
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
BRIEF RESEARCH REPORT article
Front. Mar. Sci., 30 June 2022
Sec. Marine Molecular Biology and Ecology
Volume 9 - 2022 | https://doi.org/10.3389/fmars.2022.904835
This article is part of the Research TopicMarine Biological Materials: Functional Mechanisms and Environmental Impacts from the Molecular to the Macro-ScaleView all 11 articles
 Jeana L. Drake1†
Jeana L. Drake1† Tali Mass1,2*
Tali Mass1,2*Black corals, ecologically important cnidarians found from shallow to deep ocean depths, form a strong yet flexible skeleton of sclerotized chitin and other biomolecules including proteins. The structure and mechanical properties of the chitin component of the skeleton have been well-characterized. However, the protein component has remained a mystery. Here we used liquid chromatography-tandem mass spectrometry to sequence proteins extracted from two species of common Red Sea black corals following either one or two cleaning steps. We detected hundreds of proteins between the two corals, nearly 70 of which are each other’s reciprocal best BLAST hit. Unlike stony corals, only a few of the detected proteins were moderately acidic (biased toward aspartic and/or glutamic acid residues) suggesting less of a role for these types of proteins in black coral skeleton formation as compared to stony corals. No distinct chitin binding domains were found in the proteins, but proteins annotated as having a role in protein and chitin modifications were detected. Our results support the integral role of proteins in black coral skeleton formation, structure, and function.
Antipatharians (black corals) are colonial cnidarians easily recognized by their black or golden brown chitinous skeletons. They are found from shallow to deep depths and in all oceans as well as the Mediterranean and Red Seas (Opresko, 2001; Wagner et al., 2012; Morgulis et al., 2022), although they face increasing pressure from harvesting (legal and illegal), invasive species, and contaminant introductions (Grigg, 2004; Todinanahary et al., 2016; Ruiz-Ramos et al., 2017). Their flexible skeletons, typically displaying small protrusions (spines), are covered by animal tissue with each polyp displaying six-fold symmetry (Opresko, 2001). At photic depths, they tend to grow in low light locations and are most frequently observed in locations with strong currents (Wagner et al., 2012; Morgulis et al., 2022).
Black corals are ecologically important as food, habitat/protection, and camouflage for a variety of invertebrates, fish, and mammals, particularly where the corals grow in high density (e.g., Parrish et al., 2002; Boland and Parrish, 2005; De Clippele et al., 2019), although they are not reef builders. Black corals can be home to a high density of organisms; in one study in the Philippines some black coral forests were associated with over 8,000 individual invertebrates per m2 (Suarez et al., 2015). Further, in areas with black coral forests, some fish taxa are found only within the colonies (e.g., Boland and Parrish, 2005). However, colonies can be damaged by a variety of human activities including bottom fishing and climate change (Deidun et al., 2015; Godefroid et al., 2022), and at least one species of black coral is considered endangered (IUCN, 2021).
While they have historically been used by humans as money (Tescione, 1973), jewelry (Gress and Andradi-Brown, 2018), and curios (Grigg, 1993), black corals also display a variety of medicinal properties. As suggested by their name (antipathes is Greek for “against diseases”) black corals and their extracts have historically been used in medical treatments including antibacterial activity, reducing fever, pain, and swelling, treating ulcers, and curing lung conditions (Narchi et al., 2015; Peña Moreno, 2017). From a pharmacological perspective, chitin can be deacetylated in alkaline conditions or enzymatically converted to chitosan, a stable non-toxic polymer; this chitosan and its derivatives then displays antibacterial, antifungal, and antiviral properties (review by Nuc and Dobrzycka-Krahel, 2021). Laboratory studies have supported that several of these historical medicinal uses are biochemically founded. For example, mice exposed to cigarette smoke and black coral extract displayed higher superoxide dismutase and malondialdehyde activity and lower lung tissue inflammation than mice exposed to cigarette smoke alone (Bai et al., 2011).
The skeletons of black corals are formed of chitin and other biomolecules (Goldberg, 1976). No minerals are present in the sclerotized skeleton (Goldberg et al., 1994), although the anions iodine and bromine are abundant (Juárez-de la Rosa et al., 2007). Epithelial cells secrete the chitin and other biomolecules that form the extracellular skeleton as concentric layers with a hollow core within the skeleton stalk (Daly et al., 2007). While the chitinous component of the skeleton has been well-characterized in a number of black coral taxa (e.g., Kim et al., 1992; Nowak et al., 2009; Bo et al., 2012; Juárez-de la Rosa et al., 2012), much less is known about the proteinaceous component. Proteins make up around 50% of the skeleton (Goldberg et al., 1994). Glycine, alanine, and histidine are the most abundant amino acids, and treating skeletons with alkaline solutions results in protein loss and shifts in the relative abundance of amino acids (Goldberg et al., 1994).
It has been suggested that chitin provides a viscous-elastic response while the protein component provides a plastic response, yielding the high degree of flexibility of the skeleton (Juárez-de la Rosa et al., 2012). The arrangement of the chitin fibrils in a helical pattern layer by layer may also contribute to the flexibility and reduced buckling under stresses induced by the high water flow environments in which black corals are frequently found (Kim et al., 1992). Further, the spines on the skeleton are cemented to the skeleton creating fixed points to reduce delamination as the skeletons bend and twist in the current (Kim et al., 1992). These mechanical properties may be useful in industry when synthesizing new structural materials. Additional bio-inspiration to be taken from black coral skeleton could focus on the chitin and protein content of black corals, yielding nanocomposites with industrial and medicinal uses (João et al., 2017; Zheng et al., 2019).
To address the gaps in knowledge about particular biomolecules involved in formation of their chitinous skeleton, we use liquid chromatography-tandem mass spectrometry to characterize the protein composition of ground skeleton from two black coral species, Antipathes griggi and Stichopathes sp., two commonly occurring black corals in the northern Red Sea (Figures 1A, B) (Morgulis et al., 2022). We detected hundreds of proteins in the chitin skeleton of both species. Unlike stony corals, there are few proteins strongly biased toward the acidic amino acids. Gene ontology analysis and domain searching did not reveal any proteins with known chitin binding domains, although several proteins contained an abundance of amino acids associated with chitin binding. Nearly 70 proteins that are each other’s reciprocal best BLAST hit were observed in both species, suggesting that, like stony corals, black corals may share a consensus skeletal ‘toolkit’ with a long evolutionary history (Zaquin et al., 2021). Our findings will be important in conservation, pharmaceutical development, and synthesizing materials inspired by strong yet flexible black coral skeletons.
