
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mar. Sci., 31 May 2022
Sec. Aquatic Physiology
Volume 9 - 2022 | https://doi.org/10.3389/fmars.2022.891706
This article is part of the Research TopicGenetic Adaption and Metabolic Response of Aquatic Animals to Diverse Water Environment ParametersView all 13 articles
Sterol regulatory-element binding proteins (SREBPs), sirtuin (SIRT1), and liver X receptor α (LXRα) play important roles in regulating cholesterol metabolism in mammals. However, little is known about the relationship between cholesterol metabolism and SIRT1, LXRα, and SREBP-1 in fish. In addition, knowledge of the effects of salinity on hepatic cholesterol metabolism in euryhaline teleosts is fragmented. This study revealed that hepatic cholesterol content was significantly different between fresh water (FW)- and seawater (SW)-acclimated Indian medaka. Gene expression analysis indicated srebp-1, lxrα, and sirt1 transcripts were not affected by changes in ambient salinity. However, SREBP-1, but not LXRα and SIRT1 protein expression, was significantly induced in the liver of FW-acclimated medaka. When SREBP-1 Vivo-MO inhibited SREBP-1 translation, hepatic cholesterol content was predominantly downregulated in FW- and SW-acclimated medaka. This is the first study to show that SREBP-1 is involved in cholesterol biosynthesis in fish. Furthermore, SREBP-1 knockdown had different effects on the expression of hmgcr and fdps, which encode the key enzymes involved in cholesterol biosynthesis. This study further enhances our knowledge of cholesterol metabolism in the livers of euryhaline teleosts during salinity acclimation.
Cholesterol is an essential component of all animal cell membranes and functions as a precursor to fat-soluble vitamins and steroid hormones (Lee, 2020). Mammalian physiology is usually influenced by the modification of cholesterol metabolism (Goedeke and Fernández-Hernando, 2012). Both mammals and fish acquire cholesterol from the diet (exogenous cholesterol) and de novo synthesis (endogenous cholesterol) (Babin and Vernier, 1989). Cholesterol is largely a product of metabolism in animals. The liver synthesizes more cholesterol than any other organ (Engelking, 2015). It could be absorbed by the intestine or released from the liver into the bloodstream (Khan, 2005). When cholesterol is supplemented in diets, it can significantly improve the growth of Atlantic salmon, catfish, rainbow trout, Nile tilapia, and turbot (Farrell et al., 1986; Twibell and Wilson, 2004; Yun et al., 2011; Yun et al., 2012; Xu et al., 2018). Fish were found to grow well with exogenous cholesterol supplementation might because the energy used to synthesize cholesterol was saved for growth (Kemski et al., 2020).
The cholesterol contents in the tissues of aquatic animals varied with environmental factors including temperature (Hassett and Crockett, 2009), pollutants (Mohamed et al., 2019), and toxicity (Binukumari and Vasanthi, 2014). Environmental salinities were also reported to affect cholesterol levels in aquatic animals. Serum cholesterol levels were significantly higher in the 0 and 10 ‰ groups than the 20‰ group of pufferfish (Takifugu fasciatus, Wen et al., 2021). Cholesterol levels were also higher in hepatopancreases of the mud crab (Scylla paramamosainduring) of the 4 and 12 ‰ groups than the 25 ‰ group after overwintering. The adaptive significance of these changes in cholesterol contents may contribute to the building blocks necessary for modifying the membrane properties in response to an environmental challenge (Hazel and Williams, 1990). Cholesterol in excess of dietary requirements could facilitate lipid mobilization and storage in the hepatopancreas of the shrimp (Litopenaeus vannamei) to play important roles in increasing their adaptability to environments by improving osmoregulatory capacity, leading to better survival and growth under low salinity conditions (Roya et al., 2006). In addition, 0.4% dietary cholesterol was found to increase branchial Na+, K+-ATPase activity, and serum cortisol content of juvenile Nile tilapia to improve their ability for hyperosmotic adaptation (Xu et al., 2018).
In addition to dietary uptake, de novo synthesis is the other source of cholesterol. Cholesterol biosynthesis is tightly regulated by sterol regulatory element-binding proteins (SREBPs, Amemiya-Kudo et al., 2002; Horton et al., 2002). Mammalian cells produce three SREBP isoforms, SREBP-1a, SREBP-1c, and SREBP-2. Among them, SREBP-1c is the dominant isoform of SREBP-1 in the livers of adult mammals and is involved in the activation of genes related to the synthesis of fatty acids, but not cholesterol. No alternatively spliced isoforms of the srebp-1 gene (e.g., srebp-1a and 1c in mammals) were found in fish (Minghetti et al., 2011; Thomas et al., 2013; Dong et al., 2015). According to a comparison of the deduced amino acid sequences, fish SREBP-1 is more similar to human SREBP-1a than to human SREBP-1c (Dong et al., 2015). On the other hand, SREBP-2 was found to specifically transactivate cholesterol synthesis genes (Amemiya-Kudo et al., 2002; Horton et al., 2002). In mammals, SREBP-2 plays a vital role in the mevalonate pathway of cholesterol synthesis, involving the action of more than 20 enzymes (Horton et al., 2002; Xue et al., 2020). 3-hydroxy-3-methylglutaryl CoA reductase (HMGCR) and farnesyl diphosphate synthase (FDPS, also known as farnesyl pyrophosphate synthase; FPPS) among these enzymes are involved in highly synchronized sterol and cholesterol synthesis (Horton et al., 2002; Xue et al., 2020). HMGCR is a rate-limiting enzyme in cholesterol biosynthesis (Smith et al., 1988) and is induced by sterol depletion and repressed in response to sterol accumulation (Goldstein and Brown, 1990; Wang et al., 1994). In contrast, FDPS catalyzes the formation of farnesyl diphosphate, a key intermediate in the synthesis of cholesterol and isoprenylated cellular metabolites (Ishimoto et al., 2010). Feeding diets containing tuna fish oil for two weeks decreased the serum cholesterol concentration by 50% in mice, and the hepatic mRNA levels of fdps and hmgcr by 70% and 40%, respectively, in rats (Corcos et al., 2005).
In fish, Zhu et al. (2018) reported that the plasma cholesterol levels of rainbow trout decreased after being fed a plant-based diet. Meanwhile, the gene expression of srebp-2 and hmgcr was significantly increased in livers of the rainbow trout. In addition, when cholesterol was supplemented in the plant-based diet to feed Atlantic salmon, the gene expression of srebp-2 and fdps was suppressed, hmgcr was not altered, and srebp-1 was induced in the liver (Kortner et al., 2014). Thus, srebp-2 in trout and salmon was suggested to have similar roles in cholesterol metabolism in mammals. Cholesterol biosynthesis in fish may also be regulated by ambient salinity. The gene expression of srebp-2, hmgcr, and fdps was stimulated in livers of the tongue sole (Cynoglossus semilaevis) after transfer from 30 ‰ to 15 ‰ SW for 60 days (Si et al., 2018). In livers of the rabbitfish fed with the vegetable oil diet reared at 10 ‰ salt-water, srebp-1 displayed higher expression levels and increased biosynthesis ability of long-chain polyunsaturated fatty acid, respectively, rather than those in the 32 ‰ seawater group (Zhang et al., 2016). Differential responses of SREBP-1 in fish could assist in adapting to different ambient salinity (Dong et al., 2017). Nevertheless, it remains unclear whether SREBP-1 is involved in cholesterol metabolism in fish.
When oxysterols increase because of cholesterol overload, the liver X receptors (LXRs) which are the oxysterol receptors could act as the cholesterol sensor to protect cells (Zhao and Dahlman-Wright, 2010). Two isoforms of LXR (Bertrand et al., 2004; Cruz-Garcia et al., 2009), LXRα (NR1H3) and LXRβ (NR1H2) were identified in this nuclear receptor superfamily (Peet et al., 1998). LXRα is abundantly expressed in the liver, intestine, adipose tissue, kidneys, and immune macrophages, whereas LXRβ is ubiquitously expressed in mice (Schultz et al., 2000). LXRs can promote hepatic lipogenesis by activating the transcriptional program of fatty acid synthesis, especially by increasing the transcription of srebp-1c, the key lipogenic activator, in mice (Repa et al., 2000). LXRα is vital for cholesterol metabolism through the increased expression of genes involved in bile acid synthesis and cholesterol excretion in mouse livers (Schultz et al., 2000). LXRs in fish is also involved in cholesterol metabolism (Kortner et al., 2014; Zhu et al., 2018). After feeding with a plant-based diet, hepatic lxrα expression as well as plasma cholesterol levels of rainbow trout decreased (Zhu et al., 2018). The expression of lxr was also induced when cholesterol was supplemented into the plant-based diet to feed Atlantic salmon (Kortner et al., 2014). On the other hand, transcriptome analysis revealed salinity effects on expression of genes related to lipid metabolism including lxrα and srebp-1 in livers of the turbot (Scophthalmus maximus) and showed a significant downward trend under low salinity stress (Liu et al., 2021).
In addition to SREBP-1 and LXRα, sirtuin 1 (SIRT1) also plays a beneficial role in cholesterol metabolism by deacetylating both LXR and SREBP-1/2 (Walker et al., 2010). Sirtuins (SIRTs) are members of a protein family of NAD-dependent deacetylases (Grozinger et al., 2001; Albani et al., 2010; Vassilopoulos et al., 2011). After deacetylation, SIRT1 destabilizes SREBP-1/2 (Ponugoti et al., 2010; Walker et al., 2010). Therefore, SIRT1 can modulate hepatic cholesterol metabolism by mediating LXR (Kemper et al., 2013) and SREBP-1/2 activities. In SIRT1 knockout mice, decreased expression of LXR downstream target genes involved in cholesterol metabolism and hepatic cholesterol accumulation was reported (Feige and Auwerx, 2007; Li et al., 2017). These results are consistent with the abnormal cholesterol accumulation observed in the liver-specific SIRT1 knockout mice (Li et al., 2007).
Upon salinity challenge, the euryhaline fish are able to survive well through efficient osmoregulatory mechanisms mainly exhibited on the membrane of epithelial cells in osmoregulatory tissues (Hwang and Lee, 2007). Previous studies have indicated that in some fish cholesterol contents which were pivotal for membrane structures and functions changed with environmental salinities (Xu et al., 2018; Wen et al., 2021). On the other hand, SREBP-1/2, LXRα, and SIRT1 were reported to be crucial for regulating cholesterol metabolism in mammals (Schultz et al., 2000; Horton et al., 2002; Li et al., 2007). Although in mammalian livers SIRT1 was known to control SREBP-1 gene expression through the mechanism involving the transcription factor LXR as described above, the mechanisms in cholesterol biosynthesis and accumulation in livers of euryhaline fish in response to ambient salinity changes were not clear.
