- 1College of Fishery, Guangdong Provincial Key Laboratory of Pathogenic Biology and Epidemiology for Aquatic Economic Animals, Key Laboratory of Control for Diseases of Aquatic Economic Animals of Guangdong Higher Education Institutes, Guangdong Ocean University, Zhanjiang, China
- 2Guangdong Provincial Engineering Research Center for Aquatic Animal Health Assessment, Shenzhen Public Service Platform for Evaluation of Marine Economic Animal Seedings, Shenzhen Institute of Guangdong Ocean University, Shenzhen, China
Vibrio alginolyticus (V. alginolyticus) is one of the important epizootic pathogens in marine animals. VAGM001033 belongs to a diguanylate cyclase, responsible for the synthesis of dimeric guanosine monophosphate (c-di-GMP), a ubiquitous second messenger involved in the function of biofilm formation, motility, and virulence. This study confirmed that VAGM001033 was an active diguanylate cyclase by Congo red assay. The red-stained, dry, and rough form of colonies were observed with the increasing concentration of the L-arabinose on Congo red plates. Also, an in-frame deleted ΔVAGM001033 mutant was constructed and changes of ΔVAGM001033 mutant in physiology and pathogenicity were detected. The ΔVAGM001033 mutant displayed similar morphology and growth curve with the wild-type strain showing no significant differences. The swarming ability of the ΔVAGM001033 mutant was significantly enhanced showing bigger swarming circles, while the biofilm formation, extracellular proteases, and virulence were significantly attenuated. The results of the test for antibiotic susceptibility showed that the wild type and ΔVAGM001033 mutant had similar sensitivity or resistance to most antibiotics used in this study, except cefotaxime and nitrofurantoin. The mutant was sensitive to cefotaxime and nitrofurantoin, while the wild type was intermediate. A total of 756 differentially expressed genes (DEGs) were identified by RNA-seq, of which 109 were upregulated and 647 were downregulated. Flagellar assembly, two-component system, ATP-binding cassette (ABC) transporters, and peptidoglycan biosynthesis were significantly enriched in the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway. Moreover, the ΔVAGM001033 mutant induced high antibody titers and provided immune protectivity with a relative percent survival (RPS) of 82%. Immune-related genes of pearl gentian grouper (♀Epinephelus fuscoguttatus × ♂ Epinephelus lanceolatus), namely, IgM, MHC-Iα, interleukin-1β (IL-1β), interleukin-16 (IL-16), and tumor necrosis factor-α (TNF-α) were upregulated after vaccination. Overall, the results suggested that VAGM001033 plays a crucial role in V. alginolyticus. The ΔVAGM001033 mutant might be applied as an effective live vaccine candidate against V. alginolyticus.
Introduction
Vibrio alginolyticus (V. alginolyticus), a Gram-negative bacterium, is one of the important epizootic pathogens to marine animals. Despite the boom of fish farming in China, the farmed-marine industry has been severely hampered by fish mortality due to vibriosis diseases caused by V. alginolyticus (Lee et al., 2020). In July of 2017, there was an outbreak of vibriosis among seahorses, featured by severe ulcers on the skin, caused by the dominant causative species V. alginolyticus in Eastern China (Xie et al., 2020). Cai et al. (2018) stated that it is considered to be the major causative media of outbreak of vibriosis in fish and shellfish at the coast of South China in the recent years. V. alginolyticus infection caused tissue necrosis, skin ulceration, gastroenteritis, and inflammatory reactions in aquatic animals (Chen et al., 2020; Amenyogbe et al., 2021; Su et al., 2021).
GGDEF domain protein is a cyclic di-guanosine monophosphate (c-di-GMP)-specific diguanylate cyclase, which is responsible for the synthesis of c-di-GMP. A plethora of studies confirmed that GGDEF domain protein was critical for enzyme activity, which regulated many functions, namely, motility (Floyd et al., 2020; Webster et al., 2021), biofilm formation (Valentini and Filloux, 2019), and virulence (Joshi et al., 2020). The first GGDEF domain protein PleD was identified in Caulobacter cresenctus, which controlled the cell transition from swarms to stalks (Hecht and Newton, 1995). WspR was verified to contain a GGDEF domain, which controlled the autoaggregation in Pseudomonas aeruginosa (Hueso-Gil et al., 2020). FimX was reported to bear a GGDEF domain, which regulated the twitching motility in Pseudomonas aeruginosa (Jain et al., 2017). RocS was also reported to regulate the motility in Vibrio cholerae, which included a GGEDF domain (Wu et al., 2020). ScrC in Vibrio parahaemolyticus was reported to regulate the cell attachment and motility for containing a GGEDF domain (Kimbrough and McCarter, 2020). The gene, referred to in the article as VAGM001033, consisted of a 1,035 bp open reading frame (ORF) that encoded 344 amino acids contained a predicted GGDEF domain. However, the enzymatic activity and regulatory mechanisms of these kinds of enzymes are still relatively scarce in V. alginolyticus.
With the expansion of the fish breeding scale, increased emergence and outbreak of fish diseases posed harm to the aquaculture industry. Antibiotic abuse led to the appearance of drug-resistant strains and food safety (Gudding and Van Muiswinkel, 2013). Hence, seeking a safe and effective solution is particularly urgent in aquaculture. Vaccination is definitely the most appropriate method to prevent and control fish diseases. The earliest bony vertebrate with both innate and adaptive immune response is reported to be teleost fish (Cooper and Alder, 2006).
For the purpose to detect the role of VAGM001033 in V. alginolyticus, the ΔVAGM001033 mutant was constructed and some physiological characteristics of ΔVAGM001033 mutant were detected in this study. Furthermore, pearl gentian grouper was used to evaluate the efficacy of the mutant strain as an attenuated live vaccine candidate that will contribute to the healthy and sustainable development of the aquaculture industry.
Materials and Methods
Bacterial Strains and Fish
In this study, the bacteria, plasmids, and primers were listed in Table 1. V. alginolyticus strain HY9901 was separated from a sick maricultured grouper (Epinephelus coioides) at Zhanjiang, China, and stored in our laboratory. V. alginolyticus was grown at 28°C on Tryptic soya broth (TSB) medium supplemented with 2% NaCl (Haling, China). Escherichia coli (E. coli) strains were cultured at 37°C in Luria broth (LB) (Haling, China). Ampicillin (Amp, 100 μg/ml) or chloramphenicol (Cm, 25 μg/ml) were supplemented when the optimal antibiotics were needed.
Pearl gentian groupers (♀Epinephelus fuscoguttatus × ♂ Epinephelus lanceolate, 30.0 ± 3.0 g) were purchased from a local fish farm from Donghai island, Zhanjiang and maintained in seawater at a temperature of 28 ± 1°C in a recirculation system for 2 weeks. Fishes were considered healthy by bacteriological recovery tests as described previously (Pang et al., 2022).
Before the extraction of tissues after the cessation of the experiment, fishes were anesthetized with MS222 (tricaine methanesulfonate, 100 ng/ml). All the fish experiments were approved by the respective Animal Research and Ethics Committees of the Guangdong Provincial Key Laboratory of Pathogenic Biology and Epidemiology for Aquatic Economic Animals.
Congo Red Assay
A pair of primers pBAD-VAGM001033-F/pBAD-VAGM001033-R were designed as shown in Table 1 according to V. alginolyticus genomic sequences (GenBank number: CP072781-CP072782). PCR has conducted following the conditions: denaturation at 95°C for 5 min, followed by 38 cycles of 94°C for 30 s, 55°C for 60 s, and 72°C for 30 s in a Thermocycler (Bio-Rad, Hercules, CA, United States). The PCR product was examined on 1% agarose gel, then was purified and ligated into the pBAD-HisA vector. The recombinant plasmid was transformed into E. coli DH5α and subsequently E. coli BL21. The inserted fragment was sequenced by Sangon Biological Engineering Technology & Services Corporation, Ltd. (Guangzhou, China) using the primers pBAD-TF/pBAD-TR. Finally, the recombinant pBAD-VAGM001033 was inoculated in Congolese red LB agar plates containing different concentrations of inducing agent L-arabinose. The assay was conducted in triplicates.
Construction of a ΔVAGM001033 Mutant
The ΔVAGM001033 mutant was constructed by homologous recombination techniques described by Chen et al. (2019b). Briefly, the upstream homologous fragment of 524 bp was amplified using a primer (primers: VAGM001033-UF and VAGM001033-UR) and the downstream homologous fragment of 493 bp was amplified using a primer (primers: VAGM001033-DF and VAGM001033-DR) from V. alginolyticus HY9901 genomic DNA. Both fragments contained a 20 bp overlapping sequence were used to generate an in-frame deletion of the VAGM001033 gene by overlap extension PCR. Then, the overlap PCR product was ligated into a suicide T-vector plP12 (Luo et al., 2015), a universal genetic tool for rapid and efficient deleting mutation in Vibrio species, which is based on carrying a novel counterselectable vmi480 marker. The recombinant plP12-ΔVAGM001033 was transformed into E. coli DH5α and subsequently transformed into E. coli β2163. After homologous recombination two times, positive clones were selected and used for the conjugation with V. alginolyticus, and ΔVAGM001033 mutant was successfully constructed using a PCR identification (primers: VAGM001033-TF and VA001033-TR).
Characterization of ΔVAGM001033 Mutant
Cell Morphology
Overnight cultures were grown to the exponential phase in fresh medium. V. alginolyticus HY9901 strain and ΔVAGM001033 mutant were prepared for scanning electron microscope as the method described by Wang et al. (2007). The samples were viewed and photographed using a Hitachi XA-650 scanning electron microscope (Hitachi, Japan).
Genetic Stability of the ΔVAGM001033 Mutant
ΔVAGM001033 mutant was inoculated onto a TSA plate and passed blindly for 30 generations according to the method described by Zhou et al. (2020). In brief, a single colony was picked from Tryptic soya agar (TSA) plates and cultured in TSB (HKM, Guangzhou, China) with shaking for 12 h, and then bacteria broth culture was streaked out and cultured on a TSA plate. This process was repeated 30 times. The genetic stability of each generation was determined by PCR.
Growth Curve of Bacteria
Vibrio alginolyticus HY9901 and ΔVAGM001033 mutant were cultured in TSB overnight. The culture strains were inoculated into TSB with an initial OD600 of 0.01, respectively. Samples were measured at OD600 every hour. This procedure was repeated three times in each group.
Swarming Motility
Swarming motility was performed according to the approach explained by Mathew et al. (2001). Briefly, single colonies of V. alginolyticus HY9901 and ΔVAGM001033 mutant were selected and inoculated into TSA plates with 0.3% concentration of agar at 28°C, and swarming circle diameter was measured by Vernier calipers after 24 h incubation. The experiment was performed in triplicate.
Extracellular Protease Activity
Extracellular protease (ECPase) activity was performed as previously described by Windle and Kelleher (1997). V. alginolyticus HY9901 and ΔVAGM001033 mutant were cultured on TSA plates coated with sterile cellophane at 28°C for 24 h, washed with sterile phosphate-buffered saline (PBS), centrifuged at 4°C for 30 min, and the supernatant filtered to collect extracellular products. Inactivated sample (supernatant was boiled for 10 min) was used as a blank control. The experiment was performed in triplicate.
Biofilm Formation
Biofilm formation was assayed using crystal violet ammonium oxalate concerning the method described by Kierek and Watnick (2003). A total of 200 μl bacteria suspension (OD600 = 0.5) was inoculated in a 96-well plate with 3 replicates per sample at 28°C. Samples were taken at 24 h, fixed for 20 min with methanol, and stained with crystal violet ammonium oxalate dye for 15 min. Finally, 95% alcohol was then added and incubated at room temperature for 30 min. OD570 was determined by using a Multimode Plate Reader (PerkinElmer EnSpire, Singapore). The experiment was performed in triplicate.