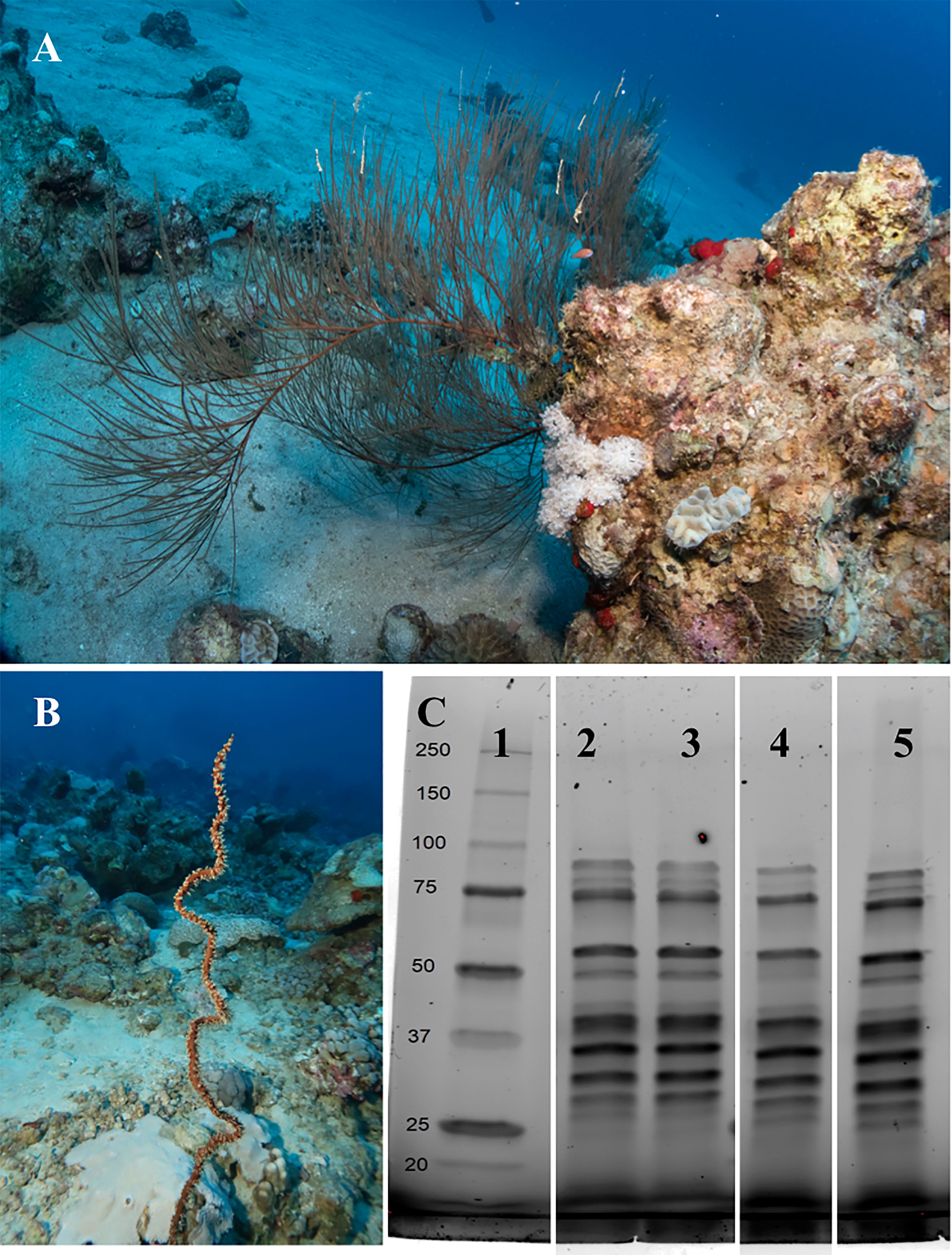
Figure 1 Antipathes griggi (A) and Stichopathes sp. (B) collected from the Red Sea adjacent to the Interuniversity Institute in Eilat, Israel, at 40-60 m depth, for proteomic sequencing of the chitinous skeleton. (C) Imaging of SDS-PAGE separation of proteins extracted by chitinase from chitinous black coral skeleton cleaned both before and after grinding. MW marker (lane 1); Antipathes cleaned only before grinding (lane 2); Stichopathes cleaned only before grinding (lane 3); Antipathes cleaned before and after grinding (lane 4); Stichopathes cleaned before and after grinding (lane 5). Photos: Hagai Nativ.
Several specimens of Antipathes griggi and Stichopathes sp. were collected at 40-60 m depth from the reef adjacent to the Interuniversity Institute in Eilat, Israel under a permit issued by the Israel Nature and Parks Authority (Figures 1A, B). Branches or coils of each species were clipped from colonies using underwater shears, segmented into 2-3 cm sections, and immediately transported to the laboratory for preservation in RNA/DNA Shield (Zymo) or 100% ethanol for protein extraction at 4°C. The next day, specimens were transported to the laboratory for processing.
Black coral segments of each species were combined and washed copiously in MilliQ water to remove the ethanol, soaked in a 10% bleach solution for 30 minutes at room temperature and for 1 hour with sonication, and then rinsed again with MilliQ and dried at 60°C. The bleaching process was repeated with 50% bleach and the sections were rinsed and re-dried. While the first wash removed most of the tissue, some tentacles of the polyps were still observed at low magnification light microscopy. The second higher concentration bleaching step appeared to remove all tissue by visual inspection. Dried skeletons were ground with a mortar and pestle; Antipathes skeletons ground to a finer powder than did Stichopathes skeletons. Ground powder was divided; half was used is-as while half was cleaned again in 50% bleach while sonicating for 2 hours, rinsed five times with MilliQ water, and dried at 60°C.
One sample of fresh black coral tissue from each species was flash frozen and then RNA was extracted using the CTAB method (Wang and Stegemann, 2010), followed by use of a Purelink RNA Mini Kit following the manufacturer’s instructions. RNA quality was examined by TapeStation (Agilent). Libraries were prepared using the INCPM-mRNA-seq (an internal protocol G-INCPM). The polyA fraction (mRNA) was purified from 500 ng of total RNA followed by fragmentation and generation of double-stranded cDNA. Then, end repair, A base addition, adapter ligation and PCR amplification steps were performed. Libraries were evaluated by Qubit (Thermo Fisher Scientific) and TapeStation (Agilent). Sequencing libraries were constructed with barcodes to allow multiplexing of samples. Around 140 million paired-end 150X2-bp reads were sequenced on an Illumina NovaSeq SP300 cyc. Poly-A/T stretches and Illumina adapters were trimmed from the reads using cutadapt; resulting reads shorter than 30 bp were discarded. Trimmed reads were used for assembly using Trinity (Trinity-v2.11.0) and likely bacterial contaminant sequences were filtered out. Highly similar sequences were clustered with CD-hit (CD-HIT version 4.5.4). Completeness was assessed (BUSCO 4.1.4), with BUSCO scores of 73.8% and 77.2% for Antipathes and Stichopathes, respectively. Sequence reads were aligned using bowtie2 (version 2.3.4.1), with alignment rates of 97.5% and 95.3% for Antipathes and Stichopathes, respectively. ORF prediction was performed in TransDecoder-v3.0.1 based on blastp-v2.7.1. Finally, sequences were characterized by InterProScan and annotated in OmicsBox. Transcriptome information and sequences are available in NCBI BioProject PRJNA809900.
Four methods were tested for protein extraction from black coral skeleton powders: Soak in 1 ml each (1) 1 M HCl at room temperature for 1 hour (modified from Shimahara and Takiguchi, 1988); (2) 6 M guanidinium hydrochloride in 8 mM imidazole, 50 mM sodium phosphate monobasic, 50 mM Tris HCl at 30°C for 1 hour (modified from Gildemeister et al., 1994); (3) 50 mM dithiothreitol (DTT) at 100°C for 15 minutes (modified from Mathys et al., 1999); or (4) 1 mg/ml chitinase in PBS 37°C for 2 hours. At the end of the incubation periods all samples were pelleted at 500 xg for 5 minutes at room temperature. Protein concentration of supernatants was quantified by bicinchoninic acid (BCA) assay (Supplementary Material Table 1), proteins in 10 μg quantified total protein were visualized by SDS-PAGE separation on Mini-Protean TGX Stain-Free precast gels (BioRad) with 5-minute UV activation, and fractions were stored separately at -80°C until further analysis. Chitinase yielded the darkest bands on UV-activated Stain-Free gels (Figure 1C); therefore, only chitinase fractions were submitted for sequencing. HCl also yielded faint bands by SDS-PAGE that were observable after computationally increasing the image contrast (Supplementary Material Figure 1A); however, these samples were not submitted for sequencing due to the low protein yield. Guanidinium hydrochloride and DTT extracts did not show any protein bands by SDS-PAGE (Supplementary Material Figures 1A, B).
