- 1Biosecurity Group, Cawthron Institute, Nelson, New Zealand
- 2Institute of Marine Science, University of Auckland, Warkworth, New Zealand
The invasive Mediterranean fanworm Sabella spallanzanii (Gmelin, 1791) is a notifiable organism under New Zealand’s Biosecurity Act and is recognized as a marine pest of particular concern, that must be reported to the Ministry for Primary Industries (MPI), New Zealand. Since its first detection in 2008, great effort and financial resources are put into surveillance and removal of individuals to contain population growth and spread. Sensitive molecular detection techniques gain great interest and are being increasingly tested for the fanworm detection in marine high-risk sites (i.e., ports and marinas) around New Zealand. However, conventional molecular detection via PCR assays from environmental DNA (eDNA) samples requires specific laboratory resources and technical expertise. This restricts the wider applicability of this approach by biosecurity practitioners or communities willing to be engaged in biosecurity surveillance. To provide end-users with a fast, easy and highly specific way to detect S. spallanzanii directly at the site of interest, a species-specific recombinase polymerase amplification (RPA) assay was designed to be read-out with lateral flow strips (RPA-LF). The RPA generates amplification within 20 minutes at 37-39°C, with a detection limit of 10 pg of the target DNA and was matching the detection limit of digital droplet PCR (ddPCR) when performed on eDNA samples. A simplified visual protocol for non-scientist users of the assay was developed and improved through independent trials with different end-user groups. The assay applicability was verified in a final validation trial with participants without scientific background resulting in 50 percent of the participants successfully detecting S. spallanzanii. Participants rated the ease of use and performance and read-out mostly as easy-to-very easy with overall positive written feedback on its usability for citizen science applications.
Introduction
The Mediterranean fanworm Sabella spallanzanii (Gmelin, 1791) is a tube-forming annelid, native to the Mediterranean Sea and Atlantic coast of Europe (Patti and Gambi, 2001). It has successfully invaded Australia in the 1990s (Clapin and Evans, 1995) and was first recorded in New Zealand in 2008 in Lyttelton Harbor (Read et al., 2011). Despite efforts to eradicate S. spallanzanii at the initial incursion area and mitigate its further spread (Fletcher, 2014), soon it was found in multiple harbors across the North and South Islands (Fletcher, 2014). Being a highly effective suspension filter feeder (Clapin, 1996) and an ecosystem engineer (O'Brien et al., 2006), the fanworm can impact abundance and recruitment patterns of invertebrate larvae (O'Brien et al., 2006; Ross et al., 2007) and alter benthic macrofauna and microbial communities adjacent to S. spallanzanii canopies (Atalah et al., 2019). Because of its high invasive potency and adverse ecological impacts, the uncontrolled spread of S. spallanzanii threatens species native to New Zealand and puts local economies at risk. In New Zealand, S. spallanzanii invasions are particularly problematic for the shellfish aquaculture sector. Dense biofouling by the fanworm, competing with mussels for food and space, may cause reduced mussel growth and compromise their attachment, therefore leading to crop losses (Bannister et al., 2019). It is estimated that S. spallanzanii could cause financial losses of roughly 14 Million NZ Dollar to the green shell mussel industry (Soliman and Inglis, 2018).
Therefore, Sabella spallanzanii is listed as a notifiable and unwanted organism in New Zealand under the Biosecurity Act 1993 (Hewitt et al., 2004; Inglis et al., 2005; Read et al., 2011) and is currently subjected to targeted surveillance as part of the national Marine High Risk Site Surveillance (MHRSS) program (Inglis et al., 2005; Wood et al., 2019b). Although the juvenile/adult stage of S. spallanzanii is morphologically distinct from most New Zealand native marine organisms and could be identified easily, when juveniles/adults are abundant in a marina, eradication or population management at this stage of invasion is near to impossible. In most cases, detecting juvenile/adult individuals at early incursion stages is crucial for effective response but very difficult to accomplish via visual surveys from the surface, i.e. when it is part of biofouling on a boat hull or harbor structures. Therefore, to effectively monitor its spread, detect incursions at the earliest instance and mitigate establishment of new populations, bi-annual diver surveys are conducted in New Zealand’s major ports and marinas (Inglis et al., 2005). Visual surveying and removal by divers can be highly efficient with a 80 – 100% detection rate for juvenile/adult individuals (Inglis et al., 2008; Inglis et al., 2009), however smaller individuals and pelagic larvae are difficult to detect and identify without a microscope and expert taxonomist (Giangrande et al., 2000; Floerl et al., 2010). Additionally, organizing and conducting underwater visual surveys are logistically complex, expensive and subject to restrictions like weather and site access and pose health and safety risks to divers in the busy port or marina environments (Bowers et al., 2021). Therefore, biosecurity authorities are interested in alternative, ideally more sensitive and cost-efficient surveillance technologies.
In recent years, molecular techniques have gained increasing popularity for bio-surveillance applications, e.g., community screening using metabarcoding or species-specific detections using targeted assays (Pochon et al., 2017; Wood et al., 2017; Zaiko et al., 2018; Hernandez et al., 2020; Gold et al., 2021). For example, a quantitative real-time polymerase chain reaction (qPCR) (Wood et al., 2017) and digital droplet polymerase chain reaction (ddPCR) assays (Wood et al., 2019a) have been developed and validated for environmental DNA (eDNA) based detection of S. spallanzanii in New Zealand’s coastal environments. These assays, however, require specific laboratory gear and technical expertise, which restricts their wide use and point-of-need applicability by non-scientist users, such as biosecurity practitioners or communities willing to be engaged in biosecurity surveillance. Engaging non-scientists to perform assays for invasive species detections could support biosecurity surveillance efforts in mitigating the establishment and spread of invasive organisms as early as possible.
To address the current need in molecular surveillance techniques that are easy, fast, specific, and usable with minimal equipment and expertise, we have designed a recombinase polymerase amplification (RPA) assay for species-specific S. spallanzanii detection. Recombinase polymerase amplification is an isothermal amplification method, that amplifies target DNA within 20 minutes at a temperature range of 25°C to 45°C (Piepenburg et al., 2006; James and Macdonald, 2015; Daher et al., 2016), which makes it possible to run assays at body temperature (~37°C), i.e., hand-held (Crannell et al., 2014). With the introduction of labelled probes and primer, RPA can be used for fluorometry-based quantitative real time amplification. Detection is also possible with lateral flow (LF) strips, that provide easy visual read-outs (Piepenburg et al., 2006). The combination of RPA and LF (RPA-LF) strip is especially valuable in resource-limited environments. RPA-LF was developed successfully for multiple disease carrying organisms/viruses (Li et al., 2019a), SARS-CoV-2 (Lau et al., 2021) or for invasive species like the Northern Pacific Seastar, Asterias amurensis (Unpublished).
Here we present the results of the successful in silico and in vitro validation of a newly designed RPA-LF assay for the Mediterranean fanworm S. spallanzanii, as well as the trialing of a simplified RPA protocol with different end-user groups.
Materials and Methods
Samples for Recombinase Polymerase Amplification (RPA) Assay Development and Validation
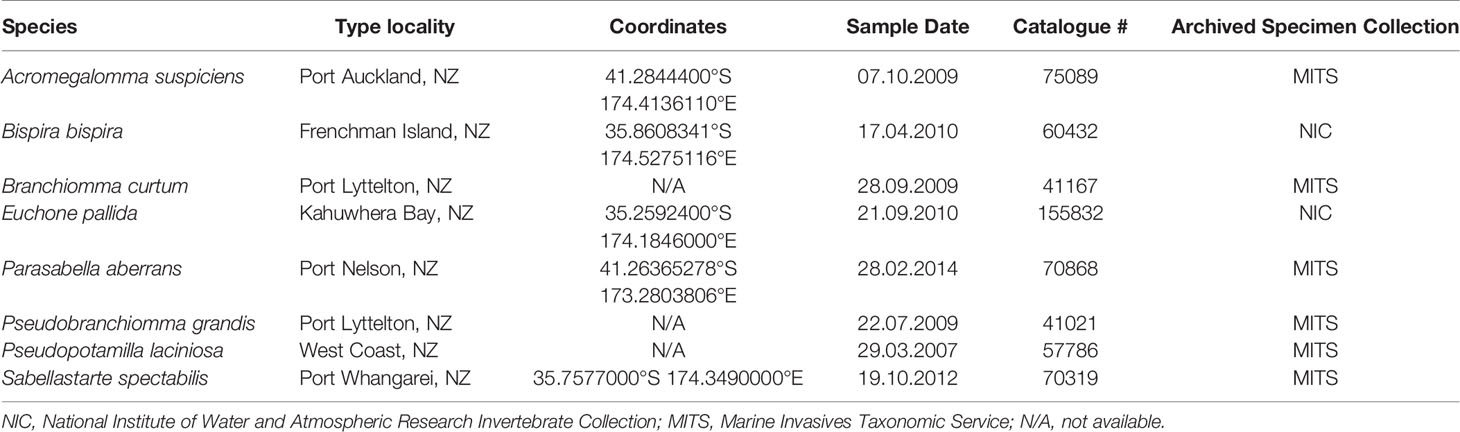
New Zealand native Sabellidae species were obtained from the NIWA Invertebrate Collection, (NIC, Wellington, New Zealand) and the Marine Invasives Taxonomic Service (MITS, Wellington, New Zealand) (Table 1), and S. spallanzanii specimens were collected by commercial divers in Tasman Bay, New Zealand. Genomic DNA was extracted from roughly 25 mg tissue of one sample of each Sabellidae species (Table 1) using the QIAamp ® DNA Mini Kit (Qiagen, Germany) with 12 h Proteinase K digest and following the manufacturer’s instructions. Additionally, five eDNA samples collected in Waitematā Harbor (New Zealand) (Supplementary Table 2) which is known to harbor S. spallanzanii populations were also included to further validate the RPA assay (von Ammon, in review). The sampling sites were a minimum of 1 km apart. We used a previously developed digital droplet PCR assay specific to S. spallanzanii (Wood et al., 2019a) to confirm that S. spallanzanii DNA was either present or absent in the environmental samples in order to verify the results obtained from our RPA assay on those same samples.