LD50 Determination
The injection concentration of V. alginolyticus HY9901 and ΔVAGM001033 mutant were 105, 106, 107, 108, and 109 colony-forming unit (CFU)/ml. A total of 330 pearl gentian groupers were randomly divided into three groups. In the V. alginolyticus HY9901 group and the ΔVAGM001033 mutant group, 100 μl bacterial suspension was intraperitoneally injected into each fish. In the PBS control group, all the fishes were injected with 100 μl PBS in the same manner. Mortalities of fish were recorded over 14 days until the mortality rate was stable. The mortality challenged by V. alginolyticus was determined and the LD50 value of the wild-type strain and ΔVAGM001033 mutant were calculated according to the modified Coriolis method (Cai et al., 2018). The experiments were conducted in triplicate.
Antimicrobial Susceptibility Test
The antimicrobial susceptibility tests were conducted using the Kirby–Bauer disc diffusion method (Bauer, 1966) on TSA nutrient agar plates according to Clinical and Laboratory Standards Institute (Clinical and Laboratory Standards Institute [CLSI], 2013) and also following manufacturer’s guidelines. The antibiotic impregnated discs were purchased from Binhe Microbial Reagent Corporation, Ltd., Hangzhou, China, namely, polymyxin B, cefoxitin, cefuroxime, kanamycin, oxacillin, gentamicin, vancomycin, cefuroxime, amikacin, cefoperazone, spectinomycin, ceftriaxone, chloramphenicol, SMZ/TMP, clarithromycin, tetracycline, norfloxacin, ofloxacin, cefixime, cefotaxime, and nitrofurantoin. Vernier calipers were used to scale the diameters of the inhibition zones. The experiments were conducted in triplicate.
Transcriptome Sequencing
For transcriptome sequencing, V. alginolyticus HY9901 and ΔVAGM003125 mutant were cultured in TSB media at 28°C overnight. According to the method of Pang et al. (2022), the bacterial cells were harvested and dissolved in TRIzol (Takara). The followed experiments, namely, RNA extraction, RNA fragmentation, cDNA synthesis, RNA-Seq libraries construction, and bioinformatics analysis were conducted by Gene Denovo Biotechnology Corporation, Ltd. (Guangzhou, China). The results of the transcriptome sequencing have been deposited at the SRA database under accession numbers PRJNA794348.
Vaccination and Bacterial Challenge
Vaccination experiments were performed to evaluate the immunoprotectivity of ΔVAGM001033 mutant as a live attenuated vaccine candidate against V. alginolyticus as the previous study of Chen et al. (2019c). Fishes were randomly divided into two groups with 90 fishes per group. Fishes in the ΔVAGM001033 mutant group were injected intraperitoneally with 100 μl 1.0 × 105 CFU/ml ΔVAGM001033 mutant. Control fishes were injected intraperitoneally with 100 μl sterile PBS. The experiment was conducted in triplicates.
Six weeks post-immunization, fishes (n = 30) were anesthetized and challenged separately by 100 μl 1 × 108 CFU/ml of V. alginolyticus HY9901 in each group. The cumulative mortality was recorded for 14 days after the challenge. The relative percent survival (RPS) was calculated according to the formula: RPS = 100% [1 – (mortality of the ΔVAGM001033 mutant group/mortality of the PBS control group)]. The bacteria were reisolated from the liver and spleen of all the dead fishes and identified by 16s rDNA. The experiment was conducted in triplicates.
Analysis of Antibody Levels
During the experimental period from 1 to 6 weeks post-vaccination, approximately 200 μl of blood from individual fish (3 fish per group) in the PBS group and the ΔVAGM001033 mutant group are collected. Serum was collected following centrifugation at 15,000 g for 5 min to measure antibody levels using ELISA as the method described by Chen et al. (2019b). Briefly, microtiter plate wells were coated with 100 μl of formalin-killed V. alginolyticus by overnight incubation at 4°C. Excess cells were discarded and wells were blocked with 100 μl of PBS containing 2% bovine serum albumin (BSA) for 3 h at 22°C. After removing the blocking solution and washing three times with PBS added with 0.05% Tween-20 (PBST), the wells were incubated for 3 h at 22°C with 100 μl of serially diluted fish serum. Antibody binding to the antigen was detected using mouse antipearle gentian grouper IgM polyclonal antibody (1:10,000), followed by rabbit antimouse IgG-HRP (Wuhan Boster, Wuhan, China) at 1:20,000 dilutions, and color was developed with a chromogenic reagent tetramethylbenzidine (TMB) (Ameresco, Ltd., Framingham, MA, United States) for 20 min with the reaction being stopped by the addition of 2.0 M H2SO4. The plates were then read at 450 nm with a microplate reader (Bio-Rad). The experiment was conducted in triplicates.
Histopathological Analysis
After 42 days post-immunization, liver and intestine tissues (3 fish per group) were taken from the PBS group, the ΔVAGM001033 mutant group, and the V. alginolyticus HY9901 group. According to the previous study (Xie et al., 2020), the tissues were fixed in Davidson’s fixative (Shanghai Tarui Bioscience, China) at room temperature for 24 h. Tissues were dehydrated with a series of different concentrations of ethanol, cleared in xylene, and embedded into paraffin. Paraffin blocks of tissues were cut at 5 μm thickness on decontaminated glass slides. After deparaffinization, samples were stained with H&E and observed under an optical microscope (Leica, Germany).
Expression Analysis of Immune-Related Gene of Grouper
According to the method described by Wei et al. (2020), liver, spleen, head, and kidney (3 fishes per group) were taken from individual fish in the PBS group and the ΔVAGM001033 mutant group at 42 days post-immunization. The real-time quantitative PCR (qPCR) was performed according to standard protocols with the use of the SYBR Green qPCR Supermix Kit (TransGen, China). The final reaction volume of 20 μl contained 1 μl cDNA, 10 μl of 2 × SYBR Green qPCR SuperMix, 1 μl of each primer (10 μM), and 7 μl of ddH2O. The reaction was performed under the following conditions: 95°C 30 s; 40 cycles for 95°C 15 s, 55°C 20 s, and 72°C 30 s in LightCycler® 96 (Roche, United States). A melting curve analysis was performed to access the amplification of specific products. The primers for interleukin-16 (IL-16), IL-1, IgM, MHC-Iα, and tumor necrosis factor-α (TNF-α) are shown in Table 1. β-actin was employed as an internal reference. The IL-16, IL-1, IgM, MHC-Iα, and TNF-α gene expressions were normalized to β-actin as relative expression values using the 2–ΔΔCt method (Pang et al., 2022).
Statistical Analysis
SPSS software version 21.0 was used for statistical analysis (SPSS Incorporation, United States). The data were analyzed using one-way ANOVA. Duncan’s test was used to test mean comparisons. For identifying significant differences between groups, data are reported as mean ± SD and analyzed by the Student’s t-test. When p < 0.05, the differences were judged significant.
Results
Congo Red Assay
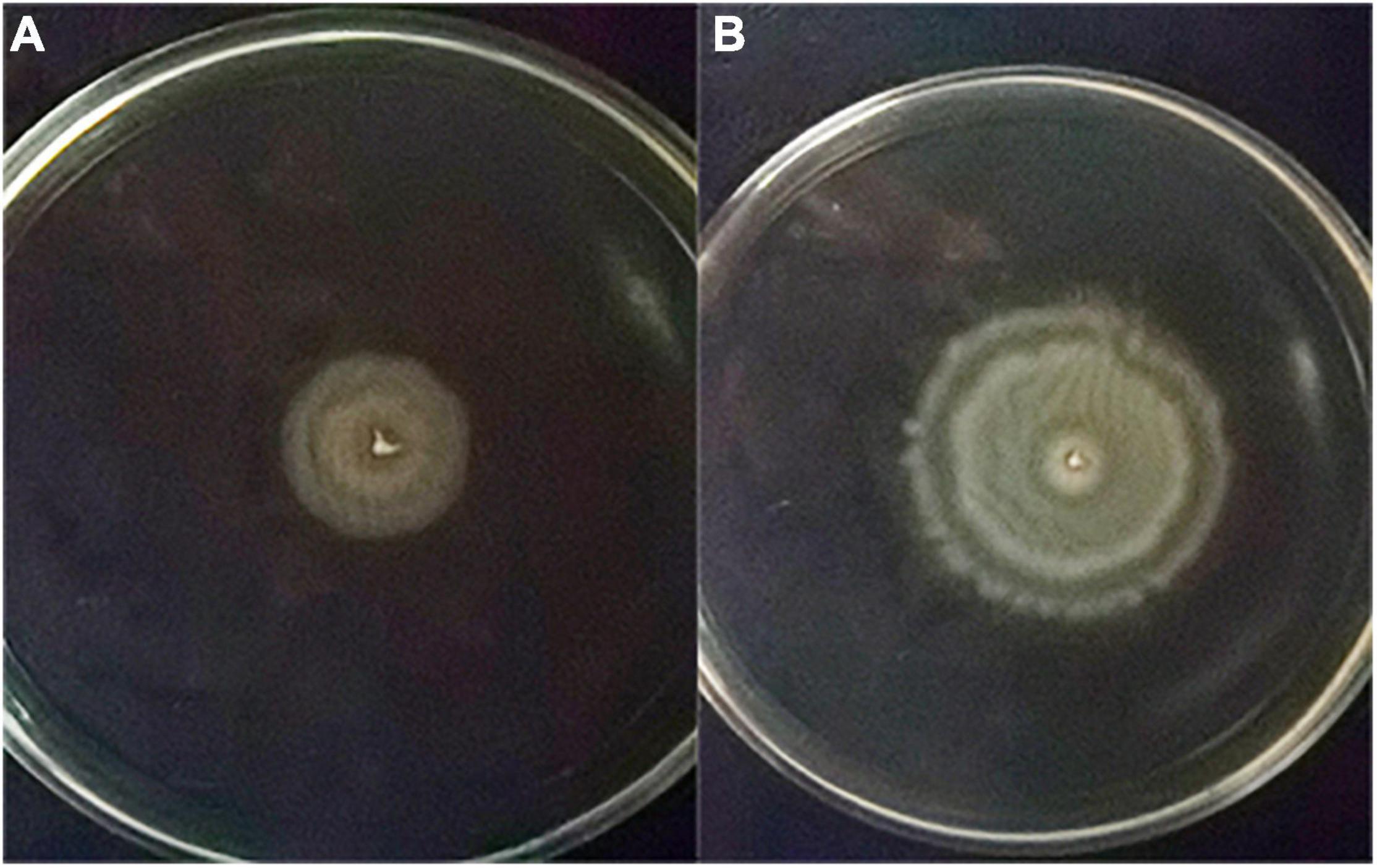
The recombinant vector pBAD-VAGM001033 was successfully constructed and confirmed by PCR identification and DNA sequencing as shown in Figure 1. The VAGM001033 gene consists of an ORF of 1,035 bp, encoding 344 amino acids with a GGDEF domain from amino acid 184–344. With the increase of the concentration of the L-arabinose, the red-stained, dry, and rough colonies were observed (Figures 2A–C), indicating that VAGM1033 synthesizes c-di-GMP as a diguanylate cyclase.
