- 1Key Laboratory of Marine Genetics and Breeding (Ministry of Education), Ocean University of China, Qingdao, China
- 2College of Marine Life Sciences, Ocean University of China, Qingdao, China
- 3Yazhou Bay Innovation Institute, Hainan Tropical Ocean University, Sanya, China
- 4Key Laboratory of Utilization and Conservation of Tropical Marine Bioresource (Ministry of Education), College of Fisheries and Life Science, Hainan Tropical Ocean University, Sanya, China
- 5Laboratory for Marine Biology and Biotechnology, Pilot National Laboratory for Marine Science and Technology (Qingdao), Qingdao, China
Neopyropia yezoensis, previously known as Pyropia yezoensis, is regarded as a model macroalgae because of its economic value and typical intertidal seaweed species. However, the genetic transformation system to introduce foreign genes into its cells is inefficient and not stable. This study developed a more efficient transformation toolbox that allows the stable expression of foreign genes in N. yezoensis cells. We constructed an efficient transformation platform with the parameters of biolistic bombardment of 6 cm target distance, 600 μg gold particles/shot, 10 μg plasmid DNA/shot, 1,350 psi of helium, and 30 mmHg vacuum pressure. Thalli at 35 days of age were the most suitable transformation conditions, in which the highest transformation efficiency was generated. The endogenous promoter pPyACT1 could control gene expression efficiently compared to pPyUBC, pPyDPE2, and pPyEF1-a, especially the exogenous promoter d35S. Finally, the foreign genes PyGUS and PyHygR were stably expressed in different generations of transformants, including monospores, gametophytes, and filamentous sporophytes. Southern blotting analysis confirmed that PyGUS was integrated into the genome of N. yezoensis transformants. Establishing an efficient gene expression toolbox provides a strong foundation for functional genomics research and molecular genetic breeding on N. yezoensis.
Introduction
Genetic transformation has become an important technique in modern molecular biology research to analyze gene functions and regulatory mechanisms, increase the tolerance of organisms to various environmental stresses, and obtain organisms that efficiently produce biofuels and medically important materials (Torney et al., 2007; Bhatnagar-Mathur et al., 2008; Rochaix, 2013). Achieving algal transformation is also crucial for investigating the function and regulation of genes and laying the foundation for genetic breeding (Walker et al., 2005; Hallmann, 2006). In microalgae, with simple cellular structures and life cycles, stable transformation systems have already been developed, such as in the model green alga Chlamydomonas reinhardtii via electroporation (Jiang et al., 2014), in the model diatom Phaeodactylum tricornutum via biolistic transformation (Kikutani et al., 2016; Slattery et al., 2018), and in Nannochloropsis sp. via electroporation (Ma et al., 2017). Macroalgae are multicellular organisms with a life cycle characterized by an alternation of generations (Bowman et al., 2016). The establishment of a transformation system in macroalgae is relatively few compared with that in microalgae. Thalli of the red alga Kappaphycus alvarezii was used in the first report of the transient transformation of seaweeds using particle bombardment (Kurtzman and Cheney, 1991). Transient transformation systems have been established in the red macroalgae Gracilaria chagii, G. gracilis, and K. alvarezii by particle bombardment (Gan et al. 2003a; Wang et al., 2010a; Huddy et al., 2012); the life cycle of the three organisms corresponds to heteromorphic alternation of generations. In the brown macroalgae, Laminaria japonica and Undaria pinnatifida, transient transformation has been described in sporophytes by particle bombardment (Daozhan et al., 2002; Jiang et al., 2003). For the green macroalgae Ulva, which has a life cycle characterized by the isomorphic alternation of generations, transient transformation has been reported in U. lactura via electroporation and U. pertusa via particle bombardment (Huang et al., 1996; Kakinuma et al., 2009). The transformation efficiency of these macroalgae is not high, while the stable transformation is immature. Recently, in the multicellular green macroalga U. mutabilis, which has two layers of cells, a molecular toolkit was developed that stably overexpresses transgenes in cell lines. It can also be used for the expression of fluorescent proteins, marker lines, and tagged endogenous proteins (Blomme et al., 2021).
Neopyropia yezoensis, one of the Bangiophyceae algae, has a life cycle of the haploid gametophyte (thalli) and diploid sporophyte (filament) and is an essential economic seaweed for its value in the food, food additives, and medicine industries. Moreover, N. yezoensis thalli, which can tolerate broad and extreme environmental stresses, is regarded as a model red macroalga for physiological and genetic research (Saga and Kitade, 2002). The completion of the N. yezoensis genome sequence allowed the prediction of 12,855 protein-coding genes, almost half of which have an unknown function (Wang et al., 2020). Therefore, it will be beneficial to develop transformation technology to accelerate the elucidation of gene function in N. yezoensis. Some studies have reported the progress of transient transformation technologies for N. yezoensis (Hirata et al., 2011; Mikami et al., 2011; Uji et al., 2013). Plastid genetic transformation based on homologous recombination technology has also been established in N. yezoensis (Kong et al., 2017). However, many bottlenecks remain in the stable and efficient transformation of macroalgae N. yezoensis, such as the lack of effective transformation methods, efficient expression vectors, and proper selection markers.
In previous studies, the use of exogenous promoters, such as those of the cauliflower mosaic virus 35S RNA (CaMV 35S) and simian virus 40 (SV40) genes, was attempted in G. changii, L. japonica, P. miniate, and N. yezoensis (Mei et al., 1998; Jiang et al., 2003; Gan et al., 2003b). However, the activity of the CaMV 35S and SV40 promoters in these algal species is very low. Strong endogenous promoters from algae show higher expression levels of exogenous genes, such as those of the β2-tubulin (β2TUB) gene, ribulose bisphosphate carboxylase small subunit 2 (RBCS2) gene, and heat shock protein 70 (HSP70) genes (Davies et al., 1992; Schroda et al., 2000; Sizova et al., 2001). The promoter of the glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene from N. yezoensis efficiently controls PyGUS in N. yezoensis cells (Fukuda et al., 2008). Thus, selecting strong endogenous promoters is critical for controlling the high expression of foreign genes. Suitable reporter genes can accelerate the selection of positive transformants. Bacterial β-galactosidase lacZ and β-glucuronidase (GUS) uidA genes in macroalgae have already been used as reporters. For foreign genes, the incompatibility of codon usage may be responsible for the poor translation efficiency of N. yezoensis cells. AT codons are abundant in bacterial GUS and lacZ. A GUS gene optimized for codon usage in N. yezoensis is efficiently expressed compared with a nonoptimized GUS gene (Fukuda et al., 2008). Fluorescent proteins, such as green protein ZsGFP, sGFP, and cyan AmCFP, can also be transiently expressed in gametophytic cells of N. yezoensis (Uji et al., 2010), which highlights their potential utilization as reporter genes in future genetic transformation research. The gene expression efficiency in these studies was relatively low. Therefore, to establish a reliable gene delivery system in N. yezoensis, it is necessary to select an efficient promoter and appropriate reporter gene for assaying gene expressions.
Gene transfer methods are critical for exogenous gene expression. In plants, Agrobacterium-mediated, electroporation, polyethylene glycol (PEG), lipofection, virus-mediated, and biolistic bombardment transformation methods have been developed (Rathus and Birch, 1992; Chen et al., 1998; Howe et al., 2006; Zhang et al., 2011; Westwood et al., 2013; Keymer et al., 2017). New transformation methods, such as nanomaterial-mediated transformation, have also been developed to achieve efficient plant genetic transformation and no transgene integration in the genome (Demirer et al., 2019; Kwak et al., 2019). The glass bead, electroporation, PEG, PEG plus electroporation (Mei et al., 1998), and Agrobacterium-mediated (Cheney et al., 2001) methods have been used for N. yezoensis; however, these transformation methods do not generate a high transformation efficiency. Particle bombardment is the most commonly used transformation technology without the limitation of host cells. Factors such as microparticle type, amount and size, helium pressure, vacuum pressure, bombardment distance (the distance between the stopping screen and the biological target), and the amount of DNA used for coating the microcarriers can influence the final transformation efficiency. Thus, for particle bombardment, the optimization of these transformation factors will improve the transformation efficiency to select an efficient gene transfer system.
Protoplasts, conchospores, gametophytic thalli, and sporophytic filaments have been used as transformation recipients for Neopyropia. For protoplast transformation, the beta-glucuronidase (GUS) and green fluorescent protein (GFP) genes driven by the ribulose-bisphosphate-carboxylase/oxygenase gene promoter can be expressed via electroporation (Mizukami et al., 2004). The transformation efficiency is low in protoplasts due to the low viability and complex preparation procedure. For conchospore transformation, lacz and egfp can be expressed under the control of the SV40 promoter by glass bead agitation in N. haitanensis (Wang et al., 2010b). However, the preparation of conchospores requires a long time, and the transformation efficiency is nearly 0.006‰. In sporophytic filaments, the PyGUS reporter gene is controlled by the PyKPA1 promoter through particle bombardment transformation (Uji et al., 2013). However, the enzymatic activity of PyGUS is lower in sporophytic cells than in gametophytic cells by particle bombardment. It is thought that the thin filamentous form of sporophytic cells makes them difficult to target by particle bombardment (Mikami et al., 2011). Gametophytic cells have been widely used for particle bombardment in N. yezoensis and N. haitanensis, by which foreign genes, such as PyGUS and sGFP reporter genes, can be transiently expressed (Takahashi et al., 2010; Hirata et al., 2011). Gametophytic thalli are an appropriate receptor to introduce foreign genes.