Proteins extracted by chitinase from ground black coral skeleton cleaned only pre-grinding as well as skeleton cleaned again post-grinding were dissolved in 5% SDS and digested with trypsin using the S-trap method overnight at room temperature. Single samples of each species for each cleaning fraction were processed. The resulting peptides were analyzed using a nanoflow ultra-performance liquid chromatograph (nanoAcquity) coupled to a high resolution, high mass accuracy mass spectrometer (Fusion Lumos) fitted with a Symmetry C18 0.18*20mm trap column (Waters, Inc) for peptide trapping and then a HSS T3 0.075*250 mm column (Waters, Inc.) for peptide separation using a gradient of 4-28% (80% acetonitrile, 0.1% Formic acid) for 150 minutes. Spray voltage was set to +2kV. The data were acquired in a Fusion Lumos by a Top Speed Data-Dependent Acquisition method with a cycle time of 3 s. An MS1 scan was performed in the Orbitrap at 120,000 resolution with a maximum injection time of 60 ms and the data were scanned between 300-1800 m/z. MS2 was selected using a monoisotopic precursor selection set to peptides, peptide charge states set to +2-+8 and dynamic exclusion set to 30 s. MS2 was performed using HCD fragmentation scanning in the Orbitrap, with the first mass set to 130 m/z at a resolution of 15,000. Maximum injection time was set to 60 ms with automatic gain control of 5x10-4 ions as a fill target. The resulting data were searched against the protein databases derived from our de novo black coral transcriptomes described above using the Byonic search engine (Protein Metrics Inc.). A first search was carried out without any false discovery rate (FDR) filtering, to generate a focused database for a second search. The second search was set to 1% FDR, allowing fixed carbamidomethylation on C and variable oxidation on MW, deamidation on NQ and protein N-terminal acetylation. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the Pride partner repository (Perez-Riverol et al., 2019), under the dataset identifier PXD032043.
The de novo transcriptomes described above for Antipathes griggi and Stichopathes sp. were used as reference peptide databases for the mass spectrometry analysis. A common contaminants database was also included. Only proteins with at least two significant peptides or at least one significant peptide with at least 10 spectra and an identification score of 250 or greater were retained. To further filter out potential human proteins inadvertently introduced during sample preparation, all sequences were BLASTed against the ‘Primates’ database in NCBI using Blast2GO 6.0 (Conesa et al., 2005). NCBI-generated sequence alignments of black coral versus Homo sapiens proteins with Blast2GO e-values lower than e-50 and percent mean similarity greater than 50%, e-values lower than e-100, and percent similarity greater than 80% were removed from our final list of proteins if manual visual examination showed that likely tryptic peptides could have been human, rather than black coral, derived (Peled et al., 2020).
For each taxon, likely coral-specific skeleton proteins were divided into those which were found in skeleton that had only been cleaned pre-grinding (cleaned once), those which were found in skeleton that were cleaned again post-grinding (cleaned twice), and those that were found in both fractions (both). Proteins in each group were then characterized for amino acid composition using SubsetDF in the CHNOSZ package in RStudio (Team, 2019), presence of a signal peptide using the Signal P-5.0 server (Armenteros et al., 2019), and completeness of predicted sequence using the annotations described above. Proteins were further examined for strong bias toward cysteine, aspartic and/or glutamic acid, or tryptophan, lysine, and/or phenylalanine. Domains for completely predicted proteins with a strong amino acid bias were predicted in the PROSITE server (Sigrist et al., 2012) and domain visualizations were created in the PROSITE MyDomains Image Creator (Hulo et al., 2007). Clustering at 30, 50, and 90% sequence similarity of black coral and scleractinian (Drake et al., 2013; Ramos-Silva et al., 2013; Takeuchi et al., 2016; Peled et al., 2020) proteins plus proteins extracted from chitinous cuticles of the mosquito Anopheles gambiae (He et al., 2007; Mastrobuoni et al., 2013; Champion et al., 2016; Zhou et al., 2016) was conducted in CD-HIT with default settings (Huang et al., 2010). Gene ontology term enrichment was performed using a two-tailed Enrichment Analysis in Blast2GO with p<0.05 (Conesa et al., 2005), which integrates the FatiGO package (Al-Shahrour et al., 2004) using the Fisher’s Exact Test. Reciprocal BLASTing was performed in the BLAST+ command line application.
Of the four protein extraction methods tested for the two black coral species - HCl, guanidine hydrochloride, DTT, and chitinase - chitinase yielded the highest amount of protein separated by SDS-PAGE with subsequent imaging (Figure 1C). While BCA assays suggested that HCl, DTT, and guanidine hydrochloride resulted in protein extraction from the chitin skeletons (Supplementary Material Table 1), it appears more likely that these treatments released chitosan, an amide derivative of chitin that can react with the BCA reagents (Kasaai et al., 2013; Gao et al., 2019). Cleaned and uncleaned fractions of black coral skeletal proteins extracted by chitinase were therefore submitted for protein sequencing.
All samples contained at least 100 returned proteins above the designated cutoffs and that were not likely human contaminants (Supplementary Material Table 2). Antipathes skeleton yielded 167 proteins that were only in the twice-cleaned fraction, 105 proteins that were only in the once-cleaned fraction, and 99 proteins in both fractions (Figure 2A). From Stichopathes skeleton, 93 proteins were only in the twice-cleaned fraction, 62 were in the once-cleaned fraction, and 73 were in both fractions (Figure 2B). Some of the proteins from the singly cleaned fractions were likely not bound to the chitinous skeleton and as such were removed during the second cleaning step as has been shown for removal of non-intracrystalline stony coral skeletal organic matter (Hendy et al., 2012). However, we cannot exclude them from the list of potential proteins involved in the chitinous black coral skeleton as they may still serve a role in skeleton strength as has been shown in squid pen which is also a composite of chitin plus proteins, lipids, and other polysaccharides (Montroni et al., 2021).
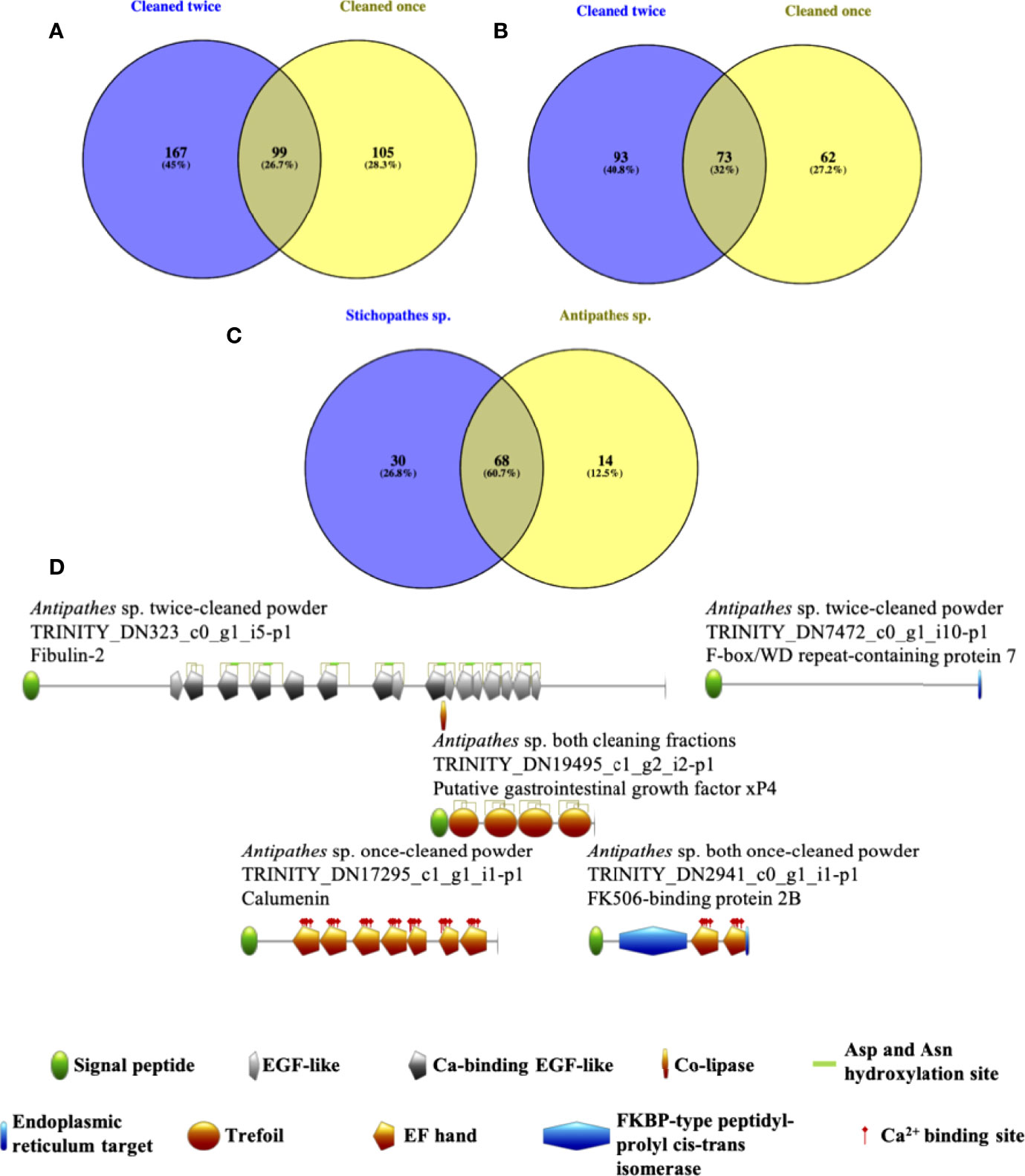
Figure 2 For both Antipathes (A) and Stichopathes (B) we observed ~30% overlap in protein detection between skeleton cleaned only before grinding (yellow) and skeleton clean both before and after grinding (purple). Unique proteins were also found in both fractions. Further, for both species, the largest number of proteins were found in the twice-cleaned fractions (purple, a & b). 60.7% of detected proteins with reciprocal best BLAST hits between the two species were observed in both species (C) across the various cleaning fractions. (D) Predicted domains in proteins extracted from black coral skeleton using chitinase. Only proteins that were completely predicted, display a proposed signal peptide, and are biased toward key amino acids were considered. Although several Stichopathes skeletal proteins meet these criteria, none are predicted to have any domains.
