- 1Key Laboratory of Tropical Aquatic Germplasm of Hainan Province, Sanya Oceanographic Institution, Ocean University of China, Sanya, China
- 2Ministry of Education (MOE) Key Laboratory of Marine Genetics and Breeding, Ocean University of China, Qingdao, China
- 3Laboratory for Marine Biology and Biotechnology, Pilot Qingdao National Laboratory for Marine Science and Technology, Qingdao, China
- 4Laboratory for Marine Fisheries Science and Food Production Processes, Pilot Qingdao National Laboratory for Marine Science and Technology, Qingdao, China
The COMMD (copper metabolism gene MuRR1 domain) gene family, highly conserved among multicellular eukaryotic organisms, plays important roles in a variety of biological processes, ranging from copper homeostasis, ionic transport, protein trafficking, NF-κB-mediated transcription, and cell proliferation. However, systematic identification, spatiotemporal expression, and stress-responsive patterns of COMMD genes remain obscure in molluscs. Here, we analyzed the characteristics of the COMMD gene family in four bivalve molluscs based on both genome and extensive transcriptomic resources. Firstly, we investigated the genomic signatures, functional domains, and phylogenetic relationships, and ten single-copy members were identified in Yesso scallop (Patinopecten yessoensis), Zhikong scallop (Chlamys farreri), Pacific oyster (Crassostrea gigas), and dwarf surf clam (Mulinia lateralis), respectively. Strong purifying selection was revealed for COMMD4. Higher expressions of most COMMDs were observed in the hepatopancreas, besides which a different tissue preference of COMMDs’ expression was found among four bivalves. Moreover, in the dwarf surf clam, the responses of COMMD members under stresses were found more sensitive in the hepatopancreas than in the gill, and MlCOMMD9 and MlCOMMD4 might be the good candidate stress indicator genes respectively for copper ion stress and V. Anguillarum infection. Our study would contribute to a better understanding for the evolution of the COMMD gene family and provide valuable information for their innate immune roles in bivalve molluscs.
Introduction
The COMMD (copper metabolism gene MuRR1 domain) family includes ten evolutionarily conserved proteins, namely, COMMD1–10, in the extreme carboxyl terminus of which they share a unique motif known as the COMM domain (Burstein et al., 2005). The COMM domain, with 70–85 amino acids in length and being rich in tryptophan, proline, and leucine, not only defines the gene family but also provides a critical interface for protein–protein interactions with each other (Maine et al., 2007). The hydrophobic residues located in the COMM domain could form two conserved nuclear export signals (NES1 and NES2) (Muller et al., 2009). Except COMMD6, other COMMD members possess an amino terminal region, which is divergent among the subfamilies but is highly conserved within the ortholog proteins (Burstein et al., 2005). COMMD1 is the first identified member, which was initially termed as Murr1 due to its proximity to the U2af1-rs1 locus in mice, and other COMMD members were identified through homologous screening (Nabetani et al., 1997; van De Sluis et al., 2002). A total of ten subfamilies were found in the vast majority of vertebrates (Burstein et al., 2005), while in invertebrates, including insects, worms, and molds, only several COMMD members were reported, and none of the COMMDs were found in unicellular eukaryotic organisms or bacteria (Nabetani et al., 1997). The wide existence as well as the highly conservative characteristic of COMMD homologues imply their critical roles during the metazoan evolution (Riera-Romo, 2018).
As the best-characterized member, COMMD1 may represent a prototype of the family (Burstein et al., 2005). COMMD1 was reported to be able to participate in two distinct activities, control of copper metabolism and regulation of the transcription factor NF-κB (Riera-Romo, 2018). Researchers found that mutations of COMMD1 are responsible for copper toxicosis in Bedlington terrier dogs, resulting in excessive copper accumulation in the liver (Tao et al., 2003). In human, biochemical findings had indicated a direct role for COMMD1 in biliary copper transport, and COMMD1 defect could impair the copper excretion (Tao et al., 2003; Riera-Romo, 2018). Besides COMMD1, various functions were found for other COMMD genes in vertebrates. COMMD4, as one of the protein kinase A targets, was able to interact with myomegalin and inhibit NF-κB activity (Uys et al., 2011). The COMMD3 and COMMD8 complex could selectively recruit GRK6, which induced GRK6-mediated phosphorylation of the receptor and activated the β-arrestin-mediated signaling (Nakai et al., 2019). COMMD3 and COMMD9, which are endogenous regulators, regulate Na+ transport through altering ENaC cell surface expression (Liu et al., 2013). COMMD5 affects cell proliferation (Solban et al., 2000), and COMMD6, 7 participates in the invasion and migration regulation of a variety of cancer (You et al., 2017; Yang et al., 2019). Research has reported that COMMD10 is related to phagosomes in murine macrophages (Dill et al., 2015), and in myeloid cells, deficiency in COMMD10 can cause increased NF-κB activation and then aggravate lipopolysaccharide systemic sepsis (Naugler and Karin, 2008; Mouhadeb et al., 2018). In comparison, only the functions of several COMMD genes were reported in invertebrates. For example, COMMD4 is found ubiquitously expressed in amphioxus, with the highest level in gonad, and lipopolysaccharide injection could induce its expression (Jin et al., 2012). Wang et al. have cloned COMMD1 in Crassostrea hongkongensis, and the transcription level of COMMD1 was increased significantly in the gill and hemolymph after salinity stimulation (Wang et al., 2017a).
As benthic filter feeders, bivalve molluscs are well adapted to highly dynamic oceans and freshwater environments since the early Cambrian. Along with intensified human activities in recent decades, the bivalve habitats are subject to various biotic/abiotic stressors, and pathogenic microbes and heavy metals are two of the main stressors. The COMMD genes play critical roles in many vital functions, and exploration of whether these genes participate in heavy-metal or bacterial resistance may help gain a better understanding for the outstanding adaptability of bivalve molluscs. In the present study, a systematic identification and characterization of COMMD genes in four bivalves were conducted, namely, Yesso scallop, Zhikong scallop, Pacific oyster, and dwarf surf clam. Detailed genic structure comparison and spatiotemporal expression analysis provided insights into the potential function of COMMD genes in bivalve molluscs. Further, transcription patterns of different COMMD members under copper ion stress and after Vibrio Anguillarum infection were investigated in the gill and hepatopancreas of M. lateralis. This is the first comprehensive research of the COMMD family genes in bivalves, which notably provided helpful information regarding the classification, evolution, and function of these genes. These findings may assist a better understanding of the bivalves’ adaption to adverse heavy-metal pollutions and bacterial challenge.
Material and Methods
Genome-Wide Identification of COMMD Genes
To identify COMMD genes, the whole genomes and transcriptomes of four bivalves (Zhang et al., 2012; Li et al., 2017; Wang et al., 2017b) were searched against the available COMMD protein sequences from representative vertebrates (Homo sapiens, Mus musculus, Xenopus laevis) and Cephalochordata (Branchiostoma belcheri). The orthologous COMMD proteins were used as query sequences for whole-genome and transcriptome-based blasts, and the threshold of the E value was 1E-5. Sequence analysis was performed by the HMM searching method (http://www.ebi.ac.uk/Tools/hmmer/search/phmmer) to ensure the integrity of COMMDs. BLASTN was used to confirm their genomic structure, and the ORF Finder program (https://www.ncbi.nlm.nih.gov/orffinder/) was used to predict the open reading frame. Further, to ensure the completeness of COMMDs, the translated sequences were submitted to the SMART tool (http://smart.embl-heidelberg.de/) and ProtParam tool (https://web.expasy.org/protparam/) to ensure the presence of the conserved COMM_domain and to predict the isoelectric point (pI), molecular weight, instability index, and grand average of hydropathicity (GRAVY) values, which were illustrated for all potential bivalve COMMDs shown in Table 1. Then, the drawing of the protein structure was done by IBS 1.0.3. Finally, we identified and counted COMMD family genes of 26 metazoan species, namely, the deuterostomes Homo sapiens, Oryzias latipes, Danio rerio, Oreochromis niloticus, Xenopus laevis, Ciona intestinalis, Branchiostoma floridae, and Strongylocentrotus purpuratus, the protostomes Drosophila melanogaster, Daphnia pulex, Caenorhabditis elegans, Tribolium castaneum, Lingula anatina, Capitella teleta, Helobdella robusta, Lottia gigantea, Elysia chlorotica, Biomphalaria glabrata, Octopus bimaculoides, Patinopecten yessoensis, Chlamys farreri, Crassostrea gigas, and Mulinia lateralis, and the non-bilaterians Nematostella vectensis, Stylophora pistillata, and Amphimedon queenslandica (Supplementary Table 1).
