
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mar. Sci., 15 July 2022
Sec. Deep-Sea Environments and Ecology
Volume 9 - 2022 | https://doi.org/10.3389/fmars.2022.869226
This article is part of the Research TopicDeep-sea Chemosynthetic Ecosystems: Living in Extreme EnvironmentsView all 13 articles
Cold seeps globally host dense unique oasis-type ecosystems, mainly fuelled by chemosynthetic microorganisms via reduced gases such as methane and hydrogen sulfide. However, the origin and pathway of nitrogen chemosynthesis in this widely distributed symbiont ecosystem remain poorly understood. Here, we explore biomarker methods (bulk stable isotope, amino acid (AA), fatty acid (FA) and compound-specific isotope analyses in gill tissues of mussels) to demonstrate the relative contributions of inorganic and organic nitrogen to symbiotic mussels at cold seeps in the South China Sea and their impact on the synthesis and metabolism of amino acids. Gigantidas platifrons (G. platifrons) symbioses with type II methanotrophs via the Serine pathway, and Bathymodiolus aduloides (B. aduloides) thrives with sulfur-oxidizing bacteria via the Calvin pathway, as revealed by bulk δ13C and δ13C of FAs. Based on the δ15N values in gill tissues of mussels, organic nitrogen from sediment is estimated as the dominant nitrogen source for B. aduloides (97-98%), in contrast, NH4+ was the main nitrogen source for G. platifrons. Different dominant nitrogen sources result in the δ15N of AAs in the gills of two mussel species having opposite trends, which might be related to synthesis and metabolism of AAs in symbiotic bacteria and host, respectively. Our findings reveal that the mechanism of nitrogen acquisition in cold seep systems is plastic and related to DIN sources/uptake and changing environmental conditions. These findings uncover novel biosynthesis of nitrogen in the deep sea, typically at cold seeps, and may have important implications for nitrogen biogeochemistry and deep-sea conservation.
Cold seeps are widespread seafloor ecosystems supported by the emission of reduced gases from seabed reservoirs (Ruff et al., 2015; Goffredi et al., 2020). Symbioses associated with bacteria have played a central role in shaping the ecosystem surrounding invertebrates in cold seeps (Rubin-Blum et al., 2019). Among the dominant animals in these ecosystems are symbiotic mussels, which live in symbiosis with thiotrophic and/or methanotrophic symbionts and derive the vast majority of their nutrition from these symbionts (Feng et al., 2015; Rubin-Blum et al., 2017). Since their first discovery, cold seeps have been recognized as important pathways of element cycling on Earth (Paull et al., 1984; Feng et al., 2018). Many studies on symbiotic relationships, especially related to carbon chemosynthetic processes, have been conducted. These findings have led to a basic understanding of seep ecosystems in terms of biodiversity and community composition (Suess, 2014) and energy and nutritional carbon sources (Brooks et al., 1987; Orphan et al., 2001). However, much more work has been focused on the carbon-fuelled symbiont system between bacteria and mussels than the nitrogen-fuelled system (Macavoy et al., 2002; Raggi et al., 2013; Rubin-Blum et al., 2017). Evidence that methane and CO2 are the main carbon sources to the symbiont ecosystem suggests that different carbon sources result in distinct δ13C values in the gills of mussels (Brooks et al., 1987), yet the nitrogen cycles in this widely distributed symbiont ecosystem remain largely unexplored (Vokhshoori et al., 2021). The nitrogen cycles can not only reveal the synthesis and metabolism of amino acids (AAs) in symbiotic mussels but also help to elucidate the interactions between fatty acids (FAs) and AAs and to understand the adaption of mussels to extreme environments and their interactions in cold seep ecosystems. Thus, understanding the nitrogen cycles in symbiotic mussels is vital to shed light on deep sea conservation.
Biochemical tracers, such as bulk stable isotopes (δ13C and δ15N), FAs, and compound-specific isotope analysis (CSIA), have been applied to bacteria and symbionts. Because of the limited kinetic isotope fractionation of carbon associated with methanotrophs, bulk δ13C values have successfully indicated the various carbon source contributions (Orphan et al., 2001; Michaelis et al., 2002). The varied δ15N values also suggest that symbionts are capable of using different nitrogen sources (Petersen et al., 2016). FA composition analysis demonstrated that type II methanotrophs were the predominant microorganism responsible for methane oxidation (Bull et al., 2000; Kellermann et al., 2012), especially in high-methane or low combined-nitrogen environments (Jahnke et al., 1999). The analysis of δ15N in AAs, however, has rarely been applied to quantitatively estimate the nutritive nitrogen sources used by symbionts in cold seeps but has emerged as a powerful approach for tracing the origins and fate of nitrogen in ecological and biogeochemical studies (Ohkouchi et al., 2017). Moreover, the combination of multiple isotope analyses, including bulk stable isotopes and stable isotopes in FAs and AAs, can help to elucidate the interactions between AAs and FAs (Riou et al., 2010; Takano et al., 2018).
In this study, we explored the biomarker methods previously used to investigate the trophic interactions of fishes (Gerringer et al., 2017; Wang et al., 2019) to delineate the biogeochemistry and nitrogen dynamics of symbiotic mussels in the Formosa Ridge (Site F) of the South China Sea (SCS). Site F is located in the northeast region of the continental slope of the SCS. It is a NNW–SSE oriented ridge that is nearly 2 km in length and rises over 100 m above the surrounding seabed (Figure 1A) (Feng et al., 2015). At site F, Gigantidas platifrons and Bathymodiolus aduloides were the most common and abundant taxa (Figure 1B) and symbiosed with methanotrophs and sulfur-oxidizing bacteria in their gills, respectively (Feng et al., 2015; Xu et al., 2019). Therefore, a comparative study of the gills of G. platifrons and B. aduloides is conducted in this research.
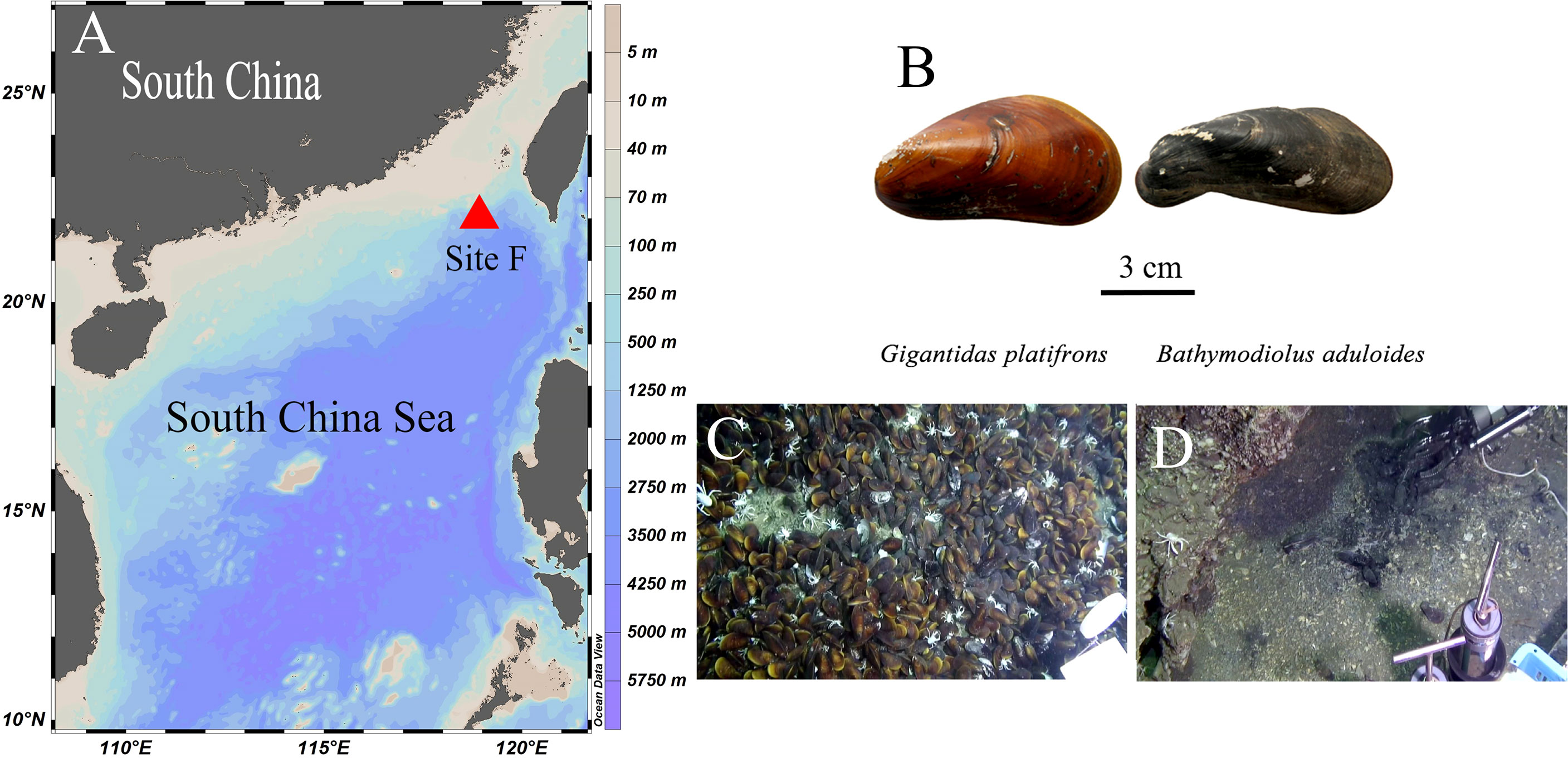
Figure 1 Figure 1 |Study site and mussel beds. (A) Site F location; (B) shells of the two mussels; (C) Gigantidas platifrons lives on carbonate pavement; (D) Bathymodiolus aduloides lives on muddy sediment; (B–D) were cited from Feng et al.4.
