
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mar. Sci., 04 May 2022
Sec. Coral Reef Research
Volume 9 - 2022 | https://doi.org/10.3389/fmars.2022.869574
This article is part of the Research TopicThe Role of Dimethylsulphide, and other Sulphur Substances, on the Climate and Ecology of Coral ReefsView all 12 articles
 Felicity W.I. Kuek1,2,3,4,5
Felicity W.I. Kuek1,2,3,4,5 Cherie A. Motti1,5†
Cherie A. Motti1,5† Jia Zhang3,4
Jia Zhang3,4 Ira R. Cooke3,4
Ira R. Cooke3,4 Jonathan D. Todd6
Jonathan D. Todd6 David J. Miller2,3,4†
David J. Miller2,3,4† David G. Bourne1,5,7†
David G. Bourne1,5,7† Jean-Baptiste Raina8*†
Jean-Baptiste Raina8*†Dimethylsulfoniopropionate (DMSP) is an important molecule in the marine sulfur cycle, produced in large amounts by corals and their dinoflagellate endosymbionts, Symbiodiniaceae. Although corals are known to harbour bacteria that can catabolise DMSP, the recent discovery of bacteria capable of producing DMSP in coastal and deep-sea environments raises the possibility of a bacterial contribution to the DMSP output of corals. Here, 157 bacteria associated with four common coral species were isolated and screened for their ability to produce DMSP by targeting dsyB, a key gene involved in DMSP biosynthesis. Approximately 9% (14 out of 157) of the bacterial isolates harboured dsyB, all being members of the Alphaproteobacteria. The ability of these isolates to produce DMSP was confirmed by liquid chromatography-mass spectrometry (LC-MS) and nuclear magnetic resonance (NMR) measurements. A dsyB-harbouring strain, Shimia aestuarii AMM-P-2, was selected for genome sequencing. This strain harbours the complete genetic machinery to (i) assimilate sulfate and synthesise the DMSP precursors, cysteine and methionine; (ii) demethylate DMSP and generate methanethiol; (iii) cleave DMSP, generating dimethyl sulfide (DMS) and acrylate; and (iv) utilise or detoxify acrylate. The impacts of varied environmental factors (temperature, salinity, light and UV radiation) on S. aestuarii AMM-P-2 DMSP biosynthesis were characterised. DMSP levels in S. aestuarii AMM-P-2 increased almost two-fold under both hypersaline conditions (40 PSU) and high UV exposure. DMSP catabolism through the cleavage pathway also increased under these conditions, producing the antioxidants DMS and acrylate, a potential response to the oxidative stress generated. Overall, our results reveal that coral-associated bacteria can synthesize DMSP and may therefore contribute to DMSP production by the coral holobiont.
Dimethylsulfoniopropionate (DMSP) is one of the most abundant organic sulfur compounds in the ocean (Sievert et al., 2007; Johnston et al., 2012), with global production estimates ranging between 12 and 103 Tmol of sulfur per year (Howard et al., 2006). Functionally speaking, this molecule is the chemical equivalent of a Swiss Army knife, being a key source of carbon and reduced-sulfur for marine microbes (Kiene et al., 2000), a potent chemoattractant affecting the behaviour of organisms ranging from bacteria to fish (DeBose et al., 2008; Seymour et al., 2010), an antioxidant (Sunda et al., 2002), a cryoprotectant (Karsten et al., 1996), a protectant against hydrostatic pressure (Zheng et al., 2020), and an osmolyte (Kirst, 1996; Stefels, 2000). In addition, its role as the main precursor of dimethyl sulfide (DMS) has received considerable attention because this highly abundant gaseous compound is released into the atmosphere where it ultimately plays a role in cloud formation, bridging marine and atmospheric sulfur cycles (Andreae et al., 1983; Bates et al., 1987; Ayers and Gras, 1991).
DMSP biosynthesis was long thought to be restricted to marine photosynthetic eukaryotes (Kiene et al., 1996). However, recent studies have demonstrated that photosynthesis is not a prerequisite for DMSP production with marine heterotrophic bacteria found in saltmarshes, the photic zone and even deep seafloor sediments producing this compound (Curson et al., 2017; Williams et al., 2019). Many marine Alphaproteobacteria belonging to the orders Rhodobacterales, Rhizobiales, and Rhodospirillales produce DMSP via the methionine transamination pathway (Curson et al., 2017). The dysB gene in these bacteria encodes the key S-adenosyl methionine-dependent methylthiohydroxybutryrate (MTHB) methyltransferase enzyme in this pathway. Furthermore, some Alphaproteobacteria and Actinobacteria have been shown to produce DMSP via a different methionine methylation pathway in which a methionine S-methyltransferase, termed MmtN, is an important enzyme (Williams et al., 2019). Approximately 0.3–0.6% of marine bacteria are predicted to produce DMSP, with the majority containing dsyB (Curson et al., 2017; Curson et al., 2018; Williams et al., 2019). Importantly, Alphaproteobacteria orders that are known to harbour dsyB can represent up to 50% of the bacterial communities associated with some reef-building coral species (Luo et al., 2021).
Coral reefs are recognised as DMSP hotspots (Jones and Trevena, 2005), thought to largely result from the photosynthetic endosymbionts of the family Symbiodiniaceae within the coral tissue that produce large amounts of the compound (Hill et al., 1995). However, the coral host itself can also produce DMSP and contain a DsyB-like MTHB S-methyltransferase, termed DSYB (Raina et al., 2013; Aguilar et al., 2017; Curson et al., 2018). Bacteria are also abundant in and around corals (Bourne et al., 2016; Pogoreutz et al., 2020) and, together with protists, fungi, archaea, and viruses, they form a metaorganism referred to as the coral holobiont (Rohwer et al., 2002). These microorganisms likely support central metabolic processes of the coral host and their microalgal partners through the fixation of carbon, cycling of nitrogen, synthesis of essential B-vitamins, and antimicrobial production (Rädecker et al., 2015; Robbins et al., 2019; Matthews et al., 2020; Ngugi et al., 2020). A large proportion of coral-associated bacteria can also catabolise DMSP, and this compound constitutes an important source of carbon and reduced sulfur for microorganisms in the holobiont (Raina et al., 2009; Raina et al., 2010; Frade et al., 2016; Tandon et al., 2020). Yet, the potential contribution of coral-associated bacteria to the production of the high DMSP concentrations measured in some reef-building corals has never been considered.
Here, bacteria associated with four common species of reef-building corals from the Great Barrier Reef (GBR) were investigated. We hypothesized that these corals harbour DMSP-producing bacteria that may contribute to DMSP concentrations measured in the holobiont. Bacterial isolates were screened for the presence of the dsyB gene, and their capacity to produce DMSP was investigated through chemical analyses. A representative DMSP-producing coral-associated bacterial isolate was exposed to abiotic stressors relevant for coral reefs and the role environmental factors may play in driving DMSP production characterised.
Coral-associated bacteria were isolated from four scleractinian coral species, Acropora millepora, Acropora tenuis, Pocillopora acuta, and Stylophora pistillata, all collected from Davies Reef (18°49’03.7”S, 147°38’39.6”E). Corals were maintained at the Australian Institute of Marine Science (AIMS) National Sea Simulator (SeaSim) and were healthy when the samples were collected, with no visual signs of bleaching or disease. Five coral fragments per colony were rinsed in sterile artificial sea water (ASW) prior to mucus collection using sterile 50 mL syringes fitted with 20-gauge hypodermic needles. Mucus samples were kept on ice and processed within an hour. In addition, two coral fragments per colony were placed in separate Whirl-Pak sterile sample bags (Nasco, United States) and immediately air-brushed with 5 mL of sterile ASW to remove coral tissue and their associated microorganisms from the coral skeleton. The tissue slurry was homogenised and transferred into sterile 50 mL centrifuge tubes, placed on ice, and processed within an hour.
To isolate coral-associated bacteria, each sample type (mucus and tissue slurry) was serially diluted in ASW (2-, 10-, 100-, and 1,000-fold; Figure 1A). Aliquots (50 µL) of each dilution were then spread onto Difco Marine Agar 2216 (MA; Becton Dickinson, United States) or modified minimal basal medium (MBM) agar enriched with mixed carbon sources [300 mM; details in Table S1; (Curson et al., 2017)], methionine (C5H11NO2S; 0.5 mM), and ammonium chloride, (NH4Cl; 20 mM) as nitrogen source. All agar plates were incubated at 28°C in the dark for one week and inspected daily for growth and the formation of morphologically distinct individual colonies. Colonies were picked using sterile 20 µL pipette tips and resuspended in 5 mL of Difco Marine Broth 2216 (MB; Becton Dickinson, United States). The isolates were incubated at 28°C and 180 RPM until growth was visible. These liquid cultures were replated on MA and this procedure repeated until pure isolates were obtained. Isolates were then cultured in MB and aliquots of each isolate stored in 20% v/v glycerol at -80°C.