Primers and Probes Design
All primers and probes used in this study were designed in Geneious Prime version 2020.2.2 (https://www.geneious.com) following the TwistDxTM (Cambridge, England) guidelines. Recombinase polymerase amplification primers and probe were designed to amplify the mitochondrial Cytochrome C Oxidase subunit 1 (COI) gene of S. spallanzanii based on a multiple sequence alignment previously generated (Wood et al., 2017). For detection on the lateral flow strip, the reverse primer was labelled with biotin at the 5’ end, the LF probe was labelled with 6-FAM (5’) and the polymerase extension blocker C3-spacer (3’) contained an internal abasic tetrahydrofuran (idSp) (Table 2). All primers and probes were checked in silico for cross-reactivity with other species using the Primer-BLAST tool (https://www.ncbi.nlm.nih.gov/tools/primer-blast/) and checked with the OligoAnalyzer™ (IDT) for in situ parameters. All primers and probes were synthesized by Integrated DNA Technologies, Inc. (IDT, Singapore).
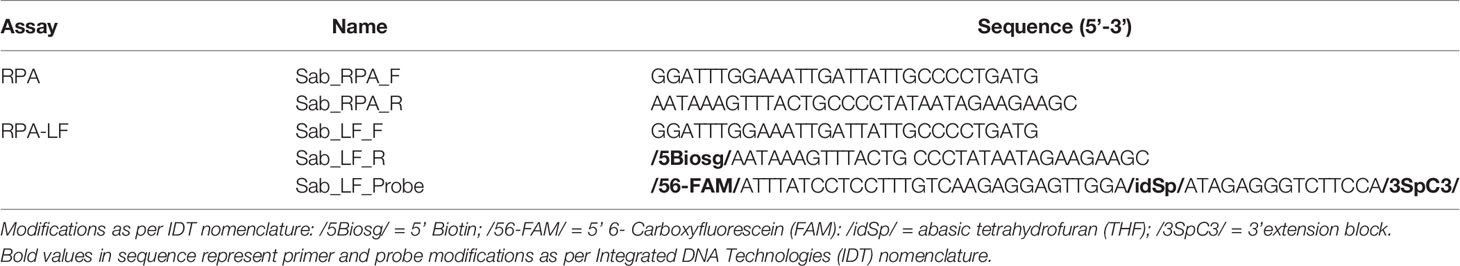
Table 2 Primer and probe sequences targeting Sabella spallanzanii, designed and tested for Recombinase Polymerase Amplification (RPA) and Recombinase Polymerase Amplification with lateral flow read-out (RPA-LF) analyses used in this study.
Recombinase Polymerase Amplification (RPA) and Droplet Digital PCR (ddPCR) Analyses
Testing of RPA primers was carried out using the TwistDxTM TwistAmp® Liquid Basic kit, with reaction volumes adapted to a total reaction volume of 20 μL. Reactions contained 10 μL 2 x reaction buffer, 2.1 μL H2O, 2 μL 10 x basic E-mix, 1 μL 20 x Core Mix, 0.9 μL dNTPs (InvitrogenTM, USA), 0.96 μL primer each (10 μM), 1 μL genomic DNA (2 ng DNA total unless stated otherwise) and 1 μL magnesium acetate to start the reaction. The reaction mixture was incubated at 37°C for 20 minutes. To ensure even distribution of reaction components, the reaction tube was flicked after 5 minutes. Amplified products were cleaned up using QIAquick® PCR Purification Kit (Qiagen, Germany) to remove any interference of RPA proteins in the electrophoresis process. The products were visualized on 1.5% agarose gel, using RedSafe™ Nucleic Acid Staining Solution (iNtRON Biotechnology, Korea).
Recombinase Polymerase Amplification Lateral Flow (RPA-LF) Reaction Mixture
Due to unavailability of the TwistDxTM Nfo kit (implications of the restricted supplies caused by SARS-CoV-2 pandemic), Endonuclease IV (EN0591, Thermofisher, USA) was supplemented into the Basic mixture to replicate the functionality of the TwistDxTM Nfo kit. We tested different concentrations of Endonuclease IV (0.2 and 0.5 U) and incubation times (5 – 40 min) at 37°C to determine the optimal reaction setup (2 ng total S. spallanzanii DNA per reaction). The resulting optimized RPA-LF reaction was: 10 μL 2x Reaction buffer, 2.1 μL H2O, 2 μL 10x basic E-mix, 1 μL 20x Core Mix, 0.9 μL dNTPs (InvitrogenTM, USA), 0.84 μL primer each (10 μM final), 0.24 μL Probe (10 μM), 0.5 U Endonuclease IV (0.25 μL of Endonuclease IV), 1 μL genomic DNA and 1 μL magnesium acetate (280mM stock concentration) to start the reaction. The reaction mixture was incubated at 37°C for 20 minutes and subsequently heat treated (5 minutes 90°C) to stop the reaction. To further increase sensitivity, we used 3 μL template (excluded H2O in the mixture) and incubated the reaction for 40 minutes (for amplification of low target DNA concentration e.g., environmental samples).
For detection of RPA products on the Milenia HybriDetect 2T lateral flow strips (Milenia Biotec GmbH, Germany), the RPA reactions were diluted 1:5 with HybriDetect 2T Assay Buffer and 10 µL of the diluted mixture was applied to the sample area. The LF strip was placed vertically into a test tube filled with 80 µL running buffer. Results were read after 5 - 10 minutes (longer incubation increases strength and clarity of bands), but within a maximum of 24h. The amplification was regarded as successful when a purple band was visible in the test-line. The run was only regarded as valid, when a purple band in the control lane (CTRL-lane) was visible, as this ensured proper functionality of the test strip itself. Assays were run repeatedly (min. 3 times) to test for consistency.
Assay Specificity
The specificity of the RPA-LF assay was tested with DNA from Sabellidae species native to New Zealand and closely related to S. spallanzanii (Table 1), using the above-described RPA-LF reaction mixture. Additionally, the environmental samples A – D containing S. spallanzanii (Supplementary Table 2) were amplified with the RPA primers Sab_RPA_F and Sab_RPA_R (Table 2). Successful amplification was confirmed using agarose gel electrophoresis (1.5% agarose gel with RedSafe™ Nucleic Acid Staining Solution [iNtRON Biotechnology, Korea]). Reactions were purified using QIAquick® PCR Purification Kit (Qiagen, Germany). Purified amplification products were bidirectionally sequenced using an ABI 3730xl DNA Analyzer at the University of Otago Genetic Analysis Services. The sequencing reads were aligned in Geneious Prime version 2020.2.2 (https://www.geneious.com). The consensus sequence of the alignment was checked against the GenBank reference database (Nucleotide collection) using the Nucleotide BLAST tool (https://blast.ncbi.nlm.nih.gov/Blast.cgi).
Limits of Detection
To assess the limit of detection, the RPA-LF assay was run on a 10-fold dilution series of S. spallanzanii DNA ranging from 1 ng to 100 fg of total DNA per reaction (0.05 ng/μl and 5 fg/μl respectively). The limit of detection was determined as the lowest concentration of S. spallanzanii DNA that produced a visible band on the lateral flow strip.
Comparative Sensitivity Assessment With the Droplet Digital PCR (ddPCR)
A ddPCR analysis specifically targeting S. spallanzanii was performed as described in (Wood et al., 2019a) for comparative assay sensitivity assessment. Briefly, the reaction mixture contained 1 × BioRad ddPCR Supermix for probes (no UDP), 450 nM of each primer and probe (Sab3F, Sab3R, Taqman probe Sab3: Wood et al., 2017), 1 μl environmental DNA and sterile water at a total volume of 21 μL. After droplet generation in a BioRad QX200 droplet generator, the reactions were amplified at 95°C for 10 min (initial denaturation), followed by 40 cycles of 94°C for 30 s, 60°C for 1 min, and a final enzyme deactivation step at 98°C for 10 min. Positive droplets were quantified using a QX200 droplet reader using the “Direct quantification” setting.
Assay Applicability Assessment by Different User Groups
To assess the wider usability of our RPA assay, we designed a simplified step-by-step protocol focused on minimizing steps that might pose difficulties to people with minimal or no background in molecular biology and laboratory procedures. This protocol was tested in two preliminary trials by groups of 5 (Preliminary Trial 1) and 4 (Preliminary Trial 2) volunteer participants, representing different educational backgrounds and occupations to improve the protocol. A final validation trial with 10 participants was performed using the improved protocol and its usability was assessed based on the final trial.
