- 1Key Laboratory of Marine Genetics and Breeding, Ministry of Education, Ocean University of China, Qingdao, China
- 2Key Laboratory of Tropical Aquatic Germplasm of Hainan Province, Sanya Oceanographic Institution, Ocean University of China, Sanya, China
- 3Hainan Yazhou Bay Seed Laboratory, Sanya, China
The origin and migration of primordial germ cells (PGCs) were regulated by miRNA during embryogenesis, but the underlying mechanism in marine teleost species was still unclear. Here, we studied the function of miR-430 in the development and migration of PGCs in Japanese flounder. By co-injecting miR-430 with eGFP-vasa 3’UTR mRNA, we found that miR-430 could cause the mis-localization of PGCs in flounder. Besides, the overexpression of miR-430 also inhibited the expression of three PGC-related genes (nanos3, piwil2 and tdrd7a), both target gene prediction and dual-luciferase reporter assay showed that miR-430 were directly binding on the canonical target site of their 3’UTRs. Significantly, the miR-430 target site of piwil2 3’UTR was necessary for PGC specific expression, where the binding seeds “GCACTTT” sequence mutation lead the distribution of eGFP-piwil2 mRNA turned from PGC-specific (wild-type) to ubiquitous (mutant). These findings will provide further evidence on the role of miR-430 in the regulation of the development and migration of PGCs.
Introduction
Primordial germ cells (PGCs) segregate from other cell lines during embryogenesis and migrate specifically toward the presumptive genital ridge that forms gonads (Wylie, 1999). The maternal germplasm was proven to play important role in the development of PGCs (Eddy, 1975; Rongo and Lehmann, 1996; Houston and King, 2000; Wang and Lin, 2004; Chen et al., 2010). Several germ cell-specific genes, such as dead end (dnd) (Weidinger et al., 2003), nanos (Subramaniam and Seydoux, 1999) and vasa (Hay et al., 1988), were expressed specifically in PGCs and the chimeric mRNA containing eGFP and the 3’UTR of them have been used for identifying and tracking PGCs (Li et al., 2015; Zhou et al., 2020).
Given that the origin and migration of PGCs during embryogenesis can be tracked by these specific markers, the migration of PGCs has been attracting increasing attention. In teleost fish, the migration of PGCs is regulated by a variety of factors. In zebrafish, the downregulation of stromal cell-derived factor 1a (sdf1a) and its receptor GPCR chemokine receptor 4b (cxcr4b) (Richardson and Lehmann, 2010) and reticulon 3 (rtn3) (Li et al., 2016a) cause clear defects in PGC migration. Besides, nanos3 has also been shown to play an important role in the migration and survival of PGCs in zebrafish (Köprunner et al., 2001). In medaka, dazl, vasa and piwi were essential for the normal migration of PGCs (Li et al., 2009; Li et al., 2012; Li et al., 2016b). Several studies showed that the participation of some microRNAs (miRNAs), such as miR-430, in the migration regulation of PGCs can cause them to be frequently mis-localized by regulating their target genes sdf1a and cxcr7 (Staton et al., 2011).
miRNAs are a group of short non-coding RNA molecules negatively controlling the target genes by binding to the 3’untranslated region (3’UTR) as well as some coding sequences of them. MiRNAs participate in the regulation of a large variety of cellular processes, and most miRNAs show tissue or developmental stage-specific expression (Lagosquintana et al., 2002; Wienholds et al., 2005). Some miRNA families play a key role in the processes of cell proliferation, development, differentiation, and apoptosis (Kloosterman and Plasterk, 2006). For example, the endogenous miR-279 regulates the formation of carbon dioxide receptors (Hartl et al., 2011). MiR-17-92 clusters have transcriptional activation which is regulated by c-Myc and E2F3 (Woods et al., 2007). miR-430 is expressed ubiquitously in early embryos and regulates hundreds of maternal mRNAs (Chen et al., 2005; Giraldez et al., 2006; Lund et al., 2009). Additionally, miR-430 is also needed to suppress PGC-specific genes, such as nanos3 (previously named nanos1) and tdrd7a in somatic cells, while the repression is not effective in germ cells (Mishima et al., 2006; Kedde et al., 2007).
In Japanese flounder, several germ cell marker genes such as tdrd7a and nanos3 have been identified, which can specifically recognize PGCs during embryonic (Li et al., 2015; Wang et al., 2019). It has been suggested that the PGCs might be formed in a unique model, which is different from those of other species. The PGCs of flounder align as two elongated lines at the posterior part of the embryonic body during the early segmentation period, then aggregate into a single loose cluster in posterior and ventral trunks, and finally migrate to the genital ridge as two clusters (Li et al., 2015). The PGC migration pattern in Japanese flounder seems to be a combination of the PGCs migration processes of medaka, herring, and ice goby. However, the underlying mechanism is unclear. This study aims to profile the function of miR-430 in the formation and migration of Japanese flounder PGCs in vivo. The PGCs were visualized by injecting green fluorescent protein (GFP) vasa 3’UTR mRNA, another germ cell-specific marker in Japanese flounder that has been studied previously (Wu et al., 2014). By co-injection of miR-430 and eGFP-vasa 3’UTR mRNA into one-cell stage embryos, we revealed that the PGCs of Japanese flounder were frequently mis-localized after the overexpression of miR-430. Finally, we investigated the effects of miR-430 on its target genes (nanos3, tdrd7a and piwil2) and proved that miR-430 repressed protein expression by binding to the 3’UTR of target genes by the dual-luciferase reporter. The current study revealed the functional association of miR-430 with the formation and migration of PGCs in flounder.
Materials and Methods
Sample Preparation
Adult Japanese flounder were obtained from a commercial hatchery in Haiyang of China and cultured in 2 m3 seawater tanks at 20°C ± 1°C under laboratory conditions for one week. Artificially fertilized eggs were incubated at 22°C ± 0.5°C in the hatching tanks, and embryos at 1-2 cell stages were used for injection. Embryos at different developmental stages from 50% epiboly to hatching were collected for observation. Each sample contained 25 embryos and was collected in triplicate. All samples were frozen immediately in liquid nitrogen and stored at −80°C for total RNA preparation. This research work was conducted in accordance with the Institutional Animal Care and Use Committee of the Ocean University of China and the China Government Principles for the Utilization and Care of Vertebrate Animals Used in Testing, Research, and Training (State Science and Technology Commission of the People’s Republic of China for No.2, October 31, 1988.http://www.gov.cn/gongbao/content/2011/content_1860757.htm).
Construction of Reporter Plasmids
The flounder vasa 3’UTR, which was produced through PCR by using pfu DNA polymerase (Thermo Fisher Scientific, Ambion, USA) with primers eGFP-vasa-Fw/Rv (Table 1), was subcloned into the BamHI and SalI sites of eGFP-tdrd7a-3’UTR plasmid (Wang et al., 2019), to replace tdrd7a 3’UTR with vasa 3’UTR for the generation of the eGFP-vasa-3’UTR plasmid using the Clone EZ Kit (GeneScript, Piscataway, USA). The eGFP-piwil2-3’UTR plasmid was constructed using the same method as that for the eGFP-vasa-3’UTR plasmid. All constructed plasmids were confirmed by sequencing.
Construction of 3’UTR -Luciferase Cassette
The flounder nanos3 3’UTR containing the miR-430 putative-recognizing sequence was cloned into the PmiRGLO dual-luciferase miRNA target expression plasmid to construct PmiRGLO-nanos3-3’UTR. The specific primer pair nanos3 3’UTR–Fw/Rv (Table 1) with restriction sites of SacI and XhoI were used to generate reporter gene constructs. The amplified PCR products were examined through 1.5% agarose gel electrophoresis and purified using the Zymoclean Gel DNA Recovery Kit (Zymo Research, Orange, CA, USA), digested and inserted into PmiRGLO dual-luciferase vector (Promega, Madison, WI, USA). PmiRGLO-piwil2-3’UTR and PmiRGLO-tdrd7a-3’UTR plasmids were constructed using the same method as that for the PmiRGLO-nanos3-3’UTR plasmid. The mutant plasmids were constructed using the 3’UTR with the putative miR-430 binding site deleted. Those plasmids were generated from the PmiRGLO-nanos3/piwil2/tdrd7a-3’UTR via PCR in accordance with the protocol of the QuikChange site directed mutagenesis kit (Agilent Technologies, Santa Clara, CA, USA). Specific mutagenic primers were designed in accordance with the cloned sequence with Integrated DNA Technologies website (http://sg.idtdna.com/Primerquest/Home/Index, Table 1). The PCR program was as follows: 95°C for 5 min and 30 cycles of 95°C for 30 s, 58°C for 30 s, and 72°C for 7 min. PCR products were subjected to DpnI (NEB) digestion to eliminate the parent vector. Besides, the dsRed control plasmid was constructed using the same method, which used the dsRed-Potdrd7b 3’UTR plasmid with the 3’UTR sequence of tdrd7b was deleted (Wang et al., 2019). All plasmids were sequenced to verify that no other mutation occurred during PCR amplification.