The euryhaline Indian medaka (Oryzias dancena, the synonym of Oryzias melastigma, Yusof et al., 2011) is an ideal model species for osmoregulatory studies (Yang et al., 2013), of which the physiological, biochemical, and molecular responses after exposure to contaminants and other environmental stressors were commonly studied due to their characteristics, i.e., small in size, short in generation time, easy to handle, and strong intolerance to environmental stress (Dong et al., 2014). Therefore, this study used this model species to investigate whether changes in ambient salinity affected cholesterol accumulation in livers of the Indian medaka through the protein and gene expression of SREBP-1, LXRα, and SIRT1. SREBP-1 knockdown was further performed by Vivo-Morpholino to reveal its role in cholesterol biosynthesis in livers of this euryhaline fish. This study will enhance our understanding of the regulatory mechanisms of cholesterol metabolism of the euryhaline teleost during salinity acclimation.
Indian medaka with a total length of 3.5 ± 0.5 cm and weight of 8.0 ± 1.0 g) were purchased from an aquarium (Taichung, Taiwan) and kept in laboratory conditions in brackish water (15 ‰) prepared by adding appropriate amounts of Blue Treasure Sea Salts (New South Wales, Australia) to aerated tap water for at least two weeks before the experiments. The medaka were then transferred into fresh water (FW; 0 ‰) and seawater (SW; 35 ‰) tanks with re-circulatory filter units for at least four weeks and 50% of the water was changed every two days. The medaka were fed to satiation with a commercial diet (A045F; Hai Feng, Nantou, Taiwan) containing 45% crude protein, 6% crude lipid, 16% ash, and 8% water. Throughout the experimental period, a 12:12 h light: dark regime (08:00 to 20:00 h light period) was maintained using timed lighting. The water temperature of the control group was maintained at 28 ± 1°C (Juo et al., 2016), and 36 to 60 fish were kept in one tank. The experimental protocol was reviewed and approved by the Institutional Animal Care and Use Committee (IACUC) of the National Chung Hsing University, Taichung, Taiwan (IACUC Approval No. 106-054).
Medaka acclimated to SW or FW were placed on ice under anesthesia, and the livers were immediately dissected. One sample (50–100 mg) from each group contained six livers collected from six individuals and was placed in an Eppendorf tube containing 200 μL of TriPure Isolation Reagent (Roche, Mannheim, Germany). The samples were frozen in liquid nitrogen and stored at -80 °C. The RNA extraction method used in this study was modified from that described by Hu et al. (2017). The RNA pellet was dissolved in sterilized diethyl pyrocarbonate (DEPC) water (20 μL). Extracted RNA integrity was verified on a 1% agarose gel (SeaKem® LE Agarose; Lonza, Basel, Switzerland) electrophoresed using SafeView™ Classic (ABM, San Jose, CA, USA). The concentration and purity of the total RNA were measured using a NanoDrop 2000 (Thermo, Wilmington, DE, USA). The purity of the RNA was evaluated by the ratio of absorbance A260/A280, with the accepted values between the ranges of 1.9–2.1.
To prepare genomic DNA-free RNA samples, purified RNA samples were further treated (10,000-25,000 units/mg) with DNase I (Thermo Fisher Scientific, Waltham, MA, USA). First-strand cDNA was synthesized using the iScript™ cDNA Synthesis Kit (Bio-Rad, Hercules, CA, USA) and 1 μg of genomic DNA-free total RNA, following the manufacturer’s instructions. After the reverse transcription reaction, real-time PCR (qPCR) was used to determine the mRNA levels using FastStart™ Universal SYBR Green Master (04913850001; Roche).
Primers used for qPCR analyses were designed according to the sequences of O. melastigma (also named O. dancena) in the National Center for Biotechnology Information (NCBI) database (www.ncbi.nlm.nih.gov). Primer and thermal cycling conditions (Applied Biosystems® Veriti® 96-Well Thermal Cycler; Thermo Fisher Scientific) were analyzed using gradient temperatures ranging from 54–62°C. Thermal cycling is performed by mixing with 1.6 μL dNTP mixture (Takara, Shiga, Japan), 2 μL 10x Ex Taq reaction buffer (Takara), 0.1 μL Ex Taq polymerase (Takara), 1 μL of cDNA (20x dilution), 0.5 μL of each forward and reverse primer (250–400 nM), and adding nucleotide, DNase, and RNase-free water (Protech, Taipei, Taiwan) to the total volume of 20 μL. The PCR product was verified by 1–2% agarose gel electrophoresis (according to the length of the target sequence). The confirmed PCR product was sent to Tri-I Biotech Company (Taipei, Taiwan) for sequence analysis. Sequencing results were confirmed against O. melastigma using the online database available in NCBI (https://www.ncbi.nlm.nih.gov).
The mRNA expression levels of sirt1, lxrα, and srebp1 were measured using a Rotor-Gene Q Real-Time PCR System (QIAGEN, Hilden, Germany) in the reaction mixture for real-time PCR containing 1 μL cDNA (20x dilution), primers (0.5–1 μL) in the concentration range from 250–400 nM (according to the primer efficiency), and 10 μL KAPA SYBR FAST qPCR Master Mix (KAPA Biosystems, Cape Town, South Africa) to a total volume of 20 μL using nucleotide, DNase, and RNase-free water (Protech). Non-specific products were confirmed by melting curve analysis and gel electrophoresis of the primers. The primer efficiency was maintained within 95–105% to verify the primer specificity (Table 1). The corresponding values of the target genes were calculated using the relative Ct method (Livak and Schmittgen, 2001) using the following formula: Relative expression= 2^- [(Ct target gene, n – Ct β-actin, n) – (Ct target gene, c – Ct β-actin,c)]. Ct corresponded to the threshold cycle number, “n” indicated each sample, and “c” indicated the control mixed with cDNA samples of all experiments (Livak and Schmittgen, 2001). Actin was used as the normalization gene.
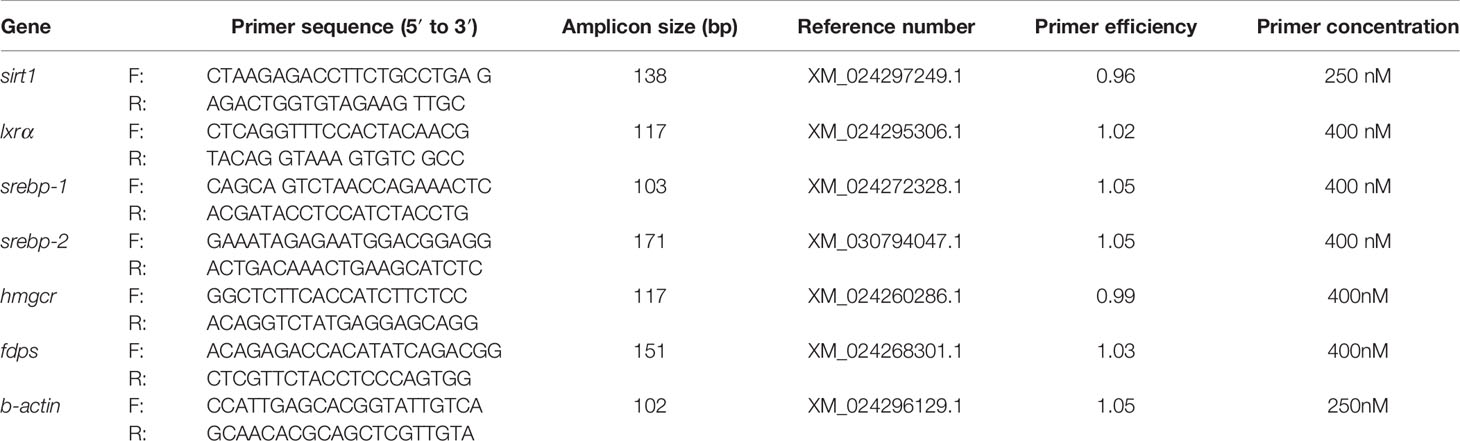
Table 1 Primer sequences used for the cDNA cloning (PCR) and expression detection (qPCR) of medaka livers.
Livers were sampled from medaka as described in the previous paragraph. After sampling, 100 mg of liver samples were ground using disposable polypropylene pellet pestles in microtubes containing 1 ml of 1x phosphate-buffered saline (PBS) using a sonicator. The ground tissues were centrifuged at 1,300 × g at 4°C for 10 min, and the supernatants were discarded. After centrifugation, the pellets were used to extract nuclear and cytoplasmic proteins using the Nuclear/Cytosol Fractionation Kit (K266-25, BioVision, Milpitas, CA, USA) according to the manufacturer’s instructions. Extracted cytoplasmic and nuclear protein samples were stored at -80°C. Protein concentration was determined using the Bradford assay (B6916, Sigma-Aldrich, St. Louis, MO, USA) and bovine serum albumin (Pierce, Hercules, CA, USA) as the standard.
Immunogens of the primary antibody were aligned with the medaka sequence to the most suitable primary antibody using multiple sequence alignments (Corpet, 1988). The primary antibodies used in this study included (1) SIRT1, a rabbit polyclonal antibody (13161-1-AP; Proteintech, Rosemont, IL, USA) raised against the residues AG3808 of human SIRT1; (2) LXRα, a mouse monoclonal antibody (sc-377260, Santa Cruz, Dallas, TX, USA) specific for epitope mapping between amino acid 433-461 of LXRα of human origin and (3) SREBP-1, a mouse monoclonal antibody (sc-13551, Santa Cruz, Dallas, Texas, USA) raised against amino acids 301–407-SREBP-1 of human origin. Primary antibodies were diluted 1:500 with antibody dilution buffer prior to immunoblotting. Secondary antibodies used in this study included the (1) goat anti-mouse IgG antibody (HRP) (GTX 213111-01, GeneTex, Irvine, CA, USA) for detecting the LXRα and SREBP-1a antibodies, and (2) goat anti-rabbit IgG antibody (HRP) (GTX 213110-01, GeneTex) for detecting the SIRT1 antibody. To test the specificity of the primary antibodies, negative controls were included using the antibody dilution buffer to replace the primary antibodies of SIRT1 (Supplementary Figure 1), LXR (Supplementary Figure 2), and SREBP-1 (Supplementary Figure 3). The antibody dilution buffer contained 1% bovine serum albumin and 0.01% sodium azide in 50 mL PBST.
