- 1CAS and Shandong Province Key Laboratory of Experimental Marine Biology, Institute of Oceanology, CAS Center for Ocean Mega-Science, Chinese Academy of Sciences, Qingdao, China
- 2Qingdao University of Science and Technology, Qingdao, China
- 3College of Environment and Safety Engineering, Qingdao University of Science and Technology, Qingdao, China
- 4Laboratory for Marine Ecology and Environmental Science, Pilot National Laboratory for Marine Science and Technology (Qingdao), Qingdao, China
- 5CAS Key Laboratory of Marine Ecology and Environmental Sciences, Institute of Oceanology, Chinese Academy of Sciences, Qingdao, China
- 6Cawthron Institute, Nelson, New Zealand
- 7School of Earth and Planetary, University of Chinese Academy of Sciences, Beijing, China
Species from the family Kareniaceae (Dinophyceae) frequently cause harmful algal blooms (HABs), with serious ecological impacts and risks to human safety and aquaculture activities in coastal waters worldwide. However, due to their small size, lack of morphological divergence, and low abundance during non-bloom periods, the diversity within this family is not well understood. By comparing the commonly used molecular markers, the Internal Transcribed Spacer (ITS) region was found to have an appropriate mutation rate to distinguish three of the most common genera (Karenia, Karlodinium, and Takayama) within the Kareniaceae family and different geographical strains of Kareniaceae. Specific primers targeting the ITS region of Karenia, and the other primers specific to the genera Karlodinium and Takayama, were designed. Specificity of the primers was tested using 17 strains of Kareniaceae species and 15 non-target species. Representative Kareniaceae species could be successfully detected even at low concentrations of target DNA template with a limit of detection of 3.2 pg. The primers were also assessed using high-throughput sequencing with two environmental samples from the South China Sea (SCS). Analysis of the reads identified as Kareniaceae species revealed a high diversity and the existence of unreported Kareniaceae species in the SCS. In conclusion, the newly developed molecular barcodes specifically detected Kareniaceae species in the field and will provide technical support for the effective warning and monitoring of Kareniaceae HABs.
Introduction
The family Kareniaceae (Gymnodiniales, Dinophyceae) was described in 2005, with the following characteristic features: a straight or sigmoid apical groove on the cell epicone and fucoxanthin and its derivatives as diagnostic pigments (Bergholtz et al., 2005). It is widely accepted that the family comprises three genera Karenia, Karlodinium, and Takayama. Recently, three genera, namely Asterodinium, Gertia, and Shimiella were added to Kareniaceae (Benico et al., 2019; Takahashi et al., 2019; Ok et al., 2021), but these additions are controversial due to morphological differences and the absence of fucoxanthin or the derivatives.
Species of Karenia, Karlodinium, and Takayama often cause harmful algal blooms (HABs), increasing in frequency, intensity, and distribution worldwide (Yang et al., 2001; Brand et al., 2012; Place et al., 2012; Li et al., 2019; Yang et al., 2021). For example, Kareniaceae HABs have occurred for more than 20 years in China since their first detection in the Pearl River Estuary in 1998 (Cui et al., 2009). More than 90 cases of Karenia mikimotoi blooms have been reported along China’s coastline (Lin et al., 2010; Liang, 2012; Lu et al., 2014; Liu et al., 2015; Lv et al., 2019). Kareniaceae species often form intense HABs with cell densities more than 106 cells L-1, and when the bloom collapses, the decomposition of the dense cell biomass causes hypoxia and threatens the health of marine life. Additionally, Kareniaceae species produce toxins and harmful substances that affect marine zooplankton, shellfish, fish, and humans. For example, K. mikimotoi produces cytotoxins, hemolytic toxins, glycolipid toxins, various polyunsaturated fatty acid toxins (Yasumoto et al., 1990; Arzul et al., 1995; Gentien et al., 2007), as well as hemolytic toxins and cytotoxins associated with massive mortalities of fish, shellfish, and invertebrates (Gentien, 1998; Mitchell and Rodger, 2007; Brand et al., 2012; Kim, 2010). Karenia brevis, another well-known HAB species, produces the neurotoxic brevetoxins that cause respiratory distress in humans (Xu et al., 2010; Errera and Campbell, 2011; Poli et al., 2000; Naar et al., 2007; Fleming et al., 2011). Karlodinium veneficum produces karlotoxins which has been associated with fish mortality (Deeds et al., 2002; Xu et al., 2012; Xu et al., 2014; Dai et al., 2014). The development of a hypoxic environment and the production of toxins mean these species can have detrimental impacts on both coastal ecosystems and marine aquaculture development. Economic losses caused by K. mikimotoi blooms were approximately US$370 million in China from 2005 to 2017 (China Marine Disaster Bulletin 2005-2017, SOAC). In 2012, a K. mikimotoi bloom decimated farmed abalone and caused financial losses exceeding US$330 million in the coastal waters of Fujian Province (Li et al., 2017).
When the family Kareniaceae was first described, 21 species were recorded (Bergholtz et al., 2005; Wang et al., 2018). To date, 37 species have been described (11 Karenia spp., 15 Karlodinium spp., seven Takayama spp., two Asterodinium spp., one Gertia sp. and one Shimiella sp.) and updated in the AlgaeBase database (https://www.algaebase.org/) (Guiry and Guiry, 2022; De Salas et al., 2008; Li and Shin, 2018; Luo et al., 2018; Benico et al., 2019; Takahashi et al., 2019; Cen et al., 2020; Ok et al., 2021). Among them, nine species of Karenia, six species of Karlodinium and one species of Takayama are considered as HAB species based on the HAB taxon list (Lundholm et al., 2009), as detailed in Table S1. There is increasing evidence that multiple Kareniaceae species co-occur or co-dominate in a bloom event. For example, in 2021 a massive HAB in the Pacific coast of eastern Hokkaido, Japan was dominated by K. selliformis, with other Kareniaceae dinoflagellates (K. longicanalis, K. mikimotoi, Karlodinium sp., Takayama cf. acrotrocha, T. tuberculata and Takayama sp.) also present (Iwataki et al., 2022). A Karenia bloom in Hong Kong waters in 2016 was dominated by the species K. mikimotoi and K. papilionacea, with K. selliformis and K. longicanalis also present in low abundance (Lin et al., 2020). Five species of Karenia (K. brevis, K. mikimotoi, K. papilionacea, and K. selliformis and one unknown Karenia species) were isolated and identified in a Karenia bloom along the west coast of Florida from 2007 to 2008 (Wolny et al., 2015). Due to the production of diverse toxins and impacts from multiple species, accurate species identification from environmental samples are crucial for early warning systems and the management of Kareniaceae blooms in coastal waters.
Morphological features are traditionally used for the identification of bloom-forming species (Richlen et al., 2008; Smayda, 2010). However, many Kareniaceae species are morphologically similar, and distinguishing them using microscopy is virtually impossible. Additionally, Kareniaceae species are unarmored and fragile, meaning they are easily destroyed during sampling and preservation processes. It is increasingly urgent and necessary to develop a reliable, sensitive method for the simultaneous detection of multiple species of Kareniaceae in the field.
DNA sequence data can provide useful auxiliary information for the classification of microalgae. Various DNA markers (e.g., the 18S ribosomal DNA (rDNA), 28S rDNA, and Internal Transcribed Spacer (ITS) regions) have been used for the detection and identification of HAB species, including Kareniaceae species. For example, using DNA sequence data, K. digitata was revised to Karl. digitatum, and recently two new species, Karl. zhouanum and Karl. elegans, have been described based on morphological characteristics, pigment composition, and 28S rDNA sequences (Luo et al., 2018; Cen et al., 2020). The ITS region has been proven as a good DNA barcode region for dinoflagellate species (Litaker et al., 2007; Stern et al., 2012); however, it cannot resolve species from some genera such as Alexandrium, Symbiodinium, Gymnodinium, Heterocapsa, Prorocentrum and Karenia (Litaker et al., 2007; Stern et al., 2012). In addition, species-specific quantitative assays, such as quantitative PCR (qPCR) and loop-mediated isothermal amplification-lateral flow dipstick (LAMP-LFD), that target the ITS region of Kareniaceae species have been developed (Yuan et al., 2012; Huang et al., 2020). However, no specific DNA marker has been developed for the whole Kareniaceae family, which has hindered the investigation of species diversity in the natural environments. Studies have shown that compared with 18S rDNA and 28S rDNA, the ITS and ribulose-bisphosphate carboxylase (rbcL) regions of the family Kareniaceae are more genetically variable, and they have been extensively used as targets for interspecific identification (Garcés et al., 2006; Al-Kandari et al., 2011; Toldrà et al., 2018). However, further studies investigating the ITS region of Kareniaceae is necessary to develop effective molecular markers for the detection of diversity within the family.
In this study, the variation rates of the 18S rDNA V1-V9 region, 28S rDNA D1-D2 region, and ITS region of Kareniaceae species were compared. The ITS region, with a relatively high variation, was selected as the target region for the design of molecular markers. Two pairs of specific primers were designed based on the ITS region of Kareniaceae species: one pair for the genus Karenia, and the other for the genera Karlodinium and Takayama. Both pairs of primers were highly sensitive with a low limit of detection. Specificity of the primers was tested using 17 strains of Kareniaceae species and 15 non-target species. The developed molecular markers were then applied using high-throughput sequencing metabarcoding to determine the diversity of Kareniaceae in the South China Sea (SCS).
Materials and Methods
Microalgal Culture
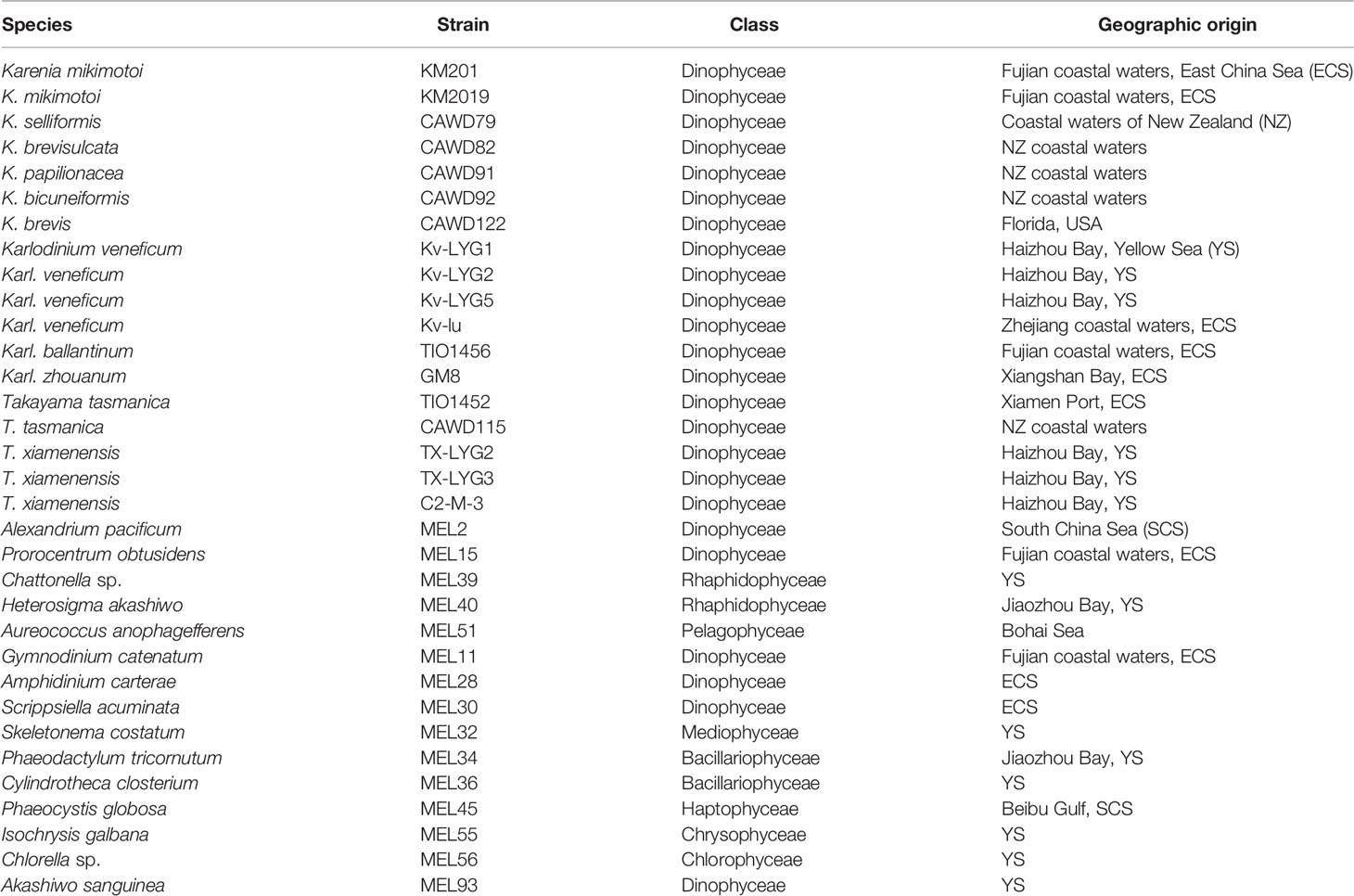
Thirty-three strains of microalgae (Table 1) were used in this study: 18 strains of Karenia, Karlodinium, and Takayama, and 15 other non-target strains. All the strains were cultured in L1 medium and maintained at 20 °C under a photon flux density of 75 μmol m-2 s-1 and a 14:10 h light-dark cycle. Approximately 50 mL of culture was collected at exponential stage from each microalgal strain and centrifuged at 5000 × g for 10 min. Cell pellets were stored at -80 °C until DNA extraction.
