- 1Key Laboratory of Healthy Mariculture for the East China Sea, Ministry of Agriculture and Rural Affairs, Jimei University, Xiamen, China
- 2Laboratory for Marine Fisheries Science and Food Production Processes, Qingdao National Laboratory for Marine Science and Technology, Qingdao, China
Sex is one of the most important scientific topics, and the existence of sex dimorphism is a conserved feature in vertebrate taxa. The research on sex-determining genes and sex chromosome evolution is a hot topic in biology. The majority of sex-determining genes expressed in somatic cells, and reciprocal interactions between germ cells and somatic cells, are important for gonadal differentiation. However, the knowledge of how signals are transmitted between somatic cells and germ cells remains unclear. In this study, we confirmed the 36 dph was a sex determination critical stage in yellow drum through transcriptome analysis at the early development stages. We further found that Kank1 participated in the germ cells’ motility process in yellow drum through KEGG pathway analysis together with protein-protein interaction network. With the dual-luciferase reporter assay, we detected that Kank1 increased the transcription of dmrt1_Luc gene in CHO cells and 293T cells. Additionally, we observed that Kank1 was not only expressed in the cytoplasm but also in the nucleus in CHO cells through a fluorescence microscope. These results suggest that Kank1 acts as a shuttling protein as in humans and may play a difunctional role at the early gonad development stage in yellow drum, and it not only participated in the germ cells’ motility but also increased the expression of dmrt1 in yellow drum.
Introduction
Sex determination is one of the most primarily biological processes, while the way of initiating female and male development exhibits remarkable diversity and variability across vertebrates (Li and Gui, 2018). The existence of sex dimorphism is a conserved feature in vertebrate taxa. It has been shown that sex can induce profound cellular and molecular dimorphism in both sexual and somatic cells of vertebrates, which is an important issue in aquaculture and medical science (Niksirat et al., 2021). Therefore, sex determination is a key issue for managing problems arising from sexual dimorphism. Importantly, various sex determination mechanisms among vertebrates suggest that it is the ideal biological event helping us understand the evolutionary conundrums underlying speciation and species diversity (Nagahama et al., 2021). The rapid development of sequencing technologies has shed light on the investigation of genomic composition in many species. Recent research advancements mainly focus on the sex chromosome evolution, screening of the master sex-determining gene,s and a part of brief gene regulatory network. A major landmark in vertebrate sex-determining genes was the discovery of the SRY gene in 1990 (Sinclair et al., 1990). Since then, considerable progress in the discovery of sex-determining genes has been made over the last three decades, such as DMY in the Oryzias latipes (Matsuda et al., 2002), amhy in the Patagonian pejerrey (Hattori et al., 2012), gsdfY in the Oryzias luzonensis (Myosho et al., 2012), sdy in the Oncorchynchus mykiss (Yano et al., 2012), and BCAR1 in the channel catfish (Bao et al., 2019), which are specifically expressed in somatic cells. The fact that sex determination of somatic cells precedes that of germ cells has been widely accepted. Furthermore, sex reversal in somatic cells leads to sex reversal in germ cells (Bowles and Koopman, 2010; Capel, 2017). However, the knowledge of how signals transmit between somatic cells and germ cells is still lacking.
Primordial germ cells (PGCs) are the stem cells of gametes, providing genome transmission to future generations (Lesch and Page, 2012), and germ cells have the potential to differentiate into eggs and spermatozoa (Golpour et al., 2016a; Golpour et al., 2016b; Golpour et al., 2017). Reciprocal interactions between germ cells and somatic cells are important for gonadal differentiation (Kocer et al., 2009). Recently, the regulatory role of germ cells during sexual development has been demonstrated. In teleost, the requirement of germ cells for gonadal development appears to be variable. Their absence leads to exclusive male development in medaka and zebrafish (Kurokawa et al., 2007; Siegfried and Nüsslein-Volhard, 2008) but not in goldfish or loach (Fujimoto et al., 2010; Goto et al., 2012). The number of PGCs likely plays an important role in teleost sexual differentiation, and females possess more germ cells than males due to their sexually dimorphic proliferation of germ cells in medaka (Saito et al., 2007), zebrafish (Siegfried and Nüsslein-Volhard, 2008) and stickleback (Lewis et al., 2008). During development, PGCs undergo specification, migration, and proliferation. These processes are regulated by transmembrane receptors that receive external chemoattractant signals, which are then translated to cytoskeletal changes by effector molecules such as phospholipids and small GTPases (Ridley et al., 2003). Meanwhile, highly regulated cell-to-cell and cell-substrate adhesion is important for PGCs motility, and localized growth factors may control PGC survival and PGC localization (Ridley et al., 2003; Richardson and Lehmann, 2010). In general, the expression pattern of genes corresponds to the developmental morphology of cells.
Yellow drum (Nibea albiflora) is an important fish species in fishery and aquaculture, widely distributed in southern Japan and East China Sea (Takita, 1974). The dmrt1 gene may be the most vital sex determing gene in yellow drum (Sun et al., submitted). In medaka, dmrt1bY expression causes a cell cycle arrest in the G2 stage and mediates a mitotic arrest in the PGCs (Herpin et al., 2007; Herpin and Schartl, 2009). It is the reason that the number of germ cells in females tends to be more than in males. In zebrafish, the dmrt1-mutated testes displayed severe testicular development defects and gradual loss of all Vasa-positive germ cells via inhibiting their self-renewal and inducing apoptosis. The dmrt1 gene might be required for the maintenance, self-renewal, and differentiation of male germ cells in zebrafish (Lin et al., 2017). In tilapia, knockdown of dmrt1 resulted in feminization of gonadal somatic cells and loss of germ cells in males (Dai et al., 2021). Therefore, it seems that the function of the dmrt1 gene in germ cell development is different in a variety of fish species. In yellow drum, two types of gonadal tissues were observed in the development stage of 40 days post-hatch (dph), which were designated as tissue type I and type II. Gonadal tissue type I showed a larger surface area and consisted of different populations of germ cells and was considered a presumptive ovary. Until 55 dph, the overall appearance of spermatogonia was similar to that of undifferentiated germ cells at 40 dph, referred to as the gonadal tissue type II (Lou et al., 2016). The different developmental stages of the germ cells in yellow drum may be associated with their different migration behavior or dimorphic proliferation. However, the connection between the sex-determining gene dmrt1 with the germ cells remained unknown.
Kank1 belongs to the Kank family. It is characterized by their unique structure, coiled-coil motifs in the N-terminal region, and ankyrin-repeats in the C-terminal region, with an additional KN motif at the N-terminus (Zhu et al., 2008; Fan et al., 2020). Kank1 was reported to play a significant role in the regulation of the action cytoskeleton (Ding et al., 2003). It can negatively regulate actin polymerization and cell motility through inhibiting the binding of Rac1 and insulin receptor substrate p53 (IRSp53) (Mhawech, 2005; Kakinuma et al., 2008; Kakinuma et al., 2009; Roy et al., 2009). In this study, we confirmed 36 dph was a sex determination critical stage in yellow drum through transcriptome analysis at the early development stages, and found that Kank1 participated in the cell motility process and interacted with Dmrt1 in yellow drum. With the dual-luciferase reporter assay, we detected that Kank1 increased the transcription of dmrt1_Luc gene in CHO cells and 293T cells. Additionally, we observed that Kank1 was not only expressed in the cytoplasm but also in the nucleus in CHO cells through a fluorescence microscope. These results suggest that Kank1 acts as a shuttling protein as in humans and may play a difunctional role at the early gonad development stage in yellow drum It not only participated in the germ cells’ motility but could increase the expression of dmrt1 in yellow drum.
Materials And Method
Ethical Approval
The sample collection and animal experiments were conducted in accordance with the regulations of the Guide for Care and Use of Laboratory Animals and were approved by the Animal Care and Use Committee of Jimei University.
Experimental Fish and Sampling, RNA Extraction and RNA Sequencing
To obtain samples for RNA-seq, artificially bred yellow drum of 26 days post-hatch (dph) purchased from a breeding farm in Zhangzhou, Fujian province, China. Fries were reared in two tanks (80 cm × 60 cm × 60 cm) with temperature at 28°C and salinity at 25 ~ 30, dissolved oxygen at 6 ppm. Fries were fed with compound feed three times a day. The sample collection at five critical stages (30 dph, 36 dph, 46 dph, 55 dph, and 61 dph) of yellow drum gonadal development according to Lou et al. (Lou et al., 2016), approximately 800 individuals were collected at each stage. Fries were anesthetized with tricaine methanesulfonate (MS-222, Shanghai Acmec Biochemical Co., China) at 300 mg/L before sampling, and the body length was recorded (Table 1). The head and tail of each fry was pruned, and the rest of the body was cut open from the abdomen and stored in RNA Later solution. Subsequently, gonads were dissected and collected from the body under a microscope for RNA extraction and the remaining part was stored at 75% ethanol for DNA extraction. The sex of each sampled fry was determined through PCR amplification with the sex specific markers (Sun et al., 2018). To obtain a sufficient quantity of RNA for sequencing, we generated three pools of gonads isolated from the fries for each sex at each stage. Total RNA was isolated using TRIZOL Reagent (Invitrogen, USA) according to the manufacturer’s instructions. The purity and integrity of the RNA were verified with agarose gel electrophoresis and Bioanalyzer 2100 (Agilent Technologies, USA). RNA integrity number (RIN) ≥ 8.0 were subjected to RNA enrichment with magnetic beads with Oligo (dT), and RNA was disrupted into short fragments by adding fragmentation buffers. These short fragments were used as templates and random hexamers as primers to synthesize the first strand of cDNA, followed by the synthesis of second strand cDNA. The AMPure XP beads were employed to purify double-strand cDNA, EB buffer was used for end-repair and A-tailing. The purified fragments were ligated with Illumina sequencing adapters, and paired-end libraries were sequenced on an Illumina NovaSeq 6000 system.