Nuclear protein fractions with 6x sample loading buffer (12% SDS, 0.06% bromophenol blue, 30% glycerol, 0.6 M dithiothreitol, and 62.5 mM Tris at pH 6.8) added were incubated at 65°C for 15 min to denature the proteins. The protein ladder (PM2500, 3-color Regular Range Protein Marker, SMOBIO, Hsinchu, Taiwan; #26616, PageRuler™ Pre-stained Protein Ladder, Thermo Fisher Scientific) loaded for immunoblotting of SIRT1, LXR, SREBP-1 were 24, 50, and 50 μg of protein per lane, respectively, and PARP1 (sc8007, Santa Cruz Biotechnology, Dallas, TX, USA) was used as a positive control. Proteins were separated on 8–10% SDS-PAGE gels using a Mini-PROTEAN® II Electrophoresis Cell (Bio-Rad). The separated proteins were transferred to a PVDF membrane (Millipore, Bedford, MA, USA) by electrophoresis on ice. After blocking with 5% (w/v) skim milk for 1 h to minimize the non-specific binding, the blots were incubated overnight at 4°C with the primary antibody. The membranes were washed three times with PBST and incubated with the secondary antibody for 1 h at room temperature. Images were developed with Immobilon™ Western Chemiluminescent HRP Substrate (Millipore) using a cooling-charge-coupled device camera (ChemiDoc XRS+, Bio-Rad) and associated software (Quantity One v 4.6.8, Bio-Rad). The bands were converted to numerical values using ImageLab 3.0 (Bio-Rad) to quantify and compare the relative protein abundance of the immunoreactive bands.
After sampling, medaka livers (16 ± 1 mg) were placed in a microtube containing 200 μL of a mixture of chloroform (Merck, 14-650-505, Phillipsburg, NJ, USA), isopropanol (#SHBK4071, Sigma-Aldrich), and NP-40 (IGEPAL® CA-630, Sigma-Aldrich) in a ratio of 7:11:0.1, followed by grinding on ice. Samples were centrifuged for 10 min at 15,000 × g at 4°C. Subsequently, the supernatants were transferred to new and air-dried at 50°C to remove chloroform. The cholesterol content in the liver was assayed as described below using the Total Cholesterol Assay Kit (STA384-192 assays, Cell Biolabs, San Diego, CA, USA) according to the manufacturer’s protocol. Cholesterol levels were measured at 570 nm using the colorimetric method with the cholesterol standard. Cholesterol standards were prepared before assays by diluting 10 mM stock solution in 1x Assay Diluent at a ratio of 1:40 to make a 250 μM working solution. The cholesterol standards were prepared promptly in serial dilutions from 0–250 μM, including 10 points, according to the manufacturer’s protocol. The formula for calculating cholesterol content are:
The translational blocking of SREBP-1 Vivo-Morpholino (5’-GATAGGCGATTCATC TCTACGGTTG-3’) was designed by Gene-Tools LLC (Philomath, OR, USA), reverse complementary to the SREBP-1 full-length sequence of the O. melastigma database (XM_024272328.1, https://www.ncbi.nlm.nih.gov/), and confirmed by the Vivo-MO sequence by BLAST search using the NCBI database. A standard Vivo-Morpholino (MO) control (5′-CCTCTTACC TCAGTTACAATTTATA-3′) was purchased from Gene-Tools.
FW- and SW-acclimated adult medaka were anesthetized with 4% MS222 made with aerated water and then injected with 0.5 mM SREBP-1 or control in 5 μL Vivo-MOs (either the original solution supplied by the vendor, 0.5 or 0.05 mM diluted with phosphate-buffered saline, pH 7.4 (PBS) using a BD insulin syringe and Ultra-Fine II needle (0.22 mm, 31G x 8 mm, Becton drive, Franklin lakes, NY, USA). Vivo-MO or control was gently injected into the region closer to the anal pore in the abdomen of the medaka. After injection, the fish were returned to their acclimated environments (i.e., FW or SW) and maintained for 24 h and 72 h. The effects of Vivo-MO were evaluated in a preliminary study using immunoblotting as previously described. The livers of the Vivo-MO and control groups were then sampled for cholesterol analysis.
Statistical analyses were done in the statistical program Minitab (Minitab® 17 Minitab Inc., PA, USA). Normality and homogeneity of variance between the groups that were statistically compared were tested (the significance level was set at P < 0.05). For the mRNA and protein abundance and cholesterol level results, data between the FW- and SW-acclimated medaka of each temperature group were compared using Student’s t-test (P < 0.05). The data of different temperature groups in FW or SW were compared using one-way analysis of variance (ANOVA) with Tukey’s test (P < 0.05). Values are expressed as mean ± standard error of the mean (SEM).
To explore the effects of ambient salinity on cholesterol accumulation in the livers of Indian medaka, fish were exposed to fresh water (FW) or seawater (SW) for two weeks. Subsequently, the livers were sampled to measure cholesterol content. Figure 1 shows that the hepatic cholesterol content in the FW group was significantly higher than that in the SW group.
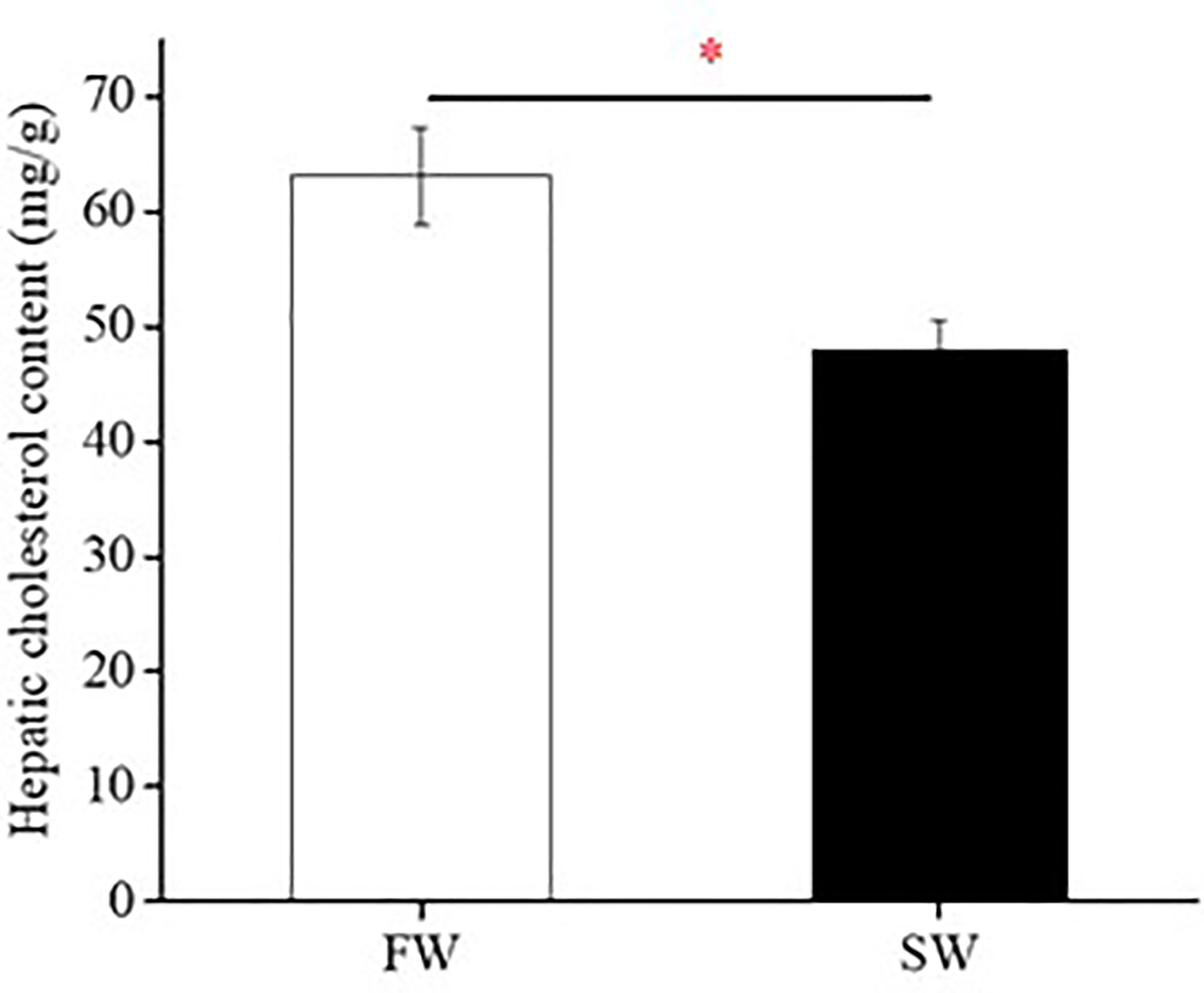
Figure 1 Hepatic cholesterol content of fresh water (FW)- and seawater (SW)-acclimated Indian medaka. Values are mean ± S.E.M. (N = 6). Student’s t-test, *p < 0.05.
Real-time PCR analyses revealed the mRNA expression of hmgcr, fdps, and srebp-1/2 in the livers of FW- and SW-acclimated medaka. Hepatic hmgcr, fdps, and srebp-1/2 transcripts were not significantly different between FW- and SW-acclimated medaka (Figure 2A). In contrast, the relative protein abundance of SREBP-1 in the liver was significantly higher in FW- than in SW-acclimated medaka (Figure 2B). Cholesterol content was approximately 7-fold higher in FW-acclimated medaka than in SW-acclimated medaka (Figure 2B).
