- 1Institute of Marine Environment and Ecology, National Taiwan Ocean University, Keelung, Taiwan
- 2Center of Excellence for the Oceans, National Taiwan Ocean University, Keelung, Taiwan
The transient impact of flooding on the community composition of marine picoeukaryotes (PEs, cell size ≤5 μm) in the East China Sea (ECS) was revealed in this study. In a summer without flooding (i.e., July 2009), photosynthetic picoeukaryotes (PPEs) were more abundant in the area covered by the Changjiang River diluted water (CDW, salinity ≤31) than in the non-CDW affected area. According to the 18S ribosomal RNA phylogeny, Alveolata (all from the superclass Dinoflagellata) was the main community component accounting for 72 to 99% of the community at each sampling station during the nonflooded summer. In addition to Dinoflagellata, diatoms or Chlorophyta also contributed a considerable proportion to the PE assemblage at the stations close to the edge of CDW coverage. In July 2010, an extreme flooding event occurred in the Changjiang River basin and led to the CDW covering nearly half of the ECS. In the flooded summer, the abundance of PPEs in the CDW-covered area decreased significantly to less than 1 × 104 cells ml-1. Compared to that during the nonflooded summer, the diversity of the PE composition was increased. While Dinophyceae still dominated the surface waters, Syndiniophyceae, which were represented by the uncultured Marine Alveolata Group (MALV)-I and MALV-II, accounted for a substantial amount in the Dinoflagellata superclass relative to this community composition in the nonflooded summer. Furthermore, a variety of plankton, including Cryptophyta, Haptophyta, Picobiliphyta, the uncultured Marine Stramenopiles (MASTs) and heterotrophic nanoflagellates, were observed. The nutrition modes of these PEs have been reported to be mixotrophic or heterotrophic. Therefore, it was inferred that the potentially mixotrophic and heterotrophic PE compositions might be favored in the marginal sea in the flooded summer.
Introduction
The distribution and succession of picoplankton (cell size ≤2 µm; recent studies defined it as ≤5 µm) are easily affected by abrupt environmental events because of their tiny cell sizes and various physiological characteristics. According to the existence of a cell nucleus, picoplankton can be divided into prokaryotic picoplankton and eukaryotic picoplankton (also known as picoeukaryotes, PEs). Picocyanobacteria (mainly Synechococcus and Prochlorococcus) are important prokaryotic picoplankton in the ocean. There have been many studies describing their responses to sudden environmental changes. For example, it has been reported that sudden environmental events, such as the passage of tropical cyclones, heavy atmospheric deposition associated with dust storms, or abundant terrestrial material inputs derived from flooding events, not only promote the transient growth of Synechococcus species but also alter their community assemblage (Chang et al., 1996; Herut et al., 2005; Chen et al., 2009; Chung et al., 2011; Chung et al., 2014; Chung et al., 2015). However, there is still a lack of relevant field studies on the relationship between PE assemblage composition and sudden environmental changes. The taxonomic assemblages of PEs are more diverse than those of picocyanobacteria. In addition, PEs exhibit versatile nutritional strategies (i.e., phototrophy, mixotrophy, and heterotrophy), allowing them to cope with changes in the surrounding environment (Worden et al., 2015). Regarding the phototrophic mode, photosynthetic picoeukaryotes (PPEs) and picocyanobacteria are important primary producers in the microbial carbon cycle in marine ecosystems. The abundance of PPEs is usually lower than that of picocyanobacteria by approximately one to two orders of magnitude; however, their contributions to total primary production are almost equivalent (Worden et al., 2004; Jardillier et al., 2010). Another large group of PEs are heterotrophic and play the role of primary consumers in the microbial food web (Stoeck et al., 2010; McKie-Krisberg and Sanders, 2014; Unrein et al., 2014; Worden et al., 2015; Leles et al., 2018). In addition to PPEs and heterotrophic PEs, mixotrophic PEs exhibit a dual nutritional strategy within single cells. These organisms shift their trophic mode from phototrophy to bacterivory when the environment is inappropriate for photosynthesis or when the cells lack nutrients (McKie-Krisberg and Sanders, 2014; Worden et al., 2015). In response to environmental changes, alterations in the composition of PEs or the conversion of their nutritional modes might further affect the microbial carbon cycle and the biogeochemical cycles of marine ecosystems (Hoegh-Guldberg and Bruno, 2010; Stoeck et al., 2010; Hilligsøe et al., 2011; Worden et al., 2015; Leles et al., 2018).
Rivers input considerable amounts of fresh water and terrestrial materials into the ocean, providing an important source of nutrients for phytoplankton growth in estuaries and adjacent oceanic areas (Justić et al., 1995; Gong et al., 2006). The Changjiang River (aka. the Yangtze River) is the longest river in Asia, and its input substantially affects the biogeochemical processes of the East China Sea (ECS) shelf. However, the completion and commissioning of the Changjiang River Dam have led to a significant decrease in the supply of fresh water and terrestrial materials to the ECS. The deficiency of the terrestrial supply of nutrients (e.g., silicate) has not only led to a decline in primary productivity but has also caused the dominant phytoplankton to shift from diatoms to dinoflagellates (Gong et al., 2006). However, the injection of large amounts of freshwater and riverine materials derived from a flood that occurred in the Changjiang River Basin stimulated a temporary massive growth of diatoms in the ECS (Gong et al., 2011). Under increasing human activities and the scenario of global environmental change, floods and droughts will likely occur often, especially in Eastern Asia (IPCC, 2021). More dynamic river discharge and loading of terrestrial materials will result in greater impacts on the marine ecosystem. Compared to microplankton (cell size ≥20 μm, e.g., diatoms), picoplankton show a relatively fast response to environmental changes due to their tiny cell size. Previous studies examining the influence of terrigenous substances on marine picoplankton ecology have often been conducted in estuaries (e.g., Paerl et al., 2020), and information from shelf and open ocean areas is still lacking. In the ECS, Tsai et al. (2010) and Lee et al. (2018) both proposed that the abundance and distribution of picoplankton were determined by the degree of the expansion of Changjiang discharge water over the shelf of the ECS. However, the community assemblages of picoplankton and their dynamics during flooding periods have not been comprehensively described.
In the summer of 2010, a devastating flood occurred in the Changjiang River Basin. The consequent freshwater inputs resulted in the expansion of the Changjiang River discharge to over half of the ECS. We have examined the influence of flooding on the ecology of prokaryotic picoplankton in our previous studies (Chung et al., 2014; Chung et al., 2015). Although PEs are larger than prokaryotic picoplankton, the PE community assemblage is difficult to determine based on microscopic observation alone due to the tiny size of these organisms. Recently, phylogenetic analyses of 18S ribosomal RNA genes have been well developed to examine PEs in the oceans at a fine taxonomic resolution (Moon-van der Staay et al., 2001; Massana, 2011). Phylogenetic information combined with hydrographic data will be of great utility in addressing the ecological roles of different PEs and inferring the impacts of environmental changes on their niches in the oceans. This study, combined with our previous findings, will provide more comprehensive information on how floods affect the succession of picoplankton in the ocean.
Materials and Methods
Sample Collection
Two voyages aboard R/V Ocean Researcher I were conducted in the ECS during the periods from 29 June to 13 July 2009 and 6 July to 17 July 2010 (Figure 1A). Temperature and salinity were measured with a conductivity/temperature/depth recorder (CTD) (SBE911 plus, Sea-Bird Electronics, Washington, USA). Water samples for determining the concentrations of nutrients and chlorophyll a and the abundance of picoplankton were collected using Teflon-coated 20-liter Niskin bottles (General Oceanics, Florida, USA) mounted on a CTD rosette sampler. A total of 24 stations located on the ECS shelf were visited. DNA samples collected from the surface (depth=5 m) at the 5 stations (S1 to S5) (Figure 1A) were used to analyze the phylogenetic diversity of 18S rRNA genes.
Determination of Chlorophyll a and Nutrient Concentrations
One liter of water to be used for chlorophyll a analysis was immediately filtered through a GF/F class filter (Whatman, Maidstone, UK), and the filter was stored at -20°C until analysis. Chlorophyll a preserved on the filter was extracted in pure acetone at 4°C overnight in the dark and was measured with a fluorometer (10-AU-005, Turner Design, California, USA) (Welschmeyer, 1994; Gong et al., 1996). To determine the concentrations of dissolved inorganic nutrients, the water sample was placed in a polypropylene bottle and immediately frozen with liquid nitrogen onboard the survey vessel. Nitrite (NO2) and nitrate (NO3) concentrations were measured via the pink azo dye method with a self-designed flow injection analyzer (Strickland and Parsons, 1972; Gong et al., 1996). The concentration of phosphate (PO4) was determined by the molybdenum blue method (Pai et al., 1990; Gong et al., 1996).
Counting of Picoplankton
Samples for picoplankton counting were fixed in seawater-buffered paraformaldehyde at a final concentration of 0.2% (w/v), frozen in liquid nitrogen and kept at -80°C until analysis. Flow cytometry (FACSAria, Becton-Dickinson Co., New Jersey, USA) was applied to distinguish different populations of picoplankton. For picocyanbacteria (i.e., Synechococcus and Prochlorococcus) and small PPEs, abundance estimates and counts were determined according to their cell size (forward- and side-scattering) and their autofluorescence in the orange (from phytoerythrin, 575 ± 15 nm) and red (from chlorophyll, >670 nm) ranges. Heterotrophic bacteria were counted in a separate subsample stained with SYBR-Green I dye (Molecular Probes Inc., Oregon, USA). A known number of fluorescent beads (TruCOUNT Tubes, Becton-Dickson Co., New Jersey, USA) was simultaneously counted to convert the flow rates for determining picoplankton abundance in the sample (Liu et al., 2002; Chung et al., 2015).
DNA Extraction From Small Planktonic Cells
Five liters of the surface water was subsequently filtered through 20 µm-pore size nylon mesh (Sefar, Heiden, Switzerland) and 5 µm-pore size polycarbonate membranes (47 mm diameter) (Nuclepore, Whatman, Maidstone, UK) to remove larger plankton. The cells in the filtrate were harvested using 0.8 µm-pore size polycarbonate membranes (47 mm diameter) (Nuclepore, Whatman, Maidstone, UK) under gentle vacuum (≤100 mmHg). The membranes were frozen in liquid nitrogen and kept at -80°C until DNA extraction. Cells (cell size between 0.8 and 5 µm) retained on the membranes were disrupted by glass beads (0.1 mm diameter) using a bead beater (BioSpec Products, Oklahoma, USA) and then treated with lysis buffer containing 0.1 M EDTA (pH 8.0), 1 mM Tris-HCl (pH 8.0), 0.25% sodium dodecyl sulfate and 0.1 mg ml-1 proteinase K (Roche, Penzberg, Germany) at 55°C overnight. After the removal of detritus and residual polysaccharides by treatment with 1% hexadecyltrimethylammonium bromide (Sigma-Aldrich, Missouri, USA), followed by chloroform extraction and an additional round of phenol/chloroform/isoamyl alcohol (v:v:v=25:24:1) extraction, DNA was precipitated by adding isopropanol. The DNA pellet was dissolved in Tris-EDTA buffer (pH 8.0), and its concentration and purity were determined using a spectrophotometer (NanoDrop, Thermo Scientific, Delaware, USA) at wavelengths of 260 and 280 nm (Chung et al., 2015).
Pyrosequencing
Variable region 4 (V4) of the 18S rRNA gene was amplified by polymerase chain reaction (PCR) with 50 ng DNA as the template. The PCR mixture contained Phusion High-Fidelity DNA polymerase (New England Biolabs, Massachusetts, USA), 1× Phusion HF buffer, a 200 μM deoxynucleotide solution mixture, and both the forward fusion primer “Primer A-MID-TAReukFWD” (5’-Primer A-MID-CCAGCASCYGCGGTAATTCC-3’, MID=multiplex identifier, S=G or C, Y=C or T) and the reverse primer “TAReukREV” (5’-ACTTTCGTTCTTGATYA-3’, Y=C or T, R=A or G) (Stoeck et al., 2010; Tamaki et al., 2011). Unidirectional pyrosequencing was performed only with Primer A (Tamaki et al., 2011). The MIDs were used to assist in sample identification after pyrosequencing. To increase specificity and sensitivity in PCR amplification, touchdown PCR was performed in a thermocycler (Model 9700, Applied Biosystems, New York, USA) under the following conditions: 1 cycle of 98°C for 30 sec; followed by 10 cycles of 98°C for 10 sec, 53°C for 30 sec, and 72°C for 30 sec; then 20 cycles of 98°C for 10 sec, 48°C for 30 sec, and 72°C for 30 sec; and finally, 1 cycle of 72°C for 10 min. The amplicons were subjected to agarose gel electrophoresis and purified using a QIAquick Gel Extraction Kit (Qiagen, California, USA). The concentrations of the purified amplicons (495 base pairs) were measured using a Qubit dsDNA High Sensitive Assay Kit (Thermo Fisher Scientific, Massachusetts, USA) and a fluorometer (Qubit 2.0) (Thermo Fisher Scientific, Massachusetts, USA). Equal molar amounts of all amplicons were combined for pyrosequencing on a Roche 454 Genome Sequencer FLX+ Instrument (454 Life Sciences, Connecticut, USA).
