
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mar. Sci. , 04 October 2022
Sec. Marine Conservation and Sustainability
Volume 9 - 2022 | https://doi.org/10.3389/fmars.2022.907714
 Lynn Drescher1†
Lynn Drescher1† Noel Jin Kiat Heng1†
Noel Jin Kiat Heng1† Min Yi Chin1†
Min Yi Chin1† Neel R. O. Karve1†
Neel R. O. Karve1† Edward Jun Wei Cheung1†
Edward Jun Wei Cheung1† Anastasia Kurniadi1†
Anastasia Kurniadi1† Mariana Quesada Urera1†
Mariana Quesada Urera1† Felipe Gabriel Waldeck1†
Felipe Gabriel Waldeck1† U. Dharshini1†
U. Dharshini1† Nirel Tze En Hoe1†
Nirel Tze En Hoe1† Joshua Song Yang Choo1†
Joshua Song Yang Choo1† Russell Fock Jun Lok1†
Russell Fock Jun Lok1† Caroline Kibat1
Caroline Kibat1 Benjamin J. Wainwright1,2*
Benjamin J. Wainwright1,2*Overfishing has significantly decreased global shark populations, with some species experiencing reductions of approximately 70% over the last 50 years. Singapore is a major shark fin transhipment hub that helps to satisfy the global demand for shark fins, which are considered status symbols and reputed to have medicinal value in Asian culture. Despite the recognised and urgent need to better protect shark populations, the success of such efforts has been limited by the difficulties associated with visually identifying the species of shark from which the fins originated. In this study, we collected 451 shark fin tissue samples from a variety of local retail markets in Singapore. Using DNA barcoding techniques, we amplified a 350 base pair fragment of the mitochondrial cytochrome c oxidase subunit I (COI) gene from each to identify the species sold in Singapore. We identified 22 shark species, of which 17 are categorised as Threatened (Critically Endangered, Endangered or Vulnerable) under the IUCN Red List. Six of these species are also listed on Appendix II of the Convention on International Trade in Endangered Species of Wild Fauna and Flora (CITES).
The global shark trade is valued at nearly US$1 billion and is responsible for the death of an estimated 100 million sharks annually (Worm et al., 2013; Steinke et al., 2017; Van Houtan et al., 2020), and shark finning is a major contributor to the decline in shark populations (Sembiring et al., 2015; Appleyard et al., 2018; Bonaccorso et al., 2021). Fins are considered a delicacy, a status symbol and are frequently served in soup at celebratory events. Additionally, fins are highly prized in Traditional Chinese Medicine (TCM) practices where they are reputed to have a host of beneficial effects, from improving general well-being, to claims promoting anti-cancer properties (Clarke et al., 2007; Dent and Clarke, 2015; Teo, 2015; Choy and Wainwright, 2022).
In efforts to prevent unsustainable fishing and international trading practices, the Convention on International Trade in Endangered Species of Wild Fauna and Flora (CITES) was established as a legally-binding framework to prevent the illegal trade of wildlife. In addition to CITES, the International Union for the Conservation of Nature (IUCN) Red List serves as a framework for identifying the extinction risk of shark species. The use of these regulatory structures is crucial to the conservation of sharks; they provide regulations through which policy can be designed, regulations upheld, and a mechanism that allows global shark populations, catches and trade to be monitored (Clarke et al., 2013).
Singapore is a consumer and key distributor of shark fins, which typically do not originate from the country’s waters (Dent and Clarke, 2015; Boon, 2017). Singapore is recognized as the world’s second-largest importer and exporter of shark fins, with an average of 2,352 tonnes imported and 2,067 tonnes exported each year (Boon, 2017). Despite recent reductions in the import and export of shark fins due to the COVID-19 pandemic and associated supply chain disruption, data from the Singapore Food Agency shows that Singapore continues to import and export large numbers of fins with 2,700 tonnes of shark fin imported and 2,400 tonnes exported in 2021 (SFA, 2022).
Highlighting the need for the improved regulation and enforcement of those that already exist, trade-regulated and endangered species of shark are frequently encountered in markets throughout the world (Cardeñosa et al., 2018). Previous studies show that IUCN red-listed and CITES trade-regulated species such as Rhynchobatus australiae and Sphyrna lewini are commonly sold in Singapore (Wainwright et al., 2018; Choo et al., 2021; Liu et al., 2021), yet these species are not typically found in the countries waters (Ip et al., 2021). Emphasising the need for more stringent enforcement and monitoring of the global shark fin trade, most of the species identified in the markets of Singapore come from populations that are acknowledged to be declining (Wainwright et al., 2018; Liu et al., 2021).