Multiple Alignment and Phylogenetic Analysis
MEGA7.0 (Sudhir et al., 2016) was used to construct phylogenetic analysis to determine which COMMD subfamily the bivalve COMMD genes belong to. The whole amino acid sequences of COMMD proteins from Human (H. sapiens), zebrafish (D. rerio), medaka fish (O. latipes), African clawed frog (X. laevis), ciona (C. intestinalis) and Stylophora (S. pistillata), were downloaded from the Ensemble genome browser database. The whole amino acid sequences of COMMD proteins from Nile tilapia (O. niloticus), notoacmea (L. gigantea), Biomphalaria (B. glabrata), and sea snail (E. chlorotica) were downloaded from the Uniport database. The whole amino acid sequences of COMMD proteins from amphioxus (B. belcheri) and octopus (O. bimaculoides) were downloaded from the NCBI database. Multi-sequence alignment was performed through ClustalW (Larkin et al., 2007) and was edited by GeneDoc software (Nicholas et al., 1997), then the phylogenetic analyses based on the neighbor-joining (NJ) method and maximum likelihood (ML) method with a bootstrap of 1,000 replicates, both including all the amino acids from the COMMDs. 154 amino acids across 16 animals were involved in this analysis. The accession numbers of 154 COMMDs are listed in Supplementary Table 2.
Selective Pressure Analysis
COMMD gene sequences were aligned based on codons, using Muscle (codons) implemented in MEGA7.0 (Sudhir et al., 2016). MEGA7.0 was used to build the alignment result into a tree file, and a Newick format file was formed. To explore selective pressure between COMMD gene sequences, after removing the gap, a strict statistical analysis was performed using the software EasyCodeML1.2 (Gao et al., 2019). Based on the Preset Site Model, the ratios of non-synonymous (dN) and synonymous (dS) substitutions for 40 COMMD genes among four bivalve were calculated. The LRT was used to test whether the selected model is significant (P < 0.05). Two likelihood ratio tests were performed to detect positively selected sites—M1a (neutral) vs. M2a (positive selection), M7 (β) vs. M8 (β and ω), and M0 (one-ratio) vs. M3 (discrete)—and the site-specific model was used for comparison. If the tests produced a significant result, then the empirical Bayes method was used to identify individual positively selected codon sites (Yang et al., 2005).
Expression Analysis
The TPM (reads per kilobase million) values were summarized from the published RNA-seq datasets of Yesso scallop (Wang et al., 2017b), Zhikong scallop (Li et al., 2017), and Pacific oyster (Zhang et al., 2012) and from our unpublished data for dwarf surf clam. During development, eleven embryo/larval developmental stages were chosen to perform expression analysis, including zygotes; multi-cells; blastula; gastrula; trochophore; D-shaped larvae; early-, mid-, and late-term umbo larvae; metamorphosis larvae; and juvenile. For adults, six tissues were chosen to perform expression analysis, namely, muscle, hepatopancreas, mantle, gill, male gonad, and female gonad. The expressional heatmaps were displayed by the heatmap package in R environment.
Copper Ion Stress and V. anguillarum Infection Experiment
Healthy adult dwarf surf clams were obtained from a laboratory breeding population. Dwarf surf clams were cultured in filtered and aerated seawater at 20°C–25°C. For the copper ion stress experimental group, the clams were acclimated in the sterilized seawater with a final copper ion concentration of 100 µg/l (from the anhydrous copper sulfate) (Zhang et al., 2012). For both control and experimental groups, five random clams were sampled at 0 h, 12 h, and 9 days, from which gill and hepatopancreas tissues were collected for RNA extraction. For the bacterial challenge group, gram-negative bacteria (Vibrio Anguillarum) were cultured in liquid 2216 E broth at 28°C to an OD600 of 0.2 and were harvested by centrifugation at 2,000 × g for 5 min. Then, the cell precipitates were suspended in filtered seawater and adjusted to 1 × 107 CFU/ml (Zhou et al., 2019) to challenge clams. For both control and experimental groups, five random clams were sampled at 0, 3, 6, 12, and 24 h, from which gill and hepatopancreas tissues were collected for RNA extraction.
RNA Isolation and Quantitative Real-Time PCR Analysis
Total mRNA was extracted from the gill and hepatopancreas of the sampled clams by using the conventional guanidinium isothiocyanate method (Chomczynski and Sacchi, 2006). The cDNA was synthesized using M-MLV Reverse Transcriptase (Promega, Madison, WI, USA). Primers of MLCOMMDs were designed using Primer Premier 5 software; the sequences of primers are listed in Supplementary Table 3. All reactions were repeated in triplicate. The transcription of target genes was standardized according to the transcription of two internal reference genes (namely, RS23 and NDUS4). For the comparisons of the COMMD transcription changes between control experimental groups, statistical analysis of the data was performed using t-test with statistical significance at P < 0.05.
Results
Identification and Characterization of COMMD Genes in Four Bivalve Molluscs
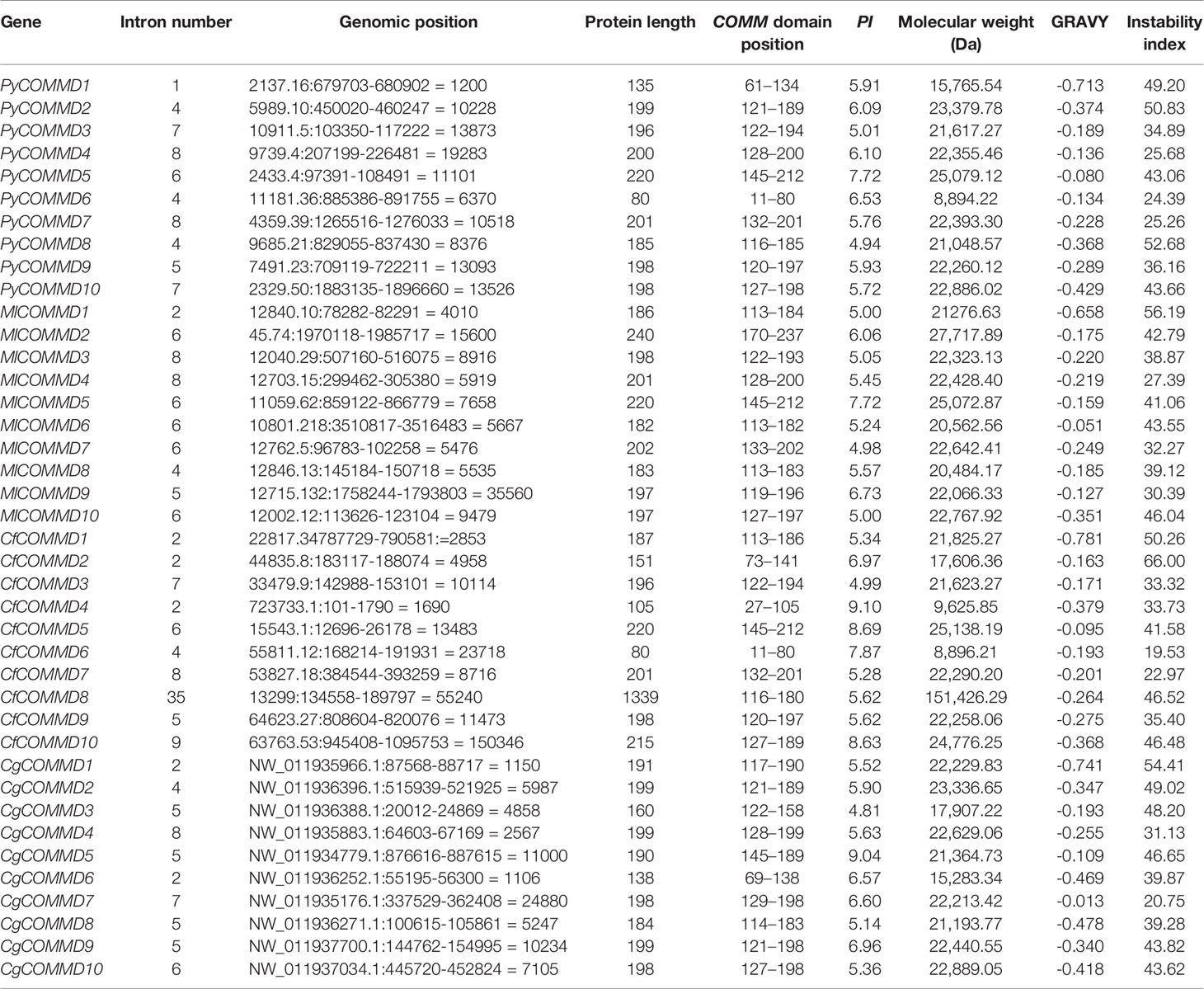
A total of 10 single-copy COMMD genes were identified in Yesso scallop, Zhikong scallop, Pacific oyster, and dwarf surf clam, respectively represented as PyCOMMDs, CfCOMMDs, CgCOMMDs, and MlCOMMDs. The length of most bivalve COMMDs ranged from 150 to 240 aa, except four relatively short COMMDs (namely, PyCOMMD1, 6, and CfCOMMD4, 6, which possess less than 135 aa) and CfCOMMD8 which was obviously longer (1,339 aa). The COMMD family shared the conserved COMM domain (Figure 1). Two highly conserved nuclear export signal regions were located at the COMM domain, namely, NES1 and NES2, which were mainly composed of the well-conserved hydrophobic amino acids L, I, V, M, and F (Figure 2). Besides, the COMMD1 proteins possessed an additional COMMD1_domain (PF17221) at the N-terminal, and a specific Glyco_hydro_15 domain (PF00723) was found at the C-terminal of CfCOMMD8 (Figure 1). Consistent with human COMMD6 (de Bie et al., 2006), the PyCOMMD6 and CfCOMMD6 lacked a variable amino terminal, while CgCOMMD6 and MlCOMMD6 contained an extended amino terminal portion. A low-complexity region was located at the amino terminal of PyCOMMD4, 8, CgCOMMD10, and MlCOMMD10. In comparison with CgCOMMDs and MlCOMMDs, a higher similarity of gene structure was revealed between COMMD orthologs from two scallops (Table 1). A subfamily-specific conserved intron number was found for scallop COMMD3, 5, 6, 7, 9, which comprised 7, 6, 4, 8, and 5 introns, respectively. Of note, the 5-intron pattern was also found in CgCOMMD9 and MlCOMMD9, making COMMD9 as the only member which showed the most conservative exon–intron structure in bivalves.