Sequence Processing, Operational Taxonomic Unit (OTU) Definition, and Diversity Analysis
Raw sequences were processed using the algorithms AmpliconNoise (Quince et al., 2011) and PyroNoise (Quince et al., 2009) to reduce the pyrosequencing error rates. Any sequence with more than one mismatch with the MID, more than two mismatches with the primers, or a PCR one-base error was excluded. Chimeras were detected and removed with UCHIME (Edgar et al., 2011). All the resultant sequences were aligned to the SILVA rRNA SEED database (release 138) and clustered into OTUs at a cutoff level of 3% divergence based on the pairwise alignment distance. The taxonomy of each OTU was annotated by the SILVA rRNA reference dataset (release 138) with the Ribosomal Database Project Classifier (Wang et al., 2007) and was subsequently confirmed using BLASTN searches against the GenBank database. The OTUs that were not classified as eukaryotic 18S rRNA genes or affiliated with larger protozoans (i.e., ciliates and radiolarians) and metazoans (e.g., copepods) were excluded from further analysis. The OTUs comprising less than 2 reads (i.e., singletons and doubletons) were also removed. The resultant OTUs were applied for the assessment of richness and diversity indices. Additionally, the OTUs with a relative contribution greater than 0.1% of the total reads in any sample were regarded as abundant OTUs and were applied to investigate the assemblage composition of PEs (Wu et al., 2017). All of the above and α- and β-diversity analyses were implemented using the Mothur software package (version 1.47.0) (Schloss et al., 2009). The phylogenetic relationship between the abundant OTUs in a taxon was interpreted by the maximum likelihood (ML) algorithm with the Kimura two-parameter model (Kimura, 1980). The effects of sampling error on tree inference and the stability of branch nodes were tested by bootstrap analyses with 1,000 repetitions. The software IQ-TREE (version 1.4.4) were used for the phylogeny-related analysis and tree construction (Nguyen et al., 2014). The vegan package in R was used in the ordination analysis.
Data Availability
All of the sequences of 18S rRNA V4 gene fragments were deposited in the Sequence Read Archive (SRA) database with accession numbers SAMN10089074 to SAMN10089083.
Results
Hydrographic Characteristics
The plume of the Changjiang River was small during the summer without flooding in 2009. The Changjiang River diluted water (CDW), which has been defined as water with a temperature ≥23°C and salinity ≤31 (Gong et al., 1996), was restricted to the inshore region of the northwestern ECS (approximately 0.4 × 105 km2). Most surfaces in the ECS were covered with high-temperature, oligotrophic waters from northward-flowing Taiwan Current Warm Water (TCWW) and the intrusion of the Kuroshio Current (Table 1, Figure 1). These conditions represent the typical hydrography of the ECS in nonflooded summers (Gong et al., 2011). In contrast, extreme flooding occurred in the Changjiang River Basin in summer 2010 and led to a large amount of fresh water being injected into the ECS. Through the extension of CDW coverage to the southern ECS (approximately 1.7 × 105 km2), more than half of the surface ECS was fertilized with terrestrial nutrients, resulting in a subsequent increase in the chlorophyll a concentration (Table 1, Figure 1).
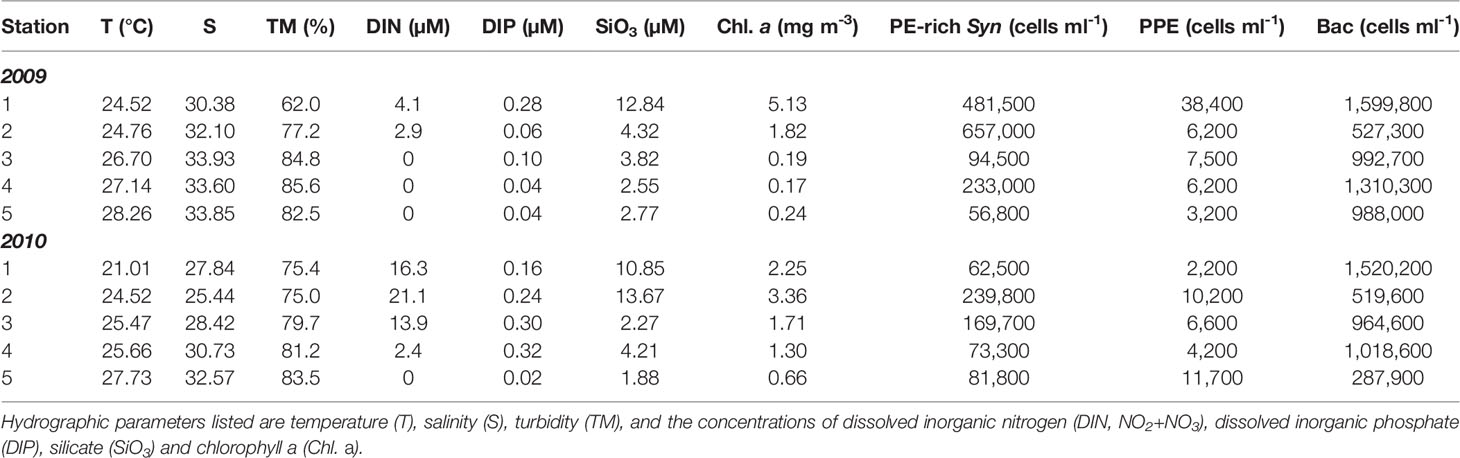
Table 1 The hydrographic properties and the cell numbers of phycoerytherin-rich Synechococcus (PE-rich Syn), photosynthetic picoeukaryotes (PPE) and heterotrophic bacteria (Bac) in the surface waters (depth=5 m) of environmental DNA sampling stations in the summers of 2009 and 2010.
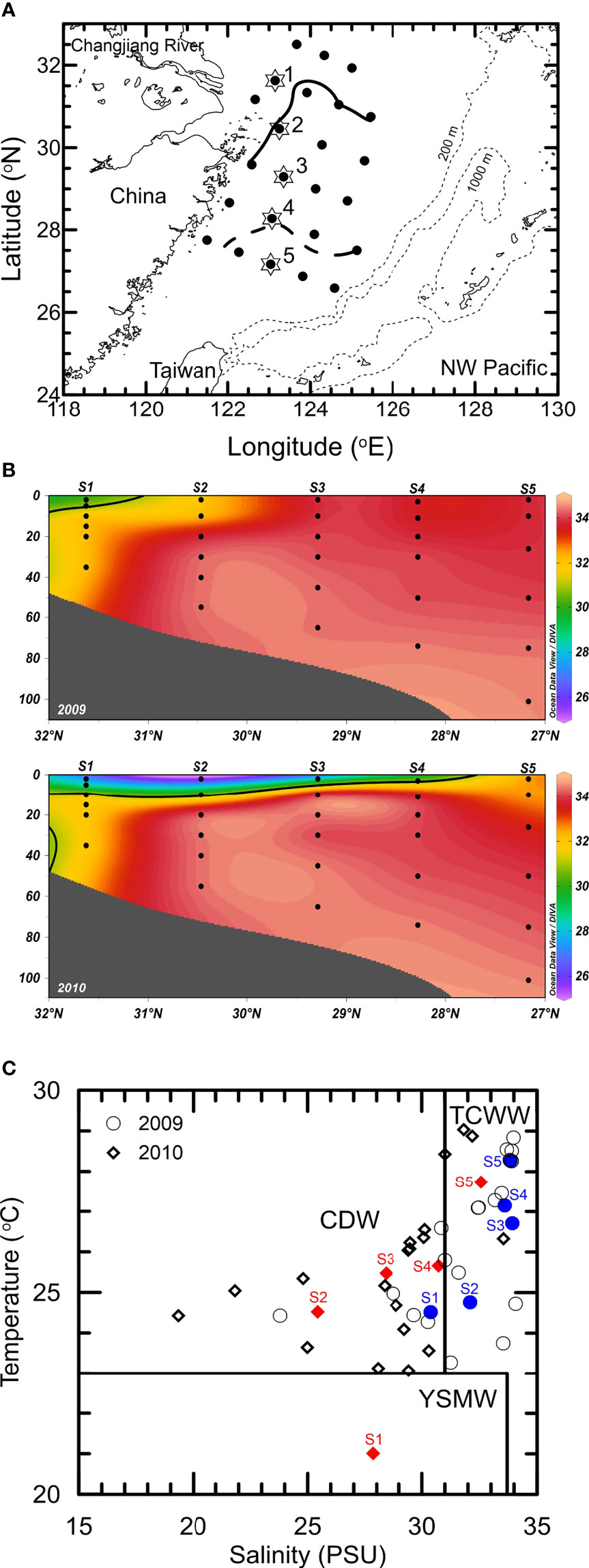
Figure 1 (A) Locations of observation stations in the East China Sea. The solid and dashed lines represent the isohaline of 31 in the summers of 2009 and 2010, respectively. A total of 24 stations were visited during the 2 cruises. The phylogenetic analysis of 18S rRNA genes was performed based on the surface samples of Stations 1 to 5 denoted by star symbols. (B) Vertical distribution of salinity at Stations 1 to 5 in the summers of 2009 and 2010. The isohaline of 31 is indicated by the solid line. (C) Temperature-salinity diagram. Characterization of different water masses, including the Changjiang diluted water (CDW), the Taiwan strait current warm water (TCWW) and the Yellow Sea mixed water (YSMW), was in accordance with the definitions of Gong et al. (1996).
Distribution of Photosynthetic Picoeukaryotes
Among PEs, only PPEs can be detected and counted by flow cytometry due to the presence of chlorophyll. The distribution of PPEs in both summers is presented in Figure 2. Regardless of whether flooding occurred, PPEs were mainly distributed in the coastal areas covered by the CDW. However, the highest count of PPEs in the nonflooded summer was greater than 2.5 × 104 cells ml-1, which was approximately twice as high as the maximum PPE count in the flooded summer. Overall, the abundance of PPEs within the CDW-influenced area in the nonflooded summer was significantly greater than that in the flooded summer (p<0.001) (Figure 2).
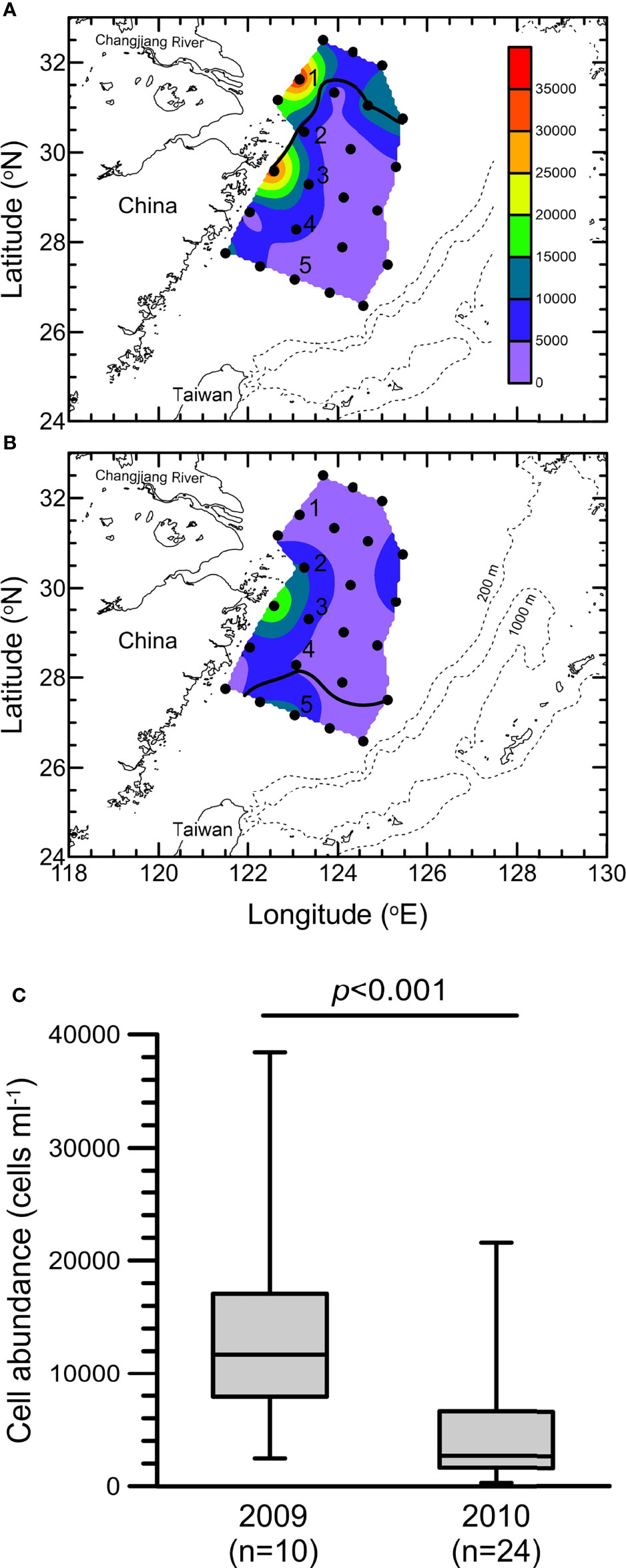
Figure 2 Distribution of photosynthetic picoeukaryotes (PPEs) (cells ml-1) at the surface on the East China Sea shelf in the summers of (A) 2009 and (B) 2010. The solid lines indicate the isohaline of 31. (C) Whisker plot of PPE abundance in the surface waters of the stations confined to the area within the isohaline of 31 (i.e., 10 stations in 2009 and 24 stations in 2010).
