- 1National Engineering Research Centre of Marine Facilities Aquaculture, Zhejiang Ocean University, Zhoushan, China
- 2Scientific Observing and Experimental Station of Fishery Resources for Key Fishing Grounds, Ministry of Agriculture and Rural Affairs of the People's Republic of China, Zhejiang Marine Fisheries Research Institute, Zhoushan, China
- 3Fishery College, Zhejiang Ocean University, Zhoushan, China
Understanding fish stock distribution is crucial in the context of sustainable fishery development. Traditional approaches such as net catching, mark recapturing, and visual counting are often time-consuming, less effective, and prone to subjective bias. Recently, environmental DNA (eDNA) technology has made up for the shortcomings of traditional fishery resource survey methods and has been widely used for species monitoring, biodiversity assessment, and biomass estimation. In this study, the eDNA method was used to analyze the potential distribution range of the black sea bream (Acanthopagrus schlegelii) in the East China Sea. The black sea bream-specific primers and probe were designed and applied in the current study. Results showed high black sea bream eDNA concentrations in Dasha fishing ground, Yangtze Estuary fishing ground, and Zhoushan fishing ground. Higher concentrations of black sea bream eDNA were found in water depths approximately 30–40 m. The eDNA hotspots were mainly concentrated at 29°N–33°N, 122°E–125°E, but no significant differences in horizontal or vertical distributions were observed. Temperature is a critical factor in the presence and black sea bream eDNA among environmental variables in the correlation analysis. In addition, a linear formula between environmental factors and the black sea bream eDNA concentration of black sea bream was predicted, which may be helpful for future fishery resource surveys. To summarize, the present paper provides a new idea for marine resource surveys. It also affords a method and theoretical basis for applying the eDNA technology in fishery surveys, which is expected to become an essential tool for fishery resource surveys in the future.
Introduction
The balance of the marine ecosystem is subject to resource over-exploitation and pollution (Pauly et al., 2005; Hader et al., 2020), resulting in severe damage to the habitats of many marine organisms (Bruno et al., 2018). The ecological functions and services of the ecosystem are also negatively affected (Hutchings et al., 2010). Accordingly, the management and protection of marine fishery resources must be strengthened. Understanding the spatial distribution and the abundance characteristics of species plays a critical role in assessing resource status, formulating strategies to conserve and restore wild stocks, and implementing marine ecological protection and marine fishery management (Gilbey et al., 2021).
Traditional approaches such as net catching, mark recapturing, and visual counting are often time-consuming (Thomsen et al., 2012; Yamamoto et al., 2016), less effective (Thomsen and Willerslev, 2015; Hänfling et al., 2016), and prone to subjective bias (Fujii et al., 2019). The environmental DNA (eDNA) approach provides a new strategy for fishery resource monitoring (Zou et al., 2020; Kume et al., 2021). Compared with traditional survey methods, the eDNA method has many advantages, such as robust sensitivity (Jerde et al., 2011), exquisite efficiency, and environmental friendliness (Goldberg et al., 2016). eDNA refers to the sum of DNA fragments released by organisms into the surrounding environment (such as soil, sediment, and water environment) (Thomsen and Willerslev, 2015). All DNA fragments are extracted from environmental samples and then undergo either high-throughput sequencing or real-time quantitative PCR (qPCR) to complete the qualitative and quantitative analyses of target organisms. The eDNA approach has been extensively used in the detection of aquatic organisms (Ficetola et al., 2008; Dejean et al., 2012; Lacoursière-Roussel et al., 2016a; Wilcox et al., 2016; Lawson Handley et al., 2019; Wang et al., 2021a). eDNA can sensitively detect the presence or the absence of fish and can be used to monitor invasive and endangered species as well as cryptic species (Ficetola et al., 2008; Dejean et al., 2012; Nester et al., 2020). Numerous studies have shown that target species’ eDNA concentration is positively correlated with biomass such that species biomass in aquatic habitats can be assessed by measuring eDNA concentrations (Takahara et al., 2012; Klymus et al., 2015; Lacoursière-Roussel et al., 2016a). When high-throughput sequencing is combined with eDNA studies, it enables rapid multispecies community monitoring and biodiversity analysis using universal primers (Jackman et al., 2021). Environmental DNA techniques have been validated in many aquatic ecosystems, including semi-enclosed or confined waters and open oceans, such as rivers, lakes, estuaries, and seas (Hänfling et al., 2016; Wilcox et al., 2016; Itakura et al., 2019; Takeuchi et al., 2019; Afzali et al., 2020; Wang et al., 2021b). However, most of the previous studies have neglected vertical distribution (Zhang et al., 2020). This has hindered a better understanding of the potential distribution of aquatic organisms. Therefore, conducting omnidirectional, deep-level investigation and research on fishery resources is imperative to evaluate fishery resources better.
China’s important fishing area, the East China Sea, has a flat seabed and multiple water masses (Zhao et al., 2015). It provides good breeding, survival, and overwintering conditions for various fish (Zhang et al., 2018). Since the 1970s, great changes have taken place in fishery resources due to the increasing intensity of marine fishing (Zhao et al., 2015). This development is manifested in the miniaturization of the fishing structure, precocious maturity, and simple age structure. The decline of fishery resources is becoming increasingly apparent (Maranghi et al., 2013). The black sea bream (Acanthopagrus schlegelii), a typical coastal warm water fish widely distributed in the north-western Pacific, is a major target for fisheries (Chen and Zhang, 2016). Over the last few years, overfishing and marine habitat environment deterioration resulted in a severe decline in black sea bream stock resources (Wang et al., 2015). The conservation, restoration, and proliferation of black sea bream resources are pressing.
In this study, specific primers and TaqMan probe are designed to monitor black sea bream resources in the East China Sea using the eDNA method. The present study aims to provide a means of assessing fishery resources in open ocean areas to facilitate future marine resource management and conservation.
Materials and methods
Field sites and sample collection
During the joint survey in the East China Sea from 14 May to 24 May 2019, a total of 44 stations (Figure 1) were designed, and 172 water samples (Figures 1 and S1) were collected using 24 × 20 L Niskin bottles controlled by the SBE 911 Plus CTD system (Sea-Bird Scientific, Bellevue, WA, USA). According to the actual water depth, three layers were defined: the first layer, which was surface water (0–10 m), the second layer (10–50 m), and the third layer (50 m to the bottom). CTD was used to record total depth, water sample depth, temperature, salinity, oxygen, and turbidity, and other environmental variables (chlorophyll, PO43−, NH4+, SiO32−, NO3− and NO2−) were determined by the staff of the First Institute of Oceanography Ministry of Natural Resources according to the Chinese national standards for each variable (AQSIQ, 1997). Seawater samples were collected using disposable sterile plastic bags, and then 1 L of seawater was immediately filtered using a vacuum pump through a 47-mm glass fiber filter with a pore size of 0.45 μm (Xingya Purification Materials Company, Shanghai, China). To avoid cross-contaminants between sampling sites, all sampling and filtering devices were soaked in 10% sodium hypochlorite solution for more than 10 min before and after each use, cleaned with tap water, and then washed with distilled water. New gloves were replaced after each sample, and 1 L of distilled water was filtered as the negative control on each day of sampling.