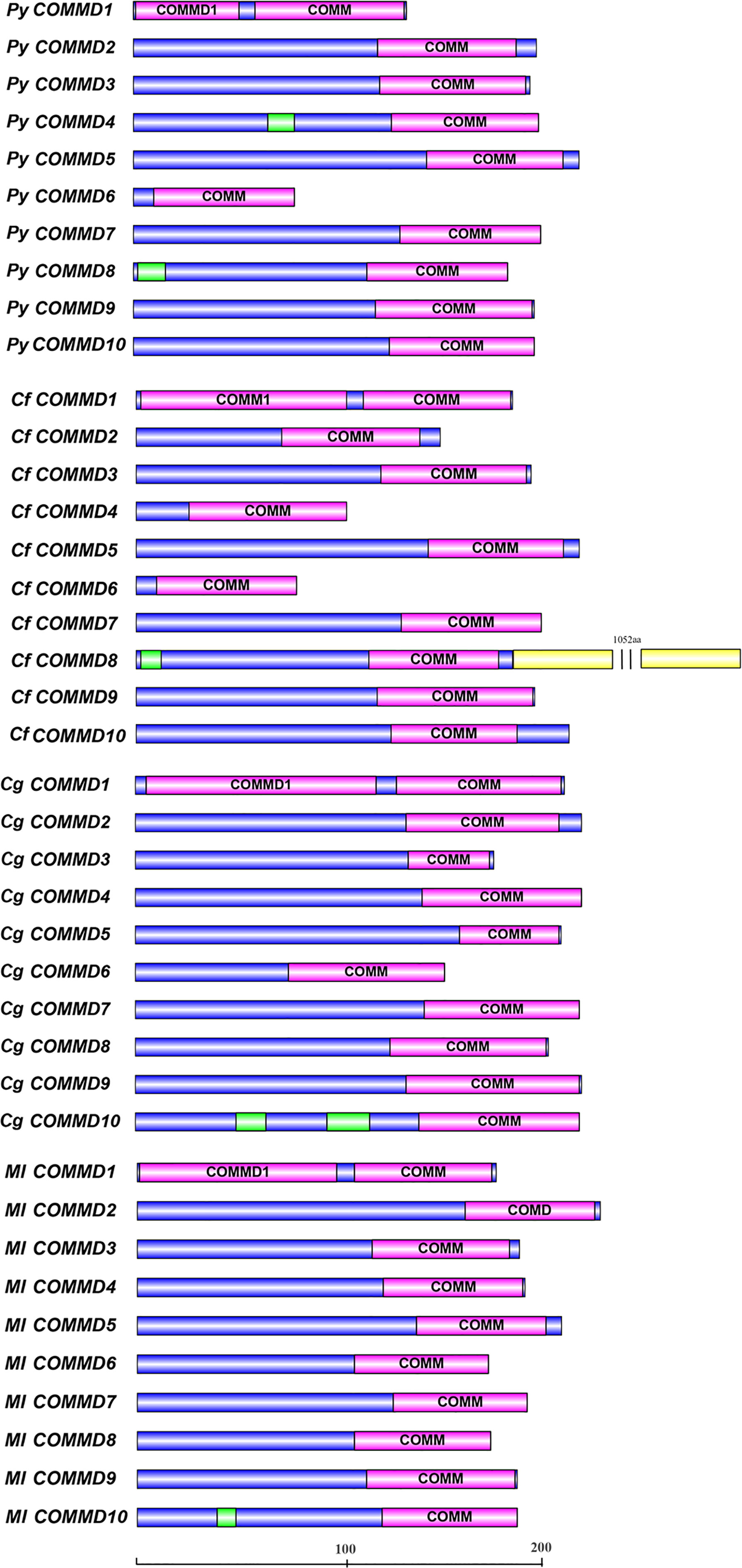
Figure 1 The structure of COMMD proteins in four bivalves. The purple boxes indicate the conserved COMM domains. The green boxes indicate the low-complexity regions, and the yellow boxes indicate the Glyco_hydro_15 Pfam domain.

Figure 2 Alignment of deduced COMM domain amino acid sequences. The two nuclear export signals (NES1 and NES2) are indicated in gray shades and by arrows pointing at conserved hydrophobic residues.
Phylogenetic and Evolutionary Analysis of the COMMD Family
In the present study, besides the COMMD family gene identification from four bivalve molluscs, we also identified COMMDs from 22 additional animal species, across the major representative groups in Metazoa (Figure 3). It revealed that five groups, namely, Deuterostomia (except Urochordata), Mollusca, Brachiopoda, Cnidaria, and Sponge, have a full set of ten COMMD subfamilies, while in Ecdysozoa, Annelida, and Urochordata, they usually lack several COMMD subfamilies (up to 9). Especially, we noted that COMMD1, 6, 9 subfamilies were absent in all investigated ecdysozoans, and within Lophotrochozoa, molluscs and brachiopods have more complete COMMD family members than annelids. We further investigated the evolutionary relationship of bivalve COMMDs. Phylogenetic analysis showed that all COMMD proteins were subdivided into ten subfamilies, including COMMD1-10, and consistent topologies were revealed based on both the NJ method (Figure 4A) and ML method (Figure 4B). For each subfamily, COMMDs from Zhikong scallop, Yesso scallop, and Pacific oyster are always grouped together first, then clustered together with COMMDs from the dwarf surf clam and other bivalves, which is in line with their assured phylogenetic relationship.
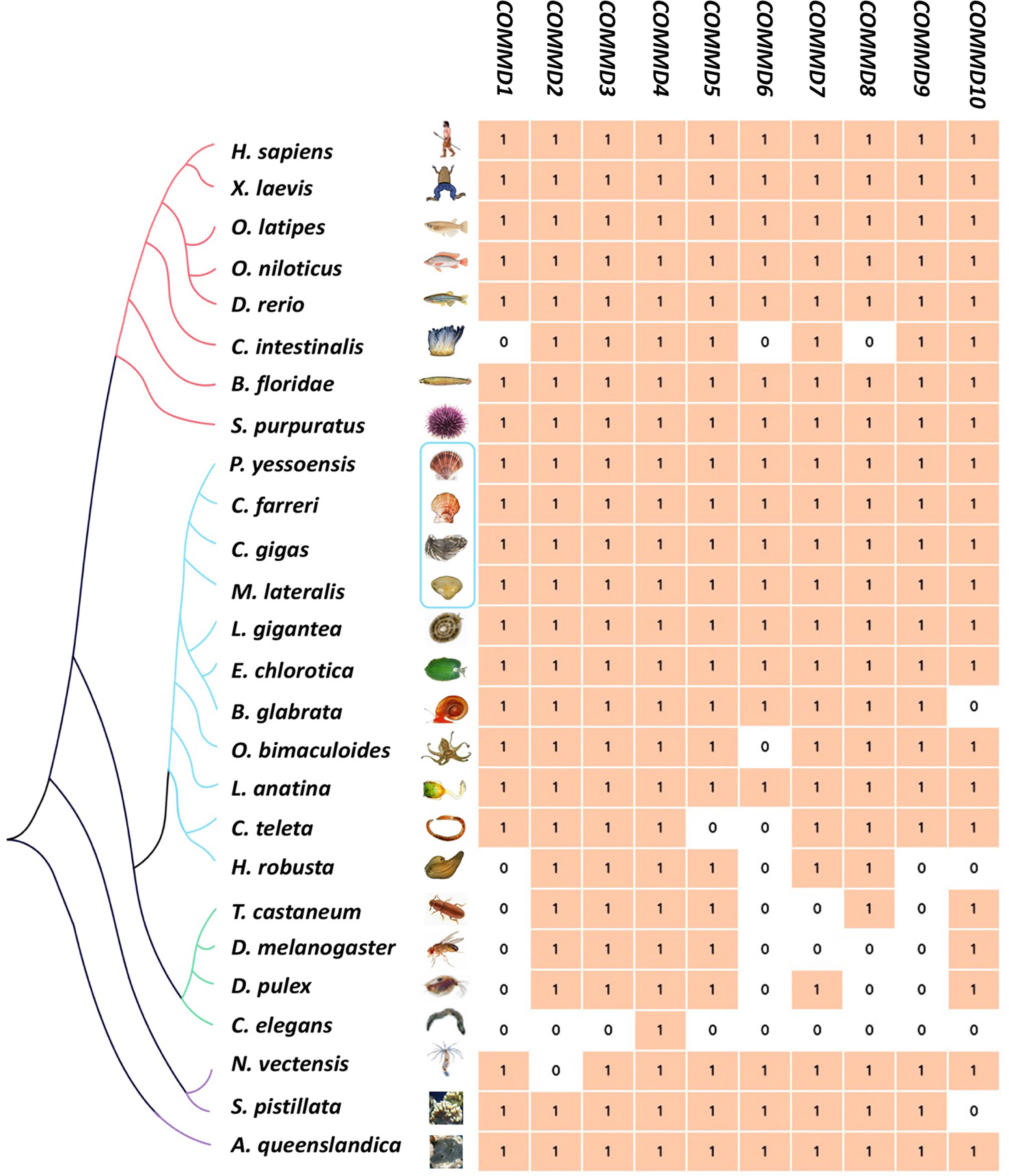
Figure 3 Genome-wide identification of COMMD gene families of 4 bivalve molluscs (in blue box) and 22 other metazoan species. Different colors of branches represent different metazoan groups (red, Deuterostomia; blue, Lophotrochozoa; green, Ecdysozoa; purple, non-Bilateria).