Based on the observation that G. platifrons and B. aduloides have obviously different biological composition characteristics, such as bulk stable isotopes and CSIA, we tested the hypothesis that G. platifrons and B. aduloides have different nitrogen-fuelled systems—fuelled by inorganic and organic nitrogen-influenced by the surrounding environment, in turn resulting in different symbiotic bacteria (Feng et al., 2015). We identified the type of methanotrophs and carbon biogeochemical cycles in mussels based on the bulk δ13C and δ13C of FAs. We also estimated the relative contributions of different nitrogen sources from δ15N values. To clarify the synthesis of AAs, we measured the δ15N of AAs and described the conversion between AAs and FAs. This multifaceted study provides a broad perspective for carbon and nitrogen transformations in symbiotic mussels in cold seeps and lays the foundation for their origin and adaptation to extreme environments.
As described in other studies (Feng et al., 2015; Sun et al., 2017), our study site, called Site F (Formosa Ridge), is located at 22°06.922´N and 119°17.130´E at a depth of 1120 m. It is an oriented ridge above the surrounding seabed and has active methane seepage on the northeastern corner of the continental slope of the South China Sea (Figure 1A). Two mussel species were collected using the manned submersible Jiaolong during the Dayang-31 expedition in July 2013 (Figure 1B). The mussels were identified as Gigantidas platifrons and Bathymodiolus aduloides by the National Center for Biotechnology Information’s gene sequence database at http://www.ncbi.nlm.nih.gov and http://www.marinespecies.org/aphia.php?p=taxdetails&id=1346726 (Wong et al., 2015; Xu et al., 2019). G. platifrons was numerically dominant and occurred mostly on carbonate pavement (Figure 1C), while B. aduloides was rare and occurred mainly on muddy sediment (Figure 1D). G. platifrons and B. aduloides harboured methanotrophic and thiotrophic symbionts, respectively. All mussels were placed in the sample baskets of the submersible. Once the deck of the ship returned, twelve individuals of G. platifrons and five individuals of B. aduloides were dissected into the mantle, gill, viscera, and foot tissue. All samples were stored at -20°C and transferred to the laboratory. In this study, the gill tissues were used to conduct the following experiments.
To compare the bulk isotopic contents in the symbiotic mussels at cold seep, the shallow-water mussels M. edulis and M. coruscus were bought from Sanggou Bay (in July 2014, 37°08´N and 122°30´E) and the aquaculture areas of Zhoushan (in June 2015, 30°32´N and 121°25´E), respectively. The species names were confirmed based on their morphological description.
All gill tissues were lyophilized in a freeze-dryer (LOC-1; Christ, Germany) and powdered using a mortar and pestle. Dried powdered samples (~0.2 mg for δ13C, ~1 mg for δ15N, three replicates) were weighed into tin cups, and stable isotopes (δ13C, δ15N) were measured using an isotope ratio mass spectrometer (Finnegan Delta plus XP; Thermo, Germany). The results were normalized to the Vienna Pee Dee Belemnite standard (V-PDB) for δ13C and to atmospheric N (N2) for δ15N. The stable isotope ratios are expressed in δ notation of units per mille as follows (Bond and Hobson, 2012):
where X =13Cor15N and R =13C/12Cor15N/14N. The precision of the stable isotope analyses was ±0.1‰.
FAs were extracted using a modified Folch method (Folch et al., 1957; Wang et al., 2019). Approximately 100 mg of the gill tissues was extracted by dichloromethane/methanol (2:1 v/v, including 0.01% BHT) and then centrifuged (3000 rpm, 10 min). The upper organic solvent layer was transferred to a flask using a pipette and evaporated to dryness under a stream of N2 at room temperature. The dried FAs were transesterified to FA methyl esters (FAMEs) in a mixture of methanol (containing 5% HCl) and n-hexane at 50 for 12 h.
The FAMEs were analysed using gas chromatography (7890A GC; Agilent, United States) equipped with a DB-FFAP capillary column (30 m in length, 0.25 mm i.d., 0.25 mm film thickness; Agilent, United States). As an internal recovery standard, 21:0 was added to the samples, and 19:0 methyl ester was added as an internal quantification standard. The injector and detector temperatures were both 250°C. Injections (1 mL) were made at 60°C, and the temperature was increased to 170°C at a rate of 30°C/min. The temperature was held constant for 5 min, increased to 220°C at 1°C/min and held at this temperature for 10 min. Nitrogen (N2) was used as the carrier gas at a flow rate of 1 mL/min. FAMEs were identified by comparing retention times with those of commercial standards (37 Component FAME Mix; Sigma, United States). The individual FAs were expressed as percentages of the total FA content based on peak areas. The FA recoveries in the analysis were >85%. If more than one sample was analysed, the data were reported as the mean ± standard deviation (SD).
To investigate the synthetic pathways of FAs from carbon sources, the δ13C values of FAs in B. aduloides and G. platifrons were measured. The compound-specific 13C FA values were analysed using a Trace Ultra GC coupled to a Delta V mass spectrometer via a GC combustion interface using helium (He) as the carrier gas (Thermo Scientific, Germany). The FAMEs were separated on a DB-5MS column (50 m, 0.32 mm i.d., 0.25 mm film thickness). The δ13C values of the FAMEs were calibrated by analysing an 18:0 FAME certified standard (certified: -23.24‰, measured: -23.34‰ ± 0.1‰) supplied by Indiana University, and the analytical precision was ≤0.3‰ (Wang et al., 2019). All samples were analysed in triplicate.
To obtain isotope data for the FAs, FAME isotopes were corrected for the isotopes of the methyl moiety originating from methanol using the following formula:
where δ13CFA is the isotope of the FA, δ13CFAME is the isotope of the FAME, δ13CMeOH is the isotope of the methanol derivatization reagent (-37.4 ± 0.1‰), and n is the number of C atoms in the FAs.
AAs were extracted from all the samples via HCl hydrolysis. Briefly, 1 mg of gill tissues was hydrolysed with 6 M HCl at 110°C for 20 h in precombusted sealed glass ampoules. After hydrolysis, the solution was neutralized to pH 8.5 with NaOH (Fitznar et al., 1999). The AAs were determined by high-performance liquid chromatography equipped with a Phenomenex™ Hyperclone column (5 µm particle diameter, BDS C18, 250 mm length, 4 mm inner diameter) (Wu et al., 2007). Eluent A was 125 mM sodium acetate with 2% methanol adjusted to pH 6.8 with acetic acid, and eluent B was 100% methanol. Gradient elution was used to separate all AAs, and external standards of the AAs (Sigma-Aldrich, Merck, USA) were used for calibration. The relative standard deviation triplicate analysis of the individual AAs was <3.5%.
The δ15N of AAs was measured by coupling instrumental analysis (ion chromatography and Precon-GC-IRMS) and oxidation-reduction of AAs (Zhang et al., 2021; Wang et al., 2022). Briefly, AAs were separated using an IC system (Dionex, ICS-5000+, Thermo Scientific, USA) equipped with an autosampler (Dionex, AS-AP) with an adjustable injection volume and coupled to an automated fraction collector (Dionex, UltiMate 3000). For the separation of AAs, the IC was equipped with an IC column (Dionex™ AminoPac™PA10, Thermo Scientific, USA), and a solvent ramp programme was used. At the end of each sample, 1 M sodium acetate was used to wash the column to remove any residue (Zhang et al., 2021). IC procedural blanks were collected after AA fractions, and were re-injected in IC system to ensure that all AAs has been collected completely. Meanwhile, the solvent blanks were also collected between Phe and Glu. These blanks were used to correct the isotopic interference from any nitrogenous compounds introduced by the analytical procedure up to this point that could be potentially converted to nitrite by NaClO [more details showed in Zhang et al. (2021)] Purified AAs were oxidized to NO2− by Strecker degradation (Schonberg and Moubacher, 1952; Zhang and Altabet, 2008). The next step was the conversion of NO2− to N2O; here, NH2OH•HCl was used as an alternative reductant (Bothner-By and Friedman, 1952; Liu et al., 2014). The stable nitrogen isotopes of the produced N2O were analysed by a Precon-GC-IRMS (Trace GC 2000, Delta-V advantage, Thermo Scientific, USA) equipped with an autosampler (Precon Automated, Thermo Scientific, USA). Standard AAs (Merck, USA; Glu, USGS40, Gly, USGS64, the Reston Stable Isotope Laboratory of the U.S. Geological Survey) were treated with the same protocol as the samples and were used to calibrate the δ15N value of the AAs. The results from this method were compared with the general GC-IRMS analysis with two lab standards (McCarthy Lab AA mixture and cyanobacteria) and only 0.6‰ deviations were found (Zhang et al., 2021).
SPSS 23.0, Primer 7.0 and MATLAB were used to perform data analysis. By SPSS 23.0, the data were tested for normality by using Shapiro–Wilk test and homogeneity of variance using Levene’s test. Cross validation using a discriminant analysis was applied to test the efficiency of FA and AA data, and linear regression was used to examine the relationship between N content and δ15N of AAs in SPSS 23.0. A cluster analysis (K-Means) based on FAs was performed using Primer 7.0. Based on the coefficients in the linear combinations of variables making up the principal components (PCs), the dominant parameters were chosen to characterize the different mussel groups. To illustrate the reliability of the data, the Spearman correlation coefficient and the p value (two-tailed) were used to evaluate the significance of the correlation.
