- 1Department of Earth and Planetary Sciences, University of California, Santa Cruz, Santa Cruz, CA, United States
- 2Department of Biological Sciences, University of Southern California, Los Angeles, CA, United States
- 3Department of Oceanography, Texas A&M University, College Station, TX, United States
- 4Agriculture and Agri-Food Canada, Swift Current Research and Development Centre, Swift Current, SK, Canada
- 5School of Marine and Environmental Sciences, University of South Alabama, Mobile, AL, United States
- 6Dauphin Island Sea Lab, Dauphin Island, AL, United States
- 7Institute of Marine Sciences, University of California, Santa Cruz, Santa Cruz, CA, United States
Our understanding of phosphorus (P) dynamics in the deep subseafloor environment remains limited. Here we investigate potential microbial P uptake mechanisms in oligotrophic marine sediments beneath the North Atlantic Gyre and their effects on the relative distribution of organic P compounds as a function of burial depth and changing redox conditions. We use metagenomic analyses to determine the presence of microbial functional genes pertaining to P uptake and metabolism, and solution 31P nuclear magnetic resonance spectroscopy (31P NMR) to characterize and quantify P substrates. Phosphorus compounds or compound classes identified with 31P NMR include inorganic P compounds (orthophosphate, pyrophosphate, polyphosphate), phosphonates, orthophosphate monoesters (including inositol hexakisphosphate stereoisomers) and orthophosphate diesters (including DNA and phospholipid degradation products). Some of the genes identified include genes related to phosphate transport, phosphonate and polyphosphate metabolism, as well as phosphite uptake. Our findings suggest that the deep sedimentary biosphere may have adapted to take advantage of a wide array of P substrates and could play a role in the gradual breakdown of inositol and sugar phosphates, as well as reduced P compounds and polyphosphates.
1 Introduction
Phosphorus (P) is a key macronutrient for life involved in the structure of nucleic acids in the form of a phosphate–ester backbone, and in the transmission of chemical energy in the form of adenosine triphosphate (ATP). Phosphorus is also a structural constituent in many cell components such as phosphoproteins, and phospholipids in cell membranes. In addition, P is involved in many cellular processes through the mechanism of phosphorylation, which alters the function and activity of proteins. Hence, P plays an important role in sustaining microbial life in the deep subseafloor environment. This environment is estimated to comprise as many prokaryotic cells as the marine water column and soils, and includes significant prokaryotic populations at depths greater than two kilometers (Roussel et al., 2008; Kallmeyer et al., 2012; Inagaki et al., 2015). Estimates based on cell counts from both ocean margins and oligotrophic ocean gyres sediments suggest that the deep sedimentary subseafloor biosphere could contain 0.18-3.6% of Earth’s total biomass and an equal amount of microbial cells as the marine water column (Kallmeyer et al., 2012). The existence and activity of deep subseafloor sediment microbial populations are likely to have profound implications on global biogeochemical cycles and our understanding of the limits of life. However, there is a substantial knowledge gap regarding the nature of P compounds sustaining life in oligotrophic open ocean sediments, and how microbial activity impacts the relative distribution of P compounds in general, especially organic P compounds, in marine sediments. Furthermore, little is known about the impact of the deep biosphere on sedimentary P cycling and P diagenetic processes. This is largely because of analytical difficulties associated with studying P cycling, particularly organic P, in deep ocean sediments that contain low concentrations of organic matter (Ruttenberg, 2014). Therefore, a better understanding of the mechanisms used by deep subseafloor microbes to obtain this essential nutrient is needed.
This study’s field site is located at North Pond, an approximately 8 × 15 km sediment-filled basin located on the western flank of the Mid-Atlantic ridge sampled during IODP Expedition 336 (Expedition 336 Scientists, 2012a). Such sediment ponds surrounded by high relief topography are common to the western flank of the Mid-Atlantic Ridge, and are thought to be representative of slow-spreading ridge flank environments (Becker et al., 2001). Sediments at North Pond are predominantly nannofossil ooze with layers of coarse foraminiferal sand and occasional pebble-sized clasts of basalt, serpentinite, gabbroic rocks, and bivalve debris (Expedition 336 Scientists, 2012a). Organic carbon and nitrogen contents in those sediments are low (Ziebis et al., 2012), and organic P was detected at low concentrations throughout the sediment column using sequential sediment extractions (Defforey and Paytan, 2015). In addition, oxygen penetrates deep in the sediment column, and has a “C” shaped depth profile due to oxygen fluxes from oxygenated seawater at the sediment-seawater interface and oxygenated fluids in the underlying basalt crust, with the middle portion of the cores being suboxic to anoxic (Orcutt et al., 2013). Deep sea, carbon-depleted sediments such as the ones sampled in this study represent approximately 70% of the seafloor (Mewes et al., 2016).
In this study, we present metagenomic data from a sediment sample collected at 59 m below seafloor (mbsf), as well as solution 31P nuclear magnetic resonance spectroscopy (31P NMR) data from two depths in the sediment column (3.2 and 56.7 mbsf). The combination of these data yields insights into putative P uptake mechanisms and transformations of P substrates by deep subseafloor microorganisms. This work also improves our knowledge of the role deep subseafloor bacteria play in P cycling and our understanding of the chemical composition of organic P in organic-lean oligotrophic sediments.
2 Materials and Methods
2.1 Sample Collection and Storage
The sediment samples used in this study were collected from Site U1382 (Hole U1382B) at North Pond using the advanced piston corer during IODP Expedition 336 in October-November 2011 (Figure 1). Site U1382 is located in the southeastern part of North Pond, where sediments are approximately 90 m thick (Expedition 336 Scientists, 2012b). The samples used for this study were U1382B 1H-3, from the oxic portion of the sediment column [average depth 3.2 mbsf, 30 μM dissolved oxygen (DO)] and U1382B 7H-5 (average depths 56.7 mbsf, 1 μM DO; Orcutt et al., 2013). Samples from both depths were used for 31P NMR spectroscopy, while sample U1382B 7H-5 from the suboxic part of the sediment column was used for metagenomic analyses due to problems with the shallow sample during sequencing. Fluorescent microspheres (Fluoresbrite Carboxylate Microspheres; Polysciences, Inc., 15700) were used in every recovered core to monitor any potential contamination (Expedition 336 Scientists, 2012a). Upon recovery, whole-round sediment cores including the sample used here for metagenomic analyses were sectioned on the catwalk, capped and stored at –80°C in sterile bags (Expedition 336 Scientists, 2012c). The sediment samples used for 31P NMR analyses were squeezed-cakes stored at –80°C after porewater collection immediately following core retrieval.
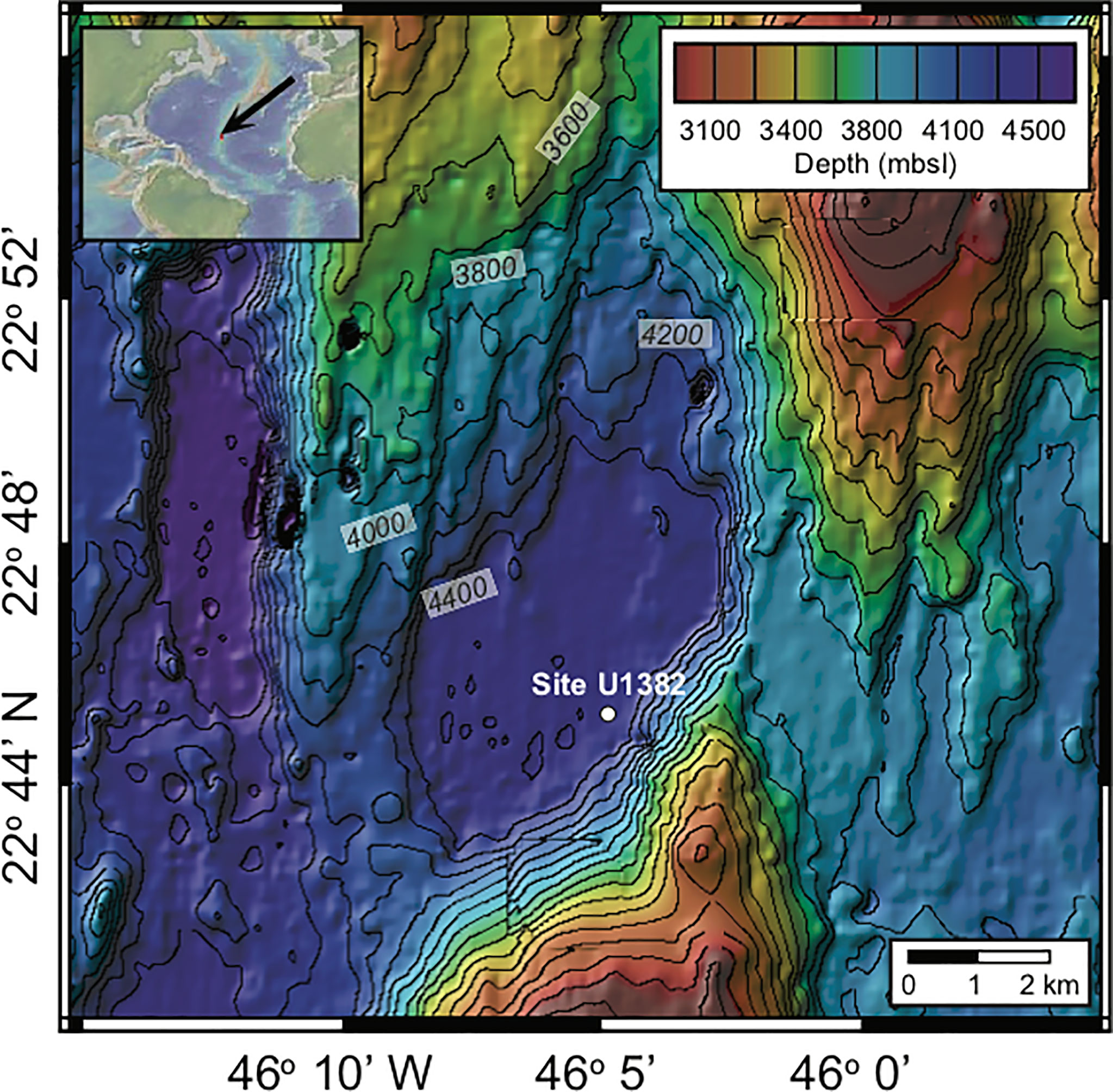
Figure 1 Location of the North Pond field site, located beneath the North Atlantic Gyre. The color scales indicate water depth for each site in meters below sea level (mbsl). Contour line intervals are 100 m for North Pond and the location of Site U1382 is indicated with a white dote. Map created using the default Global Multi-Resolution Topography Synthesis basemap (Ryan et al., 2009) in GeoMapApp version 3.6.0 (http://www.geomapapp.org).
