
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mar. Sci. , 06 May 2022
Sec. Marine Biotechnology and Bioproducts
Volume 9 - 2022 | https://doi.org/10.3389/fmars.2022.879333
This article is part of the Research Topic Biotechnology of Marine Microorganisms View all 7 articles
 Zixu Liu1†
Zixu Liu1† Biao Yuan1†
Biao Yuan1† Lingmin Zhao1
Lingmin Zhao1 Lixing Huang1
Lixing Huang1 Yingxue Qin1
Yingxue Qin1 Jiaonan Zhang2
Jiaonan Zhang2 Jiaolin Zhang2
Jiaolin Zhang2 Bing Hu2
Bing Hu2 Qingpi Yan1*
Qingpi Yan1*Pseudomonas plecoglossicida is a gram-negative pathogenic bacterium that causes visceral white spot disease in several marine and aquaculture fish species, resulting in high mortality and severe financial loss. Based on previous RNA sequencing (RNA-seq), fliK gene expression is significantly up-regulated in P. plecoglossicida during infection, indicating that fliK may contribute to its bacterial pathogenicity. To investigate the role of fliK, four specific short hairpin RNAs (shRNAs) were designed and synthesized according to the fliK gene sequence, with three of the four mutants exhibiting a significant decrease in fliK gene expression in P. plecoglossicida. The shRNA-406 mutant with the maximum silencing efficiency (97.5%) was chosen for further study. Compared with the wild-type (WT) P. plecoglossicida strain, silencing fliK in the fliK-RNA interference (RNAi) strain resulted in a significant decrease in growth, motility, chemotaxis, adhesion, and biofilm formation in P. plecoglossicida. Silencing of fliK also resulted in a 95% increase in the survival rate, a 2-day delay in the onset of death, and a significant decrease in the number of white spots on the spleen surface in infected orange-spotted groupers (Epinephelus coioides). In addition, fliK gene expression and pathogen load were significantly lower in the spleens of E. coioides infected with the fliK-RNAi strain than in those infected with the WT strain of P. plecoglossicida. RNA-seq of the spleens further revealed that fliK silencing significantly regulated the immune response of E. coioides during the pathogenic process. Compared with the WT-infected group, the differentially expressed genes (DEGs) in the fliK-RNAi-infected group were enriched in 344 and 345 KEGG pathways at 3 and 5 days post infection (dpi), respectively. Among these pathways, 21 immune system-related pathways were enriched, including the natural killer (NK) cell-mediated cytotoxicity, platelet activation, and Th17 cell differentiation signaling pathways. The NK cell-mediated cytotoxicity pathway was the most significantly enriched, which may enhance the host’s ability to remove pathogens and reduce inflammation. This study revealed the effects of the fliK gene in P. plecoglossicida pathogenicity and identified the main pathways involved in the immune response of E. coioides.
Disease has always restricted large-scale aquacultural development (Defoirdt, 2016). In recent years, visceral white spot disease has become an increasing issue in mariculture fish (Zhang et al., 2014; Li et al., 2020; He et al., 2021), resulting in high mortality and serious economic loss (Tang et al., 2020). Pseudomonas plecoglossicida, a Gram-negative proteobacterium first identified in diseased ayu (Plecoglossus altivelis) (Nishimori et al., 2000), is the causative agent of visceral white spot disease in large yellow croakers (Larimichthys crocea) and orange-spotted groupers (Epinephelus coioides) (Zhang et al., 2014; Huang et al., 2019; Li et al., 2020). While several strains of P. plecoglossicida show a strong ability to degrade pollutants and have been used in environmental pollution control (Jha et al., 2009; John et al., 2015), other strains are highly pathogenic to teleosts, leading to high death rates in a variety of farmed fish (Nishimori et al., 2000; Zhang et al., 2014; Tang et al., 2019a). Based on previous transcriptome data from our laboratory (NCBI, SRP115064), we found the fliK gene to be highly expressed in infected E. coioides (Luo et al., 2020), suggesting it may play a role in P. plecoglossicida infection.
As a multi-subunit surface organelle, flagella are present in more than 70% of bacteria (Persat et al., 2015). These flagella are either peritrichous or polar, which can affect bacterial motility (Chaban et al., 2015), chemotaxis, and virulence (Sampedro et al., 2015). Pseudomonas plecoglossicida bacteria have polar flagella, and thus exhibit excellent motility. The assembly and regulation of flagella require the participation of proteins encoded by more than 50 genes (Duan et al., 2013). The FliK protein controls flagella hook length after secretion (Waters et al., 2007; Kinoshita et al., 2020) and is considered an essential determinant of Bacillus thuringiensis virulence (Attieh et al., 2020). The fliK gene encodes the FliK protein and thus may play a role in flagella regulation and bacterial virulence; however, its role in P. plecoglossicida remains unclear.
Considering the strong pathogenicity of P. plecoglossicida to economically valuable fish and the potential impact of fliK on virulence, we silenced the fliK gene in P. plecoglossicida and analyzed differences in bacterial virulence and E. coioides immune response between wild-type (WT) and fliK-RNA interference (RNAi) strains. This study should enhance our understanding of host-pathogen interactions between E. coioides and P. plecoglossicida.
The highly pathogenic P. plecoglossicida WT strain NZBD9 was initially isolated from a L. crocea fish with visceral white spot disease. The bacteria were grown in Luria Bertani (LB) broth at 18 or 28°C with shaking at 220 rpm. Escherichia coli DH5α was purchased from TransGen Biotech (Beijing, China) and grown in LB broth (37°C, 220 rpm). Tetracycline was added to the medium when appropriate at a final concentration of 10 μg/mL.
The RNAi strains were constructed as per Darsigny et al. (2010) and Choi and Schweizer (2006), with minor modifications. Briefly, after designing four short hairpin RNA (shRNA) sequences (Supplementary Table 1) targeting the fliK gene using Thermo Fisher BLOCK-iT™ RNAi Designer (http://rnaidesigner.thermofisher.com/rnaiexpress/), the shRNAs were synthesized by Shanghai Generay Biotech Co., Ltd. (China). Tetracycline-resistant plasmid pCM130/tac was modified based on pCM130 following Luo et al. (2019). Recombinant pCM130/tac vectors were constructed by connecting shRNA to linear pCM130/tac, followed by digestion with restriction enzymes NsiI-HF and BsrGI-HF (New England Biolabs, USA) via T4 DNA ligase (Takara Biomedical Technology, Beijing, China) according to the product manual. The recombinant pCM130/tac vectors were transfected into E. coli DH5α cells by water-bathing heat shock. Plasmid DNA was extracted from the E. coli DH5α cells using an EasyPure Plasmid MiniPrep Kit (TransGen Biotech, Beijing, China) and then electroporated into P. plecoglossicida to construct the fliK-RNAi strain. Finally, the expression levels of fliK in each fliK-RNAi strain were determined by quantitative real-time polymerase chain reaction (qRT-PCR).
Optical densities (OD600) of overnight cultures of the WT and fliK-RNAi strains of P. plecoglossicida were adjusted to 0.2 ± 0.01 with fresh LB broth and diluted 4-fold. Then, 200 μL of diluted bacterial suspension was added to each well of a microtiter plate and incubated at 28°C for 24 h. The OD600 of each well was detected hourly and automatically by a SYNERGY H1 microplate reader (BioTek, USA).
Overnight cultures of the WT and fliK-RNAi strains were adjusted to OD600 = 0.3 ± 0.01 with fresh LB broth, and 1 μL of adjusted bacterial suspension was dropped onto solidified 0.4% semi-solid agar medium before incubation at 28°C for 12 h. Measurement was repeated three times.
Skin mucus was obtained from healthy E. coioides according to previously described methods (Chen et al., 2008). The protein concentration of the skin mucus was adjusted to 1 mg/mL using a Quantitative Kit (TransGen Biotech, Beijing, China). An in vitro adhesion assay was carried out following previously described methods (Jiao et al., 2021). The adhered bacteria in 20 randomly selected fields were counted under a microscope (×1 000) (Leica DM4000 B LED, Leica, Germany). Three independent biological replicates were performed per group.
Overnight cultures of the WT and fliK-RNAi strains were centrifuged, resuspended in phosphate-buffered saline (PBS, pH=7.4), and adjusted to OD600 = 0.5 ± 0.01. The bacterial suspension (0.3 mL) was then aspirated into a sterile syringe (1 mL). The open end of a capillary tube (inner diameter of 0.1 mm, sealed at one end) filled with E. coioides skin mucus was then horizontally dipped into the bacterial suspension in the syringe, and incubated at 28°C for 1 h. Finally, the number of bacteria in the mucus was determined by plate counting. Sterile PBS-EDTA in a capillary tube was used as the control. Three independent biological replicates were performed per group. The chemotactic index (CI) was calculated as CI = CFU (experimental group)/CFU (control group) (Joós et al., 2017).
Overnight cultures of the WT and fliK-RNAi strains were adjusted to OD600 = 0.3 ± 0.01 in fresh LB broth. Bacterial culture aliquots (100 μL) were added to a 12-well microtiter plate and incubated at 28°C for 24 h. After washing the wells twice with sterile PBS, the biofilm was fixed at 65°C for 45 min, then stained with 125 μL of crystal violet (0.1%) for 15 min, washed twice with PBS, and air dried. Finally, 200 μL of acetic acid (33%) was added to each well to solubilize the stained biofilm and the OD590 of each well was determined using the SYNERGY H1 microplate reader (BioTek, USA).
All infection experiments were approved by the Research Board of the Ethics Committee of Jimei University under permit number JMULAC201159.
Healthy size-matched E. coioides (body length 12 ± 2 cm) were purchased from Zhangzhou city (Fujian, China) and adaptively cultured at 18 ± 1°C for one week under specific pathogen-free laboratory conditions. The strains for infection were cultured in LB medium at 18°C for 18 h to the late stage of the logarithmic phase and then centrifuged and resuspended in PBS at 18°C.
