- 1Department of Ecosustainable Marine Biotechnology, Stazione Zoologica Anton Dohrn, Naples, Italy
- 2Department of Integrative Marine Ecology, Stazione Zoologica Anton Dohrn, Naples, Italy
- 3Dipartimento di Farmacia, Università degli Studi di Naples Federico II, Naples, Italy
Human-mediated dispersal of alien species in new biogeographic realms is one of the major drivers of biodiversity change in marine ecosystems. Among others, ascidians are invasive species spreading worldwide, thus causing ecological and economic harms in the recipient environments. An integrated taxonomic approach on selected samples allowed the identification of Botrylloides niger Herdman 1886 as a non-indigenous ascidian forming large aggregates and outcompeting native species in the Fusaro Lake (central-western Mediterranean Sea). This led to the opportunity to investigate in deep its metabolome for the first time. Untargeted mass spectrometry-based metabolomics unveiled B. niger to be a source of nutraceuticals and bioactive natural products, such as lysophospholipids, sulfonolipids, polyunsaturated fatty acids, sphingolipids, monoacylglycerols, and alkaloids. Even if causing ecosystem alterations, our results suggest that B. niger could be exploited for nutritional and/or pharmaceutical purposes, thereby turning a treat in a resource.
1 Introduction
Ascidians (Tunicata) are important members of marine benthic communities, with over 3000 species described worldwide inhabiting polar, tropical, and temperate environments, either in shallow or deeper habitats (Shenkar and Swalla, 2011). Their phylogenetic relatedness to vertebrates, coupled with a somehow fast and easy development in vitro, has attracted the attention of developmental biologists since centuries (Corbo et al., 2001; Dehal et al., 2002; Imai et al., 2006). In the last decades, ascidians also generally raised additional interest as potential alternative food sources in human consumption due to the presence of compounds with high nutritive content, such as proteins, amino acids, lipids, and secondary metabolites (Zhao and Li, 2016; Palanisamy et al., 2017; Watters, 2018; Dou and Dong, 2019), and examples of wide phenotypic and environmental plasticity, which is turn reflected in a high invasive potential (Bullard and Carman, 2009; Zhan et al., 2015; Rocha et al., 2019; Atalah et al., 2021). Moreover, they are source of a variety of bioactive chemical compounds, which include peptides, alkaloids, polyethers, macrolides, terpenes, and polysulfides (Palanisamy et al., 2017; Imperatore et al., 2019). Finally, ascidians are also emerging as model organisms for several studies including immunobiology, allorecognition, angiogenesis and whole-body regeneration, and their extensive regenerative capacity is developing huge interests in regenerative medicine and ageing research (Ballarin et al., 2001; Rinkevich et al., 2007; Gasparini et al., 2008; Franchi et al., 2011; Lauzon et al., 2013; Rinkevich et al., 2013; Voskoboynik and Weissman, 2015).
Noteworthy, these variegate interests of the scientific biological community also often merge in target species. Just to mention, the Korean common sea squirt Halocynthia roretzi (Drasche, 1884), reared and exploited as a commercial seafood in Japan and Korea (Oh et al., 1997; Hirose et al., 2009), possesses lipids that positively affected the health of diabetic/obese KK-Ay mice (Mikami et al., 2010) and ingestion of its plasmalogen enhances and keeps cognitive function through induction of neuronal growth and neuroprotective effects (Watanabe et al., 2020). The clubbed tunicate Styela clava Herdman, 1881, widely cultured in Asian countries (Ko et al., 2012), is in turn not only invading the Mediterranean Sea and nearby areas (Davis and Davis, 2010), but also possesses important compounds with variegate properties, including ACE (angiotensin-converting enzyme) inhibition (Ko et al., 2012) and antioxidant, immunomodulatory, and anticancer activities (Kim, 2011; Ju et al., 2014). Finally, specimens of the Ciona intestinalis (Linnaeus, 1767) species complex, although often reported as a sea pests in several countries worldwide, leading to conspicuous economic losses (Zhan et al., 2010; Bouchemousse et al., 2016), possess abundant phospholipids and polyunsaturated fatty acids (PUFAs) with importance to human health, thus suggesting that they could become an excellent alternative source of n-3 fatty acids (Zhao et al., 2015). Moreover, natural products (NPs) derived from ascidians have also led to the production of promising drugs already marketed for the treatment of specific cancers, such as ecteinascidine 743 or Trabectedin (Yondelis®) from Ecteinascidia turbinata Herdman, 1880 and the peptide dehydrodidemnin B or Plitidepsin (Aplidin®) from Aplidium albicans (Milne Edwards, 1841) (Ramesh et al., 2021).
Interest about ascidians is somehow eased by their ecology, as species are often abundant in shallow waters of enclosed and semi-enclosed basins such as ports, marinas, and lagoons, where they settle on artificial substrates and form high biomasses in a short time-frame (Bullard et al., 2007; Lambert, 2009; Lambert, 2019). Although this on one hand makes them a possible threat to local communities due to physical substrate dominance and release of secondary metabolites that can harm benthic assemblages (Prado et al., 2004; Minchin and Sides, 2006; Lengyel et al., 2009; Watters, 2018), on the other it guarantees easy rearing and harvesting (Davis, 1995; Manríquez and Castilla, 2007).
In this context, investigation of compounds extracted from alien ascidians may help shedding light not only in their ecological role in localities where they are introduced, but also in a possible practical use as nutritional or bioactive compounds, thus turning a treat in a resource. Charting metabolic diversity of complex natural extracts and identify novel compounds at an early stage of the research, is high challenging. Metabolomics and dereplication (i.e., early identification of already known NPs), have become more routine in the fields of NP discovery (Caso et al., 2019), environmental research (Teta et al., 2021), and ecotoxicology (De Marco et al., 2022), and may rely upon advanced and sensitive spectroscopic techniques (NMR, mass spectrometry) (Cappello, 2020). High resolution liquid chromatography coupled with tandem mass spectrometry (LC-HRMS/MS) represents a well-suited untargeted methodology to capture the entire metabolome (i.e., the chemical profile of low molecular weight metabolites) within a heterogeneous mixture, thus enabling a deep and sensitive dereplication. Indeed, high resolution metadata and acquisition of fragmentation spectra are useful in that they provide “fingerprints” of detected metabolites, thereby facilitating their identification. Many bioinformatic tools are currently available for handling such amount of metadata and provide a bird’s eye perspective of the chemical profile of a certain extract. Among these tools, to explore large MS data sets, the Feature-Based Molecular Networking represents a suitable means for fast detection, annotation and visualization of known compounds and their novel analogues as well as for discovery of completely new NPs (Nothias et al., 2020).
Herein for the first time, as far of our knowledge, we assessed the metabolome of the colonial ascidian Botrylloides niger Herdman, 1886, a renown tropical invader which already expanded its distributional range from tropical Western Atlantic to Eastern and Western Pacific (Sheets et al., 2016; Rocha et al., 2019; Nydam et al., 2021) and is now emerging as an invasive species also in the Mediterranean Sea.
2 Material and Methods
2.1 Study Area and Field Work
The Fusaro Lake (Bacoli, central Tyrrhenian Sea, central-western Mediterranean Sea) is a brackish water body situated within the Campi Flegrei Regional Park (https://www.parcodeicampiflegrei.it/) and the biggest of the four Phlegraean lagoons. The lake is connected to the Tyrrhenian Sea by three channels, although only the middle one is operative, and therefore it constitutes the only source of water exchange with the sea. During fieldwork held in June 2020 and aimed to investigate the fouling biota of the local dock wall (40.8229 N, 14.0498 E), several unidentified botryllid colonies were sampled from 0 to 2 meters. Samples were first photographed in situ (Figure 1) with a Olympus Em1 mkII camera equipped with a Zuiko 60 mm f 2.8, then scraped from hard substrates with the help of underwater knives, placed in single plastic bags filled with seawater, and brought to the Laboratory of Benthos of the Stazione Zoologica Anton Dohrn (SZN, Naples, Italy), where they were cleaned from possible contaminants (algae and other organisms adhering them) for further laboratory work.

Figure 1 Botrylloides niger Herdman, 1886 from the Fusaro Lake channel (Bacoli, central Tyrrhenian Sea, Mediterranan Sea). (A) Colony forming an aggregate on the dock wall. (B) Colony overgrowing a specimen of Styela plicata (Lesueur, 1823). (C) Colony growing amidst serpulids and other tunicates. (D) A close-up showing zooids placement and atrial openings.
2.2 Identification of the Colonies
The examination of the colonies external morphology with the help of a Zeiss Axio Zoom.V16 (Germany) microscope yielded an identification to genus level (see below). Then, since morphological identification of botryllid taxa could be deceiving, often resulting in high misidentification rates even when carried out by ascidian specialists (see discussions in Brunetti, 2009; Rocha et al., 2019; Viard et al., 2019), five colonies were randomly selected for DNA barcoding. After dissection, total genomic DNA was extracted from single zooids using the DNeasy® Blood & Tissue kit (Qiagen, Hilden, Germany), following the protocol as in Crocetta et al. (2020). Samples were then fixed in ethanol 99.9% and preserved in the collection of the Laboratory of Benthos, SZN (SZN-B-809ASC11A–813ASC11E). Partial sequences of the Cytochrome c Oxidase subunit I (COI) gene were amplified from each DNA sample using the primers designed by Folmer et al. (1994). Polymerase chain reactions (PCRs) were conducted in 25 µL volume reaction as in Tanduo et al. (2021). Amplification was performed with an initial denaturation at 95°C (5 min), followed by 39 cycles of denaturation at 95°C (1 min), annealing at 45°C (1 min), extension at 72°C (1 min), with a final extension at 72°C (5 min). The PCR products were purified and Sanger sequenced at the Molecular Biology and Sequencing Service of SZN through an Automated Capillary Electrophoresis Sequencer 3730 DNA Analyzer (Applied Biosystems, CA, USA), using the BigDye® Terminator v3.1 Cycle Sequencing Kit (Life Technologies, UK). Chromatograms for each sequence were then quality checked, assembled, and edited using Sequencher v.5.0.1 (GeneCodes, MI, USA). The identity of sequences obtained was finally checked through the Basic Local Alignment Search Tool (https://blast.ncbi.nlm.nih.gov/Blast.cgi) (Morgulis et al., 2008).
2.3 Phylogenetic Analyses
Since the BLAST results mostly reported high similarity (see below) with several sequences deposited as Botrylloides niger [or incorrectly as B. nigrum: see discussions in Ryland (2015) on Botrylloides being a masculine gender], a barely-reported alien species in the Mediterranean Sea, unpublished sequences obtained here were also included in a small phylogenetic framework as to further confirm the identification of the samples and to eventually compare them with sequences coming from other worldwide localities.
The NCBI data mining revealed the presence of twenty-eight COI partial sequences of B. niger/nigrum. However, the analysis of seven of them (MH367290, MH367289, KX650766, KX138503, KX138502, KT693199, and MH235543) revealed the presence of gaps producing misalignment, and thus they were excluded a priori. Then, by subsequently deleting identical sequences (MW858360, MT637961, MW817940, LR828514, MT232728, MT232723, KT693201, KT693200, KT693198, MW278779, KP254541, and HF548559/NC_021467), only nine sequences of B. niger were used for the subsequent phylogenetic analyses (Table 1). In addition, four sequences of Botrylloides diegensis Ritter & Forsyth, 1917 and one single sequence each of Symplegma brakenhielmi (Michaelsen, 1904) and Symplegma viride Herdman, 1886 were also added to our alignment (Table 1). Botrylloides diegensis is a species often misidentified in the Mediterranean as Botrylloides leachii (Savigny, 1816) and is known to be the sister taxon of B. niger (Nydam et al., 2021). Symplegma Herdman, 1886 taxa were chosen as outgroup of botryllids in view of recent phylogenetic studies (Perez-Portela et al., 2009; Viard et al., 2019; Nydam et al., 2021).
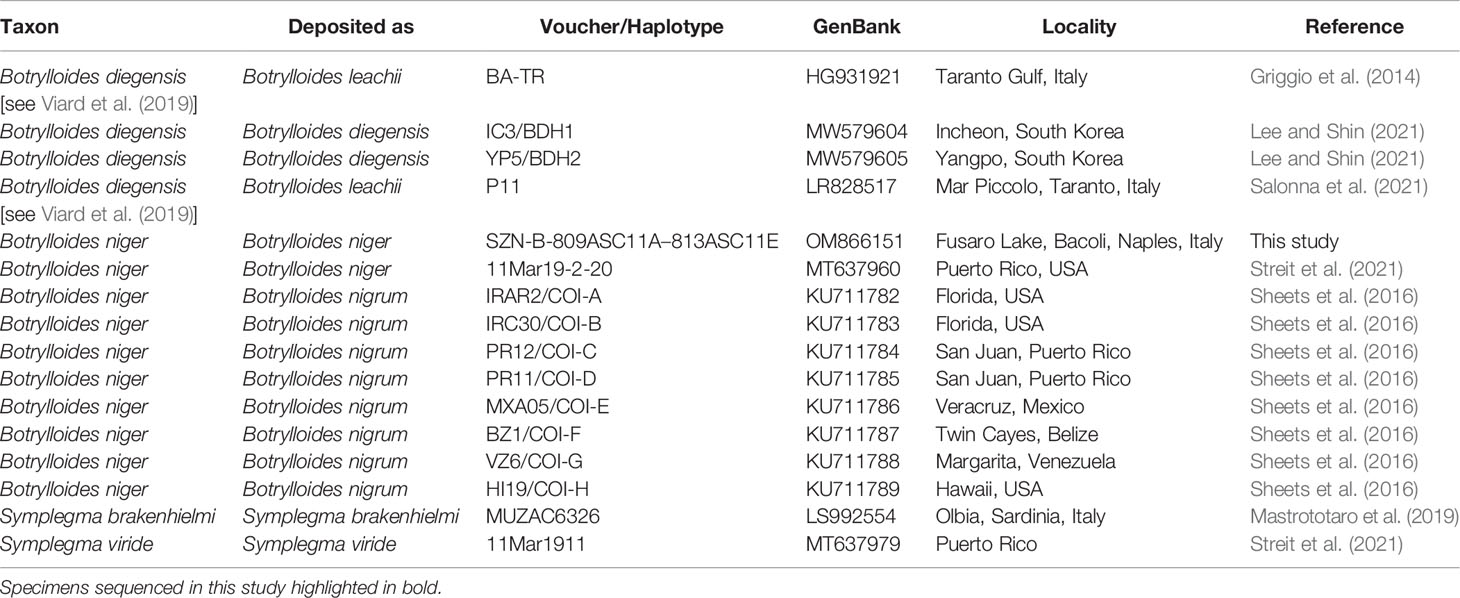
Table 1 GenBank COI partial sequences of Botrylloides and Symplegma species used in the molecular analyses and associated accession numbers and voucher data (localities obtained from GenBank and/or relevant paper/s).
Sequences were aligned using ClustalW (2.1) on the CIPRES Science Gateway (Miller et al., 2010), using default parameters. The alignment was trimmed and then used to infer phylogenies through maximum likelihood (ML) and Bayesian Inference (BI) methods. The GTR+I evolutionary model was selected through the AICc (corrected Akaike Information Criterion) algorithm, implemented in JModelTest 2 v.0.1.10 (Darriba et al., 2012), as the best-fit model for the alignment produced.
ML analysis was performed using RAxML v.2.0 (Edler et al., 2021), using 1000 rapid bootstrap pseudo-replicates under the aforementioned evolutionary model. BI was instead performed using MrBayes v.3.2.5 (Huelsenbeck and Ronquist, 2001) for 10 million generations, a sampling interval every 1000 generations, and discarding 25% of the produced trees. Tracer v1.7.1 (Rambaut et al., 2018) was used to check the convergence of Markov chain Monte Carlo (MCMC) runs. The trees obtained were checked by eye in FigTree v.1.4.4 (Rambaut, 2018) and edited in Adobe Illustrator 2019 23.1.1 (Adobe, USA).
