- College of Fisheries, Guangdong Ocean University, Zhanjiang, China
The Late Pleistocene-Holocene climate fluctuations have greatly influenced the phylogeographic structure and historical dynamics of many marine organisms in the western Pacific marginal seas. Here, we investigated the impact of Pleistocene glacial-interglacial cycles on the phylogeographic structure and demographic dynamics of Decapterus maruadsi, an economically important fish along the coast of the East China Sea (ECS) and northern South China Sea (NSCS). We obtained 430 concatenated sequences (Cyt b + control region, 1548–1554 bp) of D. maruadsi, including 246 newly sampled from the ECS and 184 previously determined from the NSCS. Genetic structure and phylogenetic analysis demonstrated a lack of significant population structure among 16 populations. Moreover, there was no significant differentiation among populations from Chinese coastal waters and northern Vietnam. Neutrality tests, unimodal mismatch distributions, Bayesian skyline plots, and the star-like haplotype networks all indicated a recent demographic expansion for D. maruadsi population during the Late Pleistocene-Holocene, explaining the low genetic diversity in D. maruadsi along the southeast coast of China. Notably, phylogenetic analyses and net genetic distances based on Cyt b jointly confirmed that 57 Cyt b haplotypes identified as D. maruadsi from the previously defined Sundaland-Rosario-Ranong clade actually represented D. russelli. These results not only reveal the complex effects of Pleistocene-Holocene climate fluctuations on the phylogeographic structure and demographic history of D. maruadsi but also provide useful genetic information for the management of genetic resources.
Introduction
Climate oscillations during the Pleistocene greatly altered the environment of marginal seas of the western Pacific (Wang and Sun, 1994; Liu et al., 2011), including the East China Sea (ECS) and South China Sea (SCS), further influencing the spatial geographic distribution and historical dynamics of many coastal marine organisms (Hewitt, 2000; Hewitt, 2004). During the glacial period, the shallow continental shelf of the Bohai Sea, Yellow Sea, and ECS was exposed and the coastline migrated about 1200 km from the coast of Bohai Bay to Okinawa Trough (Wang, 1999; Shen et al., 2011), while the SCS formed a semi-enclosed sac-shaped gulf and exposed approximately 0.7 million km2 of continental shelf (Wang and Sun, 1994; Clark et al., 2009). The coastline retreat resulted in the formation of a large land bridge from the Taiwan Strait to the Taiwan-Ryukyu (Kimura, 2000), restricting dispersal routes of marine organisms and isolating populations (Rex, 1997; Alvarado Bremer et al., 2005; Ruban, 2007). The rising sea level during the post-glaciation period resulted in the reconnection of two glacial sea basins (Okinawa Trough and the SCS), providing more habitat and an opportunity for secondary contact between isolated populations. The intermittent isolation and connection of ocean basins during repeated glacial-interglacial cycles also caused pronounced fluctuations in the ocean current pattern, upwelling intensity, and oceanic salinity, which further influenced paleo-productivity (He et al., 2013; Li et al., 2018). Moreover, Yangtze River, Pearl River, and other river systems have the potential to affect paleo-productivity in nearshore coastal waters of the ECS and SCS (Jian et al., 1999a; Jian et al., 1999b). All of these factors could significantly affect the distribution and abundance of marine organisms across the southeast coast of China and may have the profound genetic imprints on their populations (Liu et al., 2007b; He et al., 2014).
Given the substantial geographic variation and complex historical processes in the ECS and SCS, phylogeographic structure and historical demography might differ among taxa in response to coastal environmental changes during glacial periods caused by climate oscillations (Ni et al., 2014). Based on previous research, three evolutionary patterns have been identified for marine organisms along the coast of China: (1) genetic homogeneity with no lineage formation, as observed in the mud crab Scylla paramamosain (He et al., 2010), small yellow croaker Larimichthys polyactis (Wu et al., 2012), and black sea bream Acanthopagrus schlegelii (Zhao et al., 2021); (2) past lineage diversification with a current sympatric distribution and genetic homogeneity, as observed in the large yellow croaker Larimichthys crocea (Wang et al., 2013), whitespotted conger Conger myriaster (Zou et al., 2020), and spotted scat Scatophagus argus (Peng et al., 2021); and (3) past lineage diversification with an allopatric distribution and genetic heterogeneity, as observed in the redlip mullet Chelon haematocheilus (Liu et al., 2007b), lemonfish Plectorhinchus flavomaculatus (Han et al., 2008), mitten crab Eriocheir sensu stricto (Xu et al., 2009), and mottled spinefoot Siganus fuscescens (Ravago-Gotanco and Juinio-Meñez, 2010). Taken together, species-specific phylogeographic patterns and historical dynamics are expected in the ECS and SCS.
The Japanese scad Decapterus maruadsi (Temminck and Schlegel, 1844) is widely distributed across the marginal sea of the western Pacific and is an important economic fish in China, especially in the ECS and the northern SCS (NSCS) (Zheng et al., 2014; Liu et al., 2016). The annual catch of D. maruadsi in the ECS and NSCS reached approximately 5.6 × 105 tons from 1996 through 2020, with a maximum catch of about 6.3 × 105 tons in 2006 (Chinese Fishery Statistical Yearbook, 1997–2021). A recent fishery resources survey has shown that D. maruadsi can nearly always be classified as overfished in the ESC and NSCS, including in the coastal waters of southern Zhejiang (annual exploitation rate = 0.61) (Cui et al., 2020) and the Beibu Gulf (annual exploitation rate = 0.63–0.78) (Geng et al., 2018). In addition, global climate change is fundamentally altering marine environments at an alarming rate (Doney et al., 2009). Continuous overfishing and environmental changes have substantially altered the phenotypic characteristics and population structure of D. maruadsi, including miniaturization, precocious puberty, and structure simplification (Zheng et al., 2014; Geng et al., 2018). Thus, the population structure of D. maruadsi should be closely monitored in the long-term to facilitate the sustainable utilization and scientific management of fishery resources.
According to the migration patterns and ecological characteristics, the ECS D. maruadsi can be assigned to three migratory populations (i.e., West Bank of Kyushu, western ECS, and Minnan to eastern Guangdong) (Zheng et al., 2003), while the NSCS D. maruadsi can be partitioned to seven ecological populations (i.e., Minnan-Taiwan shoal, the area off Jiazi in eastern Guangdong, the Pearl River estuary, the sea off of the Pearl River estuary, western Guangdong, Hainan Qinglan, and Beibu Gulf) (Deng and Zhao, 1991). Additionally, the ECS and NSCS D. maruadsi exhibit different migratory capacities and morphological characters (Jiang et al., 2012; Wang et al., 2021). Accordingly, it is reasonable to speculate that genetic differentiation exists between the ECS and NSCS D. maruadsi if gene flow is overcome by selective pressures and genetic drift. It is, however, noteworthy that the ECS and NSCS are a relatively homogeneous and contiguous environment characterized by the near absence of physical barriers, and ocean currents further increase the connectivity. Long-distance gene flow is possible between the ECS and NSCS D. maruadsi with potential migratory ability, and this is expected to constrain population differentiation. Therefore, the level of genetic differentiation between the ECS and NSCS D. maruadsi is not clear.
Previous population genetic studies of D. maruadsi have revealed high genetic homogeneity between two populations of Fujian coastal waters based on AFLP (Zhang et al., 2010) and mitochondrial DNA control region (CR) sequences (Niu et al., 2012) and among 11 populations in the NSCS based on cytochrome b (Cyt b) (Niu et al., 2018) and CR (Niu et al., 2019). Notably, Jamaludin et al. (2020) proposed that D. maruadsi from the central Indo-West Pacific could be divided into two distinct genetic clades (Northern Vietnam and Sundaland-Rosario-Ranong) based on Cyt b (Figure 1A). The Northern Vietnam clade contained two northern Vietnam populations (Cat Ba and Nghe An), the Sundaland-Rosario-Ranong clade consisted of nine Sundaland populations, and the central Vietnam population (Khanh Hoa) was an admixed group. However, the genetic distance (0.029–0.051) between the Northern Vietnam clade and Sundaland-Rosario-Ranong clade was significantly higher than that among populations within clades (0.001–0.003) (Jamaludin et al., 2020), emphasizing the need for further molecular studies to validate these two genetic clades. Furthermore, no study to date has directly evaluated differences between populations from Chinese coastal waters and the central part of the Indo-West Pacific region.
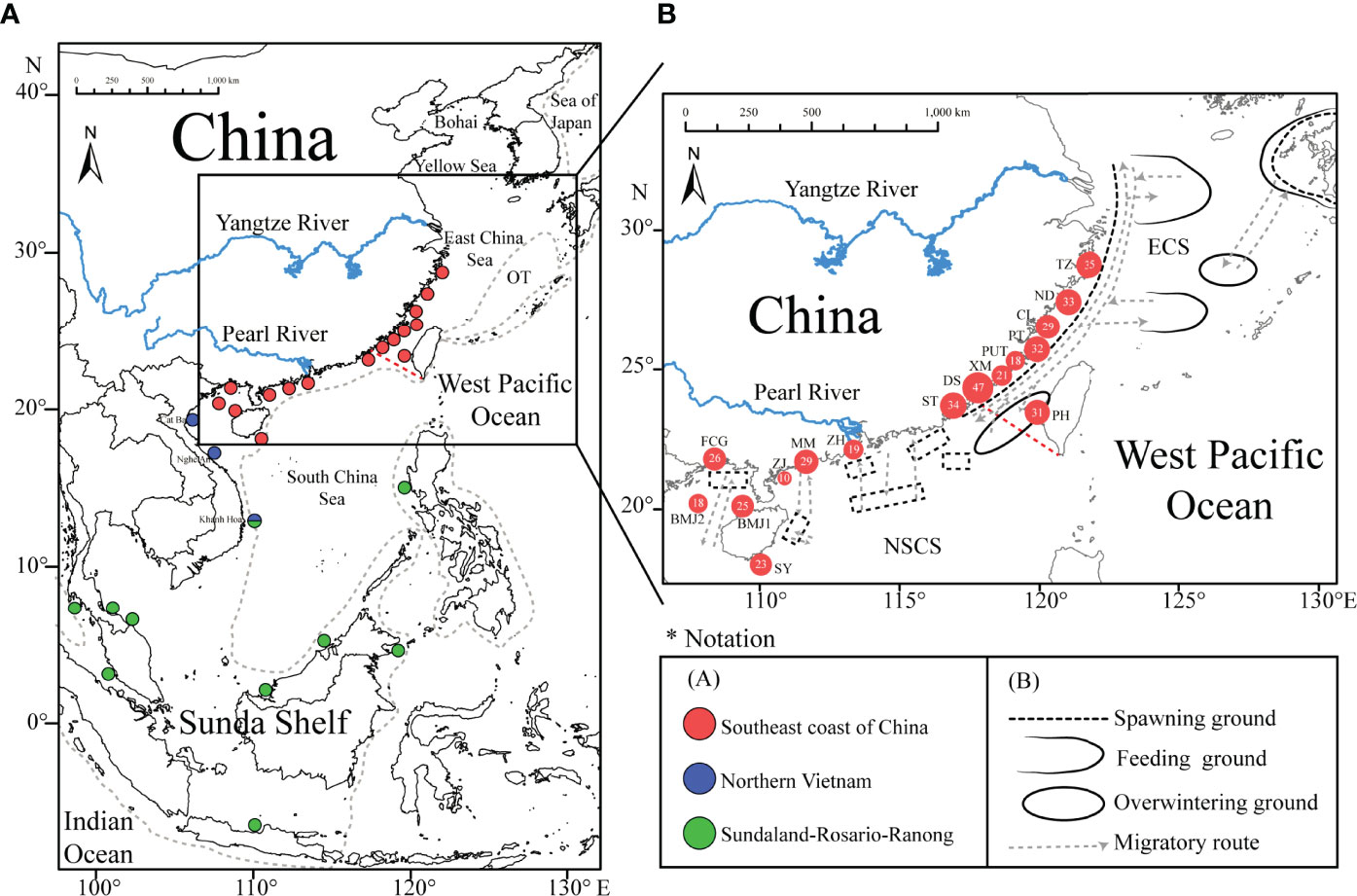
Figure 1 (A) Geographic distribution of Decapterus maruadsi populations used in this study (Niu et al., 2018; Niu et al., 2019; Jamaludin et al., 2020). The shoreline of the LGM (-120 m) is indicated by a grey dashed line and the boundary of the East China Sea (ECS) and the South China Sea (SCS) is indicated by a red dashed line. OT, Okinawa Trough. (B) Sampling locations of 430 D. maruadsi individuals along the southeast coast of China. Populations are marked by abbreviations and the size of each population’s circle indicates the number of obtained individuals. Spawning ground, feeding ground, overwintering ground, and migratory capacity of D. maruadsi are shown (Deng and Zhao, 1991; Zheng et al., 2003).