Diversity of Picoeukaryotes
According to a previous study of hydrography in the ECS (Gong et al., 1996), we defined Station 1 and Station 2 as CDW-influenced stations and Station 5 as an oceanic station. In addition to Stations 1, 2 and 5, the hydrography of Stations 3 and 4 was susceptible. In the nonflooded summer, Stations 3 and 4 were usually covered with salty, higher-temperature water. Nevertheless, during the flooded summer of 2010, the surfaces of Stations 3 and 4 were overlaid by the extended CDW (Table 1 and Figure 1). Compared to the nonflooded summer of 2009, the hydrographic conditions of the five stations were significantly affected by the occurrence of flooding in the summer of 2010. Therefore, these five stations were selected to further study the impact of flooding on the diversity of the small eukaryotic plankton assemblage.
After the removal of chimeras, singletons, doubletons, and reads belonging to larger protozoans (e.g., ciliates and radiolarians) and metazoans (e.g., copepods), we obtained a total of 960,541 high-quality reads for the 18S rRNA gene phylogenetic analysis. According to the criterion of 3% sequence divergence, these reads were categorized into 2,616 OTUs (Table 2). The rarefaction curves, except for that of Station 2, almost reached a plateau phase, indicating that the sampling efforts were enough to analyze 18S rRNA gene diversity (Supplementary Figure 1). Based on the ACE, Shannon and Pielou indices, the species richness and divergence of PEs in the flooded summer were greater than those in the nonflooded summer overall. The diversity was more pronounced at the CDW-influenced stations (i.e., Stations 1 and 2) than at the other three offshore stations (i.e., Stations 3, 4 and 5) (Table 2, Figure 3).
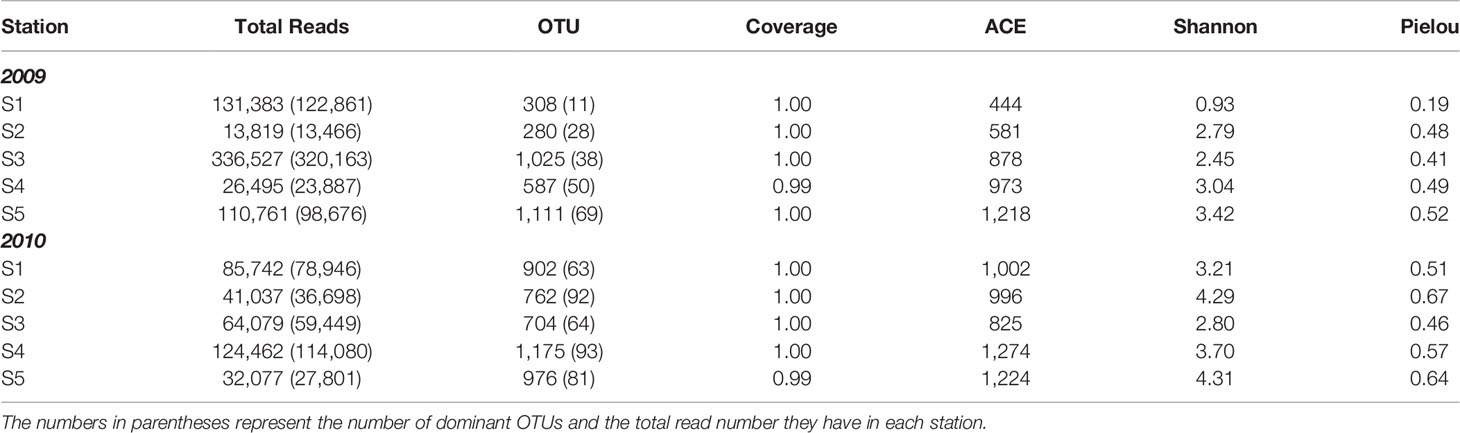
Table 2 Summary of pyrosequencing result of 18S ribosomal RNA gene V4, and the indices of richness estimate (ACE), diversity (Shannon) and evenness (Pielou) in the East China Sea in the summers of 2009 and 2010 (Figure 1).
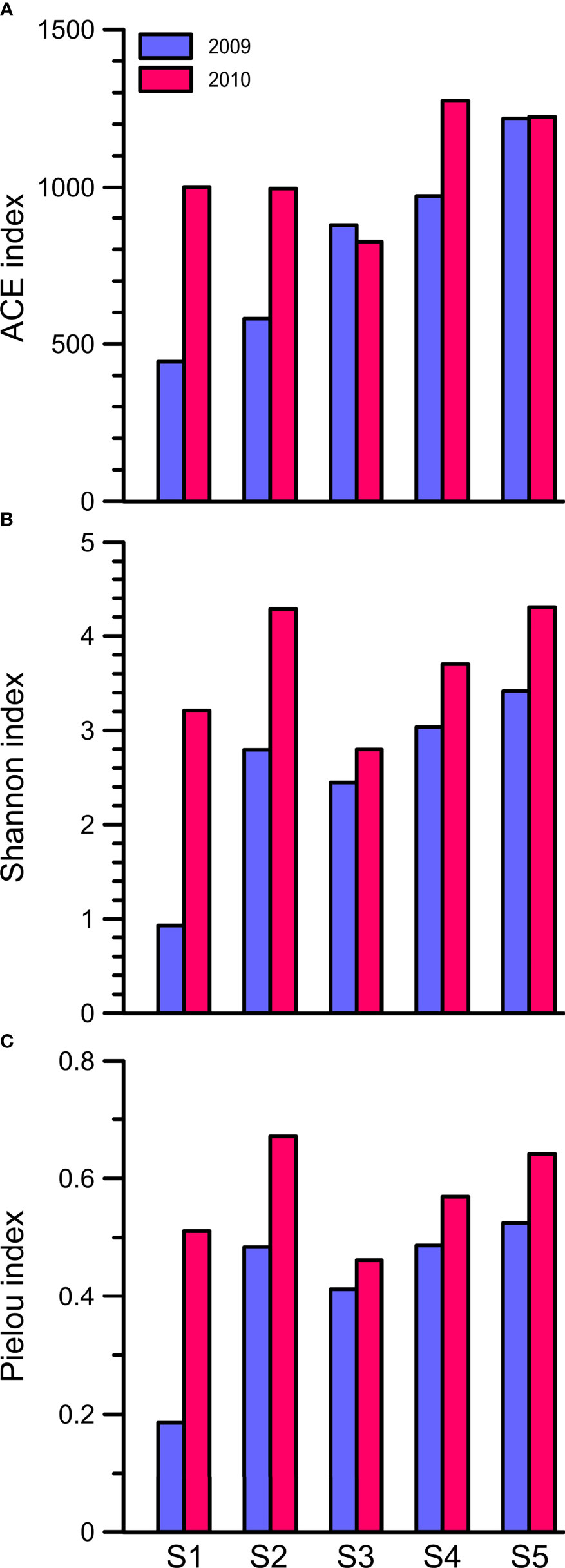
Figure 3 The (A) ACE, (B) Shannon diversity and (C) Pielou evenness indices of the community assemblages of picoeukaryotes, which were inferred from the OTU composition, at Stations 1 to 5 in the summers of 2009 and 2010.
To clarify the impact of floods on the ecology of PEs, a total of 240 abundant OTUs were selected for the subsequent analysis of the community composition. While the abundant OTUs accounted for only approximately one-tenth of the total OTUs, they comprised 902,380 reads, which accounted for 94% of the total reads (Table 2). During the nonflooded summer, the PEs at each station were almost entirely composed of the Alveolata species, except at Station 2 (Figure 4A). In this study, 63 OTUs were grouped into Alveolata. They all belonged to the phylum Dinoflagellata. Among these OTUs, 18 OTUs were grouped into the class Dinophyceae. They were all affiliated with Gymnodinium. OTU-001 accounted for the highest proportion of the community at all stations and was the most important member. The other 45 OTUs were categorized into the class Syndiniophyceae, among which 7 OTUs were affiliated with Euduboscquella spp. (the uncultured Marine Alveolata Group I, MALV-I). In addition to OTU-001, OTU-002 (i.e., Euduboscquella sp.) contributed considerable read numbers, and this group was also suggested to be the major PE at all stations (Figure 5). Euduboscquella sp. is already known to be an intracellular parasite of tintinnid ciliates (Jung et al., 2016). The uncultured Marine Alveolata Group II (MALV-II), which comprised 38 OTUs, was another dominant group within Dinoflagellata. Its abundance accounted for less than 10% of all dominant OTU sequences (Figures 4A, 5). All of the OTUs in MALV-II were Amoebophyra sp., which are syndinean dinoflagellates that parasitize free-living dinoflagellates, such as Gymnodinium (Yih and Coats, 2000). In contrast to the other 3 stations, although Dinoflagellata still accounted for most of the community assemblage at Stations 2 and 3, their proportions declined to 81% and 74%, respectively. Instead, at Station 2, Stramenopiles, mainly represented by two diatoms Fragilariopsis dollolus (OTU-49) and F. kerguelensis (OTU-101) and one uncultured Marine Stramenopile Group 13 (MAST-13) (Bicosoeca vacillans, OTU-316), jointly accounted for approximately 15% of the total reads (Figures 4A, 6). At Station 3, Chlorophyta contributed 25% of the total reads and were primarily composed of Picochlorum sp. (OTU-005) and Chloropicon roscoffensis (OTU-013), which accounted for 20% and 4%, respectively (Figures 4A, 7).
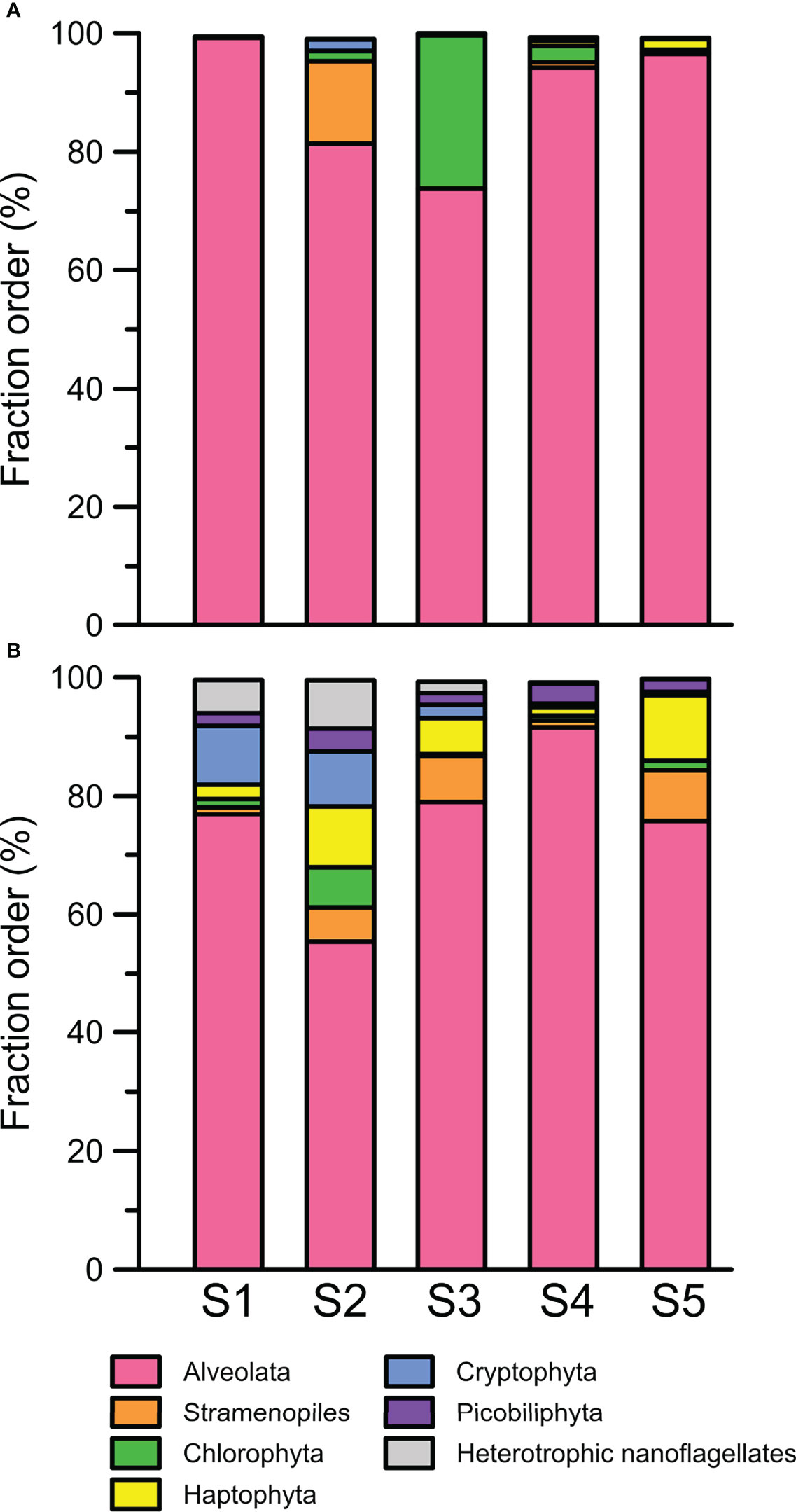
Figure 4 Community compositions of abundant picoeukaryotes indicated at the phylum level at Stations 1 to 5 in the summers of (A) 2009 and (B) 2010.