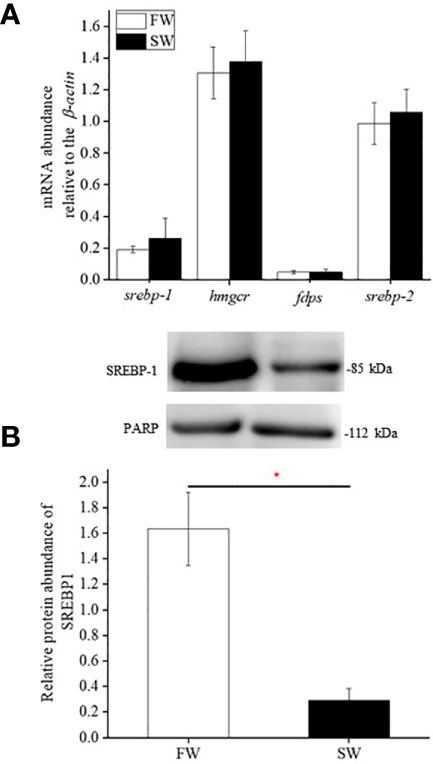
Figure 2 Relative gene (A) and protein (B) expression of SREBP-1, HMGCR, FDPS, and SREBP-2 in livers of fresh water (FW)- and seawater (SW)-acclimated Indian medaka. mRNA expression was analyzed by real-time PCR. The immunoblots were used to detect SREBP-1 protein expression of in the nuclear fractions of livers. Values are mean ± S.E.M. (N = 6). Student’s t-test, *p < 0.05.
Real-time PCR analyses revealed that the mRNA expression of lxrα and sirt1 in the liver was similar in FW- and SW-acclimated medaka (Figures 3A, 4A). Immunoblot analysis also revealed that the relative protein expression of LXRα and SIRT1 in the livers of FW- and SW-acclimated medaka was not significantly different (Figures 3B, 4B).
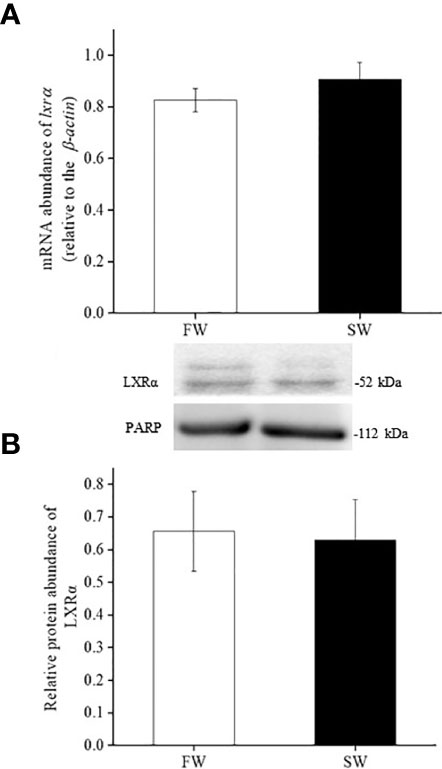
Figure 3 Relative gene (A) and protein (B) expression of LXRα in livers of fresh water (FW) and seawater (SW)- acclimated Indian medaka. mRNA expression was analyzed by real-time PCR. The immunoblots were used to detect the protein expression. Values are mean ± S.E.M. (N = 6).
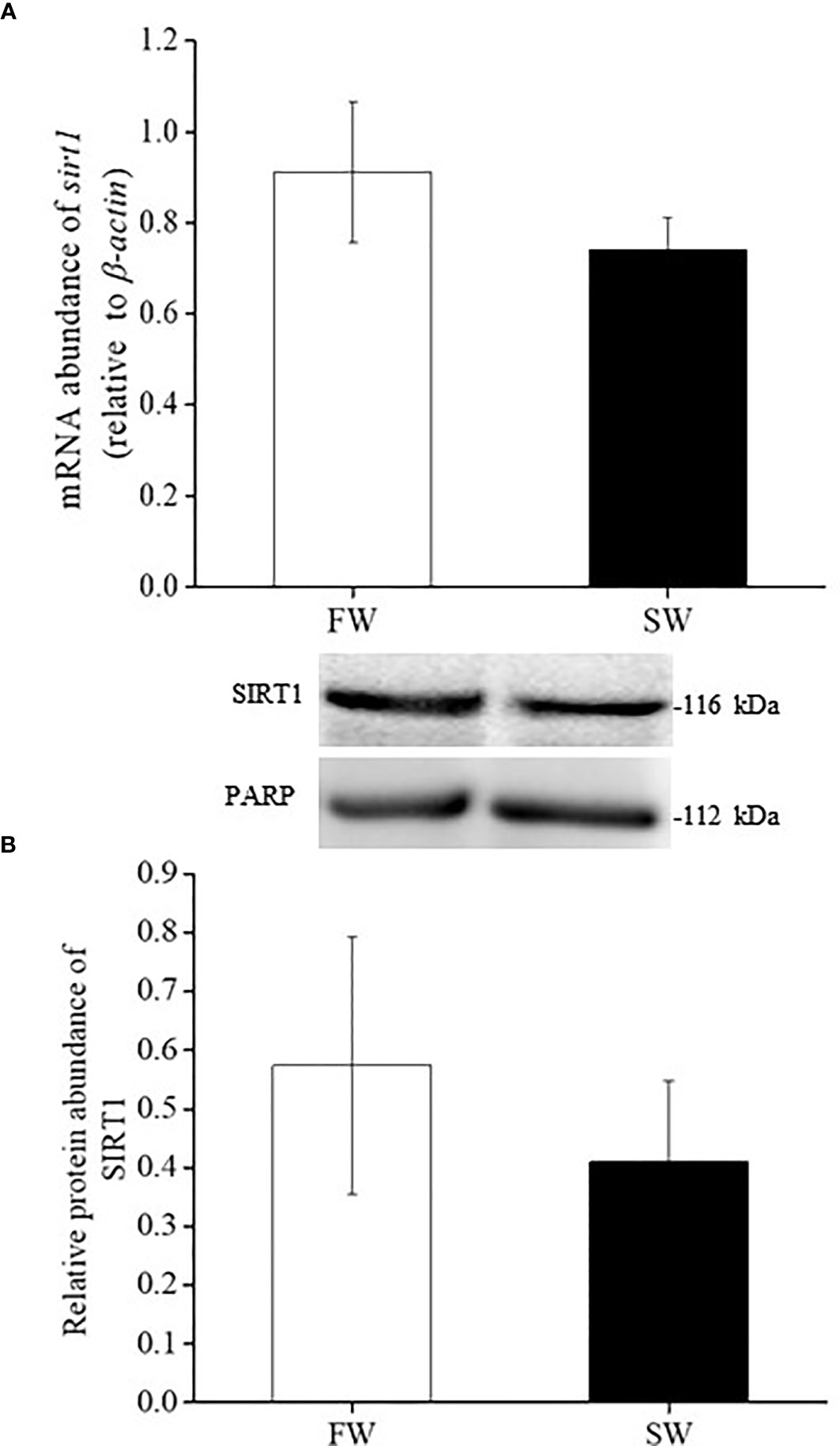
Figure 4 Relative gene (A) and protein (B) expression of SIRT1 in livers of fresh water (FW) and seawater (SW)- acclimated Indian medaka. mRNA expression was analyzed by real-time PCR. The immunoblots were used to detect the protein expression. Values are mean ± S.E.M. (N = 6).
Vivo-morpholino (Vivo-MO) of SREBP-1 was used to inhibit SREBP-1 translation in Indian medaka. Figure 5A shows that SREBP-1 Vivo-MO could inhibit SREBP-1 protein expression predominantly in the livers of Indian medaka compared to the control Vivo-MO. In contrast, SREBP-1 knockdown in FW- or SW-acclimated Indian medaka significantly inhibited hepatic cholesterol content (Figure 5B).
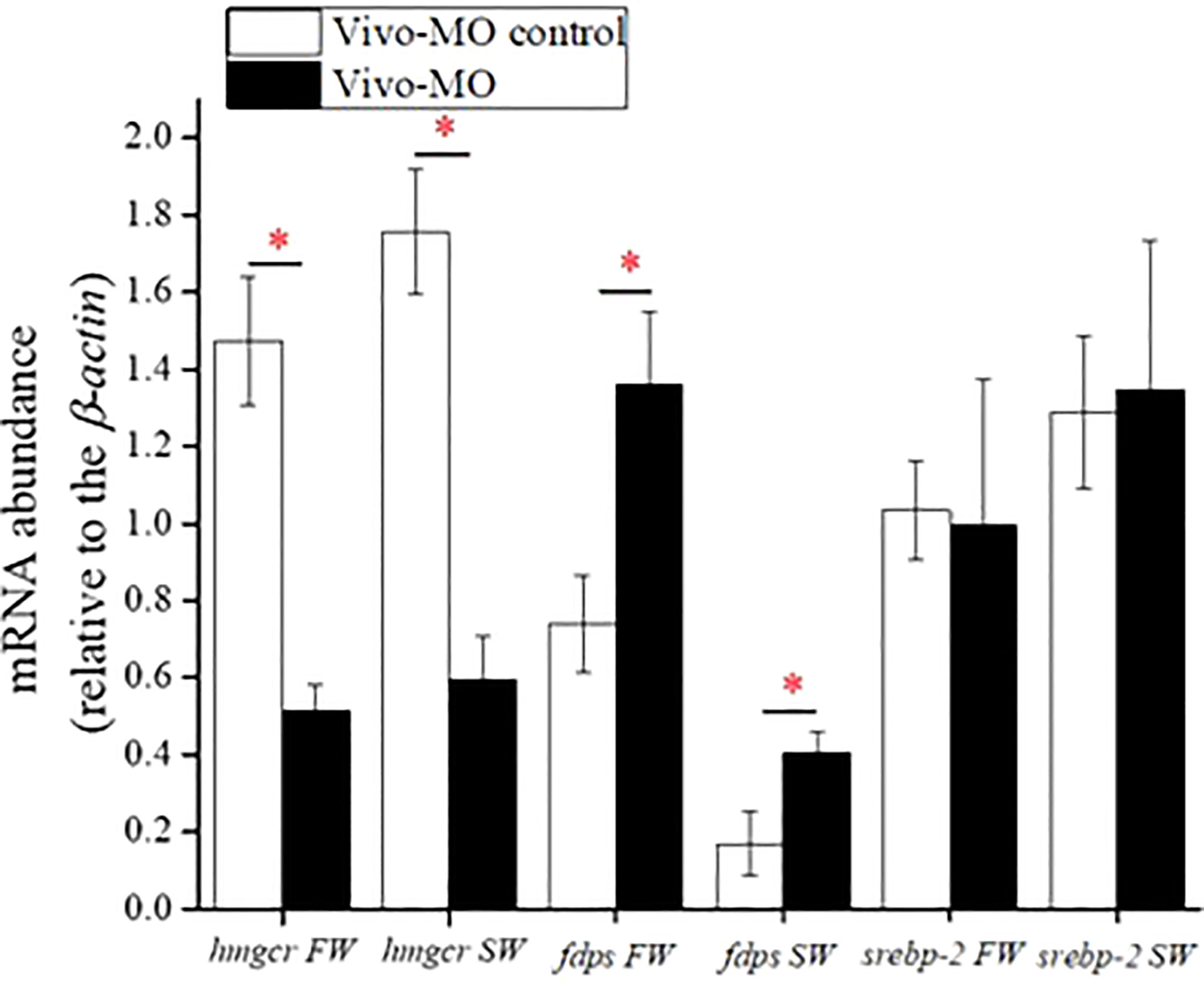
Figure 5 The effect of SREBP-1 knockdown on hepatic cholesterol content in fresh water (FW)- and seawater (SW)-acclimated Indian medaka. (A) The specificity test of SREBP-1 Vivo-MO. (B) The measurement of hepatic cholesterol content in Indian medaka with/without SREBP-1 knockdown. Values are mean ± S.E.M. (N = 6). Student’s t-test, *p < 0.05.