2.4 Preparation of Crude Extract
Approximately 30 g of sample was defrosted for the extraction, cut into small pieces, and left in water (1:20 w/v) overnight (O/N) at 20°C for removing salts. Then, the sample was extracted with methanol (1:20 w/v), O/N at 20°C. The organic phase was dried under vacuum at the rotary evaporator (R-100, BUCHI, Flawil, Switzerland) to afford about 250 mg of crude extract. The extract was fractionated by reversed-phase (RP18) column chromatography, eluted with a linear gradient of H2O/MeOH (v/v, from 50:50 to 0:100 over 1 h) yielding three fractions (50-75-100% MeOH), which were then chemically characterized by molecular networking analysis of tandem MS data. Moreover, two additional samples of B. niger, collected in different areas of the Fusaro Lake channel, were subjected to methanol extraction for qualitative assessment of reproducibility in the metabolite composition. Crude extracts from the three replicates were shown to contain a similar chemical profile by LC-HRMS/MS analysis.
2.5 Liquid Chromatography - High Resolution Tandem Mass Spectrometry (LC-HRMS2)
The RP18 eluted fractions were dissolved in methanol at a concentration of 1 mg/mL for LC-HRMS2 analyses. MS experiments were performed using a Thermo LTQ Orbitrap XL high-resolution ESI mass spectrometer equipped with a Thermo U3000 HPLC system (Thermo Fisher Scientific, Waltham, MA, USA), which included a solvent reservoir, in-line degasser, binary pump, and refrigerated autosampler. A 5-μm Kinetex C18 column (50 × 2.10 mm), maintained at room temperature, was eluted at 200 μL·min−1 with H2O (supplemented with 0.1% HCOOH) and CH3OH, using a gradient elution. The gradient program was set as follows: 30% CH3OH 1 min, 30%−100% CH3OH over 30 min, 100% CH3OH 10 min. Mass spectra were acquired in the positive ion detection mode. MS parameters were as follows: a spray voltage of 4.8 kV, a capillary temperature of 285°C, a sheath gas rate of 32 units N2 (ca. 150 mL/min), and an auxiliary gas rate of 15 units N2 (ca. 50 mL/min). Data were collected in the data-dependent acquisition mode, in which the five most intense ions of a full-scan mass spectrum were subjected to HRMS2 analysis. The m/z range for data dependent acquisition was set between 100 and 2000 amu. HRMS2 scans were obtained with CID fragmentation, an isolation width of 2.0, normalized collision energy of 35, activation Q of 0.250, and an activation time of 30 ms. HPLC profiles of the RP18 eluted fractions have been reported in the Supplementary Material (Figures S7–S13).
2.6 LC-HRMS2 Data Processing and Molecular Networking
LC-HRMS2 data from RP18 eluted fractions were processed together to generate a unique molecular network, using a previously reported method (Della Sala et al., 2020). MS raw files were imported into MZmine 2.53 (Pluskal et al., 2010). Mass detection was performed on .mzXML data and centroided masses with mass level 1 and mass level 2, by keeping the noise level at 1000 and 100, respectively. The ADAP chromatogram algorithm was used to build chromatograms setting a minimum height of 1000 and m/z tolerance of 0.05 (or 20 ppm). As it regards chromatogram deconvolution, the baseline cut-off algorithm was employed with the following settings: minimum height peak = 1000, peak duration range = 0.0–10.0 min, baseline level = 100, m/z range for MS2 scan = 0.05, retention time range = 0.5 min. Chromatogram peaks were aligned by using the Join aligner algorithm (m/z tolerance at 0.05 or 20 ppm, absolute RT tolerance at 0.5 min). [M+Na–H], [M+K–H], [M+Mg−2H], [M+NH3], [M-Na+NH4], [M+1, 13C] adducts were filtered out by setting the maximum relative height at 100%. Peaks without associated MS2 spectra were filtered out from the peak list. Processed mass data were exported to .mgf file for GNPS and the relevant chromatographic information (retention times and peak areas) were exported to a .csv file in order to generate the molecular network displayed in Figure 3, by using the Feature Based Molecular Networking (FBMN) tool (Nothias et al., 2020). FBMN parameters were set as follows: precursor ion mass tolerance = 0.02, fragment ion mass tolerances = 0.1 Da, cosine score ≥ 0.7, minimum matched fragment ions = 4. The following GNPS databases have been selected for the spectral library search: CCMS_ProteomeDatabases, CCMS_School_2019, CCMS_SpectralLibraries, RMSV000000248, and speclibs. The molecular network was visualized and analyzed in Cytoscape version 3.7.2. Chromatographic data in the .csv file were mapped to the relevant nodes in the generated network (available at https://gnps.ucsd.edu/ProteoSAFe/status.jsp?task=85d850e022d043c29ec013db633c5e91, accessed on 27/10/2021). MS tandem spectra of all compounds reported in the molecular network can be found by accessing the GNPS link.
3 Results
3.1 Morphological and Molecular Analysis of the Samples
The colonies showed morphological features peculiar to the genus Botrylloides Milne Edwards, 1841. The zooids were closely placed to each other, jointed anteriorly by elongated dorsal lip margins which created the typical zooids succession (Figure 1). The cloacal openings were present at the end of each cloacal canal, which were formed by several single atrial openings of each zooids, hence lacking atrial siphons. On the contrary, in the genus Botryllus Gaertner, 1774, the cloacal opening is formed by the jointed atrial siphons projections of each zooids which surround the opening, giving a star-shaped morphology which is typical of the genus (see Brunetti, 2009).
A 602 base pairs (bp) partial sequence of the COI gene was obtained from the five colonies, with all sequences resulting identical each other. They were deposited in GenBank under the single accession number OM866151. The sequences shared high similarity with 28 sequences deposited as Botrylloides niger (96.04–100%), including one of a specimen (NC_021467: 99.83%) from the eastern Mediterranean Sea (Achziv, Israel) formerly deposited as Botrylloides aff. leachii (Rubinstein et al., 2013; Griggio et al., 2014), that was also subsequently declared by Salonna et al. (2021) (Table 1) to be identical to specimens from Italy (Mar Piccolo, Taranto). However, it also showed high similarity with two sequences deposited as Botryllus schlosseri (Pallas, 1766) from India (KT693191: 100%; MH367291: 97.01%), and a single sequence (MG009579: 99.36%) deposited as Botrylloides aff. leachii, again from the Mediterranean coast of Israel (Reem et al., 2017). High similarities also include all worldwide sequences of B. niger deposited by Sheets et al. (2016), including two haplotypes also found in the eastern Mediterranean coast of Israel (COI-A: KU711782; COI-B: KU711783), the former of which perfectly matches (once trimmed to the same length) both HF548559/NC_021467 and our sequences. All the other taxa deposited in GenBank showed lower similarities (≤90.53%), thus excluding conspecificity. However, concerning the two mismatches listed above, there are solid morphological and molecular evidences to discriminate the two genera Botrylloides and Botryllus, suggesting that the two Indian specimens mentioned above were misidentified (see also Brunetti et al., 2017; Reem et al., 2018), whereas the paper by Reem et al. (2017) suffers of various taxonomic uncertainties, only partially solved by other authors (Viard et al., 2019; Nydam et al., 2021).
After trimming, the final alignment used for phylogenetic analyses consisted of 16 sequences of 529 bp (Supplementary Data Sheet). The ML (−lnL = 1587.16) and BI (−lnL = 1731.83 for run 1; −lnL = 1731.78 for run 2) analyses produced congruent tree topologies (Figure 2). Sequences obtained in this study fell (bootstrap, bs = 92, posterior probabilities, pp = 1) within all the B. niger haplotypes found by Sheets et al. (2016) and colleagues, and in particular clustered within the haplotypes COI-A, COI-F, and COI-H, whereas the remaining haplotypes (COI-B–COI-E, COI-G) formed an internal clade, although with a low support (bs = 56, pp = 0.98). Present results further confirm the identification of our samples as B. niger and are in agreement with the low haplotype divergence reported by Sheets et al. (2016).
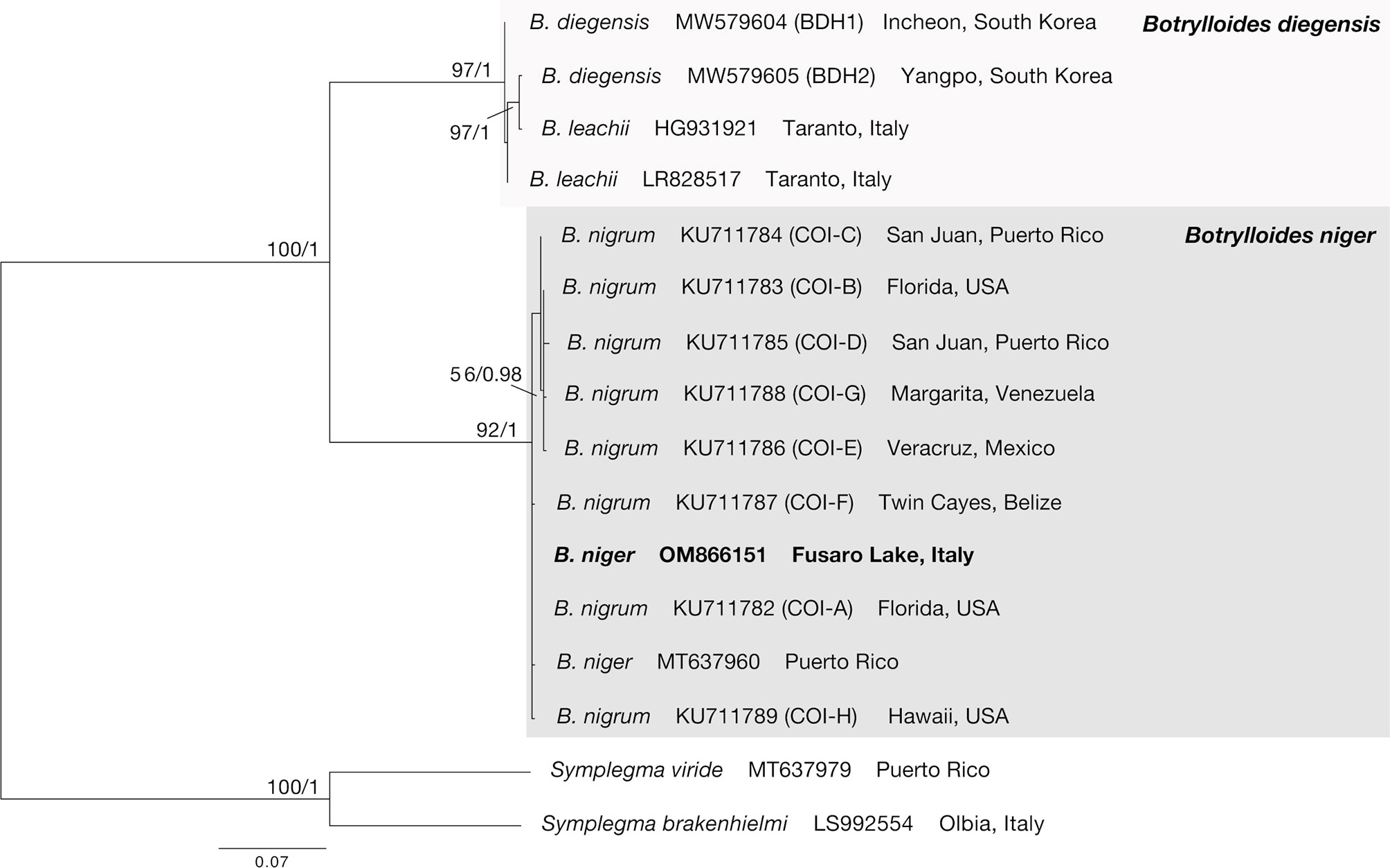
Figure 2 COI phylogenetic tree obtained from the alignment of 16 terminals. Numbers above/below branches represent bootstrap values (bs) and bayesian posterior probabilities (pp). Specimens sequenced in this study highlighted in bold.
3.2 MS-Based Molecular Networking Analysis of Botrylloides niger Metabolome
A sample of the marine tunicate B. niger was extracted with MeOH to yield the crude organic extract, which was then purified using a reversed-phase column chromatography on RP-18 silica gel.
Aiming to gain a comprehensive metabolome analysis, RP-18 eluted fractions, rather than the whole organic extract, were analysed individually by LC-HRMS2 as this approach allows to reduce the number of co-eluting metabolites and improve the quality of MS tandem spectra when untargeted fragmentation is used (Scarpato et al., 2020). After each full MS scan, the five most intense ions in the spectrum were fragmented in subsequent MS2 scans to generate data for the construction of a unique molecular network (Figure 3), representative of all RP-18 fractions, by using the FBMN tool, available on the online platform GNPS (Nothias et al., 2020). Mass spectra were acquired in the positive ion detection mode (mass accuracy ≤ 3 ppm). Based upon similarity of the MS fragmentation patterns, FBMN allows to i) group molecules with a similar chemical architecture into molecular families (molecular clusters), ii) associate these clusters with compounds reported in public available databases, and iii) identify substructures within a given molecule through detection of molecular fragments shared with known metabolites, thereby providing useful hints for structural elucidation of compounds.
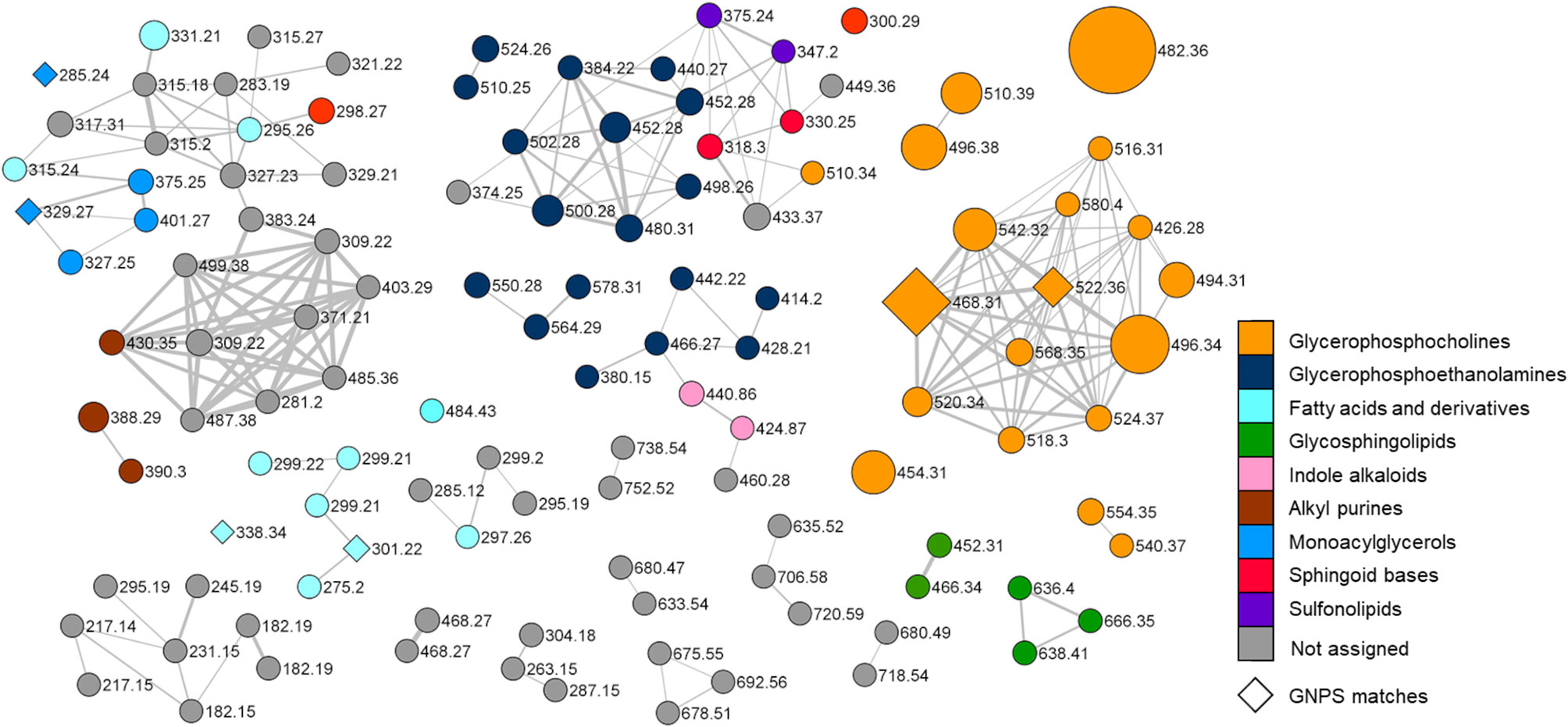
Figure 3 Molecular network of the organic extract from Botrylloides niger. Nodes are labelled with parent mass and the colour of each node is mapped to chemical class assigned by integration of molecular networking data with extensive analysis of MS tandem spectra of the metabolites. Nodes annotated by the GNPS reference database are represented as diamonds. Edge thickness reflects cosine score similarity and node size is related to metabolite amounts (peak area).