Before each trial, all participants were briefed on the protocol with a quick demonstration of how to use micropipettes. During the experiment, participants were asked to individually follow the protocol, running blinded RPA tests on 4 standardized sample types spiked with S. spallanzanii DNA to a final concentration of: 0 ng/μL [negative sample (neg)], 2 ng/μL [low concentration (low)], 20 ng/μL [medium concentration (mid)], and 200 ng/μL (high concentration [high]. Due to limited availability of necessary material at the time, participants did not perform replicate runs of each sample. Upon completion of the tests, participants were asked to report detection/non-detection of the target, based on their LF strip read-outs, and return their results in a post-experiment survey including their opinion on the usability of the assay and other suggestions on how to improve the protocol (Supplementary File “Survey and Result Sheet”). The involvement of voluntary participants for the validation trials and the survey were approved by the Cawthron Institute Ethics board (Approval number: CAW-ETH-210427). During preliminary trials (i.e., Trial 1 and 2), at least two molecular scientists were present to observe if participants struggled and supported the participants to proceed. The observations of the scientists and feedback from the participants from Preliminary Trial 1 were used to adjust the protocol before Preliminary Trial 2. Our observations and the participants feedback from Preliminary Trial 2 were in turn used to adjust the protocol before the Validation trial. During the final validation trial, no help was provided to the participants. Due to the fact that we assisted some of participants in preliminary trials 1 and 2, we decided to only use the survey and results from the final validation trial to draw conclusions on the usability of this assay as a point-of-need tool.
Results
Design and Validation of the Recombinase Polymerase Amplification With Lateral Flow Read-Out
Of the initially designed eight forward and eight reverse primers targeting the COI gene region, only six primer combinations (1 – 6) – with acceptable in silico primer parameters, checked with the IDT OligoAnalyzer™ tool – and the existing S. spallanzanii qPCR/ddPCR primers (7) (Wood et al., 2017) were chosen for further testing (Supplementary Table 1). All combinations of newly designed RPA primers produced a singular band (lane 1 – 6) corresponding to the expected length (Figure 1). The diffuse bands in the negative control are attributed to primer dimers. The existing qPCR/ddPCR primers (lane 7) produced multiple weak bands and were not considered further. Primer combination 3 (lane 3) (Figure 1, Supplementary Table 1) performed best and was chosen for the development of the RPA-LF and modified to be compatible with lateral flow read-out and an appropriate probe was designed (Table 2)
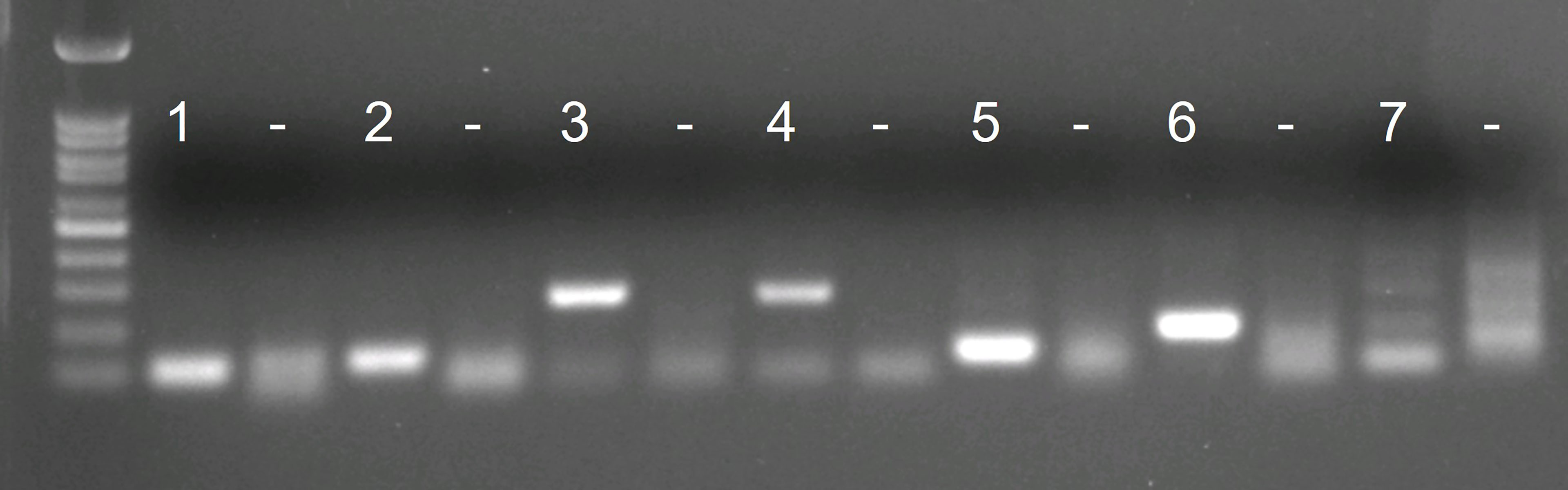
Figure 1 Agarose gel visualizations of the in vitro tests of 7 initial primer combinations. Each primer combination was tested with 2 ng of Sabella spallanzanii DNA and with water as negative control (“–”).
Different Endonuclease IV concentrations and amplification times were tested. The RPA-LF reactions were affected by the concentration of Endonuclease IV. Both tested concentrations (0.2 and 0.5 U) resulted in amplification after 20 minutes at 37°C, however the reaction containing 0.5 U produced a slightly stronger band (Figure 2A). We also observed a clear dependence of the band strength on the incubation time. A band was visible on the flow stick after 10 minutes of amplification time. The band strength increased with increasing amplification time, with maximum band strength after 40 minutes (Figure 2B).
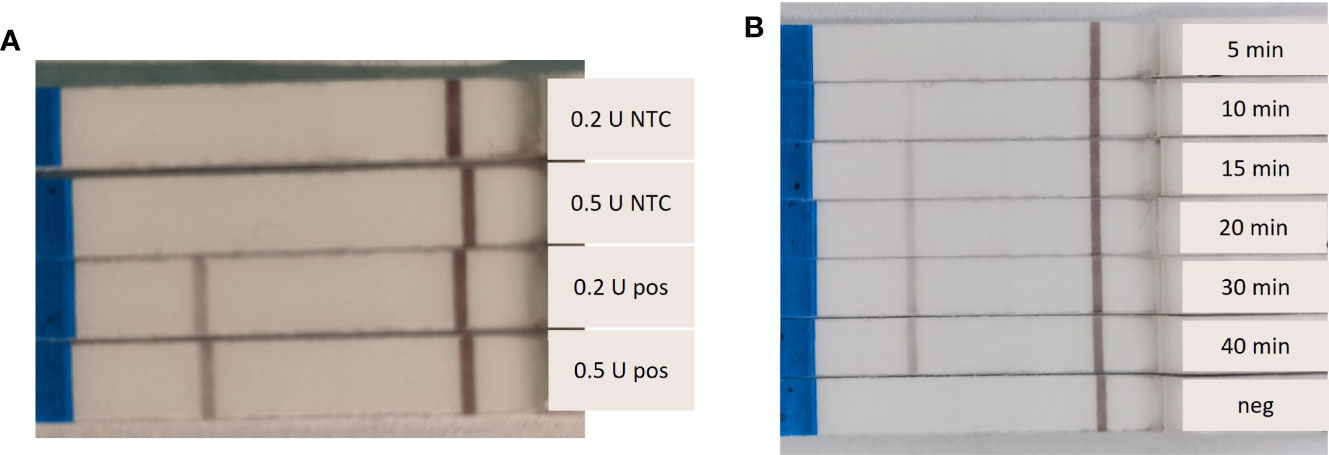
Figure 2 Dependence of the Recombinase Polymorphism Amplification-Lateral Flow (RPA-LF) amplification strength on Endonuclease IV concentration and incubation time. (A) Endonuclease IV was tested at 0.2 U and 0.5 U per reaction with (pos) and without (NTC) Sabella spallanzanii DNA. (B) amplification of 5 – 40 minutes were tested, including a negative sample (neg).
Species Specificity and Limit of Detection
Out of all tested Sabellidae species only S. spallanzanii was amplified by RPA-LF, suggesting high specificity of the designed assay (Figure 3). No false positive result in the negative control sample was detected either.
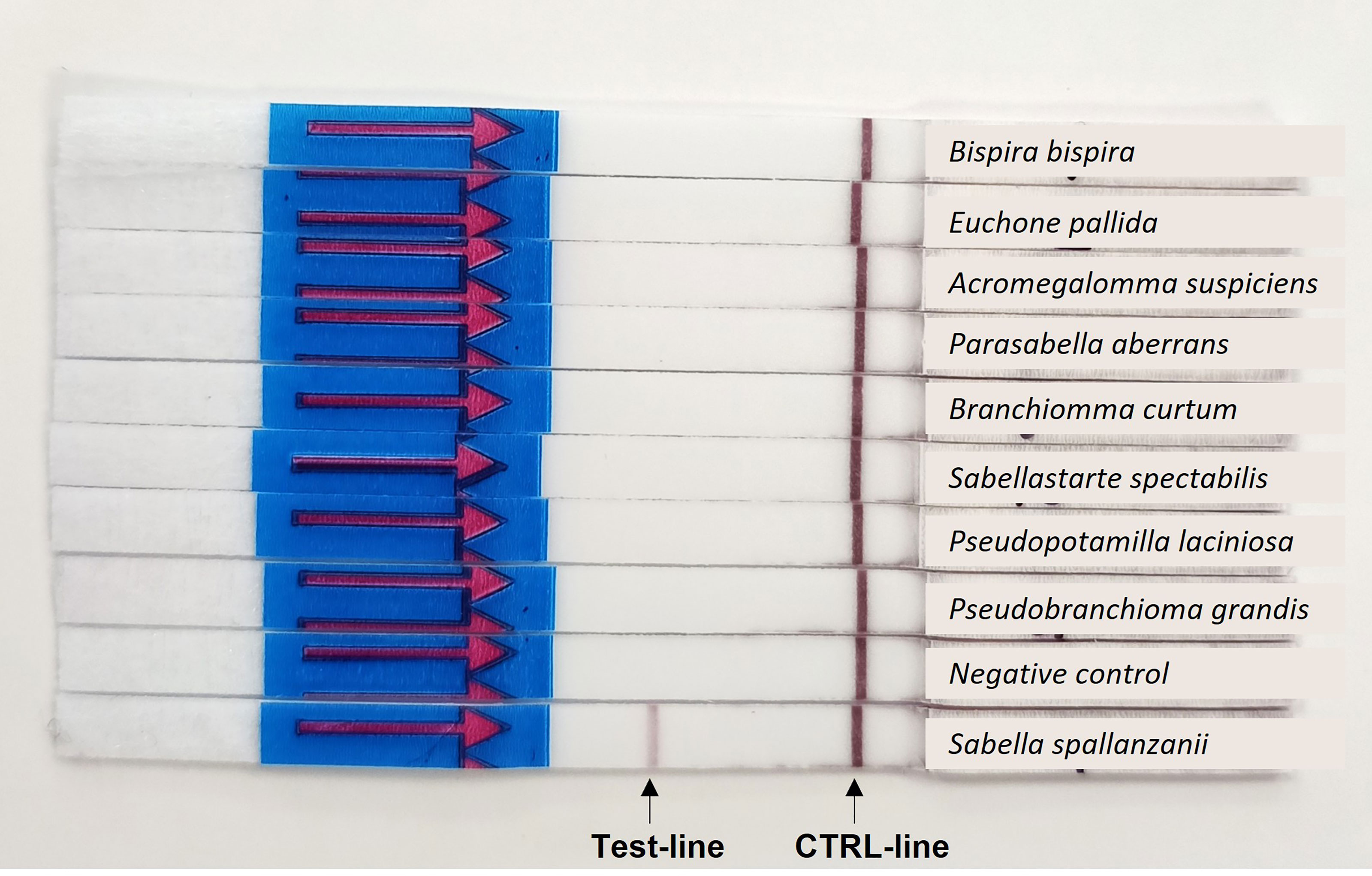
Figure 3 Results of the specificity testing: Positive amplification is indicated by a colored band in the test-line and a colored band in the control (CTRL)-line.
