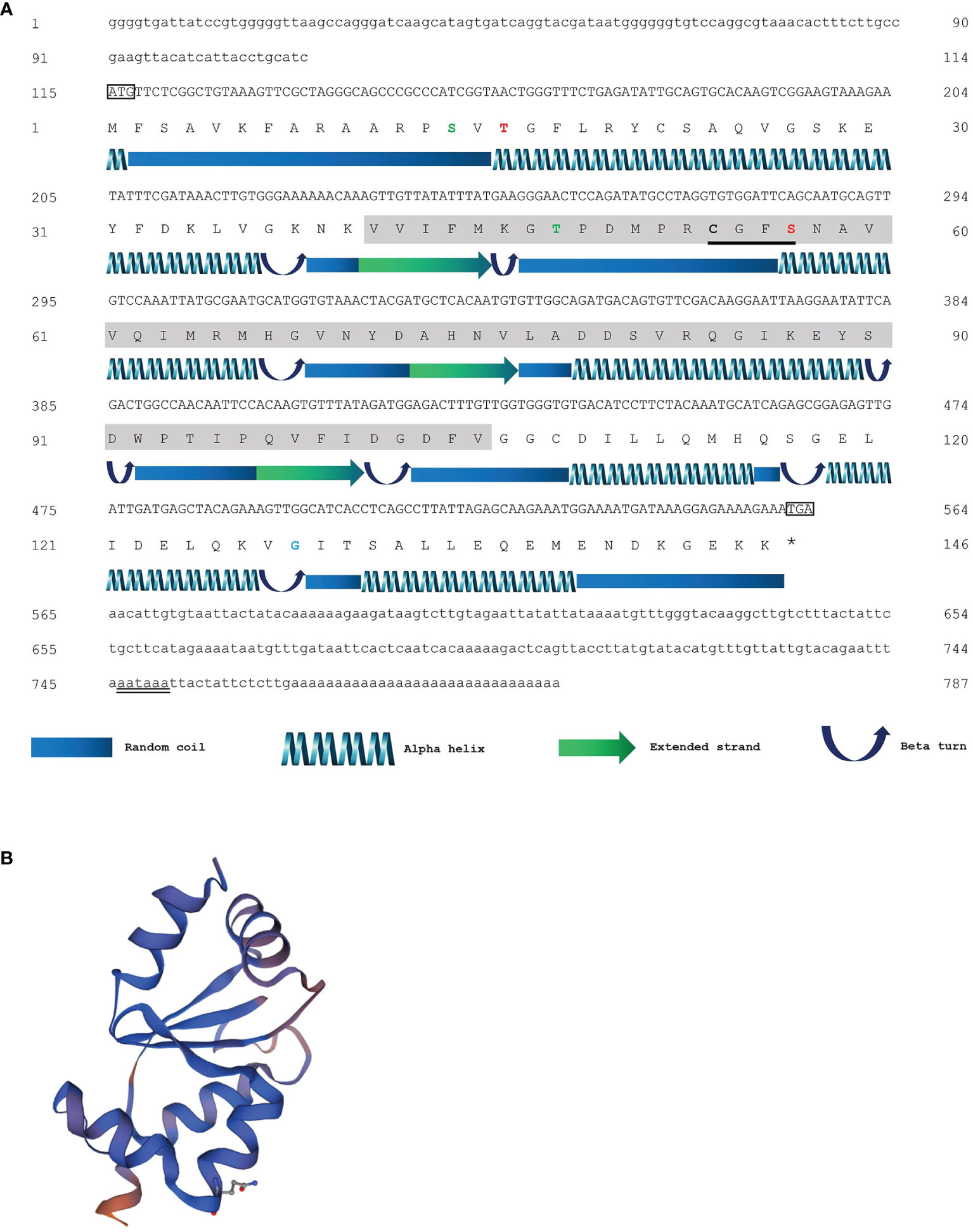
Figure 1 (A) The sequence of nucleotides, the deduced sequence of amino acids, and the putative secondary structure of PmGrx5. The initiation codon “ATG” and the termination codon “TAA” are marked in the rectangular box. The polyadenylation signal sequence (AATAAA) is marked using a double underline. The gray region indicates the predicted glutaredoxin (Grx) structural domain. The active center sequence (C-G-F-S) of the Grx domains is marked using a single underline. Cys residues are marked in bold. The phosphorylation sites, glycosylation sites, and N-myristoylation sites are highlighted in green, red, and blue, respectively. The legend of the predicted secondary structure is annotated at the bottom of the image. (B) The putative tertiary structure of PmGrx5.
As shown in Figure 2A, the Grx5 of eight arthropod species was selected for comparison with PmGrx5. The results of the multiple sequence alignment showed that in the Grx5 of all the selected species, their amino acid sequences showed a high degree of similarity. In terms of basic structure and function, their Grx5 all contained a similar Grx structural domain, and the signature active center site, C-G-F-S, was highly conserved. In addition, PmGrx5 has the highest homology to Grx5 in crustaceans. In particular, in Penaeus, PmGrx5 has 98.63% and 94.52% homology with the Grx5 of Penaeus vannamei and Penaeus japonicus, respectively. PmGrx5 has the lowest homology with the Grx5 of Armadillidium vulgare at 65.47%. In Chionoecetes opilio, Homarus americanus, Amphibalanus amphitrite, Portunus trituberculatus and Limulus polyphemus, the homology of Grx5 to PmGrx5 was 77.69%, 75.17%, 73.60%, 71.22%, and 68.99% in this order.
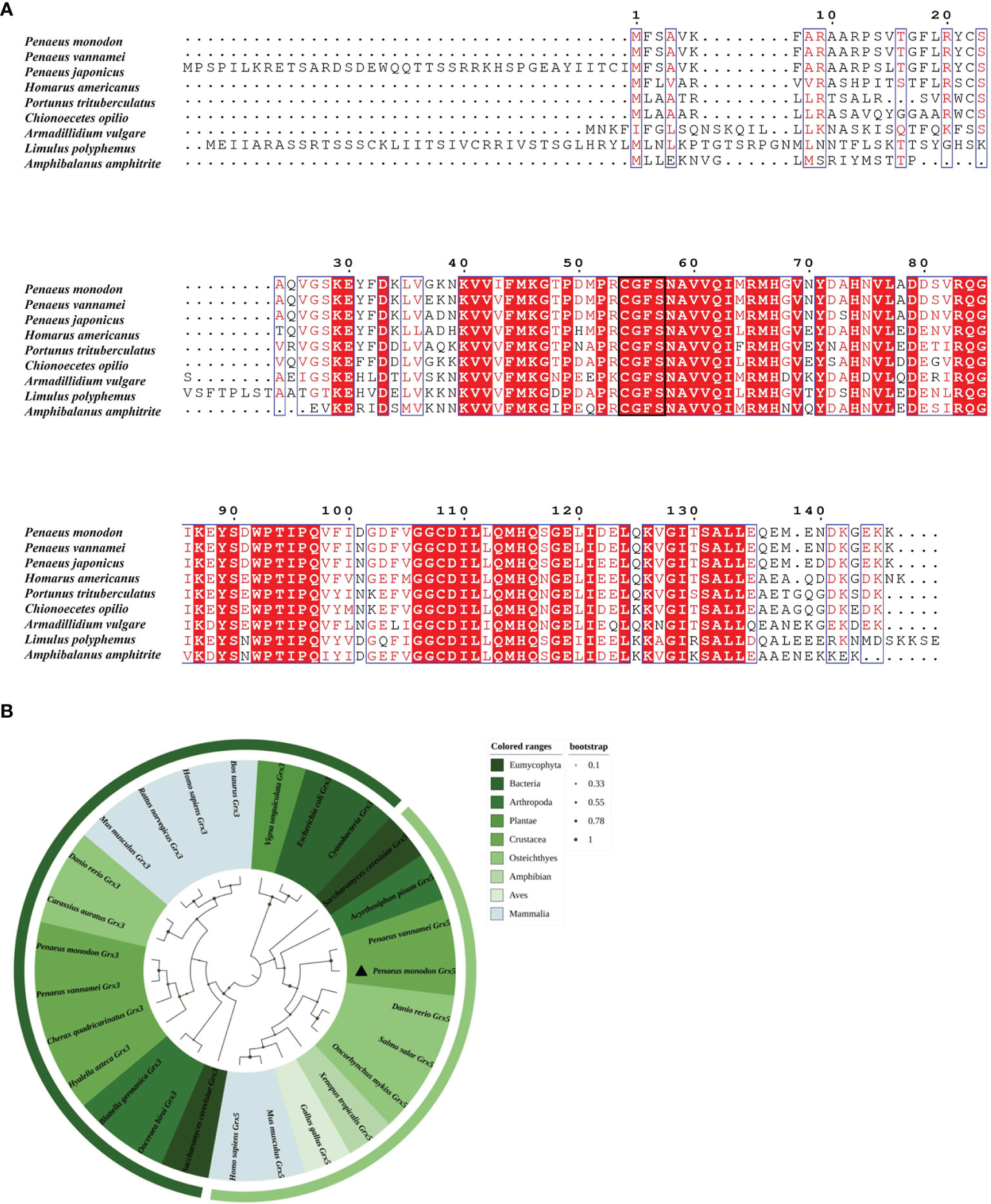
Figure 2 (A) Multiple sequence alignment of Grx5 from nine arthropod species. Red regions indicate conserved amino acid residues. Red letters indicate similar residues. Conserved active sites C-G-F-S are highlighted in a black box. The results of the amino acid sequence counts are shown at the top of each row. (B) A phylogenetic tree of mono-mercapto Grxs of various species based on amino acid sequences. Bootstrap support values corresponding to each branch are indicated by the dark green dots. The dimensions and corresponding values of the dark green dots are indicated on the right side of the picture. The corresponding species in different colors are shown on the right side of the picture. The dark green outermost arcs indicate the branches of the Grx3 protein. The light green outermost arc indicates the branch of the Grx5 protein. PmGrx5 is marked with a triangle. The scientific names of all the above species and their NCBI sequence numbers are shown below: Amphibalanus amphitrite (KAF0307822.1), Acyrthosiphon pisum (AK341132.1), Armadillidium vulgare (RXG69803.1), Blattella germanica (PSN36174.1), Bos taurus (NP_001030273.1), Carassius auratus (XP_026078075.1), Chionoecetes opilio (KAG0717517.1), Cherax quadricarinatus (AEL23128.1), Cyanobacteria (WP_190627512.1), Danio rerio (NP_001005950.1), Danio rerio (BC059659.1), Escherichia coli (SYX47894.1), Gallus gallus (AJ720261.1), Homarus americanus (XP_042230596.1), Hyalella azteca (KAA0195653.1), Homo sapiens (AAH05289.1), Homo sapiens (AB223038.1), Limulus polyphemus (XP_013786644.1), Mus musculus (NP_075629.2), Mus musculus (AK013761.1), Ooceraea biroi (EZA62809.1), Oncorhynchus mykiss (BT073697.1), Penaeus monodon (MZ827442), Penaeus japonicus (XP_042868605.1), Portunus trituberculatus (MPC08028.1), Penaeus vannamei (XP_027209771.1), Penaeus vannamei (XP_027214062.1), Penaeus vannamei (XM_027358261.1), Rattus norvegicus (AAH86381.1), Saccharomyces cerevisiae (GFP69319.1), Saccharomyces cerevisiae (NM_001183873.1), Salmo salar (BT048738.1), Vigna unguiculata (QCE13504.1), and Xenopus tropicalis (BC075374.1).