Merging molecular networking data with an in-depth investigation of MS2 spectra, led to the structural prediction of almost 70 metabolites from the organic extract of B. niger, which were assigned to nine chemical classes, namely glycerophosphocholines, glycerophosphoethanolamines, glycosphingolipids, sphingoid bases, sulfonolipids, fatty acids and derivatives, monoacylglycerols, indole alkaloids, and alkyl purines. In the network (Figure 3), the colour of each node is mapped to the relevant chemical class of the metabolite, and the size of the node is proportional to the spectral peak area of the metabolite for a visual relative quantification. Only six nodes could be annotated by comparison with the GNPS spectral database and are represented as diamonds in the network. Overall, most clusters have been shown to be related to glycerophospholipids, including lyso-glycerophosphocholines (see par 3.2.1.1), lyso-phosphatidylethanolamines (see par 3.2.1.2), and fatty acids and derivatives (see par 3.2.4). Nodes (highlighted in grey in the network) associated neither with known NPs nor with compounds predicted in this study, may indicate the presence of novel compounds, which deserve further studies to be isolated and structurally elucidated.
3.2.1 Glycerophospholipids
3.2.1.1 Lyso-Glycerophosphocholines (Lyso-GPCs)
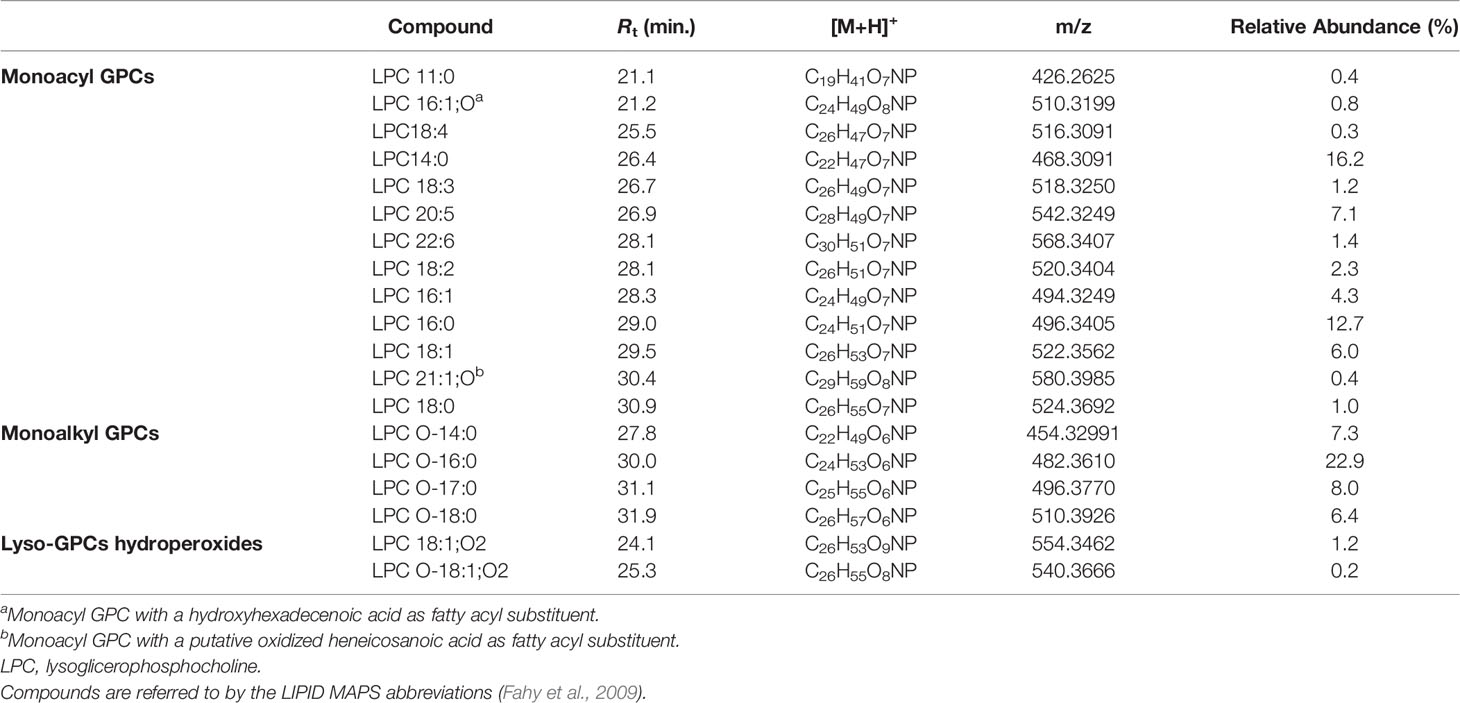
Lyso-GPCs are a class of lipids featuring a glycerophosphocholine backbone, with one free hydroxyl function, at either the sn-1 or the sn-2 position. While the sn-2 position is usually esterified with a long chain fatty acid (monoacyl GPCs), the OH at the sn-1 position may bear either a long chain acyl (monoacyl GPCs) or alkyl/alkenyl group (monoalkyl/alkenyl GPCs).
The product ion spectra generated from the [M+H]+ ions of lyso-glycerophosphocholines appeared to be dominated by the presence of a) the [M+H-H2O]+ fragment ion, derived from the neutral loss of a water molecule from the glycerol unit or the phosphate group and b) the diagnostic phosphocholine ion at m/z 184.0733 (C5H15O4NP+) (Figures 4 and S1A). Additional ions giving structural information were of low abundance but still useful for structural characterization. Fragment ions arisen from trimethylamine loss [M+H-59.0730]+ or sequential losses of water and trimethylamine [M+H-H2O-59.0730]+, were indicative of the choline moiety. Moreover, ions generated by fragmentation of the phosphocholine [M+H-183.0655]+, revealed the acyl-, alkyl, or -alkenyl glycerol backbone (Figure 4).
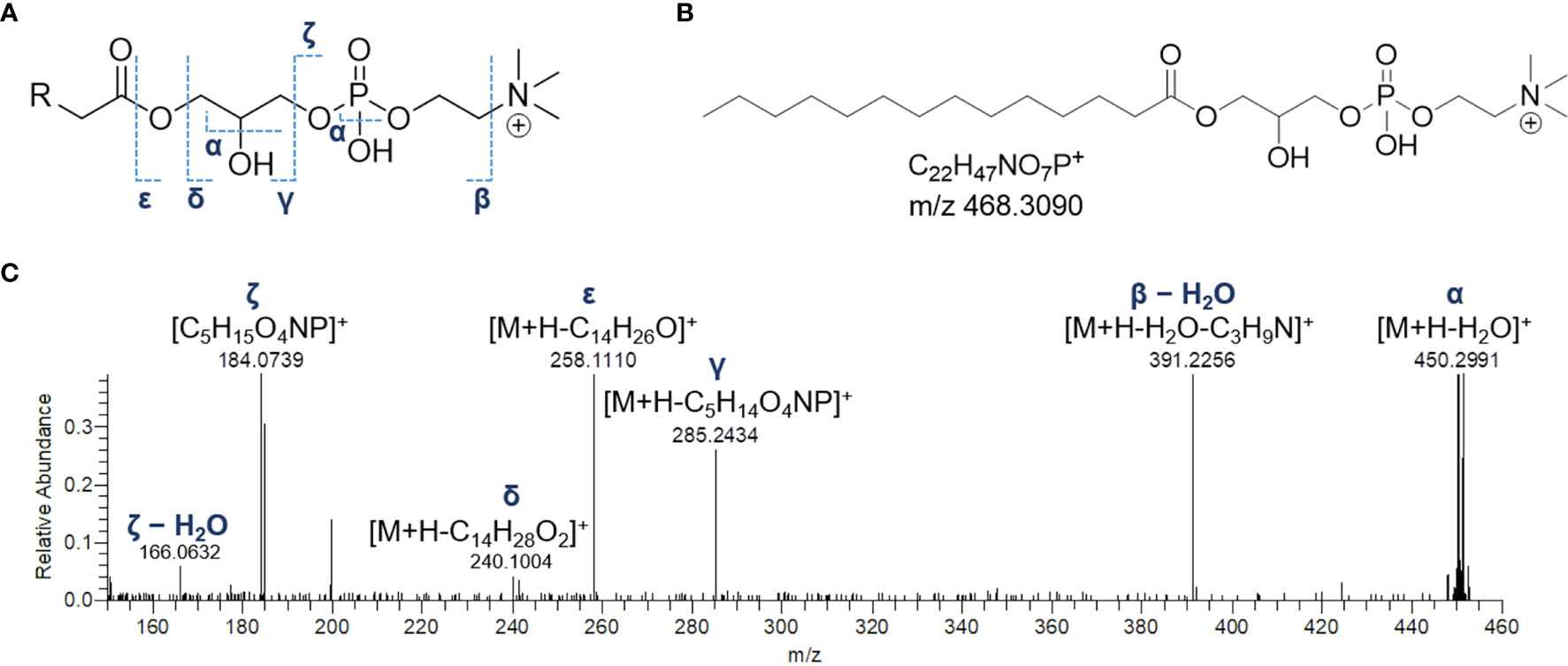
Figure 4 The fragmentation patterns of lyso-glycerophosphocholines (A). Chemical structure of a representative lyso-glycerophosphocholine from Botrylloides niger, namely 1-myristoyl-sn-glycero-3-phosphatidylcholine (LPC 14:0) (B) and HR ESI-MS2 spectrum of the relevant [M+H]+ adduct (C).
In monoacyl GPCs, also known as lysophosphatidylcholines, fatty acyl substituents could be also indirectly inferred from the presence of the glycerylphosphorylcholine fragment ion at m/z 258.1101 (C8H21O6NP+) and the corresponding dehydrated ion, as resulting from losses of the fatty acid groups as ketene and carboxylic acid, respectively (Figure 4).
Among lyso-GPCs, putative octadecenoyl GPC and octadecenyl GPC hydroperoxides were identified, as revealed by a neutral loss of 34.0055 Da, arising from fragmentation of the hydroperoxy group (Table 2).
3.2.1.2 Lyso-Phosphatidylethanolamines (Lyso-PEs)
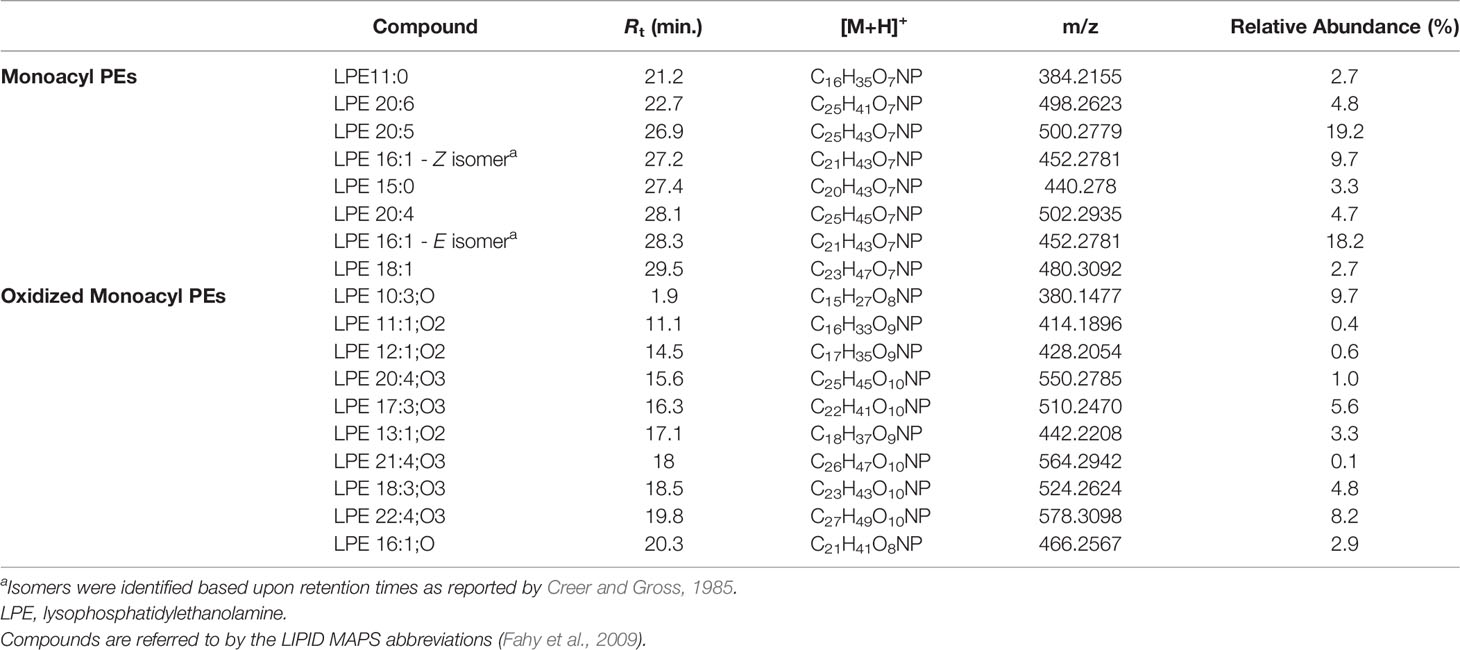
Lyso-PEs, also known as monoacylglycerophosphoethanolamines (monoacyl PEs), have a glycerophosphoethanolamine moiety with a long chain fatty acid, usually located at the sn-1 position.
Mass tandem spectra of the [M+H]+ ions of lyso-PEs displayed two abundant fragment ions, including a) the ion deriving from water loss, following the same pathway as for lyso-GPCs and b) the ion [M+H-141.0191]+ generated by elimination of the phosphoethanolamine head group via the phosphoester bond cleavage (Figure 5). Rearrangement processes leading to formation of the fragment ions a) [M+H-43.0422]+ following loss of aziridine (C2H5N), b) [M+H-61.0528]+ following loss of ethanolamine (C2H7NO), c) [M+H-59.0371]+, following loss of aminoacetaldehyde (C2H5NO) and d) [M+H-97.9769]+ and [M+H-172.0137]+ from internal losses of phosphoric acid (H3PO4) and glycerophosphophoric acid (C3H9O6P), respectively, were suggestive of the glycerophosphoethanolamine moiety and diagnostic of PEs (Figure 5) (Hsu and Turk, 2009). Differently from lyso-GPCs, acylium ions [RCO]+, together with the relevant dehydrated ions, were clearly observed in product ion spectra of lyso-PEs and were useful to characterize the fatty acyl substituents. The fragment ion at m/z 198.0531 (C5H13NO5P+), arising from the elimination of the intact carboxylic acid, was another diagnostic ion in mass tandem spectra of the [M+H]+ ions of lyso-PEs (Figure S1B). Notably, nine oxidized lyso-PEs could be detected in the organic extract of B. niger, featuring hydroxy and/or methoxy and/or oxo fatty acyl substituents (Table 3).
