- 1ICAR-Central Marine Fisheries Research Institute, Cochin, India
- 2Department of Fisheries, Port Blair, India
The genus Scomberomorus, with 18 nominal species, sustains a significant heterogeneous fishery throughout its range. The sole molecular systematic study of this genus concerned the species group S. regalis, which contains the new world taxa. The species diversity of Scomberomorus in the northern Indian Ocean has not been studied at the molecular level, often leading to misidentifications. Here, novel genetic data are provided that reconfigure species boundaries from the region. We used single and multilocus data (eight mitochondrial and three nuclear genes) to infer phylogenetic relationships, species delimitation, and the resurrection of a time-calibrated phylogenetic tree. Our aim was also to verify the hypothesis of geographical races in S. guttatus predicated on variable vertebral counts. Interestingly, all species delimitation analyses have recovered another highly cryptic species in the nominal S. guttatus previously believed to have an Indo-Pacific distribution. Scomberomorus guttatus (Bloch and Schneider, 1801) in the sensu stricto, is redeemed from its type locality based on genetic data and preliminary morphomeristic investigations and has a restricted distribution in the Bay of Bengal. The cryptic species Scomberomorus aff. guttatus which exhibits >10% genetic divergence from S. guttatus is resurrected here from the synonymy of the latter as Scomberomorus leopardus (Shaw, 1803). Widespread in the Indo-Pacific, this species contains two major molecular operational taxonomic units (MOTUs) with a divergence threshold of over 2% between them. Our analysis suggests that vertebral counts must be coupled with other features to identify the species/lineages in the nominal S. guttatus. The heterogeneity in the S. guttatus species group is discussed in relation to the ecological diversity of the region which facilitates larval recruitment and niche specialization. The results also revealed two allopatric putative species in S. commerson, found primarily in the Pacific and Indian Oceans. This study added genetic data from S. lineolatus and S. koreanus, not previously represented in the sequence repositories. Estimation of divergence time indicated that the Indo-West Pacific species group undergoes multiple diversification events besides the recent splits detected within S. leopardus.
Introduction
Scombridae represents one of the most important elements of the Pelagiaria clade (Betancur-R et al., 2017) and forms the largest and state-of-art family in Scombroidei (Collette and Russo, 1984; Fricke et al., 2021). The family includes epipelagic, commercially important, edible and recreational fishes and currently has 51 valid species in two subfamilies. Its subfamily Scombrinae is divided into four tribes, and the genus Scomberomorus, first proposed by Lacepède (1801) from Martinique Island, falls within the tribe Scomberomorini. Heretofore, the genus comprises 18 nominal species, better known as Spanish mackerels and seerfishes, with a wide distribution from tropical to temperate neritic waters within 20°C isotherms (Collette and Nauen, 1983; Collette and Russo, 1984; Collette, 2003). Although pelagic, they exhibit restricted species ranges, with the exception of Scomberomorus commerson (Lewis, 1981), indicating their limited open ocean migrations. Members of this genus are also recognized around the world as valued table fish that support diverse fisheries in their ranges (Collette and Nauen, 1983). In their magnum opus on Spanish mackerels, Collette and Russo (1984) evaluated the species relationships and systematic position of Scomberomorus within the Scombridae through a comprehensive cladistic analysis and proposed six monophyletic species groups in the genus based on synapomorphies. In 2003, Collette listed all valid species of Scomberomorus in his “Annotated checklists of fishes of family Scombridae Rafinesque 1815” which is followed globally. Their type localities, ranges, and IUCN status are shown in Table 1.

Table 1 Distribution, type locality, type specimen, and IUCN status of the nominal species in the genus Scomberomorus (Collette and Nauen, 1983; Collette and Russo, 1984; Collette, 2003; Froese and Pauly, 2022; https://www.iucnredlist.org).
Scomberomorus spp. have long been a regular subject of stock structure studies for the purpose of establishing appropriate management and monitoring plans (Welch et al., 2015), while the exclusive molecular systematics has been restricted to the Scomberomorus regalis species group, identified as the most derived clade in the genus (Banford et al., 1999). Genetic studies have primarily aimed at genetic stock identifications or phylogeography of species from major fishing areas. The species in focus were Scomberomorus semifasciatus (Australia: Broderick et al., 2011), Scomberomorus brasiliensis (Western South Atlantic: da Cunha et al., 2020), Scomberomorus sierra (Eastern Pacific: López et al., 2010), Scomberomorus concolor (Gulf of California: Magallón-Gayón et al., 2016), Scomberomorus guttatus (Persian Gulf: Abedi et al., 2011), Scomberomorus cavalla (Brazil: Santa Brígida et al., 2007), etc. Of all the species, the most extensive investigation was performed on S. commerson using a wide range of mitochondrial and nuclear molecular markers in the Pacific (e.g., Sulaiman and Ovenden, 2010; Fauvelot and Borsa, 2011; Habib and Sulaiman, 2016) and in the rim countries of the Indian Ocean (e.g., Hoolihan et al., 2006; Abedi et al., 2012; Divya et al., 2018; Vineesh et al., 2018; Johnson et al., 2021).
Biodiversity loss is one of the most serious issues with direct consequences, and species form the meter of biodiversity and conservation (Halasan et al., 2021). Spanish mackerels are harvested and exploited indiscriminately, compounded by their biological attributes (Zacharia et al., 2016; Hashemi and Doustdar, 2022). It is recommended that genetics needs to be well-integrated into the global biodiversity index while assessing conservation and risk priorities (Garner et al., 2020). Scomberomorus accounts for nearly 35% of reported Scombrids, but data-rich status regarding their genetic and life history research is limited to a few such as S. cavalla, S. commerson, and Scomberomorus maculatus (Juan-Jordá et al., 2013). The IUCN Red List (IUCN 2022. Version 2021-3, https://www.iucnredlist.org) assigns “near threatened” status to S. commerson and Scomberomorus munroi, “vulnerable” to S. concolor, and “data deficient” to S. guttatus, Scomberomorus niphonius, Scomberomorus plurilineatus, and Scomberomorus sinensis, while the remaining 11 species are of least concern. Despite shaping a cardinal transoceanic fishery, the molecular taxonomy of this group remained confined to the sole phylogenetic assessment study by Banford et al. (1999), who evaluated the effect of the Eastern Pacific/Eastern Atlantic biogeographic track (Rosen, 1975) on the diversification of new world taxa of the species group S. regalis. It may be added that certain species of Spanish mackerels are not yet represented in the GenBank (e.g., Scomberomorus lineolatus, Scomberomorus koreanus, and Scomberomorus multiradiatus). The void in the accurate identification and systematics of the Spanish mackerels necessitates detailed investigations from the respective oceanic provinces, including the Indian Ocean, which is an integral part of the speciose Central Indo-Pacific (Miller et al., 2018), for more comprehensive data coverage.
The Northern Indian Ocean, an element of the Indian Ocean covering 29% of the global ocean area (Wafar et al., 2011), is bifurcated by landmasses to the north into two intracontinental seas, viz., the Arabian Sea to the west and the Bay of Bengal to the east (Monolisha et al., 2018). In the review of “Scombrids of the world”, Collette and Nauen (1983) listed four species in Scomberomorus from India, viz., S. commerson (Lacepède, 1800), S. guttatus (Bloch and Schneider, 1801), S. lineolatus (Cuvier, 1829), and S. koreanus (Kishinouye, 1915). Silas (1964), who redescribed S. guttatus, proposed a wide distribution of the species in the Indo-Pacific while simultaneously predicting the likelihood of geographic races therein based on variations in vertebral counts cited in the earlier literature. Later, Devaraj (1976) who discovered S. koreanus in the Palk Bay (Indian Ocean) supported this observation and added some significant morphological measurements to distinguish the races. Differences in morphometry have been reported in S. guttatus distributed along the Indian coastline (Rao, 1975). Collette and Russo (1984) found no significant differences in morphometric data between populations of S. guttatus in the Arabian Sea and the Bay of Bengal, while it was evident in the races from the Pacific. They also observed a difference in the number of vertebrae between these two population clusters. Rao and Lakshmi (1993) suggested that S. lineolatus, distributed in the south-east coast of India, is a natural interspecific hybrid and not a valid species, which was refuted by Collette (1994) with a recommendation to include molecular methods to define and characterize the species in Scomberomorus. In the Indian context, taxonomic assessment supported by vanguard molecular tools remained a veritable blank, except for the classic works of the predecessors mentioned above.
Understanding evolutionary processes in marine realms where cryptic speciation is common can reveal barriers to gene flow, and species–level molecular phylogenies have been used to describe biogeographical regions (Crandall et al., 2019). Alongside this, molecular species delineation has been widely used either as a standalone technique or in combination with an integrative taxonomic approach to species identification (Luo et al., 2018). Therefore, the objectives of this study are as follows: i) generation of multilocus molecular impressions of all available species of the genus along the Indian coast and island ecosystems; ii) phylogenetic reconstruction of the genus based on globally available data for the individual loci such as the universal barcode region, i.e., the mitochondrial cytochrome c oxidase subunit 1 (COI), cytochrome b (Cyt b) fragment, along with multilocus methods using concatenated sequences of mitochondrial and nuclear markers comparable to previous studies; iii) recognition of the molecular operational taxonomic units (MOTUs) within Scomberomorus using the single-locus and multilocus species delimitation approach to assess the species diversity; and iv) resurrection of a dated phylogeny using multispecies coalescent analysis. This study also aimed to validate the hypothesis regarding the occurrence of geographic races in the Indian Ocean congeners and the results are expected to complement the universal database on this genus with glimpses into the diversity of the northern Indian Ocean.
Materials and Methods
Sampling and Overview of Materials
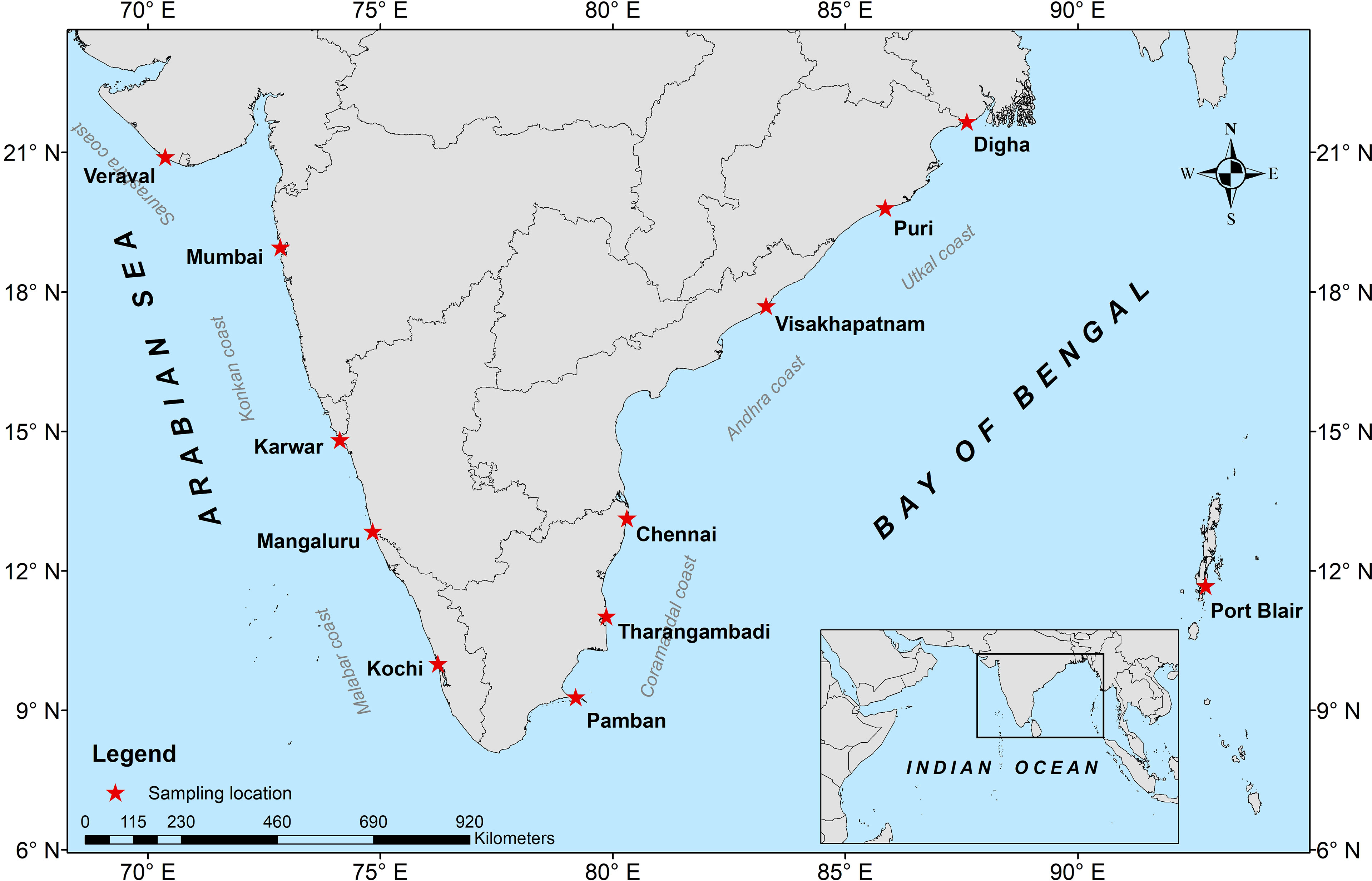
A total of 210 specimens of the genus Scomberomorus were collected in different seasons from 2019 to 2021 from different predesignated sampling sites along the coast of India, covering samples by type locality (topotypes) (Figure 1, Table 2). Pampus candidus, Brama sp., Lepidocybium flavobrunneum, and Kajikia audax were selected as outgroups for phylogenetic analysis based on the available literature (Miya et al., 2013) and digitally documented after collection (Supplementary Figure 1). All specimens were tentatively identified by morphological taxonomy (Collette and Nauen, 1983) and their fin or muscle tissues were preserved in absolute ethanol for future molecular studies. In addition, data mining of mitochondrial COI, Cyt b, NADH dehydrogenase subunit 2 (NADH2), ATPase subunits (ATPase 6 and ATPase 8), and nuclear [aldolase exon 5/intron 5 (ALD), rhodopsin (RHO), titin-like protein (Tmo-4c4)] genes were extracted from previous literature and nucleotide databases to represent the maximum species in Scomberomorus. The identity of the species underrepresented in GenBank (e.g., S. sinensis, Scomberomorus tritor, and S. multiradiatus) was deciphered by comparison, wherever possible, with either the morphological characters or the limited genetic data. Total genomic DNA (gDNA) was extracted from all tissue samples by the standard phenol–chloroform method (Sambrook et al., 1989). Isolated gDNA was visualized in 0.8% agarose gel, while NanoDrop One (Thermo Fisher Scientific, USA) determined the quality and quantity. High-quality gDNA was stored in duplicate at −80°C for further downstream processes.

Table 2 Locations of sampling (fishing harbors) and fishery information of Scomberomorus spp. along the Indian coastline including the Andaman Islands.
Polymerase Chain Reaction Amplification, Purification, and Sequencing
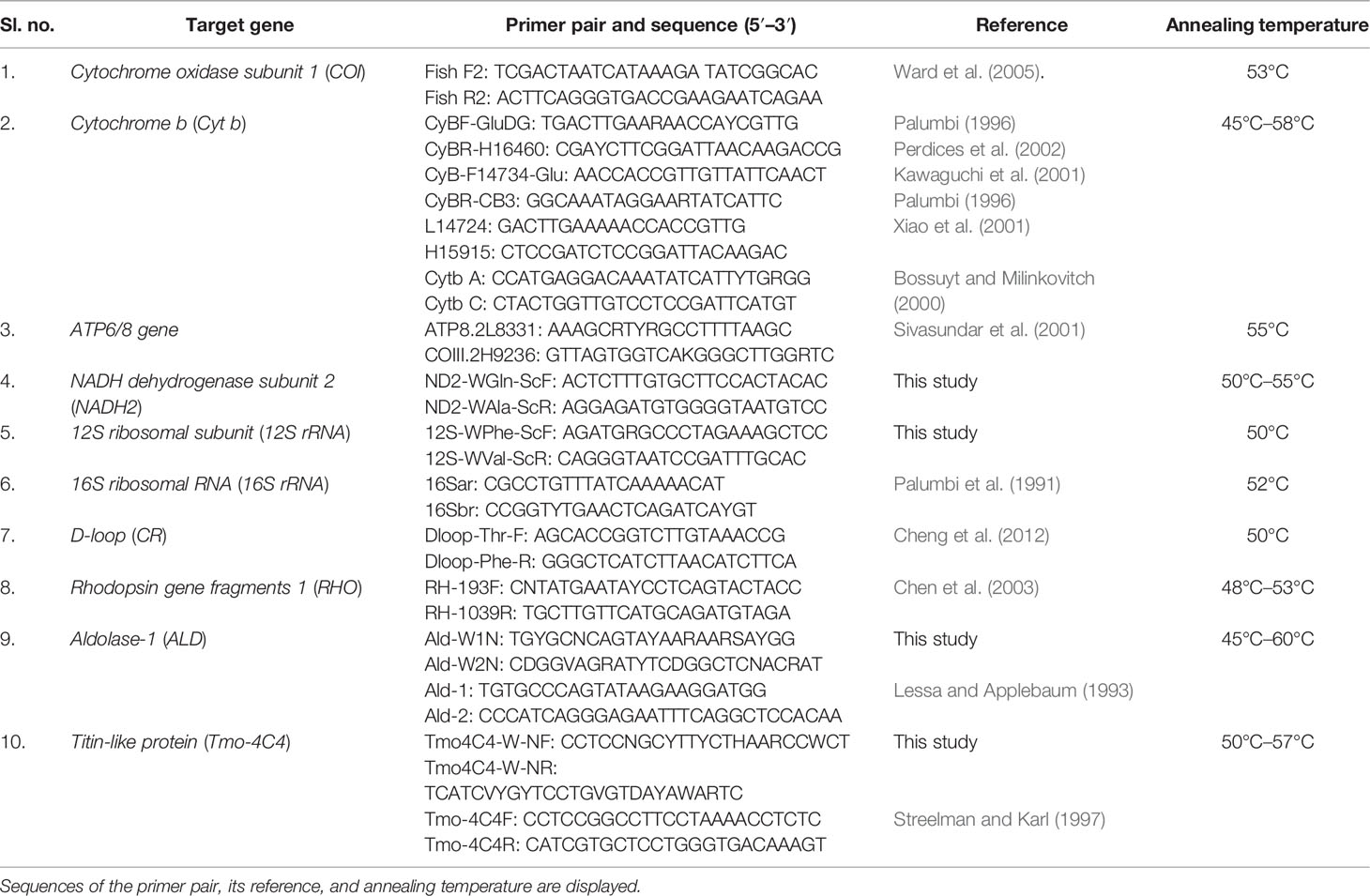
The identified specimens and the outgroups were barcoded to confirm their genetic identity. After this, no fewer than five individuals were selected for multigene amplification, taking into account collection sites and topotypes. We targeted 11 regions of DNA, eight genes from the mitochondrial (mt) genome [coding: COI, Cyt b, ATPase subunits 6 and 8, NADH2; non-coding: D-loop/Control Region (CR), 12S rRNA (12S ribosomal RNA), and 16S rRNA (16S ribosomal RNA)] and three nuclear loci (ALD, RHO, and Tmo-4C4) which, depending on their availability in GenBank, were assigned to different analyses. Amplification was carried out in a 50-µl reaction using basic polymerase chain reaction (PCR) conditions and cycling parameters following the primers and annealing temperatures (Table 3). Our analysis included multiple mt genes to increase the phylogenetic signal and to inspect the presence of accidental pseudogene amplification, while nuclear genes in particular provide regions with slower substitution rates (Banford et al., 1999). Amplified DNA fragments were visualized in 1.2%–2% agarose gel depending on the size of the product, purified, and bidirectionally sequenced.