Grx amino acid sequences from 31 different species, including Grx3 and Grx5, were selected using the minimal evolution method of MEGA-X to construct a Grx phylogenetic tree. As shown in Figure 2B, both Grx3 and Grx5 were independently aggregated. Grx3 and Grx5 of most species had one to multiple Grx structural domains and the active center site was C-G-F-S, which are typical monothiol Grxs. However, the Grx3 domains of prokaryotes and plants were clustered separately and did not have the active center site C-G-F-S, showing significant differences and signaling evolutionary distancing and separation. The phylogenetic tree showed that vertebrates, including mammals, fish, and amphibians, have Grx3 and Grx5 clustered separately. The same was true for invertebrates and fungi, including Saccharomyces cerevisiae, crustaceans, and insects, and Grx3 and Grx5 from fungi were closer to the invertebrate group. Of these, PmGrx3 was the closest phylogenetically to Grx3 from P. vannamei and clustered with Grx3 from other crustaceans. The same was observed in the phylogeny of Grx5.
The amino acid sequence of PmGrx5 was used to construct a protein interaction network based on the STRING database. As the crustacean Grx was rarely studied, the human genome was chosen as the template for the construction. As shown in Figure 3, the main proteins predicted to interact with Grx5 were BolA-like protein 1 (BOLA1), BolA-like protein 3 (BOLA3), frataxin (FXN), glutaredoxin 2 (Grx2), iron–sulfur cluster co-chaperone protein (HSCB), iron–sulfur cluster assembly factor (IBA57), iron–sulfur cluster assembly 1 (ISCA1), iron–sulfur cluster assembly 2 (ISCA2), iron–sulfur cluster assembly enzyme (ISCU), and iron–sulfur cluster scaffold protein (NFU1). The main function of these proteins was to regulate the iron–sulfur cluster and redox status in the organism.
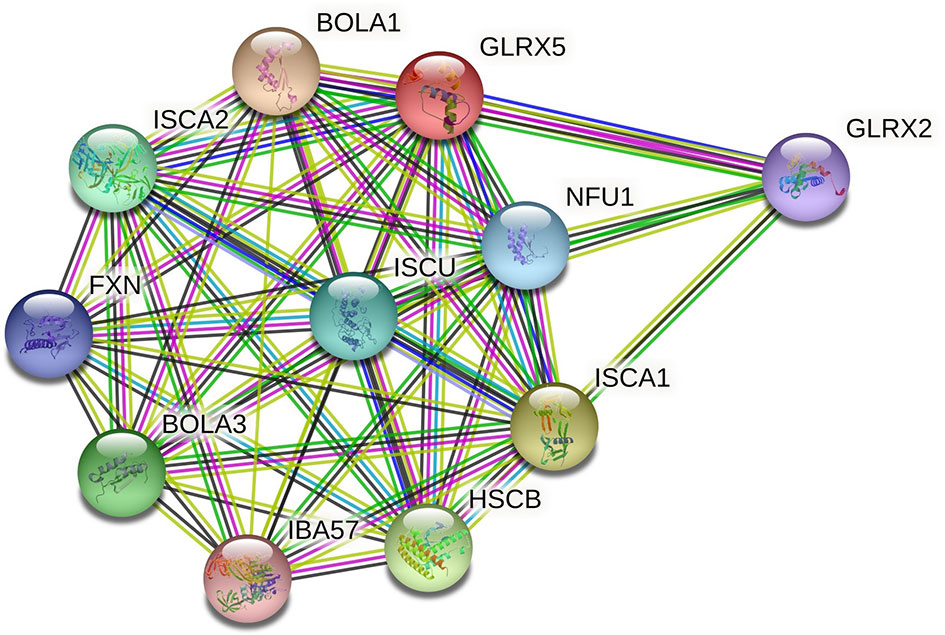
Figure 3 Grx5 protein interaction networks. The red dots represent the target protein Grx5. Other colored dots represent proteins that interact with Grx5. The names of all proteins are abbreviated to the top right of the dots.
mRNA Expression of PmGrx5 in Different Tissues
As shown in Figure 4, qRT-PCR results showed that PmGrx5 was expressed in all tissues examined. PmGrx5 expression was higher in the testis, stomach, lymphoid organ, and gill (P < 0.05). PmGrx5 expression was lower in the eyestalk nerves, brain, abdominal nerves, and ovary.
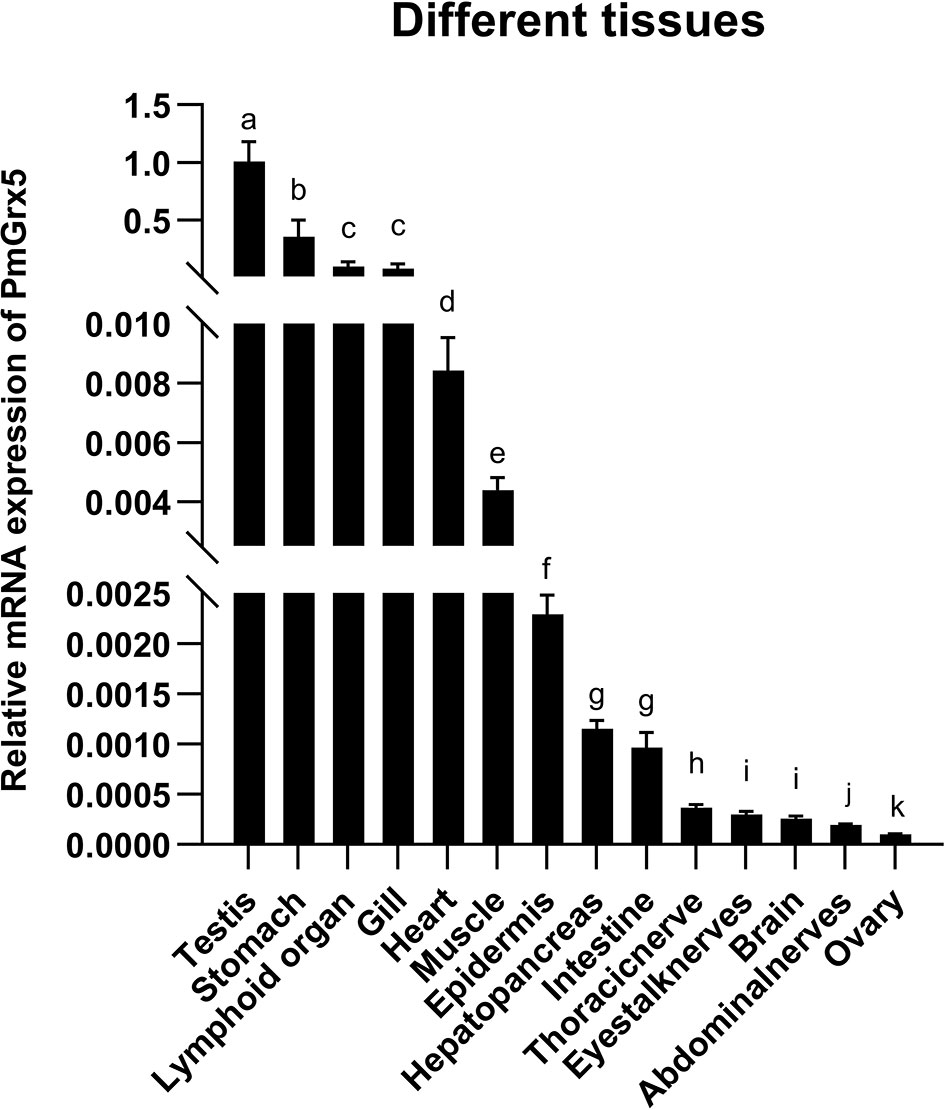
Figure 4 mRNA expression levels of PmGrx5 in different tissues. The data are presented as mean ± SD (n = 3). Different letters denote significant differences (P < 0.05).
mRNA Expression of PmGrx5 During the Developmental Period
The qRT-PCR results showed that PmGrx5 was expressed with some regularity in all early developmental stages in black tiger shrimp (Figure 5). The expression of PmGrx5 increased rapidly when development entered nauplius I. Throughout the nauplius stage, the expression of PmGrx5 showed a gradual decrease and increase and reached a peak at zoea I, which was also the highest expression value throughout the early developmental stages (P < 0.05). From mysis II until the development of post-larva, the expression of PmGrx5 showed a similar trend to the previous stage and finally peaked again at post-larva.
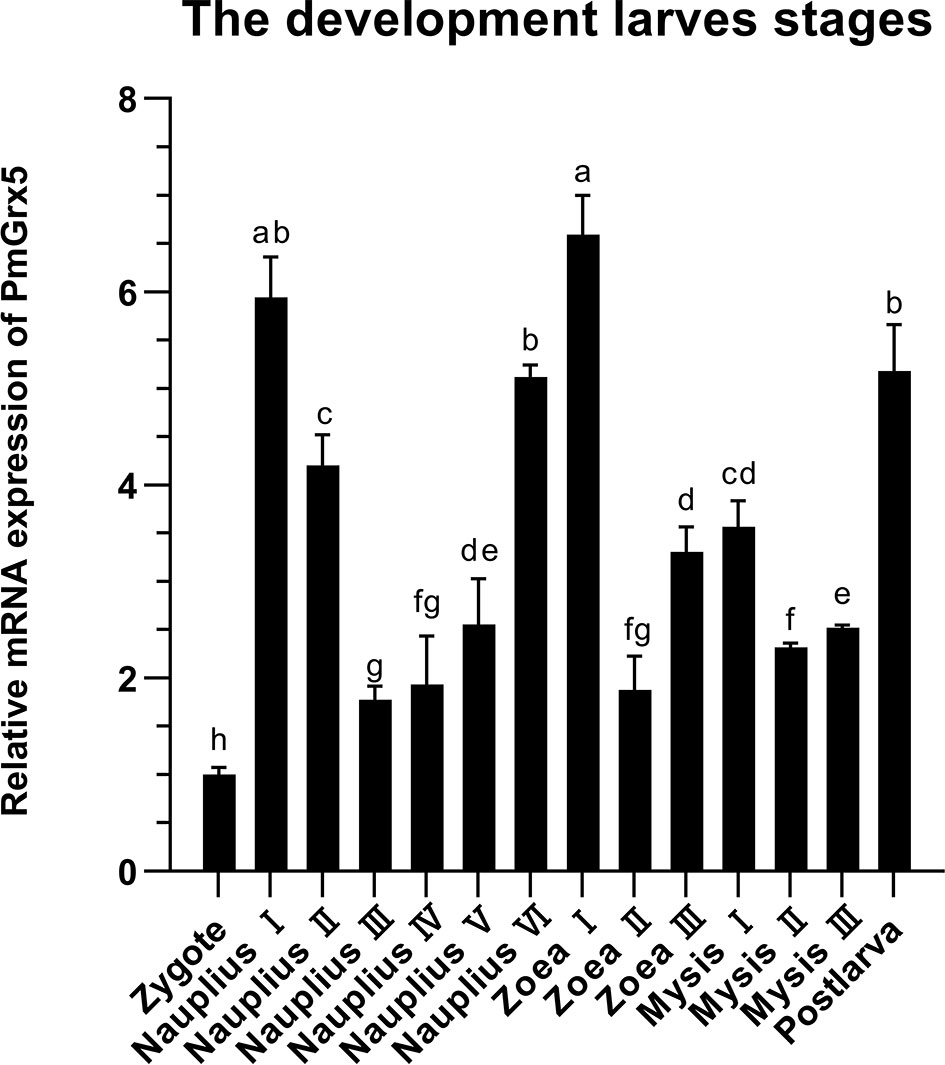
Figure 5 mRNA expression levels of PmGrx5 during the developmental period. The data are presented as mean ± SD (n = 3). Different letters denote significant differences (P < 0.05).