Transcriptome Analysis
The clean reads were obtained from raw data by filtering out adaptor contaminated reads and low-quality reads. The quality of the sequenced reads was assessed by FastQC v0.11.9 (Andrews, 2010). Clean reads were aligned to the male reference genome using STAR v2.5.3a (Dobin et al., 2013). After alignment, the transcripts were assembled using StringTie v1.3.3b (Pertea et al., 2015), and the number of reads that mapped to the genes and exons was calculated using FeatureCounts v2.0.1. The analysis of differentially expressed genes was performed with DESeq2, and the related R packages were employed for the PCA analysis. Significantly differentially expressed genes (DEG) were defined as adjust P-value < 0.05 and log102(Foldchange) > 1. Gene Ontology (GO) terms analysis of DEGs and Kyoto Encyclopaedia of Genes and Genomes (KEGG) pathway enrichment were performed using clusterProfiler R packages.
The interaction network of differential genes between females and males at 36 dph was analyzed using the STRING data [STRING: functional protein association networks (string-db.org)]. The CytoScape software was used to display the result.
Phylogenetic Analysis and Collinearity Analysis of Dmrt1
The protein sequences of 11 species including Homo sapiens (human), Gallus gallus (chicken), Lctalurus punctatus (spotted gar), Danio retio (zebrafish), Oryzia latipes (medaka), Takifugu rubripes (fugu), Tetraodon nigrovirids (spotted green pufferfish), Oreochromis niloticus (tilapia), Gasterosteus aculeatus (three spined stickleback), and Larimichthys crocea (large yellow croaker) were obtained from Ensembl website (https://asia.ensembl.org/index.html). The phylogenetic tree was constructed with MEGA 5.0 using UPGMA method. The adjacent genes with dmrt1 of the 11 species are referred to the Ensembl Genome Browser (http://www.ensembl.org).
Kank1 Gene Structure and Expression Analysis
The Kank1 gene reference sequences were extracted from transcriptome data and the amino acid sequences were translated through DNAMAN. The nuclear localization signal (NLS) was predicted using online tools NLStradamus (utoronto.ca) and the nuclear export signal (NES) was predicted using online tools LocNES NES prediction tool provided by Chook Lab (swmed.edu). The Kank1 gene expression was assessed using FPKM at different development stages in yellow drum and significant differences between the males and females were determined by t-test (p < 0.05).
Construction of Plasmid
Kank1 expression vector (pCS2-dmrt1-mCherry) was constructed and the coding sequence was obtained through PCR cloning from cDNA library of yellow drum. The pCS2- kank1-mCherry vector was double digested with BmaHI and ClaI, then pCS2-dmrt1-mCherry was constructed with dmrt1 coding sequence cloned into the digested site using ClonExpress® II One Step Cloning Kit (Vazyme, Nanjing, China). In order to distinguish with mCherry fluorescence, we amplified the EGFP coding sequences using the pEGFP-N1 vector as a template and replaced the mCherry coding sequences of the vector pCS2-dmrt1-mCherry using the ClaI and XhoI double digestion to construct the vector pCS2-dmrt1-EGFP. The dmrt1 promoter sequences were amplified from the genomic DNA of the yellow drum, the product was double digested with KpnI and HindIII, then cloned into the same site of pGL3-basic vector (Promega, USA) to construct the vector pGL3-basic-dmrt1_promoter. All the primers used in PCR cloning warelisted in Table 2.
Cell Culture
CHO (a cell line from Cricetulus griseus ovary) and HEK293 were cultured in 1640 and high glucose DMEM medium (Dulbecco’s modified eagle medium, Gibco), respectively, supplemented with 10% fetal bovine serum and 1% penicillin 100 units/ml with 100 ug/ml streptomycin, plated in 25 cm2 flask, and placed in the carbon dioxide incubator. The detailed operation steps were followed according to the reference manual.
Cell Transfection
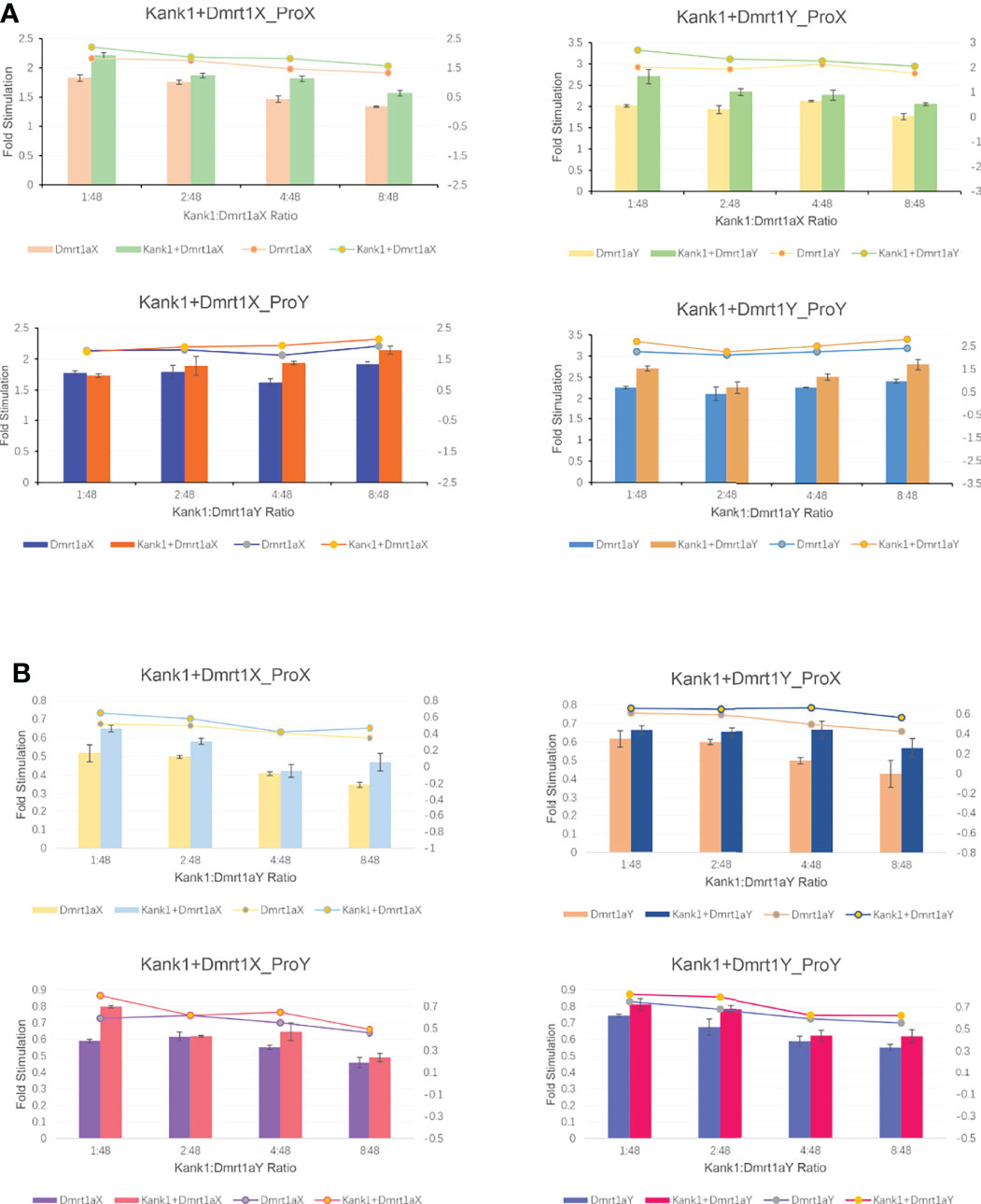
Lipo8000™ Transfection Reagent was used in vector transfection (Beyotime Biotechnology, China). Cells were plated in 12-well plates at 18 ~ 24 hours before transfection. While the cells were completely adherent and stuck to the wall, the fresh medium was replaced according to protocol. The cells were transfected with 50 μL Opti-MEM Medium, 1 μg DNA and 1.6 μL Lipo8000™. The ratio of the pGL3-basic promoter vector and the pRT-TK vector (an internal control) was about 40: 1. In the co-transfection assay, the ratio of expression vector to promoter vector was 1: 48, 2: 48, 4: 48, 8: 48, respectively. Luciferase activity was measured at 24 hours after transfection using the dual-luciferase reporter assay system (Beyotime Biotechnology, China) and LB 9508 Lumat3 (Berthold technologies, Germany) following the manufacturer’s protocol. Assays were performed in triplicate and expressed as mean ± standard error.
DAPI Dyeing and Observation
Firstly, the medium in the 12-well cell plates was discarded and washed once with 1 ml PBS. Cells were then fixed with 1 mL 4% paraformaldehyde for 10 minutes, and were permeabilized with 0.2% Tween 100 for 10 minutes and washed once again with 1 mL PBS. Nuclear staining was performed with 1 mL 0.2% DAPI for 15 minutes, and observation and photography were done with a laser confocal scanning microscope (TCSSP8, Leica).
Results
Transcriptome Analysis and the Identification of the Sex Determination Critical Stage
RNA libraries generating from the gonad of the various early stages were sequenced on an Illumina NovaSeq 6000 system. The total raw data and clean data were 318.7 Gb and 307.7 Gb, respectively. The average reads map rate was 87.9% when aligned the clean reads to the assembly of yellow drum with STAR. Transcriptome assembly obtained 23,997 transcripts from 18,596 genes (Table 3).