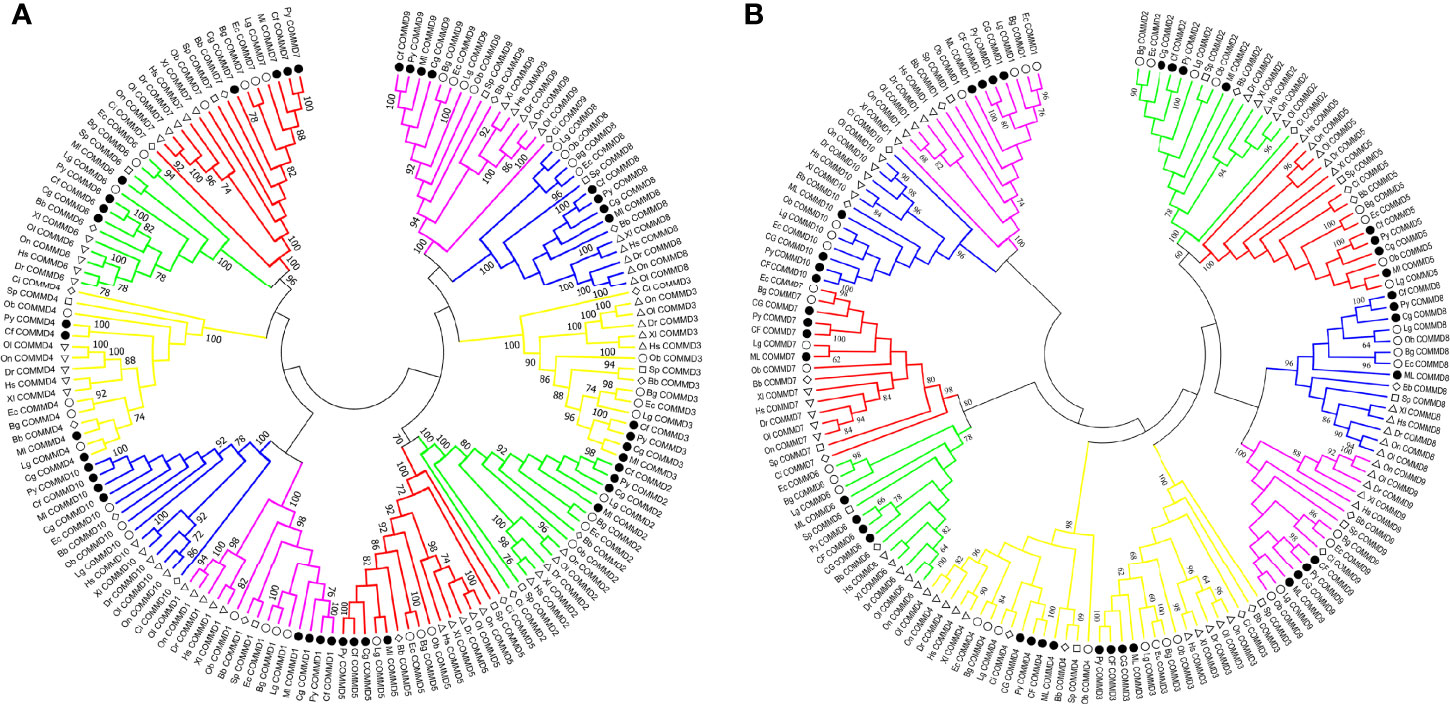
Figure 4 Phylogenetic analyses of COMMD gene families involving 154 amino acid sequences across 16 animals were conducted based on the NJ method and ML method. (A) Phylogenetic analysis of COMMD proteins using the NJ method, with a bootstrap of 1,000 replicates. (B) Phylogenetic analysis of COMMD proteins using the ML method, with a bootstrap of 1,000 replicates. Numbers in the trees represent the confidence interval. Different symbols were used to represent different clades of animals: the circles label the molluscs, the diamonds label the chordates, the triangles label the vertebrates, and the squares label the cnidarians. Four bivalve COMMDs were specially emphasized by solid color inside the symbol. Hs, Homo sapiens; Xl, Xenopus laevis; Dr, Danio rerio; Ol, Oryzias latipes; On, Oreochromis niloticus; Ci, Ciona intestinalis; Bb, Branchiostoma belcheri; Sp, Stylophora pistillata; Py, Patinopecten yessoensis; Cf, Chlamys farreri; Cg, Crassostrea gigas; Ml, Mulinia lateralis; Ec, Elysia chlorotica; Ob, Octopus bimaculoides; Lg, Lottia gigantean; Bg, Biomphalaria glabrata.
Positive Selection Analysis
To explore the selective pressure of the COMMD genes, the CODEML program in the EasyCodeML1.2 software was further used. Results show that six subfamilies were detected with positive selection sites by the M7 vs. M8 model (Table 2). According to the M8 model, COMMD4 possessed 8 positive sites, including two highly positively selected sites (P > 0.95), followed by COMMD1 (5), COMMD7 (5), and COMMD10 (4). Only one and two positive sites were respectively detected in COMMD3 and COMMD6. Overall, a total of 25 sites under potentially positive selection were identified in four bivalve COMMDs.
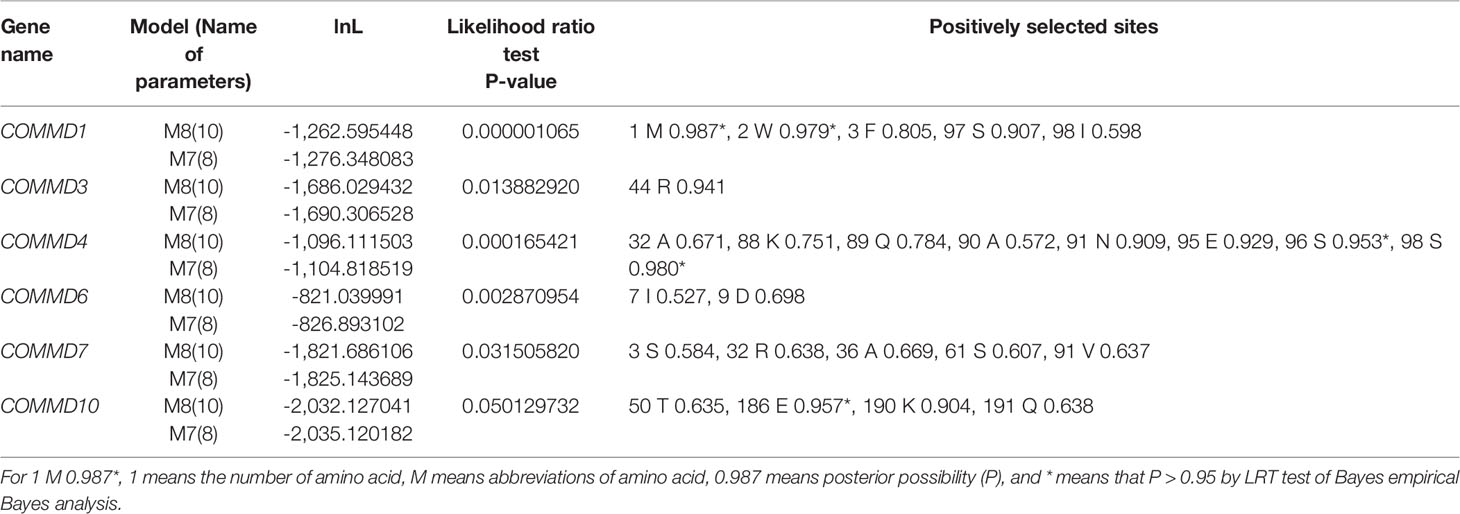
Table 2 Likelihood values and parameter estimates of computing position selection site by site model for the COMMD family members.
Temporal and Spatial Expression of the COMMDs From Four Bivalve Molluscs
The TPM (reads per kilobase million) calculated from the RNA sequence data are displayed as a heat map (Figure 5, Supplementary Tables 4 and 5). As shown in Figure 5A, the embryo expression profiles of COMMDs in four bivalves can be parted into three groups. Firstly, most of the COMMD1, 3, 4 subfamilies were detected at the beginning of fertilization and maintained high transcriptions until the multicellular stage; a similar transcription pattern was also found in PyCOMMD6, CgCOMMD2, 7, and MlCOMMD5, 8, 10, suggesting their maternal origin to play protective roles. COMMD5, 8, 9, 10 in two scallops started increasing the transcription levels during blastula, and their high transcriptions were maintained until the D-shaped veliger stage. Except the abovementioned, other members of COMMD genes, including PyCOMMD2, 7, CfCOMMD3, 7, CgCOMMD1, 3, 5, 6, 8, 9, 10, and MlCOMMD2, 6, 7, 9, were enhanced exponentially from the D-shaped veliger stage and sustained their high level of transcription during late larval development.