In this study, several endogenous strong promoters, pPyACT1, pPyUBC, pPyDPE2, and pPyEF1-a, were selected and ligated into the PBI121-GUS vector separately to compare their transcriptional activity. The antibiotic resistance hygromycin PyHygR was used as a selection marker. The constructs were transformed into N. yezoensis gametophytic thalli using biolistic bombardment, and different transforming parameters, such as target distance, gold particle/plasmid DNA content, vacuum pressure, and growing age of thalli, were optimized to improve transformation efficiency. Using these parameters, a toolbox was constructed to integrate foreign elements into the genome. This study would provide a foundation for functional genomics research and molecular genetic breeding in N. yezoensis.
Materials and Methods
Algal Strains and Culture Conditions
We selected and bred the N. yezoensis pure culture strain RZ (PYL201306-440). Gametophytic thalli and sporophytic filaments were both cultured in a PES medium made with sterilized seawater at 10°C and 20°C, respectively. Sperms and carpogonia from mature gametophytic thalli (gametophytic thalli were incubated at 20°C for approximately 30 days under stationary conditions) can inbreed to generate carpospores and further develop into sporophytic filaments. The irradiance for the gametophytic thalli was 50 μmol photons m−2 s−1 and continuous aeration with filter-sterilized air. The irradiance for the sporophytic filaments was 20 μmol photons m−2 s−1 without aeration. The photoperiod was 12 L:12 D. PES medium was renewed every 3 days and continuously aerated with filter-sterilized air. Thalli were used for particle bombardment after culturing for 5–7 weeks.
Isolation Promoter of the PyACT1, PyUBC, PyDPE2, and PyEF1-a Genes From N. yezoensis
Genomic DNA of N. yezoensis was extracted using a plant genomic DNA extraction kit (TIANGEN, China). Cis-acting elements and core promoter regions were predicted using PlantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) and Promoter 2.0. The four genes PyACT, PyUBC, PyDPE2, and PyEF1-a are housekeeping genes in N. yezoensis and encode actin, putative ubiquitin-conjugating enzyme, disproportionating enzyme type 2, and elongation factor 1-α, respectively. The four corresponding gene promoter sequences, pPyACT1, pPyUBC, pPyDPE2, and pPyEF1-a, were cloned from the N. yezoensis genome (accession number: PRJNA589917). The selected amplified promoter regions were located at around 2,000 bp upstream of each gene’s initiation codon (ATG). PCR conditions comprised an initial denaturation at 94°C for 1 min followed by 40 cycles of 94°C for 10 s and 68°C for 1 min/1 kb with the Tks Gflex DNA polymerase (Takara, Japan). The PCR amplification product was purified and ligated into the pMD19-T vector (Takara, Japan). Positive clones confirmed by sequencing were used for constructing expression vectors.
Construction of Plasmids for Genetic Transformation
To construct the vector for genetic transformation, the GUS gene was optimized based on the codon usage of the N. yezoensis genome and designated PyGUS, which was synthesized by Sangon Biotech (Shanghai, China). The promoter and PyGUS gene fragments were cloned into the vector PBI121-GUS, which replaced the 35S promoter and GUS simultaneously by multifragment homologous recombination using the ClonExpress Ultra One Step Cloning Kit (Vazyme, China), and the primers used are shown in Supplementary Table S1. Six construction vectors were generated: PBI121-pPyACT1-PyGUS, PBI121-pPyUBC-PyGUS, PBI121-pPyDPE2-PyGUS, PBI121-pPyEF1-a-PyGUS, and PBI121-d35S-PyGUS. The antibiotic resistance gene HygR was also optimized for codon usage and designated PyHygR.
Particle Bombardment and Orthogonal Experiment Design
The constructed plasmids were transformed into N. yezoensis thalli using a PDS-1000/He particle bombardment device (Bio-Rad, Foster City, CA, USA). Gold particles (30 mg, 0.6 μm diameter, Bio-Rad) were suspended in 1 ml of 70% ethanol, vortexed for 5 min, and kept on ice for 15 min. The supernatant was discarded by centrifugation at 10,000×g for 10 s, and the sample was suspended in 500 μl of 50% sterile glycerol solution and stored at −20°C until use. To coat the particles with plasmids, 50 μl gold particle mother liquor was sequentially mixed with 50 μg DNA solution by vortexing for 20 s, adding 50 μl of 2.5 M CaCl2 and vortexing an additional 20 s and 20 μl of 0.1 M spermidine by vortexing for 20 s. The mixed solution was vortexed for 1 min 10 times, kept on ice for 20 min, and finally centrifuged at 10,000×g for 10 s. The pellet was washed with 140 μl 70% ethanol and 140 μl pure ethanol and resuspended in 60 μl pure ethanol.
The orthogonal test is a multifactor and multilevel designing method that conducts tests by selecting representative points from comprehensive tests based on orthogonality (Peng et al., 2006). To achieve a high transformation efficiency, the transformation parameters were optimized by orthogonal experiment design according to the design module of Statistica 6.0 (SoftStat, 1984–2001, USA). Three relevant factors were evaluated: vacuum pressure, target distance, and gold particle/plasmid DNA content. The orthogonal table L18(3)7 (Table 1) was designed, in which interaction factors between every two factors were included, and a blank column was designated for error evaluation. Since this column is blank, the sum of the squared deviations excluded the difference between the factor levels and was only representative of the size of the experimental error; hence, repeating the experiments for error analysis was not necessary (Zhou et al., 2021). Three levels were set for each factor, and the boundary values for the levels were determined in pilot experiments to ensure an appropriate range of values. Vacuum pressure conditions were set at 28, 29, and 30 mmHg. The target distances were 3, 6, and 9 cm. The plasmid concentrations per shot were 5, 10, and 20 μg. The gold particle concentration was set at 600 μg, and the bombard pressure for the rupture disks was 1,350 psi. Thalli at 25, 30, 35, and 40 days after conchospore release were used to screen the most accessible transformation conditions. The thalli were cut and combined into 3 cm × 3 cm squares and placed in the center of 9 cm Petri dishes. After bombardment, the thalli were placed in a 96-well plate with a PES medium and incubated at 10°C for 96 h (48 h in the dark) before GUS histochemical assays.
GUS Histochemical Assay for Transformed N. yezoensis Thalli
Transformed N. yezoensis thalli individuals were assayed via GUS histochemical staining to analyze the expression of PyGUS. GUS histochemical staining of bombarded N. yezoensis thalli was performed as previously described (Jefferson et al., 1987). The thalli were incubated with a staining solution, which consisted of 2 mM X-gluc, 5 mM potassium ferricyanide, 5 mM potassium ferrocyanide, 0.5% Triton X-100, 1.5 M sorbitol, and 50 mM sodium phosphate, pH 7.0, and incubated in 37°C for 6 h. Thalli were then placed on a glass slide and observed under an optical microscope (Olympus BX53, Japan). The number of PyGUS-expressing cells was counted using a hand tally cell counter (BERM, China).
Screening and Subculture of the N. yezoensis Transformants
Experiments were conducted to prepare the gametophytic thalli individuals for the hygromycin resistance assay. The bombarded gametophytic thalli were cultured in a PES medium under nonselective conditions for 1 week. Subsequently, the medium was replaced with the PES medium containing 2 mg/ml hygromycin B. Hygromycin B concentration was confirmed by the sensitivity of N. yezoensis to the drug. After incubating for 2 weeks in an antibiotic-containing medium, N. yezoensis thalli were transferred into a normal medium. When continuously cultivated for approximately 30 days, most of the thalli cells die, and live cells can grow into a new thallus through cell division and proliferation. After cultivation for approximately 30 days, regenerated thalli were further screened using GUS histochemical staining to select successful transformants. The positive transformants released monospores using the physical sectioning method, which further grew into next-generation thalli (Chen et al., 2020). Monospores, small thalli, and filaments of positive transformants were again stained with GUS.
Southern Blot Analysis of N. yezoensis Transformants
Southern blot analysis was made to analyze whether the foreign gene was integrated into the genome in transformed thalli. Genomic DNA was extracted from the wild-type and transformants using the HP Plant DNA Kit (OMEGA, USA). DNA (10 μg) was digested with restriction enzyme BamHI and SmaI, run on an agarose gel, and transferred to a nylon membrane through the capillary siphon imprinting method. A 1,500-bp fragment of PyGUS was labeled with DIG High Prime and used as a probe. The following protocols were performed using the DIG High Prime Lab/Detection K1 Kit (Roche, Switzerland).