Sample Collection
Two phytoplankton samples were collected from the sampling sites (ZN1-3 and S30) in the South China Sea (SCS) in February 2019 and June 2020, respectively (Figure 1). Approximately 1 L of surface phytoplankton sample was filtered using sieves (mesh size 200 μm) to remove zooplankton and then filtered using an HTTP membrane (pore diameter 0.40 μm, Millipore, USA) under a vacuum of 40 kPa. The filtered membranes were stored in a freezer at -80 °C until DNA extraction.
DNA Extraction, PCR Amplification, and Sequencing
Total genomic DNA of microalgal cultures and environmental samples was extracted using the CTAB method (Winnepenninckx et al., 1993) with the following modifications: incubation with CTAB buffer at 65 °C for 1 h; centrifugation at 4 °C and 12,500 g; and isopropyl alcohol to precipitate the DNA. The extracted DNA was stored at -80 °C until further analysis. The 18S rDNA V1-V9 region and 28S rDNA D1-D2 region of five strains (Kv-LYG1, Kv-LYG5, Kv-lu, KM2019, KM201) of Kareniaceae, and ITS region of 15 strains (Kv-LYG1, Kv-LYG2, Kv-LYG5, Kv-lu, KM2019, KM201, TIO1456, GM8, TIO1452, CAWD79, CAWD82, CAWD91, CAWD92, CAWD115, CAWD122) of Kareniaceae (Table S2), were PCR amplified with universal primers for eukaryotes (Table 2). PCR was performed with a final volume of 20 μL containing 10 μL 2 × Taq PCR MasterMix (Tiangen, China), 1 μL of genomic DNA, 0.8 μL of each primer at 10 μM, and 7.4 μL sterilized deionized water. Amplifications were performed with an initial denaturation temperature of 95 °C for 3 min, followed by 35 cycles of 95 °C for 20 s; 53 °C (18S rDNA), 52 °C (28S rDNA), and 59 °C (ITS) for 30 s; 72 °C for 100 s; and a final elongation at 72 °C for 7 min. The PCR products were separated by electrophoresis on 1% agarose gel, and images were obtained using a gel imaging system (Bio-Rad, USA). Targeted DNA bands were purified by quick DNA extraction using a SteadyPure PCR DNA Purification Kit AG21003 (AG, China) and then sequenced by Sangon Biotech (Shanghai, China) and assembled using Vector NTI Advance 11.5.3 ContigExpress.
Clone Library Construction and Sequencing
Clone libraries of 18S rDNA, 28S rDNA, and ITS were constructed using purified PCR fragments to determine the polymorphism of target genes. The purified PCR fragments were cloned into pMD 18-T vectors (Takara), and the vectors were transformed into Takara Escherichia coli Competent Cells DH5α, which were placed on LB ampicillin plates for cultivation (37 °C, 16-18 h). The positive clones were screened by colony PCR, and at least three clones were sequenced by Sangon Biotech (Shanghai, China). Next, BLASTn analysis was performed on the obtained target sequences, ensuring the validity of Kareniaceae species. Finally, the obtained sequences of the Kareniaceae species in this study were submitted to GenBank (https://www.ncbi.nlm.nih.gov/genbank/).
Genetic Distance Analysis
Kareniaceae sequences (18S rDNA region, 28S rDNA region, and ITS region) were downloaded from GenBank (Table S3). Together with the sequences obtained in this study, the genetic distances of the different target sequences were estimated by MEGA7 using the pairwise distance method with bootstrap replications of 1000 (Kumar et al., 2016).
Phylogenetic Analysis of ITS Sequences of Kareniaceae Species
Sixty-two ITS sequences from the genera Karenia, Karlodinium, Takayama, Asterodinium, Gertia and Shimiella, comprising 43 sequences downloaded from GenBank (Table S3) and 19 sequences obtained in this study (Table S2), were used to construct the phylogenetic tree. ITS sequence from Gymnodinium catenatum was used as outgroup. All sequences were aligned using ClustalW, and a maximum-likelihood tree was built using the MEGA7 variance estimation method with 1000 bootstrap replications and the best-fitting nucleotide substitution models of GTR (Nei and Kumar, 2000; Kumar et al., 2016). Bayesian analysis was performed using MrBayes 3.2.6 (Ronquist et al., 2012) with generations of 20,000 and sampling frequencies of 1,000, and the best-fitting nucleotide substitution models of GTR+F+I+G4 were selected using Modelfinder (Kalyaanamoorthy et al., 2017).
Design, Optimization, and Validation of Kareniaceae Specific Primers
Design of Kareniaceae Specific Primers
Multiple alignments were performed between the obtained 19 sequences of the ITS region in this study (Table S2) and the downloaded sequences (Table S3), using Vector NTI Advance 11.5.3. Based on the alignment results, two specific primer pairs for the family Kareniaceae were designed using Primer 5: primers specific for the genus Karenia (KareF2 and KareR1) and primers specific for the genera Karlodinium and Takayama (KarlTaF1 and KarlTaR2) (Table 2). The primers were synthesized commercially by Sangon Biotech (Shanghai, China).
Optimization of PCR Reaction System
The volume of PCR reaction mixture was 20 μL: 1 μL genomic DNA (approximately 10 ng μL-1), 4 μL 5 × PCR Buffer, 1.6 μL dNTP, 0.4 μL TransStart FastPfu Fly DNA Polymerase (TransGen, China), 0.4 μL of forward and reverse primers (10 μM), and 12.2 μL sterilized deionized water. To optimize the PCR reaction, gradient PCR was first used to determine the optimal amplification temperature by using genomic DNA of K. mikimotoi, Karl. veneficum, and T. xiamenensis as templates, respectively. The reactions were performed on a TaKaRa PCR Thermal Cycler (Takara, Japan): 95°C for 2 min; followed by 35 cycles of 15 s at 95°C, 20 s at 46-61 °C, and 15 s at 72°C; and a final extension at 72°C for 5 min. The amplification products (approximately 400-440 bp) were separated using 1.5% AGE. The brightness of electrophoresis bands were used as criteria to choose the appropriate annealing temperature.
Limit of Detection
The cell concentration of microalgal cultures of K. mikimotoi, Karl. veneficum and T. xiamenensis were determined using an optical microscope and 15 mL of culture was filtered to extract total genomic DNA. DNA was quantified using a Qubit 4 fluorometer (Thermo, USA). The relationship between the cell number and DNA concertration was recorded in Table 6. The optimal assay conditions were then used to determine the primer sensitivity with 5-fold gradient diluted DNA solutions (10 ng μL-1-0.128 pg μL-1) of the K. mikimotoi, Karl. veneficum, and T. xiamenensis as DNA templates.
Test of Specificity
The specificity of the primer pairs was evaluated using the online Primer-BLAST (https://www.ncbi.nlm.nih.gov/tools/primer-blast/index.cgi?LINK_LOC=BlastHome) and by running the optimized assays with genomic DNA template from the target species (K. mikimotoi, Karl. veneficum and T. xiamenensis) and 15 non-target microalgal strains (Table 1). The resulting PCR products were analyzed using 1.5% AGE.
