
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mar. Sci. , 26 September 2022
Sec. Marine Evolutionary Biology, Biogeography and Species Diversity
Volume 9 - 2022 | https://doi.org/10.3389/fmars.2022.891236
 Lijun He1,2,3*
Lijun He1,2,3* Shasha Xu1†
Shasha Xu1† David A. Weese4†
David A. Weese4† Jie Zhang2†
Jie Zhang2† Huimin Ren1
Huimin Ren1 Fenghua Tang3
Fenghua Tang3 Zhongli Sha2,5,6,7*
Zhongli Sha2,5,6,7* Jing Zhang1
Jing Zhang1As a bio]diversity hotspot, the East Indies (Coral) Triangle possesses the highest biodiversity on the earth. However, evolutionary hypotheses around this area remain controversial; e.g., center of origin, center of accumulation, and center of overlap have been supported by different species. This study aims to answer the evolutionary influence of the Indonesian Seaway on the biodiversity of the Coral Triangle by recovering the evolutionary origins of a wide-ranging ommastrephid squid (Sthenoteuthis oualaniensis) based on integrated molecular and oceanographic clues from the Indo-Pacific. Three new clades were revealed; viz., clade I from the South China Sea, clade II from the northern East Indian Ocean, and clade III from the southern East Indian Ocean. These two Indian Ocean clades formed a monophyly closely related to clade IV from the Central-Southeast Pacific. Clade VI from the central Equatorial Pacific and clade V from the northern Eastern Pacific sit in basal positions of phylogenetic trees. Ancestral Sthenoteuthis was inferred to have originated from the Atlantic Ocean and sequentially dispersed to the northern East Pacific, central Equatorial Pacific, and West Pacific through the open Panama Seaway and being transported by westward North Equatorial Current. The East Indian Ocean was likely colonized by an ancestral population of clade IV from the Southeast Pacific. Westward South Equatorial Circulation could have promoted transoceanic migration of S. oualaniensis through the wide paleo-Indonesian Seaway. Sea level regression since the Miocene and the closure of the Indonesian Seaway at 4–3 Ma were responsible for the population genetic differentiation of S. oualaniensis in the Indo-Pacific. Therefore, the Indonesian Gateway played an important role in influencing marine organisms’ migration and population differentiation through controlling and reorganizing circulations in the Indo-Pacific.
During the past 50 million years, global marine biodiversity hotspots have gradually shifted from the West Tethyan region to the Middle East and Indo-Australia archipelago (Renema et al., 2008). The highest biodiversity of extant marine species is located in the Southeast Asian archipelago/Philippines–Malayan–Papua New Guinea region/East Indies (Coral) Triangle (Roberts et al., 2002; Hoeksema, 2007). Three main competing models/hypotheses have been invoked to explain high biodiversity in the Coral Triangle (Mora et al., 2003; Briggs, 2009), e.g., center of origin (Briggs, 2000; Briggs, 2003), center of overlap (Ekman, 1953), and center of accumulation (Bellwood and Meyer, 2009). However, none of these theories can cover all species’ evolutionary patterns (Palumbi, 1997). Thus, it is necessary to further test evolutionary models of the Coral Triangle biodiversity hotspot using wide-ranging abyssal species (e.g., squids) from the Indo-Pacific.
As large nektonic predators, ommastrephid squids play an important ecological role in open oceanic communities (Zuyev et al., 2002). Their diet consists mainly of myctophids, planktonic crustaceans, and other squids (Nesis, 1977; Chen et al., 2007). Diurnal vertical migration behavior has been observed in these squids with late juveniles and adults feeding in the photosynthetic layer (3/4 100 m) at night and returning to the deep sea (ca. 800–1,200 m) during the day (Zuyev et al., 2002). The largest and most abundant squid found in the tropical–subtropical waters of the Indian and Pacific Oceans is the purpleback flying squid, Sthenoteuthis oualaniensis (Lesson, 1830). With an estimated biomass of 8–11.2 million tons (Jereb and Roper, 2010), this species has become a species of increasing commercial interest due to its high nutritional and economical value with an annual catch production ranging from 0.3 to 0.5 million tons (Zuyev et al., 1975; Chen et al., 2007; Chen et al., 2008; Jereb and Roper, 2010). Therefore, S. oualaniensis is an ideal species to reveal the evolutionary origin of the East Indies Triangle biodiversity hotspot.
S. oualaniensis exhibits a complex and often debated population structure across the Indian and Pacific Oceans due to high levels of regional and intraspecific variations (Nesis, 1977; Snÿder, 1998; Chembian and Mathew, 2014; Jiang et al., 2015; Liu et al., 2019; Wang et al., 2019). In the West Pacific, two groups with different ecotypes and morphological characters have been described (Nesis, 1977): a small early-maturing group (EM) without dorsal photophores occupying the tropical Pacific 10°–15°N/S, and a large late-maturing group (LM) with a big dorsal mantle photophore occurring throughout the species range. The migration ability of the EM group is relatively weak as compared to the LM group, which makes long-distance migrations between productive feeding grounds and spawning areas (Nesis, 1977). Further investigation of S. oualaniensis in the Indian Ocean has suggested five groups (three main forms and two minor groups) based on morphological and ecological characters of females (e.g., size, structure of gladius, photophore, and maturing period) (Nesis, 1993) (Table S1): a small EM without photophores (dwarf), a small early-maturing middle-sized form with photophores, a late-maturing middle-sized form with single lateral axes of the rachis and photophores, a late-maturing middle-sized form with double lateral axes of the rachis and photophores, and a late-maturing giant form with photophores. However, only two groups have been observed for males: a dwarf form and a middle-sized form. These debates on the population structure may be due to phenotype plasticity in the morphological characters. For example, the early-maturing middle-sized form has a similar minimum mantle size as the dwarf form (ca. 9 cm), but they can be distinguished from each other based on the presence/absence of dorsal photophores (Table S1). However, dorsal photophores are thought to begin developing when the mantle length (MAL) reaches approximately 10 cm and before the maturation of the reproductive system. If an individual with an MAL less than 10 cm has reached maturation, the expression of photophores is inhibited (Nesis, 1977). Given this, dorsal photophores may represent unreliable environmental plastic characters for distinguishing the dwarf from the small middle-sized form. Furthermore, the giant form and middle-sized form with the single lateral axed of rachis occur sympatrically in the Red Sea, Gulf of Aden, and northern Arabian Sea (Table S1). Giant forms are thought to represent a population of delayed-maturing middle-sized females that adapt to low temperatures, low oxygen levels, and highly productive habitats with high growth rates and low metabolism rates (Nesis, 1993). Given the developing stage differences across a wide geographic range (Liu et al., 2019), morphological convergence due to similar ecological pressures (Head and Polly, 2015), and/or variation due to sexual dimorphisms (Nesis, 1993; Wang et al., 2019), morphological characters are unreliable for determining the population structure of S. oualaniensis throughout its range (Wang et al., 2019).
Molecular markers have proven highly useful in identifying species, lineages, and population structures of fishes and mollusks including squids (e.g., Carvalho et al., 1992; Santaclara et al., 2007; Wakabayashi et al., 2012; Wu et al., 2016; Yu et al., 2016; Muhammad et al., 2018). As for S. oualaniensis, four genetic clades including the East typical, Pacific typical, Central typical, and Equatorial lineages were identified in the Central Pacific and East Pacific based on the mitochondrial DNA (mtDNA) data (Staaf et al., 2010). Furthermore, in the West Pacific, three slightly divergent clades of S. oualaniensis were revealed from the Philippine Sea based on mtDNA data, and two of the clades also occurred in the northern South China and East China Seas (Liu et al., 2017). Lastly, two deeply divergent clades corresponding to dwarf and middle-sized forms of S. oualaniensis were observed from the South China Sea based on nuclear and mitochondrial genes (Li et al., 2019; Xu et al., 2020). Unfortunately, integrative studies on the evolutionary origin and colonization history of S. oualaniensis in the Indo-Pacific are still absent or unavailable because previous studies generally concentrated on local populations from restricted regions (e.g., Central-East Pacific or West Pacific) and were based on different molecular markers. Here, we integrate genetic data of samples from the East Indian Ocean, South China Sea, and Central-East Pacific Ocean to resolve the population classification and evolutionary history of S. oualaniensis. This study will provide new clues to understanding the biodiversity center of the Coral Triangle.
Specimens of S. oualaniensis were collected from the Eastern Indian Ocean (10 individuals) and South China Sea (three individuals) by using a tuck net (Figure 1; Table S2). Five grams of muscle tissue from each individual was removed and preserved in 95% ethanol. All capture and sampling of squid individuals were undertaken in accordance with an Animal Research ethics permit (W20211202) granted by the East China Normal University Animal Ethics Committee.
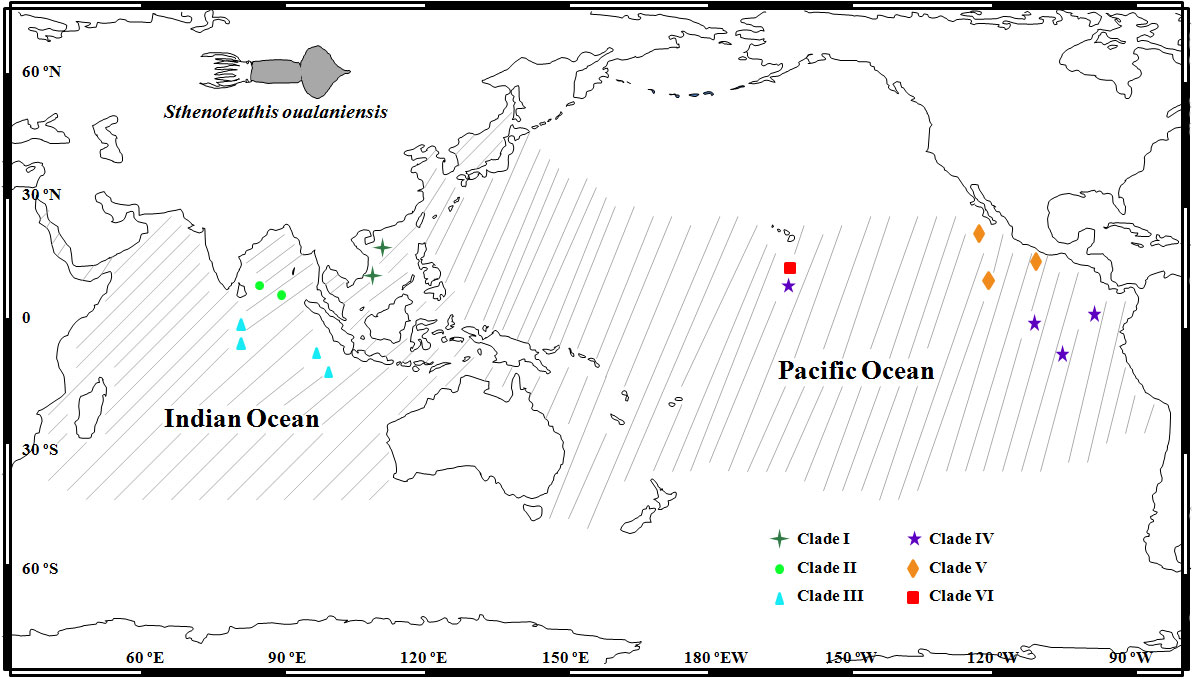
Figure 1 Sampling localities and spatial distribution of six clades in the East Indian Ocean and Pacific Ocean modified from Staaf et al. (2010). Geographic distributional range of Sthenoteuthis oualaniensis is indicated by gray lines.