Of the total 371 proteins from Antipathes skeleton and 228 proteins from Stichopathes skeleton (Additional Files S1 and S2, respectively), 112 were found in the transcriptomes of both species and 68 were each other’s reciprocal best BLAST hit (Figure 2C, Supplementary Material Table 3). Distribution of reciprocal best BLAST hit proteins between cleaning fractions between the two taxa is non-random (Fisher’s exact test, p<0.05, Supplementary Material Table 4). Orthologous proteins found in the same cleaning fractions of both species include a MAM and LDL receptor domain containing protein, three coadhesin/hemicentin/mucin proteins, and four collagen alpha proteins among others. Interestingly, these protein types are also known from stony coral skeleton, as are several protein types that were detected in both black coral skeletons but in different cleaning fractions including an EGF and laminin G domain-containing protein, a potential protocadherin, and uncharacterized skeletal organic matrix protein 5 (Drake et al., 2013; Ramos-Silva et al., 2013; Takeuchi et al., 2016; Peled et al., 2020). Only Antipathes skeleton contained a carbonic anhydrase, which is known from stony coral skeleton from multiple species (Drake et al., 2013; Ramos-Silva et al., 2013; Peled et al., 2020), where it likely supplies bicarbonate for the eventual production of the calcium carbonate exoskeleton of scleractinians (Bertucci et al., 2013; Comeau et al., 2017).
Not all proteins were completely predicted (Supplementary Material Table 2). Of 84 proteins in the twice-cleaned fraction extracted from Antipathes that were completely predicted, two displayed a disproportionate amount of aspartic and glutamic acids (‘disproportionate’ defined as >20% of total amino acids) and four had >10% cysteine; three of those with excess cysteine had signal peptides. Nearly half the number of completely predicted proteins (47) were found only in the once-cleaned fraction from Antipathes, and 20 of these displayed a signal peptide with two of this subset also showing excess aspartic and glutamic acids. Eighteen proteins observed in both fractions were both completely predicted and displayed a signal peptide; two of these were >10% cysteine.
Fewer proteins were completely predicted in Stichopathes (Supplementary Material Table 2). In the twice-cleaned fraction for this taxon, 16 of 42 completely predicted proteins have signal peptides and only one is >10% C. Similarly, 23 of 44 complete proteins found in both fractions have signal peptides and only one is >10% cysteine. No completely predicted proteins in these two fractions were biased toward acidic amino acids. The once-cleaned fraction also showed a similar breakdown of completeness and signal peptides, with 34 proteins being complete and 12 of these having a signal peptide; however, none of these were biased toward any amino acids of interest.
Bias of skeletal proteins toward acidic amino acids (aspartic and glutamic acids) is well-documented in organisms that produce biocarbonates (e.g., Nocente-McGrath et al., 1989; Gotliv et al., 2005; Mass et al., 2013) and biophosphates (He et al., 2003), and even in collagen-dominated biophosphates there are several known acidic non-collagenous proteins (Gorski, 2011). Unlike either stony coral skeletons or vertebrate bone, however, black corals’ skeletons are constructed of sclerotized chitin, a long-chained polysaccharide (Goldberg, 1976). Chitin displays a similar hierarchical organization to biocarbonates (Ehrlich, 2010), and acidic chitinases (e.g., Samac et al., 1990) have been characterized from a number of organisms. Further, an acidic chitin binding protein appears to be involved in the precipitation of calcium phosphate and then calcium carbonate in crustacean teeth (Tynyakov et al., 2015). Our results suggest that acidic proteins may be involved in black coral chitinous skeleton development; however, despite likely sharing conserved biomineralization-related genes with stony corals (review by Gilbert et al., 2022), black coral acidic proteins are perhaps not as central to the process as they appear to be in allowing stony corals biomineralize (Mass et al., 2013) and to counter the effects of ocean acidification (e.g., Drake et al., 2017).
Several amino acids are associated with chitin binding domains, including cysteine, phenylalanine, tryptophan, and tyrosine (Shen and Jacobs-Lorena, 1999). Invertebrate and plant chitin binding proteins typically display several conserved cysteines that are likely involved in disulfide bonds related to protein secondary and tertiary structure (Shen and Jacobs-Lorena, 1999). Tyrosine and phenylalanine are conserved components of the R&R consensus motif of chitin binding cuticle proteins (Rebers and Willis, 2001), and tyrosine and tryptophan are crucial conserved amino acids in the binding cleft of Bascillus chitinase. Despite a small number of completely predicted detected proteins being biased toward cysteine, and none toward phenylalanine, tryptophan, or tyrosine, some of the proteins may have chitin binding capabilities and should be tested in the future.
We also considered only those full proteins that are proposed to have a signal peptide for export to the cell membrane or extracellular space (review by von Heijne, 1990), and then queried this final group for those with predicted domains. Out of nine proteins between the two species that meet the above criteria, five are predicted to have known domains, although none are explicitly chitin binding, and no such proteins from Stichopathes were found to have predicted domains (Figure 2D). A protein annotated as fibulin-2 contains several EGF-like domains, some of which can be Ca2+ binding. EGF-like domains are found in extracellular matrix proteins and can be involved in protein-protein interactions (review by Engel, 1989; Rao et al., 1995). Trefoil domains have also been found in extracellular matrix proteins; the domain typically contains six cysteines that form disulfide bridges; these proteins may interact with mucin (Thim, 1997), which has been found in the skeleton-associated extracellular matrix of stony corals (Ramos-Silva et al., 2013; Takeuchi et al., 2016; Peled et al., 2020) and which associates with chitin in arthropod guts (Dias et al., 2018), although mucin has not been sequenced from arthropod cuticle.