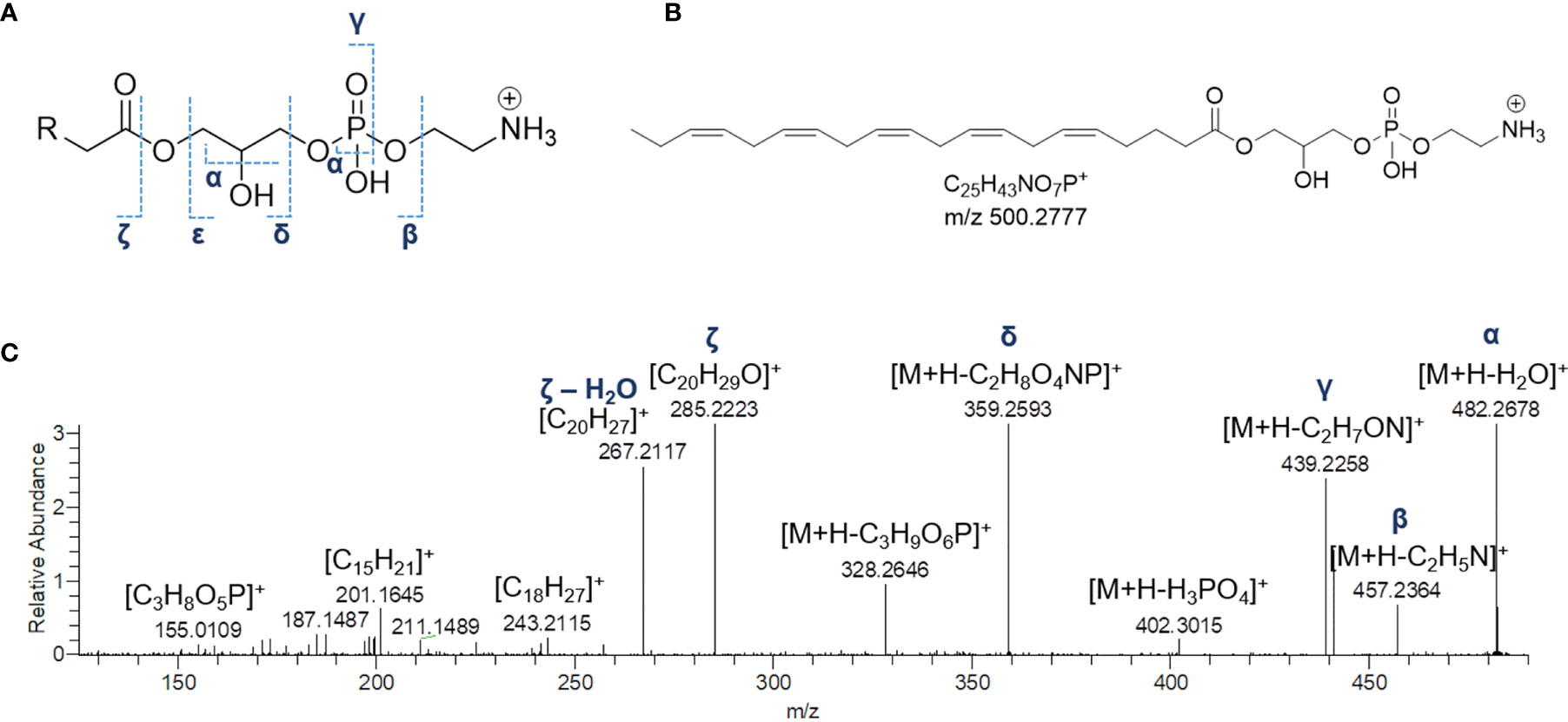
Figure 5 The fragmentation patterns of lyso-phosphatidylethanolamines (A). Chemical structure of a representative lyso-phosphatidylethanolamine from Botrylloides niger, namely 1-(5Z,8Z,11Z,14Z,17Z-eicosapentaenoyl)-glycero-3-phosphoethanolamine (LPE 20:5) (B) and HR ESI-MS2 spectrum of the relevant [M+H]+ adduct (C).
3.2.2 Sphingolipids
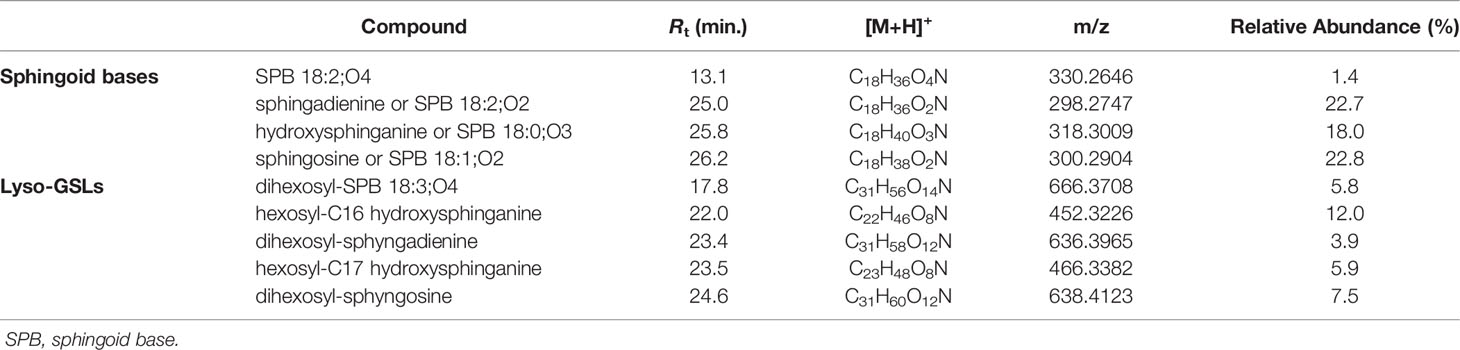
Dereplication of the organic extract of B. niger allowed the identification of nine compounds belonging to the sphingolipid class, including sphingoid bases (SPBs) and lyso-glycosphingolipids (Table 4).
Sharing similarity with previous reported ESI-MS tandem spectra (Shaner et al., 2009), the product ion spectrum of sphingosine (SPB 18:2; O2) unveiled the presence of fragments arising from losses of water, formaldehyde, water and ammonia, water and formaldehyde, and 2 water molecules and ammonia, together with minor rearrangement ions (Figure 6A). The fragmentation patterns of sphingadienine (SPB 18:2; O2) and hydroxysphinganine (SPB 18:0; O3) were quite similar to that of sphingosine, with the protonated hydroxysphinganine displaying an additional elimination of water, as expected. Interestingly, a putative novel sphingoid base, corresponding to the molecular formula C18H35O4N, was detected. As sharing almost the same fragmentation pathways with SPBs and clustering together with hydroxysphinganine in the molecular network (Figure 3), this compound (SPB 18:2; O4), has been tentatively identified as an oxidized analogue of hydroxysphinganine, featuring two degrees of unsaturation.
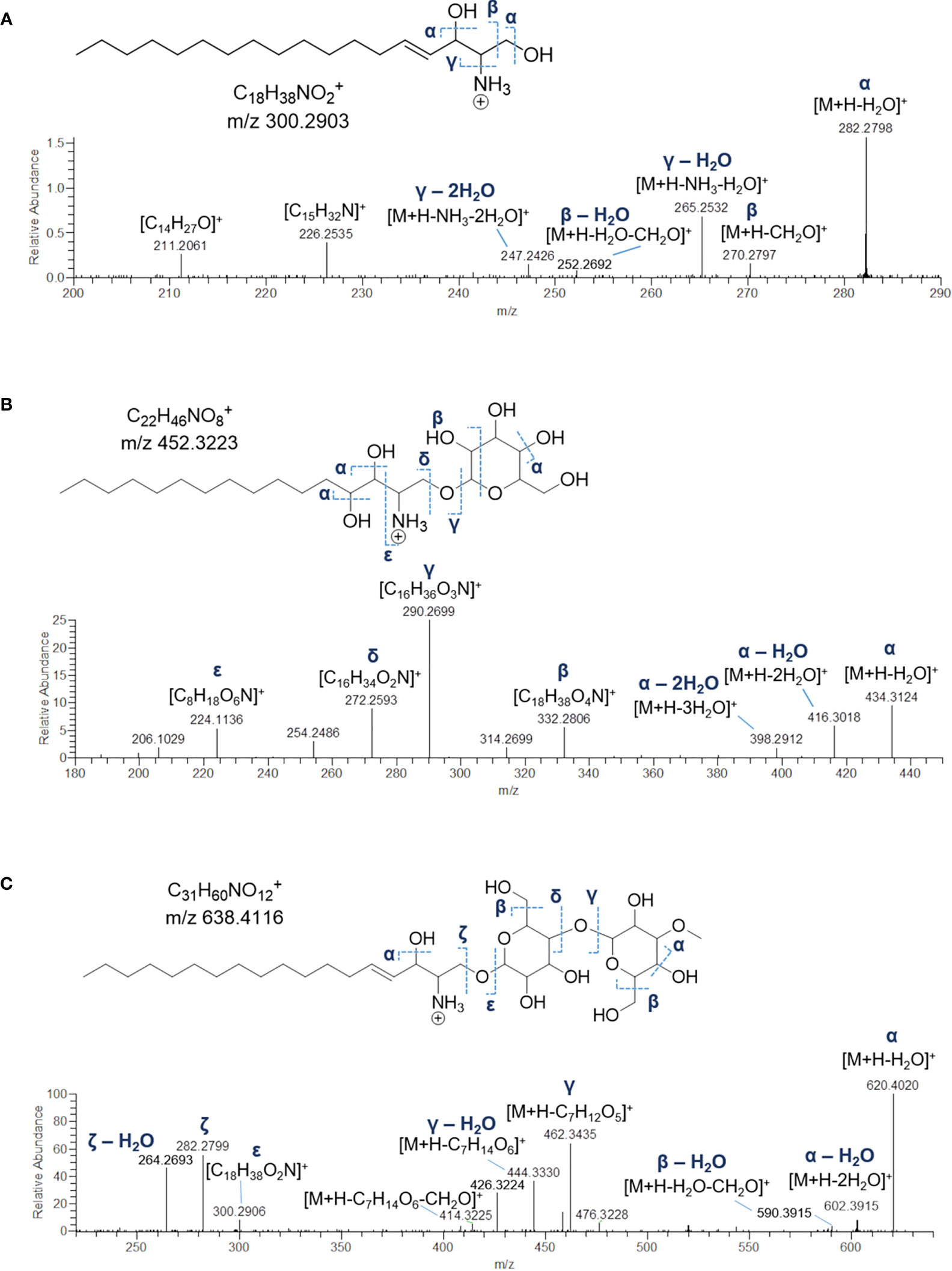
Figure 6 Fragmentation patterns and HR ESI-MS2 spectra of [M+H]+ ions of sphingosine (SPB 18:1;O2) (A), hexosyl-C16 hydroxysphinganine (B) and dihexosyl-sphyngosine (C) from Botrylloides niger.
Lyso-glycosphingolipids (lyso-GSLs) are SPBs with the primary OH group linked to a saccharidic head group (usually made up of glucose and/or galactose monomers) via a glycosidic bond, but lacking the N-acyl substituent as compared to intact glycosphingolipids (Merrill, 2011). Structural prediction of lyso-GSLs from B. niger led to the detection of two monohexosyl and three dihexosyl lyso-GSLs. Overall, the tandem mass spectra of lyso-GSL adducts contained fragment ions reflecting the long chain sphingoid base and the sugar constituents of the molecules, arising primarily from the glycosidic bond cleavage.
Fragmentations γ, δ, and ϵ indicated in Figure 6B permitted to assemble unequivocally the structures of the two monohexyl lyso-GSLs, which were shown to differ from each other as bearing a C16 and a C17 hydroxysphinganine as long chain base, respectively. In addition, the presence of the sugar unit was also suggested by fragment ions generated by a typical retro-Diels-Alder mechanism (Figure S2) involved in the fragmentation of the sugar moiety in glycosylated natural products (Demarque et al., 2016).
MS/MS spectra of the three dihexosyl lyso-GSLs displayed sequential losses of C7H12O5 (176.0679 Da) and C6H10O5 (162.0523 Da), which were consistent with a disaccharide unit composed of a putative O-methylated hexosyl starter unit linked to a hexose ring (Figure 6C). As shown in Table 4, the dihexosyl lyso-GSLs differ in degree of unsaturation and oxidation of the C18 sphingoid base.
3.2.3 Sulfonolipids
The crude extract of B. niger was shown to contain two sulfonolipids, as indicated by a) high-resolution (HR) ESI-MS spectra of their sodium adducts showing 4% intense M + 1.9957 isotope peaks suggestive of a sulphur atom and accounting for the molecular formula annotated in Table 5 and b) MS tandem spectra unveiling fragments arising from neutral losses of sulfuric and sulfurous acids. Molecular formula as well as HR MS2 spectrum of the 352-Da sulfonolipid (Figure 7A) were consistent with the structure of IOR-1 (Woznica et al., 2016), composed of a sulfonic acid head group and a C17 branched alkyl chain bearing two hydroxy groups. Acquisition of negative HR ESI-MS spectra provided more informative clues to confirm the identity of IOR-1, as displaying fragment ions at m/z 255.2305 and m/z 225.2203 arising from the neutral losses of methanesulfonic and hydroxyethanesulfonic acids, respectively (Figures 7A and S3). In the light of these findings, the 324-Da sulfonolipid, which was detected for the first time to the best of our knowledge, was tentatively identified as an inferior homologue of IOR-1, featuring a C15 alkyl chain and, therefore, indicated as C15 IOR-1 (Figure 7B). While the positive MS2 spectra were similar for both sulfonolipids, fragmentation patterns in the negative ion mode were somehow different. The ESI-MS2 spectrum of the [M-H]- ion of C15 IOR-1 unveiled the presence of the methanesulfonate fragment anion, which was consistent with a favoured fragmentation assisted by a β-hydroxy group and, therefore, indicative of the same hydroxyethylsulfonate head group as in IOR-1. However, a) the fragmentation leading to neutral loss of acetone (Figures 7B and S3) as well as b) the lack of a fragment ion homologue to the δ’ fragment in IOR-1 (Figure 7A), imply a different position for the second OH group in C15 IOR-1, which was predicted to be located on the terminal isopropyl group as shown in Figure 7B.