In both FW- and SW-acclimated Indian medaka, SREBP-1 knockdown significantly decreased hmgcr, but increased fdps expression (Figure 6). However, hepatic srebp-2 expression did not change in either FW- or SW-acclimated Indian medaka with SREBP-1 knockdown (Figure 6).
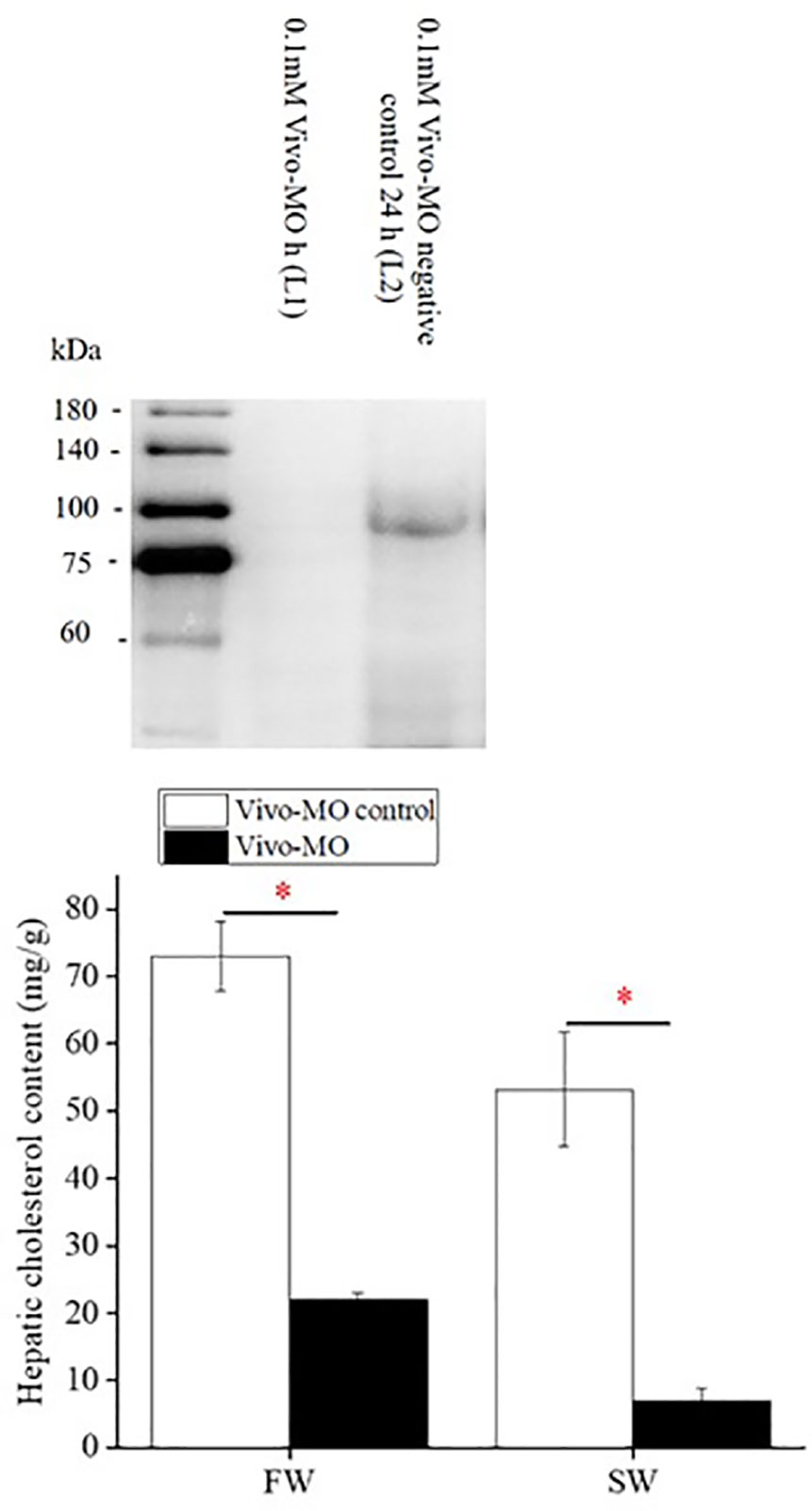
Figure 6 The effect of SREBP-1 knockdown on mRNA expression of targeted genes in livers of fresh water (FW)- and seawater (SW)-acclimated Indian medaka. mRNA expression was analyzed by real-time PCR. Values are mean ± S.E.M. (N = 6). Student’s t-test, *p < 0.05.
Steroid metabolism-related pathways, such as steroid and steroid hormone biosynthesis, are important for salinity adaptation in aquatic animals (Charmandari et al., 2005; Aruna et al., 2015). The expression of genes involved in lipid or steroid metabolism-related pathways in fish livers changes in response to ambient salinity (Si et al., 2018; Liu et al., 2021). Cholesterol is the sole precursor of steroids (Azhar et al., 2003). In this study, the cholesterol content of medaka liver, the central organ for cholesterol biosynthesis in vertebrates, was significantly higher in FW- than in SW-acclimated medaka. Cholesterol biosynthesis in the livers of Indian medaka may also be related to changes in ambient salinity, resulting in changes in hepatic cholesterol content. Kang et al. (2008) reported that plasma osmolality was significantly lower in Indian medaka under freshwater (FW) acclimation than in seawater (SW) acclimation. In contrast, the plasma osmolality was similar between the hypertonic SW and isotonic BW groups. The livers of FW-acclimated medaka may have experienced hypoosmotic stress. This is because osmotic stress can induce cell death; hence, maintenance of cell osmolality is a vital issue (Lang et al., 2000; Burg et al., 2007). Cholesterol biosynthesis in response to changes in ambient salinity is related to the physical properties of the cell membranes (Parasassi et al., 1995). The presence of cholesterol leads to a decrease in the permeability coefficients of phospholipid vesicles to Na+, K+, and Cl- (Papahadjopoulosa et al., 1972). A previous study indicated that the erythrocytes of guinea pigs containing an increased amount of cholesterol showed a dominant decrease in membrane permeability to both the active and passive components of Na+ efflux (Kroes and Ostwald, 1971). Therefore, the increased cholesterol content in the livers of FW-acclimated medaka may help protect fish against extracellular hypoosmotic stress.
Under salinity stress, the gill is the predominant organ for osmoregulation in euryhaline teleosts. Na+/K+-ATPase (NKA) is expressed in mitochondria-rich cells (i.e., ionocytes) of the gill that actively transports Na+ from and K+ into cells and can provide a primary driving force to activate other ion transport systems involved in osmoregulation (Hwang et al., 2011; Lin C. H. et al., 2021). NKA comprises α- and β-subunits. The α-subunit is considered the catalytic center of NKA, with binding sites for cations and ATP (Scheiner-Bobis, 2002). Lipid rafts (LRs) are plasma membrane microdomains enriched in cholesterol. Previous mammalian studies have shown that LRs may play a crucial role in ion exchange. In milkfish and tilapia, the branchial NKA α-subunit was mainly expressed in LR (Lin Y. T. et al., 2021). In contrast, experiments on the mammalian kidney epithelial cell line (LLC-PK1) revealed that plasma membrane cholesterol positively affects NKA α1 expression (Chen et al., 2011; Lambropoulos et al., 2016; Zhang et al., 2020). A previous study indicated that FW-acclimated Indian medaka have higher branchial NKA α1 protein expression than SW-acclimated medaka (Kang et al., 2008). The liver is the central organ involved in vertebrate cholesterol biosynthesis. Therefore, the increased liver cholesterol content in FW-acclimated medaka may increase the delivery of cholesterol to the gill and increase membrane NKA α1 protein expression.
Sterol regulatory element-binding protein-1 and 2 (SREBP-1/2) are vital regulators of cholesterol metabolism, and 3-Hydroxy-3-Methylglutaryl CoA reductase (HMGCR) and farnesyl diphosphate synthase (FDPS) are key enzymes involved in cholesterol biosynthesis (Amemiya-Kudo et al., 2002; Horton et al., 2002; Xue et al., 2020). HMGCR is an important enzyme that controls the first rate-limiting step of the cholesterol synthesis pathway (Sharpe and Brown, 2013). In mammals, hmgcr is the transcriptional target of SREBP-1a and SREBP-2 (Horton et al., 2002; Sundqvist et al., 2005; Xue et al., 2020). Si et al. (2018) indicated that the gene expression of srebp-2, hmgcr, and fdps was upregulated in the livers of Cynoglossus semilaevis under long-term hypotonic stress. In this study, we found that the hepatic gene expression of srebp-1/2, hmgcr, and fdps was not different between FW- and SW-acclimated medaka. In contrast, in this study, we found that the nuclear protein expression of SREBP-1 was significantly upregulated in FW-acclimated medaka. When SREBP-1 translation was knocked down by intraperitoneal injection of SREBP-1 Vivo-MO, we found that SREBP-1 knockdown caused a dominant decrease in hepatic cholesterol content in both FW- and SW-acclimated medaka. The decreased percentage of hepatic cholesterol content was approximately 70-80% in SREBP-1 knockdown group compared to the control group. These results revealed that SREBP-1 is positively related to cholesterol biosynthesis in the liver of teleosts. Thus, higher SREBP-1 protein expression in the liver of FW-acclimated medaka may contribute to the upregulation of cholesterol content. In addition, we found that SREBP-1 knockdown caused a dominant downregulation in the transcription of hmgcr in the livers of FW- and SW-acclimated medaka. There are two SREBP-1 proteins in mammals, SREBP-1a and 1c, in mammals (Horton et al., 2002). SREBP-1a is universal for fatty acid and cholesterol biosynthesis, but SREBP-1c is restricted to fatty acid biosynthesis (Horton et al., 2002). In teleosts, several studies have indicated that only one SREBP-1 form was translated, and the analysis of amino acid sequences showed that teleostean SREBP-1 was close to mammalian SREBP-1a (Minghetti et al., 2011; Thomas et al., 2013; Dong et al., 2015). Dong et al. (2015) reported that the gene expression of SREBP-1 in the liver of Japanese seabass was decreased by dietary polyunsaturated fatty acids compared to dietary saturated or monounsaturated fatty acids. Hence, fish SREBP-1 may be involved in lipid metabolism. This study indicated that the knockdown of SREBP-1 caused a decrease in hepatic cholesterol content in Indian medaka. Taken together, teleost SREBP-1 may function universally in lipid metabolism, similar to mammalian SREBP-1a.