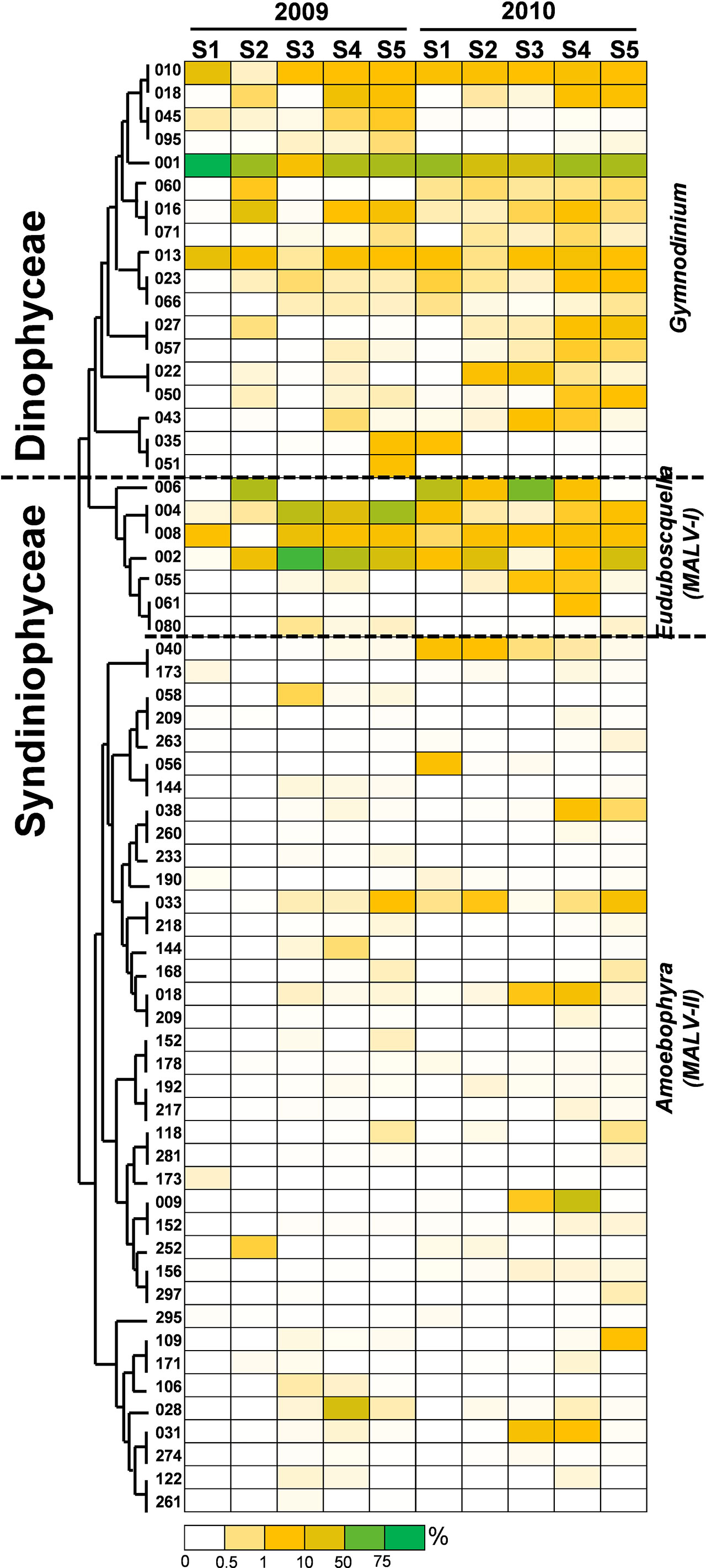
Figure 5 Heatmap representing the relative contribution of each abundant OTU of Alveolata during the summers of 2009 and 2010. All of the abundant OTUs of Alveolata identified in this study were grouped into the phylum Dinoflagellata. After phylogenetic analysis, these OTUs were further assigned to the classes Dinophyceae and Syndiniophyceae. This figure shows that the OTUs of the Dinophyceae were all affiliated with Gymnodinium. The other OTUs of the Syndiniophyceae belonged to Euduboscquella (MALV-I) and Amoebophyra (MALV-II). The relative contribution indicates the percentage of the number of 18S rRNA gene V4 sequences in each OTU. The phylogenetic tree in the left position was constructed using the maximum likelihood algorithm. The labels at the bottom position indicate the color codes of the different percentages.
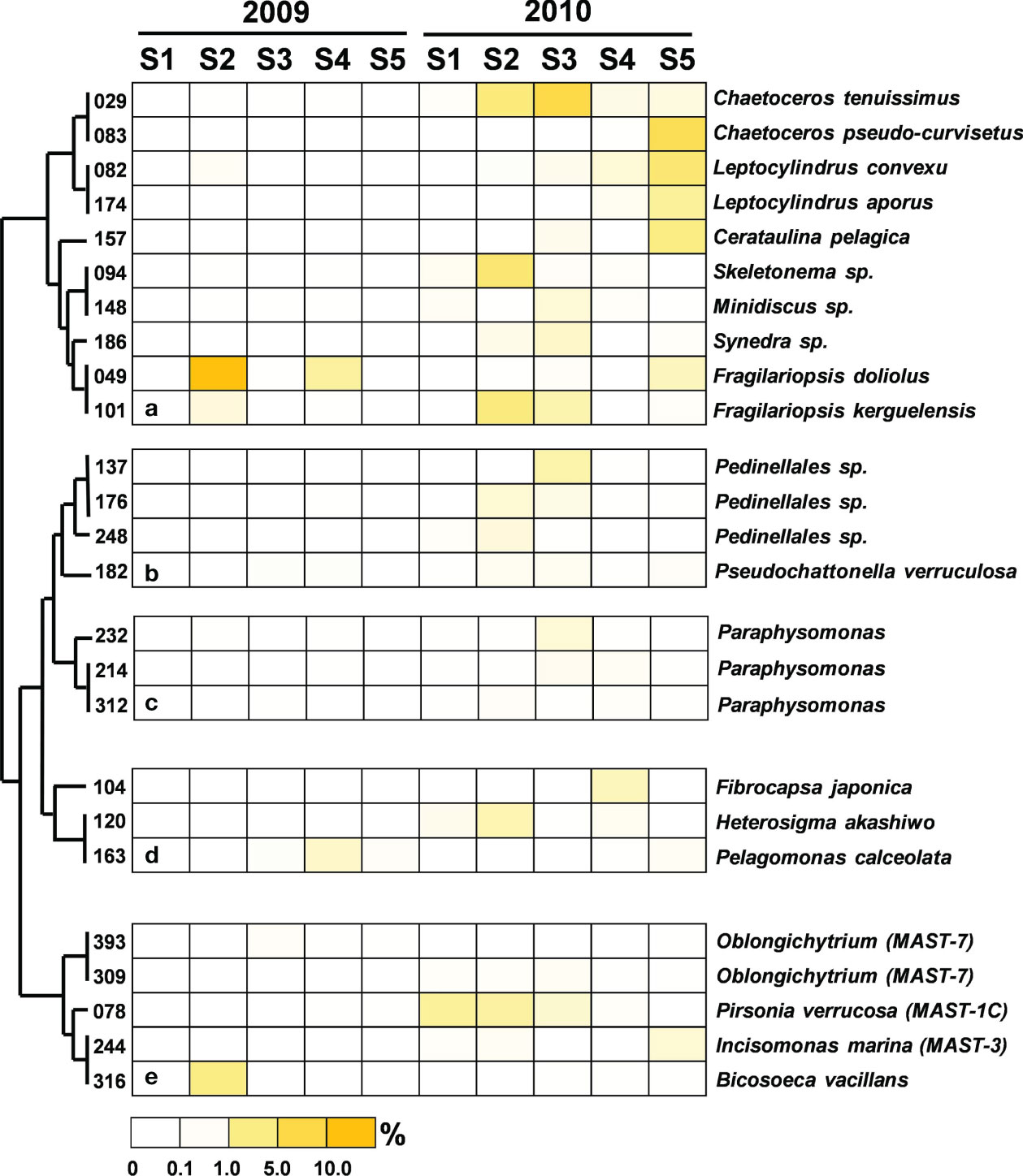
Figure 6 Heatmap representing the relative contribution of each abundant OTU of Stramenopiles during the summers of 2009 and 2010. All of the abundant Stramenopile OTUs identified in this study were grouped into (A) Bacillariophyceae, (B) Dictyochophyceae, (C) Chryosophyceae, (D) other (i.e., F. japonica belonged to Chloromonadophyceae, H. akashiwo belonged to Raphidophyceae, and P. caceolata belonged to Pelagophyceae) or (E) marine Stramenopile lineages (MAST). The relative contribution indicates the percentage of the number of 18S rRNA gene V4 sequences in each OTU. The phylogenetic tree in the left position was built using the maximum likelihood algorithm. The labels at the bottom position indicate the color codes of the different percentages.
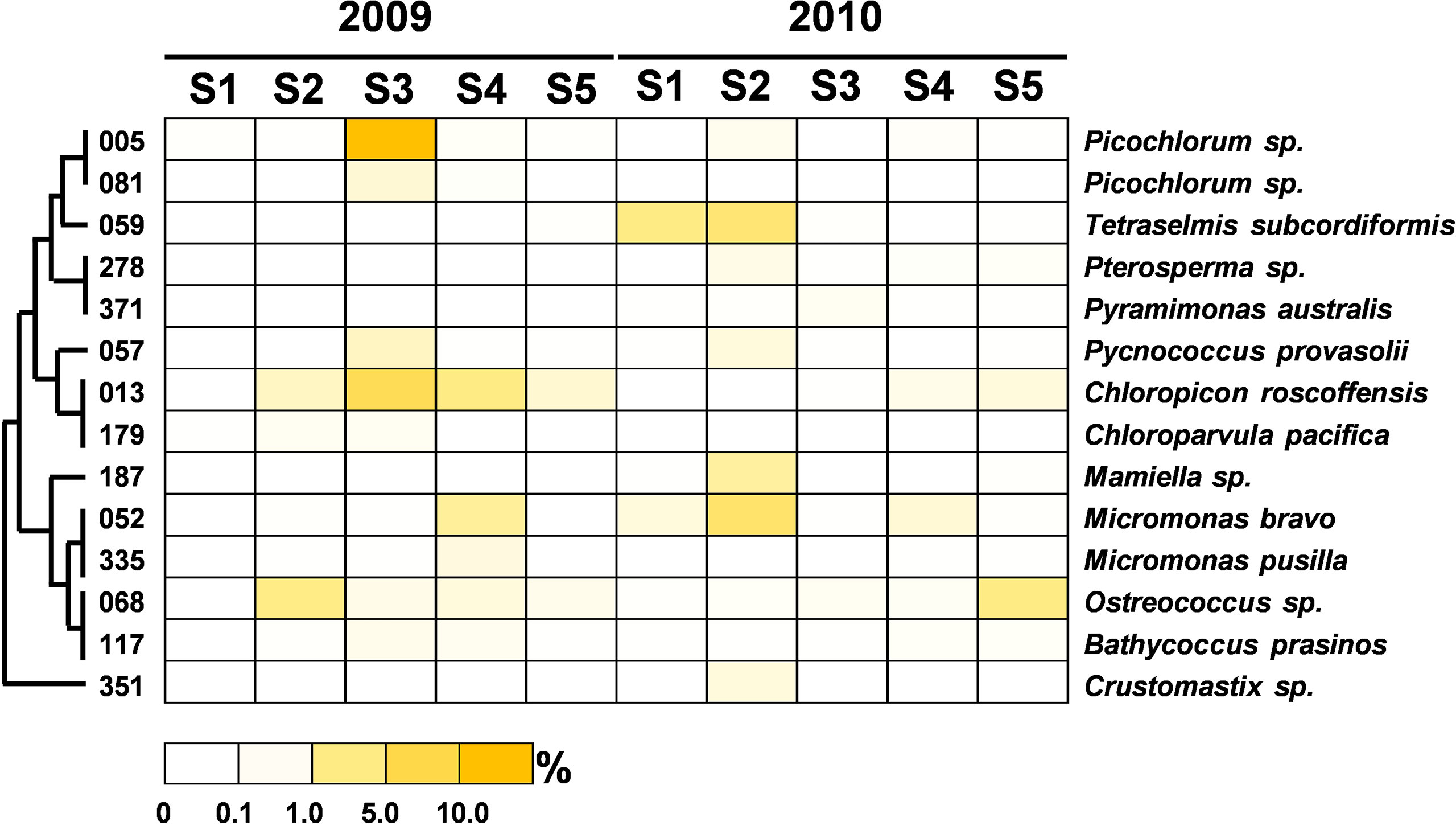
Figure 7 Heatmap representing the relative contribution of each abundant OTU of Chlorophyta during the summers of 2009 and 2010. The relative contribution indicates the percentage of the number of 18S rRNA gene V4 sequences in each OTU. The phylogenetic tree in the left position was built using the maximum likelihood algorithm. The labels at the bottom position indicate the color codes of the different percentages.