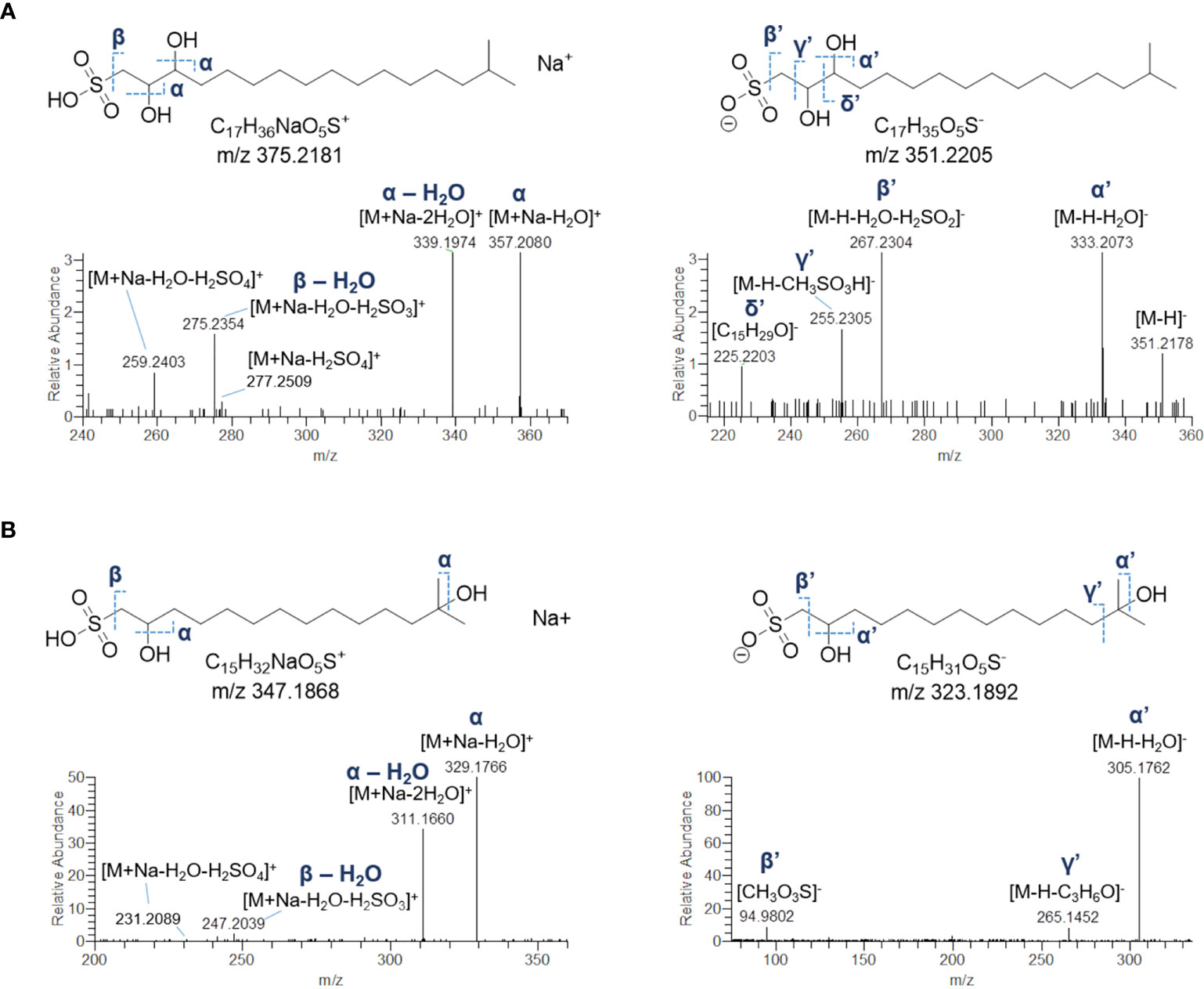
Figure 7 HR ESI-MS2 spectrum of the [M+Na]+ (left panel) and [M-H]- (right panel) ions of the putative sulfonolipid from Botrylloides niger annotated as IOR-1 (A). Predicted structure and HR ESI-MS2 spectrum of the [M+Na]+ (left panel) and [M-H]- (right panel) ions of the putative sulfonolipid C15 IOR-1 (B).
3.2.4 Monoacylglycerols and Fatty Acids and Derivatives
Monoacylglycerols (MGs) are glycerol esters, in which one hydroxy function is esterified with a long chain fatty acid, at the sn-1 (or sn-3) or the sn-2 position. However, it is widely recognised that 2-monoacylglycerols may undergo spontaneous isomerization to the corresponding 1-(or 3-) monoacyl isomers. Molecular networking analysis of MS2 data from the extract of B. niger led to the annotation of 5 monoacylglycerols (Table 6). Fragmentation spectra of the pseudomolecular ion [M+H]+of MGs was dominated by the acylium ion [RCO]+ resulting from neutral loss of glycerol (C3H8O3, 92.0473 Da), due to inductive cleavage of the ester bond assisted by the adjacent carbonyl function (Figure 8A). Moreover, the carbonyl group likely removes a proton from the terminal primary OH, thereby facilitating elimination of a lactone ring and formation of another diagnostic fragment, i.e. [M+H-C3H6O2]+ (Figure S3). MS/MS spectra of MGs were also characterized by the presence of the dehydrated ions of [M+H]+and [M+H-C3H8O3]+ fragments (Figure 8A). Notably, two MGs from B. niger displayed unusual polyunsaturated fatty acyl substituents, i.e. putative docosaheptaenoic and eicosahexaenoic acids (Table 6).
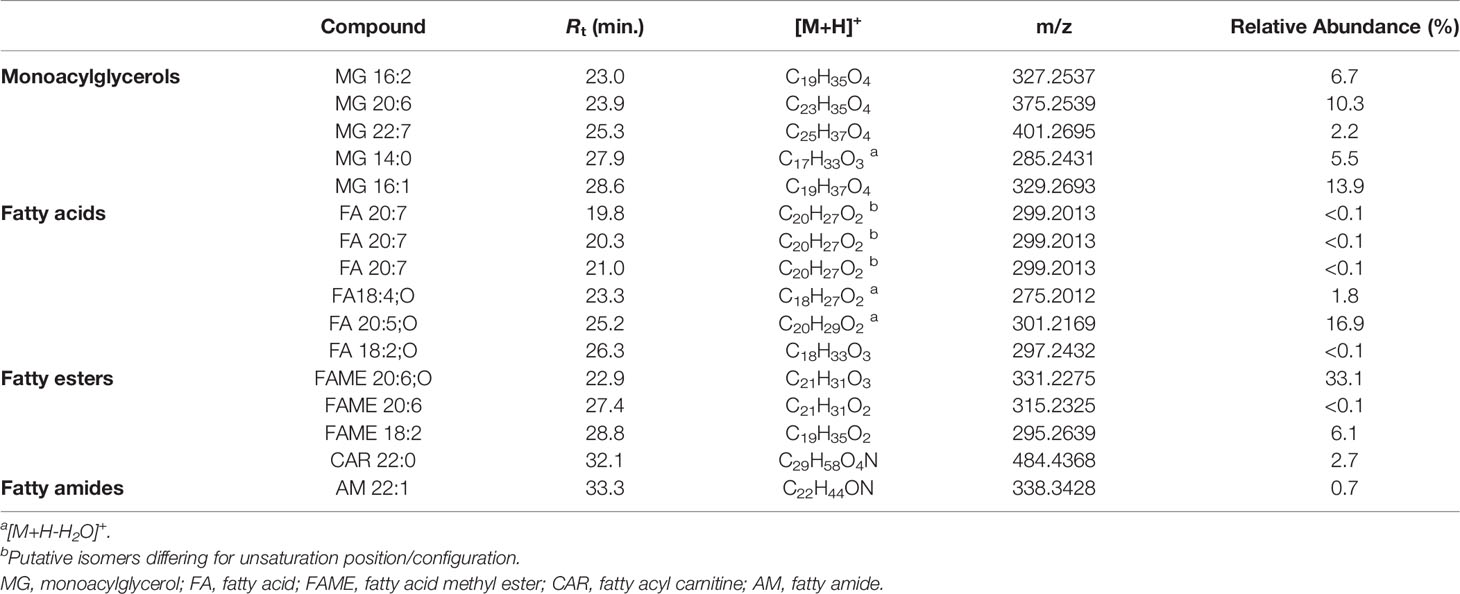
Table 6 Monoacylglycerols and fatty acids and derivatives identified in the organic extract from B. niger.
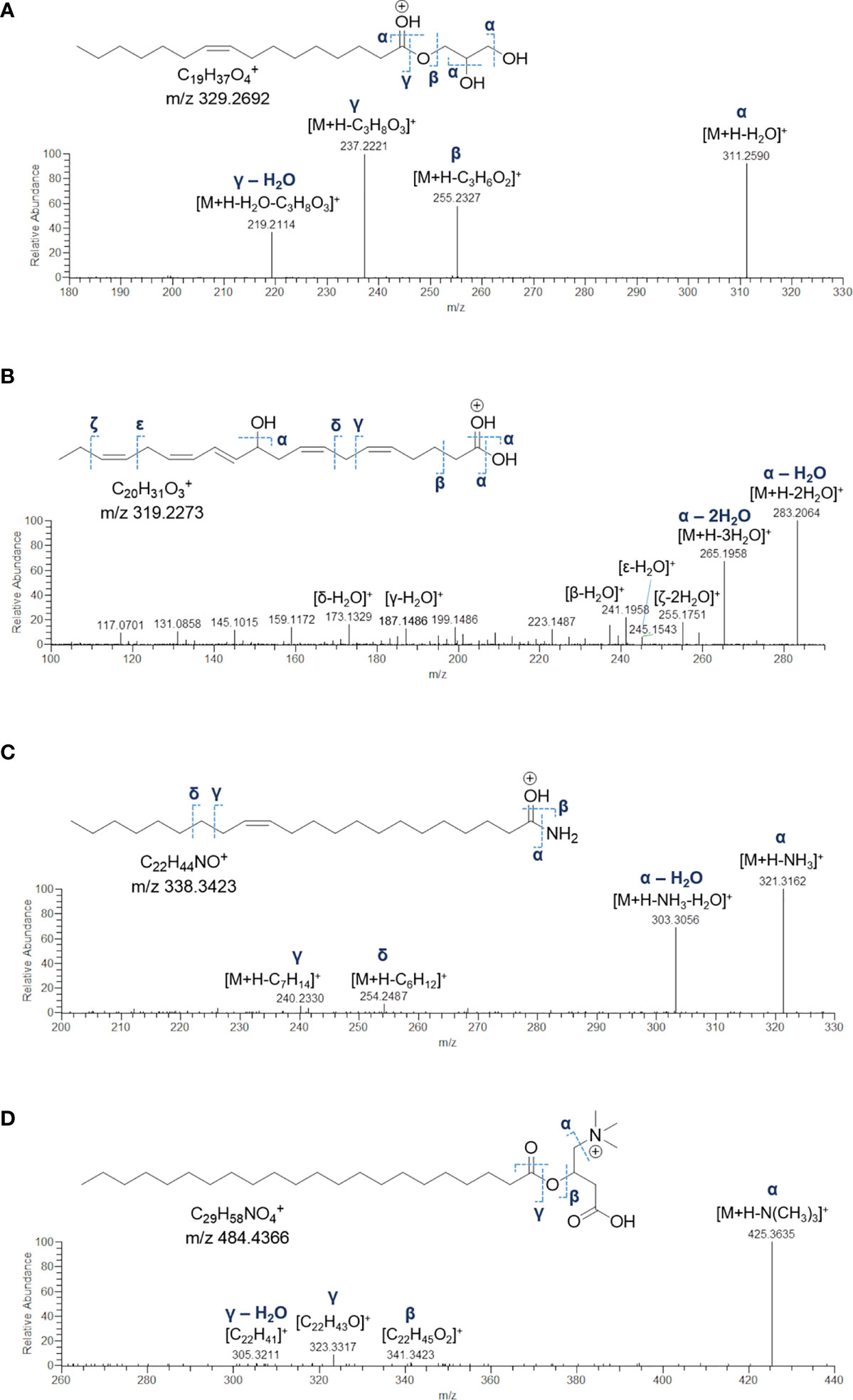
Figure 8 Fragmentation patterns and HR ESI-MS2 spectrum of the [M+H]+ ion of 1-(9Z-octadecenoyl)-sn-glycerol (MG 16:1) (A), the [M+H-H2O]+ ion of 11-hydroxy-5Z,8Z,12E,14Z,17Z-eicosapentaenoic acid (FA 20:5;O) (B), the [M+H]+ ion of 13Z-docosenamide (AM 22:1) (C), and the [M+H]+ ion of the docosanoylcarnitine (CAR 22:0) (D).
Eleven nodes in the molecular network (Figure 3) were annotated as fatty acids and derivatives, including a) a small cluster of five PUFAs, b) three fatty acid methyl ester derivatives (putative artefacts originating during extraction with methanol), c) 13Z-docosenamide (AM 22:1), and d) docosanoylcarnitine (CAR 22:0) (Table 6). Among these compounds, the 11-hydroxy-5Z,8Z,12E,14Z,17Z-eicosapentaenoic acid (FA 20:5;O), AM 22:1, and CAR 22:0 were identified by comparing their mass spectra with the GNPS spectral database (Figures 8B–D). Overall, the diagnostic product ion observed in MS/MS spectra of [M+H]+ adducts of PUFA (and [M+H-H2O]+for OH-PUFA) was the acylium ion, which in turn underwent water loss during fragmentation (Figure 8B). A series of unsaturated hydrocarbon ions with lower intensities occurred during ESI MS/MS of PUFA, due to extensive hydride shifts, thereby hampering unambiguous localization of the double bonds (Figure 8B). As it regards fatty acid methyl esters (FAMEs), mass fragmentation showed [M+H-32.0262]+ as the base peak, corresponding to the ion arising from the loss of CH3OH.
3.2.5 Indole and Purine Alkaloids
The HR ESI mass spectra of the [M+H]+ ion peaks at m/z 422.8711 (Rt = 10.9 min) and m/z 438.8659 (Rt = 12.1 min) defined the molecular formulas of these metabolites as C12H13Br3N2 and C12H13Br3N2O, respectively, as suggested by the observed isotope pattern peculiar of tribrominated compounds, showing four peaks (each separated by two mass units) of approximate intensity 1:4:4:1. In the light of these findings, the 422 Da- and 438-Da compounds were identified as 2,5,6-tribromo-1-methylgramine and the relevant N-oxide derivative, as their MS/MS spectra were exactly the same and characterized by the fragment ion [C10H7Br3]+, generated by elimination of dimethylamine (45.0578 Da) and dimethylhydroxylamine (61.0528 Da), respectively (Figure 9). In addition, the fragment [C10H7NBr3]+ undergoes a) loss of a radical bromine and b) loss of molecular bromine, thus giving the [C10H7NBr2]•+ radical cation at m/z 298.8950 and the [C10H7NBr]+ ion at m/z 219.9763, likely due to homolytic cleavage of carbon-bromine bonds (radical fragmentation) (Figures 9 and S4). Even if being unusual in CID (collision-induced dissociation)-type fragmentations, generation of odd-electron species may occur when a) unpaired electrons can occupy a delocalized antibonding orbital and b) chemical bonds are hard to be cleaved (Levsen et al., 2007), such in the case of the high conjugated indole ring in gramine alkaloids.
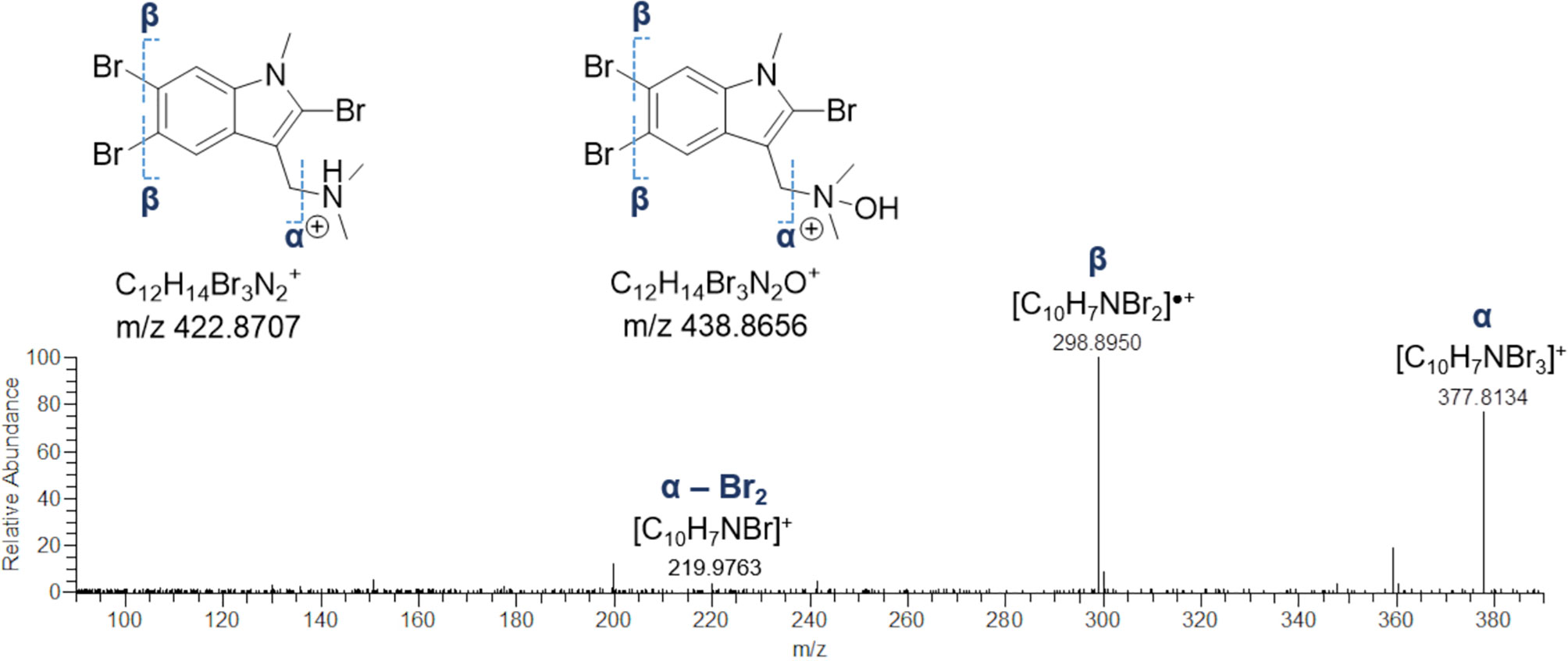
Figure 9 HR ESI-MS2 spectrum of the fragment ion at m/z 377.8134, generated by neutral loss of dimethylamine and dimethylhydroxylamine from 2,5,6-tribromo-1-methylgramine and 2,5,6-tribromo-1-methylgramine N-oxide, respectively.