To determine if invertebrates that produce a chitinous skeleton display skeletal proteome similarities across divergent phyla, we compared the black coral skeletal proteome with that of mosquito cuticle. No clustering of black coral and Anopheles gambiae cuticular proteins was observed at 30, 50, or 90% sequence similarity. While no clustering of black and stony coral skeletal proteins was observed at 90% similarity, several clusters were observed at 50 (7 clusters) and 30% (11 clusters) similarity. Clustered black and stony coral proteins are annotated as collagenase like, CUB and peptidase domain containing, digestive cysteine proteins like, EGF and laminin G domain containing, fibronectin type III domain containing, kielin/chordin like, MAM and LDL receptor domain containing, protein MLP like, uncharacterized skeletal organic matrix protein 5, ZP domain containing, and uncharacterized in stony corals (Table 1).
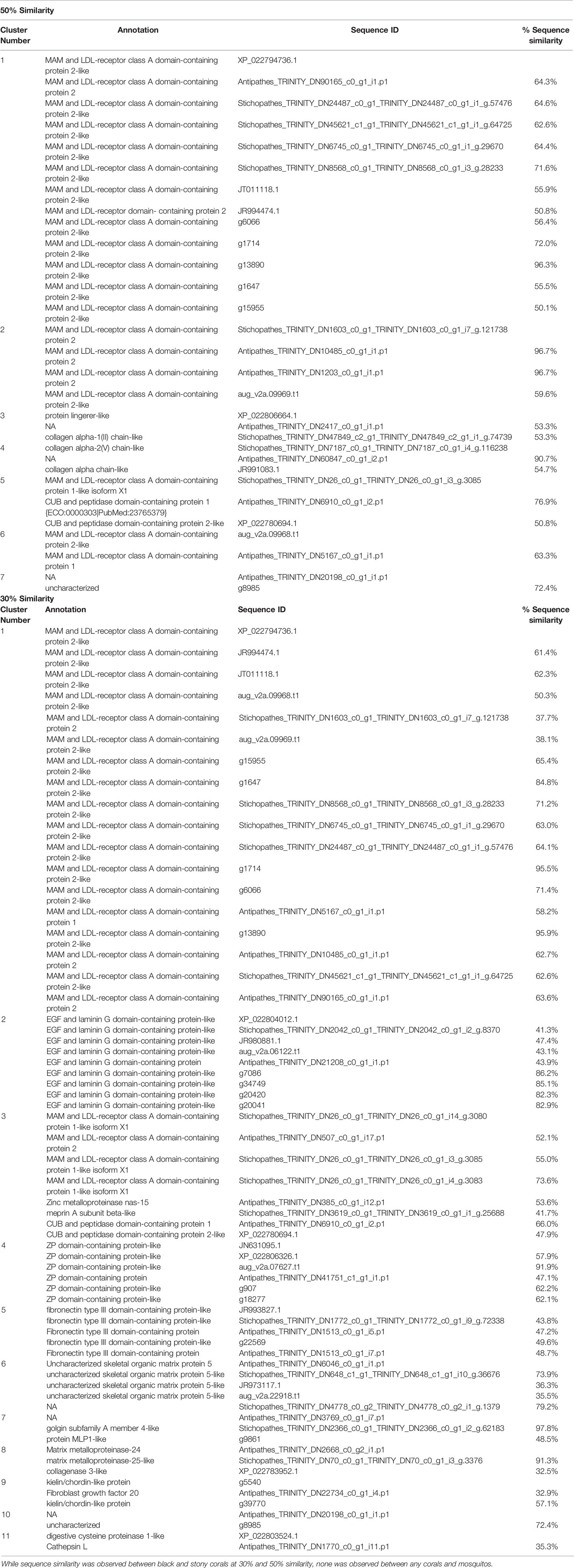
Table 1 Black coral, stony coral, and mosquito exoskeleton proteins clustered by sequence similarity in CD-Hit.
Antipathes skeletal proteins sequenced in this work are over-enriched in GO terms related to binding, the cell cortex/cytoskeleton/membrane, cell differentiation, enzymes, extracellular location, metabolism, organelles, regulation, signaling/stimulus response, and vesicles/transport (Figure 3). Similarly, Stichopathes skeletal proteins are over-enriched in GO terms for binding, enzymes, extracellular location, and regulation; additionally, they are enriched for neurons and protein interaction (Figure 3). The only GO term cluster that was under-enriched was observed for Stichopathes and included terms related to binding: organic cyclic compound binding [GO:0097159] and heterocyclic compound binding [GO:1901363]. Proteins with extracellular location and membrane GO terms may be important as black coral chitin formation and sclerotization are extracellular processes (Wagner et al., 2012). As vesicular transport of ions and organic molecules that form the skeleton appears to be important in stony corals (Schmidt et al., 2022), vesicle and transport proteins may also play a role in black coral skeleton formation. Proteins which function as enzymes may also be important to the formation and modification of the chitinous skeleton; these include a carbonic anhydrase in Antipathes, a calmodulin in Stichopathes, and several proteases and chitinases in both species.
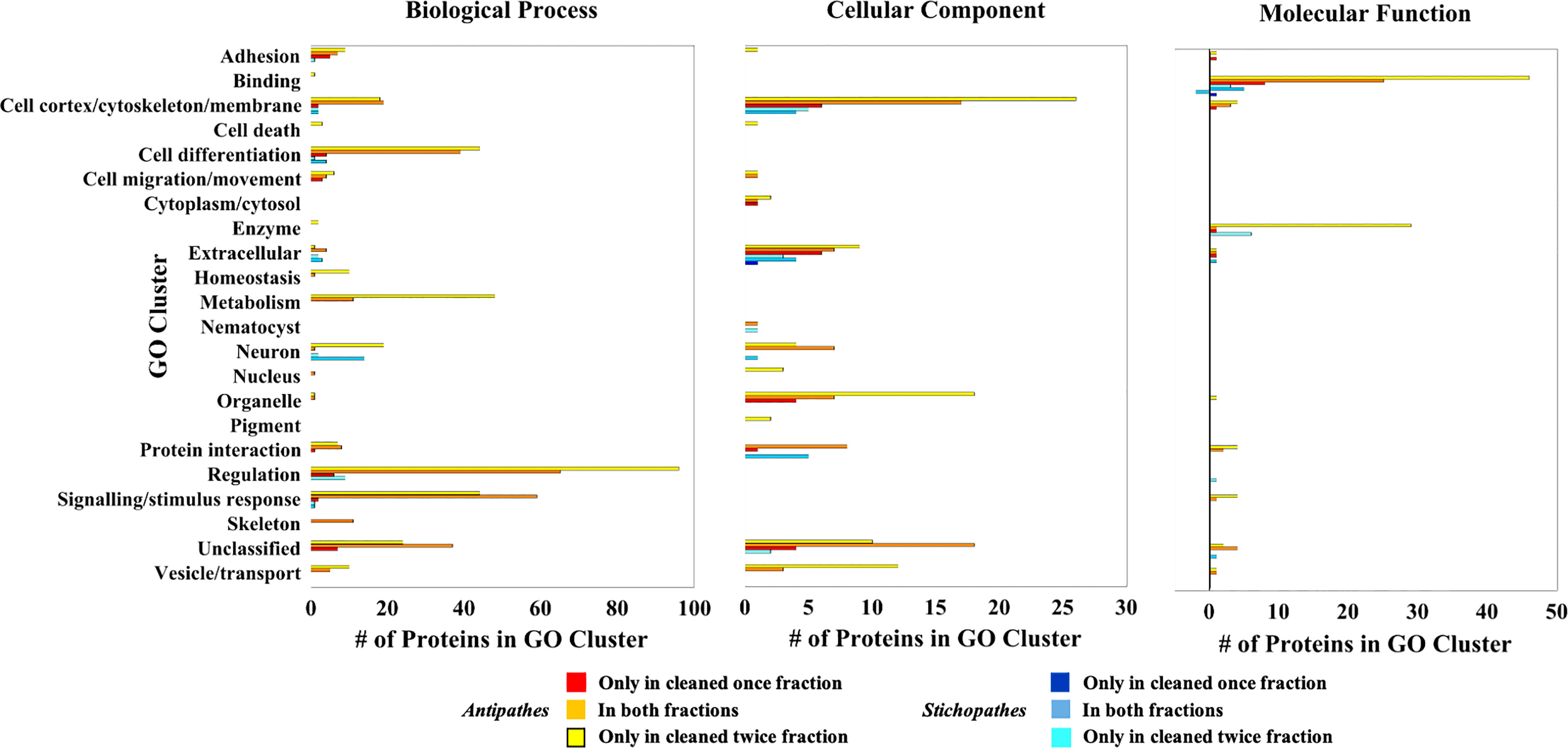
Figure 3 Clustered skeletal proteins with enriched GO terms relative to each species’ transcriptome. Bars greater than zero are the number of proteins with over-enriched GO terms in the skeletal proteomes whereas those less than zero have under-enriched GO terms. Note the different ranges in the numbers of proteins in each GO cluster.