The community composition of PEs in the flooded summer was more diverse than that in the nonflooded summer. The proportion of Alveolata declined at all stations except Station 4, falling to 80% at Stations 1 and 5 and approximately 50% at Station 2 (Figure 4B). Although the composition of the Alveolata to that in the nonflooded summer was similar, the ratio of MALV-II at each station increased to 10% (Figure 5). In addition to Alveolata, the other phyla found at all stations were Stramenopiles, Chlorophyta, Haptophyta and Picobiliphyta. Chlorophyta were mainly composed of Tetraselmis subcordiformis (OTU-059), Mamiella sp. (OTU-187), Micromonas bravo (OTU-05) and Ostreococcus sp. (OTU-068). They were widely distributed in various stations with a proportion of less than 10%. Picochlorum and Chloropicon, which were found at Station 3 in the nonflooded summer, almost disappeared in the flooded summer. Moreover, Cryptophyta and heterotrophic nanoflagellates only appeared at the CDW-influenced stations (i.e., Stations 1 and 2) (Figure 4B). Regarding nutritional strategies, the PEs that were found in the flooded summer were suggested to be heterotrophic or mixotrophic. For example, OTU-036 (Chrysochromulina simplex, Haptophyta) (Figure 8) and OTU-019 and OTU-032 (Teleaulax amphioxeia, Cryptophyta) (Figure 9) were mixotrophic. OTU-250 (Picomonas judraskeda, Picobiliphyta) (Figure 10) and several nanoflagellates, such as OTU-063 (Protaspis grandis) and OTU-113 (Cryothecomonas aestivalis) (Figure 11), were heterotrophic. Additionally, OTU-078 (Pirsonia verrucose, MAST-1C, Stramenopiles) and OTU-244 (Incisomonas marina, MAST-3, Stramenopiles) were suggested as parasites for hosting centric diatoms (Figure 6E). The new appearance of heterotrophic and mixotrophic PEs in the flooded summer implied that the microbial cycle should be different from that in the summer without flooding.
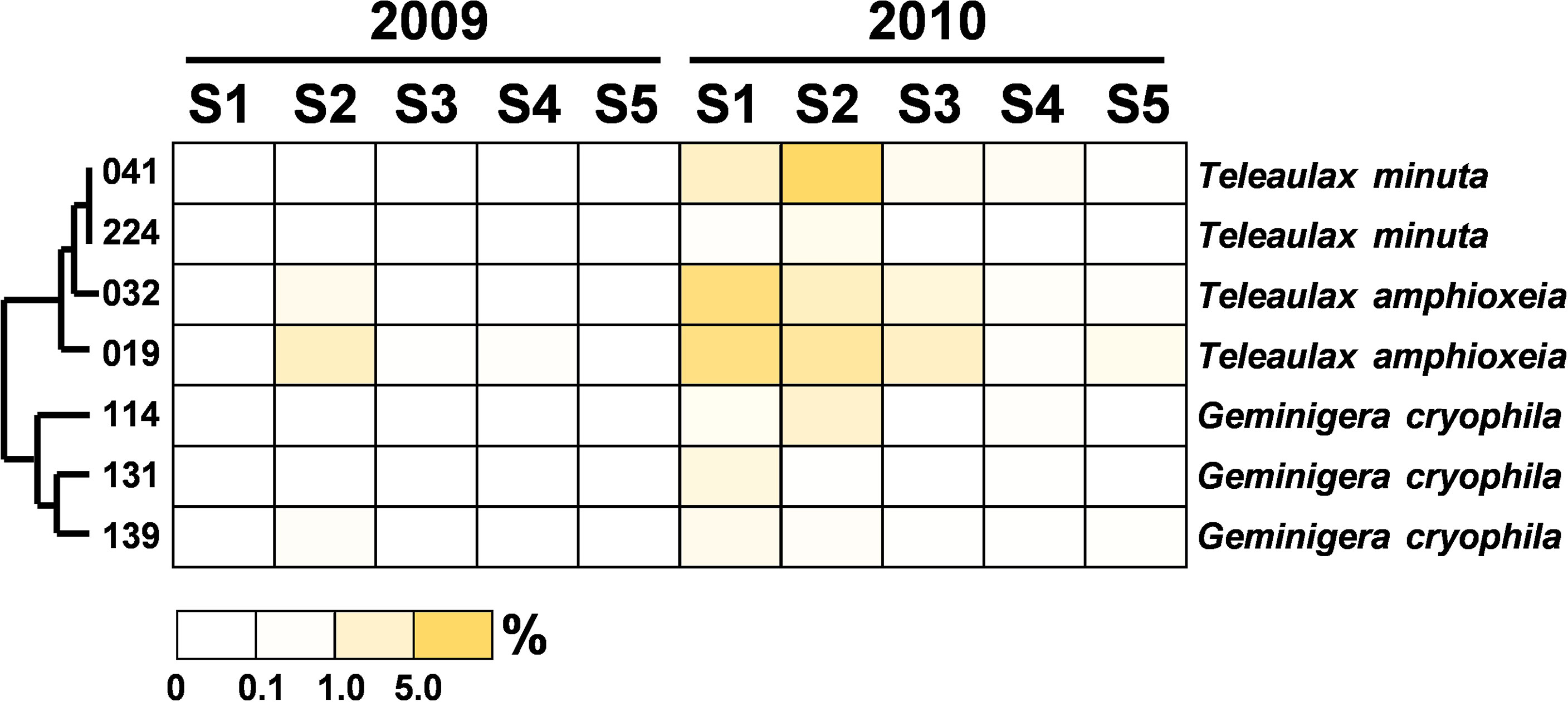
Figure 8 Heatmap representing the relative contribution of each abundant OTU of Haptophyta during the summers of 2009 and 2010. The relative contribution indicates the percentage of the number of 18S rRNA gene V4 sequences in each OTU. The phylogenetic tree in the left position was built using the maximum likelihood algorithm. The labels at the bottom position indicate the color codes of the different percentages.
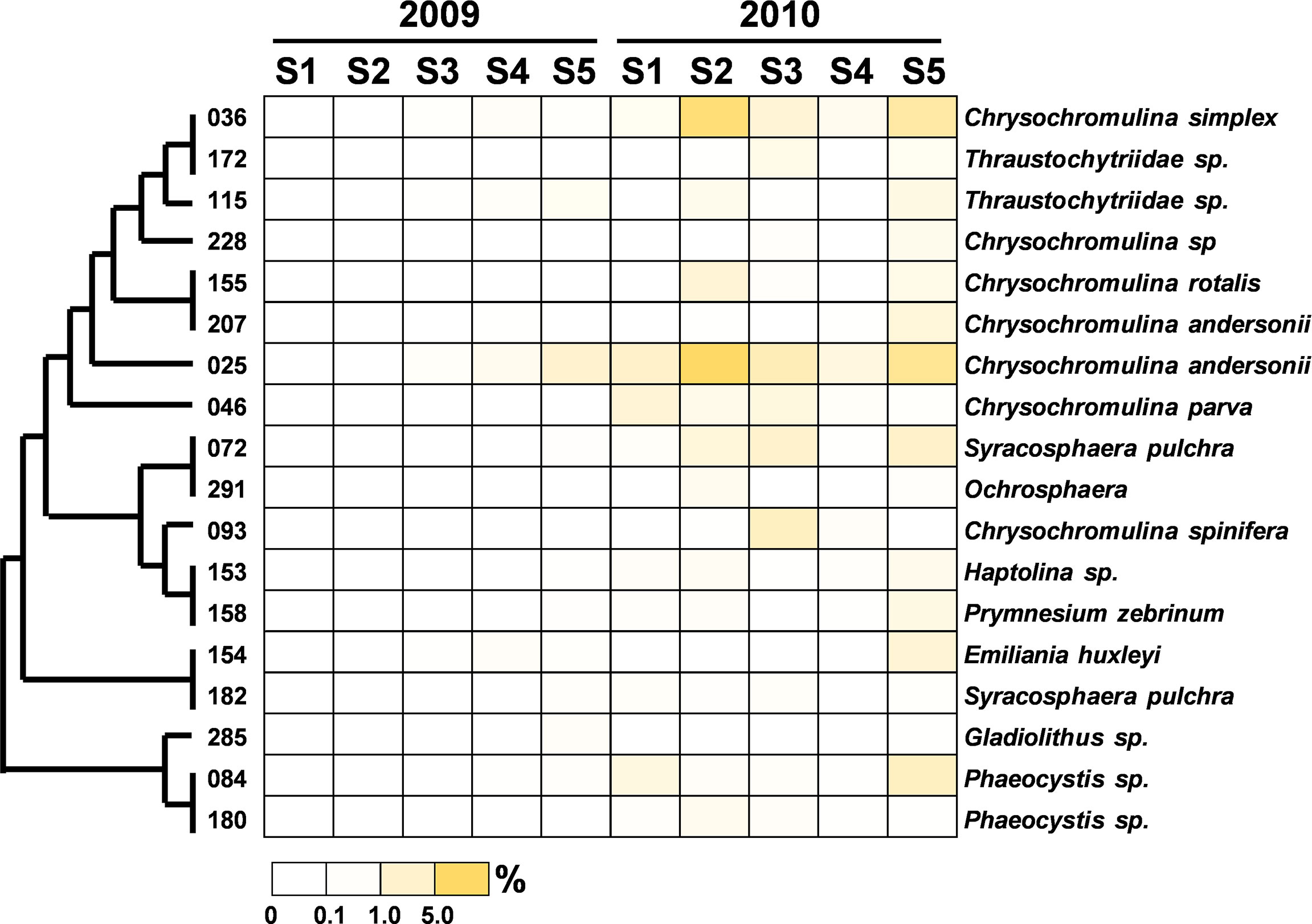
Figure 9 Heatmap representing the relative contribution of each abundant OTU of the Cryptophyta during the summers of 2009 and 2010. The relative contribution indicates the percentage of the number of 18S rRNA gene V4 sequences in each OTU. The phylogenetic tree in the left position was built using the maximum likelihood algorithm. The labels at the bottom position indicate the color codes of the different percentages.
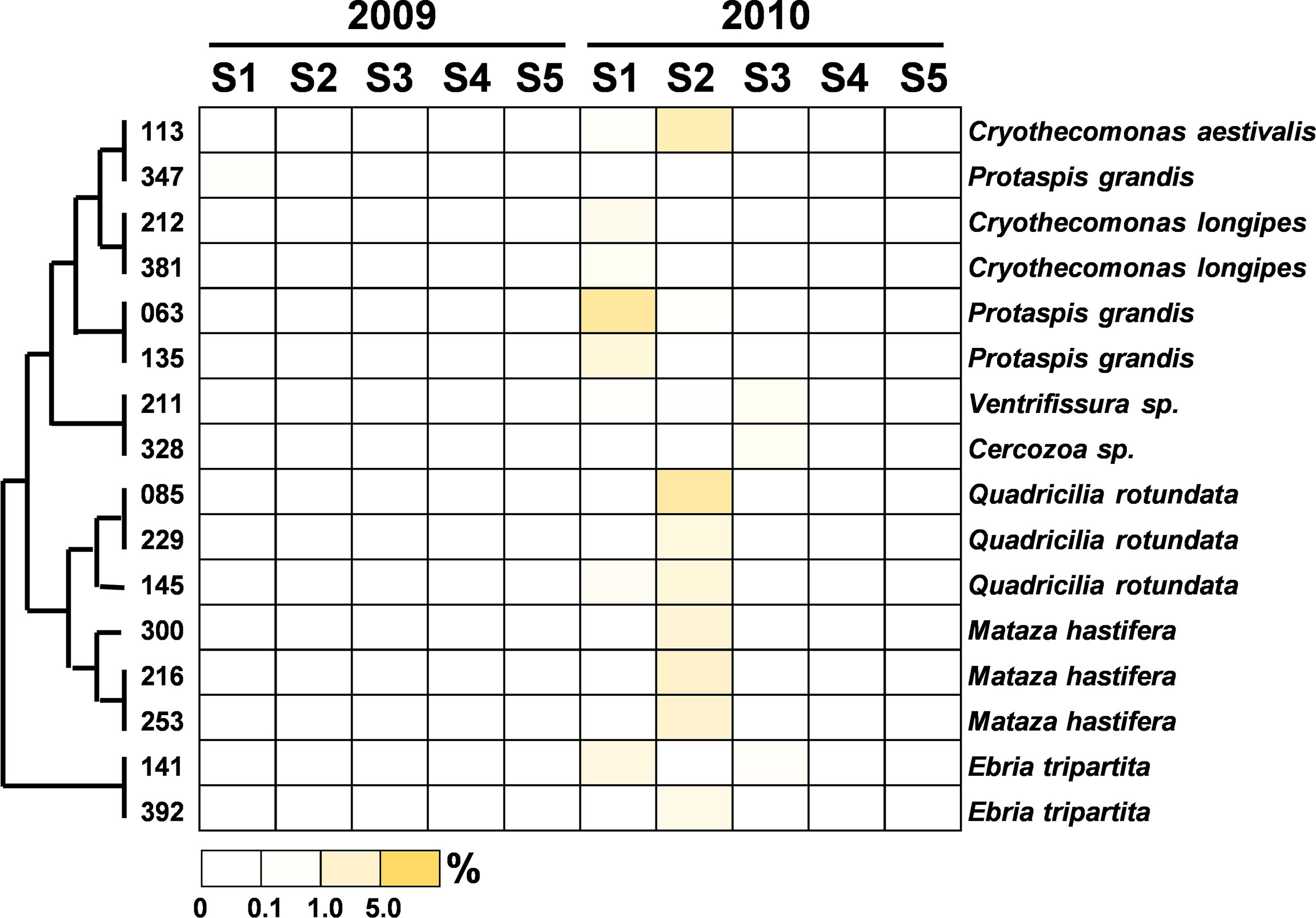
Figure 10 Heatmap representing the relative contribution of each abundant OTU of Picobiliphyta during the summers of 2009 and 2010. The relative contribution indicates the percentage of the number of 18S rRNA gene V4 sequences in each OTU. The phylogenetic tree in the left position was built using the maximum likelihood algorithm. The labels at the bottom position indicate the color codes of the different percentages.