Microinjection in Zebrafish and Japanese Flounder Embryos
The miR-430a (5’ UAAGUGCUAUUUGUUGGGGUAG 3’), miR-430c (5’ UAAGU GCUUCUCUUUGGGGUAG 3’) and control (ctrl) miRNA (5’ TAACACGTCTATA CGCCCA 3’) were synthesized by the GenePharma Company (GenePharma, Shanghai, China). Due to the core region of miR-430a and miR-430c being the same, so equal amounts of them were mixed and diluted to 10μM, then stored at -20°C. Chimeric RNA composed of the eGFP-vasa-3’UTR was synthesized in vitro using the MESSAGE mMACHINE Sp6 Kit (Thermo Fisher Scientific, Ambion, USA). eGFP-tdrd7a-3’UTR was linearized using XbaI. Each embryo was co-injected 20 pg mRNA with an equal volume of 5 μM miRNA (mir-430/ctrl miRNA) at the one-cell stage, respectively. A total of 200 embryos were injected for each group, due to the short spawning time of flounder and the low survival rate after injection, we observed about 50 flounder embryos in each group. The injected zebrafish or Japanese flounder eggs were cultured at 28.0 ± 0.5°C or 22.0 ± 0.5°C, respectively, and observed under a fluorescence microscope for eGFP and dsRed at different developmental stages.
Cell Line Culture
The flounder embryo cell (FEC) line was obtained from the cell culture of blastula embryos of Japanese flounder and maintained using a previous method (Tong et al., 1997). The monolayer cultures of cells exhibited an epithelioid morphology and subcultured for more than 200 passages. All cells were maintained in Dulbecco’s modified Eagle’s medium, DMEM/F12 (1:1) (Gibco, Carlsbad, USA) and supplemented with 10% fetal bovine serum (Gibco, Carlsbad, USA) at 24°C in an atmosphere of air (Na et al., 2009). The cell culture medium contained 100 U/mL penicillin and 100 μg/mL streptomycin.
Transient Transfection
One day before transfection, cells were seeded into 24-well cell culture plates (NEST Biotechnology, Wuxi, China) with 2 × 105 cells/well and cultured in 0.5 mL DMEM at 24°C. Transient cell transfection was performed using the Lipofectamine 3000 (Invitrogen, Carlsbad, CA, USA) in accordance with the manufacturer’s protocol when the cells grew to 80% – 90% confluence.
For the dual-luciferase reporter assay, each well was co-transfected with 500 ng PmiRGLO-nanos3/tdrd7a/piwil2-3’UTR or deletion mutation constructs alone, or together with 5 μM miR-430. Mixtures were added with 1 μL of P3000 and 1.5 μL of Lipo3000. The control cells were transfected with the PmiRGLO basic plasmid which was a 3’UTR less vector. The medium of each well was replaced with 500 μL of growth media containing 10% FBS after 6 h transfection. The transfection was maintained for 24 h, and the cells were washed with phosphate-buffered saline and lysed for 15 min with 100 μL Passive Lysis Buffer provided in a the dual-luciferase reporter Assay System (Promega, Madison, WI, USA). Twenty microliters of cell lysate was transferred into the EP tube containing 100 μL of Luciferase Assay Reagent II, then added with 100 μL of Stop & Glo®Reagent, the Renilla luciferase activities were read. The luciferase activity was measured using the GloMax 20/20 Luminometer. The luminometer was programmed to perform a 2s premeasurement delay followed by a 10s measurement period for each reporter assay. The luciferase activities were obtained by the ratio of firefly to the Renilla luciferase. All data were obtained from three independent measurements.
Target Gene Prediction
The online target site prediction software TargetScan (http://www.targetscan.org/) and MicroCosm Targets (http://www.ebi.ac.uk/enrightsrv/microcosm/htdocs/targets/v5/) were used to search potential target genes for miR-430. miR-430 mature sequence and the 3’UTR of Japanese flounder as the input files predicted the target sites by TargetScan and MicroCosm Targets. The 3’UTR of Japanese flounder was identified from its genome (NCBI accession number: PRJNA352095) and transcriptome database (NCBI accession number: SRX500343). The screening conditions were as follows: perfect complementarity between the seed sequence of miRNA and mRNA 3’UTR (free energy ≤ −20 kcal/mol).
RNA Isolation and cDNA Synthesis
The total RNA was extracted from samples using Trizol Reagent (Invitrogen, CA, USA) according to the manufacturer’s protocol, treated with RNase-free DNase I (TaKaRa, Dalian, China) to degrade genomic DNA, and then frozen at −80°C. The quality and quantity of total RNA were evaluated through 1.5% (w/v) agarose gel electrophoresis and spectrophotometry using Nano Photometer Pearl (Implen, Munich, Germany). The cDNA was transcribed with 1 μg of total RNA using the reverse transcriptase M-MLV Kit (TaKaRa, Dalian, China) and random primers (NNNNNN) according to the manufacturer’s instructions.
Quantitative Real-Time PCR
All qRT-PCR analyses were performed in a 20 µL reaction volume containing 1 μL of cDNA template (10 ng/μL), 0.4 μL of each primer (10 μM), 10 μL of 2×SYBR Green qPCR Master Mix (US Everbright Inc., Suzhou China) and 8.2 μL nuclease-free water through the LightCycler480 (Roche Applied Science, Mannheim, Germany). The expression levels of nanos3, piwil2 and tdrd7a were detected at 50% epiboly stage after injecting miR-430. The primers for qRT-PCR were designed according to their sequences at the Integrated DNA Technologies website (http://sg.idtdna.com/Primerquest/Home/Index, Table 1), β-actin was selected as the reference gene (Zheng and Sun, 2011). Verified primers were then used to perform the qPCR with 95°C (5 min) for pre-incubation, followed by 45 cycles at 95°C (15 s) and 60°C (45 s). The reaction of each sample was performed in triplicate. Melting curves were generated after the amplification reaction completion to confirm the specificity of amplicons. All samples were run in triplicate, and data were calculated using 2−ΔΔCt method.
Statistical Analysis
Expression data were analyzed using the SPSS 20.0 software. Differences were tested via one-way ANOVA and considered significant at p < 0.05. All data were expressed as mean ± standard error of the mean (SEM).
Results
Visualizing PGCs Using Flounder Vasa 3’UTR
The synthesized eGFP-vasa 3’UTR was injected into zebrafish embryos, to determine whether Japanese flounder vasa 3’UTR could specifically label PGCs. As shown in Figure 1A, in embryos injected with eGFP-vasa 3’UTR, the eGFP signal was observed in a limited number of cells in the position of putative PGCs. The position of putative PGCs showed strong eGFP expression commencing at the 9 hours post fertilization (hpf) stage (Figure 1A). In addition, PGCs showed stronger eGFP signals than other cell types. During the somitogenesis stage at 16 hpf and 36 hpf, the eGFP signals in the embryo body diminished except in cells at the position of putative PGCs. Then, the synthesized eGFP-vasa-3’UTR injected into the one cell stage embryos to recognize the germ cells of flounder. At hatching stage (72 hpf), in the embryos injected with eGFP-vasa-3’UTR mRNA, the eGFP signal was observed in a limited number of cells in the position of putative PGCs (Figure 1B).