As described in previous studies, NH4+ was the main inorganic nitrogen source to G. platifrons and methane-oxidizing bacteria (Sun et al., 2017), and particulate organic matter (POM) only supply 5% of food sources to G. platifrons by filter-feeding (Wang, 2018). Thus, we assume that NH4+-derived nitrogen is considered the sole inorganic nitrogen source in the mussel gill of G. platifrons and contribute 95% of nitrogen, thereby particulate organic matter (POM)-derived nitrogen contributing 5% by the filter feeding of G. platifrons. A simple two-source mixing model in MATLAB was used to calculate the δ15Nammonium values in the surrounding environment. The following equations were used for this calculation:
where ƒammonium is the relative contribution of NH4+ (95%), ƒOM is the relative contribution of OM (5%), δ15Nammonium is the δ15N of NH4+, Δ15Nammonium is the 15N enrichment of ammonium assimilation, δ15NPOM is the δ15N of POM, Δ15NPOM is the δ15N enrichment of POM assimilation, and δ15Ngill is the δ15N of mussel gill.
The Δ15Nammonium of ammonium assimilation is supposed as -4‰ when ambient ammonium is low (<1 mM) at the cold seeps (Liao et al., 2014). In site F, the concentration of NH4+ in sediment is about 10 mg/kg (Jing et al., 2020), which is lower than 1 mM. Thus, the Δ15Nammonium is estimated as -4‰ in this study. As measured in previous study, the δ15N of sediment at site F was 3.7‰ (Jing et al., 2020). Based on the δ15N relationship between the bulk sediment and organic nitrogen in the SCS, we estimated that δ15NPOM was 3.7‰ (Kienast et al., 2005). In symbiotic organisms, the host ingested particulate detrital or living nitrogen sources (PON), there was no δ15N enrichment observed (Ferrier-Pages and Leal, 2019), in turn Δ15NOM is supposed as zero. By the two-source mixing model, the δ15N of NH4+ was then calculated (4.5 ± 0.4‰). Furthermore, we applied δ15Nammonium value to calculate the relative contributions of inorganic and organic nitrogen sources in B. aduloides. There are no genomics studies of B. aduloides so far. Thus, the relative contributions of inorganic nitrogen (including nitrate and ammonium) are unknown. We assume that nitrate, ammonium and POM jointly determine the δ15Ngill of B. aduloides. The following equations were used for this calculation:
where ƒnitrate is the relative contribution of nitrate, δ15Nnitrate is the δ15N of nitrate, Δ15Nnitrate is the δ15N enrichment of nitrate assimilation.
To investigate the symbiotic relationship of cold seep mussels with their symbiotic bacteria and the turnover of organic and inorganic nitrogen between the mussels and the surrounding environment, we collected 47 samples (gill tissues) from cold seeps and shallow water and analysed them for FAs, AAs, bulk δ13C and δ15N, δ13C of FAs, and δ15N of AAs (Supplementary Data).
In the principal component analysis (PCA) of FAs, all gill tissues of mussels were separated into four groups (Figure 2). PC1 (79.2%) distinguished mussels into cold seep areas and shallow water areas. 16:1n-7, 18:1n-7, 20:4n-6, 20:5n-3, and 22:6n-3 were the main loading factors contributing to PC1. PC2 (5.3%) also separated the mussels, and the main loading factors were 18:1n-7 and 20:4n-6. The PCA results revealed that symbiotic mussels and filter-feeding mussels had different FA components (Supplementary Figures 1, 2). The filter-feeding mussels (Mytilus edulis, n=15; M. coruscus, n=15) had higher n-3 polyunsaturated fatty acids (n-3 PUFAs) than symbiotic mussels in cold seeps. B. aduloides (n=5) harbouring sulfur-oxidizing bacteria had a low n-3 PUFA percentage, and G. platifrons (n=12), which thrived with methanotrophs, was not detected (Supplementary Figure 1). In contrast, B. aduloides and G. platifrons had higher 16:1n-7 and 18:1n-7 levels than filter-feeding mussels (Supplementary Figure 2). Moreover, 18:1n-7 was not detected in M. edulis and M. coruscus.
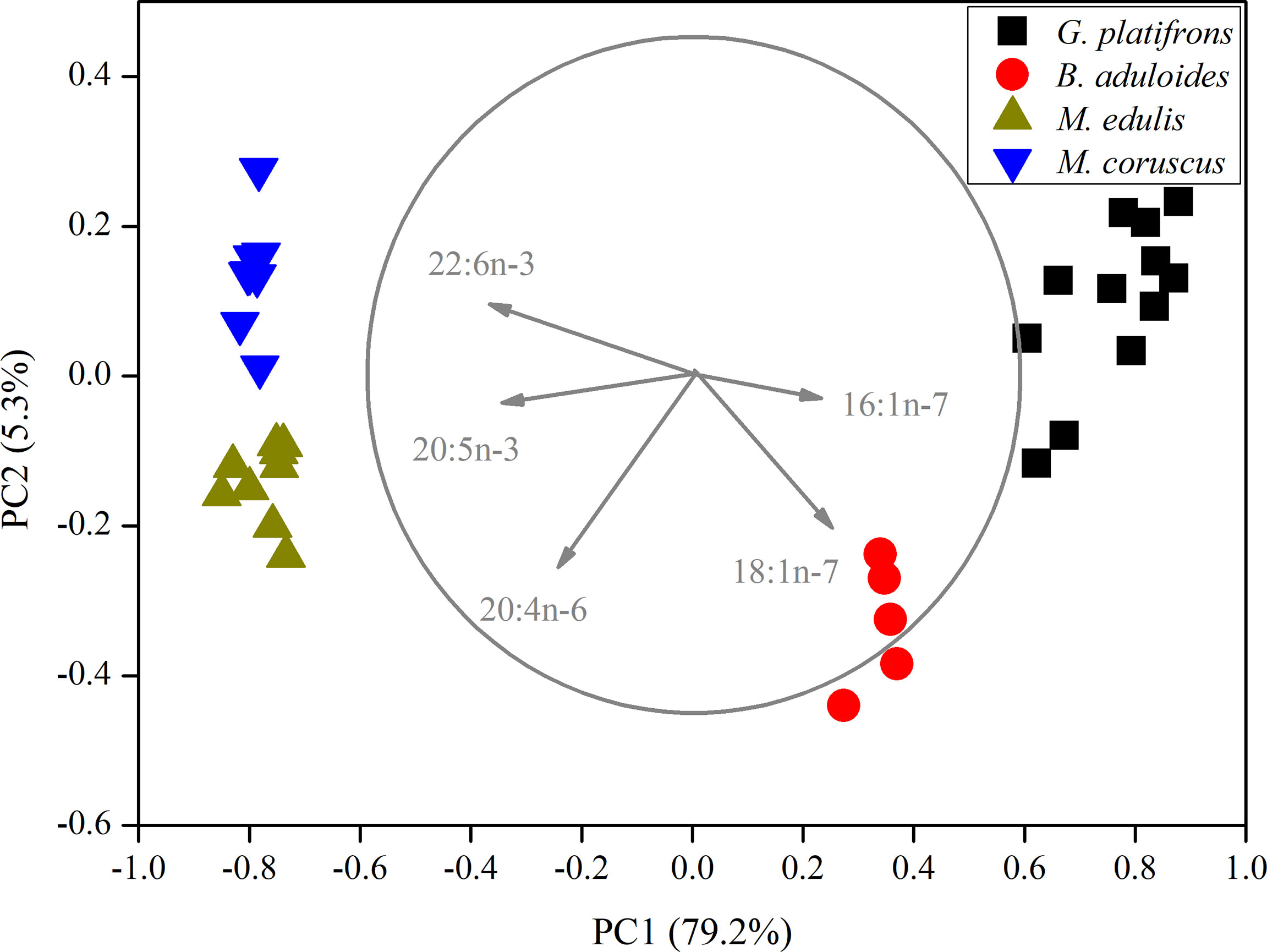
Figure 2 Principal component analysis of mussels based on fatty acids. The length of vectors represents their contributions to axis.
The bulk stable isotopes of mussels in cold seeps and shallow water differed (Figure 3). G. platifrons had more negative δ13C (-69.8 ± 1.7‰) and δ15N (0.7 ± 0.5‰) values than other mussel species, which was similar with symbiotic mussels in the cold seeps (Vokhshoori et al., 2021). The δ13C and δ15N values of B. aduloides were -34.7 ± 2.0‰ and 3.6 ± 0.3‰, respectively. M. edulis and M. coruscus had similar δ13C (-20.7 ± 0.8‰, -21.2 ± 0.5‰; P<0.05) values; however, the δ15N values significantly differed (9.7 ± 0.3‰, 2.8 ± 1.0‰; P<0.01).
δB. aduloides (-49.2 to -38.5‰) had more positive δ13C of FAs than G. platifrons (-78.4 to -70.4‰). Considering the carbon content of FAs, the average δ13C values of FAs in B. aduloides and G. platifrons were -41.3 ± 0.6‰ and -76.8 ± 0.4‰, respectively (Figure 4). However, the δ13C of 18:0 and 18:1n-7 in G. platifrons was more positive than other FAs, which was inconsistent with the results for B. aduloides (Figure 4 and Supplementary Figures 3, 4).
