- 1The Key Laboratory of Healthy Mariculture for the East China Sea, Ministry of Agriculture, Fisheries College, Jimei University, Xiamen, China
- 2Xiamen Key Laboratory for Feed Quality Testing and Safety Evaluation, Fisheries College, Jimei University, Xiamen, China
- 3Engineering Research Center of the Modern Technology for Eel Industry, Ministry of Education, Xiamen, China
A complex and dynamic microbiota exists in the intestine of fish and plays vital roles in host growth and health. However, the interactions between host originated beneficial bacteria/probiotics with gut microbiota are still largely unknown. The aim of the study is to investigate the impact of two host-derived probiotics, Lactococcus petauri LF3 and Bacillus siamensis LF4, on the intestinal microbiota of juvenile Japanese seabass (Lateolabrax japonicus). Fish were fed the control diet (C), L. petauri LF3 (LF3) and B. siamensis LF4 (LF4) supplemented diets for 6 weeks, and the intestinal microbial composition and function were evaluated by using high-throughput sequencing technology. The relative abundances of certain phyla changed significantly in the probiotic-supplemented groups, Fusobacteria and Proteobacteria decreased, while Firmicutes and Bacteroidetes increased apparently in the groups LF3 and LF4. The apparently increased relative abundances of intestinal possible beneficial Christensenellaceae_R-7_group and Lactobacillus were observed in the groups LF3 and LF4, especially in the group LF4. Compared with the control group (C), Chao1, Ace and Shannon indices enhanced remarkably in groups LF3 and LF4. Intestinal microbiota was determined to have more similarity and lower individual differences based on beta diversity analysis including PCoA, NMDS and UPGMA clustering tree in groups LF3 and LF4 compared with the control group (C). Additionally, Firmicutes as a significant biomarker emerged in the groups LF3 and LF4 compared with the control. The intestinal microbial functions (amino acid metabolism, carbohydrate metabolism, energy metabolism, membrane transport, etc.) did not alter among all groups based on level-2 KEGG pathways. In summary, host- derived probiotic L. petauri LF3 and B. siamensis LF4 shape the intestinal microbial composition, but not function in juvenile Japanese seabass (L. japonicus).
Introduction
The fish intestinal microbiota plays vital roles in host growth and health, and has co-evolved with the host in a mutually positive way (Wang et al., 2018). Intestinal microbiota is primarily affected by host phylogeny (Cornejo-Granados et al., 2017) and partly associated with growth stages, ambient condition and diet (Wang et al., 2018). With the rapid development of the modern aquaculture industry, intestinal microbial dysbiosis, immune deficiency and mass outbreak of diseases (e.g., enteritis and red-skin disease) are frequently appeared in fish because of the accumulation of opportunistic pathogens and/or other viruses under inteansive farming model (Chen et al., 2021). Previous studies have shown that maintaining the intestinal microbial homeostasis will greatly benefit the growth, immunity, and disease resistance in fish (Hoseinifar et al., 2018; Wang et al., 2019; Yang et al., 2019). Thus, exploring useful approaches to maintain the intestinal microbial homeostasis in fish will be conducive to improve the production, and effectively decrease the abuse of antibiotics in aquaculture.
Dietary supplementation of probiotics could modulate intestinal microbiota, improve production performance, immune response and disease resistance in many fish species, such as Asian seabass larvae (Lates calcarifer) (Masduki et al., 2020), grouper (Epinephelus coioides) (Yang et al., 2019), common carp (Cyprinus carpio L.) (Meng et al., 2021), tilapia (Oreochromis niloticus) (Li et al., 2019), snakehead (Channa argus) (Kong et al., 2021), and so on. Recently, probiotics as means of intestinal health maintain and diseases control in aquaculture have been reviewed extensively (Lazado et al., 2015; Hoseinifar et al., 2018; Doan et al., 2020; Teame et al., 2020; Zhang et al., 2020; Simon et al., 2021). The precise mechanism by which the positive performance of probiotics in aquaculture have must be more comprehensive and have not been well studied. So far as we know, one of the most recognized mechanisms is that probiotics can improve fish intestinal homeostasis and health via re-shaping the intestinal microbiota (Dawood et al., 2019; Doan et al., 2020; Simon et al., 2021; Guo et al., 2022). Nevertheless, information pertaining to interactions between host-derived probiotics and intestinal microbiota (including microbial composition and function) are still limited in teleost fish (Kong et al., 2021).
Japanese seabass, Lateolabrax japonicus, is one of the most economically important marine fish in China, and the annual production has reached 195, 246 tons in 2020 (China Fishery and Statistics Yearbook, 2021). Previous study has evaluated the application of five possible host-derived probiotic strains (Bacillus siamensis LF4, Lactococcus petaurid LF3, Micrococcus yunnanensis LD5, Micrococcus aloeverae LC3, Mitsuaria chitosanitabida LD3) in juvenile Japanese seabass (L. japonicus). The results demonstrated that probiotic L. petauri LF3 and B. siamensis LF4 could effectively improve growth performance, innate immunity, antioxidant activity and ammonia tolerance (Yang et al., 2021). To the best of our knowledge, limited information is still available about the impact of host-derived probiotics on intestinal microbiota in Japanese seabass. In this study, therefore, the intestinal microbial community and putative function of juvenile Japanese seabass in response to host-derived probiotic L. petauri LF3 and B. siamensis LF4 were analyzed and characterized by using high-throughput sequencing technology.
Materials and Methods
Probiotic Strains and Experimental Diets Preparation
Two host-derived probiotics L. petauri LF3 (GenBank number MZ618882) and B. siamensis LF4 (GenBank number MZ683191) isolated from the gut of Japanese seabass (L. japonicus) were selected and applied for this study. The control diet was formulated with ingredients as described by our previous study (Yang et al., 2021), For probiotic-supplemented diets preparation, two probiotic strains L. petauri LF3 and B. siamensis LF4 were inoculated into nutrient broth liquid, the culture was incubated at 30°C for 24 h. Bacterial cells were centrifuged, washed, and re-suspended in sterile phosphate buffered saline (PBS) before being surface spray to the control diet and air-dried for 24 h to obtain approximately 1 × 108 CFU g−1 of diets. All diets were stored at 4°C until use. To ensure the viability of dietary probiotics in the whole feeding trial (6 weeks), new diets were prepared every 3 weeks.
Experimental Fish and Sampling
Healthy Japanese seabass, L. japonicus, were transferred from a commercial farm to the Aquaculture Research Aquarium of Jimei University, China. After acclimation for two weeks, fish (average weight 23 g) were divided into three groups with triplicate (three tanks per group) and hand-fed control diet (C), L. petauri LF3 (LF3) and B. siamensis LF4 (LF4) supplemented diets to apparent satiation at 08:30 a.m. and 17:30 p.m. for 6 weeks. Fish husbandry management was in line with Yang et al. (2021).
At the end of the feeding trial, seabass was anaesthetized with buffered MS-222 (CAS:886-86-2) after 24 h of starvation. Subsequently, four seabass per tank were collected randomly under sterile environment, thus total twelve seabass of each group were collected. Intestines were separated and washed with sterile PBS (P7208B; Beijing LABLEAD, Inc., Beijing, China). Then, two intestines were pooled together as one sample to decrease individual variation, thus total six samples of each group were collected into 5.0 mL storage cryogenic tubes and immediately put into liquid nitrogen for subsequent DNA extraction.
Intestinal Microbiota DNA Extraction
Total DNA were extracted from each intestinal sample following the guideline of Bacterial Genomic DNA Extraction Kit (D2100; Beijing Solarbio Science & Technology Co., Ltd., China). The quality, integrity and concentration of extracted DNA were determined with 1% agarose gels and NanoDrop One (Thermo Fisher Scientific, USA) (Yang et al., 2019).