Results
Cloning and Analysis of Endogenous Promoters From N. yezoensis
PyUBC, PyACT, PyEF1-a, and PyDPE2, housekeeping genes in N. yezoensis, are stably and highly expressed under temperature stress or dehydration stress (Gao et al., 2018). The gene IDs py04835.t1 (PyUBC), py08823.t1 (PyACT), py06023.t1 (PyEF1-a), and py11282.t1 (PyDPE2) were retrieved from the N. yezoensis genome sequence. The core promoter element TATA box and some cis-acting elements were identified in the upstream region of the transcription start site (TSS) in the four genes. The results of cis-acting element analysis are presented in Supplementary Figure S1. Based on sequence analysis, pPyACT1, pPyUBC, pPyDPE2, and pPyEF1-a were cloned from the upstream sequences to the initiation codon (ATG) of the genes via PCR amplification (Supplementary Figure S2). The total lengths of pPyUBC, pPyACT1, pPyEF1-a, and pPyDPE2 were 3,620, 3,073, 2,555, and 2,852 bp, respectively.
Optimization of Particle Bombardment Parameters
The orthogonal test design results of bombardment factor effects on PyGUS expression are shown in Table 1. The number of cells expressing PyGUS was set to compare the effects of different factors on transformation efficiency. Extreme difference analysis was aimed at clarifying the significance levels of the different influencing factors. The most significant factors were identified based on the results of the range analysis. The larger the and R values, the greater the significance of the level or factor in the results. Based on the extreme difference analysis, the order of significance of the factors was vacuum pressure, DNA content, and bombardment target distance. The optimal conditions for the three factors were the third level of vacuum pressure, the second level of DNA content, and the second level of target distance. The interactive factors target distance × vacuum pressure, target distance × DNA content, and vacuum pressure × DNA content were included in the orthogonal test. The and R values of the two interactive factors were smaller than those of the single factors; therefore, the interaction between the two factors was not significant. Finally, according to the extreme difference analysis, the optimal vacuum pressure level was 30 inHg, the bombardment distance was 6 cm, and the DNA content was 10 μg.
The sum of squares of deviation (SS), degree of freedom (df), and mean-squared deviation (MS) of the number of cells expressing PyGUS were determined and are summarized in Table 2. The F-value of a factor is the ratio of the MS value to the error line. By comparing the obtained F-value with the theoretical value of a specific level and the df, the significance level can be determined for each factor. As shown in Table 2, ANOVA with vacuum pressure had a significant impact on the number of cells expressing PyGUS (p < 0.05), and DNA content significantly affected the number of cells expressing PyGUS (p < 0.1). In addition, bombardment distance had no significant effect on expression efficiency, with significant interaction between any two factors of target distance, vacuum pressure, or DNA content.
The developmental stages of thalli play an important role in the transformation efficiency of particle bombardment (Son et al., 2012). Leafy gametophytes of different ages, such as 25, 30, 35, and 40 days, were transformed and compared by estimating the number of cells expressing PyGUS (Figure 1). The highest transformation efficiency was observed in the thalli at 35 days (p < 0.01). Lower numbers of cells expressing PyGUS were observed after transformation with young thalli at 25 days of age, and the transformation efficiency at 30 days was almost similar to that at 40 days of age. Therefore, 35-day-age thalli can be used as transformation recipients in further genetic experiments.
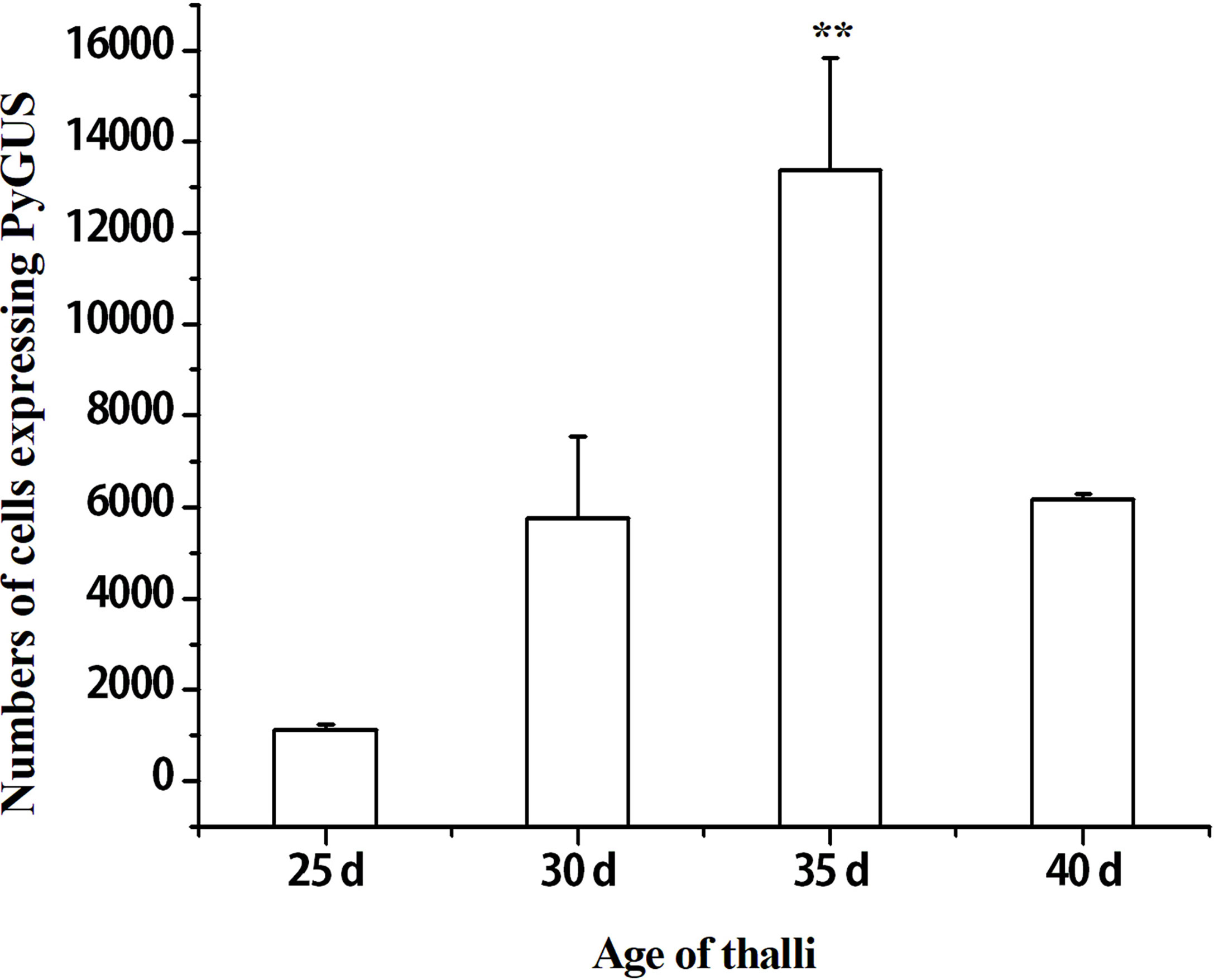
Figure 1 Effects of different thalli ages on the number of cells expressing PyGUS in N. yezoensis. The thalli age of 25, 30, 35, and 40 days correspond to the time after conchospores release. Data are expressed as mean ± SD (n = 3). **P < 0.01 by analysis of variance.
Comparison of pPyACT1, pPyUBC, pPyDPE2, pPyEF1-a, and d35S Promoter Activity
To identify the higher promoter efficiency, the promoters of four housekeeping genes (pPyACT1, pPyUBC, pPyDPE2, and pPyEF1-a) and d35S were used to control PyGUS gene expression in the PBI121 vector. Plasmids PBI121-pPyACT1-PyGUS, PBI121-pPyUBC-PyGUS, PBI121-pPyDPE2-PyGUS, PBI121-pPyEF1-a-PyGUS, and PBI121-d35S-PyGUS (a schematic representation of the plasmids is shown in Supplementary Figure S3) were transformed into N. yezoensis thalli under the optimized transformation conditions. PyGUS was expressed in all thalli after transformation with plasmids under the control of the four different endogenous promoters (Figure 2). Few cells expressing PyGUS were observed after transforming the plasmid with the pPyUBC promoter, while no cells expressing PyGUS were observed when the d35S-PyGUS plasmid was introduced into N. yezoensis. The number of cells expressing PyGUS was analyzed 96 h post bombardment (Figure 2G). The number of cells expressing PyGUS driven by the pPyACT1 promoter was the highest, with more than 20,000 cells, indicating that the pPyACT1 promoter was the strongest among the selected promoters. The number of cells expressing PyGUS driven by pPyEF1-A and pPyDPE2 ranked second and did not show a significant difference. PPyUBC was the weakest, and only a few cells expressed PyGUS. It is worth noting that no cells expressing PyGUS driven by the exogenous promoter d35S were observed in the transformed cells, indicating that the activity of the exogenous promoter in thalli was low. We conclude that the endogenous promoter pPyACT1 was most suitable for genetic transformation.
