
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mar. Sci. , 18 July 2022
Sec. Marine Megafauna
Volume 9 - 2022 | https://doi.org/10.3389/fmars.2022.900107
 Margaret A. McClain1
Margaret A. McClain1 Neil Hammerschlag2,3
Neil Hammerschlag2,3 Austin J. Gallagher2,4
Austin J. Gallagher2,4 J. Marcus Drymon5,6
J. Marcus Drymon5,6 R. Dean Grubbs7
R. Dean Grubbs7 Tristan L. Guttridge8,9
Tristan L. Guttridge8,9 Matthew J. Smukall8
Matthew J. Smukall8 Bryan S. Frazier10
Bryan S. Frazier10 Toby S. Daly-Engel11*
Toby S. Daly-Engel11*Understanding dispersal in large marine fauna is necessary for conservation, but movement patterns often vary widely by sex and life stage. In sharks, genetic studies have shown evidence of widespread male-biased dispersal, though tagging and tracking studies on the same populations show both sexes using site fidelity, including philopatry, and moving similar distances. We used a suite of microsatellite loci and DNA samples from 362 previously-tagged tiger sharks (Galeocerdo cuvier) in the northwestern Atlantic, including a large number of residential juveniles, to evaluate reproductive dispersal in light of demographic and published tracking data. We found that lumping size classes together resulted in genetic panmixia across sites, but systematic removal of large individuals showed significant population-level differentiation and three separate population clusters among juveniles less than 260 cm total length. Tests for relatedness found that 8.9% of our sample set was composed of first-order related pairs (N = 16), including several full siblings from different litters, a sign of multi-cycle genetic monogamy which carries implications for effective population size. By mapping genetic assignments of juveniles, we identified a signature of fine-scale genetic structure suggesting broad biparental site fidelity to reproductive habitat in the northeast Gulf of Mexico, which is concordant with both genetic and tracking data. Taken together, these findings demonstrate how lumping individuals from different life stages in genetic studies may obscure fine-scale genetic structure, confounding future conservation efforts.
Species of large marine predators including whales, dolphins, and fishes commonly occupy global ranges (Gaither et al., 2016) and migrate long distances (>1000 km) to pursue optimal foraging and reproductive habitat (Luschi et al., 2003; Hays et al., 2016). Such highly migratory taxa form physical linkages between disparate ecosystems (Afonso et al., 2017), which contributes to ecosystem resilience and can buffer against extinction risk over time (Doughty et al., 2016). Though belonging to disparate vertebrate groups, marine megafauna all employ a similarly-high energetic investment in reproduction, a trait that make them vulnerable to both overexploitation and the effects of climate change (Dulvy et al., 2014).
In marine mammals and elasmobranchs (sharks, skates, and rays), vagility correlates positively with body size: large species can move farther and occupy larger home ranges than small species (Musick et al., 2004). Some of the largest taxa have circumglobal distributions and home ranges that may span ocean basins (Gaither et al., 2016). However, like other megafauna, many sharks exhibit site fidelity to certain regions for feeding and reproduction (Chapman et al., 2015), which effectively limits their high-use areas to just a fraction of their home range (Lee et al., 2007; Jorgensen et al., 2009). If reproductive site fidelity is common, physical dispersal may be disconnected from gene flow, and sites regularly visited by wide ranging individuals may be reproductively isolated. Conversely, high vagility can maintain gene flow over global scales when reproduction accompanies physical movement (Chapman et al., 2015). In highly vagile sharks, studies have found more fine-scale genetic structure than expected, a pattern that is regularly attributed to female natal philopatry (when females return to their own birthplace to reproduce), a type of reproductive investment (Chapman et al., 2015; Phillips et al., 2021).
Most shark species are viviparous, with females giving birth to large-bodied young after long (4.5 month-2 year) gestations and 1-3 year reproductive cycles, which in many species includes the reoccurring use of specific habitats for parturition (Grubbs, 2010; Conrath and Musick, 2012; Chapman et al., 2015; Heupel et al., 2019). Philopatry is often inferred from detection of tagged adult females returning to the same pupping habitat over multiple reproductive cycles (Hammerschlag et al., 2011; Papastamatiou et al., 2013). The pattern for males based on tagging data is less clear, with tagged males exhibiting regional philopatry but limited association with pupping habitats (Hammerschlag et al., 2011). While tagging data can shed valuable light on reproductive behavior, patterns of reproductive dispersal and the impact on regional diversity must be verified by examining patterns of gene flow between populations (reviewed in Phillips et al., 2021). Often, data from biparentally-inherited nuclear genes (nDNA) is compared with maternally-inherited mitochondrial genes (mtDNA), with reproductive philopatry indicated by significant structure between regions at one marker type (Goudet et al., 2002; Phillips et al., 2021).
Genetic studies on sharks have regularly shown significant mtDNA structure with contrasting panmixia at nDNA, which is commonly interpreted as female natal philopatry against a background of basin-wide male dispersal (Daly-Engel et al., 2012; Veríssimo et al., 2012; Phillips et al., 2021). This contrasting pattern of female philopatry against a background of male-biased dispersal is now the prevailing view for many sharks, including large species with global ranges (e.g. Daly-Engel et al., 2012; Bernard et al., 2016; Guttridge et al., 2017). But these conclusions do not address findings from telemetry on the same populations, which provide evidence of both males and females using site fidelity and consistently show female sharks moving equivalent or longer distances than males, as vagility would predict (Chapman et al., 2015; Guttridge et al., 2017; Ajemian et al., 2020). In sexually-reproducing taxa, both sexes must pursue mating opportunities, but because mating has been observed in a very few shark species, the degree to which male dispersal potential is determined by the need to find receptive females is not yet known.
Investigating gene flow in marine megafauna is challenging given the logistical difficulties of sampling, especially for frequency-based genetic analyses, which require high sample numbers (Selkoe and Toonen, 2006). For large sharks, the opportunistic nature by which data sets are assembled often forces investigators to lump individuals together by catch location without considering age class. But this ignores the fact that most sharks undergo an ontogenetic shifts that causes their movement patterns to change at a specific point in their development (Grubbs, 2010), such that the possibility of an animal being a non-reproducing migrant appears only after this shift. The result is a mixed assemblage of non-migratory juvenile and migratory subadult and adult sharks, which when analyzed together may incorrectly present as evidence of genetic panmixia (Phillips et al., 2021).
Genetic studies on sharks have largely supported the conclusion of male-biased dispersal, though most do not parse samples by size class (Bernard et al., 2016; Santos and Coelho, 2018; Carmo et al., 2019; Klein et al., 2019; Weideli et al., 2019). A recent study on the sand tiger shark (Carcharias taurus) illustrated how including just a few genetic migrants in assignment testing could mask fine-scale structure: investigators found panmixia with biparental nDNA when size classes were lumped together, but the same analyses performed on non-migratory juvenile and young-of-the year sharks showed significant population structure between sites (Klein et al., 2019).
The complex relationship between physical and reproductive dispersal is especially apparent in tiger sharks (Galeocerdo cuvier), a well-studied species (Holland et al., 2019) in which conclusions about dispersal from tracking data have previously contrasted with genetic results (Phillips et al., 2021). The tiger shark can reach >5.5 meters total length (TL) and migrate thousands of kilometers, making this species one of a relatively few fishes with a true circumglobal distribution (Gaither et al., 2016). Their habitat is coastal-pelagic, and in general, tiger sharks have shown a similar ontogenetic habitat shift to other requiem sharks (Grubbs, 2010): larger individuals (older juveniles and adults) are seasonally migratory, while small immature individuals are residential in reproductive habitats that are broader and more diffuse than many other shark species, but still spatially limited (Driggers et al., 2008; Lea et al., 2015; Heupel et al., 2019). The size range in ontogeny at which most sharks begin to disperse is unknown and likely to vary between populations, but Lea et al. (2015) observed via tracking that tiger sharks >270 cm in Bermuda undertook seasonal long-distance migrations, while smaller, immature individuals were year ‘round residents.