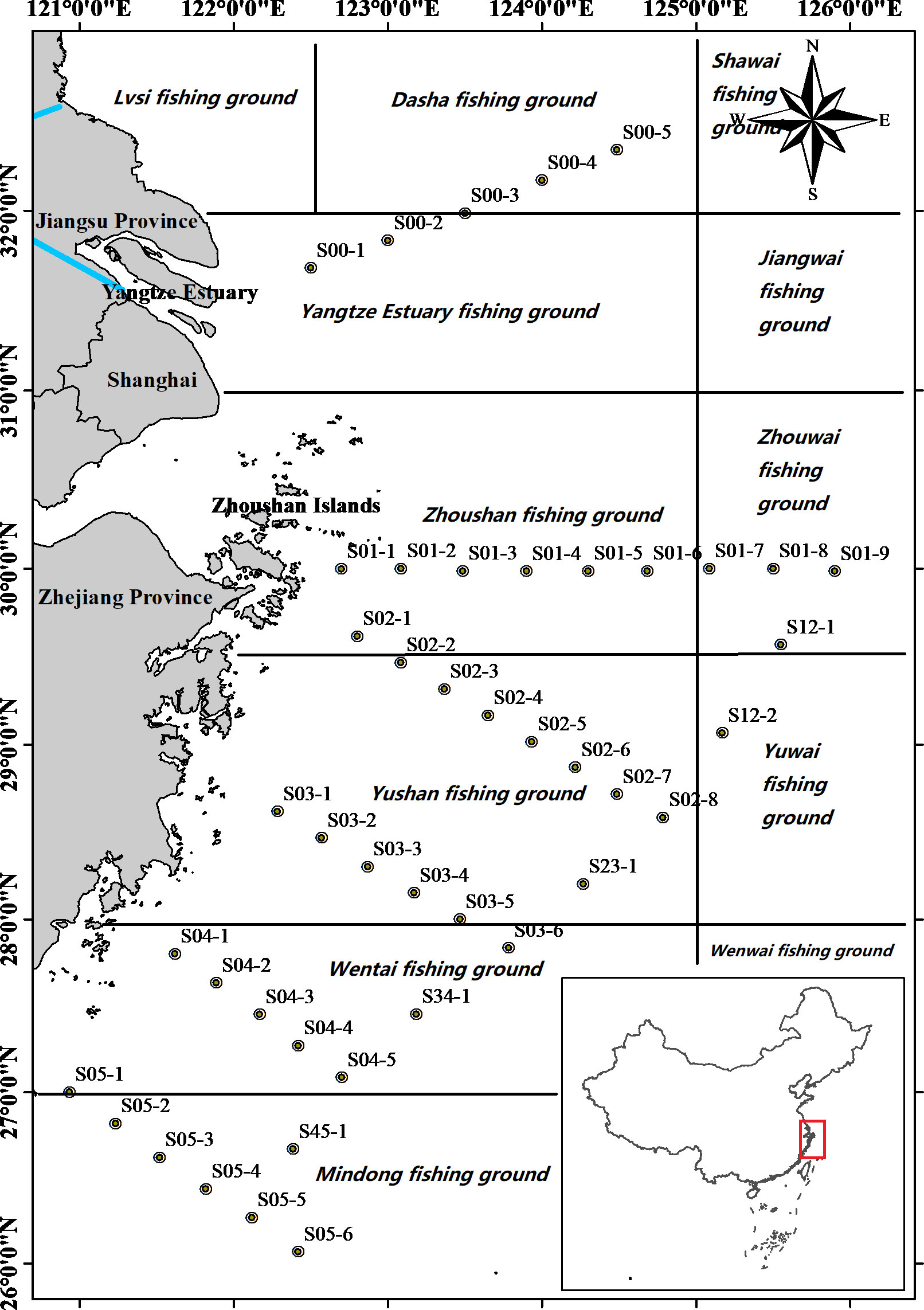
Figure 1 Map of sampling stations (black dots) of the East China Sea with adjacent provinces and fishing grounds. The black points represent sampling sites. The blue line represents the Yangtze River.
After filtration, each filter was folded individually and wrapped in aluminum foil, frozen in liquid nitrogen at −196°C, taken back to the laboratory, and stored at −80°C for further eDNA extraction.
Environmental DNA extraction
eDNA was extracted from the filters using DNeasy Blood and Tissue Kit (Qiagen, Hilden, Germany). Inhibitors were removed using OneStep PCR Inhibitor Removal Kit (Zymo Research, Irvine, CA, USA) to improve the quality of eDNA. The purity and concentrations of eDNA were measured by spectrophotometer (NanoDrop 2000, Thermo Fisher Scientific, Waltham, MA, USA).
Black sea bream-specific primers and probe design
The Cytb gene sequences of the black sea bream and its taxonomy-related species, such as Acanthopagrus latus, Acanthopagrus butcheri, Diplodus capensis, Diplodus argenteus, Menticirrhus undulatus, Sparus aurata, Sebastes owstoni, and Sebastes taczanowskii, were used to design black sea bream-specific primers and probe (Figure 2). Specific primers and TaqMan probe of the black sea bream were designed using SeqMan, Primer Express 3.0.1, AlleleID 6 software, and primer design tool (https://www.ncbi.nlm.nih.gov/tools/primer-blast). The specificity of the designed primers and probe was verified in silico (primer design tool in National Center for Biotechnology Information (NCBI)) and in vitro (i.e., performing real-time PCR using genomic DNA of black sea bream and its close relatives as the template and DNA extracted from black sea bream pond water as a template).

Figure 2 Sequences of designed black sea bream-specific primers and probe and the results of comparison with other closely related species.
Plasmid standard preparation
Black sea bream DNA was extracted by the standard phenol–chloroform–isoamyl alcohol method and amplified using the designed primers. The amplification conditions were as follows: predenaturation at 94°C for 3 min, denaturing at 94°C for 30 s, annealing at 60°C for 30 s and extended at 72°C for 1 min, and 72°C complex extensions for 10 min, for a total of 35 cycles. A 25 μl PCR system was used: 2× San Taq PCR Mix (with Blue Dye) 12.5 μl (Sangon Biotech, Shanghai, China), forward and reverse primers (0.2 μmol/L) each at 1 μl, template DNA 1 μl, and ddH2O 9.5 μl.
Plasmid standard was prepared by inserting amplified black sea bream PCR products into a pESI-T vector. Then the ligation product was transformed into Escherichia coli DH5α competent cells and cultured at 37°C for 30 min at 180 r/min. Bacterial solution measuring 100 μl was evenly coated on Luria broth (LB) solid culture medium containing ampicillin (100 μg/ml). Inverted culture was performed at 37°C overnight. After blue-white selection, PCR was performed on the bacterial fluid, and the positive results were sent to Sangon Biotech (Shanghai, China) for sequencing. The sequencing results were analyzed and compared for homology. After 24 h of expanded culture, the plasmids were extracted using a high-purity plasmid extraction kit. The purity and concentration of the extracted plasmids were measured by a spectrophotometer (NanoDrop 2000, Thermo Fisher Scientific), diluted to 108 copies/μl of standard, and stored at −80°C.
Black sea bream detection using environmental DNA
To avoid contamination, the PCR setup, including preparation and addition of the standard, was performed in one room and qPCR cycling in another. Real-time PCR was performed with a 7300Plus Real-Time PCR System (ABI, Foster City, CA, USA). The amplification conditions were as follows: denaturation at 94°C for 5 s, and annealing, extension, and data acquisition at 60°C for 30 s, for a total of 40 cycles. The reaction system (20 μl) contained TaqMan™ Fast Advanced Master Mix (Thermo Fisher Scientific) 10 μl, each primer (10 μM) 0.4 μl, TaqMan probe (10 μM) 0.2 μl, 4 μl eDNA extracted, and PCR-grade water 5 μl (Sangon, Shanghai, China). Each experiment was repeated three times, and three negative controls (pure water as the template) were set. Plasmid DNA was diluted from 108 copies/μl in a 10-fold concentration gradient to 101 copies/μl as a standard for quantitative PCR. Series dilutions of plasmid standards were used to determine the limit of detection (LOD) and limit of quantification (LOQ) of the whole method.
Statistical analysis
All statistical analyses were performed using R software version 3.5.3. Since the data between water layers and stations did not meet the normality and homogeneity of variance, statistical differences in black sea bream eDNA concentration among the three layers and stations were assessed using the Kruskal–Wallis test. The average eDNA concentration was calculated for each water layer. It is calculated as follows: the average for each layer is the sum of all eDNA concentrations in that water layer divided by the number of stations.
The relationship between environmental variables and log-transformed black sea bream eDNA concentrations (i.e. Ln(eDNA + 1)) was calculated using the 'pheatmap' package.