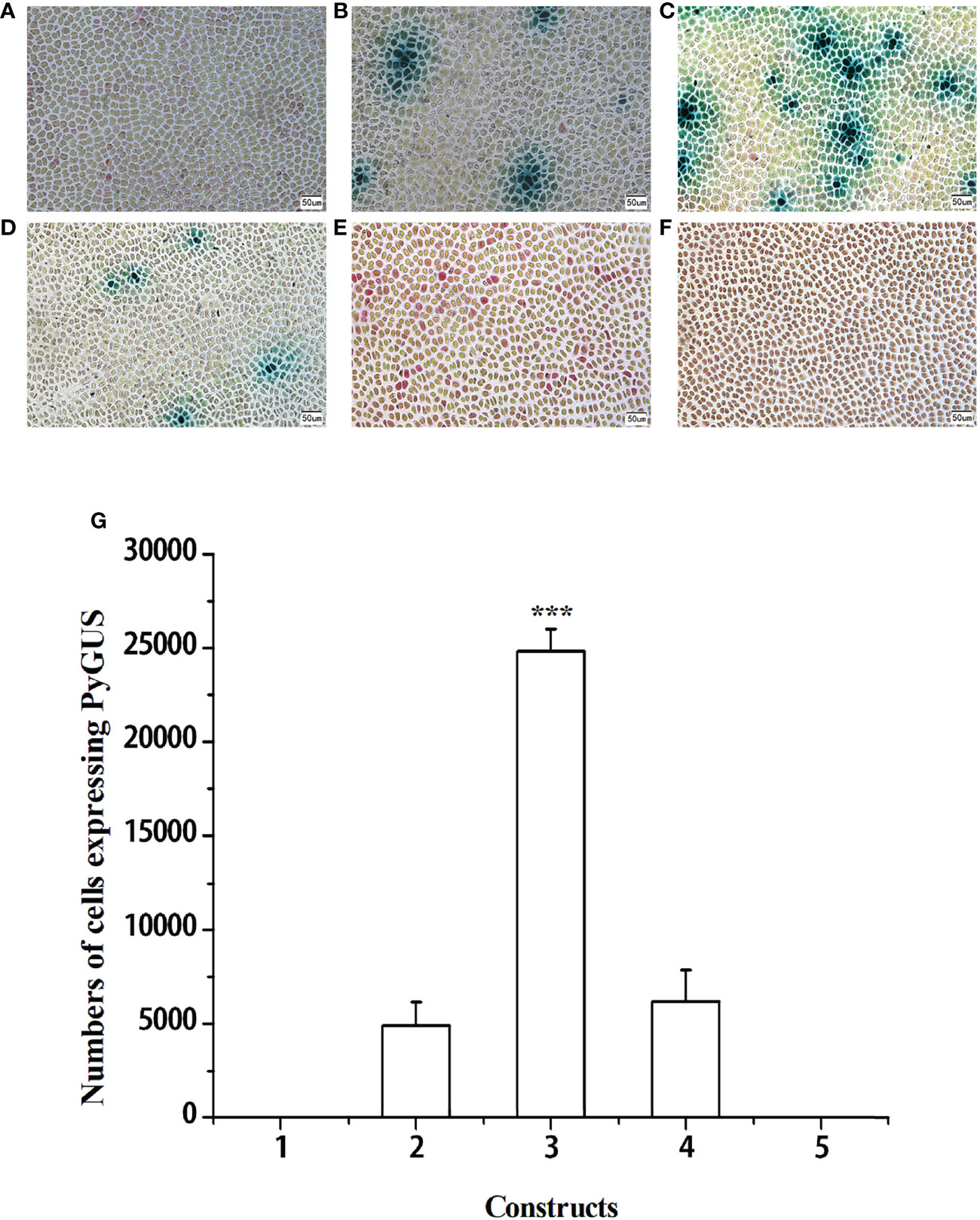
Figure 2 Expression of PyGUS in gametophytic cells of N. yezoensis. Gametophytic cells of N. yezoensis were transformed with expression plasmids PBI121-pPyUBC-PyGUS (A), PBI121-pPyEF1-a-PyGUS (B), PBI121-pPyACT1-PyGUS (C), PBI121-pPyDPE2-PyGUS (D), PBI121-d35S-PyGUS (E), and without transformation of plasmid (F). Blue spots indicate cells expressing PyGUS. (G) Comparison of the numbers of cells expressing PyGUS driven by different promoters in N. yezoensis. Plasmids with different promoters used for transformation were as follows: (1) PBI121-pPyUBC-PyGUS, (2) PBI121-pPyEF1-a-PyGUS, (3) PBI121-pPyACT1-PyGUS, (4) PBI121-pPyDPE2-PyGUS, and (5) PBI121-d35S-PyGUS. Scale bars indicate 50 μm. Data are expressed as mean ± SD (n = 3). ***P < 0.001 by analysis of variance.
Determination of the Active Promoter Region of pPyACT1
As mentioned above, the pPyACT1 promoter possessed the strongest ability to control exogenous PyGUS expression. To further determine the shortest-length functional region of the pPyACT1 promoter, different length regions of the upstream sequence from the initiation codon of the PyACT gene were used to control PyGUS to compare their activity. The different regions were denoted pPyACT1 (3,073 bp), pPyACT2 (2,316 bp), pPyACT3 (1,858 bp), and pPyACT4 (1,125 bp). The results revealed that all four constructs could be expressed in thalli cells (Figure 3), and the number of cells expressing PyGUS was not significantly different among the four constructs with different promoter lengths (Figure 3G). This indicates that the different upstream regions of pPyACT1 did not affect PyGUS expression. Bioinformatic analysis showed that the core sequence of pPyACT4 was located within the region from −1,125 bp to ATG, and the TSS was located 530 bp upstream of ATG. The 1,125-bp sequence contains the core promoter regions such as TAAT-box and CAAT box. A longer sequence did not increase promoter activity.
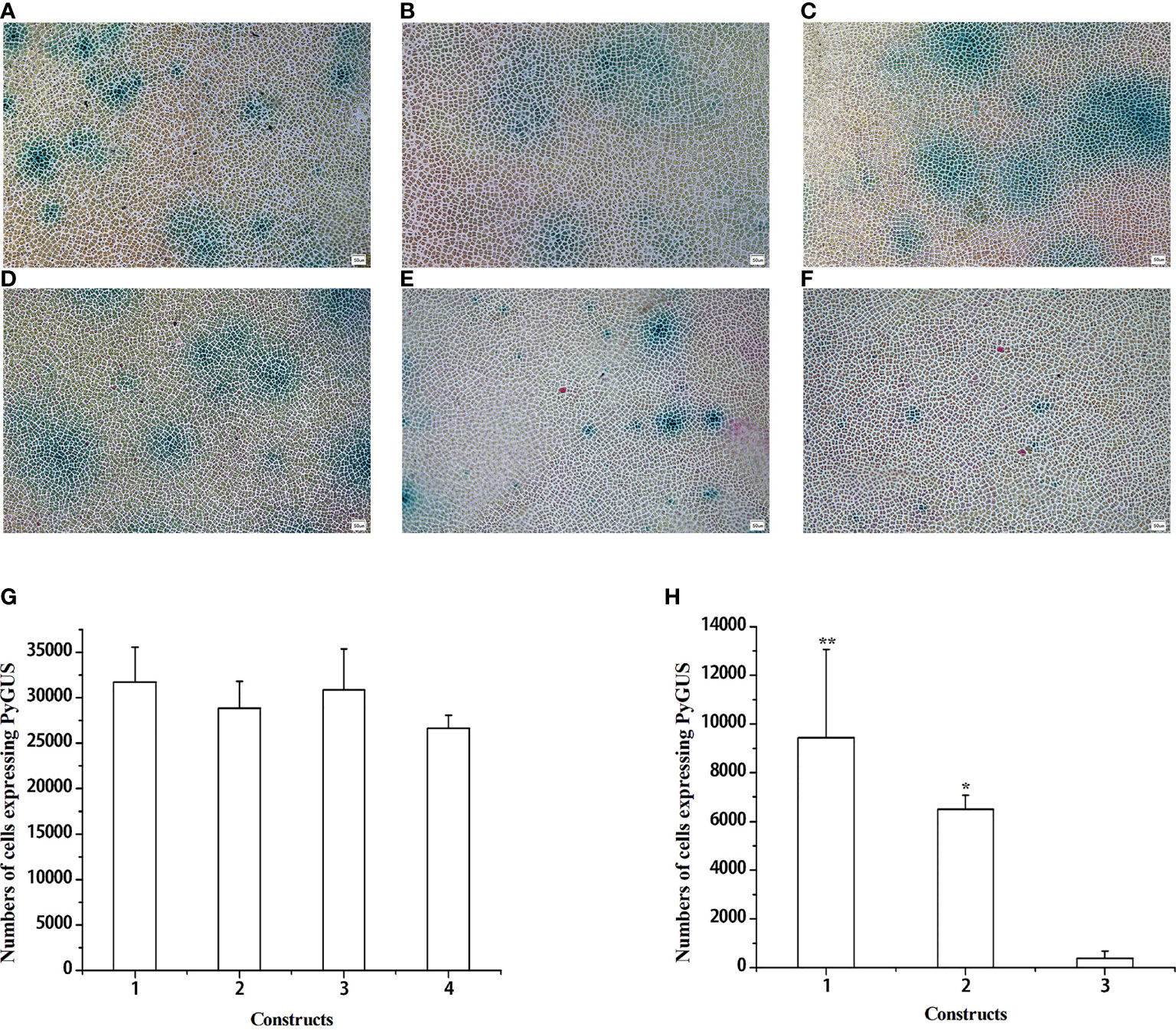
Figure 3 Expression of PyGUS driven by different promoters in gametophytic cells of N. yezoensis. Gametophytic cells of N. yezoensis were transformed with expression plasmids using different promoters. PBI121-pPyACT1-PyGUS (A), PBI121-pPyACT2-PyGUS (B), PBI121-pPyACT3-PyGUS (C), PBI121-pPyACT4-PyGUS (D), PBI121-pPyACT5-PyGUS (E), and PBI121-pPyACT6-PyGUS (F). Blue cell spots indicate cells expressing PyGUS. (G, H) Comparison of the numbers of cells expressing PyGUS with different lengths of pPyACT promoters in N. yezoensis. (G) Plasmids with different lengths of pPyACT promoters for transformation were as follows: (1) PBI121-pPyACT1-PyGUS, (2) PBI121-pPyACT2-PyGUS, (3) PBI121-pPyACT3-PyGUS, and (4) PBI121-pPyACT4-PyGUS. (H) Plasmids with different lengths of pPyACT promoters for transformation were as follows: (1) PBI121-pPyACT4-PyGUS, (2) PBI121-pPyACT5-PyGUS, and (3) PBI121-pPyACT6-PyGUS. Scale bars indicate 50 mm. Data are expressed as mean ± SD (n = 3). *P < 0.05; **P < 0.01 by analysis of variance.