Though some sex-based differences in movement have been observed, tracking studies on tiger sharks have consistently shown that both males and females move similar distances, occupy overlapping ranges, and exhibit repeated site fidelity (Papastamatiou et al., 2013; Sulikowski et al., 2016; Ajemian et al., 2020). In contrast, genetic data on the same populations have consistently shown fine-scale structure attributed to male-biased dispersal and female philopatry (Bernard et al., 2016; Carmo et al., 2019). Global genetic studies have shown deep evolutionary partitioning in tiger sharks between the Atlantic and Indo-Pacific, but biparental structure within basins is largely absent (Bernard et al., 2016; Pirog et al., 2019; Andrade et al., 2021; Bernard et al., 2021). Though no studies have examined the effect of size class, previous genetic work on tiger sharks in the northwest Atlantic identified structure at maternally-inherited mitochondrial DNA, including a divergent lineage located in the Caribbean (Carmo et al., 2019). Biparentally-inherited nuclear DNA showed either panmixia or gave inconclusive results with few indications of structure, leading to the general conclusion of male-biased dispersal (Bernard et al., 2016; Pirog et al., 2019; Bernard et al., 2021). In contrast, physical tracking data from the same population of tiger sharks found no significant differences between male and female dispersal patterns, but did identify spatial overlap between adult males and females at some areas in the Gulf of Mexico (Ajemian et al., 2020). To resolve this conflict and shed light on the patterns of fine-scale reproductive dispersal in northwest Atlantic tiger sharks, we measured evolutionary connectivity and contemporary genetic kinship using 11 nuclear microsatellite loci on a sample set of 362 animals, including individuals whose physical movement patterns were previously characterized by Ajemian et al. (2020).
Tissue samples were collected from live, free-swimming sharks captured via a variety of fishery-independent methods. Approximately 1 cm3 of fin or muscle tissue was stored in 1.5 mL of >70% ethanol or NaCl-saturated DMSO buffer for storage prior to DNA extraction with a DNeasy Blood and Tissue Kit (Qiagen, Valencia, California). We used 12 previously-published, unlinked microsatellite loci, all but one of which was species-specific (Keeney and Heist, 2003; Bernard et al., 2015). Unlabeled reverse primers and tailed forward primers were obtained from Integrated DNA Technologies, Inc. (Coralville, IA), while M13 primer tags were obtained from ThermoFisher (Waltham, MA). Polymerase chain reactions (PCR) were completed using the touch-down protocol described in Holmes et al. (Holmes et al., 2017) on a C1000 ThermoCycler from BioRad (Hercules, CA). Products were visualized with the microsatellite plugin for Geneious v7.1.8 (Kearse et al., 2012), and quality control was conducted following Davis et al. (2019). We used Arlequin v3.5.2.2 (Excoffier and Lischer, 2010) to test for Hardy-Weinberg Equilibrium and calculate diversity statistics, and calculated the likelihood of error due to mistyping, null alleles, and allele dropout in Microchecker (van Oosterhout et al., 2004).
Relatedness can be difficult to pinpoint with high accuracy using microsatellite data, with the strong possibility of overestimation (Attard et al., 2018). To be conservative, we first identified kin relationships via sibling and parental genotype reconstruction using both overall and pairwise maximum likelihood methods, then employed estimates of relatedness (r) to validate our results using a panel of model estimators. First, we tested for kinship with 95% confidence in Colony v2.0 (Jones and Wang, 2010), which accommodates incomplete sampling and varying mating strategies, but can overestimate relatedness via overall likelihood. We therefore verified the results from Colony with the more conservative log-likelihood ratio (LOD score) estimated in Cervus v3.0.7 (Kalinowski et al., 2007; Koch et al., 2008; García-Navas et al., 2014). When a related pair was identified with high confidence by both Colony and Cervus, we calculated the r value (below) and used demographic data to determine the biological relationship: for individuals related by ≥50%, an adult was assumed to be the parent of a paired juvenile rather than an older sibling, and two siblings close in size were assumed to share a litter. Half of each pair of first-order related individuals were removed from the sample set before assignment testing.
To generate relatedness values (r) and to rule out the possibility that our results reflected population-level inbreeding rather than true kinship, we measured pairwise relatedness and inbreeding using Coancestry V1.0.1.9 (Wang, 2011), which offers seven estimator models to calculate r and four methods to estimate individual inbreeding via internal relatedness (IR). We compared r values between the most commonly-used Queller-Goodnight (Q-G) estimator (Queller and Goodnight, 1989) alongside Wang’s unbiased estimator (Wang, 2017), which have complementary strengths. Q-G is robust to the occurrence of rare alleles that can lead to overestimation of relatedness, and is appropriate for populations where high relatedness is possible; in test simulations (Gonzalez et al., 2014), the Q-G showed the fewest differences between observed and expected relatedness in large datasets. Wang’s unbiased estimator is appropriate for any sample size and particularly suited for populations with possible inbreeding (Wang, 2017), while both estimators are robust to deviations from Hardy-Weinberg Equilibrium due to low heterozygosity (Wang, 2007). Individual inbreeding coefficients, a measure of internal relatedness (IR), were calculated using LynchRD in Coancestry (Wang, 2011). Coancestry runs were conducted both with and without first-order kin, accounting for inbreeding, and using 1000 reference individuals and an error rate of 0.005 (Feldheim et al., 2017). A z-test for means was used to test for differences in kinship values between groups (Glass et al., 1972).
We tested the effect of animal size on population genetic tests by systematically removing the largest individuals in 10 cm batches and performed repeated analyses. We calculated pairwise population FST between sites in Arlequin v3.5.2.2 (Excoffier and Lischer, 2010) using a Benjamini-Hochberg FDR correction for multiple tests (Benjamini and Hochberg, 1995) and 100,000 permutations. We used the program Structure 2.3 (Pritchard et al., 2000) to assign individuals to discrete genetic populations or “clusters” (K) with no a priori assumptions, first for all samples together, then systematically parsed by size. Structure’s Bayesian clustering employs an admixture model, correlated alleles, a 10,000 burn-in period, 20 replicates per K, and 300,000 MCMC repetitions (Gilbert et al., 2012). Runs were performed with a 10,000 step burn-in followed by 100,000 MCMC steps to test K = 1–10 with 20 repetitions each. Results were collated with Structure Harvester 0.9.94 (Earl and vonHoldt, 2012) and input into CLUster Matching and Permutation Program (CLUMPP; Jakobsson and Rosenberg, 2007). Structure Harvester was also used to identify the most likely number of discrete genetic groups (K) represented among our samples. For this we employed the simulation method advanced by Evanno et al. (2005) that uses an ad hoc statistic to calculate both Pr(LnK) and the second order rate of change of the log probability LnP(D) for all possible K values (ΔK), in this case K=1-9.
Among 362 sharks, we observed a change in the results of our tests for individual assignment and population connectivity after animals ≥260 cm TL were removed from our data set. But because the size at which sharks first disperse is variable, for size-based analyses we categorized animals ≥260 cm as being potentially-dispersive genetic migrants (hereafter called “dispersive”), and individuals <260 cm as likely residents (“non-dispersive”). In Structure analyses, these individuals were analyzed alone separated by site, while individuals >300 cm TL were included as their own group without site label to represent the potential reproductive migrants in the system (“adults”). Individuals between 260 and 300 cm TL were included in pairwise FST but not assigned in Structure, as they could be dispersive but were unlikely to have contributed genetically to the current population of young.
We obtained DNA samples collected from 362 tiger sharks from six sites between 2010 and 2019 (Table 1): 159 from the Gulf of Mexico (GoM; 55 from Dauphin Island, Alabama; 60 from the Big Bend of Florida; and 44 from the Florida Keys) and 203 from the Atlantic (31 from South Florida; 117 from Nassau, Little Bahama Bank, and Bimini, hereafter referred to as Bahamas; and 55 from South Carolina). After removing related individuals, our groupings of 328 individuals consisted of 217 non-dispersives <260 cm and 111 dispersives ≥260 cm, which were further divided into adults ≥300 cm (N = 55) (Branstetter et al., 1987; Whitney and Crow, 2007) and dispersive juveniles 260-299 cm (N = 56; Table 1).
Globally, a number of our loci deviated from Hardy-Weinberg Equilibrium, but when samples were grouped by catch location, significant deviation was rare (Table S1), an indication of possible genetic structure across the range. Microchecker found no evidence of significant genotyping errors, and while all loci were previously published and underwent quality testing, Cli106 was cut from final analysis after producing >2 alleles in some individuals. Nine loci amplified at all sites, while Cli103 and TGR943 failed to amplify in some locations, so while including these loci did not significantly impact our results, we excluded them from population-level analyses to reduce the chances of Type I error. No samples failed to amplify at more than two loci. We calculated the level of missing data at 1.9%, with 1.8% of that coming from locus TGR1157, which amplified poorly in some samples. Removing this locus did not impact our results, however, so it was retained in the final analysis.