The RPA-LF limit of detection was established after the standard 20 minutes and extended 40 minutes amplifications. The limit of detection for S. spallanzanii DNA, that resulted in a visible band after 20 minutes, was 10 pg (0.5 pg/μL). After 40 minutes amplification time, the lowest concentration that produced a visible band was still 10 pg, however the band was slightly brighter (Figure 4). The ddPCR assay performed on the same dilution series, consistently produced positive signals at the 100 fg concentration but also returned a positive result in the 1 fg sample for one out of the two runs (Supplementary Figure 1).
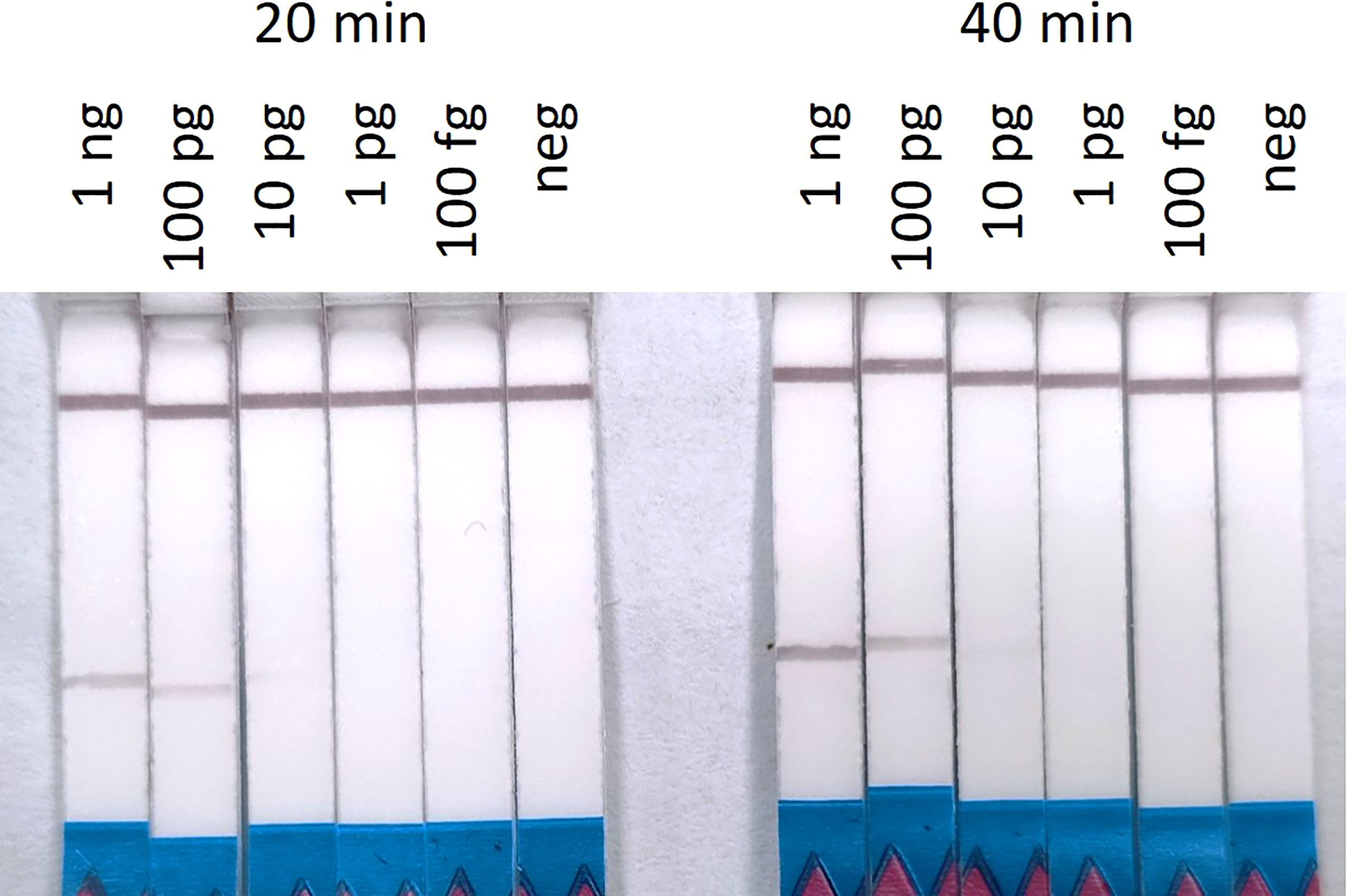
Figure 4 Recombinase Polymorphism Amplification-Lateral Flow (RPA-LF) assay on 10-fold serial dilution of Sabella spallanzanii ranging from 1 ng to 100 fg after 20 min and 40 min of amplification. Positive signals could be seen down to 10 pg of DNA.
Validation on Environmental Samples
When tested on five environmental samples (Supplementary Table 2), the RPA-LF assay was able to detect S. spallanzanii at ddPCR detection signals as low as 0.49 copies/µL, producing overall consistent results with the ddPCR assay. Only the lowest reported concentration (sample “A”, 0.06 copies/µL) failed to produce a visible band on the flow strip. Sample “E” was negative for both the ddPCR assay and the RPA-LF (Figure 5).
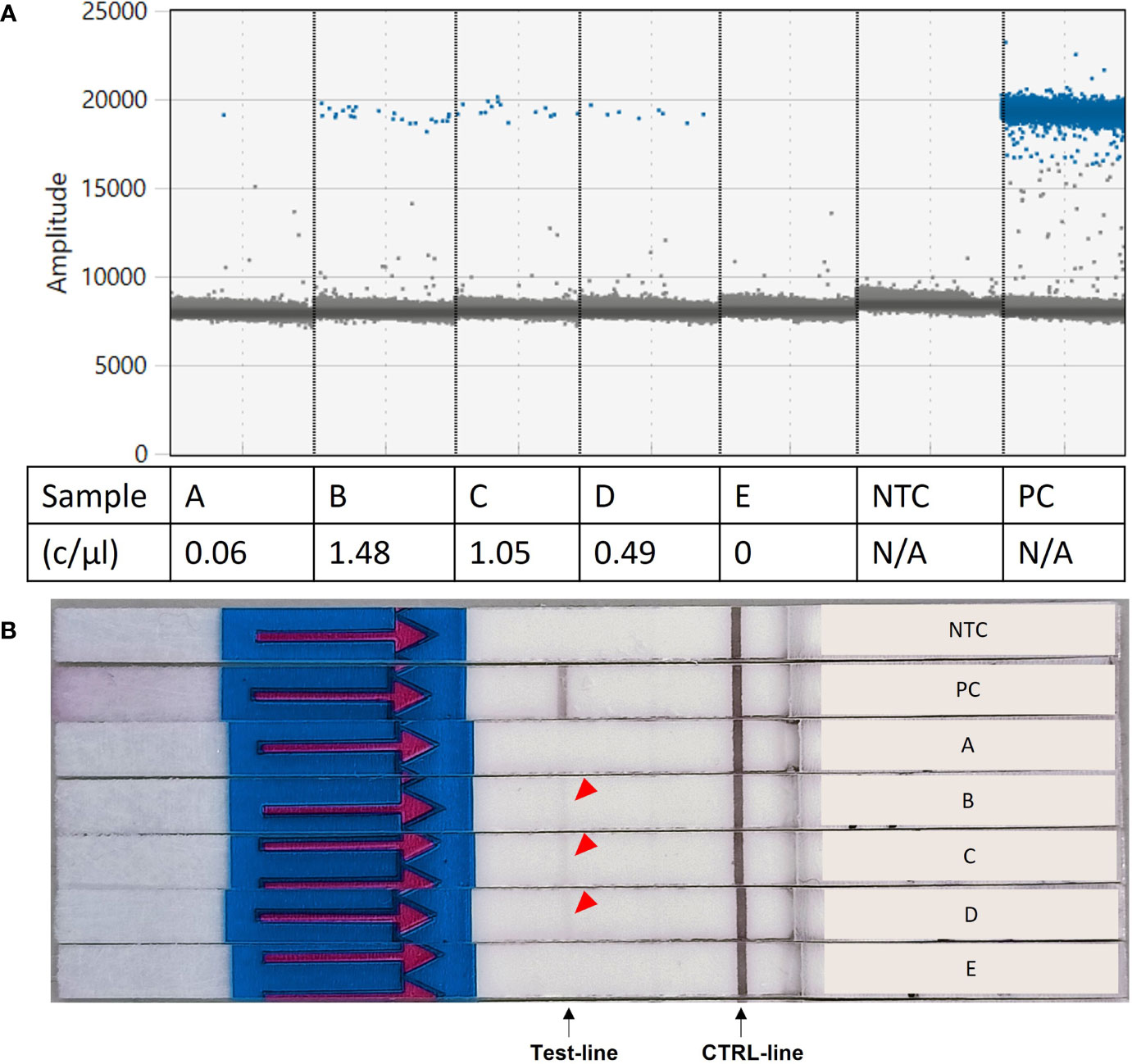
Figure 5 Results of the Recombinase Polymorphism Amplification-Lateral Flow (RPA-LF) assay test on environmental samples. (A) Environmental samples tested for Sabella spallanzanii using digital droplet PCR and reported concentrations. (B) Results of the RPA-LF assay performed on the same environmental samples, showing amplification for samples B–D (red arrow). c/mL,copies per mL; NTC, no template control; PC, positive control; N/A, not applicable.