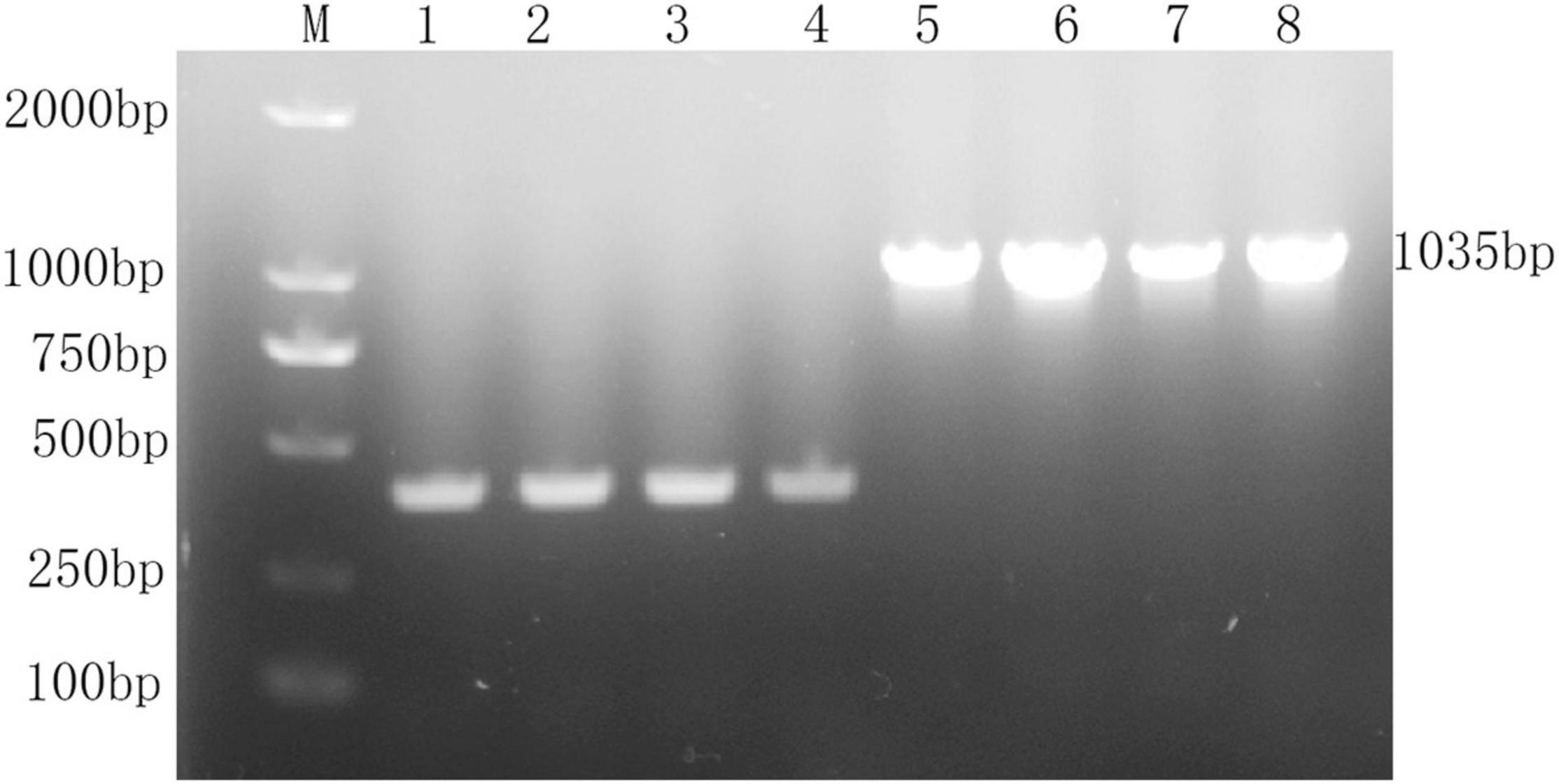
Figure 1. Construction of pBAD-VAGM001033. M: DL 2,000 marker; lane 1–4: pBAD-HisA; lane 5–8: pBAD-HisA-VAGM001033.
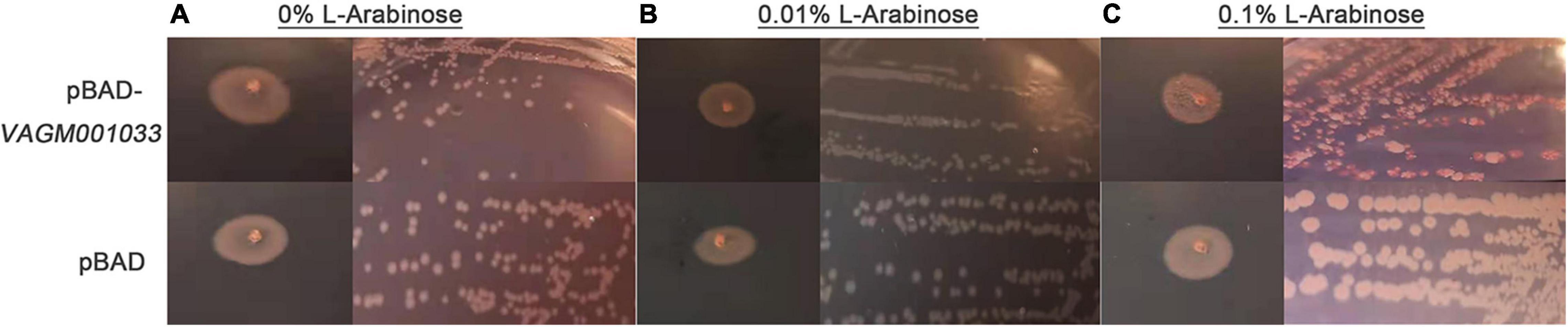
Figure 2. Phenotypic characterization on Congo red plates. With the increase of the concentration of the inducing agent L-arabinose, the phenotype of the colony displays more obvious, the red-stained, dry, and rough form (A: 0% L-arabinose; B: 0.01% L-arabinose; and C: 0.1% L-arabinose).
Identification and Characteristics of the ΔVAGM001033 Mutant
The VAGM001033 gene (locus in Chr1: 1104559-1105593) consists of an ORF of 1,035 bp, encoding 344 amino acids with a predicted GGDEF domain from amino acid 184-344. The ΔVAGM001033 mutant was constructed by deleting the total ORF of the ΔVAGM001033 gene based on genetic tools. The result of the successfully constructed ΔVAGM001033 mutant was achieved without altering the remaining sequences by a PCR identification (Figure 3) and direct DNA sequencing.
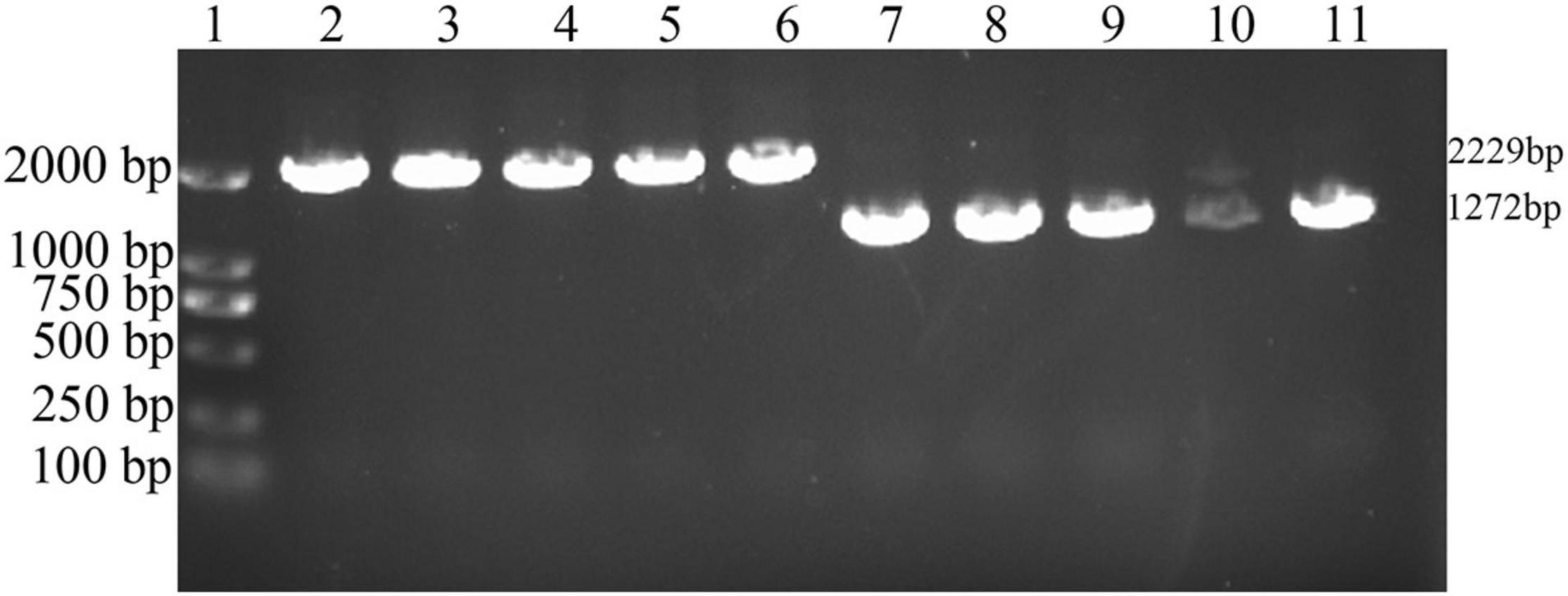
Figure 3. PCR identification of ΔVAGM001033 mutant. 1: DNA marker; 2–6: PCR amplifications with wild-type HY9901 genomic DNA (2,229 bp); 7–11: PCR amplifications with ΔVAGM00133 mutant genomic DNA (1,272 bp).
Morphology was observed by transmission electron microscope (TEM) and no discernible morphological difference was observed between the wild type and ΔVAGM001033 mutant. Both the wild type and ΔVAGM001033 mutant had a rod shape and a single polar flagellum (Figure 4). ΔVAGM001033 mutant showed a similar growth curve with the wild-type strain when cultured in the TSB medium (Figure 5). Wild type and ΔVAGM001033 mutant grew in the exponential growth phase for 0–6 h and reached the stationary phase at 15 h (OD600 ≈ 2.0).
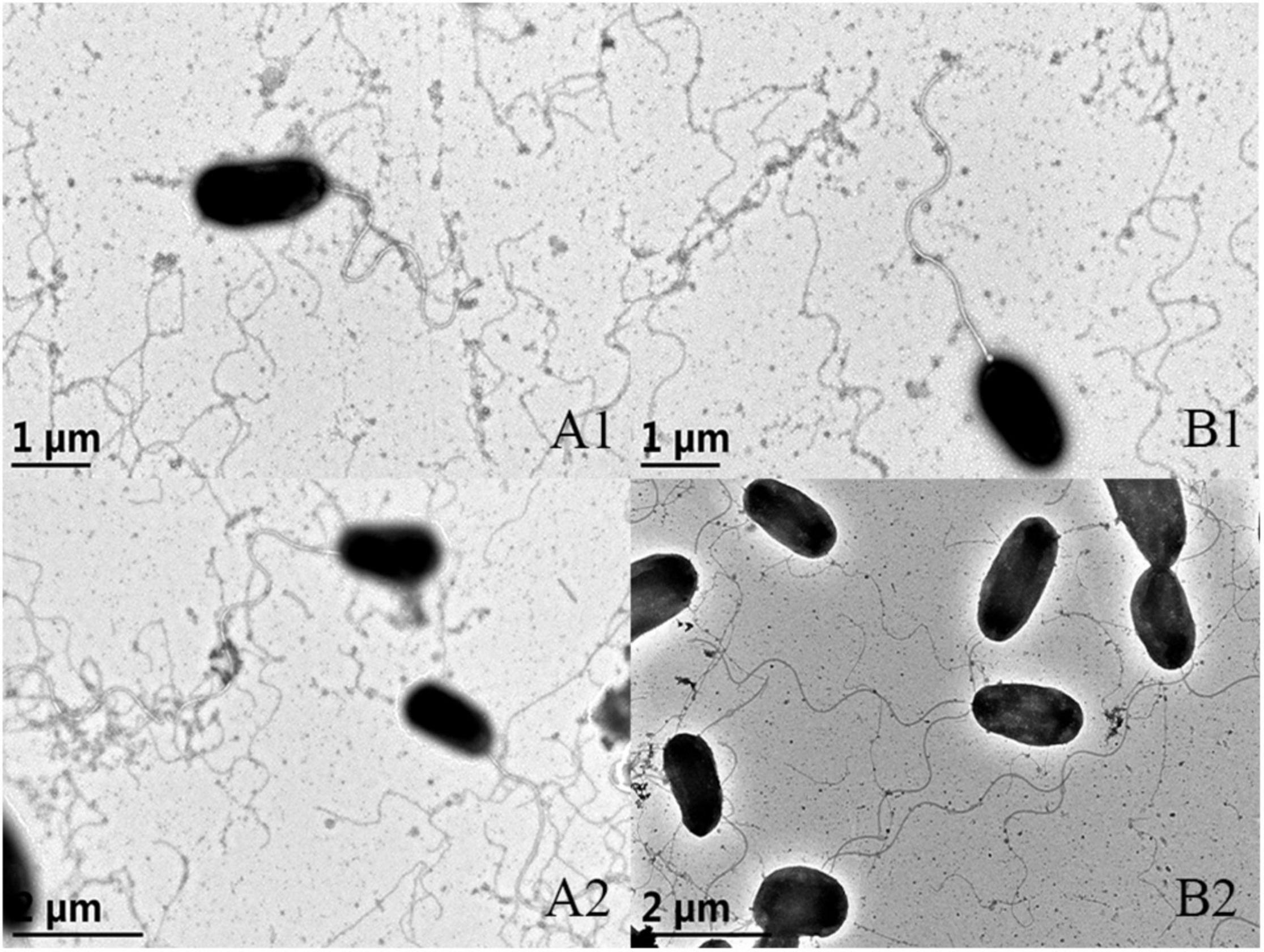
Figure 4. Morphological characteristics of the wild-type strain (A1 × 25,000, A2 × 20,000) and the ΔVAGM001033 mutant (B1 × 25,000, B2 × 20,000) via transmission electron microscope.