While the structure and mechanical properties of black coral skeleton chitin have been well-characterized, to date, their protein composition has received less attention. To address this research gap, we sequenced the skeletal proteome of two species of common Red Sea black corals under two steps of strong oxidative cleaning. Our analyses reveal hundreds of proteins in the skeletons of each species, with ~70 being each other’s reciprocal best BLAST hit, potentially representing a conserved toolkit. A small number of proteins can be considered moderately acidic and only a few are biased toward amino acids known to be involved in chitin binding. Instead, our GO analysis shows that terms related to vesicles and transport, membrane structure, and enzymatic functionality are enriched. This initial work suggests that more research is required to fully comprehend the functions of black coral skeletal proteins and their interactions with chitin.
The datasets generated for this study can be found in the following online repositories: (1) Transcriptome data are available under NCBI BioProject PRJNA809900. (2) The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (Perez-Riverol et al.) partner repository with the dataset identifier PXD032043 and 10.6019/PXD032043.
Conceptualization, investigation, writing—original draft preparation, JD and TM. Formal analysis, data curation, visualization, JD. Resources, supervision, funding acquisition, TM. All authors have read and agreed to the published version of the manuscript. All authors contributed to the article and approved the submitted version.
This project has received funding from GIF, the German-Israeli Foundation for Scientific Research and Development by the Israeli Science Foundation (Award #Grant I-1496-302.8) to TM. JD was supported by the Zuckerman STEM Leadership Postdoctoral program.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We thank the InterUniversity Institute in Eilat, Israel for accommodations during sampling; Hagai Nativ for photography; the Morris Khan Research Station diving team for SCUBA assistance; David Morgenstern at the de Botton Institute for Protein Profiling at Nancy and Stephen Grand Israel National Center for Personalized Medicine at the Weizmann Institute of Science for proteomic sequencing and analysis; the Nancy and Stephen Grand Israel National Center for Personalized Medicine at the Weizmann Institute of Science for transcriptome sequencing and analysis; and Tal Zaquin and Ricardo Almuly at the University of Haifa for laboratory assistance.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2022.904835/full#supplementary-material
Al-Shahrour F., Díaz-Uriarte R., Dopazo J. (2004). FatiGO: A Web Tool for Finding Significant Associations of Gene Ontology Terms With Groups of Genes. Bioinformatics 20, 578–580. doi: 10.1093/bioinformatics/btg455
Armenteros J. J. A., Tsirigos K. D., Sønderby C. K., Petersen T. N., Winther O., Brunak S., et al. (2019). SignalP 5.0 Improves Signal Peptide Predictions Using Deep Neural Networks. Nat. Biotechnol. 37, 420. doi: 10.1038/s41587-019-0036-z
Bai X., Chen Y., Chen W., Lei H., Gao F., Qin Y., et al. (2011). The Effect of Black Coral Extraction on Acute Lung Inflammation Induced by Cigarette Smoke in Mice. Exp. Lung Res. 37, 627–632. doi: 10.3109/01902148.2011.627084
Bertucci A., Moya A., Tambutté S., Allemand D., Supuran C. T., Zoccola D. (2013). Carbonic Anhydrases in Anthozoan Corals—A Review. Bioorganic Medicinal Chem. 21, 1437–1450. doi: 10.1016/j.bmc.2012.10.024
Bo M., Bavestrello G., Kurek D., Paasch S., Brunner E., Born R., et al. (2012). Isolation and Identification of Chitin in the Black Coral Parantipathes Larix (Anthozoa: Cnidaria). Int. J. Biol. Macromolecules 51, 129–137. doi: 10.1016/j.ijbiomac.2012.04.016
Boland R. C., Parrish F. A. (2005). A Description of Fish Assemblages in the Black Coral Beds Off Lahaina, Maui, Hawai ‘I. Pacific Sci. 59, 411–420. doi: 10.1353/psc.2005.0032
Champion M. M., Sheppard A. D., Rund S. S., Freed S. A., O’Tousa J. E., Duffield G. E. (2016). “"Qualitative and Quantitative Proteomics Methods for the Analysis of the Anopheles Gambiae Mosquito Proteome,",” in Short Views on Insect Genomics and Proteomics (Springer) Berlin, Germany, 37–62.
Comeau S., Tambutté E., Carpenter R., Edmunds P., Evensen N., Allemand D., et al. (2017). Coral Calcifying Fluid pH is Modulated by Seawater Carbonate Chemistry Not Solely Seawater pH. Proc. R. Soc B 284, 20161669. doi: 10.1098/rspb.2016.1669
Conesa A., Götz S., García-Gómez J. M., Terol J., Talón M., Robles M. (2005). Blast2GO: A Universal Tool for Annotation, Visualization and Analysis in Functional Genomics Research. Bioinformatics 21, 3674–3676. doi: 10.1093/bioinformatics/bti610
Daly M.arymegan, Brugler M. R., Cartwright P., Collins A. G., Dawson M. N., Fautin D. G., et al. (2007). The Phylum Cnidaria: A Review of Phylogenetic Patterns and Diversity 300 Years After Linnaeus. In Linnaeus Tercentenary: Progress in Invertebrate Taxonomy, Zhang Z.-Q., Shear W. A., editors. Zootaxa 1668:127–182.