In this study, we use Cyt b and CR sequences to investigate the phylogeographic structure and demographic history of D. maruadsi in the ECS and NSCS. The goals of the study were (a) to determine whether repeated glacial-interglacial climate cycles during the Pleistocene influenced the phylogeographic structure of D. maruadsi, (b) to characterize demographic history, and (c) to evaluate the molecular phylogenetic relationship between the Northern Vietnam clade and Sundaland-Rosario-Ranong clade defined by Jamaludin et al. (2020). Our results provide insight into the evolutionary history of D. maruadsi along the coastal areas of the ECS and NSCS, which is critical for species conservation.
Materials and Methods
Ethics Statement
The experimental animal protocols in the present study were reviewed and approved by the Animal Experimental Ethics Committee of Guangdong Ocean University, China.
ECS Sample Collection and Sequencing
A total of 246 D. maruadsi individuals were collected from eight geographic populations (Taizhou, Ningde, Changle, Pingtan, Putian, Xiamen, Dongshan, and Penghu) in the ECS from July 2013 to September 2018 (Figure 1 and Table 1). After morphological identification, muscle tissues were obtained from individual fish and preserved in 95% ethanol at -20°C until DNA extraction.
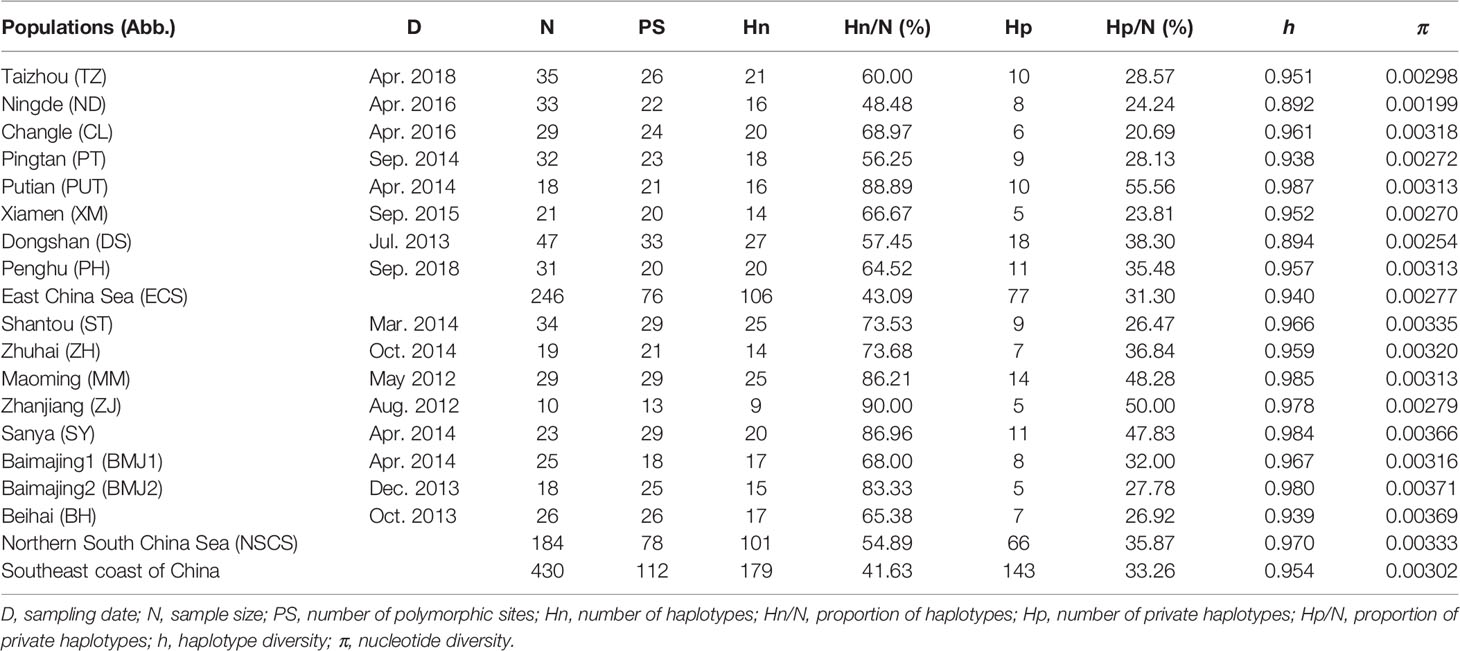
Table 1 Sampling information and genetic diversity indices of Decapterus maruadsi based on concatenated sequences (Cyt b + CR).
Total genomic DNA was extracted from muscle tissues using a standard phenol-chloroform extraction protocol (Huang et al., 2002). PCRs were implemented to amplify partial fragments of mitochondrial Cyt b and CR sequences using previously described primers, L14724 (5′-GAC TTG AAA AAC CAC CGT-3′) and H15915 (5′-CTC CGA TCT CCG GAT TTAC AAG AC-3′) for Cyt b (Xiao et al., 2001) and LF (5′-AAC TCC CAA AGC TAG GAT TCT-3′) and LR (5′-TTT GTG CTT GCG GGG CTT T-3′) for CR (Niu et al., 2012). PCR amplification was carried out in a reaction volume of 50 μL, including 39.5 μL of ddH2O, 5 μL of 10× EasyTaq buffer (containing Mg2+), 200 nM each primer, 200 μM dNTPs, 2.5 U of Taq DNA polymerase (TransGen Biotech Co., Ltd., Beijing, China), and 40–100 ng of template DNA. PCRs of Cyt b and CR were performed according to our previously described procedures (Niu et al., 2018; Niu et al., 2019). Each PCR amplification contained a negative control, which included all reagents except the template DNA. PCR products were separated by electrophoresis on a 1% agarose gel along with the Trans 2K DNA Marker (TransGen Biotech Co., Ltd.) for identifying the target gene size and sent to Sangon Biotech Co. Ltd. (Shanghai, China) for sequencing.
NSCS Samples and Sequence Integration
Cyt b (722 bp) and CR (826–832 bp) sequences of 184 D. maruadsi individuals sampled from eight localities across the NSCS determined in our previous studies were also evaluated (Niu et al., 2018; Niu et al., 2019). Thus, a total of 430 Cyt b and CR sequences of 16 geographic populations from the southeast coast of China were included in analyses. Detailed information on these samples is provided in Figure 1 and Table 1. After sequencing, the sequenced peak maps of all samples firstly were adjusted manually by Chromos software (http://technelysium.com.au/wp/chromas/) to remove the low-quality sequences. Then, partial sequences of Cyt b and CR were manually edited and checked using DNAStar 7.1 (DNASTAR, Inc.). Multiple alignments were generated using ClustalX 1.81 (Thompson et al., 1997). The sequences of Cyt b and CR were manually trimmed to the same length using MEGA 7.0 (Kumar et al., 2016). Sequence alignment found that the trimming Cyt b (722 bp) and CR (826–832 bp) sequences was located at 20–741 base and 14–843 base of their full-length (1141 bp for Cyt b and 843 bp for CR, GenBank accession number: MT818506), respectively. The trimming sequences and their concatenated sequence set (5’-Cyt b + CR-3’) were used for subsequent analyses.
Genetic Diversity and Population Structure Analysis
Population genetic diversity and differentiation among 16 geographic populations (430 individuals) of D. maruadsi collected from the ECS (246 individuals) and the NSCS (184 individuals) were assessed. All analyses described below were performed using Arlequin 3.5 (Excoffier and Lischer, 2010) and considered all sites including insertions/deletions (indels) data. Haplotype diversity (h), nucleotide diversity (π), and other molecular diversity indices were evaluated, while number of haplotypes (Hn) and number of private haplotypes (Hp) were calculated. Pairwise FST values between populations were evaluated with 10,000 permutations and all P values were adjusted by the Benjamini-Hochberg false discovery rate (FDR) (Benjamini and Hochberg, 1995). Analysis of molecular variance (AMOVA) was also performed with 10,000 permutations to characterize the population structure and genetic variation at two hierarchical levels: (1) all populations were classified into one group according to pairwise FST values and (2) 16 populations were assigned to the ECS group and the NSCS group according to sample source to detect differentiation and geographical barriers (e.g., Taiwan Strait).
Phylogenetic Analysis
The evolutionary models for each marker separately and the concatenated sequence set including and excluding outgroups were evaluated by the lowest Bayesian information criterion (BIC) score using jModelTest v2.1.10 (Darriba et al., 2012). Bayesian inference tree was constructed under Hasegawa-Kishino-Yano model (HKY + I for Cyt b, HKY + I + G for CR, and HKY + I + G for Cyt b + CR) using MrBayes v3.2.6 (Ronquist et al., 2012) with three independent runs. Each run included four chains and each chain was run for 100 million generations with a sampling frequency of 100 generations and a diagnostic frequency of 1000 generations until the average standard deviation of split frequencies fell below 0.01 (Hall, 2016). The first 10% of trees were discarded as burn-in while subsequent trees were used to produce a phylogram. The tree was visualized using FigTree (v1.4.4; http://tree.bio.ed.ac.uk/software/figtree/). Furthermore, the haplotype network was generated with the TCS network using PopART 1.7 (Leigh and Bryant, 2015). The net genetic distance was calculated under the Tamura-Nei model (TN93 + G, G = 0.75) model using MEGA 7.0. It is worth mentioning that all sites with insertions/deletions were incorporated in the phylogenetic analysis.
Demographic Analysis
Historical demography of D. maruadsi for the pooled population were inferred using three different approaches. First, we conducted neutrality tests by calculating Tajima’s D (Tajima, 1989a; Tajima, 1989b) and Fu’s Fs (Fu, 1997) with 10,000 permutations using Arlequin 3.5. Second, nucleotide mismatch distributions with 10,000 bootstrap replicates were also implemented in Arlequin 3.5 to detect historical population dynamics (Rogers and Harpending, 1992). The sum of squared deviations (SSD) and Harpending’s raggedness index (Hri) were calculated to examine the departure between observed and expected distributions. The parameter τ gained from the mismatch distribution was used to estimate the time of the population expansion with the equation t = τ/2u, where u represents a neutral mutation rate of the entire sequence in each generation calculated as u = 2μk, where μ is the nucleotide mutation rate and k is the length of the nucleotide sequence (Schneider and Excoffier, 1999). Finally, Bayesian skyline plot (BSP) was implemented under the HKY model (HKY + I for Cyt b, I = 0.76; HKY + I + G for CR, I = 0.84, G = 0.87; and HKY + I + G for Cyt b + CR, I = 0.85, G = 0.86) using BEAST v1.7.5 to identify the changes in effective population size (Ne) over time (Drummond et al., 2012). These analyses were run using the parameters as followed: 1.2 billion generations for Cyt b, 1.3 billion generations for CR, and 1.5 billion generations for Cyt b + CR, respectively. The effective sample sizes (ESS) of all runs were at least 200, and the first 10% were discarded as burn-in. These independent runs were visualized using Tracer v1.7.1 (Rambaut et al., 2018). As the divergence rate of Cyt b reported as 2% per million years (myr) in multiple bony fishes (Bermingham et al., 1997; Bowen et al., 2001; Durand et al., 2002) and that of CR used as 5–10% per myr for D. maruadsi in the previous studies (Niu et al., 2019), we took 1%, 3.75%, and 2.5% per myr as the putative nucleotide mutation rates for Cyt b, CR, and concatenated sequences (Cyt b + CR), respectively.
Reassessment of Molecular Phylogenetic Relationships Between the Northern Vietnam Clade and Sundaland-Rosario-Ranong Clade
To confirm the Northern Vietnam clade and Sundaland-Rosario-Ranong clade defined by Jamaludin et al. (2020) and investigate genetic differentiation between populations from the southeast coast of China and the central part of the Indo-West Pacific region, 80 Cyt b haplotypes (GenBank accession numbers: KX212002–KX212079, KY440109–KY440110) previously identified as D. maruadsi from two northern Vietnam populations (Cat Ba and Nghe An), one central Vietnam population (Khanh Hoa), and nine Sundaland populations were included in a phylogenetic analysis with our Cyt b sequence data. The Cyt b sequences of congeneric species Decapterus russelli from the NSCS (GenBank accession number: OM479430; the sample was collected in our previous fishery survey and was identified when fresh, and the sequence was also uploaded by our research team, ensuring the accuracy of the identification result) and the coastal waters of India (GenBank accession number: MN711693, mitochondrion complete genome), Omani Sohar (GenBank accession number: KX512731) (Damerau et al., 2018), and Indonesian Ambon (GenBank accession number: LC646650) (Kimura et al., 2022), Decapterus macarellus (GenBank accession number: KM986880) (Zou et al., 2016), and Decapterus macrosoma (GenBank accession number: KF841444) (Li et al., 2016) from the SCS were downloaded from NCBI as outgroups. Thus, a collective data set of Cyt b sequences containing 124 sequences of taxa previously identified as D. maruadsi (44 defined in our study plus 80 from NCBI) and six sequences of three congeneric species (outgroups) was formed. These sequences were truncated to target the same Cyt b fragments (606 bp) and were then used to construct a phylogenetic tree under the HKY + G model and TCS network and to calculate net genetic distances under the TN93 + G model (G = 0.29). Other parameters were the same as the methods described above.