We performed PCA analysis with all the samples (Figure 1). Clustering discovered that the samples were similarly grouped into two sections by the sex (dimension 1, Figure 1A) and the stage (dimension 2, Figure 1B). This result suggests that the samples before 36 dph formed a group and the samples after 36 dph formed another group, indicated 36 dph (body length = 24.4 ± 2.2 mm) was a critical stage. Subsequently, we analyzed the different expression genes between females and males at each stage (Figure 2A) and counted the genes of up- or down-regulated (Figure 2B), and found that the number of genes with up- and down-regulated at 36 dph were more than at other stages. This could be due to the sex biased genes expressed in two kinds of gonad before 36 dph, and switched into sex-specific expression patterns once the sex-determining key gene was expressed. A previous study showed that the critical period of sex differentiation in the yellow drum occurred at the stage of 36 dph (total length = 21.2 ± 3.9 mm) (Lou et al., 2016). Our results imply the 36 dph is a sex determination critical stage, which is inconsistent with the previous result.
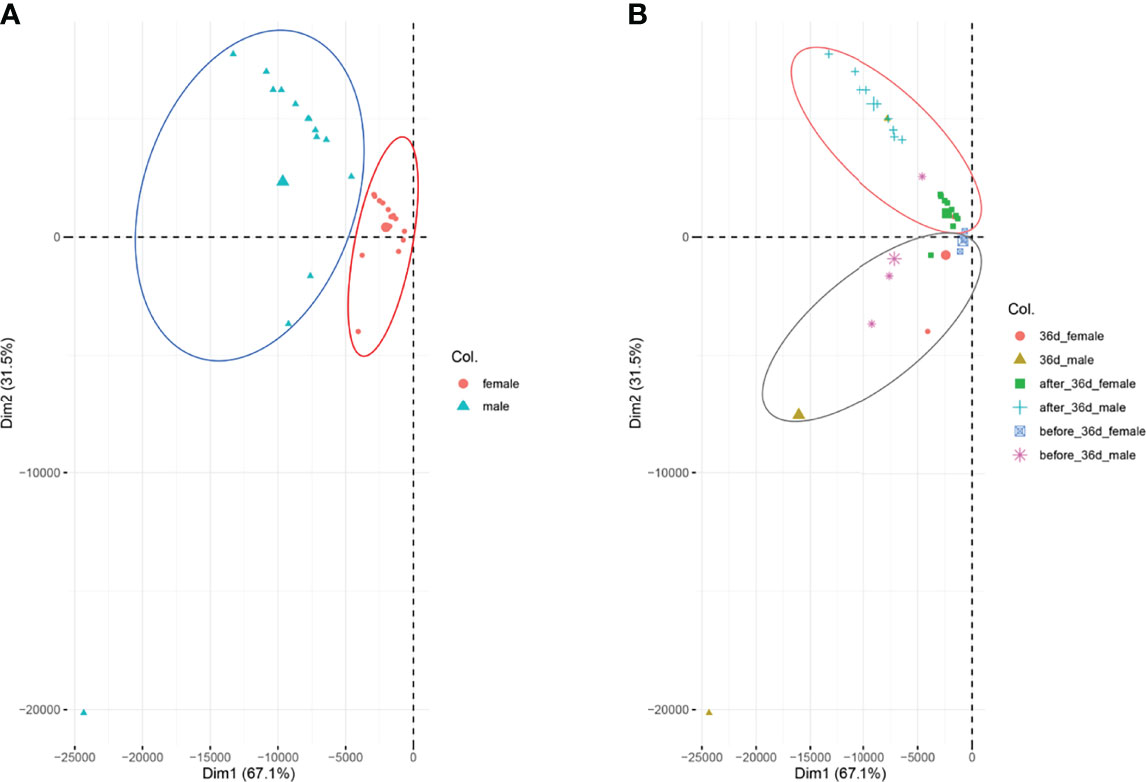
Figure 1 PAC analysis for all the samples. (A) The samples were analyzed according to sex, females are indicated in red and males are indicated in blue-green. (B) The samples were analyzed according to the stages.
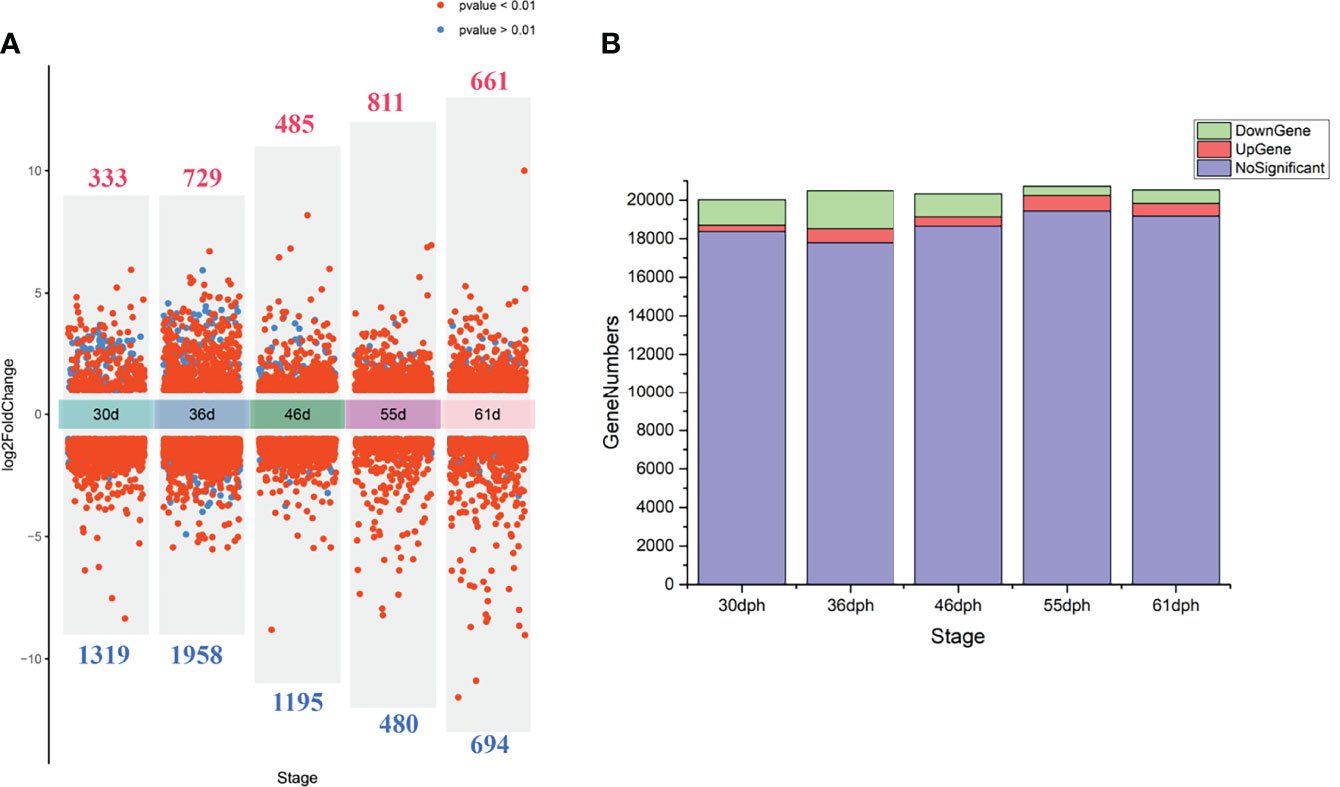
Figure 2 (A) The volcano plot demonstrates the differential expression genes between females and males at each stage, the numbers of differential expression genes are indicated in red (up-regulated) and blue (down-regulated). (B) The stacking histogram shows gene numbers of up-regulated, down-regulated and not significant at different stages.
Gene Function Enrichment of DEGs at the Sex Determination Critical Stage (36 dph)
To investigate the function of these differential genes, we performed GO and KEGG analyses between females and males at 36 dph (Figure 3). In females, the upregulated genes mainly participated in the biological processes associated with development process, reproductive process, gamete generation, germ cell development, and cellular process involved in reproduction in multicellular organisms (Figure 3A). Of note, the cell cycle and Wnt signaling pathway were primarily enriched (Figure 3C). In males, upregulated genes were mainly involved in transmembrane receptor protein tyrosine kinase signaling pathway, cell migration, cell motility, and tube morphogenesis (Figure 3B). The KEGG pathway analysis suggested that Focal adhesion and TGF-beta signaling pathway played important roles in males at 36 dph (Figure 3D). According to the general process of sexual development, we speculated that the primordial germ cells had migrated to the place of primitive gonad and began to proliferate and develop in females. However, the gonad development was relatively late in males. The primordial germ cells may be undergoing the cell migration process. In addition, we collected genes that have been reported to express in germ cells, somatic cells (Matsuda et al., 2002; Richardson and Lehmann, 2010). Comparing the genes expressed in germ cells, somatic cells, and the genes involved in the biological processes of cell cycle and cell migration at different stages (Figure 4), we found that the genes participating in the cell migration process expressed higher in males than in females. However, the genes expressed in germ cells involved in the cell cycle expressed higher in females. The gene expression trend was consistent with the results of GO and KEGG enrichment.