De Clippele L. H., Huvenne V. A., Molodtsova T. N., Roberts J. M. (2019). The Diversity and Ecological Role of non-Scleractinian Corals (Antipatharia and Alcyonacea) on Scleractinian Cold-Water Coral Mounds. Front. Mar. Sci. 6, 184. doi: 10.3389/fmars.2019.00184
Deidun A., Andaloro F., Bavestrello G., Canese S., Consoli P., Micallef A., et al. (2015). First Characterisation of a Leiopathes Glaberrima (Cnidaria: Anthozoa: Antipatharia) Forest in Maltese Exploited Fishing Grounds. Ital. J. Zoology 82, 271–280. doi: 10.1080/11250003.2014.986544
Dias R., Cardoso C., Pimentel A., Damasceno T., Ferreira C., Terra W. (2018). The Roles of Mucus-Forming Mucins, Peritrophins and Peritrophins With Mucin Domains in the Insect Midgut. Insect Mol. Biol. 27, 46–60. doi: 10.1111/imb.12340
Drake J. L., Mass T., Haramaty L., Zelzion E., Bhattacharya D., Falkowski P. G. (2013). Proteomic Analysis of Skeletal Organic Matrix From the Stony Coral Stylophora Pistillata. Proc. Natl. Acad. Sci. 110, 3788–3793. doi: 10.1073/pnas.1301419110
Drake J. L., Schaller M. F., Mass T., Godfrey L., Fu A., Sherrell R. M., et al. (2017) 63(1):107–121. Molecular and Geochemical Perspectives on the Influence of CO2 on Calcification in Coral Cell Cultures. Limnol. Oceanog. doi: 10.1002/lno.10617
Ehrlich H. (2010). Chitin and Collagen as Universal and Alternative Templates in Biomineralization. Int. Geology Rev. 52, 661–699. doi: 10.1080/00206811003679521
Engel J. (1989). EGF-Like Domains in Extracellular Matrix Proteins: Localized Signals for Growth and Differentiation? FEBS Lett. 251, 1–7. doi: 10.1016/0014-5793(89)81417-6
Gao C., Fisher Z. B., Edgar K. J. (2019). Azide Reduction by DTT or Thioacetic Acid Provides Access to Amino and Amido Polysaccharides. Cellulose 26, 445–462. doi: 10.1007/s10570-018-2195-3
Gilbert P. U., Bergmann K. D., Boekelheide N., Tambutté S., Mass T., Marin F., et al. (2022). Biomineralization: Integrating Mechanism and Evolutionary History. Sci. Adv. 8, eabl9653. doi: 10.1126/sciadv.abl9653
Gildemeister O. S., Zhu B. C., Laine R. A. (1994). Chitovibrin: A Chitin-Binding Lectin From Vibrio Parahemolyticus. Glycoconjugate J. 11, 518–526. doi: 10.1007/BF00731302
Godefroid M., Hédouin L., Mercière A., Dubois P., Consortium U. T. P. (2022). Thermal Stress Responses of the Antipatharian Stichopathes Sp. From the Mesophotic Reef of Mo'orea, French Polynesia. Sci. Total Environ., 820 153094. doi: 10.1016/j.scitotenv.2022.153094
Goldberg W. M. (1976). Comparative Study of the Chemistry and Structure of Gorgonian and Antipatharian Coral Skeletons. Mar. Biol. 35, 253–267. doi: 10.1007/BF00396873
Goldberg W. M., Hopkins T. L., Holl S. M., Schaefer J., Kramer K. J., Morgan T. D., et al. (1994). Chemical Composition of the Sclerotized Black Coral Skeleton (Coelenterata: Antipatharia): A Comparison of Two Species. Comp. Biochem. Physiol. Part B: Comp. Biochem. 107, 633–643. doi: 10.1016/0305-0491(94)90197-X
Gorski J. P. (2011). Biomineralization of Bone: A Fresh View of the Roles of non-Collagenous Proteins. Front. Biosci. (Landmark edition) 16, 2598. doi: 10.2741/3875
Gotliv B.-A., Kessler N., Sumerel J. L., Morse D. E., Tuross N., Addadi L., et al. (2005). Asprich: A Novel Aspartic Acid-Rich Protein Family From the Prismatic Shell Matrix of the Bivalve Atrina Rigida. ChemBioChem 6, 304–314. doi: 10.1002/cbic.200400221
Gress E., Andradi-Brown D. A. (2018). Assessing Population Changes of Historically Overexploited Black Corals (Order: Antipatharia) in Cozumel, Mexico. PeerJ 6, e5129. doi: 10.7717/peerj.5129
Grigg R. W. (1993). Precious Coral Fisheries of Hawaii and the US Pacific Islands. Mar. Fish. Rev. 55, 50–60.
Grigg R. W. (2004). Harvesting Impacts and Invasion by an Alien Species Decrease Estimates of Black Coral Yield Off Maui, Hawai'i. Pacific Sci. 58, 1–6. doi: 10.1353/psc.2004.0006
He N., Botelho J. M., McNall R. J., Belozerov V., Dunn W. A., Mize T., et al. (2007). Proteomic Analysis of Cast Cuticles From Anopheles Gambiae by Tandem Mass Spectrometry. Insect Biochem. Mol. Biol. 37, 135–146. doi: 10.1016/j.ibmb.2006.10.011
He G., Dahl T., Veis A., George A. (2003). Dentin Matrix Protein 1 Initiates Hydroxyapatite Formation In Vitro. Connective Tissue Res. 44, 240–245. doi: 10.1080/03008200390181726
Hendy E. J., Tomiak P. J., Collins M. J., Hellstrom J., Tudhope A. W., Lough J. M., et al. (2012). Assessing Amino Acid Racemization Variability in Coral Intra-Crystalline Protein for Geochronological Applications. Geochimica cosmochimica Acta 86, 338–353. doi: 10.1016/j.gca.2012.02.020
Huang Y., Niu B., Gao Y., Fu L., Li W. (2010). CD-HIT Suite: A Web Server for Clustering and Comparing Biological Sequences. Bioinformatics. 26(5):680–682. doi: 10.1093/bioinformatics/btq003
Hulo N., Bairoch A., Bulliard V., Cerutti L., Cuche B. A., De Castro E., et al. (2007). The 20 Years of PROSITE. Nucleic Acids Res. 36, D245–D249. doi: 10.1093/nar/gkm977
IUCN (2021). The IUCN Red List of Threatened Species. Version 2021-3. https://www.iucnredlist.org. Accessed May 2022
João C. F., Echeverria C., Velhinho A., Silva J. C., Godinho M. H., Borges J. P. (2017). Bio-Inspired Production of Chitosan/Chitin Films From Liquid Crystalline Suspensions. Carbohydr. Polymers 155, 372–381. doi: 10.1016/j.carbpol.2016.08.039
Juárez-de la Rosa B., Ardisson P.-L., Azamar-Barrios J., Quintana P., Alvarado-Gil J. (2007). Optical, Thermal, and Structural Characterization of the Sclerotized Skeleton of Two Antipatharian Coral Species. Materials Sci. Engineering: C 27, 880–885. doi: 10.1016/j.msec.2006.10.006
Juárez-de la Rosa B., Munoz-Saldana J., Torres-Torres D., Ardisson P.-L., Alvarado-Gil J. (2012). Nanoindentation Characterization of the Micro-Lamellar Arrangement of Black Coral Skeleton. J. Struct. Biol. 177, 349–357. doi: 10.1016/j.jsb.2011.12.009
Kasaai M. R., Arul J., Charlet G. (2013). Fragmentation of Chitosan by Acids. Sci. World J. 11. doi: 10.1155/2013/508540
Kim K., Goldberg W. M., Taylor G. T. (1992). Architectural and Mechanical Properties of the Black Coral Skeleton (Coelenterata: Antipatharia): A Comparison of Two Species. Biol. Bull. 182, 195–209. doi: 10.2307/1542113
Mass T., Jeana L., Haramaty L., Kim J. D., Zelzion E., Bhattacharya D., et al. (2013). Cloning and Characterization of Four Novel Coral Acid-Rich Proteins That Precipitate Carbonates In Vitro. Curr. Biol. 23, 1126–1131. doi: 10.1016/j.cub.2013.05.007
Mastrobuoni G., Qiao H., Iovinella I., Sagona S., Niccolini A., Boscaro F., et al. (2013). A Proteomic Investigation of Soluble Olfactory Proteins in Anopheles Gambiae. PloS One 8, e75162. doi: 10.1371/journal.pone.0075162
Mathys S., Evans T. C. Jr., Chute I. C., Wu H., Chong S., Benner J., et al. (1999). Characterization of a Self-Splicing Mini-Intein and its Conversion Into Autocatalytic N-And C-Terminal Cleavage Elements: Facile Production of Protein Building Blocks for Protein Ligation. Gene 231, 1–13. doi: 10.1016/S0378-1119(99)00103-1
Montroni D., Sparla F., Fermani S., Falini G. (2021). Influence of Proteins on Mechanical Properties of a Natural Chitin-Protein Composite. Acta Biomaterialia 120, 81–90. doi: 10.1016/j.actbio.2020.04.039
Morgulis M., Martinez S., Almuly R., Einbinder S., Zaslansky P., Mass T. (2022). Black Corals (Antipatharia) of the Northern Red Sea: Ancient Predators of the Mesophotic Reef. Mar. Ecol. Prog. Ser. 688, 33–47. doi: 10.3354/meps14022
Narchi N. E., Marlett C. M., Bertsch H. (2015). “"Corals and Coralline Organisms in Seri Culture: Traditional and Modern Uses,",” in Ethnobiology of Corals and Coral Reefs (Springer) Switzerland, 87–102.