16S rRNA Amplification and High-Throughput Sequencing
Qualified DNA were amplificated and then performed high-throughput sequencing by Biomarker Technologies (Beijing, China). In brief, the V3–V4 region of 16S rRNA gene was amplificated using primers 338F (5’-ACTCCTACGGGAGGCAGCA-3) and 806R (5’-GGACTACHVGGGTWTCTAAT-3’). The steps of PCR amplification as follows: one pre-denaturation step at 98°C for 2 min, followed by 30 cycles at 98°C for 30 s, 50°C for 30 s and 72°C for 60 s, then an extension step at 72°C for 5 min, followed by a final step at 4°C for ∞. The amplified products were purified and quantified, and then high-throughput sequencing based on Illumina HiSeq 2500 platform (Biomarker Biotechnology Co., Ltd., Beijing, China). Small fragment libraries were established for 16sRNA sequencing using the paired-end sequencing method.
Analyzation of Microbial Composition and Diversity
Information of high-throughput sequencing were pre-treated via trimming the low-quality sequences and adaptors to obtain effective tags. The final data were clustered in same operational taxonomic units (OTUs) under 97% similarity. Normalization of the OTUs abundance data was treated based on taxonomic database of Silva and Unite.
For each sample, alpha diversity and beta diversity were determined to exploring the effects of probiotic diets. To assess the changes in microbial community structure, differentially abundant taxa were identified in all groups (C, LF3, and LF4). In order to estimate the response of intestinal microbiota to diets, the differences of microbial abundance between the control group (C) and the probiotic groups (LF3 and LF4) were compared by the linear discriminatory analysis (LDA) effect size (LEfSe) method. Microbial composition and diversity analysis were performed on the platform BMKCloud (www.biocloud.net).
Intestinal Microbiota Function Prediction
By comparison of high-throughput sequencing data in different groups, the functional general composition could be found, and then variations of function could be analyzed and predicted. Detailly, the OTUs-table were normalized, and then KEGG and COG family information corresponded with OUTs could be obtained, resulting in that each OTUs existed a corresponding Greengene ID. Finally, relative abundances of different function could be calculated according to information in KEGG database. All above steps were performed with PICRUSt 2 software.
Statistical Analysis
Alpha diversity parameters (Chao1, Ace, Shannon and Simpson indices) were analyzed by one-way analysis of variance followed by Duncan’s multiple-range comparison in SPSS (Version 26.0; IBM Corporation, USA). The significant difference was considered when P value < 0.05. All results were exhibited as mean ± standard error.
Results
Overview of High-Throughput Sequencing
A total of 1,437,027 PE Reads and 1,407,310 raw tags were generated from the 18 intestinal samples, and 1,364,142 effective tags were obtained after dislodging adaptors. The effective rate was ranged from 87.94% to 97.01% across the 18 intestinal samples. The rarefied curves for observed species number tended to approach the saturation plateau (Figure S1), indicating that sequencing depth was sufficient and sequencing data was credible. The sequencing information have been submitted into Sequence Read Archive (Accession Number, PRJNA759698).
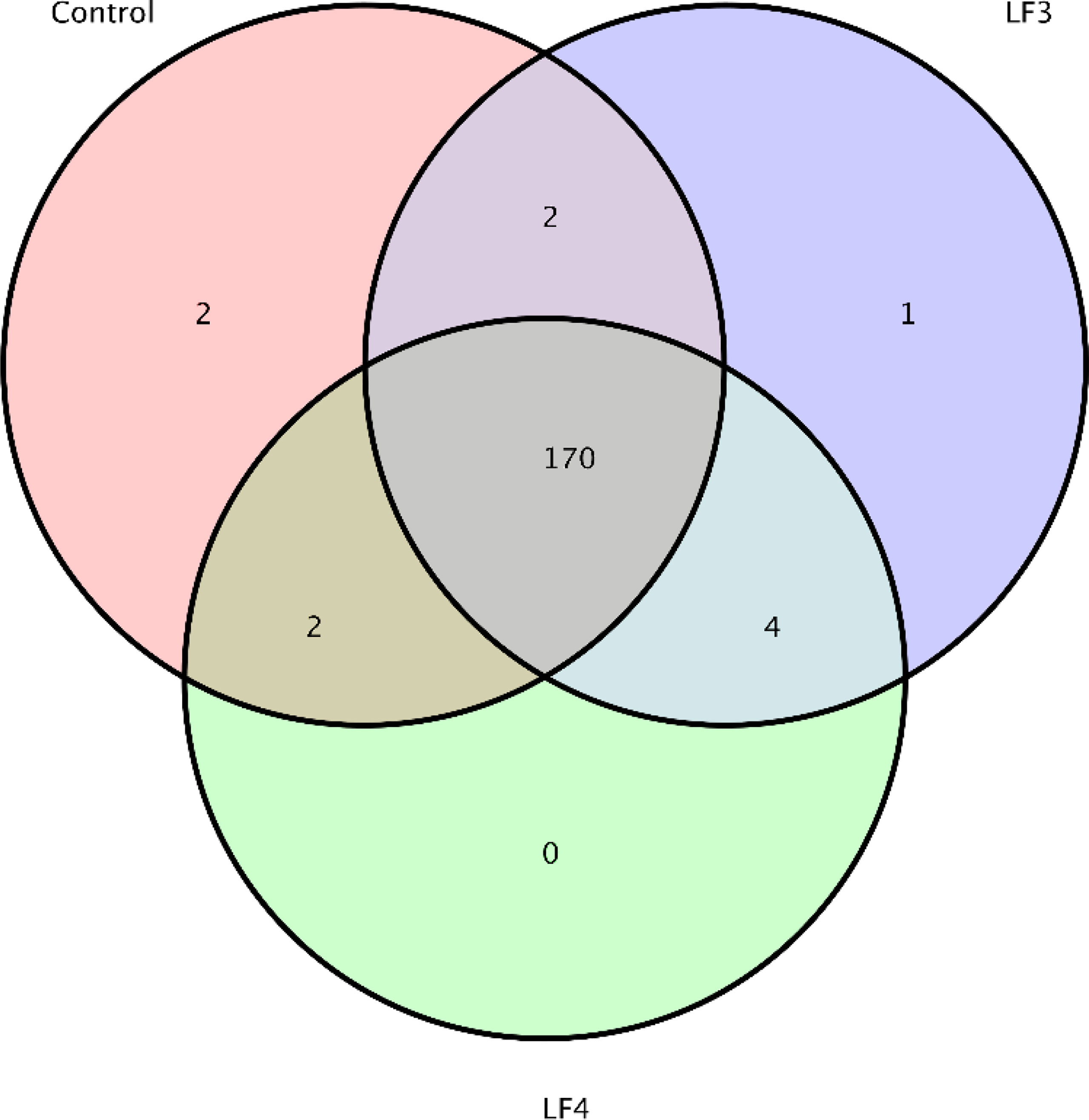
To facilitate downstream analyzation, all effective tags were normalized, and all normalized tags in 18 intestinal samples were clustered into 2420 OTUs based on 97% similar degree. The Venn diagram displayed that 170 OTUs were shared by the three groups, and the numbers of unique OTUs in groups C, LF3 and LF4 were 2, 1 and 0, respectively (Figure 1).
Alpha Diversity
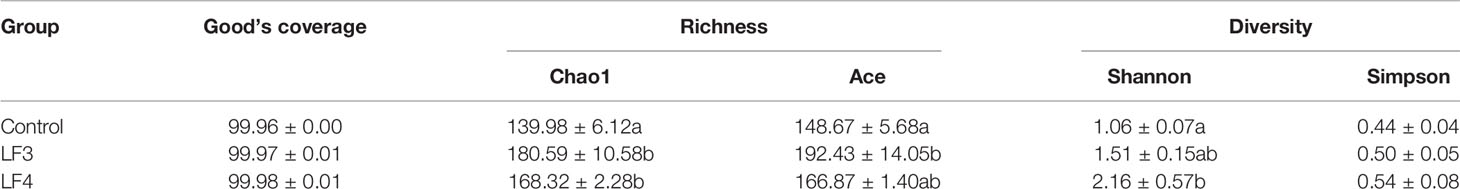
The Good’s coverage values of all the samples were over 99.96%, suggesting the adequate sampling of data and high sequencing quality (Table 1). Compared with the control group (C), intestinal microbiota richness (Chao1 and Ace) enhanced remarkably in the probiotic treated groups LF3 and LF4 (P < 0.05), except Ace indice in group LF4. Administration of host-derived probiotics resulted in improvement of Shannon indice, and significant improvement were noticed in group LF4 (P < 0.05). No significant variation in Simpson indice was documented between all groups (P > 0.05), although Simpson indices exhibited apparent enhancement in two probiotic groups.