For the survival assay, 180 fish were randomly divided into three groups (WT strain infection group, fliK-RNAi strain infection group, and PBS injection group), with each group consisting of three tanks. Each fish in the infection groups was intraperitoneally injected with the corresponding strain at a dose of 5 × 104 CFU/fish. Sixty fish injected with 200 μL of PBS were used as the negative control. The status and mortality of the injected fish were recorded twice a day. Seawater temperature was maintained at 18 ± 1°C throughout the whole experiment.
For spleen sampling, 120 fish were randomly divided into two groups (i.e., WT and fliK-RNAi strain infection groups) and intraperitoneally injected with the corresponding strain at a dose of 5 × 104 CFU/fish. Six spleens from E. coioides infected with the WT or fliK-RNAi strain of P. plecoglossicida were randomly sampled at 1, 2, 3, 4, 5, and 6 days post infection (dpi). Two spleens from the same group were randomly mixed into one sample. Spleen samples from 1, 2, 3, 4, 5, and 6 dpi were subjected to fliK expression and pathogen load assays. Spleen samples from 3 and 5 dpi were sent to Shanghai Majorbio Bio-Pharm Technology Co., Ltd. (China) for transcriptome sequencing.
Total RNA from bacteria was extracted using a TransZol Up Kit (TransGen Biotech, Beijing, China). Total RNA from fish spleens was extracted using an Easystep Super Total RNA Extraction Kit (Promega, Beijing, China). The cDNA was synthesized using TransScript All-in-One First-Strand cDNA Synthesis SuperMix for qRT-PCR (One-Step gDNA Removal) (TransGen Biotech, Beijing, China) according to the product manual.
RNA quality was analyzed by agarose gel electrophoresis and absorbance value.
qRT-PCR was implemented using a QuantStudio 6 Flex Real-Time PCR system (Life Technologies, USA). Primer sequences (Supplementary Table 2) were designed using Primer Premier v5.0. The P. plecoglossicida and E. coioides gene expression levels were normalized according to 16S rDNA and β-actin, respectively (Wang et al., 2020). The P. plecoglossicida pathogen load in each sample was estimated using the copy number of the housekeeping gene gyrB (Izumi et al., 2007). Three technical replicates were performed for each treatment. The 2−ΔΔCt method was used to calculate the relative expression levels of the genes (Luo et al., 2020).
Bacterial DNA was extracted from the infected spleens of E. coioides using an EasyPure Marine Animal Genomic DNA Kit (TransGen Biotech, Beijing, China) according to the manufacturer’s instructions. The bacterial load of P. plecoglossicida was calculated using the gyrB gene copy number, which was determined by qRT-PCR (Tang et al., 2020). Three biological replicates were carried out.
The RNA-seq libraries were prepared using protocols supplied with the Illumina TruSeq™ RNA Sample Prep Kit (San Diego, CA, USA). RNA concentration and purity were detected using the NanoDrop 2000 instrument (NanoDrop Technologies). RNA integrity number was detected using the Agilent 2100 system (Agilent Technologies, USA), with only high-quality RNA samples (1.9 ≤ A260/280 ≤ 2.0, RIN ≥ 6.5, 28 S:18 S ≥ 1.0, total amount > 2 μg) used to construct individual sequencing libraries. The mRNA was fragmented by fragmentation buffer and reverse transcribed into cDNA, which was then synthesized using a SuperScript Double-Stranded cDNA Synthesis Kit (Invitrogen, Carlsbad, CA, USA). After blunt end repair, phosphorylation, and poly (A) addition to the 3’ end, the cDNA library was amplified in 15 cycles using Phusion DNA polymerase (NEB, USA). DNA was quantified using a TBS380 Fluorometer (Turner Bio Systems, USA). The RNA-seq libraries were sequenced on the Illumina NovaSeq 6000 sequencing platform at Shanghai Majorbio Bio-Pharm Technology Co., Ltd (China).
Statistical analysis and quality control of the raw Illumina reads were performed using fastx_toolkit_0.0.14 (http://hannonlab.cshl.edu/fastx_toolkit/) (Wu et al., 2021), SeqPrep (https://github.com/jstjohn/SeqPrep), and Sickle (https://github.com/najoshi/sickle) to ensure the accuracy of subsequent analysis (Rangan et al., 2021).
All clean data were assembled from scratch using Trinity (https://github.com/trinityrnaseq/trinityrnaseq/wiki) (Marazzi et al., 2020). The sequence obtained from de novo assembly was filtered and optimized with TransRate (http://hibberdlab.com/transrate/) (Weng, 2016). Transcriptome integrity was evaluated by BUSCO (http://busco.ezlab.org) (Seppey et al., 2019). Clean reads were aligned with the reference sequence obtained by the Trinity assembly to obtain mapping results (Sherwood et al., 2019).
We performed molecular annotation to describe the biological processes related to the differentially expressed transcripts of E. coioides. The screening standards for significant DEGs were as follows: |log2FC| ≥ 1 and false discovery rate (FDR) < 0.05. Gene Ontology (GO) annotation was completed using GOATOOLS (Klopfenstein et al., 2018). Metabolic pathways were analyzed using Kyoto Encyclopedia of Genes and Genomes (KEGG) (Kanehisa et al., 2017; Lee et al., 2017), and Fisher’s exact test was used for enrichment analysis (Yang et al., 2018). Visualization of the E. coioides transcriptome data was implemented using the clusterProfiler R package and RStudio (Loraine et al., 2015).
All data are expressed as means ± standard deviation (SD) from at least three sets of independent experiments. Data analysis was implemented using IBM SPSS Statistics v22.0 (New York, USA) and one-way analysis of variance (ANOVA) with Dunnett’s test. P-values < 0.05 were considered statistically significant.
The RNA-seq data were deposited in the NCBI SRA database under accession numbers SRP315483 and SRP315268.
Four shRNAs targeting the fliK gene were designed using Thermo Fisher BLOCK-iT™ RNAi Designer. After synthesis by Shanghai Generay Biotech Co., Ltd. (China), the four shRNAs were successfully ligated to plasmid pCM130/tac to construct the recombinant pCM130/tac vectors. After the recombinant plasmid was transferred into P. plecoglossicida, four RNAi mutant strains of P. plecoglossicida were successfully constructed. The qRT-PCR results showed that the expression levels of fliK in all four RNAi mutant strains of P. plecoglossicida were lower than that in the WT strain. Although all four shRNAs led to fliK gene silencing, they showed different silencing efficiency (62.2%, 97.5%, 4.6%, and 57.2%) (Figure 1). The strain containing pCM130/tac-fliK-shRNA-406 (hereafter called the fliK-RNAi strain) exhibited the best silencing efficiency (97.5%) and was chosen for further study.
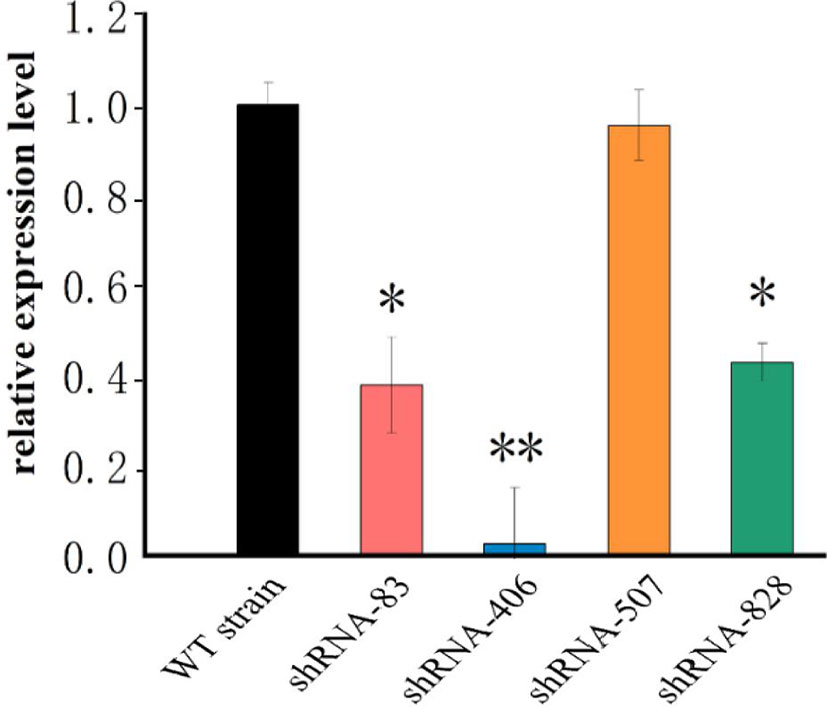
Figure 1 mRNA levels of fliK gene in WT and mutant strains of P. plecoglossicida. *P < 0.05, **P < 0.01.
Figure 2A shows the growth curves of the fliK-RNAi and WT strains of P. plecoglossicida. The growth and maximum growth rates of the fliK-RNAi strain were lower than those of the WT strain. After 11 h of incubation, the OD600 value of the fliK-RNAi strain was significantly lower than that of the WT strain.