Measures of pairwise FST (α = 0.016) between collection sites among all 362 sharks showed some indications of weak genetic structure, but after performing the same tests among the 111 dispersive individuals alone, we observed no significant differences between sites (Table 2). In contrast, comparisons among 217 non-dispersives showed significant FST values, primarily between the Big Bend (BB) region of Florida and all other sites except the Florida Keys (FK). Dauphin Island (DI) showed significant differentiation from the Florida Keys (FK) and the Bahamas (BA), but not South Florida (SF) or South Carolina (SC; Table 2). Some significance tests may have been impacted by low sample sizes: DI and SF had low numbers of dispersive individuals, for example (N = 8 at each site), while among non-dispersives, SF and SC had only 23 samples and 16 samples each (Table 1), which may explain the lack of significance between these sites and the Big Bend. Other comparisons show significance despite no change or a decrease in FST values (such as the comparison between DI and both BB and FK), another possible effect of variation in sample size, making these results somewhat ambiguous.
Bayesian assignment testing in Structure (Pritchard et al., 2000) to identify the best K for all unrelated sharks together using both the Pr(LnK) and ΔK methods (Evanno et al., 2005) showed a high likelihood of two genetic populations, hereafter known as “clusters” (K =2, Figures S1A, B). One cluster contained 92.8% of the population, with the remaining 7.2% composed of individuals that showed evidence of shared ancestry with another genetic type and could not be assigned to a single cluster. These were distributed evenly across the sampling range (Figure 1A), with little to no distinction between sampling sites. In contrast, assignment testing of unrelated, non-dispersive small tiger sharks (<260 cm) indicated three discrete genetic clusters (K = 3, Figures S1C, D) represented in our data, with 59.5% of individuals assigning to Cluster 1, 5.5% to Cluster 2, 19.4% to Cluster 3, and 15.7% unassigned. Of the 55 adults, 87.3% assigned to Cluster 1, 1.8% assigned to Cluster 2, 0% assigned to Cluster 3, and 10.9% were unassigned (Figure 1B). Cluster distribution was geographically asymmetric, with Cluster 1 being prevalent in the northern Gulf and Atlantic on either end of our sampling range, and Cluster 3 dominating the center and southern regions (Figure 2).
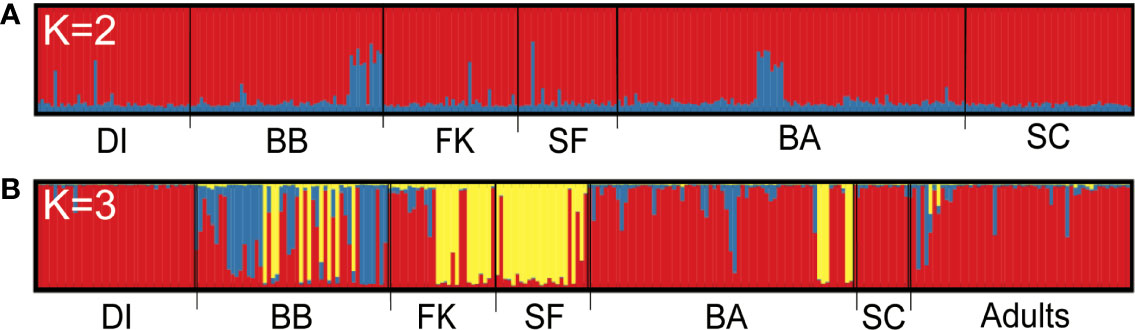
Figure 1 Assignment bar plots showing the effect of partitioning samples by size. Each vertical bar represents an individual, grouped by catch location and shaded by proportion of population ancestry from Structure. (A) All unrelated individuals; (B) All non-dispersive individuals <260 cm, with adults ≥300 cm TL separated into their own group with no location. K, number of unique genetic clusters or populations identified across the sampling range; DI, Dauphin Island; BB, Big Bend, Florida; FK, Florida Keys; SF, South Florida; BA, Bahamas; SC, South Carolina.
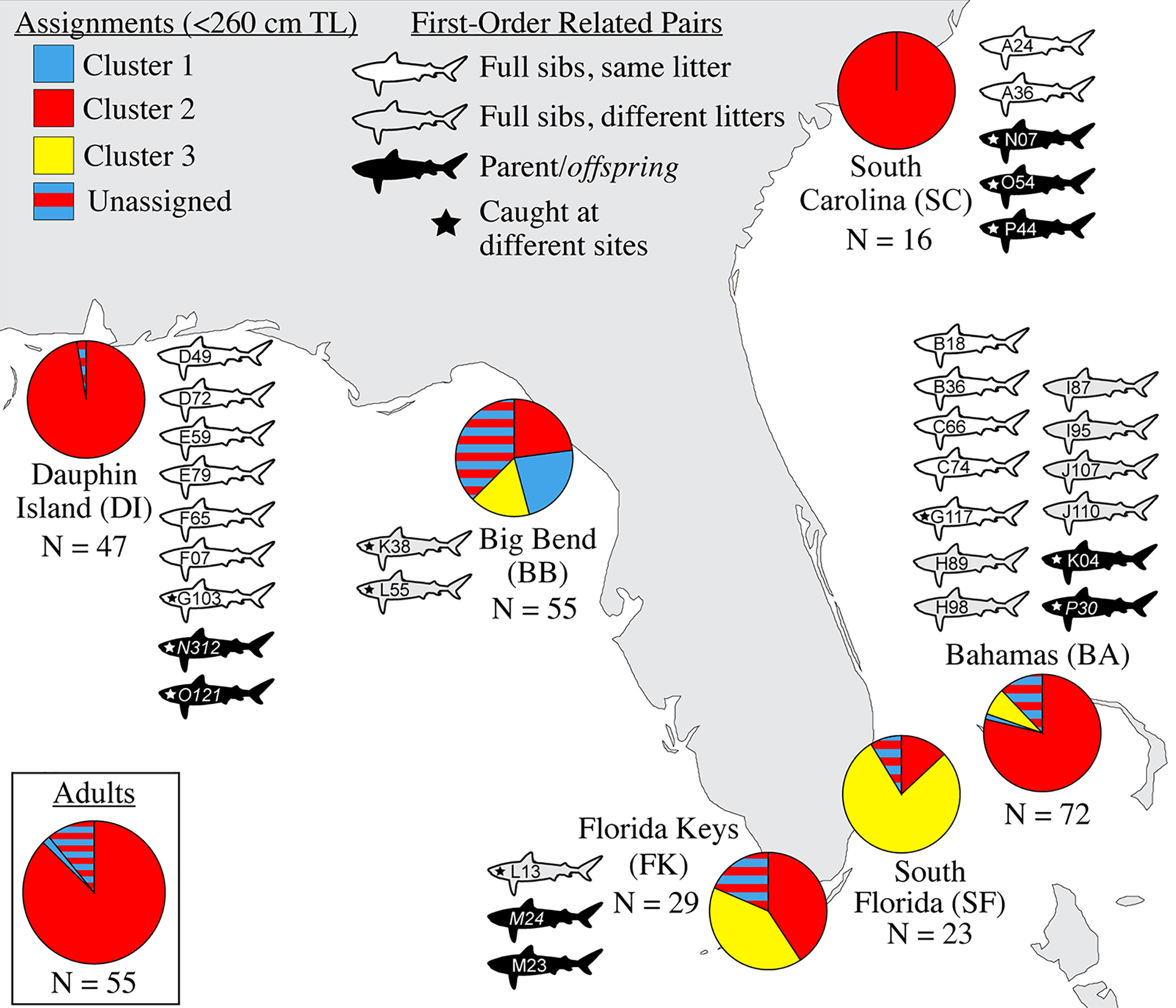
Figure 2 Genetic assignments and related pairs, by sampling site. Pie charts at sampling sites indicate the relative proportion of population ancestry among individuals <260 cm TL that assigned strongly to three genetic clusters (C1-C3), with the remaining individuals that could not be strongly assigned forming a separate “unassigned” group. The inset pie chart shows genetic assignments of potential reproductive migrants ≥300 cm TL for all sites together. Shark icons represent the catch location of individuals related by ~50% (first-order kin: full sibling or parent-offspring); the letters and numbers identify them as the related pairs described in Table 3. N, sample size.