Results
Genetic Diversity and Population Structure
A total of 430 Cyt b and CR sequences were obtained from 16 geographic populations along the ECS and the NSCS. Partial Cyt b sequences (722 bp) had 39 polymorphic sites with no insertions/deletions, and partial CR sequences (826–832 bp) contained 73 polymorphic sites and 10 insertions/deletions (Tables S1, S2). These variant sites defined 44 Cyt b haplotypes (CH1–CH44) and 134 CR haplotypes (CR1–CR134), respectively. Two mtDNA fragments were combined to obtain a concatenated sequence of 1548–1554 bp (Cyt b + CR) for each individual for subsequent analyses.
As listed in Table 1, 430 concatenated sequences (Cyt b + CR) had 112 polymorphic sites and 179 haplotypes, designated as H1–H179 (GenBank accession numbers: OM728655–OM728833). Details for all D. maruadsi haplotypes obtained in this study were shown in Table S3. The overall h and π values for the 16 populations were 0.954 and 0.00302, respectively, indicating low levels of genetic diversity in D. maruadsi along the southeast coast of China. Furthermore, high haplotype diversities (0.892–0.987) in concurrence with low nucleotide diversities (0.00199–0.00371) were also detected in each population. The average h and π values for the ECS group were 0.940 and 0.00277, respectively, and those of the NSCS group were 0.970 and 0.00333, respectively, indicating that the level of genetic diversity was lower in the ECS group than in the NSCS group. The proportion of haplotypes and proportion of private haplotypes were also lower in the ECS group (Hn/N, 43.09%; Hp/N, 31.30%) than in the NSCS group (Hn/N, 54.89%; Hp/N, 35.87%). All the results of genetic diversity based on each marker separately (Tables S1, S2) were consistent with concatenated sequences (Cyt b + CR).
Pairwise FST values showed non-significant differentiation (FDR-adjusted P values > 0.05) among 16 geographic populations (Tables 2, S4, S5). The AMOVA with all populations set as a single group revealed that the majority of the molecular variance (> 99.50%) was partitioned within populations, while a minor fraction of variance (< 0.50%) existed among populations (Table 3). When all populations were divided into the ECS group and the NSCS group, more than 99.00% of genetic variation was explained by individuals within populations and only a small fraction of variation (< 1.00%) was attributable to the difference between groups (Table 3). These results indicated that no obvious genetic structure was found among 16 populations or between the ECS group and the NSCS group in D. maruadsi.
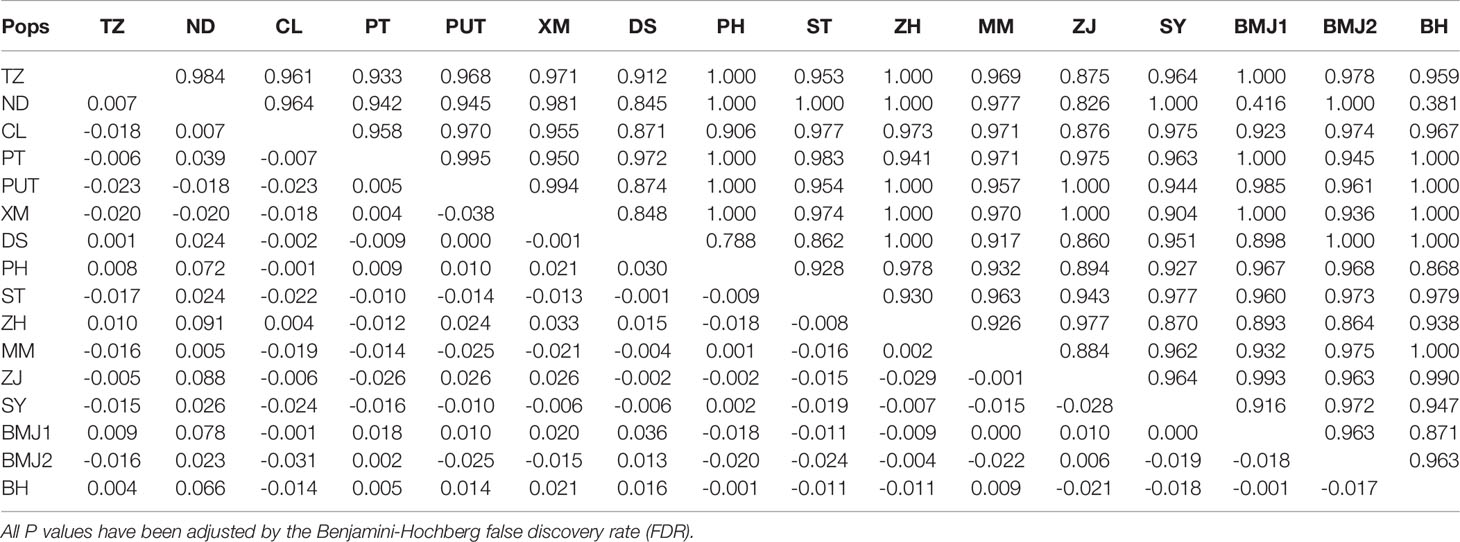
Table 2 Pairwise FST (below diagonal) and FDR-adjusted P values (upper diagonal) between 16 populations of D. maruadsi using concatenated sequences (Cyt b + CR).
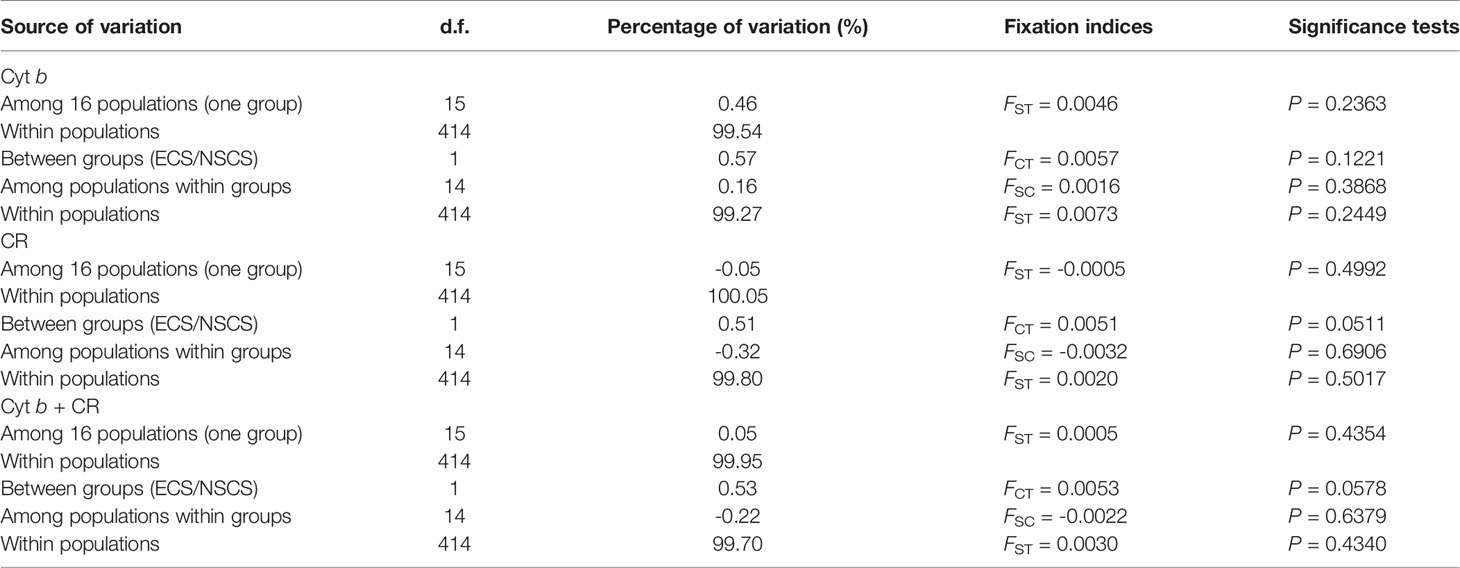
Table 3 Hierarchical analyses of molecular variance (AMOVA) testing genetic subdivision of D. maruadsi populations.
Phylogenetic Relationships
The phylogenetic trees constructed using 44 Cyt b haplotypes (Figure 2A) and 134 CR haplotypes (Figure 2B) revealed that the haplotypes of each population were scattered throughout the trees and no pronounced clusters corresponded to the sampling sites across the southeast coast of China. Interestingly, three main haplogroups (labeled A, B, and C) with low node support values (0.56 and 0.57) were detected in the Bayesian inference tree based on concatenated sequences (Cyt b + CR) (Figure 2C). Haplogroups A, B, and C included 47, 85, and 47 haplotypes comprising 104, 230, and 96 individuals, respectively. Three haplogroups were broadly sympatric across the southeast coast of China and there were no appreciable differences in their frequencies of along the ECS and NSCS. The net genetic distances among three haplogroups were 0.0007–0.0027.
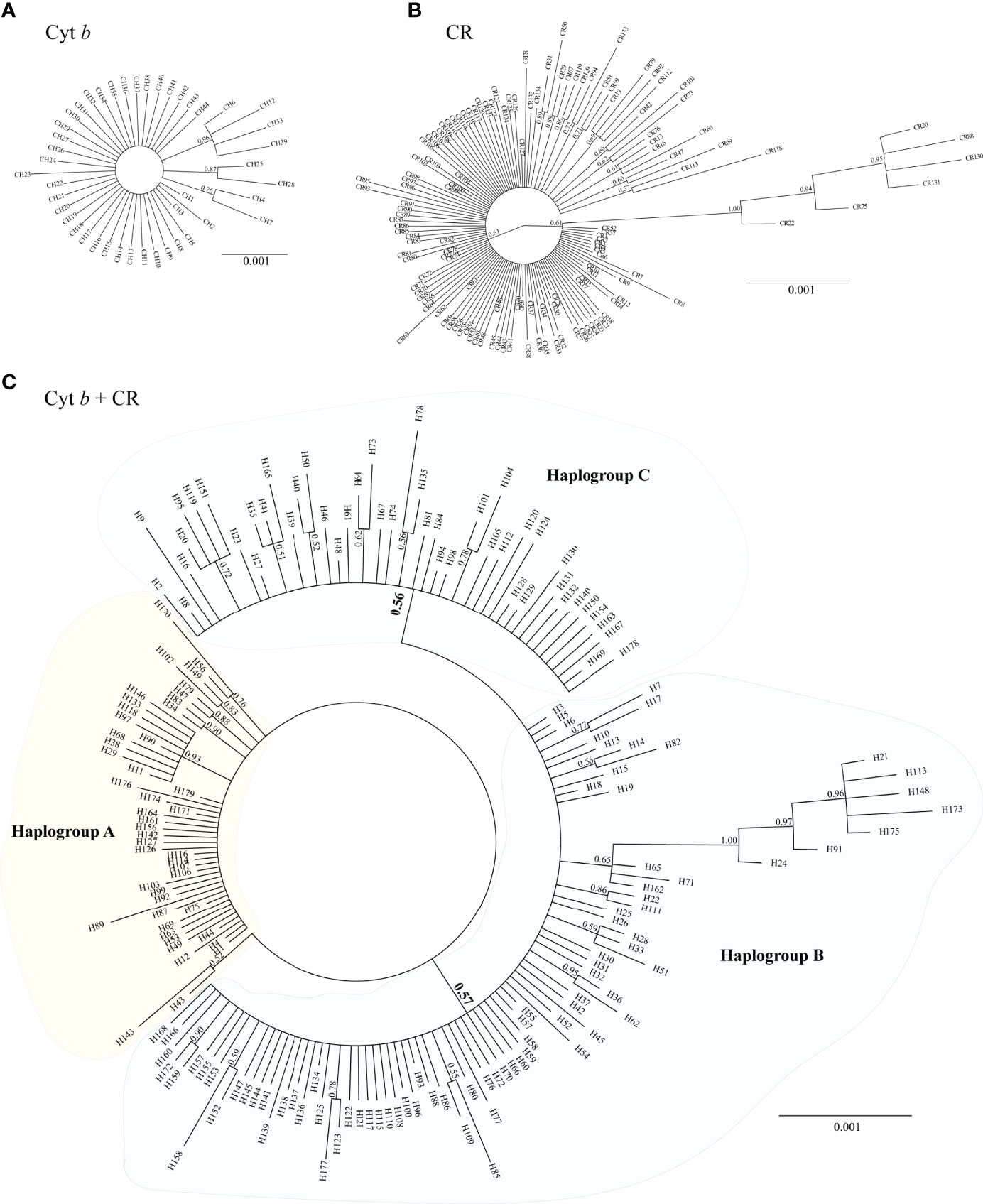
Figure 2 Bayesian inference tree for (A) 44 Cyt b, (B) 134 CR, and (C) 179 Cyt b + CR haplotypes of D. maruadsi. Numbers close to branches indicate posterior probability values above 0.5.