Analysis of PmGrx5 Transcription for the Hepatopancreas and Gill After Bacterial Challenge
Figure 6 shows the qRT-PCR results for the PmGrx3 expression in the hepatopancreas and gill at different time intervals after multiple pathogenic bacteria injections. In the hepatopancreas, the PmGrx5 expression was highly significantly upregulated 6 h after S. aureus injection compared to the control (P < 0.05). A peak was reached 72 h after injection (P < 0.05). The PmGrx5 expression was significantly upregulated, then significantly downregulated, and finally significantly upregulated again within 72 h after V. harveyi injection (P < 0.05). Within 72 h of V. anguillarum injection, the PmGrx5 expression was only significantly upregulated at 3 h post-injection compared to the control group (P < 0.05), with no significant changes at other time points. In the gill, the PmGrx5 expression was highly significantly upregulated (P < 0.05) at 3 h after S. aureus injection compared to the control, then plateaued and was significantly upregulated again after 24 h of injection (P < 0.05). The expression pattern of PmGrx5 in the gill was similar to that in the hepatopancreas within 72 h after V. harveyi or V. anguillarum injection.
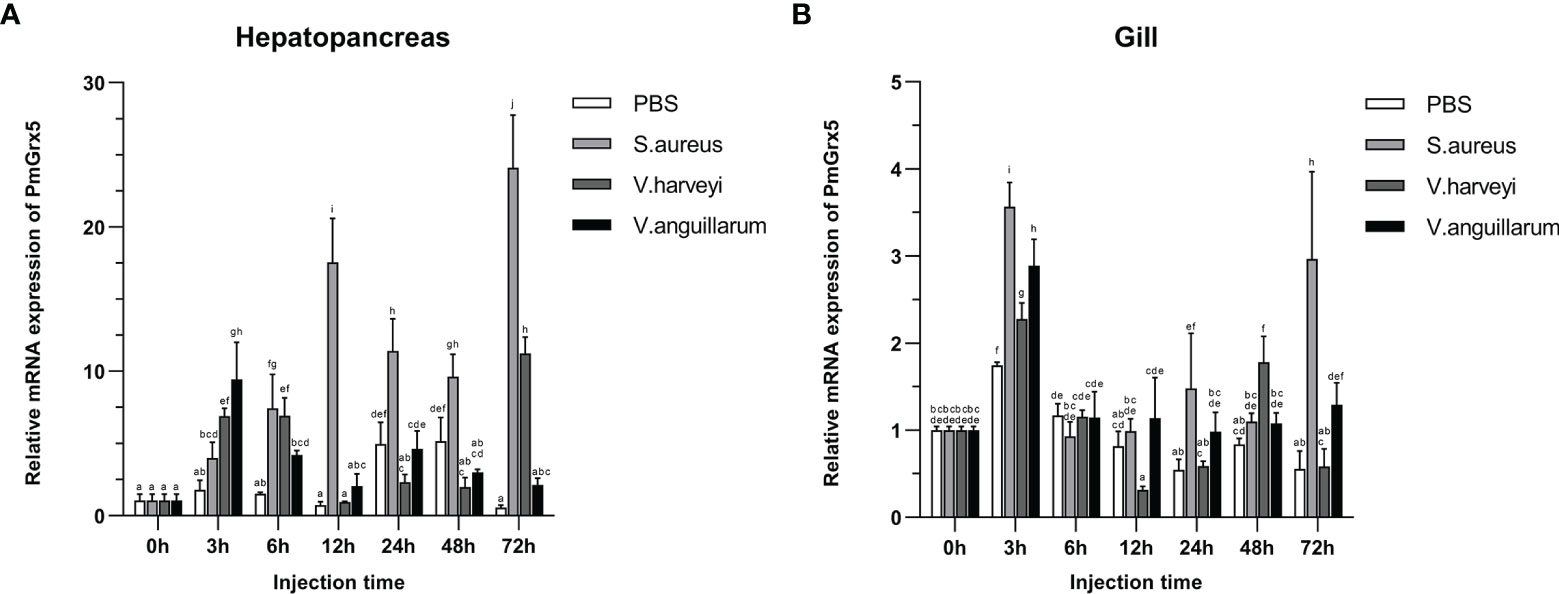
Figure 6 mRNA expression levels of PmGrx5 in the hepatopancreas (A) and gill (B) at different time intervals after multiple pathogenic bacteria injections. The data are presented as mean ± SD (n = 3). Different letters denote significant differences (P < 0.05).
Analysis of PmGrx5 Transcription for Hepatopancreas and Gill Under Ammonia-N Stress
Figure 7 shows the qRT-PCR results of PmGrx5 expression in the hepatopancreas and gill at different time intervals under different levels of ammonia-N stress. From the results, it can be seen that the expression of PmGrx5 in the hepatopancreas and gill had a similar pattern at 96 h SC, with a gradual increase followed by a decrease over 96 h after ammonia stress. PmGrx5 peaked at 12 h in the hepatopancreas and at 24 h in the gill, and both produced a fluctuation in elevated expression at 72 h (P < 0.05). At 96-h LC50, the trend in PmGrx5 expression in the hepatopancreas was similar to that at 96-h SC concentrations, except that the response was not as strong as in the former. In the gill, however, the situation was completely different. At 96-h LC50, the expression of PmGrx5 was not significantly higher in the gill compared to the control and was even consistently lower than in the control and the 96-h SC group.
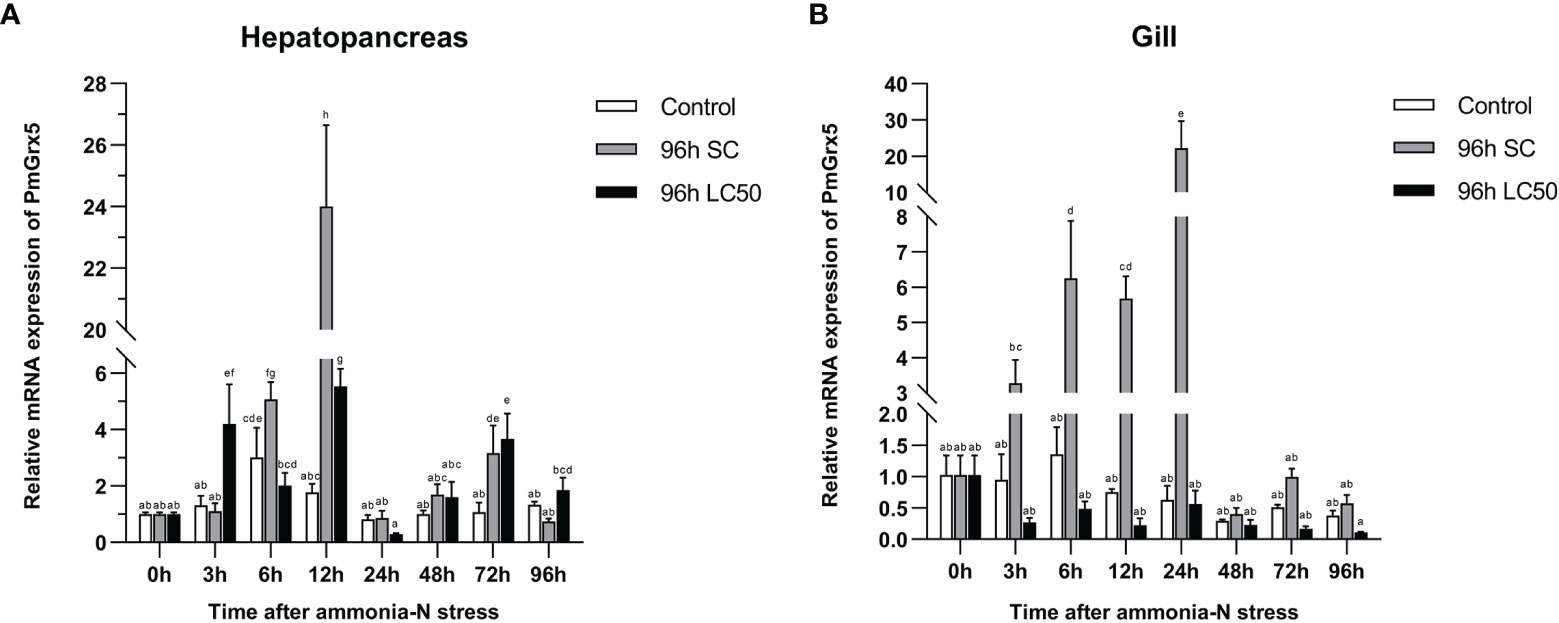
Figure 7 mRNA expression levels of PmGrx5 in the hepatopancreas (A) and gill (B) at different time intervals under different concentrations of ammonia-N stress. The data are presented as mean ± SD (n = 3). Different letters denote significant differences (P < 0.05).
The Interference Efficiency of dsGrx5 in the Hepatopancreas
As shown in Figure 8A, dsGrx5 could significantly inhibit the PmGrx5 expression in the hepatopancreas (P < 0.05), satisfying the requirements of this experiment. As shown in Figure 8B, ammonia-N stress was applied to all experimental groups 24 h after dsRNA injection. The PmGrx5 expression in the dsGFP-injected group was significantly higher than that in the dsGrx5-injected group within 48 h after ammonia-N stress (P < 0.05), which demonstrated that dsGrx5 could significantly exert its inhibitory effect within 72 h after injection.