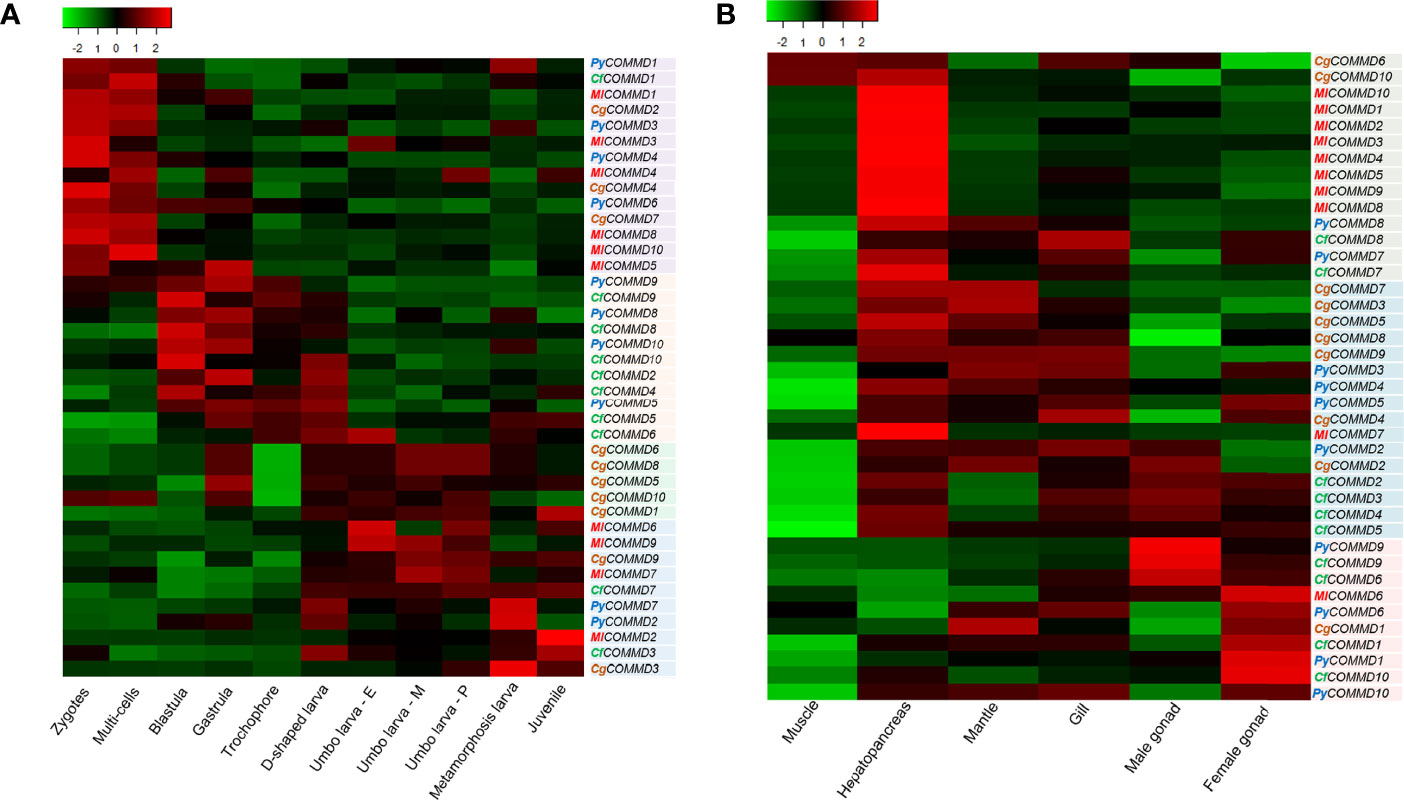
Figure 5 Heatmap of COMMD expression profiles (TPM) in embryonic developmental stages and different tissues in four bivalves. The color varies from green to red, representing the scale of the relative expression level. (A) Expression of COMMDs during embryonic development. (B) Expression of COMMDs in adult tissues of four bivalves.
According to the spatial expression pattern of COMMDs in six organs/tissues (Figure 5B), we found that most of the COMMD2, 3, 4, 5, 7, 8, 10 subfamilies were predominantly expressed in the hepatopancreas in four bivalves. Besides, most of the COMMD1, 6 members were highly expressed in the female gonads, and COMMD2, 9 of two scallops were detected with high levels in male gonads. Moreover, we noticed that COMMDs’ transcription showed a certain tissue preference among different bivalves. For example, most MlCOMMDs and CfCOMMDs were highly expressed in the hepatopancreas and gonad, respectively, while most CgCOMMDs and PyCOMMDs were highly expressed in the hepatopancreas, mantle, and gill. Overall, organ/tissue transcription patterns of COMMD genes in four bivalves may imply their diverse cellular functions.
Responses of MlCOMMD Genes Under Copper Ion Stress and Bacterial Stress
Taking the advantages of laboratory-standardized breeding and cultivation, we investigate the potentially biological functions and defensive mechanism of COMMD genes in the dwarf surf clam. First, we explored the responses of MlCOMMD genes in the gill and hepatopancreas under copper ion stress (Figure 6). After the copper ion stress, MlCOMMD1, 4, 9 showed significant responses in both tissues, with MlCOMMD9 being remarkably upregulated, while MlCOMMD1 and MlCOMMD4 were significantly suppressed. Of note, a more acute and intensive response of MlCOMMD9 was shown in the hepatopancreas (12 h, >6-fold, P < 0.001) than in the gill (day 9, >2-fold, P < 0.05). Besides, MlCOMMD4 and MlCOMMD7 were found significantly upregulated on day 9 respectively in the hepatopancreas and gill. In the next scenario, the temporal responses of MlCOMMD genes after V. Anguillarum infection were as shown in Figure 7. In the gill, only three COMMD members showed significant responses after infection, with MlCOMMD4, 8 being found to be significantly upregulated after 12 h and MlCOMMD7 being suppressed at 6 h. In comparison, responses of COMMDs in the hepatopancreas seems more ubiquitous, with six members showing significant transcription alternations. After infection, transcription of MlCOMMD4 was significantly elevated after 6 and 12 h, and MlCOMMD7 was significantly upregulated at 6 h. In the meantime, MlCOMMD5 was found to be acutely suppressed after 3 h and MlCOMMD6, 8, 9 showed a significantly lower transcription at 6 h. Notably, MlCOMMD4 was the only member that showed consistent significant induction in both the gill and hepatopancreas after infection, and similar to copper ion stress, a more acute and intensive response of MlCOMMD4 was found in the hepatopancreas (6 h, >9-fold, P < 0.05) than in the gill (12 h, >3-fold, P < 0.05). Above all, we found that responses of COMMD members were more sensitive in the hepatopancreas than the gill, in which MlCOMMD9 and MlCOMMD4 might be good candidate stress indicator genes respectively for copper ion stress and V. Anguillarum infection.
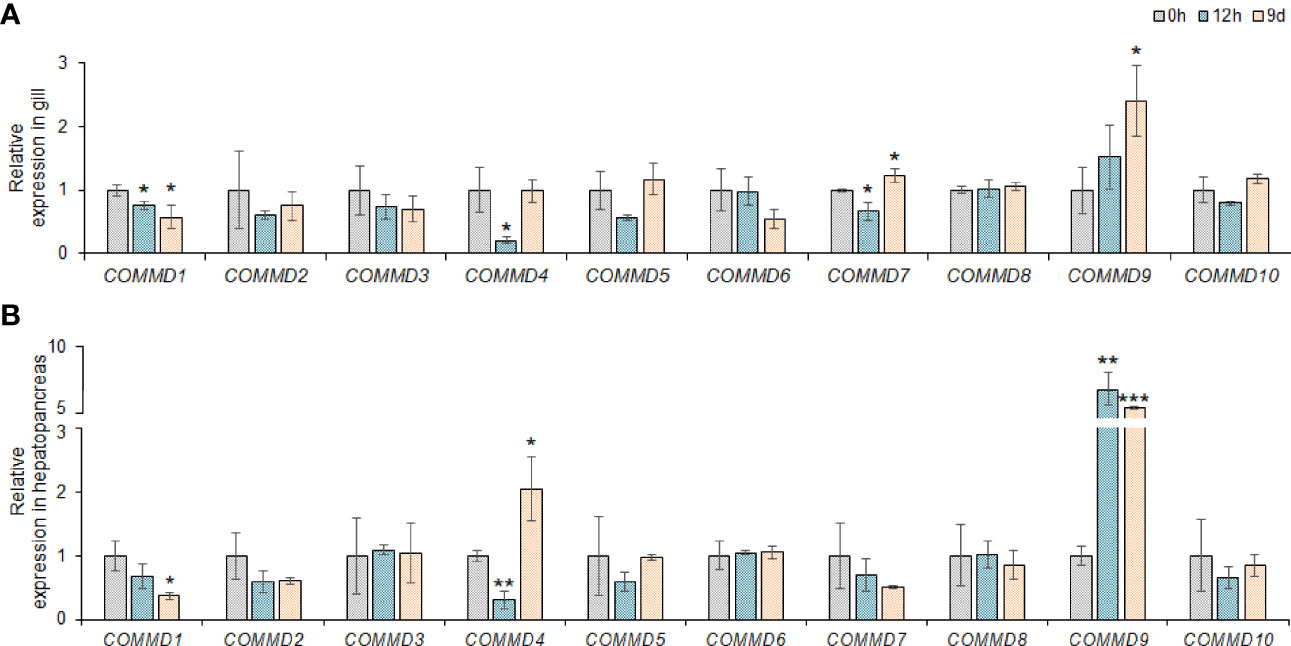
Figure 6 Relative expression of COMMD genes in M. lateralis gill and hepatopancreas under copper ion stress. (A) Relative expression of MLCOMMDs in gill under copper ion stress. (B) Relative expression of MLCOMMDs in hepatopancreas under copper ion stress. The fold changes compared with the control group for each test point are shown as a bar chart (significance: ***P < 0.001; **P < 0.01; *P < 0.05). The gray, light blue and light orange boxes respectively indicate 0 h, 9h and 9d under copper ion stress.