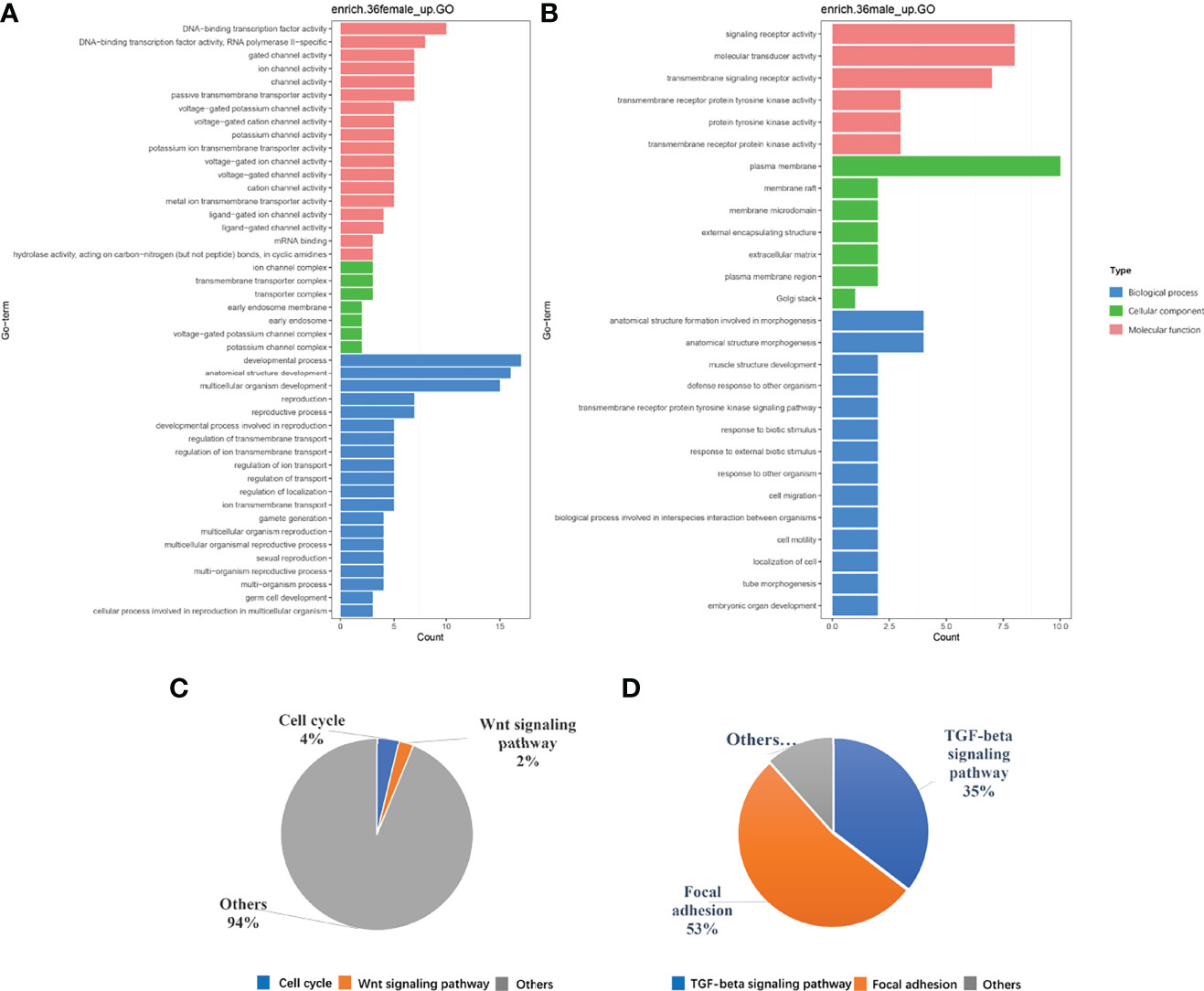
Figure 3 (A, B) are the GO enrichment results. (C, D) are the results of KEGG pathway analysis, respectively in females and in males at the 36 dph.
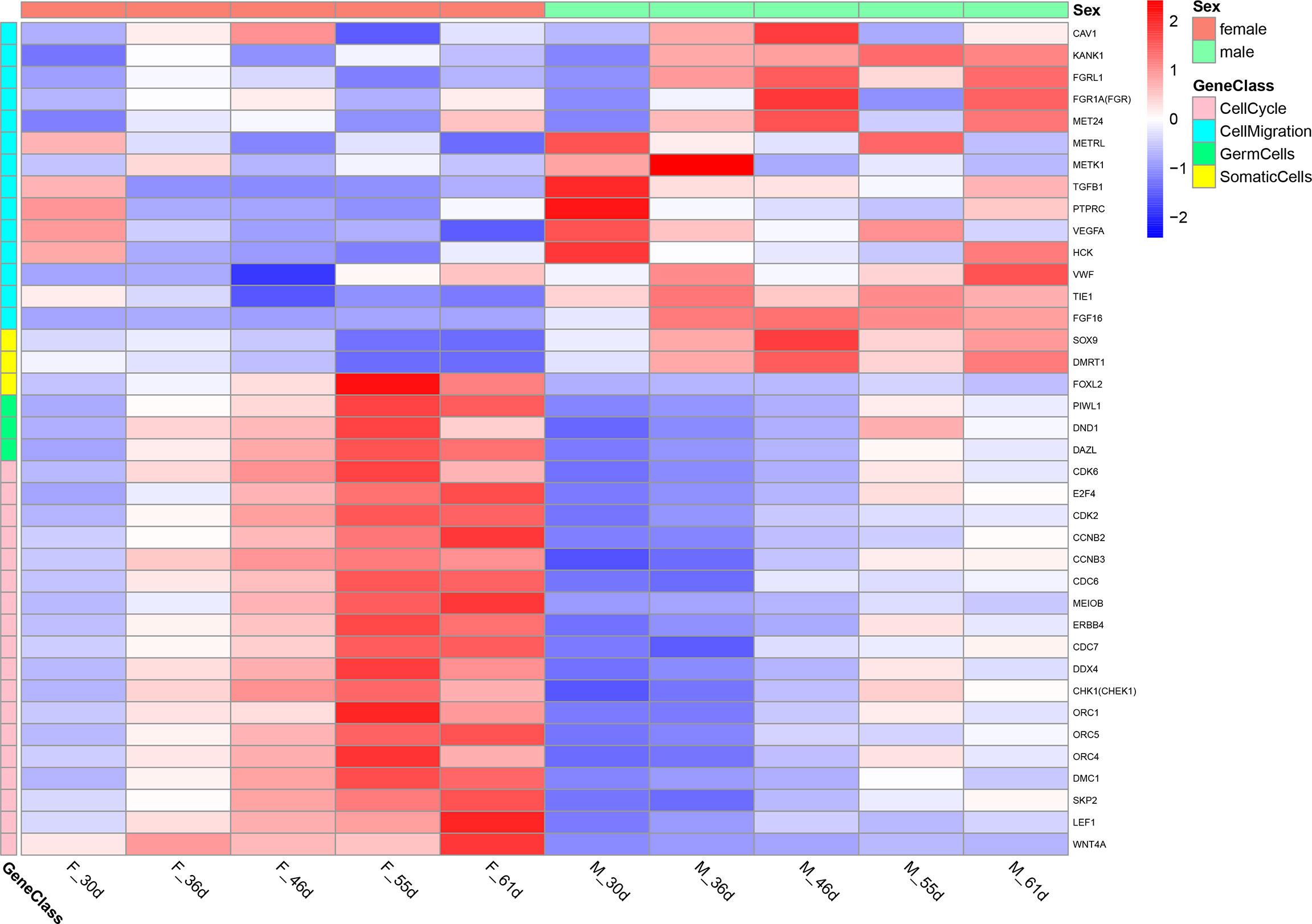
Figure 4 The heatmap of the genes expressed in germ cells, somatic cells and the genes involved in the biological processes of cell cycle and cell migration at different stage.
DEGs Interaction Network Analysis
The differential genes pathway analysis provided a clue that mabe the different behavior of the primordial germ cells caused the differences in gonad development between males and females. Given dmrt1 plays a pivotal role in the sex determination of yellow drum, we wanted to understand the link between master sex determining gene Dmrt1 and the development of primordial germ cells. We analyzed the protein-protein interaction networks with the differential genes associated with the behavior of primordial germ cells from the pathway analysis (Figure 5). The result implies that the proteins interaction mainly involved in the cell motility process and in the cell cycle in males and females, respectively. The network of proteins interaction was complex. We paid close attention to which proteins may interact with Dmrt1 in yellow drum, including Sox8, Sox9, Fgf6, Bmpr2, Kank1, Foxl2, Wnt4a, and Orc1. However, it seemed that only Kank1 could act upon Dmrt1 directly among the various genes. This result indicated that the Kank1 might take part in the germ cells migration process and could interact with Dmrt1 in yellow drum. Previous studies reported that Kank1 negatively regulated actin polymerization and cell motility through inhibiting the binding of Rac1 and insulin receptor substrate p53 (IRSp53) (Mhawech, 2005; Kakinuma et al., 2008; Kakinuma et al., 2009; Roy et al., 2009). Therefore, we suspected that Kank1 might inhibit the germ cells migration at the 36 dph in male individuals of yellow drum. However, the relationship between Dmrt1 and Kank1 is unclear, and has not been reported in previous related studies.