The TCS networks constructed using Cyt b, CR, and concatenated sequences (Cyt b + CR) all showed star-like topologies structure (Figure 3), which contained three (CH1, 15.12%; CH3, 66.98%; and CH6, 4.19%), five (CR1, 7.91%; CR2, 10.93%; CR3, 8.37%; CR5, 19.07%; and CR28, 3.72%), and five (H1, 6.51%; H3, 7.21%; H5, 16.05%; H8, 9.07%; and H55, 2.23%) dominant haplotypes at high frequencies, respectively. These high-frequency haplotypes were distributed throughout most of the sampling area and were located in the centre of the haplotype networks. All other low-frequency haplotypes were separated from these dominant haplotypes by only one or low number of mutational steps. Overall, phylogenetic analysis further indicated that there was no obvious genetic structure but a highly diverse within the species D. maruadsi across the southeast coast of China.
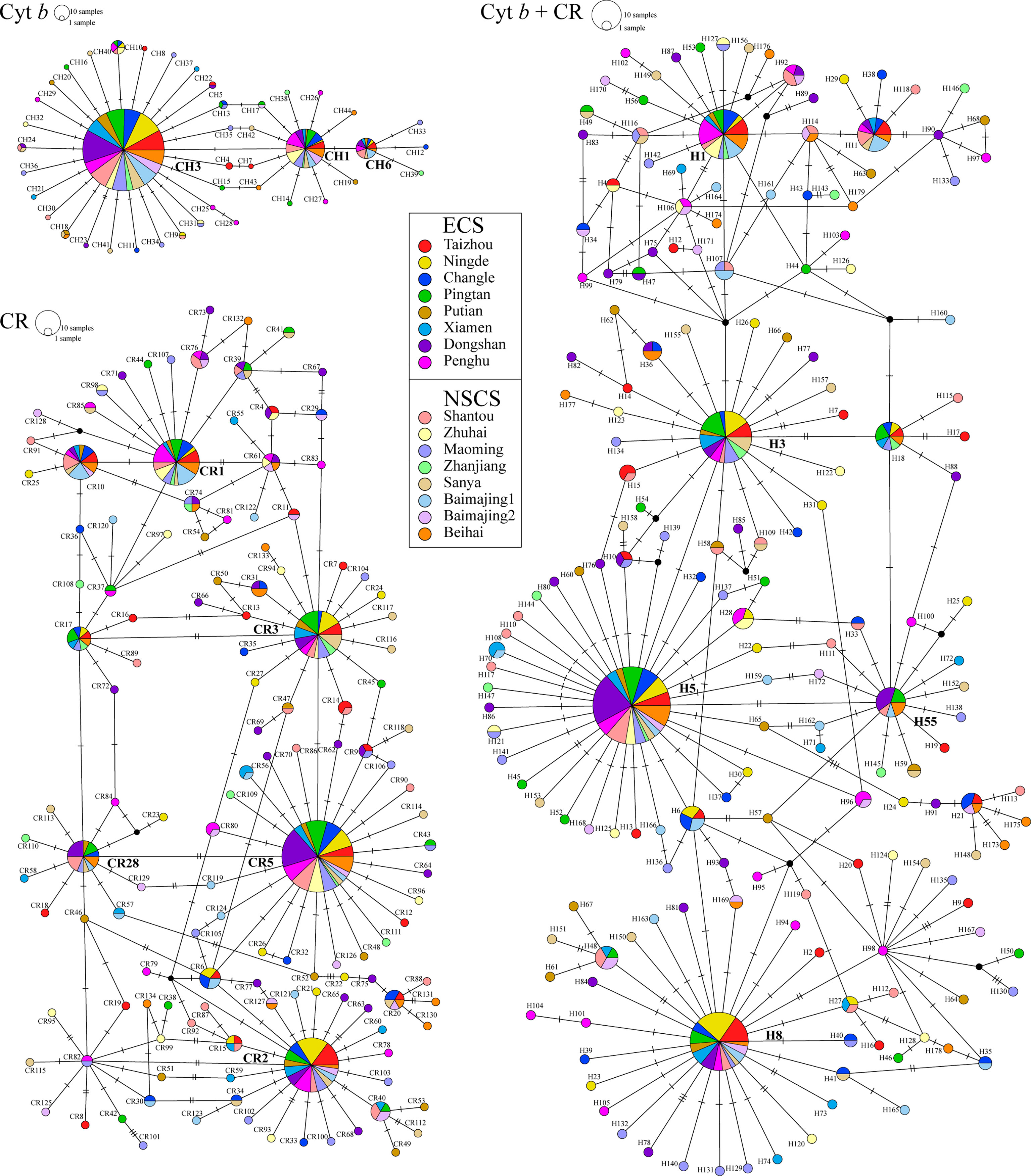
Figure 3 Unrooted TCS haplotype network for Cyt b, CR, and concatenated sequences (Cyt b + CR) haplotypes of D. maruadsi.
Demographic History
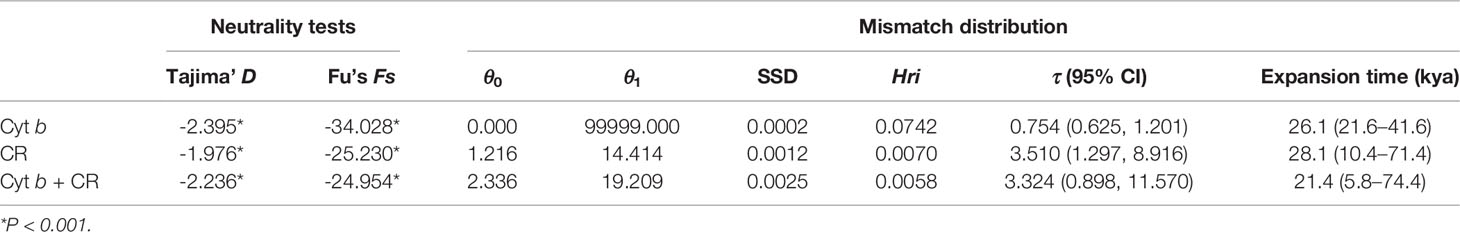
Tajima’s D and Fu’s FS values were highly significantly negative for the pooled population (Table 4), suggesting that there were population expansion events for D. maruadsi along the southeast coast of China. The nucleotide mismatch distributions of the pooled population were unimodal (Figure 4), further indicating a demographic expansion of D. maruadsi population. Both SSD and Hri values were low and not significant (Table 4), demonstrating a lack of significant deviation between the observed and expected distributions and supporting sudden expansion models. Based on the expansion parameter τ and the putative nucleotide mutation rates (1%, 3.75%, and 2.5% per myr for Cyt b, CR, and the concatenated sequences, respectively), the expansion times of D. maruadsi population were dated back to the Late Pleistocene to Holocene period (Table 4).
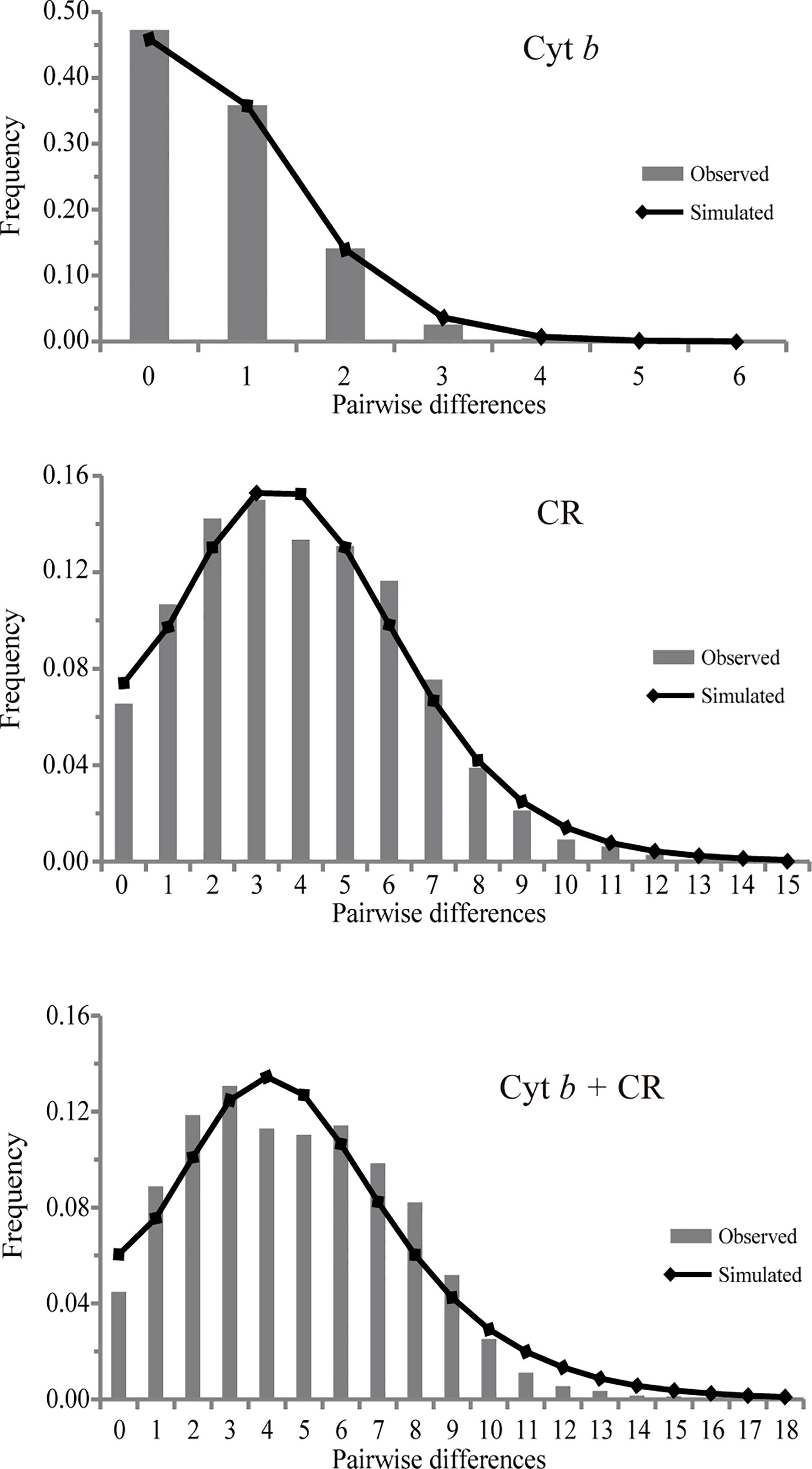
Figure 4 Mismatch distribution for the pooled population of D. maruadsi based on Cyt b, CR, and concatenated sequences (Cyt b + CR).
Bayesian skyline plots revealed a more elaborate demographic history of D. maruadsi along the southeast coast of China (Figure 5). The pooled population showed no pronounced demographic changes from 183–262 kya (coalescence times) to 18–20 kya and experienced a rapid population expansion from 18-20 kya to 3 kya (3–20 kya for Cyt b and Cyt b + CR, 3–18 kya for CR). From 3 kya to present, the effective population size remained relatively stable for the pooled population. After expansion, the effective population sizes of D. maruadsi based on Cyt b, CR, and Cyt b + CR increased about 550-, 350-, and 500-fold, respectively.