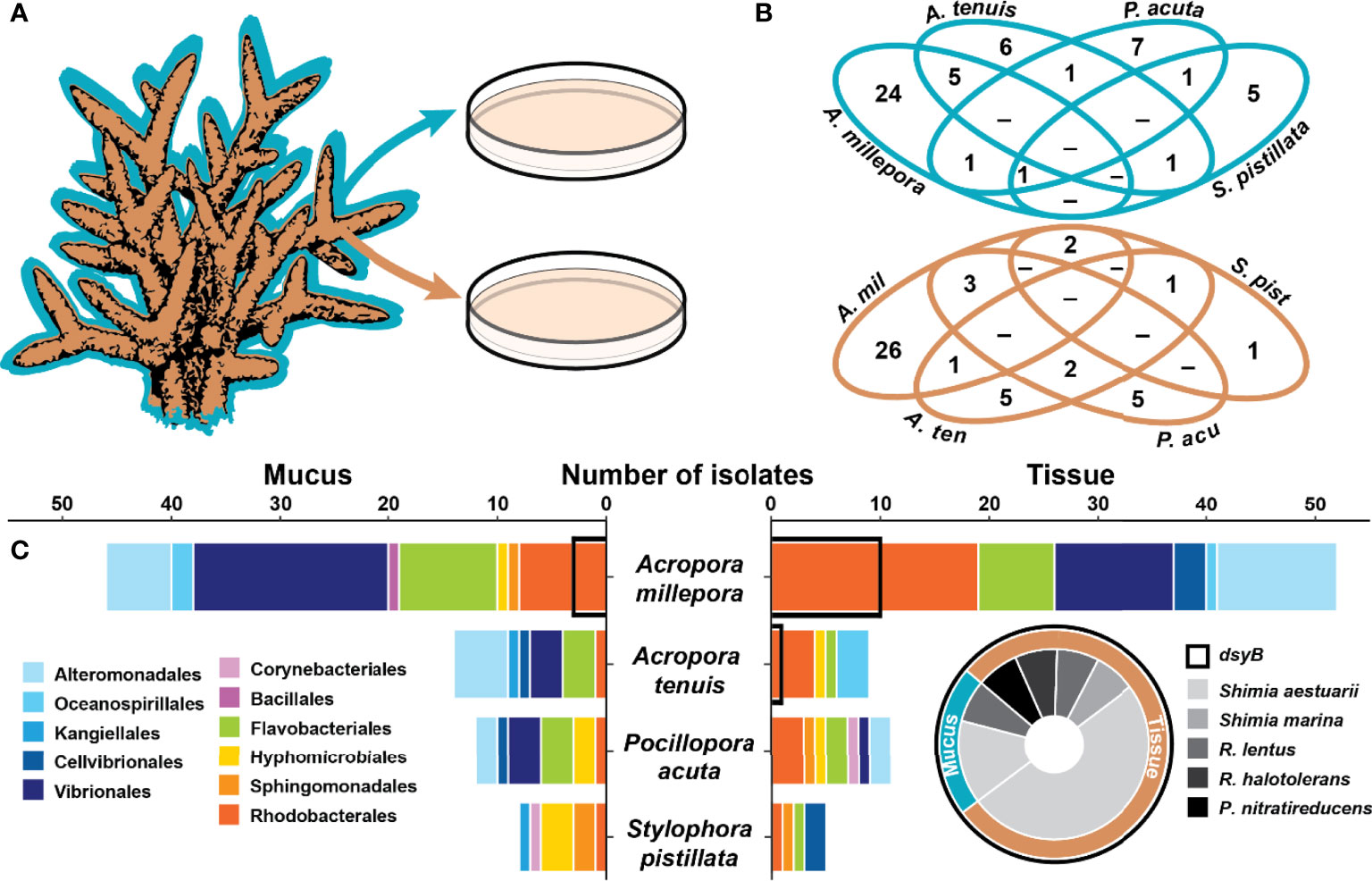
Figure 1 Coral associated bacteria harbour the dsyB gene. (A) Isolation of 157 bacteria from the mucus (blue) and tissue (brown) of four coral species. (B) Venn diagrams for the mucus (blue) and tissue (brown) of the four species showing the overlap in the isolated bacterial species (identified by their 16S rRNA gene) between the four coral species. (C) Taxonomic composition of the isolates (order level) between the four coral species for the mucus (left panel) and the tissue (right panel). Isolates harbouring dsyB (14 of 157) are indicated by a black outline on the bar graph and their taxonomic composition is presented as a pie chart (for more information, see Table S2). The genera Roseivivax and Pseudooceanicola are abbreviated.
A 5 mL liquid culture of each isolate was grown overnight in MB at 28°C with agitation (orbital shaker at 180 RPM) before being centrifuged for 5 min at 10,000 g and the supernatant decanted. DNA extraction was performed using the DNeasy UltraClean Microbial Kit (Qiagen, Germany) according to the manufacturer’s instructions. Extracted DNA was resuspended in 20 µL of UltraPure DNase/RNase-Free Distilled Water (Invitrogen, United States) and quantified by spectrophotometry (NanoDrop ND-1000, ThermoFisher, United States). Aliquots of extracted DNA were diluted with sterile Milli-Q water to 10 ng µL-1 and stored at -20°C until required.
Extracted DNA was used as template in PCR with the universal 16S rRNA genes primers 27F and 1492R (Lane, 1991), as well as the dsyB specific primers dsyB_deg1F and dsyB_deg2R that amplify a 246 bp region of the gene (Williams et al., 2019). Each PCR reaction mixture contained 1× reaction buffer, 2 mM of MgCl2 solution, 1 mM of deoxyribonucleotide triphosphate (dNTP) mix, 0.4 µM of each primer, 0.5 µL of BIOTAQ DNA Polymerase (Bioline, United Kingdom), 1 ng µL-1 of template DNA, and adjusted to a final volume of 25 µL with UltraPure DNase/RNase-free distilled water (Thermo Fisher Scientific, United States). PCR amplifications were as follows: (i) dsyB_deg1F/dsyB_deg2R: initial step at 95°C for 5 min; 30 cycles at 95°C for 30 s, 61°C for 1 min and 72°C for 15 s; and a final extension step at 72°C for 5 min; (ii) 27F/1492R: as described by Bourne and Munn (2005). PCR products were purified with the Wizard SV Gel and PCR Clean-Up System (Promega, United States) and visualised via electrophoresis on a 1% agarose gel stained with ethidium bromide.
PCR products were sequenced at Macrogen Inc. (Seoul, South Korea). The forward and reverse 16S rRNA and dsyB amplicon sequences were paired and the overlapping fragments were merged using Geneious Prime 2019.2.3 (Biomatters, New Zealand). Some dsyB amplicon sequences were too short (or of poor quality) to be merged and only one sequence (forward or reverse) was used.
To identify the closest taxonomic-relative of each isolate, BLAST searches (Altschul et al., 1990) were conducted through the National Center for Biotechnology Information (NCBI). 16S rRNA gene sequences were then aligned using MAFFT (Multiple Alignment using Fast Fourier Transform) v7 (Katoh et al., 2002; Katoh and Standley, 2013) with default settings, then trimmed using trimAl v1.4 (Capella-Gutiérrez et al., 2009) to remove sites with more than 50% missing or degraded data. Following the Bayesian Information Criterion (Schwarz, 1978), the maximum likelihood phylogeny for each sample niche was calculated. Phylogenetic trees were constructed using IQ-TREE v1.6.12, with 1,000 ultrafast bootstrap replicates (Minh et al., 2013), and formatted using the ggtree package (Yu et al., 2017) in R (R Core Team, 2020).
The dsyB amplicons were translated into protein sequences and searches to find regions of local similarity were performed using BLASTP. These translated sequences were also aligned with DsyB from Labrenzia aggregata (AOR83342) (Curson et al., 2017) to assess their similarity using the Needleman-Wunsch algorithm (Needleman and Wunsch, 1970). A multiple sequence alignment (MSA) of the prokaryotic DsyB protein sequences was visualised to show conserved residues, conservative mutations, and divergence between the different homologues. The MSA was conducted using T-Coffee v11.00 (Notredame et al., 2000; Di Tommaso et al., 2011) with default settings and formatted using Boxshade v3.21. Conserved domains within the predicted DsyB sequence were detected using CD-Search (Marchler-Bauer and Bryant, 2004) against the Conserved Domain Database (CDD) v3.18 (Lu et al., 2019).
To confirm the DMSP biosynthesis capability of bacteria harbouring dsyB, each strain was cultured in 500 mL of either MB, yeast tryptone sea salts [YTSS; (González et al., 1996)], MBM broth or methionine-enriched MBM broth (final concentration 0.5 mM). The cultures were incubated at 28°C with agitation (orbital shaker at 180 RPM) for 24 hours before being harvested by centrifugation (3,000 g for 15 min at 4°C). The clarified medium was discarded and the remaining cell pellets snap-frozen with liquid nitrogen, lyophilised overnight (Dynavac freeze dryer, Massachusetts, United States; model FD12) and stored at -20°C until required.
The freeze-dried cell pellets were resuspended in 1 mL of deuterated methanol (CD3OD; Cambridge Isotope Laboratories, Massachusetts, United States), vortexed at maximum speed for 5 min, and sonicated for 5 min at room temperature. A further 1 mL of CD3OD and 666 µL of deuterium oxide (D2O; Cambridge Isotope Laboratories, Massachusetts, United States) were added into the mixture (for a final CD3OD to D2O ratio of 3:1), which were then vortexed at maximum speed for 5 min and sonicated for 10 min at room temperature. Bacterial extracts were subsequently centrifuged at 3,000 g for 5 min. The particulate-free extracts (final volume of ~2.6 mL) were then used for subsequent analyses on the LC-MS and NMR.
LC-MS was used to assess the presence of intracellular DMSP in dsyB-positive bacterial isolates. Particulate-free bacterial extracts were analysed on an Agilent 1100 series high performance liquid chromatograph coupled to a Bruker Esquire 3000 quadrupole ion trap mass spectrometer (LC-MS; Bruker Daltonics, Massachusetts, United States) equipped with an electrospray ionisation interface (ESI). Extracts (5 µL) were separated on a reverse-phase Luna 3 μm HILIC column (Phenomenex, California, United States; 150 × 3 mm, with a particle size of 3 µm) maintained at 25°C. Separation was achieved using a programmed step gradient consisting of solvent A: 0.1% formic acid (HCOOH) in Milli-Q water and solvent B: methanol (CH3OH, HPLC grade OmniSolv), at a flow rate of 0.5 mL min-1. The column was pre-equilibrated at 60% B for 10 min prior to injection. The programmed step gradient was t = 0 min, 60% B; t = 12 min, 10% B; t = 14 min, 10% B; t = 15 min, 60% B; t = 20 min, 60% B; t = 22 min, 60% B. The ESI was operated in positive mode and the target mass of m/z 135, corresponding to the [M+H]+ of DMSP, monitored (established from a DMSP standard).