In zebrafish, aplexone treatment disturbs the transcription of enzymes in the HMGCR pathway and reduces cellular cholesterol levels. This causes a feedback loop to induce fdps gene expression (Choi et al., 2011). This study revealed that fdps gene expression was predominantly upregulated in SREBP-1 knockdown medaka. The decreased hepatic cholesterol content caused by SREBP-1 knockdown in Indian medaka may also cause a feedback effect that induces hepatic fdps gene expression. In mice, the target disruption of the srebp-1 gene caused the upregulation of the srebp-2 transcript and nuclear SREBP-2 protein expression, resulting in increased cholesterol synthesis in the liver (Shimano et al., 1997). In contrast, these results indicated that hepatic srebp-2 gene expression was not changed in Indian medaka with SREBP-1 knockdown. Additionally, SREBP-1 knockdown did not positively affect hepatic cholesterol synthesis in Indian medaka. The regulatory mechanism for cholesterol homeostasis may have subtle differences between fish and mammals. Therefore, the impaired expression of SREBP-1 caused divergent SREBP-2 expression and cholesterol synthesis. SREBP-2 plays an essential role in regulating cholesterol synthesis in mammalian experiments (Amemiya-Kudo et al., 2002; Horton et al., 2002). Previous studies have indicated that dietary cholesterol depletion and supplementation induced a positive and negative feedback effect on hepatic srebp-2 gene expression in fish (Kortner et al., 2014; Zhu et al., 2018). This study indicated that the gene expression level of srebp-2 was approximately 5-fold higher than that of srebp-1 in the livers of Indian medaka. Therefore, hepatic srebp-2 gene expression was not significantly different between FW- and SW-acclimated medaka and SREBP-1 knockdown did not change SREBP-2 gene expression. The importance of SREBP-2 in cholesterol metabolism in fish cannot be neglected.
Liver X receptor α (LXRα) plays a key role in controlling cholesterol metabolism in mammalian livers (Zhao and Dahlman-Wright, 2010; Jakobsson et al., 2012). To maintain cholesterol homeostasis, LXRα induces the expression of a range of genes involved in cholesterol transport and catabolism in the target tissues (Zhao and Dahlman-Wright, 2010; Jakobsson et al., 2012). In turbot (Scophthalmus maximus), lxrα gene expression was significantly decreased 24 and 48 h post-transfer from SW to FW (Liu et al., 2021). On the contrary, our results indicated that both gene and nuclear protein expression of LXRα were unchanged in the liver of Indian medaka acclimated to FW and SW for one week. Additionally, we found that sirtuin 1 (SIRT1) gene and nuclear protein expression in the liver of Indian medaka were not significantly different between FW and SW acclimation. SIRT1 is an NAD+-dependent histone/protein deacetylase that can deacetylate LXRs and increase their activity. Increased activity of LXRs has a positive effect on cholesterol catabolism in target tissues (Li et al., 2007). According to these results, ambient salinity did not affect the nuclear protein expression of LXRα and STIR1. Cholesterol elimination in the liver may not change in Indian medaka after acclimation to FW and SW for one week. However, FW-acclimated medaka showed higher nuclear protein expression of SREBP-1, which may contribute to the elevation of hepatic cholesterol biosynthesis. Finally, these phenomena may result in elevated cholesterol levels in the liver of FW-acclimated medaka.
In conclusion, these results showed that the knockdown of SREBP-1 decreased the expression of cholesterol metabolism-related genes and cholesterol content in the liver of Indian medaka. This is the first study to show SREBP-1 may have a positive effect on hepatic cholesterol metabolism in teleosts. Thus, the SREBP-1 protein expression was upregulated in FW-acclimated medaka, which may contribute to the higher hepatic cholesterol content compared to that in SW-acclimated medaka. We found that the gene and protein expression of LXRα and SIRT1 did not differ between the FW and SW groups in Indian medaka, implying that cholesterol catabolism and other lipid metabolic reactions might not be regulated in Indian medaka under salinity stress. Further experiments will help verify these hypotheses in the future.
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
The animal study was reviewed and approved by Institutional Animal Care and Use Committee (IACUC). Written informed consent was obtained from the owners for the participation of their animals in this study.
NR, C-HL, and T-HL designed the experiments. NR performed the experiments and analyzed the data. NR, C-HL, and T-HL wrote the paper. All authors have read and approved the final manuscript.
This study was partly supported by research projects (108-2313-B-005-006-MY3) from the Ministry of Science and Technology (MOST), Taiwan, and in part by the iEGG and Animal Biotechnology Center from The Feature Area Research Center Program within the framework of the Higher Education Sprout Project by the Ministry of Education (MOE), Taiwan (MOE 109-S-0023-A) to T-HL.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2022.891706/full#supplementary-material
Supplementary Figure 1 | Immunoblotting of SIRT1 in Indian medaka (O. melastigma) livers detected with a polyclonal antibody (Proteintech-13161-1-AP). (A) One immunoreactive band with a molecular mass of about 116 kDa was detected. (B) The negative control did not indicate an immunoreactive band with the secondary antibody. The arrow indicated the immunoreactive band. M, marker; N, nuclear fraction.
Supplementary Figure 2 | Immunoblotting of LXRα in Indian medaka (O. melastigma) livers detected with a monoclonal antibody (Santa Cruz sc-377260). (A) One immunoreactive band with a molecular mass of about 52 kDa was detected. (B) The negative control did not indicate an immunoreactive band with the secondary antibody. The arrow indicated the immunoreactive band. M, marker; N, nuclear fraction.
Supplementary Figure 3 | Immunoblotting of SREBP-1 in Indian medaka (O. melastigma) livers detected with a monoclonal antibody (Santa Cruz sc-13551). (A) The major immunoreactive band had a molecular mass of about 85 kDa. (B) Negative control did not indicate an immunoreactive band with the secondary antibody. The arrow indicated the immunoreactive band. M, marker; N, nuclear fraction.
Albani D., Polito L., Forloni G. (2010). Sirtuins as Novel Targets for Alzheimer's Disease and Other Neurodegenerative Disorders: Experimental and Genetic Evidence. J. Alzheimers Dis. 19 (1), 11–26. doi: 10.3233/JAD-2010-1215
Amemiya-Kudo M., Shimano H., Hasty A. H., Yahagi N., Yoshikawa T., Matsuzaka T., et al. (2002). Transcriptional Activities of Nuclear SREBP-1a, -1c, and–2 to Different Target Promoters of Lipogenic and Cholesterogenic Genes. J. Lipid Res. 43 (8), 1220–1235. doi: 10.1194/jlr.M100417-JLR200
Aruna A., Nagarajan G., Chang C. F. (2015). The Acute Salinity Changes Activate the Dual Pathways of Endocrine Responses in the Brain and Pituitary of Tilapia. Gen. Comp. Endocrinol. 211, 154–164. doi: 10.1016/j.ygcen.2014.12.005
Azhar S., Leers-Sucheta S., Reaven E. (2003). Cholesterol Uptake in Adrenal and Gonadal Tissues: The SR-BI and 'Selective' Pathway Connection. Rev. Front. Biosci. 8, s998–1029. doi: 10.2741/1165
Babin P. J., Vernier J. M. (1989). Plasma Lipoproteins in Fish. J. Lipid Res. 30 (4), 467–489. doi: 10.1016/S0022-2275(20)38342-5
Bertrand S., Brunet F. G., Escriva H., Parmentier G., Laudet V., Robinson-Rechavi M. (2004). Evolutionary Genomics of Nuclear Receptors: From Twenty-Five Ancestral Genes to Derived Endocrine Systems. Mol. Biol. Evol. 21 (10), 1923–1937. doi: 10.1093/molbev/msh200
Binukumari S., Vasanthi J. (2014). Changes in Cholesterol Content of the Freshwater Fish, Labeo Rohita Due to the Effect of an Insecticide ‘Encounter’ (Herbal Plant Extract). Int. J. Pharm. Sci. 5 (2), 397–399. doi: 10.13040/IJPSR.0975-8232
Burg M. B., Ferraris J. D., Dmitrieva N. I. (2007). Cellular Response to Hyperosmotic Stresses. Physiol. Rev. 87 (4), 1441–1474. doi: 10.1152/physrev.00056.2006