Sequence Analysis and BLAST Annotation
Amplified sequences were aligned using the ClustalW algorithm (Thompson et al., 1994), which was manually edited using BioEdit v7.1.9 (Hall, 1999) to remove gaps and severe base pair mismatches. Curated coding regions were translated into amino acids to check for the presence of pseudogenes. All generated sequences were compared to the sequences downloaded from nucleotide databases using the search tool BLAST (Altschul et al., 1990). All amplified sequences were deposited in GenBank and the accession numbers with details were given along with the sequences retrieved from GenBank and BOLD (Supplementary Tables 1, 2 and Table 4).
Table 4 Code to Scomberomorus spp. individuals sequenced and accession numbers from GenBank generated in this study used for multilocus phylogenetic analysis are exhibited.
Phylogenetic Analyses
Initially, phylogenetic analysis using a single locus (COI and Cyt b with a truncated length of 632 and 763 bp, respectively) was implemented to incorporate maximal sequences representing the nominal species in Scomberomorus, while multigene analysis focused on increasing the phylogenetic signals, thereby verifying the validity of the primary investigation results. Three hundred and seventy-eight sequences belonging to 15 nominal species (Supplementary Table 1) and 122 sequences from available species (Supplementary Table 2) of the genus were included in the COI and Cyt b-based analyses, respectively. COI contained sequences from all species (except S. sinensis, S. multiradiatus, and S. tritor) along with those generated in this study (n = 148). We also evaluated the possible genealogical relationships between haplotypes in the putative species complex containing cryptics through a haplotype network with a maximal connectivity limit of 95%, mapped with their COI barcodes using the TCS algorithm implemented in POPART version 1.7 (Leigh et al., 2015). For the multilocus dataset, five sequences per species were selected from our samples. It included alignments of COI, Cyt b, ATP8, ATP6, NADH2, RHO, ALD, and TMO resulting in 632, 1,032, 168, 684, 1,047, 762, 410, and 499 base pairs, respectively. These gene fragments were selected to contain the maximum number of congeneric species, and only those with better data coverage/mitogenomes were included in the analysis. The saturation in each codon of each gene was tested using DAMBE v.7 (Xia, 2018) with no evidence of saturation detected at any of the codon positions. The concatenation resulted in a final length of 5,235 bp (Table 4).
Mitochondrial data were retrieved from the complete mitogenomes of the outgroups (except for P. candidus) after the species were confirmed with barcodes (accession nos. OM436012–OM436014). The individual and concatenated matrices were analyzed under the Bayesian inference (BI) and maximum likelihood (ML) method. The best-fit model (GTR) and partition for the datasets were identified using PartitionFinder v1.1.1 (Lanfear et al., 2017) under the parameters greedy algorithm, unlinked branch length, and Bayesian information criterion (BIC). BI analysis was performed through Markov chain Monte Carlo (MCMC) sampling implemented in MrBayes v3.2 (Ronquist et al., 2012) with 5 million iterations in two independent runs, sampling every 1,000 generations and 10% burn-in. Tracer v1.7 (Rambaut et al., 2018) evaluated the convergence of analyses and ensured that the effective sample size (ESS) values were above 200. Finally, the ML analysis was performed in IQ-TREE by 1,000 standard bootstrap with a given best-fit model and partition. The BI and ML phylogram topologies and their respective statistical supports—posterior probability (PP) and bootstrap support (BS) scores—were visualized in FigTree v.1.4.3.
Species Delineation Analyses
The delineation analysis was performed at two levels with previously used data from COI and Cyt b fragments, followed by a final validation using a multiple gene data matrix, considered here as one locus. Three species delineation analyses [two tree-based methods: the Generalized Mixed Yule Coalescent model (GMYC; Pons et al., 2006; Fujisawa and Barraclough, 2013) and multirate Poisson tree processes (mPTP; Kapli et al., 2017); and a distance-based method: Automatic Barcode Gap Discovery (ABGD; Puillandre et al., 2012)] were used to delineate and discover diversity among genera. The ultrametric trees generated separately for single and multiple genes by MCMC sampling in BEAST v2.6.4 (Bouckaert et al., 2019) were used as input files for single (sGMYC) and multiple-threshold (mGMYC) models, respectively. In BEAST analyses, independent runs were performed on single and multilocus data incorporating a relaxed clock log-normal and a partition and best-fit model akin to phylogenetic analysis for 10 million generation sampling every 1,000 generations. After assessing convergence between runs and ESS scores, both sGMYC and mGMYC analyses were performed using SPLIT packages implemented in R 4.0.2 (R Core Team, 2020). ML phylogram for mPTP analysis executed in Python v.3 (Van Rossum and Drake, 2009) with PTP package (Zhang et al., 2013) was generated in IQ-TREE v1.6.12 (Nguyen et al., 2015), with provided partition and 1,000 standard bootstrap. A distance-based ABGD analysis was performed following the arguments Pmin = 0.001, Pmax = 0, steps = 20, relative gap width = 1, Nb bins (for distance distribution = 20), and genetic distance as simple distance.
An additional confirmatory species delineation for the cryptic species complex using concatenated gene data was performed using the Bayesian Phylogenetics and Phylogeography program (BP&P; Yang and Rannala, 2010). This is a robust method that models the evolution of multilocus data and is widely used for explicit taxon identification and in the analyses of taxon evolution and divergence (Luo et al., 2018). The analysis was carried out with a user-specified guide tree which estimates the probability that each node represents a speciation event (i.e., a split between two distinct species) in bpp v4.4.1 (Yang, 2015). A total of 27 sequences with the concatenated dataset (Table 4) were used for the construction of the guide tree [L, ((T,K),(E,(V,W)))], where L = S. lineolatus, T = S. guttatus, K = S. koreanus, and E, V, and W = S. aff. guttatus, in BEASTv 2.4. rjMCMC algorithm 1 with fine-tuning parameter epsilon = 2 and species model prior m = 1, mutation rate across locus = 1, and an independent rates clock model were chosen to run the analysis. An inverse gamma prior IG (alpha, beta) for ancestral population size (theta) and root age (tau) were estimated using assigned prior values. The assigned alpha and beta values for theta prior were 6 and 0.002, whereas the tau prior values were 4 and 0.06, respectively.
Genetic Distance
Intra- and interspecific sequence divergence (uncorrected p-distance) of nominal species and MOTUs were calculated after species delineation analysis using the above COI, Cyt b, and concatenated gene sequences under uniform rates in MEGA 11 (Tamura et al., 2021). In MEGA 11, the criterion for the complete deletion of missing data was adopted, resulting in 3,316 nucleotides (all mitochondrial and nuclear ALD fragments used for multilocus phylogenies) being required to calculate genetic distance.
Divergence Time Estimation
The estimation of divergence time of 10 nominal species was performed using the sequences of eight mitochondrial genes (COI, Cyt b, ATP6 and 8, NADH2, 12S rRNA, 16S rRNA, and D-loop) with a total length of 5,554 bp. Of these, the sequences of six species were retrieved from GenBank (https://www.ncbi.nlm.nih.gov/genbank/), while the remaining four were gathered from this study. To include maximum molecular data and to ensure full coverage of sequences without any data loss from all the species, only mitochondrial genes were used for this analysis. The concluding data matrix for the analysis contained only one individual per species (Table 5). The estimation of evolutionary rates and approximate time to most recent common ancestors (TMRCA) of each species included in the analysis were assessed by BEAST v2.6.4 (Bouckaert et al., 2019). The analysis was conducted with an uncorrelated relaxed clock model using the GTR/GTR+I+G nucleotide substitution model recovered from PartitionFinder v1.1.1. Speciation for the tree prior was selected as the Yule model and alternative calibration points were incorporated with three fossils under the normal distribution. Firstly, the root age of the basal node of the phylogram was calibrated with a lower and upper boundary of 79.66–98 million years ago (mya) (Santini et al., 2013). The second calibration point was fixed to the Acanthocybium solandri–Gymnosarda unicolor clade with a minimum age of 52 mya and an upper boundary of 79.66 mya to calibrate its stem age (Purdy et al., 2001; Santini et al., 2013). Also, we constrain the crown node of the Scomberomorus genus with a Paleocene fossil between 56 and 59.2 mya (Sepkoski, 2002). Input file prepared by BEAUti in the BEAST package. The MCMC run for 100 million generations was executed twice, thinning every 1,000 trees to recover time-stamped phylogenies, and 25% of the initial samples were discarded as burn-in by TreeAnnotator v2.6.4. Tracer v1.7 evaluated the convergence of analysis and ensured that the ESS scores for all parameters were above 200. The chronogram after analysis was viewed in FigTree.
Table 5 Accession numbers retrieved from GenBank and generated in this study for molecular divergence estimation.
Estimation of Vertebral Counts
Since some of the aforementioned literature discussed the variation in the number of vertebrae in S. guttatus, this meristic trait was checked for 25 specimens each of S. guttatus, collected from the type locality, east coast (Odisha and West Bengal) and west coast (Gujarat) of India, along with five specimens from other Scomberomorus species. Vertebral counts were determined both from X-radiographs taken at the respective sites and by manual counting after preparing a dry vertebral column following James (2008). Specimens were boiled to 110°C to separate the meat from the bone and the bones were brushed under running water after boiling.
Results
BLAST Results and Identity of the Species
Identity of COI barcodes for Scomberomorus spp. generated in this study were verified with all available species (n = 13) in nucleotide databases. The reliable barcodes for S. lineolatus and S. koreanus were also generated here and their incorrectly labeled sequences in various nucleotide databases were thus recognized. The identity of S. koreanus could be confirmed using the published Tmo-4C4 sequence (DQ388095; Orrell et al., 2006). Some species like S. tritor and S. sinensis were exclusively represented by the Cyt b gene in NCBI. Search results employing Cyt b were correct identifications in most cases, apart from some errors (see Supplementary Tables 1, 2). Although there is no genetic information for S. multiradiatus, morphological evidence and a geographic distribution restricted to the Gulf of Papua (Collette and Russo, 1984) isolated it from the others.
Phylogenetic Analyses Using COI Sequences and the Multilocus Data
The COI-based Bayesian phylogeny and IQ tree contained two major clades (Figure 2). Major clade I branched into two subclades with a Bayesian posterior probability (BPP) of 0.75. Subclade I was further bifurcated into two groups with high BPP (0.97). Species morphologically identified as S. guttatus have been resolved into distinct barcode clusters representing diverged species in phylogenetic analysis. In this study, sequences identical to those of S. guttatus collected and identified from the type locality (Tranquebar, India) and adjacent areas are considered to represent S. guttatus (Bloch and Schneider, 1801) in sensu stricto (see Discussion). The cryptic species with indistinguishable morphological resemblance to S. guttatus is referred to as S. aff. guttatus in the phylograms and in the manuscript. Thus, group I included S. guttatus, S. aff. guttatus, S. koreanus, and S. plurilineatus. Also, S. aff. guttatus contained two reciprocally monophyletic lineages tentatively designated as S. aff. guttatus 1 and S. aff. guttatus 2. The latter had two haplogroups, hereinafter referred to as S. aff. guttatus 2a and S. aff. guttatus 2b in the manuscript. Scomberomorus aff. guttatus 2a was represented by a larger number of sequences, while the other had only a limited number. Sequence analysis indicated a restricted distribution of S. guttatus in the Bay of Bengal, Bangladesh, and Myanmar. Scomberomorus aff. guttatus 1 included individuals from the Bay of Bengal and the western Pacific Ocean (South China Sea) and some from the Arabian Sea, while S. aff. guttatus 2 comes exclusively from the Arabian Sea. Scomberomorus plurilineatus clusters with this cryptic species, while S. koreanus and S. guttatus, clustering around S. plurilineatus, occupy an interchangeable position with respect to BI and IQ-based phylograms. In addition, S. munroi and S. niphonius form sister clade to the cryptic species + S. plurilineatus + S. koreanus + S. guttatus. Subclade II included Scomberomorus queenslandicus and S. commerson, the latter of which was further subdivided into two groups tentatively designated as S. cf. commerson 1 and S. cf. commerson 2 residing mostly in the Pacific and Indian Oceans, respectively.
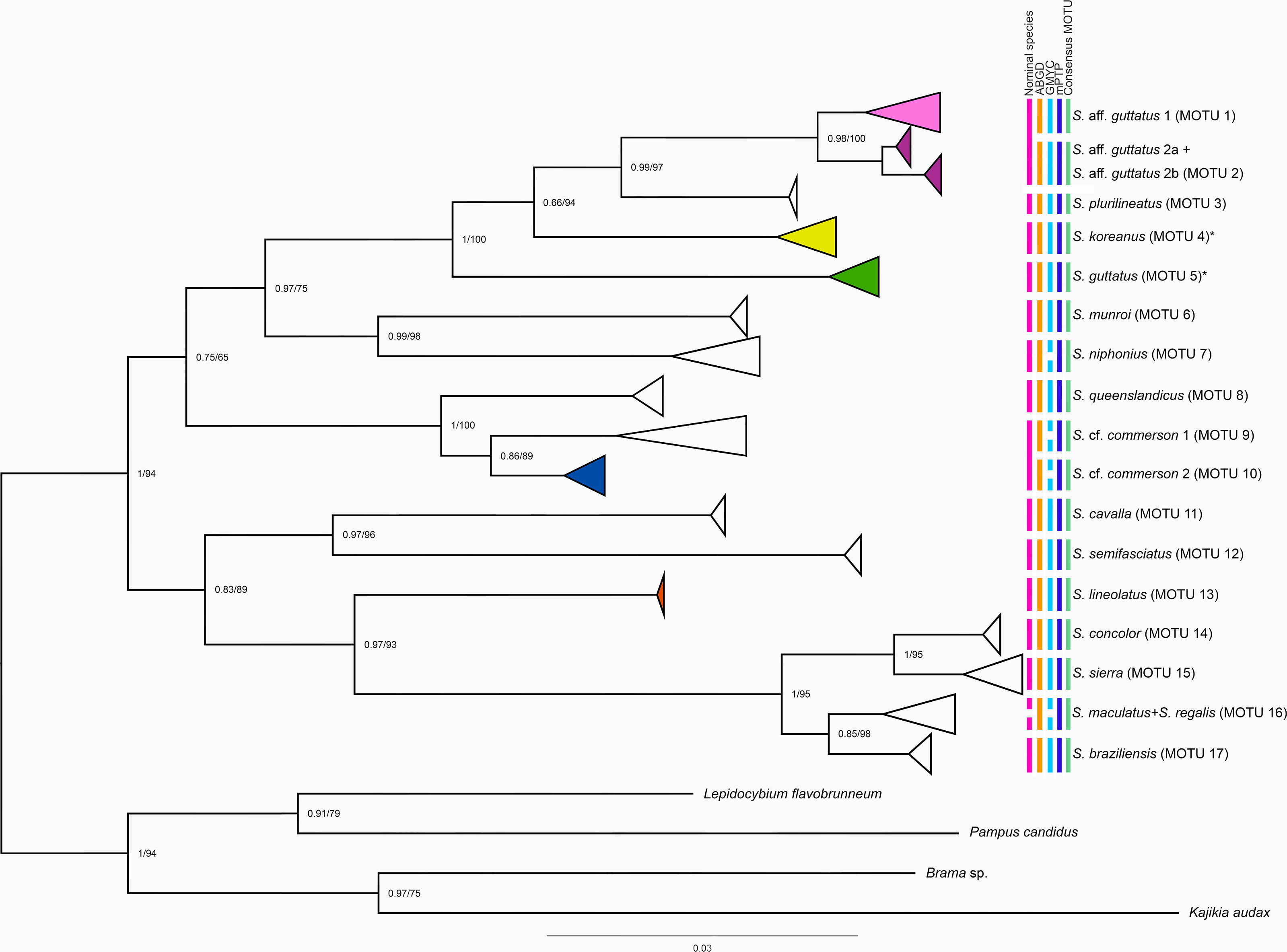
Figure 2 Phylogenetic tree depicting the relationships between Scomberomorus spp. based on Bayesian inference (BI) superimposed on maximum likelihood (ML) analysis of partial COI sequences (Supplementary Table 1). The node numbers specify the posterior probability and bootstrap support values, respectively. Leaves are collapsed to indicate the well-supported clades and species from India are highlighted in colors. The nominal species, results of delineation methods, and consensus MOTUs are indicated by different colors to the right of the phylogram.