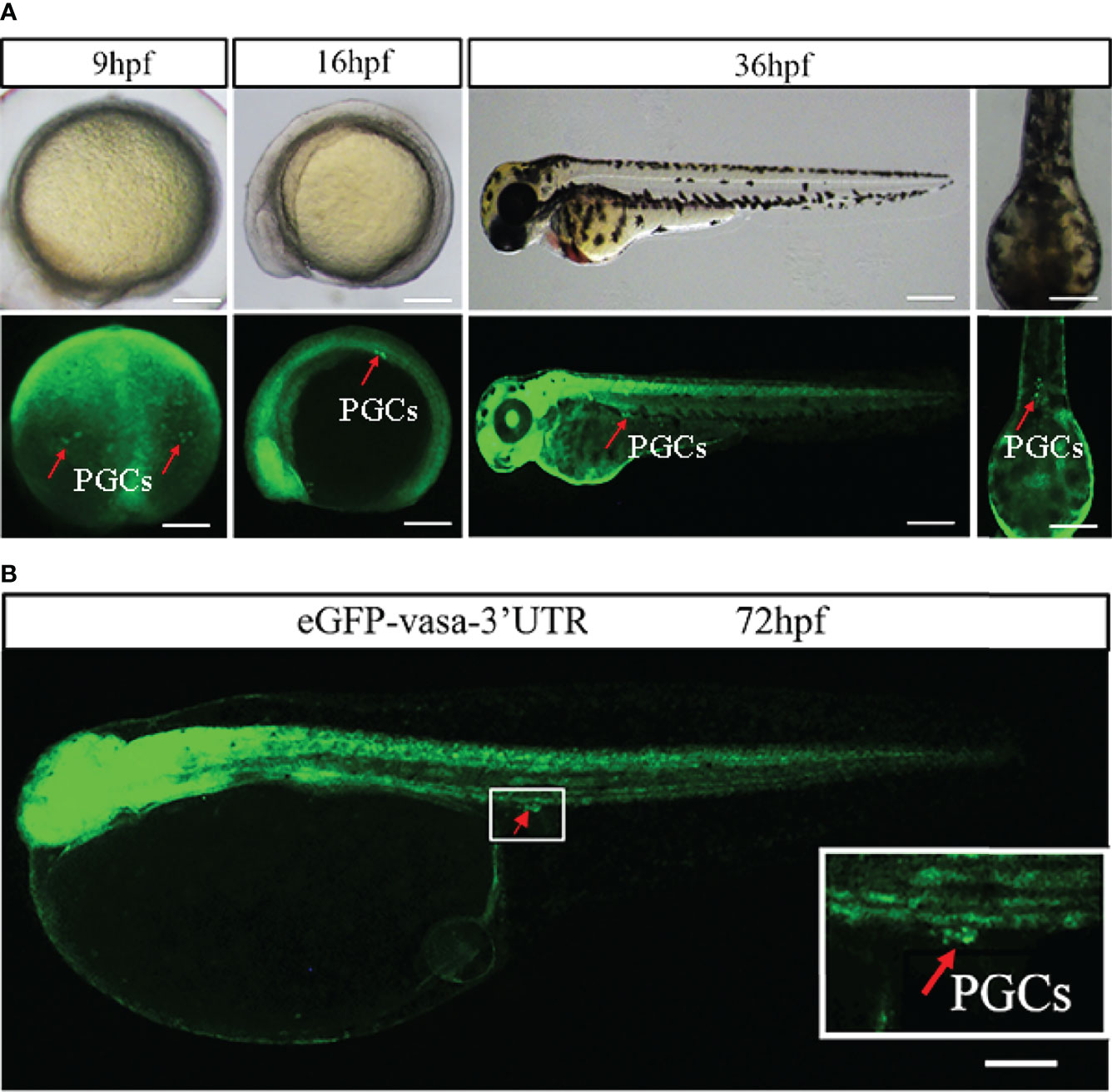
Figure 1 Visualization of PGCs by vasa. (A) Lateral views of zebrafish embryos during three different developmental stages: 9 hpf, 16 hpf and 36 hpf. At these three stages, all PGCs showed stronger eGFP signals in the position of putative PGCs than other cells. Scale bar = 250 mm. (B) Fluorescence showed eGFP (green) expression in 72 hpf embryos injected with eGFP-vasa 3’UTR mRNA. At the position of putative PGCs, eGFP signals was clearly observed. Arrows indicate the eGFP-positive cells. Scale bar = 100 mm.
Regulation of the PGCs Migration by miR-430
To analyze the effect of miR-430 on the PGCs of Japanese flounder, we co-injected the miR-430 with eGFP-vasa-3’UTR mRNA at one cell stages, because PGCs could be labeled by vasa 3’UTR (Figure 1). As shown in Figure 2A, All PGCs of co-injected with control miRNA appeared at the presumptive genital ridge region at the hatching stage (72 hpf). In the group co-injected with miR-430, a part of eGFP positive PGCs was evidently mis-localized, thereby scattering all over the posterior body Figure 2.
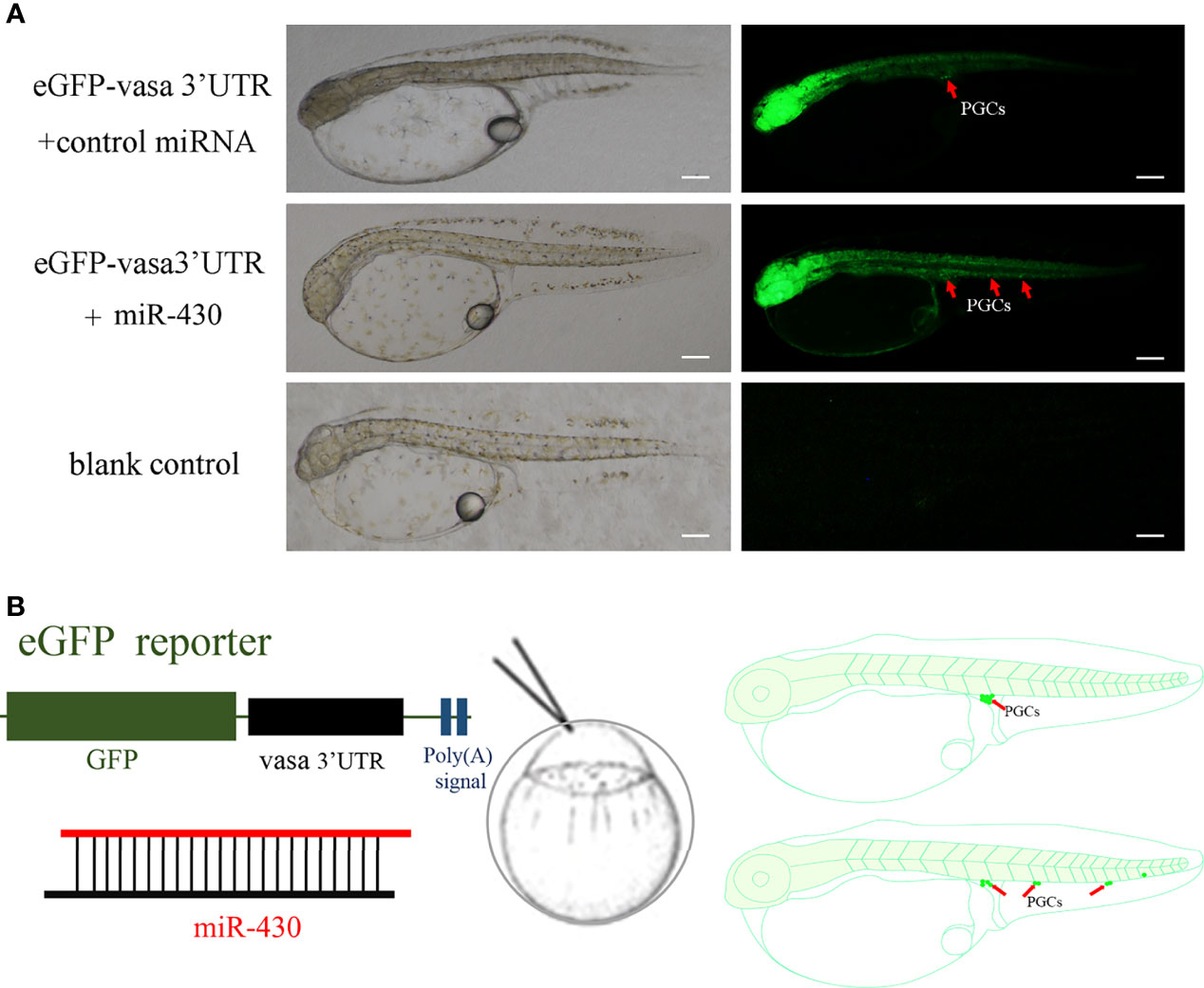
Figure 2 miR-430 regulates PGC migration in Japanese flounder. (A) Normally localized PGCs at the hatching stage (72 hpf) in the group co-injected with eGFP-vasa-3’UTR and control miRNA; Mis-localized PGCs at the hatching stage (72 hpf) in group co-injected with miR-430; The blank control group have no injected. Arrows indicate the location of PGCs. Scale bar = 100 μm. (B) Experimental approach. Vasa eGFP reporter mRNA was co-injected with miRNA into one-cell stage Japanese flounder embryos, causing the mis-localization of PGCs in the embryo.