We detected 16 pairs (32 individuals) sharing ≥50% DNA with >95% confidence which were concordant between Cervus and Colony (Table 3). Of first-order pairs, 12 were likely full siblings, and four were likely parent-offspring, based on demographic data. Of the 12 sibling pairs, we estimated the siblings in seven pairs shared a litter, and the siblings in five came from litters at least two years apart. (Table 3). All parent-offspring pairs were caught at least one year apart. We did not detect any half-siblings with >95% likelihood, though several pairs were identified with 90-95% confidence, suggesting that our dataset has low power to distinguish lower-order kin when no pedigree data are available. We therefore limited our analysis to first-order kin only. The number of related individuals per locality was not correlated with the total number of individuals caught at each site. Results from relatedness analyses showed no change with or without accounting for inbreeding, and we found no difference between the two relatedness estimators within or between any groups (p = 0.447). Average inbreeding across the entire sample set, including first order kin, was IR = 0.075 ± 0.042, while average relatedness was RW = -0.041 ± 0.061/RQG = 0.004 ± 0.054 (Figure 3). Both relatedness and inbreeding were highest in South Florida (SF), while relatedness was lowest in the Bahamas and inbreeding was lowest in South Carolina (SC; Figure 3A). Both R and IR were equivalent between the sexes (Figure 3B), and dispersive vs. non-dispersive individuals (Figure 3C), though no significant differences were detected between groups, likely due to high variance.
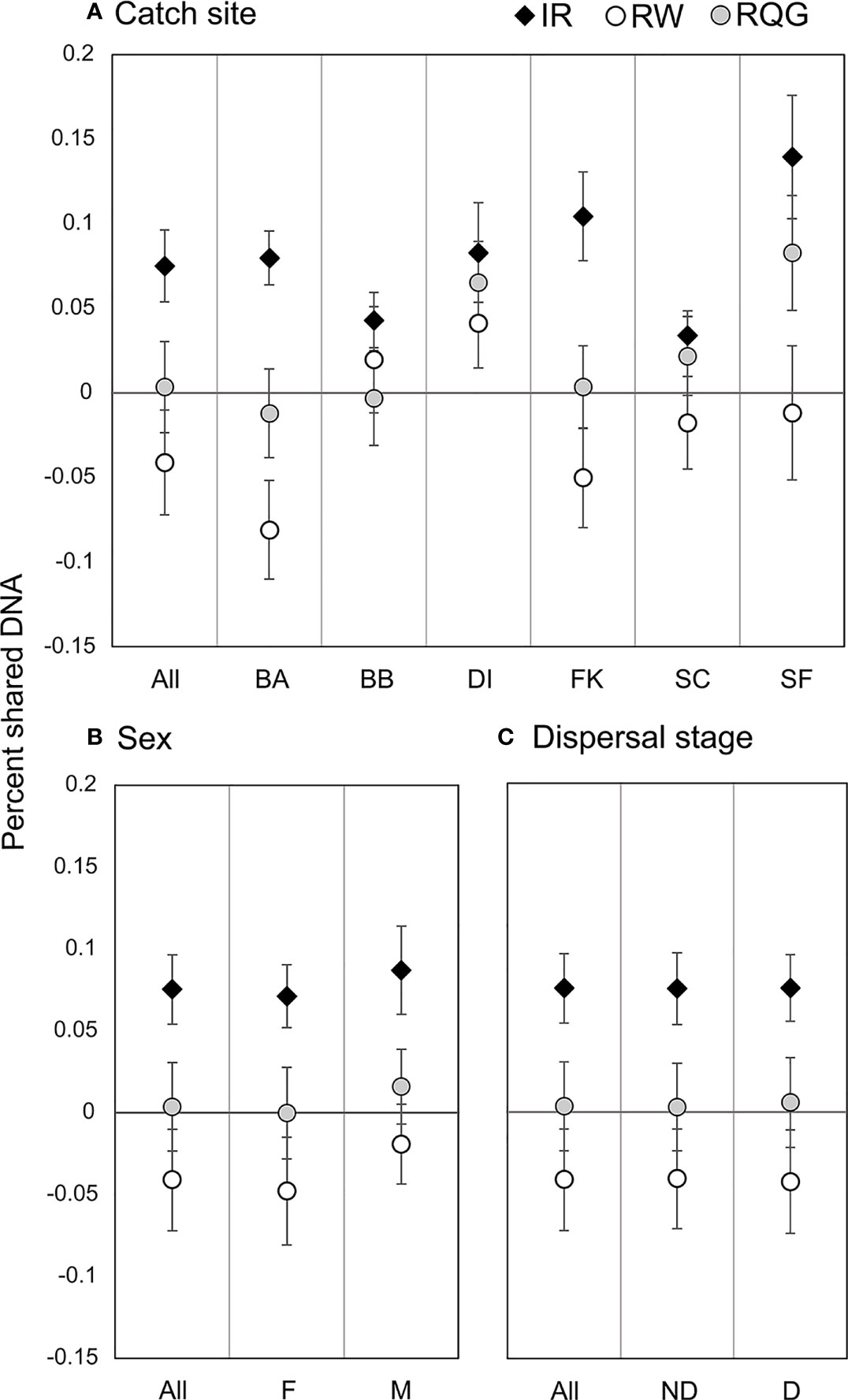
Figure 3 Pairwise relatedness and inbreeding. Circles represent the average pairwise relatedness for the Wang unbiased (RW) and Queller-Goodnight (RQG) estimators, and diamonds represent average individual inbreeding coefficient (IR) by (A) catch site (abbreviations from Table 1); (B) sex (Male/Female); and (C) dispersal stage (D, Dispersive individuals ≥260 cm TL; ND, Non-Dispersive individuals <260cm TL).
In our analysis of reproductive dispersal among tiger sharks in the northwestern Atlantic, we found that including individuals large enough to be genetic migrants in our data set of polymorphic nuclear loci had a notable impact on the results, especially on Bayesian clustering (Figure 1). When we tested individuals of all sizes together, we found no evidence of population structure, and no variation in genetic assignments between sites. Systematic removal of large individuals showed a contrasting pattern in smaller sharks <260 cm TL, namely slight but significant genetic structure and, more markedly, the presence of three distinct population clusters. The pattern of fine-scale population differentiation observed here suggests that some male tiger sharks are investing in site fidelity to particular habitats where reproduction occurs, a strategy that was assumed to be largely absent in male sharks (Phillips et al., 2021).
Evidence of biparental gene flow between habitat where sharks are likely breeding (Driggers et al., 2008) in the northern Gulf of Mexico (GoM) and Atlantic was found in the strong presence of Cluster 1 individuals (red in Figure 2) across the range, showing that Cluster 1 is the predominant genetic “type” in the broader region. We identified no significant structure between Dauphin Island, Alabama (DI) and South Carolina (SC), an indication of historical connectivity, though the relatively low sample size at SC (N = 16) means that this conclusion should be considered tentative. When mapped to the areas where these young were caught, assignment testing shows an incursion of genetic Clusters 2 and 3 (blue and yellow in Figure 2, respectively) into the center of our sampling range, especially in the Big Bend (BB), the Florida Keys (FK), and South Florida (SF). Cluster 3 was the second-most common and the dominant genetic type in the southern end of the range, suggesting it may be more frequent in the Caribbean, similar to the findings of Carmo et al., 2019. Though heavily represented among juveniles, no Cluster 3 adults were caught among the 55 included here (Figure 2). This may reflect the random chance associated with the low numbers of mature specimens we obtained, or a true scarcity of resident Cluster 3 adults in the region. If so, the presence of Cluster 3 young in the Big Bend could result from repeated northward reproductive migration into the eastern GoM by non-resident, Cluster 3 adults.
Broad site fidelity to specific habitats for reproduction can drive genetic isolation over time by increasing the likelihood of within-cluster pairings, but the considerable admixture we observed in our study indicates some degree of ongoing interbreeding between clusters. The connectivity between DI and SC is concordant with results from telemetry that show animals of both sexes migrating between the GoM and Atlantic, though such movements are rare compared with within-basin movements (Hammerschlag et al., 2012; Lea et al., 2015; Kohler and Turner, 2019; Ajemian et al., 2020). Two individuals in our study, a male tagged in 2017 and a female tagged in 2018, were observed following the same path one year apart from SF to DI, mirroring the track of a should read young-of-the-year (YOY) shark tagged in Tampa Bay that was captured nine years later in SC (unpub. data). We also found evidence of gene flow between the GoM and northwestern Atlantic in the form of related individuals caught at different sites, including two mature males caught in SC that sired pups caught in DI, and a female caught in BA that was a sibling to a male in DI (Table 3). Though it is unknown which population was their birthplace, the fact that pairwise FST show significant evolutionary differences between these two sites further highlights the occurrence of physical dispersal without mating.