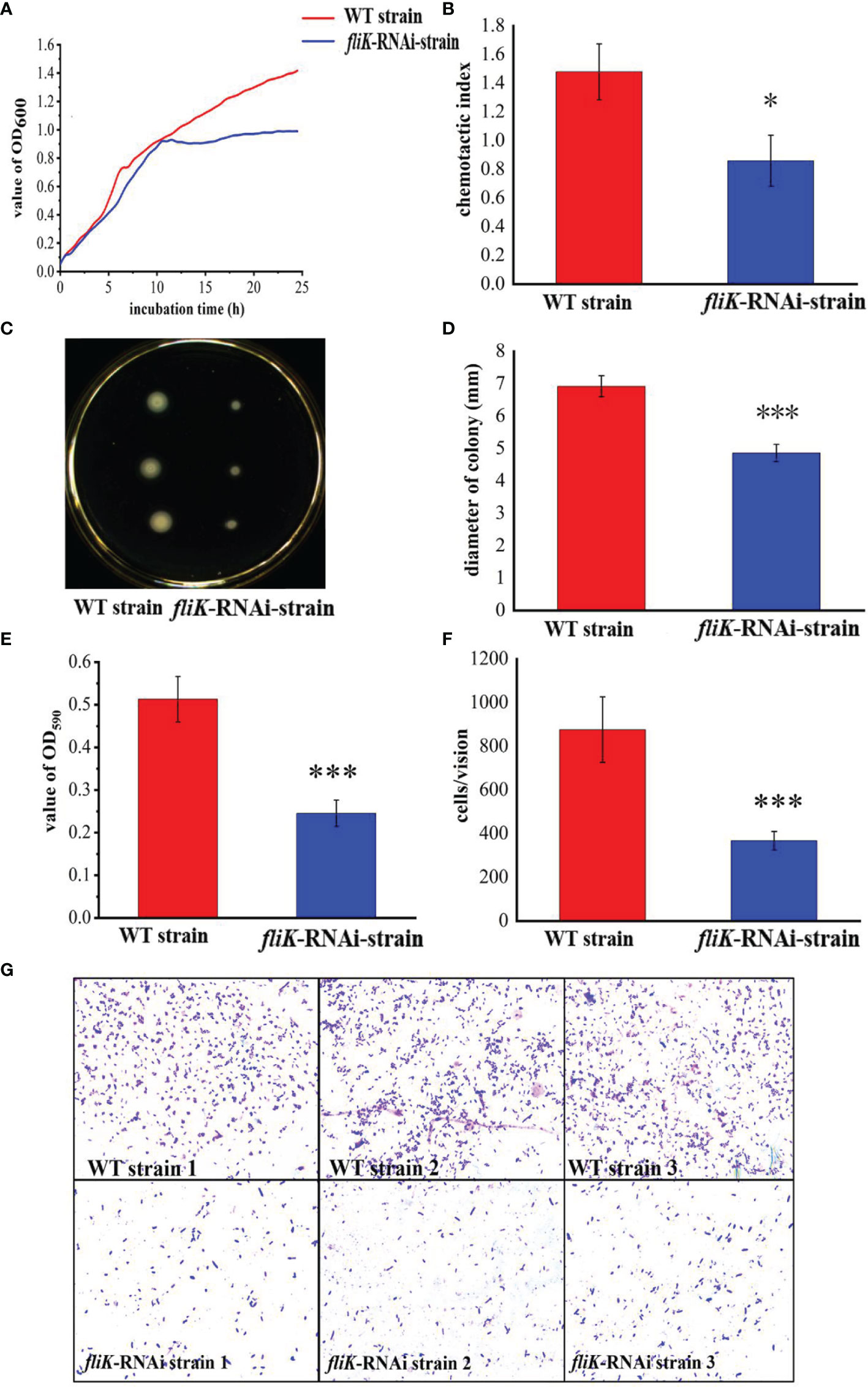
Figure 2 Comparison of biological characteristics between WT and fliK-RNAi strains of P. plecoglossicida. (A) Growth curve of WT and fliK-RNAi strains. (B) Colonies of WT and fliK-RNAi strains on semi-solid agar. (C) Colony diameter of WT and fliK-RNAi strains. (D) Adherent bacterial cells of WT and fliK-RNAi strains under light microscope (×1 000, crystal violet staining). (E) Number of cells of WT and fliK-RNAi strains attached to fish mucus. (F) Chemotaxis index of fish mucus against WT and fliK-RNAi strains. (G) Level of biofilm formation in WT and fliK-RNAi strains. Data are means ± SD from three independent biological replicates. *P < 0.05, ***P < 0.001.
Figure 2B shows the colonies of the fliK-RNAi and WT strains of P. plecoglossicida on a semi-solid LB agar plate. The colonies of the fliK-RNAi strain were significantly smaller than those of the WT strain. Colony diameter of the fliK-RNAi strain (4.86 mm) was significantly lower than that of the WT strain (6.92 mm; P < 0.001) (Figure 2C).
Figure 2D shows the number of bacterial cells of the WT and fliK-RNAi strains that adhered to the fish mucus. As detected under the microscope (×1 000), there were significantly fewer adherent cells of the fliK-RNAi strain (367 cells/vision) than the WT strain (876 cells/vision; P < 0.001; Figure 2E).
Both the WT and fliK-RNAi strains showed positive tropism to the skin mucus of E. coioides. The CI of the fliK-RNAi strain (CI = 0.86) was significantly lower than that of the WT strain (CI = 1.43) (Figure 2F). The OD of the fliK-RNAi strain biofilm stained with 0.1% crystal violet was significantly lower (47.8%) than that of the WT strain (Figure 2G).
The WT strain of P. plecoglossicida exhibited high virulence to E. coioides. Notably, E. coioides infected with 5 × 104 CFU/fish began to die at 2 dpi, and the mortality rate reached 100% at 8 dpi. The E. coioides infected with the same dose of the fliK-RNAi strain began to die at 4 dpi and the mortality rate remained at 5% thereafter. The control group injected with PBS did not die during the experiment (Figure 3A). At 6.5 dpi, the spleen surface of E. coioides infected by the WT strain was covered with numerous white spots. However, there were almost no white spots on the spleen surface of E. coioides injected with the fliK-RNAi strain or PBS (Figure 3B).
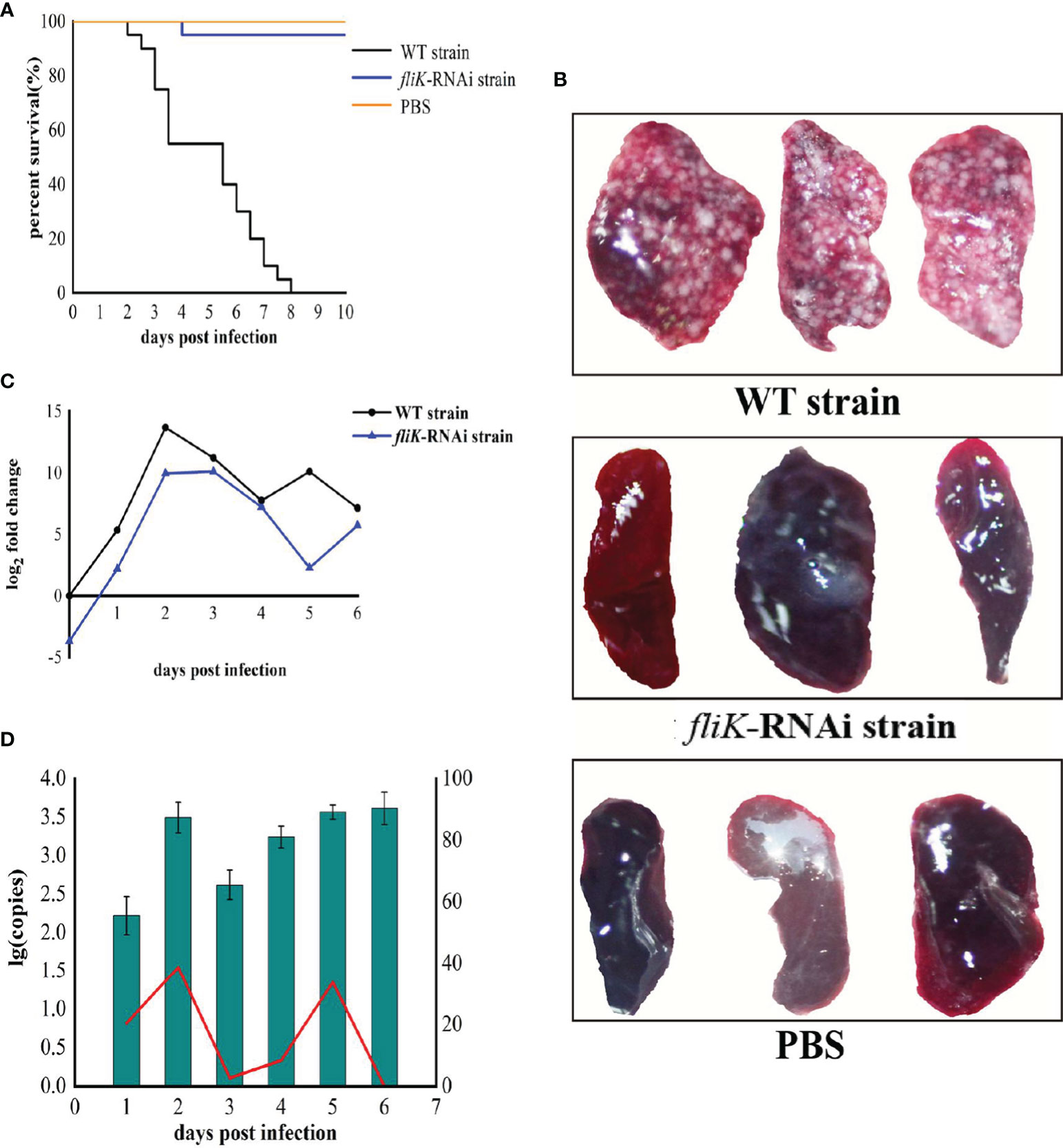
Figure 3 Comparison of virulence between WT and fliK-RNAi strains of P. plecoglossicida. (A) Survival rate of E. coioides post infection. (B) Symptoms in spleens of E. coioides injected with P. plecoglossicida or PBS. (C) Relative expression level of fliK gene in P. plecoglossicida. (D) Pathogen load of P. plecoglossicida in spleens of E. coioides during infection. Bar chart represents pathogen load of fliK-RNAi strain of P. plecoglossicida, represented by copy number of gyrB gene. Line graph represents relative pathogen load of fliK-RNAi strain to WT strain, represented by copy number of gyrB gene of fliK-RNAi strain/copy number of gyrB gene of WT strain × 100%.