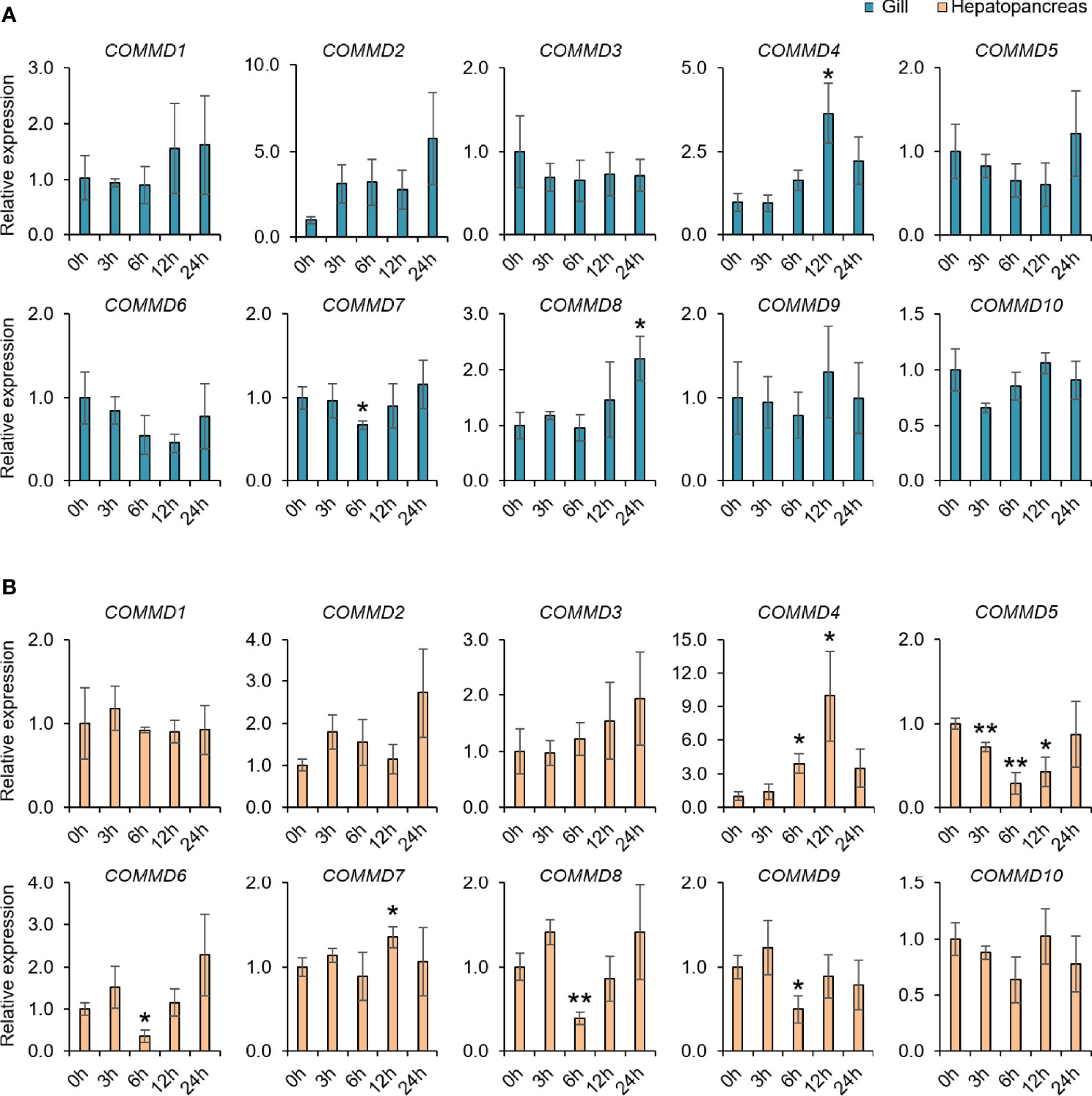
Figure 7 Relative COMMD genes expression in M. lateralis gill and hepatopancreas after V. anguillarum infection. (A) Relative expression of MLCOMMDs in gill after V. anguillarum infection. (B) Relative expression of MLCOMMDs in hepatopancreas after V. anguillarum infection. The relative fold changes compared with control group for each test point are shown as a bar chart (significance: ** p < 0.01; * p < 0.05). The blue boxes indicate gill and the light orange boxes indicate hepatopancreas.
Discussion
In this study, the COMMD family in four bivalve molluscs was identified and characterized based on the genomic and transcriptomic data. Similar to the human COMMD proteins, these bivalve COMMD proteins contain the conserved COMM domain in the extreme carboxyl terminus, which could mediate the interaction of COMMD-COMMD proteins and the formation of the COMMD polymer (Burstein et al., 2005). The amino terminal of COMMDs shares a low homology among members of the family but has highly conserved sequences with their ortholog proteins, which may contribute to the functional diversity of different subfamilies (Maine and Burstein, 2007). Besides four bivalve molluscs in our study, a 5-intron pattern of COMMD9 was also discovered in human and zebrafish, which may imply that COMMD9 has retained the ancestral exon–intron structure during the evolution. Other COMMD subfamilies of four bivalve molluscs have inconsistent intron patterns. Previously, researchers have found that intron insertion and loss may be associated with selective splicing, encoding the untranslated RNAs and enhancing the levels of mRNA transcription (Jin et al., 2012). The frequent occurrence of intron insertion and deletion among the COMMD family except for COMMD9 may be to a certain extent related to the function of introns.
Phylogenetic analysis showed that four bivalve mollusc COMMD genes always clustered together in the invertebrate clades, consisting of their evolution status and conservativeness. Previous findings show that COMMD genes are highly conserved throughout vertebrate evolution, and COMMD1 and COMMD9 seem to be restricted to vertebrates (Burstein et al., 2005). In the present study, we further retrieved COMMD genes from 26 metazoan species (Figure 3). We found that both COMMD1 and COMMD9 could be identified in the Lophotrochozoa, Deuterostomia, Cnidaria, and Sponge groups, while they were absent in the Ecdysozoa group. Besides COMMD1, 9, COMMD6, 7, 8 were also absent in most ecdysozoans and that nematodes only have the COMMD4 subfamily, which may attribute to the fact that the Ecdysozoa genomes are rapidly evolving (Telford et al., 2008). Besides, COMMD1, 6, 8 have been lost in ciona (Jin et al., 2018), while 10 intact single-copy COMMD genes were found in amphioxus as well as in most vertebrates. Species in the Mollusca, Deuterostomia (except Urochordata), Brachiopoda, Cnidaria, and Sponge groups almost have ten intact single-copy COMMD family genes; only several species were found to have loss of one the COMMD subfamily members. However, it remains unclear whether this phenomenon is due to genome assembly fragmentation or they have been lost during evolution. An across-Metazoa comparison implied that COMMD members diverged from each other at early stages of evolution and the integrated COMMD family may already exist in the metazoan last common ancestors.
Evolutionary analysis have shown that purifying selection dominated the evolution of COMMD genes (Jin et al., 2018). For four bivalves, a total of 25 sites under potentially positive selection were identified which may provide a support for the structural and functional diversity of the COMMD family members. Similarly, the selective pressure analyses of COMMD family genes in amphioxus showed that there were 16 positive selective sites detected, although the COMMD family genes have undergone very strong purifying selection during evolution (Jin et al., 2018).