Finally, three nodes in the network appeared to be related to unknown alkyl purine alkaloids (Figure S6). Analysis of the mass tandem spectra of these molecules unveiled a diagnostic fragment at m/z 150.0780 corresponding to the methyladenine ion, together with its related fragments deriving from NH3 and HCN losses (Figure S6). Therefore, these metabolites were predicted to be methyladenine derivatives bearing different alkyl/acyl substituents. However, their structures remain unsolved and require further studies to be elucidated.
4 Discussion
Human-mediated introduction of alien species in new biogeographic realms is a phenomenon that dates back centuries (Branch and Steffani, 2004; Provan et al., 2008). However, this seems to be furthermore amplified in the recent decades, with many ascidians spreading worldwide and interfering with native benthic communities and habitats by creating consistent environmental and even economic damages (Lambert and Lambert, 1998; Zhan et al., 2015). In the present case, when B. niger was locally discovered, it already formed large aggregates in the investigated channel, and its presence was also noted in additional lagoons of the area (Virgili et al., 2022). This suggests that the early eradication phase, often highlighted to limit the spread of alien species (Willan et al., 2000; Giakoumi et al., 2019), is already far to be potentially applied, and that local communities are presumably enriched by the presence of B. niger since at least years. Absence of field studies but mostly taxonomic impediments may be at the basis of such a result. Indeed, contrary to other localities where the presence of B. niger is well acknowledged (Rocha et al., 2012; Sheets et al., 2016; Streit et al., 2021), the arrival of this colonial ascidian in the area, but in general in the Mediterranean Sea, was presumably overlooked due to rarefaction of studies in zoology and confusion with the congeneric species B. leachii (see Griggio et al., 2014; Virgili et al., 2022). No certainties also occur regarding a possible pathway of arrival in the area. The Fusaro Lake is already known as a hub for the introduction of alien species of different phyla (e.g. Bianchi, 1983; Villani and Martinez, 1993; Crocetta et al., 2012; Crocetta et al., 2013; Hanson et al., 2013), and it also hosts a conspicuous mussel farm that may constitute the most likely source of introduction for this species in the lake. This seems to be in agreement with other studies worldwide, that suggested a species spread at a small scale based on recruitment preferences for mussel beds (Sheets et al., 2016; Rocha et al., 2019). However, another possibility is that this species is spreading locally favoured by the natural currents that exchange the lake waters through the channel, and most likely coming from the nearby Miseno Lake, an area recently acknowledged as a hotspot of non-indigenous species (NIS) ascidians, including B. niger (Virgili et al., 2022).
Although the dominance observed during sampling activities is worrying from an ecological point of view, our results confirm the interest of these alien species as they represent today a potential huge biological resource for nutritional or functional purposes, reducing, in this way, the economic losses caused by these invasive organisms.
Recent studies have shown that ascidians, in particular parts of their inner body tissues, are used to flavour foods intended for human consumption. In fact, all inner body tissues are rich in proteins, mainly collagens with a high essential amino acid index and high delicious amino acid (DAA) content. Moreover, they can supply the body with essential lipid components, including high contents of good-quality fatty acids (Zhao and Li, 2016). Lipids are a class of biomolecules involved in a huge number of different functions in biological systems, which make them fundamental for development and growth, and act as powerful signalling agents during metabolic disruption due to diseases, such as neurological disorders, autoimmune diseases, and cancer (Wymann and Schneiter, 2008; Cheng et al., 2016; Brown et al., 2017).
In this work, the employment of the Molecular Networking approach followed by a careful manual curation of HR ESI-MS2 data allowed us to study the metabolome of the alien species B. niger for the first time, and shed light on the presence of novel NPs, several of them remaining still unknown. FBMN was shown to be an efficient tool for fast dereplication of complex organic mixtures and support LC-MS2 data analysis for high-confidence structural prediction of detected metabolites.
Our results showed the presence of different classes of indispensable lipids, namely glycerophospholipids, glycerolipids, sphingolipids, fatty acids and derivatives, and sulfonolipids.
Among them, glycerophospholipids were the most represented lipid class, including lyso-glycerophosphocholines and lyso-phosphatidylethanolamines.
Similarly, Hou et al. (2021) recently reported the characterization of the lipid profile of C. intestinalis, H. roretzi, and S. clava, and, among the thirteen major lipid subclasses identified, glycerophospholipids and glycerolipids were the dominant components (66.30–90.60% of total lipids).
Lyso-GPCs and lyso-PEs are representatives of a class of mono-acylated/alkylated glycerophospholipids, commonly referred to as lysophospholipids, playing a key role as structural lipid constituents of cellular membranes and biological signalling molecules in eukaryotes and bacteria, including marine species. Lysophospholipids may regulate fundamental cellular functions such as cell growth, differentiation, survival, migration, adhesion, invasion, and morphogenesis, by interacting with their cognate receptors and/or modelling composition and fluidity of lipid rafts. Indeed, lysophospholipids have been reported to regulate cell motility in budding tunicates (Arai et al., 2004) as well as to enhance multicellular development in the choanoflagellate Salpingoeca rosetta Dayel et al., 2011 (Woznica et al., 2016). In the latter case, lyso-PEs and the sulfonolipids RIFs and IOR-1 are produced by the choanoflagellate endosymbiont Algoriphagus machipongonensis Alegado et al., 2013 and proposed as actors of a metabolic interplay in which multiple bacterial cues regulate the cellular growth of S. rosetta. Considering that a) lysophospholipids and sulfonolipids (IOR-1 and C15 IOR-1) co-occur in the organic extract of B. niger and that b) bacteria live in symbiosis with several Botrylloides species, the notion that a network of bacterial lipids regulate multicellular development and cell motility can be extended to B. niger, which is expected to orchestrate a complex signalling network to control seasonal dynamics of its blooms (Rinkevich et al., 1993; Oricchio and Muniz Dias, 2020; Ramalhosa et al., 2021).
To date, there has been a growing interest in studying the biological functions and metabolism of lysophospholipids, because they could represent a good starting point for the development of new therapeutics for several diseases (Varandas et al., 2019). Indeed, these molecules can affect many biological processes, such as neurogenesis, angiogenesis, wound healing, immunity, and carcinogenesis (Ishii et al., 2004).
Noteworthy, our results showed that B. niger contains also lyso-GPCs and lyso-PEs bearing PUFAs [including eicosapentaenoic acid (C20:5n-3, EPA), and docosahexaenoic acid (C22:6n-3, DHA)], which accounted for the 40% of the detected lyso-glycerophospholipids. Interestingly, glycerophospholipids enriched in PUFAs were also mainly present in the inner body tissues of different ascidians, indicating that these marine organisms could potentially be used for health-promoting food for humans (Hou et al., 2021). Marine sources containing EPA/DHA-enriched glycerophospholipids are receiving increasing attention thanks to their emerging health benefit (as reviewed in Zhang et al., 2019 and Ahmmed et al., 2020). Zhou and collaborators showed that different fatty acids composition of glycerophospholipids were involved in decreasing cognitive decline and biological damage and in brain protection, and these beneficial effects were partly enhanced in presence of EPA and DHA (Zhou et al., 2016).
In addition, unusual PUFAs (Table 6) either linked to the glycerol backbone in monoacylglycerols or as free fatty acids and methyl ester derivatives, were also identified by molecular networking analysis of tandem MS data from B. niger crude extract. In tunicates unsaturated fatty acids, including PUFAs, have been shown to exert mitogenic activity and promote cell proliferation (Arai et al., 2004). Moreover, PUFAs have attracted great attention due to their enormous benefits. They can reduce or prevent the severity of several diseases, such as hyperlipidemia, diabetes, cancers, inflammation and heart and neurodegenerative diseases (reviewed in Zhang et al., 2019).
Therefore, similarly to other marine organisms, B. niger could potentially be used for health-promoting food for humans.
Dereplication of the organic extract from B. niger shed light on the presence of lyso-glycosphingolipids and free sphingoid bases. Over the years, several sphingolipids have been identified from natural sources, including microorganisms, tunicates, sponges, corals and algae. In the past, they were merely considered as components of cellular membranes, but they demonstrated to be involved in several cellular phases (Morales et al., 2007; Ponnusamy et al., 2010), and to be effective as antiproliferative drugs against different tumors (Schmelz et al., 2000; García-Barros et al., 2014; Rethna Priya et al., 2019).
Besides the presence of different lipid classes, our findings unveiled B. niger to be a source of indole and purine alkaloids. In fact, ascidians are prolific sources of nitrogenated metabolites, and more than 300 alkaloids have been reported from these organisms (Nathani et al., 2020) till now. Noteworthy, many of these have been shown to have important activities, including antimicrobial, anticancer, and antiviral, whereby several therapeutics have been isolated or inspired from tunicates-derived NPs.
Among the 12 marine-derived NPs approved by the FDA to date, 2 molecules are native to tunicates, including the alkaloid ecteinascidine (Yondelis®) from E. turbinata and its synthetic derivative Lurbinectedin (Zepsyre®) (Phase III) used to treat different types of cancer (Della Sala et al., 2018; McCauley et al., 2020; Ramesh et al., 2021).
Particularly, two brominated indole derivatives were identified in B. niger and predicted as 2,5,6-tribromo-1-methylgramine and its N-oxide analogue. Even if ascidians have been largely acknowledged as a reservoir of brominated alkaloids, these two metabolites have never been described in tunicates before, as being only isolated from the marine bryozoan Amathia verticillata (delle Chiaje, 1822). Particularly, 2,5,6-tribromo-1-methylgramine was found to exert antifouling activity against the barnacle Amphibalanus amphitrite (Darwin, 1854) and the blue mussel Mytilus edulis Linnaeus, 1758 (Sato and Fenical, 1983; Kon-Ya et al., 1994). In addition, three novel purine alkaloids, i.e. methyladenine derivatives, were found in the metabolome of B. niger, thus their structures remaining unsolved. Alkyl purines as well as brominated indoles play a key role in the antifouling mechanism of marine organisms. Detection of such bioactive molecules could at least partially explain the invasiveness of B. niger, which is able to prevent larvae of other marine organisms from settlement and growing on its bodies (Hiebert et al., 2019).
Indeed, by competing for space and food, a congener of B. niger, namely Botrylloides violaceus Oka, 1927, is known to displace other fouling organisms (Berman et al., 1992), including native and introduced tunicates (Dijkstra et al., 2007), bryozoans, barnacles, and mussels (Dijkstra and Harris, 2009), indicating strong competitive ability (Lambert and Lambert, 2003).
In conclusion, the alien species B. niger represents a prolific source of several valuable bioactive compounds, probably developed to adapt and compete with native species. It is also rich of a huge variety of lipid species that improve human health, including lipids rarely found in other foods as PUFAs. The presence of these molecules can undoubtedly transform this alien species from damage to the ecosystem into a precious bio-resource, easily available. Therefore, B. niger together with other invasive ascidians, although concerned from an ecological point of view, could find an important economic role to benefit society, including good opportunities in the food and pharmaceutical industry for the development of functional products based on ascidians.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
GDS, DC, and FC designed and directed the research. GDS, DC, RV, GV, VT, RT, and FC performed the experiments and analysed the data. DdP proceeded to funding acquisition. GDS, DC, and FC wrote the manuscript. All authors contributed to the editing and revision of the manuscript, and read and approved the final manuscript. All authors agree to be accountable for the content of the work.
Funding
Work supported by the following projects: (i) PO FEAMP Campania 2014–2020. DRD n.35 of 15th March 2018. Innovazione, sviluppo e sostenibilità nel settore della pesca e dell’acquacoltura per la regione Campania. Misura 2.51. WP5. Task 5.5. Presenza e distribuzione di specie non indigene del macrozoobenthos e del necton in Campania; (ii) POR Campania FESR 2014–2020. Technology Platform for Therapeutic Strategies against Cancer. Antitumor Drugs and Vaccines from the Sea (ADViSE). CUP B43D18000240007–SURF 17061BP000000011. WP1. Task 1.1. Tassonomia, distribuzione, abbondanza e stagionalità dei principali organismi macrobentonici con bioattività presenti nel Golfo di Napoli.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
Centro Ittico Campano S.p.A. (Bacoli, Napoli) allowed sampling and offered support.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2022.865751/full#supplementary-material
References
Ahmmed M. K., Ahmmed F., Tian H., Carne A., Bekhit A. E. D. (2020). Marine Omega-3 (N-3) Phospholipids: A Comprehensive Review of Their Properties, Sources, Bioavailability, and Relation to Brain Health. Compr. Rev. Food Sci. Food Saf. 19, 64–123. doi: 10.1111/1541-4337.12510
Arai K., Yoshida S., Nakatani M., Fujiwara S., Yubisui T., Kawamura K. (2004). Phospholipids and Their Derivatives as Mitogen and Motogen of Budding Tunicates. J. Biochem. 135, 71–78. doi: 10.1093/jb/mvh008
Atalah J., Fletcher L. M., Forrest B. M. (2021). Impacts of a Putative Invasive Ascidian on Rocky Shore Communities. Mar. Environ. Res. 105308. doi: 10.1016/j.marenvres.2021.105308
Ballarin L., Franchini A., Ottaviani E., Sabbadin A. (2001). Morula Cells as the Major Immunomodulatory Hemocytes in Ascidians: Evidences From the Colonial Species Botryllus Schlosseri. Biol. Bull. 201, 59–64. doi: 10.2307/1543526
Berman J., Harris L., Lambert W., Buttrick M., Dufresne M. (1992). Recent Invasions of the Gulf of Maine: Three Contrasting Ecological Histories. Conserv. Biol. 6, 435–441. doi: 10.1046/j.1523-1739.1992.06030435.x
Bianchi C. (1983). Serpuloidea (Annelida, Polychaeta) Delle Lagune Costiere Laziali E Campane. Anal. Museo Civico di Storia Naturale di Genova 84, 231–243.
Bouchemousse S., Bishop J. D., Viard F. (2016). Contrasting Global Genetic Patterns in Two Biologically Similar, Widespread and Invasive Ciona Species (Tunicata, Ascidiacea). Sci. Rep. 6, 1–15. doi: 10.1038/srep24875
Branch G. M., Steffani C. N. (2004). Can We Predict the Effects of Alien Species? A Case-History of the Invasion of South Africa by Mytilus Galloprovincialis (Lamarck). J. Exp. Mar. Biol. Ecol. 300, 189–215. doi: 10.1016/j.jembe.2003.12.007
Brown H. A., Thomas P. G., Lindsley C. W. (2017). Targeting Phospholipase D in Cancer, Infection and Neurodegenerative Disorders. Nat. Rev. Drug Discov. 16, 351–367. doi: 10.1038/nrd.2016.252
Brunetti R. (2009). Botryllid Species (Tunicata, Ascidiacea) From the Mediterranean Coast of Israel, With Some Considerations on the Systematics of Botryllinae. Zootaxa 2289, 18–32. doi: 10.11646/zootaxa.2289.1.2
Brunetti R., Manni L., Mastrototaro F., Gissi C., Gasparini F. (2017). Fixation, Description and DNA Barcode of a Neotype for Botryllus Schlosseri (Palla)(Tunicata, Ascidiacea). Zootaxa 4353, 29–50. doi: 10.11646/zootaxa.4353.1.2
Bullard S. G., Carman M. R. (2009). Current Trends in Invasive Ascidian Research. Invasive Species: Detect Impact Control, 57–79.