Compared with the expression of fliK in P. plecoglossicida cultured at 18°C in vitro, the fliK gene showed high and time-dependent expression in vivo. In the early stages of infection, fliK gene expression in the WT strain gradually increased, reached a maximum at 2 dpi, and then gradually decreased. In the fliK-RNAi strain, fliK gene expression was similar to but always lower than that of the WT strain (Figure 3C).
Both the fliK-RNAi and WT strains of P. plecoglossicida persisted in the E. coioides spleen post injection. The fliK-RNAi strain pathogen load in the infected E. coioides spleens was relatively low at the early stages of infection but was high at the later stages. The fliK-RNAi strain pathogen load was always lower than that of the WT strain over the entire infection period. The relative pathogen load of the fliK-RNAi strain to WT strain increased to 38.7% at 2 dpi and decreased to 0.03% at 6 dpi (Figure 3D).
The spleens of E. coioides infected with the fliK-RNAi or WT strain of P. plecoglossicida or injected with PBS were subjected to RNA-seq. The base content distribution was balanced, and N% was within the normal range (Supplementary Figure 1). The quality of the sequence data met the requirements for subsequent analysis (Supplementary Figure 2). The percentage of bases with quality values ≥ 20 (Q20) exceeded 96% for each sample, and the base error rates of the WT and fliK-RNAi strain-infected groups were < 0.1% (Supplementary Figure 3). The reproducibility and principal component analysis (PCA) results of the three biological replicates were satisfactory (Supplementary Figure 4 & Supplementary Figure 5).
The DEGs were analyzed using the online Majorbio Cloud Platform (http://www.majorbio.com).
Spleen transcriptome data from E. coioides infected with the fliK-RNAi or WT strain were compared (hereafter referred to as the first transcriptome), and changes in expression level that met certain criteria (FDR < 0.05 and |log2FC| ≥1) were deemed statistically significant. Compared with the transcriptome of the WT strain-infected E. coioides spleen, 446 and 917 DEGs were obtained from the fliK-RNAi strain-infected E. coioides at 1 and 2 dpi, respectively. At 3 and 5 dpi, 76 208 DEGs (10 653 up-regulated and 65 555 down-regulated) and 53 102 DEGs (8 872 up-regulated and 44 230 down-regulated) were identified, respectively (Figure 4A). According to the log2FC, the top 30 up-regulated and down-regulated DEGs in the fliK-RNAi- and WT-infected groups (at 3 dpi) were selected, as shown in Figure 4B. Among the up-regulated DEGs, DN7446_c0_g1 (log2FC = 11.7) showed the greatest fold-change. Among the down-regulated DEGs, DN21377_c0_g1 (log2FC = -13.69) showed the greatest fold-change. The top 30 up-regulated and down-regulated DEGs in the fliK-RNAi- and WT-infected groups (at 5 dpi) were selected, as shown in Figure 4C. Among the up-regulated DEGs, DN14867_c1_g1 (log2FC = 11.78) showed the greatest fold-change. Among the down-regulated DEGs, DN21377_c0_g1 (log2FC = -16.91) showed the greatest fold-change.
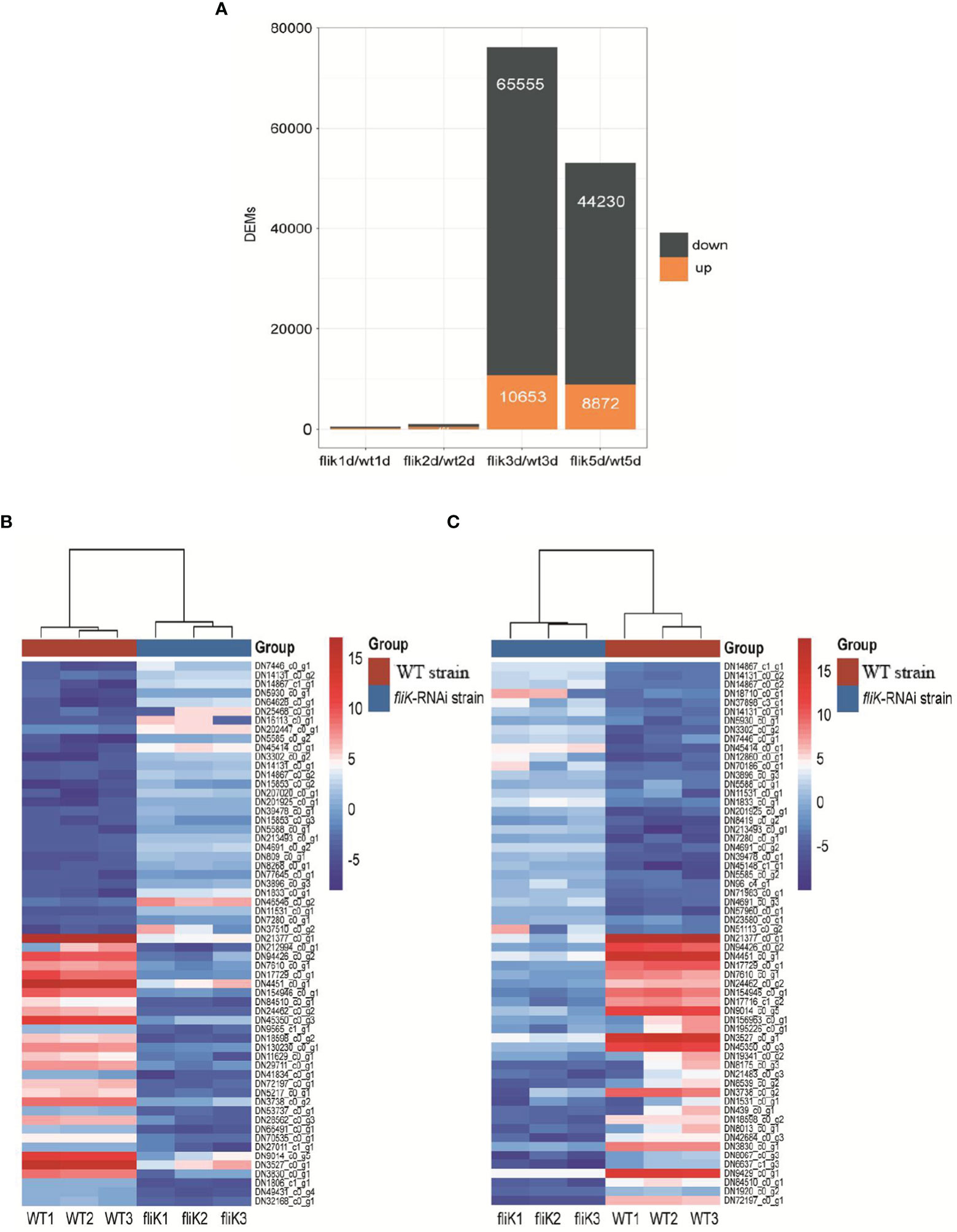
Figure 4 Comparative transcriptome analysis of E. coioides spleens. (A) Statistics of comparative transcriptome DEGs (fliK-RNAi and WT strain infection groups). (B) Heat map of top 30 up-regulated and 30 down-regulated mRNAs from transcriptome of fliK-RNAi and WT strain infection groups (3d). (C) Heat map of top 30 up-regulated and 30 down-regulated mRNAs from transcriptome of fliK-RNAi and WT strain infection groups (5 dpi).
Seven up-regulated genes and seven down-regulated genes were selected from the host transcriptomes, respectively, and qRT-PCR was performed to verify the reliability of the transcriptome data, which were found to be consistent (Supplementary Figure 6).
A total of 52 and 51 annotated GO terms were obtained at 3 and 5 dpi, respectively, based on the transcriptomes of E. coioides spleens infected with the fliK-RNAi and WT strains of P. plecoglossicida. Figure 5A shows the top 10 annotated GO terms containing the greatest number of DEGs, with “Binding” showing the highest enrichment. Figure 5B shows the number of DEGs annotated with GO terms in the “Immune System Process” between different comparison pairs. The fliK/WT(3d) and fliK/WT(5d) pair showed the most shared GO terms (Figure 5B). The distribution of DEGs in “Immune System Process” was analyzed, with DN1276_c1_g1 (log2FC = 9.26) found to be the most up-regulated and CD74 (log2FC = -9.5) found to be the most down-regulated mRNA (Figure 5C).