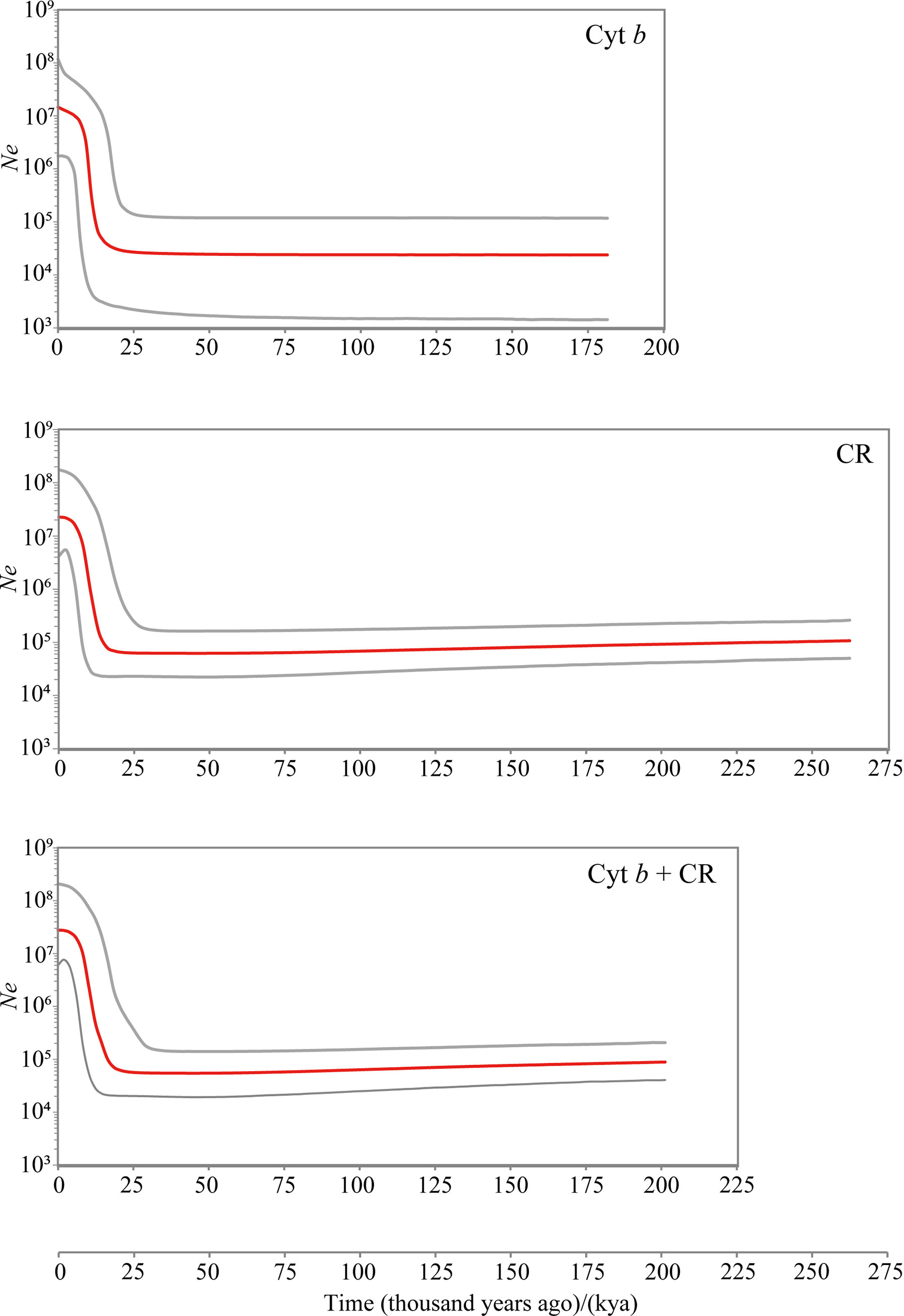
Figure 5 Bayesian skyline plots (BSP) for the pooled population of D. maruadsi based on Cyt b, CR, and concatenated sequences (Cyt b + CR). Intermediate red line represents the median of effective population size (Ne), and the gray lines indicate the upper and lower 95% highest posterior density (HPD) interval.
Molecular Phylogenetic Relationships of the Northern Vietnam Clade and Sundaland-Rosario-Ranong Clade
Phylogenetic relationships between the Northern Vietnam clade and Sundaland-Rosario-Ranong clade were further analyzed based on a collective Cyt b data set containing 124 sequences (606 bp) of named D. maruadsi and six sequences of three outgroup taxa (D. russelli, D. macarellus, and D. macrosoma). As illustrated in Figure 6A, the phylogenetic tree revealed two distinct clades (labeled I and II) with high node confidence values (1.00). The TCS network (Figure 6B) also showed two divergent clades (labeled I and II) separated by a number of mutational steps (25 steps), corresponding to the clades in the phylogenetic tree. Within clade I, 44 D. maruadsi haplotypes defined in this study (labeled red, CH1–CH44) clustered together with 23 haplotypes from two northern Vietnam populations (Cat Ba and Nghe An) and one central Vietnam population (Khanh Hoa) (blue mark, GenBank accession numbers: KX212002, KX212004, KX212051–KX212058, KX212060–KX212065, KX212068–KX212069, and KX212072–KX212076). Interestingly, within clade II, 57 Cyt b haplotypes from nine Sundaland populations and Khanh Hoa population (green mark, GenBank accession numbers: KX212003, KX212005–KX212050, KX212059, KX212066–KX212067, KX212070–KX212071, KX212077–KX212079, and KY440109–KY440110) previously identified as D. maruadsi clustered together with four D. russelli haplotypes (outgroups) from the NSCS and the coastal waters of India, Omani Sohar, and Indonesian Ambon (brown mark, GenBank accession numbers: OM479430, MN711693, KX512731, and LC646650, respectively). The net genetic distances based on Cyt b were as follows: clade I/II, 0.0594; clade I/D. macarellus, 0.1420; clade I/D. macrosoma, 0.1945; clade II/D. macarellus, 0.1256; clade II/D. macrosoma, 0.1573; and D. macarellus/D. macrosoma, 0.1350. However, the net genetic distances within clade I and II were 0.0042 and 0.0087, respectively, which were much lower than those between clades or species mentioned above. Based on these results, we conclude that Cyt b haplotypes within clade II belong to D. russelli, rather than D. maruadsi. In addition, we did not detect significant genetic differentiation among populations from Chinese coastal waters and three Vietnam populations within clade I.
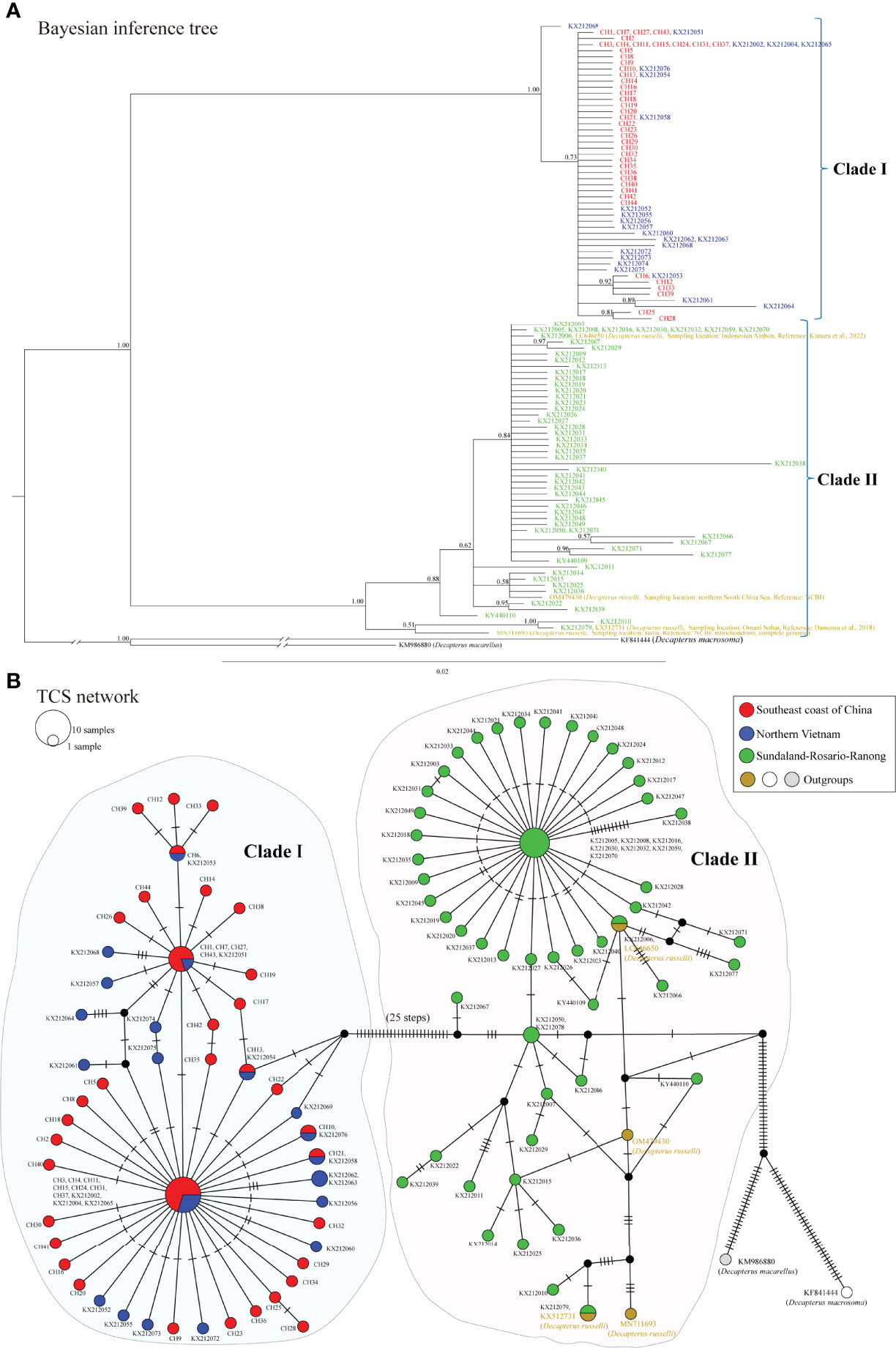
Figure 6 Bayesian inference tree (A) and TCS haplotype network (B) constructed using 124 Cyt b haplotypes (606 bp) of named D. maruadsi from the southeast coast of China, northern Vietnam, and Sundaland-Rosario-Ranong. The congeneric species Decapterus russelli (GenBank accession numbers: OM479430, KX512731, LC646650, and MN711693), Decapterus macarellus (GenBank accession number: KM986880), and Decapterus macrosoma (GenBank accession number: KF841444) Cyt b sequences were used as outgroups. Numbers near branches show posterior probability values above 0.5.
Discussion
Demographic Expansion
Neutrality tests, mismatch distribution, BSP analysis, and haplotype network all indicated a recent demographic expansion event for D. maruadsi along the southeast coast of China since the last glacial period, which may be related to a series of unique oceanographic and ecological changes from the Late Pleistocene to Holocene. During the glaciation, the East Asian monsoon prevailed over the southeast coast of China, leading to strong sea water mixing, improving shallow and mid-water nutrient transport, and accelerating nutrient recycling in the marine ecosystem (Steinke et al., 2011; Huang and Tian, 2012). These changes eventually resulted in a substantial increase in primary productivity. Based on Marine Isotope Stages (MIS), previous studies have shown that the mass accumulation rates (MARs) of total phytoplankton in the NSCS were 62.5 × 10-12 mg·cm-2·kyr-1 and 109.5 × 10-12 mg·cm-2·kyr-1 in MIS 3 (25–59 kya) and MIS 2 (11–25 kya), respectively (He et al., 2013), strongly indicating that paleo-productivity was significantly higher during MIS 2 than during past periods. Also, the total content of all marine phytoplankton biomarkers, used as a total productivity indicator, revealed a higher productivity from the ESC during MIS 2–3 (11–59 kya) than MIS 4 (59–73 kya) (Xing et al., 2008; Vats et al., 2020; Vats et al., 2021). Therefore, it is postulated that the expansion of D. maruadsi from the Last Glacial Maximum (LGM) to the early Holocene is related to increased primary productivity. After the LGM, sea levels have risen 120–140 m (Lambeck et al., 2002; Waelbroeck et al., 2002), providing substantial niche space for many marine organisms (including D. maruadsi) and accommodating extensive terrigenous nutrients rapidly from the shallow continental shelves of Chinese coastal waters. The sediment from the estuarian regions of the Pearl River and Yangtze River also showed that high levels of terrigenous nutrients were transported into the ECS and the NSCS ecosystems through the sources of fluvial runoff during deglaciation (Jian et al., 1999b; Liu et al., 2007a; Qu and Huang, 2019; Vats et al., 2020). These factors further explain the rapid growth of D. maruadsi from the deglaciation to the early Holocene. Over thousands of years of rapid growth, the effective population size of D. maruadsi increased approximately 350- to 550-fold (each marker separately and the concatenated sequences for the pooled population). Along with the increase in the effective population size, intraspecific competition should theoretically increase after the early Holocene. Moreover, the MARs of total phytoplankton in the NSCS were 22.2 × 10-12 mg·cm-2·kyr-1 in MIS 1 (0–11 kya) (He et al., 2013), revealing a decrease in paleo-productivity during the Holocene. Also, compared with the level of paleo-productivity from the LGM to early Holocene, biomarkers showed that for the ECS, total productivity and individual diatom, dinoflagellate, and coccolithophorid productivity were all lower during the Holocene (Xing et al., 2008; Vats et al., 2020; Vats et al., 2021). The increasing intraspecific competition, along with the decrease of paleo-productivity, may have limited recent D. maruadsi population growth and maintained a relatively stable effective population size within the last few thousand years.