The presence of intracellular DMSP in dsyB-positive bacterial isolates was also assessed using NMR. A 700 µL aliquot of each bacterial particulate-free extract was transferred into a 5 mm Norell 509-UP NMR tube (North Carolina, United States) and analysed immediately using quantitative NMR (qNMR) via the ERETIC method (Electronic REference To access In vivo Concentrations) (Akoka and Trierweiler, 2002) to measure the concentration of DMSP, as described in Tapiolas et al. (2013).
NMR spectra of the bacterial extracts were recorded on a Bruker Avance 600 MHz NMR spectrometer (Bruker BioSpin, United States) with a triple resonance cryoprobe (TXI), referenced using CD3OD (δH 3.31). 1H NMR spectra were acquired as outlined in Tapiolas et al., (2013) using a standard Bruker solvent suppression pulse sequence. 2D NMR spectra were also acquired to confirm the assignment of DMSP. All spectra were referenced to residual 1H and 13C resonances in CD3OD. In addition, one extract was spiked with 14 µL of 50 mM DMSP to confirm the position of the methyl singlet, as NMR signals can shift as the sample matrix changes.
A phenol:chloroform extraction method, outlined in detail in Raina et al. (2016), was used to extract high-molecular weight DNA from a representative bacterial isolate producing DMSP (called AMM-P-2 hereafter). Extracted DNA was sent to the Ramaciotti Centre for Genomics (Sydney, Australia) for library preparation using the Nextera XT DNA Library Preparation Kit (Illumina, United States) and sequenced on the Illumina MiSeq system using V2 with 2×250 bp paired-end reads.
The MiSeq read set was trimmed, assembled, and error-corrected using Trimmomatic 0.38 (Bolger et al., 2014), SPAdes v3.13.0 (Bankevich et al., 2012), and Pilon v1.23 (Walker et al., 2014), respectively. All were implemented through Shovill v1.0.4 using default settings. Prediction of coding regions and annotation were performed with Prokka v1.14.6 (Seemann, 2014) using standard databases (i.e., ISfinder, NCBI Bacterial Antimicrobial Resistance Reference Gene Database, and UniProtKB (SwissProt)) and default settings.
To confirm the presence of dsyB within the genome of AMM-P-2, we used a reciprocal BLAST approach between the protein sequence reported by Curson et al. (2017) and the AMM-P-2 genome (E-value ≥ 1 e-50). Similarly, to explore the genomic potential of the bacterium to utilise and metabolise other sulfur compounds (e.g., sulfate, cysteine), target genes were obtained from KEGG (Kyoto Encyclopedia of Genes and Genomes) (Kanehisa et al., 2004) and compared against the AMM-P-2 genome. The orthology of the highest scoring match (E-value ≥ 1 e-50) was confirmed by conducting BLASTP analyses against the NCBI NR (non-redundant) database, followed by BLASTP analyses of retrieved best match against the AMM-P-2 predicted proteins.
Isolate AMM-P-2 was grown in MBM broth with methionine (final concentration 0.5 mM) (Curson et al., 2017), and the culture incubated at 27°C, 180 RPM, ambient lighting, and 35 practical salinity units (PSU). After three days, a 1:10 dilution of this starter culture was inoculated into 60 mL replicate cultures and incubated under different environmental conditions simulating stress experienced by tropical corals: (i) high temperature (32°C; T32), (ii) low temperature (22°C; T22), (iii) high UV (through a combination of Deluxlite Black Light Blue (18W) and Reptile One UVB 5.0 (18W) with an average total radiation in the incubator of 1.328 mW cm-2 measured using a Solarmeter Model 5.0 UVA + UVB meter (Solar Technology, Pennsylvania, United States)), (iv) complete darkness, (v) high salinity (40 PSU), and (vi) low salinity (25 PSU). Three biological replicates were grown for each of the described conditions, including control conditions maintained at ambient lighting, and 35 PSU.
Culture density was monitored through time using spectrophotometry (2 µL was measured at 600 nm (OD600) on a NanoDrop 1000 spectrophotometer; Thermo Fisher Scientific, United States). Cultures were sampled over four time points, corresponding to the mid-exponential (24 h), late exponential (28 h), early stationary (32 h), and late stationary (36 h) growth stages of the bacterium, as previously established by a standard growth curve. At each time point, one culture per treatment was removed and samples were taken for quantitative nuclear magnetic resonance (qNMR) analysis (50 mL; centrifuged for 5 min at 3,000 g, pellet snap frozen and stored at -20°C until analysis).
To convert the optical density data recorded with spectrophotometry into bacterial cell numbers, OD600 and flow cytometry counts were carried out simultaneously on isolate AMM-P-2 grown under standard conditions in triplicate (27°C at 180 RPM, ambient lighting, 35 PSU, in modified MBM). After each OD600 measurement, 100 µL of cells were fixed for 15 min in 2% glutaraldehyde for subsequent flow cytometry analysis. Samples were then stained with SYBR Green (1:10,000 final dilution; ThermoFisher, Massachusetts, United States), incubated for 15 min in the dark and analysed on a CytoFLEX S flow cytometer (Beckman Coulter, California, United States) with filtered MilliQ water as the sheath fluid. For each sample, forward scatter (FSC), side scatter (SSC), and green (SYBR) fluorescence were recorded. The samples were analysed at a flow rate of 25 µL min-1. Microbial populations were characterized according to SSC and SYBR Green fluorescence (Marie et al., 1997) and cell abundances were calculated by running a standardized volume (50 µL) per sample.
Statistics were performed using IBM SPSS Statistics v27.0.1. qNMR signals associated with DMSP and acrylate were normalised to cell density and tested for significance using repeated-measures ANOVA, with Greenhouse-Geisser correction applied (a correction for sphericity). A simple main effect test, applied following significant interactions between treatments, was used to determine the difference between treatments at each time point for both DMSP and acrylate. A Pearson product-moment correlation was used to determine the relationship between DMSP and acrylate concentrations in AMM-P-2.
A total of 157 isolates were recovered from coral mucus (51%) and tissue (49%) (Figure 1B and Table S2). The highest proportion of these were members of the Gammaproteobacteria (49%), followed by Alphaproteobacteria (32%) and Flavobacteriia (17%) (Table S2). The taxonomic composition of the isolates in each compartment was different (Figure 1 and Table S2), and few isolates were shared between coral species (Figure 1B).
From the 157 bacterial isolates screened, 14 harboured the dsyB gene (9% of total isolates). These strains all belong to the Alphaproteobacteria class, specifically the family Rhodobacteraceae, and include members of the Shimia (n=10), Roseivivax (n=3), and Pseudooceanicola (n=1) genera (Figure 1C and Table S2). All of these dsyB-harbouring strains were isolated from Acropora millepora and Acropora tenuis, with 3 strains derived from the mucus and 11 from the tissue. More specifically, bacteria harbouring dsyB represented 19% (10 of 52) of the isolates from A. millepora tissue. The average sequence identity of the predicted (partial) DsyB amino acid sequences derived from the isolates to the protein from Labrenzia aggregata was ~65% (Figure S1) (Curson et al., 2017; Curson et al., 2018). Interestingly, three different Labrenzia strains were isolated from corals, including two strains with more than 98% 16S rRNA gene sequence identity to L. aggregata, but dsyB could not be amplified from these isolates.
The 14 dsyB-harbouring strains were assessed for their ability to produce DMSP. The isolates were initially grown in MB and YTSS, but DMSP could not be detected in any of the cultures using either LC-MS (Figure S2) or NMR (Figure S3). However, LC-MS did detect a peak at m/z 135 and retention time of 5.9 min, consistent with DMSP, in all isolates grown in methionine-enriched MBM (Figure S4). 1H NMR analysis established the presence of a well-resolved diagnostic singlet at δH 2.95 ppm (2 × CH3) (Tapiolas et al., 2013) (Figure 2) and ¹H-¹H correlation spectroscopy (COSY) revealed two coupled methylene groups, S-CH2- (δH 3.45, t) and -CH2-CO2H (δH 2.70, t) (Figure S5). In addition, a 1H-13C heteronuclear multiple bond correlation (HMBC) revealed long-range chemical shift correlations between: (i) the protons of the two methyl groups (S-CH3) and the carbon of the S-methylene group (δH 2.94 - δC 43.21); (ii) the carboxyl-methylene protons of the carboxyl carbon (δH 2.69 - δC 172.03) and the S-methylene carbon (δH 2.69 - δC 43.21) (Figure S6). Together, these COSY and HMBC correlations confirmed the structure of DMSP, thus verifying the presence of DMSP in the bacterial extracts.