The Nucleotide BLAST of the amplified environmental samples returned the S. spallanzanii COI region as 100% sequence identity and query cover with an e-value of 2e-100 (highest hit: MN184708.1, Supplementary Table 3). Two other species matched the submitted consensus sequences with similar value were Sabella pavonina (Sequence identity: 100%; Query cover: 100.00%; e-value: 2e-100) and Hermodice carunulata (Sequence identity: 99.02%; Query cover: 100.00%; e-value: 4e-97, GenBank Accession number: AY495947.1).
Assay Trials With Different User Groups
The participants of the preliminary assay trials and the validation trial represented different groups of potential users: biosecurity practitioners, scientists, high-school students, indigenous communities, and public representatives. None of the nine preliminary trial participants had molecular laboratory background. Five participants reported some experience operating a micropipette at least once in the past. Similarly, none of the participants in the validation trial had a background in molecular biology and only 2 out of 10 participants reported having a scientific background and used pipettes before (Figure 6). In the post-experiment surveys of the preliminary trials, participants mainly suggested to include a section about materials and an induction on micropipette usage. The issue regarding micropipette usage was consistent with our observations during the preliminary trials; therefore, we incorporated all observations and feedback from the participants into the final version of the simplified protocol (Supplementary Figure 2) and included a pipetting guide (Supplementary Figure 3) and provided these updated guides to the participants of the validation trial.
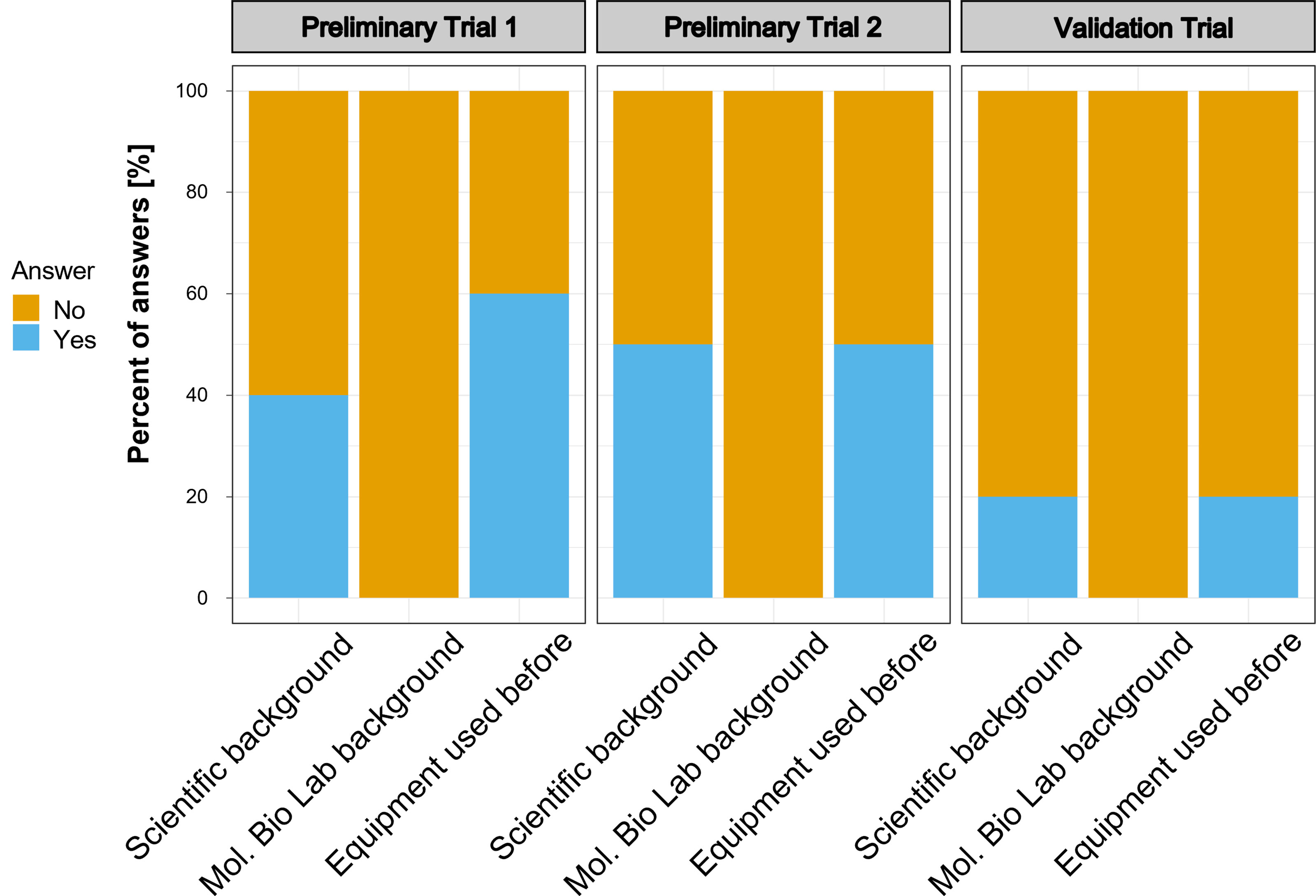
Figure 6 Background of the participants of the trial studies. Number of participants in the trials were as follows: Preliminary Trial 1 – n=4; Preliminary Trial 2 – n = 5; Validation Trial – n=10.
The assay results reported by the participants of the validation trial showed that three participants detected a positive band in the negative control. Also, two participants reported no positive band in any sample (Figure 7A). Looking only at valid sets – by removing the five participants, that had either no band at all or a band in the negative control – all participants detected S. spallanzanii in the ‘high’ concentration range. Two out of 5 did not detect S. spallanzanii in the ‘low’ sample and only one out of 5 did not detect it in the ‘mid’ concentration range (Figure 7B).
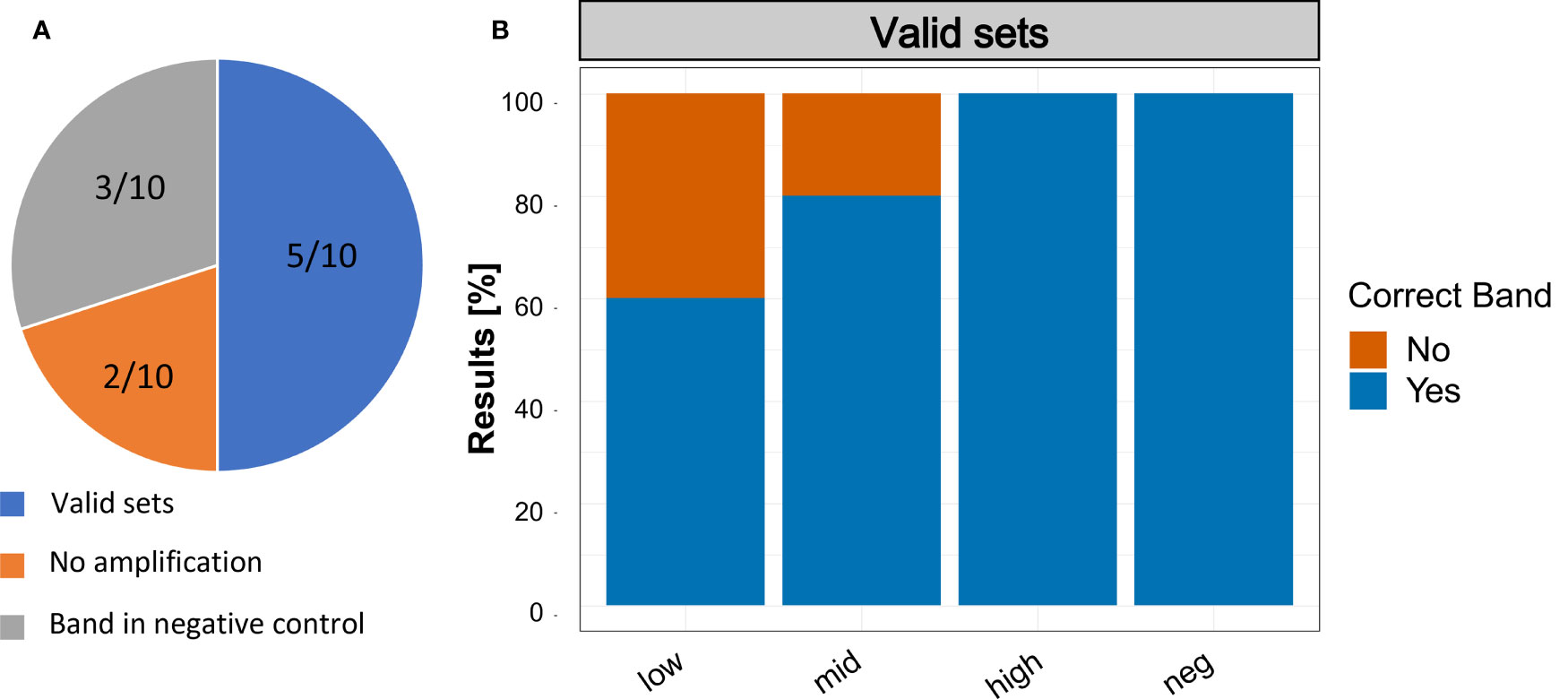
Figure 7 Results of the Recombinase Polymerase Amplification trials with non-scientist users. (A) Summary of the results reported by the participants. Valid sets: Participants that had no band in the negative control and a band in at least one of the three samples with Sabella spallanzanii DNA; No Amplification: Participants that did not report an amplification in any of the samples; Band in negative control: Participants that reported a band in the negative control. (B) Reported results of participants with ‘Valid sets’.