Shark fins come from a variety of sources, including legal fisheries, by-catch, illegal, unreported, and unregulated fisheries (IUU). Finning is the process of removing a shark’s fins and subsequently discarding the carcass (Biery and Pauly, 2012). This process is wasteful and does not utilise the entire shark, but discarding the comparatively large and low value carcass means more storage space is available for the high value fins, resulting in overfishing and contributing to the documented declines in shark populations (Biery and Pauly, 2012). Shark fins are then processed and dried before being sold to consumers in the food and beverage industry or to TCM practices. During processing and drying, key morphological features of shark fins are removed (Van Houtan et al., 2020) making species identification by visual methods alone challenging and in some cases impossible (Figure 1). These challenges hinder the accurate monitoring of the species involved in the trade and make it much more likely that endangered, or trade-regulated species inevitably end up in supply chains. In efforts to mitigate the challenges that finning presents in species identification, calls for the implementation of a Fins Naturally Attached (FNA) policy are gaining traction in numerous countries (Passantino, 2013; Ferretti et al., 2020), and have been implemented in 19 of the 43 recognised shark fishing nations (Government of India Ministry of Environment and Forests, 2013; Marine Stewardship Council)
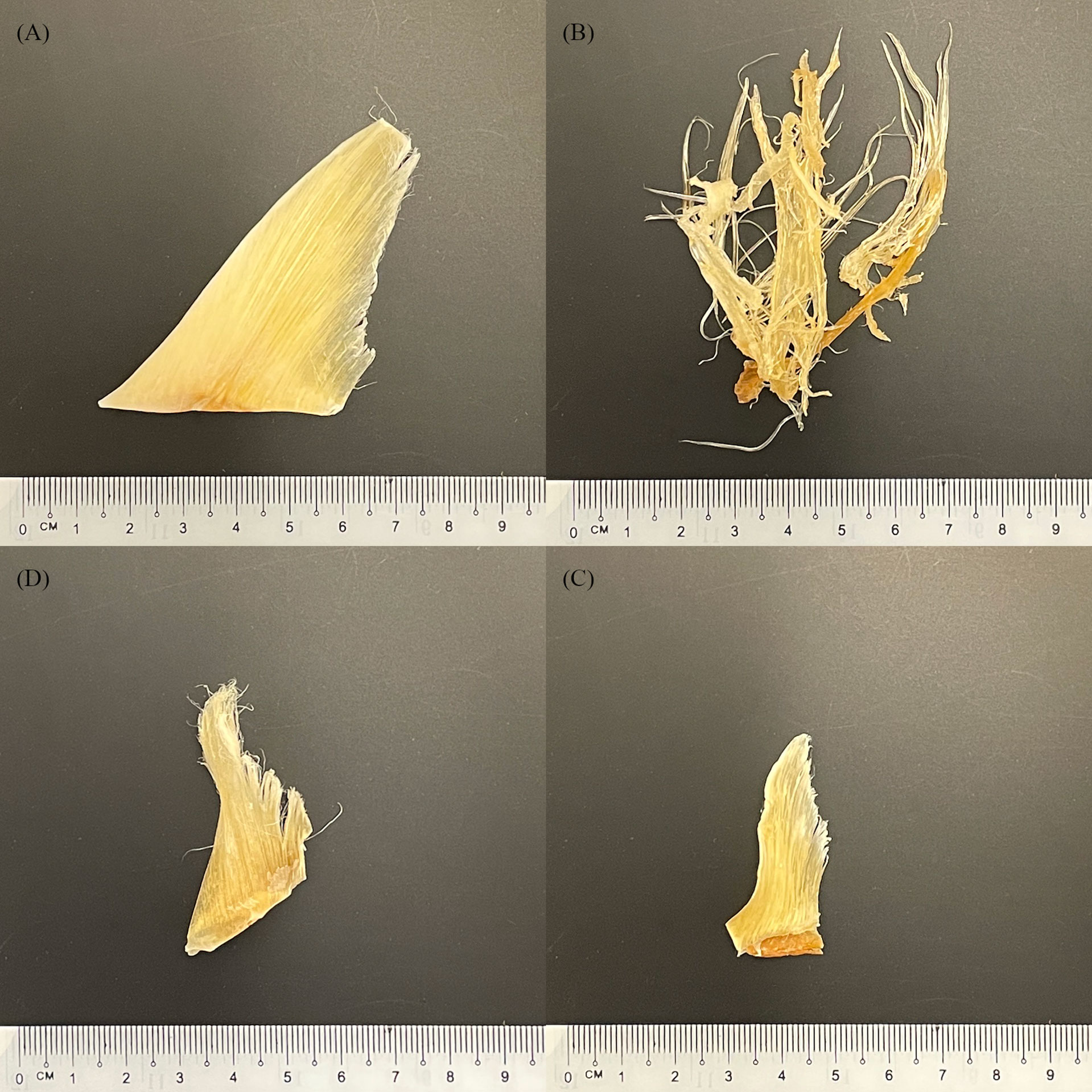
Figure 1 Selection of purchased dried shark fins (Barcoded as: (A, B) Sphyrna mokarran, (C, D) Rhizoprionodon oligolinx).
The utility of DNA barcoding in species identification is well-established (Hebert et al., 2003). It has been used to good effect throughout the world to help prevent seafood fraud and mislabelling, and it has extensive applications in the monitoring of illegal wildlife and the shark fin trade (Rehman et al., 2015; French and Wainwright, 2022; Neo et al., 2022). The technique uses species-specific differences in DNA sequence and the subsequent matching of this sequence to a known species in publicly accessible databases (e.g., GenBank and BOLD). The official animal DNA barcode is an approximate 650 bp fragment of the mitochondrial gene coding for cytochrome c oxidase 1 (COI) (Hebert et al., 2003). However, due to the processed nature of dried fins, DNA is often degraded to such an extent that it is impossible to amplify the full 650 bp region. Consequently, mini-barcoding approaches have been developed to allow DNA amplification from challenging or degraded samples (Cardeñosa et al., 2017). DNA barcoding provides an efficient, reliable and accurate method to identify the species of shark that a fin came from. Once this identification has been made, the species conservation status and any trade regulations imposed upon it can be readily determined with online resources (e.g., CITES and IUCN databases).
A total of 451 tissue samples were collected from Traditional Chinese Medicine (TCM) and retail shops across Chinatown in Singapore in January 2022, collections were made in the same way as Liu et al. (2021). Briefly, a list of TCM shops was compiled and each shop was assigned a number, this number was used to randomly pick retailers to visit. From each of 22 shops, the proprietor haphazardly selected a minimum of 20 fins from large, mixed and unmarked containers.