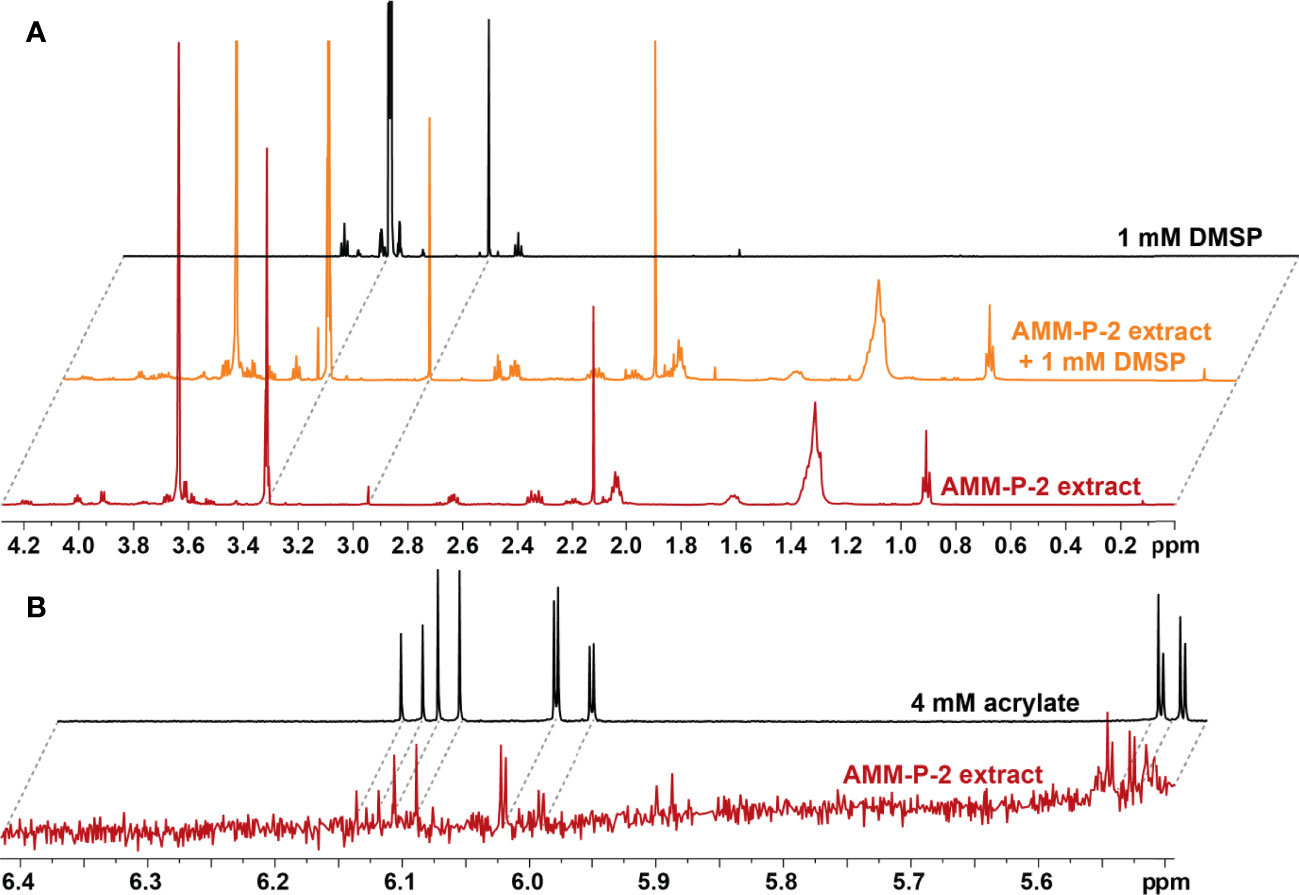
Figure 2 Isolates harbouring dsyB produce dimethylsulfoniopropionate (DMSP). (A) 1H NMR spectra of the DMSP region identifying diagnostic peaks (δH 2.95 ppm) in deuterated methanol (CD3OD) extract of Shimia AMM-P-2 cultured in methionine-enriched minimal basal medium; the same Shimia AMM-P-2 cell extract spiked with 10 µl of 1 mM DMSP standard; and 1 mM DMSP standard in CD3OD. (B) 1H NMR spectra of the acrylate region identifying diagnostic peaks (δH 5.71, 6.13 and 6.20 ppm) in CD3OD cell extracts of Shimia AMM-P-2 cultured in methionine-enriched minimal basal medium; 4 mM acrylate standard in CD3OD.
To further characterise the genomic underpinnings of DMSP production in coral-associated bacteria, isolate AMM-P-2 was selected for whole-genome sequencing (Table 1). This isolate belongs to the Shimia genus, which accounted for 71% (or 10 of 14) of dsyB-positive bacteria isolated. This strain exhibited >97% similarity with Shimia aestuarii based on its full 16S rRNA gene sequence.
Comparison of the predicted amino acid sequence of DsyB from S. aestuarii AMM-P-2 with previously characterised homologues representing the diversity of this protein family (n=14) revealed the presence of two conserved domains which are common to all dsyB orthologues (Figure S7): (i) an S-adenosylmethionine-dependent methyltransferase (AdoMet-MTase) class I superfamily domain (pfam00891; E-value 4.02 e-16) and (ii) a dimerization2 superfamily domain (pfam16864; E-value 4.13 e-7). As in several other Rhodobacterales strains (Curson et al., 2017), isc [iron sulfur cluster] or suf [sulfur formation] gene clusters were present 5’ of dsyB in S. aestuarii AMM-P-2 (Figure S8). However, in the region 3’ of dsyB, only limited synteny was observed between S. aestuarii AMM-P-2 and other Rhodobacterales.
To determine the source of sulfur used by S. aestuarii AMM-P-2 for DMSP biosynthesis, its sulfur metabolic potential was assessed. Distinct orthologues of all the enzymes in the sulfate reduction pathway were identified (Figure 3 and Table S3), confirming S. aestuarii AMM-P-2 has the genetic machinery required to uptake and convert extracellular inorganic sulfate to sulfide (Table S3). Following sulfide production, S. aestuarii AMM-P-2 can produce cysteine (through serine or homocysteine) and ultimately the DMSP precursor methionine (via aspartate; Figure 3).
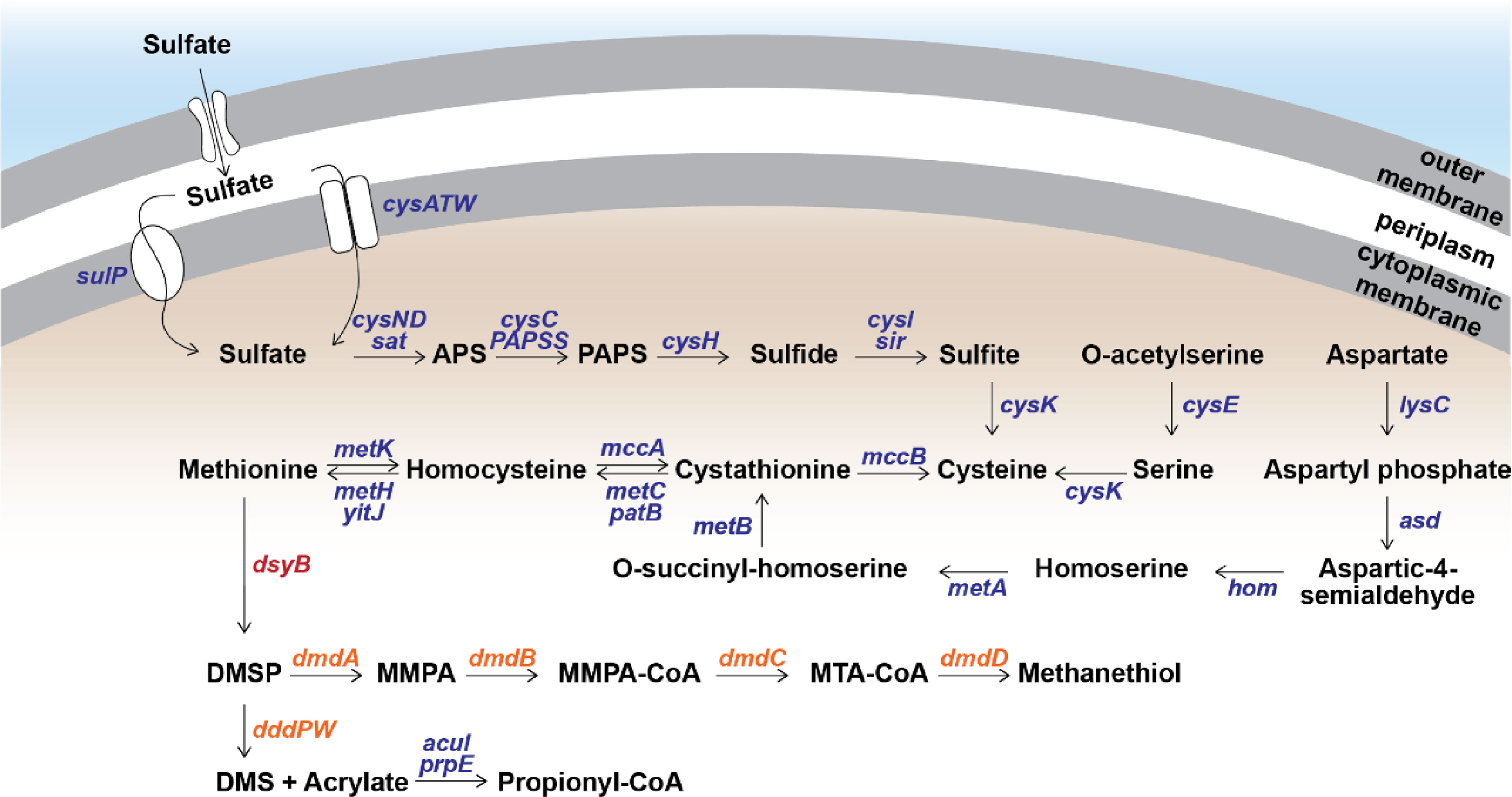
Figure 3 Metabolism of methylated sulfur compounds in Shimia aestuarii AMM-P-2. Orthologous genes for each step are presented in blue. The DMSP production gene dsyB is presented in red, while the catabolism genes are in orange. E-values and additional information can be found in Table S3.
Turning the attention to DMSP catabolism, S. aestuarii AMM-P-2 has the genetic potential to both demethylate and cleave this sulfur compound (Figure 3 and Table S3). S. aestuarii AMM-P-2 has the entire dmdABCD suite of genes enabling conversion of DMSP to MeSH (Zhang et al., 2019). This bacterium also contains the DMSP lyase genes dddP and dddW, whose products liberate equimolar amounts of DMS and acrylate from DMSP (Zhang et al., 2019). It is also able to metabolise acrylate to propionyl-CoA through acuI, acuH and prpE genes (Wang et al., 2017). Significant matches to dddA and dddC (E-values ≥ 1.39 e-93) which catabolise 3-hydroxypropionyl-CoA (3HP) to acetyl-CoA (Wang et al., 2017) (Table S3) were also identified.
To assess the influence of environmental factors on DMSP production by S. aestuarii AMM-P-2, cultures were grown in methionine-enriched MBM under six different conditions and sampled four times over a 48-hour incubation period. The conditions tested are known to elicit stress responses in corals (high and low temperature, high and low salinity, high UV, and darkness), and were compared to controls. Intracellular DMSP levels per cell within each treatment were not statistically different through time (repeated-measure ANOVA, p = 0.078; see Table S4) and no significant interaction was identified between time and treatments (repeated-measure ANOVA, p = 0.204; Table S4).