Nocente-McGrath C., Brenner C. A., Ernst S. G. (1989). Endo16, a Lineage-Specific Protein of the Sea Urchin Embryo, is First Expressed Just Prior to Gastrulation. Dev. Biol. 136, 264–272. doi: 10.1016/0012-1606(89)90147-4
Nowak D., Florek M., Nowak J., Kwiatek W., Lekki J., Chevallier P., et al. (2009). Morphology and the Chemical Make-Up of the Inorganic Components of Black Corals. Materials Sci. Engineering: C 29, 1029–1038. doi: 10.1016/j.msec.2008.08.028
Nuc Z., Dobrzycka-Krahel A. (2021). From Chitin to Chitosan - a Potential Natural Antimicrobial Agent. Prog. Chem. Appl. Chitin its Derivatives 26, 23–40. doi: 10.15259/PCACD.26.003
Opresko D. M. (2001). Revision of the Antipatharia (Cnidaria: Anthozoa). Part I. Establishment of a New Family, Myriopathidae. Zoologische Mededelingen 75, 343–370. urn:nbn:nl:ui:19-217449
Parrish F. A., Abernathy K., Marshall G. J., Buhleier B. M. (2002). Hawaiian Monk Seals (Monachus Schauinslandi) Foraging in Deep-Water Coral Beds. Mar. Mammal Sci. 18, 244–258. doi: 10.1111/j.1748-7692.2002.tb01031.x
Peled Y., Drake J. L., Malik A., Almuly R., Lalzar M., Morgenstern D., et al. (2020). Optimization of Skeletal Protein Preparation for LC–MS/MS Sequencing Yields Additional Coral Skeletal Proteins in Stylophora Pistillata. BMC Materials 2, 8. doi: 10.1186/s42833-020-00014-x
Peña Moreno D. V. (2017). Antimicrobial Activity of Culturable Microorganisms Associated With Caribbean Black Corals (Antipatharia) ('Universidad de los Andes, Bogota, Colombia).
Perez-Riverol Y., Csordas A., Bai J., Bernal-Llinares M., Hewapathirana S., Kundu D. J., et al. (2019). The PRIDE Database and Related Tools and Resources in 2019: Improving Support for Quantification Data. Nucleic Acids Res. 47, D442–D450. doi: 10.1093/nar/gky1106
Ramos-Silva P., Kaandorp J., Huisman L., Marie B., Zanella-Cleon I., Guichard N., et al. (2013). The Skeletal Proteome of the Coral Acropora Millepora: The Evolution of Calcification by Cooption and Domain Shuffling. Mol. Biol. Evol. 30(9):2099–2112. doi: 10.1093/molbev/mst109
Rao Z., Handford P., Mayhew M., Knott V., Brownlee G. G., StuartZ D. (1995). The Structure of a Ca2+-Binding Epidermal Growth Factor-Like Domain: Its Role in Protein-Protein Interactions. Cell 82, 131–141. doi: 10.1016/0092-8674(95)90059-4
Rebers J. E., Willis J. H. (2001). A Conserved Domain in Arthropod Cuticular Proteins Binds Chitin. Insect Biochem. Mol. Biol. 31, 1083–1093. doi: 10.1016/S0965-1748(01)00056-X
Ruiz-Ramos D. V., Fisher C. R., Baums I. B., Thomsen L. (2017). Stress Response of the Black Coral Leiopathes Glaberrima When Exposed to Sub-Lethal Amounts of Crude Oil and Dispersant. Elementa: Sci. Anthropocene 5. doi: 10.1525/elementa.261
Samac D. A., Hironaka C. M., Yallaly P. E., Shah D. M. (1990). Isolation and Characterization of the Genes Encoding Basic and Acidic Chitinase in Arabidopsis Thaliana. Plant Physiol. 93, 907–914. doi: 10.1104/pp.93.3.907
Schmidt C. A., Stifler C. A., Luffey E. L., Fordyce B. I., Ahmed A., Barreiro Pujol G., et al. (2022). Faster Crystallization During Coral Skeleton Formation Correlates With Resilience to Ocean Acidification. J. Am. Chem. Soc. 144(3):1332–1341. doi: 10.1021/jacs.1c11434
Shen Z., Jacobs-Lorena M. (1999). Evolution of Chitin-Binding Proteins in Invertebrates. J. Mol. Evol. 48, 341–347. doi: 10.1007/PL00006478
Shimahara K., Takiguchi Y. (1988). “"Preparation of Crustacean Chitin,",” in Methods in Enzymology (Elsevier), 417–423. Academic Press. New York, NY
Sigrist C. J., De Castro E., Cerutti L., Cuche B. A., Hulo N., Bridge A., et al. (2012). New and Continuing Developments at PROSITE. Nucleic Acids Res. 41, D344–D347. doi: 10.1093/nar/gks1067
Suarez H. N., Dy D. T., Violanda R. R. (2015). Density of Associated Macrofauna of Black Corals (Anthozoa: Antipatharia) in Jagna, Bohol, Central Philippines. Philipp J. Sci. 144, 107–115.
Takeuchi T., Yamada L., Shinzato C., Sawada H., Satoh N. (2016). Stepwise Evolution of Coral Biomineralization Revealed With Genome-Wide Proteomics and Transcriptomics. PloS One 11, e0156424. doi: 10.1371/journal.pone.0156424
Thim L. (1997). Trefoil Peptides: From Structure to Function. Cell. Mol. Life Sci. CMLS 53, 888–903. doi: 10.1007/s000180050108
Todinanahary G., Terrana L., Lavitra T., Eeckhaut I. (2016). First Records of Illegal Harvesting and Trading of Black Corals (Antipatharia) in Madagascar. Madagascar Conserv. Dev. 11:1–6. doi: 10.4314/mcd.v11i1.5
Tynyakov J., Bentov S., Abehsera S., Yehezkel G., Roth Z., Khalaila I., et al. (2015). A Crayfish Molar Tooth Protein With Putative Mineralized Exoskeletal Chitinous Matrix Properties. J. Exp. Biol. 218, 3487–3498. doi: 10.1242/jeb.123539
Wagner D., Luck D. G., Toonen R. J. (2012). The Biology and Ecology of Black Corals (Cnidaria: Anthozoa: Hexacorallia: Antipatharia). Adv. Mar. Biol. 63, 67–132. doi: 10.1016/B978-0-12-394282-1.00002-8
Wang L., Stegemann J. P. (2010). Extraction of High Quality RNA From Polysaccharide Matrices Using Cetlytrimethylammonium Bromide. Biomaterials 31, 1612–1618. doi: 10.1016/j.biomaterials.2009.11.024
Zaquin T., Malik A., Drake J. L., Putnam H. M., Mass T. (2021). Evolution of Protein-Mediated Biomineralization in Scleractinian Corals. Front. Genet. 12, 52. doi: 10.3389/fgene.2021.618517
Zheng C., Liu X., Luo X., Zheng M., Wang X., Dan W., et al. (2019). Development of a Novel Bio-Inspired “Cotton-Like” Collagen Aggregate/Chitin Based Biomaterial With a Biomimetic 3D Microstructure for Efficient Hemostasis and Tissue Repair. J. Materials Chem. B 7, 7338–7350. doi: 10.1039/C9TB02028D
Keywords: antipatharians, chitin, chitinase, skeletal proteins, LC-MS/MS protein sequencing
Citation: Drake JL and Mass T (2022) Proteomic Profiling of Black Coral (Antipatharia) Skeleton Reveals Hundreds of Skeleton-Associated Proteins Across Two Taxa. Front. Mar. Sci. 9:904835. doi: 10.3389/fmars.2022.904835
Received: 25 March 2022; Accepted: 30 May 2022;
Published: 30 June 2022.
Edited by:
Rebecca Metzler, Colgate University, United StatesReviewed by:
Anderson B. Mayfield, Coral Reef Diagnostics, United StatesCopyright © 2022 Drake and Mass. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tali Mass, dG1hc3NAdW5pdi5oYWlmYS5lZHU=
†Present address: Jeana L. Drake, Earth, Planetary, and Space Sciences Department, University of California, Los Angeles, United States
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.