DNA extraction was performed using a Qiagen DNeasy® Blood and Tissue Kit with the following modification: DNA was eluted in 50 μL of AE buffer. Other than this, all steps followed the manufacturer’s instructions. Polymerase Chain Reaction (PCR) was performed in a 25 μL volume, with each reaction containing 12.5 μL GoTaq mastermix green (Promega), 1 μL forward primer at 10μM, 1 μL reverse primer 10 μM, 1 uL Bovine Serum Albumin (BSA), 7.5 μL of nuclease-free water and 2 μL of undiluted DNA template. The forward primer mlCOIintF (5′-GGW ACW GGW TGA ACW GTW TAY CCY CC-3′) (Leray et al., 2013) and reverse primer LoboR1 (5′-TAA ACY TCW GGR TGW CCR AAR AAY CA-3′) (Lobo et al., 2013) were used to amplify a 350bp fragment of mitochondrial DNA under the following thermal cycling profile: 5 repeats of 94°C for 30 s, 48°C for 2 min, 72°C for 1 min, then 35 repeats of 94°C for 30 s, 54°C for 2 min, 72°C for 1 min, then 72°C for 5 min (Wainwright et al., 2018).
Successful PCR amplification was verified on a 1% agarose gel, and bi-directional Sanger sequencing and enzymatic cleaning was performed by Bio-Basic Asia Inc. Sequences were viewed and quality checked with Geneious Prime (v2022.0.2) (Kearse et al., 2012), we considered sequences high quality if chromatograms showed clear, well defined peaks and contained no ambiguous base calls. Sequences were then referenced against The Barcode of Life Data System (BOLD) and GenBank. We deemed a species identification positive and unambiguous if BOLD returned a 100% match and indicated no matches with other closely related congeneric species, and if the same species was the top match in GenBank (Choy and Wainwright, 2022)
Of the 451 tissues collected, 294 samples were successfully identified at the species or genus level while 157 samples returned low quality DNA sequences (i.e., multiple peaks) or did not amplify. We identified a total of 22 different shark species across 11 genera. Six species are listed under CITES Appendix II (n = 119), and the IUCN consider 17 of them as threatened. Of these 17, four species are listed as critically endangered (n = 37), five species as endangered (n = 28), and eight species as vulnerable (n = 95). Only five of those identified at the species level were listed as not threatened by the IUCN (n = 52), and we were only able to identify 82 fins to the level of genus. These 82 all belonged to the genus Carcharhinus and matched with 100% identity against two or more of the following candidate species; Carcharhinus leiodon, Carcharhinus amblyrhynchoides, Carcharhinus tilstoni, or Carcharhinus amboinensis. (Figure 2 and Table 1). The top 5 most frequently observed shark species from seven previous barcoding studies performed between 2015 and 2022 were compared (Figure 3), Carcharhinus falciformis, and Prionace glauca were frequently in the top three most encountered species found in studies performed until 2022. In this study, Prionace glauca was completely absent in the 294 samples we identified to the species or genus level, and Carcharhinus falciformis was still commonly encountered.
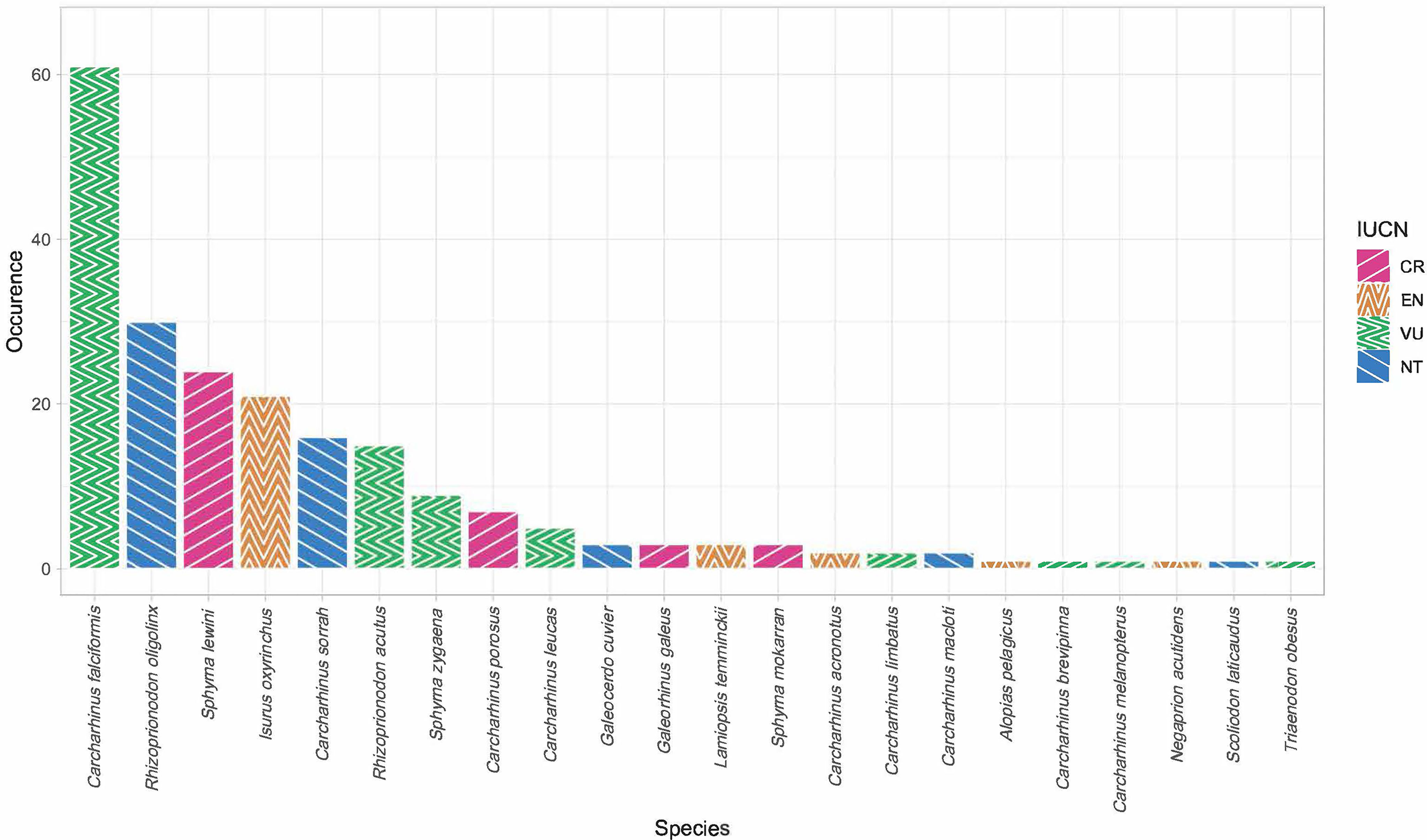
Figure 2 Bar chart showing species occurrence and their respective IUCN Red List status (CR, critically endangered, EN, endangered, VU, vulnerable, NT, near threatened). Only samples identified to the species level are included.