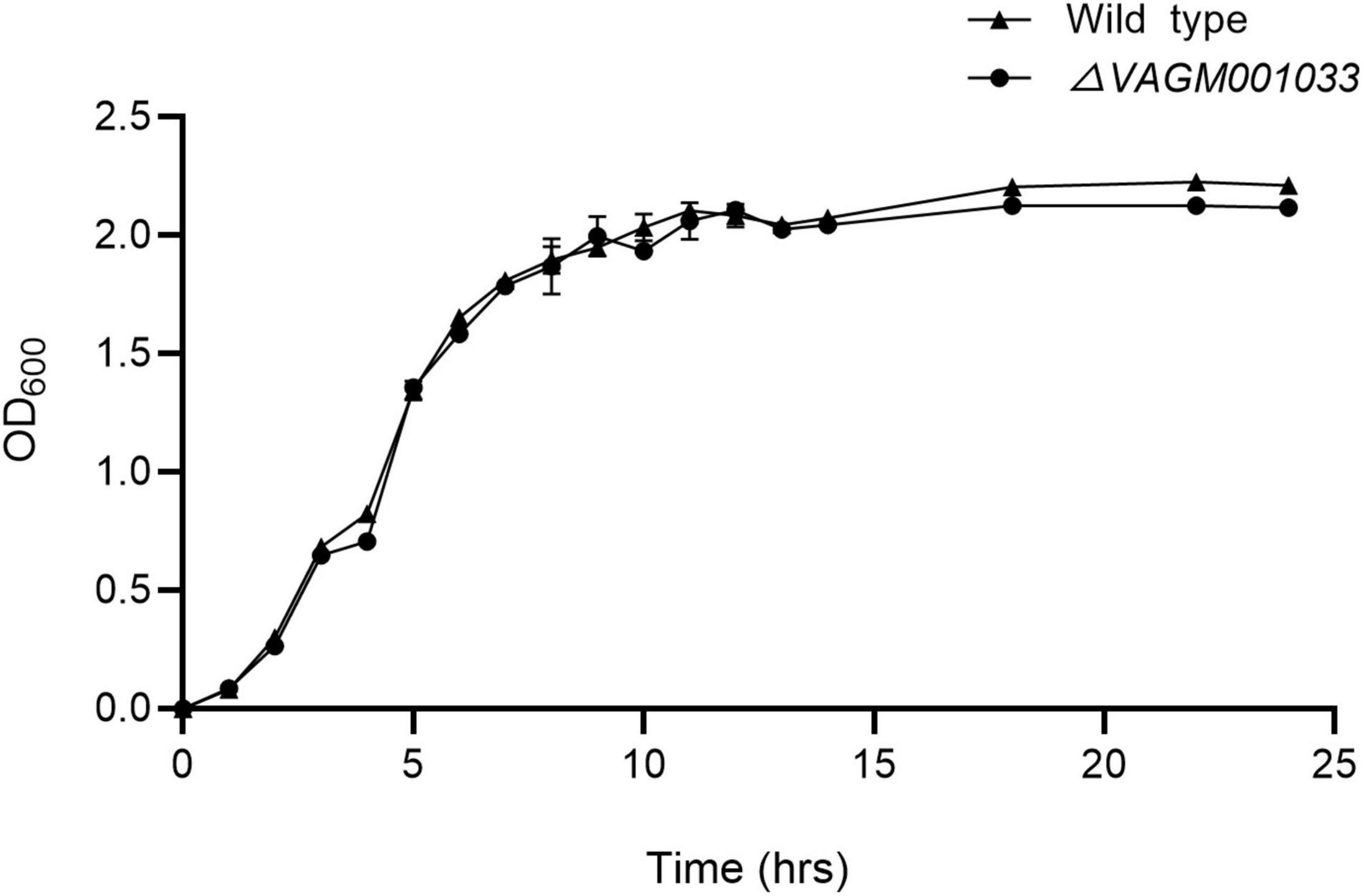
Figure 5. The growth curve of the wild-type strain and the ΔVAGM001033 mutant. At various time intervals, aliquots of cell culture were obtained and cell density was measured at OD600.
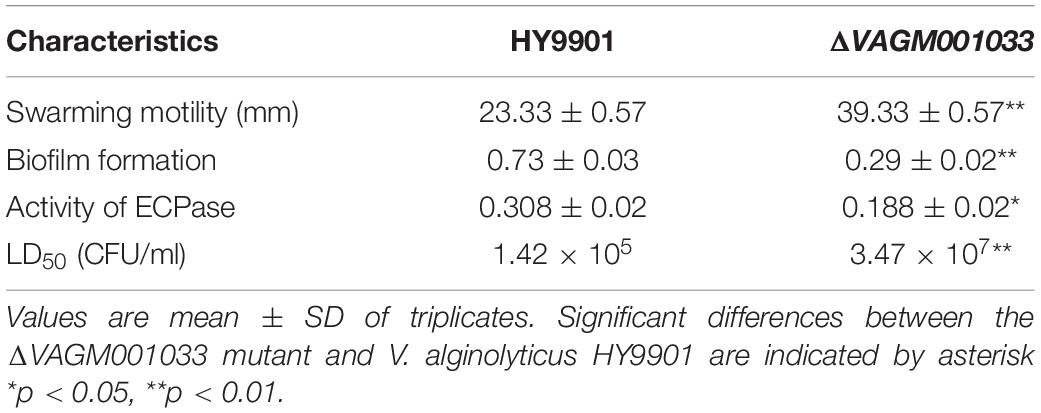
The swarming circle diameter of ΔVAGM001033 mutant (39.33 ± 0.57 mm) was significantly bigger than that in the wild type (Figure 6 and Table 2), indicating that the swarming ability of ΔVAGM001033 mutant was significantly enhanced (p < 0.01). However, biofilm formation of VAGM001033 mutant (0.29 ± 0.02) had significantly decreased when compared with that of the wild type (p < 0.01) (Table 2). The extracellular protease activity of ΔVAGM001033 mutant (0.188 ± 0.02) was significantly lower than that of the wild type (0.308 ± 0.02) (p < 0.05) (Table 2). The LD50 value of ΔVAGM001033 mutant was 100-fold higher than that of the wild type (Table 2), indicating that the virulence of ΔVAGM001033 mutant was significantly decreased (p < 0.01). All the dead groupers revealed symptoms of vibriosis specifically characterized by hemorrhaging, skin lesions, and swelling in the liver and kidney. Re-isolated bacteria taken from the liver and spleen tissues were identified as V. alginolyticus by 16S rDNA. None of the fish died or were diseased in the control group.
Antibiotic Susceptibility
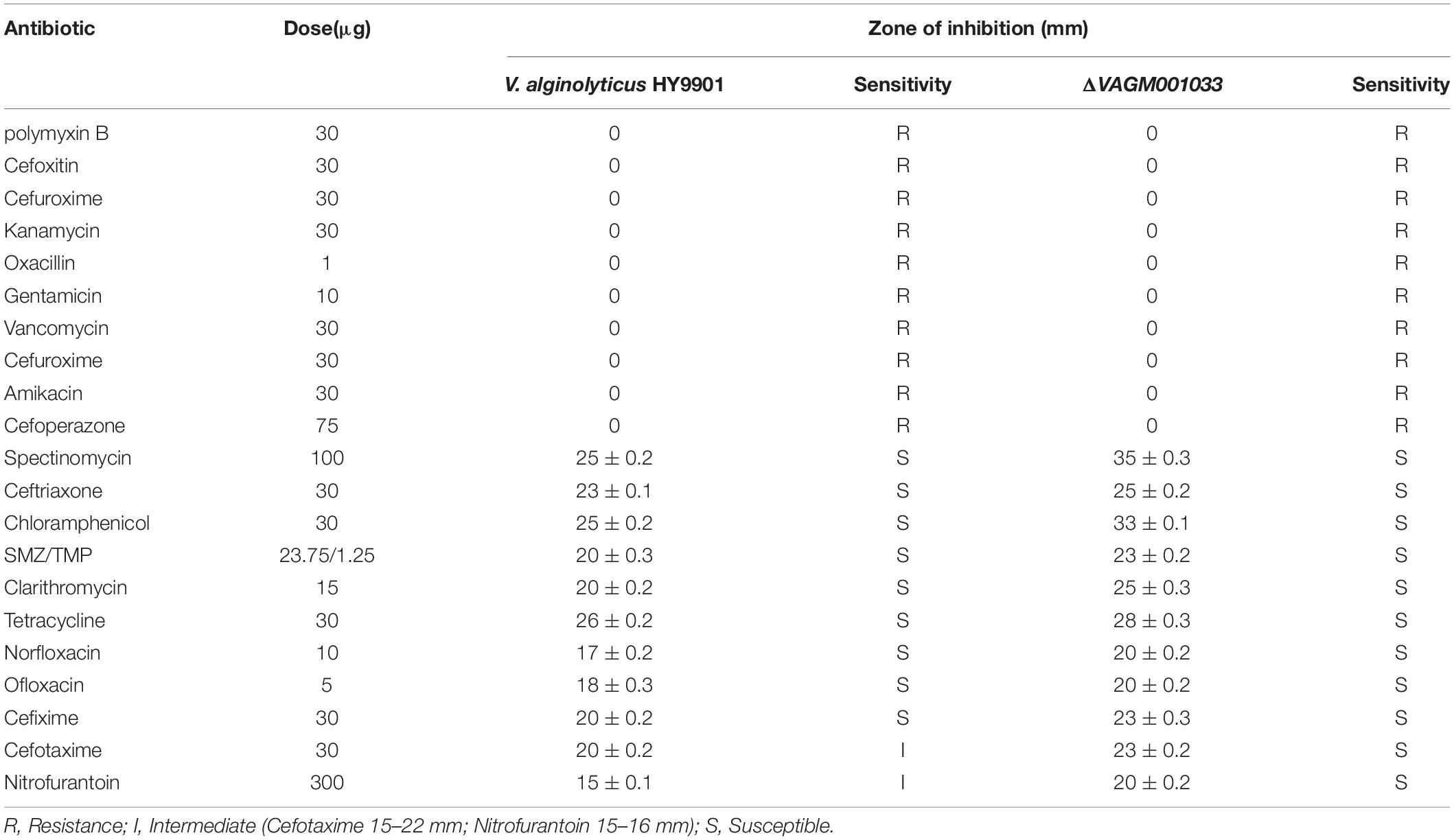
The results of the antimicrobial susceptibility are given in Table 3. Both the wild type and ΔVAGM001033 mutant were sensitive to spectinomycin, ceftriaxone, chloramphenicol, SMZ/TMP, clarithromycin, tetracycline, norfloxacin, ofloxacin, and cefixime but resistant to polymyxin B, cefoxitin, cefuroxime, kanamycin, oxacillin, gentamicin, vancomycin, amikacin, and cefoperazone. In contrast, the mutant was sensitive to cefotaxime and nitrofurantoin, while the wild type was intermediate.