The length of the promoter influences the size of the constructed vector, which adds to the difficulty of transforming the constructs. Therefore, we further determined the shortest active domain of pPyACT1, and pPyACT4 was divided into two fragments according to TSS: pPyACT5 and pPyACT6. A schematic representation of the plasmid constructs is shown in Supplementary Figure S4. The number of cells expressing PyGUS (Figure 3H) after transformation with pPyACT4-PyGUS was the highest, followed by pPyACT5-PyGUS, and pPyACT6-PyGUS was the lowest. Therefore, pPyACT4 was the smallest functional unit with promoter activity, and the pPyACT5 and pPyACT6 regions are essential components.
Screening Positive N. yezoensis Transformants and Subculture
To obtain positive transformants, the bombarded gametophytic thalli were first selected and cultured in a PES medium with 2 mg/ml hygromycin B for 2 weeks and then transferred to normal seawater with a PES medium. Approximately 30 days later, new thalli were gradually regenerated. After cultivation for approximately 30 days, the thalli were cut into small pieces and stained with GUS. All cells of the two thalli in all 156 thalli are shown in blue. PyGUS was also expressed in the released monospores and small thalli (Figure 4) originating from these two transformants. PyGUS was also expressed in sporophytic filaments (Figure 4D).
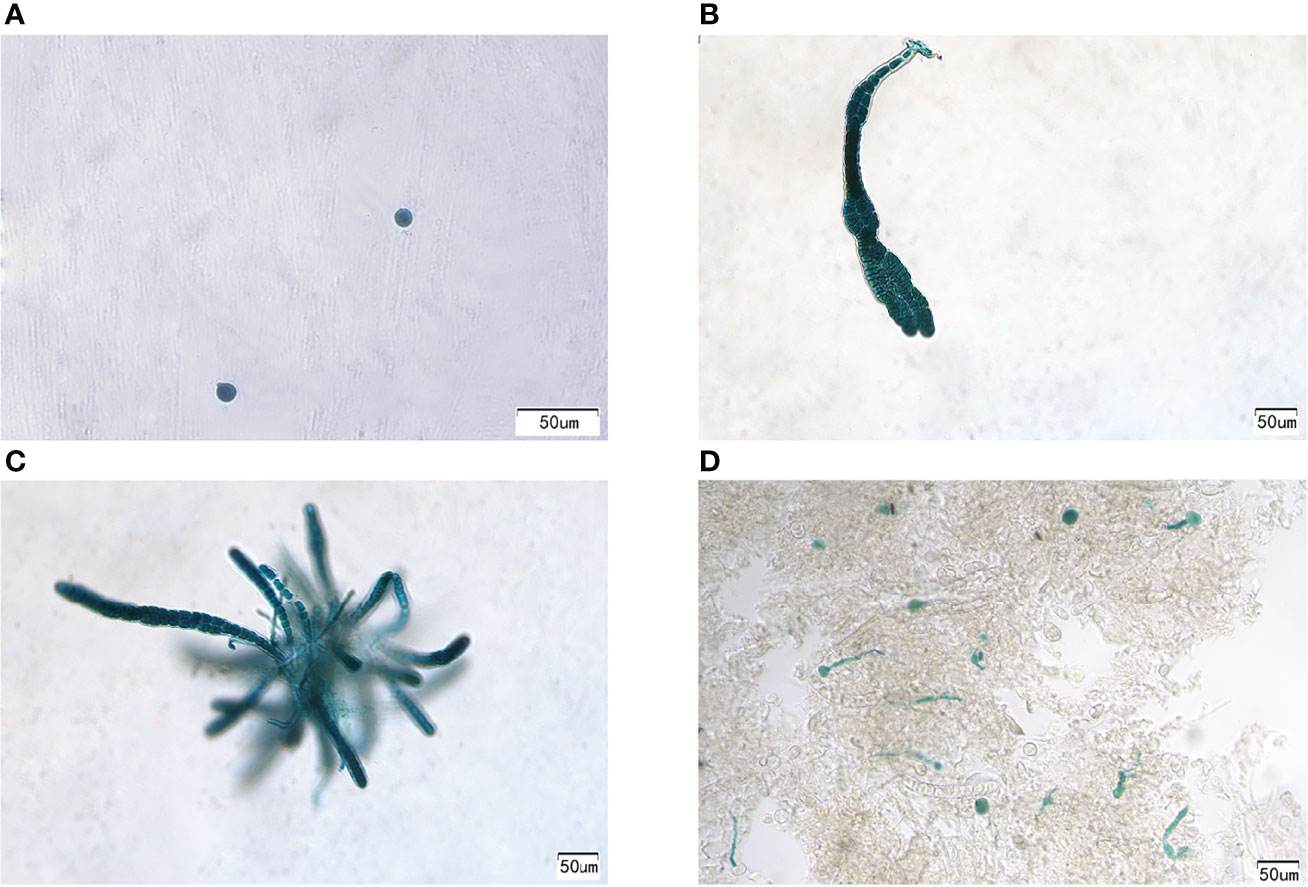
Figure 4 Expression of PyGUS in gametophytes and sporophytes from the transformants. PyGUS was expressed in monospores (A), thalli (B, C), and filaments (D). Scale bars indicate 50 μm.
Southern Blotting Analysis of N. yezoensis Transformants
PyGUS was stably expressed in different generations through asexual subculture and sexual propagation, suggesting that PyGUS was integrated into the genome. To verify this, Southern blotting was performed, which showed multiple DNA segments of different sizes in the genome of all the examined transformants, confirming that PyGUS was indeed randomly integrated into the genome (Supplementary Figure S5). Thus, PyGUS can be stably expressed in thalli through cell division and generation. However, it should be noted that similarly sized bands were observed on the nylon membrane compared with the plasmid, which indicated that the transformed plasmid was also present in thalli cells.