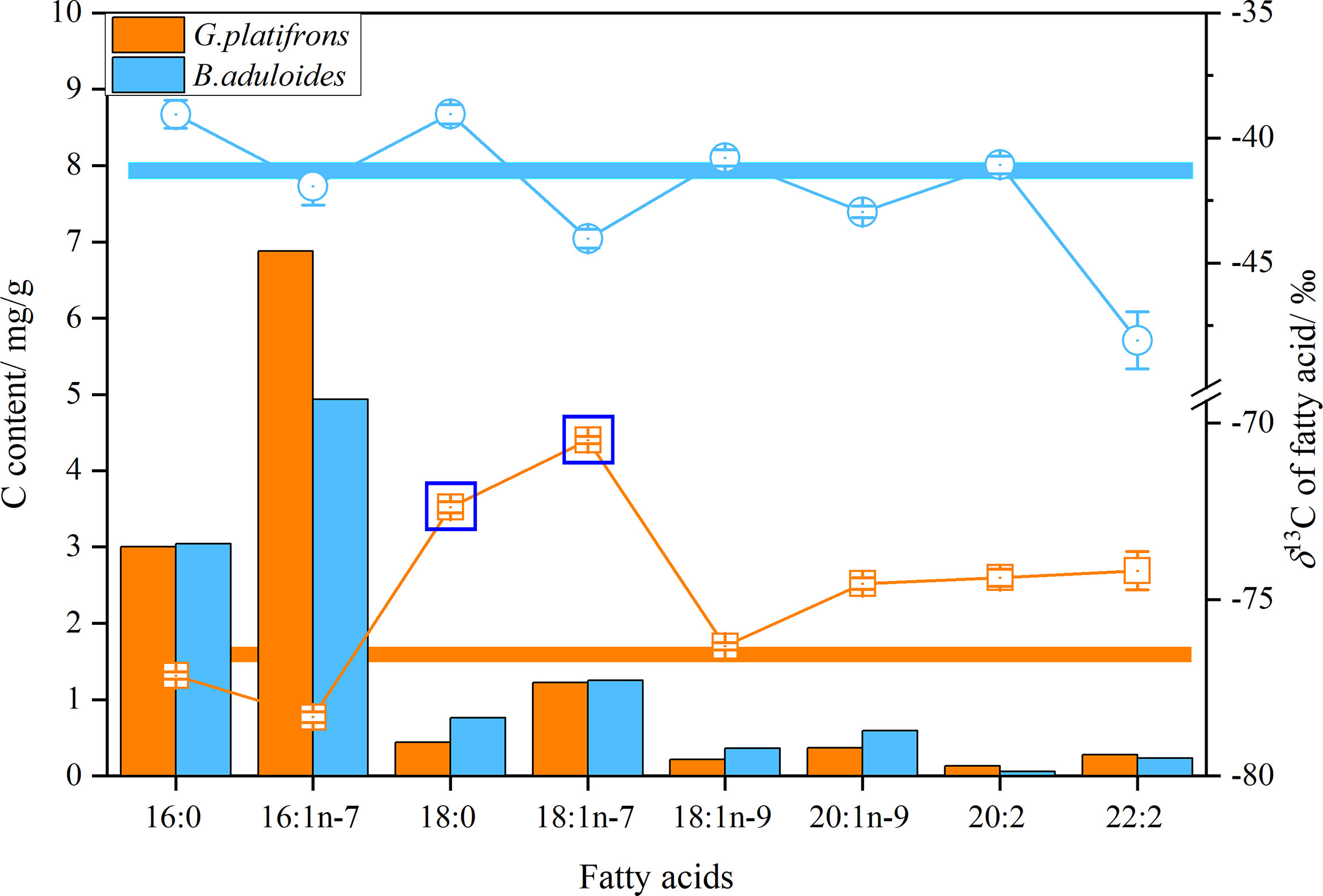
Figure 4 δ13C and carbon content of fatty acids in symbiotic mussels. Histograms represent the carbon content of fatty acids in tissues (dry weight), points represent the δ13C of a single fatty acid, and straight lines represent the average δ13C of fatty acids.
To characterize the turnover of organic and inorganic nitrogen in cold seep mussels, the δ15N values of AAs in B. aduloides and G. platifrons were used to trace the nitrogen flow between the symbiont and host. In G. platifrons, the N content was positively related to the δ15N of AAs (y=0.25x+6.52, R2 = 0.67, p<0.05) (Figure 5). In contrast, the N content in B. aduloides was negatively related to the δ15N of AAs (y=-0.35x+12.36, R2 = 0.64, p<0.05), which has not been reported to our knowledge. Glycine (Gly) and threonine (Thr) were also measured in this study (Supplementary Figures 5, 6) but these results are not shown in Figure 5 due to the Gly cleavage system (Yamaguchi et al., 2017).
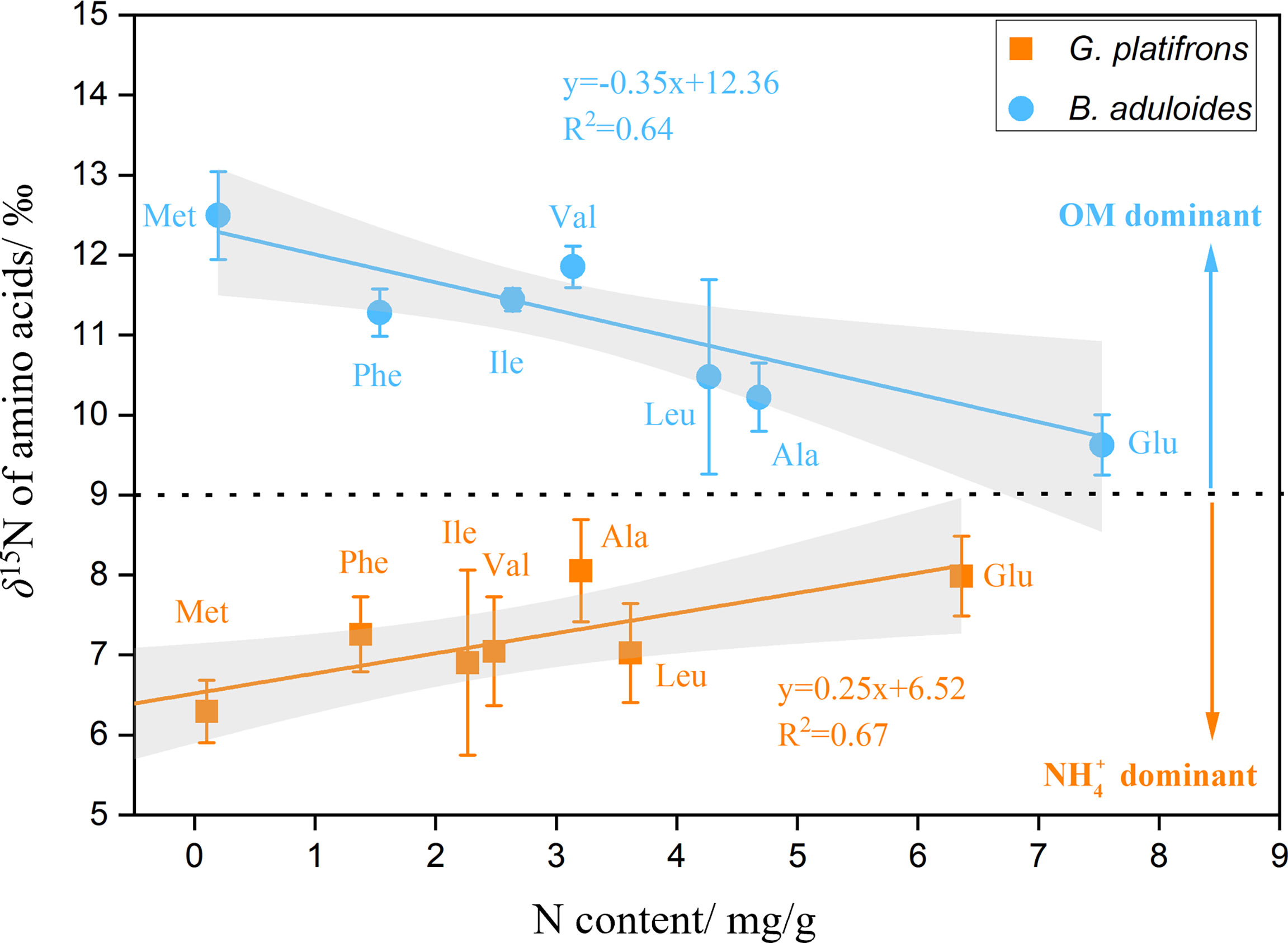
Figure 5 Nitrogen content and δ15N of amino acids in symbiotic mussels. OM, organic matter; above the black dotted line represents OM as the main nitrogen source for mussels; below the black dotted line represents nitrate as the main nitrogen source for mussels; the grey area represents the 95% confidence interval.