Charmandari E., Tsigos C., Chrousos G. (2005). Endocrinology of the Stress Response. Annu. Rev. Physiol. 67, 259–284. doi: 10.1146/annurev.physiol.67.040403.120816
Chen Y., Li X., Ye Q., Tian J., Jing R., Xie Z. (2011). Regulation of Alpha1 Na/K-ATPase Expression by Cholesterol. J. Biol. Chem. 286 (17), 15517–15524. doi: 10.1074/jbc.M110.204396
Choi J., Mouillesseaux K., Wang Z., Fiji H. D., Kinderman S. S., Otto G. W., et al. (2011). Aplexone Targets the HMG-CoA Reductase Pathway and Differentially Regulates Arteriovenous Angiogenesis. Development. 138 (6), 1173–1181. doi: 10.1242/dev.054049
Corcos C. L. J., Gonthier C., Zaghini I., Logette E., Shechter I., Bournot P. (2005). Hepatic Farnesyl Diphosphate Synthase Expression is Suppressed by Polyunsaturated Fatty Acids. Biochem. J. 385 (3), 787–794. doi: 10.1042/BJ20040933
Corpet F. (1988). Multiple Sequence Alignment With Hierarchical Clustering. Nucleic Acids Res. 16 (22), 10881–10890. doi: 10.1093/nar/16.22.10881
Cruz-Garcia L., Minghetti M., Navarro I., Tocher D. R. (2009). Molecular Cloning, Tissue Expression and Regulation of Liver X Receptor (LXR) Transcription Factors of Atlantic Salmon (Salmo Salar) and Rainbow Trout (Oncorhynchus Mykiss). Comp. Biochem. Physiol. 153 (1), 81–88. doi: 10.1016/j.cbpb.2009.02.001
Dong S., Kang M., Wu X., Ye T. (2014). Development of a Promising Fish Model (Oryzias Melastigma) for Assessing Multiple Responses to Stresses in the Marine Environment. BioMed. Res. Int. 2014, 563131. doi: 10.1155/2014/563131
Dong X., Tan P., Cai Z., Xu H., Li J., Ren W., et al. (2017). Regulation of FADS2 Transcription by SREBP-1 and PPAR-α Influences LC-PUFA Biosynthesis in Fish. Sci. Rep. 7, 40024. doi: 10.1038/srep40024
Dong X., Xu H., Mai K., Xu W., Zhang Y., Ai Q. (2015). Cloning and Characterization of SREBP-1 and PPAR-Alpha in Japanese Seabass Lateolabrax Japonicus, and Their Gene Expressions in Response to Different Dietary Fatty Acid Profiles. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 180, 48–56. doi: 10.1016/j.cbpb.2014.10.001
Engelking L. R. (2015). “In Textbook of Veterinary Physiological Chemistry,” in Cholesterol, 3rd ed., Waltham, MA, USA: Elsavier doi: 10.1016/C2010-0-66047-0
Farrell A. P., Saunders R. L., Freeman H. C., Mommsen T. P. (1986). Arteriosclerosis in Atlantic Salmon: Effects of Dietary Cholesterol and Maturation. J. Am. Heart Assoc. 6 (4), 453–461. doi: 10.1161/01.ATV.6.4.453
Feige N. J., Auwerx J. (2007). DisSIRTing on LXR and Cholesterol Metabolism. Cell Metab. 6 (5), 343–348. doi: 10.1016/j.cmet.2007.10.003
Goedeke L., Fernández-Hernando C. (2012). Regulation of Cholesterol Homeostasis. Cell Mol. Life Sci. 69 (6), 915–930. doi: 10.1007/s00018-011-0857-5
Goldstein J. L., Brown M. S. (1990). Regulation of the Mevalonate Pathway. Nature. 343 (6257), 425–430. doi: 10.1038/343425a
Grozinger C. M., Chao E. D., Blackwell H. E., Moazed D., Schreiber S. L. (2001). Identification of a Class of Small Molecule Inhibitors of the Sirtuin Family of NAD-Dependent Deacetylases by Phenotypic Screening. J. Biol. Chem. 276 (42), 38837–38843. doi: 10.1074/jbc.M106779200
Hassett R. P., Crockett E. L. (2009). Habitat Temperature Is an Important Determinant of Cholesterol Contents in Copepods. J. Exp. Biol. 212 (1), 71–77. doi: 10.1242/jeb.020552
Hazel J. R., Williams E. E. (1990). The Role of Alterations in Membrane Lipid-Composition in Enabling Physiological Adaptation of Organisms to Their Physical-Environment. Prog. Lipid Res. 29 (3), 167–227. doi: 10.1016/0163-7827(90)90002-3
Horton J. D., Goldstein J. L., Brown M. S. (2002). SREBPs: Activators of the Complete Program of Cholesterol and Fatty Acid Synthesis in the Liver. J. Clin. Investig. 109 (9), 1125–1131. doi: 10.1172/JCI200215593
Hu Y. C., Chu K. F., Yang W. K., Lee T. H. (2017). Na+, K+-ATPase β1 Subunit Associates With α1 Subunit Modulating a “Higher-NKA-In-Hyposmotic Media” Response in Gills of Euryhaline Milkfish, Chanos Chanos. J. Comp. Physiol. 187 (7), 995–1007. doi: 10.1007/s00360-017-1066-9
Hwang P. P., Lee T. H. (2007). New Insights Into Fish Ion Regulation and Mitochondrion-Rich Cells. Comp. Biochem. physiol. Mol. A Integr. Physiol. 148 (3), 479–497. doi: 10.1016/j.cbpa.2007.06.416
Hwang P. P., Lee T. H., Lin L. Y. (2011). Ion Regulation in Fish Gills: Recent Progress in the Cellular and Molecular Mechanisms. Am. J. Physiol. Regul. Integr. Comp. Physiol. 301 (1), R28–R47. doi: 10.1152/ajpregu.00047.2011
Ishimoto K., Tachibana K., Hanano I., Yamasaki D., Nakamura H., Kawai M., et al. (2010). Sterol-Regulatory-Element-Binding Protein 2 and Nuclear Factor Y Control Human Farnesyl Diphosphate Synthase Expression and Affect Cell Proliferation in Hepatoblastoma Cells. Biochem. 429 (2), 347–357. doi: 10.1042/BJ20091511
Jakobsson T., Treuter E., Gustafsson J., Steffensen K. R. (2012). Liver X Receptor Biology and Pharmacology: New Pathways, Challenges and Opportunities. Trends Pharmacol. Sci. 33 (7), 394–404. doi: 10.1016/j.tips.2012.03.013
Juo J. J., Kang C. K., Yang W. K., Yang S. Y., Lee T. H. (2016). A Stenohaline Medaka, Oryzias Woworae, Increases Expression of Gill Na+, K+-ATPase and Na+, K+, 2Cl– Cotransporter 1 to Tolerate Osmotic Stress. Zool. Sci. 33, 414–425. doi: 10.2108/zs150157
Kang C. K., Tsai S. C., Lee T. H., Hwang P. P. (2008). Differential Expression of Branchial Na+/K+-ATPase of Two Medaka Species, Oryzias Latipes and Oryzias Dancena, With Different Salinity Tolerances Acclimated to Fresh Water, Brackish Water and Seawater. CBP. A Mol. Integr. Physiol. 151 (4), 566–575. doi: 10.1016/j.cbpa.2008.07.020
Kemper J. K., Choi S. E., Kim D. H. (2013). Sirtuin 1 Deacytylase: A Key Regulator of Hepatic Lipid Metabolism. Vitam. Horm. 91, 385–404. doi: 10.1016/B978-0-12-407766-9.00016-X
Kemski M. M., Rappleye C. A., Dabrowski K., Bruno R. S., Wick M. (2020). Transcriptomic Response to Soybean Meal-Based Diets as the First Formulated Feed in Juvenile Yellow Perch (Perca Flavescens). Sci. Rep. 10 (1), 3998. doi: 10.1038/s41598-020-59691-z
Kortner T. M., Björkhem I., Krasnov A., Timmerhaus G., Krogdahl A. (2014). Dietary Cholesterol Supplementation to a Plant-Based Diet Suppresses the Complete Pathway of Cholesterol Synthesis and Induces Bile Acid Production in Atlantic Salmon (Salmo Salar L.). Br. J. Nutr. 111 (12), 2089–2103. doi: 10.1017/S0007114514000373
Kroes J., Ostwald R. (1971). Erythrocyte Membranes–Effect of Increased Cholesterol Content on Permeability. BBA. 249 (2), 647–650. doi: 10.1016/0005-2736(71)90147-7
Lambropoulos N., Garcia A., Clarke R. ,. J. (2016). Stimulation of Na+,K+-ATPase Activity as a Possible Driving Force in Cholesterol Evolution. J. Membr Biol. 249 (3), 251–259. doi: 10.1007/s00232-015-9864-z
Lang F., Ritter M., Gamper N., Huber S., Fillon S., Tanneur V., et al. (2000). Cell Volume in the Regulation of Cell Proliferation and Apoptotic Cell Death. Cell. Physiol. Biochem. 10, 417–428. doi: 10.1159/000016367
Lee G. M. D. (2020). Disorders of Lipid Metabolism (Goldman-Cecil Medicine), 1346. New York, USA: Elsavier
Li S., Monroig O., Wang T., Yuan Y., Navarro J. C., Hontoria F., et al. (2017). Functional Characterization and Differential Nutritional Regulation of Putative Elovl5 and Elovl4 Elongases in Large Yellow Croaker (Larimichthys Crocea). Sci. Rep. 7 (1), 3514–3528. doi: 10.1038/s41598-017-02646-8
Lin Y. T., Hu Y. C., Wang Y. C., Hsiao M. Y., Lorin-Nebel C., Lee T. H. (2021). Differential Expression of Two ATPases Revealed by Lipid Raft Isolation From Gills of Euryhaline Teleosts With Different Salinity Preferences. Comp. Biochem. Physiol. - B Biochem. Mol. Biol. 253, 110562. doi: 10.1016/j.cbpb.2021.110562
Lin C. H., Yeh P. L., Wang Y. C., Lee T. H. (2021). Dynamic Regulation of Ions and Amino Acids in Adult Asian Hard Clams Meretrix Lusoria Upon Hyperosmotic Salinity. Front. Mar. Sci. 8. doi: 10.3389/fmars.2021.749418
Liu Z., Ma A., Yuan C., Zhao T., Chang H., Zhang J. (2021). Transcriptome Analysis of Liver Lipid Metabolism Disorders of the Turbot Scophthalmus Maximus in Response to Low Salinity Stress. Aquaculture 534, 736273. doi: 10.1016/j.aquaculture.2020.736273
Livak K. J., Schmittgen T. D. (2001). Analysis of Relative Gene Expression Data Using Real Time Quantitative PCR and the 2-ΔΔct. Method. 25 (4), 402–408. doi: 10.1006/meth.2001.1262