Applicability of the Two Molecular Markers
Seventeen strains of Karenia (seven strains, six spp.), Karlodinium (six strains, three spp.), and Takayama (four strains, two spp.) (Table 1), and two field samples (S30 and ZN1-3) collected from the SCS were selected to test the applicability of the developed method. The genomic DNA of the Kareniaceae strains and field samples was extracted using the method described above. PCR reactions were conducted under the optimised conditions.
Application of the Barcodes in the South China Sea
To further verify the reliability of the developed molecular markers, the amplified products of the field samples (S30 and ZN1-3) were sequenced using high-throughput Illumina sequencing using the Novaseq 6000 system with 250 bp paired-end reads. The target DNA fragments were purified by quick DNA extraction using a SteadyPure PCR DNA Purification Kit AG21003 (AG, China), and the purified PCR products of each sample were quantified using a Qubit 4 fluorometer (Thermo, USA). Four sequencing libraries (two samples, two primer pairs) were constructed using the VAHTS Universal DNA Library Prep Kit for IlluminaV3 (ND607) and VAHTSTM DNA Adapters set1 (N801). Paired-end raw reads were preprocessed to generate high-quality clean reads by using fastp software (Version 0.20.1), including removal of barcode and adapter sequences, reads less than 200 bp, and low-quality filtering. Cleaned reads were assembled to generate high-quality tags (Tag refers to a contig of an F read and the associated R read, with the length around 440-450bp) using QIIME2 (Version 2021.4).
Two hundred and forty-eight ITS sequences from Kareniaceae species downloaded from GenBank and nineteen ITS sequences obtained in this study were used to create a reference database. The GenBank database was searched using each species of Kareniaceae and the full name or abbreviation of the target gene (e.g., 18S rRNA, ITS1, 5.8S rRNA, ITS2, 28S rRNA). Sequences were then aligned and trimmed to obtain the target region. Sequences with obvious alignment errors and only partial ITS sequences were removed. The homologous sequences were searched using local BLAST, with each clean tag sequence using the reference database. Homologous sequences with a similarity above 98% were clustered into an operational taxonomic unit (OTU) using QIIME2 (Version 2021.4). The mapping rarefaction curve for sampling depth verification to evaluate whether the sequencing volume was sufficient to cover all groups in the sample, and the diversity of the two samples, was analyzed with QIIME2. OTUs obtained by clustering were annotated to obtain detailed information of the two samples.
Results
Selecting the Target Region for the Molecular Marker
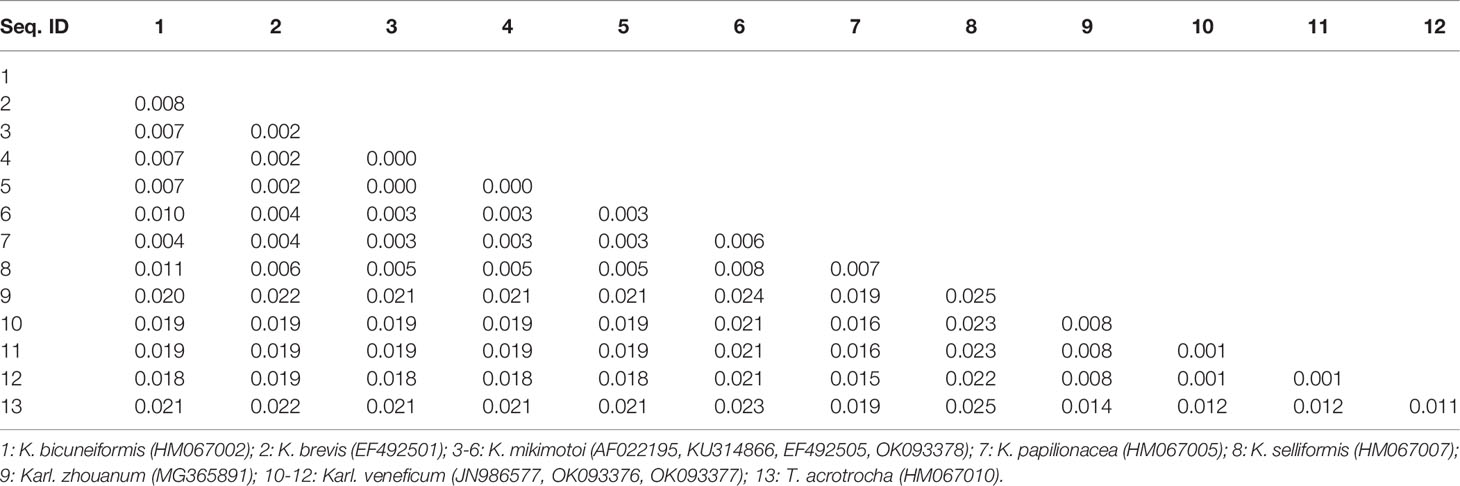
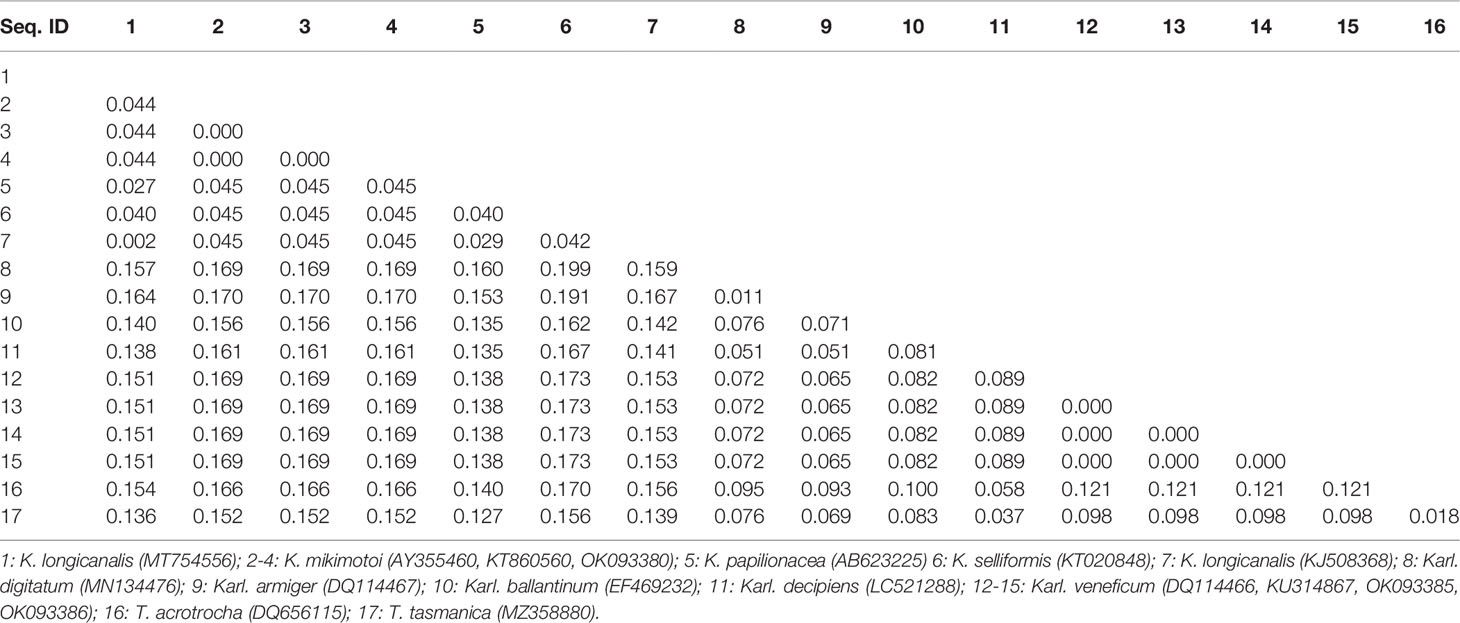
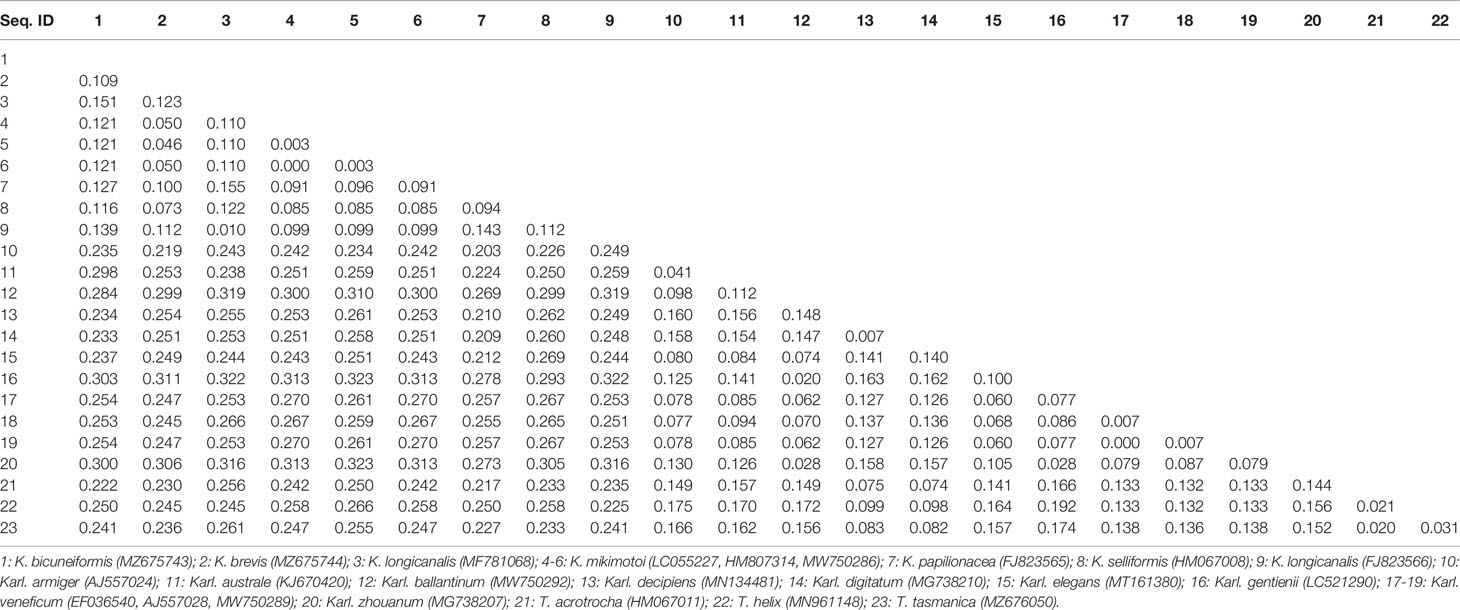
Sequences of the ITS region were successfully amplified from 15 strains of K. mikimotoi, K. brevisulcata, K. selliformis, K. bicuneiformis, K. brevis, K. papilionacea, Karl. veneficum, Karl. ballantinum, Karl. zhouanum, and T. tasmanica. Sequences of the 18S rDNA V1-V9 and 28S rDNA D1-D2 regions were successfully amplified from five strains of K. mikimotoi and Karl. veneficum. All sequences obtained in this study were deposited in GenBank (accession numbers: MW750282-MW750293, MZ323882, MZ675742-MZ675744, MZ676047-MZ676050, and OK093375-OK093386; Table S2). The genetic distances of sequences of the 18S rDNA V1-V9, 28S rDNA D1-D2, and ITS regions of Kareniaceae species obtained from this study and GenBank were analyzed.
The genetic distances between the sequences of the 18S rDNA V1-V9, 28S rDNA D1-D2, and ITS regions were 0.000-0.025, 0.000-0.199, and 0.000-0.323, respectively (Tables 3–5). The interspecific genetic distances of 18S rDNA V1-V9 and 28S rDNA D1-D2 were relatively small and were lower than those of ITS. The 18S rDNA and 28S rDNA sequences from different geographical strains of K. mikimotoi and Karl. veneficum were almost identical within a species, and the intraspecific distances of the ITS varied between 0.003 and 0.007. These results suggested that the ITS region was better suited as a molecular marker for the family Kareniaceae than the 18S rDNA and 28S rDNA.
Phylogenetic Relationship of Different Strains of Kareniaceae Species
Kareniaceae species were divided into two reciprocal sister groups, one contained Karenia and Asterodinium, and the other Karlodinium, Takayama, Gertia and Shimiella. (Figure 2). Within the latter, Shimiella formed a separate clade from Karlodinium, Takayama and Gertia. The latter clade was further divided into two subclades, and one subclade contained Gertia, Takayama, and Karl. decipiens, and the other the remaining Karlodinium species. Several Kareniaceae species showed intraspecific variation in their ITS marker, for example, the nine strains of Karl. veneficum showed slight variation (Figure 2). The results suggested that Karlodinium and Takayama have closer phylogenetic relationships than Karenia, and demonstrated variation between strains at species level. The phylogenetic analyses could distinguish between and within species of the family Kareniaceae.
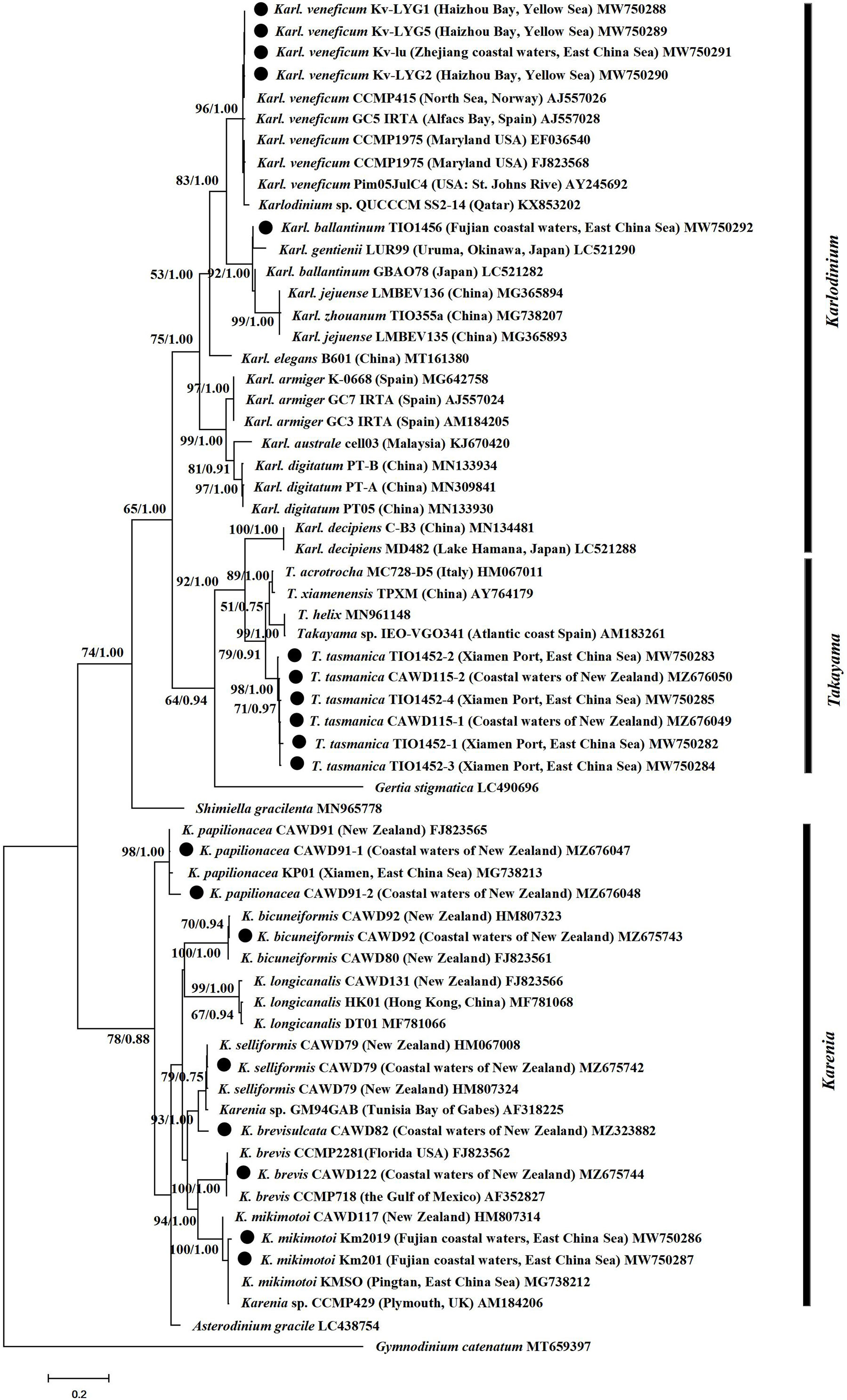
Figure 2 Phylogenetic tree of Kareniaceae species based on ITS sequences. Sequences with a circle were obtained in this study. Values at nodes suggests statistical support values (ML bootstrap values/Bayesian posterior probability). Only bootstrap values >50% and posterior probabilities >0.5 are shown.
Design, Optimization, and Verification of the Primers
Based on the alignment results of the ITS sequences from Kareniaceae species, developing a primer pair that would amplify all species from the whole family was difficult. Because the genera Karlodinium and Takayama had a close phylogenetic relationship, the Karlodinium-Takyama-specific primer pair (KarlTaF1 and KarlTaR2) was designed in a relatively conserved region covering Karlodinium and Takayama ITS sequences, whereas Karenia-specific primer pair (KareF2 and KareR1) was designed to target the ITS region of Karenia spp. independently. Genomic DNA extractions from K. mikimotoi, Karl. veneficum, and T. xiamenensis (approximately 10 ng μL-1) were used as DNA templates to optimize the annealing temperature ranging from 46 to 61°C. The PCR results showed that 49°C was the most suitable annealing temperature for both primer pairs (Figure 3A).