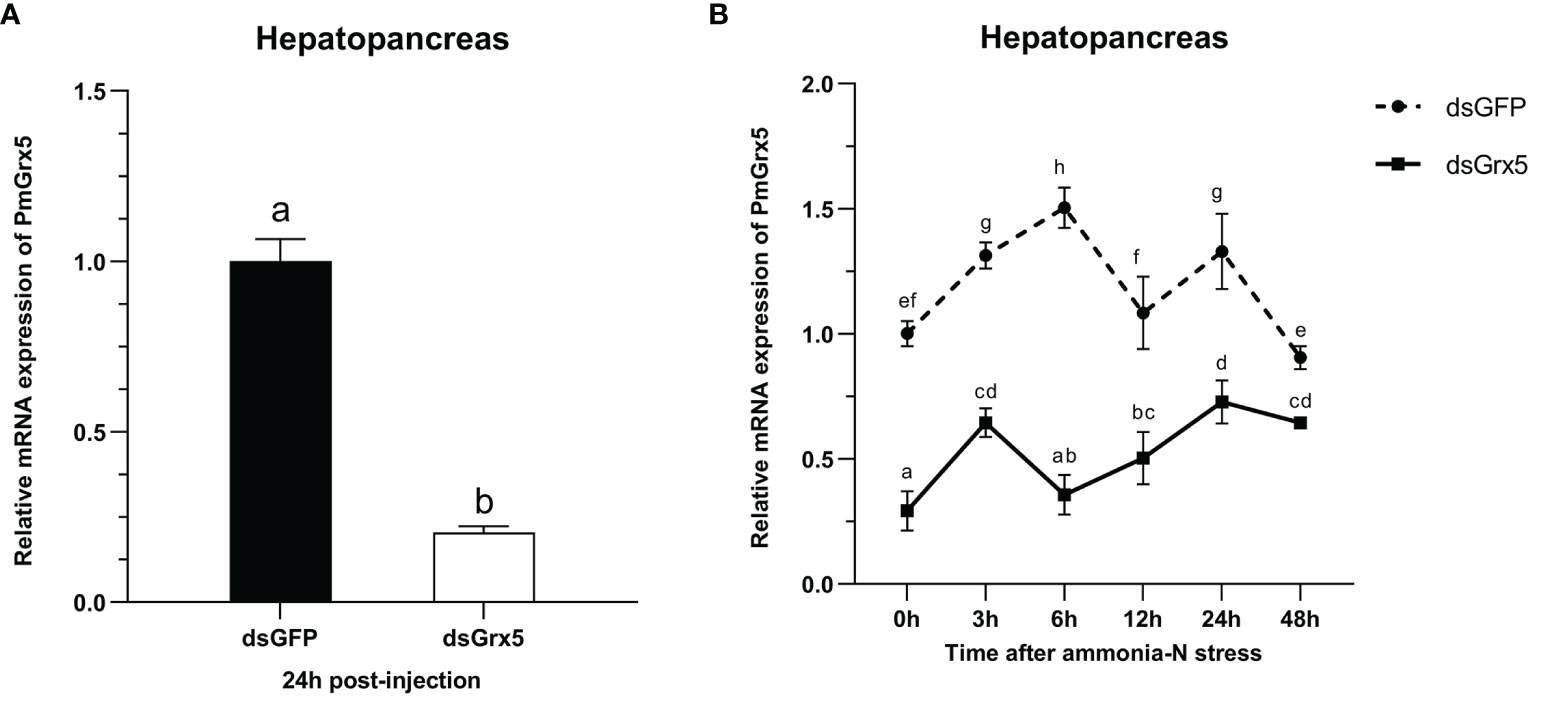
Figure 8 (A) mRNA expression levels of PmGrx5 in the hepatopancreas at 24 h after dsRNA injection. (B) Expression profile of PmGrx5 in the hepatopancreas of dsRNA-injected shrimps within 48 h under ammonia-N stress. The data are presented as mean ± SD (n = 3). Different letters denote significant differences (P < 0.05).
mRNA Expression of Genes in the Hepatopancreas of dsRNA-Injected Shrimps Under Ammonia-N Stress
As shown in Figure 9A, the PmTrx expression in the dsGFP-injected group was gradually upregulated and then downregulated within 48 h under ammonia-N stress, reaching a peak at 12 h (P < 0.05). Compared with the control group, the PmTrx expression in the PmGrx5-interfered group showed a similar trend, but its response was stronger, showing significant up- or downregulation (P < 0.05).
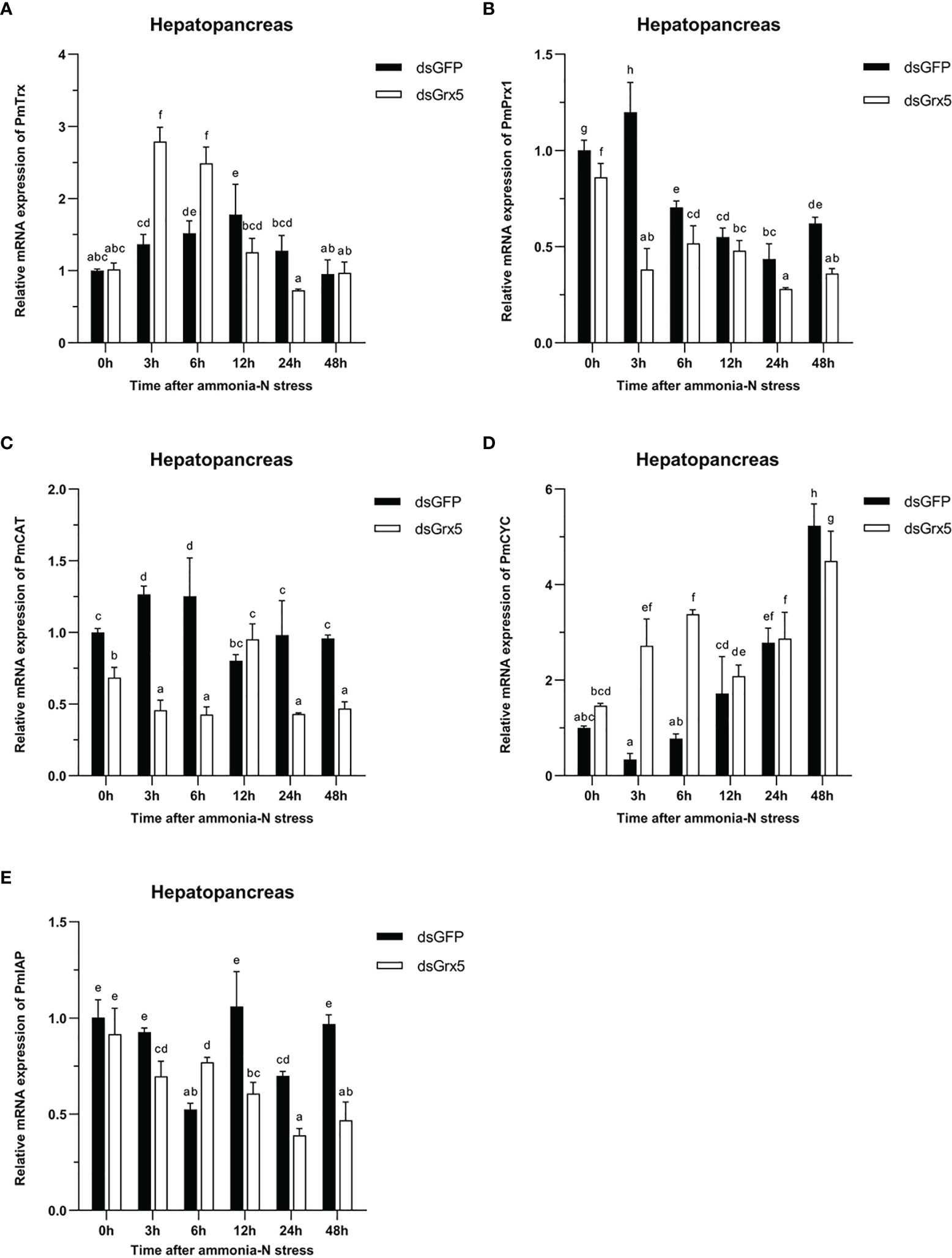
Figure 9 Expression profiles of five genes [PmTrx (A), PmPrx1 (B), PmCAT (C), PmCYC (D), and PmIAP (E)] in the hepatopancreas of dsRNA-injected shrimps under ammonia-N stress. The data are presented as mean ± SD (n = 3). Different letters denote significant differences (P < 0.05).
As shown in Figure 9B, the PmPrx1 expression in the dsGFP-injected group was significantly higher at 3 h after ammonia-N stress (P < 0.05), then gradually decreased and finally significantly upregulated again at 48 h after ammonia-N stress (P < 0.05). The expression trend of PmPrx1 in the PmGrx5-interfered group was similar to that in the control group from 3 to 48 h after ammonia-N stress. However, the expression level of PmPrx1 in the PmGrx5-interfered group was significantly lower than that in the control group at all time points (P < 0.05), except at 12 h after ammonia-N stress, when it was also lower but not significant.
As shown in Figure 9C, the PmCAT expression in the dsGFP-injected group was significantly increased within 6 h under ammonia-N stress (P < 0.05). Its expression decreased significantly at 12 h after ammonia-N stress (P < 0.05) and then stabilized. The PmCAT expression in the PmGrx5-interfered group showed significant upregulation only at 12 h after ammonia nitrogen stress (P < 0.05). At the rest of the time points, its expression level was always significantly lower than the initial level and that in the control (P < 0.05).
As shown in Figure 9D, the PmCYC expression in the dsGFP-injected group gradually increased 3 h after ammonia-N stress and reached a maximum at 48 h (P < 0.05). The PmCYC expression in the PmGrx5-interfered group was first significantly upregulated within 48 h after ammonia-N stress and significantly decreased at 12 h, then gradually increased again, also reaching a peak at 48 h (P < 0.05). The PmCYC expression in the PmGrx5-interfered group was significantly higher than that in the control group at 3–6 h after ammonia-N stress (P < 0.05). The expression pattern of PmCYC in the control and experimental groups was similar during the 12–48-h period after ammonia-N stress.
As shown in Figure 9E, the PmIAP expression in the dsGFP-injected group was significantly reduced only at 6 and 24 h after ammonia-N stress (P < 0.05). In contrast, the PmIAP expression level in the PmGrx5-interfered group was gradually reduced overall, accompanied by non-significant fluctuations. The PmIAP expression level in the PmGrx5-interfered group was significantly lower than that in the control group except for 6 h after ammonia-N stress (P < 0.05).
GSH Content in the Hepatopancreas of dsRNA-Injected Shrimps Under Ammonia-N Stress
As shown in Figure 10, GSH content in the hepatopancreas of dsGFP-injected shrimps remained essentially flat with no significant changes within 24 h after ammonia-N stress and finally decreased significantly at 48 h (P < 0.05). The trend in GSH content in the hepatopancreas of PmGrx5-interfered shrimps was similar to that of dsGFP-injected shrimps, but it was always significantly lower than that of dsGFP-injected shrimps (P < 0.05).
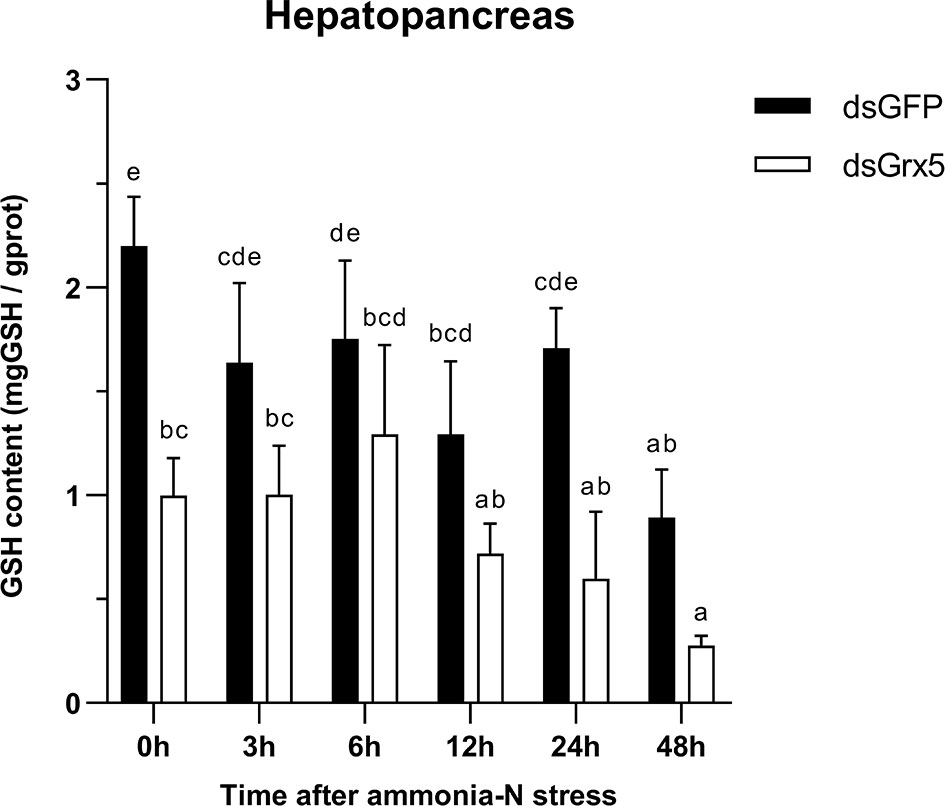
Figure 10 Changes in GSH content in the hepatopancreas of dsRNA-injected shrimps under ammonia-N stress. The data are presented as mean ± SD (n = 3). Different letters denote significant differences (P < 0.05).