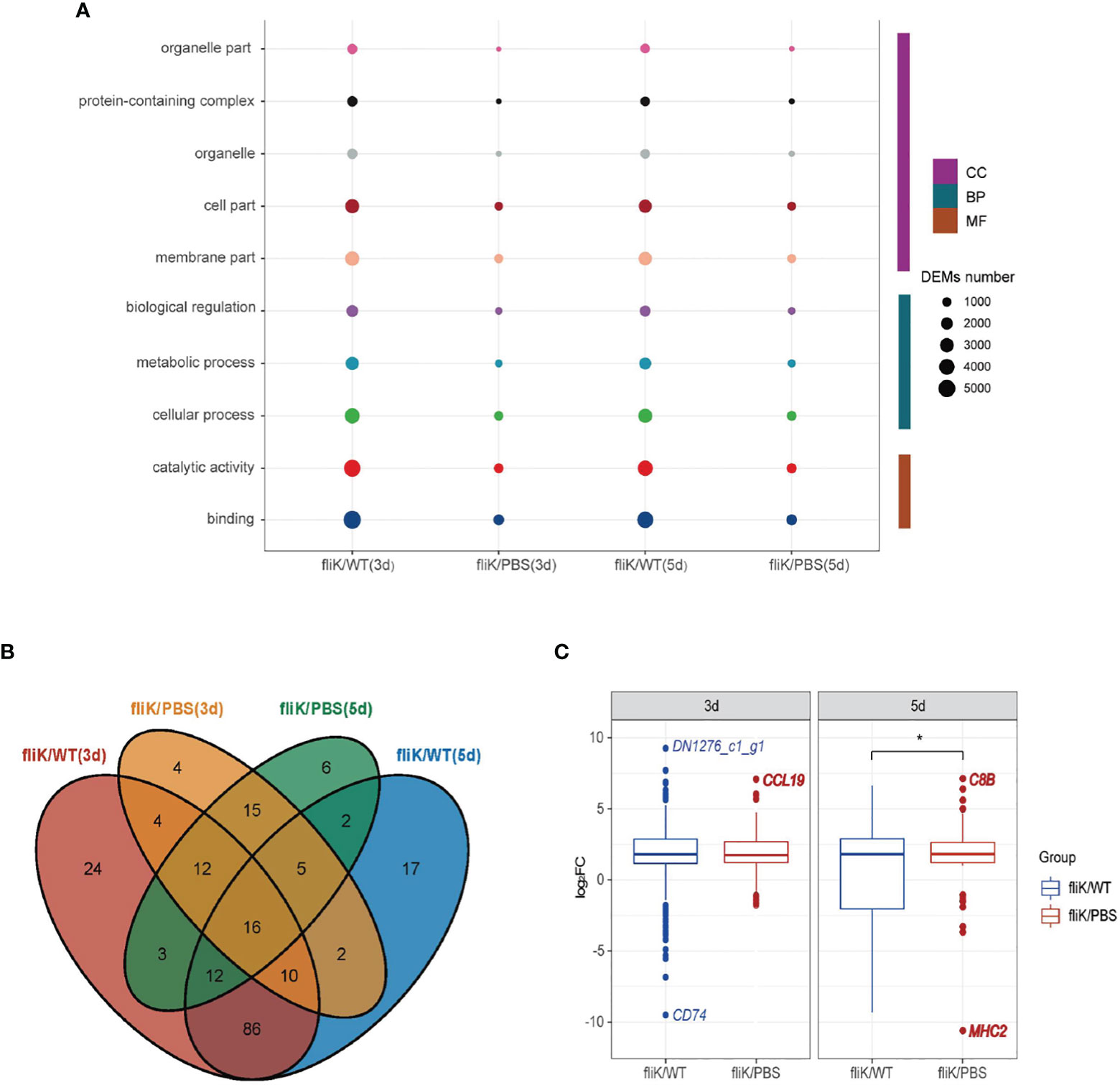
Figure 5 GO annotation of comparative transcriptome data of fliK-RNAi and WT strain infection groups (3 and 5 dpi). (A) Top 10 GO terms with number of annotated DEGs. (B) Number of DEGs annotated to “Immune System Process” in different comparison groups. (C) Distribution of expression level of DEGs annotated to “Immune System Process” (upper, middle, and lower horizontal lines in box represent upper quartile, median, and lower quartile, respectively. Upper- and lower-line segments represent maximum and minimum values of data, respectively; points outside boxes represent outliers. Blue indicates most up-regulated mRNA in fliK-RNAi and WT strain infection groups, red indicates most up-regulated mRNA in fliK-RNAi strain infection and PBS injection groups). *P < 0.05.
For the transcriptome data of the fliK-RNAi- and WT-infected groups (at 3 dpi), DEGs were enriched in 344 KEGG pathways, including 21 immune system-related pathways (Figure 6A). The DEGs enriched in the immune system-related pathways accounted for 23% of total DEGs. Among these pathways, the NOD-like receptor signaling pathway (ko04621) was enriched in the most DEGs (Figure 6A). Among the 231 DEGs enriched in the immune system-related pathways, htpG (log2FC = 6.89) was the most up-regulated and Ripk3 (log2FC = -7.94) was the most down-regulated (Figure 6B). For the transcriptome data of the fliK-RNAi- and WT-infected groups (at 5 dpi), DEGs were enriched in 345 KEGG pathways, including 21 related to the immune system. The DEGs enriched in the immune system-related pathways accounted for 24.96% of total DEGs. Among these pathways, the natural killer (NK) cell-mediated cytotoxicity (ko04650), platelet activation (ko04611), and Th17 cell differentiation (ko04659) signaling pathways were all significantly enriched (Figure 6C). DEM expression levels in these three signaling pathways were analyzed, and the most up-regulated DEGs in each pathway are shown in Figure 6D. In addition, up-regulation of 2–32 times enriched the most number of DEGs, and down-regulation of 2–32 times enriched the second large number of DEGs.
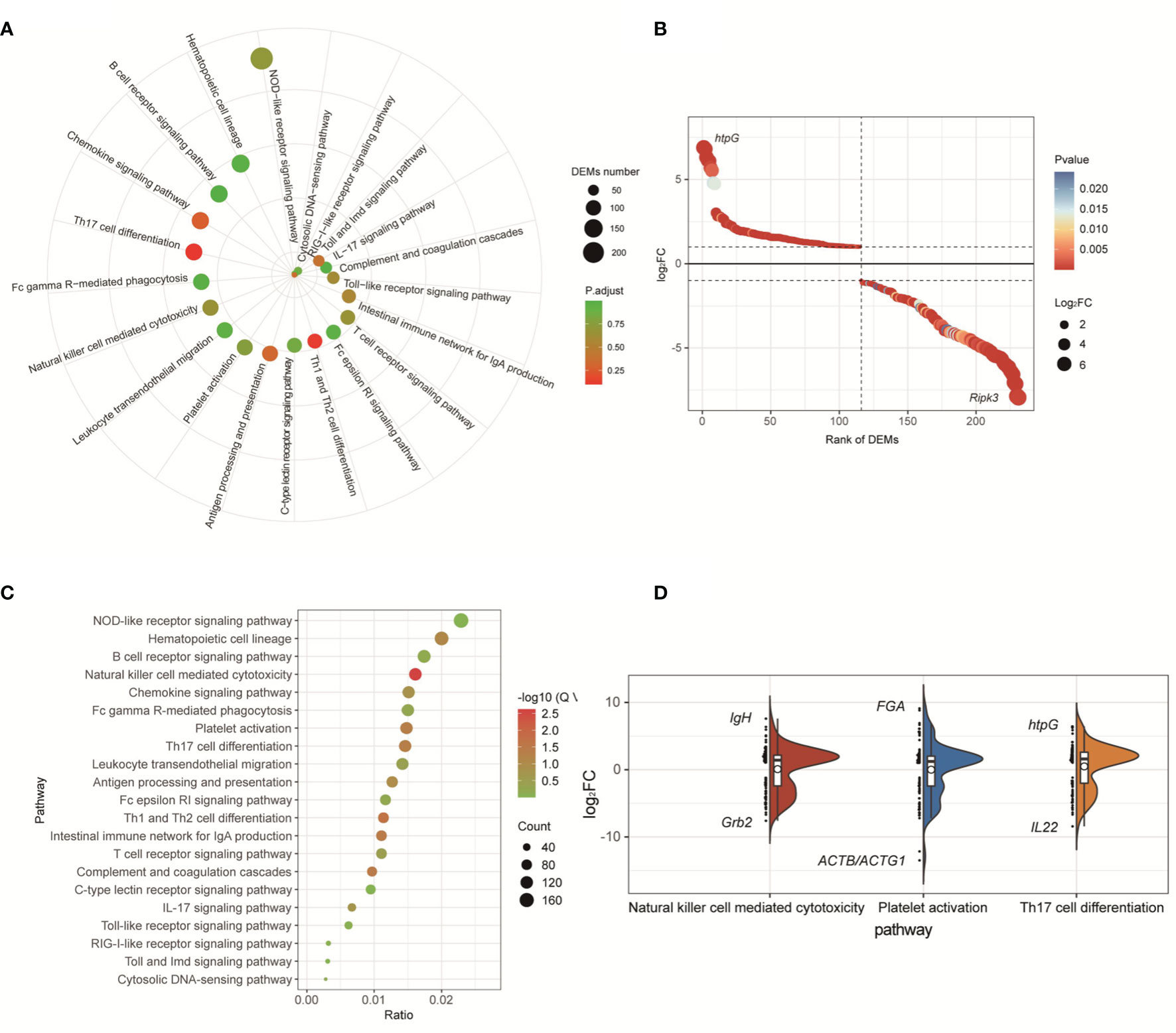
Figure 6 KEGG enrichment analysis of comparative transcriptome of fliK-RNAi and WT strain infection groups (3 and 5 dpi). (A) Signaling pathways related to immune system in comparative transcriptomes of fliK-RNAi and WT strain infection groups (3 dpi). (B) Expression level of DEGs in NOD-like receptor signaling pathway. (C) Signaling pathways related to immune system in comparative transcriptomes of fliK-RNAi and WT strain infection groups (5 dpi). (D) Expression level of DEGs in NK cell-mediated cytotoxicity, platelet activation, and Th17 cell differentiation (black horizontal lines in boxes show median expression level of each mRNA. Upper and lower frames in white box represent upper and lower quartiles in dataset, respectively. Left side reflects distribution of data, right side reflects data density, black mark indicates most up-regulated and down-regulated mRNA expression level in each pathway).
The DEGs showed gene expression in NK cell-mediated cytotoxicity. Analysis indicated that mRNAs of the oncogene Vav, related to A and C kinase (Rac), mitogen-activated protein kinase kinase (MEK1/2), perforin, factor-related apoptosis ligand (FASL), and its receptor FAS were up-regulated. While the mRNAs of phosphoinositide 3-kinase (PI3K), oncogene Raf, extracellular regulated protein kinases (ERK1/2), and tumor necrosis factor-alpha (TNF-α) were down-regulated (Figure 7).