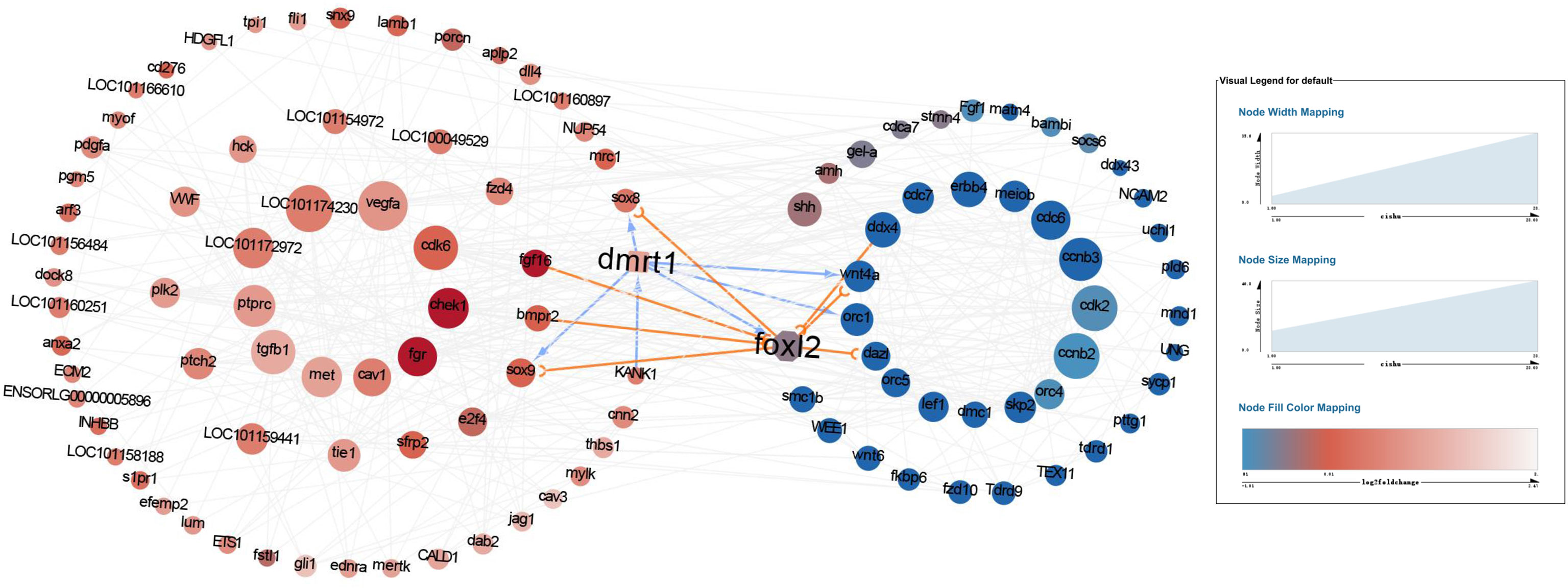
Figure 5 The protein-protein interaction networks with the differential genes detected from pathway analysis. The red and blue cycles represent the upregulated proteins in males and females, respectively. The darker the dot, the more significantly upregulated of the protein. And the bigger the dot, the more interactions.
Phylogenetic and Collinearity Analyses of Kank1 and Dmrt1
We found that the kank1 gene os located in the sex chromosome near dmrt1. Previous studies have shown that sex chromosomes usually evolved from autosomes, and once the sex determining gene fixed in the chromosome, the sexually antagonistic allele genes would be located in the surroundings (Charlesworth et al., 2005). It seemed the kank1 gene may be the sexually antagonistic allele gene. We then constructed the phylogenetic trees of dmrt1, and analyzed the collinearity between kank1 and dmrt1 in different species (Figure 6). The result suggests that the collinearity between kank1 and dmrt1 was conserved from fishes to humans. This result suggested that the kank1 gene might act as an important role in the sex determination in yellow drum, and it should be very meaningful to study the gene function of kank1.
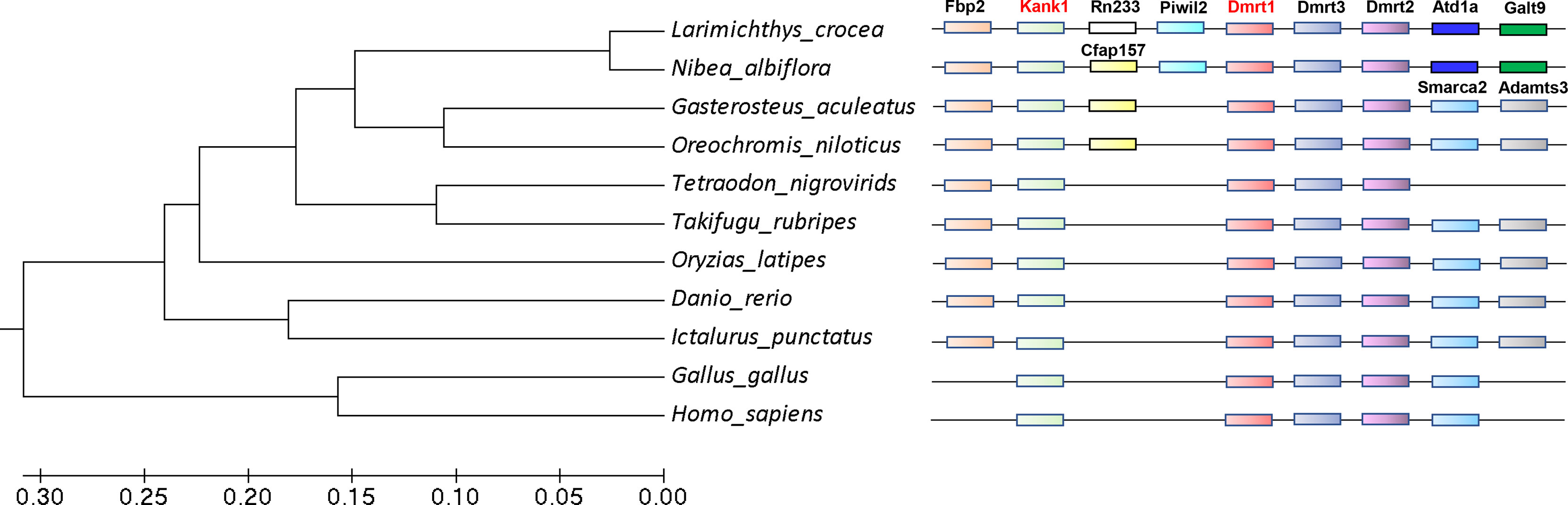
Figure 6 The phylogenetic trees of dmrt1 and collinearity analysis between dmrt1 and kank1. The different genes are represented by different color boxes.
Kank1 Gene Cloning and Expression Analysis
The ORF of kank1 consisted of 3993 bp encoding 1330 amino acid residues in yellow drum (Figure 7A). A KN motif and an Ank repeat were predicted in the C-terminal and N-terminal, respectively. The nuclear localization signal (NLS) and the nuclear export signal (NES) were predicted (Figure 7A), implying Kank1 may be a shuttling protein. The kank1 gene expressed significantly higher in males than females from 46 dph to 55 dph, which was the stage that the cytological characteristics of male gonad differentiation were gradually observed (Figure 7B). This result suggests that the kank1 gene might act as an indispensable role in the male gonad differentiation.
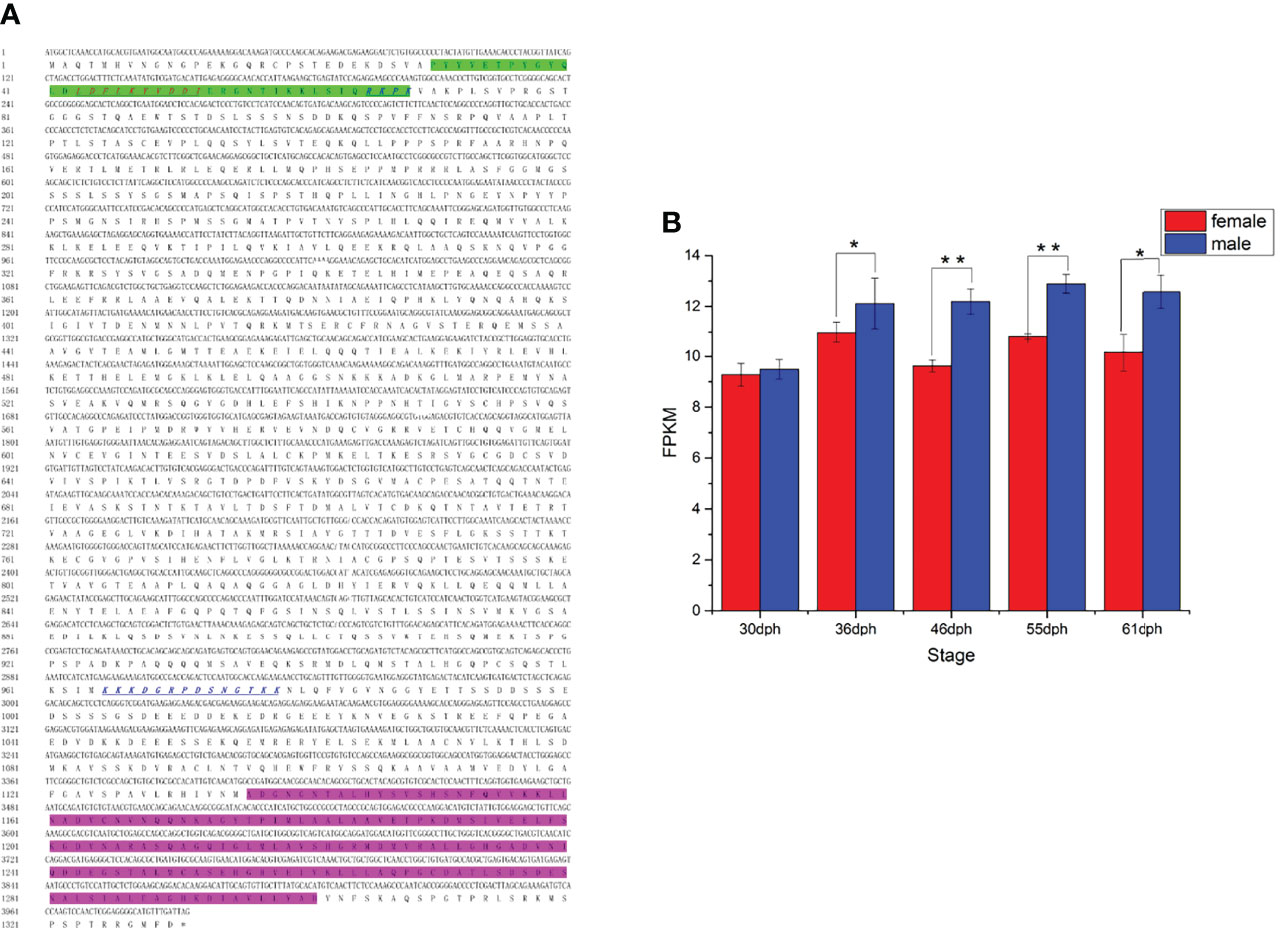
Figure 7 (A) The cDNA and deduced amino acid sequence of kank1 gene. The Kank1 KN N-terminal motif (KN) motif (green shadow) and ankyrin repeat domains from the C-terminal are marked. The sequences of NLS and NES motifs identified in Kank1 are underlined by blue italics and red italics, respectively. (B) The FPKM of kank1 gene in females and males at different stages (p < 0.01). *p < 0.05 and **p < 0.01.