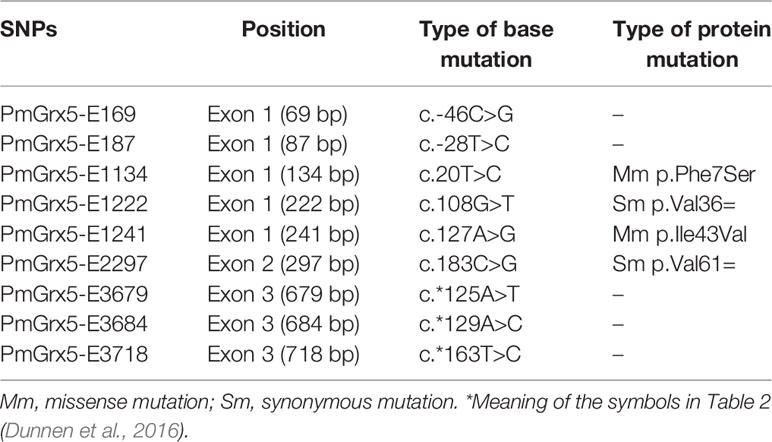
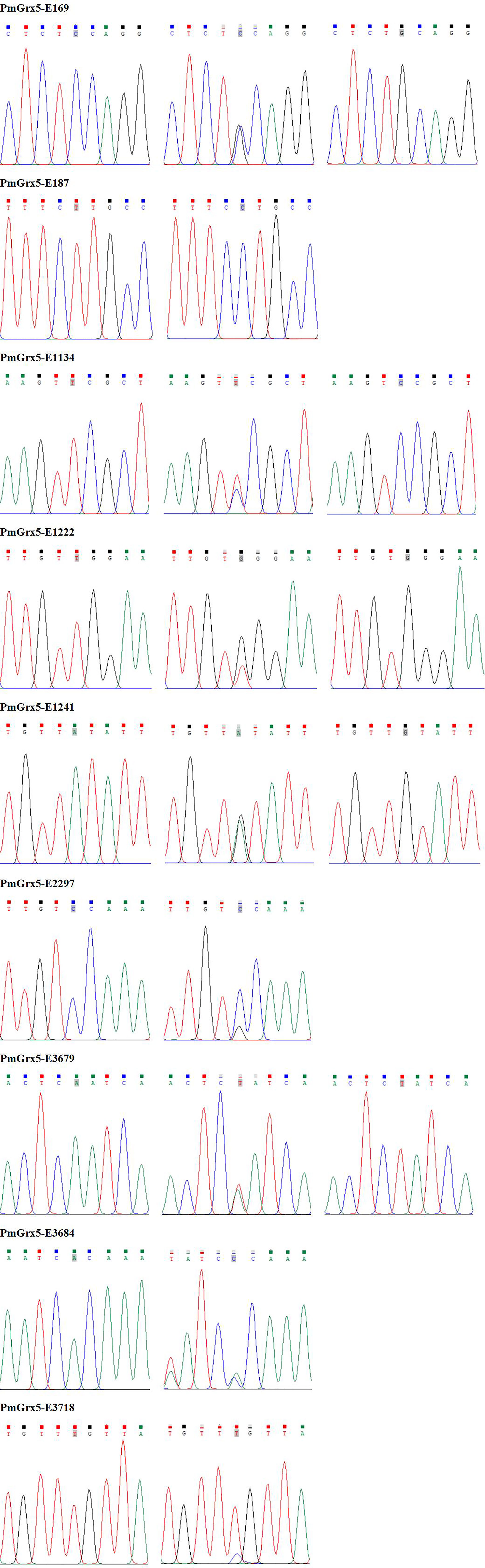
Correlation Analysis Between the SNPs of PmGrx5 and Ammonia-N Stress Tolerance Trait
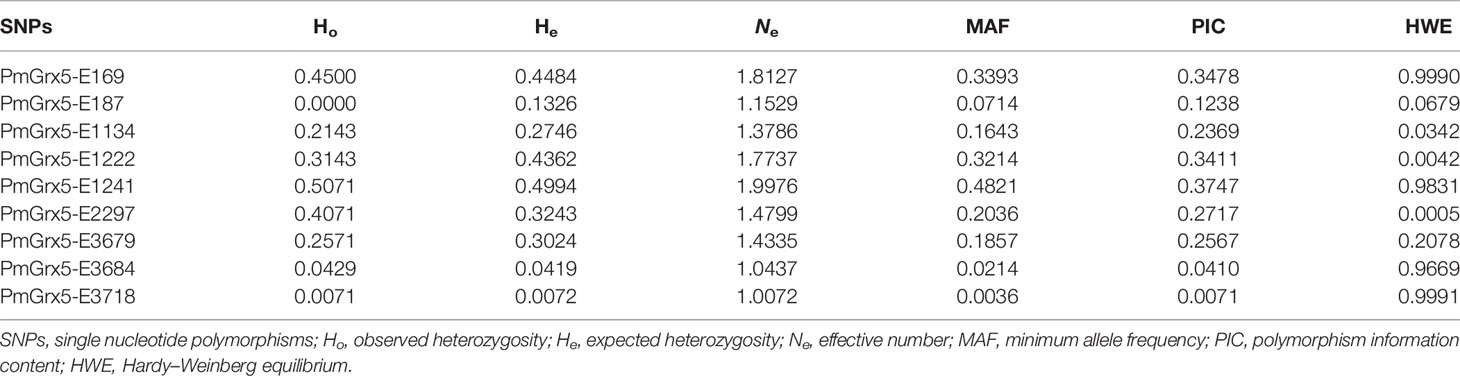
In this study, a total of nine SNPs were identified on the exons of PmGrx5. The specific information of the SNPs of PmGrx5 is shown in Table 2. Sequencing maps of the SNPs of PmGrx5 are shown in Figure 11. Analysis of the polymorphic parameters (Table 3) showed that Ho for the nine SNPs ranged from 0.0000 to 0.4500, He from 0.0072 to 0.4994, Ne from 1.0072 to 1.9976, and MAF from 0.0036 to 0.4821. PmGrx5-E187, PmGrx5-E1134, PmGrx5-E3684, and PmGrx5-E3718 showed low polymorphisms (PIC < 0.2500), and the remaining five SNPs showed moderate polymorphisms (0.2500 < PIC < 0.5000). HWE results showed that PmGrx5-E1134 significantly (P < 0.05) deviated from HWE, PmGrx5-E1222 and PmGrx5-E2297 were highly significant (P < 0.01) deviating from HWE, and the remaining six SNPs were in HWE.
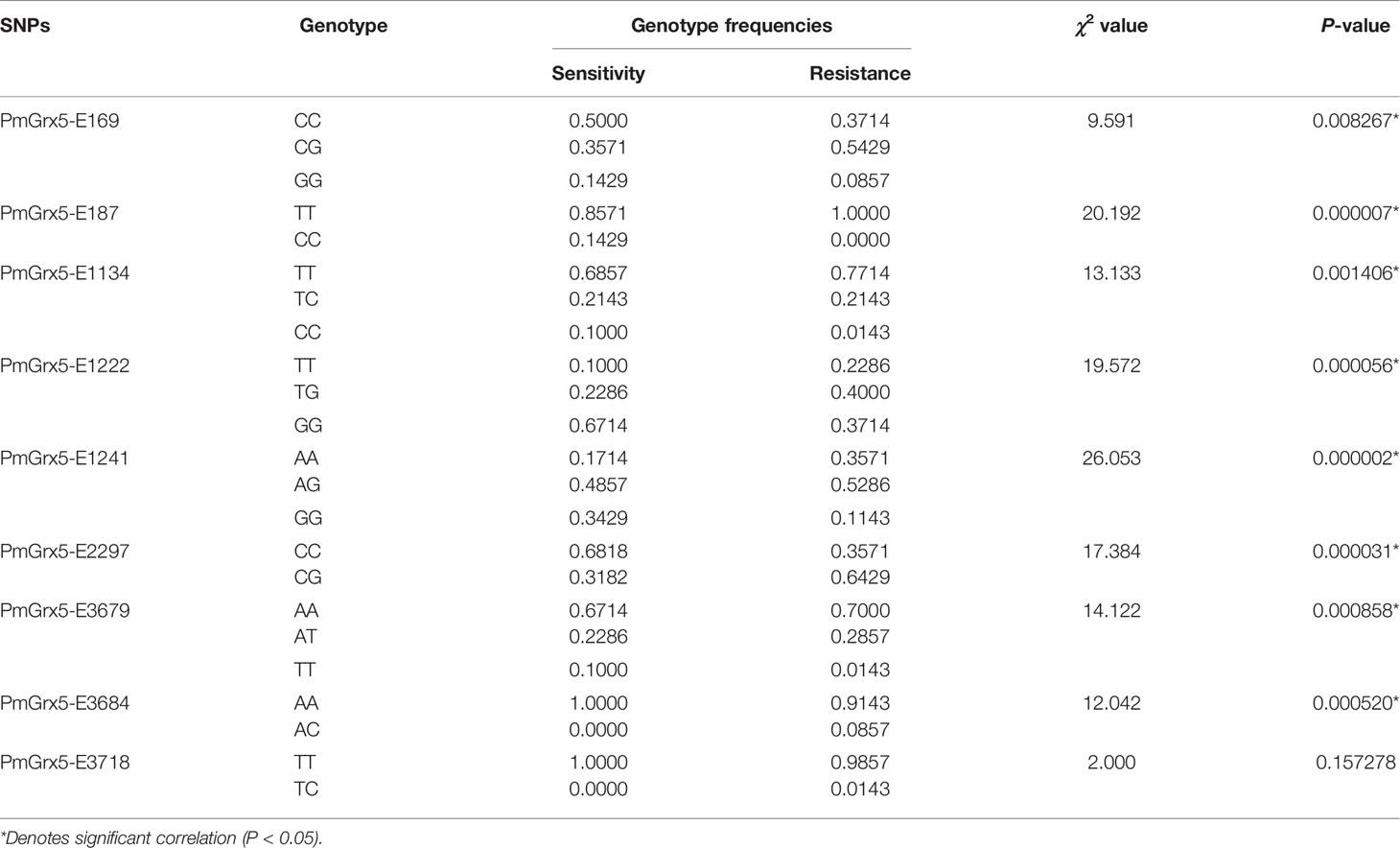
Correlation analysis between the SNPs of PmGrx5 and ammonia-N stress tolerance trait is shown in Table 4. Except for PmGrx5-E3718, which was not significantly correlated with the ammonia-N stress tolerance trait, the allele distribution of the remaining SNPs was significantly different between the sensitive and resistant groups (P < 0.05).