Major clade II also had two subclades: one subclade included S. cavalla and S. semifasciatus, while the other included S. lineolatus and the rest of the species. Scomberomorus concolor and S. sierra formed a strongly supported group in this, while S. maculatus + S. regalis and S. brasiliensis formed another. The Cyt b gene generated a tree topology (Supplementary Figure 2) that was incongruent with the COI-based phylogram.
The TCS haplotype network based on mutational steps revealed a clear grouping pattern with three major haplogroups in the cryptic complex of S. guttatus (Supplementary Figure 3A) and two haplogroups in S. commerson (Supplementary Figure 4A). The geographical distribution of these haplogroups was also depicted (Supplementary Figures 3B–D, 4B). The analysis showed a clear genetic differentiation between the haplotypes of S. aff. guttatus 1 and S. aff. guttatus 2 from S. guttatus. The first two mentioned were 10 mutational steps apart, and when taken together, they were 61 mutational steps away from the third. Similarly, the two groups in S. commerson were 18 mutational steps away from each other.
The concatenated multilocus phylogeny depicted the relationships between the 10 nominal species (Figure 3). The Bayesian and IQ phylogenetic trees showed the same topology. There were two major clades (BPP-1) and the first one had two strongly supported subclades: one with (S. aff. guttatus 1 + S. aff. guttatus 2) + (S. guttatus + S. koreanus), while the second subclade contained S. lineolatus and S. cf. commerson 2. Major clade II clustered S. cavalla outside the group of S. concolor and S. sierra and another group of S. brasiliensis, S. maculatus, and S. regalis.
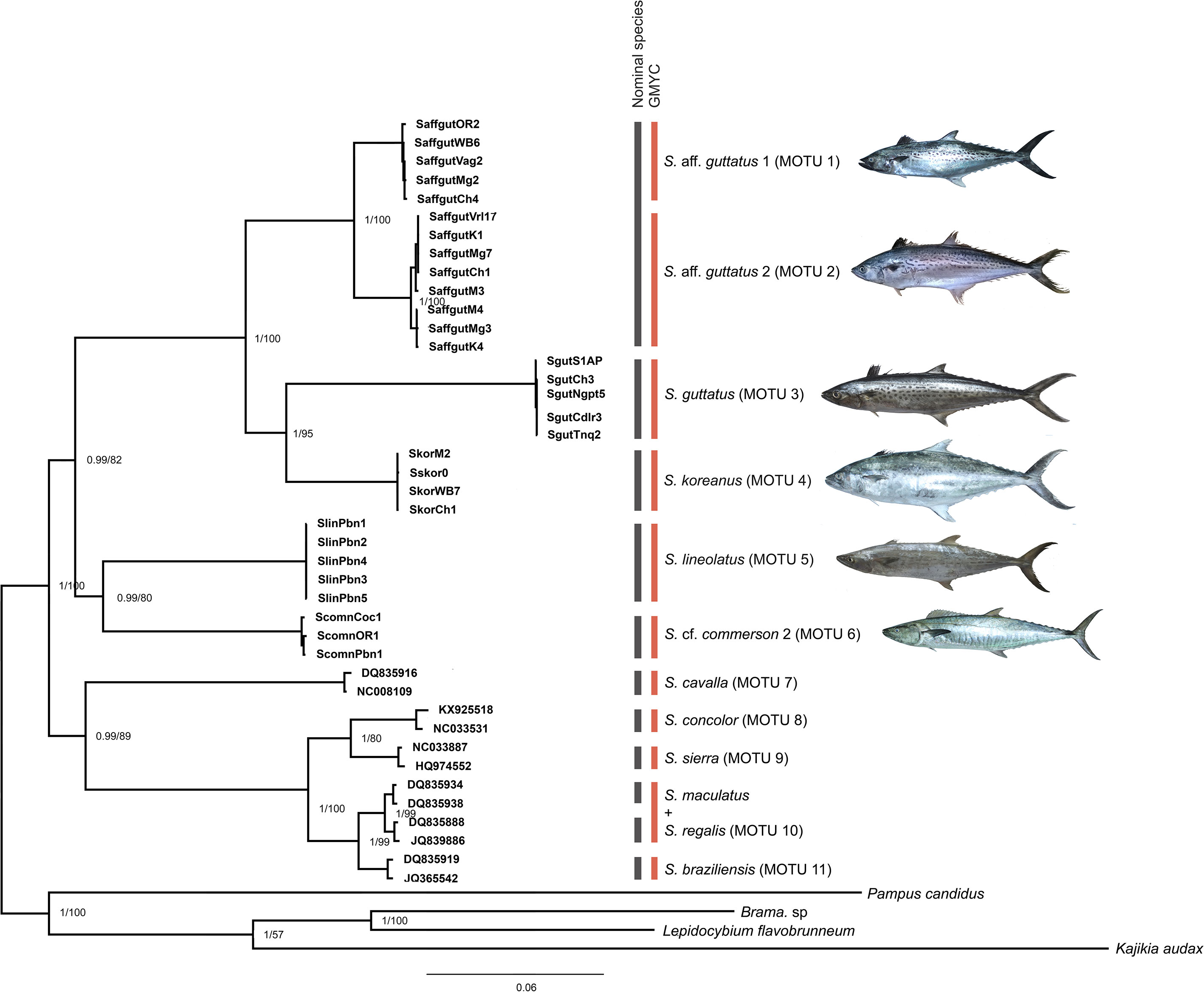
Figure 3 Bayesian inference (BI) phylogram superimposed on maximum likelihood (ML) based on concatenated multilocus data is represented (Table 4). The node numbers specify the posterior probability and bootstrap support values, respectively. The nominal species and the GMYC entities from the GMYC delineation method are indicated by different colors to the right.
Species Delimitation and Evolutionary Divergence Based on COI Sequences
The ABGD analysis detected a barcode gap at 1.7% identifying 17 hypothetical species (MOTUs) in the initial partition. The GMYC and mPTP analyses detected 21 and 17 MOTUs, respectively. The results of the species delineation analyses are given in Figure 2. Of the consensus MOTUs (n = 17), 12 nominal species, viz., S. cavalla (Cuvier, 1829); S. commerson (Lacepède, 1800); S. concolor (Lockington, 1879); S. koreanus (Kishinouye, 1915); S. lineolatus (Cuvier, 1829); S. munroi Collette and Russo, 1980; S. plurilineatus Fourmanoir, 1966; S. queenslandicus Munro, 1943; S. brasiliensis Collette, Russo and Zavala-Camin, 1978; S. niphonius (Cuvier, 1832); S. semifasciatus (Macleay, 1883); and S. sierra Jordan and Starks, 1895 were recovered by all the analyses. The two nominal species S. regalis (Bloch, 1793) and S. maculatus (Mitchill, 1815) were identified as a single MOTU in all analyses except GMYC. Interestingly, three consensus MOTUs or hypothetical species could be identified in nominal S. guttatus with all delimitation methods. Apart from S. guttatus from the type locality, two MOTUs, i.e., S. aff. guttatus 1 and S. aff. guttatus 2, have been identified on the Indian coast including the island ecosystems (Andaman and Nicobar Islands). Scomberomorus aff. guttatus 2a had a smaller intercladal divergence (0.59%) with S. aff. guttatus 2b. However, the maximum genetic divergence was found between S. aff. guttatus 1 and S. aff. guttatus 2b. Sequence identity searches based on the BLAST results of various genes and species delimitation analyses indicated that certain COI barcodes of S. guttatus are classified as S. plurilineatus. Some others actually represented S. aff. guttatus 1 and S. aff. guttatus 2, a misidentification primarily due to the deceptive spotting pattern in the S. guttatus complex.
Scomberomorus commerson also branched into two allopatric putative species in all analyses, though GMYC analysis delimited two more entities (Figure 2). Scomberomorus niphonius (Cuvier, 1832) formed a single MOTU except in GMYC. Scomberomorus lineolatus and S. koreanus, previously not represented in nucleotide databases, represented two different MOTUs.
The COI-based genetic distance values between the recognized nominal species and consensus MOTUs containing novel hypothetical species are shown in Tables 6A, B, respectively. The nominal species on analysis showed a maximum intraspecific divergence of 10.60% (S. guttatus), followed by S. commerson (4.43%), while the minimum intraspecific distance was 0.00% in S. plurilineatus and S. lineolatus. The maximum inter-MOTU distance was 12.35% (between S. semifasciatus and S. cavalla), while the minimum distance was 0.44% between S. maculatus and S. regalis. The p-distance values between S. guttatus and S. aff. guttatus was 10.26%–10.41%, while it was 2.28% between S. aff. guttatus 1 and S. aff. guttatus 2. Between S. cf. commerson 1 and S. cf. commerson 2, it was 3.74%.

Table 6 COI-based genetic distance (uncorrected p-distance) values between the (A) nominal species and (B) consensus MOTUs containing novel hypothetical species are presented.
Species Delimitation Using Cyt b Sequences
GMYC analysis of the Cyt b gene amid the limited number of proxies from each geographic region revealed 18 MOTUs representing the species records in GenBank and identified here (Supplementary Table 2; Supplementary Figure 2). Despite the absence of S. plurilineatus, S. queenslandicus, and S. regalis that were present in the COI-based analysis, the Cyt b data revealed two additional GMYC entities representing two other nominal species, viz., S. tritor and S. sinensis. Interestingly, another GMYC species of uncertain taxonomic status represented as S. guttatus in GenBank from Taiwan and as Scomberomorus sp. here was evident in Cyt b-based analyses, while their COI barcodes were not available. The observed p-distance between S. aff. guttatus 1 and S. aff. guttatus 2 was 1.82%, while Scomberomorus sp. showed a divergence of 10.42%–10.55% from above. These three showed a divergence of 10.38% to 12.57% from S. guttatus. The p-distance was 4.31% between S. cf. commerson 1 and S. cf. commerson 2. These values are comparable to the COI-based distances between the above MOTUs. A divergence of 6.61% was observed between Scomberomorus sp. and its nearest neighbor S. koreanus (Supplementary Table 3). Cyt b sequences from S. sierra of the eastern Pacific (JQ434072–JQ434074) were seen to represent S. concolor. Similarly, sequences such as DQ497884 (Taiwan) identified as S. koreanus and AY986968 identified as S. commerson (China) represented S. aff. guttatus 1.
Validation of Species Boundaries Using Multilocus Data
GMYC analysis of the multilocus data revealed 10 nominal species or 11 MOTUs (Figure 3). The genetic distance values between the nominal species and consensus MOTUs are given in Tables 7A, B. Concordant results with the previous two single-locus analyses indicating three MOTUs in nominal S. guttatus were demonstrated here. The maximum intraspecific divergence (12.2%) was observed in the nominal species S. guttatus. The p-distance values between S. guttatus and S. aff. guttatus 1 and S. aff. guttatus 2 were 11.69% and 11.99%, respectively. The p-distance was 2.14% between S. aff. guttatus 1 and S. aff. guttatus 2. The maximum inter-MOTU distance was 13.11% (between S. cavalla and S. cf. commerson 2), while the minimum was 0.60% between S. maculatus + S. regalis and S. brasiliensis. Similar to the COI-based analysis, S. maculatus and S. regalis formed a single GMYC entity here.

Table 7 Concatenated genetic distance (uncorrected p-distance) values between the (A) nominal species and (B) consensus MOTUs containing novel hypothetical species as well are shown.
According to rjMCMC algorithms 1 and 0, the BP&P analysis considered S. lineolatus, S. guttatus, S. koreanus, and three lineages in S. aff. guttatus as distinct species, with all nodes resolved with a posterior probability of 1.0, that corroborated with the multilocus tree topologies from phylogenetic analyses. The sister relationship of S. guttatus with S. koreanus was recovered here. The status of the species in S. aff. guttatus is restored similarly to all the tree topologies. In contrast to the phylogenetic and other species delineation analyses, BP&P recognized S. aff. guttatus 2a and S. aff. guttatus 2b into two separate species (Supplementary Figure 5).
Divergence Dating
The topology of the chronogram (Figure 4) mostly corroborated with both single and multilocus phylograms. The crown age of the Scomberomorus genus has been estimated at 57.3 mya in the late Paleocene [highest posterior density (HPD): 56.3–58.4 mya] and diversification in the Eocene. The regalis group species, S. concolor and S. sierra, split from S. cavalla by 46 mya (HPD: 41–49.5 mya), whereas S. concolor and S. sierra recently separated by 8.5 mya (HPD: 7.2–9.9 mya). The probability of diversification of Indo-West Pacific species was predicted in the early Eocene, i.e., 54.4 mya (HPD: 51.9–56.5 mya). The predicted recent common ancestral ages between S. commerson–S. lineolatus and S. munroi and S. niphonius have been estimated to be 45 mya (HPD: 37–45.5 mya) and 41.9 mya (HPD: 38.5–45.6 mya), respectively. The crown age of S. guttatus–S. koreanus species has been estimated at 24.9 mya (HPD: 22.5–27.5 mya), while S. koreanus split from S. aff. guttatus group by 20.2 mya (HPD: 17.8–22.8 mya). Split in S. aff. guttatus into three lineages was observed at 3.7 mya (HPD: 3–4.5 mya) and 0.91 mya (HPD: 0.6–1.2 mya).
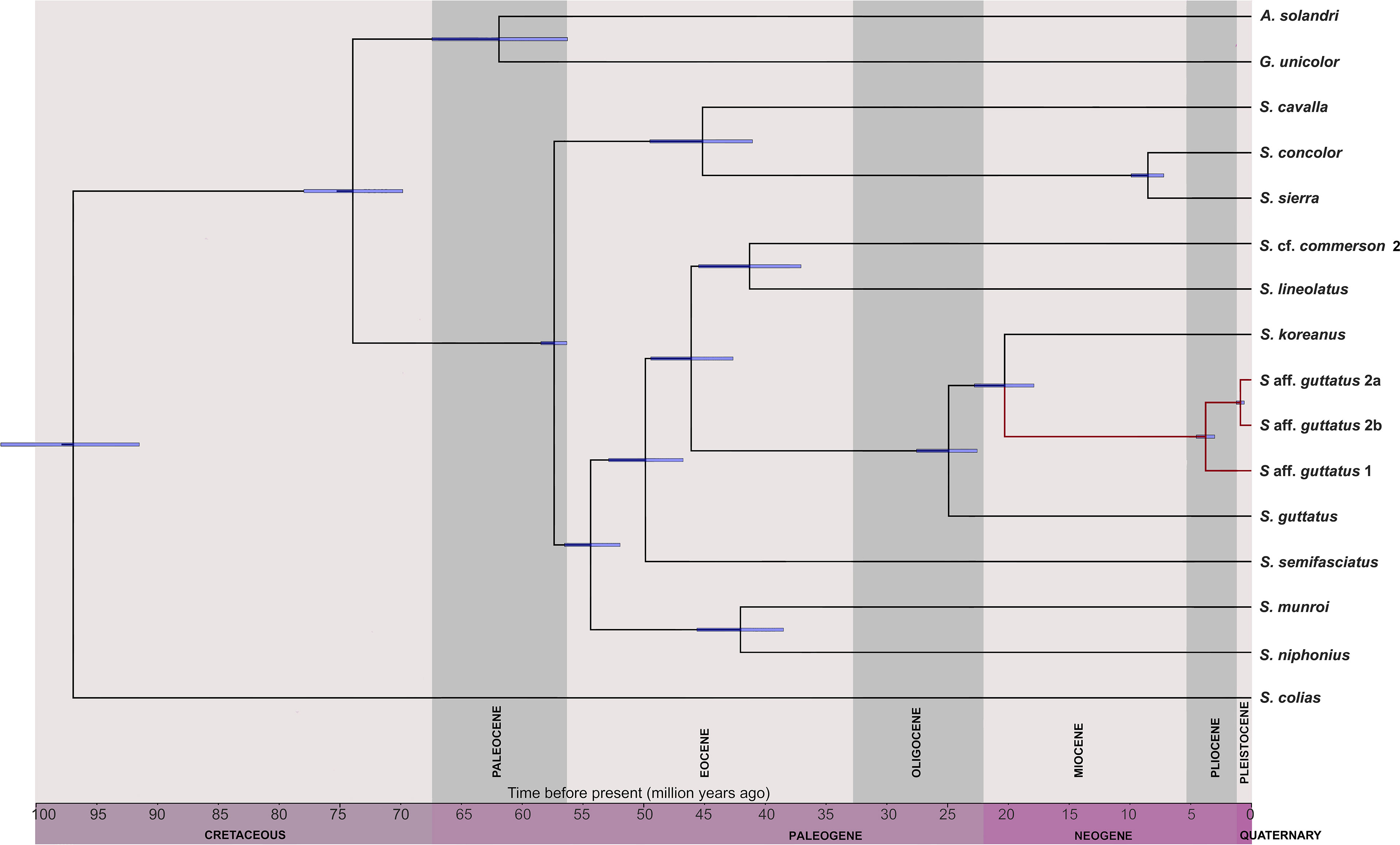
Figure 4 An ultrametric tree of Scomberomorus species (with maximum molecular data) scaled to geographical time is illustrated. Blue bars at the nodes indicate 95% confidence interval of internal nodes, and the clade highlighted in red color indicates S. guttatus and its cryptics.