RNA-seq
To compare the transcriptomic profiles of the wild-type strain and ΔVAGM001033 mutant, RNA-seq was conducted. A total of 756 differentially expressed genes (DEGs) were identified between these two strains. Of these DEGs (sequence accession number PRJNA794348), 109 were upregulated and 647 were downregulated in ΔVAGM001033 mutant (p < 0.05, more than 2-fold enrichment, Figure 7A). Genes associated with flagellar assembly were identified as the most enriched pathway according to the Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis (Figure 7B). Two-component system, ATP-binding cassette (ABC) transporters, and peptidoglycan biosynthesis were also significantly enriched (Figure 7B). Transcriptomic profiling analysis indicated that VAGM001033 regulated multiple biological pathways, namely, flagellar motility, two-component system, ABC transporters, and peptidoglycan biosynthesis.
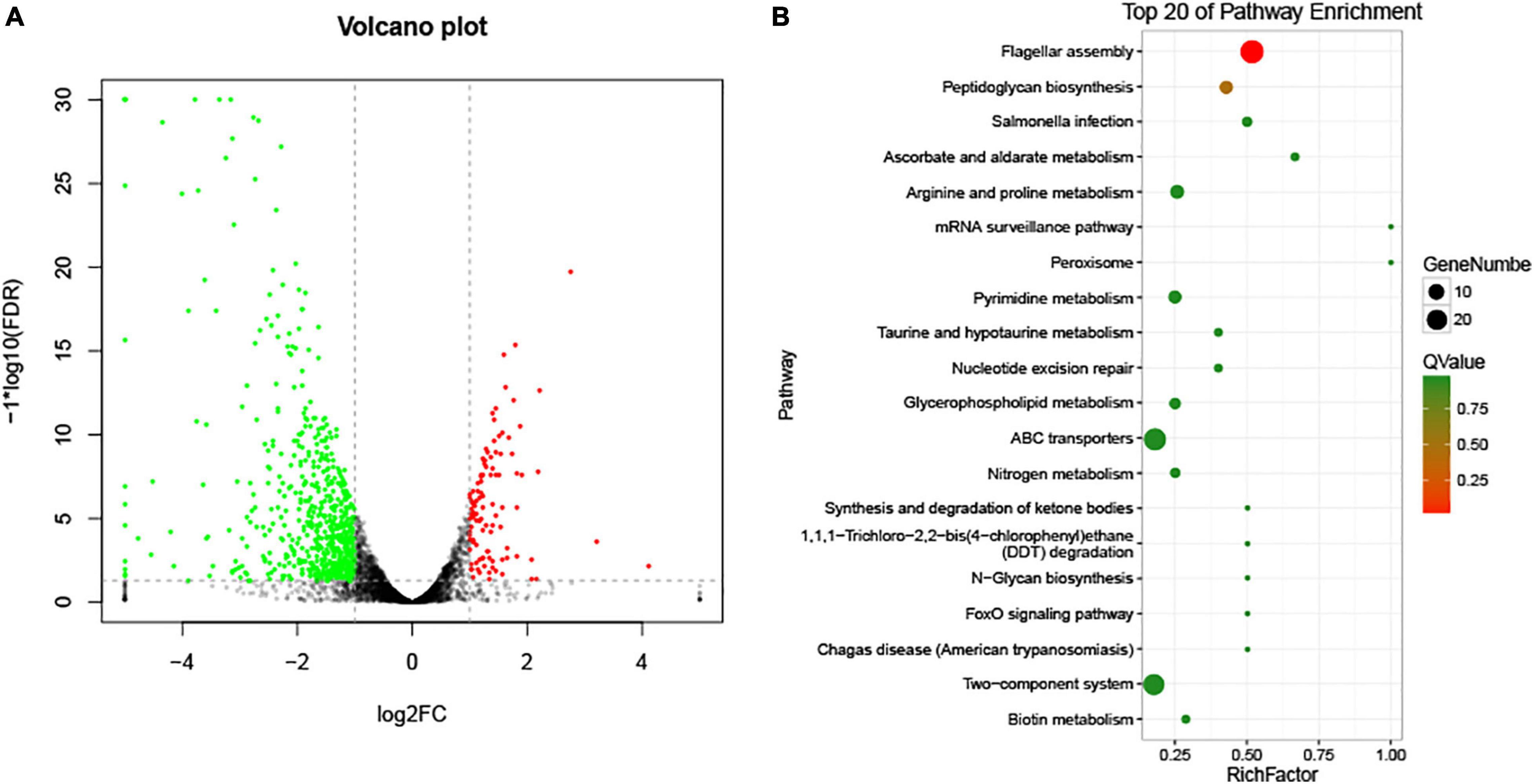
Figure 7. RNA-seq results. (A) Volcano Plot; the significantly upregulated genes are represented by red dots, the significantly downregulated genes are represented by green dots, and the black and gray dots represent no significant differences. (B) KEGG pathway enrichment.
Analysis of Antibody Levels
Enzyme-linked immunosorbent assay experiment was performed to measure the specific antibody titer of V. alginolyticus in fish of ΔVAGM001033 mutant and the PBS group. The result showed that specific antibodies in the ΔVAGM001033 mutant group could be detected at the first week after the vaccination. The specific antibody titers of fish immunized with ΔVAGM001033 mutant were significantly higher than those in the control group. During weeks 1 to 4 post-vaccination, log2 (antibody titers) in the sera of ΔVAGM001033 mutant group all reached above 4.0 and the maximum reached 9.0, while that of the PBS group was always only 2.0 to 3.0. Comparison of the means of log2 (antibody titers) between ΔVAGM001033 mutant and PBS control groups revealed a statistically significant difference (p < 0.05) (Figure 8).
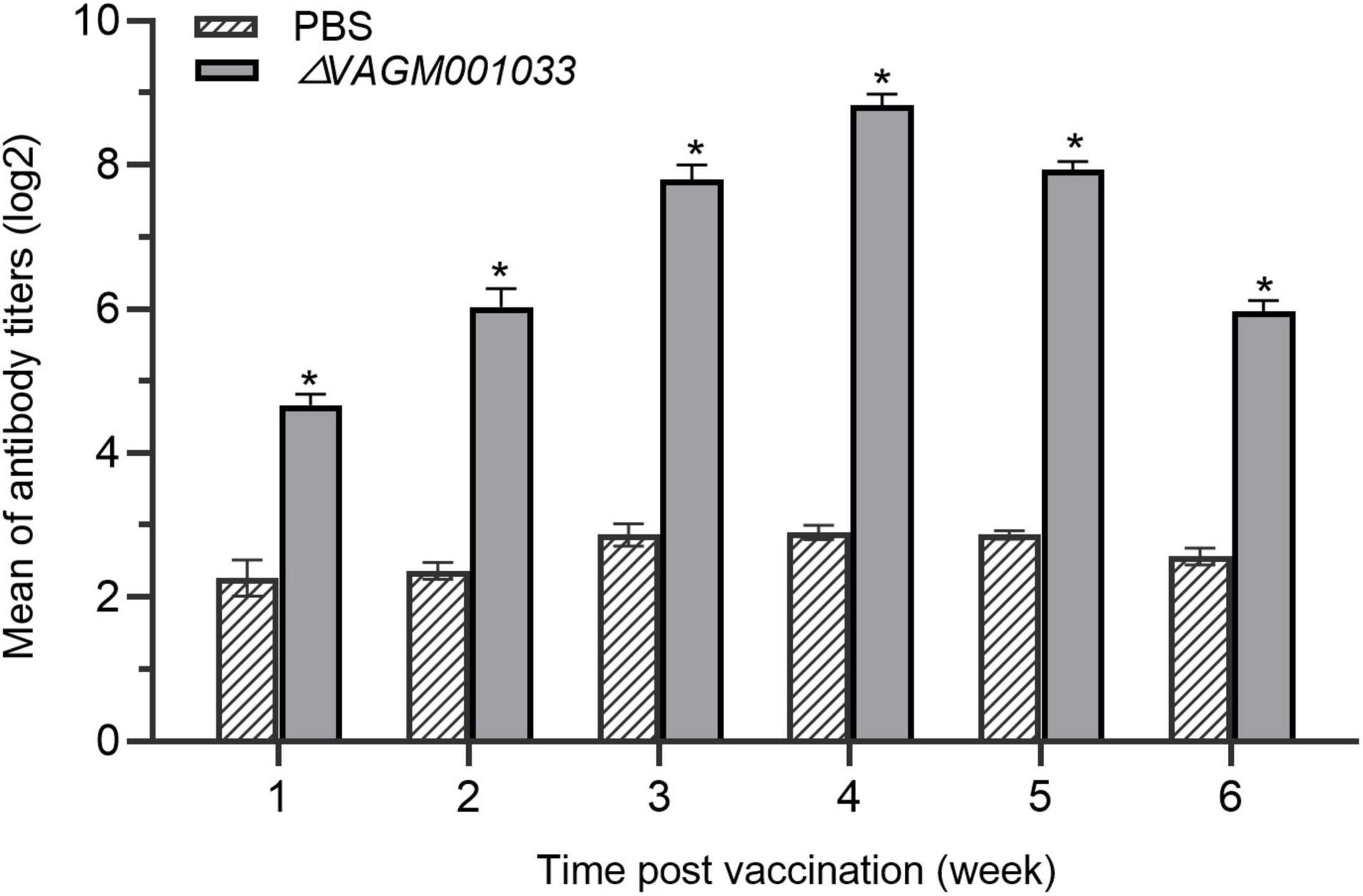
Figure 8. ELISA evaluation of antibody levels in grouper vaccinated with PBS or ΔVAGM001033 mutant. Sera samples were taken from week 1 to week 6 following immunization. The mean of log2 (antibody titers) was represented in each datum column, along with a standard error bar. The asterisks denoted significant changes between the vaccinated and control groups (p < 0.05).
Histopathological Analysis
Groupers injected with V. alginolyticus exhibit sluggish behavior, inactive movements, curve or twirl their body. No clinical signs were observed in the ΔVAGM001033 group and the PBS group. Histopathological analysis was conducted for the further observation that no histological abnormality was detected between the ΔVAGM001033 mutant group and the PBS group. Nevertheless, distinct histological changes were observed in the V. alginolyticus HY9901 group (Figure 9). The intestine section of the V. alginolyticus HY9901 group revealed degeneration and necrosis of the mucosal epithelium, destruction of the intestinal microvilli, and cell debris in the lumen (Figure 9A3). The liver section of the V. alginolyticus HY9901 group showed vacuolation, hepatocyte disruption, and hepatic necrosis (Figure 9B3).