Discussion
Grx5, a small molecule of monothiol Grx, has been previously studied in bacteria, yeast, Arabidopsis thaliana, zebrafish, and human, mainly (Camaschella et al., 2007; Oh et al., 2012; Shakamuri et al., 2012; Mapolelo et al., 2013; Petry et al., 2018). In this study, a new full-length cDNA for Grx5, PmGrx5, was successfully cloned for the first time in crustaceans. In the predicted amino acid sequence, there is a unique Grx structural domain containing an active center sequence C-G-F-S, which is typical of the monothiol Grx domain. There are two main types of monothiol Grxs: one contains only one Grx structural domain and the other contains an N-terminal Trx structural domain and one to three Grx structural domains. PmGrx5 belongs to the former group, while PmGrx3, which is also a monothiol Grx, belongs to the latter group. The difference in the number and complexity of the structural domains in their amino acid sequences predicts their evolutionary differences and potentially different functions. The amino acid sequences of Grx5 from eight species were selected for multiple comparisons with PmGrx5. The Grx5 of all nine species was a monothiol Grx and all had the active center sequence C-G-F-S, indicating that C-G-F-S is highly conserved between species. Phylogenetic analysis showed that PmGrx5 clustered with PvGrx5 with 98.63% amino acid sequence homology, which is highly conserved and predicts that Grx5 may play a similar role in crustaceans. The main function of the protein predicted to interact with Grx5 is to regulate the iron–sulfur cluster and redox state in the organism. Iron–sulfur clusters are a class of cofactors essential to living organisms and have a variety of functions such as transferring and catalyzing electrons (Glaser and Glaser, 2020). Therefore, iron–sulfur clusters are also an important component in the regulation of the redox state of cells. It has been demonstrated in several species that monosulfhydryl glutathione with the active site CGFS possesses a good ability to bind iron–sulfur clusters (Herrero and de la Torre-Ruiz, 2007), and it is considered to be an intermediate carrier for the delivery of iron–sulfur clusters to a variety of apolipoprotein target proteins (Shakamuri et al., 2012). Furthermore, inhibition of Grx5 expression in zebrafish results in the inability to synthesize iron–sulfur clusters properly in fish cells. In contrast, injection of Grx5 from other species into zebrafish, such as yeast, mice, and human, respectively, restored the normal synthesis of iron–sulfur clusters in their cells (Wingert et al., 2005). This result suggests that, at least in the physiological processes involved in iron–sulfur cluster synthesis, the function of Grx5 is conserved across multiple species.
In this study, the tissue distribution of PmGrx5 in crustaceans was examined for the first time. PmGrx5 was expressed in all tissues, indicating a wide range of biological functions. However, the expression of PmGrx5 varied greatly in different tissues, with the highest expression in the testis, stomach, lymphoid organ, and gill. In mice, the expression of Grx3, a monothiol Grx, was also the highest in the testis (Cheng et al., 2011). Meanwhile, the expression of dithiol Grx2 was increased during human sperm development. In addition, Grx2b and Grx2c were protein isoforms specific to the human testis (Lönn et al., 2008). Although their exact mechanism and role had not been proven, Grxs were closely associated with the development of the mammalian testis and sperm, and it was hypothesized that PmGrx5 performed a similar function. The stomach, lymphoid organ, and gill were all important components of the crustacean immune system. The stomach and gill were the first barriers for crustaceans against external environmental stressors and pathogens (Rowley, 2016). Among crustaceans, the lymphoid organ, currently found only, was considered to be the main immune organ that exerts phagocytosis (Rusaini, 2010). This suggested that PmGrx5 may play an important role in the immune role played by shrimps.
PmGrx5 was expressed continuously throughout the early developmental period in P. monodon. It was easy to see from Figure 6 that the expression of PmGrx5 increased each time the shrimps developed to a new stage and that the highest expression was found in the early stage of each stage. The greatest expression of PmGrx5 was found in nauplius I and zoea I. Early development was a very complex and rapid process, and as a eukaryote that did not rely on photosynthesis, this process required mitochondrial energy supply. The iron–sulfur cluster was one of the key components of the energy production process of the mitochondrial respiratory chain (Beinert, 2000). It is inferred that as a monothiol Grx, PmGrx5 may be involved in the assembly and transfer of iron–sulfur clusters (Bandyopadhyay et al., 2008) during the early development of P. monodon, thus promoting smooth development. In mammals, the monothiol Grx has been shown to be essential for embryonic development and post-embryonic growth (Cheng et al., 2011). In plants, the Grxs17 homolog (a monothiol Grx) is also required for post-embryonic growth (Knuesting et al., 2015).
To verify whether PmGrx5 was involved in the immune response of P. monodon to bacterial infection, one Gram-positive (S. aureus) and two Gram-negative (V. harveyi and V. anguillarum) strains were selected for in-vivo injection in this study. PmGrx5 produced varying degrees of immune response to all three bacteria within 72 h of injection compared to the control group. At 3 h after injection, the expression of PmGrx5 was significantly upregulated in all three treatment groups in the gill, while the expression of PmGrx5 was significantly upregulated in the group injected with Gram-negative bacteria only in the hepatopancreas. This phenomenon suggested that PmGrx5 was more sensitive to Gram-positive bacteria in the gill. However, the relative expression of PmGrx5 in the hepatopancreas was higher than that in the gill in all three treatment groups, suggesting that PmGrx5 in the hepatopancreas played a more important role in the defense of shrimp against pathogens than in the gill, the outer barrier organ of immunity. The same phenomenon was found in Litopenaeus vannamei, where the response of LvGrx3 (a monothiol Grx) was more sensitive in the gill than in the hepatopancreas after LPS injection, but the highest relative expression of lvgrx3 was detected in the hepatopancreas (Zheng et al., 2021).
Ammonia-N is one of the most common aquatic environmental stressors. Ammonia-N in water consists of non-ionic ammonia nitrogen (NH3-N) and ionic ammonia nitrogen (-N) (Randall and Tsui, 2002). Excess ammonia-N induces the production of ROS/RNS, which exacerbates oxidative damage in the organism (Ching et al., 2009; Bu et al., 2017). Excess ammonia-N in the environment is toxic to aquatic organisms, and crustaceans are no exception. It had been shown that prolonged ammonia-N stress could lead to oxidative damage and inflammation in aquatic organisms, which in turn would lead to reduced immunity and increased susceptibility to pathogens and mortality (Cheng et al., 2015; Zhang et al., 2015; Shi et al., 2019; Liu et al., 2021). The hepatopancreas is the most important organ for ammonia and nitrogen metabolism. The gill is the first immune defense barrier for shrimp in direct contact with the aquatic environment (Shi et al., 2019). In the present study, the expression of PmGrx5 was significantly upregulated in the hepatopancreas and gill of the 96-h SC group relative to the control group during 96 h of ammonia-N stress. The 96-h LC50 group also showed significant upregulation of PmGrx5 in the hepatopancreas, but the relative expression was not as strong as that of the 96-h SC group. However, the expression of PmGrx5 in the hepatopancreas of the 96-h LC50 group was even consistently lower than that of the control and the 96-h SC group. The above results suggested that both high and low concentrations of ammonia-N induced PmGrx5 in response to ammonia-N stress in shrimp and resisted ammonia-N stress by significantly upregulating PmGrx5 expression. However, as the gill was directly exposed to the environment, a high concentration of ammonia-N may severely disrupt the normal function of the gill. Also, the response of PmGrx5 in the hepatopancreas under a high concentration of ammonia-N was much less sensitive than that under a low concentration of ammonia-N. Similar results were reported in PmChi4 and PmGrx3 in P. monodon and lvGrx2 in L. vannamei (Zhou et al., 2017; Zheng et al., 2018; Fan et al., 2021). The above results suggested that within a certain period of time, shrimps can use their own defense system to counteract the adverse effects of ammonia-N stress, but the normal functions of the gill and hepatopancreas of shrimps could be impaired under a high concentration of ammonia-N.
Furthermore, to better validate the defense mechanism of PmGrx5 in shrimp coping with ammonia stress in the marine environment, we significantly inhibited the expression of PmGrx5 using dsRNA. The results showed that dsRNA could significantly exert its disrupting effect at least 72 h after injection. We selected PmTrx and PmPrx1, which are in the same Trx superfamily as PmGrx5, and analyzed their expression after PmGrx5 expression was suppressed. Within 6 h of ammonia-N stress, the expression of PmTrx in the PmGrx5-interfered group showed significant upregulation compared to the dsGFP-injected group. It had been previously demonstrated that Grx compensated for the functional defects of E. coli Trx mutant strains, and therefore, Grx and Trx overlapped in some functions in the cell (Holmgren, 1979). Shrimps may replace the absence of PmGrx5 by upregulating PmTrx during the initial phase of ammonia-N stress. The opposite was true for PmPrx1, which was significantly less expressed in the PmGrx5-interfered group than in the dsGFP-injected group. It was hypothesized that there was a dependence of PmPrx1 on PmGrx5 and that the inability of PmPrx1 to be expressed properly exacerbated oxidative damage in the organism (Zhao and Wang, 2012). A similar situation was observed in PmCAT, where the dsGFP-injected group upregulated PmCAT (a peroxidase) after 3 h under ammonia-N stress to counteract oxidative damage, while the PmGrx5-interfered group failed to do so. CYC was an enzyme that played a crucial role in apoptosis (Hu and Yao, 2016). Initially, ammonia-N stress activated the apoptotic pathway and induced massive expression of PmCYC in the PmGrx5-interfered group. With the prolongation of ammonia-N stress, PmCYC showed a gradual upregulation trend in both groups, while PmIAP (an anti-apoptotic enzyme) was significantly inhibited in the PmGrx5-interfered group. At the same time, the GSH content was also significantly reduced. The above results suggested that when PmGrx5 was inhibited, the organism would upregulate similar function genes to protect itself under ammonia-N stress. However, as the stress time increased, the expression of antioxidant enzymes was inhibited, and the expression of apoptotic genes was increased. The inhibition of PmGrx5 led to a greater risk of oxidative damage to the shrimp.
In this study, direct sequencing was applied for the first time in PmGrx5 for locus typing, and nine SNPs were successfully obtained. Of the nine SNPs, PmGrx5-E1161, PmGrx5-E1249, and PmGrx5-E2324 deviated significantly from the Hardy–Weinberg equilibrium (P < 0.05). On the one hand, artificial selection affects allele frequencies; on the other hand, the samples in this study are extreme populations, which also have an impact on the allele frequency distribution. PmGrx5-E1134, PmGrx5-E1222, PmGrx5-E1241, and PmGrx5-E2297 are located on the ORF. PmGrx5-E1134 and PmGrx5-E1241 are missense mutations that result in mutations from phenylalanine (Phe) and isoleucine (Ile) to serine (Ser) and valine (Val). Phe, Ile, and Val are non-polar amino acids, whereas Ser is a polar amino acid. This may be one of the factors contributing to the altered tolerance to ammonia-N stress in shrimp. PmGrx5-E169, PmGrx5-E187, PmGrx5-E3679, PmGrx5-E3684, and PmGrx5-E3718 are located in the UTR. Although the UTR does not directly alter the structure and function of the protein, promoters, enhancers, and transcription factor binding sites are often located in the untranslated region and, therefore, may affect the translation efficiency or stability of the mRNA (Zhang et al., 2021). In the present study, except for PmGrx5-E3745, the remaining SNPs were significantly correlated with the strength of ammonia nitrogen stress tolerance in shrimp (P < 0.05), and the exact mode of influence needs to be further investigated.