Total genomic DNA was extracted from each specimen using a standard phenol–chloroform extraction method (Zhang and Hewitt, 1998). The two mitochondrial DNA segments cytochrome b (Cyt b) and cytochrome c oxidase subunit 1 (COI) genes have proven to be good or very good at recovering interspecific and intraspecific phylogenetic relationships or population structure (Miya and Nishida, 2000). Thus, Cyt b and COI were amplified using the following two pairs of primers through polymerase chain reactions (PCRs): CytbF (5′-GAACCTCGACAMCCACCAACATG-3′) and CytbR (5′-GCTCTTGTAGATTTACCACCTGCTCC-3′) for Cyt b (Liu et al., 2017); COIF (5′-GTATTTGGGCAGGACTTCTAGGGA-3′) and COIR (5′-TAATGGCAAATATGGCGTGATCATGG-3′) for COI (Li et al., 2019). Amplifications were conducted in 50-μl volumes containing 5 μl of template DNA, 25 μl of 2× EasyTaq PCR SuperMix, 200 nM each of the appropriate primers (CytbF/CytbR, and COIF/COIR), and 18 μl of ddH2O. PCR was performed in the following cycle parameters: 1) for Cyt b, denaturation at 94°C for 4 min, followed by 35 cycles of 94°C for 30 s, 60°C for 30 s, 72°C for 60 s, and a final extension of 72°C for 10 min; 2) for COI, denaturation at 94°C for 4 min, followed by 35 cycles of 94°C for 30 s, 58°C for 30 s, 72°C for 60 s, and a final extension of 72°C for 10 min. Amplified products were visualized on a 1.5% agarose gel and then purified with the Gel Extraction Mini Kit (Watson BioTechnologies, Shanghai, China). Purified DNA products were sequenced in both directions using primer pairs of CytbF/CytbR and COIF/COIR on an ABI Prism 3730 automatic sequencer. All nucleotide sequences were deposited in GenBank (Cyt b, MZ735375–MZ735387; COI, MZ712955–MZ712967). To infer the evolutionary history of S. oualaniensis, these sequences were supplemented with Cyt b and COI sequences obtained from three complete mitochondrial genomes of S. oualaniensis (EU658923, EU660576, and EU660577) from the Central-East Pacific Ocean. Intraspecific phylogenetic relationships of the sibling species Sthenoteuthis pteropus were also reconstructed based on COI sequences including MW817786–MW817789, OM988016, OM988017, MG591279, MF980608, MF980594, MF980595, GU145073, MW255556, and AB270942 sampled from the Atlantic Ocean (Figure S1). Furthermore, sequences from S. pteropus (MW255556) and an additional squid species, Dosidicus gigas (EU068697), were utilized as outgroups in the following phylogenetic tree reconstructions of S. oualaniensis. All haplotypes and sampling information of S. oualaniensis, S. pteropus, and D. gigas used in this study are summarized in Table S2.
Cyt b and COI sequences amplified from the same individual were concatenated and aligned in Cluster X (Thompson et al., 1997). Modeltest 3.6 (Posada and Crandall, 1998) was used to select the appropriate model of evolution for the COI (TrN+I), Cyt b (TIM+I), and concatenated (TrN+I) datasets by the Akaike information criterion (AIC). The model parameters of three datasets were listed as follows: 1) COI, base frequencies with A (0.2993), C (0.2124), and G (0.1253); rate matrix with R [A–C] = 1.0000, R [A–G] = 90.4377, R [A–T] = 1.0000, R [C–G] = 1.0000, R [C–T] = 13.3904, and R [G–T] = 1.0000; proportion of invariable sites (I) = 0.7282. 2) Cyt b, base frequencies with A (0.4438), C (0.2013), and G (0.1220); rate matrix with R [A–C] = 1.0000, R [A–G] = 63.8296, R [A–T] = 0.0106, R [C–G] = 0.0106, R [C–T] = 21.7894, and R [G–T] = 1.0000; proportion of invariable sites (I) = 0.7300. 3) Concatenation, base frequencies with A (0.3539), C (0.2086), and G (0.1236); rate matrix with R [A–C] = 1.0000, R [A–G] = 109.4730, R [A–T] = 1.0000, R [C–G] = 1.0000, R [C–T] = 20.6656, and R [G–T] = 1.0000; proportion of invariable sites (I) = 0.7322.
Phylogenetic trees were inferred using the maximum likelihood (ML) (Felsenstein, 1981) methodology in PHYML 3.0 (Guindon et al., 2010). Nodal supports of the ML tree were estimated using bootstrap values (Felsenstein, 1985) with 10,000 pseudo-repetitions. A Bayesian inference (BI) (Huelsenbeck and Ronquist, 2001) tree was further recovered in MrBayes 3.2 (Ronquist et al., 2012) with an evolutionary model (TrN+I). The model parameters of BI tree were estimated in the program based on BIC criteria using the following settings: ngen = 25,000,000, samplefreq = 100, burnin = 62,500, Nchains = 4, and Nruns = 2. The convergence of independent runs was achieved when white noise was seen in the overlay plot of generation versus the log-likelihood probability for both runs with the potential scale reduction factor (PSRF) approaching 1, a lower average standard deviation of split frequencies (0.000871 < 0.01). The nodal stability of the BI tree was assessed using posterior probability (PP) with a 60% consensus rule. Moreover, median-joining (MJ) network (Bandelt et al., 1999) was used to clarify the intraspecific and interspecific evolutionary relationships of S. oualaniensis and S. pteropus based on concatenation data set and COI segment in NETWORK (version 10200; fluxusengineering, 2021), respectively. A single haplotype from each clade of S. oualaniensis was added to COI sequences of S. pteropus to overcome questions of insufficient haplotypes in network reconstruction.
Species trees for COI and Cyt b data sets were also estimated in BEAST v.1.8.2 package (Drummond and Rambaut, 2007). Unlinked models of sequence evolution of each dataset were implemented using BEAUTi (TrN+I for COI, a similar GTR+I to TIM+I for Cyt b). A strict clock model and the species tree prior to a Yule Process were used. Four independent analyses with the same parameters were performed using Markov chain Monte Carlo (MCMC) simulations for a length of chain 100 million and a sampling frequency of every 10,000 generations in BEAST. Convergences of these analyses were inspected using Tracer 1.5 (Rambaut and Drummond, 2009), and the first 10% of the trees were discarded as burn-in. A single maximum clade credibility tree was reconstructed using TreeAnnotator v1.8.2 (Drummond and Rambaut, 2007) and FigTree v. 1.4.4 (Rambaut, 2009).
In order to distinguish ambiguous internal phylogenetic relationships among four clades from the northern East Indian Ocean, southern East Indian Ocean, South China Sea, and Central-Southeast Pacific, a likelihood mapping analysis was performed in TREE-PUZZLE version 5.2 (Schmidt et al., 2002). Likelihood mapping triangle diagrams were used to compare the competing relationship between the four clusters and to assess the support values of internal branches (Strimmer and von Haeseler, 1996; Strimmer and von Haeseler, 1997).
Pairwise net genetic distances between and within different lineages were inferred using Dnet = Dxy − 1/2 * (Dx + Dy) (Nei, 1987) in MEGA X (Kumar et al., 2018), where Dxy, Dx, and Dy are the average numbers of nucleotide difference based on the Kimura 2 parameters correction (Kimura, 1980) between or within clades x and y (Nei and Li, 1979; Nei, 1987).
The divergence time between each pair of the S. oualaniensis lineages was estimated in BEAST v.1.8.2 based on the concatenation dataset. The TrN+I evolution model was used with empirically determined base frequencies. For the tree prior parameter, the Speciation : Birth–Death process (Gernhard, 2008) was used and modeled with an uncorrelated lognormal relaxed clock (Drummond et al., 2006). The following calibration node was constrained using lognormal distribution priors: 28.43 million years ago (Ma), coalescent time of S. oualaniensis and S. pteropus based on four fossils calibrated points (Pardo-Gandarillas et al., 2018). Three independent MCMC runs of 100 million generations with a sampling frequency of every 10,000 generations were performed. The log files were combined in Tracer to summarize posterior divergence times and 95% highest posterior density limits. Only runs where the ESS values over 200 for all marginal parameters were used, after discarding 10% of the first trees as burn-in.
The Bayesian binary method (BBM) implemented in RASA 4 (Yu et al., 2020) was used to reconstruct the ancestral area. Biogeographical data are based on the present distribution of S. oualaniensis and S. pteropus (Figure 1; Figure S1). The BBM analysis was run for 50 million cycles with 10 chains and a sampling frequency of every 1,000 cycles. The temperature was set at 0.1, and a fixed JC model was used. The maximum number of areas for each node was four. The information of each node was summarized and plotted as a pie chart.
Similar topologies were recovered using different tree-building methodologies under the concatanated data set (Figures 2 and S2) and species tree reconstruction (Figure 3). Three new clades (I, II, and III) of S. oualaniensis, with reliable nodal supports (BP/PP > 90/0.98), were consistently recovered by both of the phylogenetic methods (ML/BI). Clade I from the South China Sea is composed of two subclades diverging at ca. 0.27 (0.90–0.05) Ma (Dnet = 0.21%) (Table 1). Clade II and clade III distribute in the northern East Indian Ocean and southern East Indian Ocean, respectively. Based on the branching order, two basal clades (V and VI) were consistently observed with strong statistical support (BP/PP = 84/0.97, 100/1.0) (Figures 2 and S2). Clade VI/Equatorial, from the central Equatorial Pacific, was recovered to be basal to all other clades of S. oualaniensis coalescing at 14.27 (22.83–5.95) Ma (Dnet = 10.67%). Clade V/Eastern Typical from the northern East Pacific is the second most basal clade diverging from the remaining lineages (I–IV) at 3.40 (7.38–0.83) Ma (Dnet = 5.16%). A monophyletic East Indian Ocean branch grouping the two East Indian Ocean clades (II and III, Dnet = 1.99%) at 0.88 (1.9–0.25) Ma was also successfully recovered in ML and BI trees with low statistical support (BP/PP = 0.45/0.72) (Figures 2 and S2). Furthermore, this ancestral East Indian Ocean branch and clade IV/Pacific Typical coalesced at ca. 1.06 (2.48–0.31) Ma with medium-to-strong support (BP/PP = 0.69/0.97). The divergence time of clade I and clades II–IV was inferred to be 1.3 (3.0–0.4) Ma. The species tree reconstruction provided a similar topology to the ML and BI trees: among the four clades (I–IV), clade I was closely related to clade IV (Figure 3).