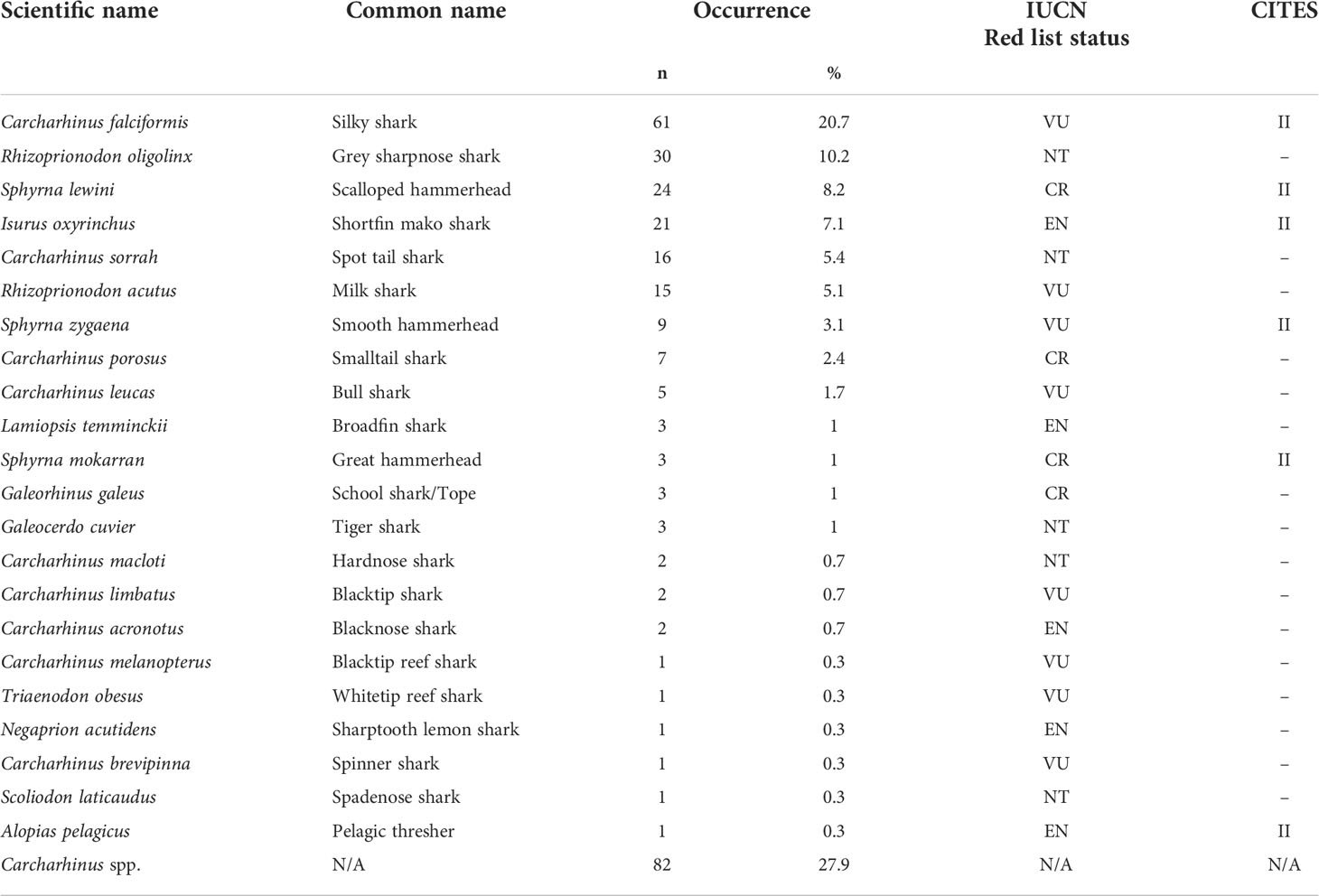
Table 1 Details of species identification, common names, occurrence, the IUCN Red List status (CR, critically endangered; EN, endangered; VU, vulnerable; NT, near threatened; LC, least concern), and CITES appendix listing.