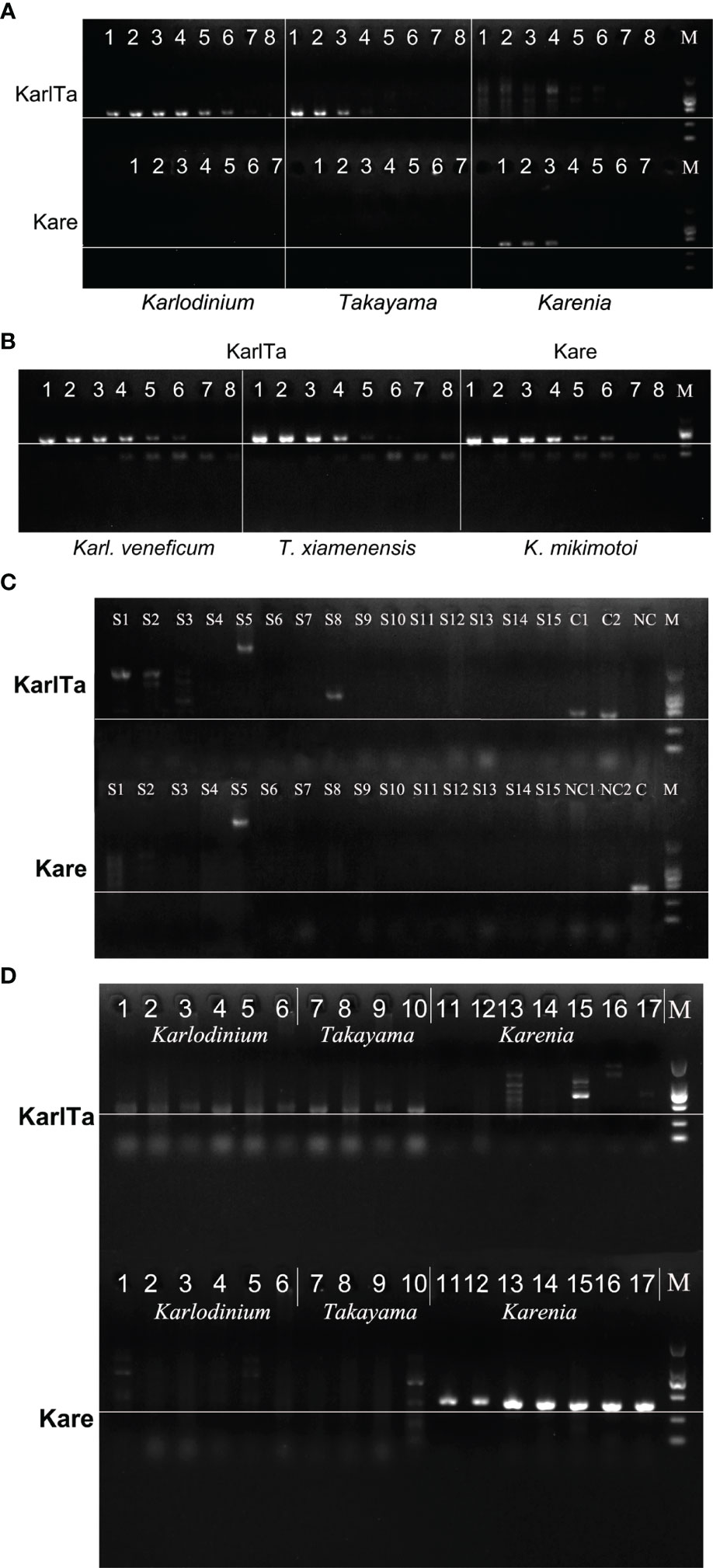
Figure 3 Sensitivity and specificity of KarlTa and Kare primer pairs. (A): Gel electrophoresis of annealing temperature gradient of KarlTa and Kare primer pairs (KarlTa, 1: 47 °C; 2: 49 °C; 3: 51 °C; 4: 53 °C; 5: 55 °C; 6: 57 °C; 7: 59 °C; 8: 61 °C; M: DL2000 DNA marker; Kare, 1: 46 °C; 2: 48 °C; 3: 50 °C; 4: 52 °C; 5: 54 °C; 6: 56 °C; 7: 58 °C; M: DL2000 DNA marker. The horizontal line indicates the correct size of target bands). (B): Gel electrophoresis of sensitivity of KarlTa and Kare primer pairs (1: 10 ng, 2:2 ng, 3: 0.4 ng, 4: 0.08 ng, 5: 16 pg, 6: 3.2 pg, 7: 0.64 pg, 8: 0.128 pg of DNA template content; M: DL2000 DNA marker. The horizontal line indicates the correct size of target bands). (C): Gel electrophoresis of specificity of KarlTa and Kare primer pairs (S1: A.pacificum; S2: P. obtusidens; S3: Chattonella sp.; S4: H.akashiwo; S5: A.anophagefferens; S6: G.catenatum; S7: A.carterae; S8: S. acuminata; S9: S. costatum; S10: P. tricornutum; S11: C.closterium; S12: P. globosa; S13: I.galbana; S14: Chlorella sp.; S15: A.sanguinea; C1 (KarlTa) & NC1 (Kare): Karl. veneficum; C2 (KarlTa) & NC2 (Kare): T. xiamenensis; NC (KarlTa) & C (Kare): K.mikimotoi; M: DL2000 DNA marker. The horizontal line indicates the correct size of target bands). (D): Validation results of KarlTa and Kare primers with laboratory samples (Karlodinium (1-6): 1-4: Karl. veneficum Kv-LYG1, Kv-LYG2, Kv-LYG5, and Kv-lu; 5: Karl. ballantinum TIO1456; 6: Karl. zhouanum GM8; Takayama (7-10): 7-9: T. xiamenensis TX-LYG2, TX-LYG3, C2-M-3; 10: T. tasmanica CAWD115; Karenia (11-17): 11-12: K.mikimotoi KM2019, KM201; 13: K.selliformis CAWD79; 14: K.brevisulcata CAWD82; 15: K.papilionacea CAWD91; 16: K.bicuneiformis CAWD92; 17: K.brevis CAWD122; M: DL2000 DNA marker. The horizontal line indicates the correct size of target bands).
Total genomic DNA was extracted from 15 mL of cultures of K. mikimotoi, Karl. veneficum, and T. xiamenensis, respectively (Table 6). The sensitivity of the designed primers was verified using serially diluted genomic DNA as DNA template. The PCR results showed that the amplified bands weakened as the DNA content decreased. The presence of target microalgae could be detected when PCR tubes contained at least 3.2 pg of target DNA, corresponding to 0.17 cells of T. xiamenensis, 0.26 cells of K. mikimotoi, and 0.60 cells of Karl. veneficum, demonstrating that the assays were sensitive and had a low detection limit (Figure 3B).
Primer-BLAST was used to evaluate the specificity of the primers and the results showed that the designed primers have good specificity. Then, strains of K. mikimotoi, Karl. veneficum, T. xiamenensis, and 15 non-target microalgal cultures were used as positive and negative controls, respectively, to test the specificity of the designed primers under the optimized experimental conditions. PCR results showed that only the genomic DNA of K. mikimotoi, Karl. veneficum, and T. xiamenensis (positive controls) had amplification bands at the expected size 400-440 bp (Figure 3C, lane 16-18), and other microalgae (negative controls) had no amplification bands at approximately 400-440 bp (Figure 3C). The KarlTa primer pair amplified only Karlodinium and Takayama, but not Karenia and the Kare primer pair only amplified Karenia, but not Karlodinium/Takayama. The findings indicated that both primer pairs were specific to their respective targets, and the combination of both primer pairs were specific to all species from the family Kareniaceae.
Applicability of the Two Primer Pairs
The results of PCR reactions using 17 unialgal strains of Kareniaceae species as DNA templates showed that the KarlTa primer pair could amplify all species of Takayama and Karlodinium in this study (samples No. 1-10, 10 strains, 5 spp.), comprising Karl. veneficum, Karl. ballantinum, Karl. zhouanum, T. tasmanica, and T. xiamenensis, but could not amplify the seven strains of Karenia (Figure 3D). By contrast, the Kare primer pair could amplify all species of Karenia in this study (samples No. 11-17, 7 strains, 6 spp.), including K. mikimotoi, K. selliformis, K. brevisulcata, K. papilionacea, K. bicuneiformis, and K. brevis, but could not amplify the 10 strains of Takayama and Karlodinium. The results implied that both molecular markers were perfectly applicable to their respective target genera and had the potential to be used for natural phytoplankton communities.
Application of the Metabarcodes for the Detection Of Field Sample
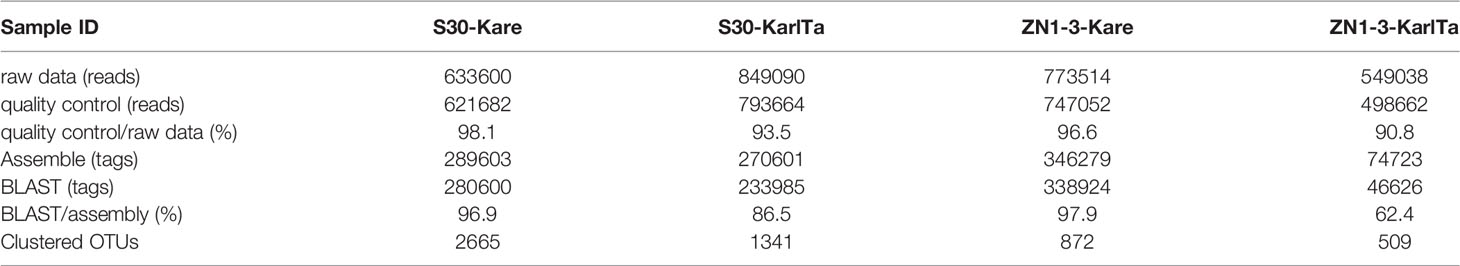
The two primer pairs were further validated for use with field samples, and both the SCS samples could be specifically amplified with the two primer pairs (Figure 4). The high-throughput sequencing results of the SCS samples using the two primer pairs as barcode were shown in Table 7. The raw data of the four libraries constructed from the SCS samples were 633,600, 849,090, 773,514, and 549,038 reads, and after quality filtering more than 90% of the reads remained, indicating that the obtained sequencing data were reliable and could be used for subsequent analyses.
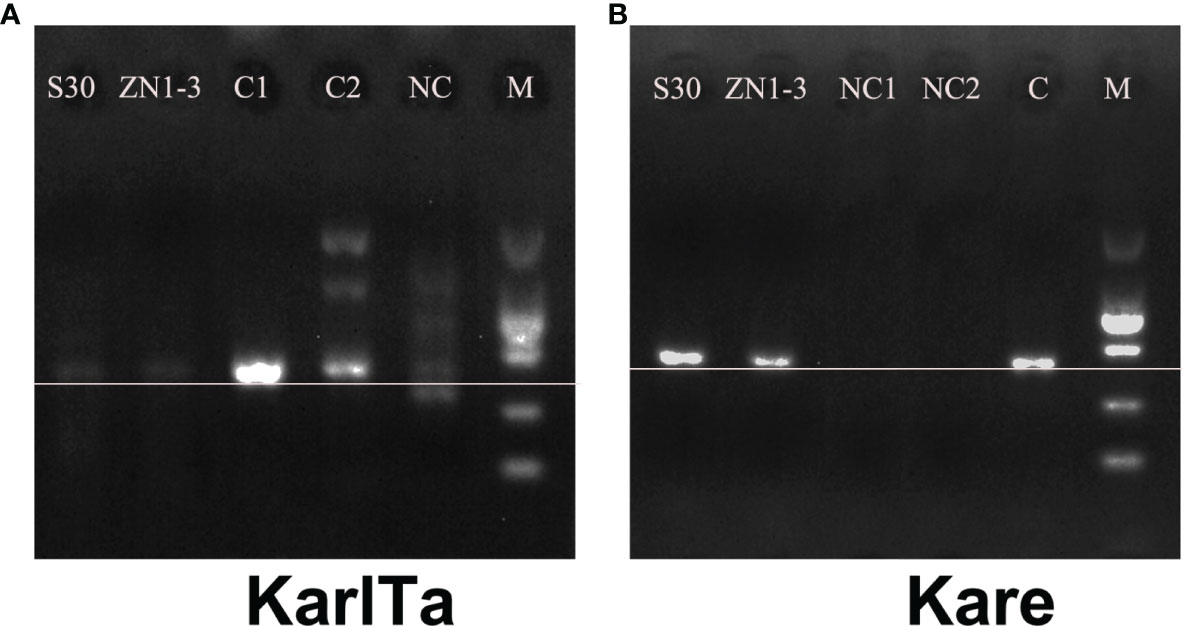
Figure 4 PCR results of specific primers in field samples (A: KarlTa; B: Kare). (Field sample S30; Field sample ZN1-3; C1 (A) & NC1 (B): Karl. veneficum; C2 (A) & NC2 (B): T. xiamenensis; NC (A) & C (B): K. mikimotoi; M: DL2000 DNA marker. The horizontal line indicates the correct size of target bands).
After bioinformatic processing, the assembled high-quality tags were 289,603, 270,601, 346,279, and 74,723. The homologous ITS sequences of Kareniaceae species obtained through BLAST analysis of the self-built reference database of Kareniaceae species were 280,600, 233,985, 338,924, and 46,626, accounting for 96.9%, 86.5%, 97.9%, and 62.4% of the assembled high-quality tags. These homologous sequences were then clustered into 2665, 1341, 872, and 509 OTUs with a similarity above 98%. The rarefaction curves of the OTUs were prepared for all samples (Figure S1), and the number of OTUs in each sample flattened with an increase in sequencing quantity, indicating that the sequencing depth was sufficient to reflect the species diversity in the samples.
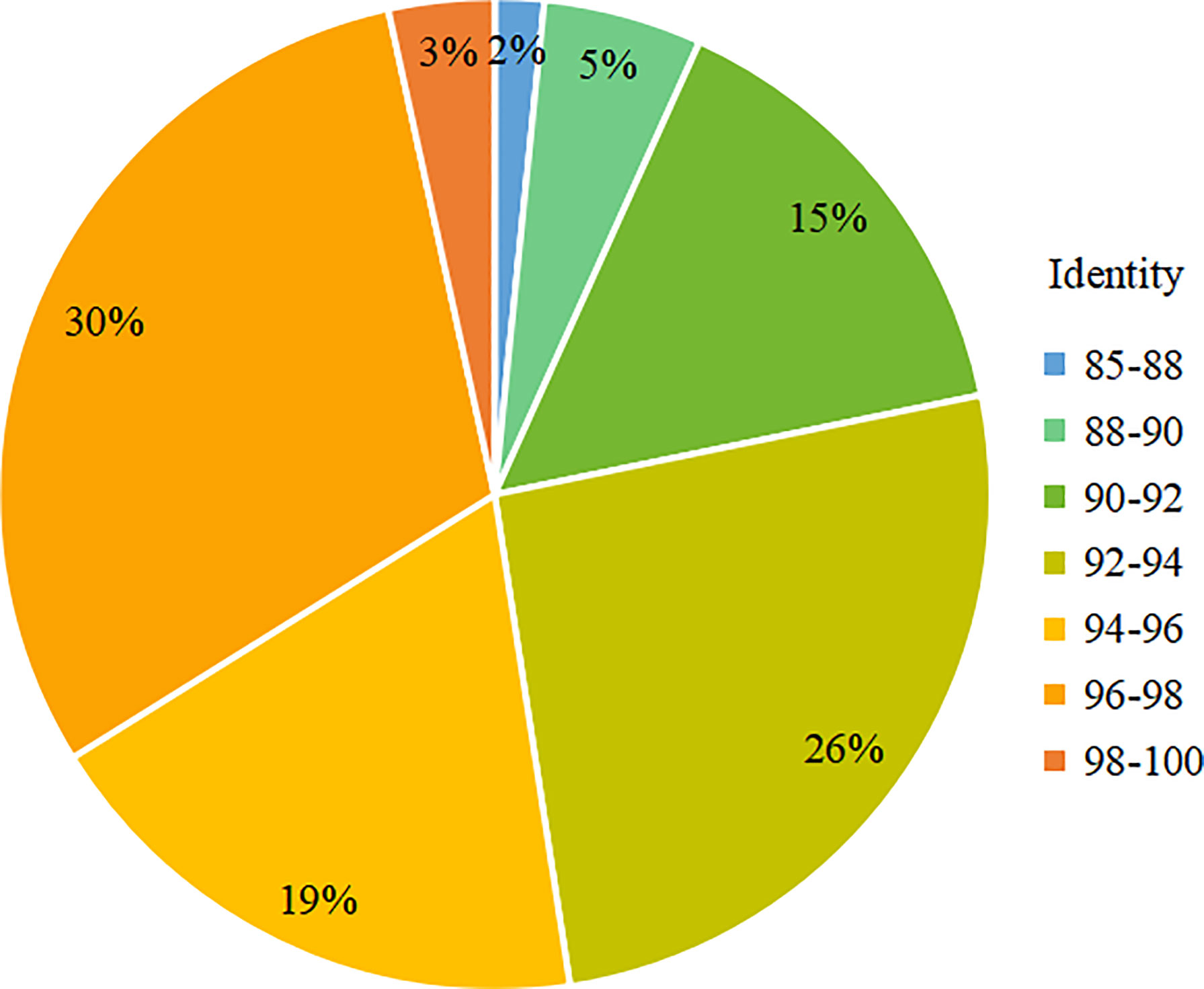
For the analysis of the different Kareniaceae species in the two SCS samples, the OTU annotation results were analysed according to different identity levels (Figure 5). Differences between the known genera of Kareniaceae were within 90% sequence identity (Table S4), so sequences with identity great than 90% were considered as homologs of the Kareniaceae family. 93% of OTUs had an identity greater than 90%, demonstrating reads from the family Kareniaceae dominated the two samples. 3% of the OTUs had identity greater than 98% and a total of fifteen known Kareniaceae species and one Brachidinium species were identified with high certainty. Sample S30 contained seven Karenia spp., five Karlodinium spp., and two Takayama spp. Sample ZN1-3 contained six Karenia spp., four Karlodinium spp., two Takayama spp., and one Brachidinium sp. (Figures 6A, B). These results dispalyed that Kareniaceae species composition was different between the two samples and the species diversity in sample S30 was slightly higher than that in ZN1-3. The dominant species in sample S30 were K. papilionacea (13.0%) and Karl. veneficum (4.4%), and those in the coastal sample ZN1-3 were K. papilionacea (10.8%), K. mikimotoi (5.6%), B. capitatum (6.6%) and Karl. veneficum (2.0%) (Table S5).