Discussion
The objective of this study was to establish an efficient and stable gene expression toolbox for N. yezoensis. In biolistic bombardment experiments, transformation efficiency is influenced by many factors, such as bombardment devices, samples, and operators. Vacuum pressure and target distance have the most significant effects on the transformation efficiency (Dhir et al., 2010). Fukuda et al. (2008) described the optimal transformation conditions using the IDERA particle acceleration device as follows: device parameters of 6 cm target distance, 700 mmHg vacuum, and 0.6 MPa helium in leafy gametophytes of N. yezoensis (strain TU-1). Son et al. (2012) reported the following optimal conditions: 3 cm target distance, 28 mmHg vacuum, and 900 psi helium. The gold-to-DNA content ratio is also important for transformation efficiency. The gold particle and plasmid DNA contents can affect the precipitation of DNA onto gold particles and determine the amount of DNA delivered into tissues (Dhir et al., 2010). Too little DNA precipitates inefficiently onto gold particles, while too much leads to a surplus. A total of 600 μg gold particles with 3.3 μg plasmid DNA and 250 μg gold particles with 20 μg plasmid DNA were employed by different groups (Fukuda et al., 2008; Son et al., 2012). Furthermore, interactions among different parameters can affect transformation efficiency (Meng et al., 2021). Previously, only single factors have been analyzed while not considering the interactions among factors. Therefore, in our study, these factors were analyzed together, and the interaction was considered using an orthogonal design. The optimized conditions were as follows: 6 cm target distance, 1,350 psi helium, 30 mmHg vacuum, bombardment particle of 600 μg gold particles/shot, and 10 μg plasmid DNA/shot. The number of cells expressing PyGUS (around 104 cells per shot) in the present study was higher than that in the above reports (around 102 to 103 cells per shot) in thalli of N. yezoensis. The growth status of recipient materials is also crucial for transformation efficiency in particle bombardment (Takumi and Shimada, 1996). Some studies suggest that different transformed materials will affect the transformation efficiency. The transient expression efficiency by particle bombardment in embryo-derived calli was higher than that in embryonic axes, immature embryos, cotyledons, shoot tips, and leaves in peaches (Ye et al., 1994). Meanwhile, higher levels of transient transformation efficiency were obtained in younger thalli of 1–3 cm in N. yezoensis leafy gametophytes (Son et al., 2012; Lim et al., 2013). The age of the N. yezoensis gametophytes used in other studies was not stated (Mikami et al., 2009; Hirata et al., 2011). However, the length range of leafy gametophytes was affected by culture conditions and could not reflect the physiological conditions of thalli. In the present study, different thalli age days were used to compare differences in the transformation efficiency of foreign genes. Finally, thalli at 35 days of age can be an ideal state for genetic transformation.
The promoter is also an essential element for vector expression during genetic transformation. The CaMV 35S promoter is widely used in heterologous plant cells because of its robust, constitutive, and nontissue-specific activity (Louis et al., 2010). In the present study, no cells expressing PyGUS were obtained after transformation with the PBI121-d35S-PyGUS construct. The expression levels of the four selected housekeeping genes were usually higher than those of most other genes in the genome (Kong et al., 2015; Gao et al., 2018). Thus, the promoters of these four genes were selected, and their ability to control exogenous gene expression was assessed. The results show that pPyACT1 had the highest activity among the selected promoters. Based on RNA-seq data from different stress factors and developmental stages, the gene expression level of PyACT was the highest, followed by PyEF1-a, pPyUBC, and PyDPE2. The activity of pPyACT1 was the highest, and the expression levels of related genes were also high (unpublished data). The actin gene promoter in D. salina has also been used to drive the expression of resistance genes for nuclear transformation (Jiang et al., 2005). The 5’ flanking region of Upactin1 of U. prolifera is a much stronger promoter than the foreign CaMV 35S (Wu et al., 2018).
The core region and length of the promoter influence the activity and complexity of the vector construction. In the present study, we analyzed the activity of different regions of pPyACT1. The promoter activities of the pPyACT5 and pPyACT6 regions were lower than that of pPyACT4. The activity of pPyACT6 was significantly lower than that of pPyACT5. According to sequence analysis, pPyACT5 was located approximately 500 bp from the TSS, which contained core elements such as CAAT box, TATA box, and G-box, and thus it could be inferred that pPyACT5 was the core region of the promoter. PPyACT6 has some responsive elements involved in light, abscisic acid, auxin, drought, and the CAT-box is involved in meristem-specific expression; hence, it may be an upstream regulatory region. The pPyACT5 and pPyACT6 regions of pPyACT4 are essential. The fragment pPyACT4 with a short length can be used to construct an expression vector for gene function research in N. yezoensis.
The PBI121 vector is a binary vector generated via Agrobacterium-mediated transformation in plants, and T-DNA can incorporate the inserted fragment into the host genome at random sites through the T-DNA LB and RB borders in the plant (Chen et al., 2003). Herein, foreign genes were inserted between the LB and RB borders in the PBI121 vector. Southern blotting analysis confirmed that the PyGUS gene was randomly integrated into the genome by transformation with particle bombardment. Few reports are available on the Agrobacterium-mediated transformation of N. yezoensis. Thus, particle bombardment could be an efficient method for delivering the vector with T-DNA LB and RB borders into the N. yezoensis genome, which would result in stable genetic transformation. This finding also suggests the possibility of developing an insertion mutant library for N. yezoensis. We also found that the entire circular plasmids with PyGUS might be stably maintained in transformed cells and inherited by subsequent generations. Interestingly, some studies have reported that plasmid DNA has been isolated from red macroalgae, including N. tenera and N. yezoensis (Goff and Coleman, 1990; Uji et al., 2014). Therefore, the vector used in the present study was possibly present in cells as an entire circular structure, except when integrated into the nuclear genome. The specific integration site will be investigated using resequencing technology in future studies.
In conclusion, the activity of the pPyACT1, pPyUBC, pPyDPE2, and pPyEF1-a promoters were compared, and pPyACT1 could be used in the genetic transformation of N. yezoensis for the highest expression levels. High transformation efficiency was achieved by optimizing the transformation parameters of the particle bombardment of N. yezoensis and thalli age, and PyHygR-resistant genes provide efficient selection markers for stable transformation. This optimized genetic transformation toolbox can deliver foreign fragments for integration in the nuclear genome and generate stable transformants, which will provide a foundation for functional genomics research and molecular genetic breeding of N. yezoensis.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/genbank/, PRJNA589917.
Author Contributions
XC, FK, and YM conceived the study. XC, BS, JY, and HR performed the experiments. XC, FK, DW,GD and YM analyzed the results. XC, HY, CY, and XT analyzed the data. HY, CY contributed to the software operation. XC wrote the manuscript. FK, XC, YM revised the manuscript. XT, GD, DW contributed to the revised version of the prof. All authors listed have made a substantial, direct, and intellectual contribution to the work and approved it for publication.
Funding
This manuscript and described work have been financed by the Shandong Province Key Research and Development Program (Grant No. 2019GHY112008), Shandong Province Key Research and Development Program (Grant No: 2021LZGC004), National Key Research and Development Program of China (Grant No: 2018YFD0900106), Natural Science Natural Science Foundation of China (Grant No. 41976146, Grant No. 31672641), National Natural Science Foundation of China (Grant No.32060829), National Key R&D Program of China (2020YFD0901101),the 2020 Research Program of Sanya Yazhou Bay Science and Technology City (No. SKJC-2020-02-009) and the Innovation Platform for Academicians of Hainan Province.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2022.856790/full#supplementary-material