G. platifrons mainly use methane-based energy sources as their carbon sources (Orphan et al., 2001; Michaelis et al., 2002), and in support of this, the δ13C of G. platifrons is consistent with the δ13C of methane (Michaelis et al., 2002; Feng et al., 2015). In contrast, the δ13C values of B. aduloides indicate a -25‰ fractionation from the measured δ13C of CO2 in seawater, which reveals that sulfur-oxidizing bacteria utilize the Calvin cycle for carbon dioxide fixation (Ponnudurai et al., 2017). Meanwhile. this species was consistent with the chemosymbiotic species from the Gulf of Cadiz, which inferred a thiotrophic mode of nutrition (Rodrigues et al., 2013). Moreover, the low content of n-3 PUFAs and high contents of 16:1n-7 and 18:1n-7 in symbiotic mussels also demonstrate that energy derived from photosynthesis has an extremely low impact on the symbiotic mussels (Ackman, 1989). The high contents of 16:1n-7 and 18:1n-7 in the mussels indicate that G. platifrons and B. aduloides symbiose with type II methanotrophs (Serine pathway) and sulfur-oxidizing bacteria, respectively (Supplementary Figure 2), which is consistent with the genome analysis (Sun et al., 2017). Type II methanotrophs are favoured in high-methane environments and utilize methane as the sole carbon and energy source (Jahnke et al., 1999). This is also the reason why G. platifrons occurs mostly on carbonate pavement, where methane is abundant. In the muddy sediment of cold seeps, sulfate reduction is achieved by sedimentary microbial mats with high metabolic rates (Tobler et al., 2016). Thus, sulfur-oxidizing bacteria provide B. aduloides with nutrition through the fixation of CO2 into biomass using produced sulfide compounds as an energy source (Volland et al., 2018). These factors result in B. aduloides living mainly on muddy sediment.
The bulk δ13C and δ13C of FAs differed substantially between the two symbiotic mussels, which is consistent with the assimilation of different carbon sources, as discussed above. Compared with the bulk δ13C, the average δ13C values of FAs were depleted by approximately 7–9‰ in B. aduloides and 2–7‰ in G. platifrons, suggesting de novo synthesis from dietary carbon sources (Deniro and Epstein, 1977). However, the different carbon isotope fractionation of δ13C in mussels might be a result of the energy supply, with 12C being the more favourable carbon source when energy is limited (Hugler and Sievert, 2011). In carbonate pavement, the abundant methane supplies enough energy to the type II methanotrophs symbiosing with G. platifrons, even though Serine pathway organisms operate in a less energetically favourable way (Anthony, 1982; Jahnke et al., 1999). Different from type II methanotrophs, sulfur-oxidizing bacteria use reduced sulfide from the anaerobic oxidation of methane as the energy source (Feng et al., 2015; Tobler et al., 2016). Instead of methane, B. aduloides use seawater CO2 fixed via sulfur-oxidizing bacteria, which agrees well with the measured δ13C values of CO2 dissolved in seawater above the mussel beds (Feng et al., 2015). Thus, different carbon sources result to various carbon isotope fractionations. In addition, in both specimens, the differences in the δ13C of FAs are also indicative of different biochemical pathways (Macavoy et al., 2002; Riou et al., 2010). In B. aduloides, unsaturated FAs are depleted relative to saturated FAs, indicating de novo transformations (Fang et al., 1993). In contrast, 18:1n-7 in G. platifrons shows more positive δ13C values than 18:0, which provides additional evidence for a biochemical pathway involving FAs in the symbiosis of type II methanotrophs and mussels. A previous study suggested that host cells can assimilate AAs directly into FAs (Riou et al., 2010). Thus, the positive δ13C of 18:1n-7 in type II methanotrophs demonstrates that this process might occur in bacteria (Bull et al., 2000; Takano et al., 2018).
Bulk nitrogen isotopes, similar to bulk carbon isotopes, reflect the nitrogen diet sources of mussels. For example, the high δ15N values of M. edulis illustrate their trophic interaction with POM in Sanggou Bay (Mahmood et al., 2016), while the low δ15N values of M. coruscus suggest that their diets different with M. edulis. In a previous study, the lower δ15N values of phytoplankton suggested that the primary producer in the East China Sea depend mainly on atmospheric nitrogen fixation (Minagawa and Wada, 1984). Thus, the low δ15N values of M. coruscus might be caused by atmospheric nitrogen indirectly. In extreme environments, these aspects have not been sufficiently explored; however, it is clear that the symbiotic mussels in cold seeps must use alternative nitrogen sources, such as ammonium, nitrate or organic nitrogen, in the sediment (Petersen et al., 2016; Ferrier-Pagès and Leal, 2018), which are rarely quantitatively estimated.
In order to estimate the various sources of nitrogen to B. aduloides, we have to delineate the contribution of ammonia, nitrate and organic nitrogen. δ15Ninorg in the SCS had a range of 3.1-4.8‰, and with the δ15N of Ninorg generally being significantly isotopically lighter than bulk sedimentary δ15N (3.7‰ in this study) (Kienast et al., 2005; Jing et al., 2020). Thus, the δ15Nnitrate in this study was assumed between 3.1and 3.7‰. The average values of δ15Nnitrate were used in this study (3.4 ± 0.3‰). During assimilation of nitrate by microbes, Δ15Nnitrate is supposed as 5‰ when nitrate is not limiting (Liao et al., 2014). In the three-source mixing model, the relative contributions of NH4+, NO3-, and OM cannot be calculated directly. Thus, we calculated the ranges of inorganic nitrogen contributions when we assumed NH4+or NO3- had the contribution to B. aduloides. When the NH4+ was the inorganic nitrogen source to B. aduloides, the OM had a relative contribution of 97%. Otherwise, the NO3- was the inorganic nitrogen source to B. aduloides, the OM had a relative contribution of 98%. In other words, POM was the main nitrogen source to B. aduloides (97-98%). As suggested in another study, filter feeding selectively on a nitrogen-rich suspension might be an important component of the nutritional requirements of B. childressi, which is consistent with our results (Pile and Young, 1999). This observation is also consistent with the regional differences, in which G. platifrons lives mainly on carbonate pavement and B. aduloides occurs mainly on muddy sediment. Moreover, these results illustrated that the limited sulfide might not satisfy their energy needs for the synthesis of all types of matter, and filter feeding OM from the surrounding environment might play a more prominent role in B. aduloides (Ponnudurai et al., 2020).
Based on the different contributions of inorganic and organic nitrogen to the symbiotic mussels, more details about the synthesis and metabolism of AAs can be revealed. The large 15N depletion of Gly relative to other AAs is caused by the Gly cleavage system in organisms, which favours 15N degradation and causes 14N depletion in the remaining Gly molecules (Yamaguchi et al., 2017). Thr is also impacted by the Gly cleavage system because of irreversible transamination reactions (Mcmahon and Mccarthy, 2016). The peculiar fractionation patterns of Thr and Gly are completely different from those of other AAs, routinely exhibiting significant depletion of 15N (Mccarthy et al., 2013; Mcmahon and Mccarthy, 2016). The degradation pattern of “heavy” AAs might produce metabolic fuel for de novo FA biosynthesis (Berg et al., 2002). Thus, the products of Thr and Gly degradation might result in positive δ13C values in 18:1n-7 in G. platifrons. However, the degradation pathways of nitrogen in AAs are independent of the synthesis of carbon in FAs. The complex carbon flows in symbiotic mussels might cause the differences in the δ13C of FAs between G. platifrons and B. aduloides (Riou et al., 2010).
In addition to Gly and Thr, the δ15N values of AAs can illustrate the conversion between AAs. The opposite linear relationships between δ15N values of AAs and nitrogen content of AAs might indicate the different synthesis and degradation of AAs in two symbiotic mussels. δIn a previous study, the nitrogen contents of AAs in bacteria show increases in the abundance of less energetically costly AAs (Akashi and Gojobori, 2002). Thus, Glu is the less energetically costly AA and have high content, Met, Phe is the highest energetic cost for biosynthesis and have low content, and other AAs (including Ile, Leu, Val, Ala) occupy the middle position (Yamaguchi et al., 2017). In G. platifrons, NH4+ was used as the inorganic nitrogen to de novo Glu synthesis. Glu, which is typically the most abundant AA in organisms, is often considered a “Glu pool” to form other AAs (Bender, 2012). In G. platifrons, both transamination and deamination favour the lighter stable isotope (14N) and leave the residual “Glu pool” 15N-enriched (Macko et al., 1986; Mcmahon and Mccarthy, 2016). Compared with that of Glu, the biosynthesis of other AAs requires more energy and favours lighter nitrogen (Wagner, 2005; Yamaguchi et al., 2017). Thus, the δ15N values of AAs in G. platifrons reflected the order of the energy cost required to form different AAs. A previous study showed that the order of the energy cost was Ile>Leu> Val> Ala. The different order in G. platifrons (Ile>Val>Ala>Leu) may be caused by the mixture of de novo AA synthesis from inorganic nitrogen coupled with AA incorporation from OM (Ohkouchi et al., 2017). Met is considered as the high energetic cost for biosynthesis, it may be reasonable for the microbes to use incorporated Met directly via salvage incorporation and not to degrade it (Yamaguchi et al., 2017). Thus, Met had lowest δ15N values in G. platifrons.
In contrast, B. aduloides has a negative correlation between N content and δ15N values of AAs, which is consistent with the assimilation of different nitrogen sources. The filter feeding of B. aduloides might play a prominent role in its ingestion of POM from the surrounding sediment (97-98% from surrounding POM) (Ponnudurai et al., 2020). B. aduloides assimilates OM and degrades it into small molecules such as free AAs (Hoppe et al., 2002) and then transports the Glu to symbiotic bacteria (Hosie et al., 2002; Ponnudurai et al., 2020). Bacteria can use Glu by metabolizing it as carbon and nitrogen sources for resynthesis of AAs or by salvage incorporation (Davies and Humphrey, 1978; Ponnudurai et al., 2020). This process results in the remaining AAs being theoretically enriched in 15N relative to the reacting AAs (Chikaraishi et al., 2007). However, the 14N of metabolized AAs is not excreted as a waste product but is recycled by the symbiont and as resynthesized AAs (Ferrier-Pagès and Leal, 2018). Of the AAs, the Glu in the “Glu pool” was most affected in terms of its 14N levels, as it incurred the lowest energy cost and was most abundant. Compared with Glu, Met is the most expensive AAs in terms of consumed moles of ATP per molecule (Kaleta et al., 2013). The symbiotic mussels use incorporated Met directly via salvage incorporation and not to degrade it as discussed above (Yamaguchi et al., 2017). Meanwhile, Met is rarely affected by the 14N of metabolized AAs, and retain the highest δ15N values in AAs.