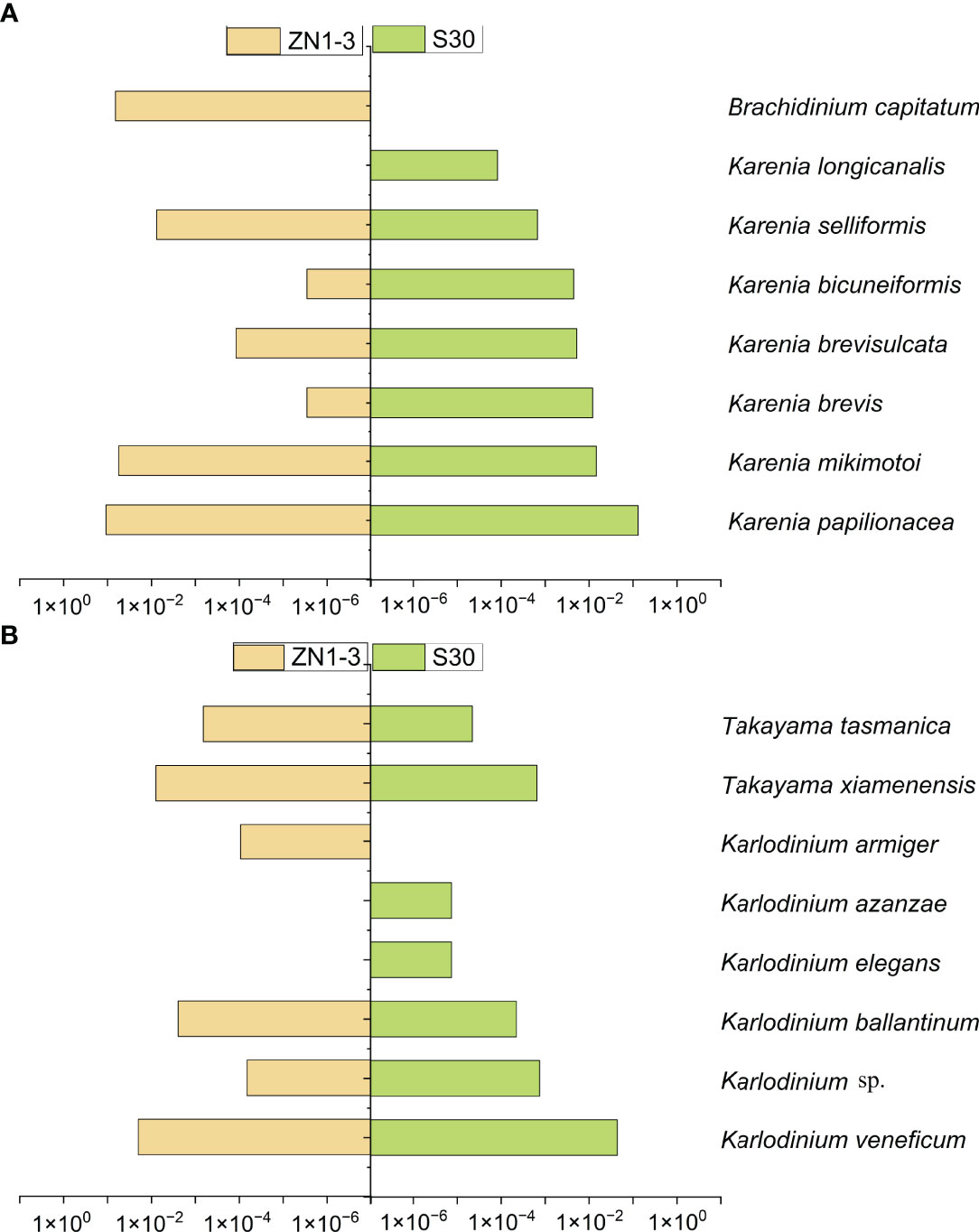
Figure 6 Comparison of the statistics OTUs annotation results between sample ZN1-3 and S30. (A): the tags proportion of the annotated as Karenia species in the two SCS samples. (B): the tags proportion of the annotated as Karlodinium and Takayama species in the two SCS samples. (Column length represents the tags proportion of the annotated Kareniaceae species with more than 98% identity in the total assembled Kareniaceae tags).
The representative sequences of the 329 OTUs (7%) with identity ranging from 80 to 90% were subjected to additional BLAST analyses in the GenBank Database, and sequences with the highest identity for OTU were pooled and analyzed (Table 8). The results showed that 266 OTUs had the closest relationship with the species from the family Kareniaceae. For the other 60 unknown hits (Uncultured eukaryotes), the second and third highest identities were Kareniaceae species (Table S6).
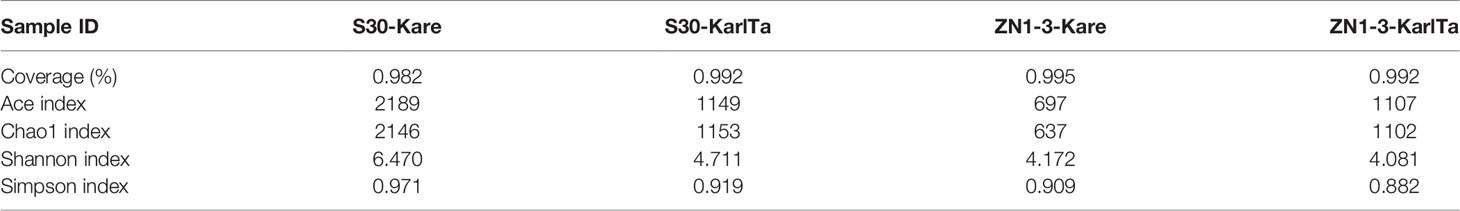
The alpha-diversity indices of the two SCS samples were analyzed using QIIME2. The coverage rates of sequencing results were greater than 98.2%, indicating that the sequencing results were effective and reliable. The Ace and Chao1 indices and the Simpson and Shannon indices reflected the species abundance and species diversity in the samples, respectively. The four indices in sample S30 were higher than those in sample ZN1-3 (Table 9), suggesting that the abundance and diversity of Kareniaceae species in sample S30 were higher than those in sample ZN1-3.
Discussion
Kareniaceae species are widely distributed in global oceans, and many form recurrent HABs in coastal waters (De Salas et al., 2008; Brand et al., 2012). Kareniaceae species can produce harmful compounds and toxins that can threaten coastal ecosystems and aquaculture (Mooney et al., 2007; Zhang et al., 2008; Brand et al., 2012; Holland et al., 2012; Fowler et al., 2015). Due to difficulties with morphological identification including small and fragile cells and the absence of distinguishable features, accurately identifying species in the family Kareniaceae from the environment is difficult. In this study, we developed a molecular method for the rapid detection of Kareniaceae species with high sensitivity and specificity. This technique shows great promise for understanding the diversity of Kareniaceae and for the early warning and monitoring of HABs.
Feasibility of the Specific Barcodes for Kareniaceae Species Detection
To date, molecular identification and detection methods for HABs have largely focused on species-specific molecular assays (Gray et al., 2003; Zhang et al., 2008; Al-Kandari et al., 2011; Toldrà et al., 2018; Huang et al., 2020; Wang et al., 2020) or universal eukaryotic molecular markers (Scorzetti et al., 2009; Zheng et al., 2009a; Marie et al., 2010; Yuan et al., 2012; Wu et al., 2015; Li et al., 2018). Kareniaceae species are generally present at a low cell density before a bloom occurs, which is usually undetectable or underestimated using universal eukaryotic primers (Gentien et al., 2005; Liu et al., 2020). Notably, as new species of Kareniaceae are regularly discovered, and the tendency that Kareniaceae HABs are not limited to a single species is becoming increasingly apparent, meaning that the successful application of species-specific primers for the early warning and monitoring of Kareniaceae HABs can be difficult and of limited use. Recently, some researchers have reported that family-level specific primers were useful to reveal the diversity, distribution and abundance of Dinophyceae species (Wietkamp et al., 2019), suggesting a type of “universal” barcode specific to the family Kareniaceae could be developed to identify various potential HAB species from environmental water samples.
Various molecular markers have been reported to detect and identify Kareniaceae HAB species, generally based on the 28S rDNA, 18S rDNA, and ITS regions (Zheng et al., 2009a; Yuan et al., 2012; Luo et al., 2018; Huang et al., 2020). These genes are arranged in tandem high copy number and thus are easily obtained using PCR amplification. In addition, many sequences from these regions are available from public sequence databases, making it easy to compare relative sequences of target species for the development of molecular methods. In this study, the comparison of genetic distances between sequences from the 18S rDNA, 28S rDNA, and ITS regions of Kareniaceae species showed that the ITS had greater variation than 18S rDNA and 28S rDNA regions. Phylogenetic analyses of the ITS sequences showed that currently classified Kareniaceae species were clustered into three clades that included the three core genera of Kareniaceae. Variation in the ITS region which corresponds to different geographical isolates is a common phenomenon in Kareniaceae species. For example, fourteen different geographical strains of K. mikimotoi clustered into four groups based on differences in the ITS region (Al-Kandari et al., 2011). Therefore, the ITS region could be used as a tool for geographical and taxonomic inference at both the intraspecific and the interspecific level. Additionally, according to the phylogenetic analyses in this study, the genera Karlodinium and Takayama clustered together and were quite distinct from the Karenia genus, which has also been reported in other studies (Huang et al., 2017; Toldrà et al., 2018; Elleuch et al., 2020), indicating the difficulty in developing a single molecular marker covering the whole family. However, we were able to design two primer pairs targeting the ITS region, one for Karenia and the other for Karlodinium and Takayama. The simultaneous use of both primer pairs allowed the successful amplification and detection of Kareniaceae species.
The specificity of molecular tools for the detection of harmful algae from environmental samples is crucial. Some detection methods based on pigment types and optical techniques have been developed to identify and detect Kareniaceae species, each with apparent advantages and disadvantages. Monitoring using pigment analysis by high-performance liquid chromatography (HPLC) has been proposed based on the biomarker pigment gyroxanthin-diester (Richardson and Pinckney, 2004), but this pigment is found in all Kareniaceae species (De Salas et al., 2003; De Salas et al., 2005) and is not exclusive to Kareniaceae, therefore, species-specific detection is not provided. An optical plankton detector (OPD, BreveBuster) developed for K. brevis relies on the unique absorption to detect its presence in a mixed phytoplankton community (Kirkpatrick et al., 2000), which exhibits high specificity for K. brevis, but cannot detect other Kareniaceae species. Current monitoring programs often use light microscopy to detect and enumerate toxic microalgae, but this method is time-consuming, requires substantial taxonomic expertise, and is based on morphological characteristics, which, in some cases, are insufficient for identification at the genus or species level. For example, identification of cells from the genus Karlodinium is particularly difficult and cells can easily be misidentified (e.g., as Gymnodinium, Karenia, Heterocapsa, and Ansanella) due to their unarmored morphology and a lack of distinct features (Shao et al., 2004; Bergholtz et al., 2005; Garcés et al., 2006; Jeong et al., 2014). The two assays designed in this study were specific for Kareniaceae species and could easily detect Karenia and Karlodinium/Takayama species. The specificity of the assays was verified against strains of other common HAB species, including dinoflagellate species closely related to Kareniaceae present in the coastal waters of China, and demonstrated their high specificity to Kareniaceae species (Table 1).
The primer pairs were also tested against environmental samples collected from the SCS using a high-throughput sequencing approach. Bioinformatic analyses of the Kare primers showed that 96.7% (S30) and 97.9% (ZN1-3) of high-quality tags were identified as belonging to the genus Karenia, indicating the high specificity of primers. To successfully amplify Karlodinium and Takayama species from the environmental samples (potentially due to low cell densities), the number of PCR cycles was increased and the number of reads that could be identified as Karlodinium/Takayama was much lower (85% for S30 and 62.4% for ZN1-3). Further BLASTN analyses were carried out on sequences that could not be successfully classified in the ZN1-3-KarlTa library, showed that only a small number could be aligned with the sequences in GenBank and most of them failed to match any sequences, which could be caused by low cell concentrations and the excess PCR cycles.