Changes in salinity affected DMSP levels per cell (Figure 4A). Levels of DMSP in cultures grown under hyposaline conditions (25 PSU) were 63% lower than the controls (35 PSU) over all four time points. In contrast, the highest DMSP levels were recorded under hypersaline conditions (40 PSU), which were 80% higher than the controls. DMSP levels also increased when cells were grown under high UV (65% higher) or in complete darkness (43% higher) compared to control ambient light conditions. Finally, at higher temperature (32°C), DMSP levels were 42% lower than the controls, while the low temperature treatment (22°C) had no effect throughout the experiment.
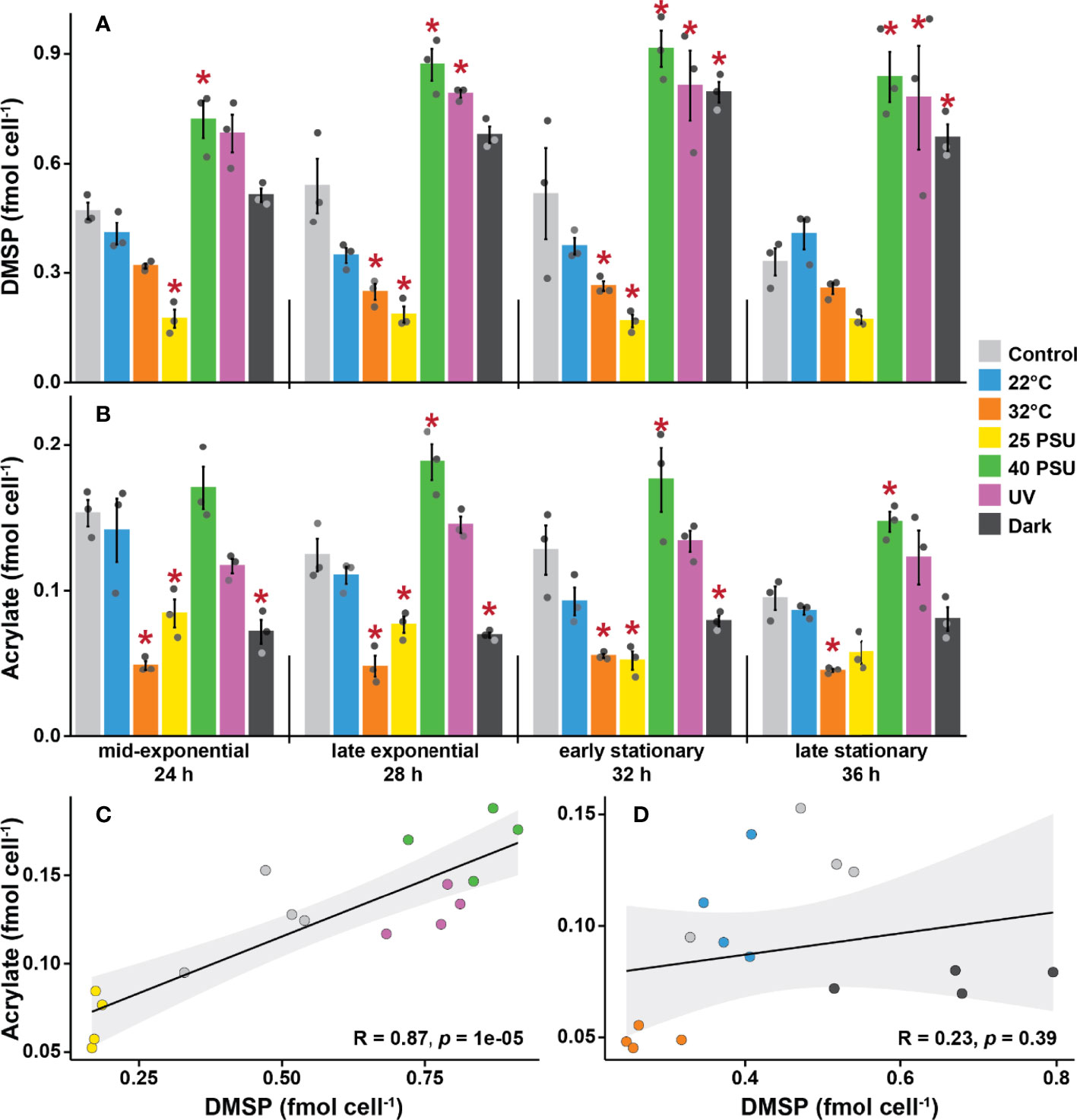
Figure 4 Dimethylsulfoniopropionate (DMSP) and acrylate levels are strongly influenced by environmental stressors in Shimia aestuarii AMM-P-2. Levels of (A) DMSP and (B) acrylate in bacterial cells exposed to high and low temperatures (32 and 22°C), high and low salinity (40 and 25 PSU), constant darkness, and high UV. Controls were grown at 27°C under ambient light in a modified MBM media adjusted to 35 PSU and supplemented with 0.5 mM methionine. Error bars indicate standard error (n = 3). Experimental conditions marked with an asterisk are significantly different (p < 0.05) from the controls (Repeated-measure ANOVA). Correlation between DMSP and acrylate levels in Shimia aestuarii AMM-P-2 under conditions (C) likely to stress the cells (high-low salinity and high UV), (D) unlikely to stress the cells (22-32°C and darkness). Pearson correlation coefficient and associated p values are displayed on each graph.
Although it was not possible to quantify DMS with qNMR due to the volatility of this compound, DMSP lysis releases equimolar concentrations of acrylate, which was detected in every sample. Overall, the level of acrylate per cell was approximately one order of magnitude lower than that of DMSP (Figure 4B). These measurements confirm that a fraction of the synthesized DMSP is channelled through the cleavage pathway. A significant interaction between time and treatments was identified for acrylate levels per cell (ANOVA, p = 0.047; Table S5). However, the differences between control conditions and the different treatments were not as pronounced for acrylate compared to DMSP. The 32°C treatment exhibited the lowest levels of acrylate across all timepoints (with 68% less acrylate on average than the control), while the high salinity treatment gave rise to the highest concentrations (with 36% more acrylate).
The six treatments were divided into two categories: (i) those likely to elicit a stress response in bacteria (i.e., high UV, low and high salinity), and (ii) those unlikely to be stressors (i.e., darkness, 22 and 32°C), and the level of correlation of the intracellular levels of DMSP and acrylate between these assessed. No significant correlation was identified between DMSP and acrylate levels for the mild conditions (Pearson’s R = 0.23, p = 0.39; Figure 4D). However, a strong positive correlation was identified for the stress treatments (Pearson’s R = 0.87, p < 0.001; Figure 4C).
Of the 157 bacteria isolated from the mucus and tissues of four coral species, approximately 9% harboured the dsyB gene (14 out of 157). Although three genera (four species) of corals were screened, dysB-positive bacteria were only isolated from the two Acropora species, which correlates with the high DMSP production by members of this genus (Tapiolas et al., 2013). Note, it is possible that other DMSP-producing bacteria were present in the corals sampled, potentially containing mmtN or other unknown DMSP synthesis genes (Williams et al., 2019). All bacteria with dsyB belonged to the Rhodobacterales order (Curson et al., 2017), with Shimia species representing 71% of these isolates (10 out of 14). Shimia are metabolically versatile members of the Roseobacter clade (Choi and Cho, 2006), and are abundant in the water column, marine sediments, and commonly associated with eukaryotic hosts (Lenk et al., 2012; Luo and Moran, 2014), particularly phytoplankton (Ajani et al., 2018; Behringer et al., 2018) and reef-building corals (Chen et al., 2011; Zhang et al., 2021). The other dsyB-harbouring bacteria isolated in our screen belonged to the genera Pseudooceanicola – which are common DMSP producers in the water column (Zheng et al., 2020) – and Roseivivax – which have been isolated from corals (Chen et al., 2012) and can induce coral larval settlement (Sharp et al., 2015).
DMSP was unambiguously identified in extracts of dsyB-harbouring bacteria when the cells were grown in methionine-supplemented MBM. In MBM lacking methionine, DMSP signal intensity was close to or below the detection limits of the LC-MS and NMR. Addition of pathway intermediates, including methionine, have been shown to enhance DMSP production in bacteria, but most dsyB-harbouring strains can also produce methionine de novo (through sulfate assimilation) (Curson et al., 2017). This study therefore reveals the presence of bacteria capable of DMSP production in corals and shows that these microorganisms are likely to contribute to the DMSP production by coral holobionts.
Three strains of Labrenzia, the bacterial genus from which the dsyB gene was initially identified (Curson et al., 2017), were isolated from Pocillopora acuta and Stylophora pistillata. The 16S rRNA genes of one of the strains was 98.6% similar to Labrenzia alba, while of the other two were more than 98% similar to L. aggregata. However, the dsyB gene was not detected in any of the three strains. This is consistent with the previously-reported lack of de novo DMS production from a Symbiodiniaceae-associated Labrenzia strain (Lawson et al., 2020), suggesting that the ability to produce DMSP is not conserved across the Labrenzia genus.
To investigate the potential of a representative DMSP-producing bacterium from corals to metabolise sulfur compounds, the genome of S. aestuarii AMM-P-2 was fully sequenced. S. aestuarii is capable of assimilating sulfate from seawater to produce sulfide and sulfur-containing amino acids. Although all the genes required for methionine synthesis were present, its growth on methionine-enriched media indicates that it can also use exogenous methylated sulfur compounds, enhancing the pool of reduced sulfur intermediates available for the synthesis of DMSP. The S. aestuarii AMM-P-2 genome also encodes both the demethylation and the cleavage pathways, allowing it to produce the sulfurous gases methanethiol and DMS. Given that metabolic interdependencies between different partners are common in symbiotic systems, it is important to note that S. aestuarii AMM-P-2 has all the required genes to produce DMSP de novo.