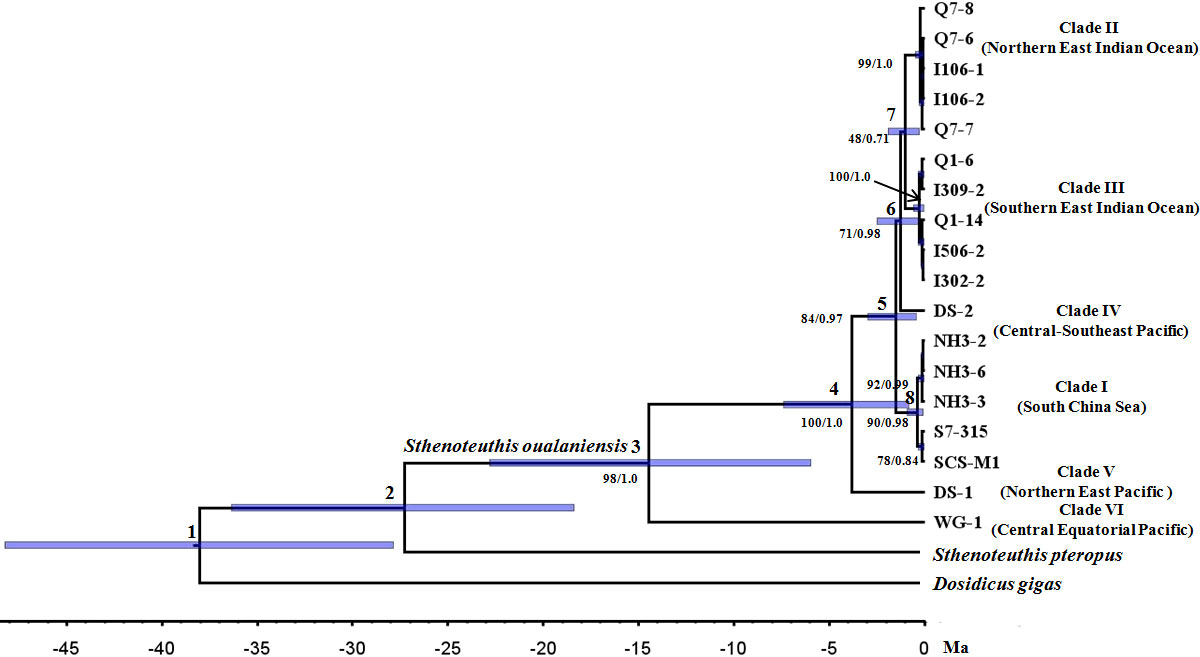
Figure 2 Bayesian relaxed clock cladogram based on concatenated mitochondrial cytochrome c oxidase subunit 1 (COI) and cytochrome b (Cyt b) sequences. Numbers above branches show ancestral nodes, and numbers below branches indicate nodal support values measured as bootstrap values (BP) and posterior probability (PP) inferred by maximum likelihood (ML) and Bayesian inference (BI) methodologies, respectively.
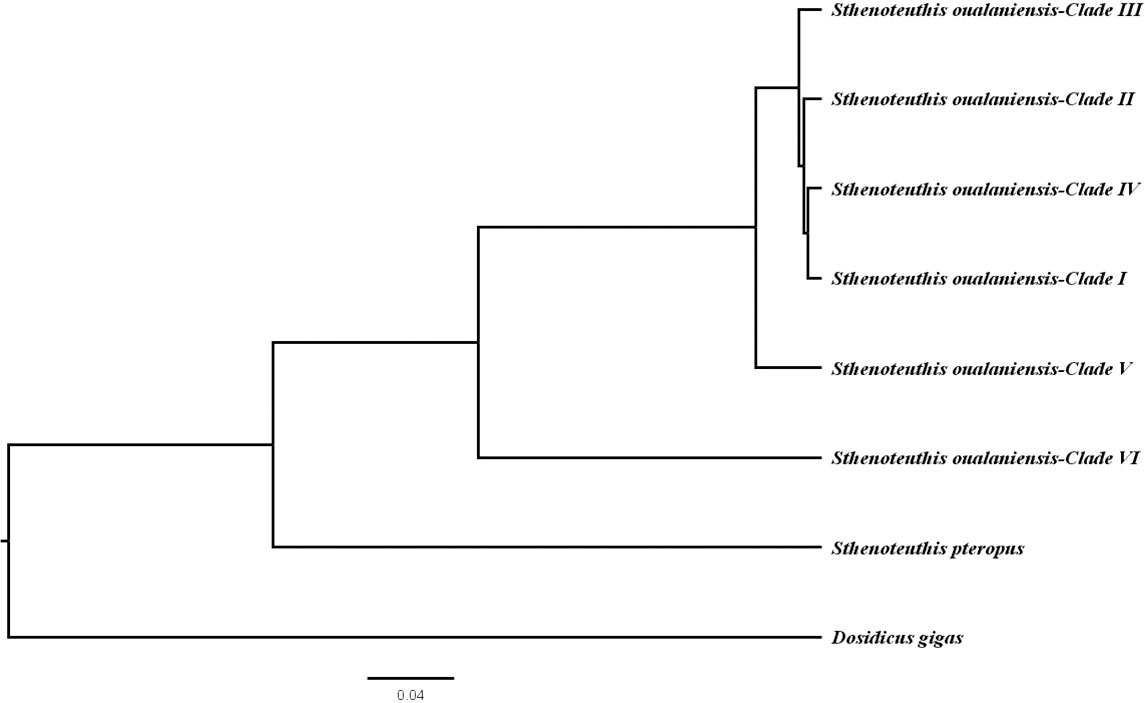
Figure 3 Species tree of the genus Sthenoteuthis showing the phylogenetic relationship of the species.
In order to further resolve the ambiguous phylogenetic relationships among clades I–IV, a quartet topology was reconstructed based on the concatenation dataset (Figure 4). The ancestral branch, grouping southern and northern East Indian Ocean clades (II and III) in Figure 4A, was strongly supported with the highest posterior weights (88.8% quartets; Figure 4D) relative to two other trees (Figures 4B, C). Thus, this ancestral East Indian Ocean branch was confirmed with reliable statistical support.
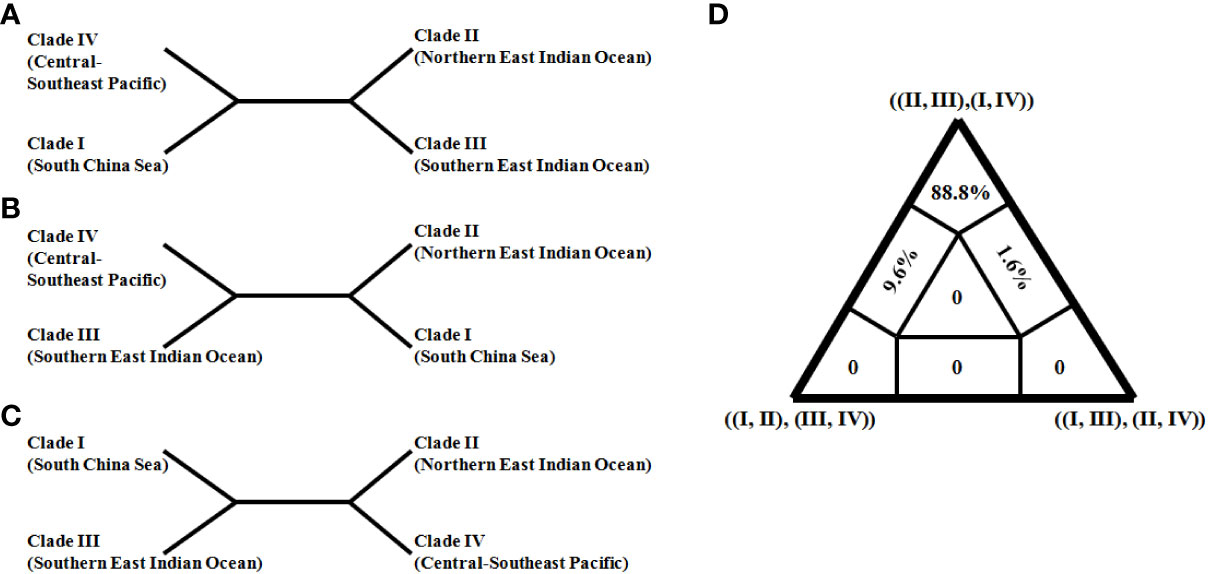
Figure 4 Three competing evolutionary relationships (A–C) among four clades (I–IV) of Sthenoteuthis oualaniensis and likely mapping triangle diagram (D). The East Indian Ocean monophylitic combination of ((II, III) and (I, IV)) obtained the highest support probability (88.8%).
The six lineages (clades I–VI) of S. oualaniensis with different divergence levels were also recovered in the network (Figure S3). However, relative to the deep divergence observed in S. oualaniensis, only one clade with two closely related haplotypes (SP1 and SP2) of S. pteropus was revealed (Figure S4).
The BBM revealed eight events of dispersal and seven events of vicariance along the phylogeny of D. gigas and Sthenoteuthis (Figure 5). Estimation of ancestral locality for the basal node (39) indicated that the recent common ancestor of D. gigas and Sthenoteuthis likely originated from the East Pacific (Southeast Pacific, PP = 0.47; northern East Pacific, PP = 0.31) at ca. 38 Ma. Node 38 showed that the Atlantic Ocean has a higher probability (PP = 0.57) to be the ancestral distribution of Sthenoteuthis relative to the northern East Pacific (PP = 0.20). Node 37 showed a mixed ancestral origin of S. oualaniensis in either the northern East Pacific (PP = 0.41) or central Equatorial Pacific (PP = 0.40). Node 36 indicated an ancestral distribution of clades I–V in the northern East Pacific (PP = 0.80). For the ancestral node 35 of clades I–IV, the South China Sea origin was indicated (PP = 0.58). Node 30 showed an ancestral origin of clades II–IV in the Central-Southeast Pacific (PP = 0.68). Node 29 suggested that the Indian Ocean ancestral population of clades II–III likely originated from the southern East Indian Ocean (PP = 0.54).
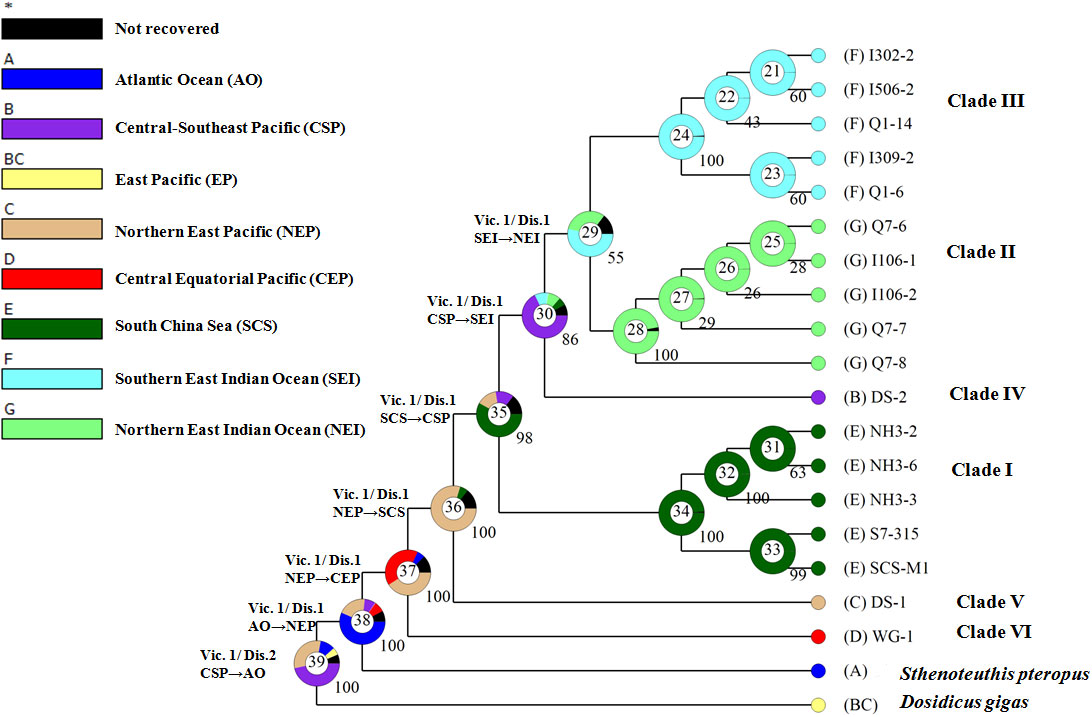
Figure 5 Historical biogeographic reconstruction of genus Sthenoteuthis and Dosidicus. Pie charts at each node show posterior probabilities of alternative ancestral distribution using different colors. Arrows show dispersal direction. Vic, vicariance; DIS, dispersal. Star "*" indicates lumped ranges with hidden probability less than 5%.