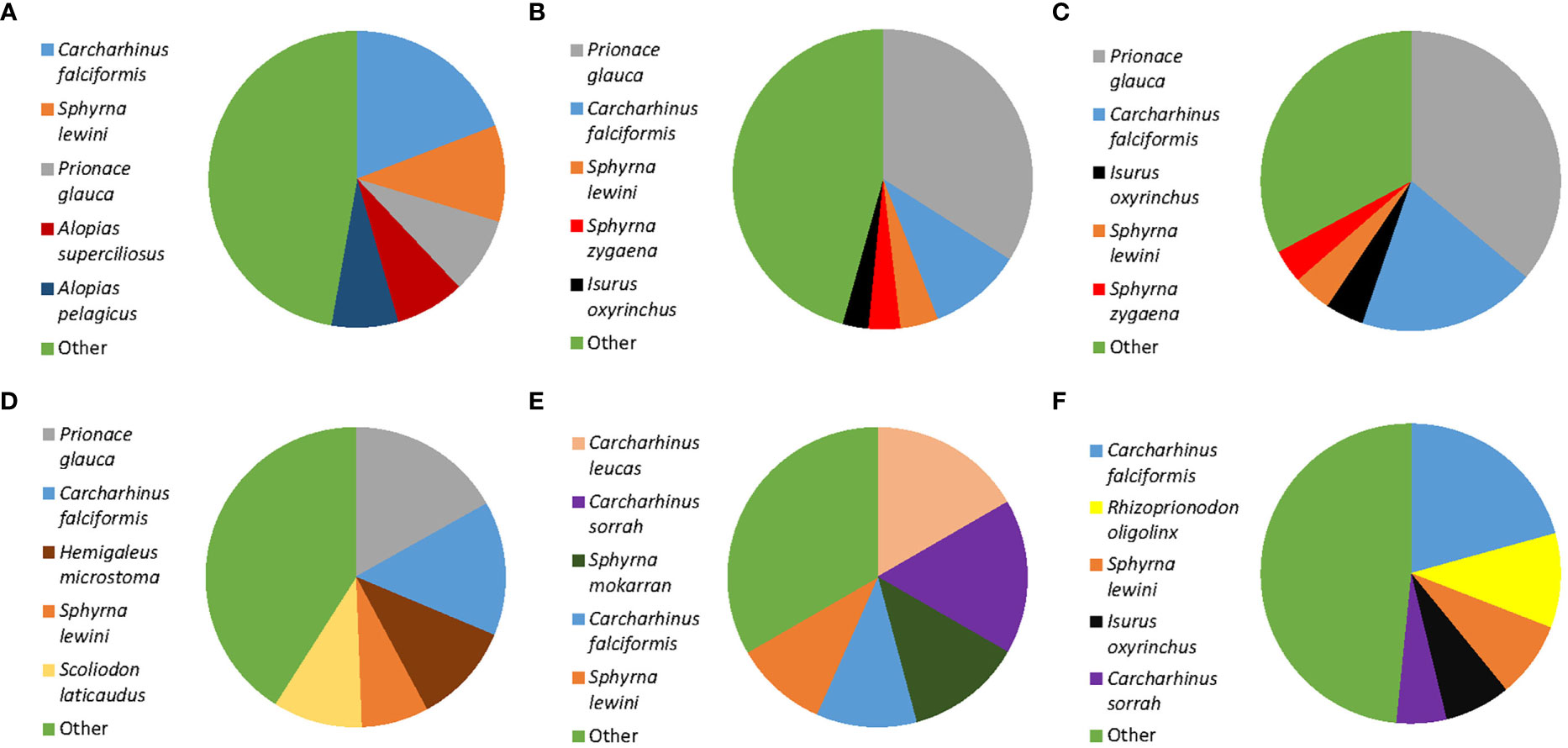
Figure 3 Comparison of shark fin DNA barcoding projects showing the top five most frequently occurring shark species in each study. Only identifications to the species level are included. (A) Sembiring et al., 2015, Indonesia. (B) Fields et al., 2018, Hong Kong. (C) Wainwright et al., 2018, Singapore. (D) Cardeñosa et al., 2020, Mainland China. (E) Seah et al., 2022, Malaysia. (F) This study, Singapore. The category ‘Other’ includes all species that are found at a low frequency, see cited studies for exact proportions of the species included in this category.
We successfully identified 294 samples to the species or genus level. It is likely samples failing to amplify or those that returned low quality sequences contained DNA that was degraded when the fins were processed, with DNA undergoing further degradation when stored in conditions that are not optimal for preservation (e.g., room temp). Of the 22 species level identifications, 17 are considered threatened by the IUCN (Critically Endangered n = 4, Endangered n = 5, or Vulnerable n = 8), and six species are listed under CITES Appendix II. These findings are in agreement with previous work performed in Singapore that shows endangered species are frequently available for purchase in public markets and retail shops (Wainwright et al., 2018; Choo et al., 2021; Liu et al., 2021).
Carcharhinus falciformis, the silky shark, was the most commonly identified species in our study (n = 61), and this species is consistently one of the most frequently encountered in retail shops and trade hubs throughout Asia (Sembiring et al., 2015; Cardeñosa et al., 2018; Fields et al., 2018; Wainwright et al., 2018). Because of rapid population declines, this species has been listed as vulnerable on the IUCN Red List since 2017 and has been included on the CITES Appendix II list since 2016. C. falciformis tends to track tuna aggregations, making it vulnerable to accidental bycatch (Poisson et al., 2014; Hutchinson et al., 2015). Additionally, the relatively long gestation and sexual maturation period of the species limits the potential for stock recovery (Branstetter, 1987). Despite regulations to prevent the trade of this species, there is still a high occurrence of it in the markets of Singapore, Malaysia and China (Fields et al., 2018; Wainwright et al., 2018; Seah et al., 2022). It should be noted that dried shark fin can last for several years if processed and stored correctly. Therefore, depending upon the turnover of inventory, we could be seeing fins that were collected before CITES and IUCN listings were implemented for this species. However, its high prevalence in markets make it very likely that at least some of these C. falciformis fins represent recent catches.