Multiple linear regression (MLR) was used to determine the best fitting models for black sea bream eDNA concentrations based on environmental variables. A preliminary analysis was first carried out to ensure that the assumptions of normality, linearity, and homoscedasticity were not violated. The eDNA concentrations were logarithm transformed to conform to a normal distribution prior to analysis. Within the MLR model, the 12 environmental variables (i.e., total depth, water sampling depth, temperature, salinity, oxygen, turbidity, chlorophyll, PO43−, NH4+, SiO32−, NO3−, and NO2−) were included as independent variables, and Ln([eDNA]+1) was included as the dependent variables. The MLR was carried out utilizing all subset regression methods of the ‘Leaps’ package. The adjusted Akaike information criterion (AIC) and the ‘gvlma’ package were used to validate the MLR model. Co-linearity between predictive variables was assessed using variance inflation factors (VIFs), and covariates with VIF greater than 5 were removed.
The sampling stations were visualized with ArcGIS 10.1 software. Gridding of black sea bream eDNA concentrations data at each sampling station was performed using Kriging interpolation and culling of gridded data from the study area using mask extraction.
The spatial distribution of black sea bream was visualized with Ocean Data view 4 software. This was performed as follows: black sea bream log transformed eDNA concentrations at each sampling site were gridded by the DIVA gridding method, adjusting the scale lengths to cover the entire study area. Other parameters were selected as default values and applied to all graphs.
Result
Specificity of primers and probe
A set of black sea bream-specific primers and a probe were developed to amplify a 146-bp sequence based on Cytb. The sequence was as follows: black sea bream-cytb-F: CTTCTCGGTCTCTGCTTAATCTCC, black sea bream-cytb-R GCGTGGAGGTTTCGGATTAG, black sea bream-cytb-P: ACTTCCGACATTGCCAC (Figure 2). The initial concentration of the plasmid standard was 2.78 × 108. LOD and LOQ were 27.80 and 84.24, respectively. In the results of qPCR, the Ct value of the plasmid standard was 13.3–32. The correlation coefficient, R2, of the curve was 99.99%. Amplification efficiency was 107.074%. Within 40 cycles, only samples from black sea bream DNA as the template by using a detection limit of 27.8 were detected, while other species and negative controls were not detected. The results showed a good linear relationship within the standard range of dilution plasmid, and the standard curve established correctly reflected the amplification of Cytb gene in black sea bream.
Distribution of black sea bream
The black sea bream eDNA was detected in 35 samples (20.35%) at only eight stations (18.18%) of all the sites sampled. There is no significant difference in the black sea bream eDNA concentration among 44 sampling sites, indicating no significant difference in the horizontal distribution of black sea bream eDNA. The average concentration of eDNA was 1.238 ± 1.359 copies/L. Among the three horizontal water layers, the eDNA concentration of Layer 2 (1.481 ± 2.426 copies/L) was higher than that of the two other water layers (Layer 1, 0.883 ± 1.655 copies/L; water Layer 3, 1.257 ± 1.878 copies/L). There was no significant difference between the different water layers. The eDNA hotspots were mainly concentrated in the Yangtze Estuary fishing ground (S00-2), the Dasha fishing ground (S00-3 and S00-4), and the Zhoushan fishing ground (S01-2 and S01-3) (Figures 3).
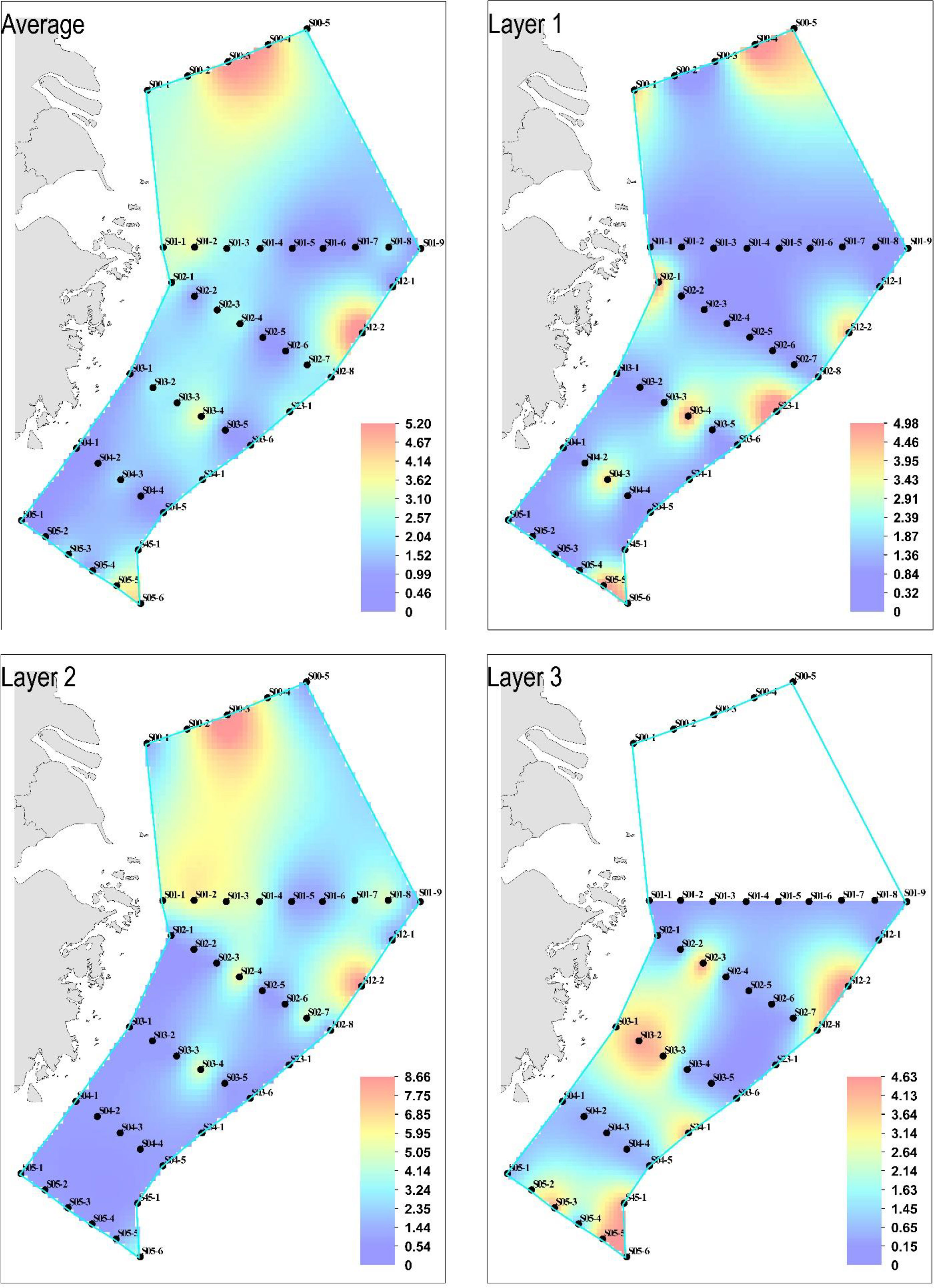
Figure 3 Horizontal distributions of black sea bream in the East China Sea. Black dots indicate sampling stations. The x-axis and the y-axis represent the longitude and the latitude, respectively. Concentrations of environmental DNA (eDNA) are depicted with gradient colors. Color bar scales indicate the log-transformed eDNA concentrations. Layer 1 (surface), Layer 2 (10–50 m), and Layer 3 (50–104 m).
Although no significant difference was observed in the vertical (difference between layers) black sea bream eDNA concentration distribution either, a higher concentration of eDNA was detected in water depths approximately 30–40 m, such as S00-1, Layer 2; S01-2, Layer 2; S01-3, Layer 2; S02-4, Layer 2; and S03-4, Layer 2 (Figure 4). For different longitudes, the eDNA hotspots were mainly concentrated at 29°N–33°N, within which the average concentration was 19.7541 copies/L. The latitude section shows that the eDNA hotspots were mainly in 122°E–125°E, and a significant difference existed between the sampling points in this range and those outside (p = 0.004212 < 0.01).