Based on morphological and behavioral characteristics, previous studies have identified two male forms (dwarf and middle-sized stocks) and five female forms (dwarf, early-maturing middle-sized form with double axes of rachis, and middle-sized form with single axes of rachis and giant form) of S. oualaniensis in the Indo-Pacific Ocean (Table S1; Nesis, 1977; Nesis, 1993). However, recent molecular investigations have suggested a more complicated population structure for S. oualaniensis throughout its range. For example, Staaf et al. (2010) recovered four clades/lineages from the Central-East Pacific, and Liu et al. (2017) identified three lineages/clades inhabiting the South China and Philippine Seas, while Xu et al. (2020) revealed two lineages/clades from the South China Sea and one lineage from the northern East Pacific (viz., clade V/Eastern Typical in this study). Here, we identified six well-supported genetic lineages with considerable population structure and genetic divergence distributed throughout the Indo-Pacific range of S. oualaniensis (Figures 1–3).
The deepest genetic divergence (Dnet = 10.67%) based on our concatenated COI and Cyt b datasets was observed between clade VI (dwarf form) from the central Equatorial Pacific and all other clades (I–V) (Figures 2, S3 and Table 1), which is similar to the divergence found between dwarf and middle-sized forms in previous studies (Li et al., 2019; Xu et al., 2020). The second deepest genetic divergence (Dnet = 5.16%) between clade V and clades I–IV is also similar to that of middle-sized forms between the East Pacific Ocean and South China Sea based on COI gene sequences (4.85%; Xu et al., 2020). These inter-clade divergences between clade VI and clades I–V or clade V and clades I–IV of S. oualaniensis are much higher than the mean level of mollusk intraspecific distance (<3%) (Yu et al., 2016). In general, a standard threshold greater than a 10-fold difference between interspecific and intraspecific genetic divergences has been used for species identification (Hebert et al., 2003; Hebert et al.,2004; Ward et al., 2005). Following this criterion, the dwarf form of clade VI found in the central Equatorial Pacific and the middle-sized form of clade V from the northern East Pacific may represent separate species or subspecies (Clarke, 1966; Li et al., 2019; Xu et al., 2020). Although no dwarf forms were presently sampled from the East Indian Ocean and the South China Sea, this form has been observed in the South China Sea (Li et al., 2019; Xu et al., 2020), suggesting ongoing transoceanic gene flow between the central Equatorial Pacific and the West Pacific.
Two morphological populations of S. oualaniensis, viz., dwarf form and middle-sized form, have been identified in the South China Sea (Liu et al., 2019; Wang et al., 2019). Based on Cyt b gene sequences, Liu et al. (2017) found two genetic lineages of S. oualaniensis (minor A and dominant B) inhabiting the South China, East China, and Philippine Seas. The genetic divergence (>1.67%) found between the two lineages (A and B) corresponds to the inter-clade divergence levels between I, II, III, and IV (1.41%–2.16%) of the middle-sized form found here (Table S3). Although no published data are available for comparison, the dominant cluster B could be analogous to clade I from the South China Sea in this study inferred from its high population abundance and sampled probability. Inferred from COI and H3 gene, the dwarf form and the dominant middle-sized form observed from the South China Sea by Xu et al. (2020) correspond well to clade VI and clade I in this study, respectively. Thus, at least three lineages including middle-sized form A, middle-sized form B/clade I, and dwarf form/clade VI have been revealed in the South China Sea, which also provide direct evidence for the three populations that were identified based on dorsal photophores by Jiang et al. (2015) (i.e., the middle-sized, dwarf-sized, and X3-sized forms). Two subclades of clade I with reliable nodal supports were further revealed from our limited sampling in the South China Sea (Figures 2, S2, S3), which may be indicative of high diversity and frequent population/lineages differentiation of S. oualaniensis in the Indo-West Pacific.
A divergence level of 1.99% was observed between clade II from the northern East Indian Ocean and clade III from the southern East Indian Ocean, which is far lower than that (10.33%–10.81%) observed between the dwarf form (clade VI) and middle-sized form (clades I–V), suggesting that both of these East Indian Ocean clades likely belong to different lineages of the middle-sized form in the Indian Ocean. Although the dominant middle-sized form and the dwarf form were found north of 10°N and south of 7°N along the neighboring southwest coast of India (Chembian and Mathew, 2014), no dwarf form was observed in the present sampling from the East Indian Ocean. Further studies will be needed to further distinguish these populations including the three middle-sized forms in the Indo-Pacific and the giant form in the Arabian Sea (Nesis, 1993; Snÿder, 1998) through integrating molecular and morphological data on a larger spatial scale.
Compared to deeply divergent lineages observed in S. oualaniensis from the Indo-Pacific, a single lineage was observed in S. pteropus from the Atlantic Ocean (Figure S4). Three possibilities can be invoked to explain the low divergence of S. pteropus: 1) recent range expansion and ongoing high gene flow transported by circulation likely have caused a homogeneous population structure of S. pteropus (He et al., 2010). 2) Population bottlenecks likely have erased other lineages of S. pteropus due to historical disaster events (Hewitt, 1996). 3) Insufficient sampling likely caused to artificially underestimate diversity of S. pteropus (Yan and Zhang, 2004). Further sampling on a larger spatial scale and more sample size will be helpful to uncover the population structure of S. pteropus.
Sthenoteuthis is thought to have originated in the Atlantic Ocean and subsequently migrated eastward into the Indian and Pacific Oceans (Zuyev et al., 1975; Zuyev et al., 2002). During the late Pliocene and early Pleistocene, low sea levels are hypothesized to have led to the genetic differentiation and speciation of Sthenoteuthis due to isolation between the Atlantic and Indo-Pacific Ocean.
Present molecular data, fossil records, simulated paleo-ocean circulations, and geologic clues partly support the abovementioned evolutionary origin of Sthenoteuthis. Biogeographical analysis based on a concatenated dataset suggested that the common ancestor of D. gigas and Sthenoteuthis likely originated from the East Pacific with a high probability of 0.78 (Central-Southeast Pacific, PP = 0.47; northern East Pacific, PP = 0.31) at ca. 38 Ma (Figure 5), and the ancestral origin of Sthenoteuthis was further inferred to be from the Atlantic Ocean with a probability of 0.57. Thus, the common ancestor of D. gigas and Sthenoteuthis was inferred to disperse from the Southeast Pacific to the Atlantic Ocean, and the ancestral population of Sthenoteuthis subsequently dispersed back to the northern East Pacific from the Atlantic Ocean.
The ancestral origin of D. gigas and Sthenoteuthis in the Pacific Ocean was also consistent with fossil clues of Ommastrephidae. The mean coalescent time of Ommastrephidae has been inferred to have occurred during the late Cretaceous (87.1 Ma; Pardo-Gandarillas et al., 2018), which preceded the earliest known Ommastrephidae fossils from the coastal northeast Atlantic Ocean, which are from the Middle Eocene (47.8–37.8 Ma) (Neige et al., 2016). However, the inferred coalescent age of Ommastrephidae is consistent with the lower jaw fossil of Teuthida from the Upper Cretaceous strata in Hokkaido, Japan (Tanabe et al., 2015). A large amount of the earliest known fossils and biodiversity hotspots of marine species including mangrove, corals, mollusk, and coral reef fish assemblages were observed in the proto-Pacific and Tethys Oceans (Piccoli et al., 1987; Bellwood, 1996; Renema et al., 2008; Neige et al., 2016). The common ancestor of D. gigas and Sthenoteuthis likely dispersed from the East Pacific to the Atlantic Ocean through the open Central American Seaway at 48–28 Ma followed by genetic divergence and speciation due to vicariance between the Pacific Ocean and Atlantic Ocean (Scotese, 2004).
Surface sea currents before 5 Ma promoted promoted the transoceanic migration of marine organisms between the Pacific Ocean and Atlantic Ocean. The North Equatorial Countercurrent extended eastward from the Pacific Ocean to the Caribbean Sea through the southern Panama Seaway during the Oligocene and Miocene (Maier-Reimer et al., 1990; Nisancioglu et al., 2003; Haug et al., 2004; von der Heydt and Dijkstra, 2006; Schneider and Schmittner, 2006; Sepulchre et al., 2014) (Figure 6A). However, the surface water of the upper 75 m flowed westward from the Atlantic Ocean to the East Pacific through the northern part of the open Central American Seaway. These bidirectional circulations might have carried larvae of marine organisms between the Pacific Ocean and Atlantic Ocean. Some transisthmus sister species pairs of octopus, hake (Merluccius spp.), and octopuses (Muusoctopus) from the East Pacific and Atlantic Oceans also shared a recent common ancestor from the East Pacific Ocean or the Atlantic Ocean (Grant and Leslie, 2001; Ibáñez et al., 2016; Lima et al., 2020).