Regulation of PGC-specific gene expression by miR-430
In present study, the target genes of miR-430 were predicted using the TargetScan and MicroCosm Targets. The 3’UTR of three putative target genes (nanos3, tdrd7a and piwil2) containing miR-430 binding site, are have been reported to be PGC-related genes in various species (Makoolati et al., 2016; Sun et al., 2017; Wang et al., 2019). The miR-430 seed region on the 3’UTR of these three genes was labeled blue (Figure 3A). The quantitative expression of nanos3, piwil2 and tdrd7a were detected by qRT-PCR after overexpression of miR-430 to investigate the regulation effects of miR-430 on the expression of these three genes. As shown in Figure 3B, expression of nanos3 was downregulated, and the piwil2 and tdrd7a mRNA levels were significantly inhibited with the overexpression of miR-430 (p < 0.01).
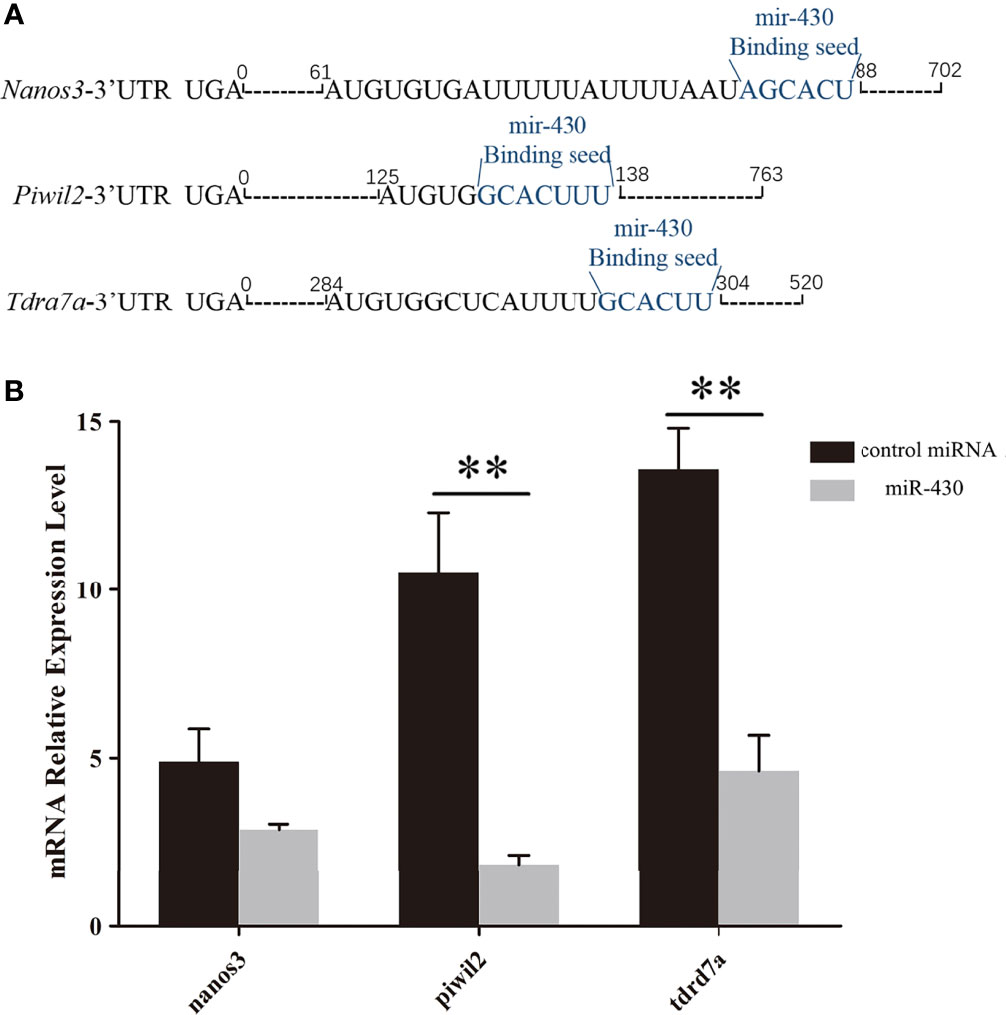
Figure 3 Quantitative expression of nanos3, piwil2 and tdrd7a by qPCR. (A) Distribution of miR-430 target seeds in nanos3, piwil2 and tdrd7a 3’UTR. Blue letters represent the miR-430 binding position. (B) Changes in the endogenous levels of nanos3, piwil2 and tdrd7a after miR-430 injection. Data are shown as mean ± SEM (n = 3). Statistical significance was marked by an asterisk (**p < 0.01).
Validation of miR-430 and Target Gene Interactions
A total of three reporter plasmids containing putative target sites (pmiRGLO-nanos3 3’UTR wt, pmiRGLO-piwil2 3’UTR wt, pmiRGLO-tdrd7a 3’UTR wt) and 3 miRNA (miR-430a, miR-430c and ctrl miRNA) were constructed for dual-luciferase assays to validate the interaction between miR-430 and its putative target genes. In order to further validate the regulating relationships between miR-430 and the three target genes (nanos3, piwil2 and tdrd7a), three mutant vectors, pmiRGLO-nanos3 3’UTR mut, pmiRGLO-piwil2 3’UTR mut and pmiRGLO-tdrd7a 3’UTR mut were constructed (Figure 4A). Three kinds of wild type plasmid pmiRGLO-3’UTR wt were co-transfected with miR-430 or ctrl miRNA to the flounder embryo cell (FEC) line, and the expression levels of nanos3 (p < 0.01), piwil2 (p < 0.05) and tdrd7a (p <0.01) were significantly decreased, respectively. By contrast, when the mutant type plasmids pmiRGLO-3’UTR mut were transfected, no evident difference in expression between the mutant vectors and the negative controls were observed (Figures 4B–D). Results strongly suggested that nanos3, piwil2 and tdrd7a were directly regulated by miR430 in Japanese flounder.