In previous studies, the expression profile of COMMD family genes has been reported in vertebrates (van De Sluis et al., 2002; Klomp et al., 2003). However, such research remains lacking in invertebrates. To better understand the characteristic and function of COMMD genes in molluscs, extensive transcriptome resources were used to profile the temporal and spatial expression patterns of COMMD genes in four bivalve molluscs (Figure 5). The COMMDs were reported to play a vital role during mouse embryonic development, and COMMD-knockout mice are embryonically lethal and die at different stages of embryogenesis (Semenova et al., 2003; van de Sluis et al., 2007; Bartuzi et al., 2013). Besides, researchers found that COMMD1 has a regulatory role in the cell cycle of HEK293 cells (Jiang et al., 2019). Our results showed that bivalve COMMD1 is highly expressed during multicellular cleavage and its transcription level declines rapidly from blastula. It may also implicate that COMMD1 is involved in the regulation of cell proliferation. Bivalve COMMD7s were found to enhance their transcription exponentially from the D-shaped veliger stage and sustained a high level of transcription during late larval development. Previous studies have found that COMMD7 promoted cell proliferation, migration, and invasion processes but suppressed cell apoptosis (Devlin et al., 2003; Zheng et al., 2018). Therefore, we speculate that COMMD7 may be involved in the regulation of organogenesis during embryonic larval formation. Among different adult tissues, high transcription levels of most COMMDs were found in the hepatopancreas. As filter-feeding animals that mainly feed on microalgae, bivalves could accumulate hazardous substance produced through diet, and the hepatopancreas is the main organ for processing and accumulating the incoming hazardous substance (Lian et al., 2019). Therefore, the relatively higher transcription levels of COMMDs in the hepatopancreas may assist with toxin tolerance in bivalves. COMMD1, 9, 10 are involved in the regulation of cell proliferation, migration, and cell-cycle progression (Yang et al., 2017; Zhan et al., 2017). We found that COMMD1, 6, 10 from two scallops were highly expressed in the female gonads, suggesting that these three COMMD members may contribute to the ovarian cell homeostasis maintenance to assist with oogenesis in scallops.
The COMMD genes play key roles in regulating copper homeostasis and innate immune response (Bartuzi et al., 2013; Jin et al., 2018; Mouhadeb et al., 2018). To investigate the possibly biological functions of the MlCOMMDs, their mRNA expression levels were measured at different time points under copper ion stress and bacterial stress. MlCOMMD1, 4, 9 transcriptions exhibited a significant alteration after copper ion stress in both assayed organs of dwarf surf clam, and MlCOMMD9 showed the most drastic upregulation, suggesting their functional relation with cellular copper ion metabolism. It was reported that COMMD1 regulates the endosomal sorting of the copper transporter (Phillips-Krawczak et al., 2015), and COMMD9 may be an endogenous regulator of the epithelial sodium channel (ENaC) to regulate Na+ transport, which could indirectly alter intracellular Cu flux (Handy et al., 2002; Liu et al., 2013). Besides, it was previously found that deficiency of COMMD1 or COMMD9 can result in hepatic copper accumulation under high-copper diets (Singla et al., 2021). Our V. Anguillarum infection experimental results revealed that MlCOMMD4 was significantly upregulated in both the gill and hepatopancreas of dwarf surf clam. Previous studies reported that COMMD4 has the ability to inhibit NF-κB, the key regulator of both innate and adaptive immune responses (de Bie et al., 2006; Maine and Burstein, 2007; Hayden and Ghosh, 2008; Naugler and Karin, 2008);, while whether bivalve COMMD4 can regulate NF-κB needs further more detailed studies.
Conclusions
In this study, 10 COMMD genes were respectively identified from the four bivalves, namely, Yesso scallop, Zhikong scallop, Pacific oyster, and dwarf surf clam. They possessed conserved COMM domains and comprised ten subfamilies. Purifying selection of six subfamilies was revealed, with the strongest selection on COMMD4. The expression profiling during embryonic development and in adult organs provided valuable implications for exploring the function of the bivalve COMMD gene. After exposure to two different stresses, MLCOMMDs exhibited different regulation patterns in different tissues or organs. The responses of COMMD members under stresses were found more sensitive in the hepatopancreas than in the gill, and MlCOMMD9 and MlCOMMD4 might be the good candidate stress indicator genes respectively for copper ion stress and V. Anguillarum infection. This study comprehensively describes the first genome-wide characterization of the COMMD gene family in bivalves, and our work will be helpful in better understanding the function and evolution of COMMD family bivalve molluscs.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
Author Contributions
SL and JH conceived and designed the study. CX and NH performed the experiments. LL and FS participated in the data analysis. XC, SL, LZ, SW, and ZB wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
We acknowledge the grant support from the Project of Sanya Yazhouwan Science and Technology City Management Foundation (SKJCKJ-2019KY01), the Key R&D Project of Shandong Province (2020ZLYS10, 2021ZLGX03), and the China Agriculture Research System of MOF and MARA.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2022.884991/full#supplementary-material
References
Bartuzi P., Hofker M. H., van de Sluis B. (2013). Tuning NF-kappaB Activity: A Touch of COMMD Proteins. Biochim. Biophys. Acta 1832, 2315–2321. doi: 10.1016/j.bbadis.2013.09.014
Burstein E., Hoberg J. E., Wilkinson A. S., Rumble J. M., Csomos R. A., Komarck C. M., et al. (2005). COMMD Proteins, a Novel Family of Structural and Functional Homologs of MURR1. J. Biol. Chem. 280, 22222–22232. doi: 10.1074/jbc.M501928200
Chomczynski P., Sacchi N. (2006). The Single-Step Method of RNA Isolation by Acid Guanidinium Thiocyanate-Phenol-Chloroform Extraction: Twenty-Something Years on. Nat. Protoc. 1, 581–585. doi: 10.1038/nprot.2006.83
de Bie P., van de Sluis B., Burstein E., Duran K. J., Berger R., Duckett C. S., et al. (2006). Characterization of COMMD Protein-Protein Interactions in NF-kappaB Signalling. Biochem. J. 398, 63–71. doi: 10.1042/BJ20051664
Devlin A. M., Solban N., Tremblay S., Gutkowska J., Schürch W., Orlov S. N., et al. (2003). HCaRG is a Novel Regulator of Renal Epithelial Cell Growth and Differentiation Causing G(2)M Arrest. Am. J. Physiol. Renal Physiol. 284, F753–F762. doi: 10.1152/ajprenal.00252.2002
Dill B. D., Gierlinski M., Hartlova A., Arandilla A. G., Guo M., Clarke R. G., et al. (2015). Quantitative Proteome Analysis of Temporally Resolved Phagosomes Following Uptake via Key Phagocytic Receptors. Mol. Cell Proteom. 14, 1334–1349. doi: 10.1074/mcp.M114.044594
Gao F., Chen C., Arab D. A., Du Z., He Y., Ho S. Y. W. (2019). EasyCodeML: A Visual Tool for Analysis of Selection Using CodeML. Ecol. Evol. 9, 3891–3898. doi: 10.1002/ece3.5015
Handy R., Eddy F., Baines H. (2002). Sodium-Dependent Copper Uptake Across Epithelia: A Review of Rationale With Experimental Evidence From Gill and Intestine. Biochim. Biophys. Acta 1566, 104–115. doi: 10.1016/s0005-2736(02)00590-4
Hayden M. S., Ghosh S. (2008). Shared Principles in NF-kappaB Signaling. Cell 132, 344–362. doi: 10.1016/j.cell.2008.01.020
Jiang Z., Yuan Y., Zheng H., Cui H., Sun X., Zhao W., et al. (2019). COMMD1 Regulates Cell Proliferation and Cell Cycle Progression by Modulating P21 Cip1 Levels. Biosci. Biotechnol. Biochem. 83, 845–850. doi: 10.1080/09168451.2019.1569497
Jin P., Gao Y., Chen L., Ma F. (2012). Cloning and Characterization of a COMMD4 Gene From Amphioxus (Branchiostoma Belcheri): An Insight Into the Function and Evolution of COMMD4. Immunol. Lett. 148, 110–116. doi: 10.1016/j.imlet.2012.10.008