Such kind of knowledge is the first time to be explored in seep ecosystem (Figure 6), it will better constrain the synthesize and metabolism of key elements governing the carbon, nitrogen and other element cycles in extreme environment. These studies will propose relationships between groups of fatty acids and amino acids, which allow testable predictions about biogeochemical processing in symbiotic mussels. An enhanced understanding of genomes and metabolic processes of microbials involved in those organic matter production and consumption in symbiotic mussels will contribute a significant relevance to this topic.
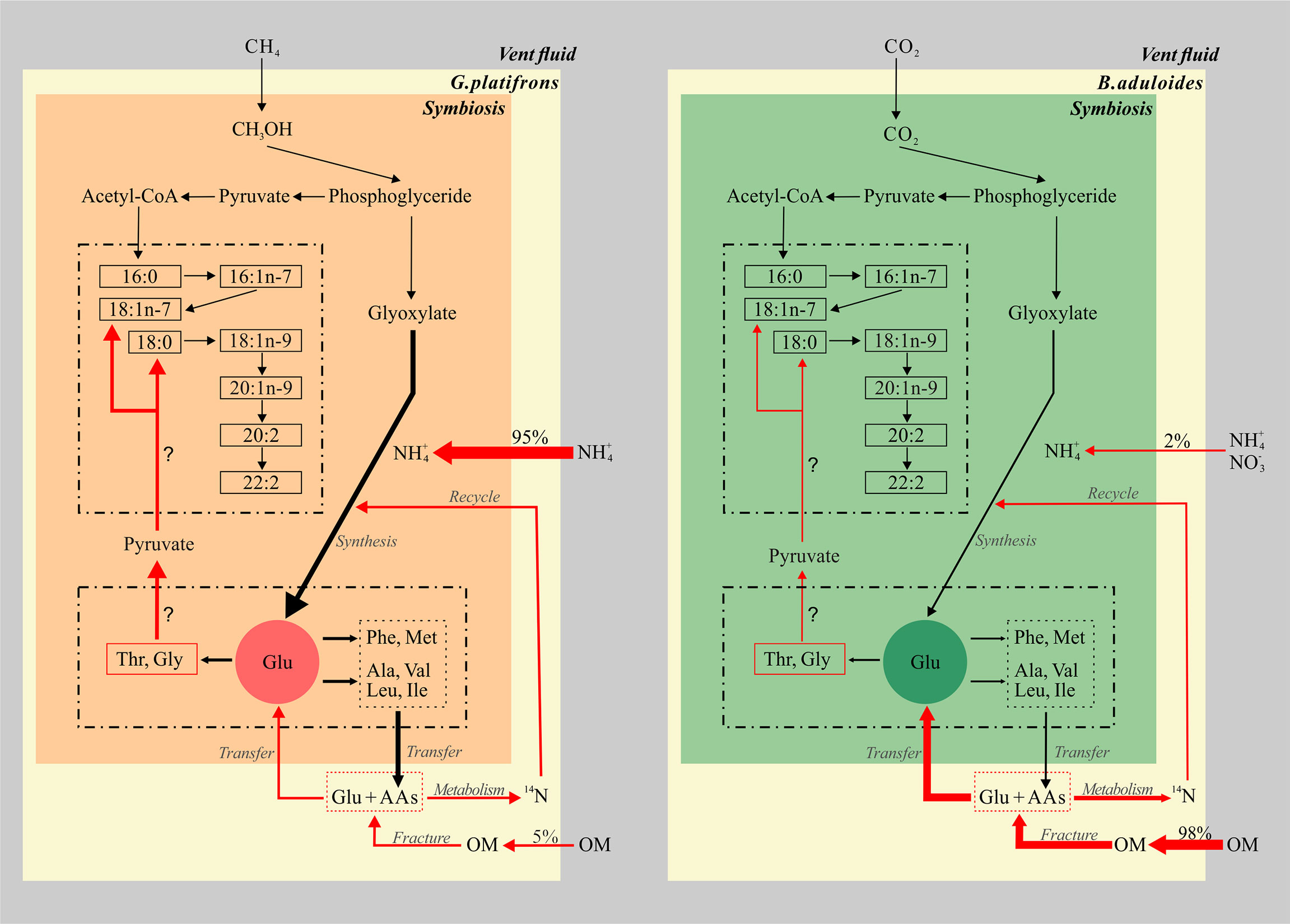
Figure 6 Conceptual model of fatty acids and amino acids in symbiotic mussels. The left part represents the symbiont of Gigantidas platifrons, and the right part represents the symbiont of Bathymodiolus aduloides; OM, organic matter; the thickness of arrows represents the relative contributions of inorganic and organic nitrogen sources.
The biomarker evidence from this study supports the hypothesis that inorganic and organic nitrogen make different contributions to the nitrogen sources of G. platifrons and B. aduloides, respectively (Figure 6). We demonstrate that methane and CO2 from the surrounding environment are the main carbon sources for G. platifrons and B. aduloides and impact the δ13C values of FAs in these mussels. Their unique symbiotic relationships lead to obvious bacterial FA signatures in the mussels, which can represent methanotrophic and thiotrophic symbionts, respectively (Bull et al., 2000; Kellermann et al., 2012). The differences in AA δ15N values, which showed opposite trends in the symbiotic mussels, might provide some evidence that the symbiotic mussels use different nitrogen sources. Inorganic nitrogen is the dominant nitrogen source for G. platifrons and OM is the dominant nitrogen source for B. aduloides, which are consistent with the nitrogen contributions from inorganic nitrogen and OM. The simple mixing model presented here provides baseline information for future studies aiming to quantify the different contributions of dietary sources in unique oasis-type ecosystems on the seafloor and to reveal the carbon and nitrogen flows in symbionts that are impacted by their surrounding environment in cold seeps.
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
FW and YW contributed for conceptualization, and writing original draft preparation. DF contributed for sample preparation, data management, and draft modification. All authors contributed to the article and approved the submitted version.
This work was supported by the Natural Science Foundation of China (41876074, 42106049) and National Key Program for Basic Research (973 program, Grant No. 2014CB441502), and funded by Pilot National Laboratory for Marine Science and Technology (Qingdao) (No. JCZX202007). The authors declare no competing financial interests.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We greatly appreciate help of the crews of the Xiangyanghong 09 as well as the Jiaolong team in helping us during the cruise, J. Sun (Hong Kong Baptist University) for help with mussel identification, L. Zhang (SCSIO, CAS) for sample preparation. This work was supported by the Natural Science Foundation of China (41876074, 42106049) and National Key Program for Basic Research (973 program, Grant No. 2014CB441502), and funded by Pilot National Laboratory for Marine Science and Technology (Qingdao) (No. JCZX202007). The authors declare no competing financial interests.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2022.869226/full#supplementary-material
AA, amino acid; Ala, alanine; Arg, arginine; Asn, asparaginate; Asp, aspartic acid; CSIA, compound-specific isotope analysis; FA,fatty acid; FAME, fatty acid methyl ester; Glu,glutamic; Gly, glycine; Ile, isoleucine; Leu, leucine; Lys, lysine; Met, methionine; OM, organic matter; Phe, phenylalanine; POM, particulate organic matter; PUFA, polyunsaturated fatty acid; SCS,South China Sea, Ser, serine; Thr, threonine; Tyr, tyrosine; Val, valine.
Ackman R. G. (1989). “Fatty Acids” in Marine Biogenic Lipids, Fats and Oils, ed. Ackman R. G. CRC press (Basel: Marcel Dekker), 103–138
Akashi H., Gojobori T. (2002). Metabolic Efficiency and Amino Acid Composition in the Proteomes of Escherichia Coli and Bacillus Subtilis. P. Natl. Acad. Sci. 99, 3695–3700. doi: 10.1073/pnas.062526999
Berg J., Tymoczko J., Stryer L. (2002). Carbon Atoms of Degraded Amino Acids Emerge as Major Metabolic Intermediates. Biochemistry 2002, 5
Bond A.L, Hobson K.A 2012. Reporting Stable-Isotope Ratios in Ecology: Recommended Terminology, Guidelines and Best Practices.Waterbirds 35. doi: 10.1675/063.035.0213.