In conclusion, the cDNA of PmGrx5 was cloned and identified as a member of the Trx superfamily, and PmGrx5 was expressed in all tissues and throughout the developmental stages of P. monodon. Both ammonia-N stress and bacterial infection significantly induced the mRNA expression level of PmGrx5 in the hepatopancreas and gill of the shrimp, suggesting that PmGrx5 plays an important role in the defense mechanism of the shrimp against marine environmental stress and pathogen infection. Inhibition of PmGrx5 expression under ammonia-N stress significantly affected the expression levels of other antioxidant enzymes and apoptotic genes, leading to a greater risk of oxidative damage in shrimp. In addition, this study identified SNPs in the exonic region of PmGrx5 and analyzed their correlation with ammonia-N stress tolerance trait in shrimp. This study will lay the foundation for further research on the role of Grx5 in crustacean innate immunity and molecular marker breeding.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, ON086315.
Ethics Statement
The animal study was reviewed and approved by the Animal Care and Use Committee of the South China Sea Fisheries Research Institute.
Author Contributions
RF, SGJ, YL, and FZ: conceptualization. RF, QY, SJ, JH, LY, and XC: methodology. RF and FZ: data curation. RF: writing—original draft preparation. FZ: writing—review and editing. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Research and Development Projects in Key Areas of Guangdong Province (2021B0202003); China Agriculture Research System of MOF and MARA (CARS‐48); Central Public-interest Scientific Institution Basal Research Fund, CAFS (No. 2020TD30); and Central Public Interest Scientific Institution Basal Research Fund, South China Sea Fisheries Research Institute, CAFS (Nos. 2020ZD01, 2021SD13).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Alday V., Roque A., Turnbull J. F. (2002). Clearing Mechanisms of Vibrio Vulnificus Biotype I in the Black Tiger Shrimp Penaeus Monodon. Dis. Aquat. Organism. 48, 91–99. doi: 10.3354/dao048091
Amornrat T., Montip T. (2010). Boonsirm Withyachumnarnkul. Characterization of Candidate Genes Involved in Growth of Black Tiger Shrimp Penaeus Monodon. Aquaculture 307, 150–156. doi: 10.1016/j.aquaculture.2010.07.008
Arun K. D., Michelle M. R., Kurt R. K. (2002). Quantitative Assay for Measuring the Taura Syndrome Virus and Yellow Head Virus Load in Shrimp by Real-Time RT-PCR Using SYBR Green Chemistry. J. Virol. Methods 104, 69–82. doi: 10.1016/S0166-0934(02)00042-3
Bandyopadhyay S., Gama F., Molina-Navarro M. M., Gualberto J. M., Claxton R., Naik S. G., et al. (2008). Chloroplast Monothiol Glutaredoxins as Scaffold Proteins for the Assembly and Delivery of [2Fe-2S] Clusters. EMBO J. 27, 1122–1133. doi: 10.1038/emboj.2008.50
Beinert H. (2000). Iron-Sulfur Proteins: Ancient Structures, Still Full of Surprises. J. Biol. Inorgan. Chem. 5, 2–15. doi: 10.1007/s007750050002
Berndt C., Poschmann G., Stühler K., Holmgren A., Bräutigam L. (2014). Zebrafish Heart Development is Regulated via Glutaredoxin 2 Dependent Migration and Survival of Neural Crest Cells. Redox Biol. 2, 673–678. doi: 10.1016/j.redox.2014.04.012
Bräutigam L., Johansson C., Kubsch B., McDonough M. A., Bill E., Holmgren A., et al. (2013). An Unusual Mode of Iron-Sulfur-Cluster Coordination in a Teleost Glutaredoxin. Biochem. Biophys. Res. Commun. 436, 491–496. doi: 10.1016/j.bbrc.2013.05.132
Bräutigam L., Schütte L. D., Godoy J. R., Prozorovski T., Gellert M., Hauptmann G., et al. (2011). Vertebrate-Specific Glutaredoxin is Essential for Brain Development. Proc. Natl. Acad. Sci. U. S. A. 108, 20532–20537. doi: 10.1073/pnas.1110085108
Bu R., Wang P., Zhao C., Bao W., Qiu L. (2017). Gene Characteristics, Immune and Stress Responses of PmPrx1 in Black Tiger Shrimp (Penaeus Monodon): Insights From Exposure to Pathogenic Bacteria and Toxic Environmental Stressors. Dev. Comp. Immunol. 77, 1–16. doi: 10.1016/j.dci.2017.07.002
Camaschella C., Campanella A., De F. L., Boschetto L., Merlini R., Silvestri L., et al. (2007). The Human Counterpart of Zebrafish Shiraz Shows Sideroblastic-Like Microcytic Anemia and Iron Overload. Blood 110, 1353–1358. doi: 10.1182/blood-2007-02-072520
Carvalho A. P., Fernandes P. A., Ramos M. J. (2006). Similarities and Differences in the Thioredoxin Superfamily. Prog. Biophys. Mol. Biol. 91, 229–248. doi: 10.1016/j.pbiomolbio.2005.06.012
Chen S. M., Chen J. C. (2002). Effects of pH on Survival, Growth, Molting and Feeding of Giant Freshwater Prawn Macrobrachium Rosenbergii. Aquaculture 218, 613–623. doi: 10.1016/S0044-8486(02)00265-X
Cheng N. (2008). AtGRX4, an Arabidopsis Chloroplastic Monothiol Glutaredoxin, is Able to Suppress Yeast Grx5 Mutant Phenotypes and Respond to Oxidative Stress. FEBS Lett. 582, 848–854. doi: 10.1016/j.febslet.2008.02.006
Cheng C., Yang F., Ling R., Liao S., Miao Y., Ye C., et al. (2015). Effects of Ammonia Exposure on Apoptosis, Oxidative Stress and Immune Response in Pufferfish (Takifugu Obscurus). Aquat. Toxicol. 164, 61–71. doi: 10.1016/j.aquatox.2015.04.004
Cheng N., Zhang W., Chen W., Jin J., Cui X., Butte N. F., et al. (2011). A Mammalian Monothiol Glutaredoxin, Grx3, is Critical for Cell Cycle Progression During Embryogenesis. FEBS J. 278, 2525–2539. doi: 10.1111/j.1742-4658.2011.08178.x
Chen J. C., Lin M. N., Ting Y. Y., Lin J. N. (1994). Survival, Haemolymph Osmolality and Tissue Water of Penaeus Chinensis Juveniles Acclimated to Different Salinity and Temperature Levels. Comp. Biochem. Physiol. Part A. Physiol. 110, 253–258. doi: 10.1016/0300-9629(94)00164-O
Ching B., Chew S. F., Wong W. P., Ip Y. K. (2009). Environmental Ammonia Exposure Induces Oxidative Stress in Gills and Brain of Boleophthalmus Boddarti (Mudskipper). Aquat. Toxicol. 95, 203–212. doi: 10.1016/j.aquatox.2009.09.004
Chi C., Tang Y., Zhang J., Dai Y., Abdalla M., Chen Y., et al. (2018). Structural and Biochemical Insights Into the Multiple Functions of Yeast Grx3. J. Mol. Biol. 430, 1235–1248. doi: 10.1016/j.jmb.2018.02.024
Couturier J., Jacquot J., Rouhier N. (2009). Evolution and Diversity of Glutaredoxins in Photosynthetic Organisms. Cell. Mol. Life Sci. 66, 2539–2557. doi: 10.1007/s00018-009-0054-y
Dunier M., Siwicki A. K. (1993). Effects of Pesticides and Other Organic Pollutants in the Aquatic Environment on Immunity of Fish: A Review. Fish. Shellfish. Immunol. 3, 423–438. doi: 10.1006/fsim.1993.1042
Dunnen J., Dalgleish R., Maglott D., Hart R., Greenblatt M., Jordan J., Roux A., Smith T., Antonarakis S., Taschner P, et al. (2016). HGVS Recommendations for the Description of Sequence Variants: 2016 Update. Human Mutation, 37, 564–569. doi: 10.1002/humu.22981
Eklund H., Cambillau C., Sjöberg B. M., Holmgren A., Jörnvall H., Höög J. O., et al. (1984). Conformational and Functional Similarities Between Glutaredoxin and Thioredoxins. EMBO J. 3, 1443–1449. doi: 10.1002/j.1460-2075.1984.tb01994.x
Fan R., Li Y., Jiang S., Jiang S., Yang Q., Yang L., et al. (2021). cDNA Cloning and Expression Analysis of Glutaredoxin 3 in Black Tiger Shrimp Penaeus Monodon. Aquacul. Int. 29, 2661–2679. doi: 10.1007/S10499-021-00774-7
FAO (2020). State of World Fishneries and Aquaculture 2020: Sustainability in Action (Rome). doi: 10.4060/ca9229en
Glaser J. R., Glaser E. M. (2020). Stereology, Morphometry, and Mapping: The Whole is Greater Than the Sum of Its Parts. J. Chem. Neuroanat. 20, 115–126. doi: 10.1016/S0891-0618(00)00073-9
Gürel T. (2005). The Larval Development of Penaeus semisulcatus (de Hann, 1850) (Decapoda: Penaeidae). J. Fisheries Aquatic Sci. 22, 195–199. doi: 10.12714/egejfas.2005.22.1.5000156908
Herrero E., de la Torre-Ruiz M. A. (2007). Monothiol Glutaredoxins: A Common Domain for Multiple Functions. Cell. Mol. Life Sci.: CMLS. 64, 1518–1530. doi: 10.1007/s00018-007-6554-8
Hiroaki M., Yoshito I., Hajime N., Junji Y., Koji S., Takahito K. (2003). Glutaredoxin Exerts an Antiapoptotic Effect by Regulating the Redox State of Akt. J. Biol. Chem. 278, 50226–50233. doi: 10.1074/jbc.m310171200
Holmgren A. (1979). Glutathione-Dependent Synthesis of Deoxyribonucleotides. Characterization of the Enzymatic Mechanism of Escherichia Coli Glutaredoxin. J. Biol. Chem. 254, 3672–3678. doi: 10.1016/S0021-9258(18)50814-0
Holmgren A., Johansson C., Berndt C., Lönn M. E., Hudemann C., Lillig C. H. (2005). Thiol Redox Control via Thioredoxin and Glutaredoxin Systems. Biochem. Soc. Trans. 33, 1375–1377. doi: 10.1042/bst20051375
Hu W., Yao C. (2016). Molecular and Immune Response Characterizations of a Novel AIF and Cytochrome C in Litopenaeus Vannamei Defending Against WSSV Infection. Fish. Shellfish. Immunol. 56, 84–95. doi: 10.1016/j.fsi.2016.06.050
Jie Y., Luo Z., Xie J., Cheng C., Ma H., Liu G., et al. (2022). Characterization of Phosphofructokinase (PFK) From Mud Crab Scylla Paramamosain and its Role in Mud Crab Dicistrovirus-1 Proliferation. Fish. Shellfish. Immunol. 124, 39–46. doi: 10.1016/J.FSI.2022.03.042
Knuesting J., Riondet C., Maria C., Kruse I., Bécuwe N., König N., et al. (2015). Arabidopsis Glutaredoxin S17 and its Partner, the Nuclear Factor Y Subunit C11/negative Cofactor 2α, Contribute to Maintenance of the Shoot Apical Meristem Under Long-Day Photoperiod. Plant Physiol. 167, 1643–1658. doi: 10.1104/pp.15.00049
Levin A., Nair D., Qureshi A. R., Bárány P., Heimburger O., Anderstam B., et al. (2018). Serum Glutaredoxin Activity as a Marker of Oxidative Stress in Chronic Kidney Disease: A Pilot Study. Nephron 140, 249–256. doi: 10.1159/000492500
Lillig C. H., Berndt C., Holmgren A. (2008). Glutaredoxin Systems. BBA-General. Subj. 1780, 1304–1317. doi: 10.1016/j.bbagen.2008.06.003
Liu M., Guo H., Liu B., Zhu K., Guo L., Liu B., et al. (2021). Gill Oxidative Damage Caused by Acute Ammonia Stress was Reduced Through the HIF-1α/NF-κb Signaling Pathway in Golden Pompano (Trachinotus Ovatus). Ecotoxicol. Environ. Saf. 222, 112504. doi: 10.1016/J.ECOENV.2021.112504
Li Y., Yang Q., Su T., Zhou F., Yang L., Huang J. (2012). The toxicity of ammonia-N on Penaeus monodon and immune parameters. J. Shanghai Ocean Univ. 21, 358–362.