Beta Diversity
Beta diversity parameters, PCoA, NMDS and UPGMA clustering tree at genus level based on binary_jaccard were used to analyze the intestinal microbial similarity among three groups (Figure 2). Regarding PCoA, the clusters of samples in the group C apparently scattered on the coordinate axis, whereas the clusters of samples in the groups LF3 and LF4 gathered on the lower left, especially that in the group LF4 (Figure 2A). Similar phenomenon was also observed in NMDS analysis (Figure 2B), and the Stress = 0.1163 (< 0.2), indicating that the NMDS data is credible. UPGMA clustering tree based on genus level showed that a remarkable separation between the group C and probiotic supplemented groups, especially in group LF4 (Figure 2C).
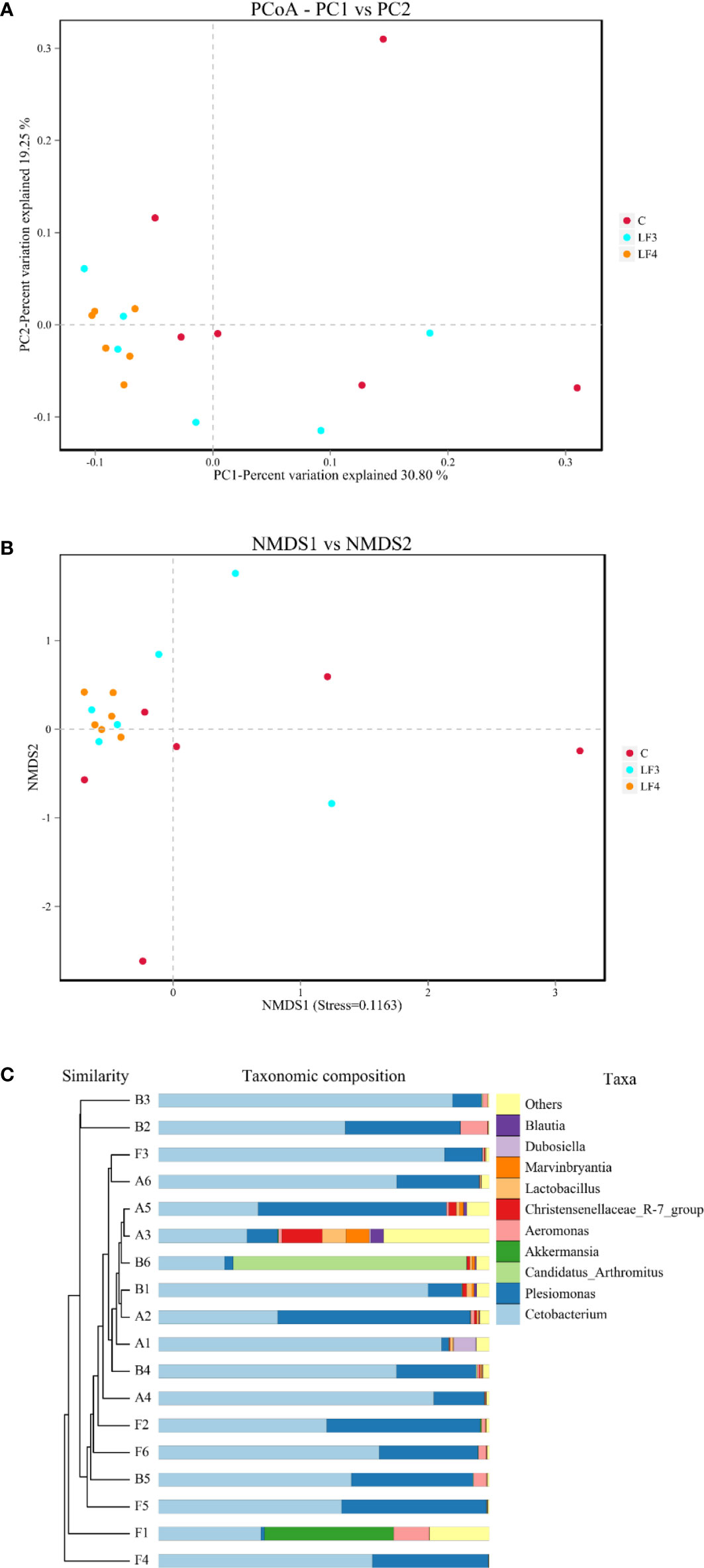
Figure 2 Beta diversity of intestinal microbiota based on genus level. (A) Principal Coordinate Analysis (PCoA) against PC1 vs PC2 axes; (B) Non-Metric Multi-Dimensional Scaling (NMDS) against NMDS1 vs NMDS2 axes; (C) UPGMA clustering trees. F1, F2, F3, F4, F5 and F6 are the six replicates of group C; B1, B2, B3, B4, B5 and B6 are the six replicates of the group LF3; A1, A2, A3, A4, A5 and A6 are the six replicates of the group LF4.
Intestinal Microbial Composition
At phylum level, Fusobacteria, Proteobacteria and Firmicutes were the three most dominant bacterial phyla, which accounted for 99.76%, 99.47% and 98.13% of the relative abundance in the groups C, LF3 and LF4, respectively (Figure 3A). The other phyla concurrently existed in the three groups, including Actinobacteria, Bacteroidetes, Verrucomicrobia, Cyanobacteria, Spirochaetes and Patescibacteria. The relative abundances of Fusobacteria and Proteobacteria were lower in the groups LF3 and LF4 compared with the group C, while apparently higher relative abundances of Firmicutes, Actinobacteria and Bacteroidetes were observed in the groups LF3 and LF4 (Figure 3B).