2.2 DNA Extraction and Sequencing
2.2.1 DNA Extraction
The method described in Mills et al. (2012) as modified by B. Kiel Reese for nucleic acids in low biomass open ocean sediments (Kiel Reese et al., 2018) was used to extract DNA. The interior of the whole-round core for sample U1382B 7H-5 was subsampled using sterile techniques and was divided into 25 splits, each weighing ~0.5 g. Initial cell lysis was achieved using five cycles of freeze (liquid nitrogen), thaw (55°C water bath) and vortex steps while stabilizing nucleic acids in a Tris–EDTA–glucose buffer. This step was followed by the addition of lysozyme to the buffer and incubation at 30°C for 10 minutes. Samples were then treated twice with buffered phenol, chloroform and isoamyl alcohol (25:24:1; pH 8.0), and sodium dodecyl sulfate to dissolve the cell membrane and solubilize both high and low molecular weight proteins. Nucleic acids within the aqueous layer above the phenol–chloroform layer were then precipitated in an ethanol and sodium acetate solution. The ethanol solution was decanted following centrifugation of samples at 4°C. Lastly, DNA pellets were resuspended in sterile water, combined into one sample. We quantified the DNA content using a Quibit fluorometer (Thermo Scientific, Waltham, MA, USA) and assessed its quality on a NanoDrop 1000 Spectrophotometer (Thermo Scientific, Waltham, MA, USA) prior to sequencing at the Marine Biological Laboratory Keck facility (Woods Hole, MA, USA). Extraction blanks were included with each sample extraction and yielded no measurable DNA on NanoDrop or Qubit, or observable DNA following PCR and gel electrophoresis.
2.2.2 Sequencing
Metagenome library construction and sequencing was performed through the Deep Carbon Observatory’s Census of Deep Life program on an Illumina HiSeq platform. The metagenome sequences can be accessed through the Marine Biological Lab’s Visualization and Analysis of Microbial Population Structures database (Huse et al., 2014).
2.3 Bioinformatic Analyses
Raw reads were quality controlled using cutadapt (-e 0.08 –overlap=3; v1.7.1) and Trimmomatic (SLIDINGWINDOW:10:28 MINLEN:75; v0.33). The QCed sequences were searched for SSU rRNA fragments using Meta-RNA (v.H3). SSU rRNA fragments were assembled using EMIRGE (v.1.3) utilizing SILVA SSU as the reference database using the SINA web portal aligner (Ludwig et al., 2004; Capella-Gutiérrez et al., 2009; Pruesse et al., 2012). For sample U1382B 7H-5, 23 full-length SSU rRNAs were reconstructed. Taxonomy was assigned to these assemblies using the mothur (v.1.3.7) commands align.seqs and classify.seqs and SILVA SSU as the reference database. The quality-checked (QCed) sequence library was aligned to its assembled SSU rRNAs using Bowtie2 (v2.2.5). Based on the total number of QCed sequences that aligned to the SSU rRNAs, a length-normalized relative abundance value was determined. Results showed that the sample contained some degree of human-related contamination, both from human DNA (~2%) and human microbiome DNA (~6.7%). In order to verify that the functional genes related to P metabolism were not from human microbiome DNA sequences, we constructed a 16S rRNA gene phylogenetic tree from the metagenome to help determine the origin of microbial taxa (see methods below; Supplementary Figure 1). Those most closely related to environmental taxa were considered valid, while those similar to known contaminants, such as common components of the human skin microbiome, were considered contaminants. We also excluded any sequences from the known human microbiome (Propionobacterium, Microbacteriaceae), as well as sequences without high-level taxonomic assignment from the Proteobacteria (that could include Enterobacteriaceae). The QCed sequence library was assembled using IDBA-UD (v.1.1.1), but the overall quality of these assemblies was low (contigs: 49614 n 50: 467 max: 48419 mean: 461). A second round of assembly using all of the contigs from the IDBA-UD output was performed using the Geneious De Novo assembler (v.6.3.8; Kearse et al., 2012). This step generated a total of approximately 130 contigs >2.5kb in length. This number was too small for genome binning analysis. To identify genes within the assembled dataset related to P metabolism, all putative coding DNA sequences (CDSs) were determined using Prodigal (v2.6.3), using the metagenome option (-p meta). The identified CDS were compared to the FOAM KEGG database (Prestat et al., 2014) using hmmsearch (Eddy, 2011; v3.1b2; parameters, –cut_tc –notextw) and P-related KO matches were extracted from the output. Additionally, a collection of proteins related to P metabolism collected from UniProtKB were compared to the predicted CDSs using BLASTP (Camacho et al., 2009; v 2.2.30+, parameters, e-value of 1×10-20, max_target_seqs 1).
2.4 16S rRNA Phylogenetic Tree
Full-length 16S rRNA gene sequences were generated using EMIRGE (as described above) and were greater than 1,000 bp in length, with the exception of ID 973 due to poor resolution within the sequence. The sequences were aligned to the SILVA SSU reference database using the SINA web portal aligner (https://www.arb-silva.de/aligner/; Ludwig et al., 2004; Capella-Gutiérrez et al., 2009; Pruesse et al., 2012). These alignments were loaded in to ARB (v6.0.3), manually assessed and added to the non-redundant 16S rRNA gene database (SSURef123 NR99) using ARB Parsimony (Quick) tool (parameters: default). We identified a selection of the nearest neighbors to the sediment metagenome sequences to be used to construct a 16S rRNA gene phylogenetic tree. We also identified additional sequences related to the environmental 16S rRNA gene using a BLAST search against the NCBI RefSeq and non-redundant databases. Environment and reference 16S rRNA gene sequences were aligned using MUSCLE (v3.8.31; parameters: -maxiters 8), processed by the automated trimming program trimAL (v1.2rev59; parameters: -automated1), and aligned for a second time using MUSCLE (parameters: -maxiters 8). The alignment was used to construct an approximate maximum-likelihood phylogenetic tree using FastTree (v2.1.3; parameters: -nt -gtr –gamma; Price et al., 2010).
2.5 Solution 31P NMR Extraction
2.5.1 Sample Extraction
Two sediment samples, U1382B 1H-3 (3.2 mbsf) and 7H-5 (56.7 mbsf), were processed following the method described in Defforey et al. (2017). This method quantitatively removes orthophosphate prior to the alkaline extraction for solution 31P NMR to amplify the signal of low-abundance P compounds, including organic P, relative to that of orthophosphate so that these P forms in oligotrophic sediments may be resolved without the need for extremely long (>24 h) NMR experiments. Briefly, each freeze-dried, ground and sieved sample (<125 μm) was divided into eight splits (1.5 g each). Sample splits were extracted in a citrate–dithionite–bicarbonate buffer to solubilize P bound to iron (Fe) oxyhydroxides (8 h; pH 7.6), after which the sediment residues were washed with ultrapure water (18.2 MΩ·cm deionized water; 2 h). The sediment residues were then extracted in a sodium acetate buffer (6 h; pH 4.0) to solubilize P bound to carbonate fluorapatite, and washed with ultrapure twice (2 h each). After this step, the 8 splits for each sample were combined and extracted in 0.25 M NaOH + 0.05 M Na2EDTA for 6 h and then centrifuged. Sediment residues were ashed at 550°C and extracted in 0.5 M sulfuric acid (16 h) to determine residual P content, and the NaOH-EDTA extracts were frozen and lyophilized. Prior to solution 31P NMR analysis, lyophilized extracts were redissolved in 500 μL D2O, H2O, NaOH–EDTA and 10 M NaOH.