Bullard S. G., Lambert G., Carman M. R., Byrnes J., Whitlatch R. B., Ruiz G., et al. (2007). The Colonial Ascidian Didemnum Sp. A: Current Distribution, Basic Biology and Potential Threat to Marine Communities of the Northeast and West Coasts of North America. J. Exp. Mar. Biol. Ecol. 342, 99–108. doi: 10.1016/j.jembe.2006.10.020
Cappello T. (2020). NMR-Based Metabolomics of Aquatic Organisms. eMagRes 9, 81–100. doi: 10.1002/9780470034590.emrstm1604
Caso A., Esposito G., Della Sala G., Pawlik J. R., Teta R., Mangoni A., et al. (2019). Fast Detection of Two Smenamide Family Members Using Molecular Networking 618. doi: 10.20944/preprints201910.0091.v1
Cheng M., Bhujwalla Z. M., Glunde K. (2016). Targeting Phospholipid Metabolism in Cancer. Front. Oncol. 6, 266. doi: 10.3389/fonc.2016.00266
Corbo J. C., Di Gregorio A., Levine M. (2001). The Ascidian as a Model Organism in Developmental and Evolutionary Biology. Cell 106, 535–538. doi: 10.1016/S0092-8674(01)00481-0
Creer M. H., Gross R. W. (1985). Separation of Isomeric Lysophospholipids by Reverse Phase HPLC. Lipids 20, 922–928. doi: 10.1007/BF02534778
Crocetta F., Macali A., Furfaro G., Cooke S., Villani G., Valdés A. (2013). Alien Molluscan Species Established Along the Italian Shores: An Update, With Discussions on Some Mediterranean “Alien Species” Categories. ZooKeys 277, 91–108. doi: 10.3897/zookeys.277.4362
Crocetta F., Tanduo V., Osca D., Turolla E. (2020). The Chinese Mitten Crab Eriocheir Sinensis H. Milne Edward(Crustacea: Decapoda: Varunidae) Reappears in the Northern Adriatic Sea: Another Intrusion Attempt or the Trace of an Overlooked Population? Mar. Pollut. Bull. 156, 111221. doi: 10.12681/mms.313
Crocetta F., Thessalou-Legaki M., Aydogan O., Bekas P., Bilge G., Boyaci Y. O., et al. (2012). New Mediterranean Biodiversity Records (December 2012). Mediterr. Mar. Sci. 13, 312–327. doi: 10.12681/mms.313
Darriba D., Taboada G., Doallo R., Posada D. (2012). Jmodeltest 2: More Models, New Heuristics and Parallel Computing. Nat. Meth 9, 772. Nature Publishing Group. doi: 10.1038/nmeth.2109
Davis A. R. (1995). Over-Exploitation of Pyura Chilensis (Ascidiacea) in Southern Chile: The Urgent Need to Establish Marine Reserves. Rev. Chil. Hist. Natural 68, 7–1.
Davis M., Davis M. (2010). The Impact of the Ascidian Styela Clava Herdman on Shellfish Farming in the Bassin De Thau, France. J. Appl. Ichthyolo. 26, 12–18. doi: 10.1111/j.1439-0426.2010.01496.x
Dehal P., Satou Y., Campbell R. K., Chapman J., Degnan B., De Tomaso A., et al. (2002). The Draft Genome of Ciona Intestinalis: Insights Into Chordate and Vertebrate Origins. Science 298, 2157–2167. doi: 10.1126/science.1080049
Della Sala G., Agriesti F., Mazzoccoli C., Tataranni T., Costantino V., Piccoli C. (2018). Clogging the Ubiquitin-Proteasome Machinery With Marine Natural Products: Last Decade Update. Mar. Drugs 16, 467. doi: 10.3390/md16120467
Della Sala G., Mangoni A., Costantino V., Teta R. (2020). Identification of the Biosynthetic Gene Cluster of Thermoactinoamides and Discovery of New Congeners by Integrated Genome Mining and MS-Based Molecular Networking. Front. Chem. 8, 397. doi: 10.3389/fchem.2020.00397
De Marco G., Brandão F., Pereira P., Pacheco M., Cappello T. (2022). Organ-Specific Metabolome Deciphering Cell Pathways to Cope With Mercury in Wild Fish (Golden Grey Mullet Chelon Auratus). Animals 12, 79. doi: 10.3390/ani12010079
Demarque D. P., Crotti A. E., Vessecchi R., Lopes J. L., Lopes N. P. (2016). Fragmentation Reactions Using Electrospray Ionization Mass Spectrometry: An Important Tool for the Structural Elucidation and Characterization of Synthetic and Natural Products. Natural Product Rep. 33, 432–455. doi: 10.1039/C5NP00073D
Dijkstra J. A., Harris L. G. (2009). Maintenance of Diversity Altered by a Shift in Dominant Species: Implications for Species Coexistence. Mar. Ecol. Prog. Ser. 387, 71–80. doi: 10.3354/meps08117
Dijkstra J., Harris L. G., Westerman E. (2007). Distribution and Long-Term Temporal Patterns of Four Invasive Colonial Ascidians in the Gulf of Maine. J. Exp. Mar. Biol. Ecol. 342, 61–68. doi: 10.1016/j.jembe.2006.10.015
Dou X., Dong B. (2019). Origins and Bioactivities of Natural Compounds Derived From Marine Ascidians and Their Symbionts. Mar. Drugs 17, 670. doi: 10.3390/md17120670
Edler D., Klein J., Antonelli A., Silvestro D. (2021). raxmlGUI 2.0: A Graphical Interface and Toolkit for Phylogenetic Analyses Using RAxML. Methods Ecol. Evol. 12, 373–377. doi: 10.1111/2041-210X.13512
Fahy E., Subramaniam S., Murphy R. C., Nishijima M., Raetz C. R., Shimizu T., et al. (2009). Update of the LIPID MAPS Comprehensive Classification System for Lipids1. J. Lipid Res. 50, S9–S14. doi: 10.1194/jlr.R800095-JLR200
Folmer O., Hoeh W., Black M., Vrijenhoek R. (1994). Conserved Primers for PCR Amplification of Mitochondrial DNA From Different Invertebrate Phyla. Mol. Mar. Biol. Biotechnol. 3, 294–299.
Franchi N., Schiavon F., Carletto M., Gasparini F., Bertoloni G., Tosatto S. C., et al. (2011). Immune Roles of a Rhamnose-Binding Lectin in the Colonial Ascidian Botryllus Schlosseri. Immunobiology 216, 725–736. doi: 10.1016/j.imbio.2010.10.011
García-Barros M., Coant N., Truman J.-P., Snider A. J., Hannun Y. A. (2014). Sphingolipids in Colon Cancer. Biochim. Biophys. Acta (BBA)-Molecular Cell Biol. Lipids 1841, 773–782. doi: 10.1016/j.bbalip.2013.09.007
Gasparini F., Burighel P., Manni L., Zaniolo G. (2008). Vascular Regeneration and Angiogenic-Like Sprouting Mechanism in a Compound Ascidian Is Similar to Vertebrates. Evol. Dev. 10, 591–605. doi: 10.1111/j.1525-142X.2008.00274.x
Giakoumi S., Katsanevakis S., Albano P. G., Azzurro E., Cardoso A. C., Cebrian E., et al. (2019). Management Priorities for Marine Invasive Species. Sci. Total Environ. 688, 976–982. doi: 10.1016/j.scitotenv.2019.06.282
Griggio F., Voskoboynik A., Iannelli F., Justy F., Tilak M.-K., Xavier T., et al. (2014). Ascidian Mitogenomics: Comparison of Evolutionary Rates in Closely Related Taxa Provides Evidence of Ongoing Speciation Events. Genome Biol. Evol. 6, 591–605. doi: 10.1093/gbe/evu041
Hanson D., Cooke S., Hirano Y., Malaquias M. A., Crocetta F., Valdés Á. (2013). Slipping Through the Cracks: The Taxonomic Impediment Conceals the Origin and Dispersal of Haminoea Japonica, an Invasive Species With Impacts to Human Health. PloS One 8, e77457. doi: 10.1371/journal.pone.0077457
Hiebert L. S., Vieira E. A., Dias G. M., Tiozzo S., Brown F. D. (2019). Colonial Ascidians Strongly Preyed Upon, Yet Dominate the Substrate in a Subtropical Fouling Community. Proc. R. Soc. B 286, 20190396. doi: 10.1098/rspb.2019.0396
Hirose E., Ohtake S. I., Azumi K. (2009). Morphological Characterization of the Tunic in the Edible Ascidian, Halocynthia Roretzi (Drasche), With Remarks on ‘Soft Tunic Syndrome’ in Aquaculture. J. Fish Dis. 32, 433–445. doi: 10.1111/j.1365-2761.2009.01034.x
Hou Q., Huang Y., Jiang L., Zhong K., Huang Y., Gao H., et al. (2021). Evaluation of Lipid Profiles in Three Species of Ascidians Using UPLC-ESI-Q-TOF-MS-Based Lipidomic Study. Food Res. Int. 110454. doi: 10.1016/j.foodres.2021.110454
Hsu F.-F., Turk J. (2009). Electrospray Ionization With Low-Energy Collisionally Activated Dissociation Tandem Mass Spectrometry of Glycerophospholipids: Mechanisms of Fragmentation and Structural Characterization. J. Chromatogr. B 877, 2673–2695. doi: 10.1016/j.jchromb.2009.02.033
Huelsenbeck J., Ronquist F. (2001). Mrbayes, Version 3.0 B4: Bayesian Influence of Phylogeny. Bioinformatics 17, 754–755. doi: 10.1093/bioinformatics/17.8.754
Imai K. S., Levine M., Satoh N., Satou Y. (2006). Regulatory Blueprint for a Chordate Embryo. Science 312, 1183–1187. doi: 10.1126/science.1123404
Imperatore C., Della Sala G., Casertano M., Luciano P., Aiello A., Laurenzana I., et al. (2019). In Vitro Antiproliferative Evaluation of Synthetic Meroterpenes Inspired by Marine Natural Products. Mar. Drugs 17, 684. doi: 10.3390/md17120684
Ishii I., Fukushima N., Ye X., Chun J. (2004). Lysophospholipid Receptors: Signaling and Biology. Annu. Rev. Biochem. 73, 321–354. doi: 10.1146/annurev.biochem.73.011303.073731
Ju B., Chen B., Zhang X., Han C., Jiang A. (2014). Purification and Characterization of Bioactive Compounds From Styela clava. J. Chem. doi: 10.1155/2014/525141
Kim S. M. (2011). Antioxidant and Anticancer Activities of Enzymatic Hydrolysates of Solitary Tunicate (Styela Clava). Food Sci. Biotechnol. 201075. doi: 10.1007/s10068-011-0146-y
Ko S.-C., Kim D. G., Han C.-H., Lee Y. J., Lee J.-K., Byun H.-G., et al. (2012). Nitric Oxide-Mediated Vasorelaxation Effects of Anti-Angiotensin I-Converting Enzyme (ACE) Peptide From Styela Clava Flesh Tissue and Its Anti-Hypertensive Effect in Spontaneously Hypertensive Rats. Food Chem. 134, 1141–1145. doi: 10.1016/j.foodchem.2012.02.210
Kon-Ya K., Shimidzu N., Adachi K., Miki W. (1994). 2, 5, 6-Tribromo-L-Methylgramine, an Antifouling Substance From the Marine Bryozoan Zoobotryon Pellucidum. Fish. Sci. 60, 773–775. doi: 10.2331/fishsci.60.773
Lambert G. (2009). Adventures of a Sea Squirt Sleuth: Unraveling the Identity of Didemnum Vexillum, a Global Ascidian Invader. Aquat. Invasions 4, 5–28. doi: 10.3391/ai.2009.4.1.2
Lambert G. (2019). Fouling Ascidians (Chordata: Ascidiacea) of the Galápagos: Santa Cruz and Baltra Islands. Aquat. Invasions 14, 132–149. doi: 10.3391/ai.2019.14.1.05
Lambert C. C., Lambert G. (1998). Non-Indigenous Ascidians in Southern California Harbors and Marinas. Mar. Biol. 130, 675–688. doi: 10.1007/s002270050289
Lambert C. C., Lambert G. (2003). Persistence and Differential Distribution of Nonindigenous Ascidians in Harbors of the Southern California Bight. Mar. Ecol. Prog. Ser. 259, 145–161. doi: 10.3354/meps259145
Lauzon R. J., Brown C., Kerr L., Tiozzo S. (2013). Phagocyte Dynamics in a Highly Regenerative Urochordate: Insights Into Development and Host Defense. Dev. Biol. 374, 357–373. doi: 10.1016/j.ydbio.2012.11.006
Lee T., Shin S. (2021). First Record of Colonial Ascidian, Botrylloides Diegensis Ritter and Forsyt(Ascidiacea, Stolidobranchia, Styelidae), in South Korea. Water 13, 2164. doi: 10.3390/w13162164
Lengyel N. L., Collie J. S., Valentine P. C. (2009). The Invasive Colonial Ascidian Didemnum Vexillum on Georges Bank-Ecological Effects and Genetic Identification. Aquat. Invasions 4, 143–152. doi: 10.3391/ai.2009.4.1.15
Levsen K., Schiebel H. M., Terlouw J. K., Jobst K. J., Elend M., Preiss A., et al. (2007). Even-Electron Ions: A Systematic Study of the Neutral Species Lost in the Dissociation of Quasi-Molecular Ions. J. Mass Spectrom. 42, 1024–1044. doi: 10.1002/jms.1234
Manríquez P. H., Castilla J. C. (2007). Roles of Larval Behaviour and Microhabitat Traits in Determining Spatial Aggregations in the Ascidian Pyura Chilensis. Mar. Ecol. Prog. Ser. 332, 155–165. doi: 10.3354/meps332155
Mastrototaro F., Montesanto F., Salonna M., Grieco F., Trainito E., Chimienti G., et al. (2019). Hitch-Hikers of the Sea: Concurrent Morphological and Molecular Identification of Symplegma Brakenhielmi (Tunicata: Ascidiacea) in the Western Mediterranean Sea. Mediterr. Mar. Sci. 20, 197–207. doi: 10.12681/mms.19390
Mccauley E. P., Piña I. C., Thompson A. D., Bashir K., Weinberg M., Kurz S. L., et al. (2020). Highlights of Marine Natural Products Having Parallel Scaffolds Found From Marine-Derived Bacteria, Sponges, and Tunicates. J. Antibiotics 73, 504–525. doi: 10.1038/s41429-020-0330-5
Merrill (2011). Sphingolipid and Glycosphingolipid Metabolic Pathways in the Era of Sphingolipidomics. Chem. Rev. 111, 6387–6422. doi: 10.1021/cr2002917
Mikami N., Hosokawa M., Miyashita K. (2010). Effects of Sea Squirt (Halocynthia Roretzi) Lipids on White Adipose Tissue Weight and Blood Glucose in Diabetic/Obese KK-Ay Mice. Mol. Med. Rep. 3, 449–453. doi: 10.3892/mmr_00000278
Miller M., Pfeiffer W. T., Schwartz T. (2010). Creating the CIPRES Science Gateway for Inference of Large Phylogenetic Trees.