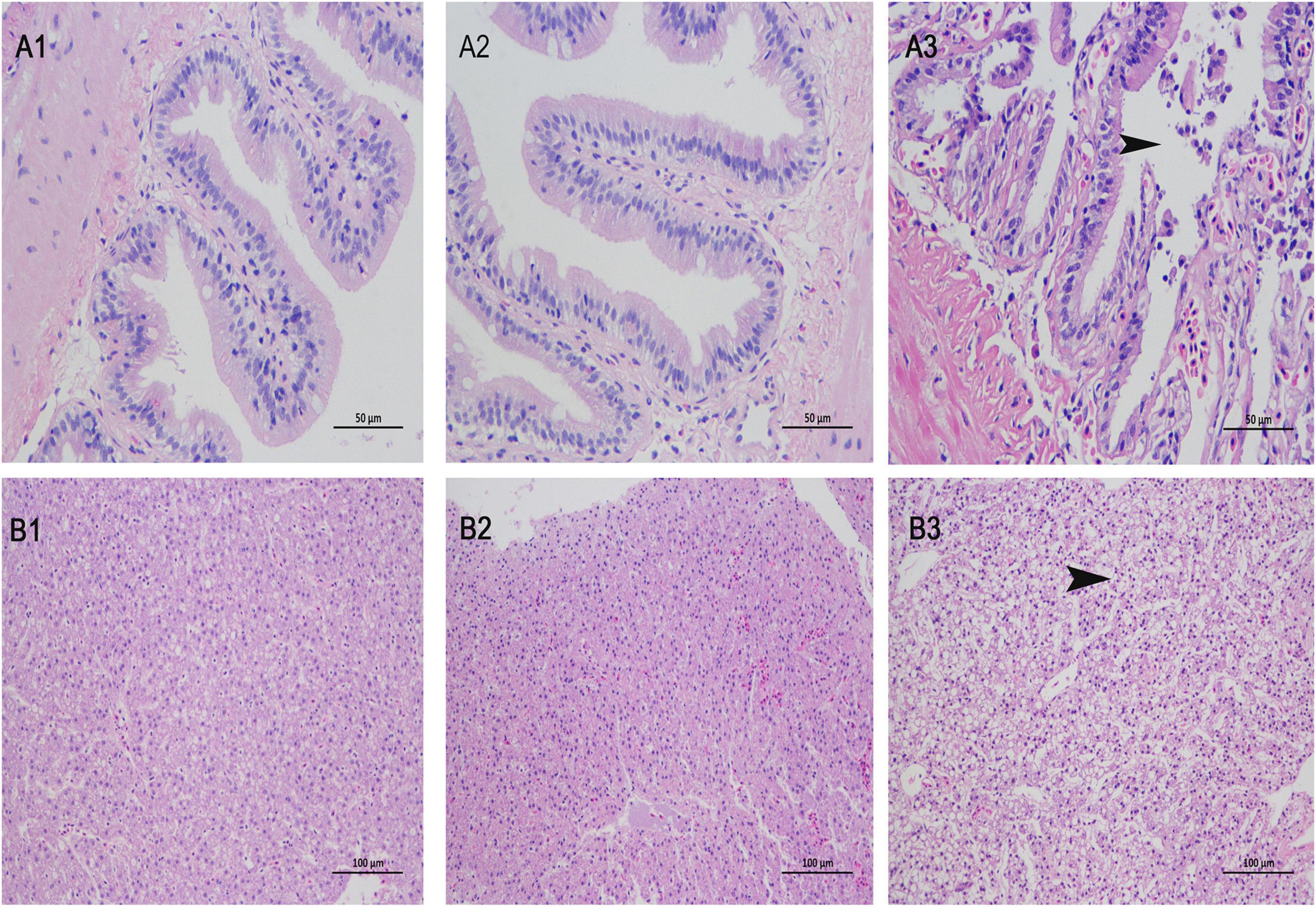
Figure 9. Histological changes of intestine (A1–3) and liver (B1–3) stained with H&E. (A1,B1), phosphate-buffered saline (PBS) group; (A2,B2), ΔVAGM001033 mutant group and (A3,B3), V. alginolyticus HY9901 group. (A1,2): intestine of the PBS group and the mutant group show intact intestinal mucosal epithelium, well-organized microvilli and no cell debris in the lumen (H&E staining; 400X). (A3): intestine of the V. alginolyticus HY9901 group show hemorrhage, degeneration, and necrosis of the mucosal epithelium, destruction of the intestinal microvilli, and cell debris in the lumen (H & E staining; 400 ×). (B1,2): liver of the PBS group and the mutant group show normal hepatocytes structure and organization (H&E staining; 200X). (B3): liver of the V. alginolyticus HY9901 group show vacuolation, hepatocyte disruption, and hepatic necrosis (H&E staining; 200X).
Immune Protective Effects of ΔVAGM001033 in Pearl Gentian Grouper
Grouper were challenged with V. alginolyticus HY9901 at 42 days post-vaccination. The results revealed that the survival rate in the ΔVAGM001033 mutant group was 84% with an RPS of 82% (Figure 10), which was significantly higher than as observed in the PBS group (p < 0.01).
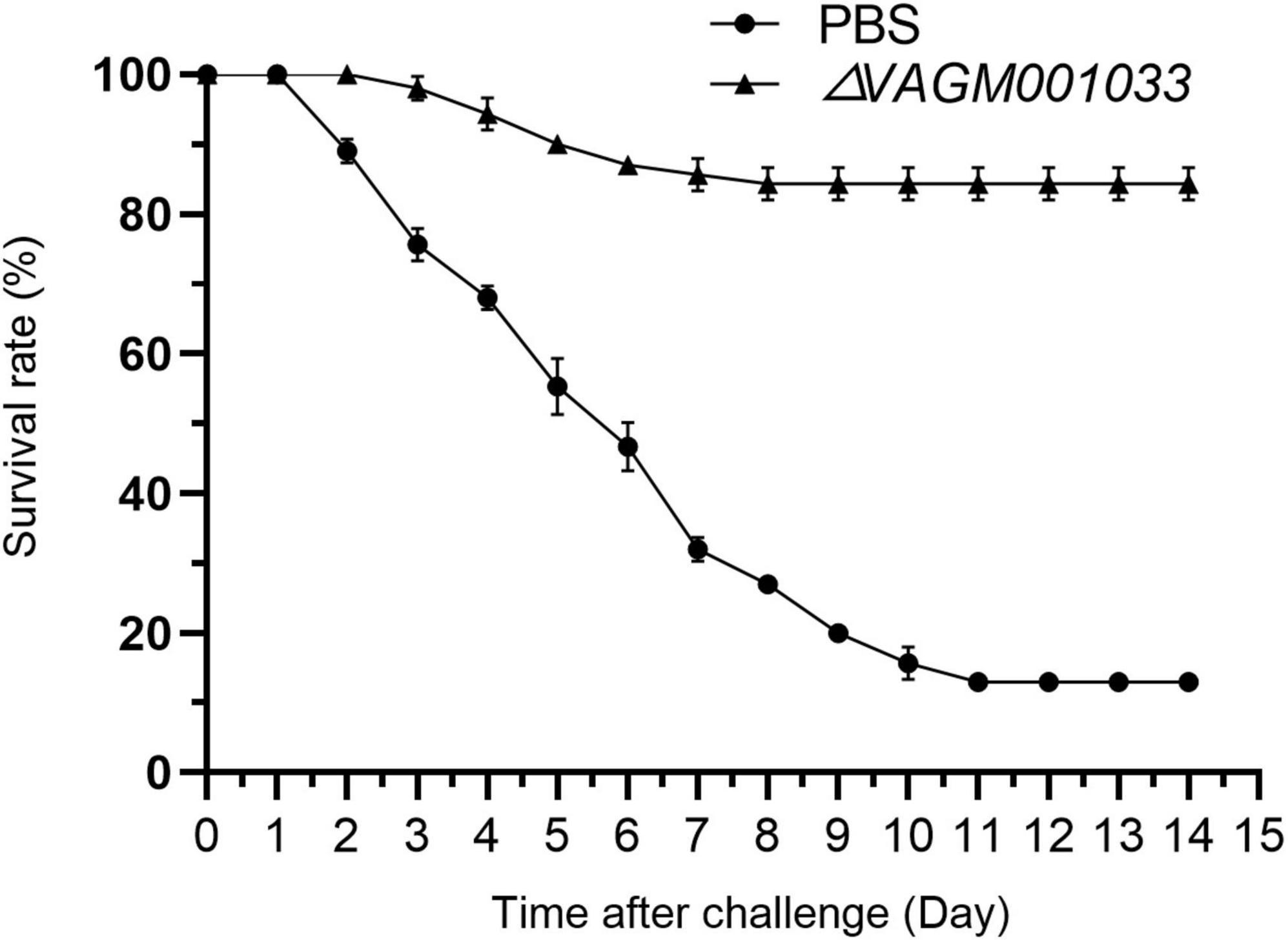
Figure 10. Survival rate of grouper after challenge with V. alginolyticus HY9901. Control fish was injected with PBS (•). Fish was vaccinated with ΔVAGM 001033 (▲).
Immune-Related Gene Expression Analysis
In this study, qPCR was conducted to evaluate the transcription levels of the immune-related genes. The immune-related genes of pearl gentian grouper, namely, IgM, MHC-Iα, IL-1β, IL-16, and TNF-α was significantly upregulated in the ΔVAGM001033 group compared with those in the control group after vaccination (Figure 11).
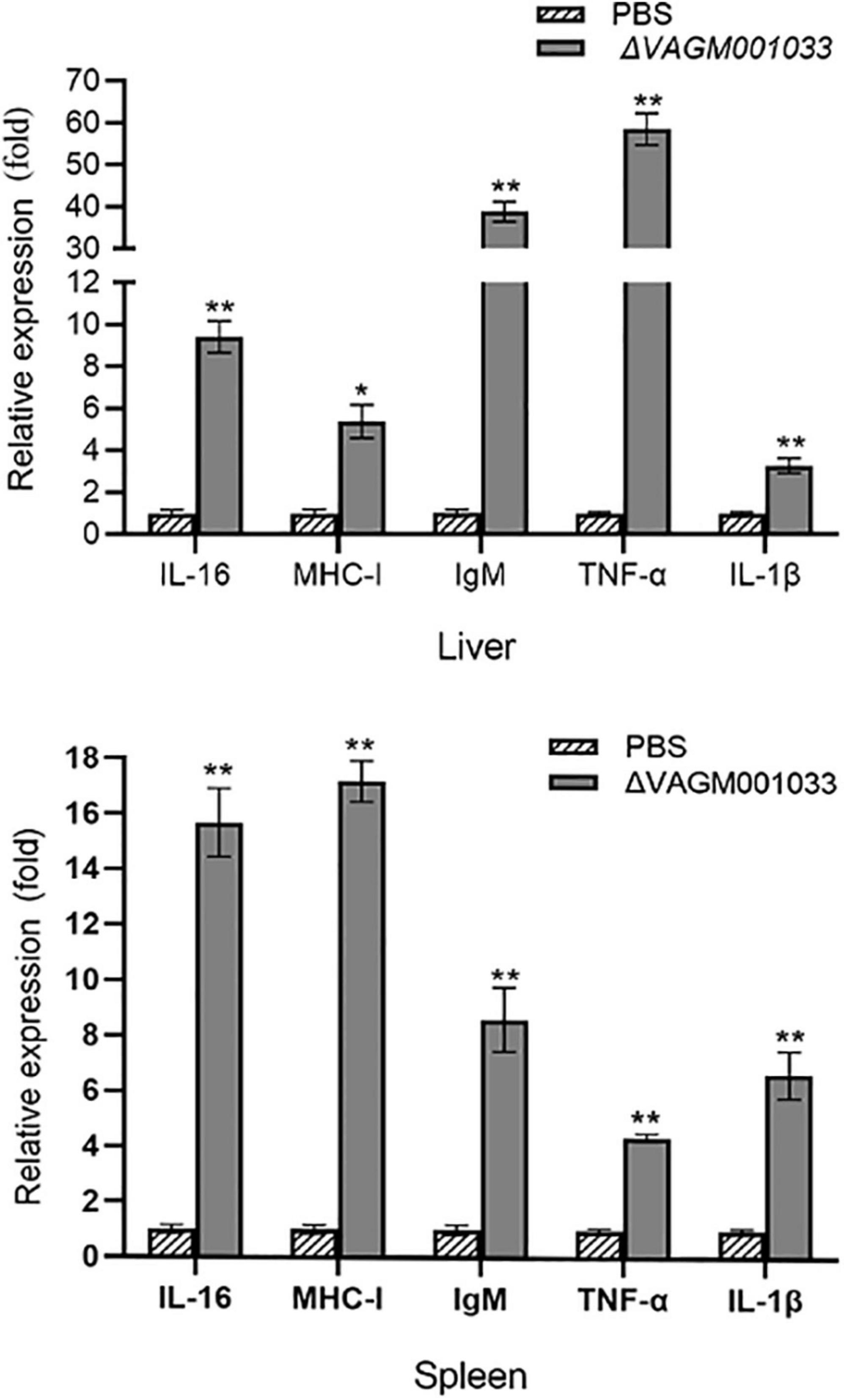
Figure 11. Expression of immune-related genes in liver and spleen. The liver and spleen of pearl gentian grouper were sampled at 42 days post-immunization. Each immune-related gene’s transcription level was compared to that of β-actin. The error bars showed the SD and the bars show the three biological replicates of mean relative expression. *p < 0.5; **p < 0.01.