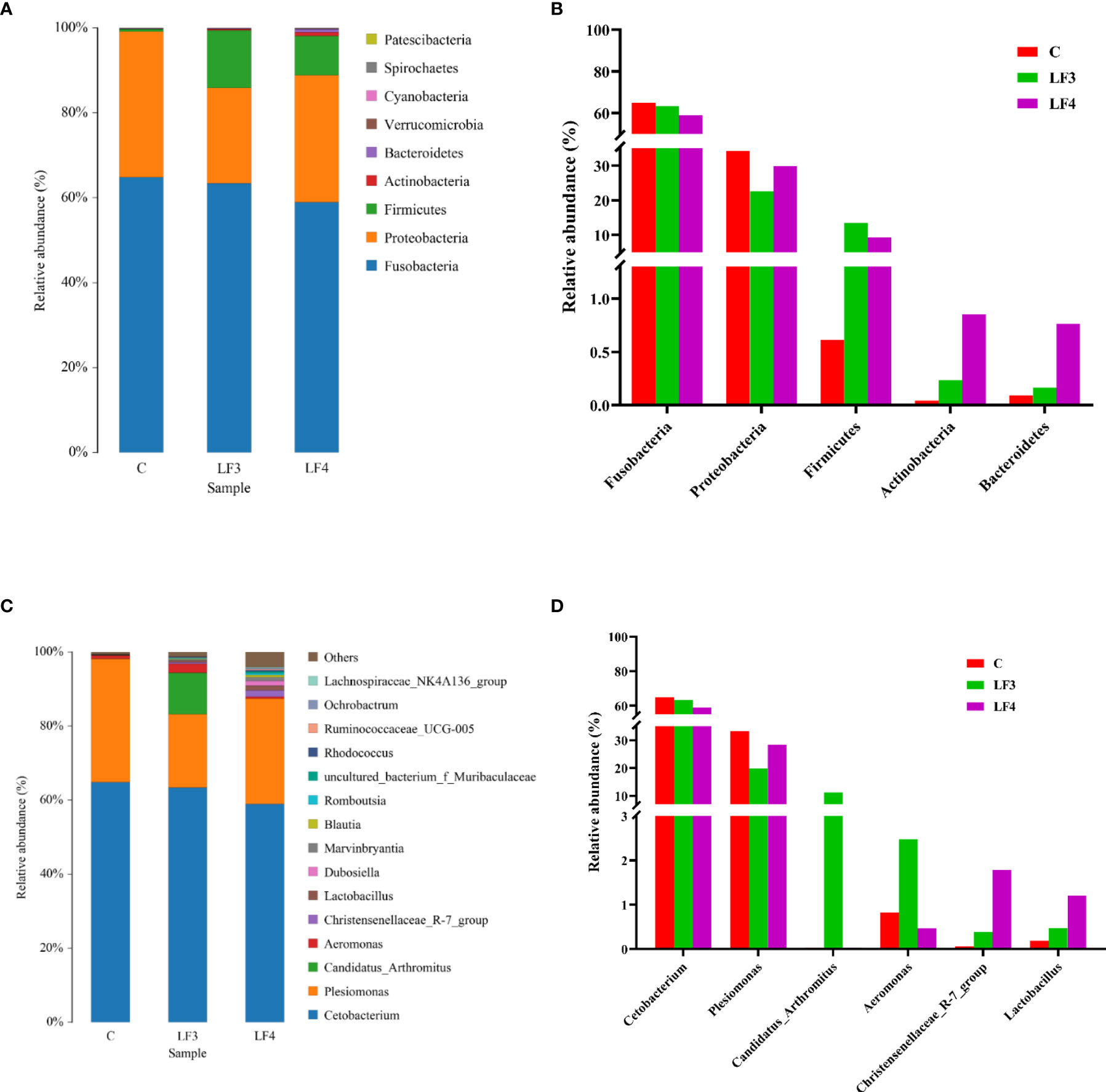
Figure 3 Relative abundances of intestinal microbiota at phyla (A) and genus (C) taxonomic levels in C, LF3 and LF4 groups. The changes of relative abundances of dominant phyla (B) and genus (D).
At genus level, the top 15 bacterial genera in the intestine of seabass were present in Figure 3C. Cetobacterium, Plesiomonas, Candidatus_Arthromitus, Aeromonas, Christensenellaceae_R-7_group, and Lactobacillus were considered as core genera with the relative abundances over 1%. As seen in Figure 3D, groups LF3 and LF4 showed a slight reduction in the relative abundances of Cetobacterium and Plesiomonas compared with the control (C). On the contrary, enhanced relative abundances of Candidatus_Arthromitus and Aeromonas was documented in the group LF3 compared with the group C. Additionally, the relative abundances of Christensenellaceae_R-7_group and Lactobacillus increased markedly in the groups LF3 and LF4, particularly in group LF4.
LEfSe Analyses
LEfSe analyses with LDA > 4 as the threshold were performed to determine whether there were significant changes in taxon abundance between control (C) group and probiotic groups LF3 or LF4 (Figure 4). For classification of phylum, the biomarkers emerging for the LF3 group indicated Firmicutes was biomarkers for the group C, and family Clostridiaceae_1, belonging to the phylum Firmicutes, had the highest LDA score (5.08, Figure 4A). On the other hand, the biomarkers emerging for the LF4 group also indicated Firmicutes was biomarkers for the group C, and the highest LDA score of phylum Firmicutes was 5.09 (Figure 4B).
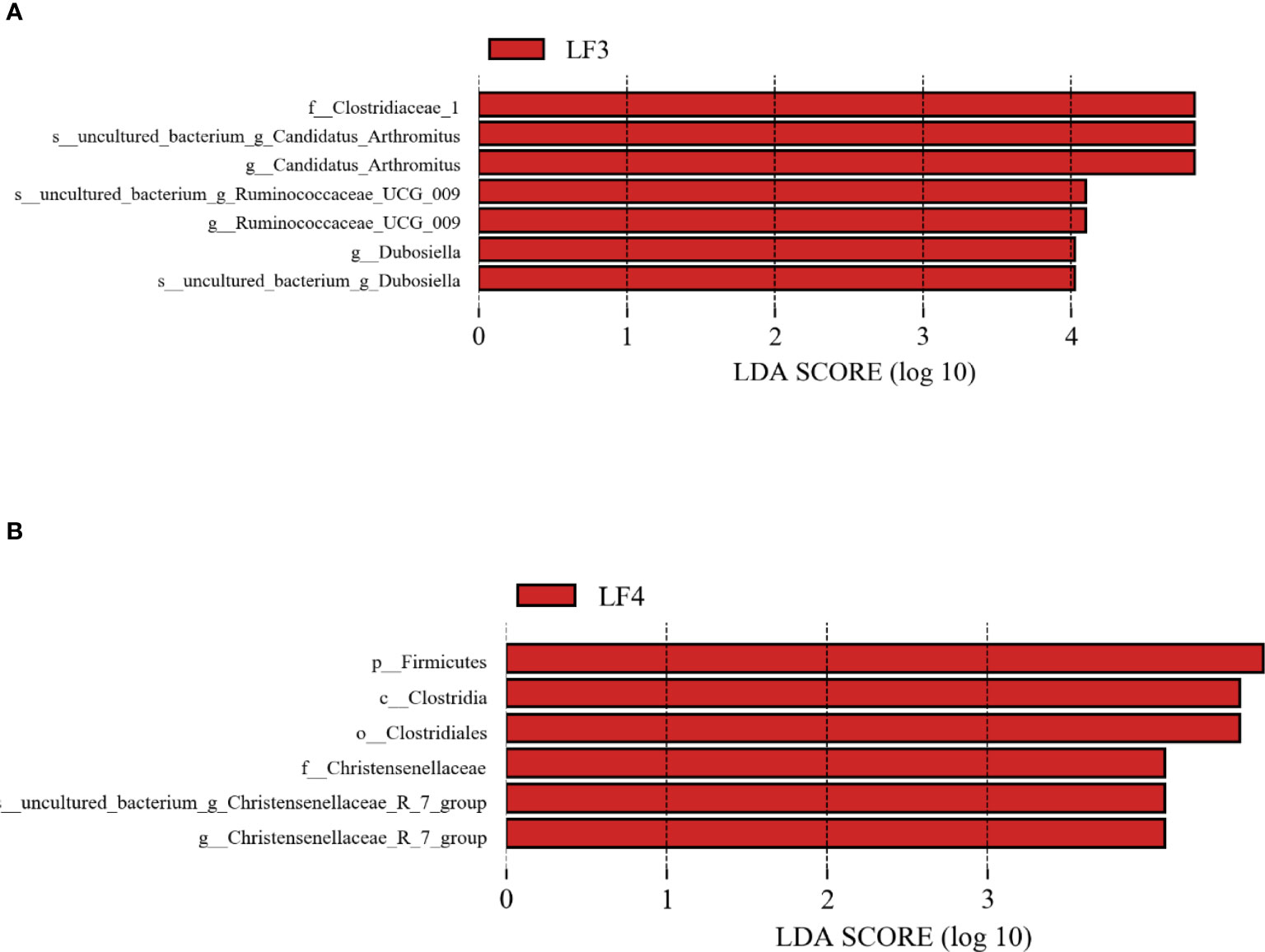
Figure 4 Lineages with linear discriminant analysis (LDA) score of the abundance of taxa. Only the taxa that LDA value above 4.0 are shown. (A) group C vs group LF3; (B) group C vs group LF4.
Functional Prediction
Changed level-2 KEGG pathways with relative abundances over 1% were displayed in Figure 5. Among predicted functions, Amino acid metabolism, Carbohydrate metabolism, Global and overview maps, energy metabolism, membrane transport, metabolism of cofactors and vitamin, and nucleotide metabolism were the most dominant. Unexpectedly, there were only slight alteration in the functions of intestinal microbiota in the groups C, LF3 and LF4.