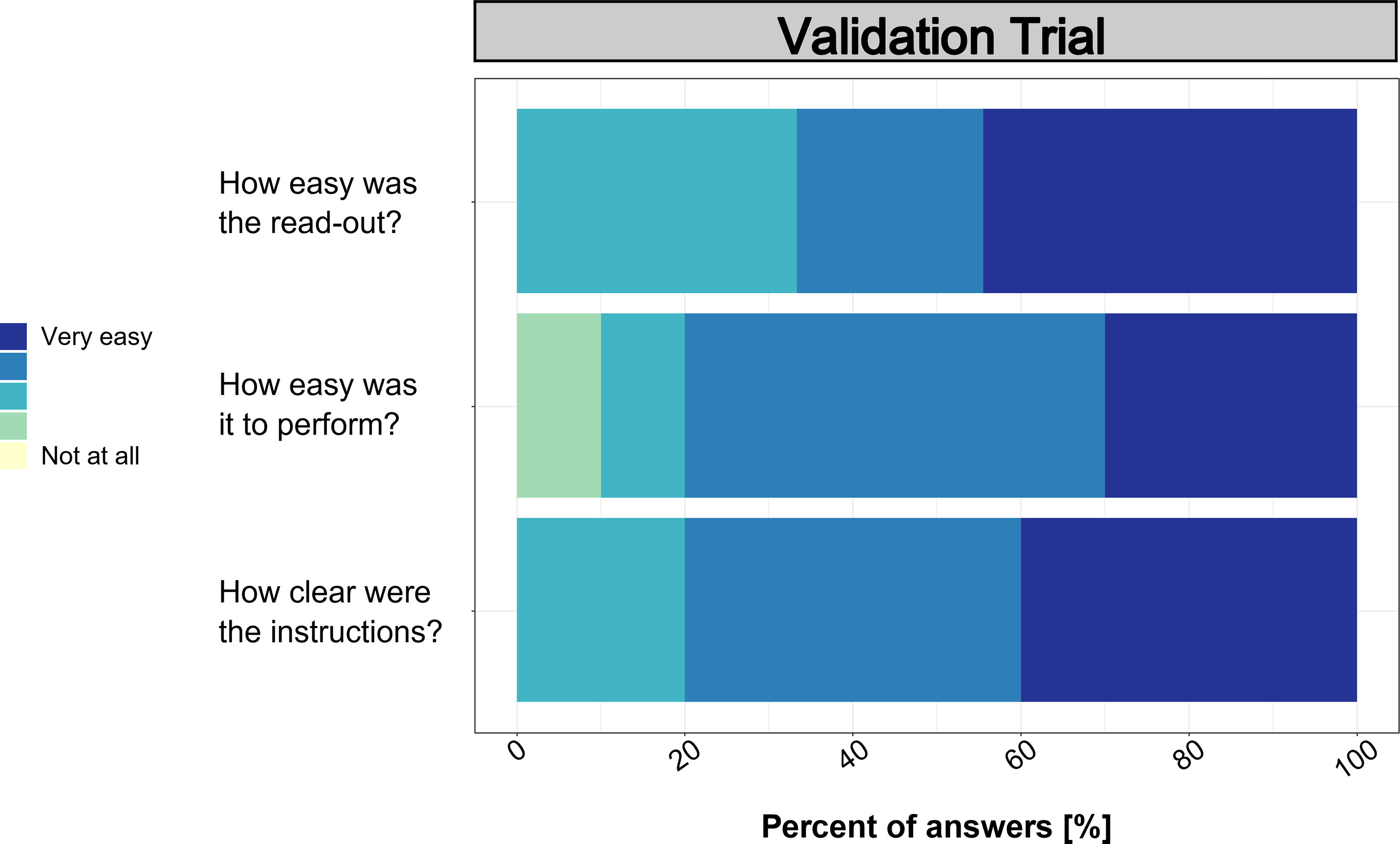
Based on the useability assessment performed after the validation trial (Figure 8), the clarity and ease of use values ranged mainly between 3 and 5 (i.e., medium to very easy/clear). Only one participant evaluated the ease of use with a 2 (not easy). However, written feedback was mainly positive and suggested that the procedure was easy and clear to follow. Still, pipetting was mentioned by multiple participants as difficult as well as the feedback to include a video description.
Discussion
In this study we developed and validated an RPA-LF assay for detecting the Mediterranean fanworm Sabella spallanzanii. Our assay showed no cross-reactivity with other Sabellidae species native to New Zealand. When we sequenced the amplification product obtained from the RPA reaction run on environmental samples and checked the sequence against GenBank using BLAST our top match was indeed S. spallanzanii. However, one closely matching polychaete species, Sabella pavonina could be amplified with our assay but is not reported to occur in New Zealand. The other close match – AY495947.1 – which was published as H. carunculata on GenBank, was later found to be a sample mix-up and indeed corresponded to S. spallanzanii and not H. carunculata (Ahyong et al., 2017). Therefore, we consider that the current assay can be robustly applied for targeted S. spallanzanii detection in New Zealand marine ecosystems but might require additional cross-reactivity validation when intended for other regions where S. pavonina might be present.
The detection limit established for our RPA-LF was 10 pg per reaction, which is higher than those reported for other published RPA-LF assays (Li et al., 2019b; Ravindran et al., 2020). We attribute this variation to the replicated reaction mixture used in this study due to unavailability of the standard RPA (TwistDx) kit. The detection limit of the newly developed RPA (10 pg/reaction) is also higher than the existing ddPCR assay for S. spallanzanii with a detection limit of 100 fg per reaction. However, when tested on environmental samples, RPA-LF consistently detected S. spallanzanii even at concentrations close to the ddPCR detection limit and failed to amplify only one ambiguously positive sample, which returned a single droplet above the amplitude threshold in the ddPCR assay. The RPA-LF assay has a higher limit of detection compared to the ddPCR assay when run on the dilution series of pure S. spallanzanii DNA but has comparable sensitivity when run on environmental samples. Considering, that detection of S. spallanzanii from eDNA is the main intended application, we conclude that the detection limit of the RPA-assay is acceptable.
In order to use the RPA-LF assay for ‘point-of-need’ applications by a wide range of potential end-users, the execution of the assay needs to be easy enough for people with no or limited laboratory background, while ensuring robust and reliable results (Larson et al., 2020; Bowers et al., 2021). We found that our visually designed step-by-step protocol was generally easy to follow by participants. One of the biggest hurdles was the use of micropipettes by inexperienced participants, which we tried to overcome by adding a visual explanation on pipette usage in the final version of the protocol. However, to make it accessible to citizens of any background, one needs to find ways around micropipettes e.g. by using so called Exact Volume transfer pipettes – disposable Pasteur pipettes that can be used to transfer a specific amount of liquid. Overall, based on the trial results and feedback provided by the participants, we can conclude that our protocol and RPA-LF assay has the potential to be used as a ‘point-of-need’ surveillance tool, e.g. for regional councils or similar institutions that already have pipettes and some scientific background.
Half of the ten participants were able to detect S. spallanzanii in at least one of the samples, while having no band in the negative control. Among those five, three participants reported all three samples containing S. spallanzanii as positive. However, three participants reported a band in the negative, and two did not report a band in any of the samples. Although the results that participants reported were overall promising, we do acknowledge that a 50% failure rate is rather high and further adjustments to the protocol or a more extensive training of volunteers is warranted. A bigger test group and replication to assess the consistency of the results might be also recommended. Providing a video instruction could give additional visual aid for participants and could be implemented as a standardized training resource for future users. Also, very faint bands, such as seen in the test on environmental samples (Figure 5) might pose difficulties for untrained users and could lead to false negative reports. This user-bias might need to be offset by higher sampling effort and replication.