Rhizoprionodon oligolinx, the grey sharpnose shark, was the second-most commonly identified species in our samples (n = 30). While this species has been identified in previous shark DNA barcoding work in Asia, it usually occurs at a much lower frequency in comparison to that observed here (Sembiring et al., 2015; Cardeñosa et al., 2018; Fields et al., 2018; Wainwright et al., 2018). R. oligolinx was recently re-categorised by the IUCN from Least Concern to Near Threatened due to population declines of 20-29% over the past three generations (Rigby et al., 2021), but it has yet to be listed under any CITES appendices. The increasing incidence of this species as demonstrated by this work supports this IUCN re-classification, and may warrant its inclusion on the CITES list in the future. The increased occurrence of R. oligolinx fins may be an outcome of more stringent regulations placed on other shark species (e.g., Carcharhinus falciformis, and all three species of thresher sharks that were added to CITES appendix II in 2016). These regulations could shift fishing pressure from one species to another, or it could simply be the case that some shark populations have reached such low levels that they are less frequently encountered meaning other species (e.g., R. oligolinx) are caught to make up for the shortfall.
Sphyrna lewini, the scalloped hammerhead, is the third most commonly encountered shark in this work. This high prevalence is consistent with a range of other work performed in Southeast Asia (Sembiring et al., 2015; Wainwright et al., 2018; Liu et al., 2021; Choy and Wainwright, 2022; Seah et al., 2022), and this species is recognised as one of the top four most traded in international shark fin markets (Cardeñosa et al., 2018; Fields et al., 2018). As previously suggested (Liu et al., 2021), the schooling behaviour that S. lewini engages in could mean it is easier to catch and therefore preferentially targeted.
A 2018 study showed that fins from guitarfish and wedgefish in the genus Rhynchobatus were readily available for purchase in the markets of Singapore (Wainwright et al., 2018). At the 2019 CITES CoP18 summit held in Geneva, critically endangered rays, guitarfish, and wedgefish were listed under CITES Appendix II. Since this regulation was introduced, two studies in Singapore using similar methods have uncovered no processed fins belonging to wedgefish or guitarfish (Liu et al., 2021; this study). It is still too early to attribute this finding to their recent CITES listing, but it is encouraging that additional surveys using similar methodologies with similar sample sizes did not uncover their presence. Repeated monitoring is required to assess whether this is a sampling artefact, a consequence of regulations or if this absence is a result of population collapse.
Despite the frequent occurrence of blue sharks, Prionace glauca, in previous work performed in Singapore and East Asia (Chuang et al., 2016; Wainwright et al., 2018; Liu et al., 2021), it was notable by its absence in the 294 fins we identified to the genus or species level in this survey (Figure 3). This could be a cause for concern, blue sharks are one of the most heavily exploited shark species in the world. They account for up to 90% of all oceanic elasmobranch catches (da Silva et al., 2021) and are frequently discarded as bycatch in long line fisheries (Campana et al., 2002; da Silva et al., 2021). Scientific evidence indicates that catches of this species have exceeded maximum sustainable yields, and recommendations that their catches be capped have been made (Simpfendorfer and Dulvy, Nicholas, 2017; da Silva et al., 2021). Yet, despite this acknowledged overfishing and observed population declines, blue sharks are not CITES regulated and are only listed as near threatened on the IUCN Red List. The evidence collected, along with the observed population declines suggest that this species would be an ideal candidate for inclusion on the CITES appendices, or the IUCN Red list. In fact, members of the Genus Prionace (e.g., blue sharks) have been proposed for listing on CITES Appendix II at the nineteenth Conference of the Parties (CITES CoP19 Prop. 37).
DNA barcoding studies are now becoming increasingly routine, and they serve an important role in monitoring the trade of shark fins throughout the world’s retail hubs and markets. Without molecular identifications, many of these fins would likely remain unidentified. Recent work shows that markets from different regions throughout East Asia and Southeast Asia have distinct species compositions (Cardeñosa et al., 2020; French and Wainwright, 2022; Seah et al., 2022; Figure 3). This suggests a number of supply chains, hubs and fisheries supply the demand for shark fin. Without routine and repeated monitoring of the trade at numerous locations, it becomes very difficult to assess whether policy and regulations are having the intended effects. As DNA sequencing technology continues to advance and the associated costs fall, it will become more feasible for governments to apply large scale and rapid testing of fins, for example, it is now possible to make accurate IDs within 60 minutes with minimal equipment (But et al., 2020). If further declines in shark populations are to be avoided, it is necessary that countries take their CITES trade obligations seriously, and sufficient resources are directed towards the enforcement of these regulations. Without it, there will be little to no impact on the conservation of shark populations.
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethical review and approval was not required for the animal study because samples came from processed fins available for sale in public retail locations.
BW conceived and designed of the study. LD, NHJK, MC, NROK, EC, AK, MU, FW, UD, NH, JC, RF, and CK performed lab work and collected samples. LD, NHJK, MC, NROK, EC, AK, MU, FW, UD, NH, JC, and RF performed analysis, and wrote the first draft of the manuscript. All authors contributed to the manuscript, read, and approved the submitted version.