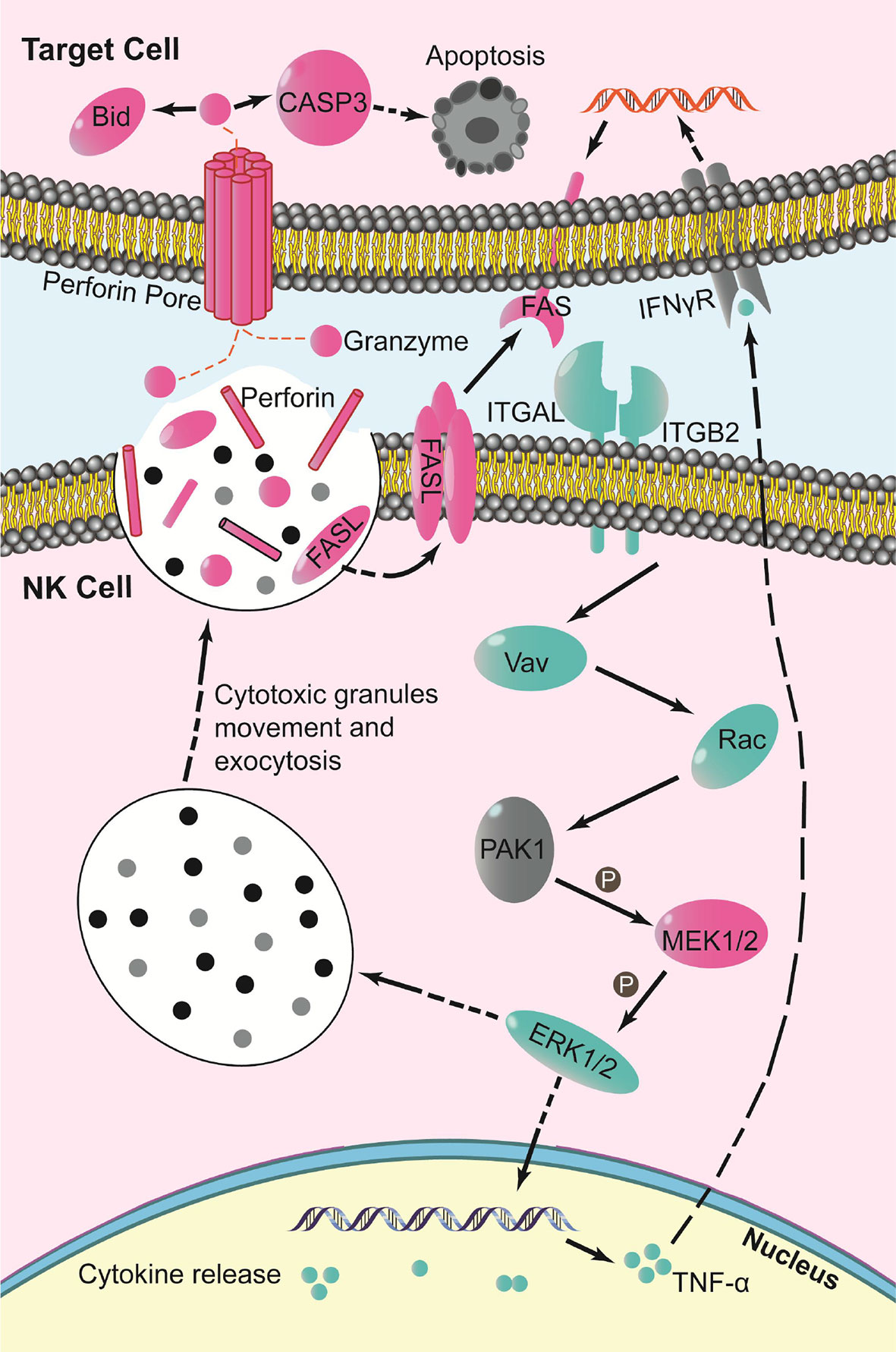
Figure 7 Schematic of key changes in NK cell-mediated cytotoxicity pathway (blue indicates down-regulation, red indicates up-regulation, gray indicates no significant change, P indicates phosphorylation).
Virulence genes play important roles in the process of pathogenic host infection (Hu et al., 2021). In recent years, several virulence-related genes have been identified and implicated in aquatic pathogen infection (He et al., 2021), including virulence genes of P. plecoglossicida. Virulence genes can affect the host immune response and are critical to pathogen-host interactions (Luo et al., 2020). At present, however, no studies have reported on the effects of the fliK gene in P. plecoglossicida pathogenicity or on host immune response.
The application of RNAi technology can reduce the expression levels of genes in different aquatic pathogens (Jiao et al., 2021) and is achieved by shRNA silencing of genes (Zhang et al., 2019). In the present study, four shRNAs were designed to silence the fliK gene. Here, fliK-shRNA-406 showed the highest silencing efficiency (97.5%), superior to that achieved for other P. plecoglossicida genes (Luo et al., 2019). Gene silencing stability is crucial for studying the influence of genes on bacterial virulence (Luo et al., 2020). Our results showed that fliK gene expression in the fliK-RNAi strain of P. plecoglossicida was always lower than that in the WT strain during infection, indicating that fliK was persistently silenced over the whole infection process. These results indicated that the RNAi technique was reliable in this study and provided the foundation for subsequent research.
In the WT and fliK-RNAi strains of P. plecoglossicida, fliK gene expression was higher in the infected E. coioides than in the in vitro cultures, suggesting that fliK may play a role in infection. Infection with the fliK-RNAi strain (5 × 104 CFU/fish) resulted in only 5% mortality in the infected E. coioides, whereas infection with the WT strain (same dose) resulted in 100% mortality at 8 dpi. Thus, silencing of the fliK gene resulted in a marked decrease in the pathogenicity of P. plecoglossicida, further suggesting that fliK is a virulence gene, as confirmed by the spleen symptoms, pathogen loads, and fliK gene expression levels. Several genes are associated with P. plecoglossicida virulence, the silencing of which results in lower or no mortality in infected fish (Tang et al., 2019b; Wang et al., 2019; Liu et al., 2020; Xin et al., 2020).
In the current study, silencing of the fliK gene resulted in a decrease in the growth rate and final concentration of P. plecoglossicida. However, whether a decrease in the growth rate is associated with a decrease in virulence of the fliK-RNAi strain remains to be verified. Motility, chemotaxis, adhesion, and biofilm formation are associated with bacterial virulence and are controlled by flagella (Luo et al., 2016). In the present study, these factors were all significantly lower in the fliK-RNAi strain than in the WT strain, indicating that fliK is associated with the regulation of motility, chemotaxis, adhesion, and biofilm formation in P. plecoglossicida. These results further suggest that fliK is a virulence gene in P. plecoglossicida.
Pathogen invasion elicits an immune response in the host and can impact the host’s transcriptome (Tang et al., 2020; Thompson et al., 2021). And several P. plecoglossicida genes has been known to affect the host transcriptomic response (Luo et al., 2019; Wang et al., 2020). In this study, we found the fliK affected the transcriptomic response of E. coioides based on the DEGs between the different P. plecoglossicida strain-infected fish, especially at 3 and 5 dpi. These DEGs mainly enriched in the immune-related GO terms and 21 KEGG pathways, including the natural killer (NK) cell-mediated cytotoxicity, platelet activation, and Th17 cell differentiation signaling pathways. NK cell-mediated cytotoxicity plays an important role in the host immune response to infection (Ma et al., 2015; Pan et al., 2015). In this pathway, ITGAL binds to ITGB2 and is down-regulated, which may reduce protein adhesion between leukocytes. The inflammatory response relating with MAPK signaling pathway (Pan et al., 2017; Habtetsion et al., 2018; Yang et al., 2019; Sharif et al., 2020) also was activated in the present study. Compared with the WT strain-infected E.coioides, the weakened autoimmune response of that infected with fliK-RNAi P.plecoglossicida seems reflect in the down-regulated secretion of TNF-α produced by NK cells. In addition, during the process of toxic granule exocytosis in NK cells to target cells: Perforin/Granzyme mediated apoptosis pathway, which plays a role in removing microorganisms and immune surveillance (Woodsworth et al., 2017; Kotov et al., 2018; Pitabut et al., 2020), FAS trimers are formed on the surface of target cells, clustering the death domains of cytoplasm, and ultimately leading to the death of target cells (Mao et al., 2021). Platelet activation signaling plays a critical role in platelet function in hemostasis and thrombosis (Estevez and Du, 2017). Platelet activation is induced by collagen or soluble platelet agonists that bind to G-protein-coupled receptors, leading to activation of platelet adhesion receptors (Estevez and Du, 2017). Various platelet adhesion receptors are critical for mediating immune or inflammatory responses in leukocytes and in the coupling between thrombosis and inflammation (Estevez and Du, 2017). Th17 cells are a new subset of IL-17-producing T helper cells that mediate immune defense against fungi and extracellular bacteria and tissue inflammation in autoimmune diseases (Huang et al., 2012). TH17 cell differentiation can be initiated by transforming growth factor-beta (TGF-b) in the presence of the inflammatory cytokines IL-6 or IL-21 and further reinforced by IL-23. TH17 cells produce several signature cytokines, e.g., IL-17, IL-17F, and IL-22, which trigger various inflammatory responses, such as neutrophilia, tissue remodeling, and antimicrobial protein production (Korn et al., 2009). Consistent with the relative pathogen loads in fish, up-regulation of the NK cell-mediated cytotoxicity, platelet activation, and Th17 cell differentiation signaling pathways in E. coioides infected with the fliK-RNAi strain compared with that infected with WT strain indicated an enhanced immunity and ability to remove microorganisms.
Our study showed that fliK is an important virulence gene of P. plecoglossicida and contributes to the pathogenicity of P. plecoglossicida in E. coioides. The silencing of fliK weakens P. plecoglossicida motility, chemotaxis, adhesion, and biofilm formation, as well as its pathogenicity. Gene silencing also significantly affects the immune response of E. coioides to P. plecoglossicida infection, with significant enrichment in the NK cell-mediated cytotoxicity pathway.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/genbank/, SRP315483 https://www.ncbi.nlm.nih.gov/genbank/, SRP315268.
The animal study was reviewed and approved by Research Board of the Ethics Committee of Jimei University under permit number JMULAC201159.