Kank1 Enhanced Dmrt1 Gene Expression
A previous study showed Dmrt1 was different in the X and Y chromosome, respectively named as Dmrt1X and Dmrt1Y. Dmrt1, as a transcription factor, can bind to its own promoter and regulate the expression of itself (submitted, Sun et al.). We constructed the expression vector of Dmrt1 and Kank1, and co-transfected them into the CHO cells. The result suggests that Dmrt1 expressed in cell nucleus and Kank1 expressed in karyoplasm (Figure 8), indicating Kank1 was really a shuttling protein. With the dual-luciferase reporter assay, we detected the transcription level of dmrt1_Luc gene was higher when transfecting the pCS2-dmrt1-EGFP and pCS2-kank1-mCherry with pGL3-basic-dmrt1_promoter simultaneously than only transfecting the pCS2-dmrt1-EGFP with pGL3-basic-dmrt1_promoter in CHO cells and 293T cells (Figures 9A, B). This result demonstrates that Kank1 together with Dmrt1 enhanced the expression of dmrt1, possibly through its interaction with Dmrt1 protein to act on the promoter of dmrt1. Previous work has shown that Kank1 negatively regulated actin polymerization and cell motility (Kakinuma et al., 2009). We speculated that Kank1 might take part in the sex determination through inhibiting the primordial germ cells migration at the cell development and increased the expression of sex-determining gene - dmrt1- in yellow drum.
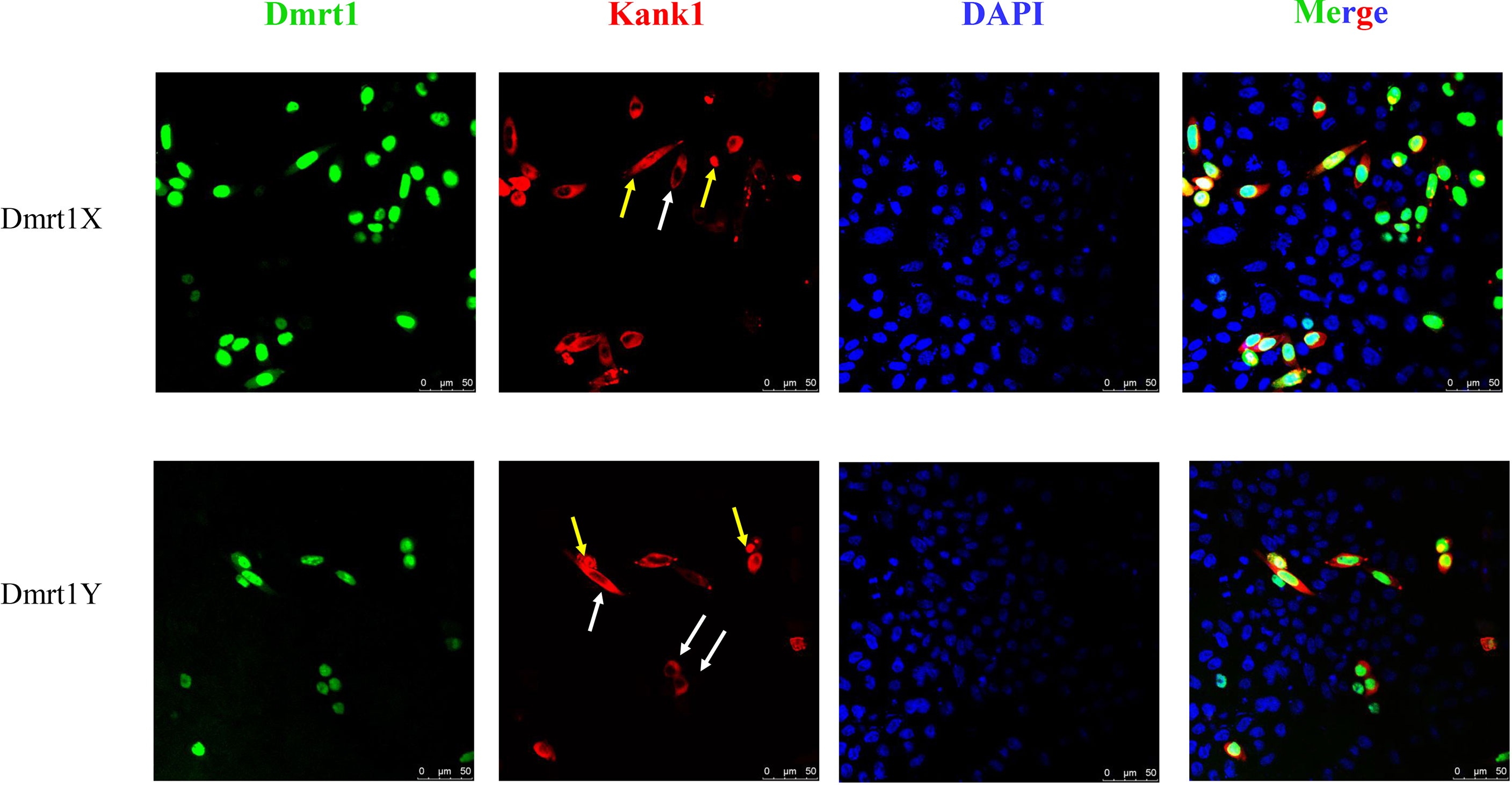
Figure 8 Co-transfection pCS2-dmrt1-EGFP and pCS2-kank1-mCherry into the CHO cells. Kank1 expressed in cytoplasm (white arrows) and in cell nucleus (yellow arrows). Scale bars: 50 μm.
Discussion
Dmrt1 Impacted the Germ Cells Migration and Development
Gonad is constituted by somatic cells and germ cells. And sex determination includes somatic sex determination and germ cell sex determination (Nagahama et al., 2021). It has long been believed that gonadal somatic cells instruct germ cells to adopt their sexual fates, and somatic signals decide germ cells whether they reside in a male or female body. The sex-determining genes that have been reported mostly express in somatic cells and the mutation of these genes result in sex reversal, such as Sry (Sinclair et al., 1990), Dmy (Matsuda et al., 2002), and Amhy (Hattori et al., 2012). Sex reversal of genetic females (XX) could be induced by the transplantation of XY somatic cells in the medaka, Oryzias latipes (Shinomiya et al., 2002). Nishimura et al. found that foxl3 was a germ cell-intrinsic factor involved in sperm-egg fate decision in medaka. XX medaka with disrupted foxl3 developed functional sperm in the expanded germinal epithelium of a histologically functional ovary (Nishimura et al., 2015). A recent report showed that the sexual fate of tilapia germline was determined by the antagonistic interaction of dmrt1 and foxl3 (Dai et al., 2021). The conventional idea is challenged by these recent studies. These results also indicate that the crosstalk between germ cells and gonadal somatic cells is critical for sex differentiation of the gonads. To address the question, the relevant research in this study was carried out.
In this study, we confirmed the 36 dph was a sex determination critical stage in yellow drum through transcriptome analysis at the early stages. Subsequently, we analyzed the differential genes and the signaling pathways they participated in at 36 dph. We discovered that the signaling pathways in females and males were different at sex determination critical stage. In females, the cell cycle and Wnt signaling pathway were mainly enriched and the most biological process was about germ cell development and gamete generation. This result suggests the germ cells in the differentiation process in females. Given germ cells confront two major cell fate decisions as they move from an immature state into sexuality, one decision is entering into a germline-specific cell cycle (i.e., meiosis), and the other is the commitment to differentiate as spermatozoa or eggs (Kimble and Page, 2007), and the cell cycle entering into meiosis is considered as a hallmark of feminization of germ cells (Kikuchi et al., 2020). We suspected that the germ cells sex determination would progress toward to the females when it was in cell cycle and reproduction. Dmrt1 has been cloned and was demonstrated as the most possible sex determination gene in yellow drum (Sun et al., submitted). In medaka, the dmrt1 gene resulted in negative regulation of germ cells proliferation and mediated a mitotic arrest of germ cells (Herpin et al., 2007). In yellow drum, the dmrt1 gene almost exclusively expressed in males, implying it was a barrier for proliferation and feminization of germ cells, and an intrigue for the gonad masculinization. In males, the main enrichment signaling pathway included TGF-beta signaling and Focal adhesion, the main biological process involved cell migration, cell motility, and tube morphogenesis. So, another possibility was the dmrt1 gene may negatively regulate the germ cells migrated to the primordial gonadial ridge and caused two types of gonadal tissues to be observed in the development stage of 40 days post-hatch (dph), which were designated as tissue type I and type II (Lou et al., 2016). Unfortunately, we failed to investigate the numbers of germ cells at early stage using histological experiments in yellow drum. Future research would pay more attention to this question.