2.5.2 Solution 31P NMR Analysis and Post-Experimental Processing
Spectra were acquired immediately following sample preparation on a Varian Unity INOVA 600 MHz spectrometer equipped with a 10 mm broadband probe [operated by the Stanford Magnetic Resonance Laboratory at Stanford University]. The analytical parameters used were: 20°C, 90° pulse, 0.48 s acquisition time, 4.52 s delay time, 5600 scans (8 h experiments), no spin and an external H3PO4 standard. No proton decoupling was used out of concern for sample degradation (Cade-Menun and Liu, 2014). We used the ratio of P concentration to Fe plus manganese (Mn) concentrations (P/Fe+Mn) as a proxy to estimate spin-lattice relaxation times (T1) to ensure adequate delays between pulses and thus quantitative spectra (McDowell et al., 2006; Cade-Menun and Liu, 2014). We used 5 s recycle delays, which correspond to three to five times the calculated T1 values. Peak identification was based on spiking with known compounds (α- and β-glycerophosphate) and on literature values (Cade-Menun, 2015). Monoester 1 refers to unidentified orthophosphate monoesters located between 7.5–6.1 ppm, while monoesters 2 and 3 include peaks between 5.7–3.5 ppm and 3.5–2.5 ppm respectively (Cade-Menun, 2017). We processed 31P NMR data using the NMR Utility Transform software (NUTS, Acorn NMR) and calculated peak areas by integration of spectra processed with a 7 Hz line broadening for the full spectrum and 2 Hz line broadening for the monoester region following baseline correction, peak picking and phasing.
2.5.3 Supernatant P Analyses
We measured total P concentrations in sediment extracts using inductively coupled plasma optical emission spectroscopy (ICP-OES) [Perkin-Elmer Optima 4300 DV Inductively Coupled Plasma Optical Emission Spectrometer operated by the University of California, Santa Cruz]. Standards were prepared with the same solutions as those used for the extraction procedure and ranged from 0 – 320 μM P. The detection limit for P on this instrument for both wavelengths is 0.4 μM. We measured molybdate reactive P concentrations (MRP, which includes primarily free orthophosphate) on a QuikChem 8000 automated ion analyzer. Standards were prepared with the same solutions used for the extraction step to minimize matrix effects on P measurements, and ranged from 0 – 30 μM P. The detection limit for P on this instrument is 0.2 μM. We derived molybdate unreactive P concentrations (MUP, which includes primarily organic P and polyphosphates) by subtracting MRP from total P concentrations.
3 Results and Discussion
3.1 Bioinformatics Data
A target subset of P-related genes was identified in sample U1382B 7H-5 (59.1 mbsf), consisting of 23 genes of interest including phosphate utilization (transport, pstABS; metabolism, phoABDEHR), phosphonate utilization (transport and metabolism; phnACDGJ), phosphite utilization (ptxABCD), glycerol 3-phosphate transport (ugpBC), polyphosphate metabolism (ppKX), and phosphatase activity (appAB; Table 1; Supplementary Table 1). This set of genes was split into two groups. Genes in group 1 (pstABS, ppX, ugpBC, ptxA, phoDR) were present in the FOAM database (accessed April 2016), a curated set of hidden Markov models (HMMs) generated using the KEGG ontology database that could be used to search conserved motifs within the environmental sequences. For FOAM searchable genes of interest (group 1), environmental sequences (excluding phoR and upgC) were searched using hmmsearch (v3.1b2; parameters, –cut_tc –notextw) with a bit score cutoff of 72 (approximately equivalent to an e-value of 1×10-20). Gene sequences for phoR and upgC represented protein subunits with generally highly conserved regions: sensor histidine kinases and ATP-binding motifs for ABC-type transporters, respectively. As such, these matches could not be trusted at the same cutoff enforced for the other genes within group 1, and a more stringent cutoff of 105 bit score (approximate e-value equivalent 1×10-50) was used. The results from the FOAM database identified the presence of ppX (exopolyphosphatase) and pstAB, the ATP-binding and permease subunits of the phosphate ABC-type transporter system. However, the crucial pstS, solute-binding subunit, was not identified. Furthermore, even with the more stringent cutoff values, 23 and 38 copies of upgC and phoR were identified. This large number suggests that even at this cutoff value, a more diverse group of these gene families was identified. The remaining set of genes of interest (group 2), including ptxABCD, ppK, phoABED and phnACD were used to search the remaining putative CDS using BLASTP (v 2.2.30+, parameters, e-value of 1×10-20, max_target_seqs 1). A total of 187 matches to the environmental sequences were identified. However, the problematic nature of the conserved region within the ATP-binding subunit of the ABC-type transporters resulted in 107 matches to environmental sequences. These were deemed uninformative and were excluded from further analysis. Matches were identified for appAB, phoD, phnADGJ, ptxCD, ppK. A large portion of the BLAST matches (after ATP-binding subunits) belonged to phoB (n = 37), a phosphate regulon response regulator, but this protein family is diverse and multiple types of regulon genes may be identified in this manner. Lastly, for each of the genes identified as related to P uptake and metabolism (both FOAM and BLAST databases), a putative taxonomy was assigned. Putative P-related environmental genes were queried using BLASTP (parameters, e-value 1×10-5, -max_target_seqs 5 -outfmt 5) against the GenBank RefSeq (v75) database. The MEGAN (v4) Last Common Ancestor (LCA) algorithm was used to assign a putative taxonomy based on the top five BLAST matches (Table 1). Most P related gene sequences belonged to Alphaproteobacteria, or to a combination of Alphaproteobacteria, Betaproteobacteria, Actinobacteria and unclassified microorganisms (Table 1). The presence of these microorganisms is further confirmed by 16S rRNA sequence data. The metagenomic analyses performed in this study only yield information regarding the presence of functional genes pertaining to P metabolism, and these may represent DNA from active and inactive or dormant microorganisms. Indeed, most phylogenetic groups identified in North Pond sediments have been shown to be inactive (Kiel Reese et al., 2018). However, active Alphaproteobacteria (mainly from the Caulobacteraceae family and Brevundimonas genus) have been found throughout the sediment column at Hole U1382B, as well as some active Betaproteobacteria (Achromobacter genus) and Gammaproteobacteria (Pseudomonas genus, and the genera Stentrophomonas and Methylophaga; Kiel Reese et al., 2018). This suggests that the functional genes reported in our study are present at depths where microorganisms are active and utilizing P and may thus be impacting sedimentary organic matter remineralization through their metabolic activity.
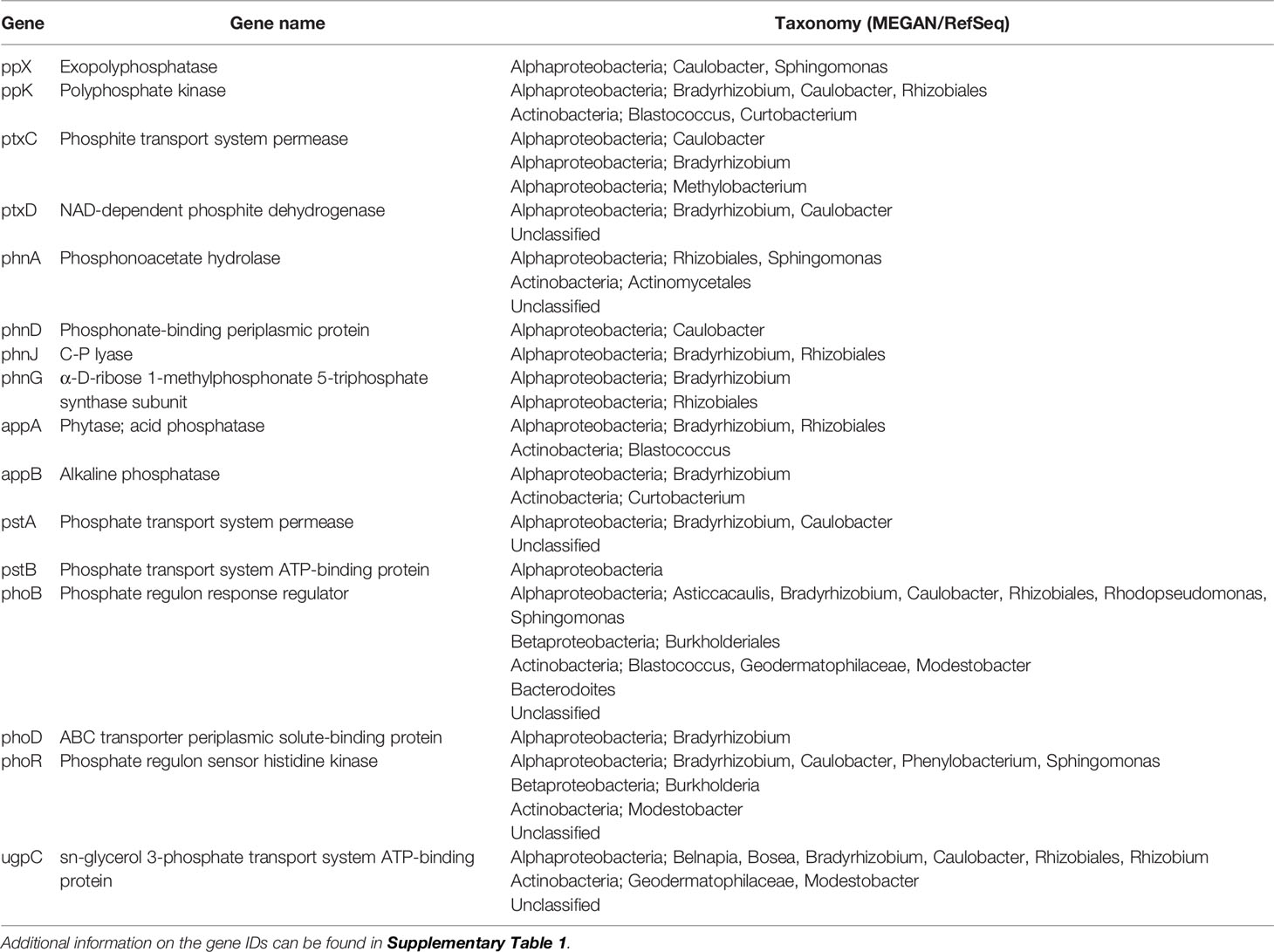
Table 1 Genes related to P uptake and metabolism identified using metagenomic analyses of sample U1382B 7H-5 (59.1 mbsf).