As in other Rhodobacterales (Curson et al., 2017), dsyB in S. aestuarii AMM-P-2 is located downstream of multiple genes (iscRS, sufBCDS) encoding iron-sulfur cluster (Isc) proteins involved in the formation of Fe-S clusters and cellular defence against oxidative stress. Specifically, the cysteine desulfurase IscS provides the sulfur that is then incorporated into Fe-S clusters and its deletion renders the cells hypersensitive to oxidative stress (Ayala-Castro et al., 2008; Rybniker et al., 2014). In addition, the IscR protein is a transcriptional regulator of the suf operon (Yeo et al., 2006) and both the suf and isc operons are highly induced by oxidative stress (Yeo et al., 2006). Therefore, the tight linkage of dsyB to the suf and isc operons suggests that DMSP transcription may be directly affected by oxidants (Sunda et al., 2002; Curson et al., 2017).
DMSP levels in S. aestuarii cells were affected by changes in salinity. Fluctuations in salinity are known to affect DMSP levels in corals (Gardner et al., 2016; Aguilar et al., 2017), while also impacting DMSP degradation in bacteria (Salgado et al., 2014; Liu et al., 2018). The nearly 5-fold increase in intracellular DMSP under hypersaline compared to hyposaline conditions reported here is consistent with previous reports in algae (Vairavamurthy et al., 1985; Kirst et al., 1991; Trossat et al., 1998), and bacteria (Curson et al., 2017), and supports the proposed function of this molecule as an osmolyte (Kirst et al., 1991).
UV exposure and complete darkness both caused significant increases in DMSP levels in S. aestuarii cells. The level of UV radiation applied here was approximately 50% higher than values previously measured at 1 m depth on the Great Barrier Reef (Nordborg et al., 2018), and is therefore likely to cause oxidative stress in the S. aestuarii cells. Indeed, because of their lack of pigmentation and low internal self-shading due to small cell volume, heterotrophic bacteria are amongst the most UV-sensitive organisms (Ruiz-Gonzalez et al., 2013). Similar increases in DMSP levels triggered by UV exposure have been reported in phytoplankton (Sunda et al., 2002; Slezak and Herndl, 2003) and have been attributed to the antioxidant capacity of DMSP (Sunda et al., 2002). More surprising was the increase in DMSP levels in cells grown in darkness. Recent studies have revealed that bacteria in aphotic environments, such as the deep ocean as well as coastal and deep sediments, produce substantial amounts of DMSP (Williams et al., 2019; Zheng et al., 2020). Although the effect of sunlight on bacterioplankton has been extensively studied at the community level (Ruiz-Gonzalez et al., 2013), the impact of prolonged darkness on the physiology of heterotrophic bacteria has received little attention to date. Our results suggest that the absence of light influences sulfur metabolism in S. aestuarii, and further investigation should aim to identify the mechanism driving the production of DMSP in aphotic conditions.
Intracellular acrylate levels in S. aestuarii AMM-P-2 were almost one order of magnitude lower than those of DMSP and decreased significantly over time in all growth conditions. Acrylate and DMSP levels were strongly and positively correlated under conditions likely to stress the bacteria (e.g., salinity, UV), but were decoupled under conditions that are stressful for the coral host but not necessarily for the bacteria (e.g., small temperature variations, darkness). This suggests that under conditions stressful for the bacteria, more DMSP is channelled towards the DMSP cleavage pathway, which generates equal amounts of DMS and acrylate, rather than the demethylation pathway (Gao et al., 2020). DMS and acrylate are efficient scavengers of hydroxyl radicals produced by the cells during stress (Sunda et al., 2002), which may explain why the cleavage pathway is preferred during stressful conditions.
This study demonstrates that some coral-associated bacteria can produce the important organosulfur compound DMSP, implying that, along with the animal host and Symbiodiniaceae, bacterial communities may contribute to DMSP production by the coral holobiont. It has recently been shown that bacteria can also use a second (dsyB-independent) pathway to produce DMSP, involving the mmtN gene (Williams et al., 2019). Since this gene is also present in Gammaproteobacteria and Actinobacteria, it is likely that some DMSP-producing bacteria were overlooked by the dsyB-centric screening approach and thus that the proportion of DMSP-producing bacteria in corals might be greater than estimated here. Analysis of the genome of a Shimia strain, representing the most abundant dsyB-harbouring genus of those isolated from corals, revealed that this bacterium has the genetic machinery to assimilate sulfate, to synthesise the sulfur-based amino acids cysteine and methionine, to catabolise DMSP to methanethiol, DMS and acrylate, and to utilise or detoxify acrylate. Furthermore, DMSP production in S. aestuarii AMM-P-2 is regulated by specific environmental conditions, some of which are not necessarily tied to coral stress. The capacity of coral-associated bacteria to produce DMSP not only adds this trait to the functional repertoire of prokaryotes associated with corals, but also indicates that bacteria may contribute to the large DMSP pool produced by this metaorganism.
The datasets presented in this study can be found in online repositories. The 16S rRNA gene amplicon sequences of the isolated bacteria have been deposited in GenBank (https://www.ncbi.nlm.nih.gov/genbank/) under the accession numbers MW828351 to MW828429 and MW828507 to MW828582. The assembled genome of isolate AMM-P-2 has been deposited in GenBank under the accession number PRJNA810763.
CM, DM, DB, FK and J-BR designed the experiments. FK collected the samples and performed all laboratory work and data analysis with the assistance of CM, DM, DB, JT and J-BR. FK, JZ, IC and DM analysed the genomic data. FK, CM, DM, DB and J-BR wrote the manuscript, with input from all authors before submission. All authors contributed to the article and approved the submitted version.
FWIK was supported through scholarships and funding from the AIMS@JCU Division of Research & Innovation and the College of Public Health, Medical and Veterinary Sciences at James Cook University. This research was funded in part by the Pilot Research Awards from AIMS@JCU.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The coral colonies were collected under permit G14/36802 issued by the Great Barrier Reef Marine Park Authority. The authors thank the staff of AIMS SeaSim for providing coral samples as needed, and Aurélie Moya, Sara Bell, Peter Thomas-Hall, and Nahshon Siboni for discussions and assistance in the lab. The authors acknowledge the Traditional Owners of the land on which the research was conducted, the Wulgurukaba, Bindal and Gadigal people; we recognise their connection to land, sea, and community, and pay our respects to Elders past, present and emerging. Finally, a special thanks goes to Libby Evans-Illidge (AIMS@JCU) for her unwavering support throughout the project.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2022.869574/full#supplementary-material
Aguilar C., Raina J.-B., Motti C. A., Fôret S., Hayward D. C., Lapeyre B., et al. (2017). Transcriptomic Analysis of the Response of Acropora Millepora to Hypo-Osmotic Stress Provides Insights Into DMSP Biosynthesis by Corals. BMC Genomics 18, 612. doi: 10.1186/s12864-017-3959-0
Ajani P. A., Kahlke T., Siboni N., Carney R., Murray S. A., Seymour J. R. (2018). The Microbiome of the Cosmopolitan Diatom Leptocylindrus Reveals Significant Spatial and Temporal Variability. Front. Microbiol. 9. doi: 10.3389/fmicb.2018.02758
Akoka S., Trierweiler M. (2002). Improvement of the ERETIC Method by Digital Synthesis of the Signal and Addition of a Broadband Antenna Inside the NMR Probe. Instrum. Sci. Technol. 30, 21–29. doi: 10.1081/CI-100108768
Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. (1990). Basic Local Alignment Search Tool. J. Mol. Biol. 215, 403–410. doi: 10.1016/S0022-2836(05)80360-2
Andreae M. O., Barnard W. R., Ammons J. M. (1983). The Biological Production of Dimethylsulfide in the Ocean and Its Role in the Global Atmospheric Sulfur Budget. Ecol. Bull. 35, 167–177. doi: jstor.org/stable/20112852
Ayala-Castro C., Saini A., Outten F. W. (2008). Fe-S Cluster Assembly Pathways in Bacteria. Microbiol. Mol. Biol. Rev. 72, 110–125. doi: 10.1128/MMBR.00034-07
Ayers G. P., Gras J. L. (1991). Seasonal Relationship Between Cloud Condensation Nuclei and Aerosol Methanesulphonate in Marine Air. Nature 353, 834–835. doi: 10.1038/353834a0
Bankevich A., Nurk S., Antipov D., Gurevich A. A., Dvorkin M., Kulikov A. S., et al. (2012). SPAdes: A New Genome Assembly Algorithm and Its Applications to Single-Cell Sequencing. J. Comput. Biol. 19, 455–477. doi: 10.1089/cmb.2012.0021
Bates T. S., Charlson R. J., Gammon R. H. (1987). Evidence for the Climatic Role of Marine Biogenic Sulphur. Nature 329, 319–321. doi: 10.1038/329319a0
Behringer G., Ochsenkühn M. A., Fei C., Fanning J., Koester J. A., Amin S. A. (2018). Bacterial Communities of Diatoms Display Strong Conservation Across Strains and Time. Front. Microbiol. 9. doi: 10.3389/fmicb.2018.00659