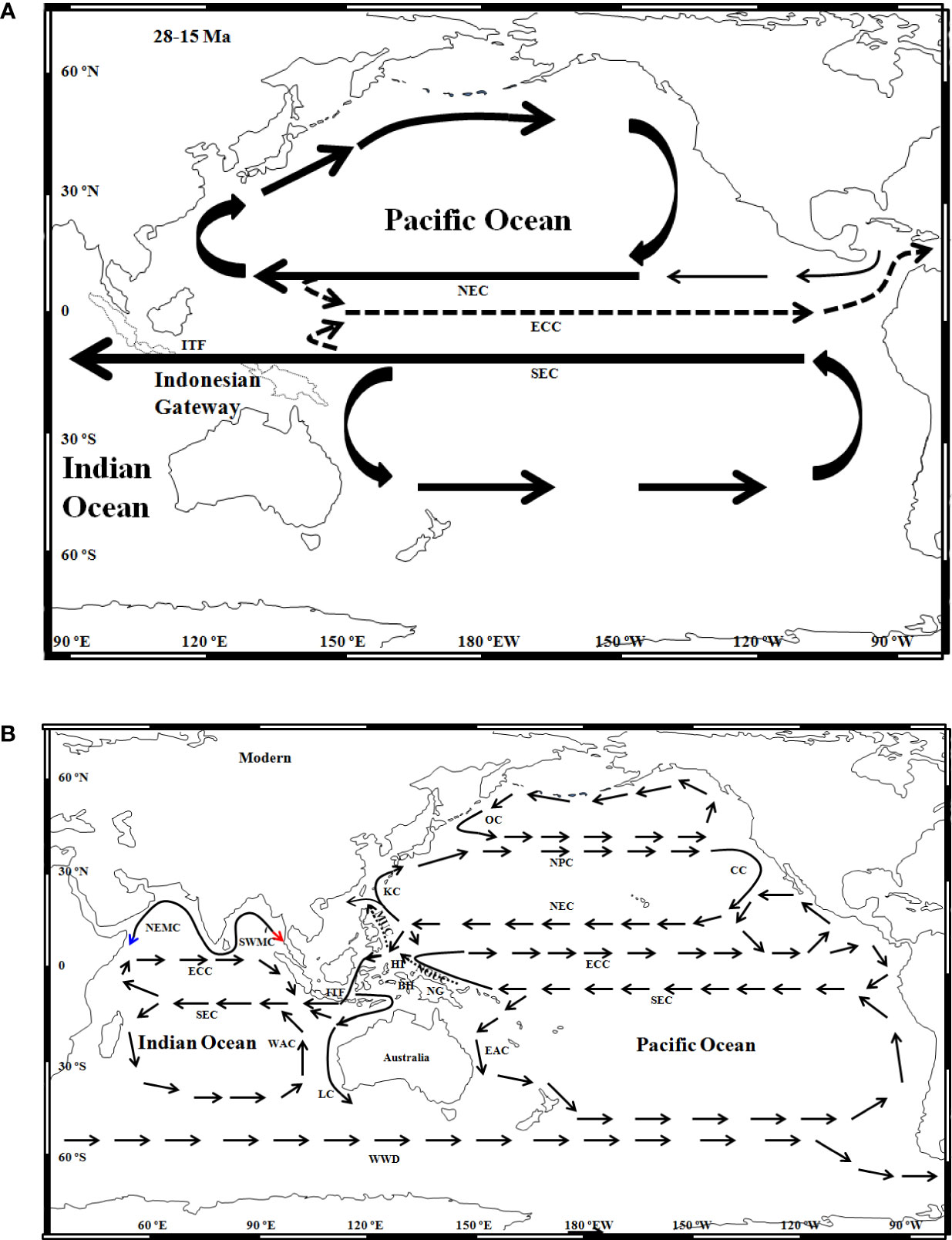
Figure 6 Circulation patterns in the Indo-Pacific Ocean during (A) the late Oligocene–Early Miocene (modified from Kennett et al., 1985; Sclater et al., 1985; von der Heydt and Dijkstra, 2006) and (B) modern (modified from Prell et al., 1980; Pollock, 1990; Pollock, 1992; Lukas et al., 1996; Morey et al., 1999; Kuhnt et al., 2004). BH, Bird Head; CC, California Current; EAC, East Australia Current; ECC, Equatorial Countercurrent; HI, Halmahera Island; ITF, Indonesian Throughflow; KC, Kuroshio Current; LC, Leeuwin Current; MUC, Mindanao Undercurrent; NG, New Guinea; NGUC, New Guinea Undercurrent; NEC, North Equatorial Current; NPC, North Pacific Current; NEMC, Northeast Monsoon Current; OC, Oyashio Current; SEC, South Equatorial Current; SWMC, Southwest Monsoon Current; WAC, West Australia Current; WWD, West Wind Drift. Blue and red arrows indicate directions of Northeast Monsoon circulation and Southwest Monsoon circulation, respectively. Dotted lines indicate undercurrents.
Lastly, the geologic history of the Central American Seaway may have also influenced speciation processes between the East Pacific and Atlantic oceans. The divergence times of D. gigas × Sthenoteuthis and S. oualaniensis × S. pteropus were traced to ca. 38 (48–28) Ma and 28 (40–17 Ma), respectively (Pardo-Gandarillas et al., 2018) (Figure 2). The western arc segment of Central America began a counterclockwise rotation between 38 and 28 Ma followed by a clockwise rotation of the central arc segment between 28 and 25 Ma, and the Central American Seaway narrowed ca. 25 Ma due to tectonic collision between Panama and South America (Farris et al., 2011; Montes et al., 2012). Moreover, an abrupt sea level regression of over 150 m occurred 35–30 Ma (Haq et al., 1987), which would have caused the exposure of shallow seas (Schaaf, 1996) and restricted water mass exchange between the East Pacific and West Atlantic Ocean preceding the near closure of the Central American Seaway (Montes et al., 2012). Low extinction and peak speciation rates have been observed in marine animals including neritic mollusks during the early Oligocene (McGowran, 1990). D. gigas × ancestral Sthenoteuthis and S. oualaniensis × S. pteropus were likely separated from each other due to habitat fragmentation and reduced gene flow (Hewitt, 1996) between the Pacific Ocean and the Atlantic Ocean during Oligocene. However, ancestral Sthenoteuthis origin should further be confirmed using some earlier fossils from the Atlantic Ocean because fossil statoliths from an extinct, potentially ancestral species (Symplectoteuthis pedroensis) of genus Sthenoteuthis have been found from Pliocene deposits in a coastal East Pacific locality (California, USA) (Clarke and Fitch, 1979).
Different from the aforementioned eastward migration pathway of ancestral Sthenoteuthis from the Atlantic Ocean to the Pacific Ocean through the Indian Ocean (Zuyev et al., 1975; Zuyev et al., 2002), multiple westward dispersal events were indicated by present molecular data (Figure 5): ancestral population of S. oualaniensis was inferred to firstly colonize the northern East Pacific from the Atlantic through the open Panama Seaway during late Oligocene (28–23 Ma), and the northern East Pacific further acted as dispersal origin of S. oualaniensis to central Equatorial Pacific and the South China Sea. Moreover, the ancestral Indian Ocean population was also inferred to have originated from Central-Southeast Pacific (clade IV) based on phylogenetic topologies and biogeographical analysis (Figures 2, 5). Westward migrations from the East Pacific to West Pacific and from the West Pacific to the East Indian Ocean were also observed in different marine species including fringehead blennies in the genus Neoclinus (Hongjamrassilp et al., 2020) and intertidal barnacle (Tetraclita squamosa) (Chan et al., 2022), respectively.
These inferred westward colonization routes correspond well to the paleo-circulation directions in the Indo-Pacific Ocean during the late Oligocene–Miocene (Kennett et al., 1985) (Figure 6A). Westward warm North Pacific Equatorial Current likely transported larvae of ancestral S. oualaniensis from the northern East Pacific to the Hawaii seamount or atolls. The population in the central Equatorial Pacific (clade VI) was firstly isolated from the remaining population (clades I–V) at ca. 14.3 (22.8–6.0) Ma. Similar population differentiation or speciation around the Hawaii island chain was also observed in other marine organisms, e.g., Panulirus marginatus (Pollock, 1992). These Hawaii seamounts/island chains could have served as a barrier to surface circulation and impeded larval gene flow between the eastern and western populations due to sea level regression during 15–6 Ma (Pollock, 1992; Woodruff, 2003).
Clade V from the northern East Pacific was subsequently separated from other clades (I–IV) at 3.4 (7.4–0.82) Ma (Figures 2; Tables 1), which indicates a possible evolutionary influence of the East Pacific Barrier (Ekman, 1953; Briggs, 1974) and seaway closure in the Pacific (Kennett et al., 1985). Panama Gateway was closed at ca. 3.5–3 Ma (O’Dea et al., 2016), which interrupted the westward water mass from the Atlantic Ocean to the East Pacific. With moving northward of the Australian Plate, the Indonesian Gateway became narrower than before and impeded South Pacific Equatorial Circulation into the Indian Ocean at ca. 3 Ma (Cane and Molnar, 2001). These seaways’ closures strengthened gyral circulations in the Indo-Pacific (Supplementary Material). As an endemic lineage, larvae of clade V were likely retained in the region by a counterclockwise circulation along the coast of Central America and could not drift into the westward North Equatorial Current (Figure 6B).
Clade I in the South China Sea was inferred to derive from the ancestral population in the northern East Pacific (Figure 5). The westward North Equatorial Current likely transported the ancestral population of clades I–V from the northern East Pacific to the West Pacific and South China Sea (Figure 6A). Furthermore, transoceanic larval exchange between the northern East Pacific and the South China Sea continued till the closure of the Panama and Indonesian seaways at ca. 3.4 Ma (Figure 2). Eastward Equatorial Countercurrent and Equatorial Undercurrent had been strengthened since these seaways’ closure in the Pacific (Kennett et al., 1985), which would be helpful to carry larvae of S. oualaniensis from the South China Sea to central Equatorial Pacific and Southeast Pacific (Figure 5). Marine species including crustaceans, fishes, and molluscan fauna in the South China Sea or Northwest Pacific could be transported to the central Equatorial Pacific and Southeast Pacific through the North Pacific Current, North Equatorial Current, and Equatorial Countercurrent or through southward coastal current along the east of the Philippines and the Equatorial Countercurrent (Randall et al., 1985; Scheltema, 1988; Ding and Bai, 1993; He et al., 2021). These alternative migration pathways can be distinguished through future sampling in a larger spatial scale from the Philippines Sea, central Equatorial Pacific, and Southwest Pacific. The South China Sea population was then isolated from the Central-Southeast Pacific population due to the glacial low sea level and semi-closed sea basin during the early Pleistocene (ca. 1.3 Ma) (Wang, 1990; Woodruff, 2003).
Based on the close evolutionary relationship between the monophyletic East Indian Ocean clades II–III and Pacific Typical clade (IV) (Figures 2, S2), an counterclockwise South Equatorial Circulation could have carried larvae of the ancestral clade IV from the Southeast Pacific to the West Pacific and Indian Oceans through the open Palaeo-Indonesian Gateway (Figures 5, 6A). The Indonesian Seaway and Indonesian Throughflow (ITF) were closely linked to the topography and bathymetry in this area (Supplementary Material). Closure of the Indonesian Gateway may have occurred more than once due to repeated tectonic events and sea level fluctuations since Miocene (Srinivasan and Sinha, 2000; von der Heydt and Dijkstra, 2006). Even if the deep-water exchange between the Pacific and Indian Oceans was restricted, surface circulation likely continued through narrower and shallower Indonesian seaway. The main composition of the ITF may have also gradually shifted from the warm South Equatorial Circulation to the cool North Equatorial water resulting from the closure of the Indonesian Seaway since 5–3 Ma (Cane and Molnar, 2001).