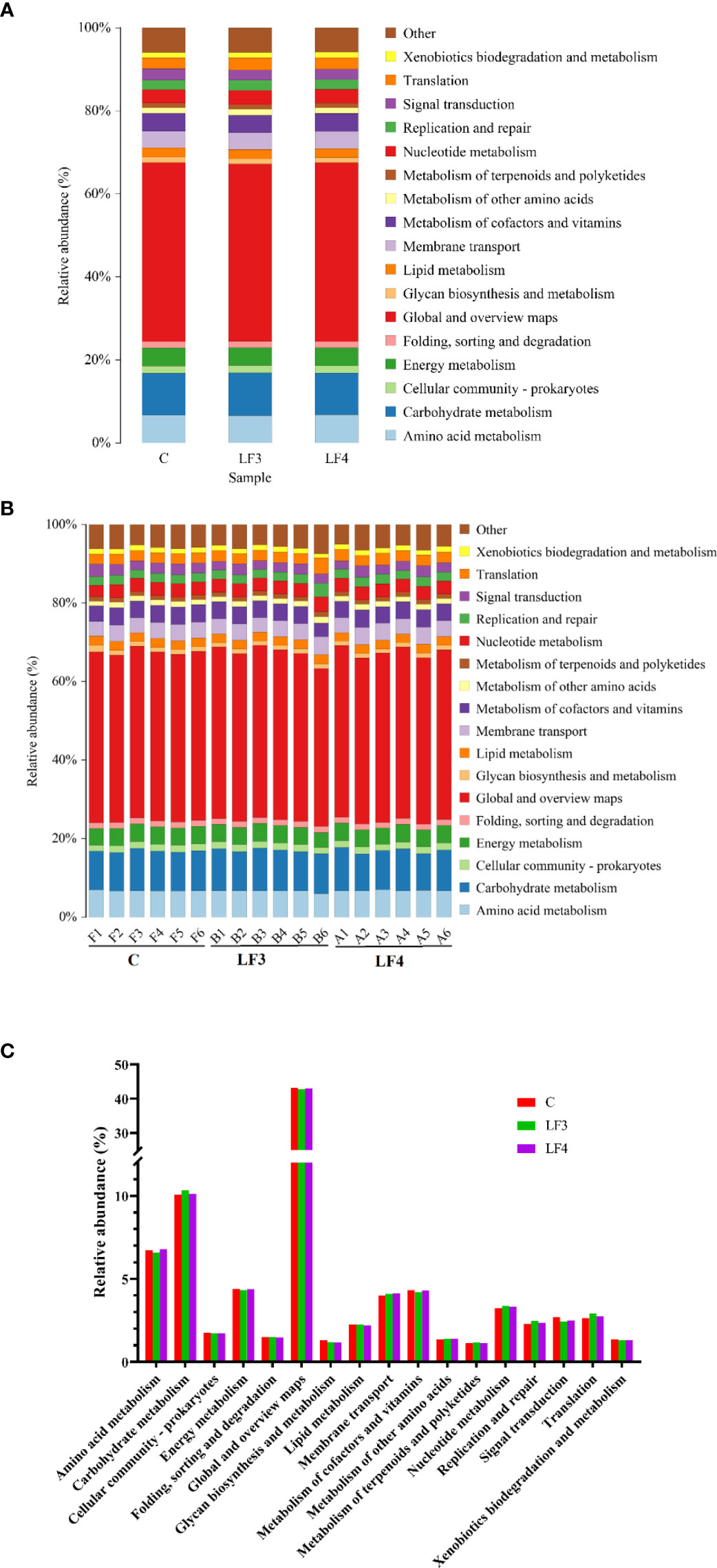
Figure 5 Predicted functions of intestinal microbiota in groups C, LF3 and LF4. (A) mean abundances of three groups; (B) abundance of each sample; (C) level-2 KEGG pathways with relative abundances over 1%.
Discussion
The application of probiotics in fish aquaculture has been extensively reported, while limited information is available about the interactions between host-derived probiotics and intestinal microbiota (Kong et al., 2021). In this study, the impact of host-derived probiotic L. petauri LF3 and B. siamensis LF4 on intestinal microbiota in juvenile Japanese seabass were analyzed by using high-throughput sequencing technology. The intestinal microbiota of Japanese seabass (L. japonicus) fed control or probiotic diets shared three dominant phyla (Fusobacteria, Proteobacteria and Firmicutes), which had been also reported as dominant phyla in the intestines of other teleost fish, such as European seabass (Dicentrarchus labrax) (Kokou et al., 2020; Wassef et al., 2020; Serra et al., 2021), Asian seabass (Lates calcarifer) (Apper et al., 2016), grass carp (Ctenopharyngodon idellus) (Shi F. et al., 2021), tilapia (O. niloticus) (Adeoye et al., 2016; Li et al., 2019), orange-spotted grouper (Epinephelus coioides) (Yang et al., 2019), and so on. Overabundances of certain taxa led to intestinal microbiota turning from balance to dysbiosis status, and then threaten host well-being (Li et al., 2019; Yang et al., 2019; Serra et al., 2021). In this study, the relative abundances of Fusobacteria and Proteobacteria were decreased in Japanese seabass fed probiotic diets. In line with the present study, decreased abundances of Fusobacteria and/or Proteobacteria following probiotics application were documented in Asian seabass (L. calcarifer) (Masduki et al., 2020) and orange-spotted grouper (E. coioides) (Yang et al., 2019). Inhibiting opportunistic pathogens Fusobacteria and Proteobacteria overgrowth would create redundant niches, which might benefit the growth of other beneficial microbes (Xun et al., 2019; Tan and Sun, 2020; Shi F. et al., 2021). On the other hand, the supplementation of host-derived probiotics in diet (groups LF3 and LF4) apparently enhanced the relative abundances of Firmicutes, Actinobacteria and Bacteroidetes in this study. Firmicutes and Bacteroidetes have been recorded previously as primarily beneficial phyla in healthy Japanese seabass (L. japonicus) (Wang et al., 2017), European sea bass (D. labrax) (Parma et al., 2019; Rimoldi et al., 2020), common carp (Cyprinus carpio L.) (Meng et al., 2021), and grouper (E. coioides) (Yang et al., 2019). Bacteria of phylum Actinobacteria could produce a wide range of exoenzymes and secondary metabolites (Ventura et al., 2007). In particular, the genus Bifidobacterium, belonging to phylum Actinobacteria, is an important lactic acid producer, which was used as probiotic in aquaculture (Jha et al., 2015). This may illustrate the better growth performance and feed utilization in Japanese seabass fed probiotic L. petauri LF3 and B. siamensis LF4 (Yang et al., 2021).
At the genus level, Japanese seabass fed with Lc. petaurid LF3 and B. siamensis LF4 containing diets showed lower relative abundances of opportunistic pathogens such as Cetobacterium and Plesiomonas, which might be partly contributed to the improvement of growth performance and general health in juvenile Japanese seabass (L. japonicus) (Yang et al., 2021). In line with the present study, previous reports have showed that lower abundances of Cetobacterium and Plesiomonas existed in the intestine of healthier fish and benefited to host health due to cooperative activity among commensal microbes (Miller et al., 2006; Behera et al., 2018; Liu et al., 2021; Zhang et al., 2021). On the other hand, higher abundances of intestinal possible beneficial Christensenellaceae_R-7_group and Lactobacillus in the phylum Firmicutes, which have been reported as beneficial bacteria to improve growth performance, immunity, antioxidant capability and other welfares in aquatic animals (Van Doan et al., 2017; Zheng et al., 2017; Hoseinifar et al., 2018; Wang et al., 2019), were presented in seabass fed with host-derived probiotics, especially with B. siamensis LF4. The genus Lactobacillus can produce high level of lactic acid, and then are able to improve feed efficiency and alleviate metabolic damage by positively modulating gut microbiota (Hoseinifar et al., 2018; Wang et al., 2019). In line with the current study, significantly elevated abundances of beneficial microorganisms have been also reported in probiotic Clostridium butyricum fed tilapia (O. niloticus) (Li et al., 2019) and common carp (C. carpio L.) (Meng et al., 2021). These results evidenced that beneficial microorganisms were usually common members of the normal microbiota in the fish gut, and under certain circumstances, the higher abundances of these microorganisms could be partly responsible for mucosal homeostasis and avoiding disease happen (Hoseinifar et al., 2018; Yu et al., 2021). Interestingly, seabass fed with Lc. petaurid LF3 demonstrated a sharp increase in the relative abundances of Aeromonas, which is a traditionally opportunistic pathogen in the intestines of aquatic animals (Jiravanichpaisal et al., 2009; Zhu et al., 2016; Chen et al., 2021). However, several studies have evidenced that Aeromonas could be use as probiotic and improve growth, immunity, and disease resistance in certain aquatic animals, such as common carp (C. carpio) (Chi et al., 2014), grass carp (Ctenopharyngodon idella) (Hao et al., 2017), Pacific white shrimp (Litopenaeus vannamei) (Hao et al., 2014), and so on. In general, these results illustrated that host-derived Lc. petaurid LF3 and B. siamensis LF4 can positively remodel intestinal microbial composition.