Minchin D., Sides E. (2006). Appearance of a Cryptogenic Tunicate, A Didemnum Sp. Fouling Marina Pontoons and Leisure Craft in Ireland. Aquat. Invasions 1, 143–147. doi: 10.3391/ai.2006.1.3.8
Morales A., Lee H., Goñi F. M., Kolesnick R., Fernandez-Checa J. C. (2007). Sphingolipids and Cell Death. Apoptosis 12, 923–939. doi: 10.1007/s10495-007-0721-0
Morgulis A., Coulouris G., Raytselis Y., Madden T. L., Agarwala R., Schäffer A. A. (2008). Database Indexing for Production MegaBLAST Searches. Bioinformatics 24, 1757–1764. doi: 10.1093/bioinformatics/btn322
Nathani N. M., Mootapally C., Gadhvi I. R., Maitreya B., Joshi C. G (2020). Marine Niche: Applications in Pharmaceutical Sciences: Translational Research. Springer Nature. doi: 10.1007/978-981-15-5017-1
Nothias L.-F., Petras D., Schmid R., Dührkop K., Rainer J., Sarvepalli A., et al. (2020). Feature-Based Molecular Networking in the GNPS Analysis Environment. Nat. Methods 17, 905–908. doi: 10.1038/s41592-020-0933-6
Nydam M. L., Lemmon A. R., Cherry J. R., Kortyna M. L., Clancy D. L., Hernandez C., et al. (2021). Phylogenomic and Morphological Relationships Among the Botryllid Ascidians (Subphylum Tunicata, Class Ascidiacea, Family Styelidae). Sci. Rep. 11, 1–13. doi: 10.1038/s41598-021-87255-2
Oh K.-S., Kim J.-S., Heu M.-S. (1997). Food Constituents of Edible Ascidians Halocynthia Roretzi and Pyura Michaelseni. Korean J. Food Sci. Technol. 29, 955–962.
Oricchio F. T., Muniz Dias G. (2020). Predation and Competition Interact to Determine Space Monopolization by Non-Indigenous Species in a Sessile Community From the Southwestern Atlantic Ocean. Aquat. Invasions 15. doi: 10.3391/ai.2020.15.1.09
Palanisamy S. K., Rajendran N., Marino A. (2017). Natural Products Diversity of Marine Ascidians (Tunicates; Ascidiacea) and Successful Drugs in Clinical Development. Natural Prod. Bioprospecting 7, 1–111. doi: 10.1007/s13659-016-0115-5
Perez-Portela R., Bishop J., Davis A. R., Turon X. (2009). Phylogeny of the Families Pyuridae and Styelidae (Stolidobranchiata, Ascidiacea) Inferred From Mitochondrial and Nuclear DNA Sequences. Mol. Phylogenet. Evol. 50, 560–570. doi: 10.1016/j.ympev.2008.11.014
Pluskal T., Castillo S., Villar-Briones A., Oresic M. (2010). MZmine 2: Modular Framework for Processing, Visualizing, and Analyzing Mass Spectrometry-Based Molecular Profile Data. BMC Bioinf. 11, 395. doi: 10.1186/1471-2105-11-395
Ponnusamy S., Meyers-Needham M., Senkal C. E., Saddoughi S. A., Sentelle D., Selvam S. P., et al. (2010). Sphingolipids and Cancer: Ceramide and Sphingosine-1-Phosphate in the Regulation of Cell Death and Drug Resistance. Future Oncol. 6, 1603–1624. doi: 10.2217/fon.10.116
Prado M. P. D., Torres Y., Berlinck R., Desiderá C., Sanchez M., Craveiro M., et al. (2004). Effects of Marine Organisms Extracts on Microtubule Integrity and Cell Cycle Progression in Cultured Cells. J. Exp. Mar. Biol. Ecol. 313, 125–137. doi: 10.1016/j.jembe.2004.08.008
Provan J., Booth D., Todd N. P., Beatty G. E., Maggs C. A. (2008). Tracking Biological Invasions in Space and Time: Elucidating the Invasive History of the Green Alga Codium Fragile Using Old DNA. Diversity Distrib. 14, 343–354. doi: 10.1111/j.1472-4642.2007.00420.x
Ramalhosa P., Gestoso I., Rocha R. M., Lambert G., Canning-Clode J. (2021). Ascidian Biodiversity in the Shallow Waters of the Madeira Archipelago: Fouling Studies on Artificial Substrates and New Records. Reg. Stud. Mar. Sci. 43, 101672. doi: 10.1016/j.rsma.2021.101672
Rambaut A. (2018). FigTree V.1.4.4. Available at: http://tree.bio.ed.ac.uk/software/figtree/ (Accessed January 27, 2022).
Rambaut A., Drummond A. J., Xie D., Baele G., Suchard M. A. (2018). Posterior Summarization in Bayesian Phylogenetics Using Tracer 1.7. Syst. Biol. 67, 901. doi: 10.1093/sysbio/syy032
Ramesh C., Tulasi B. R., Raju M., Thakur N., Dufossé L. (2021). Marine Natural Products From Tunicates and Their Associated Microbes. Mar. Drugs 19, 308. doi: 10.3390/md19060308
Reem E., Douek J., Paz G., Katzir G., Rinkevich B. (2017). Phylogenetics, Biogeography and Population Genetics of the Ascidian Botryllus Schlosseri in the Mediterranean Sea and Beyond. Mol. Phylogenet. Evol. 107, 221–231. doi: 10.1016/j.ympev.2016.10.005
Reem E., Douek J., Rinkevich B. (2018). Ambiguities in the Taxonomic Assignment and Species Delineation of Botryllid Ascidians From the Israeli Mediterranean and Other Coastlines. Mitochondrial DNA Part A 29, 1073–1080. doi: 10.1080/24701394.2017.1404047
Rethna Priya E., Ravichandran S., Gobinath T., Tilvi S., Devi S. P. (2019). Functional Characterization of Anti-Cancer Sphingolipids From the Marine Crab Dromia Dehanni. Chem. Phys. Lipids 221, 73–82. doi: 10.1016/j.chemphyslip.2019.03.010
Rinkevich Y., Paz G., Rinkevich B., Reshef R. (2007). Systemic Bud Induction and Retinoic Acid Signaling Underlie Whole Body Regeneration in the Urochordate Botrylloides Leachi. PloS Biol. 5, e71. doi: 10.1371/journal.pbio.0050071
Rinkevich B., Shlemberg Z., Lilker-Levav T., Goren M., Fishelson L. (1993). Life History Characteristics of Botrylloides (Tunicata) Populations in Akko Bay, Mediterranean Coast of Israel. Israel J. Ecol. Evol. 39, 197–212. doi: 10.1080/00212210.1993.10688712
Rinkevich Y., Voskoboynik A., Rosner A., Rabinowitz C., Paz G., Oren M., et al. (2013). Repeated, Long-Term Cycling of Putative Stem Cells Between Niches in a Basal Chordate. Dev. Cell 24, 76–88. doi: 10.1016/j.devcel.2012.11.010
Rocha R. M., Salonna M., Griggio F., Ekins M., Lambert G., Mastrototaro F., et al. (2019). The Power of Combined Molecular and Morphological Analyses for the Genus Botrylloides: Identification of a Potentially Global Invasive Ascidian and Description of a New Species. Syst. Biodivers. 17, 509–526. doi: 10.1080/14772000.2019.1649738
Rocha R. M. D., Zanata T. B., Moreno T. R. (2012). Keys for the Identification of Families and Genera of Atlantic Shallow Water Ascidians. Biota Neotropica 12, 269–303. doi: 10.1590/S1676-06032012000100022
Rubinstein N. D., Feldstein T., Shenkar N., Botero-Castro F., Griggio F., Mastrototaro F., et al. (2013). Deep Sequencing of Mixed Total DNA Without Barcodes Allows Efficient Assembly of Highly Plastic Ascidian Mitochondrial Genomes. Genome Biol. Evol. 5, 1185–1199. doi: 10.1093/gbe/evt081
Ryland J. S. (2015). Gender of the Genus Botrylloides Milne Edward)[Tunicata: Ascidiacea]. Zootaxa 3973, 398–400. doi: 10.11646/zootaxa.3973.2.13
Salonna M., Gasparini F., Huchon D., Montesanto F., Haddas-Sasson M., Ekins M., et al. (2021). An Elongated COI Fragment to Discriminate Botryllid Species and as an Improved Ascidian DNA Barcode. Sci. Rep. 11, 1–19. doi: 10.1038/s41598-021-83127-x
Sato A., Fenical W. (1983). Gramine-Derived Bromo-Alkaloids From the Marine Bryozoan Zoobotryon Verticillatum. Tetrahedron Lett. 24, 481–484. doi: 10.1016/S0040-4039(00)81442-2
Scarpato S., Teta R., Della Sala G., Pawlik J. R., Costantino V., Mangoni A. (2020). New Tricks With an Old Sponge: Feature-Based Molecular Networking Led to Fast Identification of New Stylissamide L From Stylissa Caribica. Mar. Drugs 18, 443. doi: 10.3390/md18090443
Schmelz E. M., Sullards M. C., Dillehay D. L., Merrill J. A. H. (2000). Colonic Cell Proliferation and Aberrant Crypt Foci Formation Are Inhibited by Dairy Glycosphingolipids in 1, 2-Dimethylhydrazine-Treated CF1 Mice. J. Nutr. 130, 522–527. doi: 10.1093/jn/130.3.522
Shaner R. L., Allegood J. C., Park H., Wang E., Kelly S., Haynes C. A., et al. (2009). Quantitative Analysis of Sphingolipids for Lipidomics Using Triple Quadrupole and Quadrupole Linear Ion Trap Mass Spectrometers [s]. J. Lipid Res. 50, 1692–1707. doi: 10.1194/jlr.D800051-JLR200
Sheets E. A., Cohen C. S., Ruiz G. M., Da Rocha R. M. (2016). Investigating the Widespread Introduction of a Tropical Marine Fouling Species. Ecol. Evol. 6, 2453–2471. doi: 10.1002/ece3.2065
Shenkar N., Swalla B. J. (2011). Global Diversity of Ascidiacea. PloS One 6, e20657. doi: 10.1371/journal.pone.0020657
Streit O. T., Lambert G., Erwin P. M., López-Legentil S. (2021). Diversity and Abundance of Native and Non-Native Ascidians in Puerto Rican Harbors and Marinas. Mar. Pollut. Bull. 167, 112262. doi: 10.1016/j.marpolbul.2021.112262
Tanduo V., Osca D., Crocetta F. (2021). A Bycatch Surprise: Scyllarus Subarctus Crosnie(Decapoda: Achelata: Scyllaridae) in the Mediterranean Sea. J. Crustacean Biol. 41, 1–5. doi: 10.1093/jcbiol/ruab010
Teta R., Della Sala G., Esposito G., Stornaiuolo M., Scarpato S., Casazza M., et al. (2021). Monitoring Cyanobacterial Blooms During the COVID-19 Pandemic in Campania, Italy: The Case of Lake Avernus. Toxins 13, 471. doi: 10.3390/toxins13070471
Varandas P. A., Cobb A. J., Segundo M. A., Silva E. M. (2019). Emergent Glycerophospholipid Fluorescent Probes: Synthesis and Applications. Bioconjugate Chem. 31, 417–435. doi: 10.1021/acs.bioconjchem.9b00660
Viard F., Roby C., Turon X., Bouchemousse S., Bishop J. (2019). Cryptic Diversity and Database Errors Challenge Non-Indigenous Species Surveys: An Illustration With Botrylloides Spp. In the English Channel and Mediterranean Sea. Front. Mar. Sci. 6, 615. doi: 10.3389/fmars.2019.00615
Villani G., Martinez E. (1993). Some Observations on the Opisthobranch Fauna From the Fusaro Lake, A Brackish-Water Lagoon Near Naples. Boll. Malacologico 29, 201–209.
Virgili R., Tanduo V., Katsanevakis S., Terlizzi F., Villani G., Fontana A., et al. (2022). The Miseno Lake (Central-Western Mediterranean Sea): An Overlooked Reservoir of Non-Indigenous and Cryptogenic Ascidians in a Marine Reserve. Front. Mar. Sci. 9, 866906. doi: 10.3389/fmars.2022.866906
Voskoboynik A., Weissman I. L. (2015). Botryllus schlosseri, an Emerging Model for the Study of Aging, Stem Cells, and Mechanisms of Regeneration. Invertebr. Reprod. Dev. 59, 33–38. doi: 10.1080/07924259.2014.944673
Watanabe H., Okawara M., Matahira Y., Mano T., Wada T., Suzuki N., et al. (2020). The Impact of Ascidian (Halocynthia Roretzi)-Derived Plasmalogen on Cognitive Function in Healthy Humans: A Randomized, Double-Blind, Placebo-Controlled Trial. J. Oleo Sci. ess20167. doi: 10.5650/jos.ess20167
Watters D. J. (2018). Ascidian Toxins With Potential for Drug Development. Mar. Drugs 16, 162. doi: 10.3390/md16050162
Willan R. C., Russell B. C., Murfet N. B., Moore K. L., Mcennulty F. R., Horner S. K., et al. (2000). Outbreak of Mytilopsis Sallei (Reclu)(Bivalvia: Dreissenidae) in Australia. Molluscan Res. 20, 25–30. doi: 10.1080/13235818.2000.10673730
Woznica A., Cantley A. M., Beemelmanns C., Freinkman E., Clardy J., King N. (2016). Bacterial Lipids Activate, Synergize, and Inhibit a Developmental Switch in Choanoflagellates. Proc. Natl. Acad. Sci. 113, 7894–7899. doi: 10.1073/pnas.1605015113
Wymann M. P., Schneiter R. (2008). Lipid Signalling in Disease. Nat. Rev. Mol. Cell Biol. 9, 162–176. doi: 10.1038/nrm2335
Zhan A., Briski E., Bock D. G., Ghabooli S., Macisaac H. J. (2015). Ascidians as Models for Studying Invasion Success. Mar. Biol. 162, 2449–2470. doi: 10.1007/s00227-015-2734-5
Zhang T.-T., Xu J., Wang Y.-M., Xue C.-H. (2019). Health Benefits of Dietary Marine DHA/EPA-Enriched Glycerophospholipids. Prog. Lipid Res. 75, 100997. doi: 10.1016/j.plipres.2019.100997
Zhan A., Macisaac H. J., Cristescu M. E. (2010). Invasion Genetics of the Ciona Intestinalis Species Complex: From Regional Endemism to Global Homogeneity. Mol. Ecol. 19, 4678–4694. doi: 10.1111/j.1365-294X.2010.04837.x
Zhao Y., Li J. (2016). Ascidian Bioresources: Common and Variant Chemical Compositions and Exploitation Strategy – Examples ofSp. And” Zeitschrift Für Naturforschung C 71. doi: 10.1515/znc-2016-0012
Zhao Y., Wang M., Lindström M. E., Li J. (2015). Fatty Acid and Lipid Profiles With Emphasis on N-3 Fatty Acids and Phospholipids From Ciona Intestinalis. Lipids 50, 1009–1027. doi: 10.1007/s11745-015-4049-1
Keywords: alien ascidians, fouling communities, lipids, alkaloids, mass spectrometry, molecular networking, natural products
Citation: Della Sala G, Coppola D, Virgili R, Vitale GA, Tanduo V, Teta R, Crocetta F and de Pascale D (2022) Untargeted Metabolomics Yields Insights Into the Lipidome of Botrylloides niger Herdman, 1886, An Ascidian Invading the Mediterranean Sea. Front. Mar. Sci. 9:865751. doi: 10.3389/fmars.2022.865751
Received: 30 January 2022; Accepted: 07 March 2022;
Published: 04 April 2022.
Edited by:
Francesco Tiralongo, University of Catania, ItalyReviewed by:
Tiziana Cappello, University of Messina, ItalyAgustinus R. Uria, Hokkaido University, Japan
Copyright © 2022 Della Sala, Coppola, Virgili, Vitale, Tanduo, Teta, Crocetta and de Pascale. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Gerardo Della Sala, Z2VyYXJkby5kZWxsYXNhbGFAc3puLml0
†These authors have contributed equally to this work and share first authorship
 Gerardo Della Sala
Gerardo Della Sala Daniela Coppola
Daniela Coppola Riccardo Virgili
Riccardo Virgili Giovanni Andrea Vitale
Giovanni Andrea Vitale Valentina Tanduo
Valentina Tanduo Roberta Teta
Roberta Teta Fabio Crocetta
Fabio Crocetta Donatella de Pascale
Donatella de Pascale