3.2 P NMR Data
Solution 31P NMR spectra for both samples are shown in Figure 2 and Supplemental Figure 2, and chemical shifts of peaks for compounds and compound classes are shown in Supplementary Table 2. The 2–step pretreatment preceding the alkaline extraction removed 95% of orthophosphate in sample U1382B 1H-3, and 98% in sample U1382B 7H-5 with no measurable MUP removal (Table 2; Figure 3). For both samples, the 5-s recycle delays were long enough to produce quantitative spectra (Table 3). Metal content was similar in both NMR extracts, but P content was lower for the deeper sample (Table 3). The shallower, oxic sample (U1382B 1H-3, 3.2 mbsf, 30 μM DO) contained a wider variety of P compounds than the deeper, suboxic sample (U1382B 7H-5, 56.7 mbsf, ~1 μM DO), and a greater concentration of Organic P detected in NMR spectra (Table 4). The most abundant P compounds in both samples identified using solution 31P NMR after the 2–step pretreatment were inorganic orthophosphate, pyrophosphate and polyphosphates (Table 4; Figure 2). Polyphosphates are chains of phosphates linked by anhydrous bonds; pyrophosphates are polyphosphates with only two phosphate groups. Both polyphosphates and pyrophosphates can be associated with living organisms, and can be inorganic or organic; ATP is an example of an organic polyphosphate (Heinonen, 2001; Condron et al., 2005). Pyrophosphate and polyphosphates have been identified by solution 31P NMR in culture-grown algae (Cade-Menun and Paytan, 2010) in estuary and river sediments (Shan et al., 2016; Watson et al., 2018), and in marine sediments by solid-state and solution 31P NMR (Sannigrahi and Ingall, 2005; Prüter et al., 2020). We also measured small amounts of organic P compounds, including phosphonates (reduced organic P compounds with a direct C–P bond), and orthophosphate monoesters and diesters (Table 4; Figure 2). Orthophosphate monoesters were the most abundant forms of organic P in both samples, which is consistent with numerous studies that have shown them to generally be the dominant group of organic P compounds in both marine and terrestrial environments (Ingall et al., 1990; Carman et al., 2000; Paytan et al., 2003; Sannigrahi and Ingall, 2005; Turner et al., 2005; Shan et al., 2016; Watson et al., 2018; Prüter et al., 2020). In the shallower sample, we were able to resolve individual peaks in the orthophosphate monoester region and identify a number of different monoester compounds (Table 4). Among them, the most abundant monoester compounds identified were the inositol hexakisphosphate (IHP) stereoisomers myo- and scyllo-IHP. Inositol phosphates form the dominant class of organic P compounds in soils, and are abundant in aquatic systems (Turner et al., 2002). Of all IHP stereoisomers, myo-IHP is the most abundant, but scyllo-, D-chiro- and neo-IHP have also been measured in soils and marine sediments (White and Miller, 1976; Turner et al., 2012). Peaks consistent with D-chiro- and neo-IHP were detected in the Monoester 1 region of these spectra (Supplementary Table 1), but were not specifically identified due to uncertainties about peak identification and low concentrations. Peaks for α- and β–glycerophosphate were also identified in the spectrum of the shallower sample. Although these are included with orthophosphate monoesters, they are artifacts from degradation during extraction and analysis, from phospholipids in the original samples, which are orthophosphate diesters (Cade-Menun, 2015). As such, they were included in the calculation for total orthophosphate diesters rather than total orthophosphate monoesters (Table 4). Peaks for DNA were identified in the orthophosphate diester region of spectra for both depths (Table 4). Other peaks in the orthophosphate diester region are consistent with phospholipids (Supplementary Table 2; Cade-Menun, 2015), but were not specifically identified.
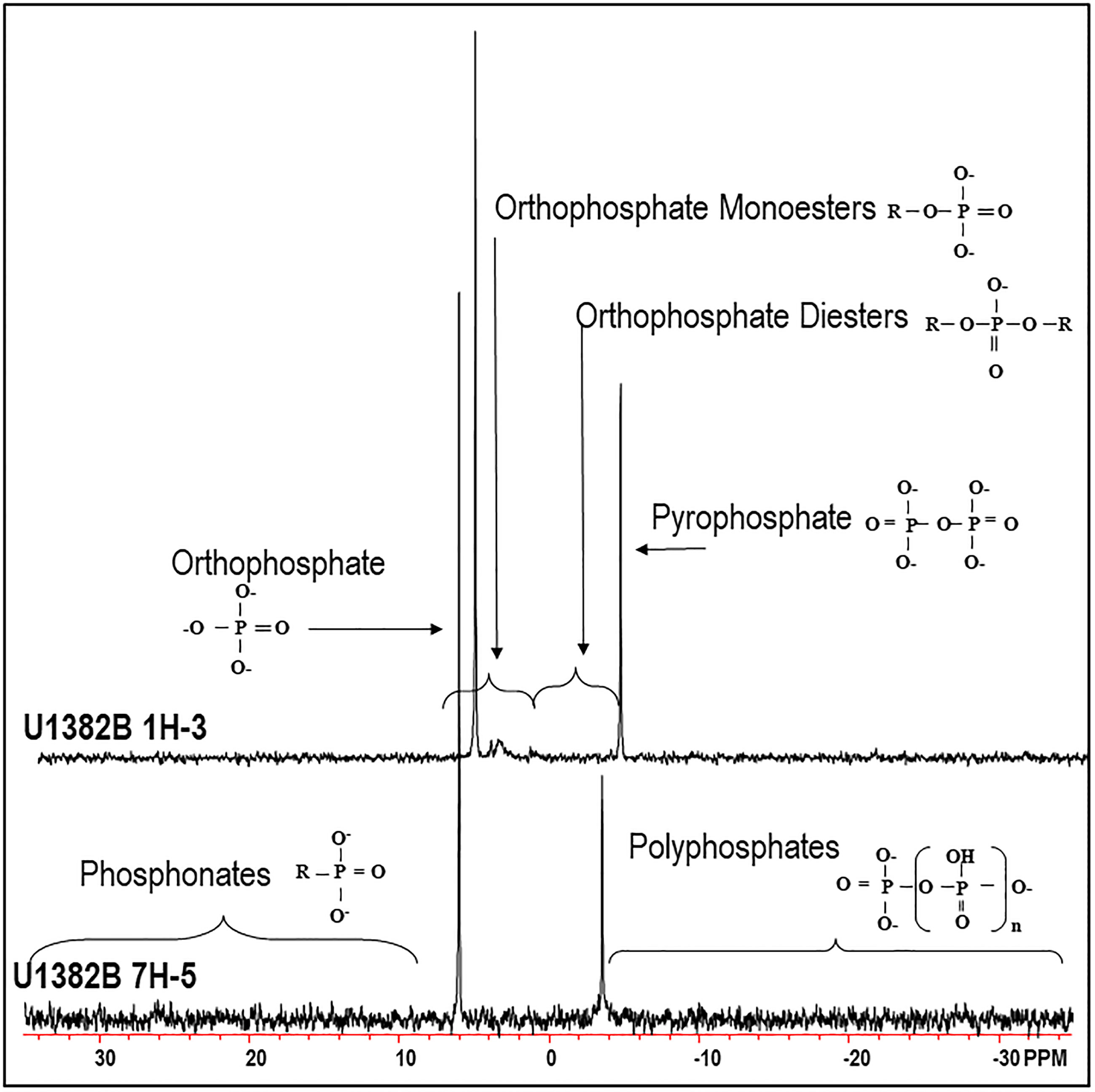
Figure 2 Solution 31P NMR spectrum of sample U1382B 1H-3 (3.2 mbsf, top), and U1382B 7H-5 (56.7 mbsf, bottom). Spectra were processed with 7 Hz line-broadening, and are plotted with the orthophosphate peak at the same height. See Supplementary Figure 2 for more details of the orthophosphate monoester region.