In recent years, studies have described new genera in the Kareniaceae family, including Asterodinium, Gertia and Shimiella (Benico et al., 2019; Takahashi et al., 2019; Ok et al., 2021), as well as Brachidinium, of which the classification is still under debate. Alignments of ITS sequences from the new genera and the two molecular markers showed that the Kare primer pair differed by only 1-2 bases from the ITS sequences of Asterodinium and Brachidinium, but differed by 4-11 bases from those of Gertia and Shimiella, implying that the Kare primer pair can potentially amplify Asterodinium and Brachidinium, but is not likely to amplifiy Gertia and Shimiella. The KarlTa primers are 5-10 bases different from the ITS sequences these four genera, and are specific to Karlodinium and Takayama. High-throughput sequencing analyses verified the existence of at least fifteen known Kareniaceae species and one Brachidinium species, and homologous sequences to Asterodinium with low identity, indicating that the barcodes developed in this study can likely distinguish species of Karenia, Karlodinium, Takayama, Brachidinium and Asterodinium from the environmental samples.
The sensitivity of molecular assays is another critical parameter that must be evaluated for field detection because it determines the applicability of a method to the early detection and timely warning for HABs. The detection limit for the developed gyroxanthin-based approach by HPLC was below 5000 cells L−1 of K. brevis in estuarine waters (Richardson and Pinckney, 2004), and the OPD method had no clear minimum detection limit (Kirkpatrick et al., 2000). Molecular detection methods have been developed for specific species of Kareniaceae, including real-time nucleic acid sequence-based amplification with internal control RNA (IC-NASBA) (Ulrich et al., 2010) and qPCR assays (Yuan et al., 2012) for K. mikimotoi. These methods had a detection limit of one and five cells, respectively. In this study, the lower limit of detection for the developed detection methods was 3.2 pg genomic DNA which corresponded to 0.17-0.60 cells of target species. Although the copy number of the ITS gene may vary between different species or strains, which may create difficulty for absolute quantification, methods targeting ribosomal regions including the ITS provide a very low limit of detection, especially compared with the detection method based on pigment or optical technology (Toldrà et al., 2018; Elleuch et al., 2020).
With the rapid development of DNA sequencing technology and the continuous improvement of bioinformatics analysis methods, metabarcoding analyses has significantly improved and are widely used in ecological studies of phytoplankton communities. The global distribution of marine chlorophyte algae was analyzed by the international Ocean Sampling Day Consortium through simultaneous sampling of 141 sites worldwide and subsequent sequencing of 18S rDNA V4 region (Tragin and Vaulot, 2018). In addition, the Earth Microbiome Project, an international cooperative project, has carried out many metabarcoding analyses of marine phytoplankton communities (Thompson et al., 2017). Metabarcoding has promoted the study of HABs in various marine ecosystems in China since its development (Lin et al., 2014; Chen et al., 2016; Zhang et al., 2017; Stern et al., 2018) and revealed many previously overlooked nano- and pico-sized HAB species, which cannot be adequately identified and distinguished by light microscopy. For example, metabarcoding analysis based on the amplification and sequencing of the 18S rDNA V4 region confirmed that the causative species of brown tide in the Bohai Sea was Aureococcus anophagefferens (Zhang et al., 2012). Metabarcoding analysis of rbcL gene revealed the phytoplankton biodiversity in Jiaozhou Bay (Liu et al., 2004), and metabarcoding analysis of ITS2 revealed the diversity of symbiotic algae in coral reefs (Qin et al., 2019; Chen et al., 2019). However, Kareniaceae species are rarely or only occasionally detected by metabarcoding analyses, potentially due to their low abundance before bloom development. For instance, only Karl. veneficum was detected among eight species of harmful dinoflagellates in Jiaozhou Bay via metabarcoding analysis (Liu et al., 2020), meaning the diversity of Kareniaceae species if often underestimated. In our study, at least 18 known species of Kareniaceae were detected in one sample by metabarcoding using the developed specific ITS primers. This method allowed for the rapid and accurate detection of low-density Kareniaceae species from field samples and as an early warning system for monitoring of HABs caused by Kareniaceae species.
Application of the Developed Molecular Markers in the SCS and its Implications
The developed detection method was applied to environmental samples collected in two areas of the SCS. Both samples were amplified using the two designed primer pairs, indicating that species of different genera of Kareniaceae existed simultaneously in these two areas of the SCS. The PCR productions of the two samples were further verified by high-throughput sequencing, and 4531 OTUs with identity greater than 90% were obtained from the two samples, implying substantial diversity in the family Kareniaceae from the SCS. A total of 15 known Kareniaceae species were detected in the two samples. Among them, nine species have been previously recorded in the coastal waters of China. New records for the region were K. brevisulcata, K. bicuneiformis, Karl. ballantinum, Karl. azanzae, and Karl. armiger (Table S7). The results suggest primers developed in this study can contribute to the discovery of Kareniaceae species that have not been reported previously in the coastal waters of China and would advance early warning and monitoring systems for Kareniaceae HABs.
High-throughput sequencing of amplicons derived from two SCS samples demonstrated that significant differences were observed in species composition between the sample S30 and sample ZN1-3. More known Kareniaceae species were found in the sample S30 and some possible reasons for this phenomenon are variations in hydrologic, chemical, and ecological environments. Notably, sea surface temperature and salinity differed between the two sampling sites, which were 30.1 °C and 33.94 in the sample S30, while 23.1°C and 33.20 in the sample ZN1-3. Higher temperatures could stimulate the growth of various dinoflagellate species (Gobler et al., 2017). Concentrations of different nutrients were relatively low and stable at site S30 in the SCS central waters, which was suitable for the co-existence of multiple species. By contrast, the environmental conditions of coastal waters are relatively unstable and changeable under the impact of human activities on the coastline, which can lead to the proliferation of one or some specific species. This hypothesis seems to be consistent with the previous results, that is, eutrophication processes in inshore areas caused by nutrient pollution from terrestrial inputs not only affects the scale of algal blooms but also changes the diversity and dominant groups (Anderson et al., 2002; Beusen et al., 2013; Glibert, 2017). Additionally, some species of Karenia, Karlodinium, and Takayama have been confirmed to be mixotrophs (organisms can combine phototrophy and phagotrophy) (Berge et al., 2012; Brand et al., 2012; Zhang et al., 2013; Jeong et al., 2016; Lim et al., 2018). To some degree, mixotrophic species can be beneficial in the coastal waters containing a high abundance of dissolved organic matter, and pico- and nano-phytoplankton.
With recent increased research on the causative species of HABs, many new Kareniaceae species have been described and identified, and some known species have been revised. For example, Karl. corsicum was revised from Gyr. corsicum (Siano et al., 2009), and several new species, including T. xiamenensis, Karl. gentienii, and Karl. elegans were published by Gu et al. (2013); Nézan et al. (2014) and Cen et al. (2020), respectively. In this study, all obtained sequences from the SCS field samples and the sequences of known Kareniaceae species were analyzed using phylogenetic methods, with Amphidinium carterae as the outgroup (Figure S3). The phylogenetic tree showed that all environmental sequences clustered together, and were far away from A. carterae. These sequences constituted 4860 OTUs, including 4531 OTUs with identity greater than 90% to sequences from species in the family Kareniaceae and 329 OTUs with identity between 80-90% to the family Kareniaceae (Table 8, S6). The identity of sequences between Kareniaceae genera was approximately 90% (Table S4), indicating that the 329 OTUs with 80-90% identity do not belong to the known Kareniaceae genera. Representative sequences of seven OTUs within the 329 OTUs were randomly selected and aligned with the ITS sequences of known Kareniaceae species and other non-Kareniaceae species in the order Gymnodiniales. The both ends of OTU sequences were highly variable over 10 to 20 bases and the middle region was highly conserved (Figure S2). These selected OTUs were more similar to known Kareniaceae species in GenBank than non-Kareniaceae species, even when compared to the related Gymnodinium and Gyrodinium species in the order Gymnodiniales. These results strongly suggest the existence of unknown diversity within the family Kareniaceae.
Conclusions
In this study, two molecular assays with a low detection limit and strong specificity were developed for the rapid detection of Kareniaceae species. The advantages of this method for monitoring of Kareniaceae blooms are its simple operation, short detection time, and easy-to-obtain results. Moreover, high-throughput sequencing metabarcoding using the two primer pairs revealed the existence of several Kareniaceae species and the high diversity of Kareniaceae in the SCS. These approaches will enable further research into the diversity and geographical distribution of Kareniaceae along the coast of China and worldwide.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: NCBI [accession: MW750282-MW750293, MZ323882, MZ675742-MZ675744, MZ676047-MZ676050, and OK093375-OK093386].
Author Contributions
QZ and LQ conceived and designed the study. WZ and QZ finished the experiment and draft of this manuscript. LQ and KS revised the manuscript. QZ and KS provided the algae. QL assisted in DNA extraction and experiment preparations. CL and XY assisted in sample collection and results analysis. All authors contributed to the article and approved the submitted version. There is no conflict of interest in submission of this manuscript, which has been approved by all authors for publication.
Funding
This study was supported by the National Science and Technology Basic Resources Investigation Program of China (grant number 2018FY100206), the National Key Research and Development Plan Project of China (grant number 2019YFC1407902), and the New Zealand Ministry for Business, Innovation and Employment (contract numbers CAWX1801 and CAWX0902).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We express our gratitude to Dr. Hae Jin Jeong (Seoul National University), Haifeng Gu (Third Institute of Oceanography, Ministry of Natural Resources of China), and Zhaohui Wang and Jingyi Cen (Jinan University) for the free provision of the cells, DNA solutions, molecular sequences of Kareniaceae species. Field samples and data were collected onboard of R/V “Kehai 68” implementing the open research cruise NORC2020-07 supported by NSFC Shiptime Sharing Project. Special appreciation is expressed to Oceanographic Data Center, IOCAS for molecular data analysis supported by Oceanographic Data Center, IOCAS.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2022.851605/full#supplementary-material
References
Al-Kandari M. A., Highfield A. C., Hall M. J., Hayes P., Schroeder D. C. (2011). Molecular Tools Separate Harmful Algal Bloom Species, Karenia Mikimotoi, From Different Geographical Regions Into Distinct Sub-Groups. Harmful Algae. 10, 636–643. doi: 10.1016/j.hal.2011.04.017
Anderson D. M., Glibert P. M., Burkholder J. M. (2002). Harmful Algal Blooms and Eutrophication: Nutrient Sources, Composition, and Consequences. Estuaries. 25, 704–726. doi: 10.1007/BF02804901
Arzul G., Erard-Le Denn E., Belin C., Nézan E. (1995). Ichthyotoxic Events Associated With Gymnodinium Cf. Nagasakiense on the Atlantic Coast of France. Harmful Algae News. 12, 8–9.