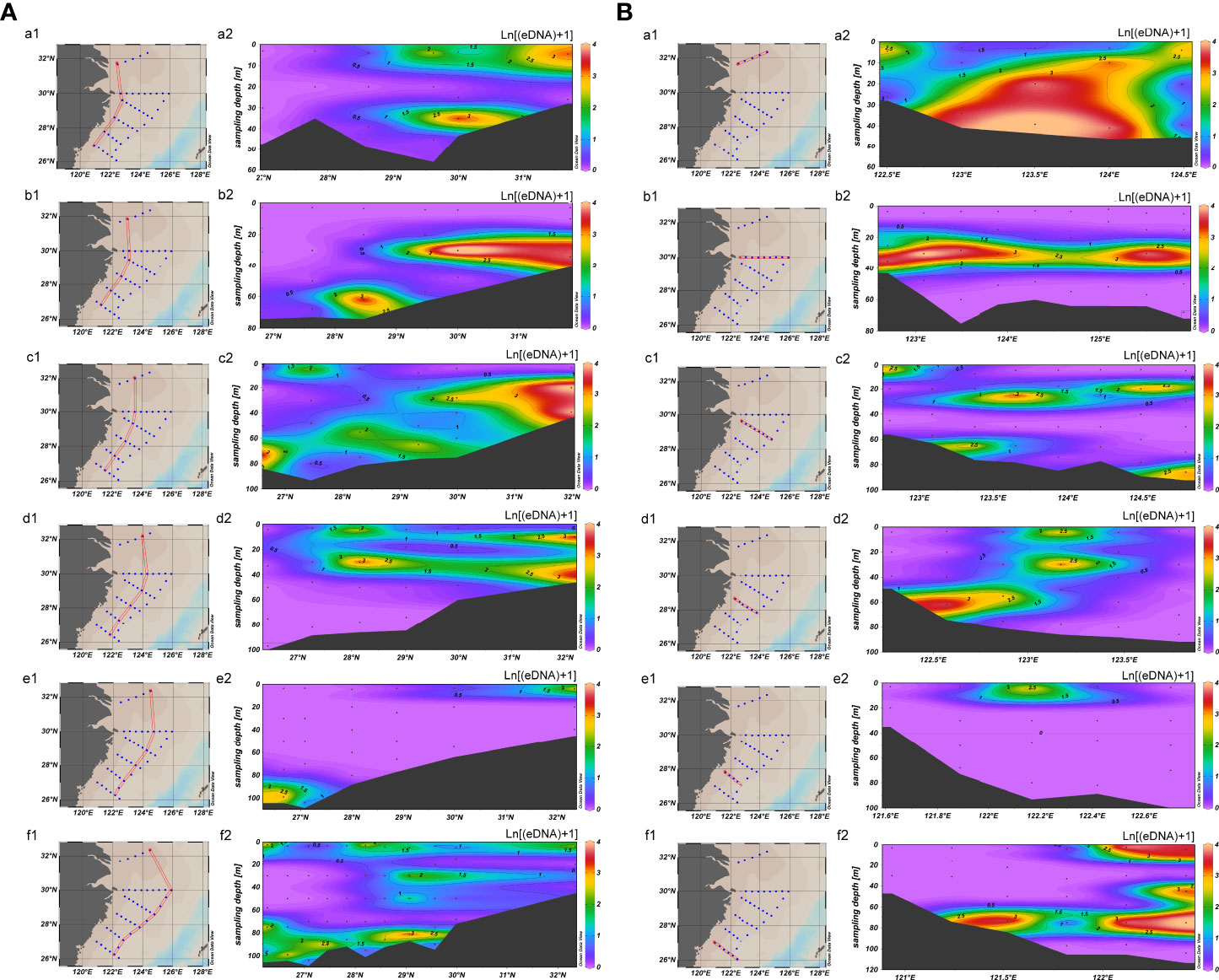
Figure 4 Vertical distributions of black sea bream in the East China Sea. (A) Vertical distributions of black sea bream along different longitude sections. The x-axis and the y-axis indicate the longitude and the depth, respectively. Black dots indicate sampling stations. Log transformed eDNA concentrations (copies/L) are shown with gradual colors. (B) Vertical distributions of black sea bream along different latitude sections. The x-axis and the y-axis indicate the latitude and the depth, respectively. Black dots indicate sampling stations. Concentrations of eDNA (copies/L) are shown with gradual colors.
Relationship between environmental factors and black sea bream environmental DNA
The correlation analysis of the 12 environmental variables at the measured sites revealed that black sea bream eDNA presence or absence and concentration are positively correlated with the temperature of sampling stations at a significant level (p < 0.05), as shown in Figure 5.
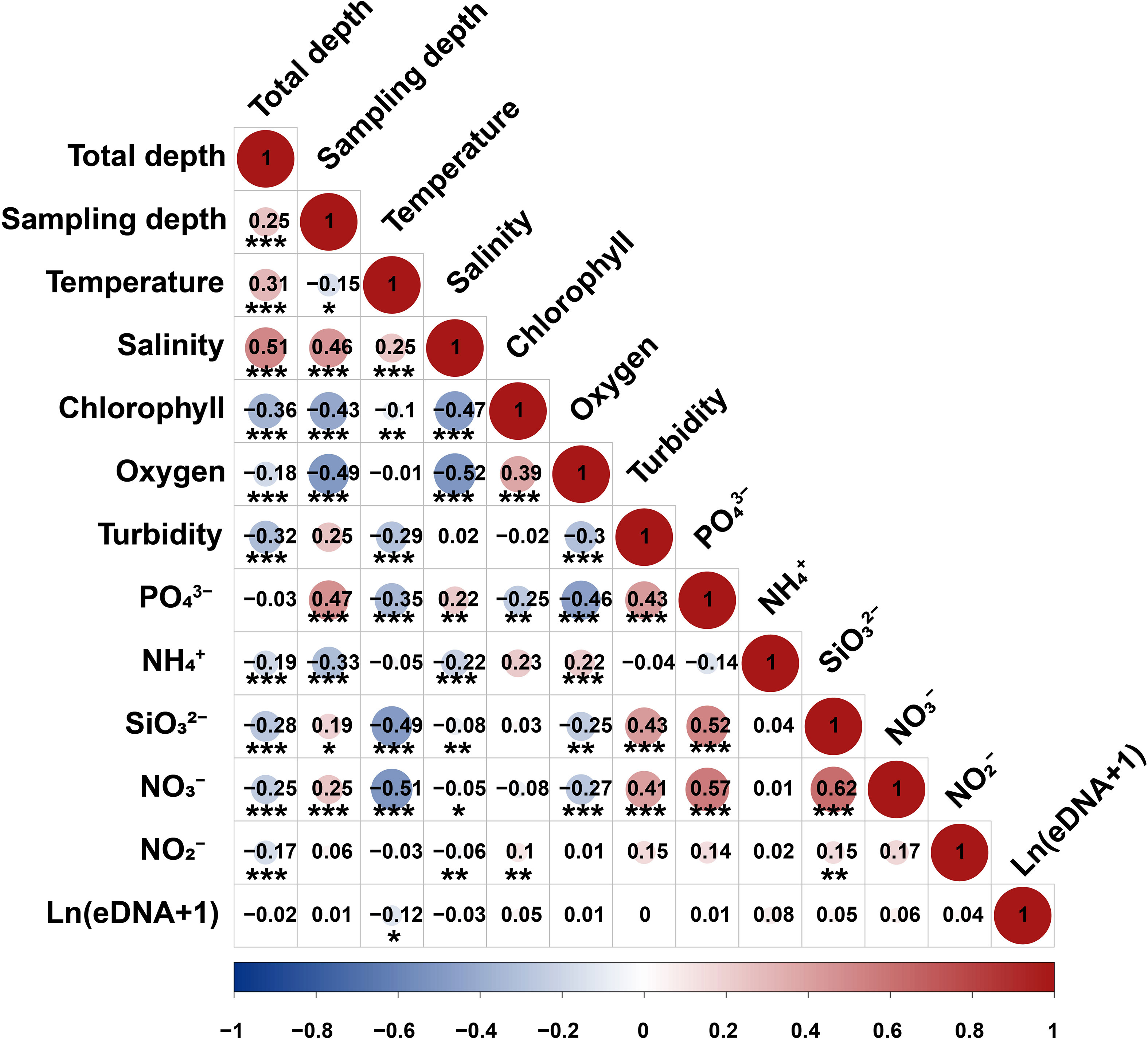
Figure 5 Correlation analysis between environmental variables and black sea bream environmental DNA (eDNA) concentrations (copies/L) as well as eDNA presence. The significance level is marked with * (p-value < 0.1), ** (0.05 < p-value < 0.1), and *** (p-value < 0.01).