Vertebral Count
Vertebral counts and X-radiographs of all available species from Indian waters are provided for comparison (Supplementary Table 4; Figure 5). Scomberomorus koreanus, S. commerson, and S. lineolatus showed a total vertebral count of 46, 44, and 46, respectively. The vertebral count of S. guttatus was 47–49 (usually 48), while it was 49–51 (usually 50) for S. aff. guttatus 1 and S. aff. guttatus 2 (Supplementary Figure 6).
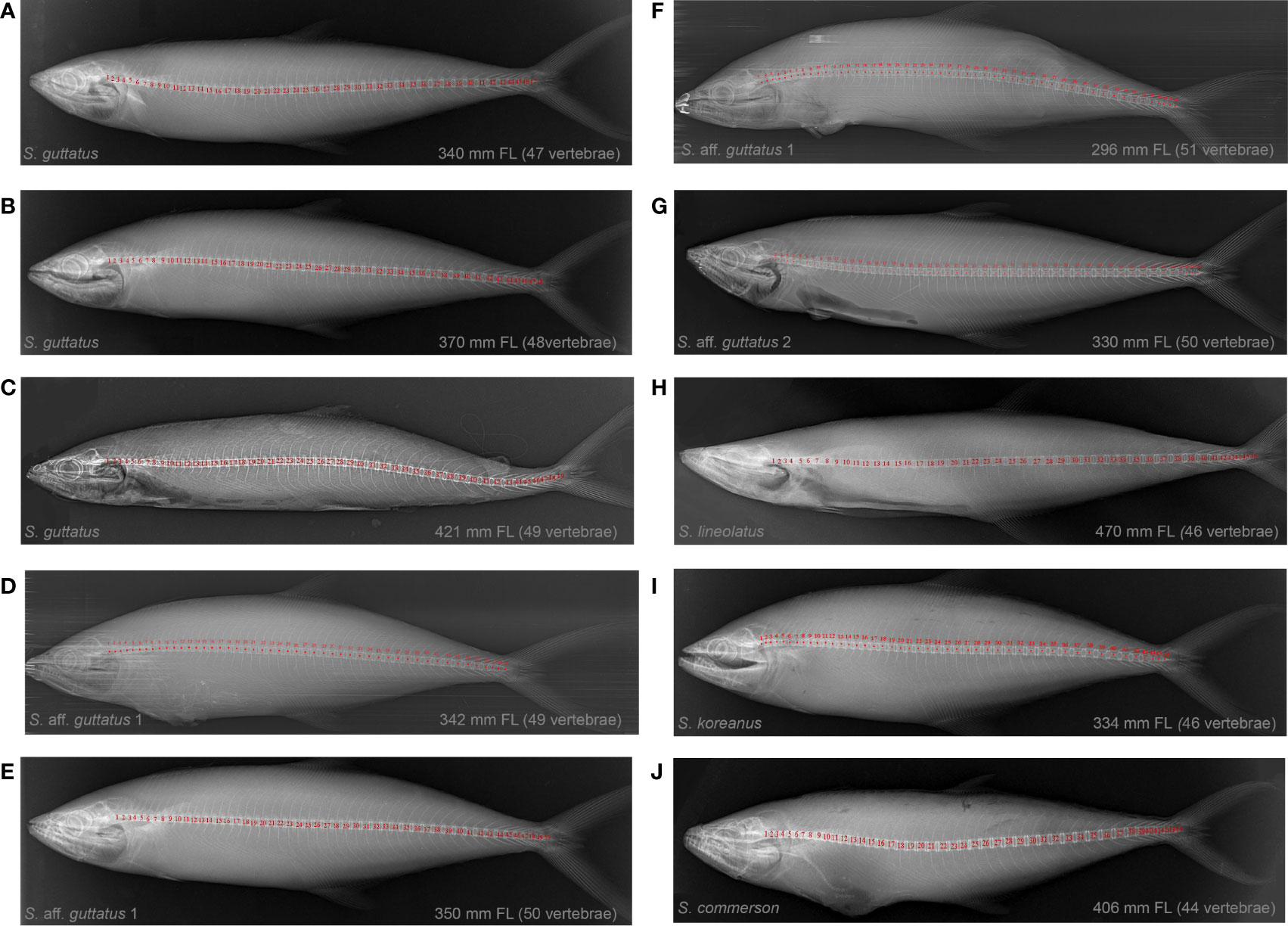
Figure 5 Digital X-ray images of Scomberomorus species (A–J) from the Northern Indian Ocean revealing the diversity, fork length (FL), and number of vertebrae.
Discussion
Phylogenetic Analyses
COI and Cyt b fragments generated incongruent gene trees in this study, which is not unexpected in Scombridae, where the phylogenetic signal varies with genes (Miya et al., 2013). The low BPP support for the subclade containing S. queenslandicus and S. commerson to the major clade I in the COI-based phylogram indicates their rather fragile affiliation within this major clade. Similarly, different levels of resolution for the species–level relationships could be observed in numerous groups of fish in individual gene trees (Clements et al., 2003; Wright et al., 2012). The incongruence in gene trees arises due to differences in their evolutionary history and various other events like incomplete lineage sorting, gene duplications, extinctions, introgression or hybridization, etc. (Som, 2015). MtDNA introgression occurs between species in Scomberomorus (e.g., Morgan et al., 2013), and the possibility of incomplete lineage sorting in the youngest lineages in Scombridae has been suggested (Ollé et al., 2022).
Collette and Russo (1984), based on a cladistic analysis of 58 characters, identified six species groups in Scomberomorus, viz., the “sinensis group,” which contains only S. sinensis; the “commerson species group” with S. commerson, S. niphonius, S. queenslandicus, and S. cavalla; the monotypic “munroi species group”; the “semifasciatus species group” with S. semifasciatus, S. plurilineatus, and S. lineolatus; the “guttatus species group” with S. guttatus, S. multiradiatus, and S. koreanus; and the “regalis species group” with S. regalis, S. tritor, S. maculatus, S. concolor, S. sierra, and S. brasiliensis. They considered S. tritor to be the basal primitive species in the regalis species group and considered S. sierra, S. brasiliensis, and S. regalis to be the three most advanced species.
The “guttatus species complex” in the COI-based phylogenetic tree contained S. aff. guttatus 1, S. aff. guttatus 2 (2a and 2b), S. koreanus, S. guttatus, and S. plurilineatus, and the grouping of the last one to this complex deviated from the “guttatus species group” proposed by Collette and Russo (1984). The TCS network showed consistent genealogical relationships. A similar genetic structure consistent with the phylogenies has been observed in the TCS network topology of the cryptic S. guttatus complex and S. commerson. Overall, S. aff. guttatus has two distinct haplogroups with a star-like topology with the predominance of a single haplotype, suggesting recolonization events through founder effects or population expansions after a bottleneck event (Allcock and Strugnell, 2012). A similar situation was observed in S. commerson. A star pattern with radial distribution was also shared in S. guttatus. The formation of haplogroups in S. aff. guttatus 2 can be an outcome of incomplete lineage sorting due to recent divergence (Van Velzen et al., 2012).
In the coalescent species tree, the clustering of S. munroi with S. niphonius and the placement of the new world taxa (Banford et al., 1999) in a highly supported clade with S. cavalla outgroup agree with Santini et al. (2013). Species–level relationships deduced from molecular data were generally inconsistent with the previously published morphological groupings by Collette and Russo (1984).
Phylogenies illustrate the historical relationships within a group of species and help study the processes of cladogenesis, and the shape of a phylogenetic tree provides clues to patterns of speciation and extinction within the clade. Taxon sampling has a strong impact on the accuracy of phylogenetic reconstruction methods, and reduced taxon sampling leads to an increase in phylogenetic tree imbalance (Heath et al., 2008; Nabhan and Sarkar, 2012). Increased sampling of taxa allows more accuracy, as exemplified in Salmonidae (Crête-Lafrenière et al., 2012). It is expected that the strength of the phylogenetic signal inferred from the concatenated gene sequences will be useful as the number of taxa increases, and therefore, we recommend global sampling and inclusion of all available species to elucidate the systematic relationships in the genus Scomberomorus.
Species Diversity Using Delimitation Analyses
Through delimitation analyses of COI and Cyt b gene fragments, we confirmed the identity of all valid species except S. maculatus and S. regalis. The latter two formed a single MOTU, an observation which is not surprising in light of the previous reports pointing to mitochondrial introgression in Scomberomorus (Banford et al., 1999; Paine et al., 2007; Morgan et al., 2013) and other scombrids (Puncher et al., 2015). Banford et al. (1999) observed that these two readily distinguishable inhabitants of the Western Atlantic are seasonally sympatric, and hypothesized the probability of ephemeral hybridization and subsequent mitochondrial introgression and recommended a comprehensive geographic and molecular assessment. Although the single-locus species delimitation approach can sometimes lead to an over- and/or underestimation of species diversity (Hofmann et al., 2019), its authenticity was confirmed here with the concordant result of the GMYC analysis of multilocus data.
The incongruence in the number of MOTUs from various molecular species delimitation methods can be due to their difference in a number of aspects. ABGD, GMYC, and PTP were developed for the analysis of single-locus data, although the latter two are often applied to concatenations of multilocus data. ABGD is sensitive to singleton sequences, while GMYC showed tendencies for oversplitting the number of species when there are violations of the model (Pentinsaari et al., 2017). Moreover, PTP and GMYC are sensitive to gene flow (Luo et al., 2018). Here, we used simple rules to find consensus between discordant MOTU delimitations using the best fit between species and MOTUs.
The Indo-Pacific king mackerel, S. guttatus, is dominant next to S. commerson in FAO Fishing Area 51 (Siddeek, 1995; Devaraj and Mohamad Kasim, 1998; Secretariat, IOTC, 2015). It is noteworthy that the nominal S. guttatus showed high intraspecific divergence, an indication of cryptic speciation events (Ge et al., 2021), and contained three consensus MOTUs with morphological similitude in all analyses except in BP&P, which produced four Bayesian species. BP&P is believed to be more accurate than other multilocus coalescent methods, showing lower rates of species overestimation and underestimation, and is generally robust to various potential confounders (Luo et al., 2018). Nevertheless, this method tends to delimit only the population structure and not the number of species, and hence, validation of genome-based results with multiple data types such as phenotypic and ecological information is required (Sukumaran and Knowles, 2017).
The species distribution along the Indian coast derived from our extensive sampling is depicted in Figure 6. Analysis of specimens from the type locality of S. guttatus and regions up to Chennai (Cuddalore, Kovalam, Nagapattinam, Kovalam) indicated the occurrence of only one haplogroup from these areas on the Coromandel Coast. The same formed a fraction of samples from other sites of fishery (Chennai, Visakhapatnam, Odisha, and West Bengal) located in the Bay of Bengal. The species having a genetic identity with the above deserves the status to be adorned as the original S. guttatus identified by Bloch and Schneider in (1801), as it is the only available species in type locality. It differed from the other two MOTUs in the nominal species by high genetic divergence. The lean and slightly cylindrical body shape of this species resembles the only existing type specimen collected from Tranquebar, India (Figure 7A: ZMB 8759: lectotype, stuffed, 361 mm SL) although its meristic trait such as vertebrae is not visible (Figure 7B). The cryptic MOTUs, viz., S. aff. guttatus 1 and S. aff. guttatus 2, hitherto hidden among the nominal species, were widespread in the collections. The Scomberomorus sp. from Taiwan recovered in Cyt b-based analysis and clades with S. koreanus represent a species with an uncertain taxonomic status that cannot be verified with the current level of information.
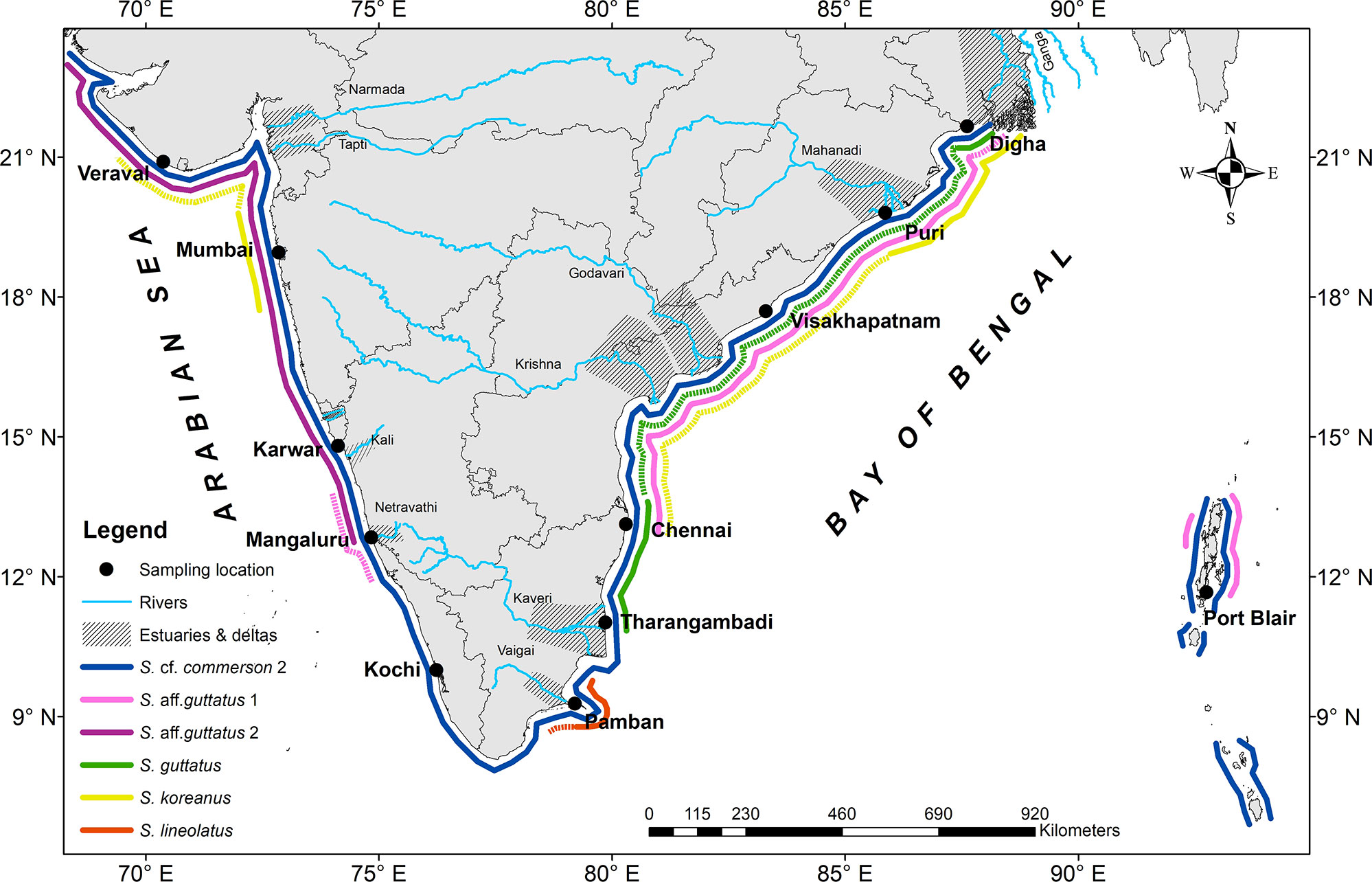
Figure 6 Geographical distribution of Scomberomorus spp. in the northern Indian Ocean based on the present study. The intense and sparse distribution of each MOTU is indicated by thick and dotted lines, respectively.

Figure 7 Lectotype of Scomberomorus guttatus (ZMB 8759) collected from Tranquebar, India. (A) Dry, stuffed specimen (373 mm FL and 361 mm SL), and (B) Computed Tomography (CT) Scan.
The allopatric putative species in S. commerson, i.e., S. cf. commerson 1 and S. cf. commerson 2, were consistent with some previous studies using mtDNA CR (Fauvelot and Borsa, 2011) with extensive geographic coverage across oceans. Scomberomorus commerson has a typically pelagic lifestyle at all developmental stages (Thangaraja and Al-Aisry, 2001) and is considered to be more migratory than all other congeners (Collette, 2001). Incidental occurrence of eggs/larvae of the Pacific Ocean lineage in the Indian Ocean (Supplementary Table 1) supports this observation. Significant differences in morphometry and spawning season between the Indo-Pacific populations were observed by Collette and Russo (1984) and Kaymaram (2010), respectively. Fauvelot and Borsa (2011) stated that the samples from the northwestern Indian Ocean, consisting of the Persian Gulf and the Oman Sea (identical to S. cf. commerson 2) and those from the Western Pacific, i.e., Southeast Asia and Oceania (S. cf. commerson 1), represent two cryptic species with a deep phylogeographic split. The additional GMYC entities observed in this study also support previous observations of geographic differentiation in this species (e.g., Sulaiman and Ovenden, 2010; Vineesh et al., 2018).
However, S. lineolatus, another valid species which is least abundant in the seerfishes in India (CMFRI FRAD, 2020), showed a limited distribution in Pamban up to Tuticorin (personal observation of the authors), although the literature describes its occurrence from Mumbai, Malabar to Palk Bay and Chennai (Collette and Russo, 1984). The shift in the fishery for S. guttatus and S. lineolatus from their former areas of frequent occurrence (Southern Gulf of Mannar and Visakhapatnam, respectively) is reminiscent of a similar trend observed for S. concolor (Fitch and Flesching, 1949). It is interesting to note the restricted distribution pattern of the nominal S. guttatus and S. koreanus along the Indian coast with an apparent break in the marine regions adjacent to peninsular India, the reasons for which will be discussed in the upcoming session.