The Big Bend (BB) region of Florida, where Driggers et al. (2008) found a evidence of non-dispersive, YOY animals, showed significant differentiation from sites in the northern GoM and west Atlantic, but connectivity with SF and FK. The BB also showed the highest observed heterozygosity of any site, and the highest proportion of non-dispersives of any site except DI (Table 1). Driggers et al. (2008) suggested that tiger shark parturition in the GoM takes place near habitat where YOY and juveniles are resident, a pattern consistent with female philopatry to areas like the Big Bend. Our observation of significant genetic structure at nuclear loci indicates that this behavior is at least partially biparental rather than just maternal.
The idea that it takes a relatively few reproductive migrants per generation to reduce polymorphism between subpopulations, as originally advanced by Wright (1931), has been well-explored over the years in a conservation context (e.g. Mills and Allendorf, 1996; Wang, 2004; Linck and Battey, 2019). It is now broadly accepted that significant differentiation can occur in the presence of gene flow (reviewed in Waples and Gaggiotti, 2006; Lowe and Allendorf, 2010). The genetic structure we observe in Atlantic tiger sharks indicates that reproductive migration is more constrained between some sites than others, and more constrained than physical dispersal patterns suggest. If this reproductive migration was exclusively male-mediated, then we would expect nuclear DNA to show high connectivity within a single population, as opposed to the fine-scale structure and multiple subpopulations (clusters) identified here. Given the high adult vagility and observed long-distance movement patterns in this species, we hypothesize that this system shows sympatry in action: both males and females from different genetic subpopulations mixing within a shared geographic range, but returning seasonally to breed in separate general areas, leading to reproductive isolation over time.
Philopatry by males is common in bony fishes, turtles, and birds, where it is thought to enhance fitness by increasing mating opportunities through increased encounters with conspecifics (Quinn and Dittman, 1990; FitzSimmons et al., 1997; Payevsky, 2016). In sharks, males are philopatric to feeding areas, but are not thought to undertake specific migrations to nurseries to eat young, which would defeat the purpose of such habitat. Though males may occasionally consume neonates, mating opportunities are more likely driving reproductive dispersal, a factor that is known to limit male fitness (Bateman, 1948).
Male site fidelity to sites at or near seasonally-specific habitats where reproduction occurs could enhance mating success through increased frequency and/or predictability of encountering a receptive female at a specific time of year, compared with the likelihood of randomly meeting one in the open ocean. Pratt (1993) noted that copulation occurs soon after parturition in some shark species, and unpublished observations of species like scalloped hammerheads (Sphyrna lewini), bonnetheads (Sphyrna tiburo), and Atlantic sharpnose sharks (Rhizoprionodon terraenovae) show that males aggregate near habitat used for parturition in the late summer, putting them in contact with post-partum females (C. Bangley, B. Frazier, and M. Giresi, pers. comm). Though the habitat that tiger sharks use for reproduction is open and diffuse compared with the well-defined nurseries used by the species above (Driggers et al., 2008), it is believed that mating in South Carolina occurs in late fall when both males and females are abundant nearshore, and resident neonates and juveniles are offshore (unpub. data). In the Gulf of Mexico, Ajemian et al. (2020) found that while more adult females than males were caught near areas where pups were present, males were sometimes detected in the area at the same time as females, and sex was not a significant predictor of adult distribution in general.
While it is generally unknown where and when mating occurs, aspects of their mating strategy suggest that reproductive opportunities may be more limited in tiger sharks than in other species. When female sharks copulate with more than one male over the breeding season (polyandry), a common strategy in elasmobranchs (Fitzpatrick et al., 2012), the first male to mate can sire the majority of the litter (Orr and Brennan, 2015). This reproductive skew should increase male competition to access females early in their reproductive cycle (Bateman, 1948), but polyandry appears rare or absent in tiger sharks: studies on Indian and Pacific Ocean populations indicate that tiger sharks may be one of a relatively few species to utilize genetic monogamy (Holmes et al., 2018; Pirog et al., 2020), especially over multiple cycles (Lamarca et al., 2020). In practice, a limit on receptive females would increase the selective pressure on male tiger sharks to be the first to encounter such individuals, promoting the evolution of reproductive site fidelity.
We found that 8.9% of our sample set was made up of first order (≥50% related) kin, either full siblings or parent-offspring pairs, whose distribution is shown in Figure 2. In cases of siblings that shared a litter (43.8% of related individuals, or 7/16 pairs; Table 3), all except one were caught prior to dispersal, so spatial and temporal proximity at birth may have increased the likelihood of capture. Among the related individuals were six pairs of full siblings caught 1-4 years apart, at sizes that indicate they came from different litters (Table 3). This repeated genetic monogamy indicates that females are likely storing sperm over multiple reproductive cycles, thought to occur every 3 years (Castro, 2009).
Many sharks including tiger sharks can store viable sperm, potentially for years (Pratt, 1993; Conrath and Musick, 2012), and though it is possible for females to mate repeatedly with the same male over subsequent years, behavioral monogamy is thus far unobserved in sharks (but would be more likely in cases of biparental philopatry). Average pairwise relatedness in all 362 animals was close to zero and average IR was 7.5% (Figure 3), so our kin identifications are unlikely to reflect historical inbreeding. Instead, these findings suggest that a relative few large reproductive females may be contributing disproportionately to the population. This phenomenon has been well-documented in bony fishes, wherein BOFFFFs (big, old, fat, fertile, female fish) are thought to sustain some fisheries (Hixon et al., 2014), but until now had not been observed in elasmobranchs. For all fishes, most mortality occurs in the juvenile phase (Werner and Gilliam, 1984), so it is possible to imagine an impacted shark population being dominated by a few large, high-value reproducers as female size increases indeterminately over time. Given that tiger sharks may grow >5.5m TL and reach ages of 50+ years (Afonso et al., 2012; Meyer et al., 2014), such high-fecundity reproducers would indeed be of value to the population.
The population overlap and mixing among non-dispersive juveniles in Florida’s Big Bend highlights the importance of this region as critical habitat within the northwestern Atlantic, and for preserving regional genetic diversity. Significant fine-scale genetic structure in the absence of physical barriers indicates long-term behavioral barriers to gene flow (e.g. philopatry), which can have a dampening effect on genetic diversity. The population differentiation detected in this study takes thousands of years of site fidelity and restricted gene flow to establish, indicating the potential for population-level consequences if even one primary parturition site is disturbed or modified by human activities. Further, the presence of “shark BOFFFFs” indicates that there may be high variation in reproductive success among females, which would lower diversity and effective population size along with the evolutionary potential of this group (Hedrick, 2005).
Our data support Lea et al. (2015) observations that vagility and body size can explain the ontogenetic shift in juvenile tiger sharks ≥260 cm TL, resulting in size-dependent genetic structure. Many studies that document long-distance migration have attributed shark movements to sea surface temperature and other abiotic features (Lea et al., 2015; Ajemian et al., 2020; Hammerschlag et al., 2022), but the potential for biparental site fidelity and the importance of size class presented here may justify a closer examination of reproductive drivers as a focus of future studies. Though size-related dispersal is well known from telemetry findings, genetics have only been used once to examine the ontogenetic shift in a shark (Klein et al., 2019), and never in a circumglobal species. Our result of similar size-dependent structure across populations of tiger sharks in the northwestern Atlantic suggests that size-based management, with the goal of prioritizing protections for high-use areas that harbor relatively non-dispersive young, would be an effective conservation strategy for this species.
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
The animal study was reviewed and approved by IACUC, Florida Institute of Technology and by separate governing bodies at authors' institutions at the time of sampling.
MM and TD designed research. NH, AG, JD, RG, TG, MS, and BF provided samples and movement data. MM performed research. MM and TD analyzed data. MM and TD wrote the paper with input from all authors. All authors contributed to the article and approved the submitted version.