Phylogeographic Structure
The previous studies have demonstrated that the alternation of historical glaciation and deglaciation had profound impacts on phylogeographical structures of many marine fishes in the western Pacific (Liu et al., 2007b; Ni et al., 2014; Gao et al., 2020). During the Late Pleistocene glacial periods, the sea level of the western Pacific dropped approximately 120 m (Lambeck et al., 2002; Waelbroeck et al., 2002), and the shallow coastal continental shelves of ESC and SCS were mostly exposed, leading to a large loss of habitat for D. maruadsi and the formation of a land bridge. Surviving D. maruadsi may be isolated, with separate populations in semi-closed SCS, Okinawa Through, as well as in the additional potential glacial refugia. Long-term glacial geographic isolation may have contributed to the formation of lineages. However, in this study, only Bayesian inference tree based on concatenated sequences (Cyt b + CR) revealed three main haplogroups with low node support values (0.56 and 0.57), which was similar to the phylogeographic pattern of Lateolabrax japonicus (Liu et al., 2006). Pairwise FST values and AMOVA indicated genetic homogeneity with no lineage formation among 16 populations along the southeast coast of China. This point was also supported by the phylogenetic analysis based on Cyt b, CR, and the concatenated sequences (no obvious clustering of haplotypes corresponding to localities was observed). In addition, the phylogenetic tree and haplotype network based on Cyt b sequences showed a lack of differentiation between 16 populations along the southeast coast of China and three populations within the Northern Vietnam clade (belonging to the NSCS) defined by Jamaludin et al. (2020). In general, D. maruadsi along the coast of the ECS and the NSCS exhibited no significant genetic structure. Similar genetic structure has been found in the cutlassfish Trichiurus japonicus (He et al., 2014) and amphibious mudskipper Periophthalmus modestus (He et al., 2015) between the ECS and the NSCS.
Avise et al. (1987) concluded that mtDNA phylogenetic continuities in the absence of current spatial separation could emerge in species having relatively extensive and recent historical interconnections through gene flow. This appears to hold true for D. maruadsi. After the last glaciation, rising sea levels opened the Taiwan Strait, which provided the opportunity for gene flow among D. maruadsi populations along the southeast coast of China. Moreover, D. maruadsi, especially the ECS population, possesses great dispersal capabilities, either as pelagic larvae, juveniles, and/or adults (Zheng et al., 2003). Adult D. maruadsi exhibit highly migratory connectivity across the species range (e.g., overlapping spawning time and spawning ground, long-distance spawning, overwintering, and preying migration), and the planktonic eggs and larvae could travel great distances on many annual or semi-annual ocean currents (Deng and Zhao, 1991; Niu et al., 2019). For instance, the southward currents (i.e., China Coastal Current and northeast monsoon drift) may carry the ECS sourced individuals toward the NSCS, whereas the northward currents (i.e., southwest monsoon drift and Taiwan Warm Current) may carry the NSCS sourced individuals toward the ECS (Li et al., 2000; Qiao, 2012). Previous studies have suggested that if there are no firm and longstanding physical barriers between sampled locations, a high level of gene flow is possible for marine species with high dispersal ability, such as mud crab S. paramamosain (He et al., 2010), small yellow croaker L. polyactis (Wu et al., 2012), Chinese beard eel Cirrhimuraena chinensis Kaup (Li et al., 2014), and black sea bream A. schlegelii (Zhao et al., 2021). In this study, we detected a high level of gene flow among 16 populations, confirming that the dispersal ability of D. maruadsi is an important determinant of the no significant genetic structure among all surveyed populations. One more point is that our BSP analysis showed that D. maruadsi recolonized the exposed continental shelf of the ECS and NSCS from glacial refugia with rising sea levels, which illustrated the species did not reach an equilibrium due to the lack of sufficient evolution time (Slatkin, 1993). Given all that, genetic homogeneity of D. maruadsi along the coast of the ECS and NSCS could mainly be explained by a combination of historical biogeography and the dispersal ability of the species.
Although the genetic differentiation between the ECS and NSCS group was not significant, a relatively higher level of genetic diversity was detected in the NSCS group than in the ECS group and the groups exhibited different migratory capacities and population compositions. The ECS group exhibits annual long-distant migration, including spawning migration, feeding migration, and overwintering migration (Figure 1B) (Zheng et al., 2003). For example, the western ECS population can migrate from the center of Taiwan Strait (24°N) to Jeju Island (30°N) coastal regions (Zheng et al., 2003). However, the NSCS group only shows north-south or deep-shallow short-distant migration across the coastal waters (Figure 1B) (Deng and Zhao, 1991). Furthermore, recently studies have indicated that the dominant body lengths in the ECS group and NSCS group vary, e.g., in the spring, the dominant body lengths for larvae and adults of D. maruadsi in the ECS are 50–110 mm and 200–290 mm (Jiang et al., 2012), while those of the species in the NSCS are 101–110 mm and 181–200 mm (Wang et al., 2021), respectively. Overall, the population genetic structure (genetic homogeneity) of D. maruadsi along the coast of the ECS and NSCS was inconsistent with patterns of genetic diversity and biological characteristics (differences), emphasizing the importance of identifying genetic resources and management units of this species with caution and accounting for this information in the future studies.
Fisheries Management Implications
It is of vital significance to correctly understand the evolutionary history, genetic diversity, population structure, and ecological distribution of a species, in particular of a population with wide distribution, for the management and diversity conservation of important economic fishes (Utter, 1991; Grant et al., 2017). D. maruadsi is widely distributed in the coastal waters of China, and its effective population size may not be small. However, a low levels of genetic diversity was detected in D. maruadsi along the southeast coast of China, especially the ECS population. Moreover, recent fishery resources survey showed that D. maruadsi was once one of the most abundant fishes, but it has suffered a considerable catch reduction in some coastal waters of China, such as Yellow Sea, Bohai, and Beibu Gulf (Chinese Fishery Statistical Yearbook, 1997–2021; Geng et al., 2018), possibly due to overfishing, environmental changes, or other factors. Genetic characterization and current status of D. maruadsi resource indicate that its germplasm resources has presented downdraft phenomena. Therefore, in order to ensure the sustainable utilization of fishery resources, D. maruadsi should be closely monitored in the long-term, and conservation and management of its germplasm resources should also be strengthened to protect the genetic diversity of this fish as much as possible. In this study, D. maruadsi along the coast of the ECS and NSCS exhibited no significant genetic structure, indicating that the ECS and NSCS populations could be considered as a management unit for conservation. But we think that the ECS and NSCS D. maruadsi cannot be entirely classified into a management unit, due to the potential mismatch among genetic structure, genetic diversity, and biological characteristics, and weak genetic evolution signal. Given the maternally inherited characteristics of mtDNA, the population structure of D. maruadsi need to further investigate based on higher resolution nuclear genetic markers, such as microsatellites and single nucleotide polymorphisms, which would be of significant help to determine more accurate and refined management units and then design an effective management policy.
Reidentification of D. russelli and D. maruadsi
Many previous studies showed that Cyt b sequences have sufficient resolution and can effectively distinguish many different group of fishes including Carangidae, which is very similar with that of COI and has been used to identify many related species (Reed et al., 2001; Kartavtsev et al., 2007; Bektas and Belduz, 2008; Xia et al., 2008; Khosravi et al., 2020). Based on phylogenetic analysis of Cyt b sequences, we detected two highly divergent clades (I and II), which were monophyletic with high confidence (1.00). The genetic distance (0.0594) between the two clades was about 14- and 7-fold higher than that within clade I (0.0042) and clade II (0.0087), respectively. In fact, the genetic distances based on Cyt b between the Northern Vietnam clade (corresponding to our clade I) and Sundaland-Rosario-Ranong clade (corresponding to our clade II) (0.0290–0.0510) computed by Jamaludin et al. (2020) were also significantly greater than that among populations within clades (0.001–0.003), consistent with the ten-fold rule between species and genera (Ward et al., 2005), which confirmed that each clade was a valid species. The genetic distances between two clades were fairly comparable to that between D. maruadsi and D. russelli (0.0245 and 0.0310) (Hou et al., 2020; Zhang et al., 2020) and the low values reported between species within the family Carangidae (0.0213–0.1680) (Reed et al., 2001; Bektas and Belduz, 2008) or other marine fishes (0.0370–0.1300) (Johns and Avise, 1998), but were higher than the accepted threshold to delimit species (0.02) according to COI barcode sequences (Hebert et al., 2003). Accordingly, we can infer that the two clades defined by Jamaludin et al. (2020) are actually a cluster of two closely related species within the genus Decapterus, rather than significant geographic structure within D. maruadsi. Furthermore, we confirmed that 23 Cyt b haplotypes of the Northern Vietnam clade recorded in Jamaludin et al. (2020) were indeed D. maruadsi because they clustered together with 430 individuals of D. maruadsi collected in our study from the southeast coast of China to form clade I. However, 57 Cyt b haplotypes of the Sundaland-Rosario-Ranong clade defined by Jamaludin et al. (2020) clustered together with four D. russelli haplotypes (outgroups) and were assigned to clade II, indicating that they are D. russelli rather than D. maruadsi.
Both D. russelli and D. maruadsi are economically important pelagic fish in the family Carangidae, and are often mistakenly identified due to the overlapping distributions and similar morphological characteristics. D. russelli and D. maruadsi are widely distributed along the coastal waters of the Indo-West Pacific and western Pacific, respectively, with extensive overlap in the SCS (Smith-Vaniz, 1984; Liu et al., 2016; FishBase, https://www.fishbase.se/search.php; GBIF, https://www.gbif.org/; TaiBNET, https://taibnet.sinica.edu.tw/). Moreover, most of the countable characteristics, such as dorsal fin, pectoral fin, pelvic fin, anal fin, gill rakes, scute, and vertebrae, largely overlap between D. russelli and D. maruadsi (Table S6) (Cheng and Zheng, 1987; Chen and Zhang, 2015; Wu and Zhong, 2021), making it impossible to distinguish the two species. There are several major morphological diagnostic characteristics to distinguish D. russelli and D. maruadsi, including color of fins, shape of the posterior end of the maxilla, scales before dorsal fin, etc (Table S7). For example, D. maruadsi has a white spot above the anterior part of the second dorsal fin but D. russelli does not; the caudal fins, dorsal fin, and pectoral fin of the latter are yellow-orange or scarlet, while those of the former are blue-gray or silver (Table S7) (Cheng and Zheng, 1987; Chen and Zhang, 2015; Wu and Zhong, 2021). However, these different diagnostic characteristics may become difficult to distinguish due to long distance transport and prolonged oxidation, which may result in species misidentification. Collectively, the accurate identification of D. russelli and D. maruadsi based on only morphology is difficult, and future studies should therefore pay more attention to this issue.
Conclusions
The present study reveals that Late Pleistocene-Holocene climate fluctuations had profound impacts on the population dynamics and phylogeographic structure of D. maruadsi in the ECS and NSCS. With respect to historical dynamics, D. maruadsi experienced recent population expansions due to increasing paleo-productivity and niche space during the LGM to early Holocene. Concerning genetic structure, there was no significant differentiation among populations from the ECS and the NSCS, which probably reflected widespread and recent historical interconnections during the post-glaciation. It is noteworthy that the population genetic structure (genetic homogeneity) of D. maruadsi was inconsistent with genetic diversity and biological characteristics, suggesting that it is necessary to identify genetic resources and management units carefully. However, further studies with larger sample range and more molecular markers are required to resolve the apparent mismatch among population genetic structure, genetic diversity, and biological characteristics, which would be of significant help to determine more accurate and refined population structure and management units. In addition, 57 Cyt b haplotypes of the Sundaland-Rosario-Ranong clade should belong to the species D. russelli rather than D. maruadsi, suggesting that morphological characters are insufficient for accurate identification of the two fishes. Overall, this study will provide useful reference and genetic information for supervision of germplasm resources, formulation of effective management policy, and taxonomic identification and evolutionary relationship research of species in the genus Decapterus.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, OM728655–OM728833.
Ethics Statement
The animal study was reviewed and approved by Animal Experimental Ethics Committee of Guangdong Ocean University, China.
Author Contributions
Q-HW, S-FN, and R-XW: conceptualization, validation, resources, writing original draft preparation, writing review and editing, and visualization. Q-HW and R-XW: methodology and software and data curation. Q-HW and S-FN: formal analysis. Q-HW, Z-LL, YZ, MH, and BL: investigation. S-FN and R-XW: supervision, project administration, and funding acquisition. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the PhD Start-up Fund of Guangdong Provincial Natural Science Foundation (NO.2016A030310329), the Special Program for Outstanding Young Teachers of Guangdong Ocean University (NO.HDYQ2017002), and the Program for Scientific Research Start-up Funds of Guangdong Ocean University (NO.R17040).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2022.878506/full#supplementary-material
References
Alvarado Bremer J. R., Viñas J., Mejuto J., Ely B., Pla C. (2005). Comparative Phylogeography of Atlantic Bluefin Tuna and Swordfish: The Combined Effects of Vicariance, Secondary Contact, Introgression, and Population Expansion on the Regional Phylogenies of Two Highly Migratory Pelagic Fishes. Mol. Phylogenet. Evol. 36, 169–187. doi: 10.1016/j.ympev.2004.12.011
Avise J. C., Arnold J., Ball R. M., Bermingham E., Lamb T., Neigel J. E., et al (1987). Intraspecific Phylogeography: The Mitochondrial DNA Bridge Between Population Genetics and Systematics. Annu. Rev. Ecol. Syst. 18, 489–522. doi: 10.1146/annurev.es.18.110187.002421
Bektas Y., Belduz A. O. (2008). Molecular Phylogeny of Turkish Trachurus Species (Perciformes: Carangidae) Inferred From Mitochondrial DNA Analyses. J. Fish Biol. 73, 1228–1248. doi: 10.1111/j.1095-8649.2008.01996.x
Benjamini Y., Hochberg Y. (1995). Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc B 57, 289–300. doi: 10.1111/j.2517-6161.1995.tb02031.x
Bermingham E., Mccafferty S. S., Martin A. P. (1997). Molecular Systematics of Fishes (San Diego: Academic Press).