Divergence Time
Although the phylogenetic investigation in the regalis species group has been performed, a comprehensive investigation on divergence time estimation for the Scomberomorus genus is yet to be undertaken. To procure the most reliable information from fossil-calibrated molecular data, molecular dating was carried out with species having at least 70% of the genes selected for the analysis. The results derived from our study differ from all previous investigations (Banford et al., 1999; Santini et al., 2013). The first split from the most recent common ancestor of Scomberomorus divided the Western Atlantic (S. cavalla)–Eastern Pacific species (S. concolor and S. sierra) with the Indo-West Pacific species group (S. munroi, S. niphonius, S. semifasciatus, S. commerson, S. lineolatus, S. guttatus, S. koreanus, and S. aff. guttatus). The recent split of the regalis species group suggested by previous nuclear sequence data (Banford et al., 1999) was also supported in this study, though the divergence time varies, which can be rectified if more species from the regalis group are added for divergence dating. The Indo-West Pacific species group has undergone multiple diversification events; of these events, the most interesting divergence was perceived in the crown group, comprising S. guttatus–S. koreanus and encircling cryptic species. Scomberomorus koreanus and S. aff. guttatus diverged from the common ancestor (S. guttatus) during the Miocene followed by population expansion of S. aff. guttatus during the Pliocene. Both divergence and expansion of the latter were probably materialized due to climatic or oceanographic changes that occurred during the Miocene–Pliocene periods that might have changed the population connectivity leading to range expansions (Renema et al., 2008). The early Miocene period is characterized by the redistribution of salt and heat resulting in significant changes in flow and climate patterns in the Indian Ocean (Bialik et al, 2019). Furthermore, the diversification of S. aff. guttatus into three lineages in the Pliocene could be accelerated by the increase in productivity associated with the biogenic bloom, and shifts in monsoon activity occurred in the Miocene period in the Indian Ocean (Karatsolis et al., 2022). The superimposed climatic changes with local adaptations followed by reproductive aggregation possibly led to sympatric speciation in this genus (Qvarnström et al., 2016; Beal et al., 2020).
Validation of Geographical Races Based on Vertebral Counts
The variations in vertebral counts, both in the precaudal and caudal regions, are observed in a number of species in Scomberomorus. The vertebral count observed for S. commerson, S. lineolatus, and S. koreanus agrees with previous observations on these species (Collette and Russo, 1984). The wide range (47–51) in nominal S. guttatus is discussed here. Jones and Silas (1961) observed 48 or 49 vertebrae in S. guttatus examined from the peninsular (Vizhinjam) and south-east coast (Tuticorin) of India. They indicated that Kishinouye (1923) observed the vertebral count of 51 for Cybium guttatum from Formosan waters and observed an insightful range of variation of this meristic character in S. guttatus. Subsequently, in 1964, Silas redescribed the species from specimens collected in the southern part of the Gulf of Mannar (Tuticorin, India). Later, Devaraj (1976) observed vertebral counts of 49–50 (mostly 50) in samples collected from the northern part of the Gulf of Mannar, which he considered to be the typical S. guttatus. He suggested that the S. guttatus from the southern Gulf of Mannar reported by Silas (1964), as well as the specimens of the contiguous Arabian Sea, characterized by shortening of the precaudal region and elongation of the caudal region, may belong to a different race suspected to be S. guttatus. It should be noted that the occurrence of S. guttatus in the southern Gulf of Mannar and Pamban is currently very rare and the authors were only able to collect very few specimens during the period of this study (2019–2021), contrary to previous observations (Jones and Silas, 1961; Devaraj, 1976; Devaraj, 1998). However, the species shares the fishery with S. commerson in the northern part of the Gulf (from Tranquebar to Nagapattinam), from where the authors were able to collect only one species with 47–49 (usually 48) vertebrae.
The cryptic species S. aff. guttatus in the Indian Ocean has 49–51 vertebrae (usually 50), while its vertebral count in the Western Pacific is usually 51 (Lee and Yang, 1983). Collette and Russo (1984) reported that the Indian Ocean population has a mode of 50 vertebrae, while populations in the East Indies, Gulf of Thailand, and China have 51 as mode. We examined the X-rays of lectotype (MNHN A.5771, Java, 1,075 mm FL, vertebrae 51) and paralectotype (MNHN A. 5715, Mumbai, 1,145 mm FL, vertebrae 50) of Cybium kuhlii (http://n2t.net/ark:/65665/3b5aba155-8948-4776-b613-ff753884c14d and http://n2t.net/ark:/65665/3cf8430ff-c20c-4d82-9424-cf17063a007b), currently synonymized with S. guttatus, to confirm our observations. The lectotype of C. kuhlii from Java and the paralectotype from Mumbai represent the Pacific and Indian Ocean lineages, respectively.
The number of vertebrae is often cited as a meristic character of diagnostic importance in fish taxonomy. The count sometimes differs between populations of a species along with its geographical range, or across the ranges of related species (e.g., Roul et al., 2021), similar to the observations here. Parallel differences in vertebral counts can also occur within the species (Hubbs, 1922). The vertebral number is influenced by a number of factors like ambient temperatures during the ontogeny of individual fish (Jordan, 1892), inheritance, etc., and the separation of environmental and genetic influences is a complex process (McDowall, 2008). Therefore, this character can be considered only in combination with other features for diagnostic purposes.
History of Seerfishes in the Northern Indian Ocean
The historical provenance of Spanish mackerels from the Indian subcontinent begins with S. guttatus, originally described as Scomber guttatus by Bloch and Schneider (1801) from Tranquebar (Tharangambadi), Tamil Nadu, India. Two years later, Russell (1803) described the seerfishes vernacularly known as “Wingeram” and “Konam” from Vizagapatam (present-day Visakhapatnam) on the Coromandel coast of India as Scomber Wingeram and Scomber Konam. They added that the latter was rarer and of inferior quality compared with the English seerfish (Wingeram) of the same size. Later that year, Shaw (1803) from the same locality, based on Russell’s description and figures of Wingeram and Konam, described them as Scomber leopardus and Scomber maculosus, respectively. Cuvier then described in Cuvier and Valenciennes, 1831, Cybium lineolatum from Malabar, Cybium interruptum from Pondicherry (Puducherry), C. kuhlii from Bombay, and C. commerson from Pondicherry and Malabar in India. Day (1878) also reported five species of seerfishes from the Indian Seas under the genus Cybium, viz., C. kuhlii, C. interruptum, C. guttatum, C. commersonii, and C. lineolatum. Jones and Silas (1961), in examining their taxonomy from India, refined all to three valid species, S. guttatus, S. lineolatus, and S. commerson. They synonymized Day’s C. kuhlii and C. guttatum with S. guttatus, C. interruptum and C. lineolatum with S. lineolatus, and C. commersonii with S. commerson. Because the original description of Scomber guttatus in Bloch and Schneider (1801) is sparse, Silas (1964) redescribed S. guttatus. He considered “Wingeram” defined by Russell (1803), later described as Scomber leopardus by Shaw (1803), identical to S. guttatus. Considering the main characters, he also considered Cybium koreanum Kishinouye, 1915 as a subspecies of S. guttatus and divided the latter into two subspecies, viz., S. guttatus guttatus (Bloch and Schneider, 1801) and S. guttatus koreanus (Kishinouye, 1915). However, Devaraj (1976) proved that S. koreanus is distinct from S. guttatus and not a subspecies of the latter, unlike Silas (1964).
Resurrection of Cryptic Species Based on Genetic Evidence
Scomberomorus aff. guttatus in the cryptic complex with two MOTUs showed a p-distance of 10.26%–10.4% with S. guttatus. There exists a divergence of more than 2% between the two MOTUs, which is widely accepted as the heuristic limit for species delineation using COI barcodes (Pereira et al., 2013). Although geographical populations of conspecifics can exhibit genetic distances in excess of 2% (Ward et al., 2005), the likelihood that these MOTUs represent two potentially cryptic species of recent origin cannot be ignored. Albeit, their divergence is not reflected in the aldolase exon 5/intron 5 regions, although the distance estimates of this nuclear gene are high among closely related taxa in the S. regalis complex that have been dated to a recent origin (Banford et al., 1999). This is due to insufficient time for the accumulation of mutations in nuclear markers (Allio et al., 2017). Microsatellite-based analysis of S. guttatus revealed a unit stock from the Persian Gulf (Abedi et al., 2011), and mtDNA sequences in GenBank from this region were identical to S. aff. guttatus 2.
Scomberomorus guttatus is characterized by the fine auxiliary branches radiating on either side of the anterior third of the lateral line, intestine with 2 folds and 3 limbs, and 47–52 vertebrae (Collette and Nauen, 1983). The first two features are common to S. guttatus and the cryptic species identified in this study, while differing in their morphometry and modes of vertebral counts. The occurrence of doppelganger in the S. guttatus species complex which is too cryptic to be identified by external morphology and its geographic populations also add to the difference in vertebral numbers noted by previous researchers. However, it can be observed that typical S. guttatus has a lesser body depth compared to the above. Their cryptic nature can be attributed to their morphological convergence in response to analogous selective regimes (Ingram and Mahler, 2013) which are discussed subsequently. Hence, based on the genetic results of specimens from type locality, vertebral counts of 47–49 (usually 48), and examination of lectotype (ZMB 8759; Figure 7), the identity of S. guttatus (Bloch and Schneider, 1801) is confirmed (Figure 8A) and its morphological redescription is strongly recommended.

Figure 8 Images of (A) Scomberomorus guttatus, collected from Tranquebar, India; (B, C) Scomberomorus leopardus (= S. aff. guttatus 1 and S. aff. guttatus 2, respectively) resurrected in this study. FL indicates fork length of the specimen.
It may be added that Shaw (1803) described Scomber leopardus, which was later synonymized with S. guttatus (Bloch and Schneider, 1801), based on the image and the original description of “Scomber Wingeram” provided by Russell (1803) from Visakhapatnam on the coast of Coromandel. Our study showed the occurrence of both species on the Coromandel coast, but since the type locality contains only S. guttatus which is less abundant in Visakhapatnam, the available senior synonym Scomber leopardus Shaw, 1803 (= Scomberomorus leopardus) merits the distinction to be accepted for the cryptic species. Based on the barcode criterion of mtDNA clustering and critical examination of Russell’s drawing (33.2 cm long), we here resurrect Scomberomorus leopardus (Shaw, 1803) (Figures 8B, C; Supplementary Figure 7) from the synonymy of S. guttatus as a valid species, which needs to be redescribed. The vertebral counts of this species have a mode of 50 for the Indian Ocean lineage, while it is reported to be 51 for the Western Pacific lineage.
It can be added that some junior synonyms, viz., Cybium interruptum and C. kuhlii, are also available in the literature (synonymized with S. guttatus) originally described by Cuvier and Valenciennes (1831) from Pondicherry and Bombay, respectively. Although C. interruptum was successively included in the same manuscript by Cuvier and Valenciennes (1831), we believe that C. interruptum cannot be applied to S. aff. guttatus 1 due to the availability of a senior synonym, Scomber leopardus Shaw, 1803 from the same geographic region. If the Arabian Sea counterpart of S. leopardus (i.e., S. aff. guttatus 2; Figure 8C) and the counterpart from the Bay of Bengal and Western Pacific (S. aff. guttatus 1; Figure 8B) turn out to be distinct on further analysis, the senior name Cybium kuhlii [= Scomberomorus kuhlii (Cuvier in Cuvier and Valenciennes, 1831)] will be applicable to the former. S. leopardus can be retained for the latter since its genesis lies in the Bay of Bengal region. In general, naming/reviving a species generally requires multiple lines of evidence along with the “Generalized Lineage Species Concept” (Coates et al., 2018), and therefore, a detailed study on the apparent phenotypic differentiation in the cryptic species and the redescription of S. guttatus are in progress.
Similarly, for the Indian Ocean complement of S. commerson (i.e., S. cf. commerson 2) which has been shown to be distinct on the basis of detailed molecular analysis, the name Scomber maculosus Shaw, 1803 (= Scomberomorus maculosus) described by Shaw (1803), based on the Konam of Russell (1803), will probably be applicable. However, in the absence of a type specimen of S. commerson, originally described as Scomber commerson after a figure from Commerson’s manuscripts, and the absence of any indication of type locality in the original description, the identity of the species can only be established by designation of a type based on the Rules of the International Code of Zoological Nomenclature.
Ecological Adaptations in the Scomberomorus guttatus Species Group
The S. guttatus species group contains S. guttatus, S. koreanus, and S. multiradiatus (Collette and Russo, 1984). The first two are available in the northern Indian Ocean. Scomberomorus guttatus, a neritic species thought to be less migratory than S. commerson, inhabits the coastal waters at depths between 15 and 200 m and is generally encountered in turbid waters with lower salinity (Fischer and Whitehead, 1974; Collette, 2001) which corroborate with the collection data from this study (Table 2). The habitat of S. guttatus is captured from river mouths and estuaries in Indonesia (Hardenberg, 1936), West Bengal in India (Hora, 1953; Dutta et al., 2021), etc., indicating their tendency to ascend estuaries of large rivers. Similarly, S. koreanus exhibited a remarkable ability to ascend brackish parts of rivers (Kishinouye, 1923). Species range boundaries indicate a species’ ecological niche in space (Sexton et al., 2009), and species morphology is partially related to its ecological niche (Schluter, 2000).
Habitat use, diet, and environmental tolerance are usually measured niche subsets (Slatyer et al., 2013). Selection on habitat preferences can be a strong reproductive barrier that promotes divergence with gene flow (Berner and Thibert-Plante, 2015). The niche breadth and size of the geographical range of S. guttatus is of lesser magnitude in India limited to the east coast, with more aggregations in Tamil Nadu and West Bengal and also in Bangladesh, while the niche expanse is wider for S. leopardus. The range of all Scomberomorus spp. in India is shown in Figure 6. The west coast of India is a coast of submergence (except for the Malabar Coast; Figure 1) with a steep slope and narrow width with only a few west-flowing rivers, forming estuaries while draining into the Arabian Sea. In contrast, the east coast is an emergent coast (Swathi Lekshmi, 2020) with the Bay of Bengal receiving discharge from many east-flowing rivers, fostering larger estuaries and major deltas. The estuaries on the west coast lie mainly between Gujarat and Karnataka, while the extensive east coast estuarine system is in the stretch from West Bengal to Tamil Nadu (Faruque and Ramachandran, 2014a; Faruque et al., 2014b) (Figure 6). The intensity of the seasonal monsoons and the precipitation patterns show a great difference in the Indian subcontinent, and the latter along with other factors creates fluctuations in salinity by freshwater runoff and determines the hydrography of the aforementioned monsoonal estuaries. Tamil Nadu receives more annual precipitation during the winter monsoon (October to November) than most other parts of the country which get it during the southwest monsoon (June–September) (Venkataramana et al., 2021). The Ganges/Brahmaputra and Irrawaddy/Salween River systems transport huge amounts of water and sediment to the northeastern Indian Ocean during and after the rainy season (June to August), resulting in the formation of freshwater pools and seasonal salinity differences (Jöhnck et al., 2020). Deltas and estuarine systems formed by the river Ganges in the northern part of the Bay of Bengal (Gangetic delta in Bangladesh) and the Southern Bay of Bengal (Hoogly-Matlah estuary forming the Sunderbans delta in India) form a highly productive marine ecosystem (Mukhopadhyay et al., 2006) that maintains a salinity gradient due to seasonal freshwater discharge and regular tidal influxes. Scomberomorus guttatus, S. leopardus, and S. koreanus have a preference for coastal and estuarine habitats which results in their adherence to these areas.
As Jordan (1922) pointed out in one of his ecogeographical rules, the nearest relative of any given form often occurs in adjacent areas usually across some barrier to distribution. This holds true for the S. guttatus complex. This occurrence represents an early declaration of the concept of allopatric speciation or perhaps vicariance (McDowall, 2008). The peninsular region of India can act as an apparent geographical barrier due to its steep slope and non-availability of suitable habitats for larval settlement. Besides, Scomberomorus spp. have pelagic eggs and larvae (Vijayaraghavan, 1955; Jones, 1961; Collette, 1986; da Cunha et al., 2020). Although information on larval dispersal with monsoon currents is sparse for the S. guttatus complex, the range overlap between the MOTUs of the lineages in S. aff. guttatus (= S. leopardus) near Mangaluru (southern Karnataka) indicates that the larvae entering the currents and circulations in the southern peninsula of the Indian subcontinent (Rao and Sreenivas, 2020) settle only in their most appropriate niches.
Recruitment and population dynamics of fish are mainly driven by their feeding ecology and dietary plasticity (Sui et al., 2021). Vijayaraghavan (1955) suggested that the dietary component of S. guttatus changes with habitat and season. The gut content analysis of S. guttatus from the north-eastern Arabian Sea showed that their major food is Acetes sp. followed by clupeids (Chellappan et al., 2018). This area is rich in extremely euryhaline Acetes spp., which provide food for many predators (Deshmukh, 2007; Metillo et al., 2015). This contradicts the feeding pattern observed on the south and south-east coasts of India, where the prey is Sardinella spp. and whitebaits (Devaraj, 1998). Piscine prey abundance is expected to affect larval survival and recruitment of Scomberomorus, as illustrated by the larval survival strategy with an early switching of diet from planktivory to piscivory, which reduces the duration of the larval stage (Shoji et al., 2005). Chale-Matsau (1996) posited that the abundance of S. plurilineatus on the KwaZulu-Natal coastline was related to the appearance of the inshore marine prey species Sardinops sagax along the coast rather than to spawning. It can be observed that the highly productive areas along the Indian coast where S. guttatus spp. inhabit provide crustacean and piscine preys that can aid in their larval recruitment.