How Kank1 Participated in the Sex Determination Process
We found the Kank1 protein contained a nuclear localization signal (NLS) and a nuclear export signal (NES), and expressed in karyoplasm in CHO cells. These results were consistent with the Kank protein in humans (Wang et al., 2006), and suggest that Kank1 of yellow drum was also a shuttling protein. Nuclei-cytoplasmic transportation of proteins plays an important role in the regulation of many cellular processes, including signal transduction, cell cycle progression and cell proliferation (Gama-Carvalho and Carmo-Fonseca, 2001). Furthermore, studies have demonstrated that Kank could bind to β-catenin and promote the nuclear localization of β-catenin, which results in the activation of β-catenin-mediated transcription (Wang et al., 2006). The β-catenin is an important component in the Wnt signaling pathway which is involved in various differentiation events during embryonic development (Lustig and Behrens, 2003). Evidence in mice and humans implied WNT4 and R-SPONDIN-1 (RSPO1) promoted the ovarian fate and blocked testis development, and activation of β-catenin was sufficient to block the testis pathway (Maatouk et al., 2008). In protandrous black porgy (Acanthopagrus schlegeli), wnt4 has important roles in late ovarian growth and natural sex change, such as oogonia proliferation, structure of ovarian lamellae, and vitellogenic oocytes (Wu and Chang, 2009). A previous study reported that rspo1 expressed more strongly in ovaries than in testes in zebrafish (Zhang et al., 2011). In Nile tilapia (Oreochromis niloticus), β-catenin1 and β-catenin2 played roles in ovarian differentiation and maintenance of female pathway (Wu et al., 2016). However, the roles of the RSPO1/Wnt/β-catenin signaling pathway in the sex determination in fish is still not clear. In teleost, the sequential hermaphroditism is not unusual (Colombo and Grandidr, 1996; Helfman et al., 1997), protogynous fishes develop into females first and become males later during fry stage, such as zebrafish (Maack and Segner, 2003), lager yellow croaker (You et al., 2012), and yellow drum (Lou et al., 2016). In the female-determination process of yellow drum, it is not clear whether Kank1 could activate the β-catenin-mediated transcription and exert the same function as in mice and humans, given that kank1 was not differentially expressed between genetic females and males before 36 dph.
The kank1 gene was expressed significantly higher in males than females at sex-determination critical stages, indicating kank1 was more favorable in male sex determination. With the protein network interaction analysis, we uncovered that Kank1 took part in the germ cells migration process and interacted with Dmrt1. Kank1 might act as a shuttling protein as in humans and play difunctional roles at the early gonadal development stages, i.e., not only participated in the germ cells motility, but also increased the expression of dmrt1 in yellow drum. Although the histological location of Kank1 has not been confirmed, we speculate it should at least express in germ cells based on the role of Kank1 in the cell migration in yellow drum. Kank1 enhanced the expression of dmrt1 in yellow drum, and dmrt1 is the master sex-determining gene and triggered the testis development. These results indicate kank1 might act as an upstream factor of the dmrt1 gene. Of note, the sex battle and antagonism between female and male are from beginning to end. So, the balance of these factors leading to either male or female development is important. Once the balance is destroyed, the sex reversal will happen.
Conclusion
We confirmed the 36 dph was a sex determination critical stage in yellow drum through transcriptome analysis at the early development stages. We further found that Kank1 participated in the cell motility process and interacted with Dmrt1 in yellow drum. With the dual-luciferase reporter assay, we detected that Kank1 increased the transcription of dmrt1_Luc gene in CHO cells and 293T cells. In addition, we observed Kank1 was not only expressed in the cytoplasm but also in the nucleus in CHO cells through fluorescence microscope. These results suggest that Kank1 acts as a shuttling protein as in humans and might play a difunctional role at the early gonad development stage - participating in the germ cells motility and increasing the expression of dmrt1 in yellow drum.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: NCBI (accession: PRJNA817837).
Ethics Statement
The animal study was reviewed and approved by the Animal Care and Use Committee of Jimei University.
Author Contributions
SS, WL and ZW designed the experiments and wrote the paper. SS, JL, SZ, QH, WH, KY, and FH performed the research. SS and WL analyzed the data. All authors read and approved the final version of the manuscript.
Funding
This work was supported by the National Key Research and Development Program of China (grant number 2018YFD0900202); the National Natural Science Foundation of China (grant numbers 31872562, U1705231, 32072969).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Bao L., Tian C., Liu S., Zhang Y., Elaswad A., Yuan Z., et al. (2019). The Y Chromosome Sequence of the Channel Catfish Suggests Novel Sex Determination Mechanisms in Teleost Fish. BMC Biol. 17, 6. doi: 10.1186/s12915-019-0627-7
Bowles J., Koopman P. (2010). Sex Determination in Mammalian Germ Cells: Extrinsic Versus Intrinsic Factors. Reprod. (Cambr. Engl.) 139, 943–958. doi: 10.1530/rep-10-0075
Capel B. (2017). Vertebrate Sex Determination: Evolutionary Plasticity of a Fundamental Switch. Nat. Rev. Genet. 18, 675–689. doi: 10.1038/nrg.2017.60
Charlesworth D., Charlesworth B., Marais G. (2005). Steps in the Evolution of Heteromorphic Sex Chromosomes. Heredity 95, 118–128. doi: 10.1038/sj.hdy.6800697
Colombo G., Grandidr G. (1996). Histological Study of the Development and Sex Differentiation of the Gonad in the European Eel. J. Fish Biol. 48, 493–512. doi: 10.1111/j.1095-8649.1996.tb01443.x
Dai S., Qi S., Wei X., Liu X., Li Y., Zhou X., et al. (2021). Germline Sexual Fate Is Determined by the Antagonistic Action of Dmrt1 and Foxl3/Foxl2 in Tilapia. Development 148, 1–14. doi: 10.1242/dev.199380
Ding M., Goncharov A., Jin Y., Chisholm A. D. (2003). C. Elegans Ankyrin Repeat Protein Vab-19 Is a Component of Epidermal Attachment Structures and Is Essential for Epidermal Morphogenesis. Development 130, 5791–5801. doi: 10.1242/dev.00791
Dobin A., Davis C. A., Schlesinger F., Drenkow J., Zaleski C., Jha S., et al. (2013). Star: Ultrafast Universal Rna-Seq Aligner. Bioinformatics 29, 15–21. doi: 10.1093/bioinformatics/bts635
Fan H., Tian H., Cheng X., Chen Y., Liang S., Zhang Z., et al. (2020). Aberrant Kank1 Expression Regulates Yap to Promote Apoptosis and Inhibit Proliferation in Oscc. J. Cell Physiol. 235, 1850–1865. doi: 10.1002/jcp.29102
Fujimoto T., Nishimura T., Goto-Kazeto R., Kawakami Y., Yamaha E., Arai K. (2010). Sexual Dimorphism of Gonadal Structure and Gene Expression in Germ Cell-Deficient Loach, a Teleost Fish. Proc. Natl. Acad. Sci. U. S. A. 107, 17211–17216. doi: 10.1073/pnas.1007032107
Gama-Carvalho M., Carmo-Fonseca M. (2001). The Rules and Roles of Nucleocytoplasmic Shuttling Proteins. FEBS Lett. 498, 157–163. doi: 10.1016/S0014-5793(01)02487-5
Golpour A., Pšenička M., Niksirat H. (2016a). Ultrastructural Localization of Intracellular Calcium During Spermatogenesis of Sterlet (Acipenser Ruthenus). Microsc. Microanal. 22, 1155–1161. doi: 10.1017/S1431927616011958
Golpour A., Pšenička M., Niksirat H. (2016b). Subcellular Localization of Calcium Deposits During Zebrafish (Danio Rerio) Oogenesis. Micron 80, 6–13. doi: 10.1016/j.micron.2015.09.004
Golpour A., Pšenička M., Niksirat H. (2017). Subcellular Distribution of Calcium During Spermatogenesis of Zebrafish, Danio Rerio. J. Morphol 278, 1149–1159. doi: 10.1002/jmor.20701
Goto R., Saito T., Takeda T., Fujimoto T., Takagi M., Arai K., et al. (2012). Germ Cells Are Not the Primary Factor for Sexual Fate Determination in Goldfish. Dev. Biol. 370, 98–109. doi: 10.1016/j.ydbio.2012.07.010
Hattori R. S., Murai Y., Oura M., Masuda S., Majhi S. K., Sakamoto T., et al. (2012). A Y-Linked Anti-Müllerian Hormone Duplication Takes Over a Critical Role in Sex Determination. Proc. Natl. Acad. Sci. U. S. A. 109, 2955–2959. doi: 10.1073/pnas.1018392109
Helfman G. S., Collette B. B., Facey D. E. (1997). The Diversity of Fishes (Boston, Mass.: Blackwell Science).