In the present study, the intestinal microbiota of fish fed probiotics L. petauri LF3 and B. siamensis LF4 demonstrated higher species diversity based on alpha diversity data (Chao1, Ace, Shannon and Simpson indices), and more similarity and lower individual differences based on beta diversity data (PCoA, NMDS and UPGMA clustering tree). The results are in line with other researches in snakehead fish (Channa argus) fed with two lactic acid bacteria (Kong et al., 2021), and common carp (C. carpio L.) fed with Clostridium butyricum (Meng et al., 2021). Generally, high intestinal bacterial diversity reflect a well-development of the host, as the increase of commensal microbial diversity and/or species richness might lead to attenuated colonization resistance against invading opportunistic pathogens, which have the potential to cause infection of the intestine (Rimoldi et al., 2020). In the current study, it is worth noting that probiotic L. petauri LF3 and B. siamensis LF4 can improve intestinal microbial diversities and meanwhile reduce individual differences, which might partly explain the previous results that the two probiotic strains facilitated growth performance, immune response, and ammonia tolerance in juvenile Japanese seabass (L. japonicus) (Yang et al., 2021). Similarly, higher and stabler intestinal microbial diversities were detected in probiotics-treated fish with improving growth, immunity and disease resistance (Kong et al., 2021); (Meng et al., 2021).
In the present study, compared to the control group, supplementation of host-derived Lc. petaurid LF3 and B. siamensis LF4 led to biomarkers emerging, especially phylum Firmicutes, as selected using LEfSe. In general, phylum Firmicutes is a well recognized class of beneficial microorganisms. Early researches have showed that bacteria (i.e., genus Bacillus, Lactobacillus, Lactococcus, Enterococcus, and Clostridium) in phylum Firmicutes can secretion of various enzymes and decreasing the intestinal pH by producing short-chain fatty acids (Dawood et al., 2019), which can inhibit the proliferation of pathogenic bacteria and improve intestinal microecology of aquatic animals (Hao et al., 2017; Hoseinifar et al., 2018; Dawood et al., 2019; Li et al., 2019; Wang et al., 2019; Yang et al., 2019; Meng et al., 2021). The results of phylum Firmicutes significantly enriched by dietary supplementation of host-derived probiotics again proved that probiotics can increase the beneficial bacteria to achieve the purpose of positively modulating intestinal microbiota of aquatic animals (Hoseinifar et al., 2018; Wang et al., 2019; Yang et al., 2019; Doan et al., 2020; Kong et al., 2021).
The NSTI values (0.19 ± 0.03) for seabass intestinal samples in the present study were in line with those for red swamp crayfish, Procambarus clarkii (0.171 ± 0.023) and pacific white shrimp, Litopenaeus vannamei (0.18 ± 0.02) (Zeng et al., 2017; Chen et al., 2021). It has been validated that PICRUSt 2 could accurately predict the functions of seabass intestinal microbiota (Chen et al., 2021). In this study, the dominated function of Amino acid metabolism, Carbohydrate metabolism and Global and overview maps were accurately predicted using PICRUSt 2, which are associated strongly with seabass digestion and absorption. Unexpectedly, the intestinal microbial functions were stable among all groups, although dietary supplementation with host-derived probiotics remarkably changed the intestinal microbial community. In agreement with the present study, the intestinal microbial functions (including carbrohydrate transport and metabolism, lipid transport and metabolism, and so on) were determined to be almost similar in snakehead fish (C. argus) fed with a basal diet or diets supplemented with 1.0 × 108 CFU/g of host-derived probiotics (L. lacti L19, E. faecalis W24, L. lacti L19 + E. faecalis W24 (Kong et al., 2021). However, these results contradict to previous studies that the change of intestinal microbial composition was associated with function change in aquatic animals, such as zebrafish (Danio rerio) (Shi Y. Y. et al., 2021) and turbot (Scophthalmus maximus) (Guo et al., 2022). Guo et al. (2022) has reported that several pathways related to glycan, lipid and amino acid metabolism upregulated in turbots (S. maximus) fed with probiotic (strain B. coagulans G1902, unspecified source). We hypothesized that this paradox might be caused by the different probiotic species and source, and rearing environment. Additionally, this study supported the hypothesis that when the host is sufficiently healthy, the function of the intestinal microbiota might maintain relative stability rather than emerge significant changes (Fan and Pedersen, 2021).
Conclusion
Host-derived probiotic L. petauri LF3 and B. siamensis LF4 decreased opportunistic pathogenic Fusobacteria, Proteobacteria, and Plesiomonas and apparently increased beneficial Firmicutes, Bacteroidetes, and Lactobacillus in juvenile Japanese seabass (L. japonicus). Dietary supplementation of host-derived probiotic L. petauri LF3 and B. siamensis LF4 significantly improve the intestinal microbial diversities and led to Firmicutes as a significant biomarker emerge. However, the intestinal microbial functions (i.e., amino acid metabolism, carbohydrate metabolism, energy metabolism, etc.) did not alter by host-derived probiotics. These results lay the necessary theoretical basis for the development and application of host-associated probiotics in Japanese seabass.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: NCBI [accession: PRJNA759698].
Ethics Statement
The animal study was reviewed and approved by the Animal Care and Use Committee of Jimei University.
Author Contributions
H-LY investigation and original draft.: Z-YL: data analysis and writing. J-TJ: investigation and data analysis. Z-XL: feed preparation and animal trial. J-DY: review and editing. Y-ZS: supervision, project administration, funding acquisition, review and editing. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National natural science foundation of China (32072990), Science and Technology Major/Special Project of Fujian Province (2021NZ029022), Xiamen Marine and Fisheries Development Fund (19CZP018HJ04) and the China Agriculture Research System (CARS-47-G14).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2022.878633/full#supplementary-material
References
Adeoye A. A., Jaramillo-Torres A., Fox S. W., Merrifield D. L., Davies S. J. (2016). Supplementation of Formulated Diets for Tilapia (Oreochromis niloticus) With Selected Exogenous Enzymes: Overall Performance and Effects on Intestinal Histology and Microbiota. Anim. Feed Sci. Technol. 215, 133–143. doi: 10.1016/j.anifeedsci.2016.03.002
Apper E., Weissman D., Respondek F., Guyonvarch A., Baron F., Boisot P., et al. (2016). Hydrolysed Wheat Gluten as Part of a Diet Based on Animal and Plant Proteins Supports Good Growth Performance of Asian Seabass (Lates calcarifer), Without Impairing Intestinal Morphology or Microbiota. Aquaculture 453, 40–48. doi: 10.1016/j.aquaculture.2015.11.018
Behera B. K., Bera A. K., Paria P., Das A., Parida P. K., Kumari S., et al. (2018). Identification and Pathogenicity of Plesiomonas shigelloides in Silver Carp. Aquaculture 493, 314–318. doi: 10.1016/j.aquaculture.2018.04.063
Chen H. L., Wang Y. J., Zhang J., Bao J. J. (2021). Intestinal Microbiota in White Spot Syndrome Virus Infected Red Swamp Crayfish (Procambarus clarkii) at Different Health Statuses. Aquaculture 542, 736826. doi: 10.1016/j.aquaculture.2021.736826
Chi C., Jiang B., Yu X. B., Liu T. Q., Xia L., Wang G. X. (2014). Effects of Three Strains of Intestinal Autochthonous Bacteria and Their Extracellular Products on the Immune Response and Disease Resistance of Common Carp, Cyprinus carpio. Fish Shellfish Immunol. 36, 9–18. doi: 10.1016/j.fsi.2013.10.003
China Fishery Statistics Yearbook (2021). Bureau of Fisheries. Ministry of Agriculture (Beijing, China: China Agriculture Press).