Bothner-By A., Friedman L. (1952). The Reaction of Nitrous Acid With Hydroxylamine. J. Chem. Phys. 20, 459–462. doi: 10.1063/1.1700442
Brooks J. M., Kennicutt M., Fisher C., Macko S., Cole K., Childress J., et al. (1987). Deep-Sea Hydrocarbon Seep Communities: Evidence for Energy and Nutritional Carbon Sources. Science 238, 1138–1142. doi: 10.1126/science.238.4830.1138
Bull I. D., Parekh N. R., Hall G. H., Ineson P., Evershed R. P. (2000). Detection and Classification of Atmospheric Methane Oxidizing Bacteria in Soil. Nature 405, 175–178. doi: 10.1038/35012061
Chikaraishi Y., Kashiyama Y., Ogawa N. O., Kitazato H., Ohkouchi N. (2007). Metabolic Control of Nitrogen Isotope Composition of Amino Acids in Macroalgae and Gastropods: Implications for Aquatic Food Web Studies. Mar. Ecol. Prog. Ser. 342, 85–90. doi: 10.3354/meps342085
Davies D. D., Humphrey T. J. (1978). Amino Acid Recycling in Relation to Protein Turnover. Plant Physiol. 61, 54–58. doi: 10.1104/pp.61.1.54
Deniro M. J., Epstein S. (1977). Mechanism of Carbon Isotope Fractionation Associated With Lipid Synthesis. Science 197, 261–263. doi: 10.1126/science.327543
Fang J., Abrajano T. A., Comet P. A., Brooks J. M., Sassen R., Macdonald I. R. (1993). Gulf of Mexico Hydrocarbon Seep Communities: XI. Carbon Isotopic Fractionation During Fatty Acid Biosynthesis of Seep Organisms and its Implication for Chemosynthetic Processes. Chem. Geo. 109, 271–279. doi: 10.1016/0009-2541(93)90074-S
Feng D., Cheng M., Kiel S., Qiu J.-W., Yang Q., Zhou H., et al. (2015). Using Bathymodiolus Tissue Stable Carbon, Nitrogen and Sulfur Isotopes to Infer Biogeochemical Process at a Cold Seep in the South China Sea. Deep-Sea Res. Pt I. 104, 52–59. doi: 10.1016/j.dsr.2015.06.011
Feng D., Qiu J.-W., Hu Y., Peckmann J., Guan H., Tong H., et al. (2018). Cold Seep Systems in the South China Sea: An Overview. J. Asian Earth Sci. 168, 3–16. doi: 10.1016/j.jseaes.2018.09.021
Ferrier-Pagès C., Leal M. C. (2018). Stable Isotopes as Tracers of Trophic Interactions in Marine Mutualistic Symbioses. Ecol. Evol. 9, 723–740. doi: 10.1002/ece3.4712
Ferrier-Pages C., Leal M. C. (2019). Stable Isotopes as Tracers of Trophic Interactions in Marine Mutualistic Symbioses. Ecol. Evol. 9, 723–740. doi: 10.1002/ece3.4712
Fitznar H. P., Lobbes J. M., Kattner G. (1999). Determination of Enantiomeric Amino Acids With High-Performance Liquid Chromatography and Pre-Column Derivatisation With O-Phthaldialdehyde and N-Isobutyrylcysteine in Seawater and Fossil Samples (Mollusks). J.Chromatogr. A. 832, 123–132. doi: 10.1016/S0021-9673(98)01000-0
Folch J., Lees M., Sloane Stanley G. H. (1957). A Simple Method for the Isolation and Purification of Total Lipids From Animal Tissues. J. Biol. Chem. 226, 497–509. doi: 10.1016/S0021-9258(18)64849-5
Gerringer M. E., Popp B. N., Linley T. D., Jamieson A. J., Drazen J. C. (2017). Comparative Feeding Ecology of Abyssal and Hadal Fishes Through Stomach Content and Amino Acid Isotope Analysis. Deep Sea Res. I Oceanogr. Res. Pap. 121, 110–120. doi: 10.1016/j.dsr.2017.01.003
Goffredi S. K., Tilic E., Mullin S. W., Dawson K. S., Keller A., Lee R. W., et al. (2020). Methanotrophic Bacterial Symbionts Fuel Dense Populations of Deep-Sea Feather Duster Worms (Sabellida, Annelida) and Extend the Spatial Influence of Methane Seepage. Sci. Adv. 6, eaay8562. doi: 10.1126/sciadv.aay8562
Hoppe H. G., Arnosti C., Herndl G. F. (2001). “Ecological significance of bacterial enzymes in the marine environment,” in Enzymes in the Environment, eds Burns R. G., Dick R. P. (Basel: Marcel Dekker), 73–107.
Hosie A. H., Allaway D., Galloway C. S., Dunsby H. A., Poole P. S. (2002). Rhizobium Leguminosarum has a Second General Amino Acid Permease With Unusually Broad Substrate Specificity and High Similarity to Branched-Chain Amino Acid Transporters (Bra/LIV) of the ABC Family. J. Bacteriol. 184, 4071–4080. doi: 10.1128/jb.184.15.4071-4080.2002
Hugler M., Sievert S. M. (2011). Beyond the Calvin Cycle: Autotrophic Carbon Fixation in the Ocean. Anuu. Rev. Mar. Sci. 3, 261–289. doi: 10.1146/annurev-marine-120709-142712
Jahnke L. L., Summons R. E., Hope J. M., Des Marais D. J. (1999). Carbon Isotopic Fractionation in Lipids From Methanotrophic Bacteria II: The Effects of Physiology and Environmental Parameters on the Biosynthesis and Isotopic Signatures of Biomarkers. Geochim. Cosmochim. Ac. 63, 79–93. doi: 10.1016/S0016-7037(98)00270-1
Jing H., Wang R., Jiang Q., Zhang Y., Peng X. (2020). Anaerobic Methane Oxidation Coupled to Denitrification is an Important Potential Methane Sink in Deep-Sea Cold Seeps. Sci. Total Environ. 748, 142459. doi: 10.1016/j.scitotenv.2020.142459
Kaleta C., Schauble S., Rinas U., Schuster S. (2013). Metabolic Costs of Amino Acid and Protein Production in Escherichia Coli. Biotechnol. J. 8, 1105–1114. doi: 10.1002/biot.201200267
Kellermann M. Y., Schubotz F., Elvert M., Lipp J. S., Birgel D., Prieto-Mollar X., et al. (2012). Symbiont–host Relationships in Chemosynthetic Mussels: A Comprehensive Lipid Biomarker Study. Org. Geochem. 43, 112–124. doi: 10.1016/j.orggeochem.2011.10.005
Kienast M., Higginson M. J., Mollenhauer G., Eglinton T. I., Chen M. T., Calvert S. E. (2005). On the Sedimentological Origin of Down-Core Variations of Bulk Sedimentary Nitrogen Isotope Ratios. Paleoceanography 20, 2. doi: 10.1029/2004pa001081
Liao L., Wankel S. D., Wu M., Cavanaugh C. M., Girguis P. R. (2014). Characterizing the Plasticity of Nitrogen Metabolism by the Host and Symbionts of the Hydrothermal Vent Chemoautotrophic Symbioses Ridgeia Piscesae. Mol. Ecol. 23, 1544–1557. doi: 10.1111/mec.12460
Liu D., Fang Y., Tu Y., Pan Y. (2014). Chemical Method for Nitrogen Isotopic Analysis of Ammonium at Natural Abundance. Anal. Chem. 86, 3787–3792. doi: 10.1021/ac403756u
Macavoy S., Macko S., Joye S. (2002). Fatty Acid Carbon Isotope Signatures in Chemosynthetic Mussels and Tube Worms From Gulf of Mexico Hydrocarbon Seep Communities. Chem. Geo. 185, 1–8. doi: 10.1016/S0009-2541(01)00394-1
Macko S. A., Estep M. L. F., Engel M. H., Hare P. (1986). Kinetic Fractionation of Stable Nitrogen Isotopes During Amino Acid Transamination. Geochim. Cosmochim. Ac. 50, 2143–2146. doi: 10.1016/0016-7037(86)90068-2
Mahmood T., Fang J., Jiang Z., Zhang J. (2016). Carbon and Nitrogen Flow, and Trophic Relationships, Among the Cultured Species in an Integrated Multi−Trophic Aquaculture (IMTA) Bay. Aquacult. Env. Interac. 8, 207–219. doi: 10.3354/aei00152
Mccarthy M. D., Lehman J., Kudela R. (2013). Compound-Specific Amino Acid δ15n Patterns in Marine Algae: Tracer Potential for Cyanobacterial vs. Eukaryotic Organic Nitrogen Sources in the Ocean. Geochim. Cosmochim. Ac. 103, 104–120. doi: 10.1016/j.gca.2012.10.037
Mcmahon K. W., Mccarthy M. D. (2016). Embracing Variability in Amino Acid δ15n Fractionation: Mechanisms, Implications, and Applications for Trophic Ecology. Ecosphere 7, e01511. doi: 10.1002/ecs2.1511
Michaelis W., Seifert R., Nauhaus K., Treude T., Thiel V., Blumenberg M., et al. (2002). Microbial Reefs in the Black Sea Fueled by Anaerobic Oxidation of Methane. Science 297, 1013–1015. doi: 10.1126/science.1072502
Minagawa M., Wada E. (1984). Stepwise Enrichment of 15N Along Food Chains: Further Evidence and the Relation Between δ15n and Animal Age. Geochim. Cosmochim. Ac. 48, 1135–1140. doi: 10.1016/0016-7037(84)90204-7
Ohkouchi N., Chikaraishi Y., Close H. G., Fry B., Larsen T., Madigan D. J., et al. (2017). Advances in the Application of Amino Acid Nitrogen Isotopic Analysis in Ecological and Biogeochemical Studies. Org. Geochem. 113, 150–174. doi: 10.1016/j.orggeochem.2017.07.009