Bolger A. M., Lohse M., Usadel B. (2014). Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Bourne D. G., Morrow K. M., Webster N. S. (2016). Insights Into the Coral Microbiome: Underpinning the Health and Resilience of Reef Ecosystems. Annu. Rev. Microbiol. 70, 317–340. doi: 10.1146/annurev-micro-102215-095440
Bourne D. G., Munn C. B. (2005). Diversity of Bacteria Associated With the Coral Pocillopora Damicornis From the Great Barrier Reef. Environ. Microbiol. 7, 1162–1174. doi: 10.1111/j.1462-2920.2005.00793.x
Capella-Gutiérrez S., Silla-Martínez J. M., Gabaldón T. (2009). trimAl: A Tool for Automated Alignment Trimming in Large-Scale Phylogenetic Analyses. Bioinformatics 25, 1972–1973. doi: 10.1093/bioinformatics/btp348
Chen M.-H., Sheu S.-Y., Chen C. A., Wang J.-T., Chen W.-M. (2011). Shimia Isoporae Sp. Nov., Isolated From the Reef-Building Coral Isopora Palifera. Int. J. Syst. Evol. Microbiol. 61, 823–827. doi: 10.1099/ijs.0.022848-0
Chen M.-H., Sheu S.-Y., Chen C. A., Wang J.-T., Chen W.-M. (2012). Roseivivax Isoporae Sp. Nov., Isolated From a Reef-Building Coral, and Emended Description of the Genus Roseivivax. Int. J. Syst. Evol. Microbiol. 62, 1259–1264. doi: 10.1099/ijs.0.032961-0
Choi D. H., Cho B. C. (2006). Shimia Marina Gen. Nov., Sp. Nov., a Novel Bacterium of the Roseobacter Clade Isolated From Biofilm in a Coastal Fish Farm. Int. J. Syst. Evol. Microbiol. 56, 1869–1873. doi: 10.1099/ijs.0.64235-0
Curson A. R. J., Liu J., Bermejo Martínez A., Green R. T., Chan Y., Carrión O., et al. (2017). Dimethylsulfoniopropionate Biosynthesis in Marine Bacteria and Identification of the Key Gene in This Process. Nat. Microbiol. 2, 17009. doi: 10.1038/nmicrobiol.2017.9
Curson A. R. J., Williams B. T., Pinchbeck B. J., Sims L. P., Martínez A. B., Rivera P. P. L., et al. (2018). DSYB Catalyses the Key Step of Dimethylsulfoniopropionate Biosynthesis in Many Phytoplankton. Nat. Microbiol. 3, 430–439. doi: 10.1038/s41564-018-0119-5
DeBose J. L., Lema S. C., Nevitt G. A. (2008). Dimethylsulfoniopropionate as a Foraging Cue for Reef Fishes. Science 319, 1356–1356. doi: 10.1126/science.1151109
Di Tommaso P., Moretti S., Xenarios I., Orobitg M., Montanyola A., Chang J.-M., et al. (2011). T-Coffee: A Web Server for the Multiple Sequence Alignment of Protein and RNA Sequences Using Structural Information and Homology Extension. Nucleic Acids Res. 39, W13–W17. doi: 10.1093/nar/gkr245
Frade P. R., Schwaninger V., Glasl B., Sintes E., Hill R. W., Simó R., et al. (2016). Dimethylsulfoniopropionate in Corals and its Interrelations With Bacterial Assemblages in Coral Surface Mucus. Environ. Chem. 13, 252–265. doi: 10.1071/EN15023
Gao C., Fernandez V. I., Lee K. S., Fenizia S., Pohnert G., Seymour J. R., et al. (2020). Single-Cell Bacterial Transcription Measurements Reveal the Importance of Dimethylsulfoniopropionate (DMSP) Hotspots in Ocean Sulfur Cycling. Nat. Commun. 11, 1942. doi: 10.1038/s41467-020-15693-z
Gardner S. G., Nielsen D. A., Laczka O., Shimmon R., Beltran V. H., Ralph P. J., et al. (2016). Dimethylsulfoniopropionate, Superoxide Dismutase and Glutathione as Stress Response Indicators in Three Corals Under Short-Term Hyposalinity Stress. Proc. R. Soc. B.: Biol. Sci. 283, 20152418. doi: 10.1098/rspb.2015.2418
González J. M., Whitman W. B., Hodson R. E., Moran M. A. (1996). Identifying Numerically Abundant Culturable Bacteria From Complex Communities: An Example From a Lignin Enrichment Culture. Appl. Environ. Microbiol. 62, 4433–4440. doi: 10.1128/aem.62.12.4433-4440.1996
Hill R. W., Dacey J. W. H., Krupp D. A. (1995). Dimethylsulfoniopropionate in Reef Corals. Bull. Marine. Sci. 57, 489–494.
Howard E. C., Henriksen J. R., Buchan A., Reisch C. R., Bürgmann H., Welsh R., et al. (2006). Bacterial Taxa That Limit Sulfur Flux From the Ocean. Science 314, 649–652. doi: 10.1126/science.1130657
Johnston A., Todd J., Curson A. (2012). Microbial Origins and Consequences of Dimethyl Sulfide. Microbe 7, 181–185. doi: 10.1128/microbe.7.181.1
Jones G. B., Trevena A. J. (2005). The Influence of Coral Reefs on Atmospheric Dimethylsulphide Over the Great Barrier Reef, Coral Sea, Gulf of Papua and Solomon and Bismarck Seas. Marine. Freshwater. Res. 56, 85–93. doi: 10.1071/MF04097
Kanehisa M., Goto S., Kawashima S., Okuno Y., Hattori M. (2004). The KEGG Resource for Deciphering the Genome. Nucleic Acids Res. 32, D277–D280. doi: 10.1093/nar/gkh063
Karsten U., Kück K., Vogt C., Kirst G. O. (1996). “Dimethylsulfoniopropionate Production in Phototrophic Organisms and its Physiological Functions as a Cryoprotectant,” in Biological and Environmental Chemistry of DMSP and Related Sulfonium Compounds. Eds. Kiene R. P., Visscher P. T., Keller M. D., Kirst G. O. (Boston, MA: Springer US), 143–153.
Katoh K., Misawa K., i. Kuma K., Miyata T. (2002). MAFFT: A Novel Method for Rapid Multiple Sequence Alignment Based on Fast Fourier Transform. Nucleic Acids Res. 30, 3059–3066. doi: 10.1093/nar/gkf436
Katoh K., Standley D. M. (2013). MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 30, 772–780. doi: 10.1093/molbev/mst010
Kiene R. P., Linn L. J., Bruton J. A. (2000). New and Important Roles for DMSP in Marine Microbial Communities. J. Sea. Res. 43, 209–224. doi: 10.1016/S1385-1101(00)00023-X
Kiene R. P., Visscher P. T., Keller M. D., Kirst G. O. (1996). Biological and Environmental Chemistry of DMSP and Related Sulfonium Compounds (Boston, MA: Springer).
Kirst G. O. (1996). “Osmotic Adjustment in Phytoplankton and MacroAlgae,” in Biological and Environmental Chemistry of DMSP and Related Sulfonium Compounds. Eds. Kiene R. P., Visscher P. T., Keller M. D., Kirst G. O. (Boston, MA: Springer US), 121–129.
Kirst G. O., Thiel C., Wolff H., Nothnagel J., Wanzek M., Ulmke R. (1991). Dimethylsulfoniopropionate (DMSP) in Icealgae and its Possible Biological Role. Marine. Chem. 35, 381–388. doi: 10.1016/S0304-4203(09)90030-5
Lane D. (1991). “16s/23s rRNA Sequencing,” in Nucleic Acid Techniques in Bacterial Systematics. Eds. Stackebrandt E., Goodfellow M. (New York, NY, USA: John Wiley & Sons), 115–175.
Lawson C. A., Seymour J. R., Possell M., Suggett D. J., Raina J.-B. (2020). The Volatilomes of Symbiodiniaceae-Associated Bacteria Are Influenced by Chemicals Derived From Their Algal Partner. Front. Marine. Sci. 7. doi: 10.3389/fmars.2020.00106
Lenk S., Moraru C., Hahnke S., Arnds J., Richter M., Kube M., et al. (2012). Roseobacter Clade Bacteria are Abundant in Coastal Sediments and Encode a Novel Combination of Sulfur Oxidation Genes. ISME. J. 6, 2178–2187. doi: 10.1038/ismej.2012.66
Liu J., Liu J., Zhang S.-H., Liang J., Lin H., Song D., et al. (2018). Novel Insights Into Bacterial Dimethylsulfoniopropionate Catabolism in the East China Sea. Front. Microbiol. 9. doi: 10.3389/fmicb.2018.03206
Luo H., Moran M. A. (2014). Evolutionary Ecology of the Marine Roseobacter Clade. Microbiol. Mol. Biol. Rev. 78, 573–587. doi: 10.1128/MMBR.00020-14
Luo D., Wang X., Feng X., Tian M., Wang S., Tang S.-L., et al. (2021). Population Differentiation of Rhodobacteraceae Along With Coral Compartments. ISME. J. 15, 3286–3302. doi: 10.1038/s41396-021-01009-6
Lu S., Wang J., Chitsaz F., Derbyshire M. K., Geer R. C., Gonzales N. R., et al. (2019). CDD/SPARCLE: The Conserved Domain Database in 2020. Nucleic Acids Res. 48, D265–D268. doi: 10.1093/nar/gkz991
Marchler-Bauer A., Bryant S. H. (2004). CD-Search: Protein Domain Annotations on the Fly. Nucleic Acids Res. 32, W327–W331. doi: 10.1093/nar/gkh454
Marie D., Partensky F., Jacquet S., Vaulot D. (1997). Enumeration and Cell Cycle Analysis of Natural Populations of Marine Picoplankton by Flow Cytometry Using the Nucleic Acid Stain SYBR Green I. Appl. Environ. Microbiol. 63, 186–193. doi: 10.1128/aem.63.1.186-193.1997
Matthews J. L., Raina J.-B., Kahlke T., Seymour J. R., van Oppen M. J. H., Suggett D. J. (2020). Symbiodiniaceae-Bacteria Interactions: Rethinking Metabolite Exchange in Reef-Building Corals as Multi-Partner Metabolic Networks. Environ. Microbiol. 22, 1675–1687. doi: 10.1111/1462-2920.14918
Minh B. Q., Nguyen M. A. T., von Haeseler A. (2013). Ultrafast Approximation for Phylogenetic Bootstrap. Mol. Biol. Evol. 30, 1188–1195. doi: 10.1093/molbev/mst024
Needleman S. B., Wunsch C. D. (1970). A General Method Applicable to the Search for Similarities in the Amino Acid Sequence of Two Proteins. J. Mol. Biol. 48, 443–453. doi: 10.1016/0022-2836(70)90057-4
Ngugi D. K., Ziegler M., Duarte C. M., Voolstra C. R. (2020). Genomic Blueprint of Glycine Betaine Metabolism in Coral Metaorganisms and Their Contribution to Reef Nitrogen Budgets. iScience 23, 101120. doi: 10.1016/j.isci.2020.101120
Nordborg F. M., Flores F., Brinkman D. L., Agustí S., Negri A. P. (2018). Phototoxic Effects of Two Common Marine Fuels on the Settlement Success of the Coral Acropora Tenuis. Sci. Rep. 8, 8635. doi: 10.1038/s41598-018-26972-7
Notredame C., Higgins D. G., Heringa J. (2000). T-Coffee: A Novel Method for Fast and Accurate Multiple Sequence Alignment. J. Mol. Biol. 302, 205–217. doi: 10.1006/jmbi.2000.4042
Pogoreutz C., Voolstra C. R., Rädecker N., Weis V., Cardenas A., Raina J.-B. (2020). “The Coral Holobiont Highlights the Dependence of Cnidarian Animal Hosts on Their Associated Microbes,” in Cellular Dialogues in the Holobiont. Eds. Bosch T. C. G., Hadfield M. G. (CRC Press: Taylor & Francis Group), 91–118.