The spawning season observed for S. guttatus on the south-east coast of India is April to July with a spawning peak in July (Krishnamoorthi, 1958), while Devaraj et al. (1987) suggested an extended spawning season from January to August with a peak from April to May. Collette and Nauen (1983) indicated it from April to July around Rameswaram Island between India and Sri Lanka. On the east coast of India (Andhra Pradesh), the peak spawning period is between November and April (Uma Mahesh et al., 2017), while on the west coast along the northern Arabian Sea, it is May (Chellappan et al., 2018). Season is longer with a peak in January to August in Indonesian waters (Noegroho et al., 2018), but the peak recruitment is from May to July in Bangladesh (Rashid et al., 2010). The difference in spawning times in S. guttatus can be due to differences in biotic and abiotic parameters in the area (Roa-Ureta et al., 2019) coupled with life-history traits of the nominal S. guttatus which is now revealed to contain cryptic species.
Fidelity to spawning sites (probably upwelling areas) rather than adaptations to oceanographic regimes has been postulated as one of the reasons for the incidence of genetically discrete groups in S. sierra (López et al., 2010). In the current context of limited information on spawning grounds along the coastline, except for the reports by Devaraj (1987) and Jones (1961), which were limited to the south-east and south-west coasts of India respectively, the above postulate cannot be verified. Recent studies evidence that marine species with pelagic larval stages are subjected to high levels of local recruitment, which can lead to reduced gene flow, niche specialization through local adaptation, and sexual selection, leading to divergence and speciation (Milá et al., 2017). Fišer et al. (2018) pointed out that cryptic species show differences in their requirements along their Grinnellian niche. Speciation is a protracted process and a continuum of reproductive isolation (Stankowski and Ravinet, 2021), which are effects from the chance fixation of diverse, epistatic incompatibilities in discrete populations subjected to identical selective pressures (Rosser et al., 2019). Though many factors point to the probability that the distribution pattern of S. guttatus species complex in India is more connected with ecological niche and feeding regime, further studies are recommended to establish the relevant eco–evo interactions (Lowe et al., 2017).
Conclusion
The congeneric species in Scomberomorus from the northern Indian Ocean have not yet been subjected to molecular taxonomic studies. Our study aimed to generate multilocus data comparable to previous studies for phylogenetic reconstruction, species delimitation, resurrection of a dated phylogeny, and validation of the hypothesis of geographic races in S. guttatus. We have collected all the nominal species available along the Indian coast, and our analysis showed that the nominal S. guttatus contains a highly cryptic species. The latter contains two geographically separated lineages distributed mainly on the east and west coasts. Based on genetic detection of specimens from the type locality and subsequent confirmatory analyses, S. guttatus (Bloch and Schneider, 1801) is redeemed and the cryptic species S. leopardus (Shaw, 1803) is resurrected. The two geographical races of S. leopardus that cross the cutoff of species delimitation may also represent two cryptic species for which further morphomeristic studies are recommended. The Indo-Pacific king mackerel, S. guttatus, thought to have a panoceanic distribution between the Persian Gulf and the Sea of Japan, is now restricted to the Bay of Bengal, while the cryptic species enjoys a wider distribution. This information on species composition is highly significant for management as its fishery is “collapsed” in certain regions (Ju et al., 2020). Reliable molecular data were generated here and included in the global database for the underrepresented species such as S. lineolatus and S. koreanus. The phylogenetic position of Indian counterparts has been ascertained and their divergence time was estimated. The geographical distribution of available species and doppelgangers was discussed in terms of their habitat preference, diet, and biological information. Our study demonstrated the necessity for global data from all species in order to arrive at a thorough conclusion about the systematics of this genus to verify its concordance with morphological groupings in the previous literature. Recognition of species in catch is essential to avoid overexploitation and to assess the potential impact of fisheries on fish populations, particularly for species with a restricted geographical range (akin to many members of Scomberomorus) that are more vulnerable to climate change (Dineshbabu et al., 2020). It is extremely important to conduct niche breadth and range size studies with a wide sampling to predict their vulnerability to altering climatic conditions.
Data Availability Statement
All gene sequences generated in this study are deposited with the NCBI under accession numbers MZ955278-MZ955299, MZ955300-MZ955315, MZ955327-MZ955334, MZ955335-MZ955342, MZ955343-MZ955361, MZ962383-MZ962389, OL362234- OL362241, OM363524-OM363528, OM363529-OM363541, OM363542-OM363555, OM363556-OM363578, OM363579-OM363600, OM363601-OM363617, OM417804- OM417807, OM417808-OM417811, OM436012-OM436014, OK086368, ON193432, ON166824, OK137200-OK137203, OK137205, OK137206-OK137207, OK041506-OK041511, MZ927323-MZ927330, OM348536, MZ922475, MZ922477-MZ922478, MZ923800-MZ923804, MZ923799, MZ927331-MZ927338, MZ854022-MZ854034, MZ914677-MZ914678, ON150870, MZ854035- MZ854038, ON128513, MZ854039- MZ854053, MZ914449-MZ914459, MZ914681, ON127539-ON127549, ON129805-ON129822, ON204167-ON204170, MZ871761-MZ871765, MZ872464-MZ872504, ON207807-ON207814, OM343183, MZ927112, OM416532-OM416538, and are accessible at https://www.ncbi.nlm.nih.gov/nuccore.
Ethics Statement
Ethical review and approval was not required for the animal study because this work does not contain any experimental studies with live animals. Fish samples caught in various gears were collected from the fishing harbors/landing centers and used for the genetic and morphological/anatomical investigations.
Author Contributions
NJ conceptualized the work, analyzed the data, and wrote the manuscript. SR carried out the lab work, submitted and analyzed the molecular data, and assisted in draft preparation. SKR incorporated the taxonomic information, reviewed the manuscript, and provided all samples from the north-east coast of India. PA provided samples from the north-west coast and prepared the GIS-based maps. Specimens were given by RV and AM from the south-east coast, HM from Visakhapatnam, SS and EA from Kerala, and EN from Andaman. PR provided samples from Mangalore and gave extensive support to this work. AG coordinated the work. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We would like to thank the Director of ICAR-CMFRI, Kerala, India, for providing facilities. We thank A. Anuraj, Joe Kizhakudan, Ajay D. Nakhawa, C. Anulekshmi, K. M. Rajesh and Y. Gladston for their participation in sampling additional materials. Pradeep from Cuddalore is thanked for assisting with the collections in the area. We also thank V. Mahesh for the sample and image provided. Our thanks go to Edda Aßel, Collection Manager, Ichthyology, and Dr. Peter Bartsch, Sc. Head of Ichthyology Collection, Museum für Naturkunde, Berlin who provided photographs, CT-image and information on the lectotype. Special thanks to Geethu Gopinath for the help in molecular lab work. The authors acknowledge the partial funding they received from the in-house project of the Marine Biotechnology Division, CMFRI (MBT/GNM/25) to carry out part of the molecular work.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2022.888463/full#supplementary-material
References
Abedi E., Mohammadi M., Qasemi A. (2011). Genetic Stock Structure of Scomberomorus Guttatus Using Microsatellite Markers in the Persian Gulf. J. Oceanogr. 2 (6), 15–21. doi; 10.5829/idosi.aejaes.2012.12.10.656
Abedi E., Zolgharnein H., Salari M. A., Qasemi A. (2012). Genetic Differentiation of Narrow-Barred Spanish Mackerel (Scomberomorus Commerson) Stocks Using Microsatellite Markers in the Persian Gulf. Am. Eurasian J. Agric. Environ. Sci. 12 (10), 1305–1310. doi; 10.5829/idosi.aejaes.2012.12.10.656
Allcock A. L., Strugnell J. M. (2012). Southern Ocean Diversity: New Paradigms From Molecular Ecology. Trends Ecol. Evol. 27, 520–528. doi: 10.1016/j.tree.2012.05.009
Allio R., Donega S., Galtier N., Nabholz B. (2017). Large Variation in the Ratio of Mitochondrial to Nuclear Mutation Rate Across Animals: Implications for Genetic Diversity and the Use of Mitochondrial DNA as a Molecular Marker. Mol. Boil. Evol. 34 (11), 2762–2772. doi: 10.1093/molbev/msx197
Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. (1990). Basic Local Alignment Search Tool. J. Mol. Biol. 215 (3), 403–410. doi: 10.1016/S0022-2836(05)80360-2
Banford H. M., Bermingham E., Collette B. B., McCafferty S. S., Stackebrandt E., Goodfellow M. (1999). Phylogenetic Systematics of the Scomberomorus Regalis (Teleostei: Scombridae) Species Group: Molecules, Morphology and Biogeography of Spanish Mackerels. Copeia, (3), 596–613. doi: 10.2307/1447593
Beal L. M., Vialard J., Roxy M. K., Li J., Andres M., Annamalai H., et al. (2020). A Road Map to IndOOS-2: Better Observations of the Rapidly Warming Indian Ocean. Bull. Am. Meteorol. Soc 101 (11), E1891–E1913. doi: 10.1175/BAMS-D-19-0209.1
Berner D., Thibert-Plante X. (2015). How Mechanisms of Habitat Preference Evolve and Promote Divergence With Gene Flow. J. Evol. Biol. 28 (9), 1641–1655. doi: 10.1111/jeb.12683
Betancur-R R., Wiley E. O., Arratia G., Acero A., Bailly N., Miya M., et al. (2017). Phylogenetic Classification of Bony Fishes. BMC Evol. Biol. 17, 162. doi: 10.1186/s12862-017-0958-3
Bialik O. M., Frank M., Betzler C., Zammit R., Waldmann N. D. (2019). Two-Step Closure of the Miocene Indian Ocean Gateway to the Mediterranean. Sci. Rep. 9, (1–10). doi: 10.1038/s41598-019-45308-7
Bloch M. E., Schneider J. G. (1801). “M.E. Blochii, Systema Ichthyologiae Iconibus Cx Ilustratum,” in Post Obitum Auctoris Opus Inchoatum Absolvit, Correxit, Interpolavit. Ed. Schneider J. G. (Berolini, Berlin: Saxo Berolini).
Bossuyt F., Milinkovitch M. C. (2000). Convergent Adaptive Radiations in Madagascan and Asian Ranid Frogs Reveal Covariation Between Larval and Adult Traits. Proc. Natl. Acad. Sci. 97, 6585–6590. doi: 10.1073/pnas.97.12.6585
Bouckaert R., Vaughan T. G., Barido-Sottani J., Duchêne S., Fourment M., Gavryushkina A., et al. (2019). Beast 2.5: An Advanced Software Platform for Bayesian Evolutionary Analysis. PloS Comput. Biol. 15, e1006650. doi: 10.1371/journal.pcbi.1006650
Broderick D., Ovenden J. R., Buckworth R. C., Newman S. J., Lester R. J. G., Welch D. J. (2011). Genetic Population Structure of Grey Mackerel Scomberomorus Semifasciatus in Northern Australia. J. Fish Biol. 79 (3), 633–661. doi: 10.1111/j.1095-8649.2011.03055.x
Chale-Matsau J. R. (1996). Age and Growth of the Queen Mackerel (Scomberomorus Plurilineatus) and Seventy-four (Polysteganus Undulosus) of KwaZulu Natal, South Africa. Dissertation (University of Natal, Durban, South Africa).
Chellappan A., Sarang J. D., Kamble S. D., Akhilesh K. V., Deshmukh V. D., Singh V. V. (2018). Biological Aspects of Spotted Seerfish Scomberomorus Guttatus (Bloch and Schneide) (Scombridae) From North-Eastern Arabian Sea. Indian J. Fish. 65 (2), 42–49.
Chen W. J., Bonillo C., Lecointre G. (2003). Repeatability of Clades as a Criterion of Reliability: A Case Study for Molecular Phylogeny of Acanthomorpha (Teleostei) With Larger Number of Taxa. Mol. Phylogenet. Evol. 26 (2), 262–288. doi: 10.1016/S1055-7903(02)00371-8
Cheng Y. Z., Xu T. J., Jin X. X., Tang D., Wei T., Sun Y. Y., et al. (2012). Universal Primers for Amplification of the Complete Mitochondrial Control Region in Marine Fish Species. Mol. Biol. 46 (5), 727–730. doi: 10.1134/S0026893312040024
Clements K. D., Gray R. D., Choat J. H. (2003). Rapid Evolutionary Divergences in Reef Fishes of the Family Acanthuridae (Perciformes: Teleostei). Mol. Phylogenet. Evol. 26 (2), 190–201. doi: 10.1016/S1055-7903(02)00325-1
CMFRI, FRAD (2020). Marine Fish Landings in India 2019 (Kochi: Technical Report. ICAR-Central Marine Fisheries Research Institute).
Coates D. J., Byrne M., Moritz C. (2018). Genetic Diversity and Conservation Units: Dealing With the Species-Population Continuum in the Age of Genomics. Front. Ecol. Evol. 6. doi: 10.3389/fevo.2018.00165
Collette B. B. (1986). “Scombridae (Including Thunnidae, Scomberomoridae, Gasterochismatidae and Sardidae),” in Fishes of the North-Eastern Atlantic and the Mediterranean, Volume 2. Eds. Whitehead P. J. P., Bauchot M. L., Hureau J. C., Nielsen J., Tortonese E. (Paris: UNESCO), 981– 997.
Collette B. B. (1994). Scomberomorus Lineolatus is a Valid Species of Spanish Mackerel, Not an Interspecific Hybrid: A Reply. J. Nat. Hist. 28 (5), 1205–1208. doi: 10.1080/00222939400770601
Collette B. B. (2001). “Scombridae. Tunas (Also, Albacore, Bonitos, Mackerels, Seerfishes, and Wahoo),” in FAO Species Identification Guide for Fishery Purposes. The Living Marine Resources of the Western Central Pacific. Vol. 6. Bony Fishes Part 4 (Labridae to Latimeriidae), Estuarine Crocodiles. Eds. Carpenter K. E., Niem V. (Rome: FAO), 3721–3756.
Collette B. B. (2003). Family Scombridae Rafinesque 1815-Mackerels, Tunas, and Bonitos, Annotated Checklists of Fishes, 19 (San Francisco, California: California Academy Sciences), 28.
Collette B. B., Nauen C. E. (1983). Scombrids of the World: An Annotated and Illustrated Catalogue of Tunas, Mackerels, Bonitos, and Related Species Known to Date. V. 2. FAO Fish. Synop. 125, vii137.
Collette B. B., Russo J. L. (1984). Morphology, Systematics, and Biology of the Spanish Mackerels (Scomberomorus, Scombridae). Fish. Bull. 82 (4), 545–692.
Crandall E. D., Riginos C., Bird C. E., Liggins L., Treml E., Beger M., et al. (2019). The Molecular Biogeography of the Indo-Pacific: Testing Hypotheses With Multispecies Genetic Patterns. Glob. Eco. Biogeogr. 28 (7), 943–960. doi: 10.1111/geb.12905
Crête-Lafrenière A., Weir L. K., Bernatchez L. (2012). Framing the Salmonidae Family Phylogenetic Portrait: A More Complete Picture From Increased Taxon Sampling. PloS One 7 (10), e46662. doi: 10.1371/journal.pone.0046662
Cuvier G. (1829). Le Règne Animal, Distribué D’après Son Organisation, Pour Servir De Base À L’histoire Naturelle Des Animaux Et D’introduction À L’anatomie Comparée, Vol. 2. v .2. Chez Díterville, Paris, i–xv 1–406 .
Cuvier G., Valenciennes A. (1831). HistoireNaturelle Des Poissons. Tome Huitième. Livre Neuvième. Des Scombéroïdes, Vol. v. 8. Levrault, Paris i–xix 5 pp. 1–509Pls. 209–245.
da Cunha D. B., Rodrigues-Filho L. F. S., de Luna Sales J. B., Rêgo P., Queiroz C., Sampaio I., et al. (2020). Molecular Inferences on Scomberomorus Brasiliensis, From the Western South Atlantic, Based on Two Mitochondrial Genes. Front. Mar. Sci. 7. doi: 10.3389/fmars.2020.558902
Day F. (1878). The Fishes of India; Being a Natural History of the Fishes Known to Inhabit the Seas and Fresh Waters of India, Burma, and Ceylon. Bernard Quaritch, London i–xx553–779 139–195.
Deshmukh V. D. (2007). Predators of Non-Penaeid Prawns of Mumbai Coast. J. Bombay Nat. Hist. Soc 104 (3), 266–274.
Devaraj M. (1976). Discovery of the Scombrid Scomberomorus Koreanus (Kishinouye) in India, With Taxonomic Discussion on the Species. Jpn. J. Ichthyol. 23 (2), 79–87.
Devaraj M. (1987). Maturity, Spawning and Fecundity of the Spotted Seer, Scomberomorus Guttatus, in the Gulf of Mannar and Palk Bay. Indian J. Fish. 34, 48–77.
Devaraj M. (1998). Food and Feeding Habits of the Spotted Seer, Scomberomorus Guttatus (Bloch and Schneider), in the Gulf of Mannar and Palk Bay. Mar. Biol. Assoc. India. 40, 105–124.
Devaraj M., Mohamad Kasim H. (1998). Seerfish Fishery of the World. Mar. Biol. Assoc. India. 40, 51–68.