Li X., Zhang S., Blander G., Tse J. G., Krieger M., Guarente L. (2007). SIRT1 Deacetylates and Positively Regulates the Nuclear Receptor LXR. Nat. Rev. Mol. Cell Biol. 28 (1), 91–106. doi: 10.1016/j.molcel.2007.07.032
Minghetti M., Leaver M. J., Tocher D. R. (2011). Transcriptional Control Mechanisms of Genes of Lipid and Fatty Acid Metabolism in the Atlantic Salmon (Salmo Salar L.) Established Cell Line, SHK-1. Int. J. Biochem. 811 (3), 194–202. doi: 10.1016/j.bbalip.2010.12.008
Mohamed A. S., Md A. E., Desoky N. S. G. (2019). The Changes in Triglyceride and Total Cholesterol Concentrations in the Liver and Muscle of Two Fish Species From Qarun Lake, Egypt. Fish. Oceanogr. 9 (4), 86–90. doi: 10.19080/OFOAJ.2019.09.555770
Papahadjopoulosa D., Nira S., Ohki S. (1972). Permeability Properties of Phospholipid Membranes: Effect of Cholesterol and Temperature. BBA. 266 (3), 561–583. doi: 10.1016/0006-3002(72)90001-7
Parasassi T., Giusti A. M., Raimondi M., Ravagnan G., Sapora O., Gratton E. (1995). Cholesterol Protects the Phospholipid Bilayer From Oxidative Damage. Free Radic. Biol. Med. 19 (4), 511–516. doi: 10.1016/0891-5849(95)00038-y
Peet D. J., Janowski B. A., Mangelsdorf D. J. (1998). The LXRs: A New Class of Oxysterol Receptors. Curr. Opin. Genet. Dev. 8 (5), 571–575. doi: 10.1016/S0959-437X(98)80013-0
Ponugoti B., Kim D. H., Xiao Z., Smith Z., Miao J., Zang M., et al. (2010). SIRT1 Deacetylates and Inhibits SREBP-1C Activity in Regulation of Hepatic Lipid Metabolism. J. Biol. Chem. 285 (44), 33959–33970. doi: 10.1074/jbc.M110.122978
Repa J. J., Liang G., Ou J., Bashmakov Y., Lobaccaro J. M. (2000). Regulation of Mouse Sterol Regulatory Element-Binding Protein-1c Gene (SREBP-1c) by Oxysterol Receptors, Lxrα and Lxrβ. Genes Dev. 14 (22), 2819–2830. doi: 10.1101/gad.844900
Roya L. A., Davisa D. A., Saoud A. P. (2006). Effects of Lecithin and Cholesterol Supplementation to Practical Diets for Litopenaeus Vannamei Reared in Low Salinity Waters. Aquac. 257, 446–452. doi: 10.1016/j.aquaculture.2006.02.059
Scheiner-Bobis G. (2002). The Sodium Pump. Its Molecular Properties and Mechanics of Ion Transport. Eur. J. Biochem. 269, 2424–2433. doi: 10.1046/j.1432-1033.2002.02909.x
Schultz J. R., Tu H., Luk A., Repa J. J., Medina J. C., Li L., et al. (2000). Role of LXRs in Control of Lipogenesis. Genes Dev. 14 (22), 2831–2838. doi: 10.1101/gad.850400
Sharpe L. J., Brown A. J. (2013). Controlling Cholesterol Synthesis Beyond 3-Hydroxy-3-Methylglutaryl-CoA Reductase (HMGCR). J. Biol. Chem. 288, 18707–18715. doi: 10.1074/jbc.R113.479808
Shimano H., Shimomura I., Hammer R. E., Herz J., Goldstein J. L., Brown M. S., et al. (1997). Elevated Levels of SREBP-2 and Cholesterol Synthesis in Livers of Mice Homozygous for a Targeted Disruption of the SREBP-1 Gene. J. Clin. Invest. 100 (8), 2115–2124. doi: 10.1172/JCI119746
Si Y., Wen H., Li Y., He F. H., Li J., Li S., et al. (2018). Liver Transcriptome Analysis Reveals Extensive Transcriptional Plasticity During Acclimation to Low Salinity in Cynoglossus Semilaevis. BMC Genom. 19 (1), 464. doi: 10.1186/s12864-018-4825-4
Smith J. R., Osborne T. F., Brown M. S., Goldstein J. L., Gil G. (1988). Multiple Sterol Regulatory Elements in Promoter for Hamster 3-Hydroxy-3-Methylglutaryl-Coenzyme A Synthase. J. Biol. Chem. 263 (34), 18480–18487. doi: 10.1016/S0021-9258(19)81383-2
Sundqvist A., Bengoechea-Alonso M. T., Ye X., Lukiyanchuk V., Jin J., Harper J. W., et al. (2005). Control of Lipid Metabolism by Phosphorylation-Dependent Degradation of the SREBP Family of Transcription Factors by SCFFbw. Cell Metab. 1 (6), 379–391. doi: 10.016/j.cmet.2005.04.010
Thomas J. K., Wiseman S., Giesy J. P., Janz D. M. (2013). Effects of Chronic Dietary Selenomethionine Exposure on Repeat Swimming Performance, Aerobic Metabolism and Methionine Catabolism in Adult Zebrafish (Danio Rerio). Aquat. Toxicol. 130–131, 112–122. doi: 10.1016/j.aquatox.2013.01.009
Twibell R. G., Wilson R. P. (2004). Preliminary Evidence That Cholesterol Improves Growth and Feed Intake of Soybean Meal-Based Diets Inaquaria Studies With Juvenile Channel Catfsh, Ictalurus Punctatus. Aquat 236 (1-4), 539–546. doi: 10.1016/j.aquaculture.2003.10.028
Vassilopoulos A., Fritz K. S., Petersen D. R., Gius D. (2011). The Human Sirtuin Family: Evolutionary Divergences and Functions. Hum. Genomics 5 (5), 485–496. doi: 10.1186/1479-7364-5-5-485
Walker A. K., Yang F., Jiang K., Ji J. Y., Jennifer L., Watts J. L., et al. (2010). Conserved Role of SIRT1 Orthologs in Fasting-Dependent Inhibition of the Lipid/Cholesterol Regulator SREBP. Genes Dev. 24 (13), 1403–1417. doi: 10.1101/gad.1901210
Wang X., Sato R., Brown M. S., Hua X., Goldstein J. L. (1994). SREBP-1, a Membrane-Bound Transcription Factor Released by Sterol-Regulated Proteolysis. Cell. 77 (1), 53–62. doi: 10.1016/0092-8674(94)90234-8
Wen X., Peng C., Xu J., Wei X., Fu D., Wang T., et al. (2021). Combined Effects of Low Temperature and Salinity on the Immune Response, Antioxidant Capacity and Lipid Metabolism in the Pufferfish (Takifugu Fasciatus). Aquac. 531, 735866. doi: 10.1016/j.aquaculture.2020.735866
Xue L., Qi H., Zhang H., Ding L., Huang Q., Zhao D., et al. (2020). Targeting SREBP-2-Regulated Mevalonate Metabolism for Cancer Therapy. Front. Oncol. 21 (10). doi: 10.3389/fonc.2020.01510
Xu C., Li E., Xu Z., Su Y., Lu M., Qin J. G., et al. (2018). Growth and Stress Axis Responses to Dietary Cholesterol in Nile Tilapia (Oreochromis Niloticus) in Brackish Water. Front. Physiol. 26 (9), 254. doi: 10.3389/fphys.2018.00254
Yang W. K., Kang C. K., Chang C. H., Hsu A. D., Lee T. H., Hwang P. P. (2013). Expression Profiles of Branchial FXYD Proteins in the Brackish Medaka Oryzias Dancena: A Potential Saltwater Fish Model for Studies of Osmoregulation. PloS One 8 (1), e55470. doi: 10.1371/journal.pone.0055470
Yun B., Ai Q., Mai K., Xu W., Qi G., Luo Y. (2012). Synergistic Efects of Dietary Cholesterol and Taurine on Growth Performance and Cholesterol Metabolism in Juvenile Turbot (Scophthalmus Maximus L.) Fed High Plant Protein Diets. Aquac. 324 (325), 85–91. doi: 10.1016/j.aquaculture.2011.10.012
Yun B., Mai K., Zhang W., Xu W. (2011). Effects of Dietary Cholesterol on Growth Performance, Feed Intake and Cholesterol Metabolism in Juvenile Turbot (Scophthalmus Maximus L.) Fed High Plant Protein Diets. Aquac. 495, 443–451. doi: 10.1016/j.aquaculture.2018.06.002
Yusof S., Ismail A., Koito T., Kinoshita M., Inoue K. (2011). Occurrence of Two Closely Related Ricefishes, Javanese Medaka (Oryzias Javanicus) and Indian Medaka (O. Dancena) at Sites With Different Salinity in Peninsular Malaysia. Environ. Biol. Fishes. 93 (1), 43–49. doi: 10.1007/s10641-011-9888-x
Zhang J., Li X., Yu H., Larre I., Dube P. R., Kennedy D., et al. (2020). Regulation of Na/K-ATPase Expression by Cholesterol: Isoform Specificity and the Molecular Mechanism. Am. J. Physiol. Cell Physiol. 319 (6), C1107–C1119. doi: 10.1152/ajpcell.00083.2020
Zhang Q., You C., Liu F., Zhu W., Wang S., Xie D., et al. (2016). Cloning and Characterization of Lxr and Srebp1, and Their Potential Roles in Regulation of LC-PUFA Biosynthesis in Rabbitfish Siganus Canaliculatus. Lipids 51, 1051–1063. doi: 10.1007/s11745-016-4176-3
Zhao C., Dahlman-Wright K. (2010). Liver X Receptor in Cholesterol Metabolism. J. Endocrinol. 204 (3), 233–240. doi: 10.1677/JOE-09-0271
Keywords: SREBP-1, cholesterol, salinity, liver, medaka
Citation: Ranasinghe N, Lin C-H and Lee T-H (2022) Cholesterol Accumulation in Livers of Indian Medaka, Oryzias dancena, Acclimated to Fresh Water and Seawater. Front. Mar. Sci. 9:891706. doi: 10.3389/fmars.2022.891706
Received: 08 March 2022; Accepted: 28 April 2022;
Published: 31 May 2022.
Edited by:
Yu-Hung Lin, National Pingtung University of Science and Technology, TaiwanReviewed by:
Zhen-Yu Du, East China Normal University, ChinaCopyright © 2022 Ranasinghe, Lin and Lee. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chia-Hao Lin, Y2gxMjNAbmt1c3QuZWR1LnR3; Tsung-Han Lee, dGhsZWVAZW1haWwubmNodS5lZHUudHc=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.