Li Y., Zhou F., Huang J., Yang L., Jiang S., Yang Q., et al. (2018). Transcriptome Reveals Involvement of Immune Defense, Oxidative Imbalance, and Apoptosis in Ammonia-Stress Response of the Black Tiger Shrimp (Penaeus Monodon). Fish. Shellfish. Immunol. 83, 162–170. doi: 10.1016/j.fsi.2018.09.026
Lönn M. E., Hudemann C., Berndt C., Cherkasov V., Capani F., Holmgren A., et al. (2008). Expression Pattern of Human Glutaredoxin 2 Isoforms: Identification and Characterization of Two Testis/Cancer Cell-Specific Isoforms. Antiox. Redox Signaling 10, 547–557. doi: 10.1089/ars.2007.1821
Mapolelo D. T., Zhang B., Randeniya S., Albetel A., Li H., Couturier J., et al. (2013). Monothiol Glutaredoxins and A-Type Proteins: Partners in Fe-S Cluster Trafficking. Dalton. Trans. 42, 3107–3115. doi: 10.1039/C2DT32263C
Mondal S., Kumar V., Singh S. P. (2019). Phylogenetic Distribution and Structural Analyses of Cyanobacterial Glutaredoxins (Grxs). Comput. Biol. Chem. 84, 107141. doi: 10.1016/j.compbiolchem.2019.107141
Nathan C., Cunningham-Bussel A. (2013). Beyond Oxidative Stress: An Immunologist’s Guide to Reactive Oxygen Species. Nat. Rev. Immunol. 13, 349–361. doi: 10.1038/nri3423
Oh Y., Hong S., Yeon J., Cha M., Kim I. (2012). Interaction Between Saccharomyces Cerevisiae Glutaredoxin 5 and SPT10 and Their In Vivo Functions. Free Radical Biol. Med. 52, 1519–1530. doi: 10.1016/j.freeradbiomed.2012.01.032
Omeka W. K. M., Liyanage D. S., Yang H., Lee J. (2019). Glutaredoxin 2 From Big Belly Seahorse (Hippocampus Abdominalis) and its Potential Involvement in Cellular Redox Homeostasis and Host Immune Responses. Fish. Shellfish. Immunol. 95, 411–421. doi: 10.1016/j.fsi.2019.09.071
Pan J. L., Bardwell J. C. (2006). The Origami of Thioredoxin-Like Folds. Protein Sci. 15, 2217–2227. doi: 10.1110/ps.062268106
Petry S. F., Sun L. M., Knapp A., Reinl S., Linn T. (2018). Distinct Shift in Beta-Cell Glutaredoxin 5 Expression Is Mediated by Hypoxia and Lipotoxicity Both In Vivo and In Vitro. Front. Endocrinol. 9. doi: 10.3389/fendo.2018.00084
Potamitou F. A., Arne H. (2004). Glutaredoxins: Glutathione-Dependent Redox Enzymes With Functions Far Beyond a Simple Thioredoxin Backup System. Antiox. Redox Signaling 6, 63–74. doi: 10.1089/152308604771978354
Qin Y., Jiang S., Huang J., Zhou F., Yang Q., Jiang S., et al. (2019). C-type lectin response to bacterial infection and ammonia nitrogen stress in tiger shrimp (Penaeus monodon). Fish and Shellfish Immunology 90, 188–198. doi: 10.1016/j.fsi.2019.04.034
Rőszer T. (2014). The Invertebrate Midintestinal Gland (“Hepatopancreas”) is an Evolutionary Forerunner in the Integration of Immunity and Metabolism. Cell Tissue Res. 358, 685–695. doi: 10.1007/s00441-014-1985-7
Randall D. J., Tsui T. K. N. (2002). Ammonia Toxicity in Fish. Mar. pollut. Bull. 45, 17–23. doi: 10.1016/S0025-326X(02)00227-8
Rowley A. F. (2016). The Immune System of Crustaceans. Encyclo. Immunobiol. 1, 437–453. doi: 10.1016/B978-0-12-374279-7.12005-3
Rusaini L. O. (2010). Insight Into the Lymphoid Organ of Penaeid Prawns: A Review. Fish. Shellfish. Immunol. 29, 367–377. doi: 10.1016/j.fsi.2010.05.011
Shakamuri P., Zhang B., Johnson M. K. (2012). Monothiol Glutaredoxins Function in Storing and Transporting [Fe2S2] Clusters Assembled on IscU Scaffold Proteins. J. Am. Chem. Soc. 134, 15213–15216. doi: 10.1021/JA306061X
Shi M., Jiang S., Li Y., Yang Q., Jiang S., Yang L., et al. (2019). Comprehensive Expression Analysis of the Beta Integrin From Penaeus Monodon Indicating its Participation in Innate Immunity and Ammonia Nitrogen Stress Response. Fish. Shellfish. Immunol. 98, 887–898. doi: 10.1016/j.fsi.2019.11.049
Soonthornchai W., Rungrassamee W., Karoonuthaisiri N., Jarayabhand P., Klinbunga S., Söderhäll K., et al. (2010). Expression of Immune-Related Genes in the Digestive Organ of Shrimp, Penaeus Monodon, After an Oral Infection by Vibrio Harveyi. Dev. Comp. Immunol. 34, 19–28. doi: 10.1016/j.dci.2009.07.007
Verma P. K., Verma S., Tripathi R. D. (2021). Pandey Nalini,Chakrabarty Debasis. CC-Type Glutaredoxin, OsGrx_C7 Plays a Crucial Role in Enhancing Protection Against Salt Stress in Rice. J. Biotechnol. 329, 192–203. doi: 10.1016/j.jbiotec.2021.02.008
Wang Z., Xing S., Birkenbihl R. P., Zachgo S. (2009). Conserved Functions of Arabidopsis and Rice CC-Type Glutaredoxins in Flower Development and Pathogen Response. (Special. Issue.: Redox Biol. I.). Mol. Plant 2, 323–335. doi: 10.1093/mp/ssn078
Wingert R. A., Galloway J. L., Barut B., Foott H., Fraenkel P., Axe J. L., et al. (2005). Deficiency of Glutaredoxin 5 Reveals Fe-S Clusters are Required for Vertebrate Haem Synthesis. Nature 436, 1035–1039. doi: 10.1038/nature03887
Xing S., Rosso M. G., Zachgo S. (2005). ROXY1, a Member of the Plant Glutaredoxin Family, is Required for Petal Development in Arabidopsis Thaliana. Development 132, 1555–1565. doi: 10.1242/dev.01725
Zhang L., Li Y., Wu M., Ouyang H., Shi R. (2021). The SNP Polymorphisms Associated With WSSV-Resistance of Prophenoloxidase in Red Swamp Crayfish (Procambarus Clarkii) and Its Immune Response Against White Spot Syndrome Virus (WSSV). Aquaculture 530, 735787. doi: 10.1016/j.aquaculture.2020.735787
Zhang Y., Ye C., Wang A., Zhu X., Chen C., Xian J., et al. (2015). Isolated and Combined Exposure to Ammonia and Nitrite in Giant Freshwater Pawn (Macrobrachium Rosenbergii): Effects on the Oxidative Stress, Antioxidant Enzymatic Activities and Apoptosis in Haemocytes. Ecotoxicology 24, 1601–1610. doi: 10.1007/s10646-015-1477-x
Zhao F., Wang Q. (2012). The Protective Effect of Peroxiredoxin II on Oxidative Stress Induced Apoptosis in Pancreatic β-Cells. Cell Biosci. 2, 22. doi: 10.1186/2045-3701-2-22
Zheng P., Wang L., Wang A., Zhang X., Ye J., Wang D., et al. (2018). cDNA Cloning and Expression Analysis of Glutaredoxin (Grx) 2 in the Pacific White Shrimp Litopenaeus Vannamei. Fish. Shellfish. Immunol. 86, 662–671. doi: 10.1016/j.fsi.2018.12.011
Zheng P., Zhang X., Wang D., Li J., Zhang Z., Lu Y., et al. (2021). Molecular Characterization and Expression Analysis of a Novel Glutaredoxin 3 Gene in Pacific White Shrimp (Litopenaeus Vannamei). Front. Mar. Sci. 8. doi: 10.3389/FMARS.2021.687377
Zhou K., Zhou F., Huang J., Yang Q., Jiang S., Qiu L., et al. (2017). Characterization and Expression Analysis of a Chitinase Gene (PmChi -4) From Black Tiger Shrimp (Penaeus Monodon) Under Pathogen Infection and Ambient Ammonia Nitrogen Stress. Fish. Shellfish. Immunol. 62, 31–40. doi: 10.1016/j.fsi.2017.01.012
Keywords: glutaredoxin, Penaeus monodon, ammonia-N stress, innate immunity, SNPs
Citation: Fan R, Jiang S, Li Y, Yang Q, Jiang S, Huang J, Yang L, Chen X and Zhou F (2022) Molecular Characterization and Expression Analysis of Glutaredoxin 5 in Black Tiger Shrimp (Penaeus monodon) and Correlation Analysis Between the SNPs of PmGrx5 and Ammonia-N Stress Tolerance Trait. Front. Mar. Sci. 9:909827. doi: 10.3389/fmars.2022.909827
Received: 31 March 2022; Accepted: 16 May 2022;
Published: 15 June 2022.
Edited by:
Xiangli Tian, Ocean University of China, ChinaCopyright © 2022 Fan, Jiang, Li, Yang, Jiang, Huang, Yang, Chen and Zhou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Falin Zhou, emhvdWZhbGluMDkyNUAxNjMuY29t
 Rui Fan
Rui Fan Shigui Jiang
Shigui Jiang Yundong Li
Yundong Li Qibin Yang1,3
Qibin Yang1,3 Song Jiang
Song Jiang