Dineshbabu A. P., Zacharia P. U., Sujitha T., Shoba J. K., Rajesh K. M., Vivekanandan E., et al. (2020). Assessment of Stock Vulnerability of Indian Marine Fishes to Past Changes in Climate and Options for Adaptation. Clim. Res. 79 (3), 175–192. doi: 10.3354/cr01586
Divya P. R., Vineesh N., Kathirvelpandian A., Basheer V. S., Gopalakrishnan A. (2018). Population Structure of Spanish Mackerel Scomberomorus Commerson (Lacepede 1800) in the Northern Indian Ocean Determined Using Microatellite Markers. Aquat. Living Resour. 31 (22), 1–10. doi: 10.1051/alr/2018011
Dutta S., Orlov A., Hazra S. (2021). Population Biology and Exploitation Status of Four Commercially Important Marine Fishes of the Northern Bay of Bengal, India. Iran J. Fish. Sci. 20 (1), 62–83.
Faruque B. M., Ramachandran K. V. (2014a). “The Continental Shelf of Western India,” in Continental Shelves of the World: Their Evolution During the Last Glacio-Eustatic Cycle, vol. 41 . Eds. Chiocci F. L., Chivas A. R. (London, Memoirs: Geological Society), 213–220.
Faruque B. M., Vaz G. G., Mohapatra G. P. (2014b). “The Continental Shelf of Eastern India,” in Continental Shelves of the World: Their Evolution During the Last Glacio-Eustatic Cycle, vol. 41 . Eds. Chiocci F. L., Chivas A. R. (London, Memoirs: Geological Society), 221–229.
Fauvelot C., Borsa P. (2011). Patterns of Genetic Isolation in a Widely Distributed Pelagic Fish, the Narrow-Barred Spanish Mackerel (Scomberomorus Commerson). Biol. J. Linn. Soc 104 (4), 886–902. doi: 10.1111/j.1095-8312.2011.01754.x
Fischer W., Whitehead P. J. P. (1974). “Eastern Indian Ocean Fishing Area 57 and Western Central Pacific Fishing Area 71,” in FAO Species Identification Sheets for Fishery Purposes, vol. IV, (Rome: Food and Agriculture Organization of the United Nations). pp 196.
Fišer C., Robinson C. T., Malard F. (2018). Cryptic Species as a Window Into the Paradigm Shift of the Species Concept. Mol. Ecol. 27 (3), 613–635. doi: 10.1111/mec.14486
Fitch E. J., Flesching A. O. (1949). A Brief Account of the Monterey Spanish Mackerel (Scomberomorus Concolor). California Fish Game. 35, 275–280.
Froese R., Pauly D. (Eds.) (2022). Fish Base World Wide Web Electronic Publication. Available at: http://www.fishbase.org
Fricke R., Eschmeyer W. N., van der Laan R. (2021) Eschmeyer's Catalog of Fishes: Genera, Species, References. Available at: http://researcharchive.calacademy.org/research/ichthyology/catalog/fishcatmain.asp (Accessed 28 April 2021).
Fujisawa T., Barraclough T. G. (2013). Delimiting Species Using Single-Locus Data and the Generalized Mixed Yule Coalescent Approach: A Revised Method and Evaluation on Simulated Data Sets. Syst. Biol. 62, 707–724. doi: 10.1093/sysbio/syt033
Garner B. A., Hoban S., Luikart G. (2020). Iucn Red List and the Value of Integrating Genetics. Conserv. Genet. 21 (5), 795–801. doi: 10.1007/s10592-020-01301-6
Ge Y., Xia C., Wang J., Zhang X., Ma X., Zhou Q. (2021). The Efficacy of DNA Barcoding in the Classification, Genetic Differentiation, and Biodiversity Assessment of Benthic Macroinvertebrates. Ecol. Evol. 11 (10), 5669–5681. doi: 10.1002/ece3.7470
Habib A., Sulaiman Z. (2016). High Genetic Connectivity of Narrow-Barred Spanish Mackerel (Scomberomorus Commerson) From the South China, Bali and Java Seas. Zool. Ecol. 26 (2), 93–99. doi: 10.1080/21658005.2016.1161121
Halasan L. C., Geraldino P. J. L., Lin H. C. (2021). First Evidence of Cryptic Species Diversity and Population Structuring of Selaroides Leptolepis in the Tropical Western Pacific. Front. Mar. Sci 8, 756163. doi: 10.3389/fmars.2021.756163
Hall T. A. (1999). BioEdit: A User-Friendly Biological Sequence Alignment Editor and Analysis Program for Windows 95/98/Nt. Nucleic Acids Symposium Ser. 41, 95–98.
Hardenberg J. D. F. (1936). On a Collection of Fishes From the Estuary and the Lower and Middle Course of the River Kapuas (W. Borneo). Treubia Buitzenzorg 15, 225–254.
Hashemi S. A. R., Doustdar M. (2022). Fishery Status of Narrow-Barred Spanish Mackerel (Scomberomorus Commerson, Lacepede 1800) in the Northern Waters of the Oman Sea, Iran. Iran. J. Ichthyol. 8 (4), 311–321. doi: 10.22034/iji.v8i4.669
Heath T. A., Hedtke S. M., Hillis D. M. (2008). Taxon Sampling and the Accuracy of Phylogenetic Analyses. J. System. Evol. 46 (3), 239–257. doi: 10.3724/SP.J.1002.2008.08016
Hofmann E. P., Nicholson K. E., Luque-Montes I. R., Köhler G., Cerrato-Mendoza C. A., Medina-Flores M., et al. (2019). Cryptic Diversity, But to What Extent? Discordance Between Single-Locus Species Delimitation Methods Within Mainland Anoles (Squamata: Dactyloidae) of Northern Central America. Front. Genet., 10, 11. doi: 10.3389/fgene.2019.00011
Hoolihan J. P., Anandh P., van Herwerden L. (2006). Mitochondrial DNA Analyses of Narrow-Barred Spanish Mackerel (Scomberomorus Commerson) Suggest a Single Genetic Stock in the ROPME Sea Area (Arabian Gulf, Gulf of Oman, and Arabian Sea). ICES J. Mar. Sci. 63, 1066–1074. doi: 10.1016/j.icesjms.2006.03.012
Hora S. L. (1953). Rising Salinity of the Hooghly River-Fishes Provide Evidences. Indian J. Pwr. Riv. Vall. Dev. 3 (6), 12–15.
Hubbs C. L. (1922). Variations in the Number of Vertebrae and Other Meristic Characters of Fishes Correlated With the Temperature of Water During Development. Am. Nat. 56, 360–372. doi: 10.1086/279875
Ingram T., Mahler D. L. (2013). SURFACE: Detecting Convergent Evolution From Comparative Data by Fitting Ornstein-Uhlenbeck Models With Stepwise Akaike Information Criterion. Methods Ecol. Evol. 4 (5), 416–425. doi: 10.1111/2041-210X.12034
James P. S. B. R. (2008). Osteo-Taxonomic Distinction of Fishes of the Family Leiognathidae. Indian J. Fish. 55 (4), 305–310.
Jöhnck J., Kuhnt W., Holbourn A., Andersen N. (2020). Variability of the Indian Monsoon in the Andaman Sea Across the Miocene-Pliocene Transition. Paleoceanogr. Paleoclimatol. 35 (9), e2020PA003923. doi: 10.1029/2020PA003923
Johnson M. G., Mgaya Y. D., Shaghude Y. W. (2021). Analysis of the Genetic Stock Structure and Phylogenetic Relationship of Narrow-Barred Spanish Mackerel Scomberomorus Commerson (Lacépède 1800) Along the Northern Tanzanian Coastal Waters Using Mitochondrial DNA. Reg. Stud. Mar. Sci. 101862. doi: 10.1016/j.rsma.2021.101862
Jones S. (1961). Notes on Eggs, Larvae and Juveniles of Fishes From Indian Waters VIII. Scomberomorus Guttatus (Bloch and Schneider), Ix. Scomberomorus Commerson (Lacepede) and X. Scomberomorus Lineolatus (Cuvier). Indian J. Fish. 8 (1), 107–120.
Jones S., Silas E. G. (1961). On Fishes of the Subfamily Scomberomorinae (Family Scombridae) From Indian Waters. Indian J. Fish. 8 (1), 189–206.
Jordan D. S. (1892). Relations of Temperature to Vertebrae Among Fishes. Proc. U. S. Natl. Mus. 1891, 107–120.
Jordan D. S. (1922). The Days of a Man: Being Memories of a Naturalist, Teacher and Minor Prophet of Democracy. Two Volumes (Yonkers on Hudson, NY: World Book Company).
Juan-Jordá M. J., Mosqueira I., Freire J., Dulvy N. K. (2013). The Conservation and Management of Tunas and Their Relatives: Setting Life History Research Priorities. PloS One 8 (8), e70405. doi: 10.1371/journal.pone.0070405
Ju P., Tian Y., Chen M., Yang S., Liu Y., Xing Q., et al. (2020). Evaluating Stock Status of 16 Commercial Fish Species in the Coastal and Offshore Waters of Taiwan Using the CMSY and BSM Methods. Front. Mar. Sci. 7, 618. doi: 10.3389/fmars.2020.00618
Kapli P., Lutteropp S., Zhang J., Kobert K., Pavlidis P., Stamatakis A., et al. (2017). Multi-Rate Poisson Tree Processes for Single-Locus Species Delimitation Under Maximum Likelihood and Markov Chain Monte Carlo. Bioinf. 33 (11), 1630–1638. doi: 10.1093/bioinformatics/btx025
Karatsolis B.-T., Lougheed B. C., De Vleeschouwer D., Henderiks J. (2022). Abrupt Conclusion of the Late Miocene-early Pliocene Biogenic Bloom at 4.6-4.4 Ma. Nat. Commun. 13, 353. doi: 10.1038/s41467-021-27784-6
Kawaguchi A., Miya M., Nishida M. (2001). Complete Mitochondrial DNA Sequence of Aulopus Japonicus (Teleostei: Aulopiformes), a Basal Eurypterygii: Longer DNA Sequences and Higher-Level Relationships. Ichthyol. Res. 48, 213–223. doi: 10.1007/s10228-001-8139-0
Kaymaram F., Hossainy S. A., Darvishi M., Talebzadeh S. A., Sadeghi M. S. (2010). Reproduction and Spawning Patterns of the Scomberomorus Commerson in the Iranian Coastal Waters of the Persian Gulf and Oman Sea. Iran. J. Fish. Sci. 9 (2), 233–244.
Kishinouye K. (1915). Contributions to the Study of Mackerels, Seerfishes, and Tunnies. Proc. Sci. Fish. Assoc. (Suisan Gakkai Ho) 1, 1–24.
Kishinouye K. (1923). Contributions to the Comparative Study of the So-Called Scombroid Fishes. J. Coll. Agricult Imperial Univ. Tokyo. 8 (3), 293–476.
Krishnamoorthi B. (1958). Observations on the Spawning Season and the Fisheries of the Spotted Seer, Scomberomorus Guttatus (Bloch and Schneider). Indian J. Fish. 5, 270–281.
Lanfear R., Frandsen P. B., Wright A. M., Senfeld T., Calcott B. (2017). PartitionFinder 2: New Methods for Selecting Partitioned Models of Evolution for Molecular and Morphological Phylogenetic Analyses. Mol. Biol. Evol. 34, 772–773. doi: 10.1093/molbev/msw260
Lee S. C., Yang H. C. (1983). Fishes of the Suborder Scombroidei of Taiwan. Bull. Inst. Zool. Acad. Sin. 22 (2), 217–242.
Leigh J. W., Bryant D., Nakagawa S. (2015). Popart: Full-Feature Software for Haplotype Network Construction. Methods Ecol. Evol. 6, 1110–1116. doi: 10.1111/2041-210X.12410
Lessa E., Applebaum G. (1993). Screening Techniques for Detecting Variation in DNA Sequences. Mol. Ecol. 2, 121–129. doi: 10.1111/j.1365-294X.1993.tb00006.x
Lewis A. (1981). Population Genetics, Ecology and Systematic of Indo-Australian Scombrid Fishes, With Particular Reference to Skipjack Tuna (Katsuwonus Pelamis). Ph.D. Thesis (Canberra: Australian National University), 314.
López M. D., Alcocer M. U., Jaimes P. D. (2010). Phylogeography and Historical Demography of the Pacific Sierra Mackerel (Scomberomorus Sierra) in the Eastern Pacific. BMC Genet. 11 (1), 1–12. doi: 10.1186/1471-2156-11-34
Lowe W. H., Kovach R. P., Allendorf F. W. (2017). Population Genetics and Demography Unite Ecology and Evolution. Trends Ecol. Evol. 32 (2), 141–152. doi: 10.1016/j.tree.2016.12.002
Luo A., Ling C., Ho S. Y., Zhu C. D. (2018). Comparison of Methods for Molecular Species Delimitation Across a Range of Speciation Scenarios. Syst. Biol. 67 (5), 830–846. doi: 10.1093/sysbio/syy011
Magallón-Gayón E., Diaz-Jaimes P., Uribe-Alcocer M. (2016). Spatial and Temporal Genetic Homogeneity of the Monterey Spanish Mackerel, Scomberomorus Concolor, in the Gulf of California. PeerJ 4, e2583. doi: 10.7717/peerj.2583
McDowall R. M. (2008). Jordan’s and Other Ecogeographical Rules, and the Vertebral Number in Fishes. J. Biogeogr. 35 (3), 501–508. doi: 10.1111/j.1365-2699.2007.01823.x
Metillo E. B., Cadelinia E. E., Hayashizaki K. I., Tsunoda T., Nishida S. (2015). Feeding Ecology of Two Sympatric Species of Acetes (Decapoda: Sergestidae) in Panguil Bay, the Philippines. Mar. Freshwater Res. 67 (10), 1420–1433. doi: 10.1071/MF15001
Milá B., Van Tassell J. L., Calderón J. A., Rüber L., Zardoya R. (2017). Cryptic Lineage Divergence in Marine Environments: Genetic Differentiation at Multiple Spatial and Temporal Scales in the Widespread Intertidal Goby Gobiosoma Bosc. Ecol. Evol. 7 (14), 5514–5523. doi: 10.1002/ece3.3161
Miller E. C., Hayashi K. T., Song D., Wiens J. J. (2018). Explaining the Ocean's Richest Biodiversity Hotspot and Global Patterns of Fish Diversity. Proc. R. Soc 285 (1888), 20181314. doi: 10.1098/rspb.2018.1314
Miya M., Friedman M., Satoh T. P., Takeshima H., Sado T., Iwasaki W., et al. (2013). Evolutionary Origin of the Scombridae (Tunas and Mackerels): Members of a Paleogene Adaptive Radiation With 14 Other Pelagic Fish Families. PloS One 8 (9), e73535. doi: 10.1371/journal.pone.0073535
Monolisha S., Platt T., Sathyendranath S., Jayasankar J., George G., Jackson T. (2018). Optical Classification of the Coastal Waters of the Northern Indian Ocean. Front. Mar. Sci. 5. doi: 10.3389/fmars.2018.00087
Morgan J. A., Macbeth M., Broderick D., Whatmore P., Street R., Welch D. J., et al. (2013). Hybridisation, Paternal Leakage and Mitochondrial DNA Linearization in Three Anomalous Fish (Scombridae). Mitochondrion. 13 (6), 852–861. doi: 10.1016/j.mito.2013.06.002
Mukhopadhyay S. K., Biswas H. D. T. K., De T. K., Jana T. K. (2006). Fluxes of Nutrients From the Tropical River Hooghly at the Land–Ocean Boundary of Sundarbans, Ne Coast of Bay of Bengal, India. J. Mar. Syst. 62 (1-2), 9–21. doi: 10.1016/j.jmarsys.2006.03.004
Nabhan A. R., Sarkar I. N. (2012). The Impact of Taxon Sampling on Phylogenetic Inference: A Review of Two Decades of Controversy. Brief. Bioinform. 13 (1), 122–134. doi: 10.1093/bib/bbr014
Nguyen L. T., Schmidt H. A., Von Haeseler A., Minh B. Q. (2015). Iq-TREE: A Fast and Effective Stochastic Algorithm for Estimating Maximum-Likelihood Phylogenies. Mol. Biol. Evol. 32 (1), 268–274. doi: 10.1093/molbev/msu300
Noegroho T., Boer M., Adrianto L. (2018). Size Structure and Population Dynamics of Indo-Pacific King Mackerel (Scomberomorus Guttatus) in Kepulauan Riau's Water, Indonesia. Aquacult Aquarium Conserv. Legislation. 11 (4), 1081–1088.
Ollé J., Vilà-Valls L., Alvarado-Bremer J., Cerdenares G., Duong T. Y., Hajjej G., et al. (2022). Population Genetics Meets Phylogenetics: New Insights Into the Relationships Among Members of the Genus Euthynnus (Family Scombridae). Hydrobiologia 849 (1), 47–62. doi: 10.1007/s10750-021-04707-6
Orrell T. M., Collette B. B., Johnson G. D. (2006). Molecular Data Support Separate Scombroid and Xiphioid Clades. Bull. Mar. Sci. 79 (3), 505–519.
Paine M. A., McDowell J. R., Graves J. E. (2007). Specific Identification of Western Atlantic Ocean Scombrids Using Mitochondrial DNA Cytochrome C Oxidase Subunit I (COI) Gene Region Sequences. Bull. Mar. Sci. 80 (2), 353–367.
Palumbi S. R. (1996). “Nucleic Acids II: The Polymerase Chain Reaction,” in Molecular Systematics. Eds. Hillis D. M., Mortiz C., Mable B. K. (Sunderland, MA: Sinauer), 205–248.
Palumbi S. R., Martin A. P., Romano S., Mcmilan W. O., Stice L., Grabowski G. (1991). A Simple Fool’s Guide to PCR (Honolulu, HI: University of Hawaii Press).