False positive results in eDNA-based assays represent a major issue for biosecurity surveillance, as false detections may trigger unnecessary response measures and compromise the credibility of molecular detection techniques for routine monitoring (Sepulveda et al., 2020; Jerde, 2021). False positive results can occur due to a lack of specificity or cross-contamination between samples (Ficetola et al., 2016). It is quite common that undesired amplification of negative controls happens in eDNA studies conducted by established molecular laboratories (Maruyama et al., 2014; Sepulveda et al., 2019), despite implementation of widely accepted guidelines to minimize/prevent cross-contaminations (Goldberg et al., 2016). To rule out false positives in the RPA-LF in a citizen science setting, a positive detection of S. spallanzanii should be further confirmed through qPCR/ddPCR. Although we acknowledge, that further improvements and a higher number of participants would need to be involved to make such assays reliably usable by non-scientists, we believe that our protocol and validation with non-scientists serve as a proof-of-concept that such assays can be run by non-scientists.
The workflow for eDNA-based detection of aquatic pests comprises several critical steps, including sample collection and DNA extraction. Traditionally, both steps require specialized equipment and trained personnel, and are therefore restricted to a very small group of dedicated specialists. To fully harness the utility of the RPA-LF assay as a ‘point-of-need’ tool and for application by non-scientist users, inexpensive, easy-to-use, yet robust and efficient sampling and DNA extraction methods are crucial. Some promising technologies are emerging in that space, that would greatly decrease the complexity of the workflow and make eDNA-based molecular biosecurity surveillance more accessible for a wide range of end-users. For example, the sampling routine can be greatly simplified by using passive sampling devices, that capture eDNA onto a sampling matrix while being deployed in the water (Kirtane et al., 2020; Bessey et al., 2021). As an alternative to time-consuming bulk/plankton-net water sampling and subsequent filtration (von Ammon et al., 2020; Bowers et al., 2021), in situ filtration systems with self-preserving filter packs are now available on the market (Thomas et al., 2019). For the DNA extraction step some promising tools are reported in the literature to be fast, produce high-quality DNA extracts and utilize equipment that can be battery-operated such as the Pretty Damn Quick Extractor (PDQeX, MicroGem), which has been successfully used in low-resource settings for pathogen detection (Boykin et al., 2019). Another commercially available solution, the SpeedXtract kit (Qiagen) has already been coupled with RPA assays (Mondal et al., 2016). Still, their efficiency on different sample types as well as compatibility with different molecular detection assays should be validated, and cost/time efficiency formally assessed through scientific and end-users’ trials.
We anticipate, that easy-to-follow molecular detection tools can be also utilized for educational purposes. Provided they are affordable and easily accessible, those tools could be used in classrooms as a hands-on experience accompanying theoretical knowledge on marine ecology, molecular biology, biodiversity and biosecurity concepts. Showcasing molecular detection assays in classrooms or public seminars as educational components would contribute to raising the public awareness about non-indigenous marine species, and ultimately help establish citizen-led biosecurity surveillance programs that effectively enable the early detection of unwanted invasive species at unprecedented spatial scales.
Conclusion
This study designed, tested and validated a novel RPA-LF assay for detection of the invasive Mediterranean fanworm S. spallanzanii from environmental samples. Levels of detection using the lateral flow stick approach were similar to those afforded by ddPCR diagnostics, but sensitivity could presumably be increased when using our primers with the fit-for-purpose TwistDx Nfo kit. Our simplified protocol is a promising means to make molecular tools more accessible to non-scientist users, although we acknowledge that some basic laboratory training would be beneficial for this purpose. Based on this experience, we recommend an iterative process whereby molecular protocols are first trialed with dedicated end-users to optimize useability and uptake, especially when application by citizen-scientists is anticipated. We believe that the combination of RPA-LF assay and the simplified protocol developed in this study can help minimizing resources and time effort for surveying the invasive Mediterranean fanworm in coastal ecosystems. More importantly, it allows bringing eDNA-based diagnostics tools closer to end-users (environmental managers, biosecurity practitioners or citizen scientists), fast-tracking the implementation of point-of-need tools in biosecurity surveillance programs.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Ethics Statement
The studies involving human participants were reviewed and approved by Cawthron Institute Ethics board (Approval number: CAW-ETH-210427). The patients/ participants provided their written informed consent toparticipate in this study.
Author Contributions
MZ designed and carried out experiments, prepared figures and wrote the manuscript. UA, XP, and AZ provided critical input into experimental design and the manuscript. All authors approved the submitted version.
Funding
This study was supported by New Zealand Ministry of Business, Innovation and Employment funding (CAWX1904 – A toolbox to underpin and enable tomorrow’s marine biosecurity system).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors greatly appreciate the help of all voluntary participants in the validation trials, including Trish Simpson, Carolina G. de Sauza, Ngairo Tahere, Zoltan McComb, Theo Wheatley, Neli Zaiko, Caleb Eason, Louis Rowell Williams, Jessica Verkuil, Krishna Priya Nimmagadda, Jemimah Olive, Richard Meffan and Grace Dowling who wished to be mentioned by name in written outputs. We also thank Aless Smith and Kaeden Leonard from the Northland Regional Council for their help in organizing our first trial with participants. We also thank Dr. Nathan Bott (RMIT) for inspiring discussions and helpful advice on RPA assay development. Figures 7 and 8 were created with BioRender.com.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2022.861657/full#supplementary-material
Supplementary Data Sheet 1 | Supplementary tables and figures.
Supplementary Data Sheet 2 | Survey and result sheet.
References
Ahyong S. T., Kupriyanova E., Burghardt I., Sun Y., Hutchings P. A., Capa M., et al (2017). Phylogeography of the Invasive Mediterranean Fan Worm, Sabella Spallanzanii (Gmelin 1791), in Australia and New Zealand. J. Mar. Biol. Assoc. U. K. 97, 985–991. doi: 10.1017/S0025315417000261
Atalah J., Floerl O., Pochon X., Townsend M., Tait L., Lohrer A. M. (2019). The Introduced Fanworm, Sabella Spallanzanii, Alters Soft Sediment Macrofauna and Bacterial Communities. Front. Ecol. Evol. 7. doi: 10.3389/fevo.2019.00481
Bannister J., Sievers M., Bush F., Bloecher N. (2019). Biofouling in Marine Aquaculture: A Review of Recent Research and Developments. Biofouling 35, 631–648. doi: 10.1080/08927014.2019.1640214
Bessey C., Neil Jarman S., Simpson T., Miller H., Stewart T., Kenneth Keesing J., et al (2021). Passive eDNA Collection Enhances Aquatic Biodiversity Analysis. Commun. Biol. 4, 236. doi: 10.1038/s42003-021-01760-8
Bowers H. A., Pochon X., von Ammon U., Gemmell N., Stanton J.-A. L., Jeunen G.-J., et al (2021). Towards the Optimization of eDNA/eRNA Sampling Technologies for Marine Biosecurity Surveillance. Water 13, 1113. doi: 10.3390/w13081113
Boykin L. M., Sseruwagi P., Alicai T., Ateka E., Mohammed I. U., Stanton J.-A. L., et al (2019). Tree Lab: Portable Genomics for Early Detection of Plant Viruses and Pests in Sub-Saharan Africa. Genes 10, 632. doi: 10.3390/genes10090632
Clapin G. (1996). The Filtration Rate, Oxygen Consumption and Biomass of the Introduced Polychaete Sabella Spallanzanii Gmelin Within Cockburn Sound : Can It Control Phytoplankton Levels and Is It an Efficient Filter Feeder? Available at: https://ro.ecu.edu.au/theses_hons/321.
Clapin G., Evans D. R. (1995). The Status of the Introduced Marine Fanworm Sabella Spallanzanii in Western Australia: A Preliminary Investigation. Hobart, Tas: CSIRO Division of Fisheries.
Crannell Z. A., Rohrman B., Richards-Kortum R. (2014). Equipment-Free Incubation of Recombinase Polymerase Amplification Reactions Using Body Heat. PloS One 9, e112146. doi: 10.1371/journal.pone.0112146
Daher R. K., Stewart G., Boissinot M., Bergeron M. G. (2016). Recombinase Polymerase Amplification for Diagnostic Applications. Clin. Chem. 62, 947–958. doi: 10.1373/clinchem.2015.245829
Ficetola G. F., Taberlet P., Coissac E. (2016). How to Limit False Positives in Environmental DNA and Metabarcoding? Mol. Ecol. Resour. 16, 604–607. doi: 10.1111/1755-0998.12508
Fletcher L. (2014). Background Information on the Mediterranean Fanworm Sabella Spallanzanii to Support Regional Response Decisions (Marlborough District Council). Nelson, New Zealand: Cawthron Institute, 35.
Floerl O., Wilkens S., Woods C. (2010). “Temporal Development of Biofouling Assemblages,” in Report Prepared for the Australian Department of Agriculture, Fisheries and Forestry. Christchurch, New Zealand: NIWA.
Giangrande A., Licciano M., Pagliara P., Gambi M. C. (2000). Gametogenesis and Larval Development in Sabella Spallanzanii (Polychaeta : Sabellidae) From the Mediterranean Sea. Mar. Biol. 136, 847–861. doi: 10.1007/s002279900251
Goldberg C. S., Turner C. R., Deiner K., Klymus K. E., Thomsen P. F., Murphy M. A., et al (2016). Critical Considerations for the Application of Environmental DNA Methods to Detect Aquatic Species. Methods Ecol. Evol. 7, 1299–1307. doi: 10.1111/2041-210X.12595
Gold Z., Sprague J., Kushner D. J., Zerecero Marin E., Barber P. H. (2021). eDNA Metabarcoding as a Biomonitoring Tool for Marine Protected Areas. PloS One 16, e0238557. doi: 10.1371/journal.pone.0238557
Hernandez C., Bougas B., Perreault-Payette A., Simard A., Côté G., Bernatchez L. (2020). 60 Specific eDNA qPCR Assays to Detect Invasive, Threatened, and Exploited Freshwater Vertebrates and Invertebrates in Eastern Canada. Environ. DNA 2, 373–386. doi: 10.1002/edn3.89
Hewitt C. L., Willing J., Bauckham A., Cassidy A. M., Cox C. M. S., Jones L., et al (2004). New Zealand Marine Biosecurity: Delivering Outcomes in a Fluid Environment. N. Z. J. Mar. Freshwater Res. 38, 429–438. doi: 10.1080/00288330.2004.9517250
Inglis G. J., Floerl O., Seaward K., Woods C., Read G., Peacock L. (2009). Sabella Local Elimination Programme - Phase 1. Final Assess. Report Prepared for MAF Biosecurity NZ Research Project MAF 11199 (Christchurch: NIWA).