Bowen W., Bass A. L., Rocha L. A., Grant W. S., Robertson D. R. (2001). Phylogeography of the Trumpetfishes (Aulostomus): Ring Species Complex on a Global Scale. Evolution 55, 1029–1039. doi: 10.1554/0014-3820(2001)055[1029:pottar]2.0.co;2
Chinese Fishery Statistical Yearbook (1997–2021a). Bureau of Fisheries, Ministry of Agriculture (Beijing: China Agriculture Press).
Clark P. U., Dyke A. S., Shakun J. D., Carlson A. E., Clark J., Wohlfarth B., et al (2009). The Last Glacial Maximum. Science (New York N. Y.) 325, 710–714. doi: 10.1126/science.1172873
Cui M. Y., Chen W. F., Dai L. B., Ma Q. Y. (2020). Growth Heterogeneity and Natural Mortality of Japanese Scad in Offshore Waters of Southern Zhejiang. J. Fish. Sci. China 27, 1427–1437.
Damerau M., Freese M., Hanel R. (2018). Multi-Gene Phylogeny of Jacks and Pompanos (Carangidae), Including Placement of Monotypic Vadigo Campogramma Glaycos. J. Fish Biol. 92, 190–202. doi: 10.1111/jfb.13509
Darriba D., Taboada G. L., Doallo R., Posada D. (2012). Jmodeltest 2: More Models, New Heuristics and Parallel Computing. Nat. Methods 9, 772. doi: 10.1038/nmeth.2109
Doney S. C., Fabry V. J., Feely R. A., Kleypas J. A. (2009). Ocean Acidification: The Other CO2 Problem. Annu. Rev. Mar. Sci. 1, 169–192. doi: 10.1146/annurev.marine.010908.163834
Drummond A. J., Suchard M. A., Xie D., Rambaut A. (2012). Bayesian Phylogenetics With BEAUti and the BEAST 1.7. Mol. Biol. Evol. 29, 1969–1973. doi: 10.1093/molbev/mss075
Durand J. D., Tsigenopoulos C. S., Unlü E., Berrebi P. (2002). Phylogeny and Biogeography of the Family Cyprinidae in the Middle East Inferred From Cytochrome B DNA-Evolutionary Significance of This Region. Mol. Phylogenet. Evol. 22, 91–100. doi: 10.1006/mpev.2001.1040
Excoffier L., Lischer H. E. (2010). Arlequin Suite Ver 3.5: A New Series of Programs to Perform Population Genetics Analyses Under Linux and Windows. Mol. Ecol. Resour. 10, 564–567. doi: 10.1111/j.1755-0998.2010.02847.x
Fu Y. X. (1997). Statistical Tests of Neutrality of Mutations Against Population Growth, Hitchhiking and Background Selection. Genetics 147, 915–925. doi: 10.1093/genetics/147.2.915
Gao T., Ying Y., Yang Q., Song N., Xiao Y. (2020). The Mitochondrial Markers Provide New Insights Into the Population Demographic History of Coilia Nasus With Two Ecotypes (Anadromous and Freshwater). Front. Mar. Sci. 7. doi: 10.3389/fmars.2020.576161
Geng P., Zhang K., Chen Z. Z., Xu Y. W., Sun M. S. (2018). Interannual Change in Biological Traits and Exploitation Rate of Decapterus Maruadsi in Beibu Gulf. South China Fish. Sci. 14, 1–9.
Grant W. S., Jasper J., Bekkevold D., Adkison M. (2017). Responsible Genetic Approach to Stock Restoration, Sea Ranching and Stock Enhancement of Marine Fishes and Invertebrates. Rev. Fish. Biol. Fish. 27, 615–649. doi: 10.1007/s11160017-9489-7
Han Z. Q., Li Y., Chen G., Gao T. X. (2008). Population Genetic Structure of Coral Reef Species Plectorhinchus Flavomaculatus in South China Sea. Afr. J. Biotechnol. 7, 1774–1781. doi: 10.5897/AJB08.204
Hebert P. D., Cywinska A., Ball S. L., Dewaard J. R. (2003). Biological Identifications Through DNA Barcodes. Proc. R. Soc B Biol. Sci. 270, 313–321. doi: 10.1098/rspb.2002.2218
He L., Mukai T., Hou Chu K., Ma Q., Zhang J. (2015). Biogeographical Role of the Kuroshio Current in the Amphibious Mudskipper Periophthalmus Modestus Indicated by Mitochondrial DNA Data. Sci. Rep. 5, 15645. doi: 10.1038/srep15645
Hewitt G. (2000). The Genetic Legacy of the Quaternary Ice Ages. Nature 405, 907–913. doi: 10.1038/35016000
Hewitt G. (2004). Genetic Consequences of Climatic Oscillations in the Quaternary. Philos. Trans. R. Soc. B Biol. Sci. 359, 183–195. doi: 10.1098/rstb.2003.1388
He L., Zhang A., Weese D., Li S., Li J., Zhang J. (2014). Demographic Response of Cutlassfish (Trichiurus Japonicus and T. Nanhaiensis) to Fluctuating Palaeo-Climate and Regional Oceanographic Conditions in the China Seas. Sci. Rep. 4, 6380. doi: 10.1038/srep06380
He L., Zhang A., Weese D., Zhu C., Jiang C., Qiao Z. (2010). Late Pleistocene Population Expansion of Scylla Paramamosain Along the Coast of China: A Population Dynamic Response to the Last Interglacial Sea Level Highstand. J. Exp. Mar. Biol. Ecol. 385, 20–28. doi: 10.1016/j.jembe.2010.01.019
He J., Zhao M., Wang P., Li L., Li Q. (2013). Changes in Phytoplankton Productivity and Community Structure in the Northern South China Sea During the Past 260 Ka. Palaeogeogr. Palaeoclimatol. Palaeoecol. 392, 312–323. doi: 10.1016/j.palaeo.2013.09.010
Hou G., Wang J., Chen Z., Zhou J., Huang W., Zhang H. (2020). Molecular and Morphological Identification and Seasonal Distribution of Eggs of Four Decapterus Fish Species in the Northern South China Sea: A Key to Conservation of Spawning Ground. Front. Mar. Sci. 7. doi: 10.3389/fmars.2020.590564
Huang E., Tian J. (2012). Sea-Level Rises at Heinrich Stadials of Early Marine Isotope Stage 3: Evidence of Terrigenous N-Alkane Input in the Southern South China Sea. Glob. Planet. Change 94-95, 1–12. doi: 10.1016/j.gloplacha.2012.06.003
Huang P. T., Wang J. X., Zhu H. C., Zhang Z. S., Chen H. P., Fan M., et al (2002). Molecular Cloning: A Laboratory Manual. 3rd edition (Beijing: Science Press).
Jamaludin N. A., Mohd-Arshaad W., Mohd Akib N. A., Zainal Abidin D. H., Nghia N. V., Nor S. M. (2020). Phylogeography of the Japanese Scad, Decapterus Maruadsi (Teleostei; Carangidae) Across the Central Indo-West Pacific: Evidence of Strong Regional Structure and Cryptic Diversity. Mitochondrial DNA A 31, 298–310. doi: 10.1080/24701394.2020.1799996
Jiang R. J., Xu H. X., Jin H. W., Zhou Y. D., He Z. T. (2012). Feeding Habits of Blue Mackerel Scad Decapterus Maruadsi Temminck Et Schlegel in the East China Sea. J. Fish. China 36, 216–227.
Jian Z., Wang L., Kienast M. (1999a). Late Quaternary Surface Paleoproductivity and Variations of the East Asian Monsoon in the South China Sea. Quat. Sci. 3, 2–40.
Jian Z., Wang L., Kienast M., Sarnthein M., Kuhnt W., Lin H., et al (1999b). Benthic Foraminiferal Paleoceanography of the South China Sea Over the Last 40,000 Years. Mar. Geol. 156, 159–186. doi: 10.1016/S0025-3227(98)00177-7
Johns G. C., Avise J. C. (1998). Tests for Ancient Species Flocks Based on Molecular Phylogenetic Appraisals of Sebastes Rockfishes and Other Marine Fishes. Evolution 52, 1135–1146. doi: 10.1111/j.1558-5646.1998.tb01840.x
Kartavtsev Y. P., Park T. J., Vinnikov K. A., Ivankov V. N., Sharina S. N., Lee J. S. (2007). Cytochrome B (Cyt-B) Gene Sequence Analysis in Six Flatfish Species (Teleostei, Pleuronectidae), With Phylogenetic and Taxonomic Insights. Mar. Biol. 152, 757–773. doi: 10.1007/s00227-007-0726-9
Khosravi M., Abdoli A., Ahmadzadeh F., Saberi-Pirooz R., Rylkova K., Kiabi B. H. (2020). Toward a Preliminary Assessment of the Diversity and Origin of Cyprinid Fish Genus Carassius in Iran. J. Appl. Ichthyol. 36, 422–430. doi: 10.1111/jai.14039
Kimura S., Takeuchi S., Yadome T. (2022). Generic Revision of the Species Formerly Belonging to the Genus Carangoides and its Related Genera (Carangiformes: Carangidae). Ichthyol. Res. 1–55. doi: 10.1007/s10228-021-00850-1
Kumar S., Stecher G., Tamura K. (2016). MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Lambeck K., Esat T. M., Potter E. K. (2002). Links Between Climate and Sea Levels for the Past Three Million Years. Nature 419, 199–206. doi: 10.1038/nature01089
Leigh J. W., Bryant D. (2015). Popart: Full-Feature Software for Haplotype Network Construction. Methods Ecol. Evol. 6, 1110–1116. doi: 10.1111/2041-210X.12410
Li D. W., Chang Y. P., Li Q., Zheng L. W., Ding X. D., Kao S. J. (2018). Effect of Sea-Level on Organic Carbon Preservation in the Okinawa Trough Over the Last 91 Kyr. Mar. Geol. 399, 148–157. doi: 10.1016/j.margeo.2018.02.013
Li H., Lin H., Li J., Ding S. (2014). Phylogeography of the Chinese Beard Eel, Cirrhimuraena Chinensis Kaup, Inferred From Mitochondrial DNA: A Range Expansion After the Last Glacial Maximum. Int. J. Mol. Sci. 15, 13564–13577. doi: 10.3390/ijms150813564
Liu J. X., Gao T. X., Wu S. F., Zhang Y. P. (2007b). Pleistocene Isolation in the Northwestern Pacific Marginal Seas and Limited Dispersal in a Marine Fish, Chelon Haematocheilus (Temminck & Schlegel 1845). Mol. Ecol. 16, 275–288. doi: 10.1111/j.1365-294X.2006.03140.x
Liu J. X., Gao T. X., Yokogawa K., Zhang Y. P. (2006). Differential Population Structuring and Demographic History of Two Closely Related Fish Species, Japanese Sea Bass (Lateolabrax Japonicus) and Spotted Sea Bass (Lateolabrax Maculatus) in Northwestern Pacific. Mol. Phylogenet. Evol. 39, 799–811. doi: 10.1016/j.ympev.2006.01.009
Liu J. X., Tatarenkov A., Beacham T. D., Gorbachev V., Wildes S., Avise J. C. (2011). Effects of Pleistocene Climatic Fluctuations on the Phylogeographic and Demographic Histories of Pacific Herring (Clupea Pallasii). Mol. Ecol. 20, 3879–3893. doi: 10.1111/j.1365-294X.2011.05213.x
Liu J. P., Xu K. H., Li A. C., Milliman J. D., Velozzi D. M., Xiao S. B., et al (2007a). Flux and Fate of Yangtze River Sediment Delivered to the East China Sea. Geomorphology 85, 208–224. doi: 10.1016/j.geomorph.2006.03.023
Li M., Zhang P., Sun D., Chen T., Fan J., Zou K., et al (2016). Characterization of the Mitochondrial Genome of the Shortfin Scad Decapterus Macrosoma (Perciformes: Carangidae). Mitochondrial DNA A 27, 82–83. doi: 10.3109/19401736.2013.873910
Li N. S., Zhao S. L., Wasiliev B. (2000). Geology of Marginal Sea in the Northwest Pacific (Harbin: Heilongjiang Education Press).