Funding for this work came from the University of West Florida and the Department of Ocean Engineering and Marine Sciences at the Florida Institute of Technology, including student funding to MM Sampling was conducted with funding and permits awarded to individual investigators Publication of this article was funded in part by the Open Access Subvention Fund and the John H. Evans Library at Florida Tech.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We thank Alex Carlson, Jeff Eble, Emily Seubert, Matt Davis, Ariel Egan, Cody Nash, Yannis Papastamatiou, Brian Bowen, and busy peer reviewers for their support and advice on this manuscript. This represents contribution 854 from the SCDNR Marine Resources Research Institute.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2022.900107/full#supplementary-material
Afonso A. S., Garla R., Hazin F. H. V. (2017). Tiger Sharks can Connect Equatorial Habitats and Fisheries Across the Atlantic Ocean Basin. PLos One 12 (9), e0184763. doi: 10.1371/journal.pone.0184763
Afonso A. S., Hazin F. H. V., Barreto R. R., Santana F. M., Lessa R. P. (2012). Extraordinary Growth in Tiger Sharks Galeocerdo Cuvier From the South Atlantic Ocean. J. Fish. Biol. 81 (6), 2080–2085. doi: 10.1111/j.1095-8649.2012.03455.x
Ajemian M. J., Drymon J. M., Hammerschlag N., Wells R. J. D., Street G., Falterman B., et al. (2020). Movement Patterns and Habitat Use of Tiger Sharks (Galeocerdo Cuvier) Across Ontogeny in the Gulf of Mexico. PLos One 15 (7), e0234868. doi: 10.1371/journal.pone.0234868
Andrade F. R. S., Afonso A. S., Hazin F. H. V., Mendonça F. F., Torres R. A. (2021). Population Genetics Reveals Global and Regional History of the Apex Predator Galeocerdo Cuvier (Carcharhiniformes) With Comments on Mitigating Shark Attacks in North-Eastern Brazil. Mar. Ecol. 42 (2), e12640. doi: 10.1111/maec.12640
Attard C. R., Beheregaray L. B., Möller L. M. (2018). Genotyping-By-Sequencing for Estimating Relatedness in Nonmodel Organisms: Avoiding the Trap of Precise Bias. Mol. Ecol. Resour. 18 (3), 381–390. doi: 10.1111/1755-0998.12739
Bateman A. J. (1948). Lntra-Sexual Selection in Drosophila. Heredity 2 (3), 349–368. doi: 10.1038/hdy.1948.21
Benjamini Y., Hochberg Y. (1995). Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc.: Ser. B (Methodological) 57 (1), 289–300. doi: 10.1111/j.2517-6161.1995.tb02031.x
Bernard A. M., Feldheim K. A., Heithaus M. R., Wintner S. P., Wetherbee B. M., Shivji M. S. (2016). Global Population Genetic Dynamics of a Highly Migratory, Apex Predator Shark. Mol. Ecol. 25 (21), 5312–5329. doi: 10.1111/mec.13845
Bernard A. M., Feldheim K. A., Shivji M. S. (2015). Isolation and Characterization of Polymorphic Microsatellite Markers From a Globally Distributed Marine Apex Predator, the Tiger Shark (Galeocerdo Cuvier). Conserv. Genet. Resour. 7 (2), 509–511. doi: 10.1007/s12686-014-0408-0
Bernard A. M., Finnegan K. A., Pavinski Bitar P., Stanhope M. J., Shivji M. S. (2021). Genomic Assessment of Global Population Structure in a Highly Migratory and Habitat Versatile Apex Predator, the Tiger Shark (Galeocerdo Cuvier). J. Heredity. 112 (6), 497–507. doi: 10.1093/jhered/esab046
Branstetter S., Musick J. A., Colvocoresses J. A. (1987). A Comparison of the Age and Growth of the Tiger Shark, Galeocerdo Cuvieri, From Off Virginia and From the Northwestern Gulf of Mexico. Fish. Bull. 85 (2), 269–279. https://scholarworks.wm.edu/vimsarticles/621.
Carmo C. B., Ferrette B. L. S., Camargo S. M., Roxo F. F., Coelho R., Garla R. C., et al. (2019). A New Map of the Tiger Shark (Galeocerdo Cuvier) Genetic Population Structure in the Western Atlantic Ocean: Hypothesis of an Equatorial Convergence Centre. Aquat. Conserv.: Mar. Freshw. Ecosyst. 29 (5), 760–772. doi: 10.1002/aqc.3029
Castro J. I. (2009). Observations on the Reproductive Cycles of Some Viviparous North American Sharks. Aqua 15 (4), 205–222.
Chapman D. D., Feldheim K. A., Papastamatiou Y. P., Hueter R. E. (2015). There and Back Again: A Review of Residency and Return Migrations in Sharks, With Implications for Population Structure and Management. Annu. Rev. Mar. Sci. 7, 547–570. doi: 10.1146/annurev-marine-010814-015730
Conrath C. L., Musick J. A. (2012). “Reproductive Biology of Elasmobranchs, in Biology of Sharks and Their Relatives, 2nd. Eds. Carrier J. C., Musick J. A., Heithaus M. R. (Boca Raton, FL, USA: CRC Press), 291–311.
Daly-Engel T. S., Seraphin K. D., Holland K. N., Coffey J. P., Nance H. A., Toonen R. J., et al. (2012). Global Phylogeography With Mixed-Marker Analysis Reveals Male-Mediated Dispersal in the Endangered Scalloped Hammerhead Shark (Sphyrna Lewini). PLos One 7 (1), e29986. doi: 10.1371/journal.pone.0029986
Davis M. M., Suarez-Moo P., Daly-Engel T. S. (2019). Genetic Structure and Congeneric Range Overlap Among Sharpnose Sharks (Genus Rhizoprionodon) in the Northwest Atlantic Ocean. Can. J. Fish. Aquat. Sci. 76 (7), 1203–1211. doi: 10.1139/cjfas-2018-0019
Doughty C. E., Roman J., Faurby S., Wolf A., Haque A., Bakker E. S., et al. (2016). Global Nutrient Transport in a World of Giants. Proc. Natl. Acad. Sci. 113 (4), 868–873. doi: 10.1073/pnas.1502549112
Driggers W. B., Ingram G. W. Jr., Grace M. A., Gledhill C. T., Henwood T. A., Horton C. N., et al. (2008). Pupping Areas and Mortality Rates of Young Tiger Sharks Galeocerdo Cuvier in the Western North Atlantic Ocean. Aquat. Biol. 2 (2), 161–170. doi: 10.3354/ab00045
Dulvy N. K., Fowler S. L., Musick J. A., Cavanagh R. D., Kyne P. M., Harrison L. R., et al. (2014). Extinction Risk and Conservation of the World’s Sharks and Rays. Elife 3, e00590. doi: 10.7554/eLife.00590
Earl D. A., vonHoldt B. M. (2012). STRUCTURE HARVESTER: A Website and Program for Visualizing STRUCTURE Output and Implementing the Evanno Method. Conserv. Genet. Resour. 4 (2), 359–361. doi: 10.1007/s12686-011-9548-7
Evanno G., Regnaut S., Goudet J. (2005). Detecting the Number of Clusters of Individuals Using the Software STRUCTURE: A Simulation Study. Mol. Ecol. 14 (8), 2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x
Excoffier L., Lischer H. E. L. (2010). Arlequin Suite Ver 3.5: A New Series of Programs to Perform Population Genetics Analyses Under Linux and Windows. Mol. Ecol. Resour. 10 (3), 564–567. doi: 10.1111/j.1755-0998.2010.02847.x
Feldheim K. A., Fields A. T., Chapman D. D., Scharer R. M., Poulakis G. R. (2017). Insights Into Reproduction and Behavior of the Smalltooth Sawfish Pristis Pectinata. Endangered Species Res. 34, 463–471. doi: 10.3354/esr00868
Fitzpatrick J. L., Kempster R. M., Daly-Engel T. S., Collin S. P., Evans J. P. (2012). Assessing the Potential for Post-Copulatory Sexual Selection in Elasmobranchs. J. Fish. Biol. 80 (5), 1141–1158. doi: 10.1111/j.1095-8649.2012.03256.x
FitzSimmons N. N., Limpus C. J., Norman J. A., Goldizen A. R., Miller J. D., Moritz C. (1997). Philopatry of Male Marine Turtles Inferred From Mitochondrial DNA Markers. Proc. Natl. Acad. Sci. 94 (16), 8912–8917. doi: 10.1073/pnas.94.16.8912
Gaither M. R., Bowen B. W., Rocha L. A., Briggs J. C. (2016). Fishes That Rule the World: Circumtropical Distributions Revisited. Fish. Fish. 17 (3), 664–679. doi: 10.1111/faf.12136
García-Navas V., Ferrer E. S., Bueno-Enciso J., Barrientos R., Sanz J. J., Ortego J. (2014). Extrapair Paternity in Mediterranean Blue Tits: Socioecological Factors and the Opportunity for Sexual Selection. Behav. Ecol. 25 (1), 228–238. doi: 10.1093/beheco/art111
Gilbert K. J., Andrew R. L., Bock D. G., Franklin M. T., Kane N. C., Moore J. S., et al. (2012). Recommendations for Utilizing and Reporting Population Genetic Analyses: The Reproducibility of Genetic Clustering Using the Program STRUCTURE. Mol. Ecol. 21 (20), 4925–4930. doi: 10.1111/j.1365-294X.2012.05754.x
Glass G. V., Peckham P. D., Sanders J. R. (1972). Consequences of Failure to Meet Assumptions Underlying the Fixed Effects Analyses of Variance and Covariance. Rev. Educ. Res. 42 (3), 237–288. doi: 10.3102/00346543042003237
Gonzalez E. G., Cerón-Souza I., Mateo J. A., Zardoya R. (2014). Island Survivors: Population Genetic Structure and Demography of the Critically Endangered Giant Lizard of La Gomera, Gallotia Bravoana. BMC Genet. 15 (1), 1–17. doi: 10.1186/s12863-014-0121-8
Goudet J., Perrin N., Waser P. (2002). Tests for Sex-Biased Dispersal Using Bi-Parentally Inherited Genetic Markers. Mol. Ecol. 11 (6), 1103–1114. doi: 10.1046/j.1365-294X.2002.01496.x
Grubbs R. D. (2010). “Ontogenetic Shifts in Movements and Habitat Use,” in Sharks and Their Relatives II: Biodiversity, Adaptive Physiology, and Conservation (Boca Raton, Florida: CRC Press), 319–350.