In MLR analysis, environmental factors and eDNA concentration were predicted as follows: ln[(eDNA) + 1] =0.0012 + 0.87 × Temperature + 1.15 × NO3−+ 1.37 × Salinity + 0.15 × PO43− (PO43−, p <0.01,Temperature, p < 0.05, NO3−, p < 0.05, Salinity, p < 0.05).
Discussion
Distribution of black sea bream
This study assessed the potential distribution of black sea bream in the East China Sea by measuring the eDNA presence/absence and concentrations at different sampling stations. From the perspective of horizontal distribution, black sea breams are mainly distributed in the Yangtze Estuary fishing ground, the Dasha fishing ground, and the Zhoushan fishing ground. The largest estuary in China and the western Pacific, the Yangtze estuary, has large seasonal variations in environmental factors such as temperature and salinity and has high primary productivity (Zhang et al., 2017). A total of 1,390 islands and more than 3,850 reefs are distributed in the sea area of the Zhoushan Archipelago. Many sessile organisms inhabit these rocky reefs (Saito et al., 2008; Bissegger et al., 2014). The excellent natural conditions of these sea areas provide an ideal habitat for black sea bream. Therefore, black sea bream is concentrated in the areas of the Yangtze River estuary and the Zhoushan Archipelago (Yamada et al., 2007; Liang et al., 2010).
Monitoring eDNA concentration enables predicting a target species’ potential distribution in the target area, reducing workload and research costs, and enabling more effective ecological monitoring (Erickson et al., 2016; Buxton et al., 2017). The spawning seasons of black sea bream populations vary between different sea areas. Black sea bream spawning times are as follows: from December to mid-March of the following year in Guangdong and Taiwan areas, from November to March of the following year in Hong Kong and its adjacent area (Law and Sadovy de Mitcheson, 2017), mid-March to early May in Fujian (Urbatzka et al., 2012), from early May to early June in Shandong (Zhang et al., 1980), and from April to May in Jiangsu (Cai, 1983). The water samples were collected in mid-May, close to the spawning time of black sea bream in Jiangsu, and the results showed that the black sea bream eDNA signals were high in the surrounding area of Jiangsu, which strongly suggests the presence of spawning fish. Previous studies have confirmed the ability of aquatic species spawning ground detection using eDNA (Takeuchi et al., 2019; Wang et al., 2020), for example, the use of environmental DNA monitoring to confirm the results of changes in spawning grounds of small yellow croaker (Larimichthys polyactis) that were first identified by traditional methods (Wang et al., 2020). The mystery spawning region of the Japanese eel Anguilla japonica was eventually discovered and validated by the eDNA approach (Takeuchi et al., 2019). In this study, environmental DNA hotspots coincided in time and extent, with black sea bream spawning grounds documented by traditional surveys (Yamada et al., 2007). We therefore speculated that the areas around S00-2 and S00-3 were the spawning sites of black sea bream in the sea near Jiangsu. Although this speculation will require more detailed field recruitment work, the use of the eDNA approach has greatly improved the efficiency of species monitoring.
The vertical distribution of eDNA depends on the location of organisms and their behavior (Murakami et al., 2019b). By investigating and researching the vertical distribution, species distribution and habitat preferences can be more comprehensively mastered. For example, changes in the eDNA concentration in the surface and deep layers during dawn and dusk can be used to estimate the times when organisms begin and end daily migrations (Govindarajan et al., 2021). eDNA concentration changes reflect the fish seasonal variation in thermal stratification and habitat preferences (Littlefair et al., 2021). Studies have shown that even when organisms are migrating, there is a residual eDNA signal at the major inhabited by the species and that this signal does not disappear completely (Allan et al., 2021). The black sea bream eDNA is mainly concentrated in the shallow zone of 20–40 m, which may be related to the fact that black sea bream live and breed mainly in this depth range (Wang et al., 2015). Black sea bream eggs are pelagic, and the planktonic life of the eggs and larvae estimated at about 20 days suggests that black sea bream can spread far from the current (Jean et al., 1998). In the sea area around the Yangtze Estuary, the distribution pattern of early fish larvae is more in the north and less in the south (Li et al., 2020). The high-density area of larvae is often on the offshore side of the spawning ground (Jiang et al., 2020), which can explain the high eDNA concentrations in surface water detected near the estuary of the Yangtze River. More eDNA was detected in the surface layer from the Mindong fishing ground (S05-5 and S05-6), which may be under the influence of the Taiwan warm current from March to May (Lu and Lee, 2014). Although the results of eDNA monitoring can be explained by the habits of black sea bream, the lack of traditional resource survey results prevents simultaneous validation of the correlation between environmental DNA and black sea bream resources. In addition, eDNA is unable to distinguish between the size, age, and survival status of target organisms (Nevers et al., 2018; Deutschmann et al., 2019). eDNA can also be dispersed and degraded by complex environmental factors in the ocean, such as currents (Yamamoto et al., 2016; Murakami et al., 2019a), water temperature (Jane et al., 2015; Lacoursière-Roussel et al., 2016b), pH, and UV (Hansen et al., 2018). However, research on these issues has focused more on closed and semi-closed environments. Fewer studies have been conducted on large open areas (Yamamoto et al., 2016; Murakami et al., 2019a). Therefore, given the obvious knowledge gaps that still exist regarding the longevity and influencing factors of eDNA signaling, eDNA studies should be seen as complementary rather than a ‘stand-alone tool’, especially in the flowing sea area. More studies are needed to further improve our knowledge of the parameters and processes that could potentially affect the eDNA distribution in the ocean.
Among the three water layers, the highest average concentration of eDNA was found in the second layer. This difference is inextricably related to halocline, pycnocline, and eDNA degradation (Hansen et al., 2018; Takasu et al., 2019; Jeunen et al., 2020). In addition, water temperature and bait distribution are influential factors affecting the vertical distribution of fish (Gibson, 1994). The results of water sample display at a sampling site may not necessarily represent the biological condition of the site (Nakagawa et al., 2018). Considering target species habitat preference and the water column environment differences, both the horizontal and vertical distributions of organisms need to be taken into account in future surveys of other marine organisms to better assess fishery resources.
Environmental variables correlated with black sea bream environmental DNA
The survival and growth of fish are inextricably linked to the marine environment. Thus, environmental factors play pivotal factors in the investigation of fishery resources (Mukherjee et al., 2012). In the predicted regression equation, in addition to the previously mentioned temperature factors that would influence black sea bream eDNA concentrations, PO43−, NO3−, and salinity may influence eDNA concentration too. The concentration and the presence or absence of eDNA of black sea bream are also significantly correlated with temperature (p < 0.05). The black sea bream’s most suitable growing temperature is 17°C–25°C, and the spawning temperature is 16°C–21°C (Jiang et al., 2012). The average temperature, 15.51°C, at the sampling stations near Jiangsu is close to the spawning temperature, resulting in more eDNA being detected. The concentrations of PO43−, in a few ranges where eDNA hotspots showed up, are between 0 and 0.5 mg/L, while the concentrations of NO3− are within the extent of 0–12 mg/L. The concentrations of both PO43− and NO3− are at normal levels. A particular concentration of PO43− and NO3− is conducive to algae growth, increasing primary productivity and providing bait directly or indirectly (Sarre et al., 2000). Salinity is also an essential factor affecting the distribution and abundance of black sea bream. Although this species is a broad-salinity fish, only the sea area with high salinity can promote its gonad maturity and spawning (Murata et al., 1997). The spatial distribution of fish stocks and their relationship with the environment has always been a research hot spot in fishery ecology (Hu et al., 2020). Consequently, we propose that in future surveys of fishery resources in the open ocean, consideration should be given to inferring the distribution patterns under different environmental conditions by predicting the regression equations between environmental factors and fish, so as to obtain the spatial distribution characteristics and influencing factors of target fish and conduct surveys with more specificity. This proposal will provide theoretical reference and support for the development of management strategies, such as the sampling design of fish communities and the establishment and optimization of fish nature reserves.