Cornejo-Granados F., Lopez-Zavala A. A., Gallardo-Becerra L., Mendoza-Vargas A., Sanchez F., Vichido R., et al. (2017). Microbiome of Pacific Whiteleg Shrimp Reveals Differential Bacterial Community Composition Between Wild, Aquacultured and AHPND/EMS Outbreak Conditions. Sci. Rep. 7, 2360–2368. doi: 10.1038/s41598-017-11805-w
Dawood M. A. O., Koshio S., Abdel-Daim M. M., Van Doan H. (2019). Probiotic Application for Sustainable Aquaculture. Rev. Aquacult. 11, 907–924. doi: 10.1111/raq.12272
Doan H. V., Hoseinifar S. H., Ringo E., Esteban M. A., Dadar M., Dawood M. A. O., et al. (2020). Host-Associated Probiotics: A Key Factor in Sustainable Aquaculture. Rev. Fish Sci. Aquac. 28, 16–42. doi: 10.1080/23308249.2019.1643288
Fan Y., Pedersen O. (2021). Gut Microbiota in Human Metabolic Health and Disease. Nat. Rev. Microbiol. 19, 55–71. doi: 10.1038/s41579-020-0433-9
Guo G. X., Li K. X., Zhu Q. H., Zhao C. Y., Li C., He Z. L., et al. (2022). Improvements of Immune Genes and Intestinal Microbiota Composition of Turbot (Scophthalmus maximus) With Dietary Oregano Oil and Probiotics. Aquaculture, 547, 737442. doi: 10.1016/j.aquaculture.2021.737442
Hao K., Liu J. Y., Ling F., Liu X. L., Lu L., Xia L., et al. (2014). Effects of Dietary Administration of Shewanella haliotis D4, Bacillus Cereus D7 and Aeromonas bivalvium D15, Single or Combined, on the Growth, Innate Immunity and Disease Resistance of Shrimp, Litopenaeus vannamei. Aquaculture 428, 141–149. doi: 10.1016/j.aquaculture.2014.03.016
Hao K., Wu Z. Q., Li D. L., Yu X. B., Wang G. X., Ling F. (2017). Effects of Dietary Administration of Shewanella xiamenensis A-1, Aeromonas veronii A-7, and Bacillus subtilis, Single or Combined, on the Grass Carp (Ctenopharyngodon idella) Intestinal Microbiota. Probiotics Antimicro. 9, 386–396. doi: 10.1007/s12602-017-9269-7
Hoseinifar S. H., Sun Y. Z., Wang A. R., Zhou Z. G. (2018). Probiotics as Means of Diseases Control in Aquaculture, a Review of Current Knowledge and Future Perspectives. Front. Microbiol. 9. doi: 10.3389/fmicb.2018.02429
Jha D. K., Bhujel R. C., Anal A. K. (2015). Dietary Supplementation of Probiotics Improves Survival and Growth of Rohu (Labeo rohita Ham.) Hatchlings and Fry in Outdoor Tanks. Aquaculture 435, 475–479. doi: 10.1016/j.aquaculture.2014.10.026
Jiravanichpaisal P., Roos S., Edsman L., Liu H. P., Soderhall K. (2009). A Highly Virulent Pathogen, Aeromonas hydrophila, From the Freshwater Crayfish Pacifastacus leniusculus. J. Invertebr. Pathol. 101, 56–66. doi: 10.1016/j.jip.2009.02.002
Kokou F., Sasson G., Mizrahi I., Cnaani A. (2020). Antibiotic Effect and Microbiome Persistence Vary Along the European Seabass Gut. Sci. Rep. 10, 12. doi: 10.1038/s41598-020-66622-5
Kong Y. D., Li M., Chu G. S., Liu H. J., Shan X. F., Wang G. Q., et al. (2021). The Positive Effects of Single or Conjoint Administration of Lactic Acid Bacteria on Channa argus: Digestive Enzyme Activity, Antioxidant Capacity, Intestinal Microbiota and Morphology. Aquaculture 531, 735852. doi: 10.1016/j.aquaculture.2020.735852
Lazado C. C., Caipang C. M. A., Estante E. G. (2015). Prospects of Host-Associated Microorganisms in Fish and Penaeids as Probiotics With Immunomodulatory Functions. Fish Shellfish Immunol. 45, 2–12. doi: 10.1016/j.fsi.2015.02.023
Liu Z. Y., Yang H. L., Hu L. H., Yang W., Ai C. X., Sun Y. Z. (2021). Dose-Dependent Effects of Histamine on Growth, Immunity and Intestinal Health in Juvenile Grouper (Epinephelus coioides). Front. Mar. Sci. 8. doi: 10.3389/fmars.2021.685720
Li H. Q., Zhou Y., Ling H. Y., Luo L., Qi D. S., Feng L. (2019). The Effect of Dietary Supplementation With Clostridium butyricum on the Growth Performance, Immunity, Intestinal Microbiota and Disease Resistance of Tilapia (Oreochromis niloticus). PloS One 14, e0223428. doi: 10.1371/journal.pone.0223428
Masduki F., Zakaria T., Min C. C., Karim M. (2020). Evaluation of Enterococcus hirae LAB3 as Potential Probiont Against Vibrio harveyi in Artemia nauplii and Asian Seabass Larvae (Lates calcarifer) Cultures. J. Environ. Biol. 41, 1153–1159. doi: 10.22438/jeb/41/5(SI)/MS_06
Meng X. L., Wu S. K., Hu W. P., Zhu Z. X., Yang G. K., Zhang Y. M., et al. (2021). Clostridium butyricum Improves Immune Responses and Remodels the Intestinal Microbiota of Common Carp (Cyprinus carpio L.). Aquaculture 530, 735753. doi: 10.1016/j.aquaculture.2020.735753
Miller W. A., Miller M. A., Gardner I. A., Atwill E. R., Byrne B. A., Jang S., et al. (2006). Salmonella spp., Vibrio spp., Clostridium Perfringens, and Plesiomonas Shigelloides in Marine and Freshwater Invertebrates From Coastal California Ecosystems. Microb. Ecol. 52, 198–206. doi: 10.1007/s00248-006-9080-6
Parma L., Yufera M., Navarro-Guillen C., Moyano F. J., Soverini M., D’Amico F., et al. (2019). Effects of Calcium Carbonate Inclusion in Low Fishmeal Diets on Growth, Gastrointestinal Ph, Digestive Enzyme Activity and Gut Bacterial Community of European Sea Bass (Dicentrarchus labrax L.) Juveniles. Aquaculture 510, 283–292. doi: 10.1016/j.aquaculture.2019.05.064
Rimoldi S., Torrecillas S., Montero D., Gini E., Makol A., Valdenegro V. V., et al. (2020). Assessment of Dietary Supplementation With Galactomannan Oligosaccharides and Phytogenics on Gut Microbiota of European Sea Bass (Dicentrarchus labrax) Fed Low Fishmeal and Fish Oil Based Diet. PloS One 15, e0231494. doi: 10.1371/journal.pone.0231494
Serra C. R., Oliva-Teles A., Enes P., Tavares F. (2021). Gut Microbiota Dynamics in Carnivorous European Seabass (Dicentrarchus labrax) Fed Plant-Based Diets. Sci. Rep. 11, 447. doi: 10.1038/s41598-020-80138-y
Shi Y. Y., Cao X. Y., Ye Z. D., Xu Y. Y., Wang Y. M., Li Z. P., et al. (2021). Role of Dietary Schizochytrium Sp. In Improving Disease Resistance of Zebrafish Through Metabolic and Microbial Analysis. Aquaculture 539, 736631. doi: 10.1016/j.aquaculture.2021.736631