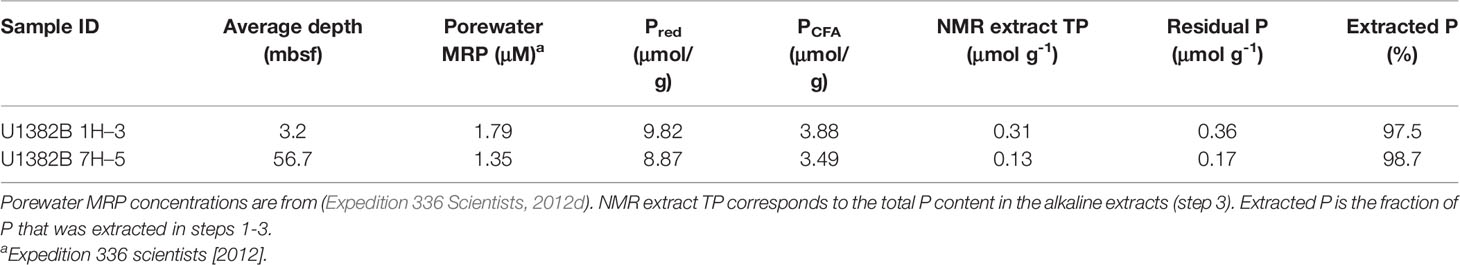
Table 2 Extraction yields for each step. Pred refers to P solubilized during the reductive step (step 1), PCFA refers to P released during the sodium acetate step (step 2).
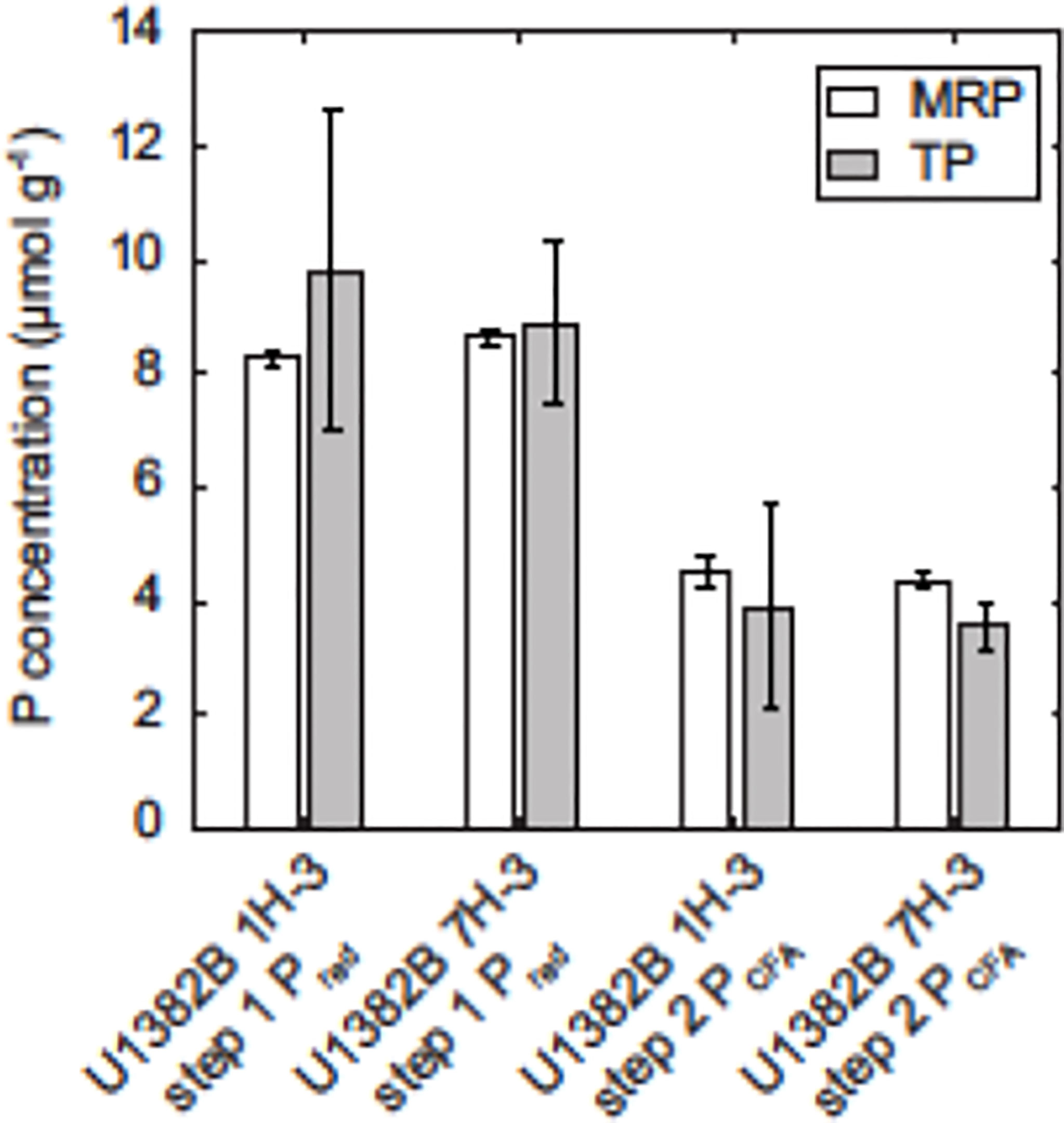
Figure 3 Mean molybdate-reactive P (MRP) and total P content in pretreatment extracts from steps 1 (Pred, citrate–dithionite–bicarbonate and water extracts) and 2 (PCFA, sodium acetate and water extracts), along with standard deviations derived from replicate sediment sample splits. Total P concentrations were measured on an ICP-OES, while MRP concentrations were measured colorimetrically on a QuikChem 8000 automated ion analyzer.
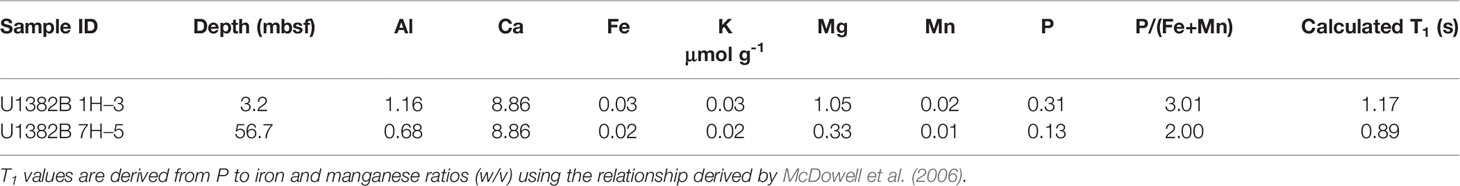
Table 3 Aluminum, calcium, iron, potassium, magnesium, manganese and phosphorus content in 31P NMR extracts.
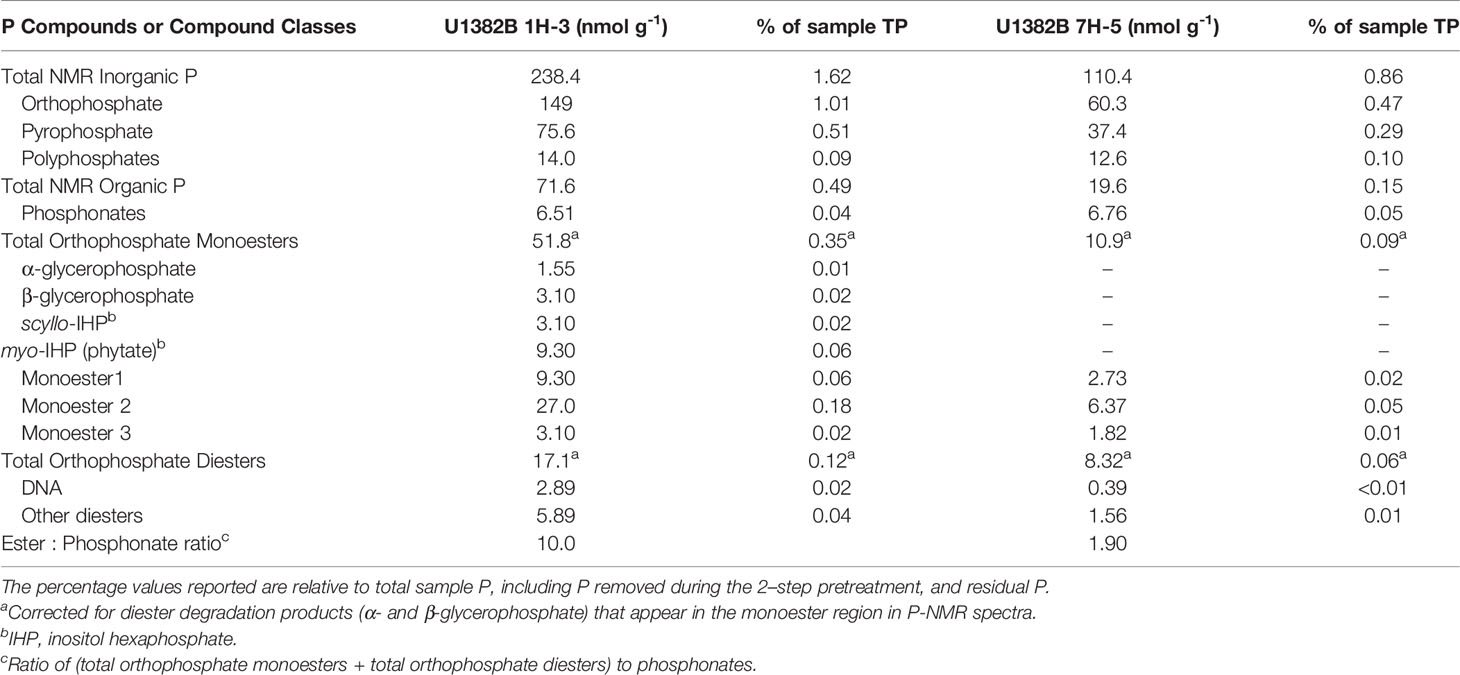
Table 4 Phosphorus compounds or compound classes identified from 31P NMR spectral analyses of samples U1382B 1H-3 (3.2 mbsf) and 7H-5 (56.7 mbsf).