Jin P., Lv C., Peng S., Cai L., Zhu J., Ma F., et al. (2018). Genome-Wide Organization, Evolutionary Diversification of the COMMD Family Genes of Amphioxus (Branchiostoma Belcheri) With the Possible Role in Innate Immunity. Fish. Shellf Immunol. 77, 31–39. doi: 10.1016/j.fsi.2018.03.019
Klomp A. E., van de Sluis B., Klomp L. W., Wijmenga C. (2003). The Ubiquitously Expressed MURR1 Protein is Absent in Canine Copper Toxicosis. J. Hepatol. 39, 703–709. doi: 10.1016/s0168-8278(03)00380-5
Larkin M. A., Blackshields G., Brown N. P., Chenna R., Mcgettigan P. A., Mcwilliam H., et al. (2007). Clustal W and Clustal X Version 2.0. Bioinformatics 23, 2947–2948. doi: 10.1093/bioinformatics/btm404
Lian S., Zhao L., Xun X., Lou J., Li M., Li X., et al. (2019). Genome-Wide Identification and Characterization of SODs in Zhikong Scallop Reveals Gene Expansion and Regulation Divergence After Toxic Dinoflagellate Exposure. Mar. Drugs 17, 700. doi: 10.3390/md17120700
Li Y., Sun X., Hu X., Xun X., Zhang J., Guo X., et al. (2017). Scallop Genome Reveals Molecular Adaptations to Semi-Sessile Life and Neurotoxins. Nat. Commun. 8, 1721. doi: 10.1038/s41467-017-01927-0
Liu Y., Swart M., Ke Y., Ly K., McDonald F. J. (2013). Functional Interaction of COMMD3 and COMMD9 With the Epithelial Sodium Channel. Am. J. Physiol. Ren. Physiol. 305, F80–F89. doi: 10.1152/ajprenal.00158.2013
Maine G. N., Burstein E. (2007). COMMD Proteins: COMMing to the Scene. Cell. Mol. Life Sci. 64, 1997–2005. doi: 10.1007/s00018-007-7078-y
Maine G. N., Mao X., Komarck C. M., Burstein E. (2007). COMMD1 Promotes the Ubiquitination of NF-κb Subunits Through a Cullin-Containing Ubiquitin Ligase. EMBO J. 26, 436–447. doi: 10.1038/sj.emboj.7601489
Mouhadeb O., Ben Shlomo S., Cohen K., Farkash I., Gruber S., Maharshak N., et al. (2018). Impaired COMMD10-Mediated Regulation of Ly6C(hi) Monocyte-Driven Inflammation Disrupts Gut Barrier Function. Front. Immunol. 9. doi: 10.3389/fimmu.2018.02623
Muller P. A., van de Sluis B., Groot A. J., Verbeek D., Vonk W. I., Maine G. N., et al. (2009). Nuclear-Cytosolic Transport of COMMD1 Regulates NF-kappaB and HIF-1 Activity. Traffic 10, 514–527. doi: 10.1111/j.1600-0854.2009.00892.x
Nabetani A., Hatada I., Morisaki H., Oshimura M., Mukai T. (1997). Mouse U2af1-Rs1 is a Neomorphic Imprinted Gene. Mol. Cell Biol. 17, 789–798. doi: 10.1128/MCB.17.2.789
Nakai A., Fujimoto J., Miyata H., Stumm R., Narazaki M., Schulz S., et al. (2019). The COMMD3/8 Complex Determines GRK6 Specificity for Chemoattractant Receptors. J. Exp. Med. 216, 1630–1647. doi: 10.1084/jem.20181494
Naugler W. E., Karin M. (2008). NF-kappaB and Cancer-Identifying Targets and Mechanisms. Curr. Opin. Genet. Dev. 18, 19–26. doi: 10.1016/j.gde.2008.01.020
Nicholas K., Nicholas H., Deerfield D. (1997). GeneDoc: Analysis and Visualization of Genetic Variation. Embnew. News 4, 14. doi: 10.11118/actaun201361041061
Phillips-Krawczak C. A., Singla A., Starokadomskyy P., Deng Z., Osborne D. G., Li H., et al. (2015). COMMD1 is Linked to the WASH Complex and Regulates Endosomal Trafficking of the Copper Transporter ATP7A. Mol. Biol. Cell. 26, 91–103. doi: 10.1091/mbc.E14-06-1073
Riera-romo M. (2018). COMMD1: A Multifunctional Regulatory Protein. J. Cell Biochem. 119, 34–51. doi: 10.1002/jcb.26151
Semenova E., Wang X., Jablonski M. M., Levorse J., Tilghman S. M. (2003). An Engineered 800 Kilobase Deletion of Uchl3 and Lmo7 on Mouse Chromosome 14 Causes Defects in Viability, Postnatal Growth and Degeneration of Muscle and Retina. Hum. Mol. Genet. 12, 1301–1312. doi: 10.1093/hmg/ddg140
Singla A., Chen Q., Suzuki K., Song J., Fedoseienko A., Wijers M., et al. (2021). Regulation of Murine Copper Homeostasis by Members of the COMMD Protein Family. Dis. Model. Mech. 14, dmm045963. doi: 10.1242/dmm.045963
Solban N., Jia H. P., Richard S., Tremblay S., Devlin A. M., Peng J., et al. (2000). HCaRG, a Novel Calcium-Regulated Gene Coding for a Nuclear Protein, is Potentially Involved in the Regulation of Cell Proliferation. J. Biol. Chem. 275, 32234–32243. doi: 10.1074/jbc.M001352200
Sudhir K., Glen S., Koichiro T. (2016). MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Tao T. Y., Liu F., Klomp L., Wijmenga C., Gitlin J. D. (2003). The Copper Toxicosis Gene Product Murr1 Directly Interacts With the Wilson Disease Protein. J. Biol. Chem. 278, 41593–41596. doi: 10.1074/jbc.C300391200
Telford M. J., Bourlat S. J., Economou A., Papillon D., Rota-Stabelli O. (2008). The Evolution of the Ecdysozoa. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 363, 1529–1537. doi: 10.1098/rstb.2007.2243
Uys G. M., Ramburan A., Loos B., Kinnear C. J., Korkie L. J., Mouton J., et al. (2011). Myomegalin is a Novel A-Kinase Anchoring Protein Involved in the Phosphorylation of Cardiac Myosin Binding Protein C. BMC Cell Biol. 12, 18. doi: 10.1186/1471-2121-12-18
van de Sluis B., Muller P., Duran K., Chen A., Groot A. J., Klomp L. W., et al. (2007). Increased Activity of Hypoxia-Inducible Factor 1 Is Associated With Early Embryonic Lethality in Commd1 Null Mice. Mol. Cell Biol. 27, 4142–4156. doi: 10.1128/MCB.01932-06
van De Sluis B., Rothuizen J., Pearson P. L., van Oost B. A., Wijmenga C. (2002). Identification of a New Copper Metabolism Gene by Positional Cloning in a Purebred Dog Population. Hum. Mol. Genet. 11, 165–173. doi: 10.1093/hmg/11.2.165
Wang F., Xiao S., Xiang Z., Yu Z. (2017a). Molecular Cloning and Expression Analysis of Commd1 Under Salinity Stress in Crassostrea Hongkongensis. J. Oceanogr. 36, 48–55. doi: 10.11978/2016022
Wang S., Zhang J., Jiao W., Li J., Xun X., Sun Y., et al. (2017b). Scallop Genome Provides Insights Into Evolution of Bilaterian Karyotype and Development. Nat. Ecol. Evol. 1, 120. doi: 10.1038/s41559-017-0120
Yang M., Huang W., Sun Y., Liang H., Chen M., Wu X., et al. (2019). Prognosis and Modulation Mechanisms of COMMD6 in Human Tumours Based on Expression Profiling and Comprehensive Bioinformatics Analysis. Br. J. Cancer 121, 699–709. doi: 10.1038/s41416-019-0571-x
Yang S., Li X., Yang M., Ren X., Hu J., Zhu X., et al. (2017). FMNL2 Destabilises COMMD10 to Activate NF-κ B Pathway in Invasion and Metastasis of Colorectal Cancer. Br. J. Cancer 117, 1164–1175. doi: 10.1038/bjc.2017.260
Yang Z., Wong W. S. W., Rasmus N. (2005). Evolution, Bayes Empirical Bayes Inference of Amino Acid Sites Under Positive Selection. Mol. Biol. Evol. 22, 1107–1118. doi: 10.1093/molbev/msi097
You N., Li J., Huang X. B., Wu K., Tang Y. C., Wang L., et al. (2017). COMMD7 Promotes Hepatocellular Carcinoma Through Regulating CXCL10. Biomed. Pharmacother. 88, 653–657. doi: 10.1016/j.biopha.2017.01.046
Zhang G., Fang X., Guo X., Li L., Luo R., Xu F., et al. (2012). The Oyster Genome Reveals Stress Adaptation and Complexity of Shell Formation. Nature 490, 49–54. doi: 10.1038/nature11413
Zhan W., Wang W., Han T., Xie C., Zhang T., Gan M., et al. (2017). COMMD9 Promotes TFDP1/E2F1 Transcriptional Activity via Interaction With TFDP1 in non-Small Cell Lung Cancer. Cell. Signal. 30, 59–66. doi: 10.1016/j.cellsig.2016.11.016
Zheng L., You N., Huang X., Gu H., Wu K., Mi N., et al. (2018). COMMD7 Regulates NF-κb Signaling Pathway in Hepatocellular Carcinoma Stem-Like Cells. Mol. Ther. Oncolyt. 12, 112–113. doi: 10.1016/j.omto.2018.12.006
Keywords: bivalve, COMMD, expression profiling, phylogenetic analysis, innate immune
Citation: Chen X, Hu N, Lian S, Li L, Sun F, Zhang L, Wang S, Bao Z and Hu J (2022) Genome-Wide Identification and Expression Profiling of the COMMD Gene Family in Four Bivalve Molluscs. Front. Mar. Sci. 9:884991. doi: 10.3389/fmars.2022.884991
Received: 27 February 2022; Accepted: 21 March 2022;
Published: 20 April 2022.
Edited by:
Zhongming Huo, Dalian Ocean University, ChinaReviewed by:
Qiong Shi, Beijing Genomics Institute (BGI), ChinaLusheng Xin, Chinese Academy of Fishery Sciences (CAFS), China
Junxia Mao, Dalian Ocean University, China
Copyright © 2022 Chen, Hu, Lian, Li, Sun, Zhang, Wang, Bao and Hu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shanshan Lian, bGlhbnNoYW5zaGFuQG91Yy5lZHUuY24=; Jingjie Hu, aHVqaW5namllQG91Yy5lZHUuY24=
†These authors have contributed equally to this work and share first authorship
 Xiaomei Chen1,2†
Xiaomei Chen1,2† Shanshan Lian
Shanshan Lian Lingling Zhang
Lingling Zhang Shi Wang
Shi Wang Zhenmin Bao
Zhenmin Bao