Guttridge T. L., Van Zinnicq Bergmann M. P. M., Bolte C., Howey L. A., Finger J. S., Kessel S. T., et al. (2017). Philopatry and Regional Connectivity of the Great Hammerhead Shark, Sphyrna Mokarran in the US and Bahamas. Front. Mar. Sci. 4, 3. doi: 10.3389/fmars.2017.00003
Hammerschlag N., Gallagher A. J., Lazarre D. M. (2011). A Review of Shark Satellite Tagging Studies. J. Exp. Mar. Biol. Ecol. 398 (1-2), 1–8. doi: 10.1016/j.jembe.2010.12.012
Hammerschlag N., Gallagher A. J., Wester J., Luo J., Ault J. S. (2012). Don’t Bite the Hand That Feeds: Assessing Ecological Impacts of Provisioning Ecotourism on an Apex Marine Predator. Funct. Ecol. 26 (3), 567–576. doi: 10.1111/j.1365-2435.2012.01973.x
Hammerschlag N., McDonnell L. H., Rider M. J., Street G. M., Hazen E. L., Natanson L. J., et al. (2022). Ocean Warming Alters the Distributional Range, Migratory Timing, and Spatial Protections of an Apex Predator, the Tiger Shark (Galeocerdo Cuvier). Global Change Biol. 00, 1–16. doi: 10.1111/gcb.16045
Hays G. C., Ferreira L. C., Sequeira A. M. M., Meekan M. G., Duarte C. M., Bailey H., et al. (2016). Key Questions in Marine Megafauna Movement Ecology. Trends Ecol. Evol. 31 (6), 463–475. doi: 10.1016/j.tree.2016.02.015
Hedrick P. (2005). Large Variance in Reproductive Success and the Ne/N Ratio. Evolution 59 (7), 1596–1599. doi: 10.1111 j.0014-3820.2005.tb01809.x
Heupel M. R., Kanno S., Martins A. P. B., Simpfendorfer C. A. (2019). Advances in Understanding the Roles and Benefits of Nursery Areas for Elasmobranch Populations. Mar. Freshw. Res. 70 (7), 897–907. doi: 10.1071/MF18081
Hixon M. A., Johnson D. W., Sogard S. M. (2014). BOFFFFs: On the Importance of Conserving Old-Growth Age Structure in Fishery Populations. ICES J. Mar. Sci. 71 (8), 2171–2185. doi: 10.1093/icesjms/fst200
Holland K. N., Anderson J. M., Coffey D. M., Holmes B. J., Meyer C. G., Royer M. A. (2019). A Perspective on Future Tiger Shark Research. Front. Mar. Sci. 6, 37. doi: 10.3389/fmars.2019.00037
Holmes B. J., Pope L. C., Williams S. M., Tibbetts I. R., Bennett M. B., Ovenden J. R. (2018). Lack of Multiple Paternity in the Oceanodromous Tiger Shark (Galeocerdo Cuvier). R. Soc. Open Sci. 5 (1), 171385. doi: 10.1098/rsos.171385
Holmes B. J., Williams S. M., Otway N. M., Nielsen E. E., Maher S. L., Bennett M. B., et al. (2017). Population Structure and Connectivity of Tiger Sharks (Galeocerdo Cuvier) Across the Indo-Pacific Ocean Basin. R. Soc. Open Sci. 4 (7), 170309. doi: 10.1098/rsos.170309
Jakobsson M., Rosenberg N. A. (2007). CLUMPP: A Cluster Matching and Permutation Program for Dealing With Label Switching and Multimodality in Analysis of Population Structure. Bioinformatics 23 (14), 1801–1806. doi: 10.1093/bioinformatics/btm233
Jones O. R., Wang J. (2010). COLONY: A Program for Parentage and Sibship Inference From Multilocus Genotype Data. Mol. Ecol. Resour. 10 (3), 551–555. doi: 10.1111/j.1755-0998.2009.02787.x
Jorgensen S. J., Reeb C. A., Chapple T. K., Anderson S., Perle C., Van Sommeran S. R., et al. (2009). Philopatry and Migration of Pacific White Sharks. Proc. R. Soc. London B: Biol. Sci. 277 (1682), rspb20091155. doi: 10.1098/rspb.2009.1155
Kalinowski S. T., Taper M. L., Marshall T. C. (2007). Revising How the Computer Program CERVUS Accommodates Genotyping Error Increases Success in Paternity Assignment. Mol. Ecol. 16 (5), 1099–1106. doi: 10.1111/j.1365-294X.2007.03089.x
Kearse M., Moir R., Wilson A., Stones-Havas S., Cheung M., Sturrock S., et al. (2012). Geneious Basic: An Integrated and Extendable Desktop Software Platform for the Organization and Analysis of Sequence Data. Bioinformatics 28 (12), 1647–1649. doi: 10.1093/bioinformatics/bts199
Keeney D. B., Heist E. J. (2003). Characterization of Microsatellite Loci Isolated From the Blacktip Shark and Their Utility in Requiem and Hammerhead Sharks. Mol. Ecol. Notes 3, 501–504. doi: 10.1046/j.1471-8286.2003.00492.x
Klein J. D., Bester-van der Merwe A. E., Dicken M. L., Mmonwa K. L., Teske P. R. (2019). Reproductive Philopatry in a Coastal Shark Drives Age-Related Population Structure. Mar. Biol. 166 (3), 26. doi: 10.1007/s00227-019-3467-7
Koch M., Hadfield J. D., Sefc K. M., Sturmbauer C. (2008). Pedigree Reconstruction in Wild Cichlid Fish Populations. Mol. Ecol. 17 (20), 4500–4511. doi: 10.1111/j.1365-294X.2008.03925.x
Kohler N. E., Turner P. A. (2019). Distributions and Movements of Atlantic Shark Species: A 52-Year Retrospective Atlas of Mark and Recapture Data. Mar. Fish. Rev. 81 (2), 1–94. doi: 10.7755/MFR.81.2.1
Lamarca F., Carvalho P. H., Vilasboa A., Netto-Ferreira A. L., Vianna M. (2020). Is Multiple Paternity in Elasmobranchs a Plesiomorphic Characteristic? Environ. Biol. Fish. 103 (12), 1463–1470. doi: 10.1007/s10641-020-01034-y
Lea J. S. E., Wetherbee B. M., Queiroz N., Burnie N., Aming C., Sousa L. L., et al. (2015). Repeated, Long-Distance Migrations by a Philopatric Predator Targeting Highly Contrasting Ecosystems. Sci. Rep. 5, 11202. doi: 10.1038/srep11202
Lee P. L. M., Luschi P., Hays G. C. (2007). Detecting Female Precise Natal Philopatry in Green Turtles Using Assignment Methods. Mol. Ecol. 16 (1), 61–74. doi: 10.1111/j.1365-294X.2006.03115.x
Linck E., Battey C. J. (2019). On the Relative Ease of Speciation With Periodic Gene Flow. bioRxiv 19(3)639–647.