Pentinsaari M., Vos R., Mutanen M. (2017). Algorithmic Single-Locus Species Delimitation: Effects of Sampling Effort, Variation and Nonmonophyly in Four Methods and 1870 Species of Beetles. Mol. Ecol. Resour. 17 (3), 393–404. doi: 10.1111/1755-0998.12557
Perdices A., Bermingham E., Montilla A., Doadrio I. (2002). Evolutionary History of the Genus Rhamdia (Teleostei: Pimelodidae) in Central America. Mol. Phylogenet. Evol. 25, 172–189. doi: 10.1016/S1055-7903(02)00224-5
Pereira L. H., Hanner R., Foresti F., Oliveira C. (2013). Can DNA Barcoding Accurately Discriminate Megadiverse Neotropical Freshwater Fish Fauna? BMC Genet. 14 (1), 1–14. doi: 10.1186/1471-2156-14-20
Pons J., Barraclough T. G., Gomez-Zurita J., Cardoso A., Duran D. P., Hazell S., et al. (2006). Sequence-Based Species Delimitation for the DNA Taxonomy of Undescribed Insects. Syst. Biol. 55, 595–609. doi: 10.1080/10635150600852011
Puillandre N., Lambert A., Brouillet S., Achaz G. J. M. E. (2012). ABGD, Automatic Barcode Gap Discovery for Primary Species Delimitation. Mol. Ecol. 21 (8), 1864–1877. doi: 10.1111/j.1365-294X.2011.05239.x
Puncher G. N., Arrizabalaga H., Alemany F., Cariani A., Oray I. K., Karakulak F. S., et al. (2015). Molecular Identification of Atlantic Bluefin Tuna (Thunnus Thynnus, Scombridae) Larvae and Development of a DNA Character-Based Identification Key for Mediterranean Scombrids. PloS One 10 (7), e0130407. doi: 10.1371/journal.pone.0130407
Purdy R. W., Schneider V. P., Applegate S. P., McLellan J. H., Meyer R. L., Slaughter B. H. (2001). The Neogene Sharks, Rays, and Bony Fishes From Lee Creek Mine, Aurora, North Carolina. Smithson. Contrib. 90, 71–202.
Qvarnström A., Ålund M., McFarlane S. E., Sirkiä P. M. (2016). Climate Adaptation and Speciation: Particular Focus on Reproductive Barriers in Ficedula Flycatchers. Evol. Appl. 9 (1), 119–134. doi: 10.1111/eva.12276
Rambaut A., Drummond A. J., Xie D., Baele G., Suchard M. A. (2018). Posterior Summarisation in Bayesian Phylogenetics Using Tracer 1.7. Syst. Biol. 67 (5), 901–904. doi: 10.1093/sysbio/syy032
Rao K. S. (1975). Biometric Comparison of the Spotted Seer, Scomberomorus Guttatus (Bloch and Schneider) From Five Localities Along the Indian Coast. Matsya 1, 63–78.
Rao K. S., Lakshmi K. (1993). Scomberomorus Lineolatus (Cuvier), an Interspecific Natural Hybrid (s. Commerson (Lacépède)× S. Guttatus (Bloch and Schneider)) Off Visakhapatnam, India. J. Nat. Hist. 27 (2), 471–491. doi: 10.1080/00222939300770231
Rao G. N., Sreenivas P. (2020). Anomalous Circulation Around the Southern Indian Peninsula Observed During Winter 2009; and the Generation Mechanisms. Ind. J. Geo Mar. Sci. 49 (03), 343–351.
Rashid H., Mustafa M. G., Dewan S. (2010). Population Dynamics and the Management of the Indo-Pacific King Mackerel Scomberomorus Guttatus From the Upper Bay of Bengal Off Bangladesh Coast. Bangladesh Veterinarian. 27 (2), 82–90. doi: 10.3329/bvet.v27i2.7558
R Core Team (2020). R: A Language and Environment for Statistical Computing R Foundation for Statistical Computing, (Vienna: R Foundation for Statistical Computing URL). Available at: https://www.R-project.org/.
Renema W., Bellwood D. R., Braga J. C., Bromfield K., Hall R., Johnson K. G., et al. (2008). Hopping Hotspots: Global Shifts in Marine Biodiversity. Science 321, 654–657. doi: 10.1126/science.1155674
Roa-Ureta R. H., Lin Y. J., Rabaoui L., Al-Abdulkader K., Qurban M. A. (2019). Life History Traits of the Narrow-Barred Spanish Mackerel (Scomberemorus Commerson) Across Jurisdictions of the Southeast Arabian Peninsula: Implications for Regional Management Policies. Reg. Stud. Mar. Sci. 31, 100797. doi: 10.1016/j.rsma.2019.100797
Ronquist F., Teslenko M., van der Mark P., Ayres D. L., Darling A., Höhna S. (2012). MrBayes 3.2: Efficient Bayesian Phylogenetic Inference and Model Choice Across a Large Model Space. Syst. Biol. 61, 539–542. doi: 10.1093/sysbio/sys029
Rosen D. E. (1975). A Vicariance Model of Caribbean Biogeography. Syst. Zool. 24, 431–464. doi: 10.2307/2412905
Rosser N., Freitas A. V., Huertas B., Joron M., Lamas G., Mérot C., et al. (2019). Cryptic Speciation Associated With Geographic and Ecological Divergence in Two Amazonian Heliconius Butterflies. Zool. J. Linn. Soc 186 (1), 233–249. doi: 10.1093/zoolinnean/zly046
Roul S. K., Jeena N. S., Kumar R., Vinothkumar R., Rahangdale S., Rahuman S., et al. (2021). Postulating the Modality of Integrative Taxonomy in Describing the Cryptic Congener Pampus Griseus (Cuvier) and Systematics of the Genus Pampus (Perciformes: Stromateidae). Front.Mar. Sci. 8. doi: 10.3389/fmars.2021.778422
Russell P. (1803). Descriptions and Figures of Two Hundred Fishes; Collected at Vizagapatam on the Coast of Coromandel (London: W. Bulmer and Co). 2 vols. v. 1–2:i–vii 78 pp. 85 pp., 197pls.
Sambrook J., Fritish E. F., Maniatis T. (1989). Molecular Cloning: A Laboratory Manual (Cold Spring Harbor: Cold Spring Harbor Laboratory Press).
Santa Brígida E. L., Cunha D. B. D., Rego P. S. D., Sampaio I., Schneider H., Vallinoto M. (2007). Population Analysis of Scomberomorus Cavalla (Cuvie) (Perciformes, Scombridae) From the Northern and Northeastern Coast of Brazil. Braz. J. Biol. 67 (4), 919–924. doi: 10.1590/S1519-69842007000500016
Santini F., Carnevale G., Sorenson L. (2013). First Molecular Scombrid Timetree (Percomorpha: Scombridae) Shows Recent Radiation of Tunas Following Invasion of Pelagic Habitat. Ital. J. Zool. 80 (2), 210–221. doi: 10.1080/11250003.2013.775366
Schluter D. (2000). Ecological Character Displacement in Adaptive Radiation. Am. Nat. 156, S4–S16. doi: 10.1086/303412
Secretariat, IOTC (2015) Assessment of Indian Ocean Indo-Pacific King Mackerel (Scomberomorus Guttatus) Using Data Poor Catch-Based Methods. Available at: http://www.iotc.org.
Sepkoski J. J. Jr (2002). A Compendium of Fossil Marine Animal Genera. Bull. Am. Paleontol. 363, 1–560.
Sexton J. P., McIntyre P. J., Angert A. L., Rice K. J. (2009). Evolution and Ecology of Species Range Limits. Annu. Rev. Ecol. Evol. Syst. 40, 415–436. doi: 10.1146/annurev.ecolsys.110308.120317
Shaw G. (1803). General Zoology or Systematic Natural History Pisces (London: G. Kearsley), 1800–1826. Pisces in vol. 4(1803) and vol. 5 (1804);(pt. 2), i–xi 187–632, Pls. 26–92.
Shoji J., Masuda R., Yamashita Y., Tanaka M. (2005). Effect of Low Dissolved Oxygen Concentrations on Behavior and Predation Rates on Red Sea Bream Pagrus Major Larvae by the Jellyfish Aurelia Aurita and by Juvenile Spanish Mackerel Scomberomorus Niphonius. Mar. Biol. 147 (4), 863–868. doi: 10.1007/s00227-005-1579-8
Siddeek M. S. M. (1995). Review of Fisheries Biology of Scomberomorus and Acanthocybium Species in the Western Indian Ocean (FAO Area 51). in Proceedings. IPTP collective volume no. 9, Colombo (Sri Lanka)
Silas E. G. (1964). “Cybium Crockewitti Bleeker, (1850) and Cybium Koreanum Kishinouye, (1915) Considered Synonyms of Scomberomorus Guttatus (Bloch and Schneider) With a Rediscription and Annotated Bibliography of S. Guttatus,” in Proceedings of Symposium on Scombroid Fishes, Marine Biological Association of India, Mandapam Camp, Symposium Series 1, 309–342.
Sivasundar A., Bermingham E., Ortí G. (2001). Population Structure and Biogeography of Migratory Freshwater Fishes (Prochilodus: Characiformes) in Major South American Rivers. Mol. Ecol. 10, 407–417. doi: 10.1046/j.1365-294x.2001.01194.x
Slatyer R. A., Hirst M., Sexton J. P. (2013). Niche Breadth Predicts Geographical Range Size: A General Ecological Pattern. Ecol. Lett. 16 (8), 1104–1114. doi: 10.1111/ele.12140
Som A. (2015). Causes, Consequences and Solutions of Phylogenetic Incongruence. Brief. Bioinform. 16 (3), 536–548. doi: 10.1093/bib/bbu015
Stankowski S., Ravinet M. (2021). Defining the Speciation Continuum. Evol. 75 (6), 1256–1273. doi: 10.1111/evo.14215
Streelman J. T., Karl S. A. (1997). Reconstructing Labroid Evolution With Single-Copy Nuclear DNA. Proc. R. Soc London B 264, 1011–1020. doi: 10.1098/rspb.1997.0140
Sui H., Xue Y., Li Y., Xu B., Zhang C., Ren Y. (2021). Feeding Ecology of Japanese Spanish Mackerel (Scomberomorus Niphonius) Along the Eastern Coastal Waters of China. Acta Oceanol. Sin. 40 (8), 98–107. doi: 10.1007/s13131-021-1796-0
Sukumaran J., Knowles L. L. (2017). Multispecies Coalescent Delimits Structure, Not Species. Proc. Natl. Acad. Sci. 114 (7), 1607–1612. doi: 10.1073/pnas.1607921114
Sulaiman Z. H., Ovenden J. R. (2010). Population Genetic Evidence for the East–West Division of the Narrow-Barred Spanish Mackerel (Scomberomorus Commerson, Perciformes: Teleostei) Along Wallace’s Line. Biodivers. Conserv. 19, 563–574. doi: 10.1007/s10531-009-9699-y
Swathi Lekshmi P. S. (2020). “Features of Indian Coastal Area in the Backdrop of Coastal Security,” in India’s National Security: A Maritime Security Perspective. Ed. Suresh R., Vij Books India Private Limited, New Delhi,142–149.
Tamura K., Stecher G., Kumar S. (2021). Mega 11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 38, (7), 3022–7 doi: 10.1093/molbev/msab120
Thangaraja M., Al-Aisry A. (2001). “Studies on the Occurrence and Abundance of Fish Eggs and Larvae in the Waters of Sultanate of Oman,” in Proceedings of 1st International Conference on Fisheries, Aquaculture and Environment in NW Indian Ocean (Muscat, Sultanate of Oman: Sultan Qaboos University), 13–36.
Thompson J. D., Higgins D. G., Gibson T. J. (1994). Clustal W: Improving the Sensitivity of Progressive Multiple Sequence Alignment Through Sequence Weighting, Position-Specific Gap Penalties and Weight Matrix Choice. Nucleic Acids Res. 22, 4673–4680. doi: 10.1093/nar/22.22.4673
Uma Mahesh V., Ghosh S., Sreeramulu K., Rao M. V., Satish Kumar M. (2017). Fishery, Biology and Stock Assessment of Spotted Seer, Scomberomorus Guttatus (Bloch and Schneider) Off Andhra Pradesh. Mar. Biol. Assoc. India. 59 (1), 67–72. doi: 10.6024/jmbai.2017.59.1.1934-10
Van Velzen R., Weitschek E., Felici G., Bakker F. T. (2012). DNA Barcoding of Recently Diverged Species: Relative Performance of Matching Methods. PloS One 7 (1), e30490. doi: 10.1371/journal.pone.0030490
Venkataramana V., Sarma V. V. S. S., Gawade L., Reddy A. M. (2021). Magnitude of River Discharge Determines the Food Habit and Community Composition of Zooplankton in the Indian Estuaries During Monsoon. Estuar. Coast. Shelf. Sci. 262, 107601. doi: 10.1016/j.ecss.2021.107601
Vijayaraghavan P. (1955). Life-History and Feeding Habits of the Spotted Seer Scomberomorus Guttatus (Block and Schneider). Indian J. Fish. 2 (2), 360–372.
Vineesh N., Divya P. R., Kathirvelpandian A., Mohitha C., Shanis C. P. R., Basheer V. S., et al. (2018). Four Evolutionarily Significant Units Among Narrow-Barred Spanish Mackerel (Scomberomorus Commerson) in the Indo-West Pacific Region. Mar. Biodivers. 48 (4), 2025–2032. doi: 10.1007/s12526-017-0714-3
Wafar M., Venkataraman K., Ingole B., Ajmal Khan S., LokaBharathi P. (2011). State of Knowledge of Coastal and Marine Biodiversity of Indian Ocean Countries. PloS One 6 (1), e14613. doi: 10.1371/journal.pone.0014613
Ward R. D., Zemlak T. S., Innes B. H., Last P. R., Hebert P. D. N. (2005). DNA Barcoding Australia’s Fish Species. Philos. Trans. R. Soc. B: Biol. Sci. 360, 1847–1857. doi: 10.1046/10.1098%2Frstb.2005.1716
Welch D. J., Newman S. J., Buckworth R. C., Ovenden J. R., Broderick D., Lester R. J., et al. (2015). Integrating Different Approaches in the Definition of Biological Stocks: A Northern Australian Multi-Jurisdictional Fisheries Example Using Grey Mackerel, Scomberomorus Semifasciatus. Mar. Policy 55, 73–80. doi: 10.1016/j.marpol.2015.01.010
Wright J. J., David S. R., Near T. J. (2012). Gene Trees, Species Trees, and Morphology Converge on a Similar Phylogeny of Living Gars (Actinopterygii: Holostei: Lepisosteidae), an Ancient Clade of Ray-Finned Fishes. Mol. Phylogenet. Evol. 63 (3), 848–856. doi: 10.1016/j.ympev.2012.02.033
Xia X. (2018). Dambe7: New and Improved Tools for Data Analysis in Molecular Biology and Evolution. Mol. Biol. Evol. 35 (6), 1550–1552. doi: 10.1093/molbev/msy073
Xiao W. H., Zhang Y. P., Liu H. Z. (2001). Molecular Systematics of Xenocyprinae (Teleostei: Cyprinidae): Taxonomy, Biogeography, and Coevolution of a Special Group Restricted in East Asia. Mol. Phylogenet. Evol. 18, 163–173. doi: 10.1006/mpev.2000.0879
Yang Z. (2015). The BPP Program for Species Tree Estimation and Species Delimitation. Curr. Zool. 61 (5), 854–865. doi: 10.1093/czoolo/61.5.854
Yang Z., Rannala B. (2010). Bayesian Species Delimitation Using Multilocus Sequence Data. Proc. Natl. Acad. Sci. U. S. A. 107 (20), 9264–9269. doi: 10.1073/pnas.0913022107
Zacharia P. U., Dineshbabu A. P., Thomas S., Kizhakudan S. J., Vivekanandan E., Pillai S. L., et al. (2016). Relative Vulnerability Assessment of Indian Marine Fishes to Climate Change Using Impact and Adaptation Attributes (Kochi, India: CMFRI Special Publication No.125, (CMFRI-NICRA Publication No. 5), Central Marine Fisheries Research Institute), pp. 192.
Keywords: Scomberomorus, phylogeny, species delimitation, divergence, resurrection, northern Indian Ocean, ecology
Citation: Jeena NS, Rahuman S, Roul SK, Azeez PA, Vinothkumar R, Manas HM, Nesnas EA, Rathinam AMM, Surya S, Rohit P, Abdussamad EM and Gopalakrishnan A (2022) Resolved and Redeemed: A New Fleck to the Evolutionary Divergence in the Genus Scomberomorus Lacepède, 1801 (Scombridae) With Cryptic Speciation. Front. Mar. Sci. 9:888463. doi: 10.3389/fmars.2022.888463
Received: 02 March 2022; Accepted: 21 April 2022;
Published: 09 June 2022.
Edited by:
Christian Marcelo Ibáñez, Andres Bello University, ChileReviewed by:
Claudio F. Cornejo, University of Concepcion, ChileMathias Hüne, Centro de Investigación para la Conservación de los Ecosistemas Australes (ICEA), Chile
Copyright © 2022 Jeena, Rahuman, Roul, Azeez, Vinothkumar, Manas, Nesnas, Rathinam, Surya, Rohit, Abdussamad and Gopalakrishnan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: N. S. Jeena, amVlbmFuc0ByZWRpZmZtYWlsLmNvbQ==
 N. S. Jeena
N. S. Jeena Summaya Rahuman
Summaya Rahuman Subal Kumar Roul
Subal Kumar Roul P. Abdul Azeez
P. Abdul Azeez R. Vinothkumar1
R. Vinothkumar1 S. Surya
S. Surya A. Gopalakrishnan
A. Gopalakrishnan