Conceptualization: JNZ, QY. Data curation: LH, YQ, JNZ. Formal analysis: LZ, JLZ, BH. Funding acquisition: QY. Investigation: ZL, BY, LZ. Methodology: LH, YQ, JLZ. Project administration: QY. Supervision: QY. Writing - original draft: ZL, BY. Writing - review and editing: QY. All authors contributed to the article and approved the submitted version.
This work was supported by the Natural Science Foundation of Fujian Province (No. 2021J01828 and 2019J06020), National Natural Science Foundation of China (No. 31972836), and Open Fund of Fujian Province Key Laboratory of Special Aquatic Formula Feed (No. TMKJZ2101).
Authors JNZ, JLZ, and BH are employed by Fujian Tianma Technology Company Limited.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2022.879333/full#supplementary-material
Attieh Z., Mouawad C., Rejasse A., Jehanno I., Perchat S., Hegna I. K., et al. (2020). The fliK Gene Is Required for the Resistance of Bacillus Thuringiensis to Antimicrobial Peptides and Virulence in Drosophila Melanogaster. Front. Microbiol. 11. doi: 10.3389/fmicb.2020.611220
Chaban B., Hughes H. V., Beeby M. (2015). The Flagellum in Bacterial Pathogens: For Motility and a Whole Lot More. Semin. Cell Dev. Biol. 46, 91–103. doi: 10.1016/j.semcdb.2015.10.032
Chen Q., Yan Q., Wang K., Zou W., Zhuang Z., Wang X. (2008). Portal of Entry for Pathogenic Vibrio alginolyticus into Pseudosciaene crocea, and Characteristic of Bacterial Adhesion to the Mucus. Dis. Aquat. Org. 80, 181–188. doi: 10.3354/dao01933
Choi K. H., Schweizer K. H. (2006). Mini-Tn7 Insertion in Bacteria With Single Atttn7 Sites: Example Pseudomonas Aeruginosa. Nat. Protoc. 1 (1), 153–161. doi: 10.1038/nprot.2006.24
Darsigny M., Babeu J. P., Seidman E. G., Gendron F. P., Levy E., Carrier J., et al. (2010). Hepatocyte Nuclear Factor-4alpha Promotes Gut Neoplasia in Mice and Protects Against the Production of Reactive Oxygen Species. Cancer Res. 70 (22), 9423–9433. doi: 10.1158/0008-5472.CAN-10-1697
Defoirdt T. (2016). Implications of Ecological Niche Differentiation in Marine Bacteria for Microbial Management in Aquaculture to Prevent Bacterial Disease. PloS Pathog. 12 (11), e1005843. doi: 10.1371/journal.ppat.1005843
Duan Q., Zhou M., Zhu L., Zhu G. (2013). Flagella and Bacterial Pathogenicity. J. Basic Microbiol. 53 (1), 1–8. doi: 10.1002/jobm.201100335
Estevez B., Du X. (2017). New Concepts and Mechanisms of Platelet Activation Signaling. Physiology 32, 162–177. doi: 10.1152/physiol.00020.2016
Habtetsion T., Ding Z. C., Pi W., Li T., Lu C., Chen T., et al. (2018). Alteration of Tumor Metabolism by CD4+ T Cells Leads to TNF-α-Dependent Intensification of Oxidative Stress and Tumor Cell Death. Cell Metab. 28 (2), 228–242.e6. doi: 10.1016/j.cmet.2018.05.012
He L., Wang L., Zhao L. M., Zhuang Z. X., Wang X. R., Huang H. B., et al. (2021). Integration of RNA-Seq and RNAi Reveals the Contribution of znuA Gene to the Pathogenicity of Pseudomonas Plecoglossicida and to the Immune Response of Epinephelus Coioides. J. Fish Dis. 44 (11), 1831–1841. doi: 10.1111/jfd.13502
Huang G., Wang Y., Chi H. (2012). Regulation of TH17 Cell Differentiation by Innate Immune Signals. Cell. Molec. Immunol. 9, 287–295. doi: 10.1038/cmi.2012.10
Huang L. X., Zuo Y. F., Jiang Q. L., Su Y. Q., Qin Y. X., Xu X. J., et al. (2019). A Metabolomic Investigation Into the Temperature-Dependent Virulence of Pseudomonas Plecoglossicida From Large Yellow Croaker (Pseudosciaena Crocea). J. Fish Dis. 42 (3), 431–446. doi: 10.1111/jfd.12957
Hu L. F., Zhao L. M., Zhuang Z. X., Wang X. R., Fu Q., Huang H. B., et al. (2021). The Effect of tonB Gene on the Virulence of Pseudomonas Plecoglossicida and the Immune Response of Epinephelus Coioides. Front. Microbiol. 12. doi: 10.3389/fmicb.2021.720967
Izumi S., Yamamoto M., Suzuki K., Aranishiet F. (2007). Identification and Detection of Pseudomonas Plecoglossicida Isolates With PCR Primers Targeting the gyrB Region. J. Fish Dis. 30 (7), 391–397. doi: 10.1111/j.1365-2761.2007.00820.x
Jha B. K., Pragash M. G., Cletus J., Raman G., Sakthivel N. (2009). Simultaneous Phosphate Solubilization Potential and Antifungal Activity of New Fluorescent Pseudomonad Strains, Pseudomonas Aeruginosa, P. Plecoglossicida and P. Mosselii. World J. Microbiol. Biotechnol. 25, 573–581. doi: 10.1007/s11274-008-9925-x
Jiao J. P., Zhao L. M., Huang L. X., Qin Y. X., Su Y. Q., Zheng W. Q., et al. (2021). The Contributions of fliG Gene to the Pathogenicity of Pseudomonas Plecoglossicida and Pathogen-Host Interactions With Epinephelus Coioides. Fish Shellfish Immunol. 119, 238–248. doi: 10.1016/j.fsi.2021.09.032
John E. M., Rebello S., Asok A. K., Jisha M. S. (2015). Pseudomonas Plecoglossicida S5, a Novel Nonpathogenic Isolate for Sodium Dodecyl Sulfate Degradation. Environ. Chem. Lett. 13, 117–123. doi: 10.1007/s10311-015-0493-7
Joós G., Jákim J., Kiss B., Szamosi R., Papp T., Felszeghy S., et al. (2017). Involvement of Adenosine A3 Receptors in the Chemotactic Navigation of Macrophages Towards Apoptotic Cells. Immunol. Lett. 183, 62–72. doi: 10.1016/j.imlet.2017.02.002
Kanehisa M., Furumichi M., Tanabe M., Sato Y., Morishima K. (2017). KEGG: New Perspectives on Genomes, Pathways, Diseases and Drugs. Nucleic Acids Res. 45 (D1), D353–D361. doi: 10.1093/nar/gkw1092
Kinoshita M., Tanaka S., Inoue Y., Namba K., Aizawa S. I., Minamino T. (2020). The Flexible Linker of the Secreted FliK Ruler is Required for Export Switching of the Flagellar Protein Export Apparatus. Sci. Rep. 10 (1), 838. doi: 10.1038/s41598-020-57782-5
Klopfenstein D. V., Zhang L. S., Pedersen B. S., Ramírez F., Vesztrocy A. W., Naldi A., et al. (2018). GOATOOLS: A Python Library for Gene Ontology Analyses. Sci. Rep. 8 (1), 10872. doi: 10.1038/s41598-018-28948-z
Korn T., Bettelli E., Oukka M., Kuchroo V. K. (2009). IL-17 and Th17 Cells. Annu. Rev. Immunol. 27, 485–517. doi: 10.1146/annurev.immunol.021908.132710
Kotov D. I., Kotov J. A., Goldberg M. F., Jenkins M. K. (2018). Many Th Cell Subsets Have Fas Ligand-Dependent Cytotoxic Potential. J. Immunol. 200 (6), 2004–2012. doi: 10.4049/jimmunol.1700420
Lee S., Park Y., Kim S. (2017). MIDAS: Mining Differentially Activated Subpaths of KEGG Pathways From Multi-Class RNA-Seq Data. Methods 124, 13–24. doi: 10.1016/j.ymeth.2017.05.026
Liu Z. X., Zhao L. M., Huang L. X., Qin Y. X., Zhang J. N., Zhang J. L., et al. (2020). Integration of RNA-Seq and RNAi Provides a Novel Insight Into the Immune Responses of Epinephelus Coioides to the impB Gene of Pseudomonas plecoglossicida. Fish Shellfish Immunol. 105, 135–143. doi: 10.1016/j.fsi.2020.06.023
Li C. W., Wang S. L., Ren Q. L., He T. L., Chen X. H. (2020). An Outbreak of Visceral White Nodules Disease Caused by Pseudomonas Plecoglossicida at a Water Temperature of 12°C in Cultured Large Yellow Croaker (Larimichthys Crocea) in China. J. Fish Dis. 43 (11), 1353–1361. doi: 10.1111/jfd.13206
Loraine A. E., Blakley I. C., Jagadeesan S., Harper J., Miller G., Firon N. (2015). Analysis and Visualization of RNAseq Expression Data Using RStudio, Bioconductor, and Integrated Genome Browser. Meth. Mol. Biol. 1284, 481–501. doi: 10.1007/978-1-4939-2444-8_24
Luo G., Huang L., Su Y., Qin Y., Xu X., Zhao L., et al. (2016). Flra, Flrb, and flrC Regulate Adhesion by Controlling the Expression of Critical Virulence Genes in Vibrio Alginolyticus. Emerg. Microb. Infect. 5, e85. doi: 10.1038/emi.2016.82
Luo G., Sun Y. J., Huang L. X., Su Y. Q., Zhao L. M., Qin Y. X., et al. (2020). Time-Resolved Dual RNA-Seq of Tissue Uncovers Pseudomonas Plecoglossicida Key Virulence Genes in Host-Pathogen Interaction With Epinephelus Coioides. Environ. Microbiol. 22 (2), 677–693. doi: 10.1111/1462-2920.14884