3.3 Combining Metagenomic and P NMR Data
The combined use of metagenomic analyses and solution 31P NMR spectroscopy yields valuable information regarding P cycling by deep sedimentary subseafloor microorganisms, as well as microbial P uptake mechanisms. While we do not have a metagenome for the shallower sediments at Hole U1382B, the decrease in abundance of organic and biogenic P compounds (Table 4) suggests that deep sedimentary subseafloor microorganisms are utilizing those compounds as a P source, and possibly as an organic C source in this C-depleted environment (Heath, 2005; Ziebis et al., 2012). Decreases in organic P may also scale with a decrease in cell density; further research is warranted.
In the shallow sample, myo-IHP can adsorb on the same sites of Fe and aluminum (Al) oxides as phosphate anions since adsorption occurs through the phosphate groups via ligand exchange with surrounding porewater and hydroxide groups on mineral surfaces (Goldberg and Sposito, 1985; Baldwin et al., 2002; Celi and Barberis, 2005; Gérard, 2016). Little is known regarding possible differences in the behavior of other stereoisomeric forms of IHP, but they may adsorb on oxide surfaces in a similar manner as myo-IHP. Once sorbed on short-range ordered Al precipitates, myo-IHP and other P compounds, including sugar phosphates such as glucose 6-phosphate have been shown to be poorly available to microorganisms (Shang et al., 1996), which would allow them to persist at depth in marine sediments. However, surface properties of oxides are modified when associated with other minerals such as clay minerals, which affect their interactions with organic P and may weaken the complex formed with oxides (Celi et al., 2003), thus increasing their bioavailability. In addition to sorption on Fe and Al oxides, myo-IHP can form soluble complexes with calcium in calcareous sediments, which may enhance the interactions between myo-IHP and calcium-bearing minerals and further stabilize it (Celi and Barberis, 2005). The bioavailability of organic P compounds is primarily dependent on the stability of the phosphate surface complexes rather than by the total amount of phosphate adsorbed (Shang et al., 1996). Varying redox conditions have been shown to strongly regulate IHP remineralization, and lead to enhanced IHP decomposition under anaerobic conditions (Suzumura and Kamatani, 1995), and the presence of the appA gene, associated with phytase (Liu et al., 2018) is consistent with mineralization of IHP compounds. The observed decrease in orthophosphate monoester abundance with depth was also reported by Prüter et al. (2020) for sediments of the Baltic Sea, who also reported a decrease in orthophosphate monoesters with distance from the coast. These decreases could be due to decreases in cell density, or to an increase in bioavailability of those compounds resulting from the transition to suboxic conditions in the middle parts of the sediment column (Zhang et al., 1994; Table 4, Figure 2). Indeed, bacteria at this depth possess genes encoding for phosphomonoesterases, (appA, appB, phoD; Vershinina and Znamenskaya, 2002) and could utilize those deeply buried P compounds as they become more susceptible to enzymatic hydrolysis.
Orthophosphate diesters were also present at both depths as DNA in both samples, phospholipid degradation products in the shallow sample, and other diesters in both samples (Table 4, Figure 2). These compounds may originate from living organisms, and DNA may also be retained by sorption. The sorption of nucleic acids on mineral surfaces is affected by the molecular weight of DNA, the presence of montmorillonite, pH and redox conditions (Greaves and Wilson, 1969; Celi and Barberis, 2005; Condron et al., 2005). The complexed nucleic acid forms with montmorillonite are weaker than those with myo-IHP due to the presence of a single hydroxide group available for ligand exchange (Greaves and Wilson, 1969). This makes nucleic acids more vulnerable to microbial enzymatic hydrolysis (Greaves and Wilson, 1970). The decrease in orthophosphate diester abundance with depth and the presence of genes such as phoD and ugpC suggest putative mechanisms for the uptake of orthophosphate diesters (phoD) and glycerophosphoryl diesters (ugpC) at Site U1382 (Wanner, 1993; Vershinina and Znamenskaya, 2002).
Other potential sources of P, and possibly C, to the deep biosphere are phosphonates, which we measured both in shallow and deeper sediments (Table 4, Figure 2). While previously thought to be refractory, these compounds have been shown to be as bioavailable as phosphate esters, especially under anoxic conditions (Benitez-Nelson et al., 2004) and represent a potential source of both P and organic C to the deep biosphere (Sosa et al., 2019). Indeed, a number of genes pertaining to phosphonate utilization were identified in our sample: phnA (phosphonoacetate hydrolase), phnD (phosphonate-binding periplasmic protein), phnJ (component of C-P lyase complex) and phnG (component of C-P lyase complex; Table 1). The comparable phosphonate abundance (6.51 and 6.76 nmol g-1 at both depths), the high ester to phosphonate ratios that decrease with depth, the presence of genes related to phosphonate utilization and redox conditions shown to enhance phosphonate remineralization (i.e., reducing conditions) suggest that phosphonates represent a likely substrate for deep subseafloor microorganisms at that depth in the sediment column. This is consistent with the results of Sosa et al. (2019) showing that C-P lyase was more prevalent in ocean regions featuring sustained periods of phosphate depletion.
Small amounts of polyphosphates were measured in both spectra, and genes for exopolyphosphatase (ppX) were present in the metagenome (Table 1, Figure 2). This enzyme catalyzes the hydrolysis of the terminal phosphate group in polyphosphates (Jones et al., 2016, p. 2). We also identified gene sequences for polyphosphate kinase (ppK), which catalyzes the reversible synthesis of polyphosphate from nucleoside triphosphates (Jones et al., 2016). Polyphosphates are synthesized by all cells, and play an important role in nutrient storage, regulatory functions and metal chelation (Rao et al., 2009). They also play a key role in nucleating the growth of authigenic carbonate fluorapatite in marine sediments, which has profound implications for reactive P burial (Diaz et al., 2008).
An unexpected result from the metagenomic analyses was the presence of two genes related to phosphite metabolism: ptxC and ptxD. Phosphite utilization genes have been identified in diverse marine microorganisms, and their abundance is higher in marine environments with low P (Martínez et al., 2012). Furthermore, it was shown that phosphite and phosphonate production in surface waters could contribute to a global P redox cycle (Van Mooy et al., 2015). While we did not measure phosphite in our samples, and, to our knowledge, phosphite has not been measured in open ocean sediments, the presence of reduced organic P in the form of phosphonates and genes related to phosphite utilization raises the possibility of this compound potentially playing a role in P sediment cycling.
4 Conclusions
In this study, we presented a combination of metagenomics analysis and solution 31P-NMR for studying putative P uptake mechanisms and metabolism, as well as the distribution of organic and biogenic P substrates in deep subseafloor sediments. We demonstrated that diverse organic P compounds persist at depth in sediments underlying oligotrophic marine settings, and that changes in redox conditions may play an important role in increasing their bioavailability. We identified a wide variety of potential P sources to the deep biosphere, as well as putative enzymatic pathways for P uptake and utilization. While this study only provides potential P uptake mechanisms, it sets the stage for future work to investigate the expression of functional genes pertaining to P uptake and metabolism and raises interesting questions about microbial transformations of sedimentary organic and biogenic P.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.bco-dmo.org/project/664073, 664073.
Author Contributions
AP: Conceptualization, Project funding, Data interpretation, Writing. DD: Conceptualization, Analytical work, Data curation, Methodology, Visualization, Writing original draft. BT and JS: Bioinformatics, Metagenomic interpretation, Data curation. BC-M: Methods development, NMR data interpretation, Editing. BKR and LZ: Molecular data analysis and interpretation. All authors have read and approved the final version of this manuscript prior to submission.
Funding
Funding was provided by a graduate student fellowship from the Center for Dark Energy Biosphere Investigations (NSF C-DEBI) to DD (award # 157598), a student research grant from the Geological Society of America to DD, a C-DEBI Research and Travel Exchange Program grant to DD, a research grant from C-DEBI to AP (award # 15626), sequencing grant to AP, DD, JS and BKR from the Census of Deep Life and internal AAFC funding (J-002238) to BC-M.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank the Integrated Ocean Drilling Program and the shipboard scientists and technical staff involved in IODP expedition 336 who collected the sediment samples used in this study. We also thank Dr. Corey Liu, Rob Franks, Michael Kong and Richard Kevorkian for their assistance with this project.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2022.907527/full#supplementary-material
References
Baldwin D. S., Mitchell A. M., Olley J. M. (2002). “Pollutant-Sediment Interactions: Sorption, Reactivity and Transport of Phosphorus,” in Agriculture, Hydrology and Water Quality. Eds. Haygarth P. M., Jarvis S. C. (Wallingford, UK: CABI Publ), 265–280.