Rädecker N., Pogoreutz C., Voolstra C. R., Wiedenmann J., Wild C. (2015). Nitrogen Cycling in Corals: The Key to Understanding Holobiont Functioning? Trends Microbiol. 23, 490–497. doi: 10.1016/j.tim.2015.03.008
Raina J.-B., Dinsdale E. A., Willis B. L., Bourne D. G. (2010). Do the Organic Sulfur Compounds DMSP and DMS Drive Coral Microbial Associations? Trends Microbiol. 18, 101–108. doi: 10.1016/j.tim.2009.12.002
Raina J.-B., Tapiolas D. M., Foret S., Lutz A., Abrego D., Ceh J., et al. (2013). DMSP Biosynthesis by an Animal and its Role in Coral Thermal Stress Response. Nature 502, 677–680. doi: 10.1038/nature12677
Raina J.-B., Tapiolas D., Motti C. A., Foret S., Seemann T., Tebben J., et al. (2016). Isolation of an Antimicrobial Compound Produced by Bacteria Associated With Reef-Building Corals. PeerJ 4, e2275. doi: 10.7717/peerj.2275
Raina J.-B., Tapiolas D., Willis B. L., Bourne D. G. (2009). Coral-Associated Bacteria and Their Role in the Biogeochemical Cycling of Sulfur. Appl. Environ. Microbiol. 75, 3492–3501. doi: 10.1128/AEM.02567-08
R Core Team (2020). R: A Language and Environment for Statistical Computing (Vienna, Austria: R Foundation for Statistical Computing).
Robbins S. J., Singleton C. M., Chan C. X., Messer L. F., Geers A. U., Ying H., et al. (2019). A Genomic View of the Reef-Building Coral Porites Lutea and its Microbial Symbionts. Nat. Microbiol. 4, 2090–2100. doi: 10.1038/s41564-019-0532-4
Rohwer F., Seguritan V., Azam F., Knowlton N. (2002). Diversity and Distribution of Coral-Associated Bacteria. Marine. Ecol. Prog. Ser. 243, 1–10. doi: 10.3354/meps243001
Ruiz-Gonzalez C., Simo R., Sommaruga R., Gasol J. M. (2013). Away From Darkness: A Review on the Effects of Solar Radiation on Heterotrophic Bacterioplankton Activity. Front. Microbiol. 4. doi: 10.3389/fmicb.2013.00131
Rybniker J., Pojer F., Marienhagen J., Kolly G. S., Chen J. M., van Gumpel E., et al. (2014). The Cysteine Desulfurase IscS of Mycobacterium Tuberculosis is Involved in Iron–Sulfur Cluster Biogenesis and Oxidative Stress Defence. Biochem. J. 459, 467–478. doi: 10.1042/BJ20130732
Salgado P., Kiene R., Wiebe W., Magalhães C. (2014). Salinity as a Regulator of DMSP Degradation in Ruegeria Pomeroyi DSS-3. J. Microbiol. 52, 948–954. doi: 10.1007/s12275-014-4409-1
Schwarz G. (1978). Estimating the Dimension of a Model. Ann. Stat 6, 461–464. doi: 10.1214/aos/1176344136
Seemann T. (2014). Prokka: Rapid Prokaryotic Genome Annotation. Bioinformatics 30, 2068–2069. doi: 10.1093/bioinformatics/btu153
Seymour J. R., Simó R., Ahmed T., Stocker R. (2010). Chemoattraction to Dimethylsulfoniopropionate Throughout the Marine Microbial Food Web. Science 329, 342–345. doi: 10.1126/science.1188418
Sharp K. H., Sneed J. M., Ritchie K. B., McDaniel L., Paul V. J. (2015). Induction of Larval Settlement in the Reef Coral Porites Astreoides by a Cultivated Marine Roseobacter Strain. Biol. Bull. 228, 98–107. doi: 10.1086/BBLv228n2p98
Sievert S. M., Kiene R. P., Schulz-Vogt H. N. (2007). The Sulfur Cycle. Oceanography 20, 117–123. doi: 10.5670/oceanog.2007.55
Slezak D., Herndl G. J. (2003). Effects of Ultraviolet and Visible Radiation on the Cellular Concentrations of Dimethylsulfoniopropionate (DMSP) in Emiliania Huxleyi (Strain L). Marine. Ecol. Prog. Ser. 246, 61–71. doi: 10.3354/meps246061
Stefels J. (2000). Physiological Aspects of the Production and Conversion of DMSP in Marine Algae and Higher Plants. J. Sea. Res. 43, 183–197. doi: 10.1016/S1385-1101(00)00030-7
Sunda W., Kieber D. J., Kiene R. P., Huntsman S. (2002). An Antioxidant Function for DMSP and DMS in Marine Algae. Nature 418, 317–320. doi: 10.1038/nature00851
Tandon K., Lu C.-Y., Chiang P.-W., Wada N., Yang S.-H., Chan Y.-F., et al. (2020). Comparative Genomics: Dominant Coral-Bacterium Endozoicomonas Acroporae Metabolizes Dimethylsulfoniopropionate (DMSP). ISME. J. 14, 1290–1303. doi: 10.1038/s41396-020-0610-x
Tapiolas D. M., Raina J.-B., Lutz A., Willis B. L., Motti C. A. (2013). Direct Measurement of Dimethylsulfoniopropionate (DMSP) in Reef-Building Corals Using Quantitative Nuclear Magnetic Resonance (qNMR) Spectroscopy. J. Exp. Marine. Biol. Ecol. 443, 85–89. doi: 10.1016/j.jembe.2013.02.037
Trossat C., Rathinasabapathi B., Weretilnyk E. A., Shen T.-L., Huang Z.-H., Gage D. A., et al. (1998). Salinity Promotes Accumulation of 3-Dimethylsulfoniopropionate and Its Precursor S-Methylmethionine in Chloroplasts. Plant Physiol. 116, 165–171. doi: 10.1104/pp.116.1.165
Vairavamurthy A., Andreae M. O., Iverson R. L. (1985). Biosynthesis of Dimethylsulfide and Dimethylpropiothetin by Hymenomonas Carterae in Relation to Sulfur Source and Salinity Variations. Limnol. Oceanogr. 30, 59–70. doi: 10.4319/lo.1985.30.1.0059
Walker B. J., Abeel T., Shea T., Priest M., Abouelliel A., Sakthikumar S., et al. (2014). Pilon: An Integrated Tool for Comprehensive Microbial Variant Detection and Genome Assembly Improvement. PloS One 9, e112963. doi: 10.1371/journal.pone.0112963
Wang P., Cao H.-Y., Chen X.-L., Li C.-Y., Li P.-Y., Zhang X.-Y., et al. (2017). Mechanistic Insight Into Acrylate Metabolism and Detoxification in Marine Dimethylsulfoniopropionate-Catabolizing Bacteria. Mol. Microbiol. 105, 674–688. doi: 10.1111/mmi.13727
Williams B. T., Cowles K., Bermejo Martínez A., Curson A. R. J., Zheng Y., Liu J., et al. (2019). Bacteria are Important Dimethylsulfoniopropionate Producers in Coastal Sediments. Nat. Microbiol. 4, 1815–1825. doi: 10.1038/s41564-019-0527-1
Yeo W.-S., Lee J.-H., Lee K.-C., Roe J.-H. (2006). IscR Acts as an Activator in Response to Oxidative Stress for the Suf Operon Encoding Fe-S Assembly Proteins. Mol. Microbiol. 61, 206–218. doi: 10.1111/j.1365-2958.2006.05220.x
Yu G., Smith D. K., Zhu H., Guan Y., Lam T. T.-Y. (2017). Ggtree: An R Package for Visualization and Annotation of Phylogenetic Trees With Their Covariates and Other Associated Data. Methods Ecol. Evol. 8, 28–36. doi: 10.1111/2041-210X.12628
Zhang X.-H., Liu J., Liu J., Yang G., Xue C.-X., Curson A. R. J., et al. (2019). Biogenic Production of DMSP and its Degradation to DMS—their Roles in the Global Sulfur Cycle. Sci. China Life Sci. 62, 1296–1319. doi: 10.1007/s11427-018-9524-y
Zhang Y., Yang Q., Zhang Y., Ahmad M., Ling J., Tang X., et al. (2021). Shifts in Abundance and Network Complexity of Coral Bacteria in Response to Elevated Ammonium Stress. Sci. Total. Environ. 768, 144631. doi: 10.1016/j.scitotenv.2020.144631
Keywords: DMSP, sulfur cycle, coral-associated bacteria, holobiont, acrylate
Citation: Kuek FWI, Motti CA, Zhang J, Cooke IR, Todd JD, Miller DJ, Bourne DG and Raina J-B (2022) DMSP Production by Coral-Associated Bacteria. Front. Mar. Sci. 9:869574. doi: 10.3389/fmars.2022.869574
Received: 04 February 2022; Accepted: 06 April 2022;
Published: 04 May 2022.
Edited by:
Gabrielle Nevitt, University of California, Davis, CA, United StatesReviewed by:
Chuya Shinzato, The University of Tokyo, JapanCopyright © 2022 Kuek, Motti, Zhang, Cooke, Todd, Miller, Bourne and Raina. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jean-Baptiste Raina, SmVhbi1CYXB0aXN0ZS5SYWluYUB1dHMuZWR1LmF1
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.