In the present context, the data obtained from quantitative eDNA monitoring methods remain questionable due to several complex environmental factors (Hinlo et al., 2018; Tillotson et al., 2018; Antognazza et al., 2021). Thus, despite the relatively high eDNA detection rates in this study, information on the spatial resolution of these detections (distribution and biomass) is inconclusive. More studies are needed in the future to validate and complement the data provided by eDNA methods, such as traditional fishery surveys for confirmation at eDNA signal hotspots.
Conclusion
This paper explored the utility of the eDNA-based approach to conduct fishery resource monitoring and provided technical support and reference for future fishery protection and resource reservation. Specific primers and probe for black sea bream were designed to characterize its vertical and horizontal distribution in the East China Sea. The relationship between the distribution of black sea bream and the environmental variables was explored. In addition, the regression prediction models for black sea bream and environmental variables were established. In conclusion, this work indicates the feasibility of using eDNA to monitor fishery resources, providing an important, effective source of information for developing fishery management and conservation policies, with significant implications for fish resource conservation and recovery. It is worth emphasizing, nevertheless, that the information provided by eDNA monitoring still needs to be validated with higher resolution information. Therefore, the eDNA approach cannot currently be used as an alternative to traditional techniques, and this needs to be acknowledged.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Author contributions
HBZ: data curation, formal analysis, writing—original draft, methodology, and validation. YZ: writing—reviewing and editing. HLZ: writing—reviewing and editing. TG: writing—reviewing and editing. XW: supervision, software, methodology, writing—reviewing and editing, funding acquisition, and project administration. All authors contributed to the article and approved the submitted version.
Funding
This paper was funded by the National Key Research and Development Programme of China (2019YFD0901303), the National Natural Science Foundation of China (41806180, 41776171), the Zhejiang Provincial Key Research and Development Programme (2021C02047), the Science and Technology Project of Zhoushan (2022C41010), and the Shared Voyage Programme of the National Natural Science Foundation of China in the East China Sea (NORC2019-02, Xiangyanghong 18).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2022.848950/full#supplementary-material
References
Afzali S. F., Bourdages H., Laporte M., Mérot C., Normandeau E., Audet C., et al. (2020). Comparing environmental metabarcoding and trawling survey of demersal fish communities in the gulf of st. Lawrence, Canada. Environ. DNA 3 (1), 22–42. doi: 10.1002/edn3.111
Allan E. A., DiBenedetto M. H., Lavery A. C., Govindarajan A. F., Zhang W. G. (2021). Modeling characterization of the vertical and temporal variability of environmental DNA in the mesopelagic ocean. Sci. Rep. 11 (1), 21273. doi: 10.1038/s41598-021-00288-5
Antognazza C. M., Britton J. R., De Santis V., Kolia K., Turunen O. A., Davies P., et al. (2021). Environmental DNA reveals the temporal and spatial extent of spawning migrations of European shad in a highly fragmented river basin. Aquat. Conservation: Mar. Freshw. Ecosyst. 31 (8), 2029–2040. doi: 10.1002/aqc.3601
AQSIQ (1997). Sea Water Quality Standard (GB 3097-1997) (Beijing: Administration of Quality Supervision, Inspection and Quarantine: Standard Press of China).
Bissegger S., Martyniuk C. J., Langlois V. S. (2014). Transcriptomic profiling in silurana tropicalis testes exposed to finasteride. Gen. Comp. Endocrinol. 203, 137–145. doi: 10.1016/j.ygcen.2014.01.018
Bruno J. F., Bates A. E., Cacciapaglia C., Pike E. P., Amstrup S. C., Hooidonk R. V., et al. (2018). Climate change threatens the world’s marine protected areas. Nat. Climate Change 8 (5330), 499–503. doi: 10.1038/s41558-018-0149-2
Buxton A. S., Groombridge J. J., Zakaria N. B., Griffiths R. A. (2017). Seasonal variation in environmental DNA in relation to population size and environmental factors. Sci. Rep. 7 (1), 46294. doi: 10.1038/srep46294
Cai X. (1983). Preliminary observation on the natural spawning and breeding of black sea bream (Sparus macrocephalus). J. Jiangsu Ocean Fish (5), 63–71.
Dejean T., Valentini A., Miquel C., Taberlet P., Bellemain E., Miaud C. (2012). Improved detection of an alien invasive species through environmental DNA barcoding: the example of the American bullfrog Lithobates catesbeianus. J. Appl. Ecol. 49 (4), 953–959. doi: 10.1111/j.1365-2664.2012.02171.x
Deutschmann B., Muller A. K., Hollert H., Brinkmann M. (2019). Assessing the fate of brown trout (Salmo trutta) environmental DNA in a natural stream using a sensitive and specific dual-labelled probe. Sci. Total Environ. 655, 321–327. doi: 10.1016/j.scitotenv.2018.11.247
Erickson R. A., Rees C. B., Coulter A. A., Merkes C. M., McCalla S. G., Touzinsky K. F., et al. (2016). Detecting the movement and spawning activity of bigheaded carps with environmental DNA. Mol. Ecol. Resour. 16 (4), 957–965. doi: 10.1111/1755-0998.12533
Ficetola G. F., Miaud C., Pompanon F., Taberlet P. (2008). Species detection using environmental DNA from water samples. Biol. Lett. 4 (4), 423–425. doi: 10.1098/rsbl.2008.0118
Fujii K., Doi H., Matsuoka S., Nagano M., Sato H., Yamanaka H. (2019). Environmental DNA metabarcoding for fish community analysis in backwater lakes: A comparison of capture methods. PloS One 14 (1), e0210357. doi: 10.1371/journal.pone.0210357
Gibson R. N. (1994). Impact of habitat quality and quantity on the recruitment of juvenile flatfishes. Netherlands J. Sea Res. 32 (2), 191–206. doi: 10.1016/0077-7579(94)90040-X
Gilbey J., Carvalho G., Castilho R., Coscia I., Coulson M. W., Dahle G., et al. (2021). Life in a drop: Sampling environmental DNA for marine fishery management and ecosystem monitoring. Mar. Policy 124, 104331. doi: 10.1016/j.marpol.2020.104331
Goldberg C. S., Turner C. R., Deiner K., Klymus K. E., Thomsen P. F., Murphy M. A., et al. (2016). Critical considerations for the application of environmental DNA methods to detect aquatic species. Methods Ecol. Evol. 7 (11), 1299–1307. doi: 10.1111/2041-210x.12595
Govindarajan A. F., Francolini R. D., Jech J. M., Lavery A. C., Llopiz J. K., Wiebe P. H., et al. (2021). Exploring the use of environmental DNA (eDNA) to detect animal taxa in the mesopelagic zone. Front. Ecol. Evol. 9, 574877. doi: 10.3389/fevo.2021.574877
Hader D. P., Banaszak A. T., Villafane V. E., Narvarte M. A., Gonzalez R. A., Helbling E. W. (2020). Anthropogenic pollution of aquatic ecosystems: Emerging problems with global implications. Sci. Total Environ. 713, 136586. doi: 10.1016/j.scitotenv.2020.136586
Hänfling B., Lawson Handley L., Read D. S., Hahn C., Li J., Nichols P., et al. (2016). Environmental DNA metabarcoding of lake fish communities reflects long-term data from established survey methods. Mol. Ecol. 25 (13), 3101–3119. doi: 10.1111/mec.13660
Hansen B. K., Bekkevold D., Clausen L. W., Nielsen E. E. (2018). The sceptical optimist: challenges and perspectives for the application of environmental DNA in marine fisheries. Fish Fish 19 (5), 751–768. doi: 10.1111/faf.12286
Hinlo R., Lintermans M., Gleeson D., Broadhurst B., Furlan E. (2018). Performance of eDNA assays to detect and quantify an elusive benthic fish in upland streams. Biol. Invasions 20 (11), 3079–3093. doi: 10.1007/s10530-018-1760-x
Hutchings J. A., Taylor E., Minto C., Ricard D., Baum J. K., Jensen O. P. (2010). Trends in the abundance of marine fishes. Can. J. Fish Aquat. Sci. 67 (8), 1205–1210. doi: 10.1139/f10-081
Hu J., Tsai W., Cheng S., Chang F. (2020). Explore the relationship between fish community and environmental factors by machine learning techniques. Environ. Res. 184, 109262. doi: 10.1016/j.envres.2020.109262
Itakura H., Wakiya R., Yamamoto S., Kaifu K., Sato T., Minamoto T. (2019). Environmental DNA analysis reveals the spatial distribution, abundance, and biomass of Japanese eels at the river-basin scale. Aquat. Conservation: Mar. Freshw. Ecosyst. 29 (3), 361–373. doi: 10.1002/aqc.3058
Jackman J. M., Benvenuto C., Coscia I., Oliveira Carvalho C., Ready J. S., Boubli J. P., et al. (2021). eDNA in a bottleneck: Obstacles to fish metabarcoding studies in megadiverse freshwater systems. Environ. DNA 3 (4), 837–849. doi: 10.1002/edn3.191
Jane S. F., Wilcox T. M., McKelvey K. S., Young M. K., Schwartz M. K., Lowe W. H., et al. (2015). Distance, flow and PCR inhibition: eDNA dynamics in two headwater streams. Mol. Ecol. Resour. 15 (1), 216–227. doi: 10.1111/1755-0998.12285
Jean C. T., Lee S. C., Chen C. T. (1998). Variation in mitochondrial DNA sequences of black porgy, Acanthopagrus schlegeli, in the coastalwaters of Taiwan. Zoolog Stud. 37 (1), 22–30.