This paper was the result of a class project: Wildlife Forensics and the Shark Fin Trade (YID3221) at Yale-NUS College, Singapore.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Appleyard S. A., White W. T., Vieira S., Sabub B. (2018). Artisanal shark fishing in Milne bay province, Papua new Guinea: Biomass estimation from genetically identified shark and ray fins. Sci. Rep. 8, 6693. doi: 10.1038/s41598-018-25101-8
Biery L., Pauly D. (2012). A global review of species-specific shark-fin-to-body-mass ratios and relevant legislation. J. Fish Biol. 80 (5), 1643–1677. doi: 10.1111/j.1095-8649.2011.03215.x
Bonaccorso E., Ordóñez-Garza N., Pazmiño D. A., Hearn A., Páez-Rosas D., Cruz S., et al. (2021). International fisheries threaten globally endangered sharks in the Eastern tropical pacific ocean: The case of the fu Yuan yu leng 999 reefer vessel seized within the galápagos marine reserve. Sci. Rep. 11 (1), 1–11. doi: 10.1038/s41598-021-94126-3
Branstetter S. (1987). Age, growth and reproductive biology of the silky shark, carcharhinus falciformis, and the scalloped hammerhead, sphyrna lewini, from the northwestern gulf of Mexico. Environ. Biol. Fishes 19 (3), 161–173. doi: 10.1007/bf00005346
But G. W. C., Wu H. Y., Shao K. T., Shaw Pang-chu (2020). rapid detection of CITES-listed shark fin species by loop-mediated isothermal amplification assay with potential for field use. Sci. Rep. 10, 4455. doi: 10.1038/s41598-020-61150-8
Campana S., Gonzalez P., Joyce W., Marl L. (2002). Catch, bycatch and landings of blueshark (Prionace glauca) in the Canadian Atlantic (Accessed April 2022).
Cardeñosa D., Fields A., Abercrombie D., Feldheim K., Shea S. K. H., Chapman D. D. (2017). A multiplex PCR mini-barcode assay to identify processed shark products in the global trade. PloS One 12, e0185368. doi: 10.1371/journal.pone.0185368
Cardeñosa D., Fields A. T., Babcock E. A., Shea S. K., Feldheim K. A., Chapman D. D. (2020). Species composition of the largest shark fin retail-market in mainland China. Sci. Rep. 10 (1), 1–10.
Cardeñosa D., Fields A. T., Babcock E. A., Zhang H., Feldheim K., Shea S. K., et al. (2018). CITES-listed sharks remain among the top species in the contemporary fin trade. Conserv. Lett. 11 (4), e12457. doi: 10.1111/conl.12457
Choo M. Y., Choy P. P. C., Ip Y. C. A., Rao M., Huang D. (2021). Diversity and origins of giant guitarfish and wedgefish products in Singapore. Aquat. Conserv. doi: 10.1002/aqc.3553
Choy P. P. C., Wainwright B. J. (2022). What is in your shark fin soup? probably an endangered shark species and a bit of mercury. Animals. doi: 10.3390/ani12070802
Chuang P. S., Hung T. C., Chang H. A., Huang C. K., Shiao J. C. (2016). The species and origin of shark fins in taiwan’s fishing ports, markets, and customs detention: A DNA barcoding analysis. PloS One 11 (1), e0147290. doi: 10.1371/journal.pone.0147290
CITES What is CITES?Available at: https://cites.org/eng/disc/what.php (Accessed March 16, 2022)
CITES CoP19 Prop. 37 Nineteenth meeting of the conference of the parties Panama city (Panama), 14 – 25 November 2022. Available at: https://cites.org/sites/default/files/documents/E-CoP19-Prop-37.pdf (Accessed 18 Aug 2022).
Clarke S. C., Harley S. J., Hoyle S. D., Rice J. S. (2013). Population trends in pacific oceanic sharks and the utility of regulations on shark finning. Conserv. Biol. 27 (1), 197–209. doi: 10.1111/j.1523-1739.2012.01943.x
Clarke S., Milner-Gulland E. J., Bjørndal T. (2007). Social, economic, and regulatory drivers of the shark fin trade. Mar. Resource Econ. 22 (3), 305–327. doi: 10.1086/mre.22.3.42629561
da Silva T. E. F., Lessa R., Santana F. M. (2021). Current knowledge on biology, fishing and conservation of the blue shark (Prionace glauca). Neotrop. Biol. Conserv. 16 (1), 71–88. doi: 10.3897/neotropical.16.e58691
Dent F., Clarke S. (2015). State of the global market for shark products. FAO Fish. Aquac. Tech. Pap. 590).
Ferretti F., Jacoby D. M. P., Pfleger M. O., White T. D., Dent F., Micheli F., et al. (2020). Shark fin trade bans and sustainable shark fisheries. Conserv. Lett. 13 (3). doi: 10.1111/conl.12708
Fields A. T., Fischer G. A., Shea S. K., Zhang H., Abercrombie D. L., Feldheim K. A., et al. (2018). Species composition of the international shark fin trade assessed through a retail-market survey in Hong Kong. Conserv. Biol. 32 (2), 376–389. doi: 10.1111/cobi.13043
French I., Wainwright B. J. (2022). DNA Barcoding identifies endangered sharks in pet food sold in Singapore. Front. Mar. Sci. 9. doi: 10.3389/fmars.2022.836941
Government of India Ministry of Environment and Forests (2013) Policy on prohibition of “finning” of sharks in the Sea. Available at: https://police.py.gov.in/Wildlife%20Ministry%20of%20environment%20and%20Forests/Policy%20on%20prohibition%20of%20(finning)%20of%20shark%20fins%20in%20the%20sea%20dt.25th%20august%202013.pdf (Accessed 19 Aug 2022).
Hebert P. D. N., Cywinska A., Ball S. L., deWaard J. R. (2003). Biological identifications through DNA barcodes. Proc. Biol. Sci. 270, 313–321. doi: 10.1098/rspb.2002.2218
Hutchinson M. R., Itano D. G., Muir J. A., Holland K. N. (2015)Post-release survival of juvenile silky sharks captured in a tropical tuna purse seine fishery (Accessed March 16, 2022).