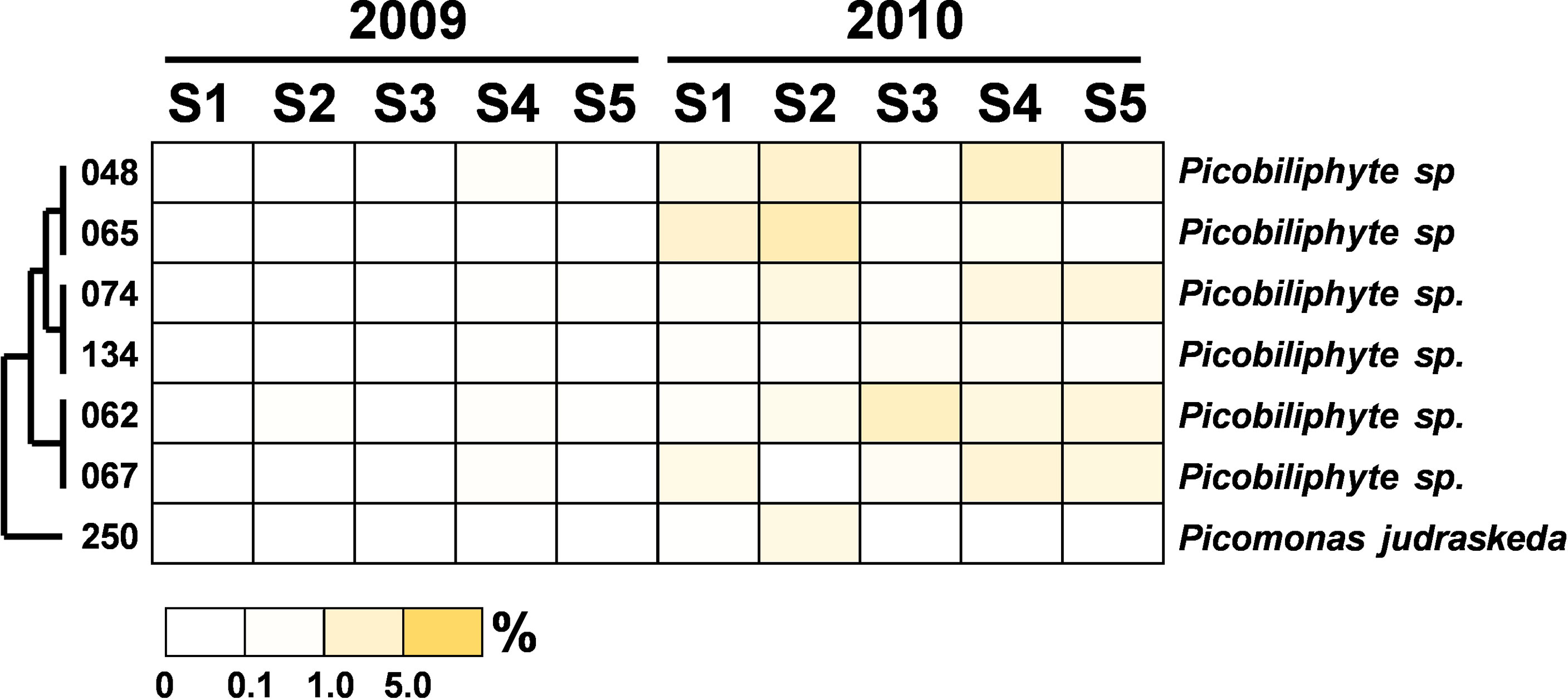
Figure 11 Heatmap representing the relative contribution of each abundant OTU of the heterotrophic nanoflagellates during the summers of 2009 and 2010. The relative contribution indicates the percentage of the number of 18S rRNA gene V4 sequences in each OTU. The phylogenetic tree in the left position was built using the maximum likelihood algorithm. The labels at the bottom position indicate the color codes of the different percentages.
Discussion
The clustering analysis of the PE assemblage composition at each station and the correlation with environmental parameters are shown in Figure 12. The correlation results indicate that salinity reduction and the input of terrestrial dissolved inorganic nitrogen (DIN) were possible factors promoting the transient growth of Cryptophyta and HNFs at the inshore stations (i.e., Stations 1 and 2). The rise in turbidity resulted in increases in Picobiliphyta and MALV-II at the offshore stations (i.e., Stations 3, 4 and 5). Moreover, the abundance of Stramenopiles (e.g., MAST-1C) and Haptophyta (e.g., Chrysochromulina simplex) both increased to account for considerable proportions of the total PE composition at each station in the flooded summer. Although there were obvious differences in the species composition at each station in the nonflooded and flooded summers, no consistent inference was obtained based on the correlation analysis between the species composition and various environmental factors (Figure 12). The term “picoeukaryotes” generally refers to eukaryotic plankton with cell sizes smaller than 2 or 5 μm. Their community composition is very complicated, and each species possesses unique biological characteristics. It is still difficult to elucidate the relationships among their succession, nutritional modes and ambient parameters (Worden et al., 2015). Our results in this study also supported this interpretation. The combination of systematic studies of single species with more comprehensive sampling of marine PE biodiversity in diverse marine environments will enable significant progress in the comparison and clarification of the relationships between PE activities and environmental factors (Worden et al., 2015).
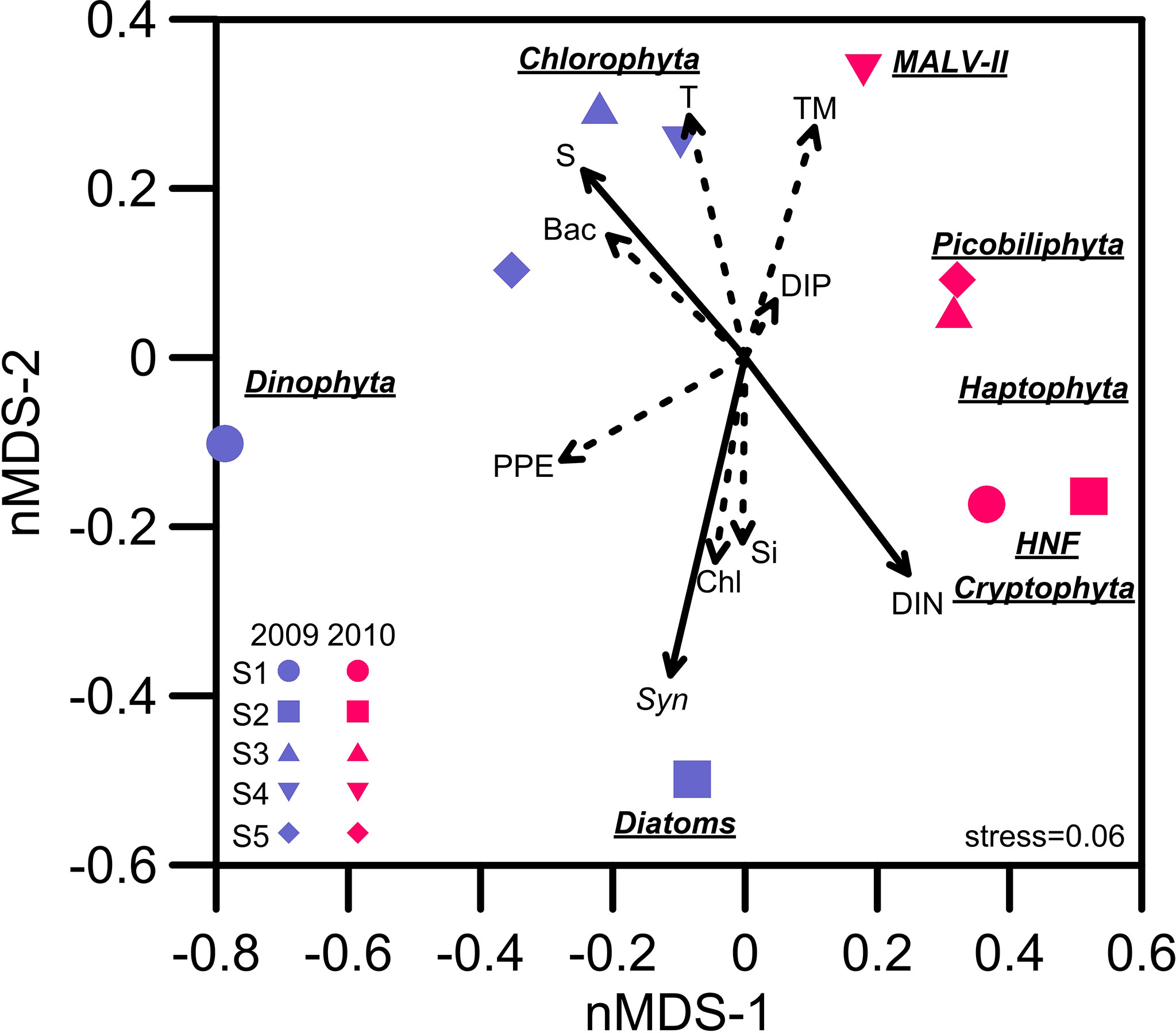
Figure 12 Nonmetric multidimensional scaling (nMDS) ordination plot of the dominant picoeukaryote assemblages at Stations 1 to 5 in the summers of 2009 and 2010. The correlation relationships between the picoeukaryote assemblages and environmental factors among all stations were assessed with the function “envfit” (number of permutations=999) in the R package “vegan”. The solid and dashed lines indicate significance (p value) at the ≤0.01 and ≤0.05 levels, respectively. S, salinity; T, temperature; TM, turbidity; DIP, dissolved inorganic phosphate; DIN, dissolved inorganic nitrogen (NO2+NO3); Si, silicate; Chl, chlorophyll a; Syn, Synechococcus; PPE, photosynthetic picoeukaryotes; Bac, bacteria.
Picoplankton succession responds quickly to changes in the environment. Therefore, when a sudden environmental event occurs, picoplankton composition shows a temporarily higher degree of divergence (e.g., Herut et al., 2005; Chen et al., 2009; Chung et al., 2011; Chung et al., 2015). The newly emerging PE populations in the flooded summer were more diverse and possessed versatile nutrition modes, mostly tending toward heterotrophy or mixotrophy. We found that Teleaulax amphioxeia was the main member of Cryptophyta at the inshore stations after the flood (Figure 9). It has been reported that T. amphioxeia is a mixotroph that shifts its nutritional mode from photosynthesis to predation when the abundance of Synechococcus or bacteria increases (Yoo et al., 2017). Accordingly, grazing by T. amphioxeia was one of the possible factors contributing to the decrease in the number of Synechococcus in the flooded summer (Chung et al., 2014; Chung et al., 2015). T. amphioxeia also served as an excellent prey for nanoflagellates and ciliates (Nishitani et al., 2010; Hernández-Urcera et al., 2018). Compared to that occurring in the nonflooded summer, the PE niches in the microbial food web were more differentiated, and the process of the pelagic ecosystem should have been more complicated when the flooding occurred.
Mixotrophic haptophytes have been considered important bacterivores in oligotrophic coastal waters (Unrein et al., 2014; Chan et al., 2018). We found that haptophytes occupied a significant proportion of the picoplankton assemblage at each station in the flooded summer. Among them, Chrysochromulina spp. were the major members. Chrysochromulina have been identified as mixotrophic haptophytes and showed high phagotrophic ability in phosphate-starved or low-irradiance conditions (Kawachi et al., 1991; Jones et al., 1993). Søgaard et al. (2021) reported that an under-ice algal bloom dominated by mixotrophic haptophytes (i.e., Chrysochromulina and Prymnesium) occurred in the early spring in the Arctic. The authors deduced that freshwater input derived from the melting ice, subsequently resulting in nutrient dilution and low light intensity, jointly led to the thriving of Chrysochromulina and Prymnesium. However, previous studies on Chrysochromulina were all carried out in high-latitude areas. Relevant research works on these species have not been performed in subtropical oceans. Based on the results of the correlation analysis (Figure 12) and the previous studies mentioned above, it was inferred that the salinity decline derived from the flooding led to the emergence of Chrysochromulina spp.