Benico G., Takahashi K., Lum W. M., Iwataki M. (2019). Morphological Variation, Ultrastructure, Pigment Composition and Phylogeny of the Star-Shaped Dinoflagellate Asterodinium Gracile (Kareniaceae, Dinophyceae). Phycologia 58, 405–418. doi: 10.1080/00318884.2019.1601948
Berge T., Poulsen L. K., Moldrup M., Daugbjerg N., Juel Hansen P. (2012). Marine Microalgae Attack and Feed on Metazoans. ISME J. 6, 1926–1936. doi: 10.1038/ismej.2012.29
Bergholtz T., Daugbjerg N., Moestrup Ø., Fernández-Tejedor M. (2005). On the Identity of Karlodinium Veneficum and Description of Karlodinium Armiger Sp. Nov. (Dinophyceae), Based on Light and Electron Microscopy, Nuclear-Encoded LSU rDNA, and Pigment Composition. J. Phycology. 42, 170–193. doi: 10.1111/j.1529-8817.2006.00172.x
Beusen A. H. W., Slomp C. P., Bouwman A. F. (2013). Global Land-Ocean Linkage: Direct Inputs of Nitrogen to Coastal Waters via Submarine Groundwater Discharge. Environ. Res. Letters. 8, 34035. doi: 10.1088/1748-9326/8/3/034035
Brand L. E., Campbell L., Bresnan E. (2012). Karenia: The Biology and Ecology of a Toxic Genus. Harmful Algae. 14, 156–178. doi: 10.1016/j.hal.2011.10.020
Cen J. Y., Wang J. Y., Huang L. F., Ding G. M., Qi Y. Z., Cao R. B., et al. (2020). Who is the "Murderer" of the Bloom in Coastal Waters of Fujian, China, in 2019? J. Oceanol. Limnol. 38, 722–732. doi: 10.1007/s00343-019-9178-6
Chen L., Wu L., Liu Y., Xu H. G. (2016). Application of Environmental DNA Metabarcoding in Ecology. Acta Ecologica Sinica. 36, 4573–4582. doi: 10.5846/stxb201501150125
Chen B., Yu K. F., Liang J. Y., Huang W., Wang G. H., Su H. F., et al. (2019). Latitudinal Variation in the Molecular Diversity and Community Composition of Symbiodiniaceae in Coral From the South China Sea. Front. Microbiol. 10:1278. doi: 10.3389/fmicb.2019.01278
Cui W. M., Yang W. D., Liu J. S., Li H. Y. (2009). Hemolytic Properties of Hemolytic Extracts From Karenia Mikimotoi Hasen. J. Trop. Subtropical Botany. 17, 237–241. doi: 10.3969/j.issn.1005-3395.2009.03.005
Dai X. F., Lu D. D., Guan W. B., Wang H. X., He P. X., Xia P., et al. (2014). Newly Recorded Karlodinium Veneficum Dinoflagellate Blooms in Stratified Water of the East China Sea. Deep Sea Res. Part II: Topical Stud. Oceanography. 101, 237–243. doi: 10.1016/j.dsr2.2013.01.015
Deeds J. R., Terlizzi D. E., Adolf J. E., Stoecker D. K., Place A. R. (2002). Toxic Activity From Cultures of Karlodinium Micrum (=Gyrodinium Galatheanum) (Dinophyceae)—a Dinoflagellate Associated With Fish Mortalities in an Estuarine Aquaculture Facility. Harmful Algae. 1, 169–189. doi: 10.1016/S1568-9883(02)00027-6
De Salas M. F., Bolch C. J. S., Botes L., Nash G., Wright S. W., Hallegraeff G. M. (2003). Takayama Gen. Nov. (Gymnodiniales, Dinophyceae), a New Genus of Unarmored Dinoflagellates With Sigmoid Apical Grooves, Including the Description of Two New Species. J. Phycology. 39, 1233–1246. doi: 10.1111/j.0022-3646.2003.03-019.x
De Salas M. F., Laza-Martínez A., Hallegraeff G. M. (2008). Novel Unarmored Dinoflagellates From the Toxigenic Family Kareniaceae (Gymnodiniales): Five New Species of Karlodinium and One New Takayama From the Australian Sector of the Southern Ocean. J. Phycology. 44, 241–257. doi: 10.1111/j.1529-8817.2007.00458.x
De Salas M. F., Rhodes L. L., Mackenzie L. A., Adamson J. E. (2005). Gymnodinoid Genera Karenia and Takayama (Dinophyceae) in New Zealand Coastal Waters. New Z. J. Mar. Freshw. Res. 39, 135–139. doi: 10.1080/00288330.2005.9517296
Elleuch J., Barkallah M., Smith K. F., Ben Neila I., Fendri I., Abdelkafi S. (2020). Quantitative PCR Assay for the Simultaneous Identification and Enumeration of Multiple Karenia Species. Environ. Sci. pollut. Res. 27, 36889–36899. doi: 10.1007/s11356-020-09739-4
Errera R. M., Campbell L. (2011). Osmotic Stress Triggers Toxin Production by the Dinoflagellate Karenia Brevis. Proc. Natl. Acad. Sci. 108, 10597–10601. doi: 10.1073/pnas.1104247108
Fleming L. E., Kirkpatrick B., Backer L. C., Walsh C. J., Nierenberg K., Clark J., et al. (2011). Review of Florida Red Tide and Human Health Effects. Harmful Algae. 10, 224–233. doi: 10.1016/j.hal.2010.08.006
Fowler N., Tomas C., Baden D., Campbell L., Bourdelais A. (2015). Chemical Analysis of Karenia Papilionacea. Toxicon. 101, 85–91. doi: 10.1016/j.toxicon.2015.05.007
Garcés E., Fernandez M., Penna A., Van Lenning K., Gutierrez A., Camp J., et al. (2006). Characterization of NW Mediterranean Karlodinium Spp. (Dinophyceae) Strains Using Morphological, Molecular, Chemical, and Physiological Methodologies. J. Phycology. 42, 1096–1112. doi: 10.1111/j.1529-8817.2006.00270.x
Gentien P. (1998). Bloom Dynamics and Ecophysiology of the Gymnodinium Mikimotoi Species Complex. Physiol. Ecol. Harmful Algal Blooms. 41, 155–173.
Gentien P., Donaghay P., Yamazaki H., Raine R., Reguera B., Osborn T. R. (2005). Harmful Algal Blooms in Stratified Environments. Oceanography. 16, 172–183. doi: 10.5670/oceanog.2005.52
Gentien P., Lunven M., Lazure P., Youenou A., Crassous M. P. (2007). Motility and Autotoxicity in Karenia Mikimotoi (Dinophyceae). Philos. Trans. R. Soc. B: Biol. Sci. 362, 1937–1946. doi: 10.1098/rstb.2007.2079
Glibert P. M. (2017). Eutrophication, Harmful Algae and Biodiversity-Challenging Paradigms in a World of Complex Nutrient Changes. Mar. pollut. Bulletin. 124, 591–606. doi: 10.1016/j.marpolbul.2017.04.027
Gobler C. J., Doherty O. M., Hattenrath-Lehmann T. K., Griffith A. W., Kang Y., Litaker R. W. (2017). Ocean Warming Since 1982 has Expanded the Niche of Toxic Algal Blooms in the North Atlantic and North Pacific Oceans. Proc. Natl. Acad. Sci. 114, 4975–4980. doi: 10.1073/pnas.1619575114
Gray M., Wawrik B., Paul J., Casper E. (2003). Molecular Detection and Quantitation of the Red Tide Dinoflagellate Karenia Brevis in the Marine Environment. Appl. Environ. Microbiol. 69, 5726–5730. doi: 10.1128/AEM.69.9.5726-5730.2003
Guillou L., Nézan E., Cueff V., Erard-Le Denn E., Cambon-Bonavita M. A., Gentien P., et al. (2002). Genetic Diversity and Molecular Detection of Three Toxic Dinoflagellate Genera (Alexandrium, Dinophysis, and Karenia) From French Coasts. Protist. 153, 223–238. doi: 10.1078/1434-4610-00100
Guiry M. D., Guiry G. M. (2022). “AlgaeBase,” in World-Wide Electronic Publication (Galway: National University of Ireland). Available at: https://www.algaebase.org.
Gu H. F., Luo Z. H., Zhang X. D., Xu B., Fang Q. (2013). Morphology, Ultrastructure and Phylogeny of Takayama Xiamenensis Sp. Nov. (Gymnodiniales, Dinophyceae) From the East China Sea. Phycologia. 52, 256–265. doi: 10.2216/12-090.1
Holland P. T., Shi F., Satake M., Hamamoto Y., Ito E., Beuzenberg V., et al. (2012). Novel Toxins Produced by the Dinoflagellate Karenia Brevisulcata. Harmful Algae. 13, 47–57. doi: 10.1016/j.hal.2011.10.002
Huang H. L., Gao W. F., Zhu P., Zhou C. X., Qiao L. L., Dang C. Y., et al. (2020). Molecular Method for Rapid Detection of the Red Tide Dinoflagellate Karenia Mikimotoi in the Coastal Region of Xiangshan Bay, China. J. Microbiological Methods 168, 105801. doi: 10.1016/j.mimet.2019.105801
Huang H. L., Zhu P., Zhou C. X., He S., Yan X. J. (2017). The Development of Loopmediated Isothermal Amplification Combined With Lateral Flow Dipstick for Detection of Karlodinium Veneficum. Harmful Algae. 62, 20–29. doi: 10.1016/j.hal.2016.11.022
Iwataki M., Lum W. M., Kuwata K., Takahashi K., Arima D., Kuribayashi T., et al. (2021). Morphological Variation and Phylogeny of Karenia Selliformis (Gymnodiniales, Dinophyceae) in an Intensive Cold-Water Algal Bloom in Eastern Hokkaido, Japan. Harmful Algae. 114, 102204. doi: 10.1016/j.hal.2022.102204
Jeong H. J., Jang S. H., Moestrup O., Kang N. S., Lee S. Y., Potvin E., et al. (2014). Ansanella Granifera Gen. Et Sp Nov (Dinophyceae), a New Dinoflagellate From the Coastal Waters of Korea. Algae. 29, 75–99. doi: 10.4490/algae.2014.29.2.075
Jeong H. J., Ok J. H., Lim A. S., Kwon J. E., Kim S. J., Lee S. Y. (2016). Mixotrophy in the Phototrophic Dinoflagellate Takayama Helix (Family Kareniaceae): Predator of Diverse Toxic and Harmful Dinoflagellates. Harmful Algae. 60, 92–106. doi: 10.1016/j.hal.2016.10.008
Kalyaanamoorthy S., Minh B. Q., Wong T. K. F., Von Haeseler A., Jermiin L. S. (2017). ModelFinder: Fast Model Selection for Accurate Phylogenetic Estimates. Nat. Methods 14, 587–589. doi: 10.1038/nmeth.4285
Kim H. G. (2010). An Overview on the Occurrences of Harmful Algal Blooms (HABs) and Mitigation Strategies in Korean Coastal Waters. Coast. Environ. ecosyst. Issues East China Sea., 121–131.
Kirkpatrick G. J., Millie D. F., Moline M. A., Schofield O. (2000). Optical Discrimination of a Phytoplankton Species in Natural Mixed Populations. Limnol. Oceanography. 45, 467–471. doi: 10.4319/lo.2000.45.2.0467
Kumar S., Stecher G., Tamura K. (2016). MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Liang Y. B. (2012). Investigation and Evaluation of Red Tide Disasters in China, (1933-2009). Oceanpress. 136.
Li Y. T., Liu K. S., Hou Y. Y., Yu J. J., Duan Y. H., Liu X. X., et al. (2018). Unveiling Picoeukaryotic Diversity and Distribution in the Bay of Large Reservoir System. Appl. Ecol. Environ. Res. 16, 7115–7128. doi: 10.15666/aeer/1605_71157128
Lim A. S., Jeong H. J., Ok J. H., Kim S. J. (2018). Feeding by the Harmful Phototrophic Dinoflagellate Takayama Tasmanica (Family Kareniaceae). Harmful Algae. 74, 19–29. doi: 10.1016/j.hal.2018.03.009
Lin Y. R., Cen J. Y., Wang J. Y., Liang Q. Y., Lv S. H. (2020). Preliminary Study on Interspecific Relationship and Hemolytic Activity of Four Karenia Species From South China Sea. Oceanologia Limnologia Sinica. 51, 1402–1411. doi: 10.11693/hyhz20200100002
Lin F. A., Guan C. J., Lu X. W. (2010). Effects of Red Tide Events Monitoring, Existence Questions and Suggestions in Coastal Areas in Recent Years in China. Mar. Environ. Science. 29, 148–151. doi: 10.1631/jzus.A1000244
Lin S. J., Wang L., Zheng L. M., Dong Y. W., Liu S. F., Ding S. X., et al. (2014). Current Status and Future Prospect of DNA Barcoding in Marine Biology. Acta Oceanologica Sinica. 36, 1–17. doi: 10.3969/j.issn.02534193.2014.12.001
Li Z., Shin H. H. (2018). Morphology and Phylogeny of an Unarmored Dinoflagellate, Karlodinium Jejuense Sp. Nov. (Gymnodiniales), Isolated From the Northern East China Sea. Phycological Res. 66, 318–328. doi: 10.1111/pre.12334
Litaker R. W., Vandersea M. W., Kibler S. R., Reece K. S., Stokes N. A., Lutzoni F. M., et al. (2007). Recognizing Dinoflagellate Species Using ITS rDNA Sequences. J. Phycol. 43, 344–355. doi: 10.1111/j.1529-8817.2007.00320.x
Liu G. Y., Ge K., Song L., Zhang Y. Y., Wang N. B. (2015). The Research Trend of Karenia Mikimotoi. Mar. Sci. 39, 117–122. doi: 10.11759/hykx20130702002
Liu S. Y., Gibson K., Cui Z. M., Chen Y., Sun X. X., Chen N. S. (2020). Metabarcoding Analysis of Harmful Algal Species in Jiaozhou Bay. Harmful Algae. 92, 101772. doi: 10.1016/j.hal.2020.101772
Liu Y. J., Yang G. P., Guan X. J. (2004). Molecular Genetic Diversity of Phytoplankton rbcL Gene in Jiaozhou Bay. Chin. J. Appl. Ecol. 15, 1626–1632. doi: 10.1007/BF02873086
Li X. D., Yan T., Lin J. N., Yu R. C., Zhou M. J. (2017). Detrimental Impacts of the Dinoflagellate Karenia Mikimotoi in Fujian Coastal Waters on Typical Marine Organisms. Harmful Algae. 61, 1–12. doi: 10.1016/j.hal.2016.11.011
Li X. D., Yan T., Yu R. C., Zhou M. J. (2019). A Review of Karenia Mikimotoi: Bloom Events, Physiology, Toxicity and Toxic Mechanism. Harmful Algae. 90, 101702. doi: 10.1016/j.hal.2019.101702
Lundholm N., Churro C., Fraga S., Hoppenrath M., Iwataki M., Larsen J., et al. (2009) IOC-UNESCO Taxonomic Reference List of Harmful Micro Algae. Available at: https://www.marinespecies.org/hab.
Luo Z. H., Wang L., Chan L., Lv S. H., Gu H. F. (2018). Karlodinium Zhouanum, a New Dinoflagellate Species From China, and Molecular Phylogeny of Karenia Digitata and Karenia Longicanalis (Gymnodiniales, Dinophyceae). Phycologia. 57, 401–412. doi: 10.2216/17-106.1
Lu D. D., Qi Y. Z., Gu H. F., Dai X. F., Wang H. X., Gao Y. H., et al. (2014). Causative Species of Harmful Algal Blooms in Chinese Coastal Waters. Algol. Stud. 145, 145–168. doi: 10.1127/1864-1318/2014/0161
Lv S. H., Cen J. Y., Wang J. Y., Ou L. J. (2019). The Research Status Quo , Hazard, and Ecological Mechanisms of Karenia Mikimotoi Red Tide in Coastal Waters of China. Oceanologia Limnologia Sinica. 50, 487–494. doi: 10.11693/hyhz20181000255
Marie D., Shi X. L., Rigaut-Jalabert F., Vaulot D. (2010). Use of Flow Cytometric Sorting to Better Assess the Diversity of Small Photosynthetic Eukaryotes in the English Channel. FEMS Microbiol. Ecol. 72, 165–178. doi: 10.1111/j.1574-6941.2010.00842.x
Mitchell S., Rodger H. (2007). Pathology of Wild and Cultured Fish Affected by a Karenia Mikimotoi Bloom in Irelan. Bull.-Eur. Assoc. Fish. Pathol. 27, 39–42.