Becker K., Bartetzko A., Davis E. E. (2001). Leg 174B Synopsis: Revisiting Hole 395A for Logging and Long-Term Monitoring of Off-Axis Hydrothermal Processes in Young Oceanic Crust. In Becker K., Malone M. J.. (Eds.), Proc. ODP, Sci.Results, 174B, 1–13 [Online]. Available at: http://www-odp.tamu.edu/publications/174B_SR/VOLUME/SYNOPSIS/SR174BSY.PDF
Benitez-Nelson C. R., O’Neill L., Kolowith L. C., Pellechia P., Thunell R. (2004). Phosphonates and Particulate Organic Phosphorus Cycling in an Anoxic Marine Basin. Limnol. Oceanogr. 49, 1593–1604. doi: 10.4319/lo.2004.49.5.1593
Cade-Menun B. J. (2015). Improved Peak Identification in 31P-NMR Spectra of Environmental Samples With a Standardized Method and Peak Library. Geoderma 257-258, 102–114. doi: 10.1016/j.geoderma.2014.12.016
Cade-Menun B. J. (2017). Characterizing Phosphorus Forms in Cropland Soils With Solution 31P−NMR: Past Studies and Future Research Needs. Chem. Biol. Technol. Agric. 4, 19. doi: 10.1186/s40538-017-0098-4
Cade-Menun B. J., Liu C. W. (2014). Solution phosphorus-31 Nuclear Magnetic Resonance Spectroscopy of Soils From 2005 to 2013: A Review of Sample Preparation and Experimental Parameters. Soil Sci. Soc Am. J. 78, 19. doi: 10.2136/sssaj2013.05.0187dgs
Cade-Menun B. J., Paytan A. (2010). Nutrient, Temperature and Light Stress Alter Phosphorus and Carbon Forms in Culture-Grown Algae. Mar. Chem. 121, 27–36. doi: 10.1016/j.marchem.2010.03.002
Camacho C., Coulouris G., Avagyan V., Ma N., Papadopoulos J., Bealer K., et al. (2009). BLAST+: Architecture and Applications. BMC Bioinf. 10, 421. doi: 10.1186/1471-2105-10-421
Capella-Gutiérrez S., Silla-Martínez J. M., Gabaldón T. (2009). trimAl: A Tool for Automated Alignment Trimming in Large-Scale Phylogenetic Analyses. Bioinformatics 25, 1972–1973. doi: 10.1093/bioinformatics/btp348
Carman R., Edlund G., Damberg C. (2000). Distribution of Organic and Inorganic Phosphorus Compounds in Marine and Lacustrine Sediments: A 31P NMR Study. Chem. Geol. 163, 101–114. doi: 10.1016/S0009-2541(99)00098-4
Celi L., Barberis E. (2005). “Abiotic Stabilization of Organic Phosphorus in the Environment,” in Organic Phosphorus in the Environment. Eds. Turner B. L., Baldwin D. S. (Wallingford, UK: CABI Publ), 113–132.
Celi L., De Luca G., Barberis E. (2003). Effects of Interaction of Organic and Inorganic P With Ferrihydrite and Kaolinite-Iron Oxide Systems on Iron Release. Soil Sci. 168, 479–488. doi: 10.1097/01.ss.0000080333.10341.a4
Condron L. M., Turner B. L., Cade-Menun B. J. (2005). “Chemistry and Dynamics of Soil Organic Phosphorus,” in Phosphorus Agriculture and the Environment. Eds. Sims J. T., Sharpley A. N. (Madison, WI, USA: Soil Science Society of America), 87–121.
Defforey D., Cade-Menun B. J., Paytan A. (2017). A New Solution 31P NMR Sample Preparation Scheme for Marine Sediments. Limnol. Oceanogr.: Methods 15, 381–393. doi: 10.1002/lom3.10166\
Defforey D., Paytan A. (2015). Data Report: Characteristics of Sedimentary Phosphorus at North Pond, IODP Expedition 336. Proc. Integr. Ocean Drill. Program 336, 1–9. doi: 10.2204/iodp.proc.336.205.2015
Diaz J., Ingall E., Benitez-Nelson C., Paterson D., de Jonge M. D., McNulty I., et al. (2008). Marine Polyphosphate: A Key Player in Geologic Phosphorus Sequestration. Science 320, 652–655. doi: 10.1126/science.1151751
Eddy S. R. (2011). Accelerated Profile HMM Searches. PloS Comput. Biol. 7, e1002195. doi: 10.1371/journal.pcbi.1002195
Expedition 336 Scientists (2012a). Initiation of Long-Term Coupled Microbiological, Geochemical, and Hydrological Experimentation Within the Seafloor at North Pond, Western Flank of the Mid-Atlantic Ridge. IODP Proc. 336, 1–72. doi: 10.2204/iodp.pr.336.2012
Expedition 336 Scientists (2012b). Site U1382. Proc. Integr. Ocean Drill. Program 336, 1–141. doi: 10.2204/iodp.proc.336.104.2012
Expedition 336 Scientists (2012c). Methods. Proc. Integr. Ocean Drill. Program 336, 1–87. doi: 10.2204/iodp.proc.336.102.2012
Expedition 336 Scientists (2012d). Sediment and Basement Contact Coring. Proc. Integr. Ocean Drill. Program 336, 1–46. doi: 10.2204/iodp.proc.336.106.2012
Gérard G. (2016). Clay Minerals, Iron/Aluminum Oxides, and Their Contribution to Phosphate Sorption in Soils – A Myth Revisited. Geoderma 262, 213–226. doi: 10.1016/j.geoderma.2015.08.036
Goldberg S., Sposito G. (1985). On the Mechanism of Specific Phosphate Adsorption by Hydroxylated Mineral Surfaces: A Review. Commun. Soil Sci. Plant Anal. 16, 801–821. doi: 10.1080/00103628509367646
Greaves M. P., Wilson M. J. (1969). The Adsorption of Nucleic Acids by Montmorillonite. Soil Biol. Biochem. 1, 317–323. doi: 10.1016/0038-0717(69)90014-5
Greaves M. P., Wilson M. J. (1970). The Degradation of Nucleic Acids and Montmorillonite-Nucleic-Acid Complexes by Soil Microorganisms. Soil Biol. Biochem. 2, 257–268. doi: 10.1016/0038-0717(70)90032-5
Heath R. T. (2005). “Microbial Turnover of Organic Phosphorus in Aquatic Systems,” in Organic Phosphorus in the Environment. Eds. Turner B. L., Frossard E., Baldwin D. S. (Wallingford, UK: CABI Publ), 185–203.