Inglis G., Hurren H., Gust N., Oldman J., Fitridge I., Floerl O. (2005). Surveillance Design for Early Detection of Unwanted Exotic Marine Organisms in New Zealand, Paper Presented at the Biosecurity New Zealand, New Zealand. Christchurch, New Zealand: NIWA.
Inglis G. J., Woods C., Willis K., Read G., Seaward K. (2008). Incursion Response to the Mediterranean Fanworm, Sabella Spallanzanii (Gmelin, 1791), in the Port Oflyttelton - Interim Measures. NIWA Client Report No: CHC2008141 (New Zealand: MAF-Biosecurity New Zealand).
James A., Macdonald J. (2015). Recombinase Polymerase Amplification: Emergence as a Critical Molecular Technology for Rapid, Low-Resource Diagnostics. Expert Rev. Mol. Diagn. 15, 1475–1489. doi: 10.1586/14737159.2015.1090877
Jerde C. L. (2021). Can We Manage Fisheries With the Inherent Uncertainty From eDNA? J. Fish Biol. 98, 341–353. doi: 10.1111/jfb.14218
Kirtane A., Atkinson J. D., Sassoubre L. (2020). Design and Validation of Passive Environmental DNA Samplers Using Granular Activated Carbon and Montmorillonite Clay. Environ. Sci. Technol. 54, 11961–11970. doi: 10.1021/acs.est.0c01863
Larson E. R., Graham B. M., Achury R., Coon J. J., Daniels M. K., Gambrell D. K., et al (2020). From eDNA to Citizen Science: Emerging Tools for the Early Detection of Invasive Species. Front. Ecol. Environ. 18, 194–202. doi: 10.1002/fee.2162
Lau Y. L., Ismail I. B., Mustapa N., Lai M. Y., Tuan Soh T. S., Haji Hassan A., et al (2021). Development of a Reverse Transcription Recombinase Polymerase Amplification Assay for Rapid and Direct Visual Detection of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2). PloS One 16, e0245164. doi: 10.1371/journal.pone.0245164
Li J., Macdonald J., von Stetten F. (2019a). Review: A Comprehensive Summary of a Decade Development of the Recombinase Polymerase Amplification. Analyst 144, 31–67. doi: 10.1039/C8AN01621F
Li T. T., Wang J. L., Zhang N. Z., Li W. H., Yan H. B., Li L., et al (2019b). Rapid and Visual Detection of Trichinella Spp. Using a Lateral Flow Strip-Based Recombinase Polymerase Amplification (LF-RPA) Assay. Front. Cell Infect. Microbiol. 9, 1. doi: 10.3389/fcimb.2019.00001
Maruyama A., Nakamura K., Yamanaka H., Kondoh M., Minamoto T. (2014). The Release Rate of Environmental DNA From Juvenile and Adult Fish. PloS One 9, e114639. doi: 10.1371/journal.pone.0114639
Mondal D., Ghosh P., Khan M. A. A., Hossain F., Böhlken-Fascher S., Matlashewski G., et al (2016). Mobile Suitcase Laboratory for Rapid Detection of Leishmania Donovani Using Recombinase Polymerase Amplification Assay. Parasit. Vectors 9, 281. doi: 10.1186/s13071-016-1572-8
O'Brien A. L., Ross D. J., Keough M. J. (2006). Effects of Sabella Spallanzanii Physical Structure on Soft Sediment Macrofaunal Assemblages. Mar. Freshwater Res. 57, 363–371. doi: 10.1071/MF05141
Patti F. P., Gambi M. C. (2001). Phylogeography of the Invasive Polychaete Sabella Spallanzanii (Sabellidae) Based on the Nucleotide Sequence of Internal Transcribed Spacer 2 (ITS2) of Nuclear rDNA. Mar. Ecol. Prog. Ser. 215, 169–177. doi: 10.3354/meps215169
Piepenburg O., Williams C. H., Stemple D. L., Armes N. A. (2006). DNA Detection Using Recombination Proteins. PloS Biol. 4, 1115–1121. doi: 10.1371/journal.pbio.0040204
Pochon X., Zaiko A., Fletcher L. M., Laroche O., Wood S. A. (2017). Wanted Dead or Alive? Using Metabarcoding of Environmental DNA and RNA to Distinguish Living Assemblages for Biosecurity Applications. PloS One 12, e0187636. doi: 10.1371/journal.pone.0187636
Ravindran V. B., Khallaf B., Surapaneni A., Crosbie N. D., Soni S. K., Ball A. S. (2020). Detection of Helminth Ova in Wastewater Using Recombinase Polymerase Amplification Coupled to Lateral Flow Strips. Water 12, 691. doi: 10.3390/w12030691
Read G., Inglis G., Stratford P., Ahyong S. (2011). Arrival of the Alien Fanworm Sabella Spallanzanii (Gmelin 1791) (Polychaeta: Sabellidae) in Two New Zealand Harbours. Aquat. Invasions 6, 273–279. doi: 10.3391/ai.2011.6.3.04
Ross D. J., Keough M. J., Longmore A. R., Knott N. A. (2007). Impacts of Two Introduced Suspension Feeders in Port Phillip Bay, Australia. Mar. Ecol. Prog. Ser. 340, 41–53. doi: 10.3354/meps340041
Sepulveda A. J., Nelson N. M., Jerde C. L., Luikart G. (2020). Are Environmental DNA Methods Ready for Aquatic Invasive Species Management? Trends Ecol. Evol. 35, 668–678. doi: 10.1016/j.tree.2020.03.011
Sepulveda A. J., Schmidt C., Amberg J., Hutchins P., Stratton C., Mebane C., et al (2019). Adding Invasive Species Biosurveillance to the U.S. Geological Survey Streamgage Network. Ecosphere 10, e02843. doi: 10.1002/ecs2.2843
Soliman T., Inglis G. J. (2018). Forecasting the Economic Impacts of Two Biofouling Invaders on Aquaculture Production of Green-Lipped Mussels Perna Canaliculus in New Zealand. Aquac. Environ. Interact. 10, 1–12. doi: 10.3354/aei00249
Thomas A. C., Nguyen P. L., Howard J., Goldberg C. S. (2019). A Self-Preserving, Partially Biodegradable eDNA Filter. Methods Ecol. Evol. 10, 1136–1141. doi: 10.1111/2041-210X.13212
von Ammon U., Jeffs A., Zaiko A., van der Reis A., Goodwin D., Beckley L. E., et al (2020). A Portable Cruising Speed Net: Expanding Global Collection of Sea Surface Plankton Data. Front. Mar. Sci. 7. doi: 10.3389/fmars.2020.615458
Wood S. A., Pochon X., Laroche O., von Ammon U., Adamson J., Zaiko A. (2019a). A Comparison of Droplet Digital Polymerase Chain Reaction (PCR), Quantitative PCR and Metabarcoding for Species-Specific Detection in Environmental DNA. Mol. Ecol. Resour. 19, 1407–1419. doi: 10.1111/1755-0998.13055
Wood S. A., Pochon X., Ming W., von Ammon U., Woods C., Carter M., et al (2019b). Considerations for Incorporating Real-Time PCR Assays Into Routine Marine Biosecurity Surveillance Programmes: A Case Study Targeting the Mediterranean Fanworm (Sabella Spallanzanii) and Club Tunicate (Styela Clava). Genome 62, 137–146. doi: 10.1139/gen-2018-0021
Wood S. A., Zaiko A., Richter I., Inglis G., Pochon X. (2017). Development of a Real-Time Polymerase Chain Reaction Assay for the Detection of the Invasive Mediterranean Fanworm, Sabella Spallanzanii, in Environmental Samples. Environ. Sci. Pollut. Res. 24, 17373–17382. doi: 10.1007/s11356-017-9357-y
Keywords: recombinase polymerase amplification (RPA), point-of-need (PON), environmental DNA (eDNA), lateral flow detection, Sabella spallanzanii
Citation: Zirngibl M, von Ammon U, Pochon X and Zaiko A (2022) A Rapid Molecular Assay for Detecting the Mediterranean Fanworm Sabella spallanzanii Trialed by Non-Scientist Users. Front. Mar. Sci. 9:861657. doi: 10.3389/fmars.2022.861657
Received: 24 January 2022; Accepted: 09 May 2022;
Published: 22 June 2022.
Edited by:
Sandra Pucciarelli, University of Camerino, ItalyReviewed by:
Elena Kupriyanova, Australian Museum, AustraliaEdwin Cruz-Rivera, University of the Virgin Islands, US Virgin Islands
Copyright © 2022 Zirngibl, von Ammon, Pochon and Zaiko. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Anastasija Zaiko, YW5hc3Rhc2lqYS56YWlrb0BjYXd0aHJvbi5vcmcubno=
 Martin Zirngibl
Martin Zirngibl Ulla von Ammon
Ulla von Ammon Xavier Pochon
Xavier Pochon Anastasija Zaiko
Anastasija Zaiko