Ni G., Li Q., Kong L., Yu H. (2014). Comparative Phylogeography in Marginal Seas of the Northwestern Pacific. Mol. Ecol. 23, 534–548. doi: 10.1111/mec.12620
Niu S. F., Su Y. Q., Wang J., Zhang L. Y. (2012). Population Genetic Structure Analysis of Decapterus Maruadsi From Fujian Coastal Waters. J. Xiamen Univ. (Nat. Sci.) 51, 759–766.
Niu S. F., Wu R. X., Zhai Y., Zhang H. R., Li Z. L., Liang Z. B., et al (2019). Demographic History and Population Genetic Analysis of Decapterus Maruadsi From the Northern South China Sea Based on Mitochondrial Control Region Sequence. PeerJ 7, e7953. doi: 10.7717/peerj.7953
Niu S. F., Wu R. X., Zhang L. Y., Zhang H. R., Liang R., Li Z. L. (2018). Genetic Diversity Analysis of Decapterus Maruadsi From Northern South China Sea Based on Mitochondrial DNA Cyt B Sequence. J. Appl. Oceanogr. 37, 263–273.
Peng M., Zhu W., Yang C., Yao J., Chen H., Jiang W., et al (2021). Genetic Diversity of Mitochondrial D-LOOP Sequences in the Spotted Scat (Scatophagus Argus) From Different Geographical Populations Along the Northern Coast of the South China Sea. J. Appl. Ichthyol. 37, 73–82. doi: 10.1111/jai.14121
Qiao F. L. (2012). Regional Oceanography of China Seas—Physical Oceanography (Beijing: China Ocean Press).
Qu H. X., Huang B. Q. (2019). Paleoclimate Change Reflected by Element Ratios of Terrigenous Sediments From Deep-Sea Oxygen Isotope MIS6 to MIS5 at MD12-3432 Station in Northern South China Sea. Earth Sci. Front. 26, 236–242.
Rambaut A., Drummond A. J., Xie D., Baele G., Suchard M. A. (2018). Posterior Summarization in Bayesian Phylogenetics Using Tracer 1.7. Syst. Biol. 67, 901–904. doi: 10.1093/sysbio/syy032
Ravago-Gotanco R. G., Juinio-Meñez M. A. (2010). Phylogeography of the Mottled Spinefoot Siganus Fuscescens: Pleistocene Divergence and Limited Genetic Connectivity Across the Philippine Archipelago. Mol. Ecol. 19, 4520–4534. doi: 10.1111/j.1365-294X.2010.04803.x
Reed D. L., Degravelle M. J., Carpenter K. E. (2001). Molecular Systematics of Selene (Perciformes: Carangidae) Based on Cytochrome B Sequences. Mol. Phylogenet. Evol. 21, 468–475. doi: 10.1006/mpev.2001.1023
Rex M. A. (1997). An Oblique Slant on Deep-Sea Biodiversity. Nature 385, 577–578. doi: 10.1038/385577a0
Rogers A. R., Harpending H. (1992). Population Growth Makes Waves in the Distribution of Pairwise Genetic Differences. Mol. Biol. Evol. 9, 552–569. doi: 10.1093/oxfordjournals.molbev.a040727
Ronquist F., Teslenko M., van der Mark P., Ayres D. L., Darling A., Höhna S., et al (2012). MrBayes 3.2: Efficient Bayesian Phylogenetic Inference and Model Choice Across a Large Model Space. Syst. Biol. 61, 539–542. doi: 10.1093/sysbio/sys029
Ruban D. A. (2007). Jurassic Transgressions and Regressions in the Caucasus (Northern Neotethys Ocean) and Their Influences on the Marine Biodiversity. Palaeogeogr. Palaeoclimatol. Palaeoecol. 251, 422–436. doi: 10.1016/j.palaeo.2007.04.008
Schneider S., Excoffier L. (1999). Estimation of Past Demographic Parameters From the Distribution of Pairwise Differences When the Mutation Rates Vary Among Sites: Application to Human Mitochondrial DNA. Genetics 152, 1079–1089. doi: 10.1093/genetics/152.3.1079
Shen K. N., Jamandre B. W., Hsu C. C., Tzeng W. N., Durand J. D. (2011). Plio-Pleistocene Sea Level and Temperature Fluctuations in the Northwestern Pacific Promoted Speciation in the Globally-Distributed Flathead Mullet Mugil Cephalus. BMC Evol. Biol. 11, 83. doi: 10.1186/1471-2148-11-83
Slatkin M. (1993). Isolation by Distance in Equilibrium and Non-Equilibrium Populations. Evolution 47, 264–279. doi: 10.1111/j.1558-5646.1993.tb01215.x
Smith-Vaniz W. F. (1984). FAO Species Identification Sheets for Fishery Purposes. Western Indian Ocean (Fishing Area 51) (Rome: FAO, United Nations).
Steinke S., Glatz C., Mohtadi M., Groeneveld J., Li Q., Jian Z. (2011). Past Dynamics of the East Asian Monsoon: No Inverse Behaviour Between the Summer and Winter Monsoon During the Holocene. Global Planet. Change 78, 170–177. doi: 10.1016/j.gloplacha.2011.06.006
Tajima F. (1989a). The Effect of Change in Population Size on DNA Polymorphism. Genetics 123, 597–601. doi: 10.1093/genetics/123.3.597
Tajima F. (1989b). Statistical Method for Testing the Neutral Mutation Hypothesis by DNA Polymorphism. Genetics 123, 585–595. doi: 10.1093/genetics/123.3.585
Thompson J. D., Gibson T. J., Plewniak F., Jeanmougin F., Higgins D. G. (1997). The CLUSTAL_X Windows Interface: Flexible Strategies for Multiple Sequence Alignment Aided by Quality Analysis Tools. Nucleic Acids Res. 25, 4876–4882. doi: 10.1093/nar/25.24.4876
Utter F. M. (1991). Biochemical Genetics and Fishery Management: An Historical Perspective. J. Fish Biol. 39, 1–20. doi: 10.1111/j.1095-8649.1991.tb05063.x
Vats N., Mishra S., Singh R. K., Gupta A. K., Pandey D. K. (2020). Paleoceanographic Changes in the East China Sea During the Last ~400 Kyr Reconstructed Using Planktic Foraminifera. Global Planet. Change 189, 103173. doi: 10.1016/j.gloplacha.2020.103173
Vats N., Singh R. K., Das M., Holbourn A., Gupta A. K., Gallagher S. J., et al (2021). Linkages Between East China Sea Deep-Sea Oxygenation and Variability in the East Asian Summer Monsoon and Kuroshio Current Over the Last 400,000 Years. Paleoceanogr. Paleoclimatol. 36, e2021PA004261. doi: 10.1029/2021PA004261
Waelbroeck C., Labeyrie L., Michel E., Duplessy J. C., Mcmanus J. F., Lambeck K., et al (2002). Sea-Level and Deep Water Temperature Changes Derived From Benthic Foraminifera Isotopic Records. Quat. Sci. Rev. 21, 295–305. doi: 10.1016/S0277-3791(01)00101-9
Wang P. (1999). Response of Western Pacific Marginal Seas to Glacial Cycles: Paleoceanographic and Sedimentological Features. Mar. Geol. 156, 5–39. doi: 10.1016/S0025-3227(98)00172-8
Wang K. L., Chen Z. Z., Xu Y. W., Sun M. S., Wang H. H., Cai Y. C., et al (2021). Biological Characteristics of Decapterus Maruadsi in the Northern South China Sea. Mar. Fish. 43, 12–21. doi: 10.3724/SP.J.1004-2490.2021.0102
Wang L., Shi X., Su Y., Meng Z., Lin H. (2013). Genetic Divergence and Historical Demography in the Endangered Large Yellow Croaker Revealed by mtDNA. Biochem. Syst. Ecol. 46, 137–144. doi: 10.1016/j.bse.2012.09.021
Wang P. X., Sun X. J. (1994). Last Glacial Maximum in China: Comparison Between Land and Sea. Catena 23, 341–353. doi: 10.1016/0341-8162(94)90077-9
Ward R. D., Zemlak T. S., Innes B. H., Last P. R., Hebert P. D. (2005). DNA Barcoding Australia's Fish Species. Philos. Trans. R. Soc. Lond. B Biol. Sci. 360, 1847–1857. doi: 10.1098/rstb.2005.1716
Wu R. X., Liang X. H., Zhuang Z. M., Liu S. F. (2012). Mitochondrial COI Sequance Variation of Silver Pomfret (Pampus Argentus) From Chinese Coastal Waters. Zool. Syst. 37, 480–488.
Wu H. L., Zhong J. S. (2021). Key to Marine and Estuarial Fishes of China (Beijing: China Agriculture Press).
Xiao W., Zhang Y., Liu H. (2001). Molecular Systematics of Xenocyprinae (Teleostei: Cyprinidae): Taxonomy, Biogeography, and Coevolution of a Special Group Restricted in East Asia. Mol. Phylogenet. Evol. 18, 163–173. doi: 10.1006/mpev.2000.0879
Xia J., Xia K., Jiang S. (2008). Complete Mitochondrial DNA Sequence of the Yellowfin Seabream Acanthopagrus Latus and a Genomic Comparison Among Closely Related Sparid Species. Mitochondrial DNA A 19, 385–393. doi: 10.1080/19401730802350998
Xing L., Zhao M. X., Zhang H. L., Shi L. X. (2008). Biomarker Reconstruction of Phytoplankton Productivity and Community Structure Changes in the Middle Okinawa Trough During the Last 15 Ka. Chin. Sci. Bull. 53, 2552–2559. doi: 10.1007/s11434-008-0231-7
Xu J., Chan T. Y., Tsang L. M., Chu K. H. (2009). Phylogeography of the Mitten Crab Eriocheir Sensu Stricto in East Asia: Pleistocene Isolation, Population Expansion and Secondary Contact. Mol. Phylogenet. Evol. 52, 45–56. doi: 10.1016/j.ympev.2009.02.007
Zhang L. Y., Su Y., Ding X. D., Niu S. F., Liu D. T., Wang J. (2010). Analysis of Genetic Diversity of Decapterus Maruadsi in the Coastal Waters of Fujian Province. J. Fish. China 34, 680–687. doi: 10.3724/SP.J.1231.2010.06731
Zhang L., Zhang J., Song P., Liu S., Liu P., Liu C., et al (2020). Reidentification of Decapterus Macarellus and D. Macrosoma (Carangidae) Reveals Inconsistencies With Current Morphological Taxonomy in China. Zookeys 995, 81–96. doi: 10.3897/zookeys.995.58092
Zhao L., Shan B., Song N., Gao T. (2021). Genetic Diversity and Population Structure of Acanthopagrus Schlegelii Inferred From mtDNA Sequences. Reg. Stud. Mar. Sci. 41, 101532. doi: 10.1016/j.rsma.2020.101532
Zheng Y. J., Chen X. Z., Cheng J. H., Wang Y. L., Shen X. Q., Chen W. Z., et al (2003). Donghai Dalujia Shengwuziyuan Yu Huanjing (Shanghai: Shanghai Science and Technology Press).
Zheng Y. J., Li J. S., Zhang Q. Y., Hong W. S. (2014). Research Progresses of Resource Biology of Important Marine Pelagic Food Fishes in China. J. Fish. China 38, 149–160.
Zou K., Chen Z., Zhang P., Li M. (2016). Mitochondrial Genome of the Mackerel Scad Decapterus Macarellus (Perciformes: Carangidae). Mitochondrial DNA A 27, 2151–2152. doi: 10.3109/19401736.2014.982601
Keywords: Decapterus maruadsi, demographic history, phylogeographic structure, genetic diversity, species reidentification
Citation: Wang Q-H, Wu R-X, Li Z-L, Niu S-F, Zhai Y, Huang M and Li B (2022) Effects of Late Pleistocene Climatic Fluctuations on the Phylogeographic and Demographic History of Japanese Scad (Decapterus maruadsi). Front. Mar. Sci. 9:878506. doi: 10.3389/fmars.2022.878506
Received: 18 February 2022; Accepted: 16 May 2022;
Published: 15 June 2022.
Edited by:
Christian Marcelo Ibáñez, Andres Bello University, ChileReviewed by:
Pamela Morales, University of Chile, ChileJuan Andrés López, University of Alaska Fairbanks, United States
Copyright © 2022 Wang, Wu, Li, Niu, Zhai, Huang and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Su-Fang Niu, d29sZjA0ODdAMTI2LmNvbQ==
†These authors have contributed equally to this work and share first authorship
 Qing-Hua Wang
Qing-Hua Wang Ren-Xie Wu
Ren-Xie Wu Zhong-Lu Li
Zhong-Lu Li Su-Fang Niu*
Su-Fang Niu*