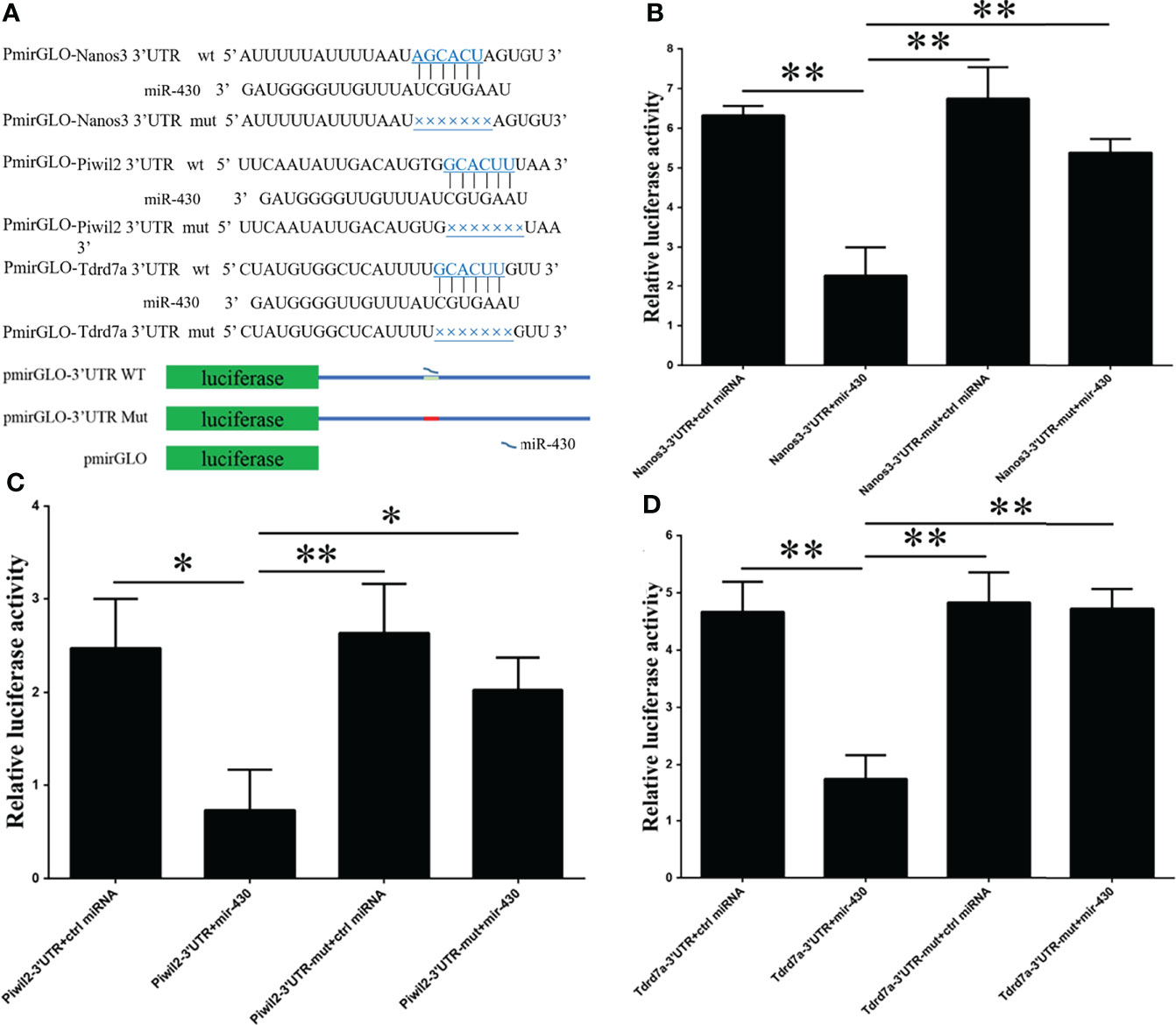
Figure 4 Schematic of the miR‐430 target sequence within the 3’UTR of nanos3, piwil2, and tard7a and activities of the dual-luciferase reporter gene linked to their 3′UTR. (A) Schematic mutant site structures in vectors PmirGLO-nanos3 3’UTR mut, PmirGLO-piwil2 3’UTR mut and PmirGLO-tdrd7a 3’UTR mut. (B–D) Dual-luciferase reporter analysis of miR-430 and target genes. The Japanese flounder embryo cell (FEC) line was co-transfected with reporter vector carrying 3’UTRs of nanos3, piwil2 and tdrd7a, and miR-430 or control miRNA. Luciferase activities were measured after 24h, and firefly luciferase activity was measured and normalized to the renilla luciferase activity. Data are shown as mean ± SEM (n = 3). Statistical significance was marked by asterisk (*p < 0.05 and **p < 0.01).
Post-Transcriptional Gene Regulation by miR-430
We constructed reporter mRNAs containing an eGFP open-reading frame and wild type or mutant 3’UTR of flounder piwil2 carrying the canonical target site for miR-430. Either the synthesized eGFP-piwil2-3’UTR wt or eGFP-piwil2-3’UTR mut mRNA was co-injected with dsRed mRNA into flounder fertilized eggs (Figure 5A). The eGFP signal was ubiquitously expressed in gastrula and neurula embryos. In the eGFP-piwil2-3’UTR wt group, eGFP showed strong signal in putative PGCs, and a weak signal in other parts of the whole body at 72 hpf (Figure 5B). However, in the group injected with eGFP-piwil2-3’UTR mut mRNA, eGFP was ubiquitously expressed all over the fry body (Figure 5B). In both groups, the co-injected dsRed was ubiquitously expressed, verifying the even distribution of injected mRNA molecules (Figure 5B).
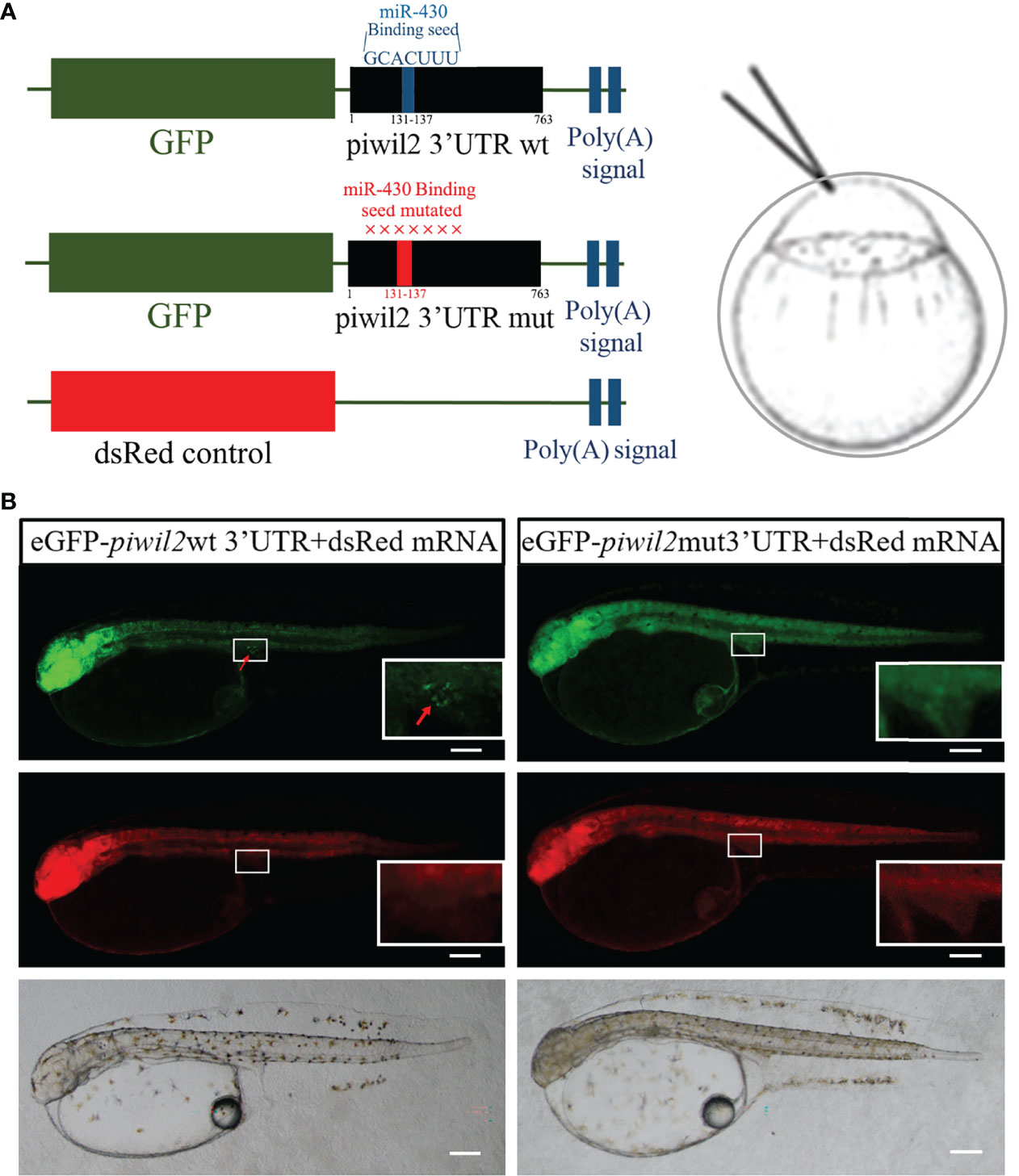
Figure 5 Piwil2 3’UTR targeted at miR-430 binding sites in Japanese flounder. (A) Schematic of the experimental set-up. The eGFP reporters with the 3’UTRs of piwil2 were used to examine regulatory the effect of miR-430 to Piwil2 in flounder embryo, dsRed mRNA lacking the target site was co-injected as a control. The miR-430 binding site in piwil2 3’UTR mutated and had a red color, the wild-type had a blue color. (B) Embryos injected with mRNA of wild-type or mutated constructs of the eGFP-piwil2-3’UTR reporter at the one-cell stage. Fluorescence microscopy showed eGFP (green) and dsRed (red) expression in 72 hpf embryos injected with eGFP reporters with piwil2 3’UTR. Endogenous miR-430 repressed the expression of reporter in wild type, and only showed strong signal in the putative PGCs, but the eGFP of mut-3’UTR reporter was highly expressed all over the fry body. The inset showed the enlargement of the gonadal region. Arrows indicate the eGFP-positive cells. Scale bar = 100 μm.