Lowe W. H., Allendorf F. W. (2010). What can Genetics Tell Us About Population Connectivity? Mol. Ecol. 19 (15), 3038–3051.doi: 10.1111/j.1365-294X.2010.04688.x
Luschi P., Hays G. C., Papi F. (2003). A Review of Long-Distance Movements by Marine Turtles, and the Possible Role of Ocean Currents. Oikos 103 (2), 293–302. doi: 10.1034/j.1600-0706.2003.12123.x
Meyer C. G., O’Malley J. M., Papastamatiou Y. P., Dale J. J., Hutchinson M. R., Anderson J. M., et al. (2014). Growth and Maximum Size of Tiger Sharks (Galeocerdo Cuvier) in Hawaii. PLos One 9 (1), e84799. doi: 10.1371/journal.pone.0084799
Mills L. S., Allendorf F. W. (1996). The One-Migrant-Per-Generation Rule in Conservation and Management. Conserv. Biol. 10 (6), 1509–1518. doi: 10.1046/j.1523-1739.1996.10061509.x
Musick J. A., Harbin M. M., Compagno L. J. V. (2004). “Historical Zoogeography of the Selachii", in Biology of Sharks and Their Relatives. Eds. Carrier J. C., Musick J. A., Heithaus M. R. (Boca Raton, FL: CRC Press).
Orr T. J., Brennan P. L. R. (2015). Sperm Storage: Distinguishing Selective Processes and Evaluating Criteria. Trends Ecol. Evol. 30 (5), 261–272. doi: 10.1016/j.tree.2015.03.006
Papastamatiou Y. P., Meyer C. G., Carvalho F., Dale J. J., Hutchinson M. R., Holland K. N. (2013). Telemetry and Random-Walk Models Reveal Complex Patterns of Partial Migration in a Large Marine Predator. Ecology 94 (11), 2595–2606. doi: 10.1890/12-2014.1
Payevsky V. A. (2016). Sex-Biased Survival and Philopatry in Birds: Do They Interact? Biol. Bull. 43 (8), 804–818. doi: 10.1134/S1062359016080136
Phillips N. M., Devloo-Delva F., McCall C., Daly-Engel T. S. (2021). Reviewing the Genetic Evidence for Sex-Biased Dispersal in Elasmobranchs. Rev. Fish. Biol. Fish. 31), 821–841. doi: 10.1007/s11160-021-09673-9
Pirog A., Jaquemet S., Ravigné V., Cliff G., Clua E., Holmes B. J., et al. (2019). Genetic Population Structure and Demography of an Apex Predator, the Tiger Shark Galeocerdo Cuvier. Ecol. Evol. 9 (10), 5551–5571. doi: 10.1002/ece3.5111
Pirog A., Magalon H., Poirout T., Jaquemet S. (2020). New Insights Into the Reproductive Biology of the Tiger Shark Galeocerdo Cuvier and No Detection of Polyandry in Reunion Island, Western Indian Ocean. Mar. Freshw. Res. 71 (10), 1301–1312. doi: 10.1071/MF19244
Pratt H. L. (1993). The Storage of Spermatozoa in the Oviducal Glands of Western North Atlantic Sharks. Environ. Biol. Fish. 38, 139–149. doi: 10.1007/BF00842910
Pritchard J. K., Stephens M., Donnelly P. (2000). Inference of Population Structure Using Multilocus Genotype Data. Genetics 155, 945–959. doi: 10.1093/genetics/155.2.945
Queller D. C., Goodnight K. F. (1989). Estimating Relatedness Using Genetic Markers. Evolution 43 (2), 258–275. doi: 10.1111/j.1558-5646.1989.tb04226.x
Quinn T. P., Dittman A. H. (1990). Pacific Salmon Migrations and Homing: Mechanisms and Adaptive Significance. Trends Ecol. Evol. 5 (6), 174–177. doi: 10.1016/0169-5347(90)90205-R
Santos C. C., Coelho R. (2018). Migrations and Habitat Use of the Smooth Hammerhead Shark (Sphyrna Zygaena) in the Atlantic Ocean. PLos One 13 (6), e0198664. doi: 10.1371/journal.pone.0198664
Selkoe K. A., Toonen R. J. (2006). Microsatellites for Ecologists: A Practical Guide for Using and Evaluating Microsatellite Markers. Ecol. Lett. 9, 615–629. doi: 10.1111/j.1461-0248.2006.00889.x
Sulikowski J. A., Wheeler C. R., Gallagher A. J., Prohaska B. K., Langan J. A., Hammerschlag N. (2016). Seasonal and Life-Stage Variation in the Reproductive Ecology of a Marine Apex Predator, the Tiger Shark Galeocerdo Cuvier, at a Protected Female-Dominated Site. Aquat. Biol. 24 (3), 175–184. doi: 10.3354/ab00648
van Oosterhout C. V., Hutchinson W., Wills D., Shipley P. (2004). MICRO-CHECKER: Software for Identifying and Correcting Genotype Errors in Microsatellite Data. Mol. Ecol. Notes 4, 535–538. doi: 10.1111/j.1471-8286.2004.00684.x
Veríssimo A., McDowell J. R., Graves J. E. (2012). Genetic Population Structure and Connectivity in a Commercially Exploited and Wide-Ranging Deepwater Shark, the Leafscale Gulper (Centrophorus Squamosus). Mar. Freshw. Res. 63 (6), 505–512. doi: 10.1071/MF11237
Wang J. (2004). Application of the One-Migrant-Per-Generation Rule to Conservation and Management. Conserv. Biol. 18 (2), 332–343. doi: 10.1111/j.1523-1739.2004.00440.x
Wang J. (2007). Triadic IBD Coefficients and Applications to Estimating Pairwise Relatedness. Genet. Res. 89 (3), 135–153. doi: 10.1017/S0016672307008798
Wang J. (2011). COANCESTRY: A Program for Simulating, Estimating and Analysing Relatedness and Inbreeding Coefficients. Mol. Ecol. Resour. 11 (1), 141–145. doi: 10.1111/j.1755-0998.2010.02885.x
Wang J. (2017). Estimating Pairwise Relatedness in a Small Sample of Individuals. Heredity 119 (5), 302–313. doi: 10.1038/hdy.2017.52
Waples R. S., Gaggiotti O. (2006). What is a Population? An Empirical Evaluation of Some Genetic Methods for Identifying the Number of Gene Pools and Their Degree of Connectivity. Mol. Ecol. 15 (6), 1419–1439. doi: 10.1111/j.1365-294X.2006.02890.x
Weideli O. C., Papastamatiou Y. P., Planes S. (2019). Size Frequency, Dispersal Distances and Variable Growth Rates of Young Sharks in a Multi-Species Aggregation. J. Fish. Biol. 94 (5), 789–797. doi: 10.1111/jfb.13968
Werner E. E., Gilliam J. F. (1984). The Ontogenetic Niche and Species Interactions in Size-Structured Populations. Annu. Rev. Ecol. Systemat. 15 (1), 393–425. doi: 10.1146/annurev.es.15.110184.002141
Whitney N. M., Crow G. L. (2007). Reproductive Biology of the Tiger Shark (Galeocerdo Cuvier) in Hawaii. Mar. Biol. 151 (1), 63–70. doi: 10.1007/s00227-006-0476-0
Keywords: philopatry, site fidelity, kinship, microsatellites, movement patterns
Citation: McClain MA, Hammerschlag N, Gallagher AJ, Drymon JM, Grubbs RD, Guttridge TL, Smukall MJ, Frazier BS and Daly-Engel TS (2022) Age-Dependent Dispersal and Relatedness in Tiger Sharks (Galeocerdo cuvier). Front. Mar. Sci. 9:900107. doi: 10.3389/fmars.2022.900107
Received: 20 March 2022; Accepted: 14 June 2022;
Published: 18 July 2022.
Edited by:
Charlie Huveneers, Flinders University, AustraliaReviewed by:
Jonathan Sandoval, Flinders University, AustraliaCopyright © 2022 McClain, Hammerschlag, Gallagher, Drymon, Grubbs, Guttridge, Smukall, Frazier and Daly-Engel. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Toby S. Daly-Engel, dGRhbHllbmdlbEBmaXQuZWR1
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.