Herpin A., Schartl M. (2009). Molecular Mechanisms of Sex Determination and Evolution of the Y-Chromosome: Insights From the Medaka Fish (Oryzias Latipes). Mol. Cell Endocrinol. 306, 51–58. doi: 10.1016/j.mce.2009.02.004
Herpin A., Schindler D., Kraiss A., Hornung U., Winkler C., Schartl M., et al. (2007). Inhibition of Primordial Germ Cell Proliferation by the Medaka Male Determining Gene Dmrt1 by. BMC Dev. Biol. 7, 99. doi: 10.1186/1471-213X-7-99
Kakinuma N., Roy B. C., Zhu Y., Wang Y., Kiyama R. (2008). Kank Regulates Rhoa-Dependent Formation of Actin Stress Fibers and Cell Migration via 14-3-3 in Pi3k-Akt Signaling. J. Cell Biol. 181, 537–549. doi: 10.1083/jcb.200707022
Kakinuma N., Zhu Y., Wang Y., Roy B. C., Kiyama R. (2009). Kank Proteins: Structure, Functions and Diseases. Cell Mol. Life Sci. 66, 2651–2659. doi: 10.1007/s00018-009-0038-y
Kikuchi M., Nishimura T., Ishishita S., Matsuda Y., Tanaka M. (2020). Foxl3, a Sexual Switch in Germ Cells, Initiates Two Independent Molecular Pathways for Commitment to Oogenesis in Medaka. Proc. Natl. Acad. Sci. U. S. A. 117, 12174–12181. doi: 10.1073/pnas.1918556117
Kimble J., Page D. C. (2007). The Mysteries of Sexual Identity. The Germ Cell's Perspective. Science 316, 400–401. doi: 10.1126/science.1142109
Kocer A., Reichmann J., Best D., Adams I. R. (2009). Germ Cell Sex Determination in Mammals. Mol. Hum. Reprod. 15, 205–213. doi: 10.1093/molehr/gap008
Kurokawa H., Saito D., Nakamura S., Katoh-Fukui Y., Ohta K., Baba T., et al. (2007). Germ Cells Are Essential for Sexual Dimorphism in the Medaka Gonad. Proc. Natl. Acad. Sci. U. S. A. 104, 16958–16963. doi: 10.1073/pnas.0609932104
Lesch B. J., Page D. C. (2012). Genetics of Germ Cell Development. Nat. Rev. Genet. 13, 781–794. doi: 10.1038/nrg3294
Lewis Z. R., McClellan M. C., Postlethwait J. H., Cresko W. A., Kaplan R. H. (2008). Female-Specific Increase in Primordial Germ Cells Marks Sex Differentiation in Threespine Stickleback (Gasterosteus Aculeatus). J. Morphol. 269, 909–921. doi: 10.1002/jmor.10608
Li X., Gui J. (2018). Diverse and Variable Sex Determination Mechanisms in Vertebrates. Sci. China Life Sci. 61, 1503–1514. doi: 10.1007/s11427-018-9415-7
Lin Q., Mei J., Li Z., Zhang X., Zhou L., Zhou L., Gui J. F. (2017). Distinct and Cooperative Roles of Amh and Dmrt1 in Self-Renewal and Differentiation of Male Germ Cells in Zebrafish. Genetics 207, 1007–1022. doi: 10.1534/genetics.117.300274
Lou B., Xu D., Geng Z., Zhang Y., Zhan W., Mao G. (2016). Histological Characterization of Gonadal Sex Differentiation in Nibea Albiflora. Aquacult Res. 47, 632–639. doi: 10.1111/are.12523
Lustig B., Behrens J. (2003). The Wnt Signaling Pathway and Its Role in Tumor Development. J. Cancer Res. Clin. Oncol. 129, 199–221. doi: 10.1007/s00432-003-0431-0
Maack G., Segner H. (2003). Morphological Development of the Gonads in Zebrafish. J. Fish Biol. 62, 895–906. doi: 10.1046/j.1095-8649.2003.00074.x
Maatouk D. M., DiNapoli L., Alvers A., Parker K. L., Taketo M. M., Capel B., et al. (2008). Stabilization of Beta-Catenin in Xy Gonads Causes Male-to-Female Sex-Reversal. Hum. Mol. Genet. 17, 2949–2955. doi: 10.1093/hmg/ddn193
Matsuda M., Nagahama Y., Shinomiya A., Sato T., Matsuda C., Kobayashi T., et al. (2002). Dmy Is a Y-Specific Dm-Domain Gene Required for Male Development in the Medaka Fish. Nature 417, 559–563. doi: 10.1038/nature751
Myosho T., Otake H., Masuyama H., Matsuda M., Kuroki Y., Fujiyama A., et al. (2012). Tracing the Emergence of a Novel Sex-Determining Gene in Medaka, Oryzias Luzonensis. Genetics 191, 163–170. doi: 10.1534/genetics.111.137497
Nagahama Y., Chakraborty T., Paul-Prasanth B., Ohta K., Nakamura M. (2021). Sex Determination, Gonadal Sex Differentiation, and Plasticity in Vertebrate Species. Physiol. Rev. 101, 1237–1308. doi: 10.1152/physrev.00044.2019
Niksirat H., Siino V., Steinbach C., Levander F. (2021). High-Resolution Proteomic Profiling Shows Sexual Dimorphism in Zebrafish Heart-Associated Proteins. J. Proteome Res. 20, 4075–4088. doi: 10.1021/acs.jproteome.1c00387
Nishimura T., Sato T., Yamamoto Y., Watakabe I., Ohkawa Y., Suyama M., et al. (2015). Sex Determination. Foxl3 Is a Germ Cell-Intrinsic Factor Involved in Sperm-Egg Fate Decision in Medaka. Science 349, 328–331. doi: 10.1126/science.aaa2657
Pertea M., Pertea G. M., Antonescu C. M., Chang T. C., Mendell J. T., Salzberg S. L. (2015). Stringtie Enables Improved Reconstruction of a Transcriptome From RNA-Seq Reads. Nat. Biotechnol. 33, 290–295. doi: 10.1038/nbt.3122
Richardson B. E., Lehmann R. (2010). Mechanisms Guiding Primordial Germ Cell Migration: Strategies From Different Organisms. Nat. Rev. Mol. Cell Biol. 11, 37–49. doi: 10.1038/nrm2815
Ridley A. J., Schwartz M. A., Burridge K., Firtel R. A., Ginsberg M. H., Borisy G., et al. (2003). Cell Migration: Integrating Signals From Front to Back. Science 302, 1704–1709. doi: 10.1126/science.1092053
Roy B. C., Kakinuma N., Kiyama R. (2009). Kank Attenuates Actin Remodeling by Preventing Interaction Between Irsp53 and Rac1. J. Cell Biol. 184, 253–267. doi: 10.1083/jcb.200805147
Saito D., Morinaga C., Aoki Y., Nakamura S., Mitani H., Furutani-Seiki M., et al. (2007). Proliferation of Germ Cells During Gonadal Sex Differentiation in Medaka: Insights From Germ Cell-Depleted Mutant Zenzai. Dev. Biol. 310, 280–290. doi: 10.1016/j.ydbio.2007.07.039
Shinomiya A., Shibata N., Sakaizumi M., Hamaguchi S. (2002). Sex Reversal of Genetic Females (Xx) Induced by the Transplantation of Xy Somatic Cells in the Medaka, Oryzias Latipes. Int. J. Dev. Biol. 46, 711–717. doi: 10.1387/ijdb.12216983
Siegfried K. R., Nüsslein-Volhard C. (2008). Germ Line Control of Female Sex Determination in Zebrafish. Dev. Biol. 324, 277–287. doi: 10.1016/j.ydbio.2008.09.025
Sinclair A. H., Berta P., Palmer M. S., Hawkins J. R., Griffiths B. L., Smith M. J., et al. (1990). A Gene From the Human Sex-Determining Region Encodes a Protein With Homology to a Conserved DNA-Binding Motif. Nature 346, 240–244. doi: 10.1038/346240a0
Sun S., Li W., Xiao S., Lin A., Han Z., Cai M., et al. (2018). Genetic Sex Identification and the Potential Sex Determination System in the Yellow Drum (Nibea Albiflora). Aquaculture 492, 253–258. doi: 10.1016/j.aquaculture.2018.03.042
Takita T. (1974). Studies on the Early Life History of Nibea Albiflora (Richardson) in Ariake Sound. Bull. Faculty Fish Nagasaki Univ. 38, 1–55. doi: 10.1007/BF02674952
Wang Y., Kakinuma N., Zhu Y., Kiyama R. (2006). Nucleo-Cytoplasmic Shuttling of Human Kank Protein Accompanies Intracellular Translocation of Beta-Catenin. J. Cell Sci. 119, 4002–4010. doi: 10.1242/jcs.03169
Wu G., Chang C. (2009). Wnt4 Is Associated With the Development of Ovarian Tissue in the Protandrous Black Porgy, Acanthopagrus Schlegeli. Biol. Reprod. 81, 1073–1082. doi: 10.1095/biolreprod.109.077362
Wu L., Wu F., Xie L., Wang D., Zhou L. (2016). Synergistic Role of β-Catenin1 and 2 in Ovarian Differentiation and Maintenance of Female Pathway in Nile Tilapia. Mol. Cell. Endocrinol. 427, 33–44. doi: 10.1016/j.mce.2016.03.002
Yano A., Guyomard R., Nicol B., Jouanno E., Quillet E., Klopp C., et al. (2012). An Immune-Related Gene Evolved Into the Master Sex-Determining Gene in Rainbow Trout, Oncorhynchus Mykiss. Curr. Biol. 22, 1423–1428. doi: 10.1016/j.cub.2012.05.045
You X., Cai M. Y., Jiang Y., Wang Z. (2012). Histological Observation on Gonadal Sex Differentiation in Large Yellow Croaker (Larimichthys Crocea). J. Fish China 36, 1057. doi: 10.3724/SP.J.1231.2012.27858
Zhang Y., Li F., Sun D., Liu J., Liu N., Yu Q. (2011). Molecular Analysis Shows Differential Expression of R-Spondin1 in Zebrafish (Danio Rerio) Gonads. Mol. Biol. Rep. 38, 275–282. doi: 10.1007/s11033-010-0105-3
Keywords: RNAseq, kank1, dmrt1, sex determination, yellow drum
Citation: Sun S, Liu J, Zhang S, He Q, Han W, Ye K, Han F, Li W and Wang Z (2022) A Shuttling Protein Kank1 Plays Important Role in the Sex Determination in Yellow Drum. Front. Mar. Sci. 9:897004. doi: 10.3389/fmars.2022.897004
Received: 15 March 2022; Accepted: 11 April 2022;
Published: 04 May 2022.
Edited by:
Hadiseh Dadras, South Bohemia Research Center of Aquaculture and Biodiversity of Hydrocenoses, CzechiaReviewed by:
Jatindra Nath Mohanty, Siksha O Anusandhan University, IndiaFan Lin, Shantou University, China
Copyright © 2022 Sun, Liu, Zhang, He, Han, Ye, Han, Li and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wanbo Li, bGkud2FuYm9Aam11LmVkdS5jbg==; Zhiyong Wang, enl3YW5nQGptdS5lZHUuY24=
 Sha Sun
Sha Sun Jieping Liu1
Jieping Liu1 Wanbo Li
Wanbo Li Zhiyong Wang
Zhiyong Wang