Discussion
The biological mechanism of PGCs migration has been reported in many species (Li et al., 2012; Li et al., 2015; Li et al., 2016a). The functions of miR-430 during the development of PGCs have been studied in different teleost, such as zebrafish (Staton et al., 2011), goldfish (Jie et al., 2014) and medaka (Tani et al., 2010). In the present study, we provide evidence that the miR-430 in Japanese flounder regulates the migration of PGCs, and repress the expression of several genes with PGC-specific expression.
Given that PGCs contribute to the transmission of genetic information from one generation to the next, the study of PGCs has been attracting attention. While PGCs specification in different species occurs via different mechanisms (Mclaren, 1999; Sasado et al., 2008; Li et al., 2015), the germplasm components required for early PGC development in different organisms, such as dnd (Weidinger et al., 2003), nanos (Subramaniam and Seydoux, 1999), and vasa (Hay et al., 1988), are common. Among the transcripts involved in PGC specification, vasa were one of the most conserved elements and universal studied marker of PGC (Lasko and Ashburner, 1988; Ikenishi and Tanaka, 1997; Kuznicki et al., 2000; Hartung et al., 2014; Reitzel et al., 2016). Vasa homologs are characterized in many organisms, such as fruit fly (Schupbach and Wieschaus, 1986), zebrafish (Yoon et al., 1997), European sea bass (Blázquez et al., 2011), Atlantic salmon (Nagasawa et al., 2013) and catfish (Raghuveer and Senthilkumaran, 2010), as a specific germline transcript. Our present results showed that vasa could label PGCs of flounder by injecting the eGFP-vasa-3’UTR mRNA, this finding was consistent with previous study that vasa is a germ cell-specific marker (Wu et al., 2014). Furthermore, the function of miR-430 was analyzed during the migration of PGCs in flounder by co-injecting the vasa reporter mRNA and miR-430 (Figure 2). The overexpression of miR-430 caused PGCs to mis-localization frequently, this finding was also observed in previous studies (Staton et al., 2011), which suggested a similar function during the early development of PGCs in different organisms.
miRNAs are a group of short non-coding RNA molecules that post-transcriptionally regulate the expression of large numbers of genes in metazoans, which controlling the target genes by binding to the 3’UTR as well as some coding sequences of them (Lewis et al., 2005; Stark et al., 2005). Previous studies have proved that miR-430 was expressed when zygotic transcription, and accelerated the deadenylation and decay of maternal mRNAs (Giraldez et al., 2006; Staton et al., 2011). In zebrafish, multiple target genes of miR-430 are identified as PGC-related genes. Among these genes, nanos3 and tdrd7a mRNA have been experimentally proven to be post-transcriptionally downregulated (Mishima et al., 2006; Kedde et al., 2007). Besides, the 3’UTR of them also contains cis-regulatory sequences, such as “GUUC”, which is required for repressing deadenylation. RNA-binding protein DAZL could direct binding to cis-regulatory sequences and is sufficient for the PGC-specific activation of miR-430 target mRNA (Mishima et al., 2006; Takeda et al., 2009). Similarly, 3’UTR of another PGC-related gene tdrd7a was also targeted by miR-430 (Tani et al., 2010). Besides, another RNA-binding protein, DND could also protect the expression of these genes in PGCs from repression by miRNAs, as exemplified by miR-430 (Kedde et al., 2007). Thus, miR-430 was suggested to suppress the PGC-related genetic cascade in somatic cells. Our present study showed that the expression levels of nanos3, piwil2 and tdrd7a were evidently downregulated after the overexpression of miR-430 in flounder embryos. Further, the dual-luciferase reporter analysis showed that miR-430 regulated nanos3, piwil2 and tdrd7a by directly binding to the 3’UTRs of these three genes in Japanese flounder, thereby affecting their roles in PGC specification, migration, and gametogenesis.
Previous studies have shown that piwil2 and piRNA have close interactions for germ-cell maintenance (Aravin et al., 2006; Grivna et al., 2006). In the present study, we provided evidence that miR-430 regulates piwil2 directly through binding to its target site of the 3’UTR. The regulation mechanism of miR-430 in the piwil2 of flounder was quite similar to those in nanos3 and tdrd7 in zebrafish (Kedde et al., 2007; Takeda et al., 2009; Chen et al., 2010). The mRNAs of this gene did not completely disappear, due to the repression was not effective in germ cells (Mishima et al., 2006; Kedde et al., 2007).
Many maternal factors that control early embryo development have been identified by maternal-effect mutant screening in zebrafish (Abrams and Mullins, 2009). Some of these factors, such as vasa, nanos and piwi, have been demonstrated to be required for PGCs formation (Köprunner et al., 2001; Hashimoto et al., 2004; Li et al., 2009). Moreover, miR-430 was shown to control PGCs development in zebrafish, carp and medaka (Tani et al., 2010; Staton et al., 2011). Furthermore, several maternal factors involved in early embryo development, such as nanos, dazl, tdrd7 and piwi, were also confirmed to be targeted by miR-430. In the future, elucidating the relationship between miR-430 and maternal mRNA in Japanese flounder is interesting.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics Statement
The animal study was reviewed and approved by Institutional Animal Care and Use Committee of the Ocean University of China.
Author Contributions
BW contributed to the experimental process, data analysis, and manuscript writing. QZ and JQ conceived and designed the experiments. FY, and CJ contributed to the experimental process. JH contributed to the acquisition of experiment samples, and all authors have read and approved the final manuscript. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by the National Key Research and Development Program of China (Grant No. 2018YFD0901205 and 2020YFD091002) and the Hainan Yazhou Bay Seed Laboratory (B21Y10802).