References
Bhatnagar-Mathur P., Vadez V., Sharma K. K. (2008). Transgenic Approaches for Abiotic Stress Tolerance in Plants: Retrospect and Prospects. Plant Cell Rep. 27 (3), 411–424. doi: 10.1007/s00299-007-0474-9
Blomme J., Liu X., Jacobs T. B., De Clerck O. (2021). A Molecular Toolkit for the Green Seaweed Ulva Mutabilis. Plant Physiol. 186 (3), 1442–1454. doi: 10.1093/plphys/kiab185
Bowman J. L., Sakakibara K., Furumizu C., Dierschke T. (2016). Evolution in the Cycles of Life. Annu. Rev. Genet. 50 (1), 133–154. doi: 10.1146/annurev-genet-120215-035227
Cheney D., Metz B., Stiller J. (2001). Agrobacterium-Metiated Genetic Transformation in the Macroscopic Marine Red Alga Porphyra Yezoensis. J. Phycol. 37 (s3), 11–11. doi: 10.1111/j.1529-8817.2001.jpy37303-22.x
Chen N., Tang L., Guan X., Chen R., Cao M., Mao Y., et al. (2020). Thallus Sectioning as an Efficient Monospore Release Method in Pyropia Yezoensis (Bangiales, Rhodophyta). J. Appl. Phycol. 32 (4), 2195–2200. doi: 10.1007/s10811-019-01992-6
Chen P.-Y., Wang C.-K., Soong S.-C., To K.-Y. (2003). Complete Sequence of the Binary Vector Pbi121 and its Application in Cloning T-DNA Insertion From Transgenic Plants. Mol. Breed. 11 (4), 287–293. doi: 10.1023/A:1023475710642
Chen L., Zhang S., Beachy R. N., Fauquet C. M. (1998). A Protocol for Consistent, Large-Scale Production of Fertile Transgenic Rice Plants. Plant Cell Rep. 18 (1), 25–31. doi: 10.1007/s002990050526
Daozhan Y., Song Q., Guoqiong S., Chengkui Z. (2002). Transient Expression of lacZ Reporter Gene in the Economic Seaweed Undaria Pinnatifida. High Technol. Lett. 12 (8), 93–95. doi: 10.2753/CSH0009-4633350347
Davies J. P., Weeks D. P., Grossman A. R. (1992). Expression of the Arylsulfatase Gene From the β2 -Tubulin Promoter in Chlamydomonas Reinhardtii. Nucleic Acids Res. 20 (12), 2959–2965. doi: 10.1093/nar/20.12.2959
Demirer G. S., Zhang H., Matos J. L., Goh N. S., Cunningham F. J., Sung Y., et al. (2019). High Aspect Ratio Nanomaterials Enable Delivery of Functional Genetic Material Without DNA Integration in Mature Plants. Nat. Nanotechnol. 14 (5), 456–464. doi: 10.1038/s41565-019-0382-5
Dhir S., Knowles K., Pagan C., Mann J., Dhir S. (2010). Optimization and Transformation of Arundo Donax L. Using Particle Bombardment. Afr. J. Biotechnol. 9, 6460–6469. doi: 10.5897/AJB09.776
Fukuda S., Mikami K., Uji T., Park E.-J., Ohba T., Asada K., et al. (2008). Factors Influencing Efficiency of Transient Gene Expression in the Red Macrophyte Porphyra Yezoensis. Plant Sci. 174 (3), 329–339. doi: 10.1016/j.plantsci.2007.12.006
Gan S. Y., Qin S., Othman R. Y., Phang S. M. (2003a) Development of a Transformation System for Gracilaria Changii (Gracilariales, Rhodophyta), a Malaysian Red Alga via Microparticle Bombardment. J Appl Phycol 15 (4), 345–349. doi: 10.1023/A:1025141913156
Gan S., Qin S., Othman D. R., Yu D., Phang S.-M. (2003b). Transient Expression of lacZ in Particle Bombarded Gracilaria Changii (Gracilariales, Rhodophyta). J. Appl. Phycol. 15, 345–349. doi: 10.1023/A:1025141913156
Gao D., Kong F., Sun P., Bi G., Mao Y. (2018). Transcriptome-Wide Identification of Optimal Reference Genes for Expression Analysis of Pyropia Yezoensis Responses to Abiotic Stress. BMC Genomics 19 (1), 251. doi: 10.1186/s12864-018-4643-8
Goff L. J., Coleman A. W. (1990). Red Algal Plasmids. Curr. Genet. 18 (6), 557–565. doi: 10.1007/BF00327028
Hirata R., Takahashi M., Saga N., Mikami K. (2011). Transient Gene Expression System Established in Porphyra Yezoensis Is Widely Applicable in Bangiophycean Algae. Mar. Biotechnol. 13 (5), 1038–1047. doi: 10.1007/s10126-011-9367-6
Howe A., Sato S., Dweikat I., Fromm M., Clemente T. (2006). Rapid and Reproducible Agrobacterium-Mediated Transformation of Sorghum. Plant Cell Rep. 25 (8), 784–791. doi: 10.1007/s00299-005-0081-6
Huang X., Weber J. C., Hinson T. K., Mathieson A. C., Minocha S. C. (1996). Transient Expression of the GUS Reporter Gene in the Protoplasts and Partially Digested Cells of Ulva Lactuca L. (Chlorophyta). Botanica Mar. 39 (1-6), 467–474. doi: 10.1515/botm.1996.39.1-6.467
Huddy S., Meyers A., Coyne V. (2012). Transformation of lacZ Using Different Promoters in the Commercially Important Red Alga, Gracilaria Gracilis. Afr. J. OF. Biotechnol. 11, 1879–1885. doi: 10.5897/AJB11.2641
Jefferson R. A., Kavanagh T. A., Bevan M. W. (1987). GUS Fusions: Beta-Glucuronidase as a Sensitive and Versatile Gene Fusion Marker in Higher Plants. EMBO J. 6 (13), 3901–3907. doi: 10.1002/j.1460-2075.1987.tb02730.x
Jiang W., Brueggeman A. J., Horken K. M., Plucinak T. M., Weeks D. P. (2014). Successful Transient Expression of Cas9 and Single Guide RNA Genes in Chlamydomonas Reinhardtii. Eukary. Cell 13 (11), 1465–1469. doi: 10.1128/ec.00213-14
Jiang G.-Z., Lü Y.-M., Niu X.-L., Xue L.-X. (2005). The Actin Gene Promoter-Driven Bar as a Dominant Selectable Marker for Nuclear Transformation of Dunaliella Salina. Yi. Chuan Xue. Bao. 32, 424–433.
Jiang P., Qin S., Tseng C. K. (2003). Expression of the lacZ Reporter Gene in Sporophytes of the Seaweed Laminaria Japonica (Phaeophyceae) by Gametophyte-Targeted Transformation. Plant Cell Rep. 21 (12), 1211–1216. doi: 10.1007/s00299-003-0645-2
Kakinuma M., Ikeda M., Coury D. A., Tominaga H., Kobayashi I., Amano H. (2009). Isolation and Characterization of the rbcS Genes From a Sterile Mutant of Ulva Pertusa (Ulvales, Chlorophyta) and Transient Gene Expression Using the rbcS Gene Promoter. Fish. Sci. 75 (4), 1015–1028. doi: 10.1007/s12562-009-0101-5
Keymer A., Pimprikar P., Wewer V., Huber C., Brands M., Bucerius S. L., et al. (2017). Lipid Transfer From Plants to Arbuscular Mycorrhiza Fungi. eLife 6, e29107. doi: 10.7554/eLife.29107
Kikutani S., Nakajima K., Nagasato C., Tsuji Y., Miyatake A., Matsuda Y. (2016). Thylakoid Luminal θ-Carbonic Anhydrase Critical for Growth and Photosynthesis in the Marine Diatom Phaeodactylum Tricornutum. Proc. Natl. Acad. Sci. 113, 9828 (35), 9828–9833. doi: 10.1073/pnas.1603112113
Kong F., Cao M., Sun P., Liu W., Mao Y. (2015). Selection of Reference Genes for Gene Expression Normalization in Pyropia Yezoensis Using Quantitative Real-Time PCR. J. Appl. Phycol. 27 (2), 1003–1010. doi: 10.1007/s10811-014-0359-6
Kong F., Zhao H., Liu W., Li N., Mao Y. (2017). Construction of Plastid Expression Vector and Development of Genetic Transformation System for the Seaweed Pyropia Yezoensis. Mar. Biotechnol. 19 (2), 147–156. doi: 10.1007/s10126-017-9736-x
Kurtzman A. M., Cheney D. P. (1991). Direct Gene Transfer and Transient Expression in a Marine Red Alga Using the Biolistic Method. J. Phycol. 27, 42.
Kwak S.-Y., Lew T. T. S., Sweeney C. J., Koman V. B., Wong M. H., Bohmert-Tatarev K., et al. (2019). Chloroplast-Selective Gene Delivery and Expression in Planta Using Chitosan-Complexed Single-Walled Carbon Nanotube Carriers. Nat. Nanotechnol. 14 (5), 447–455. doi: 10.1038/s41565-019-0375-4
Lim J.-M., Ahn J.-W., Hwangbo K., Choi D.-W., Park E.-J., Hwang M. S., et al. (2013). Development of Cyan Fluorescent Protein (CFP) Reporter System in Green Alga Chlamydomonas Reinhardtii and Macroalgae Pyropia Sp. Plant Biotechnol. Rep. 7 (3), 407–414. doi: 10.1007/s11816-013-0274-3
Louis J., Lorenc-Kukula K., Singh V., Reese J., Jander G., Shah J. (2010). Antibiosis Against the Green Peach Aphid Requires the Arabidopsis Thaliana MYZUS PERSICAE-INDUCED LIPASE1 Gene. Plant J. 64 (5), 800–811. doi: 10.1111/j.1365-313X.2010.04378.x
Ma X., Yao L., Yang B., Lee Y. K., Chen F., Liu J. (2017). RNAi-Mediated Silencing of a Pyruvate Dehydrogenase Kinase Enhances Triacylglycerol Biosynthesis in the Oleaginous Marine Alga Nannochloropsis Salina. Sci. Rep. 7 (1), 11485. doi: 10.1038/s41598-017-11932-4
Mei K., Su-juan W., Yao L., Da-leng S., Cheng-kui Z. (1998). Transient Expression of Exogenous Gus Gene in Porphyra Yezoensis (Rhodophyta). Chin. J. Oceanol. Limnol. 16 (1), 56–61. doi: 10.1007/BF02849081
Meng S., Chen X., Wang J., Fan L., Qiu L., Zheng Y., et al. (2021). Interaction Effects of Temperature, Light, Nutrients, and pH on Growth and Competition of Chlorella Vulgaris and Anabaena Sp. Strain PCC. Front. Environ. Sci. 9. doi: 10.3389/fenvs.2021.690191
Mikami K., Hirata R., Takahashi M., Uji T., Saga N. (2011). “Transient Transformation of Red Algal Cells: Breakthrough Toward Genetic Transformation of Marine Crop Porphyra Species,” in Genetic Transformation, Japan: InTech. 241–258.