Orphan V. J., House C. H., Hinrichs K.-U., Mckeegan K. D., Delong E. F. (2001). Methane-Consuming Archaea Revealed by Directly Coupled Isotopic and Phylogenetic Analysis. Science 293, 484–487. doi: 10.1126/science.1061338
Paull C. K., Hecker B., Commeau R., Freeman-Lynde R. P., Neumann C., Corso W. P., et al. (1984). Biological Communities at the Florida Escarpment Resemble Hydrothermal Vent Taxa. Science 226, 965–967. doi: 10.1126/science.226.4677.965
Petersen J. M., Kemper A., Gruber-Vodicka H., Cardini U., van der Geest M., Kleiner M., et al. (2016). Chemosynthetic Symbionts of Marine Invertebrate Animals are Capable of Nitrogen Fixation. Nat. Microbiol. 2, 16195. doi: 10.1038/nmicrobiol.2016.195
Pile A. J., Young C. M. (1999). Plankton Availability and Retention Efficiencies of Cold-Seep Symbiotic Mussels. Limnol. Oceanogr. 44, 1833–1839. doi: 10.4319/lo.1999.44.7.1833
Ponnudurai R., Heiden S. E., Sayavedra L., Hinzke T., Kleiner M., Hentschker C., et al. (2020). Comparative Proteomics of Related Symbiotic Mussel Species Reveals High Variability of Host-Symbiont Interactions. ISME. J. 14, 649–656. doi: 10.1038/s41396-019-0517-6
Ponnudurai R., Kleiner M., Sayavedra L., Petersen J. M., Moche M., Otto A., et al. (2017). Metabolic and Physiological Interdependencies in the Bathymodiolus Azoricus Symbiosis. ISME. J. 11, 463–477. doi: 10.1038/ismej.2016.124
Raggi L., Schubotz F., Hinrichs K. U., Dubilier N., Petersen J. M. (2013). Bacterial Symbionts of Bathymodiolus Mussels and Escarpia Tubeworms From Chapopote, an Asphalt Seep in the Southern Gulf of Mexico. Environ. Microbiol. 15, 1969–1987. doi: 10.1111/1462-2920.12051
Riou V., Bouillon S., Serrão Santos R., Dehairs F., Colaço A. (2010). Tracing Carbon Assimilation in Endosymbiotic Deep-Sea Hydrothermal Vent Mytilid Fatty Acids by 13C-Fingerprinting. Biogeosciences 7, 2591–2600. doi: 10.5194/bg-7-2591-2010
Rodrigues C. F., Hilário A., Cunha M. R. (2013). Chemosymbiotic Species From the Gulf of Cadiz (NE Atlantic): Distribution, Life Styles and Nutritional Patterns. Biogeosciences 10, 2569–2581. doi: 10.5194/bg-10-2569-2013
Rubin-Blum M., Antony C. P., Borowski C., Sayavedra L., Pape T., Sahling H., et al. (2017). Short-Chain Alkanes Fuel Mussel and Sponge Cycloclasticus Symbionts From Deep-Sea Gas and Oil Seeps. Nat. Microbiol. 2, 17093. doi: 10.1038/nmicrobiol.2017.93
Rubin-Blum M., Antony C. P., Sayavedra L., Martinez-Perez C., Birgel D., Peckmann J., et al. (2019). Fueled by Methane: Deep-Sea Sponges From Asphalt Seeps Gain Their Nutrition From Methane-Oxidizing Symbionts. ISME. J. 13, 1209–1225. doi: 10.1038/s41396-019-0346-7
Ruff S. E., Biddle J. F., Teske A. P., Knittel K., Boetius A., Ramette A. (2015). Global Dispersion and Local Diversification of the Methane Seep Microbiome. P. Natl. Acad. Sci. 112, 4015–4020. doi: 10.1073/pnas.1421865112
Schonberg A., Moubacher R. (1952). The Strecker Degradation of α-Amino Acids. Chem. Rev. 50, 261–277. doi: 10.1021/cr60156a002
Suess E. (2014). Marine Cold Seeps and Their Manifestations: Geological Control, Biogeochemical Criteria and Environmental Conditions. Int. J. Earth Sci. 103, 1889–1916. doi: 10.1007/s00531-014-1010-0
Sun J., Zhang Y., Xu T., Zhang Y., Mu H., Zhang Y., et al. (2017). Adaptation to Deep-Sea Chemosynthetic Environments as Revealed by Mussel Genomes. Nat. Ecol. Evol. 1, 121. doi: 10.1038/s41559-017-0121
Takano Y., Chikaraishi Y., Imachi H., Miyairi Y., Ogawa N. O., Kaneko M., et al. (2018). Insight Into Anaerobic Methanotrophy From 13C/12C- Amino Acids and 14C/12C-ANME Cells in Seafloor Microbial Ecology. Sci. Rep. 8, 14070. doi: 10.1038/s41598-018-31004-5
Tobler M., Passow C. N., Greenway R., Kelley J. L., Shaw J. H. (2016). The Evolutionary Ecology of Animals Inhabiting Hydrogen Sulfide–Rich Environments. Annu. Rev. Ecol. Evol. S. 47, 239–262. doi: 10.1146/annurev-ecolsys-121415-032418
Vokhshoori N. L., Mccarthy M. D., Close H. G., Demopoulos A. W. J., Prouty N. G. (2021). New Geochemical Tools for Investigating Resource and Energy Functions at Deep-Sea Cold Seeps Using Amino Acid δ15n in Chemosymbiotic Mussels (Bathymodiolus Childressi). Geobiology 19(6), 601–617. doi: 10.1111/gbi.12458
Volland J. M., Schintlmeister A., Zambalos H., Reipert S., Mozetic P., Espada-Hinojosa S., et al. (2018). NanoSIMS and Tissue Autoradiography Reveal Symbiont Carbon Fixation and Organic Carbon Transfer to Giant Ciliate Host. ISME. J. 12, 714–727. doi: 10.1038/s41396-018-0069-1
Wagner A. (2005). Energy Constraints on the Evolution of Gene Expression. Mol. Biol. Evol. 22, 1365–1374. doi: 10.1093/molbev/msi126
Wang X. (2018). Nutritional Sources Analysis and the Heavy-Metal Enrichment of the Macrofauna From Deep-Sea Chemotrophic Ecosystem Qingdao: (Institute of Oceanology, Chinese Academy of Sciences).
Wang F., Wu Y., Chen Z., Zhang G., Zhang J., Zheng S., et al. (2019). Trophic Interactions of Mesopelagic Fishes in the South China Sea Illustrated by Stable Isotopes and Fatty Acids. Front. Mar. Sci. 5. doi: 10.3389/fmars.2018.00522
Wang F., Wu Y., Zhang L., Jin J., Chen Z., Zhang J., et al. (2022). Improved Method for Measuring the δ15n Compound-Specific Amino Acids: Application on Mesopelagic Fishes in the South China Sea. Acta Oceanol. Sin. 41, 30–38. doi: 10.1007/s13131-021-1812-4
Wong Y. H., Sun J., He L. S., Chen L. G., Qiu J. W., Qian P. Y. (2015). High-Throughput Transcriptome Sequencing of the Cold Seep Mussel Bathymodiolus Platifrons. Sci. Rep. 5, 16597. doi: 10.1038/srep16597
Wu Y., Dittmar T., Ludwichowski K.-U., Kattner G., Zhang J., Zhu Z. Y., et al. (2007). Tracing Suspended Organic Nitrogen From the Yangtze River Catchment Into the East China Sea. Mar. Chem. 107, 367–377. doi: 10.1016/j.marchem.2007.01.022
Xu T., Feng D., Tao J., Qiu J.-W. (2019). A New Species of Deep-Sea Mussel (Bivalvia: Mytilidae: Gigantidas) From the South China Sea: Morphology, Phylogenetic Position, and Gill-Associated Microbes. Deep-Sea Res. Pt I. 146, 79–90. doi: 10.1016/j.dsr.2019.03.001
Yamaguchi Y. T., Chikaraishi Y., Takano Y., Ogawa N. O., Imachi H., Yokoyama Y., et al. (2017). Fractionation of Nitrogen Isotopes During Amino Acid Metabolism in Heterotrophic and Chemolithoautotrophic Microbes Across Eukarya, Bacteria, and Archaea: Effects of Nitrogen Sources and Metabolic Pathways. Org. Geochem. 111, 101–112. doi: 10.1016/j.orggeochem.2017.04.004
Zhang L., Altabet M. A. (2008). Amino-Group-Specific Natural Abundance Nitrogen Isotope Ratio Analysis in Amino Acids. Rapid Commun. Mass Sp. 22, 559–566. doi: 10.1002/rcm.3393
Zhang L., Lee W. M., Kreider-Mueller A., Kuhnel E., Baca J., Ji C., et al. (2021). High-Precision Measurement of Phenylalanine and Glutamic Acid δ15N by Coupling Ion-Exchange Chromatography and Purge-and-Trap Continuous-Flow Isotope Ratio Mass Spectrometry. Rapid Commun. Mass Sp. 35, e9085. doi: 10.1002/rcm.9085
Keywords: symbiotic mussels, cold seeps, nitrogen sources, biomarker methods, δ13C of fatty acids, δ15N of amino acids
Citation: Wang F, Wu Y and Feng D (2022) Different Nitrogen Sources Fuel Symbiotic Mussels at Cold Seeps. Front. Mar. Sci. 9:869226. doi: 10.3389/fmars.2022.869226
Received: 04 February 2022; Accepted: 13 June 2022;
Published: 15 July 2022.
Edited by:
Clara F. Rodrigues, University of Aveiro, PortugalReviewed by:
Ulisse Cardini, Stazione Zoologica Anton Dohrn Napoli, ItalyCopyright © 2022 Wang, Wu and Feng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ying Wu, d3V5aW5nQHNrbGVjLmVjbnUuZWR1LmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.