The operation of the Three Gorges Dam drastically decreased territorial silicate loading into the ECS, resulting in a regime shift in the dominant phytoplankton species from diatoms to dinoflagellates (Gong et al., 2006). A similar distribution of phytoplankton was observed in this study. Regarding the relationship between Stramenopiles distribution and flooding, during the nonflooded period, only a higher proportion of diatoms could be found at Station 2. By contrast, diatoms prevailed at every station in the flooded summer. 18S rRNA sequences from large chain-form diatoms (e.g., Chaetoceros tenuissimus, Leptocylindrus convexu and Cerataulins pelagica, etc.) found in our picoplankton dataset might be attributed to cell rupture during filtration. However, this also suggested that more diatoms should be present in the waters after the input of terrestrial materials (Chung et al., 2012). In addition to diatoms, some heterotrophic lineages of MAST (i.e., MAST-1C, MAST-3 and MAST-7) (Logares et al., 2012; Labarre et al., 2021) were found after flooding. Among them, Pirsonia verrucosa (OTU-076) (MAST-1C) and the diatom C. tenuissimus appeared concurrently at Stations 1 to 3 (Figure 6). Pirsonia are parasitic flagellates that are specifically hosted by marine-centric diatoms (Kühn et al., 1996; Kühn et al., 2004). A similar co-occurrence of MAST-3 and diatoms was observed at Station 5 (Figure 6). MAST-3 has poor mobility, and it was speculated that epiphytism or parasitism should be one of its lifestyles (Cavalier-Smith and Scoble, 2013; Gómez et al., 2010; Massana et al., 2014; Seeleuthner et al., 2018). Solenicola setigera (MAST-3I) has been confirmed to be parasitic to diatoms (Gómez et al., 2011). Although relative evidence is still lacking, Incisomonas marina (OTU-244) (MAST-3J) was suggested to also follow the same life strategy (Cavalier-Smith and Scoble, 2013; Seeleuthner et al., 2018). The cooccurrence of MAST-1C or MAST-3 accompanied diatoms implied that diatom abundance in the flooded summer should be controlled by these parasitic flagellates at this time.
Due to its high growth rate and tolerance to environmental changes, Picochlorum (Chlorophyta) has been widely used in the algae-related biotechnology industry (Foflonker et al., 2018; Gonzalez-Esquer et al., 2019; Krishnan et al., 2021). However, the ecology of Picochlorum in the oceans has not been studied. Genomic analysis conducted with multiple Picochlorum isolates revealed that allelic diversity and horizontal gene transfer might be the mechanisms for shaping their adaptation to variable environments (Foflonker et al., 2018). Picochlorum sp., occupied approximately 20% of the total reads at Station 3 in the nonflooded summer. A similar distribution of Picochlorum in the ECS was also found in summer 2019, when the Changjiang River plume was small. The emergence of Picochlorum should be attributed to the relatively stable hydrography (high temperature, high salinity and low DIN) in the summers without disturbances. This situation seems to be at odds with the high adaptability of Picochlorum to fluctuating environmental conditions. Pang et al. (2022) first confirmed that Picochlorum sp. (GLMF1 strain) was mixotrophic. Its bacterivory activity increased when the ambient conditions became more suitable for photosynthesis. The operation of a dual trophic strategy (phago-phototrophy) might benefit Picochlorum growth on the ECS shelf in the interference-free summer.
The development of high-throughput sequencing in the past decade has allowed us to comprehensively analyze 18S rRNA gene diversity so that we are able to explore the community composition of PEs in depth. However, with regard to protists, large differences in the copy numbers of rRNA genes among various species may lead to deviations in the assessment of PE abundance and the contribution of various PEs to the ecosystem (Zhu et al., 2005). The cell debris of larger plankton or small particles attached to their nuclear DNA fragments mixed in the sample will contribute to mischaracterization of the PE composition (Sørensen et al., 2013; Chan et al., 2020). Moreover, it is difficult to determine the taxonomic identity of PEs and simultaneously evaluate their nutritional modes. In response to these problems, high-throughput 18S rRNA gene sequencing combined with fluorescent in situ hybridization (FISH) can more accurately assess PE assemblages in the field (Unrein et al., 2014). Alternatively, the application of other index genes (e.g., rbcL) provides a feasible way to assess the community composition of PEs in the field (Meakin and Wyman, 2011).
The quantity of nutrients and freshwater carried by river flows affects the phytoplankton composition and primary productivity in marginal seas (Smith and Demaster, 1966; Rabalais et al., 1996; Gong et al., 2006; Gong et al., 2011). Under the scenario of global warming and increasing human activities, the entry of riverine materials into seas will become more dynamic, in turn affecting biogeochemical cycles on the continental shelf (Gong et al., 2006; Gong et al., 2011; Chou et al., 2013; IPCC, 2021). While various in situ studies of the impact of extreme weather events on phytoplankton ecology in the northwest Pacific have been performed (Chang et al., 1996; Chen et al., 2009; Gong et al., 2011; Chung et al., 2014; Chung et al., 2015), our knowledge of how marine ecosystems respond to global climate changes and increasing human activities is still insufficient. During the flooded summer of 2010, we observed that PE diversity increased and that more 18S rRNA reads from bacterivores appeared, although the detailed mechanism still needs to be elucidated. It might be beneficial to deliver organic carbon from the microbial cycle to the grazing food chain (Tsai et al., 2010; Stibor et al., 2019; Gilbert and Mitra, 2022). Our findings provide insight into the change in the community composition of PEs between the dry period and flooding period over the large continental shelf system in the NW Pacific. This information will facilitate a complete assessment of the impact of freshwater discharge on changing the energy flow in the microbial food web.
Data Availability Statement
All of the sequences of 18S rRNA V4 gene fragments were deposited in the Sequence Read Archive (SRA) Database with accession numbers SAMN10089074 to SAMN10089083.
Author Contributions
C-CC and G-CG designed the cruises and the sampling work. C-CC and C-WH performed the high-throughput sequencing. C-CC, G-CG, and Y-CL analyzed the hydrographic and sequencing data. C-CC wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by the Ministry of Science and Technology of Taiwan (MOST109-2611-M-019-004 and MOST110-2611-M-019-004) and the Center of Excellence for the Oceans of National Taiwan Ocean University. Y-CL was supported by the Research Scholar fellowship (MOST110-2611-M-019-018) from the Ministry of Science and Technology of Taiwan.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank the captain and crew of R/V Ocean Researcher I for their assistance. We also thank the National Core Facility for Biopharmaceuticals (NCFB) and the National Center for High-performance Computing (NCHC) of National Applied Research Laboratories (NARLabs) for providing computational resources.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2022.853847/full#supplementary-material
References
Cavalier-Smith T., Scoble J. M. (2013). Phylogeny of Heterokonta: Incisomonas marina, A Uniciliate Gliding Opalozoan Related to Solenicola (Nanomonadea), and Evidence That Actinophryida Evolved From Raphidophytes. Eur. J. Protistol. 49, 328–353. doi: 10.1016/j.ejop.2012.09.002
Chan Y.-F., Chung C.-C., Gong G.-C., Hsu C.-W. (2020). Spatial Variation of Abundant Picoeukaryotes in the Subtropical Kuroshio Current in Winter. Mar. Ecol. 41, e12579. doi: 10.1111/maec.12579
Chan Y.-F., Chiang K.-P., Ku Y., Gong G.-C. (2019). Abiotic and Biotic Factors Affecting the Ingestion Rates of Mixotrophic Nanoflagellates (Haptophyta). Microb. Ecol. 77, 607–615. doi: 10.1007/s00248-018-1249-2
Chang J., Chung C.-C., Gong G.-C. (1996). Influences of Cyclones on Chlorophyll a Concentration and Synechococcus Abundance in a Subtropical Western Pacific Coastal Ecosystem. Mar. Ecol. Prog. Ser. 140, 199–205. doi: 10.3354/meps140199
Chen Y.-L. L., Chen H.-Y., Jan S., Tuo S.-H. (2009). Phytoplankton Productivity Enhancement and Assemblage Change in the Upstream Kuroshio After Typhoons. Mar. Ecol. Prog. Ser. 385, 111–126. doi: 10.3354/meps08053
Chou C., Chiang J. C. H., Lan C.-W., Chung C.-H., Liao Y.-C., Lee C.-J. (2013). Increase in the Range Between Wet and Dry Season Precipitation. Nat. Geosci. 6, 263–267. doi: 10.1038/ngeo1744
Chung C.-C., Chang J., Gong G.-C., Hsu S.-C., Chiang K.-P., Liao C.-W. (2011). Effects of Asian Dust Storms on Synechococcus Populaitons in the Subtropical Kuroshio Current. Mar. Biotechnol. 13, 751–763. doi: 10.1007/s10126-010-9336-5
Chung C.-C., Gong G.-C., Huang C.-Y., Lin J.-Y., Lin Y.-C. (2015). Changes in the Synechococcus Assemblage Composition at the Surface of the East China Sea Due to Flooding of the Changjiang River. Microb. Ecol. 70, 677–688. doi: 10.1007/s00248-015-0608-5
Chung C.-C., Gong G.-C., Hung C.-C. (2012). Effect of Typhoon Morakot on Microphytoplankton Population Dynamics in the Subtropical Northwest Pacific. Mar. Ecol. Prog. Ser. 448, 39–49. doi: 10.3354/meps09490
Chung C.-C., Huang C.-Y., Gong G.-C., Lin Y.-C. (2014). Influence of the Changjiang River Flood on Synechococcus Ecology in the Surface Waters of the East China Sea. Microb. Ecol. 67, 273–285. doi: 10.1007/s00248-013-0299-8
Edgar R. C., Haas B. J., Clemente J. C., Quince C., Knight R. (2011). UCHIME Improves Sensitivity and Speed of Chimera Detection. Bioinformatics 27, 2194–2200. doi: 10.1093/bioinformatics/btr381
Foflonker F., Mollegard D., Ong M., Yoon H. S., Bhattacharya D. (2018). Genomic Analysis of Picochlorum Species Reveals How Microalgae may Adapt to Variable Environments. Mol. Biol. Evol. 35, 2702–2711. doi: 10.1093/molbev/msy167
Gilbert P. M., Mitra A. (2022). From Webs, Loops, Shunts, and Pumps to Icrobial Multitasking: Evolving Concepts of Marine Microbial Ecology, the Mixoplankton Paradigm, and Implications for a Future Ocean. Limnol. Oceanogr. 67, 585–597. doi: 10.1002/lno.12018
Gómez F., Moreira D., Benzerara K., López-García P. (2011). Solenicola setigera Is the First Characterized Member of the Abundant and Cosmopolitan Uncultured Marine Stramenopile Group MAST-3. Environ. Microbiol. 13, 193–202. doi: 10.1111/j.1462-2920.2010.02320.x
Gong G.-C., Chang J., Chiang K.-P., Hsiung T.-M., Hung C.-C., Duan S.-W., et al. (2006). Reduction of Primary Production and Changing of Nutrient Ratio in the East China Sea: Effect of the Three Gorges Dam? Geophys. Res. Lett. 33, L07610. doi: 10.1029/2006GL025800
Gong G.-C., Chen Y.-L. L., Liu K.-K. (1996). Chemical Hydrography and Chlorophyll a Distribution in the East China Sea in Summer: Implications in Nutrient Dynamics. Cont. Shelf Res. 16, 1561–1590. doi: 10.1016/0278-4343(96)00005-2
Gong G.-C., Liu K.-K., Chiang K.-P., Hsiung T.-M., Chang J., Chen C.-C., et al. (2011). Yangtze River Floods Enhance Coastal Ocean Phytoplankton Biomass and Potential Fish Production. Geophy. Res. Lett. 38, L13603. doi: 10.1029/2011GL047519
Gonzalez-Esquer C. R., Wright K. T., Sudasinghe N., Carr C. K., Sanders C. K., Turmo A., et al. (2019). Demonstration of the Potential of Picochlorum soloecismus as a Microalgal Platform for the Production of Renewable Fuels. Algal Res. 43, 101658. doi: 10.1016/j.algal.2019.101658
Hernández-Urcera J., Rial P., García-Portela M., Lourés P., Kilcoyne J., Rodríguez F., et al. (2018). Notes on the Cultivation of Two Mixotrophic Dinophysis Species and Their Ciliate Prey Mesodinium Rubrum. Toxins 10, 505. doi: 10.3390/toxins10120505