Mikami K., Uji T., Li L., Takahashi M., Yasui H., Saga N. (2009). Visualization of Phosphoinositides via the Development of the Transient Expression System of a Cyan Fluorescent Protein in the Red Alga Porphyra Yezoensis. Mar. Biotechnol. 11 (5), 563–569. doi: 10.1007/s10126-008-9172-z
Mizukami Y., Hado M., Kito H., Kunimoto M., Murase N. (2004). Reporter Gene Introduction and Transient Expression in Protoplasts of Porphyra Yezoensis. J. Appl. Phycol. 16 (1), 23–29. doi: 10.1023/B:JAPH.0000019050.54703.14
Peng J., Dong F., Xu Q., Xu Y., Qi Y., Han X., et al. (2006). Orthogonal Test Design for Optimization of Supercritical Fluid Extraction of Daphnoretin, 7-Methoxy-Daphnoretin and 1,5-Diphenyl-1-Pentanone From Stellera Chamaejasme L. And Subsequent Isolation by High-Speed Counter-Current Chromatography. J. Chromatogr. A. 1135 (2), 151–157. doi: 10.1016/j.chroma.2006.08.099
Rathus C., Birch R. G. (1992). Optimization of Conditions for Electroporation and Transient Expression of Foreign Genes in Sugarcane Protoplasts. Plant Sci. 81 (1), 65–74. doi: 10.1016/0168-9452(92)90025-H
Rochaix J. D. (2013). “Chlamydomonas Reinhardtii,”. Brenner’s. Ency. Genet. (Second. Edition). Eds. S. Maloy. K. Hughes. (San Diego: Acad. Press). 521-524.
Saga N., Kitade Y. (2002). Porphyra: A Model Plant in Marine Sciences. Fish. Sci. 68 (sup2), 1075–1078. doi: 10.2331/fishsci.68.sup2_1075
Schroda M., Blöcker D., Beck C. F. (2000). The HSP70A Promoter as a Tool for the Improved Expression of Transgenes in Chlamydomonas. Plant J. 21 (2), 121–131. doi: 10.1046/j.1365-313x.2000.00652.x
Sizova I., Fuhrmann M., Hegemann P. (2001). A Streptomyces Rimosus aphVIII Gene Coding for a New Type Phosphotransferase Provides Stable Antibiotic Resistance to Chlamydomonas Reinhardtii. Gene 277 (1), 221–229. doi: 10.1016/S0378-1119(01)00616-3
Slattery S. S., Diamond A., Wang H., Therrien J. A., Lant J. T., Jazey T., et al. (2018). An Expanded Plasmid-Based Genetic Toolbox Enables Cas9 Genome Editing and Stable Maintenance of Synthetic Pathways in Phaeodactylum Tricornutum. ACS Synthet. Biol. 7 (2), 328–338. doi: 10.1021/acssynbio.7b00191
Son S. H., Ahn J.-W., Uji T., Choi D.-W., Park E.-J., Hwang M. S., et al. (2012). Development of an Expression System Using the Heat Shock Protein 70 Promoter in the Red Macroalga, Porphyra Tenera. J. Appl. Phycol. 24 (1), 79–87. doi: 10.1007/s10811-011-9652-9
Takahashi M., Uji T., Saga N., Mikami K. (2010). Isolation and Regeneration of Transiently Transformed Protoplasts From Gametophytic Blades of the Marine Red Alga Porphyra Yezoensis. Electron. J. Biotechnol. 13 (2), 8–9. doi: 10.2225/vol13-issue2-fulltext-7
Takumi S., Shimada T. (1996). Production of Transgenic Wheat Through Particle Bombardment of Scutellar Tissues: Frequency is Influenced by Culture Duration. J. Plant Physiol. 149 (3), 418–423. doi: 10.1016/S0176-1617(96)80143-X
Torney F., Moeller L., Scarpa A., Wang K. (2007). Genetic Engineering Approaches to Improve Bioethanol Production From Maize. Curr. Opin. Biotechnol. 18 (3), 193–199. doi: 10.1016/j.copbio.2007.03.006
Uji T., Hirata R., Fukuda S., Mizuta H., Saga N. (2014). A Codon-Optimized Bacterial Antibiotic Gene Used as Selection Marker for Stable Nuclear Transformation in the Marine Red Alga Pyropia Yezoensis. Mar. Biotechnol. 16 (3), 251–255. doi: 10.1007/s10126-013-9549-5
Uji T., Mizuta H., Saga N. (2013). Characterization of the Sporophyte-Preferential Gene Promoter From the Red Alga Porphyra Yezoensis Using Transient Gene Expression. Mar. Biotechnol. 15 (2), 188–196. doi: 10.1007/s10126-012-9475-y
Uji T., Takahashi M., Saga N., Mikami K. (2010). Visualization of Nuclear Localization of Transcription Factors With Cyan and Green Fluorescent Proteins in the Red Alga Porphyra Yezoensis. Mar. Biotechnol. 12 (2), 150–159. doi: 10.1007/s10126-009-9210-5
Walker T. L., Collet C., Purton S. (2005). ALGAL TRANSGENICS IN THE GENOMIC Era1. J. Phycol. 41 (6), 1077–1093. doi: 10.1111/j.1529-8817.2005.00133.x
Wang D., Yu X., Xu K., Bi G., Cao M., Zelzion E., et al (2020). Pyropia yezoensis Genome Reveals Diverse Mechanisms of Carbon Acquisition in the Intertidal Environment. Nat. Commun. 11, 4028. doi: 10.1038/s41467s-020-17689-1
Wang J., Jiang P., Cui Y., Deng X., Li F., Liu J., et al. (2010a). Genetic Transformation in Kappaphycus Alvarezii Using Micro-Particle Bombardment: A Potential Strategy for Germplasm Improvement. Aquacul. Int. 18 (6), 1027–1034. doi: 10.1007/s10499-010-9320-0
Wang J., Jiang P., Cui Y., Guan X., Qin S. (2010b). Gene Transfer Into Conchospores of Porphyra Haitanensis (Bangiales, Rhodophyta) by Glass Bead Agitation. Phycologia 49 (4), 355–360. doi: 10.2216/09-91.1
Westwood J. H., Groen S. C., Du Z., Murphy A. M., Anggoro D. T., Tungadi T., et al. (2013). A Trio of Viral Proteins Tunes Aphid-Plant Interactions in Arabidopsis Thaliana. PloS One 8 (12), e83066. doi: 10.1371/journal.pone.0083066
Wu C., Jiang P., Guo Y., Liu J., Zhao J., Fu H. (2018). Isolation and Characterization of Ulva Prolifera Actin1 Gene and Function Verification of the 5′ Flanking Region as a Strong Promoter. Bioengineered 9 (1), 124–133. doi: 10.1080/21655979.2017.1325041
Ye B. S., Scorza R., cordts, Sanford (1994). Genetic Transformation of Peach Tissues by Particle Bombardment. J. Am. Soc. Hortic. Sci. 119, 367–373. doi: 10.21273/JASHS.119.2.367
Zhang Y., Su J., Duan S., Ao Y., Dai J., Liu J., et al. (2011). A Highly Efficient Rice Green Tissue Protoplast System for Transient Gene Expression and Studying Light/Chloroplast-Related Processes. Plant Methods 7 (1), 30–30. doi: 10.1186/1746-4811-7-30
Keywords: Neopyropia yezoensis, endogenous promoters, selection marker, stable transformation, particle bombardment
Citation: Cao X, Kong F, Sun B, Yin J, Ren H, Yue H, Yu C, Tang X, Du G, Wang D and Mao Y (2022) A Toolbox for Constructing a Stable Genetic Transformation Platform Allowing Foreign Fragment Integration in the Genome of Neopyropia yezoensis. Front. Mar. Sci. 9:856790. doi: 10.3389/fmars.2022.856790
Received: 17 January 2022; Accepted: 28 March 2022;
Published: 13 May 2022.
Edited by:
Hao Chen, Institute of Oceanology (CAS), ChinaReviewed by:
Wenlei Wang, Jimei University, ChinaKyle J. Lauersen, King Abdullah University of Science and Technology, Saudi Arabia
Henry Taunt, University College London, United Kingdom
Copyright © 2022 Cao, Kong, Sun, Yin, Ren, Yue, Yu, Tang, Du, Wang and Mao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Fanna Kong, Zm5rb25nQG91Yy5lZHUuY24=; Yunxiang Mao, eXhtYW9AaG50b3UuZWR1LmNu
 Xuexue Cao
Xuexue Cao Fanna Kong
Fanna Kong Bin Sun
Bin Sun Jiqiang Yin1,2
Jiqiang Yin1,2 Yunxiang Mao
Yunxiang Mao