Conflict of Interest
The reviewer [ZQ] declared a shared affiliation with the authors to the handling editor at the time of review.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abrams E. W., Mullins M. C. (2009). Early Zebrafish Development: It's in the Maternal Genes. Curr. Opin. Genet. Dev. 19 (4), 396–403. doi: 10.1016/j.gde.2009.06.002
Aravin A., Gaidatzis D., Pfeffer S., Lagosquintana M., Landgraf P., Iovino N., et al. (2006). A Novel Class of Small RNAs Bind to MILI Protein in Mouse Testes. Nature 442 (7099), 203–207. doi: 10.1038/nature04916
Blázquez M., González A., Mylonas C. C., Piferrer F. (2011). Cloning and Sequence Analysis of a Vasa Homolog in the European Sea Bass (Dicentrarchus Labrax): Tissue Distribution and mRNA Expression Levels During Early Development and Sex Differentiation. Gen. Comp. Endocrinol. 170 (2), 322–333. doi: 10.1016/j.ygcen.2010.10.007
Chen P. Y., Manninga H., Slanchev K., Chien M., Russo J. J., Ju J., et al. (2005). The Developmental miRNA Profiles of Zebrafish as Determined by Small RNA Cloning. Genes Dev. 19 (11), 1288–1293. doi: 10.1101/gad.1310605
Chen S., Zeng M., Sun H., Deng W., Lu Y., Tao D., et al. (2010). Zebrafish Dnd Protein Binds to 3'UTR of Geminin mRNA and Regulates its Expression. Bmb Rep. 43 (6), 438–444. doi: 10.5483/BMBRep.2010.43.6.438
Eddy E. M. (1975). Germ Plasm and the Differentiation of the Germ Cell Line. Intrevcytol 43, 229–280. doi: 10.1016/s0074-7696(08)60070-4
Giraldez A. J., Mishima Y., Rihel J., Grocock R. J., Van Dongen S., Inoue K., et al. (2006). Zebrafish MiR-430 Promotes Deadenylation and Clearance of Maternal mRNAs. Science 312 (5770), 75–79. doi: 10.1126/science.1122689
Grivna S. T., Pyhtila B., Lin H. (2006). MIWI Associates With Translational Machinery and PIWI-Interacting RNAs (piRNAs) in Regulating Spermatogenesis. Proc. Natl. Acad. Sci. U.S.A. 103 (36), 13415–13420. doi: 10.1073/pnas.0605506103
Hartl M., Loschek L. F., Stephan D., Siju K. P., Knappmeyer C., Kadow I. C. (2011). A New Prospero and microRNA-279 Pathway Restricts CO2 Receptor Neuron Formation. J. Neurosci. 31 (44), 15660–15673. doi: 10.1523/JNEUROSCI.2592-11.2011
Hartung O., Forbes M. M., Marlow F. L. (2014). Zebrafish Vasa is Required for Germ-Cell Differentiation and Maintenance. Mol. Reprod. Dev. 81 (10), 946–961. doi: 10.1002/mrd.22414
Hashimoto Y., Maegawa S., Nagai T., Yamaha E., Suzuki H., Yasuda K., et al. (2004). Localized Maternal Factor are Required for Zebrafish Germ Cell Formation. Dev. Biol. 268 (1), 152–161. doi: 10.1016/j.ydbio.2003.12.013
Hay B., Jan L. Y., Jan Y. N. (1988). A Protein Component of Drosophila Polar Granules is Encoded by Vasa and has Extensive Sequence Similarity to ATP-Dependent Helicases. Cel 55 (4), 577–587. doi: 10.1016/0092-8674(88)90216-4
Houston D. W., King M. L. (2000). Germ Plasm and Molecular Determinants of Germ Cell Fate. Curr. Topics Dev. Biol. 50, 155–181. doi: 10.1016/S0070-2153(00)50008-8
Ikenishi K., Tanaka T. S. (1997). Involvement of the Protein of Xenopus Vasa Homolog (Xenopus Vasa-Like Gene 1, XVLG1) in the Differentiation of Primordial Germ Cells. Dev. Growth Differ. 39 (5), 625–633. doi: 10.1046/j.1440-169X.1997.t01-4-00010.x
Jie M., Yue H. M., Zhi L., Bo C., Zhong J. X., Cheng D., et al. (2014). C1q-Like Factor, a Target of miR-430, Regulates Primordial Germ Cell Development in Early Embryos of Carassius Auratus. Int. J. Biol. Sci. 10 (1), 15–24. doi: 10.7150/ijbs.7490
Kedde M., Strasser M. J., Boldajipour B., Vrielink J. A. F. O., Slanchev K., Sage C. L., et al. (2007). RNA-Binding Protein Dnd1 Inhibits MicroRNA Access to Target mRNA. Cell 131 (7), 1273–1286. doi: 10.1016/j.cell.2007.11.034
Kloosterman W. P., Plasterk R. H. (2006). The Diverse Functions of MicroRNAs in Animal Development and Disease. Dev. Cell 11 (4), 441–450. doi: 10.1016/j.devcel.2006.09.009
Köprunner M., Thisse C., Thisse B., Raz E. (2001). A Zebrafish Nanos-Related Gene is Essential for the Development of Primordial Germ Cells. Genes Dev. 15 (21), 2877–2885. doi: 10.1101/gad.212401
Kuznicki K. A., Smith P. A., Leung-Chiu W. M., Estevez A. O., Scott H. C., Bennett K. L. (2000). Combinatorial RNA Interference Indicates GLH-4 can Compensate for GLH-1; These Two P Granule Components are Critical for Fertility in C. Elegans. Development 127 (13), 2907–2916. doi: 10.1242/dev.127.13.2907
Lagosquintana M., Rauhut R., Yalcin A., Meyer J., Lendeckel W., Tuschl T., et al. (2002). Identification of Tissue-Specific MicroRNAs From Mouse. Curr. Biol. 12 (9), 735–739. doi: 10.1016/S0960-9822(02)00809-6
Lasko P. F., Ashburner M. (1988). The Product of the Drosophila Gene Vasa is Very Similar to Eukaryotic Initiation Factor-4A. Nature 335 (6191), 611–617. doi: 10.1038/335611a0
Lewis B. P., Burge C. B., Bartel D. P. (2005). Conserved Seed Pairing, Often Flanked by Adenosines, Indicates That Thousands of Human Genes are MicroRNA Targets. Cell 120 (1), 15–20. doi: 10.1016/j.cell.2004.12.035
Li M., Hong N., Gui J., Hong Y. (2012). Medaka Piwi is Essential for Primordial Germ Cell Migration. Curr. Mol. Med. 12 (8), 1040–1049. doi: 10.2174/156652412802480853
Li M., Hong N., Xu H., Yi M., Li C., Gui J., et al. (2009). Medaka Vasa is Required for Migration But Not Survival of Primordial Germ Cells. Mech. Dev. 126 (5-6), 366–381. doi: 10.1016/j.mod.2009.02.004
Li H., Rong L., Lu Y., Wang M., Li Z. (2016a). RTN3 Regulates the Expression Level of Chemokine Receptor CXCR4 and is Required for Migration of Primordial Germ Cells. Int. J. Mol. Sci. 17 (4), 382. doi: 10.3390/ijms17040382
Li M., Tan X., Jiao S., Wang Q., Wu Z., You F., et al. (2015). A New Pattern of Primordial Germ Cell Migration in Olive Flounder (Paralichthys Olivaceus) Identified Using Nanos3. Dev. Genes Evol. 225 (4), 195–206. doi: 10.1007/s00427-015-0503-6
Li M., Zhu F., Li Z., Hong N., Hong Y. (2016b). Dazl is a Critical Player for Primordial Germ Cell Formation in Medaka. Sci. Rep. 6, 28317. doi: 10.1038/srep28317
Lund E., Liu M., Hartley R. S., Sheets M. D., Dahlberg J. E. (2009). Deadenylation of Maternal mRNAs Mediated by miR-427 in Xenopus Laevis Embryos. Rna 15 (12), 2351–2363. doi: 10.1261/rna.1882009
Makoolati Z., Movahedin M., Forouzandeh-Moghadam M. (2016). Proliferation in Culture of Primordial Germ Cells Derived From Embryonic Stem Cell: Induction by Retinoic Acid. Bioscience Rep. 36 (6), e00428. doi: 10.1042/BSR20160441
Mishima Y., Giraldez A. J., Takeda Y., Fujiwara T., Sakamoto H., Schier A. F., et al. (2006). Differential Regulation of Germline mRNAs in Soma and Germ Cells by Zebrafish miR-430. Curr. Biol. 16 (21), 2135–2142. doi: 10.1016/j.cub.2006.08.086