Ip Y. C. A., Chang J. J. M., Lim K. K. P., et al. (2021). Seeing through sedimented waters: environmental DNA reduces the phantom diversity of sharks and rays in turbid marine habitats. BMC Ecol. Evo. 21, 166. doi: 10.1186/s12862-021-01895-6
Kearse M., Moir R., Wilson A., Stones-Havas S., Cheung M., Sturrock S., et al. (2012). Geneious basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28, 1647–1649. doi: 10.1093/bioinformatics/bts199
Leray M., Yang J. Y., Meyer C. P., Mills S. C., Agudelo N., Ranwez V., et al. (2013). A new versatile primer set targeting a short fragment of the mitochondrial COI region for metabarcoding metazoan diversity: application for characterizing coral reef fish gut contents. Front. Zool. 10 (1), 1–14. doi: 10.1186/1742-9994-10-34
Liu C. J., Neo S., Rengifo N. M., French I., Chiang S., Ooi M., et al. (2021). Sharks in hot soup: DNA barcoding of shark species traded in Singapore. Fish. Res. 241, 105994. doi: 10.1016/j.fishres.2021.105994
Lobo J., Costa P. M., Teixeira M. A., Ferreira M. S., Costa M. H., Costa F. O. (2013). Enhanced primers for amplification of DNA barcodes from a broad range of marine metazoans. BMC Ecol. 13 (1), 1–8. doi: 10.1186/1472-6785-13-34
Marine Stewardship Council Ending shark finning. Available at: https://www.msc.org/what-we-are-doing/ending-shark-finning (Accessed 19 Aug 2022).
Neo S., Kibat C., Wainwright B. J. (2022). Seafood mislabelling in Singapore. Food Control 135, 108821. doi: 10.1016/j.foodcont.2022.108821
Passantino A. (2013). The EU shark finning ban at the beginning of the new millennium: The legal framework. ICES J. Mar. Sci. 71 (3), 429–434. doi: 10.1093/icesjms/fst190
Poisson F., Filmalter J. D., Vernet A. L., Dagorn L. (2014). Mortality rate of silky sharks (Carcharhinus falciformis) caught in the tropical tuna purse seine fishery in the Indian ocean. Can. J. Fish. Aquat. Sci. 71 (6), 795–798. doi: 10.1139/cjfas-2013-0561
Rehman A., Jafar S., Raja N. A., Mahar J. (2015). Use of DNA barcoding to control the illegal wildlife trade: A CITES case.
Rigby C. L., Bin Ali A., Derrick D., Fernando D., Haque A. B., Maung A. (2021) Rhizoprionodon oligolinx. the IUCN red list of threatened species 2021. Available at: https://dx.doi.org/10.2305/IUCN.UK.2021-2.RLTS.T41851A173435874.en (Accessed March 11, 2022).
Seah Y. G., Kibat C., Hew S., Wainwright B. J. (2022)DNA Barcoding of traded shark fins in peninsular Malaysia Rev. Fish Biol. Fish. doi: 10.1007/s11160-022-09713-y
Sembiring A., Pertiwi N. P. D., Mahardini A., Wulandari R., Kurniasih E. M., Kuncoro A. W., et al. (2015). DNA Barcoding reveals targeted fisheries for endangered sharks in Indonesia. Fish. Res. 164, 130–134. doi: 10.1016/j.fishres.2014.11.003
SFA (2022). Available at: https://www.sfa.gov.sg/tools-and-resources?topicIds=&personIds=&categoryIds=&typeIds=&page=2 (Accessed 23 March 2022).
Simpfendorfer C. A., Dulvy, Nicholas K. (2017). Bright spots of sustainable shark fishing. Curr. Biol. 27 (3), R97–R98. doi: 10.1016/j.cub.2016.12.017
Steinke D., Bernard A. M., Horn R. L., Hilton P., Hanner R., Shivji M. S. (2017). DNA Analysis of traded shark fins and mobulid gill plates reveals a high proportion of species of conservation concern. Sci. Rep. 7 (1), 1–6. doi: 10.1111/jiec.13189
Teo L. G. P. (2015). Man eating shark: Unravelling the debate on the (Un) ethical consumption of shark’s fin in Singapore (Singapore: National University of Singapore). Available at: https://core.ac.uk/download/pdf/48811915.pdf.
Van Houtan K. S., Gagné T. O., Reygondeau G., Tanaka K. R., Palumbi S. R., Jorgensen S. J. (2020). Coastal sharks supply the global shark fin trade. Biol. Lett. 16 (10), 20200609. doi: 10.1098/rsbl.2020.0609
Wainwright B. J., Ip Y. C. A., Neo M. L., Chang J. J. M., Gan C. Z., Clark-Shen N., et al. (2018). DNA Barcoding of traded shark fins, meat and mobulid gill plates in Singapore uncovers numerous threatened species. Conserv. Genet. 19 (6), 1393–1399. doi: 10.1007/s10592-018-1108-1
Keywords: conservation, CITES, DNA barcoding, IUCN, shark fins, Singapore
Citation: Drescher L, Heng NJK, Chin MY, Karve NRO, Cheung EJW, Kurniadi A, Urera MQ, Waldeck FG, Dharshini U, Hoe NTE, Choo JSY, Lok RFJ, Kibat C and Wainwright BJ (2022) Blood in the water: DNA barcoding of traded shark fins in Singapore. Front. Mar. Sci. 9:907714. doi: 10.3389/fmars.2022.907714
Received: 30 March 2022; Accepted: 22 August 2022;
Published: 04 October 2022.
Edited by:
Maria Grazia Pennino, Spanish Institute of Oceanography (IEO), SpainReviewed by:
Nelson Jurandi Rosa Fagundes, Federal University of Rio Grande do Sul, BrazilCopyright © 2022 Drescher, Heng, Chin, Karve, Cheung, Kurniadi, Urera, Waldeck, Dharshini, Hoe, Choo, Lok, Kibat and Wainwright. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Benjamin J. Wainwright, QmVuLldhaW53cmlnaHRAWWFsZS1OVVMuZWR1LnNn
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.