The divergence time of 1.1 (2.5–0.3) Ma between the East Indian Ocean clades (II–III) and Pacific Typical clade (IV) suggests an evolutionary influence of the early Pleistocene sea level fluctuation and closure of the Indonesian Seaway. Gene flow promoted by surface circulation between the Central-Southeast Pacific and East Indian Ocean could not have been interrupted completely until 1.1 Ma because a low proportion of the South Pacific water mass still kept flowing into the Indian Ocean through the Halmahera Sea and Banda Sea (Gordon and Fine, 1996). Nevertheless, frequent glacial sea level fluctuations over −120 m during the Pleistocene likely exposed shallow Indonesian seas and impeded gene flow between the Pacific and Indian Oceans (Woodruff, 2003). Similar speciation and lineage divergence due to ocean basins vicariance between the Indian Ocean and the Pacific Ocean were also observed in some other sister species pairs (Gaither and Rocha, 2013) and wide-ranging marine species including coconut crab (Birgus Zatro) (Lavery et al., 1996), green tiger prawn (Penaeus semisulcatus) (Abdul Halim et al., 2021), barnacle (Tetraclita kuroshioensis) (Chan et al., 2022), gastropod (Nerita albicilla) (Crandall et al., 2008), butterflyfishes species complex (Chaetodon punctatofasciatus and Chaetodon rhombochaetodon) (Mcmillan and Palumbi, 1995), and tropical reef fish (Chlorurus sordidus) (Bay et al., 2004). Thus, the closure of the Indonesian Seaway could have acted as a physical barrier to restrict transoceanic gene flow and promote population differentiation or speciation of widespread species between the Indian Ocean and the Pacific Ocean since the Miocene (Rocha et al., 2007).
Larvae of S. oualaniensis from the Central-Southeast Pacific firstly colonized the southern East Indian Ocean and then dispersed into the northern East Indian Ocean (Figure 5). Monsoon circulations and Equatorial Countercurrent in Bengal Bay (Schott et al., 2009) subsequently separated the ancestral East Indian Ocean population into two lineages (II and III) (Figure 6B). A similar pattern has been inferred for the north–south population differentiation of D. gigas and S. oualaniensis, which are hypothesized to have been influenced by Equatorial Countercurrents in the East Pacific (Staaf et al., 2010) due to strengthened gyral circulations in the Indian and Pacific Oceans after the closure of the Indonesian Gateway (Kennett et al., 1985). Thus, oceanographic circulation systems could be responsible for population differentiation or speciation by impeding the gene flow of wide-ranging marine species (Tsang et al., 2012).
In summary, at least eight evolutionary lineages including seven middle-sized forms (clades II and III from the East Indian Ocean, cluster B/clade I and cluster A from the South China Sea, clade IV from the Central-Southeast Pacific, clade V from the northern East Pacific, and Central Typical clade from the central Equatorial Pacific) and a dwarf form (clade VI) from the central Equatorial Pacific and the South China Sea have been identified in S. oualaniensis through comparing present and previous studies based on molecular data (Staaf et al., 2010; Liu et al., 2017; Xu et al., 2020). Tectonic events and oceanic circulations played important roles in influencing migration pathways, population differentiation, and speciation of Sthenoteuthis (Figure 7). The open Panama Seaway and Indonesian Gateway probably served as migration corridors of marine species transported by transoceanic circulations from the East Pacific to the West Atlantic or West Pacific and Indian Ocean (Figure 7A). Subsequently, gradually weakened circulations likely caused population differentiation or speciation due to the closure of these inter-continental seaways (Figure 7B). This study provided lineage divergence evidence to the model “center of overlap” in the Coral Triangle. However, more attention should further be paid to evolutionary links among geological, physical, or chemical oceanographic conditions and marine organisms to explain the origin of the biodiversity hotspot in the East Indies Triangle.
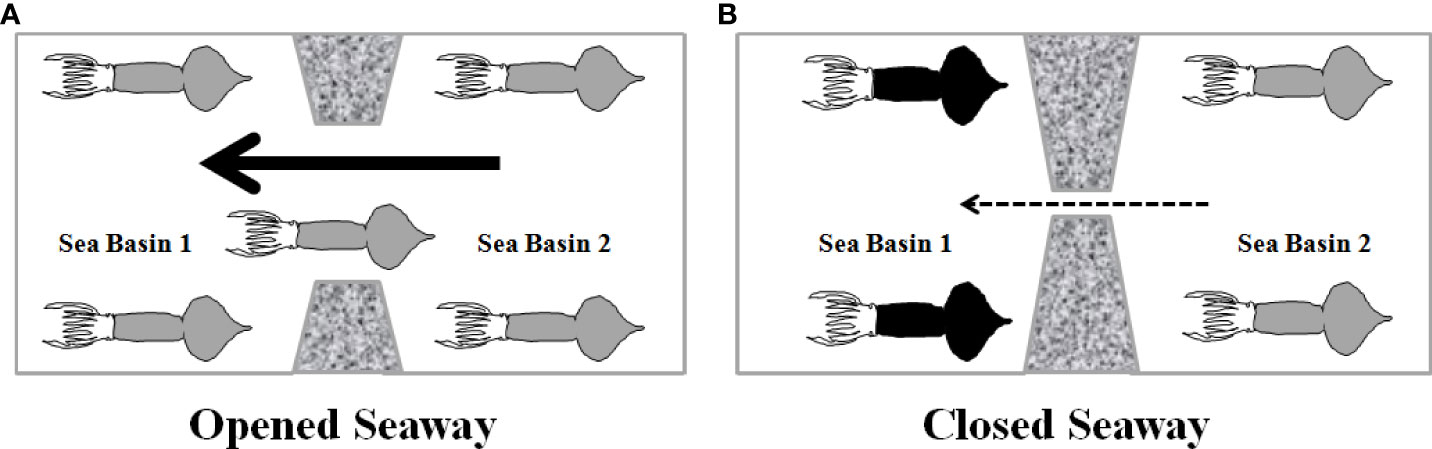
Figure 7 Biogeographic role of seaway in marine organisms: (A) genetic homogenous population between two oceans due to high migration rates carried by strong throughflow through open seaway. (B) Habitat fragmentation and population divergence between two sea basins attributed to weak throughflow and reduced gene flow through closed seaway. Solid and broken arrows indicate strong and weak throughflows, respectively.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) of the DNA sequences can be found in the article/Supplementary Material.
The animal study was reviewed and approved by East China Normal University Animal Ethics Committee (W20211202).
LJH, ZLS, and JingZ conceived and designed the research and experiments. JieZ, FHT, and JingZ collected samples. LJH, SSX, and HMR performed the experiments. LJH and SSX analyzed the data. LJH wrote the paper. DW and JingZ contributed reagents/materials/analysis tools. LJH, DW, and JingZ revised the manuscript. All authors contributed to the article and approved the submitted version.
This study was financially supported by the National Key R&D Program of China (2018YFD0900904, 2018YFD0900902), National Natural Science Foundation of China (42090043, 42025603), Strategic Priority Research Program of the Chinese Academy of Sciences (XDB42030203), and “One Belt and One Road” International Cooperation Project of Shanghai (18230743200).
We sincerely thank Xiaoyan Wu, Ying Wu, and Jinzhou Du for their kind encouragement and support during the study and manuscript preparation. Jinlong Dai, Yixue Zhang, Yao Wang, Zhengbo Liu, Xin Shi, and Xiaohui Zhang provided great help in the field sampling.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2022.891236/full#supplementary-material
Abdul Halim S. A. A., Othman A. S., Akib N. A. M., Jamaludin N. A., Esa Y., Nor S. A. M. (2021). Mitochondrial markers identify a genetic boundary of the green tiger prawn (Penaeus semisulcatus) in the Indo-Pacific Ocean. Zool. Stud. 60, 8. doi: 10.6620/ZS.2021.60-08.
Bandelt H. J., Forster P., Ro¨hl A. (1999). Median-joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 16, 37–48. doi: 10.1093/oxfordjournals.molbev.a026036
Bay L. K., Choat J. H., van Herwerden L., Robertson D. R. (2004). High genetic diversities and complex genetic structure in an Indo-Pacific tropical reef fish (Chlorurus sordidus): evidence of an unstable evolutionary past? Mar. Biol. 144, 757–767. doi: 10.1007/s00227-003-1224-3
Bellwood D. R. (1996). The Eocene fishes of Monte bolca: the earliest coral reef fish assemblage. Coral Reefs 15, 11–19. doi: 10.1007/BF01626074
Bellwood D. R., Meyer C. P. (2009). Searching for heat in a marine biodiversity hotspot. J. Biogeogr. 36, 569–576. doi: 10.1111/j.1365-2699.2008.02029.x
Briggs J. C. (2000). Centrifugal speciation and centres of origin. J. Biogeogr. 27, 1183–1188. doi: 10.1046/j.1365-2699.2000.00459.x
Briggs J. C. (2003). Marine centres of origin as evolutionary engines. J. Biogeogr. 30, 1–18. doi: 10.1046/j.1365-2699.2003.00810.x
Briggs J. C. (2009). Diversity, endemism and evolution in the Coral Triangle. J. Biogeogr. 36, 2005–2012. doi: 10.1111/j.1365-2699.2009.02146.x
Cane M. A., Molnar P. (2001). Closing of the Indonesian seaway as a precursor to east African aridification around 3–4 million years ago. Nature 411, 157–162. doi: 10.1038/35075500
Carvalho G. R., Thompson A., Stoner A. L. (1992). Genetic diversity and population differentiation of the shortfin squid Illex argentinus in the south-west Atlantic. J. Exp. Mar. Biol. Ecol. 158, 105–121. doi: 10.1016/0022-0981(92)90311-W
Chan B. K. K., Tsao Y. F., Wangkulangkul K., Amjud K., Sukparangsi W. (2022). Biogeography and biodiversity of the intertidal barnacle Tetraclita species in the Gulf of Thailand and Andaman Sea – influences of oceanographic currents and Pleistocene glaciations. Front. Mar. Sci. 8, 774041. doi: 10.3389/fmars.2021.774041
Chembian A. J., Mathew S. (2014). Population structure of the purpleback squid Sthenoteuthis oualaniensis (Lesson 1830) along the south-west coast of India. Indian J. Fish 61 (3), 20–28. Retrieved from https://epubs.icar.org.in/index.php/IJF/article/view/23110.
Chen X. J., Liu B. L., Chen Y. (2008). A review of the development of Chinese distant-water squid jigging fisheries. Fish. Res. 89, 211–221. doi: 10.1016/j.fishres.2007.10.012
Chen X. J., Liu B. L., Tian S. Q., Qian W. G., Zhao X. H. (2007). Fishery biology of purpleback squid, Sthenoteuthis oualaniensis, in the northwest Indian Ocean. Fish. Res. 83, 98–104. doi: 10.1016/j.fishres.2006.09.005
Clarke M. R. (1966). A review of the systematics and ecology of oceanic squids. Adv. Mar. Biol. 4, 91–300. doi: 10.1016/S0065-2881(08)60314-4
Clarke M. R., Fitch J. E. (1979). Statoliths of Cenozoic teuthoid Cephalopods from North America. Palaeontology 22 (2), 479–511.