Mooney B. D., Nichols P. D., De salas M. F., Hallegraeff G. M. (2007). Lipid, Fatty Acid, and Sterol Composition of Eight Species of Kareniaceae (Dinophyta): Chemotaxonomy and Putative Lipid Phycotoxins. J. Phycology. 43, 101–111. doi: 10.1111/j.1529-8817.2006.00312.x
Naar J. P., Flewelling L. J., Lenzi A., Abbott J. P., Granholm A., Jacocks H. M., et al. (2007). Brevetoxins, Like Ciguatoxins, are Potent Ichthyotoxic Neurotoxins That Accumulate in Fish. Toxicon. 50, 707–723. doi: 10.1016/j.toxicon.2007.06.005
Nézan E., Siano R., Boulben S., Six C., Bilien G., Chèze K., et al. (2014). Genetic Diversity of the Harmful Family Kareniaceae (Gymnodiniales, Dinophyceae) in France, With the Description of Karlodinium Gentienii Sp. Nov.: A New Potentially Toxic Dinoflagellate. Harmful Algae. 40, 75–91. doi: 10.1016/j.hal.2014.10.006
Ok J. H., Jeong H. J., Lee S. Y., Park S. A., Noh J. H. (2021). Shimiella Gen. Nov. And Shimiella Gracilenta Sp. Nov. (Dinophyceae, Kareniaceae), a Kleptoplastidic Dinoflagellate From Korean Waters and its Survival Under Starvation. J. Phycol. 57, 70–91. doi: 10.1111/jpy.13067
Place A. R., Bowers H. A., Bachvaroff T. R., Adolf J. E., Deeds J. R., Sheng J. (2012). Karlodinium Veneficum—The Little Dinoflagellate With a Big Bite. Harmful Algae. 14, 179–195. doi: 10.1016/j.hal.2011.10.021
Poli M. A., Musser S. M., Dickey R. W., Eilers P. P., Hall S. (2000). Neurotoxic Shellfish Poisoning and Brevetoxin Metabolites: A Case Study From Florida. Toxicon. 38, 981–993. doi: 10.1016/S0041-0101(99)00191-9
Qin Z. J., Yu K. F., Chen B., Wang Y. H., Liang J. Y., Luo W. W., et al. (2019). Diversity of Symbiodiniaceae in 15 Coral Species From the Southern South China Sea: Potential Relationship With Coral Thermal Adaptability. Front. Microbiol. 10. doi: 10.3389/fmicb.2019.02343
Richardson T. L., Pinckney J. L. (2004). Monitoring of the Toxic Dinoflagellate Karenia Brevis Using Gyroxanthin-Based Detection Methods. J. Appl. Phycology. 16, 315–328. doi: 10.1023/B:JAPH.0000047788.31312.4f
Richlen M. L., Morton S. L., Barber P. H., Lobel P. S. (2008). Phylogeography, Morphological Variation and Taxonomy of the Toxic Dinoflagellate Gambierdiscus Toxicus (Dinophyceae). Harmful Algae. 7, 614–629. doi: 10.1016/j.hal.2007.12.020
Ronquist F., Teslenko M., van der Mark P., Ayres D. L., Darling A., Höhna S., et al. (2012). MrBayes 3.2: Efficient Bayesian Phylogenetic Inference and Model Choice Across a Large Model Space. Syst. Biol. 61, 539–542. doi: 10.1093/sysbio/sys029
Scholin C. A., Herzog M., Sogin M., Anderson D. M. (1994). Identification of Group and Strain-Specific Genetic Markers for Globally Distributed Alexandrium (Dinophyceae). II. Sequence Analysis of a Fragment of the LSU rRNA Gene. J. Phycology. 30, 999–1011. doi: 10.1111/j.0022-3646.1994.00999.x
Scorzetti G., Brand L. E., Hitchcock G. L., Rein K. S., Sinigalliano C. D., Fell J. W. (2009). Multiple Simultaneous Detection of Harmful Algal Blooms (HABs) Through a High Throughput Bead Array Technology, With Potential Use in Phytoplankton Community Analysis. Harmful Algae. 8, 196–211. doi: 10.1016/j.hal.2008.05.003
Shao P., Chen Y. Q., Zhou H., Yuan J., Qu L. H., Zhao D., et al. (2004). Genetic Variability in Gymnodiniaceae ITS Regions: Implications for Species Identification and Phylogenetic Analysis. Mar. Biol. 144, 215–224. doi: 10.1007/s00227-003-1157-x
Siano R., Kooistra W. H. C. F., Montresor M., Zingone A. (2009). Unarmoured and Thin-Walled Dinoflagellates From the Gulf of Naples, With the Description of Woloszynskia Cincta Sp. Nov. (Dinophyceae, Suessiales). Phycologia. 48, 44–65. doi: 10.2216/08-61.1
Smayda T. J. (2010). Adaptations and Selection of Harmful and Other Dinoflagellate Species in Upwelling Systems 1. Morphology and Adaptive Polymorphism. Prog. Oceanography. 85, 53–70. doi: 10.1016/j.pocean.2010.02.004
Stern R. F., Andersen R. A., Jameson I., Küpper F. C., Coffroth M. A., Vaulot D., et al. (2012). Evaluating the Ribosomal Internal Transcribed Spacer (ITS) as a Candidate Dinoflagellate Barcode Marker. PloS One 7, e42780. doi: 10.1371/journal.pone.0042780
Stern R., Kraberg A., Bresnan E., Kooistra W. H. C. F., Lovejoy C., Montresor M., et al. (2018). Molecular Analyses of Protists in Long-Term Observation Programmes–Current Status and Future Perspectives. J. Plankton Res. 40, 519–536. doi: 10.1093/plankt/fby035
Takahashi K., Benico G., Lum W. M., Iwataki M. (2019). Gertia Stigmatica Gen. Et Sp. Nov. (Kareniaceae, Dinophyceae), a New Marine Unarmored Dinoflagellate Possessing the Peridinin-Type Chloroplast With an Eyespot. Protist 170, 125680. doi: 10.1016/j.protis.2019.125680
Thompson L. R., Sanders J. G., McDonald D., Amir A., Ladau J., Locey K. J., et al. (2017). A Communal Catalogue Reveals Earth’s Multiscale Microbial Diversity. Nature. 551, 457–463. doi: 10.1038/nature24621
Toldrà A., Andree K. B., Fernández-Tejedor M., Diogène J., Campàs M. (2018). Dual Quantitative PCR Assay for Identification and Enumeration of Karlodinium Veneficum and Karlodinium Armiger Combined With a Simple and Rapid DNA Extraction Method. J. Appl. Phycology. 30, 2435–2445. doi: 10.1007/s10811-018-1446-x
Tragin M., Vaulot D. (2018). Green Microalgae in Marine Coastal Waters: The Ocean Sampling Day (OSD) Dataset. Sci. Rep. 8, 14020. doi: 10.1038/s41598-018-32338-w
Ulrich R. M., Casper E. T., Campbell L., Richardson B., Heil C. A., Paul J. H. (2010). Detection and Quantification of Karenia Mikimotoi Using Real-Time Nucleic Acid Sequence-Based Amplification With Internal Control RNA (IC-NASBA). Harmful Algae. 9, 116–122. doi: 10.1016/j.hal.2009.08.010
Wang J. Y., Cen J. Y., Li S., Lv S. H., Moestrup Ø., Chan K. K., et al. (2018). A Re-Investigation of the Bloom-Forming Unarmored Dinoflagellate Karenia Longicanalis (Syn. Karenia Umbella) From Chinese Coastal Waters. J. Oceanol. Limnol. 36, 2202–2215. doi: 10.1007/s00343-019-7191-4
Wang L., Zhang C. Y., Chen G. F., Wang Y. Y., Fu M. Q. (2020). Development of a Rapid Screening Test for Karenia Mikimotoi by Using Loop-Mediated Isothermal Amplification and Lateral Flow Dipstick. J. Appl. Phycology. 32, 3143–3155. doi: 10.1007/s10811-020-02203-3
Wietkamp S., Krock B., Gu H. F., Voß D., Klemm K., Tillmann U. (2019). Occurrence and Distribution of Amphidomataceae (Dinophyceae) in Danish Coastal Waters of the North Sea, the Limfjord and the Kattegat/Belt Area. Harmful Algae. 88, 101637. doi: 10.1016/j.hal.2019.101637
Winnepenninckx B., Backeljau T., Wachter R. D. (1993). Extraction of High Molecular Weight DNA From Molluscs. Trends Genet. Tig. 9, 407. doi: 10.1016/0168-9525(93)90102-N
Wolny J. L., Scott P. S., Tustison J., Brooks C. R. (2015). Monitoring the 2007 Florida East Coast Karenia Brevis (Dinophyceae) Red Tide and Neurotoxic Shellfish Poisoning (NSP) Event. Algae. 30, 49–58. doi: 10.4490/algae.2015.30.1.049
Wu W. X., Wang L., Liao Y., Huang B. Q. (2015). Microbial Eukaryotic Diversity and Distribution in a River Plume and Cyclonic Eddy-Influenced Ecosystem in the South China Sea. Microbiologyopen. 4, 826–840. doi: 10.1002/mbo3.282
Xu Y. R., He S., Zhou C. X., Yan X. J. (2014). A Review of Karlotoxins. Mar. Sci. 38, 113–118. doi: 10.11759/hykx20130819001
Xu C. Y., Huang M. Z., Du Q. (2010). Ecological Characteristics of Important Red Tide Species in Fujian Coastal Waters. J. Appl. Oceanography. 29, 434–441. doi: 10.3724/SP.J.1035.2010.01223
Xu N., Pang S. J., Liu F. (2012). Molecular Identification of a Bloom-Forming Species Isolated From Sanggou Bay in Shandong Province. Mar. Sci. 36, 13–18.
Yang Z. B., Hodgkiss I. J., Hansen G. (2001). Karenia Longicanalis Sp. Nov. (Dinophyceae): A New Bloom-Forming Species Isolated From Hong Kong, May 1998. Bot. Mar. 44, 67–74. doi: 10.1515/BOT.2001.009
Yang H. J., Hu Z. X., Tang Y. Z. (2021). Plasticity and Multiplicity of Trophic Modes in the Dinoflagellate Karlodinium and Their Pertinence to Population Maintenance and Bloom Dynamics. J. Mar. Sci. Eng. 9, 51. doi: 10.3390/jmse9010051
Yasumoto T., Underdal B., Aune T., Hormazabal V., Skulberg O., Oshima Y. (1990). Screening for Hemolytic and Ichthyotoxic Components of Chrysochromulina Polylepis and Gyrodinium Aureolum From Norwegian Coastal Waters. Toxic Mar. Phytoplankton, 436–440.
Yuan J., Mi T. Z., Zhen Y., Yu Z. G. (2012). Development of a Rapid Detection and Quantification Method of Karenia Mikimotoi by Real-Time Quantitative PCR. Harmful Algae. 17, 83–91. doi: 10.1016/j.hal.2012.03.004
Zhang H., Litaker W., Vandersea M. W., Tester P., Lin S. J. (2008). Geographic Distribution of Karlodinium Veneficum in the US East Coast as Detected by ITS-Ferredoxin Real-Time PCR Assay. J. Plankton Res. 30, 905–922. doi: 10.1093/plankt/fbn047
Zhang Q. C., Qiu L. M., Yu R. C., Kong F. Z., Wang Y. F., Yan T., et al. (2012). Emergence of Brown Tides Caused by Aureococcus Anophagefferens Hargraves Et Sieburth in China. Harmful Algae. 19, 117–124. doi: 10.1016/j.hal.2012.06.007
Zhang Q. C., Song J. J., Yu R. C., Yan T., Wang Y. F., Kong F. Z., et al. (2013). Roles of Mixotrophy in Blooms of Different Dinoflagellates: Implications From the Growth Experiment. Harmful Algae. 30, 10–26. doi: 10.1016/j.hal.2013.08.003
Zhang W. W., Xie Y. W., Yang J. H., Yang Y. N., Li D., Zhang Y., et al. (2017). Applications and Prospects of Metabarcoding in Environmental Monitoring of Phytoplankton Community. Asian J. Ecotoxicol. 12, 15–24. doi: 10.7524/AJE.1673-5897.20160729002
Zheng J. B., Zhang F. Y., Ma L. B., Lu Y. N. (2009a). Sequence Analysis of Karenia Mikimotoi Based on Ribosomal 18S rDNA and Internal Transcribed Space Regions. J. Shanghai Ocean Univ. 18, 680–685.
Keywords: Karenia, Karlodinium, DNA barcode, species diversity, Takayama, ITS region
Citation: Zhang W, Zhang Q, Smith KF, Qiu L, Liu C, Yin X and Liu Q (2022) Development of Specific DNA Barcodes for the Dinophyceae Family Kareniaceae and Their Application in the South China Sea. Front. Mar. Sci. 9:851605. doi: 10.3389/fmars.2022.851605
Received: 10 January 2022; Accepted: 30 May 2022;
Published: 29 June 2022.
Edited by:
Giovanna Romano, Anton Dohrn Zoological Station, ItalyReviewed by:
Haifeng Gu, State Oceanic Administration, ChinaSolenn Mordret, National Research Council Canada (NRC-CNRC), Canada
Copyright © 2022 Zhang, Zhang, Smith, Qiu, Liu, Yin and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Limei Qiu, cWl1bGltZWlAcWRpby5hYy5jbg==
†These authors have contributed equally to this work and share first authorship
 Weiqian Zhang
Weiqian Zhang Qingchun Zhang
Qingchun Zhang Kirsty F. Smith
Kirsty F. Smith Limei Qiu
Limei Qiu Chao Liu4,5,7
Chao Liu4,5,7