Discussion
Diguanylate cyclase is a class of essential enzymes responsible for the synthesis of cyclic di-GMP in bacteria. Numerous studies have linked GGDEF domain proteins to processes involved in motility, biofilm formation, and virulence (Ryjenkov et al., 2005). Despite many publications concerning GGDEF domains, detailed function analyses and potential mechanisms in V. alginolyticus are relatively scarce. The rising concentration of c-di-GMP caused an increased secretion of polysaccharides, which led to binding with Congo red to form a red-stained, dry, and rough colony morphology. Polysaccharides are important metabolites secreted by bacteria during their growth and metabolism (Kenne and Lindberg, 1983). In this study, L-arabinose was a chemical inducer that involved the regulation of the L-arabinose operon of E. coli. The arabinose system was often for practical use in protein expression systems because the ara promoter acted positively to stimulate transcription in the presence of arabinose (Schleif, 2010, 2000). It was observed that with the increase of the concentration of the L-arabinose, more secretion of polysaccharide is detected and the red-stained, dry, and rough form of the colony gets more obvious.
Motility was a necessity for bacteria to detect and pursue nutrients and to reach and maintain their appropriate niches for colonization (Wadhwa and Berg, 2021). In comparison with the wild-type strain, the mutant displayed enhanced swarming motility with a bigger swarm circle diameter. Biofilm is a structured consortium that causes chronic infections due to increased tolerance to antibiotics and disinfectant chemicals, and resisting phagocytosis and the host’s immune defense system (Sharma et al., 2019; Vestby et al., 2020). Previous studies confirmed that HmsT in Yersinia pesits regulated biofilm formation by control of poly-β1,6-N-acetylglucosamine synthesis (Bobrov et al., 2011). Biofilm formation was significantly decreased in ΔVAGM001033 mutant in this study.
Antimicrobial usage in aquaculture can result in residues of antimicrobials in seafood, which was a hazard to public health (Aly and Albutti, 2014). ΔVAGM001033 mutant was sensitive to cefotaxime and nitrofurantoin, in contrast, the wild type was intermediate. Especially, the zone of inhibition of ΔVAGM001033 mutant was bigger than that in the wild type, indicating that ΔVAGM001033 mutant was more sensitive to the antibiotics. The results of antibiotic susceptibility experiments suggest the diguanylate cyclase VAGM001033 could regulate the antibiotic susceptibility through controlling the concentration of c-di-GMP and some receptors of c-di-GMP perform some important functions in the formation mechanism of drug resistance in V. alginolyticus.
A total of 756 DEGs were identified between the wild type and the mutant by RNA-seq. Flagellar assembly, two-component system, ABC transporters, and peptidoglycan biosynthesis were significantly enriched in the KEGG pathway, which was a database resource providing all knowledge about genomes and their relationships to biological systems such as cells and whole organisms and their interactions with the environment (Aoki-Kinoshita and Kanehisa, 2007). These results of RNA-seq suggest that VAGM001033 has controlled the biological phenotype such as swarming, biofilm forming, extracellular protease activity, virulence, and antibiotic susceptibility through regulating the above pathway. A previous study demonstrated that YedQ, a diguanylate cyclase, affected c-di-GMP-associated pathways, namely, flagellar assembly, siderophore pathway, exopolysaccharide biosynthesis pathway, and oxidative stress resistance in Pseudomonas syringae via RNA-sequencing (Wang et al., 2019). Taken together, the present data indicated that VAGM1033 might involve in regulating motility, biofilm formation, and virulence as diguanylate cyclase.
To evaluate the potential of the ΔVAGM001033 mutant as a live attenuated vaccine candidate, a series of experiments were conducted. A previous study reported that the accessory colonization factor acfA deletion could induce a high antibody titer, upregulated the immune-relative genes, reached the RPS value of 81.1% without showing clinical symptoms and histopathological changes (Chen et al., 2019a). A study also confirmed a superoxide dismutase sodB deletion provided an effective immunoprotection with an RPS of 86.5% after vaccination, and also induced a high antibody titer (Chen et al., 2019b). More studies confirmed that live attenuated vaccines could provide significant immunoprotectivity against Vibrio, such as ΔtyeA mutant (Zhou et al., 2020), Δhop mutant (Pang et al., 2018), ΔclpP mutant (Chen et al., 2020), and ΔvscB mutant (Pang et al., 2022). In this study, the live attenuated strain of VAGM001033 deletion could induce a high titer of specific IgM and provide the RPS of 82% in pearl gentian grouper without any side effect, suggesting ΔVAGM001033 mutant is an excellent candidate for the live attenuated vaccine.
The mRNA transcription levels of the immune-related genes, namely, IgM, MHC-Iα, IL-1β, IL-6, and TNF-α were conducted to analyze the underlying immune responses. IgM is the pioneering force against infection which is produced earliest during the adaptive immune response (Watts et al., 2001). Some previous studies reported that it was a major component of humoral immune response in teleosts and induced high expression in the spleen of vaccinated fish (Watts et al., 2001; Kim et al., 2007; Castro et al., 2013). ΔVAGM001033 mutant significantly increased IgM expression levels in this study, explaining why higher serum IgM titers were recorded in the vaccinated fish. MHC-Iα is responsible for combining with the endogenous antigen peptide, and presenting antigen to CD8+ T cells, which is distributed on all the surfaces of nucleated cells, platelets, and reticulocytes (Dijkstra et al., 2001; Šimková et al., 2006). Compared to the control group, the mRNA transcription level of MHC-Iα was significantly upregulated, indicating the recognition and presentation of V. alginolyticus could be involved in the MHC pathway. Cytokines are synthesized and secreted by immune cells and tissue cells, a class of soluble small molecules, namely, IL, IFN, TNF, CSF, GF, and chemokine. Cytokines can bind to the corresponding receptors to regulate the cellular activity, immune response, and inflammation (Secombes et al., 2001; Zou and Secombes, 2016; Sakai et al., 2021). In this study, IL-1β, IL-6, and TNF-α were upregulated in different organs, indicating the activation of immune responses.
Conclusion
In summary, an in-frame deletion strain of ΔVAGM001033 mutant was constructed successfully. The results showed that VAGM1033 was a key diguanylate cyclase in V. alginolyticus, which is involved in the regulation of motility, biofilm formation, and virulence. These results provided an essential reference for further study about the diguanylate cyclase in V. alginolyticus. Moreover, ΔVAGM001033 mutant provided significant immunoprotection to grouper against V. alginolyticus by inducing a high antibody titer and upregulating immune-related genes without histopathological abnormality. The total data indicated that ΔVAGM001033 mutant could be applied as a live attenuated vaccine to prevent and control fish diseases caused by V. alginolyticus.
Data Availability Statement
The data presented in the study are deposited in the (https://www.ncbi.nlm.nih.gov/genbank/) repository, accession number PRJNA794348.
Ethics Statement
The animal study was reviewed and approved by Epidemiology for Aquatic Economic Animals Ethics Committee of Guangdong Ocean University.
Author Contributions
HT conceived and designed the study, performed the experiment, generated the data and figures, and wrote the original draft. FD edited the data and reviewed the manuscript. GL and XW constructed the plasmids. JJ and SC made some critically contribution during the revision of the manuscript and also, they helped in the submission of sequences online. All authors contributed to the article and approved the submitted version.
Funding
This study was supported and funded by the National Natural Science Foundation of China (No. U20A2065), the Natural Science Foundation of Guangdong Province (No. 2021A1515010532), and the Natural Science Foundation of Shenzhen City (JCYJ20190813114409506).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Aly, S. M., and Albutti, A. (2014). Antimicrobials use in aquaculture and their public health impact. J. Aquac. Res. Dev. 5:247. doi: 10.4172/2155-9546.1000247
Amenyogbe, E., Huang, J., Chen, G., and Wang, W. (2021). Probiotic potential of indigenous (Bacillus sp. RCS1, Pantoea agglomerans RCS2, and Bacillus cereus strain RCS3) isolated from cobia fish (Rachycentron canadum) and their antagonistic effects on the growth of pathogenic Vibrio alginolyticus, Vibrio harvey. Front. Mar. Sci. 8:672213. doi: 10.3389/fmars.2021.672213
Aoki-Kinoshita, K. F., and Kanehisa, M. (2007). “Gene annotation and pathway mapping in KEGG,” in Comparative Genomics, ed. N. H. Bergman (Totowa: Humana Press), 71–91. doi: 10.1007/978-1-59745-515-2_6
Bauer, A. W. (1966). Antibiotic susceptibility testing by a standardized single disc method. Am. J. Clin. Pathol. 45, 149–158. doi: 10.1093/ajcp/45.4_ts.493
Bobrov, A. G., Kirillina, O., Ryjenkov, D. A., Waters, C. M., Price, P. A., Fetherston, J. D., et al. (2011). Systematic analysis of cyclic di-GMP signalling enzymes and their role in biofilm formation and virulence in Yersinia pestis. Mol. Microbiol. 79, 533–551. doi: 10.1111/j.1365-2958.2010.07470.x
Cai, S., Cheng, H., Pang, H., Jian, J., and Wu, Z. (2018). AcfA is an essential regulator for pathogenesis of fish pathogen Vibrio alginolyticus. Vet. Microbiol. 213, 35–41. doi: 10.1016/j.vetmic.2017.11.016
Castro, R., Jouneau, L., Pham, H.-P., Bouchez, O., Giudicelli, V., Lefranc, M.-P., et al. (2013). Teleost fish mount complex clonal IgM and IgT responses in spleen upon systemic viral infection. PLoS Pathog. 9:e1003098. doi: 10.1371/journal.ppat.1003098
Chen, Y., Wu, F., Pang, H., Tang, J., Cai, S., and Jian, J. (2019b). Superoxide dismutase B (sodB), an important virulence factor of Vibrio alginolyticus, contributes to antioxidative stress and its potential application for live attenuated vaccine. Fish Shellfish Immunol. 89, 354–360. doi: 10.1016/j.fsi.2019.03.061
Chen, Y., Wu, F., Wang, Z., Tang, J., Cai, S., and Jian, J. (2019c). Construction and evaluation of Vibrio alginolyticus ΔclpP mutant, as a safe live attenuated vibriosis vaccine. Fish Shellfish Immunol. 98, 917–922.
Chen, Y., Cai, S., and Jian, J. (2019a). Protection against Vibrio alginolyticus in pearl gentian grouper (♀Epinephelus fuscoguttatus × ♂ Epinephelus lanceolatu) immunized with an acfA-deletion live attenuated vaccine. Fish Shellfish Immunol. 86, 875–881. doi: 10.1016/j.fsi.2018.12.030
Chen, Y., Wu, F., Wang, Z., Tang, J., Cai, S., and Jian, J. (2020). Construction and evaluation of Vibrio alginolyticus ΔclpP mutant, as a safe live attenuated vibriosis vaccine. Fish Shellfish Immunol. 98, 917–922. doi: 10.1016/j.fsi.2019.11.054
Clinical and Laboratory Standards Institute [CLSI] (2013). Performance standards for Antimicrobial Susceptibility Testing; Twenty-Third Informational Supplement (M100- S23-2013). Wayne: Clinical and Laboratory Standards Institute.