Crandall E. D., Frey M. A., Grosberg R. K., Barber P. H. (2008). Contrasting demographic history and phylogeographical patterns in two Indo-Pacific gastropods. Mol. Ecol. 17, 611–626. doi: 10.1111/j.1365-294X.2007.03600.x
Drummond A. J., Ho S. Y., Phillips M. J., Rambau T. A. (2006). Relaxed phylogenetics and dating with confidence. PloS Biol. 4 (5), e88. doi: 10.1371/journal.pbio.0040088
Drummond A. J., Rambaut A. (2007). BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 7 (1), 214. doi: 10.1186/1471-2148-7-214
Farris D. W., Jaramillo C., Bayona G., Restrepo-Moreno S. A., Montes C., Cardona A., et al. (2011). Fracturing of the Panamanian isthmus during initial collision with south America. Geology 39, 1007–1010. doi: 10.1130/G32237.1
Felsenstein J. (1981). Evolutionary trees from DNA sequences: A maximum likelihood approach. J. Mol. Evol. 17, 368–376. doi: 10.1007/BF01734359
Felsenstein J. (1985). Confidence limits on phylogenies: An approach using the bootstrap. Evolution 39, 783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x
Gaither M. R., Rocha L. A. (2013). Origins of species richness in the Indo-Malay-Philippine biodiversity hotspot: evidence for the centre of overlap hypothesis. J. Biogeogr. 40, 1638–1648. doi: 10.1111/jbi.12126
Gernhard T. (2008). The conditioned reconstructed process. J. Theor. Biol. 253, 769–778. doi: 10.1016/j.jtbi.2008.04.005
Gordon A. L., Fine R. (1996). Pathways of water between the Pacific and Indian oceans in the Indonesian seas. Nature 379 (6561), 146–149. doi: 10.1038/379146a0.
Grant W. S., Leslie R. W. (2001). Inter-ocean dispersal is an important mechanism in the zoogeography of hakes (Pisces: Merluccius spp.). J. Biogeogr. 28, 699–721. doi: 10.1046/j.1365-2699.2001.00585.x
Guindon S., Dufayard J. F., Lefort V., Anisimova M., Hordijk W., Gascuel O. (2010). New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst. Biol. 59, 307–321. doi: 10.1093/sysbio/syq010
Haq B. U., Hardenbol J., Vail P. R. (1987). Chronology of fluctuating sea levels since the Triassic. Science 235, 1156–1167. doi: 10.1126/science.235.4793.1156
Haug G. H., Tiedemann R., Keigwin L. D. (2004). How the Isthmus of Panama put ice in the Arctic. Oceanus 42 (2), 1–4.
Head J. J., Polly P. D. (2015). Evolution of the snake body form reveals homoplasy in amniote hox gene function. Nature 520 (7545), 86–89. doi: 10.1038/nature14042.
Hebert P. D. N., Cywinska A., Ball S. L. (2003). Biological identifications through DNA barcodes. P. R. Soc. B: Biol. Sci. 270 (1512), 313–321. doi: 10.1098/rspb.2002.2218.
Hebert P. D. N., Stoeckle M. Y., Zemlak T. S., Francis C. M. (2004). Identification of birds through DNA barcodes. PloS Biol. 2, e312. doi: 10.1371/journal.pbio.0020312
He L. J., Ren H. M., Xu S. S., Zhang J. (2021). Phylogeographic pattern of marine fauna in the Indo-West Pacific. Oceanol. Limnol. Sin. 52 (2), 468–485. doi: 10.11693/hyhz20200900260.
Hewitt G. M. (1996). Some genetic consequences of ice ages, and their role in divergence and speciation. Biol. J. Linn. Soc. 58, 247–276. doi: 10.1006/bijl.1996.0035
He L. J., Zhang A. B., Weese D., Zhu C. D., Jiang C. J., Qiao Z. G. (2010). Late pleistocene expansion of Scylla paramamosain along the coast of China: a population dynamic response to the last interglacial sea level highstand. J. Exp. Mar. Biol. Ecol. 385, 20–28. doi: 10.1016/j.jembe.2010.01.019
Hoeksema B. W. (2007). “Delineation of the Indo-Malayan Centre of Maximum Marine Biodiversity: The Coral Triangle, in Biogeography, Time, and Place: Distributions, Barriers, and Islands, vol. 29 . Ed. Renema W. (Berlin:Springer), 117–178.
Hongjamrassilp W., Murase A., Miki R., Hastings P. A. (2020). Journey to the west: trans-pacific historical biogeography of fringehead blennies in the genus Neoclinus (Teleostei: Blenniiformes). Zool. Stud. 59, 9. doi: 10.6620/ZS.2020.59-09.
Huelsenbeck J. P., Ronquist F. (2001). MRBAYES: Bayesian inference of phylogeny. Bioinformatics, 17(8), 754–755. doi: 10.1093/bioinformatics/17.8.754
Ibáñez C. M., Pardo-Gandarillas M. C., Peña F., Gleadall I. G., Poulin E., Sellanes J. (2016). Phylogeny and biogeography of Muusoctopus (Cephalopoda: Enteroctopodidae). Zool. Scripta. 45, 494–503. doi: 10.1111/zsc.12171
Jereb P., Roper C. F. E. (2010). Cephalopods of the world. An annotated and illustrated catalogue of species known to date. Myopsid and Oegopsid Squids. FAO Species Catalogue for Fishery Purposes. 2. Rome, FAO
Jiang Y. E., Zhang P., Lin Z. J., Qiu Y. S., Fang Z. Q., Chen Z. Z. (2015). Statolith morphology of purpleback flying squid (Sthenoeuthis oualaniensis) in the offshore south China Sea. S. China Fish. Sci. 11 (5), 27–37. doi: 10.3969/j.issn.2095-0780.2014.04.014.
Kennett J. P., Keller G., Srinivasan M. S. (1985). Miocene planktonic foraminiferal biogeography and paleogeographic development of the Indo-Pacific region, the Miocene ocean: Paleogeography and biogeography. Geol. Soc Am. Mem 163, 197–236. doi: 10.1130/mem163-p197.
Kimura M. (1980). A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16, 111–120. doi: 10.1007/BF01731581
Kuhnt W., Holbourn A., Hall R., Zuvela M., Käse R. (2004). “Neogene history of the Indonesian Throughflow,” in Continent-ocean interactions within the East Asian marginal seas, vol. 149 . Eds. Clift P., Kuhnt W., Wang P.X., Hayes D. (Florida AVE NW, Washington: American Geophysical Union) 299–320.
Kumar S., Stecher G., Li M., Knyaz C., Tamura K. (2018). MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Lavery S., Moritz C., Fielder D. R. (1996). Indo-Pacific population structure and evolutionary history of the coconut crab Birgus latro. Mol. Ecol. 5, 557–570. doi: 10.1111/j.1365-294X.1996.tb00347.x
Lima F. D., Strugnell J. M., Leite T. S., Lima S. M. Q. (2020). A biogeographic framework of octopod species diversification: the role of the Isthmus of Panama. PeerJ 8, e8691. doi: 10.7717/peerj.8691
Liu B. L., Chen X. J., Wang X. H., Du F. Y., Fang Z., Xu L. L. (2019). Geographic, intraspecific and sexual variation in beak morphology of purpleback flying squid (Sthenoteuthis oualaniensis) throughout its distribution range. Mar. Freshw. Res. 70, 417–425. doi: 10.1071/MF18078
Liu L. W., Zhou Y. D., Lu H. J., Liu B. L., Chen X. J. (2017). Genetic structure of Sthenoteuthis oualaniensis population in the Northwest Pacific Ocean. J. Fish. China 41 (9), 1355–1364. doi: 10.11964/jfc20160910527.
Li M., Zhang P., Zhang J., Zhang K., Chen Z. Z. (2019). Genetic differentiation of the purpleback flying squid, Sthenoteuthis oualaniensis, in the South China Sea: population or species divergence. J. Fish. Sci. China 26 (1), 133–140. doi: 10.3724/SP.J.1118.2019.18193.
Lukas R., Yamagata T., McCreary J. P. (1996). Pacific low-latitude western boundary currents and the Indonesian throughflow. J. Geophys. Res-Oceans 101 (C5), 12209–12216. doi: 10.1029/96JC01204
Maier-Reimer E., Mikolajewicz U., Crowley T. (1990). Ocean general circulation model sensitivity experiment with an open Central American Isthmus. Paleoceanography 5 (3), 349–366. doi: 10.1029/PA005i003p00349
Mcmillan W. O., Palumbi S. R. (1995). Concordant evolutionary patterns among Indo-West Pacific butterflyfishes. P. R. Soc B. 260, 229–236. doi: 10.1098/rspb.1995.0085
Miya M., Nishida M. (2000). Use of mitogenomic information in teleostean molecular phylogenetics: A tree-based exploration under the maximum-parsimony optimality criterion. Mol. Phylogenet. Evol. 17 (3), 437–455. doi: 10.1006/mpev.2000.0839
Montes C., Bayona G., Cardona A., Buchs D. M., Silva C. A., Morón S., et al. (2012). Arc-continent collision and orocline formation: Closing of the Central American seaway. J. Geophys. Res. 117, B04105. doi: 10.1029/2011JB008959
Mora C., Chittaro P. M., Sale P. F., Kritzer J. P., Ludsin S. A. (2003). Patterns and processes in reef fish diversity. Nature 421, 933–936. doi: 10.1038/nature01393
Morey S. L., Shriver J. F., OʼBrien J. J. (1999). The effects of Halmahera on the Indonesian throughflow. J. Geophys. Res. 104 (C10), 23281–23296. doi: 10.1029/1999JC900195
Muhammad F., Lü Z. M., Liu L. Q., Gong L., Du X., Shafi M., et al. (2018). Genetic structure of Octopus minor around Chinese waters as indicated by nuclear DNA variations (Mollusca, Cephalopoda). ZooKeys 775, 1–14. doi: 10.3897/zookeys.775.24258
Neige P., Lapierre H., Merle D. (2016). New Eocene coleoid (Cephalopoda) diversity from statolith remains: Taxonomic assignation, fossil record analysis, and new data for calibrating molecular phylogenies. PloS One 11 (5), e0154062. doi: 10.1371/journal.pone.0154062
Nei M., Li W. (1979). Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc. Natl. Acad. Sci. U.S.A. 76, 5269–5273. doi: 10.1073/pnas.76.10.5269
Nesis K. N. (1977). Population structure of the squid Sthenoteuthis oualaniensis (Lesson 1930) (Ommastrephidae) in the tropical Western Pacific. Trudy Inst. Okeanol. P.P. Shirshov 107, 15–29.
Nesis K. N. (1993). “Population structure of oceanic ommastrephids, with particular reference to sthenoteuthis oualaniensis: a review,” in Recent advances in fisheries biology, vol. 375–383 . Eds. Okutani T., O’Dor R. K., Kubodera T. (Tokyo:Takai University Press).
Nisancioglu K. H., Raymo M. E., Stone P. H. (2003). Reorganization of Miocene deep water circulation in response to the shoaling of the Central American seaway. Paleoceanography 18 (1), 1006. doi: 10.1029/2002PA000767
O’Dea A., Lessios H. A., Coates A. G., Eytan R. I., Restrepo-Moreno S. A., Cione A. L. (2016). Formation of the Isthmus of Panama. Sci. Adv. 2, e1600883. doi: 10.1126/sciadv.1600883
Palumbi S. R. (1997). Molecular biogeography of the Pacific. Coral Reefs 16 (Sl), S47–S52. doi: 10.1007/s003380050241
Pardo-Gandarillas M. C., Torres F. I., Fuchs D., Ibáñez C. M. (2018). Updated molecular phylogeny of the squid family ommastrephidae: Insights into the evolution of spawning strategies. Mol. Phylogenet. Evol. 120, 212–217. doi: 10.1016/j.ympev.2017.12.014
Piccoli G., Santori S., Franchino A. (1987). “Benthic molluscs of shallow tethys and their destiny. shallow tethys 2,” in Proceedings of the International Symposium on Shallow Tethys 2. Ed. McKenzie K. G. (Rotterdam: Balkema). 333–373.