Huse S. M., Welch D. B. M., Voorhis A., Shipunova A., Morrison H. G., Eren A. M., et al. (2014). Vamps: A Website for Visualization and Analysis of Microbial Population Structures. BCM Bioinf. 14, 41. doi: 10.1186/1471-2105-15-41
Inagaki F., Hinrichs K.-U., Kubo Y. (2015). Exploring Deep Microbial Life in Coal-Bearing Sediment Down to ~2.5 Km Below the Ocean Floor. Science 349, 420–424. doi: 10.1126/science.aaa6882
Ingall E. D., Schroeder P. A., Berner R. A. (1990). The Nature of Organic Phosphorus in Marine Sediments: New Insights From 31P NMR. Geochim. Cosmochim. Acta 54, 2617–2620. doi: 10.1016/0016-7037(90)90248-J
Jones D. S., Flood B. E., Bailey J. V. (2016). Metatranscriptomic Insights Into Polyphosphate Metabolism in Marine Sediments. ISME J. 10, 1015–1019. doi: 10.1038/ismej.2015.169
Kallmeyer J., Pockalny R., Adhikari R. R., Smith D. C., D’Hondt S. (2012). Global Distribution of Microbial Abundance and Biomass in Subseafloor Sediment. Proc. Natl. Acad. Sci. 109, 16213–16216. doi: 10.1073/pnas.1203849109
Kearse M., Moir R., Wilson A., Stones-Havas S., Cheung M., Sturrock S., et al. (2012). Geneious Basic: An Integrated and Extendable Desktop Software Platform for the Organization and Analysis of Sequence Data. Bioinformatics 28, 1647–1649. doi: 10.1093/bioinformatics/bts199
Kiel Reese B., Zinke L. A., Sobol M. S., LaRoue D. E., Orcutt B. N., Zhang X., et al (2018). Nitrogen Cycling of Active Bacteria Within Oligotrophic Sediment of the Mid-Atlantic Ridge Flank. Geomicrobiol. J. 35, 468–483. doi: 10.1080/01490451.2017.1392649
Liu J., Cade-Menun B. J., Bainard L., Yang J., Hu Y. F., Liu C. W., et al. (2018). Long-Term Land Use Affects Phosphorus Speciation and the Composition of Phosphorus Cycling Functional Genes in Agricultural Soils. Front. Microbiol. 9. doi: 10.3389/fmicb.2018.01643
Ludwig W., Strunk O., Westram R., Richter L., Meier H., Yadhukumar, et al. (2004). ARB: A Software Environment for Sequence Data. Nucleic Acids Res. 32, 1363–1371. doi: 10.1093/nar/gkh293
Martínez A., Osburne M. S., Sharma A. K., DeLong E. F., Chisholm S. W. (2012). Phosphite Utilization by the Marine Picocyanobacterium Prochlorococcus Mit9301: Phosphite Utilization by Prochlorococcus Mit9301. Environ. Microbiol. 14, 1363–1377. doi: 10.1111/j.1462-2920.2011.02612.x
McDowell R. W., Stewart I., Cade-Menun B. J. (2006). An Examination of Spin–Lattice Relaxation Times for Analysis of Soil and Manure Extracts by Liquid State phosphorus-31 Nuclear Magnetic Resonance Spectroscopy. J. Environ. Qual. 35, 293. doi: 10.2134/jeq2005.0285
Mewes K., Mogollón J. M., Picard A., Rühlemann C., Eisenhauer A., Kuhn T., et al. (2016). Diffusive Transfer of Oxygen From Seamount Basaltic Crust Into Overlying Sediments: An Example From the Clarion–Clipperton Fracture Zone. Earth Planet. Sci. Lett. 433, 215–225. doi: 10.1016/j.epsl.2015.10.028
Mills H. J., Kiel Reese B., Peter C. S. (2012). Characterization of Microbial Population Shifts During Sample Storage. Front. Microbiol. 3. doi: 10.3389/fmicb.2012.00049
Orcutt B. N., Wheat C. G., Rouxel O., Hulme S., Edwards K. J., Bach W. (2013). Oxygen Consumption Rates in Subseafloor Basaltic Crust Derived From a Reaction Transport Model. Nat. Commun. 4, 2539. doi: 10.1038/ncomms3539
Paytan A., Cade-Menun B. J., McLaughlin K., Faul K. L. (2003). Selective Phosphorus Regeneration of Sinking Marine Particles: Evidence From 31P NMR. Mar. Chem. 82, 55–70. doi: 10.1016/S0304-4203(03)00052-5
Prestat E., David M. M., Hultman J., Taş N., Lamendella R., Dvornik J., et al. (2014). Foam (Functional Ontology Assignments for Metagenomes): A Hidden Markov Model (HMM) Database With Environmental Focus. Nucleic Acids Res. 42, e145. doi: 10.1093/nar/gku702
Price M. N., Dehal P. S., Arkin A. P. (2010). FastTree 2 – Approximately Maximum-Likelihood Trees for Large Alignments. PloS One 5, e9490. doi: 10.1371/journal.pone.0009490
Pruesse E., Peplies J., Glöckner F. O. (2012). Sina: Accurate High-Throughput Multiple Sequence Alignment of Ribosomal RNA Genes. Bioinformatics 28, 1823–1829. doi: 10.1093/bioinformatics/bts252
Prüter J., Leipe T., Michalik D., Klysubun W., Leinweber P. (2020). Phosphorus Speciation in Sediments From the Baltic Sea, Evaluated by a Multi-Method Approach. J. Soils Sediments 20, 1676–1691. doi: 10.1007/s11368-019-02518-w
Rao N. N., Gómez-García M. R., Kornberg A. (2009). Inorganic Polyphosphate: Essential for Growth and Survival. Annu. Rev. Biochem. 78, 605–647. doi: 10.1146/annurev.biochem.77.083007.093039
Roussel E. G., Bonavita M.-A. C., Querellou J., Cragg B. A., Webster G., Prieur D., et al. (2008). Extending the Sub-Sea-Floor Biosphere. Science 320, 1046–1046. doi: 10.1126/science.1154545
Ruttenberg K. C. (2014). “The Global Phosphorus Cycle,” in Treatise on Geochemistry. 2nd Ed. Holland H., Turekian K. (Amsterdam: Elsevier), 499–558.
Ryan W. B. F., Carbotte S. M., Coplan J. O. (2009). Global Multi-Resolution Topography Synthesis. Geochem. Geophys. Geosystems 10, Q03014. doi: 10.1029/2008GC002332
Sannigrahi P., Ingall E. (2005). Polyphosphates as a Source of Enhanced P Fluxes in Marine Sediments Overlain by Anoxic Waters: Evidence From P-31 Nmr. Geochem. Trans. 6, 52–59. doi: 10.1063/1.1946447
Shang C., Caldwell D. E., Stewart J. W. B., Tiessen H., Huang P. M. (1996). Bioavailability of Organic and Inorganic Phosphates Adsorbed on Short-Range Ordered Aluminum Precipitate. Microb. Ecol. 31, 29–39. doi: 10.1007/BF00175073
Shan B., Li J., Zhang W. Q., Di Z., Jin X. (2016). Characteristics of Phosphorus Components in the Sediments of Main Rivers Into the Bohai Sea. Ecol. Engineer. 97, 426–433. doi: 10.1016/j.ecoleng.2016.10.042
Sosa O. A., Repeta D. J., DeLong E. F., Ashkezari M. D., Karl D. M. (2019). Phosphate-Limited Ocean Regions Select for Bacteria Populations Enriched in the Carbon-Phosphorus Lyase Pathway for Phosphonate Degradation. Environ. Microbiol. 21, 2402–2412. doi: 10.1111/1462-2920.14628
Suzumura M., Kamatani A. (1995). Mineralization of Inositol Hexaphosphate in Aerobic and Anaerobic Marine Sediments: Implications for the Phosphorus Cycle. Geochim. Cosmochim. Acta 59, 1021–1026. doi: 10.1016/0016-7037(95)00006-2
Turner B. L., Cade-Menun B. J., Condron L. M., Newman S. (2005). Extraction of Soil Organic Phosphorus. Talanta 66, 294–306. doi: 10.1016/j.talanta.2004.11.012
Turner B. L., Cheesman A. W., Godage H. Y., Riley A. M., Potter B. V. L. (2012). Determination of Neo- and D-Chiro-Inositol Hexakisphosphate in Soils by Solution 31P NMR Spectroscopy. Environ. Sci. Technol. 46, 4994–5002. doi: 10.1021/es204446z
Turner B. L., Papházy M. J., Haygarth P. M., Mckelvie I. D. (2002). Inositol Phosphates in the Environment. Philos. Trans. R. Soc B. Biol. Sci. 357, 449–469. doi: 10.1098/rstb.2001.0837
Van Mooy B. A. S., Krupke A., Dyhrman S. T. (2015). Major Role of Planktonic Phosphate Reduction in the Marine Phosphorus Redox Cycle. Science 348, 783–785. doi: 10.1126/science.aaa8181
Vershinina O. A., Znamenskaya L. V. (2002). The Pho Regulons of Bacteria. Microbiology 71, 497–511. doi: 10.1023/A:1020547616096
Wanner B. L. (1993). Gene Regulation by Phosphate in Enteric Bacteria. J. Cell. Biochem. 51, 47–54. doi: 10.1002/jcb.240510110
Watson S. J., Cade-Menun B. J., Needoba J. A., Peterson T. D. (2018). Phosphorus Forms in Sediments of a River-Dominated Estruary. Front. Mar. Sci. 5. doi: 10.3389/fmars.2018.00302
White R. H., Miller S. L. (1976). Inositol Isomers: Occurrence in Marine Sediments. Science 193, 885–886. doi: 10.1126/science.193.4256.885
Zhang Y. S., Werner W., Scherer H. W., Sun X. (1994). Effect of Organic Manure on Organic Phosphorus Fractions in Two Paddy Soils. Biol. Fertil. Soils 17, 64–68. doi: 10.1007/BF00418674
Keywords: Marine Sediments, P-NMR, deep biosphere, P substrate, metagenome
Citation: Defforey D, Tully BJ, Sylvan JB, Cade-Menun BJ, Kiel Reese B, Zinke L and Paytan A (2022) Potential Phosphorus Uptake Mechanisms in the Deep Sedimentary Biosphere. Front. Mar. Sci. 9:907527. doi: 10.3389/fmars.2022.907527
Received: 29 March 2022; Accepted: 26 April 2022;
Published: 02 June 2022.
Edited by:
Travis Blake Meador, Academy of Sciences of the Czech Republic (ASCR), CzechiaReviewed by:
Beverly E. Flood, University of Minnesota Twin Cities, United StatesMatthew John Harke, Gloucester Marine Genomics Institute (GMGI), United States
Copyright © 2022 Delphine Defforey, Benjamin Tully, Jason Sylvan, Brandi Kiel Reese, Laura Zinke, Adina Paytan and Her Majesty the Queen in Right of Canada, as represented by the Minister of Agriculture and Agri-Food Canada for the contribution of Barbara Cade-Menun. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Delphine Defforey, ZGVmZm9yZXlAZ21haWwuY29t; Adina Paytan, YXBheXRhbkB1Y3NjLmVkdQ==
 Delphine Defforey1*
Delphine Defforey1* Benjamin J. Tully
Benjamin J. Tully Jason B. Sylvan
Jason B. Sylvan Barbara J. Cade-Menun
Barbara J. Cade-Menun Brandi Kiel Reese
Brandi Kiel Reese Adina Paytan
Adina Paytan