Cooper, M. D., and Alder, M. N. (2006). The evolution of adaptive immune systems. Cell 124, 815–822. doi: 10.1016/j.cell.2006.02.001
Dijkstra, J. M., Fischer, U., Sawamoto, Y., Ototake, M., and Nakanishi, T. (2001). Exogenous antigens and the stimulation of MHC class I restricted cell-mediated cytotoxicity: possible strategies for fish vaccines. Fish Shellfish Immunol. 11, 437–458. doi: 10.1006/fsim.2001.0351
Floyd, K. A., Lee, C. K., Xian, W., Nametalla, M., Valentine, A., Crair, B., et al. (2020). c-di-GMP modulates type IV MSHA pilus retraction and surface attachment in Vibrio cholerae. Nat. Commun. 11:1549. doi: 10.1038/s41467-020-15331-8
Gudding, R., and Van Muiswinkel, W. B. (2013). A history of fish vaccination: science-based disease prevention in aquaculture. Fish Shellfish Immunol. 35, 1683–1688. doi: 10.1016/j.fsi.2013.09.031
Hecht, G. B., and Newton, A. (1995). Identification of a novel response regulator required for the swarmer-to-stalked-cell transition in Caulobacter crescentus. J. Bacteriol. 177, 6223–6229. doi: 10.1128/jb.177.21.6223-6229.1995
Hueso-Gil, Á., Calles, B., and de Lorenzo, V. (2020). The Wsp intermembrane complex mediates metabolic control of the swim-attach decision of Pseudomonas putida. Environ. Microbiol. 22, 3535–3547. doi: 10.1111/1462-2920.15126
Jain, R., Sliusarenko, O., and Kazmierczak, B. I. (2017). Interaction of the cyclic-di-GMP binding protein FimX and the Type 4 pilus assembly ATPase promotes pilus assembly. PLoS Pathog. 13:e1006594. doi: 10.1371/journal.ppat.1006594
Joshi, A., Mahmoud, S. A., Kim, S.-K., Ogdahl, J. L., Lee, V. T., Chien, P., et al. (2020). c-di-GMP inhibits LonA-dependent proteolysis of TfoY in Vibrio cholerae. PLoS Genet. 16:e1008897. doi: 10.1371/journal.pgen.1008897
Kenne, L., and Lindberg, B. (1983). “Bacterial polysaccharides,” in The Polysaccharides, ed. G. O. Aspinall (New York: Academic Press), 287–363. doi: 10.1016/B978-0-12-065602-8.50010-8
Kierek, K., and Watnick, P. I. (2003). Environmental determinants of Vibrio cholerae biofilm development. Appl. Environ. Microbiol. 69, 5079–5088. doi: 10.1128/AEM.69.9.5079-5088.2003
Kim, W. S., Nishizawa, T., and Yoshimizu, M. (2007). Non-specific adsorption of fish immunoglobulin M (IgM) to blocking reagents on ELISA plate wells. Dis. Aquat. Organ. 78, 55–59. doi: 10.3354/dao01843
Kimbrough, J. H., and McCarter, L. L. (2020). Identification of three new GGDEF and EAL domain-containing proteins participating in the Scr surface colonization regulatory network in Vibrio parahaemolyticus. J. Bacteriol. 203, e00409–20. doi: 10.1128/JB.00409-20
Lee, P.-T., Liao, Z.-H., Huang, H.-T., Chuang, C.-Y., and Nan, F.-H. (2020). β-glucan alleviates the immunosuppressive effects of oxytetracycline on the non-specific immune responses and resistance against Vibrio alginolyticus infection in Epinephelus fuscoguttatus × Epinephelus lanceolatus hybrids. Fish Shellfish Immunol. 100, 467–475. doi: 10.1016/j.fsi.2020.03.046
Luo, P., He, X., Liu, Q., and Hu, C. (2015). Developing universal genetic tools for rapid and efficient deletion mutation in vibrio species based on suicide t-vectors carrying a novel counterselectable marker, vmi480. PLoS One 10:e0144465. doi: 10.1371/journal.pone.0144465
Mathew, J. A., Tan, Y. P., Srinivasa Rao, P. S., Lim, T. M., and Leung, K. Y. (2001). Edwardsiella tarda mutants defective in siderophore production, motility, serum resistance and catalase activity. Microbiology 147, 449–457. doi: 10.1099/00221287-147-2-449
Pang, H., Chang, Y., Zheng, H., Tan, H., Zhou, S., Zeng, F., et al. (2022). A live attenuated strain of HY9901ΔvscB provides protection against Vibrio alginolyticus in pearl gentian grouper (♀Epinephelus fuscoguttatus × ♂ Epinephelus lanceolatu). Aquaculture 546:737353. doi: 10.1016/j.aquaculture.2021.737353
Pang, H., Qiu, M., Zhao, J., Hoare, R., Monaghan, S. J., Song, D., et al. (2018). Construction of a Vibrio alginolyticus hopPmaJ (hop) mutant and evaluation of its potential as a live attenuated vaccine in orange-spotted grouper (Epinephelus coioides). Fish Shellfish Immunol. 76, 93–100. doi: 10.1016/j.fsi.2018.02.012
Ryjenkov, D. A., Tarutina, M., Moskvin, O. V., and Gomelsky, M. (2005). Cyclic diguanylate is a ubiquitous signaling molecule in bacteria: insights into biochemistry of the GGDEF protein domain. J. Bacteriol. 187, 1792–1798. doi: 10.1128/JB.187.5.1792-1798.2005
Sakai, M., Hikima, J., and Kono, T. (2021). Fish cytokines: current research and applications. Fish. Sci. 87, 1–9. doi: 10.1007/s12562-020-01476-4
Schleif, R. (2000). Regulation of the L-arabinose operon of Escherichia coli. Trends Genet. 16, 559–565. doi: 10.1016/S0168-9525(00)02153-3
Schleif, R. (2010). AraC protein, regulation of the l-arabinose operon in Escherichia coli, and the light switch mechanism of AraC action. FEMS Microbiol. Rev. 34, 779–796. doi: 10.1111/j.1574-6976.2010.00226.x
Secombes, C. J., Wang, T., Hong, S., Peddie, S., Crampe, M., Laing, K. J., et al. (2001). Cytokines and innate immunity of fish. Dev. Comp. Immunol. 25, 713–723. doi: 10.1016/s0145-305x(01)00032-5
Sharma, D., Misba, L., and Khan, A. U. (2019). Antibiotics versus biofilm: an emerging battleground in microbial communities. Antimicrob. Resist. Infect. Control 8:76. doi: 10.1186/s13756-019-0533-3
Šimková, A., Ottová, E., and Morand, S. (2006). MHC variability, life-traits and parasite diversity of European cyprinid fish. Evol. Ecol. 20, 465–477.
Su, Y.-L., Jiang, Y.-F., Chen, H.-J., Ye, S., Zhou, W.-H., Liu, H.-P., et al. (2021). Molecular characterization, expression and function analysis of Epinephelus coioides MKK4 response to SGIV and Vibrio alginolyticus infection. Dev. Comp. Immunol. 119:104020. doi: 10.1016/j.dci.2021.104020
Valentini, M., and Filloux, A. (2019). Multiple roles of c-di-GMP signaling in bacterial pathogenesis. Annu. Rev. Microbiol. 73, 387–406. doi: 10.1146/annurev-micro-020518-115555
Vestby, L. K., Grønseth, T., Simm, R., and Nesse, L. L. (2020). Bacterial biofilm and its role in the pathogenesis of disease. Antibiotics 9:59. doi: 10.3390/antibiotics9020059
Wadhwa, N., and Berg, H. C. (2021). Bacterial motility: machinery and mechanisms. Nat. Rev. Microbiol. doi: 10.1038/s41579-021-00626-4 [Epub ahead of print].
Wang, T., Cai, Z., Shao, X., Zhang, W., Xie, Y., Zhang, Y., et al. (2019). Pleiotropic effects of c-di-GMP content in Pseudomonas syringae. Appl. Environ. Microbiol. 85, e00152–19.
Wang, X., Du, Y., Yang, J., Tang, Y., and Luo, J. (2007). Preparation, characterization, and antimicrobial activity of quaternized chitosan/organic montmorillonite nanocomposites. J. Biomed. Mater. Res. A 84, 384–390. doi: 10.1002/jbm.a
Watts, M., Munday, B. L., and Burke, C. M. (2001). Immune responses of teleost fish. Aust. Vet. J. 79, 570–574. doi: 10.1111/j.1751-0813.2001.tb10753.x
Webster, S. S., Lee, C. K., Schmidt, W. C., Wong, G. C. L., and O’Toole, G. A. (2021). Interaction between the type 4 pili machinery and a diguanylate cyclase fine-tune c-di-GMP levels during early biofilm formation. Proc. Natl. Acad. Sci. U. S. A. 118:e2105566118. doi: 10.1073/pnas.2105566118
Wei, G., Tan, H., Ma, S., Sun, G., Zhang, Y., Wu, Y., et al. (2020). Protective effects of β-glucan as adjuvant combined inactivated Vibrio harveyi vaccine in pearl gentian grouper. Fish Shellfish Immunol. 106, 1025–1030. doi: 10.1016/j.fsi.2020.09.027
Windle, H. J., and Kelleher, D. (1997). Identification and characterization of a metalloprotease activity from Helicobacter pylori. Infect. Immun. 65, 3132–3137. doi: 10.1128/iai.65.8.3132-3137.1997
Wu, D. C., Zamorano-Sánchez, D., Pagliai, F. A., Park, J. H., Floyd, K. A., Lee, C. K., et al. (2020). Reciprocal c-di-GMP signaling: incomplete flagellum biogenesis triggers c-di-GMP signaling pathways that promote biofilm formation. PLoS Genet. 16:e1008703.
Xie, J., Bu, L., Jin, S., Wang, X., Zhao, Q., Zhou, S., et al. (2020). Outbreak of vibriosis caused by Vibrio harveyi and Vibrio alginolyticus in farmed seahorse Hippocampus kuda in China. Aquaculture 523:735168. doi: 10.1016/j.aquaculture.2020.735168
Zhou, S., Tu, X., Pang, H., Hoare, R., Monaghan, S. J., Luo, J., et al. (2020). A T3SS regulator mutant of Vibrio alginolyticus affects antibiotic susceptibilities and provides significant protection to Danio rerio as a live attenuated vaccine. Front. Cell. Infect. Microbiol. 10:183. doi: 10.3389/fcimb.2020.00183
Keywords: Vibrio alginolyticus, VAGM001033, diguanylate cyclase, live attenuated vaccine, mutant
Citation: Tan H, Da F, Lin G, Wan X, Jian J and Cai S (2022) Characterization of a Diguanylate Cyclase VAGM001033 of Vibrio alginolyticus and Protective Efficacy as a Live Attenuated Vaccine Candidate in Pearl Gentian Grouper. Front. Mar. Sci. 9:841564. doi: 10.3389/fmars.2022.841564
Received: 22 December 2021; Accepted: 31 January 2022;
Published: 21 March 2022.
Edited by:
Carlo C. Lazado, Norwegian Institute of Food, Fisheries and Aquaculture Research (Nofima), NorwayReviewed by:
Rolando Pakingking Jr., Southeast Asian Fisheries Development Center, PhilippinesEugenio O. Spencer, University of Santiago, Chile
Qingchao Wang, Huazhong Agricultural University, China
Ana Maria Sandino, University of Santiago, Chile
Copyright © 2022 Tan, Da, Lin, Wan, Jian and Cai. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shuanghu Cai, Y2Fpc2hAZ2RvdS5lZHUuY24=
 Huiming Tan
Huiming Tan Fan Da1
Fan Da1