Luo G., Zhao L., Xu X., Qin Y., Huang L., Su Y., et al. (2019). Integrated Dual RNA-Seq and Dual iTRAQ of Infected Tissue Reveals the Functions of a Diguanylate Cyclase Gene of Pseudomonas Plecoglossicida in Host-Pathogen Interactions With Epinephelus Coioides. Fish Shellfish Immunol. 95, 481–490. doi: 10.1016/j.fsi.2019.11.008
Mao M. G., Xu J., Liu R. T., Ye L. (2021). Fas/FasL of Pacific Cod Mediated Apoptosis. Dev. Comp. Immunol. 119, 104022. doi: 10.1016/j.dci.2021.104022
Marazzi L., Kohlib P., Eastman D. (2020). Transcriptome Dataset for RNA-Seq Analysis of Axolotl Embryonic Oropharyngeal Endoderm Explants. Data Brief 32, 106126. doi: 10.1016/j.dib.2020.106126
Ma J. Y., Zhao D., Wu Y. Q., Xu C., Zhang F. Q. (2015). Cyclic Stretch Induced Gene Expression of Extracellular Matrix and Adhesion Molecules in Human Periodontal Ligament Cells. Arch. Oral. Biol. 60 (3), 447–455. doi: 10.1016/j.archoralbio.2014.11.019
Nishimori E., Kita-Tsukamoto K., Wakabayashi H. (2000). Pseudomonas Plecoglossicida Sp. Nov., the Causative Agent of Bacterial Haemorrhagic Ascites of Ayu, Plecoglossus Altivelis. Intern. J. Syst. Evol. Microbiol. 50 (1), 83–89. doi: 10.1099/00207713-50-1-83
Pan D., Amison R. T., Riffo-Vasquez Y., Spina D., Cleary S. J., Wakelam M. J., et al. (2015). P-Rex and Vav Rac-GEFs in Platelets Control Leukocyte Recruitment to Sites of Inflammation. Blood 125 (7), 1146–1158. doi: 10.1182/blood-2014-07-591040
Pan R. Q., Ruvolo V., Mu H., Leverson J. D., Nichols G., Reed J. C., et al. (2017). Synthetic Lethality of Combined Bcl-2 Inhibition and P53 Activation in AML: Mechanisms and Superior Antileukemic Efficacy. Cancer Cell 32 (6), 748–760.e6. doi: 10.1016/j.ccell.2017.11.003
Persat A., Nadell C. D., Kim M. K., Ingremeau F., Siryaporn A., Drescher K., et al. (2015). The Mechanical World of Bacteria. Cell 161 (5), 988–997. doi: 10.1016/j.cell.2015.05.005
Pitabut N., Dhepakson P., Sakurada S., Keicho N., Khusmith S. (2020). Coordinated In Vitro Release of Granulysin, Perforin and IFN-γ in TB and HIV/TB Co-Infection Associated With Clinical Outcomes Before and After Anti-TB Treatment. Pathogens 9 (8), 655. doi: 10.3390/pathogens9080655
Rangan P., Furtado A., Henry R., Gaikwad A. (2021). “Development of Transcriptome Analysis Methods,” in Compreh. Foodom. Comprehensive Foodo. Amsterdam, Netherlands: Elsevier. pp. 462–471. doi: 10.1016/B978-0-08-100596-5.22752-2
Sampedro I., Parales R. E., Krell T., Hill J. E. (2015). Pseudomonas Chemotaxis. FEMS Microb. Rev. 39 (1), 17–46. doi: 10.1111/1574-6976.12081
Seppey M., Manni M., Zdobnov E. M. (2019). BUSCO: Assessing Genome Assembly and Annotation Completeness. Meth. Molec. Biol. 1962, 227–245. doi: 10.1007/978-1-4939-9173-0_14
Sharif P. M., Jabbaria P., Razia S., Keshavarz M., Rezaeiet N. (2020). Importance of TNF-Alpha and its Alterations in the Development of Cancers. Cytokine 130, 155066. doi: 10.1016/j.cyto.2020.155066
Sherwood T. A., Main K. L., Wetzel D. L. (2019). De Novo Assembly and Transcriptome Dataset of Liver, Testis and Head Kidney From Red Drum (Sciaenops Ocellatus). Data Brief 22, 934–939. doi: 10.1016/j.dib.2019.01.011
Tang R., Luo G., Zhao L., Huang L., Qin Y., Xu X., et al. (2019a). The Effect of a LysR-Type Transcriptional Regulator Gene of Pseudomonas Plecoglossicida on the Immune Responses of Epinephelus Coioides. Fish Shellfish Immunol. 89, 420–427. doi: 10.1016/j.fsi.2019.03.051
Tang Y., Sun Y., Zhao L., Xu X., Huang L., Qin Y., et al. (2019b). Mechanistic Insight Into the Roles of Pseudomonas Plecoglossicida clpV Gene in Host-Pathogen Interactions With Larimichthys Crocea by Dual RNA-Seq. Fish Shellfish Immunol. 93, 344–353. doi: 10.1016/j.fsi.2019.07.066
Tang Y., Xin G., Zhao L. M., Huang L. X., Qin Y. X., Su Y. Q., et al. (2020). Novel Insights Into Host-Pathogen Interactions of Large Yellow Croakers (Larimichthys Crocea) and Pathogenic Bacterium Pseudomonas Plecoglossicida Using Time-Resolved Dual RNA-Seq of Infected Spleens. Zool. Res. 41 (3), 314–327. doi: 10.24272/j.issn.2095-8137.2020.035
Thompson D., Watt J. A., Brissette C. A. (2021). Host Transcriptome Response to Borrelia Burgdorferi Sensu Lato. Ticks Tick Borne Dis. 12 (2), 101638. doi: 10.1016/j.ttbdis.2020.101638
Wang L. Y., Liu Z. X., Zhao L. M., Huang L. X., Qin Y. X., Su Y. Q., et al. (2020). Dual RNA-Seq Provides Novel Insight Into the Roles of dksA From Pseudomonas Plecoglossicida in Pathogen-Host Interactions With Large Yellow Croakers (Larimichthys Crocea). Zool. Res. 41 (4), 410–422. doi: 10.24272/j.issn.2095-8137.2020.048
Wang L. Y., Sun Y. J., Zhao L. M., Xu X. J., Huang L. X., Qin Y. X., et al. (2019). Dual RNA-Seq Uncovers the Immune Response of Larimichthys Crocea to the secY Gene of Pseudomonas Plecoglossicida From the Perspective of Host-Pathogen Interactions. Fish Shellfish Immunol. 93, 949–957. doi: 10.1016/j.fsi.2019.08.040
Waters R. C., O'Toole P. W., Ryan K. A. (2007). The FliK Protein and Flagellar Hook-Length Control. Prot. Sci. 16 (5), 769–780. doi: 10.1110/ps.072785407
Weng C. H. (2016). Identifying Association Rules of Specific Later-Marketed Products. Appl. Soft Comput. 38, 518–529. doi: 10.1016/j.asoc.2015.09.047
Woodsworth D. J., Dreolini L., Abraham L., Holt R. A. (2017). Targeted Cell-To-Cell Delivery of Protein Payloads via the Granzyme-Perforin Pathway. Molec. Ther. Meth. Clin. Dev. 7, 132–145. doi: 10.1016/j.omtm.2017.10.003
Wu Y. F., Tian Y., Tan J., Zhao S., Zhou W. Z., Luo R., et al. (2021). Differential Metabolism of Juvenile Hormone III Between Diapause and non-Diapause of Aspongopus Chinensis Dallas (Hemiptera: Dinidoridae) Revealed by Transcriptome Sequencing. J. Asia Pac. Entomol. 24 (2), 199–204. doi: 10.1016/j.aspen.2021.02.009
Xin G., Wang F., Zhao L. M., Qin Y. X., Huang L. X., Yan Q. P. (2020). Integration of RNA-Seq and RNAi Provides a Novel Insight Into the Effect of pvdE Gene to the Pathogenic of Pseudomonas Plecoglossicida and on the Immune Responses of Orange-Spotted Grouper (Epinephelus Coioides). Aquaculture 529, 735695. doi: 10.1016/j.aquaculture.2020.735695
Yang Y., Adeola A. C., Xie H. B., Zhang Y. P. (2018). Genomic and Transcriptomic Analyses Reveal Selection of Genes for Puberty in Bama Xiang Pigs. Zool. Res. 39 (6), 424–430. doi: 10.24272/j.issn.2095-8137
Yang X., Zheng Y. T., Rong W. (2019). Sevoflurane Induces Apoptosis and Inhibits the Growth and Motility of Colon Cancer In Vitro and In Vivo via Inactivating Ras/Raf/MEK/ERK Signaling. Life Sci. 239, 116916. doi: 10.1016/j.lfs.2019.116916
Zhang M. M., Qin Y. X., Huang L. X., Yan Q. P., Mao L. L., Xu X. J., et al. (2019). The Role of sodA and sodB in Aeromonas Hydrophila Resisting Oxidative Damage to Survive in Fish Macrophages and Escape for Further Infection. Fish Shellfish Immunol. 88, 489–495. doi: 10.1016/j.fsi.2019.03.021
Keywords: immune response, Epinephelus coioides, Pseudomonas plecoglossicida, fliK, RNA-seq
Citation: Liu Z, Yuan B, Zhao L, Huang L, Qin Y, Zhang J, Zhang J, Hu B and Yan Q (2022) Function of the fliK Gene in Pseudomonas plecoglossicida Pathogenicity and Epinephelus coioides’ Immune Response. Front. Mar. Sci. 9:879333. doi: 10.3389/fmars.2022.879333
Received: 19 February 2022; Accepted: 13 April 2022;
Published: 06 May 2022.
Edited by:
Sophia Letsiou, Agricultural University of Athens, GreeceReviewed by:
Minglan Guo, Chinese Academy of Sciences, ChinaCopyright © 2022 Liu, Yuan, Zhao, Huang, Qin, Zhang, Zhang, Hu and Yan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qingpi Yan, eWFucXBAam11LmVkdS5jbg==
†These authors share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.