Nagasawa K., Fernandes J. M. O., Yoshizaki G., Miwa M., Babiak I. (2013). Identification and Migration of Primordial Germ Cells in Atlantic Salmon, Salmo Salar: Characterization of Vasa, Dead End, and Lymphocyte Antigen 75 Genes. Mol. Reprod. Dev. 80 (2), 118–131. doi: 10.1002/mrd.22142
Na N., Guo H. R., Zhang S. C., Li Z. J., Yin L. C. (2009). In Vitro and In Vivo Acute Toxicity of Fenpyroximate to Flounder Paralichthys Olivaceus and its Gill Cell Line FG. Aquat. Toxicol. 92 (2), 76–85. doi: 10.1016/j.aquatox.2008.12.006
Raghuveer K., Senthilkumaran B. (2010). Cloning and Differential Expression Pattern of Vasa in the Developing and Recrudescing Gonads of Catfish, Clarias Gariepinus. Comp. Biochem. Physiol. Part A: Physiol. 157 (1), 79–85. doi: 10.1016/j.cbpa.2010.04.017
Reitzel A., Pang K., Martindale M. Q. (2016). Developmental Expression of “Germline”- and “Sex Determination”-Related Genes in the Ctenophore Mnemiopsis Leidyi. Evodevo 7, 17. doi: 10.1186/s13227-016-0051-9
Richardson B. E., Lehmann R. (2010). Mechanisms Guiding Primordial Germ Cell Migration: Strategies From Different Organisms. Nat. Rev. Mol. Cell Biol. 11 (1), 37–49. doi: 10.1038/nrm2815
Rongo C., Lehmann R. (1996). Regulated Synthesis, Transport and Assembly of the Drosophila Germ Plasm. Trends Genet. 12 (3), 102–109. doi: 10.1016/0168-9525(96)81421-1
Sasado T., Yasuoka A., Abe K., Mitani H., Furutaniseiki M., Tanaka M., et al. (2008). Distinct Contributions of CXCR4b and CXCR7/RDC1 Receptor Systems in Regulation of PGC Migration Revealed by Medaka Mutants Kazura and Yanagi. Dev. Biol. 320 (2), 328–339. doi: 10.1016/j.ydbio.2008.05.544
Schupbach T., Wieschaus E. (1986). Germline Autonomy of Maternal-Effect Mutations Altering the Embryonic Body Pattern of Drosophila. Dev. Biol. 113 (2), 443–448. doi: 10.1016/0012-1606(86)90179-X
Stark A., Brennecke J., Bushati N., Russell R. B., Cohen S. M. (2005). Animal MicroRNAs Confer Robustness to Gene Expression and Have a Significant Impact on 3'UTR Evolution. Cell 123 (6), 1133–1146. doi: 10.1016/j.cell.2005.11.023
Staton A. A., Knaut H., Giraldez A. J. (2011). miRNA Regulation of Sdf1 Chemokine Signaling Provides Genetic Robustness to Germ Cell Migration. Nat. Genet. 43 (3), 204–211. doi: 10.1038/ng.758
Subramaniam K., Seydoux G. (1999). Nos-1 and Nos-2, Two Genes Related to Drosophila Nanos, Regulate Primordial Germ Cell Development and Survival in Caenorhabditis Elegans. Development 126 (21), 4861–4871. doi: 10.1242/dev.126.21.4861
Sun Z. H., Wang Y., Lu W. J., Li Z., Liu X. C., Li S. S., et al. (2017). Divergent Expression Patterns and Function Implications of Four Nanos Genes in a Hermaphroditic Fish, Epinephelus Coioides. Int. J. Mol. Sci. 18 (4), 685. doi: 10.3390/ijms18040685
Takeda Y., Mishima Y., Fujiwara T., Sakamoto H., Inoue K. (2009). DAZL Relieves miRNA-Mediated Repression of Germline mRNAs by Controlling Poly(A) Tail Length in Zebrafish. PloS One 4 (10), e7513. doi: 10.1371/journal.pone.0007513
Tani S., Kusakabe R., Naruse K., Sakamoto H., Inoue K. (2010). Genomic Organization and Embryonic Expression of miR-430 in Medaka (Oryzias Latipes): Insights Into the Post-Transcriptional Gene Regulation in Early Development. Gene 449 (1-2), 41–49. doi: 10.1016/j.gene.2009.09.005
Tong S. L., Li H., Miao H. Z. (1997). The Establishment and Partial Characterization of a Continuous Fish Cell Line FG-9307 From the Gill of Flounder Paralichthys Olivaceus. Aquaculture 156 (3-4), 327–333. doi: 10.1016/S0044-8486(97)00070-7
Wang B., Du X., Wang H., Jin C., Gao C., Liu J., et al. (2019). Comparative Studies on Duplicated Tdrd7 Paralogs in Teleosts: Molecular Evolution Caused Neo-Functionalization. Comp. Biochem. Physiol. Part D Genomics Proteomics 6 (30), 347–357. doi: 10.1016/j.cbd.2019.04.006
Wang Z., Lin H. (2004). Nanos Maintains Germline Stem Cell Self-Renewal by Preventing Differentiation. Science 303 (5666), 2016–2019. doi: 10.1126/science.1093983
Weidinger G., Stebler J., Slanchev K., Dumstrei K., Wise C., Lovell-Badge R., et al. (2003). Dead End, a Novel Vertebrate Germ Plasm Component, Is Required for Zebrafish Primordial Germ Cell Migration and Survival. Curr. Biol. 13 (16), 1429–1434. doi: 10.1016/S0960-9822(03)00537-2
Wienholds E., Kloosterman W. P., Miska E., Alvarez-Saavedra E., Berezikov E., De B. E., et al. (2005). MicroRNA Expression in Zebrafish Embryonic Development. Science 309 (5732), 310–311. doi: 10.1126/science.1114519
Woods K., Thomson J. M., Hammond S. M. (2007). Direct Regulation of an Oncogenic Micro-RNA Cluster by E2F Transcription Factors. J. Biol. Chem. 282 (4), 2130–2134. doi: 10.1074/jbc.C600252200
Wu X., Wang Z., Jiang J., Gao J., Wang J., Zhou X., et al. (2014). Cloning, Expression Promoter Analysis of Vasa Gene in Japanese Flounder (Paralichthys Olivaceus). Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 1 (167), 41–50. doi: 10.1016/j.cbpb.2013.06.004
Yoon C., Kawakami K., Hopkins N. (1997). Zebrafish Vasa Homologue RNA is Localized to the Cleavage Planes of 2- and 4-Cell-Stage Embryos and is Expressed in the Primordial Germ Cells. Development 124 (16), 3157–3165. doi: 10.1242/dev.124.16.3157
Zheng W. J., Sun L. (2011). Evaluation of Housekeeping Genes as References for Quantitative Real Time RT-PCR Analysis of Gene Expression in Japanese Flounder (Paralichthys Olivaceus). Fish &. Shellfish Immunol. 30 (2), 638–645. doi: 10.1016/j.fsi.2010.12.014
Zhou L., Wang X., Du S., Wang Y., Zhao H., Du T., et al. (2020). Germline Specific Expression of a Vasa Homologue Gene in the Viviparous Fish Black Rockfish (Sebastes Schlegelii) and Functional Analysis of the Vasa 3' Untranslated Region. Front. Cell Dev. Biol. 8, 575788. doi: 10.3389/fcell.2020.575788
Keywords: Japanese flounder, microinjection, primordial germ cells, migration, miR-430
Citation: Wang B, Yang F, Jin C, Hu J, Qi J and Zhang Q (2022) Migration of Primordial Germ Cells Is Regulated by miR-430 During Embryonic Development of Japanese Flounder. Front. Mar. Sci. 9:904376. doi: 10.3389/fmars.2022.904376
Received: 25 March 2022; Accepted: 24 May 2022;
Published: 14 June 2022.
Edited by:
Libin Zhang, Institute of Oceanology, Chinese Academy of Sciences (CAS), ChinaReviewed by:
Mingyou Li, Shanghai Ocean University, ChinaZhenkui Qin, Ocean University of China, China
Jitendra Kumar Sundaray, Indian Council of Agricultural Research, India
Ashis Saha, Central Institute of Freshwater Aquaculture (ICAR), India
Copyright © 2022 Wang, Yang, Jin, Hu, Qi and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jie Qi, cWlqaWVAb3VjLmVkdS5jbg==; Quanqi Zhang, cXpoYW5nQG91Yy5lZHUuY24=
 Bo Wang
Bo Wang Fan Yang1
Fan Yang1 Chaofan Jin
Chaofan Jin Jie Qi
Jie Qi Quanqi Zhang
Quanqi Zhang