Jerde C. L., Mahon A. R., Chadderton W. L., Lodge D. M. (2011). “Sight-unseen” detection of rare aquatic species using environmental DNA. Conserv. Lett. 4 (2), 150–157. doi: 10.1111/j.1755-263X.2010.00158.x
Jeunen G. J., Miles D. L., Michael K., Spencer H. G., Taylor H. R., Stat M., et al. (2020). Water stratification in the marine biome restricts vertical environmental DNA (eDNA) signal dispersal. Environ. DNA 2 (1), 33–111. doi: 10.1002/edn3.49
Jiang S., Fang D., Xu D. (2020). Transcriptome analysis of Takifugu obscurus liver in response to acute retene exposure. J. Environ. Sci. Health Part A-toxic/hazardous Substances Environ. Eng. 55 (10), 1188–1200. doi: 10.1080/10934529.2020.1780852
Jiang S., Su T., Xia J., Li J., Qu Y., Liu H, et al (2012). “Germplasm resources and utilization of sparidae fishes in China,” (Beijing: China Ocean Press).
Klymus K. E., Richter C. A., Chapman D. C., Paukert C. (2015). Quantification of eDNA shedding rates from invasive bighead carp Hypophthalmichthys nobilis and silver carp Hypophthalmichthys molitrix. Biol. Conserv. 183, 77–84. doi: 10.1016/j.biocon.2014.11.020
Kume M., Lavergne E., Ahn H., Terashima Y., Kadowaki K., Ye F., et al. (2021). Factors structuring estuarine and coastal fish communities across Japan using environmental DNA metabarcoding. Ecol. Indic. 121, 107276. doi: 10.1016/j.ecolind.2020.107216
Lacoursière-Roussel A., Côté G., Leclerc V., Bernatchez L., Cadotte M. (2016a). Quantifying relative fish abundance with eDNA: a promising tool for fisheries management. J. Appl. Ecol. 53 (4), 1148–1157. doi: 10.1111/1365-2664.12598
Lacoursière-Roussel A., Rosabal M., Bernatchez L. (2016b). Estimating fish abundance and biomass from eDNA concentrations: variability among capture methods and environmental conditions. Mol. Ecol. Resour. 16 (6), 1401–1414. doi: 10.1111/1755-0998.12522
Law C. S. W., Sadovy de Mitcheson Y. (2017). Reproductive biology of black seabream Acanthopagrus schlegelii, threadfin porgy Evynnis cardinalis and red pargo Pagrus major in the northern south China Sea with consideration of fishery status and management needs. J. Fish Biol. 91 (1), 101–125. doi: 10.1111/jfb.13331
Lawson Handley L., Read D. S., Winfield I. J., Kimbell H., Johnson H., Li J., et al. (2019). Temporal and spatial variation in distribution of fish environmental DNA in england’s largest lake. Environ. DNA 1 (1), 26–39. doi: 10.1002/edn3.5
Liang J., Wang W., Lin G., Xu K., Guo A. (2010). Effect and assessment of enhancement release of nibea japonica and sparus macrocephalus in artificial reef habitat waters of zhoushan, zhejiang. J. Fish Sci. China 17 (5), 1075–1084.
Li L., Gao F., Jian Y., Wang X., Wang X., Pan L., et al. (2020). Transcriptomic analysis of liver tissue in fat greenling (Hexagrammos otakii) exposed to elevated ambient ammonia. Front. Mar. Sci. 7, 00418. doi: 10.3389/fmars.2020.00418
Littlefair J. E., Hrenchuk L. E., Blanchfield P. J., Rennie M. D., Cristescu M. E. (2021). Thermal stratification and fish thermal preference explain vertical eDNA distributions in lakes. Mol. Ecol. 30 (13), 3083–3096. doi: 10.1111/mec.15623
Lu H., Lee H. (2014). Changes in the fish species composition in the coastal zones of the kuroshio current and China coastal current during periods of climate change: Observations from the set-net fishery (1993–2011). Fish Res. 155, 103–113. doi: 10.1016/j.fishres.2014.02.032
Maranghi F., Tassinari R., Moracci G., Altieri I., Rasinger J. D., Carroll T. S., et al. (2013). Dietary exposure of juvenile female mice to polyhalogenated seafood contaminants (HBCD, BDE-47, PCB-153, TCDD): comparative assessment of effects in potential target tissues. Food Chem. Toxicol. 56, 443–449. doi: 10.1016/j.fct.2013.02.056
Mukherjee S., Chaudhuri A., Kundu N., Mitra S., Homechaudhuri S. (2012). Comprehensive analysis of fish assemblages in relation to seasonal environmental variables in an estuarine river of Indian sundarbans. Estuaries Coasts 36 (1), 192–202. doi: 10.1007/s12237-012-9558-z
Murakami H., Yoon S., Kasai A., Minamoto T., Yamamoto S., Sakata M. K., et al. (2019a). Correction to: Dispersion and degradation of environmental DNA from caged fish in a marine environment. Fish Sci. 85 (6), 1109–1109. doi: 10.1007/s12562-019-01341-z
Murakami H., Yoon S., Kasai A., Minamoto T., Yamamoto S., Sakata M. K., et al. (2019b). Dispersion and degradation of environmental DNA from caged fish in a marine environment. Fish Sci. 85 (2), 327–337. doi: 10.1007/s12562-018-1282-6
Murata O., Kato K., Ishitani Y., Nasu T., Miyashita S., Yamamoto S., et al. (1997). Gonadal maturation of Pagrus major×acanthopagrus schlegeliand and Pagrus major×sparus sarba sea bream hybrids. Aquacult Sci. 45 (1), 75–80.