Pollock D. E. (1990). Palaeoceanography and speciation in the spiny lobster genus Jasus. B. Mar. Sci. 46 (2), 387–405.
Pollock D. E. (1992). Palaeoceanography and speciation in the spiny lobster genus Panulirus in the Indo-Pacific. B. Mar. Sci. 51 (2), 135–146.
Posada D., Crandall K. A. (1998). Modeltest: testing the model of DNA substitution. Bioinformatics 14, 817–818. doi: 10.1093/bioinformatics/14.9.817
Prell W. L., Hutson W. H., Williams D. F., Bé A. W. H., Geitzenauer K., Molfino B. (1980). Surface circulation of the Indian Ocean during the last glacial maximum, approximately 18000 yr B.P. Quat. Res. 14, 309–336. doi: 10.1016/0033-5894(80)90014-9
Rambaut A. (2009). FigTree v1.4. Available at: http://tree.bio.ed.ac.uk/software/tracer.
Rambaut A., Drummond A. J. (2009). Tracer v.1.5. Available at: http://beast.bio.ed.ac.uk/LogCombiner.
Randall J. E., Lobel P. S., Chave E. H. (1985). Annotated checklist of the fishes of Johnston Island. Pac. Sci. 39, 24–79.
Renema W., Bellwood D. R., Braga J. C., Bromfield K., Hall R., Johnson K. G., et al. (2008). Hopping hotspots: Global shifts in marine biodiversity. Science 321, 654–657. doi: 10.1126/science.1155674
Roberts C. M., McClean C. J., Veron J. E. N., Hawkins J. P., Allen G. R., McAllister D. E., et al. (2002). Marine biodiversity hotspots and conservation priorities for tropical reefs. Science 295, 1280–1284. doi: 10.1126/science.1067728
Rocha L. A., Craig M. T., Bowen B. W. (2007). Phylogeography and the conservation of coral reef fishes. Coral Reefs 26, 501–512. doi: 10.1007/s00338-007-0261-7
Ronquist F., Teslenko M., Mark P. V. D., Ayres D. L., Darling A., Hohna S., et al. (2012). MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61, 539–542. doi: 10.1093/sysbio/sys029
Santaclara F. J., Espiñeira M., Vieites J. M. (2007). Genetic identification of squids (families Ommastrephidae and Loliginidae) by PCR–RFLP and FINS methodologies. J. Agr. Food Chem. 55, 9913–9920. doi: 10.1021/jf0707177
Schaaf A. (1996). Sea Level changes, continental shelf morphology, and global paleoecological constraints in the shallow benthic realm: a theoretical approach. Palaeogeogr. Palaeoclimatol. Palaeoecol. 121, 259–271. doi: 10.1016/0031-0182(95)00085-2
Scheltema R. S. (1988). Initial evidence for the transport of teleplanic larvae of benthic invertebrates across the East Pacific barrier. BioI. Bull. 174, 145–152. doi: 10.2307/1541781
Schmidt H. A., Strimmer K., Vingron M., von Haeseler A. (2002). TREE-PUZZLE: maximum likelihood phylogenetic analysis using quartets and parallel computing. Bioinformatics 18 (3), 502–504. doi: 10.1093/bioinformatics/18.3.502
Schneider B., Schmittner A. (2006). Simulating the impact of the Panamanian seaway closure on ocean circulation, marine productivity and nutrient cycling. Earth Planet. Sci. Lett. 246, 367–380. doi: 10.1016/j.epsl.2006.04.028
Schott F. A., Xie S. P., McCreary J. P. (2009). Indian Ocean circulation and climate variability. Rev. Geophys. 47, RG1002. doi: 10.1029/2007RG000245
Sclater J. G., Meinke L., Bennett A., Murphy C. (1985). “The depth of the ocean through the Neogene,” in The Miocene ocean: Paleogeography and biogeography, Kennett J. P. (Boulder, CO: Geological Society of America) vol. 163, 1–20.
Sepulchre P., Arsouze T., Donnadieu Y., Dutay J. C., Jaramillo C., Bras J. L., et al. (2014). Consequences of shoaling of the Central American seaway determined from modeling Nd isotopes. Paleoceanography 29, PA002501. doi: 10.1002/2013PA002501
Snÿder R. (1998). Aspects of the biology of the giant form of Sthenoteuthis oualaniensis (Cephalopoda: Ommastrephidae) from the Arabian Sea. J. Mollus. Stud. 64, 21–34. doi: 10.1093/mollus/64.1.21
Srinivasan M. S., Sinha D. K. (2000). Ocean circulation in the tropical Indo-Pacific during early Pliocene (5.6-4.2 ma): Paleobiogeographic and isotopic evidence. Proc. Indian Acad. Sci. (Earth Planet. Sci.) 109 (3), 315–328. doi: 10.1007/bf03549815
Staaf D. J., Ruiz-Cooley R. I., Elliger C., Lebaric Z., Campos B., Markaida U., et al. (2010). Ommastrephid squids Sthenoteuthis oualaniensis and Dosidicus gigas in the eastern Pacific show convergent biogeographic breaks but contrasting population structures. Mar. Ecol. Prog. Ser. 418, 165–178. doi: 10.3354/meps08829
Strimmer K., von Haeseler A. (1996). Quartet puzzling: A quartet maximum–likelihood method for reconstructing tree topologies. Mol. Biol. Evol. 13, 964–969. doi: 10.1093/oxfordjournals.molbev.a025664
Strimmer K., von Haeseler A. (1997). Likelihood–mapping: A simple method to visualize phylogenetic content of a sequence alignment. Proc. Natl. Acad. Sci. U.S.A. 94, 6815–6819. doi: 10.1073/pnas.94.13.6815
Tanabe K., Misaki A., Ubukata T. (2015). Late Cretaceous record of large soft-bodied coleoids based on lower jaw remains from Hokkaido, Japan. Acta Palaeontol. Pol. 60, 27–38. doi: 10.4202/app.00052.2013.
Thompson J. D., Gibson T. J., Plewniak F., Jeanmougin F., Higgins D. G. (1997). The clustal X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25, 4876–4882. doi: 10.1093/nar/25.24.4876
Tsang L. M., Achituv Y., Chu K. H., Chan B. K. K. (2012). Zoogeography of intertidal communities in the West Indian Ocean as determined by ocean circulation systems: Patterns from the Tetraclita barnacles. PloS One 7 (9), e45120. doi: 10.1371/journal.pone.0045120
von der Heydt A., Dijkstra H. A. (2006). Effect of ocean gateways on the global ocean circulation in the late Oliocene and early Miocene. Paleoceanography 21, PA1011. doi: 10.1029/2005PA001149
Wakabayashi T., Wada S., Ochi Y., Ichii T., Sakai M. (2012). Genetic differentiation of the neon flying squid Ommastrephes bartramii between North Pacific and South Atlantic populations. Nippon Suisan Gakk 78 (2), 198–203. doi: 10.2331/suisan.78.198
Wang P. X. (1990). The ice-age China Sea–status and problems. Quaternary Sicences 2, 111–124. doi: 10.3321/j.issn:1001-7410.1990.02.002.
Wang X. H., Liu B. L., Qiu Y. S., Zhang P., Du F. Y., Chen X. J., et al. (2019). Regional and intraspecific variation in Sthenoteuthis oualaniensis (Cephalopoda: Ommastrephidae) morphology from the western central Pacific Ocean. B. Mar. Sci. 95 (1), 1–12. doi: 10.5343/bms.2017.1173
Ward R. D., Zemlak T. S., Innes B. H., Last P. R., Hebert P. D. N. (2005). DNA Barcoding australia’s fish species. Philos. T. R. Soc B Biol. Sci. 360 (1462), 1847–1857. doi: 10.1098/rstb.2005.1716.
Woodruff D. S. (2003). Neogene marine transgressions, palaeogeography and biogeographic transitions on the Thai–Malay Peninsula. J. Biogeo. 30, 551–567. doi: 10.1046/j.1365-2699.2003.00846.x
Wu T. H., Tsang L. M., Chen I. S., Chu K. H. (2016). Multilocus approach reveals cryptic lineages in the goby Rhinogobius duospilus in Hong Kong streams: Role of paleodrainage systems in shaping marked population differentiation in a city. Mol. Phylogenet. Evol. 104, 112–122. doi: 10.1016/j.ympev.2016.07.014
Xu L., Liu P., Wang X. H., Damme K. V., Du F. Y. (2020). Phylogenetic relationships and cryptic species in the genus Sthenoteuthis (Cephalopoda: Ommastrephidae) in the South China Sea. Mol. Phylogenet. Evol. 149, 106846. doi: 10.1016/j.ympev.2020.106846
Yan L. N., Zhang D. X. (2004). Effects of sample size on various genetic diversity measures in population genetic study with microsatellite DNA markers. Acta Zool. Sin. 50 (2), 279–290. doi: 10.3969/j.issn.1674-5507.2004.02.019.
Yu Y., Blair C., He X. J. (2020). RASP 4: ancestral state reconstruction tool for multiple genes and characters. Mol. Biol. Evol. 37 (2), 604–606. doi: 10.1093/molbev/msz257
Yu H., Kong L. F., Li Q. (2016). Evaluation of the efficacy of twelve mitochondrial protein-coding genes as barcodes for mollusk DNA barcoding. Mitochondrial. DNA Part A 27 (2), 1336–1339. doi: 10.3109/19401736.2014.945579
Zhang D. X., Hewitt G. M. (1998). “Isolation of animal cellular total DNA,” in Molecular tools for screening biodiversity: Plants and animals. Eds. Karp A., Isaac P. G., Ingram D. S.(London:Chapman & Hall), 5–9.
Zuyev G. V., Nesis K. N., Nigmatullin C. M. (1975). Systematics and evolution of the genera Ommastrephes and Symplectoteteuthis (Cephalopoda, Ommastrephidae). Zool. Zh 54 (10), 1468–1479.
Keywords: transoceanic migration, equatorial circulation, biodiversity hotspot, Indonesian gateway, lineages diversification, ommastrephid
Citation: He L, Xu S, Weese DA, Zhang J, Ren H, Tang F, Sha Z and Zhang J (2022) Biogeographic role of the Indonesian Seaway implicated by colonization history of purpleback flying squid, Sthenoteuthis oualaniensis (Lesson, 1830), in the Indo-Pacific Ocean. Front. Mar. Sci. 9:891236. doi: 10.3389/fmars.2022.891236
Received: 07 March 2022; Accepted: 29 August 2022;
Published: 26 September 2022.
Edited by:
Benny K. K. Chan, Academia Sinica, TaiwanReviewed by:
Mariana Díaz-Santana-Iturrios, Andres Bello University, ChileCopyright © 2022 He, Xu, Weese, Zhang, Ren, Tang, Sha and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lijun He, dGlnZXIwMmpAaG90bWFpbC5jb20=; Zhongli Sha, c2hhemxAcWRpby5hYy5jbg==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.