Shi F., Lu Z. J., Yang M. X., Li F., Zhan F. B., Zhao L. J., et al. (2021). Astragalus Polysaccharides Mediate the Immune Response and Intestinal Microbiota in Grass Carp (Ctenopharyngodon idellus). Aquaculture 534, 12. doi: 10.1016/j.aquaculture.2020.736205
Simon R., Docando F., Nunez-Ortiz N., Tafalla C., Diaz-Rosales P. (2021). Mechanisms Used by Probiotics to Confer Pathogen Resistance to Teleost Fish. Front. Immunol. 12, 653025. doi: 10.3389/fimmu.2021.653025
Tan X. H., Sun Z. Z. (2020). Dietary Dandelion Extract Improved Growth Performance, Immunity, Intestinal Morphology and Microbiota Composition of Golden Pompano Trachinotus Ovatus. Aquacult. Rep. 18, 9. doi: 10.1016/j.aqrep.2020.100491
Teame T., Wang A. R., Xie M. X., Zhang Z., Yang Y. L., Ding Q. W., et al. (2020). Paraprobiotics and Postbiotics of Probiotic Lactobacilli, Their Positive Effects on the Host and Action Mechanisms: A Review. Front. Nutr. 7, 570344. doi: 10.3389/fnut.2020.570344
Van Doan H., Hoseinifar S. H., Dawood M. A. O., Chitmanat C., Tayyamath K. (2017). Effects of Cordyceps militaris Spent Mushroom Substrate and Lactobacillus plantarum on Mucosal, Serum Immunology and Growth Performance of Nile Tilapia (Oreochromis niloticus). Fish Shellfish Immunol. 70, 87–94. doi: 10.1016/j.fsi.2017.09.002
Ventura M., Canchaya C., Tauch A., Chandra G., Fitzgerald G. F., Chater K. F., et al. (2007). Genomics of Actinobacteria: Tracing the Evolutionary History of an Ancient Phylum. Microbiol. Mol. Biol. Rev. 71, 495–548. doi: 10.1128/MMBR.00005-07
Wang A. R., Ran C., Ringo E., Zhou Z. G. (2018). Progress in Fish Gastrointestinal Microbiota Research. Rev. Aquacult. 10, 626–640. doi: 10.1111/raq.12191
Wang A. R., Ran C., Wang Y. B., Zhang Z., Ding Q. W., Yang Y. L., et al. (2019). Use of Probiotics in Aquaculture of China-A Review of the Past Decade. Fish Shellfish Immunol. 86, 734–755. doi: 10.1016/j.fsi.2018.12.026
Wang J., Tao Q. Y., Wang Z., Mai K. S., Xu W., Zhang Y. J., et al. (2017). Effects of Fish Meal Replacement by Soybean Meal With Supplementation of Functional Compound Additives on Intestinal Morphology and Microbiome of Japanese Seabass (Lateolabrax japonicus). Aquac. Res. 48, 2186–2197. doi: 10.1111/are.13055
Wassef E. A., Saleh N. E., Abdel-Meguid N. E., Barakat K. M., Abdel-Mohsen H. H., El-bermawy N. M. (2020). Sodium Propionate as a Dietary Acidifier for European Seabass (Dicentrarchus labrax) Fry: Immune Competence, Gut Microbiome, and Intestinal Histology Benefits. Aquac. Int. 28, 95–111. doi: 10.1007/s10499-019-00446-7
Xun P. W., Lin H. Z., Wang R. X., Huang Z., Zhou C. P., Yu W., et al. (2019). Effects of Dietary Vitamin B-1 on Growth Performance, Intestinal Digestion and Absorption, Intestinal Microflora and Immune Response of Juvenile Golden Pompano (Trachinotus ovatus). Aquaculture 506, 75–83. doi: 10.1016/j.aquaculture.2019.03.017
Yang H. L., Liu Z. Y., Jian J. T., Ye J. D., Sun Y. Z. (2021). Host-Associated Bacillus Siamensis and Lactococcus petauri Improved Growth Performance, Innate Immunity, Antioxidant Activity and Ammonia Tolerance in Juvenile Japanese Seabass (Lateolabrax japonicus). Aquacult. Nutr. 27, 2739–2748. doi: 10.1111/anu.13399
Yang H. L., Sun Y. Z., Hu X., Ye J. D., Lu K. L., Hu L. H., et al. (2019). Bacillus pumilus SE5 Originated PG and LTA Tuned the Intestinal TLRs/MyD88 Signaling and Microbiota in Grouper (Epinephelus coioides). Fish Shellfish Immunol. 88, 266–271. doi: 10.1016/j.fsi.2019.03.005
Yu Y. Y., Ding L. G., Huang Z. Y., Xu H. Y., Xu Z. (2021). Commensal Bacteria-Immunity Crosstalk Shapes Mucosal Homeostasis in Teleost Fish. Rev. Aquacult. 13, 2322–2343. doi: 10.1111/raq.12570
Zeng S. Z., Huang Z. J., Hou D. W., Liu J., Weng S. P., He J. G. (2017). Composition, Diversity and Function of Intestinal Microbiota in Pacific White Shrimp (Litopenaeus vannamei) at Different Culture Stages. Peerj 5, e3986. doi: 10.7717/peerj.3986
Zhang H. L., Ran C., Teame T., Ding Q. W., Hoseinifar S. H., Xie M. X., et al. (2020). Research Progress on Gut Health of Farmers Teleost Fish: A Viewpoint Concerning the Intestinal Mucosal Barrier and the Impact of Its Damage. Rev. Fish Biol. Fisher. 30, 569–586. doi: 10.1007/s11160-020-09614-y
Zhang Y., Zhang P. J., Shang X. C., Lu Y. T., Li Y. H. (2021). Exposure of Lead on Intestinal Structural Integrity and the Diversity of Gut Microbiota of Common Carp. Comp. Biochem. Phys. C 239, 108877. doi: 10.1016/j.cbpc.2020.108877
Zheng X. T., Duan Y. F., Dong H. B., Zhang J. S. (2017). Effects of Dietary Lactobacillus plantarum in Different Treatments on Growth Performance and Immune Gene Expression of White Shrimp Litopenaeus vannamei Under Normal Condition and Stress of Acute Low Salinity. Fish Shellfish Immun. 62, 195–201. doi: 10.1016/j.fsi.2017.01.015
Keywords: host-derived probiotics, intestinal microbiota, diversity, functions, Lateolabrax japonicus
Citation: Yang H-L, Liu Z-Y, Jian J-T, Liu Z-X, Ye J-D and Sun Y-Z (2022) Host-Derived Probiotics Shape the Intestinal Microbial Composition, but not Putative Function in Juvenile Japanese Seabass (Lateolabrax japonicus). Front. Mar. Sci. 9:878633. doi: 10.3389/fmars.2022.878633
Received: 18 February 2022; Accepted: 23 March 2022;
Published: 19 April 2022.
Edited by:
Ana Maulvault, Portuguese Institute for Sea and Atmosphere (IPMA), PortugalReviewed by:
Christopher Marlowe Arandela Caipang, University of San Agustin, PhilippinesZhigang Zhou, Feed Research Institute (CAAS), China
Copyright © 2022 Yang, Liu, Jian, Liu, Ye and Sun. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yun-Zhang Sun, am11c3VueXVuemhhbmdAMTYzLmNvbQ==
†These authors have contributed equally to this work and share first authorship
 Hong-Ling Yang1,2†
Hong-Ling Yang1,2† Zi-Yan Liu
Zi-Yan Liu Yun-Zhang Sun
Yun-Zhang Sun