Herut B., Zohary T., Krom M. D., Mantoura R. F. C., Pitta P., Psarra S., et al. (2005). Response of East Mediterranean Surface Water to Saharan Dust: on-Board Microcosm Experiment and Field Observations. Deep Sea Res. II 52, 3024–3040. doi: 10.1016/j.dsr2.2005.09.003
Hilligsøe K. M., Richardson K., Bendtsen J., Sørensen L. L., Nielsen T. G., Lyngsgaard M. M. (2011). Linking Phytoplankton Community Size Composition With Temperature, Plankton Food Web Structure and Sea-Air CO Flux. Deep-Sea Res. I 58, 826–838. doi: 10.1016/j.dsr.2011.06.004
Hoegh-Guldberg O., Bruno J. F. (2010). The Impact of Climate Change on the World’s Marine Ecosystems. Science 328, 1523–1528. doi: 10.1126/science.1189930
IPCC (2021). Climate Change 2021: The Physical Science Basis. Contribution of Working Group I to the Sixth Assessment Report of the Intergovernmental Panel on Climate Change In: Masson-Delmotte V., Zhai P. A., Pirani S.L., Connors C., Péan S., Berger N., et al (Cambridge University Press)
Jardillier L., Zubkov M. V., Pearman J., Scanlan D. J. (2010). Significant CO2 Fixation by Small Prymnesiophytes in the Subtropical and Tropical Northeast Atlantic Ocean. ISME J. 4, 1180–1192. doi: 10.1038/ismej.2010.36
Jones H. L. J., Leadbeater B. S. C., Green J. (1993). Mixotrophy in Marine Species of Chrysochromulina (Prymnesiophyceae): Ingestion and Digestion of a Small Green Flagellate. J. Mar. Biol. Assoc. U. K. 73, 283–296. doi: 10.1017/S0025315400032859
Jung J.-H., Choi J. M., Coats D. W., Kim Y.-O. (2016). Euduboscquella Costata N. Sp. (Dinoflagellata, Syndinea), an Intracellular Parasite of the Ciliate Schmidingerella Arcuata: Morphology, Molecular Phylogeny, Life Cycle, Prevalence, and Infection Intensity. J. Eukaryot. Microbiol. 63, 3–15. doi: 10.1111/jeu.12231
Justić D., Rabalais N. N., Turner R. E., Dortch Q. (1995). Changes in Nutrient Structure of River-Dominated Coastal Waters: Stoichiometric Nutrient Balance and Its Consequences. Estuar. Coast. Shelf Sci. 40, 339–356. doi: 10.1016/S0272-7714(05)80014-9
Kawachi M., Inouye I., Maeda O., Chihara M. (1991). The Haptonema as a Food-Capturing Device: Observations on Chrysochromulina Hirta (Prymnesiophyceae). Phycologia 30, 563–573. doi: 10.2216/i0031-8884-30-6-563.1
Kimura M. (1980). A Simple Method for Estimating Evolutionary Rates of Base Substitutions Through Comparative Studies of Nucleotide Sequences. J. Mol. Evol. 16, 111–120. doi: 10.1007/BF01731581
Krishnan A., Likhogrud M., Cano M., Edmundson S., Melanson J. B., Huesemann M., et al. (2021). Picochlorum Celeri as a Model System for Robust Outdoor Algal Growth in Seawater. Sci. Rep. 11, 1–13. doi: 10.1038/s41598-021-91106-5
Kühn S., Drebes G., Schnepf E. (1996). Five New Species of the Nanoflagellate Pirsonia in German Bight, North Sea, Feeding on Planktic Diatoms. Helgoländer meeresunters. 50, 205–222. doi: 10.1007/BF02367152
Kühn S., Medlinb L., Eller G. (2004). Phylogenetic Position of the Parasitoid Nanoflagellate Pirsonia Inferred From Nuclear-Encoded Small Subunit Ribosomal DNA and a Description of Pseudopirsonia N. Gen. And Pseudopirsonia mucosa (Drebes) Comb. Nov. Protist 155, 143–156. doi: 10.1078/143446104774199556
Labarre A., López-Escardó D., Latorre F., Leonard G., Bucchini ,. F., Obiol A., et al. (2021). Comparative Genomics Reveals New Functional Insights in Uncultured MAST Species. ISME J. 15, 1767–1781. doi: 10.1038/s41396-020-00885-8
Lee Y., Yang E.-J., Youn S., Choi J. K. (2018). Influence of the Changjiang Diluted Waters on the Nanophytoplankton Distribution in the Northern East China Sea. J. Mar. Biolog. Assoc. U. K. 98, 1535–1545. doi: 10.1017/S0025315417001163
Leles S. G., Polimene L., Bruggeman J., Blackford J., Ciavatta S., Mitra A., et al. (2018). Modelling Mixotrophic Functional Diversity and Implications for Ecosystem Function. J. Plankton Res. 40, 627–642. doi: 10.1093/plankt/fby044
Liu H., Suzuki K., Minami C., Saino T., Watanabe M. (2002). Picoplankton Community Structure in the Subarctic Pacific Ocean and the Bering Sea During Summer 1999. Mar. Ecol. Prog. Ser. 237, 1–14. doi: 10.3354/meps237001
Logares R., Audic S., Santini S., Pernice M. C., de Vargas C., Massana R. (2012). Diversity Patterns and Activity of Uncultured Marine Heterotrophic Flagellates Unveiled With Pyrosequencing. ISME J. 6, 1823–1833. doi: 10.1038/ismej.2012.36
Massana R. (2011). Eukaryotic Picoplankton in Surface Oceans. Annu. Rev. Microbiol. 65, 91–110. doi: 10.1146/annurev-micro-090110-102903
Massana R., del Campo J., Sieracki M. E., Audic S., Logares R. (2014). Exploring the Uncultured Microeukaryote Majority in the Oceans: Reevaluation of Ribogroups Within Stramenopiles. ISME J. 8, 854–866. doi: 10.1038/ismej.2013.204
McKie-Krisberg Z. M., Sanders R. W. (2014). Phagotrophy by the Picoeukaryotic Green Alga Micromonas: Implications for Arctic Oceans. ISEM J. 10, 1953–1961
Meakin N. G., Wyman M. (2011). Rapid Shifts in Picoeukaryote Community Structure in Response to Ocean Acidification. ISME J. 5, 1397–1405. doi: 10.1038/ismej.2011.18
Moon-van der Staay S. Y., De Wachter R., Vaulot D. (2001). Oceanic 18s rDNA Sequences From Picoplankton Reveal Unsuspected Eukaryotic Diversity. Nature 409, 607–610. doi: 10.1038/35054541
Nguyen L.-T., Schmidt H. A., von Haeseler A., Minh B. Q. (2014). IQ-TREE: A Fast and Effective Stochastic Algorithm for Estimating Maximum-Likelihood Phylogenies. Mol. Biol. Evol. 32, 268–274. doi: 10.1093/molbev/msu300
Nishitani G., Nagai S., Baba K., Kiyokawa S., Kosaka Y., Miyamura K., et al. (2010). High-Level Congruence of Myrionecta Rubra Prey and Dinophysis Species Plastid Identities as Revealed by Genetic Analyses of Isolates From Japanese Coastal Waters. Appl. Environ. Microbiol. 76, 2791–2798. doi: 10.1128/AEM.02566-09
Paerl R. W., Venezia R. E., Sanchez J. J., Paerl H. W. (2020). Picophytoplankton Dynamics in a Large Temperate Estuary and Impacts of Extreme Storm Events. Sci. Rep. 10, 22026. doi: 10.1038/s41598-020-79157-6
Pai S.-C., Yang C.-C., Riley J. P. (1990). Effects of Acidity and Molybdate Concentration on the Kinetics of the Formation of the Phosphoantimonylmolybdenum Blue Complex. Anal. Chim. Acta 229, 115–120. doi: 10.1016/S0003-2670(00)85116-8
Pang M., Liu K., Liu H. (2022). Evidence for Mixotrophy in Pico-Chlorophytes From a New Picochlorum (Trebouxiophyceae) Strain. J. Phycol. 58, 80–91. doi: 10.1111/jpy.13218
Quince C., Lanzén A., Curtis T. P., Davenport R. J., Hall N., Head I. M., et al. (2009). Accurate Determination of Microbial Diversity From 454 Pyrosequencing Data. Nat. Methods 6, 639–641. doi: 10.1038/nmeth.1361
Quince C., Lanzen A., Davenport R. J., Turnbaugh P. J. (2011). Removing Noise From Pyrosequenced Amplicons. BMC Bioinf. 12, 38. doi: 10.1186/1471-2105-12-38
Rabalais N. N., Turner R. E., JustiĆ D., Dortch Q., Wiseman W. J., Sen Gupta B. K. (1996). Nutrient Changes in the Mississippi River and System Responses on the Adjacent Continental Shelf. Estuaries 19, 386–407. doi: 10.2307/1352458
Søgaard D. H., Sorrell B. K., Sejr M. K., Andersen P., Rysgaard S., Hansen P. J., et al. (2021). An Under−Ice Bloom of Mixotrophic Haptophytes in Low Nutrient and Freshwater−Influenced Arctic Waters. Sci. Rep. 11, 2915. doi: 10.1038/s41598-021-82413-y
Sørensen N., Daugbjerg N., Richardson K. (2013). Choice of Pore Size can Introduce Artefacts When Filtering Picoeukaryotes for Molecular Biodiversity Studies. Microb. Ecol. 65, 964–968. doi: 10.1007/s00248-012-0174-z
Schloss P. D., Westcott S. L., Ryabin T., Hall J. R., Hartmann M., Hollister E. B., et al. (2009). Introducing Mothur: Open-Source, Platform-Independent, Community-Supported Software for Describing and Comparing Microbial Communities. Appl. Environ. Microbiol. 75, 7537–7541. doi: 10.1128/AEM.01541-09
Seeleuthner Y., Mondy S., Lombard V., Carradec Q., Pelletier E., Wessner M., et al. (2018). Single-Cell Genomics of Multiple Uncultured Stramenopiles Reveals Underestimated Functional Diversity Across Oceans. Nat. Commun. 9, 310. doi: 10.1038/s41467-017-02235-3
Smith J. W. O., Demaster D. J. (1966). Phytoplankton Biomass and Productivity in the Amazon River Plume: Correlation With Seasonal River Discharge. Cont. Shelf Res. 16, 291–319. doi: 10.1016/0278-4343(95)00007-N
Stibor H., Stockenreiter M., Nejstgaard J. C., Ptacnik R., Sommer U. (2019). Trophic Switches in Pelagic Systems. Curr. Opin. Syst. Biol. 13, 108–114. doi: 10.1016/j.coisb.2018.11.006
Stoeck T., Bass D., Nebel M., Christen R., Jones M. D. M., Breiner H.-W., et al. (2010). Multiple Marker Parallel Tag Environmental DNA Sequencing Reveals a Highly Complex Eukaryotic Community in Marine Anoxic Water. Mol. Ecol. 19, 21–31. doi: 10.1111/j.1365-294X.2009.04480.x
Strickland J. D. H., Parsons T. R. (1972). A Practical Handbook of Seawater Analysis (Ottawa, Canada: Fisheries Research Board of Canada).
Tamaki H., Wright C. L., Li X., Lin Q., Hwang C., Wang S., et al. (2011). Analysis of 16S rRNA Amplicon Sequencing Options on the Roche/454 Next-Generation Titanium Sequencing Platform. PloS One 6 (9), e25263–e25263. doi: 10.1371/journal.pone.0025263
Tsai A.-Y., Gong G.-C., Sanders R. W., Wang C.-J., Chiang K.-P. (2010). The Impact of the Changjiang River Plume Extension on the Nanoflagellate Community in the East China Sea. Estuar. Coast. Shelf Sci. 89, 21–30. doi: 10.1016/j.ecss.2010.05.005
Unrein F., Gasol J. M., Not F., Forn I., Massana R. (2014). Mixotrophic Haptophytes Are Key Bacterial Grazers in Oligotrophic Coastal Waters. ISME J. 8, 164–176. doi: 10.1038/ismej.2013.132
Wang Q., Garrity G. M., Tiedje J. M., Cole J. R. (2007). Naive Bayesian Classifier for Rapid Assignment of rRNA Sequences Into the New Bacterial Taxonomy. Appl. Environ. Microbiol. 73, 5261–5267. doi: 10.1128/AEM.00062-07
Welschmeyer N. A. (1994). Fluorometric Analysis of Chlorophyll a in the Presence of Chlorophyll B and Pheopigments. Limnol. Oceanogra. 39, 1985–1992. doi: 10.4319/lo.1994.39.8.1985
Worden A. Z., Follows M. J., Giovannoni S. J., Wilken S., Zimmerman A. E., Keeling P. J. (2015). Rethinking the Marine Carbon Cycle: Factoring in the Multifarious Lifestyles of Microbes. Science 347, 1257594. doi: 10.1126/science.1257594
Worden A. Z., Nolan J. K., Palenik B. (2004). Assessing the Dynamics and Ecology of Marine Picophytoplankton: The Importance of the Eukaryotic Component. Limnol. Oceanogra. 49, 168–179. doi: 10.4319/lo.2004.49.1.0168
Wu W., Logares R., Huang B., Hsieh C. h. (2017). Abundant and Rare Picoeukaryotic Sub-Communities Present Contrasting Patterns in the Epipelagic Waters of Marginal Seas in the Northwestern Pacific Ocean. Environ. Microbiol. 19, 287–300. doi: 10.1111/1462-2920.13606
Yih W., Coats D. W. (2000). Infection of Gymnodinium Sanguineum by the Dinoflagellate Amoebophrya Sp.: Effect of Nutrient Environment on Parasite Generation Time, Reproduction, and Infectivity. J. Eukary. Microbiol. 47, 504–510. doi: 10.1111/j.1550-7408.2000.tb00082.x
Yoo Y. D., Seong K. A., Jeong H. J., Yih W., Rho J.-R., Nam S. W., et al. (2017). Mixotrophy in the Marine Red-Tide Cryptophyte Teleaulax Amphioxeia and Ingestion and Grazing Impact of Cryptophytes on Natural Populations of Bacteria in Korean Coastal Waters. Harmful Algae 68, 105–117. doi: 10.1016/j.hal.2017.07.012
Keywords: picoeukaryote, 18S rRNA phylogeny, flooding, global climate change, East China Sea
Citation: Chung C-C, Gong G-C, Lin Y-C and Hsu C-W (2022) Differences in the Composition of Abundant Marine Picoeukaryotes in the Marginal Sea Derived from Flooding. Front. Mar. Sci. 9:853847. doi: 10.3389/fmars.2022.853847
Received: 13 January 2022; Accepted: 11 April 2022;
Published: 18 May 2022.
Edited by:
Hiroaki Saito, The University of Tokyo, JapanReviewed by:
Shunyan Cheung, University of California, Santa Cruz, United StatesYasuhide Nakamura, Shimane University, Japan
Copyright © 2022 Chung, Gong, Lin and Hsu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chih-Ching Chung, Y2h1bmdjY0BtYWlsLm50b3UuZWR1LnR3
 Chih-Ching Chung
Chih-Ching Chung Gwo-Ching Gong
Gwo-Ching Gong Yun-Chi Lin
Yun-Chi Lin Ching-Wei Hsu1
Ching-Wei Hsu1