Nakagawa H., Yamamoto S., Sato Y., Sado T., Minamoto T., Miya M. (2018). Comparing local- and regional-scale estimations of the diversity of stream fish using eDNA metabarcoding and conventional observation methods. Freshw. Biol. 63 (6), 569–580. doi: 10.1111/fwb.13094
Nester G. M., De Brauwer M., Koziol A., West K. M., DiBattista J. D., White N. E., et al. (2020). Development and evaluation of fish eDNA metabarcoding assays facilitate the detection of cryptic seahorse taxa (family: Syngnathidae). Environ. DNA 2 (4), 614–626. doi: 10.1002/edn3.93
Nevers M. B., Byappanahalli M. N., Morris C. C., Shively D., Przybyla-Kelly K., Spoljaric A. M., et al. (2018). (eDNA): A tool for quantifying the abundant but elusive round goby (Neogobius melanostomus). PloS One 13 (1), e0191720. doi: 10.1371/journal.pone.0191720
Pauly D., Beddington J. R., Kirkwood G. P., Watson R., Alder J. (2005). Global trends in world fisheries: impacts on marine ecosystems and food security. Philos. Trans. R. Soc. B: Biol. Sci. 360 (1453), 5–12. doi: 10.1098/rstb.2004.1574
Saito H., Nakanishi Y., Shigeta T., Umino T., Kawai K., Imabayashi H. (2008). Effect of predation of fishes on oyster spats in Hiroshima bay. Nippon Suisan Gakk 74 (5), 809–815. doi: 10.2331/suisan.74.809
Sarre G. A., Platell M. E., Potter I. C. (2000). Do the dietary compositions of acanthopagrus butcheri in four estuaries and a coastal lake vary with body size and season and within and amongst these water bodies? J. Fish Biol. 56 (1), 103–122. doi: 10.1111/j.1095-8649.2000.tb02089.x
Takahara T., Minamoto T., Yamanaka H., Doi H., Kawabata Z. (2012). Estimation of fish biomass using environmental DNA. PloS One 7 (4), e35868. doi: 10.1371/journal.pone.0035868
Takasu H., Inomata H., Uchino K., Tahara S., Mori K., Hirano Y., et al. (2019). Spatio-temporal distribution of environmental DNA derived from Japanese sea nettle jellyfish Chrysaora pacifica in Omura Bay, Kyushu, Japan. Plankton Benthos Res. 14 (4), 320–323. doi: 10.3800/pbr.14.320
Takeuchi A., Watanabe S., Yamamoto S., Miller M. J., Fukuba T., Miwa T., et al. (2019). First use of oceanic environmental DNA to study the spawning ecology of the Japanese eel Anguilla japonica. Mar. Ecol. Prog. Ser. 609, 187–196. doi: 10.3354/meps12828
Thomsen P. F., Kielgast J., Iversen L. L., Moller P. R., Rasmussen M., Willerslev E. (2012). Detection of a diverse marine fish fauna using environmental DNA from seawater samples. PloS One 7 (8), e41732. doi: 10.1371/journal.pone.0041732
Thomsen P. F., Willerslev E. (2015). Environmental DNA – an emerging tool in conservation for monitoring past and present biodiversity. Biol. Conserv. 183, 4–18. doi: 10.1016/j.biocon.2014.11.019
Tillotson M. D., Kelly R. P., Duda J. J., Hoy M., Kralj J., Quinn T. P. (2018). Concentrations of environmental DNA (eDNA) reflect spawning salmon abundance at fine spatial and temporal scales. Biol. Conserv. 220, 1–11. doi: 10.1016/j.biocon.2018.01.030
Urbatzka R., Rocha E., Reis B., Cruzeiro C., Monteiro R. A., Rocha M. J. (2012). Effects of ethinylestradiol and of an environmentally relevant mixture of xenoestrogens on steroidogenic gene expression and specific transcription factors in zebrafish. Environ. Pollut. 164, 28–35. doi: 10.1016/j.envpol.2012.01.018
Wang X., Lu G., Zhao L., Du X., Gao T. (2021b). Assessment of fishery resources using environmental DNA: The large yellow croaker (Larimichthys crocea) in the East China Sea. Fish Res. 235, 105813. doi: 10.1016/j.fishres.2020.105813
Wang X., Lu G., Zhao L., Yang Q., Gao T. (2020). Assessment of fishery resources using environmental DNA: Small yellow croaker (Larimichthys polyactis) in East China Sea. PloS One 15 (12), e0244495. doi: 10.1371/journal.pone.0244495
Wang S., Yan Z., Hanfling B., Zheng X., Wang P., Fan J., et al. (2021a). Methodology of fish eDNA and its applications in ecology and environment. Sci. Total Environ. 755 (2), 142622. doi: 10.1016/j.scitotenv.2020.142622
Wang H., Zhang W., Li C., Lv Z., Jin C. (2015). Identification and characterization of a novel foxo transcription factors in Apostichopus japonicus. Fish Shellfish Immunol. 44 (1), 164–171. doi: 10.1016/j.fsi.2015.02.006
Wilcox T. M., McKelvey K. S., Young M. K., Sepulveda A. J., Shepard B. B., Jane S. F., et al. (2016). Understanding environmental DNA detection probabilities: A case study using a stream-dwelling char Salvelinus fontinalis. Biol. Conserv. 194, 209–216. doi: 10.1016/j.biocon.2015.12.023
Yamada U., Tokimura M., Hiroshi H., Nakabo T. (2007). Fishes and Fisheries of the East China and Yellow Seas (Kanagawa: Tokai University Press).
Yamamoto S., Minami K., Fukaya K., Takahashi K., Sawada H., Murakami H., et al. (2016). Environmental DNA as a ‘Snapshot’ of fish distribution: a case study of Japanese jack mackerel in maizuru bay, Sea of Japan. PloS One 11 (3), e0149786. doi: 10.1371/journal.pone.0149786
Zhang X., He G., Sha X. (1980). Morphological studies of the eggs, larvae and young fish of the black porgy, Sparus macrocephalus (Basilewsky). Acta Zoologica Sin. 26 (4), 331–336.
Zhang Y., Pavlovska M., Stoica E., Prekrasna I., Yang J., Slobodnik J., et al. (2020). Holistic pelagic biodiversity monitoring of the black Sea via eDNA metabarcoding approach: From bacteria to marine mammals. Environ. Int. 135, 105307. doi: 10.1016/j.envint.2019.105307
Zhang J., Zhang C., Sun P., Huang M., Fan M., Liu M. (2017). RNA-Sequencing and pathway analysis reveal alteration of hepatic steroid biosynthesis and retinol metabolism by tributyltin exposure in male rare minnow (Gobiocypris rarus). Aquat. Toxicol. 188, 109–118. doi: 10.1016/j.aquatox.2017.03.015
Zhang K., Zhang J., Xu Y., Sun M., Chen Z., Yuan M. (2018). Application of a catch-based method for stock assessment of three important fisheries in the East China Sea. Acta Oceanol Sin. 37 (2), 102–109. doi: 10.1007/s13131-018-1173-9
Zhao S., Lu B., Li R., Zhu A., Wu C. (2015). Analysis of the causes of the decline of fishery resources in the East China Sea in the context of species extinction. Scientia Sinica(Terrae) 45 (11), 1628–1640. doi: 10.1007/s11430-015-5193-4
Keywords: East China Sea, environmental DNA, black sea bream, fishery resource assessment, distribution
Citation: Zhang H, Zhou Y, Zhang H, Gao T and Wang X (2022) Fishery resource monitoring of the East China Sea via environmental DNA approach: a case study using black sea bream (Acanthopagrus schlegelii). Front. Mar. Sci. 9:848950. doi: 10.3389/fmars.2022.848950
Received: 19 January 2022; Accepted: 18 July 2022;
Published: 19 August 2022.
Edited by:
Wei Huang, Ministry of Natural Resources, ChinaReviewed by:
Alessia Cariani, University of Bologna, ItalyOlivier Laroche, Cawthron Institute, New Zealand
Copyright © 2022 Zhang, Zhou, Zhang, Gao and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaoyan Wang, d2FuZ3hpYW95YW5AempvdS5lZHUuY24=
 Haobo Zhang
Haobo Zhang Yongdong Zhou2
Yongdong Zhou2 Tianxiang Gao
Tianxiang Gao Xiaoyan Wang
Xiaoyan Wang